SUMMARY
In his 2001 article, “Translation: in retrospect and prospect,” the late Carl Woese made a prescient observation that there was a need for the then-current view of translation to be “reformulated to become an all-embracing perspective about which 21st century Biology can develop” (RNA 7:1055–1067, 2001, https://doi.org/10.1017/s1355838201010615). The quest to decipher the origins of life and the road to the genetic code are both inextricably linked with the history of the ribosome. After over 60 years of research, significant progress in our understanding of how ribosomes work has been made. Particularly attractive is a model in which the ribosome may facilitate an ∼180° rotation of the CCA end of the tRNA from the A-site to the P-site while the acceptor stem of the tRNA would then undergo a translation from the A-site to the P-site. However, the central question of how the ribosome originated remains unresolved. Along the path from a primitive RNA world or an RNA-peptide world to a proto-ribosome world, the advent of the peptidyl transferase activity would have been a seminal event. This functionality is now housed within a local region of the large-subunit (LSU) rRNA, namely, the peptidyl transferase center (PTC). The PTC is responsible for peptide bond formation during protein synthesis and is usually considered to be the oldest part of the modern ribosome. What is frequently overlooked is that by examining the origins of the PTC itself, one is likely going back even further in time. In this regard, it has been proposed that the modern PTC originated from the association of two smaller RNAs that were once independent and now comprise a pseudosymmetric region in the modern PTC. Could such an association have survived? Recent studies have shown that the extant PTC is largely depleted of ribosomal protein interactions. It is other elements like metallic ion coordination and nonstandard base/base interactions that would have had to stabilize the association of RNAs. Here, we present a detailed review of the literature focused on the nature of the extant PTC and its proposed ancestor, the proto-ribosome.
KEYWORDS: PTC, peptidyl transferase center, accretion model, evolutionary history, protein synthesis, ribosomes, translation
INTRODUCTION
The ribosome has a unique place in efforts to understand the origin and early evolution of life (1). This reflects the fact that its history predates the last universal common ancestor (LUCA) and may “have evolved in an RNA world before the evolution of the genetic code and proteins” (2). The ribosome houses an activity center that catalyzes peptidyl transfer during protein synthesis. The general location where this activity is located is referred to as the peptidyl transferase center (PTC). The present review will focus on recent developments in our understanding of the evolution of the PTC, which is where peptide bonds are formed without the direct involvement of proteins.
The modern ribosome is a complex molecular machine that is responsible for coded protein synthesis. It is comprised of a small subunit (SSU), which contains the decoding center, and the large subunit (LSU) that contains the PTC (3), which in turn is responsible for peptide bond synthesis (4–8). The subunits are ribonucleoprotein complexes comprised of rRNA and ribosomal proteins (rProteins) (9). Ribosome function is directly facilitated by tRNAs and proteins such as elongation factors and initiation factors and indirectly facilitated by proteins such as the aminoacyl-tRNA synthetases that attach amino acids to the tRNAs. Since many of the core components are universally found, a functional ribosome is considered to be an important component of LUCA (10–16). Secondary structures deduced by comparative sequence analysis initially revealed that the LSU rRNA has six domains (17, 18). An additional domain referred to as domain 0 was recognized later (19). Atomic resolution studies show that domain 5 houses the PTC region (Fig. 1) (6, 20, 21), which includes an RNA pore (22, 23). The pore serves as the entrance to the exit tunnel through which a growing peptide leaves the ribosome.
FIG 1.
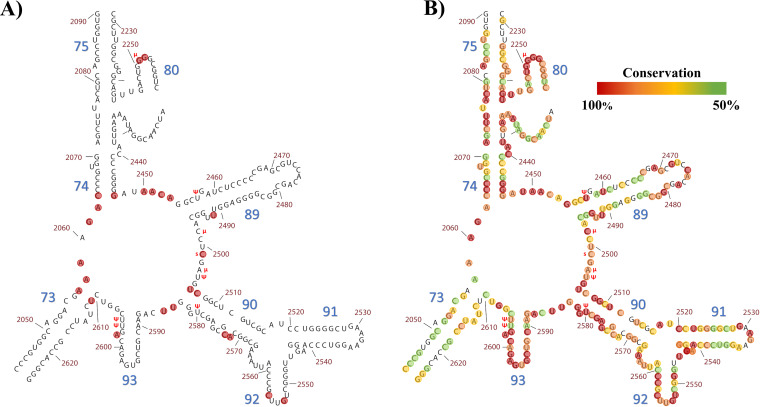
Fragment from the complete secondary structure of the LSU of Thermus thermophilus derived from crystallographic data (19), which contains the nucleotides within and around the PTC. Modified bases described in the text are marked with a red symbol in front of the corresponding base, “Ψ” for pseudouridine, “μ” for methylation, and “s” for 2-thiocytidine. Helix numbers are in large blue font, while position numbers are in smaller red font. (A) PTC residues mentioned throughout the text with a previously described functional role are highlighted within red circles. Details on functional role and/or implication for the ribosome activity are summarized in Table 1. As can be seen, PTC functional residues are not represented by a single contiguous chain of residues; instead, the PTC is formed from a diverse collection of distant residues that, when the RNA is folded into their tridimensional shape, act together as a catalytic unit, the PTC. (B) Conservation of the nucleobases within this fragment according to crystallographic data and corresponding alignments from Bernier et al. (275). This fragment comprises 282 nucleotides (nt). Red circles highlight those with 100% conservation (78 nt). Orange circles highlight those with 90 to 99.9% conservation (68 nt). Yellow and green circles highlight those with 70 to 89.9% and 50 to 69.9% conservation, respectively (52 and 49 nt). Thirty-five nucleotides from this fragment have shown less than 50% conservation.
THE PEPTIDYL TRANSFERASE CENTER: LOCATION AND COMPOSITION
PTC is a somewhat vague term that refers to the region of the ribosome formed by approximately 180 nucleotide residues of the LSU rRNA. Of these, at least 18 noncontiguous residues (24) facilitate the PTC activity. Collectively, these residues and their nearest neighbors are the most conserved and therefore likely the oldest sections of the LSU rRNA (25) (Fig. 1). Together they form a molecular platform for the ideal relative positioning of the reaction substrates (26), ensuring the efficient accommodation of the aminoacyl-tRNA 3′ end into the 50S A-site (27–29) and thereby enabling the transpeptidation activity (30–33). It is noteworthy that the structures of the PTC in the 50S subunit and in intact ribosomes are very similar (34, 35).
MUTATIONAL STUDIES AND RIBOSOMAL MUTATIONAL FLEXIBILITY
The PTC region is positioned to accommodate the acceptor ends of the tRNAs (36) (Fig. 2). This may in fact be far more vital than the nature of the conserved residues themselves, as demonstrated by mutation experiments. For example, individual mutations at positions C2063, G2447, A2450, A2451, A2453, and U2585 and the combination of A2451U and A2602G did not eliminate the transpeptidation activity (26, 37–45). Positions A2451, U2506, U2585, and A2602 were defined as forming part of an “inner shell” of the PTC. Mutations at these positions affected the peptide release significantly but did not equally affect the PTC activity (44). Three of these residues, namely, A2451, U2506, and U2585, along with G2252, are important for the binding of peptidyl-tRNA to the P-site of the large ribosomal subunit (38). Likewise, A2572 is in the immediate neighborhood of the aminoacyl-tRNA in the A-site (46) and is thought to be a sensor of conformational changes in the PTC (47, 48). It was shown to be important for PTC activity (49). The next adjacent base, C2573 (in collaboration with U2492 and C2556), facilitates the placement of the aminoacyl-tRNA at the A-site, tentatively labeled the “accommodation gate” (50–52). Mutations at A2572 and C2573 affect the peptide release but not the PTC activity (52). Several highly conserved residues are particularly important for their roles in positioning the tRNA at the PTC center. G2251 is essential for positioning the P-site tRNA during peptidyl transfer, and its mutation severely affects the PTC activity (53). Mutations of G2252, which base pairs with C74 of the P-site tRNA (54), can severely impact peptide release while not affecting the PTC activity itself (55). G2553 makes base-pairing interactions with C75 of the A-site tRNA (56), and its mutation reduces the PTC activity (57).
FIG 2.
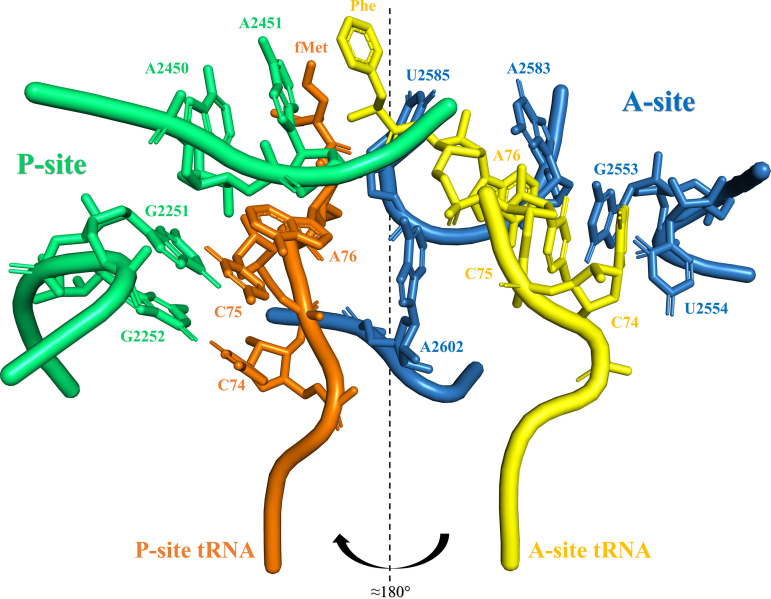
Closeup view of PTC with P-site and A-site tRNAs associated. View of some of the nucleotides that coordinate the CCA end and the amino acid residue that is carried by the P-site and A-site tRNAs. P-site rRNA chain and residues are colored green, and A-site rRNA chain and residues are colored blue. P-site tRNA is colored orange, while A-site tRNA is colored yellow. The CCA end of P-site tRNA is mainly coordinated by the Watson and Crick (WC) base pairs of G2252:C74 and G2251:C75, while an A-minor interaction of A76 with A2450 from the P-site tRNA is established. Both P-site residues A2450 and A2451 are considered important to the transpeptidation reaction. The CCA end of A-site tRNA is mainly stabilized by WC base pair G2553:C75, while U2554 establishes a hydrogen bond with C74. A2583 creates an A-minor interaction with A76 from the A-site tRNA. The “rotatory motion” mechanism implies that the 3′ end of the tRNA passage moves in a helical motion of almost 180° coupled with peptide bond formation. The tRNA transitions from the A- to the P-site along a rotation axis and around U2585 and A2602. This figure was created from the crystal structure NCBI ID 4WPO, using the PyMOL Molecular Graphics System, version 2.0 (Schrödinger, LLC).
In addition, there are nine modified residues of interest in the PTC. However, these residues are not universally conserved at their respective locations. Six pseudouridine (Ψ) residues are found around the PTC of bacterial and eukaryotic ribosomes, with roles in A-site tRNA positioning, PTC activity, translocation, initiation, termination, and tunnel composition (6, 58, 59) (Fig. 1). In this context, only 2 of these 9 modified residues are found among the 41 modified residues in the cytoplasmic LSU rRNA of Euglena gracilis (60). Furthermore, mutational studies of Ψ2504, Ψ2580, Ψ2604, and Ψ2605 (A-site) and Ψ2457 (P-site) suggest that they are not necessary for the overall activity (61). Other modified nucleotides in the PTC (Fig. 1) are reported as having some function. Methylated guanosine (G2251) is useful for positioning the P-site tRNA (53), while 2-methyladenosine (A2503) and Ψ2504 are integral parts of the tunnel entrance near the A-site (62). 2′-O-methylated C2498 is thought to help stabilize the PTC loop (62, 63). Finally, 2-thiocytidine (C2501) was found to be the only partially modified base in the PTC (64). Overall, of all the posttranscriptional modifications on RNAs, N6-methyladenosine (m6A) is shared by all the RNA types (namely, mRNA, rRNA, tRNA, and snRNA [small nucleolar RNA]) (65). The PTC is enriched in modified residues in most cases, and yet the variation in the type of modified residues is clearly evident. Although these modifications are useful in extant ribosomes, they are likely the outcome of evolutionary improvement. None of them seem to be essential for peptide bond formation.
In a more recent study (66), a detailed mutational analysis employing a single nucleotide mutation(s) within the PTC region was performed. More than 85% of the PTC nucleotides were found to be mutationally flexible. The residues classified as the least mutationally flexible, namely, C2063, A2450, and C2501, are in very close proximity to the P-site tRNA. These residues form a spatial configuration with G2252 and G2251 of the P-loop of the 23S rRNA, both of which interact with the CCA end of the P-site tRNA (53, 54) (Fig. 2). A2450 and C2063 form a wobble pair and along with C2501 form a base triple setup, which forms stacking interactions with another base triple formed by G2447-A2451-G2061 (67). Furthermore, C2063 forms base stacking interactions with G2061, A2059, A2058, and A2503 (68). In silico simulation experiments have shown that C2063 forms an intricate network of H bonds with water molecules (69). C2063 (along with C2064 and A2435) is also part of helix 74. Helix 74 is an important contact region for the 3′-CCA end of the tRNA in the conformational rearrangements during translocation (70). Meanwhile, G2447 forms noncanonical interactions with U2504 and C2452 (68). Thus, the complex noncanonical interaction network involving C2063, A2450, and C2501 likely contributes to their very limited mutational flexibility. The importance of C2063 for the PTC is further demonstrated by the observation that its interaction with A76 of the P-site tRNA is disrupted by an antibiotic like bactobolin A. The antibiotic displaces the CCA backbone of the P-site (71). Likewise, C2507 in H90 interacts with multiple partners. It base pairs with G2582 (H90) and interacts with G2553. Most importantly, the 2′OH of C2507 forms hydrogen bonds with the 2′OH and O2 of C75 in the tRNA (72). This four-base interaction is thought to stabilize the 3′ end of the tRNA in the A-site. Its importance was first established when a mutation at C2507 was found to have an adverse effect on PTC activity (73). This was further corroborated by the findings of d’Aquino et al. in 2020 (66).
In 2012, 15 residues were reported to define the entrance to the exit pore (22). When placing these residues in the context of the mutational flexibility scale described by d’Aquino et al. (66), the following was observed. P-site residues, namely, G2061, C2063, A2450, and A2451, and two A-site residues, namely, U2506 and U2585, would rank as the least flexible. U2506 interacts with residue A76 (74). The 2′OH and the O2 of U2506 hydrogen bond with the 2′OH of A76 in the A-site (72). Its mutation severely affects PTC activity (44), and its lack of mutational flexibility was further established by d’Aquino et al. (66).
On the other hand, three P-site residues, A2062, A2451, and C2452, and three A-site residues, G2505, U2586, and A2587, are the most flexible of the 15 residues lining the exit pore. Overall, the 23S rRNA residues that are the most mutationally flexible are found near the entrance to the exit tunnel (66, 75). The mutational flexibility seen was directly proportional to the distance from the PTC region that surrounds the aminoacyl end of the P-site tRNA (66). This work corroborated the findings from several earlier lines of inquiry, including a chemical footprinting assay mapping the interaction sites of macrolide antibiotics on the 23S rRNA (76). In that study by Vester and Douthwaite (76), residues A2062, A2057, A2058, A2059, C2611, and C2452 were found to confer macrolide resistance via mutations, thus establishing their mutational flexibility (Table 1).
TABLE 1.
Functional implications/roles of nucleobases that form the PTC (E. coli numbering system)a
| Positionb | Functional role/implication |
|---|---|
| A2057 | Mutation confers macrolide resistance |
| A2058 | Mutation confers macrolide resistance |
| A2059 | Mutation confers macrolide resistance |
| G2061 | Mutation reduces transpeptidation activity |
| A2062 | Mutation confers macrolide resistance |
| C2063* | Mutation reduces transpeptidation activity; involved in peptide bond formation |
| G2251 | Involved in accommodation of the P-site tRNA during peptidyl transfer; mutation reduces transpeptidation activity |
| G2252 | Binding of P-site tRNA; mutation impacts peptide release |
| G2447 | Mutation reduces transpeptidation activity |
| A2450 | Mutation reduces transpeptidation activity |
| A2451* | Has role in peptide release; binding of P-site tRNA; mutation reduces transpeptidation activity; involved in peptide bond formation |
| C2452 | Mutation confers macrolide resistance |
| A2453 | Mutation reduces transpeptidation activity |
| U2492 | Involved in accommodation of the A-site tRNA |
| C2501 | Mutation reduces transpeptidation activity |
| U2506 | Has role in peptide release; binding of P-site tRNA; mutation reduces transpeptidation activity |
| C2507 | Mutation reduces transpeptidation activity |
| G2553 | Interacts with A-site tRNA; mutation reduces transpeptidation activity |
| C2556 | Involved in accommodation of the A-site tRNA |
| G2572 | Sensor of conformational changes at the PTC; involved in transpeptidation activity; has role in peptide release |
| C2573 | Involved in accommodation of the A-site tRNA; has role in peptide release |
| U2584* | Involved in peptide bond formation |
| U2585 | Has role in peptide release; binding of P-site tRNA; mutation reduces transpeptidation activity |
| A2602* | Has role in peptide release; involved in transpeptidation activity; involved in peptide bond formation |
| C2611 | Mutation confers macrolide resistance |
Assuming that the mutational flexibility of these residues was similar in the proto-ribosome, the pore at the PTC that would eventually evolve into the exit tunnel was likely formed from residues of varying mutational flexibility. Thus, the “plasticity” observed in the PTC in extant ribosomes could well be the signature of a phenomenon with ancient origins.
In one study on yeast ribosomes, the rRNA mutations involving universally or highly conserved bases in the PTC region resulted in subtle structural rearrangements (51). However, these changes had a minimal effect on the overall translational accuracy, aminoacyl-tRNA binding, or PTC activity. Thus, the extant PTC rRNA could accommodate a certain level of plasticity without affecting the overall PTC function (51). Such plasticity could well be attributed to the functional groups and likely be a clue to its functionally generalized pasts. Several significant studies highlight this “plasticity.” Two studies have shown that it was not base A2451 but rather the 2′-hydroxyl group that was important for the reaction to occur (45, 77). Likewise, in the case of A2602, the role of the ribose moiety at that location in aiding peptide release has been demonstrated (78). This “plasticity” in effect confers functional “versatility” to the PTC. This can be seen in its demonstrated ability to support different substrates such as esters and thioesters (79, 80). It has been suggested that this capability may be reminiscent of the PTC’s ancient abilities, when it may have had to accommodate multiple substrates (81). This could also be traced to the fact that the extant ribosomes can accommodate 20 different amino acids from aminoacyl-tRNA carriers. Overall, the structural organization of the extant PTC is characterized by a property defined as an “induced-fit mechanism” (conformational change) (82). This is an extension of the induced-fit mechanism originally proposed to explain general enzyme-substrate specificity (83). Such a mechanism, in turn, is thought to confer an extraordinary ability to the ribosome to handle substrates of various sizes (84).
RIBOSOMES ARE HETEROGENOUS WITH THE PTC CORE UNCHANGED OVER TIME
In parallel with cellular evolution, the proto-ribosome evolved to give rise to the present-day ribosome that is much larger in size and complexity than its ancestral state. During this process, it also became structurally and functionally heterogenous. In several small bacterial genomes, the loss of rRNA segments (thus leading to reduction in rRNA size and complexity) has been shown to be coupled with the loss of ribosomal proteins (85). The deletions in the 23S rRNAs in these genomes were commonly located in helices H10, H63, H79, and H98, thus preserving the PTC core. Diversity and specialization have both been observed at the interspecies and intraorganismal levels (86). Both eukaryotic cells and eukaryotic cellular organelles (mitochondria) show substantial heterogeneity in their overall ribosomal structure and composition (87–91). The evolutionary origins of the mitochondria are thought to be rooted in alphaproteobacteria through endosymbiosis (92–94). Seldom discussed is the fact that this would place mitochondrial origins well after LUCA. In fact, mitochondria possess ribosomes, which show substantial divergence from their roots (95). The differences in their structures, functions, and compositions have been described in detail. Mitoribosomes are diverse in size among different lineages (96–100). Substantial sections of the rRNA have been replaced by additional protein sections (101–105). In the (55S) mitoribosome (which is specific to animals), the rRNA has been shown to have been reduced to an inner core retaining the functionally important sites, namely, the PTC active site and the decoding site (106). A detailed structural study of the yeast mitoribosome suggests that a base pairing between U2877 (C2610 in Escherichia coli) and A1958 (A2058 in E. coli) causes the narrowing of the exit tunnel entrance (107). However, this narrowing is far removed from the actual PTC activity site. The key regions of interactions between the 3′ CCA ends of the tRNAs and the PTC in the mitoribosomes, which are vital for the peptide bond formation, are not different from what is observed in the corresponding bacterial and eukaryotic (cytosolic) ribosomes (102, 108–110). In one study on the human cytosolic ribosome, cross-linking methods were used to show the proximity of an rProtein to the P-site tRNA. The rProtein rpL36a-like (rpL36AL) was found to be directly contacting the CCA 3′-terminal adenosine of the P-site tRNA (111). Otherwise, the arrangements of the PTC regions found in the eukaryotic nuclear ribosomes are not much different than those found in the ribosomes of bacteria (112, 113).
THE PTC HAS BEEN “FOSSILIZED” IN THE RIBOSOMAL LARGE SUBUNIT SINCE PRE-LUCA TIMES
Early efforts to understand ribosome evolution were inspired by the observation that the rRNAs are far too large to have emerged in a single step. Thus, some portions of the rRNAs must precede others (114), but how can we tell which are the oldest? A proposed solution was to identify timing events that provide insight into the relative age of different regions (115). The first example that was utilized was connectivity. It was argued that highly connected regions of the rRNA are likely older than less-connected regions. With this in mind, the number of long-range interactions between domains was identified (115). This approach was extended to individual helical elements by looking at A-minor interactions where adenines interact with a necessarily preexisting helical element (116–118). In the onion model (119), the PTC was assumed to be the oldest region and then other regions were dated by their distance from the PTC.
A significant breakthrough came with the realization that there was evidence of past accretion events even in the conserved regions of the rRNA (120, 121). From structural comparisons, it was inferred that new RNA was added to “older” RNA (120). Using this accretion approach, the various regions of the rRNA have been used to create a chronological hierarchy (12, 15, 120–125). This accretion model provides a detailed helix-by-helix model of LSU rRNA evolution in which each addition is assigned to one of six phases. In this model, the PTC is thought to have been assembled in phases one and two. Thus, much of the evolution of the rRNAs likely occurred well before LUCA. This approach has been extended to include the SSU rRNA as well (126).
The accretion model can also be explained and related to the function of the extant ribosome. The ribosome has been defined as a ratcheting motor (127, 128) and a Brownian machine (129) that undergoes several intermediate stages of motion described as “hybrid” states (130–134). These states include the rotation of the 30S (SSU) with respect to the 50S (LSU) (127, 135–138), translocation of the transfer RNAs and mRNA during the reverse rotation of the 30S subunit by the elongation factor G (EF-G) (139–146). It is now fairly well established that just before peptide bond formation, the ribosome is “locked” in the nonrotated state. Thus, the entire ribosomal evolution and accretion around the PTC have occurred in such a manner to accommodate all these motions associated with the extant ribosome, without in any way compromising the oldest part of the ribosome, the PTC. Thus, these subtle motions are paused during that “locked” nonrotated state, ostensibly to promote catalysis at the PTC (147). In fact, it is peptidyl transfer itself which is thought to induce the rotated conformation (136, 138, 148). At the molecular level, these various motions are associated with weak sites such as nonstandard base-base interactions and three-way junctions in the rRNAs (51, 149, 150). A comprehensive search for pivot points associated with EF-G and EF-Tu binding to the ribosome has been reported (151, 152). The importance of weak sites was found to be a general rule.
COEVOLUTION OF THE PTC AND THE tRNA
The origins of the tRNA are also linked with the origins of the PTC. The choice of the amino acid that were likely involved in early PTC activity has implications for the origins of coding. The ancestral small RNAs of extant tRNAs were aminoacylated based on the amino acid affinity of the CCA stem. This was most likely facilitated by an ancestral operational genetic code (153–155). Evidence for the same can be found in the examples of minihelix RNA comprising the acceptor stem that could be charged (156, 157) by extant aminoacyl-tRNA synthetases without the need for the anticodon. The modern tRNA has the universal CCA sequence at its 3′ end, with the anticodon segment of the tRNA at a molecular distance of around 70 Å. Depending on which tRNA is associated with the PTC, the CCA end carries the incoming amino acid or the growing peptide chain. During the initial stages of evolution, the PTC and the CCA end of the tRNA likely coevolved together (155). Recently, an ancient insertion fingerprint in the tRNA was discovered that gives credence to the idea of the addition of the anticodon section (along with the D arm) to the older fragment comprising the CCA stem (and the T arm) (120).
PROTEINS STABILIZE THE RIBOSOMES BUT ARE NOT ESSENTIAL FOR PTC ACTIVITY
Ribosome stability is achieved through interactions between bases, metal ion coordination, and the interaction with rProteins. RNA-protein interactions are significant in the ribosome as a whole, as well as in the PTC. However, none of the rProteins alone seem to be indispensable for peptide bond formation (4). There are several rProteins, such as uL2, uL3, and uL4, that have unstructured segments which dive deep into the ribosome in areas near the PTC (6, 158–160). Other rProteins, namely, uL6, L10e, uL14, bL27, and L15e/L31, are also found in the general vicinity of the PTC (6, 21, 161). Structural studies on bacterial ribosomes suggest that P-site tRNA likely interacts with the tails of rProteins such as uL5, uL16, bL27, uS9, and uS13 (34, 161). In a photoaffinity labeling experiment, it was shown that while L1 and L33 are close to the 3′ end of the deacylated tRNA at the E-site, bL27 likely interacts with the 3′ end (A76) of the P-site tRNA (162). Deletion of only three residues of the N-terminal tail of bL27 resulted in the reduction of the PTC activity (159), as does the deletion of uL16 (163, 164). However, in both cases, the PTC activity is not abolished entirely. It should also be noted that bL27 is a bacterium-specific protein with no homolog in Archaea or Eukaryota and hence not available to facilitate peptidyl transfer until after LUCA. It has been shown that uL16 is important for the proper placement of the tRNA in the A-site (34). Furthermore, in a more recent study, when 5S rRNA is depleted, uL16 is not incorporated into the LSU, which in turn disrupts the correct assembly of the PTC region in the modern ribosomes (165). However, this may not have been a necessity in the rudimentary stages of the PTC, when it was far from the excellent protein factory, which it later evolved to be. Thus, all of these rProteins are possibly the product of ribosome evolution in a protein-dominated world (158, 166, 167). Therefore, it is natural to consider other more elemental forces, like base-base interactions and metal ion coordination, that likely played supportive roles at the very early stages of PTC evolution.
METAL IONS AT THE PTC IN EXTANT RIBOSOMES
It is well established that metal ions such as magnesium, calcium, sodium, and potassium are biologically important in general and particularly for RNA (168). Among monovalent cations, potassium (K+) seems to be the most common and the most effective in stabilizing RNA structures in early folding stages or in the absence of divalent cations (169). It has been suggested that K+ ions play a major role in RNA condensation because they promote the formation of secondary structure (168). This most likely occurs because early neutralization of the intrinsic negative charge from the phosphate backbone allows RNA molecules to collapse into more compact structures. In the context of the ribosome, the importance of K+ ions was established when mammalian ribosomes were observed to lose polymerizing activity in the absence of potassium (170, 171). The specific occurrence of K+ ions in the PTC region was first reported in the 50S subunit of the archaeon Haloarcula marismortui. In this peculiar case, the K+ ion at the PTC region showed planar coordination with ion base distances that are consistent with the presence of other K+ ions (35). Recently, the relative abundance of K+ ions in the Thermus thermophilus ribosome and within its PTC has been determined using long-wavelength X-ray crystallography (172). Two independent studies, one by Klein et al. (166) and another by Rozov et al. (172), arrived at similar inferences about K+ ions. One K+ ion was assigned to the PTC (coordinating to G2061, G2447, C2501, and U2503), and another was assigned to the H11 of the 23S rRNA (coordinating to C192, U193, and A202) (166, 172). Overall, K+ ions interact with either the carbonyl groups on bases or the hydroxyl group of a ribose (173).
Among divalent cations, magnesium (Mg2+) is thought to be the most suitable for neutralizing the intrinsic negative charge of the RNA phosphate backbone due to its abundant intracellular concentrations (174). In addition, Mg2+ ions possess the highest charge density amid biologically available ions, a feature that seems to enhance its bonding to RNA (175, 176). Two types of magnesium bonding to RNA have been described, and they represent the current basis for the understanding of the Mg2+ ion interaction with rRNA (166, 168). The first type involves “diffuse binding,” which provides long-range charge screening that overcomes electrostatic repulsion between the RNA phosphate backbone and the hexahydrate Mg2+, essentially following Coulomb’s law. The second type is “site binding,” which involves specific coordination of anionic ligands to a particular dehydrated state of Mg2+. Site binding involves two subtypes. In the first, the “site-bound, outer sphere” subcategory, Mg2+ ions interact with the RNA but do not form direct contacts. Instead, the water ligands around Mg2+ ions interact with the bases and the phosphate backbone to stabilize specific RNA motifs. The second, the “site-bound, inner sphere” subcategory, is where Mg2+ ions directly interact with the bases, the sugar, and the backbone of RNA.
The role of magnesium in the structure and function of the ribosome has been well documented (177). As early as in the 1950s, it was reported that the association of a functional ribosome depended heavily on Mg2+ concentrations (178, 179). Magnesium was next established as indispensable for the cellular ribosomal maintenance (180), the structural integrity of the ribosome (181), and eventually for the PTC reaction (182). The presence of hydrated Mg2+ ions was first reported in the Deinococcus radiodurans (bacterium) 50S subunit (74). In a further analysis of ribosome crystallographic structures, the role of Mg2+ ions in assisting the interaction of distal segments of rRNA by coordination of phosphate groups and its ability to mediate protein interaction with rRNA was recognized (183). Mg2+ ions are particularly abundant in the region surrounding the PTC, where they form magnesium microclusters (184). In one model featuring the structure of the E. coli (bacterial) ribosome at 3.5-Å resolution, 170 Mg2+ ions per ribosome were reported (185). Overall, metal ions like Mg2+ are concentrated in functional regions, such as the PTC, where they stabilize RNA chains at places that rProteins cannot access (166) (Fig. 3). Recently, the conservation of these Mg2+ interactions in and around the PTC across diverse phylogenetic groups has been confirmed, and a plausible role of these Mg2+ interactions in the early evolution of the PTC has been proposed (68). Going forward, a more judicious use of stereochemical principles applied to structures with higher resolution is expected to better resolve the nature and identity of the ions in ribosomes (186).
FIG 3.
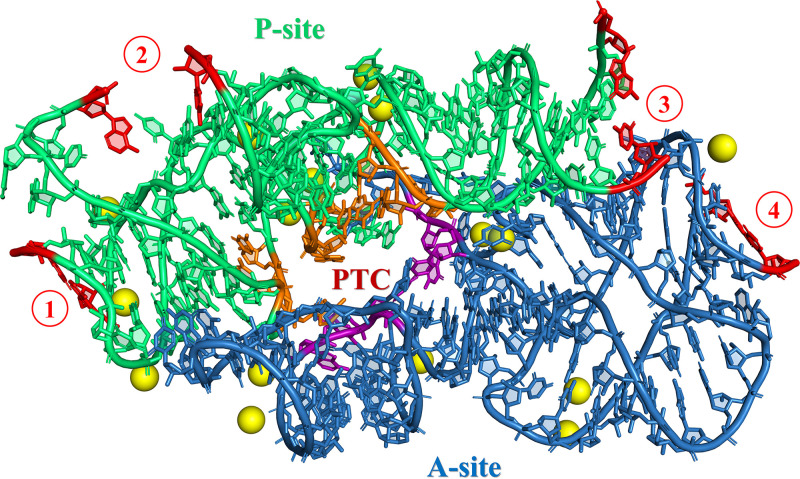
Three-dimensional representation of the pseudosymmetrical region (SymR; Agmon et al. [250]) extracted from the Thermus thermophilus crystallographic structure (NCBI ID 4WPO) using PyMOL (The PyMOL Molecular Graphics System, version 2.0; Schrödinger, LLC). The residues from the P-site and the A-site are colored green and blue, respectively. Residues delineating the entrance to the PTC pore are colored orange (P-site) and purple (A-site), as defined by Fox et al. (22). Sixteen conserved magnesium ions that are around the contact area of the SymR residues are shown as yellow spheres, as described by Rivas and Fox (68). Regions where the addition of fragments is required for the ligation of smaller fragments are highlighted by red numbers within red circles at the end of each helix, three at the P-site and one at the A-site. Also, the last residues of each ligation point are colored red. A secondary structure depiction of these elements is presented in Fig. 4.
METALS AND THE PROTO-RIBOSOME ON THE EARLY EARTH
Although Mg2+ is recognized as an essential cofactor with diverse structural and functional roles within the modern ribosome, conditions on the early Earth may have favored other ions. In particular, the ancient Earth would have provided environments with low O2 concentrations (187–189) and high ferrous iron (Fe2+) and manganese (Mn2+) availability (190, 191). By replacing Mg2+ with Fe2+, the catalytic abilities of several RNA molecules are expanded, including a previously uncharacterized ability to catalyze single-electron transfer (192). Under anaerobic conditions, Mg2+ can degrade RNA (193). Evidence that the role in folding and catalysis of Mg2+ can be replaced by Fe2+ has been presented from both quantum mechanical calculations and experiments with several ribozymes (194). Fe2+ and Mn2+ not only could aid rRNA folding but also could replace Mg2+ as the cation during translation under experimental conditions (195). These results suggest that within the anoxic environment of the early Earth, other metallic ions like Fe2+ could have been interacting with earlier versions of the ribosome, including its catalytic core, the PTC. Irrespective of the metal involved, what matters is the positioning of the various players, including the CCA end of the tRNAs and the highly conserved residues of the PTC. If experimental evidence supports the notion that the PTC activity is flexible with its metal occupants, then it would provide further support for the idea that the PTC was present during the anoxic chemical era of early Earth. It would also show that its metal-accommodating ability is a function rooted in its early origins.
THE PTC IS A DYNAMIC PEPTIDE SYNTHESIS MACHINE
Multiple lines of evidence have established that the extant PTC region is an RNA “engine” powering the peptide bond formation reaction (196–198). The PTC has not only virtually survived unchanged over 4 billion years of evolution but hasn also remained central to modern biology (15, 24, 119, 125, 199, 200). The modern PTC is postulated to act as an entropy trap platform, which ensures a certain three-dimensional placement/orientation of the substrate molecules (201, 202).
Molecular biology has been described as “a brilliantly staged molecular theater” (203). If so, the PTC sets the stage for a molecular “theater” of a kind, involving the following molecules, namely, the incoming tRNA charged with an amino acid and the nascent peptide linked to the 3′ hydroxyl end of the 3′ terminal ribose of the P-site by an ester bond (24, 204). The nucleophilic α-amino group of the A-site aminoacyl-tRNA breaks the electrophilic ester carbonyl carbon of the peptidyl-tRNA (aminolysis) (205). This results in the transfer of the peptidyl chain to the aminoacyl-tRNA at the A-site. This process is thought to involve the movement of three protons in the PTC along a proton wire (206). An initial deprotonation of the α-NH3+ of the A-site tRNA creates the α-NH2. Then, a proton is further removed from this amine along the proton wire, and at the end of the reaction, another proton is made available to the 3′-oxygen of the A76 P-site tRNA ribose.
In a significant study using quantum ab initio calculations, supported by crystal structure data, conserved water molecules “trapped” in the PTC region have been shown to be important players as well. One water molecule plays a mediatory role in this proton shuttle, while another stabilizes the negative charge on the substrate oxyanion (207). Water molecules at the PTC are thus thought to be important for maintaining the thermodynamic parameters of the reaction. Another player is the 2′-OH of the peptidyl-tRNA substrate, which is thought to ensure an efficient proton movement via a network of hydrogen bonds (208, 209). The P-site A76 2′-OH is placed in close proximity to the attacking α-amine and the O3′ leaving group and is thus optimally positioned to enable efficient catalysis (69, 82, 210, 211) (Fig. 2). This 2′-OH is important, irrespective of whether the LSU is by itself or is part of the whole ribosome unit along with the SSU (209, 212, 213). While the 2′-OH of the A76 ribose is essential for maintaining the reaction rate, the catalysis itself is attributed to the reduction of the activation free energy, achieved via electrostatic effects (214). A76 is also essential for peptide release and EF-Tu binding (215, 216).
This entire process goes on cyclically (3), and the peptidyl transferase reaction is thus pushed forward, resulting in the fairly rapid synthesis of 15 to 50 peptide bonds/s in a modern organism (217). This continues until a stop codon is reached, at which point ester hydrolysis breaks the bond between the P-site tRNA and the peptide. At each step, the growing peptide is released into an RNA pore (22, 23), which forms the entrance of the exit tunnel (218) that ultimately allows the finished peptide to get out of the way and leave the ribosome. Thus, the nanopore at the PTC acts like a molecular womb, where not only is the peptide formed, but it is also given an exit. The uniqueness of this pore at the PTC was demonstrated in a study which examined multiple “pores” formed and found in rRNAs as well as other RNAs (23). The nanopore at the PTC region is formed by RNA residues in which the nitrogenous bases are exposed. The phosphate backbone is twisted away from the lumen of the pore. Significantly, in contrast with the PTC pore, other known pores feature the nitrogenous bases twisting away, leaving the phosphate backbone to line the lumen of the pore (23, 219). Although much progress has been made in understanding ribosome structure and function, the actual chemistry of the PTC continues to be deciphered (215, 220–224).
In the modern ribosome when there are obstacles in the tunnel, peptide growth is inhibited. The coordinated train of movement of the peptide then stalls and prevents the PTC from functioning. This has been amply demonstrated with antibiotics (225–232). Antibiotic-enabled disruption of any of the players in the ribosome PTC arena, including their respective three-dimensional positioning of acceptor or donor substrates, results in a stalled or dysfunctional ribosome (24, 233, 234). Growing nascent peptides can stall the PTC activity by interacting with the exit tunnel (234–240). In its early stages, an evolving PTC would not have the fully grown ribosome with an exit tunnel to contend with. Therefore, its efficiency in making peptides would have been centered on a relatively “simple” process of a “substrate in” and a “product out.” However, when accretion events occurred, the exit tunnel might have been blocked, thereby preventing peptide synthesis. Thus, selection would favor mutations that would accommodate the peptide chain’s exit, through any obstructive expansion segments. The exit tunnel’s interactions with the growing peptide and the related stalling are thus likely consequences of ribosomal evolution (241). As the genetic code was established, the nature of the nascent peptide as well as the quality of the mRNA also played vital roles in stalling events (242–245). In order to solve the problems arising from such stalling, systems like ribosome rescue may have evolved (243, 246–249). The early PTC likely did not have to deal with such problems.
DISSECTING THE STRUCTURE OF THE PTC REVEALS ITS ORIGINS FROM SIMPLER FORMS
Close examination of the high-resolution structure of the entire domain 5 revealed the presence of a region of pseudosymmetry now known as SymR (250, 251) (Fig. 3 and 4). The two LSU rRNA fragments that define this region correspond to the A-site and P-site segments of the extant PTC (250, 251). These segments are listed as occurring in phase 1 and phase 2 in the accretion model (120). In addition, within the ribosome, the complex forms an RNA pore (22, 23). Overall, the level of conservation observed among the residues of the extant PTC (112, 252, 253) is at many locations extremely high (Fig. 1).
FIG 4.
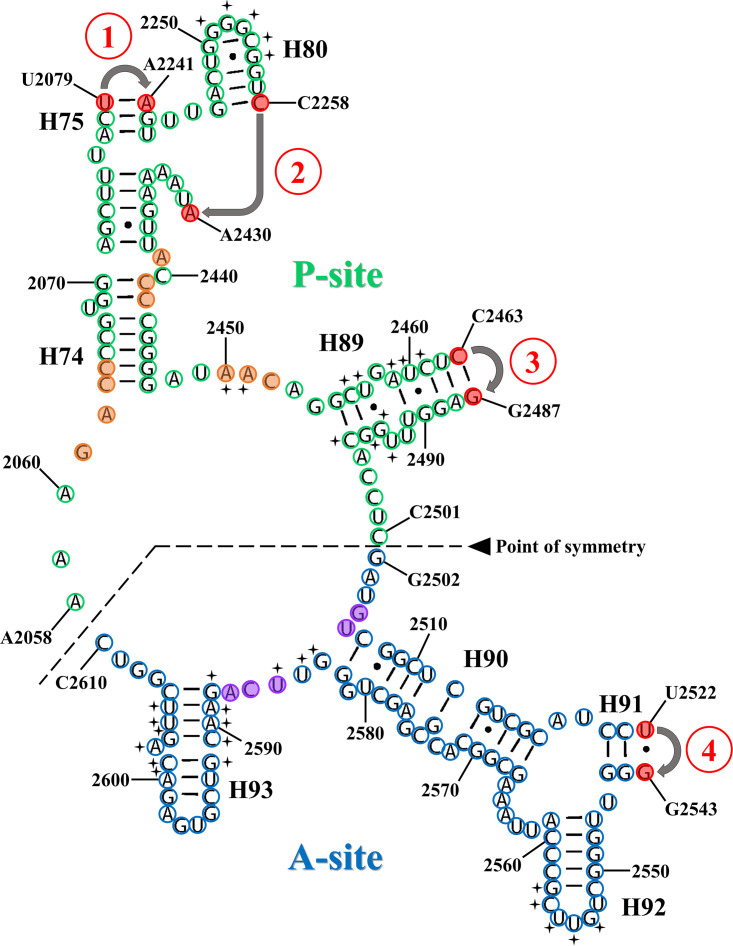
Secondary structure of the pseudosymmetrical region (SymR; Agmon et al. [250]), derived from the LSU secondary structure of Thermus thermophilus (Petrov et al. [19]). A black dotted line indicates the boundary between the P-site and the A-site. Bases from the P-site are highlighted by green circles, and bases from the A-site are highlighted by blue circles. The orange (P-site) and purple (A-site) colored circles highlight residues that delineate the entrance to the exit pore of the PTC, as defined by Fox et al. (22). In order to dissect SymR from the extant ribosomal structure, one must artificially cut and anneal the four sites highlighted by red numbers within red circles, three at the P-site (H75, H80, and H89) and one at the A-site (H91). Also, the last residues of each ligation point are highlighted by red circles. Bases involved in the “rotatory motion” mechanism from the front and rear walls and the P- and A-loops are marked by a black plus symbol in front of each base according to Agmon et al. (251) and Bashan et al. (74, 256).
This initial observation immediately led to the hypothesis that an ancient duplication and dimerization event had created a functional proto-ribosome (2, 250, 251, 254, 255) that was “frozen” and is now preserved in the modern PTC. In an elegant series of papers (250, 251, 254, 255), the Yonath group have proposed a detailed model of how the modern ribosome works. Grounded on the analysis of several ribosomal crystallographic structures, they proposed a “rotatory motion” mechanism that describes the tRNA passage from the A-site to the P-site. This requires two independent motions: (i) a helical rotation of ∼180° of the A-site tRNA-3′ coupled with the peptide bond formation (Fig. 2) and (ii) an A-to-P-site translation of the tRNA acceptor steam (74, 256). As stated by Yonath in 2003 (257), the most significant biological implication of the rotatory motion seems to be the establishment of favorable stereochemistry for peptide bond formation without drastic conformational rearrangements at the PTC. Due to its helical nature, the entrance of the nascent peptide into the ribosomal exit tunnel is ensured. Peptide bond formation, translocation, and nascent protein progression seemed to be the result of this rotatory motion, although it is not clear if peptide bond formation is triggered by the motion or the motion is triggered by the reaction (257). Yonath and coworkers have recognized the participation of several nucleotides in the proposed rotatory motion, most of which are contained within SymR (74, 250, 251, 256, 257) (Fig. 4).
From an evolutionary perspective, the rotation model depends strongly on the ability of the two fragments to anneal to one another. Each fragment by itself is stabilized by several standard base pairs and Mg2+ ions (Fig. 3). However, a cursory observation fails to find standard base pairs that could stabilize the proposed association of the two pieces. Likely as a result of this and the difficulty until recently of synthesizing RNAs in the 80–100-nucleotide size range, the hypothesis has largely been ignored. However, in fact, six conserved regions of nonstandard interactions have been identified that could facilitate association of the two RNAs (68, 250, 254). Among these, long-range interactions seem to be a dominant theme. Most of the interactions are represented by splayed-apart bases that due to their peculiar nature are able to establish several hydrogen bond contacts between the A and P fragments. In addition, there are several highly conserved Mg2+ ions that interact with the two fragments (Fig. 3). This includes two conserved Mg2+ ions that connect with both regions and that might have played a role in the association and stabilization of the two segments. Even further, one of these Mg2+ ions is in a position where it not only fixed the end of the P-site chain but can also have played a role as a potential catalytic element that enhanced the ligation of the two aboriginal fragments (68). It has been proposed that together, these interactions can stabilize an association between the two fragments. To test this hypothesis, computational studies of such an RNA pair were conducted using quantum kernel energy parameters. It was found that the two fragments could indeed be expected to hybridize (2). Adding to the optimism is the fact that examples exist in extant biology, where the LSU rRNA is expressed as multiple fragments that come together to assemble the ribosome (258, 259). This includes the PTC being formed by the association of two independent RNA pieces in both the Euglena gracilis cytosolic ribosome (260) and the Chlamydomonas reinhardtii mitochondrial ribosome (261).
Like its modern version, the hypothetical proto-ribosome would actually perform two tasks, not just peptide bond formation. In the prebiotic world, after the initial task of formation of the first peptide dimer, the question that follows is whether it will be extended to a tripeptide or be released as a dipeptide. Extension seems favorable, as the rate of synthesis is higher than the rate of peptide release (218). However, the ancient machine would soon clog unless there was a mechanism to keep the growing peptide out of the way. Thus, after a peptide bond is formed between two starter amino acids, the newly made dipeptide might begin to enter the RNA pore that is formed by the association of the two segments of the pseudosymmetrical region and then move along via the pore as synthesis continues. Thus, in the proto-ribosome and its modern descendants, the process would again not be just synthesis but polymerization as well. The primitive “entropy trap” would only need activated amino acids to make peptide bonds to extend the peptide.
However, as they exist in the modern ribosome, the two fragments are connected to the remainder of the LSU rRNA. To experimentally test the dimerization theory, it is necessary to synthesize RNAs that in effect remove the two hypothesized regions from the PTC rRNA as two independent RNAs. This requires the addition of three small RNA connectors to the P-site RNA in order to shorten helixes 75 and 89 and to connect H80 to H74 and one addition to the A-site RNA to shorten H91 (Fig. 3 and 4). By shortening H75, one removes the L1 stalk, and by dissecting and connecting H80 to H74, one removes the rRNA that corresponds to the central protuberance. Deletion of the whole helix 73 would be the final step in dissecting the SymR from the LSU rRNA. It is not clear how critical the specific choice of the additions and cuts would be.
CAN THE PTC BE BUILT FROM ITS SIMPLER COMPONENTS WHILE RETAINING ACTIVITY?
Recently, Xu and Wang have shown that a slightly larger version of two fragments encompassing the proto-ribosome can in fact form a stable complex (262). Subsequently, a seven-residue fragment ending with CCA and carrying lysine at its 3′ end was incubated with the two-piece RNA construct. A nine-residue polylysine was apparently formed (263). If this is correct, in essence, both peptide bond formation and polymerization had occurred as in the modern ribosome (263). If these findings survive scrutiny, our understanding of the prebiotic world will be enhanced. There is, however, concern that products of shorter or longer size were not observed.
Having previously shown that the large subunit alone could catalyze peptide bond formation (31), Noller pointedly observed in 1993 that the intricately intertwined RNA-protein “cage” in the large subunit would make further progress toward separating the RNA from the protein and still make the former retain its catalytic activity, a worthy challenge (264). This has proven to be true, as several groups have reportedly made multiple attempts with at best partial success. However, with the exception of the effort of Xu and Wang (262), they have not reached the mainstream literature. In terms of future studies, much depends on a convincing demonstration that the still hypothetical proto-ribosome would have been functional. This would, for example, strengthen arguments for the operational code (153, 154, 265). Also, the menu of likely follow-on studies will have to be directed toward understanding how two initially independent fragments came to be covalently linked and when, in the larger scheme of ribosomal evolution, this occurred. Such linkage would likely be an important step in stabilizing the evolving proto-ribosome. It has been suggested that translation evolved to extend the structural and functional capabilities of the hypothetical RNA world (167) and, during the course of its evolution, might have terminated the RNA world (219). How does the evolving system accrete what ultimately becomes the ribosome? And, of course, can one simplify the system further?
The existence of an active proto-ribosome would also set the stage to inquire about the potential role of the peptides produced. Nothing dramatic is thought to have been associated with early peptide functionality, and a compelling argument might be that the proto-ribosome would simply make peptides because that is what it did, in a random “nonutilitarian”/“nonspecific” fashion, and not because they were useful. But they might have been. Recent studies have shown that even extremely small peptides can perform a large variety of functions (266, 267). At the least, peptides made by the proto-ribosome might simply have augmented the ancestral PTC. For example, this could have been accomplished by building layers (7) and using the peptides to stabilize the emerging PTC. Thus, the activity would represent the possible beginnings of a transition from an “RNA world” to an “RNA/peptide world.” Studies of the ribosomal protein structures support this, as they become increasingly more complex as one proceeds from the older regions of the ribosome to the newer (268–271).
In summary, all of this reminds us that there must have been a predecessor to the modern PTC. This premise is based on the nature of RNA structure itself, since small RNAs derived from larger RNAs can still maintain their local structures (7). Thus, in the prebiotic context, simpler, smaller RNAs could have given rise to the PTC through association (272, 273). Sulima and Dinman in 2019 referred to the evolution of a proto-ribosome, forming the primordial ribosomal core, approximately 4 billion years ago as the “Big Bang” of the riboverse (86). Understanding the origin and properties of that ancestor will be a major advance and should be obtainable if the community continues to pursue it. Quoting Darwin “… from so simple a beginning endless forms most beautiful and most wonderful have been, and are being, evolved …” (274).
ACKNOWLEDGMENTS
This work was supported in part by NASA contract 80NSSC18K1139 under the Center for the Origin of Life, Georgia Institute of Technology, to George E. Fox. Mario Rivas Medrano’s research was supported by an appointment to the NASA Postdoctoral Program at the NASA Astrobiology Institute, administered by Universities Space Research Association under contract with NASA.
We declare no conflicts of interest.
Author order was determined on the basis of contribution. M.R.T., M.R., and G.E.F. designed the structure and content of the review. M.R.T., M.R., and Q.T. performed literature research. M.R.T. and G.E.F. wrote the manuscript with substantial contributions from M.R. M.R. generated the figures. All authors critically reviewed and revised manuscript and read and approved the final manuscript.
Biographies

Madhan R. Tirumalai is a Research Assistant Professor at the University of Houston, TX. He obtained his Ph.D. from Goa University, India, in 2005. He worked at the Institute of Bioinformatics in Bengaluru, India, before receiving a NASA Planetary Biology internship to work in the group of Dr. George E. Fox at the University of Houston. Continuing as a postdoc in the same group, he worked on the genomes of the spacecraft assembly facility isolates Bacillus pumilus SAFR-032, Bacillus safensis F036b, and B. safensis JPL-MERTA-8-2. The radiation- and desiccation-resistant spores produced by these organisms are a significant planetary protection concern. Following that, he completed an experimental study of the long-term effects of simulated microgravity on microorganisms. He is now part of an effort towards understanding the origins and evolution of the modern protein synthesis machinery, namely, the ribosomes. He also contributes to education and public outreach activities.

Mario Rivas received his Bachelor’s degree in biology and Ph.D. in biomedicine from The National Autonomous University of Mexico (UNAM), Mexico. He is currently a NASA Postdoctoral Fellow associated with the NASA Astrobiology Institute (NAI) and the Center for the Origin of Life (COOL) based at the Fox lab in the Biology and Biochemistry Department of the University of Houston, Texas, USA. He has been a member of the International Society for the Study of the Origin of Life (ISSOL) since 2011 and was a NASA Planetary Biology Intern in 2012. His research interest focuses on the origin and early evolution of life from the perspective of molecular evolution. The scope of his research ranges from the early evolution of the metabolic pathways to the origin and early evolution of the ribosome.

Quyen Tran received his B.Sc. and Ph.D. from the University of Houston and is currently conducting his postdoctoral work under George E. Fox. He is interested in bioinformatics, specifically the meta-analysis of changes in bacterial genomes, and dynamical chemical systems that can lead to the emergence of information and propagation and storage of said information. Notably, he was part of the collaborative effort by JPL to catalog the microbial burden and populational dynamics of the International Space Station (ISS). Most recently, he also participated in a project whereby a special biochemical model was used to model RNA sequence space exploration in prebiotic environments. The model utilized the dynamical process of ligation and hydrolysis carried out by T4-RNA ligase and Benzonase, respectively, on an initial population of short RNAs to combinatorially generate diversity.

George E. Fox earned his B.Sc. and Ph.D. in chemical engineering at Syracuse University. As a postdoctoral fellow with Carl R. Woese at the University of Illinois, he was the codiscoverer of the Archaea, the progenote concept, and the use of comparative sequence analysis to predict RNA secondary structure. In 1977, Fox joined the University of Houston, where he remains an Emeritus Moores Professor of Biology and Biochemistry. In 1980, Fox, Woese, and collaborators published the first comprehensive tree of bacterial relationships. In “How Close Is Close: 16S rRNA Sequence Identity May Not Be Sufficient To Guarantee Species Identity” (G. E. Fox, J. D. Wisotzkey, and P. Jurtshuk, Jr., Int J Syst Evol Microbiol 42:166–170, 1992, https://doi.org/10.1099/00207713-42-1-166), Fox recognized the limitations of 16S rRNA sequence comparisons. His current research is focused on ribosome function and evolution. He is an elected fellow of the American Academy of Microbiology, the American Association for the Advancement of Science, the American Institute for Medical and Biological Engineering, and the International Astrobiology Society. He chaired the 2010 Origin of Life Gordon Conference and has over 180 peer-reviewed publications.
REFERENCES
- 1.Fox GE. 2010. Origin and evolution of the ribosome. Cold Spring Harb Perspect Biol 2:a003483. 10.1101/cshperspect.a003483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang L, Krupkin M, Bashan A, Yonath A, Massa L. 2013. Protoribosome by quantum kernel energy method. Proc Natl Acad Sci USA 110:14900–14905. 10.1073/pnas.1314112110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Steitz TA. 2008. A structural understanding of the dynamic ribosome machine. Nat Rev Mol Cell Biol 9:242–253. 10.1038/nrm2352. [DOI] [PubMed] [Google Scholar]
- 4.Cech TR. 2000. Structural biology. The ribosome is a ribozyme. Science 289:878–879. 10.1126/science.289.5481.878. [DOI] [PubMed] [Google Scholar]
- 5.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 289:905–920. 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 6.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289:920–930. 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 7.Noller HF. 2012. Evolution of protein synthesis from an RNA world. Cold Spring Harb Perspect Biol 4:a003681. 10.1101/cshperspect.a003681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steitz TA, Moore PB. 2003. RNA, the first macromolecular catalyst: the ribosome is a ribozyme. Trends Biochem Sci 28:411–418. 10.1016/S0968-0004(03)00169-5. [DOI] [PubMed] [Google Scholar]
- 9.Green R, Noller HF. 1997. Ribosomes and translation. Annu Rev Biochem 66:679–716. 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 10.Woese C. 1998. The universal ancestor. Proc Natl Acad Sci USA 95:6854–6859. 10.1073/pnas.95.12.6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harris JK, Kelley ST, Spiegelman GB, Pace NR. 2003. The genetic core of the universal ancestor. Genome Res 13:407–412. 10.1101/gr.652803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bernier CR, Petrov AS, Kovacs NA, Penev PI, Williams LD. 2018. Translation: the universal structural core of life. Mol Biol Evol 35:2065–2076. 10.1093/molbev/msy101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mirkin BG, Fenner TI, Galperin MY, Koonin EV. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 3:2. 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delaye L, Becerra A, Lazcano A. 2005. The last common ancestor: what's in a name? Orig Life Evol Biosph 35:537–554. 10.1007/s11084-005-5760-3. [DOI] [PubMed] [Google Scholar]
- 15.Bowman JC, Petrov AS, Frenkel-Pinter M, Penev PI, Williams LD. 2020. Root of the tree: the significance, evolution, and origins of the ribosome. Chem Rev 120:4848–4878. 10.1021/acs.chemrev.9b00742. [DOI] [PubMed] [Google Scholar]
- 16.Ranea JA, Sillero A, Thornton JM, Orengo CA. 2006. Protein superfamily evolution and the last universal common ancestor (LUCA). J Mol Evol 63:513–525. 10.1007/s00239-005-0289-7. [DOI] [PubMed] [Google Scholar]
- 17.Cannone JJ, Subramanian S, Schnare MN, Collett JR, D'Souza LM, Du Y, Feng B, Lin N, Madabusi LV, Muller KM, Pande N, Shang Z, Yu N, Gutell RR. 2002. The comparative RNA web (CRW) site: an online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3:2. 10.1186/1471-2105-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brimacombe R. 2000. The bacterial ribosome at atomic resolution. Structure 8:R195–R200. 10.1016/S0969-2126(00)00510-4. [DOI] [PubMed] [Google Scholar]
- 19.Petrov AS, Bernier CR, Hershkovits E, Xue Y, Waterbury CC, Hsiao C, Stepanov VG, Gaucher EA, Grover MA, Harvey SC, Hud NV, Wartell RM, Fox GE, Williams LD. 2013. Secondary structure and domain architecture of the 23S and 5S rRNAs. Nucleic Acids Res 41:7522–7535. 10.1093/nar/gkt513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292:883–896. 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 21.Smith TF, Lee JC, Gutell RR, Hartman H. 2008. The origin and evolution of the ribosome. Biol Direct 3:16. 10.1186/1745-6150-3-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox GE, Tran Q, Yonath A. 2012. An exit cavity was crucial to the polymerase activity of the early ribosome. Astrobiology 12:57–60. 10.1089/ast.2011.0692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rivas M, Tran Q, Fox GE. 2013. Nanometer scale pores similar in size to the entrance of the ribosomal exit cavity are a common feature of large RNAs. RNA 19:1349–1354. 10.1261/rna.038828.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Polacek N, Mankin AS. 2005. The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol 40:285–311. 10.1080/10409230500326334. [DOI] [PubMed] [Google Scholar]
- 25.Prosdocimi F, José MV, de Farias ST. 2021. The theory of chemical symbiosis: a Margulian view for the emergence of biological systems (origin of life). Acta Biotheor 69:67–78. 10.1007/s10441-020-09388-7. [DOI] [PubMed] [Google Scholar]
- 26.Beringer M, Bruell C, Xiong L, Pfister P, Bieling P, Katunin VI, Mankin AS, Bottger EC, Rodnina MV. 2005. Essential mechanisms in the catalysis of peptide bond formation on the ribosome. J Biol Chem 280:36065–36072. 10.1074/jbc.M507961200. [DOI] [PubMed] [Google Scholar]
- 27.Sanbonmatsu KY. 2006. Alignment/misalignment hypothesis for tRNA selection by the ribosome. Biochimie 88:1075–1089. 10.1016/j.biochi.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 28.Wohlgemuth I, Beringer M, Rodnina MV. 2006. Rapid peptide bond formation on isolated 50S ribosomal subunits. EMBO Rep 7:699–703. 10.1038/sj.embor.7400732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wohlgemuth I, Brenner S, Beringer M, Rodnina MV. 2008. Modulation of the rate of peptidyl transfer on the ribosome by the nature of substrates. J Biol Chem 283:32229–32235. 10.1074/jbc.M805316200. [DOI] [PubMed] [Google Scholar]
- 30.Belousoff MJ, Davidovich C, Zimmerman E, Caspi Y, Wekselman I, Rozenszajn L, Shapira T, Sade-Falk O, Taha L, Bashan A, Weiss MS, Yonath A. 2010. Ancient machinery embedded in the contemporary ribosome. Biochem Soc Trans 38:422–427. 10.1042/BST0380422. [DOI] [PubMed] [Google Scholar]
- 31.Noller HF, Hoffarth V, Zimniak L. 1992. Unusual resistance of peptidyl transferase to protein extraction procedures. Science 256:1416–1419. 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 32.Spirin AS. 1999. Elongation cycle, step II: transpeptidation (peptide bond formation), p 195–211. In Ribosomes, 1st ed. Springer, Boston, MA. 10.1007/978-1-4615-7817-8_11. [DOI] [Google Scholar]
- 33.Beringer M, Rodnina MV. 2007. The ribosomal peptidyl transferase. Mol Cell 26:311–321. 10.1016/j.molcel.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 34.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. 2009. Insights into substrate stabilization from snapshots of the peptidyl transferase center of the intact 70S ribosome. Nat Struct Mol Biol 16:528–533. 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simonović M, Steitz TA. 2008. Cross-crystal averaging reveals that the structure of the peptidyl-transferase center is the same in the 70S ribosome and the 50S subunit. Proc Natl Acad Sci USA 105:500–505. 10.1073/pnas.0711076105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Noller HF. 1993. tRNA-rRNA interactions and peptidyl transferase. FASEB J 7:87–89. 10.1096/fasebj.7.1.8422979. [DOI] [PubMed] [Google Scholar]
- 37.Muth GW, Ortoleva-Donnelly L, Strobel SA. 2000. A single adenosine with a neutral pKa in the ribosomal peptidyl transferase center. Science 289:947–950. 10.1126/science.289.5481.947. [DOI] [PubMed] [Google Scholar]
- 38.Bocchetta M, Xiong L, Mankin AS. 1998. 23S rRNA positions essential for tRNA binding in ribosomal functional sites. Proc Natl Acad Sci USA 95:3525–3530. 10.1073/pnas.95.7.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thompson J, Kim DF, O'Connor M, Lieberman KR, Bayfield MA, Gregory ST, Green R, Noller HF, Dahlberg AE. 2001. Analysis of mutations at residues A2451 and G2447 of 23S rRNA in the peptidyltransferase active site of the 50S ribosomal subunit. Proc Natl Acad Sci USA 98:9002–9007. 10.1073/pnas.151257098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polacek N, Gaynor M, Yassin A, Mankin AS. 2001. Ribosomal peptidyl transferase can withstand mutations at the putative catalytic nucleotide. Nature 411:498–501. 10.1038/35078113. [DOI] [PubMed] [Google Scholar]
- 41.Polacek N, Swaney S, Shinabarger D, Mankin AS. 2002. SPARKA novel method to monitor ribosomal peptidyl transferase activity. Biochemistry 41:11602–11610. 10.1021/bi026040s. [DOI] [PubMed] [Google Scholar]
- 42.Polacek N, Gomez MJ, Ito K, Xiong L, Nakamura Y, Mankin A. 2003. The critical role of the universally conserved A2602 of 23S ribosomal RNA in the release of the nascent peptide during translation termination. Mol Cell 11:103–112. 10.1016/s1097-2765(02)00825-0. [DOI] [PubMed] [Google Scholar]
- 43.Beringer M, Adio S, Wintermeyer W, Rodnina M. 2003. The G2447A mutation does not affect ionization of a ribosomal group taking part in peptide bond formation. RNA 9:919–922. 10.1261/rna.5600503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Youngman EM, Brunelle JL, Kochaniak AB, Green R. 2004. The active site of the ribosome is composed of two layers of conserved nucleotides with distinct roles in peptide bond formation and peptide release. Cell 117:589–599. 10.1016/s0092-8674(04)00411-8. [DOI] [PubMed] [Google Scholar]
- 45.Erlacher MD, Lang K, Shankaran N, Wotzel B, Huttenhofer A, Micura R, Mankin AS, Polacek N. 2005. Chemical engineering of the peptidyl transferase center reveals an important role of the 2'-hydroxyl group of A2451. Nucleic Acids Res 33:1618–1627. 10.1093/nar/gki308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joseph S, Noller HF. 1996. Mapping the rRNA neighborhood of the acceptor end of tRNA in the ribosome. EMBO J 15:910–916. 10.1002/j.1460-2075.1996.tb00425.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bayfield MA, Dahlberg AE, Schulmeister U, Dorner S, Barta A. 2001. A conformational change in the ribosomal peptidyl transferase center upon active/inactive transition. Proc Natl Acad Sci USA 98:10096–10101. 10.1073/pnas.171319598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cruz-Vera LR, Gong M, Yanofsky C. 2006. Changes produced by bound tryptophan in the ribosome peptidyl transferase center in response to TnaC, a nascent leader peptide. Proc Natl Acad Sci USA 103:3598–3603. 10.1073/pnas.0600082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Triman KL. 1999. Mutational analysis of 23S ribosomal RNA structure and function in Escherichia coli. Adv Genet 41:157–195. 10.1016/s0065-2660(08)60153-4. [DOI] [PubMed] [Google Scholar]
- 50.Sanbonmatsu KY, Joseph S, Tung C-S. 2005. Simulating movement of tRNA into the ribosome during decoding. Proc Natl Acad Sci USA 102:15854–15859. 10.1073/pnas.0503456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rakauskaite R, Dinman JD. 2011. Mutations of highly conserved bases in the peptidyltransferase center induce compensatory rearrangements in yeast ribosomes. RNA 17:855–864. 10.1261/rna.2593211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Burakovsky DE, Sergiev PV, Steblyanko MA, Kubarenko AV, Konevega AL, Bogdanov AA, Rodnina MV, Dontsova OA. 2010. Mutations at the accommodation gate of the ribosome impair RF2-dependent translation termination. RNA 16:1848–1853. 10.1261/rna.2185710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Green R, Samaha RR, Noller HF. 1997. Mutations at nucleotides G2251 and U2585 of 23 S rRNA perturb the peptidyl transferase center of the ribosome. J Mol Biol 266:40–50. 10.1006/jmbi.1996.0780. [DOI] [PubMed] [Google Scholar]
- 54.Samaha RR, Green R, Noller HF. 1995. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature 377:309–314. 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 55.Feinberg JS, Joseph S. 2006. A conserved base-pair between tRNA and 23 S rRNA in the peptidyl transferase center is important for peptide release. J Mol Biol 364:1010–1020. 10.1016/j.jmb.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 56.Brunelle JL, Youngman EM, Sharma D, Green R. 2006. The interaction between C75 of tRNA and the A loop of the ribosome stimulates peptidyl transferase activity. RNA 12:33–39. 10.1261/rna.2256706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim DF, Green R. 1999. Base-pairing between 23S rRNA and tRNA in the ribosomal A site. Mol Cell 4:859–864. 10.1016/s1097-2765(00)80395-0. [DOI] [PubMed] [Google Scholar]
- 58.Bakin A, Lane BG, Ofengand J. 1994. Clustering of pseudouridine residues around the peptidyltransferase center of yeast cytoplasmic and mitochondrial ribosomes. Biochemistry 33:13475–13483. 10.1021/bi00249a036. [DOI] [PubMed] [Google Scholar]
- 59.Decatur WA, Fournier MJ. 2002. rRNA modifications and ribosome function. Trends Biochem Sci 27:344–351. 10.1016/s0968-0004(02)02109-6. [DOI] [PubMed] [Google Scholar]
- 60.Schnare MN, Gray MW. 2011. Complete modification maps for the cytosolic small and large subunit rRNAs of Euglena gracilis: functional and evolutionary implications of contrasting patterns between the two rRNA components. J Mol Biol 413:66–83. 10.1016/j.jmb.2011.08.037. [DOI] [PubMed] [Google Scholar]
- 61.Ofengand J, Del Campo M. 2004. Modified nucleosides of Escherichia coli ribosomal RNA. EcoSal Plus 1. 10.1128/ecosalplus.4.6.1. [DOI] [PubMed] [Google Scholar]
- 62.Sergiev PV, Golovina AY, Prokhorova IV, Sergeeva OV, Osterman IA, Nesterchuk MV, Burakovsky DE, Bogdanov AA, Dontsova OA. 2011. Modifications of ribosomal RNA: from enzymes to function, p 97–110. In Rodnina MV, Wintermeyer W, Green R (ed), Ribosomes: structure, function, and dynamics Springer, Vienna, Austria. 10.1007/978-3-7091-0215-2_9. [DOI] [Google Scholar]
- 63.Purta E, O'Connor M, Bujnicki JM, Douthwaite S. 2009. YgdE is the 2′-O-ribose methyltransferase RlmM specific for nucleotide C2498 in bacterial 23S rRNA. Mol Microbiol 72:1147–1158. 10.1111/j.1365-2958.2009.06709.x. [DOI] [PubMed] [Google Scholar]
- 64.Andersen TE, Porse BT, Kirpekar F. 2004. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA 10:907–913. 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Oerum S, Meynier V, Catala M, Tisné C. 2021. A comprehensive review of m6A/m6Am RNA methyltransferase structures. Nucleic Acids Res 49:7239–7255. 10.1093/nar/gkab378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.d'Aquino AE, Azim T, Aleksashin NA, Hockenberry AJ, Krüger A, Jewett MC. 2020. Mutational characterization and mapping of the 70S ribosome active site. Nucleic Acids Res 48:2777–2789. 10.1093/nar/gkaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hesslein AE, Katunin VI, Beringer M, Kosek AB, Rodnina MV, Strobel SA. 2004. Exploration of the conserved A+C wobble pair within the ribosomal peptidyl transferase center using affinity purified mutant ribosomes. Nucleic Acids Res 32:3760–3770. 10.1093/nar/gkh672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rivas M, Fox GE. 2020. Further characterization of the pseudo-symmetrical ribosomal region. Life (Basel) 10:201. 10.3390/life10090201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trobro S, Aqvist J. 2005. Mechanism of peptide bond synthesis on the ribosome. Proc Natl Acad Sci USA 102:12395–12400. 10.1073/pnas.0504043102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Whitford PC, Sanbonmatsu KY. 2013. Simulating movement of tRNA through the ribosome during hybrid-state formation. J Chem Phys 139:121919. 10.1063/1.4817212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Amunts A, Fiedorczuk K, Truong TT, Chandler J, Greenberg EP, Ramakrishnan V. 2015. Bactobolin A binds to a site on the 70S ribosome distinct from previously seen antibiotics. J Mol Biol 427:753–755. 10.1016/j.jmb.2014.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sato NS, Hirabayashi N, Agmon I, Yonath A, Suzuki T. 2006. Comprehensive genetic selection revealed essential bases in the peptidyl-transferase center. Proc Natl Acad Sci USA 103:15386–15391. 10.1073/pnas.0605970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Spahn CMT, Schäfer MA, Krayevsky AA, Nierhaus KH. 1996. Conserved nucleotides of 23 S rRNA located at the ribosomal peptidyltransferase center. J Biol Chem 271:32857–32862. 10.1074/jbc.271.51.32857. [DOI] [PubMed] [Google Scholar]
- 74.Bashan A, Agmon I, Zarivach R, Schluenzen F, Harms J, Berisio R, Bartels H, Franceschi F, Auerbach T, Hansen HAS, Kossoy E, Kessler M, Yonath A. 2003. Structural basis of the ribosomal machinery for peptide bond formation, translocation, and nascent chain progression. Mol Cell 11:91–102. 10.1016/s1097-2765(03)00009-1. [DOI] [PubMed] [Google Scholar]
- 75.d'Aquino A, Azim T, Aleksashin N, Hockenberry A, Hoang K, DeFoe A, Jewett M. 2019. Mutating the ribosome peptidyl transferase center in vitro. FASEB J 33:628.1. 10.1096/fasebj.2019.33.1_supplement.628.1. [DOI] [Google Scholar]
- 76.Vester B, Douthwaite S. 2001. Macrolide resistance conferred by base substitutions in 23S rRNA. Antimicrob Agents Chemother 45:1–12. 10.1128/AAC.45.1.1-12.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Erlacher MD, Lang K, Wotzel B, Rieder R, Micura R, Polacek N. 2006. Efficient ribosomal peptidyl transfer critically relies on the presence of the ribose 2'-OH at A2451 of 23S rRNA. J Am Chem Soc 128:4453–4459. 10.1021/ja0588454. [DOI] [PubMed] [Google Scholar]
- 78.Amort M, Wotzel B, Bakowska-Zywicka K, Erlacher MD, Micura R, Polacek N. 2007. An intact ribose moiety at A2602 of 23S rRNA is key to trigger peptidyl-tRNA hydrolysis during translation termination. Nucleic Acids Res 35:5130–5140. 10.1093/nar/gkm539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu CC, Schultz PG. 2010. Adding new chemistries to the genetic code. Annu Rev Biochem 79:413–444. 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 80.Kawakami T, Murakami H. 2012. Genetically encoded libraries of nonstandard peptides. J Nucleic Acids 2012:713510. 10.1155/2012/713510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rodnina MV. 2013. The ribosome as a versatile catalyst: reactions at the peptidyl transferase center. Curr Opin Struct Biol 23:595–602. 10.1016/j.sbi.2013.04.012. [DOI] [PubMed] [Google Scholar]
- 82.Schmeing TM, Huang KS, Strobel SA, Steitz TA. 2005. An induced-fit mechanism to promote peptide bond formation and exclude hydrolysis of peptidyl-tRNA. Nature 438:520–524. 10.1038/nature04152. [DOI] [PubMed] [Google Scholar]
- 83.Koshland DE. 1958. Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA 44:98–104. 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lehmann J. 2017. Induced fit of the peptidyl-transferase center of the ribosome and conformational freedom of the esterified amino acids. RNA 23:229–239. 10.1261/rna.057273.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nikolaeva DD, Gelfand MS, Garushyants SK. 2021. Simplification of ribosomes in bacteria with tiny genomes. Mol Biol Evol 38:58–66. 10.1093/molbev/msaa184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sulima SO, Dinman JD. 2019. The expanding riboverse. Cells 8:1205. 10.3390/cells8101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Norris K, Hopes T, Aspden JL. 2021. Ribosome heterogeneity and specialization in development. Wiley Interdiscip Rev RNA 12:e1644. 10.1002/wrna.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Poitevin F, Kushner A, Li X, Dao Duc K. 2020. Structural heterogeneities of the ribosome: new frontiers and opportunities for cryo-EM. Molecules 25:4262. 10.3390/molecules25184262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li D, Wang J. 2020. Ribosome heterogeneity in stem cells and development. J Cell Biol 219:e202001108. 10.1083/jcb.202001108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ford D. 2020. Ribosomal heterogeneity—a new inroad for pharmacological innovation. Biochem Pharmacol 175:113874. 10.1016/j.bcp.2020.113874. [DOI] [PubMed] [Google Scholar]
- 91.Petrov AS, Wood EC, Bernier CR, Norris AM, Brown A, Amunts A. 2019. Structural patching fosters divergence of mitochondrial ribosomes. Mol Biol Evol 36:207–219. 10.1093/molbev/msy221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sagan L. 1967. On the origin of mitosing cells. J Theoret Biol 14:225–274. 10.1016/0022-5193(67)90079-3. [DOI] [PubMed] [Google Scholar]
- 93.Woese CR. 1977. Endosymbionts and mitochondrial origins. J Mol Evol 10:93–96. 10.1007/BF01751802. [DOI] [PubMed] [Google Scholar]
- 94.Yang D, Oyaizu Y, Oyaizu H, Olsen GJ, Woese CR. 1985. Mitochondrial origins. Proc Natl Acad Sci USA 82:4443–4447. 10.1073/pnas.82.13.4443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.O'Brien TW. 2002. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene 286:73–79. 10.1016/S0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- 96.Greber BJ, Ban N. 2016. Structure and function of the mitochondrial ribosome. Annu Rev Biochem 85:103–132. 10.1146/annurev-biochem-060815-014343. [DOI] [PubMed] [Google Scholar]
- 97.Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, Young IG. 1981. Sequence and organization of the human mitochondrial genome. Nature 290:457–465. 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 98.van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, Frauenfeld J, Berninghausen O, Neupert W, Herrmann JM, Beckmann R. 2015. Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol 7:1235–1251. 10.1093/gbe/evv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ott M, Herrmann JM. 2010. Co-translational membrane insertion of mitochondrially encoded proteins. Biochim Biophys Acta 1803:767–775. 10.1016/j.bbamcr.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 100.Tomal A, Kwasniak-Owczarek M, Janska H. 2019. An update on mitochondrial ribosome biology: the plant mitoribosome in the spotlight. Cells 8:1562. 10.3390/cells8121562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. 2001. The large subunit of the mammalian mitochondrial ribosome: ANALYSIS OF THE COMPLEMENT OF RIBOSOMAL PROTEINS PRESENT*. J Biol Chem 276:43958–43969. 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- 102.Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. 2001. Structural compensation for the deficit of rRNA with proteins in the mammalian mitochondrial ribosome: systematic analysis of protein components of the large ribosomal subunit from mammalian mitochondria. J Biol Chem 276:21724–21736. 10.1074/jbc.M100432200. [DOI] [PubMed] [Google Scholar]
- 103.Waltz F, Soufari H, Bochler A, Giege P, Hashem Y. 2020. Cryo-EM structure of the RNA-rich plant mitochondrial ribosome. Nat Plants 6:377–383. 10.1038/s41477-020-0631-5. [DOI] [PubMed] [Google Scholar]
- 104.Waltz F, Nguyen TT, Arrive M, Bochler A, Chicher J, Hammann P, Kuhn L, Quadrado M, Mireau H, Hashem Y, Giege P. 2019. Small is big in Arabidopsis mitochondrial ribosome. Nat Plants 5:106–117. 10.1038/s41477-018-0339-y. [DOI] [PubMed] [Google Scholar]
- 105.Agrawal RK, Sharma MR, Yassin A, Lahiri I, Spremulli iL. 2011. Structure and function of organellar ribosomes as revealed by cryo-EM, p 83–96. In Rodnina MV, Wintermeyer W, Green R (ed), Ribosomes: structure, function, and dynamics Springer, Vienna, Austria. 10.1007/978-3-7091-0215-2_8. [DOI] [Google Scholar]
- 106.Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. 2003. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell 115:97–108. 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- 107.Amunts A, Brown A, Bai X-c, Llácer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V. 2014. Structure of the yeast mitochondrial large ribosomal subunit. Science 343:1485–1489. 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL. 2001. The small subunit of the mammalian mitochondrial ribosome: identification of the full complement of ribosomal proteins present. J Biol Chem 276:19363–19374. 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- 109.Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. 2014. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 515:283–286. 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 110.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. 2015. Ribosome. The structure of the human mitochondrial ribosome. Science 348:95–98. 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Baouz S, Woisard A, Sinapah S, Le Caer J-P, Argentini M, Bulygin K, Aguié G, Hountondji C. 2009. The human large subunit ribosomal protein L36A-like contacts the CCA end of P-site bound tRNA. Biochimie 91:1420–1425. 10.1016/j.biochi.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 112.Prosdocimi F, Zamudio GS, Palacios-Pérez M, Torres de Farias S, V José M. 2020. The ancient history of peptidyl transferase center formation as told by conservation and information analyses. Life (Basel, Switzerland) 10:134. 10.3390/life10080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Graifer D, Karpova G. 2012. Structural and functional topography of the human ribosome. Acta Biochim Biophys Sin (Shanghai) 44:281–299. 10.1093/abbs/gmr118. [DOI] [PubMed] [Google Scholar]
- 114.Clark CG. 1987. On the evolution of ribosomal RNA. J Mol Evol 25:343–350. 10.1007/BF02603119. [DOI] [PubMed] [Google Scholar]
- 115.Hury J, Nagaswamy U, Larios-Sanz M, Fox GE. 2006. Ribosome origins: the relative age of 23S rRNA domains. Orig Life Evol Biosph 36:421–429. 10.1007/s11084-006-9011-z. [DOI] [PubMed] [Google Scholar]
- 116.Bokov K, Steinberg SV. 2009. A hierarchical model for evolution of 23S ribosomal RNA. Nature 457:977–980. 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- 117.Nissen P, Ippolito JA, Ban N, Moore PB, Steitz TA. 2001. RNA tertiary interactions in the large ribosomal subunit: the A-minor motif. Proc Natl Acad Sci USA 98:4899–4903. 10.1073/pnas.081082398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Noller HF. 2005. RNA structure: reading the ribosome. Science 309:1508–1514. 10.1126/science.1111771. [DOI] [PubMed] [Google Scholar]
- 119.Hsiao C, Mohan S, Kalahar BK, Williams LD. 2009. Peeling the onion: ribosomes are ancient molecular fossils. Mol Biol Evol 26:2415–2425. 10.1093/molbev/msp163. [DOI] [PubMed] [Google Scholar]
- 120.Petrov AS, Gulen B, Norris AM, Kovacs NA, Bernier CR, Lanier KA, Fox GE, Harvey SC, Wartell RM, Hud NV, Williams LD. 2015. History of the ribosome and the origin of translation. Proc Natl Acad Sci USA 112:15396–15401. 10.1073/pnas.1509761112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Petrov AS, Bernier CR, Hsiao C, Norris AM, Kovacs NA, Waterbury CC, Stepanov VG, Harvey SC, Fox GE, Wartell RM, Hud NV, Williams LD. 2014. Evolution of the ribosome at atomic resolution. Proc Natl Acad Sci USA 111:10251–10256. 10.1073/pnas.1407205111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hsiao C, Lenz TK, Peters JK, Fang PY, Schneider DM, Anderson EJ, Preeprem T, Bowman JC, O'Neill EB, Lie L, Athavale SS, Gossett JJ, Trippe C, Murray J, Petrov AS, Wartell RM, Harvey SC, Hud NV, Williams LD. 2013. Molecular paleontology: a biochemical model of the ancestral ribosome. Nucleic Acids Res 41:3373–3385. 10.1093/nar/gkt023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lanier KA, Athavale SS, Petrov AS, Wartell R, Williams LD. 2016. Imprint of ancient evolution on rRNA folding. Biochemistry 55:4603–4613. 10.1021/acs.biochem.6b00168. [DOI] [PubMed] [Google Scholar]
- 124.Petrov AS, Bernier CR, Gulen B, Waterbury CC, Hershkovits E, Hsiao C, Harvey SC, Hud NV, Fox GE, Wartell RM, Williams LD. 2014. Secondary structures of rRNAs from all three domains of life. PLoS One 9:e88222. 10.1371/journal.pone.0088222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Bowman JC, Hud NV, Williams LD. 2015. The ribosome challenge to the RNA world. J Mol Evol 80:143–161. 10.1007/s00239-015-9669-9. [DOI] [PubMed] [Google Scholar]
- 126.Gulen B, Petrov AS, Okafor CD, Vander Wood D, O'Neill EB, Hud NV, Williams LD. 2016. Ribosomal small subunit domains radiate from a central core. Sci Rep 6:20885. 10.1038/srep20885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Frank J, Agrawal RK. 2000. A ratchet-like inter-subunit reorganization of the ribosome during translocation. Nature 406:318–322. 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 128.Woese C. 1970. Molecular mechanics of translation: a reciprocating ratchet mechanism. Nature 226:817–820. 10.1038/226817a0. [DOI] [PubMed] [Google Scholar]
- 129.Frank J, Gonzalez RL, Jr. 2010. Structure and dynamics of a processive Brownian motor: the translating ribosome. Annu Rev Biochem 79:381–412. 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Bretscher MS. 1968. Translocation in protein synthesis: a hybrid structure model. Nature 218:675–677. 10.1038/218675a0. [DOI] [PubMed] [Google Scholar]
- 131.Moazed D, Noller HF. 1989. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342:142–148. 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 132.Schmeing TM, Seila AC, Hansen JL, Freeborn B, Soukup JK, Scaringe SA, Strobel SA, Moore PB, Steitz TA. 2002. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat Struct Biol 9:225–230. 10.1038/nsb758. [DOI] [PubMed] [Google Scholar]
- 133.Chen J, Tsai A, O'Leary SE, Petrov A, Puglisi JD. 2012. Unraveling the dynamics of ribosome translocation. Curr Opin Struct Biol 22:804–814. 10.1016/j.sbi.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Petrov A, Chen J, O'Leary S, Tsai A, Puglisi JD. 2012. Single-molecule analysis of translational dynamics. Cold Spring Harb Perspect Biol 4:a011551. 10.1101/cshperspect.a011551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Sharma H, Adio S, Senyushkina T, Belardinelli R, Peske F, Rodnina MV. 2016. Kinetics of spontaneous and EF-G-accelerated rotation of ribosomal subunits. Cell Rep 16:2187–2196. 10.1016/j.celrep.2016.07.051. [DOI] [PubMed] [Google Scholar]
- 136.Marshall RA, Dorywalska M, Puglisi JD. 2008. Irreversible chemical steps control intersubunit dynamics during translation. Proc Natl Acad Sci USA 105:15364–15369. 10.1073/pnas.0805299105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Ermolenko DN, Majumdar ZK, Hickerson RP, Spiegel PC, Clegg RM, Noller HF. 2007. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol 370:530–540. 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 138.Chen J, Petrov A, Tsai A, O'Leary SE, Puglisi JD. 2013. Coordinated conformational and compositional dynamics drive ribosome translocation. Nat Struct Mol Biol 20:718–727. 10.1038/nsmb.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Tourigny DS, Fernández IS, Kelley AC, Ramakrishnan V. 2013. Elongation factor G bound to the ribosome in an intermediate state of translocation. Science 340:1235490. 10.1126/science.1235490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Brilot AF, Korostelev AA, Ermolenko DN, Grigorieff N. 2013. Structure of the ribosome with elongation factor G trapped in the pretranslocation state. Proc Natl Acad Sci USA 110:20994–20999. 10.1073/pnas.1311423110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Ermolenko DN, Noller HF. 2011. mRNA translocation occurs during the second step of ribosomal intersubunit rotation. Nat Struct Mol Biol 18:457–462. 10.1038/nsmb.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ramrath DJF, Lancaster L, Sprink T, Mielke T, Loerke J, Noller HF, Spahn CMT. 2013. Visualization of two transfer RNAs trapped in transit during elongation factor G-mediated translocation. Proc Natl Acad Sci USA 110:20964–20969. 10.1073/pnas.1320387110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Guo Z, Noller HF. 2012. Rotation of the head of the 30S ribosomal subunit during mRNA translocation. Proc Natl Acad Sci USA 109:20391–20394. 10.1073/pnas.1218999109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Zhou J, Lancaster L, Donohue JP, Noller HF. 2014. How the ribosome hands the A-site tRNA to the P site during EF-G-catalyzed translocation. Science 345:1188–1191. 10.1126/science.1255030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Noller HF, Lancaster L, Mohan S, Zhou J. 2017. Ribosome structural dynamics in translocation: yet another functional role for ribosomal RNA. Q Rev Biophys 50:e12. 10.1017/S0033583517000117. [DOI] [PubMed] [Google Scholar]
- 146.Noller HF, Lancaster L, Zhou J, Mohan S. 2017. The ribosome moves: RNA mechanics and translocation. Nat Struct Mol Biol 24:1021–1027. 10.1038/nsmb.3505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. 2003. Locking and unlocking of ribosomal motions. Cell 114:123–134. 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 148.Aitken CE, Puglisi JD. 2010. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol 17:793–800. 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Harvey SC, McCammon JA. 1981. Intramolecular flexibility in phenylalanine transfer RNA. Nature 294:286–287. 10.1038/294286a0. [DOI] [PubMed] [Google Scholar]
- 150.Mohan S, Donohue JP, Noller HF. 2014. Molecular mechanics of 30S subunit head rotation. Proc Natl Acad Sci USA 111:13325–13330. 10.1073/pnas.1413731111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Paci M, Fox GE. 2015. Major centers of motion in the large ribosomal RNAs. Nucleic Acids Res 43:4640–4649. 10.1093/nar/gkv289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Paci M, Fox GE. 2016. Centers of motion associated with EF-Tu binding to the ribosome. RNA Biol 13:524–530. 10.1080/15476286.2015.1114204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Shaul S, Berel D, Benjamini Y, Graur D. 2010. Revisiting the operational RNA code for amino acids: ensemble attributes and their implications. RNA 16:141–153. 10.1261/rna.1745910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Schimmel P, Giege R, Moras D, Yokoyama S. 1993. An operational RNA code for amino acids and possible relationship to genetic code. Proc Natl Acad Sci USA 90:8763–8768. 10.1073/pnas.90.19.8763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Schimmel P, Ribas de Pouplana L. 1995. Transfer RNA: from minihelix to genetic code. Cell 81:983–986. 10.1016/s0092-8674(05)80002-9. [DOI] [PubMed] [Google Scholar]
- 156.Francklyn C, Schimmel P. 1989. Aminoacylation of RNA minihelices with alanine. Nature 337:478–481. 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- 157.Francklyn C, Schimmel P. 1990. Enzymatic aminoacylation of an eight-base-pair microhelix with histidine. Proc Natl Acad Sci USA 87:8655–8659. 10.1073/pnas.87.21.8655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Klein DJ, Moore PB, Steitz TA. 2004. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J Mol Biol 340:141–177. 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- 159.Maguire BA, Beniaminov AD, Ramu H, Mankin AS, Zimmermann RA. 2005. A protein component at the heart of an RNA machine: the importance of protein L27 for the function of the bacterial ribosome. Mol Cell 20:427–435. 10.1016/j.molcel.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 160.Mailliot J, Garreau de Loubresse N, Yusupova G, Meskauskas A, Dinman JD, Yusupov M. 2016. Crystal structures of the uL3 mutant ribosome: illustration of the importance of ribosomal proteins for translation efficiency. J Mol Biol 428:2195–2202. 10.1016/j.jmb.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Selmer M, Dunham CM, Murphy FVt, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. 2006. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 313:1935–1942. 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 162.Kirillov SV, Wower J, Hixson SS, Zimmermann RA. 2002. Transit of tRNA through the Escherichia coli ribosome: cross-linking of the 3′ end of tRNA to ribosomal proteins at the P and E sites. FEBS Lett 514:60–66. 10.1016/S0014-5793(02)02302-5. [DOI] [PubMed] [Google Scholar]
- 163.Kazemie M. 1976. Binding of aminoacyl-tRNA to reconstituted subparticles of Escherichia coli large ribosomal subunits. Eur J Biochem 67:373–378. 10.1111/j.1432-1033.1976.tb10701.x. [DOI] [PubMed] [Google Scholar]
- 164.Moore VG, Atchison RE, Thomas G, Moran M, Noller HF. 1975. Identification of a ribosomal protein essential for peptidyl transferase activity. Proc Natl Acad Sci USA 72:844–848. 10.1073/pnas.72.3.844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Huang S, Aleksashin NA, Loveland AB, Klepacki D, Reier K, Kefi A, Szal T, Remme J, Jaeger L, Vázquez-Laslop N, Korostelev AA, Mankin AS. 2020. Ribosome engineering reveals the importance of 5S rRNA autonomy for ribosome assembly. Nat Commun 11:2900. 10.1038/s41467-020-16694-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Klein DJ, Moore PB, Steitz TA. 2004. The contribution of metal ions to the structural stability of the large ribosomal subunit. RNA 10:1366–1379. 10.1261/rna.7390804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Noller HF. 2004. The driving force for molecular evolution of translation. RNA 10:1833–1837. 10.1261/rna.7142404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Pyle A. 2002. Metal ions in the structure and function of RNA. J Biol Inorg Chem 7:679–690. 10.1007/s00775-002-0387-6. [DOI] [PubMed] [Google Scholar]
- 169.Shiman R, Draper DE. 2000. Stabilization of RNA tertiary structure by monovalent cations. J Mol Biol 302:79–91. 10.1006/jmbi.2000.4031. [DOI] [PubMed] [Google Scholar]
- 170.Näslund PH, Hultin T. 1970. Effects of potassium deficiency on mammalian ribosomes. Biochim Biophys Acta 204:237–247. 10.1016/0005-2787(70)90507-1. [DOI] [PubMed] [Google Scholar]
- 171.Näslund PH, Hultin T. 1971. Structural and functional defects in mammalian ribosomes after potassium deficiency. Biochim Biophys Acta 254:104–116. 10.1016/0005-2787(71)90117-1. [DOI] [PubMed] [Google Scholar]
- 172.Rozov A, Khusainov I, El Omari K, Duman R, Mykhaylyk V, Yusupov M, Westhof E, Wagner A, Yusupova G. 2019. Importance of potassium ions for ribosome structure and function revealed by long-wavelength X-ray diffraction. Nat Commun 10:2519. 10.1038/s41467-019-10409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Leonarski F, D'Ascenzo L, Auffinger P. 2019. Nucleobase carbonyl groups are poor Mg2+ inner-sphere binders but excellent monovalent ion binders—a critical PDB survey. RNA 25:173–192. 10.1261/rna.068437.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Wacker WE. 1969. The biochemistry of magnesium. Ann N Y Acad Sci 162:717–726. 10.1111/j.1749-6632.1969.tb13003.x. [DOI] [PubMed] [Google Scholar]
- 175.Misra VK, Draper DE. 1999. The interpretation of Mg(2+) binding isotherms for nucleic acids using Poisson-Boltzmann theory. J Mol Biol 294:1135–1147. 10.1006/jmbi.1999.3334. [DOI] [PubMed] [Google Scholar]
- 176.Misra VK, Draper DE. 2001. A thermodynamic framework for Mg2+ binding to RNA. Proc Natl Acad Sci USA 98:12456–12461. 10.1073/pnas.221234598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Parnell KM, Strobel SA. 2003. HIS & HERS, magic magnesium and the ballet of protein synthesis. Curr Opin Chem Biol 7:528–533. 10.1016/j.cbpa.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 178.Chao F-C, Schachman HK. 1956. The isolation and characterization of a macromolecular ribonucleoprotein from yeast. Arch Biochem Biophys 61:220–230. 10.1016/0003-9861(56)90334-4. [DOI] [PubMed] [Google Scholar]
- 179.Chao FC. 1957. Dissociation of macromolecular ribonucleoprotein of yeast. Arch Biochem Biophys 70:426–431. 10.1016/0003-9861(57)90130-3. [DOI] [PubMed] [Google Scholar]
- 180.McCarthy BJ. 1962. The effects of magnesium starvation on the ribosome content of Escherichia coli. Biochim Biophys Acta 55:880–889. 10.1016/0926-6550(62)90345-6. [DOI] [Google Scholar]
- 181.Gesteland RF. 1966. Unfolding of Escherichia coli ribosomes by removal of magnesium. J Mol Biol 18:356–371. 10.1016/S0022-2836(66)80253-X. [DOI] [PubMed] [Google Scholar]
- 182.Monro RE. 1967. Catalysis of peptide bond formation by 50 S ribosomal subunits from Escherichia coli. J Mol Biol 26:147–151. 10.1016/0022-2836(67)90271-9. [DOI] [PubMed] [Google Scholar]
- 183.Petrov AS, Bernier CR, Hsiao C, Okafor CD, Tannenbaum E, Stern J, Gaucher E, Schneider D, Hud NV, Harvey SC, Williams LD. 2012. RNA-magnesium-protein interactions in large ribosomal subunit. J Phys Chem B 116:8113–8120. 10.1021/jp304723w. [DOI] [PubMed] [Google Scholar]
- 184.Hsiao C, Williams LD. 2009. A recurrent magnesium-binding motif provides a framework for the ribosomal peptidyl transferase center. Nucleic Acids Res 37:3134–3142. 10.1093/nar/gkp119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JHD. 2005. Structures of the bacterial ribosome at 3.5 Å resolution. Science 310:827–834. 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 186.Auffinger P, Ennifar E, D'Ascenzo L. 2021. Deflating the RNA Mg2+ bubble: stereochemistry to the rescue! RNA 27:243–252. 10.1261/rna.076067.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zimorski V, Mentel M, Tielens AGM, Martin WF. 2019. Energy metabolism in anaerobic eukaryotes and Earth's late oxygenation. Free Radic Biol Med 140:279–294. 10.1016/j.freeradbiomed.2019.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kasting J. 1993. Earth's early atmosphere. Science 259:920–926. 10.1126/science.11536547. [DOI] [PubMed] [Google Scholar]
- 189.Holland HD. 2006. The oxygenation of the atmosphere and oceans. Philos Trans R Soc Lond B Biol Sci 361:903–915. 10.1098/rstb.2006.1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hong Enriquez RP, Do TN. 2012. Bioavailability of metal ions and evolutionary adaptation. Life (Basel) 2:274–285. 10.3390/life2040274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Fausto Da Silva J, Williams R. 1997. The biological chemistry of the elements: the inorganic chemistry of life. Oxford University Press, Oxford, United Kingdom. [Google Scholar]
- 192.Hsiao C, Chou IC, Okafor CD, Bowman JC, O'Neill EB, Athavale SS, Petrov AS, Hud NV, Wartell RM, Harvey SC, Williams LD. 2013. RNA with iron(II) as a cofactor catalyses electron transfer. Nat Chem 5:525–528. 10.1038/nchem.1649. [DOI] [PubMed] [Google Scholar]
- 193.Guth-Metzler R, Bray MS, Frenkel-Pinter M, Suttapitugsakul S, Montllor-Albalate C, Bowman JC, Wu R, Reddi AR, Okafor CD, Glass JB, Williams LD. 2020. Cutting in-line with iron: ribosomal function and non-oxidative RNA cleavage. Nucleic Acids Res 48:8663–8674. 10.1093/nar/gkaa586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Athavale SS, Petrov AS, Hsiao C, Watkins D, Prickett CD, Gossett JJ, Lie L, Bowman JC, O'Neill E, Bernier CR, Hud NV, Wartell RM, Harvey SC, Williams LD. 2012. RNA folding and catalysis mediated by iron (II). PLoS One 7:e38024. 10.1371/journal.pone.0038024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Bray MS, Lenz TK, Haynes JW, Bowman JC, Petrov AS, Reddi AR, Hud NV, Williams LD, Glass JB. 2018. Multiple prebiotic metals mediate translation. Proc Natl Acad Sci USA 115:12164–12169. 10.1073/pnas.1803636115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wower IK, Wower J, Zimmermann RA. 1998. Ribosomal protein L27 participates in both 50 S subunit assembly and the peptidyl transferase reaction. J Biol Chem 273:19847–19852. 10.1074/jbc.273.31.19847. [DOI] [PubMed] [Google Scholar]
- 197.Krupkin M, Matzov D, Tang H, Metz M, Kalaora R, Belousoff MJ, Zimmerman E, Bashan A, Yonath A. 2011. A vestige of a prebiotic bonding machine is functioning within the contemporary ribosome. Philos Trans R Soc Lond B Biol Sci 366:2972–2978. 10.1098/rstb.2011.0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Maracci C, Wohlgemuth I, Rodnina MV. 2015. Activities of the peptidyl transferase center of ribosomes lacking protein L27. RNA 21:2047–2052. 10.1261/rna.053330.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Leung EKY, Suslov N, Tuttle N, Sengupta R, Piccirilli JA. 2011. The mechanism of peptidyl transfer catalysis by the ribosome. Annu Rev Biochem 80:527–555. 10.1146/annurev-biochem-082108-165150. [DOI] [PubMed] [Google Scholar]
- 200.Agmon I, Davidovich C, Bashan A, Yonath A. 2009. Identification of the prebiotic translation apparatus within the contemporary ribosome. Nat Prec 10.1038/npre.2009.2921.1. [DOI] [Google Scholar]
- 201.Sievers A, Beringer M, Rodnina MV, Wolfenden R. 2004. The ribosome as an entropy trap. Proc Natl Acad Sci USA 101:7897–7901. 10.1073/pnas.0402488101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Schroeder GK, Wolfenden R. 2007. The rate enhancement produced by the ribosome: an improved model. Biochemistry 46:4037–4044. 10.1021/bi602600p. [DOI] [PubMed] [Google Scholar]
- 203.Buchachenko AL. 2020. The beauty and fascination of science, p 95–111. Springer Singapore, Singapore. 10.1007/978-981-15-2592-6_6. [DOI] [Google Scholar]
- 204.Hansen JL, Schmeing TM, Moore PB, Steitz TA. 2002. Structural insights into peptide bond formation. Proc Natl Acad Sci USA 99:11670–11675. 10.1073/pnas.172404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Vesper O, Nierhaus KH. 2004. Translation elongation in bacteria, p 214–223. In Lennarz WJ, Lane MD (ed), Encyclopedia of biological chemistry. Elsevier, New York, NY. 10.1016/B0-12-443710-9/00530-5. [DOI] [Google Scholar]
- 206.Polikanov YS, Steitz TA, Innis CA. 2014. A proton wire to couple aminoacyl-tRNA accommodation and peptide-bond formation on the ribosome. Nat Struct Mol Biol 21:787–793. 10.1038/nsmb.2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Wallin G, Åqvist J. 2010. The transition state for peptide bond formation reveals the ribosome as a water trap. Proc Natl Acad Sci USA 107:1888–1893. 10.1073/pnas.0914192107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Weinger JS, Strobel SA. 2006. Participation of the tRNA A76 hydroxyl groups throughout translation. Biochemistry 45:5939–5948. 10.1021/bi060183n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Weinger JS, Parnell KM, Dorner S, Green R, Strobel SA. 2004. Substrate-assisted catalysis of peptide bond formation by the ribosome. Nat Struct Mol Biol 11:1101–1106. 10.1038/nsmb841. [DOI] [PubMed] [Google Scholar]
- 210.Schmeing TM, Huang KS, Kitchen DE, Strobel SA, Steitz TA. 2005. Structural insights into the roles of water and the 2' hydroxyl of the P site tRNA in the peptidyl transferase reaction. Mol Cell 20:437–448. 10.1016/j.molcel.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 211.Trobro S, Aqvist J. 2006. Analysis of predictions for the catalytic mechanism of ribosomal peptidyl transfer. Biochemistry 45:7049–7056. 10.1021/bi0605383. [DOI] [PubMed] [Google Scholar]
- 212.Krayevsky AA, Kukhanova MK. 1979. The peptidyltransferase center of ribosomes. Prog Nucleic Acid Res Mol Biol 23:1–51. 10.1016/s0079-6603(08)60130-0. [DOI] [PubMed] [Google Scholar]
- 213.Dorner S, Panuschka C, Schmid W, Barta A. 2003. Mononucleotide derivatives as ribosomal P‐site substrates reveal an important contribution of the 2′‐OH to activity. Nucleic Acids Res 31:6536–6542. 10.1093/nar/gkg842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sharma PK, Xiang Y, Kato M, Warshel A. 2005. What are the roles of substrate-assisted catalysis and proximity effects in peptide bond formation by the ribosome? Biochemistry 44:11307–11314. 10.1021/bi0509806. [DOI] [PubMed] [Google Scholar]
- 215.Zaher HS, Shaw JJ, Strobel SA, Green R. 2011. The 2'-OH group of the peptidyl-tRNA stabilizes an active conformation of the ribosomal PTC. EMBO J 30:2445–2453. 10.1038/emboj.2011.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Brunelle JL, Shaw JJ, Youngman EM, Green R. 2008. Peptide release on the ribosome depends critically on the 2′ OH of the peptidyl–tRNA substrate. RNA 14:1526–1531. 10.1261/rna.1057908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Katunin VI, Muth GW, Strobel SA, Wintermeyer W, Rodnina MV. 2002. Important contribution to catalysis of peptide bond formation by a single ionizing group within the ribosome. Mol Cell 10:339–346. 10.1016/s1097-2765(02)00566-x. [DOI] [PubMed] [Google Scholar]
- 218.Zavialov AV, Mora L, Buckingham RH, Ehrenberg M. 2002. Release of peptide promoted by the GGQ motif of class 1 release factors regulates the GTPase activity of RF3. Mol Cell 10:789–798. 10.1016/s1097-2765(02)00691-3. [DOI] [PubMed] [Google Scholar]
- 219.Fox GE. 2016. Origins and early evolution of the ribosome, p 31–60. In Hernández G, Jagus R (ed), Evolution of the protein synthesis machinery and its regulation. Springer International Publishing, Cham, Switzerland. 10.1007/978-3-319-39468-8_3. [DOI] [Google Scholar]
- 220.Moore PB, Steitz TA. 2003. After the ribosome structures: how does peptidyl transferase work? RNA 9:155–159. 10.1261/rna.2127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Świderek K, Marti S, Tuñón I, Moliner V, Bertran J. 2015. Peptide bond formation mechanism catalyzed by ribosome. J Am Chem Soc 137:12024–12034. 10.1021/jacs.5b05916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Rodnina MV, Beringer M, Wintermeyer W. 2007. How ribosomes make peptide bonds. Trends Biochem Sci 32:20–26. 10.1016/j.tibs.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 223.Rodnina MV, Wintermeyer W. 2003. Peptide bond formation on the ribosome: structure and mechanism. Curr Opin Struct Biol 13:334–340. 10.1016/s0959-440x(03)00065-4. [DOI] [PubMed] [Google Scholar]
- 224.Carrasco N, Hiller DA, Strobel SA. 2011. Minimal transition state charge stabilization of the oxyanion during peptide bond formation by the ribosome. Biochemistry 50:10491–10498. 10.1021/bi201290s. [DOI] [PubMed] [Google Scholar]
- 225.Schlunzen F, Zarivach R, Harms J, Bashan A, Tocilj A, Albrecht R, Yonath A, Franceschi F. 2001. Structural basis for the interaction of antibiotics with the peptidyl transferase centre in eubacteria. Nature 413:814–821. 10.1038/35101544. [DOI] [PubMed] [Google Scholar]
- 226.Polikanov YS, Aleksashin NA, Beckert B, Wilson DN. 2018. The mechanisms of action of ribosome-targeting peptide antibiotics. Front Mol Biosci 5:48. 10.3389/fmolb.2018.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Svetlov MS, Syroegin EA, Aleksandrova EV, Atkinson GC, Gregory ST, Mankin AS, Polikanov YS. 2021. Structure of Erm-modified 70S ribosome reveals the mechanism of macrolide resistance. Nat Chem Biol 17:412–420. 10.1038/s41589-020-00715-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Svetlov MS, Vazquez-Laslop N, Mankin AS. 2017. Kinetics of drug-ribosome interactions defines the cidality of macrolide antibiotics. Proc Natl Acad Sci USA 114:13673–13678. 10.1073/pnas.1717168115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Mondal S, Pathak BK, Ray S, Barat C. 2014. Impact of P-Site tRNA and antibiotics on ribosome mediated protein folding: studies using the Escherichia coli ribosome. PLoS One 9:e101293. 10.1371/journal.pone.0101293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sothiselvam S, Liu B, Han W, Ramu H, Klepacki D, Atkinson GC, Brauer A, Remm M, Tenson T, Schulten K, Vazquez-Laslop N, Mankin AS. 2014. Macrolide antibiotics allosterically predispose the ribosome for translation arrest. Proc Natl Acad Sci USA 111:9804–9809. 10.1073/pnas.1403586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Lambert T. 2012. Antibiotics that affect the ribosome. Rev Sci Tech 31:57–64. 10.20506/rst.31.1.2095. [DOI] [PubMed] [Google Scholar]
- 232.Kannan K, Mankin AS. 2011. Macrolide antibiotics in the ribosome exit tunnel: species-specific binding and action. Ann N Y Acad Sci 1241:33–47. 10.1111/j.1749-6632.2011.06315.x. [DOI] [PubMed] [Google Scholar]
- 233.Vázquez-Laslop N, Mankin AS. 2018. Context-specific action of ribosomal antibiotics. Annu Rev Microbiol 72:185–207. 10.1146/annurev-micro-090817-062329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Ramu H, Vázquez-Laslop N, Klepacki D, Dai Q, Piccirilli J, Micura R, Mankin AS. 2011. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol Cell 41:321–330. 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 235.Seidelt B, Innis CA, Wilson DN, Gartmann M, Armache JP, Villa E, Trabuco LG, Becker T, Mielke T, Schulten K, Steitz TA, Beckmann R. 2009. Structural insight into nascent polypeptide chain-mediated translational stalling. Science 326:1412–1415. 10.1126/science.1177662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ito K, Chiba S. 2013. Arrest peptides: cis-acting modulators of translation. Annu Rev Biochem 82:171–202. 10.1146/annurev-biochem-080211-105026. [DOI] [PubMed] [Google Scholar]
- 237.Deutsch C. 2014. Tunnel vision: insights from biochemical and biophysical studies, p 61–86. In Ito K (ed), Regulatory nascent polypeptides. Springer Japan, Tokyo. 10.1007/978-4-431-55052-5_4. [DOI] [Google Scholar]
- 238.Wilson DN, Beckmann R. 2011. The ribosomal tunnel as a functional environment for nascent polypeptide folding and translational stalling. Curr Opin Struct Biol 21:274–282. 10.1016/j.sbi.2011.01.007. [DOI] [PubMed] [Google Scholar]
- 239.Guzel P, Yildirim HZ, Yuce M, Kurkcuoglu O. 2020. Exploring allosteric signaling in the exit tunnel of the bacterial ribosome by molecular dynamics simulations and residue network model. Front Mol Biosci 7:586075. 10.3389/fmolb.2020.586075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Huter P, Arenz S, Bock LV, Graf M, Frister JO, Heuer A, Peil L, Starosta AL, Wohlgemuth I, Peske F, Nováček J, Berninghausen O, Grubmüller H, Tenson T, Beckmann R, Rodnina MV, Vaiana AC, Wilson DN. 2017. Structural basis for polyproline-mediated ribosome stalling and rescue by the translation elongation factor EF-P. Mol Cell 68:515–527.e6. 10.1016/j.molcel.2017.10.014. [DOI] [PubMed] [Google Scholar]
- 241.Kovacs NA, Petrov AS, Lanier KA, Williams LD. 2017. Frozen in time: the history of proteins. Mol Biol Evol 34:1252–1260. 10.1093/molbev/msx086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Woolstenhulme CJ, Parajuli S, Healey DW, Valverde DP, Petersen EN, Starosta AL, Guydosh NR, Johnson WE, Wilson DN, Buskirk AR. 2013. Nascent peptides that block protein synthesis in bacteria. Proc Natl Acad Sci USA 110:E878–E887. 10.1073/pnas.1219536110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Buskirk AR, Green R. 2017. Ribosome pausing, arrest and rescue in bacteria and eukaryotes. Philos Trans R Soc Lond B Biol Sci 372:20160183. 10.1098/rstb.2016.0183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hayes CS, Bose B, Sauer RT. 2002. Proline residues at the C terminus of nascent chains induce SsrA tagging during translation termination. J Biol Chem 277:33825–33832. 10.1074/jbc.M205405200. [DOI] [PubMed] [Google Scholar]
- 245.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. 2004. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem 279:15368–15375. 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 246.Huter P, Müller C, Arenz S, Beckert B, Wilson DN. 2017. Structural basis for ribosome rescue in bacteria. Trends Biochem Sci 42:669–680. 10.1016/j.tibs.2017.05.009. [DOI] [PubMed] [Google Scholar]
- 247.Ayyub SA, Gao F, Lightowlers RN, Chrzanowska-Lightowlers ZM. 2020. Rescuing stalled mammalian mitoribosomes—what can we learn from bacteria? J Cell Sci 133:jcs231811. 10.1242/jcs.231811. [DOI] [PubMed] [Google Scholar]
- 248.Desai N, Yang H, Chandrasekaran V, Kazi R, Minczuk M, Ramakrishnan V. 2020. Elongational stalling activates mitoribosome-associated quality control. Science 370:1105–1110. 10.1126/science.abc7782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Müller C, Crowe-McAuliffe C, Wilson DN. 2021. Ribosome rescue pathways in bacteria. Front Microbiol 12:652980. 10.3389/fmicb.2021.652980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Agmon I, Bashan A, Zarivach R, Yonath A. 2005. Symmetry at the active site of the ribosome: structural and functional implications. Biol Chem 386:833–844. 10.1515/BC.2005.098. [DOI] [PubMed] [Google Scholar]
- 251.Agmon I, Auerbach T, Baram D, Bartels H, Bashan A, Berisio R, Fucini P, Hansen HA, Harms J, Kessler M, Peretz M, Schluenzen F, Yonath A, Zarivach R. 2003. On peptide bond formation, translocation, nascent protein progression and the regulatory properties of ribosomes. Derived on 20 October 2002 at the 28th FEBS Meeting in Istanbul. Eur J Biochem 270:2543–2556. 10.1046/j.1432-1033.2003.03634.x. [DOI] [PubMed] [Google Scholar]
- 252.Mears JA, Cannone JJ, Stagg SM, Gutell RR, Agrawal RK, Harvey SC. 2002. Modeling a minimal ribosome based on comparative sequence analysis. J Mol Biol 321:215–234. 10.1016/s0022-2836(02)00568-5. [DOI] [PubMed] [Google Scholar]
- 253.Chirkova A, Erlacher MD, Clementi N, Zywicki M, Aigner M, Polacek N. 2010. The role of the universally conserved A2450-C2063 base pair in the ribosomal peptidyl transferase center. Nucleic Acids Res 38:4844–4855. 10.1093/nar/gkq213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Agmon I. 2009. The dimeric proto-ribosome: structural details and possible implications on the origin of life. Int J Mol Sci 10:2921–2934. 10.3390/ijms10072921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Agmon IC. 2016. Could a proto-ribosome emerge spontaneously in the prebiotic world? Molecules 21:1701. 10.3390/molecules21121701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Bashan A, Zarivach R, Schluenzen F, Agmon I, Harms J, Auerbach T, Baram D, Berisio R, Bartels H, Hansen HA, Fucini P, Wilson D, Peretz M, Kessler M, Yonath A. 2003. Ribosomal crystallography: peptide bond formation and its inhibition. Biopolymers 70:19–41. 10.1002/bip.10412. [DOI] [PubMed] [Google Scholar]
- 257.Yonath A. 2003. Ribosomal tolerance and peptide bond formation. Biol Chem 384:1411–1419. 10.1515/BC.2003.156. [DOI] [PubMed] [Google Scholar]
- 258.Matzov D, Taoka M, Nobe Y, Yamauchi Y, Halfon Y, Asis N, Zimermann E, Rozenberg H, Bashan A, Bhushan S, Isobe T, Gray MW, Yonath A, Shalev-Benami M. 2020. Cryo-EM structure of the highly atypical cytoplasmic ribosome of Euglena gracilis. Nucleic Acids Res 48:11750–11761. 10.1093/nar/gkaa893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Gray MW, Gopalan V. 2020. Piece by piece: building a ribozyme. J Biol Chem 295:2313–2323. 10.1074/jbc.REV119.009929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Schnare MN, Gray MW. 1990. Sixteen discrete RNA components in the cytoplasmic ribosome of Euglena gracilis. J Mol Biol 215:73–83. 10.1016/S0022-2836(05)80096-8. [DOI] [PubMed] [Google Scholar]
- 261.Boer PH, Gray MW. 1988. Scrambled ribosomal RNA gene pieces in Chlamydomonas reinhardtii mitochondrial DNA. Cell 55:399–411. 10.1016/0092-8674(88)90026-8. [DOI] [PubMed] [Google Scholar]
- 262.Xu D, Wang Y. 2021. smFRET study of rRNA dimerization at the peptidyl transfer center. Biophys Chem 277:106657. 10.1016/j.bpc.2021.106657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Xu D, Wang Y. 2021. Protein-free ribosomal RNA scaffolds can assemble poly-lysine oligos from charged tRNA fragments. Biochem Biophys Res Commun 544:81–85. 10.1016/j.bbrc.2021.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Noller HF. 1993. Peptidyl transferase: protein, ribonucleoprotein, or RNA? J Bacteriol 175:5297–5300. 10.1128/jb.175.17.5297-5300.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Hipps D, Shiba K, Henderson B, Schimmel P. 1995. Operational RNA code for amino acids: species-specific aminoacylation of minihelices switched by a single nucleotide. Proc Natl Acad Sci USA 92:5550–5552. 10.1073/pnas.92.12.5550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Gazit E. 2018. Reductionist approach in peptide-based nanotechnology. Annu Rev Biochem 87:533–553. 10.1146/annurev-biochem-062917-012541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Makam P, Gazit E. 2018. Minimalistic peptide supramolecular co-assembly: expanding the conformational space for nanotechnology. Chem Soc Rev 47:3406–3420. 10.1039/c7cs00827a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Korobeinikova AV, Garber MB, Gongadze GM. 2012. Ribosomal proteins: structure, function, and evolution. Biochemistry (Mosc) 77:562–574. 10.1134/S0006297912060028. [DOI] [PubMed] [Google Scholar]
- 269.Ramakrishnan V, White SW. 1998. Ribosomal protein structures: insights into the architecture, machinery and evolution of the ribosome. Trends Biochem Sci 23:208–212. 10.1016/s0968-0004(98)01214-6. [DOI] [PubMed] [Google Scholar]
- 270.Pilla SP, Bahadur RP. 2019. Residue conservation elucidates the evolution of r-proteins in ribosomal assembly and function. Int J Biol Macromol 140:323–329. 10.1016/j.ijbiomac.2019.08.127. [DOI] [PubMed] [Google Scholar]
- 271.Vishwanath P, Favaretto P, Hartman H, Mohr SC, Smith TF. 2004. Ribosomal protein-sequence block structure suggests complex prokaryotic evolution with implications for the origin of eukaryotes. Mol Phylogenet Evol 33:615–625. 10.1016/j.ympev.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 272.Welch M, Majerfeld I, Yarus M. 1997. 23S rRNA similarity from selection for peptidyl transferase mimicry. Biochemistry 36:6614–6623. 10.1021/bi963135j. [DOI] [PubMed] [Google Scholar]
- 273.Zhang B, Cech TR. 1998. Peptidyl-transferase ribozymes: trans reactions, structural characterization and ribosomal RNA-like features. Chem Biol 5:539–553. 10.1016/s1074-5521(98)90113-2. [DOI] [PubMed] [Google Scholar]
- 274.Darwin C. 1859. On the origin of species by means of natural selection, or preservation of favoured races in the struggle for life. John Murray, London, England. [PMC free article] [PubMed] [Google Scholar]
- 275.Bernier CR, Petrov AS, Waterbury CC, Jett J, Li F, Freil LE, Xiong X, Wang L, Migliozzi BL, Hershkovits E, Xue Y, Hsiao C, Bowman JC, Harvey SC, Grover MA, Wartell ZJ, Williams LD. 2014. RiboVision suite for visualization and analysis of ribosomes. Faraday Discuss 169:195–207. 10.1039/c3fd00126a. [DOI] [PubMed] [Google Scholar]