Abstract
In the last 2 decades, lipidomics has become one of the fastest expanding scientific disciplines in biomedical research. With an increasing number of new research groups to the field, it is even more important to design guidelines for assuring high standards of data quality. The Lipidomics Standards Initiative is a community-based endeavor for the coordination of development of these best practice guidelines in lipidomics and is embedded within the International Lipidomics Society. It is the intention of this review to highlight the most quality-relevant aspects of the lipidomics workflow, including preanalytics, sample preparation, MS, and lipid species identification and quantitation. Furthermore, this review just does not only highlights examples of best practice but also sheds light on strengths, drawbacks, and pitfalls in the lipidomic analysis workflow. While this review is neither designed to be a step-by-step protocol by itself nor dedicated to a specific application of lipidomics, it should nevertheless provide the interested reader with links and original publications to obtain a comprehensive overview concerning the state-of-the-art practices in the field.
Supplementary key words: lipidomics, metabolomics, MS, chromatography, ion mobility spectrometry, phospholipids, sphingolipids, LC-MS, lipid identification
Abbreviations: DTIMS, drift tube ion mobility spectrometry; FAIMS, field asymmetric waveform ion mobility spectrometry; HILIC, hydrophilic interaction liquid chromatography; HRMS, high-resolution MS; ICR, ion cyclotron resonance; ILS, International Lipidomics Society; IMS, ion mobility spectrometry; IS, internal standard; LCF, lipid class-specific fragment; LPA, lysophosphatidic acid; LSI, Lipidomics Standards Initiative; PC, phosphatidylcholine; QqQ, triple quadrupole; QTOF, quadrupole TOF; RP, reverse-phase; RT, retention time; S1P, sphingosine-1-phosphate; SPE, solid phase extraction; SRM, selected reaction monitoring; TG, triglyceride; TIMS, trapped ion mobility spectrometry; TWIMS, traveling wave ion mobility spectrometry; UHPLC, ultrahigh-performance liquid chromatography; UHPSFC, ultrahigh-performance supercritical fluid chromatography
Lipidomics is the science based on large-scale lipid determination by bioanalytical methods, including the interpretation of these data in a broader biological context. Because of a combination of high sensitivity and specificity, MS is the method of choice (1, 2). This review article focuses on ESI because according to Web of Science, more than two-thirds of the lipidomics publications since 2010 used this ionization technique. The other two ionization techniques worth mentioning are MALDI and electron impact/chemical ionization in combination with GC-MS. Since MALDI is overwhelmingly used for MS imaging, which is beyond the scope of this article, and GC-MS does not provide comprehensive lipidomic data but only lipid classes like fatty acyl and sterols, these two ionization techniques are not covered in this review. When coupled to separation techniques like chromatography and ion mobility, ESI-based mass spectrometric analysis can even increase its analytical performance. While in the very beginning, only small sets of lipids were determined by GC-MS, the invention of ESI enabled the acquisition of many lipid classes in one analytical run (3). This development led from overwhelmingly direct infusion MS techniques in the mid-90s (4) to the emerging of a novel discipline called lipidomics from the early new millennium onward (5, 6). Nowadays, the analytical bases of this science are coarsely divided into direct infusion (shotgun) lipidomics (7) and separation coupled lipidomics (8, 9). It has been recognized within the last decade that the steps after data acquisition are crucial for the quality of reported studies: software-assisted data processing, including not only identification and quantitation of lipids but also further biological interpretation of the resulting large datasets (10) and integration with other omics data (11). It has further been recognized in the last decade that the field desperately needs some sort of quality assurance. The first step was the design of a shorthand nomenclature for lipids, which reflects the experimental evidence for the existence of a lipid (12, 13). The take-home message conveyed by this group of authors was: “Report only what is experimentally proven, and clearly state where assumptions were made.” Within the following years, this project evolved into the Lipidomics Standards Initiative (LSI) (https://lipidomics-standards-initiative.org/), a platform concerned by coordination of quality issues in the lipidomics field (10). But since the quality issues within the field soon expanded beyond nomenclature aspects and data acquisition, some core members of the LSI group decided to found a scientific society, the International Lipidomics Society (ILS) (https://lipidomicssociety.org/). Having ILS as scientific society is not only beneficial for developing analytical quality standards but also covers other aspects of interest for the community. Divided into several interest groups, ILS coordinates ring trials in clinical lipidomics, the availability of reference materials, ontology issues, ring trials in bioinformatics, and it is the interface between the community and instrument developers (vendors). But above all, ILS is the communication platform needed by the community, which connects scientists by organizing conferences (International Lipidomics Conference), virtual seminar series, and so on.
This review article aims at giving an overview about the state of the art in the lipidomics field with particular emphasis on recommended best practice examples guiding newcomers and less experienced researchers in the field. Therefore, it will include all aspects of the lipidomics workflow from preanalytics to data analysis, as depicted in Fig. 1.
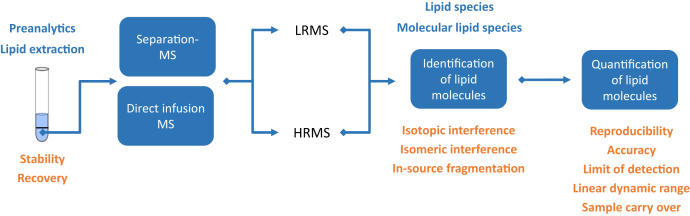
Fig. 1.
Lipidomic workflow. Typical lipidomic workflows including lipid extraction, direct infusion, or separation-based mass spectrometric analysis by nominal resolution, that is, low-resolution MS or HRMS. Data analysis includes lipid identification at respective structure level, for example, lipid species or lipid molecular species level and their quantification. Important considerations to achieve high-quality results are depicted in orange and are discussed in the respective sections.
Preanalytics and sample preparation
The first and clearly a critical step in any lipidomics workflow is sampling and subsequent sample processing. Especially, tissues should always be immediately frozen in liquid nitrogen, whereas biofluids like plasma should be either immediately processed or frozen at −80°C. If samples are kept for too long at room temperature, enzymatic and chemical degradation processes could result in lipid peroxidation or hydrolysis (14, 15, 16). It was even observed that samples, which were kept at room temperature and pH >6 for longer periods, showed scrambling of fatty acyls in lysophospholipids, resulting in the loss of regioisomeric specificity (17). Inappropriate preanalytic conditions may change the concentration dramatically like for lysolipids, lysophosphatidic acid (LPA) and sphingosine-1-phosphate (S1P), which are generated instantly after drawing blood samples (15) and therefore require special precautions to preserve in vivo concentrations (18). Lipolytic activity (19), which may continue even after the addition of organic solvents like alcohol and could be monitored by lipid class ratios reflecting degradation (20), is highly undesirable in lipidomics. Generally, the analysis of potential degradation products like oxidized lipids, lysophospholipids, or phosphatidic acid needs a particular precaution to ensure that they do not just arise artificially from sample processing. If possible, it is always good advice to process samples immediately. If unprocessed or processed samples are stored at −80°C for a longer period, then the stability should be verified for the studied lipid species.
Liquid-liquid extraction is the gold standard in lipidomic sample preparation. The historically most widely used methods rely on biphasic chloroform-methanol-water mixtures, such as the methods described by Lebaron and Folch (21) and Bligh and Dyer (22) (Fig. 2). While the Bligh and Dyer protocol has a higher proportion of methanol and is thus slightly more suitable for polar lipids (e.g., glycerophospholipids), the Folch protocol has a higher content of nonpolar chloroform in its organic phase, which increases the solubility of nonpolar lipids (e.g., triglyceride [TG]). Within the last decade, the supremacy of these two classical methods was challenged by a new liquid-liquid extraction method based on methyl-tert-butyl ether for reasons of reduced toxicity and improved sample handling (23). When polar anionic lipids, such as LPA or S1P, are of interest, an acidified Bligh and Dyer protocol should be the extraction method of choice (24). Using this extraction, it is of utmost importance to strictly keep to the protocol concerning the concentration of HCl and the extraction time, because otherwise acid-sensitive hydrolysis effects could cause significant alternations of natural concentrations of these sensitive lipid classes. Particularly for large-scale lipidomics studies, biphasic liquid-liquid extraction protocols are often replaced by monophasic methods based on the protein precipitation only because they are simpler in handling and thus are more suitable for workflow automation (25, 26). Nevertheless, precaution needs to be taken to avoid precipitation of lipid classes like TG or cholesteryl esters (unpublished observation).
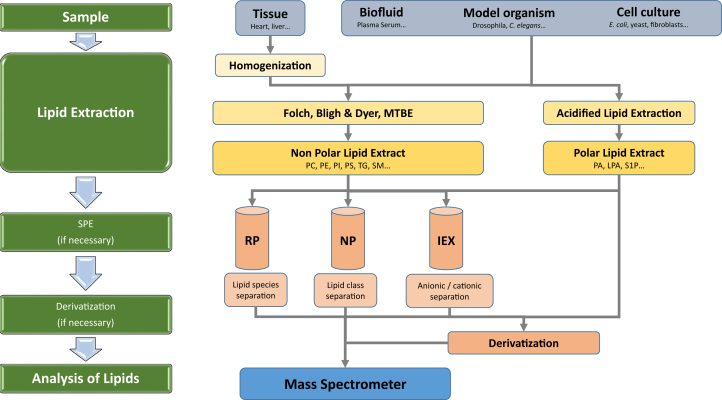
Fig. 2.
Modular sample preparation workflow. Various sample types (powder blue) are extracted by liquid-liquid extraction (yellow) and eventually enriched by SPE or derivatized (reddish) before they are subjected to mass spectrometric analysis (blue).
While the extraction of fluidic samples is straightforward, solid samples like tissues need homogenization prior to extraction. This step needs careful evaluation since lipid recovery could be compromised by various factors, including sample concentration, solvent, and the method applied for homogenization (27). Thus, inappropriate conditions may lead to a selective loss of either nonpolar or ionic lipid classes. Furthermore, it should be emphasized that each type of sample (e.g., solid vs. fluidic, urine vs. blood, blood plasma vs. serum, etc.) will eventually require dedicated considerations (which are however beyond the scope of this review) (28).
In summary, it should be noted that each extraction protocol has its inherent merits and drawbacks, and none of them fits equally well for all lipid categories and classes, which makes it necessary to choose the protocol best suited for the lipids under investigation and the individual needs of the study (29). Solid phase extraction (SPE) can be a good enrichment step when only a few lipids are of interest in a targeted approach (30) (Fig. 2). SPE is beneficial for further fractionation of total lipid extracts, allowing separation of phospholipids or sphingolipids even into individual classes (31, 32, 33). Another application of SPE in lipidomics is the selective enrichment of low-abundance lipids like steroids (34), N-acyl phosphatidylethanolamines (35), long-chain base phosphates (36), or glycosphingolipids (37). If necessary, further analytical enhancement of individual lipid classes is possible by derivatization, which results in an enhancement of ionization efficiency and/or introduction of characteristic fragments (38, 39, 40) (Fig. 2). It is advised that whatever method is used for lipid sample preparation, it is always better to add internal standards (ISs) prior extraction to the samples for internal control and quantification (41).
Direct infusion—MS
In the early days of lipidomic analysis, direct infusion systems coupled to a triple quadrupole (QqQ) mass spectrometer were the state of the art (Fig. 3). Worth mentioning in this respect are the instrumental settings proposed by the groups of Han and Gross (3, 7) and Liebisch et al. (14, 42). The former used a setup relying on either a syringe pump or a static nanoESI source (Nanomate), whereas the latter used a HPLC-based flow injection system. In such a setting, the QqQ is typically utilized for a combination of precursor ion and neutral loss scans. This method is particularly useful if characteristic head group fragments or neutral losses are available, like it is the case for most phospholipids (43, 44, 45, 46, 47) and sphingolipids (48, 49, 50, 51). In addition, precursor ion or neutral loss scans on specific fatty acyl fragments/neutral losses complement the overall picture of a given lipid structure. If all these pieces of information are properly put together, it is possible to identify not only the lipid class and the cumulative number of fatty acyl carbons and double bonds by its corresponding head group fragmentation (e.g., phosphatidylcholine [PC] 38:4) but eventually also the constituent fatty acyls by their specific scans (e.g., PC 18:0_20:4) (7). Since any chromatographic information and separation are missing, it is good advice to include as much fragment information as possible for increasing the selectivity of the analytical system such as multidimensional MS (52). For improved selectivity, alternatively, contemporary direct infusion platforms use high-resolution mass spectrometers, with the gold standard nowadays being Orbitrap MS (53), often coupled to a Nanomate nanoESI chip (Fig. 3) (54, 55, 56). Achieving a mass accuracy of better than 3 ppm enables this mass analyzer to pin down the elemental composition of precursor and fragment masses alike (57, 58). The idea of this setup is the generation of full scan spectra of intact lipid adduct ions and MS/MS spectra of each detected lipid species, both at a mass resolution of 100,000 or higher (54, 55, 59, 60, 61). Furthermore, the nanoESI chip offers another set of advantages compared with conventional direct infusion systems: 1) It relies on one nanoESI needle per sample, and therefore, any carryover from the injection system is abolished (62). 2) NanoESI significantly enhances signal intensity resulting in improved limits of detection (63). Generally, direct infusion MS methods benefit from a steady ionization environment, generating very robust quantitative data. Consequently, just one or eventually two nonendogenous ISs for each lipid class are sufficient for quantitation, which is in sharp contrast to the requirements concerning the number of IS for lipid species separation-based methods (7, 41, 53, 62). Nevertheless, it is important to strictly control the concentration of the infused lipids because at concentrations above 10 μM, aggregation of lipids starts to distort quantitative data (4, 64), and at above 100 μM (multiply charged) multimers (e.g., [3M + 2Na]2+ or [2M + 3Na]+) start to be the dominant forms (3). Furthermore, it is important for quantitation to allow the formation of only one adduct, preferably the one with maximal ionization efficiency. This is the reason why ammonia salts are often used, because they result in PC and SM to be detected mainly as protonated compounds (4), they increase the ionization efficiency of TG as NH4+ adducts and anionic lipid classes, such as phosphatidylglycerol, phosphatidylinositol, phosphatidylserine, phosphatidic acid, and cardiolipin as deprotonated species (63). Another issue, which could hamper quantitation, would be multiply charged compounds, distributing the intact lipids intensity over several charge states. Since lipids are rather small molecules with a mass below 1,000 Da, this phenomenon is just observed for cardiolipin, phosphatidylinositol phosphate, or gangliosides, which are not the bulk lipid classes. In these cases, the existence of several charge states has of course to be factored in for quantitative aspects. The drawback of direct infusion MS methods is the inherently limited ionization capacity of electrospray leading to ion suppression effects, which results in detection problems for minor lipid constituents of certain nonpolar lipid classes when, for example, compared with ultrahigh-performance supercritical fluid chromatography (UHPSFC) (9). This physical limitation could be overcome either by the use of chromatography or by selective enhancement/suppression techniques, where ionization of specific lipid classes is enhanced by the addition of certain additives, for example, NH4OH or LiOH (63, 65). Another drawback of any direct infusion MS approach is the lack of chromatographic information (retention time [RT]) for identification. This can be a particular challenge with lipid extracts rich in overlapping isomeric and isobaric species because isomers and isobars result in mixed MS/MS spectra and tend to complicate data interpretation, although strategies to overcome this issue have been proposed recently (60).
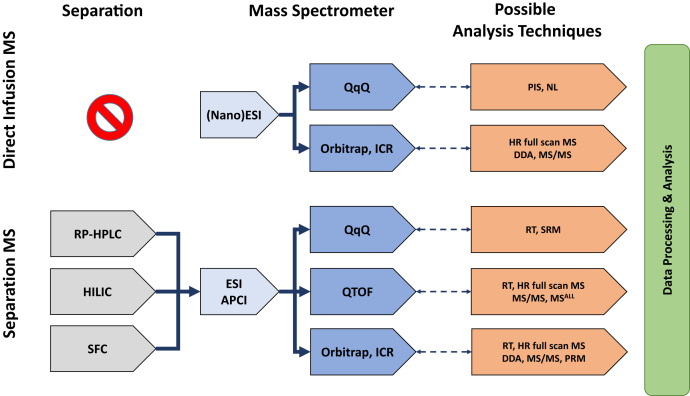
Fig. 3.
Frequently used instrumental setups. This figure shows an overview of frequently used combinations of chromatography, MS (ion sources and analyzers), and the possible associated analytical techniques, which are provided by the corresponding instrumental setup.
Separation—MS
Chromatography
By enabling molecular separation, chromatography opens up another analytical dimension for the analysis of lipids by MS (Fig. 3). On the one hand, the separation of total lipid extracts reduces the complexity of mass spectra, whereas on the other hand, RT provides valuable information about the lipid identity. On the downside, chromatography often needs more demanding and sophisticated quantitation strategies, when compared with direct infusion MS. Reverse-phase (RP)-HPLC, the most widely used separation technique in lipidomics, separates lipid species by their hydrophobic fatty acyl moieties according to the equivalent carbon number model (66). When coupled to QqQ, one specific mass transition in selected reaction monitoring (SRM) for each lipid species is essential for unambiguous identification and further quantitation. This is even possible in a validated clinical setting as has been shown for the analysis of ceramides (67). When using the Orbitrap option for such a platform, molecular species are underpinned by high mass accuracy. Depending on the instrument type and availability of parallel and/or sequential acquisition modes, full MS/MS spectra are either obtained in high or in nominal mass resolution (68). When using a Q-Exactive, parallel reaction monitoring, a product ion scan of a precursor mass selected in Q1 and fragmented in the higher-energy collision dissociation cell, can be used to increase the duty cycle when compared with SRM on this kind of instrument (69, 70). Furthermore, the use of a nanoHPLC system could even further increase the coverage by a factor of three (69). In contrast to RP-HPLC, hydrophilic interaction liquid chromatography (HILIC) separates lipids according to their polar head group into lipid classes. Because of the coelution of all molecular species of a lipid class in a very narrow RT window, this kind of chromatography is very convenient for quantitation, because similarly to shotgun lipidomics, one IS per lipid class could be considered (71). HILIC is often used for the analysis of polar phosphorylated lipids (e.g., LPA, S1P, and bis(monoacylglycerol)phosphate), either coupled with QqQ in SRM mode (15, 72, 73), with an Fourier transform-ion cyclotron resonance-MS (ICR-MS) in all ion fragmentation mode (74), with a Q-Exactive in data-dependent MS/MS mode (75), or to an Orbitrap generating high-resolution full scan spectra and nominal resolution linear ion trap MS/MS spectra in parallel (24). In a HILIC/Orbitrap MS setup, it was even possible to selectively monitor the 13C labeling enrichment status of the glycerol backbones of all individual lipid species of a lipid class within just one cumulative MS/MS spectrum per lipid class (76). The merits of HILIC separation running on an ultrahigh-performance liquid chromatography (UHPLC)/quadrupole TOF (QTOF)-MS system in high-resolution full scan MS/MS mode have recently resulted in the highest number of gangliosides being reported so far in one chromatographic run (37). The big advantage of this system is its high duty cycle caused by the speed of both the QTOF and the UHPLC. UHPSFC has recently become a very promising separation technique. It excels by its high chromatographic resolution and its speed. The coupling of UHPSFC running in HILIC-like mode to a QTOF mass spectrometer resulted, for example, in separation of 25 lipid classes in just 6 min runtime (77). When compared with UHPLC, it became evident that UHPSFC has the potential to increase the coverage of the lipidome by a factor of 3.4 with a decrease of analysis time by 40% (9).
Ion mobility spectrometry
Ion mobility spectrometry (IMS) separations coupled with MS (IMS-MS) have recently been added to many lipidomics workflows to investigate and separate isomeric and isobaric lipid classes and species. IMS enables rapid gas phase-based size separations by employing different electric field and buffer gas introduction combinations. IMS-based techniques include field asymmetric waveform ion mobility spectrometry (FAIMS), drift tube ion mobility spectrometry (DTIMS), traveling wave ion mobility spectrometry (TWIMS), and trapped ion mobility spectrometry (TIMS) (78, 79). FAIMS (or differential ion mobility spectrometry) is one of the most commonly used IMS-based techniques in current lipidomic studies since its baseline separates lipids of different classes in <1 s (80) and provides trend line groupings (81). DTIMS, TWIMS, and TIMS are also widely used in lipidomic analyses since they can be easily coupled with LC separations. The drift time versus m/z plots for these separations illustrate trend lines for each lipid class similar to the FAIMS results (82, 83, 84, 85). Moreover, it has been shown that DTIMS with a 4 Torr and 1 m IMS drift region separated lipid isomers such as sn-1/sn-2 positional, cis/trans double bond and stereochemical isomers (e.g., R vs. S) (82). While these were very promising results for standards, many of the isomers were only partially separated, limiting the utility of this DTIMS platform for full lipid species separations in complex mixtures and sample types. To obtain better DTIMS separations, both the pressure and length of drift tubes have been increased and advanced multiplexing techniques employed, illustrating greatly improved lipid separation (86, 87). Advances in the other IMS techniques are also showing exciting lipid separation enhancements including long trapping times in TIMS (88), FAIMS devices constructed with higher voltages than commercial versions (89), and the structures for lossless ion manipulation TWIMS platform having a compact serpentine ion drift path ranging from 16 to 500 m (90, 91), and enabling ultrahigh resolving power IMS separations (92, 93).
Lipid identification
A major challenge in lipidomics is the correct identification of lipid species. Misidentifications were discussed for several studies and represent an ongoing issue (10, 94, 95, 96). Identification and annotation of lipid molecules is based on a hierarchical concept related to the structural details provided by the analysis. Annotation of lipid molecules should mirror the structural resolution, as supported by MS-derived evidence, and follow the recently updated shorthand notation for MS-derived lipid structures (13). The fundamental principle for most lipid classes is the detection of lipid class-specific fragments (LCFs) (see https://lipidomics-standards-initiative.org/resources/lipid-class-specific-fragments) and ions specific for variable components, typically hydrocarbon chains, the so-called molecular lipid species-specific fragments (see https://lipidomics-standards-initiative.org/resources/lipid-molecular-species-fragments) (97) (Fig. 4). Consequently, the detection of only LCF justifies the annotation of lipid species, that is, the sum composition of carbon atoms and double bond equivalents of the hydrocarbon chains. Additional detection of molecular lipid species-specific fragments approves the annotation of hydrocarbon chains, that is, the annotation of molecular lipid species. The determination of sn positions requires additional data, for example, application of fatty acyl chain ratios either experimentally determined for specific molecules (98) or included into algorithms (99) as well as response models (60). The identification of double bond positions or other structural details requires sophisticated methods like ozone-mediated (100) or radical-mediated (101) cleavage, derivatization (102), gas-phase chemistry (103), or charge remote fragmentations (104, 105, 106) to generate structure-specific ions.
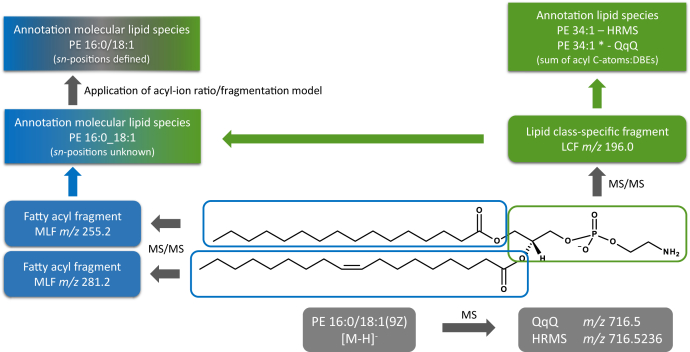
Fig. 4.
Hierarchical lipid annotation. Annotation levels of PE 16:0/18:1(9Z) are related to mass spectrometric data. Annotation at lipid species level requires precursor mass and LCF ion: Without separation of isobaric PE O-35:1 ([M − H]−m/z 716.5600) by HRMS, annotation of PE 34:1 ([M − H]−m/z 716.5236) is based on (∗) the assumption that only even carbon number fatty acyls are present. Molecular lipid species annotation needs the LCF and both molecular lipid fragments (molecular lipid species-specific fragment, here fatty acyl product ions) but does not specify sn-positions PE of 16:0_18:1. Annotation at sn-position level PE 16:0/18:1 demands further data, for example, the application of fatty acyl ion ratios (98, 99) or fragmentation models (60).
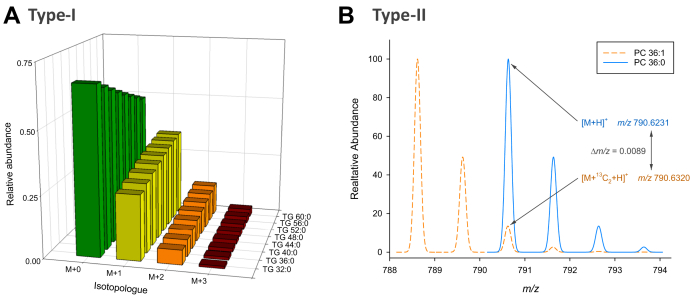
Another important aspect to achieve correct identification is the consideration of isomeric (see https://lipidomics-standards-initiative.org/resources/isomeric-overlap) and isobaric overlap (see https://lipidomics-standards-initiative.org/resources/isobaric-overlap). Thus, PC 33:1 is isomeric with PE 36:1, that is, their sum formula is identical. Therefore, in the positive ion mode, [M + H]+ or [M + Na]+ ions of these species have identical m/z, which requires an additional analytical dimension like MS/MS and formation of distinct LCFs for separation. Isobaric overlap may occur from variations in bond types, for example, ether bond containing PE O-35:1 ([M − H]− m/z 716.5600) has the same nominal m/z as diacyl PE 34:1 ([M − H]− m/z 716.5236). Such kind of isobaric overlap may require assumptions based on biological knowledge, like the presence of only even carbon number fatty acyls (Fig. 4), to annotate species when high-resolution MS (HRMS) is not available. Another isobaric interference inherent in lipidomes is the so-called type II overlap (107) occurring in double bond series within the lipid class (Fig. 5B). This interference on monoisotopic peaks results from the second isotopic peak of a species with one additional double bond, for example, [PC 32:1 + H + 13C2]+ overlaps with [PC 32:0 + H]+). These isobaric peaks only have a mass difference of 8.94 mDa. Therefore, when confident separation of these species is not ensured, type II overlap needs to be evaluated. In nominal resolution MS (low-resolution MS), this effect is typically corrected by calculating isotope patterns in both direct infusion (107, 108) and lipid class separation (72). In contrast, sufficient resolving power in HRMS (e.g., Orbitrap or ICR) can resolve this isobaric overlap (54). Finally, in-source fragmentation could lead to misidentification (see https://lipidomics-standards-initiative.org/resources/in-source-fragmentation) and should be evaluated when ion source parameters are optimized (109, 110).
Fig. 5.
Isotopic effects. A: Type I isotopic effect illustrated for triacylglycerols with increasing number of carbon atoms in all fatty acyl chains. The fraction (abundance is normalized to sum of all isotopologues) of M + 0 decreases, and higher isotopologues increase with the number of carbon atoms. B: Type II isotopic overlap occurs in DB series, that is, within a lipid class, the M + 2-isotopologue of an unsaturated species overlaps M + 0 of species with one double bond less. The overlap of PC 36:1 with PC 36:0 is shown (isotopologue abundance is normalized to the monoisotopic peak M + 0).
In general, the identification principles discussed previously apply to all types of lipidomics approaches. When additional separations, such as chromatography, are utilized, this extra dimension can reduce isomeric and isobaric overlaps. Here, the RT should be used as an additional criterion for lipid species identification. Thus, for lipid class separation like HILIC, the observed RT is similar for the respective lipid class and follows polarity shifts, for example, by hydrocarbon chain length (37, 77) or introduction of additional hydroxyl groups (111). In RP-HPLC, the separation is based on the hydrophobic interaction, which is related to the length of hydrocarbon chains, the number of double bonds, and other structural features, such as polar functional groups. These structural elements directly relate to the RT of the lipid molecules and therefore could be used in mathematical models to justify the correct lipid molecule annotation (66, 112).
Quantification of lipids
The lipid composition of cell membranes or organelles defines their biophysical properties and function (1, 113). Only an accurate quantification of lipid classes and their species as a basis for lipid composition will permit a detailed understanding of the functional role of lipid species. Moreover, repositories of lipid species for any kind of biological material are only valuable in molar concentrations but not in arbitrary units. Molar concentrations make the determination of mass or ratio changes possible and meaningful for studying lipid metabolism. Finally, the confirmation and validation of lipid biomarkers in different laboratories using concentrations is straightforward and will speed up the transition of such markers into clinical diagnostics (10).
The vast majority of lipidomic methods is based on ESI. It is a fundamental principle of this type of ionization that solvent composition, additives, and other components influence the ionization efficiency (52, 64). This influence, also termed matrix effect, could be compensated by addition of IS exposed to the identical ionization conditions (41). Accurate compensation will only be possible if the molecule used as IS is chemically similar (preferably identical) to the analyte and coionizes with the target molecule (114). Therefore, at least one exogenous IS should be added per lipid class as a minimum requirement for reliable comparison.
The classical way for accurate quantification by application of calibration curves using authentic standards and matching stable isotope-labeled IS is only feasible for targeted analysis of a few analytes (67). In lipidomics, structure-response relationships within each lipid class do vary to a certain extent, with some lipid classes being predominantly dependent on the polar head group, whereas others are rather affected by their fatty acyl composition. It is well known that some lipid classes like PC and SM, which are analyzed in positive ion mode using fragment ion m/z 184, are slightly affected by structural modifications, double bond content, and fatty acyl chains because of their inherent head group charge. Some lipid classes like nonpolar cholesteryl ester are substantially affected by the number of carbon atoms and particular double bonds in their fatty acyl chains (53). When fatty acyl fragments are used for glycerophospholipid quantification, beside the number and position of double bonds, carbon number and also the sn positions determine their responses (60). These effects can be addressed by the application of response factor models (53, 60). One factor pertaining to all lipid classes is the so-called type I isotopic effect (107) that describes the decrease of the proportion of monoisotopic peaks for increasing number of carbon atoms (Fig. 5A). This effect could be simply corrected based on the calculated isotopic pattern (107). As discussed previously for identification, when type II overlap is present, it needs to be corrected also for accurate quantification (Fig. 5B). Substantial overlap may occur, for example, in nominal resolution direct infusion analysis when PC and SM M + 1 isotopologues overlap with monoisotopic peaks in positive ion mode precursor ion scans of m/z 184 (108). In HRMS using Orbitrap or ICR-MS, the partial overlap of type II isobaric peaks may affect accurate quantification because of peak interference. This phenomenon requires evaluation and resolution-dependent data analysis (115).
Besides appropriate data handling, other factors like background signal or preanalytic conditions could substantially influence lipid molecule concentrations as discussed previously. Too high or even false reporting may originate from a chemical background. For example, saturated fatty acids like palmitic and stearic acid are ubiquitous in substantial concentrations (116), and therefore, blank samples should monitor such background especially when a limited amount of sample is subjected to analysis.
Data reporting
So far, most of the published lipidomics studies report lipid lists using several dialects of lipid shorthand names (117) and custom formatting of lipid quantities and study variables or other proprietary formats. However, data in such semistructured formats may not be portable because of number encoding issues, and it may be inaccessible to computational approaches because of the absence of an underlying scheme describing the structure of the data. Data repositories such as MetaboLights (118) already require the deposition of both raw data and experimental metadata in structured and accessible formats to allow for better reproducibility and transparency of experimental results. However, for MS data from small molecules in general and lipids in particular, additional information that provides normalized and unambiguous shorthand lipid names, appropriate to the level of structural resolution, as well as details on the m/z signals used for identification and quantification in addition to preanalytical study parameters such as RT and CCS values, is vital to support reported identifications and quantities. This additional evidence facilitates crosschecking for unfounded identifications and typical analytical or data analysis issues (see aforementioned). The data format mzTab-M for metabolomics addresses these issues (119). As a tab-delimited and spreadsheet-like format, mzTab-M is easily accessible by both humans and machines, based on a well-defined pattern and encoding. It captures experimental metadata, summary information on small molecules across assays, MS features as a basis for quantitation, and evidence to support reporting of individual lipid or feature group identifications. The format was developed with widespread consultation of different approaches taken in the field and involvement of software teams from academic research groups as well as industry. The standard has undergone a rigorous peer-review process by both the Metabolomics Standardization Initiative and the Human Proteome Organization's Proteomics Standards Initiative to ensure that the resulting specification is of high quality and mature. In addition, a web application for conveniently validating mzTab-M files by a graphical user interface and a command line validator are available (120). Support for mzTab-M as an output format has already been included in LDA 2 (121), MS-DIAL 4 (122) and MZmine 3 (unpublished), whereas support for mzTab-M as an input format is available in GNPS (123) and MetaboAnalyst (124).
Conclusion
Good lipidomic practice should consider the entire lipid workflow starting from sample collection, lipid extraction, mass spectrometric analysis, identification, and quantification of lipid molecules (Fig. 1). To ensure data quality, the workflow should be evaluated before its application. The depth of method validation could be limited when research questions are addressed based on semiquantitative data, but clinical studies or biomarker validation require quantitative data generated with fully validated methods. As a guidance for researchers and reviewers, LSI currently refines the guidelines for lipidomics including a checklist for quality reporting (10). Other currently ongoing projects should further support and promote the development of lipocentric hierarchical databases like LIPID MAPS (125), SwissLipids (http://www.swisslipids.org/#/) (126), comprehensive lipid compendia, and literature collections, like ”The LipidWeb” (https://www.lipidmaps.org/resources/lipidweb/lipidweb_html/index.html) and the introduction of mzTab-M as a small molecule MS transcommunity data format (119). The aim of these developments is to improve the quality of reported lipidomics data to take lipidomics research to the next level, that is, to facilitate a better understanding of lipid molecule functions in health and disease.
Data availability
All data are contained within the article.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the Austrian Federal Ministry of Education, Science and Research (grant number: BMWFW-10.420/0005-WF/V/3c/2017), the Deutsche Forschungsgemeinschaft (grant numbers: LI 923/4-1 and LI 923/11-1), the Czech Science Foundation (grant number: 21-20238S), the National Institute on Aging (grant numbers: RF1 AG061729 and RF1 AG061872), and the National Institutes of Health (grant numbers: P30 ES025128, P42 ES027704, and P42 ES031009). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author contributions
H. C. K., G. L., and E. S. B. writing–original draft; R. A., K. E., X. H., N. H., M. H., and M. R. W. writing–review and editing.
Funding and additional information
H. C. K. gratefully acknowledges funding from the Austrian Federal Ministry of Education, Science and Research (grant number: BMWFW-10.420/0005-WF/V/3c/2017). M. H. acknowledges the support of grant project no. 21-20238S sponsored by the Czech Science Foundation. X. H. acknowledges the partial support of National Institute on Aging grants (RF1 AG061729 and RF1 AG061872). E. S. B. acknowledges grants from the National Institutes of Health (P30 ES025128, P42 ES027704, and P42 ES031009). G. L. acknowledges funding by the Deutsche Forschungsgemeinschaft (grant numbers: LI 923/4-1 and LI 923/11-1). N. H. acknowledges funding from the Federal Ministry of Education and Research in Germany (BMBF) (grant number: FKZ 031A532B). R. A. acknowledges funding from the Austrian Science Foundation (FWF) (grant number: P33298-B).
Contributor Information
Harald C. Köfeler, Email: harald.koefeler@medunigraz.at.
Gerhard Liebisch, Email: gerhard.liebisch@klinik.uni-regensburg.de.
References
- 1.Wenk M.R. Lipidomics: new tools and applications. Cell. 2010;143:888–895. doi: 10.1016/j.cell.2010.11.033. [DOI] [PubMed] [Google Scholar]
- 2.Holcapek M., Liebisch G., Ekroos K. Lipidomic analysis. Anal. Chem. 2018;90:4249–4257. doi: 10.1021/acs.analchem.7b05395. [DOI] [PubMed] [Google Scholar]
- 3.Han X., Gross R.W. Electrospray ionization mass spectroscopic analysis of human erythrocyte plasma membrane phospholipids. Proc. Natl. Acad. Sci. U. S. A. 1994;91:10635–10639. doi: 10.1073/pnas.91.22.10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brugger B., Erben G., Sandhoff R., Wieland F.T., Lehmann W.D. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 1997;94:2339–2344. doi: 10.1073/pnas.94.6.2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han X., Gross R.W. Global analyses of cellular lipidomes directly from crude extracts of biological samples by ESI mass spectrometry: a bridge to lipidomics. J. Lipid Res. 2003;44:1071–1079. doi: 10.1194/jlr.R300004-JLR200. [DOI] [PubMed] [Google Scholar]
- 6.Spener F., Lagarde M., Géloên A., Record M. Editorial: what is lipidomics? Eur. J. Lipid Sci. Technol. 2003;105:481–482. [Google Scholar]
- 7.Han X., Gross R.W. Shotgun lipidomics: electrospray ionization mass spectrometric analysis and quantitation of cellular lipidomes directly from crude extracts of biological samples. Mass Spectrom. Rev. 2005;24:367–412. doi: 10.1002/mas.20023. [DOI] [PubMed] [Google Scholar]
- 8.Fauland A., Kofeler H., Trotzmuller M., Knopf A., Hartler J., Eberl A., Chitraju C., Lankmayr E., Spener F. A comprehensive method for lipid profiling by liquid chromatography-ion cyclotron resonance mass spectrometry. J. Lipid Res. 2011;52:2314–2322. doi: 10.1194/jlr.D016550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lisa M., Cifkova E., Khalikova M., Ovcacikova M., Holcapek M. Lipidomic analysis of biological samples: comparison of liquid chromatography, supercritical fluid chromatography and direct infusion mass spectrometry methods. J. Chromatogr. A. 2017;1525:96–108. doi: 10.1016/j.chroma.2017.10.022. [DOI] [PubMed] [Google Scholar]
- 10.Liebisch G., Ahrends R., Arita M., Arita M., Bowden J.A., Ejsing C.S., Griffiths W.J., Holcapek M., Köfeler H.C., Mitchell T.W., Wenk M.R., Ekroos K. Lipidomics needs more standardization. Nat. Metab. 2019;1:745–747. doi: 10.1038/s42255-019-0094-z. [DOI] [PubMed] [Google Scholar]
- 11.Worheide M.A., Krumsiek J., Kastenmuller G., Arnold M. Multi-omics integration in biomedical research - a metabolomics-centric review. Anal. Chim. Acta. 2021;1141:144–162. doi: 10.1016/j.aca.2020.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liebisch G., Vizcaino J.A., Kofeler H., Trotzmuller M., Griffiths W.J., Schmitz G., Spener F., Wakelam M.J.O. Shorthand notation for lipid structures derived from mass spectrometry. J. Lipid Res. 2013;54:1523–1530. doi: 10.1194/jlr.M033506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liebisch G., Fahy E., Aoki J., Dennis E.A., Durand T., Ejsing C.S., Fedorova M., Feussner I., Griffiths W.J., Kofeler H., Merrill A.H., Jr., Murphy R.C., O'Donnell V.B., Oskolkova O., Subramaniam S., et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J. Lipid Res. 2020;61:1539–1555. doi: 10.1194/jlr.S120001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebisch G., Drobnik W., Lieser B., Schmitz G. High-throughput quantification of lysophosphatidylcholine by electrospray ionization tandem mass spectrometry. Clin. Chem. 2002;48:2217–2224. [PubMed] [Google Scholar]
- 15.Scherer M., Schmitz G., Liebisch G. High-throughput analysis of sphingosine 1-phosphate, sphinganine 1-phosphate, and lysophosphatidic acid in plasma samples by liquid chromatography-tandem mass spectrometry. Clin. Chem. 2009;55:1218–1222. doi: 10.1373/clinchem.2008.113779. [DOI] [PubMed] [Google Scholar]
- 16.Kim J., Hoppel C.L. Comprehensive approach to the quantitative analysis of mitochondrial phospholipids by HPLC-MS. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2013;912:105–114. doi: 10.1016/j.jchromb.2012.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Okudaira M., Inoue A., Shuto A., Nakanaga K., Kano K., Makide K., Saigusa D., Tomioka Y., Aoki J. Separation and quantification of 2-acyl-1-lysophospholipids and 1-acyl-2-lysophospholipids in biological samples by LC-MS/MS. J. Lipid Res. 2014;55:2178–2192. doi: 10.1194/jlr.D048439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kano K., Matsumoto H., Kono N., Kurano M., Yatomi Y., Aoki J. Suppressing postcollection lysophosphatidic acid metabolism improves the precision of plasma LPA quantification. J. Lipid Res. 2021;62:100029. doi: 10.1016/j.jlr.2021.100029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulmer C.Z., Koelmel J.P., Jones C.M., Garrett T.J., Aristizabal-Henao J.J., Vesper H.W., Bowden J.A. A review of efforts to improve lipid stability during sample preparation and standardization efforts to ensure accuracy in the reporting of lipid measurements. Lipids. 2021;56:3–16. doi: 10.1002/lipd.12263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krautbauer S., Blazquez R., Liebisch G., Hoering M., Neubert P., Pukrop T., Burkhardt R., Sigruener A. Application of lipid class ratios for sample stability monitoring-evaluation of murine tissue homogenates and SDS as a stabilizer. Metabolites. 2021;11:277. doi: 10.3390/metabo11050277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lebaron F.N., Folch J. The effect of pH and salt concentration on aqueous extraction of brain proteins and lipoproteins. J. Neurochem. 1959;4:1–8. doi: 10.1111/j.1471-4159.1959.tb13168.x. [DOI] [PubMed] [Google Scholar]
- 22.Bligh E.G., Dyer W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 23.Matyash V., Liebisch G., Kurzchalia T.V., Shevchenko A., Schwudke D. Lipid extraction by methyl-tert-butyl ether for high-throughput lipidomics. J. Lipid Res. 2008;49:1137–1146. doi: 10.1194/jlr.D700041-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Triebl A., Trotzmuller M., Eberl A., Hanel P., Hartler J., Kofeler H.C. Quantitation of phosphatidic acid and lysophosphatidic acid molecular species using hydrophilic interaction liquid chromatography coupled to electrospray ionization high resolution mass spectrometry. J. Chromatogr. A. 2014;1347:104–110. doi: 10.1016/j.chroma.2014.04.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lofgren L., Stahlman M., Forsberg G.B., Saarinen S., Nilsson R., Hansson G.I. The BUME method: a novel automated chloroform-free 96-well total lipid extraction method for blood plasma. J. Lipid Res. 2012;53:1690–1700. doi: 10.1194/jlr.D023036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhao Z., Xu Y. An extremely simple method for extraction of lysophospholipids and phospholipids from blood samples. J. Lipid Res. 2010;51:652–659. doi: 10.1194/jlr.D001503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Höring M., Krautbauer S., Hiltl L., Babl V., Sigruener A., Burkhardt R., Liebisch G. Accurate lipid quantification of tissue homogenates requires suitable sample concentration, solvent composition, and homogenization procedure—a case study in murine liver. Metabolites. 2021;11:365. doi: 10.3390/metabo11060365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burla B., Arita M., Arita M., Bendt A.K., Cazenave-Gassiot A., Dennis E.A., Ekroos K., Han X., Ikeda K., Liebisch G., Lin M.K., Loh T.P., Meikle P.J., Oresic M., Quehenberger O., et al. MS-based lipidomics of human blood plasma: a community-initiated position paper to develop accepted guidelines. J. Lipid Res. 2018;59:2001–2017. doi: 10.1194/jlr.S087163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reis A., Rudnitskaya A., Blackburn G.J., Mohd Fauzi N., Pitt A.R., Spickett C.M. A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL. J. Lipid Res. 2013;54:1812–1824. doi: 10.1194/jlr.M034330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pati S., Nie B., Arnold R.D., Cummings B.S. Extraction, chromatographic and mass spectrometric methods for lipid analysis. Biomed. Chromatogr. 2016;30:695–709. doi: 10.1002/bmc.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fauland A., Trotzmuller M., Eberl A., Afiuni-Zadeh S., Kofeler H., Guo X., Lankmayr E. An improved SPE method for fractionation and identification of phospholipids. J. Sep. Sci. 2013;36:744–751. doi: 10.1002/jssc.201200708. [DOI] [PubMed] [Google Scholar]
- 32.Pernet F., Pelletier C.J., Milley J. Comparison of three solid-phase extraction methods for fatty acid analysis of lipid fractions in tissues of marine bivalves. J. Chromatogr. A. 2006;1137:127–137. doi: 10.1016/j.chroma.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 33.Bodennec J., Koul O., Aguado I., Brichon G., Zwingelstein G., Portoukalian J. A procedure for fractionation of sphingolipid classes by solid-phase extraction on aminopropyl cartridges. J. Lipid Res. 2000;41:1524–1531. [PubMed] [Google Scholar]
- 34.Wong C.H., Leung D.K., Tang F.P., Wong J.K., Yu N.H., Wan T.S. Rapid screening of anabolic steroids in horse urine with ultra-high-performance liquid chromatography/tandem mass spectrometry after chemical derivatisation. J. Chromatogr. A. 2012;1232:257–265. doi: 10.1016/j.chroma.2011.12.095. [DOI] [PubMed] [Google Scholar]
- 35.Triebl A., Weissengruber S., Trotzmuller M., Lankmayr E., Kofeler H. Quantitative analysis of N-acylphosphatidylethanolamine molecular species in rat brain using solid-phase extraction combined with reversed-phase chromatography and tandem mass spectrometry. J. Sep. Sci. 2016;39:2474–2480. doi: 10.1002/jssc.201600172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayanaswamy P., Shinde S., Sulc R., Kraut R., Staples G., Thiam C.H., Grimm R., Sellergren B., Torta F., Wenk M.R. Lipidomic “deep profiling”: an enhanced workflow to reveal new molecular species of signaling lipids. Anal. Chem. 2014;86:3043–3047. doi: 10.1021/ac4039652. [DOI] [PubMed] [Google Scholar]
- 37.Hajek R., Jirasko R., Lisa M., Cifkova E., Holcapek M. Hydrophilic interaction liquid chromatography-mass spectrometry characterization of gangliosides in biological samples. Anal. Chem. 2017;89:12425–12432. doi: 10.1021/acs.analchem.7b03523. [DOI] [PubMed] [Google Scholar]
- 38.Griffiths W.J., Gilmore I., Yutuc E., Abdel-Khalik J., Crick P.J., Hearn T., Dickson A., Bigger B.W., Wu T.H., Goenka A., Ghosh A., Jones S.A., Wang Y. Identification of unusual oxysterols and bile acids with 7-oxo or 3beta,5alpha,6beta-trihydroxy functions in human plasma by charge-tagging mass spectrometry with multistage fragmentation. J. Lipid Res. 2018;59:1058–1070. doi: 10.1194/jlr.D083246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Honda A., Yamashita K., Miyazaki H., Shirai M., Ikegami T., Xu G., Numazawa M., Hara T., Matsuzaki Y. Highly sensitive analysis of sterol profiles in human serum by LC-ESI-MS/MS. J. Lipid Res. 2008;49:2063–2073. doi: 10.1194/jlr.D800017-JLR200. [DOI] [PubMed] [Google Scholar]
- 40.Lee J.C., Byeon S.K., Moon M.H. Relative quantification of phospholipids based on isotope-labeled methylation by nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry: enhancement in cardiolipin profiling. Anal. Chem. 2017;89:4969–4977. doi: 10.1021/acs.analchem.7b00297. [DOI] [PubMed] [Google Scholar]
- 41.Wang M., Wang C., Han X. Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-what, how and why? Mass Spectrom. Rev. 2017;36:693–714. doi: 10.1002/mas.21492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liebisch G., Drobnik W., Reil M., Trumbach B., Arnecke R., Olgemoller B., Roscher A., Schmitz G. Quantitative measurement of different ceramide species from crude cellular extracts by electrospray ionization tandem mass spectrometry (ESI-MS/MS) J. Lipid Res. 1999;40:1539–1546. [PubMed] [Google Scholar]
- 43.Hsu F.F., Turk J. Characterization of phosphatidylinositol, phosphatidylinositol-4-phosphate, and phosphatidylinositol-4,5-bisphosphate by electrospray ionization tandem mass spectrometry: a mechanistic study. J. Am. Soc. Mass Spectrom. 2000;11:986–999. doi: 10.1016/S1044-0305(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 44.Hsu F.F., Turk J. Charge-remote and charge-driven fragmentation processes in diacyl glycerophosphoethanolamine upon low-energy collisional activation: a mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000;11:892–899. doi: 10.1016/S1044-0305(00)00159-8. [DOI] [PubMed] [Google Scholar]
- 45.Hsu F.F., Turk J. Charge-driven fragmentation processes in diacyl glycerophosphatidic acids upon low-energy collisional activation. A mechanistic proposal. J. Am. Soc. Mass Spectrom. 2000;11:797–803. doi: 10.1016/S1044-0305(00)00151-3. [DOI] [PubMed] [Google Scholar]
- 46.Hsu F.F., Turk J. Electrospray ionization/tandem quadrupole mass spectrometric studies on phosphatidylcholines: the fragmentation processes. J. Am. Soc. Mass Spectrom. 2003;14:352–363. doi: 10.1016/S1044-0305(03)00064-3. [DOI] [PubMed] [Google Scholar]
- 47.Hsu F.F., Turk J. Characterization of phosphatidylethanolamine as a lithiated adduct by triple quadrupole tandem mass spectrometry with electrospray ionization. J. Mass Spectrom. 2000;35:595–606. doi: 10.1002/(SICI)1096-9888(200005)35:5<595::AID-JMS965>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 48.Hsu F.F., Turk J. Structural determination of sphingomyelin by tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2000;11:437–449. doi: 10.1016/S1044-0305(99)00150-6. [DOI] [PubMed] [Google Scholar]
- 49.Hsu F.F., Turk J. Structural determination of glycosphingolipids as lithiated adducts by electrospray ionization mass spectrometry using low-energy collisional-activated dissociation on a triple stage quadrupole instrument. J. Am. Soc. Mass Spectrom. 2001;12:61–79. doi: 10.1016/S1044-0305(00)00194-X. [DOI] [PubMed] [Google Scholar]
- 50.Hsu F.F., Turk J. Characterization of ceramides by low energy collisional-activated dissociation tandem mass spectrometry with negative-ion electrospray ionization. J. Am. Soc. Mass Spectrom. 2002;13:558–570. doi: 10.1016/S1044-0305(02)00358-6. [DOI] [PubMed] [Google Scholar]
- 51.Hsu F.F., Turk J., Stewart M.E., Downing D.T. Structural studies on ceramides as lithiated adducts by low energy collisional-activated dissociation tandem mass spectrometry with electrospray ionization. J. Am. Soc. Mass Spectrom. 2002;13:680–695. doi: 10.1016/S1044-0305(02)00362-8. [DOI] [PubMed] [Google Scholar]
- 52.Han X., Yang K., Gross R.W. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom. Rev. 2012;31:134–178. doi: 10.1002/mas.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Horing M., Ejsing C.S., Hermansson M., Liebisch G. Quantification of cholesterol and cholesteryl ester by direct flow injection high-resolution Fourier transform mass spectrometry utilizing species-specific response factors. Anal. Chem. 2019;91:3459–3466. doi: 10.1021/acs.analchem.8b05013. [DOI] [PubMed] [Google Scholar]
- 54.Almeida R., Pauling J.K., Sokol E., Hannibal-Bach H.K., Ejsing C.S. Comprehensive lipidome analysis by shotgun lipidomics on a hybrid quadrupole-orbitrap-linear ion trap mass spectrometer. J. Am. Soc. Mass Spectrom. 2015;26:133–148. doi: 10.1007/s13361-014-1013-x. [DOI] [PubMed] [Google Scholar]
- 55.Schuhmann K., Almeida R., Baumert M., Herzog R., Bornstein S.R., Shevchenko A. Shotgun lipidomics on a LTQ Orbitrap mass spectrometer by successive switching between acquisition polarity modes. J. Mass Spectrom. 2012;47:96–104. doi: 10.1002/jms.2031. [DOI] [PubMed] [Google Scholar]
- 56.Linden D., William-Olsson L., Ahnmark A., Ekroos K., Hallberg C., Sjogren H.P., Becker B., Svensson L., Clapham J.C., Oscarsson J., Schreyer S. Liver-directed overexpression of mitochondrial glycerol-3-phosphate acyltransferase results in hepatic steatosis, increased triacylglycerol secretion and reduced fatty acid oxidation. FASEB J. 2006;20:434–443. doi: 10.1096/fj.05-4568com. [DOI] [PubMed] [Google Scholar]
- 57.Schuhmann K., Srzentic K., Nagornov K.O., Thomas H., Gutmann T., Coskun U., Tsybin Y.O., Shevchenko A. Monitoring membrane lipidome turnover by metabolic (15)N labeling and shotgun ultra-high-resolution orbitrap Fourier transform mass spectrometry. Anal. Chem. 2017;89:12857–12865. doi: 10.1021/acs.analchem.7b03437. [DOI] [PubMed] [Google Scholar]
- 58.Zullig T., Kofeler H.C. High resolution mass spectrometry in lipidomics. Mass Spectrom. Rev. 2021;40:162–176. doi: 10.1002/mas.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Almeida R., Berzina Z., Arnspang E.C., Baumgart J., Vogt J., Nitsch R., Ejsing C.S. Quantitative spatial analysis of the mouse brain lipidome by pressurized liquid extraction surface analysis. Anal. Chem. 2015;87:1749–1756. doi: 10.1021/ac503627z. [DOI] [PubMed] [Google Scholar]
- 60.Schuhmann K., Moon H., Thomas H., Ackerman J.M., Groessl M., Wagner N., Kellmann M., Henry I., Nadler A., Shevchenko A. Quantitative fragmentation model for bottom-up shotgun lipidomics. Anal. Chem. 2019;91:12085–12093. doi: 10.1021/acs.analchem.9b03270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hu C., Wang C., He L., Han X. Novel strategies for enhancing shotgun lipidomics for comprehensive analysis of cellular lipidomes. Trends Analyt. Chem. 2019;120:115330. doi: 10.1016/j.trac.2018.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwudke D., Oegema J., Burton L., Entchev E., Hannich J.T., Ejsing C.S., Kurzchalia T., Shevchenko A. Lipid profiling by multiple precursor and neutral loss scanning driven by the data-dependent acquisition. Anal. Chem. 2006;78:585–595. doi: 10.1021/ac051605m. [DOI] [PubMed] [Google Scholar]
- 63.Hsu F.F. Mass spectrometry-based shotgun lipidomics - a critical review from the technical point of view. Anal. Bioanal. Chem. 2018;410:6387–6409. doi: 10.1007/s00216-018-1252-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Koivusalo M., Haimi P., Heikinheimo L., Kostiainen R., Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. J. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- 65.Han X., Yang K., Yang J., Fikes K.N., Cheng H., Gross R.W. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J. Am. Soc. Mass Spectrom. 2006;17:264–274. doi: 10.1016/j.jasms.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 66.Ovcacikova M., Lisa M., Cifkova E., Holcapek M. Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography-electrospray ionization mass spectrometry. J. Chromatogr. A. 2016;1450:76–85. doi: 10.1016/j.chroma.2016.04.082. [DOI] [PubMed] [Google Scholar]
- 67.Kauhanen D., Sysi-Aho M., Koistinen K.M., Laaksonen R., Sinisalo J., Ekroos K. Development and validation of a high-throughput LC-MS/MS assay for routine measurement of molecular ceramides. Anal. Bioanal. Chem. 2016;408:3475–3483. doi: 10.1007/s00216-016-9425-z. [DOI] [PubMed] [Google Scholar]
- 68.Triebl A., Trotzmuller M., Hartler J., Stojakovic T., Kofeler H.C. Lipidomics by ultrahigh performance liquid chromatography-high resolution mass spectrometry and its application to complex biological samples. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2017;1053:72–80. doi: 10.1016/j.jchromb.2017.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Danne-Rasche N., Coman C., Ahrends R. Nano-LC/NSI MS refines lipidomics by enhancing lipid coverage, measurement sensitivity, and linear dynamic range. Anal. Chem. 2018;90:8093–8101. doi: 10.1021/acs.analchem.8b01275. [DOI] [PubMed] [Google Scholar]
- 70.Schott H.F., Krautbauer S., Horing M., Liebisch G., Matysik S. A validated, fast method for quantification of sterols and gut microbiome derived 5alpha/beta-stanols in human feces by isotope dilution LC-high-resolution MS. Anal. Chem. 2018;90:8487–8494. doi: 10.1021/acs.analchem.8b01278. [DOI] [PubMed] [Google Scholar]
- 71.Cifkova E., Holcapek M., Lisa M., Ovcacikova M., Lycka A., Lynen F., Sandra P. Nontargeted quantitation of lipid classes using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry with single internal standard and response factor approach. Anal. Chem. 2012;84:10064–10070. doi: 10.1021/ac3024476. [DOI] [PubMed] [Google Scholar]
- 72.Scherer M., Schmitz G., Liebisch G. Simultaneous quantification of cardiolipin, bis(monoacylglycero)phosphate and their precursors by hydrophilic interaction LC-MS/MS including correction of isotopic overlap. Anal. Chem. 2010;82:8794–8799. doi: 10.1021/ac1021826. [DOI] [PubMed] [Google Scholar]
- 73.Yu X., Chen K., Li S., Wang Y., Shen Q. Lipidomics differentiation of soft-shelled turtle strains using hydrophilic interaction liquid chromatography and mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2019;1112:11–15. doi: 10.1016/j.jchromb.2019.02.025. [DOI] [PubMed] [Google Scholar]
- 74.Ventura G., Bianco M., Calvano C.D., Losito I., Cataldi T.R.I. HILIC-ESI-FTMS with all ion fragmentation (AIF) scans as a tool for fast lipidome investigations. Molecules. 2020;25:2310. doi: 10.3390/molecules25102310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.da Costa E., Azevedo V., Melo T., Rego A.M., Evtuguin D.V., Domingues P., Calado R., Pereira R., Abreu M.H., Domingues M.R. High-resolution lipidomics of the early life stages of the red seaweed Porphyra dioica. Molecules. 2018;23:187. doi: 10.3390/molecules23010187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Leithner K., Triebl A., Trotzmuller M., Hinteregger B., Leko P., Wieser B.I., Grasmann G., Bertsch A.L., Zullig T., Stacher E., Valli A., Prassl R., Olschewski A., Harris A.L., Kofeler H.C., et al. The glycerol backbone of phospholipids derives from noncarbohydrate precursors in starved lung cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2018;115:6225–6230. doi: 10.1073/pnas.1719871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lisa M., Holcapek M. High-throughput and comprehensive lipidomic analysis using ultrahigh-performance supercritical fluid chromatography-mass spectrometry. Anal. Chem. 2015;87:7187–7195. doi: 10.1021/acs.analchem.5b01054. [DOI] [PubMed] [Google Scholar]
- 78.Kliman M., May J.C., McLean J.A. Lipid analysis and lipidomics by structurally selective ion mobility-mass spectrometry. Biochim. Biophys. Acta. 2011;1811:935–945. doi: 10.1016/j.bbalip.2011.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dodds J.N., Baker E.S. Ion mobility spectrometry: fundamental concepts, instrumentation, applications, and the road ahead. J. Am. Soc. Mass Spectrom. 2019;30:2185–2195. doi: 10.1007/s13361-019-02288-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Baker P.R., Armando A.M., Campbell J.L., Quehenberger O., Dennis E.A. Three-dimensional enhanced lipidomics analysis combining UPLC, differential ion mobility spectrometry, and mass spectrometric separation strategies. J. Lipid Res. 2014;55:2432–2442. doi: 10.1194/jlr.D051581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shvartsburg A.A., Isaac G., Leveque N., Smith R.D., Metz T.O. Separation and classification of lipids using differential ion mobility spectrometry. J. Am. Soc. Mass Spectrom. 2011;22:1146–1155. doi: 10.1007/s13361-011-0114-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kyle J.E., Zhang X., Weitz K.K., Monroe M.E., Ibrahim Y.M., Moore R.J., Cha J., Sun X., Lovelace E.S., Wagoner J., Polyak S.J., Metz T.O., Dey S.K., Smith R.D., Burnum-Johnson K.E., et al. Uncovering biologically significant lipid isomers with liquid chromatography, ion mobility spectrometry and mass spectrometry. Analyst. 2016;141:1649–1659. doi: 10.1039/c5an02062j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Paglia G., Angel P., Williams J.P., Richardson K., Olivos H.J., Thompson J.W., Menikarachchi L., Lai S., Walsh C., Moseley A., Plumb R.S., Grant D.F., Palsson B.O., Langridge J., Geromanos S., et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 2015;87:1137–1144. doi: 10.1021/ac503715v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Paglia G., Kliman M., Claude E., Geromanos S., Astarita G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015;407:4995–5007. doi: 10.1007/s00216-015-8664-8. [DOI] [PubMed] [Google Scholar]
- 85.Paglia G., Astarita G. Metabolomics and lipidomics using traveling-wave ion mobility mass spectrometry. Nat. Protoc. 2017;12:797–813. doi: 10.1038/nprot.2017.013. [DOI] [PubMed] [Google Scholar]
- 86.Groessl M., Graf S., Knochenmuss R. High resolution ion mobility-mass spectrometry for separation and identification of isomeric lipids. Analyst. 2015;140:6904–6911. doi: 10.1039/c5an00838g. [DOI] [PubMed] [Google Scholar]
- 87.May J.C., Knochenmuss R., Fjeldsted J.C., McLean J.A. Resolution of isomeric mixtures in ion mobility using a combined demultiplexing and peak deconvolution technique. Anal. Chem. 2020;92:9482–9492. doi: 10.1021/acs.analchem.9b05718. [DOI] [PubMed] [Google Scholar]
- 88.Silveira J.A., Ridgeway M.E., Park M.A. High resolution trapped ion mobility spectrometery of peptides. Anal. Chem. 2014;86:5624–5627. doi: 10.1021/ac501261h. [DOI] [PubMed] [Google Scholar]
- 89.Bowman A.P., Abzalimov R.R., Shvartsburg A.A. Broad separation of isomeric lipids by high-resolution differential ion mobility spectrometry with tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 2017;28:1552–1561. doi: 10.1007/s13361-017-1675-2. [DOI] [PubMed] [Google Scholar]
- 90.Deng L., Ibrahim Y.M., Baker E.S., Aly N.A., Hamid A.M., Zhang X., Zheng X., Garimella S.V.B., Webb I.K., Prost S.A., Sandoval J.A., Norheim R.V., Anderson G.A., Tolmachev A.V., Smith R.D. Ion mobility separations of isomers based upon long path length structures for lossless ion manipulations combined with mass spectrometry. ChemistrySelect. 2016;1:2396–2399. doi: 10.1002/slct.201600460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ibrahim Y.M., Hamid A.M., Deng L., Garimella S.V., Webb I.K., Baker E.S., Smith R.D. New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst. 2017;142:1010–1021. doi: 10.1039/c7an00031f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wojcik R., Webb I.K., Deng L., Garimella S.V., Prost S.A., Ibrahim Y.M., Baker E.S., Smith R.D. Lipid and glycolipid isomer analyses using ultra-high resolution ion mobility spectrometry separations. Int. J. Mol. Sci. 2017;18:183. doi: 10.3390/ijms18010183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wormwood Moser K.L., Van Aken G., DeBord D., Hatcher N.G., Maxon L., Sherman M., Yao L., Ekroos K. High-defined quantitative snapshots of the ganglioside lipidome using high resolution ion mobility SLIM assisted shotgun lipidomics. Anal. Chim. Acta. 2021;1146:77–87. doi: 10.1016/j.aca.2020.12.022. [DOI] [PubMed] [Google Scholar]
- 94.Liebisch G., Ejsing C.S., Ekroos K. Identification and annotation of lipid species in metabolomics studies need improvement. Clin. Chem. 2015;61:1542–1544. doi: 10.1373/clinchem.2015.244830. [DOI] [PubMed] [Google Scholar]
- 95.Koelmel J.P., Ulmer C.Z., Jones C.M., Yost R.A., Bowden J.A. Common cases of improper lipid annotation using high-resolution tandem mass spectrometry data and corresponding limitations in biological interpretation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2017;1862:766–770. doi: 10.1016/j.bbalip.2017.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kofeler H.C., Eichmann T.O., Ahrends R., Bowden J.A., Danne-Rasche N., Dennis E.A., Fedorova M., Griffiths W.J., Han X., Hartler J., Holcapek M., Jirasko R., Koelmel J.P., Ejsing C.S., Liebisch G., et al. Quality control requirements for the correct annotation of lipidomics data. Nat. Commun. 2021;12:4771. doi: 10.1038/s41467-021-24984-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pauling J.K., Hermansson M., Hartler J., Christiansen K., Gallego S.F., Peng B., Ahrends R., Ejsing C.S. Proposal for a common nomenclature for fragment ions in mass spectra of lipids. PLoS One. 2017;12 doi: 10.1371/journal.pone.0188394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ekroos K., Ejsing C.S., Bahr U., Karas M., Simons K., Shevchenko A. Charting molecular composition of phosphatidylcholines by fatty acid scanning and ion trap MS3 fragmentation. J. Lipid Res. 2003;44:2181–2192. doi: 10.1194/jlr.D300020-JLR200. [DOI] [PubMed] [Google Scholar]
- 99.Wozny K., Lehmann W.D., Wozny M., Akbulut B.S., Brugger B. A method for the quantitative determination of glycerophospholipid regioisomers by UPLC-ESI-MS/MS. Anal. Bioanal. Chem. 2019;411:915–924. doi: 10.1007/s00216-018-1517-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Marshall D.L., Criscuolo A., Young R.S.E., Poad B.L.J., Zeller M., Reid G.E., Mitchell T.W., Blanksby S.J. Mapping unsaturation in human plasma lipids by data-independent ozone-induced dissociation. J. Am. Soc. Mass Spectrom. 2019;30:1621–1630. doi: 10.1007/s13361-019-02261-z. [DOI] [PubMed] [Google Scholar]
- 101.Pham H.T., Ly T., Trevitt A.J., Mitchell T.W., Blanksby S.J. Differentiation of complex lipid isomers by radical-directed dissociation mass spectrometry. Anal. Chem. 2012;84:7525–7532. doi: 10.1021/ac301652a. [DOI] [PubMed] [Google Scholar]
- 102.Ma X., Chong L., Tian R., Shi R., Hu T.Y., Ouyang Z., Xia Y. Identification and quantitation of lipid C=C location isomers: a shotgun lipidomics approach enabled by photochemical reaction. Proc. Natl. Acad. Sci. U. S. A. 2016;113:2573–2578. doi: 10.1073/pnas.1523356113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Randolph C.E., Blanksby S.J., McLuckey S.A. Toward complete structure elucidation of glycerophospholipids in the gas phase through charge inversion ion/ion chemistry. Anal. Chem. 2020;92:1219–1227. doi: 10.1021/acs.analchem.9b04376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frankfater C., Jiang X., Hsu F.F. Characterization of long-chain fatty acid as N-(4-aminomethylphenyl) pyridinium derivative by MALDI LIFT-TOF/TOF mass spectrometry. J. Am. Soc. Mass Spectrom. 2018;29:1688–1699. doi: 10.1007/s13361-018-1993-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Jensen N.J., Gross M.L. Mass spectrometry methods for structural determination and analysis of fatty acids. Mass Spectrom. Rev. 1987;6:497–536. [Google Scholar]
- 106.Pittenauer E., Allmaier G. The renaissance of high-energy CID for structural elucidation of complex lipids: MALDI-TOF/RTOF-MS of alkali cationized triacylglycerols. J. Am. Soc. Mass Spectrom. 2009;20:1037–1047. doi: 10.1016/j.jasms.2009.01.009. [DOI] [PubMed] [Google Scholar]
- 107.Han X., Gross R.W. Quantitative analysis and molecular species fingerprinting of triacylglyceride molecular species directly from lipid extracts of biological samples by electrospray ionization tandem mass spectrometry. Anal. Biochem. 2001;295:88–100. doi: 10.1006/abio.2001.5178. [DOI] [PubMed] [Google Scholar]
- 108.Liebisch G., Lieser B., Rathenberg J., Drobnik W., Schmitz G. High-throughput quantification of phosphatidylcholine and sphingomyelin by electrospray ionization tandem mass spectrometry coupled with isotope correction algorithm. Biochim. Biophys. Acta. 2004;1686:108–117. doi: 10.1016/j.bbalip.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 109.Gathungu R.M., Larrea P., Sniatynski M.J., Marur V.R., Bowden J.A., Koelmel J.P., Starke-Reed P., Hubbard V.S., Kristal B.S. Optimization of electrospray ionization source parameters for lipidomics to reduce misannotation of in-source fragments as precursor ions. Anal. Chem. 2018;90:13523–13532. doi: 10.1021/acs.analchem.8b03436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hu C., Luo W., Xu J., Han X. Recognition and avoidance of ion source-generated artifacts in lipidomics analysis. Mass Spectrom. Rev. September 30, 2020 doi: 10.1002/mas.21659. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Horejsi K., Jirasko R., Chocholouskova M., Wolrab D., Kahoun D., Holcapek M. Comprehensive identification of glycosphingolipids in human plasma using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry. Metabolites. 2021;11:140. doi: 10.3390/metabo11030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vankova Z., Peterka O., Chocholouskova M., Wolrab D., Jirasko R., Holcapek M. Retention dependences support highly confident identification of lipid species in human plasma by reversed-phase UHPLC/MS. Anal. Bioanal. Chem. July 9, 2021 doi: 10.1007/s00216-021-03492-4. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 113.Ernst R., Ejsing C.S., Antonny B. Homeoviscous adaptation and the regulation of membrane lipids. J. Mol. Biol. 2016;428:4776–4791. doi: 10.1016/j.jmb.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 114.Krautbauer S., Buchler C., Liebisch G. Relevance in the use of appropriate internal standards for accurate quantification using LC-MS/MS: tauro-conjugated bile acids as an example. Anal. Chem. 2016;88:10957–10961. doi: 10.1021/acs.analchem.6b02596. [DOI] [PubMed] [Google Scholar]
- 115.Horing M., Ejsing C.S., Krautbauer S., Ertl V.M., Burkhardt R., Liebisch G. Accurate quantification of lipid species affected by isobaric overlap in Fourier-transform mass spectrometry. J. Lipid Res. 2021;62:100050. doi: 10.1016/j.jlr.2021.100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Gallego S.F., Hermansson M., Liebisch G., Hodson L., Ejsing C.S. Total fatty acid analysis of human blood samples in one minute by high-resolution mass spectrometry. Biomolecules. 2018;9:7. doi: 10.3390/biom9010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kopczynski D., Hoffmann N., Peng B., Ahrends R. Goslin: a grammar of succinct lipid nomenclature. Anal. Chem. 2020;92:10957–10960. doi: 10.1021/acs.analchem.0c01690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Haug K., Cochrane K., Nainala V.C., Williams M., Chang J., Jayaseelan K.V., O'Donovan C. MetaboLights: a resource evolving in response to the needs of its scientific community. Nucleic Acids Res. 2020;48:D440–D444. doi: 10.1093/nar/gkz1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Hoffmann N., Rein J., Sachsenberg T., Hartler J., Haug K., Mayer G., Alka O., Dayalan S., Pearce J.T.M., Rocca-Serra P., Qi D., Eisenacher M., Perez-Riverol Y., Vizcaino J.A., Salek R.M., et al. mzTab-M: a data standard for sharing quantitative results in mass spectrometry metabolomics. Anal. Chem. 2019;91:3302–3310. doi: 10.1021/acs.analchem.8b04310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Hoffmann N., Hartler J., Ahrends R. jmzTab-M: a reference parser, writer, and validator for the proteomics standards initiative mzTab 2.0 metabolomics standard. Anal. Chem. 2019;91:12615–12618. doi: 10.1021/acs.analchem.9b01987. [DOI] [PubMed] [Google Scholar]
- 121.Hartler J., Triebl A., Ziegl A., Trotzmuller M., Rechberger G.N., Zeleznik O.A., Zierler K.A., Torta F., Cazenave-Gassiot A., Wenk M.R., Fauland A., Wheelock C.E., Armando A.M., Quehenberger O., Zhang Q., et al. Deciphering lipid structures based on platform-independent decision rules. Nat. Methods. 2017;14:1171–1174. doi: 10.1038/nmeth.4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tsugawa H., Ikeda K., Takahashi M., Satoh A., Mori Y., Uchino H., Okahashi N., Yamada Y., Tada I., Bonini P., Higashi Y., Okazaki Y., Zhou Z., Zhu Z.J., Koelmel J., et al. A lipidome atlas in MS-DIAL 4. Nat. Biotechnol. 2020;38:1159–1163. doi: 10.1038/s41587-020-0531-2. [DOI] [PubMed] [Google Scholar]
- 123.Wang M., Carver J.J., Phelan V.V., Sanchez L.M., Garg N., Peng Y., Nguyen D.D., Watrous J., Kapono C.A., Luzzatto-Knaan T., Porto C., Bouslimani A., Melnik A.V., Meehan M.J., Liu W.T., et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat. Biotechnol. 2016;34:828–837. doi: 10.1038/nbt.3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Pang Z., Chong J., Zhou G., de Lima Morais D.A., Chang L., Barrette M., Gauthier C., Jacques P.E., Li S., Xia J. MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021;49:W388–W396. doi: 10.1093/nar/gkab382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sud M., Fahy E., Cotter D., Brown A., Dennis E.A., Glass C.K., Merrill A.H., Jr., Murphy R.C., Raetz C.R., Russell D.W., Subramaniam S. LMSD: LIPID MAPS structure database. Nucleic Acids Res. 2007;35:D527–D532. doi: 10.1093/nar/gkl838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Aimo L., Liechti R., Hyka-Nouspikel N., Niknejad A., Gleizes A., Gotz L., Kuznetsov D., David F.P., van der Goot F.G., Riezman H., Bougueleret L., Xenarios I., Bridge A. The SwissLipids knowledgebase for lipid biology. Bioinformatics. 2015;31:2860–2866. doi: 10.1093/bioinformatics/btv285. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data are contained within the article.