Abstract
The PTEN gene is a tumor suppressor localized in the frequently altered chromosomal region 10q23. The tumor suppressor function of the PTEN protein (PTEN) has been linked to its ability to dephosphorylate the lipid second-messenger phosphatidylinositol 3,4,5-trisphosphate and phosphatidylinositol 3,4-bisphosphate and, by doing so, to antagonize the phosphoinositide 3-kinase pathway. The PTEN protein consists of an amino-terminal phosphatase domain, a lipid binding C2 domain, and a 50-amino-acid C-terminal domain (the “tail”) of unknown function. A number of studies have shown that the tail is dispensable for both phosphatase activity and blocking cell growth. Here, we show that the PTEN tail is necessary for maintaining protein stability and that it also acts to inhibit PTEN function. Thus, removing the tail results in a loss of stability but does not result in a loss of function because the resultant protein is more active. Furthermore, tail-dependent regulation of stability and activity is linked to the phosphorylation of three residues (S380, T382, and T383) within the tail. Therefore, the tail is likely to mediate the regulation of PTEN function through phosphorylation.
The PTEN gene was cloned as a candidate tumor suppressor gene from the chromosome 10q23 region, a locus frequently targeted for genetic loss in tumors (24, 26, 42). Somatic inactivation of both PTEN alleles and loss of heterozygosity have been demonstrated in a number of tumors including glioblastoma, melanoma, and prostate, breast, and endometrial carcinomas (reviewed in reference 46). Germ line PTEN mutations are associated with the development of the related dominantly inherited disorders known as Cowden disease and Bannayan-Zonana syndrome (28–30, 34). These disorders are characterized by the presence of benign hamartomas of the skin, intestinal tract, and central nervous system and by an increased incidence of cancers of the thyroid and breast (28, 29, 34). Similarly, heterozygous PTEN mice develop a variety of tumors and proliferative lesions of multiple tissues (10, 11, 37, 43).
Reconstitution of PTEN expression to certain PTEN null cells results in an increase in the population of cells in the G1 phase of the cell cycle (13, 23, 39); in other PTEN null cells it results in the induction of apoptosis or anoikis (9, 25, 32). Accumulating evidence suggests that these functions are linked to the lipid phosphatase activity of PTEN, which allows PTEN to antagonize the phosphatidylinositol 3-kinase (PI3K) pathway (reviewed in references 4 and 46). A number of downstream targets of phosphatidylinositol 3,4,5-triphosphate and phosphatidylinositol 3,4-bisphosphate including the serine-threonine kinase Akt, BTK, SGK, and p70S6K, have been identified (6, 12, 18–20, 27). Akt, in particular, appears to play a role in both proliferative and apoptotic signals. Constitutive activation of Akt has been found in cells that lack functional PTEN, and PTEN can inhibit Akt kinase activity in cells. A number of downstream targets of Akt have been described and include GSK3, BAD, caspase-9, IKKα, and the forkhead transcription factors FKHR, FKHRL1, and AFX (2, 3, 5, 7, 8, 21, 36, 44). Our group has recently found that forkhead transcription factors are inactive in PTEN null cells and that reconstitution of FKHR activity, in the absence of PTEN, can induce both cell cycle arrest and apoptosis in susceptible PTEN null cells (N. Nakamura, S. Ramaswamy, F. Vazquez, and W. Sellers, submitted for publication).
Each molecular constituent of the PI3K pathway, such as receptor tyrosine kinases, PI3K, and Akt, is subjected to regulation of its activity. Likewise, it has been speculated that PTEN might be regulated, but to date evidence of such regulation has remained elusive (4). In keeping with the idea that PTEN might be regulated, protein phosphatases in general are regulated by a number of mechanisms including phosphorylation, second messengers, regulatory subunits, subcellular localization, dimerization, and binding to inhibitory proteins (reviewed in references 1 and 17).
The PTEN protein contains the signature motif (HCXXGXXR) of the family of protein tyrosine phosphatases and dual-specificity phosphatases. The PTEN crystal structure shows that PTEN consists of an amino-terminal phosphatase domain (PD; residues 7 to 185), which includes the phosphatase signature motif, and a lipid binding C2 domain that extends from residues 186 to 351. C2 domains, named for homology to a domain found in protein kinase C (PKC), have been identified in a number of proteins involved in signal transduction or membrane trafficking such as PKC, cPLA2, phospholipase Cs, and synaptotagmins (reviewed in reference 40). C2 domains can play a role in mediating Ca2+-dependent lipid interactions. However, the C2 domain of PTEN is unlikely to bind Ca2+, and its in vitro binding to lipids is independent of Ca2+ (22). The last 50 amino acid residues (354 to 403) (referred to herein as the “tail”) were not crystallized, and structural prediction programs fail to identify regions of secondary structure. The function of this domain and its relationship in three dimensions to the remainder of the protein remain unknown.
The PTEN tail is dispensable for PTEN phosphatase activity and for activity in a number of cellular assays including soft-agar colony suppression assays (14, 22; S. Ramaswamy and W. R. Sellers, unpublished data). Here we show that the tail is necessary for maintaining PTEN stability. However, deletion of the tail also results in an increase in activity as measured by the ability of PTEN to induce a G1 arrest or to induce the transcriptional activity of FKHR. Thus, deletion of the tail does not result in a loss of PTEN function because, while unstable, the resultant protein is more active. We further demonstrate that the tail is a site for PTEN phosphorylation and that phosphorylation of the tail regulates both PTEN stability and activity.
MATERIALS AND METHODS
Plasmids.
pCD19, pSG5L, pSG5L-HA-PTEN, pSG5L-HA-PTEN;1-393, pSG5L-HA-PTEN;1-373, pSG5L-HA-PTEN;1-353, pSG5L-HA-PTEN;1-343, pSG5L-HA-PTEN;1-336; pcDNA3-Flag-FKHR, and pGL3-promoter-FasL were described previously (39, 41, 44, 45; Nakamura et al., submitted).
pSG5L-HA-PTEN;360ΔA, pSG5L-HA-PTEN;S370A, pSG5L-HA-PTEN;A4, pSG5L-HA-PTEN;S380A, pSG5L-HA-PTEN;T382A, pSG5L-HA-PTEN;T383A, pSG5L-HA-PTEN;S385A, pSG5L-HA-PTEN;A3, and pSG5L-HA-PTEN;D3 were generated by site-directed mutagenesis using single-stranded DNA generated from pSG5L-HA-PTEN (Muta-Gene; Bio-Rad). The oligonucleotides used for site-directed mutagenesis are the following: 5′-CACCAGATGTggccGACAATGAAC-3′ (PTEN;S370A), 5′-GATCATTATAGATATgCTGACgCCgCgGACgCaGATCCAGAGAATGAAC-3′ (PTEN;A4), 5′-CTGATCATTATcGaTATgCaGACACaACTGACTCTG-3′ (PTEN; S380A), 5′-GATCATTATcgaTATTCTGACgcaACTGACTCTG-3′ (PTEN;T382A), 5′-GATATTCTGACACCgcgGACTCTGATC-3′ (PTEN;T383A), 5′-GATATTCTGACACtACaGACgcaGATCCAGAG-3′ (PTEN:S385A), 5′-GATCATTATAGATATgCTGACgCCgCgGACTCTGATCCAGAG-3′ (PTEN;A3), 5′-GATCATTATcGATATgaTGACgaCgaTGACTCTGATCCAG-3′ (PTEN;D3).
Antibodies and immunoblotting.
HA-11 (Babco), antihemagglutinin (HA) antibody was used for immunoblotting at 1:1,000; C54 anti-PTEN serum was previously described and was used at 1:1,000 dilution (39).
Cells were washed in phosphate-buffered saline, and cellular proteins were extracted in TNN buffer (150 mM NaCl, 50 mM Tris [pH 7.4], 0.5% NP-40) for 20 min at 4°C. Lysates were cleared by centrifugation, and proteins were separated by gel electrophoresis. Immunoblots were obtained essentially as described previously (39). Briefly, membranes were blocked in Tris-buffered saline–0.05% Triton X-100 (TBS-T)–4% (wt/vol) milk for 1 h at room temperature (RT). Membranes were then incubated with primary antibodies diluted in TBS-T–4% (wt/vol) milk for 1 h at RT. Subsequently, membranes were washed with TBS-T and incubated with horseradish peroxidase secondary antibody (1:20,000; Pierce Chemicals) diluted in TBS-T–4% (wt/vol) milk. Membranes were washed in TBS-T, and bound antibody was detected by enhanced chemiluminescence (Pierce Supersignal).
Cell lines, cell culture, and transfection.
786-0 and ACHN renal carcinoma cells and U2-OS osteosarcoma cells were maintained in Dulbecco's modified Eagle medium (DMEM) containing 4,500 mg of glucose/ml, 2 mM l-glutamine, 10% fetal clone (HyClone), and penicillin and streptomycin and were maintained at 37°C in a humidified 10% CO2 atmosphere. 786-0 cells were transfected using Fugene reagent (Boehringer Mannheim), and U2-OS cells were transfected using calcium phosphate (BBS method), as previously described (39).
Pulse-chase labeling.
786-0 cells were transfected with various pSG5L-HA-PTEN plasmids and split into p60 plates. Forty hours after transfection, cells were washed twice with methionine-free DMEM and then incubated for 45 min in methionine-free DMEM with 10% dialyzed fetal bovine serum (DFBS) (Gibco BRL). Cells were then incubated for 45 min with methionine-free DMEM–10% DFBS containing [35S]methionine (150 μCi/ml) (NEN Life Science Products). The medium was then replaced with complete medium. HA epitope-tagged proteins were isolated by anti-HA immunoprecipitation and resolved on a 7.5% polyacrylamide gel. The labeled protein present at each time point was quantified by phosphorimaging and normalized to the amount of protein present at the zero-time point.
Cell cycle assays.
Cell cycle assays were performed essentially as previously described (39). Briefly, 786-0 cells were cotransfected with 4 μg of pCD19 plasmid along with pSG5L vector or the relevant pSG5L-HA-PTEN wild-type or mutant plasmid. Forty hours after transfection the cells were harvested with trypsin, stained with fluorescein isothiocyanate-conjugated anti-CD19 antibody (CalTag), fixed in 70% ethanol, and stained with propidium iodide in the presence of RNase A. The cell cycle profile of the CD19-positive cells was determined by two-color fluorescence-activated cell sorting (FACS). Data are shown as the percentages of increase in the G1 population. This was determined by dividing the absolute percentage difference between the vector control and the experimental data point by the percentage of G1 cells in the vector and then multiplying by 100.
Luciferase reporter assays.
FKHR transactivation assays were performed essentially as described previously (Nakamura et al., submitted). Briefly, cells were transfected in 12-well plates with 0.25 μg of the FasL luciferase and β-galactosidase reporter plasmids, 0.5 μg of pCDNA3-FHKR, and various amounts of pSG5L-HA-PTEN. Cells were lysed 40 h after transfection using 1× reporter lysis buffer by following the manufacturer's instructions (Promega). Luciferase and β-galactosidase activities were measured as described previously (41). Luciferase activity was normalized to β-galactosidase activity. Fold activation was calculated by dividing the normalized luciferase activity by the normalized activity obtained in the presence of the vector and reporter plasmid alone.
Metabolic labeling, proteolytic digestions, and phosphoamino acid analysis.
ACHN or transfected U2-OS cells were washed twice with phosphate-free DMEM. Then the medium was changed to a mixture of phosphate-free DMEM, 10% DFBS, and 1 mCi (endogenous) or 200 μCi (transfected) of [32P]orthophosphate (NEN Life Science Products)/ml, and the cells were incubated from 2 to 4 h. Labeled proteins were isolated by immunoprecipitation using HA-11 (transfected) or C54 (endogenous) antibodies, resolved by sodium dodecyl sulfate–7.5% polyacrylamide gel electrophoresis, and transferred to a nitrocellulose membrane. The phosphorylated proteins were visualized by autoradiography.
Proteolytic digestions were done essentially as described previously (38). Membrane pieces containing phosphoproteins were excised, washed with double-distilled water (ddH20), and blocked with 0.5% polyvinylpyrrolidone MW360 (PVP-360) in 100 mM acetic acid for 30 min at 37°C. Digestion was performed with 5 μg of sequencing-grade trypsin (Promega) overnight at 37°C. Peptides were twice lyophilized to dryness and washed with ddH20. Peptides were then resuspended in a small volume of Laemmli sample buffer and then resolved in 16.5% Tris-Tricine gels.
For phosphoamino acid analysis a fraction of the tryptic digested peptide was hydrolyzed in 5.7 N HCl. The resulting amino acids were then lyophilized, washed with ddH20, and resuspended in a small volume of ddH20. Samples were then spotted on thin-layer chromatography (TLC) plates (EM Science) together with 1 μg of cold phosphoamino acid standards (Sigma). Phosphoamino acids were then resolved by electrophoresis at pH 1.9 in a buffer containing 2.5% (vol/vol) formic acid (88%) and 7.8% (vol/vol) glacial acetic acid for 45 min at 800 V in the first dimension and by chromatography in the second dimension in a buffer containing 70% (vol/vol) 2-propanol and 15% (vol/vol) HCl. Cold phosphoamino acid standards were visualized by developing the TLC plates with 0.2% (wt/vol) ninhydrin in acetone and baking them at 65°C until color developed.
RESULTS
The PTEN tail modulates PTEN stability.
Recently the crystal structure of PTEN has been solved from residues 7 to 353 (eliminating an internal loop of residues 286 to 309). This truncated protein has in vitro lipid phosphatase activity comparable to that of full-length PTEN (PTEN;WT) and can induce apoptosis in LNCaP cells to the same extent as the wild type (14, 22). Similarly, our group mapped the minimal in vivo functional domain of PTEN by C-terminal and N-terminal deletion mutations (S. Ramaswamy and W. R. Sellers, unpublished data). We found that a truncated PTEN protein of residues 10 to 353 retained protein and lipid phosphatase activity in vitro and was able to induce a G1 arrest in cells. Furthermore, PTEN;1-353 was comparable to PTEN;WT in suppressing soft-agar colony formation in PTEN null renal carcinoma cells (786-0 cells) (S. Ramaswamy and W. R. Sellers, unpublished data). These results indicate that the last 50 amino acids of PTEN are not necessary for lipid or protein phosphatase activity or for its ability to inhibit proliferation or induce apoptosis. For simplicity we refer to these last 50 residues as the PTEN tail (Fig. 1A).
FIG. 1.
The PTEN tail is required for protein stability. (A) Schematic representation of the PTEN protein. Dark gray box, phosphatase signature motif (HCXXGXXR); light gray box, PD; white box, C2 domain; black box, C-terminal 50-residues (PTEN tail). The minimal domain that is functional as a growth suppressor is shown. (B) Pulse-chase analysis of PTEN;WT and PTEN;1-353. 786-0 cells were metabolically labeled with [35S]methionine for 45 min. The medium was then replaced with complete growth medium at time zero, and cells were harvested at the indicated times. HA-PTEN and HA-PTEN;1-353 were immunoprecipitated from labeled cell extracts, separated by gel electrophoresis, and detected by autoradiography. (C) Log plot of the percentage of the time-zero protein remaining at the times indicated on the x axis. [35S]methionine-labeled, immunoprecipitated protein (B) was quantified by phosphorimager analysis.
In our experiments we noted that PTEN;1-353 was produced at markedly reduced steady-state levels compared to PTEN;WT. To determine whether the changes in the steady-state protein levels were related to changes in protein stability, 786-0 cells (PTEN null) transiently transfected with plasmids encoding PTEN;WT or PTEN;1-353 were pulse-labeled with [35S]methionine for 45 min. HA-PTEN and HA-PTEN;1-353 were recovered by immunoprecipitation and detected by autoradiography. In these experiments the half-life of PTEN;1-353 was found to be reduced by more than fourfold compared to that of PTEN;WT (Fig. 1B and C). In keeping with these results, it was recently reported that the steady-state level of PTEN;1-351 is reduced compared to that of PTEN;WT when the protein is produced by transfection in COS-7 cells (14). Together these data suggest that the PTEN tail, while not required for the functional activity of the protein, is required for maintaining stability.
The tail domain modulates PTEN biological activity.
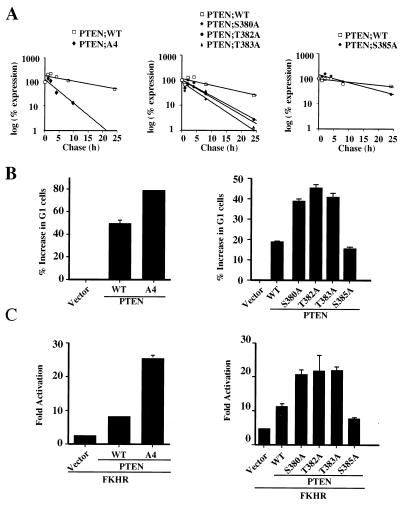
As a result of the reduced half-life, PTEN;1-353 is produced at significantly lower levels than PTEN;WT. In order to determine whether the resulting decrease in protein production results in reduced activity, PTEN;1-353 and PTEN;WT were compared in a cell cycle arrest assay that reflects the ability of PTEN to act as a lipid phosphatase (39). 786-0 cells were transfected with increasing doses of plasmids encoding PTEN;WT and PTEN;1-353 along with a plasmid encoding the cell surface marker pCD19. The cell cycle distribution of the CD19-positive cells (as a marker of transfection) was determined by staining with fluorescein isothiocyanate-conjugated anti-CD19 and propidium iodide followed by two-color FACS. Surprisingly, at equivalent input plasmid concentrations PTEN;1-353 reproducibly induced a greater increase in G1 than PTEN;WT (data not shown). Next, the activities of PTEN;WT and PTEN;1-353 were compared when the proteins were produced at similar steady-state levels. Plasmid titration indicated that equivalent protein levels were obtained at 2 μg of PTEN;1-353 and 0.5 μg of PTEN;WT (Fig. 2A and B). At these levels PTEN;1-353 induced a significantly more robust G1 arrest (Fig. 2C).
FIG. 2.
The PTEN tail is an inhibitory domain. (A) Steady-state protein levels of PTEN;WT and PTEN;1-353. 786-0 cells were transfected with the indicated amounts of plasmids encoding HA-PTEN;WT or HA-PTEN;1-353. Forty hours after transfection, cell lysates were prepared and separated by gel electrophoresis. HA-PTEN and HA-PTEN;1-353 were detected by anti-HA immunoblotting. (B) Quantification of the immunoblot shown in panel A. The radiograph was digitized, and the relative protein quantities were determined using ImageQuant software. The results are expressed as a percentages of PTEN;WT at 2 μg of input plasmid DNA. (C) Induction of G1 arrest by PTEN;WT and PTEN;1-353. 786-0 cells were cotransfected with a plasmid encoding the cell surface marker CD19 (pCD19) along with plasmids encoding PTEN;WT (0.5 μg) or PTEN;1-353 (2 μg). Forty hours after transfection, the cell cycle distribution of the CD19-positive cells was determined by FACS analysis. Shown are the means and standard errors of duplicate experiments. These data are representative of three independent experiments. (D) Induction of FKHR transcriptional activity by PTEN;WT and PTEN;1-353. 786-0 cells were transfected with a FasL promoter luciferase reporter plasmid and a plasmid encoding FKHR in combination with the indicated amounts of plasmid encoding PTEN; WT or PTEN;1-353. Forty hours after transfection, luciferase activity was determined as described in Materials and Methods. Shown are the means and standard errors of the fold activation relative to the activity obtained with the reporter alone.
Forkhead transcription factors are targets of Akt regulation both in mammalian cells and in Caenorhabditis elegans (2, 3, 15, 21, 33, 35). Akt phosphorylation creates 14-3-3 binding sites and results in the cytoplasmic localization of forkhead proteins. Furthermore, recent data in our laboratory have shown that in PTEN null cells FKHRL1 and exogenously expressed FKHR are retained in the cytoplasm and that exogenously expressed FKHR fails to activate transcription. Coexpression of wild-type PTEN, but not a PTEN mutant lacking lipid phosphatase activity, with FKHR restores cytoplasmic localization and FKHR transactivation as measured with a 3XIRS promoter or FasL promoter luciferase reporters (Nakamura et al., submitted). Thus, FKHR transcriptional activity requires PTEN function. The induction of FKHR transcriptional activity by PTEN is also dose dependent, as shown in Fig. 2D.
We next compared PTEN;WT and PTEN;1-353 in a FKHR transcriptional activation assay. Consistent with the results obtained in the cell cycle assay (Fig. 2C), the ability of PTEN;1-353 to induce FKHR transcriptional activity was enhanced compared to that of PTEN;WT. Furthermore, at every DNA plasmid concentration tested, PTEN;1-353 induced FKHR activation more efficiently than PTEN;WT, although protein levels were reduced by more than fourfold. These results suggest that the PTEN tail not only plays a role in maintaining its protein stability but also in regulating its biological activity. Specifically, these data suggest that the tail acts to restrict or inhibit PTEN function.
PTEN is a phosphoprotein.
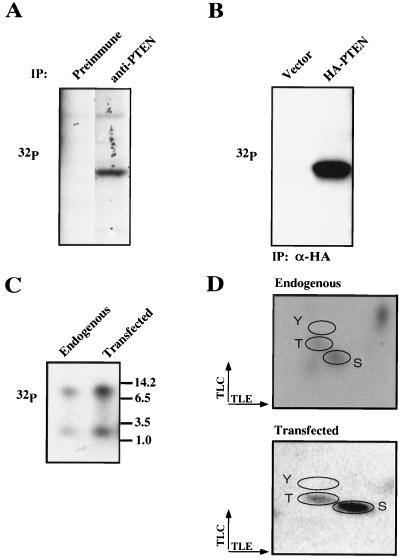
The PTEN tail is rich in serine and threonine (28% of the residues) and contains consensus phosphorylation sites for GSK3, PKA, CK1, and CK2. In order to determine whether regulation of PTEN stability and activity might be linked to phosphorylation, we determined whether endogenous PTEN is phosphorylated. To this end, ACHN renal carcinoma cells that contained PTEN were metabolically labeled with [32P]orthophosphate. Labeled lysates were incubated with an anti-PTEN antibody (C54) or a preimmune control. Bound proteins were separated by gel electrophoresis and detected by autoradiography. A 32P-labeled protein of the same molecular weight as PTEN was detected in anti-PTEN immunoprecipitates but not in the preimmune control (Fig. 3A). In separate experiments in which the labeled proteins were transferred to nitrocellulose, this 32P-labeled species comigrated with PTEN, as detected by immunoblotting. These data suggest that endogenous PTEN is phosphorylated. Next, PTEN plus U2-OS cells were transfected with either the empty vector (pSG5L) or pSG5L-HA-PTEN and metabolically labeled with orthophosphate. Labeled cells were lysed, and epitope-tagged proteins were immunoprecipitated with anti-HA antibody. In parallel, endogenous PTEN was immunoprecipitated from lysates prepared from untransfected orthophosphate-labeled U2-OS cells. A phosphorylated protein of 58 kDa was detected in the anti-HA immunoprecipitates from cells expressing HA-PTEN but not in the vector-transfected cells (Fig. 3B). Next, phosphorylated endogenous PTEN and exogenously produced HA-PTEN were digested with trypsin. As shown in Fig. 3C, an identical pattern of phosphotryptic peptides was observed when phosphopeptides were separated by Tris-Tricine gel electrophoresis (16.5% acrylamide). Phosphoamino acid analysis of both endogenous and transfected PTEN proteins showed phosphorylation of serine and threonine residues, while tyrosine phosphorylation was not detected (Fig. 3D). These data suggest that endogenous PTEN is a phosphoprotein, that HA-PTEN produced by transfection is a phosphoprotein, and that these proteins are phosphorylated on the same peptides, predominantly on serine and threonine.
FIG. 3.
PTEN is a phosphoprotein. (A) Phosphorylation of endogenous PTEN. Asynchronously growing ACHN cells were metabolically labeled with [32P]orthophosphate for 4 h. Protein extracts were prepared and immunoprecipitated with either preimmune or anti-PTEN (C54) antibody. Bound proteins were resolved by gel electrophoresis, transferred to a nitrocellulose membrane, and detected by autoradiography. Data shown are from the same exposure of the same gel; the lanes were rearranged for clarity. (B) Phosphorylation of exogenous HA-PTEN. U2-OS cells were transfected with the backbone vector or pSG5L-HA-PTEN. Forty hours after transfection cells were labeled with 32P-orthophosphate for 2 h. Protein extracts were prepared and immunoprecipitated (IP) with anti-HA antibody. Bound proteins were resolved and detected as for panel A. (C) Tryptic phosphopeptides of endogenous PTEN and exogenously produced HA-PTEN. The bands corresponding to endogenous PTEN or HA-PTEN (A and B) were excised, digested with trypsin, and resolved on a 16.5% Tris-Tricine gel. Phosphopeptides were detected by autoradiography. (D) Phosphoamino acid analysis of endogenous PTEN and exogenously produced HA-PTEN. Tryptic digest products of in vivo-labeled endogenous PTEN or HA-PTEN were hydrolyzed with acid, and the resulting phosphoamino acids were resolved by two-dimensional thin-layer electrophoresis and detected by autoradiography. Phosphoserine (S), phosphothreonine (T), and phosphotyrosine (Y) standards were visualized by ninhydrin staining.
PTEN is phosphorylated within the tail domain.
To determine the exact sites of phosphorylation, U2-OS cells were transfected with plasmids encoding a series of PTEN C-terminal deletion mutants and labeled with orthophosphate. 32P-labeled HA-tagged proteins were recovered by anti-HA immunoprecipitation and detected by autoradiography. These experiments revealed that deletion of residues 354 to 403 (the tail) abrogated most PTEN phosphorylation (Fig. 4A). In addition, gel electrophoretic separation of peptides generated by cyanogen bromide cleavage of orthophosphate-labeled HA-PTEN revealed a single phosphorylated peptide consistent in size with the predicted CNBr peptide containing the PTEN tail (data not shown). Next, every serine and threonine in the tail was mutated either singly or in clusters to alanine. These PTEN mutants were transfected into U2-OS cells and labeled with orthophosphate. No single-amino-acid substitution abrogated or significantly reduced the total phosphorylation of PTEN (data not shown). However, the substitution of a serine/threonine cluster, S380, T382, T383, and S385 (the A4 mutant), did significantly alter total PTEN phosphorylation. In addition, mutation of these sites led to the loss of the more slowly migrating tryptic phosphopeptide (Fig. 4B). This peptide therefore is likely to be peptide 2 (Fig. 4D). In contrast, substitution of alanines for the serine/threonine cluster beginning at S360 (which contains a GSK3 consensus phosphorylation site) had no effect on either the total phosphate incorporated into PTEN or the phosphorylation of the two phosphopeptides detected in Tris-Tricine gels (Fig. 4B). Next, the A4 mutation was combined with a single alanine substitution at a consensus CK2 site (S370) found in peptide 1 of the tail to give the A5 mutation (Fig. 4D). When PTEN;A5 was expressed in U2-OS cells and tested for phosphorylation, 32P labeled protein was not detected despite adequate protein expression (Fig. 4C). Phosphopeptide analysis was not possible because no labeled protein could be excised from the gel. These results indicate that most, if not all, of the PTEN tail phosphorylation occurs on serine 370 and one or more sites of the A4 cluster (S380, T382, T383, and S385).
FIG. 4.
PTEN is phosphorylated within the tail. (A) Deletion of the tail impairs PTEN phosphorylation. Plasmids encoding either wild-type (WT) HA-PTEN or the indicated C-terminal truncation mutants were transfected into U2-OS cells. Twenty-four hours after transfection, cells were split into T25 flasks and 35-mm plates. Forty hours after transfection, T25 flasks were metabolically labeled with [32P]orthophosphate; anti-HA immunoprecipitates of protein extracts were prepared, and bound labeled proteins were detected by autoradiography (top). In parallel, whole-cell extracts prepared from the 35-mm plates were separated by gel electrophoresis, transferred to nitrocellulose, and immunoblotted with anti-HA antibody (bottom). (B [left] and C) Alanine mutations in the PTEN tail impair phosphorylation. U2-OS cells were transfected with plasmids encoding PTEN;WT or the indicated alanine substitution mutants. Transfected cells were split, replated, grown overnight, and used in parallel for metabolic labeling and for the preparation of whole-cell extracts. [32P]orthophosphate labeling, immunoprecipitation, and autoradiography (top) were performed as for panel A. Anti-HA immunoblotting (bottom) was performed as for panel A. (B [right]) Tryptic phosphopeptides of PTEN tail substitution mutants. Phosphopeptides resulting from the tryptic digestion of either HA-PTEN;WT or the indicated mutant proteins were resolved on Tris-Tricine gels and detected as for Fig. 3C. (D) Schematic representation of PTEN tail substitution mutants used. Dashes indicate amino acids that were not altered. The predicted tryptic peptides of the PTEN tail are shown as peptide 1 and peptide 2. PHD, phosphatase domain; C2D, C2 domain.
To directly identify the PTEN phosphorylation sites, 2 μg of HA-PTEN isolated by anti-HA immunoaffinity purification was digested with trypsin and analyzed by LC/MSMS. Here, peptide 1 with a phosphoserine at residue 370 was identified. However, peptide 2 was not detectable in either a phosphorylated or unphosphorylated state. No other phosphopeptides were identified (data not shown).
Mutation of the phosphorylation sites in the tail alter PTEN stability and biological activity.
To determine whether phosphorylation of one or more of the amino acid residues delineated above has a role in modulating PTEN stability, the relevant phosphorylation site mutants were produced in U2-OS cells by transient transfection and the steady-state levels of HA-PTEN and the phosphorylation mutants were determined by immunoblot analysis. While mutation of serine 370 did not change the steady-state level of PTEN (data not shown), mutation of the S/T cluster (PTEN;A4) resulted in a marked decrease in the steady-state protein level (data not shown). Furthermore, pulse-chase labeling experiments revealed a marked reduction in the PTEN;A4 half-life (Fig. 5A, left).
FIG. 5.
Mutation of phosphoacceptor sites in the PTEN tail alters protein stability and activity. (A) Protein half-life of PTEN;A4 (left) and single-substitution mutants (middle and right). 786-0 cells were transfected with plasmids encoding HA-PTEN;WT and HA-PTEN;A4 or the indicated single-substitution mutants, metabolically labeled with [35S]methionine, and chased for the indicated times. Data are shown as in Fig. 1C. (B) Increased activity of PTEN;A4 (left) and mutants with at single substitution of S380, T382, or T383 (right) in the cell cycle arrest assay. 786-0 cells were cotransfected with a plasmid encoding CD19 (pCD19) along with plasmids encoding PTEN;WT, PTEN;A4, or single-substitution mutants at concentrations resulting in equivalent protein production (0.5 μg of pSG5L-HA-PTEN;WT and pSG5L-HA-PTEN;S385A and 2.0 μg of pSG5L-HA-PTEN; A4, pSG5L-HA-PTEN;S380A, pSG5L-HA-PTEN;T382A, and pSG5L-HA-PTEN;T383A). Forty hours after transfection the cell cycle distribution of the CD19-positive cells was analyzed as for Fig. 2C. These data are representative of three independent experiments. (C) FKHR transcriptional activity induced by PTEN;A4 (left) and single-substitution mutants (right). 786-0 cells were transfected with a plasmid encoding FKHR along with either the backbone vector or plasmids encoding PTEN;WT, PTEN;A4, or single-substitution mutants as indicated. Forty hours after transfection luciferase activity was measured and the fold activation of FKHR relative to the activity obtained with the reporter alone was calculated as for Fig. 2D. Shown are the means and standard errors of experimental duplicates. These data are representative of two independent experiments.
As deletion of the tail led to an increase in PTEN activity, we next asked if the PTEN;A4 mutant was similarly more active in biological assays. In keeping with the data for the PTEN tail, the PTEN;A4 mutant, while expressed at lower levels, was more active in both inducing a G1 arrest in PTEN null 786-0 cells (Fig. 5B, left) and inducing FKHR transcriptional activation (Fig. 5C, left). These data suggest that phosphorylation within the A4 cluster is required to maintain stability and is linked to an inhibitory activity of the PTEN tail.
Next, the individual point mutations within the A4 cluster were tested in the same assays of protein half-life and activity. While replacement of serine 385 with alanine did not alter the steady-state PTEN protein levels, mutation of S380, T382, and T383 each reduced both the steady-state protein levels and the protein half-life (Fig. 5A, right, and data not shown). More specifically, the half-life of the PTEN;A4 mutant was reduced more than six-fold compared to that of PTEN;WT. Similarly, mutating serine 380 reduced the half-life by more than fivefold. Mutation of threonines 382 and 383 reduced PTEN half-life by 2.7- to 3-fold. Furthermore, the individual phosphorylation mutants with substitutions S380A, T382A, and T383A, but not S385A, were again more active in inducing a G1 arrest and in inducing FKHR transcriptional activation (Fig. 5B and C, right). Taken together, these data show that the increased activity associated with deletion of the tail is entirely mimicked by mutations within the A4 cluster, specifically S380, T382, or T383.
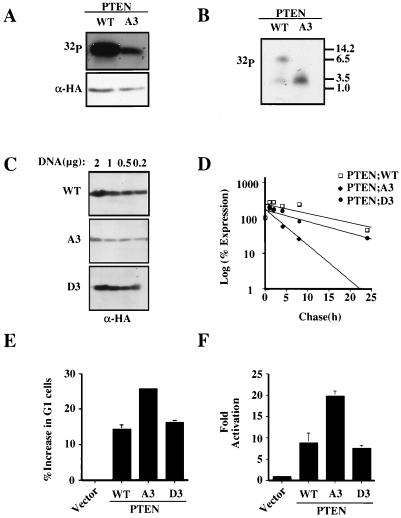
The above data raised the possibility that phosphorylation of these three specific residues (S380, T382, and T383) might be required to maintain PTEN in a stable yet relatively inactive state. While mutation of each of these individual residues did not alter total incorporation of 32P into the PTEN protein (data not shown) when the S380, T382, and T383 residues were mutated to alanine (PTEN;A3) incorporation of 32P into PTEN during orthophosphate labeling was reduced (Fig. 6A) and the most slowly migrating tryptic phosphopeptide (peptide 2) was not detectable (Fig. 6B). In keeping with the data for PTEN;A4 and for the individual phosphorylation site mutants (with mutations S380A, T382A, and T383A), PTEN;A3 was found to have a reduced protein half-life, to be expressed at lower steady-state levels, and to be more active than wild-type PTEN in biological assays (Fig. 6C to F).
FIG. 6.
Aspartic acid substitutions of serine 380, threonine 382, and threonine 383 restore PTEN expression levels and half-life. (A) Substitution mutations of serine 380, threonine 382, and threonine 383 impair PTEN phosphorylation. Plasmids encoding PTEN;WT and PTEN;A3 were transfected into U2-OS cells, and phosphorylation of the resulting HA-PTEN proteins was determined as for Fig. 4A. (B) Phosphorylated tryptic peptides of the PTEN tail substitution mutants. Phosphopeptides resulting from the tryptic digestion of HA-PTEN;WT or HA-PTEN;A3 were resolved on Tris-Tricine gels and detected by autoradiography. (C) Aspartic acid substitution restores HA-PTEN steady-state protein levels. 786-0 cells were transfected with the indicated amounts of plasmids encoding HA-PTEN;WT and HA-PTEN;A3 and HA-PTEN;D3 mutants. Forty hours after transfection whole-cell extracts were analyzed by immunoblotting with anti-HA antibody. (D) Aspartic acid substitution restores PTEN stability. 786-0 cells were transfected with plasmids encoding HA-PTEN;WT, HA-PTEN;A3, or HA-PTEN;D3, metabolically labeled with [35S]methionine, and chased for the indicated time. Data are shown as in Fig. 1C. (E) Cell cycle arrest induced by PTEN;WT or the indicated substitution mutants. 786-0 cells were transfected with plasmids encoding PTEN;WT or the indicated PTEN mutants. Forty hours after transfection the cell cycle distribution of the CD19-positive 786-0 cells was determined as for Fig. 2C. Data are the means and standard errors of experimental duplicates. The data are representative of three independent experiments. (F) FKHR transcriptional activation. 786-0 cells were transfected with the FasL promoter luciferase reporter plasmid and a plasmid encoding FKHR alone or in combination with a plasmid encoding PTEN;WT or the indicated PTEN substitution mutants. Forty hours after transfection the fold activation of reporter activity was determined as for Fig. 2D. The data are the means and standard errors of experimental duplicates. These data are representative of two independent experiments.
Aspartic acid substitutions at the phosphorylation sites in the PTEN tail lead to a recovery of PTEN stability.
A reasonable interpretation of these results is that the serine/threonine-to-alanine substitutions of PTEN block phosphorylation and thereby alter the stability and activity of PTEN in cells. On the other hand, it is formally possible that mutation of these residues might result in these changes independent of the changes in PTEN tail phosphorylation. To distinguish these possibilities, conversion of the serines/threonines to aspartic acid was used to try and mimic phosphorylation of these residues. As we had noted that mutation of any one of the putative phosphorylation sites altered both stability and activity, it appeared that phosphorylation of all three residues might be required for maintaining PTEN stability. Therefore, we generated a PTEN;D3 cDNA in which codons 380, 382, and 383 encoded aspartic acid.
In contrast to the results obtained with PTEN;A3, PTEN;D3 recovered the expression levels of PTEN;WT, suggesting that a negative charge was enough to maintain protein stability (Fig. 6C). To test this possibility, we performed a pulse-chase experiment to determine if aspartic acid substitution could restore PTEN stability. As shown in Fig. 6D, PTEN;D3 recovered the stability of PTEN;A3 and was similar to PTEN;WT. As expected, when analyzed after orthophosphate labeling, PTEN;D3 like PTEN;A3 was found not to contain phosphopeptide 1 (data not shown). These data argue that the D3 mutation does not restore stability simply be restoring phosphorylation of PTEN at other sites but rather that phosphorylation of the S380 cluster is required for appropriate stability.
Next, we asked whether aspartic acid substitution led to a change in the activity of PTEN in the cell cycle assay and the FKHR transactivation assay. As would be predicted if loss of phosphorylation was responsible for the changes seen in PTEN activity, PTEN;D3 recovered the activity of PTEN;WT and its activity was reduced compared to that of PTEN;A3 (Fig. 6E-F).
DISCUSSION
Recently it was shown that PTEN;1-351 has in vitro lipid phosphatase activity, can inhibit Akt activation, and can suppress anchorage-independent growth (14). In accordance with these results, we found that PTEN;1-353 inhibits the growth of PTEN null 786-0 cells in soft agar and induces apoptosis in LNCaP cells (S. Ramaswamy and W. R. Sellers, unpublished data). Furthermore, the PTEN crystal structure shows that these residues include the entire PD and a C2 lipid binding domain (22). Together these data suggest that the tail (residues 354 to 403) is not required for biological activity of the protein.
Here, we have shown that deletion of the PTEN tail results in a loss of protein stability. One might expect that loss of stability would lead to a loss of PTEN function; however, as stated above, no requirement for the tail was found in multiple assays of PTEN function. This paradox can be explained by the concomitant loss of an inhibitory activity that maps to the tail region. Thus, the tail contains sequences that are required for protein stability and for negative regulation of PTEN function. This linkage between stability and activity argues for a model in which PTEN is normally found in a stable yet relatively inactive state. In such a model, activation of PTEN would be accompanied by a decrease in protein half-life, presumably reflecting degradation of the active molecule (Fig. 7). Such a mechanism would prevent the untimely or promiscuous activation of PTEN. The linkage between protein activation and protein instability is a common theme in molecular biology. Examples of proteins where the activated form of the protein is unstable include Src, EGFR, PDGFR, and the nuclear receptors RAR and RXR (16, 31, 47).
FIG. 7.
Model for PTEN regulation by phosphorylation of the tail. The black extension to PTEN represents the tail, and the circles labeled P represent phosphorylated residues within the tail. PTEN phosphorylation on the tail would restrict PTEN activity. Dephosphorylation of the tail would result in an increase in PTEN activity and in its rapid degradation.
The tail is also required for PTEN phosphorylation. In vitro mutagenesis revealed specific phosphorylation of S380, T382, and T383. Mutation of these sites led to a loss of stability and a gain in PTEN function. Conversely, aspartic acid mutations of these same residues preserved the protein half-life and the function of PTEN. Together, these data argue strongly that phosphorylation of these residues is required for stability and that the changes in stability are not simply the result of a misfolding secondary to changes in the amino acid residues. Recently others have shown that mutations in the C2 domain can reduce expression levels and protein half-life (14). The mutants are, however, found to be functionally inactive or significantly impaired in biological assays; thus the instability of these mutants might indeed arise as a consequence of protein misfolding (14, 22). On the other hand, the PTEN;A3 mutant, while unstable, is more active, arguing strongly against protein misfolding as a mechanism for instability in this instance.
What is the mechanism by which PTEN phosphorylation regulates protein stability? As pointed out by Georgescu et al. (14) the tail contains two putative PEST sequences (residues 350 to 375 and 379 to 386) implicated in targeting proteins for proteolytic degradation. We have found that the second PEST region is the site of three phosphorylation sites (S380, T382, T383), raising the possibility that this sequence is normally masked by phosphorylation. Arguing against this model, however, is the fact that deletion of the entire tail also results in a shorter half-life. Secondary structure prediction and the results of proteolytic digestion experiments suggest that the tail is a relatively unstructured and presumably flexible region. Thus, one model is that the tail can mask a degradation signal present elsewhere in the PTEN protein when phosphorylated; this signal would be unmasked by dephosphorylation leading to a shift in the position of the tail (Fig. 7).
As an alternative model, the tail might regulate PTEN localization through interactions with the adjacent C2 domain. If so, then stability might simply reflect the localization of PTEN to a subcellular compartment where PTEN degradation can take place. To date we have not seen an obvious effect on the membrane localization of the relevant C-terminal truncations or phosphorylation site mutants (data not shown). Surprisingly, PTEN;1-353 and PTEN;A4 manifestly increased localization to the nucleus (data not shown). Whether this apparent change in localization results from a more rapid degradation of the cytoplasmic component of these mutants or from a true shift in localization is not yet clear. Nonetheless, regulation of localization by tail phosphorylation might play an important role in the regulation of PTEN.
What is the mechanism through which PTEN function is regulated by the tail? There are a number of mechanisms that could account for the inhibitory activity of the tail on PTEN. First, the phosphatase activity itself could be regulated through an allosteric or steric mechanism. To date, however, we have seen no effects of phosphorylation site mutations on intrinsic phosphatase activity (data not shown). Second, the ability of PTEN to gain access to a substrate could be altered either through changes in localization (see above), through changes in the position of the tail, or perhaps through inhibitory interactions with tail-associated proteins. With respect to the last idea, the PTEN tail contains a PDZ binding domain. Given that the PDZ binding sequence (along with the entire tail) is dispensable for the biological function of PTEN, it would seem likely that this domain is linked to the role for the tail that we have put forth. Specifically, the interaction of a PTEN with a PDZ domain-containing protein might be regulated through PTEN phosphorylation.
Are the PTEN phosphorylation events that we have identified constitutive or regulated? Our data are most consistent with the idea that PTEN exists in a predominantly phosphorylated state and that dephosphorylation of the S380 cluster is a regulated event. While PTEN runs as a single band under standard sodium dodecyl sulfate-gel electrophoresis conditions, on two-dimensional gels multiple isoforms of PTEN can be distinguished based on differences in the isoelectric point (data not shown). While it is likely that these forms represent different PTEN phosphoisoforms, how these forms are related to each phosphorylation site or to PTEN activity or stability is not yet known. Our model suggests that PTEN function is regulated by the balance between a kinase and a phosphatase. It is possible that the kinase constitutively phosphorylates PTEN and that a phosphatase regulates the activity. An intriguing possibility is that PTEN activates itself through autodephosphorylation maintaining a constant loop of activity. Identification of the kinase and phosphatase activities responsible for the regulation of these sites should provide further insights into how regulation of PTEN is achieved.
ACKNOWLEDGMENTS
This work was supported by the grants from the Gillette Women's Cancer Program, the Department of Defense (DAMD17-98-1-8596), NIH (K11CA65594), the American Cancer Society (RPG-00-113-01), and the CaPCURE foundation to W.R.S. and from the Department of Defense (PC990016) to F.V.
We thank John Alberta for his help with the phosphoamino acid analysis and Thomas Roberts, Alan D'Andrea, and Charles Stiles for their critical reading of the manuscript.
REFERENCES
- 1.Barford D, Das A K, Egloff M P. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu Rev Biophys Biomol Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 2.Biggs W H, III, Meisenhelder J, Hunter T, Cavenee W K, Arden K C. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc Natl Acad Sci USA. 1999;96:7421–7426. doi: 10.1073/pnas.96.13.7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunet A, Bonni A, Zigmond M J, Lin M Z, Juo P, Hu L S, Anderson M J, Arden K C, Blenis J, Greenberg M E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857–868. doi: 10.1016/s0092-8674(00)80595-4. [DOI] [PubMed] [Google Scholar]
- 4.Cantley L C, Neel B G. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cardone M H, Roy N, Stennicke H R, Salvesen G S, Franke T F, Stanbridge E, Frisch S, Reed J C. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318–1321. doi: 10.1126/science.282.5392.1318. [DOI] [PubMed] [Google Scholar]
- 6.Chung J, Grammer T C, Lemon K P, Kazlauskas A, Blenis J. PDGF- and insulin-dependent pp70S6k activation mediated by phosphatidylinositol-3-OH kinase. Nature. 1994;370:71–75. doi: 10.1038/370071a0. [DOI] [PubMed] [Google Scholar]
- 7.Cross D A, Alessi D R, Cohen P, Andjelkovich M, Hemmings B A. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 8.Datta S R, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg M E. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231–241. doi: 10.1016/s0092-8674(00)80405-5. [DOI] [PubMed] [Google Scholar]
- 9.Davies M A, Koul D, Dhesi H, Berman R, McDonnell T J, McConkey D, Yung W K, Steck P A. Regulation of Akt/PKB activity, cellular growth, and apoptosis in prostate carcinoma cells by MMAC/PTEN. Cancer Res. 1999;59:2551–2556. [PubMed] [Google Scholar]
- 10.Di Cristofano A, Kotsi P, Peng Y F, Cordon-Cardo C, Elkon K B, Pandolfi P P. Impaired fas response and autoimmunity in Pten(+/−) mice. Science. 1999;285:2122–2125. doi: 10.1126/science.285.5436.2122. [DOI] [PubMed] [Google Scholar]
- 11.Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi P P. Pten is essential for embryonic development and tumour suppression. Nat Genet. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- 12.Franke T F, Yang S I, Chan T O, Datta K, Kazlauskas A, Morrison D K, Kaplan D R, Tsichlis P N. The protein kinase encoded by the Akt proto-oncogene is a target of the PDGF-activated phosphatidylinositol 3-kinase. Cell. 1995;81:727–736. doi: 10.1016/0092-8674(95)90534-0. [DOI] [PubMed] [Google Scholar]
- 13.Furnari F B, Huang H J, Cavenee W K. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 1998;58:5002–5008. [PubMed] [Google Scholar]
- 14.Georgescu M M, Kirsch K H, Akagi T, Shishido T, Hanafusa H. The tumor-suppressor activity of PTEN is regulated by its carboxyl-terminal region. Proc Natl Acad Sci USA. 1999;96:10182–10187. doi: 10.1073/pnas.96.18.10182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo S, Rena G, Cichy S, He X, Cohen P, Unterman T. Phosphorylation of serine 256 by protein kinase B disrupts transactivation by FKHR and mediates effects of insulin on insulin-like growth factor-binding protein-1 promoter activity through a conserved insulin response sequence. J Biol Chem. 1999;274:17184–17192. doi: 10.1074/jbc.274.24.17184. [DOI] [PubMed] [Google Scholar]
- 16.Harris K F, Shoji I, Cooper E M, Kumar S, Oda H, Howley P M. Ubiquitin-mediated degradation of active Src tyrosine kinase. Proc Natl Acad Sci USA. 1999;96:13738–13743. doi: 10.1073/pnas.96.24.13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hunter T. Signaling—2000 and beyond. Cell. 2000;100:113–127. doi: 10.1016/s0092-8674(00)81688-8. [DOI] [PubMed] [Google Scholar]
- 18.James S R, Downes C P, Gigg R, Grove S J, Holmes A B, Alessi D R. Specific binding of the Akt-1 protein kinase to phosphatidylinositol 3,4,5-trisphosphate without subsequent activation. Biochem J. 1996;315:709–713. doi: 10.1042/bj3150709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klippel A, Kavanaugh W M, Pot D, Williams L T. A specific product of phosphatidylinositol 3-kinase directly activates the protein kinase Akt through its pleckstrin homology domain. Mol Cell Biol. 1997;17:338–344. doi: 10.1128/mcb.17.1.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kobayashi T, Cohen P. Activation of serum- and glucocorticoid-regulated protein kinase by agonists that activate phosphatidylinositide 3-kinase is mediated by 3-phosphoinositide-dependent protein kinase-1 (PDK1) and PDK2. Biochem J. 1999;339:319–328. [PMC free article] [PubMed] [Google Scholar]
- 21.Kops G J, de Ruiter N D, De Vries-Smits A M, Powell D R, Bos J L, Burgering B M. Direct control of the Forkhead transcription factor AFX by protein kinase B. Nature. 1999;398:630–634. doi: 10.1038/19328. [DOI] [PubMed] [Google Scholar]
- 22.Lee J O, Yang H, Georgescu M M, Di Cristofano A, Maehama T, Shi Y, Dixon J E, Pandolfi P, Pavletich N P. Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association. Cell. 1999;99:323–334. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 23.Li D, Sun H. PTEN/MMAC1/TEP1 suppresses the tumorigenicity and induces G1 cell cycle arrest in human glioblastoma cells. Proc Natl Acad Sci USA. 1998;95:15406–15411. doi: 10.1073/pnas.95.26.15406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li D M, Sun H. TEP1, encoded by a candidate tumor suppressor locus, is a novel protein tyrosine phosphatase regulated by transforming growth factor beta. Cancer Res. 1997;57:2124–2129. [PubMed] [Google Scholar]
- 25.Li J, Simpson L, Takahashi M, Miliaresis C, Myers M P, Tonks N, Parsons R. The PTEN/MMAC1 tumor suppressor induces cell death that is rescued by the AKT/protein kinase B oncogene. Cancer Res. 1998;58:5667–5672. [PubMed] [Google Scholar]
- 26.Li J, Yen C, Liaw D, Podsypanina K, Bose S, Wang S I, Puc J, Miliaresis C, Rodgers L, McCombie R, Bigner S H, Giovanella B C, Ittmann M, Tycko B, Hibshoosh H, Wigler M H, Parsons R. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–1947. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 27.Li Z, Wahl M I, Eguinoa A, Stephens L R, Hawkins P T, Witte O N. Phosphatidylinositol 3-kinase-gamma activates Bruton's tyrosine kinase in concert with Src family kinases. Proc Natl Acad Sci USA. 1997;94:13820–13825. doi: 10.1073/pnas.94.25.13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liaw D, Marsh D J, Li J, Dahia P L, Wang S I, Zheng Z, Bose S, Call K M, Tsou H C, Peacocke M, Eng C, Parsons R. Germline mutations of the PTEN gene in Cowden disease, an inherited breast and thyroid cancer syndrome. Nat Genet. 1997;16:64–67. doi: 10.1038/ng0597-64. [DOI] [PubMed] [Google Scholar]
- 29.Marsh D J, Dahia P L, Coulon V, Zheng Z, Dorion-Bonnet F, Call K M, Little R, Lin A Y, Eeles R A, Goldstein A M, Hodgson S V, Richardson A L, Robinson B G, Weber H C, Longy M, Eng C. Allelic imbalance, including deletion of PTEN/MMACI, at the Cowden disease locus on 10q22-23, in hamartomas from patients with Cowden syndrome and germline PTEN mutation. Genes Chromosomes Cancer. 1998;21:61–69. doi: 10.1002/(sici)1098-2264(199801)21:1<61::aid-gcc8>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 30.Marsh D J, Kum J B, Lunetta K L, Bennett M J, Gorlin R J, Ahmed S F, Bodurtha J, Crowe C, Curtis M A, Dasouki M, Dunn T, Feit H, Geraghty M T, Graham J M, Jr, Hodgson S V, Hunter A, Korf B R, Manchester D, Miesfeldt S, Murday V A, Nathanson K L, Parisi M, Pober B, Romano C, Tolmie J L, et al. PTEN mutation spectrum and genotype-phenotype correlations in Bannayan-Riley-ruvalcaba syndrome suggest a single entity with Cowden syndrome. Hum Mol Genet. 1999;8:1461–1472. doi: 10.1093/hmg/8.8.1461. [DOI] [PubMed] [Google Scholar]
- 31.Moghal N, Sternberg P W. Multiple positive and negative regulators of signaling by the EGF-receptor. Curr Opin Cell Biol. 1999;11:190–196. doi: 10.1016/s0955-0674(99)80025-8. [DOI] [PubMed] [Google Scholar]
- 32.Myers M P, Pass I, Batty I H, van der Kaay J, Stolarov J P, Hemmings B A, Wigler M H, Downes C P, Tonks N K. The lipid phosphatase activity of PTEN is critical for its tumor supressor function. Proc Natl Acad Sci USA. 1998;95:13513–13518. doi: 10.1073/pnas.95.23.13513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nakae J, Park B C, Accili D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J Biol Chem. 1999;274:15982–15985. doi: 10.1074/jbc.274.23.15982. [DOI] [PubMed] [Google Scholar]
- 34.Nelen M R, van Staveren W C, Peeters E A, Hassel M B, Gorlin R J, Hamm H, Lindboe C F, Fryns J P, Sijmons R H, Woods D G, Mariman E C, Padberg G W, Kremer H. Germline mutations in the PTEN/MMAC1 gene in patients with Cowden disease. Hum Mol Genet. 1997;6:1383–1387. doi: 10.1093/hmg/6.8.1383. [DOI] [PubMed] [Google Scholar]
- 35.Ogg S, Paradis S, Gottlieb S, Patterson G I, Lee L, Tissenbaum H A, Ruvkun G. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C. elegans. Nature. 1997;389:994–999. doi: 10.1038/40194. [DOI] [PubMed] [Google Scholar]
- 36.Ozes O N, Mayo L D, Gustin J A, Pfeffer S R, Pfeffer L M, Donner D B. NF-kappaB activation by tumor necrosis factor requires the Akt serine-threonine kinase. Nature. 1999;401:82–85. doi: 10.1038/43466. [DOI] [PubMed] [Google Scholar]
- 37.Podsypanina K, Ellenson L H, Nemes A, Gu J, Tamura M, Yamada K M, Cordon-Cardo C, Catoretti G, Fisher P E, Parsons R. Mutation of Pten/Mmac1 in mice causes neoplasia in multiple organ systems. Proc Natl Acad Sci USA. 1999;96:1563–1568. doi: 10.1073/pnas.96.4.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Potter L R, Hunter T. Identification and characterization of the major phosphorylation sites of the B-type natriuretic peptide receptor. J Biol Chem. 1998;273:15533–15539. doi: 10.1074/jbc.273.25.15533. [DOI] [PubMed] [Google Scholar]
- 39.Ramaswamy S, Nakamura N, Vazquez F, Batt D B, Perera S, Roberts T M, Sellers W R. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/Akt pathway. Proc Natl Acad Sci USA. 1999;96:2110–2115. doi: 10.1073/pnas.96.5.2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rizo J, Sudhof T C. C2-domains, structure and function of a universal Ca2+-binding domain. J Biol Chem. 1998;273:15879–15882. doi: 10.1074/jbc.273.26.15879. [DOI] [PubMed] [Google Scholar]
- 41.Sellers W R, Novitch B G, Miyake S, Heith A, Otterson G A, Kaye F J, Lassar A B, Kaelin W G., Jr Stable binding to E2F is not required for the retinoblastoma protein to activate transcription, promote differentiation, and suppress tumor cell growth. Genes Dev. 1998;12:95–106. doi: 10.1101/gad.12.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Steck P A, Pershouse M A, Jasser S A, Yung W K, Lin H, Ligon A H, Langford L A, Baumgard M L, Hattier T, Davis T, Frye C, Hu R, Swedlund B, Teng D H, Tavtigian S V. Identification of a candidate tumour suppressor gene, MMAC1, at chromosome 10q23.3 that is mutated in multiple advanced cancers. Nat Genet. 1997;15:356–362. doi: 10.1038/ng0497-356. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki A, de la Pompa J L, Stambolic V, Elia A J, Sasaki T, del Barco Barrantes I, Ho A, Wakeham A, Itie A, Khoo W, Fukumoto M, Mak T W. High cancer susceptibility and embryonic lethality associated with mutation of the PTEN tumor suppressor gene in mice. Curr Biol. 1998;8:1169–1178. doi: 10.1016/s0960-9822(07)00488-5. [DOI] [PubMed] [Google Scholar]
- 44.Tang E D, Nunez G, Barr F G, Guan K L. Negative regulation of the forkhead transcription factor FKHR by Akt. J Biol Chem. 1999;274:16741–16746. doi: 10.1074/jbc.274.24.16741. [DOI] [PubMed] [Google Scholar]
- 45.Tedder T F, Isaacs C M. Isolation of cDNAs encoding the CD19 antigen of human and mouse B lymphocytes. J Immunol. 1989;143:712–717. [PubMed] [Google Scholar]
- 46.Vazquez F, Sellers W R. The PTEN tumor suppressor protein: an antagonist of phosphoinositide 3-kinase signaling. Biochim Biophys Acta. 2000;1470:M21–M35. doi: 10.1016/s0304-419x(99)00032-3. [DOI] [PubMed] [Google Scholar]
- 47.Zhu J, Gianni M, Kopf E, Honore N, Chelbi-Alix M, Koken M, Quignon F, Rochette-Egly C, de The H. Retinoic acid induces proteasome-dependent degradation of retinoic acid receptor alpha (RARalpha) and oncogenic RARalpha fusion proteins. Proc Natl Acad Sci USA. 1999;96:14807–14812. doi: 10.1073/pnas.96.26.14807. [DOI] [PMC free article] [PubMed] [Google Scholar]