Abstract
Mammalian chromosomes terminate with a 3′ tail which consists of reiterations of the G-rich repeat, d(TTAGGG). The telomeric tail is the primer for replication by telomerase, and it may also invade telomeric duplex DNA to form terminal lariat structures, or T loops. Here we show that the ubiquitous and highly conserved mammalian protein hnRNP D interacts specifically with the G-rich strand of the telomeric repeat. A single gene encodes multiple isoforms of hnRNP D. All isoforms bind comparably to the G-rich strand, and certain isoforms can also bind tightly and specifically to the C-rich telomeric strand. G-rich telomeric sequences readily form structures stabilized by G-G pairing, which can interfere with telomere replication by telomerase. We show that hnRNP D binding to the G-rich strand destabilizes intrastrand G-G pairing and that hnRNP D interacts specifically with telomerase in human cell extracts. This biochemical analysis suggest that hnRNP D could function in vivo to destabilize structures formed by telomeric G-rich tails and facilitate their extension by telomerase.
Telomeres are regions of specialized sequence, structure, and function located at both ends of each linear eukaryotic chromosome. Telomeres are of particular interest because they regulate cellular life span. Telomeres undergo programmed shortening as an individual ages, and telomere shortening over time provides a clock that limits the number of cell generations (20; reviewed in references 16, 17, and 47). Tumor cells must overcome this built-in senescence by either reactivating telomerase or turning on alternative mechanisms that maintain telomere length.
Essentially all eukaryotic telomeres consist of repeats of G-rich sequence motifs. In humans and other mammals, the telomeric repeat is d(TTAGGG)n. Telomeres in human somatic cells contain 3 to 18 kb of duplex DNA and terminate with a single-stranded G-rich tail (G tail), which is the primer for telomere extension by telomerase. Mammalian telomeric G tails are approximately 130 to 210 nucleotides (nt) in length (38, 44, 64). Because telomeric tails are G rich and single stranded, they have the potential to form structures stabilized by G-G pairing (21, 62). This is a common property of G-rich eukaryotic telomeres, and it may be important for telomere function. Nonetheless, formation of such G-G paired structures has the potential to interfere with the ability of telomerase to replicate telomeres (12, 65).
Telomeric DNA in vivo is complexed with specific proteins, and some of these proteins function to regulate telomere length and maintain telomeric sequence (27, 47; reviewed in references 3 and 61). In Saccharomyces cerevisiae, duplex telomeric DNA is bound by the protein Rap1p, which regulates telomere length (39; reviewed in references 15, 25, 52, and 53). Other S. cerevisiae proteins have been shown to interact with single-stranded G-rich telomeric tails. The S. cerevisiae protein Cdc13p functions to protect the telomeric ends from degradation, prevent single-stranded ends from activating the Rad9 cell cycle checkpoint, and regulate telomere length (10, 14, 33, 46). Another S. cerevisiae protein, Est1p, is essential for telomere maintenance (37), coprecipitates with telomerase (32, 55), and binds G-rich single-stranded DNA (32, 55, 59). In mammals, telomeric duplex DNA is bound by TRF1, which can be visualized on the telomeres of metaphase and interphase chromosomes and functions at least in part to regulate telomere length (1, 5, 67; reviewed in reference 54). A closely related mammalian protein, TRF2, binds to telomeric duplex repeats and prevents end-to-end chromosomal fusion and loss of G tails (58).
Several highly conserved mammalian proteins were identified as candidate telomere binding proteins in a screen which used DNA affinity chromatography to isolate proteins that recognized the mammalian telomeric repeat as single-stranded DNA (23). One protein identified by this affinity screen was hnRNP A1, a nuclear protein known to be involved in regulation of alternative splicing (19, 42) and to function in mRNA transport (49) and packaging (reviewed in references 26 and 43). The N-terminal fragment of hnRNP A1, referred to as UP1, binds the telomeric G strand and interacts with telomerase; the CB3 murine erythroleukemia line, which is deficient in hnRNP A1, contains shortened telomeres, similar to cells in which telomerase is not active (27). This affinity screen also identified another hnRNP family member, hnRNP D (23). hnRNP D is a highly conserved protein (human and mouse polypeptides are 97% identical and 99% similar [7]), consistent with one or more critical cellular functions. The HNRPD gene consists of eight coding exons, two of which are regulated by alternative splicing, and it encodes four distinct isoforms of hnRNP D, with apparent molecular masses of 37 to 45 kD (7, 24) (Fig. 1). All isoforms of hnRNP D contain two canonical RNA binding domains (RBDs; also called RNA recognition motifs), structural domains which are common among proteins that interact with RNA or single-stranded DNA and which are found in many hnRNP family proteins, including hnRNP A1 (reviewed in references 2, 4, and 60). hnRNP D was originally identified as associating with hnRNA in the mammalian nucleus, but this association is quite loose (9, 13, 48). hnRNP D (also known as AUF1 [66]) has been reported to regulate the stability of specific mRNAs containing AUUUA repeats (30, 35).
FIG. 1.
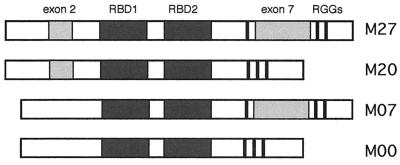
Isoforms of hnRNP D. Alternative splicing of hnRNP D exons 2 and 7 produces four distinct forms of hnRNP D, referred to as M27, M20, M07, and M00 (7). M27 contains a 19-residue region encoded by alternative exon 2 and a 49-residue region encoded by alternative exon 7 (light shading); M20 contains the region encoded by exon 2; M07 contains the region encoded by exon 7; M00 contains neither of the regions encoded by exons 2 and 7. RBD1 and RBD2 (dark shading) indicate the conserved RBDs, and RGG indicates the three Arg-Gly-Gly motifs.
The G-rich telomeric repeats can spontaneously form G-G paired structures in vitro (51, 56, 62), and we have recently found that hnRNP D binds tightly (KD = 0.5 nM) to G-G paired DNA (6). This property, and the results of telomeric affinity chromatography (23) described above, led us to study possible interactions between hnRNP D, telomeres, and telomerase. Here we report that hnRNP D binds with high affinity and in sequence-specific fashion to single-stranded repeats of the telomeric G strand, d(TTAGGG); that certain isoforms of hnRNP D also interact well with the C-rich strand (C strand); and that hnRNP D interacts specifically with telomerase. We show that a synthetic oligonucleotide bearing the mammalian telomeric repeat, (TTAGGG)4, spontaneously forms G-G paired structures in vitro and that binding by hnRNP D destabilizes such G-G paired structures, while binding by hnRNP A1 produces a canonical pattern of protection. A cocrystal of hnRNP A1 with telomeric repeats has recently been reported (8), and comparison of the hnRNP D and hnRNP A1 sequences shows that essentially all of the amino acids residues that make sequence-specific contacts with telomeric DNA are conserved between hnRNP D and hnRNP A1. These in vitro data are consistent with the possibility that hnRNP D may function in telomere maintenance in vivo.
MATERIALS AND METHODS
Protein purification.
Murine hnRNP D cDNAs (7) were subcloned into the pET30 vector (Novagen) to create N-terminal fusions with the His6 tag. His-tagged recombinant protein was produced either from expression clones that produced full-length His-tagged recombinant protein or from expression clones which used an internal methionine as a start codon, truncating a 29-amino-acid N-terminal region rich in glycine and alanine; similar results were obtained from both. Protein expression was induced by addition of isopropyl-β-d-thiogalactopyranoside to a log-phase culture, and protein was purified by nickel chelate chromatography (Novagen) followed by Mono S chromatography. Protein concentration was determined by the Bradford assay, and purity was assessed by silver staining of sodium dodecyl sulfate (SDS)-polyacrylamide gels.
Gel mobility shift assays.
Synthetic oligonucleotides were synthesized at the Keck Center for Biotechnology, Yale Medical School, and 5′ end labeled with [γ-32P]ATP (NEN) and T4 polynucleotide kinase (New England Biolabs). A typical 15-μl binding reaction mixture contained 10 fmol of DNA (approximately 104 cpm), 100 mM NaCl, 10 mM Tris (pH 7.5), 1 mM EDTA, 35 ng of poly(dI-dC) nonspecific competitor, and various amounts of protein. Binding reaction mixtures were incubated for 30 min at 37°C and then resolved on a 10% polyacrylamide Tris-borate-EDTA (TBE) gel. Gels were dried, and radioactivity was quantified by phosphoimaging.
Footprint analysis.
The d(TTAGGG)4 oligonucleotide was gel purified and 32P labeled, heated to 90°C for 10 min, and then cooled to room temperature. Then, 500 fmol of DNA was incubated in a 50-μl binding reaction with or without protein (500 nM unless otherwise specified) for 30 min at 37°C. The hnRNP A1 protein used in footprinting was provided by Kenneth R. Williams, Yale University School of Medicine. After incubation, 5 μl of the reaction was removed to assay binding on a native gel. For dimethyl sulfate (DMS) footprinting, the remainder was made up to 250 μl containing 0.4% DMS, 50 mM sodium cacodylate, and 1 mM EDTA (pH 8.0), incubated for 5 min at room temperature, and then quenched with 40 μl of 1.5 M sodium acetate (pH 7.0)–1 M β-mercaptoethanol. Following two ethanol precipitations, the DNA pellet was dried, resuspended in 1 M piperidine, and incubated for 30 min at 90°C. Piperidine was removed by speed vacuum drying, and the DNA was resuspended in water and dried again. For P1 nuclease footprinting, 0.1 μg of P1 (Roche Pharmaceuticals) was added to the binding reaction mixture, quickly mixed, and then immediately quenched by adding 1/25 volume (2 μl) of 0.25 M EDTA–0.125 M EGTA. Footprinting reactions were analyzed on 10% acrylamide–8 M urea gels in TBE.
Pull-down and TRAP telomerase activity assays.
A cell pellet containing 106 HT1080 cells was lysed in 200 μl of 1× CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate} buffer, and 2 μl was tested for telomerase activity using a TRAP assay kit as directed by the manufacturer (Intergen). In pull-down experiments (e.g., Fig. 4B), 100 pmol of recombinant His6-tagged hnRNP D or HuR (see Results) was bound in batch to 40 μl of nickel resin, rinsed three times with 1× CHAPS buffer, and then incubated with 50 μl of HT1080 cell extract and 50 μl of 1× CHAPS buffer for 30 min at 37°C. Sonicated salmon sperm competitor DNA was preincubated with protein-loaded beads and also included during incubation with cell extracts, at a final concentration of 10 μg/ml. Following incubation, the beads were rinsed three times with 1× CHAPS buffer and resuspended in 40 μl of 1× CHAPS buffer; 10 μl was tested for telomerase activity.
FIG. 4.
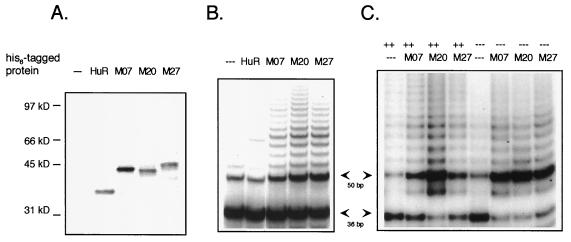
hnRNP D interacts with telomerase. (A) SDS-PAGE analysis of uncoated nickel beads (−−−) or beads coated with hnRNP D M07, M20, or M27 or with HuR. (B) PAGE analysis of products of TRAP assays of telomerase activity bound to beads. Arrows mark the positions of 36-bp control and 50-bp product DNAs. (C) As for panel B except that assays were carried out in the absence (−−−) or presence (++) of sonicated salmon sperm DNA as nonspecific competitor, as indicated on the top line.
RESULTS
hnRNP D binds the telomeric repeat as single-stranded but not duplex DNA.
hnRNP D is produced in distinct isoforms, related by alternative splicing of two conserved coding exons (7). The hnRNP D isoforms are referred to as hnRNP D M00, M07, M20, and M27, to reflect the presence or absence of the regions encoded by alternative exons 2 and 7 (Fig. 1). We assayed the ability of each hnRNP D isoform to bind a synthetic oligonucleotide carrying four iterations of the telomeric DNA repeat, d(TTAGGG)4. An example of a mobility shift performed with recombinant hnRNP D M07 is shown in Fig. 2A. From this and other data, we calculate that hnRNP D M07 binds the d(TTAGGG)4 oligonucleotide with a dissociation constant of approximately 30 nM. Similar results were obtained with the other hnRNP D isoforms (data not shown).
FIG. 2.
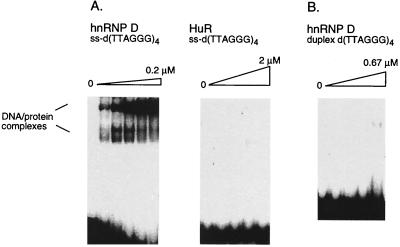
hnRNP D binds specifically to the single-stranded (ss) G-rich telomeric repeat. (A) Gel mobility shift analysis of binding reactions containing 32P-labeled telomeric oligonucleotide d(TTAGGG)4 and hnRNP D M07 (left) or HuR (right). DNA-protein complexes are indicated. The oligonucleotide was incubated with hnRNP D at 0.025, 0.05, 0.1, and 0.2 μM and with HuR at 0.20, 0.67, 1, and 2 μM. Other hnRNP D isoforms gave results similar to those for hnRNP D M07. (B) Gel mobility shift analysis of binding reactions containing 32P-labeled duplex d(TTAGGG)4 and hnRNP D M07 at 0.09, 0.17, 0.35, and 0.67 μM.
Like many eukaryotic proteins that interact with single-stranded nucleic acids, hnRNP D contains conserved RBDs. We tested whether binding to the d(TTAGGG)4 telomeric repeat is a property of all proteins that contain RBDs by assaying binding by HuR, an Elav family protein that contains three RBDs and binds AU-rich elements in mRNA to regulate transcript stability (11, 13, 31, 45). Recombinant HuR (a generous gift of C. Fan and J. Steitz, Yale Medical School) does not bind to the telomeric G-rich repeat even at very high protein concentrations (KD > 2 μM) (Fig. 2A).
hnRNP D does not recognize duplex d(TTAGGG) repeats.
Telomeres contain both single-stranded and duplex DNA. We tested the ability of hnRNP D to bind telomeric sequences as duplex DNA in gel mobility shift assays. As shown in Fig. 2B, recombinant hnRNP D M07 did not interact with the telomeric duplex 4-mer (KD >> 0.67 μM). Similar results were obtained in experiments that analyzed binding by all other hnRNP D isoforms (data not shown).
Binding affinity is length dependent.
Telomeric G tails in normal human cells are 130 to 210 nt in length (38, 44, 64). We examined whether binding affinities were dependent on length of the single-stranded G tail by assaying binding to a panel of synthetic oligonucleotides bearing different numbers of d(TTAGGG) repeats. Because many nucleic acid binding proteins can bind their specific sites only within a minimum length of DNA, the oligonucleotides used in this experiment carried nontelomeric flanking sequences so that the shortest oligonucleotide assayed was a 16-mer carrying one telomeric repeat. In other experiments, the presence of nontelomeric sequence at the 5′ or 3′ end of repeats has been shown not to influence hnRNP D binding (see below; also data not shown). As summarized in Table 1, hnRNP D did not bind the oligonucleotide bearing only a single d(TTAGGG) repeat (KD > 400 nM), bound to some extent to oligonucleotides bearing two (KD = 150 nM) or three (KD = 60 nM) repeats, and bound best to oligonucleotides carrying four or more repeats (KD ≤ 29 nM). Therefore, at least four repeats are required for optimal binding.
TABLE 1.
Effect of repeat length on hnRNP D bindinga
| DNA sequence | Length (nt) | KD (nM) |
|---|---|---|
| GACGTC(TTAGGG)1CGAT | 16 | >400 |
| GACGTC(TTAGGG)2CGAT | 22 | 150 |
| GACGTC(TTAGGG)3CGAT | 28 | 60 |
| GACGTC(TTAGGG)4CGAT | 34 | 29 |
| GACGTC(TTAGGG)5CGAT | 40 | 25 |
| GACGTC(TTAGGG)6CGAT | 46 | 21 |
hnRNP D was incubated with the indicated radiolabeled oligonucleotides, and the complexes were resolved by gel electrophoresis.
hnRNP D binding to d(TTAGGG)4 is sequence specific.
We determined sequence specificity of the interaction of hnRNP D with telomeric G-rich repeats by assaying binding of hnRNP D to a panel of DNAs in which C had been substituted for the naturally occurring base at each of the six positions in the d(TTAGGG)4 repeat. These data are summarized in Table 2. In almost all cases, single base mutations diminished binding from 5-fold to more than 20-fold. Mutation of either of the last two guanines in the repeat had the most deleterious effect, diminishing binding of all isoforms to less than 7% of the wild-type level. The effect of mutation on binding was essentially identical for all hnRNP D isoforms tested. The interaction of hnRNP D with the G-rich telomeric repeat is therefore sequence specific.
TABLE 2.
Sequence specificity of hnRNP D interaction with telomere repeatsa
| Repeat | % Bound
|
||
|---|---|---|---|
| M07 | M20 | M27 | |
| (TTAGGG)4 | 100 | 100 | 100 |
| (C-----)4 | 5 | 15 | 12 |
| (-C----)4 | 16 | 10 | 9 |
| (--C---)4 | 14 | 14 | 41 |
| (---C--)4 | 11 | 11 | 18 |
| (----C-)4 | 2 | 4 | 3 |
| (-----C)4 | 6 | 7 | 4 |
| (CCCTAA)4 | 100 | 4 | 5 |
| (-A----)4 | 140 | 23 | 23 |
| (--A---)4 | 50 | 6 | 9 |
| (---A--)4 | 3 | 2 | 3 |
| (----T-)4 | 20 | 61 | 85 |
hnRNP D (67 nM) was bound to radiolabeled oligonucleotides, and the bound complex was resolved by gel electrophoresis. The percentage bound was normalized for (TTAGGG)4 binding.
Specific isoforms of hnRNP D have high affinity for the C strand.
We assayed the ability of each hnRNP D isoform to bind a synthetic oligonucleotide bearing the wild-type telomeric C-strand sequence, d(CCCTAA)4, and a panel of DNAs carrying single base changes in the C-strand repeat. hnRNP D M07 bound to d(CCCTAA)4 with affinity comparable to that of hnRNP D (all isoforms) binding to the G-strand sequence, d(TTAGGG)4. Mutational analysis showed that hnRNP D M07 binding was impaired by changes at some but not all positions. hnRNP D M00 binding to the C strand was comparable to that of hnRNP D M07 (data not shown), while hnRNP D M27 and hnRNP D M20 bound to the telomeric C strand with affinities lower than those of hnRNP D M07 and hnRNP D M00 (Table 2). Thus, sequences encoded by alternative exon 2 appear to interfere somewhat with binding to the single-stranded C-rich telomeric repeat. hnRNP A1 does not bind the telomeric C strand (data not shown).
hnRNP D destabilizes G-G paired structures formed by telomeric G-tails.
We carried out footprint analyses to determine how hnRNP D interacts with telomeric G tails. Recombinant hnRNP D was incubated with the telomeric G-tail oligonucleotide d(TTAGGG)4, and free DNA or DNA-protein complexes were then footprinted. G-rich telomeric repeats readily form structures stabilized by G-G pairing (51, 56, 62). G-G pairing involves the N7 of guanine, and its experimental hallmark is resistance of guanine bases to methylation by DMS (50). It was therefore not surprising that in the absence of protein, most guanines in the G-tail oligonucleotides were not fully accessible to DMS probing (Fig. 3A), as this is typical of molecules that have formed structures stabilized by G-G pairing. Remarkably, binding by hnRNP D dramatically increased the sensitivity of the guanines to methylation (Fig. 3A). Thus, hnRNP D binding appeared to destabilize structures formed spontaneously by the telomeric oligonucleotide.
FIG. 3.
hnRNP D destabilizes G-G paired structures spontaneously formed by telomeric G tails. (A) DMS footprint of hnRNP D M07, M27, and M20 bound to 32P-labeled (TTAGGG)4. (B) P1 nuclease footprint of hnRNP D M07, M27, and M20 bound to 32P-labeled (TTAGGG)4. (C) DMS footprint of hnRNP D and hnRNP A1 bound to 32P-labeled (TTAGGG)4. Protein concentrations are 40, 80, and 160 nM. (D) DMS footprint of hnRNP D bound to 32P-labeled (TTAGGG)4, (AGGGTT)4N7, or N7(TTAGGG)4, where N7 corresponds to the sequence ATGACGA. (E) DMS footprint of hnRNP D bound to 32P-labeled (TTAGGG)8 or N7(TTAGGG)8 where N7 corresponds to ATGACGA. —, lane with no protein.
We also carried out footprint analysis with endonuclease P1, which cleaves single-stranded DNA. In the absence of protein, the telomeric oligonucleotide was resistant to P1 digestion, while incubation with hnRNP D rendered the telomeric oligonucleotide sensitive to digestion (Fig. 3B). Nuclease protection analysis thus further supports the notion that hnRNP D destabilizes structures formed by the G-rich telomeric repeat.
To determine whether hnRNP D and hnRNP A1 interact similarly with the G-rich telomeric repeat, we compared their DMS footprints at various protein concentrations. As shown in Fig. 3C, the methylation enhancement produced by hnRNP D binding contrasted with results observed with hnRNP A1. Recombinant hnRNP A1 (a generous gift of Kenneth R. Williams) binding to the (TTAGGG)4 repeat did not render all G's sensitive to methylation; instead, two G's are protected and the third G in each repeat is sensitive. We note that the cocrystal structure of the UP1 derivative of hnRNP A1 complexed to the 12-mer TTAGGGTTAGGG suggested that the protein makes base-specific contacts with two of three G's in each repeat, but that the third G in each repeat was not in close proximity with the protein (8). This is consistent with the observed DMS accessibility of only the third G in each repeat. Like hnRNP D, hnRNP A1 may unstructure the repeat upon binding; but in the hnRNP A1 complex, some contacts with the DNA may preclude accessibility to DMS.
Binding by the different isoforms of hnRNP D produced essentially identical patterns of methylation enhancement (Fig. 3C and data not shown). In addition, the presence of nontelomeric sequences at either the 5′ or 3′ terminus did not alter methylation enhancement (Fig. 3D). Methylation enhancement was similarly apparent upon hnRNP D binding to substrates containing eight rather than four iterations of the TTAGGG repeat (compare Fig. 3E and D; also data not shown).
hnRNP D interacts with telomerase.
Essentially all eukaryotic telomeres contain runs of three or more guanines in the strand that generates the 3′ tail. Single-stranded nucleic acid molecules containing runs of guanines have the potential to become spontaneously structured by G-G pairing, as visualized by the footprints in Fig. 3. If analogous structuring occurred in vivo, it could interfere with use of the 3′ tail as a primer for telomere extension by telomerase. The ability of hnRNP D to bind to the telomeric G strand and to destabilize G-G pairing within this region suggested that hnRNP D might function in telomere replication to facilitate telomerase interaction with telomeric tails. This prompted us to ask if hnRNP D can interact with telomerase. Recombinant hnRNP D, expressed as a His6-tagged fusion protein, was bound to nickel resin and assayed for the ability to pull down telomerase activity from a cell extract. Beads loaded with HuR (which does not bind telomeric repeats [Fig. 2A]) were also tested as a control for specificity. SDS-polyacrylamide gel electrophoresis (PAGE) analysis of coated beads verified comparable loading with each recombinant protein (Fig. 4A). Coated beads were incubated with extracts of HT1080, a human fibrosarcoma line, and washed extensively to remove any protein that was nonspecifically bound to the resin. Bound telomerase activity was then assayed by adding primer, deoxynucleoside triphosphates, and buffer appropriate for telomerase extension to the resuspended beads. After incubation for 30 min at 30°C, samples were heated to 94°C for 4 min, and extended products were amplified by PCR in the TRAP assay (Intergen). As shown in Fig. 4B, beads coated with hnRNP D M07, hnRNP D M20, and hnRNP D M27 all pulled down telomerase activity from the extract. In contrast, uncoated beads or beads coated with HuR did not. (M00 also pulls down telomerase activity [data not shown].) The results of this experiment therefore show that hnRNP D interacts specifically with a component of the telomerase complex.
When two nucleic acid binding proteins interact nonspecifically with the same nucleic acid molecule, they can become tethered to one another, enabling them to interact in a pull-down even though this interaction is not specific. One way to prevent tethering is to destroy nucleic acids by treating extracts with RNase and/or DNase; however, this was not possible for the experiments shown in Fig. 4B because the RNA component of telomerase is sensitive to RNase, and the PCR step of the TRAP assay is sensitive to DNase. Instead, we examined whether the hnRNP D-telomerase interaction was sensitive to competition by a nonspecific oligonucleotide by carrying out the pull-down in the presence of sonicated salmon sperm DNA. As shown in Fig. 4C, preincubation of hnRNP D-coated beads with this competitor did not affect the ability of hnRNP D to pull down telomerase activity.
Contacts between hnRNP A1 and telomere repeats are conserved in the hnRNP D sequence.
Both hnRNP D and hnRNP A1 contain conserved RBD structural motifs. The cocrystal structure of the N-terminal fragment of hnRNP A1 (UP1) bound to d(TTAGGG)2 was recently solved, and the amino acid residues that make specific contacts with DNA were inferred by proximity (8). It was therefore of interest to compare the conservation of amino acid residues of hnRNP D and hnRNP A1 at contact positions identified in the cocrystal.
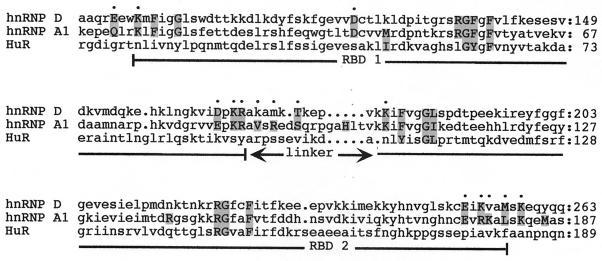
Figure 5 shows an alignment of the RBDs of hnRNP D and hnRNP A1; HuR, which does not bind the telomeric repeats (Fig. 2A) or interact with telomerase (Fig. 4B), is included for comparison. The protein-DNA contacts in the UP1 cocrystal were classified as either sequence specific (if they depended on a specific interaction between a base and an amino acid side chain) or non-sequence specific (if they involved interactions with amino acid main chain atoms, base stacking, or van der Waals interactions). Strikingly, 12 of 15 (80%) of the sequence-specific contacts are conserved between hnRNP A1 and hnRNP D. Moreover, two of the three nonconserved contacts are in the linker region, where hnRNP proteins are highly variable and the structure is thought to be flexible. In contrast, HuR, which contains RBDs but does not interact with either telomere DNA or telomerase, has no residues similar to those in either hnRNP A1 or hnRNP D at any of the positions that appear to mediate base-specific contacts. At positions that appear to be involved in non-sequence-specific contacts, hnRNP D and hnRNP A1 are similar at 13 of 17 positions (76%), while hnRNP A1 and HuR are similar at 10 of 17 positions (59%). The conservation between hnRNP D and hnRNP A1 at the sequence-specific contacts may well reflect the ability of these two proteins to interact with the same nucleic acid sequence in vivo.
FIG. 5.
ClustalW sequence alignment of hnRNP D, hnRNP A1, and HuR. Only the regions corresponding to the RBDs are shown; the first two RBDs of HuR are included in the alignment, and the third (and least conserved) RBD is omitted. Positions of contacts are in uppercase letters and shaded, and the remainder of the sequence is in lowercase letters; base-specific contacts are indicated with dots on the uppermost line, while non-sequence-specific contacts are not marked. RBD1, RBD2, and the linker region between them are indicated.
DISCUSSION
We have shown that the highly conserved mammalian protein hnRNP D binds in vitro to the single-stranded G-rich telomeric repeat TTAGGG and that this binding is sequence specific. We have further shown that hnRNP D destabilizes intramolecular G-G base pairs which form spontaneously within the G-rich telomeric strand in solution and that hnRNP D can interact with telomerase. These properties suggest that hnRNP D may enhance telomere extension by telomerase in vivo, by guiding telomerase to telomeric tails and facilitating their use as primers.
Telomeres in essentially all eukaryotes are G-rich repeats. The repeat in mammals is TTAGGG; in Oxytricha nova, it is T4G4; and in S. cerevisiae, it is TG1–3. Single-stranded DNAs containing runs of three or more G's spontaneously form intramolecular structures stabilized by G-G pairing (29, 50, 51, 56, 62, 63). G-G paired DNAs form rapidly and spontaneously in solution and once formed are quite stable; also, G-G pairing has been shown to interfere with telomere extension by telomerase (12, 65). Inefficient extension of a G-G paired telomeric tail probably reflects inefficient base pairing of the G-rich DNA strand with the RNA template of telomerase or inability of the 3′ end of the tail to gain access to the active site of the enzyme and prime extension. In either case, destabilization of G-G paired regions by hnRNP D would overcome this inhibition. It is also possible that hnRNP D does not function in telomere extension but is involved in telomere maintenance, by protecting the G-rich tail or by participating in forming or stabilizing the lariat-shaped T loops recently visualized at the telomeric termini (18).
hnRNP D has also been reported to regulate stability of mRNAs containing AU-rich regions (30, 35), but the mechanism by which this occurs is not established. While hnRNP D was first identified in association with hnRNA, it is not one of the core proteins in the stable 40S hnRNP particle and is readily removed during low-salt washes (9, 48, 57). Moreover, in contrast to HuR, another factor implicated in stabilization of mRNAs containing AU-rich regions (11, 45), hnRNP D does not associate with polysomes or cross-link to poly(A)+ mRNA (13). A telomeric function of hnRNP D is also consistent with biochemical fractionation carried out by another laboratory: when nuclei were extracted with detergent to produce a nuclear matrix fraction, hnRNP D, telomeric DNA, and the telomere duplex DNA binding protein TRF1 all cofractionated with the nuclear matrix (36). This fractionation pattern contrasts with that of hnRNP A2/B1, a protein which is part of the hnRNP particle core and is involved in splice site selection (22): hnRNP A2/B1 partitions with the unextracted nuclei and not the nuclear matrix fraction (36).
Another hnRNP family protein, hnRNP A1, may also function in telomere maintenance in mammalian cells (27). hnRNP A1 is known to regulate alternative splicing (42) and to function in mRNA transport (49) and packaging (reviewed in references 26, 43). Evidence for telomeric function was provided by experiments showing that the murine erythroleukemia cell line CB3, which does not express hnRNP A1, has short telomeres which become elongated upon transfection with constructs which express hnRNP A1 or its UP1 derivative. Redundancy of hnRNP A1 function is suggested by the observation that the CB3 line does proliferate in the absence of hnRNP A1. The sequence conservation between hnRNP D and hnRNP A1 at residues critical for interaction with the telomeric repeat (Fig. 5) and the shared ability of hnRNP D and hnRNP A1 (UP1) to interact with telomerase suggest that hnRNP D could perform a redundant or complementary role. Alternatively, the potential telomeric functions of hnRNP D and hnRNP A1 are opposing, with one protein stimulating and the other inhibiting telomere elongation or maintenance.
Alternative splicing produces distinct isoforms of hnRNP D which differ in the presence or absence of sequences encoded by exons 2 and 7 (Fig. 1). All hnRNP D isoforms tested bound in sequence-specific fashion to the G-rich repeat TTAGGG, but only the hnRNP D isoforms which lack exon 2 bound well to the C-rich repeat CCCTAA. Exon 2 is a short (57-nt) exon just upstream of the exon that encodes RBD1 (Fig. 1), and the 19-amino-acid region encoded by exon 2 may alter the region of hnRNP D that interacts with the C strand. These observations suggest that interactions with the C strand and G strand involve distinctive protein-nucleic acid contacts. Several different activities have been reported to interact with the telomeric C strand in vitro (28, 40, 41). Two of these have been identified as hnRNP K and ASF/SF2, both of which contain RBDs (28). The identity of the other has not been established, but its molecular mass is estimated to be 40 kDa (41), raising the possibility that it may be hnRNP D.
Parallels between yeast and mammalian cells may be a valuable guide to learning more about the possible telomeric function of hnRNP D. Genetic experiments in S. cerevisiae identified the EST1 gene as necessary to prevent progressive telomere shortening (37). Est1p was subsequently shown to be associated with the telomerase complex (32, 55) and to bind telomeric tails, although its binding affinity (KD = 250 nM) is lower than that of hnRNP D (59). Quite recently it was reported that Est1p contains an RBD which mediates its interaction with telomerase, apparently via the RNA component of the enzyme (68). The possible structural homology between Est1p and hnRNP D raises the very interesting and testable possibility that these proteins may also be functional homologs.
ACKNOWLEDGMENTS
We thank Cynthia Fan and Joan Steitz for providing recombinant HuR, and we thank Kenneth R. Williams for providing recombinant hnRNP A1. We are grateful to George Miller, David Schatz, and Joan Steitz for helpful suggestions and to Laurie Dempsey, David Hesslein, Michelle Duquette, and Yilun Liu for valuable experimental advice.
This research was supported by NCI P01 16038 to N.M. A.E. was supported by NIH predoctoral training grant T32 GM07223.
REFERENCES
- 1.Bianchi A, Smith S, Chong L, Elias P, de Lange T. TRF1 is a dimer and bends telomeric DNA. EMBO J. 1997;16:1785–1794. doi: 10.1093/emboj/16.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birney E, Kumar S, Krainer A R. Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21:5803–5816. doi: 10.1093/nar/21.25.5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bryan T M, Cech T R. Telomerase and the maintenance of chromosome ends. Curr Opin Cell Biol. 1999;11:318–324. doi: 10.1016/S0955-0674(99)80043-X. [DOI] [PubMed] [Google Scholar]
- 4.Burd C G, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- 5.Chong L, van Steensel B, Broccoli D, Erdjument-Bromage H, Hanish J, Tempst P, de Lange T. A human telomeric protein. Science. 1995;270:1663–1667. doi: 10.1126/science.270.5242.1663. [DOI] [PubMed] [Google Scholar]
- 6.Dempsey L A. G4 DNA binding by LR1 and its subunits, nucleolin and hnRNP D: a role for G-G pairing in immunoglobin switch recombination. J Biol Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 7.Dempsey L A, Li M-J, DePace A, Bray-Ward P, Maizels N. The human HNRPD locus maps to 4q21 and encodes a highly conserved protein. Genomics. 1998;49:378–384. doi: 10.1006/geno.1998.5237. [DOI] [PubMed] [Google Scholar]
- 8.Ding J, Hayashi M K, Zhang Y, Manche L, Krainer A R, Xu R M. Crystal structure of the two-RRM domain of hnRNP A1 (UP1) complexed with single-stranded telomeric DNA. Genes Dev. 1999;13:1102–1115. doi: 10.1101/gad.13.9.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dreyfuss G, Matunis M J, Piñol-Roma S, Burd C G. hnRNP proteins and the biogenesis of mRNA. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 10.Evans S K, Lundblad V. Est1 and cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 11.Fan X C, Steitz J A. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher T M, Sun D, Salazar M, Hurley L H. Effect of DNA secondary structure on human telomerase activity. Biochemistry. 1998;37:5536–5541. doi: 10.1021/bi972681p. [DOI] [PubMed] [Google Scholar]
- 13.Gallouzi I-E, Brennan C M, Steinberg M G, Swanson M S, Eversole A, Maizels N, Steitz J A. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci USA. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gotta M, Cockell M. Telomeres, not the end of the story. Bioessays. 1997;19:367–370. doi: 10.1002/bies.950190503. [DOI] [PubMed] [Google Scholar]
- 16.Greider C W. Telomerase activity, cell proliferation, and cancer. Proc Natl Acad Sci USA. 1998;95:90–92. doi: 10.1073/pnas.95.1.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greider C W. Telomeres and senescence: the history, the experiment, the future. Curr Biol. 1998;8:R178–R181. doi: 10.1016/s0960-9822(98)70105-8. [DOI] [PubMed] [Google Scholar]
- 18.Griffith J D, Comeau L, Rosenfield S, Stansel R M, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 19.Hanamura A, Caceres J F, Mayeda A, Franza B R, Jr, Krainer A R. Regulated tissue-specific expression of antagonistic pre-mRNA splicing factors. RNA. 1998;4:430–444. [PMC free article] [PubMed] [Google Scholar]
- 20.Harley C B. Telomere loss: mitotic clock or genetic time bomb? Mutat Res. 1991;256:271–282. doi: 10.1016/0921-8734(91)90018-7. [DOI] [PubMed] [Google Scholar]
- 21.Henderson E R. Telomere DNA structure. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1995. [Google Scholar]
- 22.Huang M, Rech J E, Northington S J, Flicker P F, Mayeda A, Krainer A R, LeStourgeon W M. The C-protein tetramer binds 230 to 240 nucleotides of pre-mRNA and nucleates the assembly of 40S heterogeneous nuclear ribonucleoprotein particles. Mol Cell Biol. 1994;14:518–533. doi: 10.1128/mcb.14.1.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa F, Matunis M J, Dreyfuss G, Cech T R. Nuclear proteins that bind the pre-mRNA 3′ splice site sequence r(UUAG/G) and the human telomeric DNA sequence d(TTAGGG)n. Mol Cell Biol. 1993;13:4301–4310. doi: 10.1128/mcb.13.7.4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kajita Y, Nakayama J, Aizawa M, Ishikawa F. The UUAG-specific RNA binding protein, heterogeneous nuclear ribonucleoprotein D0. J Biol Chem. 1995;270:22167–22175. doi: 10.1074/jbc.270.38.22167. [DOI] [PubMed] [Google Scholar]
- 25.Konig P, Rhodes D. Recognition of telomeric DNA. Trends Biochem Sci. 1997;22:43–47. doi: 10.1016/s0968-0004(97)01008-6. [DOI] [PubMed] [Google Scholar]
- 26.Krecic A M, Swanson M S. hnRNP complexes: composition, structure, and function. Curr Opin Cell Biol. 1999;11:363–371. doi: 10.1016/S0955-0674(99)80051-9. [DOI] [PubMed] [Google Scholar]
- 27.LaBranche H, Dupuis S, Ben-David Y, Bani M-R, Wellinger R J, Chabot B. Telomere elongation by hnRNP A1 and a derivative that interacts with telomeric repeats and telomerase. Nat Genet. 1998;19:1–4. doi: 10.1038/575. [DOI] [PubMed] [Google Scholar]
- 28.Lacroix L, Lienard H, Labourier E, Djavaheri-Mergny M, Lacoste J, Leffers H, Tazi J, Helene C, Mergny J-L. Identification of two human nuclear proteins that recognise the cytosine-rich strand of human telomeres in vitro. Nucleic Acids Res. 2000;28:1564–1575. doi: 10.1093/nar/28.7.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laporte L, Thomas G J., Jr A hairpin conformation for the 3′ overhang of Oxytricha nova telomeric DNA. J Mol Biol. 1998;281:261–270. doi: 10.1006/jmbi.1998.1938. [DOI] [PubMed] [Google Scholar]
- 30.Laroia G, Cuesta R, Brewer G, Schneider R J. Control of mRNA decay by heat shock-ubiquitin-proteosome pathway. Science. 1999;284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 31.Levine T D, Gao F, King P H, Andrews L G, Keene J D. Hel-N1: an autoimmune RNA-binding protein with specificity for 3′ uridylate-rich untranslated regions of growth factor mRNAs. Mol Cell Biol. 1993;13:3494–3504. doi: 10.1128/mcb.13.6.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin J J, Zakian V A. An in vitro assay for Saccharomyces telomerase requires EST1. Cell. 1995;81:1127–1135. doi: 10.1016/s0092-8674(05)80017-0. [DOI] [PubMed] [Google Scholar]
- 33.Lin J J, Zakian V A. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lingner J, Cech T R, Hughes T R, Lundblad V. Three Ever Shorter Telomere (EST) genes are dispensable for in vitro yeast telomerase activity. Proc Natl Acad Sci USA. 1997;14:11190–11195. doi: 10.1073/pnas.94.21.11190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loflin P, Chyi-Ying C, Shyu A-B. Unraveling a cytoplasmic role for hnRNP D in the in vivo mRNA destabilization directed by the AU-rich element. Genes Dev. 1999;13:1884–1897. doi: 10.1101/gad.13.14.1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luderus M E E, van Steensel B, Chong L, Sibon O C M, Cremers F F M, de Lange T. Structure, subnuclear distribution, and nuclear matrix association of the mammalian telomeric complex. J Cell Biol. 1996;135:867–881. doi: 10.1083/jcb.135.4.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lundblad V, Szostak J W. A mutant with a defect in telomere elongation leads to senescence in yeast. Cell. 1989;57:633–643. doi: 10.1016/0092-8674(89)90132-3. [DOI] [PubMed] [Google Scholar]
- 38.Makarov V L, Hirose Y, Langmore J P. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 39.Marcand S, Gilson E, Shore D. A protein-counting mechanism for telomere length regulation in yeast. Science. 1997;275:986–990. doi: 10.1126/science.275.5302.986. [DOI] [PubMed] [Google Scholar]
- 40.Marsich E, Piccini A, Xodo L E, Manzini G. Evidence for a HeLa nuclear protein that binds specifically to the single-stranded d(CCCTAA)n telomeric motif. Nucleic Acids Res. 1996;24:4029–4033. doi: 10.1093/nar/24.20.4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marsich E, Xodo L E, Manzini G. Widespread presence in mammals and high binding specificity of a nuclear protein that recognises the single-stranded telomeric motif (CCCTAA)n. Eur J Biochem. 1998;258:93–99. doi: 10.1046/j.1432-1327.1998.2580093.x. [DOI] [PubMed] [Google Scholar]
- 42.Mayeda A, Krainer A R. Regulation of alternative pre-mRNA splicing by hnRNP A1 and splicing factor SF2. Cell. 1992;68:365–375. doi: 10.1016/0092-8674(92)90477-t. [DOI] [PubMed] [Google Scholar]
- 43.McAfee J G, Huang M, Soltaninassab S, Rech J E, Iyengar S, LeStourgeon W M. The packaging of pre-mRNA. In: Krainer A R, editor. Eukaryotic mRNA processing. Oxford, United Kingdom: IRL Press; 1997. pp. 68–102. [Google Scholar]
- 44.McElligott R, Wellinger R J. The terminal DNA structure of mammalian chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Myer V E, Fan X C, Steitz J A. Identification of HuR as a protein implicated in AUUUA-mediated RNA decay. EMBO J. 1997;16:2130–2139. doi: 10.1093/emboj/16.8.2130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nugent C I, Hughes T R, Lue N F, Lundblad V. Cdc13p: a single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 47.Nugent C I, Lundblad V. The telomerase reverse transcriptase: components and regulation. Genes Dev. 1998;12:1073–1085. doi: 10.1101/gad.12.8.1073. [DOI] [PubMed] [Google Scholar]
- 48.Piñol-Roma S, Choi Y D, Matunis M J, Dreyfuss G. Immunopurification of heterogeneous nuclear ribonucleoprotein particles reveals an assortment of RNA-binding proteins. Genes Dev. 1988;2:215–227. doi: 10.1101/gad.2.2.215. [DOI] [PubMed] [Google Scholar]
- 49.Piñol-Roma S, Dreyfuss G. Shuttling of pre-mRNA binding proteins between nucleus and cytoplasm. Nature. 1992;355:730–732. doi: 10.1038/355730a0. [DOI] [PubMed] [Google Scholar]
- 50.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 51.Sen D, Gilbert W. Novel DNA superstructures formed by telomere-like oligomers. Biochemistry. 1992;31:65–70. doi: 10.1021/bi00116a011. [DOI] [PubMed] [Google Scholar]
- 52.Shore D. RAP1: a protean regulator in yeast. Trends Genet. 1994;10:408–412. doi: 10.1016/0168-9525(94)90058-2. [DOI] [PubMed] [Google Scholar]
- 53.Shore D. Telomere length regulation: getting the measure of chromosome ends. J Biol Chem. 1997;378:591–597. [PubMed] [Google Scholar]
- 54.Smith S, de Lange T. TRF1, a mammalian telomeric protein. Trends Genet. 1997;13:21–26. doi: 10.1016/s0168-9525(96)10052-4. [DOI] [PubMed] [Google Scholar]
- 55.Steiner B R, Hidaka K, Futcher B. Association of the Est1 protein with telomerase activity in yeast. Proc Natl Acad Sci USA. 1996;93:2817–2821. doi: 10.1073/pnas.93.7.2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sundquist W L, Klug A. Telomeric DNA dimerizes by formation of guanine tetrads between hairpin loops. Nature. 1989;342:825–829. doi: 10.1038/342825a0. [DOI] [PubMed] [Google Scholar]
- 57.Swanson M S, Dreyfuss G. RNA binding specificity of hnRNP proteins: a subset bind to the 3′ end of introns. EMBO J. 1988;7:3519–3529. doi: 10.1002/j.1460-2075.1988.tb03228.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Steensel B, Smogorzewska A, De Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–413. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
- 59.Virta-Pearlman V, Morris D K, Lundblad V. Est1 has the properties of a single-stranded telomere end-binding protein. Genes Dev. 1996;10:3094–3104. doi: 10.1101/gad.10.24.3094. [DOI] [PubMed] [Google Scholar]
- 60.Weighardt F, Biamonti G, Riva S. The roles of heterogeneous nuclear ribonucleoproteins (hnRNP) in RNA metabolism. Bioessays. 1996;18:747–756. doi: 10.1002/bies.950180910. [DOI] [PubMed] [Google Scholar]
- 61.Weilbaecher R G, Lundblad V. Assembly and regulation of telomerase. Curr Opin Chem Biol. 1999;3:573–577. doi: 10.1016/s1367-5931(99)00011-3. [DOI] [PubMed] [Google Scholar]
- 62.Williamson J R. G-quartet structures in telomeric DNA. Annu Rev Biophys Biomol Struct. 1994;23:703–730. doi: 10.1146/annurev.bb.23.060194.003415. [DOI] [PubMed] [Google Scholar]
- 63.Williamson J R, Raghuraman M K, Cech T R. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 64.Wright W E, Tesmer V M, Huffman K E, Levene S D, Shay J W. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zahler A M, Williamson J R, Cech T R, Prescott D M. Inhibition of telomerase by G-quartet DNA structures. Nature. 1991;350:718–720. doi: 10.1038/350718a0. [DOI] [PubMed] [Google Scholar]
- 66.Zhang W, Wagner B J, Ehrenman K, Schaefer A W, DeMaria C T, Crater D, DeHaven K, Long L, Brewer G. Purification, characterization, and cDNA cloning of an AU-rich element RNA-binding protein, AUF1. Mol Cell Biol. 1993;13:7652–7665. doi: 10.1128/mcb.13.12.7652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhong Z, Shiue L, Kaplan S, de Lange T. A mammalian factor that binds telomeric TTAGGG repeats in vitro. Mol Cell Biol. 1992;13:4834–4843. doi: 10.1128/mcb.12.11.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou J, Hidaka K, Futcher B. The Est1 subunit of yeast telomerase binds the Tlc1 telomerase RNA. Mol Cell Biol. 2000;20:1947–1955. doi: 10.1128/mcb.20.6.1947-1955.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]