Abstract
Motivation
It is essential to study bacterial strains in environmental samples. Existing methods and tools often depend on known strains or known variations, cannot work on individual samples, not reliable, or not easy to use, etc. It is thus important to develop more user-friendly tools that can identify bacterial strains more accurately.
Results
We developed a new tool called mixtureS that can de novo identify bacterial strains from shotgun reads of a clonal or metagenomic sample, without prior knowledge about the strains and their variations. Tested on 243 simulated datasets and 195 experimental datasets, mixtureS reliably identified the strains, their numbers and their abundance. Compared with three tools, mixtureS showed better performance in almost all simulated datasets and the vast majority of experimental datasets.
Availability and implementation
The source code and tool mixtureS is available at http://www.cs.ucf.edu/˜xiaoman/mixtureS/.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
It is imperative to reconstruct bacterial strain genomes from shotgun reads of clonal samples of individual species or metagenomic samples of many species (Luo et al., 2015; Pulido-Tamayo et al., 2015). Bacterial genomes are constantly evolving, where mutations are accumulated in different copies of a species genome that result in different strain genomes of the same species mixed in a sample (Zolfo et al., 2017). To identify bacterial strains in a sample, shotgun sequencing is routinely used to generate short DNA segments from mixed strain genomes in a sample, which are called reads and approximate the full DNA content and abundance of the mixed strain genomes in the sample (Nayfach et al., 2016). To reconstruct the strain genomes from these reads is thus crucial for our understanding of the bacterial diversity, evolution, function, drug resistance and so on (Nayfach et al., 2016; Pulido-Tamayo et al., 2015; Truong et al., 2017; Zolfo et al., 2017). More than a dozen methods are available for strain studies. The vast majority of them depend on known strains and/or known variations in strains, or intent to identify only variations in the species genome or a portion of the strain genomes, which cannot be generally applied or cannot de novo reconstruct the entire strain genomes (Ahn et al., 2015; Albanese and Donati, 2017; Hong et al., 2014; Luo et al., 2015; Nayfach et al., 2016; Quince et al., 2017; Roosaare et al., 2017; Truong et al., 2017; Zolfo et al., 2017). This leaves only a few methods that can de novo reconstruct bacterial strain genomes from reads in individual samples (Li et al., 2019; Pulido-Tamayo et al., 2015; Smillie et al., 2018). Moreover, to our knowledge, the performance of these remaining methods is still suboptimal. In addition, some tools are difficult to use by general biologists. We thus create a new tool called mixtureS that have better accuracy and are more user-friendly.
2 Methods and implementation
As previous studies (Li et al., 2019; Pulido-Tamayo et al., 2015; Smillie et al., 2018), mixtureS assumes that different strains of a species are likely to have different abundance. It also assumes that there are two types of nucleotides at a true polymorphic site, because almost all polymorphic sites in microbial genomes are biallelic (Foster et al., 2020). The first assumption makes the separation of the polymorphic sites in different strains possible, and the second one enables a simpler solution as shown below. Note that different strains are still allowed to share polymorphic sites.
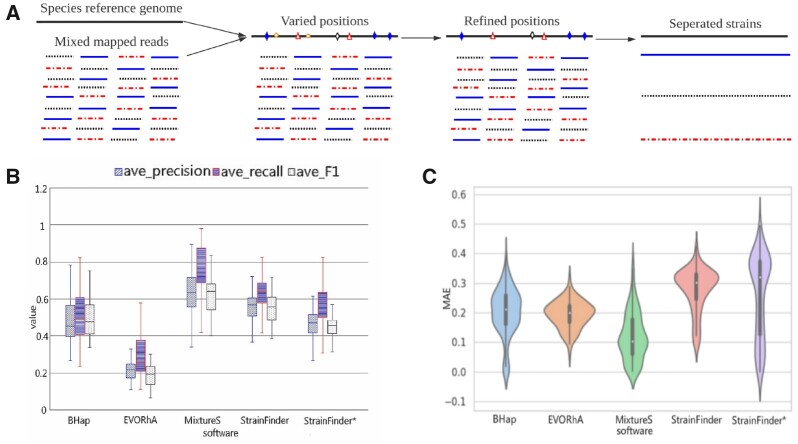
Starting from the mapped reads to a species genome, mixtureS infers the strain genomes and their abundance in three main steps (Fig. 1A). First, it identifies all genome positions with varied nucleotides in the mapped reads. Second, it refines the identified positions by removing positions with low-coverage (<10% of the average coverage of the genome) and positions with variations highly likely due to sequencing errors. Finally, mixtureS applies an expectation maximization (EM) algorithm to infer the strains from the remaining polymorphic positions. EM algorithms have shown good performance previously (Li and Waterman, 2003; Smillie et al., 2018; Wang et al., 2015, 2016, 2017).
Fig. 1.
The mixtureS tool and its performance. (A) The three main steps in mixtureS. (B) Performance of mixtureS and other tools on simulated data. (C) Performance of mixtureS and other tools on experimental data. MAE on the y-axis is the average absolute difference between the predicted abundance of a predicted strain and the corresponding known abundance of the corresponding known strain across strains and samples
In brief, assume that there are n remaining polymorphic positions, which are from m strains, and the frequency of the wild-type nucleotide and the mutated nucleotide at the ith position are and , respectively. Assume the relative abundance of jth strain is πj and the probability that a mutated nucleotide at a position belongs to the jth strain is αj. We have the expectation of the missing data at the E-step calculated as , where the missing data means the mutated nucleotide at the ith position is from the jth strain, and . We have the parameter estimation and at the M-step. Since m is unknown, mixtureS starts from m = 1 and applies the Bayesian information criterion to adjust m and identify the best m. See details in Supplementary Material S1.
We implement the above pipeline into the mixtureS tool package. This package provides the tool in both Linux and Windows versions. It also includes a script to process the FASTQ raw reads, to map reads to the reference genome of interest, and to generate a four by n matrix as the input to the above pipeline. The readme, source code, information about simulated and experimental datasets, together with example test datasets, are also provided. Compared with existing tools (Pulido-Tamayo et al., 2015; Smillie et al., 2018; Truong et al., 2017), it is much easier to set up the running environment for mixtureS and simpler to interpret the output of mixtureS, which makes it easy to apply mixtureS for strain studies.
3 Results
We tested mixtureS on 243 simulated datasets (Supplementary Material S2). In each dataset, we randomly generated shotgun reads for 2–4 strains of a bacterial species and mixed these reads together. We tested mixtureS together with three other tools, BHap, EVORhA and strainFinder on the mixed reads (Li et al., 2019; Pulido-Tamayo et al., 2015; Smillie et al., 2018). MixtureS predicted the correct strain numbers in 202 datasets, while BHap, EVORhA and strainFinder did it in 40, 46 and 0 datasets, respectively. Because strainFinder had trouble to find the right strain number, we input the correct strain number to run strainFinder, which was called strainFinder*. Even with this advantage, in terms of the strain abundance, mixtureS predicted at least 2.96, 1.74, 7.68 and 3.71 times closer to the true abundance than BHap, EVORhA, strainFinder and strainFinder*, respectively (the corresponding standard deviation as 8, 4, 40.70 and 18.50, respectively). In addition, the predicted polymorphic sites by mixtureS was much more accurate (Fig. 1B).
We also tested mixtureS on 195 experimental datasets (Supplementary Material S3) (Sobkowiak et al., 2018). There were two strains of Mycobacterium tuberculosis with known abundance in each dataset, while the polymorphic sites in the two strains were unknown. We compared how well the four methods predicted the number of strains and their abundance. BHap, EVORhA, strainFinder and mixtureS predicted two strains in 22, 0, 0 and 84 datasets, respectively. As to the strain abundance, mixtureS had a much accurate estimate than other tools, including strainFinder* (Fig. 1C).
4 Discussion
We demonstrated the usage of mixtureS on samples of individual species. For metagenomic samples with multiple species, users can map reads to the species genome of interest first and then apply mixtureS. mixtureS can infer strains more accurately than existing tools and is fast (Supplementary Material S4), which makes it a valuable addition to study bacterial strains.
Supplementary Material
Acknowledgements
The authors thank Saidi Wang and Amlan Talukder for their help with the revision.
Funding
This work was supported by the National Science Foundation [1661414, 2015838] and the National Institutes of Health [R15GM123407].
Conflict of Interest: none declared.
Contributor Information
Xin Li, Department of Computer Science.
Haiyan Hu, Department of Computer Science.
Xiaoman Li, Burnett School of Biomedical Science, College of Medicine, University of Central Florida, Orlando, FL 32816, USA.
References
- Ahn T.-H. et al. (2015) Sigma: strain-level inference of genomes from metagenomic analysis for biosurveillance. Bioinformatics, 31, 170–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albanese D., Donati C. (2017) Strain profiling and epidemiology of bacterial species from metagenomic sequencing. Nat. Commun., 8, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J.T. et al. (2020) Ricin forensics: comparisons to microbial forensics. In: Budowle, S. (eds) Microbial Forensics. Acedemia Press, pp. 315–326. [Google Scholar]
- Hong C. et al. (2014) Pathoscope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome, 2, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Waterman M.S. (2003) Estimating the repeat structure and length of dna sequences using -tuples. Genome Res., 13, 1916–1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. (2019) BHap: a novel approach for bacterial haplotype reconstruction. Bioinformatics, 35, 4624–4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C. et al. (2015) Constrains identifies microbial strains in metagenomic datasets. Nat. Biotechnol., 33, 1045–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayfach S. et al. (2016) An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res., 26, 1612–1625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Tamayo S. et al. (2015) Frequency-based haplotype reconstruction from deep sequencing data of bacterial populations. Nucleic Acids Res., 43, e105–e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C. et al. (2017) DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol., 18, 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roosaare M. et al. (2017) Strainseeker: fast identification of bacterial strains from raw sequencing reads using user-provided guide trees. PeerJ, 5, e3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smillie C.S. et al. (2018) Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe, 23, 229–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobkowiak B. et al. (2018) Identifying mixed Mycobacterium tuberculosis infections from whole genome sequence data. BMC Genomics, 19, 613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong D.T. et al. (2017) Microbial strain-level population structure and genetic diversity from metagenomes. Genome Res., 27, 626–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2015) MBBC: an efficient approach for metagenomic binning based on clustering. BMC Bioinformatics, 16, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2016) MBMC: an effective markov chain approach for binning metagenomic reads from environmental shotgun sequencing projects. Omics J. Integr. Biol., 20, 470–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. et al. (2017) rrnafilter: a fast approach for ribosomal RNA read removal without a reference database. J. Comput. Biol., 24, 368–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolfo M. et al. (2017) MetaMLST: multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res., 45, e7–e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.