Abstract
Much of our understanding of GH’s action stems from animal models and the generation and characterization of genetically altered or modified mice. Manipulation of genes in the GH/IGF1 family in animals started in 1982 when the first GH transgenic mice were produced. Since then, multiple laboratories have altered mouse DNA to globally disrupt Gh, Ghr, and other genes upstream or downstream of GH or its receptor. The ability to stay current with the various genetically manipulated mouse lines within the realm of GH/IGF1 research has been daunting. As such, this review attempts to consolidate and summarize the literature related to the initial characterization of many of the known gene-manipulated mice relating to the actions of GH, PRL and IGF1. We have organized the mouse lines by modifications made to constituents of the GH/IGF1 family either upstream or downstream of GHR or to the GHR itself. Available data on the effect of altered gene expression on growth, GH/IGF1 levels, body composition, reproduction, diabetes, metabolism, cancer, and aging are summarized. For the ease of finding this information, key words are highlighted in bold throughout the main text for each mouse line and this information is summarized in Tables 1, 2, 3 and 4. Most importantly, the collective data derived from and reported for these mice have enhanced our understanding of GH action.
Keywords: Growth hormone, Prolactin, Insulin-like growth factor 1, Transgenic mice, Knockout mice, Metabolism, Cancer, Aging
Introduction
Growth hormone (GH) helps regulate and coordinate growth and other physiological processes, including metabolism, fluid balance, immunity, and aging. The investigation of GH’s actions has an extensive history. The growth-promoting activity of GH was recognized in 1921 when chronic administration of extracts from bovine pituitary glands resulted in enhanced weight gain when injected into rats [1, 2]. Then in 1936, Houssay demonstrated both the diabetogenic activity of anterior pituitary extracts and the decreased severity of diabetes in anterior-hypophysectomized dogs [3]. The protein responsible, GH, was first purified from bovine (b) pituitary extracts in 1944 [4]. Human (h) GH was purified in 1956 from cadaver pituitary glands [5], and its efficacy was established in the treatment of pediatric GH-deficient patients [6]. Purified hGH was approved for use in the US in 1958 and became the standard treatment until recombinant human GH was approved for use in 1986 [7–9]. The interesting history of GH discoveries, both basic and clinical, has been recently reviewed [2, 10].
In humans, a GH-related gene cluster is located in a 78 k base pair portion of chromosome 17 [11] and contains five tandemly linked GH-related genes, in which one (GH1) present at the 5’ end of the cluster is expressed in the anterior pituitary. Three of the other genes are expressed in the placenta, and one is a non-expressed pseudogene. GH1 encodes a 22 kDa protein consisting of 191 amino acids following cleavage of the 26-amino acid secretory signal peptide. It contains four antiparallel α helices and has significant structural homology with prolactin (PRL) and placental lactogen [12].
GH exerts its actions by binding to a specific cell surface receptor (R). The hGH receptor (GHR) gene is located on chromosome 5, encodes a single-chain transmembrane glycoprotein composed of 638 amino acids, and is a member of the type I cytokine receptor family. After removal of its 18-amino acid secretory signal peptide, hGHR is composed of a N-terminal, 246-amino acid extracellular domain; a 24-amino acid transmembrane domain; and a C-terminal, 350-amino acid intracellular domain [13, 14]. The extracellular domain contains three disulfide bonds; two of which are essential for ligand binding [15]. The cytoplasmic domain contains two highly conserved sequences among cytokine receptors, Box 1 and Box 2. Box 1 contains nine amino acids with proline-rich and hydrophobic residues and acts as a binding site for a signal-transducing Janus kinase 2 (JAK2). The elegant work of Waters et al. provided a mechanistic model for this initiation of GH-GHR-induced intracellular signal transduction via JAK2 activation [14, 16]. That is, GHRs exist as preformed dimers in the absence of ligands [17]. Two JAK2 molecules, each bound to a GHR, are closely located; however, trans-interaction of the kinase domain of one JAK2 molecule and the pseudokinase domain of the other JAK2 inhibit each other, and the JAK2 stays inactive. Upon GH binding, the relative position of GHRs changes, resulting in JAK2 activation [16]. Activated JAK2 further phosphorylates multiple tyrosine residues on the intracellular domain of the GHR [18–20], which serves as binding sites for proteins possessing SH2 domains. The most common and best described of the GH induced intracellular signaling pathways involves signal transducer and activator of transcription (STAT) 5a and 5b molecules. STAT5 molecules are recruited to the phosphotyrosine residues on the GHR and become activated through tyrosine-phosphorylation by JAK2. Tyrosine phosphorylation of STAT molecules results in the dissociation of the STAT molecules from the receptor followed by homo- or heterodimerization and translocation to the nucleus, where they regulate the expression of GH target genes [21].
One of the negative regulators of the JAK-STAT signaling pathway is the suppressor of the cytokine signaling (SOCS) protein family. SOCS1-3 and cytokine-inducible SH2-containing protein (CISH) are implicated in the negative regulation of GH action, of which SOCS2 appears to play a major role [22]. All SOCS proteins are able to direct the ubiquitination of SH2 and N-terminal bound substrates for degradation [23]. Additionally, SOCS3 has been shown to directly inhibit the enzyme activity of JAK2 by its kinase-inhibitory region [24].
Manipulation of GH genes in animals started in 1982 when the first GH transgenic mouse was produced by Palmiter et al. using a fusion gene consisting of the promoter/enhancer of the mouse metallothionein-1 (Mt1) gene and the rat (r) Gh gene [25]. The fusion gene was microinjected into the pronuclei of fertilized mouse eggs and gave rise to giant mice, featured on the cover of a 1982 issue of Nature [25]. Usually, mice generated in this manner (microinjection of cloned DNA in fertilized mouse eggs) are termed hemizygous, signifying random incorporation of the injected DNA into the mouse genome. Breeding of hemizygous mice can result in new mouse strains containing two or more copies/alleles of the injected DNA.
Our group has employed a structure/function experimental design using transgenic mice for the past three decades. During this time, we discovered that substitution of one amino acid, Gly119 in bGH (Gly120 in hGH), by several amino acids (except Ala) resulted in a competitive antagonist of the GHR [26–28]. Expression of this GHR antagonist in vivo resulted in dwarf mice [26]. At that time, we predicted that GH interacted with a secondary target protein to explain the mechanism of GHR antagonism [26]. Later, Cunningham et al. demonstrated that, indeed, one GH molecule interacted with two GHR molecules to initiate signal transduction [29].
We and others have also employed gene disruption, or ‘knockout’ (KO) technology, to globally disrupt Gh, Ghr, or other genes upstream or downstream of GH or its receptor. Our group has focused on Ghr gene disruptions. Ghr-/- (also called GHRKO or GHR-/-) mice are dwarf and obese, with low insulin-like growth factor 1 (IGF1) and high GH levels [30]. Importantly, since the mice lack functional GHRs, they are GH insensitive or resistant and, thus, more insulin sensitive than wild-type (WT) littermates [31]. They are also resistant to high-fat diet (HFD)-induced type 2 diabetes (T2D) [32] and cancer [33–36]. Surprisingly, GHR-/- mice have a longer lifespan than WT mice [37]. One GHR-/- mouse lived a week short of five years and set the standard for the Methuselah Mouse Prize as the world’s longest-lived laboratory mouse (http://reason.com/archives/2004/08/18/methuselah-mouse). Since then, our group, as well as others, have developed many tissue-specific GHRKO mice, which will be described below [38–65].
Internally, the task of ‘keeping up’ with the various genetically manipulated mouse lines within the GH/IGF1 family has been daunting. Thus, in this review, we have critically reviewed the literature related to the initial characterization of many natural and gene-manipulated mice related to the actions of GH, prolactin (PRL), and IGF1. We acknowledge that additional phenotypic/biochemical/endocrine data may exist for these mouse lines but consider this beyond the scope of this review.
Below, we have organized the mouse lines by modifications made to constituents of the GH/IGF family either upstream or downstream of GHR or to the GHR itself. Throughout this review, we define global homozygous null (-/-) mice as knockouts (KOs) and heterozygotes as +/− . For all mouse lines discussed, we recognize the individual(s) who generated the mice along with the date and laboratory name. Mice with ‘upstream’ modifications include GH transgenic, GH-/-, GH releasing hormone (GHRH) transgenic, GHRH-/-, GHRHR-/-, GHR antagonist, PRL-/-, PRLR-/-, and PRLR antagonist transgenic mouse lines. GHR modifications include global GHRKO (GHR-/-), various tissue-specific GHRKOs, and temporal GHRKOs. Modifications downstream of GHR include those made to several signal transduction molecules including JAK2 and STAT5, IGF1 and IGF1R, IGF binding proteins (BPs), ALS and PAPP-A transgenic, and KOs. Importantly, available data on the effect of altered gene expression on growth, GH/IGF1 levels, body composition, reproduction, diabetes, metabolism, cancer, and aging are summarized for each mouse line and in Tables 1, 2, 3, and 4. For ease of finding this information, key words are shown in bold throughout the main text. Also, to aid the reader, we have divided each section into “origin” and “phenotype”. To provide additional context, Fig. 1 illustrates the mouse lines with alteration in the GH/IGF family and relevant upstream and downstream constituents referred to in this review. Figure 2 compares several transgenic and null mouse lines related to GH action for adiposity, metabolism, cancer incidence, and longevity. Figure 3 provides a timeline of when the mouse lines were generated. Overall, we hope this review will provide a comprehensive reference to investigators by collating numerous results and references relating to specific mouse lines within the GH/IGF1 family. Importantly, the collective data derived and reported for these mice have enhanced our understanding of GH action.
Table 1.
Mouse lines upstream of GHR
| Mouse line | Discovery (year/lab/1st author) | Expression control | Serum GH | Serum IGF1 | Size / weight | Body composition | Insulin sensitivity | Reproductive capacity | Cancer incidence | Lifespan | Original references |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Snell (Pit1−/−) | 1929 Snell | spontaneous mutation in Pou1f1 | ↓ | ↓ | ↓ | ↑ muscle at 3 mo, though lower quality | ↑ | ↓ | ↓ | ↑ | [67] |
| Ames (Prop1−/−) | 1961 Schaible and Gowen | spontaneous mutation in Prop1 | ↓ | ↓ | ↓ | ↑ fat | ↑ | ↓ | ↓ | ↑ | [83] |
| hGHRH | 1994 Hyde (Moore) | Mt1 | ↑ | ↑ | n/a | n/a | n/a | n/a | n/a | n/a | [99] |
| GHRH-/- | 2004 Salvatori (Alba) | NeoR replacing amino acid residues 1–42 | ↓ | ↓ |
↓ size; – weight |
↑ fat | ↑ | ↓ | n/a | ↑ | [108] |
| lit/lit (Ghrhr−/−) | 1976 Eicher (Beamer) | spontaneous mutation in Ghrhr | ↓ | ↓ | ↓ | ↑ fat | ↑ | ↓ | ↓ | ↑ | [115, 116] |
| MT1-hGH | 1983 Brinster (Palmiter) | Mt1 | ↑ | ↑ | ↑ | ↑ soleus weight | n/a | ↓ | ↑♀ | n/a | [124] |
| hGH | 1991 Isaksson (Tornell) | Mt1 | ↑ | n/a | ↑ size | n/a | n/a | n/a | ↑♀ | n/a | [132] |
| 171hGH/CS-TG | 2009 Cattini (Jin) | LCR | n/a | n/a | – | n/a | n/a | n/a | n/a | n/a | [135] |
| hGH | 1991 Brem (Gunzburg) | Wap | n/a | n/a | – | ↑ heart weight ♂ | n/a | n/a | n/a | n/a | [139] |
| hGH |
1994 Houdebine (Devinoy) |
(Rabbit) Wap | n/a | n/a | n/a | n/a | n/a | ↓ | n/a | n/a | [140] |
| GHv | 1988 Goodman (Selden) | Mt1 | n/a | ↑ | ↑ | n/a | n/a | ↓ | n/a | n/a | [128] |
| bGH MT | 1985 Palmiter (Hammer) | Mt1 | ↑ | ↑ | ↑ weight | ↑ lean; ↓ fat | dysregulated | ↓ | ↑ | ↓ | [145] |
| bGH PEPCK | 1988 Handon (McGrane) | Pck1 | ↑ | ↑ | ↑ weight | ↑ lean; ↓ fat | ↓serum glucose | ↓ | n/a | ↓ | [155] |
| GH-/- | 2019 Kopchick (List) | VelociGene KOMP null allele/ZEN-UB1 reporter | ↓ | ↓ | ↓ | ↑ fat; ↓ lean | ↑; glucose intolerant | n/a | n/a | n/a | [167] |
| AOiGHD | 2011 Kineman (Luque) | rGh/Cre with iDTR | ↓ | ↓ | – | ↑ fat (HFD) | ↑ | n/a | n/a | n/a | [168] |
| GHA | 1990-mGHA; 1991-hGHA Kopchick (Chen) | Mt1 | ↑ | ↓ | ↓ | ↑ fat; ↓ lean | ↑ | ↓ | ↓ | – | [26, 169, 170] |
“—” indicates no change; n/a indicates not available; “mo” indicates months of age; ♂ indicates males; ♀ indicates females
Table 2.
Global, temporal and tissue-specific GHRKO mouse lines
| Mouse line | Discovery (year/lab/1st author) | Expression control | Serum GH | Serum IGF1 | Size / weight | Body composition | Insulin sensitivity | Lifespan | Original references | |
|---|---|---|---|---|---|---|---|---|---|---|
| Global GHRKO | GHR-/- | 1997 Kopchick (Zhou) | NeoR replacing exon4 of Ghr | ↑ | ↓ | ↓ | ↑ fat; ↓ lean | ↑; glucose intolerant | ↑ | [30, 31] |
| Global Adult Onset | aGHRKO | 2016 Kopchick (Junnila) | ROSA26/Cre | ↑ | ↓ | ↓ | ↑ fat; ↓ lean | ↑; glucose intolerant | ↑ maximal lifespan ♀ | [60] |
| Liver-specific KO | GHRLD | 2009 Sperling (Fan) | albumin/Cre | ↑ | ↓ | – | – | ↓; glucose intolerant | n/a | [38] |
| LiGHRKO | 2014 Kopchick (List) | albumin/Cre | ↑ | ↓ | ↓ after 5 mo | ↑ fat at early ages; ↓ adulthood | ↓ glucose homeostasis | – | [48] | |
| aLivGHRkd | 2015 Kineman (Cordoba) | thyroxin-binding promotor/Cre | ↑♂ | ↓ | n/a | n/a | n/a | n/a | [55] | |
| Li-GHRKO | 2016 Yakar (Liu) | albumin/Cre | n/a | ↓ | – | ↑ fat | ↓; increased blood glucose | n/a | [59] | |
| L-Ghr-/- | 2019 Liang (Fang) | albumin/Cre | ↑ | ↓ | – | – | hypoglycemic under CR | n/a | [63] | |
| Muscle-specific KO | ΔGHR | 2010 Clemens (Mavalli) | fem-2c-73 k/Cre | – | – | ↑ weight | ↑ fat | ↓ | n/a | [40] |
| mGHRKO | 2012 LeRoith (Vijayakumar) | Ckmm/Cre | – | – | – | ↓ lean | – | n/a | [42] | |
| MuGHRKO | 2015 Kopchick (List) | Ckmm/Cre | – | – | – | – | ↑ glucose homeostasis ♂ | ↑ | [54] | |
| Brain-specific KO | LeprEYFPΔGHR | 2017 Sadagurski (Cady) | leptin receptor/Cre | – | – | – | – | ↓ glucose homeostasis | n/a | [61] |
| AgRP-IRES-Cre | 2019 Donato (Furigo) | AgRP IRES/Cre | n/a | n/a | – | – | – | n/a | [64] | |
| LepR-IRES-Cre | 2019 Donato (Furigo) | LepR IRES/Cre | n/a | n/a | ↑ | ↓ fat | n/a | n/a | [64] | |
| Nestin-Cre | 2019 Donato (Furigo) | Nestin/Cre | n/a | n/a | ↑ | ↑ lean | n/a | n/a | [64] | |
| Fat-specific KO | FaGHRKO | 2013 Kopchick (List) | aP2/Cre (aka, FABP4/Cre) | – | ↑♂ | ↑ weight | ↑ fat, ↑fluid; ↑ lean mass ♀ | – | ↓ | [44] |
| AdGHRKO | 2019 Kopchick (List) | adiponectin/Cre | – | – | – | ↑ fat; ↑brown AT ♀ | ↑ | n/a | [220] | |
| Fat-Ghr-/- | 2019 Liang (Fang) | adiponectin/Cre | n/a | n/a | n/a | – | n/a | n/a | [63] | |
| Macrophage/monocyte-specific KO | GHRMacD | 2010 Menon (Lu) | Lyzs/Cre | n/a | n/a | – | ↑ epididymal fat on HFD | –; ↓ on HFD | n/a | [39, 47] |
| Beta cell-specific KO | βGHRKO | 2011 LeRoith (Wu) | insulin/Cre | – | – | – | – | n/a | n/a | [41] |
| HSC-specific KO | Ghrfl/fl;Vav1Cre/+ | 2014 Rossi (Stewart) | Vav1/Cre | n/a | n/a | n/a | n/a | n/a | n/a | [50] |
| Bone-specific KO | DMP-GHRKO | 2016 Yakar (Liu) | Dmp1/Cre | ↑ 8 wk, – 12 wk | – | – | – | n/a | n/a | [224] |
| Heart-specific KO | iC-GHRKO | 2016 Kopchick (Jara) | Myh6/Cre | n/a | –; ↓12.5 mo | – | ↓ fat; ↑ lean | ↑ 6.5 mo; ↓ 12.5 mo | n/a | [58] |
| Intestine-specific KO | IntGHRKO | 2019 Kopchick (Young) | villin/Cre | n/a | n/a | – | – | ↓♀ | n/a | [65] |
“—” indicates no change; n/a indicates not available. “wk” indicates weeks of age, “mo” indicates months of age; ♂ indicates males; ♀ indicates females
Table 3.
Global and tissue-specific IGF1 transgenic and IGF1(R) KO mouse lines
| Mouse lines | Discovery (year/lab/first) | Expression control | Serum GH | Serum IGF1 | Size / weight | Body composition | Insulin sensitivity | Reproductive capacity | Cancer incidence | Lifespan | Original references | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global | IGF1 | 1988 Palmiter (Mathews) | Mt1 + human IGFI cDNA | ↓ | ↑ | ↑ weight | n/a | n/a | – | n/a | n/a | [257] |
| IGF2+/− | 1990 Robertson (DeChiara) | NeoR replacing exon 2 of Igf2 | n/a | n/a | ↓ | n/a | n/a | – | n/a | n/a | [259] | |
| IGF1-/- | 1993 Efstratiadis (Liu) | deleting exon 4 of Igf1 | n/a | ↓ | ↓ | n/a | n/a | n/a | n/a | neonatal lethality; ↓ | [260] | |
| IGF1R-/- | 1993 Efstratiadis (Liu) | deleting exon 3 of Igf1r | n/a | n/a | ↓ | n/a | n/a | n/a | n/a | complete neonatal lethality | [260] | |
| Liver | TTR-IGF-1 | 2006 Xu (Liao) | transthyretin | ↓ | ↑ | ↑ | ↑ lean | ↑ glucose tolerance | – | – | n/a | [263] |
| HIT |
2009 LeRoith (Wu) |
transthyretin | – | ↑ | ↑;↑L,K,S | ↓fat | – | – | n/a | n/a | [265] | |
| KO-HIT |
2009 LeRoith (Wu) |
transthyretin + IGF1 null | – | ↑ | ↓at birth; – at 16 wk | ↓fat | – | –♂;↓♀ | n/a | No prenatal lethailty | [265] | |
| GHRKO-HIT | 2013 Yakar (Wu) | transthyretin | n/a | – | ↓; ↓K,S,H | ↓ muscle; ↑brown AT | – | n/a | n/a | n/a | [267] | |
| LID | 1999 LeRoith (Yakar) | albumin/Cre | ↑ | ↓ | – | – | ↓ | – | ↓ | ↑ ♀ | [268] | |
| LI-IGF1-/- |
1999 Ohlsson (Sjögren) |
Mx/Cre induced at ~ 1 mo | ↑ | ↓ | –; ↓K, ↑L at 3mo; ↓ wight at 13 mo | ↓ femur length at 3 mo; ↓ fat at 13 mo | ↓ | – | n/a | ↑ mean lifespan ♀ | [270] | |
| Adipose | IGF-1RaP2Cre | 2008 Blüher (Kloting) | aP2/Cre | – | ↑ | ↑ | ↑ fat | – | n/a | n/a | n/a | [273] |
| F-IGFRKO | 2016 Kahn (Boucher) | Adipo/Cre | n/a | ↑ | – | ↓ fat | – | n/a | n/a | n/a | [277] | |
| Brain | bIGF1RKO+/− | 2008 Holzenberger (Kappeler) | nestin/Cre | ↓ | ↓ | ↓ | ↑ fat | ↓ | infertile (-/-) | ↓ | ↑ mean lifespan (+/−); – (-/-) | [278] |
| Muscle | Skeletal Muscle IGF1 | 1995 Schwartz (Coleman) | avian skeletal α-actin driving human IGF1 | n/a | – | – | ↑ superficial gluteus muscle mass | n/a | n/a | n/a | n/a | [279] |
| MKR | 2001 LeRoith (Fernandez) | MCK + dominant-negative IGF1R | n/a | n/a | ↓ weight | n/a | ↓ | n/a | n/a | n/a | [280] | |
| MIGIRKO | 2015 Kahn (O'Neil) | ACTA1/Cre | n/a | n/a | ↓ | ↓ muscle | – | n/a | n/a | ↓ | [281] | |
| M-IGF1R KO | 2016 Kahn (O'Neil) | ACTA1/Cre | n/a | n/a | – | – | n/a | n/a | n/a | ↓ | [281, 282] | |
| Cardiac | IGF1 | 1996 Anversa (Reiss) | α-MHC + human IGF1 | n/a | ↑ | – at birth, ↑ on day 210; ↑ organ weights | – | n/a | ↓ using both Tg mice as breeders | n/a | – | [283] |
| Cardiomyocyte IGF-1 | 2007 Rosenthal (Santini) | α-MHC + rat mIGF1 | n/a | n/a | n/a | n/a | n/a | Hets as breeders | n/a | n/a | [284] | |
| CIGF1RKO | 2008 Abel (Kim) | Myosin 6/Cre | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | [285] | |
| iCMIGF-IRKO | 2012 Gödecke (Moellendorf) | 4-OHTX inducible Myosin 6/Cre at 3&11 mo | n/a | ↑ | ↓ size in newborns | n/a | – | n/a | n/a | n/a | [286] | |
| Endothelial | hIGFREO | 2012 Kearney (Imrie) | TIE2 + hIGF1R | n/a | n/a | – | n/a | – | n/a | n/a | n/a | [287] |
| EC IGF-1RKO | 2011 Kearney (Abbas) | TIE2/Cre, male 3–5 mo | n/a | n/a | n/a | n/a | – | n/a | n/a | n/a | [288] | |
| Endothelial IGF-1RKO | 2015 Cheng (Liang) | VE-Cadherin/Cre, male 3–4 mo | n/a | n/a | – | n/a | n/a | – | n/a | n/a | [289] | |
| Myeloid (Macrophage) | MIKO | 2016 Dixit (Spadaro) | LysM/Cre | n/a | n/a | n/a | ↑ fat | ↓ on HFD | n/a | n/a | ↑ pro-longevity effects | [290] |
| MΦ-IGF1RKO | 2016 Delafontaine (Higashi) | LysM/Cre x IGF1R/APOE-/- FLOX | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | [292] | |
| Bone | OC-IGF-1 Tg | 2000 Clemens (Zhao) | osteocalcin + rat IGF1 | n/a | – | – | n/a | n/a | n/a | n/a | n/a | [293] |
| Osteoblast IGF1 Tg | 2006 Kream (Jiang) | rat Col1a1 + murine Igf1 | n/a | ↑ | ↑ weight ♂ | n/a | n/a | n/a | n/a | n/a | [294] | |
|
Osteoblast IGF1R KO |
2002 Clemens (Zhang) | osteocalcin/Cre | n/a | n/a | – | n/a | n/a | n/a | n/a | n/a | [296] | |
| OBIGF1R−/− | 2015 Bikle (Wang) | Col1α1/Cre | n/a | n/a | – | n/a | n/a | n/a | n/a | n/a | [298] | |
| Chondrocyte IGF1 KO | 2007 Mohan (Govoni) | Col2α1/Cre | n/a | – | ↓ length | ↓ bone size, weight | n/a | n/a | n/a | n/a | [299] | |
| Osteocyte IGF1 KO | 2013 Lau (Sheng) | Dmp-1/Cre | n/a | – | n/a | ↓ femur length | n/a | n/a | n/a | n/a | [301] | |
| DMP-IGF-1R KO | 2016 Yakar (Liu) | DMP-1/Cre | ↑ at 8 wk; – at 16 wk | – | – | n/a | n/a | n/a | n/a | n/a | [224] | |
| Ovarian granulosa cells | IGF1Rgcko | 2017 Stocco (Baumgarten) | ESR2 + CYp19 | n/a | n/a | n/a | n/a | – | infertile | n/a | n/a | [303] |
| Pancreatic beta cells | β cell IGF-1R KO |
2002 Efstratiadis (Xuan); Kahn (Kulkarni) |
rat insulin/Cre | n/a | n/a | n/a | n/a | glucose intolerant; ↓ insulin secretion | n/a | n/a | n/a | [304, 305] |
| Steroidogenic cells | Steroidogenic cells IGF-1R KO | 2018 Nef (Neirijnck) | human P450SCC/Cre | n/a | n/a | – | ↓ testicular weight | n/a | – | n/a | n/a | [307] |
| Somatotroph | SIGFRKO | 2010 Radovick (Romero) | rGHpCre | ↑ | ↑ | ↓ weight;—length; ↑ L, S | ↓ fat | – | n/a | n/a | n/a | [308] |
| Thyrocyte | Thyrocyte specific IGF-1R KO | 2011 Krohn (Muller) | thyroglobulin/Cre | n/a | – | ↑weight in males; ↓ in females | alteration in perigonadal fat mass | ↑ males | n/a | n/a | n/a | [310] |
“—” indicates no change; n/a indicates not available. “wk” indicates weeks of age, “mo” indicates months of age. “Tg” indicates transgenic mice. “ +/− “ indicates heterozygous; “-/- “ indicates homozygous. ♂ indicates males; ♀ indicates females; K, kidney, L, liver, S, spleen, H, heart.
Table 4.
IGFBP transgenic and KO mouse lines
| Mouse lines | Discovery (Year/Lab/first) | Expression control | Serum GH | Serum IGF1 | Size / weight | Body composition | Insulin sensitivity | Reproductive capacity | Cancer incidence | Lifespan | Original references | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| IGFBP1 | human IGFBP1 transgene | 1995 Dai (D'Ercole) | mouse Mt1 | n/a | n/a | ↓ weight | not consistent | ↓ | ↓ ♀ | n/a | n/a | [312] |
| IGFBP1 KO | 2003 Taub (Leu) | NeoR replacement of promotor and exons 1–2 | n/a | ↑ before 4 mo | – | n/a | – | n/a | ↓ prostate tumor size; ↓ proliferation, but not incidence | n/a | [318] | |
| IGFBP2 | IGFBP2 transgene | 1999 Wolf (Hoeflich) | CMV | – | – | – | ↑ fat | ↑ sensitivity; ↓ serum insulin | n/a | ↓ colorectal tumor growth with induced carcinogenesis | n/a | [320] |
| IGFBP2 KO | 2000 Pintar (Wood) | NeoR replacement of exon 3 | – | – |
–weight;↑L; ↓S,H,K |
n/a | – | – | n/a | n/a | [323] | |
| IGFBP3 | human IGFBP3 transgene | 1995 Molnar (Murphy) | mouse Mt1 | n/a | n/a | ↑ S,L,H | ↑ fat | n/a | – | n/a | n/a | [325] |
| IGFBP3 KO | 2006 Pintar (Ning) | NeoR replacement of exon1-3 | n/a | n/a | ↑ weight until 22 wk; – afterward | – | –; impaired glucose homeostasis on HFD | n/a | ↑ lung cancer tumorigenesis | n/a | [328] | |
| IGFBP4 | IGFBP4 transgene | 1998 Fagin (Wang) | murine cDNA driven by α-actin | n/a | n/a | ↓ thymus | n/a | n/a | n/a | n/a | n/a | [331] |
| IGFBP4 KO | 2006 Pintar (Ning) | NeoR replacement of exon1 | n/a | – | ↓ | ↓ fat, femur length | n/a | – | n/a | n/a | [328] | |
| IGFBP5 | IGFBP5 transgene | 2002 Flint (Tonner) | β-lactoglobulin | n/a | ↑ | ↓ weight | ↓ lean | n/a | ↓ fertility ♀ | n/a | ↑ neonatal mortality | [335] |
| IGFBP5 KO | 2006 Pintar (Ning) | NeoR replacement of exon1 | n/a | – | – | ↑ fat | mild glucose intolerance | – | n/a | n/a | [328] | |
| IGFBP6 | IGFBP6 transgene | 2004 Babajko (Bienvenu) | glial fibrillary acidic protein promoter/enhancer | n/a | ↓ 15 d; – 1, 3 mo | ↓ weight; growth retardation up to 3 mo | n/a | mild insulin resistance w/ diet-induced obesity | ↓ | n/a | n/a | [340] |
“—” indicates no changes; n/a indicates not available; “d” indicates days of age, “wk” indicates weeks of age, “mo” indicates months of age; ♀ indicates female. K, kidney, L, liver, S, spleen, H, heart
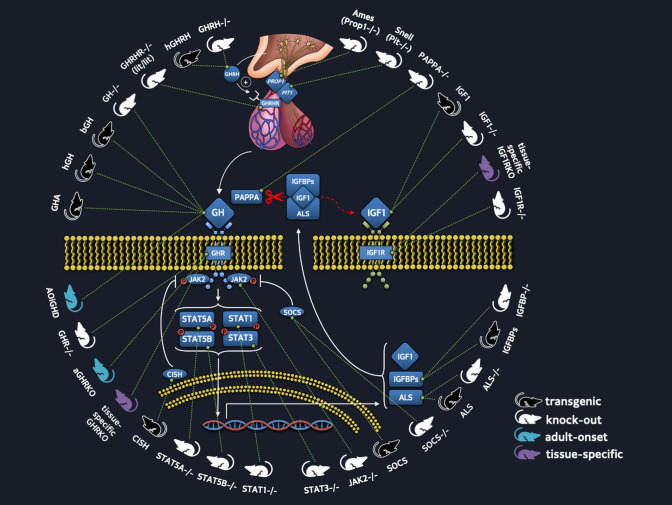
Fig. 1.
Summary of transgenic and knockout mouse lines with altered GH/IGF action. The diagram shows proteins involved in the regulation of GH secretion, GH induced intracellular signaling, and the production of IGF1, ALS, IGFBP3. The different mouse colors represent mice with a transgene overexpressed (black), mice with genes that have been knocked out globally (white), adult-onset knockouts (blue) or tissue-specific knockouts (purple) (Color figurre online)
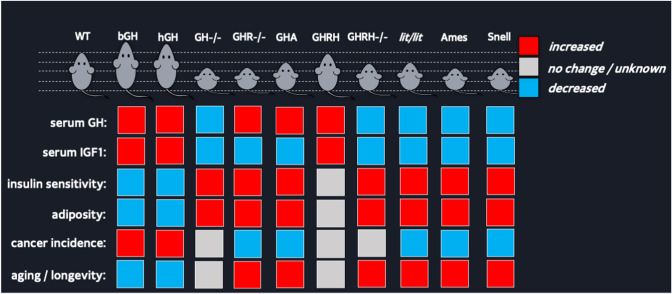
Fig. 2.
Phenotypic comparison among several transgenic and knockout mouse lines with altered GH action. The mice compared are depicted at the top of the figure along with their relative size and the altered genes. The red box indicates increased growth, the blue box indicates decreased growth, and the grey box indicates no change in growth relative to WT controls (Color figure online)
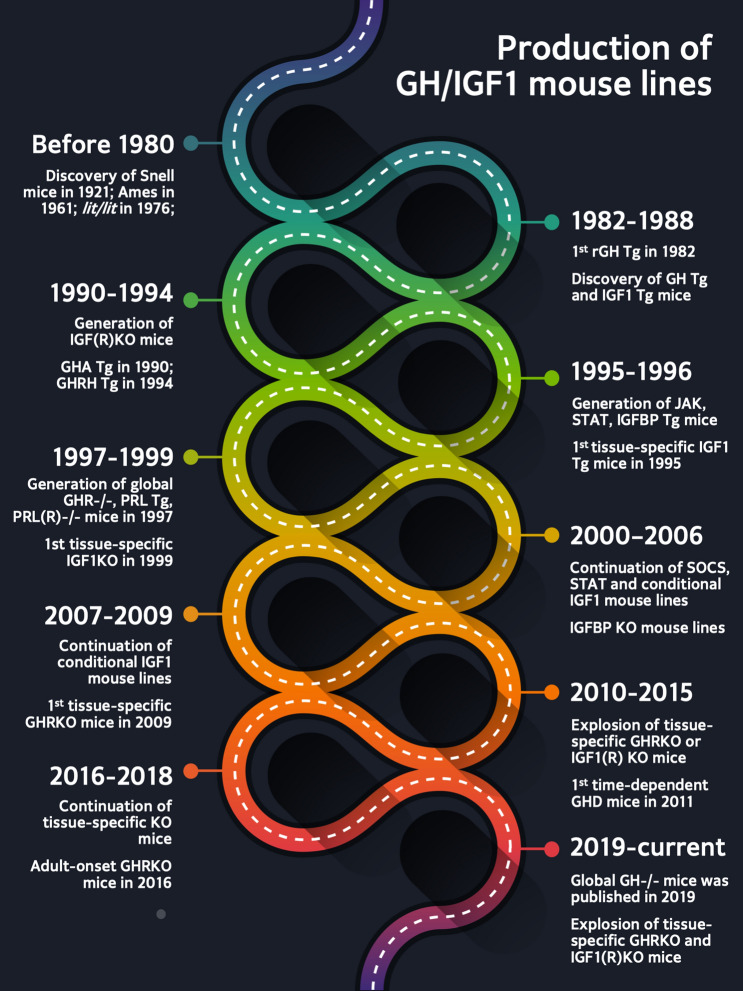
Fig. 3.
Production timeline of GH/IGF1 mouse lines. Before 1980, only three mouse lines related to the GH/IGF family had been discovered (Snell, Ames and lit/lit mice). Through transgenic and ‘knock-out’ technology, generation of different mouse lines with altered GH/IGF family signaling increased. The 1990s were mainly dedicated to transgenic and global knockouts associated with altered GHR, IGF1, PRL and JAK/STAT. The first tissue-specific mouse line was generated in 1999, foretelling more than two decades of additional conditional GHR and IGF1 knockouts. To date, there are 137 mouse lines dedicated to exploring the effects of the GH/IGF family; all these mouse lines contribute to a deeper understanding of the impact of GH and IGF1 on health and disease. Tg transgenic mouse lines, KO knockout mouse lines
Mouse lines upstream of GHR
Circulating GH is produced by the acidophilic somatotroph cells of the anterior pituitary gland. The transcription factors Prophet of Pit-1 (PROP1, gene product of Prop1), pituitary-specific transcription factor 1 (PIT1; gene product of Pou1f1), and GH releasing hormone receptor (Ghrhr) are sequentially expressed in the developing pituitary and are together responsible for the regulation of GH production. PROP1 is critical for both the development of anterior pituitary cell types (somatotrophs, gonadotrophs, lactotrophs, thyrotropes) and in inducing PIT1 expression. PIT1 regulates further differentiation of the pituitary cell lineages, as well as the expression of the Ghrhr gene, which in turn, promotes the clonal expansion of these cells [66]. Pituitary GH production is positively regulated by hypothalamic GHRH and gastric ghrelin, and negatively by hypothalamic somatostatin (SRIF) and endocrine IGF1. Each of these proteins binds to its cognate receptors − GHRH receptor (GHRHR), ghrelin receptor / GH secretagogue receptor (GHS-R), SRIF receptor subtypes, and IGF1R, in order to elicit their actions [66]. Mouse lines discovered or engineered to focus on each of these regulatory nodes of GH production have allowed us to understand developmental regulation and downstream physiological effects in a new light. Each will be discussed briefly below and is summarized in Table 1.
Snell (Pit1-/-) and Ames (Prop1-/-)
The earliest mouse lines discovered to have a somatotrophic deficiency in GH production resulting in distinctive phenotypes were Snell and Ames mice. These mice have been at the center of hundreds of published research reports since their discovery and are essential in the current understanding of the action of GH.
Snell dwarf mice (dw/dw; Pit1-/-; Pou1f1-/-)
Origin In 1921, George Snell (Nobel Prize, 1980) observed a new recessive Mendelian genotype of dwarfism in his mouse stock [67]. This Snell dwarf mouse (also termed dw/dw or Pit1-/- or Pou1f1-/-) represented the second case of hereditary dwarfism in rodents, following a previous report in guinea pigs [67].
Phenotype Snell reported the length of this mature dwarf mouse reaches that of a 16–17-day-old ‘normal’ mouse. The Snell dwarf mice also weigh only one fourth of their wild-type counterparts [67]. Snell mice have since been part of numerous studies worldwide and are characterized by pituitary hypoplasia, with a combined deficiency of GH, PRL, and thyroid-stimulating hormone (TSH), which later was found to be due to a spontaneous point mutation in the Pou1f1 gene [68]. This mutation abrogates the interaction of the PIT1, a POU family transcription factor, with its target transcriptional regulatory sequence. This, in turn, leads to improper formation and dysfunction of the pituitary somatotrophs, lactotrophs, and thyrotrophs [68], as well as nearly undetectable levels of serum IGF1 [69]. The severely suppressed growth of Snell dwarf mice [70] is partially restored following thyroxine and GH replacement therapy [71].
A ‘diabetogenic’ effect of GH has been known since 1930s [72]. Subsequent studies have revealed that GH induces insulin resistance primarily in peripheral tissues [73] via (i) elevated free fatty acid (FFA) from increased lipolysis leading to increase of diacylglycerol and ceramides and suppression of IRS1 activation in liver and skeletal muscle; (ii) elevated FFA induced increase of acetyl-CoA, leading to increased gluconeogenesis in liver and kidney; (iii) upregulation of PI3K regulatory p85a subunit in mouse white adipose tissues (AT); and (iv) upregulated SOCS expression [74]. Insulin resistance is an important metabolic hallmark in patients with acromegaly [75] while congenital GH insensitivity in Laron Syndrome (LS) individuals is associated with improved insulin sensitivity [76]. In agreement, GH-deficient Snell mice display a low utilization of circulating glucose, reduced serum insulin levels, and increased insulin sensitivity, as well as decreased free radical-induced damage (lower protein carbonyl content) [77].
Snell mice show an increase in lifespan compared to WT mice, with a 50% and 29% increase in males and females, respectively [52, 70] and are protected from a number of age-related pathophysiologies, including neurological decline [78], collagen denaturation [70], cataract development, glomerular damage and cancer [79]. However, these dwarf mice have defects in hearing, musculature, immunity, and reproduction. At three months of age, Snell mice have more muscle mass as compared to WT but also a compromised muscle quality and poor fatigue recovery [80]. Defects in reproductive capacity of Snell mice include sterility and delayed testicular growth [81]. Hormone replacement (GH + thyroxine + TSH) restores fertility in male mice but not in females [71]. Congenital deafness due to a lack of TSH is partially rescued in these mice by thyroid hormone treatment [82]. Overall, results from the Snell mouse were the first to strongly implicate GH in lifespan determination.
Ames mice (Prop1-/-)
Origin The Ames mouse was first reported in 1961 by Schaible and Gowen [83]. These mice have a spontaneous recessive mutation in the Prop1 gene, necessary for expression of PIT1, which results in the lack of somatotrophs, lactotrophs, and thyrotrophs similar to that seen in Snell mice.
Phenotype Ames mice have a severe lack of GH, PRL, and TSH, and very low circulating IGF1 [84]. Ames mice are small with a low body weight [85] and are one-third the body size of WT mice. Although Ames mice have increased adiposity, they exhibit lower circulating blood glucose and enhanced insulin sensitivity due to the lack of GH’s diabetogenic effect [86, 87]. These dwarf mice are protected from HFD-induced insulin resistance unlike age-matched WT mice [88]. Additional distinct physiological characteristics of Ames mice include significantly higher brown AT [89], lower resting core body temperature [90], and a reduced senescent cell burden in white AT [91]. Similar to Snell mice, Ames mice are also markedly resistant to standard oxidative stress inducers like paraquat and diaquat, even at older ages [92]. Moreover, Ames dwarf mice have a significantly lower incidence of fatal neoplasms, including lung adenocarcinoma [93]. Similar to Snell mice, Ames mice also exhibit a markedly increased lifespan, with males living an average of ~ 50% longer and females living > 60% longer than their WT littermates [94, 95]. Interestingly, a further extension of lifespan is observed in these mice when subjected to caloric restriction (CR), which indicates that the anti-aging effects exhibited via CR and the Prop1 gene mutation occur through independent mechanisms [96, 97]. Despite these positive attributes, Ames mice suffer from a number of reproductive deficiencies. They are hypogonadal with decreased levels of gonadotropin and testosterone [98]. Although some male Ames mice remain fertile, all females are sterile. Further, Ames mice suffer from auditory deficits but, unlike in the Snell mice, are almost completely rescued by early life thyroid hormone treatment [82].
Overall, Ames and Snell mice present similar deficiencies in three pituitary hormones (GH, PRL, TSH), resulting in similar phenotypes of extended lifespan, improved oxidative stress response, improved insulin sensitivity, and reduced incidence of cancer. Numerous studies surrounding them have deepened our understanding of the endocrine control of specific aspects of health, disease, and lifespan.
Growth hormone releasing hormone (GHRH) and its receptor (GHRHR)
GHRH and its cognate receptor, GHRH receptor (GHRHR), promote GH release primarily along the hypothalamus-pituitary axis. Human patients with isolated GH deficiency (IGHD) are often found to have inactivating mutations in the GHRHR or GHRHR gene locus. Therefore, a better understanding of this ligand-receptor pair in modulating the physiological effects of GH has clinical relevance. Below we discuss three mouse lines associated with the GHRH-GHRHR pair.
Human GHRH transgenic mice
Origin In order to study the effects of GHRH in modulating the GH/IGF axis, Hyde and colleagues developed a human GHRH transgenic mouse (hGHRH) in 1994 using the Mt1 gene promoter/enhancer to drive expression of the hGHRH gene [99].
Phenotype GHRH transgenic mice have increased serum concentrations of mouse GH, PRL and IGF1 and are significantly larger in body size than WT mice [99]. As such, they are a mouse model of pituitary associated acromegaly. Also, endogenous hypothalamic GHRH levels are significantly suppressed, while levels of somatostatin (SST or SRIF) and SST receptor subtypes are elevated compared to littermate controls [100]. The upregulation of GHRH action in mice leads to massive hyperplasia of mammosomatotrophs observable at 8-months of age [101]. In adulthood (16–24-months age), pituitary adenomas immunoreactive for GH and PRL are often observed [102–104]. Transgenic GHRH mice were employed to study the effect of GH in regulating the production of neuropeptides from the anterior pituitary. In the anterior pituitary of GHRH transgenic mice, the tachykinins (substance-P and neurokinin A) are markedly increased in males and females [105]. Tachykinins, found in nearly all vertebrates, are one of the largest family of neuropeptides involved in neuronal excitation, behavioral response, vasodilation, and regulation of smooth muscle contraction. Also, the hyperpolarizing neuropeptide galanin, found in human central nervous system (CNS) and gut, are known to be produced by pituitary cells following GH stimulation in vitro [106]. In the pituitaries of male GHRH transgenic mice, galanin mRNA and peptide contents are also highly upregulated [99]. However, the levels of the neuropeptide, vasoactive intestinal polypeptide, in the anterior pituitary of male hGHRH transgenic mice are half of that of nontransgenic animals [107]. No data on the lifespan of these hGHRH transgenic mice are available.
GHRHKO mice (Ghrh-/- or GHRH-/-)
Origin The GHRHKO mouse (Ghrh-/- or GHRH-/-) was generated as a new mouse line of congenital GH deficiency in 2004 by Alba and Salvatori. Amino acid residues 1–42 of the Ghrh gene were replaced by a neomycin resistance (NeoR) gene [108].
Phenotype GHRH-/- (Ghrh-/-) mutant mice exhibit highly reduced levels of pituitary Gh mRNA and protein and reduced liver Igf1 mRNA and serum IGF1 [108]. Growth retardation in the null animals is first detected at 3 weeks of age, and null mice are 60% the body size of either Ghrh +/+ or Ghrh +/− littermates by 12-weeks [108]. GHRH analogs, acting as agonists of the cognate receptor, improve body length and body weight [109]. GHRH-/- mice have increased intra-abdominal and subcutaneous fat depots, concomitant with an increase in food intake [110, 111]. Increased body temperature, intrascapular brown AT, and thermogenesis is observed in GHRH-/- mice, which could be a function of the increased metabolic rate of a smaller sized mouse to maintain body-temperature [111, 112]. Adiponectin levels are suppressed in both intra-abdominal and subcutaneous white AT depots, while it is elevated in the serum of these animals [110]. Despite an increased adiposity, insulin sensitivity is markedly improved in null mice and is found to be associated with decreased TOR signaling in white AT [113]. These GHRH-/- dwarf mice are long-lived with median lifespan increased in males and females by 50% and 43%, respectively. Maximal lifespan is increased by 18% in males and 33% in females [113]. CR also significantly increases overall survival along with both relative and maximal lifespan, indicating an additive effect especially in females [113]. Microarray analysis reveals several differentially regulated genes in the liver of GHRH-/- mice compared to WT littermates, wherein expression of multiple xenobiotic detoxification genes are dramatically increased [113]. The reported reproductive deficiencies in these null mice include suppressed rates of apoptosis and lipid peroxidation in testes of adult GHRH-/- mice compared to controls [114]. As another model of congenital GH/IGF1 deficiency, GHRH-/- mice share multiple phenotypes of the Snell and Ames mice.
GHRHRKO mice (little; lit/lit; Ghrhr-/- or GHRHR-/-)
Origin In 1976, Beamer and Eicher first reported the ‘little’ (or lit/lit) mouse, a new dwarf mouse deficient in GH and PRL due to a homozygous missense mutation in the Ghrhr gene [115, 116].
Phenotype This dwarf had very low levels of GH and, consequently, IGF1 [117]. The serum GH levels in these mice are only 1% of those of WT controls [118], and serum IGF1 and IGFBP3 are also highly reduced [118, 119], while IGFBP1, 2, and 4 remain unaffected [119]. Low serum leptin [70] and reduced PRL levels are observed in the lit/lit mice [115]. The body weight of these mice is about 2/3rd that of WT mice [70, 118], along with reduced levels of body fluid, protein and minerals. The lit/lit mice exhibit abnormally larger AT, especially in males [119]. Numerous results show that the growth of several cancers, including sarcoma and prostate tumor implants [120, 121], is reduced in this GH deficient mouse. Also, MCF7 breast cancer xenograft growth is reduced by almost half in lit/lit mice compared to WT controls [122]. The femoral lengths, periosteal circumference, and bone mineral density (BMD) are reduced in the lit/lit mice [123], and these mice have an extension in lifespan by 23% in males and 25% in females[70]. Thus, both GHRH-/- and GHRHR-/- mice have significantly suppressed GH/IGF action, resulting in considerably smaller body size, increased adiposity, reduced cancer growth, and extended lifespan.
Growth Hormone (GH)
The clinical relevance of GH treatment for GH deficient children and adults, as well as the extended lifespans of both Ames and Snell mice, fueled interest in the study of GH action in genetically altered mice. The first GH transgenic mouse with the rat Gh gene expressed under the mouse Mt1 promoter/enhancer developed by Palmiter et al. in 1982, grew almost twice as large as the littermate controls [25] and opened up a transformative scope of studying human conditions in laboratory mice. Beginning there, several mouse lines, transgenic for both human (h) and mouse (m) GH have been produced, which partially recapitulated several features of the human condition of GH excess found in patients with acromegaly.
Human GH transgenic mice (hGH)
MT1-hGH transgenic mice
Origin The first hGH transgenic mouse was generated by Palmiter and Brinster in 1983 using the Mt1 promoter/enhancer driving hGH expression [124].
Phenotype Zn or Cd treatment of the MT1-bGH mice further increase the Mt1-promoter/enhancer activity by up to tenfold [124]. The serum hGH levels in these mice are reported to be as high as 3000–900,000 ng/mL [125]. Expectedly, serum IGF1 levels in hGH mice are also significantly higher than those of WT mice [124]. In addition, the serum PRL level is reduced [126] while hypothalamic somatostatin expression is twofold higher than normal [127]. These MT1-hGH transgenic mice are larger in body size the wild type (WT) littermates [128] with markedly increased body weight and greater muscle mass with more and larger type-1 and type-2 fibers [129]. However, hGH transgenic mice suffer from reproductive defects, including a dramatic decrease in ability of males to impregnate females possibly due to the lactogenic effects of ectopically expressed hGH. This occurs despite enlarged testes and seminal vesicles [130]. Likewise, female mice are sterile, possibly due to a dysregulated PRL axis. Daily progesterone injections as well as PRL-secreting ectopic pituitary transplants from WT female mice reverses this reproductive defect [126]. Other abnormalities include severe kidney lesions, glomerular hypertrophy with sclerosis, and hyalinosis associated with tubule-interstitial changes [125]. Transgenic female mice also have a markedly higher incidence of malignant mammary tumors at 27–43 weeks of age [131]. No reports on the lifespan of hGH mice are available.
MT1-hGH transgenic mice
Origin A second transgenic mouse line expressing hGH under the mouse Mt1 promoter/enhancer was produced in 1991 by Tornell and Isaksson [132].
Phenotype These transgenic mice resemble those produced by Palmiter and Brinster with larger body size and higher levels of circulating hGH than WT mice. Female transgenic mice also have markedly higher levels of spontaneous mammary carcinomas similar to those described above [131, 132]. This high frequency of spontaneous mammary carcinomas is probably due to hGH-mediated activation of the mouse PRLRs rather than GHRs [133, 134]. This finding is later clarified by the same group via generation of bovine (b) GH transgenic mice in the same genetic background as the hGH mice, which did not exhibit spontaneous mammary carcinomas, as only hGH binds and activates both the GHR and PRLR [133].
171hGH/CS mice
Origin Cattini and colleagues in 2009 generated a third hGH transgenic mouse line named 171hGH/CS [135] to analyze the pituitary regulation of human GH production. These 171hGH/CS-TG mice contain a 171-kb DNA fragment containing the intact hGH / chorionic somatomammotropin (GH/CS) gene locus, along with the locus control region (LCR) from chromosome-17, including sequences required for pituitary specific expression [135].
Phenotype Both pituitary and placental expression of hCS-A, hCS-B, and placental hGH-variant are detected in these transgenic mice during gestation, in proportions comparable to that in the human placenta, along with high hGH levels [135, 136]. Corticosteroid treatments increase both human and mouse GH levels as well as the Ghrhr mRNA in primary pituitary cells from 171hGH/CS-TG mice [136]. Studies using these mice reveal that hGH production is impacted by the circadian rhythm via direct binding of circadian transcription factors at an enhancer motif in the hGH promoter locus. GH production is suppressed in these mice by acute sleep deprivation [137] and by HFD feeding only during the light (inactive) stage of daily cycle [138].
Wap-hGH mice
Origin In a fourth transgenic mouse line expressing human GH generated by Gunzburg et al. in 1991, the mammary specific whey acidic protein (Wap) promoter/enhancer was used to drive ectopic expression of hGH in mouse milk [139]. Another attempt at producing hGH in the milk of transgenic mice driven by a 6.3 kb long 5’-flanking region of the rabbit WAP promoter/enhancer was undertaken in 1994 by Houdebine and colleagues [140]. These models highlight the important lactogenic effect of human GH, given its unique ability to bind to and activate both GH and PRL receptors [141].
Phenotype Male transgenic mice from Gunzburg have higher plasma LDL-cholesterol and lipid peroxides and increased heart weights and lipid accumulation in liver compared to WT counterparts [142], suggesting a potential cardiac risk for male mice chronically exposed to hGH via the mammary gland. The body size of these mice does not differ from controls. The second Wap-hGH mouse from Houdebine produce up to 22 mg/ml of hGH in the milk but the lactogenic activity of hGH induces multiple dysfunctions including sterility in some of the transgenic females [140]. The same group generated another mouse line using the same transcriptional regulatory system, which express up to 16 mg/mL of bGH in the milk [143].
MT1-GHv mice
Origin A fifth transgenic mouse line, expressing the human placental GH-variant (GH2, or GH-V; GHv) under the mouse Mt1 promoter/enhancer was created by Selden and colleagues in 1988 [128].
Phenotype These mice, similar to the MT1-hGH animals, have a larger body size than normal with elevated IGF1 levels and present a range of reproductive defects including small litter size (significantly lower than the expected at 50%), reduced fetal growth, increased pre- and post-natal mortality, as well as a 20% infertility rate in females[144]. Male MT1-hGHv mice are unable to impregnate the females in most cases and have increased testes and seminal vesicle weights like the MT1-hGH mice [130]. In both the MT1-hGH and MT1-GHv mice, spermatogenesis is unaffected [130].
Bovine GH transgenic mice (MT1-bGH and PEPCK-bGH)
MT1-bGH mice
Origin The first mouse overexpressing bGH (bGH) driven by the Mt1 promoter/enhancer was generated by Hammer et al. in 1985 [145].
Phenotype In the MT1-bGH mice, bGH concentrations are 40- to 400-fold those of WT mice (m) GH, and the transgene is expressed in almost all tissues [145, 146]. Serum IGF1 as well as somatostatin levels are markedly upregulated [127, 147]. The MT1-bGH mice weigh significantly more than controls with increased organ weights and higher lean mass and reduced fat mass [146, 148, 149]. They have larger body size and also model the human condition of acromegaly. These transgenic mice exhibit dysregulated insulin sensitivity as they are hyperinsulinemic at young ages but hypoinsulinemic and hypoglycemic at older ages [150]. Interestingly, both male and female mice also have increased adiposity in early life but switch to a leaner than normal phenotype at four (males) to six (females) months of age [146]. While GH is known to increase gluconeogenesis, MT1-bGH mice surprisingly exhibit suppressed glucose production following a pyruvate challenge, which could be confounded by higher insulin levels [151]. On HFDs, they are resistant to diet-induced obesity but develop dyslipidemia and diabetes [152]. Further, a dysregulated adipokine profile with decreased adiponectin and increased inflammatory IL-6, TNFα, and increased serum cholesterol have been reported [153, 154].
PEPCK-bGH mice
Origin McGrane et al. developed a second bGH mouse line in 1988, employing the phosphoenolpyruvate carboxykinase (PEPCK; Pck1) transcriptional regulatory region ligated upstream to the bGH gene [155].
Phenotype PEPCK-bGH mice have serum bGH levels higher than that of MT1-bGH mice [127], reaching up to 2300 ng/mL, and cAMP administration causes a further twofold increase in bGH levels. As the bGH transgene in these mice is driven by the PEPCK promoter/enhancer, interventions such as a high carbohydrate diet that can suppress PEPCK mRNA, might also suppress the bGH transgene expression. Accordingly, a carbohydrate-rich diet intake by these mice does result in suppressed gluconeogenesis and hence PEPCK expression and in turn, suppresses GH expression by 90%, while increasing serum insulin levels. The PEPCK-bGH animals have a twofold higher growth rate despite the transgene being expressed in the liver and kidney, indicating an endocrine effect of the bGH transgene. The pituitary weight of PEPCK-bGH mice is elevated, with smaller Golgi in pituitary somatotrophs. Serum IGF1 concentrations of these transgenic mice range between 2–threefold higher than those of WT mice [156] along with upregulated somatostatin levels similar to the MT1-bGH transgenic animals [127]. PEPCK-bGH mice weigh approximately 1.5-times more than WT mice [156], with increased lean mass[146, 148] and increased weights of internal organs, including kidney, liver, and heart [149]. In addition, seven-month-old PEPCK-bGH mice display improved glucose clearance, and lower blood glucose and HbA1c levels, while glucose and insulin sensitivities are comparable to WT mice [156]. These mice also develop inflammatory arthritis with production of autoantibodies [157].
Similar to hGH transgenic mice, a range of reproductive disorders are observed in the females of both MT1- and PEPCK-bGH mice, including an increased interval between pairing with a male and conception, increased interval between litters, reduced number of litters, reduced fetal growth, increased pre- and postnatal mortality and alterations in sex ratio [144]. More than 60% of the PEPCK-bGH and 20% of the MT1-bGH female mice are infertile, concomitant with the higher level of circulating bGH in PEPCK compared to MT1 animals [144]. Male bGH transgenic mice (both MT1 and PEPCK) have significantly higher weight of the testes and seminal vesicles but spermatogenesis or fertility is unaffected [130].
There is a significant decrease in the lifespan of both MT1- and PEPCK- driven bGH transgenic, giant mice. MT1-bGH mice have a maximal lifespan of 24-months and a 1-year survival-rate of 44%, while PEPCK-bGH mice have a maximal lifespan of only 18-months and a 1-year survival-rate of 25% [158, 159]. The early morbidity of bGH mice recapitulates several factors underlying the shortened lifespan observed in untreated human patients with acromegaly. MT1-bGH mice suffer from renal and cardiac defects [160]. These mice exhibit renal disorders like hypercellular glomeruli early in life, advancing to increased glomerular size and progressive glomerulosclerosis at adulthood [160]. A significantly increased heart mass concomitant with impaired systolic function and a decreased energy reserve in the myocardium is also observed [149]. An increased mitogenic action of excess GH on its main target organ – the liver – leads to a number of hepatic abnormalities in both bGH transgenic mouse lines. For example, in both MT1- and PEPCK-bGH mice, hepatomegaly is observed as early as 2 weeks of age and progresses maximally into young adulthood, with an enhanced expression of proto-oncogenes and activation of multiple mitogenic signaling intermediates like c-SRC, mTOR, STAT3, GSK3, NFkB, c-fos, c-jun, and c-myc [153, 154, 161]. Additionally, pro-tumorigenic hepatocellular events, including upregulation of tumorigenic galectin-1 [162], and elevated oncogenic signaling pathways, are observed in the livers of both male and female PEPCK-bGH mice [163]. Both MT1- and PEPCK-bGH mice are known to develop spontaneous liver tumors [164]. In both mouse lines, a sustained hepatic hypertrophy and inflammation lead to a significantly higher rate of spontaneous hepatocellular carcinogenesis compared to WT controls [164–166].
In summary, human and bovine GH transgenic mice have a decreased fat mass, with increased body size and lean mass. However, these mice have fertility defects, exhibit kidney and cardiovascular dysfunction, and have elevated neoplasm incidence along with a decreased lifespan.
GH-/- mice (Gh-/- or GHKO)
Origin In order to investigate the effects of GH absence and GH replacement on phenotypic variables, GH-/- mice were generated in the Kopchick laboratory in 2019, using a VelociGene KOMP definitive null allele that replaces the Gh gene with a ZEN-UB1 selectable reporter [167].
Phenotype Circulating GH in GH-/- mice is reduced to an undetectable level compared with that of WT controls [167]. Serum IGF1 levels are also significantly reduced (~ 90%). Disruption of the Gh gene significantly reduces nasal-anal body length (> 30%), and body composition is significantly altered in both sexes, with body weight and lean mass significantly decreased and fat mass significantly increased relative to controls. GH-/- mice of both sexes demonstrate greatly enhanced insulin sensitivity probably due to the lack of GH’s diabetogenic effect. However, GH-/- mice are significantly glucose intolerant (although greater in males than females), which is attributed to their decreased pancreatic islet size. Liver, kidney, heart, spleen, gastrocnemius, soleus, and quadriceps masses are also significantly decreased, whereas AT mass and relative brain weight are significantly increased. Liver triglyceride content and adipocyte size in the subcutaneous depot are elevated in both male and female GH-/- mice. White AT fibrosis is significantly decreased in the subcutaneous white AT depot of both sexes compared to controls, suggesting depot-specific effects of GH. In summary, GH-/- mice show similar phenotypes as other mouse lines that lack GH action, although their cancer incidence and lifespan have not been reported at the time of this publication.
Adult onset-isolated GH deficiency mice (AOiGHD)
Origin To better understand the metabolic effects of somatopause – the progressive decline of hormones in the hypothalamic-pituitary-somatotrophic axis with age – a mouse line of adult onset-isolated GH deficiency (AOiGHD) was created by Kineman and colleagues in 2011 by breeding the inducible monkey diphtheria toxin receptor mice (iDTR) with mice having a rat Gh promoter/enhancer driven Cre recombinase [168].
Phenotype The adult Cre+/−iDTR+/− offspring are treated with diptheria toxin (DT) to selectively ablate somatotroph cells expressing diphtheria toxin receptor at 10–12 weeks of age, resulting in a ~ 50% decrease in circulating GH and IGF1 levels [168]. These mice also have lower fasting insulin levels and improved whole-body insulin sensitivity when fed either low-fat or HFD relative to WT littermates. Indirect calorimetry suggested that these mice utilize mainly carbohydrates for energy metabolism. Furthermore, detrimental physiological effects are seen only in HFD animals including increased fat mass, decreased hepatic lipids, and impaired glucose clearance and insulin output. The AOiGHD mice also have decreased liver weight accompanied with reduced liver triglyceride content. Overall, the mouse line shows that reduction in circulating GH and IGF1 levels with age improves insulin sensitivity and prevents metabolic dysfunction under moderated caloric intake.
GHR antagonist transgenic mice (GHA)
Origin To understand some of the effects of pharmacological perturbations to GH action, a transgenic mouse line that expresses a mutated-bovine GH gene that effectively antagonizes endogenous GH action was created. These transgenic GHR antagonist (GHA) mice were generated in a C57BL/6 J background in the Kopchick laboratory between 1990 and 1991 via the fusion of the mutated GH transgene downstream of the mouse Mt1 promoter/enhancer [26, 169, 170]. The mutated bovine GH gene differs from its WT counterpart in that it encodes a single amino acid substitution at position 119. The glycine that typically occupies this position, found in the third alpha-helix of bGH (G119 in bovine GH; G120 in human GH), is critical for the successful activation of the GHR [171]. When glycine 119 is substituted with arginine, the resulting molecule competitively inhibits the association of mouse GH with the GHR [172]. Similarly, when a lysine is substituted for the glycine at position 120 of the human GH gene, an effective human GHR antagonist is produced. Following these discoveries, Kopchick and colleagues went on to develop the novel drug, SOMAVERT® (Pegvisomant for injection), which is a GHR antagonist that inhibits the interaction of endogenous GH with GHR and is now used world-wide for the treatment of patients with acromegaly [173].
Phenotype As a result of the overexpression of the GHR antagonist, GHA mice have smaller body size and show a 30% lower mean growth ratio [26, 170] with significantly reduced body weight [147], wherein lean mass is reduced, and body fat is increased compared to controls [174]. GHA mice have increased lipid storage in the inguinal subcutaneous white AT depot and a relative increase in extra-peritoneal to intra-peritoneal white AT [174]. Additionally, GHA mice have markedly lower serum IGF1 [26, 147, 170, 172], lower serum IGFBP3 [37], and higher pituitary mouse (m) GH levels than nontransgenic littermates [26, 170]. The pituitary weight of GHA mice is about half that of controls, with moderate to sparsely granulated somatotrophs compared to those densely granulated in WT mice [147]. Despite increased obesity [175], GHA mice are more insulin sensitive than controls [174]. Increased brown AT mass accompanied by higher expression of thermogenic factors has also been reported [175]. On a HFD, although GHA mice gain more weight than WT controls (males > females), they are protected from HFD-induced glucose intolerance and hyperinsulinemia [176]. Additionally, GHA mice are protected from streptozotocin-induced diabetic kidney lesions [175] and from cancer. For example, after treatment with the mammary carcinogen DMBA, ~ 66% of GHA mice remain tumor-free compared to only 1/3rd of the controls and have less tumors and a smaller tumor burden [172]. Although no significant difference in lifespan between GHA and WT mice has been reported [158], female GHA mice tend to live longer than controls.
In summary, the phenotypes observed in GH transgenic mice with elevated GH action contrast significantly to those seen in the GH-/- or GHA mice. All these observations strongly suggest that GH plays a critical role in promoting growth, body size, lean mass, glucose intolerance, and reproductive deficiency, while the absence or deficiency of GH improves glucose homeostasis, adiposity, cancer resistance, and longevity.
Prolactin and prolactin receptor (PRL and PRLR)
Prolactin (PRL) is a protein secreted from the lactotrophs of the anterior pituitary gland [177] and has a structure similar to that of GH. PRL secretion is stimulated by PRL releasing factors such as thyrotropin releasing hormone, oxytocin and neurotensin [178]. On the contrary, PRL secretion is inhibited by dopamine and somatostatin [179] and induced by gamma-aminobutyric acid [180]. PRL binds to PRL receptors (PRLR), which are a member of cytokine receptors that lack intrinsic kinase domains but possess JAK2 associating regions; thus, PRL resembles the GHR and transduces similar intracellular signals. Human PRLR can bind at least three ligands including PRL, placental lactogen and hGH. Like GHR, PRLR consists of an extracellular domain for ligand binding, a helical transmembrane portion and an intracellular region. However, alternative precursor mRNA splicing leads to different isoforms of the PRLR with identical extracellular domains while the intracellular domains differ in size (referred to as ‘long’ or ‘short’ PRLR) [177]. The receptor homodimer is constitutively expressed on cell surfaces in a ligand-independent manner in several tissues and peripheral organs including the breast, prostate, brain, pituitary gland, heart, uterus and skin [178]. Although hundreds of actions of PRL have been reported [181, 182], a main function of PRL is to promote both growth of the mammary gland and to induce and maintain lactation. In the following section, PRL transgenic, PRL-/-, PRLR-/-, PRLR variants, and PRLR antagonist mice will be discussed.
MT1-PRL transgenic mice
Origin To study prostate hyperplasia, Tornell and colleagues in 1997 generated MT1-PRL transgenic mouse lines which overexpressed PRL [183].
Phenotype These transgenic mice have ubiquitous expression of rat PRL (rPrl) under the control of Mt1 promoter/enhancer. Three mouse lines generated, L1, L2 and L3, have an increase in serum rat PRL by ~ 250 ng/ml, 15 ng/ml, and 100 ng/ml respectively [183]. The endogenous mouse PRL serum levels are not reported though mouse PRL mRNA is detected in all parts of the prostate glands. The three PRL transgenic mouse lines exhibit enlarged prostates due to increased PRL secretion along with increased prostate weight and hyperplasia compared to the controls. Interestingly, these mice also have elevated IGF1 levels close to that of bGH mice although the GH levels are not reported. L1 and L2 mice remain fertile, while L3 mice, with the highest PRL levels, are infertile [183].
Local prostate specific prolactin expression: Pb-PRL transgenic mice
Origin To assess the role of PRL in abnormal prostate growth in transgenic animals that overexpress PRL, Kindblom et al. in 2003, developed a Pb-PRL transgenic mouse line, which locally produce PRL in the prostate [184].
Phenotype In this mouse line, the minimal probasin (Pb) promoter/enhancer is used to direct rPrl expression in the epithelial cells of dorsolateral, ventral, and anterior of prostate lobes. Marked enlargement of prostate glands is observed in the transgenic males, which is also observed in the MT1-PRL mice. Though both MT1-PRL and Pb-PRL have marked ductal dilation and elongation, MT1-PRL mice have significantly elevated ductal branching points and tips while Pb-PRL mice have normal branching points. The data suggest that PRL action can differentially impact a variety of prostate cells. The heterozygous Pb-PRL animals remain fertile [184].
Mammary epithelial PRL overexpressing mice (NRL-PRL)
Origin PRL is crucial in development and differentiation of the mammary gland. Many epidemiological studies have linked PRL with increased risk of estrogen receptor positive (ERα +) breast tumors [185]. To specifically study the role of PRL in breast cancer, Schuler and colleagues developed a PRL transgenic mouse line in 2003 [186]. This mouse line called NRL-PRL has locally overexpressed rPrl transgene in mammary epithelia driven by a hormonally nonresponsive promoter/enhancer – neu-related lipocalin (NRL).
Phenotype The NRL-PRL females develop mammary pathology and ERα + and ERα- carcinomas [186, 187]. Overall, breast cancer development in NRL-PRL mice strongly implicates PRL in development of ERα + cancers.
PRL knockout mice (Prl-/- or PRL-/-)
Origin To determine the effects of a lack of PRL, Nelson Horseman et al. generated the PRL-/- mouse line in 1997 through a targeted insertion of a NeoR gene into the region of the PRL gene encoding the second α helix [188].
Phenotype Although no detectable effect on growth or adiposity at any age is observed [188], male PRL-/- mice exhibit impaired glucose tolerance at 4 weeks of age [189]. Also, higher leptin concentrations are found in PRL-/- mice on normal chow compared to WT mice [189]. Females are sterile, indicating that PRL is essential for female fertility, whereas males remain reproductively viable [188]. Since PRL has been found to influence the immune system, it was expected that these mice would be immunocompromised. However, no significant difference is reported in the number of B- and T-cells in PRL-/- mice compared to controls [188].
PRLR knockout mice (Prlr-/- or PRLR-/-)
Origin To determine the effects of a lack of PRL action, Ormandy et al. in the laboratory of Paul Kelly generated a prolactin receptor knockout mouse line (PRLR-/-) in 1997 [190].
Phenotype These mice present features like those noted in PRL-/- mice. Female PRLR-/- mice are sterile and show changes in estrous cyclicity when compared to WT mice [190]. Heterozygous female mice are fertile but display abnormal maternal behavior including decreased pup retrieval, leaving pups unattended or scattering them around the cage [190]. However, males are ‘partially infertile’ [190], with 20% of all tested males exhibiting delayed fertility [181]. Both male and female PRLR-/- mice experience a significant decrease in bone formation and a reduction in bone mineral density compared to controls [191]. In terms of glucose homeostasis, PRLR-/- mice have reduced pancreatic islet density and β-cell mass, as well as reduced pancreatic insulin mRNA levels in both sexes [192]. There is also a marked reduction in abdominal fat mass in both sexes. Importantly, PRLR-/- mice are protected from prostate carcinogenesis [193], suggesting that abrogated PRL action might be protective against prostate cancer.
PRLR variants
PRLR is expressed ubiquitously with various proportions of long and short isoforms in different tissues. In mice, four PRLR variants have been classified as one long (LPRLR) and three short forms (S1PRLR, S2PRLR, S3PRLR). S1PRLR and S2PRLR forms are mouse specific while LPRLR and S3PRLR are homologous in other species [194]. Only LPRLR has been shown to induce transcription of milk producing genes while both LPRLR and S1PRLR have been shown to modulate cell proliferation. Similarly, rat PRLR has a long (LPRLR), a variant (Nb2), and a short (F3-SPRLR) form. Interestingly, F3-SPRLR results in formation of inactive heterodimer resulting in absence of downstream signaling in vitro. Several mouse lines have been generated to study the individual effects of each type of isoforms.
F3-SPRLR mice
Origin To assess the dominant negative effects of SPRLR and role of PRL in normal mammary gland development, Saunier et al. developed a transgenic mouse line in which the F3-short form of the rat PRLR (F3-SPRLR) was expressed in mouse mammary epithelium driven by mouse mammary tumor virus-long terminal repeat (MMTV-LTR) in 2003 [194].
Phenotype Mice with low levels of transgene expression exhibit phenotypes similar to WT animals while mice expressing high levels of transgene show impaired mammary gland development and lactation although fertility is unaffected [194]. Hence, locally blocking PRL/PRLR at the mammary gland hinders mammary gland development indicating the crucial role of PRLR signaling in mammary tumors.
PR-1 mice
Origin To assess the signal transduction of the short PRLR isoform, Binart et al. in the Kelly laboratory developed a mouse line with overexpression of the short isoform of the mouse PRLR (originally called PR-1, also known as S1PRLR) in 2003 [195]. The Pr1 gene is expressed in heterozygous Prlr+/− mice driven by the elongation factor 1α (EF1A) promoter/enhancer.
Phenotype Previous studies have shown that heterozygote Prlr+/− mice exhibit severe defects in lactation after the first pregnancy [190]. Interestingly, introducing the short form of the gene (Pr1) in Prlr+/− mice results in normal mammary ductal development and the ability to lactate after the first pregnancy. The results from this study strongly indicate that the short form of PRLR is specifically involved in mammary stem cell formation.
Tg-RL and CL-RL mice
Origin PRL is involved in corpus luteum (CL) formation and progesterone production crucial in embryo implantation and maintenance of pregnancy. To delineate the role of PRLR long form in CL function, Le et al. in 2012 developed two transgenic mouse lines expressing only PRLR long form—one ubiquitously expressed and named Tg-RL driven by the EF1A promoter/enhancer, and the other in CL-specific manner and named CL-RL driven by the transcriptional regulatory region of the hydroxysteroid 17-beta dehydrogenase 7 (hsd17b7) CL-specific gene [196].
Phenotype Both mouse lines have normal follicular development and ovulation rates. An interesting malformation of vasculature is observed in both mouse lines, which can be attributed to lack of PRLRs (short form) function [196].
PRLR antagonist transgenic mice
Origin The rational design for competitive PRLR antagonist where it competes with endogenous PRL and binds but does not activate the PRLR was based on the pioneering work on the GHR antagonist (Pegvisomant) by the Kopchick laboratory. Goffin and colleagues in 2003 generated the first PRLR antagonist by replacing the glycine in the 3rd PRL α-helix. This glycine, when replaced with arginine at position 129 (G129R), resulted in a strong antagonist of the PRLR [197]. Also, deleting the first nine residues (Δ1–9) at the N-terminus in the G129R-hPRL proved to enhance the effectiveness of the antagonist [197]. To study the effects of blocking the PRL action in prostate tumorigenesis, Rouet et al. in 2010 developed the Δ1–9-G129R–hPRL transgenic mouse line driven by Mt1 promoter/enhancer for ubiquitous expression of the antagonist [198].
Phenotype These mice express about 200 ng/ml of circulating PRL antagonist. No prostate hypertrophy is observed in these mice. However, increased pituitary weight is observed in both sexes [199, 200]. Inhibition of lactotroph cell proliferation and increased apoptosis are also observed when mice are treated with dopamine agonist (D2R) and then treated with PRL [200]. In 2010, the latter team also generated a double transgenic mouse by crossing Pb-PRL (rat Prl expressed only in the prostates) with Δ1–9-G129R-hPRL mice. The weight of dorsal prostate in these mice is reduced at 6-month of age as compared to Pb-PRL mice. These mice also had a stark reduction in STAT5 phosphorylation in dorsal prostates and reduced tumorigenesis. Overall, these findings point to the role of endocrine PRLR antagonists in preventing early prostate tumorigenesis [198].
In summary, PRL is closely related to GH as both belong to the same cytokine family, have approximately the same mass, similar quaternary structures, bind to a homo-dimerized cognate receptor like GHR, and activate STAT5 in their downstream signaling. PRL-PRLR axis plays important physiological roles especially in lactation and in maintaining fertility. Also, blocking PRL can retard/inhibit prostate tumorigenesis.
Global, temporal and tissue-specific GHRKO mice
For GH to elicit a response in cells, it must bind to its cognate receptor, the GHR, which is a pre-formed single membrane spanning dimer and a member of the cytokine family receptors—all lacking a kinase domain. After GH binds to the preformed GHR homodimer, the intracellular domain associated JAK2 kinases then phosphorylate one another and begin the process of GH induced GHR signal transduction. Inactivating mutations in the GHR or down-stream signaling intermediates lead to GH insensitivity. In humans this condition is called Laron Syndrome (LS). LS is characterized by low IGF1, elevated GH, short stature, obesity, and resistance to cancer [76]. Furthermore, the Ecuadorian cohort which is the largest cohort of individuals with LS, exhibit extreme insulin sensitivity and resistance to cancer and diabetes. In mice, GHR gene disruption (GHR-/-) produces a similar phenotype to humans with LS [31]. To date, GHR-/- mice have been used in over 130 published studies that have greatly enhanced our knowledge of GH action in vivo. In addition to global GHR-/- mice, temporal and tissue-specific GHR gene disrupted mice have been generated (Table 2) as will be discussed in the subsequent section.
Global GHRKO
GHR knockout mice (Ghr-/-; GHR-/- or GHRKO)
Origin To determine the effects of a lack of GH action, the GHR null or GHR-/- or GHRKO mouse line was developed by Zhou et al. in the Kopchick laboratory in 1997 through a targeted mutation in which a NeoR gene was used to replace a major portion of exon 4 of the Ghr along with ~ 500 bp of intron 4/5 [30].
Phenotype The resulting homozygous null mice are dwarf with decreased body length and weight. These mice experience delayed sexual maturation and decreased litter sizes [30, 201]. GHR-/- mice have ~ 50–100 fold increase in serum GH and a ~ 90% decrease in serum IGF1 levels [30]. In regard to body composition, these mice have increased fat mass and decreased lean mass [202]. Surprisingly, the largest increase in adiposity occurs in the subcutaneous white AT depot. Although obese, these mice show improved insulin sensitivity and decreased serum insulin [31]. However, GHR -/- mice have impaired glucose tolerance due to decreased pancreatic islet size and function [203, 204]. Additionally, these mice have normal to high levels of serum leptin [148, 205, 206] and adiponectin [148, 205, 207] with normal to low levels of cholesterol [206, 208] and T3 and T4 [209]. GHR-/- mice have increased oxygen consumption and lower respiratory quotient values, which indicate a shift towards fat oxidation [210, 211]. Additionally, these mice show 23–26% greater neuron density in the somatosensory cortex of the brain along with improved memory retention and reduced memory loss with age [212]. Remarkably, these mice display resistance to several disease states, including the development of certain types of cancer [33–35], nephropathy when type 1 diabetes is induced [213], resistance to T2D when placed on a HFD [32] and age-related loss of grip strength [214]. Finally, these mice have increased longevity [37, 215] and hold a world record for the longest-lived laboratory mouse [31].
Temporal GHRKO
Global adult onset—aGHRKO mice
Origin To investigate the physiological effects of disrupting GH action in adulthood, Junnila et al. in the Kopchick laboratory in 2016 generated a mouse line with ablated GHR at 1.5 months of age using the Cre gene transcriptionally driven by ROSA26 gene promotor/enhancer [60].
Phenotype Adult-onset GHRKO (aGHRKO) mice have a variable but significant decrease in tissue specific GHR gene expression, with liver and AT showing the greatest reduction, and skeletal muscle and heart, the least [60]. In terms of phenotype, the aGHRKO mice have reduced circulating IGF1 and elevated circulating GH when compared to control mice. These mice have reduced body weight and body size (5–10%), with an increase in fat mass and a decrease in lean mass when compared to controls. Despite the increased adiposity, both male and female aGHRKO mice show increased insulin sensitivity and decreased circulating insulin levels. Similar to the germline GHR-/- mice, aGHRKO mice have decreased glucose tolerance in comparison to controls. The adipokine profile is altered in these mice with increased adiponectin but no difference in leptin levels. Changes in circulating IGFBPs were also seen in the aGHRKO mice when compared to WT mice. That is, similar to GHR-/- mice, aGHRKO mice exhibit a decrease in IGFBP3 and an increase in IGFBP1, 2 and 6. Therefore, while IGFBP3 is known to be positively associated with GH action, IGFBP1, 2, and 6 appear to be negatively associated with it. Finally, longevity studies show that aGHRKO females have an increased maximal lifespan when compared to female controls.
Tissue-specific GHRKO
Liver-specific GHR knockout mice
Liver is one of the most important organs in the GH/IGF1 axis since it is the site where the majority of circulating GH-stimulated IGF1 is produced. It is estimated that 75–90% of circulating IGF1 is produced from the liver [48, 216]. Indicative of this importance, there are five liver-specific GHR knockout mouse lines that have been independently produced by different laboratories between 2009–2019.
GHRLD
Origin In 2009, Fan et al. produced the first liver-specific GHR knockout (GHRLD) mouse in the laboratory of Mark Sperling [38]. To produce these mice, an albumin promoter/enhancer was used to drive Cre recombinase in liver hepatocytes.
Phenotype These mice have decreased serum IGF1 and elevated serum GH levels [38]. Despite the reduction to circulating IGF1, these mice show no change in body weight, body length, tibia length or body composition. Several organs are altered in size in these mice including increased liver weight and decreased kidney weight. Glucose homeostasis in these mice is negatively affected, as GHRLD mice are glucose intolerant and insulin resistant. Additionally, male mice exhibit increased liver steatosis. Finally, these mice have increased hepatic fibrosis, circulating inflammatory cytokines and decreased bone density.
LiGHRKO
Origin In 2014, List et al. in the Kopchick laboratory produced the second liver-specific GHR knockout mouse (LiGHRKO) [48]. To produce these mice, an albumin promoter/enhancer was used to drive Cre recombinase specifically in the liver hepatocytes.
Phenotype The resulting mice are significantly smaller with decreased body weight and body length at 6 months of age [48]. Analysis of body composition shows a higher percentage of body fat at early ages followed by a lower percentage in adulthood similar to the body composition profile of bGH mice that results from elevated GH levels. In some sense, these animals could be considered mice with ‘extrahepatic acromegaly’. For example, liver IGF1 mRNA is quite low yet the levels are increased in skeletal muscle and AT. Interestingly, there is a male-specific development of fatty liver. Similar to GHRLD, LiGHRKO mice have impaired glucose homeostasis with an increase in several adipokines, including leptin, resistin and adiponectin, and increased inflammatory cytokines (IL-6 and MCP-1). These null mice also have increased grip strength compared to controls. Additionally, LiGHRKO mice have smaller kidneys and spleens and increased liver, heart and lung mass relative to body weight. Aging studies at two separate institutions reveal that liver-specific disruption of the GHR does not alter lifespan in LiGHRKO mice [52] despite severe reductions to circulating IGF1 [48]. We suspect that the benefits of lower circulating IGF1 in LiGHRKO— which normally favors lifespan extension— were offset by impaired glucose homeostasis and elevated circulating GH, that in turn increased local IGF1 in non-hepatic tissues. To date, no other liver-specific mouse lines have been evaluated for lifespan.
aLivGHRkd
Origin To investigate the role of GH in hepatic fat production and accumulation, Cordoba et al. produced an adult-onset (induction at 10–12 weeks of age), liver GHR knockdown mouse (aLivGHRkd) in the laboratory of Rhonda Kineman in 2015 [55]. These mice were generated utilizing a Cre system driven by the thyroxine-binding promoter/enhancer.
Phenotype Both male and female mice have reduced circulating IGF1 and hepatic Igf1 mRNA levels, although the reduction is less pronounced in females [55]. There is also an increase in GH, GHRHR and the ghrelin receptor (previously known as the GH secretagogue receptor 1a) in male mice. These mice have increased liver weight, hepatic de novo lipogenesis, triglycerides, and glycolysis-driving factors, such as glucokinase and fructose 2,6-bisphosphate.
Li-GHRKO
Origin To investigate the role of hepatic GH on lipid and carbohydrate metabolism, Liu et al. produced the liver-specific GHR deletion mouse (Li-GHRKO) in the laboratory of Shoshana Yakar in 2016 [59]. These mice were produced using a Cre system driven by an albumin promoter/enhancer.
Phenotype There is no change in body weight of these mice, but there is an increase in fat mass, as seen before in other mouse lines [59]. Similarly, these mice have reduced serum IGF1 levels with increased blood glucose and serum insulin, as well as impaired insulin tolerance. They also have increased serum triglycerides, cholesterol, FFAs and leptin levels. Furthermore, the liver weight of these mice is increased, as well as hepatic triglyceride and fatty acid content. Finally, hepatic glycogen is increased, as well as enzyme markers for gluconeogenesis (i.e., glucokinase, PCK1).
L-Ghr-/-
Origin To investigate the role of liver-specific GH on CR, Fang et al. produced the L-Ghr-/- mouse using a Cre system driven by an albumin promoter/enhancer in the laboratory of Guosheng Liang in 2019 [63].
Phenotype There is no resulting change in body weight or body composition [63]. Additionally, these mice have blood glucose levels comparable to controls. However, when placed on a CR diet, the mice have decreased blood glucose resulting in a hypoglycemic state. They also have an increase in plasma GH and ghrelin. Differing from previous findings in the other liver-specific GHR KO mouse lines, the livers of these mice have decreased triglycerides and reduction in autophagic vacuoles.
Overall, physiological data obtained from these five liver-specific Ghr gene disrupted mouse lines are in agreement with only a few discrepancies. Most notably, Fan et al. reports that deletion of GHR in liver does not affect body composition or growth as measured by total body weight and body length. In contrast, List et al. found that LiGHRKO mice have a higher percentage of adiposity at a young age, then a lower percentage in adulthood when compared to controls. Furthermore, List et al. found that body weight and body length are all significantly decreased in LiGHRKO mice compared to controls. While the precise reason for the inconsistencies is unknown, we suspect that they may be due to the age at which these measures are recorded, and/or the numbers of mice used in each study. Specifically, Fan et al. evaluated these parameters at 16 weeks of age using a n of 6 to 8, while List et al. measured growth factors at 6 months of age using a n of 15 to 16 and body composition over time up to 22 months of age using a n of 13 to 19. Importantly, List et al. observed no changes in weight until later in life, which may explain why Fan et al. observed no differences in growth.
Muscle-specific GHRKO mice
Since GH has significant anabolic effects on muscle, three muscle-specific GHR knockout mouse lines have been generated independently to understand the roles of the GH-axis on muscle size, fiber type, metabolism, glucose homeostasis and longevity.
ΔGHR
Origin In 2010, Mavelli et al. created a muscle-specific GHR knockout mouse (ΔGHR) in the laboratory of Thomas Clemens [40]. These mice were produced using the Mef-2c-73 k promoter/enhancer to drive Cre expression in muscle. However, off target expression is reported for this Cre line (described below in comparison of the three muscle-specific knockout mouse lines).
Phenotype These mice show no change in either serum GH or IGF1 levels [40]. In terms of phenotype, these mice reveal an increase in body weight over controls, starting at 12 weeks of age. Body composition analysis shows that these mice also have increased fat mass compared to controls. Additionally, ΔGHR mice have increased glucose and triglyceride levels, indicating the development of insulin resistance.
mGHRKO
Origin In 2012, Vijayakumar et al. produced the muscle GHRKO mouse (mGHRKO) in the laboratory of Derek LeRoith [42].
Phenotype These mice were produced utilizing the Cre system driven by the mouse muscle creatine kinase (Ckmm) transcriptional regulatory region [42]. No difference is seen in GH and IGF1 levels. While body size is comparable to WT controls, the lean mass of the mGHRKO mice is significantly decreased. Additionally, both subcutaneous and gonadal AT are significantly reduced along with an increase in serum adiponectin levels.
MuGHRKO
Origin In 2015, to understand the effects of muscle GHR on glucose homeostasis and aging, List et al. in the Kopchick laboratory produced the MuGHRKO mouse [54]. These mice were produced utilizing the Cre system driven by the mouse muscle creatine kinase (Ckmm) promoter/enhancer, which is specifically expressed in skeletal and cardiac muscle.
Phenotype No changes to the GH/IGF1 axis are found [54]. Body length and weight are comparable to controls, and no difference is observed in fat or lean mass as measured over time. Male MuGHRKO mice have enhanced insulin sensitivity and increased lifespan although this increase does not recapitulate that seen in global GHR-/- mice.
Comparison of the three muscle-specific Ghr gene disrupted mouse lines shows conflicting results. Mavalli et al. [40] report that muscle-specific disruption of the GHR in male mice produces increased adiposity with insulin resistance and glucose intolerance. In contrast, both List et al. and Vijayakumar et al. report reduced adiposity and overall improvement in glucose homeostasis [42, 54]. The difference among Mavalli’s results [40] and those of the two other laboratories [42, 54] likely reflects the use of different promoter/enhancers driving Cre expression. Both List et al. and Vijayakumar et al. used muscle creatine kinase (Ckmm) promoter/enhancer [42, 54], which drives Cre expression in postnatal skeletal and cardiac muscle [217] while Mavalli et al. used the mef-2c promoter/enhancer, which directs Cre expression in postnatal skeletal muscle [44]. Unfortunately, while mef-2c Cre expression was thought to exclusively target skeletal muscle, more recently it has been shown that it is an important regulator of brain, bone, lymphocyte, blood vessel, endothelium, neural crest, craniofacial, and melanocyte development [218, 219]. Therefore, it is likely that unanticipated expression of Cre by the mef-2c promoter/enhancer in tissues other than muscle accounts for the differences between mice generated by Mavalli et al. versus other two mouse lines.
Brain-specific GHRKO mice
To understand the roles of GH axis on brain, four independent GHR brain-specific mouse lines have been generated between 2017–2019.
LeprEYFPΔGHR
Origin To comprehend the role of GHR signaling on the CNS, Cady et al. produced the LeprEYFPΔGHR mouse in the laboratory of Marianna Sadagurski in 2017 [61]. A Cre/loxP system was used to ablate Ghr in the leptin receptor-expressing neurons.
Phenotype No changes to body weight, length or composition are observed, and there is no change in serum IGF1 or GH levels [61]. LeprEYFPΔGHR mice do show impaired glucose homeostasis when compared to controls but normal insulin tolerance. This impaired glucose homeostasis may be due to the observed increase in hepatic gluconeogenesis.
AgRP-IRES-Cre
Origin To investigate the role of brain-specific GH action on energy homeostasis, Furigo et al. produced an agouti-related protein (AgRP) GHR knockout (AgRP-IRES-Cre) mouse in the laboratory of Jose Donato in 2019 [64]. These mice were produced by crossing mice carrying loxP-flanked Ghr alleles with AgRP-IRES-Cre mouse (Agrptm1(cre)Lowl/J).
Phenotype There is no change in glucose tolerance or insulin sensitivity, body weight, length or composition of these mice [64]. Also, no changes are observed in leptin sensitivity, ghrelin-induced food intake or ghrelin-induced c-Fos expression in the arcuate nucleus (ARH) of these mice. While there is no change in number of AgRP cells of the ARH, there is a reduction in c-Fos positive cells in a food-deprived state. These mice also show an attenuated neuroendocrine response that normally aids in energy conservation, when under food deprivation. Moreover, in this state, these mice show increased weight loss and decreased blood glucose compared to controls.
LepR-IRES-Cre
Origin In the same publication from the laboratory of Jose Donato (mentioned above) for AgRP-IRES-Cre mice, Furigo et al. reported the generation of a leptin receptor-presenting-cell-specific GHR knockout (LepR-IRES-Cre) mouse [64]. These mice were produced by crossing mice carrying loxP-flanked Ghr alleles with LepR-IRES-Cre mouse (B6.129-Leprtm2(cre)Rck/J).
Phenotype These mice have an increase in body weight and body length, as well as a reduction in body fat mass [64]. There are no changes to food intake, leptin sensitivity or energy expenditure, but there is a decrease in serum leptin levels. Under food deprivation, these mice have increased weight loss with some mice becoming lethargic.
Nestin-Cre
Origin Furigo et al. produced an entire brain GHR knockout (Nestin-Cre) mouse in their 2019 publication [64]. As the name implies, these mice were produced using a Cre system driven by the nestin promoter/enhancer.
Phenotype Nestin-Cre mice have increased body weight and body length with an increase in lean mass [64]. There is also an upregulation of GHRH expression in the hypothalamus. While there is no change in food intake, leptin sensitivity or energy expenditure, an increase in weight loss is observed during food deprivation in these mice compared to controls.
Disruption of GHR in the brain has helped establish that GH has a role in neurological processes. By targeting Ghr in various regions of the brain, researchers have established that hypothalamic GHR controls hepatic glucose production in nutrient-sensing, leptin receptor-expressing neurons [61], and GH regulates responses to weight loss in AgRP neurons [64]. Given the intricacies of the brain and the vast number of cell populations, we anticipate that many more brain-specific GHR knockout mice will be generated and evaluated.
Fat-specific GHRKO mice
GH plays an important role in AT catabolism. To understand how the GH-axis in AT affects glucose homeostasis and longevity, three fat-specific GHR knockout mice have been independently generated and characterized.
FaGHRKO
Origin The first fat-specific GHR knockout mouse line (FaGHRKO) was produced by List et al. in the Kopchick laboratory in 2013, utilizing the Cre/LoxP system driven by aP2, also known as Fabp4 promotor/enhancer [44].
Phenotype These mice show increased body weight with a 96% increase in total fat mass and an overall increase in body fluid when compared to controls [44]. Additionally, female mice show an 8% increase in lean mass. Both brown AT and all white AT depots are significantly increased in these mice. While no change is seen in insulin sensitivity, female mice show an increase in IGFBP5, IL-6 and leptin. Both male and female mice show a decrease in adipsin, with male mice displaying an additional decrease in adiponectin and IGF1 levels. Finally, these mice have a shortened lifespan when compared to WT controls.
AdGHRKO
Origin Later, expression of the aP2 promoter/enhancer was found in non-ATs, interfering with the interpretation of results seen in the FaGHRKO mice. To use a more reliable and robust model to investigate the direct effects of GH on AT, List et al. produced the AdGHRKO mouse, an adipocyte-specific GHR knockout mouse driven by adipoenctin/Cre [220].
Phenotype These mice exhibit no change in body length or body weight, though they have increased fat mass [220]. More specifically, all white AT depots had increased mass in female mice, and all but the perigonadal depot are increased in males. There is also an increase in brown AT in female mice. Adipocyte size is increased in these mice, with the only exception, again, being the perigonadal depot in male mice. These mice have improved glucose homeostasis with an increase in insulin sensitivity and no change in glucose tolerance. Furthermore, there is no change in serum GH, IGF1 or fasting blood glucose, but there is a decrease in total insulin in male mice. These mice also have a reduction in liver triglycerides. Overall, the more recent AdGHRKO mouse line has an AT profile remarkably like the previously reported FaGHRKO produced in the same laboratory.
Fat-Ghr-/-
Origin To investigate the role of adipocyte-specific GH-action on CR, Fang et al. produced the Fat-Ghr-/- mouse in the laboratory of Guosheng Liang in 2019 using the Cre/LoxP system driven by an adiponectin promoter/enhancer [63].
Phenotype Differing from AdGHRKO mice, Fat-Ghr-/- mice have no change in body fat mass. When placed on CR, there is no change in blood glucose, plasma ghrelin or plasma GH levels [63].
Three distinct fat-specific GHRKO mouse lines have been created. FaGHRKO and AdGHRKO lines generated in the same laboratory by List et al. have a similar AT profile, with increased adiposity resulting in an overall increase in percent body fat. In contrast, Fat-Ghr-/- mice generated by Fang et al. have no phenotypic change in any parameter including percent body fat. The difference between these mouse lines is unknown but may result from incomplete disruption of the GHR in AT. Genetic background is likely not a factor since all three lines were produced in a mixed C57BL/6 N x C57BL/6 J background, where floxed mice were generated in C57BL/6 N then crossed to Cre mice in a C57BL/6 J background. It should be noted that both FaGHRKO and AdGHRKO mouse lines were generated using the same floxed mouse – generated in the Kopchick laboratory, while Fat-Ghr-/- mice were generated using floxed mice generated in the Liang laboratory. Thus, it is possible that differences in the floxed mouse lines may exist.
Other GHRKO mouse lines
In the following section, we will discuss several individual mouse lines generated to explore the tissue-specific effects of GH and GHR on the heart, bone and intestines or cell types such as macrophages, beta-cells and hematopoietic stem cells.
Macrophage—GHRMacD
Origin While GHRs are expressed on macrophages, little is known about the role of GH in macrophage function. Accordingly, in 2010, Lu et al. produced the GHRMacD mouse in the laboratory of Ram Menon [39]. These mice were produced using the Cre/LoxP system driven by the Lyzs locus, expressed specifically in macrophages, monocytes and granulocytes (neutrophils, basophils, etc.).
Phenotype In vivo characterization is not described for this mouse line in this initial paper; however, in vitro studies show that cultured media collected from primary macrophages in the stromal vascular compartment (SVC) of AT from GHRMacD mice have an inhibitory effect on preadipocyte differentiation when placed on 3T3-L1 cells [39]. This finding indicates that intact GH-action in primary macrophages increases preadipocyte differentiation. However, GH does not increase IGF1 expression in macrophages. There is no difference between IGF1 levels in GHRMacD macrophages and control macrophages when treated with GH. In a follow up study in live mice, the Menon laboratory showed that GHRMacD mice (also called MacGHR KO mice in this paper) have no observable phenotypic changes except when challenged with a HFD [47]. When fed a HFD, GHRMacD mice had increased macrophage abundance in AT resulting in increased AT crown like structures and increased expression of pro-inflammatory cytokines (IL-1β, TNF-α, IL-6, and osteopontin) in AT stromal vascular fraction. These results support the possibility that GH may have beneficial effects on diet induced obesity related chronic inflammation.
Beta cell—βGHRKO
Origin To determine the role of the GHR in β-cell mass and function, Wu et al. created the β-cell GHR knockout mouse (βGHRKO) in the laboratory of Derek LeRoith in 2011 [41]. βGHRKO mice were generated by crossing GHR floxed mice with a rat insulin 2 promoter (RIP)/Cre/hGH mouse line constructed by Pedro Herrera [221].
Phenotype When fed a standard chow diet, βGHRKO mice show no difference in body weight, body composition or IGF1 and insulin levels compared to controls [221]. However, these mice show a significant decrease in islet cell size and number, suggesting that GH stimulates the growth and proliferation of islet cells. On a HFD, βGHRKO mice show a significant decrease in β-cell mass and higher glucose levels.
It should be noted that there is controversy surrounding the Cre line used in this study as it inadvertently expresses hGH, thus results obtained by studies using βGHRKO mice are difficult to interpret. More specifically, multiple laboratories have demonstrated that fusion genes containing the hGH minigene used to enhance transgene expression and thought to not be transcribed or translated do in fact produce significant amounts of hGH [222, 223]. Furthermore, GH expressed in pancreatic islets can bind to the PRLR thus augmenting phenotypic factors such as beta cell mass and insulin content [222].
Hematopoietic stem cells (HSC)
Origin To investigate the impact of GH signaling on hematopoietic stem cells (HSC), Stewart et al. produced the Ghrfl/fl;Vav1Cre/+mouse in the laboratory of Rossi in 2014, using the Cre/LoxP system driven by vav1 gene transcriptional regulatory sequences [50].
Phenotype Ex vivo analyses conducted using primary hematopoietic stem cells from these mice show no significant ‘blood cell’ differences apart from a decrease in number of platelets [50]. Also, there are no changes in progenitor compartments, progenitor cell action or in peripheral blood engraftment following the primary and secondary competitive transplants. These results suggest that GH signaling is dispensable for HSC function.
Bone—DMP-GHRKO
Origin To investigate the role of GHR action on bone growth, Liu et al. produced the DMP-GHRKO mouse in the laboratory of Shoshana Yakar in 2016, using the Cre/LoxP system driven by a dentin matrix protein 1 (Dmp1) promoter/enhancer [224].
Phenotype These mice show no change in body weight or composition [224]. Also, there is no change in osteocyte morphology or serum IGF1 levels. Serum GH is increased at 8 weeks and is similar to controls by 16 weeks of age. The DMP-GHRKO mice have decreased lacunae and cross-sectional area, resulting in a slender bone phenotype. Additionally, female mice have similar cortical bone thickness to controls but decreased bone marrow area. Males, however, have decreased cortical bone thickness and increased marrow area. DMP-GHRKO mice also have reduced levels of parathyroid hormone. To further understand the role of autocrine/paracrine IGF1 in bones, a DMP-IGF1RKO mouse, as more thoroughly described below, was produced. These mice have increased cortical bone cross-sectional area and reduced bone thickness and marrow area. Thus, the authors suggest that IGF1R and GHR may have overlapping as well as distinct effects on osteocytes [224].
Heart—iC-GHRKO
Origin To study the role of GH-action on the heart, Jara et al. produced the adult-inducible cardiac-specific GHR knockout mouse (iC-GHRKO) in the Kopchick laboratory in 2016, using a Cre/LoxP system driven by myosin heavy chain 6 promoter/enhancer [58].
Phenotype These mice show no change in body weight or length; however, they do have changes in body composition [58]. That is, the knockout mice have reduced fat mass and increased lean mass when compared to controls. There is no change in circulating insulin, with a decrease in circulating IGF1 only at 12.5 months of age. At 6.5 months, there is no change in glucose tolerance, but an increase in insulin sensitivity is observed. At 12.5 months, however, these mice have decreased glucose tolerance and increased insulin resistance. The iC-GHRKO mice have no changes in cardiac dimension but have decreased cardiac wall thickness. Additionally, blood pressure is unaltered in iC-GHRKO mice compared to age matched controls. Thus, taken together, removal of GHR in cardiac tissue specifically, has no observable effect on cardiac physiology but results in a decreased cardiac wall thickness and altered whole body glucose homeostasis.
Intestine—IntGHRKO
Origin In 2019, to investigate the effect of GH on the intestines, Young et al. produced the intestinal epithelial cell-specific GHR knockout mouse (IntGHRKO) in the Kopchick laboratory, utilizing the Cre/LoxP system driven by a villin promoter/enhancer [65].
Phenotype These mice have comparable body weights to controls, with no persistent body composition differences [65]. In male mice, there is a decrease in large intestine length. Also, there is a trend, albeit not significant, towards shorter villi in the small intestine, as well as decreased crypt depth in both small and large intestines. Female mice have decreased glucose tolerance and show insulin resistance, while males do not. In terms of intestinal permeability measurements, male mice have increased expression of occludin and females have decreased fecal albumin, indicating that there is a modest improvement to barrier function. Finally, males present with decreased fat absorption. These results demonstrate that removal of GH-action in the intestinal epithelial cells has modest and sex-specific effects on intestinal morphology and function.
Mouse lines downstream of GHR
GH induced intracellular signaling molecules downstream of the GHR have been manipulated in mice and include Janus kinases (JAK), signal transducers and activators of transcription (STAT), suppressors of cytokine signaling (SOCS), acid-labile subunits (ALS), IGF1 and IGFBPs. These molecules play critical roles in growth and development, glucose homeostasis and other physiological processes; thus, mouse lines with alterations in the levels or actions of these molecules are of interest and will be discussed below. Results related to some of these mouse lines are summarized in Tables 3 and 4.
JAKs, STATs, SOCSs
The canonical GH intracellular signaling pathway, through JAK2 and STAT5b phosphorylation, has been targeted in addition to the other JAK and STAT proteins. In fact, almost every member of the JAK family and the STAT family has been knocked out in a mouse line, and a transgenic line overexpressing STAT4 has also been reported. Importantly, the JAK/STAT pathway is shared among many different hormones and cytokines; as such, disruption of genes in this pathway generally results in impaired immune response and decreased growth and are difficult to attribute solely to GH action. Further downstream from JAK/STAT are SOCS proteins that serve as important inhibitors of this signaling pathway. Specific phenotypes of each gene disruption or transgenic mouse will be discussed below. All of the knockouts discussed in this section were generated using homologous recombination; for detailed description of the methods used for each mouse line, the reader is referred to the original publications.
JAK family knockout mice
Global JAK knockout mice
Origin Janus kinase proteins are intracellular tyrosine kinases that transduce signals of many cytokines. There are four members of the family: JAK1, JAK2, JAK3 and Tyrosine Kinase 2 (TYK2), each of which has been disrupted in a mouse line. Jak1-/- mice were first reported in 1998 by Rodig et al. [225]. Jak2-/- mice were produced in 1998 by both the Pfeffer laboratory and Ihle laboratory [226, 227]. JAK3 expression is more limited than JAK1 or JAK2, specific to hematopoietic cells and epithelial cells, so the creation of Jak3-/- mice was driven in part by the desire to develop a new mouse line of immunodeficiency. Jak3-/- mice were produced by Park et al. in 1995 [228]. TYK2 is ubiquitously expressed, and its disruption in a mouse was first reported by Shimoda et al. in 2000 [229].
Phenotype Jak1-/- mice have decreased size compared to controls and an impaired immune response [225]. They also have a failure to nurse, leading to death within days of birth, indicating a broad range of cytokine signaling disruptions. In contrast, Jak2-/- mice die in utero, presumably due to their impaired erythropoiesis, as stem cells from Jak2-/- mice respond to interferon α but not to erythropoietin or interferon γ [226, 227]. Jak3-/- mice are born in the expected Mendelian ratio (when heterozygous mice are bred, 25% of the resultant offspring are Jak3-/-) and survive to adulthood but have impaired lymphocyte development [228]. Specifically, they have decreased B and T cells and lack peripheral lymph nodes, natural killer cells and γδ T cells in the skin and intestines. Tyk2-/- mice develop normally, but have impaired IFNα signaling and their response to interleukin (IL)-12 is completely disrupted [229]. Interestingly, these mice also develop obesity and glucose intolerance due to abnormal BAT development [230].
Mice with tissue specific disruption of JAK2
Origin Although systemic JAK2 gene disruption is fatal, at least two tissue-specific JAK2 gene disrupted mice with direct relevance to GH’s metabolic effects have been reported. Liver-specific disruption of JAK2 (JAK2L mice) was first reported by Sos et al. in 2011, using the Cre/LoxP system with albumin promoter/enhancer to drive Cre [231]. To further explore the relationship between JAK2 and metabolism, the same laboratory developed an adipose-specific JAK2 disrupted (JAK2A) mouse line (first reported in 2013), also using the Cre/LoxP system with adiponectin promoter/enhancer driving Cre expression [232].
Phenotype JAK2L mice exhibit impaired lipid metabolism, with increased liver triglycerides and serum free fatty acids [231]. JAK2A mice have decreased lipolysis and increased body fat, as one would expect when GH signaling is disrupted in fat [232]. Interestingly, when the JAK2L and JAK2A mice are crossed to produce JAK2LA mice, those with JAK2 disruption in both tissues show the same increase in body fat and decreased lipolysis, but without interfering with liver lipid metabolism seen in JAK2L mice, indicating that the regulation of lipid metabolism through JAK2 involves coordination among multiple tissues [232].
STAT transgenic and STAT knockout mice
The STAT family are proteins that lie downstream of JAKs in various cytokine signaling pathways. The STAT family consists of 7 members—STAT1, STAT2, STAT3, STAT4, STAT5a, STAT5b, and STAT6—each of which has been disrupted in a mouse line; STAT4 transgenic mice have also been reported.
STAT1-/- mice
Origin Because STAT1 is a central factor in interferon signaling and involved in the signaling of other cytokines, Stat1-/- mice were generated in 1996 by Meraz et al. and Durbin et al. to determine whether STAT1 is necessary for all interferon-induced signaling, as well as signaling of other cytokines [233, 234].
Phenotype STAT1 deficient mice have normal reproduction [233, 234]. Despite GH’s ability to activate STAT1, this mouse line has no change in body size and responds normally to GH administration, gaining the same amount of weight as vehicle-treated controls. Stat1-/- mice also have normal responses to epidermal growth factor (EGF), IL-10, and IL-6. The most prominent phenotype of Stat1-/- mice is their complete lack of responsiveness to interferon (α and γ) [233, 234], leading to a strong susceptibility to infection by both bacteria and viruses, despite normal immune cell populations. The specificity of interferon signaling disruption in Stat1-/- mice has led to their common use as a model of interferon deficiency.
STAT2-/- and STAT3-/- mice
Origin STAT2, in contrast to STAT1, is specific to type 1 interferon (α and β) signaling. Stat2-/- mice were first reported by Park et al. in 2000 [235]. STAT3 was initially identified as a downstream effector of IL-6, but later found to be activated in response to other cytokines. In an attempt to clarify the role of STAT3 in cytokine signaling, Stat3-/- mice were developed in 1997 by Takeda et al. [236].
Phenotype As expected, Stat2-/- animals have increased susceptibility to infection but have unique deficiencies in T cells and macrophages as well as decreased STAT1 expression in some tissues [235]. STAT3 knockout mice die early in embryogenesis, thus limiting the utility of this mouse line [236].
STAT4 transgenic and STAT4-/- mice
Origin STAT4 is predominantly associated with IL-12 signaling, and both STAT4 transgenic and null mice were generated to confirm this specificity. STAT4 transgenic mice were first reported in 1999 by Wirtz et al. using the cytomegalovirus (CMV) promoter/enhancer to drive expression of murine Stat4 cDNA [237]. Stat4-/- mice were first reported in 1996 by Thierfelder et al. and Kaplan et al. [238, 239].
Phenotype Although no transgenic Stat4 mRNA is initially detected in the colon, STAT4 expression is induced by injecting dinitrophenyl-keyhole limpet hemocyanin, and upon this treatment, colitis developed in the transgenic mice [237]. This phenotype agrees with the finding that IL-12 is associated with Crohn’s disease in humans. In contrast to STAT4 transgenic mice, Stat4-/- mice also have normal growth and reproduction but an impaired immune system [238, 239]. STAT4 ablation in mice did result in disrupted IL-12 signaling, which causes decreased interferon-γ secretion, decreased T cell proliferation, decreased natural killer cell toxicity, and a shift from Th1 to Th2 cell differentiation.
STAT5a-/-, STAT5b-/- and STAT5a-/-5b-/- mice
Origin STAT5 denotes two highly similar proteins, STAT5a and STAT5b, which have unique and overlapping functions, and may work together through the formation of heterodimers. As such, each has been knocked out in mice individually, as well as jointly. Stat5a-/- mice were first reported in 1997 by Liu et al. [240], while Stat5b-/- mice were reported in the same year by Udy et al. [241], and Stat5a-/-5b-/- were reported the following year (1998) by Teglund et al. [242].
Phenotype Stat5a-/- mice exhibit normal size, weight, and fertility, but they are unable to lactate, indicating a probable disruption of PRL signaling [240]. These mice also exhibit an impaired IL-2 response in T cells that can be overcome by IL-2 administration. STAT5b is part of the canonical GH signaling pathway and thus, disruption of the Stat5b gene, yields an expected decrease in growth [241]. Unexpectedly, this growth deficit is limited to males. The ablation of STAT5b also results in a sex-specific pattern of gene expression in the liver (e.g. CYP and MUP). IL-2 resistance is more pronounced in Stat5b-/- mice, as excess IL-2 does not ameliorate this resistance. Stat5b-/- mice also exhibit IL-15 resistance. When STAT5a and STAT5b are knocked out in combination, a stronger phenotype is observed [242]. These double null mice have decreased lymphocytes in circulation and are infertile due to impaired corpus luteum formation. Similar to GHR-/- animals, Stat5a-/-5b-/- mice are dwarf and have low serum IGF1 levels and decreased epididymal fat. Interestingly, about 1/3 of the double knockout mice in the initial study died within 48 h of birth. The results from these three mouse lines underscore the importance of STAT5a and STAT5b in growth, lactation, and reproduction.
STAT6-/- mice
Origin STAT6 is considered a key component in IL-4 signaling. To examine this relationship, Takeda et al. generated Stat6-/- mice [243].
Phenotype: Stat6-/- mice are similar in phenotype to many of the other STAT null mice and are also relatively “normal”, with no reported change to body length, body weight, or reproduction. As expected, Stat6-/- mice experience disrupted IL-4 signaling, resulting in decreased MHC class II and CD23 expression. Stat6-/- animals also have impaired immunoglobin class switching, lymphocyte proliferation, and Th2 cell development [243]. Thus, STAT6 is important in adaptive and humoral immunity.
SOCSs transgenic and SOCS-/- mice
Further downstream of the GHR are SOCS proteins. As their name implies, SOCS proteins inhibit cytokine signaling in the JAK-STAT pathway. There are eight members of the SOCS family: SOCS1-SOCS7 and CISH, and each has been disrupted in a mouse line (except for SOCS4 and CISH). Transgenic models overexpressing CISH, SOCS1, SOCS2, SOCS3, SOCS5, and SOCS6 have also been reported. For more detail on SOCS family transgenic and null mouse lines, see a previous review on the subject [244].
SOCS1 transgenic and SOCS1-/- mice
Origin The SOCS1 protein has been shown to inhibit GHR signaling [245], and thus Socs1-/- mice were generated by Starr et al. in 1998 [246]. SOCS1 transgenic mice were generated by expressing the transgene in the T cell lineage of mice via fusion of the cDNA to the lck tyrosine kinase proximal promoter /enhancer (first reported by Fujimoto et al. in 2000) [247].
Phenotype Socs1-/- mice are normal size at birth but show decreased growth and die before weaning [246]. The role of SOCS1 in immune development is further substantiated by the principal phenotypes of both SOCS1 transgenic and null animals. In the T-cell specific SOCS1 transgenic mice, impaired T cell development is observed. In addition, Socs1-/- mice present phenotypic differences associated with alterations to the interferon gamma (IFNγ) pathway, ranging from lymphopenia to monocyte infiltration into organs [246]. When IFNγ is knocked out alongside SOCS1, chronic inflammation and perturbed T cell development is detected, along with polycystic kidneys [248].
SOCS2, SOCS3 transgenic and SOCS2-/-, SOCS3-/- mice
Origin SOCS2 and SOCS3 were among the earliest SOCS proteins discovered, and both transgenic and null animals were generated for each gene/protein. Socs2-/- mice were reported by Metcalf et al., in 2000 [22], while SOCS2 transgenic mice were generated by Greenhalgh et al. in 2002 using the UBC promoter to drive gene expression [249]. SOCS3 transgenic and Socs3-/- mice were reported by Marine et al. in 1999 [250].
Phenotype Due to the role of SOCS2 in inhibiting the GH axis, Socs2-/- mice display gigantism [22]. These mice also show increased collagen deposition in their skin, another indication of increased GH action, and decreased levels of major urinary protein (MUP) in the urine. Interestingly, in SOCS2 transgenic mice, a counterintuitive result is observed; that is, the mice are giant [249]. These results suggest that excess or deficit of SOCS2 activate the GH/IGF axis, while moderate levels inhibit GH action. It is hypothesized that this activation of the GH/IGF axis is due to SOCS2 outcompeting SOCS3 (a more potent GHR inhibitor) for GHR-binding at high concentrations. The status of SOCS3 as a more potent GHR inhibitor is demonstrated by the more extreme phenotype seen when SOCS3 is altered. Because of embryonic lethality, no growth-associated phenotypes could be assessed in SOCS3 null and transgenic mice [250].
SOCS5 transgenic and SOCS5-/- mice
Origin Another member of the SOCS family, SOCS5 is also believed to be involved in immune development, but knowledge of its association lags that surrounding other SOCS proteins. To help rectify this, SOCS5 transgenic and null mice were developed, in 2002 by Seki et al. [251] and in 2004 by Brender et al. [252], respectively. In the transgenic mice, a FLAG tagged SOCS5 protein is expressed in mice under the control of the lck proximal promoter/enhancer.
Phenotype Alterations to SOCS5 seem to have milder phenotypes than those seen with SOCS1 manipulation, which may explain why relatively little was known about SOCS5. In SOCS5 transgenic mice, the phenotype is limited to decreased Th2 cell differentiation [251]. Socs5-/- mice, on the other hand, have no alteration in phenotype [252].
SOCS6, CISH transgenic and SOCS6-/-, SOCS7-/- mice
Origin Transgenic mice that overexpress SOCS6 were generated by Li et al. in 2004 using the elongation factor 1 (EF1) promoter/enhancer to drive Socs6 expression [253], and CISH transgenic mice were generated by Matsumoto et al. in 1999 using the β-actin promoter to drive Cis1 expression [254]. The SOCS6 and SOCS7 genes have also been disrupted in mouse lines, with Socs6-/- mice being reported by Krebs et al. in 2002 [255] and Socs7-/- mice reported in 2005 by Banks et al. [256].
Phenotype SOCS6 and SOCS7 manipulation results in phenotypes marked by alterations in glucose metabolism. Specifically, in SOCS6 transgenic mice, an improvement in glucose metabolism is observed [253]. Socs7-/- exhibit increased pancreatic islet size and improved glucose metabolism [256]. Socs6 gene disruption (Socs6-/-), on the other hand, causes mild dwarfism with no reported change to glucose metabolism [255]. CISH transgenic mice phenotypically resemble Stat5-/- mice with normal development but with a defect in GH signaling [254]. Features of CISH transgenic mice include lactation deficiencies, indicating prolactin inhibition, as well as decreased body size, indicating the inhibition of the GH axis. However, CISH transgenic mice have normal fertility, differentiating them from Stat5-/- mice. CISH transgenic mice also have alterations to their T cells [decreased γδ T cells, natural killer (NK) cells, and NK T Cells and a shift in Th1/Th2 differentiation towards Th2 cells], further illustrating the many roles of CISH.
The strong phenotypes of some of the molecules downstream of the GHR demonstrate the complex regulation of GH signaling even before the main effector of GH action, IGF1, is taken into account.
IGF1, IGF1R, and tissue-specific KO
As one of the most important products of GH action, IGF1 and its receptor have been manipulated in numerous mouse lines to study its endocrine, autocrine and paracrine effects both globally and in specific tissues or cells. The IGFs are synthesized by almost all tissues and are important mediators of cell growth, differentiation, and transformation. IGFs have a fundamental role in both prenatal and postnatal development and exert their physiologic effects by binding to the IGF receptors or, albeit with less affinity, the insulin receptor. In addition, IGF1’s effects are modulated by multiple IGF binding proteins (BP). In the following section, we will summarize the transgenic and knockout mouse lines relating to both IGF1 and its receptor. Details regarding each mouse line can also be found in Table 3.
IGF1 transgenic mice
Origin In 1988, Palmiter’s laboratory generated IGF1 transgenic mice containing a fusion chimeric gene with Mt1 promoter/enhancer, a sequence encoding the rat somatostatin secretory signal sequence to allow for secretion, the human IGF1 cDNA, and a sequence containing the human GH 3'-RNA processing signals [257].
Phenotype These mice express 1.5 times higher circulating IGF1 levels than controls and, as expected, decreased GH levels [257]. No phenotypic differences are evident until 6–8 weeks of age. Overall, IGF1 transgenic mice display 1.3 times higher weight gain compared to WT mice though no increase in skeletal growth is observed. The spleen, pancreas, kidneys, and brain display increased growth. Also, fertility is not affected. Notably, changes in kidney structure have been identified in IGF1 transgenic mice by Striker’s laboratory [258]. That is, IGF1 transgenic mice have enlarged glomeruli without glomerulosclerosis, in contrast to GH transgenic mice that display enlarged glomeruli with sclerosis. This implies that GH plays a direct role in the formation of kidney sclerosis while IGF1 stimulates increased glomerular size.
IGF2+/− mice (Igf2+/−)
Origin IGF2+/− (or Igf2+/−) mice were generated in 1990 by T. DeChiara in Robertson’s laboratory by deleting a portion of exon 2 of the mouse Igf2 gene [259].
Phenotype No homozygous Igf2-/- pups survive [259]. Heterozygous Igf2+/− pups display considerably smaller body size (60% of normal size). Genotyping of heterozygous embryos reveals that the mutant allele exerts its effect in the early embryonic stage (earlier than day 16) and maintains its effect in post-natal growth. Despite their diminutive size, the heterozygous mice appear normal and display normal reproductive capacity. Interestingly, this was the first study to identify the presence of imprinted genes (paternal), verifying previous hypotheses regarding this epigenetic phenomenon.
IGF1-/- and IGF1R-/- mice, and associated double mutants
IGF1-/- mice (Igf1-/-)
Origin In 1993, the Efstratiadis laboratory reported the generation of the IGF1-/- (Igf1-/-) mouse generated via the deletion of exon 4 of the mouse Igf1 gene [260].
Phenotype Igf1-/- mice experience increased neonatal lethality, although the rate of survivability is 10–68%, which is dependent on genetic background [260]. At birth, Igf1-/- mice display decreased body mass (65% of normal size). Post-natal effects include a progressively decreased growth rate, displaying 30% of control mouse size in adulthood [261]. The heterozygous Igf1+/− progeny do not display any obvious phenotypic difference from control littermates.
IGF1R-/- mice (Igf1r-/-)
Origin In the same 1993 publication for the generation of IGF1-/- mice, the Efstratiadis group also reported generation of the Igf1r-/- mice via the deletion of exon 3 in the gene encoding Igf1r [260].
Phenotype These mice display severe growth deficiency with a body mass reduction of 45% compared to WT mice at birth. The mutant neonates, however, are not viable due to respiratory issues, and unlike IGF1-/- mice, lethality appears independent of the genetic background strain of the mice. In addition, mutant IGF1R-/- mice exhibit delayed ossification of bones in the extremities and trunk by 1–2 days post-birth.
IGF1-/- with IGF1R-/- mice
Origin This same paper by Estratiadis also describes double mutants (Igf1-/- with Igf1r-/-) [260].
Phenotype The phenotype of the double knockout does not differ from the IGF1R-/- mice [260].
Overall, the role of IGF1/IGF1R in mouse embryonic development appears essential for viability, and the absence of which shows a considerable impact on bone development, muscle development and growth.
IGF1R-/- with GHR-/- mice
Origin In 2001, the Efstratiadis laboratory also reported the crossing of mutant mice lacking either IGF1, GHR or both simultaneously to examine the impact of GH and IGF1 in controlling postnatal growth [262]. Note that GHR null mice were generated using a targeting vector that replace exons 7, 8a, and 8, distinct from that reported by Zhou et al., which is described above, but with a similar growth phenotype [30].
Phenotype With respect to growth, these studies estimated that 17% of body weight is attributed to processes unrelated to GH or IGF1 while IGF1 accounts for 35% of growth and 14% for GH [262]. Importantly, the study reveals that 34% of growth is associated with overlapping functions of GH and IGF1. This study also assesses chondrocytes and bone ossification and reports that GH and IGF1 have independent and overlapping functions in chondrocytes since the phenotype of double mutants is more severe than that manifested in either class of single mutant. Thus, these mutants provide conclusive evidence of the importance of both of these hormones acting independently and in concert to support body growth.
Tissue-specific IGF1 and IGF1R manipulation
To understand the role of IGF1 in specific tissues and cell types, IGF1 and IGF1R have been either knocked in or out in specific tissues and cell types. In the following section, we describe numerous tissue-specific mouse lines and provide additional details about each in Table 3.
Liver-specific IGF1 transgenic and KO mice
(i) Hepatic IGF1 transgenic (TTR-IGF-I) mice
Origin In 2006, Xu’s laboratory created hepatic IGF1 transgenic (TTR-IGF-I) mice using a fusion gene consisting of the promoter/enhancer of the transthyretin (TTR) gene, the mouse Igf1 cDNA, and the SV40t polyadenylation-signal [263]. Note that the TTR promoter/enhancer targets transgene expression specifically to the liver, and the authors estimate approximately three copies of the TTR-IGF1 transgene in these mice.
Phenotype As expected with increased circulating levels of IGF1, these mice show decreased levels of GH and increased IGFBP3 levels [263]. As Pegvisomant treatment does not alter IGFBP3 levels in WT mice, these results collectively indicate that IGFBP3 is not a direct target of the GH signaling pathway. The authors suggest that liver-expressed IGF1 can stimulate IGFBP3 expression and stabilize IGF1 under GH-deficient conditions. These mice display a larger body size and organ weight, presumably due to the higher circulating IGF1 levels. When TTR-IGF-I mice are bred with MMTV-ErbB2 mice to investigate the effect of elevated IGF1 on ErbB2 driven mammary carcinogenesis, the high levels of systemic IGF1 appear to have no effect on promoting ErbB2 driven mammary carcinogenesis [264].
(ii) Hepatic IGF1 transgenic (HIT) mice and KO-HIT mice
Origin In 2009, LeRoith and colleagues, developed the hepatic IGF1 transgenic (HIT) mice, which overexpresses the rat Igf1 transgene in the liver of mice, as well as KO-HIT mice, in which only the liver produces IGF1 (i.e. mice that have a null Igf1 gene in all tissues but overexpress a rat Igf1 transgene specifically in the liver) [265].
Phenotype HIT mice have increased IGF1, unaffected GH levels, increased body mass, organ sizes and skeletal sizes, but decreased adiposity [265]. In contrast, KO-HIT mice have total absence of tissue IGF1, but elevated levels of serum IGF1, which can support normal body size and weight at puberty and postpubertal ages. Early deficits in skeletal structure of KO-HIT mice are restored by adulthood [266]. Insulin sensitivity is not altered by elevated levels of serum IGF1. Female KO-HIT mice have insufficient tissue IGF1 to fully support the female reproductive system, while male mice reproductive function is not affected. Overall, KO-HIT mice show that most autocrine/paracrine actions of IGF1 related to tissue growth and function can be offset by elevated levels of endocrine IGF1 although autocrine/paracrine IGF1 appears critical for neonatal development.
(iii) GHRKO-HIT mice
Origin In 2013 Yakar et al. combined the GHRKO mouse with the HIT mouse to generate the GHRKO-HIT mouse [267].
Phenotype The results with GHRKO-HIT suggest that, with the absence of GH-GHR mediated action, serum IGF1 is not sufficient to restore body and skeletal size, but sufficient to restore impaired glucose tolerance in GHRKO mice [267].
(iv) LID mice
Origin The first liver specific IGF1 KO (LID) mouse line was produced by Yakar and LeRoith in 1999 via crossing albumin Cre mice with Igf1 floxed mice.
Phenotype LID mice have increased GH and decreased IGF1 levels due to IGF1 ablation in the liver [268]. Their body weight, selected organ weights (kidney, fat, muscle, spleen, and heart), body length and femur length are not different from WT controls. LID mice exhibit decreased insulin sensitivity and display normal reproductive capacity. Interestingly, when treated with GH, female LID mice exhibit an accelerated growth rate compared to males [216]. LID mice also show decreased cancer incidence and an increased lifespan in females compared to controls [21, 269] presumably attributing to lowered levels of circulating IGF1. These results challenged the idea that circulating IGF1 is critical for normal growth and development and suggest that growth is preserved even when IGF1 is absent from the liver and/or the importance of the autocrine/paracrine role of IGF1.
(v) Conditional liver IGF1KO mice (LI-IGF-I-/-)
Origin In 1999, Sjögren et al. produced conditional liver IGF1KO mice (referred to in the paper as LI-IGF-I-/- mice) by crossing mice with a Mx Cre (Mx dynamin-like GTPase 1) promoter/enhancer, which is activated in an interferon-dependent manner, to Igf1 floxed mice [270].
Phenotype Similar to LID mice, these mice have increased GH levels, decreased IGF1 levels in serum (~ 75%) and exhibit no changes in postnatal growth with induction of interferon at ~ 1 month and measurements at ~ 2 months after induction. Interestingly, kidneys are slightly smaller and the livers larger in LI-IGF-I-/- mice than in controls [270]. At 13 months of age, these mice have decreased fat mass and become insulin resistant [271]. The female mice also have an increased mean lifespan [272].
In summary, these results suggest that decreased endocrine IGF1 has a critical role in decreasing cancer incidence and extending lifespan, but it does not affect growth and development significantly. These findings are in contrast to what is observed in LiGHRKO mice in which lifespan is not altered and body size is decreased [48]. In these cases, GH and local IGF1 may be able to sustain growth of the whole organism and organs. On the other hand, increased IGF1 levels could further increase body size, organ weight and glucose tolerance.
Adipose-specific IGF1R KO mice
(i) aP2 adipose-specific IGF1R KO mice
Origin Different transcriptional regulators have been used to determine the physiological role of the IGF1R signaling in AT. Initially, an aP2 promoter/enhancer-driven Cre was utilized by Kloting et al. in 2008 [273].
Phenotype These ap2 adipose-specific IGF1R KO mice have a marked increase in somatic growth with increases in both body weight and body length [273]. They also have elevated circulating IGF1 and IGFBP3 levels with no change in GH. The authors suggest that the ~ 20% increase in circulating IGF1 is responsible for the increased growth. Other notable metabolic features in these mice include elevated glucose levels and suppressed adiponectin levels, despite normal glucose and insulin tolerance. Regarding their AT phenotype, these mice have increased fat mass, more prominent in the gonadal region versus the subcutaneous region, and significant increases in adipocyte size. The increase in lipid accumulation is attributed to an increase in IRs and insulin-stimulated glucose uptake into adipocytes with the deletion of the IGF1R. Importantly, these authors reveal a decrease in IGF1R protein not only in AT but also in the brain. More recently, other groups have confirmed the promiscuity of the ap2 promoter in several tissues including regions of the brain [274–276]. The “leaky” nature of this promoter sheds doubts on whether the phenotype observed in these mice is due to a deletion of IGF1R AT or other tissues. Regardless, the authors conclude that IGF1R signaling in adipocytes is not crucial for the development and differentiation of AT/adipocytes but does seem to participate in regulating circulating IGF1 levels.
(ii) Adiponectin adipose-specific IGF1R KO mice
Origin To uncover the specific role of IGF1R in adipocytes, a second adipocyte-specific IGF1R KO mouse was created using the adiponectin promoter/enhancer by Ron Kahn’s group in 2016 [277].
Phenotype These mice have a distinct phenotype as compared to the first mouse line made with the ap2 Cre. The adipo-Cre IGF1R KO mice have modest reductions in both white AT and brown AT mass (~ 25%), despite a 73% increase in circulating IGF1 levels [277]. They also have reduced expression of lipogenic genes in intra-abdominal fat depots, reduced levels of circulating leptin and adiponectin [277] with no change in ectopic fat deposition. However, these mice have no appreciable changes in response to a glucose or insulin challenge or basal insulin or glucose levels. In comparison, insulin receptor adipocyte-specific KO results in a severe lipodystrophic state, severely impaired glucose metabolism (higher basal glucose and insulin, impaired GTT and ITT), and increased ectopic fat deposition than IGF1R KO mice. Thus, the authors conclude that insulin and IGF1 signaling play essential but distinct roles in the development and function of white and brown fat.
Brain-specific IGF1R KO mice (bIGF1RKO +/− and bIGF1RKO −/−)
Origin In 2008, Holzenberger’s laboratory generated brain-specific IGF1R KO mice by crossing Igf1 floxed females with Nestin-Cre transgenic males (flox/+; NesCre +/0) [278]. Nestin driven Cre recombinase is specific to neural and glial precursors early in neural development.
Phenotype Homozygous double mutants express no IGF1R on CNS neurons or glia [278]. The homozygous animals have microcephaly with severe growth retardation; they are also infertile and exhibit abnormal behavior (e.g. male KO mice have impaired exploratory behavior and are less anxious) but have normal lifespans. On the other hand, heterozygotes, whose IGF1R levels are depleted by half in the neurons and glia, exhibit healthier aging (delayed mortality and longer mean lifespan) and behave normally. By 90 days, heterozygote adults weigh 90% of WT controls and are 5% shorter in length. They have normal IGF1 levels in peripheral tissues but lower plasma GH and IGF1 levels. Adult pituitaries are 30–40% smaller with markedly fewer somatotrophs, and most other organs are smaller in adult bIGF1RKO +/− mice with the exception of AT, which is significantly increased in both adult males and females. Adult heterozygous males also have significantly higher circulating lipid levels (triglyceride, HDL, total cholesterol and free fatty acid) compared to WT animals. Both sexes of heterozygous mice have impaired glucose tolerance. Like homozygous mice, there is no change in maximum lifespan of heterozygous mice; however, heterozygous mice do have an increase in mean lifespan, which is attributed to fewer degenerative diseases as well as tumors compared to WT. Overall, the authors conclude that partially lowered GH/IGF1 signaling in the brain favors lifespan extension and that the ability to alter somatotropic function in stressful environments allows the organism to decelerate growth and preserve resources, and thereby improve health span.
Muscle-specific IGF1 transgenic and IGF1R KO mice
(i) Skeletal muscle IGF1 transgenic Mice
Origin Striated muscle-specific IGF1 transgenic mice were created in 1995 by the Schwartz laboratory using the avian skeletal α-actin gene proximal promoter/enhancer appended to the human IGF1 gene [279].
Phenotype Striated muscle-specific IGF1 transgenic mice have no changes in serum IGF1 levels or body weight [279]. However, concentrations of IGF1 in muscle are 47-fold greater in transgenic mice compared to WT controls, causing myofiber hypertrophy with a change in overall fiber types and increased superficial gluteus muscle.
(ii) MKR Mice
Origin In 2001, Le Roith’s laboratory generated skeletal muscle-specific transgenic mice by overexpressing a dominant-negative IGF1R (MKR mice) via fusion of mutant IGF1R (KR-hIGF1R) cDNA downstream of the muscle-creatine kinase (MCK) promoter/enhancer [280]. In these mice, the mutated gene encodes a protein that has lysine at position 1003 changed to arginine (KR mutant), which abolishes the ATP-binding within the β-subunit of the human IGF1R cDNA.
Phenotype In these mice, expression at the protein level results in the formation of hybrid receptors between mutant and endogenous IGF1R and IRs, abrogating their normal function and resulting in a marked decrease in glucose uptake upon stimulation with either IGF1 or insulin [280]. Although normal glucose tolerance is maintained, peripheral insulin resistance and pancreatic beta cell dysfunction develop by seven to twelve weeks of age in MKR mice, contributing to a chronic hyperglycemic state. Overall, body glucose disposal, glycolysis and glycogen synthesis are significantly reduced in MKR mice. In the skeletal muscle and brown AT of these mice, glucose transport activity is reduced by 50%. There is also a marked increase in the number of glycogen deposits, FFAs, and triglycerides in the livers consistent with an aggravation of the insulin-resistant state. MKR mice also exhibit a 10–20% reduction in body weight relative to WT controls.
(iii) MIGIRKO Mice
Origin A double knockout mouse (MIGIRKO), which has a loss of both IR and IGF1R signaling reported by O’Neill et al., in 2015, was generated via the use of a Cre/LoxP system using a skeletal muscle actin promoter/enhancer [281].
Phenotype MIGIRKO mice exhibit a 60% decrease in muscle mass, accompanied by loss of both muscle strength and endurance, and a shortened lifespan (6 months) due to atrophy of the diaphragm [281]. These mice have normal glucose and insulin tolerance but lower fasting glucose levels and increased basal glucose uptake. The alteration in glucose metabolism is due to increased membrane localization of glucose transporters (Glut 4 and Glut 1) as a result of decreased TBC1D1, a protein critical to the regulation of glucose transport in muscle cells.
(iv) M-IGF1R KO Mice
Origin In 2016, O’Neil et al. also reported the characterization of muscle-specific IGF1R KO mice (M-IGF1R KO) [282].
Phenotype M-IGF1R KO has no significant reduction in muscle mass in contrast to MIGIRKO, most likely due to compensation on behalf of functional IR signaling [282]. Overall, these mice do not display a dramatic phenotype resulting from disruption of solely IGF1 action in muscle, again, likely due to compensation via functional IR signaling.
Cardiac-specific IGF1 transgenic and IGF1R KO mice
(i) Cardiac-specific IGF1 transgenic mice
Origin In 1996, IGF1 transgenic mice were generated by the Anversa laboratory using human IGF1 cDNA placed under transcriptional control of rat α-myosin heavy chain promoter/enhancer [283].
Phenotype These transgenic mice have increased serum IGF1 despite cardiomyocytes being the only source of transgenic IGF1 [283]. This finding is similar to what is reported above for cardiac-specific GHR disruption by Jara et al. [58] and emphasizes the significant contribution of cardiomyocytes to endocrine IGF1. These mice have significantly greater total heart mass, liver, brain, spleen and kidney due to the increase in IGF1. The enlarged hearts are attributed to overexpression of IGF1-induced myocyte proliferation, suggesting that local and endocrine IGF1 increase organ sizes and promote myocyte proliferation.
(ii) Cardiomyocyte IGF1 transgenic mice
Origin Another cardiomyocyte IGF1 transgenic mouse line was created using mouse α-myosin heavy chain promoter/enhancer by the Rosenthal group in 2007 [284].
Phenotype In these mice, local IGF1 expression results in accelerated postnatal cardiac growth and greater heart size [284]. These mice have the capacity to repair their hearts more efficiently both morphologically and functionally in response to injuries induced by cardiotoxin or ligation.
(iii) CIGF1RKO mice
Origin In 2008, the laboratory of Abel developed a constitutive cardiac-specific IGF1R knockout mouse (CIGF1RKO) [285].
Phenotype These mice are resistant to exercise-induce cardiac hypertrophy, implicating IGF1 in this process [285].
(iv) iCMIGF-IRKO mice
Origin Adult heart, tamoxifen-inducible, cardiomyocyte-specific IGF1R KO mice (iCMIGF-IRKO) were reported in 2012 by Gödecke et al. [286]. Mice with tamoxifen induction at 3 months and 11 months of age, with measurements taken 6 weeks after gene deletion, are described.
Phenotype Younger induction (3 months) results in no functional or structural consequences; however, induction at the older age (11 months) results in cardiac dysfunction without structural abnormality [286].
In summary, these studies show that autocrine/paracrine IGF1 promotes heart repair in response to injury and conservation of cardiac function. However, the absence of IGF1 signaling in cardiomyocytes does not affect the morphology or function of hearts significantly, unless induction occurs at a later age (11-month-old). Similarly, the removal of GH action in heart at adult age (4-month-old) affects neither the local IGF1 levels nor the function of hearts [58] even though endocrine IGF1 levels are altered.
Endothelial IGF1R transgenic and KO mice
(i) Endothelial IGF1R transgenic mice (hIGFREO)
Origin Generated by Kearney et al. team in 2012, the endothelial IGF1R transgenic mice (hIGFREO) were produced by overexpressing human IGF1R following the Tie2 (mouse endothelial-specific receptor tyrosine kinase) promoter/enhancer [287].
Phenotype These transgenic mice exhibit no change in size/weight or glucose homeostasis [287]. Reduced basal and insulin-stimulated eNOS activity is reported in these mice. As for cardiac function, no difference in endothelial cell eNOS is observed with only enhanced aortic constriction in response to phenylephrine. These mice have normal blood pressure and aortic response to acetylcholine (ACH) and nitroprusside but increased endothelial cell migration and regeneration.
(ii) EC IGF-1R KO mice
Origin Also reported by Kearney et al. in 2011, opposite results are observed for the endothelium-specific IGF1R KO mouse (EC IGF-1R KO) produced by Tie2 Cre [288]. A second endothelial cell (EC)-specific IGF1-R KO mouse line was generated by Cheng and colleague in 2015 within the context of chronic kidney disease (CDK)-induced pathology via vascular epithelial (VE)-cadherin-Cre [289].
Phenotype Kearney et al. report that male EC IGF-1R KO mice show normal glucose homeostasis with enhanced basal and insulin-stimulated eNOS phosphorylation[288]. As for cardiac function, there is blunted aortic constriction in response to phenylephrine and enhanced aortic constriction in response to l-NG-nitro-l-arginine methyl ester (L-NMMA). No difference in endothelial cell eNOS is observed. The EC specific IGF1-R KO mice produced by Cheng et al. have no changes in overall body size, weight, or reproductive capacity [289]. However, these KO mice display significantly more severe tubular injury and interstitial collagen deposition in obstructed kidneys compared to WT. The phosphorylation state of VE-cadherin, correlating with the disassembly of EC junctions, is significantly higher, along with markedly increased platelet accumulation and vascular permeability in null animals.
Collectively, these results support an important role for IGF1R within a physiological range in regulating nitric oxide bioavailability and vascular repair, which are hallmarks of several human diseases involving tissue growth and vascularization.
Myeloid and macrophage-specific IGF1R KO mice
(i) MIKO mice
Origin In 2016, Dixit and colleagues at Yale University created myeloid-specific IGF1R KO mice (MIKO) with Cre driven by LysM promotor/enhancer [290, 291].
Phenotype MIKO mice have decreased NLRP3 (NOD-, LRR- and pyrin domain-containing protein 3) inflammasome activation in aging macrophages [290]. They also exhibit increased adiposity, with fewer macrophages in the stromal vascular fraction of visceral AT and a decrease in M2 macrophage activation, unlike the increase in visceral AT M2 macrophage polarization reported in GHRKO mice [291]. Interestingly, these mice show delayed resolution from helminth infection (which induces an adaptive immune response characterized by a distinct T helper cell driven cellular and cytokine profile) and have increased insulin resistance when placed on a HFD.
(ii) MΦ-IGF1RKO mice
Origin As autocrine/paracrine action of IGF1 plays an important role in increasing macrophage activities [290], in 2016, Delafontaine and colleagues created a monocyte/macrophage-specific IGF1R KO mouse (MΦ-IGF1RKO), bred on an apolipoprotein E-deficient genetic background [292].
Phenotype These mice show increased atherosclerotic lesion formation with less stable plaques and marked by increased macrophage content [292]. Plaque-associated macrophages exhibit increased inflammatory responses to stimulation, as well as increased expression of antioxidant genes. Production of cytokines or chemokines such as IL-1α, IL-6, TNFα, MPC1 and fractalkine (an unusual chemokine encoded by the gene CX3CL that can act as either a soluble or membrane-bound mediator), are associated with increases in NFκB activity. These macrophages also demonstrate decreased expression of ATP-binding cassette transporters ABCA1 and ABCG1 and, therefore, a significant reduction in HDL-dependent cholesterol efflux, which leads to atherogenesis.
In conclusion, these studies suggest that macrophage IGF1 signaling exerts anti-atherogenic effects through suppressing macrophage activities, atherosclerotic lesion formation, and reducing plaque vulnerability.
Bone-specific IGF1 transgenic and IGF1(R) KO mice
(i) Osteoblast-specific IGF1 transgenic mice
Origin In 2000, Clemens’ Laboratory created the osteoblast-specific IGF1 transgenic by fusing the rIgf1 cDNA transgene to the human osteocalcin (OC) promoter/enhancer [293]. Another osteoblast-lineage IGF1 transgenic mouse reported by Kream’s group in 2006 utilized the upstream regulatory sequence of rat Col1a1 gene followed by murine Igf1 [294].
Phenotype The mice from Clemens’ group have increased bone formation rate and cortical and trabecular bone mass density [295]. The mice generated by Kream’s group show increased bone formation and resorption; male transgenic mice have increased serum IGF1 levels and body weight [294].
(ii) Osteoblast IGF1R KO mice and OBIGF1R-/- mice
Origin Osteoblast IGF1R KO mice were generated by Clemens’s group in 2002 (human osteocalcin promoter/enhancer) [296–298] and OBIGF1R-/- mice (Col1α1 promotor/enhancer) by Bikle’s laboratory in 2015 [296–298].
Phenotype Both mice have decreased bone formation rate and cancellous bone volume/connectivity with normal body size [296–298]. These mice also have increased trabecular bone separation with a decrease in trabecular number.
(iii) Chondrocyte IGF1 KO mice
Origin Chondrocyte IGF1 KO mice originated using the procollagen Col2α1 gene promoter/enhancer driven Cre by Mohan’s laboratory in 2007 [299, 300].
Phenotype These mice exhibit decreased bone mineral content, bone mineral density, bone size, weight and body length when compared to WT [299, 300].
(iv) Osteocyte IGF1 KO mice
Origin In 2013, Lau and colleagues created the osteocyte IGF1 KO mice using dentin matrix protein 1 (Dmp1) driven Cre [301].
Phenotype These mice have significantly smaller periosteal diameter of femurs, shorter femur lengths, reduction in bone mineral contents, bone formation and bone turnover [301].
(v) DMP-IGF-1R KO mice
Origin Osteocyte IGF1R KO mice (DMP-IGF-1R KO) were generated by Yakar et al. in 2016 [224].
Phenotype DMP-IGF-1R KO mice show an increase in total cross-sectional areas of femora with reductions in bone area but significant increases to marrow area. DMP-IGF-1R KO mice also exhibit cortical bone thickness with enlarged marrow area, which indicates increased endosteal resorption [224]. When DMP-GHR KO mice are compared with DMP-IGF-1R KO, there is a decrease in bone accrual for both [224]. These results imply that GH and IGF1 share some overlapping yet distinct effects on osteocytes.
In summary, the GH/IGF1 axis controls skeletal growth through an endocrine and autocrine/paracrine fashion. Studies above clearly show that IGF1 signaling regulates bone length, radial bone growth, cortical and trabecular bone properties through chondrocyte, osteoblast and osteocyte function. Detailed information can be found in the review from Yakar et al., 2018 [302].
Ovarian granulosa cells IGF1R KO mice
Origin In 2017, Stocco’s laboratory generated ovarian granulosa cell specific IGF1R KO mice (IGF1Rgcko) using Cre driven by the estrogen receptor β (Esr2) and the aromatase (Cyp19) promoter/enhancers [303].
Phenotype These mice do not possess antral follicles, even with gonadotropin stimulation. They are sterile and have smaller ovaries [303]. Serum estradiol levels are decreased by 90% compared to controls, while follicle stimulation hormone (FSH) receptor expression is not altered. Their insulin sensitivity is unchanged in comparison to control mice. Activation of AKT is significantly dampened, and apoptosis levels in follicles from primary to secondary stages are increased. Overall, these data suggest that IGF1R has an essential role in granulosa cell function and, as a result, in female fertility.
Pancreatic β Cell IGF1R KO mice
(i) β cell IGF-1R KO mice
Origin In 2002, Efstratiadis’ laboratory generated β Cell IGF-1R KO mice using Cre driven by the rat insulin promoter/enhancer (InsPr-Cre) [304]. Another β Cell IGF1R KO mouse was generated in Kahn’s laboratory in 2002, also using InsPr-Cre [305].
Phenotype The lack of IGF1R in the mice produced by Efstratiadis’s laboratory does not affect β cell mass but does lead to age-dependent glucose intolerance and decreased insulin secretion in response to arginine and glucose [304]. With these results, the authors suggest that IGF1R signaling is a requirement in regulating insulin secretion. The finding from Kahn’s laboratory corroborates those of the Efstratiadis laboratory, except that they report normal insulin secretion in vivo in response to arginine [305]. Kahn’s mice also have a decrease in insulin secretion in response to glucose.
(ii) βDKO mice
Origin βDKO mice with disruptions in both IR and IGF1R in β cells were generated using an Ins/Pr-Cre [306].
Phenotype These double KO mice have low insulin levels with high levels of glucagon and are highly glucose-intolerant [306]. Both the mass and insulin content of pancreatic β cells are decreased. While β cell-specific IGF-1R KO and IR KO are both glucose intolerant, a more severe intolerance is observed in βDKO mice, probably due to the overlapping functions of IGF1 and insulin.
Steroidogenic cell IGF1R KO mice
(i) Steroidogenic cell IGF1R KO mice
Origin In 2018, Nef’s laboratory developed IGF1R KO mice with IGF1R deleted in steroidogenic cells using Cre driven by the human P450SCC promoter/enhancer [307].
Phenotype IGF1R KO in steroidogenic cells result in mice that grow normally, have normal adrenal gland development, and no change in corticoid synthesis [307]. Male KO mice have significantly decreased testicular weight in comparison to control mice, but seminal vesicle size and anogenital index are unchanged. Leydig cells, which are steroidogenic cells that produce androgens, are found to have decreased responsiveness to human chorionic gonadotropin (hCG). The authors indicate that IGF1R signaling is necessary for the development of Leydig cells and their steroidogenic activity as adults. They also noted that disruption of IGF1R signaling did not have a significant effect on adrenal gland development or function.
(ii) Steroidogenic cell IGF1R;IR KO mice
Origin Nef’s laboratory also developed IGF1R;IR KO mice with both IR and IGF1R deleted in steroidogenic cells using Cre driven by the human P450SCC promoter/enhancer [307].
Phenotype The phenotype of IGF1R;IR double KO mouse is dramatic [307]. That is, Leydig cells fail to mature, resulting in impaired steroidogenic function, decreased steroidogenic cells and serum testosterone levels. These mice have a substantial reduction in the size of the adrenal cortex and testis and are infertile. After weaning, survival rate of these mice is significantly reduced due to disparity in salt and water metabolism.
Somatotroph IGF1R KO mice
(i) SIGFRKO mice
Origin In 2010, Radovick’s laboratory generated somatotroph-specific IGF1R KO mice (SIGFRKO) using Cre driven by the GH promoter/enhancer (rGHpCre) [308].
Phenotype SIGFRKO mice do not respond to feedback by IGF1 [308]. By 14 weeks of age, these mice grow normally but weigh significantly less than controls; and body length is unchanged in comparison to control mice at all life stages. These mice have increases in the levels of fasting serum GH and IGF1 and have altered body composition with decreased fat mass but no change in lean mass. Average weights of many tissues (brain, heart, lungs, and kidney) are unchanged although liver and spleen mass are increased. Additionally, IGFBP3 levels are unchanged and ALS levels increased. There are also decreased mRNA levels of GHRH and increased mRNA levels of somatostatin in pituitary tissue, likely contributing to the growth deficiency observed by 14 weeks of age. Glucose and insulin tolerance are both unchanged in SIGFRKO mice [308].
(ii) HiGH mice
Origin In 2011, Kineman’s laboratory generated somatotroph IGF1R and IR KO mice (HiGH) with somatotroph specific inactivation of both the IR and IGF1R using the rat Gh promoter/enhancer driving Cre [309].
Phenotype HiGH mice are characterized by increased levels of GH and IGF1 [309]. From birth to 3 weeks of age, these mice are the same size as control mice, but their weight is modestly increased in adult life. The increase in GH promotes a lean phenotype but has minimal effects on adiposity in males, even in response to HFD. These mice have decreased insulin sensitivity and elevated insulin levels. HiGH mice also have a mild elevation in the GH/IGF1 axis and provide a means to understand the role of the GH/IGF1 axis within more physiological levels than transgenic GH mice, which have extraordinarily high levels of GH.
Thyrocyte-specific IGF1R KO mice
Origin In 2011, Müller et al. generated thyrocyte-specific IGF-1R KO mice using Cre driven by the thyroid-specific thyroglobulin promoter/enhancer [310]. Mice lacking one or two alleles of the Igf1r (Igf1r+/− or Igf1r-/-) were characterized.
Phenotype These mice have no difference in thyroid weights; however, both Igf1rflox/wt and Igf1rflox/flox mice exhibit a more abnormally large thyroid follicles than controls [310]. They also have a greater number of papillary structures resembling papillary thyroid hyperplasia, increased thyroid-stimulating hormone (TSH) levels, and normal thyroid hormone synthesis. Igf1rflox/flox males exhibit increases in body weight consistent with latent hypothyroidism. Conversely, the weights of female Igf1rflox/flox remain lower compared to WT. There is also a sex- and age-dependent alteration in perigonadal fat mass. Both Igf1rflox/wt and Igf1rflox/flox mice of both sexes retain normal glucose tolerance, though male Igf1rflox/flox experience lower insulin resistance. Overall, specific ablation of IGF1R in thyrocytes does not affect thyroid hormones synthesis, but it does affect thyroid homeostasis and systemic alterations in metabolism.
IGF Binding Proteins (IGFBPs) and Acid Labile Subunit (ALS)
IGFBPs transgenic and knockout mice
In circulation and in tissues, most IGF molecules are bound by one of the six distinct members of the IGF-binding protein family (IGFBP) designated as IGFBP1 through IGFBP6 [311]. IGFBPs bind to IGF molecules with high affinity, regulating their bioavailability and functions. In addition, several IGFBPs have been reported to have cellular actions that are independent of their IGF binding. To determine the specific function of each IGFBP in vivo, different mouse lines have been generated. Mice with IGFBP 1, 2, 3, 4, and 5 expressed as transgenes or knockouts have been reported. For IGFBP6, data are only published for transgenic mice. Due to the overlapping functions of IGFBPs, some phenotypes of transgenic or null mice are mild. More details for the mice described in this section are provided in Table 4.
Human IGFBP1 transgenic and IGFBP1-/- mice
(i) IGFBP1 transgenic mice
Origin The hIGFBP1 transgenic mice were created by D’Ercole et al. in 1995 in which gene expression was controlled by the mouse Mt1 promoter/enhancer [312]. The inserted transgene was a full-length human IGFBP1 (hIGFBP1) cDNA, which was truncated at the 3’ untranslated (3’UT) region.
Phenotype IGFBP1 is expressed ubiquitously in these animals [312] whereas liver is the major site of expression in nontransgenic mice [313]. Transgenic hIGFBP1 mice have lower body weight, smaller brains, as well as smaller and sometimes dysmorphic bone structure [312, 314]. Mice display insulin resistance in skeletal muscle, hyperglycemia and impaired glucose tolerance with advancing age [315], reduced fertility in the female mice due to changes in follicular growth [316], and increased extracellular matrix deposition and glomerulosclerosis in kidneys [317].
(ii) IGFBP1-/- mice
Origin Igfbp1-/- mice, reported by the Taub laboratory in 2003, used a SpeI restriction enzyme to insert a NeoR gene that disrupted the Igfbp1 gene [318].
Phenotype These null mice present with increased serum IGF1 levels that normalizes by 4-months of age [318]. These mice show no major alterations in their metabolic phenotype or insulin sensitivity. When crossed with c-Myc transgenic mice to induce prostate cancer, there is no significant difference in the incidence of cancer though the prostate tumor size tends to be smaller, and proliferation is decreased [319]. Overall, the impact of lost IGFBP1 action appears to be minimal.
IGFBP2 transgenic and IGFBP2-/- mice
(i) IGFBP2 transgenic mice
Origin IGFBP2 transgenic mice were generated via the CMV promoter/enhancer fused to the Igfbp2 cDNA by Hoeflich et al. in 1999 [320].
Phenotype The CMV transcriptional regulatory region is known to direct expression in multiple cell types with transgene expression being highest in the pancreas and stomach, whereas IGFBP2 is normally produced primarily in the liver and kidneys in adult nontransgenic mice [313]. IGFBP2 transgenic mice display no changes in circulating levels of GH or IGF1, total body weight, or bone size; however, a reduction in body length and a significant increase in fat mass in males is observed [321]. These mice also have reduced serum insulin levels and increased insulin sensitivity as well as lower systolic blood pressure [321, 322]. When subjected to HFD, IGFBP2 mice are more resistant to obesity when compared to WT controls and have decreased leptin levels, increased glucose sensitivity, and lower blood pressure. Surprisingly, IGFBP2 transgene expression has a protective effect against colon cancer due to decreased cell proliferation and is protective against metabolic diseases [321, 322].
(ii) IGFBP2-/- mice
Origin Igfbp2-/- mice were generated by the Pintar group in 2000 via deletion of exon 3 in the Igfbp2 gene [323].
Phenotype Igfbp2-/- mice have no noticeable changes in circulating GH, IGF1 or body weight when compared with WT controls [323]. However, organ specific differences are observed, with a notable increase in liver size and a decrease in the size of the spleen, heart, and kidneys. There are no differences in insulin sensitivity, other metabolic parameters or fertility. Blocking IGFBP2 action can improve cancer outcomes, at least in mice susceptible to glioblastoma due to reduced immunosuppression caused by IGFBP2 [324]. This indicates that IGFBP2 has tissue dependent effects on susceptibility to cancers although a comprehensive analysis of cancer incidence for Igfbp2-/- mice are not reported.
Human IGFBP3 transgenic and IGFBP3-/- mice
(i) IGFBP3 transgenic mice
Origin IGFBP3 transgenic mice were generated by Murphy et al. in 1995 using a Mt1 promoter/enhancer and the cDNA of the human IGFBP3 transgene [325]. IGFBP3 transgenic mice created using the CMV promotor/enhancer or the phosphoglycerate kinase (PGK) promoter/enhancer have also been reported (called CMVBP-3 and PGKBP-3 mice) [326].
Phenotype IGFBP3 is normally expressed predominantly in the kidneys of adult nontransgenic mice and is also highest in the kidneys of transgenic mice as well [313, 325]. Transgenic mouse using Mt1 promotor/enhancer exhibits greater spleen, liver, heart, and fat weight. Other detectable changes include reduced alveoli size and a significant age-related decrease in pancreatic beta cell mass [313, 325, 327]. Although not assessed in IGFBP3 mice made with the Mt1 promoter/enhancer, other IGFBP3 transgenic mice (CMVBP-3 or PGKBP-3) show no change in fertility for males or females [326].
(ii) IGFBP3-/- mice
Origin Igfbp3-/- mice were generated by the laboratory of Pintar in 2006 [328] using a NeoR cassette inserted between exon 1 and 3 of the Igfbp3 gene.
Phenotype These mice have no change in body weight or size [328, 329]. The initial analysis of these animals reveals a decreased metabolic rate and reduced plasma triglyceride and adiponectin levels [329]. When challenged with a HFD, Igfbp3-/- animals also maintain hepatic insulin sensitivity despite impaired fasting glucose. Null mice show an increase in lung tumorigenesis due to IGFBP3’s influence on IGF1 signaling [330]. Changes in reproductive capabilities are not reported. In summary, gene disruption of IGFBP3 creates a mouse that has some positive effects on metabolism, but a negative effect as it relates to at least one type of cancer.
IGFBP4 transgenic and IGFBP4-/- mice
(i) IGFBP4 transgenic mice
Origin In 1998 and as first reported by Fagin’s group, IGFBP4 transgenic mice were generated via microinjection of murine Igfbp4 cDNA cloned downstream of α-actin 5′-flanking region [331].
Phenotype IGFBP4 is expressed mainly in the adult kidney, liver, and spleen of nontransgenic mice although transgene expression is highest in the bladder and the aorta of transgenic animals [313, 331]. The expression of the transgene negatively affects cellular proliferation in lymphoid tissues although total lymphocyte development is not inhibited. Further, growth of the thymus is limited via the increased stimulation of apoptosis in the thymocytes. No data about the fertility of IGFBP4 transgenic mice are reported.
(ii) IGFBP4-/- mice
Origin Igfbp4-/- mice were also generated by Pintar’s group in 2006 [328].
Phenotype These mice have no significant changes in metabolic parameters or serum levels of any other IGFBPs or IGF1. Although these mice exhibit a standard growth rate in later life, they never catch up to achieve full WT size [332] and have reductions in fat mass, total body length and femur length [333, 334]. Igfbp4-/- mice are reproductively viable, though pups show decreased growth in utero and are 10–15% smaller than WT controls through 14 weeks of age.
IGFBP5 transgenic and IGFBP5-/- mice
(i) IGFBP5 transgenic mice
Origin Expression of IGFBP5 in transgenic mice was directed to the mammary gland via the β-lactoglobulin promoter/enhancer. This mouse line was first reported by Tonner et al. in 2002 [335]. WT mice normally have highest IGFBP5 expression in the kidney, muscle, ovaries among other tissues [313].
Phenotype After birth, IGFBP5 transgenic mice exhibit a decrease in total body weight as compared to controls. A reduction in total muscle mass and a transient decrease in bone volume and mineral density through 8 weeks of age is reported [336, 337]. Females show reduced fertility with an increase in the mortality of neonates.
(ii) IGFBP5-/- mice
Origin In 2006, Pintar’s group reported on Igfbp5-/- mice, which were achieved by insertion of a NeoR cassette into exon 1 of the Igfbp5 gene [328].
Phenotype Null mice have a similar total body size, with a modest increase in lung weight [338], an increase in adiposity, a mild glucose intolerance and increased susceptibility to diet-induced obesity as compared to controls [339]. No impact on fertility of null mice is reported.
IGFBP6 transgenic mice
Origin IGFBP6 transgenic mice were developed by Bienvenu et al. in 2004 using human IGFBP6 cDNA with a glial fibrillary acidic protein (GFAP) promoter/enhancer [340]. Igfbp6-/- mice have not been reported.
Phenotype Transgene expression in these mice is high in the CNS [340], while nontransgenic mice normally express the highest levels of IGFBP6 in the lungs and heart with variable amounts in other tissues [313]. Transgenic mice exhibit increased levels of IGFBP6 between 3 and 15 days of age, along with decreased detected plasma IGF1 levels at 15 days [340]. This change is transient with IGF1 levels being the same as the WT group at both 1 and 3 months of age. Mice have reduced litter size, with sterility in 5–20% of females. They also exhibit growth retardation as neonates through three months of age. These mice have a reduction in the size of the cerebellum [340] along with cerebellar abnormalities although changes in cognition or behavior are not reported. With diet-induced obesity, these mice develop mild insulin resistance and obesity. They also show a decrease in brown AT UCP-1 expression, along with an increase in plasma levels of glucose, insulin, and leptin [341].
ALS transgenic and ALS-/- mice (or Igfals-/-)
Origin The acid-labile subunit (ALS) is component of the IGF1 ternary complex along with IGF1 and IGFBP3, mediating the stability and bioavailability of IGF1. ALS transgenic mice were generated in 2001 by a group led by Murphy using the ALS cDNA driven by the CMV promoter/enhancer [342]. Igfals-/- mice were reported in 2000 by the Boisclair group and were made by replacing the Igfals gene with a neomycin phosphotransferase gene [343].
Phenotype ALS transgenic mice have decreased body size but no change in circulating IGF1 or IGFBP3 levels [342]. Therefore, it is hypothesized that the decreased body size is due to altered tissue availability of IGF1. The authors also report decreased litter size in ALS transgenic mothers, suggested to be due to IGF1’s role in ovarian follicular development. Igfals-/- mice have decreased serum IGF1 and IGFBP3 levels, with a corresponding decrease in body size [343]. Interestingly, there is no significant change in Igf1 or Igfbp3 mRNA expression, indicating that the decrease in protein level is due to decreased stability in the serum, as is often seen when IGFs are not sequestered in ternary complexes. A later study published by Yakar’s group in 2010 reported the same decrease in body size and serum IGF1 levels but with an additional skeletal phenotype [344], i.e. Igfals-/- mice had a sex- and age-dependent decrease in the periosteum formation around the femur leading to decreased bone formation. This decrease in bone thickness is compensated with an increase in the endosteal surface inside the bone that covers the bone marrow. Thus, the outer layer of the bones becomes thinner but the inner layer increases in thickness. Overall, it appears that disruption of ALS in mice decreases size in mice and affects skeletal shape.
PAPP-A-/- (Pregnancy-associated plasma protein-A)
Pregnancy-associated plasma protein A (PAPP-A) modulates the activity and bioavailability of IGF1 by cleaving IGFBP2, IGFBP4, and IGFBP5. Importantly, while IGFBP2 and IGFPB5 may be cleaved by other proteases, the proteolysis of IGFBP4 seems to be limited to PAPP-A [345].
Origin Germline Pappa-/- (PAPP-A-/- or PAPP-A-null) mice were reported in 2004 by Conover et al. [346]. To evaluate the phenotype of PAPP-A disruption at an adult age, a separate mouse line was reported by Bale et al., in 2017 [346]. This mouse line was generated using the tamoxifen-inducible Cre/LoxP system in which the Pappa gene was disrupted at five months of age (fPAPP-A/pos).
Phenotype Pappa-/- mice are dwarf with a 40% body size reduction and have compromised fertility, with an 80% reduction in litter size [346, 347]. Pappa-/- mice have normal circulating GH and IGF1 levels compared to controls. They also have significantly increased longevity, with males showing a 33% and females a 41% lifespan extension [348]. Although these mice do not show changes in glucose metabolism, food intake, and total energy expenditure and resting energy [349], they do show a decrease in the prevalence and severity of age-related diseases, such as cardiomyopathy, nephropathy, and cancer [346, 350, 351]. Treadmill experiments provide evidence of improved skeletal muscle function with a decrease in fatigue and an increase in endurance in PAPP-A-null mice [352]. Postnatal ablation of the Pappa gene in fPAPP-A/pos mice results in a significant extension of lifespan with an increase in median lifespan of 21% compared to control mice [351, 353]. As tamoxifen can induce scrotal enlargement and subsequent complications in male mice, only female mice are used in the longevity study. Thus, although germline disruption of Pappa shows positive results in terms of aging and some age-related diseases in both male and female mice, it is unknown if adult disruption of the Pappa gene will also lead to lifespan extension in male mice. It is interesting to note that disruption of GHR at an adult age also leads to lifespan extension in females [60]. As both strategies – PAPP-A and GHR ablation – lead to reduced IGF1 action, it is of interest to test if the PAPP-A system is also regulated by GH. To that end, AT of GHR-/- and bGH mice show no change in gene expression of Igfbp4 and Pappa when compared to WT mice, although protein levels of IGFBP4 are increased in bGH mice compared to GHR-/- mice. Furthermore, the C terminal-IGFBP4 fragment, which is generated after PAPP-A-cleavage, does not differ among bGH, GHR-/-, and WT mice [354]. Despite no change in AT, PAPP-A is expressed in different tissues [345]; thus, it is possible that PAPP-A expression and activity is modulated by GH in other tissues.
Concluding remarks
The above review characterizes 137 mouse strains in which genes in the GH/IGF1 family have been altered. A few of these alterations are via ‘natural gene mutations;’ however, the majority are via genetic manipulations, namely (1) generation of transgenic mice that express a gene or cDNA encoding a component of the family or (2) disruption of specific genes within the family. A summary of the published data is presented, including authors, date of publication, corresponding references as well as salient physiological consequences of the gene alterations. Also, we had a ‘personal laboratory reason’ for generating this review; namely, we often need a consolidated document that can be easily used and referenced when preparing new manuscripts that refer to subsets of these mice. The reader is encouraged to use this review in a similar manner and, when doing so, a few new tidbits of information may be acquired. Although 137 different mouse lines are described, we are sure we missed some, and for that we apologize. Also, we tried to extract the salient physiological points from the published data when describing these mice. Again, if we missed some of these points or mistakenly described them, we are very sorry. Finally, I (JJK) am indebted to the authors of this review, which includes Ohio University faculty, staff of the Edison Biotechnology Institute, post-doctoral fellows, graduate students, undergraduate students, medical students, and technicians. Without their effort, this review would not have been accomplished.
Acknowledgements
We thank Delaney Geitgey, Jaycie Kuhn, Savannah McKenna, Cole Smith, and Grace Lach for their support and contribution to this work. Most of this review was written during the pandemic. Thus, we dedicate it to those who suffered with, fought against, or died from COVID-19.
Funding
This work was supported, in part, by the State of Ohio’s Eminent Scholar Program that includes a gift from Milton and Lawrence Goll to J.J.K., NIH-R01AG059779.
Declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
The authors of this review have abided by and are compliant with all typical ethical standards.
Research involving human and animal rights
This review only documents mice with alterations in the GH/IGF family. Thus, no research was discussed or cited related to humans. Also, we believe that each research group involved in generation of the various mouse strains used animal use and care procedures that were approved by their individual university/hospital/institute.
Data availability
Not applicable.
Code availability
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Evans HM, Long JA. The effect of the anterior lobe administered intraperitoneally upon growth, maturity and estrous cycles of the rat. Anat Rec. 1921;21:62–63. [Google Scholar]
- 2.Buchman M, Bell S, Kopchick JJ. Growth hormone discovery and structure. Pediatr Endocrinol Rev. 2018;16(Suppl 1):2–10. doi: 10.17458/per.vol16.2018.bbk.ghdiscoverystructure. [DOI] [PubMed] [Google Scholar]
- 3.Houssay BA. The hypophysis and metabolism. N Engl J Med. 1936;214:961–971. doi: 10.1056/nejm193605142142001. [DOI] [Google Scholar]
- 4.Li CH, Evans HM. The isolation of pituitary growth hormone. Science. 1944;99(2566):183–184. doi: 10.1126/science.99.2566.183. [DOI] [PubMed] [Google Scholar]
- 5.Li CH, Papkoff H. Preparation and properties of growth hormone from human and monkey pituitary glands. Science. 1956;124(3235):1293–1294. doi: 10.1126/science.124.3235.1293. [DOI] [PubMed] [Google Scholar]
- 6.Frasier SD. The not-so-good old days: working with pituitary growth hormone in North America, 1956 to 1985. J Pediatr. 1997;131(1 Pt 2):S1–4. doi: 10.1016/s0022-3476(97)70001-5. [DOI] [PubMed] [Google Scholar]
- 7.Flodh H. Human growth hormone produced with recombinant DNA technology: development and production. Acta Paediatr Scand Suppl. 1986;325:1–9. doi: 10.1111/j.1651-2227.1986.tb10356.x. [DOI] [PubMed] [Google Scholar]
- 8.Laron Z. The era of cadaveric pituitary extracted human growth hormone (1958–1985): biological and clinical aspects. Pediatr Endocrinol Rev. 2018;16(Suppl 1):11–16. doi: 10.17458/per.vol16.2018.la.hghcadavericpituitary. [DOI] [PubMed] [Google Scholar]
- 9.Raben MS. Treatment of a pituitary dwarf with human growth hormone. J Clin Endocrinol Metab. 1958;18(8):901–903. doi: 10.1210/jcem-18-8-901. [DOI] [PubMed] [Google Scholar]
- 10.Ranke MB, Wit JM. Growth hormone—past, present and future. Nat Rev Endocrinol. 2018;14(5):285–300. doi: 10.1038/nrendo.2018.22. [DOI] [PubMed] [Google Scholar]
- 11.Barsh GS, Seeburg PH, Gelinas RE. The human growth hormone gene family: structure and evolution of the chromosomal locus. Nucleic Acids Res. 1983;11(12):3939–3958. doi: 10.1093/nar/11.12.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fiddes JC, Seeburg PH, DeNoto FM, Hallewell RA, Baxter JD, Goodman HM. Structure of genes for human growth hormone and chorionic somatomammotropin. Proc Natl Acad Sci USA. 1979;76(9):4294–4298. doi: 10.1073/pnas.76.9.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dehkhoda F, Lee CMM, Medina J, Brooks AJ. The growth hormone receptor: mechanism of receptor activation, cell signaling, and physiological aspects. Front Endocrinol (Lausanne) 2018;9:35. doi: 10.3389/fendo.2018.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brooks AJ, Dai W, O'Mara ML, Abankwa D, Chhabra Y, Pelekanos RA, Gardon O, Tunny KA, Blucher KM, Morton CJ, Parker MW, Sierecki E, Gambin Y, Gomez GA, Alexandrov K, Wilson IA, Doxastakis M, Mark AE, Waters MJ. Mechanism of activation of protein kinase JAK2 by the growth hormone receptor. Science. 2014;344(6185):1249783. doi: 10.1126/science.1249783. [DOI] [PubMed] [Google Scholar]
- 15.van den Eijnden MJ, Lahaye LL, Strous GJ. Disulfide bonds determine growth hormone receptor folding, dimerisation and ligand binding. J Cell Sci. 2006;119(Pt 15):3078–3086. doi: 10.1242/jcs.03036. [DOI] [PubMed] [Google Scholar]
- 16.Waters MJ, Brooks AJ. JAK2 activation by growth hormone and other cytokines. Biochem J. 2015;466(1):1–11. doi: 10.1042/bj20141293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ross RJ, Leung KC, Maamra M, Bennett W, Doyle N, Waters MJ, Ho KK. Binding and functional studies with the growth hormone receptor antagonist, B2036-PEG (pegvisomant), reveal effects of pegylation and evidence that it binds to a receptor dimer. J Clin Endocrinol Metab. 2001;86(4):1716–1723. doi: 10.1210/jcem.86.4.7403. [DOI] [PubMed] [Google Scholar]
- 18.Hansen LH, Wang X, Kopchick JJ, Bouchelouche P, Nielsen JH, Galsgaard ED, Billestrup N. Identification of tyrosine residues in the intracellular domain of the growth hormone receptor required for transcriptional signaling and Stat5 activation. J Biol Chem. 1996;271(21):12669–12673. doi: 10.1074/jbc.271.21.12669. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Darus CJ, Xu BC, Kopchick JJ. Identification of growth hormone receptor (GHR) tyrosine residues required for GHR phosphorylation and JAK2 and STAT5 activation. Mol Endocrinol. 1996;10(10):1249–1260. doi: 10.1210/mend.10.10.9121492. [DOI] [PubMed] [Google Scholar]
- 20.Xu BC, Wang X, Darus CJ, Kopchick JJ. Growth hormone promotes the association of transcription factor STAT5 with the growth hormone receptor. J Biol Chem. 1996;271(33):19768–19773. doi: 10.1074/jbc.271.33.19768. [DOI] [PubMed] [Google Scholar]
- 21.Aaronson DS, Horvath CM. A road map for those who don't know JAK-STAT. Science. 2002;296(5573):1653–1655. doi: 10.1126/science.1071545. [DOI] [PubMed] [Google Scholar]
- 22.Metcalf D, Greenhalgh CJ, Viney E, Willson TA, Starr R, Nicola NA, Hilton DJ, Alexander WS. Gigantism in mice lacking suppressor of cytokine signalling-2. Nature. 2000;405(6790):1069–1073. doi: 10.1038/35016611. [DOI] [PubMed] [Google Scholar]
- 23.Linossi EM, Babon JJ, Hilton DJ, Nicholson SE. Suppression of cytokine signaling: the SOCS perspective. Cytokine Growth Factor Rev. 2013;24(3):241–248. doi: 10.1016/j.cytogfr.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kershaw NJ, Murphy JM, Liau NP, Varghese LN, Laktyushin A, Whitlock EL, Lucet IS, Nicola NA, Babon JJ. SOCS3 binds specific receptor-JAK complexes to control cytokine signaling by direct kinase inhibition. Nat Struct Mol Biol. 2013;20(4):469–476. doi: 10.1038/nsmb.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, Birnberg NC, Evans RM. Dramatic growth of mice that develop from eggs microinjected with metallothionein-growth hormone fusion genes. Nature. 1982;300(5893):611–615. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen WY, Wight DC, Wagner TE, Kopchick JJ. Expression of a mutated bovine growth hormone gene suppresses growth of transgenic mice. Proc Natl Acad Sci USA. 1990;87(13):5061–5065. doi: 10.1073/pnas.87.13.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen WY, Wight DC, Mehta BV, Wagner TE, Kopchick JJ. Glycine 119 of bovine growth hormone is critical for growth-promoting activity. Mol Endocrinol. 1991;5(12):1845–1852. doi: 10.1210/mend-5-12-1845. [DOI] [PubMed] [Google Scholar]
- 28.Okada S, Chen WY, Wiehl P, Kelder B, Goodman HM, Guller S, Sonenberg M, Kopchick JJ. A growth hormone (GH) analog can antagonize the ability of native GH to promote differentiation of 3T3-F442A preadipocytes and stimulate insulin-like and lipolytic activities in primary rat adipocytes. Endocrinology. 1992;130(4):2284–2290. doi: 10.1210/endo.130.4.1547740. [DOI] [PubMed] [Google Scholar]
- 29.Cunningham BC, Ultsch M, De Vos AM, Mulkerrin MG, Clauser KR, Wells JA. Dimerization of the extracellular domain of the human growth hormone receptor by a single hormone molecule. Science. 1991;254(5033):821–825. doi: 10.1126/science.1948064. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y, Xu BC, Maheshwari HG, He L, Reed M, Lozykowski M, Okada S, Cataldo L, Coschigamo K, Wagner TE, Baumann G, Kopchick JJ. A mammalian model for Laron syndrome produced by targeted disruption of the mouse growth hormone receptor/binding protein gene (the Laron mouse) Proc Natl Acad Sci USA. 1997;94(24):13215–13220. doi: 10.1073/pnas.94.24.13215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.List EO, Sackmann-Sala L, Berryman DE, Funk K, Kelder B, Gosney ES, Okada S, Ding J, Cruz-Topete D, Kopchick JJ. Endocrine parameters and phenotypes of the growth hormone receptor gene disrupted (GHR-/-) mouse. Endocr Rev. 2011;32(3):356–386. doi: 10.1210/er.2010-0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Berryman DE, List EO, Kohn DT, Coschigano KT, Seeley RJ, Kopchick JJ. Effect of growth hormone on susceptibility to diet-induced obesity. Endocrinology. 2006;147(6):2801–2808. doi: 10.1210/en.2006-0086. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Prins GS, Coschigano KT, Kopchick JJ, Green JE, Ray VH, Hedayat S, Christov KT, Unterman TG, Swanson SM. Disruption of growth hormone signaling retards early stages of prostate carcinogenesis in the C3(1)/T antigen mouse. Endocrinology. 2005;146(12):5188–5196. doi: 10.1210/en.2005-0607. [DOI] [PubMed] [Google Scholar]
- 34.Zhang X, Mehta RG, Lantvit DD, Coschigano KT, Kopchick JJ, Green JE, Hedayat S, Christov KT, Ray VH, Unterman TG, Swanson SM. Inhibition of estrogen-independent mammary carcinogenesis by disruption of growth hormone signaling. Carcinogenesis. 2007;28(1):143–150. doi: 10.1093/carcin/bgl138. [DOI] [PubMed] [Google Scholar]
- 35.Ikeno Y, Hubbard GB, Lee S, Cortez LA, Lew CM, Webb CR, Berryman DE, List EO, Kopchick JJ, Bartke A. Reduced incidence and delayed occurrence of fatal neoplastic diseases in growth hormone receptor/binding protein knockout mice. J Gerontol A. 2009;64(5):522–529. doi: 10.1093/gerona/glp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Basu R, Qian Y, Kopchick JJ. MECHANISMS IN ENDOCRINOLOGY: lessons from growth hormone receptor gene-disrupted mice: are there benefits of endocrine defects? Eur J Endocrinol. 2018;178(5):R155–R181. doi: 10.1530/eje-18-0018. [DOI] [PubMed] [Google Scholar]
- 37.Coschigano KT, Holland AN, Riders ME, List EO, Flyvbjerg A, Kopchick JJ. Deletion, but not antagonism, of the mouse growth hormone receptor results in severely decreased body weights, insulin, and insulin-like growth factor I levels and increased life span. Endocrinology. 2003;144(9):3799–3810. doi: 10.1210/en.2003-0374. [DOI] [PubMed] [Google Scholar]
- 38.Fan Y, Menon RK, Cohen P, Hwang D, Clemens T, DiGirolamo DJ, Kopchick JJ, Le Roith D, Trucco M, Sperling MA. Liver-specific deletion of the growth hormone receptor reveals essential role of growth hormone signaling in hepatic lipid metabolism. J Biol Chem. 2009;284(30):19937–19944. doi: 10.1074/jbc.M109.014308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lu C, Kumar PA, Fan Y, Sperling MA, Menon RK. A novel effect of growth hormone on macrophage modulates macrophage-dependent adipocyte differentiation. Endocrinology. 2010;151(5):2189–2199. doi: 10.1210/en.2009-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mavalli MD, DiGirolamo DJ, Fan Y, Riddle RC, Campbell KS, van Groen T, Frank SJ, Sperling MA, Esser KA, Bamman MM, Clemens TL. Distinct growth hormone receptor signaling modes regulate skeletal muscle development and insulin sensitivity in mice. J Clin Invest. 2010;120(11):4007–4020. doi: 10.1172/jci42447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Liu C, Sun H, Vijayakumar A, Giglou PR, Qiao R, Oppenheimer J, Yakar S, LeRoith D. Growth hormone receptor regulates beta cell hyperplasia and glucose-stimulated insulin secretion in obese mice. J Clin Invest. 2011;121(6):2422–2426. doi: 10.1172/jci45027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vijayakumar A, Wu Y, Sun H, Li X, Jeddy Z, Liu C, Schwartz GJ, Yakar S, LeRoith D. Targeted loss of GHR signaling in mouse skeletal muscle protects against high-fat diet-induced metabolic deterioration. Diabetes. 2012;61(1):94–103. doi: 10.2337/db11-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vijayakumar A, Wu Y, Buffin NJ, Li X, Sun H, Gordon RE, Yakar S, LeRoith D. Skeletal muscle growth hormone receptor signaling regulates basal, but not fasting-induced, lipid oxidation. PLoS ONE. 2012;7(9):e44777. doi: 10.1371/journal.pone.0044777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.List EO, Berryman DE, Funk K, Gosney ES, Jara A, Kelder B, Wang X, Kutz L, Troike K, Lozier N, Mikula V, Lubbers ER, Zhang H, Vesel C, Junnila RK, Frank SJ, Masternak MM, Bartke A, Kopchick JJ. The role of GH in adipose tissue: lessons from adipose-specific GH receptor gene-disrupted mice. Mol Endocrinol. 2013;27(3):524–535. doi: 10.1210/me.2012-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vijayakumar A, Buffin NJ, Gallagher EJ, Blank J, Wu Y, Yakar S, LeRoith D. Deletion of growth hormone receptors in postnatal skeletal muscle of male mice does not alter muscle mass and response to pathological injury. Endocrinology. 2013;154(10):3776–3783. doi: 10.1210/en.2013-1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X, Bartke A, Berryman DE, Funk K, Kopchick JJ, List EO, Sun L, Miller RA. Direct and indirect effects of growth hormone receptor ablation on liver expression of xenobiotic metabolizing genes. Am J Physiol Endocrinol Metab. 2013;305(8):E942–950. doi: 10.1152/ajpendo.00304.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu C, Kumar PA, Sun J, Aggarwal A, Fan Y, Sperling MA, Lumeng CN, Menon RK. Targeted deletion of growth hormone (GH) receptor in macrophage reveals novel osteopontin-mediated effects of GH on glucose homeostasis and insulin sensitivity in diet-induced obesity. J Biol Chem. 2013;288(22):15725–15735. doi: 10.1074/jbc.M113.460212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.List EO, Berryman DE, Funk K, Jara A, Kelder B, Wang F, Stout MB, Zhi X, Sun L, White TA, LeBrasseur NK, Pirtskhalava T, Tchkonia T, Jensen EA, Zhang W, Masternak MM, Kirkland JL, Miller RA, Bartke A, Kopchick JJ. Liver-specific GH receptor gene-disrupted (LiGHRKO) mice have decreased endocrine IGF-I, increased local IGF-I, and altered body size, body composition, and adipokine profiles. Endocrinology. 2014;155(5):1793–1805. doi: 10.1210/en.2013-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan Y, Fang X, Tajima A, Geng X, Ranganathan S, Dong H, Trucco M, Sperling MA. Evolution of hepatic steatosis to fibrosis and adenoma formation in liver-specific growth hormone receptor knockout mice. Front Endocrinol (Lausanne) 2014;5:218. doi: 10.3389/fendo.2014.00218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stewart MH, Gutierrez-Martinez P, Beerman I, Garrison B, Gallagher EJ, LeRoith D, Rossi DJ. Growth hormone receptor signaling is dispensable for HSC function and aging. Blood. 2014;124(20):3076–3080. doi: 10.1182/blood-2014-05-575308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gesing A, Wang F, List EO, Berryman DE, Masternak MM, Lewinski A, Karbownik-Lewinska M, Kopchick JJ, Bartke A. Expression of apoptosis-related genes in liver-specific growth hormone receptor gene-disrupted mice is sex dependent. J Gerontol A. 2015;70(1):44–52. doi: 10.1093/gerona/glu008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dominick G, Berryman DE, List EO, Kopchick JJ, Li X, Miller RA, Garcia GG. Regulation of mTOR activity in Snell dwarf and GH receptor gene-disrupted mice. Endocrinology. 2015;156(2):565–575. doi: 10.1210/en.2014-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zawada I, Masternak MM, List EO, Stout MB, Berryman DE, Lewinski A, Kopchick JJ, Bartke A, Karbownik-Lewinska M, Gesing A. Gene expression of key regulators of mitochondrial biogenesis is sex dependent in mice with growth hormone receptor deletion in liver. Aging (Albany NY) 2015;7(3):195–204. doi: 10.18632/aging.100733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.List EO, Berryman DE, Ikeno Y, Hubbard GB, Funk K, Comisford R, Young JA, Stout MB, Tchkonia T, Masternak MM, Bartke A, Kirkland JL, Miller RA, Kopchick JJ. Removal of growth hormone receptor (GHR) in muscle of male mice replicates some of the health benefits seen in global GHR-/- mice. Aging (Albany NY) 2015;7(7):500–512. doi: 10.18632/aging.100766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cordoba-Chacon J, Majumdar N, List EO, Diaz-Ruiz A, Frank SJ, Manzano A, Bartrons R, Puchowicz M, Kopchick JJ, Kineman RD. Growth hormone inhibits hepatic de novo lipogenesis in adult mice. Diabetes. 2015;64(9):3093–3103. doi: 10.2337/db15-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sadagurski M, Landeryou T, Cady G, Kopchick JJ, List EO, Berryman DE, Bartke A, Miller RA. Growth hormone modulates hypothalamic inflammation in long-lived pituitary dwarf mice. Aging Cell. 2015;14(6):1045–1054. doi: 10.1111/acel.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Y, Knop E, Knop N, Sullivan DA, List EO, Kopchick JJ, Kam WR, Ding J. Growth hormone influence on the morphology and size of the mouse meibomian gland. J Ophthalmol. 2016;2016:5728071. doi: 10.1155/2016/5728071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jara A, Liu X, Sim D, Benner CM, Duran-Ortiz S, Qian Y, List EO, Berryman DE, Kim JK, Kopchick JJ. Cardiac-specific disruption of GH receptor alters glucose homeostasis while maintaining normal cardiac performance in adult male mice. Endocrinology. 2016;157(5):1929–1941. doi: 10.1210/en.2015-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, Cordoba-Chacon J, Kineman RD, Cronstein BN, Muzumdar R, Gong Z, Werner H, Yakar S. Growth hormone control of hepatic lipid metabolism. Diabetes. 2016;65(12):3598–3609. doi: 10.2337/db16-0649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Junnila RK, Duran-Ortiz S, Suer O, Sustarsic EG, Berryman DE, List EO, Kopchick JJ. Disruption of the GH receptor gene in adult mice increases maximal lifespan in females. Endocrinology. 2016;157(12):4502–4513. doi: 10.1210/en.2016-1649. [DOI] [PubMed] [Google Scholar]
- 61.Cady G, Landeryou T, Garratt M, Kopchick JJ, Qi N, Garcia-Galiano D, Elias CF, Myers MG, Jr, Miller RA, Sandoval DA, Sadagurski M. Hypothalamic growth hormone receptor (GHR) controls hepatic glucose production in nutrient-sensing leptin receptor (LepRb) expressing neurons. Mol Metab. 2017;6(5):393–405. doi: 10.1016/j.molmet.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.List EO, Berryman DE, Jensen EA, Kulkarni P, McKenna S, Kopchick JJ. New insights of growth hormone (GH) actions from tissue-specific GH receptor knockouts in mice. Arch Endocrinol Metab. 2019;63(6):557–567. doi: 10.20945/2359-3997000000185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fang F, Shi X, Brown MS, Goldstein JL, Liang G. Growth hormone acts on liver to stimulate autophagy, support glucose production, and preserve blood glucose in chronically starved mice. Proc Natl Acad Sci USA. 2019;116(15):7449–7454. doi: 10.1073/pnas.1901867116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Furigo IC, Teixeira PDS, de Souza GO, Couto GCL, Romero GG, Perello M, Frazao R, Elias LL, Metzger M, List EO, Kopchick JJ, Donato J., Jr Growth hormone regulates neuroendocrine responses to weight loss via AgRP neurons. Nat Commun. 2019;10(1):662. doi: 10.1038/s41467-019-08607-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Young JA, Jensen EA, Stevens A, Duran-Ortiz S, List EO, Berryman DE, Kopchick JJ. Characterization of an intestine-specific GH receptor knockout (IntGHRKO) mouse. Growth Horm IGF Res. 2019;46–47:5–15. doi: 10.1016/j.ghir.2019.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kato Y, Murakami Y, Sohmiya M, Nishiki M. Regulation of human growth hormone secretion and its disorders. Intern Med. 2002;41(1):7–13. doi: 10.2169/internalmedicine.41.7. [DOI] [PubMed] [Google Scholar]
- 67.Snell GD. Dwarf, a new mendelian recessive character of the house mouse. Proc Natl Acad Sci USA. 1929;15(9):733–734. doi: 10.1073/pnas.15.9.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li S, Crenshaw EB, 3rd, Rawson EJ, Simmons DM, Swanson LW, Rosenfeld MG. Dwarf locus mutants lacking three pituitary cell types result from mutations in the POU-domain gene pit-1. Nature. 1990;347(6293):528–533. doi: 10.1038/347528a0. [DOI] [PubMed] [Google Scholar]
- 69.Brown-Borg HM, Bartke A. GH and IGF1: roles in energy metabolism of long-living GH mutant mice. J Gerontol A. 2012;67(6):652–660. doi: 10.1093/gerona/gls086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Flurkey K, Papaconstantinou J, Miller RA, Harrison DE. Lifespan extension and delayed immune and collagen aging in mutant mice with defects in growth hormone production. Proc Natl Acad Sci USA. 2001;98(12):6736–6741. doi: 10.1073/pnas.111158898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bartke A. The response of two types of dwarf mice to growth hormone, thyrotropin, and thyroxine. Gen Comp Endocrinol. 1965;5(4):418–426. doi: 10.1016/0016-6480(65)90102-4. [DOI] [PubMed] [Google Scholar]
- 72.Levine R, Luft R. The relation between the growth and diabetogenic effects of the so-called growth hormone of the anterior pituitary. Diabetes. 1964;13:651–655. doi: 10.2337/diab.13.6.651. [DOI] [PubMed] [Google Scholar]
- 73.Bratusch-Marrain PR, Smith D, DeFronzo RA. The effect of growth hormone on glucose metabolism and insulin secretion in man. J Clin Endocrinol Metab. 1982;55(5):973–982. doi: 10.1210/jcem-55-5-973. [DOI] [PubMed] [Google Scholar]
- 74.Kim SH, Park MJ. Effects of growth hormone on glucose metabolism and insulin resistance in human. Ann Pediatr Endocrinol Metab. 2017;22(3):145–152. doi: 10.6065/apem.2017.22.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vila G, Jorgensen JOL, Luger A, Stalla GK. Insulin resistance in patients with acromegaly. Front Endocrinol (Lausanne) 2019;10:509. doi: 10.3389/fendo.2019.00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Guevara-Aguirre J, Balasubramanian P, Guevara-Aguirre M, Wei M, Madia F, Cheng CW, Hwang D, Martin-Montalvo A, Saavedra J, Ingles S, de Cabo R, Cohen P, Longo VD. Growth hormone receptor deficiency is associated with a major reduction in pro-aging signaling, cancer, and diabetes in humans. Sci Transl Med. 2011;3(70):70ra13. doi: 10.1126/scitranslmed.3001845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brooks NL, Trent CM, Raetzsch CF, Flurkey K, Boysen G, Perfetti MT, Jeong YC, Klebanov S, Patel KB, Khodush VR, Kupper LL, Carling D, Swenberg JA, Harrison DE, Combs TP. Low utilization of circulating glucose after food withdrawal in Snell dwarf mice. J Biol Chem. 2007;282(48):35069–35077. doi: 10.1074/jbc.M700484200. [DOI] [PubMed] [Google Scholar]
- 78.Tallaksen-Greene SJ, Sadagurski M, Zeng L, Mauch R, Perkins M, Banduseela VC, Lieberman AP, Miller RA, Paulson HL, Albin RL. Differential effects of delayed aging on phenotype and striatal pathology in a murine model of Huntington disease. J Neurosci. 2014;34(47):15658–15668. doi: 10.1523/JNEUROSCI.1830-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Vergara M, Smith-Wheelock M, Harper JM, Sigler R, Miller RA. Hormone-treated snell dwarf mice regain fertility but remain long lived and disease resistant. J Gerontol A. 2004;59(12):1244–1250. doi: 10.1093/gerona/59.12.1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rader EP, Naimo MA, Ensey J, Baker BA. VCAM-1 upregulation accompanies muscle remodeling following resistance-type exercise in Snell dwarf (Pit1(dw/dw) ) mice. Aging Cell. 2018;17(5):e12816. doi: 10.1111/acel.12816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hochereau-de Reviers MT, de Reviers MM, Monet-Kuntz C, Perreau C, Fontaine I, Viguier-Martinez MC. Testicular growth and hormonal parameters in the male Snell dwarf mouse. Acta Endocrinol (Copenh) 1987;115(3):399–405. doi: 10.1530/acta.0.1150399. [DOI] [PubMed] [Google Scholar]
- 82.Karolyi IJ, Dootz GA, Halsey K, Beyer L, Probst FJ, Johnson KR, Parlow AF, Raphael Y, Dolan DF, Camper SA. Dietary thyroid hormone replacement ameliorates hearing deficits in hypothyroid mice. Mamm Genome. 2007;18(8):596–608. doi: 10.1007/s00335-007-9038-0. [DOI] [PubMed] [Google Scholar]
- 83.Schaible R, Gowen JW. A new dwarf mouse. Abstr. Genetics. 1961;46:896. [Google Scholar]
- 84.Bartke A, Brown-Borg H. Life extension in the dwarf mouse. Curr Top Dev Biol. 2004;63:189–225. doi: 10.1016/s0070-2153(04)63006-7. [DOI] [PubMed] [Google Scholar]
- 85.Wang Z, Al-Regaiey KA, Masternak MM, Bartke A. Adipocytokines and lipid levels in Ames dwarf and calorie-restricted mice. J Gerontol A. 2006;61(4):323–331. doi: 10.1093/gerona/61.4.323. [DOI] [PubMed] [Google Scholar]
- 86.Dominici FP, Hauck S, Argentino DP, Bartke A, Turyn D. Increased insulin sensitivity and upregulation of insulin receptor, insulin receptor substrate (IRS)-1 and IRS-2 in liver of Ames dwarf mice. J Endocrinol. 2002;173(1):81–94. doi: 10.1677/joe.0.1730081. [DOI] [PubMed] [Google Scholar]
- 87.Borg KE, Brown-Borg HM, Bartke A. Assessment of the primary adrenal cortical and pancreatic hormone basal levels in relation to plasma glucose and age in the unstressed Ames dwarf mouse. Proc Soc Exp Biol Med. 1995;210(2):126–133. doi: 10.3181/00379727-210-43931. [DOI] [PubMed] [Google Scholar]
- 88.Hill CM, Fang Y, Miquet JG, Sun LY, Masternak MM, Bartke A. Long-lived hypopituitary Ames dwarf mice are resistant to the detrimental effects of high-fat diet on metabolic function and energy expenditure. Aging Cell. 2016;15(3):509–521. doi: 10.1111/acel.12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Darcy J, McFadden S, Fang Y, Huber JA, Zhang C, Sun LY, Bartke A. Brown adipose tissue function is enhanced in long-lived. Male Ames Dwarf Mice Endocrinol. 2016;157(12):4744–4753. doi: 10.1210/en.2016-1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hunter WS, Croson WB, Bartke A, Gentry MV, Meliska CJ. Low body temperature in long-lived Ames dwarf mice at rest and during stress. Physiol Behav. 1999;67(3):433–437. doi: 10.1016/s0031-9384(99)00098-0. [DOI] [PubMed] [Google Scholar]
- 91.Darcy J, McFadden S, Bartke A. Altered structure and function of adipose tissue in long-lived mice with growth hormone-related mutations. Adipocyte. 2017;6(2):69–75. doi: 10.1080/21623945.2017.1308990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bokov AF, Lindsey ML, Khodr C, Sabia MR, Richardson A. Long-lived ames dwarf mice are resistant to chemical stressors. J Gerontol A. 2009;64(8):819–827. doi: 10.1093/gerona/glp052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ikeno Y, Bronson RT, Hubbard GB, Lee S, Bartke A. Delayed occurrence of fatal neoplastic diseases in ames dwarf mice: correlation to extended longevity. J Gerontol A. 2003;58(4):291–296. doi: 10.1093/gerona/58.4.b291. [DOI] [PubMed] [Google Scholar]
- 94.Bartke A, Coschigano K, Kopchick J, Chandrashekar V, Mattison J, Kinney B, Hauck S. Genes that prolong life: relationships of growth hormone and growth to aging and life span. J Gerontol A. 2001;56(8):B340–349. doi: 10.1093/gerona/56.8.b340. [DOI] [PubMed] [Google Scholar]
- 95.Brown-Borg HM, Borg KE, Meliska CJ, Bartke A. Dwarf mice and the ageing process. Nature. 1996;384(6604):33. doi: 10.1038/384033a0. [DOI] [PubMed] [Google Scholar]
- 96.Bartke A, Wright JC, Mattison JA, Ingram DK, Miller RA, Roth GS. Extending the lifespan of long-lived mice. Nature. 2001;414(6862):412. doi: 10.1038/35106646. [DOI] [PubMed] [Google Scholar]
- 97.Ikeno Y, Hubbard GB, Lee S, Dube SM, Flores LC, Roman MG, Bartke A. Do Ames dwarf and calorie-restricted mice share common effects on age-related pathology? Pathobiol Aging Age Relat Dis. 2013 doi: 10.3402/pba.v3i0.20833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chandrashekar V, Bartke A. Induction of endogenous insulin-like growth factor-I secretion alters the hypothalamic-pituitary-testicular function in growth hormone-deficient adult dwarf mice. Biol Reprod. 1993;48(3):544–551. doi: 10.1095/biolreprod48.3.544. [DOI] [PubMed] [Google Scholar]
- 99.Moore JP, Jr, Morrison DG, Hyde JF. Galanin gene expression is increased in the anterior pituitary gland of the human growth hormone-releasing hormone transgenic mouse. Endocrinology. 1994;134(5):2005–2010. doi: 10.1210/endo.134.5.7512494. [DOI] [PubMed] [Google Scholar]
- 100.Peng XD, Park S, Gadelha MR, Coschigano KT, Kopchick JJ, Frohman LA, Kineman RD. The growth hormone (GH)-axis of GH receptor/binding protein gene-disrupted and metallothionein-human GH-releasing hormone transgenic mice: hypothalamic neuropeptide and pituitary receptor expression in the absence and presence of GH feedback. Endocrinology. 2001;142(3):1117–1123. doi: 10.1210/endo.142.3.8005. [DOI] [PubMed] [Google Scholar]
- 101.Asa SL, Kovacs K, Stefaneanu L, Horvath E, Billestrup N, Gonzalez-Manchon C, Vale W. Pituitary mammosomatotroph adenomas develop in old mice transgenic for growth hormone-releasing hormone. Proc Soc Exp Biol Med. 1990;193(3):232–235. doi: 10.3181/00379727-193-3-rc1. [DOI] [PubMed] [Google Scholar]
- 102.Asa SL, Kovacs K, Stefaneanu L, Horvath E, Billestrup N, Gonzalez-Manchon C, Vale W. Pituitary adenomas in mice transgenic for growth hormone-releasing hormone. Endocrinology. 1992;131(5):2083–2089. doi: 10.1210/endo.131.5.1425411. [DOI] [PubMed] [Google Scholar]
- 103.Donangelo I, Melmed S. Implication of pituitary tropic status on tumor development. Front Horm Res. 2006;35:1–8. doi: 10.1159/000094259. [DOI] [PubMed] [Google Scholar]
- 104.Luque RM, Soares BS, Peng XD, Krishnan S, Cordoba-Chacon J, Frohman LA, Kineman RD. Use of the metallothionein promoter-human growth hormone-releasing hormone (GHRH) mouse to identify regulatory pathways that suppress pituitary somatotrope hyperplasia and adenoma formation due to GHRH-receptor hyperactivation. Endocrinology. 2009;150(7):3177–3185. doi: 10.1210/en.2008-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Debeljuk L, Wright JC, Phelps C, Bartke A. Transgenic mice overexpressing the growth-hormone-releasing hormone gene have high concentrations of tachykinins in the anterior pituitary gland. Neuroendocrinology. 1999;70(2):107–116. doi: 10.1159/000054465. [DOI] [PubMed] [Google Scholar]
- 106.Hyde JF, Moore JP, Cai A (1998) Galanin in normal and hyperplastic anterior pituitary cells. From pituitary tumor cell lines to transgenic mice. Ann N Y Acad Sci 863:48–55. 10.1111/j.1749-6632.1998.tb10682.x [DOI] [PubMed]
- 107.Hyde JF, Morrison DG, Drake KW, Moore JP, Jr, Maley BE. Vasoactive intestinal polypeptide mRNA and peptide levels are decreased in the anterior pituitary of the human growth hormone-releasing hormone transgenic mouse. J Neuroendocrinol. 1996;8(1):9–15. doi: 10.1111/j.1365-2826.1996.tb00681.x. [DOI] [PubMed] [Google Scholar]
- 108.Alba M, Salvatori R. A mouse with targeted ablation of the growth hormone-releasing hormone gene: a new model of isolated growth hormone deficiency. Endocrinology. 2004;145(9):4134–4143. doi: 10.1210/en.2004-0119. [DOI] [PubMed] [Google Scholar]
- 109.Fintini D, Alba M, Schally AV, Bowers CY, Parlow AF, Salvatori R. Effects of combined long-term treatment with a growth hormone-releasing hormone analogue and a growth hormone secretagogue in the growth hormone-releasing hormone knock out mouse. Neuroendocrinology. 2005;82(3–4):198–207. doi: 10.1159/000092520. [DOI] [PubMed] [Google Scholar]
- 110.Recinella L, Shohreh R, Salvatori R, Orlando G, Vacca M, Brunetti L. Effects of isolated GH deficiency on adipose tissue, feeding and adipokines in mice. Growth Horm IGF Res. 2013;23(6):237–242. doi: 10.1016/j.ghir.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 111.Leone S, Chiavaroli A, Shohreh R, Ferrante C, Ricciuti A, Manippa F, Recinella L, Di Nisio C, Orlando G, Salvatori R, Vacca M, Brunetti L. Increased locomotor and thermogenic activity in mice with targeted ablation of the GHRH gene. Growth Horm IGF Res. 2015;25(2):80–84. doi: 10.1016/j.ghir.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 112.Bartke A, Westbrook R. Metabolic characteristics of long-lived mice. Front Genet. 2012;3:288. doi: 10.3389/fgene.2012.00288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sun LY, Spong A, Swindell WR, Fang Y, Hill C, Huber JA, Boehm JD, Westbrook R, Salvatori R, Bartke A. Growth hormone-releasing hormone disruption extends lifespan and regulates response to caloric restriction in mice. Elife. 2013;2:e01098. doi: 10.7554/eLife.01098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Matzkin ME, Miquet JG, Fang Y, Hill CM, Turyn D, Calandra RS, Bartke A, Frungieri MB. Alterations in oxidative, inflammatory and apoptotic events in short-lived and long-lived mice testes. Aging (Albany NY) 2016;8(1):95–110. doi: 10.18632/aging.100875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Beamer WH, Eicher EM. Stimulation of growth in the little mouse. J Endocrinol. 1976;71(1):37–45. doi: 10.1677/joe.0.0710037. [DOI] [PubMed] [Google Scholar]
- 116.Eicher EM, Beamer WG. Inherited ateliotic dwarfism in mice. Characteristics of the mutation, little, on chromosome 6. J Hered. 1976;67(2):87–91. doi: 10.1093/oxfordjournals.jhered.a108682. [DOI] [PubMed] [Google Scholar]
- 117.Godfrey P, Rahal JO, Beamer WG, Copeland NG, Jenkins NA, Mayo KE. GHRH receptor of little mice contains a missense mutation in the extracellular domain that disrupts receptor function. Nat Genet. 1993;4(3):227–232. doi: 10.1038/ng0793-227. [DOI] [PubMed] [Google Scholar]
- 118.Liang H, Masoro EJ, Nelson JF, Strong R, McMahan CA, Richardson A. Genetic mouse models of extended lifespan. Exp Gerontol. 2003;38(11–12):1353–1364. doi: 10.1016/j.exger.2003.10.019. [DOI] [PubMed] [Google Scholar]
- 119.Donahue LR, Beamer WG. Growth hormone deficiency in 'little' mice results in aberrant body composition, reduced insulin-like growth factor-I and insulin-like growth factor-binding protein-3 (IGFBP-3), but does not affect IGFBP-2, -1 or -4. J Endocrinol. 1993;136(1):91–104. doi: 10.1677/joe.0.1360091. [DOI] [PubMed] [Google Scholar]
- 120.Deitel K, Dantzer D, Ferguson P, Pollak M, Beamer W, Andrulis I, Bell R. Reduced growth of human sarcoma xenografts in hosts homozygous for the lit mutation. J Surg Oncol. 2002;81(2):75–79. doi: 10.1002/jso.10136. [DOI] [PubMed] [Google Scholar]
- 121.Takahara K, Tearle H, Ghaffari M, Gleave ME, Pollak M, Cox ME. Human prostate cancer xenografts in lit/lit mice exhibit reduced growth and androgen-independent progression. Prostate. 2011;71(5):525–537. doi: 10.1002/pros.21268. [DOI] [PubMed] [Google Scholar]
- 122.Yang XF, Beamer WG, Huynh H, Pollak M. Reduced growth of human breast cancer xenografts in hosts homozygous for the lit mutation. Cancer Res. 1996;56(7):1509–1511. [PubMed] [Google Scholar]
- 123.Mohan S, Richman C, Guo R, Amaar Y, Donahue LR, Wergedal J, Baylink DJ. Insulin-like growth factor regulates peak bone mineral density in mice by both growth hormone-dependent and -independent mechanisms. Endocrinology. 2003;144(3):929–936. doi: 10.1210/en.2002-220948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Palmiter RD, Norstedt G, Gelinas RE, Hammer RE, Brinster RL. Metallothionein-human GH fusion genes stimulate growth of mice. Science. 1983;222(4625):809–814. doi: 10.1126/science.6356363. [DOI] [PubMed] [Google Scholar]
- 125.Wolf E, Kahnt E, Ehrlein J, Hermanns W, Brem G, Wanke R. Effects of long-term elevated serum levels of growth hormone on life expectancy of mice: lessons from transgenic animal models. Mech Ageing Dev. 1993;68(1–3):71–87. doi: 10.1016/0047-6374(93)90141-d. [DOI] [PubMed] [Google Scholar]
- 126.Bartke A, Steger RW, Hodges SL, Parkening TA, Collins TJ, Yun JS, Wagner TE. Infertility in transgenic female mice with human growth hormone expression: evidence for luteal failure. J Exp Zool. 1988;248(1):121–124. doi: 10.1002/jez.1402480116. [DOI] [PubMed] [Google Scholar]
- 127.Hurley DL, Bartke A, Wagner TE, Wee BE, Phelps CJ. Increased hypothalamic somatostatin expression in mice transgenic for bovine or human GH. J Neuroendocrinol. 1994;6(5):539–548. doi: 10.1111/j.1365-2826.1994.tb00617.x. [DOI] [PubMed] [Google Scholar]
- 128.Selden RF, Wagner TE, Blethen S, Yun JS, Rowe ME, Goodman HM. Expression of the human growth hormone variant gene in cultured fibroblasts and transgenic mice. Proc Natl Acad Sci USA. 1988;85(21):8241–8245. doi: 10.1073/pnas.85.21.8241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Dudley GA, Portanova R. Histochemical characteristics of soleus muscle in hGH transgenic mice. Proc Soc Exp Biol Med. 1987;185(4):403–408. doi: 10.3181/00379727-185-42561. [DOI] [PubMed] [Google Scholar]
- 130.Bartke A, Naar EM, Johnson L, May MR, Cecim M, Yun JS, Wagner TE. Effects of expression of human or bovine growth hormone genes on sperm production and male reproductive performance in four lines of transgenic mice. J Reprod Fertil. 1992;95(1):109–118. doi: 10.1530/jrf.0.0950109. [DOI] [PubMed] [Google Scholar]
- 131.Tornell J, Carlsson B, Pohjanen P, Wennbo H, Rymo L, Isaksson O. High frequency of mammary adenocarcinomas in metallothionein promoter-human growth hormone transgenic mice created from two different strains of mice. J Steroid Biochem Mol Biol. 1992;43(1–3):237–242. doi: 10.1016/0960-0760(92)90213-3. [DOI] [PubMed] [Google Scholar]
- 132.Tornell J, Rymo L, Isaksson OG. Induction of mammary adenocarcinomas in metallothionein promoter-human growth hormone transgenic mice. Int J Cancer. 1991;49(1):114–117. doi: 10.1002/ijc.2910490121. [DOI] [PubMed] [Google Scholar]
- 133.Wennbo H, Gebre-Medhin M, Gritli-Linde A, Ohlsson C, Isaksson OG, Tornell J. Activation of the prolactin receptor but not the growth hormone receptor is important for induction of mammary tumors in transgenic mice. J Clin Invest. 1997;100(11):2744–2751. doi: 10.1172/jci119820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Wennbo H, Tornell J. The role of prolactin and growth hormone in breast cancer. Oncogene. 2000;19(8):1072–1076. doi: 10.1038/sj.onc.1203349. [DOI] [PubMed] [Google Scholar]
- 135.Jin Y, Lu SY, Fresnoza A, Detillieux KA, Duckworth ML, Cattini PA. Differential placental hormone gene expression during pregnancy in a transgenic mouse containing the human growth hormone/chorionic somatomammotropin locus. Placenta. 2009;30(3):226–235. doi: 10.1016/j.placenta.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Vakili H, Jin Y, Nagy JI, Cattini PA. Transgenic mice expressing the human growth hormone gene provide a model system to study human growth hormone synthesis and secretion in non-tumor-derived pituitary cells: differential effects of dexamethasone and thyroid hormone. Mol Cell Endocrinol. 2011;345(1–2):48–57. doi: 10.1016/j.mce.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 137.Jarmasz JS, Jin Y, Vakili H, Cattini PA. Sleep deprivation and diet affect human GH gene expression in transgenic mice in vivo. Endocr Connect. 2020;9(12):1135–1147. doi: 10.1530/EC-20-0354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Vakili H, Jin Y, Cattini PA. Evidence for a circadian effect on the reduction of human growth hormone gene expression in response to excess caloric intake. J Biol Chem. 2016;291(26):13823–13833. doi: 10.1074/jbc.M116.722744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Gunzburg WH, Salmons B, Zimmermann B, Muller M, Erfle V, Brem G. A mammary-specific promoter directs expression of growth hormone not only to the mammary gland, but also to Bergman glia cells in transgenic mice. Mol Endocrinol. 1991;5(1):123–133. doi: 10.1210/mend-5-1-123. [DOI] [PubMed] [Google Scholar]
- 140.Devinoy E, Thepot D, Stinnakre MG, Fontaine ML, Grabowski H, Puissant C, Pavirani A, Houdebine LM. High level production of human growth hormone in the milk of transgenic mice: the upstream region of the rabbit whey acidic protein (WAP) gene targets transgene expression to the mammary gland. Transgenic Res. 1994;3(2):79–89. doi: 10.1007/bf01974085. [DOI] [PubMed] [Google Scholar]
- 141.Bartke A, Kopchick JJ. The forgotten lactogenic activity of growth hormone: important implications for rodent studies. Endocrinology. 2015;156(5):1620–1622. doi: 10.1210/en.2015-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Naraoka H, Ito K, Suzuki M, Naito K, Tojo H. Analysis of gender difference of cardiac risk biomarkers using hGH-transgenic mice. Exp Anim. 2006;55(1):1–9. doi: 10.1538/expanim.55.1. [DOI] [PubMed] [Google Scholar]
- 143.Thepot D, Devinoy E, Fontaine ML, Stinnakre MG, Massoud M, Kann G, Houdebine LM. Rabbit whey acidic protein gene upstream region controls high-level expression of bovine growth hormone in the mammary gland of transgenic mice. Mol Reprod Dev. 1995;42(3):261–267. doi: 10.1002/mrd.1080420302. [DOI] [PubMed] [Google Scholar]
- 144.Naar EM, Bartke A, Majumdar SS, Buonomo FC, Yun JS, Wagner TE. Fertility of transgenic female mice expressing bovine growth hormone or human growth hormone variant genes. Biol Reprod. 1991;45(1):178–187. doi: 10.1095/biolreprod45.1.178. [DOI] [PubMed] [Google Scholar]
- 145.Hammer RE, Brinster RL, Palmiter RD. Use of gene transfer to increase animal growth. Cold Spring Harb Symp Quant Biol. 1985;50:379–387. doi: 10.1101/sqb.1985.050.01.048. [DOI] [PubMed] [Google Scholar]
- 146.Palmer AJ, Chung MY, List EO, Walker J, Okada S, Kopchick JJ, Berryman DE. Age-related changes in body composition of bovine growth hormone transgenic mice. Endocrinology. 2009;150(3):1353–1360. doi: 10.1210/en.2008-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Asa SL, Coschigano KT, Bellush L, Kopchick JJ, Ezzat S. Evidence for growth hormone (GH) autoregulation in pituitary somatotrophs in GH antagonist-transgenic mice and GH receptor-deficient mice. Am J Pathol. 2000;156(3):1009–1015. doi: 10.1016/s0002-9440(10)64968-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Berryman DE, List EO, Coschigano KT, Behar K, Kim JK, Kopchick JJ. Comparing adiposity profiles in three mouse models with altered GH signaling. Growth Horm IGF Res. 2004;14(4):309–318. doi: 10.1016/j.ghir.2004.02.005. [DOI] [PubMed] [Google Scholar]
- 149.Bollano E, Omerovic E, Bohlooly-y M, Kujacic V, Madhu B, Tornell J, Isaksson O, Soussi B, Schulze W, Fu ML, Matejka G, Waagstein F, Isgaard J. Impairment of cardiac function and bioenergetics in adult transgenic mice overexpressing the bovine growth hormone gene. Endocrinology. 2000;141(6):2229–2235. doi: 10.1210/endo.141.6.7486. [DOI] [PubMed] [Google Scholar]
- 150.Ding J, Berryman DE, Kopchick JJ. Plasma proteomic profiles of bovine growth hormone transgenic mice as they age. Transgenic Res. 2011;20(6):1305–1320. doi: 10.1007/s11248-011-9499-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hjortebjerg R, Berryman DE, Comisford R, Frank SJ, List EO, Bjerre M, Frystyk J, Kopchick JJ. Insulin, IGF-1, and GH receptors are altered in an adipose tissue depot-specific manner in male mice with modified GH action. Endocrinology. 2017;158(5):1406–1418. doi: 10.1210/en.2017-00084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Olsson B, Bohlooly YM, Fitzgerald SM, Frick F, Ljungberg A, Ahren B, Tornell J, Bergstrom G, Oscarsson J. Bovine growth hormone transgenic mice are resistant to diet-induced obesity but develop hyperphagia, dyslipidemia, and diabetes on a high-fat diet. Endocrinology. 2005;146(2):920–930. doi: 10.1210/en.2004-1232. [DOI] [PubMed] [Google Scholar]
- 153.Wang Z, Masternak MM, Al-Regaiey KA, Bartke A. Adipocytokines and the regulation of lipid metabolism in growth hormone transgenic and calorie-restricted mice. Endocrinology. 2007;148(6):2845–2853. doi: 10.1210/en.2006-1313. [DOI] [PubMed] [Google Scholar]
- 154.Quaife CJ, Mathews LS, Pinkert CA, Hammer RE, Brinster RL, Palmiter RD. Histopathology associated with elevated levels of growth hormone and insulin-like growth factor I in transgenic mice. Endocrinology. 1989;124(1):40–48. doi: 10.1210/endo-124-1-40. [DOI] [PubMed] [Google Scholar]
- 155.McGrane MM, de Vente J, Yun J, Bloom J, Park E, Wynshaw-Boris A, Wagner T, Rottman FM, Hanson RW. Tissue-specific expression and dietary regulation of a chimeric phosphoenolpyruvate carboxykinase/bovine growth hormone gene in transgenic mice. J Biol Chem. 1988;263(23):11443–11451. doi: 10.1016/S0021-9258(18)37977-8. [DOI] [PubMed] [Google Scholar]
- 156.McGrane MM, Yun JS, Moorman AF, Lamers WH, Hendrick GK, Arafah BM, Park EA, Wagner TE, Hanson RW. Metabolic effects of developmental, tissue-, and cell-specific expression of a chimeric phosphoenolpyruvate carboxykinase (GTP)/bovine growth hormone gene in transgenic mice. J Biol Chem. 1990;265(36):22371–22379. doi: 10.1016/S0021-9258(18)45715-8. [DOI] [PubMed] [Google Scholar]
- 157.Ogueta S, Olazabal I, Santos I, Delgado-Baeza E, Garcia-Ruiz JP. Transgenic mice expressing bovine GH develop arthritic disorder and self-antibodies. J Endocrinol. 2000;165(2):321–328. doi: 10.1677/joe.0.1650321. [DOI] [PubMed] [Google Scholar]
- 158.Junnila RK, List EO, Berryman DE, Murrey JW, Kopchick JJ. The GH/IGF-1 axis in ageing and longevity. Nat Rev Endocrinol. 2013;9(6):366–376. doi: 10.1038/nrendo.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bartke A. Can growth hormone (GH) accelerate aging? Evidence from GH-transgenic mice. Neuroendocrinology. 2003;78(4):210–216. doi: 10.1159/000073704. [DOI] [PubMed] [Google Scholar]
- 160.Doi T, Striker LJ, Quaife C, Conti FG, Palmiter R, Behringer R, Brinster R, Striker GE. Progressive glomerulosclerosis develops in transgenic mice chronically expressing growth hormone and growth hormone releasing factor but not in those expressing insulinlike growth factor-1. Am J Pathol. 1988;131(3):398–403. [PMC free article] [PubMed] [Google Scholar]
- 161.Martinez CS, Piazza VG, Gonzalez L, Fang Y, Bartke A, Turyn D, Miquet JG, Sotelo AI. Mitogenic signaling pathways in the liver of growth hormone (GH)-overexpressing mice during the growth period. Cell Cycle. 2016;15(5):748–759. doi: 10.1080/15384101.2016.1148844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bacigalupo ML, Piazza VG, Cicconi NS, Carabias P, Bartke A, Fang Y, Sotelo AI, Rabinovich GA, Troncoso MF, Miquet JG. Growth hormone upregulates the pro-tumorigenic galectin 1 in mouse liver. Endocr Connect. 2019;8(8):1108–1117. doi: 10.1530/ec-19-0292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Miquet JG, Freund T, Martinez CS, Gonzalez L, Diaz ME, Micucci GP, Zotta E, Boparai RK, Bartke A, Turyn D, Sotelo AI. Hepatocellular alterations and dysregulation of oncogenic pathways in the liver of transgenic mice overexpressing growth hormone. Cell Cycle. 2013;12(7):1042–1057. doi: 10.4161/cc.24026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Snibson KJ, Bhathal PS, Hardy CL, Brandon MR, Adams TE. High, persistent hepatocellular proliferation and apoptosis precede hepatocarcinogenesis in growth hormone transgenic mice. Liver. 1999;19(3):242–252. doi: 10.1111/j.1478-3231.1999.tb00042.x. [DOI] [PubMed] [Google Scholar]
- 165.Snibson KJ. Hepatocellular kinetics and the expression of growth hormone (GH) in the livers and liver tumours of GH-transgenic mice. Tissue Cell. 2002;34(2):88–97. doi: 10.1016/s0040-8166(02)00012-5. [DOI] [PubMed] [Google Scholar]
- 166.Kong X, Wu W, Yuan Y, Pandey V, Wu Z, Lu X, Zhang W, Chen Y, Wu M, Zhang M, Li G, Tan S, Qian P, Perry JK, Lobie PE, Zhu T. Human growth hormone and human prolactin function as autocrine/paracrine promoters of progression of hepatocellular carcinoma. Oncotarget. 2016;7(20):29465–29479. doi: 10.18632/oncotarget.8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.List EO, Berryman DE, Buchman M, Jensen EA, Funk K, Duran-Ortiz S, Qian Y, Young JA, Slyby J, McKenna S, Kopchick JJ. GH Knockout Mice have increased subcutaneous adipose tissue with decreased fibrosis and enhanced insulin sensitivity. Endocrinology. 2019;160(7):1743–1756. doi: 10.1210/en.2019-00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Luque RM, Lin Q, Cordoba-Chacon J, Subbaiah PV, Buch T, Waisman A, Vankelecom H, Kineman RD. Metabolic impact of adult-onset, isolated, growth hormone deficiency (AOiGHD) due to destruction of pituitary somatotropes. PLoS ONE. 2011;6(1):e15767. doi: 10.1371/journal.pone.0015767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Kopchick JJ, McAndrew SJ, Shafer A, Blue WT, Yun JS, Wagner TE, Chen WY. In-vitro mutagenesis of the bovine growth hormone gene. J Reprod Fertil Suppl. 1990;41:25–35. [PubMed] [Google Scholar]
- 170.Chen WY, White ME, Wagner TE, Kopchick JJ. Functional antagonism between endogenous mouse growth hormone (GH) and a GH analog results in dwarf transgenic mice. Endocrinology. 1991;129(3):1402–1408. doi: 10.1210/endo-129-3-1402. [DOI] [PubMed] [Google Scholar]
- 171.Wang YD, Wood WI. Amino acids of the human growth hormone receptor that are required for proliferation and Jak-STAT signaling. Mol Endocrinol. 1995;9(3):303–311. doi: 10.1210/mend.9.3.7539888. [DOI] [PubMed] [Google Scholar]
- 172.Chen WY, Chen NY, Yun J, Wagner TE, Kopchick JJ. In vitro and in vivo studies of antagonistic effects of human growth hormone analogs. J Biol Chem. 1994;269(22):15892–15897. doi: 10.1016/S0021-9258(17)40764-2. [DOI] [PubMed] [Google Scholar]
- 173.Kopchick JJ. Discovery and mechanism of action of pegvisomant. Eur J Endocrinol. 2003;148(Suppl 2):S21–25. doi: 10.1530/eje.0.148s021. [DOI] [PubMed] [Google Scholar]
- 174.Comisford R, Lubbers ER, Householder LA, Suer O, Tchkonia T, Kirkland JL, List EO, Kopchick JJ, Berryman DE. Growth hormone receptor antagonist transgenic mice have increased subcutaneous adipose tissue mass, altered glucose homeostasis and no change in white adipose tissue cellular senescence. Gerontology. 2016;62(2):163–172. doi: 10.1159/000439050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Li Y, Knapp JR, Kopchick JJ. Enlargement of interscapular brown adipose tissue in growth hormone antagonist transgenic and in growth hormone receptor gene-disrupted dwarf mice. Exp Biol Med (Maywood) 2003;228(2):207–215. doi: 10.1177/153537020322800212. [DOI] [PubMed] [Google Scholar]
- 176.Yang T, Householder LA, Lubbers ER, List EO, Troike K, Vesel C, Duran-Ortiz S, Kopchick JJ, Berryman DE. Growth hormone receptor antagonist transgenic mice are protected from hyperinsulinemia and glucose intolerance despite obesity when placed on a HF diet. Endocrinology. 2015;156(2):555–564. doi: 10.1210/en.2014-1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Bernard V, Young J, Binart N. Prolactin—a pleiotropic factor in health and disease. Nat Rev Endocrinol. 2019;15(6):356–365. doi: 10.1038/s41574-019-0194-6. [DOI] [PubMed] [Google Scholar]
- 178.Freeman ME, Kanyicska B, Lerant A, Nagy G. Prolactin: structure, function, and regulation of secretion. Physiol Rev. 2000;80(4):1523–1631. doi: 10.1152/physrev.2000.80.4.1523. [DOI] [PubMed] [Google Scholar]
- 179.Enjalbert A, Bertrand P, Le Dafniet M, Epelbaum J, Hugues JN, Kordon C, Moyse E, Peillon F, Shu C. Somatostatin and regulation of prolactin secretion. Psychoneuroendocrinology. 1986;11(2):155–165. doi: 10.1016/0306-4530(86)90050-8. [DOI] [PubMed] [Google Scholar]
- 180.Willoughby JO, Jervois PM, Menadue MF, Blessing WW. Activation of GABA receptors in the hypothalamus stimulates secretion of growth hormone and prolactin. Brain Res. 1986;374(1):119–125. doi: 10.1016/0006-8993(86)90400-2. [DOI] [PubMed] [Google Scholar]
- 181.Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19(3):225–268. doi: 10.1210/edrv.19.3.0334. [DOI] [PubMed] [Google Scholar]
- 182.Goffin V, Bernichtein S, Touraine P, Kelly PA. Development and potential clinical uses of human prolactin receptor antagonists. Endocr Rev. 2005;26(3):400–422. doi: 10.1210/er.2004-0016. [DOI] [PubMed] [Google Scholar]
- 183.Wennbo H, Kindblom J, Isaksson OG, Tornell J. Transgenic mice overexpressing the prolactin gene develop dramatic enlargement of the prostate gland. Endocrinology. 1997;138(10):4410–4415. doi: 10.1210/endo.138.10.5461. [DOI] [PubMed] [Google Scholar]
- 184.Kindblom J, Dillner K, Sahlin L, Robertson F, Ormandy C, Tornell J, Wennbo H. Prostate hyperplasia in a transgenic mouse with prostate-specific expression of prolactin. Endocrinology. 2003;144(6):2269–2278. doi: 10.1210/en.2002-0187. [DOI] [PubMed] [Google Scholar]
- 185.Tworoger SS, Hankinson SE. Prolactin and breast cancer etiology: an epidemiologic perspective. J Mammary Gland Biol Neoplasia. 2008;13(1):41–53. doi: 10.1007/s10911-008-9063-y. [DOI] [PubMed] [Google Scholar]
- 186.Rose-Hellekant TA, Arendt LM, Schroeder MD, Gilchrist K, Sandgren EP, Schuler LA. Prolactin induces ERalpha-positive and ERalpha-negative mammary cancer in transgenic mice. Oncogene. 2003;22(30):4664–4674. doi: 10.1038/sj.onc.1206619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Arendt LM, Rugowski DE, Grafwallner-Huseth TA, Garcia-Barchino MJ, Rui H, Schuler LA. Prolactin-induced mouse mammary carcinomas model estrogen resistant luminal breast cancer. Breast Cancer Res. 2011;13(1):R11. doi: 10.1186/bcr2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Horseman ND, Zhao W, Montecino-Rodriguez E, Tanaka M, Nakashima K, Engle SJ, Smith F, Markoff E, Dorshkind K. Defective mammopoiesis, but normal hematopoiesis, in mice with a targeted disruption of the prolactin gene. EMBO J. 1997;16(23):6926–6935. doi: 10.1093/emboj/16.23.6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.LaPensee CR, Horseman ND, Tso P, Brandebourg TD, Hugo ER, Ben-Jonathan N. The prolactin-deficient mouse has an unaltered metabolic phenotype. Endocrinology. 2006;147(10):4638–4645. doi: 10.1210/en.2006-0487. [DOI] [PubMed] [Google Scholar]
- 190.Ormandy CJ, Camus A, Barra J, Damotte D, Lucas B, Buteau H, Edery M, Brousse N, Babinet C, Binart N, Kelly PA. Null mutation of the prolactin receptor gene produces multiple reproductive defects in the mouse. Genes Dev. 1997;11(2):167–178. doi: 10.1101/gad.11.2.167. [DOI] [PubMed] [Google Scholar]
- 191.Kelly PA, Binart N, Lucas B, Bouchard B, Goffin V. Implications of multiple phenotypes observed in prolactin receptor knockout mice. Front Neuroendocrinol. 2001;22(2):140–145. doi: 10.1006/frne.2001.0212. [DOI] [PubMed] [Google Scholar]
- 192.Freemark M, Avril I, Fleenor D, Driscoll P, Petro A, Opara E, Kendall W, Oden J, Bridges S, Binart N, Breant B, Kelly PA. Targeted deletion of the PRL receptor: effects on islet development, insulin production, and glucose tolerance. Endocrinology. 2002;143(4):1378–1385. doi: 10.1210/endo.143.4.8722. [DOI] [PubMed] [Google Scholar]
- 193.Robertson FG, Harris J, Naylor MJ, Oakes SR, Kindblom J, Dillner K, Wennbo H, Tornell J, Kelly PA, Green J, Ormandy CJ. Prostate development and carcinogenesis in prolactin receptor knockout mice. Endocrinology. 2003;144(7):3196–3205. doi: 10.1210/en.2003-0068. [DOI] [PubMed] [Google Scholar]
- 194.Saunier E, Dif F, Kelly PA, Edery M. Targeted expression of the dominant-negative prolactin receptor in the mammary gland of transgenic mice results in impaired lactation. Endocrinology. 2003;144(6):2669–2675. doi: 10.1210/en.2002-221038. [DOI] [PubMed] [Google Scholar]
- 195.Binart N, Imbert-Bollore P, Baran N, Viglietta C, Kelly PA. A short form of the prolactin (PRL) receptor is able to rescue mammopoiesis in heterozygous PRL receptor mice. Mol Endocrinol. 2003;17(6):1066–1074. doi: 10.1210/me.2002-0181. [DOI] [PubMed] [Google Scholar]
- 196.Le JA, Wilson HM, Shehu A, Mao J, Devi YS, Halperin J, Aguilar T, Seibold A, Maizels E, Gibori G. Generation of mice expressing only the long form of the prolactin receptor reveals that both isoforms of the receptor are required for normal ovarian function. Biol Reprod. 2012;86(3):86. doi: 10.1095/biolreprod.111.095927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Bernichtein S, Kayser C, Dillner K, Moulin S, Kopchick JJ, Martial JA, Norstedt G, Isaksson O, Kelly PA, Goffin V. Development of pure prolactin receptor antagonists. J Biol Chem. 2003;278(38):35988–35999. doi: 10.1074/jbc.M305687200. [DOI] [PubMed] [Google Scholar]
- 198.Rouet V, Bogorad RL, Kayser C, Kessal K, Genestie C, Bardier A, Grattan DR, Kelder B, Kopchick JJ, Kelly PA, Goffin V. Local prolactin is a target to prevent expansion of basal/stem cells in prostate tumors. Proc Natl Acad Sci USA. 2010;107(34):15199–15204. doi: 10.1073/pnas.0911651107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Ferraris J, Boutillon F, Bernadet M, Seilicovich A, Goffin V, Pisera D. Prolactin receptor antagonism in mouse anterior pituitary: effects on cell turnover and prolactin receptor expression. Am J Physiol Endocrinol Metab. 2012;302(3):E356–364. doi: 10.1152/ajpendo.00333.2011. [DOI] [PubMed] [Google Scholar]
- 200.Ferraris J, Zarate S, Jaita G, Boutillon F, Bernadet M, Auffret J, Seilicovich A, Binart N, Goffin V, Pisera D. Prolactin induces apoptosis of lactotropes in female rodents. PLoS ONE. 2014;9(5):e97383. doi: 10.1371/journal.pone.0097383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Danilovich N, Wernsing D, Coschigano KT, Kopchick JJ, Bartke A. Deficits in female reproductive function in GH-R-KO mice; role of IGF-I. Endocrinology. 1999;140(6):2637–2640. doi: 10.1210/endo.140.6.6992. [DOI] [PubMed] [Google Scholar]
- 202.Berryman DE, List EO, Palmer AJ, Chung MY, Wright-Piekarski J, Lubbers E, O'Connor P, Okada S, Kopchick JJ. Two-year body composition analyses of long-lived GHR null mice. J Gerontol A. 2010;65(1):31–40. doi: 10.1093/gerona/glp175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Liu JL, Coschigano KT, Robertson K, Lipsett M, Guo Y, Kopchick JJ, Kumar U, Liu YL. Disruption of growth hormone receptor gene causes diminished pancreatic islet size and increased insulin sensitivity in mice. Am J Physiol Endocrinol Metab. 2004;287(3):E405–413. doi: 10.1152/ajpendo.00423.2003. [DOI] [PubMed] [Google Scholar]
- 204.Guo Y, Lu Y, Houle D, Robertson K, Tang Z, Kopchick JJ, Liu YL, Liu JL. Pancreatic islet-specific expression of an insulin-like growth factor-I transgene compensates islet cell growth in growth hormone receptor gene-deficient mice. Endocrinology. 2005;146(6):2602–2609. doi: 10.1210/en.2004-1203. [DOI] [PubMed] [Google Scholar]
- 205.Al-Regaiey KA, Masternak MM, Bonkowski M, Sun L, Bartke A. Long-lived growth hormone receptor knockout mice: interaction of reduced insulin-like growth factor i/insulin signaling and caloric restriction. Endocrinology. 2005;146(2):851–860. doi: 10.1210/en.2004-1120. [DOI] [PubMed] [Google Scholar]
- 206.Egecioglu E, Bjursell M, Ljungberg A, Dickson SL, Kopchick JJ, Bergstrom G, Svensson L, Oscarsson J, Tornell J, Bohlooly YM. Growth hormone receptor deficiency results in blunted ghrelin feeding response, obesity, and hypolipidemia in mice. Am J Physiol Endocrinol Metab. 2006;290(2):E317–325. doi: 10.1152/ajpendo.00181.2005. [DOI] [PubMed] [Google Scholar]
- 207.Lubbers ER, List EO, Jara A, Sackman-Sala L, Cordoba-Chacon J, Gahete MD, Kineman RD, Boparai R, Bartke A, Kopchick JJ, Berryman DE. Adiponectin in mice with altered GH action: links to insulin sensitivity and longevity? J Endocrinol. 2013;216(3):363–374. doi: 10.1530/JOE-12-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Bartke A, Peluso MR, Moretz N, Wright C, Bonkowski M, Winters TA, Shanahan MF, Kopchick JJ, Banz WJ. Effects of Soy-derived diets on plasma and liver lipids, glucose tolerance, and longevity in normal, long-lived and short-lived mice. Horm Metab Res. 2004;36(8):550–558. doi: 10.1055/s-2004-825796. [DOI] [PubMed] [Google Scholar]
- 209.Hauck SJ, Hunter WS, Danilovich N, Kopchick JJ, Bartke A. Reduced levels of thyroid hormones, insulin, and glucose, and lower body core temperature in the growth hormone receptor/binding protein knockout mouse. Exp Biol Med (Maywood) 2001;226(6):552–558. doi: 10.1177/153537020122600607. [DOI] [PubMed] [Google Scholar]
- 210.Longo KA, Berryman DE, Kelder B, Charoenthongtrakul S, Distefano PS, Geddes BJ, Kopchick JJ. Daily energy balance in growth hormone receptor/binding protein (GHR -/-) gene-disrupted mice is achieved through an increase in dark-phase energy efficiency. Growth Horm IGF Res. 2010;20(1):73–79. doi: 10.1016/j.ghir.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Westbrook R, Bonkowski MS, Strader AD, Bartke A. Alterations in oxygen consumption, respiratory quotient, and heat production in long-lived GHRKO and Ames dwarf mice, and short-lived bGH transgenic mice. J Gerontol A Biol Sci Med Sci. 2009;64(4):443–451. doi: 10.1093/gerona/gln075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Kinney BA, Coschigano KT, Kopchick JJ, Steger RW, Bartke A. Evidence that age-induced decline in memory retention is delayed in growth hormone resistant GH-R-KO (Laron) mice. Physiol Behav. 2001;72(5):653–660. doi: 10.1016/s0031-9384(01)00423-1. [DOI] [PubMed] [Google Scholar]
- 213.Bellush LL, Doublier S, Holland AN, Striker LJ, Striker GE, Kopchick JJ. Protection against diabetes-induced nephropathy in growth hormone receptor/binding protein gene-disrupted mice. Endocrinology. 2000;141(1):163–168. doi: 10.1210/endo.141.1.7284. [DOI] [PubMed] [Google Scholar]
- 214.Arum O, Rickman DJ, Kopchick JJ, Bartke A. The slow-aging growth hormone receptor/binding protein gene-disrupted (GHR-KO) mouse is protected from aging-resultant neuromusculoskeletal frailty. Age (Dordr) 2014;36(1):117–127. doi: 10.1007/s11357-013-9551-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Coschigano KT, Clemmons D, Bellush LL, Kopchick JJ. Assessment of growth parameters and life span of GHR/BP gene-disrupted mice. Endocrinology. 2000;141(7):2608–2613. doi: 10.1210/endo.141.7.7586. [DOI] [PubMed] [Google Scholar]
- 216.Liu JL, Yakar S, LeRoith D. Mice deficient in liver production of insulin-like growth factor I display sexual dimorphism in growth hormone-stimulated postnatal growth. Endocrinology. 2000;141(12):4436–4441. doi: 10.1210/endo.141.12.7825. [DOI] [PubMed] [Google Scholar]
- 217.Caplan AI, Fiszman MY, Eppenberger HM. Molecular and cell isoforms during development. Science. 1983;221(4614):921–927. doi: 10.1126/science.6348946. [DOI] [PubMed] [Google Scholar]
- 218.Agarwal P, Verzi MP, Nguyen T, Hu J, Ehlers ML, McCulley DJ, Xu SM, Dodou E, Anderson JP, Wei ML, Black BL. The MADS box transcription factor MEF2C regulates melanocyte development and is a direct transcriptional target and partner of SOX10. Development. 2011;138(12):2555–2565. doi: 10.1242/dev.056804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Potthoff MJ, Olson EN. MEF2: a central regulator of diverse developmental programs. Development. 2007;134(23):4131–4140. doi: 10.1242/dev.008367. [DOI] [PubMed] [Google Scholar]
- 220.List EO, Berryman DE, Buchman M, Parker C, Funk K, Bell S, Duran-Ortiz S, Qian Y, Young JA, Wilson C, Slyby J, McKenna S, Jensen EA, Kopchick JJ. Adipocyte-specific GH receptor-null (AdGHRKO) mice have enhanced insulin sensitivity with reduced liver triglycerides. Endocrinology. 2019;160(1):68–80. doi: 10.1210/en.2018-00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Herrera PL. Adult insulin- and glucagon-producing cells differentiate from two independent cell lineages. Development. 2000;127(11):2317–2322. doi: 10.1242/dev.127.11.2317. [DOI] [PubMed] [Google Scholar]
- 222.Brouwers B, de Faudeur G, Osipovich AB, Goyvaerts L, Lemaire K, Boesmans L, Cauwelier EJ, Granvik M, Pruniau VP, Van Lommel L, Van Schoors J, Stancill JS, Smolders I, Goffin V, Binart N, in't Veld P, Declercq J, Magnuson MA, Creemers JW, Schuit F, Schraenen A (2014) Impaired islet function in commonly used transgenic mouse lines due to human growth hormone minigene expression. Cell Metab 20(6): 979–990. 10.1016/j.cmet.2014.11.004 [DOI] [PMC free article] [PubMed]
- 223.Baan M, Kibbe CR, Bushkofsky JR, Harris TW, Sherman DS, Davis DB. Transgenic expression of the human growth hormone minigene promotes pancreatic beta-cell proliferation. Am J Physiol Regul Integr Comp Physiol. 2015;309(7):R788–794. doi: 10.1152/ajpregu.00244.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Liu Z, Kennedy OD, Cardoso L, Basta-Pljakic J, Partridge NC, Schaffler MB, Rosen CJ, Yakar S. DMP-1-mediated Ghr gene recombination compromises skeletal development and impairs skeletal response to intermittent PTH. Faseb J. 2016;30(2):635–652. doi: 10.1096/fj.15-275859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Rodig SJ, Meraz MA, White JM, Lampe PA, Riley JK, Arthur CD, King KL, Sheehan KC, Yin L, Pennica D, Johnson EM, Jr, Schreiber RD. Disruption of the Jak1 gene demonstrates obligatory and nonredundant roles of the Jaks in cytokine-induced biologic responses. Cell. 1998;93(3):373–383. doi: 10.1016/s0092-8674(00)81166-6. [DOI] [PubMed] [Google Scholar]
- 226.Parganas E, Wang D, Stravopodis D, Topham DJ, Marine JC, Teglund S, Vanin EF, Bodner S, Colamonici OR, van Deursen JM, Grosveld G, Ihle JN. Jak2 is essential for signaling through a variety of cytokine receptors. Cell. 1998;93(3):385–395. doi: 10.1016/s0092-8674(00)81167-8. [DOI] [PubMed] [Google Scholar]
- 227.Neubauer H, Cumano A, Muller M, Wu H, Huffstadt U, Pfeffer K. Jak2 deficiency defines an essential developmental checkpoint in definitive hematopoiesis. Cell. 1998;93(3):397–409. doi: 10.1016/s0092-8674(00)81168-x. [DOI] [PubMed] [Google Scholar]
- 228.Park SY, Saijo K, Takahashi T, Osawa M, Arase H, Hirayama N, Miyake K, Nakauchi H, Shirasawa T, Saito T. Developmental defects of lymphoid cells in Jak3 kinase-deficient mice. Immunity. 1995;3(6):771–782. doi: 10.1016/1074-7613(95)90066-7. [DOI] [PubMed] [Google Scholar]
- 229.Shimoda K, Kato K, Aoki K, Matsuda T, Miyamoto A, Shibamori M, Yamashita M, Numata A, Takase K, Kobayashi S, Shibata S, Asano Y, Gondo H, Sekiguchi K, Nakayama K, Nakayama T, Okamura T, Okamura S, Niho Y. Tyk2 plays a restricted role in IFN alpha signaling, although it is required for IL-12-mediated T cell function. Immunity. 2000;13(4):561–571. doi: 10.1016/s1074-7613(00)00055-8. [DOI] [PubMed] [Google Scholar]
- 230.Derecka M, Gornicka A, Koralov SB, Szczepanek K, Morgan M, Raje V, Sisler J, Zhang Q, Otero D, Cichy J, Rajewsky K, Shimoda K, Poli V, Strobl B, Pellegrini S, Harris TE, Seale P, Russell AP, McAinch AJ, O'Brien PE, Keller SR, Croniger CM, Kordula T, Larner AC. Tyk2 and Stat3 regulate brown adipose tissue differentiation and obesity. Cell Metab. 2012;16(6):814–824. doi: 10.1016/j.cmet.2012.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sos BC, Harris C, Nordstrom SM, Tran JL, Balazs M, Caplazi P, Febbraio M, Applegate MA, Wagner KU, Weiss EJ. Abrogation of growth hormone secretion rescues fatty liver in mice with hepatocyte-specific deletion of JAK2. J Clin Invest. 2011;121(4):1412–1423. doi: 10.1172/JCI42894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nordstrom SM, Tran JL, Sos BC, Wagner KU, Weiss EJ. Disruption of JAK2 in adipocytes impairs lipolysis and improves fatty liver in mice with elevated GH. Mol Endocrinol. 2013;27(8):1333–1342. doi: 10.1210/me.2013-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Meraz MA, White JM, Sheehan KC, Bach EA, Rodig SJ, Dighe AS, Kaplan DH, Riley JK, Greenlund AC, Campbell D, Carver-Moore K, DuBois RN, Clark R, Aguet M, Schreiber RD. Targeted disruption of the Stat1 gene in mice reveals unexpected physiologic specificity in the JAK-STAT signaling pathway. Cell. 1996;84(3):431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 234.Durbin JE, Hackenmiller R, Simon MC, Levy DE. Targeted disruption of the mouse Stat1 gene results in compromised innate immunity to viral disease. Cell. 1996;84(3):443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 235.Park C, Li S, Cha E, Schindler C. Immune response in Stat2 knockout mice. Immunity. 2000;13(6):795–804. doi: 10.1016/s1074-7613(00)00077-7. [DOI] [PubMed] [Google Scholar]
- 236.Takeda K, Noguchi K, Shi W, Tanaka T, Matsumoto M, Yoshida N, Kishimoto T, Akira S. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94(8):3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Wirtz S, Finotto S, Kanzler S, Lohse AW, Blessing M, Lehr HA, Galle PR, Neurath MF. Cutting edge: chronic intestinal inflammation in STAT-4 transgenic mice: characterization of disease and adoptive transfer by TNF- plus IFN-gamma-producing CD4+ T cells that respond to bacterial antigens. J Immunol. 1999;162(4):1884–1888. [PubMed] [Google Scholar]
- 238.Thierfelder WE, van Deursen JM, Yamamoto K, Tripp RA, Sarawar SR, Carson RT, Sangster MY, Vignali DA, Doherty PC, Grosveld GC, Ihle JN. Requirement for Stat4 in interleukin-12-mediated responses of natural killer and T cells. Nature. 1996;382(6587):171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 239.Kaplan MH, Sun YL, Hoey T, Grusby MJ. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 1996;382(6587):174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 240.Liu X, Robinson GW, Wagner KU, Garrett L, Wynshaw-Boris A, Hennighausen L. Stat5a is mandatory for adult mammary gland development and lactogenesis. Genes Dev. 1997;11(2):179–186. doi: 10.1101/gad.11.2.179. [DOI] [PubMed] [Google Scholar]
- 241.Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94(14):7239–7244. doi: 10.1073/pnas.94.14.7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Teglund S, McKay C, Schuetz E, van Deursen JM, Stravopodis D, Wang D, Brown M, Bodner S, Grosveld G, Ihle JN. Stat5a and Stat5b proteins have essential and nonessential, or redundant, roles in cytokine responses. Cell. 1998;93(5):841–850. doi: 10.1016/s0092-8674(00)81444-0. [DOI] [PubMed] [Google Scholar]
- 243.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Essential role of Stat6 in IL-4 signalling. Nature. 1996;380(6575):627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 244.Trengove MC, Ward AC. SOCS proteins in development and disease. Am J Clin Exp Immunol. 2013;2(1):1–29. [PMC free article] [PubMed] [Google Scholar]
- 245.Ram PA, Waxman DJ. SOCS/CIS protein inhibition of growth hormone-stimulated STAT5 signaling by multiple mechanisms. J Biol Chem. 1999;274(50):35553–35561. doi: 10.1074/jbc.274.50.35553. [DOI] [PubMed] [Google Scholar]
- 246.Starr R, Metcalf D, Elefanty AG, Brysha M, Willson TA, Nicola NA, Hilton DJ, Alexander WS. Liver degeneration and lymphoid deficiencies in mice lacking suppressor of cytokine signaling-1. Proc Natl Acad Sci U S A. 1998;95(24):14395–14399. doi: 10.1073/pnas.95.24.14395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Fujimoto M, Naka T, Nakagawa R, Kawazoe Y, Morita Y, Tateishi A, Okumura K, Narazaki M, Kishimoto T. Defective thymocyte development and perturbed homeostasis of T cells in STAT-induced STAT inhibitor-1/suppressors of cytokine signaling-1 transgenic mice. J Immunol. 2000;165(4):1799–1806. doi: 10.4049/jimmunol.165.4.1799. [DOI] [PubMed] [Google Scholar]
- 248.Kaplan DH, Shankaran V, Dighe AS, Stockert E, Aguet M, Old LJ, Schreiber RD. Demonstration of an interferon gamma-dependent tumor surveillance system in immunocompetent mice. Proc Natl Acad Sci USA. 1998;95(13):7556–7561. doi: 10.1073/pnas.95.13.7556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Greenhalgh CJ, Metcalf D, Thaus AL, Corbin JE, Uren R, Morgan PO, Fabri LJ, Zhang JG, Martin HM, Willson TA, Billestrup N, Nicola NA, Baca M, Alexander WS, Hilton DJ. Biological evidence that SOCS-2 can act either as an enhancer or suppressor of growth hormone signaling. J Biol Chem. 2002;277(43):40181–40184. doi: 10.1074/jbc.C200450200. [DOI] [PubMed] [Google Scholar]
- 250.Marine JC, McKay C, Wang D, Topham DJ, Parganas E, Nakajima H, Pendeville H, Yasukawa H, Sasaki A, Yoshimura A, Ihle JN. SOCS3 is essential in the regulation of fetal liver erythropoiesis. Cell. 1999;98(5):617–627. doi: 10.1016/s0092-8674(00)80049-5. [DOI] [PubMed] [Google Scholar]
- 251.Seki Y, Hayashi K, Matsumoto A, Seki N, Tsukada J, Ransom J, Naka T, Kishimoto T, Yoshimura A, Kubo M. Expression of the suppressor of cytokine signaling-5 (SOCS5) negatively regulates IL-4-dependent STAT6 activation and Th2 differentiation. Proc Natl Acad Sci USA. 2002;99(20):13003–13008. doi: 10.1073/pnas.202477099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Brender C, Columbus R, Metcalf D, Handman E, Starr R, Huntington N, Tarlinton D, Odum N, Nicholson SE, Nicola NA, Hilton DJ, Alexander WS. SOCS5 is expressed in primary B and T lymphoid cells but is dispensable for lymphocyte production and function. Mol Cell Biol. 2004;24(13):6094–6103. doi: 10.1128/mcb.24.13.6094-6103.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Li L, Gronning LM, Anderson PO, Li S, Edvardsen K, Johnston J, Kioussis D, Shepherd PR, Wang P. Insulin induces SOCS-6 expression and its binding to the p85 monomer of phosphoinositide 3-kinase, resulting in improvement in glucose metabolism. J Biol Chem. 2004;279(33):34107–34114. doi: 10.1074/jbc.M312672200. [DOI] [PubMed] [Google Scholar]
- 254.Matsumoto A, Seki Y, Kubo M, Ohtsuka S, Suzuki A, Hayashi I, Tsuji K, Nakahata T, Okabe M, Yamada S, Yoshimura A. Suppression of STAT5 functions in liver, mammary glands, and T cells in cytokine-inducible SH2-containing protein 1 transgenic mice. Mol Cell Biol. 1999;19(9):6396–6407. doi: 10.1128/mcb.19.9.6396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Krebs DL, Uren RT, Metcalf D, Rakar S, Zhang JG, Starr R, De Souza DP, Hanzinikolas K, Eyles J, Connolly LM, Simpson RJ, Nicola NA, Nicholson SE, Baca M, Hilton DJ, Alexander WS. SOCS-6 binds to insulin receptor substrate 4, and mice lacking the SOCS-6 gene exhibit mild growth retardation. Mol Cell Biol. 2002;22(13):4567–4578. doi: 10.1128/mcb.22.13.4567-4578.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Banks AS, Li J, McKeag L, Hribal ML, Kashiwada M, Accili D, Rothman PB. Deletion of SOCS7 leads to enhanced insulin action and enlarged islets of Langerhans. J Clin Invest. 2005;115(9):2462–2471. doi: 10.1172/jci23853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Mathews LS, Hammer RE, Behringer RR, D'Ercole AJ, Bell GI, Brinster RL, Palmiter RD. Growth enhancement of transgenic mice expressing human insulin-like growth factor I. Endocrinology. 1988;123(6):2827–2833. doi: 10.1210/endo-123-6-2827. [DOI] [PubMed] [Google Scholar]
- 258.Doi T, Striker LJ, Gibson CC, Agodoa LY, Brinster RL, Striker GE. Glomerular lesions in mice transgenic for growth hormone and insulinlike growth factor-I. I. Relationship between increased glomerular size and mesangial sclerosis. Am J Pathol. 1990;137(3):541–552. [PMC free article] [PubMed] [Google Scholar]
- 259.DeChiara TM, Efstratiadis A, Robertson EJ. A growth-deficiency phenotype in heterozygous mice carrying an insulin-like growth factor II gene disrupted by targeting. Nature. 1990;345(6270):78–80. doi: 10.1038/345078a0. [DOI] [PubMed] [Google Scholar]
- 260.Liu JP, Baker J, Perkins AS, Robertson EJ, Efstratiadis A. Mice carrying null mutations of the genes encoding insulin-like growth factor I (Igf-1) and type 1 IGF receptor (Igf1r) Cell. 1993;75(1):59–72. [PubMed] [Google Scholar]
- 261.Baker J, Liu JP, Robertson EJ, Efstratiadis A. Role of insulin-like growth factors in embryonic and postnatal growth. Cell. 1993;75(1):73–82. doi: 10.1016/S0092-8674(05)80085-6. [DOI] [PubMed] [Google Scholar]
- 262.Lupu F, Terwilliger JD, Lee K, Segre GV, Efstratiadis A. Roles of growth hormone and insulin-like growth factor 1 in mouse postnatal growth. Dev Biol. 2001;229(1):141–162. doi: 10.1006/dbio.2000.9975. [DOI] [PubMed] [Google Scholar]
- 263.Liao L, Dearth RK, Zhou S, Britton OL, Lee AV, Xu J. Liver-specific overexpression of the insulin-like growth factor-I enhances somatic growth and partially prevents the effects of growth hormone deficiency. Endocrinology. 2006;147(8):3877–3888. doi: 10.1210/en.2005-1537. [DOI] [PubMed] [Google Scholar]
- 264.Dearth RK, Kuiatse I, Wang YF, Liao L, Hilsenbeck SG, Brown PH, Xu J, Lee AV. A moderate elevation of circulating levels of IGF-I does not alter ErbB2 induced mammary tumorigenesis. BMC Cancer. 2011;11:377. doi: 10.1186/1471-2407-11-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wu Y, Sun H, Yakar S, LeRoith D. Elevated levels of insulin-like growth factor (IGF)-I in serum rescue the severe growth retardation of IGF-I null mice. Endocrinology. 2009;150(9):4395–4403. doi: 10.1210/en.2009-0272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Elis S, Courtland HW, Wu Y, Rosen CJ, Sun H, Jepsen KJ, Majeska RJ, Yakar S. Elevated serum levels of IGF-1 are sufficient to establish normal body size and skeletal properties even in the absence of tissue IGF-1. J Bone Miner Res. 2010;25(6):1257–1266. doi: 10.1002/jbmr.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Wu Y, Sun H, Basta-Pljakic J, Cardoso L, Kennedy OD, Jasper H, Domene H, Karabatas L, Guida C, Schaffler MB, Rosen CJ, Yakar S. Serum IGF-1 is insufficient to restore skeletal size in the total absence of the growth hormone receptor. J Bone Miner Res. 2013;28(7):1575–1586. doi: 10.1002/jbmr.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Yakar S, Liu JL, Stannard B, Butler A, Accili D, Sauer B, LeRoith D. Normal growth and development in the absence of hepatic insulin-like growth factor I. Proc Natl Acad Sci USA. 1999;96(13):7324–7329. doi: 10.1073/pnas.96.13.7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Novosyadlyy R, Leroith D. Insulin-like growth factors and insulin: at the crossroad between tumor development and longevity. J Gerontol A. 2012;67(6):640–651. doi: 10.1093/gerona/gls065. [DOI] [PubMed] [Google Scholar]
- 270.Sjogren K, Liu JL, Blad K, Skrtic S, Vidal O, Wallenius V, LeRoith D, Tornell J, Isaksson OG, Jansson JO, Ohlsson C. Liver-derived insulin-like growth factor I (IGF-I) is the principal source of IGF-I in blood but is not required for postnatal body growth in mice. Proc Natl Acad Sci U S A. 1999;96(12):7088–7092. doi: 10.1073/pnas.96.12.7088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Sjogren K, Wallenius K, Liu JL, Bohlooly YM, Pacini G, Svensson L, Tornell J, Isaksson OG, Ahren B, Jansson JO, Ohlsson C. Liver-derived IGF-I is of importance for normal carbohydrate and lipid metabolism. Diabetes. 2001;50(7):1539–1545. doi: 10.2337/diabetes.50.7.1539. [DOI] [PubMed] [Google Scholar]
- 272.Svensson J, Sjogren K, Faldt J, Andersson N, Isaksson O, Jansson JO, Ohlsson C. Liver-derived IGF-I regulates mean life span in mice. PLoS ONE. 2011;6(7):e22640. doi: 10.1371/journal.pone.0022640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Kloting N, Koch L, Wunderlich T, Kern M, Ruschke K, Krone W, Bruning JC, Bluher M. Autocrine IGF-1 action in adipocytes controls systemic IGF-1 concentrations and growth. Diabetes. 2008;57(8):2074–2082. doi: 10.2337/db07-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lee KY, Russell SJ, Ussar S, Boucher J, Vernochet C, Mori MA, Smyth G, Rourk M, Cederquist C, Rosen ED, Kahn BB, Kahn CR. Lessons on conditional gene targeting in mouse adipose tissue. Diabetes. 2013;62(3):864–874. doi: 10.2337/db12-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Krueger KC, Costa MJ, Du H, Feldman BJ. Characterization of Cre recombinase activity for in vivo targeting of adipocyte precursor cells. Stem Cell Reports. 2014;3(6):1147–1158. doi: 10.1016/j.stemcr.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Martens K, Bottelbergs A, Baes M. Ectopic recombination in the central and peripheral nervous system by aP2/FABP4-Cre mice: implications for metabolism research. FEBS Lett. 2010;584(5):1054–1058. doi: 10.1016/j.febslet.2010.01.061. [DOI] [PubMed] [Google Scholar]
- 277.Boucher J, Softic S, El Ouaamari A, Krumpoch MT, Kleinridders A, Kulkarni RN, O'Neill BT, Kahn CR. Differential roles of insulin and IGF-1 receptors in adipose tissue development and function. Diabetes. 2016;65(8):2201–2213. doi: 10.2337/db16-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Kappeler L, De Magalhaes Filho C, Dupont J, Leneuve P, Cervera P, Perin L, Loudes C, Blaise A, Klein R, Epelbaum J, Le Bouc Y, Holzenberger M. Brain IGF-1 receptors control mammalian growth and lifespan through a neuroendocrine mechanism. PLoS Biol. 2008;6(10):e254. doi: 10.1371/journal.pbio.0060254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Coleman ME, DeMayo F, Yin KC, Lee HM, Geske R, Montgomery C, Schwartz RJ. Myogenic vector expression of insulin-like growth factor I stimulates muscle cell differentiation and myofiber hypertrophy in transgenic mice. J Biol Chem. 1995;270(20):12109–12116. doi: 10.1074/jbc.270.20.12109. [DOI] [PubMed] [Google Scholar]
- 280.Fernandez AM, Kim JK, Yakar S, Dupont J, Hernandez-Sanchez C, Castle AL, Filmore J, Shulman GI, Le Roith D. Functional inactivation of the IGF-I and insulin receptors in skeletal muscle causes type 2 diabetes. Genes Dev. 2001;15(15):1926–1934. doi: 10.1101/gad.908001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.O'Neill BT, Lauritzen HP, Hirshman MF, Smyth G, Goodyear LJ, Kahn CR. Differential role of insulin/IGF-1 receptor signaling in muscle growth and glucose homeostasis. Cell Rep. 2015;11(8):1220–1235. doi: 10.1016/j.celrep.2015.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.O'Neill BT, Lee KY, Klaus K, Softic S, Krumpoch MT, Fentz J, Stanford KI, Robinson MM, Cai W, Kleinridders A, Pereira RO, Hirshman MF, Abel ED, Accili D, Goodyear LJ, Nair KS, Kahn CR. Insulin and IGF-1 receptors regulate FoxO-mediated signaling in muscle proteostasis. J Clin Invest. 2016;126(9):3433–3446. doi: 10.1172/jci86522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Reiss K, Cheng W, Ferber A, Kajstura J, Li P, Li B, Olivetti G, Homcy CJ, Baserga R, Anversa P. Overexpression of insulin-like growth factor-1 in the heart is coupled with myocyte proliferation in transgenic mice. Proc Natl Acad Sci U S A. 1996;93(16):8630–8635. doi: 10.1073/pnas.93.16.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Santini MP, Tsao L, Monassier L, Theodoropoulos C, Carter J, Lara-Pezzi E, Slonimsky E, Salimova E, Delafontaine P, Song YH, Bergmann M, Freund C, Suzuki K, Rosenthal N. Enhancing repair of the mammalian heart. Circ Res. 2007;100(12):1732–1740. doi: 10.1161/circresaha.107.148791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Kim J, Wende AR, Sena S, Theobald HA, Soto J, Sloan C, Wayment BE, Litwin SE, Holzenberger M, LeRoith D, Abel ED. Insulin-like growth factor I receptor signaling is required for exercise-induced cardiac hypertrophy. Mol Endocrinol. 2008;22(11):2531–2543. doi: 10.1210/me.2008-0265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Moellendorf S, Kessels C, Peiseler L, Raupach A, Jacoby C, Vogt N, Lindecke A, Koch L, Bruning J, Heger J, Kohrer K, Godecke A. IGF-IR signaling attenuates the age-related decline of diastolic cardiac function. Am J Physiol Endocrinol Metab. 2012;303(2):E213–222. doi: 10.1152/ajpendo.00538.2011. [DOI] [PubMed] [Google Scholar]
- 287.Imrie H, Viswambharan H, Sukumar P, Abbas A, Cubbon RM, Yuldasheva N, Gage M, Smith J, Galloway S, Skromna A, Rashid ST, Futers TS, Xuan S, Gatenby VK, Grant PJ, Channon KM, Beech DJ, Wheatcroft SB, Kearney MT. Novel role of the IGF-1 receptor in endothelial function and repair: studies in endothelium-targeted IGF-1 receptor transgenic mice. Diabetes. 2012;61(9):2359–2368. doi: 10.2337/db11-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Abbas A, Imrie H, Viswambharan H, Sukumar P, Rajwani A, Cubbon RM, Gage M, Smith J, Galloway S, Yuldeshava N, Kahn M, Xuan S, Grant PJ, Channon KM, Beech DJ, Wheatcroft SB, Kearney MT. The insulin-like growth factor-1 receptor is a negative regulator of nitric oxide bioavailability and insulin sensitivity in the endothelium. Diabetes. 2011;60(8):2169–2178. doi: 10.2337/db11-0197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Liang M, Woodard LE, Liang A, Luo J, Wilson MH, Mitch WE, Cheng J. Protective role of insulin-like growth factor-1 receptor in endothelial cells against unilateral ureteral obstruction-induced renal fibrosis. Am J Pathol. 2015;185(5):1234–1250. doi: 10.1016/j.ajpath.2015.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Spadaro O, Goldberg EL, Camell CD, Youm YH, Kopchick JJ, Nguyen KY, Bartke A, Sun LY, Dixit VD. Growth hormone receptor deficiency protects against age-related NLRP3 inflammasome activation and immune senescence. Cell Rep. 2016;14(7):1571–1580. doi: 10.1016/j.celrep.2016.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Spadaro O, Camell CD, Bosurgi L, Nguyen KY, Youm YH, Rothlin CV, Dixit VD. IGF1 shapes macrophage activation in response to immunometabolic challenge. Cell Rep. 2017;19(2):225–234. doi: 10.1016/j.celrep.2017.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Higashi Y, Sukhanov S, Shai SY, Danchuk S, Tang R, Snarski P, Li Z, Lobelle-Rich P, Wang M, Wang D, Yu H, Korthuis R, Delafontaine P. Insulin-like growth factor-1 receptor deficiency in macrophages accelerates atherosclerosis and induces an unstable plaque phenotype in apolipoprotein E-deficient mice. Circulation. 2016;133(23):2263–2278. doi: 10.1161/circulationaha.116.021805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zhao G, Monier-Faugere MC, Langub MC, Geng Z, Nakayama T, Pike JW, Chernausek SD, Rosen CJ, Donahue LR, Malluche HH, Fagin JA, Clemens TL. Targeted overexpression of insulin-like growth factor I to osteoblasts of transgenic mice: increased trabecular bone volume without increased osteoblast proliferation. Endocrinology. 2000;141(7):2674–2682. doi: 10.1210/endo.141.7.7585. [DOI] [PubMed] [Google Scholar]
- 294.Jiang J, Lichtler AC, Gronowicz GA, Adams DJ, Clark SH, Rosen CJ, Kream BE. Transgenic mice with osteoblast-targeted insulin-like growth factor-I show increased bone remodeling. Bone. 2006;39(3):494–504. doi: 10.1016/j.bone.2006.02.068. [DOI] [PubMed] [Google Scholar]
- 295.Rutter MM, Markoff E, Clayton L, Akeno N, Zhao G, Clemens TL, Chernausek SD. Osteoblast-specific expression of insulin-like growth factor-1 in bone of transgenic mice induces insulin-like growth factor binding protein-5. Bone. 2005;36(2):224–231. doi: 10.1016/j.bone.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 296.Zhang M, Xuan S, Bouxsein ML, von Stechow D, Akeno N, Faugere MC, Malluche H, Zhao G, Rosen CJ, Efstratiadis A, Clemens TL. Osteoblast-specific knockout of the insulin-like growth factor (IGF) receptor gene reveals an essential role of IGF signaling in bone matrix mineralization. J Biol Chem. 2002;277(46):44005–44012. doi: 10.1074/jbc.M208265200. [DOI] [PubMed] [Google Scholar]
- 297.Kubota T, Elalieh HZ, Saless N, Fong C, Wang Y, Babey M, Cheng Z, Bikle DD. Insulin-like growth factor-1 receptor in mature osteoblasts is required for periosteal bone formation induced by reloading. Acta Astronaut. 2013;92(1):73–78. doi: 10.1016/j.actaastro.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Wang T, Wang Y, Menendez A, Fong C, Babey M, Tahimic CG, Cheng Z, Li A, Chang W, Bikle DD. Osteoblast-Specific Loss of IGF1R Signaling Results in Impaired Endochondral Bone Formation During Fracture Healing. J Bone Miner Res. 2015;30(9):1572–1584. doi: 10.1002/jbmr.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 299.Govoni KE, Lee SK, Chung YS, Behringer RR, Wergedal JE, Baylink DJ, Mohan S. Disruption of insulin-like growth factor-I expression in type IIalphaI collagen-expressing cells reduces bone length and width in mice. Physiol Genomics. 2007;30(3):354–362. doi: 10.1152/physiolgenomics.00022.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Govoni KE, Wergedal JE, Florin L, Angel P, Baylink DJ, Mohan S. Conditional deletion of insulin-like growth factor-I in collagen type 1alpha2-expressing cells results in postnatal lethality and a dramatic reduction in bone accretion. Endocrinology. 2007;148(12):5706–5715. doi: 10.1210/en.2007-0608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sheng MH, Zhou XD, Bonewald LF, Baylink DJ, Lau KH. Disruption of the insulin-like growth factor-1 gene in osteocytes impairs developmental bone growth in mice. Bone. 2013;52(1):133–144. doi: 10.1016/j.bone.2012.09.027. [DOI] [PubMed] [Google Scholar]
- 302.Yakar S, Werner H, Rosen CJ. Insulin-like growth factors: actions on the skeleton. J Mol Endocrinol. 2018;61(1):T115–T137. doi: 10.1530/jme-17-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Baumgarten SC, Armouti M, Ko C, Stocco C. IGF1R expression in ovarian granulosa cells is essential for steroidogenesis, follicle survival, and fertility in female mice. Endocrinology. 2017;158(7):2309–2318. doi: 10.1210/en.2017-00146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Xuan S, Kitamura T, Nakae J, Politi K, Kido Y, Fisher PE, Morroni M, Cinti S, White MF, Herrera PL, Accili D, Efstratiadis A. Defective insulin secretion in pancreatic beta cells lacking type 1 IGF receptor. J Clin Invest. 2002;110(7):1011–1019. doi: 10.1172/jci15276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Kulkarni RN, Holzenberger M, Shih DQ, Ozcan U, Stoffel M, Magnuson MA, Kahn CR. beta-cell-specific deletion of the Igf1 receptor leads to hyperinsulinemia and glucose intolerance but does not alter beta-cell mass. Nat Genet. 2002;31(1):111–115. doi: 10.1038/ng872. [DOI] [PubMed] [Google Scholar]
- 306.Ueki K, Okada T, Hu J, Liew CW, Assmann A, Dahlgren GM, Peters JL, Shackman JG, Zhang M, Artner I, Satin LS, Stein R, Holzenberger M, Kennedy RT, Kahn CR, Kulkarni RN. Total insulin and IGF-I resistance in pancreatic beta cells causes overt diabetes. Nat Genet. 2006;38(5):583–588. doi: 10.1038/ng1787. [DOI] [PubMed] [Google Scholar]
- 307.Neirijnck Y, Calvel P, Kilcoyne KR, Kuhne F, Stevant I, Griffeth RJ, Pitetti JL, Andric SA, Hu MC, Pralong F, Smith LB, Nef S. Insulin and IGF1 receptors are essential for the development and steroidogenic function of adult Leydig cells. Faseb J. 2018;32(6):3321–3335. doi: 10.1096/fj.201700769RR. [DOI] [PubMed] [Google Scholar]
- 308.Romero CJ, Ng Y, Luque RM, Kineman RD, Koch L, Bruning JC, Radovick S. Targeted deletion of somatotroph insulin-like growth factor-I signaling in a cell-specific knockout mouse model. Mol Endocrinol. 2010;24(5):1077–1089. doi: 10.1210/me.2009-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.Gahete MD, Cordoba-Chacon J, Anadumaka CV, Lin Q, Bruning JC, Kahn CR, Luque RM, Kineman RD. Elevated GH/IGF-I, due to somatotrope-specific loss of both IGF-I and insulin receptors, alters glucose homeostasis and insulin sensitivity in a diet-dependent manner. Endocrinology. 2011;152(12):4825–4837. doi: 10.1210/en.2011-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Muller K, Fuhrer D, Mittag J, Kloting N, Bluher M, Weiss RE, Many MC, Schmid KW, Krohn K. TSH compensates thyroid-specific IGF-I receptor knockout and causes papillary thyroid hyperplasia. Mol Endocrinol. 2011;25(11):1867–1879. doi: 10.1210/me.2011-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Bach LA. IGF-binding proteins. J Mol Endocrinol. 2018;61(1):T11–T28. doi: 10.1530/JME-17-0254. [DOI] [PubMed] [Google Scholar]
- 312.D'Ercole AJ, Ye P, Dai Z. Human insulin-like growth factor binding protein-1 (hIGFBP-1) transgenic mice: insights into hIGFBP-1 regulation and actions. Prog Growth Factor Res. 1995;6(2–4):417–423. doi: 10.1016/0955-2235(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 313.Schneider MR, Lahm H, Wu M, Hoeflich A, Wolf E. Transgenic mouse models for studying the functions of insulin-like growth factor-binding proteins. Faseb J. 2000;14(5):629–640. doi: 10.1096/fasebj.14.5.629. [DOI] [PubMed] [Google Scholar]
- 314.Ben Lagha N, Seurin D, Le Bouc Y, Binoux M, Berdal A, Menuelle P, Babajko S. Insulin-like growth factor binding protein (IGFBP-1) involvement in intrauterine growth retardation: study on IGFBP-1 overexpressing transgenic mice. Endocrinology. 2006;147(10):4730–4737. doi: 10.1210/en.2006-0171. [DOI] [PubMed] [Google Scholar]
- 315.Rajkumar K, Krsek M, Dheen ST, Murphy LJ. Impaired glucose homeostasis in insulin-like growth factor binding protein-1 transgenic mice. J Clin Invest. 1996;98(8):1818–1825. doi: 10.1172/jci118982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 316.Froment P, Seurin D, Hembert S, Levine JE, Pisselet C, Monniaux D, Binoux M, Monget P. Reproductive abnormalities in human IGF binding protein-1 transgenic female mice. Endocrinology. 2002;143(5):1801–1808. doi: 10.1210/endo.143.5.8815. [DOI] [PubMed] [Google Scholar]
- 317.Doublier S, Seurin D, Fouqueray B, Verpont MC, Callard P, Striker LJ, Striker GE, Binoux M, Baud L. Glomerulosclerosis in mice transgenic for human insulin-like growth factor-binding protein-1. Kidney Int. 2000;57(6):2299–2307. doi: 10.1046/j.1523-1755.2000.00090.x. [DOI] [PubMed] [Google Scholar]
- 318.Leu JI, Crissey MA, Craig LE, Taub R. Impaired hepatocyte DNA synthetic response posthepatectomy in insulin-like growth factor binding protein 1-deficient mice with defects in C/EBP beta and mitogen-activated protein kinase/extracellular signal-regulated kinase regulation. Mol Cell Biol. 2003;23(4):1251–1259. doi: 10.1128/mcb.23.4.1251-1259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Gray A, Aronson WJ, Barnard RJ, Mehta H, Wan J, Said J, Cohen P, Galet C. Global Igfbp1 deletion does not affect prostate cancer development in a c-Myc transgenic mouse model. J Endocrinol. 2011;211(3):297–304. doi: 10.1530/JOE-11-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 320.Hoeflich A, Wu M, Mohan S, Foll J, Wanke R, Froehlich T, Arnold GJ, Lahm H, Kolb HJ, Wolf E. Overexpression of insulin-like growth factor-binding protein-2 in transgenic mice reduces postnatal body weight gain. Endocrinology. 1999;140(12):5488–5496. doi: 10.1210/endo.140.12.7169. [DOI] [PubMed] [Google Scholar]
- 321.Wheatcroft SB, Kearney MT, Shah AM, Ezzat VA, Miell JR, Modo M, Williams SC, Cawthorn WP, Medina-Gomez G, Vidal-Puig A, Sethi JK, Crossey PA. IGF-binding protein-2 protects against the development of obesity and insulin resistance. Diabetes. 2007;56(2):285–294. doi: 10.2337/db06-0436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322.Diehl D, Hessel E, Oesterle D, Renner-Muller I, Elmlinger M, Langhammer M, Gottlicher M, Wolf E, Lahm H, Hoeflich A. IGFBP-2 overexpression reduces the appearance of dysplastic aberrant crypt foci and inhibits growth of adenomas in chemically induced colorectal carcinogenesis. Int J Cancer. 2009;124(9):2220–2225. doi: 10.1002/ijc.24193. [DOI] [PubMed] [Google Scholar]
- 323.Wood TL, Rogler LE, Czick ME, Schuller AG, Pintar JE. Selective alterations in organ sizes in mice with a targeted disruption of the insulin-like growth factor binding protein-2 gene. Mol Endocrinol. 2000;14(9):1472–1482. doi: 10.1210/mend.14.9.0517. [DOI] [PubMed] [Google Scholar]
- 324.Liu Y, Song C, Shen F, Zhang J, Song SW. IGFBP2 promotes immunosuppression associated with its mesenchymal induction and FcgammaRIIB phosphorylation in glioblastoma. PLoS ONE. 2019;14(9):e0222999. doi: 10.1371/journal.pone.0222999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Murphy LJ, Rajkumar K, Molnar P. Phenotypic manifestations of insulin-like growth factor binding protein-1 (IGFBP-1) and IGFBP-3 overexpression in transgenic mice. Prog Growth Factor Res. 1995;6(2–4):425–432. doi: 10.1016/0955-2235(95)00026-7. [DOI] [PubMed] [Google Scholar]
- 326.Modric T, Silha JV, Shi Z, Gui Y, Suwanichkul A, Durham SK, Powell DR, Murphy LJ. Phenotypic manifestations of insulin-like growth factor-binding protein-3 overexpression in transgenic mice. Endocrinology. 2001;142(5):1958–1967. doi: 10.1210/endo.142.5.8165. [DOI] [PubMed] [Google Scholar]
- 327.Nguyen KH, Yao XH, Erickson AG, Mishra S, Nyomba BL. Glucose intolerance in aging male IGFBP-3 transgenic mice: differential effects of human IGFBP-3 and its mutant IGFBP-3 devoid of IGF binding ability. Endocrinology. 2015;156(2):462–474. doi: 10.1210/en.2014-1271. [DOI] [PubMed] [Google Scholar]
- 328.Ning Y, Schuller AG, Bradshaw S, Rotwein P, Ludwig T, Frystyk J, Pintar JE. Diminished growth and enhanced glucose metabolism in triple knockout mice containing mutations of insulin-like growth factor binding protein-3, -4, and -5. Mol Endocrinol. 2006;20(9):2173–2186. doi: 10.1210/me.2005-0196. [DOI] [PubMed] [Google Scholar]
- 329.Yamada PM, Mehta HH, Hwang D, Roos KP, Hevener AL, Lee KW. Evidence of a role for insulin-like growth factor binding protein (IGFBP)-3 in metabolic regulation. Endocrinology. 2010;151(12):5741–5750. doi: 10.1210/en.2010-0672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330.Wang YA, Sun Y, Palmer J, Solomides C, Huang LC, Shyr Y, Dicker AP, Lu B. IGFBP3 modulates lung tumorigenesis and cell growth through IGF1 signaling. Mol Cancer Res. 2017;15(7):896–904. doi: 10.1158/1541-7786.mcr-16-0390. [DOI] [PubMed] [Google Scholar]
- 331.Wang J, Niu W, Witte DP, Chernausek SD, Nikiforov YE, Clemens TL, Sharifi B, Strauch AR, Fagin JA. Overexpression of insulin-like growth factor-binding protein-4 (IGFBP-4) in smooth muscle cells of transgenic mice through a smooth muscle alpha-actin-IGFBP-4 fusion gene induces smooth muscle hypoplasia. Endocrinology. 1998;139(5):2605–2614. doi: 10.1210/endo.139.5.5986. [DOI] [PubMed] [Google Scholar]
- 332.Ning Y, Schuller AG, Conover CA, Pintar JE. Insulin-like growth factor (IGF) binding protein-4 is both a positive and negative regulator of IGF activity in vivo. Mol Endocrinol. 2008;22(5):1213–1225. doi: 10.1210/me.2007-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Maridas DE, DeMambro VE, Le PT, Mohan S, Rosen CJ. IGFBP4 is required for adipogenesis and influences the distribution of adipose depots. Endocrinology. 2017;158(10):3488–3500. doi: 10.1210/en.2017-00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334.Maridas DE, DeMambro VE, Le PT, Nagano K, Baron R, Mohan S, Rosen CJ. IGFBP-4 regulates adult skeletal growth in a sex-specific manner. J Endocrinol. 2017;233(1):131–144. doi: 10.1530/joe-16-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Tonner E, Barber MC, Allan GJ, Beattie J, Webster J, Whitelaw CB, Flint DJ. Insulin-like growth factor binding protein-5 (IGFBP-5) induces premature cell death in the mammary glands of transgenic mice. Development. 2002;129(19):4547–4557. doi: 10.1242/dev.129.19.4547. [DOI] [PubMed] [Google Scholar]
- 336.Salih DA, Tripathi G, Holding C, Szestak TA, Gonzalez MI, Carter EJ, Cobb LJ, Eisemann JE, Pell JM. Insulin-like growth factor-binding protein 5 (Igfbp5) compromises survival, growth, muscle development, and fertility in mice. Proc Natl Acad Sci U S A. 2004;101(12):4314–4319. doi: 10.1073/pnas.0400230101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337.Devlin RD, Du Z, Buccilli V, Jorgetti V, Canalis E. Transgenic mice overexpressing insulin-like growth factor binding protein-5 display transiently decreased osteoblastic function and osteopenia. Endocrinology. 2002;143(10):3955–3962. doi: 10.1210/en.2002-220129. [DOI] [PubMed] [Google Scholar]
- 338.Ning Y, Hoang B, Schuller AG, Cominski TP, Hsu MS, Wood TL, Pintar JE. Delayed mammary gland involution in mice with mutation of the insulin-like growth factor binding protein 5 gene. Endocrinology. 2007;148(5):2138–2147. doi: 10.1210/en.2006-0041. [DOI] [PubMed] [Google Scholar]
- 339.Gleason CE, Ning Y, Cominski TP, Gupta R, Kaestner KH, Pintar JE, Birnbaum MJ. Role of insulin-like growth factor-binding protein 5 (IGFBP5) in organismal and pancreatic beta-cell growth. Mol Endocrinol. 2010;24(1):178–192. doi: 10.1210/me.2009-0167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 340.Bienvenu G, Seurin D, Grellier P, Froment P, Baudrimont M, Monget P, Le Bouc Y, Babajko S. Insulin-like growth factor binding protein-6 transgenic mice: postnatal growth, brain development, and reproduction abnormalities. Endocrinology. 2004;145(5):2412–2420. doi: 10.1210/en.2003-1196. [DOI] [PubMed] [Google Scholar]
- 341.Bienvenu G, Seurin D, Le Bouc Y, Even P, Babajko S, Magnan C. Dysregulation of energy homeostasis in mice overexpressing insulin-like growth factor-binding protein 6 in the brain. Diabetologia. 2005;48(6):1189–1197. doi: 10.1007/s00125-005-1767-6. [DOI] [PubMed] [Google Scholar]
- 342.Silha JV, Gui Y, Modric T, Suwanichkul A, Durham SK, Powell DR, Murphy LJ. Overexpression of the acid-labile subunit of the IGF ternary complex in transgenic mice. Endocrinology. 2001;142(10):4305–4313. doi: 10.1210/endo.142.10.8427. [DOI] [PubMed] [Google Scholar]
- 343.Ueki I, Ooi GT, Tremblay ML, Hurst KR, Bach LA, Boisclair YR. Inactivation of the acid labile subunit gene in mice results in mild retardation of postnatal growth despite profound disruptions in the circulating insulin-like growth factor system. Proc Natl Acad Sci U S A. 2000;97(12):6868–6873. doi: 10.1073/pnas.120172697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 344.Courtland HW, DeMambro V, Maynard J, Sun H, Elis S, Rosen C, Yakar S. Sex-specific regulation of body size and bone slenderness by the acid labile subunit. J Bone Miner Res. 2010;25(9):2059–2068. doi: 10.1002/jbmr.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 345.Oxvig C. The role of PAPP-A in the IGF system: location, location, location. J Cell Commun Signal. 2015;9(2):177–187. doi: 10.1007/s12079-015-0259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Conover CA, Bale LK, Overgaard MT, Johnstone EW, Laursen UH, Fuchtbauer EM, Oxvig C, van Deursen J. Metalloproteinase pregnancy-associated plasma protein A is a critical growth regulatory factor during fetal development. Development. 2004;131(5):1187–1194. doi: 10.1242/dev.00997. [DOI] [PubMed] [Google Scholar]
- 347.Nyegaard M, Overgaard MT, Su YQ, Hamilton AE, Kwintkiewicz J, Hsieh M, Nayak NR, Conti M, Conover CA, Giudice LC. Lack of functional pregnancy-associated plasma protein-A (PAPPA) compromises mouse ovarian steroidogenesis and female fertility. Biol Reprod. 2010;82(6):1129–1138. doi: 10.1095/biolreprod.109.079517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 348.Conover CA, Bale LK. Loss of pregnancy-associated plasma protein A extends lifespan in mice. Aging Cell. 2007;6(5):727–729. doi: 10.1111/j.1474-9726.2007.00328.x. [DOI] [PubMed] [Google Scholar]
- 349.Conover CA, Mason MA, Levine JA, Novak CM. Metabolic consequences of pregnancy-associated plasma protein-A deficiency in mice: exploring possible relationship to the longevity phenotype. J Endocrinol. 2008;198(3):599–605. doi: 10.1677/joe-08-0179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 350.Conover CA, Bale LK, Oxvig C. Targeted inhibition of pregnancy-associated plasma protein-a activity reduces atherosclerotic plaque burden in mice. J Cardiovasc Transl Res. 2016;9(1):77–79. doi: 10.1007/s12265-015-9666-9. [DOI] [PubMed] [Google Scholar]
- 351.Conover CA, Oxvig C. PAPP-A: a promising therapeutic target for healthy longevity. Aging Cell. 2017;16(2):205–209. doi: 10.1111/acel.12564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Conover CA, Bale LK, Nair KS. Comparative gene expression and phenotype analyses of skeletal muscle from aged wild-type and PAPP-A-deficient mice. Exp Gerontol. 2016;80:36–42. doi: 10.1016/j.exger.2016.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Bale LK, West SA, Conover CA. Inducible knockdown of pregnancy-associated plasma protein-A gene expression in adult female mice extends life span. Aging Cell. 2017;16(4):895–897. doi: 10.1111/acel.12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Hjortebjerg R, Berryman DE, Comisford R, List EO, Oxvig C, Bjerre M, Frystyk J, Kopchick JJ. Depot-specific and GH-dependent regulation of IGF binding protein-4, pregnancy-associated plasma protein-A, and stanniocalcin-2 in murine adipose tissue. Growth Horm IGF Res. 2018;39:54–61. doi: 10.1016/j.ghir.2018.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.