Summary
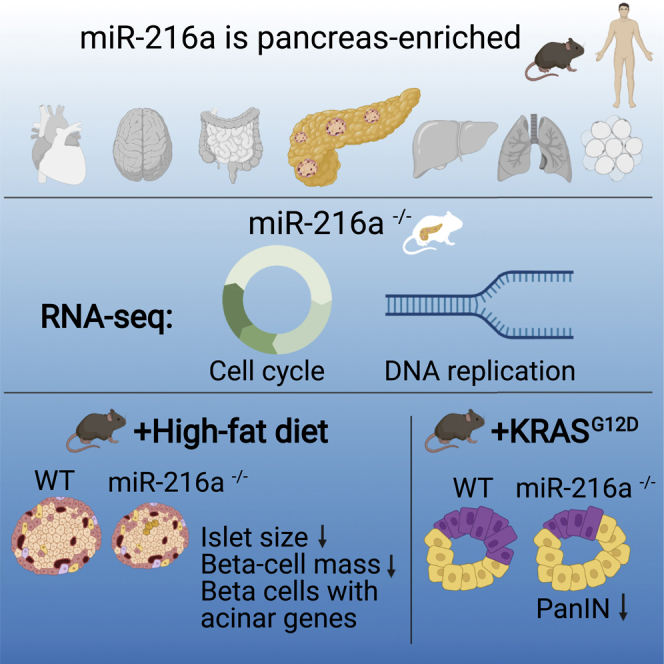
miRNAs have crucial functions in many biological processes and are candidate biomarkers of disease. Here, we show that miR-216a is a conserved, pancreas-specific miRNA with important roles in pancreatic islet and acinar cells. Deletion of miR-216a in mice leads to a reduction in islet size, β-cell mass, and insulin levels. Single-cell RNA sequencing reveals a subpopulation of β-cells with upregulated acinar cell markers under a high-fat diet. miR-216a is induced by TGF-β signaling, and inhibition of miR-216a increases apoptosis and decreases cell proliferation in pancreatic cells. Deletion of miR-216a in the pancreatic cancer-prone mouse line KrasG12D;Ptf1aCreER reduces the propensity of pancreatic cancer precursor lesions. Notably, circulating miR-216a levels are elevated in both mice and humans with pancreatic cancer. Collectively, our study gives insights into how β-cell mass and acinar cell growth are modulated by a pancreas-specific miRNA and also suggests miR-216a as a potential biomarker for diagnosis of pancreatic diseases.
Keywords: miR-216a, β-cell mass, diabetes, pancreatic cancer, PDAC, biomarker, micro-RNA, circulating miRNA
Graphical abstract
Highlights
miR-216a is highly abundant and enriched in mouse and human pancreas
Deletion of miR-216a alters pancreatic cell cycle and DNA replication pathways
miR-216a deletion decreases susceptibility to pancreatic neoplasia
miR-216a levels are elevated in mice and humans with pancreatic cancer
Erener et al. identify miR-216a as a pancreas-specific miRNA affecting pancreatic pathologies. Under a high-fat diet, deletion of miR-216a reduces beta cell mass and induces acinar gene expression. In cancer-prone mice, miR-216a deletion decreases pancreatic neoplasia frequency. Circulating miR-216a levels are elevated in mice and humans with pancreatic cancer.
Introduction
Perturbations in the function of pancreatic endocrine and exocrine cells leads to the development of life-burdening and/or life-threatening diseases such as diabetes and pancreatic cancer. Although diabetes is characterized by hyperglycemia resulting from defects in insulin secretion, insulin action, or both, pancreatic ductal adenocarcinoma (PDAC), a major form of pancreatic cancer in humans, results from the abnormal proliferation of the exocrine cells in response to oncogenic mutations.1,2 To date many protein coding genes necessary for pancreas development and function have been identified, but the role of the majority of small RNAs is still unclear.
MicroRNAs (miRNAs) are short (∼21–22 nt long) non-coding RNAs that have emerged in the past two decades as buffers of signaling pathways to maintain normal tissue development and function.3 Mature miRNAs function as evolutionarily conserved post-transcriptional gene regulators that mainly decrease the stability or inhibit translation of messenger RNAs (mRNAs) through binding to complementary sequences.4 A single miRNA can affect the regulation of hundreds of genes with multiple targets within cellular networks that enable modulation of entire pathways in the context of an individual biological process.5,6 In the pancreas, conditional deletion of the miRNA processing endonuclease Dicer at the onset of pancreatic development (e9.5) using a Pdx1-Cre strain results in defects in all pancreatic lineages with dramatic reduction in the ventral pancreas as well as a reduction in the overall epithelial contribution to the dorsal pancreas at e18.5.7 Postnatal Dicer ablation in β-cells using the conditional RIP2-Cre or Pdx1-CreER strain impairs islet architecture, insulin secretion, and β-cell mass, while deletion of Dicer in acinar cells using Mist1-CreERT mice promotes epithelial to mesenchymal transition accompanied by acinar-to-ductal metaplasia.8, 9, 10, 11 Although previous findings demonstrated important roles for Dicer in the pancreas, in vivo studies investigating the role of individual miRNAs in both endocrine and exocrine function are limited. Furthermore, the majority of investigated miRNAs do not have pancreas-specific expression.
Tissue-specific patterns of gene expression play fundamental roles in tissue development and function.12 Although the majority of miRNAs are ubiquitously expressed, some miRNAs exhibit tissue-specific or developmental stage-specific expression patterns and contribute to maintaining normal tissue identity and function.13,14 Tissue-specific miRNAs are also found in mouse and human plasma and serum at surprisingly high concentrations.15 In the present study, we hypothesized that pancreas-enriched miRNAs might have critical roles for endocrine and exocrine cell function and serve as biomarkers for diagnosing pancreatic diseases. We find that miR-216a is a pancreas-specific miRNA and contributes to β-cell and acinar cell function by modulating cell growth. We further show that circulating miR-216a levels are altered in both mice and humans with pancreatic tumors. Our data unveil the potential of tissue-specific miRNAs to contribute to disease mechanisms and to serve as biomarkers.
Results
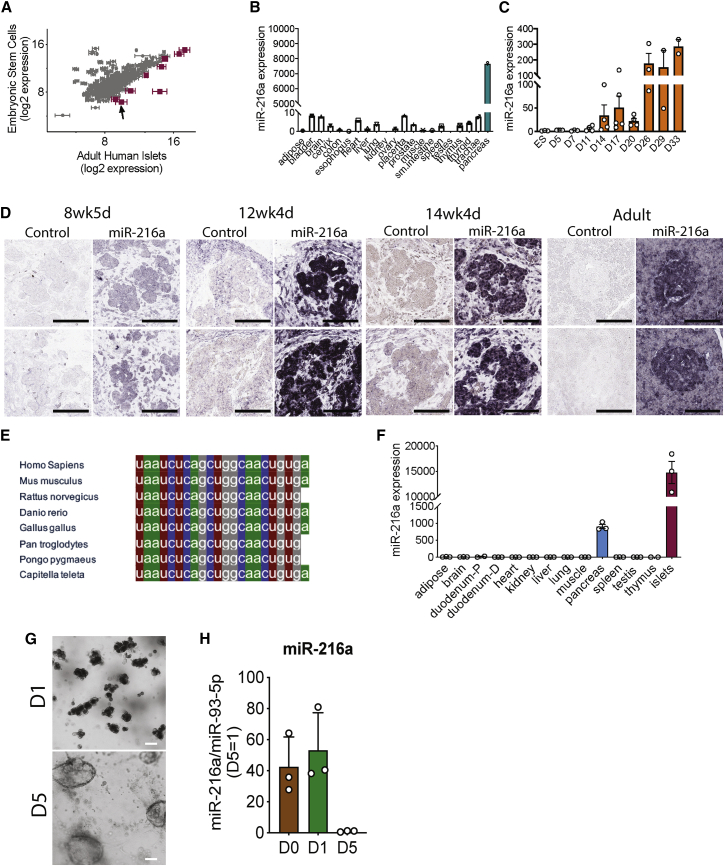
To identify the miRNAs that are enriched in pancreatic islets, we performed miRNA profiling from adult human islets and compared it with embryonic stem cells (ESCs). We identified nine miRNAs that showed greater than 3-fold expression in human islets compared with ESCs (Figure 1A). Among the nine miRNAs identified, only miR-216a showed a pancreas-specific expression pattern (Figure 1B), while the other miRNAs were ubiquitously expressed in the analyzed tissues (Figure S1A). qRT-PCR analysis of miR-216a levels in multiple tissues showed that miR-216a was expressed in human islets and whole pancreas to comparable levels (Figure S1B). To examine whether miR-216a levels are changed during endocrine cell development, we differentiated human ESCs to pancreatic-like cells. When comparing sequential time points, the single largest increase in levels of miR-216a occurred on day 14 (Figure 1C) of the differentiation protocol, which marks the generation of PDX1+/NKX6.1+ pancreatic endocrine progenitor cells.16 miR-216a levels further increased during differentiation, reaching the highest levels at the final stage of differentiation (days 26–33), correlating with the presence of pancreatic endocrine cells. To further investigate miR-216a expression during development, we performed in situ hybridization with human fetal pancreatic tissue using DIG-labeled miR-216a-LNA probes. There was faint staining in the pancreatic tissue at the gestational age 8 weeks, 4 days (Figure 1D; Figure S2), during the time when insulin and glucagon double-positive cells emerge.17 However, we detected strong miR-216a staining in the branching pancreatic epithelium that harbor PDX1+/NKX6.1+ pancreatic endocrine progenitor cells.17 Analysis of human adult pancreas also revealed strong pancreatic miR-216a staining with further enrichment in pancreatic islets (Figure 1D; Figure S2). To further investigate the specificity of miR-216a for pancreatic tissue, we performed in situ hybridization on kidney capsule grafts obtained from mice and rats implanted with pancreatic progenitor cells that had developed to endocrine cells.18 There was strong reactivity for miR-216a in the grafts, whereas neighboring kidney sections had no detectable staining (Figure S1C). We next examined the miR-216a sequence across diverse species and found it to be highly conserved (Figure 1E), suggestive of a functional importance. A thorough tissue expression analysis from C57BL/6 mice showed that miR-216a levels were specifically expressed in the pancreas with further enrichment in pancreatic islets (Figure 1F). To further investigate the expression of miR-216a in ductal cells, we differentiated mouse exocrine cells into ductal cells and quantified miR-216a expression by qRT-PCR. Expression of miR-216a was significantly downregulated as the cells differentiated into ductal cells (Figures 1G and 1H). Overall, these data suggest a role for miR-216a miRNA in pancreatic acinar cell and islet development and/or function.
Figure 1.
miR-216a is enriched in pancreatic islets and is highly conserved among various species
(A) miRNA profiling of adult human islets compared with human embryonic stem cells. miRNAs with greater than 3-fold increase in the islets are shown in magenta. Arrow points to miR-216a. n = 3 human islet donors. Data represent mean log2 signals ± SEM (from human islets).
(B) Equal amounts of RNA from various human tissues (each a pool of three tissue donors) was reverse-transcribed, and miR-216a expression was determined using qRT-PCR. Threshold cycle 33 (Ct = 33) was arbitrarily set as 1.
(C) Human embryonic stem cells (ESCs) were differentiated to pancreatic endocrine cells for the indicated days, and miR-216a expression was measured using qRT-PCR and expressed relative to levels in undifferentiated ESCs. In ESCs, Ct = 27.
(D) Fetal and adult human pancreata were probed with DIG-labeled miR-216a and scrambled control miRNA probes at the indicated gestational weeks. Purple color indicates presence of miRNA expression. Scale bar, 100 μm.
(E) Comparison of mature miR-216a sequences in different species.
(F) Same as in (B) except that the tissues were harvested from 8-week-old C57BL/6 male mice. n = 3 mice. Threshold cycle 33 (Ct = 33) was arbitrarily set as 1. Individual data points are shown in (C) and (F), and data represent mean ± SEM.
(G) Bright-field images of acinar cells differentiated toward ductal cells at day 1 (D1) and day 5 (D5) of differentiation. Scale bar, 100 μm.
(H) miR-216a expression was measured using qRT-PCR and normalized to miR-93-5p levels. Expression at day 5 is arbitrarily set as 1. At day 0, miR-216a Ct = ∼18.
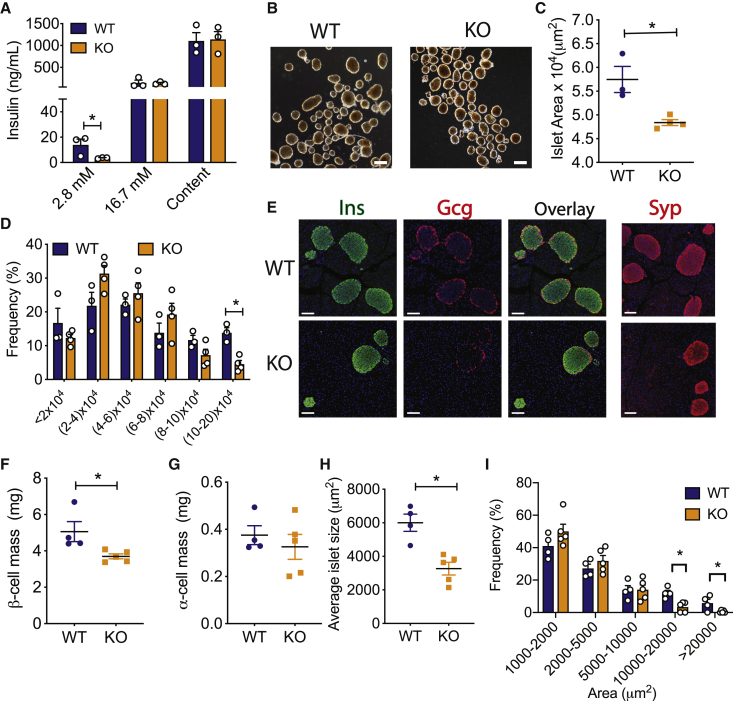
To explore the role of miR-216a in pancreatic islet function in vivo, we generated miR-216a knockout (KO) mice in which the precursor sequence of miR-216a (pre-miR-216) was deleted by homologous recombination.19 miR-216a KO mice were viable, and qRT-PCR analysis from isolated islets of miR-216a KO mice confirmed miR-216a was efficiently deleted (Figure S3A). Body weight, fasting blood glucose levels, and glucose tolerance were comparable between miR-216a KO and wild-type (WT) littermate controls when fed with a chow diet (Figures S3B–S3G). Pancreas weight and pancreatic cell size were unchanged (Figures S3H and S3I). To investigate the effect of miR-216a deletion on endocrine pancreas, we isolated islets from miR-216a KO mice and WT littermates and analyzed islet function. During glucose-stimulated insulin secretion (GSIS) assays, miR-216a KO islets secreted less insulin at basal glucose concentration (2.8 mM) (Figure 2A). To determine whether the observed effects on insulin secretion were cell autonomous and direct, we transfected rat INS-1 cells with miR-216a mimetics and performed GSIS assays. Inhibition of miR-216a did not change insulin secretion at low or high glucose concentrations (Figure S2J). Analysis of isolated islets revealed that miR-216a KO were smaller than the WT islets (Figure 2B), with miR-216a KO mice lacking bigger islets and trending toward having an increased number of smaller islets (Figures 2C and 2D). Immunostaining of pancreas from adult mice with insulin and glucagon antibodies demonstrated that β-cell mass was significantly reduced in the miR-216a KO mice compared with controls, while α-cell mass was unchanged (Figures 2E–2G). Islet circularity and the location of α-cells were unchanged (Figures S3K and S3L). Consistent with the isolated islet data, the area of islets determined by synaptophysin immunostaining was significantly smaller in miR-216a KO mice (Figures 2E and 2H), and miR-216a KO pancreatic sections harbored less of the larger islets, compared with WT controls (Figure 2I). In 1-day-old miR-216a KO pups and WT littermates, β-cell and α-cell area were not different (Figures S4A–S4C). Similarly, average islet size was comparable, with no statistical difference in various islet size groups (Figures S4A, S4D, and S4E). Islet circularity and peripheral α-cells were also comparable (Figures S4F and S4G), suggesting a postnatal role for miR-216a in regulating β-cell mass and islet size.
Figure 2.
miR-216a knockout (KO) mice have smaller islets and reduced β-cell mass
(A) Insulin secretion and content from islets isolated from male WT and KO mice exposed to 2.8 and 16.7 mM glucose. n = 3 mice.
(B) Representative images of isolated islets from 10-week-old male WT and KO mice. Scale bar, 100 μm.
(C and D) Average size of isolated islets (C) and the distribution of islet size (D). n = 3 or 4 mice. A two-tailed Student’s t test was performed to assess significance.
(E–I) Pancreata from 21-week-old male WT and KO mice were immunostained for insulin (Ins), glucagon (Gcg), and synaptophysin (Syp). (E) Representative images of insulin, glucagon, and synaptophysin immunostaining, along with an overlay of Ins and Gcg. Nuclei were identified with DAPI (blue). Scale bars, 100 μm. (F) β-cell mass, (G) α-cell mass, (H) average islet size, and (I) islet size distribution. n = 4 or 5 mice.
Individual data points are shown in ( (A), (C), (D), and (F–I). Data represent mean ± SEM. ∗p < 0.05.
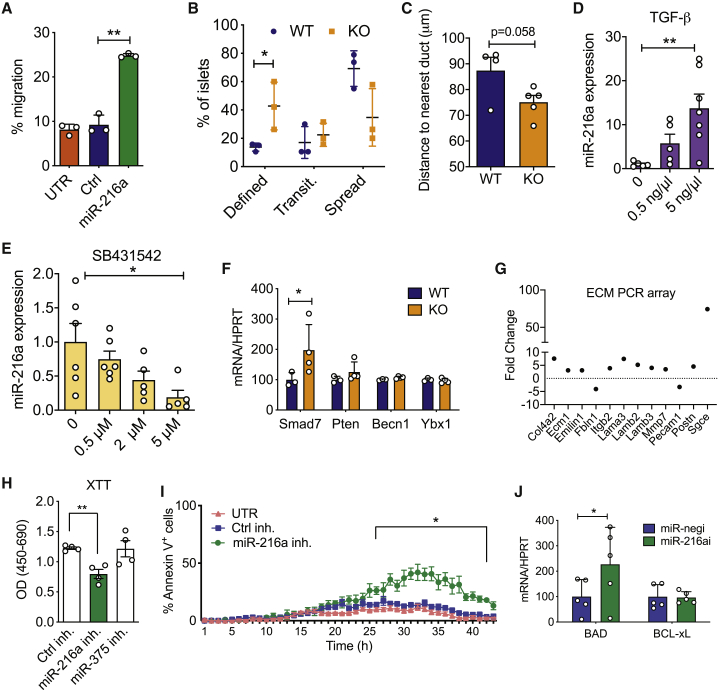
Regulation of islet size is complex and involves cellular processes such as fusion, fission, growth, and migration.20 Cell migration is critical for both islet formation and the movement of islets away from ducts.21,22 To test whether miR-216a alters cell migration and/or proliferation, we transfected PDAC PANC-1 cells that have very low miR-216a levels (not shown) and migration ability with control (ctrl) or miR-216a mimetics and performed a migration assay using transwell chambers. Quantification of the cells that traversed the Boyden chambers demonstrated that miR-216a more than doubled the number of migrating cells compared with untransfected and control miRNA-transfected wells (Figure 3A). We next assessed the migration ability of miR-216a KO islet cells ex vivo. We coated cell culture plates with the matrix secreted by 804G cells and monitored the spreading of islets by light microscopy. Five days post-seeding, more WT islets spread compared with miR-216a KO islets, while miR-216a KO islets had more defined borders, suggestive of less spreading (Figure 3B). To further explore the possibility that miR-216a KO alters islet migration, we quantified the distance of islets from the ducts in both WT and KO pancreata. Islets from miR-216a KO mice were closer to the ducts compared with WT islets (p = 0.058) (Figure 3C).
Figure 3.
miR-216a regulates cell migration and apoptosis
(A) Percentage of calcein-stained PANC-1 cells migrating through a transwell following transfection with miR-216a or control (Ctrl) mimetics or untransfected (UTR). n = 3 technical replicates.
(B) Percentage of islets isolated from 10-week-old male WT or KO mice that appeared as having defined, transitionary (transit.), or spread boundaries 3 days after plating on collagen wells. n = 3 mice.
(C) Distance to duct was quantified using the H&E stains of WT and miR-216a KO adult pancreata.
(D and E) Human EndoC-βH1 cells were treated with varying concentrations of TGF-β or TGF-β inhibitor SB431542, and miR-216a levels were quantified using qRT-PCR. n = 6 technical replicates. Ct levels for miR-216a in untreated human EndoC-βH1 cells = ∼21.
(F) Islets from 10-week-old male WT and KO mice were isolated and expression of Smad7, Pten, Becn1, and Ybx1 was quantified with qRT-PCR. n = 3 or 4 mice.
(G) MIN6 cells were transfected with miR-216a and control mimetics and expression of 84 ECM-related genes was quantified using qRT-PCR. Genes displaying >2-fold difference are shown. Each value is the mean of three independent transfections.
(H and I) INS1-E cells were transfected with the indicated miRNA inhibitors (inh.) or with a scrambled control miRNA inhibitor (Ctrl inh.), and cell viability was assessed using XTT (2,3-bis-[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide) assay (H) or live cell imaging using Hoechst and Alexa 647 annexin V (I). TNF-α, IFN-γ, and IL-1β were added to media prior to imaging cells at 37°C and 5% CO2 in an ImageXpress Micro. n = 4 technical replicates.
(J) Mouse islets were transfected with a control miRNA (miR-negi) and miR-216a inhibitor. Expression of Bad and Bcl-xl was quantified using qRT-PCR. n = 5 mice.
A two-tailed Student’s t test (A–F, H, and J) or two-way ANOVA with Bonferroni’s multiple-comparison post-test (I) was performed to assess significance. ∗p < 0.05 and ∗∗p < 0.01. Individual data points are shown in (B)–(F), (H), and (J). Data represent mean ± SEM.
TGF-β signaling regulates pancreatic epithelium branching and cell migration to form islet clusters.23 It has been previously shown that miR-216a expression is regulated by TGF-β signaling and alters expression of Pten and Smad7.24,25 To investigate the potential signaling pathways regulating miR-216a expression in the pancreas, we treated EndoC-βH1 cells, which have high endogenous miR-216a levels, with a TGF-β agonist TGF-β1 and the inhibitor SB431542 and measured miR-216a levels by qPCR. TGF-β1 treatment significantly increased miR-216a expression and inhibition of TGF signaling with SB431542 significantly decreased miR-216a levels (Figures 3D and 3E). We next examined mRNA levels of potential miR-216a target genes in the pancreatic islets of WT and miR-216a KO mice. qRT-PCR analysis of islets of WT and miR-216a KO mice indicated that Smad7 expression was significantly upregulated in the miR-216a KO islets, whereas Pten, Becn1, and Ybx1 levels were unchanged (Figure 3F). We also examined expression of key hormones and transcription factors regulating endocrine cell identity. Insulin, Glucagon, Pdx1, and Nkx6.1 levels were not changed in islets from miR-216a KO mice (Figure S5). TGF-β signaling and Smad7 are known regulators of cell migration, and their mode of action is thought to be via altering the expression of genes involved in regulating extracellular matrix (ECM) composition.26 We analyzed the levels of ECM genes in miR-216a transfected cells using an ECM gene array. miR-216a altered the expression of 12 of the 87 genes analyzed (Figure 3G). Levels of genes involved in basement membrane, such as Col4a2, Ecm1, Fbln1, Lama3, Lamb2, and Lamb3, were upregulated in the miR-216a transfected cells, suggesting that TGF-β-induced miR-216a can increase cell migration by altering the ECM integrity of basement membrane. We next investigated whether inhibition of miR-216a can affect cell proliferation and cell death by transfecting INS-1E β-cells with miR-216a and a control scrambled miRNA inhibitor. INS-1E β-cells have higher levels of miR-216a compared with other rodent β-cell lines such as MIN6 (not shown) and thus were chosen for miR-216a inhibition experiments. Inhibition of miR-216a significantly reduced cell proliferation compared with control miRNA inhibitors (Figure 3H). Analysis of cell death using live cell imaging indicated that inhibition of miR-216a in the presence of cytotoxic factors (TNF-α, IFN-γ, IL-1β) increased the rate of apoptosis (Figure 3I). Similarly, inhibition of miR-216a in mouse islets increased expression of the pro-apoptotic gene Bad (Figure 3J).
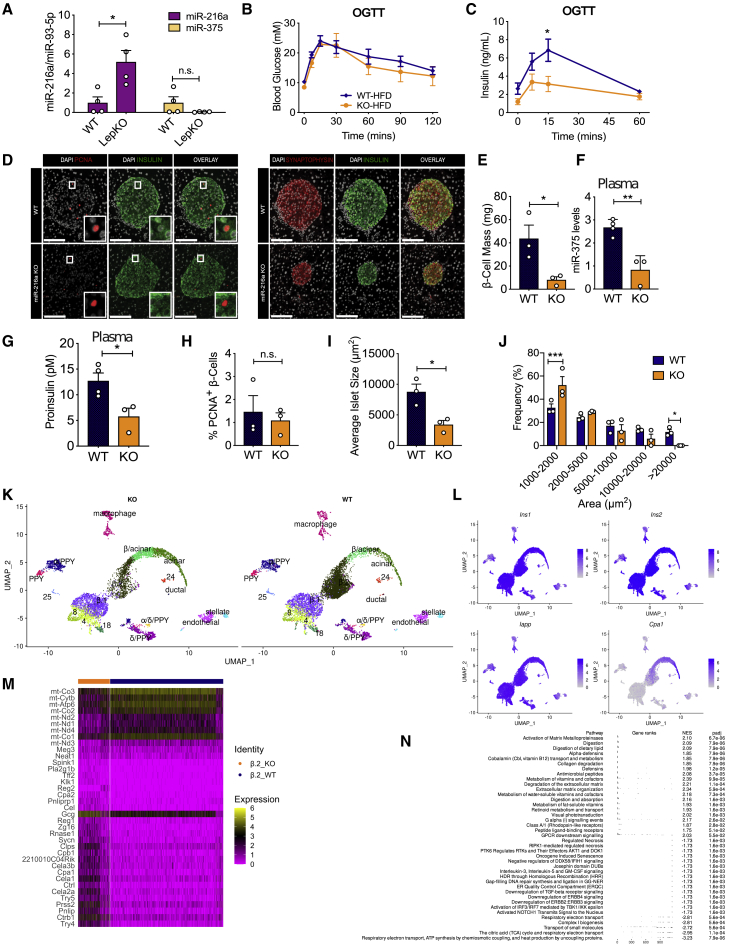
Many miRNAs regulated by metabolic stressors coordinate the adaptive responses of islet cells to metabolic stresses during obesity and diabetes.27 We sought to investigate whether deletion of miR-216a would change insulin secretion when challenged with metabolic overload. We first explored miR-216a levels in the islets of leptin KO (LepKO) rats, a model of obesity with disturbed glucose and energy metabolism and larger islets.28 miR-216a levels were significantly higher in the LepKO rat islets, whereas levels of another well-studied islet enriched miRNA, miR-375, were unchanged (Figure 4A). Notably, miR-216a levels were also reported to be increased in human islets from subjects with T2D.29 When placed on a 60% high-fat diet (HFD) for 8 weeks, fasting body weight and blood glucose levels were comparable between miR-216a KO mice and littermate WT controls (Figures S6A and S6B). However, an oral glucose tolerance test performed 2 weeks after HFD revealed that miR-216a KO mice secreted less insulin in response to glucose administration despite no significant differences in blood glucose levels (Figures 4B and 4C). Insulin tolerance tests indicated similar insulin sensitivity of miR-216a KO and WT mice (Figure S6C), suggesting compromised islet function in miR-216a KO mice under HFD. Consistent with the reduced insulin secretion from miR-216a KO mice in vivo, pancreata from miR-216a KO mice had significantly reduced β-cell mass compared with controls after HFD feeding (Figures 4D and 4E). Furthermore, miR-216a KO mice had decreased circulating miR-375 and proinsulin levels compared with WT after 8 weeks of HFD feeding (Figures 4F and 4G). The degree of cell proliferation was comparable in the WT and miR-216a KO β-cells as assessed by PCNA immunostaining (Figure 4H), and miR-216a KO mice fed the HFD also had significantly smaller islets (Figure 4I), with increased frequency of smaller islets and decreased occurrence of larger islets (Figure 4J). Overall, these data indicate that under metabolically stressed condition of 60% HFD, miR-216a KO mice secrete lower insulin during a glucose challenge and have decreased islet size with reduced β-cell mass and proinsulin protein levels.
Figure 4.
miR-216a KO mice have decreased insulin secretion, β-cell mass, and islet size on a high-fat diet (HFD)
(A) Islets were isolated from WT and leptin knockout (LepKO) male rats, and expression of miRNAs was quantified using qRT-PCR and expressed relative to levels in WT cells.
(B–J) WT and miR-216a KO male mice were fed with a 60% HFD for 8 weeks. Oral glucose tolerance tests (OGTT) were performed 2 weeks post-HFD; blood glucose levels are shown in (B) and plasma insulin concentrations in (C). (D) Pancreata from WT and miR-216a KO mice were fixed and stained with the indicated antibodies 8 weeks post-HFD (n = 3 mice/group). Representative images are shown. Scale bars, 100 μm. Insets are enlarged 4×. Quantifications of insulin (E), PCNA (H), and synaptophysin (I and J) immunoreactivity. (F) qRT-PCR analysis for miR-375 from plasma of WT and miR-216a KO mice, 8 weeks post-HFD; n = 3 or 4 mice. (I) Average islet size and (J) islet size distribution on the basis of synaptophysin immunostaining.
(K–N) Single-cell RNA-seq was carried out on mouse pancreatic islets. (K) UMAP plot of cells from KO and WT animals labeled by cell type, identified by expression of canonical genes, showing the presence of all major cell types in both genotypes in approximately equal proportions. (L) Normalized expression values of Ins1, Ins2, and Iapp shows likely contamination across all cells but particularly high expression of all three canonical β-cell genes in the β.1 and β.2 clusters. Normalized expression of Cpa1, a gene differentially expressed in acinar cells, is highest in the acinar cluster but has increased expression in the β.2 cluster compared with the β.1 cluster. (M) A heatmap showing normalized gene expression for the statistically significantly up- or downregulated genes within the β.2 cluster comparing between genotypes, further analyzed in (N), showing gene set enrichment analysis on all differentially expressed genes, reveals an increased acinar phenotype in cells within this cluster from KO animals compared with WT.
A two-tailed Student’s t test (A, E–J) or two-way ANOVA with Sidak’s multiple-comparison post-test (C) was performed to assess significance. ∗p < 0.05, ∗p < 0.01, and ∗∗∗p < 0.001. Individual data points are shown in (A) and (E)–(J). Data represent mean ± SEM.
For an unbiased investigation of islet dysfunction, we performed single-cell RNA sequencing (RNA-seq) analysis from the islets of WT and miR-216a KO mice exposed to 60% HFD for 9 weeks. Cell counts and viability assessments prior to sequencing did not indicate any differences between WT and KO islet cell preparations (Table S1). After sequencing, we assigned cell clusters to specific cell types for further analysis on the basis of differential expression of canonical genes (Tables S2 and S3). Some clusters contained multiple cell populations (Figure 4K), and some cell types were contained in multiple clusters. The proportion of cells in each cluster were similar, and all clusters were represented in both genotypes (Figure 4K). β-cells were restricted to those that had statistically significantly upregulated expression of Ins1, Ins2, and Iapp genes (Figure 4L). Many clusters had no (7 of 30 clusters) or fewer than 10 (15 of 30 clusters) differentially expressed genes between genotypes (Table S4), suggesting that overall, most pancreatic cells were similar between genotypes. There appeared to be two major subpopulations of β-cells (Figures 4K and 4L); one population contained clusters with upregulation of genes typical for mature β-cells, such as Unc3 and Mafa (β.1; Table S3), whereas the other population (β.2) lacked these genes and had higher expression of Cpa1, a canonical acinar gene, as well as being closer to the acinar population in local UMAP (uniform manifold approximation and projection) space (Figure 4L) compared with cluster β.1. There were no or few differentially expressed genes in the β.1 cluster between the genotypes, but many genes were differentially expressed in the β.2 cluster when comparing WT and KO cells (Figure 4M; Table S4). Similarly, many genes were differentially expressed in the acinar cell cluster (Figure S6D; Table S4). To further investigate the differentially expressed genes, we performed gene set enrichment analysis for the two most different clusters, β.2 and acinar cells (Figure 4N; Figure S6E). For the β.2 cluster, many genes related to digestion were upregulated, suggesting that islets from KO mice had more characteristics of acinar-like cells than those of WT mice. Additionally, the TGF-β receptor signaling pathway was downregulated in KO cells compared with WT. Many pathways that were significantly different between WT and KO cells in the β.2 cluster, including “activation of matrix metalloproteinases,” “digestion,” and “collagen degradation,” were also different in the acinar cell clusters. Interestingly, Gcg was statistically significantly upregulated in many of the KO clusters.
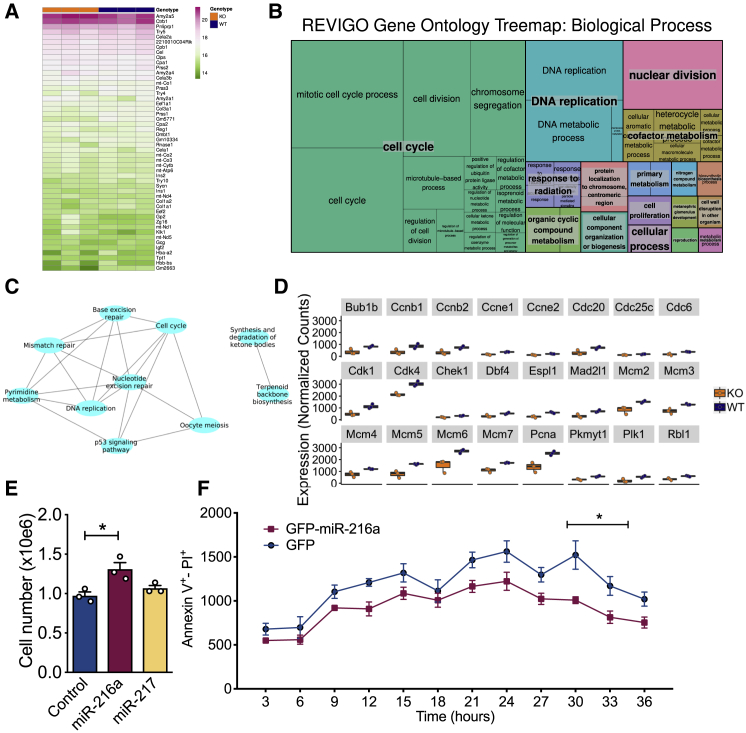
As miR-216a is expressed in both endocrine and exocrine pancreas, we sought to perform a global analysis to further explore signaling pathways targeted by miR-216a in the whole pancreas. We performed RNA-seq from the pancreata of 1-day-old WT and miR-216a KO mice. The RNA integrity numbers (RINs) obtained from the pancreata of miR-216a KO mice were all above 8 and suitable for RNA-seq (not shown). We examined the top 50 most abundant genes expressed in WT and miR-216a KO pancreata and observed the expected abundance for genes encoding amylase, trypsin, and insulin, but they were not significantly different between WT and miR-216a KO mice (Figure 5A). Differential gene expression analysis from all the transcripts using an adjusted p value < 0.05 revealed that 409 genes were differentially expressed between WT and KO pancreata (Table S5). To identify more globally the affected biological processes in the miR-216a KO pancreata, we next performed a Gene Ontology (GO) analysis and found cell cycle to be most significantly altered (Figure 5B). Similarly, KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis indicated that cell cycle, DNA replication, and repair pathways were statistically different in the miR-216a KO versus WT pancreata (Figure 5C). Next, we compared the expression of genes involved in cell cycle pathways between WT and miR-216a KO pancreata and observed that many cyclin-dependent kinases (e.g., Cdk4, Cdk1) and cyclins (e.g., Ccne1) as well as DNA replication genes (e.g., Pcna, Mcm2, Mcm5) were decreased in the pancreas of miR-216a KO mice (Figure 5D). This was consistent with our observation that inhibition of miR-216a decreases cell proliferation in INS-1E β-cells (Figure 3H) and the presence of reduced β-cell mass and islet size in the miR-216a KO mice. A map of the cell cycle pathway with genes significantly regulated in miR-216a KO pancreata is shown in (Figure S7). To investigate the effect of miR-216a on cell proliferation in cells with exocrine origin, we transfected human PANC-1 cells with the miR-216a mimetic and another miRNA, miR-217. Similar to INS1-1E β-cells, miR-216a transfection significantly increased the number of cells (Figure 5E), and decreased the rate of apoptosis (Figure 5F), thus confirming the role of miR-216a in regulating cell proliferation in another pancreatic cell type.
Figure 5.
RNA-seq analysis of pancreata from miR-216a-KO mice
(A) RNA from the pancreata of 1-day old male WT and miR-216a KO mice was isolated and subjected to RNA sequencing and top 50 most abundant genes are shown by heatmap.
(B) A treemap plot, combining statistically significant GO terms in the biological processes category into similar terms. The size of the square increases with a decreasing p value, and the color of the square indicates the grouping of like terms (labeled in larger text with a gray background). Statistically significant terms were identified using hypergeometric tests with a false discovery rate of 0.1.
(C) All statistically significant KEGG terms are shown in a network map, with nodes representing KEGG terms and edges connecting nodes representing differentially expressed genes in common between KEGG terms. Statistically significant terms were identified using hypergeometric tests with a false discovery rate of 0.1.
(D) Normalized gene expression data for key genes of interest. All genes shown are differentially expressed, with adjusted p values < 0.05 (adjusted by the Benjamini-Hochberg correction).
(E and F) PANC-1 cells were transfected with the indicated miRNAs, and 48 h later TGF-β was added to cell culture media and cell number was counted (E). Individual data points are shown. n = 3 technical replicates. (F) Cell death was assessed with live cell imaging using Hoechst and Alexa 647 annexin V. Cells were imaged at 37°C and 5% CO2 in an ImageXpress Micro. n = 5 technical replicates.
A two-tailed Student’s t test (E) or two-way ANOVA with Bonferroni’s multiple-comparison post-test (F) was performed to assess significance. ∗p < 0.05. Data represent mean ± SEM.
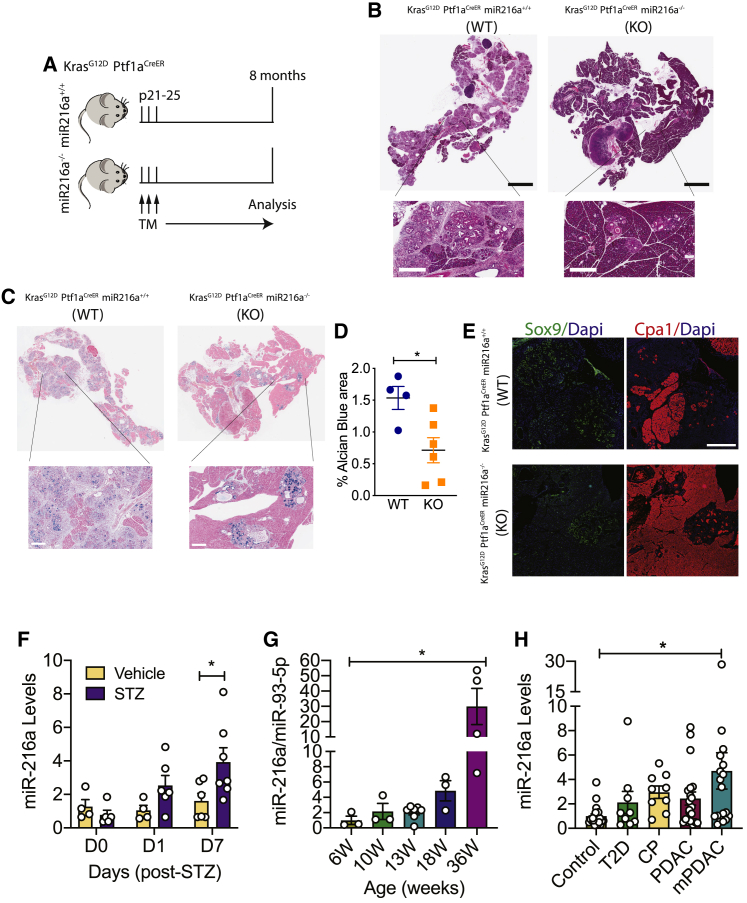
We next investigated whether the absence of miR-216a alters the progression of a pancreatic pathology related to cell cycle/proliferation in acinar cells. Oncogenic KRAS can induce PDAC precursor lesions from pancreatic acinar cells.30,31 A number of studies have shown that PDAC develops from abnormally proliferating cells in the precursor lesions termed pancreatic intraepithelial neoplasia (PanIN), the most common precursor lesions observed in humans.32 A mutation in KRAS oncogene is currently considered as the initiating factor in pancreatic cancer.33 Expression of constitutively active KrasG12D allele in mice induces PanINs and, after a significant latency period, PDAC.34 We crossed miR-216a KO mice with the pancreatic cancer-prone KrasG12D;Ptf1aCreER mice (Figure 6A) and analyzed the frequency of PanINs in the pancreata of offspring. As expected, H&E staining from the pancreata of KrasG12D;Ptf1aCreER;miR-216a(+/+) mice revealed widespread lesions with columnar to cuboidal cells with varying degrees of cytological and architectural atypia (Figure 6B). In contrast, the pancreata of mice lacking miR-216a had limited neoplastic lesions. Further histological analysis with Alcian blue staining confirmed the histological characteristic of high acidic mucin content of PanINs in the WT mice, whereas miR-216a KO pancreata had decreased PanIN frequency (Figure 6C). In KrasG12D;Ptf1aCreER;miR-216a(−/−) mice, there was more than 2-fold reduction in the Alcian blue+ area compared to KrasG12D;Ptf1aCreER;miR-216a(+/+) mice (Figure 6D). We assessed acinar-to-ductal metaplasia by immunostaining the pancreas using the acinar and ductal cell markers Cpa1 and Sox9, respectively. Pancreata from the KrasG12D;Ptf1aCreER ;miR-216a(+/+) mice had increased Sox9 and reduced CpaI expression indicative of ductal metaplasia, with widespread acinar cell loss. Pancreata from KrasG12D;Ptf1aCreER;miR-216a(−/−) mice maintained abundant acinar cell content with little ductal-like cells (Figure 6E). These data indicate that acinar cells from the miR-216a(−/−) mice have less propensity than miR-216a(+/+) mice to form PanINs and acinar-to-ductal metaplasia in response to oncogenic KRAS.
Figure 6.
miR-216a-KO mice have a lower incidence of pancreatic intraepithelial (PanIN) lesions
(A) Schematic showing the design of the study. Expression of KrasG12D was induced in exocrine cells by three tamoxifen injections on alternating days when the mice were 21–25 days old. Pancreata were harvested 8 months later.
(B) Pancreatic H&E staining. Scale bars (black), 3 mm; inset scale bars (white), 400 μm.
(C) Pancreatic Alcian blue staining. Insets are 5× enlarged; inset scale bars, 400 μm.
(D) Alcian blue-positive area. Individual data points are shown. n = 4–6 mice. A two-tailed Student’s t test was performed to assess significance. Data represent mean ± SEM. ∗p < 0.05.
(E) Representative images of Cpa1 and Sox9 immunostaining. Scale bar, 500 μm.
(F) C57BL/6 mice were treated with a single high dose (180 mg/kg) of the β-cell toxin STZ or vehicle injection. Total RNA was isolated from the plasma on the indicated days (D0, D1, and D7), and miR-216a levels were measured using qRT-PCR. n = 4–7 mice.
(G) Total RNA was isolated from the plasma of LSL KrasG12D;Ptf1a-Creex1 mice at the indicated weeks. miR-216a levels were measured using qRT-PCR and normalized to miR-93-5p levels. n = 3–6 mice.
(H) Total RNA was isolated from the sera of patients with type 2 diabetes (T2D), chronic pancreatitis (CP), pancreatic ductal adenocarcinoma (PDAC), and pancreatic ductal adenocarcinoma with metastasis (mPDAC) and control subjects. miR-216a levels were measured using qRT-PCR. n = 9–20 human subjects. Ct for controls = ∼32.
A two-way ANOVA (F) or one-way ANOVA (G and H) with Bonferroni’s multiple-comparison post-test was performed to assess significance. ∗p < 0.05. Data represent mean ± SEM.
Finally, to explore whether circulating miR-216a levels are changed in diabetes and pancreatic cancer, we used STZ-induced diabetes and LSL-KrasG12D;Ptf1a-Creex1 pancreatic cancer-prone mouse models, respectively. Plasma miR-216a levels were increased by 1 day post-STZ, reaching significant levels 7 days post-injection (Figure 6F). In the pancreatic cancer mouse model, we detected a significant increase in plasma miR-216a levels as the mice developed tumors, reaching significance at 36 weeks of age (Figure 6G). To investigate the relevance in humans, we measured miR-216a levels from the sera of patients with type 2 diabetes (T2D), chronic pancreatitis (CP), PDAC, or PDAC with metastasis (mPDAC). Age and gender information of the donors is provided in Table S6. miR-216a levels were not elevated in T2D patients (Figure 6H). Yet there was a trend toward an increase in miR-216a levels in all exocrine pancreas-related diseases, with a significant increase in sera from patients with PDAC with metastasis. These data suggest a clinical opportunity to use circulating miR-216a as a biomarker for exocrine-related pancreatic diseases.
Discussion
We predicted that pancreas-enriched miRNAs likely have specific, critical roles for pancreas function and hence sought to identify miRNAs enriched in pancreatic cells. Using human and mouse tissue panels, fetal and adult human pancreas and human endocrine cell transplant models, we demonstrated that miR-216a is specific to pancreatic tissue. It was highly expressed in the islets and acinar cells, with little expression in the ducts. Therefore, a whole-body miR-216a KO mouse was used to investigate the role of miR-216a in both endocrine and exocrine compartments in vivo. Islets isolated from the miR-216a KO mice were smaller and secreted less insulin than control islets. Similarly, analysis of pancreatic tissue confirmed the presence of smaller islets and reduced β-cell mass in the miR-216a KO adult mice. Interestingly, pancreata from the miR-216a KO newborn mice had a trend toward increased islet size. Islet growth is size dependent during development, with preferential expansion of smaller islets and fission of large interconnected islet-like structures occurring most actively at approximately 3 weeks of age at the time of weaning.20 The ROCK-nmMyoII pathway was shown to coordinate epithelial cell morphogenesis by narrowing the apical surface of the cell, basalward cell movement and cell-rear detachment from the apical lumen, enabling clustering as nascent islets under the basement membrane.35 More recently, it was shown that as endocrine progenitors differentiate, they migrate in cohesion and form bud-like islet precursors.36 As we observed a major difference in islet size in the miR-216a KO mice postnatally, our data support a postnatal role for miR-216a in islet cell expansion and migration. Indeed, we showed miR-216a increases cell migration and proliferation in vitro. Islets from miR-216a KO mice were also closer to the ducts compared with WT islets. Further studies are required to examine the spatiotemporal expression of miR-216a within the epithelial plexus.
Levels of miR-216a were regulated by TGF-β signaling and deletion of miR-216a led to an increase in Smad7 levels in the miR-216a KO islets. This result is in alignment with published data showing that forced expression of miR-216a increases epithelial-to-mesenchymal transition (EMT) and migration with metastatic ability in epithelial hepatic cellular carcinoma (HCC) cells by targeting Pten and Smad7.25 Although we did not observe any differences in Pten, Becn1, and Ybx1 mRNA levels, we found an increase in Smad7 levels in islets of miR-216a KO mice. We also observed changes in the expression of genes involved in EMT and migration in MIN6 and PANC-1 cells. Lack of Pten, Becn1, and Ybx1 regulation by miR-216a in our study could be explained by differences in target gene selection under physiological versus overexpression conditions. Alternatively, the selection of studied cell types (pancreatic islets versus renal mesangial cells or endothelial cells) could lead to differences in miRNA targets.
miRNAs can mediate various stress responses in diseases.27 Although miR-216a KO mice had smaller islets and reduced β-cell mass, it was insufficient to cause glucose intolerance even when placed on 60% HFD, suggesting absence of miR-216a did not affect glucose homeostasis in vivo under metabolic stress. In rodents, hyperglycemia is reported to develop when ∼80% of β-cell mass is lost.37 The extent of β-cell loss in patients with overt T2D is estimated at ∼65%.38 Type 1 diabetes is diagnosed when patients lose the majority of β-cells (>70%).39 The degree of β-cell mass reduction in the KO mice was not sufficient to significantly impair glucose tolerance. However, miR-216a KO mice had smaller islets, reduced β-cell mass, and diminished insulin secretion. A recent study showed that miR-216a-targeting theranostic nanoparticles could promote proliferation of insulin-secreting cells in vivo.40 These results are in line with our data and provide additional evidence for a role of miR-216a in regulating β-cell mass. Single-cell RNA sequencing from the islets of HFD fed WT and KO mice revealed no major differences in the proportions of different cell types. However, interestingly, we observed a subpopulation of β-cells with upregulated acinar cell markers in the KO islets, suggesting a dysfunctional β-cell subpopulation may contribute to the islet dysfunction in the KO mice.
Analysis of RNA-seq results from the pancreata indicated that cell cycle pathways were the most significantly enriched GO term in the miR-216a KO mice. Similarly, DNA replication, nuclear division, and cell proliferation were among the most altered biological processes. As miRNAs affect the expression of multiple genes, identifying signaling pathways targeted by miRNAs rather than minor changes in individual genes is more meaningful to interpret the role of miRNAs in the cellular and physiological context. By opting to isolate pancreatic RNA from 1-day-old pups, we focused on the earliest biological changes that occurred in the miR-216a KO mice postnatally, thereby revealing the potential causal mechanisms for the observed phenotype. On the basis of the expression of miR-216a in the acinar tissue as well as on the altered pathways in miR-216a KO pancreata, we hypothesized miR-216a may have a functional role in pancreatic cancer. Our data suggest that deletion of miR-216 in the presence of oncogenic signals reduces incidence of neoplastic lesions in pancreatic cancer. In contrast to our findings, ectopic miR-216 expression in PANC-1 cells inhibited tumor growth in mouse xenografts.41,42 However, these studies are considerably different from ours. We induced pancreatic cancer in the pancreatic acinar cells in vivo by crossing the pancreatic cancer-prone mouse line KrasG12D with Ptf1CreER mice. Thus, in our studies, miR-216a was fully absent, including during embryonic development. In addition, pancreatic cancer was induced in our studies in the acinar pancreatic tissue, whereas in the aforementioned studies, PANC-1 cells were implanted subcutaneously, such that the findings may not be directly comparable with ours. Engineered exosomes carrying short hairpin RNA specific to oncogenic KrasG12D targeted oncogenic KRAS with enhanced efficacy and suppressed cancer in multiple mouse models of pancreatic cancer.43 It remains to be investigated whether inhibition of miR-216a either alone or in combination with KRAS can lead to a greater attenuation of tumor growth and an increase in overall survival in PDAC.
Cancer is characterized by uncontrolled proliferation resulting from aberrant activity of cell cycle proteins, so many cell cycle regulators are important targets in cancer therapy.44 A hallmark genetic event in PDAC is loss of the CDKN2A/2B tumor suppressor locus, which encodes Cdk4/6 inhibitors that are particularly important for KRAS driven tumors, such as PDAC.45, 46, 47 Our RNA-seq analysis identified downregulation of many cell cycle and proliferation genes, such as Cdk1, Cdk4, and Pcna, in the miR-216a KO mice, and we found reduced PanIN formation in the presence of oncogenic KRAS, both of which suggest partial protection from pancreatic cancer initiation. TGF-β signaling pathway is another important and commonly deregulated signaling pathway in pancreatic carcinomas.48 Activation of TGF-β leads to phosphorylation of Smad complexes to translocate to the nucleus and activate their target genes involved in cell cycle progression.48 We showed that miR-216a levels are regulated by TGF-β signaling and miR-216a deletion increases levels of Smad7, which is a known inhibitor of cell cycle progression. Our results support a model whereby TGF-β-induced miR-216 directly reduces Smad7 levels and increases the expression of genes involved in cell cycle progression, which in the presence of oncogenic mutations such as KRAS give rise to PanIN lesions. Whether there are other direct miR-216a targets mitigating this effect needs further evaluation. RNA-interference-based therapies are becoming more attractive because of their stability and sequence specificity and the relative ease of synthesis.49
miR-375 was the first reported islet-“specific” miRNA with a role in maintaining normal α- and β-cell mass.50,51 Although we also observed an enrichment for miR-375 in human and mouse islets, it is also expressed at high levels in other tissues, and non-pancreatic roles for this miRNA have been identified, such as in the regulation of gut mucosal immunity and gastric tumorigenesis.52,53 Specificity of miR-216a for the pancreas suggests clinical opportunities to exploit this miRNA as a biomarker in pancreatic pathologies. Indeed, in humans, we show a significant increase in serum miR-216a levels in patients with metastatic PDAC. Previously, Goodwin et al.54 showed that circulating miR-216 levels are increased in a mouse model of acute pancreatitis. Given the pancreas specificity of miR-216a, it is not surprising for miR-216a to be elevated in pancreatic cell damage mouse models. Our human data also point to a trend toward elevated miR-216a levels in CP. Given the complexity of accessing pancreatic tissue for biopsy, a blood-based biomarker that can diagnose pancreatic pathologies would be of tremendous value for clinical use. At the very least, circulating miR-216a could be useful as a biomarker for the noninvasive monitoring of PDAC progression in mice.
In summary, our results reveal a pancreas-specific miRNA (miR-216a) with physiological roles in both endocrine and exocrine cells and reveal how dysregulation of miR-216a levels affects β-cell mass and pancreatic cancer initiation. Given the specificity of miR-216a for the pancreas and elevated blood levels in pancreatic diseases, our findings also provide biomarker opportunities to evaluate disease status or progression.
Limitations of study
In this study, we investigated the function of miR-216a using whole-body miR-216a KO mice. Although miR-216a is expressed mainly in the pancreas, we cannot exclude the possibility that some of the effects we observed resulted from miR-216a deletion in other organs. The role of miR-216a in pancreatic cancer was studied in a mouse model in which a common KrasG12D mutation seen in pancreatic cancer was induced in acinar cells. Yet pancreatic cancer can originate in acinar or ductal cells, and pancreatic tumors accumulate other mutations such as TP53, CDKN2A, and SMAD4. It remains to be investigated how miR-216a affects tumor progression in tumors with ductal origin and in the presence of multiple driver mutations. We observed an increase in serum miR-216a levels in pancreatic pathologies, especially in pancreatic cancer. Further studies are required to evaluate the utility of miR-216a as a serum biomarker of pancreatic pathologies in larger cohorts and whether it can distinguish among different forms of pancreatic cancer and/or pancreatitis.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit α-Insulin | Cell Signaling | Cell Signaling Technology Cat# 3014; RRID: AB_2126503 |
| Guinea-pig α-Insulin | Thermo Scientific | Thermo Fisher Scientific Cat# PA1-26938; RRID: AB_794668 |
| Mouse α-Glucagon | Sigma Aldrich | Sigma-Aldrich Cat# G2654; RRID: AB_259852 |
| Mouse α-Synaptophysin | Monosan | Cell Sciences Cat# MON9013; RRID: AB_419808 |
| Rabbit α-Synaptophysin | Novus | Novus Cat# NB300-653; RRID: AB_10126406 |
| Mouse α-PCNA | BD Transduction | BD Biosciences Cat# 610665; RRID: AB_397992 |
| Rabbit α-Sox9 | Millipore | Millipore Cat# AB5535; RRID: AB_2239761 |
| Goat α-Cpa1 | R&D systems | R and D Systems Cat# AF2765; RRID: AB_2085841 |
| Biological samples | ||
| Human Total RNA Survey panel | Thermo Fisher Scientific | #AM6000 |
| Human islets | Irving K. Barber Human Islet Isolation Laboratory (Vancouver, British Columbia) | N/A |
| Human fetal pancreas | Riedel et al., 201117 Provided by Dr. Renian Wang, University of Western Ontorio, London, Canada | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Type XI collagenase | Sigma Aldrich | #C7657 |
| FBS | Sigma Aldrich | #F1051 |
| Sodium pyruvate | Sigma Aldrich | #S8636 |
| TGF-β | R&D systems | #240-B-002 |
| SB431542 | Selleck Chemicals | #S1067 |
| TNF-α | R&D systems | #510-RT-010 |
| IL-1β | Cederlane | #CLR01 |
| IFN-γ | Ray Biotech | #228-10827-2 |
| Critical commercial assays | ||
| ECM PCR array | QIAGEN | PAMM-013ZC |
| Migration assay | Trevigen | #3465-096-K |
| Proinsulin ELISA | Mercodia | #10-1232-01 |
| Insulin ELISA | Alpco | #80-INSMSU-E01 |
| miRNeasyRNA isolation kit | QIAGEN | #217004 |
| miRCURY RNA isolation kit | Exiqon (now QIAGEN) | #301110 |
| Universal cDNA Synthesis kit | Exiqon (now QIAGEN) | #203300 |
| ExiLENT SYBR® Green master mix | Exiqon (now QIAGEN) | #203403 |
| iScript cDNA Synthesis kit | Bio-Rad Laboratories | #1725035 |
| Ssofast EvaGreen Supermix | Bio-Rad Laboratories | #1725201 |
| In situ hybridization kit | Biochain | #K2191050 |
| Deposited data | ||
| Single-cell RNA-seq data | This paper | GEO: GSE159844 |
| RNA-seq data | This paper | GEO: GSE159970 |
| Experimental models: Cell lines | ||
| miR-216a KO ES cell clones (4H7 and 4H9) | Prosser et al., 201119 Wellcome Trust Sanger Institute | Mutant Mouse Resource and Research Center (USA) |
| CA1S | Caron et al., 201355 Provided by Dr. James Piret | N/A |
| Human EndoC-βH1 | Ravassard et al., 201156 Provided by Drs. Raphael Scharfmann, and Philippe Ravassard | N/A |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | The Jackson Laboratory | Stock No: 000664 |
| Mouse: miR-216a−/− | This paper | N/A |
| Mouse: B6.129S4-Krastm4Tyj/J | The Jackson Laboratory | Stock No: 008179 |
| Mouse: B6.129S6(Cg)-Ptf1atm2 (cre/ESR1)Cvw/J |
The Jackson Laboratory | Stock No: 036577 |
| Oligonucleotides | ||
| Insulin_fwd: CATCAGCAAGCAGGTCATTG | This paper | N/A |
| Insulin_rev: GCTCCCAGAGGGCAAG | This paper | N/A |
| Glucagon_fwd: CGCAGGCACGCTGATG | This paper | N/A |
|
Glucagon_rev: ACGAGATGTTGTGAAGA TGGTTG |
This paper | N/A |
|
Smad 7_fwd: CTCATTGGAAATTTAGCAT TGTAGTGC |
This paper | N/A |
|
Smad 7_rev: CACATTGCACAATACATAA ATAATGATGC |
This paper | N/A |
| Pten_fwd: GGTGGAACGGCTGACAGCTA | This paper | N/A |
| Pten_rev: GGATGCTTCATGTGCTGCCTG | This paper | N/A |
|
Becn1_fwd: CAGCCTCTGAAACTGGAC ACGA |
Primer sequences are from Origene | #MP201300 |
|
Becn1_rev: CTCTCCTGAGTTAGCCTC TTCC |
Primer sequences are from Origene | #MP201300 |
| miR-216a, LNA PCR primer set, UniRT | Exiqon (now QIAGEN) | #204167 |
| miR-375, LNA PCR primer set, UniRT | Exiqon (now QIAGEN) | #204362 |
| miR-93, LNA PCR primer set, UniRT | Exiqon (now QIAGEN) | #204715 |
| DIG-labeled hsa-miR-216a miRCURY LNA detection probe | Exiqon (now QIAGEN) | #38495-15 |
| DIG-labeled scrambled miRNA detection probe | Exiqon (now QIAGEN) | #99004-15 |
| Recombinant DNA | ||
| pScamp EF1a-premir-216a-EGFP | This paper | N/A |
| Software and algorithms | ||
| GraphPad Prism 7 | GraphPad Software | GraphPad Prism, RRID: SCR_002798 |
| ImageJ | ImageJ | ImageJ, RRID: SCR_003070 |
| MetaXpress 5.3.0.5 Software | Molecular Devices | MetaXpress, RRID: SCR_016654 |
| R (v3.6.0) | R Core | R Project for Statistical Computing, RRID: SCR_001905 |
| Cell Ranger pipeline (v3.1.0 and 4.0.0) | 10x Genomics | Cell Ranger, RRID: SCR_017344 |
| Seurat v3.2.1 | Stuart et al., 201957 Hafemeister and Satija 201958 | SEURAT, RRID: SCR_007322 |
| ggplot2 v.3.3.2 | Wickham et al., 201659 | ggplot2, RRID: SCR_014601 |
| fgsea v1.14.0 | Korotkevich et al., 201960 | fgsea, RRID: SCR_020938 |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Timothy J. Kieffer (tim.kieffer@ubc.ca).
Materials availability
miR-216a KO ES cell clones (4H7 and 4H9) are available for purchase from the Mutant Mouse Resource and Research Center (USA). pScamp EF1a-premir-216a-EGFP expression plasmid generated in this study is available upon request.
Experimental model and subject details
Cell culture
The INS-1 rat insulinoma cell line was cultured in RPMI-1640 containing 11.2 mM glucose and 2 mM L-glutamine. The medium was supplemented with 10% fetal bovine serum, 1 mM sodium pyruvate (#S8636, Sigma-Aldrich), 10 mM HEPES, 50 μM 2-mercaptoethanol (Sigma Aldrich), 100 U/mL penicillin (Sigma Aldrich), and 100 μg/mL streptomycin (Sigma Aldrich). PANC-1 cells were cultured in DMEM media (Life Technologies) containing 4 mM L-glutamine, 4500 mg/L glucose, 1 mM sodium pyruvate, and 1500 mg/L sodium bicarbonate supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. EndoC-βH1 cells provided by (Drs. R Scharfmann, and P. Ravassard) were cultured on ECM-fibronectin–coated (1% and 2 μg/ml, respectively) (Sigma-Aldrich) culture wells and maintained in DMEM (Sigma Aldrich) that contained 5.6 mM glucose, 2% BSA fraction V (Roche Diagnostics), 50 μM 2-mercaptoethanol, 10 mM nicotinamide (Sigma-Aldrich), 5.5 μg/ml transferrin (Sigma-Aldrich), 6.7 ng/ml selenite (Sigma-Aldrich), 100 U/ml penicillin, and 100 μg/ml streptomycin.56 For transfections, cells were plated on 6-well plates (unless stated otherwise), and the next day 50%–70% confluent cells were transfected with miR-216a (50 nM) and miRNA controls (50 nM) using RNAimax (Life Technologies) following manufacturer’s instructions. To control for transfection effects, at least 3 wells were left untransfected. To differentiate CA1S human ES cells into pancreatic endocrine cells, we used our previously published 33-day, 7-stage differentiation protocol.16,55 Briefly, CA1S cells were cultured in mTesR1 media (Stem Cell Technologies, Vancouver BC, Canada) on 1:30 Matrigel coated surfaces (Stem Cell Technologies). Upon ∼90%–95% confluency, hESCs were directed in MCDB 131 media through 7 stages of pancreatic development in the presence of growth factors and small molecules as described in detail before.16 Briefly following compounds were added to induce differentiation: Stage 1 (Day1): GDF-8 (100 ng/mL) and MCK-928 (2.5 μM); Stage 1 (Days 2-4): GDF-8 (100 ng/mL)); Stage 2 (Days 5-6): FGF-7 (50 ng/mL); Stage 3 (Days 7-10): FGF-7 (50 ng/mL), Activin A (20 ng/mL); SANT-1 (0.25 μM), retinoic acid (2 μM), retinoic acid (2 μM), LDN193189 (200 nM); Stage 4 (Days 11-13): SANT-1 (0.25 μM), LDN193189 (200 nM), TBP (500 nM), CYP26a inhibitor (100 nM); Stage 5 (Days 14-16): LDN193189 (200 nM), ALK5 inhibitor (1 μM), Cyp26a inhibitor (100 nM); Stage 6 (Days 17-19): LDN193189 (200 nM), ALK5 inhibitor (1 μM); Stage 7 (Days 20-33): LDN193189 (200 nM), ALK5 inhibitor (1 μM), Vitamin A (100 nM).
Mouse models
The miR-216a KO ES cell line was generated by the Wellcome Trust Sanger Institute19 and the miR-216a−/− (miR-216a KO) mouse line was generated by the Centre for Phenogenomics in Toronto, Canada. Briefly, two frozen miR-216a KO ES cell (JM8A3 derived from C57BL/6N) clones (4H7 and 4H9) were purchased from the Mutant Mouse Resource and Research Center (USA) and were expanded and aggregated with diploid CD1 embryos. Coat color chimerism was scored and chimeras were bred with albino B6N (B6N-Tyrc-Brd/BrdCrCrl) females to test germline transmission. Successful germline transmission was confirmed with TaqMan qPCR of DNA prepared from tail clips, using the primers as indicated below in the experimental details.
miR-216a−/− mice were crossed with WT C57BL/6 mice for at least 3 generations before use in experiments and maintained on a C57BL/6 background obtained from the University of British Columbia Animal Care Facility. Mice were fed a chow diet (2918, Research Diets). Mice were housed with a 12-h:12-h light-dark cycle with constant temperature and humidity and ad libitum access to food and water. miR-216a−/− male mice were sacrificed at 21-weeks of age for tissue analysis. In all experiments, aged-matched littermates of miR-216a KO mice were used as controls.
For high-fat-diet (HFD) studies, C57BL/6 wild-type littermates and miR-216a KO male mice were fed a 60% HFD diet (D12492i, Research Diets, Cedarlane, Burlington, Canada) starting at 6-8 weeks of age for 8 weeks. Pancreata were analyzed from 14-16 weeks old male mice. For STZ studies, 8-week old C57BL/6 male mice were rendered diabetic with a single intraperitoneal injection of streptozotocin (STZ) at a dose of 180 mg/kg. Blood samples were collected 2 hours, 24 hours and 7 days after STZ injection. LSL-KrasG12D (KrasG12D) and Ptf1aCreER have been described previously.30,61, 62, 63 Briefly, the LSL-KrasG12D strain carries a Lox-Stop-Lox (LSL) sequence followed by the Kras G12D point mutation and the Ptf1aCreER strain carries a knock-in allele to abolish the function of the Ptf1a gene function and express tamoxifen-inducible CreER protein from the Ptf1a promoter/enhancer elements. Expression of Kras G12D was induced in KrasG12D x Ptf1aCreER x miR-216a−/− and KrasG12D x Ptf1aCreER x miR-216a+/+ mice by 3 subcutaneous tamoxifen injections at a dose of 125 μg/g body weight on alternating days when the mice were 21-25 days old. Pancreata were harvested 8 months later from both male and female mice for analysis. Leptin knock-out rats harbored a 151 bp deletion within Exon 1 of Leptin gene on chromosome 4 and were on Sprague-Dawley background strain.28 Leptin knock-out and littermate control rats used for islet isolation and the subsequent miRNA analysis in this study were female and 20-weeks old. Kidney capsule grafts were obtained from mice and rats implanted with pancreatic progenitor cells. Briefly, male 8- to 10-week-old SCID-beige mice (C.B-Igh-1b/GbmsTac-Prkdcscid-LystbgN7) and male 8- to 10-week-old nude rats (Crl:NIH-Foxn1rnu) were transplanted with pancreatic progenitor cells under the left kidney capsule as we described before.18 Engrafted kidneys were harvested at 22 weeks post-transplantation.
Ptf1a-Creex1 mice carry Cre recombinase in place of the exon 1 (ex1) of the Ptf1a locus.64 Murine serum samples from mutant LSL-KrasG12D;Ptf1a-Creex1 mice were collected at between 6- and 36-weeks of age (at different stages of tumor development) from both male and female mice. All procedures with animals were approved by the local Animal Care Committee and carried out in accordance with the national Animal Care guidelines (Canadian Council on Animal Care guidelines or German Animal Protection Law).
Human samples
Human fetal pancreata were kindly provided by Dr. Renian Wang, University of Western Ontario, London, Canada.17 Briefly, Human fetal pancreata (8-week to 14-week) were fixed in 4% (wt/vol.) paraformaldehyde, embedded in paraffin and sectioned for histological analysis (Wax-it Histology Services, Vancouver, BC, Canada). Human fetal pancreata were collected according to protocols approved by the Health Sciences Research Ethics Board at the University of Western Ontario with informed consent to use for research purposes. Based on the ethics protocol, we are unable to obtain gender information of these samples. Human adult islets (Donor 1: male, 54 years; donor 2: male, 22 years; donor 3: male, 46 years) were provided by the Irving K. Barber Human Islet Isolation Laboratory (Vancouver, British Columbia) with informed consent to use for research purposes.
Human serum samples were selected for analysis on the basis of the following criteria: 1) patients with chronic pancreatitis (CP), 2) patients who were newly diagnosed and previously untreated (PDAC), 3) patients with metastatic pancreatic cancer, previously untreated (mPDAC), 4) control samples with no current diabetes or pancreas cancer diagnosis (Control) and, 5) patients diagnosed with type 2 diabetes (T2D). Serum samples from patients (1-3) were collected at the Second Department of Internal Medicine and Surgery, Klinikum rechts der Isar at the Technical University Munich and all patients accepted and signed an informed consent that had been approved by the local ethics committee (license number: 1926/07). Serum samples (4-5) were collected at the Department of Medicine of University of Leipzig. The study was approved by the Ethics Committee of the University of Leipzig (approval numbers: 363-10-13122010 159-12-21052012), and performed in accordance to the declaration of Helsinki. All subjects gave written informed consent to use their data in anonymized form for research purposes. The age and gender of the donors can be found in Table S6 and sample size is included in the legend of Figure 6. All serum samples were frozen at −80°C until used.
Method details
Mouse model experimental details
Successful germline transmission was confirmed with TaqMan qPCR of DNA prepared from tail clips, using the primers in two separate reactions (KO and WT allele). The KO allele assay yields a 125 bp product. Forward: 5′AGT TCC TAT TCC GAA GTT CCT ATT C 3′. Reverse: 5′ AGA GGT TGA GGA CAG ACA GTA 3′. Probe (sense); TGG TCA TAG CTG TTT CCT GAA CAC CA. Cycling conditions: 98°C, 30 s; 40 cycles of 98°C, 5 s; 56°C, 10 s. The WT allele assay yields a 135 bp product. Forward: 5′ GGC TAT GAG TTG GTT TA 3′. Reverse: 5′ GGA AAT TGC TCT GTT TAG 3′. Probe (antisense): CTG TGA GGA ATG ATA GGG AC. Cycling conditions: 95°C, 30 s; 35 cycles of 95°C, 5 s; 59°C, 10 s. Reactions were performed using the iTaq Universal Probes Supermix (Bio-Rad #1725131, Hercules, California, USA) on the CFX96 Touch Thermal Cycler (Bio-Rad 1855195). Both probes were labeled with 6’-FAM.
Body weight and blood glucose were measured after a 4-hour morning fast. Blood was collected from the saphenous vein with heparin coated capillary tubes.
Metabolic assessments
All metabolic analyses were performed in conscious mice that were restrained during blood sampling. Blood glucose values were determined with a OneTouch Ultra 2 glucometer (LifeScan, Inc., Burnaby, Canada) measured from saphenous vein blood. Fasting (4 hours) glucose and body weight were monitored weekly. For glucose tolerance tests with chow-diet or HFD fed mice, 6 hour fasted mice were given 2 g/kg glucose either by intraperitoneal injection or oral gavage, and blood was collected into heparin-coated capillary tubes at indicated times following glucose administration. For insulin tolerance tests, 4 hour fasted mice were injected intraperitoneally with 0.75 U/kg insulin (Novolin ge Toronto, Novo Nordisk Canada, Mississauga, Canada), with glucose measures at indicated times following glucose administration.
Islet isolation
Mouse islets were isolated as previously described using Type XI collagenase (1000 units/mL; #C7657, Sigma-Aldrich, St. Louis, MO).65 Following digestion and filtration, islets were picked with a pipette in three rounds to > 95% purity. Islets were cultured overnight in RPMI-1640 (Sigma-Aldrich, St. Louis, MO) supplemented with 10% FBS (#F1051, Sigma-Aldrich, St. Louis, MO), 100 units/mL penicillin and 100 μg/mL streptomycin. For glucose stimulated insulin secretion assays, the next day after isolation 30 islets per mouse were placed in 24-well plates with KRBB buffer (129 mM NaCl, 4.8 mM KCl, 1.2 mM MgSO4, 1.2 mM KH2PO4, 2.5 mM CaCl2, 5.0 mM NaHCO3, 10 mM HEPES, 0.5% BSA, pH: 7.4) supplemented with 2.8 mM Glucose (low-Glucose). After one hour of incubation at 37°C, media was discarded. Islets were then serially placed in low Glucose (2.8 mM Glucose) KRBB buffer, high Glucose (16.7 mM) KRBB buffer and 20 mM KCl for 1 hour at 37°C. At the end of each incubation media was collected, centrifuged and stored at −20°C. Following the last media collection, islets were lysed in 0.18 M HCl/70% ethanol, homogenized, then incubated again at −20°C overnight. Following centrifugation, the aqueous solution was neutralized 1:2 with 1 M Tris, pH 7.5.
Isolation of pancreatic acinar cells and 3D-culture
Primary pancreatic acinar cells were prepared as previously described.66 In brief, the pancreas was removed, washed three times with ice-cold Hank’s balanced salt solution (HBSS), minced to ∼1 mm and digested with collagenase I at 37 °C with gentle shaking. The digested pancreatic pieces were washed twice with HBSS containing 5% FBS and then pipetted through 100 μm filters. The supernatant of the cell suspension containing acinar cells was added dropwise to HBSS containing 30% FBS. Acinar cells were then centrifuged and re-suspended in Waymouth media supplemented with 1% FBS, 0.1 mg ml−1 trypsin inhibitor, 1 μg ml−1 dexamethasone. For the 3D acinar culture, cell culture plates were coated with collagen I in Waymouth media. Isolated primary pancreatic acinar cells were added as a 1:1 mixture with collagen I/Waymouth media on the top of this layer. The cells were cultured in the presence of TGF-α (50 ng/mL). Further, Waymouth complete media with 10% FBS, 100 units/mL penicillin and 100 μg/mL streptomycin, 0.1 mg ml−1 trypsin inhibitor, 1 μg ml−1 dexamethasone was added on top of the cell/gel mixture, replaced on days 1 and 3. At days 1 and 5 formation of ducts were visualized under a microscope.
Cloning
miRNA-GFP expression vectors were generated by initially creating a EGFP expression vector, pScamp EF1a-EGFP, which contains the 5′ end of the human EF1A gene (a genomic PCR amplicon containing 1407 bp of proximal promoter, exon 1, intron 1, and the UTR of exon 2), followed by the EGFP ORF and SV40 poly(A) signal, all cloned into pBluescript. This was subsequently modified into a “miRtron” expression vector, pScamp EF1a-premir-216a-EGFP, by removing 450 bp of the middle EF1A intron with restriction endonucleases BfuAI and XhoI, and replacing it with a compatible BsmBI-SalI restriction fragment of a 575-bp amplicon∗ consisting of the full hsa-premir-216a sequence, flanked on either side by roughly 200 bp of endogenous genomic sequences. Cells transfected with this plasmid produce a chimeric EF1A-EGFP mRNA that gets translated into a fluorescent protein, while the excised intron is processed into mature hsa-miR-216a.∗ miRtron hsa-premir-216a cloning primers (BsmBI and XhoI 5′ overhangs are underlined). Forward: 5′ gggcacacaCGTCTCGACGCAGATTATACTTTTATGACATTACATGCAATATAGC 3′; Reverse: 5′ cacacaGTCGACCCAAGTAGCACTGAAGGAGCG 3′.
Pancreas immunohistochemistry and histology
Pancreata were fixed overnight in 4% paraformaldehyde (PFA) and then stored in 70% EtOH prior to paraffin embedding and sectioning (5 μm thickness; Wax-it Histology Services; Vancouver, BC). Immunofluorescent staining was performed as previously described.67 Briefly, slides were deparaffinized in xylene and hydrated by graded ethanol washes. Slides were then washed in phosphate-buffered saline (PBS) followed by 10–15 min incubation in 10 mM sodium citrate/0.05% Tween 20, pH 6.0 at 95°C. Slides were incubated with DAKO Protein Block Serum-Free (Agilent Technologies, Inc, Santa Clara, CA) at room temperature for 10 minutes and then with the primary antibodies diluted in Dako Antibody Diluent (Agilent Technologies, Inc.) overnight at 4°C. The following primary antibodies were used: insulin (Cell Signaling, # 3014 at 1:200), glucagon (Sigma Aldrich, #G2654 at 1:1000), synaptophysin (Monosan, #MON9013 at 1:10), and PCNA (BD Transduction, #610665 at 1:100). The next day slides were serially washed with PBS and were incubated in appropriate Alexa Fluor 488 or 594 secondary antibodies (Thermo Fisher Scientific, Waltham, MA) diluted in Dako Antibody Diluent. Following serial PBS washes, slides were coverslipped with VECTASHIELD HardSet Mounting Medium with DAPI (Vector Laboratories, Burlingame, CA). Images were captured using a ImageXpress Micro™ Imaging System and analyzed using MetaXpress 5.3.0.5 Software (Molecular Devices Corporation, Sunnyvale, CA, USA). β-cell mass was analyzed in three pancreas sections per mouse, at least 200 μm apart, on insulin-labeled sections. Islet size was quantified as the synaptophysin-positive area per islet. For the α-cell analysis, glucagon-immunoreactive cells were counted using the ImageJ Cell Counter tool. Only islets with visible DAPI stained nuclei and at least 40 cells and 10 or more non-β cells were used to determine periphery or core localization of α-cells. α-cells were considered to be in the periphery of the islet if they were within 2 outermost cell layers of the islet or the core if they were deeper than the 2 outermost cell layers of the islet. For the islet circularity analysis, islets stained with synaptophysin and DAPI were used. Only islets with at least 40 cells and 10 or more non-β cells were used to determine islet circularity. All images were converted to 8-bit and non-islet nuclei were erased using ImageJ outline and clear function. Islet circularity was determined using Analyze Particles with threshold set until the entire area of islet was filled. H&E staining and Alcian blue staining on pancreatic sections were performed by Wax-it Histology Services Inc. (Vancouver, Canada). Distance to ducts was quantified on the H&E stains. Distance from islet to nearest duct was defined with islets as minimum of 3 cells with a cell shape and eosinophilic appearance of endocrine cells.
RNA isolation and quantitative RT-PCR
Tissues were homogenized with an Ultra-Turrax and total RNA was isolated using the miRCURY RNA isolation kit (QIAGEN) as per the manufacturer’s instructions including a DNase (Life Technologies) treatment step. mRNA was reverse transcribed with iScript cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA) and quantitative RT-PCR was performed using Ssofast EvaGreen Supermix (Bio-Rad). Primer sequences to amplify Pdx1, Nkx6.1, and Hprt transcripts were described previously.68 A human RNA panel was purchased from ThermoFisher Scientific (#AM6000). miRNA was reverse transcribed with a Universal cDNA synthesis kit (QIAGEN and qRT-PCR was performed using SYBR Green master mix (QIAGEN) with LNA-based miRNA primers (QIAGEN). Relative values were calculated by the quantified by the 2−ΔΔCT method. RNA from the pancreata of 1-day old mice was isolated using a protocol specific for pancreatic RNA isolations.69
Extracellular RNA was isolated from 50 μL mouse plasma or 200 μL human serum (centrifuged for 5 minutes at 3000 × g) in the presence of an MS2 carrier RNA (Roche, Laval, Quebec) using a miRNeasy kit (QIAGEN, Mississauga, Ontario, Canada). RNA was reverse transcribed with a Universal cDNA synthesis kit (QIAGEN Quantitative RT-PCR (qRT-PCR) was performed using SYBR Green master mix (QIAGEN) with LNA-based miRNA primers (QIAGEN). Relative values were calculated by the quantified by the 2−ΔΔCT method.
In situ hybridization
Human fetal pancreata were kindly provided by Dr. Renian Wang, University of Western Ontorio, London, Canada and were described elsewhere.17 Human adult pancreas was provided by the Irving K. Barber Human Islet Isolation Laboratory (Vancouver, British Columbia) with consent to use for research purposes. All samples were fixed overnight in 4% PFA, embedded in paraffin and sectioned (5 μm thickness; Wax-it Histology Services; Vancouver, BC). Slides were de-paraffinized with three consecutive xylene washes, rehydrated with graded ethanol washes (100% x 3, 95%, 70%, each 5 minutes), and washed in diethylpyrocarbonate (DEPC)-treated water. In situ hybridization was carried out with a IsHyb In Situ Hybridization kit (Biochain, San Francisco, California). Sections were fixed with 4% PFA for 20 minutes, washed twice with DEPC-treated PBS, and treated with 10 μg/mL Proteinase K (Sigma Aldrich) at 37°C for 15 minutes. Slides were washed with DEPC-treated PBS, fixed again with PFA for 15 minutes, and washed with DEPC-treated water. Sections were incubated for 4 hours at 50°C in pre-hybridization solution (Biochain). Afterward, pancreatic sections were incubated with DIG-labeled hsa-miR-216a miRCURY LNA detection probe (Exiqon#38495-15- now QIAGEN) or control scrambled miRNA probe (Exiqon#99004-15, now QIAGEN) at 0.25 ng/μL in hybridization solution (Biochain) for 14 hours at 45°C. Then, slides were washed in SSC buffer (Biochain) as follows: 2X SSC buffer, 2x10 min, 45°C; 1.5X SSC buffer, 1x10min, 45°C; 2x SSC buffer, 2x20 minutes, 37°C. After washing steps, sections were incubated with 1X blocking solution (Biochain) in PBS for 1 hour. Slides were incubated overnight with alkaline-phosphate conjugated anti-digoxinogen antibody (diluted 1:500 in PBS) (Biochain) at 4°C. The following day, slides were washes three times with PBS for 10 minutes, twice with alkaline phosphatase buffer (Biochain) for 5 minutes, and incubated with nitro-blue tetrazolium chloride (NBT) and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt (BCIP) solution (6.6 μL NBT and 3.3 μL BCIP were diluted in 1 mL alkaline phosphatase buffer) (Biochain) for 20 hours at room temperature. Slides were scanned using a ScanScope CS system (Aperio; Vista, CA).
Assays
Insulin was measured in plasma, islets and cell culture media using a Mouse Ultrasensitive Insulin ELISA (ALPCO, Salem, NH). Proinsulin was measured from plasma collected from cardiac blood using Rat/Mouse Proinsulin ELISA (Mercodia, Sweden). To perform migration assay with cultured cell lines, PANC-1 cells were transfected with the miR-216a and control scrambled miRNA mimetics (Dharmacon, Lafayette, CO) and 24 hours later, seeded on trans-well migration chambers (Trevigen, Gaithersburg, MD) in media without FBS. Cells were allowed to migrate for 16 hours and the number of cells transversing the boyer chamber was quantified by incubating the cells in the bottom chamber with Calcein-AM for one hour and measuring the fluorescence at 485 nm excitation, 520 nm emission with a Tecan Plate Reader. For the XTT (2,3-bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide) assay, cells were transfected with miR-216a and the control miRNAs and an XTT assay was performed 3 days post-transfection in 96-well plates. XTT (Life Technologies) was dissolved in pre-warmed 37°C cell culture media at 1 mg/mL and stock Phenazine methosulfate (PMS) solution was prepared in PBS at 10 mM. PMS was mixed with the XTT solution immediately before labeling the cells and 25 μL of XTT/PMS solution was directly added to each well containing 100 μL cell culture media. Cells were incubated for two hours at 37°C in a CO2 incubator. Absorbance was read at 450 nm using a Tecan Plate Reader. For live cell imaging, INS-1 cells seeded on 96-well plates were transfected with control or miR-216a expressing plasmids or left untreated. Two days after transfection, cells were incubated with 50 ng/mL Hoechst and 1:500 diluted Alexa647 annexinV 30 min prior to imaging. TNF-α, IFN-ϒ and IL-1β (10 ng/mL each) was added to media and cells were imaged every 2 hours at 37°C and 5% CO2 in an ImageXpress Micro™ (Molecular Devices).
Single-cell RNA sequencing and analysis
C57BL/6 wild-type littermates and miR-216a KO mice were fed a 60% HFD diet (D12492i, Research Diets, Cedarlane, Burlington, Canada) starting at 6-8 weeks of age for 9 weeks. Mouse islet were isolated by ductal inflation, followed by collagenase incubation, filtration and hand-picking to ∼95% purity as described above. Islets were resuspended in RPMI 1640 (Thermo Fisher Scientific #11875-093), 10% FBS, 1% Pen/Strep) then picked into PBS in microcentrifuge tubes (Eppendorf, DNA LoBind, 022-43-104-8). Islets were centrifuged at 290 g for 5 minutes at 4°C then transferred to TrypLE, resuspended, then incubated at 37°C for 13.5 minutes in total, pipetting up and down five times every 2 minutes. FACS buffer was added to stop the dissociation process, then single cells were centrifuged again using the same parameters. Islets were resuspended in fresh FACS buffer then filtered into a fresh microcentrifuge tube. Cell concentration was counted and viability measured using a NucleoCounter-200 (ChemoMetec).
All samples were processed according to 10X Genomics Chromium Single Cell 3′ Reagent Guidelines v3.1 Chemistry as per the manufacturer’s protocol. RNA sequencing was provided by the UBC Biomedical Research Centre Core Facility. Fastq files were generated using the cellranger pipeline (version 3.1.0), then version 4.0.0 were used for all subsequent steps. Prior to alignment, files were trimmed using cutadapt version 2.10 with default options except for a maximum error rate of 0.1. Trimmed files were aligned to GRCm38 release 101 and genes counted using cellranger count with default options. Counts were read into R using Seurat version 3.2.1 for quality control, dimensionality reduction, cell clustering, and differential expression analysis.57 Cells were filtered from the analysis if they had less than 200 or greater than 10,000 features or greater than 5% mitochondrial genes. Libraries were integrated using the Seurat SCTransform pipeline and donor effects regressed out during normalization using the variance stabilizing transform methodology with 3000 variable features.58 Unsupervised clustering was done with the original Louvain algorithm using the FindNeighbors and FindClusters functions with a resolution of 1.2. Assignment of cell clusters to cell types was based on differentially expressed canonical genes for each cluster detected in a minimum of 5% of cells enriched by a minimum log fold change threshold of 0.15 in populations compared (Table S2) using the Wilcoxon Rank Sum test.70 Clusters marked as more than one cell type had more than one canonical marker differentially expressed compared to the rest of the dataset, suggesting that the cluster contains a mix of cell types. Differentially expressed genes within clusters or cell types between genotypes were identified in a similar manner. Heatmap plotting top marker genes for clusters and between genotypes were generated using the DoHeatmap function with thematic modifications using ggplot2 v.3.3.2.59
Gene set enrichment analysis was performed using the fgsea package version 1.14.0 using Reactome pathways, with genes included if they were expressed in a minimum of 5% of cells enriched by a minimum log fold change threshold of 0.05 to speed up calculations.60,71
RNA sequencing and analysis
RNA sequencing was performed by the Biomedical Research Center Genomics facility at the University of British Columbia, Vancouver, Canada. Sample quality control was assessed using an Agilent 2100 Bioanalyzer. Qualifying samples (samples with RNA integrity numbers > 8) were then prepared following the standard protocol for the NEBnext Ultra ii Stranded mRNA (New England Biolabs). Sequencing was performed on the Illumina NextSeq 500 with Paired End 42bp × 42bp reads. Sequencing data were demultiplexed using Illumina’s bcl2fastq2.
Fastq files were retrieved from Illumina Basespace and aligned with salmon (version 0.13.1) to the most current Ensembl Mus musculus genome (as of 2019-05-08) using the gcBias, validateMappings, and rangeFactorizationBins (with 4 as the binning parameter) options. Quality of the Fastq files was assessed using FastQC (version 0.11.8).72, 73, 74 All fastq files passed quality control. All further analyses were performed in R (version 3.6.0).75 Counts were imported using the tximport package (version 1.12.0) and the biomaRt package (version 0.8.0).76, 77, 78 Analysis of differentially expressed genes was performed using the DESeq2 (version 1.24.0).79 Genes with less than 5 counts in 2 samples or fewer were dropped from the analysis. Gene Ontology analysis was performed on genes with an IQR greater than 0.5 across all samples and an adjusted p value < 0.05 (adjusted by Benjamini-Hochberg method) between wild-type and control mice. Significantly enriched terms for Biological Process, Cellular Components, and Molecular Functions were identified using the categoryCompare package (version 1.28.0) and visualized using Cytoscape via the RCy3 package (version 2.4.0).80,81 Further KEGG network analyses were performed using the gage (version 2.34.0) and pathview (version 1.24.0) packages.82,83 Finally, GO terms were summarized using the REViGO methodology and plotted using the treemap (version 2.4.2) package.84,85
Quantification and statistical analysis
Statistical analyses were performed using GraphPad Prism 7, with significance defined as p < 0.05. Unless specified in the figure legend, Student’s t tests were used for normally distributed data (two-tailed, unpaired). Bonferroni correction was used to correct for multiple comparisons, where applicable. Data are represented by bar plots with individual values indicated, individual values only, or line graphs. Results show mean±SEM.
Acknowledgements
We thank Dr. Anna D’Souza for providing islets from LepKO rats and Alexandra Berninger for providing plasma from LSL-KrasG12D;Ptf1a-Creex1 mice. T.J.K. gratefully acknowledges funding from the JDRF and the Canadian Institutes for Health Research for this research. S.E. is a recipient of a JDRF advanced postdoctoral fellowship. A.R. gratefully acknowledges studentship support from the CIHR (Vanier Canada Graduate Scholarship) and Vancouver Coastal Health (CIHR-UBC MD/PhD Studentship). We would also like to thank Dr. Ziliang Ao and Dr. Garth L. Warnock from the Irving K. Barber Human Islet Isolation Laboratory (Vancouver, BC, Canada) for providing human islets. The graphical abstract was created using BioRender.com.
Author contributions
S.E. designed and coordinated the studies, performed experiments, analyzed the data, and wrote the manuscript with input from all co-authors. C.E.E. analyzed data. A.R., M.M.G., S.O., S.P., T.W., J.P., J.E.B., M.J.R., R.K.B., and T.D.W. performed experiments. M.L., M.B., H.A., J.L.K., and S.H. provided reagents and clinical samples. S.E., C.E.E., J.L.K., and T.J.K. interpreted results. S.E. and T.J.K. supervised the studies.
Declaration of interests
The authors declare no competing interests.
Published: November 16, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.xcrm.2021.100434.
Supplemental information
Data and code availability
Single-cell RNA-seq and bulk RNaseq data generated during the current study are available in the NCBI’s Gene Expression Omnibus database (single cell RNA-seq: GEO: GSE159844, bulk RNA-seq: GEO: GSE159970). Code related to the bulk and single cell RNA-seq analyses can be found at https://github.com/caraee/miR216a_KO.
References
- 1.American Diabetes Association Diagnosis and classification of diabetes mellitus. Diabetes Care. 2010;33(Suppl 1):S62–S69. doi: 10.2337/dc10-S062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz P. Acinar cell plasticity and development of pancreatic ductal adenocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2017;14:296–304. doi: 10.1038/nrgastro.2017.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 4.Carthew R.W., Sontheimer E.J. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Godard P., van Eyll J. Pathway analysis from lists of microRNAs: common pitfalls and alternative strategy. Nucleic Acids Res. 2015;43:3490–3497. doi: 10.1093/nar/gkv249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akhtar M.M., Micolucci L., Islam M.S., Olivieri F., Procopio A.D. Bioinformatic tools for microRNA dissection. Nucleic Acids Res. 2016;44:24–44. doi: 10.1093/nar/gkv1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lynn F.C., Skewes-Cox P., Kosaka Y., McManus M.T., Harfe B.D., German M.S. MicroRNA expression is required for pancreatic islet cell genesis in the mouse. Diabetes. 2007;56:2938–2945. doi: 10.2337/db07-0175. [DOI] [PubMed] [Google Scholar]
- 8.Mandelbaum A.D., Melkman-Zehavi T., Oren R., Kredo-Russo S., Nir T., Dor Y., Hornstein E. Dysregulation of Dicer1 in beta cells impairs islet architecture and glucose metabolism. Exp. Diabetes Res. 2012;2012:470302. doi: 10.1155/2012/470302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kalis M., Bolmeson C., Esguerra J.L., Gupta S., Edlund A., Tormo-Badia N., Speidel D., Holmberg D., Mayans S., Khoo N.K., et al. Beta-cell specific deletion of Dicer1 leads to defective insulin secretion and diabetes mellitus. PLoS ONE. 2011;6:e29166. doi: 10.1371/journal.pone.0029166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez-Sanchez A., Nguyen-Tu M.S., Rutter G.A. DICER inactivation identifies pancreatic β-cell “disallowed” genes targeted by microRNAs. Mol. Endocrinol. 2015;29:1067–1079. doi: 10.1210/me.2015-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y.J., McAllister F., Bailey J.M., Scott S.G., Hendley A.M., Leach S.D., Ghosh B. Dicer is required for maintenance of adult pancreatic acinar cell identity and plays a role in Kras-driven pancreatic neoplasia. PLoS ONE. 2014;9:e113127. doi: 10.1371/journal.pone.0113127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Niehrs C., Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. doi: 10.1038/990025. [DOI] [PubMed] [Google Scholar]
- 13.Londin E., Loher P., Telonis A.G., Quann K., Clark P., Jing Y., Hatzimichael E., Kirino Y., Honda S., Lally M., et al. Analysis of 13 cell types reveals evidence for the expression of numerous novel primate- and tissue-specific microRNAs. Proc. Natl. Acad. Sci. U S A. 2015;112:E1106–E1115. doi: 10.1073/pnas.1420955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun K., Lai E.C. Adult-specific functions of animal microRNAs. Nat. Rev. Genet. 2013;14:535–548. doi: 10.1038/nrg3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L., et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc. Natl. Acad. Sci. U S A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruin J.E., Erener S., Vela J., Hu X., Johnson J.D., Kurata H.T., Lynn F.C., Piret J.M., Asadi A., Rezania A., et al. Characterization of polyhormonal insulin-producing cells derived in vitro from human embryonic stem cells. Stem Cell Res. 2014;12:194–208. doi: 10.1016/j.scr.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Riedel M.J., Asadi A., Wang R., Ao Z., Warnock G.L., Kieffer T.J. Immunohistochemical characterisation of cells co-producing insulin and glucagon in the developing human pancreas. Diabetologia. 2012;55:372–381. doi: 10.1007/s00125-011-2344-9. [DOI] [PubMed] [Google Scholar]
- 18.Bruin J.E., Asadi A., Fox J.K., Erener S., Rezania A., Kieffer T.J. Accelerated maturation of human stem cell-derived pancreatic progenitor cells into insulin-secreting cells in immunodeficient rats relative to mice. Stem Cell Reports. 2015;5:1081–1096. doi: 10.1016/j.stemcr.2015.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Prosser H.M., Koike-Yusa H., Cooper J.D., Law F.C., Bradley A. A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat. Biotechnol. 2011;29:840–845. doi: 10.1038/nbt.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jo J., Kilimnik G., Kim A., Guo C., Periwal V., Hara M. Formation of pancreatic islets involves coordinated expansion of small islets and fission of large interconnected islet-like structures. Biophys. J. 2011;101:565–574. doi: 10.1016/j.bpj.2011.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miettinen P.J., Huotari M., Koivisto T., Ustinov J., Palgi J., Rasilainen S., Lehtonen E., Keski-Oja J., Otonkoski T. Impaired migration and delayed differentiation of pancreatic islet cells in mice lacking EGF-receptors. Development. 2000;127:2617–2627. doi: 10.1242/dev.127.12.2617. [DOI] [PubMed] [Google Scholar]
- 22.Greiner T.U., Kesavan G., Ståhlberg A., Semb H. Rac1 regulates pancreatic islet morphogenesis. BMC Dev. Biol. 2009;9:2. doi: 10.1186/1471-213X-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ritvos O., Tuuri T., Erämaa M., Sainio K., Hildén K., Saxén L., Gilbert S.F. Activin disrupts epithelial branching morphogenesis in developing glandular organs of the mouse. Mech. Dev. 1995;50:229–245. doi: 10.1016/0925-4773(94)00342-k. [DOI] [PubMed] [Google Scholar]
- 24.Kato M., Putta S., Wang M., Yuan H., Lanting L., Nair I., Gunn A., Nakagawa Y., Shimano H., Todorov I., et al. TGF-beta activates Akt kinase through a microRNA-dependent amplifying circuit targeting PTEN. Nat. Cell Biol. 2009;11:881–889. doi: 10.1038/ncb1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xia H., Ooi L.L., Hui K.M. MicroRNA-216a/217-induced epithelial-mesenchymal transition targets PTEN and SMAD7 to promote drug resistance and recurrence of liver cancer. Hepatology. 2013;58:629–641. doi: 10.1002/hep.26369. [DOI] [PubMed] [Google Scholar]
- 26.Xu J., Lamouille S., Derynck R. TGF-beta-induced epithelial to mesenchymal transition. Cell Res. 2009;19:156–172. doi: 10.1038/cr.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.LaPierre M.P., Stoffel M. MicroRNAs as stress regulators in pancreatic beta cells and diabetes. Mol. Metab. 2017;6:1010–1023. doi: 10.1016/j.molmet.2017.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D’souza A.M., Asadi A., Johnson J.D., Covey S.D., Kieffer T.J. Leptin deficiency in rats results in hyperinsulinemia and impaired glucose homeostasis. Endocrinology. 2014;155:1268–1279. doi: 10.1210/en.2013-1523. [DOI] [PubMed] [Google Scholar]
- 29.Kameswaran V., Bramswig N.C., McKenna L.B., Penn M., Schug J., Hand N.J., Chen Y., Choi I., Vourekas A., Won K.J., et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014;19:135–145. doi: 10.1016/j.cmet.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopp J.L., von Figura G., Mayes E., Liu F.F., Dubois C.L., Morris J.P., 4th, Pan F.C., Akiyama H., Wright C.V., Jensen K., et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guerra C., Schuhmacher A.J., Cañamero M., Grippo P.J., Verdaguer L., Pérez-Gallego L., Dubus P., Sandgren E.P., Barbacid M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell. 2007;11:291–302. doi: 10.1016/j.ccr.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 32.Hezel A.F., Kimmelman A.C., Stanger B.Z., Bardeesy N., Depinho R.A. Genetics and biology of pancreatic ductal adenocarcinoma. Genes Dev. 2006;20:1218–1249. doi: 10.1101/gad.1415606. [DOI] [PubMed] [Google Scholar]
- 33.Kanda M., Matthaei H., Wu J., Hong S.M., Yu J., Borges M., Hruban R.H., Maitra A., Kinzler K., Vogelstein B., Goggins M. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology. 2012;142:730–733.e9. doi: 10.1053/j.gastro.2011.12.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 35.Bankaitis E.D., Bechard M.E., Gu G., Magnuson M.A., Wright C.V.E. ROCK-nmMyoII, Notch and Neurog3 gene-dosage link epithelial morphogenesis with cell fate in the pancreatic endocrine-progenitor niche. Development. 2018;145:dev162115. doi: 10.1242/dev.162115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sharon N., Chawla R., Mueller J., Vanderhooft J., Whitehorn L.J., Rosenthal B., Gürtler M., Estanboulieh R.R., Shvartsman D., Gifford D.K., et al. A peninsular structure coordinates asynchronous differentiation with morphogenesis to generate pancreatic islets. Cell. 2019;176:790–804.e13. doi: 10.1016/j.cell.2018.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonner-Weir S., Trent D.F., Weir G.C. Partial pancreatectomy in the rat and subsequent defect in glucose-induced insulin release. J. Clin. Invest. 1983;71:1544–1553. doi: 10.1172/JCI110910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Butler A.E., Janson J., Bonner-Weir S., Ritzel R., Rizza R.A., Butler P.C. Beta-cell deficit and increased beta-cell apoptosis in humans with type 2 diabetes. Diabetes. 2003;52:102–110. doi: 10.2337/diabetes.52.1.102. [DOI] [PubMed] [Google Scholar]
- 39.Cnop M., Welsh N., Jonas J.C., Jörns A., Lenzen S., Eizirik D.L. Mechanisms of pancreatic beta-cell death in type 1 and type 2 diabetes: many differences, few similarities. Diabetes. 2005;54(Suppl 2):S97–S107. doi: 10.2337/diabetes.54.suppl_2.s97. [DOI] [PubMed] [Google Scholar]
- 40.Wang P., Liu Q., Zhao H., Bishop J.O., Zhou G., Olson L.K., Moore A. miR-216a-targeting theranostic nanoparticles promote proliferation of insulin-secreting cells in type 1 diabetes animal model. Sci. Rep. 2020;10:5302. doi: 10.1038/s41598-020-62269-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hou B.H., Jian Z.X., Cui P., Li S.J., Tian R.Q., Ou J.R. miR-216a may inhibit pancreatic tumor growth by targeting JAK2. FEBS Lett. 2015;589:2224–2232. doi: 10.1016/j.febslet.2015.06.036. [DOI] [PubMed] [Google Scholar]
- 42.Wang S., Chen X., Tang M. MicroRNA-216a inhibits pancreatic cancer by directly targeting Janus kinase 2. Oncol. Rep. 2014;32:2824–2830. doi: 10.3892/or.2014.3478. [DOI] [PubMed] [Google Scholar]
- 43.Kamerkar S., LeBleu V.S., Sugimoto H., Yang S., Ruivo C.F., Melo S.A., Lee J.J., Kalluri R. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503. doi: 10.1038/nature22341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Otto T., Sicinski P. Cell cycle proteins as promising targets in cancer therapy. Nat. Rev. Cancer. 2017;17:93–115. doi: 10.1038/nrc.2016.138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feldmann G., Beaty R., Hruban R.H., Maitra A. Molecular genetics of pancreatic intraepithelial neoplasia. J. Hepatobiliary Pancreat. Surg. 2007;14:224–232. doi: 10.1007/s00534-006-1166-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.LaPak K.M., Burd C.E. The molecular balancing act of p16(INK4a) in cancer and aging. Mol. Cancer Res. 2014;12:167–183. doi: 10.1158/1541-7786.MCR-13-0350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schutte M., Hruban R.H., Geradts J., Maynard R., Hilgers W., Rabindran S.K., Moskaluk C.A., Hahn S.A., Schwarte-Waldhoff I., Schmiegel W., et al. Abrogation of the Rb/p16 tumor-suppressive pathway in virtually all pancreatic carcinomas. Cancer Res. 1997;57:3126–3130. [PubMed] [Google Scholar]
- 48.Shen W., Tao G.Q., Zhang Y., Cai B., Sun J., Tian Z.Q. TGF-β in pancreatic cancer initiation and progression: two sides of the same coin. Cell Biosci. 2017;7:39. doi: 10.1186/s13578-017-0168-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Smith B., Agarwal P., Bhowmick N.A. MicroRNA applications for prostate, ovarian and breast cancer in the era of precision medicine. Endocr. Relat. Cancer. 2017;24:R157–R172. doi: 10.1530/ERC-16-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Poy M.N., Eliasson L., Krutzfeldt J., Kuwajima S., Ma X., Macdonald P.E., Pfeffer S., Tuschl T., Rajewsky N., Rorsman P., Stoffel M. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 51.Poy M.N., Hausser J., Trajkovski M., Braun M., Collins S., Rorsman P., Zavolan M., Stoffel M. miR-375 maintains normal pancreatic alpha- and beta-cell mass. Proc. Natl. Acad. Sci. U S A. 2009;106:5813–5818. doi: 10.1073/pnas.0810550106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Biton M., Levin A., Slyper M., Alkalay I., Horwitz E., Mor H., Kredo-Russo S., Avnit-Sagi T., Cojocaru G., Zreik F., et al. Epithelial microRNAs regulate gut mucosal immunity via epithelium-T cell crosstalk. Nat. Immunol. 2011;12:239–246. doi: 10.1038/ni.1994. [DOI] [PubMed] [Google Scholar]
- 53.Kang W., Huang T., Zhou Y., Zhang J., Lung R.W.M., Tong J.H.M., Chan A.W.H., Zhang B., Wong C.C., Wu F., et al. miR-375 is involved in Hippo pathway by targeting YAP1/TEAD4-CTGF axis in gastric carcinogenesis. Cell Death Dis. 2018;9:92. doi: 10.1038/s41419-017-0134-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goodwin D., Rosenzweig B., Zhang J., Xu L., Stewart S., Thompson K., Rouse R. Evaluation of miR-216a and miR-217 as potential biomarkers of acute pancreatic injury in rats and mice. Biomarkers. 2014;19:517–529. doi: 10.3109/1354750X.2014.944217. [DOI] [PubMed] [Google Scholar]
- 55.Caron N.J., Gage B.K., O’Connor M.D., Eaves C.J., Kieffer T.J., Piret J.M. A human embryonic stem cell line adapted for high throughput screening. Biotechnol. Bioeng. 2013;110:2706–2716. doi: 10.1002/bit.24936. [DOI] [PubMed] [Google Scholar]
- 56.Ravassard P., Hazhouz Y., Pechberty S., Bricout-Neveu E., Armanet M., Czernichow P., Scharfmann R. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 2011;121:3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wickham H., Navarro D., Pedersen T.L. Springer-Verlag; New York: 2009. ggplot2: Elegant Graphics for Data Analysis. [Google Scholar]
- 60.Korotkevich G., Sukhov V., Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2019 doi: 10.1101/060012. [DOI] [Google Scholar]
- 61.Tuveson D.A., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5:375–387. doi: 10.1016/s1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 62.Johnson L., Mercer K., Greenbaum D., Bronson R.T., Crowley D., Tuveson D.A., Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- 63.Kawaguchi Y., Cooper B., Gannon M., Ray M., MacDonald R.J., Wright C.V. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- 64.Lesina M., Wörmann S.M., Morton J., Diakopoulos K.N., Korneeva O., Wimmer M., Einwächter H., Sperveslage J., Demir I.E., Kehl T., et al. RelA regulates CXCL1/CXCR2-dependent oncogene-induced senescence in murine Kras-driven pancreatic carcinogenesis. J. Clin. Invest. 2016;126:2919–2932. doi: 10.1172/JCI86477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Salvalaggio P.R., Deng S., Ariyan C.E., Millet I., Zawalich W.S., Basadonna G.P., Rothstein D.M. Islet filtration: a simple and rapid new purification procedure that avoids ficoll and improves islet mass and function. Transplantation. 2002;74:877–879. doi: 10.1097/00007890-200209270-00023. [DOI] [PubMed] [Google Scholar]
- 66.Liou G.Y., Döppler H., Braun U.B., Panayiotou R., Scotti Buzhardt M., Radisky D.C., Crawford H.C., Fields A.P., Murray N.R., Wang Q.J., et al. Protein kinase D1 drives pancreatic acinar cell reprogramming and progression to intraepithelial neoplasia. Nat. Commun. 2015;6:6200. doi: 10.1038/ncomms7200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Asadi A., Bruin J.E., Kieffer T.J. Characterization of antibodies to products of proinsulin processing using immunofluorescence staining of pancreas in multiple species. J. Histochem. Cytochem. 2015;63:646–662. doi: 10.1369/0022155415576541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Glavas M.M., Hui Q., Tudurí E., Erener S., Kasteel N.L., Johnson J.D., Kieffer T.J. Early overnutrition reduces Pdx1 expression and induces β cell failure in Swiss Webster mice. Sci. Rep. 2019;9:3619. doi: 10.1038/s41598-019-39177-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Azevedo-Pouly A.C., Elgamal O.A., Schmittgen T.D. RNA isolation from mouse pancreas: a ribonuclease-rich tissue. J. Vis. Exp. 2014;(90):e51779. doi: 10.3791/51779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Baron M., Veres A., Wolock S.L., Faust A.L., Gaujoux R., Vetere A., Ryu J.H., Wagner B.K., Shen-Orr S.S., Klein A.M., et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 2016;3:346–360.e4. doi: 10.1016/j.cels.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A., Sidiropoulos K., Cook J., Gillespie M., Haw R., et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patro R., Duggal G., Love M.I., Irizarry R.A., Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J., Billis K., Cummins C., Gall A., Girón C.G., et al. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Andrews S. 2010. FastQC: a quality control tool for high throughput sequence data.https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 75.R Core Team . R Foundation for Statistical Computing; Vienna, Austria: 2019. A language and environment for statistical computing. [Google Scholar]
- 76.Soneson C., Love M.I., Robinson M.D. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521. doi: 10.12688/f1000research.7563.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 78.Durinck S., Spellman P.T., Birney E., Huber W. Mapping identifiers for the integration of genomic datasets with the R/Bioconductor package biomaRt. Nat. Protoc. 2009;4:1184–1191. doi: 10.1038/nprot.2009.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Flight R.M., Harrison B.J., Mohammad F., Bunge M.B., Moon L.D., Petruska J.C., Rouchka E.C. categoryCompare, an analytical tool based on feature annotations. Front. Genet. 2014;5:98. doi: 10.3389/fgene.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ono K., Muetze T., Kolishovski G., Shannon P., Demchak B. CyREST: Turbocharging Cytoscape Access for External Tools via a RESTful API. F1000Res. 2015;4:478. doi: 10.12688/f1000research.6767.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Luo W., Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29:1830–1831. doi: 10.1093/bioinformatics/btt285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Luo W., Friedman M.S., Shedden K., Hankenson K.D., Woolf P.J. GAGE: generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of Gene Ontology terms. PLoS ONE. 2011;6:e21800. doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tennekes M. 2017. treemap: Treemap Visualization. R package version 2.4-2.https://cran.r-project.org/web/packages/treemap/index.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Single-cell RNA-seq and bulk RNaseq data generated during the current study are available in the NCBI’s Gene Expression Omnibus database (single cell RNA-seq: GEO: GSE159844, bulk RNA-seq: GEO: GSE159970). Code related to the bulk and single cell RNA-seq analyses can be found at https://github.com/caraee/miR216a_KO.