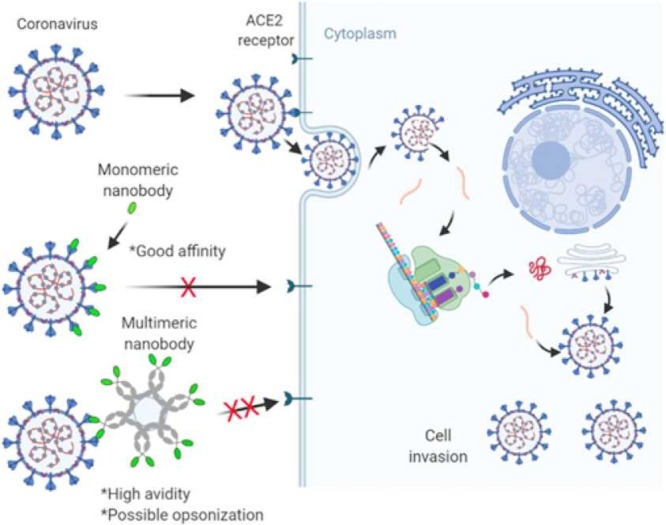
Graphical Abstract
Keywords: Coronavirus, Nanobody, Affinity, Avidity, Multivancy, Nanoparticles, Therapy
Abstract
In the past two decades, the emergence of coronavirus diseases has been dire distress on both continental and global fronts and has resulted in the search for potent treatment strategies. One crucial challenge in this search is the recurrent mutations in the causative virus spike protein, which lead to viral escape issues. Among the current promising therapeutic discoveries is the use of nanobodies and nanobody-like molecules. While these nanobodies have demonstrated high-affinity interaction with the virus, the unpredictable spike mutations have warranted the need for avidity-inspired therapeutics of potent inhibitors such as nanobodies. This article discusses novel approaches for the design of anti-SARS-CoV-1 and −2 nanobodies to facilitate advanced innovations in treatment technologies. It further discusses molecular interactions and suggests multivalent protein nanotechnology and chemistry approaches to translate mere molecular affinity into avidity.
Introduction
The past two decades have witnessed the emergence of many deadly coronavirus-associated (CoV) diseases on both epi- and pandemic levels in forms such as severe acute respiratory syndrome (SARS) and the middle east respiratory syndrome (MERS). The disease, especially in the present decade, has warranted the search for potent antidotes such as vaccines [1], [2], [3] and therapeutics [4], [5], [6] to help combat the seemingly unending life loss. Antibodies and soluble angiotensin-converting enzyme 2 (ACE2)- the virus receptor on the cell- are the most popular biologics suggested for coronavirus treatment. Also, several nanobodies have been reported as potent inhibitors of coronaviruses and present advantages in terms of size, solubility and stability. However, with the emergence of mutated virus strains capable of escaping inhibition by many monovalent blockers [7], it is presently highly expedient to probe into the design of potential inhibitors towards creating “avidity-inspired” therapeutics. We define an avidity-inspired therapeutic as one generated via the utilization of multivalent protein engineering and nanotechnology approaches that translate mere molecular affinity into avidity of functional interactions for optimum cumulative performance.
This article provides a general overview of coronaviruses, emphasizing SARS-CoV-1 and −2 and their spike glycoproteins. The article follows to discuss nanobodies and their molecular architecture, highlighting key benefits. It henceforward presents the impressive nanobodies developed against the spike glycoprotein to date and suggests interesting multivalent approaches based on protein engineering and particulate nanochemistry to guide the development of treatment technologies against the challenges of spike mutations.
Coronavirus disease: an overview of components, functions and mechanism of infection
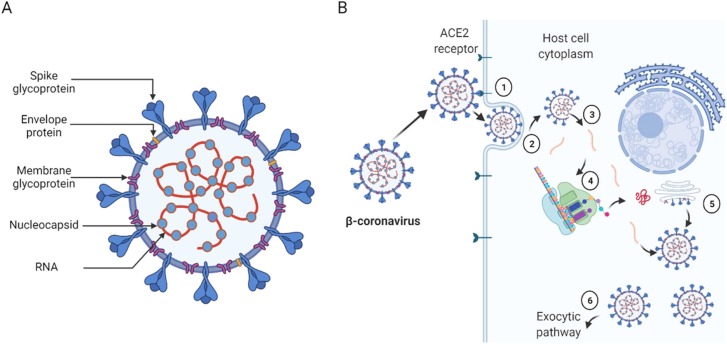
Coronavirus is a positive-strand RNA virus bound with a well-defined envelope. The virus belongs to the Coronaviridae family and is sub-classified into four genera: α-, β-, γ-, and δ-coronavirus [8]. The α- and β-coronaviruses appreciably infect mammals, whereas the γ- and δ-coronaviruses infect birds, fish and, at times, mammals. SARS-CoV-1 and −2 are β-coronaviruses and, thus, share similar structural and molecular architecture. As depicted in Fig. 1A, the β-coronaviruses possess protruding spike proteins that serve for receptor attachment and fusion to membranes. Their membrane glycoproteins shape the virion and support the nucleocapsid, whereas the envelope protein functions in viral assembly, release and pathogenesis [9], [10]. As expected, the viral genomic RNA harbours the replicative identity of the virus, whereas the nucleocapsid encapsidates the genome into the virus [9], [10]. During infection (Fig. 1B), the spike protein interacts with the cell-surface ACE2 receptor and fuses with the cell membrane with the assistance of host cell proteases such as cathepsin L and TMPRSS2.
Fig. 1.
Schematic description of β-coronaviruses (A) and their simplified cell entry and replication mechanism. Before cell invasion, the spike protein interacts with the ACE2 cell surface receptor (1) and fuses the cell membrane aided by enzymatic proteolysis (2). Next, the virus enters the cytosol and releases the positive-strand RNA (3), which adopts the host cell replicating and translation machinery (4) to propagate and package the virus (5). Finally, the matured virus exits through the exocytic pathway (6).
Interestingly, the host invasion and virus propagation cycle would putatively be inefficient without the spike protein. Consequently, cell entry is preceded by viral RNA release, where the host transcription and translation machinery produce all components required for viral replication and packaging. The packaged virus then plies the exocytic pathway out of the cell to infect other healthy cells to continue the cycle.
Spike glycoprotein: the molecular and structural similarities and differences
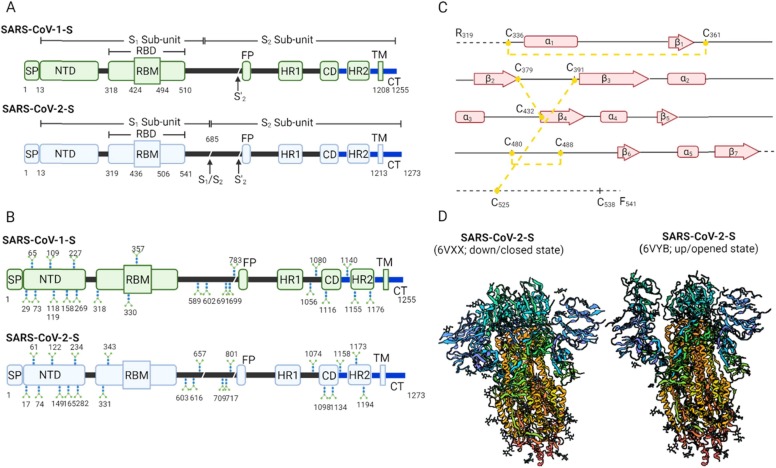
The critical role of the spike (S) glycoprotein in the virus's life cycle is important for vaccine and therapy development, especially by understanding its molecular and structural characteristics. In general, the SARS-CoV-1 and −2-S glycoproteins are multidomain proteins consisting of 1255aa and 1273aa, respectively ( Fig. 2A). The multidomain structure composes a signal peptide (SP) and S1 and S2 sub-units from the N-terminus, respectively. In the native state, the structure forms a homotrimer. The S1 subunit harbours a rigid N-terminal domain (NTD) and a receptor-binding domain (RBD) responsible for receptor attachment. On the other hand, the S2 sub-unit is known for host cell membrane fusion and thus comprises a fusion peptide (FP), heptapeptide repeat sequence (HR 1–2), and connecting domain (CD), transmembrane (TM) and cytoplasmic (CP) domains. One key distinguishing feature in SARS-CoV-2-S is the presence of a novel multibasic protease cleavage site (S1/S2) located at the junction of the S1 and S2 sub-units. Unlike the S1 cleavage site processible by cathepsin and TMPRSS2, the S1/S2 site has expanded proteolytic activation machinery, including Furin-like proteases [11], [12]. The protease expansion is putatively liable for the stability and transmission of SARS-CoV-2.
Fig. 2.
Spike glycoprotein characteristics. A: Schematic comparison of the multidomain spike glycoprotein structure of SARS-CoV-1 and SARS-CoV-2. B: Schematic representation of glycosylated site differences between SARS-CoV-1-S and SARS-CoV-2-S. C: β-sheets, α-helices and disulfide bond connections in SARS-CoV-2-S RBD. D: Closed and opened states of SARS-CoV-2-S ([17]; Reproducible under Creative Commons Attribution license CC0 1.0) Signal peptide (SP), N-terminal domain (NTD), receptor-binding domain (RBD), receptor-binding motif (RBM), protease cleavage sites (S1/S2, S’1), fusion peptide (FP), heptad repeat 1&2 (HR1&2), connecting domain (CD), transmembrane domain (TM) and C-terminal domain (CT).
Notably, the spike proteins of SARS-CoV-1 and − 2 exhibit differences in glycosylation levels (Fig. 2B). These glycans are host-derived and putatively play a role in protein folding, viral tropism and immune escape [13], [14]. SARS-CoV-1-S has 23 N-acetylglucosamines (NAGs) per protomer vis-a-vis 22 NAGs per protomer for SARS-CoV-2-S [15], [16]. In SARS-CoV-2-S, 20 out of 22 glycans are conserved and distributed across both S1 sub-unit (i.e., 9 out of 13 NAGs conserved) and S2 sub-unit (all the 9 NAGs conserved) [16], [17]. Thus, the presence of the glycans has the penchant for causing specific epitope shielding that may weaken the neutralization abilities of potential blockers and cause viral immune escape.
Notwithstanding, viral immune escape could also be caused by epitope-associated residue substitutions, deletions and insertions that lead to an altered epitope conformation, increased receptor binding affinity and overall antigenicity [18], [19], [20], [21]. The concept of immune escape [22] and mutations of SARS-CoV-2 [23] have been excellently reviewed in recent papers for readers reference.
Focusing on the RBDs, both SARS-CoV-1 and − 2 SRBDs compose of five-stranded anti-parallel β-sheets (β1–β4 and β7) core that is linked together by short connecting α-helices and loops and stabilized by three pairs of disulfide bonds [9], [24], [25]. Within this RBD structure lies the receptor-binding motif (RBM) positioned between β4 and β7. The RBM comprises a pair of β-strands (i.e., β5 and β6) and α-helices (i.e., α4 and α5) stabilized by a single disulfide bond at the distal end [24], [25]. Interestingly, the RBD contains most of the receptor contacting residues and is thus essential for vaccine and therapy development. There are about 16 and 17 receptors contacting residues in SARS-CoV-1-S and SARS-CoV-2-S, respectively [24]. Fourteen of the contacting residues share similarities. The entire individual contacting residues interact with 20 ACE2 residues, where 17 of the ACE2 contact points are similar for SARS-CoV-1 and − 2 [24]. Fig. 2C describes the β-sheets, α-helices and disulfide bond connections in SARS-CoV-2S RBD.
In terms of conformation dynamics, both RBDs show similar changes, as represented in Fig. 2D. For instance, they assume an up promotor (receptor accessible; metastable) and down promoter (receptor inaccessible; stable) states in the similitude of a hinge [15], [16], [17], [26]. However, the conformation of SARS-CoV-1 SRBD packs tightly against the N-terminal domain of the neighbouring promotor, whereas SARS-CoV-2 SRBD angles closer to the central cavity of the trimer [16]. To this end, the cryptic characteristics of the spike protein open up interest in nanobody therapy due to its ability to target cryptic epitopes and viral canyons.
Nanobodies: molecular and structural characteristics
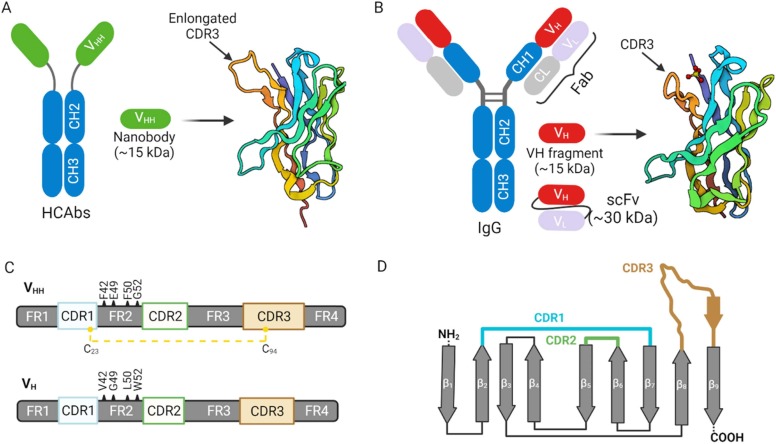
Nanobodies are single-domain, antigen-specific fragments derived from the heavy-chain-only antibodies (HCAbs) of camelids, such as alpacas, llamas and dromedaries. These heavy-chain-only antibodies are a unique class of antigen binders serendipitously discovered during the retrieval of a single-chain variable fragment (scFv) from naive immunoglobulin (IgG) libraries [27]. Unlike canonical antibodies, which are heterotetrameric, the structure of HCAbs is homodimeric and consists of two heavy chains fused to the constant fragment lacking the first constant domain (CH1) ( Fig. 3A-B). In other words, the HCAbs structure is devoid of the light chain and the characteristic first constant domain (CH1) of the heavy chain polypeptide of canonical antibodies. The absence of these domains does not impair the specificity and affinity of HCAbs [28]. However, these characteristics make HCAbs interestingly monovalent and smaller than conventional antibodies; thus, a promising candidate for disease diagnostics and therapeutics.
Fig. 3.
Characteristics of nanobodies and nanobody-like structures. A: Schematic description of camelid heavy-chain only antibody (HCAbs) demonstrating the nanobody (VHH) section and its cartoon. The HCAbs lacks the CH1 domain. B: Schematic description of IgG antibody showing heavy-chain (VH) section and its cartoon. C: Schematic comparison of VHH and VH, showing the complementary determining regions (CDR1–3), framework regions (FR1–4) and the hydrophilic residues (F42, E49, R50, and G52) and its hydrophobic (V42, G49, L50, and W52) counterparts in VHH and VH, respectively. The disulfide bond linkage in VHH is also shown. D: Schematic arrangement of the CDR loops within the nine β-stranded (β1–9) core architecture of VHH. Variable light (VL), constant chains (CH1-CH3), antigen-binding fragment (Fab); and single-chain variable fragment (scFv).
Of note, nanobodies represent the smallest (~15 kDa; 2.5 nm width and 4.2 nm height), detachable, functional antigen binders that form the variable (VHH) domain of the HCAbs. Comparatively, antibodies (IgG) are about 150–170 kDa and with ~13.7 nm width and ~8.4 nm height [29]. Nanobodies fairly compare with antibody VH in size, affinity and biodistribution, but differs from scFv—a fusion protein consisting of antibody VH and VL (Fig. 3B).
Like canonical antibodies, nanobodies perform their binding functionalities with the help of their hypervariable complementary determining regions (CDR1-CDR3) encoded by the IGHV gene or generated via V-D-J gene recombination [30]. Both nanobodies and IgG VH have four framework structures (framework regions, FRs) which enclose the CDRs (Fig. 3C). Clearly, there is a high sequence resemblance between the human/mouse VH and camelid VHH generated for the same antigen. However, there is a pronounced presence of solvent-exposed, hydrophilic amino acid units in the FR2 of nanobodies (F42, E49, R50, and G52) which contrast with the hydrophobic residues of human VH (V42, G49, L50, and W52). This sequence variation does not cause any conformational alterations of the backbone [31]; nevertheless, the hydrophilic inclusions make nanobodies highly water-soluble [32], [33]. On the other hand, antibody VH often suffers aggregation tendencies and stickiness [34], [35], [36]; its associated scFv versions are no different since they inherit the intrinsic properties of corresponding VH-VL domains [37], [38]. Importantly, the intrinsinc hydrophilicity in nanobodies facilitates their facile production, especially in forms such as multimers to confer avidity (see subsequent discussions). As such, several groups have incorporated the hydrophilic residues of nanobodies into IgG VH with resounding outcomes [39], [40], [41].
In terms of the CDR architecture, antibody VH has short-looped CDRs, whereas nanobodies (especially CDR1 and CDR3) are known to be flexibly extended such that they are capable of accessing catalytic sites of enzymes and cryptic epitopes such as viral surface canyons [42], [43], [44], [45]. In nanobodies, the CDR1 and CDR3 are often linked by an interloop disulfide bond through Cys23 and Cys94, respectively [32], [46]. This interloop disulfide bond is conserved among all nanobodies, and it serves a dual purpose of stabilizing the overall nanobody structure and rigidifying the base of the flexible CDR3 loop to facilitate a stronger epitope interaction [47]. The presence of the disulfide bond could also be the rationale for the thermal and chemical stability of nanobodies, and its removal does not significantly compromise the stability [48]. It is worth noting that dromedary-derived nanobodies sometimes have an additional stabilizing disulfide bond linking the CDR2 and CDR3 in a solvent-exposed fashion to prevent aggregation [47], [49]. Fig. 3D shows the arrangement of the CDR loops within the nine β-stranded core architecture of a typical nanobody. The presence of the β-hairpin within the CDR3 contributes to the totemic cryptic epitope meandering.
In production terms, initial approaches for raising nanobodies shared similarity with the laborious conventional techniques for antibodies [50]. The method required the immunization of the camelid with an immunogen of interest followed by blood lymphocyte isolation and mRNA extraction for cloning and gene library construction. Positive clones were selected via panning or immunoassay techniques prior to the final soluble nanobody expression. But the expression of nanobodies from this stage has been relatively straightforward, unlike the stringent methods for antibody production, which often suffers from batch-to-batch variability, instability, specificity, and selectivity. Interestingly, recent advancements in nanobody technology has seen an immense transformation aided by high-throughput sequencing, in silico translation and LC-MS/MS techniques to expedite nanobody identification and production [51], [52]. These have resulted in a robust pipeline facile production of nanobodies in systems such as high-order eukaryotic cells [53], [54], [55], yeast [53], [54], [55], [56], Escherichia coli [57] and Aspergillus [58]. These achievements put nanobodies in a position of commercial relevance for all ailments, including coronavirus disease. Lastly, nanobodies' smaller size and stability characteristics offer high tissue penetrability advantages and allow the delivery of nanobody therapeutics through alternative routes (e.g., pulmonary via nebulization), which is hardly achievable by other products [44], [56].
Nanobody interaction with coronavirus spike
The interaction of nanobodies with the coronavirus spike protein could be classified into three main groups: receptor binding site (RBS), non-receptor binding site (nRBS), and the overlapping cahoot interactions. For the RBS category, the nanobody competitively binds to the ACE2 receptor-associated epitope on the spike RBD to prematurely trigger the fusion machinery in the absence of ACE2. The result of this decoy interaction may not alter the RBD architecture but leads to the abrogation of cell membrane fusion and virus entry. On the other hand, the nRBS class interacts with the other sections of the spike protein to distort the spike protein’s conformational liberty and thus render it defective towards cell membrane interaction. Finally, the overlapping cahoot sits between RBS and nRBS, where the nanobody-spike protein interaction occurs at the boundary between the two previous scenarios discussed. In this case, the effect is a combination of blocking and distortion of conformation liberty.
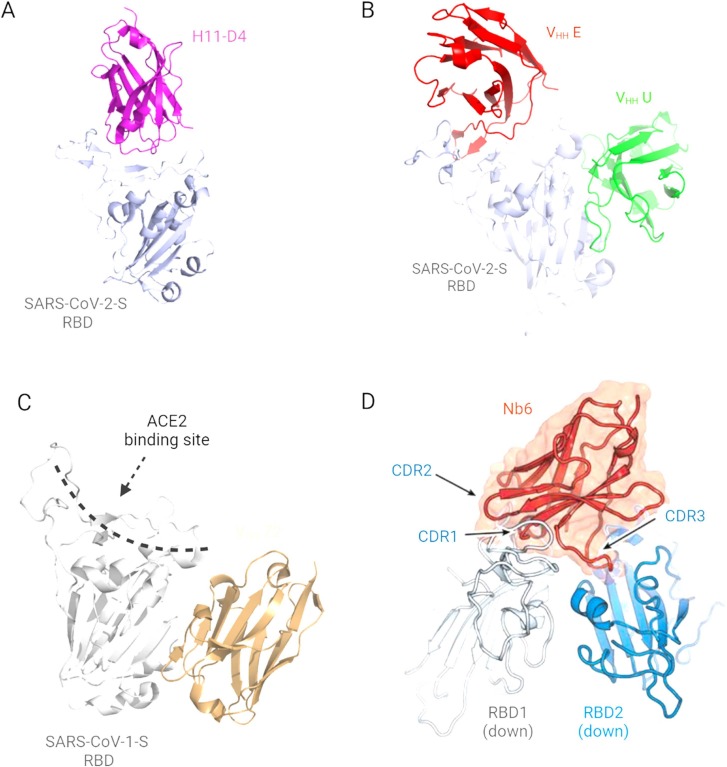
The RBS category is the most explored and reported interaction. For example, Huo et al.[59] reported two closely related nanobodies, H11-H4 and H11-D4, from naïve llama library that recognized the same epitope in the SARS-CoV-2-S RBD. The nanobodies interacted with the spike protein unevenly using all three CDRs. It is interesting to note that the structures of nanobody-spike complexes were indistinguishable, although these nanobodies have sequence variations, especially in the extended CDR3 loops. Between the two constructs, H11-H4 showed a higher binding affinity (KD) of 12 nM compared to 39 nM for H11-D4, and this was evident from the presence of bonds such as salt-bridging, π–cation and hydrogen bonds identified between the interacting surface. Also, Koenig et al.[60] isolated four RBS categorized SARS-CoV-2 nanobodies, namely VHH E, VHH U, VHH V and VHH W. Herein, the VHH E and VHH U interact with distinct epitopes on the RBD in the ‘open’ conformation state ( Fig. 4B), which leads to the induction of the premature fusion state dynamics. This characteristic makes VHH E and VHH U promising for tandem pairing (see subsequent discussion). Notably, the VHH E is llama-originated, whereas the others are alpaca derived. VHH E uses its CDR1 and extended β-hairpin containing CDR3 to interact with the ACE2 binding site via polar, hydrophobic and hydrogen bonding, sharing 16 RBD residue contact points with ACE2. Conversely, VHH U binds the RBD using all three CDRs but shares identical RBD residue contact points between SARS-CoV-1 and −2 SRBD, although it does not block the former. VHH V and VHH W were also alpaca-derived and thus shared a similar structure and binding interface with VHH U. Similarly, Hanke et al. [61] isolated an alpaca-derived RBS nanobody, Ty1, that specifically bound the individual RBDs in the ‘open’ spike trimer state. Ty1 showed an unchanged molecular orientation in their binding state, although binding to two “down” and one “up” RBD conformations within the spike trimer. Ty1 bound the RBD epitope using CDR1 and CDR3 loops that contacted “T470, V483-E484” and “Y449, F490, Q493” RBD residues, respectively. Conversely, the CDR2 was found to mainly stabilized the CDR1 conformation in the RBD complex.
Fig. 4.
Nanobody interaction with coronavirus spike. A: Receptor binding site (RBS) associated nanobodies ([59]; PDB 6YZ5; Reproducible under Creative Commons Attribution license CC0 1.0). B: Promising tandem RBS nanobody pairs ([60]; PDB 7B17; Reproducible under Creative Commons Attribution license CC0 1.0). C: Non-receptor binding site (nRBS) associated nanobody ([62]; PDB 6WAQ; Reproducible under Creative Commons Attribution license CC0 1.0). D: An example of the overlapping cahoots ([63]; Reproducible under Creative Commons Attribution license CC0 1.0).
For the nRBS associated and the overlapping cahoot nanobodies, there has not been many reports and characterizations. Dwelling on the few reported, Wrapp et al.[62] identified nanobody VHH-72 that interacts with SARS-CoV-1 SRBD away from the footprint of ACE2 binding interface (Fig. 4C). The VHH-72 is llama-derived and interacts with its CDR2 and CDR3 in such a way that its backbone groups reinforce the hydrogen bonding with the spike. Similarly, Xiang et al.[64] isolated nanobody Nb95 among several others and discovered through epitope mapping analysis that Nb95 associates with the spike NTD to possibly restrict the conformational flexibility of the spike trimer. Herein, it is opined that the Nb96 related restriction of RBD conformation contributes to a compromised membrane fusion mechanism. Also, Mast et al.[65] reported 16 nRBS nanobodies that show potent neutralization of SARS-CoV-2. However, the molecular determinants of their binding are not available yet.
Lastly, for the overlapping cahoot (Fig. 4D), Schoof et al. [63] reported an ultrapotent synthetic nanobody, Nb6. This nanobody was able to contact two adjacent RBDs in the spike trimer. All three CDRs participated in the interaction, but most of the contacting residues reside within the CDR1 and CDR2. However, it is the CDR3 that meanders its way across the two adjacent RBDs.
Avidity-inspired approaches for effective viral blocking
Protein engineering approaches
Immunoglobulin (Ig) Fc fusion approach
The fusion of molecules to Fc domains of human immunoglobulin (Ig) is not uncommon because the fused Fc passes on antibody-like characteristics such as the recruitment of effector functions, complement activation and long serum circulatory half-life to resultant molecules [66], [67], [68], [69]. Moreover, in relation to coronavirus disease, the approach multimerizes the efficiency and neutralization potency of ACE2 decoys and antibodies, as reviewed elsewhere [70], [71], [72].
In terms of nanobodies, Wrapp et al.[62] engineered a bivalent form of VHH-72 via fusion to the human IgG Fc domain. Compared to the monovalent state, the bivalent VHH-72 firmly bound SARS-CoV-2 RBD and neutralized SARS-CoV-2 pseudoviruses with a half-maximal inhibitory concentration (IC50) of approximately 13 nM. In the quest to boost the reported potency, the authors further modified VHH-72-Fc using computational modelling in a subsequent study [73]. Herein, the affinity-matured modified molecule (VHH72_S56A-Fc) neutralized SARS-CoV-1 and −2 at the subnanomolar level. Interestingly, the new design bound other serbacovirus clades and strongly prevented both SARS-CoV-2 wild type and virulent mutant variants in hamsters.
In another study involving llama and nanomice, Xu et al.[74] reported two groups of interesting nanobodies: Group 1 and 2. The Group 1 versions bound highly conserved epitopes within Serbacoviruses that are inaccessible to conventional antibodies. Also, the nanobodies bypass the antigenic drift associated with the spike variant E484K and N501Y. The Group 2 versions were strictly ACE2 inhibitors. However, they fail to neutralize SARS-CoV-2 variants bearing E484K or N501Y mutations within the ACE2-binding site of the spike RBD. Nevertheless, the Group 2 nanobodies elicited remarkable neutralization activity against these variants in their homotrimer-IgG Fc fusion state and showed an IC50 of 4–538 pM. This enhanced neutralization prowess is a result of enhanced avidity and concomitant crosslinking of multiple viral spike proteins. Also, Dong et al. engineered several humanized nanobody-Fc fusions based on isolated llama-derived nanobodies that prevent ACE2-RBD [75], [76]. Among the versions ranging from monoclonal VHH-Fc to tri-specific VHH-Fc, the tri-specific version showed significantly better blocking and neutralization of SARS-CoV-2 pseudoviruses than the others. Similar concepts were published by other groups [77], [78], where the multivalent nanobody-Fc fusions showed affinities (KD) and IC50 ranging from 59.2 to 250 pM and 43–111pM, respectively. There is also the possibility to tither both ends of the Fc domain with nanobodies via engineered linkers to circumvent notorious incorrect pairing problems that may arise with direct bi or tri-specific nanobody-Fc.
On a different dimension, polyvalent immunoglobulins Fc of IgA and IgM could also be employed in ‘avidifying’ nanobodies since they, respectively, form dimers and pentamers in the presence of the joining chain (J-chain). For instance, the IgA-Fc approach was utilized for engineering ACE2 decoy receptors, where the ACE2-IgAFc neutralized both wild type and mutated variants (N501Y, L452R; N501Y and E484K) [79]. Also, Xu et al.[80] engineered SARS-CoV-2 IgA and IgM by stitching previously identified SARS-CoV-2 IgG1 monoclonal antibodies onto IgA-Fc and IgM-Fc scaffold, respectively. Both engineered IgA and IgM strongly inhibited live SARS-CoV-2 virus better than their IgG–based counterparts. In particular, the work reported IgM-14 as the most potent neutralizer, where the enhanced avidity effect successfully neutralized the South African B.1.351, Brazilian P.1 and UK B.1.1.7 variants by 374-, 547- and 45-fold, respectively, in comparison with their IgG counterparts. Furthermore, IgM-14 neutralized other resistant viruses and 21 generated RBD mutants, of which numerous reported antibodies fail to recognize. To even make smaller pentameric versions, research can consider using the 18-amino acid secretary tailpiece of the IgM to scaffold nanobodies in a similar fashion done in other areas [69], [81], [82], [83], [84], [85], [86].
Nanobody cocktail approach
The use of protein cocktails presents a promising avidity approach to thwart SARS-CoV-2 mutational escape, as demonstrated for antibodies [87], [88], [89]. In this case, the concocted antibodies achieve the optimum neutralization benefit if they bind to distinct and non-overlapping epitopes. Also, this approach offers conformational freedom to the blocking molecule, which may be advantageous. Given the promising features of nanobodies against the variants of concern, a combinatorial mixture of multiple non-overlapping nanobodies could offer superior protection in the fight against mutational escape. For instance, Pymm et al. [90] showed that a pairwise combination of four non-competing alpaca-derived nanobodies or their Fc-fusion versions potently neutralize wild-type SARS-CoV-2 and variants with N501Y and D614G mutation. The researchers reported a 104-fold reduction in the viral load of these strains in mice studies. Similarly, Dong et al. [75], [76] showed via in-silico pairwise combinations that their llama-derived nanobodies synergistically block SARS-CoV-2.
In another study that raised llama-derived nanobodies against individual spike protein subunits such S1, S2, and non-RBD [65], the researchers demonstrated the effect of cooperativity and synergism in cocktail therapies. Herein, the work identified nanobodies 71, 16 and 26 having high-affinity interaction (KD<1 nM) with S1 RBD, S1 non-RBD region, and S2 SARS-CoV-2, respectively. Furthermore, a pairwise binary combination of these nanobodies successfully blocked escape mutants and variants of concern. These results confirmed that careful selection of non-competing nanobodies is a plausible way to combat future SARS-CoV-2 recalcitrance.
Engineered linker fusion approach
Immunoglobulin-based fusion may not be devoid of aggregation due to woolly pairing of intra- and inter-chain disulphide bonds. Also, methods to quantitatively identify proportionate cocktails may be exponentially laborious, requiring a sophisticated multidimensional synergy framework, modelling and in-silico design to achieve batch-dependent dosage estimations. Moreover, the inclusion of cocktails components targeting the same epitope may deceptively display additive effects instead of an expected synergized low-concentration benefits. However, the use of engineered linker fusions obviates these challenges and presents extra advantages in relation to molecular proximity, especially when epitopes within the same spike protein are to be targeted. Moreover, strategic linking of nanobodies into bivalent, trivalent, bispecific and tri-specific formats becomes straightforward towards avidity-inspired therapeutics. With knowledge of intra-epitope distances on SARS-CoV-2 spike protein [91] and rational linker length engineering [92], the much-needed avidity-inspired nanobodies can be engineered to crosslink spike proteins in the prevention of immunity escape. For example, Colton et al. [93] assembled synthetic non-overlapping SARS-CoV-2 nanobodies into bi-paratopic and multivalent formats with linkers and reported approximately 600- and 1400-fold increase in spike protein binding affinity and virus neutralization potency, respectively. Herein, the most potent trivalent nanobody neutralized live SARS-CoV-2 virus with an IC50 of 4 nM. Koenig et al. [60] also engineered linker based multivalent and bi-paratopic nanobodies from the four individual nanobodies already mentioned. The engineered multivalent nanobodies displayed high avidity towards the spike protein and were 100-fold more potent than their monomeric nanobodies in neutralizing the SARS-CoV-2 virus. Impressively, biparatopic nanobody fusion (EV and VE) prevented escape mutant compared to binary nanobody cocktails E + U, E + V and E + W. Also, Xiang et al. [64] isolated ultrapotent anti-SARS-CoV-2 nanobodies with pico- to femtomolar affinities to SARS-CoV-2-S and inhibited live the SARS-CoV-2 virus at sub-nanogram per millilitre concentration. The researchers further engineered linker-based homo-, heterodimeric and homotrimeric nanobodies out of Nb20 and Nb21, using flexible linkers such as (GS)25 and (EK)31. The resulting multivalent nanobodies showed exceptional avidity and ultrahigh neutralization potency against SARS-CoV-2 virus. The authors showed in a subsequent report that their homotrimeric Nb21 (PiN-21) require low-dosage (0.2–0.6 mg/kg) to protected infected Syrian hamster [94]. Many other recent studies have demonstrated and confirmed the potential of linker-based bi-paratopic or multi-paratopic nanobodies for enhanced blocking and neutralization of SARS-CoV-2 variants [63], [95], [96], [97].
Of note, the success of this approach indispensably dwells on impressive linker design and engineering techniques. As such, the nature, design and functionality of selected linkers contribute to the conformational freedom, expression yield, stability, bioactivity and pharmacokinetic profiles of the recombinant fusion protein [98], [99]. Furthermore, the length and composition of the linker could also affect the cytotoxicity of the fusion protein [99]. Over the years, selection considerations include flexible, rigid, and in vivo cleavable linkers depending on the intended application. However, flexible glycine-serine rich linkers are the most preferred options for antibody fragments engineering to date. Readers are referred to Rosmalen et al. [100] and Gräwe et al. [92] for guidance in rational flexible linker design approaches in relation to multidomain protein engineering.
Multimeric domains and non-immunoglobulin scaffolds
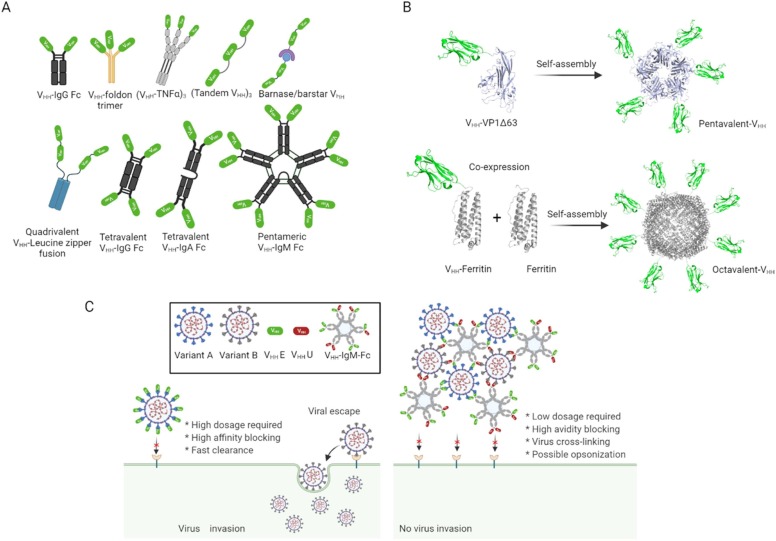
So far, it is apparent that avid spike protein binding and viral blocking are enhanced as oligomeric assemblies and valency of nanobodies increase. In line with this, protein engineers have access to several beneficial higher oligomerization domains that could confer nanobodies with high-avidity abilities for SARS-CoV-2 treatment and mutational escape prevention. In addition, some of these scaffolds also allow steric complementary binding in the correct orientation for diverse complexities, passing on much-needed stability and protease cleavage resistance to involving molecules. Notably, stability and protease cleavage resistance are important for drug delivery purposes. Interestingly, some of these oligomeric domains have been utilized for multivalent antibodies engineering in other therapeutic applications as reviewed elsewhere [70], [71]. However, despite these enormous efforts in searching for ultrapotent multivalent nanobodies against the SARS-CoV-2 virus, these oligomeric domains are yet to be utilized. Below are some straightforward oligomeric domain technologies capable of improving the valency of nanobodies from 2 through to 10.
For instance, helix-turn-helix motif leucine zipper dimerization peptide domains (Fos or Jun) can be used to generate mono-, bispecific, bivalent and tetravalent nanobodies imitating the Y-shape of antibodies [101]. Also, trivalent nanobodies can be generated by fusing a single nanobody to the N-terminal of trimeric domains such as foldon[102], triple-helical bundle, [103] viral capsid protein SHP [104], and collagen XV, VIII or triplex-forming peptide [105], [106], [107]. These resulting multivalent nanobodies could potentially bind and lock the trimeric spike protein to render the virus defective. For tetramerization, mono or bivalent nanobodies can be genetically fused to p53 protein TD [108], GCN4 amphipathic coiled-coil helix [109], and streptavidin core [110]. Moreover, biparatopic hexavalent nanobodies can be generated by fusing nanobodies to both exposed N- and C-terminus of trimeric TIEXVIII collagen domain [111].
Furthermore, pentameric nanobodies can be generated via fusion to the N-terminus of non-toxic verotoxin B-subunit [112]. Fusion to both the N- and C- terminus results in bivalent decameric molecules [113]. Lastly, heptavalent nanobodies can be exploited by N-terminus fusion with heptameric ring-like protein IMX313[114] and heptameric domain of Archaeal RNA binding protein Sm1 [115]. Besides these interesting possibilities, we anticipate that a higher-order expression system such as yeast may be required to overcome potential challenges such as incorrect domain swapping, stoichiometry mismatch and aggregation problems.
Docking ligands also offer a flexible, alternative avenue to create multimeric proteins. For instance, SnoopTagIr-DogTag [116], SpyTag-SpyCatcher [117], barnase–barstar [118] and dock-and-lock (DNL) assembly approach [119] can be exploited to design bivalent, trivalent and multi-specific tetravalent anti-SARS-CoV nanobody candidates with permanent, stable and robust pairing/association. However, in all these examples, the immunogenicity and overall safety are yet to be accessed appropriately. We, therefore, refer readers to Löfblom et al.[120] for more information on non-immunogenic scaffolds.
Protein cages and virus-like particles
Protein cages, viral-like particles (VLPs) and protein nanoparticle scaffolds serve as ideal platforms for multimerizing protein molecules. This attribute stems from their interesting properties, including optimal size and space orientation, tunable surface area and valency, biocompatibility and stability, monodispersity, and many others [121], [122]. These hallmarks make it interesting to attempt their use for avidity-inspired nanobodies therapeutics. Another hallmark of these platforms is that they can assemble and display up to 72 monomeric nanobody units if properly engineered. Thought-provokingly, since the SARS-CoV-2 viral architecture contains 24 ± 9 spike trimer units [91], this multi-affinity nanobody approach may uniquely crosslink several viruses into a defective state. In this case, protein cages such as ferritin (24-mer) [123], dihydrolipoyl acetyltransferase [124], and Lumazine synthase (60-mer)[125] could be exploited. Notably, these scaffolds have previously been used to display several proteins [126], [127], [128], [129].
In the context of coronavirus disease, Rujas et al.[130] genetically fused an already reported VHH-72 to the N-terminal light chain of human apoferritin protomer to generate an ultrapotent SARS-CoV-2 neutralizer. Herein, the modular unit successfully assembled 24 copies of VHH-72 into homogenous spherical particles with high avidity towards SARS-CoV-2-S. Remarkably, the 24-mer VHH-72 particle showed approximately 10,000-fold blocking potency in comparison to the IgG counterparts. In terms of VLPs, many useful scaffolds such as VP1, MS2, HPV, HbsAg and AP205 are available to display more than 60 copies of nanobody for SARS-CoV-2 neutralization applications. However, VLP-based super-scaffolds (72-mer) have not been applied yet in light of nanobody presentation. Nevertheless, they find applications in diverse antigen presentation as reviewed already reviewed [131]. Fig. 5 summarizes the protein engineering approaches and associated benefits. Table 1 summarizes recent high avidity and ultrapotent anti-SARS-CoV-1 and −2 nanobodies reported in literature.
Fig. 5.
Some protein engineering approaches. A: Immonoglobulin Fc and linker tandem fusions. B: Scaffolds and protein cages. VHH-VP1Δ63 spontaneously forms pentamers. The structure is capable of forming 72-mer. VHH-Ferritin scaffolds up to 24 molecules. A co-expression with unfused ferritin enhances the spatial orientation of scaffolded molecules. C: Benefits of an avidity-inspired nanobody for coronavirus treatment. Using soluble nanobodies may suffer from issues of viral escape, which can easily be prevented with an avidity-inspired design.
Table 1.
Representative high avidity and ultrapotent anti-SARS-CoV-1 and −2 nanobodies.
| Nanobody Name | Source and method | Virus type | Binding affinity (KD) | Neutralizing activity (IC50) | Mechanism of neutralization/inhibition | Protective efficacy | Ref |
|---|---|---|---|---|---|---|---|
| VHH-72 Bivalent VHH-72-Fc |
Immune library + phage display, IgG Fc fusion | SARS-CoV-1 MERS-CoV SARS-CoV-2 RBD |
36.8 nM | 13.3 nM | Recognize epitope residues (Trp100, Tyr356/494, Cys366, Phe 364,Ser358,Arg 426) on spike RBD, Blocks RBD-ACE2 interaction, Neutralized SARS-CoV-2 pseudoviruses |
N/A | [62] |
| H11-D4 H11-H4 H11-D4-Fc H11-H4-Fc |
Naïve library, IgG Fc fusions | SARS-CoV-2 RBD | 39 nM 12 nM N/A N/A |
N/A N/A 4–6 nM 18 nM |
H11-H4 recognize RBD epitope via hydrogen bonding (residues; Lys 449-Phe456, Gly482 – Ser 494), van der Waals interaction and salt bridge contact; Blocks RBD–ACE2 interaction; Neutralized SARS-CoV-2 pseudoviruses |
N/A | [132] |
| Ty1 Ty1-Fc Fu2 Fu2-Fc Fu2-Ty1 homodimer/ heterodimer |
Immune library + phage display, IgG Fc fusion, Chemical linkage (Sortase A labelling and Cu-free click-chemistry) |
SARS-CoV SARS-CoV-2 SARS-CoV-2 Variant (B.1.351) |
5–10 nM 0.12 nM |
54 nM 1 nM 7 nM 0.75 nM 0.8 nM 140 pM |
Ty1 recognize RBD epitope residues (T470, V483-E484, Y449, F490, Q493); Fu2 recognize RBD epitope residues (381 – S375) via anti-parallel β-strand hydrogen bond; Fu2 induced the formation of spike trimer-dimers; Block RBD-ACE2 interaction, Neutralized pseudotyped and live SARS-CoV-1, −2 and variant (B.1.351) |
Fu2-Ty1 prophylactically and therapeutically mice from SARS-CoV-2 challenge | [61], [95] |
| Nb-15,Nb17, Nb19 Nb56; Nb-30,Nb-12 Bivalent format Trimer format |
Immune (llama and Nanomouse) library, IgG Fc fusion |
SARS-CoV-2 Variants (B.1.1.7, B.1.351, P.1) SARS-CoV-1 |
Monomer 30–3 nM Multimeric 0.8 nM − 2.4pM |
Monomer 11.7 nM Multimeric (65–9 pM) |
Recognize conserved epitope residues different from RBD; Focus and block RBD – ACE2 interaction; Neutralized pseudotyped and live SARS-CoV-1, −2 and variant (B.1.351) |
N/A | [74] |
| Sb23 Bivalent Sb23-Fc |
Synthetic library + Phage display IgG Fc fusion |
SARS-CoV-2 | 10 nM 225 pM |
0.6 µg/ml 0.007 µg/ml |
Recognize spike RBD residues Block RBD – ACE2 interaction Neutralized pseudotyped SARS-CoV-2 virus |
N/A | [133] |
| sdAb Bivalent sdAb-Fc |
Humanized Synthetic library + phage display IgG Fc fusion |
SARS-CoV-2 | 0.99–35.5 nM | 0.0009–0.07 µg/ml 0.001–0.043 µg/ml |
Recognize spike RBD residues Block RBD – ACE2 association Neutralized pseudotyped and authentic SARS-CoV-2 virus |
N/A | [134] |
| Nb91, Nb3 Homodimer homotrimer Heterodimer |
Naïve library IgG Fc fusion |
SARS-CoV-2 | N/A | N/A 32.36–54.07 nM 4.7 – 4.89 nM 1.54 nM |
Recognize spike RBD residues Block RBD – ACE2 association Neutralized pseudotyped SARS-CoV-2 virus |
N/A | [77] |
| WNbFc2,7,15,36 WNbFc mixtures |
Immune library + phage display IgG Fc fusion Cocktail |
SARS-CoV-2 Variant N501Y D614G |
0.25–0.55 nM 0.22– 0.52 nM |
0.1 – 3.18 nM | Recognize spike RBD residues (Y109,E484,F486,Q493,N501,K417,R403) Block RBD – ACE2 interactions Neutralized both wild-type SARS-CoV-2 and N501Y D614G variant |
Nanobody-Fc mixtures prophylactically prevented Wild-type SARS-CoV-2 and N501Y D614G variant in mice | [90] |
| 1B, 3 F,2A-Fc Bispecific 1B-3 F;1B-3 F-Fc Trispecific 3 F‑1B‑2A-Fc 1B-3 F-2A-Fc |
Naïve and synthetic humanized phage libraries IgG Fc fusion Cocktail |
SARS-CoV-2 | 0.82 – 1.6 nM 0.25 nM 95 pM 47 pM |
1 nM 0.71 nM 0.74 nM |
Bind spike S1 RBD residues Block SARS-CoV-2/ACE2 interaction Neutralized pseudotyped SARS-CoV-2 virus |
N/A | [75], [76] |
| Nbs 20,34,89,95 Nb 21 Tri-Nb 20 Tri-Nb21 (PiN-21) |
Immune library + MS proteomic strategy Strategic linking |
SARS-CoV-2 | 0.102/0.133 nM 0.045 nM 4.1 pM 1.3 pM |
10.4 – 108 pM < 1 pM < 1 pM < 1 pM |
Recognize spike RBD residues and epitope closer to trimmer NTD Block SARS-CoV-2/ACE2 interaction Neutralized pseudotyped and authentic SARS-CoV-2 virus |
PiN-21 (Tri-Nb21) prophylactically and therapeutically prevent and treat SARS-CoV-2 infection in Syrian hamster | [64], [94] |
| Nb 6 mNb 6 mNb6-tri |
Synthetic library + yeast surface display Strategic linking |
SARS-CoV-2 | 210 nM 0.45 mM < 1 pM |
2 μM 6.3 nM 54–120 pM |
Recognize spike RBD residues, bind and lock spike in the inactive state Block SARS-CoV-2/ACE2 interaction Neutralized pseudotyped and authentic SARS-CoV-2 virus |
N/A | [63] |
| VH A01, B01, B02; Biparatopic (VH2A01-B01) Multivalent (VH 3 B01) |
Synthetic humanized library + phage library IgG Fc fusion Strategic linking |
SARS-CoV-2 | 23 – 113 nM 0.1 nM 0.12 nM |
33.5 nM 26.2 nM 4.0 nM |
Out-Competed ACE2 and bind RBD and spike ectodomain Neutralize pseudotyped and authentic SARS-CoV-2 virus |
N/A | [93] |
| VHH E,U,V,W Biparatopic/Bivalent (VE,EV,EE,VV) Trivalent EEE |
Immune library + phage library Strategic linking |
SARS-CoV-2 Escape mutants |
1.86 – 22 nM 84/200 pM N/A |
60 nM 0.7 – 1.32 nM 170 pM |
VHH V, U and E bind and recognize distinct ACE2 binding epitopes on spike RBD in different orientation; Nanobodies trigger activation of the fusion machinery; Block ACE2 – RBD interaction Neutralize pseudotyped and authentic SARS-CoV-2 virus |
N/A | [60] |
| VHH-72 Bivalent VHH-72-Fc VHH-hFerr |
Immune library 24-mer Apoferritin protein cage as scaffold | SARS-CoV-2 | N/A 36.8 nM < 1 pM (beyond detection limit) |
1.3 µg/ml 0.00011 µg/ml |
Recognize spike RBD epitope residues Neutralized pseudotyped SARS-CoV-2 |
N/A | [130] |
Particulate nanotechnology approaches
Herein, advancements in particulate nanotechnology in areas such as inhalable [135], [136], [137] and injectable [138], [139] nanomaterials show tremendous promises for applications in medicine. These materials include, but are not limited to, liposomes, dendrimers, polymers, fullerenes, nanoribbons, carbon nanotubes and dots ( Fig. 6A). Although these nanomaterials possess excellent bioavailability and pharmacokinetics of medicinal relevance [140], yet they remain to be fully utilized. Also, the surface area of these nanomaterials is highly available for implanting a plethora of ligands at high densities for avidity-inspired drug delivery purposes.
Fig. 6.
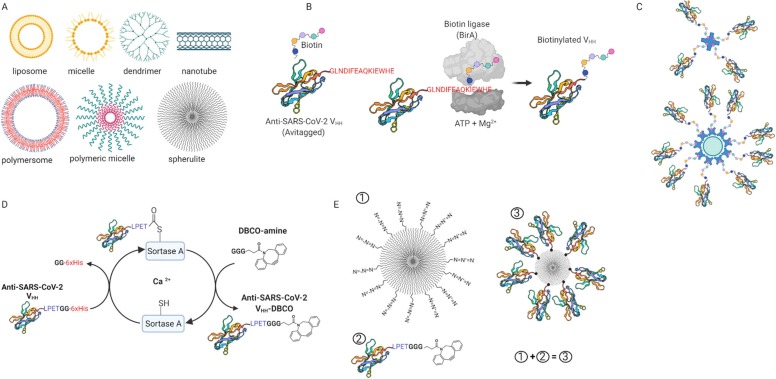
Particulate nanotechnology approaches towards avidity-inspired therapeutics. A: Some examples of medically relevant nanoparticles. B: Protein biotinylation through biotin ligase. C: Multivalent nanobodies based on biotin-streptavidin interaction. D: Sortase A transpeptidation. E: Click chemistry based multimerization, where nanoparticle with azide group (1) reacts with nanobody-DBCO (2) to produce a heptavalent molecule (3).
Focusing on inhalable nanoparticles by virtue of their potential in curbing lung-associated diseases, many incredible chemistries could be applied on these nanoparticles in tandem with anti-SARS-CoV-1 and − 2 nanobodies towards achieving avidity responses. In fact, the past two decades have seen several efforts towards achieving orthogonal, site-specific multivalent molecules through chemical exploitation of inherent reactive functional groups [141], [142], [143], [144], [145], genetic code expansion [146], [147], [148], [149] and enzymatic approaches [150], [151], [152]. There is, therefore, no right time but now to consider these options for improving the avidity of viral blocking molecules such as nanobodies. In the case of avidity-inspired nanobodies, the chemical functionalization approach will be preferable for carrier nanoparticle modification, whereas the genetic code expansion and enzymatic approaches would favour protein modification due to the mild reaction requirements. In the genetic synthesis approach, aminoacyl tRNA synthetases (aaRS) are used to recognize and replace specific codons (e.g., amber UAG) with an unnatural amino acid that bears the required orthogonal reactive handle [153]. Enzymatic approaches such as proximity-dependent biotinylation [154], [155], [156], [157] and sortase A (Srt A) transpeptidation [158], [159], [160], [161] facilely append handles necessary for multimerizing nanobodies on nanocarriers. For instance, in proximity-dependent biotinylation (Fig. 6B), enzymes such as biotin ligase (BirA) attach biotin molecules via nucleophilic interaction onto Avi-tag (GLNDIFEAQKIEWHE) bearing proteins using Mg2+ and ATP as cofactors [162]. The biotinylated protein can then be multimerized onto nanoparticle carriers exploiting the strong binding affinity between biotin and strept(avidin) (KD 4 × 10−14 M; Fig. 6C) [163], [164]. Herein, the approach has been used to produce tetrameric molecules for cancer treatment [165], [166]. In Srt A transpeptidation (Fig. 5D), the enzyme recognizes the C-terminal peptide motif (LPXTG) on target molecules, cleaving between threonine (T) and glycine (G), and ligates available molecules bearing N-terminal glycine repeat. The “X” is preferentially aspartic acid (D), glutamic acid (E), alanine (A), asparagine (N), glutamine (Q) or lysine (K). The transpeptidation mechanism is mostly Ca2+ dependent, but there is a Ca2+ independent variant available [167]. In view of this, several applications associated with protein multi-functionalization [161], protein cyclization [168], protein lipidation [169], and cell-surface labelling [170], [171] have been attempted. Modifications such as C- to N-terminal [172], [173], N- to C-terminal [174], [175], and unnatural C- to C- and N-to N-terminal [176] are available. The method could be complemented with click chemistry to load functional nanobodies onto nanoparticles to obtain avidity-inspired therapeutics, as depicted in Fig. 6E. These products can easily be formulated and delivered via the nasal route at low dosages for coronavirus and respiratory infections. In line with these approaches, Moliner‐morro et al. [177] has reported bivalent (Ty1-Ty1 and Ty1-PEG-Ty1) and 4-arm polyethylene glycol (PEG) tetrameric construct using combined Srt A transpeptidation and strain promoted azide-alkyne click chemistry. Herein, the bivalent formats yielded a similar potency as Ty1-Fc fusion, whereas the tetrameric Ty1 construct dramatically neutralized SARS-CoV-2 at IC50 = ~13 pM. The group has further reported that their bispecific nanobody induces spike trimer dimerization [95]. Having these toolsets available, it is important to note that bioconjugation approaches for medical applications must be orthogonal and site-specific, yielding safe and quantitative products. Therefore, safety verifications of the final product are mandated.
Avidity-inspired nanobody therapeutics: outlook
Recently, traditional antibody therapeutics for viral disease treatment has shown the potential to intensify viral infections or cause detrimental immunopathology via a phenomenon known as antibody-dependent enhancement (ADE). Simply put, ADE results from an antibody-Fc receptor (FcR) complex-mediated virus uptake into phagocytic cells, which results in heightened inflammation and tissue injury (reviewed in [178], [179], [180]). Whiles ADE is improbable for nanobody therapeutics bearing no Fc domains, multivalent nanobodies fused with immunoglobulin Fc may exhibit some potential danger. However, there is no direct evidence of COVID-19 or SARS-related ADE reported for nanobody-Fc fusion to date, partly because of inadequate knowledge of ADE and assessment techniques. Since the ADE effect is generally overlooked, we suggest that a careful selection, engineering and optimization of immunoglobulin Fc domains that offer improved potency and safer routes be considered. For instance, Yamin et al. [181] and Pinto et al. [182] have exceptionally demonstrated approaches to circumvent detrimental immunopathology, which may prove helpful for avidity-inspired therapeutics.
Also, with the potential of nanobodies to prevent viral escape with no autoimmune responses, the future of avidity-inspired nanobody therapeutics remains oceanic. For instance, we foresee an overflow of exploitation of de novo protein design strategies [183], [184], [185], [186] that have the potential to generate complex non-immunogenic protein scaffolds for use in avidifying nanobodies in no distant future. A case in point is reported by Divine et al. [187], where antibodies were computationally clustered within a protein super-structure for SARS-CoV-2 neutralization. Many more in line with this is anticipated.
Conclusion
Nanobody technology remains a versatile therapeutic approach of high potential in the emergence of coronavirus disease and associated variants of concern. Considering the trajectory of coronavirus, with different variants appearing quarterly, the focus now is on approaches to easily multimerized these potent inhibitors into high-avidity therapeutics. Many highlighted approaches concerning protein engineering and nanoparticle chemistry are available and ready to be explored towards advanced innovations in treatment technologies. With uncompromised safety implemented, the product can be formulated and delivered via the nasal route at low dosages to treat coronavirus and other respiratory infections.
CRediT authorship contribution statement
Eugene M. Obeng: Conceptualization, Writing – original draft, Writing – review & editing, Visualization. Christian K.O. Dzuvor: Writing – original draft, Writing – review & editing. Michael K. Danquah: Writing – review & editing, Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
EMO and CKOD acknowledge Monash Graduate Scholarship for their PhD studies. The figures were drawn and assembled with BioRender.com. The authors acknowledge all other relevant papers that were not cited due to lack of space.
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Dong Y., Dai T., Wei Y., Zhang L., Zheng M., Zhou F. A systematic review of SARS-CoV-2 vaccine candidates. Signal Transduct. Target. Ther. 2020;5:237. doi: 10.1038/s41392-020-00352-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Der Li Y., Chi W.Y., Su J.H., Ferrall L., Hung C.F., Wu T.C. Coronavirus vaccine development: from SARS and MERS to COVID-19. J. Biomed. Sci. 2020;27:1–23. doi: 10.1186/s12929-020-00695-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poland G.A., Ovsyannikova I.G., Kennedy R.B. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396:1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Taylor P.C., Adams A.C., Hufford M.M., de la Torre I., Winthrop K., Gottlieb R.L. Neutralizing monoclonal antibodies for treatment of COVID-19. Nat. Rev. Immunol. 2021;21:382–393. doi: 10.1038/s41577-021-00542-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Idris A., Davis A., Supramaniam A., Acharya D., Kelly G., Tayyar Y., West N., Zhang P., McMillan C.L.D., Soemardy C., Ray R., O’Meally D., Scott T.A., McMillan N.A.J., Morris K.V. A SARS-CoV-2 targeted siRNA-nanoparticle therapy for COVID-19. Mol. Ther. 2021;29:1–8. doi: 10.1016/j.ymthe.2021.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duan K., Liu B., Li C., Zhang H., Yu T., Qu J., Zhou M., Chen L., Meng S., Hu Y., Peng C., Yuan M., Huang J., Wang Z., Yu J., Gao X., Wang D., Yu X., Li L., Zhang J., Wu X., Li B., Xu Y., Chen W., Peng Y., Hu Y., Lin L., Liu X., Huang S., Zhou Z., Zhang L., Wang Y., Zhang Z., Deng K., Xia Z., Gong Q., Zhang W., Zheng X., Liu Y., Yang H., Zhou D., Yu D., Hou J., Shi Z., Chen S., Chen Z., Zhang X., Yang X. Effectiveness of convalescent plasma therapy in severe COVID-19 patients. Proc. Natl. Acad. Sci. 2020;117:9490–9496. doi: 10.1073/pnas.2004168117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bulutoglu B., Macazo F.C., Bale J., King N., Baker D., Minteer S.D., Banta S. Multimerization of an alcohol dehydrogenase by fusion to a designed self-assembling protein results in enhanced bioelectrocatalytic operational stability. ACS Appl. Mater. Interfaces. 2019;11:20022–20028. doi: 10.1021/acsami.9b04256. [DOI] [PubMed] [Google Scholar]
- 8.Millet J.K., Whittaker G.R. Host cell proteases: critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015;202:120–134. doi: 10.1016/j.virusres.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du L., He Y., Zhou Y., Liu S., Zheng B.-J., Jiang S. The spike protein of SARS-CoV — a target for vaccine and therapeutic development. Nat. Rev. Microbiol. 2009;7:226–236. doi: 10.1038/nrmicro2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Y., Liu Q., Guo D. Emerging coronaviruses: genome structure, replication, and pathogenesis. J. Med. Virol. 2020;92:418–423. doi: 10.1002/jmv.25681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jaimes J.A., Millet J.K., Whittaker G.R. Proteolytic cleavage of the SARS-CoV-2 spike protein and the role of the novel S1/S2 site. IScience. 2020;23 doi: 10.1016/j.isci.2020.101212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffmann M., Kleine-Weber H., Pöhlmann S. A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol. Cell. 2020;78:779–784. doi: 10.1016/j.molcel.2020.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watanabe Y., Allen J.D., Wrapp D., McLellan J.S., Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Sci. (80-. ) 2020;369:330–333. doi: 10.1126/science.abb9983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watanabe Y., Bowden T.A., Wilson I.A., Crispin M. Exploitation of glycosylation in enveloped virus pathobiology. Biochim. Biophys. Acta - Gen. Subj. 2019;1863:1480–1497. doi: 10.1016/j.bbagen.2019.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yuan Y., Cao D., Zhang Y., Ma J., Qi J., Wang Q., Lu G., Wu Y., Yan J., Shi Y., Zhang X., Gao G.F. Cryo-EM structures of MERS-CoV and SARS-CoV spike glycoproteins reveal the dynamic receptor binding domains. Nat. Commun. 2017;8:1–9. doi: 10.1038/ncomms15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C.-L., Abiona O., Graham B.S., McLellan J.S. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367:1260–1263. doi: 10.1126/science.abb2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walls A.C., Park Y.J., Tortorici M.A., Wall A., McGuire A.T., Veesler D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell. 2020;181:281–292. doi: 10.1016/j.cell.2020.02.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Andreano E., Piccini G., Licastro D., Casalino L., Johnson N.V., Paciello I., Monego S.D., Pantano E., Manganaro N., Manenti A., Manna R., Casa E., Hyseni I., Benincasa L., Montomoli E., Amaro R.E., McLellan J.S., Rappuoli R. SARS-CoV-2 escape in vitro from a highly neutralizing COVID-19 convalescent plasma. BioRxiv Prepr. Serv. Biol. 2020 doi: 10.1101/2020.12.28.424451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCarthy K.R., Rennick L.J., Nambulli S., Robinson-McCarthy L.R., Bain W.G., Haidar G., Duprex W.P. Recurrent deletions in the SARS-CoV-2 spike glycoprotein drive antibody escape. Science. 2021;371:1139–1142. doi: 10.1126/science.abf6950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McCallum M., De Marco A., Lempp F.A., Tortorici M.A., Pinto D., Walls A.C., Beltramello M., Chen A., Liu Z., Zatta F., Zepeda S., di Iulio J., Bowen J.E., Montiel-Ruiz M., Zhou J., Rosen L.E., Bianchi S., Guarino B., Fregni C.S., Abdelnabi R., Foo S.-Y.C., Rothlauf P.W., Bloyet L.-M., Benigni F., Cameroni E., Neyts J., Riva A., Snell G., Telenti A., Whelan S.P.J., Virgin H.W., Corti D., Pizzuto M.S., Veesler D. N-terminal domain antigenic mapping reveals a site of vulnerability for SARS-CoV-2. Cell. 2021;184:2332–2347. doi: 10.1016/j.cell.2021.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Starr T.N., Greaney A.J., Hilton S.K., Ellis D., Crawford K.H.D., Dingens A.S., Navarro M.J., Bowen J.E., Tortorici M.A., Walls A.C., King N.P., Veesler D., Bloom J.D. Deep mutational scanning of SARS-CoV-2 receptor binding domain reveals constraints on folding and ACE2 binding. Cell. 2020;182:1295–1310. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harvey W.T., Carabelli A.M., Jackson B., Gupta R.K., Thomson E.C., Harrison E.M., Ludden C., Reeve R., Rambaut A., Peacock S.J., Robertson D.L. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021;19:409–424. doi: 10.1038/s41579-021-00573-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X., Zhang L., Chen S., Ji W., Li C., Ren L. Recent progress on the mutations of SARS-CoV-2 spike protein and suggestions for prevention and controlling of the pandemic. Infect. Genet. Evol. 2021;93 doi: 10.1016/j.meegid.2021.104971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581:215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 25.Li F., Li W., Farzan M., Harrison S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science. 2005;309:1864–1868. doi: 10.1126/science.1116480. [DOI] [PubMed] [Google Scholar]
- 26.Yuan M., Wu N.C., Zhu X., Lee C.C.D., So R.T.Y., Lv H., Mok C.K.P., Wilson I.A. A highly conserved cryptic epitope in the receptor binding domains of SARS-CoV-2 and SARS-CoV. Science. 2020;368:630–633. doi: 10.1126/science.abb7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamers-Casterman C., Atarhouch T., Muyldermans S., Robinson G., Hammers C., Songa E.B., Bendahman N., Hammers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 28.Hassanzadeh-Ghassabeh G., Devoogdt N., De Pauw P., Vincke C., Muyldermans S. Nanobodies and their potential applications. Nanomedicine. 2013;8:1013–1026. doi: 10.2217/nnm.13.86. [DOI] [PubMed] [Google Scholar]
- 29.Tan Y.H., Liu M., Nolting B., Go J.G., Gervay-Hague J., Liu G. A nanoengineering approach for investigation and regulation of protein immobilization. ACS Nano. 2008;2:2374–2384. doi: 10.1021/nn800508f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muyldermans S. Nanobodies: natural single-domain antibodies. Annu. Rev. Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 31.Muyldermans S., Cambillau C., Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem. Sci. 2001;26:230–235. doi: 10.1016/S0968-0004(01)01790-X. [DOI] [PubMed] [Google Scholar]
- 32.Schröter C., Krah S., Zielonka S., Empting M., Kolmar H., Grzeschik J., Hock B., Könning D., Valldorf B., Sellmann C. Camelid and shark single domain antibodies: structural features and therapeutic potential. Curr. Opin. Struct. Biol. 2016;45:10–16. doi: 10.1016/j.sbi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 33.Flajnik M.F., Deschacht N., Muyldermans S. A case of convergence: why did a simple alternative to canonical antibodies arise in sharks and camels? PLoS Biol. 2011;9 doi: 10.1371/journal.pbio.1001120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dudgeon K., Rouet R., Kokmeijer I., Schofield P., Stolp J., Langley D., Stock D., Christ D. General strategy for the generation of human antibody variable domains with increased aggregation resistance. Proc. Natl. Acad. Sci. 2012;109:10879–10884. doi: 10.1073/pnas.1202866109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ewert S., Huber T., Honegger A., Plückthun A. Biophysical properties of human antibody variable domains. J. Mol. Biol. 2003;325:531–553. doi: 10.1016/S0022-2836(02)01237-8. [DOI] [PubMed] [Google Scholar]
- 36.Jespers L., Schon O., Famm K., Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat. Biotechnol. 2004;22:1161–1165. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 37.Wörn A., Plückthun A. Different equilibrium stability behavior of ScFv fragments: identification, classification, and improvement by protein engineering. Biochemistry. 1999;38:8739–8750. doi: 10.1021/bi9902079. [DOI] [PubMed] [Google Scholar]
- 38.P.S. Chowdhury, G. Vasmatzis, Engineering scFvs for Improved Stability, in: Recomb. Antibodies Cancer Ther., Humana Press, New Jersey, n.d.: pp. 237–254. doi: 10.1385/1-59259-334-8:237. [DOI] [PubMed]
- 39.Wu Y., Li C., Xia S., Tian X., Kong Y., Wang Z., Gu C., Zhang R., Tu C., Xie Y., Yang Z., Lu L., Jiang S., Ying T. Identification of human single-domain antibodies against SARS-CoV-2. Cell Host Microbe. 2020;27:891–898. doi: 10.1016/j.chom.2020.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanha J., Nguyen T.-D., Ng A., Ryan S., Ni F., MacKenzie R. Improving solubility and refolding efficiency of human VHs by a novel mutational approach. Protein Eng. Des. Sel. 2006;19:503–509. doi: 10.1093/protein/gzl037. [DOI] [PubMed] [Google Scholar]
- 41.da Silva F.A., Santa-Marta M., Freitas-Vieira A., Mascarenhas P., Barahona I., Moniz-Pereira J., Gabuzda D., Goncalves J. Camelized rabbit-derived VH single-domain intrabodies against vif strongly neutralize HIV-1 infectivity. J. Mol. Biol. 2004;340:525–542. doi: 10.1016/j.jmb.2004.04.062. [DOI] [PubMed] [Google Scholar]
- 42.Desmyter A., Transue T.R., Ghahroudi M.A., Thi M.H.D., Poortmans F., Hamers R., Muyldermans S., Wyns L. Crystal structure of a camel single-domain V(H) antibody fragment in complex with lysozyme. Nat. Struct. Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 43.Lauwereys M. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rossey I., Gilman M.S.A., Kabeche S.C., Sedeyn K., Wrapp D., Kanekiyo M., Chen M., Mas V., Spitaels J., Melero J.A., Graham B.S., Schepens B., McLellan J.S., Saelens X. Potent single-domain antibodies that arrest respiratory syncytial virus fusion protein in its prefusion state. Nat. Commun. 2017;8:16165. doi: 10.1038/ncomms16165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mahony J., Spinelli S., Blangy S., Bebeacua C., van Sinderen D., Desmyter A., Veesler D., Farenc C., Cambillau C. Viral infection modulation and neutralization by camelid nanobodies. Proc. Natl. Acad. Sci. 2013;110:E1371–E1379. doi: 10.1073/pnas.1301336110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muyldermans S., Atarhouch T., Saldanha J., Barbosa J.A.R.G., Hamers R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. "Protein Eng. Des. Sel. 1994;7:1129–1135. doi: 10.1093/protein/7.9.1129. [DOI] [PubMed] [Google Scholar]
- 47.Govaert J., Pellis M., Deschacht N., Vincke C., Conrath K., Muyldermans S., Saerens D. Dual beneficial effect of interloop disulfide bond for single domain antibody fragments. J. Biol. Chem. 2012;287:1970–1979. doi: 10.1074/jbc.M111.242818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu H., Schittny V., Nash M.A. Removal of a conserved disulfide bond does not compromise mechanical stability of a VHH antibody complex. Nano Lett. 2019;19:5524–5529. doi: 10.1021/acs.nanolett.9b02062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunz P., Zinner K., Mücke N., Bartoschik T., Muyldermans S., Hoheisel J.D. The structural basis of nanobody unfolding reversibility and thermoresistance. Sci. Rep. 2018;8:1–10. doi: 10.1038/s41598-018-26338-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Muyldermans S. Single domain camel antibodies: current status. Rev. Mol. Biotechnol. 2001;74:277–302. doi: 10.1016/S1389-0352(01)00021-6. [DOI] [PubMed] [Google Scholar]
- 51.Pardon E., Laeremans T., Triest S., Rasmussen S.G.F., Wohlkönig A., Ruf A., Muyldermans S., Hol W.G.J., Kobilka B.K., Steyaert J. A general protocol for the generation of nanobodies for structural biology. Nat. Protoc. 2014;9:674–693. doi: 10.1038/nprot.2014.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fridy P.C., Li Y., Keegan S., Thompson M.K., Nudelman I., Scheid J.F., Oeffinger M., Nussenzweig M.C., Fenyö D., Chait B.T., Rout M.P. A robust pipeline for rapid production of versatile nanobody repertoires. Nat. Methods. 2014;11:1253–1260. doi: 10.1038/nmeth.3170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomassen Y.E., Verkleij A.J., Boonstra J., Verrips C.T. Specific production rate of VHH antibody fragments by Saccharomyces cerevisiae is correlated with growth rate, independent of nutrient limitation. J. Biotechnol. 2005;118:270–277. doi: 10.1016/j.jbiotec.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 54.Thomassen Y.E., Meijer W., Sierkstra L., Verrips C.T. Large-scale production of VHH antibody fragments by Saccharomyces cerevisiae. Enzym. Microb. Technol. 2002;30:273–278. doi: 10.1016/S0141-0229(01)00497-5. [DOI] [Google Scholar]
- 55.van de Laar T., Visser C., Holster M., López C.G., Kreuning D., Sierkstra L., Lindner N., Verrips T. Increased heterologous protein production bySaccharomyces cerevisiae growing on ethanol as sole carbon source. Biotechnol. Bioeng. 2007;96:483–494. doi: 10.1002/bit.21150. [DOI] [PubMed] [Google Scholar]
- 56.Detalle L., Stohr T., Palomo C., Piedra P.A., Gilbert B.E., Mas V., Millar A., Power U.F., Stortelers C., Allosery K., Melero J.A., Depla E. Generation and characterization of ALX-0171, a potent novel therapeutic nanobody for the treatment of respiratory syncytial virus infection. Antimicrob. Agents Chemother. 2016;60:6–13. doi: 10.1128/AAC.01802-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zarschler K., Witecy S., Kapplusch F., Foerster C., Stephan H. High-yield production of functional soluble single-domain antibodies in the cytoplasm of Escherichia coli. Microb. Cell Fact. 2013;12:97. doi: 10.1186/1475-2859-12-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Joosten V., Gouka R.J., Van Den Hondel C.A.M.J.J., Verrips C.T., Lokman B.C. Expression and production of llama variable heavy-chain antibody fragments (VHHs) by Aspergillus awamori. Appl. Microbiol. Biotechnol. 2005;66:384–392. doi: 10.1007/s00253-004-1689-0. [DOI] [PubMed] [Google Scholar]
- 59.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N., Ren J., Zhou D., Harrison P.J., Weckener M., Clare D.K., Vogirala V.K., Radecke J., Moynié L., Zhao Y., Gilbert-Jaramillo J., Knight M.L., Tree J.A., Buttigieg K.R., Coombes N., Elmore M.J., Carroll M.W., Carrique L., Shah P.N.M., James W., Townsend A.R., Stuart D.I., Owens R.J., Naismith J.H. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 60.Koenig P.-A., Das H., Liu H., Kümmerer B.M., Gohr F.N., Jenster L.-M., Schiffelers L.D.J., Tesfamariam Y.M., Uchima M., Wuerth J.D., Gatterdam K., Ruetalo N., Christensen M.H., Fandrey C.I., Normann S., Tödtmann J.M.P., Pritzl S., Hanke L., Boos J., Yuan M., Zhu X., Schmid-Burgk J.L., Kato H., Schindler M., Wilson I.A., Geyer M., Ludwig K.U., Hällberg B.M., Wu N.C., Schmidt F.I. Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape. Science. 2021;371:eabe6230. doi: 10.1126/science.abe6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hanke L., Vidakovics Perez L., Sheward D.J., Das H., Schulte T., Moliner-Morro A., Corcoran M., Achour A., Karlsson Hedestam G.B., Hällberg B.M., Murrell B., McInerney G.M. An alpaca nanobody neutralizes SARS-CoV-2 by blocking receptor interaction. Nat. Commun. 2020;11:4420. doi: 10.1038/s41467-020-18174-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wrapp D., De Vlieger D., Corbett K.S., Torres G.M., Wang N., Van Breedam W., Roose K., van Schie L., Hoffmann M., Pöhlmann S., Graham B.S., Callewaert N., Schepens B., Saelens X., McLellan J.S. Structural basis for potent neutralization of betacoronaviruses by single-domain camelid antibodies. Cell. 2020;181:1004–1015. doi: 10.1016/j.cell.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schoof M., Faust B., Saunders R.A., Sangwan S., Rezelj V., Hoppe N., Boone M., Billesbølle C.B., Puchades C., Azumaya C.M., Kratochvil H.T., Zimanyi M., Deshpande I., Liang J., Dickinson S., Nguyen H.C., Chio C.M., Merz G.E., Thompson M.C., Diwanji D., Schaefer K., Anand A.A., Dobzinski N., Zha B.S., Simoneau C.R., Leon K., White K.M., Chio U.S., Gupta M., Jin M., Li F., Liu Y., Zhang K., Bulkley D., Sun M., Smith A.M., Rizo A.N., Moss F., Brilot A.F., Pourmal S., Trenker R., Pospiech T., Gupta S., Barsi-Rhyne B., Belyy V., Barile-Hill A.W., Nock S., Liu Y., Krogan N.J., Ralston C.Y., Swaney D.L., García-Sastre A., Ott M., Vignuzzi M., Walter P., Manglik A. An ultrapotent synthetic nanobody neutralizes SARS-CoV-2 by stabilizing inactive Spike. Science. 2020;1479:eabe3255. doi: 10.1126/science.abe3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xiang Y., Nambulli S., Xiao Z., Liu H., Sang Z., Duprex W.P., Schneidman-Duhovny D., Zhang C., Shi Y. Versatile and multivalent nanobodies efficiently neutralize SARS-CoV-2. Sci. (80-. ) 2020;370:eabe4747. doi: 10.1126/science.abe4747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mast F.D., Fridy P.C., Ketaren N.E., Wang J., Jacobs E.Y., Olivier J.P., Sanyal T., Molloy K.R., Schmidt F., Rutkowska M., Weisblum Y., Rich L.M., Vanderwall E.R., Dambrauskas N., Vigdorovich V., Keegan S., Jiler J.B., Stein M.E., Olinares P.D.B., Hatziioannou T., Sather D.N., Debley J.S., Fenyö D., Sali A., Bieniasz P.D., Aitchison J.D., Chait B.T., Rout M.P. Nanobody repertoires for exposing vulnerabilities of SARS-CoV-2. BioRxiv. 2021 doi: 10.1101/2021.04.08.438911. [DOI] [Google Scholar]
- 66.Strohl W.R. Fusion proteins for half-life extension of biologics as a strategy to make biobetters. BioDrugs. 2015;29:215–239. doi: 10.1007/s40259-015-0133-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saunders K.O. Conceptual approaches to modulating antibody effector functions and circulation half-life. Front. Immunol. 2019;10:1296. doi: 10.3389/fimmu.2019.01296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Schäfer A., Muecksch F., Lorenzi J.C.C., Leist S.R., Cipolla M., Bournazos S., Schmidt F., Maison R.M., Gazumyan A., Martinez D.R. Antibody potency, effector function, and combinations in protection and therapy for SARS-CoV-2 infection in vivo. J. Exp. Med. 2020;218 doi: 10.1084/jem.20201993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qureshi O.S., Rowley T.F., Junker F., Peters S.J., Crilly S., Compson J., Eddleston A., Björkelund H., Greenslade K., Parkinson M. Multivalent Fc γ-receptor engagement by a hexameric Fc-fusion protein triggers Fc γ-receptor internalisation and modulation of Fc γ-receptor functions. Sci. Rep. 2017;7:1–15. doi: 10.1038/s41598-017-17255-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nunez-Prado N., Compte M., Harwood S., Alvarez-Mendez A., Lykkemark S., Sanz L., Alvarez-Vallina L. The coming of age of engineered multivalent antibodies. Drug Discov. Today. 2015;20:588–594. doi: 10.1016/j.drudis.2015.02.013. [DOI] [PubMed] [Google Scholar]
- 71.Deyev S.M., Lebedenko E.N. Multivalency: the hallmark of antibodies used for optimization of tumor targeting by design. Bioessays. 2008;30:904–918. doi: 10.1002/bies.20805. [DOI] [PubMed] [Google Scholar]
- 72.Jing W., Procko E. ACE2-based decoy receptors for SARS coronavirus 2. Proteins Struct. Funct. Bioinforma. 2021:1–14. doi: 10.1002/prot.26140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schepens B., van Schie L., Nerinckx W., Roose K., Van Breedam W., Fijalkowska D., Devos S., Weyts W., De Cae S., Vanmarcke S. Drug development of an affinity enhanced, broadly neutralizing heavy chain-only antibody that restricts SARS-CoV-2 in rodents. BioRxiv. 2021 doi: 10.1126/scitranslmed.abi7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu J., Xu K., Jung S., Conte A., Lieberman J., Muecksch F., Lorenzi J.C.C., Park S., Schmidt F., Wang Z., Huang Y., Luo Y., Nair M., Wang P., Schulz J.E., Tessarollo L., Bylund T., Chuang G.Y., Olia A.S., Stephens T., Teng I.T., Tsybovsky Y., Zhou T., Munster V., Ho D.D., Hatziioannou T., Bieniasz P.D., Nussenzweig M.C., Kwong P.D., Casellas R. Nanobodies from camelid mice and llamas neutralize SARS-CoV-2 variants. Nature. 2021 doi: 10.1038/s41586-021-03676-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong J., Huang B., Wang B., Titong A., Kankanamalage S.G., Jia Z., Wright M., Parthasarathy P., Liu Y. Development of humanized tri-specific nanobodies with potent neutralization for SARS-CoV-2. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-74761-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong J., Huang B., Jia Z., Wang B., Gallolu Kankanamalage S., Titong A., Liu Y. Development of multi-specific humanized llama antibodies blocking SARS-CoV-2/ACE2 interaction with high affinity and avidity. Emerg. Microbes Infect. 2020;9:1034–1036. doi: 10.1080/22221751.2020.1768806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu Q., Zhang Z., Li H., Zhong K., Zhao Q., Wang Z., Wu Z., Yang D., Sun S., Yang N. Development of multivalent nanobodies blocking SARS-CoV-2 infection by targeting RBD of spike protein. J. Nanobiotechnol. 2021;19:1–12. doi: 10.1186/s12951-021-00768-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ma H., Zeng W., Meng X., Huang X., Yang Y., Zhao D., Zhou P., Wang X., Zhao C., Sun Y. Potent neutralization of sars-cov-2 by hetero-bivalent alpaca nanobodies targeting the spike receptor-binding domain. J. Virol. 2021;95:e02438–20. doi: 10.1128/JVI.02438-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tanaka S., Nelson G., Olson C.A., Buzko O., Higashide W., Shin A., Gonzalez M., Taft J., Patel R., Buta S. An ACE2 Triple Decoy that neutralizes SARS-CoV-2 shows enhanced affinity for virus variants. Sci. Rep. 2021;11:1–12. doi: 10.1038/s41598-021-91809-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ku Z., Xie X., Hinton P.R., Liu X., Ye X., Muruato A.E., Ng D.C., Biswas S., Zou J., Liu Y. Nasal delivery of an IgM offers broad protection from SARS-CoV-2 variants. Nature. 2021:1–9. doi: 10.1038/s41586-021-03673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kim M.Y., Copland A., Nayak K., Chandele A., Ahmed M.S., Zhang Q., Diogo G.R., Paul M.J., Hofmann S., Yang M. Plant‐expressed Fc‐fusion protein tetravalent dengue vaccine with inherent adjuvant properties. Plant Biotechnol. J. 2018;16:1283–1294. doi: 10.1111/pbi.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mekhaiel D.N.A., Czajkowsky D.M., Andersen J.T., Shi J., El-Faham M., Doenhoff M., McIntosh R.S., Sandlie I., He J., Hu J. Polymeric human Fc-fusion proteins with modified effector functions. Sci. Rep. 2011;1:1–11. doi: 10.1038/srep00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rowley T.F., Peters S.J., Aylott M., Griffin R., Davies N.L., Healy L.J., Cutler R.M., Eddleston A., Pither T.L., Sopp J.M. Engineered hexavalent Fc proteins with enhanced Fc-gamma receptor avidity provide insights into immune-complex interactions. Commun. Biol. 2018;1:1–12. doi: 10.1038/s42003-018-0149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sørensen V., Rasmussen I.B., Norderhaug L., Natvig I., Michaelsen T.E., Sandlie I. Effect of the IgM and IgA secretory tailpieces on polymerization and secretion of IgM and IgG. J. Immunol. 1996;156:2858–2865. [PubMed] [Google Scholar]
- 85.Teye K., Hashimoto K., Numata S., Ohta K., Haftek M., Hashimoto T. Multimerization is required for antigen binding activity of an engineered IgM/IgG chimeric antibody recognizing a skin-related antigen. Sci. Rep. 2017;7:1–12. doi: 10.1038/s41598-017-08294-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Webster G.R., van Dolleweerd C., Guerra T., Stelter S., Hofmann S., Kim M., Teh A.Y., Diogo G.R., Copland A., Paul M.J. A polymeric immunoglobulin—antigen fusion protein strategy for enhancing vaccine immunogenicity. Plant Biotechnol. J. 2018;16:1983–1996. doi: 10.1111/pbi.12932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ku Z., Xie X., Davidson E., Ye X., Su H., Menachery V.D., Li Y., Yuan Z., Zhang X., Muruato A.E. Molecular determinants and mechanism for antibody cocktail preventing SARS-CoV-2 escape. Nat. Commun. 2021;12:1–13. doi: 10.1038/s41467-020-20789-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hansen J., Baum A., Pascal K.E., Russo V., Giordano S., Wloga E., Fulton B.O., Yan Y., Koon K., Patel K. Studies in humanized mice and convalescent humans yield a SARS-CoV-2 antibody cocktail. Sci. (80-. ) 2020;369:1010–1014. doi: 10.1126/science.abd0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Baum A., Fulton B.O., Wloga E., Copin R., Pascal K.E., Russo V., Giordano S., Lanza K., Negron N., Ni M., Wei Y., Atwal G.S., Murphy A.J., Stahl N., Yancopoulos G.D., Kyratsous C.A. Antibody cocktail to SARS-CoV-2 spike protein prevents rapid mutational escape seen with individual antibodies. Sci. (80-. ) 2020;369:1014–1018. doi: 10.1126/science.abd0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Pymm P., Adair A., Chan L.-J., Cooney J.P., Mordant F.L., Allison C.C., Lopez E., Haycroft E.R., O’Neill M.T., Tan L.L. Nanobody cocktails potently neutralize SARS-CoV-2 D614G N501Y variant and protect mice. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2101918118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ke Z., Oton J., Qu K., Cortese M., Zila V., McKeane L., Nakane T., Zivanov J., Neufeldt C.J., Cerikan B. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. 2020;588:498–502. doi: 10.1038/s41586-020-2665-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gräwe A., Stein V. Linker engineering in the context of synthetic protein switches and sensors. Trends Biotechnol. 2021;39:731–744. doi: 10.1016/j.tibtech.2020.11.007. [DOI] [PubMed] [Google Scholar]
- 93.Bracken C.J., Lim S.A., Solomon P., Rettko N.J., Nguyen D.P., Zha B.S., Schaefer K., Byrnes J.R., Zhou J., Lui I., Liu J., Pance K., Azumaya C.M., Braxton J.R., Brilot A.F., Gupta M., Li F., Lopez K.E., Melo A., Merz G.E., Moss F., Paulino J., Pospiech T.H., Pourmal S., Puchades C., Rizo A.N., Smith A.M., Sun M., Thomas P.V., Wang F., Yu Z., Asarnow D., Braxton J.R., Campbell M.G., Chio C.M., Chio U.S., Dickinson M.S., Diwanji D., Faust B., Gupta M., Hoppe N., Jin M., Li F., Li J., Liu Y., Merz G.E., Nguyen H.C., Paulino J., Pospiech T.H., Pourmal S., Sangwan S., Trenker R., Trinidad D., Tse E., Zhang K., Zhou F., Azumaya C.M., Billesboelle C., Bowen A., Campbell M.G., Diwanji D., Hoppe N., Li Y.L., Nguyen P., Nowotny C., Puchades C., Safari M., Sangwan S., Schaefer K., Smith A.M., Trenker R., Tsui T.K.M., Whitis N., Zhao J., Asarnow D., Azumaya C.M., Chio C.M., Faust B., Gupta M., Kim K., Moritz M., Owens T.W., Paulino J., Peters J.K., Pourmal S., Schaefer K., Tsui T.K.M., Biel J., Deshpande I., Herrera N., Kratochvil H.T., Liu X., Schulze-Gahmen U., Young I.D., Chen J., Diallo A., Doan L., Flores S., Gupta M., Jin M., Kratochvil H.T., Lam V.L., Li Y., Lo M., Merz G.E., Paulino J., Thwin A.C., Titus E.W., Yu Z., Zhou F., Zhang Y., Bulkley D., Joves A., Joves A., McKay L., Tabios M., Tse E., Agard D.A., Cheng Y., Fraser J.S., Frost A., Jura N., Kortemme T., Krogan N.J., Manglik A., Rosenberg O.S., Southworth D.R., Stroud R.M., Verba K.A., Zhou X.X., Leung K.K., Wells J.A. Bi-paratopic and multivalent VH domains block ACE2 binding and neutralize SARS-CoV-2. Nat. Chem. Biol. 2021;17:113–121. doi: 10.1038/s41589-020-00679-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nambulli S., Xiang Y., Tilston-Lunel N.L., Rennick L.J., Sang Z., Klimstra W.B., Reed D.S., Crossland N.A., Shi Y., Duprex W.P. Inhalable nanobody (PiN-21) prevents and treats SARS-CoV-2 infections in Syrian hamsters at ultra-low doses. Sci. Adv. 2021;7:eabh0319. doi: 10.1126/sciadv.abh0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hanke L., Das H., Sheward D.J., Vidakovics L.P., Urgard E., Moliner-Morro A., Karl V., Pankow A., Kim C., Smith N.L. A bispecific monomeric nanobody induces SARS-COV-2 spike trimer dimers. BioRxiv. 2021 doi: 10.1101/2021.03.20.436243v2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.D. Li, T. Li, H. Cai, H. Yao, B. Zhou, N. Zhang, Y. Gong, Y. Zhao, Q. Shen, W. Qin, A potent synthetic nanobody targets RBD and protects mice from SARS-CoV-2 infection, (2020).
- 97.Yao H., Cai H., Li T., Zhou B., Qin W., Lavillette D., Li D. A high-affinity RBD-targeting nanobody improves fusion partner’s potency against SARS-CoV-2. PLoS Pathog. 2021;17 doi: 10.1371/journal.ppat.1009328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chen X., Zaro J.L., Shen W.C. Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev. 2013;65:1357–1369. doi: 10.1016/j.addr.2012.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Klement M., Liu C., Loo B.L.W., Choo A.B.H., Ow D.S.W., Lee D.Y. Effect of linker flexibility and length on the functionality of a cytotoxic engineered antibody fragment. J. Biotechnol. 2015;199:90–97. doi: 10.1016/j.jbiotec.2015.02.008. [DOI] [PubMed] [Google Scholar]
- 100.Van Rosmalen M., Krom M., Merkx M. Tuning the flexibility of glycine-serine linkers to allow rational design of multidomain proteins. Biochemistry. 2017;56:6565–6574. doi: 10.1021/acs.biochem.7b00902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kostelny S.A., Cole M.S., Tso J.Y. Formation of a bispecific antibody by the use of leucine zippers. J. Immunol. 1992;148:1547–1553. [PubMed] [Google Scholar]
- 102.Guo L., Bi W., Wang X., Xu W., Yan R., Zhang Y., Zhao K., Li Y., Zhang M., Cai X., Jiang S., Xie Y., Zhou Q., Lu L., Dang B. Engineered trimeric ACE2 binds viral spike protein and locks it in “Three-up” conformation to potently inhibit SARS-CoV-2 infection. Cell Res. 2021;31:98–100. doi: 10.1038/s41422-020-00438-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Fletcher J.M., Boyle A.L., Bruning M., Bartlett G.J., Vincent T.L., Zaccai N.R., Armstrong C.T., Bromley E.H.C., Booth P.J., Brady R.L. A basis set of de novo coiled-coil peptide oligomers for rational protein design and synthetic biology. ACS Synth. Biol. 2012;1:240–250. doi: 10.1021/sb300028q. [DOI] [PubMed] [Google Scholar]
- 104.Forrer P., Chang C., Ott D., Wlodawer A., Plückthun A. Kinetic stability and crystal structure of the viral capsid protein SHP. J. Mol. Biol. 2004;344:179–193. doi: 10.1016/j.jmb.2004.09.030. [DOI] [PubMed] [Google Scholar]
- 105.Fan C., Huang C., Chiu W., Lai C., Liou G., Li H., Chou M. Production of multivalent protein binders using a self‐trimerizing collagen‐like peptide scaffold. FASEB J. 2008;22:3795–3804. doi: 10.1096/fj.08-111484. [DOI] [PubMed] [Google Scholar]
- 106.Sánchez-Arévalo Lobo V.J., Cuesta Á.M., Sanz L., Compte M., García P., Prieto J., Blanco F.J., Álvarez-Vallina L. Enhanced antiangiogenic therapy with antibody-collagen XVIII NC1 domain fusion proteins engineered to exploit matrix remodeling events. Int. J. Cancer. 2006;119:455–462. doi: 10.1002/ijc.21851. [DOI] [PubMed] [Google Scholar]
- 107.Cuesta Á.M., Sánchez-Martín D., Blanco-Toribio A., Villate M., Enciso-Álvarez K., Alvarez-Cienfuegos A., Sainz-Pastor N., Sanz L., Blanco F.J., Álvarez-Vallina L. MAbs. Taylor & Francis; 2012. Improved stability of multivalent antibodies containing the human collagen XV trimerization domain; pp. 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Miller A., Leach A., Thomas J., McAndrew C., Bentley E., Mattiuzzo G., John L., Mirazimi A., Harris G., Gamage N., Carr S., Ali H., Van Montfort R., Rabbitts T. A super-potent tetramerized ACE2 protein displays enhanced neutralization of SARS-CoV-2 virus infection. Sci. Rep. 2021;11:1–13. doi: 10.1038/s41598-021-89957-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.P. Pack, K. MÜller, R. Zahn, A. Plückthun, Tetravalent Miniantibodies with High Avidity Assembling in Escherichia coli, 1995. [DOI] [PubMed]
- 110.Kipriyanov S.M., Breitling F., Little M., Dübel S. Single-chain antibody streptavidin fusions: Tetrameric bifunctional scFv-complexes with biotin binding activity and enhanced affinity to antigen. Hum. Antibodies. 1995;6:93–101. doi: 10.3233/HAB-1995-6303. [DOI] [PubMed] [Google Scholar]
- 111.Blanco-Toribio A., Sainz-Pastor N., Álvarez-Cienfuegos A., Merino N., Cuesta Á.M., Sánchez-Martín D., Bonet J., Santos-Valle P., Sanz L., Oliva B. MAbs. Taylor & Francis; 2013. Generation and Characterization of Monospecific and Bispecific Hexavalent Trimerbodies; pp. 70–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Zhang J., Tanha J., Hirama T., Khieu N.H., To R., Tong-Sevinc H., Stone E., Brisson J.-R., MacKenzie C.R. Pentamerization of single-domain antibodies from phage libraries: a novel strategy for the rapid generation of high-avidity antibody reagents. J. Mol. Biol. 2004;335:49–56. doi: 10.1016/j.jmb.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 113.Stone E., Hirama T., Tanha J., Tong-Sevinc H., Li S., MacKenzie C.R., Zhang J. The assembly of single domain antibodies into bispecific decavalent molecules. J. Immunol. Methods. 2007;318:88–94. doi: 10.1016/j.jim.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 114.Li Y., Leneghan D.B., Miura K., Nikolaeva D., Brian I.J., Dicks M.D.J., Fyfe A.J., Zakutansky S.E., de Cassan S., Long C.A. Enhancing immunogenicity and transmission-blocking activity of malaria vaccines by fusing Pfs25 to IMX313 multimerization technology. Sci. Rep. 2016;6:1–13. doi: 10.1038/srep18848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.D. Kim, Y. Yan, C.A. Valencia, R. Liu, Heptameric targeting ligands against EGFR and HER2 with high stability and avidity, (2012). [DOI] [PMC free article] [PubMed]
- 116.Andersson A.-M.C., Buldun C.M., Pattinson D.J., Draper S.J., Howarth M. SnoopLigase peptide-peptide conjugation enables modular vaccine assembly. Sci. Rep. 2019;9:1–13. doi: 10.1038/s41598-019-40985-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zakeri B., Fierer J.O., Celik E., Chittock E.C., Schwarz-Linek U., Moy V.T., Howarth M. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl. Acad. Sci. 2012;109:E690–E697. doi: 10.1073/pnas.1115485109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Deyev S.M., Waibel R., Lebedenko E.N., Schubiger A.P., Plückthun A. Design of multivalent complexes using the barnase·barstar module. Nat. Biotechnol. 2003;21:1486–1492. doi: 10.1038/nbt916. [DOI] [PubMed] [Google Scholar]
- 119.Goldenberg D.M., Rossi E.A., Sharkey R.M., McBride W.J., Chang C.H. Multifunctional antibodies by the dock-and-lock method for improved cancer imaging and therapy by pretargeting. J. Nucl. Med. 2008;49:158–163. doi: 10.2967/jnumed.107.046185. [DOI] [PubMed] [Google Scholar]
- 120.Löfblom J., Frejd F.Y., Ståhl S. Non-immunoglobulin based protein scaffolds. Curr. Opin. Biotechnol. 2011;22:843–848. doi: 10.1016/j.copbio.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 121.Bhaskar S., Lim S. Engineering protein nanocages as carriers for biomedical applications. NPG Asia Mater. 2017;9 doi: 10.1038/am.2016.128. e371–e371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Berckman E.A., Hartzell E.J., Mitkas A.A., Sun Q., Chen W. Biological assembly of modular protein building blocks as sensing, delivery, and therapeutic agents. Annu. Rev. Chem. Biomol. Eng. 2020;11:35–62. doi: 10.1146/annurev-chembioeng-101519-121526. [DOI] [PubMed] [Google Scholar]
- 123.Hempstead P.D., Yewdall S.J., Fernie A.R., Lawson D.M., Artymiuk P.J., Rice D.W., Ford G.C., Harrison P.M. Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J. Mol. Biol. 1997;268:424–448. doi: 10.1006/jmbi.1997.0970. [DOI] [PubMed] [Google Scholar]
- 124.Milne J.L.S., Shi D., Rosenthal P.B., Sunshine J.S., Domingo G.J., Wu X., Brooks B.R., Perham R.N., Henderson R., Subramaniam S. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: a multifunctional catalytic machine. EMBO J. 2002;21:5587–5598. doi: 10.1093/emboj/cdf574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang X., Meining W., Fischer M., Bacher A., Ladenstein R. X-ray structure analysis and crystallographic refinement of lumazine synthase from the hyperthermophile Aquifex aeolicus at 1.6 Å resolution: determinants of thermostability revealed from structural comparisons. J. Mol. Biol. 2001;306:1099–1114. doi: 10.1006/jmbi.2000.4435. [DOI] [PubMed] [Google Scholar]
- 126.Ji P., Zhu J., Li X., Fan W., Liu Q., Wang K., Zhao J., Sun Y., Liu B., Zhou E.M., Zhao Q. Fenobody and RANbody-based sandwich enzyme-linked immunosorbent assay to detect Newcastle disease virus. J. Nanobiotechnol. 2020;18:1–19. doi: 10.1186/s12951-020-00598-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fan K., Jiang B., Guan Z., He J., Yang D., Xie N., Nie G., Xie C., Yan X. Fenobody: a ferritin-displayed nanobody with high apparent affinity and half-life extension. Anal. Chem. 2018;90:5671–5677. doi: 10.1021/acs.analchem.7b05217. [DOI] [PubMed] [Google Scholar]
- 128.Swartz A.R., Chen W. Rapid quantification of monoclonal antibody titer in cell culture harvests by antibody-induced Z-ELP-E2 nanoparticle cross-linking. Anal. Chem. 2018;90:14447–14452. doi: 10.1021/acs.analchem.8b04083. [DOI] [PubMed] [Google Scholar]
- 129.Zhang B., Chao C.W., Tsybovsky Y., Abiona O.M., Hutchinson G.B., Moliva J.I., Olia A.S., Pegu A., Phung E., Stewart-Jones G.B.E. A platform incorporating trimeric antigens into self-assembling nanoparticles reveals SARS-CoV-2-spike nanoparticles to elicit substantially higher neutralizing responses than spike alone. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-74949-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rujas E., Kucharska I., Tan Y.Z., Benlekbir S., Cui H., Zhao T., Wasney G.A., Budylowski P., Guvenc F., Newton J.C., Sicard T., Semesi A., Muthuraman K., Nouanesengsy A., Aschner C.B., Prieto K., Bueler S.A., Youssef S., Liao-Chan S., Glanville J., Christie-Holmes N., Mubareka S., Gray-Owen S.D., Rubinstein J.L., Treanor B., Julien J.P. Multivalency transforms SARS-CoV-2 antibodies into ultrapotent neutralizers. Nat. Commun. 2021;12:1–12. doi: 10.1038/s41467-021-23825-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Nguyen B., Tolia N.H. Protein-based antigen presentation platforms for nanoparticle vaccines. npj Vaccin. 2021;6:1–11. doi: 10.1038/s41541-021-00330-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Huo J., Le Bas A., Ruza R.R., Duyvesteyn H.M.E., Mikolajek H., Malinauskas T., Tan T.K., Rijal P., Dumoux M., Ward P.N. Neutralizing nanobodies bind SARS-CoV-2 spike RBD and block interaction with ACE2. Nat. Struct. Mol. Biol. 2020;27:846–854. doi: 10.1038/s41594-020-0469-6. [DOI] [PubMed] [Google Scholar]
- 133.Custódio T.F., Das H., Sheward D.J., Hanke L., Pazicky S., Pieprzyk J., Sorgenfrei M., Schroer M.A., Gruzinov A.Y., Jeffries C.M. Selection, biophysical and structural analysis of synthetic nanobodies that effectively neutralize SARS-CoV-2. Nat. Commun. 2020;11:1–11. doi: 10.1038/s41467-020-19204-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chi X., Liu X., Wang C., Zhang X., Li X., Hou J., Ren L., Jin Q., Wang J., Yang W. Humanized single domain antibodies neutralize SARS-CoV-2 by targeting the spike receptor binding domain. Nat. Commun. 2020;11:2–8. doi: 10.1038/s41467-020-18387-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Roa W.H., Azarmi S., Al-Hallak M.H.D.K., Finlay W.H., Magliocco A.M., Löbenberg R. Inhalable nanoparticles, a non-invasive approach to treat lung cancer in a mouse model. J. Control. Release. 2011;150:49–55. doi: 10.1016/j.jconrel.2010.10.035. [DOI] [PubMed] [Google Scholar]
- 136.Sadhukha T., Wiedmann T.S., Panyam J. Inhalable magnetic nanoparticles for targeted hyperthermia in lung cancer therapy. Biomaterials. 2013;34:5163–5171. doi: 10.1016/j.biomaterials.2013.03.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Miranda M.S., Rodrigues M.T., Domingues R.M.A., Costa R.R., Paz E., Rodríguez-Abreu C., Freitas P., Almeida B.G., Carvalho M.A., Gonçalves C., Ferreira C.M., Torrado E., Reis R.L., Pedrosa J., Gomes M.E. Development of inhalable superparamagnetic iron oxide nanoparticles (SPIONs) in microparticulate system for antituberculosis drug delivery. Adv. Healthc. Mater. 2018;7:1–12. doi: 10.1002/adhm.201800124. [DOI] [PubMed] [Google Scholar]
- 138.Xu R., Zhang G., Mai J., Deng X., Segura-Ibarra V., Wu S., Shen J., Liu H., Hu Z., Chen L., Huang Y., Koay E., Huang Y., Liu J., Ensor J.E., Blanco E., Liu X., Ferrari M., Shen H. An injectable nanoparticle generator enhances delivery of cancer therapeutics. Nat. Biotechnol. 2016;34:414–418. doi: 10.1038/nbt.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pantarotto D., Partidos C.D., Hoebeke J., Brown F., Kramer E., Briand J.-P., Muller S., Prato M., Bianco A. Immunization with peptide-functionalized carbon nanotubes enhances virus-specific neutralizing antibody responses. Chem. Biol. 2003;10:961–966. doi: 10.1016/j.chembiol.2003.09.011. [DOI] [PubMed] [Google Scholar]
- 140.Gill S., Löbenberg R., Ku T., Azarmi S., Roa W., Prenner E.J. Nanoparticles: characteristics, mechanisms of action, and toxicity in pulmonary drug delivery—a review. J. Biomed. Nanotechnol. 2007;3:107–119. doi: 10.1166/jbn.2007.015. [DOI] [Google Scholar]
- 141.Rodionov V.O., Presolski S.I., Gardinier S., Lim Y.-H., Finn M.G. Benzimidazole and Related Ligands for Cu-Catalyzed Azide−Alkyne Cycloaddition. J. Am. Chem. Soc. 2007;129:12696–12704. doi: 10.1021/ja072678l. [DOI] [PubMed] [Google Scholar]
- 142.Boutureira O., Bernardes G.J.L. Advances in chemical protein modification. Chem. Rev. 2015;115:2174–2195. doi: 10.1021/cr500399p. [DOI] [PubMed] [Google Scholar]
- 143.Agouridas V., El Mahdi O., Diemer V., Cargoët M., Monbaliu J.C.M., Melnyk O. Native Chemical Ligation and Extended Methods: Mechanisms, Catalysis, Scope, and Limitations. Chem. Rev. 2019;119 doi: 10.1021/acs.chemrev.8b00712. [DOI] [PubMed] [Google Scholar]
- 144.Spicer C.D., Davis B.G. Selective chemical protein modification. Nat. Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 145.Zhang C., Dai P., Vinogradov A.A., Gates Z.P., Pentelute B.L. Site-selective cysteine-cyclooctyne conjugation. Angew. Chem. Int. Ed. 2018;57:6459–6463. doi: 10.1002/anie.201800860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Chin J.W. Expanding and reprogramming the genetic code. Nature. 2017;550:53–60. doi: 10.1038/nature24031. [DOI] [PubMed] [Google Scholar]
- 147.Liu C.C., Schultz P.G. Adding new chemistries to the genetic code. Annu. Rev. Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- 148.Young T.S., Schultz P.G. Beyond the canonical 20 amino acids: expanding the genetic lexicon. J. Biol. Chem. 2010;285:11039–11044. doi: 10.1074/jbc.R109.091306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Lang K., Davis L., Torres-Kolbus J., Chou C., Deiters A., Chin J.W. Genetically encoded norbornene directs site-specific cellular protein labelling via a rapid bioorthogonal reaction. Nat. Chem. 2012;4:298–304. doi: 10.1038/nchem.1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Walper S.A., Turner K.B., Medintz I.L. Enzymatic bioconjugation of nanoparticles: developing specificity and control. Curr. Opin. Biotechnol. 2015;34:232–241. doi: 10.1016/j.copbio.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 151.Shadish J.A., DeForest C.A. Site-selective protein modification: from functionalized proteins to functional biomaterials. Matter. 2020;2:50–77. doi: 10.1016/j.matt.2019.11.011. [DOI] [Google Scholar]
- 152.Zhang Y., Park K.-Y., Suazo K.F., Distefano M.D. Recent progress in enzymatic protein labelling techniques and their applications. Chem. Soc. Rev. 2018;47:9106–9136. doi: 10.1039/C8CS00537K. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Wals K., Ovaa H. Unnatural amino acid incorporation in E. coli: current and future applications in the design of therapeutic proteins. Front. Chem. 2014;2:1–12. doi: 10.3389/fchem.2014.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Samavarchi-Tehrani P., Samson R., Gingras A.-C. Proximity dependent biotinylation: key enzymes and adaptation to proteomics approaches. Mol. Cell. Proteom. 2020;19:757–773. doi: 10.1074/mcp.R120.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Cronan J.E. Targeted and proximity-dependent promiscuous protein biotinylation by a mutant Escherichia coli biotin protein ligase. J. Nutr. Biochem. 2005;16:416–418. doi: 10.1016/j.jnutbio.2005.03.017. [DOI] [PubMed] [Google Scholar]
- 156.Cho K.F., Branon T.C., Udeshi N.D., Myers S.A., Carr S.A., Ting A.Y. Proximity labeling in mammalian cells with TurboID and split-TurboID. Nat. Protoc. 2020;15:3971–3999. doi: 10.1038/s41596-020-0399-0. [DOI] [PubMed] [Google Scholar]
- 157.Kim D.I., Jensen S.C., Noble K.A., KC B., Roux K.H., Motamedchaboki K., Roux K.J. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 2016;27:1188–1196. doi: 10.1091/mbc.E15-12-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pishesha N., Ingram J.R., Ploegh H.L. Sortase a: a model for transpeptidation and its biological applications. Annu. Rev. Cell Dev. Biol. 2018;34:163–188. doi: 10.1146/annurev-cellbio-100617-062527. [DOI] [PubMed] [Google Scholar]
- 159.Heck T., Pham P.H., Yerlikaya A., Thöny-Meyer L., Richter M. Sortase A catalyzed reaction pathways: a comparative study with six SrtA variants. Catal. Sci. Technol. 2014;4:2946–2956. doi: 10.1039/c4cy00347k. [DOI] [Google Scholar]
- 160.Tsukiji S., Nagamune T. Sortase-mediated ligation: a gift from gram-positive bacteria to protein engineering. ChemBioChem. 2009;10:787–798. doi: 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 161.Popp M.W., Antos J.M., Grotenbreg G.M., Spooner E., Ploegh H.L. Sortagging: a versatile method for protein labeling. Nat. Chem. Biol. 2007;3:707–708. doi: 10.1038/nchembio.2007.31. [DOI] [PubMed] [Google Scholar]
- 162.Fairhead M., Howarth M. In: Anal. Biochem. Gautier A., Hinner M.J., editors. Springer New York; New York, NY: 2015. Site-specific biotinylation of purified proteins using BirA; pp. 171–184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bayer E.A., Wilchek M. Methods in Enzymology. Vol. 184. 1990. Protein biotinylation; pp. 138–160. [DOI] [PubMed] [Google Scholar]
- 164.Green N.M. Avidin and streptavidin. Methods Enzym. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 165.Schultz J., Lin Y., Sanderson J., Zuo Y., Stone D., Mallett R., Wilbert S., Axworthy D. A tetravalent single-chain antibody-streptavidin fusion protein for pretargeted lymphoma therapy. Cancer Res. 2000;60:6663–6669. [PubMed] [Google Scholar]
- 166.Kipriyanov S.M., Little M., Kropshofer H., Breitling F., Gotter S., Dübel S. Affinity enhancement of a recombinant antibody: formation of complexes with multiple valency by a single-chain Fv fragment–core streptavidin fusion. "Protein Eng. Des. Sel. 1996;9:203–211. doi: 10.1093/protein/9.2.203. [DOI] [PubMed] [Google Scholar]
- 167.Das S., Pawale V.S., Dadireddy V., Singh A.K., Ramakumar S., Roy R.P. Structure and specificity of a new class of Ca 2+ -independent housekeeping sortase from Streptomyces avermitilis provide insights into its non-canonical substrate preference. J. Biol. Chem. 2017;292:7244–7257. doi: 10.1074/jbc.M117.782037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Popp M.W., Dougan S.K., Chuang T.-Y., Spooner E., Ploegh H.L. Sortase-catalyzed transformations that improve the properties of cytokines. Proc. Natl. Acad. Sci. 2011;108:3169–3174. doi: 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Antos J.M., Miller G.M., Grotenbreg G.M., Ploegh H.L. Lipid modification of proteins through sortase-catalyzed transpeptidation. J. Am. Chem. Soc. 2008;130:16338–16343. doi: 10.1021/ja806779e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Tanaka T., Yamamoto T., Tsukiji S., Nagamune T. Site-specific protein modification on living cells catalyzed by sortase. ChemBioChem. 2008;9:802–807. doi: 10.1002/cbic.200700614. [DOI] [PubMed] [Google Scholar]
- 171.Yamamoto T., Nagamune T. Expansion of the sortase-mediated labeling method for site-specific N-terminal labeling of cell surface proteins on living cells. Chem. Commun. 2009:1022. doi: 10.1039/b818792d. [DOI] [PubMed] [Google Scholar]
- 172.Witte M.D., Theile C.S., Wu T., Guimaraes C.P., Blom A.E.M.M., Ploegh H.L. Production of unnaturally linked chimeric proteins using a combination of sortase-catalyzed transpeptidation and click chemistry. Nat. Protoc. 2013;8:1808–1819. doi: 10.1038/nprot.2013.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Guimaraes C.P., Witte M.D., Theile C.S., Bozkurt G., Kundrat L., Blom A.E.M., Ploegh H.L. Site-specific C-terminal and internal loop labeling of proteins using sortase-mediated reactions. Nat. Protoc. 2013;8:1787–1799. doi: 10.1038/nprot.2013.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Antos J.M., Chew G.-L.L., Guimaraes C.P., Yoder N.C., Grotenbreg G.M., Popp M.W.-L.L., Ploegh H.L. Site-specific N- and C-terminal labeling of a single polypeptide using sortases of different specificity. J. Am. Chem. Soc. 2009;131:10800–10801. doi: 10.1021/ja902681k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Theile C.S., Witte M.D., Blom A.E.M., Kundrat L., Ploegh H.L., Guimaraes C.P. Site-specific N-terminal labeling of proteins using sortase-mediated reactions. Nat. Protoc. 2013;8:1800–1807. doi: 10.1038/nprot.2013.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ploegh H.L., Popp M.W., Witte M.D., Cragnolini J.J., Dougan S.K., Yoder N.C. Site-specific modification of single-chain antibody fragments for bioconjugation and vascular immunotargeting. Proc. Natl. Acad. Sci. 2012;109:11993–11998. doi: 10.1073/pnas.1205427109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Moliner‐morro A., Sheward D.J., Karl V., Vidakovics L.P., Murrell B., McInerney G.M., Hanke L. Picomolar sars‐cov‐2 neutralization using multi‐arm peg nanobody constructs. Biomolecules. 2020;10:1–11. doi: 10.3390/biom10121661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Iwasaki A., Yang Y. The potential danger of suboptimal antibody responses in COVID-19. Nat. Rev. Immunol. 2020;20:339–341. doi: 10.1038/s41577-020-0321-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. doi: 10.1038/s41586-020-2538-8. [DOI] [PubMed] [Google Scholar]
- 180.Lee W.S., Wheatley A.K., Kent S.J., DeKosky B.J. Antibody-dependent enhancement and SARS-CoV-2 vaccines and therapies. Nat. Microbiol. 2020;5:1185–1191. doi: 10.1038/s41564-020-00789-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yamin R., Jones A.T., Hoffmann H.-H., Schäfer A., Kao K.S., Francis R.L., Sheahan T.P., Baric R.S., Rice C.M., Ravetch J.V., Bournazos S. Fc-engineered antibody therapeutics with improved anti-SARS-CoV-2 efficacy. Nature. 2021 doi: 10.1038/s41586-021-04017-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pinto D., Park Y.-J., Beltramello M., Walls A.C., Tortorici M.A., Bianchi S., Jaconi S., Culap K., Zatta F., De Marco A., Peter A., Guarino B., Spreafico R., Cameroni E., Case J.B., Chen R.E., Havenar-Daughton C., Snell G., Telenti A., Virgin H.W., Lanzavecchia A., Diamond M.S., Fink K., Veesler D., Corti D. Cross-neutralization of SARS-CoV-2 by a human monoclonal SARS-CoV antibody. Nature. 2020;583:290–295. doi: 10.1038/s41586-020-2349-y. [DOI] [PubMed] [Google Scholar]
- 183.Vulovic I., Yao Q., Park Y.-J., Courbet A., Norris A., Busch F., Sahasrabuddhe A., Merten H., Sahtoe D.D., Ueda G., Fallas J.A., Weaver S.J., Hsia Y., Langan R.A., Plückthun A., Wysocki V.H., Veesler D., Jensen G.J., Baker D. Generation of ordered protein assemblies using rigid three-body fusion. Proc. Natl. Acad. Sci. 2021;118 doi: 10.1073/pnas.2015037118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hsia Y., Mout R., Sheffler W., Edman N.I., Vulovic I., Park Y.-J., Redler R.L., Bick M.J., Bera A.K., Courbet A., Kang A., Brunette T.J., Nattermann U., Tsai E., Saleem A., Chow C.M., Ekiert D., Bhabha G., Veesler D., Baker D. Design of multi-scale protein complexes by hierarchical building block fusion. Nat. Commun. 2021;12:2294. doi: 10.1038/s41467-021-22276-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Bale J.B., Gonen S., Liu Y., Sheffler W., Ellis D., Thomas C., Cascio D., Yeates T.O., Gonen T., King N.P., Baker D. Accurate design of megadalton-scale two-component icosahedral protein complexes. Science. 2016;353:389–394. doi: 10.1126/science.aaf8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Ueda G., Antanasijevic A., Fallas J.A., Sheffler W., Copps J., Ellis D., Hutchinson G.B., Moyer A., Yasmeen A., Tsybovsky Y., Park Y.-J., Bick M.J., Sankaran B., Gillespie R.A., Brouwer P.J.M., Zwart P.H., Veesler D., Kanekiyo M., Graham B.S., Sanders R.W., Moore J.P., Klasse P.J., Ward A.B., King N.P., Baker D. Tailored design of protein nanoparticle scaffolds for multivalent presentation of viral glycoprotein antigens. Elife. 2020;9:1–30. doi: 10.7554/eLife.57659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Divine R., Dang H.V., Ueda G., Fallas J.A., Vulovic I., Sheffler W., Saini S., Zhao Y.T., Raj I.X., Morawski P.A., Jennewein M.F., Homad L.J., Wan Y.-H., Tooley M.R., Seeger F., Etemadi A., Fahning M.L., Lazarovits J., Roederer A., Walls A.C., Stewart L., Mazloomi M., King N.P., Campbell D.J., McGuire A.T., Stamatatos L., Ruohola-Baker H., Mathieu J., Veesler D., Baker D. Designed proteins assemble antibodies into modular nanocages. Science. 2021;372:eabd9994. doi: 10.1126/science.abd9994. [DOI] [PMC free article] [PubMed] [Google Scholar]