Highlights
-
•
Bacterial dysbiosis induces various changes in host cell machinery.
-
•
Bacterial infections trigger chronic inflammation leading to cell proliferation.
-
•
Immune response modulation and genomic instability assist tumour development.
-
•
Bacterial minicells, microswimmers, and OMVs can act as feasible drug carriers.
-
•
Bacteria acts as a promising approach for targeting the hypoxic core of the tumour.
Keywords: Bacterial infections, Chronic inflammation, Immune response modulation, Genomic instability, Metastasis, Bacteriotherapy
Abstract
Understanding various responses of cells towards change in their external environment, presence of other species and is important in identifying and correlating the mechanisms leading to malignant transformations and cancer development. Although uncovering and comprehending the association between bacteria and cancer is highly challenging, it promises excellent perspectives and approaches for successful cancer therapy. This review introduces various bacterial species, their virulence factors, and their role in cell transformations leading to cancer (particularly gastric, oral, colon, and breast cancer). Bacterial dysbiosis permutates host cells, causes inflammation, and results in tumorigenesis. This review explored bacterial-mediated host cell transformation causing chronic inflammation, immune receptor hyperactivation/absconding immune recognition, and genomic instability. Bacterial infections downregulate E-cadherin, leading to loosening of epithelial tight junction polarity and triggers metastasis. In addition to understanding the role of bacterial infections in cancer development, we have also reviewed the application of bacteria for cancer therapy. The emergence of bacteriotherapy combined with conventional therapies led to new and effective ways of overcoming challenges associated with available treatments. This review discusses the application of bacterial minicells, microswimmers, and outer cell membrane vesicles (OMV) for drug delivery applications.
.
1. Introduction
Cancer remains a prime cause of morbidity and mortality worldwide. It is a resultant of the extensive and uncontrollable proliferation of cells. These cells are heterogenous and constantly evolve evading immune responses, developing drug resistance and recurrence (Gallaher et al., 2019). An adequate understanding of the biological components provoking cancer progression can reduce the mortality rate. Extensive efforts of researchers concluded, the intrinsic risk factors (such as an error in DNA replication/spontaneous mutations) cause only 10–30% of cancer. The extrinsic risk factors (endogenous risk factors: biological ageing, genetic susceptibility, hormones, etc., and exogenous risk factors: chemical carcinogens, tumour-causing micro-organisms, unhealthy lifestyles, etc.) are the significant drivers of cancer progression (Wu et al., 2016). In 2018, about 18.1 million new cancer cases were recorded worldwide (Bray et al., 2018). In 2020 19.3 million new cancer cases and 10 million deaths have been reported, indicating the rampant cancer progression worldwide. The cancer cases are expected to increase further and reach 28.4 million by 2040 (World Health Organisation 2018; Sung et al., 2021). A better understanding of cancer-causing factors, early detection, and improvised therapy is crucial and highly required to address cancer fatality.
Various environmental factors can cause changes in the host-genetic relationship leading to cancer development. Micro-organisms like bacteria, viruses, fungi colonizing the gastrointestinal tract, mouth, skin and vagina could be vital and potential environmental factors. Infectious agents contribute to 16.1% of malignancy worldwide (Takeshima and Ushijima, 2019; Dekaboruah et al., 2020; De Martel et al., 2012). Micro-organisms can colonize inside and outside the body without harmful effects as commensals or mutuals (living with beneficial co-relation with host cells). However, an imbalance between organisms, i.e. dysbiosis, alters the normal physiological homoeostasis in host cells. This unhealthy relationship between host and microbes leads to various diseases like obesity (Turnbaugh et al., 2006), malnutrition (Blanton et al., 2016), diabetes (Al-Rawi and Al-Marzooq, 2017), chronic inflammatory diseases such as inflammatory bowel disease (IBD) (Mazmanian et al., 2008), and cancer (Polk and Peek, 2010). An average human (aged 20–30 years with a bodyweight of 70 kg and a height of 170 cm) contains about 3.8 × 1013 bacterial cells. The colon alone contains 1011 bacterial cells, and the stomach, duodenum, jejunum (upper small intestine) contains 103–104 bacterial cells (Sender et al., 2016). The interactions between the microbes and the host cells are diverse, making it hard to determine and define the exact role played by microbiota in cancer progression. The microbiota-mediated infection has been reported as a potent cancer-inducing factor that triggers cancer progression by attacking the host cell DNA, modulating the immune system, inducing inflammation, hormone secretion, lymphoproliferation, and promoting epithelial-mesenchymal transition (EMT) (Geng et al., 2020; Paciello et al., 2013; Grebowska et al., 2008; Leclercq et al., 2016; Jin et al., 2019; Rosean et al., 2019; Li et al., 2019). The microbes translocate, causes genotoxicity and affect the metabolism altering the host cell's environment. The symbiotic relations between bacteria and host cells, when disrupted, lead to translocation of bacteria into host cells, triggers inflammation leading to the development of tumours. Bacterial genotoxins like colibactin, CDTs (cytolethal distending toxin) are known to break the host cells' DNA (Li et al., 2019; Shine et al., 2018; Zhou et al., 2020). Microbiota is also known for altering the gut's bile acid homoeostasis, catalysing deconjugation and dihydroxylation (Wang et al., 2019; Li et al., 2020).
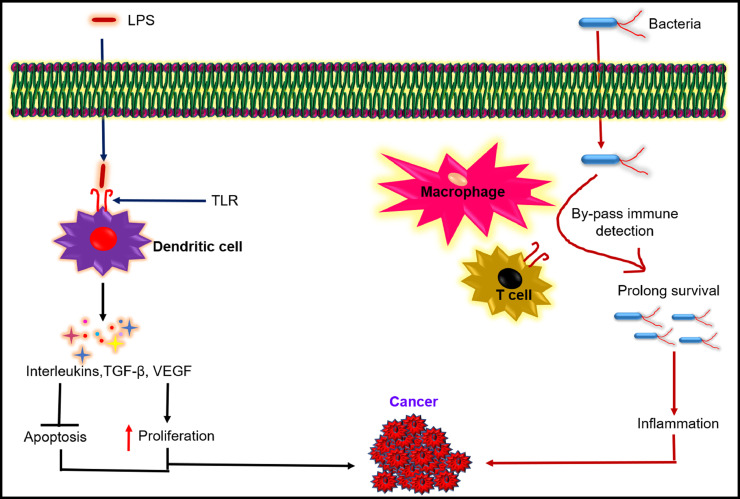
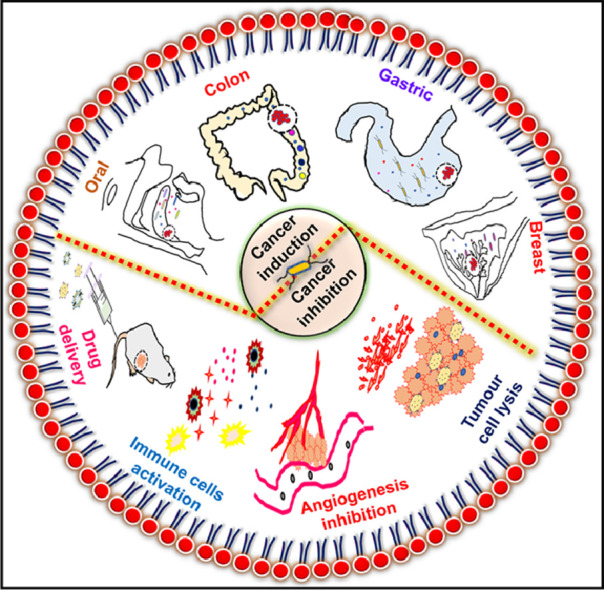
Microbiota and cancer have a bi-modal relationship, as represented in Fig. 1. Though microbiota plays a critical role in triggering and progression of malignancy, it is also an emerging cancer therapy approach (Zhang et al., 2018; Yu et al., 2012; Park et al., 2017; Park et al., 2013a). Conventional cancer therapies, like chemotherapy, surgery, radiation, etc., lack target specificity and adverse side effects. The traditional cancer drugs cannot penetrate the tumour tissues deeply, and some of the tumour tissues develop drug resistance as well (Navya et al., 2019; Maeda and Khatami, 2018). The unique internal pathophysiology of solid tumours and the limitations of conventional treatments indicate the immediate need for new therapeutic modalities. Several alternative treatments like photodynamic therapy, gene therapy, insulin potentiating therapy, human alpha‐lactalbumin made lethal to tumour cells (HAMLET), telomerase therapy, hyperthermia, dichloroacetate, non‐invasive radiofrequency cancer treatment, etc. have been explored for enhanced therapeutic efficacy (Sedighi et al., 2019). Bacteria have an advantage over all the approaches mentioned above: they can perforate into the tumour, colonize in its hypoxic core, and the body does not show any immune resistance, making it a more practical therapeutic approach (Zhang et al., 2018; Yu et al., 2012; Park et al., 2017). Application of non-pathogenic bacteria for anticancer drug delivery leads to regression of tumour growth (Zhang et al., 2018; Jiang et al., 2010; Xiong et al., 2013; Sawant et al., 2020).
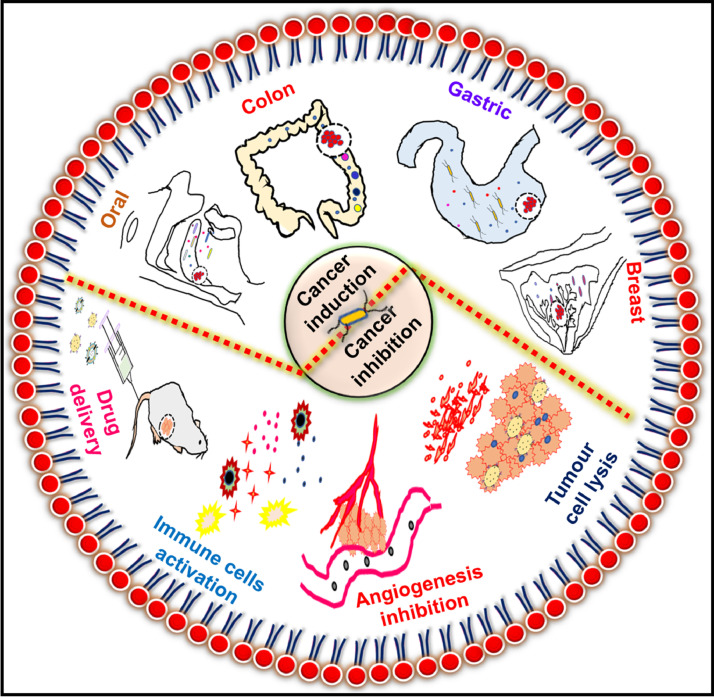
Fig. 1.
Schematic showing the role of bacteria in the induction and inhibition of cancer.
This review discusses the inter-relation between bacteria and cancer, cancer-causing virulence factors, and infections. We have also explored the various mechanisms to understand the role played by bacteria in the development and progression of cancer. Finally, the application of bacteria for cancer therapy is also discussed.
2. Understanding the mechanism of cancer progression
It is arduous to identify the particular bacterial species that causes cancer, as more than a million bacterial species are present in the human body. For example, the colon alone contains more than 100 trillion bacterial species, and the oral cavity contains over 500 bacterial species. Furthermore, a bacterial species found to be responsible for cancer progression follows different mechanisms at different sites. Example: Fusobacterium can cause both oral and colon carcinoma (Dekaboruah et al., 2020; Rubinstein et al., 2013; Harrandah et al., 2021). The mechanisms by which bacteria promotes malignancy and metastasis are discussed in detail in this section.
2.1. Chronic inflammation
The inflammatory reaction protects the host cells by fighting the foreign organisms, but inflammation/chronic inflammation persistence leads to cancer development. Several studies showed that chronic inflammation acts as a plethoric driving force in cancer development, intensifying malignant transformation, tumour growth, invasion, and metastasis. About 25% of human cancer is associated with chronic inflammation. Inflammation shows greater replicative potential and self-determination of growth factors. Furthermore, growth inhibition is also absent in inflammation conditions with a legit escape from apoptosis. All these factors lead to tumour extravasation and have great potential to enhance angiogenesis (Afrasiabi et al., 2015; Crusz and Balkwill, 2015; Gruffaz et al., 2017). The mechanism of chronic inflammation leading to cancer development and progression is not specific to bacteria. Other micro-organisms like viruses (Liu et al., 2015) and fungi (Ramirez-Garcia et al., 2013) can also promote an inflammatory pathway leading to cancer metastasis.
Inflammation is a natural immune response to protect host cells. Phagocytes recruited to the infection site secrete several pro-inflammatory cytokines like TNF-α (tumour necrosis factor-alpha), IL-23, IL-1, attracting other immune cells, enhancing the inflammatory response. These inflammatory responses activate STAT3 and NF-Kβ signalling pathways, triggering uncontrolled cell proliferation. The recruited immune cells increase reactive oxygen species (ROS) and nitrogen oxide species (NOS) by activating enzyme-like NADPH (nicotinamide adenine dinucleotide phosphate) oxidase, superoxide dismutase (SOD), nitric oxide synthase (NOS), etc. These free radicals further kill pathogens (Messex et al., 2020; Duque and Descoteaux, 2014b). However, these free radicals and derived metabolic products like HNO2, N2O3, peroxynitrite also damage proteins, DNA, and cell membranes. The damaged DNA is propagated through uncontrolled cell division, escaping the cell cycle check-point and repair system, leading to point mutations, deletion, and translocation. Hence chronic inflammation is associated with DNA damage, mutation, and aberrant cell differentiation, provoking cancer development (Habib and Moinuddin, 2005; Teshima et al., 2018; Irrazabal et al., 2020).
Fig. 2 shows the schematic illustration of how bacterial dysbiosis causes chronic inflammation and cancer. For example, H. pylori infection causes an influx of phagocytic cells (macrophage and neutrophils) in an attempt to kill the pathogens. These phagocytic cells release ROS/RNS (reactive oxyfen species/reactive nitrogen species) to clear the infection. However, the persistence of these free radicals damages the epithelial cells, leading to cancer development (den Hartog et al., 2016; Ding et al., 2007). H.pylori induces inflammation via several mechanisms, of which cagPAI mediated NF-kβ induction was discussed earlier in detail (2.1.Gastric cancer) (Chen et al., 2016; Krisch et al., 2016; Epiya-motifs, 2019; Stein et al., 2002). In addition, H.pylori also causes inflammation in gastric epithelial cells via their surface adhesion proteins like OipA (outer inflammatory protein OipA) and BabA (blood group antigen binding adhesin). These proteins aid in the longer persistence of bacteria in host epithelial cells, induces the secretion of pro-inflammatory cytokines like IL-6, IL-11, causing severe inflammation and develops stomach cancer (Sugimoto et al., 2011). F.nucleatum also has surface adhesion proteins: FadA (fusobacterium adhesin A), Fap2 (fibroblast activation protein 2), and RadD (radiation gene D). Researchers have found that FadA modulates the tumour micro-environment by downregulating the tight junction proteins: cadherin and its functional association with catenin, evoking chronic inflammation and cancer metastasis (Rubinstein et al., 2013; Kaplan et al., 2010).
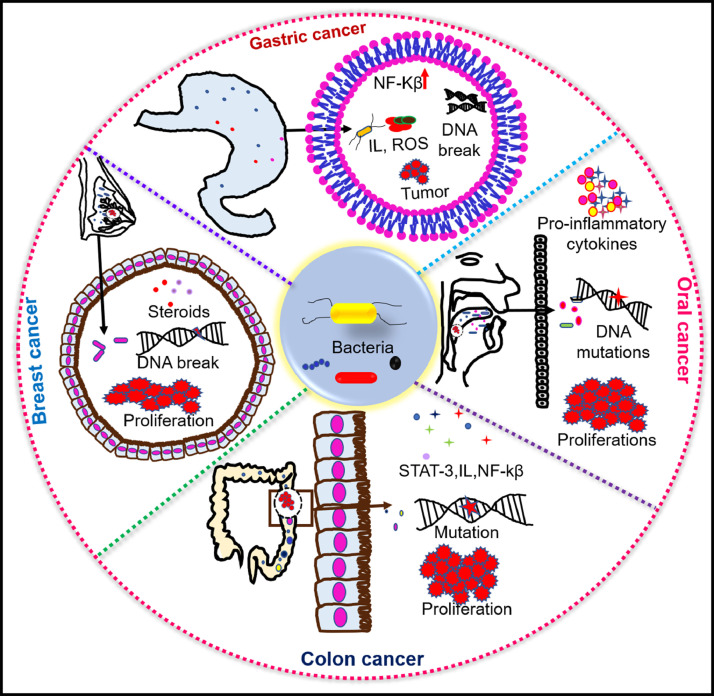
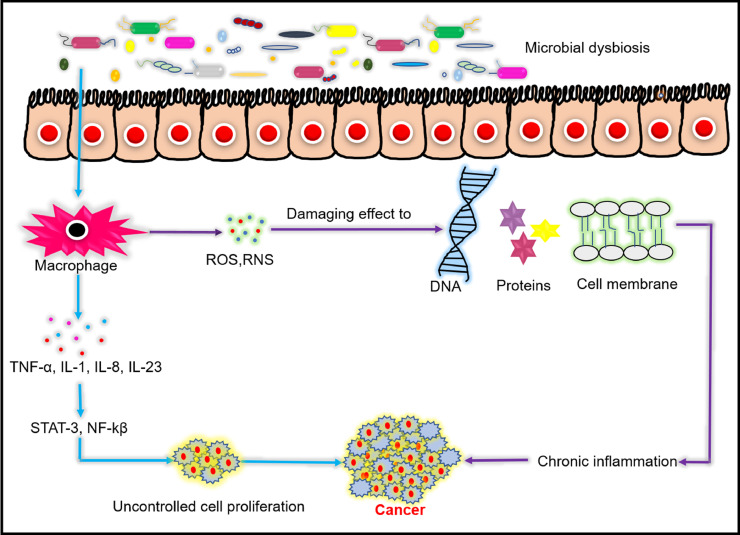
Fig. 2.
Bacterial dysbiosis causes chronic inflammation leading to cancer. Infected host immune cells produces inflammatory signalling molecules like TNF-α (tumour necrosis factor alpha), IL-1 (interleukin-1), IL-8, IL23, STAT-3 (signal transducer & activator of transcription 3), NF-κβ (nuclear factor kappa-light-chain-enhancer of activated B cells), ROS (reactive oxygen species), RNS (reactive nitrogen species), which causes chronic inflammation and leads to progression of cancer.
2.2. Cross talk between bacterial infections and immune system
The crosstalk between bacteria and immune system results in multifold interactions causing homoeostasis and pathogenesis. The commensal bacteria play an essential role in training and functionalizing major components of the host's innate and adaptive immune system. The innate immune system is composed of several pattern recognition receptors (PRRs). These PRRs have broad specificity towards antigens and can bind to many molecules with a common structural motif present in pathogens called pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) released by the damaged cells. There are several PRRs like toll-like receptors (TLRs), nucleotide-binding oligomerization domain (NOD) and leucine-rich repeats (LRR)-containing receptors (NLR), retinoic acid-inducible gene 1 (RIG-1) -like receptors (RLR) and C-type lectin receptors (CLRs). Among all these PRRs, TLRs are the most crucial pattern recognition receptors to elicit antimicrobial and inflammatory responses.
In contrast, the adaptive immune system consists of several effector immune cells like B and T cells. It recognizes the antigen by multiple antigen receptors, distributed on B and T cells. The genetic and somatic recombination of these receptors on B and T cells generates multiple repertoires of receptors. These receptors enable the identification of clonal selection of antigen-specific receptors and develop immunological memory. These receptors enable the identification of a clonal selection of antigen-specific receptors and develop immunological memory. After TLRs become activated by PAMPs/DAMPs, an immune response is transmitted to macrophages/B cells/ T cells. The activated immune cells produce pro-inflammatory cytokines like tumour-necrosis factor alpha (TNF α), interleukin-1β (IL-1β) and IL-6, neutralizing antigens. Therefore innate and adaptive immune responses commingle to kill pathogens (Amarante-Mendes et al., 2018; Chi et al., 2020).
Many bacteria present in the host's gut environment maintain a symbiotic/mutualistic relationship with the host immune system. The host-microbe mutualistic relationship operates optimally in healthy conditions and induces strong protective responses against foreign pathogens (Y. and Belkaid, 2015). Dendritic cells (DCs) and T cells are important components of innate and adaptive immune systems respectively. Dendritic cells present antigens to T cells and endow the differentiation of effector T cells. Microbes in the gut are directly and indirectly involved in DC cell activation or effector T cell differentiation/regulation preventing mucosal inflammation. It has been found that short-chain fatty acid (SCFAs)-C2, C3, C4, C5 fermented products of digestive resistant dietary fibres are produced by the anaerobic microbiota in the colon. These SCFAs involve in the regulation of DC, T cells’ function and developing protective immunity. It has been reported that SCFAs control the effector T cell function by activating or suppressing the FoxP3 Treg (forkhead box P3T regulatory) cells’ transcription. Park et al. reported that in cases of high T cell activation, SCFAs activate the histone deacetylase (HDAC), downregulating the transcription of the FoxP3 Treg gene. In contrast, under low T cell activation, SCFAs inhibit HDAC, upregulating the transcription of the FoxP3 Treg gene. Thus, microbiome-produced SCFAs can control both effector and regulatory cells function, which is crucial to boost immunity against pathogens (Furusawa et al., 2013; Park et al., 2015).
Several studies reported that alteration in host-microbe interactions/bacterial dysbiosis disrupts the host cells' homoeostasis leading to the evolution of malignancy (Sobhani et al., 2011; Weir et al., 2013). C. Xuan observed that breast tissues with microbial dysbiosis cause lower expression of immune sensors like TLR 2,5,9. Reduction in expression of antimicrobial response effectors: interleukin-12 A (IL-12A), myeloperoxidase (MPO) and bactericidal/permeability-increasing protein (BPI) was also observed, as compared to healthy breast tissues. In conclusion, bacterial dysbiosis causes inhibition of immune sensors and progression of malignancy (Xuan et al., 2014).
Researchers also reported that immune cell hyperactivation/evasion by bacteria develops cancer. Microbial ligands like LPS (lipopolysaccharides), lipopeptides, lipoteichoic acids, peptidoglycans, flagellins, etc., bind with specific TLR. Following successful binding, various cascade signalling pathways regulating cell proliferation or apoptosis, cell differentiation, secretion of inflammatory cytokines like interferons (IFNs), TNF-α, interleukins (IL1, IL6, IL8, IL12, IL16), etc., are affected (Ernst and Chandler, 2017; van Bergenhenegouwen et al., 2016; Schirbel et al., 2013). The hyper-activation of TLRs or escaping immune recognition can lead to cancer development, as illustrated in Fig. 3. Ye et al. showed that gram-negative bacteria like E.coli causes outgrowth and uncontrolled metastasis in non-small cell lung cancer (NSCLC). Overactivation of TLR4 and TLR9 related with lipid synthesis was observed in-vitro and in-vivo, suggesting immune receptor overactivation by bacterial infection (Ye et al., 2016).
Fig. 3.
Bacterial infections manipulate host immune response and stimulates pathogenesis. Bacterial LPS (lipopolysaccharides) binds to TLR (toll like receptor), activate dendritic cells, helps bacteria in bypassing the immune detection and trigger the production of interleukins, TGF-β (tumour growth factor beta), VEGF (vascular endothelial growth factor) which in turn induces cellular proliferation, inhibits apoptosis and cancer progression.
The bacterial outer surface contains complex antigenic moieties, allowing escape immune recognition (by modifying the surface antigens) and survival. For example, gram-negative bacteria like Shigella, Neisseria, etc., contain a polysaccharide-rich capsule to escape immune recognition. This immune escape mechanism protects them from phagocytosis and prevents complement activation, ensuring survival within host cells, leading to inflammation (Paciello et al., 2013; Sivagnanam et al., 2010). Bacteria can also dampen immune recognition by acquiring mutation on their surface flagellin. TLR-5 recognizes a conserved motif named αD1a. Several bacteria like Salmonella, Vibrio contains these conserved domains. It has also been reported that the highly virulent Helicobacter has a mutation in the D1 domain of its flagellin F1aA, which helps in evading immune recognition by TLR-5 and progresses gastric carcinoma (Il Yoon et al., 2012; Pachathundikandi et al., 2019). Salmonella typhimurium was also reported to have modified lipid A, inhibiting the TLR4 mediated immune activation. These immune-escaping mechanisms secure the residence within host organisms for a more extended period, allowing pathogenic bacteria modify host cell integrity and signalling cascade triggering cellular malignancies (Kawasaki et al., 2005).
In addition to innate immune evasion/hyperactivation, bacterial infections can also modulate the adaptive immune response in host cells. Adaptive immune responses become activated through secretion of cytokines, activation and proliferation of effector T cells and B cells. Cytolytic activity of effector T cell (CD8+ T cell) and antibodies produced by B cells contributes to killing bacterial pathogens. Pro-inflammatory cytokines like TNF- α, IL-1,8,12 are crucial to clear the bacterial infection from host cells (Duque and Descoteaux, 2014a). Some bacterial pathogens evolve their intracellular mechanism to modulate cytokine production and inhibit B/T cell activation inside host cells. Researchers reported that mycobacterium infected macrophages produce anti-inflammatory cytokines IL-6 inhibiting the activation of effector T cells. The secretion of immunosuppressive cytokines IL-10 was also observed. IL-10 suppresses macrophages and inhibits specific immune responses like MHC-II (major histocompatibility complex) antigen presentation, CD86 costimulatory signals. Activation of Treg cells, downregulating the immune response has also been observed. All the above observations confirm that bacterial infections manipulate innate and adaptive immune responses in the host stimulating pathogenesis (Jung et al., 2017; Giacomini et al., 2001).
2.3. Genomic instability
Genomic stability is essential for maintaining cellular integrity and protecting cells from: a) DNA replication error, b) oxidative stress (resulting from metabolic by-products), c) various exogenous carcinogenic agents causing DNA damage, etc. Genomic instability initiates and progresses carcinogenesis. The baseline mutations are insufficient for developing multiple changes in the host cell genome leading to cancer development. A spontaneous mutation obtained by the host cell induces malignancy leading to metastasis (Van Zijl et al., 2011). The bacterial genotoxins like cytolethal distending toxin (CDT, produced by certain gram-negative bacteria, e.g., Helicobacter pylori) (Toller et al., 2011), colibactin (produced by E. coli) (Cuevas-Ramos et al., 2010), and endonucleases (produced by N. gonorrhoea) (Weyler et al., 2014) induce DNA damage, favouring genomic instability and oncogenic transformation.
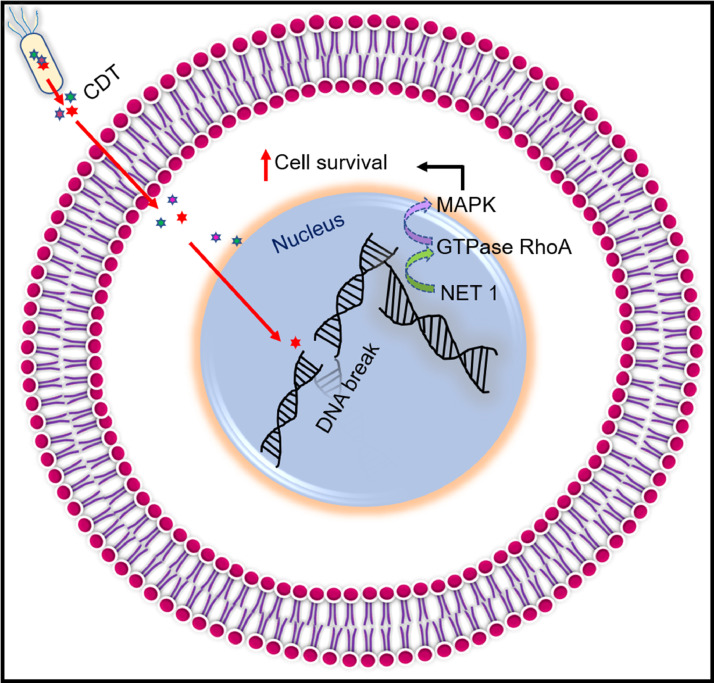
CDT is a protein toxin comprising of three subunits- CdtA, CdtB, and CdtC, encoded by different genes present in one operon. The subunits CdtA and CdtC ensure the binding of CDT to the target cell plasma membrane, and CdtB catalyzes the induction of double-stranded DNA breakage. After shielding the attachment by CdtA and CdtC, CDT endocytoses into the target cells. Following endocytosis, CdtB goes through Endosome-Golgi retrograde transport to the endoplasmic reticulum and translocates into the nucleus. CdtB then instigates double-stranded DNA break, seeking DNA damage response. This response induces G1-S and G2-M cell cycle arrest, responsible for DNA damage and prolongs the accumulation of genetic mutation. The DNA damage response also enhances the mitogen-activated protein kinase (MAPK) function by activating the neuroepithelial cell-transforming gene 1 protein (NET1). The GTPase RhoA leads to more prolonged survival of infected cells, as illustrated in Fig. 4. As a result, the infected cells acquire genetic mutation leading to malignancy (Dixon et al., 2015; Boesze-Battaglia et al., 2016; Guidi et al., 2013). In addition to CDT, certain
Fig. 4.
Bacterial genotoxins mediated DNA double stranded damage leading to cancer. CDT (cytolethal distending toxin) enters the nucleus and disrupts the DNA double helix structure. The DNA damage response intensifies MAPK (mitogen-activated protein kinase) function by activating NET 1 (neuroepithelial cell-transforming gene one protein), GTPase RhoA (guanosine triphosphatase Ras homologue family member A) protein which prolongs the cell survival.
E. coli strains secrete colibactin, which induces DNA double-stranded break, causing genomic instability. The DNA damage activates DNA damage checkpoint pathways ATM (ataxia-telangiectasia, mutated), CHK1 (check point kinase 1), and CHK2 (check point kinase 2), resulting in G2-M cell cycle arrest and chromosomal instability. All these factors lead to the development of cancer (Cuevas-Ramos et al., 2010; Bezine et al., 2014).
3. Bacterial infections mediate the progression of cancer
Bacterial inflammation is one of the crucial factors leading to cancer progression. The amplified inflammation, resulting in enhanced cell division, leads to cancer (Crusz and Balkwill, 2015; Domingo-Gonzalez et al., 2017; Korniluk et al., 2017). Researchers found bacterial infection to be carcinogenic and tumour stimulating, leading to the progression of malignancy. In addition, bacterial infections cause genomic instability and produce toxins hampering the regulatory signals associated with cell growth and proliferation (Rubinstein et al., 2013; Sugimoto et al., 2011; Cuevas-Ramos et al., 2010). Bacterial infections and their effects associated with various cancers are represented in Fig. 5. All bacterial strains are not carcinogenic, but few of them are. Examples: Helicobacter pylori cause gastric cancer (Stein et al., 2017), S. typhi develops gallbladder carcinoma (Scanu et al., 2015). Other bacterial strains are also found to be linked with cancer. Examples include Campylobacter jejuni (associated with small intestinal lymphomas) (He et al., 2019), Mycobacterium tuberculosis (associated with lung cancer) (Yamaguchi et al., 2020), and Citrobacter rodentium (associated with human colorectal cancer) (Umar, 2012).
Fig. 5.
Various cancers associated with bacterial infections. Bacteria infects the host cell and ensure their prolonged survival by modulating the host cell machinery; NF-kβ (nuclear factor kappa-light-chain-enhancer of activated B cells), IL (interleukins), ROS (reactive oxygen species), STAT-3 (signal transducer and activator of transcription 3).
Some well-known cancer-causing bacteria, major virulence factors, and alterations in the host micro-environment with infections are listed below in Table 1. Although researchers have explored the association between bacterial infections and cancer, bacteria's role in causing cancer and factors leading to metastasis are not very well understood. There is an excellent scope for future research to study how bacteria act as mediators of rapid progression and cancer metastasis.
Table 1.
Summary of cancer-causing major virulence factors of bacteria and post-infectious state in host cells.
| Name of the bacteria | Cancer-causing major virulence factors | Post-infectious changes in the host. | Developed cancer | References |
|---|---|---|---|---|
| Helicobacter pylori | cag pathogenicity island (cagPAI). membrane proteins OipA (outer inflammatory protein OipA) and BabA (blood group antigen binding adhesin) | Impaired tight junction function, high invasion, secretion of proinflammatory cytokines (IL-4, 6, 10, 11, TNF-α), STAT-3 and NF-kβ activation, cell survival, proliferation, chronic inflammation. | Gastric cancer | (Sugimoto et al., 2011; Stein et al., 2017; Dadashzadeh et al., 2017; Bounder et al., 2020; Hu et al., 2018) |
| Fusobacterium nucleatum | Surface adhesion protein FadA (fusobacterium adhesin A) | Downregulation of tight junction protein cadherin, recruitment of tumour-infiltrating immune cells (tumour-associated neutrophils and macrophages) secretion of IL-1β, IL-6, IL-8, NF-kβ1, NF-Kβ2, TNF-α, chronic inflammation, cell proliferation. | Colorectal cancer | (Rubinstein et al., 2013; Kostic et al., 2013) |
| Porphyromonas gingivalis | Lipopolysaccharides (LPS) | Altered oral epithelial interconnections, enhanced inflammatory response (IL-1β, TNF-α, and IL-6), epithelial cell proliferation, and differentiation. | Oral cancer | (Katz et al., 2011; Chen et al., 2019; Zhang et al., 2008) |
| Chlamydia pneumonia | LPS(lipopolysaccharides) and Heat shock protein 60 (HSP60) | Prolonged chronic inflammation with iNOS, IL-12, IL-23, and TNF-α. | Lung cancer | (Jupelli et al., 2013; Krüll and Suttorp, 2007; Chaturvedi et al., 2010) |
| Chlamydia Trachomatis | Plasmid encoded protein Pgp3/4 (P-glycoprotein 3/4) | Inhibit apoptosis, trigger inflammation, cell invasion, and malignancy | Cervical cancer | (Safaeian et al., 2010; Zou et al., 2019; Sigar et al., 2014; Song et al., 2013) |
| Campylobacter jejuni | HtrA (high-temperature requirement protein A) protease | Disruption of tight junction proteins (claudin, occludins) and loss of intestinal barrier function enhance cellular invasion, transmigration, proinflammatory cytokines such as TNF-α (tumour necrosis factor alpha) and IFN-γ (interferon gamma), and the matrix-degrading enzyme matrix metalloproteinase-2 (MMP-2), proliferation. | Small intestine cancer | (Harrer et al., 2019; Alzheimer et al., 2020; Abfalter et al., 2016; Heimesaat et al., 2014; Jing et al., 2021; Bartlett, 2004) |
| Campylobacter jejuni | Genotoxin, cytolethal distending toxin (CDT) | DNA double-strand breaks through the ATM (ataxia-telangiectasia mutated) -CHK2 (check point kinase 2), and ATR (ATM- and Rad3-Related) -CHK1 (check point kinase 1),, cell cycle arrest, promote the secretion of proinflammatory mediators IL-1β, IL-6, and IL-8, activate cell proliferation. | Colon cancer | (He et al., 2019; Pons et al., 2020) |
| Staphylococcus aureus | Clumping factor B (ClfB) protein | Increased expression of human β-defensin-2 (hBD-2) protein and AMP (antimicrobial peptides) causing chronic inflammation, tumour cell proliferation, and growth. | Skin carcinoma | (Madhusudhan et al., 2020; Lacey et al., 2019; Marcinkiewicz and Majewski, 2016) |
| S. Typhi | Genotoxin (typhoid toxin) | DNA damage, chronic inflammation, epithelial hyperplasia. | Gall bladder cancer | (Del Bel Belluz et al., 2016; Walawalkar et al., 2013; Gonzalez-Escobedo and Gunn, 2013) |
| Neisseria meningitidis | Outer membrane protein Opc | Enhanced bacterial transcytosis through the endothelial barrier, increase secretion of inflammatory cytokines and chemokines, activation of caspase-3. | Astrocyotmas/glioblastomas | (Cunha et al., 2010; Pereira et al., 2011) |
3.1. Gastric cancer
Gastric cancer is associated with a gram-negative bacterium: Helicobacter pylori (H. pylori), colonizing the human stomach. Approximately 1089,103 new cases and 768,793 new deaths due to stomach cancer were reported globally in 2020 (Sung et al., 2021; Mentis et al., 2019). Most of the person infected with H. pylori do not show any clinical symptoms, but the long-term infection causes inflammation of gastric epithelium. Approximately 10% of infected patients develop peptic ulcers, while 1–3% develop gastric adenocarcinoma. H. pylori infection can modulate host cells' signal transduction and metabolic pathways, including glycolysis, TCA cycle, and amino acid metabolism. The genetic heterogeneity of H. pylori influences its virulence factors which plays a crucial role in the pathogenesis of gastric cancer (Zhang et al., 2017; Sgouras et al., 2015; Matsunaga et al., 2018). Cytotoxin-associated gene A (CagA) and peptidoglycan are a few of the various virulence factors of H. pylori that penetrate the infected gastric epithelial cells and potentially trigger oncogenic pathways. CagA is a 120–140 kDa protein present in the DNA insertion element. Cag pathogenicity island (cagPAI) induces malignancy by translocating into the host cells. After translocation, CagA is phosphorylated by two tyrosine kinase Abl and Src (cytoplasmic tyrosine kinase) on tyrosine residue at four distinct glutamate-proline‑isoleucine‑tyrosine‑alanine (EPIYA) motifs present at the C-terminal region of the protein, inducing cell migration (Chen et al., 2016; Krisch et al., 2016; Epiya-motifs, 2019; Stein et al., 2002). Along with the CagA protein, peptidoglycan is also transmitted into host cells, triggering the nucleotide-binding oligomerization domain-containing protein 1 (NOD1) dependent nuclear factor kappa light chain enhancer activated B cells (NF-kβ) mediated inflammation leading to chronic inflammation in gastric epithelial cells. Inside the host cells, peptidoglycan can activate the phosphoinositide-3-kinase–protein kinase B/Akt (PI3K-PKB/Akt) signalling in an Src-EGFR (epidermal growth factor receptor) dependent pathway that induces cell proliferation, migration/metastasis, and apoptosis prevention, leading to carcinoma, (Viala et al., 2004; Suarez et al., 2015; R.C. Migration, and Apoptosis By Activation of Pi3K Signaling 2010). Cyclooxygenase-2/prostaglandin E2 pathway and IL-1β are other critical factors that induce chronic active gastritis and adenocarcinoma (Zhang et al., 2017; Echizen et al., 2016). Fig. 6 shows the schematic illustration of cancer development and progression by CagA and peptidoglycan from H. pylori.
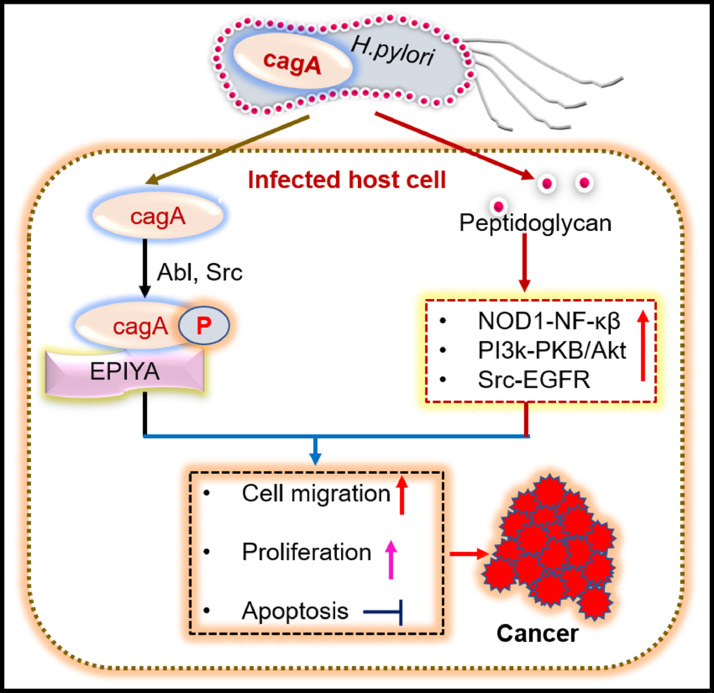
Fig. 6.
Cytotoxin-associated gene A (Cag A) and peptidoglycan of H. pylori triggers signalling pathways leading to cancer progression. Cag A, phosphorylated by Abl and Src tyrosine kinase on tyrosine motif EPIYA (glutamate-proline-isoleucine-tyrosine-alanine) induces cellular proliferation. Peptidoglycan induces proliferations, metastasis, inhibition of apoptosis by activation of NOD1- NF-kβ (nucleotide-binding oligomerization domain-containing protein one- nuclear factor kappa light chain enhancer activated B cells), PI3K-PKB/Akt(phosphoinositide-3-kinase–protein kinase B/Akt), Src-EGFR (epidermal growth factor receptor) signalling pathway.
E-cadherin is an epithelial adhesion protein that maintains proper cellular barriers associated with p120, β-catenin, and cytoskeletal components. Following H. pylori infection translocated CagA binds with E-cadherin and forms the complex E-cadherin-β-catenin. The impaired function of E-cadherin loosens the epithelial junction and enhances the nucleo-cytoplasmic accumulation of β-catenin, activating the β-catenin dependent carcinogenesis. Downregulation of β-catenin enhances the cytoplasmic/nuclear expression of p120, which further interacts with Ras-related C3 botulinum toxin substrate 1 (Rac1), p21-activated kinases (PAKs), Rho GTPases (Ras homologue family member A guanosine triphosphatase) and upgrades cellular motility and metastasis (Hu et al., 2018; Oliveira et al., 2009; Li et al., 2015). A high level of reactive oxygen species with H. pylori infection in gastric cells fuels the DDBs (DNA double-stranded breaks), DSBs (DNA single-stranded breaks), and impaired cell cycle checkpoint function, further leading to the development of malignancy (Shi et al., 2019).
3.3. Oral cancer
Oral cancer, also called oral squamous cell carcinoma (OSCC), originates from the oral mucosa. Nearly 377,713 new cases and 177,757 new deaths due to cancer in the lips and oral cavity were reported globally in 2020 (Sung et al., 2021). The oral cavity homes more than 700 bacterial species, and bacteria like Fusobacterium, Dialister, Peptostreptococcus, Filifactor, Peptococcus, Catonella, and Parvimonas were associated with oral squamous cell carcinoma (Ferlay et al., 2019; Chattopadhyay et al., 2019; Zhao et al., 2017).
The bacteria interact with oral mucosa by binding with toll-like receptors (TLRs). These bacterial-host cell interactions induce chronic inflammation in the oral cavity by secreting IL-6 (interleukin-6), NF-kβ (nuclear factor-kβ), IL-8 (interleukin-8). In addition to these, upregulation of the cell survival factors: MYC (Master Regulator of Cell Cycle Entry and Proliferative Metabolism), JAK1 (Janus kinase 1), STAT-3 (Signal transducer and activator of transcription 3) and mesenchymal transition markers: ZEB 1(zinc finger E-Box binding homeobox 1), TGF-β (transforming growth factor β)) were observed. These factors drive cell survival, proliferation, invasion, metastasis, and development of oral carcinoma (Harrandah et al., 2021; Gallimidi et al., 2015). Several causative factors, like tobacco, alcohol, etc., also alter the oral cavity's microbial composition, influencing microbial dysbiosis and making oral mucosa susceptible to carcinoma (Börnigen et al., 2017).
3.4. Colon cancer
The human gastrointestinal tract (GIT) contains a diverse micro-organisms called the intestine's normal microflora. Almost 1148,515 new cases and 576,858 new deaths due to colon cancer were reported in 2020 across the globe (Sung et al., 2021). Approximately 100 trillion bacteria are present in the human gut. These microflorae control the intestine's homoeostasis by regulating metabolic functions, developing immunity, and preventing pathogenic attacks (Gensollen et al., 2016; Claesson et al., 2012; Gagnière et al., 2016). However, when these flora's regular population alters (bacterial dysbiosis), it results in pathogenic conditions like cancer. The colon is the most heavily bacterial-populated part of GIT and is 12 folds more prone to cancer than the small intestine (Gagnière et al., 2016).
Fusobacterium nucleatum is a harmful gram-negative bacteria, generally found in the oral cavity and associated with colorectal cancer. F. nucleatum has a highly conserved adhesion protein FadA that binds to E-cadherin and activates β-catenin signalling. This further triggers the inflammatory genes NF-kβ, pro-inflammatory cytokines like IL-6, IL-8, and IL-18 in the colon epithelial cells inducing colorectal cancer (CRC) (Rubinstein et al., 2013). Furthermore, Fusobacterium nucleatum was also reported to enhance tumour multiplicity by recruiting tumour-infiltrating myeloid cells that promote colorectal malignancy (Kostic et al., 2013).
Some E.coli strains (B2 E.coli 11G5) can produce cyclomodulins/genotoxins, breaking host cells' DNA and inducing colorectal cancer. In addition, these bacterial strains can induce inflammation, cause colon epithelial damage, enhance cell proliferation, progress colonic neoplasia (Raisch et al., 2014; Buc et al., 2013). Furthermore, various other bacteria like Streptococcus gallolyticus (ex S. bovis) (Amado et al., 2015), Enterococcus faecalis (Escolà-Vergé et al., 2020), Clostridium septicum (Nanjappa et al., 2015) were also found to be associated with colorectal carcinoma.
3.5. Breast cancer
A diverse population of bacteria exists in breast tissue irrespective of lactation. Around 2261,419 new cases and 684,996 new deaths due to breast cancer were reported globally in 2020 (Sung et al., 2021). Proteobacteria and Firmicutes (mainly the class Bacilli) are the primary species found in breast cells. Breasts, being fatty tissues with wider vasculature, support bacterial growth. Bacterial composition plays an essential role in building immunity in neonates (Urbaniak et al., 2014). The commensal bacteria in the human body plays a vital role in maintaining the various physiological functions, but microbial dysbiosis induces inflammation leading to cancer. The increased bacterial population (high abundance of genus Alistipes) in the nipple was reported to progress breast cancer by modulating therapeutic responses (Chan et al., 2016).
Bacillus, Enterobacteriaceae, and Staphylococcus were found in great numbers in breast cancer tissues. These bacteria caused DNA double-stranded breaks in HeLa cells (Urbaniak et al., 2016). A strong association between gut microbiota and breast cancer has also been observed. The steroid hormone oestrogen is a prominent risk factor in breast cancer, particularly in post-menopausal women. Gut microbes are capable of encoding oestrogen deconjugating enzymes like β-glucuronidase and hydroxysteroid dehydrogenase. These enzymes can activate oestrogen metabolism, enhance their excretion, and propel them into hepatic circulation in an active form leading to hormonal imbalance and breast cancer (Rebecca et al., 2011; Yang et al., 2017).
4. Role of bacterial pathogens in epithelial to mesenchymal transition (EMT) leading to metastasis
Epithelial cells act as a functional interface between adjacent cells of our body distinguishing different organs. These cells are tightly bound to their surrounding cells and basement membrane by multiple junction proteins like tight junctions, adherens junctions, desmosomes, and hemidesmosomes (FARQUHAR and PALADE, 1963). However, the epithelial barrier loses its static phenotype during the developmental process, fibrogenesis or tumorigenesis and gains drastic migratory behaviour, leading to metastasis (Jechlinger et al., 2003). Loss of tight junction proteins cadherin/E-cadherin, cytokeratin and gain of vimentin, matrix metalloprotease, fibronectin and integrin αvβ3 are the characteristics of EMT (Lamouille et al., 2014). Multiple kinase-mediated signalling pathways like TGFβ−1/Smad2/3, Wnt/Notch, Ras>Raf>Mek>Erk>NF-κ β signalling contribute majorly to initiate the EMT and metastasis (Zhang et al., 2016; Olea-Flores et al., 2019).
Although the microbes are crucial in protecting host cells, an alteration in their composition and distribution disrupts homoeostasis in host cells. This altered relation favours inflammatory disorders promoting EMT (Y. and Belkaid, 2015). The bacterial lipopolysaccharide (LPS), a major component of gram-negative bacteria's outer membrane, induces EMT. TGF β1 is one of the most potent instigators of EMT and liver fibroblast. Smad 2/Smad 3 are the direct downstream signalling molecules in the TGF β1 transduction pathway. Activated TGF β1 receptor induces Smad2/3 phosphorylation forming a heteromeric complex with Smad 4. This complex further translocates to the nucleus for regulating specific gene expression (Schiffer et al., 2000). Zhao et al. demonstrated that stimulation of human intrahepatic biliary epithelial cells (HIBEpiCs) with LPS resulted in phenotypic changes producing bipolar cells with fibroblastic morphology. Downregulation of E-cadherin and upregulation of mRNA along witha protein expression of TGF β1 were also observed. The expression of mesenchymal markers like S100A and α-SMA (α-smooth muscle actin) was also increased significantly. Their study concluded that LPS triggers EMT via TGF β1/ Smad 2/ Smad 3 pathway (Zhao et al., 2011).
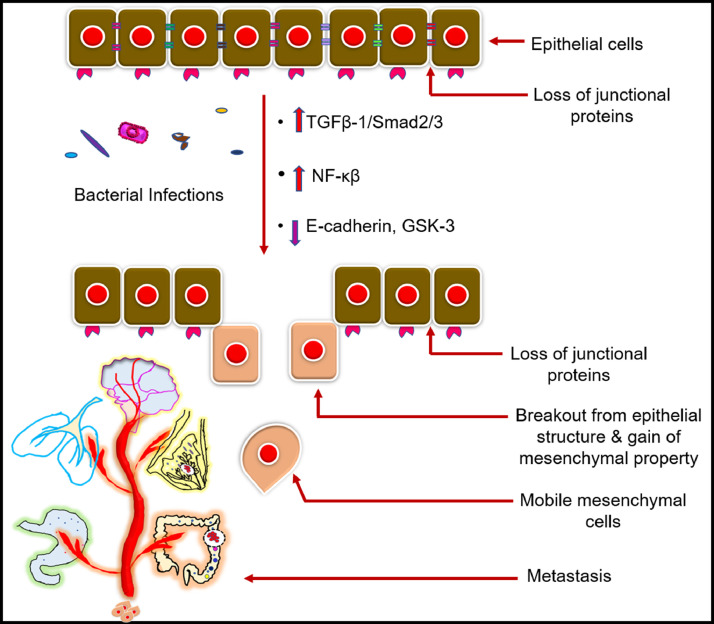
Loss of epithelial polarity is one of the major hallmarks of EMT and cancer metastasis, as illustrated in Fig. 7. Transcriptional repressor Snail acts as a potent inducer of EMT suppressing E-cadherin transcription/expression. Lee et al. showed that infections with cytotoxin-associated gene A (CagA) positive H.pylori strain 60,190 downregulate the epithelial tight junction protein E-cadherin in gastric cancer cells with increased tyrosine phosphorylation. The glycogen synthase kinase 3 (GSK 3) is a serine/threonine kinase that induces proteasomal degradation of oncogenic protein Snail, c-Myc (cellular myelocytomatosis oncogene), Mcl-1 (induced myeloid leukaemia cell differentiation protein). The CagA infections showed the destruction of GSK 3 (glycogen synthase kinase 3), confirming the stability of Snail protein and downregulation of E-cadherin, leading to EMT. The CagA induced excessive cellular transformation with cell motility, scattering, and metastasis known as ‘hummingbird phenotype’ was also observed. CagA was also found to induce in-vivo invasive progression of non-invasive MCF-7 cells. The cells crossed the basement membrane in the chick chorioallantoic membrane (CAM) assay (Lee et al., 2014).
Fig. 7.
Bacterial infections downregulate epithelial tight junction proteins leading to cancer-metastasis. Multiple kinase-mediated signalling pathways like TGFβ−1 (tumour growth factor beta one) /Smad2/3 (intracellular mediator, translocate TGF-β from cytoplasm to nucleus), GSK-3 (Glycogen synthase kinase three) are activated which downregulates the tight junction proteins E-cadherin and promotes EMT (epithelial mesenchymal transition).
Chow et al. exhibited that incubation of non-small lung cancer (NSCLC) cells with heat-inactivated E.coli elevated cell adhesion, proliferation, migration and metastasis via TLR4 (toll-like receptor 4) signalling. TLR4 is a pathogen recognition receptor. Incubation of NSCLC cells with E.coli showed the inhibition of TLR4, leading to the malignant characteristics. They also revealed that NSCLC cells have increased in vivo adhesion to hepatic sinusoids using the C57BL/6 mice model after incubation with E.coli. All these observations confirm the influence of bacterial infections in metastasis both in vitro and in vivo (Chow et al., 2015).
Wiele et.al. also deciphered certain quorum sensing peptides produced by bacteria like Phr0662 (Bacillus sp.), EntF metabolite (Enterococcus faecium), and EDF-derived (Escherichia coli) peptides interact with mammalian cells leading to cancer metastasis. It was observed from multiple confirmation assays that bacterial quorum sensing peptides activate the Ras/raf/MEK/MAPK, PI3K/Akt, STAT intracellular signalling cascades in mammalian cells, leading to genetic alterations and cancer metastasis. Hyper phosphorylation of epidermal growth factor receptors (EGFR) and activation of Smad2/Smad3 protein linked with cell cytoskeleton rearrangement and cell migration were also noticed. Histidine 4 (HIST1H4) protein is a well-known histone protein associated with increased Notch 1 stimulation, NF-kβ production, contributing to cell proliferation, angiogenesis and metastasis. In the presence of bacterial quorum sensing peptide upregulation of HIST1H4 was observed. All these observations confirm that bacterial infections can lead to uncontrolled metastasis in mammalian cells through activation of several signalling cascade mechanisms (Van De Wiele et al., 2015).
5. Application of bacteria for cancer therapy
Cancer is neither a transmissible disease nor an infectious disease, but bacterial infections increase the chances of cancer development. Though conventional cancer therapies like radiotherapy, chemotherapy, etc., has been exceptional in improving survival rates over the years, there are several drawbacks and limitations. Bacteriotherapy, the application of bacteria for therapeutic purposes, is a novel strategy to overcome the limitations of conventional therapy and offers effective cancer therapy. Bacteriotherapy can be used as a stand-alone therapy or in combination with conventional therapies. The application of bacteria for the delivery of therapeutics, genes, and drugs broadens its application in cancer therapy. Researchers demonstrated the successful application of bacteriotherapy for regression of tumours and inhibition of metastasis (Riglar and Silver, 2018; Enejiyon et al., 2020; Yan et al., 2015). Examples of few bacteria used for cancer therapy are E.coli (Yan et al., 2015), Salmonella (Gao et al., 2020), Pseudomonas (Chang et al., 2015), Bifidobacterium (Parisa et al., 2020), and Lactobacillus (Sentürk et al., 2020). Table 2 lists the summary of bacteriotherapy for cancer treatment.
Table 2.
Various bacteria used for cancer therapy and their significant outcome post-treatment.
| Bacterial strain | Treated cancer cell line | Significant outcome | References |
|---|---|---|---|
| E. coli Nissle 1917 (EcN) | MCF-7, 4T1 (Breast cancer cell). | Rapid internalization and drug release in the tumor's hypoxic region; significant tumour regression; no adverse effects. | (Zhang et al., 2018) |
| Salmonella enterica | K1735, B16F10 (murine melanoma), 4T1 (murine breast cancer). | upregulation of connexin 43 (Cx43) expressions in tumour cells; a significant increase in mitogen-activated protein kinases (MAPK); enhanced tumour-intracellular communication & chemo sensitization. | (Chang et al., 2013) |
| Salmonella enterica | B16F10 (mouse melanoma) and 4T1 (mouse breast cancer). | Reduced expression of immune tolerance helping rate-limiting enzyme indoleamine 2, 3-dioxygenase 1 (IDO), phospho-protein kinase B (P-AKT), phospho-mammalian targets of rapamycin (P-mTOR), and phospho-p70 ribosomal s6 kinase (P-p70s6K), tumour cell lysis. |
(Kuan and Lee, 2016) |
| Listeria monocytogenes | 501 Mel, A375, and SK-Mel-28 melanoma cells. | Enhanced ROS production; activated caspase-3 dependent apoptosis of tumour cells; increased cytokines (e.g., IL-2), differentiation of CD8+ T cells; and inhibited metastasis and tumour cell growth. | (Vitiello et al., 2019) |
| E. coli MG1655 | CT26 (colon), RENCA (renal), and TRAMPC1 (prostate) cancer cell | Secretion of TNF-α, IL-12, IL-6, and IL-10; inhibition of cancer cell proliferation and metastasis. | (Murphy et al., 2017) |
| Bifidobacterium breve UCC2003 and Clostridium difficile CCUG 37,780 | A549 (lung cancer cell). | Rapid delivery of hypoxic region-specific anti-cancerous nanomaterials; inhibition of epidermal growth factor receptors, and HIF-1α (hypoxia-induced factor); regression of tumour growth. | (Luo et al., 2016) |
| Clostridium perfringens | Caco-2 (colon adenocarcinoma), MDA-MB-231 (mammary adenocarcinoma), OE-33 (oesophageal adenocarcinoma), Kyse140 (oesophageal squamous carcinoma), Hela (cervix carcinoma) | Dysregulated expression of claudin-3,4 and 7; increased apoptosis; significant tumour cell killing. | (Becker et al., 2018) |
| S. typhimurium A1-R | BxPC-3, Capan-1, Hs766T, MiaPaCa-2 and Panc-1 (Pancreatic cancer cell line) | Inhibition of vascular endothelial growth factor (VEGF) and angiogenesis; prevents metastasis and tumour growth. | (Hiroshima et al., 2014) |
| Streptococcus pyogenes | HROG02, HROG05 and HROG10, ADI and Palomid 529 (glioblastoma cell) | Induction of apoptosis, inhibition of proliferation and cancer cell growth. | (Fiedler et al., 2015) |
| Lactococcus lactis | SW480 (colon cancer epithelial cell) | Cytotoxic effects on colorectal cancer cells; apoptosis induction by increasing the bax/bcl-2 ratio in both mRNA; protein levels and induction of pro-apoptotic stress protein CHAC1 (cation transport regulator-like protein 1); cell cycle arrest; tumour growth inhibition. | (Ahmadi et al., 2017) |
| Bacillus subtilis | MCF-7, MDA-MB‑231 (breast cancer cell) | Downregulation of MMP-9 (matrix metalloprotease-9) expression; activation of nuclear factor-κB (NF-κB), activator protein-1 (AP-1); inhibition of phosphorylation of Akt, extracellular signal-regulated kinase (ERK), tumour cell invasion, migration and proliferation. |
(Park et al., 2013b) |
| Clostridium butyricum MIYAIRI 588 | 253J-BV cells (human bladder cancer), H460 (human lung cancer), 786-O (human renal cancer), HEK293 (human embryonic kidney) and MBT-2 cell lines (murine bladder cancer) | Endogenous release of tumour necrosis factor-related apoptosis-inducing ligand (TRAIL), release of cytokines and chemokines, tumour cell lysis. | (Shinnoh et al., 2013) |
In this part of the review, the application of bacteria for drug delivery, immunotherapy, and combinational therapeutic approaches are discussed in detail.
5.1. Drug delivery
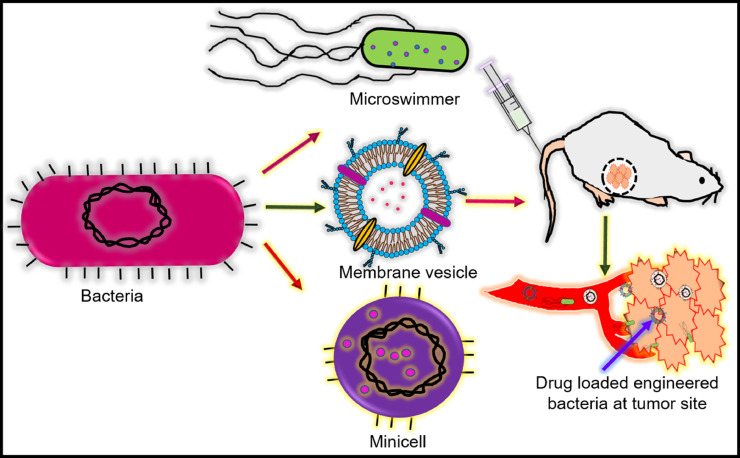
Poor vascularization and high interstitial fluid pressures are the characteristics of solid tumours, hindering the penetration of anticancer drugs into the tumour core and reducing their efficacy. Several drug carriers like liposomes, micelles, etc., accumulate in the perivascular region following their leakage from the vasculature. However, the released drug still does not penetrate the hypoxic tumour core due to the absence of arteries and high interstitial fluid pressure (Muthu et al., 2014; Ogihara, 2013). Bacteria can be one of the robust platforms for anticancer drug delivery under both in-vitro and in-vivo conditions, as shown in Fig. 8. Research shows that bacterial species like S. typhimurium (Yu et al., 2012), E. coli Nissle 1917 (EcN) (Stritzker et al., 2007) can colonize and proliferate in the tumour's hypoxic region. Therefore, bacteria and bacteria-derived cells can be emerging carriers for targeted drug delivery applications. Zhang et al. designed and developed a drug delivery vehicle by genetically engineering E. coli Nissle 1917 (EcN) with pHLIP (pH-(low)-insertion-peptide). The in-vitro and in-vivo experiments showed doxorubicin-loaded minicellspHLIP selectively targeted the tumour acidic micro-environment. MinicellspHLIP were observed to penetrate the tumour core, releasing doxorubicin in a controlled fashion, killing the cancer cells (Zhang et al., 2018).
Fig. 8.
Bacteria derived vehicles for targeted drug delivery. Bacterial microswimmer, membrane coated vesicles, minicell delivers anticancer drug to hypoxic core of tumour microenvironment and prevent the tumour reoccurrence.
The bacterial outer membrane can also be used as a drug carrier for targeted cancer therapy. Gujrati et al. used mutant E. coli strain to develop outer membrane vesicles (OMVs), loaded with siRNA (model drug) and decorated with human epidermal growth factor receptor 2 (HER2)- specific antibody (targeting ligand). In-vivo studies using siRNA loaded OMVs showed rapid extravasation and accumulation selectively in the tumour tissues. In-vivo anti-tumour efficacy of HER2-targeted, siRNA-carrying OMVs demonstrated that a small amount of siRNA was sufficient for substantial inhibition compared to the non-targeted carrier or free siRNA. Further in-vitro and in-vivo data revealed no non-specific binding, uptake, or toxicity against normal cells (Gujrati et al., 2014).
In addition to the bacterial minicells and OMVs, bacteria-driven microswimmers were developed for target-specific drug delivery. Microswimmers are hybrid microsystems integrated with bacteria that can swim like Serratia marcescens (S. marcescens), E. coli (E.coli), etc. The inherent physical and chemical features of bacteria like flagella, chemotaxis and viscoelasticity support micro swimming (Park et al., 2017; Zhuang and Sitti, 2016). Park et al. demonstrated the design and application of bacterial microswimmers for targeted drug delivery. A microswimmer was engineered by attaching a single E. coli bacterium to the surface of doxorubicin-loaded polyelectrolyte multilayer (PEM) microparticles embedding magnetic nanoparticles. The in-vitro studies demonstrated the drug delivery efficacy of these bacterial microswimmers, suggesting the valuable and significant contribution of bacteria to developing target-specific drug carriers (Park et al., 2017).
5.2. Immunotherapy
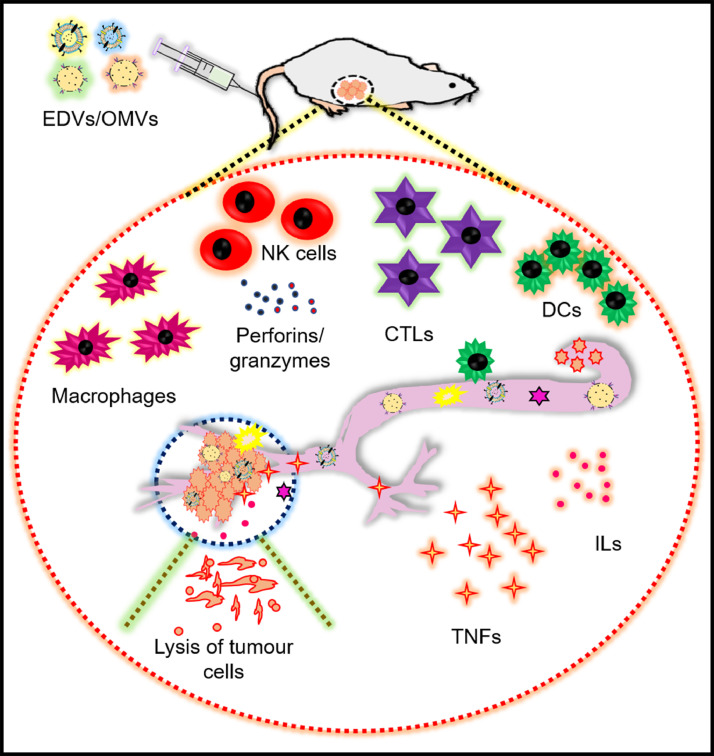
Immunotherapy is a powerful strategy to fight cancer and has greater advantages than chemotherapy or radiotherapy. However, the clinical outcome of immunotherapy for various tumours is limited by immune toxicity, recurrence, lack of initial response to therapy, etc. A successful tumour immune response is generated when a) tumour antigens are taken up by antigen-presenting cells (APC like dendritic cells), b) APCs are properly matured to be differentiated further, c) tumour-specific T cells are generated, and d) anti-tumour T cells are infiltrated into the tumour cells (Sagnella et al., 2020). Fig. 9 shows the role of bacterial vectors in activating several immune cells and tumour cell lysis.
Fig. 9.
Bacteria mediated immune cell activation promotes the lysis of tumour cells. Drug loaded EDVs (EnGeneIC Dream Vector)/OMVs (outer membrane vesicles) activates the host immune cells like DCs (dendritic cells), NK (natural killer) cells, CTLs (cytotoxic T lymphocytes and stimulate secretion of perforins/granzymes, TNFs (tumour necrosis factors), ILs (interleukins) causing lysis of tumour cells.
Sagnella et al. developed the EnGeneIC Dream Vector (EDV): a bacteria-derived delivery system conjugated with target-specific antibody and loaded with a super cytotoxic drug PNU-159,682 (682). In addition to delivering the drug to the target site, EDV also induces the tumour-specific immune response, triggers the in-vivo priming and maturation of dendritic cells within the tumour micro-environment. Up-regulation of tumour-specific CD4+ T cells, CD8+ T cell, CD80, and CD86 in response to clear cancer cells were also detected. Natural killer cells (NK) and interferon-γ (IFN-γ) have the inherent ability to kill the cancer cell by releasing perforin and granzyme. The EDV treatment also resulted in a remarkable production of NK cells, recruitment of IFNs to the target site, lysis of the cancer cells, and inhibition of tumour relapse. Thus, the bacteria-derived vector is indeed a novel and significant contribution to cancer immunotherapy (Sagnella et al., 2020).
Chen et al. designed nanoformulations with bacterial outer membrane vesicles (OMVs) to improve immunotherapy's efficacy. In this study, OMVs of attenuated Salmonella were functionalized with polyethylene glycol (PEG). These functionalized OMVs were loaded with tegafur, a fluorouracil prodrug (5-FU), and were conjugated with tumour-specific ligand Arg-Gly-Asp (RGD) peptide. This nanoformulation was observed to be immune-stimulatory, demonstrated improved blood circulation and tumour specificity. In-vivo biodistribution studies showed target-specific accumulation, and histological examinations of the liver, spleen, and kidney showed no pathological abnormalities, indicating biocompatibility. Furthermore, TNF-α, interferon-γ (IFN-γ), interleukin-12 (IL-12), IL-4, IL-17, CD4+, CD8+ T cells were upregulated, resulting in the death of cancer cells and inhibiting metastasis. This study demonstrated that bacteriotherapy could be a valuable strategy for cancer immunotherapy (Chen et al., 2020).
5.3. Combinational therapy
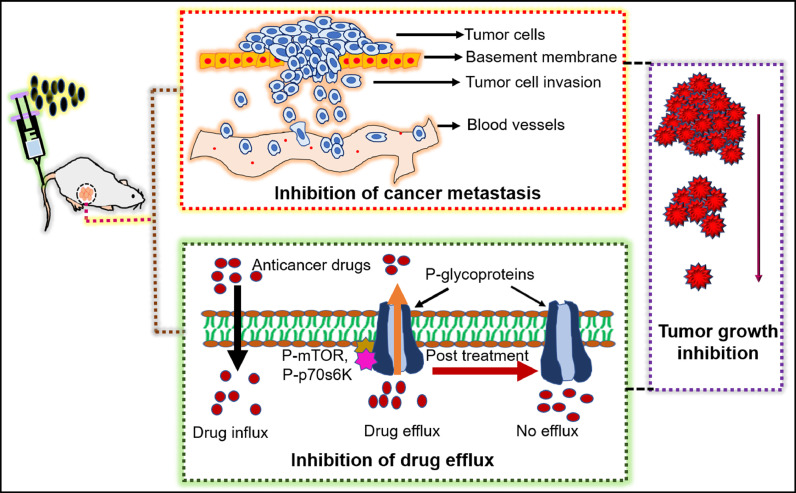
The combination of bacteriotherapy and conventional therapy has shown significant and sustained therapeutic potential. Conventional chemo/radiotherapy is associated with a relapse of tumours and adverse side effects due to their lack of specificity and systemic toxicity (D'Alterio et al., 2020). Bacteria therapy combined with chemo/radiotherapy can be a new approach to overcome these drawbacks. The combination of bacteriotherapy with conventional therapy can prevent cancer cells' growth, as demonstrated in Fig. 10.
Fig. 10.
Therapeutic benefits of combinational therapy. Bacteria therapy combined with chemo/radiotherapy prevents the multidrug resistance and tumour regrowth by inhibiting the phosphorylation, activation of kinase like P-mTOR (phospho-mammalian targets of rapamycin), P-p70s6K (phospho-p70 ribosomal s6 kinase) which are the main regulator of p-glycoprotein (protein responsible for drug resistance) expression.
The chemotherapy is limited by high dosages and low penetration, in addition to the poor vasculature and hypoxic micro-environment. Jia et al. demonstrated the improved efficacy of cyclophosphamide (CTX), a chemotherapeutic drug combined with tumour-targeting bacteria Salmonella VNP20009. This combinational therapy also showed reduced toxicity and payload of the drug. Also, more bacteria were found to be infiltrated at the tumour region. The vascular endothelial growth factor (VEGF) at the tumour site was also reduced as compared to individual treatments, suggesting the clinical importance of this combined cancer therapy (Jia et al., 2007).
Another dominant problem in chemotherapy is the development of drug resistance.
P-glycoprotein 1 (P-gp) is the multidrug resistance protein 1 (MDR1) present on the plasma membrane, fluxing out chemo drugs. The expression of P-gp is controlled by phospho-protein kinase B (P-AKT), phospho-mammalian targets of rapamycin (P-mTOR), and phospho-p70 ribosomal s6 kinase (P-p70s6K). Salmonella with a chemo drug (5-Fluorouracil) was observed to downregulate P-gp by inhibiting the protein kinases, making the cancer cells sensitive to chemo drug. This finding demonstrates that the bacteria combined with chemotherapy could prevent drug resistance and improve cancer therapy (Yang et al., 2018).
In addition to chemotherapy, the application of bacterial vectors also improved radiotherapy. Jiang et al. designed a cytotoxic protein cytolysin A (ClyA) producing E. coli strain K-12 and combined it with radiotherapy. Individual treatments resulted in reduced tumour growth, but re-growth of tumours was observed post-treatment. The novel combined therapy (E.coli-radiotherapy) reduced tumour growth and metastasis in colon cancer significantly, depicting the significance and efficacy of combinational treatment (Jiang et al., 2010).
6. Conclusions
Bacteria inhabited in our body plays a vital role in a) maintaining the digestive system's normal physiological health (intestine, colon, stomach), b) protecting the immune system and c) preventing the colonization of pathogens. However, an imbalance in these organisms, i.e., bacterial dysbiosis, modulates several host cellular signalling pathways, leading to cancer development, eventually causing death. In this review, we have discussed several inherent bacterial characteristics that can trigger carcinogenesis. H.pylori containing cagPAI can activate PI3K-Akt kinase pathway, increase oxidative stress, cause disruption of E-cadherin/β-catenin/p120 interactions, damage host cell DNA, and develop gastric cancer. Bacterial dysbiosis can be associated with several cancers (oral cancer, colon cancer, and breast cancer) due to excessive ROS generation, genomic instability, inhibition of apoptosis and excessive cell proliferation.
Researchers, over the years, have successfully understood several mechanisms like chronic inflammation, immune system evasion, and genomic instability, triggering cancer. Bacterial dysbiosis can modulate the host cell signalling by enhancing the secretion of several pro-inflammatory cytokines like NF-κβ, IL-6, IL-8, IL-10, IL-18, etc. NF-kβ is the central player of inflammation/ chronic inflammation, instigating hyper-proliferation of host cells leading to cancer. These inflammatory cytokines are also indirectly associated with a high level of free radicals and DNA mutations, enhancing chronic inflammation by damaging proteins and cell membranes. Bacterial infections also provoke malignancy by their surface modification, immune receptor activation/ evasion forming DNA double-strand break, and genomic instability by secreting genotoxins (CTD). Though researchers have linked bacterial infections and cancer, the number of bacterial-mediated host cell transformations leading to cancer development is enormous than reported. Furthermore, it is crucial to understand the association between bacterial infections and cancer to develop alternative and novel therapeutic approaches.
Bacterial infections lead to cancer progression by various mechanisms, but bacteria significantly improve cancer therapy when applied as a vector or drug carrier. Bacterial minicells, microswimmers, and outer cell membrane vesicles (OMV) deliver the drug into the tumour's hypoxic region, resulting in tumour regression. Bacterio-immunotherapy is emerging as an important therapeutic approach addressing the immunotherapy mediated toxicity and recurrence of tumours. The combinational therapy of bacteria and chemo/radiotherapy is also evolving as a dominant scheme for overcoming the limitations of conventional treatment. Furthermore, there is a great need for understanding the mechanism by which bacteria targets, penetrates, and colonizes the tumour. This understanding would lead to the development of more effective treatment strategies. Research on the application of bacteria in combinational with conventional treatment can open up a new door for researchers to develop safe, biocompatible, and target specific cancer therapies.
CRediT authorship contribution statement
Sajmina Khatun: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Tejaswini Appidi: Data curation, Formal analysis, Writing – original draft, Writing – review & editing. Aravind Kumar Rengan: Resources, Writing – review & editing.
Declaration of Competing Interest
There are no conflicts to declare.
Acknowledgements
The authors acknowledge Dept. of Science & Technology (DST), Dept. of Biotechnology (DBT), Govt. of India for research funding of DST-Inspire (DST/INSPIRE/04/2015/000377), DST-AMT (DST/TDT/AMT/2017/227(G)), DBT-NNT (BT/NNT/28/1386/2017) projects. The authors also acknowledge the financial support by Vasudha foundations, Hyderabad, India. Author SK acknowledges University Grants Commission (UGC), Govt. of India for UGC-JRF fellowship (18-340769). Author TA acknowledges Dept. of Science & Technology (DST), Govt. of India for Inspire fellowship (IF160291).
References
- Abfalter C.M., Schubert M., Götz C., Schmidt T.P., Posselt G., Wessler S. HtrA-mediated E-cadherin cleavage is limited to DegP and DegQ homologs expressed by gram-negative pathogens. Cell Commun. Signal. 2016;14:1–12. doi: 10.1186/s12964-016-0153-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afrasiabi K., Zhou Y.H., Fleischman A. Chronic inflammation: is it the driver or is it paving the road for malignant transformation. Genes Cancer. 2015;6:214–219. doi: 10.18632/genesandcancer.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadi S., Ghollasi M., Hosseini H.M. The apoptotic impact of nisin as a potent bacteriocin on the colon cancer cells. Microb. Pathog. 2017;111:193–197. doi: 10.1016/j.micpath.2017.08.037. [DOI] [PubMed] [Google Scholar]
- Al-Rawi N., Al-Marzooq F. The relation between periodontopathogenic bacterial levels and resistin in the saliva of obese type 2 diabetic patients. J. Diabetes Res. 2017:2017. doi: 10.1155/2017/2643079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- M. Alzheimer, S.L. Svensson, F. König, M. Schweinlin, M. Metzger, H. Walles, C.M. Sharma, A three-dimensional intestinal tissue model reveals factors and small regulatory RNAs important for colonization with Campylobacter jejuni, 2020. https://doi.org/10.1371/journal.ppat.1008304. [DOI] [PMC free article] [PubMed]
- Amado C., Hidalgo M.J., Sedano C., Hebel A., Porte L., Braun S., Dabanch J., Fica A. Bacteriemias por Streptococcus gallolyticus (ex S. bovis) y su relación con patología colónica o hepatobiliar y endocarditis. Rev. Chil. Infectol. 2015;32:430–434. doi: 10.4067/S0716-10182015000500009. [DOI] [PubMed] [Google Scholar]
- Amarante-Mendes G.P., Adjemian S., Branco L.M., Zanetti L.C., Weinlich R., Bortoluci K.R. Pattern recognition receptors and the host cell death molecular machinery. Front. Immunol. 2018;9:1–19. doi: 10.3389/fimmu.2018.02379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartlett J.G. Immunoproliferative small intestinal disease associated with campylobacter jejuni. Infect. Dis. Clin. Pract. 2004;12:382. doi: 10.1097/01.idc.0000144912.27311.19. [DOI] [Google Scholar]
- Becker A., Leskau M., Schlingmann-Molina B.L., Hohmeier S.C., Alnajjar S., Escobar H.M., Ngezahayo A. Functionalization of gold-nanoparticles by the Clostridium perfringens enterotoxin C-terminus for tumor cell ablation using the gold nanoparticle-mediated laser perforation technique. Sci. Rep. 2018;8:1–15. doi: 10.1038/s41598-018-33392-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bezine E., Vignard J., Mirey G. The cytolethal distending toxin effects on mammalian cells: a DNA damage perspective. Cells. 2014;3:592–615. doi: 10.3390/cells3020592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanton L.V., Barratt M.J., Charbonneau M.R., Ahmed T., Gordon J.I. Childhood undernutrition, the gut microbiota, and microbiota-directed therapeutics. Science (80-.) 2016:352. doi: 10.1126/science.aad9359. [DOI] [PubMed] [Google Scholar]
- Boesze-Battaglia K., Alexander D., Dlakić M., Shenker B.J. A journey of cytolethal distending toxins through cell membranes. Front. Cell. Infect. Microbiol. 2016;6:1–11. doi: 10.3389/fcimb.2016.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börnigen D., Ren B., Pickard R., Li J., Ozer E., Hartmann E.M., Xiao W., Tickle T., Rider J., Gevers D., Franzosa E.A., Davey M.E., Gillison M.L., Huttenhower C. Alterations in oral bacterial communities are associated with risk factors for oral and oropharyngeal cancer. Sci. Rep. 2017;7:1–13. doi: 10.1038/s41598-017-17795-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounder G., Jouimyi M.R., Boura H., Jouhadi H., Badre W., Benomar H., Kettani A., Lebrazi H., Maachi F. Association of tumor necrosis factor receptor 1 promoter gene polymorphisms (-580 A/G and-609G/T) and TNFR1 serum levels with the susceptibility to gastric precancerous lesions and gastric cancer related to H. pylori infection in a Moroccan population. Biomed Res. Int. 2020;2020 doi: 10.1155/2020/2451854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA. Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- Buc E., Dubois D., Sauvanet P., Raisch J., Delmas J., Darfeuille-Michaud A., Pezet D., Bonnet R. High prevalence of mucosa-associated E. coli producing cyclomodulin and genotoxin in colon cancer. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0056964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A.A., Bashir M., Rivas M.N., Duvall K., Sieling P.A., Pieber T.R., Vaishampayan P.A., Love S.M., Lee D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016;6:1–11. doi: 10.1038/srep28061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Liu Y., Han B., Zhou C., Bai C., Li J. Pseudomonas aeruginosa preparation plus chemotherapy for advanced non-small-cell lung cancer: a randomized, multicenter, double-blind phase III study. Med. Oncol. 2015;32:1–7. doi: 10.1007/s12032-015-0583-1. [DOI] [PubMed] [Google Scholar]
- Chang W.W., Lai C.H., Chen M.C., Liu C.F., Kuan Y.D., Lin S.T., Lee C.H. Salmonella enhance chemosensitivity in tumor through connexin 43 upregulation. Int. J. Cancer. 2013;133:1926–1935. doi: 10.1002/ijc.28155. [DOI] [PubMed] [Google Scholar]
- Chattopadhyay I., Verma M., Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol. Cancer Res. Treat. 2019;18:1–19. doi: 10.1177/1533033819867354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi A.K., Gaydos C.A., Agreda P., Holden J.P., Chatterjee N., Goedert J.J., Caporaso N.E., Engels E.A. Chlamydia pneumoniae infection and risk for lung cancer. Cancer Epidemiol. Biomarkers Prev. 2010;19:1498–1505. doi: 10.1158/1055-9965.EPI-09-1261. [DOI] [PubMed] [Google Scholar]
- Chen Q., Bai H., Wu W., Huang G., Li Y., Wu M., Tang G., Ping Y. Bioengineering bacterial vesicle-coated polymeric nanomedicine for enhanced cancer immunotherapy and metastasis prevention. Nano Lett. 2020;20:11–21. doi: 10.1021/acs.nanolett.9b02182. [DOI] [PubMed] [Google Scholar]
- Chen S.Y., Zhang R.G., Duan G.C. Pathogenic mechanisms of the oncoprotein CagA in H. pylori-induced gastric cancer (Review) Oncol. Rep. 2016;36:3087–3094. doi: 10.3892/or.2016.5145. [DOI] [PubMed] [Google Scholar]
- Chen W., Alshaikh A., Kim S., Kim J., Chun C., Mehrazarin S., Lee J., Lux R., Kim R.H., Shin K.H., Park N.H., Walentin K., Schmidt-Ott K.M., Kang M.K. Porphyromonas gingivalis impairs oral epithelial barrier through targeting GRHL2. J. Dent. Res. 2019;98:1150–1158. doi: 10.1177/0022034519865184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi X., Li Y., Qiu X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: mechanism and regulation. Immunology. 2020;160:233–247. doi: 10.1111/imm.13176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S.C. Chow, S.D. Gowing, J.J. Cools-lartigue, C.B. Chen, J. Berube, H. Yoon, C.H.F. Chan, M.C. Rousseau, F. Bourdeau, B. Giannias, L. Roussel, S.T. Qureshi, S. Rousseau, L.E. Ferri, Gram negative bacteria increase non-small cell lung cancer metastasis via toll-like receptor 4 activation and mitogen-activated protein kinase phosphorylation, 4 (2015). https://doi.org/10.1002/ijc.29111. [DOI] [PubMed]
- Claesson M.J., Jeffery I.B., Conde S., Power S.E., O’connor E.M., Cusack S., Harris H.M.B., Coakley M., Lakshminarayanan B., O’sullivan O., Fitzgerald G.F., Deane J., O’connor M., Harnedy N., O’connor K., O’mahony D., Van Sinderen D., Wallace M., Brennan L., Stanton C., Marchesi J.R., Fitzgerald A.P., Shanahan F., Hill C., Paul Ross R., O’toole P.W. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- Crusz S.M., Balkwill F.R. Inflammation and cancer: advances and new agents. Nat. Rev. Clin. Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- Cuevas-Ramos G., Petit C.R., Marcq I., Boury M., Oswald E., Nougayrède J.P. Escherichia coli induces DNA damage in vivo and triggers genomic instability in mammalian cells. Proc. Natl. Acad. Sci. USA. 2010;107:11537–11542. doi: 10.1073/pnas.1001261107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunha C.S.E., Griffiths N.J., Virji M. Neisseria meningitidis opc invasin binds to the sulphated tyrosines of activated vitronectin to attach to and invade human brain endothelial cells. PLoS Pathog. 2010;6:1–18. doi: 10.1371/journal.ppat.1000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadashzadeh K., Peppelenbosch M.P., Adamu A.I. Helicobacter pylori pathogenicity factors related to gastric cancer. Can. J. Gastroenterol. Hepatol. 2017;2017 doi: 10.1155/2017/7942489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Alterio C., Scala S., Sozzi G., Roz L., Bertolini G. Paradoxical effects of chemotherapy on tumor relapse and metastasis promotion. Semin. Cancer Biol. 2020;60:351–361. doi: 10.1016/j.semcancer.2019.08.019. [DOI] [PubMed] [Google Scholar]
- De Martel C., Ferlay J., Franceschi S., Vignat J., Bray F., Forman D., Plummer M. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13:607–615. doi: 10.1016/S1470-2045(12)70137-7. [DOI] [PubMed] [Google Scholar]
- Dekaboruah E., Suryavanshi M.V., Chettri D., Verma A.K. Human microbiome: an academic update on human body site specific surveillance and its possible role. Arch. Microbiol. 2020;202:2147–2167. doi: 10.1007/s00203-020-01931-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Bel Belluz L., Guidi R., Pateras I.S., Levi L., Mihaljevic B., Rouf S.F., Wrande M., Candela M., Turroni S., Nastasi C., Consolandi C., Peano C., Tebaldi T., Viero G., Gorgoulis V.G., Krejsgaard T., Rhen M., Frisan T. The typhoid toxin promotes host survival and the establishment of a persistent asymptomatic infection. PLoS Pathog. 2016;12:1–25. doi: 10.1371/journal.ppat.1005528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hartog G., Chattopadhyay R., Ablack A., Hall E.H., Butcher L.D., Bhattacharyya A., Eckmann L., Harris P.R., Das S., Ernst P.B., Crowe S.E. Regulation of Rac1 and reactive oxygen species production in response to infection of gastrointestinal epithelia. PLoS Pathog. 2016;12:1–20. doi: 10.1371/journal.ppat.1005382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S.Z., Minohara Y., Xue J.F., Wang J., Reyes V.E., Patel J., Dirden-Kramer B., Boldogh I., Ernst P.B., Crowe S.E. Helicobacter pylori infection induces oxidative stress and programmed cell death in human gastric epithelial cells. Infect. Immun. 2007;75:4030–4039. doi: 10.1128/IAI.00172-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon S.D., Huynh M.M., Tamilselvam B., Spiegelman L.M., Son S.B., Eshraghi A., Blanke S.R., Bradley K.A. Distinct roles for CdtA and CdtC during intoxication by cytolethal distending toxins. PLoS ONE. 2015;10:1–16. doi: 10.1371/journal.pone.0143977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo-Gonzalez R., Prince O., Cooper A., Khader S.A. Cytokines and Chemokines in Mycobacterium tuberculosis Infection. Tuberc. Tuber. Bacillus. 2017;4:33–72. doi: 10.1128/9781555819569.ch2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque G.A., Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front. Immunol. 2014;5:1–13. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duque G.A., Descoteaux A. Macrophage cytokines: involvement in immunity and infectious diseases. Front. Immunol. 2014;5:1–12. doi: 10.3389/fimmu.2014.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echizen K., Hirose O., Maeda Y., Oshima M. Inflammation in gastric cancer: interplay of the COX-2/prostaglandin E2 and Toll-like receptor/MyD88 pathways. Cancer Sci. 2016;107:391–397. doi: 10.1111/cas.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enejiyon S.O., Adabara N.U., Wuna M.M., Fasasi R.A. Salmonella typhimurium as a potential anticancer agent: a review. Sri Lankan J. Infect. Dis. 2020;10(98) doi: 10.4038/sljid.v10i2.8289. [DOI] [Google Scholar]
- N. Epiya-motifs, Tailor-made detection of individual phosphorylated, (2019). [DOI] [PMC free article] [PubMed]
- Ernst R.K., Chandler C.E. Bacterial lipids: powerful modifiers of the innate immune response. F1000Res. 2017;6:1–11. doi: 10.12688/f1000research.11388.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escolà-Vergé L., Peghin M., Givone F., Pérez-Rodríguez M.T., Suárez-Varela M., Meije Y., Abelenda G., Almirante B., Fernández-Hidalgo N. Prevalence of colorectal disease in Enterococcus faecalis infective endocarditis: results of an observational multicenter study. Rev. Española Cardiol. (English Ed. 2020;73:711–717. doi: 10.1016/j.rec.2019.07.007. [DOI] [PubMed] [Google Scholar]
- FARQUHAR M.G., PALADE G.E. Junctional complexes in various epithelia. J. Cell Biol. 1963;17:375–412. doi: 10.1083/jcb.17.2.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlay J., Colombet M., Soerjomataram I., Mathers C., Parkin D.M., Piñeros M., Znaor A., Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- Fiedler T., Strauss M., Hering S., Redanz U., William D., Rosche Y., Classen C.F., Kreikemeyer B., Linnebacher M., Maletzki C. Arginine deprivation by arginine deiminase of Streptococcus pyogenes controls primary glioblastoma growth in vitro and in vivo. Cancer Biol. Ther. 2015;16:1047–1055. doi: 10.1080/15384047.2015.1026478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furusawa Y., Obata Y., Fukuda S., Endo T.A., Nakato G., Takahashi D., Nakanishi Y., Uetake C., Kato K., Kato T., Takahashi M., Fukuda N.N., Murakami S., Miyauchi E., Hino S., Atarashi K., Onawa S., Fujimura Y., Lockett T., Clarke J.M., Topping D.L., Tomita M., Hori S., Ohara O., Morita T., Koseki H., Kikuchi J., Honda K., Hase K., Ohno H. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- Gagnière J., Raisch J., Veziant J., Barnich N., Bonnet R., Buc E., Bringer M.A., Pezet D., Bonnet M. Gut microbiota imbalance and colorectal cancer. World J. Gastroenterol. 2016;22:501–518. doi: 10.3748/wjg.v22.i2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallaher J.A., Brown J.S., Anderson A.R.A. The impact of proliferation-migration tradeoffs on phenotypic evolution in cancer. Sci. Rep. 2019;9:1–10. doi: 10.1038/s41598-019-39636-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallimidi A.B., Fischman S., Revach B., Bulvik R., Rubinstein A.M., Nussbaum G., Elkin M. Nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget. 2015;6 doi: 10.18632/oncotarget.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S., Jung J.H., Lin S.M., Jang A.Y., Zhi Y., Bum Ahn K., Ji H.J., Hyang Lim J., Guo H., Choy H.E., Lim S., Seo H.S. Development of oxytolerant salmonella typhimurium using radiation mutation technology (RMT) for cancer therapy. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-60396-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng F., Zhang Y., Lu Z., Zhang S., Pan Y. Fusobacterium nucleatum caused DNA damage and promoted cell proliferation by the Ku70/p53 pathway in oral cancer cells. DNA Cell Biol. 2020;39:144–151. doi: 10.1089/dna.2019.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gensollen T., Iyer S.S., Kasper D.L., Blumberg R.S. How colonization by microbiota in early life shapes the immune system. Science (80-.) 2016;352:539–544. doi: 10.1126/science.aad9378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giacomini E., Iona E., Ferroni L., Miettinen M., Fattorini L., Orefici G., Julkunen I., Coccia E.M. Infection of human macrophages and dendritic cells with mycobacterium tuberculosis induces a differential cytokine gene expression that modulates T cell response. J. Immunol. 2001;166:7033–7041. doi: 10.4049/jimmunol.166.12.7033. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Escobedo G., Gunn J.S. Gallbladder epithelium as a niche for chronic salmonella carriage. Infect. Immun. 2013;81:2920–2930. doi: 10.1128/IAI.00258-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebowska A., Moran A.P., Matusiak A., Bak-Romaniszyn L., Czkwianianc E., Rechciński T., Walencka M., Płaneta-Małecka I., Rudnicka W., Chmiela M. Anti-phagocytic activity of Helicobacter pylori lipopolysaccharide (LPS)–possible modulation of the innate immune response to these bacteria. Polish J. Microbiol. 2008;57:185–192. [PubMed] [Google Scholar]
- Gruffaz M., Vasan K., Tan B., Da Silva S.R., Gao S.J. TLR4-mediated inflammation promotes KSHV-induced cellular transformation and tumorigenesis by activating the STAT3 pathway. Cancer Res. 2017;77:7094–7108. doi: 10.1158/0008-5472.CAN-17-2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidi R., Guerra L., Levi L., Stenerlöw B., Fox J.G., Josenhans C., Masucci M.G., Frisan T. Chronic exposure to the cytolethal distending toxins of Gram-negative bacteria promotes genomic instability and altered DNA damage response. Cell. Microbiol. 2013;15:98–113. doi: 10.1111/cmi.12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati V., Kim S., Kim S.H., Min J.J., Choy H.E., Kim S.C., Jon S. Bioengineered bacterial outer membrane vesicles as cell-specific drug-delivery vehicles for cancer therapy. ACS Nano. 2014;8:1525–1537. doi: 10.1021/nn405724x. [DOI] [PubMed] [Google Scholar]
- Habib S., Moinuddin R.Ali. Acquired antigenicity of DNA after modification with peroxynitrite. Int. J. Biol. Macromol. 2005;35:221–225. doi: 10.1016/j.ijbiomac.2005.02.005. [DOI] [PubMed] [Google Scholar]
- Harrandah A.M., Chukkapalli S.S., Bhattacharyya I., Progulske-Fox A., Chan E.K.L. Fusobacteria modulate oral carcinogenesis and promote cancer progression. J. Oral Microbiol. 2021;13 doi: 10.1080/20002297.2020.1849493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrer A., Bücker R., Boehm M., Zarzecka U., Tegtmeyer N., Sticht H., Schulzke J.D., Backert S. Campylobacter jejuni enters gut epithelial cells and impairs intestinal barrier function through cleavage of occludin by serine protease HtrA. Gut Pathog. 2019;11:1–16. doi: 10.1186/s13099-019-0283-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Z., Gharaibeh R.Z., Newsome R.C., Pope J.L., Dougherty M.W., Tomkovich S., Pons B., Mirey G., Vignard J., Hendrixson D.R., Jobin C. Campylobacter jejuni promotes colorectal tumorigenesis through the action of cytolethal distending toxin. Gut. 2019;68:289–300. doi: 10.1136/gutjnl-2018-317200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimesaat M.M., Fischer A., Alutis M., Grundmann U., Boehm M., Tegtmeyer N., Göbel U.B., Kühl A.A., Bereswill S., Backert S. The impact of serine protease HtrA in apoptosis, intestinal immune responses and extra-intestinal histopathology during Campylobacter jejuni infection of infant mice. Gut Pathog. 2014;6:1–10. doi: 10.1186/1757-4749-6-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiroshima Y., Zhang Y., Murakami T., Maawy A., Miwa S., Yamamoto M., Yano S., Sato S., Momiyama M., Mori R., Matsuyama R., Chishima T., Tanaka K., Ichikawa Y., Bouvet M., Endo I., Zhao M., Hoffman R.M. Efficacy of tumor-targeting Salmonella typhimurium A1-R in combination with anti-angiogenesis therapy on a pancreatic cancer patient-derived orthotopic xenograft (PDOX) and cellline mouse models. Oncotarget. 2014;5:12346–12357. doi: 10.18632/oncotarget.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Guo L., Ye G. Helicobacter pylori infection impairs gastric epithelial barrier function via microRNA-100-mediated mTOR signaling inhibition. Mol. Med. Rep. 2018;18:587–594. doi: 10.3892/mmr.2018.8971. [DOI] [PubMed] [Google Scholar]
- Il Yoon S., Kurnasov O., Natarajan V., Hong M., Gudkov A.V., Osterman A.L., Wilson I.A. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;80-(335):859–864. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irrazabal T., Thakur B.K., Kang M., Malaise Y., Streutker C., Wong E.O.Y., Copeland J., Gryfe R., Guttman D.S., Navarre W.W., Martin A. Limiting oxidative DNA damage reduces microbe-induced colitis-associated colorectal cancer. Nat. Commun. 2020;11 doi: 10.1038/s41467-020-15549-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jechlinger M., Grunert S., Tamir I.H., Janda E., Lüdemann S., Waerner T., Seither P., Weith A., Beug H., Kraut N. Expression profiling of epithelial plasticity in tumor progression. Oncogene. 2003;22:7155–7169. doi: 10.1038/sj.onc.1206887. [DOI] [PubMed] [Google Scholar]
- Jia L.J., Wei D.P., Sun Q.M., Jin G.H., Li S.F., Huang Y., Hua Z.C. Tumor-targeting Salmonella typhimurium improves cyclophosphamide chemotherapy at maximum tolerated dose and low-dose metronomic regimens in a murine melanoma model. Int. J. Cancer. 2007;121:666–674. doi: 10.1002/ijc.22688. [DOI] [PubMed] [Google Scholar]
- Jiang S.N., Phan T.X., Nam T.K., Nguyen V.H., Kim H.S., Bom H.S., Choy H.E., Hong Y., Min J.J. Inhibition of tumor growth and metastasis by a combination of escherichia coli-mediated cytolytic therapy and radiotherapy. Mol. Ther. 2010;18:635–642. doi: 10.1038/mt.2009.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin C., Lagoudas G.K., Zhao C., Bullman S., Bhutkar A., Hu B., Ameh S., Sandel D., Liang X.S., Mazzilli S., Whary M.T., Meyerson M., Germain R., Blainey P.C., Fox J.G., Jacks T. Commensal microbiota promote lung cancer development via γδ T cells. Cell. 2019;176:998–1013. doi: 10.1016/j.cell.2018.12.040. e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing X., Korchagina A.A., Shein S.A., Muraoka W.T., Koroleva E., Tumanov A.V. IL-23 contributes to campylobacter jejuni-induced intestinal pathology via promoting IL-17 and IFNγ responses by innate lymphoid cells. Front. Immunol. 2021:11. doi: 10.3389/fimmu.2020.579615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung B.G., Wang X., Yi N., Ma J., Turner J., Samten B. Early secreted antigenic target of 6-kDa of mycobacterium tuberculosis stimulates IL-6 production by macrophages through activation of STAT3. Sci. Rep. 2017;7:1–14. doi: 10.1038/srep40984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jupelli M., Shimada K., Chiba N., Slepenkin A., Alsabeh R., Jones H.D., Peterson E., Chen S., Arditi M., Crother T.R. Chlamydia pneumoniae infection in mice induces chronic lung inflammation, iBALT formation, and fibrosis. PLoS ONE. 2013;8:2–10. doi: 10.1371/journal.pone.0077447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan C.W., Ma X., Paranjpe A., Jewett A., Lux R., Kinder-Haake S., Shi W. Fusobacterium nucleatum outer membrane proteins Fap2 and RadD induce cell death in human lymphocytes. Infect. Immun. 2010;78:4773–4778. doi: 10.1128/IAI.00567-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz J., Onate M.D., Pauley K.M., Bhattacharyya I., Cha S. Presence of Porphyromonas gingivalis in gingival squamous cell carcinoma. Int. J. Oral Sci. 2011;3:209–215. doi: 10.4248/IJOS11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawasaki K., Ernst R.K., Miller S.I. Inhibition of Salmonella enterica serovar typhimurium lipopolysaccharide deacylation by aminoarabinose membrane modification. J. Bacteriol. 2005;187:2448–2457. doi: 10.1128/JB.187.7.2448-2457.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korniluk A., Koper O., Kemona H., Dymicka-Piekarska V. From inflammation to cancer. Ir. J. Med. Sci. 2017;186:57–62. doi: 10.1007/s11845-016-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostic A.D., Chun E., Robertson L., Glickman J.N., Gallini C.A., Michaud M., Clancy T.E., Chung D.C., Lochhead P., Hold G.L., El-Omar E.M., Brenner D., Fuchs C.S., Meyerson M., Garrett W.S. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14:207–215. doi: 10.1016/j.chom.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krisch L.M., Posselt G., Hammerl P., Wesslera S. CagA phosphorylation in helicobacter pylori-infected B cells is mediated by the nonreceptor tyrosine kinases of the src and Abl families. Infect. Immun. 2016;84:2671–2680. doi: 10.1128/IAI.00349-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüll M., Suttorp N. Pathogenesis of Chlamydophila pneumoniae infections — epidemiology, immunity, cell biology, virulence factors. Commun.-Acquired Pneumonia. 2007:83–110. doi: 10.1007/978-3-7643-7563-8_6. [DOI] [Google Scholar]
- Kuan Y.D., Lee C.H. Salmonella overcomes tumor immune tolerance by inhibition of tumor indoleamine 2, 3-dioxygenase 1 expression. Oncotarget. 2016;7:374–385. doi: 10.18632/ONCOTARGET.6258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacey K.A., Mulcahy M.E., Towell A.M., Geoghegan J.A., McLoughlin R.M. Clumping factor B is an important virulence factor during Staphylococcus aureus skin infection and a promising vaccine target. PLoS Pathog. 2019;15:1–20. doi: 10.1371/journal.ppat.1007713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. Fakultas Psikologi Dan Sosial Budaya Universitas Islam Indonesia Yogyakarta. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758.Molecular. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leclercq S.Y., Sullivan M.J., Ipe D.S., Smith J.P., Cripps A.W., Ulett G.C. Pathogenesis of Streptococcus urinary tract infection depends on bacterial strain and β-hemolysin/cytolysin that mediates cytotoxicity, cytokine synthesis, inflammation and virulence. Sci. Rep. 2016;6:1–14. doi: 10.1038/srep29000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.G., Kim H.S., Lee Y.S., Kim S., Cha S.Y., Ota I., Kim N.H., Cha Y.H., Yang D.H., Lee Y., Park G.J., Yook J.I., Lee Y.C. Helicobacter pylori CagA promotes Snail-mediated epithelial-mesenchymal transition by reducing GSK-3 activity. Nat. Commun. 2014;5 doi: 10.1038/ncomms5423. [DOI] [PubMed] [Google Scholar]
- Li M., Liu S., Wang M., Hu H., Yin J., Liu C., Huang Y. Gut microbiota dysbiosis associated with bile acid metabolism in neonatal cholestasis disease. Sci. Rep. 2020;10:1–13. doi: 10.1038/s41598-020-64728-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q., Ma L., Shen S., Guo Y., Cao Q., Cai X., Feng J., Yan Y., Hu T., Luo S., Zhou L., Peng B., Yang Z., Hua Y. Intestinal dysbacteriosis-induced IL-25 promotes development of HCC via alternative activation of macrophages in tumor microenvironment. J. Exp. Clin. Cancer Res. 2019;38:1–13. doi: 10.1186/s13046-019-1271-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.F., Qin S.H., Ruan X.Z., Wang X. P120-catenin participates in the progress of gastric cancer through regulating the Rac1 and Pak1 signaling pathway. Oncol. Rep. 2015;34:2357–2364. doi: 10.3892/or.2015.4226. [DOI] [PubMed] [Google Scholar]
- Liu X., Ma X., Lei Z., Feng H., Wang S., Cen X., Gao S., Jiang Y., Jiang J., Chen Q., Tang Y., Tang Y., Liang X. Chronic inflammation-related HPV: a driving force speeds oropharyngeal carcinogenesis. PLoS ONE. 2015;10:1–14. doi: 10.1371/journal.pone.0133681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C.H., Huang C.T., Su C.H., Yeh C.S. Bacteria-mediated hypoxia-specific delivery of nanoparticles for tumors imaging and therapy. Nano Lett. 2016;16:3493–3499. doi: 10.1021/acs.nanolett.6b00262. [DOI] [PubMed] [Google Scholar]
- Madhusudhan N., Pausan M.R., Halwachs B., Durdević M., Windisch M., Kehrmann J., Patra V.K., Wolf P., Boukamp P., Moissl-Eichinger C., Cerroni L., Becker J.C., Gorkiewicz G. Molecular profiling of keratinocyte skin tumors links staphylococcus aureus overabundance and increased human β-defensin-2 expression to growth promotion of squamous cell carcinoma. Cancers (Basel) 2020;12 doi: 10.3390/cancers12030541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda H., Khatami M. Analyses of repeated failures in cancer therapy for solid tumors: poor tumor-selective drug delivery, low therapeutic efficacy and unsustainable costs. Clin. Transl. Med. 2018;7:1–20. doi: 10.1186/s40169-018-0185-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcinkiewicz M., Majewski S. The role of antimicrobial peptides in chronic inflammatory skin diseases. Postep. Dermatologii i Alergol. 2016;33:6–12. doi: 10.5114/pdia.2015.48066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunaga S., Nishiumi S., Tagawa R., Yoshida M. Alterations in metabolic pathways in gastric epithelial cells infected with Helicobacter pylori. Microb. Pathog. 2018;124:122–129. doi: 10.1016/j.micpath.2018.08.033. [DOI] [PubMed] [Google Scholar]
- Mazmanian S.K., Round J.L., Kasper D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- Mentis A.F.A., Boziki M., Grigoriadis N., Papavassiliou A.G. Helicobacter pylori infection and gastric cancer biology: tempering a double-edged sword. Cell. Mol. Life Sci. 2019;76:2477–2486. doi: 10.1007/s00018-019-03044-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messex J.K., Byrd C.J., Liou G.Y. Signaling of macrophages that contours the tumor microenvironment for promoting cancer development. Cells. 2020:9. doi: 10.3390/cells9040919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy C., Rettedal E., Lehouritis P., Devoy C., Tangney M. Intratumoural production of TNFα by bacteria mediates cancer therapy. PLoS ONE. 2017;12:1–13. doi: 10.1371/journal.pone.0180034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthu M.S., Leong D.T., Mei L., Feng S.S. Nanotheranostics - application and further development of nanomedicine strategies for advanced theranostics. Theranostics. 2014;4:660–677. doi: 10.7150/thno.8698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanjappa S., Shah S., Pabbathi S. Clostridium septicum gas gangrene in colon cancer: importance of early diagnosis. Case Rep. Infect. Dis. 2015;2015:1–3. doi: 10.1155/2015/694247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navya P.N., Kaphle A., Srinivas S.P., Bhargava S.K., Rotello V.M., Daima H.K. Current trends and challenges in cancer management and therapy using designer nanomaterials. Nano Converg. 2019;6 doi: 10.1186/s40580-019-0193-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogihara T. Challenges of drug delivery systems that contribute to cancer chemotherapy. Foreword., Biol. Pharm. Bull. 2013;36:691. doi: 10.1248/bpb.b13-f3605. [DOI] [PubMed] [Google Scholar]
- Olea-Flores M., Zuñiga-Eulogio M.D., Mendoza-Catalán M.A., Rodríguez-Ruiz H.A., Castañeda-Saucedo E., Ortuño-Pineda C., Padilla-Benavides T., Navarro-Tito N. Extracellular-signal regulated kinase: a central molecule driving epithelial–mesenchymal transition in cancer. Int. J. Mol. Sci. 2019;20:1–32. doi: 10.3390/ijms20122885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira M.J., Costa A.M., Costa A.C., Ferreira R.M., Sampaio P., Machado J.C., Seruca R., Mareei M., Figueiredo C. CagA associates with c-met, E-cadherin, and p120catenin in a multiproteic complex that suppresses helicobacter pylori-induced cell-invasive phenotype. J. Infect. Dis. 2009;200:745–755. doi: 10.1086/604727. [DOI] [PubMed] [Google Scholar]
- Pachathundikandi S.K., Tegtmeyer N., Arnold I.C., Lind J., Neddermann M., Falkeis-Veits C., Chattopadhyay S., Brönstrup M., Tegge W., Hong M., Sticht H., Vieth M., Müller A., Backert S. T4SS-dependent TLR5 activation by Helicobacter pylori infection. Nat. Commun. 2019;10:1–11. doi: 10.1038/s41467-019-13506-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paciello I., Silipo A., Lembo-Fazio L., Curcurù L., Zumsteg A., Noël G., Ciancarella V., Sturiale L., Molinaro A., Bernardini M.L. Proc. Natl. Acad. Sci. U. S. A. Vol. 110. 2013. Erratum: intracellular Shigella remodels its LPS to dampen the innate immune recognition and evade inflammasome activation (Proceedings of the National Academy of Sciences of the United States of America; p. 20843. (2013) 110: (E4345-E4354) DOI: 10.1073/pnas.1303641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisa A., Roya G., Mahdi R., Shabnam R., Maryam E., Malihe T. Anti-cancer effects of Bifidobacterium species in colon cancer cells and a mouse model of carcinogenesis. PLoS ONE. 2020;15:1–18. doi: 10.1371/journal.pone.0232930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park B.W., Zhuang J., Yasa O., Sitti M. Multifunctional bacteria-driven microswimmers for targeted active drug delivery. ACS Nano. 2017;11:8910–8923. doi: 10.1021/acsnano.7b03207. [DOI] [PubMed] [Google Scholar]
- Park J., Kim M., Kang S.G., Jannasch A.H., Cooper B., Patterson J., Kim C.H. Short-chain fatty acids induce both effector and regulatory T cells by suppression of histone deacetylases and regulation of the mTOR-S6K pathway. Mucosal Immunol. 2015;8:80–93. doi: 10.1038/mi.2014.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.J., Park S.H., Cho S., Kim D.M., Lee Y., Ko S.Y., Hong Y., Choy H.E., Min J.J., Park J.O., Park S. New paradigm for tumor theranostic methodology using bacteria-based microrobot. Sci. Rep. 2013;3:1–8. doi: 10.1038/srep03394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.Y., Kim J.H., Lee Y.J., Lee S.J., Kim Y. Surfactin suppresses TPA-induced breast cancer cell invasion through the inhibition of MMP-9 expression. Int. J. Oncol. 2013;42:287–296. doi: 10.3892/ijo.2012.1695. [DOI] [PubMed] [Google Scholar]
- Pereira R.F.C., Alves D.A., Jacinto R.K., de Hollanda L.M., Verinaud L.M.C. Effects of Neisseria meningitidis infection in tumor glioblastoma cell line NG97: respiratory pathogen inducing apoptosis. J. Bacteriol. Parasitol. 2011:02. doi: 10.4172/2155-9597.1000114. [DOI] [Google Scholar]
- Polk D.B., Peek R.M. Helicobacter pylori: gastric cancer and beyond. Nat. Rev. Cancer. 2010;10:403–414. doi: 10.1038/nrc2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- B. Pons, A. Pettes-Duler, C. Naylies, F. Taieb, S. Hashim, S. Tadrist, Y. Lippi, G. Mirey, J. Vignard, Cytolethal distending toxin: from mitotic DNA damage to cGAS-dependent pro-inflammatory response, (2020) 1–21. https://doi.org/10.1101/2020.06.09.141648.
- R.C. Migration, and Apoptosis By Activation of Pi3K Signaling Apoptosis. 2010;199:641–651. doi: 10.1086/596660.HELICOBACTER. [DOI] [Google Scholar]
- Raisch J., Buc E., Bonnet M., Sauvanet P., Vazeille E., de Vallée A., Déchelotte P., Darcha C., Pezet D., Bonnet R., Bringer M.A., Darfeuille-Michaud A. Colon cancer-associated B2 Escherichia coli colonize gut mucosa and promote cell proliferation. World J. Gastroenterol. 2014;20:6560–6572. doi: 10.3748/wjg.v20.i21.6560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Garcia A., Arteta B., Abad-Diaz-de-Cerio A., Pellon A., Antoran A., Marquez J., Rementeria A., Hernando F.L. Candida albicans increases tumor cell adhesion to endothelial cells in vitro: intraspecific differences and importance of the mannose receptor. PLoS ONE. 2013;8:1–10. doi: 10.1371/journal.pone.0053584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebecca E.J., Lukanova A., Dossus L., Becker S., Rinaldi S., Tjønneland A., Olsen A., Overvad K., Mesrine S., Engel P., Clavel-Chapelon F., Chang-Claude J., Vrieling A., Boeing H., Schütze M., Trichopoulou A., Lagiou P., Trichopoulos D., Palli D., Krogh V., Panico S., Tumino R., Sacerdote C., Rodríguez L., Buckland G., Sánchez M.J., Amiano P., Ardanaz E., Bueno-de-Mesquita B., Ros M.M., Van Gils C.H., Peeters P.H., Khaw K.T., Wareham N., Key T.J., Allen N.E., Romieu I., Siddiq A., Cox D., Riboli E., Kaaks R. Postmenopausal serum sex steroids and risk of hormone receptor-positive and -negative breast cancer: a nested case-control study. Cancer Prev. Res. 2011;4:1626–1635. doi: 10.1158/1940-6207.CAPR-11-0090. [DOI] [PubMed] [Google Scholar]
- Riglar D.T., Silver P.A. Engineering bacteria for diagnostic and therapeutic applications. Nat. Rev. Microbiol. 2018;16:214–225. doi: 10.1038/nrmicro.2017.172. [DOI] [PubMed] [Google Scholar]
- Rosean C.B., Bostic R.R., Ferey J.C.M., Feng T.Y., Azar F.N., Tung K.S., Dozmorov M.G., Smirnova E., Bos P.D., Rutkowski M.R. Preexisting commensal dysbiosis is a host-intrinsic regulator of tissue inflammation and tumor cell dissemination in hormone receptor–positive breast cancer. Cancer Res. 2019;79:3662–3675. doi: 10.1158/0008-5472.CAN-18-3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein M.R., Wang X., Liu W., Hao Y., Cai G., Han Y.W. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating e-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195–206. doi: 10.1016/j.chom.2013.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safaeian M., Quint K., Schiffman M., Rodriguez A.C., Wacholder S., Herrero R., Hildesheim A., Viscidi R.P., Quint W., Burk R.D. Chlamydia trachomatis and risk of prevalent and incident cervical premalignancy in a population-based cohort. J. Natl. Cancer Inst. 2010;102:1794–1804. doi: 10.1093/jnci/djq436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sagnella S.M., Yang L., Stubbs G.E., Boslem E., Martino-Echarri E., Smolarczyk K., Pattison S.L., Vanegas N., St. Clair E., Clarke S., Boockvar J., MacDiarmid J.A., Brahmbhatt H. Cyto-immuno-therapy for cancer: a pathway elicited by tumor-targeted, cytotoxic drug-packaged bacterially derived nanocells. Cancer Cell. 2020;37:354–370. doi: 10.1016/j.ccell.2020.02.001. e7. [DOI] [PubMed] [Google Scholar]
- Sawant S.S., Patil S.M., Gupta V., Kunda N.K. Microbes as medicines: harnessing the power of bacteria in advancing cancer treatment. Int. J. Mol. Sci. 2020;21:1–24. doi: 10.3390/ijms21207575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scanu T., Spaapen R.M., Bakker J.M., Pratap C.B., en Wu L., Hofland I., Broeks A., Shukla V.K., Kumar M., Janssen H., Song J.Y., Neefjes-Borst E.A., te Riele H., Holden D.W., Nath G., Neefjes J. Salmonella manipulation of host signaling pathways provokes cellular transformation associated with gallbladder carcinoma. Cell Host Microbe. 2015;17:763–774. doi: 10.1016/j.chom.2015.05.002. [DOI] [PubMed] [Google Scholar]
- Schiffer M., Von Gersdorff G., Bitzer M., Susztak K., Bottinger E.P. Smad proteins and transforming growth factor-β signaling. Kidney Int. Suppl. 2000;58:45–52. doi: 10.1046/j.1523-1755.2000.07708.x. [DOI] [PubMed] [Google Scholar]
- Schirbel A., Kessler S., Rieder F., West G., Rebert N., Asosingh K., McDonald C., Fiocchi C. Pro-angiogenic activity of TLRs and NLRs: a novel link between gut microbiota and intestinal angiogenesis. Gastroenterology. 2013;144:613–623. doi: 10.1053/j.gastro.2012.11.005. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedighi M., Zahedi Bialvaei A., Hamblin M.R., Ohadi E., Asadi A., Halajzadeh M., Lohrasbi V., Mohammadzadeh N., Amiriani T., Krutova M., Amini A., Kouhsari E. Therapeutic bacteria to combat cancer; current advances, challenges, and opportunities. Cancer Med. 2019;8:3167–3181. doi: 10.1002/cam4.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14:1–14. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sentürk M., Ercan F., Yalcin S. The secondary metabolites produced by Lactobacillus plantarum downregulate BCL-2 and BUFFY genes on breast cancer cell line and model organism Drosophila melanogaster: molecular docking approach. Cancer Chemother. Pharmacol. 2020;85:33–45. doi: 10.1007/s00280-019-03978-0. [DOI] [PubMed] [Google Scholar]
- Sgouras D.N., Trang T.T.H., Yamaoka Y. Pathogenesis of Helicobacter pylori Infection. Helicobacter. 2015;20:8–16. doi: 10.1111/hel.12251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Wang P., Guo Y., Liang X., Li Y., Ding S. Helicobacter pylori-Induced DNA damage is a potential driver for human gastric cancer AGS cells. DNA Cell Biol. 2019;38:272–280. doi: 10.1089/dna.2018.4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine E.E., Xue M., Patel J.R., Healy A.R., Surovtseva Y.V., Herzon S.B., Crawford J.M. Model Colibactins exhibit human cell genotoxicity in the absence of host bacteria. ACS Chem. Biol. 2018;13:3286–3293. doi: 10.1021/acschembio.8b00714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinnoh M., Horinaka M., Yasuda T., Yoshikawa S., Morita M., Yamada T., Miki T., Sakai T. Clostridium butyricum MIYAIRI 588 shows antitumor effects by enhancing the release of TRAIL from neutrophils through MMP-8. Int. J. Oncol. 2013;42:903–911. doi: 10.3892/ijo.2013.1790. [DOI] [PubMed] [Google Scholar]
- Sigar I.M., Schripsema J.H., Wang Y., Clarke I.N., Cutcliffe L.T., Seth-Smith H.M.B., Thomson N.R., Bjartling C., Unemo M., Persson K., Ramsey K.H. Plasmid deficiency in urogenital isolates of Chlamydia trachomatis reduces infectivity and virulence in a mouse model. Pathog. Dis. 2014;70:61–69. doi: 10.1111/2049-632X.12086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivagnanam V., Zhu X., Schlichter L.C. Dominance of E. coli phagocytosis over LPS in the inflammatory response of microglia. J. Neuroimmunol. 2010;227:111–119. doi: 10.1016/j.jneuroim.2010.06.021. [DOI] [PubMed] [Google Scholar]
- Sobhani I., Tap J., Roudot-Thoraval F., Roperch J.P., Letulle S., Langella P., Gérard C., van Nhieu J.T., Furet J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE. 2011;6 doi: 10.1371/journal.pone.0016393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Carlson J.H., Whitmire W.M., Kari L., Virtaneva K., Sturdevant D.E., Watkins H., Zhou B., Sturdevant G.L., Porcella S.F., McClarty G., Caldwell H.D. Chlamydia trachomatis plasmid-encoded pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 2013;81:636–644. doi: 10.1128/IAI.01305-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein M., Bagnoli F., Halenbeck R., Rappuoli R., Fantl W.J. A. Covacci, c-Src/Lyn kinases activate Helicobacter pylori CagA through tyrosine phosphorylation of the EPIYA motifs. Mol. Microbiol. 2002;43:971–980. doi: 10.1046/j.1365-2958.2002.02781.x. [DOI] [PubMed] [Google Scholar]
- S.C. Stein, E. Faber, S.H. Bats, T. Murillo, Y. Speidel, N. Coombs, C. Josenhans, Helicobacter pylori modulates host cell responses by CagT4SS-dependent translocation of an intermediate metabolite of LPS inner core heptose biosynthesis, 2017. https://doi.org/10.1371/journal.ppat.1006514. [DOI] [PMC free article] [PubMed]
- Stritzker J., Weibel S., Hill P.J., Oelschlaeger T.A., Goebel W., Szalay A.A. Tumor-specific colonization, tissue distribution, and gene induction by probiotic Escherichia coli Nissle 1917 in live mice. Int. J. Med. Microbiol. 2007;297:151–162. doi: 10.1016/j.ijmm.2007.01.008. [DOI] [PubMed] [Google Scholar]
- Suarez G., Romero-Gallo J., Piazuelo M.B., Wang G., Maier R.J., Forsberg L.S., Azadi P., Gomez M.A., Correa P., Peek R.M. Modification of Helicobacter pylori peptidoglycan enhances NOD1 activation and promotes cancer of the stomach. Cancer Res. 2015;75:1749–1759. doi: 10.1158/0008-5472.CAN-14-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto M., Ohno T., Graham D.Y., Yamaoka Y. Helicobacter pylori outer membrane proteins on gastric mucosal interleukin 6 and 11 expression in Mongolian gerbils. J. Gastroenterol. Hepatol. 2011;26:1677–1684. doi: 10.1111/j.1440-1746.2011.06817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA. Cancer J. Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- Takeshima H., Ushijima T. Accumulation of genetic and epigenetic alterations in normal cells and cancer risk. Npj Precis. Oncol. 2019;3:1–8. doi: 10.1038/s41698-019-0079-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima R., Hanada K., Akada J., Kawano K., Yamaoka Y. Aggregatibacter actinomycetemcomitans infection causes DNA double-strand breaks in host cells. Genes to Cells. 2018;23:264–273. doi: 10.1111/gtc.12570. [DOI] [PubMed] [Google Scholar]
- Toller I.M., Neelsen K.J., Steger M., Hartung M.L., Hottiger M.O., Stucki M., Kalali B., Gerhard M., Sartori A.A., Lopes M., Müller A. Carcinogenic bacterial pathogen Helicobacter pylori triggers DNA double-strand breaks and a DNA damage response in its host cells. Proc. Natl. Acad. Sci. USA. 2011;108:14944–14949. doi: 10.1073/pnas.1100959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Mahowald M.A., Magrini V., Mardis E.R., Gordon J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444:1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Umar S. Citrobacter infection and wnt signaling. Curr. Colorectal Cancer Rep. 2012;8:298–306. doi: 10.1007/s11888-012-0143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Cummins J., Brackstone M., Macklaim J.M., Gloor G.B., Baban C.K., Scott L., O'Hanlon D.M., Burton J.P., Francis K.P., Tangney M., Reida G. Microbiota of human breast tissue. Appl. Environ. Microbiol. 2014;80:3007–3014. doi: 10.1128/AEM.00242-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbaniak C., Gloor G.B., Brackstone M., Scott L., Tangney M., Reida G. The microbiota of breast tissue and its association with breast cancer. Appl. Environ. Microbiol. 2016;82:5039–5048. doi: 10.1128/AEM.01235-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bergenhenegouwen J., Kraneveld A.D., Rutten L., Garssen J., Vos A.P., Hartog A. Lipoproteins attenuate TLR2 and TLR4 activation by bacteria and bacterial ligands with differences in affinity and kinetics. BMC Immunol. 2016;17:1–10. doi: 10.1186/s12865-016-0180-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van De Wiele C., Burvenich C., Peremans K. Crosstalk between the microbiome and cancer cells by quorum sensing peptides. Peptides. 2015:1–9. doi: 10.1016/j.peptides.2014.12.009. [DOI] [PubMed] [Google Scholar]
- Van Zijl F., Krupitza G., Mikulits W. Initial steps of metastasis: cell invasion and endothelial transmigration. Mutat. Res. - Rev. Mutat. Res. 2011;728:23–34. doi: 10.1016/j.mrrev.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J., Chaput C., Boneca I.G., Cardona A., Girardin S.E., Moran A.P., Athman R., Mémet S., Huerre M.R., Coyle A.J., DiStefano P.S., Sansonetti P.J., Labigne A., Bertin J., Philpott D.J., Ferrero R.L. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004;5:1166–1174. doi: 10.1038/ni1131. [DOI] [PubMed] [Google Scholar]
- Vitiello M., Evangelista M., Di Lascio N., Kusmic C., Massa A., Orso F., Sarti S., Marranci A., Rodzik K., Germelli L., Chandra D., Salvetti A., Pucci A., Taverna D., Faita F., Gravekamp C., Poliseno L. Antitumoral effects of attenuated Listeria monocytogenes in a genetically engineered mouse model of melanoma. Oncogene. 2019;38:3756–3762. doi: 10.1038/s41388-019-0681-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walawalkar Y.D., Gaind R., Nayak V. Study on Salmonella Typhi occurrence in gallbladder of patients suffering from chronic cholelithiasis-a predisposing factor for carcinoma of gallbladder. Diagn. Microbiol. Infect. Dis. 2013;77:69–73. doi: 10.1016/j.diagmicrobio.2013.05.014. [DOI] [PubMed] [Google Scholar]
- Wang S., Dong W., Liu L., Xu M., Wang Y., Liu T., Zhang Y., Wang B., Cao H. Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol. Carcinog. 2019;58:1155–1167. doi: 10.1002/mc.22999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir T.L., Manter D.K., Sheflin A.M., Barnett B.A., Heuberger A.L., Ryan E.P. Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0070803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weyler L., Engelbrecht M., Forsberg M.M., Brehwens K., Vare D., Vielfort K., Wojcik A., Aro H. Restriction endonucleases from invasive Neisseria gonorrhoeae cause double-strand breaks and distort mitosis in epithelial cells during infection. PLoS ONE. 2014;9:1–23. doi: 10.1371/journal.pone.0114208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organisation Latest global cancer data : cancer burden rises to 18 . 1 million new cases and 9 . 6 million cancer deaths in 2018 Latest global cancer data : cancer burden rises to 18 . 1 million new cases and 9 . 6 million cancer deaths in 2018. Int. Agency Res. Cancer. 2018:13–15. [Google Scholar]
- Wu S., Powers S., Zhu W., Hannun Y.A. Substantial contribution of extrinsic risk factors to cancer development. Nature. 2016;529:43–47. doi: 10.1038/nature16166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong M.H., Bao Y., Du X.J., Bin Tan Z., Jiang Q., Wang H.X., Zhu Y.H., Wang J. Differential anticancer drug delivery with a nanogel sensitive to bacteria-accumulated tumor artificial environment. ACS Nano. 2013;7:10636–10645. doi: 10.1021/nn403146t. [DOI] [PubMed] [Google Scholar]
- Xuan C., Shamonki J.M., Chung A., DiNome M.L., Chung M., Sieling P.A., Lee D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE. 2014;9:1–7. doi: 10.1371/journal.pone.0083744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Y, Belkaid T.H. Role of the microbiota in immunity and inflammation Yasmine. Cell. 2015;157:121–141. doi: 10.1016/j.cell.2014.03.011.Role. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi F., Minakata T., Miura S., Suzuki T., Sagara H. Heterogeneity of latent tuberculosis infection in a patient with lung cancer. J. Infect. Public Health. 2020;13:151–153. doi: 10.1016/j.jiph.2019.07.009. [DOI] [PubMed] [Google Scholar]
- Yan L., Kanada M., Zhang J., Okazaki S., Terakawa S. Photodynamic treatment of tumor with bacteria expressing killerred. PLoS ONE. 2015;10:1–15. doi: 10.1371/journal.pone.0131518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C.J., Chang W.W., Lin S.T., Chen M.C., Lee C.H. Salmonella overcomes drug resistance in tumor through P-glycoprotein downregulation. Int. J. Med. Sci. 2018;15:574–579. doi: 10.7150/ijms.23285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Tan Q., Fu Q., Zhou Y., Hu Y., Tang S., Zhou Y., Zhang J., Qiu J., Lv Q. Gastrointestinal microbiome and breast cancer: correlations, mechanisms and potential clinical implications. Breast Cancer. 2017;24:220–228. doi: 10.1007/s12282-016-0734-z. [DOI] [PubMed] [Google Scholar]
- Ye M., Gu X., Han Y., Jin M., Ren T. Gram-negative bacteria facilitate tumor outgrowth and metastasis by promoting lipid synthesis in lung cancer patients. J. Thorac. Dis. 2016;8:1943–1955. doi: 10.21037/jtd.2016.06.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu B., Yang M., Shi L., Yao Y., Jiang Q., Li X., Tang L.H., Zheng B.J., Yuen K.Y., Smith D.K., Song E., Huang J.D. Explicit hypoxia targeting with tumor suppression by creating an “obligate” anaerobic Salmonella Typhimurium strain. Sci. Rep. 2012;2:1–10. doi: 10.1038/srep00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Chen L., Li S., Gu Z., Yan J. Lipopolysaccharide (LPS) of porphyromonas gingivalis induces IL-1β, TNF-α and IL-6 production by THP-1 cells in a way different from that of Escherichia coli LPS. Innate Immun. 2008;14:99–107. doi: 10.1177/1753425907088244. [DOI] [PubMed] [Google Scholar]
- Zhang J., Tian X.-.J., Xing J. Signal transduction pathways of EMT Induced by TGF-β, SHH, and WNT and Their Crosstalks. J. Clin. Med. 2016;5:41. doi: 10.3390/jcm5040041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X.Y., Zhang P.Y., Aboul-Soud M.A.M. From inflammation to gastric cancer: role of Helicobacter pylori. Oncol. Lett. 2017;13:543–548. doi: 10.3892/ol.2016.5506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Ji W., He L., Chen Y., Ding X., Sun Y., Hu S., Yang H., Huang W., Zhang Y., Liu F., Xia L., coli E. Nissle 1917-derived minicells for targeted delivery of chemotherapeutic drug to hypoxic regions for cancer therapy. Theranostics. 2018;8:1690–1705. doi: 10.7150/thno.21575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Chu M., Huang Z., Yang X., Ran S., Hu B., Zhang C., Liang J. Variations in oral microbiota associated with oral cancer. Sci. Rep. 2017;7:1–10. doi: 10.1038/s41598-017-11779-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L., Yang R., Cheng L., Wang M., Jiang Y., Wang S. LPS-induced epithelial-mesenchymal transition of intrahepatic biliary epithelial cells. J. Surg. Res. 2011;171:819–825. doi: 10.1016/j.jss.2010.04.059. [DOI] [PubMed] [Google Scholar]
- Zhou X., Chen C., Zhong Y.N., Zhao F., Hao Z., Xu Y., Lai R., Shen G., Yin X. Effect and mechanism of vitamin D on the development of colorectal cancer based on intestinal flora disorder. J. Gastroenterol. Hepatol. 2020;35:1023–1031. doi: 10.1111/jgh.14949. [DOI] [PubMed] [Google Scholar]
- Zhuang J., Sitti M. Chemotaxis of bio-hybrid multiple bacteria-driven microswimmers. Sci. Rep. 2016;6:1–10. doi: 10.1038/srep32135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., Lei W., Su S., Bu J., Zhu S., Huang Q., Li Z. Chlamydia trachomatis plasmid-encoded protein Pgp3 inhibits apoptosis via the PI3K-AKT-mediated MDM2-p53 axis. Mol. Cell. Biochem. 2019;452:167–176. doi: 10.1007/s11010-018-3422-9. [DOI] [PubMed] [Google Scholar]