Abstract
Saccharomyces cerevisiae MutL homologues Mlh1p and Pms1p form a heterodimer, termed MutLα, that is required for DNA mismatch repair after mismatch binding by MutS homologues. Recent sequence and structural studies have placed the NH2 termini of MutL homologues in a new family of ATPases. To address the functional significance of this putative ATPase activity in MutLα, we mutated conserved motifs for ATP hydrolysis and ATP binding in both Mlh1p and Pms1p and found that these changes disrupted DNA mismatch repair in vivo. Limited proteolysis with purified recombinant MutLα demonstrated that the NH2 terminus of MutLα undergoes conformational changes in the presence of ATP and nonhydrolyzable ATP analogs. Furthermore, two-hybrid analysis suggested that these ATP-binding-induced conformational changes promote an interaction between the NH2 termini of Mlh1p and Pms1p. Surprisingly, analysis of specific mutants suggested differential requirements for the ATPase motifs of Mlh1p and Pms1p during DNA mismatch repair. Taken together, these results suggest that MutLα undergoes ATP-dependent conformational changes that may serve to coordinate downstream events during yeast DNA mismatch repair.
The process of DNA mismatch repair (MMR) has been the focus of intense study since human MMR gene mutations were implicated in hereditary and sporadic forms of human cancer (13, 21, 39, 52). A primary role of MMR is to correct base-base mismatches and insertion-deletion loops (IDLs) resulting from DNA replication, endogenous or exogenous sources of DNA damage, and recombination (14, 36, 38, 43). In Escherichia coli, where MMR has been reconstituted in vitro by using purified proteins, a homodimer of MutS binds the mismatch, followed by the formation of an ATP-binding-dependent ternary complex with a homodimer of MutL. The latent endonuclease activity of MutH is stimulated by the ATP-dependent MutS-MutL ternary complex to nick the undermethylated strand at the nearest hemimethylated GATC site. After incision, UvrD helicase and four exonucleases excise the nascent strand some distance past the mismatch (68). The resultant single-strand gap (up to 1 kb) is filled in by DNA polymerase III, and the nick is sealed by DNA ligase (43).
In the yeast Saccharomyces cerevisiae, the mutation avoidance functions of MMR involve multiple MutS and MutL heterodimers with partially overlapping functions (36, 38). For example, a heterodimer of Msh2p and Msh6p, MutSα, appears to be involved primarily in correcting mismatches and +1 IDL heterologies (2, 3, 12, 19, 23, 32, 33, 37, 41, 42, 48, 64), whereas, an Msh2p-Msh3p heterodimer, MutSβ, functions in correction of IDLs with 1 to 14 bases (23, 37, 41, 49). The major MMR MutL activity, MutLα, in yeast is a heterodimer of Mlh1p and Pms1p (27, 51, 58, 59). An additional yeast MutL activity, MutLβ, comprised of Mlh1p and Mlh3p, appears to act in conjunction with MutSβ to correct a small fraction of IDLs (22).
An important clue to a possible biochemical activity of the MutL homologues was the appreciation of sequence similarity between the highly conserved NH2 termini of the MutLs and a new family of ATPases (11, 44). The so-called GHL ATPase family is comprised of E. coli gyrase b subunit, the Hsp90 homologues, and the MutL homologues (10, 20). The supercoiling activity of E. coli DNA gyrase is dependent on the ATPase activity of the homodimeric gyrase b subunits (65). Recently, the homodimer Hsp90 has been demonstrated to have a weak intrinsic ATPase activity required for Hsp90 function (47, 50). The crystal structures of the NH2 termini of Hsp90 and gyrase b revealed strong structural similarity within their ATPase motifs (55, 56, 71). In addition, Hsp90 and gyrase b appear to have similar ATPase cycles, which include functionally important NH2-terminal conformational changes (4, 25, 26, 55, 56, 71). The NH2-terminal conformational changes for gyrase b have been associated with dimerization of the NH2-terminal domains in the ATP-bound form (4, 55, 56, 71). Recently, the crystal structure of an NH2-terminal fragment of MutL was solved and demonstrated that MutL possesses an ATP-binding pocket homologous to the gyrase b and Hsp90 proteins. In addition, MutL appears to have the ATPase-dependent NH2-terminal dimerization cycle found in the other GHL family member. Interestingly, Ban et al. reported that the NH2-terminal-dimerized, ATP-bound form of MutL could activate the MutH endonuclease in a MutS-independent manner (9, 10).
Our previous studies have shown the importance of the NH2 terminus of yeast Mlh1p and Pms1p in MMR (51). The abovementioned findings for the GHL family of proteins now present a working paradigm for detailed studies of the ATPase motifs found in the eukaryotic MutL homologues. In this report, we investigate the function of predicted ATPase motifs in S. cerevisiae MutLα (Mlh1p-Pms1p). Our results suggest that yeast MutLα has structural and functional properties consistent with other members of the GHL family of ATPases. Specifically, genetic results suggest that the ATPase motifs of both Mlh1p and Pms1p are absolutely required for MMR in vivo. In addition, biochemical and in vivo findings suggest that ATP binding induces conformational changes in MutLα that are associated with heterodimerization between the NH2 termini of Mlh1p and Pms1p. Surprisingly, our genetic results suggest differential requirements for Mlh1p and Pms1p ATPase motifs during MMR.
MATERIALS AND METHODS
Strains and media.
E. coli strains DH5α and DH-10B were used for plasmid construction and amplification. E. coli MAX Efficiency DH-10Bac [F− mcrA D(mrr-hsdRMS-mcrBC) φ80dlacZDM15 ΔlacX74 deoR recA1 endA1 araD139 Δ(ara, leu)7697 galU galK λ− rpsL nupG/bMON14272/pMON7124] was used to produce recombinant baculoviruses as described below under “Expression and purification of yeast MutLα.” The S. cerevisiae strains used in this study are described in Table 1. Bacterial and yeast strains were grown under conditions described previously (51). Yeast transformations were performed by the polyethylene glycol-lithium acetate method (24).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Source (reference) |
|---|---|---|
| GCY35 | MATa his3Δ200 hom3-10 ura3-52 ade2-101oc trp1 met13 met4 | G. F. Crouse (45) |
| PTY100 | Same as GCY35, but mlh1::URA3 | This study |
| PTY101 | Same as GCY35, but pms1::hisG-URA3-hisG | This study |
| PTY102 | Same as GCY35, but mlh3::hisG | This study |
| PTY103 | Same as GCY35, but mlh1::URA3 mlh3::hisG | This study |
| PTY104 | Same as GCY35, but pms1::hisG-URA3-hisG mlh3::hisG | This study |
| PTY200 | Same as GCY35, but mlh1-E31A | This study |
| PTY201 | Same as GCY35, but pms1-E61A | This study |
| PTY202 | Same as GCY35, but pms1-E61A mlh3::hisG | This study |
| PTY300 | Same as GCY35, but mlh1-G98A | This study |
| PTY301 | Same as GCY35, but pms1-G128A | This study |
| PTY302 | Same as GCY35, but pms1-G128A mlh3::hisG | This study |
| PTY400 | Same as GCY35, but mlh1-E31A pms1-E61A | This study |
| PTY500 | Same as GCY35, but mlh1-E31A pms1-G128A | This study |
| PTY501 | Same as GCY35, but mlh1-G98A pms1-E61A | This study |
| PTY600 | Same as GCY35, but mlh1-G98A pms1-G128A | This study |
| L40 | MATa his3Δ200 trp1-901 leu2-3,112 ade2 LYS2::(lexAop)4-HIS3 URA3::(lexAop)8-lacZ GAL4 gal80 | S. Hollenberg (69) |
| AMR70 | MATα his3Δ200 trp1-901 leu2-3,112 URA3::(lexAop)8-lacZ GAL4 gal80 | S. Hollenberg (69) |
Deletions of mlh1Δ and pms1Δ in the GCY35 (45) background were created as described previously (51, 53).
Genomic mlh1 point mutant strains used in this study (Table 1) were created by a two-step recombination procedure. Targeting constructs pYI-mlh1-31, and -98 were digested with PstI and transformed into the appropriate strains. Purified Ura+ transformants were replica plated onto yeast extract-peptone-dextrose (YPD) plates and grown overnight. YPD replica plates were replica plated to 5-fluoroorotic acid (5-FOA)-containing plates. Purified 5-FOAR isolates were screened for retention of the mlh1 point mutant allele by the mutator replica patch test for hom3-10 reversion, and the point mutation was confirmed by sequencing a PCR amplicon of the MLH1 gene. Both alleles were screened by using the same PCR oligonucleotides: yMLH1.S (5′-CGGGATCCCTCGAGACACCATGTCTCTCAGAATAAAAGC-3′) and yMLH1-F96A anchor.R (5′-GGAGTAAACGCTGTTCAAAGCTCT-3′). Alleles mlh1-E31A and mlh1-G98A were sequenced with the oligonucleotides ymlh1-98.AS (5′-GGCTAAAGCTTCAGCTCGGAATCCATACGTTTGAATCTG-3′) and ymlh1-31.S (5′-CCCGTAAATGCTCTCAAAGCTATGATGGAGAATTCC-3′), respectively. All double point mutant strains, PTY400, -500, -501, and -600, were generated by mutation of the MLH1 gene last.
Genomic pms1 point mutant strains were created similarly by using targeting constructs pYI-pms1-61 and -128 TV II digested with MluI or XbaI. PCR oligonucleotides yPMS1-86.S (5′-GTATGTCCAGCAGTTTCCATCAG-3′) and yPMS1-1281.AS (5′-GCAAGCTTATCGGTGTATTTCCCAAGCATTC-3′) were used to amplify a portion of the PMS1 gene, and the resulting PCR product was sequenced with oligonucleotides ypms1-128.AS (5′-GAAGATAGGGCCTCAGCTCTAAACCCTAACGTCTGTACTTTAGC-3′) and ypms1-61.S (5′-ACAACTGCAGTGAAAGCTCTCGTTGATAATAGTATAGATGCG-3′) for the pms1-E61A and pms1-G128A alleles, respectively.
Disruptions of mlh3Δ were generated by transformation with XhoI- and SacI-digested pΔmlh3::hisG-URA3-hisG (1) and selection on −Ura dropout media. Targeting of mlh3 was confirmed by Southern analysis of EcoRV-digested genomic DNA with a PCR-generated probe by using oligonucleotides 5′-TGGTTCGCCGATCTTATC-3′ and 5′-AAATACACTCCCTCTCCATCAC-3′.
Plasmid construction.
All DNA manipulations were performed by standard molecular biology procedures (40). Automated DNA sequencing was done at the Vollum Institute core sequencing facilities with an ABI automated sequencer.
(i) Targeting vectors.
pYI-mlh1-31 was created as follows. The MLH1 open reading frame (ORF) and approximately 800 bp of upstream sequence were cloned into pYI-lacZ. The E31A mutation was generated in the resultant construct by using the Quikchange Site-Directed Mutagenesis kit (Stratagene) and the following oligonucleotides: ymlh1-31.S and ymlh1-31.AS (5′-GGAATTCTCCATCATAGCTTTGAGAGCATTTACGGG-3′). The desired mutations were detected by sequencing with oligonucleotide ymlh1-98.AS. An approximately 400-bp KpnI fragment containing the E31A mutation was cloned back into the parental construct to erase the potential for second site mutations elsewhere in the construct. pYI-mlh1-98 was created in a similar fashion, except the following oligonucleotides were used instead: ymlh1-98.S (5′-CAGATTCAAACGTATGGATTCCGAGCTGAAGCTTTAGCC-3′) and ymlh1-98.AS for mutagenesis and ymlh1-31.S for identification of the point mutation.
pYI-pms1-61 TV II was constructed as follows. pYI-ypms1 TV II was generated by PCR to contain 686 bp upstream of the ATG codon to position 2426 of the yPMS1 ORF in pYI-lacZ. A PstI-BspMI fragment from pFB-ypms1-61 (see below) that contained the E61A codon mutation was used to replace the wild-type PstI-BspMI fragment of pYI-ypms1 TV II to create pYI-pms1-61 TV II. pYI-pms1-128 TV II was created similarly with a PstI-BspMI fragment from pFB-ypms1-128 (see below) that contained the G128A codon mutation. pYI-ypms1-61 and -128 TV II were both shown to be free of second site mutations in the germane regions by sequencing.
pΔmlh3::hisG-URA3-hisG was a kind gift from David Jacobson (Oregon Health Sciences University, Department of Molecular and Medical Genetics).
(ii) Two-hybrid vectors.
All of the following constructs were sequenced to confirm that point mutations were present in the desired plasmids. pNBTM116 was a generous gift of Stanley Hollenberg (Oregon Health Sciences University, Department of Cell and Developmental Biology) and allows construction of two-hybrid “bait” fusions with the lexA DNA binding domain fused at the carboxy terminus of the bait protein. pNBTM-mlh1 N-354 was engineered with the oligonucleotides ymlh1 N-anchor.S (5′-CGGGATCCATGTCTCTCAGAATAAAAGCAC-3′) and ymlh1 N-354.AS (5′-AGCCTCGAGCTCTGGCTTGTTTGTTGAAATTG-3′) to generate a PCR amplicon that was cloned into pNBTM116 at the BamHI and XhoI sites. Plasmid pNBTM-mlh1-31 N-354 was generated in an identical fashion, but the PCR was performed on template DNA that contained the E31A codon mutation.
pNBTM-pms1 N-401 and pNBTM-pms1-61 N-401 were generated by using a similar procedure to pNBTM-mlh1 N-354, but with oligonucleotides ypms1 N-anchor.S (5′-CGGGATCCAAAATGTTTCACCACATCGAAAAC-3′) and ypms1 N-401.AS (5′-AGCCTCGAGTTGTGAGCACATTCTTTTGGG-3′). The pNBTM-pms1-61, -128 N-401 double point mutant was made by using the Quikchange Site-Directed Mutagenesis kit (Stratagene) with plasmid pNBTM-pms1-61 N-401 and the ypms1-128.S and -.AS oligonucleotides.
pCAD3 analogous to pNBTM116 allows fusion of the GAL4 activation domain to the carboxy terminus of the “prey” protein (54). pCAD-mlh1 N-354 and alanine point mutant version E31A were constructed by cloning a PCR product generated from oligonucleotides ymlh1 N-anchor.S and ymlh1 N-354(BamHI).AS (5′-AGCGGATCCCTCTGGCTTGTTTGTTGAAATTG-3′) into pCAD3 at a BamHI site. Plasmids with the correct insert orientation were isolated for further study. The pCAD-mlh1-31, -98 N-354 double point mutant was made by using the Quikchange Site-Directed Mutagenesis kit (Stratagene) with construct pCAD-mlh1-31 N-354 and the ymlh1-98.S and -.AS oligonucleotides.
pCAD-pms1-61 N-401 was created in a likewise fashion, except that oligonucleotides ypms1 N-anchor.S and ypms1 N-401 (BamHI).AS (5′-AGCGGATCCTTGTGAGCACATTCTTTTGGG-3′) were used.
(iii) Baculovirus plasmids.
The 6×His-MLH1 recombinant baculovirus was constructed as follows. A PCR product was generated that engineered a 6×His affinity tag in frame with the MLH1 ORF after the initiator methionine. This 6×His-encoding PCR product was cloned into the BamHI and NdeI restriction sites of pBTM-MLH1, replacing approximately 360 bp of the native gene. Automated sequencing of the construct confirmed that the 6×His tag was in frame with the MLH1 ORF and that no PCR-generated mutations arose. The 6×His-MLH1 ORF was then cloned into pFastBac DUAL (pFBD) (Life Technologies) by using polylinker sites BamHI and SalI. Based upon mutator assays, the 6×His epitope-tagged Mlh1p functionally complemented an mlh1Δ strain (data not shown).
The PMS1 recombinant baculovirus was produced as follows. The PMS1 ORF was removed from genomic clone pJH480-PMS1 by using AseI and SalI restriction enzymes and ligated to a synthetic linker containing AseI- and NcoI-compatible overhangs. This PMS1 ORF ligation product was then cloned into pEAE51 at sites NcoI and SalI, replacing the MSH6 ORF (3). The PMS1 ORF was then excised with XhoI and SalI restriction enzymes and ligated into the pFastBac1 (pFB) (Life Technologies) polylinker at an XhoI site. A pFB-PMS1 construct in the desired orientation was identified and sequenced to examine the site of the synthetic linker.
Two-hybrid analysis and β-galactosidase assays.
Protein-protein interactions were assessed by the two-hybrid technique. Bait and prey plasmids were transformed into L40 and AMR70 yeast, respectively (69). L40 bait strains were mated with AMR70 prey strains as described previously (67). Growth on −Ura −Trp −Leu (−UTL) plates indicated efficiency of mating, while growth on −Trp −His −Ura −Leu −Lys (−THULL) plates indicated bait-prey interaction. Expression of a subset of constructs was confirmed by Western blotting of L40 strains with the indicated bait or prey. Extracts were made from 10-ml saturated cultures by glass bead lysis for 30 min at 4°C in a mixture of 25 mM Tris (pH 7.5), 1 mM EDTA, 10 mM β-mercaptoethanol (β-ME), 1 mM phenylmethylsulfonyl fluoride (PMSF), and Complete Proteolysis Inhibitor (Roche Molecular Biochemicals) and centrifuged at 14,000 × g for 5 min; concentrations of soluble protein fractions were determined by Bradford (Bio-Rad). Ten to 15 μg of each extract was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on 10% polyacrylamide gels, transferred to nylon membranes (Ambion), probed with the either anti-GAL4-TA (1:200 dilution) or anti-lexA-DB (1:200 dilution) (Santa Cruz Bio. Inc.) followed by the appropriate secondary antibody, and detected by chemiluminescence.
Diploid L40/AMR70 is homozygous for a second chromosomal lexA-GAL4A reporter system, URA3::(lexAop)8-lacZ. β-Galactosidase assays were performed on −THULL plates as described previously (67). Reaction mixtures were placed at 30°C until the desired blue color development was achieved.
Measurement of mutation rates and CAN1 mutational spectrum analysis.
Briefly, strains were streak purified before the mutation rate assay, individual colonies were grown to saturation in YPD, and then various dilutions were plated onto complete synthetic medium (CSM), −Thr and +canavanine (+CAN [60 μg/ml]) plates and colonies were counted after 2 to 3 days of growth at 30°C. Rates were determined as previously described (51). Statistical analyses were performed by using a two-tailed Mann-Whitney test with Prism 2.0a software (GraphPad Software, Inc.); P values of <0.05 were considered statistically significant.
Canavanine resistance (CanR) mutations were determined from genomic preparations by using the glass bead lysis method, followed by PCR of the CAN1 gene, as described previously (66), and direct sequencing of the QIAquick (Qiagen)-purified PCR amplicon with an ABI automated sequencer.
Expression and purification of yeast MutLα.
The Bac-to-Bac Baculovirus (Life Technologies) expression system was used to express MutLα in Spodoptera frugiperda (Sf9) cells infected with recombinant baculovirus. Recombinant baculoviruses that express 6×His-Mlh1p and Pms1p were created as described in the manufacturer's instructions (Bac-to-Bac Baculovirus expression system; Life Technologies). A 200-ml culture of Sf9 cells (typically 1 × 106 to 2 × 106 cells/ml) was coinfected with recombinant baculoviruses at multiplicities of infection of 2 to 2.5 and 11 to 15 for 6×His-Mlh1p- and Pms1p-expressing baculoviruses, respectively. Cells were harvested at 44 to 48 h of coinfection, frozen as cell pellets with liquid nitrogen, and stored at −80°C.
All subsequent steps were performed at 0 to 4°C, and purification was monitored by SDS-PAGE and Western blot analysis. Western blots were probed with a 1:1,000 dilution of anti-4×His monoclonal antibody (Qiagen) or a cross-reacting 1:100 dilution of anti-hPms2p polyclonal antibody and then visualized by using a 1:2,000 dilution of anti-mouse immunoglobulin G (mIgG)-horseradish peroxidase mIgG-(HRP) (Pierce) or a 1:1,250 dilution of anti-rat IgG (rIgG) rIgG-HRP (Bio-Rad), respectively, and Enhanced Luminol reagent (NEN). All buffers included 0.5 to 1 mM PMSF (Sigma Chemicals), 4 to 10 μg of leupeptin per ml (Sigma Chemicals), and 4 to 10 μg of aprotinin per ml (Sigma Chemicals). Cell pellets were resuspended in 5 ml of Sf9 lysis buffer per gram of wet cell pellet (Sf9 lysis buffer: 50 mM Tris-HCl [pH 7.6], 5 mM β-ME, 100 mM KCl, Complete-Mini EDTA Free pills [1 pill/5 or 10 ml] [Roche Molecular Biochemicals], 1% Nonidet P-40 [NP-40] [Sigma Chemicals]). The cell lysate was then spun at 10,000 × g for 10 min. The cleared lysate was incubated in a batch with 1.0 ml of a 50% slurry of Ni-nitrilotriacetic acid agarose resin (Qiagen) in buffer H (400 mM NaCl, 25 mM Tris-HCl [pH 7.8], 20% glycerol, 5 mM β-ME) plus 0.6 M (NH4)2SO4 and 10 to 15 mM imidazole (pH 8) for 1 h. The resin was washed in a batch three times: once with 40 ml of buffer H plus 0.6 M (NH4)2SO4 and 25 mM imidazole and twice with 40 ml of buffer H plus 0.6 M (NH4)2SO4 and 50 mM imidazole. The resin was loaded onto an Econo-column (Bio-Rad) and eluted with buffer H plus 0.5 M imidazole. Peak fractions were pooled, desalted into buffer T (50 mM Tris-HCl [pH 7.8], 10% glyercol, 1 mM dithiothreitol) plus 100 mM NaCl and 0.01% NP-40 by using PD-10 desalting columns (Amersham Pharmacia Biotechnologies), and further purified on a 1-ml HiTrap heparin column by using a 20-ml gradient from 100 mM to 1 M NaCl. Peak fractions were concentrated with Vivaspin 500 (50,000 molecular weight cutoff) (Vivascience, Ltd., Brinbrook Hill, United Kingdom) as described by the manufacturers. Concentrated fractions were frozen in liquid nitrogen and stored at −80°C. MutLα protein concentration was determined by scanning densitometry of a Coomassie blue-stained gel by using bovine serum albumin standards (Pierce) and analyzed with NIH image 1.61 software.
Limited proteolysis assays.
Limited proteolysis reaction mixtures (20 μl) consisted of 150 ng of MutLα, 30 mM Tris (pH 7.6), 150 mM NaCl, 5 mM MgCl2, and 0.5 mM dithioerythritol with or without 5 mM ATP, adenosine 5′-(β,γ-imino)triphosphate (AMP-PNP), ATPγS, or ADP. Reaction mixtures were incubated for 15 min at 30°C, followed by the addition of 50 ng of modified trypsin (Promega Corp.), incubation at 30°C for a specified interval, addition of SDS-sample buffer, and boiling for 7 min. Processed reactions were separated on an SDS-PAGE (10% polyacrylamide) gel and transferred onto polyvinylidene difluoride membranes (Ambion), and Western blotting was performed with specified antibodies. Anti-4×His Western blots were performed as described above. The anti-Mlh1p polyclonal antibody was a kind gift of T. Kunkel (National Institute of Environmental Health Sciences) and was generated against a COOH-terminal peptide of yeast Mlh1p. The anti-Mlh1p polyclonal antibody was used to probe limited proteolysis blots at a 1:10,000 dilution and detected as described above.
RESULTS
Predicted ATPase residues of MutLα are necessary for MMR in vivo.
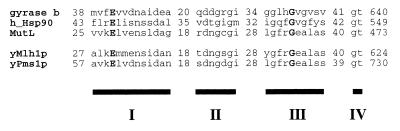
To examine the importance of putative ATPase domains of S. cerevisiae Mlh1p and Pms1p, we examined the effects of mutations introduced at two of the ATPase motifs, I and III, conserved in the GHL family (Fig. 1). We chose to examine residues E31 and E61 of Mlh1p and Pms1p, respectively, because mutations at the homologous glutamate of MutL, gyrase B, and Hsp90 have been shown to eliminate ATP hydrolysis with little or no effect on ATP binding (9, 10, 34, 47, 50). In motif III, we focused on residues G98 and G128 of Mlh1p and Pms1p, respectively, which are modeled to affect ATP binding and/or an associated conformational change induced upon ATP binding (9, 10, 26, 47, 50). For brevity, we will refer to alanine substitution mutations at E31 and E61 of Mlh1p and Pms1p, respectively, as hydrolysis mutations and the mutations G98A and G128A in Mlh1p and Pms1p, respectively, as ATP-binding mutations.
FIG. 1.
NH2-terminal ATPase domains of GHL ATPases. ATPase motifs I to IV are designated by black boxes, and sequences are shown above motif boxes. Numbers correspond to the number of amino acids preceding or following sequence alignments. Boldface letters are the absolutely conserved residues that were substituted for with alanine in Mlh1p and Pms1p.
To address whether the ATP hydrolysis and ATP-binding motifs of MutLα are necessary for mutation avoidance by MMR, double point mutants (e.g., mlh1-E31A pms1-E61A) were generated and examined for their spontaneous mutation rate (see Materials and Methods). We analyzed the effects of double hydrolysis and double binding mutants by using two mutator rate assays: reversion of hom3-10 and forward mutation at CAN1. Relative to the wild-type strain, both the double ATP hydrolysis mutant (strain PTY400) and the double ATP-binding mutant (strain PTY600) exhibited spontaneous mutation rates similar to those observed in mlh1Δ and pms1Δ strains (Table 2, compare strains PTY400 and PTY600 with PTY100 and PTY101). The mutator phenotype of a complex double mutant, mlh1 hydrolysis mutant plus pms1 binding mutant, or vice versa, was also similar to that of an MMR-deficient (e.g., mlh1Δ) strain (Table 2, compare strains PTY500 and PTY501 with PTY100 and PTY101). These data indicate that any combinations of double ATP hydrolysis and/or binding mutations affecting MutLα result in defects in the mutation avoidance functions of MMR comparable to the defects seen in mlh1Δ and pms1Δ strains.
TABLE 2.
Mutation rate effects of genomic mutations altering the NH2 termini of MutLα and MutLβ
| Straina | Relevant genotype | Biochemical deficiencyb | Fold mutator (rate ± SD)c
|
|
|---|---|---|---|---|
| hom3-10d | CanRe | |||
| GCY35 | Wild type | 1 | 1 | |
| PTY100 | mlh1Δ | 1,253 ± 301 | 29 ± 6 | |
| PTY101 | pms1Δ | 1,212 ± 79 | 28 ± 5 | |
| PTY102 | mlh3Δ | 4 ± 1 | 2 ± 1 | |
| PTY103 | mlh1Δmlh3Δ | 919 ± 13 | 20 ± 2 | |
| PTY104 | pms1Δmlh3Δ | 1,051 ± 199 | 27 ± 2 | |
| PTY400 | mlh1-E31A pms1-E61A | ATP hydrolysisf | 715 ± 291 | 26 ± 8 |
| PTY600 | mlh1-G98A pms1-G128A | ATP binding or heterodimerizationf | 831 ± 41 | 22 ± 2 |
| PTY200 | mlh1-E31A | ATP hydrolysisg | 311 ± 135 | 7 ± 2 |
| PTY201 | pms1-E61A | ATP hydrolysisg | 19 ± 8 | 1 ± 0.5 |
| PTY202 | pms1-E61A mlh3Δ | ATP hydrolysisg | 60 ± 31 | 2 ± 0.8 |
| PTY300 | mlh1-G98A | ATP binding or heterodimerizationg | 725 ± 126 | 22 ± 8 |
| PTY301 | pms1-G128A | ATP binding or heterodimerizationg | 78 ± 28 | 4 ± 1 |
| PTY302 | pms1-G128A mlh3Δ | ATP binding or heterodimerizationg | 159 ± 63 | 5 ± 0.6 |
| PTY500 | mlh1-E31A pms1-G128A | Complex effect (?)f | 681 ± 80 | 25 ± 14 |
| PTY501 | mlh1-G98A pms1-E61A | Complex effect (?)f | 651 ± 104 | 25 ± 6 |
Similar results for strains W303 (mlh1) and MW3317-21A (pms1) (unpublished data).
Experiments repeated two to five times with 5 to 11 cultures per experiment (Materials and Methods).
Wild-type GCY35 rate of 9.90 × 10−9.
Wild-type GCY35 rate of 3.01 × 10−7.
The entire MutLα heterodimer is affected.
Only one protomer of the heterodimer(s) is affected.
Mutator effects of single alterations in the putative ATPase domains of S. cerevisiae MutLα.
To examine the individual contributions of Mlh1p and Pms1p ATPase motifs to MutLα function, we examined the effect of single mutations on mutation avoidance. As shown in Table 2, the single mutations affecting the ATPase motifs had effects on mutation avoidance that were significantly less than the corresponding mlh1Δ and pms1Δ strains, with the exception of the mlh1 binding mutant (PTY300), the effect of which was only slightly less than the mlh1Δ strain (PTY100). Interestingly, the homologous ATPase mutations made in MLH1 and PMS1 had different effects on mutation avoidance. The mlh1 hydrolysis mutant (PTY200) displayed 16-fold and 7-fold higher rates of mutation at hom3-10 and CAN1, respectively, than the corresponding pms1 hydrolysis mutant strain PTY201 (P < 0.0286 for both loci). Likewise, the mlh1 binding mutant (PTY300) showed 9- and 5.5-fold higher rates of mutation at hom3-10 and CAN1, respectively, than the corresponding pms1 binding mutant strain PTY301 (P < 0.0286 for both loci). One trivial explanation for the differential effects of homologous mlh1 and pms1 mutations on mutation avoidance was that MLH3, which is involved in a minor mutation avoidance pathway (22), compensates for the pms1 mutations. However, as shown in Table 2, pms1 point mutant strains deleted for MLH3, PTY202 (pms1-E61A mlh3Δ), and PTY302 (pms1-G128A mlh3Δ) still demonstrated mutation rates significantly smaller than that observed for the respective homologous mlh1 ATPase point mutant strains PTY200 (mlh1-E31A) and PTY300 (mlh1-G98A) (P < 0.0159 for PTY200 versus PTY202 and P < 0.0286 for PTY300 versus PTY302). These results suggest that the differences seen between homologous mlh1 and pms1 ATPase point mutations with respect to mutation rate are not due to the redundant functions of MLH3, but rather infer an intrinsic asymmetry within the MutLα complex.
In addition to the differential effects of mlh1 versus pms1 mutants noted above, we also observed that ATP-binding mutations produced more severe effects on mutation avoidance than did ATP hydrolysis mutations. Both ATP-binding mutant strains PTY300 (mlh1-G98A) and PTY301 (pms1-G128A) exhibited a two- to fourfold higher rate of spontaneous mutation at hom3-10 and CAN1 relative to the hydrolysis mutants PTY200 (mlh1-E31A) and PTY201 (pms1-E61A), respectively (all comparisons had a P value of <0.0286). These results suggest that individual ATP binding mutations of Mlh1p and Pms1p produce greater effects on the mutation avoidance functions of MMR than the individual hydrolysis mutations.
To better define the effects of individual Mlh1p and Pms1p ATPase mutations on MMR, we examined the mutational spectra at the CAN1 reporter. CAN1 reports base substitutions, frameshifts, deletions, insertions, and large chromosomal rearrangements (15). As seen in Table 3, the spectra of deletion strains PTY100 (mlh1Δ), PTY101 (pms1Δ), and PTY104 (pms1Δ mlh3Δ) and point mutant strains PTY200 (mlh1-E31A) and PTY300 (mlh1-G98A) showed a preponderance of frameshift mutations (FS) relative to base substitutions (BS), similar to previously published reports for an msh2Δ strain (66). In contrast, strain PTY301 (pms1-G128A) showed a different spectrum, namely, a majority of BS, represented in Table 3 by an FS/BS ratio of 0.8. Because the mutation rate of PTY301 (pms1-G128A) for CANR is only fourfold greater than the wild-type rate (Table 2), one-quarter of the mutations seen with PTY301 represent the wild-type spectrum. Correcting for the wild-type contribution, we still observed a majority of base substitutions (10 of 18 [56%] versus 7.5 of 13.5 [56%]). Next, because MLH3 is partially redundant with PMS1 in correcting frameshift mutations, we examined the CAN1 spectrum in a pms1-G128A mlh3Δ strain (PTY302). As shown in Table 3, the pms1-G128A mlh3Δ strain (PTY302) showed a spectrum at CAN1 that was indistinguishable from that of an MMR-null strain (FS/BS ratio of 3.3). In contrast to the asymmetry observed with the mutation rates, the spectrum results indicate that the mlh1 and pms1 ATPase mutations result in the same mutational spectra.
TABLE 3.
Summary of mutation spectra at CAN1
| Strain | Relevant genotype | Result for class of mutation
|
FS/BS ratiob | |||
|---|---|---|---|---|---|---|
| FS
|
BS frequency (%) | Complex frequency (%) | ||||
| Frequency (%) | % Typea | |||||
| GCY35f | Wild type | 7/20 (35) | 86:14 | 11/20 (55) | 2/20 (10)c | 0.6 |
| PTY100 | mlh1Δ | 8/10 (80) | 100:0 | 2/10 (20) | NAd | 4.0 |
| PTY200 | mlh1-E31A | 12/20 (60) | 75:25 | 8/20 (40) | NA | 1.9e |
| PTY300 | mlh1-G98A | 17/20 (85) | 99:12 | 3/20 (15) | NA | 5.7 |
| PTY101 | pms1Δ | 8/10 (80) | 62:38 | 2/10 (20) | NA | 4.0 |
| PTY104 | pms1Δmlh3Δ | 8/10 (80) | 62:38 | 2/10 (20) | NA | 4.0 |
| PTY301 | pms1-G128A | 8/18 (44) | 87:13 | 10/18 (56) | NA | 0.8e |
| PTY302 | pms1-G128A mlh3Δ | 18/25 (72) | 94:6 | 7/25 (28) | NA | 3.3e |
Ratio of contractions to expansions.
FS/BS ratio, where a signature MMR-defective spectrum is ≥2.
Duplication events that are flanked by direct repeats.
NA, not applicable.
This value is the FS/BS ratio with the wild-type spectrum contribution subtracted.
The spectrum of an mlh3Δ strain is similar to that of the wild type (R. D. Kolodner, personal communication).
MutLα undergoes an ATP-dependent conformational change.
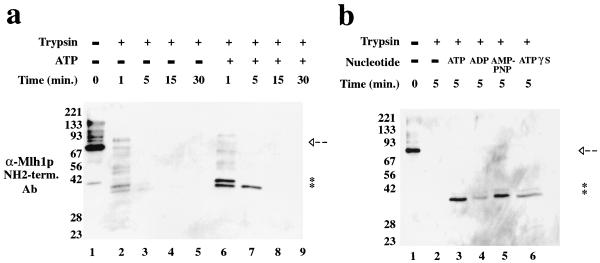
To investigate further the role of candidate ATP-binding or hydrolysis motifs in MutLα function, we used limited proteolysis to examine the effects of adenine nucleotides on the conformation of recombinant MutLα purified from insect cells (data not shown). We used an antibody directed against the 6×His tag at the NH2 terminus of Mlh1p to detect NH2-terminal fragments following limited proteolysis. As depicted in Fig. 2a, the presence of ATP led to the protection of distinct NH2-terminal fragments of Mlh1p from trypsin proteolysis. The protected NH2-terminal fragments of approximately 42 and 38 kDa coincide with the E. coli MutL LN40 thrombin proteolytic fragment that possessed the core ATPase domain (9, 10). We did not observe any differences between MutLα in the presence or absence of ATP when using a polyclonal antibody directed against the COOH terminus of Mlh1p (data not shown), suggesting that the COOH terminus of Mlh1p does not undergo an ATP-dependent conformational change. However, we did detect an approximately 30-kDa band that was resistant to proteolysis in the presence or absence of ATP even after a 30-min incubation with 750 ng of trypsin (data not shown). This highly trypsin-resistant Mlh1p COOH-terminal fragment may represent the COOH-terminal heterodimerization domain of Mlh1p. As shown in Fig. 2b, lanes 3 to 5 demonstrate that nonhydrolyzable ATP analogs AMP-PNP and ATPγS, as well as ADP, also protect the NH2 terminus of Mlh1p from trypsin proteolysis. Qualitatively, the relative levels of protection from proteolysis in the presence of nucleotide are as follows: ATP ≈ AMP-PNP > ATPγS > ADP. As demonstrated for ATP, we saw no differential protection of the COOH terminus of Mlh1p in the presence of ADP, AMP-PNP, or ATPγS by reprobing with the antibody directed against the COOH terminus of Mlh1p (data not shown). The limited proteolysis results indicate that at least the Mlh1p NH2 terminus of MutLα undergoes an ATP-binding-dependent conformational change. We were unable to address whether the NH2 terminus of Pms1p undergoes a similar ATP-dependent conformational change, because an antibody specific for the NH2 terminus of Pms1p antibody was not available. However, as described below, yeast two-hybrid results suggest that Pms1p also undergoes an ATP-dependent conformational change.
FIG. 2.
Adenine nucleotides alter trypsin sensitivity of MutLα. (a) One hundred fifty nanograms of MutLα was subjected to proteolysis with modified trypsin as described in Materials and Methods in the presence or absence of 5 mM ATP for the indicated time at 30°C. Products were treated with SDS-sample buffer, boiled, separated on an SDS-PAGE (10% polyacrylamide) gel, and detected by immunoblotting with anti-4×His antibody. Arrows denote full-length 6×His-Mlh1p, and asterisks designate NH2-terminal (term.) fragments of 6×His-Mlh1p that are protected from proteolysis in the presence of ATP. Equal loading of samples and even transfer of the blot were demonstrated by using a polyclonal antibody raised against the COOH terminus of Mlh1p (data not shown). (b) The same analysis was performed as described for panel a, but the effects of 5 mM adenine nucleotides ADP, AMP-PNP, and ATPγS were examined.
ATP binding promotes heterodimerization of the NH2 termini of MutLα in vivo.
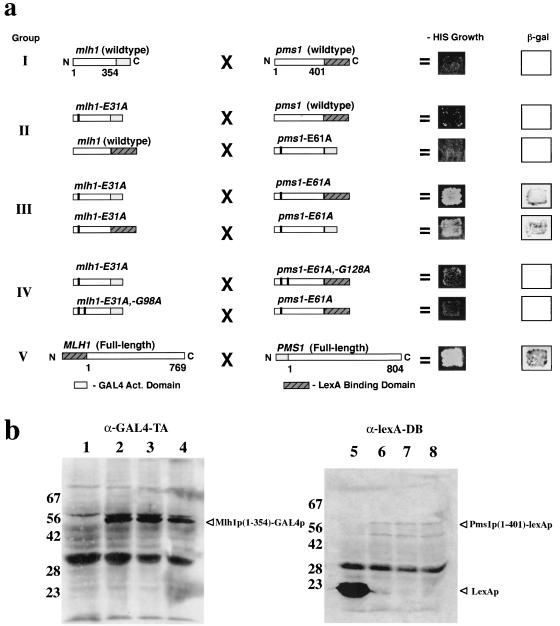
ATP-binding-dependent conformational changes in other GHL family members are associated with dimerization of their NH2-terminal ATP binding domains (9, 71). To inquire into the functional significance of the ATP-dependent conformational change data described above, we used the yeast two-hybrid system to assay interactions between wild-type and mutant NH2-terminal fragments of Mlh1p and Pms1p. Based upon sequence alignments with GHL family members, the fragments 1 to 354 and 1 to 401 of Mlh1p and Pms1p, respectively, should each contain the structural elements necessary for ATP binding and hydrolysis (9). These NH2-terminal fragments of Mlh1p and Pms1p were fused at their COOH termini to either the lexA DNA binding domain or the GAL4 activation domain (Fig. 3a). Consistent with our previous studies (51), no interaction was seen between wild-type Mlh1p and Pms1p NH2-terminal fragments (Fig. 3a, group I). As stated before, homologous ATP hydrolysis mutations in other GHL ATPases have been shown to abolish ATP hydrolysis activity with little or no effect on ATP binding (9, 10, 34, 47, 50). Interestingly, for an NH2-terminal fragment of gyrase b, ATP binding was only observed in vitro for a hydrolysis-deficient form (34). Therefore, we reasoned that ATP hydrolysis mutations in the NH2 termini of both Mlh1p and Pms1p might prolong a double ATP-bound state in vivo and allow interaction to be detected by the yeast two hybrid assay. Indeed, as shown in Fig. 3a (group III), a robust interaction was seen when both Mlh1p and Pms1p NH2-terminal fragments possessed ATP hydrolysis mutations. However, the interaction was not observed when only the Mlh1p or Pms1p fragment possessed the ATP hydrolysis mutation E31A or E61A, respectively (Fig. 3a, group II). To demonstrate that this novel interaction was dependent on the putative ATP-binding activities of Mlh1p and Pms1p, we superimposed either the mlh1-G98A or pms1-G128A ATP-binding mutation onto the hydrolysis-defective NH2-terminal fragments of mlh1p-E31A or pms1p-E61A, respectively. Supporting our hypothesis, we observed that superimposing a mutation designed to prevent the putative ATP-binding or conformational change in one fragment ablated the two-hybrid interaction (Fig. 3a, compare groups III and IV). Interestingly, the two-hybrid interaction seen in Fig. 3a, group III, was specific only for the mlh1p-E31A and pms1p-E61A NH2-terminal fusion pairs, because the NH2 terminus of mlh1p-E31A did not interact with itself (data not shown). The same observation was seen with the NH2 terminus of pms1p-E61A (data not shown), suggesting that, similar to their respective COOH-terminal domains (51), the NH2 termini of Mlh1p and Pms1p do not homodimerize. The two-hybrid results of Fig. 3a are not due to ATP hydrolysis or ATP-binding mutations grossly affecting expression or stability of the fusion proteins, because Western analysis demonstrates that all fusion proteins are expressed at similar levels (Fig. 3b). Taken together, the two-hybrid results suggest that ATP binding, but not hydrolysis, by both Mlh1p and Pms1p is necessary for MutLα NH2-terminal heterodimerization.
FIG. 3.
Two-hybrid analysis detects NH2-terminal Mlh1p and Pms1p interaction. (a) Boxes correspond to bait and prey constructs tested for interaction. The residues included in the fusions are indicated below the group I and V constructs, respectively. Amino acid substitutions designated above each construct are indicated by black bars within the construct boxes. Interaction is scored as growth on −HIS media and blue color development with the substrate X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described in Materials and Methods. Group I, wild-type NH2-terminal fusion fragments; group II, one NH2-terminal fusion fragment contains a hydrolysis point mutation; group III, both NH2-terminal fusion fragments contain hydrolysis point mutations; group IV, one NH2-terminal fusion fragment has the indicated compound mutations; and group V, positive control reaction with full-length Pms1p and Mlh1p. (b) Western analysis of L40 strains with two-hybrid constructs from panel a using anti-GAL4-TA or anti-lexA-DB monoclonal antibody as described in Materials and Methods. Lanes: 1, pCAD3 (empty vector); 2, pCAD-mlh1 N-354; 3, pCAD-mlh1-E31A N-354; 4, pCAD-mlh1-E31A, -G98A N-354; 5, pNBTM (lexA); 6, pNBTM-pms1 N-401; 7, pNBTM-pms1-E61A N-401; 8, pNBTM-pms1-E61A, -G128A N-401. Fusion products and lexAp are indicated by arrowheads. The approximately 90-kDa band in lanes 1 to 4 may be endogenous Gal4p. The other bands present in control lanes 1 and 5 and in lanes 2 to 4 and 6 to 8, respectively, represent nonspecificity by the primary and secondary antibodies. In lanes 6 to 8, the faster-migrating specific anti-lexA-DB reacting species is unknown, but may be a pms1p(1-401)-lexAp degradation product.
DISCUSSION
Although clearly crucial for MMR, little information exists on the function of the major MutL activity in yeast, MutLα, composed of Mlh1p and Pms1p. Previous studies with yeast have defined COOH-terminal domains as important for Mlh1p and Pms1p interaction (51) and conserved NH2-terminal residues as necessary for MMR activity (51, 63). Recent investigations of MutL and other members of the GHL family of ATPases have suggested guidelines for more detailed studies of MutLα function. Here, we present evidence defining yeast MutLα as a functional member of the GHL ATPase superfamily. First, residues critical for the ATPase function of GHL family members, when substituted for alanine in both Mlh1p and Pms1p, disrupt MMR. Second, adenine nucleotide binding protects the NH2 terminus of Mlh1p from trypsin proteolysis, suggesting that MutLα undergoes ATP-dependent conformational changes. Third, results from the two-hybrid system suggest that one consequence of the ATP-induced conformational changes is an interaction between the NH2 termini of Mlh1p and Pms1p. Finally, analysis of single mlh1 and pms1 ATPase motif mutants indicates a functional asymmetry within yeast MutLα.
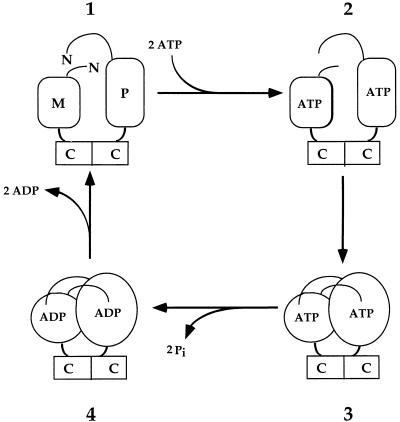
GHL family members appear to share an ATPase cycle that is highlighted by an NH2-terminal dimerized intermediate in the ATP-bound form (4, 9, 10, 55–57, 71). In the case of MutL, this ATP-binding-induced NH2-terminal dimerization activated MutH endonuclease in vitro (10). The limited proteolysis and two-hybrid analyses presented here support an ATPase cycle for MutLα, composed of at least four intermediates, that is similar to those of other GHL family members (Fig. 4). Limited proteolysis suggests that MutLα undergoes a conformational change in vitro that is dependent on ATP binding, as AMP-PNP (and to a lesser extent ATPγS) produced the same effect as ATP (Fig. 4, intermediate 2). Furthermore, we observed a specific two-hybrid interaction between Mlh1p and Pms1p NH2-terminal fragments, each containing ATP hydrolysis mutations (Fig. 4, intermediate 3). Interaction was not detected by a yeast two-hybrid assay with wild-type NH2-terminal fragments of Mlh1p and Pms1p, presumably because ATP hydrolysis renders the interaction transient. In further support of the existence of intermediate 3, our recent observations suggest that the double hydrolysis mutant form of MutLα is highly resistant to trypsin-limited proteolysis even without added ATP (unpublished observations). Also, that ADP provided some protection from limited proteolysis suggests the existence of an ADP-bound intermediate (Fig. 4, intermediate 4). Double mutant mlh1 pms1 strains with alanine substitutions at ATP hydrolysis or ATP-binding residues showed increased spontaneous mutation rates indistinguishable from those of completely MMR-defective cells. This double mutant analysis suggests that the candidate ATPase domains of both Mlh1p and Pm1p and the ATPase cycle described above are required for MutLα function in yeast MMR for mutation avoidance. It is intriguing to speculate on the function of the NH2-terminal ATP-bound MutLα intermediate, because similar findings from other GHL ATPases (25, 55–57), namely MutL (10), suggest that this MutLα intermediate may play a significant role in coordinating downstream steps with known and perhaps unidentified MMR proteins. Although our data are consistent with the ATPase cycle represented in Fig. 4, further biochemical work with MutLα is required to confirm and characterize the contribution of Mlh1p and Pms1p ATP binding and hydrolysis activities to MutLα function. Similar to earlier work from the Hsp90 field (35, 61), we have not been able to specifically assign an intrinsic ATPase activity to MutLα with our current protein preparations (unpublished observations). However, similar to what is currently known for GHL ATPases (9, 10, 34, 47, 50), our double mlh1 pms1 hydrolysis mutant phenotype and our two-hybrid results suggest a crucial role for ATP hydrolysis during mutation avoidance in yeast MMR (Table 2).
FIG. 4.
A model for the yeast MutLα ATPase cycle. Briefly, intermediate 1 is the nucleotide-free state. ATP binding induces conformational changes in the NH2 termini of Mlh1p and Pms1p, represented by a change in shape from rectangular to oval that occurs in the step(s) between intermediates 2 and 3. Intermediate 3 is heterodimerization of the NH2 termini of Mlh1p and Pms1p in the ATP-bound state. Intermediate 4 is the ADP-bound form following ATP hydrolysis. The mlh1-G98A and pms1-G128A ATP-binding mutants were constructed to affect the transition(s) from intermediate 1 to intermediate 2 and/or intermediate 2 to intermediate 3. In contrast, the ATP hydrolysis mutations, mlh1-E31A and pms1-E61A, were modeled to prevent the transition from intermediate 3 to intermediate 4. M, the NH2 terminus of Mlh1p; P, the NH2 terminus of Pms1p; C, COOH termini of Mlh1p and Pms1p. Each arrow may represent multiple distinct steps. This model, which is consistent with the studies reported here, was adapted from a model for MutL proposed by Ban and Yang (9).
Interestingly, genetic analysis revealed a functional asymmetry with respect to the two ATPase domains of the MutLα heterodimer. Specifically, alanine substitution mutations affecting the predicted ATPase motifs of Mlh1p had a greater impact on mutation avoidance than the corresponding mutations in Pms1p. Formally, our genetic results argue that Mlh1p can compensate better for ATPase mutations in Pms1p for mutation avoidance than can Pms1p for the corresponding ATPase mutations in Mlh1p. The apparent genetic asymmetry detected for MutLα may reflect at the mechanistic level a kinetic asymmetry similar to that observed in the homodimeric ATPases, topoisomerase II from S. cerevisiae, and the γ complex from E. coli (6, 28, 29). Biologically, the genetic asymmetry observed with MutLα may represent distinct but overlapping roles of Mlh1p and Pms1p during mutation avoidance, e.g., excision tracts originating 5′ versus 3′ from the mismatch or IDL (18, 46) or differential roles during strand discrimination.
MutL has been referred to as a “molecular matchmaker,” coupling the mismatch binding activity of MutS to the latent endonuclease MutH (60). One criterion of a molecular matchmaker that MutL has always appeared to lack was an intrinsic ATPase activity. Recent work has now identified this “missing” activity, and as suggested previously, it appears to be critical for MutL activity in MMR (5, 9, 10). Moreover, the MutL ATPase activity was responsible for coordinated interaction and activation of MutH in vitro (10). In this report, we have shown that the conserved ATPase motifs of MutLα are necessary for mutation avoidance by MMR in yeast. The role of the ATPase motifs of Mlh1p and Pms1p in other MMR-related functions, such as meiotic (7, 8, 30, 70) and homeologous recombination (16, 17, 31, 62), remains to be determined. Finally, as for MutL (9, 10), the ATP-dependent conformational changes in yeast MutLα are likely to facilitate interaction with downstream proteins in MMR. Our ability to produce a stable Mlh1p-Pms1p NH2-terminal interaction via the yeast two-hybrid system may provide a means to identify these proteins.
ACKNOWLEDGMENTS
This work was supported by NSF grant MCB9631061 to R.M.L. and OHSU Molecular Hematology Training grant 5-T32-HL07781 to P.T.T.
We thank Andrew Buermeyer, Suzanne Deschênes, Guy Tomer, and Betsy Ferguson for helpful comments on the manuscript and Sandra Dudley for expert technical assistance with the mutational spectra. Special thanks go to Eric Alani and Jayson Bowers for their superb technical advice on the MutLα purification.
REFERENCES
- 1.Alani E, Cao L, Kleckner N. A method for gene disruption that allows repeated use of URA3 selection in the construction of multiply disrupted yeast strains. Genetics. 1987;116:541–545. doi: 10.1534/genetics.112.541.test. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alani E, Chi N-W, Kolodner R. The Saccharomyces cerevisiae Msh2 protein specifically binds to duplex oligonucleotides containing mismatched DNA base pairs and insertions. Genes Dev. 1995;9:234–247. doi: 10.1101/gad.9.2.234. [DOI] [PubMed] [Google Scholar]
- 3.Alani E, Sokolsky T, Studamire B, Miret J J, Lahue R S. Genetic and biochemical analysis of Msh2p-Msh6p: role of ATP hydrolysis and Msh2p-Msh6p subunit interactions in mismatch base pair recognition. Mol Cell Biol. 1997;17:2436–2447. doi: 10.1128/mcb.17.5.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ali J A, Jackson A P, Howells A J, Maxwell A. The 43-kilodalton N-terminal fragment of the DNA gyrase b protein hydrolyzes ATP and binds coumarin drugs. Biochemistry. 1993;32:2717–2724. doi: 10.1021/bi00061a033. [DOI] [PubMed] [Google Scholar]
- 5.Aronshtam A, Marinus M G. Dominant negative mutator mutations in the mutL gene of Escherichia coli. Nucleic Acids Res. 1996;24:2498–2504. doi: 10.1093/nar/24.13.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baird C L, Harkins T T, Morris S K, Lindsley J E. Topoisomerase II drives DNA transport by hydrolyzing one ATP. Proc Natl Acad Sci USA. 1999;96:13685–13690. doi: 10.1073/pnas.96.24.13685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baker S, Plug A, Prolla T, Bronner C, Harris A, Yao X, Christie D-M, Monell C, Arnheim N, Bradley A, Ashley T, Liskay R. Involvement of Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet. 1996;13:336–342. doi: 10.1038/ng0796-336. [DOI] [PubMed] [Google Scholar]
- 8.Baker S M, Bronner C E, Zhang L, Plug A W, Robatzek M, Warren G, Elliott E A, Yu J, Arnheim A T N, Flavell R A, Liskay R M. Male mice defective in the DNA mismatch repair gene PMS2 exhibit abnormal chromosome synapsis in meiosis. Cell. 1995;82:309–319. doi: 10.1016/0092-8674(95)90318-6. [DOI] [PubMed] [Google Scholar]
- 9.Ban C, Junop M, Yang W. Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell. 1999;97:85–97. doi: 10.1016/s0092-8674(00)80717-5. [DOI] [PubMed] [Google Scholar]
- 10.Ban C, Yang W. Crystal structure and ATPase activity of MutL: implications for DNA repair and mutagenesis. Cell. 1998;95:541–552. doi: 10.1016/s0092-8674(00)81621-9. [DOI] [PubMed] [Google Scholar]
- 11.Bergerat A, de Massy B, Gadelle D, Varoutas P C, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 12.Bowers J, Sokolsky T, Quach T, Alani E. A mutation in the MSH6 subunit of the Saccharomyces cerevisiae MSH2-MSH6 complex disrupts mismatch recognition. J Biol Chem. 1999;274:16115–16125. doi: 10.1074/jbc.274.23.16115. [DOI] [PubMed] [Google Scholar]
- 13.Bronner C E, Baker S M, Morrison P T, Warren G, Smith L G, Lescoe M K, Kane M, Earabino C, Lipford J, Lindblom A, Tannergard P, Bollag R J, Godwin A R, Ward D C, Nordenskjold M, Fishel R, Kolodner R, Liskay R M. Mutation in the DNA mismatch repair gene homologue hMLH1 is associated with hereditary nonpolyposis colon cancer. Nature. 1994;368:258–261. doi: 10.1038/368258a0. [DOI] [PubMed] [Google Scholar]
- 14.Buermeyer A B, Deschenes S M, Baker S M, Liskay R M. Mammalian DNA mismatch repair. Annu Rev Genet. 1999;33:533–564. doi: 10.1146/annurev.genet.33.1.533. [DOI] [PubMed] [Google Scholar]
- 15.Chen C, Merrill B J, Lau P J, Holm C, Kolodner R D. Saccharomyces cerevisiae pol30 (proliferating cell nuclear antigen) mutations impair replication fidelity and mismatch repair. Mol Cell Biol. 1999;19:7801–7815. doi: 10.1128/mcb.19.11.7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen W, Jinks-Robertson S. The role of the mismatch repair machinery in regulating mitotic and meiotic recombination between diverged sequences in yeast. Genetics. 1999;151:1299–1313. doi: 10.1093/genetics/151.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Datta A, Hendrix M, Lipsitch M, Jinks-Robertson S. Dual roles for DNA sequence identity and the mismatch repair system in the regulation of mitotic crossing-over in yeast. Proc Natl Acad Sci USA. 1997;94:9757–9762. doi: 10.1073/pnas.94.18.9757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Drummond J T, Anthoney A, Brown R, Modrich P. Cisplatin and adriamycin resistance are associated with MutLalpha and mismatch repair deficiency in an ovarian tumor cell line. J Biol Chem. 1996;271:19645–19648. doi: 10.1074/jbc.271.33.19645. [DOI] [PubMed] [Google Scholar]
- 19.Drummond J T, Li G-M, Longley M J, Modrich P. Isolation of an hMSH2-p160 heterodimer that restores DNA mismatch repair to tumor cells. Science. 1995;268:1909–1912. doi: 10.1126/science.7604264. [DOI] [PubMed] [Google Scholar]
- 20.Dutta I, Inouye I. GHKL, an emergent ATPase/kinase superfamily. Trends Biochem Sci. 2000;25:24–28. doi: 10.1016/s0968-0004(99)01503-0. [DOI] [PubMed] [Google Scholar]
- 21.Fishel R A, Lescoe M K, Rao M R S, Copeland N, Jenkins N, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 22.Flores-Rozas H, Kolodner R D. The Saccharomyces cerevisiae MLH3 gene functions in MSH3-dependent suppression of frameshift mutations. Proc Natl Acad Sci USA. 1998;95:12404–12409. doi: 10.1073/pnas.95.21.12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Genschel J, Littman S J, Drummond J T, Modrich P. Isolation of MutSbeta from human cells and comparison of the mismatch repair specificities of MutSbeta and MutSalpha. J Biol Chem. 1998;273:19895–19901. doi: 10.1074/jbc.273.31.19895. [DOI] [PubMed] [Google Scholar]
- 24.Gietz R D, Schiestl R H. Applications of high efficiency lithium acetate transformation of intact yeast cells using single-stranded nucleic acids as carrier. Yeast. 1991;7:253–263. doi: 10.1002/yea.320070307. [DOI] [PubMed] [Google Scholar]
- 25.Grenert J P, Johnson B D, Toft D O. The importance of ATP binding and hydrolysis by hsp90 in formation and function of protein heterocomplexes. J Biol Chem. 1999;274:17525–17533. doi: 10.1074/jbc.274.25.17525. [DOI] [PubMed] [Google Scholar]
- 26.Grenert J P, Sullivan W P, Fadden P, Haystead T A J, Clark J, Mimnaugh E, Krutzsch H, Ochel H J, Schulte T W, Sausville E, Neckers L M, Toft D O. The amino-terminal domain of heat shock protein 90 (hsp90) that binds geldanamycin is an ATP/ADP switch domain that regulates hsp90 conformation. J Biol Chem. 1997;272:23843–23850. doi: 10.1074/jbc.272.38.23843. [DOI] [PubMed] [Google Scholar]
- 27.Habraken Y, Sung P, Prakash L, Prakash S. Enhancement of MSH2-MSH3-mediated mismatch recognition by the yeast MLH1-PMS1 complex. Curr Biol. 1997;7:790–793. doi: 10.1016/s0960-9822(06)00337-x. [DOI] [PubMed] [Google Scholar]
- 28.Harkins T T, Lewis T J, Lindsley J E. Pre-steady-state analysis of ATP hydrolysis by Saccharomyces cerevisiae DNA topoisomerase II. 2. Kinetic mechanism for the sequential hydrolysis of two ATP. Biochemistry. 1998;37:7299–7312. doi: 10.1021/bi9729108. [DOI] [PubMed] [Google Scholar]
- 29.Hingorani M M, Bloom L B, Goodman M F, O'Donnell M. Division of labor—sequential ATP hydrolysis drives assembly of a DNA polymerase sliding clamp around DNA. EMBO J. 1999;18:5131–5144. doi: 10.1093/emboj/18.18.5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter N, Borts R H. Mlh1 is unique among mismatch repair proteins in its ability to promote crossing-over during meiosis. Genes Dev. 1997;11:1573–1582. doi: 10.1101/gad.11.12.1573. [DOI] [PubMed] [Google Scholar]
- 31.Hunter N, Chambers S R, Louis E J, Borts R H. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. EMBO J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- 32.Iaccarino I, Marra G, Palombo F, Jiricny J. hMSH2 and hMSH6 play distinct roles in mismatch binding and contribute differently to the ATPase activity of hMutSalpha. EMBO J. 1998;17:2677–2686. doi: 10.1093/emboj/17.9.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iaccarino I, Palombo F, Drummond J, Totty N F, Hsuan J J, Modrich P, Jiricny J. MSH6, a Saccharomyces cerevisiae protein that binds to mismatches as a heterodimer with MSH2. Curr Biol. 1996;6:484–486. doi: 10.1016/s0960-9822(02)00516-x. [DOI] [PubMed] [Google Scholar]
- 34.Jackson A P, Maxwell A. Identifying the catalytic residue of the ATPase reaction of DNA gyrase. Proc Natl Acad Sci USA. 1993;90:11232–11236. doi: 10.1073/pnas.90.23.11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jakob U, Scheibel T, Bose S, Reinstein J, Buchner J. Assessment of the ATP binding properties of Hsp90. J Biol Chem. 1996;271:10035–10041. doi: 10.1074/jbc.271.17.10035. [DOI] [PubMed] [Google Scholar]
- 36.Jiricny J. Eukaryotic mismatch repair: an update. Mutat Res. 1998;409:107–121. doi: 10.1016/s0921-8777(98)00056-1. [DOI] [PubMed] [Google Scholar]
- 37.Johnson R E, Kovvali G K, Prakash L, Prakash S. Requirement of the yeast MSH3 and MSH6 genes for MSH2-dependent genomic stability. J Biol Chem. 1996;271:7285–7288. doi: 10.1074/jbc.271.13.7285. [DOI] [PubMed] [Google Scholar]
- 38.Kolodner R D, Marsischky G T. Eukaryotic DNA mismatch repair. Curr Opin Genet Dev. 1999;9:89–96. doi: 10.1016/s0959-437x(99)80013-6. [DOI] [PubMed] [Google Scholar]
- 39.Leach F S, Nicolaides N C, Papadopoulos N, Liu B, Jen J, Parsons R, Peltomaki P, Sistonen P, Aaltonen L A, Nystrom-Lahti M, Guan X-Y, Zhang J, Meltzer P S, Yu J-W, Kao F-T, Chen D J, Cerosaletti K M, Fournier R E K, Todd S, Lewis T, Leach R J, Naylor S L, Weissenbach J, Mecklin J-P, Jarvinen H, Petersen G M, Hamilton S R, Green J, Jass J, Watson P, Lynch H T, Trent J M, de la Chapelle A, Kinzler K W, Vogelstein B. Mutations of a mutS homolog in hereditary nonpolyposis colorectal cancer. Cell. 1993;75:1215–1225. doi: 10.1016/0092-8674(93)90330-s. [DOI] [PubMed] [Google Scholar]
- 40.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 41.Marsischky G T, Filosi M, Kane M F, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- 42.Marsischky G T, Kolodner R D. Biochemical characterization of the interaction between the Saccharomyces cerevisiae MSH2-MSH6 complex and mispaired bases in DNA. J Biol Chem. 1999;274:26668–26682. doi: 10.1074/jbc.274.38.26668. [DOI] [PubMed] [Google Scholar]
- 43.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu Rev Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 44.Mushegian A R, Bassett D E, Jr, Boguski M S, Bork P, Koonin E V. Positionally cloned human disease genes: patterns of evolutionary conservation and functional motifs. Proc Natl Acad Sci USA. 1997;94:5831–5836. doi: 10.1073/pnas.94.11.5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.New L, Liu K, Crouse G F. The yeast gene MSH3 defines a new class of eukaryotic MutS homologues. Mol Gen Genet. 1993;239:97–108. doi: 10.1007/BF00281607. [DOI] [PubMed] [Google Scholar]
- 46.Nicolaides N C, Littman S J, Modrich P, Kinzler K W, Vogelstein B. A naturally occurring hPMS2 mutation can confer a dominant negative mutator phenotype. Mol Cell Biol. 1998;18:1635–1641. doi: 10.1128/mcb.18.3.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Obermann W M J, Sondermann H, Russo A A, Pavletich N P, Hartl F U. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J Cell Biol. 1998;143:901–910. doi: 10.1083/jcb.143.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Palombo F, Gallinari P, Iaccarino I, Lettier T, Hughes M, D'Arrigo A, Truong O, Hsuan J J, Jiricny J. GTBP, a 160-kilodalton protein essential for mismatch-binding activity in human cells. Science. 1995;268:1912–1914. doi: 10.1126/science.7604265. [DOI] [PubMed] [Google Scholar]
- 49.Palombo F, Iaccarino I, Nakajima E, Ikejima M, Shimada T, Jiricny J. hMutSbeta, a heterodimer of hMSH2 and hMSH3, binds to insertion/deletion loops in DNA. Curr Biol. 1996;6:1181–1184. doi: 10.1016/s0960-9822(02)70685-4. [DOI] [PubMed] [Google Scholar]
- 50.Panaretou B, Prodromou C, Roe S M, O'Brien R, Ladbury J E, Piper P W, Pearl L H. ATP binding and hydrolysis are essential to the function of the Hsp90 molecular chaperone in vivo. EMBO J. 1998;17:4829–4836. doi: 10.1093/emboj/17.16.4829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pang Q, Prolla T A, Liskay R M. Functional domains of the Saccharomyces cerevisiae Mlh1p and Pms1p DNA mismatch repair proteins and their relevance to hereditary nonpolyposis colorectal cancer-associated mutations. Mol Cell Biol. 1997;17:4465–4473. doi: 10.1128/mcb.17.8.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Papadopoulos N, Nicolaides N C, Wei Y-F, Ruben S M, Carter K C, Rosen C A, Haseltine W A, Fleischmann R D, Fraser C M, Adams M D, Venter J C, Hamilton S R, Petersen G M, Watson P, Lynch H T, Peltomaki P, Mecklin J-P, de la Chapelle A, Kinzler K W, Vogelstein B. Mutation of a mutL homolog in hereditary colon cancer. Science. 1994;263:1625–1629. doi: 10.1126/science.8128251. [DOI] [PubMed] [Google Scholar]
- 53.Petes T D, Malone R E, Symington L S. Recombination in yeast. In: Broach J, Jones E, Pringle J, editors. The molecular and cellular biology of the yeast Saccharomyces. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1991. pp. 407–521. [Google Scholar]
- 54.Printen J A, Sprague G F., Jr Protein-protein interactions in the yeast pheromone response pathway: Ste5p interacts with all members of the MAP kinase cascade. Genetics. 1994;138:609–619. doi: 10.1093/genetics/138.3.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Prodromou C, Roe S M, O'Brien R, Ladbury J E, Piper P W, Pearl L H. Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell. 1997;90:65–75. doi: 10.1016/s0092-8674(00)80314-1. [DOI] [PubMed] [Google Scholar]
- 56.Prodromou C, Roe S M, Piper P W, Pearl L H. A molecular clamp in the crystal structure of the N-terminal domain of the yeast Hsp90 chaperone. Nat Struct Biol. 1997;4:477–482. doi: 10.1038/nsb0697-477. [DOI] [PubMed] [Google Scholar]
- 57.Prodromou C, Siligardi G, O'Brien R, Woolfson D N, Regan L, Panaretou B, Ladbury J E, Piper P W, Pearl L H. Regulation of Hsp90 ATPase activity by tetratricopeptide repeat (TPR)-domain co-chaperones. EMBO J. 1999;18:754–762. doi: 10.1093/emboj/18.3.754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Prolla T A, Christie D-M, Liskay R M. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prolla T A, Pang Q, Alani E, Kolodner R D, Liskay R M. Interactions between the Msh2, Mlh1 and Pms1 proteins during the initiation of DNA mismatch repair. Science. 1994;265:1091–1093. doi: 10.1126/science.8066446. [DOI] [PubMed] [Google Scholar]
- 60.Sancar A, Hearst J E. Molecular matchmakers. Science. 1993;259:1415–1420. doi: 10.1126/science.8451638. [DOI] [PubMed] [Google Scholar]
- 61.Scheibel T, Neuhofen S, Weikl T, Mayr C, Reinstein J, Vogel P D, Buchner J. ATP-binding properties of human Hsp90. J Biol Chem. 1997;272:18608–18613. doi: 10.1074/jbc.272.30.18608. [DOI] [PubMed] [Google Scholar]
- 62.Selva E M, New L, Crouse G F, Lahue R S. Mismatch correction acts as a barrier to homeologous recombination in Saccharomyces cerevisiae. Genetics. 1995;139:1175–1188. doi: 10.1093/genetics/139.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shcherbakova P V, Kunkel T A. Mutator phenotypes conferred by MLH1 overexpression and by heterozygosity for mlh1 mutations. Mol Cell Biol. 1999;19:3177–3183. doi: 10.1128/mcb.19.4.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Studamire B, Quach T, Alani E. Saccharomyces cerevisiae Msh2p and Msh6p ATPase activities are both required during mismatch repair. Mol Cell Biol. 1998;18:7590–7601. doi: 10.1128/mcb.18.12.7590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugino A, Cozzarelli N R. The intrinsic ATPase of DNA gyrase. J Biol Chem. 1980;255:6299–6306. [PubMed] [Google Scholar]
- 66.Tishkoff D X, Filosi N, Gaida G M, Kolodner R D. A novel mutation avoidance mechanism dependent on S. cerevisiae RAD27 is distinct from DNA mismatch repair. Cell. 1997;88:253–263. doi: 10.1016/s0092-8674(00)81846-2. [DOI] [PubMed] [Google Scholar]
- 67.Umar A, Buermeyer A B, Simon J A, Thomas D C, Clark A B, Liskay R M, Kunkel T A. Requirement for PCNA in DNA mismatch repair at a step preceding DNA resynthesis. Cell. 1996;87:65–73. doi: 10.1016/s0092-8674(00)81323-9. [DOI] [PubMed] [Google Scholar]
- 68.Viswanathan M, Lovett S T. Single-strand DNA-specific exonucleases in Escherichia coli. Roles in repair and mutation avoidance. Genetics. 1998;149:7–16. doi: 10.1093/genetics/149.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vojtek A B, Hollenberg S M, Cooper J A. Mammalian ras interacts directly with the serine/threonine kinase raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 70.Wang T F, Kleckner N, Hunter N. Functional specificity of MutL homologs in yeast: evidence for three Mlh1-based heterocomplexes with distinct roles during meiosis in recombination and mismatch correction. Proc Natl Acad Sci USA. 1999;96:13914–13919. doi: 10.1073/pnas.96.24.13914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wigley D B, Davies G J, Dodson E J, Maxwell A, Dodson G. Crystal structure of an N-terminal fragment of the DNA gyrase B protein. Nature. 1991;351:624–629. doi: 10.1038/351624a0. [DOI] [PubMed] [Google Scholar]