Abstract
Objective:
To investigate whether variations in CIDEA mRNA expression and protein levels are modulated by the pattern of abdominal fat distribution, in adolescent girls with obesity.
Methods:
We recruited 35 adolescent girls with obesity and characterized their abdominal fat distribution by MRI. Subjects had only periumbilical/abdominal (n=14) or paired abdominal and gluteal SAT biopsy (n=21). CIDEA expression were determined by RT-PCR, CIDEA protein level by western blot, and the turnover of adipose lipids and adipocytes by 2H2O labeling. In 6 girls, we performed a second abdominal SAT biopsy (after ~34.2 months) to explore the weight gain effect on CIDEA expression in abdominal SAT.
Results:
CIDEA expression decreased in abdominal SAT from subjects with high VAT/(VAT+SAT); CIDEA inversely correlated with number of small adipocytes, with the increase in pre-adipocyte proliferation, and with adipogenesis. We also found a strong inverse correlation between CIDEA protein level with the newly synthetized glycerol (r= − 0.839, p= 0.0047). Following weight gain, we observed an increase in adipocytes cell diameter with a decrease in CIDEA expression and RNA-seq transcriptomic profile typical of adipocyte dysfunction.
Conclusions:
A reduced expression of CIDEA in girls with high VAT/(VAT+SAT) is associated with adipocyte hypertrophy and insulin resistance.
Keywords: Obesity, adipocytes, subcutaneous adipose tissue, insulin resistance, Adolescents
Introduction
In the US about 20% of adolescents are obese (1). Adolescent obesity frequently persists into adulthood (2) and is associated with type 2 diabetes and fatty liver (3, 4, 5, 6). Studies from our group (3, 5, 7, 8, 9) showed that in girls with obesity, the high ratio of visceral AT depot (VAT) to abdominal subcutaneous AT depot (SAT) (High VAT/(VAT+SAT)) is associated with a profound insulin resistance (3, 8, 10), ultimately increasing the risk for type 2 diabetes (3, 11, 12). Furthermore, insulin resistance seen in these girls with the unhealthy metabolic profile, is characterized by an increased in vivo rate of lipolysis in abdominal SAT, a higher mature adipocyte turnover and a strong association between increased lipolysis and fatty liver development (13). However, the molecular changes associated with AT dysfunction in subjects with high VAT/(VAT+SAT) still remain uncharacterized.
CIDEA (Cell Death-Inducing DFFA-like Effector A), a component of the CIDE-family, is mainly located on the surface of lipid droplets (14, 15). First described as specific activator/inducer of apoptosis in mammalian cells (16, 17), more recently CIDEA’s important role in lipid droplets formation and lipid accumulation emerged (14, 17, 18, 19), being highly expressed in human white AT (20, 21, 22) and mouse beige/brown AT (20, 21, 23). In contrast to the literature reporting that high levels of CIDEA expression improve insulin sensitivity in both adults (14) and humanized mouse models (24) and protect from diet-induced obesity in a mouse model (20), little is known about the expression of CIDEA in AT from adolescents with obesity and different patterns of body fat distribution. Herein, we hypothesized that variations in CIDEA mRNA expression and protein levels would be modulated by the pattern of abdominal fat distribution, and closely partake in the development of insulin resistance. To test this hypothesis we investigated: 1) both the mRNA expression and protein levels of CIDEA in adolescent girls with obesity and different VAT/(VAT+SAT) levels, 2) explored possible correlations between CIDEA expression level with key markers of insulin resistance, cell sizing measurements, as well as the role of CIDEA in the regulation of lipolysis in SAT adipocytes; 3) assessed, by western blot, the phosphorylation of ATGL and HSL, two key enzymes for lipolysis in adipocytes (25, 26), and 4) determined the effects of natural increases in weight gain on changes in CIDEA expression, in a group of girls that consented to have a baseline and follow-up SAT biopsy after weight gain.
Methods
Study participants.
From the “Prevalence of Carbohydrate intolerance in obese children” (NCT0197849) multiethnic cohort (12), we recruited 35 adolescent girls with obesity between the ages of 12 and 20 years, who agreed to have a periumbilical SAT biopsy or a paired periumbilical and gluteal SAT biopsy (n=21). Using the median of the VAT/(VAT+SAT) ratio of the entire large cohort, the subjects were separated into two groups: low (<0.0972) and high (>0.0972) VAT/(VAT+SAT) (12). Six subjects consented for a follow-up abdominal SAT biopsy after a median of 34.2 months (95% CI: 18.8–83.9). None of the subjects received treatment with any medications. Clinical and metabolic characteristics on 11 subjects have been previously reported (13).
Oral Glucose Tolerance Test (OGTT).
After 12-hour overnight fasting, all participants underwent a 3-hour OGTT at the Yale Center for Clinical Investigation (YCCI) (7). The insulin sensitivity index (WBISI) was calculated using the formula by Matsuda (27).
Abdominal MRI and total body composition (DEXA).
Multi-slice abdominal MRI studies were performed on a Siemens Sonata 1.5 Tesla system (7). Liver fat content was measured by MRI using the Proton Density Fat Fraction method (28). Total body composition was measured by DEXA with a Hologic scanner (Hologic, Boston, MA).
Deuterated water (D2O) labeling protocol (NCT02395003).
Selected subjects were enrolled into an 8-week 70% D2O labeling protocol prior to the biopsies (13). Briefly, to achieve an enrichment around 1.0% in body water pool, each subject returned to YCCI and drank two doses of 70 mL of D2O within 3–4 hours apart. Thereafter, each subject drank 40 ml aliquots of D2O three times a day for 5 days and then twice a day for the remainder of the 8-weeks labeling period (29, 30, 31). Compliance with D2O intake was checked weekly by evaluating D2O enrichments in urine.
Biopsy of Subcutaneous Adipose Tissue (SAT) (NCT02395003).
The abdominal or paired abdominal and gluteal SAT biopsy was performed under sterile conditions at Yale YCCI-Hospital Research Unit (YCCI-HRU) after administration of local anesthesia (lidocaine without adrenaline/epinephrine) (13).
Analytical method.
Plasma glucose levels were measured using the Yellow Springs Instruments 2700 STAT Analyzer, and lipid levels using an autoanalyzer (747–200; Roche-Hitachi). Plasma insulin, adiponectin, and leptin levels were measured using radioimmunoassay (EMD Millipore).
Adipocyte size measurement.
Samples of 20–30 mg of SAT were immediately fixed in osmium tetroxide and analyzed using Multisizer 4 (Beckman Coulter). We performed a curve-fitting analysis technique as previously described (8). The diameter at which the Gaussian curve peaked was defined as the “peak diameter” of the large adipose cells. We also calculated the “% of adipose cells above” (% large cells) and “% below” (% small cells) the nadir. The number of adipocytes in the abdominal depot was estimated by the following formula: cell number volume of abdominal SAT/weighted volume per cell (8).
Real-time PCR.
Adipose tissues were homogenized in QIAzol Lysis Reagent (QIAGEN Inc.), total RNA isolated using RNeasy Mini Kit (QIAGEN Inc.), and reverse-transcribed using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, ThermoFisher Scientific). Real-time PCR was performed using SYBR Green master mix (Bio-Rad) on an Applied Biosystems 7500 Fast Real-Time PCR (ThermoFisher Scientific). Gene expression was normalized to housekeeping gene TBP. Relative gene expression levels were calculated using the 2-ΔΔCt method. The list of all primers used is in Supplemental Table S1.
RNA next-generation sequencing (RNA-seq) and analysis.
Quality and quantity of total RNA was evaluated using Agilent 2200 Bioanalyzer. RNAs with RNA integrity number > 8.0 were used to construct the cDNA library, and sequencing was subsequently performed with Illumina HiSeq 4000. The sequencing reads for each of the samples were aligned to the GRCh38 human reference genome using HISAT2 (32). Gene-level read counts were generated using the feature Counts function of Rsubread (33), based on annotations from the ENCODE v27 GTF file. Differential gene expression was performed using DESeq2 (34). The DESeq2 analysis results were submitted to the IPA software (QIAGEN Inc.) (35), and a core analysis was used to perform pathway enrichment analysis on the differentially expressed genes. RNA-Seq data are deposited into the National Center for Biotechnology Information’s Gene Expression Omnibus, accession GSE159955.
Western blotting.
SAT protein extraction was performed using RIPA buffer supplemented with phosphatase (PhosSTOP, Roche) and proteinase (cOmplete MINI, Roche) inhibitors, and protein content was quantified using BCA assay (Pierce). After denaturation, an equal amount of proteins was run in 4–12% or 16% Tris-Glycine Gel (Novex, Invitrogen). Proteins were transferred to PVDF membranes (Millipore Sigma) by semidry transfer, blocked with 3% BSA, and blotted overnight at 4°C with specific primary antibodies. Actin (#4967), ATGL (#2138), HSL (#4107), pSer660 HSL (#4126), were from Cell Signaling, pSer406 ATGL (ab135093) and CIDEA (ab151577) were from Abcam. Membranes were washed in TBS-Tween (TBS-T) 3 times and then incubated 1 hour with specific HRP-conjugated secondary antibody (Cell Signaling #7074). After 3 washings in TBS-T, the specific band was visualized using enhanced ECL chemiluminescence substrate (Pierce, Thermo Fisher Scientific). Films were developed within the linear dynamic range of signal intensity and then scanned. The intensity of the bands was measured using ImageJ software (NIH). Because of the limited amount of SAT tissue collected, CIDEA protein expression assay was performed on 12 abdominal and 11 gluteal SAT collected.
In vivo of SAT lipid fluxes and cell dynamics measurements by D2O.
In frozen SAT, we assessed TG synthesis based on the incorporation of deuterium into the glycerol moiety of TGs, followed by GCMS and application of MIDA (30). This labeling approach also allows us to measure DNL from D2O using MIDA. DNA synthesis (as index of proliferation and cell turnover) rate was measured evaluating the deuterium enrichment into the deoxyribose moiety of both pre-adipocytes and mature adipocytes isolated from SAT (13, 30).
Statistical analysis.
Where appropriate, continuous variables were log-transformed to normalize their distributions. Wilcoxon Rank Sum Test or Wilcoxon Signed Rank Test was applied for between-group and within-group comparisons, respectively. Unadjusted bivariate associations for continuous variables were summarized using Pearson correlations (using log-transformed variables) and Spearman correlations (when the sample size was limited to n<20 subjects). After adjustment for age, BMI and race, multiple linear regression was applied to identify different factors as predictors of CIDEA mRNA expression levels. Data are expressed as mean ± SEM. GraphPad Prism 8.2 and R-statistical software were used for statistical analysis.
Results
Anthropometric, clinical and metabolic characteristics of study participants
Based on a previously identified VAT/(VAT+SAT) cut-off value (Ratio=0.0972) from our multiethnic cohort of 88 girls with obesity (12, 13), we divided the enrolled adolescent girls, into a group of 21 subjects with low VAT/(VAT+SAT) and 14 subjects with high VAT/(VAT+SAT) (Table 1). Age, race/ethnicity, Tanner stage and BMI were similar in both groups. However, the high VAT/(VAT+SAT) group had higher VAT content (p<0.0001), seemingly similar SAT depot, and importantly was characterized by high content of fat in the liver (p=0.0001), greater ALT concentrations (p=0.03), an increase in HOMA-IR (p=0.043), a decrease in WBISI (p=0.038), and a greater mean 2-hour plasma glucose (p=0.036), fasting C-peptide (p=0.003), 2-hour plasma insulin concentrations (p=0.024) and AUC for the insulin (p=0.042).
Table 1.
Clinical and metabolic characteristics of adolescent subjects with obesity.
| Low VAT/(VAT+SAT) Mean (Range) (n=21) | High VAT/(VAT+SAT) Mean (Range) (n=14) | p-value | |
|---|---|---|---|
| Age (Years) | 16.6 (13–20) | 15.3 (12–19) | 0.073 |
| Race (C/H/AA/other) | 3/9/9/0 | 8/4/1/1 | 0.014 |
| Tanner stage (III/IV/V) | 3/5/13 | 6/2/6 | 0.16 |
| Anthropometrics | |||
| Weight (Kg) | 97.8 (66.4–154.5) | 101.2 (69.7–137.9) | 0.16 |
| Height (cm) | 164.4 (149–177) | 161.2 (151–172) | 0.68 |
| BMI (Kg/m2) | 36.2 (25.9–55.6) | 38.8 (28.3–56.1) | 0.38 |
| Body composition (DEXA) | |||
| % Fat Mass | 45.3 (39.0–51.3) * | 45.9 (32.5–53.7) † | 0.55 |
| Total body fat (Kg) | 44.8 (21.7–67.4) * | 45.6 (22.4–67.7) † | 0.98 |
| Lean body mass (Kg) | 52,1 (36.3–82.0) * | 50.5 (39.4–66.3) † | 0.96 |
| Body Fat Distribution (MRI) | |||
| VAT (cm2) | 52.9 (7.7–94.5) | 101.4 (42.2–172.8) | <0.0001 |
| SAT (cm2) | 636.9 (302.8–1074.1) | 576.5 (315.9–1063.8) | 0.36 |
| Superficial SAT (cm2) | 187.6 (114.8–336.3) † | 172.7 (73–401.9) | 0.27 |
| Deep SAT (cm2) | 214.9 (83.4–361.1) † | 187.6 (75–312) | 0.40 |
| VAT/(VAT+SAT) | 0.076 (0.025–0.096) | 0.153 (0.107–0.230) | <0.0001 |
| Metabolic measurements | |||
| Fasting glucose (mg/dL) | 90.5 (77–101.5) | 91.3 (83.5–112) | 0.82 |
| 2-hour glucose (mg/dL) | 112.7 (69–166) | 125.8 (87–149) | 0.036 |
| Fasting insulin (μU/mL) | 24.2 (7.5–45.5) | 33.8 (12–82.5) † | 0.16 |
| 2-hour insulin (μU/mL) | 108.5 (19–310) | 219.8 (81–560) | 0.024 |
| Fasting C-peptide (pmol/L) | 879.2 (460–1775) * | 1271.9 (770–2610) | 0.003 |
| HOMA-IR ((mg/dL)(μU/mL)) | 5.52 (1.78–11.17) | 8.80 (2.91–20.99) | 0.043 |
| WBISI (l2/(mg × μU)) | 2.66 (1.20–6.75) | 1.71 (0.49–3.30) | 0.038 |
| AUC180 Glucose (mg/dL/min) | 113.4 (86.8–131.1) | 122.5 (88.1–145.0) | 0.09 |
| AUC180 Insulin (μU/mL/min) | 104.7 (32.9–220.2) | 201.6 (61.0–522.0) | 0.042 |
| AUC180 C-peptide(pmol/L/min) | 2530 (1545–4513) | 3710 (1648–8370) | 0.07 |
| Leptin (ng/mL) | 52.2 (14.4–169-4) | 68.2 (28–188.6) † | 0.12 |
| Adiponectin (ng/ml) | 7.39 (1.78–17.20) ‡ | 6.26 (2.2–12.5) ‡ | 0.53 |
| Adiponectin/Leptin ratio | 0.228 (0.010–1.146) ‡ | 0.130 (0.025–0.446) ‡ | 0.35 |
| Lipids | |||
| Total Cholesterol (mg/dL) | 152.5 (108–242) | 147.7 (117–222) | 0.49 |
| HDL (mg/dL) | 45 (30–67) | 39.2 (27–51) | 0.12 |
| LDL (mg/dL) | 91 (48–166) ‡ | 79.4 (40–147) | 0.21 |
| TG (mg/dL) | 86.9 (22–213) | 110.2 (53–206) | 0.09 |
| FFA (μmol/L) | 0.490 (0.231–0.854) | 0.538 (0.218–0.934) | 0.44 |
| Liver | |||
| ALT (U/L) | 14.1 (6–56) † | 35 (1–194) | 0.030 |
| AST (U/L) | 17.4 (6–38) * | 28.1 (11–108) † | 0.18 |
| Fat Liver (% HFF) | 0.68 (0–6) | 10.99 (0–38.90) † | 0.0001 |
Demographic, anthropometric and metabolic characteristics are shown for 35 adolescent females enrolled for abdominal and gluteal subcutaneous adipose tissue biopsy. Wilcoxon Rank Sum test was used to compare Low vs High VAT/(VAT+SAT) groups. Data on 11 subjects have been previously reported [Ref. 13]. AA: Afro-Americans; AUC180=Area Under the Curve calculated after 180 minutes; C: Caucasians; FFA: Free Fatty Acid; H: Hispanics; HOMA-IR: Homeostatic Model Assessment of Insulin Resistance; SAT: Abdominal Subcutaneous Adipose Tissue; TG: Triglycerides; VAT: Visceral Adipose Tissue; WBISI: Whole-Body Insulin Sensitivity Index;
= 1 missing data,
= 2 missing data,
= 3 data missing.
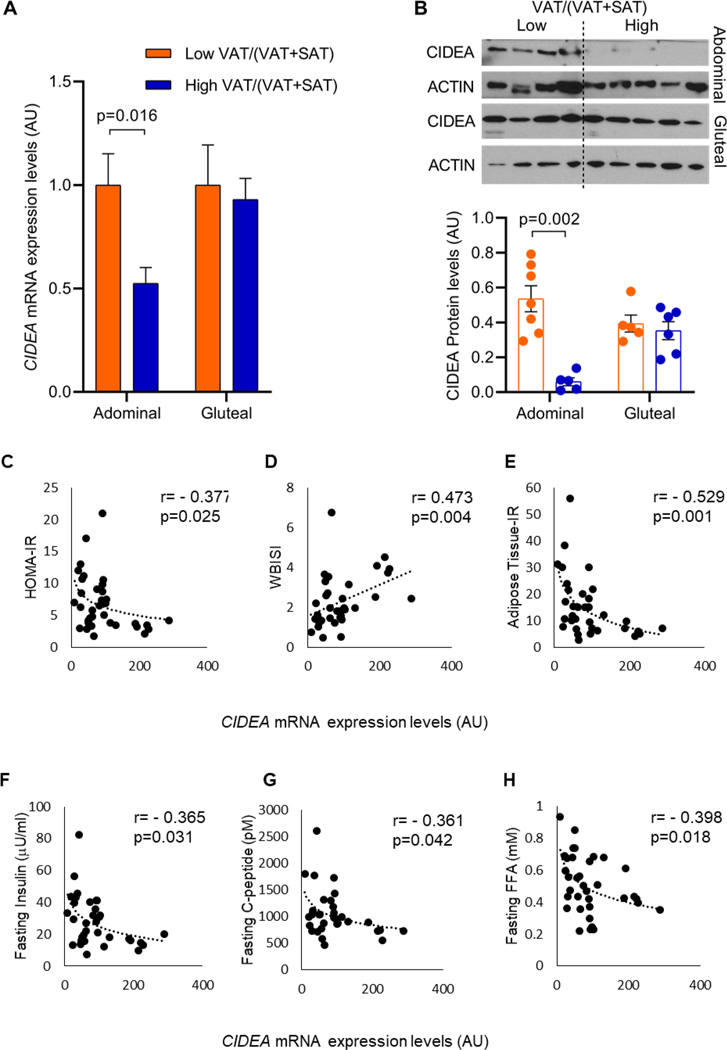
CIDEA expression and protein level are reduced in adolescent girls with high VAT/(VAT+SAT)
To identify the role of CIDEA in regulating SAT functionality, we determined CIDEA mRNA expression by RT-PCR in abdominal and gluteal depots obtained from adolescent girls with obesity with low or high VAT/(VAT+SAT). In abdominal SAT, the CIDEA mRNA expression was significantly reduced in the high VAT/(VAT+SAT) group compared to the low VAT/(VAT+SAT) group (p=0.016), but this reduction was not observed in SAT samples obtained from the gluteal depot (Figure 1A). Using Western Blot, we also confirmed these data at protein level (Figure 1B). Since we observed a modulation in CIDEA expression both at mRNA and protein levels exclusively in the abdominal SAT, we focused our attention on studying the impact of CIDEA expression in the abdominal SAT from subjects with different VAT/(VAT+SAT).
Figure 1. CIDEA mRNA expression is reduced in abdominal SAT in high VAT/(VAT+SAT) group and correlates with an increase in markers associated to insulin resistance.
(A) CIDEA mRNA expression in abdominal SAT (Low VAT/(VAT+SAT): n=21 subjects, High VAT/(VAT+SAT): n=14 subjects) and gluteal SAT (Low VAT/(VAT+SAT): n=7 subjects, High VAT/(VAT+SAT): n=14 subjects). (B) Representative blots showing CIDEA levels in abdominal and gluteal SAT in low and high VAT/(VAT+SAT) groups and CIDEA protein quantification showing significant reduction in abdominal SAT in high VAT/(VAT+SAT) group and no variation in gluteal SAT. (Abdominal low VAT/(VAT+SAT): n=7 subjects, abdominal high VAT/(VAT+SAT): n=5 subjects; gluteal low VAT/(VAT+SAT): n=5 subjects, gluteal high VAT/(VAT+SAT): n=6 subjects). Data are expressed as mean ± SEM. All data were analyzed using multiple linear regression adjusted for age, race and BMI. (C-D) CIDEA mRNA expression significantly correlates with HOMA-IR and WBISI, (E) inversely correlates with Adipose tissue Insulin Resistance, (F) fasting insulin, (G) fasting C-peptide, and (H) fasting serum FFA. Data were analyzed using Pearson’s correlation after logarithmic transformation of x and y variables and using all the subjects enrolled including both Low and High VAT/(VAT+SAT) (n=32–35 subjects).
Because CIDEA belongs to the family of browning genes, we investigated using RNA-seq the expression of other genes associated with the browning process, as well as mitochondrial markers. Interestingly, we were not able to observe differences between the 2 groups in any browning/beige genes (including UCP1, TBX1, PPARA, PGC1A, PRDM16, EVA1, DIO2), nor in mitochondrial activity marker genes (such as COX7C, COX4L1, CKMT2, CPT2). No statistical differences were observed in the expression of CIDE-C, a component of CIDE family also involved in lipid droplets enlargement (Supplemental Figure S1A–D). We were also able to confirm by qPCR that the expression of some mitochondrial and browning markers (such as PPARA, PGC1A and TFAM), did not vary between groups (Supplemental Figure S1E).
CIDEA mRNA level inversely correlates with insulin resistance and is associated with changes in adipocytes size and morphometry
As shown in Figure 1C–H, in the abdominal SAT we observed that CIDEA inversely correlated with HOMA-IR, Adipose Tissue-IR, fasting insulin, C-peptide, FFA, and directly correlated with WBISI, but did not correlate with BMI (r= −0.166, p=0.34), and fasting glucose (r= −0.172, p=0.32).
Consistent with our previous reports (13), we observed an increase in cell peak diameter of adipocytes isolated from abdominal SAT of subjects with high VAT/(VAT+SAT) compared to the adipocytes from subjects with low VAT/(VAT+SAT) (p=0.026), (Supplemental Table S2). We also found in subjects with high VAT/(VAT+SAT) a reduction in the total number of large adipocytes (p=0.038) but no changes in either percentage of small adipocytes or total number of small adipocytes.
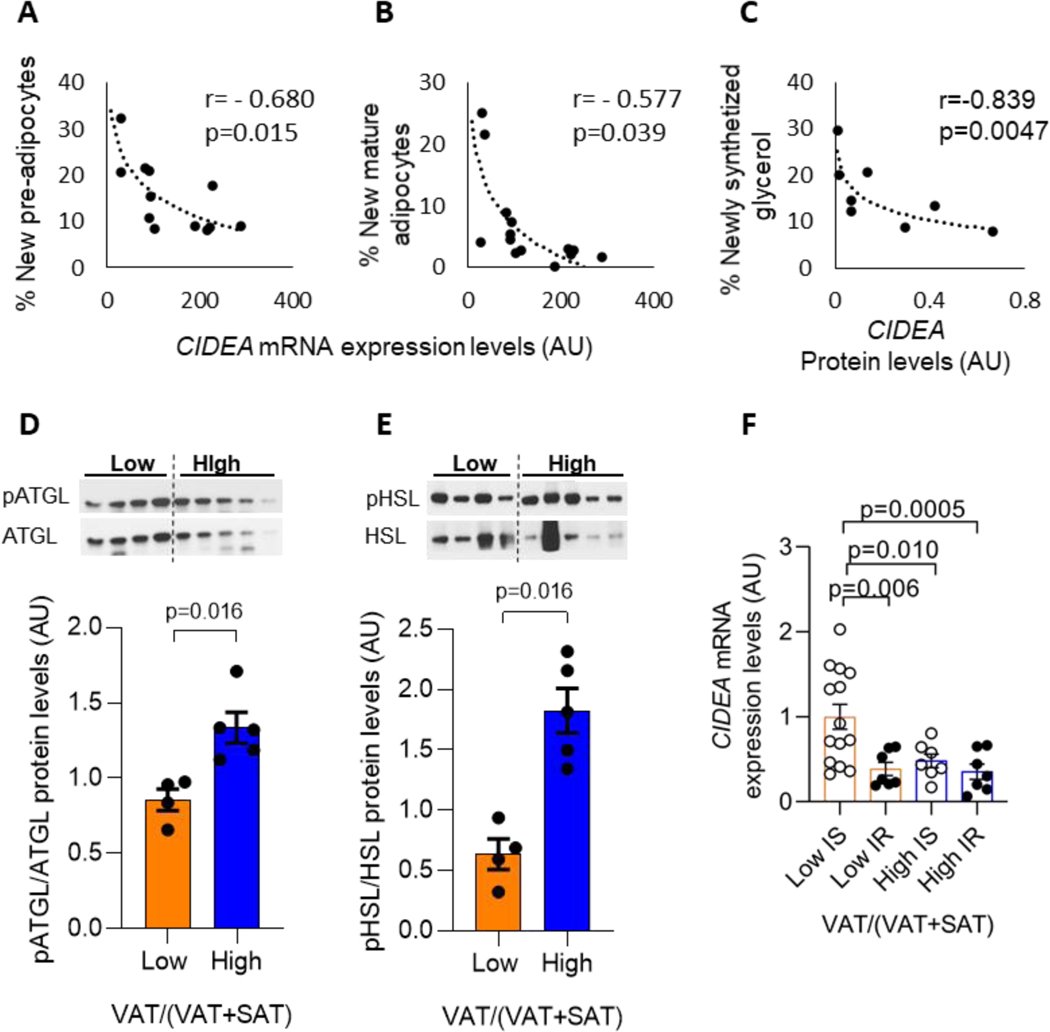
Using the deuterated water (D2O) labelling protocol in a subset of subjects (13), we analyzed the incorporation of deuterium into the DNA of preadipocytes and mature adipocytes in order to evaluate the percentage of new pre-adipocytes (indicating preadipocytes proliferation) and newly differentiated adipocytes (indicating adipogenesis) as a function of the CIDEA expression and differences in VAT/(VAT+SAT) levels.
We found a significant correlation between the decrease in CIDEA expression and the increase in pre-adipocyte’s proliferation as well as adipogenesis (Figure 2A, B), indicating that the decrease in CIDEA expression is associated with an increase in adipocytes turnover.
Figure 2. Association between CIDEA mRNA expression and insulin resistance, adipocyte measurements and lipolysis.
(A) CIDEA mRNA expression correlates with percentage of new pre-adipocytes (n=12 subjects), and (C) with the percentage of new mature adipocytes (n=13 subjects). (D) CIDEA protein level correlates with the percentage of newly synthesized glycerol as index of lipolysis (n=8 subjects). (B-D) Data were analyzed using Spearman correlation. (E-F) Blots and protein quantification showing significant increase in both ATGL and HSL phosphorylation levels in abdominal SAT in high vs low VAT/(VAT+SAT) groups (Low VAT/(VAT+SAT): n=4 subjects, high VAT/(VAT+SAT): n=5 subjects). (G) CIDEA mRNA expression in subjects grouped by VAT/(VAT+SAT) and characterized by insulin sensitivity (IS) or insulin resistance (IR). Data are expressed as mean ± SEM and analyzed using multiple linear regression adjusted by age, BMI, and race. IS Low VAT/(VAT+SAT) n=14 subjects, IR Low VAT/(VAT+SAT) n=7, IS High VAT/(VAT+SAT) n=7, IR High VAT/(VAT+SAT) n=7.
CIDEA is involved in regulating lipolysis
In a subset of subjects, we analyzed the deuterium incorporation in newly synthetized glycerol (as index of lipolysis) and found a strong inverse correlation between CIDEA protein level with the newly synthetized glycerol (r= − 0.839, p= 0.0047) (Figure 2C). Additionally, we demonstrate that the higher lipolytic rates are due to a greater phosphorylation of ATGL and HSL, two key enzymes involved in lipolysis in adipocytes (25, 26), in the SAT depot, as assessed by western blot (Figure 2D, E). These data suggest that the reduction in CIDEA expression partakes in the accelerated rate of lipolysis from the abdominal SAT.
Encouraged by the results indicating an increase in cell peak and a concomitant increase in lipolysis in SAT obtained from high VAT/(VAT+SAT), we decided to investigate if VAT/(VAT+SAT) was involved in the regulation of CIDEA expression independently from the effect of insulin as lipolytic regulator. We regrouped our subjects based on both VAT/(VAT+SAT) and insulin sensitivity levels, as reported in Figure 2F, and compared the CIDEA expression levels among these 4 subgroups. We found that in low VAT/(VAT+SAT) groups, insulin resistant (IR) subjects had reduced CIDEA expression compared to insulin sensitive (IS) subjects. In high VAT/(VAT+SAT) groups, no notable differences were observed between IS and IR subjects. We also found comparable CIDEA expression levels in IR subjects with low VAT/(VAT+SAT), IR subjects with high VAT/(VAT+SAT), and IS subjects with high VAT/(VAT+SAT). Interestingly, IS subjects showed a reduction in CIDEA expression in high VAT/(VAT+SAT) compared to low VAT/(VAT+SAT) (Figure 2F).
To further confirm these observations, we applied multiple linear regression analysis to identify if VAT/(VAT+SAT), insulin sensitivity state, and/or adipocyte cell peak were meaningful predictors of CIDEA expression, adjusting for age, BMI, and race (Table 2). We found that all these variables were independent predictors associated with changes in CIDEA expression.
Table 2.
Factors affecting CIDEA mRNA expression level in adolescent girls with obesity.
| Independent variable | Estimate (SE) | p value |
|---|---|---|
| Intercept | −2.53 (1.13) | 0.034 |
| IS/IR two-level variable | −0.88 (0.28) | 0.004 |
| VAT/(VAT+SAT) | −0.81 (0.29) | 0.009 |
| Cell peak | 0.02 (0.01) | 0.020 |
R2=0.402, p=0.003
Parameters estimated by multiple linear regression analysis adjusted for age, BMI, and race. The log of the CIDEA mRNA expression level was used as the dependent variable and VAT/(VAT+SAT), IS/IR two-level variable (insulin sensitivity/resistance was defined by WBISI cutoff=1.79), and cell peak as independent variables. IS=insulin sensitivity, IR=insulin resistance, SE=Standard Error.
The increase in body weight induces reduction in CIDEA mRNA expression and adipocytes dysfunction.
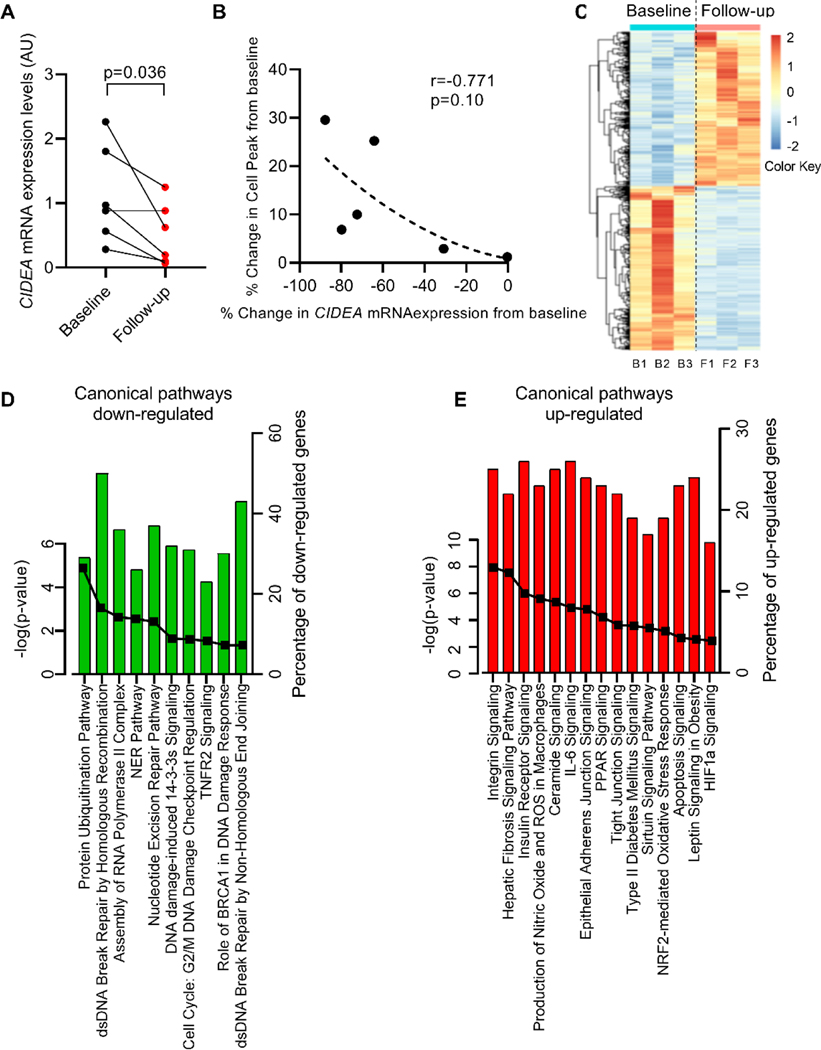
We explored the effect of further weight gain on CIDEA mRNA expression in abdominal SAT collected from a group of adolescent girls that consented to have a second abdominal SAT biopsy after a median of 34.2 months from the first biopsy.
All subjects gained weight (p=0.031) with an increased in BMI (p=0.031). These changes were reflected by changes in total fat mass and lean body mass, by an increase in visceral fat depot (p=0.062) and consequently a small increase in VAT/(VAT+SAT) (p=0.063) (Table 3). Furthermore, adipocyte cell peak (p=0.031) and percentage of small adipocytes (p=0.031), as well as in total number of adipocytes (p=0.063) all increased (Supplemental Table S3). In this group of subjects, we analyzed the CIDEA mRNA expression at baseline and at follow-up after gaining weight and found its expression to be decreased at the time of the follow-up biopsy (Figure 3A), suggesting that the increase in fat accumulation in adipocytes (evaluated by increasing of cell peak) is linked with a decrease in CIDEA mRNA expression. Interestingly, we were able to connect the change in cell sizing between baseline and follow-up biopsy with the change in CIDEA gene expression in abdominal SAT. We also found a strong inverse correlation between the change in cell peak and change in CIDEA mRNA expression (r= − 0.771, p=0.10), albeit it was based on the small number of subjects (Figure 3B). Based on these longitudinal data, we suggest that in abdominal SAT the variation in CIDEA and cell sizing are closely interconnected.
Table 3.
Baseline and follow up clinical and metabolic characteristics of adolescent subjects that returned for a second biopsy.
| Baseline (n=6)Mean (Range) | Follow-up (n=6)Mean (Range) | p-value | |
|---|---|---|---|
| Age (Years) | 14.7 (10.0–18.0) | 18.0 (14.0–24.0) | 0.031 |
| Race (C/H/AA) | 1/4/1 | ||
| Tanner stage (II/III/IV/V) | 1/1/2/2 | 0/1/0/5 | 0.23 |
| Anthropometrics | |||
| Weight (Kg) | 90.0 (66.4–116.75) | 106.7 (77.9–144.5) | 0.031 |
| Height (cm) | 160.3 (143.0–166.0) | 161.6 (150.0–167.0) | 0.50 |
| BMI (Kg/m2) | 35.1 (25.9–42.4) | 42.0 (30.1–51.8) | 0.031 |
| Body composition (TANITA) | |||
| Total body fat (Kg) | 42.2 (28.9–56.0) | 47.5 (31.9–62.6) * | 0.13 |
| Lean body mass (Kg) | 47.4 (37.3–59.6) | 50.9 (42.4–58.4) * | 0.062 |
| Body Fat Distribution (MRI) | |||
| VAT (cm2) | 56.6 (34.2–85.0) | 78.1 (37.0–129.1) * | 0.062 |
| SAT (cm2) | 554.6 (374.8–727.1) | 646.6 (424.3–969.5) * | 0.13 |
| Superficial SAT (cm2) | 176.8 (122.6–227.1) | 199.7 (133.2–258.5) * | 0.062 |
| Deep SAT (cm2) | 194.1 (122.6–293.7) | 218.6 (160.0–381.3) * | 0.63 |
| VAT/(VAT+SAT) | 0.092 (0.067–0.131) | 0.103 (0.068–0.123) * | 0.31 |
| Metabolic measurements | |||
| Fasting glucose (mg/dL) | 88.8 (86.0–93.5) | 87.9 (78.0–95.0) | 0.91 |
| 2-hour glucose (mg/dL) | 119.8 (88–149) | 130.3 (111–157.5) | 0.44 |
| Fasting insulin (μU/mL) | 27.7 (15.5–39.5) | 25.9 (15–38) | 0.81 |
| Fasting C-peptide (pmol/L) | 960.2 (585–1733) * | 916.3 (565–1463) * | 0.50 |
| WBISI (l2/(mg × μU)) | 1.79 (0.46–3.65) | 1.78 (0.87–2.86) | 0.84 |
| Lipids | |||
| Total Cholesterol (mg/dL) | 141.8 (117–163) | 144.2 (116–176) * | 0.38 |
| HDL (mg/dL) | 44.0 (29–53) | 44.8 (34–53) * | 0.50 |
| LDL (mg/dL) | 88.2 (67–118) | 83.4 (64–119) * | 0.75 |
| TG (mg/dL) | 77.5 (30–117) | 80.6 (27–125) * | 0.44 |
| Liver | |||
| ALT (U/L) | 18.8 (8–51) | 14.6 (8–20) | 0.75 |
| Fat Liver (% HFF) | 8.3 (0–32.7) | 5.7 (0.7–9.7) * | 0.18 |
Demographic, anthropometric and metabolic characteristics are shown for adolescent girls that had an abdominal SAT biopsy at baseline and after gaining weight (follow-up). Wilcoxon Signed Rank test was used to compare baseline vs follow-up distributions. AA: Afro-Americans; C: Caucasians; H: Hispanics; HOMA-IR: Homeostatic Model Assessment of Insulin Resistance; SAT: Abdominal Subcutaneous Adipose Tissue; TG: Triglycerides; VAT: Visceral Adipose Tissue; WBISI: Whole-Body Insulin Sensitivity Index;
= 1 missing data.
Figure 3. CIDEA mRNA expression significantly decreases in abdominal SAT after weight gain.
(A) CIDEA mRNA expression in SAT collected at baseline and after gaining weight (follow-up). A linear model was fitted using generalized least squares with unstructured covariance, corrected for change in age and BMI (n=6 subjects each group) (B) Inverse correlation between percent change in CIDEA mRNA expression and percent change in cell peak. Data were analyzed using Spearman correlation (n=6 subjects each group). (C) Heatmap originated from RNA-seq analysis of SAT collected at baseline and during follow-up biopsy. (D-E) Canonical pathways enrichment analysis for down-/up-regulated genes in follow up SAT biopsy compared to baseline biopsy. The bar represents the percent of genes up-/down-regulated within the specific pathways. The solid line represents the -log(p value) for each pathway (n=3 subjects each group).
To determine whether SAT showed a differential gene profile after weight gain, we used RNA-seq to compare gene expression in abdominal SAT collected at baseline and at the follow-up biopsy. A shown from heatmap plot, we identified a different pattern of gene expression between baseline and follow-up biopsy (Figure 3C).
As shown from IPA analysis, we were able to identify a general perturbation in tissue functionality in SAT collected in the follow-up biopsy after weight gain. Particularly, we observed that most of the downregulated genes in the follow-up biopsy belonged to pathways associated with ubiquitination and DNA repair processes (Figure 3D and Supplemental Table S4).
Thus, after gaining further weight, the ability of the abdominal SAT to maintain normal tissue morphometry and functionality deteriorated, as indicated through DNA repair and ubiquitin-dependent protein degradation, suggesting an accumulation of dysfunctional protein and mismatch errors in the DNA that lead to a dysfunctional adipose tissue.
In contrast, upregulated genes after gaining weight belonged to pathways associated to cell signaling (including PLC, PI3K, Akt pathways), inflammatory pathways (including IL8, IL6, IL9, IL1beta, TGF signaling), oxidative stress (including HIF1a and Nrf2 signaling), apoptosis signaling, extracellular matrix remodeling and signaling (including integrins, markers of fibrosis, cell junctions), and PPAR signaling (Figure 3E and Supplemental Table S4).
Overall, these results from follow-up biopsies suggest that gaining further weight leads to an enlargement of adipocytes which is closely linked to a decrease in CIDEA mRNA expression and a modulation of genes associated with adipocyte dysfunction.
Discussion
In this study, we demonstrated that in adolescent girls with obesity and a high VAT/(VAT+SAT), CIDEA expression was reduced at both mRNA and protein levels exclusively in abdominal SAT depot and not in gluteal depot. This reduction was not associated with reduction in any other gene markers associated to browning of white adipose tissue or mitochondrial markers. Interestingly, the reduction in CIDEA was associated with adipocyte hypertrophy and an increase in percentage of small cells, as well as increase in pre-adipocyte proliferation and differentiation into mature adipocytes. Of note, we demonstrate that the higher lipolytic rates are due to a greater phosphorylation of ATGL and HSL, two key enzymes involved in lipolysis in adipocytes (25, 26), in the SAT depot. These data suggest that the reduction in CIDEA expression partakes in the accelerated rate of lipolysis from the abdominal SAT, thus greatly contributing the insulin resistant state of these adolescents with central obesity.
Interestingly, our observations clearly showed that CIDEA mRNA/protein expression was not reduced in gluteal SAT depot from adolescent girls with obesity and with high VAT/(VAT+SAT). We also did not find any correlation between CIDEA mRNA expression and any of the common outcomes of insulin resistance or new pre-adipocyte/adipocyte turnover (data not shown), supporting the idea that CIDEA modulation is depot specific. We can speculate that the reason behind this depot specificity is based on the “limited SAT expandability” theory, therefore the excessive enlargement of abdominal adipocytes induces cell dysfunction but this phenomenon is not observed in more flexible gluteal adipocytes.
CIDEA, a member of the CIDE (cell death–inducing DFFA-like effector) family, is expressed in both brown and white adipose tissue (21, 22). Originally described as master regulator of the apoptotic process, more recently it has been reported that CIDEA plays important roles in the development of metabolic disorders (21, 36) and regulation of lipid metabolism (14).
Herein, this study investigated, for the first time, the expression of CIDEA in a group of adolescent girls with obesity and different patterns of abdominal body fat distribution. Although in the current study no direct comparison were made with adult forms of obesity, a reduction in CIDEA expression associated with markers of insulin resistance has been described in adults with obesity (14, 21, 22, 37, 38). Interestingly, in this present study we demonstrated that this phenomenon was driven not only by insulin resistance but also by VAT/(VAT+SAT) and adipocyte cell peak. In fact, using statistical analysis, we were able to identify insulin resistance, VAT/(VAT+SAT) and adipocyte cell peak as independent mediators involved in the regulation of CIDEA mRNA expression.
Although CIDEA belongs to a family of browning/beige markers, surprisingly, our data did not find a reduction in the mRNA expression of other browning/beige gene markers. This observation is in contrast with the data reported using in vitro human adipocytes and in vivo mouse model (39) which reported that the increase in CIDEA expression was associated with a parallel increase in UCP1 in in vitro differentiated SAT adipocytes, and increase in energy expenditure as well as improvement in thermoregulation using a mouse model (39). Of note, the connection between CIDEA and browning was observed in mouse models and in vitro SAT human adipocytes. Until now, there has been no evidence of the role of CIDEA in in vivo human abdominal SAT tissue.
From the cell sizing measurements, we showed that in subjects with high VAT/(VAT+SAT), the abdominal SAT has larger adipocyte cell peak but reduced number of large cells, and that the increase in CIDEA mRNA expression correlates with the decrease in the number of small adipocytes. These findings are different from what was previously reported in CIDEA-null mice (20) which showed a strong reduction in adiposity associated with reduction in adipocyte cell size independent from HSL activity modulation. It should be noted that these data were generated from a whole-body knock-down mouse focusing on the role of CIDEA in brown adipose tissue, and no translational studies in humans were reported showing similar functions in human adipose tissue.
Using an in vivo dynamic measure of the SAT cell population turnover, we demonstrated that the reduction in CIDEA gene expression was associated with an increase in both pre-adipocytes proliferation and differentiation of new mature adipocytes. Our findings are consistent with the observation published in a CIDEA humanized mouse model (24). The proliferation of new pre-adipocytes and differentiation into new mature adipocytes, supported from the increase in the number of small adipocytes, indicates an increase in adipocytes turnover probably associated to an increase in cell death.
In our experiments, we also observed an increase in lipolysis (described as increase in both TAG synthesis rate and pATGL and pHSL) associated with a reduction in CIDEA mRNA expression and increase in adipocyte cell diameter. This might suggest that in SAT from subjects with high VAT/(VAT+SAT), the adipocytes are still functional but not able to retain lipids because they have reached the maximum capacity of lipid storage (40). Therefore, lipids are spilling out, leading to accumulation in VAT and ectopic location as liver, resulting in fatty liver disease. However, the role of CIDEA in regulating the lipolytic machinery is still unclear, as previously noted (21).
To further test our observations longitudinally, we collected abdominal SAT from a small group of subjects undergoing a second biopsy after gaining weight. We found that, adipocyte cell diameter increased in parallel with a significant reduction in CIDEA mRNA expression, suggesting a possible direct association with the adipocyte diameter. Of note, the changes in adipocyte gene expression indicated by RNA-seq after gaining weight are showing a gene profile typical of adipocyte dysfunction.
Our study limitations include a small sample size for the cross-sectional studies, and an even smaller number of subjects consenting to having a repeated follow-up biopsy. Moreover, the small amount of tissue collected during the biopsies limited our ability to perform further functional assays/experiments. Another limitation to this study is that the D2O labeling approach cannot differentiate between newly synthesized fatty acids that originate from the liver and are transported to adipose tissue and fatty acids newly synthetized from adipose tissue. Finally, we should note that osmium fixation may induce an increase in adipocyte diameter, as reported by others, however the adipocyte diameter observed here were consistent with the dimensions already reported from our and other groups using the same methodology (8, 13, 41, 42).
In conclusion, this study demonstrates that the reduction in CIDEA expression is associated with a poor metabolic state, and for the first time we have demonstrated a strong connection between CIDEA mRNA expression and adipocyte cell size and functionality in SAT from adolescent girls with obesity.
Supplementary Material
Study Importance Questions:
What is already known about this subject?
In adolescent girls with obesity, the high ratio of visceral AT depot (VAT) to abdominal subcutaneous AT depot (SAT) (High VAT/(VAT+SAT)) is associated with insulin resistance, increased risk for type 2 diabetes and fatty liver
CIDEA is originally described as principal driver involved in lipid droplets formation and lipid accumulation, more recently, however, it was described to have additional functions
The increase in CIDEA expression in AT improve metabolic profile in a mouse model
What are the new findings in your manuscript?
CIDEA is reduced in SAT from subjects with High VAT/(VAT+SAT) compared to Low VAT/(VAT+SAT)
CIDEA expression inversely correlates with Insulin resistance indexes and inversely correlates with preadipocyte proliferation and differentiation
After gaining weight, CIDEA is reduced and correlates with increase in adipocyte diameter
How might your results change the direction of research or the focus of clinical practice?
A better understanding of the molecular mechanism behind the adipose tissue dysfunction in adolescent girls with obesity might open new prospectives for therapeutic intervention and reduce the risk of type 2 diabetes or other complication in adulthood
Acknowledgments
The authors thank all the volunteers and research nurses at the Yale HRU for their skillful help in the study. In memory of Bridget Pierpont, all the authors are also grateful for her extraordinary work and dedication to this project.
Funding
This study was supported by the Robert E. Leet and Clara Guthrie Patterson Trust (Mentored Research Award to JN); NIH Eunice Kennedy Shriver National Institute of Child Health and Human Development grants R01-HD-40787, R01-HD-28016, and K24-HD-01464 to SC; Clinical and Translational Science Award grant UL1-TR001863 from the National Center for Advancing Translational Science, a component of the NIH; grant R01-EB006494 (Bioimage Suite); and Distinguished Clinical Scientist Award from the American Diabetes Association (to SC), as well as grants R01 DK113984 and the Yale Diabetes Research Center P30 DK-045735 to GIS.
Footnotes
Disclosure
The authors declared no conflict of interest.
References
- 1.Hales CM, Fryar CD, Carroll MD, Freedman DS, Ogden CL. Trends in Obesity and Severe Obesity Prevalence in US Youth and Adults by Sex and Age, 2007–2008 to 2015–2016. JAMA 2018;319: 1723–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Srinivasan SR, Bao W, Wattigney WA, Berenson GS. Adolescent overweight is associated with adult overweight and related multiple cardiovascular risk factors: the Bogalusa Heart Study. Metabolism 1996;45: 235–240. [DOI] [PubMed] [Google Scholar]
- 3.Weiss R, Dufour S, Taksali SE, Tamborlane WV, Petersen KF, Bonadonna RC, et al. Prediabetes in obese youth: a syndrome of impaired glucose tolerance, severe insulin resistance, and altered myocellular and abdominal fat partitioning. Lancet 2003;362: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mayer-Davis EJ, Dabelea D, Lawrence JM. Incidence Trends of Type 1 and Type 2 Diabetes among Youths, 2002–2012. N Engl J Med 2017;377: 301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.D’Adamo E, Cali AM, Weiss R, Santoro N, Pierpont B, Northrup V, et al. Central role of fatty liver in the pathogenesis of insulin resistance in obese adolescents. Diabetes Care 2010;33: 1817–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Petersen MC, Shulman GI. Mechanisms of Insulin Action and Insulin Resistance. Physiol Rev 2018;98: 2133–2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taksali SE, Caprio S, Dziura J, Dufour S, Cali AM, Goodman TR, et al. High visceral and low abdominal subcutaneous fat stores in the obese adolescent: a determinant of an adverse metabolic phenotype. Diabetes 2008;57: 367–371. [DOI] [PubMed] [Google Scholar]
- 8.Kursawe R, Eszlinger M, Narayan D, Liu T, Bazuine M, Cali AM, et al. Cellularity and adipogenic profile of the abdominal subcutaneous adipose tissue from obese adolescents: association with insulin resistance and hepatic steatosis. Diabetes 2010;59: 2288–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weiss R, Taksali SE, Dufour S, Yeckel CW, Papademetris X, Cline G, et al. The “obese insulin-sensitive” adolescent: importance of adiponectin and lipid partitioning. J Clin Endocrinol Metab 2005;90: 3731–3737. [DOI] [PubMed] [Google Scholar]
- 10.Caprio S, Perry R, Kursawe R. Adolescent Obesity and Insulin Resistance: Roles of Ectopic Fat Accumulation and Adipose Inflammation. Gastroenterology 2017;152: 1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weiss R. Fat distribution and storage: how much, where, and how? Eur J Endocrinol 2007;157 Suppl 1: S39–45. [DOI] [PubMed] [Google Scholar]
- 12.Umano GR, Shabanova V, Pierpont B, Mata M, Nouws J, Trico D, et al. A low visceral fat proportion, independent of total body fat mass, protects obese adolescent girls against fatty liver and glucose dysregulation: a longitudinal study. Int J Obes (Lond) 2019;43: 673–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nouws J, Fitch M, Mata M, Santoro N, Galuppo B, Kursawe R, et al. Altered In Vivo Lipid Fluxes and Cell Dynamics in Subcutaneous Adipose Tissues Are Associated With the Unfavorable Pattern of Fat Distribution in Obese Adolescent Girls. Diabetes 2019;68: 1168–1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puri V, Ranjit S, Konda S, Nicoloro SM, Straubhaar J, Chawla A, et al. Cidea is associated with lipid droplets and insulin sensitivity in humans. Proc Natl Acad Sci U S A 2008;105: 7833–7838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hallberg M, Morganstein DL, Kiskinis E, Shah K, Kralli A, Dilworth SM, et al. A functional interaction between RIP140 and PGC-1alpha regulates the expression of the lipid droplet protein CIDEA. Mol Cell Biol 2008;28: 6785–6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Inohara N, Koseki T, Chen S, Wu X, Nunez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J 1998;17: 2526–2533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ito M, Nagasawa M, Omae N, Ide T, Akasaka Y, Murakami K. Differential regulation of CIDEA and CIDEC expression by insulin via Akt1/2- and JNK2-dependent pathways in human adipocytes. J Lipid Res 2011;52: 1450–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishimoto Y, Nakajima S, Tateya S, Saito M, Ogawa W, Tamori Y. Cell death-inducing DNA fragmentation factor A-like effector A and fat-specific protein 27beta coordinately control lipid droplet size in brown adipocytes. J Biol Chem 2017;292: 10824–10834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao GG, Chen FJ, Zhou LK, Su L, Xu DJ, Xu L, et al. Control of lipid droplet fusion and growth by CIDE family proteins. Bba-Mol Cell Biol L 2017;1862: 1197–1204. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Z, Yon Toh S, Chen Z, Guo K, Ng CP, Ponniah S, et al. Cidea-deficient mice have lean phenotype and are resistant to obesity. Nat Genet 2003;35: 49–56. [DOI] [PubMed] [Google Scholar]
- 21.Nordstrom EA, Ryden M, Backlund EC, Dahlman I, Kaaman M, Blomqvist L, et al. A human-specific role of cell death-inducing DFFA (DNA fragmentation factor-alpha)-like effector A (CIDEA) in adipocyte lipolysis and obesity. Diabetes 2005;54: 1726–1734. [DOI] [PubMed] [Google Scholar]
- 22.Gummesson A, Jernas M, Svensson PA, Larsson I, Glad CA, Schele E, et al. Relations of adipose tissue CIDEA gene expression to basal metabolic rate, energy restriction, and obesity: population-based and dietary intervention studies. J Clin Endocrinol Metab 2007;92: 4759–4765. [DOI] [PubMed] [Google Scholar]
- 23.Barbatelli G, Murano I, Madsen L, Hao Q, Jimenez M, Kristiansen K, et al. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am J Physiol Endocrinol Metab 2010;298: E1244–1253. [DOI] [PubMed] [Google Scholar]
- 24.Abreu-Vieira G, Fischer AW, Mattsson C, de Jong JM, Shabalina IG, Ryden M, et al. Cidea improves the metabolic profile through expansion of adipose tissue. Nat Commun 2015;6: 7433. [DOI] [PubMed] [Google Scholar]
- 25.Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science 2004;306: 1383–1386. [DOI] [PubMed] [Google Scholar]
- 26.Kraemer FB, Shen WJ. Hormone-sensitive lipase: control of intracellular tri-(di-)acylglycerol and cholesteryl ester hydrolysis. J Lipid Res 2002;43: 1585–1594. [DOI] [PubMed] [Google Scholar]
- 27.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care 1999;22: 1462–1470. [DOI] [PubMed] [Google Scholar]
- 28.Tang A, Tan J, Sun M, Hamilton G, Bydder M, Wolfson T, et al. Nonalcoholic fatty liver disease: MR imaging of liver proton density fat fraction to assess hepatic steatosis. Radiology 2013;267: 422–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.White UA, Fitch MD, Beyl RA, Hellerstein MK, Ravussin E. Differences in In Vivo Cellular Kinetics in Abdominal and Femoral Subcutaneous Adipose Tissue in Women. Diabetes 2016;65: 1642–1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Strawford A, Antelo F, Christiansen M, Hellerstein MK. Adipose tissue triglyceride turnover, de novo lipogenesis, and cell proliferation in humans measured with 2H2O. Am J Physiol Endocrinol Metab 2004;286: E577–588. [DOI] [PubMed] [Google Scholar]
- 31.Allister CA, Liu LF, Lamendola CA, Craig CM, Cushman SW, Hellerstein MK, et al. In vivo 2H2O administration reveals impaired triglyceride storage in adipose tissue of insulin-resistant humans. J Lipid Res 2015;56: 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013;41: e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15: 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kramer A, Green J, Pollard J, Jr., Tugendreich S. Causal analysis approaches in Ingenuity Pathway Analysis. Bioinformatics 2014;30: 523–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gong J, Sun Z, Li P. CIDE proteins and metabolic disorders. Curr Opin Lipidol 2009;20: 121–126. [DOI] [PubMed] [Google Scholar]
- 37.Feldo M, Kocki J, Lukasik S, Bogucki J, Feldo J, Terlecki P, et al. CIDE--A gene expression in patients with abdominal obesity and LDL hyperlipoproteinemia qualified for surgical revascularization in chronic limb ischemia. Pol Przegl Chir 2013;85: 644–648. [DOI] [PubMed] [Google Scholar]
- 38.Wu J, Zhang L, Zhang J, Dai Y, Bian L, Song M, et al. The genetic contribution of CIDEA polymorphisms, haplotypes and loci interaction to obesity in a Han Chinese population. Mol Biol Rep 2013;40: 5691–5699. [DOI] [PubMed] [Google Scholar]
- 39.Jash S, Banerjee S, Lee MJ, Farmer SR, Puri V. CIDEA Transcriptionally Regulates UCP1 for Britening and Thermogenesis in Human Fat Cells. iScience 2019;20: 73–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Virtue S, Vidal-Puig A. Adipose tissue expandability, lipotoxicity and the Metabolic Syndrome--an allostatic perspective. Biochim Biophys Acta 2010;1801: 338–349. [DOI] [PubMed] [Google Scholar]
- 41.McLaughlin T, Sherman A, Tsao P, Gonzalez O, Yee G, Lamendola C, et al. Enhanced proportion of small adipose cells in insulin-resistant vs insulin-sensitive obese individuals implicates impaired adipogenesis. Diabetologia 2007;50: 1707–1715. [DOI] [PubMed] [Google Scholar]
- 42.Liu A, McLaughlin T, Liu T, Sherman A, Yee G, Abbasi F, et al. Differential intra-abdominal adipose tissue profiling in obese, insulin-resistant women. Obes Surg 2009;19: 1564–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.