Abstract
The emergence of severe acute syndrome coronavirus 2 (SARS-CoV-2) in December 2019 has led to the global COVID-19 pandemic. Although the symptoms of most COVID-19 patients are mild or self-curable, most of severe patients have sepsis caused by cytokine storms, which greatly increases the case fatality rate. Moreover, there is no effective drug that can limit the novel coronavirus thus far, so it is more needed to develop antiviral drugs for the SARS-CoV-2. In our research, we employed the techniques of molecular docking to screen 35 flavonoid compounds among which 29 compounds have Z-scores lower than − 6. Then, ( −)-gallocatechin gallate, ( +)-gallocatechin and baicalein were identified to have potent inhibitory activity against SARS-CoV-2 Mpro with IC50 values of 5.774 ± 0.805 μM, 13.14 ± 2.081 μM and 5.158 ± 0.928 μM respectively by FRET assay. Molecular docking results also showed that ( −)-gallocatechin gallate, ( +)-gallocatechin and baicalein can non-covalently bind to Mpro through π-π stacking and hydrogen bonds in the Cys145 catalytic site. We further evaluated the effect of ( −)-gallocatechin gallate and baicalein on cytokine storms using a mouse model of sepsis. ( −)-Gallocatechin gallate and baicalein significantly reduced sepsis of mouse models on weight, murine sepsis score, and survival rate and reduced the inflammatory factor levels, such as TNF-α, IL-1α, IL-4, and IL-10. Overall, ( −)-gallocatechin gallate and baicalein show certain potential of treatment against COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s10753-021-01602-z.
KEY WORDS: SARS-CoV-2, Mpro protease, flavonoids, sepsis, cytokine storm
Introduction
The coronavirus pandemic starting in 2019 has been named by the World Health Organization (WHO) as “COVID-19” [1]. According to the WHO’s assessment, the current COVID-19 outbreak can be classified as a global pandemic and has potential of significant impacts on human health and life [2]. As of May 7, 2021, there were more than 157 million confirmed cases and 3.28 million deaths worldwide. According to the current data, the symptoms of most COVID-19 patients are mild and self-recoverable, with clinical manifestations of fever, cough, fatigue, myalgia, and diarrhea, and most patients have a good prognosis. Patients with critical conditions often develop respiratory failure, sepsis, septic shock, and multiple organ failure 1 week after the initial symptom was presented. Sepsis, caused by cytokine factor storms, greatly increases the mortality of critically ill patients [3]. At present, there is no drug with good efficacy for novel coronavirus. Therefore, there is strong a need for discovering new leads to develop a drug against SARS-CoV-2.
Coronaviruses are a large family of viruses which often lead to the illness with the symptom of colds and fever; some infections can result in severe illnesses, such as Middle East respiratory syndrome (MERS) and severe acute respiratory syndrome (SARS) [4]. The COVID-19 virus as a single plus-strand RNA virus has never been found in the human body before, which has round or elliptical shape approximately 60–140 nm in diameter [5]. The viral genome encodes replicases, four structural proteins (spines, envelopes, membranes, and nucleocapsid proteins), 16 non-structural proteins (NSPs), and nine accessory proteins. Among these proteins, non-structural proteins play very important roles in virus replication and transcription [6, 7]. NSP5 is the main protease of SARS-CoV-2, which can cut polyproteins translated by viral RNA. The main protease cleaves 12 smaller proteins from the polyprotein, which will be involved in the replication of viral RNA. Therefore, the main protease of coronavirus is an important potential drug target, which is crucial to inhibit virus replication [8].
In recent years, increasing attention has been given to natural products extracted from plants and animals. At present, many natural products have been proven to have therapeutic effects on different diseases [9]. Since the COVID-19 outbreak, scientists have performed many studies, such as screening SARS-CoV-2 inhibitors by molecular docking [10–14] and so on, but few have performed further enzymatic tests. Quercetin known as a SARS-CoV-2 3CLpro inhibitor was screened from a small chemical library consisting of 150 compounds by using a FRET assay. Quercetin has a good inhibition constant of approximately 7 μM and compares quite well to SARS-CoV-2 3CLpro inhibitors already reported, which are considered promising candidates for optimization or preclinical studies [15]. Mangiferin, glucogallin, and phlorizin are natural polyphenols with antiviral activity. They showed good affinity toward the SARS-CoV-2 main protease and interact with some common interacting residues and binding in the same pocket of Mpro in comparison to the reported inhibitor N3. Natural polyphenolic compounds may be potentially safe Mpro inhibitors that can prevent posttranslational modification of viral proteins by inhibiting the activity of the main proteases [16]. The Indian medicinal plant Withania somnifera (Ashwagandha) is widely used by the traditional medical practitioners in India for curing various diseases. Withanoside V, one of the active components of Ashwagandha, has been reported to be a potent inhibitor of SARS-CoV-2 Mpro, which Ashwagandha was proposed to treat COVID-19 [17]. In the past year, many natural compounds, such as andrographolide [18], glycyrrhizin [19], and phenylbenzopyrone of flavonoids [20], were reported to inhibit the activity of the SARS-CoV-2 main protease and stop virus replication. These studies provide inspiration for finding protease inhibitors from natural compounds. In our study, ( −)-gallocatechin gallate, ( +)-gallocatechin, and baicalein were selected from 35 natural flavonoid products as potent inhibitors of SARS-CoV-2 Mpro by molecular docking and enzyme activity screening. We also used a mouse model of fecal dilution-induced sepsis to evaluate the effects of ( −)-gallocatechin gallate and baicalein on cytokine storms. They all showed a good effect in reducing the severity of sepsis, suggesting that ( −)-gallocatechin gallate and baicalein may have potential to inhibit COVID-19.
Materials and Methods
Drugs and Reagents
The 29 extracted compounds were obtained mainly from Topscience Co. Ltd (Shanghai, China). Cefpirome sulfate was obtained from Yuanye Bio-Technology Co., Ltd (Shanghai, China). ( −)-Gallocatechin gallate and baicalein were purchased from Topscience Co., Ltd (Shanghai, China). An enzyme activity inhibitor screening kit was purchased from Beyotime Biotechnology (Shanghai, China). An ELISA kit was provided by Enzymelink Biotechnology Co., Ltd (Shanghai, China).
Molecular Modeling
The crystal structure of SARS-CoV-2 Mpro (PDB ID 6Y2F), which was resolved by Zhang et al. in 2020 [8], was downloaded from the RCSB Protein Data Bank (PDB) and prepared using the Protein Preparation Wizard module in Schrödinger 2017 to remove all crystallographic water molecules, made up the missing atoms of unseen side chains, add hydrogen atoms, and assign protonation states and partial charges with the OPLS_2005 force field. Finally, the crystal structure was minimized until the root-mean-square deviation (RMSD) of the non-hydrogen atoms reached less than 0.3 Å. The cocrystallized SARS-CoV-2 Mpro inhibitor was initially removed from the PDB complex and then redocked to the crystal structure. The RMSD between the redocked and crystal conformation of the ligand was 1.98 Å. The 22 flavonoids were prepared using the LigPrep module of the Schrödinger 2017 molecular modeling package to add hydrogen atoms, convert 2D structures to 3D, generate stereoisomers, and determine the ionization state at pH 7.0 ± 2.0 with Epik[22]. Using the prepared receptor structure, a receptor grid file was generated around the original ligand of the protein, and the 22 flavonoids were docked to the receptor using the Glide XP protocol.
Protease Activity Assay
Enzyme activity inhibitor screening employs the fluorescence resonance energy transfer method. In the reaction system, there are drugs, Mpro, and substrates with fluorescent donors and receptors. The Mpro with enzyme activity can decompose the substrate and produce fluorescence. The enzyme activity assay was performed in 96-well black flat-bottomed plates. For the screening of Mpro inhibitor and IC50 measurement, 300 nM Mpro was incubated with compounds at 37 ℃ for 5 min in assay buffer (50 mM Tris, 150 mM NaCl, 1 mM EDTA, 1% glycerol, PH 7.3), then 20 μM substrate, Dabcyl-KTSAVLQSGFRKME-Edans, was added to the reaction system and incubated for 10 min at 37 ℃ before detection. The negative control well consisted of 93 μL assay buffer, 5 μL DMSO, and 2 μL substrate. Enzyme activity control well contained 92 μL assay buffer, 1 μL Mpro, 5 μL DMSO, and 2 μL substrate; sample wells are 92 μL assay buffer, 1 μL Mpro, 5 μL compound, and 2 μL substrate. The IC50 was calculated by plotting the relative fluorescence units (RFUs) against various concentrations of Mpro inhibitors using a dose–response curve in GraphPad Prism 7 software. The results: inhibition rate (%) = (RFU 100% enzyme activity control-RFU sample) / (RFU 100% enzyme activity control-RFU negative control) × 100%.
Experimental Animals
Male C57BL/6 (6–8 weeks, 20–22 g) mice were purchased from Charles river (Beijing, China). All animal care and experimental procedures complied with guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of Nankai University (Permit No. SYXK 2014–0003). All experimental protocols were approved by the Animal Experiment Committee of Tianjin International Joint Academy of Biomedicine (approval no. SYXK (JIN) 2017–0003).
Modeling and Animal Grouping
In this experiment, a mouse sepsis model was established by intraperitoneal injection of 10% fecal diluent. The mice were randomly divided into five groups (n = 7): saline group, 10% fecal dilution only group, fecal dilution with cefpirome sulfate (CS) group (100 mg/kg), and fecal dilution with ( −)-gallocatechin gallate (50 and 100 mg/kg) treatment groups and fecal dilution with baicalein (100 and 200 mg/kg). Intraperitoneal injection of 10% fecal dilution was given to the model group and treatment group at day 0. For the treatment group, intraperitoneal injections of cefpirome sulfate (100 mg/kg), ( −)-gallocatechin gallate (50 and 100 mg/kg) and baicalein (100 and 200 mg/kg) were given 2 h before fecal dilution injection. The saline group received normal saline by the same procedure.
Weight Monitoring
The weight of each group was measured and recorded at the 0th hour, 24th hour, and 48th hour of the model.
MSS
The murine sepsis score (MSS) was tested according to the manufacturer’s introduction [23]. At 0 h, 24 h, and 48 h, the appearance and ideology of the mice were scored. In the scoring method, variables such as the degree of erection, activity, posture, behavior, chest movement, chest sound, eye opening, and weight loss were evaluated. Score 1 to 4 points.
Histological
The heart, liver, spleen, lung, and kidney were fixed with 10% formaldehyde at the 48 h. After 2 days of fixation, the sections were embedded in paraffin and sectioned into 4-μm slices. The prepared slices were dried at 60 ℃ and then dewaxed them as usual. Hematoxylin staining was performed for 3–5 min, then the slices were rinsed with ultra-pure water for 10–15 min, with hydrochloric ethanol separation for 10–20 s and dye with eosin for 3 min. After dehydration and transparency, neutral gum was added to seal the slice.
ELISA for the Detection of Inflammatory Factors
The levels of IL-1α, TNF-α, IL-4, and IL-10 in the serum of fecal dilution-induced sepsis mice were determined by enzyme-linked immunosorbent assay (ELISA) kits. The experimental process strictly followed the instructions of the ELISA kit. After collecting mouse serum, 50 μL of the standard substance and 50 μL of the test sample were placed into the reaction well, and then 50 μL of the sample was added as a thinner. Then, 50 μL biotin-labeled antibody was added immediately. After incubating for 1 h at 37 °C, the liquid in the wells was shaken off, wash solution was added to each well, and the wells were washed 3 times with wash solution; 80 μL affinity streptococcus-HRP was added to each well and incubated at 37 °C for 30 min. The cleaning was repeated 3 times. Fifty microliters each of substrate A and B was added to each well and incubated at 37 °C for 10 min. Quickly, 50 μL stop solution was add to the ELISA plate, and the OD value of each well was immediately measured at 450 nm.
Statistical Analysis
Data are presented using the Prism version 7.0 software as the means ± SD. Differences between the experimental and control groups were assessed by Student’s t test. Significant differences among multiple groups were detected by one-way ANOVA. P < 0.05 was considered as statistically significant.
Results
Molecular Docking
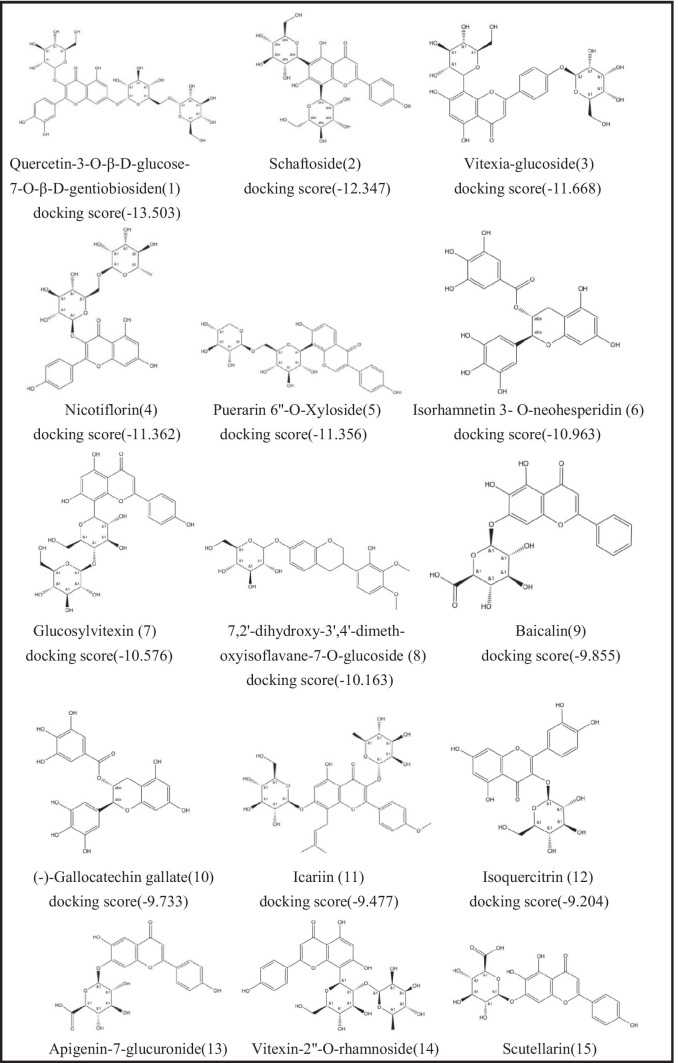
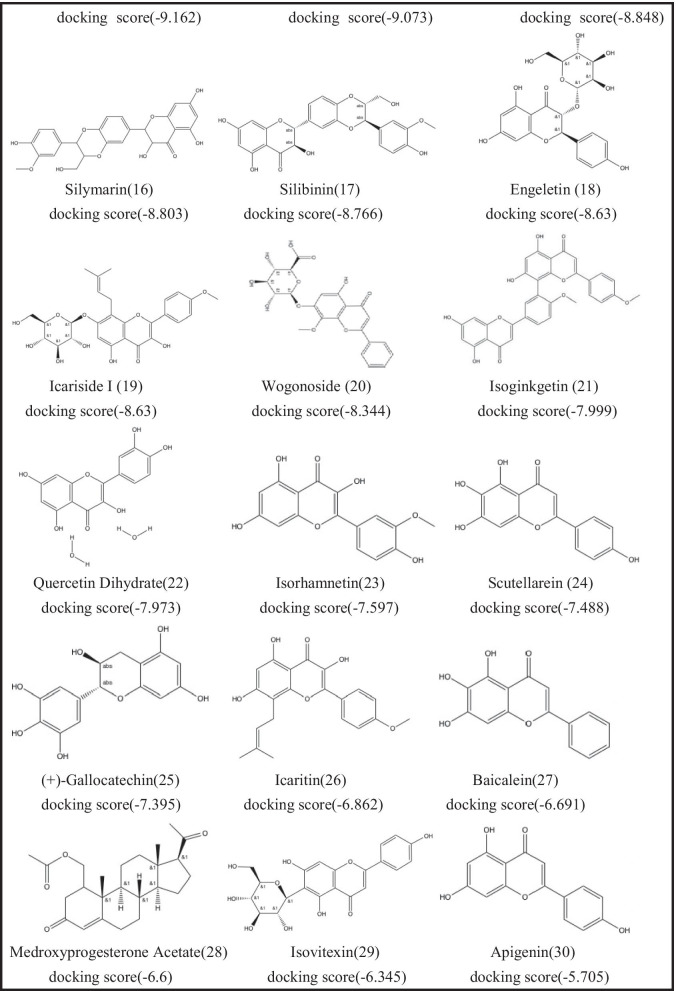
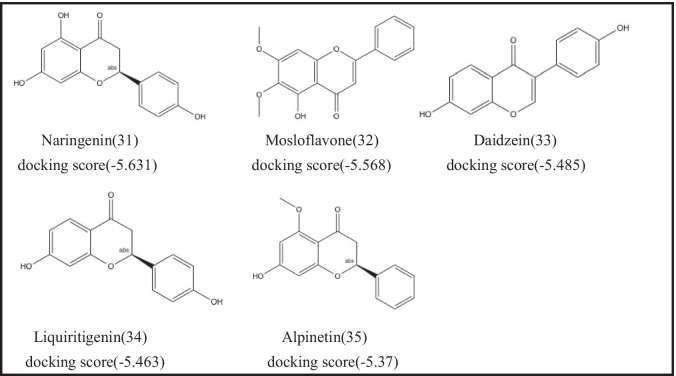
Recently, a large number of studies reported the screening results of natural compounds, especially flavonoids, as anti-SARS-CoV-2 inhibitors based on in silico drug discovery approaches. Therefore, we selected some flavonoid drugs for molecular docking with the Mpro main protease. We docked 35 flavonoids to SARS-CoV-2 Mpro. The 2D structures of the 35 flavonoids and the corresponding Glide XP docking scores are listed in Table 1. Twenty-nine of the 35 flavonoid compounds have docking scores lower than − 6.0, which indicates that these compounds may have potential to effectively inhibit SARS-CoV-2 Mpro activity. We further evaluated their activities of 29 flavonoid compounds by the following biological assays.
Table 1.
List of Drugs for Molecular Docking
FRET Assays of SARS-CoV-2 Mpro Inhibitors
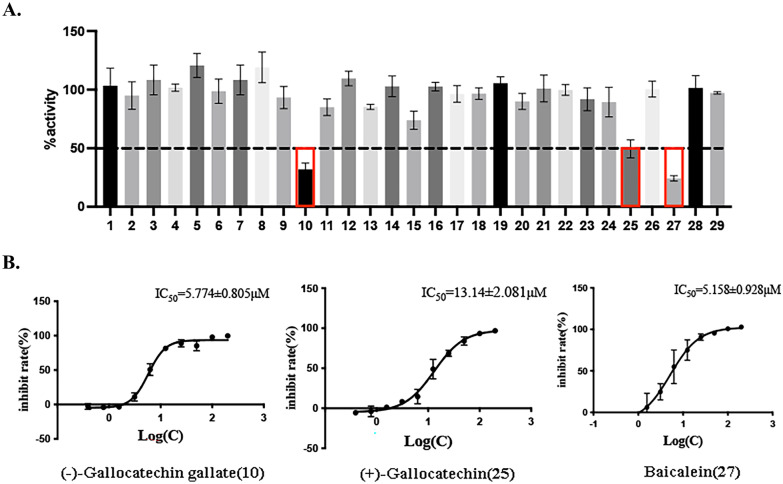
With the established FRET assay system, we screened 29 flavonoid compounds with molecular docking scores lower than − 6, to identify potential SARS-CoV-2 Mpro inhibitors. The inhibitors at a final concentrate of 20 μM were preincubated with 0.3 μM Mpro at 37 ℃ for 5 min before 20 μM FRET substrate was added. Inspiringly, three compounds, ( −)-gallocatechin gallate (10), ( +)-gallocatechin (25), and baicalein (27) had fine inhibition and inhibition rates of more than 50% against Mpro (Fig. 1A and Table S1). As negative control, other compounds that have lower docking scores, such as quercetin-3-O-β-D-glucose-7-O-β-D-gentiobiosiden, schaftoside, vitexia-glucoside, nicotiflorin, and isorhamnetin 3- O-neohesperidin, did not exhibit detectable inhibitory activity.
Fig. 1.
Screening of 29 flavonoids, docking score lower than -6, against SARS-CoV-2 Mpro using the FRET assay. A 40-μM compound (final concentration 20 μM) was pre-incubated with 0.3 μM SARS-CoV-2 M pro at 30 °C for 10 min, and then 200 μM FRET substrate was added to the reaction mixture to initiate the reaction. The excitation wavelength is 340 nm, and the emission wavelength is 490 nm for fluorescence measurement. Results: Inhibition rate (%) = (RFU100% enzyme activity control-RFU sample) / (RFU100% enzyme activity control-RFU blank control) × 100%. The results are average ± standard deviation of three repeats. B The inhibitory assay of ( −)-gallocatechin gallate, ( +)-gallocatechin, and baicalein show efficient inhibition for Mpro. Error bars: mean ± S.D. of three independent replicates.
Inhibitory Activities of SARS-CoV-2 Mpro by the ( −)-gallocatechin gallate, ( +)-gallocatechin and Baicalein and Their Structural Basis
In the enzyme activity screening assay, the results showed three potent flavonoid compounds were confirmed as SARS-CoV-2 inhibitors. To further evaluate the inhibitory activity of these three compounds, we detected their inhibition in a gradient concentrate and calculated the 50% inhibitory concentration values (IC50), respectively. The IC50 values of ( −)-gallocatechin gallate, ( +)-gallocatechin, and baicalein were 5.774 ± 0.805 μM, 13.14 ± 2.081 μM, and 5.158 ± 0.928 μM, respectively (Fig. 1B). Among these compounds, ( −)-gallocatechin gallate and baicalein showed better inhibitory effects, and the IC50 was lower than 10 μM.
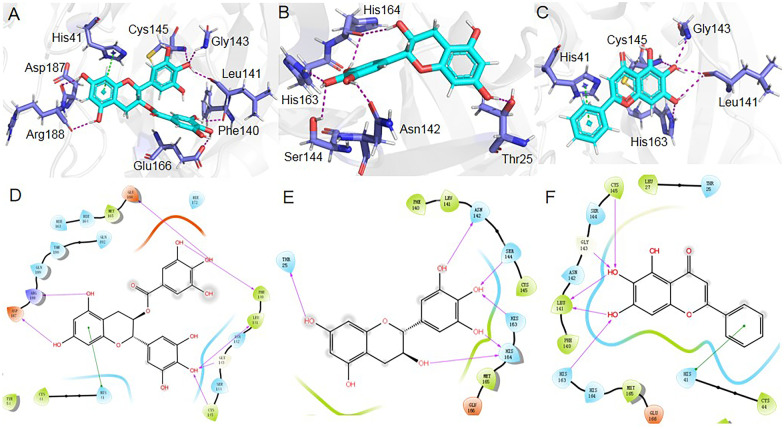
The docked binding positions and the interaction details of ( −)-gallocatechin gallate, ( +)-gallocatechin, and baicalein with SARS-CoV-2 Mpro are presented in Fig. 2. In the structure of ( −)-gallocatechin gallate bound with SARS-CoV-2 Mpro (Fig. 2A and D), the dihydrobenzopyran ring of ( −)-gallocatechin gallate interacts with the imidazole side chain of His41 through π-π stacking. The 5- and 7-hydroxyl groups of dihydrobenzopyran form hydrogen bonds with the backbone oxygen of Asp187 and Arg188. The 4-hydroxyl of trihydroxyphenyl forms hydrogen bonds with the backbone oxygen of Leu141, the backbone nitrogen of Gly143, and the backbone nitrogen of Cys145. The 3- and 4-hydroxyl groups of trihydroxybenzoate form hydrogen bonds with the backbone oxygen of Phe140 and the side chain oxygen Glu166.
Fig. 2.
Docked conformations of ( −)-gallocatechin gallate, ( +)-gallocatechin, and baicalein in SARS-CoV-2 Mpro. A–C 3D binding interactions of the compounds with surrounding amino acids. D–F 2D binding interactions of the compounds with surrounding amino acids.
In the structure of ( +)-gallocatechin bound with SARS-CoV-2 Mpro (Fig. 2B and E), the 3- and 7-hydyoxyl groups of ( +)-gallocatechin formed hydrogen bonds with the backbone oxygen of His164 and the side chain oxygen of Thr25. The 3′-, 4′- and5′-hydroxyl groups of ( +)-gallocatechin form hydrogen bonds with the backbone oxygen of His164, the imidazole side chain of His 163, the side chain oxygen Ser144, and the side chain oxygen Asn142.
In the structure of baicalein binding with SARS-CoV-2 Mpro (Fig. 2C and F), the 2-phenyl group of baicalein interacts with the imidazole side chain of His41 through π-π stacking. The 6- and 7-hydroxyl groups of baicalein form hydrogen bonds with the backbone nitrogen of Cys145, the backbone nitrogen of Gly13, the backbone oxygen of Leu141, and the imidazole side chain of His 163.
( −)-gallocatechin Gallate and Baicalein Attenuate Fecal Dilution-Induced Sepsis in Mice
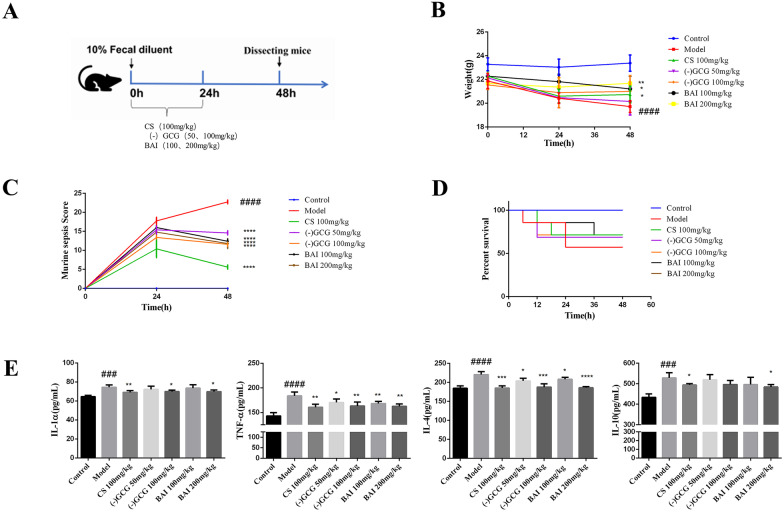
As shown in Fig. 3A, we evaluated the effect of ( −)-gallocatechin gallate and baicalein on sepsis using a mouse model. The change of body weight is displayed in Fig. 3B. Compared with the control group, the weight of the model group was significantly reduced. The weight of the mice in the treatment group was higher than the weight of the mice in the model group, and the efficacy of baicalein was better than the efficacy of ( −)-gallocatechin gallate. The mouse sepsis score (MSS) of the model group was significantly higher than the MSS of the control group at the 24 h and 48 h after the model was established, but the group treated with ( −)-gallocatechin gallate (50 and 100 mg/kg) and baicalein (100 and 200 mg/kg) attenuated the symptoms caused by sepsis (Fig. 3C). Observing the mortality of mice every 6 h, it was found that 3 mice were found to have died in the model group, and the mortality rate is 42%, while the mortality rate of the treatment group was reduced to 28% except for the low-dose ( −)-gallocatechin gallate group. The results showed that ( −)-gallocatechin gallate and baicalein can improve the survival rate of sepsis caused by fecal dilution (Fig. 3D).
Fig. 3.
Effect of ( −)-gallocatechin gallate and baicalein on fecal dilution-induced sepsis in mice. A Experimental scheme of mice sepsis induced by fecal diluent. B Changes in body weight in different groups of mice in review (control, model, 100 mg/kg cefpirome sulfate, 50 mg/kg ( −)-gallocatechin gallate, and 100 mg/kg ( −)-gallocatechin gallate, 100 mg/kg baicalein, and 200 mg/kg baicalein). C The effect of ( −)-gallocatechin gallate and baicalein on murine sepsis score (MSS) of the different groups of mice described above. D The effect of ( −)-gallocatechin gallate and baicalein on the survival time of the different groups of mice described above. E Serum IL-1α, TNF-α, IL-4, and IL-10 were detected by ELISA. Values are presented as the mean ± SEM (n = 5), ##P < 0.01; ####P < 0.0001,significantly different from control group; *P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.0001, significantly different from model groups.
( −)-Gallocatechin gallate and Baicalein Inhibit the Release of Proinflammatory Cytokines in Serum
COVID-19 and sepsis both have the pathological manifestation of cytokine storms, so we detected the levels of proinflammatory factors in the serum of septic mice. As shown in Fig. 3E, compared with the control group, the levels of the serum inflammatory factors IL-1α, TNF-α, IL-4, and IL-10 in the model group were significantly increased, while they were markedly decreased after treatment with ( −)-gallocatechin gallate and baicalein. The data suggested that ( −)-gallocatechin gallate and baicalein could downregulate the level of serum inflammatory cytokines induced by sepsis.
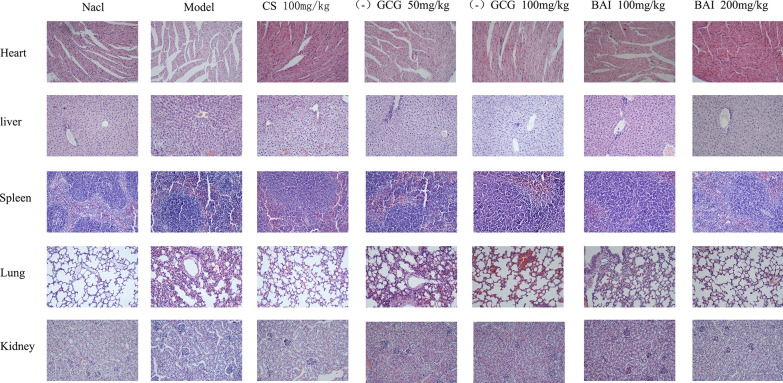
We also evaluated the effect of ( −)-gallocatechin gallate and baicalein on tissue damage induced by sepsis. In the pathological sections of the heart (Fig. 4), the myocardial tissue cells of the control group were evenly distributed, the fibers were clearly organized, and are long spindle-shaped; the myocardial cells of the model group were swollen and deformed or even ruptured, a large number of nuclei were constricted and deeply stained, and the myocardial fibers are severely disordered and deformed. In the CS group and ( −)-gallocatechin gallate group, some cardiomyocytes were swollen and deformed, a small number of nuclei were densely stained, and myocardial fibers were disordered and slightly deformed; in the baicalein group, a small number of myocardial cells were enlarged and deformed, and fewer nuclei were constricted and deeply stained, and myocardial fibers were disorderly by performance and dense structure. The structure of the alveoli in the lung tissue of the control group mice was disordered, the alveolar interval was significantly widened, and the lung interstitium had obvious congestion and edema, some atelectasis, and a large amount of neutrophil infiltration. In ( −)-gallocatechin gallate and baicalein group mouse, the alveoli structure was basically complete, the alveolar compartment was widened, the pulmonary interstitial congestion and edema were significantly improved, and the neutrophil infiltration was significantly reduced. A large amount of inflammatory cell infiltration, hepatocellular edema, and necrosis were observed in the liver tissue of the model group; the liver tissue of the positive drug group and the (-)-gallocatechin gallate and baicalein group showed different degrees of congestion and inflammatory cell infiltration, but the degree of damage was significantly less than the model group. In the model group, the spleen and kidney tissues were structurally disordered or even ruptured, with a large number of inflammatory cells appearing. After (-)-gallocatechin gallate and baicalein treatment, the tissue structure was more complete than the tissue structure of the model group, and the inflammatory cells were reduced.
Fig. 4.
Effect of ( −)-gallocatechin gallate and baicalein on serum proinflammatory cytokine expression. HE staining was used to evaluate the effect of ( −)-gallocatechin gallate and baicalein on fecal dilution-induced pathological changes (20 × magnification).
Discussion
The epidemic of COVID-19 urgently requires new treatment strategies [6, 7, 24, 25]. Currently, there is no specific drug for SARS-CoV-2, but a large number of compounds have been reported to have good binding affinity with Mpro through virtual screening of the structure [21, 13, 26, 27]. For example, a team from the University of Alberta, Canada, found that GC376, an earlier protease inhibitor prodrug used to treat feline coronavirus infection, and its mother GC373 compound were effective in inhibiting SARS-CoV and SARS-CoV-2 Mpro in the nanomolar range [28–31]. Antiretroviral drugs such as lopinavir and nefinavir for patients with immune deficiency syndrome (AIDS) and HIV-1 infection that they can bind to the main protease of novel coronavirus and inhibit virus replication [32]. However, in clinical trials, such drugs are given at a high dose, resulting in low patient compliance [33]. In China, many scientists focus their attention on natural products, such as flavonoids [34], alkaloids [35, 36], and green tea polyphenols [37], in an attempt to find drugs that are effective against novel coronavirus. Some more studies are carried out to further develop them to be potential therapeutic drugs against novel coronavirus.
In our study, the binding affinities of 35 natural flavonoid compounds with SARS-CoV-2 Mpro were screened through computer simulation of molecular docking. The results showed that ( −)-gallocatechin gallate, ( +)-gallocatechin, and baicalein have high affinity for Mpro. ( −)-Gallocatechin gallate, ( +)-gallocatechin, and baicalein also exhibited significant inhibition with IC50 values of 5.774 ± 0.805 μM, 13.14 ± 2.081 μM, and 5.158 ± 0.928 μM, respectively. Structurally, three compounds can non-covalently bind to SARS-CoV-2 Mpro through π-π stacking and hydrogen bonds in the Cys145 catalytic site.
The novel coronavirus causes an overactive inflammatory response known as a cytokine storm, followed by severe lung inflammation. Sepsis is the most common symptom in severe and critical COVID-19 patients, 42% of survived, and 100% of fatal patients had sepsis [38]. These severe and critical patients presented clinical manifestations of sepsis or septic shock, including fever, intractable hypotension, microcirculatory dysfunction, severe metabolic acidosis, and different degrees of multiple organ dysfunction. COVID-19 complications with sepsis undoubtedly increase the difficulty of treatment and seriously affect the prognosis of patients. In this study, we evaluated the inhibitory effect of ( −)-gallocatechin gallate and baicalein on SARS-CoV-2 main protease and sepsis. Both compounds showed an effective effect in relieving sepsis in mice by evaluating body weight, MSS score, and survival rate. They effectively inhibited the expression of inflammatory factors such as IL-1α, TNF-α, IL-4, and IL-10, which are main factors inducing cytokine storms in sepsis. We also compared the pathological sections of the heart, liver, spleen, lung, and kidney of each group of mice and found that in the model group, the damage to the spleen of the model group mice was serious, and the white pulp was significantly enlarged; a large amount of inflammatory infiltration was found in the heart, liver, lung, and kidney; the damage was very obvious and the cell arrangement was disordered; after (-)-gallocatechin gallate and baicalein intervention, the damages and observations mentioned above were alleviated.
Conclusion
In summary, the data indicated that ( −)-gallocatechin gallate and baicalein may be a potential targeting SARS-CoV-2 main protease inhibitor and also significantly reduces the storm of inflammatory factors and systemic inflammatory response in septic mice. ( −)-Gallocatechin gallate and baicalein may be used against COVID-19. At the same time, our study also shows that natural products of small molecular bioactivity may be a useful source of SARS-CoV-2 Mpro inhibitors and can be a powerful line of defense against COVID-19.
Supplementary Information
Below is the link to the electronic supplementary material.
Author Contribution
Data curation, Ting Xiao and Mengqi Cui; formal analysis, Shanfa Ren and Jiali Bao; funding acquisition, Honggang Zhou and Cheng Yang; investigation, Peipei Zhang and Liang Zhang; methodology, Dandi Gao; project administration, Honggang Zhou; resources, Mingjiang Li; Software, Caijuan Zheng; supervision, Jianping Lin; validation, Ming Wang; visualization, Ronghao Sun; writing — original draft, Ting Xiao; writing — review and editing, Dongmei Li. All the authors read and approved the final manuscript. All data were generated in-house, and no paper mill was used. All authors agree to be accountable for all aspects of work ensuring integrity and accuracy.
Funding
This study was supported by Tianjin Science and Technology Project (Grant No. 20ZXGBSY00050) and The Fundamental Research Funds for the Central University (Grant 735–63201241 and Grant 735–63201239).
Data Availability
Data available on request from the authors.
Declarations
Ethics Approval and Consent to Participate
All animals care and experimental procedures conformed guidelines approved by the Institutional Animal Care and Use Committee (IACUC) of Nankai University (Permit No. SYXK 2014–0003).
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Ting Xiao, Mengqi Cui, Caijuan Zheng, and Peipei Zhang contributed equally to this work.
Contributor Information
Ting Xiao, Email: xting2011@163.com.
Mengqi Cui, Email: cuimengqi2018@163.com.
Caijuan Zheng, Email: 2120191180@mail.nankai.edu.cn.
Peipei Zhang, Email: zhangpeipei6223408@126.com.
Dongmei Li, Email: dongmeili@nankai.edu.cn.
Honggang Zhou, Email: honggang.zhou@nankai.edu.cn.
Cheng Yang, Email: cheng.yang@nankai.edu.cn.
References
- 1.Ciotti M, Angeletti S, Minieri M, Giovannetti M, Benvenuto D, Pascarella S, Sagnelli C, Bianchi M, Bernardini S, Ciccozzi M. COVID-19 Outbreak: An Overview. Chemotherapy. 2019;64:215–223. doi: 10.1159/000507423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tandon PN. COVID-19: Impact on health of people & wealth of nations. Indian Journal of Medical Research. 2020;151:121–123. doi: 10.4103/ijmr.IJMR_664_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Special Expert Group for Control of the Epidemic of Novel Coronavirus Pneumonia of the Chinese Preventive Medicine A. 2020. An update on the epidemiological characteristics of novel coronavirus pneumonia (COVID-19). Zhonghua Liu Xing Bing Xue Za Zhi 41:139–144. [DOI] [PubMed]
- 4.Wang HJ, Du SH, Yue X, Chen CX. Review and Prospect of Pathological Features of Corona Virus Disease. Fa Yi Xue Za Zhi. 2020;36:16–20. doi: 10.12116/j.issn.1004-5619.2020.01.004. [DOI] [PubMed] [Google Scholar]
- 5.Gupta SP. Progress in Studies on Structural and Remedial Aspects of Newly Born Coronavirus, SARS-CoV-2. Current Topics in Medicinal Chemistry. 2020;20:2362–2378. doi: 10.2174/1568026620666200922112300. [DOI] [PubMed] [Google Scholar]
- 6.Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved alpha-ketoamide inhibitors. Science. 2020;368:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harvey AL. Natural products in drug discovery. Drug Discovery Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 10.Gentile, D., V. Patamia, A. Scala, M.T. Sciortino, A. Piperno, and A. Rescifina. 2020. Putative Inhibitors of SARS-CoV-2 Main Protease from A Library of Marine Natural Products: A Virtual Screening and Molecular Modeling Study. Marine Drugs 18. [DOI] [PMC free article] [PubMed]
- 11.Joshi T, Joshi T, Sharma P, Mathpal S, Pundir H, Bhatt V, Chandra S. In silico screening of natural compounds against COVID-19 by targeting Mpro and ACE2 using molecular docking. European Review for Medical and Pharmacological Sciences. 2020;24:4529–4536. doi: 10.26355/eurrev_202004_21036. [DOI] [PubMed] [Google Scholar]
- 12.Kandeel, M., and M. Al-Nazawi. 2020. Virtual screening and repurposing of FDA approved drugs against COVID-19 main protease. Life Science 251:117627. [DOI] [PMC free article] [PubMed]
- 13.Hakmi M, Bouricha EM, Kandoussi I, Harti JE, Ibrahimi A. Repurposing of known anti-virals as potential inhibitors for SARS-CoV-2 main protease using molecular docking analysis. Bioinformation. 2020;16:301–306. doi: 10.6026/97320630016301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ton, A.T., F. Gentile, M. Hsing, F. Ban, and A. Cherkasov. 2020. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Molecular Informatics 39:e2000028. [DOI] [PMC free article] [PubMed]
- 15.Abian O, Ortega-Alarcon D, Jimenez-Alesanco A, Ceballos-Laita L, Vega S, Reyburn HT, Rizzuti B, Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. International Journal of Biological Macromolecules. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh, R., A. Gautam, S. Chandel, A. Ghosh, D. Dey, S. Roy, V. Ravichandiran, and D. Ghosh. 2020. Protease Inhibitory Effect of Natural Polyphenolic Compounds on SARS-CoV-2: An In Silico Study. Molecules 25. [DOI] [PMC free article] [PubMed]
- 17.Tripathi MK, Singh P, Sharma S, Singh TP, Ethayathulla AS, Kaur P. Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor. Journal of Biomolecular Structure & Dynamics. 2021;39:5668–5681. doi: 10.1080/07391102.2020.1790425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi TH, Huang YL, Chen CC, Pi WC, Hsu YL, Lo LC, Chen WY, Fu SL, Lin CH. Andrographolide and its fluorescent derivative inhibit the main proteases of 2019-nCoV and SARS-CoV through covalent linkage. Biochemical and Biophysical Research Communications. 2020;533:467–473. doi: 10.1016/j.bbrc.2020.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van de Sand, L., M. Bormann, M. Alt, L. Schipper, C.S. Heilingloh, E. Steinmann, D. Todt, U. Dittmer, C. Elsner, O. Witzke, and A Krawczyk. 2021. Glycyrrhizin Effectively Inhibits SARS-CoV-2 Replication by Inhibiting the Viral Main Protease. Viruses 13. [DOI] [PMC free article] [PubMed]
- 20.Potshangbam, A.M., P. Nongdam, A.K. Kumar, and R.S. Rathore. 2021. Phenylbenzopyrone of flavonoids as a potential scaffold to prevent SARS-CoV-2 replication by inhibiting its MPRO main protease. Current Pharmaceutical Biotechnology. [DOI] [PubMed]
- 21.Dai, W., B. Zhang, X.M. Jiang, H. Su, J. Li, Y. Zhao, et al. 2020. Stucture-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 368: 1331-1335 [DOI] [PMC free article] [PubMed]
- 22.Verma P, Tiwari M, Tiwari V. In silico high-throughput virtual screening and molecular dynamics simulation study to identify inhibitor for AdeABC efflux pump of Acinetobacter baumannii. Journal of Biomolecular Structure & Dynamics. 2018;36:1182–1194. doi: 10.1080/07391102.2017.1317025. [DOI] [PubMed] [Google Scholar]
- 23.Shrum B, Anantha RV, Xu SX, Donnelly M, Haeryfar SM, McCormick JK, Mele T. A robust scoring system to evaluate sepsis severity in an animal model. BMC Research Notes. 2014;7:233. doi: 10.1186/1756-0500-7-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. New England Journal of Medicine. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Guan X, Wu P, Wang X, Zhou L, Tong Y, Ren R, Leung KSM, Lau EHY, Wong JY, et al. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus-Infected Pneumonia. New England Journal of Medicine. 2020;382:1199–1207. doi: 10.1056/NEJMoa2001316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Das, S., S. Sarmah, S. Lyndem, and A. Singha Roy. 2020. An investigation into the identification of potential inhibitors of SARS-CoV-2 main protease using molecular docking study. Journal of Biomolecular Structure and Dynamics 1–11. [DOI] [PMC free article] [PubMed]
- 27.Fischer, A., M. Sellner, S. Neranjan, M. Smiesko, and M.A. Lill. 2020. Potential Inhibitors for Novel Coronavirus Protease Identified by Virtual Screening of 606 Million Compounds. International Journal of Molecular Sciences 21. [DOI] [PMC free article] [PubMed]
- 28.Vuong W, Khan MB, Fischer C, Arutyunova E, Lamer T, Shields J, Saffran HA, McKay RT, van Belkum MJ, Joyce MA, et al. Feline coronavirus drug inhibits the main protease of SARS-CoV-2 and blocks virus replication. Nature Communications. 2020;11:4282. doi: 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Lovell S, Tiew KC, Mandadapu SR, Alliston KR, Battaile KP, Groutas WC, Chang KO. Broad-spectrum antivirals against 3C or 3C-like proteases of picornaviruses, noroviruses, and coronaviruses. Journal of Virology. 2012;86:11754–11762. doi: 10.1128/JVI.01348-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tiew KC, He G, Aravapalli S, Mandadapu SR, Gunnam MR, Alliston KR, Lushington GH, Kim Y, Chang KO, Groutas WC. Design, synthesis, and evaluation of inhibitors of Norwalk virus 3C protease. Bioorganic & Medicinal Chemistry Letters. 2011;21:5315–5319. doi: 10.1016/j.bmcl.2011.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandadapu SR, Weerawarna PM, Gunnam MR, Alliston KR, Lushington GH, Kim Y, Chang KO, Groutas WC. Potent inhibition of norovirus 3CL protease by peptidyl alpha-ketoamides and alpha-ketoheterocycles. Bioorganic & Medicinal Chemistry Letters. 2012;22:4820–4826. doi: 10.1016/j.bmcl.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Costanzo M, De Giglio MAR, Roviello GN. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Current Medicinal Chemistry. 2020;27:4536–4541. doi: 10.2174/0929867327666200416131117. [DOI] [PubMed] [Google Scholar]
- 33.Cao B, Wang Y, Wen D, Liu W, Wang J, Fan G, Ruan L, Song B, Cai Y, Wei M, et al. A Trial of Lopinavir-Ritonavir in Adults Hospitalized with Severe Covid-19. New England Journal of Medicine. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Das, P., R. Majumder, M. Mandal, and P. Basak. 2020. In-Silico approach for identification of effective and stable inhibitors for COVID-19 main protease (M(pro)) from flavonoid based phytochemical constituents of Calendula officinalis. Journal of Biomolecular Structure and Dynamics 1–16. [DOI] [PMC free article] [PubMed]
- 35.Garg, S., and A. Roy. 2020. In silico analysis of selected alkaloids against main protease (M(pro)) of SARS-CoV-2. Chemico-Biological Interactions 332:109309. [DOI] [PMC free article] [PubMed]
- 36.Ghosh, R., A. Chakraborty, A. Biswas, and S. Chowdhuri. 2020. Identification of alkaloids from Justicia adhatoda as potent SARS CoV-2 main protease inhibitors: An in silico perspective. Journal of Molecular Structure 129489. [DOI] [PMC free article] [PubMed]
- 37.Ghosh, R., A. Chakraborty, A. Biswas, and S. Chowdhuri. 2020. Evaluation of green tea polyphenols as novel corona virus (SARS CoV-2) main protease (Mpro) inhibitors - an in silico docking and molecular dynamics simulation study. Journal of Biomolecular Structure and Dynamics 1–13. [DOI] [PMC free article] [PubMed]
- 38.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, Xiang J, Wang Y, Song B, Gu X, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/S0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data available on request from the authors.