Abstract
The essential genome of a bacterium encompasses core genes associated with basic cellular processes and conditionally essential genes dependent upon environmental conditions or the genetic context. Comprehensive knowledge of those gene sets allows for a better understanding of fundamental bacterial biology and offers new perspectives for antimicrobial drug research against detrimental bacteria such as pathogens. We investigated the essential genome of Xanthomonas hortorum pv. vitians, a gammaproteobacterial plant pathogen of lettuce (Lactuca sativa L.) which belongs to the plant-pathogen reservoir genus Xanthomonas and is affiliated to the family Xanthomonadaceae . No practical means of disease control or prevention against this pathogen is currently available, and its molecular biology is virtually unknown. To reach a comprehensive overview of the essential genome of X. hortorum pv. vitians LM16734, we developed a mixed approach combining high-quality full genome sequencing, saturated transposon insertion sequencing (Tn-Seq) in optimal growth conditions, and coupled computational analyses such as comparative genomics, synteny assessment and phylogenomics. Among the 370 essential loci identified by Tn-Seq, a majority was bound to critical cell processes conserved across bacteria. The remaining genes were either related to specific ecological features of Xanthomonas or Xanthomonadaceae species, or acquired through horizontal gene transfer of mobile genetic elements and associated with ancestral parasitic gene behaviour and bacterial defence systems. Our study sheds new light on our usual concepts about gene essentiality and is pioneering in the molecular and genomic study of X. hortorum pv. vitians.
Keywords: Xanthomonas hortorum, Tn-Seq, bacterial leaf spot of lettuce, mobile genetic elements, Xanthomonadaceae, essential genome
Data Summary
All new genome and sequencing data were deposited in the corresponding databases of the National Center for Biotechnology Information (NCBI, https://www.ncbi.nlm.nih.gov), and are associated with BioProject PRJNA530964 and BioSample SAMN11340225. The Illumina and PacBio raw sequencing reads of the complete genome sequencing of strain LM16734 are accessible at the Sequence Read Archive (SRA) database under accession numbers SRR12066994–SRR12066995. The manually revised annotated assembly carried out at the MicroScope annotation platform (https://mage.genoscope.cns.fr) was deposited in the GenBank database under accession number GCA_014338485.1. The transposon insertion sequencing reads of the two duplicates were also deposited in the SRA database under accession numbers SRR12067051–SRR12067052. The GenBank assembly accession numbers for the other published genome data used in this study are given in Table S1 (available in the online version of this article). Tables S1–S7 and Figs S1–S3 are available at figshare with the following doi: https://figshare.com/s/aab51b0deba42f67e969.
Impact Statement.
The genus Xanthomonas constitutes a huge reservoir of economically and ecologically important plant-pathogenic species that pressure agroecosystems and threaten global food sustainability. Amongst these, Xanthomonas hortorum pv. vitians causes a bacterial leaf spot of lettuce. This disease attacks all varieties of lettuce around the world, and no sustainable and operational means of control has yet been found. Moreover, little is known about the genetic and molecular functioning of X. hortorum species. Previous studies conducted on other organisms have shown that transposon insertion sequencing (Tn-Seq) is a powerful tool for acquiring fundamental knowledge on cell functioning and critical cellular processes because it reliably identifies prospective targets for new antimicrobial compounds. Using the discriminative power of highly saturated genome-wide Tn-Seq and high-quality complete genome sequencing, the present study offers an extensive description and analysis of X. hortorum pv. vitians LM16734 genes required for cell survival in optimal growth conditions. Portrayal of the essential genome is accompanied by a critical reflection on the concepts underlying gene essentiality based on gene conservation, synteny and phylogenomic analyses. In this way, we discriminated between core essential genes of xanthomonads and horizontally acquired contextual essential genes in this first Xanthomonas essential genome.
Introduction
Evolutionary forces shape bacterial genomes through persistence and loss of genes in relation to the fitness cost or benefit they provide to the organism or population. However, for most genes, the conferred fitness depends on occupied environments and ecological niches, or on the lifestyle adopted by the bacterium [1, 2]. A minority of coding sequences are considered indispensable regardless of the environmental context, as they regulate critical cellular processes such as DNA replication, cell wall integrity or energy metabolism [3]. These coding sequences and structural elements as a whole are referred to in the literature as the minimal gene set [4] or essential genome [5] of a given bacterium. However, gene essentiality is usually deduced from indirect observations of lethal mutant phenotypes in a given medium. Thus, observed essentiality could either result from direct alteration of critical functions required for cell survival (e.g. DNA replication, membrane synthesis), or context-dependent effects related to the genome content or environmental conditions (e.g. toxin–antitoxin systems, auxotrophy) [6, 7]. Consequently, the essentiality of some genes varies among strains within the same species [8–10]. Therefore, the essential genome of a bacterium can be seen as a combination of ‘core’ and ‘accessory’ essential genes, and the latter depend on environmental conditions or the genetic context.
Essential genomes have been historically assessed through systematic single-gene knock-out or deletion experiments [11, 12], or identification of conserved genes by comparative genomics [13, 14]. Yet, both methods have substantial limitations. For example, computational approaches based on protein sequence homology yielded artificially small essential genomes because similar functions can be encoded by non-orthologous genes, while experimental approaches were time-consuming and subject to variability due to methodological differences [3, 15]. However, the development and extensive application of genome-wide transposon insertion sequencing – abbreviated Tn-Seq [16] – in the last decade, and of its variants referred to as INSeq [17], TraDIS [18] or HITS [19] has allowed for highly efficient and systematic identification of the essential features of cell survival in a genome. Briefly, Tn-Seq experiments use high-throughput sequencing of a pooled population of mutants covering the entire genome to assess the impact of each mutation site on overall fitness and survival. Experiments can be carried out both in optimal growth conditions to identify genes commonly considered as absolutely essential [5, 20–26], and in diverse environmental conditions (e.g. in vivo, in synthetic communities) to reveal conditionally essential genes [27–32]. Essential genomes have been so far characterized in vitro for at least 63 bacteria from various phyla (DEG database, accessed September 2020) [33]. However, research has been mostly focused on human-pathogenic bacteria (51 essential genomes out of 63) because systematic essentiality assessment is considered a promising way to identify new targets for antimicrobial drugs [34]. However, apart from the two major α-proteobacterial plant pathogens Agrobacterium fabrum [35] and Ralstonia solanacearum [36], no other essential genomes of plant pathogens, especially from the γ-proteobacterial class, have been investigated.
The genus Xanthomonas is a large reservoir of important plant-pathogenic γ-proteobacteria, as Xanthomonas species are the causative agents of diseases of almost 400 plant species, some of which are major crops worldwide (rice, cabbage, tomato, citruses, etc.) [37]. They belong to the same taxonomic family – Xanthomonadaceae – as the environmental and opportunistic pathogenic genus Stenotrophomonas and the devastating plant-pathogenic genus Xylella . While some Xanthomonas species have been extensively studied, e.g. major X. campestris or X. oryzae species [38], there still remains a lot to discover about most species. Among these, Xanthomonas hortorum , which comprises seven distinct pathovars and an increasing number of non-classified pathogenic strains attacking a wide variety of plants, has been scarcely investigated, perhaps partially because of its complex and until recently unresolved taxonomy [39]. Indeed, apart from a few studies on some type III effectors of the tomato pathogen X. hortorum pv. gardneri (formerly known as ‘ X. gardneri ’) [40, 41], and a noticeable genomic study of X. hortorum pv. carotae [42], extensive functional genomic studies and mutagenesis experiments have not yet been attempted on X. hortorum .
Our research work focuses primarily on X. hortorum pv. vitians, the causal agent of the bacterial leaf spot of lettuce disease (BLSL). This foliar disease is characterized by an outburst of small water-soaked lesions that later coalesce to form large necrotic patches [43]. Although sporadic, BLSL seems to have been re-emerging all around the world since the 1990s [39], sometimes dramatically reducing plant yield and quality, and increasing post-harvest losses [44]. Moreover, this pathogen can disseminate across the environment and persist mostly through seed infestation [45], but also in buried plant debris, irrigation water and epiphytically on a wide variety of weeds [46, 47]. It has become a concerning threat for lettuce production in the absence of efficient and practical means of disease control or prevention [48]. Understanding the architecture and evolution of the X. hortorum pv. vitians genome would certainly support the many attempts made to select resistant lettuce cultivars [49] and prevent predictable resistance by identifying virulence and resistance genes.
The present study aims at giving a comprehensive description of the essential genome of X. hortorum pv. vitians in optimal growth conditions, and provides an overview of the conservation and horizontal transfer of essential genes within the family Xanthomonadaceae . Using high-resolution complete genome sequencing and the design of a highly saturated transposon mutant library, we combined the discriminative power of genome-wide Tn-Seq experiments with critical insights into gene essentiality brought by multiple computational analyses. This innovative approach allowed us to identify core essential features conserved at various taxonomic levels, and genetic-context-dependent essential genes acquired through horizontal transfer events. This primary functional genomic study of X. hortorum pv. vitians will push forward investigations of this under-investigated species and lead to advances in both fundamental genomic understanding and disease control research.
Methods
Bacterial strains, plasmids and growth conditions
Xanthomonas hortorum pv. vitians strain LM16734 (=CFBP 8638) was isolated from a diseased lettuce in 2016 in the Rhône-Alpes region, France [39], and routinely cultured at 28 °C on 1/10th tryptic soy agar (TSA) plates or in 1/10th tryptic soy broth (TSB). Escherichia coli MFDpir/pSamEc [27] was grown at 37 °C in LB medium supplemented with diaminopimelic acid (DAP), for which it is auxotrophic, at 57 µg.ml−1. When required, the media were supplemented with 100 µg.ml−1 ampicillin and 25 µg.ml−1 kanamycin. The pSamEc plasmid is a mobilizable suicide vector carrying (i) the ampicillin resistance gene bla, (ii) a Himar1-C9 transposase gene under the control of the lac promoter and (iii) a kanamycin resistance gene flanked by mariner inverted repeat sequences containing MmeI restriction sites. The strains were kept at −80 °C in 30 % (v/v) glycerol vials for long-term storage.
Complete genome sequencing, assembly and annotation
High-quality complete genome sequencing of X. hortorum pv. vitians LM16734 was achieved using a combination of long-read and short-read technologies. Genomic DNA was extracted and purified from overnight broth cultures using the standard phenol–chloroform method [50], then sent to GATC Biotech for PacBio long-read sequencing with a PacBio RS instrument (Pacific Biosciences) and to Eurofins Genomics (Eurofins Scientific) for Illumina HiSeq 2×150 bp short-read sequencing on a NovaSeq 6000 instrument (Illumina Inc.). Hybrid assembly of raw reads from the two sequencing strategies was performed using unicycler v0.4.2 [51], and the resulting assembly was deposited and annotated on the MicroScope platform (https://mage.genoscope.cns.fr) [52]. Annotation of the genes of interest was cured manually based on homology and synteny analysis following the MicroScope recommendations. The replication origin (oriC) was identified based on its homology with the origin of the X. hortorum B07-007 genome recorded in the DoriC v10 database (DoriC ID: ORI97015374) [53]. The replication terminus sequence (dif) was detected by blast-searching the dif site of X. campestris pv. campestris 17 [54]. Both predicted oriC and dif sequences are presented in Table S7. Finally, leading and lagging strands of replication were determined by analysing the GC-skew bias computed with the GenSkew web tool (http://genskew.csb.univie.ac.at/).
Construction of the transposon library, DNA preparation and sequencing
A transposon mutant library was generated by performing a conjugation of X. hortorum pv. vitians LM16734 with a donor strain of the mariner transposon Himar1-C9. Briefly, six mixed solutions containing 15 ml of an overnight culture of strain LM16734 and Escherichia coli MFDpir/pSamEc were mixed in equal parts, centrifuged at 3344 x g for 20 min, and resuspended each in 200 µl of TSB supplemented with DAP at 57 µg ml−1. The resulting suspensions were deposited on over-dried TSA plates and incubated at 28 °C for 2 h, then resuspended in 1 ml of TSB. Conjugation efficiencies were evaluated by plating serially diluted aliquots on TSA plates supplemented with kanamycin. The remaining suspensions of the conjugation experiments were diluted to 1/5th and plated on TSA kanamycin plates, and then incubated at 28 °C for 3 days. Finally, the plates were washed with 800 µl of TSB, and colonies were resuspended with a spreader. Bacteria were subsequently mixed with 30 % (v/v) glycerol for long-term storage at −80 °C. The library theoretically consisted of 250,000 individual colonies, i.e. individual transposition events, at a concentration of 1010 CFU.ml−1.
Library DNA preparation and sequencing procedures were derived from Royet et al. [27]. The genomic DNA of the mutant library was extracted in two replicates from 125 µl aliquots using a Promega Wizard Genomic DNA Purification kit (Promega). Then, 50 µg of DNA from each sample was digested with 50 U of MmeI in a final volume of 1.2 ml at 37 °C for 1.5 h. Digestions were heat-inactivated for 20 min at 80 °C and purified with a QIAquick PCR Purification kit (Qiagen). The samples were then concentrated with a vacuum concentrator to obtain final volumes of 20 µl, which were deposited on 1 % (w/v) agarose gels. Each gel was cut out to extract the digested 1.3–1.8 kb fragments containing the transposon and adjacent genomic DNA using a QIAquick Gel Extraction kit (Qiagen). For each sample, 500 ng of digested and purified DNA was ligated to 0.44 µM of double-stranded barcoded adapters obtained as described previously [27] with 2000 U of T4 DNA ligase in a final volume of 50 µl at 16 °C overnight. Five 18-cycle PCRs were performed per sample to amplify the genomic DNA adjacent to the transposon. Each reaction mix contained 100 ng of DNA, 1 U of Q5 DNA polymerase (New England Biolabs), 1× Q5 buffer, 0.2 mM of dNTPs, and 0.4 µM of the LIB_PCR_5 and LIB_PCR_3 primers [32]. The five reaction products per sample were pooled and concentrated with a vacuum concentrator to obtain 25 µl samples that were deposited on a 1.8 % (w/v) agarose gel. The 125 bp bands were cut out and extracted with a QIAquick Gel Extraction kit (Qiagen). All gel electrophoresis and gel extraction steps were carried out on a blue-light transilluminator to avoid DNA damage, and DNA was stained with GelGreen (Biotium). Finally, DNA samples were dialysed for 4 h on 0.025 µm nitrocellulose membranes (Milipore) deposited on sterile ultra-purified water. Thirty nanograms of each DNA library were sent to MGX (CNRS Sequencing Service) for quality control assessment and high-throughput sequencing on Illumina HiSeq 2500 instruments (Illumina Inc.).
Assessment of genome essentiality
Raw reads were treated with cutadapt v1.15 [55] to retain only those containing the mariner inverted left repeat (ACAGGTTGGATGATAAGTCCCCGGTCTT) and cut the adapter sequence. Then, sequencing yield differences between the two replicates were normalized to the lowest yield by random subsampling with seqtk v1.2 (https://github.com/lh3/seqtk). Trimmed read datasets were mapped to unique TA sites on independent replicons without mismatches on the genome using a modified version of the TPP script [27] from the TRANSIT software program v2.0.2 [56]. Gene essentiality was determined by analysing the datasets with TRANSIT v2.3.4 using the Hidden Markov Model method (HMM) [57]. We performed Trimmed Total Reads (TTR normalization, summed the read counts of the two replicates, applied a LOESS correction for genome positional bias, and discarded the read counts in the 10 % N-terminus and C-terminus of the ORF that may have led to erroneous interpretations. Eventually, reciprocal blastn of genes classified as essential, growth-defect and growth-advantage were performed with blast+ v2.6.0 [58] against the genome to exclude the genes present in multiple copies on the same or different replicons, considered as false positives. Visualizations of the circular genome and local genomic regions were plotted with DNAPlotter [59].
Gene set enrichment analysis and cellular pathway mapping
Cluster of Orthologous Groups (COG)- [60] based annotation of genes was retrieved from the MicroScope platform. Gene set enrichment analyses were performed by computing the proportion of each COG category in the different gene states (i.e. non-essential, essential, growth-defect and growth-advantage) reported against their global proportion in the genome. The fold-change values were log-transformed to determine over- and under-represented COG categories depending on gene state. Kyoto Encyclopedia of Genes and Genomes (KEGG) (https://www.kegg.jp/) annotations were assigned to genes with BlastKOALA [61] and mapped to Xanthomonas hortorum reference pathways with KEGG Mapper [62].
Analysis of essential gene conservation at multiple taxonomic levels
Conservation of the essential genes identified within the genus Xanthomonas and within the family Xanthomonadaceae was investigated. First, GenBank assemblies of 72 genomes exhaustively representing the taxonomic diversity of Xanthomonas , three genomes of Stenotrophomonas maltophilia and three genomes of Xylella fastidiosa subspecies fastidiosa , multiplex and pauca were retrieved from the NCBI database. Genome accession numbers, taxonomic names and general assembly statistics are given in Table S1. Then, a whole-genome-based phylogeny was constructed using PhyloPhlan2 v0.40 (https://bitbucket.org/nsegata/phylophlan/wiki/phylophlan2) [63]. Briefly, a core of ~350 single-copy highly conserved proteins from the PhyloPhlan database were retrieved from the genomes using usearch v11.0.667 [64], aligned and concatenated with muscle v3.8.31 [65], and a phylogeny was built with FastTree v2.1.11 SSE3 [66]. Robustness was assessed locally using 1000 resamples of the Shimodaira–Hasegawa (SH) test [67]. Previously identified homologues of the essential protein-coding genes were then searched for among the 78 genomes using blastp with thresholds of 50 % coverage, 30 % amino-acid identity and an e-value <10−5. To check for potential false negatives caused by erroneous automatic annotation, sequences were also searched for using tblastn and the same thresholds, and homologous nucleotide sequences were searched for using blastn with thresholds of 50 % coverage, 70 % nucleotide identity and an e-value <10−5. Finally, essential and growth-defect genes were searched for in the DEG10 v15.2 database of essential genes (accessed June 2020) [33] with blastp using thresholds of 50 % coverage, 30 % amino-acid identity and an e-value <10−5. The phylogenetic tree and blast results were plotted together with the ggtree R package [68].
Results and discussion
Complete genome sequencing of X. hortorum pv. vitians LM16734 and design of a highly saturated transposon mutant library
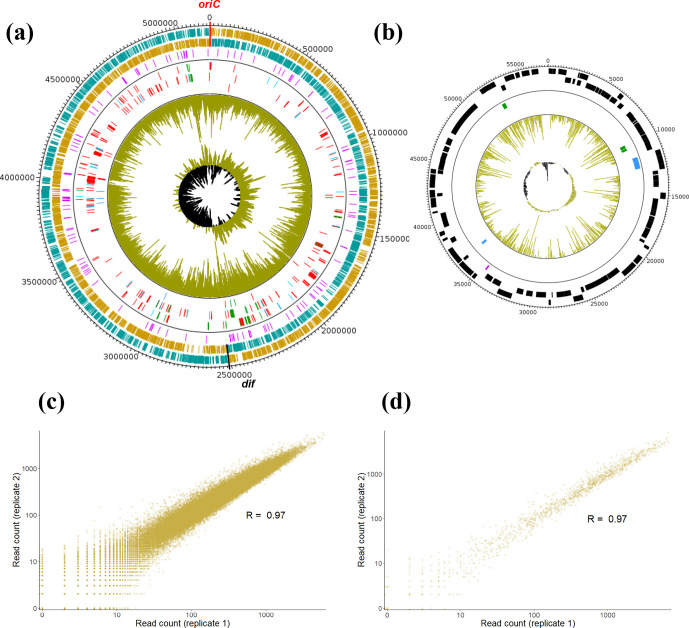
The complete genome of Xanthomonas hortorum pv. vitians LM16734 was sequenced using a combination of PacBio long-read and Illumina HiSeq short-read technologies. PacBio sequencing yielded 94,722 reads with a mean read length of 10,717 bp, while Illumina HiSeq sequencing yielded more than 7.2 million paired-end reads with a mean read length of 151 bp. Hybrid assembly constructed with Unicycler resulted in two replicons: a circular chromosome of 5,213,310 bp (Fig. 1a) and a native plasmid of 57,250 bp, designated pLM16734 (Fig. 1b), with an overall sequencing depth of 587.5×. The presence of a pLM16734-like plasmid might be a common feature among X. hortorum pv. vitians strains. Plasmid detection experiments performed on 18 strains representative of the known diversity of this lettuce pathogen indeed revealed that they all contained a single native plasmid ranging from around 40 to 60 kb [39]. The curated annotation carried out at the MicroScope platform detected 4,909 predicted protein-coding sequences (CDSs), 56 tRNAs, six rRNAs, one transfer-messenger RNA (tmRNA), seven regulatory riboswitches and 77 non-coding RNAs (ncRNAs) on the chromosome, and 89 CDSs and one ncRNA on plasmid pLM16734. The protein-coding density was therefore 87.6 % on the chromosome and 79.1 % on pLM16734, similar to values observed in the complete genome of X. hortorum pv. carotae M081 and other xanthomonads [42].
Fig. 1.
Complete genome atlas of X. hortorum pv. vitians LM16734. The localizations of essential, growth-defect and growth-advantage loci identified by Tn-Seq are indicated on (a) the circular chromosome (5, 213,310 bp) and (b) plasmid pLM16734 (57, 250 bp). The outermost ring track is a clockwise-arranged base pair (bp) ruler, with ticks every 20,000 bp for (a) and every 200 bp for (b). The next two tracks represent predicted protein-coding gene sequences on the forward and reverse strands, respectively. The chromosome replication origin (oriC) and terminus (dif) are marked as red and black ticks in (a), respectively, and protein-coding genes on the leading and lagging strands of replication are shown in orange and blue, respectively. The third (purple) track shows predicted ncRNA sequences on both strands. After the separation line, the next two tracks represent essential (red), growth-defect (blue) or growth-advantage (green) protein-coding genes on the forward and reverse strands, while the sixth track indicates important RNAs with the same colour code. Finally, the seventh track (gold) shows the mean distribution of read counts between the two technical replicates throughout the entire genome, and the eighth track depicts the GC-skew (positive in gold and negative in black). (c, d) Correlations between the two technical replicates of the Tn-Seq experiment. The log-transformed numbers of reads per TA site are plotted and compared between replicates, and Pearson correlation coefficients are indicated for (a) the circular chromosome (R=0.97) and (b) plasmid pLM16734 (R=0.97).
We designed a transposon mutant library for our Tn-Seq experiment by mating recipient strain LM16734 with an E. coli MFDpir donor strain containing a transferable Himar9 mariner transposon on the suicide vector pSamEc. Approximately 250, 000 individual transconjugants were recovered on 1/10th TSA rich medium supplemented with kanamycin and pooled. Then, two DNA libraries were prepared from two different aliquots of the mutant pool and massively sequenced by Illumina sequencing. The sequencing yielded more than 23 million reads in each replicate, which were normalized to the lowest value (Table 1). Of these 23 million reads, 87.4 % (replicate 1) and 87.6 % (replicate 2) were considered as ‘true’ TA-site-disrupted reads because they carried the Himar9 end sequence. The fact that 99.5 % of the Himar9-end-harbouring reads mapped to unique TA sites on the chromosome in both replicates certified the quasi-certain complete absence of persistence of the vector within the mutant library. This phenomenon often occurs in mariner-based Tn-Seq studies that do not use a suicide vector, and can result in a drastic reduction in the number of informative reads mapping the genome because vector sequences can be amplified during the PCR steps [9, 22]. It is worth noting that the discrepancy between the total number of Himar9-end-harbouring reads (~20.3 million) and the sum of reads mapped to the two replicons (~21.1 million) suggested that some sequences of plasmid pLM16734 were identical or highly similar to some chromosomal regions. Mapping these filtered reads to the genome also revealed that out of the 85,314 and 1,361 TA sites potentially disrupted by the transposon on the chromosome and the plasmid, respectively, 60,186 (replicate 1) to 60,715 (replicate 2) of the chromosomal TA sites and 1,197 (replicate 1) to 1,218 (replicate 2) of the plasmid TA sites contained at least one insertion (Table 1). Hence, the insertion density achieved was 70.5–71.2 % on the chromosome and 88–89.5 % on the plasmid, i.e. one insertion every 85 bp on the chromosome and one every 47 bp on the plasmid on average (Fig. 1a, b). The TA site saturation of the library is a critical parameter for statistical inference of essentiality in Tn-Seq studies [57, 69]. Saturation achieved was higher in the present study than in most mariner-based Tn-Seq studies [10, 22, 24, 57]. Furthermore, correlations between the number of reads at each TA site (i.e. the read count) of the two replicates were optimal on both replicons as asserted by Pearson correlation coefficients of 0.97 in both cases (Fig. 1c, d). In brief, previous analyses of our complete genome sequence and mutant library representativeness showed that we achieved a high quality in all key-point statistics that strongly supported further statistical gene state interpretations.
Table 1.
Transposon-insertion sequencing (Tn-Seq) statistics
|
Replicate |
Sequencing yield |
No. of reads after normalization* |
No. of Tn-end containing reads |
Replicon |
Read count (unique TA sites) |
TA hits† |
Insertion density† |
Mean read count over non-zero TA‡ |
|---|---|---|---|---|---|---|---|---|
|
1 |
23,662,661 |
23,179,825 |
20, 267, 199 |
Chromosome |
20,168,343 |
60,186 |
70.5 |
335.1 |
|
pLM16734 |
938, 617 |
1,197 |
88.0 |
784.1 |
||||
|
2 |
23,179, 825 |
23,179, 825 |
20, 309, 279 |
Chromosome |
20, 209, 37 |
60,715 |
71.2 |
332.9 |
|
pLM16734 |
92, 4 781 |
1,218 |
89.5 |
759.3 |
||||
|
|
|
|
|
|
|
|
|
|
*Numbers of reads were normalized based on the lowest sequencing yield (i.e. replicate 2).
†The complete X. hortorum pv. vitians LM16734 genome contains 85 314 TA sites in the chromosome and 1361 in plasmid pLM16734. Insertion density was calculated as the ratio of TA sites with at least one read mapped over the total number of TA sites in the replicon.
‡Mean value of mapped reads per TA site with at least one read.
In vitro identification of essential and fitness-related genes
The computational assessment of gene essentiality by Tn-Seq methods generally relies on examination of the number of disrupted sites within a given gene relative to the total number of potential transposon insertion sites (i.e. the insertion rate). This rate theoretically reflects the tolerance of the organism to the loss of the encoded function [57, 70]. Yet, other approaches for Tn-Seq data analysis use the number of reads at insertion sites (i.e. the read count) as a quantitative measure by calculating the mean read count in a sliding window to identify regions of the genome presenting lower insertion values [71]. Both methods have their drawbacks: the first tend to overestimate the number of essential genes [71] and are sensitive to insertions in the N- and C-terminal regions of genes, incorrectly concluding that they are non-essential [72], while the second have been criticized for their lack of statistical significance [56, 57]. We decided to use the HMM-based approach offered by TRANSIT because it uses both the qualitative insertion rate and quantitative read count parameters. Moreover, it assesses the probability of essentiality for each individual TA site, so that domains of essentiality can be identified in genes. It also performs a four-state classification, as genes and regions can be labelled as essential, non-essential, growth-defect or growth-advantage. The growth-defect state, also referred to as critical in other studies [10, 22], corresponds to observations that a deficit rather than complete lack of insertions within a region might be a sign that it is not directly vital for the cell, but that its disruption would impair the growth of the organism [16]. Likewise, genes or regions presenting a significantly higher read count than the rest of the genome are classified as growth-advantage, meaning that their disruption would lead to a higher growth rate, possibly because they represent an unnecessary metabolic cost in the studied conditions (e.g. secretion of an extracellular enzyme or toxin) [57].
Our HMM-based analysis conducted with TRANSIT primarily identified 571 essential chromosomal loci. Nine were subsequently considered as artefactual CDSs after manual examination on the MicroScope comparative genomics platform, and another 192 were discarded because they were duplicated in the genome, on the same or different replicons, and thus were impossible to classify according to our workflow. Nineteen loci computationally identified as growth-defect were also discarded manually because they were duplicated. Finally, 34 more loci were impossible to categorize because they did not contain TA sites. All these loci were therefore classified as ‘undetermined’ and are detailed in Table S2. Following these refinement steps, 370 chromosomal loci were positively identified as essential (Table S3), consisting of 359 protein-coding genes, eight tRNAs and three ncRNAs. Additionally, 44 protein-coding genes and six tRNAs were categorized as growth-defect (Table S4), and 77 protein-coding genes, one tRNA and one ncRNA were considered as growth-advantage after curation (Table S5). The initial analysis and subsequent manual curation identified three growth-defect and three growth-advantage genes on plasmid pLM16734 (Table S6).
Analysis of essential genes of X. hortorum pv. vitians associated with core bacterial features
A striking observation one can make about the number of essential genes in various bacteria is that most of them appear to rely on a restricted set of 300–500 essential genes despite important variability in terms of genome size, architecture or lifestyles [33]. To determine whether the essential genes we identified in X. hortorum pv. vitians were common across the bacterial kingdom, we searched for homologues in the Database of Essential Genes (DEG10 v15.2, accessed June 2020), which at the time contained 48 essential genomes from 29 Gram-negative bacteria, 17 from Gram-positive bacteria and two from Mollicutes . An overwhelming majority (93.5 %) of the essential genes we identified in X. hortorum pv. vitians had at least one putative homologue in the database (Table S3). Therefore, it seems that essential genes and functions are widely conserved across bacteria. Moreover, the comparison of our growth-defect gene set to the DEG database revealed that 86 % of them also had putative homologues in the database, suggesting that they could have been annotated as essential under more stringent experimental conditions.
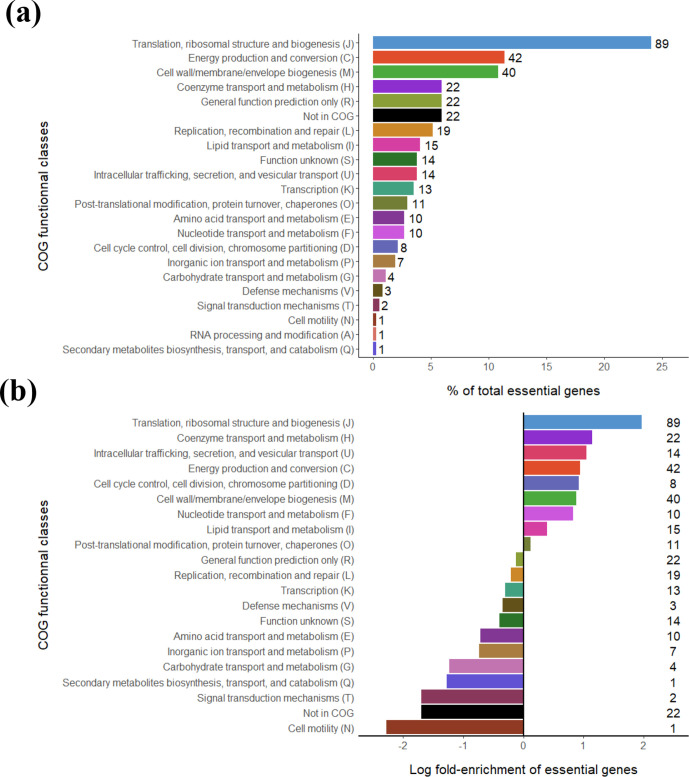
To further compare the key cellular functions of X. hortorum pv. vitians and those described in other bacteria, we classified genes into general functional categories or metabolic pathways based on the COG and KEGG databases. The COG classification of essential genes showed that almost half of them were clustered in only three categories: 24 % were related to fundamental processes of translation and ribosome biogenesis (J), 11 % were associated with energy production and conversion (C), and another 11 % were related to cell wall and membrane biogenesis (M) (Fig. 2a). However, it appeared more accurate to consider how enriched the COG categories were in the essential gene set considering their representation in the complete genome. Cellular processes presenting an enrichment in the essential gene set were associated with translation and ribosome structure (J), coenzyme metabolism (H), intracellular trafficking and secretion (U), energy production (C), cell division (D), cell wall and membrane biogenesis (M), and nucleotide metabolism (F) (Fig. 2b). Similar analyses in other bacteria also showed that translation and ribosome structure were the key essential functions of bacterial cells, while the other COG categories enriched in our gene set were usually identified as essential but varied in terms of importance and gene numbers [8, 23, 24, 26, 73]. Other COG-based analyses of non-essential, growth-defect and growth-advantage genes are not discussed here but can be found in Fig. S1. In addition, KEGG pathway clustering reached better precision in the determination of crucial cellular pathways, and underlined that most of the genes required for aminoacyl-tRNA biosynthesis, ribosome structure, protein export, DNA replication and lipopolysaccharide biosynthesis were essential (Table 2). About half of the genes involved in peptidoglycan biosynthesis, the citrate cycle, oxidative phosphorylation and RNA degradation were also required for cell survival. Unlike what was described in other organisms [10, 22, 25, 73], our analysis revealed that genes of the glucose catabolism pathways were apparently not essential for X. hortorum pv. vitians survival in rich medium (Fig. S2), even though glucose is the main carbon source in TSA medium. The only essential gene in the pentose phosphate pathway encoded the ribose-phosphate pyrophosphokinase (prsA, XHV734_3940), which catalyses the conversion of ribose-5-phosphate into 5-phosphoribosyl-1-pyrophosphate, a key compound in the biosynthesis pathways of the essential purine and pyrimidine bases [74]. More interestingly, the only essential gene associated with the glycolysis pathway encoded the phosphopyruvate hydratase, also known as enolase (eno, XHV734_2903), which converts 2-phospho-d-glycerate into phosphoenolpyruvate. Considering that this enzyme is multifunctional – it is also involved in the RNA degradosome [75], a pathway containing seven essential genes (Table 2) – this secondary function of the enolase probably accounts more for its essentiality phenotype than does its role in glycolysis. Overall, these two examples highlight the fact that some genes in our essential gene set act as metabolic hubs, and this makes it difficult to precisely determine which of their many roles is critical and responsible for the observed essentiality phenotype. The absence of essential genes in the usually key pathways of glucose catabolism is probably a reflection of the ability of xanthomonads to rely on the Entner-Doudoroff alternative glucose catabolism pathway (Fig. S2) [76]. A metabolic flux study of the utilization of 13C-labelled glucose in X. campestris revealed that it was mostly processed through the Entner-Doudoroff pathway to be converted into pyruvate; yet, it could also be marginally handled by the glycolysis and pentose phosphate pathways [77]. Therefore, we can reasonably conclude that no matter which of the three glucose catabolism pathways was disrupted in X. hortorum pv. vitians in our experiment, the others would be able to take over and prevent the occurrence of deleterious phenotypes. In a nutshell, most of the X. hortorum pv. vitians essential genome overlapped with key cellular functions described in other organisms, while displaying a few peculiar features.
Fig. 2.
Clustering of essential genes based on Cluster of Orthologous Groups (COG) functional classes. (a) Proportion of each class in the essential gene set, with the number of genes next to the bars; (b) log-fold enrichment of COG classes, calculated as the ratio of the proportion of a given class in the essential gene set to its overall proportion in the genome.
Table 2.
Essential KEGG pathways in X. hortorum pv. vitians LM16734 (non-exhaustive)
|
KEGG pathways |
Essential+Critical genes* |
Total genes† |
|---|---|---|
|
Aminoacyl-tRNA biosynthesis |
22+1 |
24 |
|
Ribosome |
48 |
54 |
|
Protein export |
10+1 |
16 |
|
DNA replication |
10 |
15 |
|
Lipopolysaccharide biosynthesis |
11 |
17 |
|
Peptidoglycan biosynthesis |
12 |
21 |
|
Citrate cycle |
13 |
24 |
|
Oxidative phosphorylation |
26 |
51 |
|
RNA degradation |
7 |
15 |
|
Fatty acid biosynthesis |
6+1 |
19 |
|
Pyrimidine metabolism |
6+2 |
25 |
|
Amino sugar and nucleotide sugar metabolism |
6 |
34 |
|
Purine metabolism |
8 |
55 |
|
Biosynthesis of amino acids |
9+3 |
105 |
*Number of genes identified as essential and growth-defect.
†Total number of genes in the model pathway in the X. campestris pv. campestris ATCC 33913T organism-specific KEGG database.
Conservation of essential genes among Xanthomonadaceae and investigations of specific essential features and mobile genetic elements
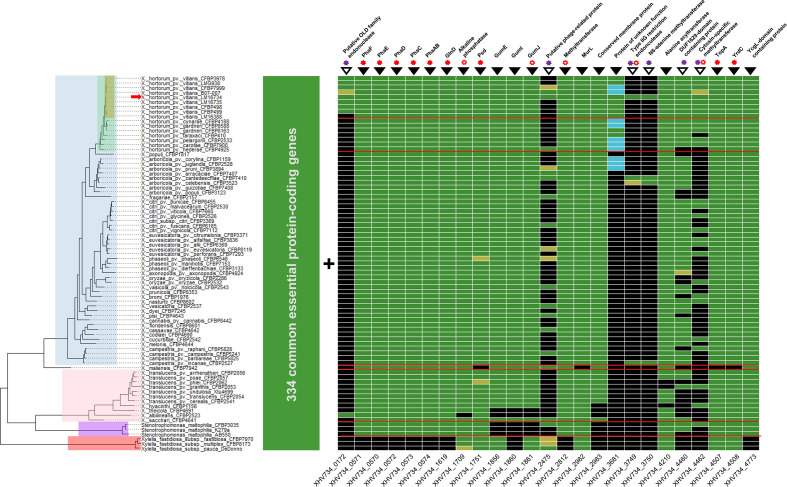
As we intended to go beyond the description of common essential bacterial functions, we further looked into the specific essential genes showing no homology in the DEG database and investigated the conservation of our essential gene set among the genus Xanthomonas , and also to some extent the family Xanthomonadaceae . To that end, we built a genome-based phylogeny of all the known species and pathovars of Xanthomonas species, Stenotrophomonas maltophilia and Xylella fastidiosa using relevant type and pathotype strain genomes (Table S1), and analysed the conservation of X. hortorum pv. vitians essential genes across the tree (Fig. 3). We deliberately chose weakly stringent coverage and identity thresholds to look at the conservation of protein functions and deal with quality heterogeneity in genome assemblies and annotations, but we checked for inconsistencies and potential artefacts manually. Hence, 334 out of 359 protein-coding genes were shared among all investigated Xanthomonadaceae (Table S3). The remaining 25 genes could be distinguished into two groups according to their distribution pattern in the phylogeny: (i) those that were mostly conserved among Xanthomonadaceae or displayed a phylogeny-related distribution (Fig. 3, filled inverse triangles), and (ii) those that showed a heterogenous distribution unrelated to phylogeny (Fig. 3, empty inverse triangles).
Fig. 3.
Conservation of the essential genes of X. hortorum pv. vitians LM16734 in the family Xanthomonadaceae . Colours on the phylogenetic tree indicate taxonomic levels: yellow = X. hortorum pv. vitians, green = X. hortorum , blue = Xanthomonas clade I, pink = Xanthomonas clade II, purple = Stenotrophomonas maltophilia, and red = Xylella fastidiosa . Taxonomic separations are also indicated by red lines on the gene conservation matrix. The position of the X.hortorum pv. vitians LM16734 genome in the phylogeny is highlighted by a red arrow. In the matrix, green boxes indicate detection with blastp (>50% coverage, >30% identity, e-value >10-5), yellow boxes detection with tblastn (> 50 % coverage, > 30 % identity, e-value > 10-5), and blue boxes detection with blastn (>50 % coverage, >70% identity, e-value >10-5). The numbers of gene loci are shown below the matrix, and gene product annotations are indicated above it. Empty and filled black inverted triangles refer to the two distribution patterns discussed in the text. Filled red asterisks indicate that at least one homologue was detected in the DEG10 v15.2 essential gene database with blastp (>50% coverage, >30% identity, e-value >10-5), while empty red asterisks indicate lower homology thresholds (>40% coverage and/or >20% identity and e-value >10-5). Finally, purple asterisks highlight the presence of mobile genetic-element-associated genes (transposase, integrase, reverse transcriptase) in the neighbourhood of the gene (<10 kb distance).
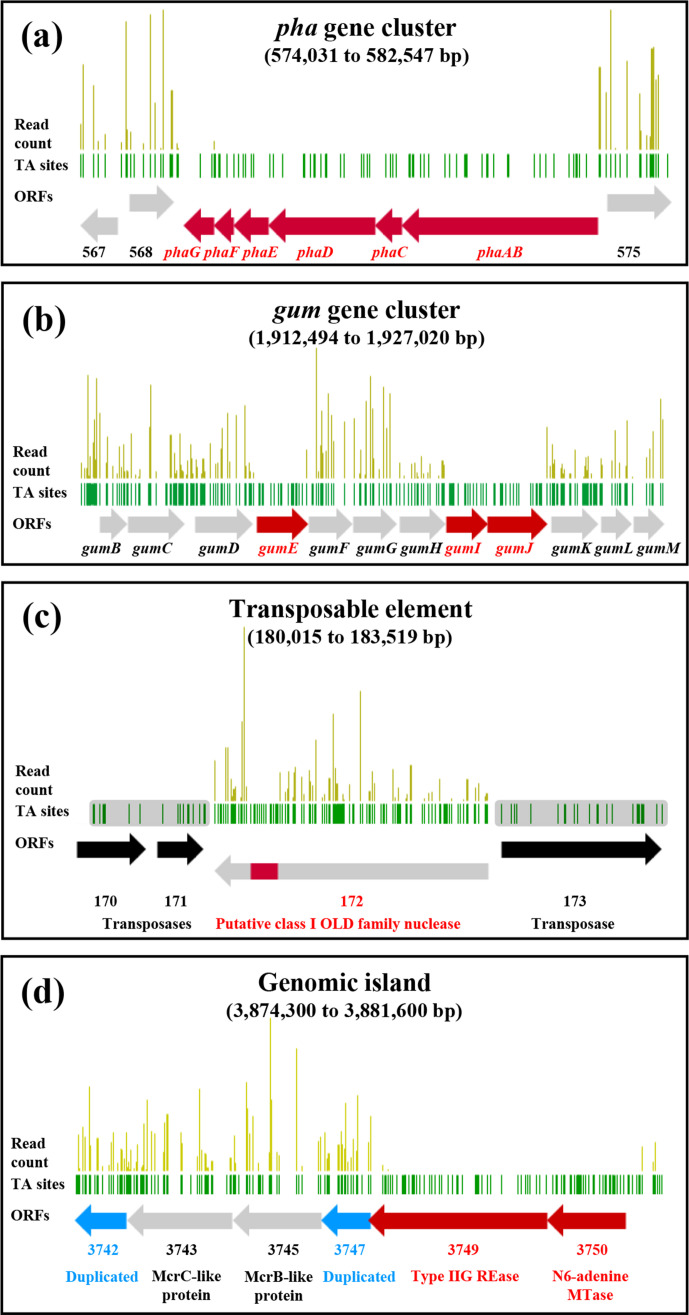
The genes included in the first category (phylogeny-related distribution) were mainly related to known genes and functions. For example, the six genes of the pha operon, which encodes the monovalent cation/proton antiporter system Pha also referred to as Mrp or Mnh, were essential according to our analysis as no in-frame transposon insertions were observed (Fig. 4a). These genes are conserved in all Xanthomonas species and S. maltophilia , but completely absent in X. fastidiosa (Fig. 3). Note that all six genes in the operon are required for the antiporter to be functional [78]. Antiporters of the Pha family are widely distributed across bacteria and usually involved in cytoplasmic pH homeostasis and efflux of toxic cations, but also in a broad range of other resistance and environmental adaptation phenotypes [79]. The pha operon is essential for a few bacteria to grow in rich medium [11, 35, 80]. Its absence in X. fastidiosa could be a consequence of the extreme genome reduction undergone by this pathogen, which made it fit for only a few restricted homeostatic environments [81], while Xanthomonas species and S. maltophilia are adapted to living and surviving in more variable and stressful environments. Thus, they might partly rely on Pha antiporters to buffer physicochemical toxicity in challenging environments (e.g. epiphytic life for Xanthomonas species).
Fig. 4.
Schematic representations of gene essentiality within the genomic context. Representations are shown for (a) the pha gene cluser (574,031…582,547 bp), (b) the gum gene cluster (1,912,494…1,927,020 bp), (c) the transposable element containing the putative OLD family endonuclease XHV734_0172 (180,015…183,519 bp) and (d) the genomic island containing the restriction-modification system composed of restriction endonuclease XHV734_3749 and methyltransferase XHV734_3750 (3,868,330…3,884,140 bp). Essential genes or domains are shown in red, non-essential genes or domains in grey, transposases in black in (c), and duplicated loci in blue in (d). Gene names or simplified loci numbers are indicated at the bottom of each box. Green bars indicate the position of TA sites, and golden bars represent the read count at each TA site. In (c), shadowed TA sites indicate unavailability of read counts because of repeated sequences.
Similarly, three components of the gum gene cluster were also essential (Fig. 4b). This set of 12 genes (gumB to gumM) encodes the synthesis and export of the well-known in planta virulence factor and biotechnologically important exopolysaccharide (EPS) xanthan. The three genes – gumE, gumI and gumJ – turned out to be conserved in most Xanthomonas species but absent in X. sacchari and X. albilineans (Fig. 3), as previously reported [82]. They had no homologues in the DEG database (Table S2), except gumJ that showed low but significant homology (85 % coverage and 25 % amino acid identity, e-value of 3.27e−20) with the succinoglycan biosynthesis transport protein ExoT of Agrobacterium tumefaciens , essential in rich medium [35]. They were also absent in S. maltophilia, in line with the inability of Stenotrophomonas to synthesize xanthan as one of the major features that led to their differentiation from the genus Xanthomonas [83]. Concerning X. fastidiosa , it was shown that the absence of gumG, I and L leads to the formation of another type of EPS [84], and independent acquisition of the gum cluster by Xanthomonas species and Xylella species may account for these differences [85]. Interestingly, a previous study by Katzen et al. [86] reported the impossibility to create insertion mutants of gumB, C, E, J and M in X. campestris pv. campestris. Yet, Kim et al. [87] later claimed that only the knock-out mutation of gumJ was lethal to X. oryzae pv. oryzae. Lethality of gum gene mutations might thus be species-specific, probably because of complex transcriptional patterns that are likely to differ among Xanthomonas species [88–90]. In our case, considering that gumJ and gumE are responsible for the translocation of the lipid-linked sugar repeat units from the cytoplasm to the periplasm and for the polymerization of these units in the periplasm, respectively [76, 91], their disruption would eventually cause EPS units to accumulate in the cytoplasm or the periplasm. Some lipid-linked intermediates of xanthan biosynthesis have been hypothesized to be toxic when they accumulate inside the cell [86], an assumption further supported by the demonstration that intermediate components of a similar extracellular EPS of Streptococcus pneumonia were indeed toxic [92]. Therefore, the toxicity of the lipid-linked repeat units of xanthan probably accounts for the lethal phenotype of gumJ and gumE mutants. However, the absence of insertions within gumI is more difficult to interpret. This gene encodes a mannosyltransferase that participates in the formation of the pentasaccharide repeat units before their translocation and polymerization in the periplasm [93]. Either its disruption also leads to accumulation of toxic intermediates, or its essentiality might be an artefact caused by the potential polar effects that possibly occurred in operons using our mutagenesis approach [27]. Nevertheless, these basic findings concerning the xanthan biosynthesis pathway of X. hortorum pv. vitians might be helpful for more applied research, as the industrial potential of X. hortorum pv. pelargonii strains for xanthan production has been investigated [94].
On the other hand, examining the conservation of X. hortorum pv. vitians LM16734 essential genes revealed a second category of genes whose distribution across the phylogeny was more chaotic (Fig. 3, empty inverted triangles). Although most of these genes were not automatically annotated, manual annotation and analysis of synteny and gene neighbourhood on the MicroScope platform revealed that they were all associated with mobile genetic elements (MGEs), i.e. prophages, transposons, genomic islands, etc. For example, locus XHV734_0172 encodes a putative ATP-dependent endonuclease of the OLD family flanked by transposase genes (Fig. 4c), suggesting transposable-element-mediated horizontal acquisition. The conservation pattern among the Xanthomonadaceae implies that it was acquired independently in X. hortorum pv. vitians and X. albilineans (Fig. 3). The absence of this element in strain LM16388 is consistent with the early divergence of this strain from the rest of the pathovar, as we previously reported [39]. According to the classification proposed by Schiltz et al. [95], this nuclease belongs to class 1 OLD nucleases, like the canonical old gene of phage P2. Exhaustive biological roles of OLD nucleases remain to be unravelled, yet they are known to play a significant role in phage exclusion by protecting lysogenized host genomes against superinfection by other phages, presumably by degrading nucleic acids from newcomers through their ribonuclease and DNA exonuclease activities [95, 96]. Intriguingly, XHV734_0172 was the only locus in our analysis that displayed only one short domain intolerant to insertions (I422 to Y476) (Fig. 4c). A comparison with the recently described structure of OLD nucleases [95] revealed that this essential domain was located in the C-ter Topoisomerase/Primase (Toprim) catalytic domain that bears the nuclease activity, and more precisely starts with the highly conserved metal A-binding aspartate motif D425xD427 (Fig. S3) [97]. Further knowledge of the in vivo biological activity of OLD nucleases would be required to understand why this segment of the Toprim domain is essential in X. hortorum pv. vitians. One possible explanation would be that transposon insertion near catalytic sites generates truncated proteins displaying uncontrolled exonuclease and ribonuclease activities, leading to cell death. Nevertheless, a genome-wide Tn-Seq screening of Salmonella enterica also showed that another OLD nuclease was conditionally essential [30], confirming that some OLD nuclease genes behave as ‘essential’ genes whose disruption results in a lethal phenotype. Furthermore, knowledge of the phage exclusion-related genes of X. hortorum pv. vitians could be of importance in the near future, as phage therapy arises as a promising way to control plant diseases caused by Xanthomonas species [98].
We identified other MGE-associated essential features related to restriction–modification (R-M) systems. R-M systems are often considered as primitive immune systems as their biological role is to protect bacterial genomes from foreign DNA integration by recognizing self and non-self DNA according to methylation profiles [99]. A typical R-M system involves a restriction endonuclease (REase) that recognizes and cleaves foreign DNA at specific sites, and a methyltransferase (MTase) that methylates the host DNA at the same specific sites to protect it from restriction. We notably identified an essential R-M system composed of a type-II restriction endonuclease (XHV734_3749) and its cognate N6-adenine methyltransferase (XHV734_3750) (Fig. 3). This system is located on a small genomic island of 7.3 kb also containing an McrBC-like R-M system and two small duplicated unknown proteins upstream of each R-M system (Fig. 4d). Interestingly, this genomic island is conserved in the few xanthomonads possessing the essential R-M system (Fig. 3), but also strongly homologous to cluster S13 of Pseudomonas protegens Cab57, yet with a completely different genomic environment [100]. From a wider perspective, this genomic island is part of a larger variable genomic region located between moaE downstream (XHV734_3751) and dnaX upstream (XHV734_3720), which seems to have undergone multiple horizontal acquisition events, as indicated by the persistence of several mobility-associated genes (XHV734_3722, 3725, 3734 and 3735). Searching for homologues of XHV734_3749 and 3750 in the restriction enzyme database REBASE [101] yielded a perfect match for an identical R-M system in X. arboricola pv. juglandis CPBF 1521 (M.Xar1521ORF31530P and Xar1521ORF31540P), with moaE downstream and an McrB-like protein-coding gene upstream. However, this genomic island was not detected in the type strain of X. arboricola pv. juglandis CFBP 2528T included in our analyses (Fig. 3). This shows some limitations of our taxonomy-based conservation analysis when it comes to MGEs. The presence of this genomic island upstream of the moa operon in some of the phylogenetically distant xanthomonads investigated suggests either early acquisition followed by loss events in most strains, or multiple independent site-specific integration events. Nevertheless, it was interesting to observe that both the REase and the cognate MTase were annotated as essential. As R-M systems are often compared to toxin/antitoxin systems because they act as genetic addiction systems [102], we would have expected the sole MTase to be essential. Yet, the similarity with Xar1521ORF31540p and domain analysis using MicroScope revealed that XHV734_3749 is a type-IIG REase, meaning that it possesses both the endonuclease and the methyltransferase domains, and that the presence of a cognate separate MTase is accessory [103]. Thus, its disruption would probably lead to post-segregational killing as the cell REase concentration would become too low to successfully protect bacterial DNA from its own endonuclease activity. Nevertheless, the co-occurrence of two distinct restriction modification systems on a small genomic island raises questions about their biological roles. The type-I R-M system McrBC was originally described as part of the so-called ‘Immigration Control Region’ in E. coli K12 [104] because it plays a role in the protection of the host genome against foreign invading DNA. On the other hand, type-II R-M systems frequently behave like ‘selfish’ genetic elements promoting their own survival to increase their relative frequency [105, 106]. Observations that type-II R-M systems are often associated with MGEs also suggest that they play an important role in the stabilization of genomic islands [99, 107], a perspective that echoes the essentiality of the type-II R-M system of the genomic island we described.
Conclusions
In this study, we fully exploited the power of genome-wide Tn-Seq essentiality screening by putting our results in perspective with the insights offered by comparative genomics, synteny analyses and phylogenomics. Combining these approaches allowed us to separate core essential genes conserved across bacterial phyla from context-dependent essential genes in the essential genome of X. hortorum pv. vitians. Among these, we identified specific Xanthomonas or Xanthomonadaceae features that were essential in vitro, but also probably play an important role in natural ecological niches. Also, we discovered through systematic synteny and phylogenomic examination that some essential genes were acquired through horizontal gene transfer events, either separately or on genomic islands, and were associated with bacterial defence systems or genetic addiction complexes. Thus, we showed that gene essentiality assessment by Tn-Seq gains significance by systematically examining genome content and gene conservation. Our complete genome sequence and high-density transposon library will reveal useful and reliable tools for the next steps of our research on this important lettuce pathogen, which will aim at discovering X. hortorum pv. vitians genes associated with virulence and survival in planta.
Supplementary Data
Funding information
L.Morinière was funded for his PhD by a grant from the French Ministry of Higher Education, Research and Innovation. This work was done as part of the Rhône Alpes rural development programme and funded by the Rhône-Alpes region and the EU FEADER programme.
Acknowledgements
The authors are grateful to Laurène Mirabel and Géraldine Effantin for their technical assistance in the development of the transposon library. The LABGeM (CEA/Genoscope and CNRS UMR8030), the France Génomique and French Bioinformatics Institute national infrastructures (funded as part of Investissement d'Avenir programme managed by Agence Nationale pour la Recherche, contracts ANR-10-INBS-09 and ANR-11-INBS-0013) are acknowledged for support within the MicroScope annotation platform. The authors would like to thank Annie Buchwalter for English proofreading and corrections.
Author contributions
F.B. supervised the manuscript, funding acquisition and project administration. E.G. also participated in supervision, methodology development and provided some of the resources. S.L. helped to perform experimental investigations and provided some of the resources used. L.M. conceptualized the study, carried out the investigations, data curation, formal analysis, visualization, and wrote the original draft. All four authors contributed to the reviewing and editing of the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: BLSL, bacterial leaf spot of lettuce; CDS, coding sequence; COG, Cluster of Orthologeous Groups; DEG, database of essential genes; EPS, exopolysaccharide; HMM, hiden markov model; KEGG, kyoto encyclopediae of genes and genomes; LB, luria-bertani; MGE, mobile genetic element; MTase, methyltransferase; OLD, overcoming lysogenization defect; PCR, polymerase chain reaction; REase, restriction endonuclease; R-M, restriction–modification; Tn-Seq, transposon insertion sequencing; TSA / TSB, tryptic soy agar / tryptic soy broth; TTR, trimmed total reads.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Seven supplementary tables and three supplementary figures are available with the online version of this article.
References
- 1.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 2.Hacker J, Carniel E. Ecological fitness, genomic islands and bacterial pathogenicity. EMBO Rep. 2001;2:376–381. doi: 10.1093/embo-reports/kve097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Juhas M, Eberl L, Glass JI. Essence of life: essential genes of minimal genomes. Trends Cell Biol. 2011;21:562–568. doi: 10.1016/j.tcb.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Koonin EV. How many genes can make a cell: the minimal-gene-set concept. Annu Rev Genomics Hum Genet. 2000;1:99–116. doi: 10.1146/annurev.genom.1.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christen B, Abeliuk E, Collier JM, Kalogeraki VS, Passarelli B, et al. The essential genome of a bacterium. Mol Syst Biol. 2011;7:528. doi: 10.1038/msb.2011.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.D’Elia MA, Pereira MP, Brown ED. Are essential genes really essential? Trends Microbiol. 2009;17:433–438. doi: 10.1016/j.tim.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Rancati G, Moffat J, Typas A, Pavelka N. Emerging and evolving concepts in gene essentiality. Nat Rev Genet. 2018;19:34–49. doi: 10.1038/nrg.2017.74. [DOI] [PubMed] [Google Scholar]
- 8.Gislason AS, Turner K, Domaratzki M, Cardona ST. Comparative analysis of the Burkholderia cenocepacia K56-2 essential genome reveals cell envelope functions that are uniquely required for survival in species of the genus Burkholderia . Microb Genomics. 2017;3 doi: 10.1099/mgen.0.000140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poulsen BE, Yang R, Clatworthy AE, White T, Osmulski SJ, et al. Defining the core essential genome of Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2019;116:10072–10080. doi: 10.1073/pnas.1900570116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Le Breton Y, Belew AT, Valdes KM, Islam E, Curry P, et al. Essential genes in the core genome of the human pathogen Streptococcus pyogenes . Sci Rep. 2015;5:9838. doi: 10.1038/srep09838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, et al. Essential Bacillus subtilis genes. Proc Natl Acad Sci. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:2006.0008. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mushegian AR, Koonin EV. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc Natl Acad Sci. 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gil R, Silva FJ, Peretó J, Moya A. Determination of the core of a minimal bacterial gene set. Microbiol Mol Biol Rev. 2004;68:518–537. doi: 10.1128/MMBR.68.3.518-537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat Rev Microbiol. 2003;1:127–136. doi: 10.1038/nrmicro751. [DOI] [PubMed] [Google Scholar]
- 16.van Opijnen T, Bodi KL, Tn-seq CA. High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods. 2009;6:767–772. doi: 10.1038/nmeth.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goodman AL, McNulty NP, Zhao Y, Leip D, Mitra RD, et al. Identifying genetic determinants needed to establish a human gut symbiont in its habitat. Cell Host Microbe. 2009;6:279–289. doi: 10.1016/j.chom.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langridge GC, Phan M-D, Turner DJ, Perkins TT, Parts L, et al. Simultaneous assay of every Salmonella Typhi gene using one million transposon mutants. Genome Res. 2009;19:2308–2316. doi: 10.1101/gr.097097.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gawronski JD, Wong SMS, Giannoukos G, Ward DV, Akerley BJ. Tracking insertion mutants within libraries by deep sequencing and a genome-wide screen for Haemophilus genes required in the lung. Proc Natl Acad Sci. 2009;106:16422–16427. doi: 10.1073/pnas.0906627106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goodall ECA, Robinson A, Johnston IG, Jabbari S, Turner KA, et al. The essential genome of Escherichia coli K-12. MBio. 2018;9:e02096–17. doi: 10.1128/mBio.02096-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Higgins S, Sanchez-Contreras M, Gualdi S, Pinto-Carbó M, Carlier A, et al. The essential genome of Burkholderia cenocepacia H111. J Bacteriol. 2017;199:e00260–17. doi: 10.1128/JB.00260-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hooven TA, Catomeris AJ, Akabas LH, Randis TM, Maskell DJ, et al. The essential genome of Streptococcus agalactiae . BMC Genomics. 2016;17:406. doi: 10.1186/s12864-016-2741-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mandal RK, Jiang T, Kwon YM. Essential genome of Campylobacter jejuni . BMC Genomics. 2017;18:616. doi: 10.1186/s12864-017-4032-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosconi F, de Vries SPW, Baig A, Fabiano E, Grant AJ. essential genes for in vitro growth of the endophyte Herbaspirillum seropedicae SmR1 as revealed by transposon insertion site sequencing. Appl Environ Microbiol. 2016;82:6664–6671. doi: 10.1128/AEM.02281-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubin BE, Wetmore KM, Price MN, Diamond S, Shultzaberger RK, et al. The essential gene set of a photosynthetic organism. Proc Natl Acad Sci. 2015;112:E6634–6643. doi: 10.1073/pnas.1519220112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wong Y-C, Abd El Ghany M, Naeem R, Lee K-W, Tan Y-C, et al. Candidate essential genes in Burkholderia cenocepacia J2315 identified by genome-wide TraDIS. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Royet K, Parisot N, Rodrigue A, Gueguen E, Condemine G. Identification by Tn-seq of Dickeya dadantii genes required for survival in chicory plants. Mol Plant Pathol. 2019;20:287–306. doi: 10.1111/mpp.12754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bishop AH, Rachwal PA, Vaid A. Identification of genes required by Bacillus thuringiensis for survival in soil by transposon-directed insertion site sequencing. Curr Microbiol. 2014;68:477–485. doi: 10.1007/s00284-013-0502-7. [DOI] [PubMed] [Google Scholar]
- 29.Cole BJ, Feltcher ME, Waters RJ, Wetmore KM, Mucyn TS, et al. Genome-wide identification of bacterial plant colonization genes. PLOS Biol. 2017;15:e2002860. doi: 10.1371/journal.pbio.2002860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Khatiwara A, Jiang T, Sung S-S, Dawoud T, Kim JN, et al. Genome scanning for conditionally essential genes in Salmonella enterica serotype Typhimurium. Appl Environ Microbiol. 2012;78:3098–3107. doi: 10.1128/AEM.06865-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.LaSarre B, Deutschbauer AM, Love CE, McKinlay JB. Covert cross-feeding revealed by genome-wide analysis of fitness determinants in a synthetic bacterial mutualism. Appl Environ Microbiol. 2020;86 doi: 10.1128/AEM.00543-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Skurnik D, Roux D, Aschard H, Cattoir V, Yoder-Himes D, et al. A comprehensive analysis of in vitro and in vivo genetic fitness of Pseudomonas aeruginosa using high-throughput sequencing of transposon libraries. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Luo H, Lin Y, Gao F, Zhang C-T, DEG ZR. 10, an update of the database of essential genes that includes both protein-coding genes and noncoding genomic elements. Nucleic Acids Res. 2014;42:D574–580. doi: 10.1093/nar/gkt1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mobegi FM, van Hijum SA, Burghout P, Bootsma HJ, de Vries SP, et al. From microbial gene essentiality to novel antimicrobial drug targets. BMC Genomics. 2014;15:958. doi: 10.1186/1471-2164-15-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Curtis PD, Brun YV. Identification of essential alphaproteobacterial genes reveals operational variability in conserved developmental and cell cycle systems. Mol Microbiol. 2014;93:713–735. doi: 10.1111/mmi.12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Su Y, Xu Y, Li Q, Yuan G, Zheng D. The essential genome of Ralstonia solanacearum . Microbiol Res. 2020;238:126500. doi: 10.1016/j.micres.2020.126500. [DOI] [PubMed] [Google Scholar]
- 37.S-Q A, Potnis N, Dow M, Vorhölter F-J, Y-Q H, et al. Mechanistic insights into host adaptation, virulence and epidemiology of the phytopathogen Xanthomonas . FEMS Microbiol Rev. 2020;44:1–32. doi: 10.1093/femsre/fuz024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ryan RP, Vorhölter F-J, Potnis N, Jones JB, Van Sluys M-A, et al. Pathogenomics of Xanthomonas: understanding bacterium–plant interactions. Nat Rev Microbiol. 2011;9:344–355. doi: 10.1038/nrmicro2558. [DOI] [PubMed] [Google Scholar]
- 39.Morinière L, Burlet A, Rosenthal ER, Nesme X, Portier P, et al. Clarifying the taxonomy of the causal agent of bacterial leaf spot of lettuce through a polyphasic approach leads to combine Xanthomonas hortorum Vauterin, et al. 1995 and Xanthomonas cynarae Trébaol 2000 emend. Timilsina, et al. 2019. Syst Appl Microbiol. 2020:126087. doi: 10.1016/j.syapm.2020.126087. [DOI] [PubMed] [Google Scholar]
- 40.Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T. Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 2008;179:546–556. doi: 10.1111/j.1469-8137.2008.02487.x. [DOI] [PubMed] [Google Scholar]
- 41.Potnis N, Minsavage G, Smith JK, Hurlbert JC, Norman D, et al. Avirulence proteins AvrBs7 from Xanthomonas gardneri and AvrBs1.1 from Xanthomonas euvesicatoria contribute to a novel gene-for-gene interaction in pepper. Mol Plant-Microbe Interactions®. 2011;25:307–320. doi: 10.1094/MPMI-08-11-0205. [DOI] [PubMed] [Google Scholar]
- 42.Kimbrel JA, Givan SA, Temple TN, Johnson KB, Chang JH. Genome sequencing and comparative analysis of the carrot bacterial blight pathogen, Xanthomonas hortorum pv. carotae M081, for insights into pathogenicity and applications in molecular diagnostics. Mol Plant Pathol. 2011;12:580–594. doi: 10.1111/j.1364-3703.2010.00694.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bull CT, Koike ST. Evaluating the efficacy of commercial products for management of bacterial leaf spot on lettuce. Plant Health Prog. 2005 [Google Scholar]
- 44.Bull CT. Genetic diversity of lettuce for resistance to bacterial leaf spot caused by Xanthomonas campestris pv. vitians . Plant Health Prog. 2007 [Google Scholar]
- 45.Koike ST, Gilbertson RL. Detect. Plant-Pathog. Bact. Seed Plant. Mater. 2nd ed. The American Phytopathological Society; 2017. Chapter 25: Detection of Xanthomonas campestris pv. vitians in lettuce seeds; pp. 173–178. [Google Scholar]
- 46.Fayette J, Jones JB, Pernezny K, Roberts PD, Raid R. Survival of Xanthomonas campestris pv. vitians on lettuce in crop debris, irrigation water, and weeds in south Florida. Eur J Plant Pathol. 2017 [Google Scholar]
- 47.Toussaint V, Benoit DL, Carisse O. Potential of weed species to serve as a reservoir for Xanthomonas campestris pv. vitians, the causal agent of bacterial leaf spot of lettuce. Crop Prot. 2012;41:64–70. [Google Scholar]
- 48.Bull CT, Koike ST. Practical benefits of knowing the enemy: modern molecular tools for diagnosing the etiology of bacterial diseases and understanding the taxonomy and diversity of plant-pathogenic bacteria. Annu Rev Phytopathol. 2015;53:157–180. doi: 10.1146/annurev-phyto-080614-120122. [DOI] [PubMed] [Google Scholar]
- 49.Sandoya GV, Maisonneuve B, Truco MJ, Bull CT, Simko I, et al. Genetic analysis of resistance to bacterial leaf spot in the Heirloom lettuce cultivar Reine des Glaces. Mol Breed. 2019;39:160. [Google Scholar]
- 50.Sambrook J, Russell DW. The Condensed Protocols from Molecular Cloning: a Laboratory Manual. CSHL Press; 2006. [Google Scholar]
- 51.Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13:e1005595. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallenet D, Calteau A, Dubois M, Amours P, Bazin A, et al. Microscope: an integrated platform for the annotation and exploration of microbial gene functions through genomic, pangenomic and metabolic comparative analysis. Nucleic Acids Res. 2020;48:D579–589. doi: 10.1093/nar/gkz926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo H, Gao F. DoriC 10.0: an updated database of replication origins in prokaryotic genomes including chromosomes and plasmids. Nucleic Acids Res. 2019;47:D74–77. doi: 10.1093/nar/gky1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yen M-R, Lin N-T, Hung C-H, Choy K-T, Weng S-F, et al. oriC region and replication termination site, dif, of the Xanthomonas campestris pv. campestris 17 chromosome. Appl Environ Microbiol. 2002;68:2924–2933. doi: 10.1128/AEM.68.6.2924-2933.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetJournal. 2011;17:10–12. [Google Scholar]
- 56.DeJesus MA, Ambadipudi C, Baker R, Sassetti C, Ioerger TR. TRANSIT - A software tool for Himar1 TnSeq analysis. PLOS Comput Biol. 2015;11:e1004401. doi: 10.1371/journal.pcbi.1004401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.DeJesus MA, Ioerger TR. A hidden Markov model for identifying essential and growth-defect regions in bacterial genomes from transposon insertion sequencing data. BMC Bioinformatics. 2013;14:303. doi: 10.1186/1471-2105-14-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics. 2009;25:119–120. doi: 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Galperin MY, Makarova KS, Wolf YI, Koonin EV. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kanehisa M, Sato Y, Morishima K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol. 2016;428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 62.Kanehisa M, Sato Y. Kegg Mapper for inferring cellular functions from protein sequences. Protein Sci. 2020;29:28–35. doi: 10.1002/pro.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Segata N, Börnigen D, Morgan XC, Huttenhower C. PhyloPhlAn is a new method for improved phylogenetic and taxonomic placement of microbes. Nat Commun. 2013;4:2304. doi: 10.1038/ncomms3304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Edgar RC. Search and clustering orders of magnitude faster than blast . Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 65.Edgar RC. Muscle: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shimodaira H, Hasegawa M. Multiple comparisons of log-likelihoods with applications to phylogenetic inference. Mol Biol Evol. 1999;16:1114–1116. [Google Scholar]
- 68.Yu G, Smith DK, Zhu H, Guan Y, TT-Y L. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8:28–36. [Google Scholar]
- 69.DeJesus MA, Nambi S, Smith CM, Baker RE, Sassetti CM, et al. Statistical analysis of genetic interactions in Tn-Seq data. Nucleic Acids Res. 2017;45:e93. doi: 10.1093/nar/gkx128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zomer A, Burghout P, Bootsma HJ, Hermans PWM, van Hijum S. Essentials: software for rapid analysis of high throughput transposon insertion sequencing data. PloS One. 2012;7:e43012. doi: 10.1371/journal.pone.0043012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Solaimanpour S, Sarmiento F, Mrázek J. Tn-seq explorer: a tool for analysis of high-throughput sequencing data of transposon mutant libraries. PLoS One. 2015;10:e0126070. doi: 10.1371/journal.pone.0126070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, et al. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis . PLOS Pathog. 2012;8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shields RC, Zeng L, Culp DJ, Burne RA. Genomewide identification of essential genes and fitness determinants of Streptococcus mutans UA159. mSphere. 2018;3 doi: 10.1128/mSphere.00031-18. 07 02 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan Z, Wang L, Sun S, Wu Y, Qian W. Genetic and proteomic analyses of a Xanthomonas campestris pv. campestris purC mutant deficient in purine biosynthesis and virulence. J Genet Genomics. 2013;40:473–487. doi: 10.1016/j.jgg.2013.05.003. [DOI] [PubMed] [Google Scholar]
- 75.Chandran V, Luisi BF. Recognition of enolase in the Escherichia coli RNA degradosome. J Mol Biol. 2006;358:8–15. doi: 10.1016/j.jmb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 76.Vorhölter F-J, Schneiker S, Goesmann A, Krause L, Bekel T, et al. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J Biotechnol. 2008;134:33–45. doi: 10.1016/j.jbiotec.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 77.Schatschneider S, Huber C, Neuweger H, Watt TF, Pühler A, et al. Metabolic flux pattern of glucose utilization by Xanthomonas campestris pv. campestris: prevalent role of the Entner–Doudoroff pathway and minor fluxes through the pentose phosphate pathway and glycolysis. Mol Biosyst. 2014;10:2663–2676. doi: 10.1039/c4mb00198b. [DOI] [PubMed] [Google Scholar]
- 78.Ito M, Guffanti AA, Wang W, Krulwich TA. Effects of nonpolar mutations in each of the seven Bacillus subtilis mrp genes suggest complex interactions among the gene products in support of Na+ and alkali but not cholate resistance. J Bacteriol. 2000;182:5663–5670. doi: 10.1128/jb.182.20.5663-5670.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ito M, Morino M, Krulwich TA. Mrp antiporters have important roles in diverse bacteria and archaea. Front Microbiol. 2017;8:2325. doi: 10.3389/fmicb.2017.02325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, et al. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol. 2008;4:174. doi: 10.1038/msb.2008.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gerlin L, Cottret L, Cesbron S, Taghouti G, Jacques M-A, et al. Genome-scale investigation of the metabolic determinants generating bacterial fastidious growth. MSystems. 2020;5:e00698–19. doi: 10.1128/mSystems.00698-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pieretti I, Pesic A, Petras D, Royer M, Süssmuth RD, et al. What makes Xanthomonas albilineans unique amongst xanthomonads? Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Palleroni NJ, Bradbury JF. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings, et al. 1983. Int J Syst Evol Microbiol. 1993;43:606–609. doi: 10.1099/00207713-43-3-606. [DOI] [PubMed] [Google Scholar]
- 84.da Silva FR, Vettore AL, Kemper EL, Leite A, Arruda P. Fastidian gum: the Xylella fastidiosa exopolysaccharide possibly involved in bacterial pathogenicity. FEMS Microbiol Lett. 2001;203:165–171. doi: 10.1111/j.1574-6968.2001.tb10836.x. [DOI] [PubMed] [Google Scholar]
- 85.Lu H, Patil P, Van Sluys M-A, White FF, Ryan RP, et al. Acquisition and evolution of plant pathogenesis–associated gene clusters and candidate determinants of tissue-specificity in Xanthomonas . PLoS ONE. 2008;3:e3828. doi: 10.1371/journal.pone.0003828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Katzen F, Ferreiro DU, Oddo CG, Ielmini MV, Becker A, et al. Xanthomonas campestris pv. campestris gum mutants: Effects on xanthan biosynthesis and plant virulence. J Bacteriol. 1998;180:1607–1617. doi: 10.1128/jb.180.7.1607-1617.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kim S-Y, Kim J-G, Lee B-M, Cho J-Y. Mutational analysis of the gum gene cluster required for xanthan biosynthesis in Xanthomonas oryzae pv oryzae . Biotechnol Lett. 2008;31:265. doi: 10.1007/s10529-008-9858-3. [DOI] [PubMed] [Google Scholar]
- 88.Alkhateeb RS, Vorhölter F-J, Rückert C, Mentz A, Wibberg D, et al. Genome wide transcription start sites analysis of Xanthomonas campestris pv. campestris B100 with insights into the gum gene cluster directing the biosynthesis of the exopolysaccharide xanthan. J Biotechnol. 2016;225:18–28. doi: 10.1016/j.jbiotec.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 89.Yoon K-H, Cho J-Y. Transcriptional analysis of the gum gene cluster from Xanthomonas oryzae pathovar oryzae . Biotechnol Lett. 2007;29:95–103. doi: 10.1007/s10529-006-9217-1. [DOI] [PubMed] [Google Scholar]
- 90.Katzen F, Becker A, Zorreguieta A, Pühler A, Ielpi L. Promoter analysis of the Xanthomonas campestris pv. campestris gum operon directing biosynthesis of the xanthan polysaccharide. J Bacteriol. 1996;178:4313–4318. doi: 10.1128/jb.178.14.4313-4318.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Galván EM, Ielmini MV, Patel YN, Bianco MI, Franceschini EA, et al. Xanthan chain length is modulated by increasing the availability of the polysaccharide copolymerase protein GumC and the outer membrane polysaccharide export protein GumB. Glycobiology. 2013;23:259–272. doi: 10.1093/glycob/cws146. [DOI] [PubMed] [Google Scholar]
- 92.Xayarath B, Yother J. Mutations blocking side chain assembly, polymerization, or transport of a Wzy-dependent Streptococcus pneumoniae capsule are lethal in the absence of suppressor mutations and can affect polymer transfer to the cell wall. J Bacteriol. 2007;189:3369–3381. doi: 10.1128/JB.01938-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Salinas SR, Bianco MI, Barreras M, Ielpi L. Expression, purification and biochemical characterization of GumI, a monotopic membrane GDP-mannose:glycolipid 4-β-D-mannosyltransferase from Xanthomonas campestris pv. campestris . Glycobiology. 2011;21:903–913. doi: 10.1093/glycob/cwr022. [DOI] [PubMed] [Google Scholar]
- 94.Niknezhad SV, Asadollahi MA, Zamani A, Biria D. Production of xanthan gum by free and immobilized cells of Xanthomonas campestris and Xanthomonas pelargonii . Int J Biol Macromol. 2016;82:751–756. doi: 10.1016/j.ijbiomac.2015.10.065. [DOI] [PubMed] [Google Scholar]
- 95.Schiltz CJ, Adams MC, Chappie JS. The full-length structure of Thermus scotoductus OLD defines the ATP hydrolysis properties and catalytic mechanism of Class 1 OLD family nucleases. Nucleic Acids Res. 2020;48:2762–2776. doi: 10.1093/nar/gkaa059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Myung H, Calendar R. The old exonuclease of bacteriophage P2. J Bacteriol. 1995;177:497–501. doi: 10.1128/jb.177.3.497-501.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schiltz CJ, Lee A, Partlow EA, Hosford CJ, Chappie JS. Structural characterization of class 2 old family nucleases supports a two-metal catalysis mechanism for cleavage. Nucleic Acids Res. 2019;47:9448–9463. doi: 10.1093/nar/gkz703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ranjani P, Gowthami Y, Gnanamanickam S, Palani P. Bacteriophages: A new weapon for the control of bacterial blight disease in rice caused by Xanthomonas oryzae . Microbiol Biotechnol Lett. 2018;46:346–359. [Google Scholar]
- 99.Vasu K, Nagaraja V. Diverse functions of restriction-modification systems in addition to cellular defense. Microbiol Mol Biol Rev. 2013;77:53–72. doi: 10.1128/MMBR.00044-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Takeuchi K, Noda N, Someya N. Complete genome sequence of the biocontrol strain Pseudomonas protegens Cab57 discovered in Japan reveals strain-specific diversity of this species. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0093683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE — A database for DNA restriction and modification: enzymes, genes and genomes. Nucleic Acids Res. 2015;43:D298–D299. doi: 10.1093/nar/gku1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mruk I, Kobayashi I. To be or not to be: regulation of restriction–modification systems and other toxin–antitoxin systems. Nucleic Acids Res. 2014;42:70–86. doi: 10.1093/nar/gkt711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Loenen WAM, Dryden DTF, Raleigh EA, Wilson GG. Type I restriction enzymes and their relatives. Nucleic Acids Res. 2014;42:20–44. doi: 10.1093/nar/gkt847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Raleigh EA. Organization and function of the mcrBC genes of Escherichia coli K-12. Mol Microbiol. 1992;6:1079–1086. doi: 10.1111/j.1365-2958.1992.tb01546.x. [DOI] [PubMed] [Google Scholar]
- 105.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Mochizuki A, Yahara K, Kobayashi I, Iwasa Y. Genetic addiction: selfish gene's strategy for symbiosis in the genome. Genetics. 2006;172:1309–1323. doi: 10.1534/genetics.105.042895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Furuta Y, Abe K, Kobayashi I. Genome comparison and context analysis reveals putative mobile forms of restriction-modification systems and related rearrangements. Nucleic Acids Res. 2010;38:2428–2443. doi: 10.1093/nar/gkp1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.