Abstract
Background and objectives:
Plakophilin-2 (PKP2) is an intracellular desmosomal anchoring protein that has been implicated in a genome-wide association study, in which genetic variants of PKP2 are associated with Porphyromonas gingivalis (P.gingivalis)-dominant periodontal dysbiosis. In this study, we compared the ex vivo PKP2 expression in periodontitis gingival biopsies to periodontitis-free subjects and assessed the in vitro role of PKP2 in gingival epithelial barrier function and the mechanism by which P.gingivalis modulates PKP2 expression.
Material and methods:
Using reverse transcription quantitative real-time PCR (RT-qPCR), we determined PKP2 mRNA expression levels in gingival biopsies collected from 11 periodontally healthy, 10 experimental gingivitis, and 10 chronic periodontitis subjects. PKP2 protein expression in gingival biopsies was detected by immunohistochemistry. We then challenged primary gingival epithelial cells with bacteria including P.gingivalis, Campylobacter rectus, and various Toll-like receptor agonists. Western blot and immunofluorescence staining were used to detect protein expression. Inhibitors blocking proteases pathways were tested for P.gingivalis-mediated PKP2 protein degradations. We also knocked down endogenous epithelial PKP2 using lentiviral short-hairpin RNA (shRNA) and evaluated cell proliferation, spreading, and barrier function.
Results:
Periodontitis gingival biopsies had approximately twofold less PKP2 mRNA than did healthy controls (p < .05). PKP2 protein was predominantly expressed in gingival epithelium. In primary gingival epithelial cells, P.gingivalis challenge increased PKP2 mRNA levels, while protein expression decreased, which suggests that P.gingivalis has a protein degradation mechanism. Cysteine proteases inhibitors greatly attenuated P.gingivalis-m ediated PKP2 protein degradation. Epithelial cells with deficient PKP2 exhibited inhibited cell proliferation and spreading and failed to form monolayers. Finally, P.gingivalis impaired gingival epithelial barrier function.
Conclusions:
PKP2 appears to be critical in maintaining gingival epithelial barrier function and is susceptible to degradation by cysteine proteases produced by P.gingivalis. Our findings have identified a mechanism by which P.gingivalis impairs epithelial barrier function by promoting PKP2 degradation.
Keywords: desmosome, epithelium, gene expression, periodontal disease, PKP2
1 |. INTRODUCTION
Maintenance of periodontal health requires intact cell junctions that form a strong gingival epithelial barrier to wall off microbial invasion. As a major cell junction complex, desmosome junctions reside throughout the oral, sulcular, and junctional epithelium.1,2 Desmoglein (DSG) and desmocollin (DSC) are desmosomal extracellular components (also known as Cadherin proteins), while plakophilin, plakoglobin, and desmoplakin are the intracellular components that connect desmosomes and intermediate filaments.3 Disruptions of desmosome junctions lead to skin blister diseases such as pemphigus and impetigo.4,5 In impetigo, desmoglein proteins are subject to proteases from Streptococcus and Staphylococcus species, leading to desmosomal disassembly that subsequently causes blisters.6–8 There is a gap in knowledge about how desmosome proteins respond to periodontal bacteria such as Porphyromonas gingivalis (P.gingivalis) and P.gingivalis-associated proteases.
Within the desmosome junction, plakophilin-2 (PKP2) is a well-characterized intracellular anchoring protein. Mutations in the PKP2 gene are etiologically associated with a severe inherited heart disease-arrhythmogenic right ventricular cardiomyopathy. pkp2 null mice experience embryonic lethality due to heart morphogenesis defects.9,10 PKP2 has a multi-faceted role in cell proliferation and migration.11,12 The interest in the role of PKP2 as it relates to P.gingivalis-associated periodontitis has emerged from a genome-wide association study (GWAS) that demonstrated a strong association between P.gingivalis-dominant periodontal dysbiosis and single nucleotide polymorphism variants at rs6488099 locus within the PKP2 gene locus.13 Results from the GWAS study suggest PKP2 as a potential candidate gene for periodontal disease among subjects with P.gingivalis-dominated dysbiosis, but the mechanism of PKP2 in periodontal disease needs further investigations.
In this study, we aimed to assess the ex vivo expression of PKP2 in periodontal disease and the in vitro role of PKP2 in gingival epithelial cells. In addition, this investigation provides novel insights into the modulation of desmosomal proteins by P.gingivalis and suggests that gingival epithelial barrier disruption is a key mechanism of P.gingivalis-mediated pathogenesis in periodontitis.
2 |. MATERIALS AND METHODS
2.1 |. Participant selection
In this study, we included analyses of clinical gingival biopsies from participants in a case-control clinical study and in vitro experiments. In the clinical portion, we recruited participants with healthy periodontium, experimentally induced gingivitis, and chronic periodontitis at the University of North Carolina at Chapel Hill (UNC-CH) Adams School of Dentistry between 2010 and 2015. All clinical components including gingival biopsies that were acquired through surgical procedures were approved by the Institutional Review Board of UNC-CH. A signed informed consent was collected from each participant. Experimental gingivitis was induced in a group of participants in accord with a well-described stent-induced biofilm overgrowth model (SIBO).14 These participants went through a 21-day gingival inflammation induction period and their gingival biopsies were collected on Day 21, according to the time frame described by Löe et al.15 Definitions of healthy and periodontitis participants were based on the 1999 International Workshop for a Classification of Periodontal Diseases and Conditions.16 Details of the exclusion and inclusion criteria for three groups of participants can be found in previously published papers.14,17 Briefly, the exclusion criteria include the following: (1) no use of antibiotics in the 1 month before the screening examinations; (2) no signs of systemic diseases with any oral manifestation at the time of the enrollment; (3) no treatment for systemic diseases 3 months prior to the collection of gingival biopsies. Periodontally healthy participants were either healthy volunteers or those scheduled for crown lengthening or tooth extraction. All periodontitis participants went through an initial non-surgical treatment phase at which point, they still required further surgical interventions. At the time of biopsy collections, we first examined six sites of the affected tooth: mesial-lingual, mid-lingual, distal-lingual, mesial-buccal, mid-buccal, and distal-buccal for clinical parameters. The average probing depth (PD) and clinical attachment level of biopsy-involved teeth were reported. We then anesthetized the tooth with 2% lidocaine with 1:100, 000 epinephrine. Incisions were made on keratinized gingival tissues around the selected tooth. A band of 0.5-to 1-mm gingival collar was mostly removed from the interproximal site where the deepest pocket was located in a periodontitis patient. Then, tissue biopsies were immediately processed with designated preservation methods for further analyses. A similar size gingival biopsy was also collected from the interproximal site from each gingivitis or periodontally healthy participant. This project was a pilot feasibility clinical investigation, and a sample size of 10 subjects per group was estimated to provide a power of 80% and an alpha of p < .05 (two-t ailed) to detect a 1.4-f old difference in PKP2 mRNA expression using variance estimates based upon changes in IL-1β expression within diseased tissues.18 A total of 31 gingival biopsies were processed in RNAlater (Thermo) and stored at −80°C until RNA was isolated. An additional six biopsies (three healthy and three periodontitis) were collected for immune-histochemistry (IHC) staining of PKP2 from six different participants in addition to the subjects mentioned above, so that we could determine in histology where PKP2 expression was mainly localized in gingival biopsies. These additional tissues were fixed in 10% neutral-buffered formalin, dehydrated with ethanol, and sent to the Histology Core Facility at UNC-CH for paraffin embedding, sectioning, and IHC staining for PKP2 protein.
2.2 |. RNA isolation, reverse transcription quantitative real-time PCR (RT-qPCR), and IHC staining
Total RNA was isolated using an RNeasy Mini kit (Qiagen). 500 ng total RNA was reverse-transcribed to cDNA (SuperScript VILO, Invitrogen). RT-qPCR was conducted in a 7500 Sequence Detection System, detecting PKP2 and Keratin 5/KRT5 gene expression using a Taqman assay (Thermo Scientific, Hs00428040 and Hs00361185, respectively). 18s ribosomal RNA was used as the housekeeping gene for normalizations (Thermo Scientific, 4319413). For IHC staining, heat-induced antigen retrieval was performed on the sliced paraffin-embedded gingival biopsies prior to staining. After blocking, slides were stained with a mouse anti-human anti-PKP2 antibody (Santa Cruz, sc-393711, C-1 clone, dilution: 1:100) overnight at 4°C. After washing, those slides were incubated for 1 hour (1 h) with a goat anti-mouse secondary antibody (Jackson ImmunoResearch, 115–065-166, dilution: 1:500) at room temperature (RT). Slides were then treated with ABC elite reagents (Vector) and DAB (Thermo) for visualization under a bright field microscope. Slides treated without a primary antibody served as the negative controls.
2.3 |. In vitro stimulation of gingival epithelial cells
Primary human gingival epithelium progenitor cells (HGEPs, single donor, CellnTec) were maintained in a CnT-Prime (CellnTec) medium without antibiotics. Cells were seeded at a density of 5 × 105 in six-well plates and stimulated, respectively, with P.ginigvalis A7436, Campylobacter rectus (C.retus) 314, and multiple Toll-l ike receptor (TLR) agonists including tumor necrosis factor-α (TNF-α , R&D 210-TA-020), P.ginigvalis strain W83-derived lipopolysaccharide/LPS (a courtesy from Salvador Nares lab at University of Illinois Chicago), P.ginigvalis strain A7436-derived LPS, and Escherichia coli (E. coli strain O55:B6) LPS for up to 24 hours (24 h). Several multiplicities of infections (MOIs) were applied, but an MOI of 50 was most frequently utilized throughout the study. Heat-killed P.ginigvalis and C.rectus were prepared in a 70°C heat block for 30 min to inactivate bacterial proteases while still keep the inflammatory stimulatory capability. The absence of colony formation was confirmed by culturing heat-treated P.ginigvalis in an anaerobic chamber. One proteasomal inhibitor—MG132 (a courtesy from Qing Zhang lab at UNC-CH)—and one lysosomal inhibitor—NH4Cl—were used in vitro. Serine protease inhibitor—Pefabloc SC (Roche 11429868001)—and cysteine protease inhibitors—Leupeptin (Roche 11017101001) and Cathepsin B inhibitor (Sigma 219385)—were incubated with P.ginigvalis in an anaerobic chamber at 37°C for 1 h before P.ginigvalis was added into the cell culture.
2.4 |. Western blot
Whole-cell lysates of HGEPs were collected with a RIPA buffer (Cell Signaling, 9806) 24 h after the challenge. We used a mouse anti-human anti-PKP2 antibody (molecular weights: 130 kDa, Santa Cruz, sc-393711, C-1 clone, dilution 1:200), a rabbit anti-human anti-DSG1 antibody (molecular weights: 150 kDa, Santa Cruz, sc-20114, H-290 clone, dilution 1:200), and a rabbit anti-human anti-β-actin antibody (molecular weights: 45 kDa, Cell Signaling, 13E5, dilution 1:1000) for western blotting. DSG1 (Desmoglein 1) is an extracellular desmosomal protein. Specific HRP-conjugated secondary antibodies (Thermo Scientific 31430, dilution 1:5000; Jackson ImmunoResearch, 111–035-144, dilution 1:10 000) were incubated for 1 h at RT for relevant blots. An ECL system (SuperSignal West Pico, Pierce) was used to detect the intensity of the bands. Densitometry analysis was conducted by ImageJ to quantify protein expression levels.
2.5 |. Calcium switch and immunofluorescence staining
As calcium plays a role in both maintaining normal desmosome structures and function and protects cells from protease cleavage,19 we switched a regular low calcium-containing medium (0.7 μM) per cell supplier to a medium containing a higher calcium concentration (1.8 mM) in some of the experiments to investigate the effect of different calcium concentrations on desmosomal protein expression levels. 5 × 104 HGEPs cells were seeded in 24-well glass-bottom plates (MatTek) that were coated with 10 μg/ml Fibronectin (Sigma). When cells were approximately 80% confluent, the cell culture medium was changed to a high-calcium concentration medium (1.8 mM). After an overnight incubation, cells were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 30 min, and then blocked with 1% BSA/PBS/0.1% Triton X-100 at RT for 1 h. Cells were either incubated with an anti-PKP2 (Santa Cruz, sc-393711, C-1 clone, dilution 1:200) or anti-DSG1 (Santa Cruz, sc-20114, H-290 clone, dilution 1:200) primary antibody overnight at 4°C. A secondary antibody (Alexa Fluor 488 for anti-PKP2, Alexa Fluor 590 for anti-DSG1) was incubated with cells at 1:200 dilution at RT for 1 h, avoiding light. A confocal microscopy (Zeiss LSM 710) was used to capture the immunofluorescence signals. In a separate experiment, live P.ginigvalis with an MOI of 50 was added to HGEPs under either a low-or high-calcium concentration to study the effect of calcium on epithelial responses to bacterial proteolysis. Western blotting was used to detect PKP2 and DSG1 protein expression levels.
2.6 |. PKP2 silencing in gingival epithelial cells
To test for the functional relevance of the reduced epithelial PKP2 expression, we knocked down PKP2 using lentiviral short-hairpin RNA (shRNA) clones and examined cellular phenotypic changes in vitro. Target sequences of five shRNAs and one scramble (scr) control RNA construct are listed in Table S1. Each shRNA lentiviral particle-containing medium was prepared at the Lentivirus Core Facility at UNC-CH. HGEPs were treated with a lentiviral particle-containing medium for 48 h, and successfully transduced cells were selected by puromycin (2 μg/ml, MP Biomedicals) for up to 7 days. We examined cell proliferation using the MTS assay (CellTiter 96 ® Aqueous) following the manufacturer’s protocol. 2 × 104 puromycin-selected cells were seeded on 96-well plates for an initial 48 h and culture medium was collected at day 0 (starting point), 1, and 3 for MTS assay. Cells were also seeded onto fibronectin-coated plate and observed under a phase-contrast microscope for cell spreading 1 h post-seeding.
2.7 |. Epithelial barrier function assay
We conducted a fluorescein isothiocyanate-dextran (FITC-Dextran, Sigma, #46945, mol wt 70 000) transwell dye exclusion assay to assess gingival epithelial barrier function. After puromycin selection, PKP2 knockdown or scramble shRNA-transduced HGEPs were seeded onto fibronectin-coated 0.4-μ m pore size transwell inserts (Sigma, CLS3470) to form a monolayer. FITC dye (1 mg/ml, final concentration) was added onto the top compartment of each insert. In a parallel experiment, regularly maintained wild-type HEGPs were seeded in a density of 1 × 105 onto transwell inserts. In addition to FITC dye, live P.ginigvalis of 109 CFU/ml with or without Leupeptin, live 108 CFU/ml, or heat-killed P.ginigvalis 109 CFU/ ml was added into the top compartments of the inserts in a final volume of 200 μl. Here, we did not follow the previously utilized MOI quantifications for P.ginigvalis since cells may become over confluence in the inserts and since the exact confluence is hard to observe given the porosity of the inserts. All inserts were immersed in culture medium in the bottom wells of a 24-well plate. Five hours later, medium from bottom wells was collected and measured for fluorescent signals at the 495 nm/527 nm excitation-emission wavelengths.
2.8 |. Cell viability assay
Human gingival epithelium progenitor cells stimulated by P.ginigvalis at different MOIs and P.ginigvalis-challenged cells used in epithelial barrier function as mentioned above were collected and washed with PBS. After washing, cells were immediately stained with a Live/ Dead Green Dead Cell Stain Kit (Thermo Fisher Scientific) for 30 min at 4°C in dark. Then, fluorescence signals were read by a BD LSR II with Violet flow cytometer (Becton Dickinson). Heat-killed cells were included as a positive control for complete cell death.
2.9 |. Statistical analysis
Statistical significance was determined by Student’s t-test (two-tailed) for two-sample comparisons or one-way ANOVA for multiple sample comparisons. Changes in gene expression were calculated by the 2−ΔΔCT method.20 Within-group differences were evaluated by Fisher’s least significant differences (LSD) test. Chi-square test was used for categorical variable data analyses. All statistical analyses were performed by GraphPad Prism version 6.0. The statistical significance threshold was set at p < .05. Three replicates were used in each in vitro experiment, and every experiment was independently repeated at least twice.
3 |. RESULTS
3.1 |. Characterizations of PKP2 expression in gingival biopsies
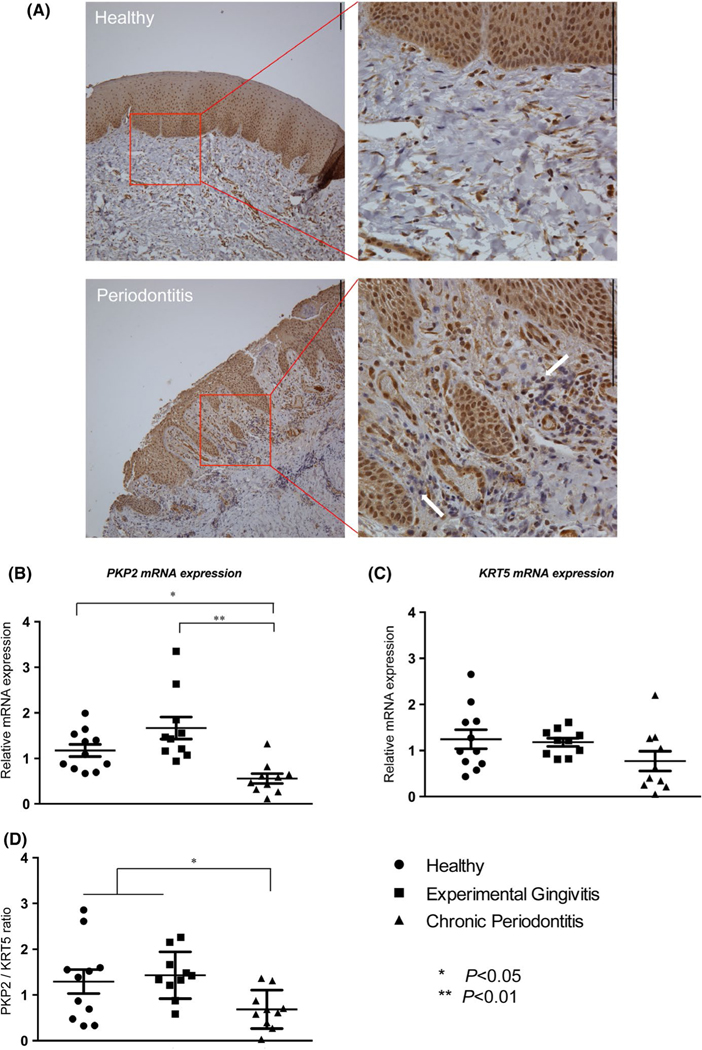
A total of 37 participants, aged 19– 64, were recruited sequentially in the clinic. Among them, six samples (three from participants who were periodontally healthy and three participants suffering from chronic periodontitis) were processed exclusively for IHC staining of PKP2 protein. In Figure 1, we observed that PKP2 expression was predominantly localized in epithelial cells in both cytosol and nuclear compartments, while endothelial cells and fibroblasts expressed moderate levels of PKP2. Compared with control samples with periodontal health, periodontitis gingival biopsy samples displayed disrupted epithelium, prominent rete pegs, and more immune cell infiltrates (Figure 1A, lower left panel). We did not quantify and compare the expression of PKP2 between healthy and periodontitis biopsies because of the limited numbers of samples (three in each group) and because of the uniform staining patterns of PKP2 in gingival epithelium in all six samples (Figure S1).
FIGURE 1.
PKP2 expression in gingiva biopsies. (A) PKP2 expression was primarily localized to gingival epithelium in gingival biopsies. Scale bar is 130 μm. Periodontitis biopsies displayed disrupted epithelium, prominent rete pegs, and more immune cell infiltrates (white arrows). PKP2 was detected in endothelial cells and fibroblast at a moderate level (lower right panel). (B) Periodontitis biopsies had about twofold less PKP2 mRNA expression than those with healthy gingival biopsies. (C) KRT5 mRNA expression was not significantly different among those three groups. (D) The fold change ratio of PKP2/KRT5 remained significantly less in periodontitis biopsies than in healthy gingiva and experimental gingivitis biopsies. * indicates p < .05; ** indicates p < .01
We utilized the remaining 31 gingival biopsy samples for exploring PKP2 mRNA transcription, focusing on the epithelial PKP2. Among them, 11 samples were from periodontally healthy subjects; ten samples were from experimental gingivitis subjects; ten samples were from chronic periodontitis subjects. Demographics and clinical parameters for the three groups are listed in Table S2. As expected, the differences in PD and attachment level at the biopsied teeth were significant among the three groups of subjects (p < .05). Chronic periodontitis subjects were significantly older than the other two groups (p < .05). Demographic parameters including gender, ethnicity, and smoking status were comparable between three groups. Chronic periodontitis subjects had approximately two-fold less PKP2 mRNA expression in gingival biopsies (Figure 1B, p < .05) than did periodontally healthy subjects. The difference of PKP2 mRNA expression between experimental gingivitis subjects and healthy controls was not statistically significant (p > .05). We used KRT5 as an internal reference for the epithelial content of each biopsy to adjust for cell type heterogeneity in tissues. No significant differences were detected in KRT5 mRNA expression among the three groups, even though there was a trend toward lower KRT5 content for subjects with chronic periodontitis (Figure 1C, p > .05). We then normalized the level of PKP2 to KRT5 by plotting a ratio for each sample from each subject. After the normalization, PKP2 mRNA expression level was still significantly lower in periodontitis biopsies than in control and experimental gingivitis samples (Figure 1D). These results suggest that PKP2 is primarily localized in epithelial cells and a decreased epithelial PKP2 mRNA expression level is associated with periodontitis.
3.2 |. PKP2 protein is specifically degraded by live P.gingivalis
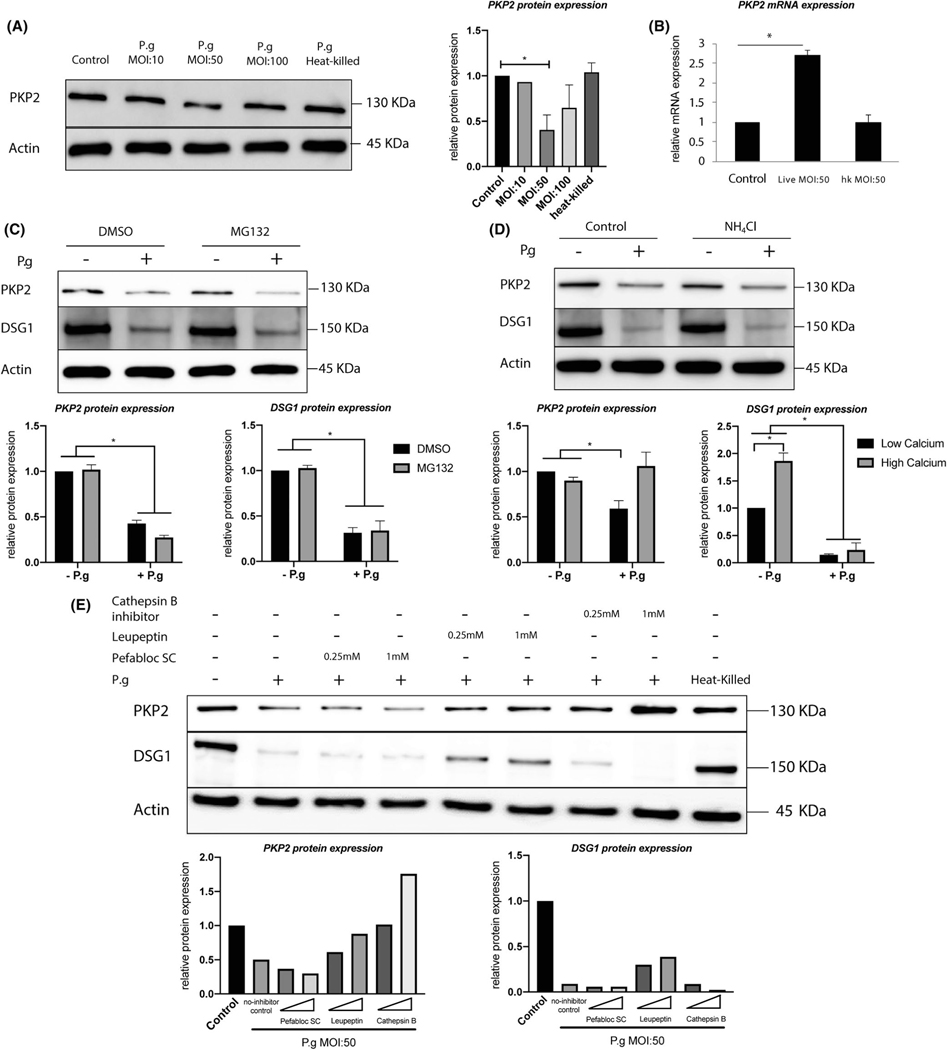
We then focused on the epithelial PKP2 and utilized human gingival epithelial progenitors (HGEPs) cells that were acquired from normal gingiva from a single donor to study the effect of P.ginigvalis on PKP2 expression. We found that HGEPs under P.ginigvalis challenge for 24 h at an MOI of 50 exhibited consistent reduction in PKP2 protein expression (Figure 2A). Although cells challenged with P.ginigvalis at an MOI of 100 exhibited reduced PKP2 protein expression, such a reduction of PKP2 expression was not consistent and, therefore, was not statistically significant in comparison with control cells with P.ginigvalis challenge based on three independent experiments (Figure 2A, right panel, densitometry, and Figure S2, densitometry). Heat-killed P.ginigvalis did not affect PKP2 protein expression, which suggests that heat-sensitive bacterial contents might be responsible for the P.ginigvalis-mediated PKP2 expression reduction. We also tested effects of different P.ginigvalis MOIs over various lengths of treatment time (2 h, 8 h, 24 h) on both PKP2 and DSG1 expression. The protein levels of both molecules were reduced in a dosage-and time-dependent manner (Figure S2). Additionally, we tested cytotoxicity of different MOIs on epithelial cells for 24 h (Figure S3). We found that more than 80% cells were alive with an MOI of 50 for 24 h, and about 75.5% cells were alive with an MOI of 100. Therefore, we decided that the cytotoxicity of P.ginigvalis under an MOI of 50 was an appropriate condition for ongoing experiments. Taken together, the treatment of HGEPs with P.ginigvalis at an MOI of 50 for 24 h was optimal and was, therefore, used for the remaining mechanistic experiments in the co-culture model with HGEPs. Interestingly, in response to a live P.ginigvalis challenge, PKP2 mRNA expression in HGEPs increased by about 2.5-fold (Figure 2B). This increase stood in contrast to the decreased PKP2 mRNA expression found in the periodontitis tissue samples (Figure 1B). The increased mRNA and decreased protein pattern also suggest a protein degradation mechanism rather than a decreased protein synthesis. To further test the hypothesis that the PKP2 protein degradation was specific to the P.ginigvalis challenge, we used another periodontal disease-associated pathogen, C.rectus (strain 314), and TLR ligands including TNF-α , LPS from E. coli and two P.ginigvalis strains (A7436 and W83) to challenge HGEPs and found none of those agents affected PKP2 or DSG1 protein expression (Figure S4). Therefore, these data suggest that PKP2 protein degradation is specifically attributed to the live P.ginigvalis challenge.
FIGURE 2.
PKP2 protein is specifically degraded by Porphyomonas gingivalis by actions of cysteine proteases. (A) Western blot results showed that 24 h of live P.ginigvalis challenge at an MOI of 50 significantly decreased PKP2 protein expression in HGEPs. Heat-killed P.ginigvalis had no effects on PKP2 protein expression. Densitometry is presented on the right panel, averaging from replicates of two or more independent experiments. Heat-killed P.ginigvalis was at an MOI of 50. (B) 24 h of live P.ginigvalis challenge at an MOI of 50 increased PKP2 mRNA. Heat-killed P.ginigvalis (MOI:50) had no effects on PKP2 mRNA. (C and D) The treatment of HGEPs with a proteasome inhibitor MG132 or lysosome inhibitor NH4Cl did not block PKP2 and DSG1 protein degradation by live P.ginigvalis. HGEPs were pre-treated with MG132 10 μM for 3 h or NH4Cl for 6 h and then challenged with live P.ginigvalis at an MOI of 50 for 24 h. Densitometry analysis is presented in lower panels. (E) P.ginigvalis were co-cultured with different protease inhibitors for 1 h anaerobically at 37°C before being applied to treat HGEPs for 24 h. Leupeptin and Cathepsin B inhibitor greatly attenuated PKP2 degradation, but only partially restored DSG1 degradation. PKP2 protein-130 kDa; DSG1 protein-150 kDa; β-actin protein-45 kDa. * indicates p < .05. Error bars represent standard deviations
3.3 |. P.ginigvalis-mediated PKP2 protein degradation is due to cysteine proteases produced by P.ginigvalis
To further explore P.ginigvalis-mediated degradation of PKP2 proteins, both intracellular and extracellular protein degradation pathways were investigated using pathway-specific inhibitors. To investigate the involvement of intracellular degradation pathway, we treated cells with the proteasome inhibitor MG132 or the lysosome inhibitor NH4Cl. We found that neither MG132 nor NH4Cl blocked P.ginigvalis-mediated PKP2 or DSG1 degradations (Figure 2C,D). To explore the extracellular degradation pathway, we found that both cysteine proteases inhibitors, Leupeptin and Cathepsin B inhibitor, restored PKP2 expression in the presence of P.ginigvalis challenge to a similar level as its endogenous expression without P.ginigvalis. However, both inhibitors only partially restored DSG1 expression in P.ginigvalis-challenged cells (Figure 2E). We, therefore, propose that desmoglein 1 (DSG1), an extracellular desmosomal protein, is more susceptible to extracellular sources of proteolytic enzymes from P.ginigvalis than PKP2. In addition, the serine protease inhibitor, Pefabloc SC, did not reverse P.ginigvalis-mediated degradation of either molecule, indicating that the degradation was specific to bacterial cysteine proteases activity. In summary, P.ginigvalis-mediated desmosomal protein degradation does not rely on epithelial intracellular degradation pathways but on the bacteria-associated cysteine proteases extracellular degradation pathway.
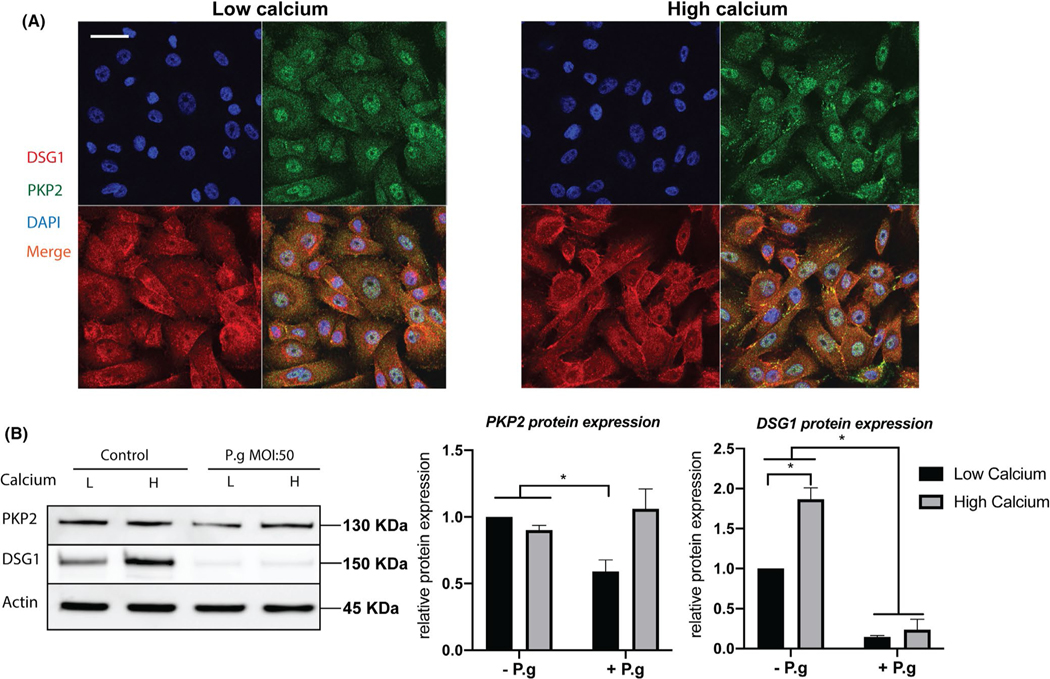
3.4 |. Calcium switch results in co-localization of PKP2 and DSG1
Since calcium levels directly affect desmosome structures and epithelial barrier homeostasis, we switched from a low concentration of calcium (0.7 μM) to a high concentration (1.8 mM) and found that calcium switch promoted the formation of punctated PKP2 and DSG1 protein expression in unstimulated HGEPs (Figure 3A). Co-l ocalization of DSG1 and PKP2 at the periphery of epithelial cells may indicate strengthened desmosome junctions. From western blotting results, we found that the high-calcium concentration (1.8 mM) increased DSG1, but not PKP2 expression (Figure 3B, left panels). With a low calcium concentration, HGEPs co-cultured with P.ginigvalis exhibited decreases in DSG1 and PKP2 protein expression levels, while the high-calcium concentration protected PKP2 from being degraded by P.ginigvalis (MOI: 50)—a bacterial load that effectively degraded PKP2 when subjected to a low concentration of calcium (Figure 3B). These results suggest that calcium strengthens desmosome junctions at the cell surface and mitigates the P.ginigvalis-mediated degradation of PKP2.
FIGURE 3.
PKP2 and DSG1 co-localize under high calcium. (A) High calcium (1.8 mM) in medium led to punctated DSG1 and PKP2 expression and enhanced their co-localization at cell junctions. Scale bar is 30 μm. (B) The high-calcium condition increased DSG1 protein expression but did not affect the overall PKP2 amount by western blotting. High calcium protected PKP2 from being degraded by live P.ginigvalis at an MOI of 50. Densitometry is presented in right panels
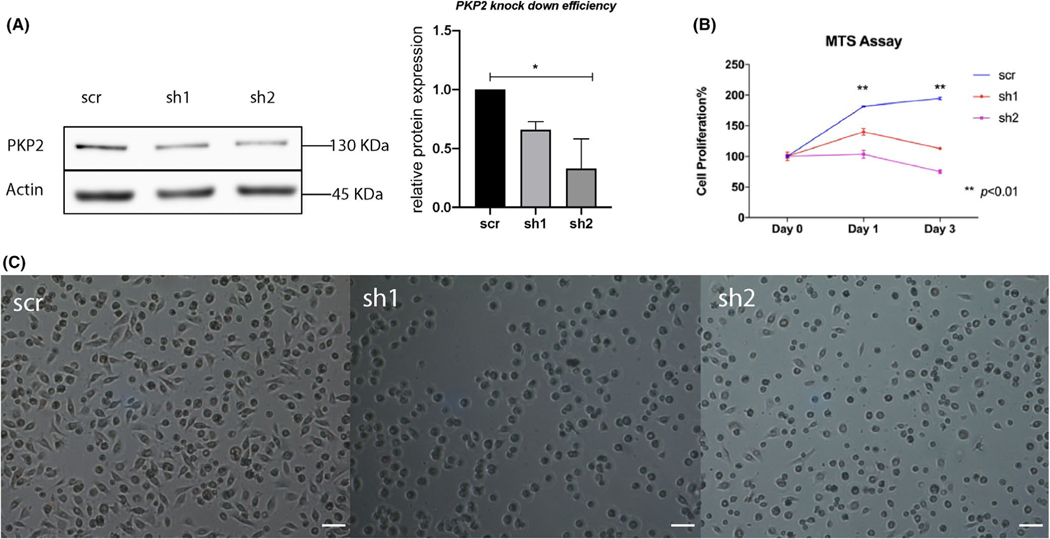
3.5 |. PKP2 deficiency inhibits cell proliferation and spreading
Among five tested shRNA constructs (Table S1), sh1, sh2, and sh4 shRNA constructs consistently knocked down PKP2 in HEGPs, especially sh2 with an average knockdown efficiency of more than 50% (Figure 4A, right panel). Phenotypic characterizations of HGEPs in which PKP2 was knocked down by sh1 and sh2 were reported here (Figure 4A), and those of sh4 were reported in a recent review article.21 Compared with scramble control, the cell proliferation of sh1-and sh2-transduced cells was significantly inhibited (Figure 4B). Scramble shRNA-transduced cells were able to spread and polarize 1 h after being seeded, while sh1-and sh2-transduced cells had round shapes that occupied less areas, which suggests an inefficient spreading (Figure 4C). We attempted to seed scramble shRNA-transduced cells and sh2-transduced cells on transwell inserts and apply FITC dye to test for cell barrier function. However, sh2-transduced cells failed to form a monolayer barrier on transwell inserts (Figure S5). This observation provides additional support to the hypothesis that PKP2 deficiency impairs cell spreading and cell-to-cell contact.
FIGURE 4.
PKP2 deficiency impairs cell proliferation and spreading. (A) PKP2 was moderately knocked down by short-hairpin 1 (sh1) and short-hairpin 2 (sh2) lentivirus-shRNA constructs in HGEPs compared to scramble control (scr), as shown by western blotting. Densitometry analysis of PKP2 protein expression in sh1 and sh2 from two separate western blots (right panel) is to demonstrate the knockdown efficiency. (B) Epithelial cells with deficient PKP2 displayed significantly inhibited cell proliferations. ** indicates p < .01. Error bars represent standard deviations. 2 × 104 puromycin-selected cells had been seeded on 96-well plates and waited for 48 h before we collected culture medium at day 0 (the first medium collection), 1, and 3 for MTS assay. (C) HGEPs with deficient PKP2 displayed impaired cell spreading 1 h after being seeded. Scale bar is 31 μm
3.6 |. P.ginigvalis impairs gingival epithelial barrier function
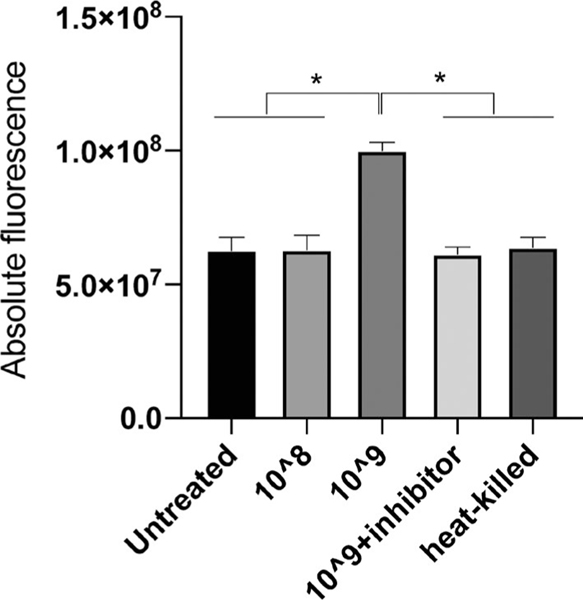
Significantly, more FITC dye, reflected by absolute fluorescence signals, was detected in wild-type HGEPs treated with live P.ginigvalis at 109 CFU/ml than in cells treated with P.ginigvalis at 108 CFU/ml, heat-killed P.ginigvalis at 109 CFU/ml, no P.ginigvalis control, or live P.ginigvalis (109 CFU/ml) together with Leupeptin (p < .05) (Figure 5). Permeation to the FITC-dextran fluorescent dye in HGEPs was dose-dependent in the live P.ginigvalis challenge, as higher fluorescence signals from bottom wells in cells treated with 109 CFU/ml P.ginigvalis than treated with 108 CFU/ml P.ginigvalis. We also conducted a cytotoxicity assay on cells treated with P.ginigvalis and found that almost all epithelial cells remained alive for 109 CFU/ ml live P.ginigvalis challenged for 5 h, suggesting that the compromised gingival epithelial barrier upon P.ginigvalis challenged was not due to cell death (Figure S6). Interestingly, heat-killed P.ginigvalis at 109 CFU/ml led to 27% cell death (Figure S6), regardless of their null impacts on epithelial barrier. The addition of cysteine proteases (Leupeptin) into live P.ginigvalis at 109 CFU/ml restored the gingival epithelial barrier function. Therefore, we can provide indirect evidence that P.ginigvalis impairs gingival epithelial barrier function through degrading PKP2, likely by actions of cysteine proteases.
FIGURE 5.
Live P.ginigvalis impairs gingival epithelial barrier function through a cysteine protease-mediated mechanism. In a transwell system, FITC dye was loaded onto the upper wells where confluent HEGPs had been seeded. Those cells were treated with live or heat-killed P.ginigvalis or P.ginigvalis in the presence of Leupeptin, a cysteine protease, for 5 h. We observed significantly more FITC dye migrating to the lower bottom wells from HGEPs treated with live P.ginigvalis at 109 CFU/ml but not P.ginigvalis at 108 CFU/ml, suggesting a dosage response of P.ginigvalis in disruption of epithelial barrier function. Heat-killed P.ginigvalis at 109 CFU/ml or leupeptin (0.25 mM) plus live P.ginigvalis at 109 CFU/ml had no significant impact on the dye exclusion. Error bars represent standard deviations. * indicates p < .05
4 |. DISCUSSION
In this investigation, we unraveled a novel mechanism of P.ginigvalis-mediated PKP2 protein degradation in periodontal disease. The data from clinical samples demonstrate a lower level of epithelial PKP2 mRNA expression in gingival biopsies from subjects with chronic periodontitis. The downregulation of PKP2 in periodontitis implies a weak epithelial barrier at the gingival surface. Three groups of participants from whom gingival biopsies were acquired for ex vivo PKP2 mRNA expression had comparable demographic parameters (gender, ethnicity, and smoking), but not age. Chronic periodontitis subjects are older than the other two groups of participants. Aging has been associated with an overall impaired epithelial barrier function in multiple organs.22 There is limited evidence about the effect of aging on epithelial barrier function. Smoking was controlled among groups, as no significant difference was present for our study participants. Nevertheless, smoking affects gingival epithelium and connective tissues in endothelial vessels and immune infiltrates.23 Smoking also increases gingival epithelium thickness.24 Therefore, it is important to control for smoking among participant groups when studying gingival epithelial barrier function.
Our ex vivo data align with previous studies reporting a dampened expression of different desmosomal components in gingiva with periodontitis. Abe et al.25 reported similar findings that DSG1 and DSC1 mRNA expression levels were reduced in gingival biopsies from periodontitis subjects. Besides desmosome, tight junction structure molecules including occludin, claudin-4 and 15, ZO-1 and 2, and JAM-A were also affected in periodontitis.26,27 In addition, the expression of E-cadherin, an adherens junction component, was significantly decreased for periodontitis gingival biopsies than it was for healthy controls.28–30 Gap junction members Connexin 26 and 43 were reduced in the pathological lining epithelium of periodontal pockets.31 Although it is anticipated that genes of desmosome junctional molecules are downregulated in periodontitis, Gursoy et al.32 reported a higher DSG1 protein expression level from periodontitis biopsies. Both DSG1 and PKP2 are considered multi-functional, and the roles of the desmosomal proteins involved in periodontal disease are complex. Therefore, more data from in vivo and in vitro experiments are needed to delineate their precise activities in periodontitis at each stage.
One major finding of this study is that PKP2 expression is highly susceptible to P.ginigvalis modulation. The interaction between PKP2 and P.ginigvalis was originally identified in a GWAS study, which associated an intronic variant rs6488099 with PKP2 gene locus with a trait characterized by a P.ginigvalis-dominant dysbiosis.13 Without functional genomic analysis, one can only infer that the variant forms of PKP2 may compromise the integrity of this barrier function-associated gene and impair host response to the bacterial insult, which contributes to dysbiosis and periodontal tissue breakdown. However, we are able to provide evidence to show how PKP2 is associated with P.ginigvalis-mediated periodontal disease.33–35
Our data clearly showed that P.ginigvalis decreased PKP2 protein expression, but increased its transcription in vitro. The increased PKP2 mRNA seems to contradict another study published by our group in 2018, in which P.ginigvalis decreased PKP2 mRNA.36 Gingival epithelial cells used in that study came from different donors than the donors in this study. Therefore, the different responses to P.ginigvalis challenge could be explained by different genetic backgrounds of the donor cells. According to a recent study by Chen et al.,37 the mRNA levels of multiple tight junction molecules were also inhibited by a P.ginigvalis challenge. It suggests that future studies utilizing primary cells should take host genetic variabilities into consideration. The increased PKP2 transcription in vitro also contrasts with the decreased ex vivo PKP2 mRNA levels from the periodontitis biopsies. This discrepancy may be explained by noting that the PKP2 transcription was examined in primary gingival epithelial cells upon single periodontal pathogen challenge for 24 h, while the mRNA assayed from biopsies came from a site in close proximity to a sustained plaque biofilm, which reflect a long-term change of PKP2 at the transcriptional level.
The discrepancy between increased PKP2 mRNA and decreased protein elicits an investigation on the potential post-transcriptional modification mechanisms, including mRNA instability, inhibition of protein translation, or protein degradation. P.ginigvalis is known to produce proteases that degrade protein extracellularly, resulting in periodontium destruction.38 In fact, we found that cysteine proteases inhibitors mitigated P.ginigvalis-mediated PKP2 protein reduction. We also ruled out two intracellular degradation pathways: (1) lysosomal degradation, which delivers structural proteins to lysosome for degradation; and (2) proteasome degradation, in which target proteins become ubiquitinated and then are transported to proteasome for degradation.39 By performing a comprehensive profiling of protein degradation pathways, we conclude that P.ginigvalis degrades PKP2 protein through an extracellular cysteine protease-specific mechanism. Cysteine proteinases of P.ginigvalis exist in the form of gingipains and the P.ginigvalis strain A7436 used in this study contains functional gingipains.40 It is unlikely that proteases secreted by P.ginigvalis can cross cell membranes to degrade PKP2 intracellularly. However, the P.ginigvalis A7436 strain used in this report can attach to cell surfaces and invade cells.41,42 Although PKP2 is an intracellular protein, the punctated pattern of PKP2 staining was clearly present on cell surfaces (see Figures 1 and 3), while some fractions still stay inside the nucleus. In our data, the fact that P.ginigvalis cannot completely abrogate PKP2 protein expression suggests that P.ginigvalis decreases PKP2 protein in a fraction that is close to the cell membrane. Both leupeptin and cathepsin B inhibitors are cysteine protease inhibitors, nevertheless demonstrating differential inhibitory effects on DSG1 recovery (see Figure 2E). Houle et al.43 have shown similar findings that leupeptin completely inhibits collagenase proteolytic activity of P. gingivalis, while cathepsin B inhibitor exhibits poor inhibitory effect.
Even when PKP2 was only moderately knocked down in gingival epithelial cells, cell physiology presented drastic changes such as inhibited proliferation and cell spreading/adherence. PKP2 is multi-functional and plays different roles depending on the cell type. PKP2’s role in cardiomyocytes has been studied most widely because a PKP2 exonic mutation is associated with a severe heart condition-arrhythmogenic right ventricular cardiomyopathy that leads to sudden death.9 A defective PKP2 causes desmosome disassembly in cardiomyocytes, compromises calcium cycling and cardiac rhythm, and reduces heart integrity.10,44 In skin keratinocytes, a deficient PKP2 decreases cell proliferation and migration through its interaction with epidermal growth factor receptor.11 Koetsier et al.12 showed that keratinocytes with deficient PKP2 exhibited smaller areas of cell attachment due to compromised focal adhesion. Their findings mirrored what we showed in Figure 4C as PKP2-knocked down cells occupied fewer areas when attached to the surface. The phenotypic changes associated with a deficient PKP2 in gingival epithelial cells will likely lead to a “leaky” epithelial barrier that permits penetration of P.ginigvalis or its metabolites into the sub-epithelial connective tissue compartment and elicits overt inflammation.45,46 Many studies have utilized FITC dye to test epithelial permeability in the presence of P.ginigvalis or other bacteria and showed that periodontal pathogenic bacteria could impair epithelial barrier function.37,47 Even though we could not provide direct in vivo evidence that P.ginigvalis impairs barrier function through degrading PKP2, we present compelling in vitro evidence to support that P.ginigvalis-associated gingival epithelial barrier destruction involves PKP2 degradation by cysteine proteases produced by P.ginigvalis (Figures 2 and 5). Meanwhile, the cytotoxicity assay in Figure S6 shows that heat-killed P.ginigvalis induces more cell death (27%) than live P.ginigvalis at 109 CFU/ml, suggesting that P.ginigvalis-mediated impaired epithelial barrier is not attributed to cell death. This result is in accordance with findings in Bugueno et al.48 In this study, heat-killed P.ginigvalis increases more apoptosis than live P.ginigvalis and live P.ginigvalis actually inhibits oral epithelial cells apoptosis. Although some studies show that live P.ginigvalis leads to cell death,49 this discrepancy may be explained by different bacterial strains, length of exposure, cell source, and cell death detection methods. Another study by Bruewer et al. shows that proinflammatory cytokines impair epithelial barrier in an apoptosis-independent mechanism, as inhibiting apoptosis does not restore barrier function. Therefore, our findings support that P.ginigvalis-mediated impaired barrier function is not associated with apoptosis/cell death.50
In summary, we propose a novel mechanism by which P.ginigvalis impairs gingival epithelial barrier function by promoting PKP2 degradation. Future functional genomic studies of the PKP2 gene will provide more insights into host-mediated periodontal dysbiosis.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Mr. Michael Tilley (Scientific Writer and Editor) of Iowa Institute for Oral Health Research for his help on editing this manuscript. This work was part of Ning Yu’s PhD thesis 2015 under the mentorship of the late Professor Dr. Steven Offenbacher, who sadly passed away during the manuscript preparation. We would like to thank the PhD committee members (Drs. Zhi Liu, Silvana Barros, Eric Everett, Asma Khan) for instructions. Both Yu and Zhang were supported by NIDCR R90DE022527 when the work was conducted. Zhang is currently supported by NIDCR R00DE027086. Dr. Offenbacher’s guidance and insightful discussion on this project and his inspiration on our scientific careers will be remembered forever.
Funding information
SZ is sponsored by an NIDCR R00DE027086 grant
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section.
CONFLICT OF INTEREST
All authors declare no conflicts of interest with respect to the authorship and/or publication of this article.
REFERENCES
- 1.Hatakeyama S, Yaegashi T, Oikawa Y, et al. Expression pattern of adhesion molecules in junctional epithelium differs from that in other gingival epithelia. J Periodontal Res. 2006;41(4):322–328. [DOI] [PubMed] [Google Scholar]
- 2.Green KJ, Gaudry CA. Are desmosomes more than tethers for intermediate filaments? Nat Rev Mol Cell Biol. 2000;1(3):208–216. [DOI] [PubMed] [Google Scholar]
- 3.Garrod D, Chidgey M. Desmosome structure, composition and function. Biochem Biophys Acta. 2008;1778(3):572–5 87. [DOI] [PubMed] [Google Scholar]
- 4.Kasperkiewicz M, Ellebrecht CT, Takahashi H, et al. Pemphigus. Nat Rev Dis Primers. 2017;3:17026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Payne AS, Hanakawa Y, Amagai M, Stanley JR. Desmosomes and disease: pemphigus and bullous impetigo. Curr Opin Cell Biol. 2004;16(5):536–543. [DOI] [PubMed] [Google Scholar]
- 6.Amagai M, Matsuyoshi N, Wang ZH, Andl C, Stanley JR. Toxin in bullous impetigo and staphylococcal scalded-skin syndrome targets desmoglein 1. Nat Med. 2000;6(11):1275–1277. [DOI] [PubMed] [Google Scholar]
- 7.Hanakawa Y, Schechter NM, Lin C, Nishifuji K, Amagai M, Stanley JR. Enzymatic and molecular characteristics of the efficiency and specificity of exfoliative toxin cleavage of desmoglein 1. J Biol Chem. 2004;279(7):5268–5277. [DOI] [PubMed] [Google Scholar]
- 8.Sumitomo T, Mori Y, Nakamura Y, et al. Streptococcal cysteine protease-mediated cleavage of desmogleins is involved in the pathogenesis of cutaneous infection. Front Cell Infect Microbiol. 2018;8:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Tintelen JP, Entius MM, Bhuiyan ZA, et al. Plakophilin-2 mutations are the major determinant of familial arrhythmogenic right ventricular dysplasia/cardiomyopathy. Circulation. 2006;113(13):1650–1658. [DOI] [PubMed] [Google Scholar]
- 10.Grossmann KS, Grund C, Huelsken J, et al. Requirement of plakophilin 2 for heart morphogenesis and cardiac junction formation. J Cell Biol. 2004;167(1):149–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arimoto K, Burkart C, Yan M, Ran D, Weng S, Zhang DE. Plakophilin-2 promotes tumor development by enhancing ligand-dependent and-independent epidermal growth factor receptor dimerization and activation. Mol Cell Biol. 2014;34(20):3843–3854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koetsier JL, Amargo EV, Todorovic V, Green KJ, Godsel LM. Plakophilin 2 affects cell migration by modulating focal adhesion dynamics and integrin protein expression. J Invest Dermatol. 2014;134(1):112–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Offenbacher S, Divaris K, Barros SP, et al. Genome-wide association study of biologically-informed periodontal complex traits offers novel insights into the genetic basis of periodontal disease. Hum Mol Genet. 2016;25(10):2113–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Offenbacher S, Barros SP, Paquette DW, et al. Gingival transcriptome patterns during induction and resolution of experimental gingivitis in humans. J Periodontol. 2009;80(12):1963–1982. [DOI] [PubMed] [Google Scholar]
- 15.Loe H, Theilade E, Jensen SB. Experimental gingivitis in man. J Periodontol. 1965;36:177–187. [DOI] [PubMed] [Google Scholar]
- 16.Armitage GC. Development of a classification system for periodontal diseases and conditions. Ann Periodontol. 1999;4(1):1–6. [DOI] [PubMed] [Google Scholar]
- 17.Zhang S, Barros SP, Niculescu MD, Moretti AJ, Preisser JS, Offenbacher S. Alteration of PTGS2 promoter methylation in chronic periodontitis. J Dent Res. 2010;89(2):133–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Divaris K, Monda KL, North KE, et al. Exploring the genetic basis of chronic periodontitis: a genome-wide association study. Hum Mol Genet. 2013;22(11):2312–2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanakawa Y, Selwood T, Woo D, Lin C, Schechter NM, Stanley JR. Calcium-dependent conformation of desmoglein 1 is required for its cleavage by exfoliative toxin. J Invest Dermatol. 2003;121(2):383–389. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. [DOI] [PubMed] [Google Scholar]
- 21.Zhang S, Yu N, Arce RM. Periodontal inflammation: integrating genes and dysbiosis. Periodontol 2000. 2020;82(1):129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parrish AR. The impact of aging on epithelial barriers. Tissue Barriers. 2017;5(4):e1343172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jalayer Naderi N, Semyari H, Elahinia Z. The impact of smoking on gingiva: a histopathological study. Iran J Pathol. 2015;10(3):214–220. [PMC free article] [PubMed] [Google Scholar]
- 24.Villar CC, de Lima AF. Smoking influences on the thickness of marginal gingival epithelium. Pesqui Odontol Bras. 2003;17(1):41–45. [DOI] [PubMed] [Google Scholar]
- 25.Abe D, Kubota T, Morozumi T, et al. Altered gene expression in leukocyte transendothelial migration and cell communication pathways in periodontitis-affected gingival tissues. J Periodontal Res. 2011;46(3):345–353. [DOI] [PubMed] [Google Scholar]
- 26.Ye P, Yu H, Simonian M, Hunter N. Expression patterns of tight junction components induced by CD24 in an oral epithelial cell-culture model correlated to affected periodontal tissues. J Periodontal Res. 2014;49(2):253–259. [DOI] [PubMed] [Google Scholar]
- 27.Choi YS, Kim YC, Ji S, Choi Y. Increased bacterial invasion and differential expression of tight-j unction proteins, growth factors, and growth factor receptors in periodontal lesions. J Periodontol. 2014;85(8):e313–322. [DOI] [PubMed] [Google Scholar]
- 28.Arun R, Hemalatha R, Arun KV, Kumar T. E-cadherin and CD1a expression in gingival epithelium in periodontal health, disease and post-treatment. Indian J Dent Res. 2010;21(3):396–4 01. [DOI] [PubMed] [Google Scholar]
- 29.Loo WT, Jin L, Cheung MN, Wang M, Chow LW. Epigenetic change in E-cadherin and COX-2 to predict chronic periodontitis. J Transl Med. 2010;8:110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagarakanti S, Ramya S, Babu P, Arun KV, Sudarsan S. Differential expression of E-cadherin and cytokeratin 19 and net proliferative rate of gingival keratinocytes in oral epithelium in periodontal health and disease. J Periodontol. 2007;78(11):2197–2202. [DOI] [PubMed] [Google Scholar]
- 31.Ye P, Chapple CC, Kumar RK, Hunter N. Expression patterns of E-cadherin, involucrin, and connexin gap junction proteins in the lining epithelia of inflamed gingiva. J Pathol. 2000;192(1):58–66. [DOI] [PubMed] [Google Scholar]
- 32.Gursoy UK, Zeidan-Chulia F, Yilmaz D, et al. Analyses of gingival adhesion molecules in periodontitis: theoretical in silico, comparative in vivo, and explanatory in vitro models. J Periodontol. 2016;87(2):193–202. [DOI] [PubMed] [Google Scholar]
- 33.Offenbacher S, Jiao Y, Kim SJ, et al. GWAS for Interleukin-1beta levels in gingival crevicular fluid identifies IL37 variants in periodontal inflammation. Nat Commun. 2018;9(1):3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang S, Divaris K, Moss K, et al. The novel ASIC2 locus is associated with severe gingival inflammation. JDR Clin Trans Res. 2016;1(2):163–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marchesan JT, Jiao Y, Moss K, et al. Common polymorphisms in IFI16 and AIM2 genes are associated with periodontal disease. J Periodontol. 2017;88(7):663–672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barros SP, Hefni E, Nepomuceno R, Offenbacher S, North K. Targeting epigenetic mechanisms in periodontal diseases. Periodontol 2000. 2018;78(1):174–184. [DOI] [PubMed] [Google Scholar]
- 37.Chen W, Alshaikh A, Kim S, et al. Porphyromonas gingivalis impairs oral epithelial barrier through targeting GRHL2. J Dent Res. 2019;98(10):1150–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Grenier D, La VD. Proteases of Porphyromonas gingivalis as important virulence factors in periodontal disease and potential targets for plant-derived compounds: a review article. Curr Drug Targets. 2011;12(3):322–331. [DOI] [PubMed] [Google Scholar]
- 39.Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17(7):1807–1819. [DOI] [PubMed] [Google Scholar]
- 40.Genco CA, Odusanya BM, Potempa J, Mikolajczyk-Pawlinska J, Travis J. A peptide domain on gingipain R which confers immunity against Porphyromonas gingivalis infection in mice. Infect Immun. 1998;66(9):4108–4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deshpande RG, Khan MB, Genco CA. Invasion of aortic and heart endothelial cells by Porphyromonas gingivalis. Infect Immun. 1998;66(11):5337–5343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khlgatian M, Nassar H, Chou HH, Gibson FC 3rd, Genco CA. Fimbria-dependent activation of cell adhesion molecule expression in Porphyromonas gingivalis-infected endothelial cells. Infect Immun. 2002;70(1):257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houle MA, Grenier D, Plamondon P, Nakayama K. The collagenase activity of Porphyromonas gingivalis is due to Arg-gingipain. FEMS Microbiol Lett. 2003;221(2):181–185. [DOI] [PubMed] [Google Scholar]
- 44.Cerrone M, Montnach J, Lin X, et al. Plakophilin-2 is required for transcription of genes that control calcium cycling and cardiac rhythm. Nat Commun. 2017;8(1):106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romanowski AW, Squier CA, Lesch CA. Permeability of rodent junctional epithelium to exogenous protein. J Periodontal Res. 1988;23(2):81–86. [DOI] [PubMed] [Google Scholar]
- 46.Squier CA. The permeability of oral mucosa. Crit Rev Oral Biol Med. 1991;2(1):13–32. [DOI] [PubMed] [Google Scholar]
- 47.Li S, Dong G, Moschidis A, et al. P. gingivalis modulates keratinocytes through FOXO transcription factors. PLoS One. 2013;8(11):e78541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bugueno IM, Batool F, Korah L, Benkirane-Jessel N, Huck O. Porphyromonas gingivalis differentially modulates apoptosome apoptotic peptidase activating factor 1 in epithelial cells and fibroblasts. Am J Pathol. 2018;188(2):404–416. [DOI] [PubMed] [Google Scholar]
- 49.Stathopoulou PG, Galicia JC, Benakanakere MR, Garcia CA, Potempa J, Kinane DF. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 2009;9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bruewer M, Luegering A, Kucharzik T, et al. Proinflammatory cytokines disrupt epithelial barrier function by apoptosis-independent mechanisms. J Immunol. 2003;171(11):6164–6172. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.