Abstract
Signal transducer and activator of transcription 1 (STAT1) mediates gene expression in response to cytokines and growth factors. Activation of STAT1 is achieved through its tyrosine phosphorylation, a process that involves Jak tyrosine kinases. Here we show that STAT1, although phosphorylated on Y701, is unable to localize in the nucleus in the absence of Jak1 or Jak1 kinase activity. In contrast, the nuclear accumulation of STAT1 in Tyk2-deficient cells remains intact. Nuclear presence of tyrosine-phosphorylated STAT1 could be restored in Jak1-deficient cells by leptomycin B, an inhibitor of nuclear export. Amino acids 197 to 205 of STAT1 were found to encode a leucine-rich nuclear export signal (NES). An L→A mutation within the NES restored nuclear retention of STAT1 in Jak1-deficient cells. Impaired binding of the transcriptional coactivator CBP to tyrosine-phosphorylated STAT1 derived from Jak1-deficient cells offers a model for the intermolecular regulation of the nuclear export sequence.
Interferons (IFNs) as well as many other cytokines and growth factors mediate their biological effects through the induction of a set of immediate-early response genes (9, 11, 14, 18, 26, 30, 34, 40, 50, 59). This process depends on the activation of a family of SH2 and SH3 domain-containing signal transducers and activators of transcription (STATs) (15, 16, 28, 29, 33, 41). Activation of latent, cytoplasmic, or membrane-associated STAT proteins is accomplished through their tyrosine phosphorylation (6, 15, 28, 29), which in most cases depends on the activity of the Janus protein-tyrosine kinases (Jaks) (5, 7, 23, 24, 36, 48, 54, 55). IFN-γ initiation of STAT1 tyrosine phosphorylation requires the activity of Jak1 and Jak2 (36, 55), whereas IFN-α/β mediates STAT1 activation through the kinases Jak1 and Tyk2 (36, 54). After its tyrosine phosphorylation, STAT1 either homodimerizes or forms heterodimers when STAT2 is activated by IFN-α/β, in order to translocate to the nucleus, where site-specific binding to enhancer elements leads to gene activation (17, 44). Tyrosine phosphorylation of STAT1 is an absolute prerequisite for its nuclear translocation and its ability to bind DNA (45, 46). We have recently shown that the SH2 domain of STAT1 is essential for its nuclear translocation (35), demonstrating the importance of dimerization over tyrosine phosphorylation alone in the process of nuclear import. Furthermore, we found the SH2 domain of STAT1 to be required for its activation by IFN-γ but dispensable for STAT1 activation by IFN-α/β (35).
Regulation of nuclear localization occurs at the level of both nuclear import and nuclear export (53). Transport across the nuclear pore is usually an energy-dependent process, and the fact that the process is saturable indicates the involvement of receptors which recognize signal sequences in the target proteins (38). Two groups of transport signals for nuclear import have been well characterized. The basic nuclear localization signal (NLS) is recognized by the α-subunit of the importin complex and is found in many nuclear proteins (8). The M9 domain, which is found in hnRNP A1 and related proteins, mediates nuclear import through recognition by transportin, an importin β-related protein (39). Analogous to the events characterizing nuclear import, nucleocytoplasmic transport also involves the recognition of signal sequences (53). In the recent past, significant progress was made in the understanding of the process of nuclear export. The existence of leucine-rich nuclear export signals (NES) was defined (12, 56); subsequently, the nuclear protein Crm1 was identified as the receptor for NES sequences, which is responsible for the GTP-dependent nuclear export of NES-containing proteins (13, 49). The antibiotic leptomycin B (LMB) was characterized as a highly specific inhibitor of Crm1-mediated nuclear export, acting through the disruption of Crm1-NES interaction (58). The subcellular localization of several signaling molecules such as IκB, c-Abl, mitogen-activated protein kinase-activated protein kinase 2, and IRF-3 has been shown to be at least partially controlled through regulated nuclear export (3, 10, 52, 60).
In this study we wanted to address the question of whether tyrosine phosphorylation and dimerization of STAT1 are sufficient for its nuclear accumulation. Sequence analysis of STAT1 does not indicate the presence of domains homologous to nuclear localization signals such as the bipartite NLS or an M9 sequence. However, in the present study we demonstrate that STAT1 contains a leucine-rich NES sequence and that the nuclear localization of STAT1 after IFN-α/β or epidermal growth factor (EGF) treatment is, at least in part, regulated through inhibition of its nuclear export. The regulation of this process requires the presence and catalytic activity of Jak1 but not Tyk2, since tyrosine-phosphorylated STAT1 translocates to the nucleus in Tyk2−/− but not Jak1−/− cells. The nuclear translocation of tyrosine-phosphorylated STAT1 can be restored in Jak1−/− cells through the addition of LMB, indicating that Jak1 controls the function of the STAT1 NES.
MATERIALS AND METHODS
Cells.
2fTGH, U3SH2, and U6A cells were described previously (32, 37), as were wild-type and Jak1−/− HeLa cells. All cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin (Irvine Scientific).
IFNs and reagents.
IFN-α, IFN-β, and IFN-γ were generous gifts from Hoffman LaRoche, Chiron, and Genentech, respectively. Sodium vanadate (50 mM) and 100 mM hydrogen peroxide were incubated in DMEM without FBS for 15 min prior to addition to cells. LMB (100 nM) was added to cells 90 min prior to stimulation.
Plasmids.
Putative NES-green fluorescent protein (GFP) fusion proteins were generated by inserting hybridized oligonucleotides corresponding to STAT1 amino acids 197 to 205 (5′-GATCTCTGTTACTCAAGAAGATGTATTTA) and 519 to 528 (5′-GATCTCTGAACATGTTGGGAGAGAAGCTTCTT) into the BglII (5′) and SmaI (3′) sites of the pEGFP-C1 vector (Clontech). Full-length STAT1-GFP fusion proteins were constructed by inserting STAT1 cDNA into the BglII and BamHI sites of the pEGFP-C1 vector (Clontech). The NES mutant was generated by site-directed mutagenesis using overlapping oligonucleotides containing the NES mutation.
Transfections.
Cells were seeded onto coverslips in six-well plates and incubated overnight at 37°C. Plasmid DNA (0.4 μg/ml) was transfected using Effectene or Superfect (Qiagen) according to the manufacturer's protocol. Cells were assayed for GFP expression 15 to 24 h after transfection.
Whole-cell extracts and EMSA.
Following treatment, cells were washed with phosphate-buffered saline (PBS) and lysed on the plates with lysis buffer (1 ml) containing 20 mM HEPES (pH 7.4), 1% Triton X-100, 100 mM NaCl, 50 mM NaF, 10 mM β-glycerophosphate, 1 mM sodium vanadate, and 1 mM phenylmethylsulfonyl fluoride (PMSF). Lysates were centrifuged at 13,000 rpm for 5 min, and protein concentration was determined by the Lowry assay (Bio-Rad protein assay). Electrophoretic mobility shift assays (EMSAs) were performed using whole-cell extracts prepared as described above and an end-labeled oligonucleotide corresponding to the GRR sequence found in the promoter sequence of the Fc gamma receptor I (FcγRI) (5′-AATTAGCATGTTTCAAGGATTTGAGATGTATTTCCCA-GAAAAG-3′) as described previously (57).
Immunoprecipitation and immunoblotting.
For coimmunoprecipitation experiments, cells were lysed in lysis buffer (1 ml) containing 100 mM NaCl, 50 mM Tris (pH 7.5), 1 mM EDTA, 0.1% Triton X-100, 10 mM NaF, 1 mM PMSF, and 1 mM vanadate. Lysates were centrifuged at 13,000 rpm for 5 min. Supernatant was collected and cleared with protein G-Sepharose for 30 min. Following the clearing, lysates were incubated with a Crm1 polyclonal antibody for 2 h and protein G-Sepharose for an additional hour. After sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transfer onto a polyvinylidene difluoride (PVDF) membrane, proteins were detected with monoclonal anti-STAT1 antibodies (Transduction Labs). Blots were developed with horseradish peroxidase-conjugated secondary antibodies and enhanced chemiluminescence (Amersham).
GST fusion protein affinity precipitations.
For affinity precipitation experiments, cells were lysed in lysis buffer (0.5 ml) containing 100 mM NaCl, 20 mM HEPES (pH 7.5), 1 mM EDTA, 10% glycerol, 0.1% NP-40, and 1 mM orthovanadate. After centrifugation, an equal volume of lysis buffer without detergent was added. Previously described glutathione S-transferase (GST) fusion proteins were incubated with the cell lysates for 12 h, and bound proteins were resolved by SDS-PAGE and analyzed by Western blot.
Immunofluorescence.
Cells were seeded onto coverslips in six-well plates and incubated overnight at 37°C in DMEM containing 10% FBS. After treatment, coverslips were rinsed with PBS followed by one wash with PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] buffer. Cells were fixed in methanol at room temperature for 6 min, and nuclei were permeabilized by incubating with 0.5% Nonidet P-40–PIPES buffer for 13 min at room temperature. Coverslips were washed three times with PBS, blocked with 10% goat serum for 35 min, and incubated with anti-STAT1 (Transduction Laboratory) for 50 min at room temperature. Cells were rinsed four times for 5 min in PBS prior to incubation with Cy3-conjugated secondary antibody for 40 min at room temperature. After washing, coverslips were mounted onto glass slides in 50% glycerol–PBS.
RNase protection assays.
Total RNA was isolated using Trizol reagent. 32P-labeled antisense riboprobes were generated by in vitro transcription using T7 or SP6 RNA polymerase. Labeled riboprobe and 10 μg of RNA were incubated in hybridization buffer (4:1 formamide and 5× stock; 5× stock was 200 mM PIPES [pH 6.4], 2 M NaCl, 5 mM EDTA) overnight at 56°C prior to digestion with T1 RNase. Protected fragments were separated by electrophoresis on a 4.5% polyacrylamide–urea gel.
RESULTS
Activation of STAT1 by IFN-α/β in Jak−/− and Tyk2−/− cells.
The phosphorylation of STAT1 on Y701 and the subsequent dimerization are obligatory for the nuclear translocation of STAT1. However, it is unclear whether these events are sufficient. It has previously been reported that incubation of cells with a combination of 1 mM hydrogen peroxide and 0.1 mM orthovanadate (H/V) results in ligand-independent activation of STAT proteins (19, 22). This treatment can activate STAT1 in the absence of the Jak tyrosine kinases that are required for STAT activation by ligands (19), indicating that H/V treatment bypasses the need for receptor-mediated signaling. In our hands, exposure to H/V resulted in the tyrosine phosphorylation of STAT1, and this was observed with wild-type 2fTGH cells and with Jak1- and Tyk2-deficient cells (data not shown). However, pretreatment with H/V prevented IFN-α from stimulating the nuclear translocation of STAT1 despite the fact that this costimulation resulted in a further increase in STAT1 tyrosine phosphorylation (data not shown).
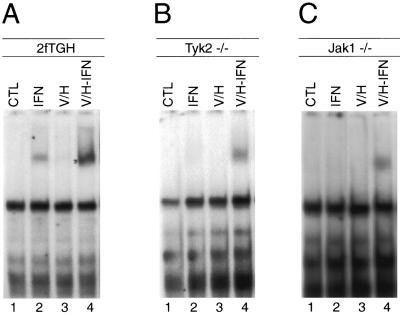
We therefore reduced the concentration of H/V to a level where it did not affect the nuclear translocation of STAT1 after stimulation of cells with IFN-α/β. A significantly lower concentration of H/V (h/v: 10 μM hydrogen peroxide and 5 μM orthovanadate) was found not to interfere with the nuclear translocation of STAT1 after IFN stimulation in wild-type cells (see Fig. 2A, top right). This h/v treatment was also unable to activate STAT1 (Fig. 1A to C, lanes 3), but it promoted the subsequent induction of STAT1 DNA binding by IFN-α/β in both Jak1−/− and Tyk2−/− cells (Fig. 1B and C, lanes 4). Western blot analysis using p(Y701)STAT-specific antibodies confirmed the results of the DNA-binding assay (Fig. 2A). It thus appears that the priming of cells with a subthreshold h/v concentration facilitates activation of STAT1 after IFN-α/β treatment in the absence of Jak1 and Tyk2 while preserving the ligand-dependent nature of the stimulation.
FIG. 2.
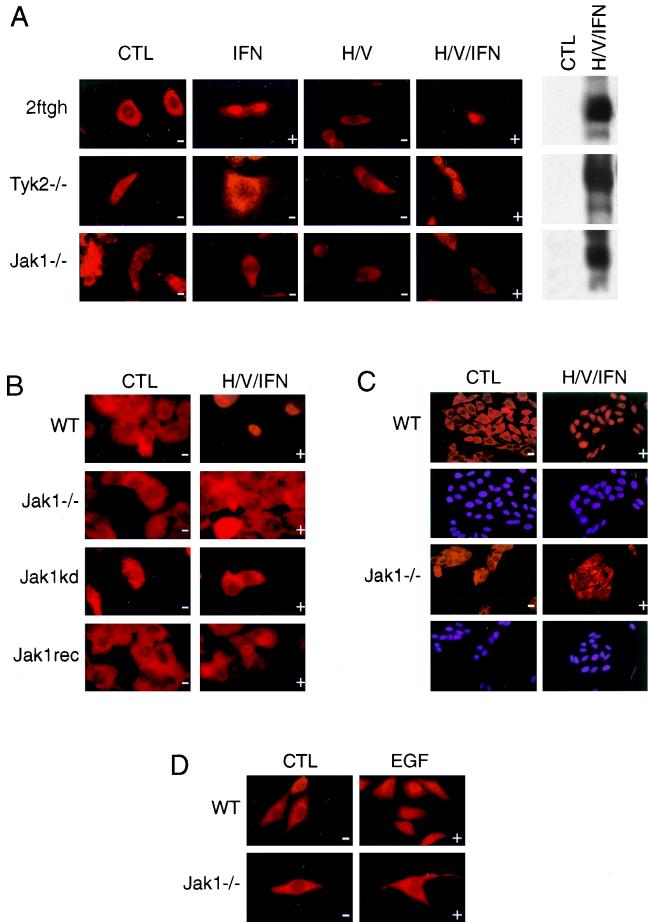
Tyrosine-phosphorylated STAT1 does not accumulate in the nucleus in Jak1−/− cells. (A) Parental 2fTGH, Tyk2−/− U1A, and Jak1−/− U4A cells were treated as in Fig. 1. Subcellular distribution of STAT1 was detected using a monoclonal antibody against STAT1. (B) Jak1−/− U4A cells reconstituted with wild-type (WT) or kinase-inactive Jak1 were treated as in Fig. 1, and subcellular distribution of STAT1 was analyzed. (C and D) Wild-type (WT) and Jak1−/− HeLa cells were stimulated with h/v and IFN-α (C) or EGF (2 ng/ml) (D) for 30 min, and the compartmentalization of STAT1 was analyzed. The + and − signs indicate the presence and absence, respectively, of STAT1 tyrosine phosphorylation after the indicated treatments.
FIG. 1.
Priming with h/v restores IFN-dependent activation of STAT1 in Jak1−/− and Tyk2−/− cells. Parental 2fTGH (A), Tyk2−/− U1A (B), and Jak1−/− U4A (C) cells were either left untreated (lanes 1), stimulated for 30 min with 500 U of IFN-α per ml (lanes 2) or pretreated with h/v for 30 min followed by an additional 30 min of incubation without (lanes 3) or with (lanes 4) IFN-α (500 U/ml). Total cell extracts (CTL) were prepared and subject to EMSA using end-labeled GRR as a probe.
Impaired nuclear translocation of STAT1 in the absence of Jak1 kinase activity.
Our primary goal was to investigate whether the tyrosine phosphorylation of STAT1 was sufficient for its nuclear translocation. We therefore used immunohistochemistry to analyze the subcellular localization of tyrosine-phosphorylated STAT1 in Tyk2−/− and Jak1−/− cells. As shown in Fig. 2A (top panel), IFN-α-activated, tyrosine-phosphorylated STAT1 translocated efficiently into the nucleus in wild-type 2fTGH cells, and h/v priming had no apparent effect on the process. In Tyk2−/− cells, STAT1 remained cytoplasmic after stimulation with IFN-α alone. Interestingly, STAT1 appeared in the nucleus of Tyk2−/− cells when appropriate tyrosine phosphorylation was achieved through h/v priming followed by IFN-α stimulation (Fig. 2A, second panel from top). In contrast, STAT1 was found exclusively in the cytoplasm after the same costimulation was applied in Jak1−/− cells (Fig. 2A, third panel from top). These results suggested that Jak1 but not Tyk2 is required in a function other than tyrosine phosphorylation to facilitate the nuclear accumulation of STAT1. Similarly, STAT1 translocated to the nucleus in Jak2−/− cells but not Jak1−/− cells when IFN-γ was used for stimulation (data not shown).
To determine if the enzymatic activity of Jak1 was required for tyrosine-phosphorylated STAT1 to accumulate in the nucleus, we performed the h/v priming experiment with Jak1−/− cells reconstituted with a kinase-inactive form of Jak1. As seen in the Jak1−/− cells, h/v and IFN-α costimulation resulted in the tyrosine phosphorylation and DNA binding of STAT1 in these cells (data not shown); however, the kinase-inactive form of Jak1 was unable to promote the nuclear presence of STAT1 (Fig. 2B). In contrast, Jak1−/− cells reconstituted with wild-type Jak1 supported IFN-α/β-stimulated STAT1 nuclear localization (Fig. 2B, bottom panel), demonstrating that the lack of STAT1 nuclear localization seen in Jak1−/− cells can indeed be attributed to the lack of Jak1.
In order to exclude that our findings were a peculiarity of the 2fTGH cells series, we repeated the experiments in wild-type and Jak1-deficient HeLa cells. Indeed, the dependence of STAT1 nuclear localization on Jak1 after IFN-α stimulation was also observed in these cells (Fig. 2C). We next wanted to explore whether our findings were restricted to stimulation by IFN-α, or if they were of a more general nature. We were particularly interested in the subcellular distribution of STAT1 after EGF stimulation. It had been shown by several laboratories that although Jak1 is activated in response to EGF, tyrosine phosphorylation of STAT1 after EGF treatment occurs independent of Jak1; rather, it requires the intrinsic kinase activity of the EGF receptor (7, 47). This unique feature allowed us to assay for the subcellular distribution of tyrosine-phosphorylated STAT1 following EGF stimulation without the need for h/v priming.
Wild-type and Jak1−/− HeLa cells were stimulated with EGF for 30 min, and the localization of STAT1 was analyzed. As shown in Fig. 2D, EGF was only able to target STAT1 into the nucleus in wild-type HeLa cells (upper panel) but not in the Jak1−/− mutant (lower panel). Thus, it appears that Jak1 is dispensable for mediating the tyrosine phosphorylation of STAT1 in response to EGF (7, 47) but, as in the case with IFN-α-activated STAT1, is essential for subsequent accumulation of the phosphorylated protein in the nucleus. It is noteworthy that EGF is still able to induce transcription of c-fos in Jak1−/− cells; however, mutation analysis of the c-fos promoter demonstrated that this event does not require the presence of the STAT-responsive element SIE (31).
Inhibition of nuclear export restores nuclear accumulation of tyrosine-phosphorylated STAT1 in Jak1−/− cells.
It becomes evident from the results described above that, although necessary, tyrosine phosphorylation of STAT1 alone is insufficient for its nuclear presence. As mentioned in the introduction, STAT1 does not appear to contain any domains resembling known conserved NLS sequences. However, analysis of the STAT1 amino acid sequence revealed three leucine-rich domains that have homology to the consensus signal sequence for nuclear export (Table 1). We therefore decided to test whether the highly specific nuclear export inhibitor LMB would influence the subcellular distribution of unphosphorylated or tyrosine-phosphorylated STAT1.
TABLE 1.
Comparison of NES consensus sequences with leucine-rich domains of STAT1
| Protein | Sequence |
|---|---|
| HIV-1 REV | LPP LERLTL |
| PKI | LALKLAGLDI |
| IκB | LQQQLGQLTL |
| MEK1 | LQKKLEELEL |
| c-Abl | LESNLRELQI |
| IRF-3 | LDELLGNMVL |
| Consensus | LX1–3LX2–3MLXIL |
| STAT1 amino acids | |
| 197–205 | LL LKKMYL? |
| 349–358 | LRL LVKLQEL? |
| 519–528 | LNM LGEKL L? |
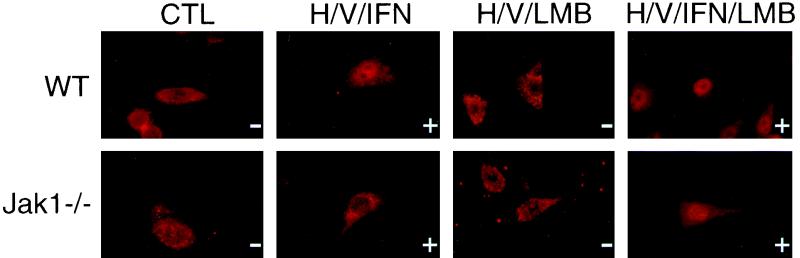
Cells were incubated with LMB for 30 min simultaneously with h/v priming. In wild-type 2fTGH cells, treatment with h/v and LMB alone had no apparent effect on localization of the unphosphorylated STAT1 protein (Fig. 3, upper panel). Even exposure to LMB for 8 h was unable to target unphosphorylated STAT1 to the nucleus. A slight enhancement of nuclear staining was observed after IFN-α stimulation when the combined treatment with h/v and LMB was applied compared to h/v priming alone (Fig. 3, top panel). Similar to the wild-type cells, LMB alone or in combination with h/v cotreatment had no effect in Jak1-deficient cells (Fig. 3, lower panel). However, LMB was able to restore nuclear localization of tyrosine-phosphorylated STAT1 after IFN-α stimulation of h/v-primed Jak1−/− cells (Fig. 3, bottom right). Thus, our results indicate that the kinase activity of Jak1 has an inhibitory effect on the nuclear export of STAT1.
FIG. 3.
Nuclear accumulation of tyrosine-phosphorylated STAT1 in Jak1−/− cells after addition of LMB. Parental 2fTGH and Jak1−/− U4A cells were primed with h/v as in Fig. 1 without or with prior addition (90 min) of 100 nM LMB. Cells were stimulated with IFN-α (500 U/ml) for 30 min and STAT1 localization was analyzed as in Fig. 2. The + and − signs indicate the presence and absence, respectively, of STAT1 tyrosine phosphorylation after the indicated treatments.
Since we had previously shown that an SH2 domain-mutated, tyrosine-phosphorylated STAT1 fails to display nuclear presence (35), we also tested the effects of LMB treatment on STAT1−/− cells reconstituted with the SH2 domain-mutated STAT1. Interestingly, LMB was unable to facilitate the nuclear translocation of the SH2-mutated STAT1 (data not shown), suggesting that this STAT mutant is impaired in LMB-insensitive nuclear import, whereas the absence of Jak1 appears to affect LMB-sensitive nuclear export.
Amino acids 197 to 205 of STAT1 encode a functional NES.
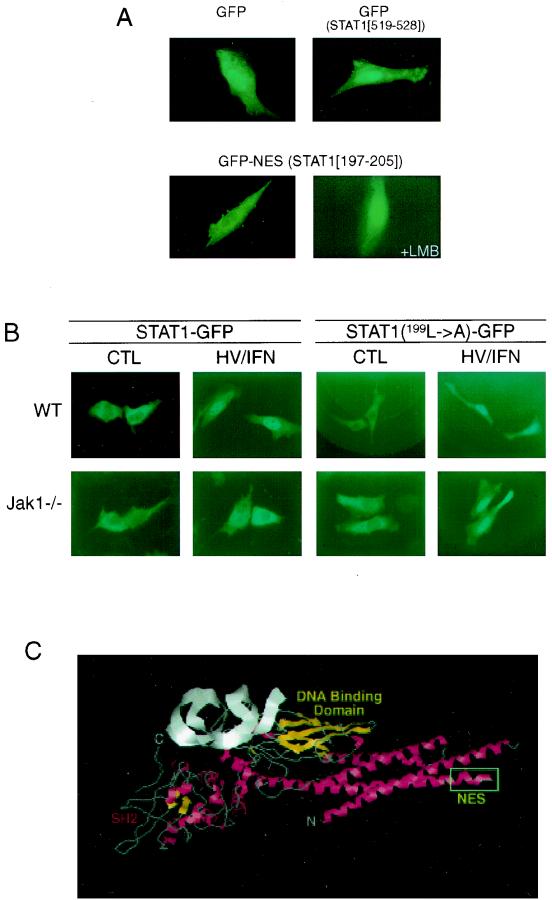
In order to investigate which, if any, of the leucine-rich domains of STAT1 function as an NES, we decided to generate fusions with GFP. GFP displays a tendency to accumulate in the nucleus when expressed ectopically. It has been shown previously that the coupling of a functional NES onto GFP causes the resulting fusion protein to be excluded from the nucleus (49). Therefore, we created individual fusion proteins of GFP and the three leucine-rich sequences of STAT1 depicted in Table 1. The constructs were transiently transfected into primary human fibroblasts, and the localization of the fusion proteins was visualized after 15 h. Parental GFP was found to accumulate in the nucleus (Fig. 4A, upper left panel), as predicted, as did the GFP fusion with the leucine-rich domain STAT1519–528 (upper right panel) and STAT1349–358 (data not shown). However, the coupling of STAT1197–205 to GFP caused the exclusion of the resulting fusion protein from the nucleus (Fig. 4A, lower left panel).
FIG. 4.
Amino acids 197 to 205 of STAT1 encode a functional NES. (A) Primary human foreskin fibroblasts were transiently transfected with either parental GFP plasmid (top left), GFP-STAT1519–528 (top right), or GFP-STAT1197–205 (bottom panels). In addition, cells expressing GFP-STAT1197–205 were also incubated with 100 nM LMB for 90 min prior to visualization of the fusion proteins (bottom right). (B) Wild-type (WT) and Jak1−/− HeLa cells were transiently transfected with either STAT1-GFP or STAT1 (L199→A)-GFP, and localization of the fusion proteins after stimulation with h/v and IFN-β (1,000 U/ml) was determined. (C) The crystal structure of tyrosine-phosphorylated STAT1 bound to DNA was downloaded from the Protein Data Bank of the Brookhaven National Laboratory (1, 4), and the positions of individual amino acids were identified using Chemscape Chime. The highly exposed NES (boxed) is located at the start of the second α-helix.
One possible concern was that not the function of an NES but unpredictable steric hindrance prevented STAT1197–205-GFP from entering the nucleus. We therefore exposed the cells expressing STAT1197–205-GFP to LMB for 60 min prior to determining the localization of the fusion protein. As would be expected for an NES-mediated nuclear exclusion, STAT1197–205- GFP was found in the nucleus in LMB-treated cells (Fig. 4A, lower right panel). These results clearly demonstrate that the leucine-rich domain encompassing amino acids 197 to 205 encodes the functional NES of STAT1. A critical regulatory event in nucleocytoplasmic transport is the binding of a nuclear export receptor such as Crm1 to the leucine-rich NES sequences of target proteins. Indeed, coimmunoprecipitation experiments revealed a specific interaction of STAT1 with Crm1 (data not shown), providing further evidence that the leucine-rich region between amino acids 197 and 205 of STAT1 encodes a functional NES sequence.
Mutation of STAT1 NES restores its nuclear accumulation in Jak1−/− cells.
To further demonstrate that the above-identified NES is the target of regulation by Jak1, we generated GFP fusion proteins containing either wild-type STAT1 or STAT1 bearing a single point mutation within the NES (L199A) that had previously been shown in other proteins to abrogate NES function (52, 56). When transfected into wild-type HeLa cells, both wild-type and NES mutant STAT1 translocated to the nucleus upon IFN stimulation (Fig. 4B, top panels). In striking contrast, only the NES-mutated STAT1 could be detected in the nucleus of Jak1−/− HeLa cells upon h/v and IFN-α costimulation (Fig. 4B, bottom right panels), whereas wild-type STAT1 remained restricted to the cytoplasm (Fig. 4B, bottom left panels). These results clearly demonstrate that Jak1 mediates additional events beyond STAT1 tyrosine phosphorylation that exert control over STAT1 nuclear export.
Crystal structure of STAT1 reveals highly exposed position of the NES.
In order for STAT1 amino acids 197 to 205 to interact with Crm1, these residues should be readily accessible. We therefore decided to determine the position of the NES within STAT1 by means of the crystal structure coordinates recently posted with the Protein Data Bank of the Brookhaven National Laboratory (1, 4). Indeed, the crystal structure of tyrosine-phosphorylated STAT1 bound to DNA reveals a highly exposed position of the NES at the start of the second α-helix (Fig. 4C). This isolated position of the strongly hydrophobic NES argues against intramolecular regulation based on a conformational change, but rather supports the notion that access to the NES may be governed by other proteins capable of interacting with STAT1 in the nucleus.
STAT1 binding to CBP is impaired in Jak1−/− cells.
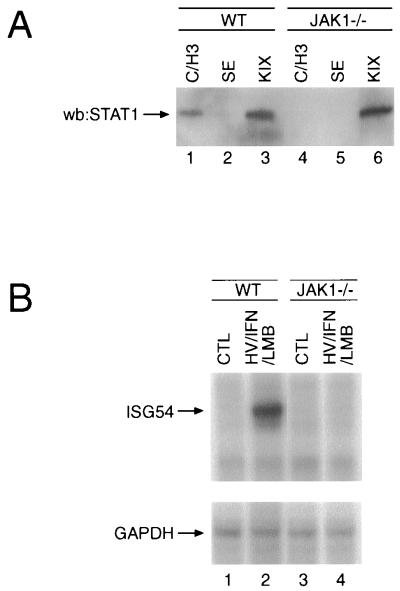
The hypothesis of intermolecular regulation of access to the NES led us to investigate the previously reported association of STAT1 with the transcriptional coactivator CBP with respect to its dependence on Jak1. Several regions of CBP have been shown to interact with STAT1 independently of its tyrosine phosphorylation (21, 27, 61). We therefore tested the ability of these CBP domains to bind STAT1 in lysates derived from either wild-type or Jak1−/− cells. GST-CBP fusion proteins bound to glutathione-agarose were incubated with lysates from h/v-primed, IFN-α-stimulated wild-type or Jak1-deficient HeLa cells, and the resulting affinity precipitates were analyzed for the presence of STAT1. A fusion protein representing the KIX domain (CBP residues 451 to 720), which has been shown to interact with STAT1, CREB, and c-Jun (21), was able to isolate STAT1 from lysates of either cell type (Fig. 5A, lanes 1 and 4). In contrast, the fusion protein encompassing the C/H3 region (CBP residues 1455 to 1891), capable of association with E1A, STAT1, or c-Fos (21), was only able to sequester STAT1 in lysates derived from wild-type cells, but failed to bind STAT1 in lysates lacking Jak1 (lanes 3 and 6). A fusion protein to the SE domain (CBP residues 1492 to 2441), which reportedly does not interact with STAT1 (21), was used as a negative control (lanes 2 and 5). Reprobing of the blot with GST antiserum verified that similar amounts of the various fusion proteins were present in each sample; in addition, STAT1 phosphorylation on Y701 and S727 was verified with phosphorylated-STAT1-specific antiserum (data not shown).
FIG. 5.
Impaired interaction of STAT1 and CBP and STAT1-mediated gene induction in Jak1−/− cells. (A) Wild-type (WT) and Jak1−/− HeLa cells were stimulated with h/v and IFN-α, and the lysates were incubated with glutathione-agarose-conjugated GST fusion proteins representing the KIX domain (lanes 1 and 4), the SE region (lanes 2 and 5), or the C/H3 domain (lanes 3 and 6) of CBP. Affinity-precipitated proteins were resolved by SDS-PAGE, transferred to polyvinylidene difluoride, and probed for the presence of STAT1. (B) Wild-type and Jak1−/− HeLa cells were stimulated with h/v and IFN-α in the presence of LMB, and ISG54 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA levels were analyzed by RNase protection assay.
It would be expected that impaired association of STAT1 with CBP leads to a lack of IFN-induced gene transcription. To test this hypothesis, wild-type and Jak1−/− HeLa cells were exposed to h/v prior to IFN-β stimulation, and the nuclear presence of STAT1 was enforced by the addition of LMB. Indeed, whereas ISG54 induction was readily detectable in wild-type HeLa cells (Fig. 5B, lanes 1 and 2), no ISG54 mRNA could be detected in Jak1−/− cells (Fig. 5B, lanes 3 and 4), despite the nuclear presence of appropriately tyrosine- and serine-phosphorylated STAT1. Furthermore, identical results were obtained when a probe corresponding to IRF1 was used in the RNase protection assays (data not shown), demonstrating that genes that are induced by either STAT1 homodimers or STAT1-STAT2 heterodimers are both affected by the impaired STAT1-CBP interaction.
DISCUSSION
Nuclear compartmentalization allows eukaryotes an additional level of gene regulation, whereby transcriptional regulators must be allowed access to their regulatory elements. Previous work demonstrated STAT1 tyrosine phosphorylation to be necessary for ligand-induced nuclear localization (45, 46). Using a STAT1 SH2 domain mutant incapable of dimerization, we have previously demonstrated that tyrosine phosphorylation is necessary but not sufficient for nuclear localization of STAT1 (35).
Although much research has focused on the activation of STAT1, little is known about STAT1 nuclear translocation. Sekimoto et al. have identified the small GTPase Ran/TC4 as a member of the nuclear import machinery that is required for nuclear import of IFN-γ-activated STAT1 (43). GTP hydrolysis by Ran is a critical step in the movement of import receptors and their cargo into the nucleus (38). Karyophilic protein cargo binds to the import receptors via an NLS. The best-characterized NLS consists of a highly basic amino acid stretch. However, no such NLS has been identified in STAT1. Interestingly, the basic NLS contained in the simian virus 40 T antigen and STAT1 can noncompetitively bind the NPI-1 import complex, indicating that each cargo binds to a distinct domain of NPI-1 and suggesting the existence of a unique, uncharacterized NLS in STAT1 (42).
Nuclear localization can also be regulated by nuclear export. A leucine-rich domain was first identified as a nuclear export sequence in the HIV-1 Rev protein (12) and protein kinase I (56). Since then, additional polypeptides such as IκB, MEK1, c-Abl, and p53 (3, 10, 51, 52, 60) have been classified as NES-containing proteins. Like nuclear import, export also is an energy-dependent process involving the GTPase activity of Ran. The chromosome maintenance (Crm1) gene was identified as the NES receptor that facilitated export (13, 49). Furthermore, Crm1 was shown to be the target of the drug LMB, which disrupts Crm1 interaction with the leucine-rich NES and thereby traps Crm1 cargo in the nucleus (58).
In this study we demonstrate that the nuclear localization of STAT1 after its tyrosine phosphorylation and dimerization is also intrinsically controlled at the level of nuclear export. Our results show that the IFN receptor-associated tyrosine kinase Jak1 but not Tyk2 is required to achieve IFN-induced nuclear accumulation of STAT1. As such, STAT1 is exclusively found in the cytoplasm in Jak1-deficient cells even when it is appropriately phosphorylated on Y701 and S727. Importantly, a kinase-dead Jak1 was unable to restore IFN-induced STAT1 nuclear localization, but Jak1−/− cells reconstituted with wild-type Jak1 were able to accumulate nuclear STAT1, indicating a distinct role for Jak1 kinase activity in STAT1 nuclear localization. Jak1 was also required for STAT1 nuclear localization mediated by EGF stimulation, where the intrinsic kinase activity of the EGF receptor rather than Jak1 is required for STAT1 tyrosine phosphorylation (7, 47). Interestingly, STAT5 nuclear localization has also been shown to be uncoupled with its tyrosine phosphorylation. Two separate residues of the prolactin receptor have been shown to be necessary for tyrosine phosphorylation and nuclear localization of STAT5 (2). Additionally, Src activation led to the tyrosine phosphorylation of both STAT5a and -b but nuclear localization of only STAT5b, demonstrating the need for an additional signal for STAT nuclear localization (25).
Sequence analysis of STAT1 indicated the existence of several potential leucine-rich NES. Fusion of STAT1 amino acids 197 to 205 to GFP led to an exclusively cytoplasmic fusion protein, whereas GFP alone or GFP fused to the other putative export sequences yielded a predominantly nucleus-localized protein. Furthermore, the addition of LMB retained the STAT1 NES-GFP fusion in the nucleus, indicating a disruption in binding between the short amino acid stretch and Crm1. Additionally, an interaction between Crm1 and the full-length STAT1 protein was detected in coimmunoprecipitation experiments, and analysis of the crystal structure of STAT1 revealed a readily accessible position of the NES within the protein.
Jak1-mediated inhibition of STAT1 nuclear export is necessary in order for STAT1 to accumulate in the nucleus and fulfill its function as a transcriptional activator. The Jak1−/− cell deficiency of nucleus-localized, tyrosine-phosphorylated STAT1 was recovered when cells were supplemented with LMB, implying a role for Jak1 in preventing the nuclear export of the active STAT1 species. Indeed, a single amino acid mutation within the STAT1 NES (L199A) was able to retain STAT1 in the nucleus of Jak1-deficient cells, demonstrating that the lack of nuclear STAT1 in Jak1−/− cells was due to nuclear export. The presence of the hydrophobic leucine-rich NES region of STAT1 within the coiled-coil domain of STAT1, a region shown by crystal structure to be quite exposed, is suggestive of continuous protein interaction. As such, the association of the C/H3 region with STAT1 in wild-type but not in Jak1-deficient cells offers a model for the functional regulation of the NES by Jak1, whereby Jak1−/− cells are unable to recruit CBP and other CBP/STAT-interacting proteins. An example of such a potential protein is Nmi, recently shown to interact with the STAT5 coiled-coil domain and to interact with STAT1. Nmi was shown to enhance the association of STAT1 with CBP and enhance IFN-γ-induced transcription (62). Without Jak1, STAT1 may be unable to recruit such proteins, disrupting the transcriptional protein scaffold and thus allowing unrestricted access of Crm1 to the NES, resulting in uninhibited nuclear export of STAT1. Intriguingly, nuclear localization is not sufficient for phosphorylated STAT1 to induce transcription, as h/v and IFN costimulation does not induce the synthesis of ISG54 or IRF1 in Jak1−/− cells even in the presence of LMB. This observation suggests that Jak1 supplies an additional signal that both enhances STAT1-mediated transcription and maintains STAT1 nuclear presence. Interestingly, we had observed that the Ser/Thr phosphatase inhibitor okadaic acid can substitute for LMB in restoring STAT1 nuclear localization in Jak1−/− cells. It is therefore interesting to speculate that Jak1 might regulate Ser/Thr phosphorylation, which is necessary for the intermolecular interactions between STAT1 and the transcriptional scaffold as well as for STAT1 nuclear retention.
The presence of a functional NES suggests that STAT1 is recycled back to the nucleus after its dephosphorylation. In fact, pulse-chase experiments have detected 35S-labeled STAT1 in the cytoplasm after nuclear residence (20). Importantly, prolonged exposure to LMB alone is insufficient to promote STAT1 nuclear localization in the absence of tyrosine phosphorylation. This demonstrates that STAT1 is not continuously shuttling between the cytoplasmic and nuclear compartments and represents further evidence that the nuclear presence of STAT1 is regulated through both nuclear import and nuclear export.
We describe here a novel aspect of STAT1 regulation that relies on the kinase activity of Jak1 for nuclear retention in addition to tyrosine phosphorylation. We have identified amino acids 197 to 205 as the NES that accounts for Jak1-regulated nuclear export and whose function is abolished upon the addition of LMB. Limited interaction of STAT1 with transcriptional coactivator complexes in Jak1−/− cells offers an intriguing possibility of NES access regulation through intermolecular masking.
ACKNOWLEDGMENTS
We thank G. Grosveld for the generous gift of Crm1 antiserum and G. Stark and R. Flavell for the mutant cell lines. CBP fusion constructs were kindly made available by C. Glass. IFN-α, IFN-β, and IFN-γ were kind gifts from Hoffman-LaRoche, Biogen, and Genentech, respectively. LMB was generously provided by B. Wolff-Winiski (Novartis).
This work was supported in part by NIH grant CA80105. M.D. is a recipient of the Sidney Kimmel Foundation for Cancer Research Scholar Award.
REFERENCES
- 1.Abola E E, Sussman J L, Prilusky J, Manning N O. Protein data bank archives of three-dimensional macromolecular structures. Methods Enzymol. 1997;277:556–571. doi: 10.1016/s0076-6879(97)77031-9. [DOI] [PubMed] [Google Scholar]
- 2.Ali S, Ali S. Prolactin receptor regulates Stat5 tyrosine phosphorylation and nuclear translocation by two separate pathways. J Biol Chem. 1998;273:7709–7716. doi: 10.1074/jbc.273.13.7709. [DOI] [PubMed] [Google Scholar]
- 3.Arenzana-Seisdedos F, Turpin P R, Thomas M D, Hay R T, Virelizier J L, Dargemont C. Nuclear localization of I kappa B alpha promotes active transport of NF-kappa B from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, Vinkemeier U, Zhao Y, Jeruzalmi D, Darnell J E, Kuriyan J. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 5.Darnell J E, Kerr I M, Stark G R. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 6.David M, Romero G, Zhang Z Y, Dixon J E, Larner A C. In vitro activation of the transcription factor ISGF3 by IFNα involves a membrane associated tyrosine phosphatase and kinase. J Biol Chem. 1993;268:6593–6599. [PubMed] [Google Scholar]
- 7.David M, Wong L, Flavell R, Thompson S, Larner A C, Johnson G. STAT activation by EGF and amphiregulin: requirement for the EGF receptor kinase, but not for tyrosine phosphorylation sites or JAK1. J Biol Chem. 1996;271:9185–9188. doi: 10.1074/jbc.271.16.9185. [DOI] [PubMed] [Google Scholar]
- 8.Dingwall C, Laskey R A. Nuclear targeting sequences—a consensus? TIBS Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 9.Driggers P H, Ennist D L, Gleason S L, Wai-Han M, Marks M S, Levi B-Z, Flanagan J R, Appella E, Ozato K. An interferon g-regulated protein that binds the interferon-inducible enhancer element of major histocompatibility class I genes. Proc Natl Acad Sci USA. 1990;87:3743–3747. doi: 10.1073/pnas.87.10.3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engel K, Kotlyarov A, Gaestel M. Leptomycin B-sensitive nuclear export of MAPKAP kinase 2 is regulated by phosphorylation. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finbloom D S, Larner A C. Induction of early response genes by interferons, interleukins, and growth factors by the tyrosine phosphorylation of latent transcription factors: implications for chronic inflammatory diseases. Arthritis Rheumatism. 1995;38:877–889. doi: 10.1002/art.1780380702. [DOI] [PubMed] [Google Scholar]
- 12.Fischer U, Huber J, Boelens W C, Mattaj I W, Luehrmann R. The HIV-1 rev activation domain is a nuclear export signal that accesses an export pathway used by specific cellular RNAs. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 13.Fornerod M, Ohno M, Yoshida M, Mattaj I. Crm1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 14.Friedman R L, Stark G R. α-Interferon-induced transcription of HLA and metallothionine genes containing homologous upstream sequences. Nature. 1985;314:637–639. doi: 10.1038/314637a0. [DOI] [PubMed] [Google Scholar]
- 15.Fu X-Y. A transcription factor with SH2 and SH3 domains is directly activated by an interferon-α induced cytoplasmic protein tyrosine kinase(s) Cell. 1992;70:323–335. doi: 10.1016/0092-8674(92)90106-m. [DOI] [PubMed] [Google Scholar]
- 16.Fu X-Y, Schindler C, Improta T, Aebersold R, Darnell J E J. The proteins of ISGF-3, the interferon α-induced transcriptional activator, define a gene family involved in signal transduction. Proc Natl Acad Sci USA. 1992;89:7840–7843. doi: 10.1073/pnas.89.16.7840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gupta S, Yan H, Wong L H, Ralph S, Krolewski J, Schindler C. The SH2 domains of Stat1 and Stat2 mediate multiple interactions in the transduction of IFN-α signals. EMBO J. 1996;15:1075–1084. [PMC free article] [PubMed] [Google Scholar]
- 18.Hallek M, Lepisto E M, Slattery K E, Griffin J D, Ernst T J. Interferon-γ increases the expression of the gene encoding the β subunit of the granulocyte-macrophage colony-stimulating factor receptor. Blood. 1992;80:1736–1742. [PubMed] [Google Scholar]
- 19.Haque S, Wu Q, Kammer W, Friedrich K, Smith J, Kerr I, Stark G, Williams B. Receptor-associated constitutive protein tyrosine phosphatase activity controls the kinase function of JAK1. Proc Natl Acad Sci USA. 1997;94:8563–8568. doi: 10.1073/pnas.94.16.8563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haspel R L, Darnell J E., Jr A nuclear protein tyrosine phosphatase is required for the inactivation of Stat1. Proc Natl Acad Sci USA. 1999;96:10188–10193. doi: 10.1073/pnas.96.18.10188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvai A E, Xu L, Korzus E, Brard G, Kalafus D, Mullen T-M, Rose D W, Rosenfeld M G, Glass C K. Nuclear integration of JAK/STAT and Ras/AP-1 signaling by CBP and p300. Proc Natl Acad Sci USA. 1997;94:1074–1079. doi: 10.1073/pnas.94.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Igarashi K, David M, Larner A C, Finbloom D S. In vitro activation of a transcription factor by gamma interferon requires a membrane-associated tyrosine kinase and is mimicked by vanadate. Mol Cell Biol. 1993;13:3984–3989. doi: 10.1128/mcb.13.7.3984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ihle J N, Witthuhn B A, Quelle F W, Yamamoto K, Thierfelder W E, Kreider B, Silvennoinen O. Signaling by the cytokine receptor superfamily: JAKS and STATS. Trends Biochem Sci. 1994;19:222–227. doi: 10.1016/0968-0004(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 24.Johnston J A, Kawamura M, Kirken R A, Chen Y-Q, Blake T B, Shibuya K, Ortaldo J R, McVicar D W, O'Shea J J. Phosphorylation and activation of the Jak-3 Janus kinase in response to interleukin-2. Nature. 1994;370:151–153. doi: 10.1038/370151a0. [DOI] [PubMed] [Google Scholar]
- 25.Kazansky A V, Kabotyanski E B, Wyszomierski S L, Mancini M A, Rosen J M. Differential effects of prolactin and src/abl kinases on the nuclear translocation of STAT5B and STAT5A. J Biol Chem. 1999;274:22484–22492. doi: 10.1074/jbc.274.32.22484. [DOI] [PubMed] [Google Scholar]
- 26.Khan K D, Shuai K, Lindwall G, Maher S E, Darnell J E, Jr, Bothwell A L M. Induction of the Ly-6A/E gene by interferon α/β and γ requires a DNA element to which a tyrosine-phosphorylated 91-kDa protein binds. Proc Natl Acad Sci USA. 1993;90:6806–6810. doi: 10.1073/pnas.90.14.6806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kurokawa R, Kalafus D, Ogliastro M-H, Kioussi C, Xu L, Torchia J, Rosenfeld M G, Glass C K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- 28.Larner A C, David M, Feldman G M, Igarashi K, Hackett R H, Webb D A S, Sweitzer S M, Petricoin III E F, Finbloom D S. Tyrosine phosphorylation of DNA binding proteins by multiple cytokines. Science. 1993;261:1730–1733. doi: 10.1126/science.8378773. [DOI] [PubMed] [Google Scholar]
- 29.Larner A C, Finbloom D S. Protein tyrosine phosphorylation as a mechanism which regulates cytokine activation of early response genes. Biochim Biophys Acta. 1995;1266:278–287. doi: 10.1016/0167-4889(95)00015-k. [DOI] [PubMed] [Google Scholar]
- 30.Larner A C, Jonak G, Cheng Y-S E, Korant B, Knight E, Darnell J E. Transcriptional induction of two genes in human cells by β interferon. Proc Natl Acad Sci USA. 1984;81:6733–6737. doi: 10.1073/pnas.81.21.6733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leaman D W, Pisharody S, Flickinger T, Commane W, M A, Schlessinger J, Levy K I M, D E, Stark G R. Roles of JAKs in activation of STAT and stimulation of c-fos gene expression by epidermal growth factor. Mol Cell Biol. 1996;16:369–375. doi: 10.1128/mcb.16.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leung S, Qureshi S, Kerr I, Darnell J, Stark G. Role of STAT2 in the alpha interferon signaling pathway. Mol Cell Biol. 1995;15:1312–1317. doi: 10.1128/mcb.15.3.1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy D E, Kessler D S, Pine R, Darnell J E., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-α-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 34.Luster A D, Unkeless J C, Ravetch J V. γ-Interferon transcriptionally regulates an early-response gene containing homology to platelet proteins. Nature. 1985;315:672–676. doi: 10.1038/315672a0. [DOI] [PubMed] [Google Scholar]
- 35.Mowen K A, David M. Role of the STAT1-SH2 domain and STAT2 in the nuclear translocation of STAT1. J Biol Chem. 1998;273:30073–30076. doi: 10.1074/jbc.273.46.30073. [DOI] [PubMed] [Google Scholar]
- 36.Muller M, Briscoe J, Laxton C, Guschin D, Ziemiecki A, Silvennoinen O, Harpur A G, Barbieri G, Withuhn B A, Schindler C, Pellegrini S, Wilks A F, Ihle J N, Stark G R, Kerr I M. The protein tyrosine kinase JAK1 complements defects in the interferon-α/β and -γ signal transduction. Nature. 1993;366:129–135. doi: 10.1038/366129a0. [DOI] [PubMed] [Google Scholar]
- 37.Muller M, Laxton C, Briscoe J, Schindler C, Improta T, Darnell J E, Jr, Stark G R, Kerr I M. Complementation of a mutant cell line: central role of the 91kDa polypeptide of ISGF3 in the interferon-α and -γ signal transduction pathways. EMBO J. 1993;12:4221–4228. doi: 10.1002/j.1460-2075.1993.tb06106.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nigg E. Nucleocytoplasmic transport: signals, mechanism and regulation. Nature. 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 39.Pollard V W, Michael M W, Nakielny S, Siomi M C, Wang F, Dreyfuss G. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 40.Revel M, Chebath J. Interferon-activated genes. Trends Biochem Sci. 1986;11:166–170. [Google Scholar]
- 41.Schindler C, Fu X-Y, Improta T, Aebersold R, Darnell J E., Jr Proteins of transcription factor ISGF-3: one gene encodes the 91- and 84-kDa ISGF-3 proteins that are activated by interferon α. Proc Natl Acad Sci USA. 1992;89:7836–7839. doi: 10.1073/pnas.89.16.7836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sekimoto T, Nakajima K, Tachibana T, Hirano T, Yoneda Y. Interferon-γ-dependent nuclear import of Stat1 is mediated by the GTPase activity of Ran/TC4. J Biol Chem. 1996;271:31017–31020. doi: 10.1074/jbc.271.49.31017. [DOI] [PubMed] [Google Scholar]
- 44.Shuai K, Horvath C M, Tsai Huang L H, Qureshi S A, Cowburn D, Darnell J E., Jr Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 45.Shuai K, Schindler C, Prezioso R V, Darnell J E., Jr Activation of transcription by IFN-γ: tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 46.Shuai K, Stark G R, Kerr I M, Darnell J E., Jr A single phosphotyrosine residue of Stat91 required for gene activation by interferonγ. Science. 1993;261:1744–1746. doi: 10.1126/science.7690989. [DOI] [PubMed] [Google Scholar]
- 47.Shuai K, Ziemiecki A, Wilks A F, Harpur A G, Sadowski H B, Gilman M Z, Darnell J E., Jr Polypeptide signalling to the nucleus through tyrosine phosphorylation of JAK and Stat proteins. Nature. 1993;366:580–583. doi: 10.1038/366580a0. [DOI] [PubMed] [Google Scholar]
- 48.Silvennoinen O, Ihle J N, Schlessinger J, Levy D E. Interferon-induced nuclear signalling by Jak protein tyrosine kinases. Nature. 1993;366:583–585. doi: 10.1038/366583a0. [DOI] [PubMed] [Google Scholar]
- 49.Stade K, Ford C, Guthrie C, Weis K. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 50.Staeheli P, Danielson P, Haller O, Sutcliffe J G. Transcriptional activation of the mouse Mx gene by type I interferon. Mol Cell Biol. 1986;6:4770–4774. doi: 10.1128/mcb.6.12.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stommel J M, Marchenko N D, Jimenez G S, Moll U M, Hope T J, Wahl G M. A leucine-rich nuclear export signal in the p53 tetramerization domain: regulation of subcellular localization and p53 activity by NES masking. EMBO J. 1999;18:1660–1672. doi: 10.1093/emboj/18.6.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taagepera S, McDonald D, Loeb J E, Whitaker L L, MacLeod K A, Wang J Y J, Hope T. Nuclear-cytoplasmic shuttling of c-Abl tyrosine kinase. Proc Natl Acad Sci USA. 1998;95:7457–7462. doi: 10.1073/pnas.95.13.7457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ullman K, Powers M, Forbes D. Nuclear export receptors: from importin to exportin. Cell. 1997;90:967–970. doi: 10.1016/s0092-8674(00)80361-x. [DOI] [PubMed] [Google Scholar]
- 54.Velazquez L, Fellous M, Stark G R, Pellegrini S. A protein tyrosine kinase in the interferon α/β signaling pathway. Cell. 1992;70:313–322. doi: 10.1016/0092-8674(92)90105-l. [DOI] [PubMed] [Google Scholar]
- 55.Watling D, Guschin D, Muller M, Silvennoinen O, Witthuhn B A, Quelle F W, Rogers N C, Schindler C, Ihle J N, Stark G R, Kerr I M. Complementation by the protein tyrosine kinase JAK2 of a mutant cell line defective in the interferon-γ signal transduction pathway. Nature. 1993;366:166–170. doi: 10.1038/366166a0. [DOI] [PubMed] [Google Scholar]
- 56.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Identification of a signal for rapid export of proteins from the nucleus. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 57.Wilson K C, Finbloom D S. Interferon γ rapidly induces in human monocytes a DNA-binding factor that recognizes the γ response region within the promoter of the gene for the high-affinity Fcγ receptor. Proc Natl Acad Sci USA. 1992;89:11964–11968. doi: 10.1073/pnas.89.24.11964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wolff B, Sanglier J J, Wang Y. Leptomycin B is an inhibitor of nuclear export: inhibition of nucleo-cytoplasmic translocation of the human immunodeficiency virus type 1 (HIV-1) Rev protein and Rev-dependent mRNA. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 59.Yan C, Sehgal P B, Tamm I. Signal transduction pathways in the induction of 2′,5′-oligoadenylate synthetase gene expression by interferon α/β. Proc Natl Acad Sci USA. 1989;86:2243–2247. doi: 10.1073/pnas.86.7.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang J J, Vinkemeier U, Gu W, Chakravarti D, Horvath K, Darnell J. Two contact regions between Stat1 and CBP/p300 in interferon γ signaling. Proc Natl Acad Sci USA. 1996;93:15092–15096. doi: 10.1073/pnas.93.26.15092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu M, John S, Berg M, Leonard W J. Functional association of Nmi with Stat5 and Stat1 in IL-2- and IFNgamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]