Abstract
The major histocompatibility complex (MHC) class II transactivator CIITA plays a pivotal role in the control of the cellular immune response through the quantitative regulation of MHC class II expression. We have analyzed a region of CIITA with similarity to leucine-rich repeats (LRRs). CIITA LRR alanine mutations abolish both the transactivation capacity of full-length CIITA and the dominant-negative phenotype of CIITA mutants with N-terminal deletions. We demonstrate direct interaction of CIITA with the MHC class II promoter binding protein RFX5 and could also detect novel interactions with RFXANK, NF-YB, and -YC. However, none of these interactions is influenced by CIITA LRR mutagenesis. On the other hand, chromatin immunoprecipitation shows that in vivo binding of CIITA to the MHC class II promoter is dependent on LRR integrity. LRR mutations lead to an impaired nuclear localization of CIITA, indicating that a major function of the CIITA LRRs is in nucleocytoplasmic translocation. There is, however, evidence that the CIITA LRRs are also involved more directly in MHC class II gene transactivation. CIITA interacts with a novel protein of 33 kDa in a manner sensitive to LRR mutagenesis. CIITA is therefore imported into the nucleus by an LRR-dependent mechanism, where it activates transcription through multiple protein-protein interactions with the MHC class II promoter binding complex.
The expression of major histocompatibility complex class II (MHC-II) molecules, which play a critical role in immune responses by presenting processed exogenous antigens to CD4+ T lymphocytes, is controlled in a highly complex manner. MHC-II molecules are expressed constitutively only on a restricted set of cell types specialized in antigen presentation, such as B lymphocytes, macrophages, and dendritic cells, whereas MHC-II gene expression can be induced and modulated on many other cell types by different stimuli, most prominently gamma interferon (IFN-γ) (16, 27). In humans, three MHC-II isotypes are known, human leukocyte antigen DR (HLA-DR), -DQ, and -DP. Expression is controlled mainly at the level of transcription via conserved upstream sequence elements in the proximal promoters, the W (S), X, X2, and Y boxes, which mediate constitutive and IFN-γ-induced expression of the MHC-II genes (reviewed in references 16 and 27).
Four essential MHC-II regulatory factors were discovered through analysis of cell lines derived from patients suffering from hereditary HLA class II deficiency (also known as bare lymphocyte syndrome [BLS]), a genetically heterogeneous disease of gene regulation, or of in vitro-generated mutant cell lines (18, 27). These factors, called RFX5 (regulatory factor binding to X box 5), RFXAP (RFX-associated protein), RFXANK (RFX protein containing ankyrin repeats; also called RFX-B), and CIITA (class II transactivator), are essential for the expression of all MHC-II genes (12, 30, 33, 44, 45). Mutations in the corresponding regulatory genes could be identified in BLS patients in all four complementation groups. RFX5, RFXAP, and RFXANK are components of the multisubunit RFX complex that binds to the X box of the MHC-II promoter (38).
The MHC-II transactivator CIITA, the first MHC-II deficiency gene identified, is the master regulator of MHC-II gene expression (46). While RFX and the other MHC-II promoter binding complexes such as NF-Y (nuclear factor binding to the Y box) and X2BP (X2 box binding protein) are also present in MHC-II-negative cells, a strictly concordant expression between CIITA and MHC-II mRNA has been observed in multiple cell lines and tissues (36, 45, 47). CIITA is the obligatory mediator of IFN-γ-induced MHC-II expression (7, 9, 47). CIITA expression is both necessary and sufficient to induce expression of all MHC-II promoter-containing genes, controlling MHC-II expression qualitatively and quantitatively, with a nearly linear correlation between CIITA and MHC-II expression over a wide range of expression levels (6, 7, 21, 36, 47).
CIITA is probably not itself a DNA binding protein and is believed to act in a coactivator-like fashion through protein-protein interactions with MHC-II promoter binding proteins (40, 42, 45, 50). Removal of the N-terminal acidic region or both the acidic and proline-, serine-, and threonine-rich regions of CIITA leads to a dominant-negative phenotype (8, 51). Efficient dominant-negative mutants have been selected through a functional approach from a library of mutants with random N-terminal deletions (5). CIITA contains a tripartite GTP binding motif, which is important for the predominantly nuclear localization (19). A second motif involved in the nuclear localization was found at amino acid positions 955 to 959 (11).
The C-terminal part of CIITA contains a region showing sequence similarity to so-called leucine-rich repeat motifs (LRRs) (24). LRRs mediate protein-protein interactions and are found in many different classes of proteins (24). We demonstrate here that the LRRs of CIITA are essential for its function. By CIITA LRR alanine scanning mutagenesis we identified a number of residues in which single alanine exchanges completely abolish CIITA function. CIITA interacts with at least four MHC-II promoter binding proteins, but these interactions are not affected by LRR mutagenesis. On the other hand, in vivo recruitment of CIITA to the MHC-II promoter binding complex is dependent on the integrity of the LRRs. Immunofluorescence analysis revealed that LRR mutagenesis leads to an impaired nuclear localization of CIITA, demonstrating that a major role of the CIITA LRRs is in protein transport. However, the observation of one particular mutant (MT1) that is still recruited to the promoter without activating MHC-II gene expression reveals that the CIITA LRRs might also have a function within the nucleus. A first indication of a CIITA LRR-interacting protein was obtained by coimmunoprecipitation.
MATERIALS AND METHODS
Cells and cell culture.
Raji (ATCC CCL-86) is a human MHC-II-positive Burkitt lymphoma cell line; RJ2.2.5 is a CIITA-deficient, MHC-II-negative cell line derived from Raji (1, 45). Both cell lines were grown in RPMI 1640 medium supplemented with 2 mM l-glutamine, 10% fetal calf serum, 10 U of penicillin/ml, and 10 μg of streptomycin/ml. HEK293-EBNA (Invitrogen) is a CIITA-negative embryonal human kidney cell line, and was grown in Dulbecco's modified Eagle medium, supplemented as described above. Cells were grown at 37°C in a humidified, 7.5% CO2 atmosphere.
Transfections.
Burkitt lymphoma cells were transfected by electroporation as described previously (45). Transfected cells were selected with hygromycin B (Calbiochem) at 200 μg/ml and were cultivated under the constant presence of hygromycin. Hygromycin-resistant cells were analyzed in bulk without sorting or cloning. HEK293-EBNA cells were transfected by calcium phosphate precipitation and analyzed 3 days after transfection.
Expression vectors.
The Epstein-Barr virus episomal expression vectors EBO-76PL, with a simian virus 40 (SV40) early promoter, and EBS-PL, with an SRα promoter (49), have been described previously (5, 43). They contain a hygromycin resistance gene for antibiotic selection in mammalian cells. The CS2+MT vector, a gift from R. Rupp, contains a cytomegalovirus IE enhancer/promoter and six N-terminal copies of the myc epitope (41).
CIITA alanine mutants.
CIITA alanine mutants were created by a two-step PCR procedure. The template (KS-BBN-CIITAshort, a full-length CIITA gene open reading frame with a deleted 3′-untranslated region) was amplified in a standard PCR with the 5′ primer F14 (CIITA gene nucleotide positions 2792 to 2812) and a mutagenic 3′ primer. The second PCR was performed with the purified PCR product as a megaprimer, a CIITA gene construct with a deleted binding site for the 5′ primer as a template, primer F14, and a 3′ primer downstream of the CIITA gene stop codon. The PCR product was isolated and subcloned, and the mutations were verified by sequencing. Sequences of primers and PCR conditions are available upon request.
cDNA expression constructs.
CS2+MT-RFX5 contains RFX5 from amino acid position 194 (44) in frame with the six myc epitopes. This RFX5 construct is similar to the one used by Scholl and colleagues (42). Full-length OBF-1 (48), a gift from M. Strubin, was cloned into EBO-76PL. EBO-RFXANK has been described (30). CS2+MT-RFXAP was created by inserting full-length RFXAP into the CS2+MT vector in frame with the six myc epitope tags (12). CS2+MT-NF-YA, -YB, and -YC were generated by reverse transcription-PCR using Raji cDNA as a template. Full-length open reading frames were cloned into the CS2+MT vector in frame with the six myc epitope tags. KEBS-PL-NLS-L102 and KEBS-PL-NLS-L335 have been described (5). EBO-76PL-EGFP and EBS-PL-EGFP were created by inserting enhanced GFP-N1 (EGFP-N1) (Clontech) into EBO-76PL and EBS-PL. EGFP-CIITA is an N-terminal fusion of EGFP to the second in-frame ATG of CIITAshort with the connecting sequence KQCATM (the last amino acids of EGFP and the first of CIITA are underlined). The CIITA part in this fusion protein corresponds to the naturally occurring form IV of CIITA (32). EGFP-CIITA shows wild-type activity for MHC-II transactivation (C. Kammerbauer, S. B. Hake, and V. Steimle, unpublished data).
Antibodies and FACS analysis.
Cell staining and fluorescence-activated cell sorter (FACS) analysis were performed as described previously (45). One million cells were stained with the following antibodies at the indicated dilutions: HLA class II DR-specific D1.12 (1), 1:250; DQ-specific SPV-L3 (Serotec), undiluted; rabbit anti-mouse fluorescein isothiocyanate-labeled secondary antibody (Serotec), 1:100. Cells transfected with EGFP fusion constructs were stained with monoclonal anti-human HLA-DR antibody coupled to Quantum Red (clone HK14; Sigma), at a dilution of 1:200. Staining and analysis were performed on live cells. Dead cells were excluded from the analysis by their forward and sideways light scatter properties and, where possible, by staining with propidium iodide.
The following antibodies for the Western blot analysis were used at the indicated concentrations. The primary antibodies were polyclonal rabbit anti-human CIITA serum 20, diluted 1:4,000 (5), polyclonal rabbit anti-human RFXANK serum, diluted 1:4,000 (30), polyclonal rabbit anti-human OBF-1 serum, diluted 1:2,000 (48), and monoclonal mouse anti-myc, diluted 1:2,000 (Santa Cruz Biotechnology). Secondary antibodies were polyclonal sheep anti-mouse horseradish peroxidase-linked antibody, diluted 1:10,000 (Amersham), and polyclonal donkey anti-rabbit horseradish peroxidase-linked antibody, diluted 1:10,000 (Amersham). The antibodies for the immunoprecipitation assay were used in the following amounts: 4 μg of monoclonal mouse anti-green fluorescent protein (GFP) (clones 7.1 and 13.1; Roche) and 10 μl of polyclonal rabbit anti-GFP (Clontech).
Protein extracts and Western blot analysis.
Total protein was extracted from 107 cells (5). Sodium dodecyl sulfate (SDS)-polyacrylamide gels (7.5 or 15%) were loaded with 100 μg of total protein. After gel electrophoresis, the proteins were semidry blotted onto a polyvinylidene difluoride membrane (Immobilon-P; Millipore). The membrane was washed once with phosphate-buffered saline (PBS) containing 0.5% Tween 20 (Sigma) and then blocked for 30 min with blocking solution (Roche). The first antibody was added, and the membrane was incubated for 45 min. The membrane was washed three times with PBS containing 0.5% Tween 20 and then blocked again and incubated with the second, peroxidase-labeled, antibody. After being washed, the membrane was incubated with ECL-Plus substrate solution (Amersham) and the proteins were detected by exposure to X-ray film (Hyperfilm; Amersham).
Immunoprecipitations. (i) Protein G-agarose.
Fifty microliters of the total protein lysates was mixed with WP-1 buffer (Roche) to a final volume of 1 ml. Packed protein G-agarose (Roche) was resuspended in 10 volumes of WP-1 buffer. For preclearing, the solution was incubated for 3 h with 100 μl of protein G-agarose. After centrifugation (13,000 × g, 4°C, 30 s), the supernatant was incubated with the appropriate antisera for 1 h. Immunocomplexes were precipitated overnight using 200 μl of protein G-agarose at 4°C. The complexes were washed three times with WP-1 for 30 min, three times with WP-2, and twice with WP-3 (Roche). Proteins were resolved by electrophoresis and revealed by Western blotting.
(ii) Dynabeads.
Dynabeads (100 μl) coated with anti-rabbit antibody (Dynal) were washed three times with WP-1 buffer and incubated overnight at 4°C with anti-GFP antibodies (Roche) in WP-1 buffer. The beads were washed twice with WP-1 buffer and incubated with 50 μl of the total protein lysates at 4°C in WP-1 buffer for 3 h. Washes were performed on a magnet; all other treatments and buffers were as described above.
Chromatin immunoprecipitation.
Chromatin immunoprecipitations were carried out, with modifications described previously (31), according to a protocol provided by J. Wells and P. Farnham (University of Wisconsin). Briefly, 3 days after transfection with the different EGFP-CIITA constructs, 70 million HEK293-EBNA cells per transfection were cross-linked with 1% formaldehyde for 5 min at room temperature. Cross-linking was stopped with 0.125 M glycine. After being washed in PBS, cells were lysed in Tris-EDTA (TE) buffer containing protease inhibitors and 0.5% NP-40. Nuclei were pelleted by centrifugation and resuspended in nucleus lysis buffer (TE buffer containing 0.5 M NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, and 0.5% sarcosyl). After centrifugation, chromatin was resuspended in 1 ml of immunoprecipitation buffer (20 mM Tris-HCl [pH 8.0], 200 mM NaCl, 2 mM EDTA, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride), sheared by sonication to an average length of 500 to 800 bp and stored in aliquots at −70°C. Immunoprecipitation was carried out with the equivalent of 107 cells per immunoprecipitation at room temperature in the presence of protease inhibitors. Input of similar amounts and quality of chromatin was verified by gel electrophoresis. For each immunoprecipitation 100 μl of rat anti-mouse immunoglobulin G-coupled magnetic beads (Dynal) was preincubated overnight with 4 μg of GFP-specific monoclonal antibodies (Roche) and washed before use. The chromatin preparation was supplemented with 1 mg of bovine serum albumin BSA, 100 μg of tRNA, and 50 μg of salmon testis DNA/ml and incubated with the bead-coupled antibodies for 3 h at room temperature. All washes were carried out on a magnet. Beads were washed three times each for 10 min with immunoprecipitation buffer, with immunoprecipitation buffer with 500 mM NaCl, and with immunoprecipitation buffer with 20 mM Tris-HCl, pH 8.0, 0.25 M LiCl, 2 mM EDTA, and 0.5% NP-40 and once in TE buffer containing 0.1% NP-40. Beads were resuspended in 200 μl of elution buffer (100 mM Tris-HCl [pH 8.0], 200 mM NaCl, 0.5% SDS, 100 mg of proteinase K/ml) and incubated for 1 h at 50°C, followed by 10 min at 65°C. Cross-linking of the chromatin supernatant was reversed overnight at 65°C. After phenol-chloroform extraction and precipitation, DNA was resuspended in 25 μl of TE and used for PCR and slot blot hybridization. DNA isolated from the unbound chromatin supernatant of the GFP transfectant after immunoprecipitation was resuspended in 250 μl of TE and used as a positive control. PCR was carried out under standard conditions with a primer pair spanning the DRA promoter (DRA-PForw., 5′CTTGATTTGTTGTTGTTGTTGTC; DRA-PRev., 5′CTTTTGGGAGTCAGTAGAGC) and with 1 μl of immunoprecipitation material per reaction. Unspecific DNA precipitation was quantified by slot blot hybridization with an Alu repeat probe.
Radioactive labeling of cells.
Forty-eight hours after transfection HEK293-EBNA cells were incubated for 2 h in modified Eagle medium (Gibco) without methionine. After being washed, the cells were incubated overnight in the same medium supplemented with 50 μCi of [35S]methionine (Amersham)/ml. The cells were then lysed, and total protein was isolated, immunoprecipitated, and resolved by SDS-polyacrylamide gel electrophoresis (PAGE) as described above. Dried gels were exposed to X-ray film (X-OMAT AR; Kodak).
Fluorescence microscopy analysis.
HEK293-EBNA cells were grown and transfected on glass coverslips. After 48 h transfected cells were fixed for 10 min in 3% paraformaldehyde-PBS and stained for 10 min in 2 μg of bis-benzimidine–methanol/ml. The stained cells were analyzed and photographed on a Zeiss Axioskop microscope equipped with a cooled charge-coupled device camera (Hamamatsu). Image processing was carried out with the OpenLab (Improvision) and Photoshop (Adobe) software packages.
RESULTS
Alanine scanning mutagenesis of CIITA LRR sequences.
The crystal structure of the RNase inhibitor (RI), which consists almost entirely of LRRs, is a horseshoe-shaped structure with a large inner surface composed of solvent-exposed parallel β-strands. The β-strands are connected via loop regions to the backbone-forming α-helices. The substrate RNase A is bound in the central cavity via numerous amino acid contacts leading to one of the strongest protein-protein bonds on record (22, 24, 25).
In an attempt to develop a rational basis for a mutational analysis of the CIITA LRRs, we asked the question whether the contact positions between the RI and its substrate (25) are found predominantly at certain positions within the individual repeats. Identification of the contact positions of the RI in an alignment of the alternating A-type and B-type repeats of the RI indicates that contact points are only found in the β-strands and loop regions of the repeats (Fig. 1A). Contact points in the β-strands are confined to positions interspersed with the highly conserved structural residues. In the last two repeats, the loop regions contribute strongly to the binding in addition to the β-strand regions. The lower part of Fig. 1A shows the alignment of the CIITA LRRs with those of the RI. CIITA shows a better homology to the B-type repeats, due to the conserved asparagine at position 10 of the repeat. By combining the contact positions of both types of RI repeats, we identified nine potential contact positions in each CIITA LRR unit.
FIG. 1.
Generation of CIITA LRR alanine mutants. (A) Alignment of RI with the LRRs of CIITA. The alternating A- and B-type repeats of porcine RI (accession no. P10775) are aligned separately on the upper left and right sides. Amino acids that are in contact with the substrate RNase A are indicated as follows: those involved in van der Waals interactions are shaded, and those participating in hydrogen bond formation are in lowercase (25). The consensus sequences of the RI repeats are shown below the RI sequence, and positions participating in substrate binding are shaded. The LRR regions of human CIITA (amino acid positions 988 to 1097) are at the bottom. Potential contact positions were combined from both types of RI repeats, and the corresponding positions in CIITA are shaded. (B) CIITA LRR multiple-alanine mutants. At the top the RI B-type repeat consensus (cons.) sequence is shown. Compiled contact positions are indicated together with the delimitation of β-strand-, loop-, and α-helical regions. The positions of the multiple alanine mutations are indicated by circles. The names of the mutants are indicated on the left. Open circles, mutations without effect on CIITA function; solid circles, mutations leading to a loss of CIITA transactivation activity (see Fig. 2A). (C) CIITA LRR single-alanine mutants. Solid circles, mutations which abolish CIITA function in single-alanine mutants (see Fig. 2B); open circles, mutations without effect in single-alanine mutants. The single alanine mutations are identified by their amino acid positions.
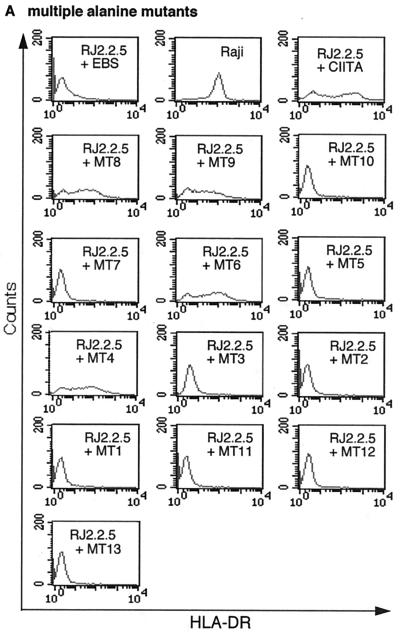
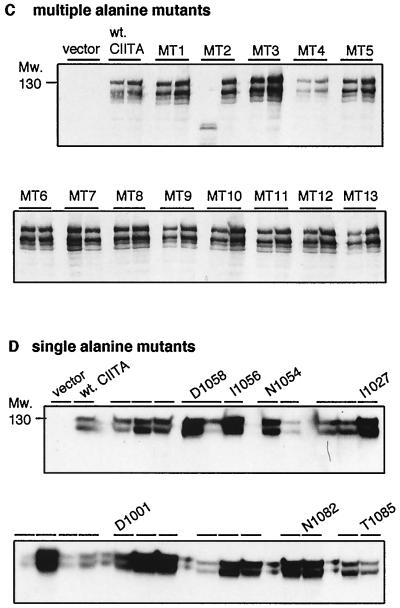
The functional significance of these consensus LRR positions was tested by alanine scanning mutagenesis of the corresponding CIITA residues. The relevant positions were first mutated in groups of two to four residues at a time (Fig. 1B). The resulting 13 CIITA mutants were stably transfected into the CIITA-deficient Burkitt lymphoma B-cell line RJ2.2.5 (1, 45). Nine of the 13 mutants completely lacked CIITA transactivation of HLA-DR, as shown by flow cytometry (Fig. 2A and 1B). Transient transfection of CIITA-negative HEK293-EBNA cells yielded similar results (data not shown). Levels of mutant protein expression were generally not affected (Fig. 2C); only mutants MT2 and MT11 showed reduced stability, especially when fused to GFP (Fig. 2C and 6A; data not shown). The multiple-alanine mutants demonstrated the functional importance of all four loop regions and of the β-regions of repeats two and four (Fig. 1B).
FIG. 2.
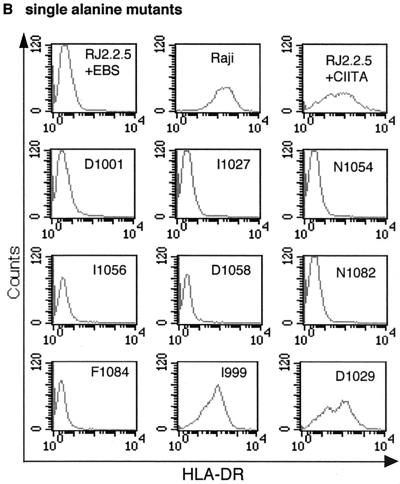
FACS analysis of CIITA LRR alanine mutants. (A) Multiple-alanine mutants. Wild-type and mutant CIITA cDNAs in the episomal expression vector EBS-PL were transfected into the CIITA-deficient cell line RJ2.2.5 and analyzed in bulk for HLA-DR expression by FACS after 2 weeks of antibiotic selection. The mutants are presented in N-terminal-to-C-terminal order (see also Fig. 1B). (B) Single-alanine mutants. All seven mutants with mutations which abolish CIITA function and two mutants with mutations without effect are shown. Mutations are identified by their amino acid positions (see also Fig. 1C). (C) Protein expression analysis of CIITA LRR multiple-alanine mutants. CIITA cDNA expression constructs were transfected into HEK293-EBNA cells, and transient protein expression was analyzed by Western blotting with a CIITA-specific antiserum. The blot shows two independent transfections for each construct. The two major bands of CIITA correspond most probably to products resulting from initiation at the first and second AUGs of the CIITA cDNA, form III (32, 45), used here. (D) Protein expression analysis of CIITA LRR single-alanine mutants. Only the mutants that have lost their transactivation potential are identified individually; the others are indicated by a horizontal bar.
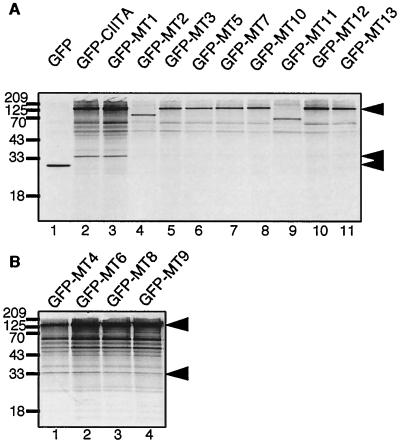
FIG. 6.
Immunoprecipitation of in vivo-labeled CIITA transfectants. Wild-type EGFP-CIITA and EGFP-CIITA LRR multiple-alanine mutants were transfected individually into HEK293-EBNA cells, and the cells were metabolically labeled overnight with [35S]methionine. EGFP-CIITA and associated proteins were immunoprecipitated with a GFP-specific polyclonal antiserum (Clontech), followed by SDS-PAGE on 7.5 or 15% gels and autoradiography. (A) A 15% gel of cells transfected with EGFP, EGFP-CIITA, and all nine EGFP-CIITA multiple-alanine mutants that abolish MHC-II transactivation. (B) Analysis of mutants that have retained their transactivation potential. Arrowheads, positions of EGFP, the CIITA-associated 33-kDa band, and EGFP-CIITA. Two of the mutants, MT2 and MT11, became unstable when fused to EGFP (lanes 4 and 9).
Next, 25 single-alanine mutants, covering all potentially important positions revealed by experiments with the multiple-alanine mutants, were generated (Fig. 1C). Transfection of these mutants into RJ2.2.5 and HEK293-EBNA cells identified seven CIITA mutants in which HLA class II activation was completely abolished (Fig. 2B and 1C; data not shown). As before, levels of expression of mutant proteins that had lost transactivation capacity were at least equal to the average level for all 25 mutants (Fig. 2D). Two of the mutations target the highly conserved asparagine at position 10 of repeats three and four (N1054, N1082), whereas the other 5 mutations are located at positions 1 or 3 of the IxD loop motif of the CIITA LRRs (Fig. 2B and 1C). In the region of the β-strands no single alanine exchange was able to abolish CIITA function. The results of the alanine mutagenesis of CIITA LRRs are summarized in Fig. 1B and C.
Interaction of CIITA with subunits of the MHC-II promoter binding complexes.
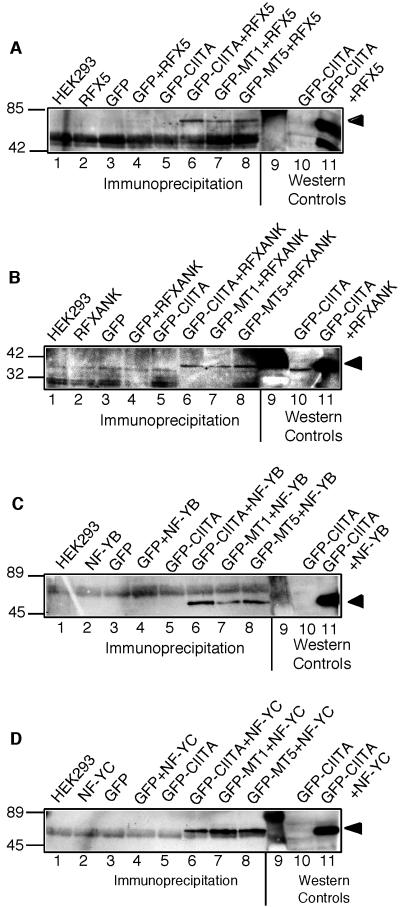
The major function of LRRs is protein-protein interaction (23, 24). We therefore wanted to know whether CIITA interacts with known MHC-II promoter binding proteins and whether the CIITA LRRs play a role in these interactions. Analysis was carried out by coimmunoprecipitation of native and epitope-tagged proteins cotransfected into HEK293-EBNA cells. Scholl and colleagues (42) had shown an interaction between CIITA and RFX5, the 75-kDa subunit of the RFX complex, in a yeast two-hybrid assay and by far-Western analysis. As shown in Fig. 3A, immunoprecipitation of CIITA leads to precipitation of a cotransfected RFX5 construct (lane 6). Precipitation of RFX5 is strictly dependent on the presence of CIITA as shown by the different controls (lanes 1 to 5). The LRR regions of CIITA do not seem to play an important role in this interaction since all CIITA multiple-alanine mutants precipitated RFX5 as well as the wild-type construct (Fig. 3A, lanes 7 and 8, and data not shown). CIITA also bound to RFXANK (Fig. 3B, lane 6), but again this interaction was not affected by CIITA LRR mutagenesis (Fig. 3B, lanes 7 and 8, and data not shown). No interaction between CIITA and RFXAP, the 36-kDa subunit of the RFX complex (12), could be detected (data not shown).
FIG. 3.
Coimmunoprecipitation of CIITA and MHC-II promoter binding proteins. (A) CIITA and RFX5. EGFP-CIITA and Myc-RFX5 were transfected individually or together into HEK293-EBNA cells and immunoprecipitated with an anti-GFP serum. Coprecipitated RFX5 was detected with a myc antibody in a Western blot analysis. Lane 6, cotransfection of CIITA and RFX5; lanes 1 to 5, controls. All CIITA multiple-alanine mutants coprecipitated RFX5 (lanes 7 and 8; data not shown). Lanes 10 and 11, Western blot controls of the material used in lanes 5 and 6, respectively; lane 9, molecular weight standard. (B) CIITA and RFXANK. Interaction between EGFP-CIITA and RFXANK was analyzed as for RFX5 with the exception that unmodified RFXANK was transfected and that an RFXANK-specific polyclonal antiserum was used for detection. Again, all CIITA multiple-alanine mutants coimmunoprecipitated RFXANK (lanes 7 and 8; data not shown). The band detected in the EGFP-CIITA Western blot control (lane 10) migrates below that of RFXANK and is therefore nonspecific. (C and D) CIITA and NF-YB and NF-YC. Immunoprecipitations were carried out here with a GFP-specific serum and Dynabeads. The calculated molecular masses for the various constructs are as follows: Myc6-RFX5, 62 kDa; RFXANK, 29 kDa; Myc6-NF-YB, 33 kDa; Myc6-NF-YC, 47 kDa. Arrowheads, positions of bands of the expected sizes.
The binding of CIITA to the three subunits of NF-Y, NF-YA, YB, and YC, was also tested. Immunoprecipitation was carried out with paramagnetic beads due to background problems of NF-Y precipitations with protein G-agarose (data not shown). Both NF-YB and NF-YC, but not NF-YA, could be coimmunoprecipitated with CIITA (Fig. 3C and D and data not shown). As before, these interactions were not influenced by any of the LRR multiple-alanine mutations (Fig. 3C and D, and data not shown). Interactions of CIITA with TAFs or CREB binding protein (CBP) were not tested. These take place in the N-terminal region of CIITA (14, 15, 26, 28) and were therefore not expected to be influenced by LRR mutagenesis.
CIITA has also been reported to interact with the B-cell-specific coactivator OBF-1 (also called Bob1 and OCA-B) (13, 48). We could not detect any interaction between these two proteins in our coimmunoprecipitation assay (data not shown). Furthermore, we could not detect any cooperativity between OBF-1 and CIITA for HLA-DR induction in cotransfection assays of HEK293-EBNA and HeLa cells, which do not express endogenous OBF-1 (S. B. Hake and V. Steimle, unpublished data).
LRR mutations abolish the dominant-negative phenotype of CIITA mutants.
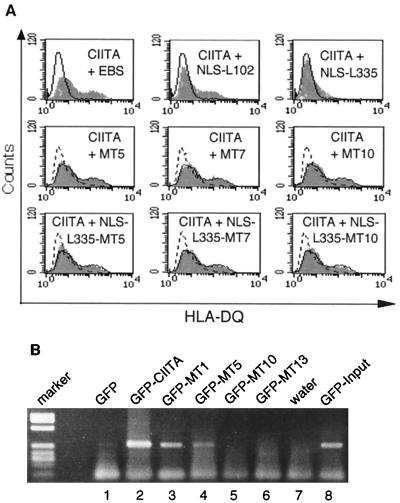
Dominant-negative CIITA mutants presumably act by competing with wild-type CIITA for interaction with MHC-II promoter binding complexes. We wanted to test whether this effect is also impaired by CIITA LRR mutagenesis. CIITA LRR and dominant-negative mutants were cotransfected with equal amounts of wild-type CIITA into HEK293-EBNA cells, and transient HLA class II expression was analyzed by flow cytometry (Fig. 4A). Dominant-negative CIITA mutants affect the mRNA expression of all three class II isotypes equally, but the effect at the cell surface is strongest for HLA-DQ (5). We therefore used an HLA-DQ-specific antibody for the analysis. A comparison with different dominant-negative CIITA mutants, which we had generated earlier (5), showed that even the effect of a weak dominant-negative mutant such as NLS-L102 can be detected in this assay (Fig. 4A, top). None of the multiple-CIITA LRR mutants showed any dominant-negative effect (Fig. 4A, middle, and data not shown). Next, the LRR mutations of CIITA-MT5, -MT7, and -MT10 were introduced into the backbone of the very strongly dominant-negative mutant with N-terminal deletions, NLS-L335 (5), and were tested in the same assay (Fig. 4A, bottom). CIITA LRR mutations completely abolished the dominant-negative phenotype of NLS-L335, indicating that the CIITA LRRs are essential not only for transactivation but also for the dominant-negative function of CIITA.
FIG. 4.
In vivo recruitment of CIITA to the MHC-II promoter. (A) CIITA LRR mutations abolish dominant-negative effect. Plasmids encoding CIITA LRR mutants were cotransfected with equal amounts of wild-type CIITA plasmid in HEK293-EBNA cells, and HLA-DQ expression was analyzed 72 h later. (Top) Open profile, results for cells transfected with empty vector alone; shaded profiles, cotransfection of CIITA wild-type cDNA either with empty vector (left; this profile is used as an overlay for comparison in the middle and bottom portions and is shown there as a solid line), a weak dominant-negative CIITA mutant, NLS-102 (middle), and a strongly dominant-negative mutant, NLS-L335 (right; this profile is shown in an overlay for comparison in the middle and lower portions as a dashed line). (Middle) CIITA-MT5, -MT7, and -MT10 (shaded profiles) were cotransfected with wild-type CIITA and analyzed as described above. (Bottom) The LRR alanine mutations of MT5, MT7, and MT10 (shaded profiles) were introduced into the N-terminal deletion mutant NLS-L335 and analyzed as described above. (B) CIITA LRR mutations impair the in vivo recruitment of CIITA to the HLA-DRA promoter. Transient transfectants of wild-type and LRR-mutated EGFP-CIITA constructs in HEK293-EBNA cells were cross-linked in vivo with formaldehyde, and cross-linked protein-chromatin complexes were immunoprecipitated with GFP-specific monoclonal antibodies. Precipitation of the HLA-DRA promoter was analyzed by PCR amplification.
In vivo recruitment of CIITA to the MHC-II promoter is dependent on the integrity of the CIITA LRRs.
Abolition of the CIITA dominant-negative phenotype by LRR mutagenesis indicated that the LRRs might be essential for recruitment of CIITA to the MHC-II promoter. This hypothesis was tested directly by chromatin immunoprecipitation (34, 35). HEK293-EBNA cells transiently transfected with EGFP or with wild-type or LRR-mutated EGFP-CIITA constructs were cross-linked in vivo with formaldehyde. All transfectants expressed similar amounts of the EGFP-CIITA fusion proteins as shown by GFP fluorescence (data not shown). Sonicated, cross-linked chromatin was immunoprecipitated with GFP-specific monoclonal antibodies, and precipitation of a DRA promoter fragment was revealed by PCR amplification (Fig. 4B). Wild-type EGFP-CIITA coimmunoprecipitated the DRA promoter DNA (Fig. 4B, lane 2) thus demonstrating that CIITA indeed contacts this promoter in vivo. In contrast, promoter binding is strongly reduced or absent in most CIITA LRR mutants (lanes 4 to 6). Only EGFP-MT1 showed a clear, if reduced, association with the promoter (lane 3). In the experiment shown here, EGFP-MT5 (lane 4) also gave rise to a weak DRA promoter signal. In a second immunoprecipitation experiment the signal for EGFP-MT1 was found to be comparable to that for wild-type CIITA, whereas EGFP-MT5-, -MT10-, and -MT13-dependent amplification was at background levels (data not shown). Levels of nonspecific DNA precipitation were analyzed by slot blot hybridization with an Alu repeat probe and were found to be similar in the different samples (data not shown). These experiments demonstrate that CIITA is physically associated with the MHC-II promoter in vivo and that this interaction is dependent on the integrity of the CIITA LRRs.
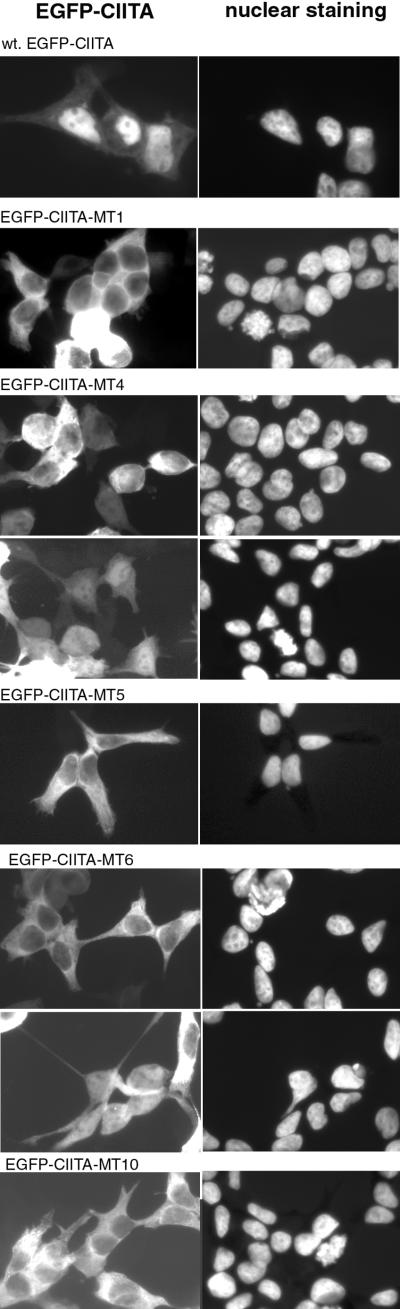
LRR function is essential for nuclear localization of CIITA.
The preceding experiments had led to an apparent contradiction: on the one hand CIITA LRRs were found to be essential for transactivation, dominant-negative function, and in vivo promoter occupation; on the other hand the multiple interactions of CIITA with MHC-II promoter binding proteins were not influenced by LRR mutagenesis. These observations could best be explained by a function of the CIITA LRRs which is only indirectly associated with promoter occupation, for example, nuclear translocation. Wild-type or LRR-mutated EGFP-CIITA constructs were therefore transfected into HEK293-EBNA cells and analyzed 48 h later by fluorescence microscopy (Fig. 5). As expected (11, 19), wild-type CIITA showed a mostly nuclear staining. In contrast, all multiple-LRR mutants showed impaired nuclear translocation, whether they were still functional or not (Fig. 5, and data not shown). The transactivation-competent mutants EGFP-CIITA-MT4 and -MT6 displayed clear nuclear staining when cells with the lowest detectable GFP expression levels were analyzed (Fig. 5), but the other two transactivation-competent mutants (EGFP-CIITA-MT8 and -MT9) were indistinguishable from the nonfunctional mutants in this respect. These results indicate that the CIITA LRRs are important for nuclear localization of the protein. However, when mutant EGFP-MT1, which was still recruited to the promoter despite having lost its transactivation potential, is taken into consideration, it is most likely that the LRRs are also of functional importance for CIITA bound to the MHC-II promoter. The localization data shown here were found to be reproducible in numerous experiments and were also confirmed with Vero and COS cells (data not shown). Mutants EGFP-MT2 and EGFP-MT11 were not analyzed because of their instability (Fig. 6).
FIG. 5.
Subcellular localization of CIITA LRR mutants. HEK293-EBNA cells were transfected with wild-type (wt) or LRR-mutated EGFP-CIITA constructs as indicated and analyzed 48 h later. Left, GFP fluorescence; right, nuclear staining with bis-benzimidine. The lower images of EGFP-MT4 and -MT6 show cells with a very low level of GFP fluorescence.
Coimmunoprecipitation of a CIITA LRR-associated protein from in vivo-labeled extracts.
The coimmunoprecipitation data and fluorescence microscopy experiments had indicated that the CIITA LRRs are involved in a function distinct from promoter binding. We therefore wanted to see whether the CIITA LRRs bind to as yet unknown proteins and whether this interaction is sensitive to LRR mutagenesis. HEK293-EBNA cells transfected with wild-type or LRR-mutated EGFP-CIITA constructs were labeled metabolically with [35S]methionine, and the CIITA proteins were immunoprecipitated via the EGFP epitope tag (Fig. 6).
Immunoprecipitations were resolved by 7.5 or 15% SDS– PAGE and analyzed by autoradiography. Figure 6A shows a 15% gel in which a CIITA-dependent immunoprecipitated protein band of 33 kDa is visible. Several additional bands are detectable in the higher-molecular-weight range, but these are present in cells expressing either wild-type CIITA or LRR alanine mutants. Resolving these immunoprecipitations on 7.5% gels did not lead to the detection of additional, differentially associated bands (data not shown). The 33-kDa protein was coimmunoprecipitated with the nonfunctional mutant EGFP-MT1 (Fig. 6A, lane 3) and, with somewhat reduced efficiency, with the still-functional CIITA LRR mutants EGFP-MT4, -MT6, -MT8, and -MT9 (Fig. 6B). On the other hand, precipitation of p33 is not at all or very faintly detectable in cells transfected with the other nonfunctional mutants and in cells transfected with EGFP alone (Fig. 6A), thus establishing a strong correlation between p33 binding and CIITA function.
DISCUSSION
In the initial sequence analysis of CIITA we had detected a weak homology in the C-terminal part to Rna1, a yeast protein rich in leucines (45). Kobe and Deisenhofer (25) noted that this region of CIITA contains two LRRs, thus identifying what we now recognize as CIITA LRRs 2 and 3 (Fig. 1). LRRs 1 and 4, which do not fit the LRR consensus sequence perfectly, were identified in a dot plot comparison of the CIITA sequence with itself (data not shown). A first indication that this region of CIITA is functionally important came from the analysis of a CIITA allele in the CIITA-deficient patient cell line BCH (4), in which most of LRR 4 of CIITA is deleted (amino acids 1079 to 1106). Both this mutant and one with a complete C-terminal deletion of the LRRs (from position 980) have no transactivation or dominant-negative potential and abolish the dominant-negative phenotype of CIITA constructs with N-terminal deletions (4, 5).
These observations encouraged us to examine the LRR-containing region of CIITA by alanine scanning mutagenesis. Analysis of the RI contact positions in the RI-RNase A complex (25) allowed us to identify potential consensus LRR contact positions (Fig. 1A). The complete loss of function of nine multiple-CIITA LRR alanine mutants indicated the functional importance of all four loop regions and of the β-strands of repeats 2 and 4 (Fig. 1B and 2A). Mutants with single-alanine mutations of the corresponding positions led to the identification of seven functionally critical residues (Fig. 1C and 2B). Five of these mutations are located at positions 1 and 3 of a conserved IxD motif in the loop regions (Fig. 1C). The locations of functionally critical CIITA LRR positions strongly suggest that CIITA is a bona fide LRR protein.
Our findings may have wider implications for the structure and function analysis of other LRR proteins. Papageorgiou and colleagues (37) analyzed the structure of the complex between human RI (hRI) and angiogenin (Ang), another member of the pancreatic RNase superfamily of proteins which are bound by RI. While 10 out of 26 RI contact positions differ in the hRI-Ang complex from those in the porcine RI-RNase A complex, they all are located within the consensus binding positions identified here. The fact that single alanine mutations can completely abolish protein-protein interaction is not astonishing. It has been shown that in most protein-protein interactions only very few contact positions, so-called hot spots, account for the majority of the binding energy (3, 10). Strategies similar to ours may be useful for the identification of LRR hot spots in other LRR proteins.
Through its overall control of MHC-II expression CIITA has a strong impact on the cellular immune response. The CIITA LRR hot spots identified here provide now precise target sites for the development of agents that inhibit MHC-II expression and thus suppress the immune response.
LRRs are known as protein-protein interaction domains; therefore we wanted to know whether CIITA interacts with MHC-II promoter binding proteins directly and whether the CIITA LRRs play a role in these interactions. We could detect coimmunoprecipitation of CIITA not only with RFX5 as shown earlier (42) but also for the first time with RXANK and with the NF-Y subunits NF-YB and NF-YC (Fig. 3). While the interactions demonstrated here are specific as shown by the stringent controls in Fig. 3, lanes 1 to 5, they are quite weak and could only be shown through overexpression of both interaction partners (data not shown). Indeed, at physiological levels of protein expression these interactions could not be demonstrated individually, but only in the form of a DNA-bound multiprotein complex (31). We could not detect any interaction of CIITA with RFXAP or NF-YA or with OBF-1 (Bob1, OCA-B), a factor which had been reported earlier to interact with CIITA (13). The fact that we do not observe indirect binding of RFXAP or NF-YA to CIITA via the two other subunits of the respective complexes is a strong indication that we are detecting direct protein-protein interactions in our coimmunoprecipitation system.
The importance of the LRRs for recruitment of CIITA to the MHC-II promoter was demonstrated indirectly through abolition of the CIITA dominant-negative function and directly through chromatin immunoprecipitation (Fig. 4). CIITA LRR alanine mutants showed no dominant-negative phenotype on their own. More importantly, the introduction of the corresponding LRR mutations into a strongly dominant-negative CIITA mutant (NLS-L335) completely abolished its dominant-negative phenotype (Fig. 4A). Chromatin immunoprecipitation experiments with in vivo-cross-linked CIITA transfectants demonstrated that wild-type CIITA is efficiently recruited to the MHC-II promoter (Fig. 4B). This interaction is severely reduced or absent for most of the nonfunctional mutants. Only MT1 behaves differently and is recruited to the HLA-DRA promoter to a substantial degree. Immune fluorescence analysis of chimeric EGFP-CIITA constructs demonstrated that the LRRs are important for the subcellular localization of CIITA. All EGFP-CIITA LRR multiple-alanine mutants showed impaired nuclear translocalization (Fig. 5). Metabolic labeling and immunoprecipitation identified a 33-kDa protein (p33) that associates with CIITA in an LRR-dependent manner (Fig. 6).
One of the main functions of the CIITA LRRs appears therefore to be nuclear import or localization of CIITA. Three distinct sequence elements which are important for nuclear localization have now been identified in CIITA, though none of them appears to be a classical nuclear localization signal (NLS). A five-amino-acid RDLKK motif just N-terminal of the LRRs (amino acid positions 955 to 959) can confer nuclear localization to GFP but cannot be replaced by a SV40 NLS, nor can it be transferred to another position in CIITA (11). Both features are hallmarks of the classical NLS. It has recently been shown that the GTP-binding domain of CIITA is also necessary for nuclear localization (19). GTP-binding proteins are essential for nucleocytoplasmic transport, and GTP hydrolysis is normally carried out by small GTP-binding proteins such as Ran (17). Our findings that the CIITA LRRs are also important for nuclear localization demonstrate that subcellular localization is even more complex than previously thought and involves regions spanning more than half of the protein. It is intriguing that one of the main Ran-binding proteins, RanGAP, binds to Ran through an LRR domain with a three-dimensional structure very similar to that of the RI (20). Indeed Rna1, the first protein discovered with homology to CIITA, has been identified as the RanGAP of yeast (2, 45). Finding two such elements involved in nucleocytoplasmic transport in the same molecule is highly unusual and points to a novel mechanism.
The situation becomes yet more complex when the nonfunctional mutant MT1 and the transactivation-competent mutants MT4, MT6, MT8, and MT9 are taken into consideration. MT1 differs from the other multiple-alanine mutants by targeting mainly residues lying outside of our consensus positions at the N-terminal end of the β-strand of LRR 4 (Fig. 1B). These residues were mutated because of their homology with the LRVxx motif at the very C-terminal end of the RI (Fig. 1A). While MT1 behaves like the other mutants in terms of transactivation, direct protein-protein interactions with promoter binding complexes, and dominant-negative effect, it shows substantial in vivo recruitment to the MHC-II promoter and coimmunoprecipitates p33 (Fig. 2 to 5).
Astonishingly, EGFP-MT1 and the functional mutants EGFP-MT4, -MT6, -MT8, and -MT9 are also excluded from the nucleus (Fig. 5). This result can best be explained by overexpression, which is necessary for the fluorescence microscopy experiments. With the help of the EGFP-CIITA chimeras we could show clearly that amounts of CIITA which are undetectable by FACS or by fluorescence microscopy are sufficient to induce substantial MHC-II expression (data not shown). In view of their phenotype, it is not astonishing that the functional mutants MT4, MT6, MT8, and MT9 are efficiently recruited to the MHC-II promoter in vivo (data not shown). MT4 and MT6 show clear nuclear staining at the lowest levels of GFP fluorescence that can be distinguished from background fluorescence, but all other mutants, including MT1 and the other two functional mutants, MT8 and MT9, are indistinguishable. It has to be assumed, therefore, that the amounts of CIITA necessary for efficient MHC-II transactivation are below the level of detection. Indeed, to date no laboratory has been able to demonstrate localization of CIITA without substantial overexpression (11, 19).
We can thus distinguish several levels of function in which the CIITA LRRs are important; all LRR mutants are impaired in nuclear localization as discussed above. Most nonfunctional mutants fail to bind to the MHC-II promoter in vivo (such as EGFP-MT5, -MT10, and -MT13; Fig. 4B), most probably due to the altered subcellular localization. EGFP-MT1 binds to the promoter in vivo (Fig. 4B) and interacts with p33 but is completely inactive in terms of transactivation (Fig. 2A). The existence of the CIITA LRR mutant MT1 is a strong indication that the CIITA LRRs are important for nuclear translocation and also play a more direct role in transactivation.
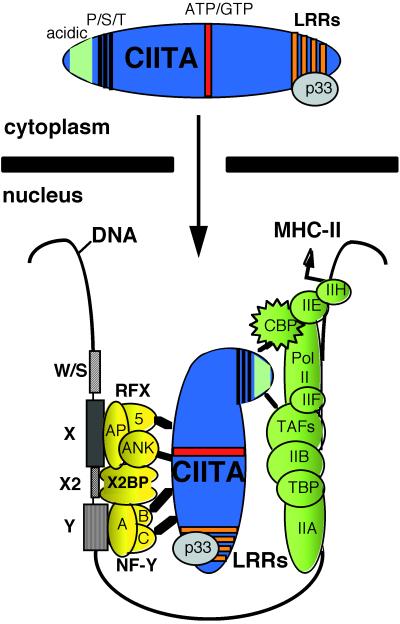
Figure 7 summarizes our findings. CIITA enters the nucleus through a mechanism which is dependent on the CIITA LRRs. A strong correlation between the binding of p33 to the CIITA LRRs, promoter recruitment, and transactivation exists. At the MHC-II promoter CIITA is essential for the formation of the MHC-II “transcriptosome.” In this case it is a split transcriptosome. At least in B cells the MHC-II promoter binding proteins are capable of forming an enhanceosome-like structure through cooperative interaction even in the absence of CIITA (reviewed in reference 39). Yet, despite the fact that at least some of the DNA binding factors contain potential transcriptional activation domains or can interact with other coactivators (29), this complex is completely inert in the absence of CIITA. Rather, the MHC-II promoter binding complex forms a landing pad for CIITA, which interacts with it through at least four discrete interactions, three of which have been shown here for the first time. CIITA bridges the silent enhanceosome and the basal transcription machinery through these multiple protein-protein interactions and thus activates transcription. The CIITA LRRs are critical for nuclear localization, promoter recruitment, and transactivation. Whether these are separate functions or different facets of the same function remains to be seen.
FIG. 7.
LRR-dependent nuclear localization of CIITA and MHC-II transactivation through multiple protein-protein interactions. CBP, CREB binding protein; TBP, TATA-binding protein; TAFs, TBP-associated factors.
ACKNOWLEDGMENTS
We thank R. Accolla (Verona, Italy) R. Rupp (Tübingen, Germany), and M. Strubin (Geneva, Switzerland) for cells, reagents, and plasmids and J. Wells and P. Farnham (University of Wisconsin) for providing us with a ChIP protocol. We thank Felix Schnappauf for help with some experiments, and Hubertus Kohler for help with the FACS analyses, Séverine Bontron, Rose Brugger, Stuart Clarkson, Lisa Denzin, Michael Reth, and Michel Strubin for critical reading of the manuscript and for discussion, and Hans-Ulrich Weltzien and the Steimle laboratory for moral support and help in many ways.
REFERENCES
- 1.Accolla R S. Human B cell variants immunoselected against a single Ia antigen subset have lost expression of several Ia antigen subsets. J Exp Med. 1983;157:1053–1058. doi: 10.1084/jem.157.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becker J, Melchior F, Gerke V, Bischoff F R, Ponstingl H, Wittinghofer A. RNA1 encodes a GTPase-activating protein specific for Gsp1p, the Ran/TC4 homologue of Saccharomyces cerevisiae. J Biol Chem. 1995;270:11860–11865. doi: 10.1074/jbc.270.20.11860. [DOI] [PubMed] [Google Scholar]
- 3.Bogan A A, Thorn K S. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 4.Bontron S, Steimle V, Ucla C, Eibl M M, Mach B. Two novel mutations in the MHC class II transactivator CIITA in a second patient from MHC class II deficiency complementation group A. Hum Genet. 1997;99:541–546. doi: 10.1007/s004390050403. [DOI] [PubMed] [Google Scholar]
- 5.Bontron S, Ucla C, Mach B, Steimle V. Efficient repression of endogenous major histocompatibility complex class II expression through dominant negative CIITA mutants isolated by a functional selection strategy. Mol Cell Biol. 1997;17:4249–4258. doi: 10.1128/mcb.17.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chang C H, Flavell R A. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med. 1995;181:765–767. doi: 10.1084/jem.181.2.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chin K C, Li G G, Ting J P. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chin K C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P. Molecular analysis of G1B and G3A IFN gamma mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii gene induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 10.Clackson T, Wells J A. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 11.Cressman D E, Chin K C, Taxman D J, Ting J P. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 12.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex, is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fontes J D, Jabrane-Ferrat N, Toth C R, Peterlin B M. Binding and cooperative interactions between two B cell-specific transcriptional coactivators. J Exp Med. 1996;183:2517–2521. doi: 10.1084/jem.183.6.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontes J D, Jiang B, Peterlin B M. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 17.Gorlich D. Transport into and out of the cell nucleus. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griscelli C, Lisowska-Grospierre B, Mach B. Combined immunodeficiency with defective expression in MHC class II genes. Immunodefic Rev. 1989;1:135–153. [PubMed] [Google Scholar]
- 19.Harton J A, Cressman D E, Chin K C, Der C J, Ting J P. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 20.Hillig R C, Renault L, Vetter I R, Drell I T, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 21.Kern I, Steimle V, Siegrist C A, Mach B. The two novel MHC class II transactivators RFX5 and CIITA both control expression of HLA-DM genes. Int Immunol. 1995;7:1295–1299. doi: 10.1093/intimm/7.8.1295. [DOI] [PubMed] [Google Scholar]
- 22.Kobe B, Deisenhofer J. Crystal-structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 23.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 24.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 25.Kobe B, Deisenhofer J. A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature. 1995;374:183–186. doi: 10.1038/374183a0. [DOI] [PubMed] [Google Scholar]
- 26.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 28.Mahanta S K, Scholl T, Yang F C, Strominger J L. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci USA. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maity S N, de Crombrugghe B. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem Sci. 1998;23:174–178. doi: 10.1016/s0968-0004(98)01201-8. [DOI] [PubMed] [Google Scholar]
- 30.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 31.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlethaler-Mottet A, Otten L A, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 34.Orlando V, Paro R. Mapping polycomb-repressed domains in the bithorax complex using in-Vivo Formaldehyde Cross-Linked Chromatin. Cell. 1993;75:1187–1198. doi: 10.1016/0092-8674(93)90328-n. [DOI] [PubMed] [Google Scholar]
- 35.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 36.Otten L A, Steimle V, Bontron S, Mach B. Quantitative control of MHC class II expression by the transactivator CIITA. Eur J Immunol. 1998;28:473–478. doi: 10.1002/(SICI)1521-4141(199802)28:02<473::AID-IMMU473>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 37.Papageorgiou A C, Shapiro R, Acharya K R. Molecular recognition of human angiogenin by placental ribonuclease inhibitor—an X-ray crystallographic study at 2.0 angstrom resolution. EMBO J. 1997;16:5162–5177. doi: 10.1093/emboj/16.17.5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Reith W, Satola S, Sanchez C H, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 39.Reith W, Steimle V, Mach B. Molecular defects in the bare lymphocyte syndrome and regulation of MHC class II genes. Immunol Today. 1995;16:539–546. doi: 10.1016/0167-5699(95)80048-4. [DOI] [PubMed] [Google Scholar]
- 40.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 41.Rupp R A, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 42.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Silacci P, Mottet A, Steimle V, Reith W, Mach B. Developmental extinction of major histocompatibility complex class II gene expression in plasmocytes is mediated by silencing of the transactivator gene CIITA. J Exp Med. 1994;180:1329–1336. doi: 10.1084/jem.180.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 45.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 46.Steimle V, Reith W, Mach B. Major histocompatibility complex class II deficiency: a disease of gene regulation. Adv Immunol. 1996;61:327–340. doi: 10.1016/s0065-2776(08)60870-6. [DOI] [PubMed] [Google Scholar]
- 47.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 48.Strubin M, Newell J W, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 49.Takebe Y, Seiki M, Fujisawa J-I, Hoy P, Yokota K, Arai K-I, Yoshida M, Arai N. SRα promoter: an efficient and versatile mammalian cDNA expression system composed of the simian virus 40 early promoter and the R-U5 segment of human T-cell leukemia virus type 1 long terminal repeat. Mol Cell Biol. 1988;8:466–472. doi: 10.1128/mcb.8.1.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 51.Zhou H, Su H S, Zhang X, Douhan III J, Glimcher L H. CIITA-dependent and -independent class II MHC expression revealed by a dominant negative mutant. J Immunol. 1997;158:4741–4749. [PubMed] [Google Scholar]