Abstract
Parvoviruses and especially the adeno-associated virus (AAV) species provide an exciting and versatile platform for the rational design or molecular evolution of human gene-therapy vectors, documented by literature from over half a century, hundreds of clinical trials, and the recent commercialization of multiple AAV gene therapeutics. For the last three decades, the power of these vectors has been further potentiated through various types of hybrid vectors created by intra- or inter-genus juxtaposition of viral DNA and protein cis elements or by synergistic complementation of parvoviral features with those of heterologous, prokaryotic, or eukaryotic viruses. Here, we provide an overview of the history and promise of this rapidly expanding field of hybrid parvoviral gene-therapy vectors, starting with early generations of chimeric particles composed of a recombinant AAV genome encapsidated in shells of synthetic AAVs or of adeno-, herpes-, baculo-, or protoparvoviruses. We then dedicate our attention to two newer, highly promising types of hybrid vectors created via (1) pseudotyping of AAV genomes with bocaviral serotypes and capsid mutants or (2) packaging of AAV DNA into, or tethering of entire vector particles to, bacteriophages. Finally, we conclude with an outlook summarizing critical requirements and improvements toward clinical translation of these original concepts.
Keywords: AAV, adeno-associated virus, bacteriophage, bocavirus, parvovirus
Graphical abstract
Gene transfer vectors based on parvoviruses hold enormous potential for human gene therapy. Here, Fakhiri and Grimm provide an overview of exciting, recently reported strategies to further increase this potential by combining features from multiple parvoviruses in a single vector or by juxtaposing parvoviruses with heterologous viruses such as bacteriophages.
Introduction
Within half a century, parvoviruses, including the adeno-associated virus (AAV) species, have evolved from a biologically interesting observation into a powerhouse and key driver in human gene therapy, for which its potential has been showcased in numerous preclinical and clinical studies all over the world. Whereas the reasons for this impressive success story are manifold, the greatest assets of these viruses with respect to their development and use as recombinant gene-therapy vectors for DNA delivery may be their small size, their simple genetic composition and structure, and the high degree of flexibility and amenability of genome and capsid to genetic engineering. Further enhancing their versatility is that both wild-type (WT) and synthetic viral components can be juxtaposed in a plug-and-play fashion, including packaging of genomes from one viral genotype into the capsid of another serotype (“pseudotyping”); cross-genera combinations of DNA, protein, and capsid; encapsidation of parvoviral DNA into further heterologous viruses; or biochemical coupling (“piggy-backing”) of transgene-encoding AAV particles to prokaryotic viruses. Surprisingly, although the concept of pseudotyping is widely known, well studied, and reported in a vast amount of original literature and review articles, hybrid parvoviral vectors have yet to gain the same popularity, despite their undisputed potential and the rapidly accumulating evidence of their promise as an alternative or add-on to more conventional vector designs.
Here, we aim to fill in this critical gap by providing an overview of the multitude of concepts for the creation of hybrid parvoviral vectors that have been reported to date, starting with a short history of genuine parvoviral vectors and traditional pseudotyping approaches before focusing on the latest original avenues and critically discussing their clinical potential. Because of the complexity of the topic and the wealth of superb reviews dedicated to parvoviral vector biology and traditional technology, we refer to this prior literature for general aspects and background information.1, 2, 3, 4, 5, 6 Here, we will rather emphasize other facets that we deem underappreciated and concurrently very promising for the future of the field, comprising innovative combinations of different AAV elements with those of mammalian bocaviruses (BoVs) and prokaryotic bacteriophages.
A brief history of genuine parvoviral vectors
When the labs of Rowe,7 Hammon,8 and Rapp9 simultaneously reported their discovery of tiny icosahedral particles in adenovirus (AdV) preparations from human and simian samples in the 1960s, no one anticipated that decades later, this would lay the foundation for a unique class of biotherapeutics, namely, recombinant AAVs (rAAVs). Around the same time, Toolan and Ledinko10 described another new (at the time) virus, called H1, that can infect and grow in human and simian cells and that would eventually be of particular promise in cancer therapy. Much more recently, in 2005, a seminal third discovery that would shape gene-therapy research was reported by Allander and colleagues11 who had cloned another small virus, nowadays known as human BoV 1 (HBoV1), from human nasopharyngeal aspirates. Today, over 50 years after the initial report of AAV, we know that all three of these viruses, AAV, H1, and HBoV1, are members of the virus family Parvoviridae and the same subfamily Parvovirinae, which comprise eight genera with >50 species that can all infect vertebrates, i.e., mammals, birds, and reptiles. In contrast, Densovirinae, the only other Parvovirinae subfamily, infect invertebrates. More specifically, AAV, together with the goose parvovirus and the Muscovy duck parvovirus, belongs to the genus dependoparvovirus, which reflects a peculiarity of the life cycle of these viruses in that they depend on co-infection with a second, unrelated helper virus for their propagation (the name-giving AdV in the case of AAV, as discovered by Atchison et al.8 and Hoggan et al.7). However, although goose and Muscovy duck parvovirus are pathogenic12,13 and share features with autonomously replicating parvoviruses (APVs; see below),14 AAV is believed to be apathogenic, including historic evidence that newborn mice and hamsters showed no pathologies 2 months post-AAV injection via different routes of administration.8 Nonetheless, we note recently emerging, conflicting evidence of a potential contribution of WT AAV integration to the formation and maintenance of hepatocellular carcinoma in humans, which remains the topic of intense and ongoing research.15, 16, 17
In contrast to AAV, H1 and HBoV1 are APVs, which comprise a total of seven genera: protoparvovirus (H1 and others), bocaparvovirus (HBoV1 and others), erythroparvovirus, amdoparvovirus, aveparvovirus, copiparvovirus, and tetraparvovirus.18 The name protoparvovirus is derived from the Greek term “proto” for “first,” as these viruses belong to the first identified and described parvoviruses,19 whereas bocaparvovirus stems from the hosts of the genus founders discovered more than four decades ago, (bo)vine parvovirus and minute virus of (ca)nines.20, 21, 22 Unlike AAV, these viruses can independently replicate their genomes in target cells. Another feature of APVs that distinguishes them from AAV is that they are lytic viruses that eventually kill the infected cell. Despite their independence of extra viral genes, APVs strictly require cellular factors expressed during the cellular S-phase and hence preferentially infect dividing cells.23, 24, 25 One notable exception is HBoV1, which can replicate in differentiated, quiescent primary human airway epithelial (pHAE) cells.26 Still, work, especially from the Rommelaere lab,27, 28, 29 showed that active cellular proliferation is not enough for efficient replication but that it is rather neoplastic transformation that fosters APV infection and cytotoxicity. This implies that transformation with oncogenic agents provides a unique environment for APV replication that is both cell-type and oncogene dependent,30,31 an observation supporting the historical notion that these parvoviruses act as tumor suppressors via their preferred replication in, and lysis of, tumor cells.10,32 Interestingly, also WT AAV was shown to provide anti-tumor effects in certain cancers by either directly reducing the expression of oncogenes33 or by interfering with oncogenic viruses, such as the human papillomavirus type 16 (HPV16).34
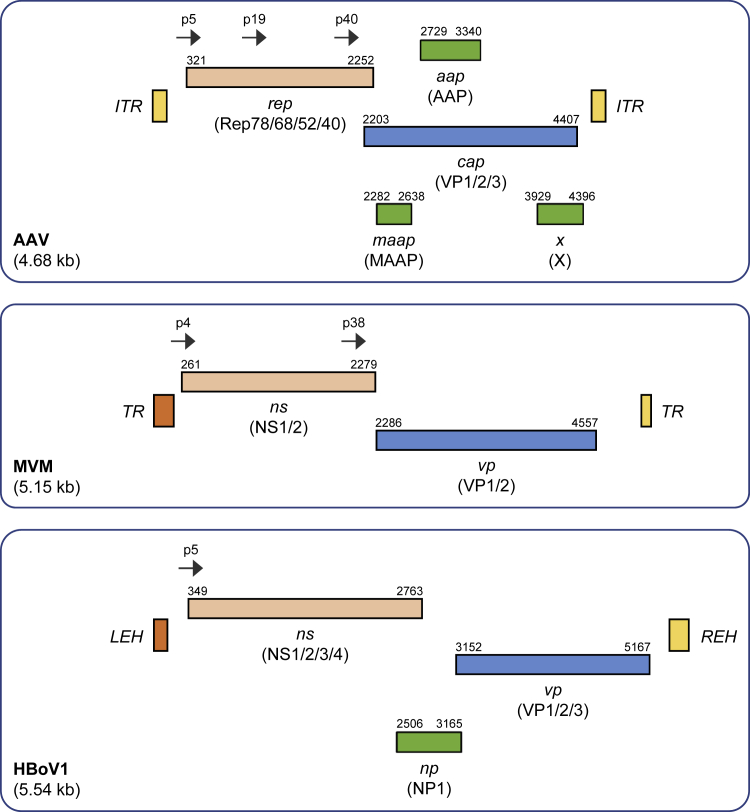
Despite the distinct differences in the life cycles among the eight genera of the Parvovirinae subfamily, a common feature that explains their enormous popularity in the gene and cell-therapy communities is their relatively simple structure (Figure 1), which in turn facilitates their engineering as recombinant, authentic, or hybrid parvoviral vectors. Typically, all parvoviruses are composed of a small (hence the name from Latin “parvus” = small), non-enveloped, T = 1 icosahedral capsid containing a single-stranded (ss), ∼4 to 6 kilobase (kb) DNA genome of plus or minus polarity. This genome encodes non-structural (NS or Rep), capsid or virion (Cap or viral protein [VP]), as well as accessory proteins, such as AAP35 or MAAP36 in the case of AAV or nuclear phosphoprotein (NP) in the case of HBoV1. Notably, whereas the AAV p5 promoter is regulated by AdV and only weakly active or silent in its absence,37,38 the p4 promoter in protoparvoviruses is constitutively active in target cells and transcribes the cytotoxic NS proteins. The latter explains why protoparvoviruses have already been studied extensively and utilized as vectors,18 especially the minute virus of mice (MVM), the rat parvovirus H1 (both rodent parvoviruses), and the closely related LuIII (whose natural host remains unknown). When engineered as gene-delivery vehicles, they combine several attractive features: (1) ability to infect a variety of cell types, (2) specificity for transformed cells,39 and (3) safety, as they are apathogenic in humans.40 In general, two main types of genuine protoparvoviral vectors have been developed to date, i.e., (1) cap replacement or (2) “gutted” vectors. In the first type, the ns open reading frame (ORF) is kept, whereas the cap ORF is partly deleted and replaced by a transgene of interest. The underlying idea is to exploit the cancer-specific and cytotoxic properties of the NS proteins and to enhance these by adding small transgene cassettes encoding immunostimulatory cytokines, e.g., interleukin (IL)-241 or cytotoxic genes, such as the herpes simplex virus (HSV) thymidine kinase.42 Indeed, these engineered protoparvoviral vectors can trigger more potent anti-neoplastic effects than the parental WT viruses.41 Such replication-competent, native, or engineered protoparvoviral vectors are nowadays applied to target a variety of cancer types, such as glioblastoma (ClinicalTrials.gov: NCT01301430), pancreatic carcinoma,43, 44, 45 or lymphoma.46
Figure 1.
Genome organization of three prototypes of Parvoviridae
Shown are (from top to bottom) schemes of the genomes of wild-type AAV, MVM, and HBoV1, as representatives of the main species of parvoviruses that are often used as the basis for the creation of hybrid parvoviral vectors. Numbers denote nucleotide positions within each genome. All elements (except for promoters; shown as arrows) are drawn to scale. ORFs are shown in italics and proteins in parentheses. For clarity, not all protein species are depicted, and introns have been omitted (e.g., AAV encodes four different Rep proteins that are produced from two promoters and via alternative splicing). The information used to generate these schemes was gathered from multiple sources including original literature (e.g., Srivastava et al.47) and the NCBI database (https://www.ncbi.nlm.nih.gov/).
Notably, although all protoparvoviruses currently under investigation are derived from rodents, three human variants have been found recently—bufavirus (BuV), tusavirus (TuV), and cutavirus (CuV)48—which may have adapted to human tissue and hence are worth studying in the future.
Next to the use in cancer therapy and vaccination,32,49 protoparvoviruses have also been engineered as gutted or “gutless” vectors that have all viral genes removed and replaced by transgenes of interest, with only the terminal repeats (TRs) remaining as the sole mandatory element for production of vectors. Importantly, since these vectors lack the ns ORF, they lose their ability to replicate in, and lyse, tumor cells. For instance, as one of the first, the Maxwell lab50 demonstrated the packaging of a recombinant LuIII genome encoding a firefly luciferase or lacZ transgene into the capsid of LuIII. As expected, this vector mediated robust but transient gene expression in a variety of human cell lines, including HeLa and HepG2.50 In these gutted vectors, the constitutive viral p4 promoter is typically replaced by heterologous, tissue-specific, or inducible promoters51 that allow restriction or control of transgene expression, respectively.
In principle, the two identical designs were also applied to, and tested with, AAV vectors soon after the cloning of the AAV serotype 2 (AAV2) genome in the early 1980s.52,53 Similar to the predominant form of APVs, the first AAV vectors maintained an intact rep ORF and were thus denoted as “rep+ cap−.”54,55 Due to the expression of the Rep proteins, these vectors are capable of site specifically integrating into the human genome at a defined locus (AAVS1 on the long arm of chromosome 19).56 However, this feature was never fully validated and explored, mostly because of the already limited cargo size of AAV of less than 5 kb of foreign DNA that restricts its use to smaller transgenes and because the inverted TRs (ITRs) are the only viral element required for vector DNA replication and packaging in cis.57 Accordingly, rep− cap− (i.e., gutless) vectors that can transiently transduce a large variety of target cells but lost their ability to site specifically integrate into the cellular chromosome were established and have become the prime AAV vector design, which is also used for the approved AAV gene therapeutics Glybera, Luxturna, and Zolgensma.57,58
Notably, conventional AAV vectors deliver a ss recombinant genome, which has to undergo second-strand DNA synthesis or annealing of two strands of opposite polarity in order to form transcription-competent double-stranded DNA in the transduced cells. Because this process can be slow and rate limiting, McCarty et al.59 from the Samulski lab have devised a so-called self-complementary (sc) or double-stranded AAV vector design in which one of the ITRs carries a small truncation that results in arrest of AAV DNA replication during vector production and packaging as a genome carrying an inverted copy of the transgene. In the transduced cells, the two copies then quickly self-anneal into expression-competent, double-stranded DNA in the absence of the aforementioned processes, explaining the faster expression kinetics often observed with these scAAV versus genuine ssAAV vectors. However, owing to the necessity to encapsidate two copies of the same transgene, packaging capacity is reduced to ∼2.4 kb of foreign DNA, which limits the use of scAAV vectors to smaller transgene expression cassettes such as those encoding short cDNA, short hairpin (sh)RNAs, or guide (g)RNAs.
Finally, also important to mention in this context is that gutless vectors differ from WT viruses in several aspects of packaging and transduction. First, vectors display a higher ratio of total viral particles to infectious units (vp:iu) than their WT counterpart. ius are the particles that enter a cell and yield efficient replication in the presence of AdV co-infection (WT viruses or recombinant vectors). For example, the infectivity of LuIII vectors is only one-tenth of the WT counterpart, resulting in a vp:iu ratio of 5,000−10,000:1 versus ∼800:1 for the WT.1,50 rAAV vectors show a similar trend, but the overall vp:iu ratio was often estimated to be lower (e.g., vp:iu ∼1:1 for WTAAV2 versus only 53−124:1 for rAAV260), a notion that further illustrates the particularly high promise of this viral vector system. Importantly, vp:iu ratios can vary depending on the type of analyzed samples (e.g., in cell lysates,61 ratios of 1:40 and 1:1.800 were reported for WTAAV2 and rAAV2, respectively, versus 1:1 for iodixanol-purified virus60), the cell types used for transduction/infection,62 abundance of host factors such as receptors, the analyzed time points,63 and the setting (in vitro or in vivo64). Also, the lack of robust biopotency assays for gutless vectors derived from protoparvoviruses hampers a direct comparison to those based on dependoparvoviruses. Thus, the higher vp:iu ratio found in protoparvoviruses might mainly result from differences in assay setup. Alternatively, biological differences may exist that impact vector performance such as a higher prevalence of empty capsids in vector stocks and/or more stringent requirements for genome:capsid interactions (as observed for canine parvovirus [CPV],65,66 immunosuppressive strain of MVM [MVMi],67 and H168). Finally, as mentioned above, the rodent protoparvoviruses that were studied as vectors so far might not infect/transduce human cells as well as the human-derived ones.
Besides the different origins and types of the parvoviral vectors, it was shown that common post-packaging and -entry barriers to transduction exist. For example, vectors based on AAV (dependoparvovirus) and BoV (bocaparvovirus) were shown to be targets of proteasomal degradation in the cytoplasm.69,70 Inhibition of proteasome activity by either chemical reagents70, 71, 72 or mutation of capsid surface residues amenable to ubiquitination73 yielded an increase in viral genome accumulation in the nucleus and higher transgene expression. Moreover, recent studies imply a direct role for the AAV capsid in promoting transcription of the viral genome after uncoating and second-strand synthesis.74,75 The effect was linked to highly conserved residues in the so-called “dead zone” of the viral capsid, as mutations in this region or physical blocking by the A20 antibody abolished transgene mRNA synthesis. Albeit the exact mechanisms remain unknown, these studies highlight the importance of post-nuclear entry steps in the processing and efficient expression of viral vector. Support comes from a recent study by Powell et al.,76 who linked a region on the VP1/VP2 interface of the AAV9 capsid to the differential expression from certain promoters. For example, when an AAV9 vector carrying a full-length chicken β-actin promoter was infused into the rat striatum, it was predominantly expressed in neurons. Yet, when six glutamate residues were inserted at the VP1/VP2 interface after amino acid 138, transgene expression shifted toward oligodendrocytes. By contrast, insertion of an alanine stretch had no effect on the expression pattern. This observation of a direct influence of a capsid region on promoter expression in a cell-type-dependent fashion is an intriguing new aspect, which again highlights the complexity of parvovirus biology and suggests that further research into the mechanisms will eventually lead to the development of better viral vectors.
Hybrid AAV vector systems based on AdV, HSV, and baculovirus
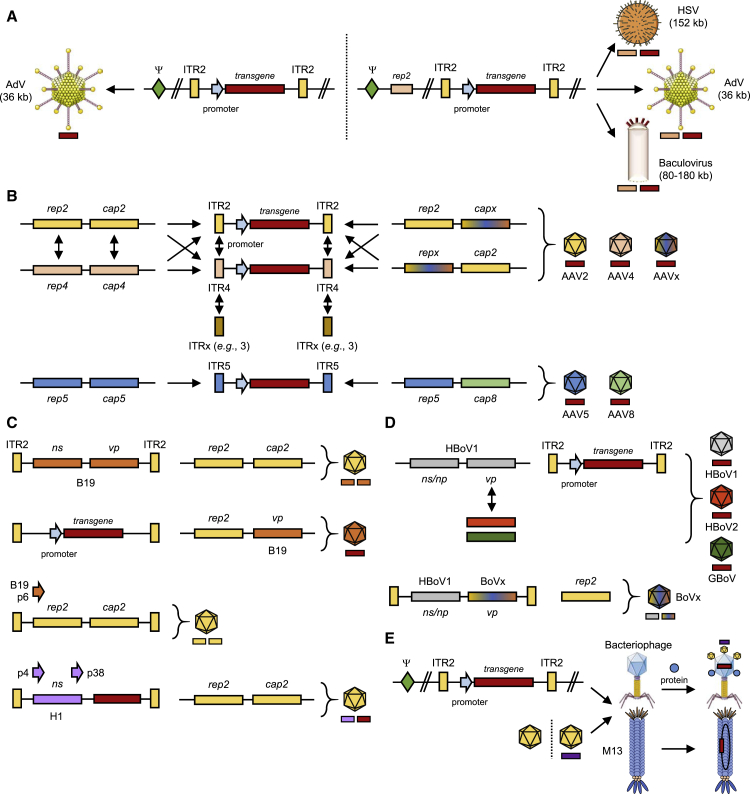
As noted, AAV depends on a heterologous helper virus for the completion of its own life cycle, such as the name-giving AdV or others including HSV, which provide essential, albeit different, helper functions for AAV replication, expression, or progeny formation. Although this dependency on a second, usually pathogenic, virus is disadvantageous from a manufacturing standpoint and thus avoided in state-of-the-art AAV production schemes, several groups purposely combined AAV and helper virus functions into hybrid vectors that aimed to merge the best of both worlds (Figure 2A). In particular, attempts were made to harness the high in vitro and in vivo efficiency of AdV, HSV, or baculoviruses (BaVs) and to concurrently overcome the extra-chromosomal nature of their genomes, which typically results in transient gene expression. Hence, the idea evolved to combine these vectors with AAV genomes that have the propensity to integrate randomly or in a site-specific manner, depending on the absence or presence of Rep proteins, respectively, hoping this would result in high-level and persistent transgene expression. However, as most of these concepts have been published about two decades ago and have not been pursued since then, we will not elaborate further on these proofs of principle. Instead, we refer the reader to the original literature77, 78, 79, 80, 81, 82, 83, 84, 85, 86, 87, 88 and to the overviews provided in Figure 2 as well as in Table 1.
Figure 2.
Different strategies for generation of hybrid parvoviral vectors
Depicted is an overview of the approaches discussed in detail in the text. In all panels, the AAV serotype is denoted after the label ITR, rep or cap (e.g., rep2 is rep from AAV2; x indicates chimeric rep or cap genes). (A) Packaging of AAV genomes into heterologous capsids from distantly related viruses. Ψ, packaging signal. (B) Intragenus pseudotyping with natural (left) or synthetic (right) elements. Note that packaging of vectors carrying the AAV5 ITRs requires helpers expressing AAV5 rep. (C) Intergenus pseudotyping. (D) AAV pseudotyping with bocaviral capsids. (E) Hybrids between AAV and bacteriophages. This figure contains clip art from Servier Medical Art (https://smart.servier.com/).
Table 1.
Overview of hybrid inter- and intragenus parvovirus vectors
| Genus | Genome | Capsid | Production | Targets | Reference | Pros | Cons |
|---|---|---|---|---|---|---|---|
| Protoparvovirus | LuIII | LuIII | NB324K | HeLa, iD5, NB324K, WI38, A375, WM1617, OVCARIII, T47D, HepG2, H929, Jurkat | 2,50,89, 90, 91 | wide diversity in nature | gutless vectors poorly characterized in terms of cell tropism, ip:vp ratio, and yields |
| H1 | NB324K | HeLa | 90,92 | non-human origin but mostly able to transduce human tissue (except FPV and CPV) | no comparative analysis to standard AAV vectors | ||
| FPV | NB324K | NB324K, CFK, A72 | 89,91,93,94 | wealth of data on wild-type (WT) virus biology that inform vector design | no in vivo studies | ||
| CPV | NB324K | CFK, A72 | 89,93 | low immunogenicity | low titers of ∼108−109 gc/mL | ||
| MVMi | NB324K | NB324K, iD5 (∼10× less efficient than MVMp) | 90,95 | compatible with peptide display94 | small packaging capacity of ∼4.7 kb | ||
| MVMp | NB324K | A9, NB324K, iD5 | 90,92,95 | no evidence of integration or genotoxicity | |||
| Dependoparvovirus | mostly AAV2 (but also several others96,97) | AAV1 to -13, chimeric/synthetic, e.g., DJ, AAV2.5T | HEK293, Sf9, stable producer cell lines | countless cell lines and primary cells | 96, 97, 98, 99, 100, 101,102, 103, 104, 105, 106, 107 | often transcend the species barrier | small packaging capacity of ∼4.7 kb |
| safe, high in vitro and in vivo activity | high seroprevalence of natural serotypes | ||||||
| can be produced to high titers (e.g., 3.3 × 1013 vg/L108) | only transient expression in dividing cells (may benefit CRISPR applications) | ||||||
| high amenability to genetic manipulations | integration into the human genome, albeit at low frequency | ||||||
| can be packaged into, and combined with, parvoviruses and distantly related viruses | |||||||
| Dependoparvovirus/erythroparvovirus | WTB19 flanked by AAV2 ITRs or WTAAV genome with p6 promoter | AAV2 | KB | NBM | 109,110 | The hybrid vector is both infectious and replication competent in B19 target cells. | no reported selectivity of B19 for cancer cells |
| The replacement of the p5 promoter by B19 p6 allows autonomous replication of AAV in human hematopoietic progenitor cells (may be beneficial for hemoglobinopathies). | Hybrid vectors were proven useful for studying the early events of virus trafficking but might not truthfully reflect the complete infectious cycle. | ||||||
| AAV2 | B19 | HEK293 | HEK293, undifferentiated and differentiated MB-02, LDBM, primary CD34+, HEL, K562, M07e, HL-60, HUVEC, NHLF | 111,112 | Restricted tissue tropism for human erythroid cell lineage allows tissue-specific transgene delivery. | low titers of ∼109 gc/mL | |
| efficient transduction and specificity even at a particle-to-cell ratio of 200:1 (versus 100,000:1 for AAV2) | Vector cannot be used to efficiently target cells from different origins. | ||||||
| compatibility with gutless ssAAV vectors | pathogenicity of B19 and strong immune response provoked by the viral capsid proteins | ||||||
| Dependoparvovirus/protoparvovirus | hybrid = WTH1 flanked by AAV2 ITRs | AAV2 | HEK293T | 113 | Vector was functional und gave higher expression in tumor cell lines. | only weak selectivity for cancer tissue in vivo | |
| Killing of transformed cells was comparable to standard AAV vectors. | |||||||
| no autonomous replication of the hybrid vector | |||||||
| Dependoparvovirus/bocaparvovirus | AAV2 | HBoV1-4, GBoV | HEK293, Sf9 | cell lines: Huh7, HeLa, HEK293, LX-2, PancI, RAW267.4, MCF-7 | 69,70,114, 115, 116 | larger packaging capacity than AAV (5.5−5.8 kb) | gutless vectors need to be boosted by proteasome inhibitors |
| primary cells: pHAE, skeletal muscle, cardiac myocytes, lung fibroblasts, saphenous vein endothelial cells, gut or lung organoids | numerous human and non-human primate serotypes | reports of triggered immune responses against the HBoV1 capsid | |||||
| high titers (>1 × 104 gc/cell, total yields > 5 × 1012 gc) | |||||||
| proven compatibility with both ss and scAAV vectors | |||||||
| amenability to DNA family shuffling (due to high homology) | |||||||
| Dependoparvovirus/mastadenovirus, simplexvirus/betabaculovirus | AdV/AAV | AdV | AdV/AAV: HEK293, 293CRE4, PER.C6 | AdV/AAV: M07e, K562, HEL, HeLa, Huh7, Hep3B, HepG2, SKHEP-1, primary amniocytes and keratinocytes | AdV/AAV: 78,79,81,84 | higher stability of intracellular extrachromosomal forms and genome integration tested in vitro and in vivo | need for specialized production protocols |
| HSV/AAV | HSV | HSV/AAV: V27 | HSV/AAV: BHK21, Hs913T, NPLC, U87 | HSV/AAV: 86,87 | larger packaging capacity than AAV | high capsid immunogenicity | |
| BaV/AAV | BaV | BaV/AAV: Sf9 | BaV/AAV: Huh7, MRC-5, HEK293, C17.2, PC12, NT-2 | BaV/AAV: 88,117 | high titers | risk of in vivo recombination (AdV and HSV) | |
| hybrid genomes | packaging of oversized rAAV by encapsidation of replication intermediates (AdV/AAV) | contamination of vector stocks with WT virus | |||||
| persistent transgene expression (AdV/AAV) | instability of in vivo transgene expression (HSV/AAV) | ||||||
| broad in vivo cell tropism (AdV) | |||||||
| Dependoparvovirus/inovirus | hybrid = phage genome including AAV ITRs and a transgene cassette | phage (M13 or T4) | purified from host E. coli culture supernatants (M13) or in vitro assembly (T4) | antigen-presenting DCs and many cancer cell lines, e.g., KS1767, HEK293(T), | 118, 119, 120, 121, 122 | targeting of tumor cells by phages displaying tumor-homing peptides | unclear scalability and ability to manufacture under GMP conditions |
| HeLa, U87, DU145, human melanoma cell lines, 9L, MCF-7, LN229, SNB19, C2C12, LNCaP | higher persistence of delivered transgenes flanked by AAV ITRs | low transduction ability in mammalian cells due to multiple entry barriers | |||||
| high cargo capacity | cellular and immune response against phage proteins is possible | ||||||
| can deliver both proteins and transgenes | |||||||
| no pre-existing immunity | |||||||
| Dependoparvovirus/tequatrovirus | phage and AAV | AAV capsids coupled to the T4 phage capsid | in vitro DNA packaging (T4) | HEK293 | 123 | can deliver both proteins and transgenes | cumbersome production process that requires several components to be purified separately in sufficient quantity and quality |
| HEK293 cells (AAV) | high cargo capacity | Scalability of production has yet to be assessed. | |||||
| co-delivery of multiple AAVs with different cargos | Cellular and immune response against phage proteins is possible. | ||||||
| in vivo data available | |||||||
| no pre-existing immunity |
Cell lines: NB324K, SV40-transformed human newborn kidney cells; HeLa, human cervical tumor cell line; iD5, a subclone of the somatic cell hybrid line Hybl/ll (a fusion of A9 ouabrll cells with EL4 cells); WI38, human embryonic lung cells; A375, WM1617, human melanoma cell lines; OVCARIII, human ovarian carcinoma cell line; T47D, human breast cancer cell line; HepG2, human liver cancer cell line; H929, human plasma cell myeloma cell line; Jurkat, human T-lymphocyte cell line; CFK, Crandell feline kidney cells; A72, canine fibroma cells; A9, ouabrll cells are a ouabain-resistant derivative of the HGPRT cell line A9; HEK293, human embryonic kidney cells; Sf9, a clonal isolate derived from the parental Spodoptera frugiperda cell line IPLB-Sf-21-AE; KB, human keratin-forming tumor cell line; NBM, normal human bone marrow cells; MB-02, megakaryocytic leukemia cell line; LDBM, low-density bone marrow cells; HEL/K562, human erythroleukemia cell lines; M07e, megakaryocytic leukemia cell line; HL-60, human promyelocytic leukemia cell line; HUVEC, human umbilical vein endothelial cell; NHLF, normal human lung fibroblasts; Huh7, human liver carcinoma cell line; LX-2, human hepatic stellate cell line; PancI, pancreatic cancer cell lines; RAW267.4, murine macrophage cell line; MCF-7, breast cancer cell line; pHAE, primary human airway epithelia; HepG2, Hep3B, human hepatoma-derived cell lines; SKHep1, human cell line of endothelial origin; 293Cre4, Cre-expressing 293 cells; PER.C6, Ad5 E1-transformed human embryonic retinoblasts; BHK21, baby hamster kidney cells; Hs913T, human lung fibroblasts; NPLC, human hepatocytes; V27, derivative of Vero cells that constitutively expresses HSV-1 ICP27; U87, human glioma cells; MRC-5, human diploid fibroblasts; C17.2, murine immortalized neuronal cell line; PC12, cell line derived from a rat pheochromocytoma; NT-2, pluripotent human testicular embryonal carcinoma cell line; KS1767, Kaposi’s sarcoma-derived cell line; DU145, prostate cancer cell line; 9L, rat glioblastoma cell line; LN229, SNB19, glioblastoma cells; C2C12, murine myoblast cell line; LNCaP, prostate cancer cell line; ip:vp, ratio of infectious particles to viral particles.
Intragenus pseudotyping of parvoviruses with natural capsids
As mentioned, the TRs are typically the only viral element that is required in cis for packaging of WT or recombinant parvoviral genomes into parvoviral capsids. This quickly led to the concept of vector pseudotyping, whereby a given recombinant parvoviral genome based on (I)TRs from one viral isolate is encapsidated into a capsid from the same genus but a different isolate, resulting in a prototype of a chimeric parvoviral vector. This strategy has been used extensively and has become a routine approach for several reasons: (1) as a universal concept, it alleviates the need to modify and adapt the ITRs every time a new capsid, natural or synthetic, is tested; (2) it facilitates investigations of natural virus properties such as tropism or immunogenicity and thus reduces time and costs; (3) the use of the same genetic backbone to encode a transgene permits a fair comparison among capsid variants by eliminating an impact of the ITRs; and (4) it concurrently enables high-throughput screening as well as the deliberate exploitation of molecularly evolved or rationally designed capsids or entire libraries thereof. The latter is particularly attractive for AAV vectors, considering that 13 natural AAV serotypes and hundreds of variants have been identified, to date, in numerous primate and non-primate species and that the latest capsid engineering technology offers manifold avenues for additional capsid diversification, stratification, and enhancement.4, 5, 6,124,125
As an example from the protoparvovirus field, a recombinant LuIII genome was packaged into multiple, closely or distantly, related species including H1 parvovirus,92 feline parvovirus (FPV), and CPV,89,93 as well as both fibrotropic and lymphotropic strains of MVM (MVMp90,92 or MVMi90). General observations, which were later also verified for AAV, were that the capsid but not the packaged genome is the major determinant of tissue tropism and that pivotal residues are located in the VP2 protein,93,95 whereas VP1 is required for particle infectivity.126 Still, exceptions were also reported. For example, the murine specificity of MVMp not only depends on the viral capsid but strictly requires viral genomes based on MVMp but not on H1.127 Interestingly, this only applies to cap replacement but not to gutless vectors; in the latter case, recombinant LuIII genomes were identical to recombinant MVM genomes.90
Today, pseudotyping is particularly popular in the AAV field and is used routinely to cross package viral genomes derived from one AAV serotype into capsids from another to study or harness distinct features such as cell selectivity or differential reaction with anti-AAV antibodies. Since AAV2 is the first isolated and best-characterized serotype, it comes as no surprise that the first packaging system was also derived from this variant; i.e., the AAV2 ITRs were used to flank a recombinant genome prior to packaging into another serotype’s capsid (Figure 2B). In one of the earliest studies, Chiorini et al.98 aimed to characterize their newly isolated AAV4 serotype by packaging a rAAV2 genome expressing lacZ into the capsid of AAV4, using the Rep proteins from AAV4. Soon after, Beck et al.128 packaged a rAAV2 genome into either the AAV2 or AAV3 capsid to study effects of vector readministration to the lung. These pioneering studies were rapidly and largely expanded by a string of publications from the Samulski129,130 and Kay and Kleinschmidt96 labs, demonstrating robust packaging of various AAV2 ITR-flanked transgenes into the capsids of AAV1 or AAV3 through AAV6, and followed by countless additional reports, to date, including natural (AAV8 to -13) and synthetic AAV capsids.99,131
Notably, despite the popularity of the basic pseudotyping system using both Rep and ITRs from AAV2, other combinations were also tested successfully. For example, it was shown that Rep proteins from AAV1 or AAV4 to -6 recognize AAV2 ITRs, replicate the genomes, and package them in the cognate viral capsid. Vice versa, ITRs from AAV1, -3, and -6 are recognized by all of the other Rep proteins and mediate DNA encapsidation.97 An exception is AAV5, for which the ITRs and Rep proteins are not interchangeable due to the unique and extended terminal resolution site (trs) in the AAV5 ITR, which is nicked by the Rep78/68 proteins during genome replication.96,132 Thus, AAV5 Rep proteins are unable to package AAV2 ITR-flanked genomes, and vice versa, AAV2 Rep proteins cannot recognize and cross package AAV5 ITR-flanked DNA.97 Typically, the original AAV2 Rep/ITR combination for pseudotyping gives comparable vectors yields96 or even outperformed other Rep/ITR combinations.133 For example, rAAV6 titers were higher when the AAV2 Rep/ITR pair was used for transgene packaging rather than AAV6 Rep/ITR.133 Also notable is that 3- to 4-fold higher vector titers were achieved when the AAV3 Rep/ITR pair was used instead of the one from AAV2.134 Similarly, AAV4 vectors gave five-fold higher titers when AAV4 Rep was used instead of AAV2 Rep. As replication of the AAV4 ITR-based genomes was not compromised with AAV2 Rep, the effect was possibly due to incompatibility between these Rep variants and the AAV4 capsid.96 Interesting in this context is an observation in the study by Rabinowitz et al.130 that titers of AAV1 and AAV3 to -5 vectors produced with the AAV2 Rep/ITR-based pseudotyping system can be increased by appending serotype-specific sequences 5′ to the start codon for the major capsid protein VP1 in the Rep-coding region. This implies that a Rep-specific, capsid-interacting domain might be required for efficient serotype-dependent encapsidation,130 albeit this small stretch might not be enough, as rAAV4 vectors still yielded lower titers than rAAV3 and rAAV5. We finally note that in most of these studies, only one transgene cassette was tested. Thus, further work is required to validate these data, as packaging efficiency can vary dramatically between different vector genome compositions.
In addition to governing vector titers, the AAV ITRs also play a pleiotropic role in rAAV transduction, which is an aspect that has been scarcely addressed in the long history of AAV vector development. In fact, they are not only crucial viral elements for rescue, replication, and packaging of viral genomes,57 but they also support second-strand DNA synthesis from the ssDNA genome in the nucleus after transduction135,136 and form substrates for genome self-circularization in cells and tissues.137,138 In general, it was shown that the capsid phenotype and not the vector genotype determines AAV vector tropism in vitro97 and in vivo.97,133 Still, the careful selection of ITRs can be of value in dual-vector, trans-splicing approaches, where two different ITRs are used to foster directional recombination that results in a functional transgene cassette.137 These vector types were used successfully in multiple studies to deliver transgenes that exceed the inherent AAV packaging capacity.139, 140, 141, 142, 143 Finally, ITRs from different serotypes were shown, including in recent work from the Samulski lab,144,145 to mediate differential cryptic transcriptional activities that can drive transgene expression in vitro146 and in vivo.
Intragenus pseudotyping of AAV genomes with synthetic AAV capsids
As indicated above, the promiscuity of the AAV2 ITRs makes them and vector genomes derived thereof amenable to packaging not only into other WTAAV variants but also into synthetic AAV capsids created through molecular evolution or rational design. Because of the long history and great diversity of these AAV capsid engineering technologies, we refer the readers to more dedicated comprehensive review articles by others and us for details.3,4,6,124,147, 148, 149 Here, it may suffice to note that AAV vectors have already been pseudotyped with, for instance, capsids that were constructed by swapping AAV cap domains, with the aim to elucidate sequences and/or structures that govern tissue and cell specificity. In one example, Shen et al.150 created hybrid capsids between AAV2 and AAV8 and used these to uncover the importance of the loop IV domain in AAV8 liver transduction. Many other groups concurrently designed and validated high-throughput methodologies that allow for the screening of large libraries of up to 108 capsid variants in parallel.151 One example is DNA family shuffling, a homology- and recombination-based approach that is technically challenging but has great potential when combined with strong selection pressure,99,152 as it can breed novel variants that not only blend the properties of the parental capsids but also ideally outperform their ancestors. One interesting AAV chimera and the prototype in the field of parvoviral cap shuffling is AAV-DJ, which combines the assets of three AAV serotypes, AAV2, AAV8, and AAV9, and shows high transduction of mouse hepatocytes in vivo and partial escape from antibody neutralization.99 We particularly highlight this technology because it has not only been used extensively to molecularly evolve hybrid AAV capsids99,153, 154, 155 but has also recently been applied to breed chimeric BoV capsids,70 illustrating its breath and usefulness in the field of hybrid parvoviral vectors. Next to shuffling, we note other powerful approaches to alter capsid sequences, including peptide display,131,156,157 fusion of epitopes or nanobodies to the capsid surface,158, 159, 160 or creation of mosaic capsids by mixing plasmids encoding VPs from different AAV serotypes during vector production.161,162
Parvoviral hybrid vectors created through intergenus packaging
Beyond encapsidating parvoviral genomes into natural or synthetic capsids from the same species or genus, or into heterologous virus capsids, hybrid vectors were also created by combining genome and capsid from two of the eight genera of Parvovirinae (Figure 2C). As this research area is very complex and as these vectors have already found manifold applications in basic and applied research, we can only scratch its surface here. The first hybrid vectors were constructed with the aim to reduce WT, replication-competent virus contaminations in vector preparations, originating from homologous recombination between helper and vector constructs.127 Subsequently, chimeric parvoviral vectors were mainly devised in attempts to increase vector titer, study biological aspects, decrease capsid recognition by neutralizing antibodies, or re-target capsids to other cell types.
As one interesting example, we highlight a hybrid between AAV and erythroparvoviruses, which encompass a small group of parvoviruses with a unique tropism for red blood progenitor cells.163 A well-studied and characterized member is the pathogenic B19 parvovirus that causes erythema infectiosum in humans. Efforts to study B19 in cell culture were initially hampered by the lack of suitable producer cell lines. Therefore, in the late 1980s, Srivastava et al.109 developed a cross-packaging approach in which the B19 genome was flanked with AAV2 ITRs. Supplying AAV2 Rep in trans then sufficed to mediate packaging into the AAV2 capsid.109 Intriguingly, this system allowed encapsidation of B19 genomes in cell culture and yielded infectious vps that behaved like a WT B19 virus in cells, i.e., the ensuing particles could replicate in, and lyse, erythroid progenitor cells in the absence of AAV2 Rep. This suggests that B19 NS proteins can recognize and resolve AAV2 ITRs, which is interesting in view of the homoterminal nature of the repeats in both viruses. Based on this pioneering work, Ponnazhagan et al.111 constructed another hybrid parvoviral vector by packaging an AAV2 ITR-flanked genome into the B19 capsid using AAV2 Rep. The resulting vectors robustly and specifically transduced red blood progenitor cells, congruent with the idea that the parvovirus capsid but not the genome determines tropism. In contrast to other hybrid parvoviral vectors (e.g., AAV2/HBoV1, see below), viral yields were low (∼1 × 109 genome copies [gc] per mL).111 Still, the high specificity of B19/AAV vectors mediated much better transduction than AAV in the erythroid fraction of primary human bone marrow cells (ratio of 200:1 vector particles to iu, as compared to 100,000:1 for rAAV),111 which highlights the promise of this viral system and supports its further development. Additionally, the recombinant vector was also used to study biological aspects of B19 tropism and cell entry.112
Another B19/AAV hybrid vector was constructed by Wang et al.110 to study the role of the B19 p6 promoter in infectivity and cell specificity. Therefore, the AAV2 p5 promoter was replaced by the p6 promoter, and the hybrid vector was packaged into an AAV2 capsid. Interestingly, the p6 promoter was sufficient to promote autonomous replication of AAV in erythroid progenitor cells but not in other cell types.110 Although the target specificity of B19 is known to be promoted by erythrocyte P-antigen receptors,111 this study revealed that, next to capsid-receptor interactions, other elements in the B19 genome are crucial to confer specificity to a target cell type.
In another approach by Krüger et al.,113 protoparvoviral genomes were combined with AAV2 capsids to broaden their tropism in human cells, while retaining their cancer-specific cytotoxicity. To this end, a hybrid AAV2/H1 vector was created by flanking an H1 genome comprising a p4 promoter-driven ns gene and a p38 promoter-driven transgene with AAV2 ITRs. Packaging with a conventional AAV2 helper plasmid resulted in hybrid vectors that permitted transformation-selective transduction of various cancer cells, owing to the preferred p4 activity in these cells.
Furthermore, hybrid vectors were devised to examine tissue tropism and cytopathic activity of rodent protoparvoviruses in human cells. For example, MVM/LuIII chimeras were constructed to define the region in LuIII that confers its ability to efficiently kill transformed human fibroblasts, which are refractory to MVM.164 This showed that the region spanning vp2 of LuIII sufficed to promote productive infection, which, akin to the experience with AAV chimeras, supports the generation and use of engineered viruses with combined and novel properties.
Additionally, pseudotyping and cross-genera packaging studies revealed interesting aspects of genome-packaging polarity. For example, it was found that genome polarity is an intrinsic property of the genome itself and not determined by the capsid used. When a LuIII genome was pseudotyped with a MVM capsid, both minus and plus strands were encapsidated with equal efficiencies, which is a feature that is determined by LuIII but not MVM.92 Likewise, when AAV2 genomes were pseudotyped with the HBoV1 capsid, the polarity of AAV2 was retained.69 The differential packaging of (−) and (+) strands depends on the TR sequences.165 In heterotelomeric viruses with two distinct TRs, one (usually the one at the 5′ end of the negative strand) is often resolved by terminal resolution during replication of the genome. This leads to a preferential displacement and packaging of negative-sense genomes.166 In contrast, the 3′ TR is resolved by a different mechanism with slower kinetics, i.e., asymmetric junction resolution.167 The effects of different TRs on parvoviral vector transduction, especially with respect to cross-genera pseudotyping, remain elusive and an interesting topic for further work. Notable in this context is a report of chimeric AAV2/HBoV1 vectors that showed no major differences in titer or transduction when recombinant HBoV1 or rAAV2 genomes were packaged into the HBoV1 capsid.69 As for polarity of viral genomes, we know from AAV that vectors exclusively packaging (−) or (+) strands perform equally well in vivo, suggesting that polarity itself does not affect transduction with a genuine parvoviral vector.168 Still, this does not rule out that synthetic (I)TRs may facilitate a better interaction with the DNA machinery responsible for second-strand DNA synthesis and may improve transduction or foster site-specific integration into the host genome.
Hybrid parvoviral vectors between AAV and BoVs
One category of hybrid parvoviral vectors that has been introduced over the last few years and that we highlight for its unique assets is based on the BoV genus, which comprises an assortment of viruses that infect primates or non-primates, including pigs,169 bats,170 sea lions,125 rodents,171 and dogs.22 The isolation of the first primate BoV (HBoV1) from respiratory samples by Allander et al.11 was followed by the discovery of numerous other BoVs in stool from humans (HBoV2 to -4172,173), gorilla (GBoV),174 or chimpanzee.175 Although many non-primate BoVs are highly pathogenic, causing reproductive failure and neonatal respiratory diseases,176 primate BoVs are associated with a diffuse phenotype and not unanimously linked to human diseases.177 Still, we note sporadic reports, including from our own group and collaborators, on severe outcomes of BoV infection, comprising a fatality in an immunodeficient boy for which HBoV1 may have been the causative agent,178 as well as evidence from the Schildgen lab179,180 and others of a link with lung fibrosis and tumorigenesis.
Efforts to engineer and utilize this parvovirus as a vector for gene transfer, especially in the human lung, were kick started with a 2012 publication by the Söderlund-Venermo, Qiu, and Engelhardt labs,181 in which a reverse genetics system for HBoV1 production and testing was established. This permitted the discovery of a unique tropism of WT HBoV1 for infection of airway epithelia from the apical side and by reporting a molecular clone of the full-length, ∼5.5-kb HBoV1 genome (i.e., ∼18% larger than AAV2), laid the foundation for the development of recombinant HBoV1 vectors. Analysis of this clone showed that, unlike AAV, the two terminal hairpins in HBoV1 flanking the predominantly minus-stranded DNA genome are asymmetric and that the genome encodes different NS proteins—NS1 to -4182 and NP1183—together with VP1 to -3, all expressed from a single p5 promoter. Also, as a typical APV, HBoV1 replicates in permissive cells without a helper virus and usually, ultimately lyses the host cell during progeny release. Interestingly, HBoV1 encodes a small non-coding (nc)RNA in its 3′ UTR (untranslated region), namely, BocaSR, which is essential for the expression of the NS proteins NS1 to -3/NP1 and indispensable for the replication of the viral genome.184
To harness the specific tissue tropism and larger size of HBoV1 for vector development, Yan et al.69 engineered a chimeric AAV2/HBoV1 vector system in which a rAAV genome was cross packaged into the HBoV1 capsid (Figure 2D). To this end, they set up a production system based on four-plasmid transfection of HEK293 cells comprising (1) the AAV2 vector, (2) an adenoviral helper, (3) an AAV2 Rep expression plasmid, and (4) a bocaviral helper encoding ns and capsid genes. This resulted in chimeric AAV2/HBoV1 vector particles at yields that were up to 20-fold lower than those of genuine AAV2 vectors but much higher than those obtained for a hybrid AAV2/B19 vector. Importantly, as shown with over-sized reporter AAV2 genomes, the AAV2/HBoV1 vector can package large ssAAV genomes of up to 5.4 kb without truncations, which exceeds the cargo limit of conventional AAV2 capsids. As hoped for, the vector enabled efficient transduction of pHAE and CuFi air-liquid-interface cultures from the apical but not the basolateral side, reflecting the infection bias seen in natural infections.181 In direct comparison, the hybrid AAV2/HBoV1 vector outperformed AAV1 or AAV2 vectors at the apical side by ∼5- to 70-fold, respectively, whereas the two authentic AAV vectors excelled from the basolateral side. A more detailed analysis of the transduced cells revealed that the majority of AAV2/HBoV1 particles are retained in the cytoplasm, tempting Yan et al.69 to speculate that endosomal processing and intracellular trafficking might be impaired and, vice versa, could be improved by treatment of the cells with proteasome inhibitors such as N-acetyl-l-leucyl-l-leucyl-l-norleucine (LLnL) or doxorubicin that are both known to enhance transduction of various AAV serotypes. Indeed, the addition of these two inhibitors increased AAV2/HBoV1 transgene expression by over 1,000-fold, especially from the apical side, implying that AAV and BoV share intracellular pathways and barriers. Finally, it was reported that the enhanced cargo capacity of the new hybrid AAV2/HBoV1 vector enables the transduction of an over-sized (with respect to AAV) 5.5-kb genome encoding the cystic fibrosis transmembrane conductance regulator (CFTR) cDNA under a strong chicken beta-actin (CBA) promoter and functional improvement in CF HAE.
More recently, these assets were confirmed and expanded in an in vivo study that showcased the potential of AAV2/HBoV1 vectors to transduce the ferret airways, which display high similarities to human lungs including CF phenotypes.114 Although vector performance in ferret airway epithelia ex vivo or in a mouse tracheal xenograft model was subpar in comparison to human target cells, the AAV2/HBoV1 vector (in the presence of doxorubicin) gave robust expression following transtracheal installation in 3- to 29-day-old ferrets, even after repeated vector administration. Yet, the reporter signals were unevenly distributed and most abundant in the lower respiratory tract including the bronchiolar airways and alveolar regions of the lungs.
In our own group, we have recently harnessed this cross-genera packaging system to study the kinetics of AAV2/HBoV1 transduction70 and to dissect the influence of naturally occurring point mutations (“singletons”) in the HBoV1 capsid on the producibility, transduction ability, and antibody escape of chimeric AAV2/HBoV1 vectors.115 Interestingly, we found the kinetics of transgene expression with bocaviral vectors to be similar to a WT infection, starting 3 days post-transduction/-infection and stabilizing at 6 to 9 days.181 Moreover, our collection of 64 HBoV1-positive patient samples permitted us to clone 29 HBoV1 capsid variants, differing from the reference strain11 in 32 nucleotides (nt) or four amino acids. Among these, most notable was a change from threonine to serine at position 590 that significantly affects virus and vector titer.115 Surprisingly, whereas a serine at this position was associated with lower viral loads in patients, it increased titers but not performance or immunoreactivity of rAAV2/HBoV1 vectors. Furthermore, via a combination of structural modeling and targeted mutagenesis, we identified several capsid residues, which, next to transduction, govern assembly and packaging and should thus inform the future design of improved AAV2/HBoV1 hybrid vectors.
Intrigued by the potential and capacity of this system, our group has recently expanded this concept to four additional bocaviral variants, namely, HBoV2 to -4 and GBoV.70 To this end, we cloned new helper plasmids expressing the respective capsid genes together with the HBoV1 NS and NP1 proteins and then used these to pseudotype various ss- or scAAV vector genomes. This enabled a comparative analysis of the natural tropism of bocaviral species, showing that besides HBoV1, also HBoV4 and GBoV effectively transduce pHAE, albeit they also require proteasome inhibitors for maximum performance. Moreover, all five BoV variants were able to transduce 3D primary human lung organoids depending on the administration route, with some performing better after mechanical organoid shearing (GBoV) and others transducing best after direct microinjection (HBoV2 and -3). Several variants, most notably HBoV4 and GBoV, also performed well in other primary cell types from the liver or the musculature, or in human CD4+ T cells, which makes them highly attractive for in vivo or ex vivo gene therapies outside the lung. Further increasing their potential is that HBoV4 and GBoV were much less inhibited by human immunoglobulins (intravenous immunoglobulin [IVIg]) in a cell culture-based transduction/inhibition assay, implying their particular usefulness in gene-therapy patients with pre-existing anti-BoV immunity. Besides, we could show that the HBoV1 capsid efficiently packages up to 6.1 kb of ss- or 3.6 kb of scAAV DNA, representing ∼130% of the DNA that fits into an AAV2 capsid. This further underscores the usefulness of hybrid AAV/HBoV vectors for encapsidation and delivery of over-sized DNA cargos that are incompatible with genuine AAV vectors. Finally, in unpublished work, we observed a high instability of BoV TR sequences in bacterial cells as compared to AAV ITRs and encountered difficulties in packaging HBoV1 TR-flanked genomes into capsids of HBoV2 to -4 but not GBoV. Whereas these preliminary notions require further investigation, they additionally inform vector development and support the use of the pseudotyping production system.
As a whole, the work on AAV/BoV vectors by multiple labs has revealed a series of features and advantages that, in our opinion, should encourage continued research into and application of this intriguing hybrid parvoviral vector. A first asset is the greatly expanded packaging capacity, which enables delivery of cassettes that would be incompatible with any other known parvoviral vectors, including all-in-one CRISPR cassettes comprising Cas9 and gRNA(s), as already demonstrated by us.70 A second advantage of pseudotyped BoV vectors is their good producibility, yielding typical titers of ∼1 × 1012 vector genomes per mL, which is below the yields of genuine AAV vectors but higher than other cross-genera parvoviral packaging systems. Importantly, an optimized production system has been reported recently that is completely devoid of BoV NS proteins and instead only relies on the use of AAV Rep and AAV genomes in HEK293185 or insect cells.116 Mechanistically, it is tempting to speculate that AAV Rep proteins recognize the BoV capsid (and the B19 capsid, as mentioned) and to package the AAV genomes, which would foster the future development of other hybrid parvoviral vector systems. Importantly, as little information is available about BoV genome packaging, other scenarios are also possible, such as the involvement of host proteins or a different mode of capsid assembly than AAV, e.g., capsid formation around the genomes and packaging via strand displacement. Third, the ease of pseudotyping by swapping BoV capsid genes in helper plasmids, akin to the process used for AAV, greatly facilitates the generation and testing of other, natural or synthetic bocaviral variants beyond those already studied, such as those from chimpanzees,175 pigs,169 or dogs.22 This is motivated by our aforementioned data on the robust performance and improved immunoreactivity of non-HBoV1 variants,70,115 and it is further stimulated by our finding of cell-type specificity within the lung, such as that of HBoV4 for basal cells.70 Last but not least, it is important to mention that despite the promise of the BoV/AAV pseudotyping approach and the multiple improvements reported so far, several hurdles still limit its applicability and are thus in the focus of current investigations. For example, the high dependency on proteasome inhibition necessitates the application of inhibitors, which are chemicals with a certain level of toxicity. Importantly, proteasome inhibitors were already successfully and safely combined with AAV and BoV vectors in vivo in multiple pre-clinical studies.72,186, 187, 188 Still, it is clear that a thorough risk/benefit assessment is needed prior to clinical translation of these vectors and strategies.
Besides chemical reagents, other strategies to shield capsids from the proteasome include modulating surface-exposed residues that are recognized by the proteasome, an approach that is used successfully in the AAV field. In addition, we envision further strategies to improve the AAV/BoV system, based on the intricacy of parvovirus biology and recent data. For example, we consider it interesting to test whether inclusion of the above-mentioned BocaSR ncRNA (∼250 nt) in the 3′ UTR that localizes to virus DNA replication centers benefits BoV vector production and performance. Finally, besides rational vector design, it is encouraging that BoVs are amenable to directed molecular evolution, as demonstrated by the rescue and selection of a replication-competent library in which AAV2 ITR-flanked BoV genomes were packaged into chimeric BoV capsids, when AAV2 Rep and AdV proteins were expressed in trans.70 These libraries can be subjected to customized and well-defined selection pressures, e.g., to allow the transduction of cell types of interest in the absence of proteasome inhibitors. This could eventually lead to the identification of novel variants that partially or fully overcome current limitations.
Hybrid vector systems based on AAV and phages
In 2006, the Arap lab118 introduced an original, intriguing, and versatile hybrid vector system called AAV/phage (AAVP) to the field, which merges cis elements from AAV with a prokaryotic, tumor-targeted fd-tet bacteriophage (a filamentous phage derived from M13) (Figure 2E). In more detail, an AAV cassette consisting of a gfp reporter driven by a cytomegalovirus (CMV) promoter and flanked by AAV2 ITRs was inserted into the genome of a phage displaying the cyclic peptide CDCRGDCFC (in short, RGD4C) on its minor pIII coat protein. This peptide targets αV integrins that are frequently expressed on the surface of tumor and of tumor vascular endothelial cells. As hoped, the resulting chimeric AAVP transduced cultured mammalian cells in a ligand-dependent manner and delivered an intact AAV genome. This was confirmed by the successful rescue of progeny AAV particles following delivery of all helper functions to the transduced cells via transfection (rep and cap) and infection (AdV). Notably, via a combination of techniques, the authors obtained evidence for the persistence of the AAV genomes in AAVP-transduced cells as head-to-tail DNA concatemers. This tempted them to postulate that the presence of the ITRs in the hybrid viruses bestows an advantage in that it alters the intracellular fate of the transgene cassette and enhances AAV maintenance. Still, neither this pioneering study nor follow-up work reported data on the exact chromosomal integration site(s), leaving unclear whether AAV genome integration was specific (e.g., in the AAVS1 locus) and safe. Importantly, the predicted tumor cell selectivity after systemic delivery mediated by the displayed cyclic RDG peptide was confirmed in multiple immunocompetent or immunocompromised mouse tumor models. These also revealed a pronounced liver detargeting, which is notable considering that this organ acts as a non-specific sink for peripherally administered vectors, including genuine AAVs. Moreover, these studies in murine models illustrated the capacity of the AAVP vector to suppress tumor growth in vivo when used to express the HSV thymidine kinase suicide gene and combined with systemic ganciclovir administration. Finally, worth noting is that the same group followed up with a detailed protocol for the cloning, production, purification, and application of AAVP, which facilitates its broader testing and use in the vector and gene-therapy communities.189
As encouraging as this pioneering work was, it also raised a series of seminal questions about the clinical potential and translation of this novel hybrid vector, including the possibility to manufacture clinical-grade AAVP under good manufacturing practice (GMP) conditions. Moreover, albeit the authors provided preliminary data that the vector continues to function in the presence of anti-phage antibodies, the immunogenicity of phages and hence the induction of a humoral and/or cellular immune response in treated humans remain a concern. Finally, despite evidence that the inclusion of AAV ITRs enhanced vector persistence, a deeper understanding of the integration mechanisms and site(s) in the host cell chromosome and of the genomic vector forms is mandatory for safety assessment. Of note, the ability to compare AAVP vectors with or without ITRs concurrently provides a useful tool to dissect the role of the ITRs for integration or concatemerization, which will, in turn, help to answer the aforementioned questions.
Another pivotal task that was addressed comprehensively in a series of more recent studies concerns experimental strategies to enhance vector efficacy in vitro and in vivo. In a first study, the Hajitou group118,189,190 investigated hurdles to vector entry and intracellular trafficking that may explain the poor vector efficiency, evidenced by the high doses needed for robust transduction and the fact that only ∼10% of transduced cells express an encoded transgene, despite almost 100% vector internalization in all cells. With the use of a battery of complementary techniques, Stoneham et al.190 could partially resolve the intracellular fate of RGD4C-displaying AAVP and show that, following αV integrin binding and uptake in a dynamin-/clathrin-dependent manner, the majority of particles are sequestered by the endo-lysosomal pathway. Also important is their discovery that lysosome disruption via chloroquine, an agent that neutralizes acidic organelles such as late endosomes, can boost AAVP transgene expression in a dose- and cell type-dependent fashion, even in cells that were refractory to the vector per se.190
An additional intracellular barrier to AAVP transduction was identified and resolved in two closely related studies by Przystal et al.191 and Tsafa et al.,119 both also from the Hajitou lab. Specifically, the two teams consistently reported that, akin to AAV and other parvoviruses,192 also AAVP is susceptible to proteasomal degradation and that proteasome inhibition enhances targeted tumor cell killing by AAVP in vitro and in vivo. Investigation of this particular pathway made sense considering that proteasome activity is frequently elevated in tumor cells,193 implying that it may pose a critical barrier for AAVP in these cells. Indeed, transduction was improved in the presence of two well-known and widely used proteasome inhibitors, MG132 and LLnL (peptide aldehyde inhibitors that block the 26S proteasome subunit).191 The effect correlated with longer vector persistence and, as expected from proteasome inhibition, polyubiquitination of the phage coat proteins. Notably, the two inhibitors also triggered dose- and cell type-dependent cytotoxicity, suggesting a need to carefully balance enhancement of AAVP transduction versus safety. Still, the combination of MG132 (the stronger of the two inhibitors) and a luciferase-encoding AAVP was well tolerated in vivo in a melanoma mouse model and shown to enhance tumor-specific transgene expression, and the same inhibitor also boosted killing of U87 human glioblastoma cells in mice with AAVP encoding HSV thymidine kinase.191 Of note, similar conclusions were drawn in the related work by Tsafa et al.,119 albeit the agent used here—the natural dietary plant product genistein—is fundamentally different. Genistein is a well-characterized and clinically evaluated, soy-derived isoflavone that exhibits pronounced inhibitory activity on various cancer types, likely via sensitization to, or induction of, apoptosis and by targeting multiple cancer-related signaling pathways. Genistein has previously also been shown to boost AAV2 transduction at doses that, unlike proteasome inhibitors, were below the cytotoxic half-maximal inhibitory concentration (IC50) doses.194 Here, genistein was found to enhance AAVP transduction of cultured cells, in a manner independent of cell attachment or entry, but mediated by inhibition of proteasomal degradation and increased nuclear accumulation of AAVP vector DNA (possibly due to genistein-mediated enlargement of nuclear pores).119 This is reminiscent of data with MG132 and LLnL,191 as well as findings with AAV2,194 and thus supports the key role of the proteasome as a natural regulator of AAVP efficacy and a means to exogenously control the latter.
Yet another possible mechanism of action of genistein could be that it fosters the conversion of incoming ssAAV(P) DNA into transcriptionally active and more stable double-stranded DNA.119 A crucial role of the AAV DNA component was also postulated by Kia et al.,195 who noted a rapid decline of gene expression in AAVP/GFP-transduced tumor cells, possibly due to promoter silencing. Treatment of the cells with inhibitors of histone deacetylases (HDACs) such as trichostatin-A or suberoylanilide hydroxamic acid significantly increased numbers of GFP-positive cells and mean fluorescence intensities. Similarly, treatment with the DNA methylation inhibitor 5-azacytidine yielded a dose-dependent increase in GFP expression. These data were corroborated by Campbell et al.196 who found a positive effect of another HDAC inhibitor, C1A, that specifically blocks HDAC6. Their additional notion that the HDAC6-selective inhibitor outperformed the pan-HDAC inhibitor sodium butyrate is encouraging, as it suggests the feasibility to boost AAVP efficiency in human patients with low toxicity and costs.
One more host cell factor that may determine the efficacy of AAVP transduction was reported by Yata et al.,120 who studied the role of the tumor extracellular matrix (ECM) in various 2D and 3D models. As the ECM can spatially block access to cell-surface receptors and thus inhibit vector transport to cells, binding, and subsequent uptake, it was speculated that its deliberate degradation and depletion may improve the performance of therapeutics including AAVP. Indeed, when the Hajitou group118,189,190 treated various tumor cell lines with collagenase, hyaluronidase, or a combination, they observed a decrease in type I collagen, fibronectin, and hyaluronic acid in cell supernatants. In turn, this modulation of the tumor cell microenvironment improved AAVP movement, internalization, and reporter gene expression, while retaining specificity for integrin-expressing tumor cells. This was corroborated with the HSV thymidine kinase gene in which its cytotoxic effect was also boosted after ECM depletion in the absence of non-specific toxicity from collagenase and/or hyaluronidase. Still, concerns regarding the clinical translation are that collagenase cannot discriminate healthy from diseased tissues and that high collagenase levels are frequently associated with increased cancer metastasis, raising a risk of toxicity and other adverse events in patients. Thus, it is encouraging that Yata et al.120 could recapitulate their findings upon cell treatment with the US Food and Drug Administration (FDA)-approved antihypertensive losartan, which inhibits collagen I de novo synthesis and offers a better safety profile in cancer patients. This drug is particularly attractive, as it may preferentially impact metabolically active tumor cells and concomitantly suppress metastasis formation, which may synergize with the relief of physical and spatial ECM barriers and thus warrants further (pre-)clinical investigation.
Finally, we highlight three interesting recent proofs of concept for the feasibility to enhance AAVP transduction by modulating the phages themselves, rather than cellular host factors or structures. In the first, Yata et al.121 coupled the inherently negatively charged phages with a synthetic cationic polymer. This yielded positively charged particle aggregates that bound better to cells than uncoupled AAVP, owing to the alleviation of electrostatic repulsion, and that yielded improved, cancer-specific gene expression and cell killing. An intriguing explanation is the high buffering capacity of the hybrid nanomaterial complexes, implying their ability to trigger osmotic endosome swelling during intracellular vector trafficking and to thereby facilitate endosomal AAVP escape. This is reminiscent of findings with traditional AAV2 vectors following their coupling with polylysine, which enhanced their in vivo efficacy.197 A concern in view of data from the same group on the limiting role of the ECM120 is that the larger size of the phage-polymer complexes could restrict particle diffusion and hamper efficacy, which remains to be studied. Notably, cancer cells often express higher levels of anionic molecules, creating a negative net charge of their membranes and in turn increasing their susceptibility to cationic entities, such as the hybrid nanocomplexes reported in this work. Taken together, the two studies by Yata and co-workers120,121 illustrate the possibility to harness natural differences between normal and tumor cells for the rational design of next-generation, synthetic gene-delivery systems including chimeric parvoviral/phage vectors for cancer gene therapy.
In a second study, Smith et al.198 combined AAVP with gold (AU) nanoparticles, yielding a “transducing matrix” that gave better transduction than genuine AAVP in cell culture. An inspiring notion is that Au nanoparticles are biocompatible and trackable by, e.g., computed tomography, which implies the possibility to monitor AAVP complexes in vivo regardless of their cargo. Moreover, it may be possible to repurpose the transducing scaffold for compatibility with magnetic particles, which could enable spatially confined or patterned gene delivery.
In the third and most comprehensive study on AAVP engineering to date, Suwan et al.199 pursued a clever strategy to simultaneously mitigate pre- and post-internalization hurdles, including adsorption by the ECM or neutralizing antibodies, as well as endo-lysosomal degradation. To this end, the authors first established a phage construct carrying two versions—WT or recombinant—of the major, multithousand-copy pVIII coat protein, permitting the production of multifunctional AAVP particles displaying the original RGD4C targeting ligand on the pIII minor coat protein in combination with other peptides with desirable attributes on pVIII. Functionality was verified by displaying both RGD4C and a well-characterized streptavidin-binding peptide (SBP) on the same particle and by showing that the resulting hybrid AAVP concurrently interacts with streptavidin and integrins. Next, SBP was replaced with the short-charged neutralizing peptide AKAS, hoping it would display zwitterionic properties and thus alleviate the adverse adsorption of plasma proteins (“protein corona”) on the phage surface. Indeed, RGD4C-AKAS-AAVP particles interacted much less with a positively charged polymer and were shifted toward lower negative ζ-potential values. Impressively, the multifunctional particles also interacted less with fibrinogen, as well as with antiphage antibodies, suggesting they had acquired escape mechanisms to circumvent non-specific adsorption and neutralization. On top, the authors displayed three different peptides expected to promote endo-lysosomal escape and found that especially one of them, a histidine-rich sequence called H5WYG, gave substantially higher transgene expression in cultured cells as well as in tumors in a mouse model than unmodified AAVP. This may be due to the “proton-sponge” effect, i.e., the protonation of imidazole groups in the histidine residues, followed by an influx of chloride ions into the endosome, their osmotic swelling and eventual rupture, and the release of the modified AAVP particles from the endo-lysosomal degradative pathway. As compared to other strategies pursuing the same aim previously reported from this group, such as the use of chloroquine,190 the design of these latest, genetically engineered, and multifunctional AAVP derivatives seems superior, especially as it can, in principle, be extended to the display of an even greater variety of peptides on the same particle and should also be compatible with AAV.199,200
As a whole, the AAVP story, thus far, perfectly highlights the tremendous potential of combining parvoviral elements (here, a rAAV2 genome) with a heterologous virus (here, a prokaryotic bacteriophage). It simultaneously illustrates the power and potential of smart vector engineering, as well as the great promise for clinical applications in gene therapy of cancer and other diseases. In fact, a wealth of additional studies has already documented the versatility and potency of AAVP in various mouse or rat models of human tumors, such as soft-tissue sarcoma,201 melanoma,202,203 glioblastoma,204, 205, 206 neuroendocrine tumors of the pancreas,207 breast cancer,208 or prostate cancer.209 Further exemplifying the flexibility of this system is the variety of other AAVP elements that were swapped and adapted to a particular application, including promoters (e.g., Grp78204,205), transgenes (e.g., tumor necrosis factor202,203,206,207), and peptides (e.g., targeting Grp78,209 somatostatin receptors,207 or IL-11 receptor201). Last but not least, several reports consistently illustrated the capacity of this hybrid vector for theranostics approaches, using the HSV thymidine kinase gene as a tumor-suicide gene and for monitoring of transduced cells via positron emission tomography.118,201,206,208,209
In addition to the work on AAVP by the Hajitou, Arap, and Pasqualini labs, we note a related, recent study by Khalili et al.,210 who further repurposed this hybrid AAV/phage vector platform and exemplified its utility for cancer immunotherapy. Therefore, the authors displayed a cell-penetrating TAT (transactivator protein from human immunodeficiency virus 1) peptide on the pVIII protein and embedded the dickkopf-1 (DKK1) gene, encoding a secretory glycoprotein with a prominent role in cancer, in the phage-encapsidated AAV2 vector genome. Intramuscular vector injection in mice triggered a robust humoral immune response, i.e., the formation of anti-DKK1 antibodies, as detected by ELISA. Although this work left a substantial number of open questions, such as on the biology of the exposed TAT peptide in this context and the target cells that were transduced by the hybrid vector in vivo, it provides an appealing proof of concept for the potential of this vector as an immunization and vaccination platform.
Lastly, we highlight a recent, highly versatile, and promising hybrid vector system reported by the Rao lab,123 which also combines AAV and bacteriophages but in a unique fashion that fundamentally differs from the AAVP design. The same group had previously already illustrated the great capacity of the bacteriophage T4 for engineering and repurposing as a gene and protein delivery vector in mammalian cells in vitro and in vivo.122 The underlying rationale was that, first, T4 phages possess one of the fastest and most powerful DNA packaging machineries, which is promiscuous and thus compatible with any linear phage or non-phage DNA. Second, they also display two non-essential proteins (Hoc, highly antigenic outer-capsid protein; Soc, small outer-capsid protein) that can be harnessed for fusion and phage decoration with heterologous proteins including targeting ligands. Together, this allows for the concurrent delivery of foreign DNA and proteins into the same cell, using a single, double-engineered T4 particle. Notably, this could enable the transfer of up to ∼170 kb DNA and ∼1,025 heterologous protein molecules, calculated from the size and structure of genuine T4 particles. Moreover, T4 is neither toxic nor infectious in mammalian cells, nor is it blocked by pre-existing immunity, which makes it very attractive as a delivery vehicle for eukaryotic gene and protein transfer. This potential was exemplified in the 2013 study by Tao et al.,122 in which the authors packaged various reporter plasmids into T4 in vitro (about 10 plasmids per head) and then arrayed a series of foreign peptides on the phage surface as recombinant Hoc or Soc fusions that were spiked into the assembly reactions and incorporated via simple first-order kinetics. For instance, delivery of a luciferase reporter with phages decorated with the cell-penetrating TAT protein (the same used in AAVP210) was found to be efficient in HEK293T cells, matching that of a potent transfection reagent (lipofectamine). As noted, this system also permits co-delivery of foreign DNA and protein into mammalian cells, as documented with particles containing GFP or luciferase plasmids and concomitantly displaying β-galactosidase fusions on the surface. Moreover, these particles were successfully targeted to dendritic cells (DCs) via additional display of DC-targeting ligands. Importantly, the engineered phages mediate effective reporter gene expression in the mouse muscle following intramuscular application, and duration of in vivo expression was extended (from 14 to at least 30 days) when the reporter cassette was flanked by AAV2 ITRs, reminiscent of the data with the AAVP system (see above). Finally, it was shown that parallel expression of the Yersinia pestis (the causative agent of plague) F1-V protein and F1-V display on the T4 surface yields a potent vaccine that elicits both a humoral and a cellular immune response in intramuscularly injected and immunized mice.
Although this pioneering work compellingly illustrated the potential of engineered T4 phages, a critical bottleneck that remained and has been addressed in a recent study123 from the same lab is the relatively poor efficiency of recombinant T4 in mammalian cells, owing to the fact that it is a prokaryotic virus that has never adapted to eukaryotes. Thus, to boost its potency in cells from higher organisms, the Rao lab devised a complex experimental scheme to tether complete AAV particles onto the T4 head via streptavidin-biotin-mediated coupling to Hoc or Soc. Hence, the AAV vectors are piggy backed onto T4 and act as drivers that mediate efficient and specific transfer of the entire complex into mammalian cells. For proof of concept, the group exploited the same reporters and transgenes as in the original 2013 study,122 as well as the AAV-DJ capsid.99 Biotinylation of AAV-DJ, coupling to T4 through Soc- or Hoc-avidin fusions carrying a hexahistidine tag, and purification via nickel chromatography resulted in hybrid T4/AAV vectors in which up to four AAV-DJ capsids were tethered to one T4 capsid. Remarkably, as compared to genuine recombinant T4 with fluorescence reporter genes, transgene expression in HEK293 cells was enhanced by up to 40,000-fold upon AAV-DJ tethering in a dose-dependent fashion, validating the key role of the AAV component in this hybrid vector. An extra benefit is the ability to co-express another transgene from the AAV vector itself, as shown with various combinations of reporters encoded in T4 or AAV-DJ. Akin to the preceding study,122 evidence was provided that this new hybrid vector can transfer combinations of foreign DNA encapsidated in the phage or the viral vector with protein fusions displayed on the T4 surface. This also revealed another asset of the AAV component, i.e., its ability to mediate efficient endosomal escape and nuclear gene expression, which, based on the application, may synergize with the inherently cytosolic activity of proteins exposed on the T4 surface. Importantly, the hybrid particles yielded long-term (at least 60 days), non-toxic reporter gene expression in the murine muscle when delivering a transgene cassette comprising AAV2 ITRs, extending prior data122 and complementing AAVP studies (see above). Lastly, this latest generation of T4/AAV hybrid vector is again promising as a recombinant vaccine, as shown by co-delivery of influenza virus hemagglutinin or Y. pestis F1-V DNA or protein into mice via intramuscular administration, resulting in long-lasting, broad, and protective immunization via induction of humoral and cellular immune responses.
Conclusions and prospects
Among all modalities for therapeutic gene transfer in humans that have been evaluated over the years, recombinant vectors derived from natural or synthetic viruses represent one of the most powerful, promising, and versatile, owing to the sheer complexity of available templates and to their often high amenability to genetic engineering and targeted repurposing. Further expanding the capacity of viruses and their range of clinical indications is the concept that was in the center of the present article, i.e., the coupling of functional DNA and protein elements from different viruses. Ideally, in the resulting hybrid particles, the two input viruses will co-opt each other’s properties without compromising their individual abilities, enabling the components to perfectly synergize and to yield an integrated chimeric particle with unique attributes and unprecedented potential for numerous applications. This is well exemplified by the T4/AAV hybrid discussed above that merges the large cargo capacity and flexibility of the phage with the potent entry and endosome escape mechanisms provided by the AAV component, as well as with the ability of the latter to deliver additional reporter or therapeutic DNA cassettes.
Notably, although the examples reviewed here were centered on the juxtaposition of parvoviral features with other viruses, the same paradigm was concurrently applied and tested with other combinations of viruses too, such as chimeras between AdV and retroviruses, SV40, or Epstein-Barr virus.211, 212, 213 Nonetheless, the vast majority of studies on hybrid viral vectors reported to date comprises at least one parvoviral component in the form of genome, protein, and/or capsid, which most likely reflects the numerous assets provided by the Parvoviridae family, such as site-specific and safe vector DNA integration or the ease of genome pseudotyping. Indeed, together with our knowledge of parvovirus biology from decades of research, this large and diverse virus family may offer an optimal assortment of distinct natural viral building blocks that can be freely combined to create superior new vector platforms.
Still, it is equally clear that substantial additional work is required to further accelerate the development of these exciting vector platforms and to ultimately justify their clinical evaluation and translation. Specifically, we envision at least three seminal advances that should be pursued by the community and that are also in our own focus during our continued work on hybrid AAV/BoV vectors and others. First and foremost, despite our previously gathered insights into many aspects of parvovirus DNA packaging, capsid structure, infectivity, and other facets of their life cycle, there are significant gaps in our understanding of the fundamental biology of AAV and APVs that hamper the rational design and enhancement of hybrid parvoviral vectors.
Second, and related to this, another prerequisite for clinical translation is improvements in technology for manufacturing of hybrid vectors, including production, purification, titration, and storage in large scale and clinical grade and according to current GMP. In fact, to date, protocols used for hybrid parvoviral vector generation are designed for smaller scales that suffice for experimental lab work but not for clinical evaluation. Fortunately, we are optimistic that the field can largely and quickly benefit from the numerous advances in up- or downstream processing of AAV vectors that were reported and reviewed over the years.214, 215, 216, 217, 218 The exemplification of these requirements and of how better insights into virus biology benefit vector manufacturing offers a string of reports on BoV from the Yan,185 Qiu,183,219 and Engelhardt220 labs, in addition to those reviewed above, which yielded important insights into, e.g., the function of the viral NS proteins or cellular host factors for capsid protein expression183,220 or cis elements in the viral DNA hairpins that govern genome replication,219 many of which have already been used to improve AAV/HBoV vector production protocols.185 Concurrently, we anticipate a leap in our understanding of parvovirus biology and an ensuing benefit for vector engineering from the expansion of capsid diversification and selection strategies beyond AAV, as illustrated by our shuffling of bocaviral capsid variants.70 Together with the rapid concomitant progress in the resolution of the capsid structures of many parvoviral species as well as the prediction of critical residues and domains particularly driven by the Agbandje-McKenna lab,68,73, 74, 75,107,115 this combination of top-down (library screening) and bottom-up (rational design) strategies promises to propel the field of hybrid parvoviral vectors fast forward, as it has done over the last decades for AAV.
Third, as our knowledge of vector biology and our manufacturing skills increase, it will be exciting and highly informative to comprehensively study hybrid parvoviral vectors in small or large animal models of human disease. This will allow more learning about their clinically relevant features such as cell and tissue biodistribution, reactivity with pre-existing or induced, cellular or humoral immunity, amenability to vector readministration, or kinetics and efficiencies of transgene expression depending on route of administration and dose, to name a few. Most important will also be to carefully monitor potential in vivo toxicity of the novel hybrid vectors, a need that is most drastically illustrated by the unexpected and dire recent outcomes of a trial assessing high AAV8 vector doses for treatment of X-linked myotubular myopathy in children.221 Along these lines, others reported AAV vector toxicity due to high-level transgene expression in dorsal root ganglia (DRG).222,223 Luckily, they concurrently identified a solution that is fully compatible with hybrid parvoviral vectors, i.e., embedding microRNA (miR)-183 binding sites in the vector DNA, resulting in vector silencing in DRG due to their high inherent levels of miR-183.224
As a whole, we are convinced that these looming studies will allow us to even better understand and subsequently tailor the mechanisms underlying specific, robust, stable, and safe gene or protein delivery with hybrid vectors in vivo and to thereby fully realize the enormous potential and plasticity of this exhilarating vector type for human gene therapy.
Acknowledgments
The authors acknowledge support from the Heidelberg Biosciences International Graduate School HBIGS at Heidelberg University. This work was funded by the German Research Foundation (DFG) (Cluster of Excellence CellNetworks, EXC81; SFB1129, TP16, project number 240245660; TRR179, TP18, project number 272983813); the Cystic Fibrosis Foundation (CFF) (GRIMM15XX0); as well as the German Center for Infection Research (DZIF, BMBF) (TTU-HIV 04.803 and 04.815).
Declaration of interests
D.G. is a co-founder, shareholder, and chief scientific officer (CSO) of the company AaviGen GmbH.
References
- 1.Corsini J., Afanasiev B., Maxwell I.H., Carlson J.O. Autonomous parvovirus and densovirus gene vectors. Adv. Virus Res. 1996;47:303–351. doi: 10.1016/s0065-3527(08)60738-1. [DOI] [PubMed] [Google Scholar]
- 2.Maxwell I.H., Terrell K.L., Maxwell F. Autonomous parvovirus vectors. Methods. 2002;28:168–181. doi: 10.1016/s1046-2023(02)00221-9. [DOI] [PubMed] [Google Scholar]
- 3.Li C., Samulski R.J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 2020;21:255–272. doi: 10.1038/s41576-019-0205-4. [DOI] [PubMed] [Google Scholar]
- 4.Grimm D., Zolotukhin S. E Pluribus Unum: 50 Years of Research, Millions of Viruses, and One Goal--Tailored Acceleration of AAV Evolution. Mol. Ther. 2015;23:1819–1831. doi: 10.1038/mt.2015.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wu Z., Asokan A., Samulski R.J. Adeno-associated virus serotypes: vector toolkit for human gene therapy. Mol. Ther. 2006;14:316–327. doi: 10.1016/j.ymthe.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Wang D., Tai P.W.L., Gao G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019;18:358–378. doi: 10.1038/s41573-019-0012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoggan M.D., Blacklow N.R., Rowe W.P. Studies of small DNA viruses found in various adenovirus preparations: physical, biological, and immunological characteristics. Proc. Natl. Acad. Sci. USA. 1966;55:1467–1474. doi: 10.1073/pnas.55.6.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atchison R.W., Casto B.C., Hammon W.M. Adenovirus-Associated Defective Virus Particles. Science. 1965;149:754–756. doi: 10.1126/science.149.3685.754. [DOI] [PubMed] [Google Scholar]
- 9.Melnick J.L., Mayor H.D., Smith K.O., Rapp F. Association of 20-Millimicron Particles with Adenoviruses. J. Bacteriol. 1965;90:271–274. doi: 10.1128/jb.90.1.271-274.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toolan H., Ledinko N. Growth and cytopathogenicity of H-viruses in human and simian cell cultures. Nature. 1965;208:812–813. doi: 10.1038/208812a0. [DOI] [PubMed] [Google Scholar]
- 11.Allander T., Tammi M.T., Eriksson M., Bjerkner A., Tiveljung-Lindell A., Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc. Natl. Acad. Sci. USA. 2005;102:12891–12896. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu Q., Huang Y., Wan C., Fu G., Qi B., Cheng L., Shi S., Chen H., Liu R., Chen Z. Genomic and pathogenic analysis of a Muscovy duck parvovirus strain causing short beak and dwarfism syndrome without tongue protrusion. Res. Vet. Sci. 2017;115:393–400. doi: 10.1016/j.rvsc.2017.07.006. [DOI] [PubMed] [Google Scholar]
- 13.Ning K., Liang T., Wang M., Dong Y., Qu S., Zhang D. Pathogenicity of a variant goose parvovirus, from short beak and dwarfism syndrome of Pekin ducks, in goose embryos and goslings. Avian Pathol. 2018;47:391–399. doi: 10.1080/03079457.2018.1459040. [DOI] [PubMed] [Google Scholar]
- 14.Qiu J., Cheng F., Yoto Y., Zádori Z., Pintel D. The expression strategy of goose parvovirus exhibits features of both the Dependovirus and Parvovirus genera. J. Virol. 2005;79:11035–11044. doi: 10.1128/JVI.79.17.11035-11044.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nault J.C., Datta S., Imbeaud S., Franconi A., Mallet M., Couchy G., Letouzé E., Pilati C., Verret B., Blanc J.F., et al. Recurrent AAV2-related insertional mutagenesis in human hepatocellular carcinomas. Nat. Genet. 2015;47:1187–1193. doi: 10.1038/ng.3389. [DOI] [PubMed] [Google Scholar]
- 16.Chandler R.J., LaFave M.C., Varshney G.K., Trivedi N.S., Carrillo-Carrasco N., Senac J.S., Wu W., Hoffmann V., Elkahloun A.G., Burgess S.M., Venditti C.P. Vector design influences hepatic genotoxicity after adeno-associated virus gene therapy. J. Clin. Invest. 2015;125:870–880. doi: 10.1172/JCI79213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan G.J., Dane A.P., Hallwirth C.V., Smyth C.M., Wilkie E.E., Amaya A.K., Zhu E., Khandekar N., Ginn S.L., Liao S.H.Y., et al. Identification of liver-specific enhancer-promoter activity in the 3′ untranslated region of the wild-type AAV2 genome. Nat. Genet. 2017;49:1267–1273. doi: 10.1038/ng.3893. [DOI] [PubMed] [Google Scholar]
- 18.Cotmore S.F., Agbandje-McKenna M., Chiorini J.A., Mukha D.V., Pintel D.J., Qiu J., Soderlund-Venermo M., Tattersall P., Tijssen P., Gatherer D., Davison A.J. The family Parvoviridae. Arch. Virol. 2014;159:1239–1247. doi: 10.1007/s00705-013-1914-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toolan H.W. Experimental production of mongoloid hamsters. Science. 1960;131:1446–1448. doi: 10.1126/science.131.3411.1446. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz D., Green B., Carmichael L.E., Parrish C.R. The canine minute virus (minute virus of canines) is a distinct parvovirus that is most similar to bovine parvovirus. Virology. 2002;302:219–223. doi: 10.1006/viro.2002.1674. [DOI] [PubMed] [Google Scholar]
- 21.Abinanti F.R., Warfield M.S. Recovery of a hemadsorbing virus (HADEN) from the gastrointestinal tract of calves. Virology. 1961;14:288–289. doi: 10.1016/0042-6822(61)90206-9. [DOI] [PubMed] [Google Scholar]
- 22.Binn L.N., Lazar E.C., Eddy G.A., Kajima M. Recovery and characterization of a minute virus of canines. Infect. Immun. 1970;1:503–508. doi: 10.1128/iai.1.5.503-508.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deleu L., Pujol A., Faisst S., Rommelaere J. Activation of promoter P4 of the autonomous parvovirus minute virus of mice at early S phase is required for productive infection. J. Virol. 1999;73:3877–3885. doi: 10.1128/jvi.73.5.3877-3885.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adeyemi R.O., Pintel D.J. Replication of minute virus of mice in murine cells is facilitated by virally induced depletion of p21. J. Virol. 2012;86:8328–8332. doi: 10.1128/JVI.00820-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muharram G., Le Rhun E., Loison I., Wizla P., Richard A., Martin N., Roussel A., Begue A., Devos P., Baranzelli M.C., et al. Parvovirus H-1 induces cytopathic effects in breast carcinoma-derived cultures. Breast Cancer Res. Treat. 2010;121:23–33. doi: 10.1007/s10549-009-0451-9. [DOI] [PubMed] [Google Scholar]
- 26.Deng X., Yan Z., Cheng F., Engelhardt J.F., Qiu J. Replication of an Autonomous Human Parvovirus in Non-dividing Human Airway Epithelium Is Facilitated through the DNA Damage and Repair Pathways. PLoS Pathog. 2016;12:e1005399. doi: 10.1371/journal.ppat.1005399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cornelis J.J., Becquart P., Duponchel N., Salomé N., Avalosse B.L., Namba M., Rommelaere J. Transformation of human fibroblasts by ionizing radiation, a chemical carcinogen, or simian virus 40 correlates with an increase in susceptibility to the autonomous parvoviruses H-1 virus and minute virus of mice. J. Virol. 1988;62:1679–1686. doi: 10.1128/jvi.62.5.1679-1686.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Spegelaere P., van Hille B., Spruyt N., Faisst S., Cornelis J.J., Rommelaere J. Initiation of transcription from the minute virus of mice P4 promoter is stimulated in rat cells expressing a c-Ha-ras oncogene. J. Virol. 1991;65:4919–4928. doi: 10.1128/jvi.65.9.4919-4928.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Salomé N., van Hille B., Duponchel N., Meneguzzi G., Cuzin F., Rommelaere J., Cornelis J.J. Sensitization of transformed rat cells to parvovirus MVMp is restricted to specific oncogenes. Oncogene. 1990;5:123–130. [PubMed] [Google Scholar]
- 30.Cornelis J.J., Chen Y.Q., Spruyt N., Duponchel N., Cotmore S.F., Tattersall P., Rommelaere J. Susceptibility of human cells to killing by the parvoviruses H-1 and minute virus of mice correlates with viral transcription. J. Virol. 1990;64:2537–2544. doi: 10.1128/jvi.64.6.2537-2544.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacroix J., Schlund F., Leuchs B., Adolph K., Sturm D., Bender S., Hielscher T., Pfister S.M., Witt O., Rommelaere J., et al. Oncolytic effects of parvovirus H-1 in medulloblastoma are associated with repression of master regulators of early neurogenesis. Int. J. Cancer. 2014;134:703–716. doi: 10.1002/ijc.28386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angelova A.L., Geletneky K., Nüesch J.P., Rommelaere J. Tumor Selectivity of Oncolytic Parvoviruses: From in vitro and Animal Models to Cancer Patients. Front. Bioeng. Biotechnol. 2015;3:55. doi: 10.3389/fbioe.2015.00055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermonat P.L. Down-regulation of the human c-fos and c-myc proto-oncogene promoters by adeno-associated virus Rep78. Cancer Lett. 1994;81:129–136. doi: 10.1016/0304-3835(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 34.Zhan D., Santin A.D., Liu Y., Parham G.P., Li C., Meyers C., Hermonat P.L. Binding of the human papillomavirus type 16 p97 promoter by the adeno-associated virus Rep78 major regulatory protein correlates with inhibition. J. Biol. Chem. 1999;274:31619–31624. doi: 10.1074/jbc.274.44.31619. [DOI] [PubMed] [Google Scholar]
- 35.Sonntag F., Schmidt K., Kleinschmidt J.A. A viral assembly factor promotes AAV2 capsid formation in the nucleolus. Proc. Natl. Acad. Sci. USA. 2010;107:10220–10225. doi: 10.1073/pnas.1001673107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ogden P.J., Kelsic E.D., Sinai S., Church G.M. Comprehensive AAV capsid fitness landscape reveals a viral gene and enables machine-guided design. Science. 2019;366:1139–1143. doi: 10.1126/science.aaw2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang L.S., Shi Y., Shenk T. Adeno-associated virus P5 promoter contains an adenovirus E1A-inducible element and a binding site for the major late transcription factor. J. Virol. 1989;63:3479–3488. doi: 10.1128/jvi.63.8.3479-3488.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Y., Seto E., Chang L.S., Shenk T. Transcriptional repression by YY1, a human GLI-Krüppel-related protein, and relief of repression by adenovirus E1A protein. Cell. 1991;67:377–388. doi: 10.1016/0092-8674(91)90189-6. [DOI] [PubMed] [Google Scholar]
- 39.Nüesch J.P., Lacroix J., Marchini A., Rommelaere J. Molecular pathways: rodent parvoviruses--mechanisms of oncolysis and prospects for clinical cancer treatment. Clin. Cancer Res. 2012;18:3516–3523. doi: 10.1158/1078-0432.CCR-11-2325. [DOI] [PubMed] [Google Scholar]
- 40.Dupont F. Risk assessment of the use of autonomous parvovirus-based vectors. Curr. Gene Ther. 2003;3:567–582. doi: 10.2174/1566523034578104. [DOI] [PubMed] [Google Scholar]
- 41.Haag A., Menten P., Van Damme J., Dinsart C., Rommelaere J., Cornelis J.J. Highly efficient transduction and expression of cytokine genes in human tumor cells by means of autonomous parvovirus vectors; generation of antitumor responses in recipient mice. Hum. Gene Ther. 2000;11:597–609. doi: 10.1089/10430340050015789. [DOI] [PubMed] [Google Scholar]
- 42.Dupont F., Avalosse B., Karim A., Mine N., Bosseler M., Maron A., Van den Broeke A.V., Ghanem G.E., Burny A., Zeicher M. Tumor-selective gene transduction and cell killing with an oncotropic autonomous parvovirus-based vector. Gene Ther. 2000;7:790–796. doi: 10.1038/sj.gt.3301161. [DOI] [PubMed] [Google Scholar]
- 43.Grekova S.P., Aprahamian M., Daeffler L., Leuchs B., Angelova A., Giese T., Galabov A., Heller A., Giese N.A., Rommelaere J., Raykov Z. Interferon γ improves the vaccination potential of oncolytic parvovirus H-1PV for the treatment of peritoneal carcinomatosis in pancreatic cancer. Cancer Biol. Ther. 2011;12:888–895. doi: 10.4161/cbt.12.10.17678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dempe S., Lavie M., Struyf S., Bhat R., Verbeke H., Paschek S., Berghmans N., Geibig R., Rommelaere J., Van Damme J., Dinsart C. Antitumoral activity of parvovirus-mediated IL-2 and MCP-3/CCL7 delivery into human pancreatic cancer: implication of leucocyte recruitment. Cancer Immunol. Immunother. 2012;61:2113–2123. doi: 10.1007/s00262-012-1279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hajda J., Lehmann M., Krebs O., Kieser M., Geletneky K., Jäger D., Dahm M., Huber B., Schöning T., Sedlaczek O., et al. A non-controlled, single arm, open label, phase II study of intravenous and intratumoral administration of ParvOryx in patients with metastatic, inoperable pancreatic cancer: ParvOryx02 protocol. BMC Cancer. 2017;17:576. doi: 10.1186/s12885-017-3604-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Angelova A.L., Aprahamian M., Balboni G., Delecluse H.J., Feederle R., Kiprianova I., Grekova S.P., Galabov A.S., Witzens-Harig M., Ho A.D., et al. Oncolytic rat parvovirus H-1PV, a candidate for the treatment of human lymphoma: In vitro and in vivo studies. Mol. Ther. 2009;17:1164–1172. doi: 10.1038/mt.2009.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Srivastava A., Lusby E.W., Berns K.I. Nucleotide sequence and organization of the adeno-associated virus 2 genome. J. Virol. 1983;45:555–564. doi: 10.1128/jvi.45.2.555-564.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Väisänen E., Fu Y., Hedman K., Söderlund-Venermo M. Human Protoparvoviruses. Viruses. 2017;9:354. doi: 10.3390/v9110354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Angelova A., Rommelaere J. Immune System Stimulation by Oncolytic Rodent Protoparvoviruses. Viruses. 2019;11:415. doi: 10.3390/v11050415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Maxwell I.H., Maxwell F., Rhode S.L., 3rd, Corsini J., Carlson J.O. Recombinant LuIII autonomous parvovirus as a transient transducing vector for human cells. Hum. Gene Ther. 1993;4:441–450. doi: 10.1089/hum.1993.4.4-441. [DOI] [PubMed] [Google Scholar]
- 51.Maxwell I.H., Spitzer A.L., Long C.J., Maxwell F. Autonomous parvovirus transduction of a gene under control of tissue-specific or inducible promoters. Gene Ther. 1996;3:28–36. [PubMed] [Google Scholar]
- 52.Samulski R.J., Berns K.I., Tan M., Muzyczka N. Cloning of adeno-associated virus into pBR322: rescue of intact virus from the recombinant plasmid in human cells. Proc. Natl. Acad. Sci. USA. 1982;79:2077–2081. doi: 10.1073/pnas.79.6.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Laughlin C.A., Tratschin J.D., Coon H., Carter B.J. Cloning of infectious adeno-associated virus genomes in bacterial plasmids. Gene. 1983;23:65–73. doi: 10.1016/0378-1119(83)90217-2. [DOI] [PubMed] [Google Scholar]
- 54.Hermonat P.L., Muzyczka N. Use of adeno-associated virus as a mammalian DNA cloning vector: transduction of neomycin resistance into mammalian tissue culture cells. Proc. Natl. Acad. Sci. USA. 1984;81:6466–6470. doi: 10.1073/pnas.81.20.6466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tratschin J.D., West M.H., Sandbank T., Carter B.J. A human parvovirus, adeno-associated virus, as a eucaryotic vector: transient expression and encapsidation of the procaryotic gene for chloramphenicol acetyltransferase. Mol. Cell. Biol. 1984;4:2072–2081. doi: 10.1128/mcb.4.10.2072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kotin R.M., Siniscalco M., Samulski R.J., Zhu X.D., Hunter L., Laughlin C.A., McLaughlin S., Muzyczka N., Rocchi M., Berns K.I. Site-specific integration by adeno-associated virus. Proc. Natl. Acad. Sci. USA. 1990;87:2211–2215. doi: 10.1073/pnas.87.6.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McLaughlin S.K., Collis P., Hermonat P.L., Muzyczka N. Adeno-associated virus general transduction vectors: analysis of proviral structures. J. Virol. 1988;62:1963–1973. doi: 10.1128/jvi.62.6.1963-1973.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Samulski R.J., Chang L.S., Shenk T. Helper-free stocks of recombinant adeno-associated viruses: normal integration does not require viral gene expression. J. Virol. 1989;63:3822–3828. doi: 10.1128/jvi.63.9.3822-3828.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McCarty D.M., Monahan P.E., Samulski R.J. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248–1254. doi: 10.1038/sj.gt.3301514. [DOI] [PubMed] [Google Scholar]
- 60.Zeltner N., Kohlbrenner E., Clément N., Weber T., Linden R.M. Near-perfect infectivity of wild-type AAV as benchmark for infectivity of recombinant AAV vectors. Gene Ther. 2010;17:872–879. doi: 10.1038/gt.2010.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Grimm D., Kern A., Pawlita M., Ferrari F., Samulski R., Kleinschmidt J. Titration of AAV-2 particles via a novel capsid ELISA: packaging of genomes can limit production of recombinant AAV-2. Gene Ther. 1999;6:1322–1330. doi: 10.1038/sj.gt.3300946. [DOI] [PubMed] [Google Scholar]
- 62.Buck T.M., Wijnholds J. Recombinant Adeno-Associated Viral Vectors (rAAV)-Vector Elements in Ocular Gene Therapy Clinical Trials and Transgene Expression and Bioactivity Assays. Int. J. Mol. Sci. 2020;21:4197. doi: 10.3390/ijms21124197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lengler J., Coulibaly S., Gruber B., Ilk R., Mayrhofer J., Scheiflinger F., Hoellriegl W., Falkner F.G., Rottensteiner H. Development of an In Vitro Biopotency Assay for an AAV8 Hemophilia B Gene Therapy Vector Suitable for Clinical Product Release. Mol. Ther. Methods Clin. Dev. 2020;17:581–588. doi: 10.1016/j.omtm.2020.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.De B.P., Chen A., Salami C.O., Van de Graaf B., Rosenberg J.B., Pagovich O.E., Sondhi D., Crystal R.G., Kaminsky S.M. In Vivo Potency Assay for Adeno-Associated Virus-Based Gene Therapy Vectors Using AAVrh.10 as an Example. Hum. Gene Ther. Methods. 2018;29:146–155. doi: 10.1089/hgtb.2017.246. [DOI] [PubMed] [Google Scholar]
- 65.Tsao J., Chapman M.S., Agbandje M., Keller W., Smith K., Wu H., Luo M., Smith T.J., Rossmann M.G., Compans R.W., et al. The three-dimensional structure of canine parvovirus and its functional implications. Science. 1991;251:1456–1464. doi: 10.1126/science.2006420. [DOI] [PubMed] [Google Scholar]
- 66.Chapman M.S., Rossmann M.G. Single-stranded DNA-protein interactions in canine parvovirus. Structure. 1995;3:151–162. doi: 10.1016/s0969-2126(01)00146-0. [DOI] [PubMed] [Google Scholar]
- 67.Llamas-Saiz A.L., Agbandje-McKenna M., Wikoff W.R., Bratton J., Tattersall P., Rossmann M.G. Structure determination of minute virus of mice. Acta Crystallogr. D Biol. Crystallogr. 1997;53:93–102. doi: 10.1107/S0907444996010566. [DOI] [PubMed] [Google Scholar]
- 68.Halder S., Nam H.J., Govindasamy L., Vogel M., Dinsart C., Salomé N., McKenna R., Agbandje-McKenna M. Structural characterization of H-1 parvovirus: comparison of infectious virions to empty capsids. J. Virol. 2013;87:5128–5140. doi: 10.1128/JVI.03416-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan Z., Keiser N.W., Song Y., Deng X., Cheng F., Qiu J., Engelhardt J.F. A novel chimeric adenoassociated virus 2/human bocavirus 1 parvovirus vector efficiently transduces human airway epithelia. Mol. Ther. 2013;21:2181–2194. doi: 10.1038/mt.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fakhiri J., Schneider M.A., Puschhof J., Stanifer M., Schildgen V., Holderbach S., Voss Y., El Andari J., Schildgen O., Boulant S., et al. Novel Chimeric Gene Therapy Vectors Based on Adeno-Associated Virus and Four Different Mammalian Bocaviruses. Mol. Ther. Methods Clin. Dev. 2019;12:202–222. doi: 10.1016/j.omtm.2019.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Douar A.M., Poulard K., Stockholm D., Danos O. Intracellular trafficking of adeno-associated virus vectors: routing to the late endosomal compartment and proteasome degradation. J. Virol. 2001;75:1824–1833. doi: 10.1128/JVI.75.4.1824-1833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duan D., Yue Y., Yan Z., Yang J., Engelhardt J.F. Endosomal processing limits gene transfer to polarized airway epithelia by adeno-associated virus. J. Clin. Invest. 2000;105:1573–1587. doi: 10.1172/JCI8317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhong L., Li B., Mah C.S., Govindasamy L., Agbandje-McKenna M., Cooper M., Herzog R.W., Zolotukhin I., Warrington K.H., Jr., Weigel-Van Aken K.A., et al. Next generation of adeno-associated virus 2 vectors: point mutations in tyrosines lead to high-efficiency transduction at lower doses. Proc. Natl. Acad. Sci. USA. 2008;105:7827–7832. doi: 10.1073/pnas.0802866105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Salganik M., Aydemir F., Nam H.J., McKenna R., Agbandje-McKenna M., Muzyczka N. Adeno-associated virus capsid proteins may play a role in transcription and second-strand synthesis of recombinant genomes. J. Virol. 2014;88:1071–1079. doi: 10.1128/JVI.02093-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Aydemir F., Salganik M., Resztak J., Singh J., Bennett A., Agbandje-McKenna M., Muzyczka N. Mutants at the 2-Fold Interface of Adeno-associated Virus Type 2 (AAV2) Structural Proteins Suggest a Role in Viral Transcription for AAV Capsids. J. Virol. 2016;90:7196–7204. doi: 10.1128/JVI.00493-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Powell S.K., Samulski R.J., McCown T.J. AAV Capsid-Promoter Interactions Determine CNS Cell-Selective Gene Expression In Vivo. Mol. Ther. 2020;28:1373–1380. doi: 10.1016/j.ymthe.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Johnson M., Huyn S., Burton J., Sato M., Wu L. Differential biodistribution of adenoviral vector in vivo as monitored by bioluminescence imaging and quantitative polymerase chain reaction. Hum. Gene Ther. 2006;17:1262–1269. doi: 10.1089/hum.2006.17.1262. [DOI] [PubMed] [Google Scholar]
- 78.Fisher K.J., Kelley W.M., Burda J.F., Wilson J.M. A novel adenovirus-adeno-associated virus hybrid vector that displays efficient rescue and delivery of the AAV genome. Hum. Gene Ther. 1996;7:2079–2087. doi: 10.1089/hum.1996.7.17-2079. [DOI] [PubMed] [Google Scholar]
- 79.Recchia A., Parks R.J., Lamartina S., Toniatti C., Pieroni L., Palombo F., Ciliberto G., Graham F.L., Cortese R., La Monica N., Colloca S. Site-specific integration mediated by a hybrid adenovirus/adeno-associated virus vector. Proc. Natl. Acad. Sci. USA. 1999;96:2615–2620. doi: 10.1073/pnas.96.6.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Steinwaerder D.S., Carlson C.A., Lieber A. Generation of adenovirus vectors devoid of all viral genes by recombination between inverted repeats. J. Virol. 1999;73:9303–9313. doi: 10.1128/jvi.73.11.9303-9313.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lieber A., Steinwaerder D.S., Carlson C.A., Kay M.A. Integrating adenovirus-adeno-associated virus hybrid vectors devoid of all viral genes. J. Virol. 1999;73:9314–9324. doi: 10.1128/jvi.73.11.9314-9324.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shayakhmetov D.M., Carlson C.A., Stecher H., Li Q., Stamatoyannopoulos G., Lieber A. A high-capacity, capsid-modified hybrid adenovirus/adeno-associated virus vector for stable transduction of human hematopoietic cells. J. Virol. 2002;76:1135–1143. doi: 10.1128/JVI.76.3.1135-1143.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gonçalves M.A., Pau M.G., de Vries A.A., Valerio D. Generation of a high-capacity hybrid vector: packaging of recombinant adenoassociated virus replicative intermediates in adenovirus capsids overcomes the limited cloning capacity of adenoassociated virus vectors. Virology. 2001;288:236–246. doi: 10.1006/viro.2001.1073. [DOI] [PubMed] [Google Scholar]
- 84.Recchia A., Perani L., Sartori D., Olgiati C., Mavilio F. Site-specific integration of functional transgenes into the human genome by adeno/AAV hybrid vectors. Mol. Ther. 2004;10:660–670. doi: 10.1016/j.ymthe.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 85.Yang C.C., Xiao X., Zhu X., Ansardi D.C., Epstein N.D., Frey M.R., Matera A.G., Samulski R.J. Cellular recombination pathways and viral terminal repeat hairpin structures are sufficient for adeno-associated virus integration in vivo and in vitro. J. Virol. 1997;71:9231–9247. doi: 10.1128/jvi.71.12.9231-9247.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnston K.M., Jacoby D., Pechan P.A., Fraefel C., Borghesani P., Schuback D., Dunn R.J., Smith F.I., Breakefield X.O. HSV/AAV hybrid amplicon vectors extend transgene expression in human glioma cells. Hum. Gene Ther. 1997;8:359–370. doi: 10.1089/hum.1997.8.3-359. [DOI] [PubMed] [Google Scholar]
- 87.Fraefel C., Jacoby D.R., Lage C., Hilderbrand H., Chou J.Y., Alt F.W., Breakefield X.O., Majzoub J.A. Gene transfer into hepatocytes mediated by helper virus-free HSV/AAV hybrid vectors. Mol. Med. 1997;3:813–825. [PMC free article] [PubMed] [Google Scholar]
- 88.Palombo F., Monciotti A., Recchia A., Cortese R., Ciliberto G., La Monica N. Site-specific integration in mammalian cells mediated by a new hybrid baculovirus-adeno-associated virus vector. J. Virol. 1998;72:5025–5034. doi: 10.1128/jvi.72.6.5025-5034.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Spitzer A.L., Maxwell F., Corsini J., Maxwell I.H. Species specificity for transduction of cultured cells by a recombinant LuIII rodent parvovirus genome encapsidated by canine parvovirus or feline panleukopenia virus. J. Gen. Virol. 1996;77:1787–1792. doi: 10.1099/0022-1317-77-8-1787. [DOI] [PubMed] [Google Scholar]
- 90.Maxwell I.H., Long C.J., Carlson J.O., Rhode S.L., 3rd, Maxwell F. Encapsidation of a recombinant LuIII parvovirus genome by H1 virus and the fibrotropic or lymphotropic strains of minute virus of mice. J. Gen. Virol. 1993;74:1175–1179. doi: 10.1099/0022-1317-74-6-1175. [DOI] [PubMed] [Google Scholar]
- 91.Maxwell I.H., Maxwell F. Parvovirus LuIII transducing vectors packaged by LuIII versus FPV capsid proteins: the VP1 N-terminal region is not a major determinant of human cell permissiveness. J. Gen. Virol. 2004;85:1251–1257. doi: 10.1099/vir.0.19490-0. [DOI] [PubMed] [Google Scholar]
- 92.Corsini J., Carlson J.O., Maxwell F., Maxwell I.H. Symmetric-strand packaging of recombinant parvovirus LuIII genomes that retain only the terminal regions. J. Virol. 1995;69:2692–2696. doi: 10.1128/jvi.69.4.2692-2696.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Spitzer A.L., Parrish C.R., Maxwell I.H. Tropic determinant for canine parvovirus and feline panleukopenia virus functions through the capsid protein VP2. J. Gen. Virol. 1997;78:925–928. doi: 10.1099/0022-1317-78-4-925. [DOI] [PubMed] [Google Scholar]
- 94.Maxwell I.H., Chapman J.T., Scherrer L.C., Spitzer A.L., Leptihn S., Maxwell F., Corsini J.A. Expansion of tropism of a feline parvovirus to target a human tumor cell line by display of an alpha(v) integrin binding peptide on the capsid. Gene Ther. 2001;8:324–331. doi: 10.1038/sj.gt.3301399. [DOI] [PubMed] [Google Scholar]
- 95.Maxwell I.H., Spitzer A.L., Maxwell F., Pintel D.J. The capsid determinant of fibrotropism for the MVMp strain of minute virus of mice functions via VP2 and not VP1. J. Virol. 1995;69:5829–5832. doi: 10.1128/jvi.69.9.5829-5832.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Grimm D., Kay M.A., Kleinschmidt J.A. Helper virus-free, optically controllable, and two-plasmid-based production of adeno-associated virus vectors of serotypes 1 to 6. Mol. Ther. 2003;7:839–850. doi: 10.1016/s1525-0016(03)00095-9. [DOI] [PubMed] [Google Scholar]
- 97.Grimm D., Pandey K., Nakai H., Storm T.A., Kay M.A. Liver transduction with recombinant adeno-associated virus is primarily restricted by capsid serotype not vector genotype. J. Virol. 2006;80:426–439. doi: 10.1128/JVI.80.1.426-439.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chiorini J.A., Yang L., Liu Y., Safer B., Kotin R.M. Cloning of adeno-associated virus type 4 (AAV4) and generation of recombinant AAV4 particles. J. Virol. 1997;71:6823–6833. doi: 10.1128/jvi.71.9.6823-6833.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Grimm D., Lee J.S., Wang L., Desai T., Akache B., Storm T.A., Kay M.A. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Xiao W., Chirmule N., Berta S.C., McCullough B., Gao G., Wilson J.M. Gene therapy vectors based on adeno-associated virus type 1. J. Virol. 1999;73:3994–4003. doi: 10.1128/jvi.73.5.3994-4003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Excoffon K.J., Koerber J.T., Dickey D.D., Murtha M., Keshavjee S., Kaspar B.K., Zabner J., Schaffer D.V. Directed evolution of adeno-associated virus to an infectious respiratory virus. Proc. Natl. Acad. Sci. USA. 2009;106:3865–3870. doi: 10.1073/pnas.0813365106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rutledge E.A., Halbert C.L., Russell D.W. Infectious clones and vectors derived from adeno-associated virus (AAV) serotypes other than AAV type 2. J. Virol. 1998;72:309–319. doi: 10.1128/jvi.72.1.309-319.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Davidson B.L., Stein C.S., Heth J.A., Martins I., Kotin R.M., Derksen T.A., Zabner J., Ghodsi A., Chiorini J.A. Recombinant adeno-associated virus type 2, 4, and 5 vectors: transduction of variant cell types and regions in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 2000;97:3428–3432. doi: 10.1073/pnas.050581197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gao G.P., Alvira M.R., Wang L., Calcedo R., Johnston J., Wilson J.M. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc. Natl. Acad. Sci. USA. 2002;99:11854–11859. doi: 10.1073/pnas.182412299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mori S., Wang L., Takeuchi T., Kanda T. Two novel adeno-associated viruses from cynomolgus monkey: pseudotyping characterization of capsid protein. Virology. 2004;330:375–383. doi: 10.1016/j.virol.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 106.Gao G., Vandenberghe L.H., Alvira M.R., Lu Y., Calcedo R., Zhou X., Wilson J.M. Clades of Adeno-associated viruses are widely disseminated in human tissues. J. Virol. 2004;78:6381–6388. doi: 10.1128/JVI.78.12.6381-6388.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Schmidt M., Govindasamy L., Afione S., Kaludov N., Agbandje-McKenna M., Chiorini J.A. Molecular characterization of the heparin-dependent transduction domain on the capsid of a novel adeno-associated virus isolate, AAV(VR-942) J. Virol. 2008;82:8911–8916. doi: 10.1128/JVI.00672-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Grieger J.C., Soltys S.M., Samulski R.J. Production of Recombinant Adeno-associated Virus Vectors Using Suspension HEK293 Cells and Continuous Harvest of Vector From the Culture Media for GMP FIX and FLT1 Clinical Vector. Mol. Ther. 2016;24:287–297. doi: 10.1038/mt.2015.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Srivastava C.H., Samulski R.J., Lu L., Larsen S.H., Srivastava A. Construction of a recombinant human parvovirus B19: adeno-associated virus 2 (AAV) DNA inverted terminal repeats are functional in an AAV-B19 hybrid virus. Proc. Natl. Acad. Sci. USA. 1989;86:8078–8082. doi: 10.1073/pnas.86.20.8078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang X.S., Yoder M.C., Zhou S.Z., Srivastava A. Parvovirus B19 promoter at map unit 6 confers autonomous replication competence and erythroid specificity to adeno-associated virus 2 in primary human hematopoietic progenitor cells. Proc. Natl. Acad. Sci. USA. 1995;92:12416–12420. doi: 10.1073/pnas.92.26.12416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ponnazhagan S., Weigel K.A., Raikwar S.P., Mukherjee P., Yoder M.C., Srivastava A. Recombinant human parvovirus B19 vectors: erythroid cell-specific delivery and expression of transduced genes. J. Virol. 1998;72:5224–5230. doi: 10.1128/jvi.72.6.5224-5230.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Weigel-Kelley K.A., Yoder M.C., Srivastava A. Recombinant human parvovirus B19 vectors: erythrocyte P antigen is necessary but not sufficient for successful transduction of human hematopoietic cells. J. Virol. 2001;75:4110–4116. doi: 10.1128/JVI.75.9.4110-4116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Krüger L., Eskerski H., Dinsart C., Cornelis J., Rommelaere J., Haberkorn U., Kleinschmidt J.A. Augmented transgene expression in transformed cells using a parvoviral hybrid vector. Cancer Gene Ther. 2008;15:252–267. doi: 10.1038/sj.cgt.7701113. [DOI] [PubMed] [Google Scholar]
- 114.Yan Z., Feng Z., Sun X., Zhang Y., Zou W., Wang Z., Jensen-Cody C., Liang B., Park S.Y., Qiu J., Engelhardt J.F. Human Bocavirus Type-1 Capsid Facilitates the Transduction of Ferret Airways by Adeno-Associated Virus Genomes. Hum. Gene Ther. 2017;28:612–625. doi: 10.1089/hum.2017.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Fakhiri J., Linse K.P., Mietzsch M., Xu M., Schneider M.A., Meister M., Schildgen O., Schnitzler P., Soderlund-Venermo M., Agbandje-McKenna M., Grimm D. Impact of Natural or Synthetic Singletons in the Capsid of Human Bocavirus 1 on Particle Infectivity and Immunoreactivity. J. Virol. 2020;94:e00170. doi: 10.1128/JVI.00170-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Deng X., Zou W., Yan Z., Qiu J. Establishment of a Recombinant AAV2/HBoV1 Vector Production System in Insect Cells. Genes (Basel) 2020;11:439. doi: 10.3390/genes11040439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wang C.Y., Wang S. Adeno-associated virus inverted terminal repeats improve neuronal transgene expression mediated by baculoviral vectors in rat brain. Hum. Gene Ther. 2005;16:1219–1226. doi: 10.1089/hum.2005.16.1219. [DOI] [PubMed] [Google Scholar]
- 118.Hajitou A., Trepel M., Lilley C.E., Soghomonyan S., Alauddin M.M., Marini F.C., 3rd, Restel B.H., Ozawa M.G., Moya C.A., Rangel R., et al. A hybrid vector for ligand-directed tumor targeting and molecular imaging. Cell. 2006;125:385–398. doi: 10.1016/j.cell.2006.02.042. [DOI] [PubMed] [Google Scholar]
- 119.Tsafa E., Al-Bahrani M., Bentayebi K., Przystal J., Suwan K., Hajitou A. The natural dietary genistein boosts bacteriophage-mediated cancer cell killing by improving phage-targeted tumor cell transduction. Oncotarget. 2016;7:52135–52149. doi: 10.18632/oncotarget.10662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Yata T., Lee E.L., Suwan K., Syed N., Asavarut P., Hajitou A. Modulation of extracellular matrix in cancer is associated with enhanced tumor cell targeting by bacteriophage vectors. Mol. Cancer. 2015;14:110. doi: 10.1186/s12943-015-0383-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Yata T., Lee K.Y., Dharakul T., Songsivilai S., Bismarck A., Mintz P.J., Hajitou A. Hybrid Nanomaterial Complexes for Advanced Phage-guided Gene Delivery. Mol. Ther. Nucleic Acids. 2014;3:e185. doi: 10.1038/mtna.2014.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Tao P., Mahalingam M., Marasa B.S., Zhang Z., Chopra A.K., Rao V.B. In vitro and in vivo delivery of genes and proteins using the bacteriophage T4 DNA packaging machine. Proc. Natl. Acad. Sci. USA. 2013;110:5846–5851. doi: 10.1073/pnas.1300867110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Zhu J., Tao P., Mahalingam M., Sha J., Kilgore P., Chopra A.K., Rao V. A prokaryotic-eukaryotic hybrid viral vector for delivery of large cargos of genes and proteins into human cells. Sci. Adv. 2019;5:eaax0064. doi: 10.1126/sciadv.aax0064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kotterman M.A., Schaffer D.V. Engineering adeno-associated viruses for clinical gene therapy. Nat. Rev. Genet. 2014;15:445–451. doi: 10.1038/nrg3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Li L., Shan T., Wang C., Côté C., Kolman J., Onions D., Gulland F.M., Delwart E. The fecal viral flora of California sea lions. J. Virol. 2011;85:9909–9917. doi: 10.1128/JVI.05026-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Tu M., Liu F., Chen S., Wang M., Cheng A. Role of capsid proteins in parvoviruses infection. Virol. J. 2015;12:114. doi: 10.1186/s12985-015-0344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Wrzesinski C., Tesfay L., Salomé N., Jauniaux J.C., Rommelaere J., Cornelis J., Dinsart C. Chimeric and pseudotyped parvoviruses minimize the contamination of recombinant stocks with replication-competent viruses and identify a DNA sequence that restricts parvovirus H-1 in mouse cells. J. Virol. 2003;77:3851–3858. doi: 10.1128/JVI.77.6.3851-3858.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Beck S.E., Jones L.A., Chesnut K., Walsh S.M., Reynolds T.C., Carter B.J., Askin F.B., Flotte T.R., Guggino W.B. Repeated delivery of adeno-associated virus vectors to the rabbit airway. J. Virol. 1999;73:9446–9455. doi: 10.1128/jvi.73.11.9446-9455.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Chao H., Liu Y., Rabinowitz J., Li C., Samulski R.J., Walsh C.E. Several log increase in therapeutic transgene delivery by distinct adeno-associated viral serotype vectors. Mol. Ther. 2000;2:619–623. doi: 10.1006/mthe.2000.0219. [DOI] [PubMed] [Google Scholar]
- 130.Rabinowitz J.E., Rolling F., Li C., Conrath H., Xiao W., Xiao X., Samulski R.J. Cross-packaging of a single adeno-associated virus (AAV) type 2 vector genome into multiple AAV serotypes enables transduction with broad specificity. J. Virol. 2002;76:791–801. doi: 10.1128/JVI.76.2.791-801.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Börner K., Kienle E., Huang L.Y., Weinmann J., Sacher A., Bayer P., Stüllein C., Fakhiri J., Zimmermann L., Westhaus A., et al. Pre-arrayed Pan-AAV Peptide Display Libraries for Rapid Single-Round Screening. Mol. Ther. 2020;28:1016–1032. doi: 10.1016/j.ymthe.2020.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Chiorini J.A., Afione S., Kotin R.M. Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J. Virol. 1999;73:4293–4298. doi: 10.1128/jvi.73.5.4293-4298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Halbert C.L., Allen J.M., Miller A.D. Adeno-associated virus type 6 (AAV6) vectors mediate efficient transduction of airway epithelial cells in mouse lungs compared to that of AAV2 vectors. J. Virol. 2001;75:6615–6624. doi: 10.1128/JVI.75.14.6615-6624.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ling C., Yin Z., Li J., Zhang D., Aslanidi G., Srivastava A. Strategies to generate high-titer, high-potency recombinant AAV3 serotype vectors. Mol. Ther. Methods Clin. Dev. 2016;3:16029. doi: 10.1038/mtm.2016.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Ferrari F.K., Samulski T., Shenk T., Samulski R.J. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J. Virol. 1996;70:3227–3234. doi: 10.1128/jvi.70.5.3227-3234.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Sanlioglu S., Duan D., Engelhardt J.F. Two independent molecular pathways for recombinant adeno-associated virus genome conversion occur after UV-C and E4orf6 augmentation of transduction. Hum. Gene Ther. 1999;10:591–602. doi: 10.1089/10430349950018661. [DOI] [PubMed] [Google Scholar]
- 137.Yan Z., Zak R., Zhang Y., Engelhardt J.F. Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol. 2005;79:364–379. doi: 10.1128/JVI.79.1.364-379.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Duan D., Sharma P., Yang J., Yue Y., Dudus L., Zhang Y., Fisher K.J., Engelhardt J.F. Circular intermediates of recombinant adeno-associated virus have defined structural characteristics responsible for long-term episomal persistence in muscle tissue. J. Virol. 1998;72:8568–8577. doi: 10.1128/jvi.72.11.8568-8577.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chamberlain K., Riyad J.M., Weber T. Expressing Transgenes That Exceed the Packaging Capacity of Adeno-Associated Virus Capsids. Hum. Gene Ther. Methods. 2016;27:1–12. doi: 10.1089/hgtb.2015.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yan Z., Zhang Y., Duan D., Engelhardt J.F. Trans-splicing vectors expand the utility of adeno-associated virus for gene therapy. Proc. Natl. Acad. Sci. USA. 2000;97:6716–6721. doi: 10.1073/pnas.97.12.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Duan D., Yue Y., Engelhardt J.F. Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther. 2001;4:383–391. doi: 10.1006/mthe.2001.0456. [DOI] [PubMed] [Google Scholar]
- 142.Koo T., Popplewell L., Athanasopoulos T., Dickson G. Triple trans-splicing adeno-associated virus vectors capable of transferring the coding sequence for full-length dystrophin protein into dystrophic mice. Hum. Gene Ther. 2014;25:98–108. doi: 10.1089/hum.2013.164. [DOI] [PubMed] [Google Scholar]
- 143.Carvalho L.S., Turunen H.T., Wassmer S.J., Luna-Velez M.V., Xiao R., Bennett J., Vandenberghe L.H. Evaluating Efficiencies of Dual AAV Approaches for Retinal Targeting. Front. Neurosci. 2017;11:503. doi: 10.3389/fnins.2017.00503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Haberman R.P., McCown T.J., Samulski R.J. Novel transcriptional regulatory signals in the adeno-associated virus terminal repeat A/D junction element. J. Virol. 2000;74:8732–8739. doi: 10.1128/jvi.74.18.8732-8739.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Earley L.F., Conatser L.M., Lue V.M., Dobbins A.L., Li C., Hirsch M.L., Samulski R.J. Adeno-Associated Virus Serotype-Specific Inverted Terminal Repeat Sequence Role in Vector Transgene Expression. Hum. Gene Ther. 2020;31:151–162. doi: 10.1089/hum.2019.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Flotte T.R., Afione S.A., Solow R., Drumm M.L., Markakis D., Guggino W.B., Zeitlin P.L., Carter B.J. Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem. 1993;268:3781–3790. [PubMed] [Google Scholar]
- 147.Choi V.W., McCarty D.M., Samulski R.J. AAV hybrid serotypes: improved vectors for gene delivery. Curr. Gene Ther. 2005;5:299–310. doi: 10.2174/1566523054064968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Weinmann J., Grimm D. Next-generation AAV vectors for clinical use: an ever-accelerating race. Virus Genes. 2017;53:707–713. doi: 10.1007/s11262-017-1502-7. [DOI] [PubMed] [Google Scholar]
- 149.Grimm D., Büning H. Small But Increasingly Mighty: Latest Advances in AAV Vector Research, Design, and Evolution. Hum. Gene Ther. 2017;28:1075–1086. doi: 10.1089/hum.2017.172. [DOI] [PubMed] [Google Scholar]
- 150.Shen X., Storm T., Kay M.A. Characterization of the relationship of AAV capsid domain swapping to liver transduction efficiency. Mol. Ther. 2007;15:1955–1962. doi: 10.1038/sj.mt.6300293. [DOI] [PubMed] [Google Scholar]
- 151.Körbelin J., Hunger A., Alawi M., Sieber T., Binder M., Trepel M. Optimization of design and production strategies for novel adeno-associated viral display peptide libraries. Gene Ther. 2017;24:470–481. doi: 10.1038/gt.2017.51. [DOI] [PubMed] [Google Scholar]
- 152.Maheshri N., Koerber J.T., Kaspar B.K., Schaffer D.V. Directed evolution of adeno-associated virus yields enhanced gene delivery vectors. Nat. Biotechnol. 2006;24:198–204. doi: 10.1038/nbt1182. [DOI] [PubMed] [Google Scholar]
- 153.Koerber J.T., Jang J.H., Schaffer D.V. DNA shuffling of adeno-associated virus yields functionally diverse viral progeny. Mol. Ther. 2008;16:1703–1709. doi: 10.1038/mt.2008.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Herrmann A.K., Bender C., Kienle E., Grosse S., El Andari J., Botta J., Schürmann N., Wiedtke E., Niopek D., Grimm D. A Robust and All-Inclusive Pipeline for Shuffling of Adeno-Associated Viruses. ACS Synth. Biol. 2019;8:194–206. doi: 10.1021/acssynbio.8b00373. [DOI] [PubMed] [Google Scholar]
- 155.Kienle E., Senís E., Börner K., Niopek D., Wiedtke E., Grosse S., Grimm D. Engineering and evolution of synthetic adeno-associated virus (AAV) gene therapy vectors via DNA family shuffling. J. Vis. Exp. 2012;2012:3819. doi: 10.3791/3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Müller O.J., Kaul F., Weitzman M.D., Pasqualini R., Arap W., Kleinschmidt J.A., Trepel M. Random peptide libraries displayed on adeno-associated virus to select for targeted gene therapy vectors. Nat. Biotechnol. 2003;21:1040–1046. doi: 10.1038/nbt856. [DOI] [PubMed] [Google Scholar]
- 157.Körbelin J., Sieber T., Michelfelder S., Lunding L., Spies E., Hunger A., Alawi M., Rapti K., Indenbirken D., Müller O.J., et al. Pulmonary Targeting of Adeno-associated Viral Vectors by Next-generation Sequencing-guided Screening of Random Capsid Displayed Peptide Libraries. Mol. Ther. 2016;24:1050–1061. doi: 10.1038/mt.2016.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Loiler S.A., Conlon T.J., Song S., Tang Q., Warrington K.H., Agarwal A., Kapturczak M., Li C., Ricordi C., Atkinson M.A., et al. Targeting recombinant adeno-associated virus vectors to enhance gene transfer to pancreatic islets and liver. Gene Ther. 2003;10:1551–1558. doi: 10.1038/sj.gt.3302046. [DOI] [PubMed] [Google Scholar]
- 159.Eichhoff A.M., Börner K., Albrecht B., Schäfer W., Baum N., Haag F., Körbelin J., Trepel M., Braren I., Grimm D., et al. Nanobody-Enhanced Targeting of AAV Gene Therapy Vectors. Mol. Ther. Methods Clin. Dev. 2019;15:211–220. doi: 10.1016/j.omtm.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Warrington K.H., Jr., Gorbatyuk O.S., Harrison J.K., Opie S.R., Zolotukhin S., Muzyczka N. Adeno-associated virus type 2 VP2 capsid protein is nonessential and can tolerate large peptide insertions at its N terminus. J. Virol. 2004;78:6595–6609. doi: 10.1128/JVI.78.12.6595-6609.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Bowles D.E., Rabinowitz J.E., Samulski R.J. Marker rescue of adeno-associated virus (AAV) capsid mutants: a novel approach for chimeric AAV production. J. Virol. 2003;77:423–432. doi: 10.1128/JVI.77.1.423-432.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Rabinowitz J.E., Bowles D.E., Faust S.M., Ledford J.G., Cunningham S.E., Samulski R.J. Cross-dressing the virion: the transcapsidation of adeno-associated virus serotypes functionally defines subgroups. J. Virol. 2004;78:4421–4432. doi: 10.1128/JVI.78.9.4421-4432.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Srivastava A., Lu L. Replication of B19 parvovirus in highly enriched hematopoietic progenitor cells from normal human bone marrow. J. Virol. 1988;62:3059–3063. doi: 10.1128/jvi.62.8.3059-3063.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Paglino J., Tattersall P. The parvoviral capsid controls an intracellular phase of infection essential for efficient killing of stepwise-transformed human fibroblasts. Virology. 2011;416:32–41. doi: 10.1016/j.virol.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cotmore S.F., Tattersall P. Parvovirus diversity and DNA damage responses. Cold Spring Harb. Perspect. Biol. 2013;5:a012989. doi: 10.1101/cshperspect.a012989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cotmore S.F., Tattersall P. Genome packaging sense is controlled by the efficiency of the nick site in the right-end replication origin of parvoviruses minute virus of mice and LuIII. J. Virol. 2005;79:2287–2300. doi: 10.1128/JVI.79.4.2287-2300.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cotmore S.F., Tattersall P. Resolution of parvovirus dimer junctions proceeds through a novel heterocruciform intermediate. J. Virol. 2003;77:6245–6254. doi: 10.1128/JVI.77.11.6245-6254.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou X., Zeng X., Fan Z., Li C., McCown T., Samulski R.J., Xiao X. Adeno-associated virus of a single-polarity DNA genome is capable of transduction in vivo. Mol. Ther. 2008;16:494–499. doi: 10.1038/sj.mt.6300397. [DOI] [PubMed] [Google Scholar]
- 169.Zhou F., Sun H., Wang Y. Porcine bocavirus: achievements in the past five years. Viruses. 2014;6:4946–4960. doi: 10.3390/v6124946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lau S.K.P., Ahmed S.S., Yeung H.C., Li K.S.M., Fan R.Y.Y., Cheng T.Y.C., Cai J.P., Wang M., Zheng B.J., Wong S.S.Y., et al. Identification and interspecies transmission of a novel bocaparvovirus among different bat species in China. J. Gen. Virol. 2016;97:3345–3358. doi: 10.1099/jgv.0.000645. [DOI] [PubMed] [Google Scholar]
- 171.Zhang C., Song F., Xiu L., Liu Y., Yang J., Yao L., Peng J. Identification and characterization of a novel rodent bocavirus from different rodent species in China. Emerg. Microbes Infect. 2018;7:48. doi: 10.1038/s41426-018-0052-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Arthur J.L., Higgins G.D., Davidson G.P., Givney R.C., Ratcliff R.M. A novel bocavirus associated with acute gastroenteritis in Australian children. PLoS Pathog. 2009;5:e1000391. doi: 10.1371/journal.ppat.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kapoor A., Simmonds P., Slikas E., Li L., Bodhidatta L., Sethabutr O., Triki H., Bahri O., Oderinde B.S., Baba M.M., et al. Human bocaviruses are highly diverse, dispersed, recombination prone, and prevalent in enteric infections. J. Infect. Dis. 2010;201:1633–1643. doi: 10.1086/652416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Kapoor A., Mehta N., Esper F., Poljsak-Prijatelj M., Quan P.L., Qaisar N., Delwart E., Lipkin W.I. Identification and characterization of a new bocavirus species in gorillas. PLoS ONE. 2010;5:e11948. doi: 10.1371/journal.pone.0011948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Brožová K., Hrazdilová K., Slaninková E., Modrý D., Černý J., Celer V. Genetic and phylogenetic characterization of novel bocaparvovirus infecting chimpanzee. Infect. Genet. Evol. 2016;37:231–236. doi: 10.1016/j.meegid.2015.11.013. [DOI] [PubMed] [Google Scholar]
- 176.Manteufel J., Truyen U. Animal bocaviruses: a brief review. Intervirology. 2008;51:328–334. doi: 10.1159/000173734. [DOI] [PubMed] [Google Scholar]
- 177.Guido M., Tumolo M.R., Verri T., Romano A., Serio F., De Giorgi M., De Donno A., Bagordo F., Zizza A. Human bocavirus: Current knowledge and future challenges. World J. Gastroenterol. 2016;22:8684–8697. doi: 10.3748/wjg.v22.i39.8684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Tabatabai J., Fakhiri J., Meyburg J., Linse K.P., Xu M., Söderlund-Venermo M., Grimm D., Schnitzler P. Severe Human Bocavirus 1 Respiratory Tract Infection in an Immunodeficient Child With Fatal Outcome. Pediatr. Infect. Dis. J. 2019;38:e219–e222. doi: 10.1097/INF.0000000000002354. [DOI] [PubMed] [Google Scholar]
- 179.Windisch W., Schildgen V., Malecki M., Lenz J., Brockmann M., Karagiannidis C., Schildgen O. Detection of HBoV DNA in idiopathic lung fibrosis, Cologne, Germany. J. Clin. Virol. 2013;58:325–327. doi: 10.1016/j.jcv.2013.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Höpken M., Förster I., Maune S., Brockmann M., Schildgen O., Schildgen V. Association of the Human Bocavirus With Tonsil Squamous Cell Carcinomas. Front. Microbiol. 2018;9:2450. doi: 10.3389/fmicb.2018.02450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Huang Q., Deng X., Yan Z., Cheng F., Luo Y., Shen W., Lei-Butters D.C., Chen A.Y., Li Y., Tang L., et al. Establishment of a reverse genetics system for studying human bocavirus in human airway epithelia. PLoS Pathog. 2012;8:e1002899. doi: 10.1371/journal.ppat.1002899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shen W., Deng X., Zou W., Cheng F., Engelhardt J.F., Yan Z., Qiu J. Identification and Functional Analysis of Novel Nonstructural Proteins of Human Bocavirus 1. J. Virol. 2015;89:10097–10109. doi: 10.1128/JVI.01374-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Zou W., Cheng F., Shen W., Engelhardt J.F., Yan Z., Qiu J. Nonstructural Protein NP1 of Human Bocavirus 1 Plays a Critical Role in the Expression of Viral Capsid Proteins. J. Virol. 2016;90:4658–4669. doi: 10.1128/JVI.02964-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Wang Z., Shen W., Cheng F., Deng X., Engelhardt J.F., Yan Z., Qiu J. Parvovirus Expresses a Small Noncoding RNA That Plays an Essential Role in Virus Replication. J. Virol. 2017;91:e02375-16. doi: 10.1128/JVI.02375-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Yan Z., Zou W., Feng Z., Shen W., Park S.Y., Deng X., Qiu J., Engelhardt J.F. Establishment of a High-Yield Recombinant Adeno-Associated Virus/Human Bocavirus Vector Production System Independent of Bocavirus Nonstructural Proteins. Hum. Gene Ther. 2019;30:556–570. doi: 10.1089/hum.2018.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Monahan P.E., Lothrop C.D., Sun J., Hirsch M.L., Kafri T., Kantor B., Sarkar R., Tillson D.M., Elia J.R., Samulski R.J. Proteasome inhibitors enhance gene delivery by AAV virus vectors expressing large genomes in hemophilia mouse and dog models: a strategy for broad clinical application. Mol. Ther. 2010;18:1907–1916. doi: 10.1038/mt.2010.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Mitchell A.M., Samulski R.J. Mechanistic insights into the enhancement of adeno-associated virus transduction by proteasome inhibitors. J. Virol. 2013;87:13035–13041. doi: 10.1128/JVI.01826-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Nathwani A.C., Cochrane M., McIntosh J., Ng C.Y., Zhou J., Gray J.T., Davidoff A.M. Enhancing transduction of the liver by adeno-associated viral vectors. Gene Ther. 2009;16:60–69. doi: 10.1038/gt.2008.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Hajitou A., Rangel R., Trepel M., Soghomonyan S., Gelovani J.G., Alauddin M.M., Pasqualini R., Arap W. Design and construction of targeted AAVP vectors for mammalian cell transduction. Nat. Protoc. 2007;2:523–531. doi: 10.1038/nprot.2007.51. [DOI] [PubMed] [Google Scholar]
- 190.Stoneham C.A., Hollinshead M., Hajitou A. Clathrin-mediated endocytosis and subsequent endo-lysosomal trafficking of adeno-associated virus/phage. J. Biol. Chem. 2012;287:35849–35859. doi: 10.1074/jbc.M112.369389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Przystal J.M., Umukoro E., Stoneham C.A., Yata T., O’Neill K., Syed N., Hajitou A. Proteasome inhibition in cancer is associated with enhanced tumor targeting by the adeno-associated virus/phage. Mol. Oncol. 2013;7:55–66. doi: 10.1016/j.molonc.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yan Z., Zak R., Luxton G.W., Ritchie T.C., Bantel-Schaal U., Engelhardt J.F. Ubiquitination of both adeno-associated virus type 2 and 5 capsid proteins affects the transduction efficiency of recombinant vectors. J. Virol. 2002;76:2043–2053. doi: 10.1128/jvi.76.5.2043-2053.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Wu W.K., Cho C.H., Lee C.W., Wu K., Fan D., Yu J., Sung J.J. Proteasome inhibition: a new therapeutic strategy to cancer treatment. Cancer Lett. 2010;293:15–22. doi: 10.1016/j.canlet.2009.12.002. [DOI] [PubMed] [Google Scholar]
- 194.Mah C., Qing K., Khuntirat B., Ponnazhagan S., Wang X.S., Kube D.M., Yoder M.C., Srivastava A. Adeno-associated virus type 2-mediated gene transfer: role of epidermal growth factor receptor protein tyrosine kinase in transgene expression. J. Virol. 1998;72:9835–9843. doi: 10.1128/jvi.72.12.9835-9843.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Kia A., Yata T., Hajji N., Hajitou A. Inhibition of histone deacetylation and DNA methylation improves gene expression mediated by the adeno-associated virus/phage in cancer cells. Viruses. 2013;5:2561–2572. doi: 10.3390/v5102561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Campbell S., Suwan K., Waramit S., Aboagye E.O., Hajitou A. Selective Inhibition of Histone Deacetylation in Melanoma Increases Targeted Gene Delivery by a Bacteriophage Viral Vector. Cancers (Basel) 2018;10:125. doi: 10.3390/cancers10040125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Moulay G., Boutin S., Masurier C., Scherman D., Kichler A. Polymers for improving the in vivo transduction efficiency of AAV2 vectors. PLoS ONE. 2010;5:e15576. doi: 10.1371/journal.pone.0015576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Smith T.L., Souza G.R., Sidman R.L., Arap W., Pasqualini R. An AAVP-based solid-phase transducing matrix for transgene delivery: potential for translational applications. Cancer Gene Ther. 2017;24:358–360. doi: 10.1038/cgt.2017.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Suwan K., Yata T., Waramit S., Przystal J.M., Stoneham C.A., Bentayebi K., Asavarut P., Chongchai A., Pothachareon P., Lee K.Y., et al. Next-generation of targeted AAVP vectors for systemic transgene delivery against cancer. Proc. Natl. Acad. Sci. USA. 2019;116:18571–18577. doi: 10.1073/pnas.1906653116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Alves-Cruzeiro J., Webster C.P., Azzouz M. The hybrid AAVP tool gets an upgrade. Proc. Natl. Acad. Sci. USA. 2019;116:18162–18164. doi: 10.1073/pnas.1912635116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Hajitou A., Lev D.C., Hannay J.A., Korchin B., Staquicini F.I., Soghomonyan S., Alauddin M.M., Benjamin R.S., Pollock R.E., Gelovani J.G., et al. A preclinical model for predicting drug response in soft-tissue sarcoma with targeted AAVP molecular imaging. Proc. Natl. Acad. Sci. USA. 2008;105:4471–4476. doi: 10.1073/pnas.0712184105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Tandle A., Hanna E., Lorang D., Hajitou A., Moya C.A., Pasqualini R., Arap W., Adem A., Starker E., Hewitt S., Libutti S.K. Tumor vasculature-targeted delivery of tumor necrosis factor-alpha. Cancer. 2009;115:128–139. doi: 10.1002/cncr.24001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Yuan Z., Syrkin G., Adem A., Geha R., Pastoriza J., Vrikshajanani C., Smith T., Quinn T.J., Alemu G., Cho H., et al. Blockade of inhibitors of apoptosis (IAPs) in combination with tumor-targeted delivery of tumor necrosis factor-α leads to synergistic antitumor activity. Cancer Gene Ther. 2013;20:46–56. doi: 10.1038/cgt.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kia A., Przystal J.M., Nianiaris N., Mazarakis N.D., Mintz P.J., Hajitou A. Dual systemic tumor targeting with ligand-directed phage and Grp78 promoter induces tumor regression. Mol. Cancer Ther. 2012;11:2566–2577. doi: 10.1158/1535-7163.MCT-12-0587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Przystal J.M., Waramit S., Pranjol M.Z.I., Yan W., Chu G., Chongchai A., Samarth G., Olaciregui N.G., Tabatabai G., Carcaboso A.M., et al. Efficacy of systemic temozolomide-activated phage-targeted gene therapy in human glioblastoma. EMBO Mol. Med. 2019;11:e8492. doi: 10.15252/emmm.201708492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Staquicini F.I., Smith T.L., Tang F.H.F., Gelovani J.G., Giordano R.J., Libutti S.K., Sidman R.L., Cavenee W.K., Arap W., Pasqualini R. Targeted AAVP-based therapy in a mouse model of human glioblastoma: a comparison of cytotoxic versus suicide gene delivery strategies. Cancer Gene Ther. 2020;27:301–310. doi: 10.1038/s41417-019-0101-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Smith T.L., Yuan Z., Cardó-Vila M., Sanchez Claros C., Adem A., Cui M.H., Branch C.A., Gelovani J.G., Libutti S.K., Sidman R.L., et al. AAVP displaying octreotide for ligand-directed therapeutic transgene delivery in neuroendocrine tumors of the pancreas. Proc. Natl. Acad. Sci. USA. 2016;113:2466–2471. doi: 10.1073/pnas.1525709113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Dobroff A.S., D’Angelo S., Eckhardt B.L., Ferrara F., Staquicini D.I., Cardó-Vila M., Staquicini F.I., Nunes D.N., Kim K., Driessen W.H.P., et al. Towards a transcriptome-based theranostic platform for unfavorable breast cancer phenotypes. Proc. Natl. Acad. Sci. USA. 2016;113:12780–12785. doi: 10.1073/pnas.1615288113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Ferrara F., Staquicini D.I., Driessen W.H.P., D’Angelo S., Dobroff A.S., Barry M., Lomo L.C., Staquicini F.I., Cardó-Vila M., Soghomonyan S., et al. Targeted molecular-genetic imaging and ligand-directed therapy in aggressive variant prostate cancer. Proc. Natl. Acad. Sci. USA. 2016;113:12786–12791. doi: 10.1073/pnas.1615400113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Khalili S., Rasaee M.J., Bamdad T., Mard-Soltani M., Asadi Ghalehni M., Jahangiri A., Pouriayevali M.H., Aghasadeghi M.R., Malaei F. A Novel Molecular Design for a Hybrid Phage-DNA Construct Against DKK1. Mol. Biotechnol. 2018;60:833–842. doi: 10.1007/s12033-018-0115-2. [DOI] [PubMed] [Google Scholar]
- 211.Feng M., Jackson W.H., Jr., Goldman C.K., Rancourt C., Wang M., Dusing S.K., Siegal G., Curiel D.T. Stable in vivo gene transduction via a novel adenoviral/retroviral chimeric vector. Nat. Biotechnol. 1997;15:866–870. doi: 10.1038/nbt0997-866. [DOI] [PubMed] [Google Scholar]
- 212.Gluzman Y., Van Doren K. Palindromic adenovirus type 5-simian virus 40 hybrid. J. Virol. 1983;45:91–103. doi: 10.1128/jvi.45.1.91-103.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Leblois H., Roche C., Di Falco N., Orsini C., Yeh P., Perricaudet M. Stable transduction of actively dividing cells via a novel adenoviral/episomal vector. Mol. Ther. 2000;1:314–322. doi: 10.1006/mthe.2000.0042. [DOI] [PubMed] [Google Scholar]
- 214.Ayuso E., Mingozzi F., Bosch F. Production, purification and characterization of adeno-associated vectors. Curr. Gene Ther. 2010;10:423–436. doi: 10.2174/156652310793797685. [DOI] [PubMed] [Google Scholar]
- 215.Clément N., Grieger J.C. Manufacturing of recombinant adeno-associated viral vectors for clinical trials. Mol. Ther. Methods Clin. Dev. 2016;3:16002. doi: 10.1038/mtm.2016.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Aponte-Ubillus J.J., Barajas D., Peltier J., Bardliving C., Shamlou P., Gold D. Molecular design for recombinant adeno-associated virus (rAAV) vector production. Appl. Microbiol. Biotechnol. 2018;102:1045–1054. doi: 10.1007/s00253-017-8670-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.O’Riordan C.R., Lachapelle A.L., Vincent K.A., Wadsworth S.C. Scaleable chromatographic purification process for recombinant adeno-associated virus (rAAV) J. Gene Med. 2000;2:444–454. doi: 10.1002/1521-2254(200011/12)2:6<444::AID-JGM132>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 218.El Andari J., Grimm D. Production, Processing, and Characterization of Synthetic AAV Gene Therapy Vectors. Biotechnol. J. 2021;16:e2000025. doi: 10.1002/biot.202000025. [DOI] [PubMed] [Google Scholar]
- 219.Shen W., Deng X., Zou W., Engelhardt J.F., Yan Z., Qiu J. Analysis of cis and trans Requirements for DNA Replication at the Right-End Hairpin of the Human Bocavirus 1 Genome. J. Virol. 2016;90:7761–7777. doi: 10.1128/JVI.00708-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang X., Xu P., Cheng F., Li Y., Wang Z., Hao S., Wang J., Ning K., Ganaie S.S., Engelhardt J.F., et al. Cellular Cleavage and Polyadenylation Specificity Factor 6 (CPSF6) Mediates Nuclear Import of Human Bocavirus 1 NP1 Protein and Modulates Viral Capsid Protein Expression. J. Virol. 2020;94:e01444. doi: 10.1128/JVI.01444-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Wilson J.M., Flotte T.R. Moving Forward After Two Deaths in a Gene Therapy Trial of Myotubular Myopathy. Hum. Gene Ther. 2020;31:695–696. doi: 10.1089/hum.2020.182. [DOI] [PubMed] [Google Scholar]
- 222.Hordeaux J., Hinderer C., Buza E.L., Louboutin J.P., Jahan T., Bell P., Chichester J.A., Tarantal A.F., Wilson J.M. Safe and Sustained Expression of Human Iduronidase After Intrathecal Administration of Adeno-Associated Virus Serotype 9 in Infant Rhesus Monkeys. Hum. Gene Ther. 2019;30:957–966. doi: 10.1089/hum.2019.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Hinderer C., Katz N., Buza E.L., Dyer C., Goode T., Bell P., Richman L.K., Wilson J.M. Severe Toxicity in Nonhuman Primates and Piglets Following High-Dose Intravenous Administration of an Adeno-Associated Virus Vector Expressing Human SMN. Hum. Gene Ther. 2018;29:285–298. doi: 10.1089/hum.2018.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Hordeaux J., Buza E.L., Jeffrey B., Song C., Jahan T., Yuan Y., Zhu Y., Bell P., Li M., Chichester J.A., et al. MicroRNA-mediated inhibition of transgene expression reduces dorsal root ganglion toxicity by AAV vectors in primates. Sci. Transl. Med. 2020;12:eaba9188. doi: 10.1126/scitranslmed.aba9188. [DOI] [PubMed] [Google Scholar]