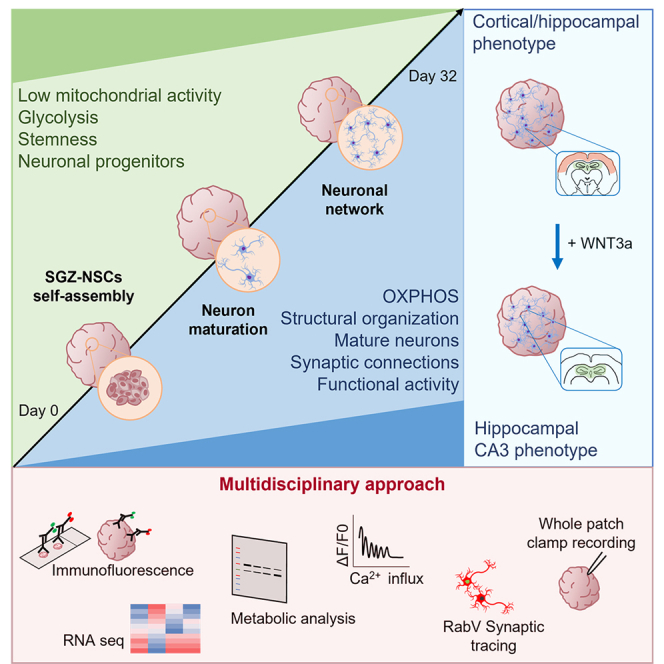
Summary
Brain organoids are in vitro three-dimensional (3D) self-organized neural structures, which can enable disease modeling and drug screening. However, their use for standardized large-scale drug screening studies is limited by their high batch-to-batch variability, long differentiation time (10–20 weeks), and high production costs. This is particularly relevant when brain organoids are obtained from human induced pluripotent stem cells (iPSCs). Here, we developed, for the first time, a highly standardized, reproducible, and fast (5 weeks) murine brain organoid model starting from embryonic neural stem cells. We obtained brain organoids, which progressively differentiated and self-organized into 3D networks of functional neurons with dorsal forebrain phenotype. Furthermore, by adding the morphogen WNT3a, we generated brain organoids with specific hippocampal region identity. Overall, our results showed the establishment of a fast, robust and reproducible murine 3D in vitro brain model that may represent a useful tool for high-throughput drug screening and disease modeling.
Subject areas: Biological sciences, Neuroscience, Cell biology, Developmental biology
Graphical abstract
Highlights
-
•
Murine embryonic NSCs are able to self-organize into brain organoids
-
•
Murine brain organoids mature in 32 days, showing dorsal forebrain identity
-
•
Brain organoids develop 3D network of functional neurons
-
•
WNT3a supplementation induces specific hippocampal brain region identity
Biological sciences; Neuroscience; Cell biology; Developmental biology
Introduction
In recent years, in vitro three-dimensional (3D) biological systems have emerged as the state-of-the-art biotechnology to investigate brain development, neurological or neuropsychiatric disorders, and to perform large-scale pharmacological studies. Drug discovery is a complex process with a low success rate (Xue et al., 2018). To ameliorate this aspect, high-throughput drug screening and drug repositioning strategies have been developed. High-throughput drug screening allows us to assay a large amount of compounds, but specific platforms are necessary to make the process reliable and fast. A crucial aspect is the possibility of using a robust and appropriate model of disease. This is particularly challenging for disorders affecting the central nervous system (CNS), given the complexity of the cerebral architecture. To overcome this aspect, 3D-based in vitro approaches, such as brain organoids, represent an appealing alternative to animal models for preclinical assays (Bedard et al., 2020). Brain organoids are in vitro 3D structures able to recapitulate the development and topological organization of the brain tissue. They are typically derived from induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs), exploiting the stem cell properties of self-renewal and self-organization (Lancaster and Knoblich, 2014; Lancaster et al., 2013).
Since the original work by Reynolds (Reynolds and Weiss, 1992), traditional 3D neuronal cultures have been identified as neurospheres or neuro-aggregates in which neuronal progenitors grow in suspension in a medium that favors their expansion maintaining their stem cell properties (Singec et al., 2006; Soares et al., 2021). With time, in vitro cultured brain organoids develop into mature neural structure, which recapitulate the development and topological organization of the in vivo brain tissue. Eiraku et al. showed that mouse ESCs could be steered to differentiate into neuroepithelial tissues when cultured as floating aggregates in a serum-free medium (Eiraku et al., 2008). Starting from this pioneering study, more sophisticated and fine-tuned culture methods have been developed to generate human brain organ-like structures, termed organoids or spheroids (Lancaster et al., 2013; Pasca et al., 2015; Bagley et al., 2017; Qian et al., 2016; Yoon et al., 2019). These structures can also generate brain region-specific organoids (Qian et al., 2018; Muguruma et al., 2015; Xiang et al., 2019; Monzel et al., 2017; Pellegrini et al., 2020; Sakaguchi et al., 2015; Winanto et al., 2019).
The intensive use of human iPSCs (hiPSCs) is prompted by the achievement of comparable results between the patient-derived organoid and the diseased tissue (Lee et al., 2017). hiPSC-derived brain organoids own relevant translational potential and several groups have recently improved the protocol for their reliable and reproducible generation (Velasco et al., 2019; Yoon et al., 2019). On the other hand, accordingly to human development timeline (Budday et al., 2015), hiPSC-derived brain organoids require long time and high production cost for their in vitro generation that may hinder their potential application in large-scale studies.
The development of highly reproducible functional murine brain organoids may overcome some of these limitations and may represent a useful supportive tool to optimize protocols for human-derived brain organoids and to perform large-scale drug discovery studies. Given the fast growth rate and short scaling time of mouse brain development, murine brain organoids can be generated in a short time and with low production costs. Moreover, the extensive amount of in vivo data available on mouse brain development, together with the possibility of obtaining genetically mutant phenotype samples, provide invaluable points of comparison to validate murine organoids as systems to study the pathophysiology of different brain disorders (Marshall and Mason, 2019). Despite early attempts to generate 3D forebrain structures from mouse and human ESCs by Sasai et al. (Eiraku et al., 2008; Watanabe et al., 2005; Nasu et al., 2012), there are no data in literature on the generation of robust and reproducible functional rodent brain organoids (Marshall and Mason, 2019).
In this work, we have generated highly standardized murine brain organoids with functional neurons in 32 d of in vitro culture. Several complementary techniques, including global transcriptomic analysis, protein expression, and metabolic analysis showed the highly reproducible and consistent progressive maturation of the brain organoids. Moreover, by combining different methods such as trans-synaptic tracing, wholemount calcium imaging, whole-cell patch clamp, and microelectrode array (MEA) recordings, we showed that brain organoids acquire functional neurons with neuronal synaptic connections organized in 3D networks. Finally, we demonstrated that murine brain organoids can be further induced to acquire brain region–specific features, namely, by adding the morphogen WNT3a we obtained brain organoids enriched in neurons with CA3 hippocampal signature.
Overall, our results showed robust and reproducible murine 3D in vitro brain model with mature neural structure and functional neurons.
Results
Murine brain organoids show reproducible development and maturation
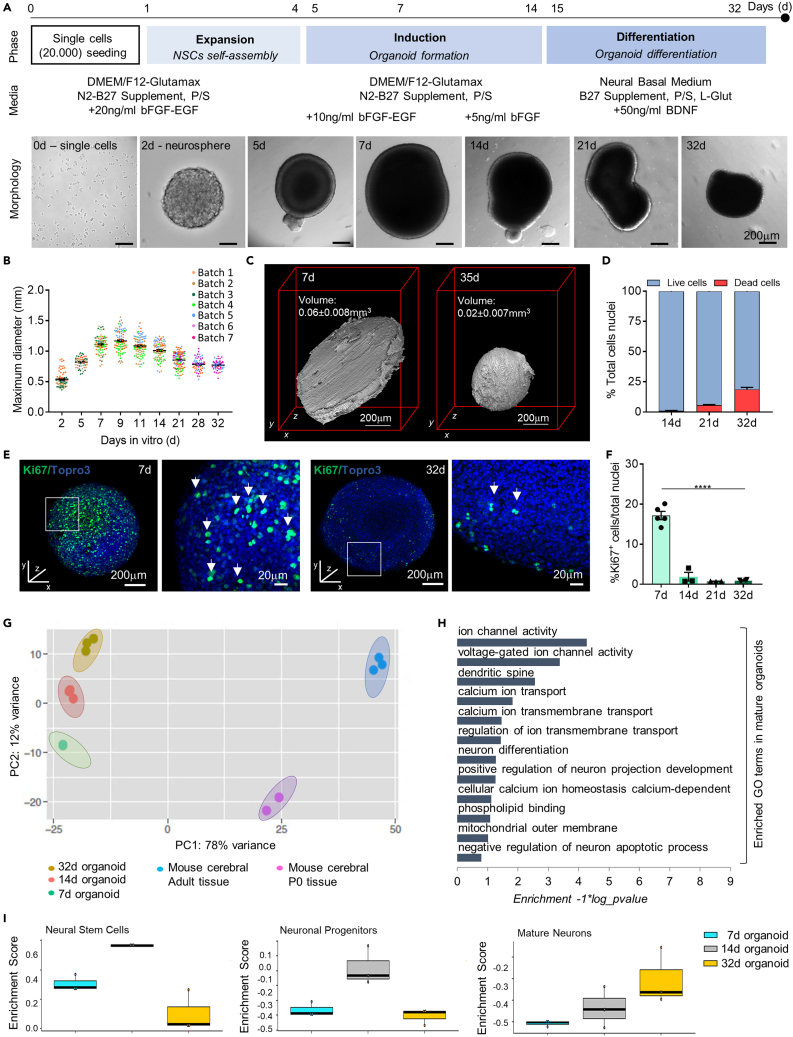
We isolated and cultured in vitro neural stem cells (NSCs) from the subgranular zone (SGZ), a well-known stem cells niche (Pino et al., 2017; Decimo et al., 2020), of mouse brain embryos (E14.5) following a three-step culture method. During the first phase of the protocol, referred to as expansion phase (days 0–4), the SGZ-derived NSCs were seeded at a specific cell density (20000 cells/well) (Lancaster and Knoblich, 2014) in each well of a 24-well plate (day 0) and kept in dynamic culture in growth media (see STAR Methods section) supplemented with 20 ng/mL of epidermal growth factor (EGF) and 20 ng/mL of basic fibroblast growth factor (bFGF). At this stage, cells started to self-assemble into neurospheres. The accurate initial cell seeding density allowed us to obtain, in each well, one organoid with defined size (maximum diameter: 0.82 ± 0.01 mm) at the end of the expansion phase (day 4, Figures 1A and 1B). To reduce cell proliferation and trigger cell differentiation, we subsequently lowered the growth factors concentration to 10 ng/mL EGF and bFGF (day 5) and then 5 ng/mL bFGF (day 7) (induction phase, days 5–14) (Figure 1A). In line with the morphology of early development human brain organoid (Lancaster and Knoblich, 2014), by day 7, we observed organoids with smooth edges, a bright and optically translucent surface, and a darker center (Figure 1A). At day 9, the organoids reached the maximum diameter of 1.7 ± 0.02 mm (n = 89) (Figure 1B).
Figure 1.
Reliability of murine brain organoid model
(A) Schematic representation of the three-phase organoid generation protocol. After single cells (20000 cells/well) seeding on a 24-well plate (day 0), the protocol starts with the initial Expansion Phase that lasts up to day 4: mouse NSCs proliferation and neurosphere formation in culture medium enriched with bFGF and EGF (20ng/ml). Induction Phase from day 5 to day 14: early organoid formation in medium supplemented with gradual decrease of bFGF and EGF (day 5 to day 6: 10ng/ml bFGF and EGF; day 7 to day 14: 5ng/ml bFGF). Differentiation Phase from day 15 to day 32: organoids maturation in differentiation medium (BDNF 50ng/ml) up to day 32. In all the three phases, organoids were maintained on an orbital shaker (dynamic culture). Pictures in (A) are representative brightfield images of single NSCs, a neurosphere and organoids at different stages of the protocol. At day 5 the early formed organoids show the presence of brightened translucent surface tissue, whereas, the central core is quite dark. Late organoids develop gradual dark and compact tissue morphology and structural remodeling.
(B) Graph representing the growth curve of murine brain organoids. An exponential growth is observed up to day 11 whereas the maximum diameter progressively diminishes as the differentiation proceeds. Data are expressed as mean ± SEM of the maximum diameter (n>30). Organoids belonging to different SGZ-NSCs batches were reported in different colors.
(C) Optical projection tomography (OPT) reconstruction of representative organoids at day 7 and day 35 showing the 3D structure of the in vitro generated organoid. The reported organoids volume was calculated for n = 2 organoids at 7d and n = 3 organoids at 35d based on the 360-degree OPT reconstructions using Avizo. Data are expressed as mean ± SEM.
(D) Graph representing the number of live and dead cells expressed in % of total cells number. The analysis was performed on n = 3 organoids/time point. Dead cells were identified as propidium iodide positive cells while the number of live cells was obtained by subtracting dead cells from the total nuclei.
(E) Representative confocal immunofluorescence maximum intensity projections of z stack images of whole mount murine brain organoids at 7 and 32 days in culture, showing the proliferative marker Ki67 (in green). Total nuclei are visible in blue (TOPRO-3). White boxes indicate a magnification of a region of the whole mount organoids, presented as single plane image. White arrows show Ki67+ cells.
(F) Graph representing the percentage of Ki67+ cells over total cell nuclei at different time points. Data are expressed as mean ± SEM. Analysis was performed on n = 3 different organoids and at least on 3 entire sections for each organoid. p < 0.05 was considered statistically significant. Statistical differences between marker expression at different time points were calculated by ordinary one-way ANOVA followed by Tukey's multiple comparison test. ∗∗∗∗p < 0.0001.
(G) Principal components analysis (PCA) of RNAseq data from organoid samples at different stages of differentiation (7 days: n = 3; 14 days: n = 3; 32 days: n = 3) and from murine cerebral tissues (P0: n = 2; adult: n = 3). PCA shows organoids and tissues cluster according to their different maturation phases.
(H) Gene ontology (GO) analysis for up-regulated genes in mature (32 days) versus early (7 days) organoids. Based on global transcriptomic analysis, GO analysis was performed in order to identify GO terms enriched in the up-regulated genes in mature compared to early organoids and to classify them based on molecular function, biological process and cellular components.
(I) Box plots of GSVA RNAseq data analysis generated evaluating specific gene lists for neural stem cells, neuronal progenitors and mature neurons in 7d, 14d and 32d organoids showing upregulation of genes related to neuronal maturation in late organoids. The Enrichment Score represents the amount to which the genes in the specific gene lists are over-represented.
To specifically induce neuronal differentiation and maturation, following the induction phase, we added the brain-derived neurotrophic factor (BDNF) 50 ng/mL to the culture media (differentiation phase, days 15–32) (Figure 1A). The BDNF belongs to the neurotrophin family, is a key molecule in neural development and function, and is required to increase the neurite number, length and complexity, as well as dendritic spine density in primary 2D NSC-derived neuronal cultures (Silva et al., 2009). During this phase, the organoids showed a thinner translucent surface with a darker core and they achieved a stationary maximum diameter of 0.79 ± 0.01 mm (n = 59) at day 28 and of 0.78 ± 0.01 mm (n = 49) at day 32 (Figure 1B). These data indicated that the organoids developed with consistent size throughout the different phases of the protocol. To assess the overall success rate of organoid formation across different murine SGZ-derived NSC batches (n = 7), we started the production of at least 54 organoids for each batch for a total of 732 generated organoids. We found a 95% rate of successful organoid formation and maintenance along the in vitro culture timing (32d) with a 5% rate of organoid disintegration after 14d or no aggregation within the first 48h (see Figure S1A).
To assess the volume of the organoids, we measured the 3D structures by performing a 3D optical projection tomography (OPT) reconstruction using in-house build system (Belay et al., 2021) (Figure 1C and Videos S1 and S2). In line with the maximum diameter measures, this analysis showed a higher organoid volume during the induction phase (0.063 ± 0.008 mm3) compared to the differentiation stage (0.023 ± 0.007 mm3) (Figure 1C). 3D reconstruction of organoid morphology showed that early organoids (7d in culture), in which a considerable volume is associated with a flat appearance, acquired a spherical shape at later maturation stage (35d in culture) (see Figure 1C and Videos S1 and S2). The different shapes and volumes observed in the mature organoid compared with the early organoid may be correlated to a progressive changing in the cellular spatial organization during the in vitro organoid maturation. Moreover, we observed high cell viability at all time points analyzed (98.97 ± 0.36% vs 94.17 ± 0.48% vs 81.21 ± 1.72% of live cells detected inside the organoids at 14, 21 and 32d respectively; n = 3 organoids/time point) (Figure 1D) indicating that the relatively small dimension of mature organoids guarantees sufficient nutrient and oxygen diffusion without substantial impact on the cell survival within the 3D structure. To assess cell proliferation, we quantified the cells expressing the cell-cycle G1/S-G2/M marker Ki67 (Figure 1E). As expected, we detected a high cell proliferation rate within the organoids during the expansion phase, which drastically decreased from the end of the induction phase (Ki67+ cells over total cell nuclei: 17.18 ± 1.03% at 7d vs 1.9 ± 1.08% at 14d; p<0.0001) to the differentiation phase (0.76 ± 0.04% and 0.98 ± 0.12% at 21d and 32d, respectively; p<0.0001) (Figure 1F). These data were in line with the morphological analysis (size – maximum diameter), suggesting a decrease in the proliferative activity, promoting cellular maturation during the induction phase.
To assess the brain organoid maturation through the developmental stages, we performed global transcriptomic analysis (RNA sequencing – RNAseq) of organoids at (1) early (day 7 – end of expansion phase), (2) intermediate (day 14 – end of induction phase) and (3) mature developmental stages (day 32 – differentiation phase) and compared them to neonatal (P0) and adult (3 months) mouse cerebral tissues. Principal component analysis (PCA) of the gene expression profiles clustered the different samples in five groups (Figure 1G): (1) organoids at 7 d; (2) organoids at 14 d; (3) organoids at 32 d; (4) P0; and (5) adult mouse cerebral tissues. These results confirmed that organoids at the same developmental stage had a similar gene expression signature, further indicating the reproducibility and robustness of the protocol. In addition, the PCA distribution of the organoid clusters suggests a progressive maturation of organoids through the different growth protocol stages (Figure 1G). The predominant source of variation was related to the nature of the samples (tissue or organoid) (first principal component – PC1, explaining 78% of the variance), whereas different organoids at distinct differentiation phases accounted for the 12% of the variance (PC2), with different samples belonging to the same maturation stage clustering close together (Figure 1G). The transcriptional differences observed between organoids and the murine tissue may be attributed to the indisputable in vitro limitations in resembling accurately the in vivo development (Mason and Price, 2016; Qian et al., 2019). Gene ontology analysis of mature organoids compared with immature organoids indicated that the most significant categories of up-regulated genes were related to (1) neuron differentiation, including dendritic spines and regulation of neuron projection, (2) ion channel, including voltage gated ion channels, calcium ion transport and development of calcium ion homeostasis, and (3) synapse structures, vesicular trafficking and glutamate- and GABA-specific receptors (Figure 1H). Gene set variation analysis (GSVA) showed that early and intermediate organoids had a higher expression of genes related to the NSCs identity (e.g., Nes, Pax6, Sox2, Sox9 and Vim), while organoids at 32 d increased the expression of genes related to mature neurons (e.g., NeuN, Map1b, Vamp1, Snap25 and Syt1) (Figure 1I and Table S1).
Overall, our data indicated the reproducibility and reliability of the brain organoid development, showing the progressive maturation and acquisition of the neuronal phenotype.
Cellular phenotype and distribution during murine brain organoid maturation
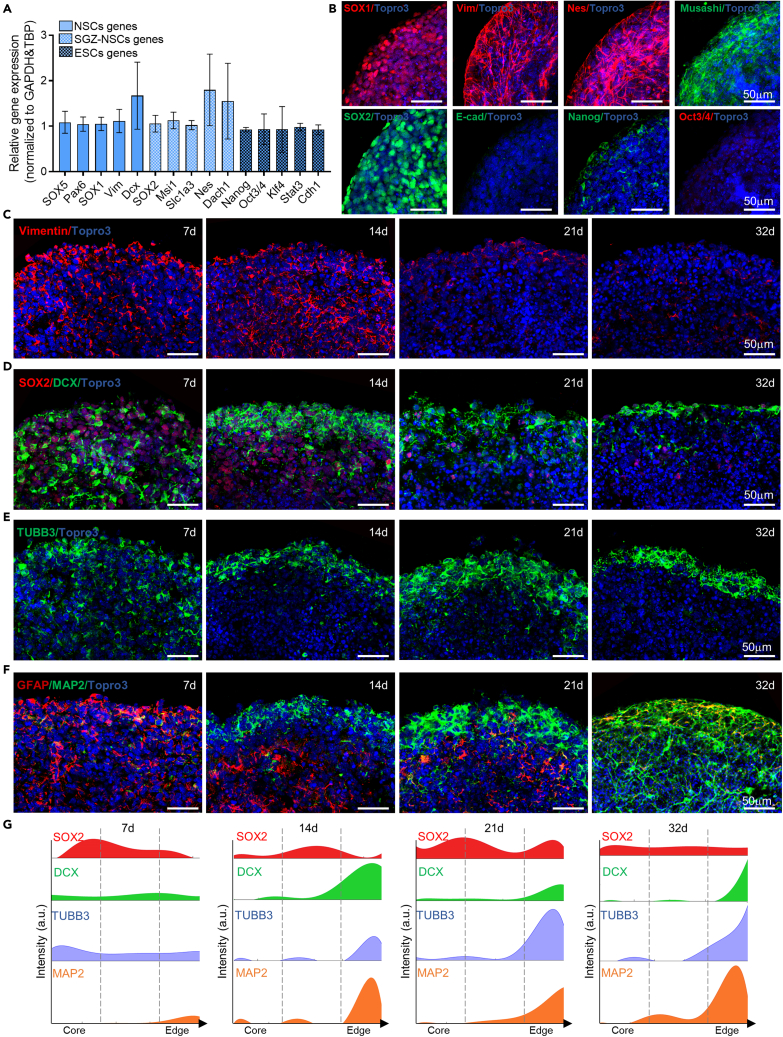
To assess the progressive brain organoid cell differentiation and distribution, we evaluated the expression and localization of NSCs, neural progenitors, neurons, and glial cells throughout the brain organoid developmental stages. First, we characterized the SGZ-derived NSC by RT-PCR and immunofluorescence analysis. As expected, we found that embryonic (E14.5) SGZ-derived neurospheres expressed the specific SGZ hippocampal derived NSC genes (Sox2, Musashi, Slc1a3, Nestin, Dach1), bona fide NSCs hallmark genes (Sox1, Sox5, Dcx, Pax6, Vimentin) and ESC genes (Nanog, Oct3/4, Klf4, Stat3, Cdh1) (Abranches et al., 2009; Kuegler et al., 2010; Palmqvist et al., 2005) (Figure 2A). The expression of SGZ hippocampal derived NSC (SOX2, Musashi, Nestin) and bona fide NSC (SOX1, Vimentin) markers was confirmed by protein expression, while little to no expression was observed for ESC markers (Oct3/4, Nanog, E-cad) (Figure 2B).
Figure 2.
Murine brain organoids show progressive cellular organization and maturation into discrete regions
(A) Graph showing Relative gene expression (normalized to GAPDH and TBP) levels of genes related to bona fide NSCs (i.e., SOX5, Pax6, Sox1, Vim, Dcx), SGZ-NSCs (Sox2, Msi1, Slc1a3, Nes, Dach1) and to ESCs (i.e., Nanog, Oct3/4, Klf4, Stat3, Cdh1) evaluated in n = 4 different SGZ-derived NSCs batches. Data are expressed as mean ± SEM.
(B) Representative confocal immunofluorescence images of SGZ-derived NSCs neurospheres at 3d in culture, showing the presence of the specific NSCs markers SOX1 and Vimentin, the SGZ-NSCs markers Musashi, SOX2 and Nestin, and little or no expression of Nanog, Oct3/4 and E-cadherin ESCs markers.
(C) Representative confocal immunofluorescence images of sliced murine brain organoids at different protocol stages, showing the stemness (Vimentin) marker. At day 7 and day 14, the distribution of Vimentin+ cells is widespread, and no evident cellular organization is observed. At a later stage of maturation (day 21 ad 32), few Vimentin+ cells were still detected.
(D) Representative confocal immunofluorescence images of sliced NSCs-derived organoids at different protocol stages, showing the stemness (SOX2) and the neuronal progenitors (DCX) markers. At day 7, all the cell populations inside the organoids do not show any type of organization but are scattered and strongly staminal. Starting from the induction phase (day 14), the immature neuronal cells spatially rearrange to cluster on the external region, while the middle layer is mainly staminal. At later time points (days 21 and 32), few SOX2+ cells were detected while DCX+ cells still populated the edge of the organoid.
(E) Representative confocal immunofluorescence images of sliced murine brain organoids at different protocol stages, showing the immature neurons (TUBB3) marker. At day 7, few immature neurons were detected within the organoid structure and progressive organization to the external layer was observed along the developmental stages. Starting from the induction phase (day 14), the immature neurons mainly cluster on the external region with a progressive thickening of the TUBB3+ cells layer.
(F) Representative confocal immunofluorescence images of sliced murine brain organoids at different protocol phases, illustrating the mature neuronal cells (MAP2) in close association with astrocytes (GFAP) cells. At later stages of culture, organoids show an ongoing neuronal maturation, mainly restricted to the outer organoid edge. A progressive thickening of the neuronal cells layers and the ability to form a neuronal network is observed from day 21 to day 32. All the pictures in B-F are maximum intensity projections of z stack images.
(G) Fluorescence intensity analysis showing expression of SOX2 (red), DCX (green), TUBB3 (blue) and MAP2 (orange) plotted versus the distance from the center (left) to the edge (right) of brain organoids at day 7, 14, 21, 32. Dashed lines delineate different zones of the organoid slice, from the core to the edge. A progressive accumulation of neuronal cells population from the core to the edge is observed during the later time point.
Then, we assessed the NSC markers expression during organoid maturation. Immunofluorescence analysis performed at day 7 (end of the expansion phase) revealed a high number of stem cell expressing Vimentin (15.45 ± 0.5% among the total cell nuclei) and sex determining region Y-box 2 (SOX2: 79.1 ± 2.95% among the total cell nuclei) (Figures 2C, 2D, S1B, and S1C). During organoid differentiation, the number of Vimentin+ and SOX2+ NSCs progressively decreased (see Figures S1B and S1C) (8.79 ± 1.02% and 7.88 ± 1.15% Vimentin+ cells at 21 and 32d, respectively, p = 0.01 and p = 0.02 respect to 7d; 9.95 ± 2.42% and 10.79 ± 0.51% SOX2+ cells at 21 and 32d, respectively, p<0.0001 respect to 7d). Qualitative analysis of cellular distribution showed an initial (day 7) widespread localization of Vimentin+ and SOX2+ NSCs inside the organoid (Figures 2C, 2D, and 2G and see Figure S1I), that maintained an intersperse distribution both in the inner and the external layer until the later stage of differentiation (Figure 2G).
During the initial stages of organoid formation, we also observed the presence of doublecortin (DCX) positive neural progenitors (10.97 ± 1.60% and 17.1 ± 2.83% at 7d and 14d, respectively) (Figure 2D), while they significantly decreased up to ∼5% at later time points (day 21: 5.77 ± 1.07%; day 32: 5.73 ± 0.9%, p = 0.01 and p = 0.009 respect to 14d) (Figures 2D and S1D). The DCX neural progenitors showed a widespread distribution within the organoid at day 7 (see Figure S1I) and, starting from the induction phase (day 14), they spatially rearranged to cluster in the external region of the organoid (Figures 2D and 2G). As expected, during brain organoid maturation, we observed a progressive statistically significant increase in the number of immature (Tubulin Beta 3 Class III -TUBB3) (17.55 ± 2.35% and 7.63 ± 1.56% at 32d and 7d, respectively; p = 0.0014) (Figure S1E) and mature (Microtubule-Associated Protein 2 - MAP2) (24.4 ± 2.08% and 7.38 ± 1.43% at 32d and 7d, respectively; p < 0.0001) (Figure S1F) neurons. Notably, starting from day 14 and more consistently in the later differentiation stage, we observed a major edge zone of TUBB3+ (Figures 2E and 2G) and MAP2+ cells (Figures 2F and 2G), which accumulated in the external layer ((Figures 2E–2G). In line with the increase in the total neuronal number and with their modified distribution in the organoid, the thickness of both TUBB3+ and MAP2+ cells outer layer increased as the maturation proceeds (TUBB3+ cells layer: 18.69 ± 0.55μm and 30.96 ± 3.99μm at 14d and 32d, respectively; MAP2+ cells layer: 17.89 ± 1.13μm and 30.76 ± 2.19μm at 14d and 32d, respectively; p = 0.0009).
These data suggested that SGZ-derived NSCs inside the brain organoid progressively differentiate into neurons, which are mostly compartmentalized at the organoid edge forming a net of neuronal cells (Figure S1J). The maturation of neuronal cells observed by immunofluorescence was further supported by RNAseq data which highlighted an increased expression in late-stage organoid of genes related to synaptic formation and plasticity (Cttn, Nsg2, Syt1, Snap25), signal transduction (Srrm3, Stmn2, Trim67) as well as genes encoding specific mature neuron proteins (Vamp1, Map1b) (Cahoy et al., 2008; Bifari et al., 2020; Zhang et al., 2014) (Figure S1K and Table S2).
To evaluate the presence of different cell populations in brain organoids, we analyzed the protein expression of glial fibrillary acidic protein (GFAP)+ astrocytes, neural/glial antigen 2 (NG2)+ oligodendrocytes, ionized calcium-binding adapter molecule 1 (Iba1)+ microglia cells and CD31+ endothelial cells. While we did not detect at any time point oligodendrocytes, microglia, nor endothelial cells (data not shown), we found a stable ∼10% of astrocytes (see Figure S1G), closely associated with neural cells (Figures 2F and S1L). Although we did not observe any change in the number of astrocytes through the organoid development, the transcriptome analyses indicated that genes related to astrocyte maturation such as Slc1a2, encoding for a transporter protein that clears the glutamate from the extracellular space at synapses, Gfap, Aqp4, Slc1a3, and Slc4a4 (Boisvert et al., 2018; Cahoy et al., 2008), were progressively up-regulated in the mature organoid stage (Figure S1M and Table S3).
Among the total number of cells detected, early organoids (7d) were mainly characterized by the presence of NSCs and neuronal progenitors (∼80%). In line with the progressive maturation stages of the organoid development, intermediate stage organoids (14d) showed an increased number of immature neurons and neural progenitors (∼32%) and at later stages (21d and 32d) ∼ 60% of the entire cellular population was represented by immature and mature neurons (Figure S1H).
Overall, these data indicated that the distribution of neural cell types in different regions of the brain organoid showed a progressive organization. Early-stage organoids were characterized by a non-organized structure, with spread proliferative and stemness markers, then they progressively showed a radial distribution of neurons which, at late stage of differentiation, increased in number and outer layer thickness.
Murine brain organoids undergo an oxidative metabolic switch supporting neuronal maturation
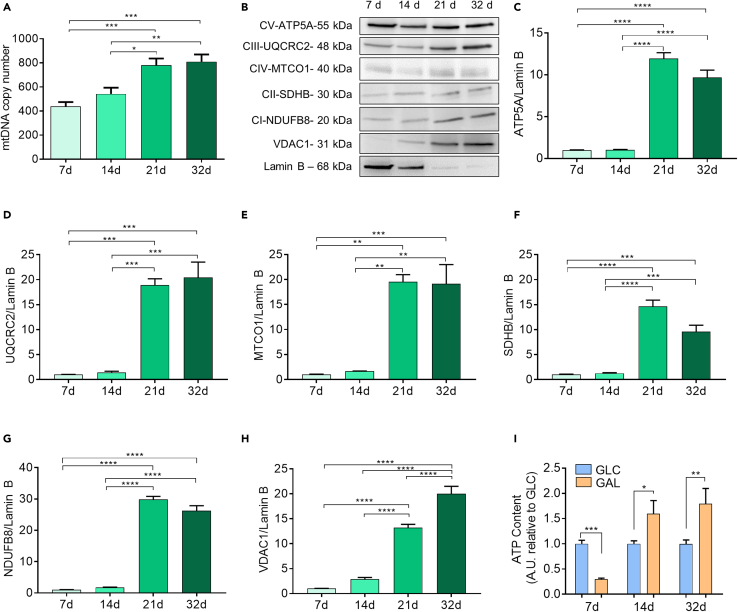
Together with changes in morphology, a switch from glycolytic to oxidative metabolism is required for neuronal differentiation (Bifari et al., 2020). To describe brain organoid progressive maturation through oxidative phosphorylation, we characterized their metabolic profile. First, we assessed the mitochondrial mass and mitochondrial respiratory chain complexes content of the organoids throughout the maturation stages. The mitochondrial DNA (mtDNA) copy number, an index of mitochondrial mass, progressively and significantly increased with time (Figure 3A), rising from 438 ± 36.47 mtDNA molecules/cell at day 7–809.7 ± 59.82 mtDNA molecules/cell at day 32 (p < 0.001), indicating an increase in the mitochondrial amount during brain organoid maturation. Therefore, we performed immunoblot analysis on total cellular extracts to evaluate the mitochondrial proteins and oxidative phosphorylation (OXPHOS) system quantity. Given the heterogeneity of cell composition of brain organoids across the developmental stages, equal amounts of protein/organoid pool at different maturation phases may not reflect the cell number. Therefore, we expressed the mitochondrial markers as fold-change of the nuclear marker Lamin B, rather than to the total protein content per lane. The voltage-dependent anion-selective channel 1 (VDAC 1) protein, a mitochondrial channel of the outer mitochondrial membrane widely used as an indicator of mitochondrial mass (Bugiardini et al., 2020), significantly increased in the organoids at different maturation times (from 1 ± 0.59 at 7d to 19.99 ± 1.52 at 32d; p < 0.0001) (Figures 3B and 3H). Mitochondria are mainly responsible for energy production through the OXPHOS process that is upregulated in mature neurons compared to NSCs (Beckervordersandforth et al., 2017; Bifari et al., 2020; Lorenz et al., 2017; Martano et al., 2019; Zheng et al., 2016; Inak et al., 2021). Therefore, we checked the amounts of the mitochondrial respiratory chain complexes I-V by immunoblot analysis (Figure 3B) of specific complex subunits. We found that the ATP5A, UQCRC2, MTCO1, SDHB, NDUFB8 proteins, structural subunits of complexes V, III, IV, II, and I, respectively, significantly increased during the maturation process (Figures 3C–3G). Overall, these data suggested that brain organoids increased their mitochondrial mass during maturation.
Figure 3.
Murine brain organoids switch from glycolysis to oxidative phosphorylation during development
(A) Mitochondrial DNA (mtDNA) in organoids progressively increases over time, suggesting an increment of mitochondrial mass from day 7 to day 32. n = 9 organoids for each time point. Data are reported as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. P < 0.05 was considered statistically significant. ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05.
(B) Representative western blotting analysis of organoids at different time points shows an increase in OXPHOS complex subunits (ATP5A, UQCRC2, MTCO1, SDHB, NDUFB8), and VDAC1 when reported to nuclear marker Lamin B, suggesting an intensification of electron transport chain activity and acquired mitochondrial mass in the cells composing mature organoids.
(C–H) Quantitative analysis of the western blotting described in (B). n>3 organoids for each time point. Data are expressed as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. P < 0.05 was considered statistically significant. ∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01.
(I) Graphs showing the total ATP content (expressed as A.U. relative to glucose) measured in organoids fed with different carbon sources (glucose – GLC; galactose – GAL) at different time points (n = ≥8 samples/time point), and normalized to cell number. Data are expressed as mean ± SEM. Statistical differences were evaluated by a two-tailed unpaired ttest for two datasets (glucose vs galactose) at each time point. P < 0.05 were considered significantly different. ∗∗∗p < 0.001; ∗∗p <0.01; ∗p <0.05.
To further confirm the metabolic development of the organoids toward oxidative phosphorylation during the process of neuronal maturation, we switched the carbon source to a fully oxidative substrate by replacing glucose with galactose in the culture medium, forcing the organoids to use OXPHOS for the ATP production. Early organoids (7d) significantly dropped their ATP content in response to galactose, demonstrating their inefficiency in producing energy with solely OXPHOS. Interestingly, a 2-fold increase in cellular ATP content was observed in intermediate organoids (14d) cultured with galactose, suggesting their capability to switch to mitochondrial metabolism, if required. Total ATP content in mature organoids (32d) was more stimulated (Figure 3I), indicating their full differentiation toward mature neurons which could easily adapt to oxidative substrates.
Accordingly, we observed a statistically significant higher expression of genes related to OXPHOS pathway (e.g., Ndufa13, Ndufa1, Ndufa3, Ndufb7, Ndufv1, Cox7a1, Ndufa6, Cox7b, Ndufa2, Ndufa7, Sdhaf2, Atp5k, Ndufb10, Sdhc, Sdha, Ndufaf7) (see Table S4) in mature organoids compared to early organoids, further suggesting a metabolic switch toward oxidative phosphorylation adaptation.
We found a significant upregulation of peroxisome proliferator-activated receptor gamma (Pparg) in the mature organoids. The PPARγ activation may influence neuronal differentiation of mouse NSCs and rat embryonic midbrain cells (Ghoochani et al., 2012; Park et al., 2004). The PPAR-agonist drug bezafibrate could improve neuronal maturation that is altered in an organoid model of genetic neurological disease (Inak et al., 2021), further suggesting that the activation of PPARG contributed to the correct neuronal development. According to the upregulation of PPARG, several genes related to peroxisomal (Acox, Lbp, Dbp, Ech1), and mitochondrial (Acad11, Acadsb, Acad12, Slc25a20, Cpt1c, Echdc2, Hadhb) (see Table S4) beta-oxidation pathways were upregulated in the mature organoid. Peroxisomal and mitochondrial beta-oxidation have different substrate specificity and respond to different environmental changes and cellular processes, including developmental stages (Poirier et al., 2006), to support energy requirements.
Taken together, these results indicated that the maturation of the organoids was accompanied by a significant upregulation of the mitochondrial biogenesis, which sustained the oxidative metabolism in differentiating neurons.
Mature brain organoids develop a 3D neuronal network with excitatory and inhibitory synapses
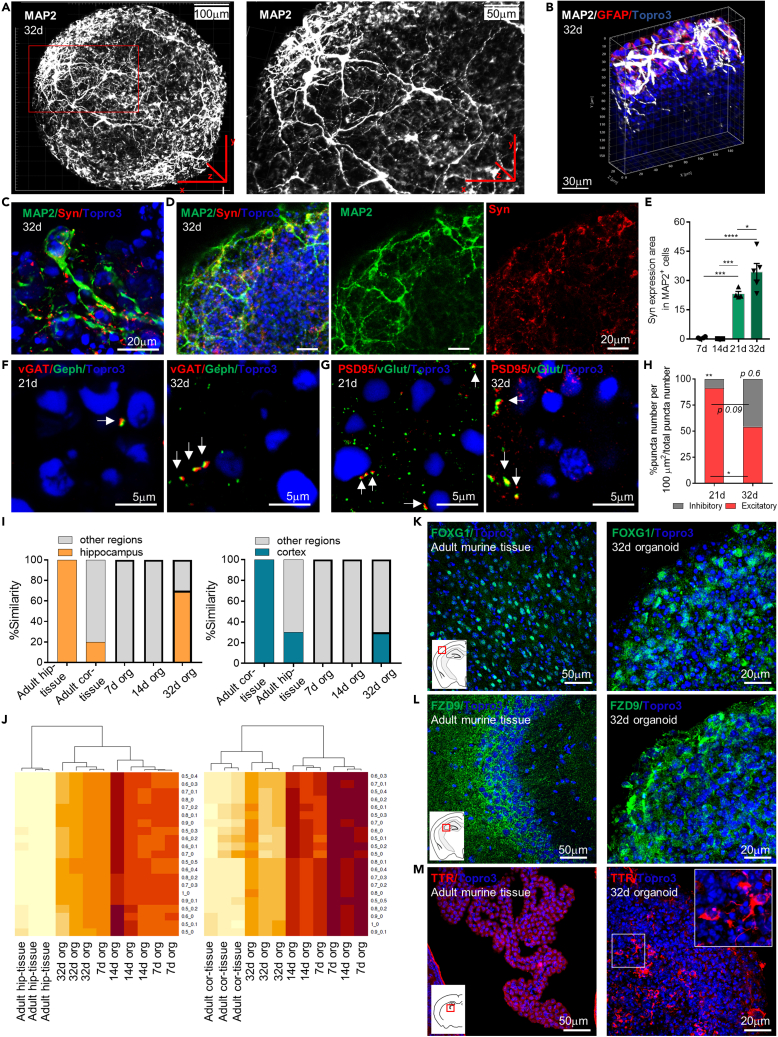
To analyze the distribution of neurons inside the organoid, we performed wholemount immunostaining and 3D imaging reconstruction. As shown in Figures 4A and 4B, mature MAP2+ neuronal cells were characterized by the formation of an intricate neuronal network with cells populating the entire organoid surface and elongating processes extending in the three dimensions across the organoid structure (see Videos S3 and S4), suggesting that they may contact both adjacent and distal cells.
Figure 4.
Neuronal network formation and synapse development within mature murine brain organoids
(A) Representative maximum Z-projection of confocal images of whole mount immunostaining performed at 32 d for the MAP2 marker, showing the formation of an extended neuronal network on the organoids surface, with the characteristic process elongation. The red box highlights the magnification of an organoid region.
(B) Representative 3D reconstruction of confocal image of a mature organoid (32d) region using IMARIS software showing the distribution of neuronal cells (MAP2+, in white) in all the spatial three-dimensions.
(C) High magnification confocal maximum intensity z stack projections of image showing a mature neuronal cell (MAP2) inside the organoid at 32d with the characteristic process elongation and the expression of synaptic density marker (Syn).
(D) Representative maximum intensity z stack projection of confocal images of whole mount immunostaining performed at 32d for the Syn and the MAP2 markers, showing colocalization between neuronal cells and the synaptic marker.
(E) Graph showing the synaptic density (% Syn+ area in MAP2+ cells area, in 100 μm2) at different time points. Data are expressed as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗∗∗p < 0.001; ∗p < 0.05.
(F and G) Representative confocal immunofluorescence maximum intensity z stack projections images of sliced murine brain organoids at 21 and 32 d for the detection of inhibitory vGat-Gephyrin (F) and excitatory PSD95-vGlut (G) synapses. White arrows indicate the co-localization of PSD95-vGlut and vGat-Gephyrin in double-positive cells, able to generate excitatory and inhibitory synaptic contacts, respectively.
(H) Graph showing the average inhibitory and excitatory synaptic puncta colocalization in 100 μm2. Note the general synaptic decrement, mainly the excitatory synapses reduction, matched with the increment of inhibitory synapses at 32 d. Analysis performed on n≥3 different organoids and at least 3 entire sections for each organoid. Data in all graphs are showed as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗∗p < 0.01; ∗p < 0.05.
(I) Graphs showing the percentage of similarity of adult murine hippocampal tissue (adult hip-tissue), adult cortical tissue (adult cor-tissue), 7d organoids (7d org), 14d organoids (14d org), and 32d organoids (32d org) to hippocampal (left graph) or cortical (right graph) tissues.
(J) Heatmaps showing the degree of similarity between adult murine hippocampal tissue (adult hip-tissue) or adult cortical tissue (adult cor-tissue) and 7d organoids (7d org), 14d organoids (14d org), and 32d organoids (32d org) considering different proportions of hippocampal and cortical components, according to gene expression profiles of selected genes.
(K–M) Representative confocal immunofluorescence maximum intensity z stack projections images of sliced adult (8 weeks) murine brain tissue and mature (32d) murine brain organoids showing the expression of FOXG1-cortical (K), FZD9-hippocampal (L) and TTR-choroid plexus (M) markers. White insets represent a sketch of coronal mouse brain sections while red boxes highlight the brain region of the murine tissue staining.
Neurons MAP2+, astrocytes GFAP+ and total nuclei (TOPRO-3) are shown in white, red and blue, respectively.
Neurons MAP2+ and total nuclei (TOPRO-3) are shown in green and blue, respectively.
To assess the organoid potential to develop neuronal connections, we performed immunofluorescence analysis evaluating the expression area of the presynaptic marker synaptophysin (Syn) in MAP2+ neurons at different developmental stages. In line with the overall increment of MAP2+ cells throughout organoid development (see Figures 2F and S1F), we observed expression of Syn marker (Figures 4C and 4D), which increased at day 32 (Syn positive area over MAP2+ area: 34.52 ± 4.26 vs 0.62 ± 0.25 at 32 and 7 d respectively, p< 0.001) (Figures 4E and S2A).
We next assessed the presence of inhibitory and excitatory synaptic punctae at later stages of organoids maturation (21d and 32d). The co-localization of presynaptic and postsynaptic punctae revealed the presence of synapses (Dzyubenko et al., 2016). In order to distinguish GABAergic and glutamatergic synapses, we assessed the co-localization of specific presynaptic and postsynaptic marker proteins (Dzyubenko et al., 2016). We used the vesicular GABA transporter vGAT and gephyrin to identify respectively the pre- and post-synaptic terminals of inhibitory synapses (Figure 4F), while the vesicular glutamate transporter VGlut and the scaffolding protein PSD95 were used to identify respectively the pre- and post-synaptic terminals of the excitatory synapses (Figure 4G). In keeping with what is observed during brain development (Levinson and El-Husseini, 2005; Cline, 2005), immunofluorescence analysis of vGLUT-PSD95 punctae revealed that excitatory synapses were mainly expressed in early mature organoids (21d) reaching about 80% of the total synaptic punctae per 100 μm2. The analysis of the vGAT-gephyrin colocalization showed an increase of inhibitory synapses during organoid maturation (about 20% and 50% over the total synaptic punctae per 100μm2 at 21d and 32d, respectively). At the latter stage (32d) of organoid development, the relative amount of excitatory and inhibitory synaptic punctae was found to be in equal proportion (Figure 4H). In addition to the observed expression of synaptic proteins, we further evaluated the presence of inhibitory (Figure S2B) and excitatory (Figure S2C) synapses by transcriptomic analysis using a selected gene panel (Hayamizu et al., 2005). Heatmap profiles and DESeq2 analysis revealed that mature (32d) rather than early (7d) organoids showed an upregulation of genes related to NMDA and cholinergic receptors (i.e. Chrm1, Chrm4), glutamate receptors and reuptake (i.e. Grin2b, Grin1, Grm4, Grik1) as well as GABA receptors and transporters (i.e. Gabra4, Gabrb1, Camk4, Gabra1, Slc32a1, Gabra5, Gabrg2, GAD2) (see Table S5). The presence of inhibitory (GAD65-67+) and excitatory (NMDA+) neurons in organoids at later stage of differentiation (Figures S2D and S2E) was also supported by immunofluorescence analysis. RNAseq data further revealed that the gene expression of the chloride extruding K+-Cl− cotransporter isoform 2 (Kcc2) was upregulated in mature compared to early organoids (padj value <0.00001; log2FoldChange: 3.11) while the chloride-importing Na+-K+-Cl− cotransporter isoform 1 (Nkcc1) was downregulated during organoid development (padj value <0.00001; log2FoldChange:−0.81) (see Table S5). The change in the expression of Nkcc1 and Kcc2 during organoid development might suggest a switch of GABAergic-signaling from excitatory-to-inhibitory which characterized in vivo neuronal brain development (Ganguly et al., 2001; Rivera et al., 2005; Ben-Ari and Spitzer, 2004).
To investigate whether the mature astrocytes found at later time points of organoid development contributed to boosting the synapses formation and specification, we stained astrocytes for glutamine synthetase (Gln Syn), a protein involved in the ATP-dependent conversion of glutamate and ammonia to glutamine. In the brain, glutamine synthetase produced by astrocytes plays an important role in regulating the glutamate/glutamine cycle between neurons and astrocytes and it has been suggested to play a pivotal role in synaptogenesis (Son et al., 2019). We found that at 21 d astrocyte expressed Gln Syn (Figure S2F), indicating their possible contribution in the neuronal network formation.
The murine cerebral organoid method described here relies on the spontaneous neuronal differentiation without any extrinsic patterning to guide the development into a specific brain region. The SGZ neural stem cells source used may have defined an intrinsic patterning toward dorsal, rather than ventral, brain regional specification. To characterize the brain regional identity and neuronal diversity of the tissue-like mature organoid generated, we analyzed RNA-seq data by evaluating the degree of similarity (percentage) between organoids (7d, 14d, 32d) and murine adult cortex and hippocampal tissue, considering selected gene lists (cortical signature: Stard8, Sytl2, Tnnc1, Myl4, Drd2, Calb2, Reln, Lhx1, Lhx5, Ebf3, Cux1, Satb2, Dok5, Ctp2, Emx1, Tiam2, Tbr1, Foxg1, Pvrl3; hippocampal signature: Zbtb20, Sipa1l3, Gpr161, Crlf1, Glis3, Dcx, Nrp2, Tgf2b, Gria1, Tnip2, Slc39a6, Prox1, Slc26a10, Trpc6, Neurod1, C1ql2, Gqrik4, Elavl2, Elavl4, Dkk3, Nectin3, Scip, Spock1). As expected, hippocampal and cortical tissue revealed a score of 100% of similarity for hippocampal and cortical gene set, respectively. This further validated the reliability of the selected genes. This analysis did not find any affinity of organoids at 7d and 14d to murine adult cortex and hippocampal tissue gene set, indicating that organoid brain regional identity is not defined at the early stage of development. Interestingly, we observed about 70% of gene expression similarity between hippocampal adult tissue and the 32d mature organoids and about 30% between cortical adult tissue and mature organoid (Figures 4I and 4J). These results further indicated that mature organoids may better reproduce dorsal forebrain with a predominant hippocampal component. However, the presence of other brain regions cannot be excluded. We then analyzed the expression of specific cortical (FOXG1), hippocampal (FZD9), choroid plexus (TTR) (Figures 4K–4M) and ganglionic (NKX2.1, GSX2) (see Figures S2G and S2H) markers (Renner et al., 2017) in adult brain murine tissue and mature organoids by immunofluorescence. We found that in all the analyzed mature organoids (32d: n = 4), both cortical and hippocampal markers were expressed mainly at the edge of the organoids as well as in the specific brain tissue region (Figures 4K and4 L). Ganglionic markers were undetectable while they were expressed in ventral brain tissue (see Figures S2G and S2H). We also found few TTR positive cells at the center of the organoid structure (Figure 4M). These data suggested that during development the murine brain organoids acquired features of dorsal forebrain tissue.
Taken together, these results suggested that the mature brain organoid display 3D neuronal network coupling via inhibitory and excitatory synapses, which is supported by mature astrocytes. Regional identity analysis revealed that murine brain organoids acquired, along the maturation, dorsal forebrain phenotype. Next, we questioned about the presence of neuronal activity.
Whole-cell patch-clamp recording reveal functional neurons inside the brain organoids
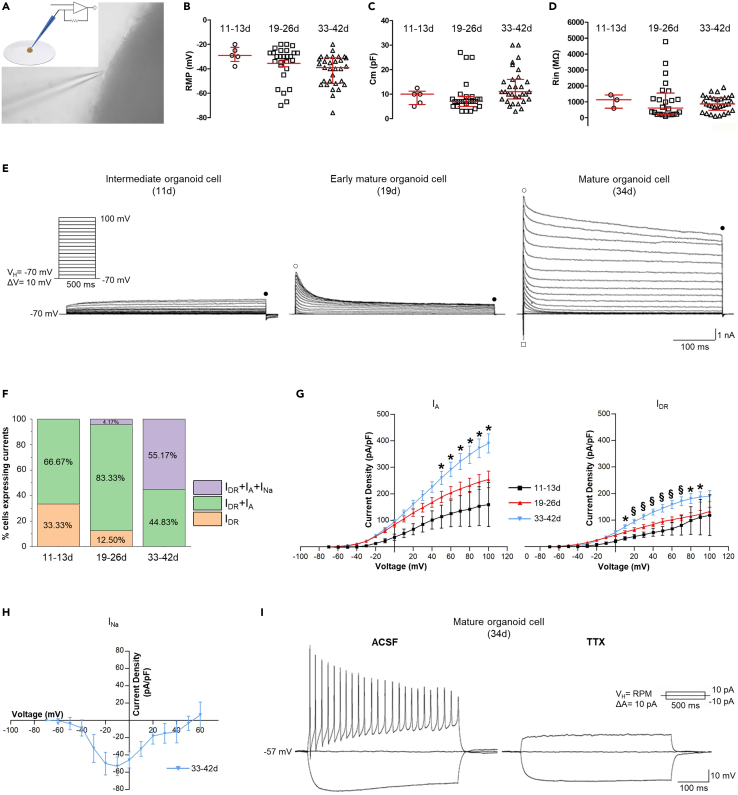
To assess the functionality of the organoids, we performed whole-cell patch-clamp recording. To preserve the cellular network and properties, we used whole-mount organoids (Figure 5A), rather than organoid slices or dissociated cells – techniques commonly used by other laboratories (Birey et al., 2017; Xiang et al., 2019). Recorded cells were primarily localized at the edge of the organoid. We investigated the passive membrane properties and the expression of voltage-gated currents (outward potassium (K+) and inward sodium (Na+) currents) in a total of 63 cells recorded from at least 56 different organoids at three different stages of development: intermediate (11–13d), early mature (19–26d), and mature (33–42d).
Figure 5.
Patch-clamp analysis of cells in intact organoids at different developmental stages show functional neurons
(A) Picture showing a glass pipette during a patch-clamp recording; note the edge position of the cell under recording. In the inset, a scheme shows the glass pipette used for patch-clamp recording of a whole mount organoid placed on a glass coverslip.
(B–D) Passive membrane properties: resting membrane potential (RMP) (B), membrane capacitance (Cm) (C), and input resistance (Rin) (D) of cells in intermediate (11–13d), early mature (19–26d) and mature (33–42d) organoids. In all graphs, the median is represented by the longest horizontal red lines, whereas the 25–75 confidence interval is marked by the vertical red bars.
(E) Representative traces of K+ and Na+ currents elicited by depolarizing voltage steps in cells recorded from the whole mount organoid. At intermediate (11d, left panel) and early (19d, middle panel) developmental stages, cells show outward currents but do not exhibit inward Na+ current, consistent with the absence of action potentials. At more mature developmental stages (34d, right panel), cell exhibits both inward Na+ and outward K+ currents. In the inset (left panel), the stimulus protocol applied is shown. • indicates the point where delayed rectifier K+ current (IDR) amplitude was measured; ○ indicates point where A-type K+ current (IA) amplitude was measured; ○ indicates point where Na+ current (INa) amplitude was measured.
(F) Graph showing the percentage of cells exhibiting IDR, IDR + IA, and IDR + IA + INa in intermediate (11–13d), early mature (19–26d) and mature (33–42d) organoids.
(G) Averaged current-voltage (I-V) relationship for IA(left) and IDR(right). Black line refers to intermediate organoid cells (11–13d); red line refers to early mature organoid cells (19–26d); light blue line refers to mature organoid cells (33–42d). Data are presented as mean of current density (pA/pF) ± SEM. ∗p<0.05, §<0.01.
(H) Averaged I-V relationship for INa. Light blue line refers to mature organoid cells (33-42d), while INa was not observed in intermediate (11–13d) and early mature (19–26d) organoids. Data are presented as mean of current density (pA/pF) ± SEM.
(I) Representative current-clamp traces evoked in a mature organoid cell (34d) by 500 ms current pulses; depolarization induces action potentials that are blocked by TTX (1 μM). In the inset (right panel), the stimulus protocol applied is shown.
Resting membrane potential (RMP) (Figure 5B) showed a trend from highly depolarized (lowest RMP:−29 mV) to less depolarized values (highest RMP:−52 mV) from intermediate toward mature organoid cells, although statistical significance was not reached due to the wide range of RMP values exhibited by the organoids at each of these stages (−29.0 [-22.5 to −34.0] mV, n= 5 at 11-13d;−30.0 [-27.0 to −40.0] mV, n = 27 at 19–26 d;−39.0 [-31.0 to −52.0] mV, n = 31 at 33–42 d; ANOVA). The membrane capacitance (Cm) (Figure 5C) was 10.0 [5.8–11.2] pF, n= 5 at 11-13d, 7.0 [5.0–9.0] pF, n = 27 at 19–26 d, and 11.0 [8–16] pF, n = 31 at 33–42 d (values not statistically different; ANOVA). Input resistance (Rin) (Figure 5D) was 1130.0 [596.3–1437.0] MΩ, n= 5 at 11-13d, 610.2 [224.1–1558.0] MΩ, n = 24 at 19-26d, and 865.1 [464.9–1266.0] MΩ, n = 31 at 33-42d (values not statistically different; ANOVA).
All the patched neurons showed outward K+ currents (Figure 5E) elicited by depolarizing voltage steps in voltage-clamp whole-cell configuration (Figure 5E, inset in left panel). In particular, while delayed rectifier K+ currents (IDR) were recorded in 100% of cells at any developmental stage (n = 3 at 11-13d, n = 24 at 19-26d, and n = 29 t 33-42d; Figures 5E and 5F), the A-type K+ current (IA) appeared only in 2 out of 3 cells recorded from intermediate organoid cells, in 87.50% (21 out of 24) cells recorded from early mature organoid (19–26d), and in 100% (n = 29) in mature organoids (Figure 5F). Organoid cells at 11–13d (n = 3) did not exhibit inward Na+ current (INa), while it was recorded in 1 out 24 cells (4.1%) at 19-26d, and in 16 out of 29 organoid cells (55.17%) at 33–42d (Figures 5E and 5F). Number of cells manifesting IDR (n = 1 at 11–13d, n = 3 at 19-26d, n = 0 at 33-42d), IDR + IA (n = 2 at 11-13d, n = 20 at 19–26d, n = 13 at 33–42d), or IDR + IA + INa (n = 0 at 11–13d, n = 1 at 19–26d, n = 16 at 33-42d) were statistically different in intermediate, early mature and mature organoids (p = 0.00003, Fisher Exact test). I-V relationships for the voltage-activated K+ currents showed an increase in current density recorded up to sub-maximal stimulus amplitude; for example, at +90 mV voltage step, IA density was 153.4 ± 73.4 pA/pF at 11–13d (n = 2), 244.4 ± 28.31 pA/pF at 19-26d (n = 19) and 371.8 ± 35.24 pA/pF at 33–42d (n = 26, p = 0.015, ANOVA), while IDR density was 110.9 ± 66.8 pA/pF at 11–13d (n = 3), 121.9 ± 14.2 pA/pF at 19–26d (n = 22) and 187.0 ± 19.4 pA/pF at 33-42d (n = 26, p = 0.0321, ANOVA) (Figure 5G). INa recorded in mature organoid cells showed typical INa I-V relationships (Figure 5H). In accordance with these results, none of the patched organoid cells recorded in current-clamp mode at 11–13d (n= 5) and 19–26d (n = 4) phases could generate action potentials when injected with positive current, while 6 out of 16 mature organoid cells (37,50%) were able to generate action potentials; both action potential and INa were completely abolished when by the sodium channel blocker tetrodotoxin (TTX, 1 μM; n = 6; Figure 5I).
In line with patch-clamp results, upregulated genes for voltage-gated ion channels were detected with gene ontology analysis (see Figure 1H). In particular, upregulation of genes related to voltage-gated K-channels (i.e., Kcnc4 and Kcna3) in mature (32d) rather than early (7d) organoids were confirmed by heatmap profiles and DESeq2 analysis (see Figure S2C).
Overall, electrophysiological data indicated that neurons within the organoid progressively mature and were able to generate action potentials at late stages of development.
Mature murine brain organoids exhibit functional synaptic connection and neuronal spontaneous activity
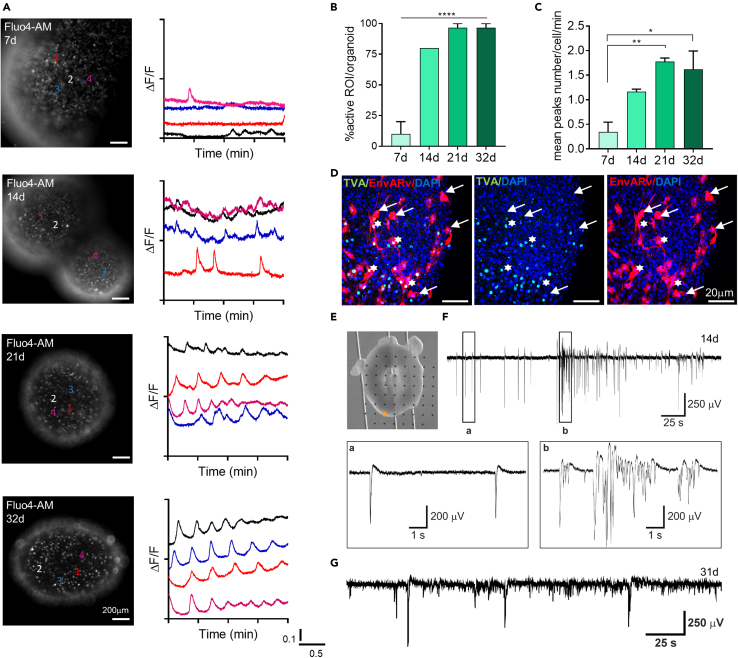
Spontaneous neuronal activity is an important step during neuronal development. The first spontaneous electrical activity in the immature brain is driven by intracellular calcium fluctuations (Rosenberg and Spitzer, 2011). Thus, we examined spontaneous calcium fluctuations by timecourse live calcium imaging using different approaches.
We examined the overall intracellular spontaneous calcium dynamics of the entire brain organoid at early (7d), intermediate (14d), and mature (21d and 32d) stages by using the fluorescent calcium dye Fluo4-AM (Figure 6A, left panels). We analyzed the cellular activity within organoids evaluating the fluorescence intensity signal of the calcium indicator in different cells (n = at least 10 cells/organoid, n= at least 3 organoids/time point) by 3 min of live-imaging acquisition. The Fluo4-AM dye exhibits an increase in fluorescence upon binding Ca2+, thus, variations of intensity correspond to intracellular calcium fluctuations (Figure 6A, right panels).
Figure 6.
Calcium imaging, transsynaptic tracing and electrophysiology on intact organoids show neuronal synaptic connections and spontaneous activity
(A) Representative fluorescence images reported in greyscale of whole mount organoids at different time points during the time-lapse Fluo4-AM dye calcium imaging assay (left side), paired to graphs of calcium oscillations of representative cells analyzed (right side). It is possible to observe an increment of cellular activity over time, as well as major peaks synchronization, from early (7d) to mature (21-28d) organoids.
(B and C) Graphs showing (B) the percentage of active cells within the organoid at different time points (n = 3 organoids/time point; n = 10 cells selected in each organoid) and (C) the average number of the peaks counted for each cell per minute at different time points (n = 3 organoids/time point; n = 10 cells selected in each organoid). Data in all graphs are shown as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p< 0.05.
(D) Representative maximum intensity z stack projection of confocal images of Pseudotyped rabies-virus (EnvARv)–based transsynaptic tracing in neural cells of whole mount organoids at 32d. White asterisks indicate the “starter” neurons (yellow); white arrows indicate the projecting neurons (red) taking synaptic contact with TVA-neurons.
(E) Optical bright-field image of a 14d organoid laying on a planar MEA and held in place by a custom hold-down anchor.
(F) Electrical activity generated by the organoid in (E) and recorded by the electrode marked by the orange dot. The insets show a representative population spike (A) and a representative population burst (B) corresponding to the boxed signal portions.
(G) Electrical activity generated by a 31d organoid (different from E), showing the persistence of the population spikes and bursts resembling those observed at the earlier organoid developmental stage shown in (F).
At early stages of organoid development (7d), almost no peaks were detected through the brain organoids (4 active cells out of a total of 40 evaluated cells). Starting from day 14, we observed an increment in the spontaneous activity with 24 cells displaying calcium flux out of a total of 30 analyzed cells (Figure 6B). The large majority of the cells (29 active cells/30 evaluated cells) analyzed both at 21 and 32 days showed spontaneous activity along the acquisition time (Figure 6B and see Videos S5 and S6). By evaluating the mean number of the detected peaks per minute of recording per organoids at different developmental stage, we observed a statistically significant higher number of calcium peaks in early mature (21d: 1.78 ± 0.07 peaks/min, p = 0.006) and mature (32d: 1.62 ± 0.37 peaks/min, p = 0.01) organoids compared to the peaks detected in early organoids (7d: 0.34 ± 0.20 peaks/min) (Figure 6C), suggesting the progressive increase of spontaneous calcium activity.
The video is reported in grayscale and 10X speed.
The video is reported in grayscale and 10X speed.
Calcium imaging analysis revealed that mature brain organoids organized in 3D networks of functional neurons with synaptic components exhibited spontaneous cellular activity. Collectively, these features set the basis for the generation of functional circuits throughout the 3D structure (Khazipov and Luhmann, 2006). Therefore, we investigated the presence of functional synaptic connection. To this aim we took advantage of the rabies virus (RABV)-based monosynaptic tracing technique (Wickersham et al., 2007). A lentiviral vector encoding GFP- and EnvA-pseudotyped DG RABV (pAAV-Syn-H2B-GFP-TVA-oG-WPRE3) was delivered to the organoids at 18d in order to drive the expression of the EnvA receptor (TVA) under synaptophysin promoter. This allowed us to confine primary RABV infection to neurons as ‘‘starter’’ population expressing GFP. 48 h later, we transduced the organoid with the rabies virus encoding for mCherry and the RABV glycoprotein (BRVenVA-1 G-deleted Rabies mCherry), which is necessary for subsequent monosynaptic transfer to their coupled neuron. Thus, we observed yellow (GFP+/mCherry+ double-positive) starter neurons which had received the G-TVA retrovirus encoding the EnvA receptor (TVA) 24 h later (Figure 6D). Only the neurons activating a synaptic contact with a starter neuron (GFP+/mCherry+ double-positive) received the viral particle and expressed exclusively mCherry (mCherry+/GFP−). One week following the transduction we observed clusters of mCherry+/GFP− neurons distributed at the edge of the organoid indicating that they had generated monosynaptic connections with the starter neurons (Figure 6D). The specificity of RABV to infect only cells in organoids that previously had received the RABV glycoprotein was confirmed by the absence of signals in organoids infected only with the BRVenVA-1 G-deleted Rabies mCherry (see Figure S2I).
These results suggested that neurons of murine brain organoid established neuronal synaptic connections.
Next, we sought to determine whether the neurons populating the organoids were forming electrically active networks. To this end, we performed MEA electrophysiology in intact organoids to evaluate their functional profile during development. Organoids exhibited spontaneous electrical activity as of day 14 and up to 4–5 weeks in culture. Figure 6E shows a 14-d organoid on a planar MEA. This organoid could generate spontaneous electrical activity (Figure 6F) consisting of a mixed pattern of population spikes (Figure 6F, inset a) and population bursts (Figure 6F, inset b). The electrical activity recorded from a 31-d organoid presented similarities with the pattern observed at day 14 (Figure 6G). Overall, we observed spontaneous electrical activity in 3 of the 30 tested organoids. This activity was generated sporadically and disappeared over time. The electrical activity of functional organoids was typically restricted to 1–2 electrodes at the edge of the organoid, out of the 15–25 electrodes covered within the 60-channel MEA. These results indicated that functional neurons self-organize in small ensembles located at the periphery of the organoid.
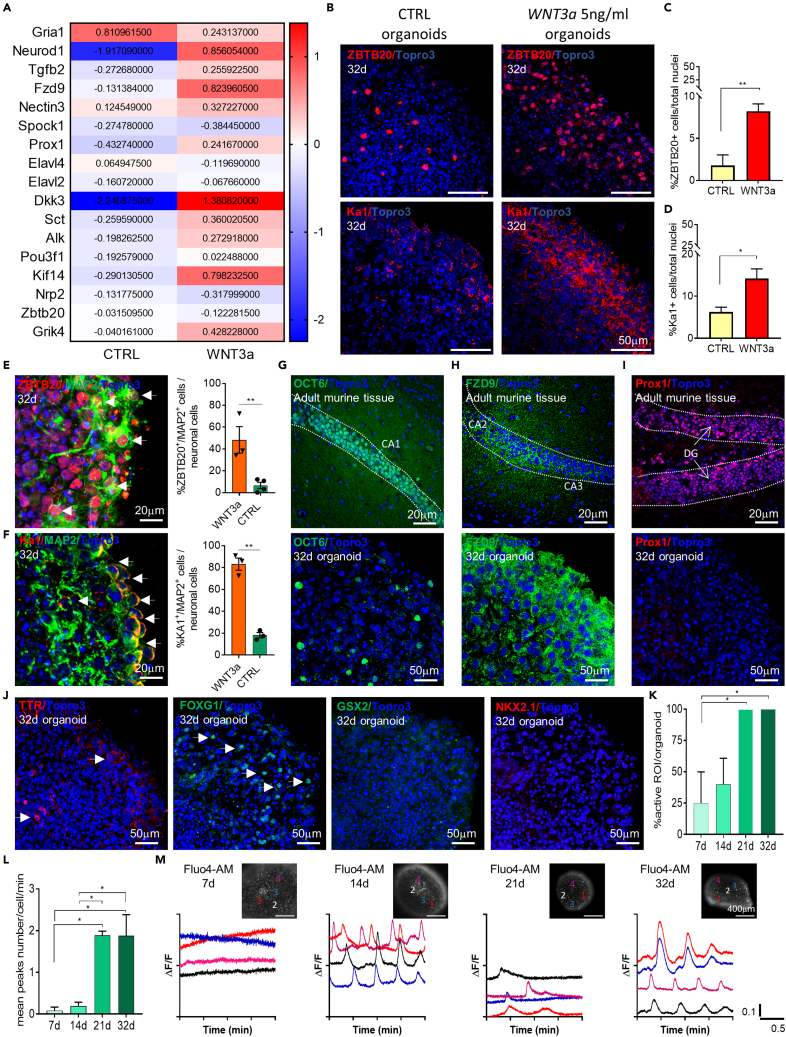
WNT3a induces organoids enriched in CA3 hippocampal neurons
The hippocampus is one of the brain regions mostly involved in the pathology progression of many neurodegenerative disorders including Alzheimer’s disease and epilepsy, therefore, an in vitro 3D structure able to replicate the hippocampal signature can be extremely useful for drug testing and neurodegenerative disease studies. Thus, we first assessed the hippocampal signature of the generated organoids by comparing their hippocampal gene expression profile with the one of the neonatal and adult mouse hippocampal tissue. This analysis revealed that brain organoids generated by NSCs originally obtained from SGZ showed a gene signature similar to the hippocampal tissue, for the upregulation of genes related to hippocampal cell growth and division (e.g., Ywhae, Bcan, Ptprs, Usp9x), regulators of proliferation and axon sprouting (e.g., Nr2e1, Uba6, Gli3, DCX, Plxna3, Btg2, Kdm6b, Hdac1) (Figure S3A and Table S6). Interestingly, we noted that murine brain organoid had a lower expression of the genes related to hippocampal neural differentiation and migration, such as WNT3a compared to hippocampal tissue (e.g., Lef1, Drd1, Dlx2, Neurod6, Alk, Dlx1, Fgf13, Wnt3a, Cdk5r2), hippocampal neurogenesis and cell fate determination (e.g., Cdk5r1, Srf, Nr4a3, Prox1) (Figure S3A and Table S6).
In order to improve the hippocampal signature in the brain organoid, we added WNT3a factor during the expansion and induction phase; the neuronal differentiation was then reached by providing the BDNF to the culture medium from day 15 (Figure S3B), as for the standard protocol (CTRL). By using a low concentration of WNT3a (5 ng/mL) we attempted to drive specifically the CA3 hippocampal neurons phenotype (Sarkar et al., 2018).
We first characterized the WNT3a-treated organoid cellular organization and phenotype specification by immunofluorescence, as previously performed for the control organoids. We followed the progressive WNT3a-treated organoid NSCs (Ki67, SOX2, Vim), neuronal progenitors (DCX), mature neurons (TUBB3, MAP2), and glial cells (GFAP) differentiation, maturation, and spatial distribution through the different developmental stages. Results showed that WNT3a supplementation did not impact the organoids spatial distribution and maturation compared with control organoids (see Figure S3).
Second, we assessed the organoid hippocampal signature following WNT3a supplementation and compared it to the standard control organoid. We performed RT-PCR analysis for specific hippocampal genes (Wheeler et al., 2015) in mature organoids samples (n = 3 pool of 6–8 organoids/pool) generated both with control and WNT3a protocol. Importantly, we found higher expression of hippocampal-related genes (e.g., Grik4, Alk, Sct, Dkk3, Nectin3, Fzd9, Tgfb2, Neurod1) in WNT3a organoids compared to control organoids (Figure 7A), suggesting that WNT3a factor is able to induce the hippocampal patterning. Then, we evaluated the protein expression of the specific pan-hippocampal marker ZBTB20 and the glutamate receptor kainate type 1 (KA1) which is typical of the cornu ammonis 3 (CA3) hippocampal pyramidal neurons (Sakaguchi et al., 2015; Sarkar et al., 2018) by immunofluorescence (Figure 7B). Results showed that mature organoids (32d) supplied with WNT3a displayed a significantly higher expression of ZBTB20 (1.8 ± 1.25% vs 8.24 ± 0.87% of ZBTB20+ cells in CTRL and WNT3a organoids, respectively; p = 0.0047) (Figure 7C) and KA1 markers (6.28 ± 1.11% and 14.2 ± 2.3% of KA1+ cells in CTRL and WNT3a organoids, respectively; p = 0.04) (Figure 7D) compared to control organoids. Moreover, we found that mature (32d) WNT3a organoids drastically increased the proportion of hippocampal ZBTB20 (Figure 7E) and KA1 (Figure 7F) neurons compared to controls (48.33 ± 12.09% and 6.59 ± 2.90% of ZBTB20+/MAP2+ cells over total number of neuronal cells in WNT3a and CTRL organoids, respectively; p = 0.0058. 83.13 ± 5.45% and 18.14 ± 2.45% of KA1+/MAP2+ cells over total number of neuronal cells in WNT3a and CTRL organoids, respectively; p = 0.001) (Figures 7E and 7F). The improved hippocampal signature of WNT3a organoid was also confirmed by the expression of the hippocampal CA1/2 region markers (OCT6, FDZ9) (Figures 7G and 7H). The expression of the DG granule neuronal marker PROX1 was not found (Figure 7I), further suggesting that the usage of low WNT3a concentration is able to specifically boost the CA3 and to a lesser extent CA1/CA2 commitment. Regional identity analysis further confirmed the specific hippocampal signature of WNT3 organoid showing rare cortical and choroid plexus marker expression and undetectable level of ganglionic eminence hallmarks (Figure 7J).
Figure 7.
Murine brain organoids develop CA3 hippocampal-specific neurons after WNT3a supplementation
(A) Heatmap showing the log2 fold change of genes related to hippocampal signature in control (CTRL) and WNT3a-treated (WNT3a) mature organoids.
(B) Representative confocal immunofluorescence images of sliced NSCs-derived control (CTRL) against WNT3a-treated organoids, at 32 d, showing cells positive for the pan-hippocampal ZBTB20 and the CA3-specific KA1 markers.
(C and D) Graphs showing the percentage of ZBTB20 (C) and Ka1 (D) positive cells over total cell nuclei at 32d in control and WNT3a-treated organoids. Analysis was performed on n≥3 different organoids and at least on 3 entire sections for each organoid. Data in all graphs are expressed as mean ± SEM. Statistical differences between experimental conditions were evaluated by two-tailed unpaired t test for two datasets (CTRL vs WNT3a) for both markers. ∗∗p<0.01; ∗p <0.05.
(E) Representative confocal immunofluorescence images of sliced WNT3a-treated organoids (32d), showing double-positive cells (highlighted by white arrows) for ZBTB20 and MAP2 mature neuronal markers. Graph on the right showing the percentage of ZBTB20+/MAP2+ cells over total neuronal cells in WNT3a and CTRL organoids at 32d. Data are expressed as mean ± SEM. Statistical differences were evaluated by two-tailed unpaired t test for two datasets (CTRL vs WNT3a). ∗∗p<0.01.
(F) Representative confocal immunofluorescence images of sliced WNT3a-treated organoids (32d), showing double-positive cells (highlighted by white arrows) for Ka1 and MAP2 mature neuronal markers. Graph on the right showing the percentage of Ka1+/MAP2+ cells over total neuronal cells in WNT3a and CTRL organoids at 32d. Data are expressed as mean ± SEM. Statistical differences were evaluated by two-tailed unpaired t test for two datasets (CTRL vs WNT3a). ∗∗p<0.01.
(G–I) Representative confocal immunofluorescence images of sliced adult (8 weeks) murine brain tissue and mature (32d) murine brain organoids showing the expression of OCT6 (G), FZD9 (H) and Prox1 (I) as CA1, CA2 and DG hippocampal markers, respectively. White insets represent a sketch of coronal mouse brain sections while red boxes highlight the brain region of the murine tissue staining.
(J) Representative confocal immunofluorescence images of mature (32d) murine WNT3a brain organoids showing the low or no expression of TTR-choroid plexus, FOXG1-cortical, GSX2 and NKX2.1 ganglionic eminences markers. All the pictures in B, E-F, G-J are maximum intensity projections of z stack images.
(K) Graph showing the percentage of active cells within the organoid at different time points (n≥3 organoids/time point; n = 10 cells selected in each organoid). Data in all graphs are shown as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗p< 0.05.
(L) Graph showing the average number of the peaks counted for each cell per minute at different time points (n≥3 organoids/time point; n = 10 cells selected in each organoid). Data in all graphs are shown as mean ± SEM. Statistical differences in graphs were calculated by ordinary one-way ANOVA. ∗p< 0.05.
(M) Time-lapse Fluo4-AM calcium imaging on whole mount WNT3a-treated organoids at different time points, paired to representative graphs of calcium flux. Note the increasing neural activity over time as well as a major peak synchronization.
Calcium imaging analysis using Fluo4-AM was lastly performed to test the cellular activity of the WNT3a organoids. Similarly, to control organoids, WNT3a treated organoids (n= at least 10 cells/organoid, n= at least 3 organoids/time point) displayed a progressive increase of spontaneous calcium activity during development (Figures 7K–7M) suggesting that WNT3a modulation preserved the spontaneous cell activity of murine brain organoid.
Overall, these data suggested that murine brain organoids can be induced to specifically acquire the neuronal signature of the CA3 hippocampus.
Discussion
In this work, we obtained robust, reproducible, and functional 3D brain organoids starting from mouse SGZ-derived NSCs. In line with the different developmental time required for human and murine brain development (Chen et al., 2017; Clancy et al., 2007; Molnar and Clowry, 2012; Semple et al., 2013), we effectively generated brain organoids in 32 d of in vitro culture, overthrowing the time necessary for human iPSCs, which requires at least 30 d for neuronal induction and further 60 to 90 d for overall differentiation (Kadoshima et al., 2013; Mariani et al., 2012; Pasca et al., 2015; Lancaster et al., 2013).
The fast production of murine brain organoids composed of functional mature neurons ensures their use as feasible, cost-effective, time-saving tools for high-throughput drug screening before the validation on human-models. Global transcriptomic analysis demonstrated the robustness of the method through the protocol phases showing similar gene profile of organoids belonging to the same developmental stage. We used principal component analysis to characterize transcriptional variance between different organoid's maturation stage and murine tissue counterparts. Different organoids at distinct differentiation phase accounted for only 12% of the variance with different samples belonging to the same developmental stage clustering close together suggesting the reproducibility of the protocol and the low variability across multiple organoid samples. This aspect (low variability) is of fundamental importance for future drug screening applications (Marshall and Mason, 2019). Moreover, taking advantages of the fast growth rate and the possibility of easily obtaining mutant lines, organoids derived from mouse embryonic NSCs may represent a valuable tool to investigate both neurodevelopmental physiological processes and disorders thanks to the lower number of mice required to generate a high number of brain organoids.
The standardization of the proposed protocol was reached by two important steps: (1) the seeding of a precise number of cells in the expansion phase (20000 cells/well), that allowed us to obtain in each well one organoid with defined size and developmental stage; (2) the use of a time-controlled growth factor media concentration, to start the cells maturation process. In addition, we demonstrated that the neurotrophin BDNF, commonly used to increase neuronal maturation in primary 2D NSCs-derived culture (Ji et al., 2005; Silva et al., 2009), was able to induce neuronal maturation also in 3D structures. In this work we demonstrated that mSGZ-derived NSCs can efficiently self-assemble and differentiate into brain tissue-like structure endowed with functional mature neurons. As already described (Abranches et al., 2009; Kuegler et al., 2010; Palmqvist et al., 2005), mSGZ-derived NSCs derived from E14.5 embryos display neuronal commitment with the expression of markers that are specific for NSCs. This pointed out an important difference with the protocol used for human-derived organoids (Lancaster and Knoblich, 2014; Pasca et al., 2015; Sakaguchi et al., 2015; Andersen et al., 2020) since no neuroepithelia induction step was required for mSGZ-derived NSC brain organoid generation.
The maturation of murine SGZ-NSCs-derived organoids was confirmed by comparative techniques, evaluating organoids morphology, transcriptomic profile, cellular organization, metabolic profile, and functionality.
In line with in vivo brain development (Seto and Eiraku, 2019) whole transcriptomic analysis showed the progressive maturation of the organoid highlighting the down-regulation of gene related to neural stem cells and the up-regulation of gene related to neuron differentiation, synapse structures, calcium ion transport, and the development of calcium ion homeostasis. We specifically observed increasing expression of gene related to NMDA and cholinergic receptors, glutamate receptor and reuptake as well as GABA receptors and transporters further supporting functional neuronal maturation. Expression of chloride (Cl−) co-transporters, such Nkcc1 and Kcc2, is important for intracellular Cl− homeostasis and for GABA reversal potential, playing a critical role in GABAergic transmission. NKCC1 is best known for its role in maintaining a high intracellular Cl−concentration in immature neurons. During the second postnatal week in rats and mice (Rivera et al., 2005), NKCC1 expression decreases while KCC2, the main Cl− extruder in the CNS, is upregulated, leading to a decrease in neuronal intracellular Cl− concentration and a shift toward hyperpolarizing GABAergic responses. Accordingly, transcriptomic analysis of the murine brain organoid showed the increase of KCC2 during organoid development, while NKCC1 expression decreased over organoid maturation, suggesting that the maturity level reached by the organoid at 32d is comparable to the maturity of a postnatal brain. However, it has to be considered that in vitro culture determines the loss of important intrinsic and morphogenic signals (Cederquist et al., 2019) thus leading to obvious differences in terms of specific cell identity commitment, differentiation, and maturation time respect to what is commonly observed in vivo.
We reported the formation of 3D structures in which the initial high abundance of stem cells and neuronal progenitors is scattered through the entire organoid. As soon as the differentiation proceeds, we observed neuronal cells mainly localized at the edge of the 3D structure. DCX+, TUBB3+ and, later, MAP2+ cells moved from the center to the edge of the organoid, acquiring a radial-like organization. Radial cell migration replicates neuronal in vivo migration from the SVZ to the cortical plate (Lange et al., 2016; Bifari et al., 2017). Interestingly, we observed a metabolic switch from glycolytic to oxidative metabolism during the maturation of the organoid. In vivo and in vitro experiments previously showed that neuronal precursors are highly glycolytic and switched toward oxidative metabolism during the migration and maturation to the upper cortical layer (Beckervordersandforth et al., 2017; Lange et al., 2016; Zheng et al., 2016). Indeed, 3D brain organoids maturation proceeded toward oxidative metabolism replicating in vivo brain development metabolic switch (Lange et al., 2016; Beckervordersandforth et al., 2017).
Neuronal cells populating the brain organoids were characterized by extending processes that converged into 3D neuronal network with synaptic connections. Proper brain development and function require the presence and a stringent balance between excitatory and inhibitory synapses formation during neural circuit assembly (Cline, 2005). The initial overproduction of excitatory synapses subsequently undergoes a pruning process which consists of an activity-dependent elimination of synapses until the synaptic density is almost stable (Neniskyte and Gross, 2017). This process ensures the right ratio between excitatory and inhibitory synapses, which in turn contribute to this balance by refining the amount of excitatory synapses, a typical feature of a maturational process. By evaluating the expression of specific excitatory and inhibitory synaptic markers, we showed that the murine brain organoids were able to undergo neuronal maturation through a process similar to the one that occurs in vivo, reaching a balance between the number of excitatory and inhibitory synapses overtime, suggesting the accomplishment of the pruning process.
Several studies showed that the synaptic development depends not only on neurotrophins and neuronal cells, but also on glia (Neniskyte and Gross, 2017), in particular revealing a key astrocytes' contribution to the synaptic pruning and equilibrium. The presence of astrocytes within human brain organoids is already disclosed (Pasca et al., 2015; Quadrato et al., 2017; Lancaster et al., 2013) and it is reported that the addition of astrocytes to neuronal cultures promotes synapses maturation and spontaneous action potential formation (Allen and Eroglu, 2017). We showed that astrocytes within the brain organoid progressively matured and were in close contact with differentiating neuronal cells suggesting they might support the synaptogenesis and the regular neuronal activity.
Currently, unguided brain organoid generation protocol starting from hiPSCs show the presence of different brain regional identities (cortex, hippocampus, ganglionic eminences, and choroid plexus). The murine brain organoids generated with our protocol presented dorsal forebrain signature with no evident expression of ganglionic components. Interestingly, a higher similarity to hippocampal rather than to cortical tissue was observed in mature SGZ-NSCs derived organoids. The lack of a ganglionic eminence identity observed in the murine SGZ-NSCs derived organoid may be explained by the absence of extrinsic pattering factors. On the other hand, possibly, the brain regional identity differences between the SGZ-NSCs derived murine brain organoids described here and the embryonic and hiPSC derived cerebral organoid may be due to the different signature of the initial stem cell source.
Calcium signaling and spontaneous oscillation of intracellular calcium concentrations are important to maintain brain development and function, playing a pivotal role in several aspects of neuronal growth and differentiation, such as synapse maturation and synaptic contact formation (Rosenberg and Spitzer, 2011). By calcium imaging analysis, we noted a developing cellular activity inside the organoids along the stages of maturation suggesting the possible existence of a cellular network composed of neurons exhibiting a functional synaptic connection. To identify an active neuronal connection, we exploited the transneuronal tracing technology of the rabies virus, which specifically propagates exclusively between connected neurons by strictly unidirectional (retrograde) transneuronal transfer (Ugolini, 2011). Thus, we demonstrated that neurons composing the 3D murine brain organoids were connected by active synapses.
Previous works reported functional characterization of neurons within the organoid and their network activity; however, they have not been entirely described (Pacitti et al., 2019; Qian et al., 2019; Song et al., 2019; Trujillo et al., 2019). A critical point about the complete evaluation of neuron activity within the organoid is represented by the 3D spheroid structure that limits the use of current electrophysiological technologies. To the best of our knowledge, acute MEA recordings were performed in few studies only (Fair et al., 2020; Trujillo et al., 2019; Renner et al., 2020; Park et al., 2021; Zafeiriou et al., 2020; Yao et al., 2020; Giandomenico et al., 2019). The large majority of brain organoid functionalities described were performed in organoid-derived cells outgrowing as a monolayer culture (Trujillo et al., 2019; Yao et al., 2020) or in organoid slices (Zafeiriou et al., 2020; Giandomenico et al., 2019). In this study, we presented a functional assessment via MEA electrophysiology performed on intact organoids, preserving their 3D architecture and neuronal connections. Although the yield of MEA recording was low, these experiments demonstrated the formation of electrically active neuronal clusters located at the periphery of the organoid, thus providing the very first evidence of functional microcircuits in intact organoids derived from murine SGZ-derived NSCs. These results were in line with the functional studies on human iPSC-derived organoids, wherein the detected activity was confined within the outer organoid layer, and, overall, in a restricted number of MEA electrodes (Giandomenico et al., 2019). The low yield reported in this study may be explained by the use of planar MEAs, not penetrating the organoid, thus, not allowing us to detect the activity possibly generated by neurons populating deeper layers of the 3D structure. Another possible explanation is that despite the presence of active neurons, as also supported by calcium imaging analysis, their activity is not robust/synchronous enough to give origin to local field potentials or multiunit activity, which are recorded with low-density planar MEAs. Further studies deploying 3D MEAs or silicon probes or high-density MEAs will help address more in depth the functionality of murine organoids.
To stabilize the organoids during patch-clamp recording and facilitate giga-ohm seal formation, we plated them on poly-D-lysine coated cover glasses, a strategy that allowed us to record from over 60 living cells. All cells recorded were located at the edge of the organoid where neurons tend to cluster, especially in the mature organoids. The patch-clamp analysis revealed the presence of functional cells as 100% of the recorded ones showed ionic currents that progressively changed in density and type over organoid maturation. Specifically, potassium currents were observed in all developmental stages and increased in density with organoid maturation, while sodium currents appeared only in the most mature phase. These results were in line with the increasing expression of genes related to voltage-gated channels in mature compared to early organoids observed by gene ontology, heatmap profiles, and DESeq2 analysis, and confirming a developmental process in which neurons in mature organoids were able to fire action potentials.
Lastly, we provided a proof of concept of the possibility to modulate the 3D model by inducing the brain organoid toward a hippocampal specific phenotype. Since WNT pathway is crucial for the hippocampal fate determination (Lie et al., 2005), we specifically prompted the hippocampal signature by supplying the culture medium with specific concentration of WNT3a factor (Sakaguchi et al., 2015; Sarkar et al., 2018) during the proliferation and induction phases of the protocol. This modulation allowed us to obtain consistent hippocampal specific neurons, expressing both the pan-hippocampal ZBTB20 marker, typical of all neurons committed to hippocampal identity and the KA1 marker which, within the hippocampus, is primarily expressed by pyramidal neurons in the CA3 subfield. Interestingly, a slight expression of other CA hippocampal subfield markers (i.e., OCT6 and FZD9) was found in mature WNT3a-treated organoids while no expression of PROX1, specific for the DG hippocampal region, was observed. The reliability and flexibility of the herein developed protocol not only enable to adopt it systematically for generating in vitro brain hippocampal replicas, but may be also further implemented to mimic different hippocampal brain subregions.
In conclusion, our results showed for the first time the effective generation of reliable and reproducible murine brain organoids. SGZ-derived NSCs progressively mature in a short time (5 weeks) forming 3D networks of functional neurons with synaptic connections able to acquire a specific hippocampal brain region signature. The accessibility of the stem cell source, the reproducibility, robustness, and functionality of the murine SGZ-NSC-derived organoids may represent valuable supporting tools for high throughput drug screening, hiPSCs-derived organoid protocol optimization and brain disease modeling.
Limitations of the study
This approach would highly benefit from further improvements including the generation of ventral forebrain cell types and the development of a defined brain region specific organization, such as the cortical layering showed in human-derived organoids. This aspect could be improved by combining ECM components or by creating assembloids, which consist of a fusion of more brain region specific organoids. In addition, the brain organoid co-culture with other cell types (i.e., microglia/oligodendrocytes) and the implementation of other strategies, such as optogenetic approaches with cell-type-specific reporter, could allow us to increase the study of functional neurons and neuronal network activity.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| SOX2 | R&D Systems | Cat# AF2018; RRID: AB_355110 |
| VIMENTIN | Millipore | Cat# AB5733; RRID: AB_11212377 |
| KI67 | Abcam | Cat# ab16667; RRID: AB_302459 |
| DCX | Cell Signaling Technology | Cat# 4604; RRID: AB_561007 |
| B3 TUBULIN | Promega | Cat# G7121; RRID: AB_430874 |
| MAP2 | Sigma-Aldrich | Cat# M1406; RRID: AB_477171 |
| ZBTB20 | Genetex | Cat# GTX121616; RRID: AB_11177870 |
| KA1 | Abcam | Cat# ab67404; RRID: AB_1140942 |
| NEUN | Genetex | Cat# GTX133127; RRID: AB_2886836 |
| GAD65/67 | Santa Cruz Biotechnology | Cat# sc-365180; RRID: AB_10710523 |
| NMDA | Santa Cruz Biotechnology | Cat# sc-365597; RRID: AB_10847218 |
| GLSYN | Santa Cruz Biotechnology | Cat# sc-74430; RRID: AB_1127501 |
| GFAP | Abcam | Cat# ab53554; RRID: AB_880202 |
| SYNAPTOPHYSIN | Synaptic Systems | Cat# 101 004; RRID: AB_1210382 |
| PSD95 | Millipore | Cat# MAB1596; RRID: AB_2092365 |
| GEPHYRIN | Synaptic Systems | Cat# 147 011; RRID: AB_887717 |
| VGAT | Synaptic Systems | Cat# 131 004; RRID: AB_887873 |
| VGLUT | Synaptic Systems | Cat# 135 303; RRID: AB_887875 |
| GSX2 | Genetex | Cat# GTX129390; RRID: AB_2885981 |
| FOXG1 | Abcam | Cat# ab18259; RRID: AB_732415 |
| NKX2.1 | Genetex | Cat# GTX34907 |
| PREALBUMIN | Genetex | Cat# GTX85112; RRID: AB_10723946 |
| FRIZZLED 9 | Genetex | Cat# GTX71581; RRID: AB_375823 |
| OCT6 | Abcam | Cat# ab272925 |
| MUSASHI | Genetex | Cat# GTX117808; RRID: AB_10722801 |
| NANOG | Genetex | Cat# GTX62742; RRID: AB_11169335 |
| OCT3/4 | Santa Cruz Biotechnology | Cat# SC-5279; RRID: AB_628051 |
| E-CADHERIN | Genetex | Cat# GTX100443; RRID: AB_10729586 |
| NESTIN | R&D System | Cat# MAB2736; RRID: AB_2282664 |
| SOX1 | R&D System | Cat# AF3369; RRID: AB_2239879 |
| Total OXPHOS Rodent WB Antibody Cocktail | Abcam | Cat# MS604-300; RRID: AB_1622581 |
| VDAC1 | Abcam | Cat# ab15895; RRID: AB_2214787 |
| LAMIN B | Abcam | Cat# ab16048; RRID: AB_443298 |
| Donkey anti-rabbit Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21206; RRID: AB_2535792 |
| Donkey anti-mouse Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-21202; RRID: AB_141607 |
| Donkey anti-guinea pig CY3 | Jackson ImmunoResearch | Cat# 706-165-148; RRID: AB_2340460 |
| Goat anti-mouse CY3 | Amersham | Cat# PA43002; RRID: AB_772235 |
| Donkey anti-goat Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11056; RRID: AB_142628 |
| Goat anti-rabbit CY3 | Amersham | Cat# PA43004; RRID: AB_772236 |
| Donkey anti-rabbit Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A10040; RRID: AB_2534016 |
| Goat anti-chicken Alexa Fluor 546 | Thermo Fisher Scientific | Cat# A-11040; RRID: AB_2534097 |
| Bacterial and virus strains | ||
| BRVenVA-1 G-deleted Rabies mcherry | Osakada et al. (2011) | Addgene plasmid #32630, #32631, #32632, #32633, #32634 |
| pAAV-Syn-H2B-GFP-TVA-oG-WPRE3 | Charitè Viral Core Facility, University of Berlin | N/A |
| Biological samples | ||
| Embryonic (E14.5) mouse neural stem cells | This paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| DMEM-F12 GlutaMAX | Gibco | Cat#31331-028 |
| DMEM, no glucose | Gibco | Cat# 11966025 |
| Neurobasal Media | Gibco | Cat#21103-049 |
| Penicillin/Streptomycin (100ml) | Gibco | Cat#15140-122 |
| N2 | Gibco | Cat#17502-048 |
| B27 | Gibco | Cat#17504-044 |
| bFGF | Peprotech | Cat#100-18B |
| EGF | Peprotech | Cat#AF-100-15 |
| BDNF | Peprotech | Cat#450-02 |
| Wnt3a | Genetex | GTX109037-Pro |
| Accutase (100 ml) | Gibco | Cat#A1105-01 |
| 0.25% Trypsin – EDTA (1X) (100 ml) | Gibco | Cat#25200-056 |
| L-Glutamine 200mM | Gibco | Cat# 25030081 |
| HBSS 10X | Gibco | Cat#14180-046 |
| HEPES | Sigma | Cat# H3375-250G |
| Clarity Western ECL Substrate | Biorad | Cat#170-5060 |
| PFA | Mondial | FM0622 |
| Sucrose | Sigma-Aldrich | Cat#84100 |
| Propidium Iodide | Sigma-Aldrich | Cat#P4170 |
| Fluorescein Diacetate | Merck-Millipore | Cat#343209 |
| Cryobloc | Diapath | Cat#070130 |
| Bovine Serum Albumine | Sigma-Aldrich | A7906-10G |
| Amido Black Staining Solution 2X | Sigma-Aldrich | A8181-1EA |
| 1,4-diazabicyclo [2.2.2] octane (DABCO) | Sigma-Aldrich | D-2522 |
| RNase A, DNase and protease-free (10 mg/mL) | Thermo Fisher Scientific | Cat#EN0531 |
| Critical commercial assays | ||
| Pierce BCA protein assay kit | Thermo Fisher Scientific | 23227 |
| TruSeq Stranded mRNA library prep kit | Illumina | 20020594 |
| RNeasy Plus Micro Kit | Qiagen | Cat#74034 |
| PowerSYBR Green PCR Master Mix | Thermo Fisher Scientific | 4367659 |
| CyQUANT Cell Proliferation Assay Kit | Thermo Fisher Scientific | C7026 |
| CellTiter-Glo 3D Cell Viability Assay | Promega | G9681 |
| Fluo-4 Direct calcium assay kit | Thermo Fisher Scientific | F10472 |
| SuperscriptTM VILOTm Master mix | Thermo Fisher Scientific | 11755050 |
| Deposited data | ||
| RNAseq | This paper | https://www.ncbi.nlm.nih.gov/bioproject/PRJNA778927 |
| Experimental models: Organisms/strains | ||
| Mouse: c57Bl/6J | Charles River | #001CD |
| Oligonucleotides | ||
| Primer for ND1 forward: 5’– CTAGCAGAAACAAACCGGGC-3’ |
Invitrogen; Quiros et al., 2017 | N/A |
| Primer for ND1 reverse: 5’– CCGGCTGCGTATTCTACGTT-3’ |
Invitrogen; Quiros et al., 2017 | N/A |
| Primer for HK2 forward: 5’-GCCAGCCTCTCCTGATTTTAGTGT-3’ |
Invitrogen; Quiros et al., 2017 | N/A |
| Primer for HK2 reverse: 5’- GGGAACACAAAAGACCTCTTCTGG-3’ |
Invitrogen; Quiros et al., 2017 | N/A |
| Software and algorithms | ||
| ImageJ | U.S. National Institutes of Health | https://imagej.nih.gov/ij/ |
| Image Lab | Biorad | https://www.bio-rad.com/it-it/product/image-lab-software?ID=KRE6P5E8Z |
| Prism | GrapPhad | https://www.graphpad.com/scientificsoftware/prism/ |
| Imaris | Bitplane | https://www.graphpad.com/scientificsoftware/prism/ |
| Avizo | ThermoFisher Scientific | http://www.fei.com/software/avizo3d/ |
| Other | ||
| ORBi shaker | Benchmark Scientific | #BT3001 |
| 96-well cell imaging plates | Eppendorf | Cat# 0030741030 |
| Confocal Microscope LSM710 | Zeiss | RRID:SCR_018063 |
| Ti Eclipse Microscope | Nikon | RRID:SCR_021242 |
| Axiovert 200M | Zeiss | RRID:SCR_020915 |
| NanoDropTM One/OneC Microvolume UV-Vis Spectrophotometer | Thermofisher Scientific | #ND-ONE-W |
Resource availability
Lead contact
Further information and requests for resources should be directed to and will be fulfilled by the lead contact, Ilaria Decimo (ilaria.decimo@univr.it)
Materials availability
This study did not generate any unique reagents.
Experimental model and subject details
Primary neural stem cells
Pregnant wild-type (WT) C57Bl/6J mice were obtained from Charles River Laboratories (Wilmington, MA). E14.5 mice subgranular zone (SGZ) was isolated to obtain NSCs and to generate organoids. Animal housing and all experimental procedures were approved by the Istituto Superiore di Sanità (I.S.S., National Institute of Health; protocol n. C46F4.N.N4E, Italy) and the Animal Ethics Committee (C.I.R.S.A.L., Centro Interdipartimentale di Servizio alla Ricerca Sperimentale) of the University of Verona (Italy).
Method details
Mouse neural stem cells (NSCs) culture
Pregnant C57Bl/6J female mice were sacrificed by cervical dislocation; NSCs were extracted from the SGZ of E14.5 embryos’ brains using fine tweezers under stereo microscope and collected into ice-cold HBSS solution (sterile water, HBSS 10X, HEPES 0.3 M, 1% Pen/Strep). After centrifugation at 300g per 1 min, HBSS was substituted with PBS 1X and then the sample was centrifuged again at 300g for 1 min. The pellet was suspended in a solution containing DNase (Sigma Aldrich) and Trypsin 1X (GIBCO) diluted 1:5; the first round of dissociation was performed using the gentleMACS™ Dissociator (Miltenyi Biotec). After incubation for 10 min at 37°C, other two rounds of dissociation were done. PBS was added to inactivate the enzymes and then the sample was filtered with a cell strainer (40 μm diameter). After 10 min final centrifugation at 300g, samples were suspended in DMEM-F12 (GIBCO) with 2% B27 (GIBCO), 1%N2 (GIBCO), 1% Pen/Strep (GIBCO) enriched with 20ng/ml bFGF and EGF. Cells were stained with Trypan blue 0,4% (Lonza BioWhittaker), counted and cultured as neurospheres in suspension in T75 flasks (Falcon) up to the second passage at 37°C in a humidified atmosphere of 5% CO2 in air. After 7–10 d, neurospheres were collected, centrifuged, mechanically dissociated to a single-cell suspension (Dolci et al., 2017) and further used for organoid generation.
Organoid generation protocol
Brain organoids were obtained starting from E14.5 murine-derived NSCs. A three-phase organoid generation protocol was developed, each stage relying on a different media composition (see section media composition of organoid generation protocol). Specifically, the protocol consisted of the (1) expansion phase, (2) induction phase and (3) differentiation phase. The expansion phase starts after the seeding of the single cells (day 0) and lasts 5 d. Briefly, 20000 cells/well were seeded into each well of 24-well plates (Thermofisher) in 500 μl of medium enriched with 20 ng/ml EGF and 20 ng/ml bFGF (Expansion medium). During the induction phase, that continues for up to day 14, the concentration of bFGF and EGF was gradually scaled down: First, at day 5, both EGF and bFGF were decreased to a concentration of 10 ng/ml (Induction medium I); second, at day 7, bFGF concentration was reduced to 5 ng/ml, while EGF was eliminated altogether (Induction medium II). During the last step (differentiation phase), starting at day 15, the organoids were cultured in the differentiation medium enriched with 50 ng/ml BDNF. Cells plates were kept in continuously agitation (65rpm) using the ORBi-SHAKER CO2 (Benchmark Scientific) at 37°C in a humidified atmosphere of 5% CO2 in air for all the culture time. Seven different batches of SGZ-NSCs belonging to seven different embryos extraction were used to generate organoids. The organoids used in this work were generated starting from single cells obtained after no more than 3/4 subsequent phases of expansion following tissue isolation.
Media composition of organoid generation protocol
Expansion medium (day 0-4): DMEM/F-12 GlutaMAX, (GIBCO), 2% B27 supplement (GIBCO), 1% N2 supplement (GIBCO), 1% Pen/Strep (GIBCO), 20 ng/ml EGF, 20 ng/ml bFGF.
Induction medium I (days 5-6): DMEM/F-12 GlutaMAX, (GIBCO), 2% B27 supplement (GIBCO), 1% N2 supplement (GIBCO), 1% Pen/Strep (GIBCO), 10 ng/ml EGF, 10 ng/ml bFGF.
Induction medium II (days 7-14): DMEM/F-12 GlutaMAX, (GIBCO), 2% B27 supplement (GIBCO), 1% N2 supplement (GIBCO), 1% Pen/Strep (GIBCO), 5 ng/ml bFGF.
Differentiation medium (days 15-42): Neurobasal medium (GIBCO), 2% B27 supplement (GIBCO), 1% Pen/Strep (GIBCO), 0,25% L-glutamine, 50 ng/ml BDNF.
RNAseq analysis
Total RNA was extracted from 14 samples (7 days organoids: n=3 pools of 3 organoids each, 14 d organoids: n=3 pools of 3 organoids each, 30 d organoids: n=3 pools of 3 organoids each, adult – 3 months – cerebral tissue: n=3, P0 cerebral tissue: n=2) using the RNeasy Plus Micro Kit (Qiagen, Cat No. 74034) according to the manufacturer's protocol and RNA integrity was evaluated using the Fragment Analyzer (Agilent Technologies). RNAseq library preparation was performed starting from 100ng high-quality total RNA using the “TruSeq Stranded mRNA library prep kit” (Illumina, San Diego, USA). Briefly, the mRNA fraction was purified from total RNA by polyA capture, fragmented and subjected to cDNA synthesis. Barcoded DNA adapters were ligated to both ends of the double-stranded cDNA and subjected to PCR amplification. The library products were evaluated using Fragment Analyzer (Agilent Technologies), then sequenced on an Illumina NextSeq500 sequencer using 75bp single-end reads, generating ∼17 million reads per sample. Quality control was conducted using the software Scythe (v0.991) and Sickle (v1.33). Transcript expression levels were then quantified in each sample by running the computer software Salmon v.1.0.0 against the reference sequence of the mouse transcriptome (Gencode M23-GRCm38).
Differential expression analysis on sequence count data was performed using negative binomial distribution models as implemented in the library DESeq2 of R software (https://www.r-project.org/), using the Benjamini-Hochberg method to adjust the p-values for multiple comparisons. Transcript expression fold-changes were estimated for each expressed transcript in every comparison. The most relevant up-regulated genes were then summarized into biological processes and molecular functions by an enrichment analysis based on gene ontologies as implemented in the library dnet of R. Gene Set Variation Analysis was performed using the package GSVA (ver 1.38.2) of R to identify functionally enriched gene sets.
Selected gene lists were adapted from literature: (i) neural stem cell dataset, neuronal progenitors (Bifari et al., 2020); (ii) mature neurons (Cahoy et al., 2008; Zhang et al., 2014; Bifari et al., 2020); (iii) astrocytes (Boisvert et al., 2018; Zhang et al., 2014); (iv) inhibitory synapses and excitatory synapses (Hayamizu et al., 2005); (v) hippocampal development (Lein et al., 2004; Wheeler et al., 2015). The possible contribution from Hippocampus (H) and Cortex (C) cells in the organoid tissue based on gene expression was estimated by investigating different proportions of H and C cells into the organoid tissue according to gene expression profiles of selected genes (cortical signature: Stard8, Sytl2, Tnnc1, Myl4, Drd2, Calb2, Reln, Lhx1, Lhx5, Ebf3, Cux1, Satb2, Dok5, Ctp2, Emx1, Tiam2, Tbr1, Foxg1, Pvrl3; hippocampal signature: Zbtb20, Sipa1l3, Gpr161, Crlf1, Glis3, Dcx, Nrp2, Tgf2b, Gria1, Tnip2, Slc39a6, Prox1, Slc26a10, Trpc6, Neurod1, C1ql2, Gqrik4, Elavl2, Elavl4, Dkk3, Nectin3, Scip, Spock1) defining either hippocampal or cortical phenotype. For each gene lists and different amounts of H and C cells, it was calculated the sum of the difference of gene expression levels (log-TMP transformed values) with the organoid sample (7 d organoids: n=3 pools of 3 organoids each, 14 d organoids: n=3 pools of 3 organoids each, 30 d organoids: n=3 pools of 3 organoids each) under investigation. The difference between samples was estimated as the normalized values of the squared differences between samples (C+H and organoid tissues). Lower overall values of differences suggested a higher confidence of the suggested proportion H and C cells in the organoid tissue. In order to validate the selected gene list, we performed the above analysis by including adult murine hippocampal and cortical tissue (adult – 3 months – cortical tissue: n=3, adult – 3 months – hippocampal tissue: n=3). A low value of differences was obtained by both hippocampal and cortical samples, thus suggesting reliability of the analysis performed. The percentage of similarity to hippocampus or cortex was evaluated based on the H/C proportion showing the higher confidence.
Optical projection tomography (OPT) and volume measurement
In-house-built 3D imaging system for Optical Projection Tomography (OPT) (Belay et al., 2021) was used to image and characterize the 3D structures of the organoids. Brightfield OPT was used to capture the 360-degree structure of the organoids. White LED source (LTCL23; Opto Engineering, Mantova, Italy) with a 0.65 μm voxel size (1 px=0.65 μm) illuminated the samples in 0.9° intervals while a 10× objective captured a total of 400 projection images simultaneously rotating the sample. The projection images were then used to reconstruct the 3D tomographic image of the sample using a filtered back-projection algorithm (Matlab; MathWorks, Natick, MA, USA) (Figueiras et al., 2014) and further edited using Fiji (ImageJ) and Avizo (ThermoFisher Scientific). All 3D images were manually segmented using Avizo, and organoid volumes, images, and rotating videos were calculated and processed based on the 360-degree reconstructions using Avizo.
Organoids maximum diameter measurements
To determine the organoids’ maximum diameter, bright field images were acquired, during the various stages of the protocol (2, 5, 7, 9, 11, 14, 21, 28, 32d; n>30) using the Axiovert 200M (Carl Zeiss, Munch, Germany) microscope. The maximum diameter was measured using the “straight line” tool of ImageJ software (U.S. National Institutes of Health). The pixel obtained value was converted in mm.
Viability assay
Organoid viability was assessed using a fluorescence staining solution containing propidium iodide (50%), fluorescein diacetate (50%) and Hoechst (1:1200), freshly prepared the day of the experiment and protected from light. The propidium iodide is an intercalating red fluorescent dye labeling dead cells, fluorescein diacetate is hydrolized to fluorescent fluoresceine in live cells, whereas Hoechst is used for total nuclei staining. Whole mount organoids (n=3 sample/time point) were washed in PBS 1X and incubated for 5 min in the staining solution. Three PBS 1X washes were performed. Then, the samples were fixed in a 4% PFA and 4% sucrose solution for 10 min, before being acquired at Zeiss LSM710 confocal microscope (Carl Zeiss, Munich, Germany). Quantification of the propidium iodide positive cells was done by counting positive cells on total nuclei. Then, the number of viable cells was obtained. The quantification was performed using a specific plugin of ImageJ software (U.S. National Institutes of Health) and a semi-automated cell count.
Organoid Immunohistochemistry
Murine NSCs-derived organoids were selected at 7, 14, 21, and 32 d after seeding and fixed for 15 min in 4% PFA/4% sucrose, then rinsed and stored in PBS 1X at 4°C. For cryosectioning, each organoid was embedded in Cryobloc (Diapath, Italy) and cut at 30 μm. Immunostaining was performed after 1 hour incubation in blocking solution (PBS 1X with 0.25% Triton X-100, 2% bovine serum albumin for cytosolic antigens and PBS 1X with 0.5% Triton X-100, 2% bovine serum albumin (BSA) for nuclear antigens) (Formaggio et al., 2010). Sections were then incubated with primary antibodies in blocking solution at 4°C for 2 hours. After rinsing 3 times for 10 min in blocking solution, appropriate secondary antibodies were applied for 1.5 h at room temperature. After final washing steps in blocking solution and then in PBS 1X, nuclear staining with TOPRO™-3 or DAPI (Invitrogen Thermo Fisher Scientific) was performed, and slides were mounted using 1,4-Diazabicyclo[2.2.2] octane (DABCO) (Sigma-Aldrich). For whole-mount immunofluorescence staining, whole organoids were incubated in blocking solution (PBS 1X with 0.5% Triton X-100, 2% bovine serum albumin) for 1 h; then, they were incubated with primary antibodies in blocking solution and kept on a shaker overnight at 4°C. After rinsing 3 times for 10 min in PBS 1X, secondary antibodies were applied for 6 h at room temperature. After final washing steps in PBS 1X, nuclear staining with TOPRO™-3 or DAPI (Invitrogen Thermo Fisher Scientific) was performed; in order to preserve the 3D structure of the organoid, an in-house method was set up, using a parafilm chamber fixed on a glass slide; the whole organoids were inserted in the chamber and mounted using DABCO.
Tissue immunofluorescence
Intact adult mice brain tissues were extracted after animal perfusion in 4% PFA then rinsed in PBS and stored in sucrose 30% at 4°C. Medio-lateral sagittal or coronal sections were cryosectioned at 35μm throughout the entire samples. Slides were stored at -20°C. Immunostaining on cryosections was performed after 30 min incubation in blocking solution (PBS 1X with 0.25% Triton X-100, 2% bovine serum albumin). Sections were then incubated with primary antibodies in blocking solution overnight at 4°C. After rinsing 6 times for 5 min in blocking solution, appropriate secondary antibodies were applied for 4 h at room temperature. After final washing steps in blocking solution and then in PBS 1X, nuclear staining with TOPRO™-3 (Invitrogen Thermo Fisher Scientific) was performed and slides were mounted using DABCO (Sigma-Aldrich).
Antibodies
The following primary antibodies were used: anti-SOX2 (goat, 1:200, R&D System, Cat#AF2018), anti-Vimentin (chicken 1:400, Millipore, Cat#AB5733), anti-Ki67 (rabbit, 1:200, Abcam, Cat#ab16667), anti-DCX (rabbit, 1:400, Cell Signaling Technology, Cat#4604), anti-β3Tubulin (mouse, 1:400, Promega, Cat#G7121), anti-MAP2 (mouse, 1:200, Sigma-Aldrich, Cat#M1406), anti-ZBTB20 (rabbit, 1:200, Genetex, Cat#GTX121616), anti-KA1 (rabbit, 1:200, Abcam, Cat#ab67404), anti-NeuN (rabbit, 1:200, Genetex, Cat#GTX121616), anti-GAD65/67 (mouse, 1:400, Santa Cruz Biotechnology, Cat#sc-365180), anti-NMDA (mouse, 1:400, Santa Cruz Biotechnology, Cat#sc-365597), anti-GLSYN (mouse, 1:400, Santa Cruz Biotechnology, Cat#sc-74430), anti-GFAP (goat, 1:200, Abcam, Cat#ab53554), anti-synaptophysin (guinea pig, 1:200, Synaptic System, Cat#101004), anti-PSD95 (mouse, 1:200, Millipore, Cat#MAB1596), anti-gephyrin (mouse, 1:200, Synaptic Systems, Cat#147011), anti-VGAT (guinea pig, 1:400, Synaptic Systems, Cat#131004), anti-VGLUT (rabbit, 1:400, Synaptic Systems, Cat#135303), anti-Total OXPHOS Rodent WB Antibody Cocktail (mouse, 1:200, Abcam, ab110413), anti-VDAC1 (rabbit, 1:200, Cat# ab15895), anti-Lamin B (rabbit, 1:200, Abcam, Cat#ab16048), anti-GSX2 (rabbit, 1:200, Genetex, Cat#GTX129390), anti-FOXG1 (rabbit, 1:200, Abcam, Cat#18259), anti-NKX2.1 (mouse, 1:200, Genetex, Cat#GTX34907), anti-PreAlbumin (chicken, 1:200, Genetex, Cat#GTX85112), anti-Frizzled9 (rabbit, 1:200, Genetex, Cat#GTX71581), anti-Musashi 2 (rabbit, 1:200, Genetex, Cat#GTX117808), anti-OCT3/4 (mouse, 1:200, Santa Cruz Biotechnology, Cat#sc-5279), anti-OCT6 (rabbit, 1:200, Abcam, Cat#ab272925), anti-E-Cadherin,(rabbit, 1:200, Genetex, Cat#GTX100443), anti-NANOG (mouse, 1:200, Genetex, Cat#GTX627421), anti-SOX1 (goat, 1:200, R&D Systems, Cat#967237), anti-Nestin (mouse, 1:200, R&D Systems, Cat#MAB2736). Appropriate secondary antibodies were used: donkey anti-rabbit Alexa Fluor 488 (donkey, 1:1000, Thermo Fisher Scientific, Cat#A21206), donkey anti-mouse Alexa Fluor 488 (donkey, 1:1000, Thermo Fisher Scientific, Cat#A21202), donkey anti-guinea pig CY3 (donkey, 1:1000, Jackson ImmunoResarch, Cat#AB2340460), goat anti-mouse CY3 (goat, 1:1000, Amersham, Cat#PA43002), donkey anti-goat Alexa Fluor 546 (donkey, 1:1000, Invitrogen by Thermo Fisher Scientific, Cat#A-11056), goat anti-rabbit CY3 (goat, 1:1000, Amersham, Cat#PA43004), donkey anti-rabbit Alexa Fluor 546 (donkey, 1:1000, Thermo Fisher Scientific, Cat#A10040), goat anti-chicken Alexa Fluor 546 (goat, 1:1000, Thermo Fisher Scientific, Cat#A11040). TO-PRO™-3 (1:3000, Molecular Probes-Thermo Fisher Scientific) or DAPI (1:2000, Invitrogen Thermo Fisher Scientific) were used to visualize the cell nuclei.
Immunofluorescence image acquisition, analysis and quantification
Immunofluorescence imaging of sliced and whole-mount organoids was performed using a Zeiss LSM710 confocal microscope (Carl Zeiss, Munich, Germany). Both reconstruction of 3D images and quantification were performed using a specific plugin of ImageJ software (U.S. National Institutes of Health) and a semi-automated cell count. Quantification of positive-immunoreactive cells for Ki67, Vimentin, SOX2, DCX, TUBB3, ZBTB20, KA1, GFAP and MAP2 was done by counting positive cells over the total number of nuclei (TOPRO™-3) per field. The number of excitatory synaptic puncta was measured by evaluating the co-localization of the expression of the presynaptic marker vGLUT and the postsynaptic marker PSD95; the presence of inhibitory GABAergic synapses was quantified by analyzing the co-localization of the presynaptic marker vGAT and the postsynaptic marker gephyrin. The colocalization analysis was performed in blind using a custom-designed macro in Image J. Synaptic density was expressed as synaptophysin-positive area against the MAP2+ neuronal cells area. For the quantification of organoid marker expression, at least 3 slices for each organoid sample were analyzed, from n ≥ 3 different organoids. For whole-mount staining, at least 10 plane sections were acquired and analyzed for each organoid. Statistical differences between marker expression at different time points were calculated by ordinary one-way ANOVA followed by Tukey’s post-hoc test. P < 0.05 was considered statistically significant. Statistical differences between two groups (CTRL vs WNT3a) for hippocampal markers expression were calculated by two-tailed unpaired t-test for two datasets. P < 0.05 was considered significantly different.
Fluorescence intensity quantification
Fluorescence intensity in cryosections was obtained using the Plot Profile command in ImageJ (Fiji). Fluorescence intensity graphs for each marker was obtained by further using the Fit Function (8th Degree Polynomial) in ImageJ.
Immunoblot analysis
Immunoblot analysis was performed as previously described (Brunetti et al., 2020). Murine NSCs-derived organoids at 7, 14, 21 and 32 d of culture were pooled (n=2 or 5 organoids/sample depending on the analyzed time point) and homogenized in modified radioimmunoprecipitation assay (RIPA) buffer [100 mM NaCl, 50 mM Tris-HCl pH 7.4, 1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate (SDS), DTT 5 mM, MgCl2 5 mM, Glycerol 3%, Phenylmethylsulfonyl fluoride (PMSF) 5 mM, and Benzonase 2 unit/μl] in the presence of protease and phosphatase inhibitors. Samples were incubated on ice for 30 min and centrifuged at 10,000 x g for 15 min at 4°C. Protein concentration was determined with the Bicinchoninic Acid (BCA) protein assay Kit (Thermo Scientific). Aliquots (20 μg each) were run through a 4–15% SDS–polyacrylamide gel electrophoresis (PAGE), electroblotted onto a PVDF membrane. Amido Black staining solution (Sigma, Cat#A8181-1EA) was used to detect total protein. After blocking, the membranes were probed by the primary antibodies overnight at +4°C and subsequently incubated with horseradish peroxidase-conjugated secondary antibodies (anti-mouse, 1:5000, Promega, Cat#W402B; anti-rabbit, 1:5000, Promega, Cat#W4018) for 2 h at room temperature. Chemiluminescence-based immunostaining (Clarity Western ECL Substrate, Biorad, Cat#170-5060) was performed.
The following primary antibodies were used: total oxidative phosphorylation (OXPHOS) rodent antibody cocktail (mouse, 1:1000, Abcam, Cat#ms604-300), VDAC-1 (rabbit, 1:5000, Abcam, Cat#ab15895), Lamin B (rabbit, 1:1000, Abcam Cat# ab16048). Images were acquired with the ImageQuant LAS 4000 apparatus (GE Healthcare); the quantification was performed using Image Lab™ software, version 6.0.1 for Windows (Bio-Rad Laboratories). For each time point n>3 organoid pools were evaluated. Statistical differences in graphs were calculated by ordinary one-way ANOVA. P < 0.05 was considered statistically significant.
DNA extraction and mitochondrial DNA quantification
Total DNA was extracted from organoids (n=9-11 for each time point) with proteinase k-based protocol. Briefly, each organoid was incubated in Lysis Buffer (Tris-HCl pH 8.5 100mM, EDTA 5mM, SDS 0.2% and NaCl 200mM) supplemented with 10 μg/ ml of proteinase K at 55°C for 2 h. After centrifugation, the DNA was precipitated with isopropyl alcohol, washed with 70% v/v ethanol, suspended in TE buffer and quantified with NanoDrop™ One/OneC Microvolume UV-Vis Spectrophotometer (ThermoFisher Scientific). Mitochondrial DNA was quantified by Sybr Green-based real time PCR (QuantStudio™ 3 Real-Time PCR System, Applied Biosystem) according to the ΔΔCt method. The mitochondrial gene MT-ND1 and the nuclear gene Hexokinase 2 (HK2) were amplified from approximately 20 ng of total DNA. Triplicate experiments were carried out for each biological replicates. For each time point, n=9 organoids were evaluated; Statistical analysis was performed by one-way ANOVA followed by Tukey’s comparison test. P < 0.05 was considered statistically significant.
Bioenergetic assessment
Organoids were kept at 37°C in a humidified atmosphere of 5% CO2 in air, on an orbital shaker, with culture medium supplemented either with glucose or galactose as carbon source. Total ATP content was evaluated in samples at different time points. The glucose media consisted of DMEM supplemented with 17mM glucose, 1 mM sodium pyruvate, 1% pen-strep, 2% B27 plus 1% N2 and 5 ng/ml bFGF or 50 ng/ml BDNF for organoids at 7 d or 14, 21, and 32 d, respectively. Galactose media consisted of DMEM supplemented with 5 mM galactose, 1 mM sodium pyruvate, 1% pen-strep, 2% B27 plus 1% N2 and 5 ng/ml bFGF or 50 ng/ml BDNF for organoids at 7 d or 14, 21, and 32 days, respectively. After 60h, the cellular ATP content was evaluated by CellTiter-Glo 3D Cell Viability Assay in n=8 (7d), n=8 (14d), n=10 (32d) organoids for each condition, following manufacturer’s instructions. To exclude any differences in cell proliferation/viability, a subset of organoids treated as above (7d: n=4, 14d: n=4 and 32d: n=6 for each media condition) was used for data normalization by evaluating the DNA content with CyQUANT Cell Proliferation Assay Kit, following manufacturer's instructions. Results were then expressed as A.U. relative to glucose and presented as mean ±SEM. Statistical differences were evaluated by two-tailed unpaired t-test for two datasets (glucose versus galactose) at each time point. P < 0.05 was considered significantly different.
Patch-clamp recordings
Organoid at different stages of development (11-13d: intermediate; 19-26d: early mature; 33-42d: mature) were plated on glass coverslips coated with poly-D-lysine (10 μg/mL; Sigma) more than 24h before recording and left in the incubator under controlled conditions (37°C, 5% CO2). Prior to recording, the coverslips with organoids were moved from a Petri dish (35 mm) to a recording chamber where the experiments were performed at room temperature in artificial cerebrospinal fluid (ACSF) composed of (in mM): NaCl (115.0), KCl (2.0), KH2PO4 (1.25), MgSO4 (1.0), CaCl2 (2.0), D-Glucose (25.0), and NaHCO3 (26.0), and bubbled in carbogen (95% O2 and 5% CO2). Borosilicate glass patch pipettes were pulled (P-97 micropipette puller, Sutter Instrument, USA) to a final resistance of 5-7 MΩ and filled with an intracellular solution composed of (in mM): K-Gluconate (120.0), NaCl (5 mM), HEPES (10.0), EGTA (10.0), MgCl2 (2.0), CaCl2 (1.0), ATP-Na2 (2.0), and GTP-Na (0.4), osmolality was adjusted at 285 mOsm with 10 mM sucrose. Resting membrane potential (RMP) was measured in whole-cell configuration immediately after disrupting the patch in current-clamp mode with null current injection. Membrane capacitance (Cm) was measured calibrating whole-cell compensation (Cslow) and series resistance compensation from a holding potential of -70 mV. Input resistance (Rin) was determined from the analysis of response to depolarizing steps with 10 mV increments applied from the holding potential of -70 mV. For voltage-clamp recordings, membrane potential was held at -70 mV and positive voltage steps (500 ms) were applied from -70 mV to 100 mV (ΔV=10 mV). For current density analysis, current amplitude was measured at the positive peak for IA (○ in Figure 5D), at the steady-state for IDR (• in Figure 5D), and at negative peak for INa (□ in Figure 5D) (Safronov, 1999). Current-clamp activity was recorded at resting membrane potential and evoked by 500 ms current pulses from -10 pA to +10 pA (ΔA=10 pA). Data were acquired with an Axopatch 1D amplifier and Digidata 1322, using pClamp 10.6 software (all from Molecular Devices, USA) and stored in a pc for offline analysis. pClamp 10 (Molecular Devices, USA) and Origin 2020 (OriginLab, USA) software were used routinely during data analysis and graph plotting. Data of passive membrane properties experiments are presented as median followed by 25-75 confidence interval. Data of current density are presented as mean±SEM. Statistical differences were evaluated by one-way ANOVA followed by post hoc Tukey’s Multiple Comparison Test if applicable (passive properties and current densities), or with Fisher exact test (number of cells variably expressing Na+ and K+ currents). P < 0.05 was considered significantly different. Drugs: the sodium channel blocker Tetrodotoxin (TTX) was purchased from Tocris Bioscience (Bio-Techne, USA), while all the other chemicals, unless otherwise stated, were obtained from Carlo Erba Reagents (Italy) or Sigma-Aldrich (Merck Life Science, Germany).
Intracellular calcium imaging
To test spontaneous calcium activity, organoids at 7 (n=4), 14 (n=3), 21 (n=3), 32 (n=3) days of maturation were assessed with Fluo-4 Direct Calcium Assay Kit (Invitrogen). WNT3a-treated organoids at different time points (7d: n=2; 14d: n=3; 21d: n=3; 32d: n=3) were likewise used. For calcium dye loading, the organoids were first incubated 10 minutes with HEPES pH 7.3 (Sigma, H3375-250G), rinsed and then with equal parts of physiological salt solution (PSS) pH 7.4 (140 mM NaCl, 5 mM KCl, 1.2 mM Na2PO4, 1.4 mM MgCl2, 1.8 mM CaCl2, 11.5 mM glucose, 10 mM HEPES) and Fluo-4 acetoxymethyl ester solutions for 3 h at 37°C. Excess dye was removed by washing with culture medium, and a 30-min incubation was done at 37°C in order to allow the complete de-esterification of the dye and avoid artifacts such as continuous increase in fluorescence intensity during the experiment. Imaging was carried out at 37°C and 5% CO2 using epifluorescence microscopy optics (Eclipse Ti Nikon Microscope), while the organoids were placed in glass bottom 96-well cell imaging plate (Eppendorf #0030741030) maintained in the appropriate culture medium (see media composition of organoid generation protocol section).
Time-lapse imaging was performed at 1 frame/100 ms for 3 min and then processed using ImageJ software. For calcium imaging analysis the intensity signal of the calcium indicator of different cells of interest (n=10/organoid) within the organoids over the entire time of acquisition was measured using the “Plot Z-axis profile” plugin of ImageJ. The fluorescence change over time was defined as ΔF/F=(F-Fbasal)/Fbasal, where F is the fluorescence of a ROI at a specific time point and Fbasal is the corresponding fluorescence of a background area. Cells were considered active when at least a change in the fluorescence intensity (peak) was detected. For each ROI corresponding to a selected cell within an organoid, the number of intensity fluorescence peaks were counted and expressed as mean of peaks per minute.
Rabies virus and adenovirus
To assess the synaptic connections between neuronal cells, organoids at 18 d (n=6) were transfected with adenoviral vector pAAV-Syn-H2B-GFP-TVA-oG-WPRE3 (MOI=50) and kept at 37°C in a humidified atmosphere of 5% CO2 in air, on an orbital shaker for the first 30 min, to promote the transfection and then in static culture for about 20 h. Fresh medium was supplied the day after transfection and the AVV cellular transfection was evaluated with a fluorescence microscope (Nikon Ti Eclipse). 48h later, BRVenVA-1 G-deleted Rabies mcherry (Osakada et al., 2011) virus (MOI=0.25) was added to the culture media. The green, red, and yellow spots were evaluated in the following days by confocal microscope (LSM510 confocal microscope).
Microelectrode array recording
To favor adhesion of the organoid to the microelectrode array (MEA), the array electrodes were first cleaned enzymatically and autoclaved, then made hydrophilic by overnight incubation with sodium phosphate buffer and subsequent overnight coating with a 50 μl drop poly-D-lysine (PDL, 50 mg/ml in sodium phosphate buffer), both at 37°C. MEAs were then thoroughly washed with MilliQ water to remove excess coating, filled with MilliQ water, sealed and stored at 4°C until use.
On the day of recording, individual intact organoids were transferred in a 50 μl drop of culture medium to the previously dried MEA. A custom hold-down anchor was then placed on top of the organoid to ensure its mechanical stability. The MEA chamber was subsequently filled with warm (∼37°C) artificial cerebrospinal fluid (ACSF) and sealed with a custom replica-moulded polydimethylsiloxane lid (Blau et al., 2009) to ensure sterility and maintenance of a physiological microenvironment throughout recordings. The ACSF was composed of (mM) NaCl 115, KCl 2, KH2PO4, 1.25, MgSO4 1, CaCl2 2, D-glucose 25, NaHCO3 26, L-Ascorbic Acid 1, equilibrated at pH = 7.4 with carbogen gas mixture (95% O2 / 5% CO2), and maintained at ∼37°C with the use of a custom-made mini-incubator placed on top the MEA head stage.
Extracellular field potentials were acquired using the Multichannel Experimenter software through an 8 x 8 planar MEA (TiN electrodes, diameter 30 mm, inter-electrode distance 200 mm, impedance < 100 kΩ) connected to a MEA2100-Mini60 head stage (all from Multichannel Systems – Reutlingen, Germany). Signals were low pass filtered at 2-10 kHz before digitization at 10 kHz, and stored in the computer hard drive for offline analysis.
Quantitative RT-PCR analysis for SGZ-NSCs characterization and hippocampal phenotype
Total RNA was extracted from 4 different SGZ-NSCs samples (500000 cells/sample) and from 6 different mature (32d) organoids sample (3 CTRL organoids: n=3 pools of 6–8 organoids each, 3 WNT3a-treated organoids: n=3 pools of 6–8 organoids each) using the RNeasy Plus Micro Kit (Qiagen, Cat No. 74034) according to the manufacturer's protocol and RNA abundance was evaluated using the NanoDrop™ One/OneC Microvolume UV-Vis Spectrophotometer (ThermoFisher Scientific). Reverse transcription was carried out using Superscript VILO Master Mix (Invitrogen, Thermo-Fisher Scientific, #11755050, Waltham, USA). Expression level of specific hippocampal or embryonic and neural stem cells genes (see Table S7) was quantified by Sybr Green-based real-time quantitative RT-PCR (7900HT Real-time PCR System, Applied Biosystems) according to the ΔΔCt method and by using Tbp and Gapdh as reference genes for data normalization.
Quantification and statistical analysis
Unless otherwise stated, n ≥ 3 samples or replicates were used for statistical analysis. Data on graphs are expressed as mean ± SEM. Statistical differences were calculated by two-tailed unpaired t-test for two datasets and ordinary one-way or two-way analysis of variance (ANOVA) test for multiple datasets using Prism (GraphPad Inc., La Jolla, CA). P < 0.05 was considered statistically significant.
∗∗∗∗p < 0.0001; ∗∗∗p < 0.001; ∗∗p < 0.01; ∗p < 0.05; ns not statistically significant.
Acknowledgments
We acknowledge Dr. Edward Callaway, Systems Neurobiology Laboratories, The Salk Institute for Biological Studies, La Jolla, CA 92037, USA, for providing the modified rabies virus particle, Margrate Anyanwu (University of Verona) for help with immunofluorescence staining, Dr. Monica Castellucci, CPT University of Verona, for technical help with RNAseq, Dr. Giacomo Pruzzo, IIT Genoa, for MEA mold design and realization of the custom mini-incubator. “Centro Interdipartimentale di Servizi per la Ricerca che utilizza Animali da Laboratorio”– C.I.R.S.A.L. and “Centro PiattaformeTecnologiche”–CPT (University of Verona) are acknowledged for services and support. European Union project FETPROACT-2018-2020 HERMES [grant number 824164], is acknowledged for the support on research provided to I.D, G.C., G.P. and G.P., J.H.; Fondazione Telethon–Italy (Grant Number GSP20004_PAsMCT8006) to I.D. Italian patient association la Colonna and GALM and University of Verona (Grant Number DDSP-FUR-6616) to I.D. and G.F.; University of Milan (Grant Number BIOMETRA15-6-3003005-1 and PSR2018_RIVA_BIFARI) to F.B; and Fondazione Telethon–Italy (Grant Number GGP19250) to F.B.
Author contributions
F.C., R.G.Z., and A.C. performed experiments, analyzed and interpreted data; E.R. performed electrophysiological experiments, analyzed and interpreted data; G.P., E.B. performed metabolic experiments, analyzed and interpreted data; A.B., M.D.C., S.D. performed immunofluorescence experiments; D.C. performed MEA recording and interpreted data; A.A., J.H. performed OPT microscopy and organoid volume analysis; G. M., G.M. analyzed and interpreted gene expression data; R.B. performed electrophysiological experiments; G.F., F.B., G.P., G.P., G.C. analyzed and interpreted data. F.C., G.C., I.D wrote the paper. All authors discussed results and commented on the manuscript. F.C., R.G.Z., I.D conceptualized the study and I.D. supervised the project and had the scientific direction.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Published: December 17, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103438.
Supplemental information
Data and code availability
-
•
The RNAseq dataset has been deposited at GenBank and is publicly available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA778927. The accession link is listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Abranches E., Silva M., Pradier L., Schulz H., Hummel O., Henrique D., Bekman E. Neural differentiation of embryonic stem cells in vitro: a road map to neurogenesis in the embryo. PLoS One. 2009;4:e6286. doi: 10.1371/journal.pone.0006286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen N.J., Eroglu C. Cell biology of astrocyte-synapse interactions. Neuron. 2017;96:697–708. doi: 10.1016/j.neuron.2017.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen J., Revah O., Miura Y., Thom N., Amin N.D., Kelley K.W., Singh M., Chen X., Thete M.V., Walczak E.M., et al. Generation of functional human 3D cortico-motor assembloids. Cell. 2020;183:1913–1929 e26. doi: 10.1016/j.cell.2020.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagley J.A., Reumann D., Bian S., Levi-Strauss J., Knoblich J.A. Fused cerebral organoids model interactions between brain regions. Nat. Methods. 2017;14:743–751. doi: 10.1038/nmeth.4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckervordersandforth R., Ebert B., Schaffner I., Moss J., Fiebig C., Shin J., Moore D.L., Ghosh L., Trinchero M.F., Stockburger C., et al. Role of mitochondrial metabolism in the control of early lineage progression and aging phenotypes in adult hippocampal neurogenesis. Neuron. 2017;93:1518. doi: 10.1016/j.neuron.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedard P., Gauvin S., Ferland K., Caneparo C., Pellerin E., Chabaud S., Bolduc S. Innovative human three-dimensional tissue-engineered models as an alternative to animal testing. Bioengineering. 2020;7:115. doi: 10.3390/bioengineering7030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belay B., Koivisto J.T., Parraga J., Koskela O., Montonen T., Kellomaki M., Figueiras E., Hyttinen J. Optical projection tomography as a quantitative tool for analysis of cell morphology and density in 3D hydrogels. Sci. Rep. 2021;11:6538. doi: 10.1038/s41598-021-85996-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Ari Y., Spitzer N.C. Nature and nurture in brain development. Trends Neurosci. 2004;27:361. doi: 10.1016/j.tins.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Bifari F., Decimo I., Pino A., Llorens-Bobadilla E., Zhao S., Lange C., Panuccio G., Boeckx B., Thienpont B., Vinckier S., et al. Neurogenic radial glia-like cells in meninges migrate and differentiate into functionally integrated neurons in the neonatal cortex. Cell Stem Cell. 2017;20:360–373.e7. doi: 10.1016/j.stem.2016.10.020. [DOI] [PubMed] [Google Scholar]
- Bifari F., Dolci S., Bottani E., Pino A., Di Chio M., Zorzin S., Ragni M., Zamfir R.G., Brunetti D., Bardelli D., et al. Complete neural stem cell (NSC) neuronal differentiation requires a branched chain amino acids-induced persistent metabolic shift towards energy metabolism. Pharmacol. Res. 2020;158:104863. doi: 10.1016/j.phrs.2020.104863. [DOI] [PubMed] [Google Scholar]
- Birey F., Andersen J., Makinson C.D., Islam S., Wei W., Huber N., Fan H.C., Metzler K.R.C., Panagiotakos G., Thom N., et al. Assembly of functionally integrated human forebrain spheroids. Nature. 2017;545:54–59. doi: 10.1038/nature22330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau A., Neumann T., Ziegler C., Benfenati F. Replica-moulded polydimethylsiloxane culture vessel lids attenuate osmotic drift in long-term cell cultures. J. Biosci. 2009;34:59–69. doi: 10.1007/s12038-009-0009-3. [DOI] [PubMed] [Google Scholar]
- Boisvert M.M., Erikson G.A., Shokhirev M.N., Allen N.J. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 2018;22:269–285. doi: 10.1016/j.celrep.2017.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunetti D., Bottani E., Segala A., Marchet S., Rossi F., Orlando F., Malavolta M., Carruba M.O., Lamperti C., Provinciali M., et al. Targeting multiple mitochondrial processes by a metabolic modulator prevents sarcopenia and cognitive decline in SAMP8 mice. Front. Pharmacol. 2020;11:1171. doi: 10.3389/fphar.2020.01171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budday S., Steinmann P., Kuhl E. Physical biology of human brain development. Front Cell Neurosci. 2015;9:257. doi: 10.3389/fncel.2015.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bugiardini E., Bottani E., Marchet S., Poole O.V., Beninca C., Horga A., Woodward C., Lam A., Hargreaves I., Chalasani A., et al. Expanding the molecular and phenotypic spectrum of truncating MT-ATP6 mutations. Neurol. Genet. 2020;6:e381. doi: 10.1212/NXG.0000000000000381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A., et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci. 2008;28:264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist G.Y., Asciolla J.J., Tchieu J., Walsh R.M., Cornacchia D., Resh M.D., Studer L. Specification of positional identity in forebrain organoids. Nat. Biotechnol. 2019;37:436–444. doi: 10.1038/s41587-019-0085-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen V.S., Morrison J.P., Southwell M.F., Foley J.F., Bolon B., Elmore S.A. Histology atlas of the developing prenatal and postnatal mouse central nervous system, with emphasis on prenatal days E7.5 to E18.5. Toxicol. Pathol. 2017;45:705–744. doi: 10.1177/0192623317728134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clancy B., Finlay B.L., Darlington R.B., Anand K.J. Extrapolating brain development from experimental species to humans. Neurotoxicology. 2007;28:931–937. doi: 10.1016/j.neuro.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline H. Synaptogenesis: a balancing act between excitation and inhibition. Curr. Biol. 2005;15:R203–R205. doi: 10.1016/j.cub.2005.03.010. [DOI] [PubMed] [Google Scholar]
- Decimo I., Dolci S., Panuccio G., Riva M., Fumagalli G., Bifari F. Meninges: a widespread niche of neural progenitors for the brain. Neuroscientist. 2020;27:506–528. doi: 10.1177/1073858420954826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolci S., Pino A., Berton V., Gonzalez P., Braga A., Fumagalli M., Bonfanti E., Malpeli G., Pari F., Zorzin S., et al. High yield of adult oligodendrocyte lineage cells obtained from meningeal biopsy. Front. Pharmacol. 2017;8:703. doi: 10.3389/fphar.2017.00703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzyubenko E., Rozenberg A., Hermann D.M., Faissner A. Colocalization of synapse marker proteins evaluated by STED-microscopy reveals patterns of neuronal synapse distribution in vitro. J. Neurosci. Methods. 2016;273:149–159. doi: 10.1016/j.jneumeth.2016.09.001. [DOI] [PubMed] [Google Scholar]
- Eiraku M., Watanabe K., Matsuo-Takasaki M., Kawada M., Yonemura S., Matsumura M., Wataya T., Nishiyama A., Muguruma K., Sasai Y. Self-organized formation of polarized cortical tissues from ESCs and its active manipulation by extrinsic signals. Cell Stem Cell. 2008;3:519–532. doi: 10.1016/j.stem.2008.09.002. [DOI] [PubMed] [Google Scholar]
- Fair S.R., Julian D., Hartlaub A.M., Pusuluri S.T., Malik G., Summerfied T.L., Zhao G., Hester A.B., Ackerman W.E.T., Hollingsworth E.W., et al. Electrophysiological maturation of cerebral organoids correlates with dynamic morphological and cellular development. Stem Cell Rep. 2020;15:855–868. doi: 10.1016/j.stemcr.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiras E., Soto A.M., Jesus D., Lehti M., Koivisto J., Parraga J.E., Silva-Correia J., Oliveira J.M., Reis R.L., Kellomaki M., Hyttinen J. Optical projection tomography as a tool for 3D imaging of hydrogels. Biomed. Opt. Express. 2014;5:3443–3449. doi: 10.1364/BOE.5.003443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formaggio E., Fazzini F., Dalfini A.C., Di Chio M., Cantu C., Decimo I., Fiorini Z., Fumagalli G., Chiamulera C. Nicotine increases the expression of neurotrophin receptor tyrosine kinase receptor A in basal forebrain cholinergic neurons. Neuroscience. 2010;166:580–589. doi: 10.1016/j.neuroscience.2009.12.073. [DOI] [PubMed] [Google Scholar]
- Ganguly K., Schinder A.F., Wong S.T., Poo M. GABA itself promotes the developmental switch of neuronal GABAergic responses from excitation to inhibition. Cell. 2001;105:521–532. doi: 10.1016/s0092-8674(01)00341-5. [DOI] [PubMed] [Google Scholar]
- Ghoochani A., Shabani K., Peymani M., Ghaedi K., Karamali F., Karbalaei K., Tanhaie S., Salamian A., Esmaeili A., Valian-Borujeni S., et al. The influence of peroxisome proliferator-activated receptor gamma(1) during differentiation of mouse embryonic stem cells to neural cells. Differentiation. 2012;83:60–67. doi: 10.1016/j.diff.2011.08.009. [DOI] [PubMed] [Google Scholar]
- Giandomenico S.L., Mierau S.B., Gibbons G.M., Wenger L.M.D., Masullo L., Sit T., Sutcliffe M., Boulanger J., Tripodi M., Derivery E., et al. Cerebral organoids at the air-liquid interface generate diverse nerve tracts with functional output. Nat. Neurosci. 2019;22:669–679. doi: 10.1038/s41593-019-0350-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayamizu T.F., Mangan M., Corradi J.P., Kadin J.A., Ringwald M. The Adult Mouse Anatomical Dictionary: a tool for annotating and integrating data. Genome Biol. 2005;6:R29. doi: 10.1186/gb-2005-6-3-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inak G., Rybak-Wolf A., Lisowski P., Pentimalli T.M., Juttner R., Glazar P., Uppal K., Bottani E., Brunetti D., Secker C., et al. Defective metabolic programming impairs early neuronal morphogenesis in neural cultures and an organoid model of Leigh syndrome. Nat. Commun. 2021;12:1929. doi: 10.1038/s41467-021-22117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., Pang P.T., Feng L., Lu B. Cyclic AMP controls BDNF-induced TrkB phosphorylation and dendritic spine formation in mature hippocampal neurons. Nat. Neurosci. 2005;8:164–172. doi: 10.1038/nn1381. [DOI] [PubMed] [Google Scholar]
- Kadoshima T., Sakaguchi H., Nakano T., Soen M., Ando S., Eiraku M., Sasai Y. Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proc. Natl. Acad. Sci. U S A. 2013;110:20284–20289. doi: 10.1073/pnas.1315710110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khazipov R., Luhmann H.J. Early patterns of electrical activity in the developing cerebral cortex of humans and rodents. Trends Neurosci. 2006;29:414–418. doi: 10.1016/j.tins.2006.05.007. [DOI] [PubMed] [Google Scholar]
- Kuegler P.B., Zimmer B., Waldmann T., Baudis B., Ilmjarv S., Hescheler J., Gaughwin P., Brundin P., Mundy W., Bal-Price A.K., et al. Markers of murine embryonic and neural stem cells, neurons and astrocytes: reference points for developmental neurotoxicity testing. ALTEX. 2010;27:17–42. doi: 10.14573/altex.2010.1.16. [DOI] [PubMed] [Google Scholar]
- Lancaster M.A., Knoblich J.A. Generation of cerebral organoids from human pluripotent stem cells. Nat. Protoc. 2014;9:2329–2340. doi: 10.1038/nprot.2014.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster M.A., Renner M., Martin C.A., Wenzel D., Bicknell L.S., Hurles M.E., Homfray T., Penninger J.M., Jackson A.P., Knoblich J.A. Cerebral organoids model human brain development and microcephaly. Nature. 2013;501:373–379. doi: 10.1038/nature12517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C., Turrero Garcia M., Decimo I., Bifari F., Eelen G., Quaegebeur A., Boon R., Zhao H., Boeckx B., Chang J., et al. Relief of hypoxia by angiogenesis promotes neural stem cell differentiation by targeting glycolysis. Embo J. 2016;35:924–941. doi: 10.15252/embj.201592372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C.T., Bendriem R.M., Wu W.W., Shen R.F. 3D brain Organoids derived from pluripotent stem cells: promising experimental models for brain development and neurodegenerative disorders. J. Biomed. Sci. 2017;24:59. doi: 10.1186/s12929-017-0362-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lein E.S., Zhao X., Gage F.H. Defining a molecular atlas of the hippocampus using DNA microarrays and high-throughput in situ hybridization. J. Neurosci. 2004;24:3879–3889. doi: 10.1523/JNEUROSCI.4710-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson J.N., El-Husseini A. Building excitatory and inhibitory synapses: balancing neuroligin partnerships. Neuron. 2005;48:171–174. doi: 10.1016/j.neuron.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Lie D.C., Colamarino S.A., Song H.J., Desire L., Mira H., Consiglio A., Lein E.S., Jessberger S., Lansford H., Dearie A.R., Gage F.H. Wnt signalling regulates adult hippocampal neurogenesis. Nature. 2005;437:1370–1375. doi: 10.1038/nature04108. [DOI] [PubMed] [Google Scholar]
- Lorenz C., Lesimple P., Bukowiecki R., Zink A., Inak G., Mlody B., Singh M., Semtner M., Mah N., Aure K., et al. Human iPSC-derived neural progenitors are an effective drug discovery model for neurological mtDNA disorders. Cell Stem Cell. 2017;20:659–674 e9. doi: 10.1016/j.stem.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Mariani J., Simonini M.V., Palejev D., Tomasini L., Coppola G., Szekely A.M., Horvath T.L., Vaccarino F.M. Modeling human cortical development in vitro using induced pluripotent stem cells. Proc. Natl. Acad. Sci. U S A. 2012;109:12770–12775. doi: 10.1073/pnas.1202944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall J.J., Mason J.O. Mouse vs man: organoid models of brain development & disease. Brain Res. 2019;1724:146427. doi: 10.1016/j.brainres.2019.146427. [DOI] [PubMed] [Google Scholar]
- Martano G., Borroni E.M., Lopci E., Cattaneo M.G., Mattioli M., Bachi A., Decimo I., Bifari F. Metabolism of stem and progenitor cells: proper methods to answer specific questions. Front Mol. Neurosci. 2019;12:151. doi: 10.3389/fnmol.2019.00151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mason J.O., Price D.J. Building brains in a dish: prospects for growing cerebral organoids from stem cells. Neuroscience. 2016;334:105–118. doi: 10.1016/j.neuroscience.2016.07.048. [DOI] [PubMed] [Google Scholar]
- Molnar Z., Clowry G. Cerebral cortical development in rodents and primates. Prog. Brain Res. 2012;195:45–70. doi: 10.1016/B978-0-444-53860-4.00003-9. [DOI] [PubMed] [Google Scholar]
- Monzel A.S., Smits L.M., Hemmer K., Hachi S., Moreno E.L., Van Wuellen T., JARAZO J., Walter J., Bruggemann I., Boussaad I., et al. Derivation of human midbrain-specific organoids from neuroepithelial stem cells. Stem Cell Rep. 2017;8:1144–1154. doi: 10.1016/j.stemcr.2017.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muguruma K., Nishiyama A., Kawakami H., Hashimoto K., Sasai Y. Self-organization of polarized cerebellar tissue in 3D culture of human pluripotent stem cells. Cell Rep. 2015;10:537–550. doi: 10.1016/j.celrep.2014.12.051. [DOI] [PubMed] [Google Scholar]
- Nasu M., Takata N., Danjo T., Sakaguchi H., Kadoshima T., Futaki S., Sekiguchi K., Eiraku M., Sasai Y. Robust formation and maintenance of continuous stratified cortical neuroepithelium by laminin-containing matrix in mouse ES cell culture. PLoS One. 2012;7:e53024. doi: 10.1371/journal.pone.0053024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neniskyte U., Gross C.T. Errant gardeners: glial-cell-dependent synaptic pruning and neurodevelopmental disorders. Nat. Rev. Neurosci. 2017;18:658–670. doi: 10.1038/nrn.2017.110. [DOI] [PubMed] [Google Scholar]
- Osakada F., Mori T., Cetin A.H., Marshel J.H., Virgen B., Callaway E.M. New rabies virus variants for monitoring and manipulating activity and gene expression in defined neural circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacitti D., Privolizzi R., Bax B.E. Organs to cells and cells to organoids: the evolution of in vitro central nervous system modelling. Front.Cell. Neurosci. 2019;13:129. doi: 10.3389/fncel.2019.00129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmqvist L., Glover C.H., Hsu L., Lu M., Bossen B., Piret J.M., Humphries R.K., Helgason C.D. Correlation of murine embryonic stem cell gene expression profiles with functional measures of pluripotency. Stem Cells. 2005;23:663–680. doi: 10.1634/stemcells.2004-0157. [DOI] [PubMed] [Google Scholar]
- Park K.S., Lee R.D., Kang S.K., Han S.Y., Park K.L., Yang K.H., Song Y.S., Park H.J., Lee Y.M., Yun Y.P., et al. Neuronal differentiation of embryonic midbrain cells by upregulation of peroxisome proliferator-activated receptor-gamma via the JNK-dependent pathway. Exp.Cell Res. 2004;297:424–433. doi: 10.1016/j.yexcr.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Park Y., Franz C.K., Ryu H., Luan H., Cotton K.Y., Kim J.U., Chung T.S., Zhao S., Vazquez-Guardado A., Yang D.S., et al. Three-dimensional, multifunctional neural interfaces for cortical spheroids and engineered assembloids. Sci. Adv. 2021;7:eabf9153. doi: 10.1126/sciadv.abf9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasca A.M., Sloan S.A., Clarke L.E., Tian Y., Makinson C.D., Huber N., Kim C.H., Park J.Y., O'rourke N.A., Nguyen K.D., et al. Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nat. Methods. 2015;12:671–678. doi: 10.1038/nmeth.3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrini L., Albecka A., Mallery D.L., Kellner M.J., Paul D., Carter A.P., James L.C., Lancaster M.A. SARS-CoV-2 infects the brain choroid plexus and disrupts the blood-CSF barrier in human brain organoids. Cell Stem Cell. 2020;27:951–961 e5. doi: 10.1016/j.stem.2020.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pino A., Fumagalli G., Bifari F., Decimo I. New neurons in adult brain: distribution, molecular mechanisms and therapies. Biochem. Pharmacol. 2017;141:4–22. doi: 10.1016/j.bcp.2017.07.003. [DOI] [PubMed] [Google Scholar]
- Poirier Y., Antonenkov V.D., Glumoff T., Hiltunen J.K. Peroxisomal beta-oxidation--a metabolic pathway with multiple functions. Biochim. Biophys. Acta. 2006;1763:1413–1426. doi: 10.1016/j.bbamcr.2006.08.034. [DOI] [PubMed] [Google Scholar]
- Qian X., Nguyen H.N., Song M.M., Hadiono C., Ogden S.C., Hammack C., Yao B., Hamersky G.R., Jacob F., Zhong C., et al. Brain-region-specific organoids using mini-bioreactors for modeling ZIKV exposure. Cell. 2016;165:1238–1254. doi: 10.1016/j.cell.2016.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Jacob F., Song M.M., Nguyen H.N., Song H., Ming G.L. Generation of human brain region-specific organoids using a miniaturized spinning bioreactor. Nat. Protoc. 2018;13:565–580. doi: 10.1038/nprot.2017.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X., Song H., Ming G.L. Brain organoids: advances, applications and challenges. Development. 2019;146:dev166074. doi: 10.1242/dev.166074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G., Nguyen T., Macosko E.Z., Sherwood J.L., Min Yang S., Berger D.R., Maria N., Scholvin J., Goldman M., Kinney J.P., et al. Cell diversity and network dynamics in photosensitive human brain organoids. Nature. 2017;545:48–53. doi: 10.1038/nature22047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner M., Lancaster M.A., Bian S., Choi H., Ku T., Peer A., Chung K., Knoblich J.A. Self-organized developmental patterning and differentiation in cerebral organoids. EMBO J. 2017;36:1316–1329. doi: 10.15252/embj.201694700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiros P.M., Goyal A., Jha P., Auwerx J. Analysis of mtDNA/nDNA ratio in mice. Curr Protoc Mouse Biol. 2017;7:47–54. doi: 10.1002/cpmo.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner H., Grabos M., Becker K.J., Kagermeier T.E., Wu J., Otto M., Peischard S., Zeuschner D., Tsytsyura Y., Disse P., et al. A fully automated high-throughput workflow for 3D-based chemical screening in human midbrain organoids. Elife. 2020;9:e52904. doi: 10.7554/eLife.52904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds B.A., Weiss S. Generation of neurons and astrocytes from isolated cells of the adult mammalian central nervous system. Science. 1992;255:1707–1710. doi: 10.1126/science.1553558. [DOI] [PubMed] [Google Scholar]
- Rivera C., Voipio J., Kaila K. Two developmental switches in GABAergic signalling: the K+-Cl- cotransporter KCC2 and carbonic anhydrase CAVII. J. Physiol. 2005;562:27–36. doi: 10.1113/jphysiol.2004.077495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S.S., Spitzer N.C. Calcium signaling in neuronal development. Cold Spring Harb. Perspect. Biol. 2011;3:a004259. doi: 10.1101/cshperspect.a004259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safronov B.V. Spatial distribution of NA+ and K+ channels in spinal dorsal horn neurones: role of the soma, axon and dendrites in spike generation. Prog. Neurobiol. 1999;59:217–241. doi: 10.1016/s0301-0082(98)00051-3. [DOI] [PubMed] [Google Scholar]
- Sakaguchi H., Kadoshima T., Soen M., Narii N., Ishida Y., Ohgushi M., Takahashi J., Eiraku M., Sasai Y. Generation of functional hippocampal neurons from self-organizing human embryonic stem cell-derived dorsomedial telencephalic tissue. Nat. Commun. 2015;6:8896. doi: 10.1038/ncomms9896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Mei A., Paquola A.C.M., Stern S., Bardy C., Klug J.R., Kim S., Neshat N., Kim H.J., Ku M., et al. Efficient generation of CA3 neurons from human pluripotent stem cells enables modeling of hippocampal connectivity in vitro. Cell Stem Cell. 2018;22:684–697 e9. doi: 10.1016/j.stem.2018.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple B.D., Blomgren K., Gimlin K., Ferriero D.M., Noble-Haeusslein L.J. Brain development in rodents and humans: identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 2013;106-107:1–16. doi: 10.1016/j.pneurobio.2013.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto Y., Eiraku M. Human brain development and its in vitro recapitulation. Neurosci. Res. 2019;138:33–42. doi: 10.1016/j.neures.2018.09.011. [DOI] [PubMed] [Google Scholar]
- Silva A., Pereira J., Oliveira C.R., Relvas J.B., Rego A.C. BDNF and extracellular matrix regulate differentiation of mice neurosphere-derived cells into a GABAergic neuronal phenotype. J. Neurosci. Res. 2009;87:1986–1996. doi: 10.1002/jnr.22041. [DOI] [PubMed] [Google Scholar]
- Singec I., Knoth R., Meyer R.P., Maciaczyk J., Volk B., Nikkhah G., Frotscher M., Snyder E.Y. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat. Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- Soares R., Ribeiro F.F., Lourenco D.M., Rodrigues R.S., Moreira J.B., Sebastiao A.M., Morais V.A., Xapelli S. The neurosphere assay: an effective in vitro technique to study neural stem cells. Neural Regen. Res. 2021;16:2229–2231. doi: 10.4103/1673-5374.310678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son H., Kim S., Jung D.H., Baek J.H., Lee D.H., Roh G.S., Kang S.S., Cho G.J., Choi W.S., Lee D.K., Kim H.J. Insufficient glutamine synthetase activity during synaptogenesis causes spatial memory impairment in adult mice. Sci. Rep. 2019;9:252. doi: 10.1038/s41598-018-36619-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L., Yuan X., Jones Z., Vied C., Miao Y., Marzano M., Hua T., Sang Q.A., Guan J., Ma T., et al. Functionalization of brain region-specific spheroids with isogenic microglia-like cells. Sci. Rep. 2019;9:11055. doi: 10.1038/s41598-019-47444-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo C.A., Gao R., Negraes P.D., Gu J., Buchanan J., Preissl S., Wang A., Wu W., Haddad G.G., Chaim I.A., et al. Complex oscillatory waves emerging from cortical organoids model early human brain network development. Cell Stem Cell. 2019;25:558–569 e7. doi: 10.1016/j.stem.2019.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini G. Rabies virus as a transneuronal tracer of neuronal connections. Adv.Virus Res. 2011;79:165–202. doi: 10.1016/B978-0-12-387040-7.00010-X. [DOI] [PubMed] [Google Scholar]
- Velasco S., Kedaigle A.J., Simmons S.K., Nash A., Rocha M., Quadrato G., Paulsen B., Nguyen L., Adiconis X., Regev A., et al. Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature. 2019;570:523–527. doi: 10.1038/s41586-019-1289-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe K., Kamiya D., Nishiyama A., Katayama T., Nozaki S., Kawasaki H., Watanabe Y., Mizuseki K., Sasai Y. Directed differentiation of telencephalic precursors from embryonic stem cells. Nat. Neurosci. 2005;8:288–296. doi: 10.1038/nn1402. [DOI] [PubMed] [Google Scholar]
- Wheeler D.W., White C.M., Rees C.L., Komendantov A.O., Hamilton D.J., Ascoli G.A. Hippocampome.org: a knowledge base of neuron types in the rodent hippocampus. Elife. 2015;4:e09960. doi: 10.7554/eLife.09960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham I.R., Finke S., Conzelmann K.K., Callaway E.M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods. 2007;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winanto, Khong Z.J., Hor J.H., Ng S.Y. Spinal cord organoids add an extra dimension to traditional motor neuron cultures. Neural Regen. Res. 2019;14:1515–1516. doi: 10.4103/1673-5374.255966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y.F., Tanaka Y., Cakir B., Patterson B., Kim K.Y., Sun P.N., Kang Y.J., Zhong M., Liu X.R., Patra P., et al. hESC-derived thalamic organoids form reciprocal projections when fused with cortical organoids. Cell Stem Cell. 2019;24:487. doi: 10.1016/j.stem.2018.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue H., Li J., Xie H., Wang Y. Review of drug repositioning approaches and resources. Int. J. Biol. Sci. 2018;14:1232–1244. doi: 10.7150/ijbs.24612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao H., Wu W., Cerf I., Zhao H.W., Wang J., Negraes P.D., Muotri A.R., Haddad G.G. Methadone interrupts neural growth and function in human cortical organoids. Stem Cell Res. 2020;49:102065. doi: 10.1016/j.scr.2020.102065. [DOI] [PubMed] [Google Scholar]
- Yoon S.J., Elahi L.S., Pasca A.M., Marton R.M., Gordon A., Revah O., Miura Y., Walczak E.M., Holdgate G.M., FAN H.C., et al. Reliability of human cortical organoid generation. Nat. Methods. 2019;16:75–78. doi: 10.1038/s41592-018-0255-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafeiriou M.P., Bao G., Hudson J., Halder R., Blenkle A., Schreiber M.K., Fischer A., Schild D., Zimmermann W.H. Developmental GABA polarity switch and neuronal plasticity in bioengineered neuronal organoids. Nat. Commun. 2020;11:3791. doi: 10.1038/s41467-020-17521-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Chen K., Sloan S.A., Bennett M.L., Scholze A.R., O'keeffe S., Phatnani H.P., Guarnieri P., Caneda C., Ruderisch N., et al. An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 2014;34:11929–11947. doi: 10.1523/JNEUROSCI.1860-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng X., Boyer L., Jin M., Mertens J., Kim Y., Ma L., Ma L., Hamm M., Gage F.H., Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Neurons MAP2+, astrocytes GFAP+ and total nuclei (TOPRO-3) are shown in white, red and blue, respectively.
Neurons MAP2+ and total nuclei (TOPRO-3) are shown in green and blue, respectively.
The video is reported in grayscale and 10X speed.
The video is reported in grayscale and 10X speed.
Data Availability Statement
-
•
The RNAseq dataset has been deposited at GenBank and is publicly available at https://www.ncbi.nlm.nih.gov/bioproject/PRJNA778927. The accession link is listed in the key resources table. All other data reported in this paper will be shared by the lead contact upon request.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.