Abstract
A tool was developed to automatically segment several subcortical limbic structures (nucleus accumbens, basal forebrain, septal nuclei, hypothalamus without mammillary bodies, the mammillary bodies, and fornix) using only a T1-weighted MRI as input. This tool fills an unmet need as there are few, if any, publicly available tools to segment these clinically relevant structures. A U-Net with spatial, intensity, contrast, and noise augmentation was trained using 39 manually labeled MRI data sets. In general, the Dice scores, true positive rates, false discovery rates, and manual-automatic volume correlation were very good relative to comparable tools for other structures. A diverse data set of 698 subjects were segmented using the tool; evaluation of the resulting labelings showed that the tool failed in less than 1% of cases. Test-retest reliability of the tool was excellent. The automatically segmented volume of all structures except mammillary bodies showed effectiveness at detecting either clinical AD effects, age effects, or both. This tool will be publicly released with FreeSurfer (surfer.nmr.mgh.harvard.edu/fswiki/ScLimbic). Together with the other cortical and subcortical limbic segmentations, this tool will allow FreeSurfer to provide a comprehensive view of the limbic system in an automated way.
Graphical Abstract
1. Introduction
The limbic system is a set of brain structures that govern the interplay between subcortical regions and association cortices. The limbic system was originally defined by Maclean (MacLean, 1949), but its composition has evolved and been debated (Kotter and Stephan, 1997; LeDoux, 2012). In cortex, the limbic lobe includes the olfactory cortex (paleocortex), hippocampus (allocortex), caudal orbitofrontal, medial frontal, temporopolar, anteroventral insular, cingulate, retrosplenial, and parahippocampal gyri. Subcortically, limbic structures include, but are not limited to, hypothalamus (including the mammillary bodies), amygdala, the extended amygdala, nucleus accumbens, ventral pallidum, association thalamic nuclei, basal forebrain, septal nuclei, cerebellum, fornix, and the reticular formation of the brainstem. The limbic system supports a wide variety of functions and behaviors, including autonomic regulation (heart rate, blood pressure, hunger thirst, sexual arousal circadian rhythm), cognitive/attentional/emotional processing, spatial memory, long term memory, fear, emotional memory, anxiety, aggression, reward, and addiction (Heimer and Van Hoesen, 2006; L. Heimer et al., 2008; Mesulam, 1985). Thus, understanding the role of the limbic system in health and disease is clinically relevant and significant.
Brain imaging (e.g., MRI and PET) can be used to enhance this understanding. However, scientists and clinicians who use neuroimaging often do not have the anatomical expertise to properly locate these anatomical structures. Further, it is a tedious, error prone, and time-consuming task to manually label structures in a whole brain image, especially in a large data set with many subjects. Accordingly, imaging scientists have developed methods that will automatically label structures of interest (Despotovic et al., 2015). Typically, this starts with an expert manually labeling a set of images; a tool is then trained using the expert labels as input; this tool is then applied to a novel image to automatically predict how an expert would have labeled the image. Performance of such methods vary depending upon the structure, method, and quality of the training and test images.
Many tools to automatically label the brain have already been developed using parametric methods, machine learning techniques, or by simply deforming label atlases to an individual via a nonlinear registration. Cortically, Fischl et al., 2004, developed a method to automatically segment cortical regions, including two limbic areas (cingulate and parahippocampal gyri), using a surface-based Bayesian method. With respect to the subcortical limbic system, several groups have created tools that include hippocampus, amygdala, thalamus, and nucleus accumbens (Billot et al., 2020b; Fischl et al., 2002; Henschel et al., 2020; Iglesias et al., 2015; Jog et al., 2019; Patenaude et al., 2011; Puonti et al., 2016; Saygin et al., 2017; Wenzel et al., 2018).
For hypothalamus, Rodrigues et al., 2020 used a U-Net (Ronnenberger et al., 2015) to segment whole hypothalamus, and Billot et al., 2020a implemented a U-Net to segment the hypothalamic subunits. Even less has been done for fornix, septal nucleus, and basal forebrain. Butler et al., 2014 and Butler et al., 2012, created a (fixed, i.e., non-probabilistic) septal nuclei label in MNI space which is then simply mapped to an individual’s brain image after non-linear registration. Teipel et al., 2014 and Cavedo et al., 2017 used a similar method to segment basal forebrain. Jin et al., 2015 developed a segmentation tool for fornix, but it requires a diffusion MRI volume.
In this manuscript, we develop, test, and validate an easy-to-use tool to automatically segment several subcortical limbic structures from T1-weighted anatomical MRIs. These structures include hypothalamus1 (HTh), mammillary bodies (MB), basal forebrain (BF), septal nuclei (SepN), fornix (Fx), and nucleus accumbens (NA) (see Fig. 1). Despite the clinical significance of the limbic system, several of these structures (MB, BF, SepN, and Fx) have few, if any, publicly available automatic segmentation tools. Unique aspects of this tool include: MB segmentation (to our knowledge, there are no other tools to segment MB), Fx segmentation from T1-weighted images, probabilistic segmentation of BF and SepN, the combination of limbic regions, ease-to-use, self-contained (but easy integration with FreeSurfer), and extensive testing. We show the clinical utility of the tool by applying it to independent aging and Alzheimer’s disease (AD) data sets. The robustness of the tool was tested on a diverse set of 698 independent images from various scanners. This tool is publicly available with FreeSurfer (surfer.nmr.mgh.harvard.edu/fswiki/ScLimbic); these segmentations can be combined with other segmentations in FreeSurfer to provide a more complete representation of the limbic system using automated methods.
Fig. 1.
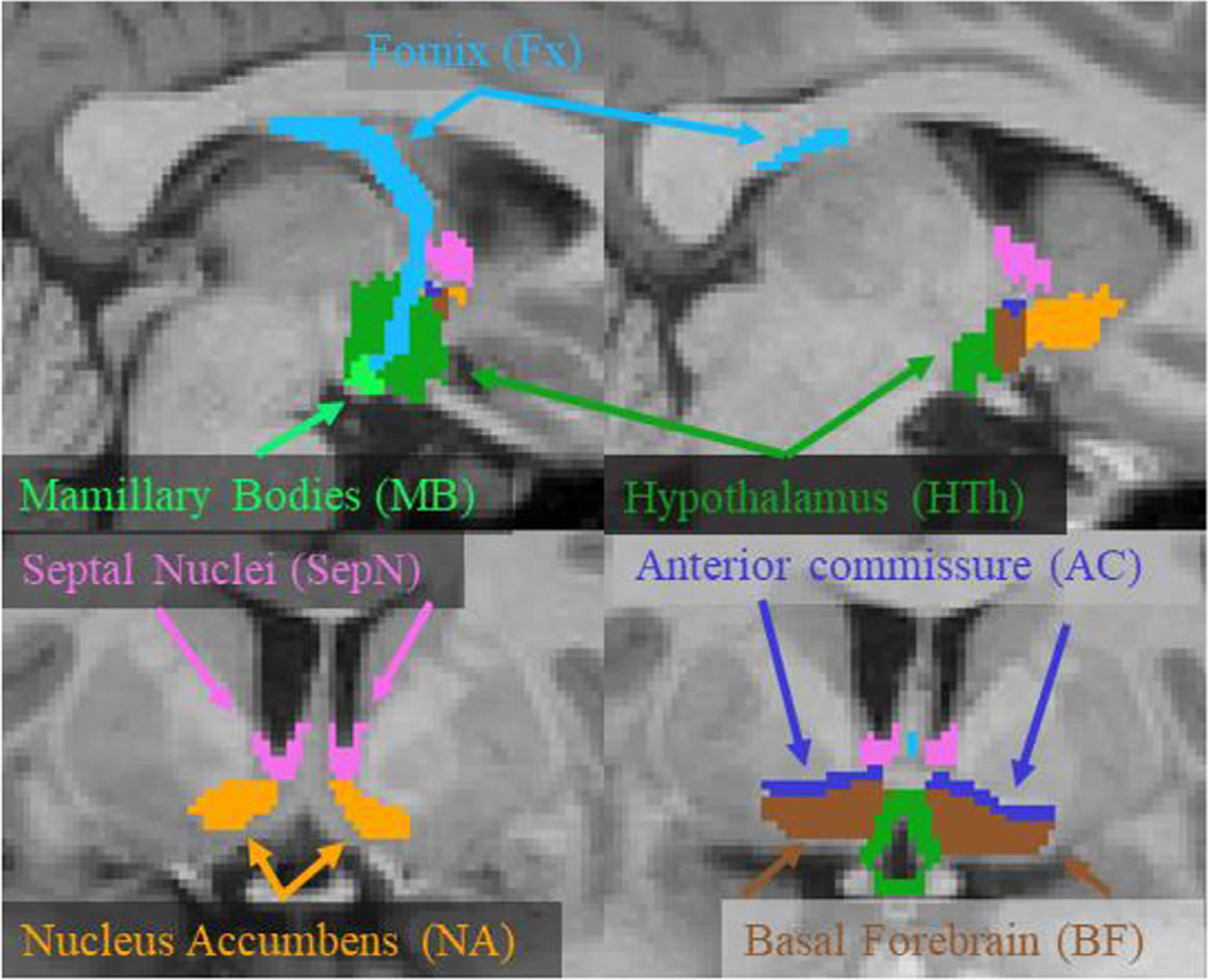
Example manual segmentations of the labels used in this study. The hypothalamus label excludes mamillary bodies, which were included as a separate label. The anterior commissure (AC) was labeled only to provide a reference for manually labeling the other structures. The upper images are sagittal slices; the bottom images are coronal slices.
2. Methods
2.1. Data sets
Several data sets were used for manual labeling and network training and validation as well as for testing robustness and clinical validation. In all cases, images were resampled into a 2563 1mm3 vol, and the intensities rescaled into an 8-bit range. These operations, known as “conforming”, are the first step in the FreeSurfer pipeline. Inputs to the tool must be 1mm3 but do not need to be 2563 or 8-bit (the tool can reslice to 1mm3 if needed).
2.2. FreeSurfer maintenance (FSM)
MRI images were acquired on 29 subjects (M = 15, F = 14), mean age 44.8 years (+/− 18.5, min = 19 max = 76). This study was approved by the Massachusetts General Hospital Internal Review Board for the protection of human subjects; all subjects gave written informed consent. Scanning was performed on a Siemens 3T Prisma with a 32-channel head coil. Two acquisitions were used for this study: a multiecho MPRAGE sequence (van der Kouwe et al., 2008) and a single echo MP2RAGE sequence (Marques et al., 2010). MPRAGE parameters were 1 mm isotropic voxel size, 256 × 256 × 176, inversion time 1250 ms, TR 2530 ms, readout flip angle 7°, time between readout pulses 9.8 ms, GRAPPA acceleration factor 2, bandwidth 650 Hz/Px, four echoes (1.69, 3.55, 5.41, and 7.27 ms). The four echoes were combined by computing the root-mean-square (RMS) of the four images yielding a single T1-weighted (T1w) volume. MP2RAGE parameters were 1 mm isotropic, 256 × 256 × 176, 1st inversion time 700 ms, 2nd inversion time 2500 ms, TR 5000 ms, readout flip angle for 1st inversion 4°, readout flip angle for 2nd inversion 5°, time between readout pulses 7.1 ms, TE 2.98 ms, GRAPPA acceleration factor 3, bandwidth 240 Hz/Px. The MP2RAGE sequence automatically produces a quantitative T1 (qT1) map.
2.3. Alzheimer’s disease neuroimaging initiative (ADNI, Weiner et al., 2010)
T1w images from 110 ADNI subjects were used. Ten subjects (5 M/5F, mean age 77y) were manually labeled; all these subjects had an AD diagnosis. The remaining 100 subjects were used to evaluate the effect of AD on the volume of the limbic structures (50 healthy controls (HC), 22 M/28F, age mean/std/min/max 75.0/4.8/62/90y; 50 diagnosed with AD 28 M/22F, age mean/std/min/max 74.3/7.2/56/88y); we refer to this set as the ADNI100.
2.4. Harvard aging brain study (HABS, Mormino et al., 2014)
Ninety-nine subjects were drawn from HABS, which had approval from the Massachusetts General Hospital Internal Review Board; all subjects gave written informed consent. Subjects were healthy and aged from 66 to 87 years (mean 73.9y, s.d. 5.8y), 44 males and 55 females. MPRAGEs were acquired on a Siemens 3T Trio. Greve et al., 2016 describes additional scanning parameter details of this cohort.
2.5. Minimal interval resonance imaging in Alzheimer’s disease (MIRIAD, Malone et al., 2013)
In this data set, we analyzed 40 subjects (20 AD, 20 HC; 18 F, 22 M; mean age 68y; GE 1.5T Signa scanner; partially defaced), each with two time points 14 days apart, to evaluate test-retest reliability. This data is publicly available from miriad.drc.ion.ucl.ac.uk.
2.6. Thousand Functional connectomes (FC1k)
We analyzed 499 cases from the FC1k data base (fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html), a public collection of anonymized MRI data. While best known for fMRI, the 1000 Functional Connectomes also has T1-weighted anatomical MRI data from which we analyzed 499 subjects from three sites: Beijing (198 subjects), Cambridge (198 subjects), and Oulu (103 subjects), all 3T scanners. The subjects ranged in age from 18 to 30y; all images were defaced. The voxel size was Beijing: 1.3 × 1 × 1 mm, Cambridge: 1.2 × 1.2 × 1.2 mm, Oulu: .94x.94 × 1 mm. We point out here that the Oulu data set was very noisy based on visual inspection.
2.7. Manual labeling
The left and right sides of six structures (HTh, MB, BF, Fx, SepN, and NA; see Fig. 1) were manually labeled for a total of 12 distinct labels on the 29 FSM subjects and the 10 ADNI subjects. The manual labeling was overseen by an experienced neuroanatomist (JCA). Parts of the anterior commissure (AC) were labeled, but only to provide a reference for manually labeling the other structures. For the FSM, qT1 images were used for manual labeling as they provided the best contrast for the boundaries of interest; T1w images were used for the ADNI subjects. A description of the anatomical labeling protocol is given in Appendix A.
2.8. U-Net architecture and training
We used the network described in Billot et al., 2020a. This is network is a simple 3D variant of the popular U-Net architecture (Ronnenberger et al., 2015). The training software (Neuron (Dalca et al., 2018) and Lab2Im (Billot et al., 2020a) Python packages) is publicly available at https://github.com/BBillot/hypothalamus_seg. Billot et al., 2020a extensively tuned the hyperparameters and showed that this network out-performed state-of-the-art multi-atlas segmentation (Artaechevarria et al., 2009). For this tool, the network architecture, augmentation, and training were identical to that of Billot et al., 2020a with the exception that we include intensity noise augmentation (i.e., the adding of white Gaussian noise to the image during training).
Briefly, the network has three resolution layers. Convolutions are performed with a 3 × 3 × 3 kernel. The first convolution has 24 output feature maps followed by a batch normalization and a max pooling step; the number of features is doubled after each max pooling and halved after each up-convolution. All layers, except the last, use an Exponential Linear Unit (ELU) activation function. The last layer has a softmax activation function. The input is always a T1w MRI. Augmentation consisted of spatial transformations (left-right flipping, affine and nonlinear transforms) and intensity transforms (multiplication by a bias field, noise augmentation, rescaling with min-max normalization, and contrast augmentation with nonlinear gamma (power law) distortion). The network was trained by optimizing the “soft” Dice score between the manual labels and the predicted labels. The first 50 epochs were trained without noise augmentation followed by 50 epochs with noise augmentation. Each epoch consisted of 1000 batches with a batch size of 1. The network easily reached convergence in this time. A batch size of 1 was used due to GPU memory limitations; as pointed out by Billot et al., 2020a, this low batch size is compensated for by using a large number of voxels (1603) to compute the loss function and gradient. The network was trained with the ADAM optimizer (Kingma, 2015). In Experiment 1, the network was trained on a subset of the 39 subjects for cross validation purposes. In the rest of the experiments, the network was trained on all 39 subjects.
2.9. Experiment 1: Cross-validation
The 39 manually labeled subjects were divided into a training group (N = 21, 10 female, 6 AD, 57y mean age) and a testing/validation group (N = 18, 11 female, 4 CE, 52y mean age). The network was trained on the training group and then applied to the (independent) test group. The manual and automatic labels were then compared in terms of Dice, correlation coefficient, true positive rate (TPR), and false discovery rate (FDR); a paired t-test was used to determine whether the manual and automatic volumes systematically differed.
2.10. Experiment 2: Robustness
The performance of machine learning tools is highly dependent on the training set and augmentation; if the tool sees an input that is somewhat different from the augmented training set, it may underlabel or fail to label at all. To evaluate the robustness of this tool, we applied it to 698 data samples from five data sets (ADNI100, HABS, Beijing, Cambridge, and Oulu) that were neither in the training or test sets and represent a variety of scanners and populations. To avoid visually inspecting each case, we used reverse classification accuracy (RCA, Robinson et al., 2019; Valindria et al., 2017) to flag individual data sets for visual scrutiny. In RCA, the test image was nonlinearly registered using ANTs (Avants et al., 2011) to each of the 39 manual labeled subjects, the manual labels were then mapped into the test image space where Dice scores were computed for each subject and label. For a given label, the maximum Dice score across the 39 was used as the quality metric, where 0 was bad and 1 was perfect. The idea here is that if one of the manually labeled subjects is anatomically close to the test subject, and this procedure will produce a reasonably good overlap between the segmentations. For the purposes of non-linear registration, the images were skull stripped and bias field corrected using FreeSurfer (Fischl, 2012); the segmentation was always applied to the raw data. Images that had a quality score of less than 0.5 on any label were flagged for manual inspection by two of the authors (DNG and DC); all labels were evaluated for a case regardless of which label was flagged. The criteria for passing were whether a given label was in about the right place with about the right shape and did not appear to be under- or over-labeled by more than 25%. The quality control reviewers were able to do this in about 2 min per case. These criteria were intentionally vague to avoid labor equivalent to the manual labeling of the flagged cases, which would have taken months or years. While a low RCA score could be an indication of a poor segmentation, it could also be the result of a poor nonlinear registration (and so not a problem with the tool per se).
2.11. Experiment 3: Test-retest reliability
The two time points from the 40 MIRIAD subjects were used to evaluate test-retest reliability. Each subject/time point was segmented using the current method. The correlation coefficient and intraclass correlation (ICC, using the ICC(3,1) from Shrout and Fleiss, 1979) of segmentation volumes across time and subject were then computed as the test-retest measure. We also computed a paired-t to test whether the two time points were systematically different.
2.12. Experiment 4: Alzheimer’s disease and aging effects
To evaluate the effect of AD on the volume of the limbic structures, the network was applied to the ADNI100 data set. The volumes were then compared across diagnosis (AD-vs-HC) using a two-sample t-test. The volumes were corrected for estimated total intracranial volume (eTIV, Buckner et al., 2004) to account for differences in head size. To further assess clinical sensitivity, we performed an analysis quantifying the changes in the volume of these structures with age. While age itself is not a clinical condition, age does impose substantial changes on the brain similar to diseases. The T1w images of the HABS data set were segmented using the present method. The eTIV-corrected volumes of the structures were then regressed against age; the null hypothesis that there was no age effect was evaluated with a t-test.
3. Results
Fig. 2 illustrates an example of the automatic segmentation for each of the structures in an individual subject withheld from the training; this subject was in the middle of the range of Dice scores. Green indicates that a voxel was in both the manual and automatic labels; from the standpoint of the automatic segmentation, these are true positives (TPs). Yellow indicates that the voxel was present in the manual label but not in the automatic segmentation (i.e., a false negative, FN); the full manual label consists of the green and yellow voxels. Red indicates that a voxel was in the automatic segmentation but not in the manual label (i.e., a false positive, FP). The full automatic label consists of green and red voxels.
Fig. 2.
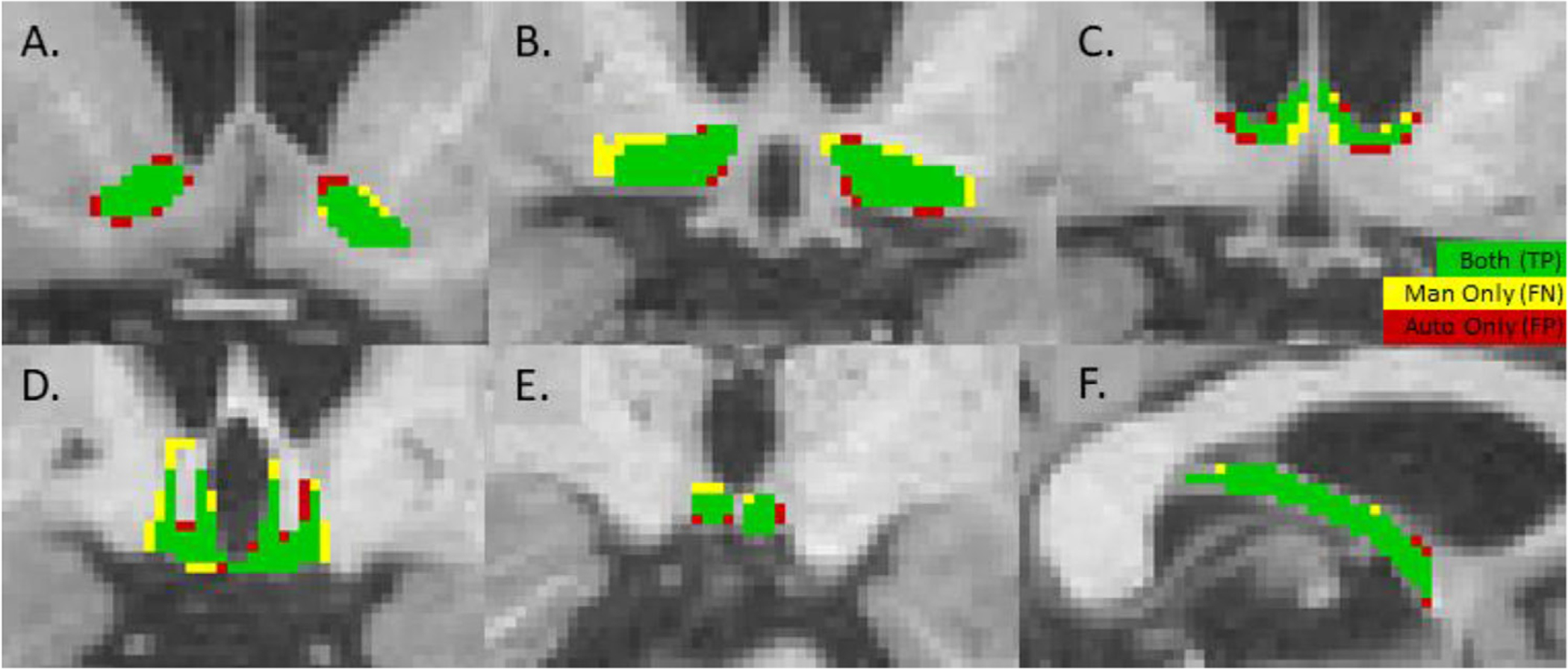
Performance of automatic segmentation on a single test subject as compared to the manual segmentation for each of the structures. Green indicates that the voxel was in both the manual and automatic segmentations (a true positive, TP). Yellow means that the voxel was only in the manual (a false negative, FN). Red means the voxel was only in the automatic (a false positive, FP). The mean Dice score for this subject was 0.78, the middle of the range for the test subjects. (A) NA, (B) BF, (C) SepN, (D) HTh, (E) MB, (F), Left Fx.
3.1. Experiment 1: Cross-validation results
Table 1 shows the cross-validation performance of the automatic segmentation. None of the automatic segmentation volumes were significantly different than that of the manual segmentations (paired t-test) indicating no systematic bias in the volume measurement. Cross-subject volume variation was also comparable. The Dice scores range from 0.69 to 0.82. Aside from MB, the correlation coefficient (CC) between the manual and automatic volumes is in the moderate to high range of 0.62 to 0.88; the MB has a relatively low CC.
Table 1.
Cross-validation performance of the automatic segmentation. Manual Vol is the mean volume of the manual segmentation in mm3; Auto Vol is the mean volume of the automatic segmentation in mm3. CC is the Pearson correlation coefficient between Manual Vol and Auto Vol; TPR: mean true positive rate; FDR: mean false discovery rate. Numbers in parentheses indicate standard deviations. NA: nucleus accumbens, BF: basal forebrain, SepN: septal nuclei, HTh: hypothalamus without mammillary bodies, MB: mammillary bodies, Fx: fornix, L: left, R: right. The table reflects only data from the 18 independent test subjects.
| Structure | Manual Vol | Auto Vol | Dice | CC | TPR | FDR |
|---|---|---|---|---|---|---|
| NA-L | 374.9 (110.9) | 404.1 (129.5) | 0.82 (0.045) | 0.88 | 0.85 (0.058) | 0.20 (0.090) |
| NA-R | 380.1 (119.8) | 422.6 (130.5) | 0.78 (0.084) | 0.72 | 0.83 (0.072) | 0.24 (0.140) |
| BF-L | 328.7 (68.8) | 304.2 (48.9) | 0.78 (0.051) | 0.63 | 0.76 (0.095) | 0.19 (0.066) |
| BF-R | 322.6 (70.3) | 318.6 (54.2) | 0.75 (0.087) | 0.70 | 0.76 (0.114) | 0.24 (0.095) |
| SepN-L | 117.5 (30.4) | 108.9 (17.5) | 0.69 (0.079) | 0.62 | 0.68 (0.093) | 0.28 (0.110) |
| SepN-R | 114.9 (31.9) | 101.1 (18.7) | 0.72 (0.074) | 0.69 | 0.69 (0.077) | 0.23 (0.130) |
| HTh-L | 439.3 (88.3) | 473.4 (68.6) | 0.81 (0.035) | 0.74 | 0.85 (0.051) | 0.21 (0.076) |
| HTh-R | 438.6 (91.6) | 471.6 (65.4) | 0.82 (0.034) | 0.78 | 0.86 (0.057) | 0.21 (0.063) |
| MB-L | 51.6 (9.2) | 50.4 (10.3) | 0.78 (0.070) | 0.50 | 0.77 (0.078) | 0.19 (0.118) |
| MB-R | 54.1 (9.9) | 51.4 (7.6) | 0.80 (0.061) | 0.37 | 0.79 (0.098) | 0.18 (0.094) |
| Fx-L | 551.9 (109.6) | 525.4 (88.3) | 0.80 (0.043) | 0.75 | 0.78 (0.076) | 0.18 (0.054) |
| Fx-R | 544.2 (127.8) | 505.6 (88.8) | 0.79 (0.040) | 0.87 | 0.77 (0.057) | 0.18 (0.064) |
The True Positive Rate (TPR, number of true positives detected by the automatic segmentation divided by the number of voxels in the manual segmentation) ranged from 0.68 to 0.86. As illustrated in Fig. 2, this value represents the number of green voxels (true positives) divided by the sum of the green and yellow voxels (number of voxels in the manual label). The False Discovery Rate (FDR, number of false positives divided by the total number of voxels in the automatic segmentation) ranged from 0.18 to 0.28. As displayed in Fig. 2, this value represents the number of red voxels (false positives) divided by the sum of the green and red voxels (total number of voxels in the automatic label).
3.2. Experiment 2: Robustness results
In the robustness test, 124/698 cases were flagged by RCA for inspection. The two raters had very similar results, agreeing 94% of the time. Of the 124, the raters agreed that 91 have no issues at all, suggesting that the RCA threshold of 0.5 was quite liberal. Issues with the 33 remaining cases all had to do with underlabeling to some degree. There were 2 cases (0.3%) where a label was simply not present (NA-L in one case and MB-R in the other), both from Oulu. There were 31 other cases (4.3%) where at least one region was underlabeled; 15 of those were fornix. Of the 18 (2.6%) remaining from the 33, the SepN were suspect in 6 subjects because of an anatomical variant (an unclosed cavum septum pellucidum, width about 10 mm); to be clear, it was not evident that SepN segmentation failed because of this, we just do not have enough experience with this variant to know that it succeeded. For the remaining 12 cases (1.7%), various regions were underlabeled. Hypothalamus failed in 2 AD cases because the portion of fornix that goes through hypothalamus was not labeled; in these cases, there was simply no contrast between HTh and Fx. Table 2 shows the underlabeling rate for each label individually averaged across the two raters. Except for fornix, the rates are all less than 1%. Of the regions, fornix incurred the most failures, some on subjects with much atrophy but a portion on young subjects with very small ventricles. One subject failed because of an extreme angle in the head position; after manual rotation, the segmentation passed. Of the 33 problematic cases, 17 came from Oulu.
Table 2.
Robustness and test-retest reliability. Underlabeling rate (UR) is the percent of the 698 subjects that had some mislabeling based on visual inspection. CC is Pearson correlation coefficient and ICC is intraclass correlation.
| Structure | UR | CC | ICC |
|---|---|---|---|
| NA-L | 0.72% | 0.94 | 0.94 |
| NA-R | 0.29% | 0.97 | 0.97 |
| BF-L | 0.29% | 0.96 | 0.96 |
| BF-R | 0.29% | 0.94 | 0.94 |
| SepN-L | 0.86% | 0.91 | 0.91 |
| SepN-R | 0.86% | 0.93 | 0.92 |
| HTh-L | 0.57% | 0.94 | 0.94 |
| HTh-R | 0.43% | 0.95 | 0.94 |
| MB-L | 0.57% | 0.90 | 0.90 |
| MB-R | 0.72% | 0.94 | 0.94 |
| Fx-L | 3.01% | 0.94 | 0.94 |
| Fx-R | 3.01% | 0.94 | 0.94 |
3.3. Experiment 3: Test-retest reliability results
The test-retest reliability across scans of the 40 MIRIAD subjects is shown in Table 2 using both correlation coefficient (CC) and intraclass correlation (ICC). The values are distributed closely around 0.95. While MB-L is the lowest, it is still high at 0.90; at 0.94, MB-R is similar to that of other labels. The time points were not significantly different when tested with a paired t-test, also suggesting good reliability.
3.4. Experiment 4: Effects of Alzheimer’s disease and aging results
The results for the effects of AD and aging are shown in Table 3. The change in volume with AD and age was always negative, indicating a loss of tissue (i.e., atrophy). All structures, except MB, show significance in either AD or age or both.
Table 3.
Effect of AD and age on the volume of the given structure. Change and Slope show the change in volume in thousandths of percent of intracranial volume. Slope is per decade. A negative Change value indicates loss of volume in AD relative to HC. A negative Slope indicates a loss in volume with age. The p-values have been corrected for 12 comparisons; those with p < 0.05 are marked with an asterisk. See Table 1 for structure abbreviations.
| Structure | AD Change | p | Age Slope | p |
|---|---|---|---|---|
| NA-L | −2.82 | 0.051142 | −2.93 | 0.000951 * |
| NA-R | −2.36 | 0.117383 | −3.15 | 0.000084 * |
| BF-L | −2.39 | 0.000641 * | −1.54 | 0.003073 * |
| BF-R | −2.38 | 0.000138 * | −1.03 | 0.032481 * |
| SepN-L | −0.47 | 0.289839 | −0.09 | 0.999968 |
| SepN-R | −0.58 | 0.007340 * | −0.05 | 1.000000 |
| HTh-L | −1.95 | 0.023953 * | −2.36 | 0.000181 * |
| HTh-R | −2.09 | 0.001621 * | −2.04 | 0.000771 * |
| MB-L | −0.20 | 0.874555 | −0.10 | 0.974076 |
| MB-R | −0.30 | 0.238399 | −0.09 | 0.998295 |
| Fx-L | −3.13 | 0.007580 * | −2.87 | 0.000475 * |
| Fx-R | −2.84 | 0.025472 * | −2.81 | 0.010283 * |
3.5. Tool usage
The tool and instructions are available from the FreeSurfer wiki at surfer.nmr.mgh.harvard.edu/fswiki/ScLimbic (“ScLimbic” is meant to abbreviate “subcortical limbic”); a diagram of the software workflow is also shown in Fig. 3. In the basic usage, one creates a folder with the T1w volumes in (NIFTI or mgz format) one wants to segment, then runs the Python script
Fig. 3.
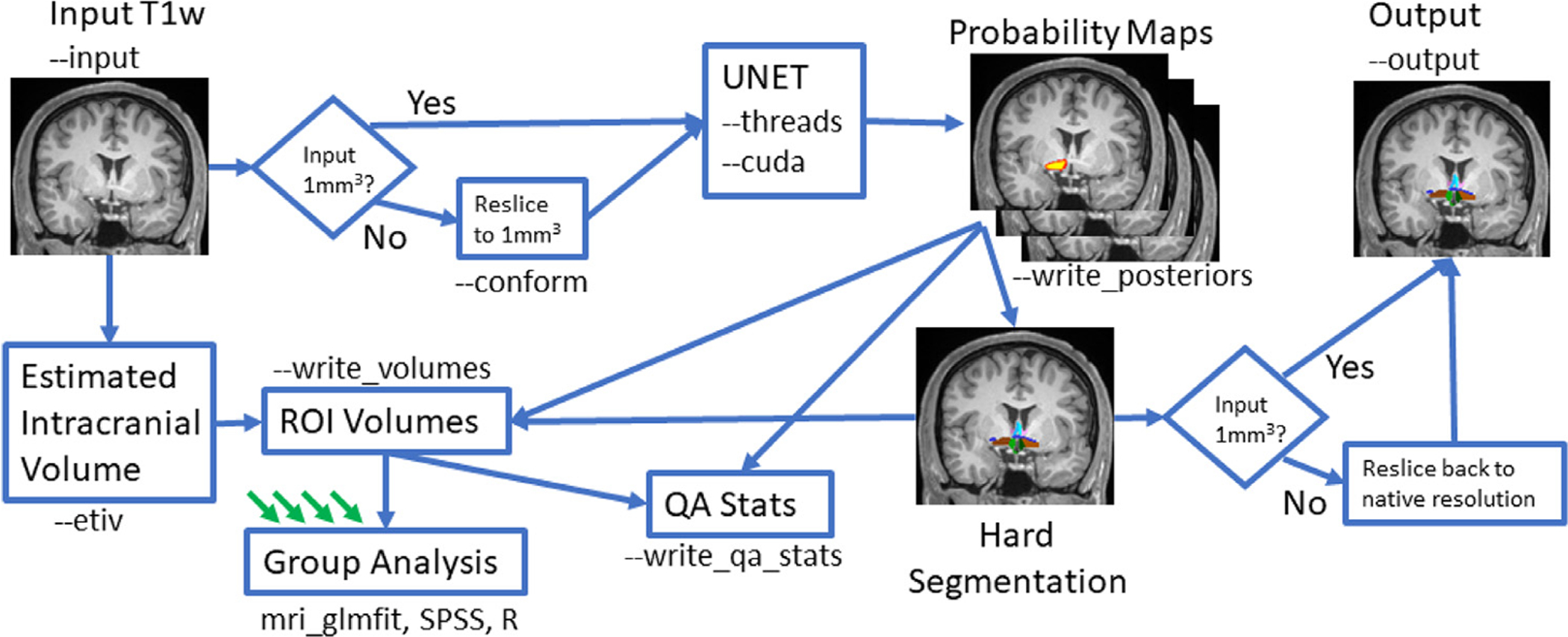
Diagram of the tool workflow showing various options and outputs. Green arrows indicate output from other subjects.
mri_sclimbic_seg --i inputfolder --o outputfolder --write_volumes
The tool will find all the input images, segment them, and write out the segmentation images into the output folder; the segmentations will resemble Fig. 1. It will also create a CSV file where each row is a case, each column is a label, and each entry is the volume of that structure in mm3. On a single threaded CPU, the program takes about 40 s to run on a single case; with 3 threads (—threads 3), the time drops to about 15 s; using a GPU (—cuda) does not reduce this significantly as much of the time is spent loading and writing. The tool uses about 20GB of memory.
If the input volume is not 1mm3, then there is an option to reslice to this resolution (—conform); the reslicing is only internal – the output segmentation is resliced back to the original resolution. Note that changing the resolution may affect the quality. If one is planning to perform a volumetric group study, then one will need to normalize by ICV. If one does not have an estimate of the ICV, then the tool can compute it using the FreeSurfer method (—etiv, Buckner et al., 2004); the ICV will be included as a column in the CSV file. Computing the ICV will increase the processing time to about 5 min for each case. The CSV file can be imported into a statistical program like SPSS or R for further processing or it can be processed using FreeSurfer’s mri_glmfit, which includes automatic application of ICV correction if ICV is in the CSV.
The user should visually inspect the segmentation output. To assist in quality control, the tool can output two additional CSV files (—write_qa_stats). One contains a z-score2 for the volume each structure based on the means and standard deviations of the manual labels. In the other, the “confidence” (mean posterior probability within the label) is reported. If the z-score is very high or the confidence is very low, then the case should be visually examined. The tool does not require knowledge of FreeSurfer; as long as FreeSurfer is installed, then the user need only understand and execute mri_sclimbic_seg.
4. Discussion
The goal of this study was to develop a deep learning segmentation tool for the following limbic structures: hypothalamus, mammillary bodies (part of hypothalamus), basal forebrain, septal nuclei, nucleus accumbens, and fornix. The tool was trained on manually labeled data and evaluated over 700 independent data sets; clinical efficacy was shown on AD and aging data sets.
4.1. Segmentation performance
The central goal of automatic segmentation is to replicate how an expert would have labeled a novel image. This capability was judged by comparing the automatic segmentation to the manual segmentation of images not included in the training. Average Dice scores ranged from 0.69 (SepN) to 0.82 (NA). This is well within the range of other studies. For example, Fischl et al., 2002 and Puonti et al., 2016 had Dice scores between 0.70 and 0.90 for much larger structures, which will generally perform better on Dice than smaller structures. For whole hypothalamus, Billot et al., 2020a had a Dice score of 0.84 and Rodrigues et al., 2020 had 0.77; our tool is comparable at 0.81. Billot et al., 2020a also had a Dice score of 0.81 for the posterior hypothalamus, the subunit closest to our definition of MB, which had a comparable Dice score of 0.78.
While the Dice score provides a good summary measure of overall accuracy, other metrics provide more meaningful evaluations in terms of how the segmentation will perform when applied for a particular purpose. The Pearson correlation coefficient (CC, Table 1) shows how the volume of the automatic segmentation scales with that of the manual label. Ideally, the volume would accurately reflect the true value; however, this is technically not necessary in studies that compare groups or correlate diagnostic parameters as long as the volume scales with the true value. This ability to scale is measured by the CC. In our study, the CCs were generally in the range of 0.62 to 0.88, except for MB, which was 0.37 and 0.50. The CC for SepN was 0.62–0.69, which exceeds the 0.34–0.66 obtained by Butler et al., 2014. The low CC score for MB indicates that the MB volume might not be a sensitive marker of cross-subject differences. Indeed, while other labels were significant in both the AD and aging studies, MB was not significant in either. The Dice for MB was a relatively high 0.78; this shows a shortcoming of the Dice score as a performance metric, which is why we tested additional metrics in this report.
4.2. Appropriateness for multimodal integration
We report the True Positive Rate (TPR) and False Discovery Rate (FDR) for each structure (Table 1 and Fig. 2). These measures are pertinent to multimodal integration studies (e.g., fMRI, dMRI, PET). For example, in a task fMRI study, the amplitude of the hemodynamic response might be averaged over the label; in a diffusion study, the label may be used as a seed region for tractography. Errors in the cross-modal analysis may result if the label does not significantly overlap the true structure or if a significant number of voxels from a neighboring structure were included. TPR measures the overlap with the true structure (sensitivity), and FDR measures the contamination for neighboring structures (specificity). In this study, the TPRs were in the range of 0.68–0.86, meaning that a large fraction of the true structure will fall within the automatically segmented label. The FDRs were in the range of 0.18–0.28, meaning that a relatively small fraction of automatically segmented voxels fall outside of the true structure. Our findings indicate that all these structures, including MB, are appropriate for cross-modal applications. We have not been able to find other automatic segmentation studies that report TPR or FDR, so there is no reference for comparison.
4.3. Robustness
we evaluated the robustness of the segmentation on 698 cases. Reverse classification accuracy (RCA, Valindria et al., 2017) liberally flagged 124 cases for visual inspection. “Failures,” as indicated by noticeable underlabeling, were found in only 33 cases. For individual labels, the failure rate was quite low (Table 2), less than 1% for all structures, except for fornix, which was 3%. While this performance is quite good, it is important to evaluate where and how the underlabeling occurs. We observed several failure modes. Six cases had large (> 10 mm) unclosed cavum septum pellucidum (CSP). A CSP is a space between the left and right septa in the lateral ventricles, very close to the septal nuclei and fornix (Born et al., 2004). The septa usually fuse shortly after birth, but closure does not occur in roughly 1–5% of the population (Chen et al., 2014). This structural irregularity can cause errors in the SepN and Fx segmentations since these structures are closely bound to the septa.
With 14 of the 32 failures, Fx was the most error prone of all the limbic labels; Fx had several failure modes. The first was the CSP cases mentioned above. The second was advanced atrophy in some cases (i.e., AD). The Fx is a white matter strand that connects HTh and hippocampus. In healthy subjects, it is clearly visible but still only a few millimeters in diameter. With aging and disease, it becomes thinner and darker, and the crus of the fornix becomes barely visible as it passes through the atrium of the lateral ventricle. This can cause the automatic segmentation to be hit-or-miss in this region. We emphasize that this was observed in only a handful of cases; the vast majority of atrophic cases had good Fx segmentations. The third Fx failure mode was in young subjects with very small ventricles in MRI with poor contrast. In such cases, the Fx tail was in near or direct contact with the corpus callosum and became indistinguishable; all of these cases were in the Oulu data set. Finally, in a two ADNI cases, the body of the Fx, which is completely surrounded by the HTh, was not segmented because there was no visible gray/white contrast; presumably, this is just part of the disease process, but we counted it as an error for both Fx and HTh.
We emphasize here that these circumstances of underlabeling mentioned above occurred in a very small fraction of cases. This robustness test probably represents a worst-case scenario as the data sets (deliberately) included low-quality data (Oulu) or data collected many years ago (ADNI). On high-quality data such as FSM, HABS, Beijing, and Cambridge datasets, virtually no errors occurred.
4.4. Test-Retest reliability
The test-retest performance of the tool was evaluated using 40 (20 AD and 20 HC) subjects scanned two weeks apart. This duration is probably too short for much true anatomical change to have occurred, so any differences are attributed to either scanning or inaccuracies of the tool. The CCs and ICCs were around 0.94, which is excellent. While the MB manual-automatic volume correlations were poor, the CC and ICC for MBs were very high (0.90 and 0.94) in test-retest. This indicates that all the structures, including MB, can be sensitive to longitudinal changes.
4.5. Clinical significance
The limbic system is especially vulnerable to Alzheimer’s disease (Mesulam, 1996; Braak and Braak, 1997; Braak and Del Tredici, 2012; Hyman et al., 1984; Terry and Katzman, 1983; Hopper and Vogel, 1976). The SepN and HTh are strongly connected to the hippocampus via the Fx, so hippocampal atrophy has substantial downstream effects on Fx, SepN and HTh. The hippocampus is a seminal structure in the staging of Alzheimer’s disease pathology (Braak and Braak, 1991, 1997) and a neuroimaging biomarker benchmark (Braskie and Thompson, 2014; Weiner et al., 2017). Our SepN label includes medial septal nuclei, while our basal forebrain label includes vertical limb (Ch2) and the horizontal limb (Ch3) of the diagonal band of Broca, and the nucleus basalis of Meynert (Ch4). The latter making up the main portion of acetylcholine input for the cerebral cortex. Acetylcholine has a large neurochemical impact on Alzheimer’s disease pathology (Ballinger et al., 2016; Geula and Mesulam, 1996; Hampel et al., 2019).
In line with this thesis, we found that BF, SepN, HTh, and Fx showed atrophy when comparing ADs to age-matched controls (Table 3). These results corroborate other studies such as Teipel et al., 2014 (BF), Butler et al., 2018 (SepN), Billot et al., 2020a (HTh), and Copenhaver et al., 2006 (Fx). NA and MB did not show an effect despite being found in other studies (Nie et al., 2017 and Copenhaver et al., 2006 respectively); NA would have been significant in this study without corrections for multiple comparisons.
While advanced age is not a clinical condition in and of itself, the aging brain undergoes many changes that are similar to clinical conditions (Salat et al., 2004). We demonstrated significant changes with age in NA, BF, HTh, and Fx, thus providing additional evidence of clinical utility.
4.6. An easy-to-use tool
the tool that performed the automatic labeling in this study is freely available via FreeSurfer along with extensive documentation for how to use it on individual and group data (see surfer.nmr.mgh.harvard.edu/fswiki/ScLimbic; for source code see github.com/freesurfer). It is self-contained and easy to use, including computing and applying corrections for ICV, if needed. The segmentation of a single case can be done on a CPU or GPU and finishes in a few minutes. The images, labels, and training code are available, so researchers can retrain the network with their own manually labeled data if desired. While we have emphasized a specific tool built on the U-Net architecture of Billot et al., 2020a, this work shows that the manual labels that we have developed are sufficient for accurately labeling these structures in the human brain, and new algorithms and architectures (e.g., Billot et al., 2020b; Isensee et al., 2021) could be employed to create new tools with even better, more robust performance.
5. Conclusion
A tool was developed to automatically segment several subcortical limbic structures (nucleus accumbens, basal forebrain, septal nuclei, hypothalamus without mammillary bodies, the mammillary bodies, and fornix) from a T1-weighted MRI. This tool fills an unmet need as there are few, if any, tools to segment these clinically relevant structures. A U-Net with spatial, intensity, contrast, and noise augmentation was trained using 39 manually labeled MRI data sets. In general, the Dice scores, true positive rates, false discovery rates, and manual-automatic volume correlation were very good relative to comparable tools for other structures. A diverse data set of 698 subjects were segmented using the tool; evaluation of the resulting labelings showed that the tool failed in less than 1% of cases. Test-retest reliability of the tool was excellent. The automatically segmented volume of all structures except mammillary bodies showed effectiveness at detecting either clinical AD effects, age effects, or both. This tool will be publicly released with FreeSurfer (surfer.nmr.mgh.harvard.edu). Together with the other cortical and subcortical limbic segmentations, this tool will allow FreeSurfer to provide a comprehensive view of the limbic system in an automated way.
Acknowledgments
Support for this research was provided in part by the National Institutes of Health (R01NS105820, R01EB023281, R01NS083534, R01AG057672, R01NS112161, R01AG059011, R01NR010827, U19AG068753, U24DA041123, R01EB019956, U01MH117023, P41EB015896, R01EB006758, R21EB018907, P41EB030006, 1R56AG064027, 1R01AG064027, 5R01AG008122, R01AG016495, R01MH123, R01MH121885, R01NS0525851, R01NS070963, 5U01NS086625, 5U24NS10059103, R01AG070988, RF1AG072056, K99HD101553, 1RF1MH123195), and was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. Additional support was provided by the NIH Blueprint for Neuroscience Research (5U01-MH093765), part of the multi-institutional Human Connectome Project. European Research Council: Starting Grant 677697, project “BUNGEE-TOOLS”; Alzheimer’s Research UK: ARUK-IRG2019A-003. BB was supported by EPSRC UCL center for Doctoral Training in Medical Imaging (EP/L016478/1). We would like to thank the contributors to and curators of the 1000 Functional Connectomes data set. The collection and sharing of the MRI data used in the group study based on ADNI was funded by the Alzheimer’s Disease Neuroimaging Initiative (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). We would like to thank the Harvard Aging Brain (HABS, habs.mgh.harvard.edu) study for supplying aging data.
Appendix A: Manual Labeling Methods
The quantitative T1 (qT1) image was used to determine the anatomical boundaries of each structure for the FSM data set and the T1w image for the ADNI data. All structures undertaken in this report – fornix, basal forebrain, hypothalamus, mammillary bodies, nucleus accumbens, septal nuclei, anterior commissure – were labeled in the coronal plane while the axial and sagittal planes served as three-dimensional checkpoints for evaluation of missing or inaccurate voxels.
Neuroanatomy structures:
We used several neuroanatomical publications and references to guide our anatomical delineation (L. Heimer, 2008; Mai et al., 2004; Ding et al., 2016; Edlow et al., 2018; Haines, 2007). Tissue slabs in educational atlases often show oblique slices, which is different than our description based on orthogonal planes. This differential – between orthogonal in MRI and oblique planes in histological references – sometimes presents as a challenge for identification. Nonetheless, the anterior, posterior, medial, lateral, superior, and inferior limits of each structure were identified based on distinct contrast and/or using the neighboring anatomical structures and manually labeled. Each structure is detailed in the following paragraphs.
Hypothalamus- The anterior slices of the hypothalamus appear when that ventral tissue connects with the optic chiasm. Continuing from anterior to posterior, the midline of the anterior commissure comes into view and to the columns of the fornix materialize just posterior to the anterior commissure. The hypothalamus occurs in the same slice with the septal nuclei or after immediately posterior to the septal nuclei (e.g., one or two millimeters), but this arrangement depends on individual case. We have observed both concurrence and adjacent slices. The posterior boundary of the hypothalamus is the posterior limit of the mammillary bodies. The superior boundary depends on the anterior-posterior slice. The midline of the anterior commissure limits hypothalamus anteriorly while the columns of the fornix pass through the hypothalamus at mid-level, and the dorsal thalamus marks the superior boundary at posterior most levels. The inferior boundary of the hypothalamus extends into the infundibular stalk that gives rise to the pituitary gland. The lateral boundary is the basal forebrain area, which house the cholinergic bands of Broca and the nucleus basal of Meynert or the sublenticular extended amygdala (included in our basal forebrain label). The third ventricle demarcated the medial boundary of the hypothalamus.
Nucleus Accumbens- The anterior boundary of the nucleus accumbens presented at the same plane coronally as the adjacent structures: posterior orbitofrontal cortex, subcallosal area, olfactory tract, and the temporal pole. The nucleus accumbens represents the ventral portion of the striatum. The midway point of the nucleus accumbens typically shows the optic tract inferior the accumbens nucleus, the olfactory tract nears its end, and temporal cortices have arrived in coronal plane (i.e., five gyri appear in temporal lobe), albeit small. Together the putamen, caudate and nucleus accumbens appear as a U-shaped structure in the coronal plane and show similar contrast in MRI. The posterior limit of the nucleus accumbens arises at same rostrocaudal level as the optic chiasm, which is located inferiorly to it.
Fornix and Anterior Commissure- the fornix and anterior commissure showed different contrast relative to neighboring gray matter contrast given they are both white matter structures. The fornix begins posteriorly, coming off the posterior hippocampus, continuing dorsally as crus of the fornix, the body of the fornix and finally descending into the basal forebrain region as the columns of the fornix. The anterior commissure appears at the midline anteriorly then arches posteriorly and laterally. Scrolling through coronal view, the anterior commissure gives the impression that it is fragmented but it is one continuous structure with each small section hitting the plane at a different slice. Image contrast and relative location provided excellent guidance to neighboring structural boundaries. Note that we only labeled the anterior commissure to aid in the labeling of the other structures.
Basal forebrain and septal nuclei- the septal nuclei occupy the region immediately inferior to the lateral ventricles at the midline. The anterior-posterior levels and the appearance of the septal nuclei depends on the particular case; it may be at the similar level as the nucleus accumbens or just posterior to it at the similar level to the hypothalamus and midline anterior commissure. The septal nuclei represent a small territory along the midline. In this work, the basal forebrain label contains the following structures: the vertical and horizontal limb of the diagonal band of Broca, ventral pallidum, some posterior remnants of ventral striatum, the extended amygdala, and the nucleus basalis of Meynert. The basal forebrain becomes evident slightly posterior to the anterior most anterior commissure. The superior boundary of the basal forebrain is either the anterior commissure or the globus pallidus while the inferior boundary goes to the ventral edge (of the brain tissue), anterior to the cerebrospinal fluid in the interpeduncular cistern. The hypothalamus bounds the basal forebrain medially. The lateral boundary of the basal forebrain is more difficult because it appears continuous (from a contrast perspective) with the medial amygdala. A fiducial marker, a fixed line, was drawn at the medial edge of the putamen, inferiorly to the tissue edge to delimit the lateral most boundary of basal forebrain. Approximately four 1 mm coronal slices were labeled per basal forebrain and each slice became smaller posteriorly.
Footnotes
CRediT authorship contribution statement
Douglas N. Greve: Conceptualization, Data Curation, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing - original draft, Supervision, Funding acquisition. Benjamin Billot: Software, Review, Methodology, Resources. Devani Cordero: Investigation, Data Curation, Review. Andrew Hoopes: Software. Malte Hoffmann: Data Curation, Review. Adrian Dalca: Software, Methodology, Review. Bruce Fischl: Conceptualization, Methodology, Review, Funding acquisition, Resources. Juan Eugenio Iglesias: Software, Methodology, Review. Jean C. Augustinack: Conceptualization, Investigation, Methodology, Validation, Review, Supervision, Writing.
The hypothalamus label here excludes the mamillary bodies since we have a separate label for those.
The z-score should only be used for quality control. It should not be interpreted as a deviation from a normal population because the subjects used for manual labeling were not chosen with this in mind.
Data Code Availability
The code to train and apply the model is available from github.com/BBillot/hypothalamus_seg. The model, code to apply it, and documentation are available at surfer.nmr.mgh.harvard.edu/fswiki/ScLimbic. Data used in the labeling can be requested from the corresponding author, subject to a data use agreement. Data from the 1000 Functional Connectomes data base is publicly available from fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html. Alzheimer’s Disease Neuroimaging Initiative (ADNI) data available from adni.loni.usc.edu. Data from the Harvard Aging Brain Study (HABS) can be requested from habs.mgh.harvard.edu/researchers/request-data.
References
- Artaechevarria X, Munoz-Barrutia A, Ortiz-de-Solorzano C, 2009. Combination strategies in multi-atlas image segmentation: application to brain MR data. IEEE Trans. Med. Imag 28, 1266–1277. [DOI] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Song G, Cook PA, Klein A, Gee JC, 2011. A reproducible evaluation of ANTs similarity metric performance in brain image registration. Neuroimage 54, 2033–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballinger EC, Ananth M, Talmage DA, Role LW, 2016. Basal forebrain cholinergic circuits and signaling in cognition and cognitive decline. Neuron 91, 1199–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot B, Bocchetta M, Todd E, Dalca AV, Rohrer JD, Iglesias JE, 2020a. Automated segmentation of the hypothalamus and associated subunits in brain MRI. Neuroimage 223, 117287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billot B, Greve DN, Van Leemput K, Fischl B, Iglesias JE, Dalca AV, 2020b. A learning strategy for contrast-agnostic MRI segmentation. MIDL: medical imaging with deep learning, arXiv:2003.01995 [cs]MIDL 2020. [Google Scholar]
- Born CM, Meisenzahl EM, Frodl T, Pfluger T, Reiser M, Moller HJ, Leinsinger GL, 2004. The septum pellucidum and its variants. An MRI study. Eur. Arch. Psychiatry Clin. Neurosci 254, 295–302. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, 1991. Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. (Berl) 82, 239–259. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E, 1997. Staging of Alzheimer-related cortical destruction. Int. Psychogeriatr 9, 257–261 discussion 269–272. [PubMed] [Google Scholar]
- Braak H, Del Tredici K, 2012. Where, when, and in what form does sporadic Alzheimer’s disease begin? Curr. Opin. Neurol 25, 708–714. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Thompson PM, 2014. A focus on structural brain imaging in the Alzheimer’s disease neuroimaging initiative. Biol. Psychiatry 75, 527–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder AZ, 2004. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. Neuroimage 23, 724–738. [DOI] [PubMed] [Google Scholar]
- Butler T, Blackmon K, Zaborszky L, Wang X, DuBois J, Carlson C, Barr WB, French J, Devinsky O, Kuzniecky R, Halgren E, Thesen T, 2012. Volume of the human septal forebrain region is a predictor of source memory accuracy. J. Int. Neuropsychol. Soc 18, 157–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T, Harvey P, Deshpande A, Tanzi E, Li Y, Tsui W, Silver C, Fischer E, Wang X, Chen J, Rusinek H, Pirraglia E, Osorio RS, Glodzik L, de Leon MJ, 2018. Basal forebrain septal nuclei are enlarged in healthy subjects prior to the development of Alzheimer’s disease. Neurobiol. Aging 65, 201–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler T, Zaborszky L, Pirraglia E, Li J, Wang XH, Li Y, Tsui W, Talos D, Devinsky O, Kuchna I, Nowicki K, French J, Kuzniecky R, Wegiel J, Glodzik L, Rusinek H, deLeon MJ, Thesen T, 2014. Comparison of human septal nuclei MRI measurements using automated segmentation and a new manual protocol based on histology. Neuroimage 97, 245–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavedo E, Grothe MJ, Colliot O, Lista S, Chupin M, Dormont D, Houot M, Lehericy S, Teipel S, Dubois B, Hampel H Hippocampus Study, G., 2017. Reduced basal forebrain atrophy progression in a randomized Donepezil trial in prodromal Alzheimer’s disease. Sci. Rep 7, 11706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JJ, Chen CJ, Chang HF, Chen DL, Hsu YC, Chang TP, 2014. Prevalence of cavum septum pellucidum and/or cavum Vergae in brain computed tomographies of Taiwanese. Acta Neurol. Taiwan 23, 49–54. [PubMed] [Google Scholar]
- Copenhaver BR, Rabin LA, Saykin AJ, Roth RM, Wishart HA, Flashman LA, Santulli RB, McHugh TL, Mamourian AC, 2006. The fornix and mammillary bodies in older adults with Alzheimer’s disease, mild cognitive impairment, and cognitive complaints: a volumetric MRI study. Psychiatry Res. 147, 93–103. [DOI] [PubMed] [Google Scholar]
- Dalca AV, Guttag J, Sabuncu M, 2018. Anatomical priors in convolutional networks for unsupervised biomedical segmentation. In: IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 9290–9299. [Google Scholar]
- Despotovic I, Goossens B, Philips W, 2015. MRI segmentation of the human brain: challenges, methods, and applications. Comput. Math. Methods Med 2015, 450341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding SL, Royall JJ, Sunkin SM, Ng L, Facer BA, Lesnar P, Guillozet-Bongaarts A, McMurray B, Szafer A, Dolbeare TA, Stevens A, Tirrell L, Benner T, Calde-jon S, Dalley RA, Dee N, Lau C, Nyhus J, Reding M, Riley ZL, Sand-man D, Shen E, van der Kouwe A, Varjabedian A, Wright M, Zollei L, Dang C, Knowles JA, Koch C, Phillips JW, Sestan N, Wohnoutka P, Zielke HR, Hohmann JG, Jones AR, Bernard A, Hawrylycz MJ, Hof PR, Fischl B, Lein ES, 2016. Comprehensive cellular-resolution atlas of the adult human brain. J. Comp. Neurol 524, 3127–3481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edlow BL, Keene CD, Perl DP, Iacono D, Folkerth RD, Stewart W, Mac Donald CL, Augustinack J, Diaz-Arrastia R, Estrada C, Flannery E, Gor-don WA, Grabowski TJ, Hansen K, Hoffman J, Kroenke C, Larson EB, Lee P, Mareyam A, McNab JA, McPhee J, Moreau AL, Renz A, Richmire K, Stevens A, Tang CY, Tirrell LS, Trittschuh EH, van der Kouwe A, Varjabedian A, Wald LL, Wu O, Yendiki A, Young L, Zollei L, Fischl B, Crane PK, Dams-O’Connor K, 2018. Multimodal characterization of the late effects of traumatic brain injury: a methodological overview of the late effects of traumatic brain injury project. J. Neurotrauma 35, 1604–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, 2012. FreeSurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Albert M, Dieterich M, Haselgrove C, Kouwe A.v.d., Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM, 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. [DOI] [PubMed] [Google Scholar]
- Fischl B, van der Kouwe A, Destrieux C, Halgren E, Segonne F, Salat DH, Busa E, Seidman LJ, Goldstein J, Kennedy D, Caviness V, Makris N, Rosen B, Dale AM, 2004. Automatically parcellating the human cerebral cortex. Cereb. Cortex 14, 11–22. [DOI] [PubMed] [Google Scholar]
- Geula C, Mesulam MM, 1996. Systematic regional variations in the loss of cortical cholinergic fibers in Alzheimer’s disease. Cereb. Cortex 6, 165–177. [DOI] [PubMed] [Google Scholar]
- Greve DN, Salat DH, Bowen SL, Izquierdo-Garcia D, Schultz AP, Catana C, Becker JA, Svarer C, Knudsen GM, Sperling RA, Johnson KA, 2016. Different partial volume correction methods lead to different conclusions: an (18)F-FDG-PET study of aging. Neuroimage 132, 334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haines D, 2007. Neuroanatomy An Atlas of Structures, Sections, and Systems, 7th ed. Lippincott Williams & Wilkins. [Google Scholar]
- Hampel H, Mesulam MM, Cuello AC, Khachaturian AS, Vergallo A, Farlow MR, Snyder PJ, Giacobini E, Khachaturian ZS, 2019. Revisiting the cholinergic hypothesis in Alzheimer’s disease: emerging evidence from translational and clinical research. J. Prev. Alzheimers Dis 6, 2–15. [DOI] [PubMed] [Google Scholar]
- Heimer L, Van Hoesen GW, 2006. The limbic lobe and its output channels: implications for emotional functions and adaptive behavior. Neurosci. Biobehav. Rev 30, 126–147. [DOI] [PubMed] [Google Scholar]
- Heimer L, Van Hoesen GW, Trimble M, Zahm DS, 2008. The New Anatomy of the Basal Forebrain and Its Implications for Neuropsychiatric Illness. Elsevier and Academic Press. [Google Scholar]
- Henschel L, Conjeti S, Estrada S, Diers K, Fischl B, Reuter M, 2020. FastSurfer - A fast and accurate deep learning based neuroimaging pipeline. Neuroimage 219, 117012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper MW, Vogel FS, 1976. The limbic system in Alzheimer’s disease. A neuropathologic investigation. Am. J. Pathol 85, 1–20. [PMC free article] [PubMed] [Google Scholar]
- Hyman BT, Van Horsen GW, Damasio AR, Barnes CL, 1984. Alzheimer’s disease: cell-specific pathology isolates the hippocampal formation. Science 225, 1168–1170. [DOI] [PubMed] [Google Scholar]
- Iglesias JE, Augustinack JC, Nguyen K, Player CM, Player A, Wright M, Roy N, Frosch MP, McKee AC, Wald LL, Fischl B, Van Leemput K Alzheimer’s Disease Neuroimaging, I., 2015. A computational atlas of the hippocampal formation using ex vivo, ultra-high resolution MRI: application to adaptive segmentation of in vivo MRI. Neuroimage 115, 117–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isensee F, Jaeger PF, Kohl SAA, Petersen J, Maier-Hein KH, 2021. nnU-Net: a self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 18, 203–211. [DOI] [PubMed] [Google Scholar]
- Jin Y, Shi Y, Zhan L, Thompson PM Alzheimer’s Disease Neuroimaging, I., 2015. Automated Multi-Atlas Labeling of the Fornix and Its Integrity in Alzheimer’s Disease. Proc. IEEE Int. Symp. Biomed. Imag 2015, 140–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jog A, Hoopes A, Greve DN, Van Leemput K, Fischl B, 2019. PSACNN: pulse sequence adaptive fast whole brain segmentation. Neuroimage 199, 553–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingma D, Ba J, 2015. Adam: a method for stochastic optimization. 3rd International Conference for Learning Representations arXiv:1412.6980.. [Google Scholar]
- Kotter R, Stephan KE, 1997. Useless or helpful? The “limbic system” concept. Rev. Neurosci 8, 139–145. [DOI] [PubMed] [Google Scholar]
- LeDoux J, 2012. Rethinking the emotional brain. Neuron 73, 653–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean P, 1949. Psychosomatic disease and the visceral brain; recent developments bearing on the Papez theory of emotion. Psychosom. Med 11, 338–353. [DOI] [PubMed] [Google Scholar]
- Mai J, Majtanik M, Paxinos G, 2004. Atlas of Teh Human Brain, 2nd ed. Elsevier. [Google Scholar]
- Malone IB, Cash D, Ridgway GR, MacManus DG, Ourselin S, Fox NC, Schott JM, 2013. MIRIAD–Public release of a multiple time point Alzheimer’s MR imaging dataset. Neuroimage 70, 33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques JP, Kober T, Krueger G, van der Zwaag W, Van de Moortele PF, Gruetter R, 2010. MP2RAGE, a self bias-field corrected sequence for improved segmentation and T1-mapping at high field. Neuroimage 49, 1271–1281. [DOI] [PubMed] [Google Scholar]
- Mesulam MM, 1985. Principles of Behavioral Neurology. Davis FA, Philadelphia, PA. [Google Scholar]
- Mesulam MM, 1996. The systems-level organization of cholinergic innervation in the human cerebral cortex and its alterations in Alzheimer’s disease. Prog. Brain Res 109, 285–297. [DOI] [PubMed] [Google Scholar]
- Mormino EC, Betensky RA, Hedden T, Schultz AP, Amariglio RE, Rentz DM, Johnson KA, Sperling RA, 2014. Synergistic effect of β-Amyloid and neurodegeneration on cognitive decline in clinically normal individuals. JAMA Neurol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie X, Sun Y, Wan S, Zhao H, Liu R, Li X, Wu S, Nedelska Z, Hort J, Qing Z, Xu Y, Zhang B, 2017. Subregional structural alterations in hippocampus and nucleus accumbens correlate with the clinical impairment in patients with Alzheimer’s disease clinical spectrum: parallel combining volume and vertex-based approach. Front. Neurol 8, 399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M, 2011. A Bayesian model of shape and appearance for subcortical brain segmentation. Neuroimage 56, 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puonti O, Iglesias JE, Van Leemput K, 2016. Fast and sequence-adaptive whole-brain segmentation using parametric Bayesian modeling. Neuroimage 143, 235–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson R, Valindria VV, Bai W, Oktay O, Kainz B, Suzuki H, Sanghvi MM, Aung N, Paiva JM, Zemrak F, Fung K, Lukaschuk E, Lee AM, Carapella V, Kim YJ, Piechnik SK, Neubauer S, Petersen SE, Page C, Matthews PM, Rueckert D, Glocker B, 2019. Automated quality control in image segmentation: application to the UK Biobank cardiovascular magnetic resonance imaging study. J. Cardiovasc. Magn. Reson 21, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues L, Rezende T, Zanesco A, Hernandez AL, Franca M, Rittner L, 2020. Hypothalamus fully automatic segmentation from MR images using a U-Net based architecture. 15th International Symposium on Medical Information Processing and Analysis. [Google Scholar]
- Ronnenberger O, Fischer P, Brox T, 2015. U-net: convolutional networks for biomedical image segmentation. In: Navab N, Wells H,J, Frangi AW, (Eds.), International Conference on Medical Image Computing and Computer-Assisted Intervention 2015. Springer, pp. 234–241. [Google Scholar]
- Salat DH, Buckner RL, Snyder AZ, Greve DN, Desikan RS, Busa E, Morris JC, Dale AM, Fischl B, 2004. Thinning of the cerebral cortex in aging. Cereb. Cortex 14, 721–730. [DOI] [PubMed] [Google Scholar]
- Saygin ZM, Kliemann D, Iglesias JE, van der Kouwe AJW, Boyd E, Reuter M, Stevens A, Van Leemput K, McKee A, Frosch MP, Fischl B, Augustinack JC Alzheimer’s Disease Neuroimaging, I., 2017. High-resolution magnetic resonance imaging reveals nuclei of the human amygdala: manual segmentation to automatic atlas. Neuroimage 155, 370–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrout PE, Fleiss JL, 1979. Intraclass correlations: uses in assessing rater reliability. Psychol. Bull 86, 420–428. [DOI] [PubMed] [Google Scholar]
- Teipel S, Heinsen H, Amaro E Jr., Grinberg LT, Krause B, Grothe M Alzheimer’s Disease Neuroimaging, I., 2014. Cholinergic basal forebrain atrophy predicts amyloid burden in Alzheimer’s disease. Neurobiol. Aging 35, 482–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry RD, Katzman R, 1983. Senile dementia of the Alzheimer type. Ann. Neurol 14, 497–506. [DOI] [PubMed] [Google Scholar]
- Valindria VV, Lavdas I, Bai W, Kamnitsas K, Aboagye EO, Rockall AG, Rueckert D, Glocker B, 2017. Reverse classification accuracy: predicting segmentation performance in the absence of ground truth. IEEE Trans. Med. Imag 36, 1597–1606. [DOI] [PubMed] [Google Scholar]
- van der Kouwe AJ, Benner T, Salat DH, Fischl B, 2008. Brain morphometry with multiecho MPRAGE. Neuroimage 40, 559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Aisen PS, Jack CR Jr., Jagust WJ, Trojanowski JQ, Shaw L, Saykin AJ, Morris JC, Cairns N, Beckett LA, Toga A, Green R, Walter S, Soares H, Snyder P, Siemers E, Potter W, Cole PE, Schmidt M Alzheimer’s Disease Neuroimaging, I., 2010. The Alzheimer’s disease neuroimaging initiative: progress report and future plans. Alzheimers Dement 6, 202–211 e207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner MW, Veitch DP, Aisen PS, Beckett LA, Cairns NJ, Green RC, Harvey D, Jack CR Jr., Jagust W, Morris JC, Petersen RC, Saykin AJ, Shaw LM, Toga AW, Trojanowski JQ Alzheimer’s Disease Neuroimaging, I., 2017. Recent publications from the Alzheimer’s Disease Neuroimaging Initiative: reviewing progress toward improved AD clinical trials. Alzheimers Dement 13, e1–e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenzel F, Meyer C, Stehle T, Peters J, Siemonsen S, Thaler C, Zagorchev L Alzheimer’s Disease Neuroimaging, I., 2018. Rapid fully automatic segmentation of subcortical brain structures by shape-constrained surface adaptation. Med. Image Anal 46, 146–161. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The code to train and apply the model is available from github.com/BBillot/hypothalamus_seg. The model, code to apply it, and documentation are available at surfer.nmr.mgh.harvard.edu/fswiki/ScLimbic. Data used in the labeling can be requested from the corresponding author, subject to a data use agreement. Data from the 1000 Functional Connectomes data base is publicly available from fcon_1000.projects.nitrc.org/fcpClassic/FcpTable.html. Alzheimer’s Disease Neuroimaging Initiative (ADNI) data available from adni.loni.usc.edu. Data from the Harvard Aging Brain Study (HABS) can be requested from habs.mgh.harvard.edu/researchers/request-data.


