Abstract
Type IV CRISPR-Cas are a distinct variety of highly derived CRISPR-Cas systems that appear to have evolved from type III systems through the loss of the target-cleaving nuclease and partial deterioration of the large subunit of the effector complex. All known type IV CRISPR-Cas systems are encoded on plasmids, integrative and conjugative elements (ICEs), or prophages, and are thought to contribute to competition between these elements, although the mechanistic details of their function remain unknown. There is a clear parallel between the compositions and likely origin of type IV and type I systems recruited by Tn7-like transposons and mediating RNA-guided transposition. We investigated the diversity and evolutionary relationships of type IV systems, with a focus on those in Acidithiobacillia, where this variety of CRISPR is particularly abundant and always found on ICEs. Our analysis revealed remarkable evolutionary plasticity of type IV CRISPR-Cas systems, with adaptation and ancillary genes originating from different ancestral CRISPR-Cas varieties, and extensive gene shuffling within the type IV loci. The adaptation module and the CRISPR array apparently were lost in the type IV ancestor but were subsequently recaptured by type IV systems on several independent occasions. We demonstrate a high level of heterogeneity among the repeats with type IV CRISPR arrays, which far exceed the heterogeneity of any other known CRISPR repeats and suggest a unique adaptation mechanism. The spacers in the type IV arrays, for which protospacers could be identified, match plasmid genes, in particular those encoding the conjugation apparatus components. Both the biochemical mechanism of type IV CRISPR-Cas function and their role in the competition among mobile genetic elements remain to be investigated.
Introduction
Class 1 CRISPR/Cas systems are divided into three types (I, III, and IV), each of which is characterized by a unique signature gene.1 The prototype of the type IV systems is the CRISPR-Cas locus of the type strain (ATCC 23270) of Acidithiobacillus ferrooxidans,2 which resides in an integrative and conjugative element (ICE)3 inserted into the chromosome of this model acidophile.4,5 This CRISPR-Cas system has been designated AFERR, and associated genes + were named Csf (CRISPR-Cas subtype as in A. ferrooxidans). Currently, type IV CRISPR-Cas systems are classified into three types (A, B, and C) that have distinct characteristic operon organizations.1 Recently, however, it has been proposed to reclassify some of these systems into new subtypes IV-D and IV-E, and to split subtype IV-A into three variants based on differences in operon organization.6
Similar to other class 1 CRISPR-Cas systems, all type IV systems encompass homologs of Cas5 (Csf3) and Cas7 (Csf2) key subunits of the effector complex involved in crRNA binding.7,8 The originally described subtypes IV-A and IV-B share a signature protein, Csf1, that has been predicted to be structurally and functionally equivalent to the large subunits of other class 1 effector complexes, although it does not share any detectable sequence similarity with the latter and is much smaller compared to the large subunits of type I and type III systems.9 The recently introduced subtype IV-C shares the Cas5 and Cas7 components with IV-A and IV-B, but also possesses a distinct large subunit fused to an active HD family nuclease domain, similarly to Cas10, the large subunit of type III systems. This observation led to the hypothesis on the origin of type IV systems from type III systems.1 The signature protein of subtype IV-A systems is DinG family helicase, whereas the majority of the type IV-B loci encode a protein predicted to be a small subunit of effector complex, Cas11_IV.1 Cas6 is encoded in most type IV-A systems, whereas in the subtype IV-B loci, cas6 genes are scarce.6 Subtype IV-B systems are strongly linked to cysH gene, the functional role of which in the context of these CRISPR-Cas systems remains unclear.10,11 Many subtype IV-A systems include CRISPR arrays but typically lack the cas1 and cas2 adaptation genes and any associated nuclease that could be implicated in the target cleavage. These distinctive features imply that type IV systems do not provide adaptive immunity in the way canonical CRISPR-Cas systems can and instead could have been recruited for non-defense functions or defense functions not involving target cleavage.11 Most type IV systems are associated with phages or plasmids and have been proposed to be involved in plasmid competition.12,13
Experimental data on type IV systems are scarce. Type IV CRISPR RNA (crRNA) production and maturation has been demonstrated in high-throughput RNAseq assays.7,8 Experiments in Escherichia coli have shown that processing of the EbN1 pre-crRNA is mediated by the cognate type IV Cas6 and that the crRNAs are specifically incorporated into the type IV effector complex.7,8 The Cas6 protein associated with type IV CRISPR-Cas from Mahella australiensis has been studied, and its structure has been solved.14 The structure of type IV-A effector complex from Aromatoleum aromaticum EbN1 was partially solved, and the predicted organization of the effector complex resembling those of type I and type III effector complexes has been confirmed.7 Most recently, a partial structure of type IV-B effector complex from Mycobacterium sp. JS623 type IV-B has been solved, and it has been shown that the type IV effector complex can be associated with heterogeneous small RNAs arranged in a pseudo-A-form configuration.8 Although the functional mode of type IV CRISPR-Cas systems remains unclear, it has been shown that the system from Pseudomonas aeruginosa strain PA83 mediates RNA-guided interference against a plasmid in vivo, both clearing and inhibiting the plasmid, and a crucial role of DinG helicase in this activity has been demonstrated.15
Several lines of evidence provide clues to potential alternative functions of the type IV CRISPR-Cas systems distinct from adaptive immunity. Most of the type IV CRISPR-Cas systems are located on plasmids,12 megaplasmids,7 ICEs,3,16 and, in several cases, prophages.6,10,11 This trend suggests that they contribute to mobile genetic elements–host interaction, and in particular could inhibit host defense, including resident CRISPR-Cas, or contribute to the maintenance and/or enhance the mobility of plasmids via as yet unknown mechanisms.6,10,11 Spacer target analysis of one such system partially supports this view. The IncHI1B/IncFIB plasmid-encoded type IV CRISPR from Klebsiella strains have been recently shown to harbor a number of spacers that match conjugational transfer genes traN and traL of IncFIIK/IncFIB(K) plasmids, suggesting a role in plasmid incompatibility.12 In general, however, the functions of type IV CRISPR-Cas systems remain poorly understood.
Here, we explore in detail the type IV CRISPR-Cas loci in several species and strains of the Acidithiobacillia class17 and reexamine the diversity, classification, and evolution of type IV systems. We demonstrate a remarkable evolutionary plasticity of type IV CRISPR-Cas, including borrowing genes from other CRISPR-Cas types and extensive shuffling of gene modules, along with the previously undetected extensive polymorphism of CRISPR repeats in type IV arrays that has no precedent in other types of CRISPR-Cas systems.
Methods
Type IV CRISPR-Cas loci
The set of genomes analyzed in this work combines sequences reported to contain previously identified type IV loci1,6 and sequences recently added to the NCBI database from Acidithiobacillia class,17 which were downloaded from NCBI FTP site (ftp://ftp.ncbi.nlm.nih.gov/genomes/all/) and are listed in Supplementary Table S1. Protein-coding genes were annotated automatically using PSI-BLAST,18 with a 10e-4 e-value cutoff and effective database size of 2 × 107. Combined profiles from NCBI CDD database19 and CRISPR-Cas profiles described previously1 were used as queries. The proteins were annotated according to the best scoring match. The HHpred program was used to identify distantly related sequences for selected genes encoded in type IV loci.20 Additionally, CRISPRCasTyper21 was used for the identification of Cas proteins and CRISPR-Cas system subtype assignments, and the annotation of several loci was manually adjusted according to the recently proposed classification.6
CRISPR array detection
CRISPR arrays were identified using the minCED tool (https://github.com/ctSkennerton/minced), which is a modification of the CRT CRISPR recognition tool22 using default parameters. Additionally, for analysis of the Acidithiobacillia class arrays, the CRISPRfinder online tool was employed,23 followed by extensive manual curation of the arrays.
Search for CRISPR protospacers
To reduce redundancy, spacers recovered from the CRISPR arrays were self-compared using BLASTN. Unique spacer sequences were analyzed using CRISPR Target24 and used as queries in BLASTN searches against the NCBI nr nucleotide database as previously described1 in order to identify candidate target protospacers. Hits with sequence identity >95% and query coverage >95% were retained for further analysis. Identical BLASTN hits mapping to spacer sequences in CRISPR arrays were removed. Candidate protospacer sequences mapping to intergenic versus intragenic regions were scored and analyzed in further detail.
Phylogenetic analysis
To select gene representatives, protein sequences for the selected genes were clustered with MMseqs225 with a sequence similarity threshold of 0.98 and a coverage threshold of 0.333. Representative sequences were aligned using MUSCLE.26 These alignments were used as input for phylogenetic tree construction using IQ-TREE27 with the TEST model-finder and 2,000 ultrafast bootstrap iterations. Combined FastTree28 and UPGMA trees were built using the previously described approach.1
Repeat polymorphism analysis
Repeat polymorphism was estimated as the sum of the number of mismatches between a repeat and the consensus repeat sequence for each repeat in an array divided by the number of repeats. Alignments for the repeats were constructed using MAFFT29 with the adjustdirectionaccurately parameter. Gaps in the alignments were not counted as mismatches and were discarded from the calculation. For each array, the repeat with the highest number of mismatches (degenerate repeat) was discarded from the calculations as well. The source data for the type IV repeats were collected from the arrays analyzed in this work, and the other CRISPR-Cas types are represented by the arrays assembled previously.1
Expression of the type IV CRISPR-Cas system
Fervidacidithiobacillus caldus ATCC 51756 (proposed nomenclatural emendation17) was cultured in mineral salts medium (MSM) with trace elements at pH 2.5 in the presence of sterile-filtered tetrathionate (5 mM) or 5 g/L of elemental sulfur. Stock solutions of tetrathionate were sterile filtered and added to the autoclaved (121°C for 15 min) MSM, whereas the finely ground ethanol-sterilized powdered sulfur was added to MSM prior to autoclaving at 105°C for 30 min. All F. caldus cultures were incubated at 40°C under aerobic conditions on a rotary shaker at 150 rpm. Cells were collected in the logarithmic growth phase (Log) 3 days after inoculation and at stationary phase (Stat) after 7 days. Sessile versus planktonic growth was achieved in 6 cm long and 2.5 cm diameter columns containing a 2:1 w/w mix of elemental sulfur and quartz. Mid-exponential sulfur-grown cultures were passed through the column at a continuous flow of 0.029 m3/(0.015 m2 × 20 seg) = 0.096 m/s for 7 days. Planktonic cells were collected from flow phase and sessile cells were separated from mineral substrate by thorough washing using a solution of MSM medium and 1% sodium dodecyl sulfate (SDS) and gentle vortexing. RNA isolation, reverse transcription (RT) polymerase chain reaction (PCR), and real-time PCR were carried out following standard protocols as described by Nieto et al.30 Briefly, cells were collected and re-suspended in ice-cold buffer TE (25:10), pH 8.0, with 1 × extraction buffer (per liter: 1% SDS, 50 mM Tris-HCl, pH 8.0, and 2 mM EDTA). After cell lysis, the suspensions were treated with TRIzol (Invitrogen), followed by two extractions with acid phenol and chloroform. RNA was precipitated with absolute ethanol overnight at −20°C, washed with 70% ethanol, and finally re-suspended in sterilized water. Samples were treated with DNase and purified with the Roche High Pure RNA Isolation Kit, following the manufacturer's recommendations. DNA-free high-quality RNA was stored at −80°C for downstream applications. Oligonucleotide primers used in the study are listed in Supplementary Table S2. Copy DNA (cDNA) was prepared from 3 μg total RNA using Superscript II reverse transcriptase (Thermo Fisher Scientific) according to the manufacturer's instructions. PCR products were amplified with proofreading DNA polymerase Dreamtaq (Thermo Fisher Scientific) in 25 μL reactions (1 × PCR buffer +1.5 mM MgCl2) containing 30 ng template cDNA, 10 μM required primers, and 0.2 mM each deoxyribonucleotides. PCR amplification conditions were as follows: initial denaturing step at 95°C for 5 min followed by 28–30 amplification cycles (denaturation at 95°C for 30 s, annealing at the appropriate temperature depending on the specific primers pairs for 30 s, and elongation at 72°C for 1 min), and a final elongation step at 72°C for 3 min. The real-time PCR reactions were performed in the RotorGene Q PCR System (Qiagen) using the KAPA SYBR FAST qPCR Kit (Roche). The 20 μL PCR reactions contained 2 μL undiluted cDNA, 200 nM each primer, and 1 × KAPA MasterMix. The cycling protocol was as follows: initial denaturation for 10 min at 95°C followed by 40 cycles of 30 s at 95°C, 15 s at 60°C, and 30 s at 72°C. Fluorescence was measured after the extension phase at 72°C. The amplification products were subjected to a melting curve analysis, and specific amplification was confirmed by a single peak in the melting curve. The reactions for each target gene were performed in triplicate and in the same PCR run, including a no template control. Amplification efficiency was calculated from a standard curve constructed by amplifying serial dilutions of genomic DNA for each gene. These values were used to obtain the fold change in expression between conditions (sessile versus planktonic) for the genes of interest normalized against rpoC gene expression levels according to Nieto et al.30
Results
Comparative genomic and phylogenetic analysis of type IV loci core genes
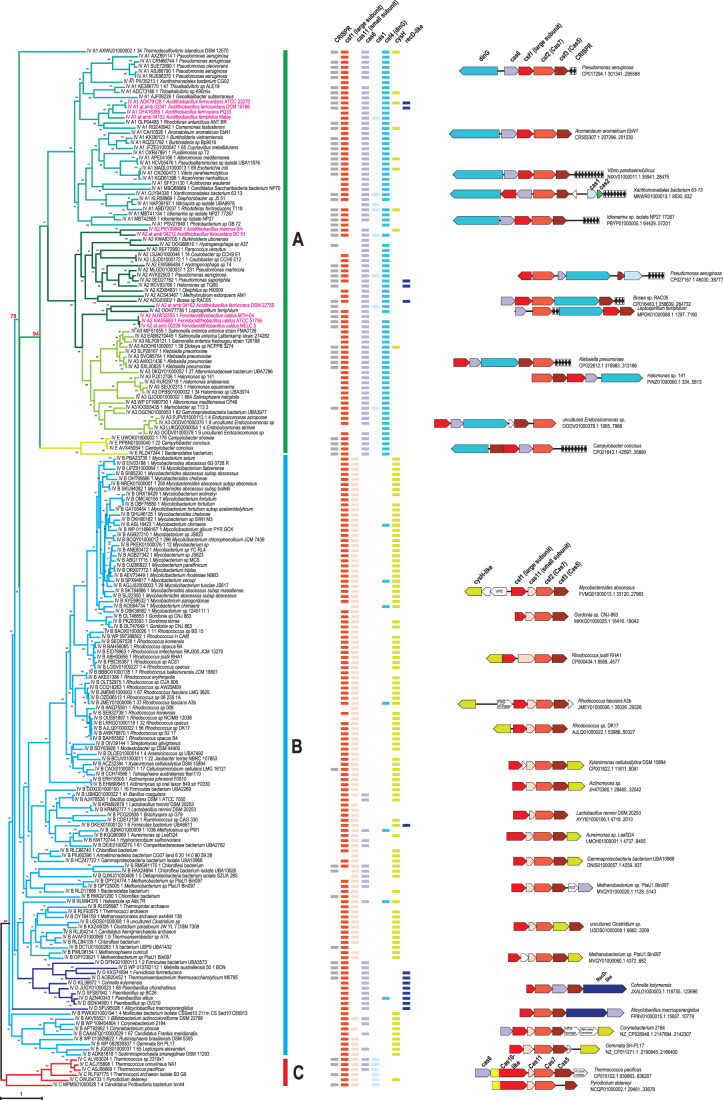
We analyzed a data set consisting of 856 type IV loci, including 12 novel loci identified in recently sequenced genomes of Acidithiobacillia class species (Supplementary Tables S1 and S3). The extended loci (30 genes upstream and downstream of the type IV core genes) were automatically annotated as described in the Methods, and selected protein families were analyzed in detail using sensitive computational methods, such as PSI-BLAST and HHpred, to search for potential homologs. First, we reconstructed phylogenetic trees for representative sets of Csf2/Cas7 and Csf3/Cas5 proteins (Fig. 1 and Supplementary Fig. S1A). Because Cas7 is the most highly conserved protein among the type IV components, Cas7 phylogeny was used as the framework to classify type IV systems and explore their evolution. The signature protein families, such as Csf4/DinG (signature of subtype IV-A), putative small subunit, Cas11 (signature of subtype IV-B), and the Cas10-like large subunit (signature of subtype IV-C), CRISPR repeats, as well as typical organizations of core genes, were mapped on the Cas7 tree (Fig. 1), revealing distinct loci configurations between the major branches. It should be emphasized that, as detailed previously1 and confirmed in the current analysis of type IV, to classify CRISPR-Cas systems unequivocally into subtypes, signature genes alone are insufficient. Multiple features, such as locus organization and presence of additional genes and CRISPR arrays, have to be taken into account, along with the phylogenies of conserved cas genes.
FIG. 1.
(A–C) Phylogenetic analysis of Cas7 (Csf2) and comparative genomic analysis of type IV systems. Phylogenetic tree for 204 representatives of Cas7 (Csf2) shown on the left was built using the IQ-TREE method as described in the Methods. The branches are colored according to the recently proposed classification of the type IV systems.6 Each leaf is denoted by subtype (IV-A1, IV-A2, IV-A3, IV-B, IV-C, IV-D, IV-E) as recently proposed,6 protein identifier, and species name. Supporting values were calculated by the IQ-TREE program. Several key values supporting monophyly of type IV-A and type IV-C are highlighted in red. The colored lines behind the tree show the amended proposal for classification of these systems based on this work as follows: green, type IV-A; blue, type IV-B; and red, type IV-C. The phyletic pattern (presence of a gene in the type IV locus in the respective genome) is shown by rectangles, which are color coded according to the legend above. Representative type IV core gene neighborhoods are shown behind the patterns and color coded according to the gene designation shown for each system once. The genes and CRISPR arrays are shown to scale, and accession for respective genomic partition, species name, and coordinates of the region are indicated on the right. A few genes that are inserted into type IV loci are shown by blank arrows, with short name (if any) indicated inside the arrow. VIP2, ADP phosphoribosyltransferase VIP2; FlhG, MinD-like ATPase involved in chromosome partitioning or flagellar assembly; HTH, helix-turn-helix, DNA binding; TGT, queuine tRNA-ribosyltransferase. Note: To visualize the details in this figure, a 200% zoom is recommended.
In the Cas7 tree, two of the current major subtypes, IV-A and IV-C, are monophyletic, whereas the subtype IV-B clade also includes the proposed new subtype IV-D (Fig. 1). Among the branches that have been proposed to be reclassified as new subtypes,6 only IV-E and IV-A3 are strictly monophyletic, although most of the loci assigned to IV-A1 and IV-D also form clades. In addition, the proposed new subtype IV-E confidently groups with the IV-A branch (Fig. 1).
In the phylogeny of Cas5, a protein that is less strongly conserved at the sequence level than Cas7, none of the major subtypes are monophyletic (although in subtypes A and C, there are only a few outliers), whereas among the proposed new subtypes and variants, E and A3 are monophyletic (Supplementary Fig. S1A). In this tree, the proposed subtype D clearly splits into two clades, one of which falls within the subtype A clade, whereas the other one groups with the proposed subtype IV-E (with support values >70%). Another example of potential gene shuffling is the confident placement of subtype IV-C Cas5 from the Candidatus Poribacteria bacterium within the IV-B branch (Fig. 1 and Supplementary Fig. S1A).
Putative large subunits (Csf1-like and Cas10-like subunits of type IV-C systems) associated with type IV systems are highly diverse and are often difficult to identify. In the previous analysis by Pinilla-Redondo et al.,6 the large subunit has not been identified in the proposed IV-A2 group, which was one of the criteria for making this group a separate variant. Using both PSI-BLAST and HHpred, we were able to identify this subunit, which is encoded divergently to the csf2 gene (Cas7), in all proposed IV-A2 systems (Fig. 1 and Supplementary Fig. S2). To analyze the relationships between the large subunits, we built a dendrogram using a combination of HHalign scores and the standard phylogenetic approach employed for clusters with alignable sequences (Supplementary Fig. S1B). None of the major subtypes—A, B, and C—formed clades in the resulting tree, although the newly proposed subtype E and the variant A3 did. Similar to the Cas5 tree, the proposed subtype D split into two branches with apparently different origins, but both with affinity to Csf1-like proteins from subtype B. Our analysis therefore indicates that the large subunit sequences of the proposed variant A1 and subtype D group together with the homologous sequences of subtype B, whereas most of the Csf1-like proteins of the proposed variants A2 and A3 are similar to each other but distant from A1 (Supplementary Fig. S1B). There are also smaller branches in this tree, which correspond to highly diverged variants of Csf1 (Supplementary Fig. S1B). Most likely, this can be explained by differences in the evolutionary rates of csf1-like genes in different groups. Also, as in the case of the Cas5 component, these observations suggest that different genes in the proposed subtype IV-D have different origins, making these loci hybrid.
In agreement with previous observations, putative small subunits were identified in the vast majority of type IV-B systems and in all IV-C systems but not in any of the type IV-A systems (Fig. 1 and Supplementary Table S3). Despite the confident placement of both Cas7 and Csf1 components of the proposed subtype IV-D within the IV-B clade, no putative small subunits were detected in the IV-D loci.
In accordance with the previous phylogenetic analysis of Cas6,6 we observed a remarkable diversity of cas6 genes associated with type IV loci. There are at least six major clades of Cas6 encoded in these loci and several smaller clades that include diverged Cas6 variants (Supplementary Fig. S3 and Table S3). Only subtypes type IV-C and the proposed subtype IV-E and variant IV-A3 were monophyletic in the Cas6 phylogeny (Supplementary Fig. S3). These observations imply a complex evolutionary history of Cas6 in type IV systems, following the apparent loss of Cas6 at the onset of type IV evolution (see discussion below).
Generally, type IV loci are evolutionarily labile, with many cases of apparent gene shuffling, inversions, and insertions of various genes next to different cas genes such that these genes might be co-transcribed with the cas genes in the respective loci (Fig. 1 and Supplementary Table S3).
In addition to the genes encoding the effector complex subunits and cas6 genes, some type IV loci, mostly of the IV-A subtype, include the adaptation genes cas1 and cas2. Here, we also analyzed Cas1 sequences from type IV loci and identified two distinct phylogenetic affinities of these genes, one with subtype I-E and the other (only in Pseudomonas species) with I-F (Supplementary Fig. S4), which is also consistent with the analysis of CRISPR repeats in the type IV loci.6 These findings suggest two independent acquisitions of adaptation module genes during the evolution of type IV systems. Considering the patchy distribution of Cas1 in type IV loci along with the apparent origin of the type IV-associated cas1 genes from a distinct clade within subtype I-E, the provenance of the type IV adaptation modules can be explained either by multiple independent losses of ancestral Cas1 or, more likely, by secondary acquisitions and multiple recombination events between type IV loci (Fig. 1).
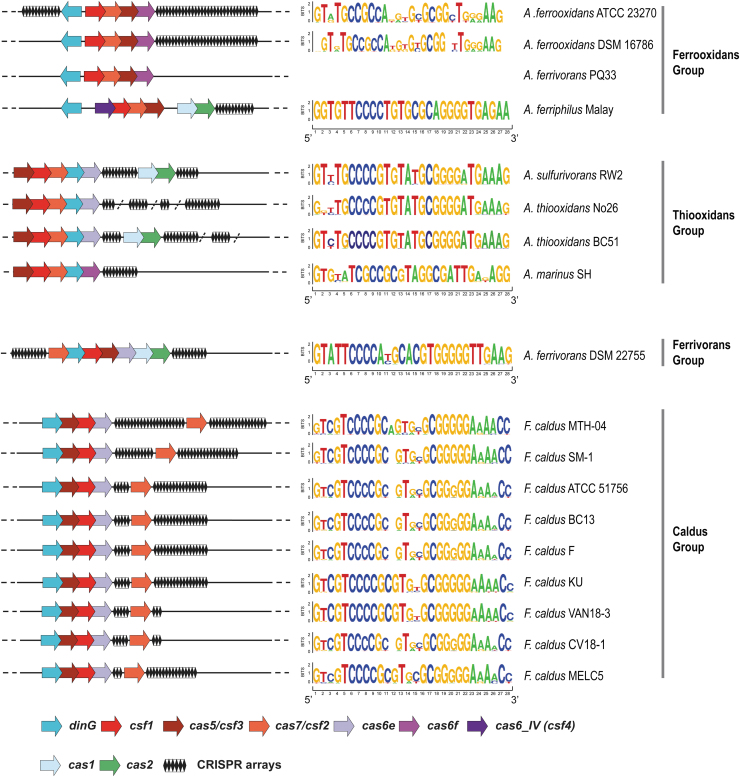
Acidithiobacillia class type IV systems are diverse and belong to four distinct groups (named after the most abundant species representative) as apparent from both the Cas7 and Cas5 phylogenies. The Caldus group, encompassing the loci from F. caldus strains and the Ferrivorans group represented by Acidithiobacillus ferrivorans DSM 22755T single loci, are close to one another in both these trees (Figs. 1 and 2 and Supplementary Fig. S1A). Type IV CRISPR-Cas systems in these groups show considerable differences with respect to the arrangement of the core genes as well as the location and size of the CRISPR arrays (Fig. 2). Moreover, even within one group, we observed extensive gene shuffling within the type IV loci. For example, the order of the cas genes in Acidithiobacillus ferriphilus Malay differs from that in the other strains in the Ferrooxidans group. Furthermore, the Cas1–Cas2 module is present in some loci but not in others. Considering that most of the cas1 genes from these loci belong to the same branch in the Cas1 phylogeny, it appears that the adaptation module is exchanged among the type IV loci (Fig. 2 and Supplementary Fig. S4). The exception is the Cas1 from A. ferriphilus Malay, which is the deepest branch in the IV-A Cas1 subtree (Supplementary Fig. S4). Interestingly, the CRISPR array in this strain is the only one in type IV systems that contains identical repeats (see below; Fig. 2). Apart from gene shuffling, the evolution of type IV system in the Acidithiobacillia was apparently affected by changes in evolutionary rates that might correspond to functional shifts. Thus, type IV loci from the Thiooxidans group encode a highly divergent Csf1 protein, suggesting rapid evolution of this gene (Fig. 2 and Supplementary Fig. S1B).
FIG. 2.
Schematic representation of the diversity of type IV CRISPR-Cas systems in Acidithiobacillia class species and strains. The core type IV-A genes are shown by arrows, and CRISPR arrays are shown as diamonds. The genes are not shown to scale. On the right side, the Seqlogos (constructed using WebLogo interface https://weblogo.berkeley.edu/logo.cgi) of the repeats in all arrays in the respective type IV-locus are shown. CRISPR arrays found elsewhere in the genome or in unassembled contigs are separated by a slash.
Genes tightly associated with type IV loci
As noticed previously, in addition to the effector complex genes, most of the subtype IV-A loci encode the DinG helicase, which is the signature of this subtype. This association was confirmed in the present analysis (Fig. 1), and indeed, it has been shown that DinG is necessary for interference against plasmids.15
The cysH-like genes that have been identified in many neighborhoods of type IV-B loci are less stably associated with the IV-B systems but are often also present in the vicinity of type IV-A systems (Fig. 1). The CysH-like proteins are members of the adenine nucleotide alpha hydrolase family, which includes homologs of 3′-phosphoadenosine-5′-phosphosulfate (PAPS)-reductase, the key enzyme of the sulfate-assimilation pathway and cysteine biosynthesis, N-type ATP PPases, and ATP sulfurilases.31 These enzymes are also components of the antiphage defense DNA phosphorothioation system32,33 and, furthermore, are also found in some phages and are thought to facilitate inorganic sulphate assimilation by their bacterial hosts.34–36
We constructed a phylogenetic tree of CysH-like proteins and reproduced our previous observations10 that CysH-like genes associated with subtype IV-B loci are largely monophyletic, but those found in the extended subtype IV-A loci are not (Supplementary Fig. S5A and Table S3). We also found that these proteins belong to a large clade that consists of CysH-like proteins that are typically encoded in mobile genetic elements (MGE), such as plasmids, phages, and other integrated elements, unlike the bona fide CysH (APS/PAPS reductase), which is involved in sulfate assimilation and forms a compact clade in the CysH tree (Supplementary Fig. S5A). Examination of the multiple alignment shows that unlike APS/PAPS reductases, most of the CysH-like proteins in this clade, including all those associated with type IV CRISPR-Cas, contain the sulfonucleotide-binding domain but lack the conserved cysteine of the active site in the C-terminal catalytic domain of the reductase (Supplementary Fig. S6).37 Thus, the type IV associated CysH-like proteins, as well as most of the other proteins in this clade, are not PAPS reductases but rather, most likely, sulfonucleotide-binding proteins. Similar observations have been recently published by Taylor et al.,38 who hypothesized that the role of the CysH-like protein in type IV systems is to stabilize the AMPylation of specific substrates through the ATP α-hydrolase activity. The actual activities and functions of the CysH-like proteins in plasmids and other MGE remain to be studied experimentally.
Another gene of interest is the RecD homolog that has been suggested as a signature for the proposed subtype IV-D.6 Indeed, this gene is found in the predicted operons of all these systems, although it is often present also within extended loci of many other type IV systems (Fig. 1 and Supplementary Tables S1 and S3). As in all of the Cas protein trees discussed above, apart from Cas7, the proposed subtype D is not monophyletic in the RecD family tree, suggesting at least three independent incorporations of the recD gene into the respective type IV operons (Supplementary Fig. S5B). Similar to CysH homologs, RecD homologs that are encoded in the type IV loci belong to a distinct clade enriched in proteins encoded in MGE, as opposed to another major clade that consists of bona fide bacterial RecD helicases, components of the exonuclease V DNA repair complex RecBCD. Furthermore, the plasmid relaxase TraA,39 a RecD homolog, belongs to the same clade as the RecD homologs from type IV loci (Supplementary Fig. S5B). The nature of the functional link between RecD and type IV systems, if any, remains obscure. However, it cannot be ruled out that RecD continues to function as a plasmid relaxase and is associated with type IV systems, since both genes are transcribed at the same time during the plasmid life cycle, a phenomenon known as gene hitch-hiking.40
Extended neighborhoods of type IV effector genes
Analysis of extended surroundings of type IV core genes revealed little conservation. This is not surprising, considering that type IV systems are encoded on a variety of plasmids, prophages, and other integrated elements as well as at least one phage, bacteriophage SPI1 from Skermania piniformis.41 All these MGE are enriched in poorly conserved and fast-evolving genes. Indeed, search of the neighboring gene products for domains from the pfam, COGs, CDD, or Cas database showed a sharp drop in the density of detected domains outside of the type IV loci core and the tightly associated genes, such as cysH (Supplementary Fig. S7). These searches detected primarily genes that are typical of MGE, such as parA and parB involved in plasmid partitioning42 and xerD-like integrases (Table 1).43 Analysis of tighter gene clusters (MMseq identity threshold 0.5) yielded an overlapping but distinct set, dominated by functionally characterized genes (Table 1). The two most abundant protein families in this list, namely Vip2-like ADP phosphoribosyltransferase (e.g., EIV03191.1) and an uncharacterized protein family (e.g., EIV03192.1), have been identified in our previous analysis of genes linked to CRISPR loci.10 Both protein sets included a MoxR-like ATPase that has been recently described as a hub in a network of functional systems implicated in various biological conflicts.44 Specifically, a three-component module that, in addition to MoxR includes a Zincin superfamily metallopeptidase fused to a vWA domain (e.g., OHT95645.1) and a putative toxin precursor (e.g., OHT95646.1), is most often present in the vicinity of type IV loci. Generally, however, the type IV loci are found in “dark matter” islands, such that the neighboring genes are present in only a few other islands, suggesting that these loci are prone to frequent gene shuffling and are not essential for the propagation of the respective MGE, but rather are more or less random clusters of genes involved in anti-defense functions and inter-MGE competition (Supplementary Table S3).
Table 1.
Protein Families Most Often Encoded in the Extended Gene Type IV Neighborhoods
| Cluster/family ID | Weighted frequency | Comment |
|---|---|---|
| MMseq 0.5 clusters | ||
| CLUSTER_52 | 28.7 | CysH-like |
| CLUSTER_53 | 28.7 | ADP phosphoribosyltransferase VIP2-like |
| CLUSTER_28 | 22.3 | MoxR-like ATPase |
| CLUSTER_141 | 21.3 | Unknown |
| CLUSTER_40 | 19.4 | MoxR associated zincin metallopeptidase fused vWFA domain |
| CLUSTER_33 | 18.8 | DNA2-like Helicase |
| CLUSTER_256 | 13.7 | Uncharacterized DUF1870 |
| CLUSTER_46 | 13.4 | CysH-like |
| CLUSTER_119 | 11.8 | MoxR associated (precursor releasing small C-terminal peptide) |
| CLUSTER_126 | 9.4 | MoxR associated (precursor releasing small C-terminal peptide) |
| CDD assignments | ||
| COG0175 | 103.17 | CysH-like |
| COG1199 | 76.0 | DinG |
| COG1674 | 33.5 | DNA segregation ATPase FtsK/SpoIIIE |
| COG1396 | 30.7 | XRE-family HTH domain |
| COG4974 | 29.7 | Site-specific recombinase XerD |
| COG1475 | 27.9 | Chromosome segregation protein Spo0J, contains ParB-like nuclease domain |
| COG0714 | 26.7 | MoxR-like ATPase |
| COG1192 | 24.4 | Chromosome segregation ATPase ParA |
| COG2801 | 24.0 | Transposase InsO |
| COG1028 | 24.0 | NAD(P)-dependent dehydrogenase |
| cd00093 | 23.3 | Helix-turn-helix XRE-family like proteins. |
| COG0582 | 20.7 | Site-specific recombinase XerC |
| pfam07510 | 20.5 | Protein of unknown function (DUF1524), predicted His-Me finger endonuclease |
| COG3864 | 20.4 | Zincin metal-dependent peptidase, MoxR associated |
Comparison of type IV CRISPR-Cas loci in Acidithiobacillia class species
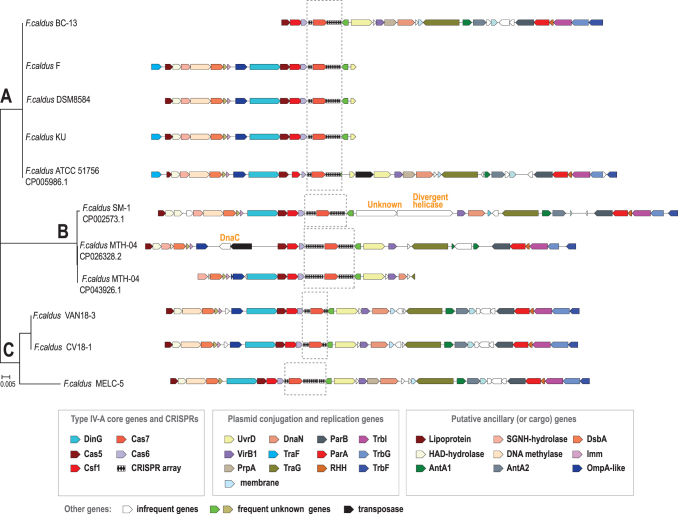
As indicated by the presence of characteristic genes of the type 4 conjugative transfer system (e.g., virB4, trb, and tra genes), all type IV CRISPR-Cas loci in Acidithiobacillia class species are located in ICEs on the bacterial chromosome (Fig. 2 and Supplementary Table S3).45–48 In A. ferrooxidans ATCC 23270T, the type IV locus occurs in ICEAfe1, a well-characterized ICE,3,43,45 whereas in F. caldus ATCC 51756T it occurs within the ICEAca1.2.16 Both ICEs are large (183–291 Kbp) and have been shown to be stably inherited and actively excised from the chromosome in response to DNA damage, and are likely prone to conjugative transfer. While ICEAfe1 is poorly conserved in Acidithiobacillus species and strains, ICEAca1.2 occurs in other F. caldus strains.45,49 The Caldus group is the largest and best represented among the available Acidithiobacillia class species, and the genome sequences can be confidently aligned on the nucleotide level. So, we selected this group for in-depth analysis of extended type IV loci and CRISPR arrays (Fig. 3 and Supplementary Table S4). The type IV systems in all these genomic islands are located upstream of the genes involved in DNA partitioning during replication (parA, parB, dnaN), in plasmid/ICE DNA rolling circle replication (uvrD), and in conjugative DNA transfer (trbE, traG, trbI, trbG, trbF). The region upstream of the uvrD-like gene, containing the type IV cas genes, shows signs of instability, with some deletions and insertions (Fig. 3). One such insertion disrupts the dinG gene in F. caldus MTH-04. Surprisingly, some of the insertions include genes that have not been previously detected in Fervidacidithiobacillus and or even the closely related Acidithiobacillales (Fig. 3). General context conservation with regional instability extends to other strains of the species (Supplementary Table S3). For example, two genes encoding very large proteins (AEK57865.1, 1,328 aa; AEK57866.1, 971 aa) were inserted in place of the uvrD gene in F. caldus SM-1, CP002573.1 (Fig. 3). One of these genes encodes a highly diverged helicase, whereas the other one shows no similarity with known protein families. The closest homologs of both proteins were detected in cyanobacteria (as indicated by top scoring hits in BLAST searches; WP_009786411.1: e-value 5e-41, 26.01% identity and WP_137908226.1: e-value 1e-07, 21.61% identity, respectively).
FIG. 3.
(A–C) Comparative analysis of extended type IV loci in Fervidacidithiobacillus caldus strains. The extended type IV-A gene neighborhoods from integrative and conjugative elements from closely related F. caldus strains are mapped to the cas6 nucleotide sequence-based tree, which was constructed using FastTree. The designations are the same as in the Figure 1. The color key and names or short descriptions of the respective genes are given underneath the schematic. A few rare genes that were inserted in the closest vicinity of type IV genes are indicated in orange above the respective blank arrows. Gray dashed lines indicate the regions that are aligned in Supplementary Figure S7.
Specific functions of most of these genes are unclear, although they typically belong to known large protein families, such as HAD and alpha/beta superfamily hydrolases, and DsbA-like thioredoxins. However, a potential functional clue comes from the observation that these genes form a putative operon with the gene coding for the superinfection immunity protein Imm (Fig. 3). This membrane protein has been identified and functionally studied in the T4 bacteriophage, and has been shown to inhibit, directly or indirectly, phage DNA injection into the host cell, so that deletion of the imm gene resulted in a phage unable to protect cells from superinfection.50 Therefore, it seems plausible that all the genes between the lipoprotein and subtype IV-A CRISPR-Cas are involved in functions related to plasmid competition and anti-defense response, clustering together in defense islands similarly to other recently described anti-MGEs genes.51–53
Polymorphic repeats in type IV CRISPR arrays of the Acidithiobacillia
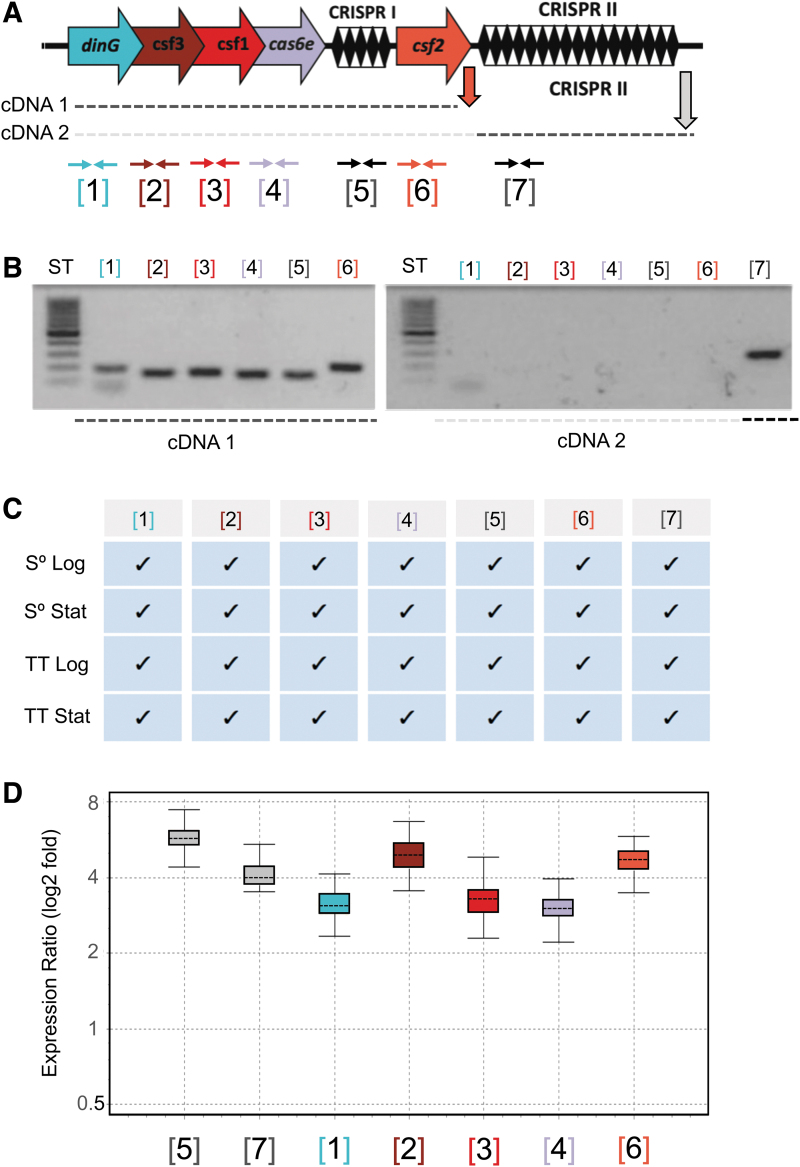
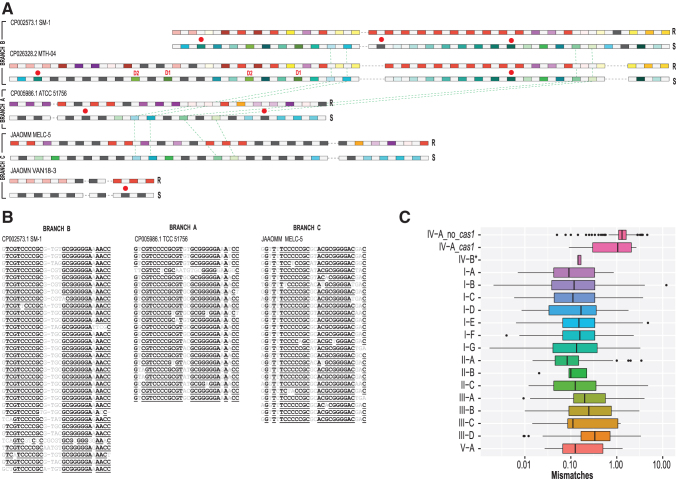
To explore the evolution of arrays and spacer acquisition dynamics, we analyzed the CRISPR arrays located upstream and/or downstream of the csf2 (cas7) gene (Fig. 3 and Supplementary Fig. S8). A tree was built from the alignment of the cas6 nucleotide sequences in order to determine the relationships between the type IV loci in the “Caldus group” (Fig. 3). In this case, the tree based on the alignment of the cas7 nucleotide sequences had the exact same topology as the cas6 tree (Supplementary Fig. S9). The three branches of this tree (A, B, and C) include five, three, and three loci, respectively. All the sequences of the entire region within the A branch were identical, but the spacer sequences differed between branches A and C, suggestive of an active process of spacer acquisition. Transcriptional expression of the A-branch loci components from representative strains (F. caldus ATCC 51756/ DSM 8584) has been demonstrated by RT-PCR, quantitative RT-PCR, and/or RNASeq under diverse growth conditions, being most prominent in attached cells (3–6 × higher) during biofilm formation (Fig. 4 and Supplementary Fig. S10).
FIG. 4.
(A–D) Expression of the type IV CRISPR-Cas system of F. caldus ATCC 51756T. Generic scheme showing the F. caldus CRISPR-Cas system, the cDNAs obtained by reverse transcription (dotted lines), and reverse transcription (RT) polymerase chain reaction (PCR; vertical arrows) and PCR primers position (horizontal arrows), numbered according to the position of the different features in the loci. Primers used in each reaction are detailed in Supplementary Table S2. (A) Analysis of the RT-PCR-amplified products by agarose gel electrophoresis. Total RNA was extracted from mid-log sulfur-grown cultures (pH 2.5; 40°C, 150 rpm; mineral salts medium) and used as template in cDNA synthesis with reverse primers indicated in panel (A). (B) Summary of RT-PCR amplification results for total RNA extracted from cells grown under different growth conditions. Cultures were grown in the presence of either sulfur (S°) or tetrathionate (TT) as energy sources, and cells were collected at two distinct growth phases, logarithmic (Log) and stationary (Stat). Positive amplification is indicated by a checkmark. (C) Fold induction of each component as assessed by quantitative RT-PCR for total RNA recovered from sessile (biofilm) versus planktonic grown cells (achieved in 6 cm long and 2.5 cm diameter columns containing elemental sulfur-quartz). Gene expression levels in each condition were normalized against a reference gene (rpoC, “housekeeping”), and normalized values were used in the calculation of the expression ratio sessile/planktonic.
To compare repeat and spacer sequences, we clustered them as follows: repeats were joined in a cluster if they were identical, and spacers were clustered if they were at least 90% identical. This clustering revealed a complex picture of array evolution. The 125 repeats in these loci fell into 42 clusters of identical sequences, demonstrating remarkable heterogeneity of repeats within arrays (Fig. 5A and Supplementary Table S4). Typically, different repeats contain several nucleotide substitutions compared to the consensus while retaining the palindromic structure (Fig. 5B). Identical repeats were often scattered along the arrays, and their location differed in the arrays from different branches in the tree. Such polymorphic repeats have not been observed in CRISPR arrays to date and seem to be poorly compatible with the preferential spacer acquisition from one end of the array, accompanied by repeat propagation, which is typically observed in other CRISPR-Cas types (Fig. 5A). Polymorphism of type IV CRISPR repeats also extends to other Acidithiobacillia class species (Supplementary Table S4).
FIG. 5.
Comparative analysis of repeats and spacers in F. caldus strains and repeat polymorphism in type IV arrays in general. (A) CRISPR arrays for five unique loci. For each CRISPR array, two lines representing repeats (above) and spacers (yellow) are shown. In the first line, repeats (R) are color coded as follows: identical repeats that occur twice or more are shown by rectangles of the same color, unique repeats are shown by dark gray rectangles, and spacers are shown by light gray rectangles. In the second line, spacers (S) are color coded as follows: spacers that are 90% identical and occur twice or more are shown by rectangles of the same color, unique spacers are shown by dark gray rectangles, and repeats are shown by light gray rectangles. Two parts of CRISPR arrays, upstream and downstream of csf2 gene and interrupted by insertions, are separated by a dashed line (see Fig. 3). Duplicated spacers are indicated by D1 and D2, respectively. Red circles indicate spacers with protospacers identified. Green dashed lines indicate spacers common between two branches of the tree shown on the Figure 3. (B) Multiple alignments of CRISPR repeats identified in three representative strains from three branches of the tree shown on Figure 3. Underlined letters indicate positions with at least 90% identity. (C) Number of repeat mismatches for CRISPR arrays per CRISPR-Cas type. The box plot shows the weighted average number of repeat mismatches versus repeat consensus per CRISPR array. CRISPR-Cas type indicated on the y-axis. For subtype IV-A, the estimates were obtained separately for loci without cas1 (IV-A_no_cas1) and with cas1 gene (IV-A_cas1). The asterisk at IV-B indicates that results were obtained only for a small branch within the subtype IV-B corresponding to the proposed subtype IV-D, where five loci with CRISPR arrays were identified. The remaining IV-B and IV-C arrays were not included because in these loci, arrays are scarce and show a highly patchy distribution (see Fig. 1). The boxes show the 25th/50th/75th percentiles, and black dots show outliers that fall above 1.5 × interquartile range. For type I, there are ∼10% outliers shown as dots.
The striking diversity of the repeats observed in the Caldus group prompted us to compare the repeats from type IV arrays with CRISPR arrays of other types in greater detail. We used the previously reported curated set of CRISPR systems1 and analyzed the arrays for nucleotide substitutions compared with the repeat consensus. This comparison showed a dramatically greater within-array variability of repeats in the type IV arrays compared to the arrays from all other CRISPR-Cas systems (Fig. 5C). Repeat sequence variability in type IV loci does not depend on the presence of adaptation modules (Cas1 and/or Cas2; Fig. 5C). It seems most likely that the repeat variability in type IV arrays is underestimated because short arrays with multiple substitutions are not identifiable by the currently available CRISPR array detection methods. Indeed, arrays were missed in several IV-A loci in Pseudomonas plasmid genomes (e.g., P. aeruginosa strain S04 90 plasmid, CP011370.1, and P. aeruginosa RW109 plasmid 2, LT969521.1) as shown by manual examination of these loci. Spacers with perfect matches to TraY from P. putida plasmid pND6-2 and a plasmid integrated in P. aeruginosa strain GIMC5019:PA52Ts1 genome were identified in these arrays (Supplementary Fig. S11) in evidence of their functionality. Furthermore, we identified RNAseq data in the SRA archive showing that the IV-A array is expressed in P. aeruginosa RW109 during exposure to antibiotics (Supplementary Fig. S11). The lack of arrays in most of the subtype IV-B systems might point to the promiscuity of these systems with respect to the crRNA identity, similar to type III effector complexes. Within the subtype IV-B, only some of the IV-D group loci contain arrays. The repeats in these arrays show much less heterogeneity than those in IV-A arrays (Fig. 5C), but the IV-D sample might be too small for a confident conclusion of a lower variability to be made.
Spacers in Acidithiobacillia type IV arrays preferentially target plasmids
The majority of the 75 unique spacers in the F. caldus arrays showed no reliable matches to any available sequences, with only six distinct spacers matching other Acidithiobacillia chromosomes or plasmids and one matching a both plasmid pACMV2JF-5 from Acidiphilium multivorum AIU301 and plasmid pACRY05 from Acidiphilium cryptum JF-5 (Fig. 5A and Supplementary Table S4). Notably, the identified protospacers came from genes known to be important for plasmid replication or conjugation, in particular, the traA gene from A. cryptum JF-5 plasmid pACRY05, mobA and a permease encoded in F. caldus MNG pTcM1, the intergenic region between repC and the primase gene in the pTcM1 plasmid from F. caldus MNG, and to a chromosomal trbB gene from the F. caldus type strain genome (pTcM1/pLAtc3) located in a putative ICE region. Interestingly, four spacers are shared by all three branches although surrounded by different repeats (Fig. 4A). Also, in CP026328.2, two spacers are duplicated. Furthermore, considering that F. caldus VAN18-3 and CV18-1 group with F. caldus MELC-5 (branch C) and F. caldus MELC-5 contains an identical region in the arrays with CP005986.1 (branch A), we can conclude that this region was present in the common ancestor of branches C and A, and was deleted from the common ancestor of F. caldus VAN18-3 and CV18-1 array (Fig. 3).
In addition to the 75 unique spacers from the F. caldus CRISPR loci shown on the Figures 3 and 5, 214 unique spacers were identified in other analyzed Acidithiobacillia strains; none of these were similar to those in the Caldus group strains. Among these 214 spacers, only seven had matches in NR and none in the viral database (Supplementary Table S4). At least three of those target ICEs or mobilizable plasmids, which are both common in Acidithiobacillia class genomes.54
Discussion
The results of in-depth analysis of type IV CRISPR-Cas systems presented here and elsewhere reveal many unusual features of this CRISPR-Cas type. The type IV systems join the expanding group of CRISPR-Cas variants that were recruited by MGE for antidefense and other functions, and in the process of this exaptation, lost their target cleavage activity. In particular, there is a clear parallel between the incorporation of type IV systems in MGE and the recruitment of defective variants of subtype I-B and I-F as well as subtype V-K by Tn7-like transposons, in which these defective CRISPR-Cas systems mediate RNA-guided transposition.11,55,56
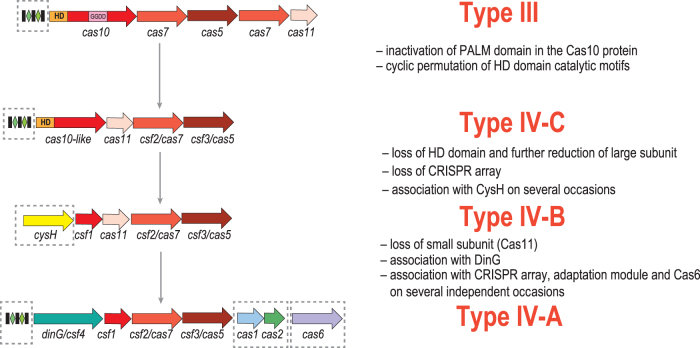
The origin and directionality of the evolution of type IV systems became more transparent with the identification of subtype IV-C systems and the first structural data on the organization of type IV effector complexes.1,7,8 These results support the previously proposed hypothesis that type IV systems originated from type III. Many subtype IV-C loci contain no CRISPR array and likely evolved from one of the numerous variants of type III that also lack CRISPR.1 At the onset of type IV evolution, the large subunit deteriorated, and the HD domain present in the type III and subtype IV-C large subunits was lost, resulting in the loss of the target cleavage capacity, and the minimalist version of the large subunit emerged, giving rise to subtype IV-B. The next major evolutionary event was the origin of subtype IV-A, which evolved by the loss of the small subunit and the recruitment of DinG, possibly, compensating for the absence of the small subunit. CRISPR arrays, Cas6 and, less frequently, adaptation modules were acquired on several independent occasions, mostly by subtype IV-A systems (Fig. 6).
FIG. 6.
A hypothetical scenario for the origins of type IV CRISPR-Cas systems. Homologous genes are shown as color-coded arrows or rectangles (for domains) and identified by a family name. The key evolutionary events are described to the right of the images. Optional genes and CRISPR arrays are denoted by dashed outlines. GGDD: key catalytic motif of the cyclase/polymerase domain of Cas10.
Type IV is exceptional among CRISPR-Cas systems in harboring highly polymorphic repeats. The biological underpinning and impact of this within-array repeat heterogeneity remain unclear, but considering that most of the type IV systems lack adaptation genes, it seems possible that they acquire new spacers via recombination between CRISPR arrays encoded on different MGEs rather than via a typical adaptation route. The few cas1 genes that are associated with type IV loci are monophyletic and originate from type I-E Cas1, with the exception of several type IV loci in Pseudomonas, which encode Cas1 apparently originating from subtype I-F. It is of interest whether these Cas1 proteins, especially the former, possess any distinct functional properties, such as an ability to insert new repeats in random positions within the CRISPR array.
Our analysis of the genes that often accompany type IV CRISPR-Cas systems showed that the most common of these genes, cysH and recD/traA, are frequently present in numerous MGE, not only within type IV CRISPR-Cas loci. However, cysH genes associated with subtype IV-B are largely monophyletic, suggesting a strong functional link. Several other functionally uncharacterized genes and apparent modules were also found to be consistently associated with type IV loci (Table 1), but the majority of the neighbors in these loci are highly diverse and belong to the genomic “dark matter.” However, analysis of the context of the Caldus group type IV loci, which encode superinfection immunity protein Imm, suggests that type IV systems, along with most of the other genes in the same neighborhoods, belong to “cargo” regions of ICEs and other MGE, and are involved in inter-MGE conflicts and, possibly, additional functions in MGE reproduction.
The results of the present analysis seem to provide little or no support for the proposed reclassification of type IV systems. Indeed, in subtype IV-A, the proposed variants A1 and A2 are not monophyletic.6 Furthermore, all proposed IV-A variants—A1, A2, and A3—share the same set of effector genes, and the sequences of the core genes Csf2 and Csf3 are closely similar. Divergence of large or small subunits of the effector complexes has not been previously considered sufficient reason to reclassify CRISPR-Cas subtypes (e.g., in subtype I-B, there are 13 distinct large subunits that share no detectable sequence similarity), and the proposed variants lack any other features that would suggest major functional differences. The proposed subtype IV-E shares the same set of effector complex genes and the ancillary gene dinG with subtype IV-A, and the fusion of the effector genes generally is not thought to justify establishing a new subtype. The E branch confidently groups with type IV-A in the Cas7 and Cas5 trees, and all the E loci encode DinG, the signature of subtype IV-A. Thus, we believe that this subfamily should remain within type IV-A.
The proposed subtype D is the most notable type IV variant. Our analysis suggests that this is a hybrid system, with Csf2 (Cas7) and the large subunit Csf1 originating from within the IV-B clade, whereas Csf3 (Cas5) is from the IV-A clade. The Csf1 and Csf3 genes are not monophyletic and apparently became independently associated with Csf2 at least twice. Most of the IV-D systems are encoded in predicted operons, together with a gene coding for a RecD/TraA family helicase. This association also appears to have evolved independently on at least three occasions. However, recD/traA genes are also found in extended loci of other systems, such as subtype IV-A. These chimeric loci present a challenge for the current classification approach. There seems to be no compelling reason to classify these loci as a separate subtype given the apparent polyphyly of all the components of the proposed IV-D system, and we therefore propose to classify these systems as type IV-B based on the Cas7 (Csf2) phylogeny. Otherwise, all the distinguishing features that were used to classify type IV systems into the A and B subtypes remain valid. So, we propose adhering to this classification, ahead of additional comparative genomic and experimental evidence that might call for modifications to the classification scheme.
Many outstanding questions regarding the functions of type IV systems remain to be answered. In particular, the source of the crRNA for type IV-B systems lacking CRISPR arrays remains to be identified. Most significantly, the specific role and the mechanism of the involvement of type IV systems in MGE interference and possibly other processes are currently unclear, and the functions of the associated genes, such as dinG, cysH, recD, and others, have also to be elucidated.
Supplementary Material
Acknowledgment
R.Q. would like to acknowledge J.F. Banfield for her contributions and inspiration at early stages of the analysis the type IV CRIPSR-Cas loci in the Acidithiobacillia class representatives.
Author Disclosure Statement
No competing financial interests exist.
Funding Information
K.S.M., Y.I.W., S.A.S., I.T., and E.V.K. are supported by the Intramural Research Program of the National Institutes of Health of the USA (National Library of Medicine). This work was supported the Agencia Nacional de Investigación y Desarrollo (ANID, Chile) under Grants FONDECYT 1181251 (R.Q.), FONDECYT 11201070 (L.G.A.), Programa de Apoyo a Centros con Financiamiento Basal AFB170004 (R.Q.), CONICYT-PFCHA/Doctorado Nacional/20171049 (A.M.-B.), CONICYT-PFCHA/Doctorado Nacional/21090398 (L.G.A.), CONICYT-PFCHA/Doctorado Nacional/21120305 (P.C.C.), CONICYT/FONDAP Grant No. 15110027 (L.G.A.), and the ANID—Millennium Science Initiative Programme—NCN17_093, granted by the Ministry of Economy, Development and Tourism from Chile (R.Q.).
Supplementary Material
References
- 1. Makarova KS, Wolf YI, Iranzo J, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol 2020;18:67–83. DOI: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Makarova KS, Haft DH, Barrangou R, et al. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol 2011;9:467–477. DOI: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bustamante P, Covarrubias PC, Levican G, et al. ICE Afe 1, an actively excising genetic element from the biomining bacterium Acidithiobacillus ferrooxidans. J Mol Microbiol Biotechnol 2012;22:399–407. DOI: 10.1159/000346669. [DOI] [PubMed] [Google Scholar]
- 4. Quatrini R, Johnson DB. Acidithiobacillus ferrooxidans. Trends Microbiol 2019;27:282–283. DOI: 10.1016/j.tim.2018.11.009. [DOI] [PubMed] [Google Scholar]
- 5. Valdes J, Pedroso I, Quatrini R, et al. Acidithiobacillus ferrooxidans metabolism: from genome sequence to industrial applications. BMC Genomics 2008;9:597. DOI: 10.1186/1471-2164-9-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pinilla-Redondo R, Mayo-Munoz D, Russel J, et al. Type IV CRISPR-Cas systems are highly diverse and involved in competition between plasmids. Nucleic Acids Res 2020;48:2000–2012. DOI: 10.1093/nar/gkz1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ozcan A, Pausch P, Linden A, et al. Type IV CRISPR RNA processing and effector complex formation in Aromatoleum aromaticum. Nat Microbiol 2019;4:89–96. DOI: 10.1038/s41564-018-0274-8. [DOI] [PubMed] [Google Scholar]
- 8. Zhou Y, Bravo JPK, Taylor HN, et al. Structure of a type IV CRISPR-Cas ribonucleoprotein complex. iScience 2021;24:102201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Makarova KS, Aravind L, Wolf YI, et al. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol Direct 2011;6:38. DOI: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shmakov SA, Makarova KS, Wolf YI, et al. Systematic prediction of genes functionally linked to CRISPR-Cas systems by gene neighborhood analysis. Proc Natl Acad Sci U S A 2018;115:E5307–E5316. DOI: 10.1073/pnas.1803440115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Faure G, Shmakov SA, Yan WX, et al. CRISPR-Cas in mobile genetic elements: counter-defence and beyond. Nat Rev Microbiol 2019;17:513–525. DOI: 10.1038/s41579-019-0204-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Newire E, Aydin A, Juma S, et al. Identification of a type IV-A CRISPR-Cas system located exclusively on IncHI1B/IncFIB plasmids in Enterobacteriaceae. Front Microbiol 2020;11:1937. DOI: 10.3389/fmicb.2020.01937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kamruzzaman M, Iredell JR. CRISPR-Cas system in antibiotic resistance plasmids in Klebsiella pneumoniae. Front Microbiol 2019;10:2934. DOI: 10.3389/fmicb.2019.02934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Taylor HN, Warner EE, Armbrust MJ, et al. Structural basis of Type IV CRISPR RNA biogenesis by a Cas6 endoribonuclease. RNA Biol 2019;16:1438–1447. DOI: 10.1080/15476286.2019.1634965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Crowley VM, Catching A, Taylor HN, et al. A type IV-A CRISPR-Cas system in Pseudomonas aeruginosa mediates RNA-guided plasmid interference in vivo. CRISPR J 2019;2:434–440. DOI: 10.1089/crispr.2019.0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Acuna LG, Cardenas JP, Covarrubias PC, et al. Architecture and gene repertoire of the flexible genome of the extreme acidophile Acidithiobacillus caldus. PLoS One 2013;8:e78237. DOI: 10.1371/journal.pone.0078237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Moya-Beltran A, Beard S, Rojas-Villalobos C, et al. Genomic evolution of the class Acidithiobacillia: deep-branching Proteobacteria living in extreme acidic conditions. ISME J 2021 May 18 [Epub ahead of print]; DOI: 10.1038/s41396-021-00995-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Altschul SF, Madden TL, Schaffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Marchler-Bauer A, Zheng C, Chitsaz F, et al. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 2013;41:D348–352. DOI: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics 2005;21:951–960. DOI: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 21. Russel J, Pinilla-Redondo R, Mayo-Munoz D, et al. CRISPRCasTyper: automated identification, annotation, and classification of CRISPR-Cas loci. CRISPR J 2020;3:462–469. DOI: 10.1089/crispr.2020.0059. [DOI] [PubMed] [Google Scholar]
- 22. Bland C, Ramsey TL, Sabree F, et al. CRISPR recognition tool (CRT): a tool for automatic detection of clustered regularly interspaced palindromic repeats. BMC Bioinformatics 2007;8:209. DOI: 10.1186/1471-2105-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Grissa I, Vergnaud G, Pourcel C. CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res 2007;35:W52–57. DOI: 10.1093/nar/gkm360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Biswas A, Gagnon JN, Brouns SJ, et al. CRISPRTarget: bioinformatic prediction and analysis of crRNA targets. RNA Biol 2013;10:817–827. DOI: 10.4161/rna.24046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Steinegger M, Soding J. MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 2017;35:1026–1028. DOI: 10.1038/nbt.3988. [DOI] [PubMed] [Google Scholar]
- 26. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nguyen LT, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 2015;32:268–274. DOI: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Price MN, Dehal PS, Arkin AP. FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 2010;5:e9490. DOI: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 2013;30:772–780. DOI: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nieto PA, Covarrubias PC, Jedlicki E, et al. Selection and evaluation of reference genes for improved interrogation of microbial transcriptomes: case study with the extremophile Acidithiobacillus ferrooxidans. BMC Mol Biol 2009;10:63. DOI: 10.1186/1471-2199-10-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bick JA, Dennis JJ, Zylstra GJ, et al. Identification of a new class of 5′-adenylylsulfate (APS) reductases from sulfate-assimilating bacteria. J Bacteriol 2000;182:135–142. DOI: 10.1128/jb.182.1.135-142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Xiong L, Liu S, Chen S, et al. A new type of DNA phosphorothioation-based antiviral system in archaea. Nat Commun 2019;10:1688. DOI: 10.1038/s41467-019-09390-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Xu T, Yao F, Zhou X, et al. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res 2010;38:7133–7141. DOI: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Farlow J, Bolkvadze D, Leshkasheli L, et al. Genomic characterization of three novel Basilisk-like phages infecting Bacillus anthracis. BMC Genomics 2018;19:685. DOI: 10.1186/s12864-018-5056-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Garcia P, Monjardin C, Martin R, et al. Isolation of new Stenotrophomonas bacteriophages and genomic characterization of temperate phage S1. Appl Environ Microbiol 2008;74:7552–7560. DOI: 10.1128/AEM.01709-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Summer EJ, Gill JJ, Upton C, et al. Role of phages in the pathogenesis of Burkholderia, or “Where are the toxin genes in Burkholderia phages?” Curr Opin Microbiol 2007;10:410–417. DOI: 10.1016/j.mib.2007.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kopriva S, Koprivova A. Plant adenosine 5′-phosphosulphate reductase: the past, the present, and the future. J Exp Bot 2004;55:1775–1783. DOI: 10.1093/jxb/erh185. [DOI] [PubMed] [Google Scholar]
- 38. Taylor HN, Laderman E, Armbrust M, et al. Positioning diverse type IV structures and functions within class 1 CRISPR-Cas systems. Front Microbiol 2021;12:671522. DOI: 10.3389/fmicb.2021.671522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yang JC, Lessard PA, Sengupta N, et al. TraA is required for megaplasmid conjugation in Rhodococcus erythropolis AN12. Plasmid 2007;57:55–70. DOI: 10.1016/j.plasmid.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 40. Rogozin IB, Makarova KS, Wolf YI, et al. Computational approaches for the analysis of gene neighbourhoods in prokaryotic genomes. Brief Bioinform 2004;5:131–149. DOI: 10.1093/bib/5.2.131. [DOI] [PubMed] [Google Scholar]
- 41. Dyson ZA, Tucci J, Seviour RJ, et al. Isolation and characterization of bacteriophage SPI1, which infects the activated-sludge-foaming bacterium Skermania piniformis. Arch Virol 2016;161:149–158. DOI: 10.1007/s00705-015-2631-8. [DOI] [PubMed] [Google Scholar]
- 42. Bignell C, Thomas CM. The bacterial ParA-ParB partitioning proteins. J Biotechnol 2001;91:1–34. DOI: 10.1016/s0168-1656(01)00293-0. [DOI] [PubMed] [Google Scholar]
- 43. Castillo F, Benmohamed A, Szatmari G. Xer site specific recombination: double and single recombinase systems. Front Microbiol 2017;8:453. DOI: 10.3389/fmicb.2017.00453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaur G, Burroughs AM, Iyer LM, et al. Highly regulated, diversifying NTP-dependent biological conflict systems with implications for the emergence of multicellularity. Elife 2020;9. DOI: 10.7554/eLife.52696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Flores-Rios R, Moya-Beltran A, Pareja-Barrueto C, et al. The type IV secretion system of ICEAfe1: formation of a conjugative pilus in Acidithiobacillus ferrooxidans. Front Microbiol 2019;10:30. DOI: 10.3389/fmicb.2019.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cabezon E, Ripoll-Rozada J, Pena A, et al. Towards an integrated model of bacterial conjugation. FEMS Microbiol Rev 2015;39:81–95. DOI: 10.1111/1574-6976.12085. [DOI] [PubMed] [Google Scholar]
- 47. Guglielmini J, Quintais L, Garcillan-Barcia MP, et al. The repertoire of ICE in prokaryotes underscores the unity, diversity, and ubiquity of conjugation. PLoS Genet 2011;7:e1002222. DOI: 10.1371/journal.pgen.1002222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wozniak RA, Waldor MK. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 2010;8:552–563. DOI: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 49. Moya-Beltran A, Rojas-Villalobos C, Diaz M, et al. Nucleotide second messenger-based signaling in extreme acidophiles of the Acidithiobacillus species complex: partition between the core and variable gene complements. Front Microbiol 2019;10:381. DOI: 10.3389/fmicb.2019.00381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lu MJ, Henning U. The immunity (imm) gene of Escherichia coli bacteriophage T4. J Virol 1989;63:3472–3478. DOI: 10.1128/JVI.63.8.3472-3478.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Koonin EV, Zhang F. Coupling immunity and programmed cell suicide in prokaryotes: life-or-death choices. Bioessays 2017;39:1–9. DOI: 10.1002/bies.201600186. [DOI] [PubMed] [Google Scholar]
- 52. Makarova KS, Wolf YI, Snir S, et al. Defense islands in bacterial and archaeal genomes and prediction of novel defense systems. J Bacteriol 2011;193:6039–6056. DOI: 10.1128/JB.05535-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Doron S, Melamed S, Ofir G, et al. Systematic discovery of antiphage defense systems in the microbial pangenome. Science 2018;359. DOI: 10.1126/science.aar4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Beard S, Ossandon FJ, Rawlings DE, et al. The flexible genome of acidophilic prokaryotes. Curr Issues Mol Biol 2021;40:231–266. DOI: 10.21775/cimb.040.231. [DOI] [PubMed] [Google Scholar]
- 55. Klompe SE, Vo PLH, Halpin-Healy TS, et al. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 2019;571:219–225. DOI: 10.1038/s41586-019-1323-z. [DOI] [PubMed] [Google Scholar]
- 56. Strecker J, Ladha A, Gardner Z, et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 2019;365:48–53. DOI: 10.1126/science.aax9181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Makarova KS, Wolf YI, Koonin EV. Classification and nomenclature of CRISPR-Cas systems: where from here? CRISPR J 2018;1:325–336. DOI: 10.1089/crispr.2018.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Makarova KS, Wolf YI, Shmakov SA, et al. Unprecedented diversity of unique CRISPR-Cas-related systems and Cas1 homologs in Asgard archaea. CRISPR J 2020;3:156–163. DOI: 10.1089/crispr.2020.0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chartron J, Carroll KS, Shiau C, et al. Substrate recognition, protein dynamics, and iron-sulfur cluster in Pseudomonas aeruginosa adenosine 5′-phosphosulfate reductase. J Mol Biol 2006;364:152–169. DOI: 10.1016/j.jmb.2006.08.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Buetti-Dinh A, Herold M, Christel S, et al. Systems biology of acidophile biofilms for efficient metal extraction. Sci Data 2020;7:215. DOI: 10.1038/s41597-020-0519-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.