Abstract
Ecological restoration can promote biodiversity conservation in anthropogenically fragmented habitats, but effectiveness of these management efforts need to be statistically validated to determine ’success.’ One such approach is to gauge the extent of recolonization as a measure of landscape permeability and, in turn, population connectivity. In this context, we estimated dispersal and population connectivity in prairie vole (Microtus ochrogaster; N = 231) and meadow vole (M. pennsylvanicus; N = 83) within five tall-grass prairie restoration sites embedded within the agricultural matrix of midwestern North America. We predicted that vole dispersal would be constrained by the extent of agricultural land surrounding restored habitat patches, spatially isolating vole populations and resulting in significant genetic structure. We first employed genetic assignment tests based on 15 microsatellite DNA loci to validate field-derived species-designations, then tested reclassified samples with multivariate and Bayesian clustering to assay for spatial and temporal genetic structure. Population connectivity was further evaluated by calculating pairwise FST, then potential demographic effects explored by computing migration rates, effective population size (Ne), and average relatedness (r). Genetic species assignments reclassified 25% of initial field identifications (N = 11 M. ochrogaster; N = 67 M. pennsylvanicus). In M. ochrogaster population connectivity was high across the study area, reflected in little to no spatial or temporal genetic structure. In M. pennsylvanicus genetic structure was detected, but relatedness estimates identified it as kin-clustering instead, underscoring social behavior among populations rather than spatial isolation as the cause. Estimates of Ne and r were stable across years, reflecting high dispersal and demographic resilience. Combined, these metrics suggest the agricultural matrix is highly permeable for voles and does not impede dispersal. High connectivity observed confirms that the restored landscape is productive and permeable for specific management targets such as voles and also demonstrates population genetic assays as a tool to statistically evaluate effectiveness of conservation initiatives.
Introduction
‘Distribution’ and ‘abundance’ are fundamental natural history aspects of species [1], but the population processes defining these ecological characteristics [2] can be easily disrupted by anthropogenic habitat fragmentation [3]. Most contemporary landscapes reflect large-scale human modifications that constrain species to persist as subdivided, often isolated populations. As a consequence, ‘dispersal’ and thus ‘population connectivity’ have become key ecological parameters that determine long-term population viability [4].
Temperate grassland biomes (>8% of the global landmass [5]) are easily transformed into human settlements and agricultural plots, and species dependent on grassland habitats have been especially impacted. The large-scale agricultural conversion of the Tall Grass Prairie in Midwestern North America [6] emphasizes the extensive human demands imposed upon these biomes. Adoption of row-crop technologies and prolongation of the growing season ([7] Table 1) have substantially enhanced this agricultural capacity [8], but also increased regional greenhouse gas emissions such that they now exceed the national average [9]. In addition, increasing demand for biofuels has further intensified agricultural practices and accelerated the removal of potential ’edge’ habitats along the margins of agricultural fields [10] that previously provided both essential connectivity among habitat fragments for prairie species as well as refugial habitat. Other consequences of large-scale habitat alterations are loss of indigenous biodiversity and increased biotic homogenization [11].
Table 1. Vole samples collected during three field seasons (2010–2012) across five SAFE sites in Illinois.
Samples were harvested non-invasively from two species, M. ochrogaster and M. pennsylvanicus, but only totals are listed. Geographic location of sites in Fig 1.
| Site | 2010 | 2011 | 2012 | Totals |
|---|---|---|---|---|
| Livingston | 27 | 4 | 10 | 41 |
| Montgomery | 0 | 56 | 41 | 97 |
| Pontiac | 21 | 1 | 9 | 31 |
| Prairie Ridge | 60 | 3 | 32 | 95 |
| Saybrook | 0 | 19 | 77 | 96 |
| Total | 108 | 83 | 169 | 360 |
Habitat restoration can reduce or even reverse the detrimental consequences of these anthropogenic impacts. In the prairie landscape of North America, numerous conservation initiatives have been implemented to improve remnant grassland parcels and increase population numbers of targeted wildlife. Earliest prairie restoration efforts date as far back as the 1930s [12] and now extend from local grass-roots efforts to governmental initiatives such as the State Acres for Wildlife Enhancement Initiative (SAFE; https://www.fsa.usda.gov/programs-and-services/conservation-programs/), a conservation program that aims at converting agricultural land into grasslands by providing annual rental pay to farmers for removing environmentally sensitive areas from production. The goal of SAFE is to promote wildlife through habitat restoration, with the primary target being high priority species designated by U.S. Fish and Wildlife Service as threatened and endangered (T&E), but with other species benefitting as well. The program is voluntary, thus specific locations and numbers of sites may fluctuate over time. In Illinois, SAFE sites are selected in proximity to permanent prairie habitats, with each site comprised of several restoration patches varying in size and time since initial restoration.
However, one critical aspect of restoration is the definition of ’success,’ and which metric(s) can gauge if such a specific goal has been reached. In this study, we evaluated the spatio-temporal genetic structure of two vole species, the prairie vole (Microtus ochrogaster) and the meadow vole (M. pennsylvanicus), as a means of quantifying the effectiveness of prairie restoration efforts within a large-scale agroecosystem. Voles can subsist in small home ranges (100m2) within prairie fragments, and are easily captured via live trapping, a technique that yields reliable abundance and movement estimates [13], an important aspect when evaluating effects of fragmentation on population dynamics and demographics [13–15].
The study area encompassed five prairie restoration SAFE sites, isolated within the fragmented row-crop landscape of Illinois and about 30–250 km apart, to test if large geographic distances separating restoration patches in this highly-modified agricultural matrix would act synergistically to curtail vole dispersal [16]. For M. ochrogaster, relatedness within and among sites was also explored, as it is a highly philopatric and socially monogamous species that often nests cooperatively [17, 18], such that social- and kin-related clusters might impact dispersal. In contrast, M. pennsylvanicus is promiscuous [19], yet can also form social groups during the non-breeding season [20].
Microsatellite DNA analysis was used to: 1) Quantify genetic diversity in both vole species within and among the five SAFE sites; and 2) Assess levels of temporal and spatial gene flow among habitat patches. Population genetic data reflect dispersal of organisms through complex environments, and hence can reveal if connectivity indeed exists amongst habitat patches widely-separated within an anthropogenically-modified landscape [21]. This approach also provides an estimate of landscape permeability, an important component of restoration programs within agroecosystems where potential habitat rehabilitation is often restricted to isolated patches, but where connectivity can be established through edge habitat within surrounding areas [22]. Our expectation was that limited connectivity among study sites would be reflected in strong population structure, particularly given the life histories of the study species, and the relative isolation of restoration patches within the agroecosystem.
Materials and methods
Sampling and DNA extraction
Voles were live-trapped from mid-May through mid-August (2010–2012) as part of a concurrent study [23] at representative SAFE sites that evaluated response of small mammal communities to prairie restoration efforts. Tissue samples were collected from five SAFE sites approximately 35–250 km apart (Fig 1B) in central/ south-central Illinois. Each site contained a mosaic of restoration patches (Fig 1C) varying in area (<2ha to >65ha), distance from other patches (0m to 8km), or time since initial restoration (1 year to >10 years) (S1 Table). The surrounding landscape was dominated by row crop agriculture, but restored patches were connected to various degrees by fencerows, roadside ditches, or grass waterways. Restoration was conducted by seeding previous cropland with either CP1 (cool-season grasses and legumes), CP2 (warm-season grasses), or mixtures [24]. However, neither patch size nor seeding type impacted vole abundance [23], but population cycles were seemingly an overriding factor. Sampling was conducted via six transects of 15 traps each set 7m apart within each restoration patch, with coverage defined by patch size. Trapping occurred over three consecutive evenings (N = 90 traps/ patch/ night; [24]), and was approved by the Institutional Animal Care and Use Committee (IACUC), University of Illinois/Urbana-Champaign.
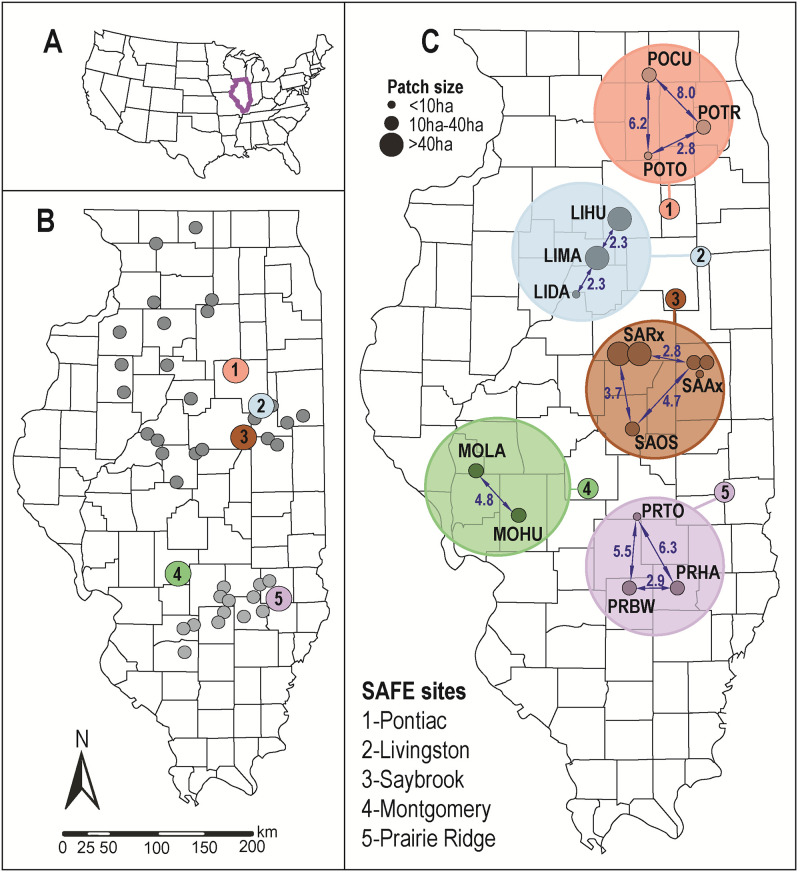
Fig 1. Map depicting State Acres for Wildlife Enhancement (SAFE) restoration sites in Illinois.
(A) Location of Illinois (purple) in the USA. (B) Distribution of SAFE sites (circles) in Illinois: color = sampled sites; dark grey sites in the Grand Prairie region; light gray = sites in the Southern Till Plain. (C) Schematic distribution of restoration patches (small circles) at sampled sites (large circles): arrows = inter-patch distances (in km); circle size = patch area (in ha). The map was generated in ArcGIS 10.4.1 using publicly available data downloaded from government institutions, with boundaries derived from Topologically Integrated Geographic Encoding and Referencing (TIGER) data via U.S. Census Bureau. Additional details on patches provided in S1 Table.
Ear tissue was sampled from each captured vole, with a maximum of 30 samples/ patch/ year and stored in 95% EtOH at -20°C for subsequent genetic analyses. Whole genomic DNA was extracted using Promega Wizard Kit (2010–2011) or Qiagen DNeasy Blood and Tissue Kit (2012) and quantified using an Implen Pearl P-300 nanophotometer.
Microsatellite DNA genotyping
Microsatellite loci previously developed for vole species were evaluated for consistent cross-amplification in M. ochrogaster and M. pennsylvanicus. A set of 23 loci was selected for genotyping and combined into six multiplex panels using fluorescently labeled forward primers (S2 Table). Amplifications were conducted in 10–15μl volume polymerase chain reactions (PCR) using approximately 10–15ng template and following standard procedures. Additional details on reaction conditions and cycling profiles are provided in S2 Table.
Fragment analysis
Microsatellite DNA fragments were resolved on an automated Applied Biosystems (ABI) Prism 3730xl GeneAnalyzer at the W. M. Keck Center, University of Illinois, Champaign. An internal size standard (Liz 500) was run with each sample, and alleles were scored using GeneMapper© 4.1 software (ABI). Genotypes were partitioned by species, site, and sampling period, then tested via Micro-Checker 2.2.3 [25] for null alleles, large allele dropout and scoring errors. All pairs of loci were tested for linkage disequilibrium (Markov Chain parameters: 10,000 dememorization steps, 500 batches, 5,000 iterations), and each locus evaluated for deviations from Hardy-Weinberg equilibrium (HWE) using exact tests (GenePop 4.0; [26]) with Bonferroni correction for multiple comparisons [27].
Assignment of individuals to species
The two vole species are morphologically similar and accurate identification can be difficult, particularly when non-lethal sampling is conducted. To verify field-based species identification, we employed the population assignment option in GenAlEx 6.50 [28] to cluster samples according to similarities in genotype. Genetic-based species assignments were then used to re-classify individuals to species and re-evaluate distinct gene pools using Bayesian clustering (Structure 2.3.4. [29]); with admixture ancestry and correlated allele frequency options selected [30].
Molecular taxonomy
To further verify species designation, we genotyped a diagnostic locus (Microtus avpr1a gene [31]) that identifies M. ochrogaster with an allele of ~600–800bp, and M. pennsylvanicus with one of ~200–300bp. Primers and PCR protocols were adapted from previous studies to test a subset of samples (N = 68) for concordance between field- and genetic-based species identification [32–34], and diagnostic alleles visualized by separating PCR products on 2.5% agarose gel stained with GelGreen (Biotium Inc., Hayward, CA) and examined on a bluelight transluminator.
Genetic diversity and structure of vole populations
Standard population genetic parameters were calculated to quantitative diversity within and among sites. These included measures of allele frequencies and heterozygosity and were estimated for each species at each site using GenAlEx. Values for allelic richness (AR) and private allelic richness (PAR) were derived from rarefaction and corrected for sample sizes (N = 23; HP-Rare v. June-6-2006; [35]). Pairwise relatedness (r) among individuals was calculated using the Ritland (1996) estimator in GenAlEx to reduce potential bias in genetic analyses by inadvertent comparison of related individuals captured in localized trapping transects (a particular concern when evaluating spatial genetic structure). To mitigate, we subsequently removed individuals at random from pairwise comparisons when r ≥ 0.25.
Multivariate clustering
To visualize genetic structure, we first performed a principal component analysis (PCA) using the dudi.pca function in the R package Adegenet [36]. Because PCA captures both among-group (i.e., ‘population structure’) and within-group genetic variation [37], a discriminant analysis of principal components (DAPC) was subsequently performed to isolate the among-group components [36]. An automated cross-validation procedure was used to select the optimal number of PCs by iteratively exploring classification error as a function of the number of retained PCs. Here, 80% of the samples served as a ‘training set’ for the remaining 20% of samples. All evaluations utilized the xvalDapc function of Adegenet [36], as implemented in a custom R script (vol_dapc.R in the Open Science Repository; doi: 10.17605/OSF.IO/K2M8W).
Bayesian clustering
To further assess distribution of gene pools within and among sampling locations assignment tests were conducted (Structure; [29]).Ten replicates were run for K-values ranging from 1–10 using a burn-in of 500,000 iterations, followed by 1,000,000 Markov Chain Monte Carlo (MCMC) replicates. To account for potential multimodality per K, results were processed in Clumpak [38]. The optimal number of clusters for each simulation (per Clumpak) was evaluated by the ad hoc statistic ‘ΔK’ [39], and also the probability by K [29]. This was done because ΔK is unable to evaluate validity of K = 1, and also has a propensity to select K = 2 [39, 40].
Genetic structure detected within localities for M. pennsylvanicus suggested the potential for aggregation of related individuals (i.e., kin structure). Therefore, family structure was evaluated (Colony v2.0.6.6; [41]) to determine if clusters corresponded with family groups. Analytical methods allowed for female polygamy as well as inbreeding and program parameters were selected as ’very long’ run length, full-likelihood analysis and ’very high’ precision.
Power analysis
To measure our ability to resolve population structure, a power analysis was conducted (PowSim; [42]) utilizing both available test options (i.e., χ2 and Fisher exact tests). The Fisher tests were performed using 1,000 dememorization steps followed by 1,000 iterations in 100 batches. Empirical allele frequencies, sample sizes, and number of populations were evaluated, while an initial effective population size of 500 was assumed. The time of divergence required to detect population differentiation was estimated by varying number of generations impacted by genetic drift (t) from 2–20. Power was assessed as the number of significant tests observed per 10,000 replicates.
Pairwise FST
Population connectivity was evaluated using pairwise FST, a standardized index that reflects average gene flow over time, with spatial/ temporal patterns estimated within years among sites, and among years within sites (GenAlEx, with 9,999 permutations and missing data interpolated). Gene flow could only be reliably assessed for M. ochrogaster, due to variability in numbers of samples obtained at sites among years for M. pennsylvanicus, as well as the uneven distribution of samples following genetic re-classification of samples.
To assay spatial connectivity, we derived pairwise FST values for M. ochrogaster at three sites in 2010 (i.e., Livingston, Pontiac, and Prairie Ridge), and between two sites in 2012 (i.e., Montgomery and Prairie Ridge). To test for potential impacts of environmental perturbations (e.g., drought in 2012; [21, 24]), we also calculated pairwise FST estimates between years at two sites (Montgomery: 2011 versus 2012; Prairie Ridge: 2010 versus 2012). Sample sizes were too small at other sites or time intervals to conduct either spatial or temporal comparisons.
Estimates of migration and effective population size
Individual movements can influence population dynamics, and two are of particular importance in this regard: Colonization (i.e., movement into an unoccupied habitat patch) and migration (i.e., immigration into occupied patches). For example, the annual replanting of crops is an agroecosystem dynamic that can alternately convert vole populations into sinks that must be potentially colonized from a source [43].
Recent migration among SAFE sites
Potential F1 descendants of migrant individuals were identified using GeneClass2 2.0; [44]) and by selecting the ’L_home’ test statistic (i.e., likelihood of obtaining the genotype of an individual from the sampled population under the assumption that not all source populations were sampled). A Monte Carlo resampling method was selected, with 10,000 bootstraps and a threshold value of 0.01, as it improves performance when identifying first generation migrants while also controlling for Type-I error rates [45].
Effective population size (Ne)
Trapping success was uneven among years, impacted by an apparent regional population decline in 2011 and a substantial drought in 2012 [23, 24]. To gauge the effects of these perturbations on vole populations, we quantified Ne (LDNE; [46]) based upon estimates of linkage disequilibrium (i.e., a non-random association of independent alleles with haplotypes occurring in unexpected frequencies; [47]). Ne reflects the rate of genetic drift (i.e., random fluctuations in allele frequencies over time; [13]), as well as the effectiveness of selection and migration. It also indicates heterozygosity loss and links strongly with demographic factors such as sex ratio, population size, and lifetime fitness.
Ne and associated 95% jackknife confidence intervals were calculated using NeEstimator v2.1 [48], with rare alleles (PCRIT) excluded from analysis [49] (i.e., PCRIT = 0.02 when N ≥ 25). Due to sample size limitations, Ne could only be calculated for M. ochrogaster at Prairie Ridge (2010 versus 2012) and Montgomery (2011 versus 2012). Mean Ne at these two sites were compared per year by implementing Welch’s t-test for unequal variances. This approach is more robust than Student’s t-test and maintains Type-I error rates despite inequalities among variances and sample sizes. The test was performed in R [50] using summary statistics and a suitably-modified web-based program: (http://stats.stackexchange.com/questions/30394/how-to-perform-two-sample-t-tests-in-r-by-inputting-sample-statistics-rather-tha).
Isolation-By-Distance (IBD)
Finally, we evaluated the degree to which dispersal limitation has contributed to spatial genetic structuring within the two species by calculating genetic differentiation as a function of geographic distance between sample sites. We first calculated pairwise genetic distances among localities (i.e., Edwards distance; [51]) using the dist.genpop function in Adegenet [36]. We then used the Mantel test to compare these data against with pairwise geographic distances derived from latitude and longitude coordinates (S1 Table), with significance computed using 999 random permutations [52].
Results
Sampling, genotyping, and species assignments
A total of 360 voles were trapped across five prairie restoration sites over three field seasons (Table 1). Of these, 194 were field-identified as M. ochrogaster, and 166 as M. pennsylvanicus. All samples were genotyped across 23 microsatellite loci, with eight subsequently removed due either to null alleles or scoring issues, leaving 15 loci for data analyses. In addition, 46 samples missing data at more than one locus were excluded, leaving 314 individual genotypes for evaluation (S1 Table). Of these, 271 had complete genotypes and 43 lacked alleles at a single locus. Linkage disequilibrium was detected but was inconsistent across temporal periods or sites, and thus attributed to demographic effects on genetic structure rather than non-independence amongst loci.
Genotype-derived species assignments were concordant with 75% of field identifications based on morphology (236 of 314). Among the remaining 78 field identifications, 11 M. ochrogaster and 67 M. pennsylvanicus were genetically reclassified (S1 Fig), resulting in 231 M. ochrogaster and 83 M. pennsylvanicus genotypes, respectively (S1 Table). A subsequent Bayesian cluster analysis using reclassified species identifications consistently allocated all 314 samples (S1 Fig). Screening with the diagnostic avpr1a locus confirmed species-level classifications for 97% of test samples (65/67), with two individuals (0.6% of 314) identified as hybrids, suggesting a rare occurrence of interbreeding between the two study species. The Bayesian assignment plot (S1 Fig) also reflected some individuals with admixed ancestry, including the two individuals of hybrid origin. Genetic species assignments rather than field identification were employed in subsequent analyses (S1 Table).
Genetic diversity and population structure
Microsatellite DNA polymorphism was high in both species, as indicated by mean numbers of alleles (NA) and observed heterozygosity (Ho) for M. ochrogaster (NA = 22.7; Ho = 0.80) and M. pennsylvanicus (NA = 16.3; Ho = 0.70), respectively (Table 2). Pairwise FST values by patch and site were non-significant for M. ochrogaster at both spatial and temporal scales, save for comparisons involving a patch in the southernmost SAFE site (i.e., Tombstone patch at Prairie Ridge (S3 Table).
Table 2. Genetic diversity in 212 M. ochrogaster (MIOC) and 75 M. pennsylvanicus (MIPE) based on 15 microsatellite loci.
| Site | N | N A | NA SE | A R | AR SE | P AR | PAR SE | H o | Ho SE | |
|---|---|---|---|---|---|---|---|---|---|---|
| MIOC | Livingston | 28 | 13.5 | 1.4 | 12.4 | 1.2 | 1.4 | 0.3 | 0.81 | 0.04 |
| Montgomery | 71 | 17.7 | 1.8 | 12.7 | 1.1 | 1.6 | 0.3 | 0.79 | 0.04 | |
| Pontiac | 23 | 13.2 | 1.1 | 13.0 | 1.1 | 1.4 | 0.3 | 0.83 | 0.04 | |
| Prairie Ridge | 85 | 16.2 | 1.7 | 12.1 | 1.1 | 1.0 | 0.3 | 0.80 | 0.05 | |
| Saybrook | 5 | 6.3 | 0.5 | - | - | - | - | 0.84 | 0.06 | |
| MIPE | Livingston | 6 | 6.3 | 0.5 | - | - | - | - | 0.63 | 0.07 |
| Saybrook | 69 | 15.7 | 1.3 | 12.0 | 0.9 | 7.15 | 0.7 | 0.70 | 0.06 |
Samples were collected from five SAFE sites. N = number of individuals genotyped; Na = mean number of alleles per locus; Na SE = standard error Na; AR = allelic richness corrected for sample size; AR SE = standard error AR; PAR = private allelic richness corrected for sample size; PAR SE = standard error PAR; Ho = observed heterozygosity; Ho SE = standard error Ho.
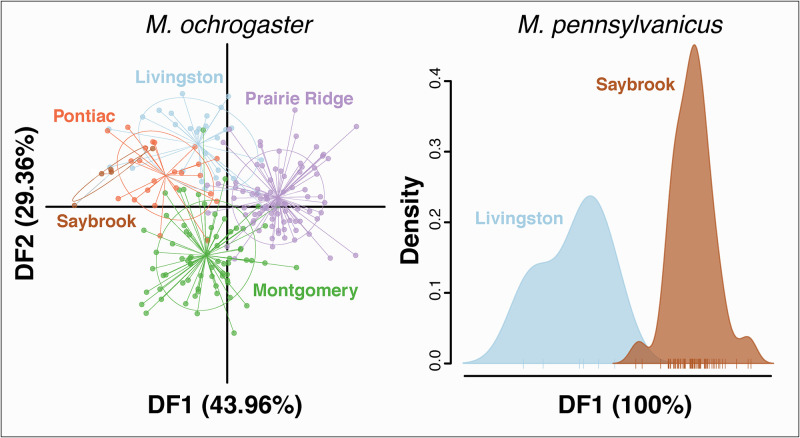
PCA revealed a greater spread within rather than among sites for both vole species, with the first two PC axes capturing 3.76% and 3.29% of variation in M. ochrogaster, and 6.12% and 4.96% of variation in M. pennsylvanicus (S2 Fig). For the DAPC (Fig 2), 60 PCs were retained for M. ochrogaster, and 30 for M. pennsylvanicus (S3 Fig). Genetic clusters in the DAPC overlapped for M. ochrogaster, but with spatial relationships generally mirroring geography, showing a slightly stronger association between the three norther-central SAFE sites (Pontiac, Livingston and Saybrook) and the two southern sites (Montgomery and Prairie Ridge), respectively (Fig 2). Clusters for M. pennsylvanicus overlapped less so, although samples were only available for two sites (i.e., Livingston and Saybrook; Fig 2).
Fig 2. Discriminant analyses of principal components (DAPC) for M. ochrogaster and M. pennsylvanicus.
Genetic clusters found at five SAFE sites: Livingston, Montgomery, Pontiac, Prairie Ridge, and Saybrook. Data derived from 15 microsatellite loci. Colors reflect unique sites and are consistent across species; individuals are represented as points for M. ochrogaster and ticks for M. pennsylvanicus. The percentage of variation from the discriminant analysis captured by each discriminant function axis (DF) is in parentheses.
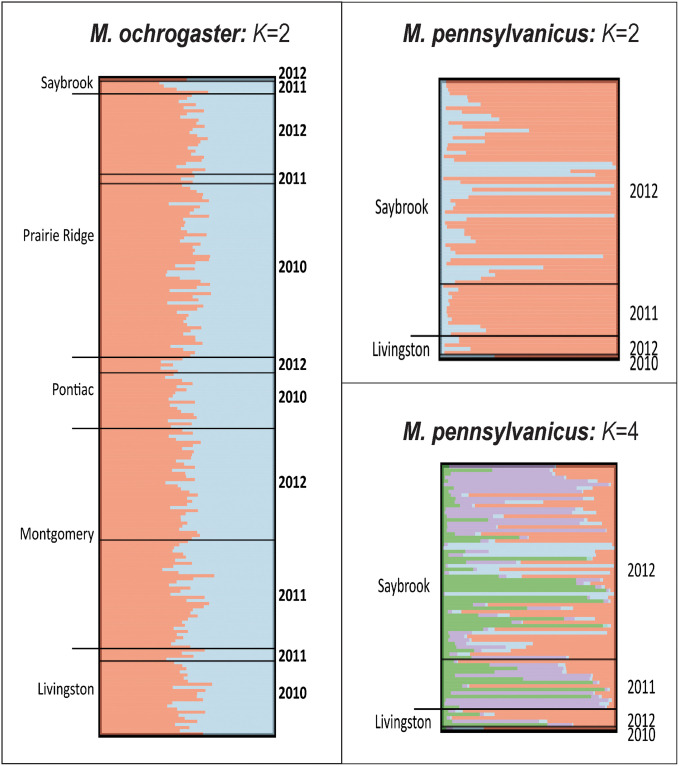
Assignment tests failed to recover distinct gene pools for M. ochrogaster, despite the selection of K = 2 (Fig 3) by the ΔK statistic (S4 Fig). Rather, the species was represented as a single homogenous population across the study area, in concordance with the selection of K = 1 (Pr(K) statistic) as the best explanation of population structure (S4 Fig). In contrast, M. pennsylvanicus (Fig 3) was represented by either two clusters (ΔK; S4 Fig) or four [Pr(K) S4 Fig]. However, one cluster in both K = 2 and K = 4 mostly corresponded to a family group recovered by Colony. Here, four (of 12) individuals (33%) in this family group were assigned fractionally to the cluster by Structure (ancestry < 0.15). We interpreted this as a consequence of the relatively low probability assigned to the family group by Colony [Pr = 0.7038; a list of family groups is provided in the Open Science Framework repository (doi:10.17605/OSF.IO/K2M8W)]. No other population or family structure was concordant among both Structure and Colony outputs. Scant evidence of spatial or temporal structure was apparent for either species.
Fig 3. Spatial and temporal genetic patterns in M. ochrogaster and M. pennsylvanicus.
Power analysis indicated adequate power to discern population clusters in both species (S5 Fig). This was true across all temporal scales in M. ochrogaster, despite an apparent lack of structure in the empirical data. However, M. pennsylvanicus shows sensitivity to divergence time in that a minimum of 15 simulated generations was required for significant population differences in >90% of replicates. This is likely a consequence of low sample size (N = 6) associated with the Livingston site.
Plots are based on Bayesian clustering (Structure v2.3.4). Colors reflect distinct gene pools and horizontal bars within plots represent individuals. The proportion of each color within an individual bar reflects the probability of ancestry. Samples are grouped by site and year (separated by black bars). Sample sizes are in Table 2.
Relatedness
We removed 19 individuals from our analyses because of relatedness (r) values >0.25. Average within-patch r-values ranged from 0.009–0.106, with higher values generally seen among individuals at the southernmost SAFE site (Prairie Ridge; S4 Table), where relatedness at one restoration patch (PRHA) differed significantly between 2010 and 2012 (t = -3.06, P<0.028), whereas those for the other patch (PRTO) did not (t = -0.54, P>0.6). An insufficient sample size prevented such a comparison for the third patch.
Average pairwise relatedness and geographic distance were significantly but inversely related across patches and sites in 2010, with relatedness diminishing as distances increased (3 sites, 6 patches; P = 0.027). However, relatedness versus distance was non-significant in 2012 (2 sites, 4 patches; P = 0.13).
Estimates of migration and effective population size
Estimates of migration (GeneClass2) indicated contemporary movements of individuals among SAFE sites (Table 3). In M. ochrogaster, five individuals (= 2%) were identified as potential migrants based on an assignment probability threshold <0.01. No potential migrants were identified for M. pennsylvanicus, but it was only captured at two patches, with only six individuals sampled from Livingston and the remaining 77 from Saybrook.
Table 3. Rates of migration versus residency for M. ochrogaster.
| 2010 | Livingston | Prairie Ridge |
| Livingston | 0.767 (0.418) | 0.291 (0.066) |
| Prairie Ridge | 0.233 (0.216) | 0.709 (0.485) |
| 2012 | Montgomery | Prairie Ridge |
| Montgomery | 0.858 (0.51) | 0.265 (0.003) |
| Prairie Ridge | 0.142 (0.0003) | 0.735 (0.341) |
Estimates were derived within and between SAFE sites for years and sites with sufficient sample sizes: 2010 (Livingston N = 24; Prairie Ridge N = 57) and 2012 {Montgomery N = 35; Prairie Ridge N = 30). Values are posterior estimates for mean and 95% highest posterior density (in parentheses). Bold values = residency estimates.
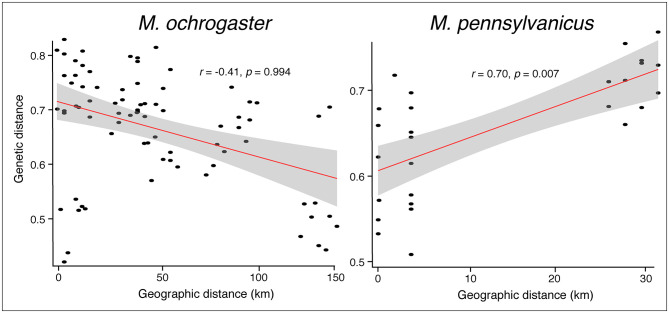
Tests for IBD notably differed between species, in a manner concordant with the migration analysis. Here, M. ochrogaster showed little evidence for dispersal limitation over the distances examined (maximum pairwise distance = 143km), with a negative but non-significant relationship between geographic and genetic distances (Mantel r = -0.41; p = 0.994; Fig 4). However, results for M. pennsylvanicus reflected an IBD pattern (Mantel r = 0.70; p = 0.007), despite a much smaller maximum observed distance (= 31.6km; Fig 4).
Fig 4. Patterns of isolation-by-distance among M. ochrogaster and M. pennsylvanicus.
For M. ochrogaster estimates are based on samples from five SAFE sites (= Livingston, Montgomery, Pontiac, Prairie Ridge, and Saybrook) and for M. pennsylvanicus on two sites (= Livingston and Saybrook). The X-axis represents pairwise geographic distances computed from latitude and longitude coordinates, whereas the Y-axis represents genetic distances computed among localities using allele frequencies for 15 microsatellite loci. Lines represent linear models, with confidence intervals in gray. Numbers above linear regression lines represent Mantel r and p-values estimated from 999 random permutations.
Population fluctuations
In M. ochrogaster Ne declined (but non-significantly) at Prairie Ridge from 148 in 2010 (95% CI = 83–479) to 58 in 2012 (95% CI = 32–197; t = 0.82; P<0.4136). At Montgomery, M. ochrogaster populations also displayed a reduced Ne estimate, with values declining from 579 (95% CI = 67—∞) in 2011 to 169 (95% CI = 67—∞) in 2012 (t = 3.06; P<0.0032). A test of significance could not be performed due to confidence intervals trending to infinity. Comparison of each confidence interval demonstrated a lack of significance, as they overlapped reciprocally with Ne estimates for each year (S6 Fig).
Discussion
Ecological restoration is an integral aspect of biodiversity conservation and wildlife management in anthropogenically fragmented habitats, but it must be quantitatively assayed to determine if ongoing efforts are indeed effective. Thus, metrics are needed to quantify how extensive and at what level restored patches are colonized and interconnected [53]. Habitat fragmentation is often best assessed by estimating connectivity among populations of small mammals [54], where level of isolation (i.e., reduced gene flow) may be ecologically more relevant than patch size, particularly when agricultural plots are converted into natural habitat [55]. However, population connectivity is less well studied within large, homogenized agricultural landscapes when compared to natural habitats, due largely to the lack of diversity in small mammal communities [56]. In addition, those studies that evaluated genetic structuring and diversity in agroecosystems reported conflicting results [57].
Our empirical evaluation of genetic structure and population connectivity in two vole species provides insights into the mechanisms by which restored patches of an agroecosystem may facilitate population persistence in target species. A lack of spatial genetic structure in our data, even over larger geographic distances, suggests the landscape mosaic dominating Illinois is highly permeable for voles and does not limit their dispersal. Despite extensive fragmentation, restored patches are sufficiently connected via edge habitat that provides corridors [58], allowing voles to rapidly colonize and supporting demographic resilience [59, 60]. Thus, our results help sustain restoration goals while documenting the effectiveness of management options.
Taxonomic uncertainty and species abundances
The correct identification of species in the field is a fundamental assumption when ecological hypotheses are tested, and when conservation and management initiatives are implemented. Species uncertainty, particularly among phenotypically-similar sibling species, can bias population estimates [61]. Microtus spp., including the two vole species in this study, are difficult to distinguish in the field, with a diagnosis often based on qualitative characters that relate to pelage, as well as differences in tooth morphology [62].
Fortunately, species-diagnostic DNA markers can unambiguously discriminate among sympatric species [63], as well as decipher potential hybridization and introgression. In our data 25% of field identifications did not match those derived genetically for M. ochrogaster and M. pennsylvanicus. Similar disparities have been observed in other studies [31].
Hybridization and resulting admixture represent another confounding factor when taxa are identified based on phenotype [64]. In our study, genotypes of two samples reflected admixture, and both were from a SAFE site (Saybrook) with a preponderance of M. pennsylvanicus. Admixture between species is often promoted by range shifts among distributions previously discrete. The construction of Illinois interstate highways have had variable impacts on wildlife [65], and seemingly facilitated a range expansion of M. pennsylvanicus from northern into central Illinois during the 1970’s. This created a local contact zone between the two species [66] and, in turn, may have facilitated opportunities for inter-species mating. These aspects underscore the necessity of molecular species identification, particularly for sympatric voles.
Field studies showed that the two vole species responded differently to the environment, particularly as it related to trophic and habitat preferences [67, 68]. While M. ochrogaster was more tolerant of sparse cover, M. pennsylvanicus preferred more dense vegetation. Habitat preference may thus explain the disparity in our sample sizes, particularly given that vegetation communities varied among study sites [23, 24]. Despite efforts to obtain equal numbers for each species, our samples were heavily skewed towards M. ochrogaster, particularly after genetically reclassifying 67 samples initially field-identified as M. pennsylvanicus (S1 Fig).
Voles are important components of grassland ecosystems [69] and they respond differentially to vegetation management. Thus, accurate identification is clearly important when the success of prairie restoration is being evaluated, and our study documents the effectiveness of genetic data in accurately diagnosing species. Species-specific assays, such as the Microtus avpr1a gene applied in this study, make molecular taxonomy easy and affordable. In addition, as demonstrated herein, appropriate analytical approaches such as Bayesian clustering can also accurately diagnose species if indeed population genetic data are available.
Dispersal capacity in agricultural landscapes
Spatially-structured habitats such as the agroecosystems dominating the midwestern plains of North America are characterized by heterogeneity in patch quality, and this serves to modulate demographic processes such as population density and dispersal rates [13]. Small mammals represent a substantial component of the biodiversity within grassland ecosystems, with deer mice (Peromyscus sp.) and vole (Microtus sp.) being both ubiquitous and linked to complex trophic interactions [69]. However, these dynamics are strongly influenced in modified agricultural landscapes by crop type, farming practices, and vegetational structure [68].
Experimental manipulations of patch quality demonstrated that food availability and predation risk indeed affected dispersal in M. ochrogaster [70], as well as intrinsic population demographics (e.g., growth rates and reproductive success; [71]), but not social structure [70, 72]. Habitat fragmentation also impacts movements, and a 7-year mark-recapture study of M. ochrogaster [15] showed less frequent dispersal occurred when habitat fragmentation was elevated, but distances so moved were greater. This frequency versus distance was interpreted as a balance between costs associated with habitat loss versus higher predation risk via distances traveled.
Given these observations, we predicted that the highly fragmented and agriculturally dominant landscape of central Illinois would impede vole dispersal, and the cumulative effects of limited gene flow would manifest themselves in genetic structure at larger spatial scales [59]. However, for M. ochrogaster we detected neither significant structure (Figs 2 and 3), nor genetic divergence among sites (S3 Table), which suggests the maintenance of connectivity across the study area. Here, the many intervening, but unsampled SAFE sites (Fig 1B) may have facilitated dispersal and persistence as a stepping-stone model with edge-habitat serving as corridors.
‘Border habitats’ such as fencerows, roadside ditches, and waterways can also promote the recolonization of restored grasslands [73], and may be especially important in maintaining vole populations [60], particularly given the rapid demographic response often observed after a population crash [74]. In this sense, restored grasslands can serve as a ‘source,’ buffering those population declines induced by the annual harvesting/ replanting of crops [23, 43]. In the context of vole population cycles, this would also promote demographic resilience [75, 76]. The spatial clustering we detected in M. pennsylvanicus is most likely related to social behavior (see below).
Genetic structure in voles
Dispersal in voles is modulated spatially and temporally [43] by landscape characteristics, population fluctuations, and social systems. Thus, gene flow is impacted by both intrinsic (life history) and extrinsic (environment) processes, and population genetic analyses can reveal which of these forces may be at play.
Structure and spatial scale
Geographic distance can be a primary factor in determining the dispersal of rodents in converted ecosystems [77]. However, we failed to detect significant spatial structure or patterns of IBD in M. ochrogaster (Fig 4). Lack of genetic structure could be due to insufficient resolution of the markers used (false negative or type II error), but our power analyses demonstrated that genetic diversity, at least in M. ochrogaster, was sufficient to detect structure, if present (S5 Fig). High levels of connectivity were also reflected in low, mostly non-significant FST values (S3 Table), as well as potential migrants between sites (Table 3). Microtus pennsylvanicus was only available from two SAFE sites, and the significant IBD pattern (Fig 4) was likely due to clustering of related individuals.
Structure and demographic cycles
Fluctuations in population sizes and densities among years can also elicit temporal shifts in genetic structure [43, 73]. Both vole species display multi-annual (but non-synchronous) population density cycles [67, 74]. Higher densities lead to increased dispersal tendencies and thus gene flow, whereas low densities reduce both, thus increasing potential population divergence via genetic drift [43]. We did not detect significant temporal shifts in population genetic parameters, with estimates of Ne and relatedness stable over time (S6 Fig).
Our sampling results were uneven among years, impacted by an apparent regional population decline (2011) and a substantial drought (2012). Most individuals (47%) were collected in 2012, fewer in 2010 (30%), and least in 2011 (23%), with but a few samples (N<5) available for three sites in 2011, and none at two sites in 2010 (Table 1). Population fluctuations occurred among years, per CPUE estimates (Catch-per-unit-effort; [23]), likely reflecting variance in population densities. However, densities may not have been similar across larger restoration patches, and trapping may have occurred in high- or low-density areas within a given patch. Genetic data are useful to infer population connectivity at large spatial scales, but they cannot inform about demographic connectivity unless direct measures of relatedness are estimated [78].
Structure and social systems
At more local scales, genetic structure in voles can be attributed to spatial clustering of kin [18], or reflect sex-specific differences in habitat use [73]. The two vole species differ in this regard, with M. ochrogaster socially monogamous with strong pair bonds and an elevated degree of philopatry and sociality [79]. Preferences for familiar peers are maintained in part by aggression toward unfamiliar individuals, as in mate partnerships [80]. Conversely, M. pennsylvanicus is promiscuous [19] and males are more likely to disperse [70], with reduced aggression toward unfamiliar conspecifics [80], and formation of social groups in the non-breeding season [20] being facilitated.
Therefore, both species seasonally gather in communal groups, and clustering of M. pennsylvanicus in our data is attributed to the aggregation of related individuals (Fig 3). Clustering of kin could also contribute to our observation of temporal stability in genetic diversity. A study involving the root vole, M. oeconomus, is consistent with this hypothesis, in that temporal stability of genetic composition was attributed in part to individuals immigrating from nearby areas that were closely-related genetically [72].
Restoration success depends upon the goal
Both patch size and connectivity (effective corridors) are essential to sustain genetically diverse populations [81], and thus underscore the success of ecological restoration. In our study, population connectivity among restoration sites was high and sustained over large geographic distances. Voles seemingly disperse through this landscape mosaic and can quickly colonize newly-restored patches, and even smaller patches appear suitable to sustain populations if indeed connectivity is maintained [23]. Thus, the SAFE model seemingly achieves restoration goals for those small mammals with early colonizing ability.
Medium- to large-sized species are more sensitive to patch size and landscape context in the prairie community [58, 82]. A comparative population genetic study of upland game birds [65] demonstrated that patch size is relevant to sustain Pheasant, Greater Prairie Chicken, and Bobwhite Quail populations, as indicated by the depressed population demographics (e.g., lower Ne, higher relatedness) found in smaller habitat patches (S5 Table).
Two models are often implemented in the effort to retain/ increase grassland habitat through conversion of agricultural land. In the land-sharing model, small patches of unfarmed natural or semi-natural vegetation are retained by embedding them within larger agricultural plots, [83]. In contrast, the land-sparing model focuses on protecting/ restoring native vegetation on existing grasslands, rather than by converting agricultural land [84]. While land-sharing seems to be a strategy that is effective for species with short generation times and natural population fluctuations such as voles (i.e., they can quickly respond to changes in the landscape due to crop rotation), land-sparing is more important for longer-lived species or those with specific habitat requirements as a component of life history (e.g., leking grounds for Greater Prairie Chicken, [85]). The SAFE model of small, time-limited restoration patches (i.e., 10–15-year enrollment intervals) works well for some biodiversity components, but the retention of larger grassland patches is essential for others and should become an integral part of ecological restoration of prairie habitats to sustain larger, more demographically resilient populations [86]. Thus, restoration ’success’ will differ across species.
Restoration beyond wildlife: A broader perspective
Benefits of ecological restoration are often aimed at providing habitat for wildlife or ensuring ecosystems functioning in an anthropogenic landscape, and are thus viewed from a biodiversity conservation stance [87, 88]. Increasingly, a broader perspective is now being applied that also considers economic impacts [89, 90] and human health [91, 92]. Financial incentives to landowners are an integral part of the SAFE initiative, but prairie habitats are also valued for their aesthetic appeal and contribution to local economies via recreation and ecotourism (e.g., bird watching, hiking, hunting) [90]. Retaining or restoring grasslands also has strong potential for natural climate mitigation [93]. Ecological restoration can benefit human health in a variety of ways, to include physiological and psychological health [92]. However, these relationships are complex and much uncertainty is involved [94].
Restoration may also have different impacts on functional connectivity of biodiversity components, including species that can contribute to spreading emerging infectious diseases (EIDs) [95, 96] In this regard, grassland rodents are recognized as important vectors and host reservoirs for EIDs, as evidenced by the contemporary spread of Hanta virus [97], as well as Lyme disease [98, 99]. In some cases, restoration reduced disease risk by diminishing the density of vectors (ticks; [100]), or pathogens (Hanta virus; [101]). In other cases, restoration practices that benefited wildlife species were also associated with higher disease risk. These can often be mitigated by targeting management of the host species (e.g., culling of deer; [102]).
In the context of grassland restoration, Lyme disease is of potential concern [103], in that the pathogen (Borrelia burgdorferi) is carried by a vector tick that relies upon vertebrate hosts (including voles) for dispersal [104]. Some studies have documented that an increase in tick density due to an increase in host prevalence did not associate with higher prevalence of the pathogen, and vice-versa [105]. In this sense, functional connectivity of the host-vector relationship may be uncoupled, and thus unpredictable. However, given the threat of Lyme disease to human health, monitoring the response of vectors to restoration efforts should be considered when benefits and risks of restoration are evaluated. An approach that coordinates agency programs, such as habitat restoration and wildlife management, can reduce potential risks [100]. Also, integrating data from different programs can inform comprehensive policies [96] and coordinate various initiatives [106].
Conclusions
Restoring agricultural plots in midwestern North America to endemic tall grass prairie is an ongoing process implemented by federal and state agencies, but often with success difficult to quantify. Molecular approaches can provide metrics to gauge if specific restoration goals have been successfully achieved, and were applied in this study to quantify connectivity among vole populations in rehabilitated prairie patches in Illinois.
Molecular approaches can complement the ecological assessment of restoration success in multiple ways, and in addition, provide insights not obtainable otherwise. As demonstrated in this study, key aspects include genetic clarification of taxonomic uncertainty, and metrics to quantify population connectivity over larger spatial and temporal scales.
Voles in this study reflected a certain degree of demographic resilience [14, 75] as indicated by temporally stable Ne and relatedness values, despite a regional population decline in 2011. Despite this, a level of caution is warranted such that genetic data is not over-interpreted in the context of ecological inferences. Genetic connectivity, as examined in this study, does not necessarily equate to demographic connectivity [78].
Advances in molecular technologies and analyses have expanded the capacity of genetic/genomic data to address questions previously refractive. Genetic data can reveal how species respond to specific management actions (e.g., invasive species control; [107]), or test if ecological methods at the local scale (e.g., mark-recapture) can be extrapolated at the regional scale.
Collaborations between molecular and restoration ecologists can invoke a more nuanced perspective of ’restoration success.’ Genetic patterns emerge as a response to different processes, and data must be carefully evaluated [108]. In our study, genetic structure in M. pennsylvanicus was a response to social behavior (clustering of kin), and not isolation (reduced geneflow), thus mirroring observations on an iconic prairie species, the Greater Prairie Chicken [85]. Ecology must be considered along with life history when interpreting genetic patterns of study species. For example, our analyses identified potential migrants amongst SAFE sites, suggesting individual dispersal across 250km of agroecosystem—a biologically improbable event.
Supporting information
Field-identification based on morphology was verified by genetic clustering, with 25% of samples reclassified.
(PDF)
Genetic clusters detected in each species at Illinois SAFE sites based on PCA applied to 15 microsatellite loci.
(PDF)
‘Optimal’ number of retained principal component (PC) axes (X-axis) based on minimum of root-mean-square-error (RMSE) assignment (Y-axis) of 20% of randomly selected samples.
(PDF)
Evaluation of 10 replicates at K values 1–10 based on the ’ad hoc statistic’ ΔK and the probability of K.
(PDF)
Ability to resolve population structure as evaluated with two statistical measures (χ2 and Fisher exact tests).
(PDF)
Comparison of Ne estimates and 95% jackknife confidence intervals between years and two SAFE sites.
(PDF)
Details on geographic location, patch area, acronyms and sample sizes based on phenotypic and genetic identifications.
(PDF)
Details on primers used to amplify 23 microsatellite loci.
(PDF)
Values calculated for 2010 and 2012 between restoration patches.
(PDF)
Estimates of within-patch average and variance by year.
(PDF)
Comparison of population genetic and life history parameters for two vole and three bird species.
(PDF)
Acknowledgments
We thank P. Wolff, S. Beyer, W. Hill, S. McLaughlin, K. Barmann, C. Griffith, B. Neece, A. Ahlers, J. Andrews, G. Spyreas, and J. Larkin for fieldwork. Illinois Department of Natural Resources (IDNR) facilitated landowner access to SAFE sites. The use of trade, product, industrial, or firm names is for informative purposes only and does not constitute an endorsement by the U.S. Government or the U.S. Fish and Wildlife Service (USFWS). Links to non-Service Web sites do not imply official USFWS endorsement of opinions or ideas expressed therein or guarantee the validity of the information provided. The findings, conclusions, and opinions expressed in this article represent those of the authors, and not necessarily that of the USFWS, Illinois Department of Natural Resources, Illinois state government, nor any other affiliated institution.
Data Availability
Data available in Open Science Framework respository under https://osf.io/k2m8w/.
Funding Statement
The U.S. Fish and Wildlife Service (USFWS) Federal Aid in Wildlife Restoration Program, as administered by the Illinois Department of Natural Resources (IDNR), provided funding for this project (awarded to MRD and MED). Additional funding was provided by endowments through University of Arkansas/ Fayetteville to MRD (Bruker Professor of Life Sciences) and MED (21st Century Chair in Global Change Biology).
References
- 1.Andrewartha HG, Birch LC. The Distribution and Abundance of Animals. Chicago: University of Chicago Press; 1954. [Google Scholar]
- 2.Chen I, Hill JK, Ohlemueller R, Roy DB, Thomas CD. Rapid range shifts of species associated with high levels of climate warming. Science. 2011; 333: 1024–1026. doi: 10.1126/science.1206432 [DOI] [PubMed] [Google Scholar]
- 3.Corlett RT. The anthropocene concept in ecology and conservation. Trends Ecol Evol. 2015; 30: 36–41. doi: 10.1016/j.tree.2014.10.007 [DOI] [PubMed] [Google Scholar]
- 4.Crooks KR, Sanjayan M, editors. Connectivity Conservation. Cambridge: Cambridge University Press; 2006. [Google Scholar]
- 5.Henwood WD. Toward a strategy for the conservation and protection of the world’s temperate grasslands. Great Plains Res. 2010; 20: 121–134. https://www.iucn.org/content/towards-conservation-strategy-worlds-temperate-grasslands [Google Scholar]
- 6.Wright CK, Wimberly MC. Recent land use change in the western corn belt threatens grasslands and wetlands. Proc Natl Acad Sci USA. 2013; 110: 4134–4139. doi: 10.1073/pnas.1215404110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tscharntke T, Klein AM, Kreuss A, Steffan-Dewenter I, Thies C. Landscape perspectives on agricultural intensification and biodiversity–ecosystem service management. Ecol Lett. 2005; 8: 857–874. doi: 10.1111/j.1461-0248.2005.00782.x [DOI] [Google Scholar]
- 8.Hibbard K, Wilson T, Avery K, Harriss R, Newmark R, Rose, et al. Ch. 10: Energy, water, and land use. In: Melillo JM, Richmond TC, Yohe GW, editors. Climate Change Impacts in the United States: The Third National Climate Assessment. Washington DC: U.S. Global Change Research Program; 2014. p. 257–285. http://nca2014.globalchange.gov/report/sectors/energy-water-and-land
- 9.Pryor SC, Scavia D, Downer C, Gaden M, Iverson L, Nordstrom R, et al. Ch. 18: Midwest. Climate Change Impacts in the United States. In: Melillo JM, Richmond TC, Yohe GW, editors. Climate Change Impacts in the United States: The Third National Climate Assessment. U.S. Washington DC: Global Change Research Program; 2014. p. 418–440. Also http://nca2014.globalchange.gov/report/regions/midwest
- 10.Gelfand I, Sahajpal R, Zhang X, Izaurralde RC, Gross KL, Robertson GP. Sustainable bioenergy production from marginal lands in the US Midwest. Nature. 2013; 493: 514–520. doi: 10.1038/nature11811 [DOI] [PubMed] [Google Scholar]
- 11.Olden JD, Douglas ME, Douglas MR. The human dimensions of biotic homogenization. Conserv Biol. 2005; 19: 2036–2038. doi: 10.1111/j.1523-1739.2005.00288.x [DOI] [Google Scholar]
- 12.Kline V, Howell E. Prairies. In: Jordan W., Gilpin M, Aver J, editors. Restoration Ecology: A Synthetic Approach to Ecological Research. New York: Cambridge University Press; 1987. p. 75–483. [Google Scholar]
- 13.Batzli GO. Habitat fragmentation, vole population fluctuations, and the ROMPA hypothesis: An experimental test using model landscapes. Integr Zool. 2016; 11: 469–482. doi: 10.1111/1749-4877.12209 [DOI] [PubMed] [Google Scholar]
- 14.Coffman CJ. The effects of corridor-linked patches on metapopulation dynamics: a field experiment with Microtus pennsylvanicus [dissertation]. Raleigh: North Carolina State University. 1997.
- 15.Diffendorfer JE, Gaines MS, Holt RD. Habitat fragmentation and movements of three small mammals (Sigmodon, Microtus, and Peromyscus). Ecology. 1995; 76: 827–839. doi: 10.2307/1939348 [DOI] [Google Scholar]
- 16.Smith JE, Batzli GO. Dispersal and mortality of prairie vole (Microtus ochrogaster) in fragmented landscapes: A field experiment. Oikos. 2006; 112: 209–217. doi: 10.1111/j.0030-1299.2006.13431.x [DOI] [Google Scholar]
- 17.Roberts RL, Williams JR, Wang AK, Carter Porges C. Cooperative breeding and monogamy in prairie voles: Influence of the sire and geographical variation. Animal Behaviour. 1998; 55: 1131–1140. doi: 10.1006/anbe.1997.0659 [DOI] [PubMed] [Google Scholar]
- 18.Keane B, Ross S, Crist TO, Solomon NG. Fine-scale spatial patterns of genetic relatedness among resident adult prairie vole. J Mammal. 2015; 96: 1194–1202. doi: 10.1093/jmammal/gyv128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boonstra R, Xia X, Pavone L. Mating system of the meadow vole, Microtus pennsylvanicus. Behav. Ecology. 1993; 4: 83–89. doi: 10.1093/beheco/4.1.83 [DOI] [Google Scholar]
- 20.Getz LL. Social structure and aggressive behavior in a population of Microtus pennsylvanicus. J. Mammal. 1972; 53: 310–317. doi: 10.2307/1379167 [DOI] [Google Scholar]
- 21.Amaral KE, Palace KEM, O’Brien KM, Fenderson LE, Kovach AI. Anthropogenic habitats facilitate dispersal of an early successional obligate: Implications for restoration of an endangered ecosystem. PLoS ONE. 2016; 11(3): e0148842. doi: 10.1371/journal.pone.0148842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson SJ, Kierepka EM, Shiwart RK, Latch OE, Rhodes OE Jr. Assessing the permeability of landscape features to animal movement: Using genetic structure to infer functional connectivity. PLoS ONE. 2015; 10(2): e0117500. doi: 10.1371/journal.pone.0117500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mulligan MP, Shooley RL, Ward MP. Effects of connectivity and regional dynamics on restoration of small mammal communities in midwestern grasslands. Restor Ecol. 2013; 21: 678–685. doi: 10.1111/rec.12039 [DOI] [Google Scholar]
- 24.Mulligan MP. Effects of connectivity and regional dynamics on restoration trajectories for small mammal communities on midwestern grasslands [thesis]. Champaign-Urbana (IL): University of Illinois. 2012.
- 25.Van Oosterhout C, Huthinson WF, Wills DMP, Shipley P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004; 4: 535–538. doi: 10.1111/j.1471-8286.2004.00684.x [DOI] [Google Scholar]
- 26.Rousset F. genepop’ 007: A complete re-implementation of the genepop software for windows and linux. Mol Ecol Resour. 2008; 8: 103–106. doi: 10.1111/j.1471-8286.2007.01931.x [DOI] [PubMed] [Google Scholar]
- 27.Rice W. Analyzing tables of statistical tests. Evolution. 1989; 43: 223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x [DOI] [PubMed] [Google Scholar]
- 28.Peakall R, Smouse PE. Genalex 6.5: Genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics. 2012; 28: 2537–2539. doi: 10.1093/bioinformatics/bts460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000; 155: 945–959. doi: 10.1093/genetics/155.2.945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Falush D, Stephens M, Pritchard J. Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics. 2003; 164: 1567–1587. doi: 10.1093/genetics/164.4.1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Henterly AC, Mabry KE, Solomon NG, Chesh AS, Keane B. Comparison of morphological versus molecular characters for discriminating between sympatric meadow and prairie voles. Am Midl Nat. 2011; 165: 412–420. doi: 10.1674/0003-0031-165.2.412 [DOI] [Google Scholar]
- 32.Hammock E, Lim M, Nair H, Young L. Association of vasopressin 1a receptor levels with a regulatory microsatellite and behavior. Genes Brain Behav. 2005; 4: 289–301. doi: 10.1111/j.1601-183X.2005.00119.x [DOI] [PubMed] [Google Scholar]
- 33.Ophir AG, Campbell P, Hanna K, Phelps SM. Field tests of cis-regulatory variation at the prairie vole avpr1a locus: Association with V1aR abundance but not sexual or social fidelity. Horm Behav. 2008; 54: 694–702. doi: 10.1016/j.yhbeh.2008.07.009 [DOI] [PubMed] [Google Scholar]
- 34.Solomon NG, Richmond AR, Harding PA, Fries A, Jacquemin S, Shaefer RL et al. Polymorphism at the avpr1a locus in male prairie voles correlated with genetic but not social monogamy in field populations. Mol Ecol. 2009; 18: 4680–4695. doi: 10.1111/j.1365-294X.2009.04361.x [DOI] [PubMed] [Google Scholar]
- 35.Kalinowski ST. HP-Rare 1.0: A computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes. 2005; 5: 187–189. doi: 10.1111/j.1471-8286.2004.00845.x [DOI] [Google Scholar]
- 36.Jombart T. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics. 2008; 24: 1403–1405. doi: 10.1093/bioinformatics/btn129 [DOI] [PubMed] [Google Scholar]
- 37.Jombart T, Devillard S, Balloux F. Discriminant analysis of principal components: a new method for the analysis of genetically structured populations. BMC Genet. 2010; 11: 94. doi: 10.1186/1471-2156-11-94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I. Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour. 2015; 15: 1179–1191. doi: 10.1111/1755-0998.12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software Structure: A simulation study. Mol Ecol. 2005; 14: 2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x [DOI] [PubMed] [Google Scholar]
- 40.Janes JK, Miller JM, Dupuis JR, Malenfant RM, Gorrell JC, Cullingham CI, et al. The K = 2 conundrum. Mol Ecol. 2017; 26: 3594–3602. doi: 10.1111/mec.14187 [DOI] [PubMed] [Google Scholar]
- 41.Jones OR, Wang J. (2010), Colony: a program for parentage and sibship inference from multilocus genotype data. Mol Ecol Resour. 2010; 10:551–555. doi: 10.1111/j.1755-0998.2009.02787.x [DOI] [PubMed] [Google Scholar]
- 42.Ryman N, Palm S. (2006), PowSim: a computer program for assessing statistical power when testing for genetic differentiation. Mol Ecol Notes. 2006; 6: 600–602. doi: 10.1111/j.1471-8286.2006.01378.x [DOI] [Google Scholar]
- 43.Gauffre B, Berthier K, Inchausti P, Chaval Y, Bretagnolle V, Cosson JF. Short-term variations in gene flow related to cyclic density fluctuations in the common vole. Mol Ecol. 2014; 23: 3214–3225. doi: 10.1111/mec.12818 [DOI] [PubMed] [Google Scholar]
- 44.Piry S, Alapetite A, Cornuet J-M, Baudoin L, Estoup A () GeneClass2: A software for [genetic assignment and first-generation migrant detection. J. Hered. 2004; 95: 536–539. doi: 10.1093/jhered/esh074 [DOI] [PubMed] [Google Scholar]
- 45.Paetkau D, Slade R, Burden M, Estoup A. Genetic assignment methods for the direct, real-time estimation of migration rate: A simulation-based exploration of accuracy power. Mol Ecol. 2004; 13: 55–65. doi: 10.1046/j.1365-294x.2004.02008.x [DOI] [PubMed] [Google Scholar]
- 46.Waples RS, Do C. LDNE: A program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour. 2008; 8: 753–756. doi: 10.1111/j.1755-0998.2007.02061.x [DOI] [PubMed] [Google Scholar]
- 47.Slatkin M. Linkage disequilibrium—Understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008; 9: 477. doi: 10.1038/nrg2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Do C, Waples RS, Peel D, Macbeth GM, Tillett BJ, Ovenden JR. NeEstimator v2: re-implementation of software for the estimation of contemporary effective population size (Ne) from genetic data. Mol Ecol Resour. 2014; 14: 209–214. doi: 10.1111/1755-0998.12157 [DOI] [PubMed] [Google Scholar]
- 49.Waples RS, Do C. Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl. 2010; 3:244–262. doi: 10.1111/j.1752-4571.2009.00104.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2008
- 51.Edwards A.W.F. (1971) Distance between populations on the basis of gene frequencies. Biometrics. 1971; 27: 873–881. [PubMed] [Google Scholar]
- 52.Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967; 27: 209–220. https://cancerres.aacrjournals.org/content/27/2_Part_1/209.long [PubMed] [Google Scholar]
- 53.Hunter-Ayad J, Hassall C. An empirical, cross-taxon evaluation of landscape-scale connectivity. Biodiv Conserv. 2020; 29:1339–1359. doi: 10.1007/s10531-020-01938-2 [DOI] [Google Scholar]
- 54.Lin YK, Batzli GO. The influence of habitat quality on dispersal demography, and population dynamics of voles. Ecol Monogr. 2001; 71: 245–275. doi: 10.1890/0012-9615(2001)071[0245:TIOHQO]2.0.CO;2 [DOI] [Google Scholar]
- 55.Oliver TH, Marshall HH, Morecroft MD, Brereton T, Prudhomme C, Huntingford C. Interacting effects of climate change and habitat fragmentation on drought-sensitive butterflies. Nature Clim Change. 2015; 5: 941–944. doi: 10.1038/NCLIMATE2746 [DOI] [Google Scholar]
- 56.Heroldová M, Bryja J, Zejda J, Tkadlec E. Structure and diversity of small mammal communities in agriculture landscape. Agr Ecosyst Environ. 2007; 120: 206–210. doi: 10.1016/j.agee.2006.09.007 [DOI] [Google Scholar]
- 57.Marchi C, Andersen LW, Damgaard C, Olsen K, Jensen TS, Loeschcke V. Gene flow and population structure of a common agricultural wild species (Microtus agrestis) under different land management regimes. Heredity. 2013; 111: 486–494. doi: 10.1038/hdy.2013.70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berry R, Schooley RL, Ward MP. Landscape context affects use of restored grasslands by mammals in a dynamic agroecosystem. Am Midl Nat. 2017; 177: 165–182. doi: 10.1674/0003-0031-177.2.165 [DOI] [Google Scholar]
- 59.Gauffre B, Estoup A, Bretagnolle V, Cosson JF. Spatial genetic structure of a small rodent in a heterogeneous landscape. Mol Ecol. 2008; 17: 4619–4629. doi: 10.1111/j.1365-294X.2008.03950.x [DOI] [PubMed] [Google Scholar]
- 60.Coffman CJ, Nichols JD, Pollock KH. Population dynamics of Microtus pennsylvanicus in corridor-linked patches. Oikos. 2001, 93: 3–21. doi: 10.1034/j.1600-0706.2001.930101.x [DOI] [Google Scholar]
- 61.Runge JP, Hines JE, Nichols JD. Estimating species-specific survival and movement when species identification is uncertain. Ecology. 2007; 88: 282–288. doi: 10.1890/0012-9658(2007)88[282:essamw]2.0.co;2 [DOI] [PubMed] [Google Scholar]
- 62.Hoffmeister DF. Mammals of Illinois. Champaign: University of Illinois Press. 2002. [Google Scholar]
- 63.Moran S, Turner PD, O’Reilly C. Non-invasive genetic identification of small mammal species using real-time polymerase chain reaction. Mol Ecol Resour. 2008; 8: 1267–1269. doi: 10.1111/j.1755-0998.2008.02324.x [DOI] [PubMed] [Google Scholar]
- 64.Frare CF, Matocq MD, Feldman CR, White AM, Manley PN, Jermstad KD, et al. Landscape disturbance and sporadic hybridization complicate field identification of chipmunks. J Wildl Manag. 2017; 81:248–258. doi: 10.1002/jwmg.21193 [DOI] [Google Scholar]
- 65.Douglas MR, Anthonysamy WJB, Mussmann SM, Davis MA, Louis W, Douglas ME. Multi-targeted management of upland game birds at the agroecosystem interface of Midwestern North America. PLoS ONE Biodiversity. 2020; 15(4): e0230735. doi: 10.1371/journal.pone.0230735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Getz L., Cole FR, Gates D. Interstate roadsides as dispersal routes for Microtus pennsylvanicus. J Mammal. 1978; 59: 208–212. doi: 10.2307/1379900 [DOI] [Google Scholar]
- 67.Getz LL, Hoffman JE, McGuire B, Dolan TW III. Twenty-five years of population fluctuations of Microtus ochrogaster and M. pennsylvanicus in three habitats in east-central Illinois. J Mammal. 2001; 82: 22–34. [Google Scholar]
- 68.Klatt BJ, Getz LL, McGuire B. Interspecific interactions and habitat use by prairie vole (Microtus ochrogaster) and meadow vole (M. pennsylvanicus). Am Midl Nat. 2015; 173: 241–252. doi: 10.1674/amid-173-02-241-252.1 [DOI] [Google Scholar]
- 69.Mérő TO, Bocz R, Polyák L. Horváth G, Lengyel S. Local habitat management and landscape-scale restoration influence small-mammal communities in grasslands. Anim Conserv. 2015; 18: 442–450. doi: 10.1111/acv.12191 [DOI] [Google Scholar]
- 70.Lin YK, Keane B, Isenhour A, Solomon NG. Effects of patch quality on dispersal and social organization of Prairie Voles: an experimental approach. J Mammal. 2006; 87: 446–453. doi: 10.1644/05-MAMM-A-201R1.1 [DOI] [Google Scholar]
- 71.Lin YK, Batzli GO. Movement of voles across habitat boundaries: Effects of food and cover. J Mammal. 2004; 85: 216–224. [Google Scholar]
- 72.Pilot M, Dabrowski M, Jancewicz E, Schtickzelle N, Gliwicz J. Temporally stable genetic variability and dynamic kinship structure in a fluctuating population of the root vole Microtus oeconomus. Mol Ecol. 2010; 19: 2800–2812. doi: 10.1111/j.1365-294X.2010.04692.x [DOI] [PubMed] [Google Scholar]
- 73.Chiappero MB, Sommaro LV, Priotto JW, Wiernes MP, Steinmann AR, Gardenal CN. Spatio-temporal genetic structure of the rodent Calomys venustus in linear, fragmented habitats. J Mammal. 2016; 97: 424–435. doi: 10.1093/jmammal/gyv186 [DOI] [Google Scholar]
- 74.Myers JH. Population cycles: Generalities, exceptions and remaining mysteries. Proc R Soc B. 2018; 285: 20172841. doi: 10.1098/rspb.2017.2841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Capedvila P, Stott I, Beger M, Salguero-Gómez S. Towards a comparative framework of demographic resilience. Trends Ecol Evol. 2020; 35: 776–786. doi: 10.1016/j.tree.2020.05.001 [DOI] [PubMed] [Google Scholar]
- 76.Thiessen Martens JR, Entz MH, Wonneck MD. Redesigning Canadian prairie cropping systems for profitability, sustainability, and resilience. Can J Plant Sci. 2015; 95:1049–1072. doi: 10.4141/CJPS-2014-173 [DOI] [Google Scholar]
- 77.Melis C, Borg AA, Jensen H, Bjorkvoll E, Ringsby TH, Saether TH. Genetic variability and structure of the water vole Arvicola amphibius across four metapopulations in northern Norway. Ecol Evol. 2013; 3: 770–778. doi: 10.1002/ece3.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Lowe WH, Allendorf FW. What can genetics tell us about population connectivity? Mol Ecol. 2010; 19:3038–3051. doi: 10.1111/j.1365-294X.2010.04688.x [DOI] [PubMed] [Google Scholar]
- 79.Getz LL, McGuire B, Pizzuto T, Hofmann JE, Fraze B. Social-organization of the prairie vole (Microtus ochrogaster). J Mammal. 1993; 74: 44–58. doi: 10.2307/1381904 [DOI] [Google Scholar]
- 80.Lee NS, Goodwin NL, Freitas KE, Beery AK. Affiliation, aggression, and selectivity of peer relationships in Meadow and Prairie Voles. Front Behav Neurosci. 2019; 13: 52. doi: 10.3389/fnbeh.2019.00052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kuparinen A, Festa-Bianchet M. Harvest-induced evolution: Insights from aquatic and terrestrial sys- tems. Phil Trans R Soc B. 2017; 372: 20160036. doi: 10.1098/rstb.2016.0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cosentino BJ, Phillips CA, Schooley RL, Lowe WH, Douglas MR. Linking extinction-colonization dynamics to the genetic structure of a salamander metapopulation. Proc R Soc B. 2012; 279: 1575–1582. doi: 10.1098/rspb.2011.1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fischer J, Abson DJ, Bustic V, Chappell MJ, Ekroos J, Hanspach J, et al. Land sparing versus land sharing: Moving forward. Conserv Lett. 2014; 7: 149–157. doi: 10.1111/conl.12084 [DOI] [Google Scholar]
- 84.Phalan BT. What have we learned from the land sparing-sharing model? Sustainability. 2018; 10:1760. doi: 10.3390/su10061760 [DOI] [Google Scholar]
- 85.Mussmann SM, Douglas MR, Anthonysamy WJB, Davis MA, Simpson SA, Wade L, et al. Genetic rescue, the Greater Prairie Chicken, and the problem of conservation-reliance in the Anthropocene. Roy Soc Open Sci. 2017; 4: e160736. doi: 10.1098/rsos.160736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matthews JW, Molano-Flores B, Ellis, Marcum PB, Handel W, Zylka J, et al. Impacts of management and antecedent site condition on restoration outcomes in a sand prairie. Rest Ecol. 2017, 25: 972–981. doi: 10.1111/rec.12525 [DOI] [Google Scholar]
- 87.Correa Ayram CA, Mendoza ME, Etter A, Salicrup DRP. Habitat connectivity in biodiversity conservation: A review of recent studies and applications. Prog Phys Geogr. 2015; 40: 7–37. doi: 10.1177/0309133315598713 [DOI] [Google Scholar]
- 88.Barral MP, Benayas JMR, Meli P, Maceira NO. Quantifying the impacts of ecological restoration on biodiversity and ecosystem services in agroecosystems: A global meta- analysis. Agr Ecosyst Environ. 2015; 202: 223–231. doi: 10.1016/j.agee.2015.01.009 [DOI] [Google Scholar]
- 89.Morlando S, Schmidt SJ, Loguidice K. Reduction in Lyme disease risk as an economic benefit of habitat restoration. Restor Ecol. 2012; 20: 498–504. doi: 10.1111/j.1526-100X.2011.00796.x [DOI] [Google Scholar]
- 90.Costanza R, d’Arge R, de Groot R, Farber S, Grasso M, Hannon B et al. The value of the world’s ecosystem services and natural capital. Nature. 1997; 387: 253–260. doi: 10.1038/387253a0 [DOI] [Google Scholar]
- 91.Aert R, Hoonay O, Van Nieuwenhuyse. Biodiversity and human health: mechanisms and evidence of the positive health effects of diversity in nature and green spaces. Brit Med Bull. 2018; 127: 5–22. doi: 10.1093/bmb/ldy021 [DOI] [PubMed] [Google Scholar]
- 92.Nabhan GP, Orlando L, Monti LS, Aronson J. Hands-on ecological restoration as a nature-based health intervention: Reciprocal restoration for people and ecosystems. Ecopsychol. 2020; 12(S1): 195–202. doi: 10.1089/eco.2020.0003 [DOI] [Google Scholar]
- 93.Fargione JE, Bassett S, Boucher T, Bridgham SD, Conant RT, Cook-Patton SC et al. Natural climate solutions for the United States. Sci Adv. 2018; 4: eaat1869. doi: 10.1126/sciadv.aat1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Speldewinde PC, Slaney D, Weinstein P. Is restoring an ecosystem good for your health? Sci Total Envir. 2015; 502: 276–279. doi: 10.1016/j.scitotenv.2014.09.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tracey JA, Bevins SN, Vanderwoude S, Crooks KR. An agent-based movement model to assess the impact of landscape fragmentation on disease transmission. Ecosphere. 2014; 5: 119. doi: 10.1890/ES13-00376.1 [DOI] [Google Scholar]
- 96.Gottdenker NL, Streicker DS, Faust CL, Carroll CR. Anthropogenic land use change and infectious diseases: A review of the evidence. Ecohealth. 2014; 11: 619–632. doi: 10.1007/s10393-014-0941-z [DOI] [PubMed] [Google Scholar]
- 97.Rubio AV, Avila-Flores R, Suzán G. Responses of small mammals to habitat fragmentation: Epidemiological considerations for rodent-borne hantaviruses in the Americas. Ecohealth. 2014; 11: 526–533. doi: 10.1007/s10393-014-0944-9 [DOI] [PubMed] [Google Scholar]
- 98.LoGuidice K, Ostfeld RS, Schmidt KA, Keesiing F. The ecology of infectious disease: Effects of host diversity and community composition on Lyme disease risk. Proc Natl Acad Sci USA. 2003; 100: 567–571. doi: 10.1073/pnas.0233733100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rydzewski JR, Mateus-Pinilla N, Warner RE, Hamer S, Weng H. Ixodes scapularis and Borrelia burgdorferi among diverse habitats within a natural area in east-central Illinois. Vector-Borne Zoonot. 2011; 11: 1351–1358. doi: 10.1089/vbz.2010.0160 [DOI] [PubMed] [Google Scholar]
- 100.Gilbert L. Can restoration of afforested peatland regulate pests and disease? J Appl Ecol. 2013; 50: 1226–1233. doi: 10.1111/1365-2664.12141 [DOI] [Google Scholar]
- 101.Priest PR, Prado A, Tambosi LR, Umetsu F, Bueno AD, Pardini R, et al. Moving to healthier landscapes: Forest restoration decreases the abundance of Hantavirus reservoir rodents in tropical forests. Sci Total Environ. 2021; 752: 141967. doi: 10.1016/j.scitotenv.2020.141967 [DOI] [PubMed] [Google Scholar]
- 102.Millins C. Gilbert L, Medlock J, Hansford K, Thompson DBA, Biek R. Effects of conservation management of landscapes and vertebrate communities on Lyme borreliosis risk in the United Kingdom. Phil Trans R Soc B. 2017; 372: 20160123. doi: 10.1098/rstb.2016.0123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kilpatrick M, Dobson ADM, Levi T, Salkeld DJ, Swei A, Ginsberg HS, et al. Lyme disease ecology in a changing world: consensus, uncertainty and critical gaps for improving control. Phil Trans R Soc B. 2017; 372: 20160117. doi: 10.1098/rstb.2016.0117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Leighton PA, Koffi JK, Pelcat Y, Lindsay LR, Ogden NH. Predicting the speed of tick invasion: An empirical model of range expansion for the Lyme disease vector Ixodes scapularis in Canada. J Appl Ecol. 2012; 49: 457–464. doi: 10.1111/j.1365-2664.2012.02112.x [DOI] [Google Scholar]
- 105.Watts AG, Saura S, Jardine C, Leighton P, Werden L, Fortin MJ. Host functional connectivity and the spread potential of Lyme disease. Landscape Ecol 2018, 33: 1925–1938. doi: 10.1007/s10980-018-0715-z [DOI] [Google Scholar]
- 106.Young HS, Wood CL, Kilpatrick M, Lafferty KD, Nunn CL, Vincent JR. Conservation, biodiversity and infectious disease: scientific evidence and policy implications. Phil Trans R Soc B. 2017; 372:20160124. doi: 10.1098/rstb.2016.0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Levine BA, Douglas MR, Yackel Adams A, Lardner B, Reed R, Savidge JA, et al. Genomic pedigree reconstruction identifies predictors of mating and reproductive success in an invasive vertebrate. Ecol Evol. 2019; 9:11863–11877. doi: 10.1002/ece3.5694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Chafin TK, Douglas MR, Martin BT, Zbinden ZD, Middaugh CM, Ballard J, et al. Age structuring and spatial heterogeneity in prion protein gene (PRNP) polymorphism in Arkansas white-tailed deer. Prion. 2020; 14: 238–248. doi: 10.1080/19336896.2020.1832947 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Field-identification based on morphology was verified by genetic clustering, with 25% of samples reclassified.
(PDF)
Genetic clusters detected in each species at Illinois SAFE sites based on PCA applied to 15 microsatellite loci.
(PDF)
‘Optimal’ number of retained principal component (PC) axes (X-axis) based on minimum of root-mean-square-error (RMSE) assignment (Y-axis) of 20% of randomly selected samples.
(PDF)
Evaluation of 10 replicates at K values 1–10 based on the ’ad hoc statistic’ ΔK and the probability of K.
(PDF)
Ability to resolve population structure as evaluated with two statistical measures (χ2 and Fisher exact tests).
(PDF)
Comparison of Ne estimates and 95% jackknife confidence intervals between years and two SAFE sites.
(PDF)
Details on geographic location, patch area, acronyms and sample sizes based on phenotypic and genetic identifications.
(PDF)
Details on primers used to amplify 23 microsatellite loci.
(PDF)
Values calculated for 2010 and 2012 between restoration patches.
(PDF)
Estimates of within-patch average and variance by year.
(PDF)
Comparison of population genetic and life history parameters for two vole and three bird species.
(PDF)
Data Availability Statement
Data available in Open Science Framework respository under https://osf.io/k2m8w/.