Abstract
Objective
Liver mitochondria adapt to high-calorie intake. We investigated how exercise alters the early compensatory response of mitochondria, thus preventing fatty liver disease as a long-term consequence of overnutrition.
Methods
We compared the effects of a steatogenic high-energy diet (HED) for six weeks on mitochondrial metabolism of sedentary and treadmill-trained C57BL/6N mice. We applied multi-OMICs analyses to study the alterations in the proteome, transcriptome, and lipids in isolated mitochondria of liver and skeletal muscle as well as in whole tissue and examined the functional consequences by high-resolution respirometry.
Results
HED increased the respiratory capacity of isolated liver mitochondria, both in sedentary and in trained mice. However, proteomics analysis of the mitochondria and transcriptomics indicated that training modified the adaptation of the hepatic metabolism to HED on the level of respiratory complex I, glucose oxidation, pyruvate and acetyl-CoA metabolism, and lipogenesis. Training also counteracted the HED-induced glucose intolerance, the increase in fasting insulin, and in liver fat by lowering diacylglycerol species and c-Jun N-terminal kinase (JNK) phosphorylation in the livers of trained HED-fed mice, two mechanisms that can reverse hepatic insulin resistance. In skeletal muscle, the combination of HED and training improved the oxidative capacity to a greater extent than training alone by increasing respiration of isolated mitochondria and total mitochondrial protein content.
Conclusion
We provide a comprehensive insight into the early adaptations of mitochondria in the liver and skeletal muscle to HED and endurance training. Our results suggest that exercise disconnects the HED-induced increase in mitochondrial substrate oxidation from pyruvate and acetyl-CoA-driven lipid synthesis. This could contribute to the prevention of deleterious long-term effects of high fat and sugar intake on hepatic mitochondrial function and insulin sensitivity.
Keywords: Exercise, Mitochondrial supercomplexes, Acetyl-CoA, MAFLD, Lipidomics, Proteomics
Highlights
-
•
High-energy diet promotes mitochondrial respiration in liver independent of training.
-
•
High-energy diet combined with training disconnects substrate oxidation from lipid synthesis.
-
•
High-energy diet combined with training reduces complex I formation in the liver.
-
•
Trained skeletal muscle unburdens the liver from substrate overload.
-
•
Comprehensive resource of mitochondrial adaptations to high-energy diet and training.
1. Introduction
Non-alcoholic fatty liver disease (NAFLD) is the most prevalent hepatic pathology associated with the global obesity pandemic. NAFLD is the leading risk factor for non-alcoholic steatohepatitis (NASH) and liver cirrhosis and is closely linked to the development of hepatocellular carcinoma [1]. Affected individuals are often insulin resistant, have reduced glucose tolerance or type 2 diabetes, and have a higher frequency of cardiovascular events and increased mortality [2]. With a prevalence of circa 25% in the global population, NAFLD is now one of the major health burdens with tremendous consequences for health care systems as well as for the individuals' quality of life [3]. To account for this increase and emphasize the connection between metabolic disturbances and excessive hepatic lipid deposition, metabolic (dysfunction) associated fatty liver disease (MAFLD) has recently been suggested as an alternative term [4].
Health organizations worldwide advocate increased regular physical activity as a potent treatment of NAFLD and for the prevention of steatohepatitis and associated metabolic comorbidities [5,6]. All kinds of exercise interventions (e.g., endurance and resistance exercise, a combination of both, or unstructured increase in daily physical activity) are effective in ameliorating NAFLD [7]. There is a clear positive correlation between weight loss and reduction in liver fat, albeit improvement of hepatic steatosis by exercise is possible in the absence of weight loss [8]. This suggests that the increased energy expenditure leads to metabolic adaptations in the liver beyond the reduction of liver fat. The additional benefit of exercise is further supported by the finding of more persistent protection against NAFLD when comparing the susceptibility after exercise training and caloric restriction [9]. The underlying mechanisms responsible for the superior potential of exercise on NAFLD beyond weight loss remain undefined.
There is accumulating evidence to suggest that steatogenic conditions responsible for the development of NAFLD first induce compensatory mechanisms in liver mitochondria. Both human and rodent studies showed that liver mitochondria can temporarily adapt to high-calorie intake and obesity by increasing mitochondrial content, mitochondrial enzyme activities, and mitochondrial oxidative capacity, resulting in enhanced fatty acid oxidation, mitochondrial respiration, and tricarboxylic acid (TCA) cycle flux [[10], [11], [12]]. It has, however, been proposed that these compensatory responses trigger a decline of mitochondrial function and are directly linked to the development of hepatic insulin resistance [13]. The upregulation of mitochondrial substrate oxidation and TCA cycle activity is accompanied by increased generation of fatty acid-derived lipids, such as diacylglycerols (DG) and ceramides (CER), and of reactive oxygen species [14,15]. Together with oxidative stress, these lipids lead to impaired hepatic insulin action and activate inflammatory responses, apoptosis, and DNA damage. Resultantly, the compensatory mechanisms fail, leading to reduced mitochondrial function, as observed in the liver of humans with advanced NAFLD and steatohepatitis [10,16,17].
Recent evidence suggests that the compensatory activation of mitochondrial substrate oxidation must be coupled to an increase in energy expenditure to counteract the side effects and to avoid the accumulation of lipids related to the induction of hepatic insulin resistance and inflammation [9]. Exercise is the most effective physiological stimulus for enhancing ATP consumption coupled to substrate oxidation. This is not restricted to the working muscle. During endurance exercise, hepatic oxygen consumption and carbon dioxide delivery can increase two-fold in humans [18], indicating higher metabolic activity and increased demand for ATP in the liver, as also reported in exercising mice [19]. It remains unknown how liver mitochondria are affected by chronically elevated levels of high-energy fuels when regular exercise training is performed and how this interaction is linked to the potency of exercise to prevent insulin resistance and mitochondrial dysfunction.
We aimed to provide a comprehensive view of the early mitochondrial adaptations when a steatogenic diet is combined with regular exercise to determine the effectiveness of exercise in preventing, ameliorating, or reversing NAFLD and its related comorbidities. Focusing on the liver and skeletal muscle, we studied the impact of regular exercise training on mitochondria and mitochondria-associated metabolic pathways in mice during the onset of hepatic steatosis. Mice were fed a high-energy diet (enriched with sugar and fat) for six weeks and they either remained untrained or underwent regular treadmill training. Sedentary and trained control groups received a matched control diet. We applied transcriptomics, proteomics, and lipidomics to elucidate the molecular alterations in mitochondria of liver and skeletal muscle and examined the functional consequences on respiration. Our in-depth analysis of alterations on protein, transcript, and lipid level provides novel hints on how to prevent the transition from a mitochondrial compensation of nutrient overload to the deterioration of mitochondrial function associated with NAFLD.
2. Results
2.1. Treadmill training ameliorates metabolic disturbances induced by HED
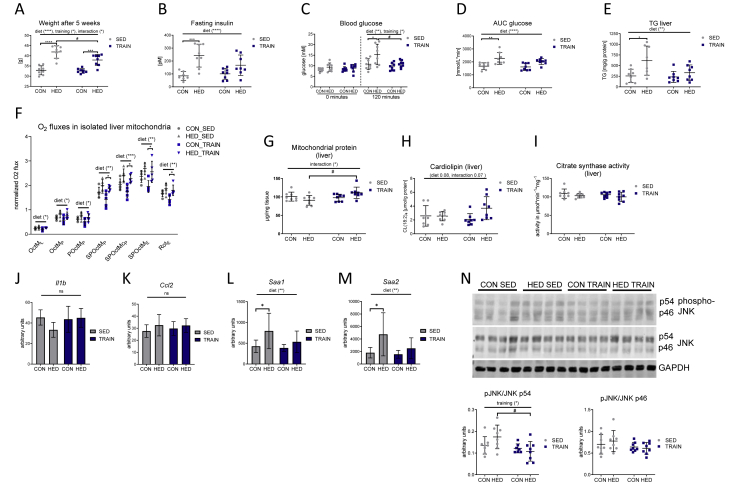
The high-energy diet (HED) contained additional sugar (10% weight sucrose) and fat (20% weight lard) and was fed to mice for six weeks to induce hepatic steatosis. At the onset of the dietary intervention, half of the HED group and of the control chow-fed group were trained three days a week for 1 h on a treadmill. Sedentary mice receiving HED gained significantly more weight than the control chow-fed mice, and the weight gain was significantly lower in the trained group (Figure 1A). HED-feeding increased fasting insulin levels and caused glucose intolerance, as evident from higher glucose levels after 120-min ipGTT (intraperitoneal glucose tolerance testing) and increased area under the curve (AUC) glucose values (Figure 1B–D and Fig. S1). HED-feeding also led to significantly higher levels of triacylglycerol (TG) in the liver (Figure 1E). These effects were lower or absent in the trained group on HED. These data indicate the suitability of the study design for investigating the impact of regular exercise training on the development of fatty liver and glucose intolerance.
Figure 1.
Training ameliorates the metabolic disturbances induced by a high-energy diet. A: Mouse body weights after 5 weeks of diet and training. B: fasting insulin levels. C: blood glucose levels before and 120 min after an intraperitoneal glucose tolerance test. D: the area under the curve (AUC) thereof. E: Triacylglycerol (TG) levels in liver samples. F: Respiratory analyses of isolated mouse liver mitochondria on an Oxygraph-2k. Oxygen fluxes in isolated liver mitochondria were normalized for the mitochondrial amount (by protein), dilution throughout the analysis and inter-day variances. Consecutive addition of malate (M), octanoylcarnitine (Oct), ADP, pyruvate (P), succinate (S), cytochrome c (c), carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP), and rotenone (Rot; complex I inhibitor); L = leak respiration (malate and octanoylcarnitine), P = phosphorylating conditions in the presence of ADP, E = uncoupled respiration using the protonophore FCCP. G: Mitochondrial protein yield from liver tissue wet weight. H: Tetralinoleoyl cardiolipin (CL) content of liver tissue. I: Citrate synthase activity in liver tissue. J–M: Hepatic transcript levels of Il1b, Ccl2, Saa1, and Saa2. N: Immunoblot and densitometric quantification of hepatic phospho-Thr-183/Tyr-185 JNK. The left side shows a representative immunoblot with 4 out of 8 tissue lysates. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training. n = 8, (n = 5 for respiratory analyses), mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 for diet, training or interaction by 2-way ANOVA as indicated. ∗p < 0.05, ∗∗p < 0.01, ∗p < 0.001, ∗p < 0.0001 for diet or #p < 0.05 for training by Tukey HSD multiple comparisons test. One outlier was excluded in the CON SED group in graph B.
2.2. HED induces an increased oxidative capacity in isolated mouse liver mitochondria
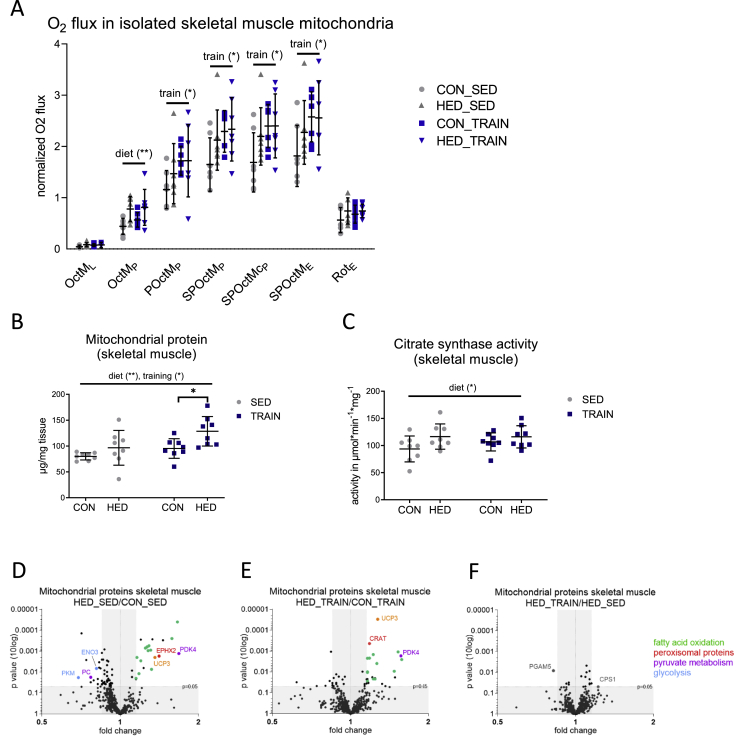
To analyze the impact of HED on mitochondrial function, high-resolution respirometry was performed. In isolated mitochondria, HED resulted in an increase in leak respiration (OctML), fatty acid oxidation capacity (OctMP, measured after addition of octanoyl carnitine, malate, and ADP), fatty acid oxidation combined with pyruvate (POctMP), maximal coupled phosphorylation after addition of succinate (SPOctMP), uncoupled phosphorylation after addition of FCCP (SPOctME) and uncoupled complex II respiration (after addition of complex I inhibitor rotenone (RotE) (Figure 1F)). The data indicate that the liver mitochondria adapted to the increased intake of carbohydrates and fat and compensated for these changes by increasing mitochondrial substrate oxidation, similar to previous findings [10,15]. While the increase in respiration of isolated mitochondria did not differ between trained and sedentary HED-fed mice (Figure 1F), the amount of mitochondrial protein in whole liver tissue was higher in trained than in sedentary HED-fed mice (Figure 1G). Livers of trained HED-fed mice also showed a trend towards a higher level of the mitochondrial lipid marker tetralinoleoyl cardiolipin ((18:2)4CL) (Figure 1H). This suggested that the combination of HED and training, but not HED alone, led to an increase in mitochondrial mass in the liver. Notably, despite the higher mitochondrial mass, citrate synthase activity did not increase in liver tissue of trained HED-fed mice (Figure 1I).
The compensatory increase in mitochondrial respiration indicated a very early stage of fatty liver, without relevant hepatic tissue inflammation. This assumption was supported by transcript data of the livers of the four groups obtained by microarray analysis. The transcripts encoding the inflammatory cytokines Tnf and Il6 were not detectable, and the transcripts of Il1b and Ccl2 did not differ between groups (Figure 1J, K). An increase in the acute phase transcripts Saa1 and Saa2A was the only indication of a slightly activated inflammatory cytokine signaling in the liver of the HED-fed mice (Figure 1L, M). However, phosphorylation of JNK in the HED-fed mice was not significantly increased (Figure 1N). The trained HED-fed mice did not show any significant increase in Saa1 and Saa2 transcripts and phospho-JNK p54 signals were lower than in sedentary HED-fed mice. Thus, the livers of HED-fed mice had only slightly elevated inflammation markers, and these were absent in the trained mice. These data confirm that the mice in our study were still at a relatively early stage of NAFLD with compensatory increased mitochondrial respiration and marginal signs of hepatic inflammation. The fact that the citrate synthase activity in liver tissue was similar despite the higher mitochondrial mass in the livers of trained HED-fed mice is a first indication that training could modify the effect of HED on mitochondrial protein composition.
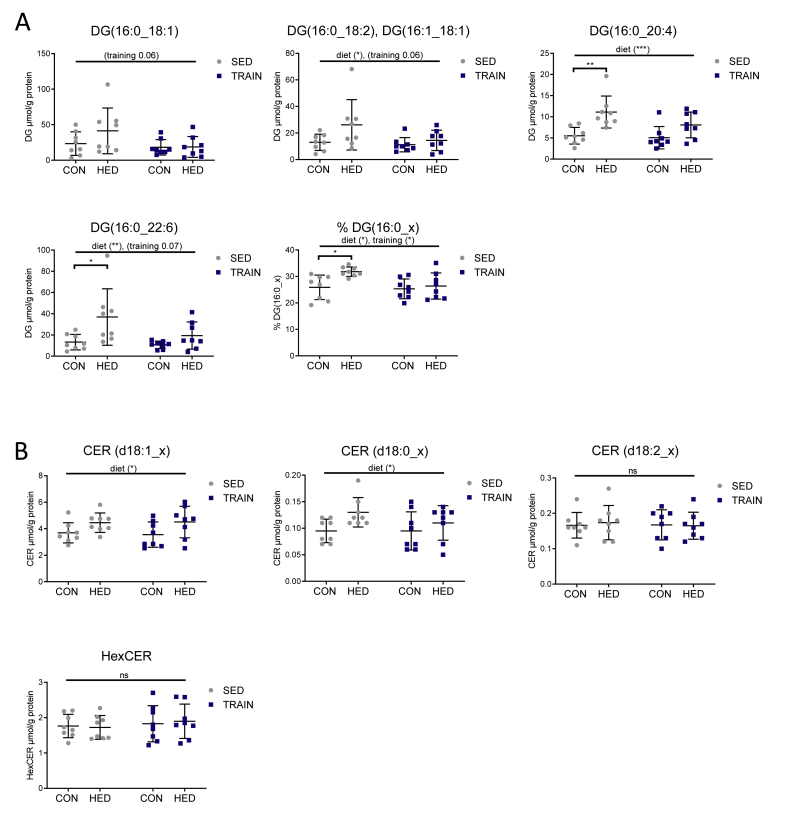
2.3. Training prevents an increase in hepatic diacylglycerols containing one palmitoyl chain in HED-fed mice
Next, we studied the effects of HED and training on hepatic lipid diacylglycerol (DG) and ceramides (CER) by ultra-high-performance liquid chromatography-mass spectrometry UHPLC-MS/MS lipidomics. The increased concentration of distinct DG and CER species has been linked to the development of hepatic insulin resistance [14]. A total of 25 DG species were detected (Table S1), most of which were induced by HED. In addition, a lowering effect of training, which was most pronounced for DG species containing one palmitoyl chain, was observed. The highly abundant DG species DG (16:0/18:1), DG (16:0/18:2)/DG (16:1/18:1), DG(16:0/20:4), and DG(16:0/22:6) showed a similar regulation by HED and training (Figure 2A). The percentage of these DG species in the sum of all DGs was increased by HED from 25.9% to 31.8% and reduced by training to 26.4% (Figure 2A). Therefore, training may have a beneficial effect on hepatic insulin action by protecting it against HED-induced elevation of distinct DG species. Contrastively, training had no effect on the abundance of CERs. Nineteen CER species were detected (7 CERs (d18:1_x), 3 dihydro-CERs (d18:0_x; d22:0_x), 7 hexosyl-CERs and 2 CERs containing two double bonds in the sphingoid base (d18:2_x)) (Table S1). HED slightly increased the sum of CERs and dihydro-CERs with no conversion by training (Figure 2B). This was also evident in the regulation of the individual CERs (Table S1). Hexosyl-CERs and CER (d18:2_x) were neither altered by HED nor by training (Figure 2B).
Figure 2.
Training prevents an increase in hepatic diacylglycerols containing one palmitoyl chain in high-energy diet-fed mice. A: C16:0 acyl-containing diacylglycerol (DG) species in μmol/g protein and as a percentage of the total sum of DGs in liver tissue obtained from sedentary and trained high-energy diet-fed and standard chow-fed mice. B: Sum of detected ceramides (CER) (d18:1_x), dihydroceramides (CER (d18:0_x)), CER (d18:2_x), and hexosyl-CER in liver tissue obtained from sedentary and trained HED-fed and control-fed mice. Individual DG and CER species are shown in Table S1. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training. n = 8, mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 for diet, training or interaction by 2-way ANOVA as indicated. ∗p < 0.05, ∗∗p < 0.01 for diet by Tukey HSD multiple comparisons test.
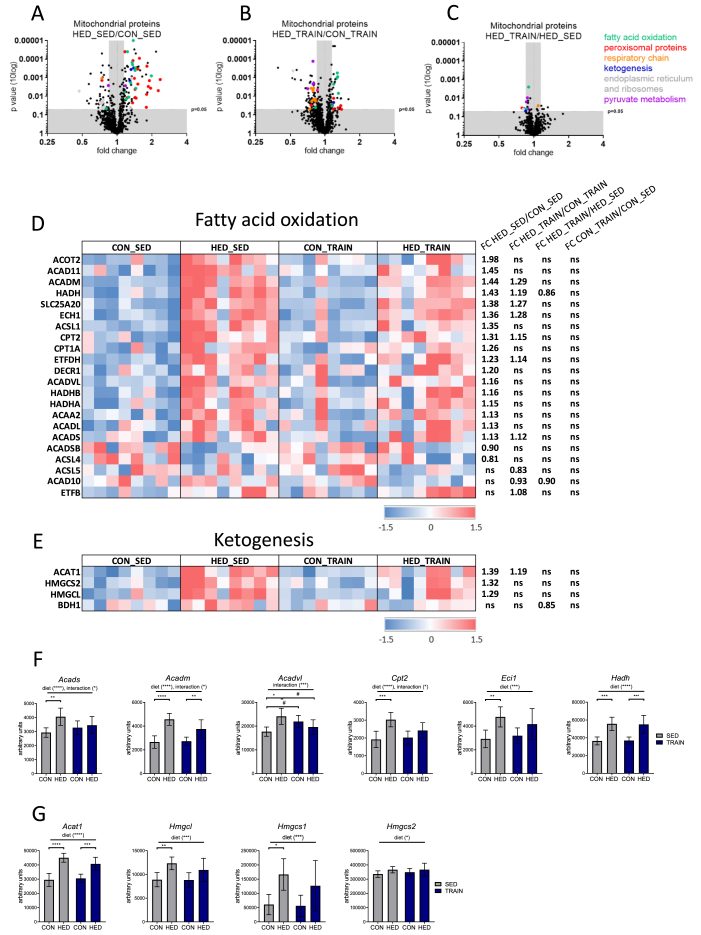
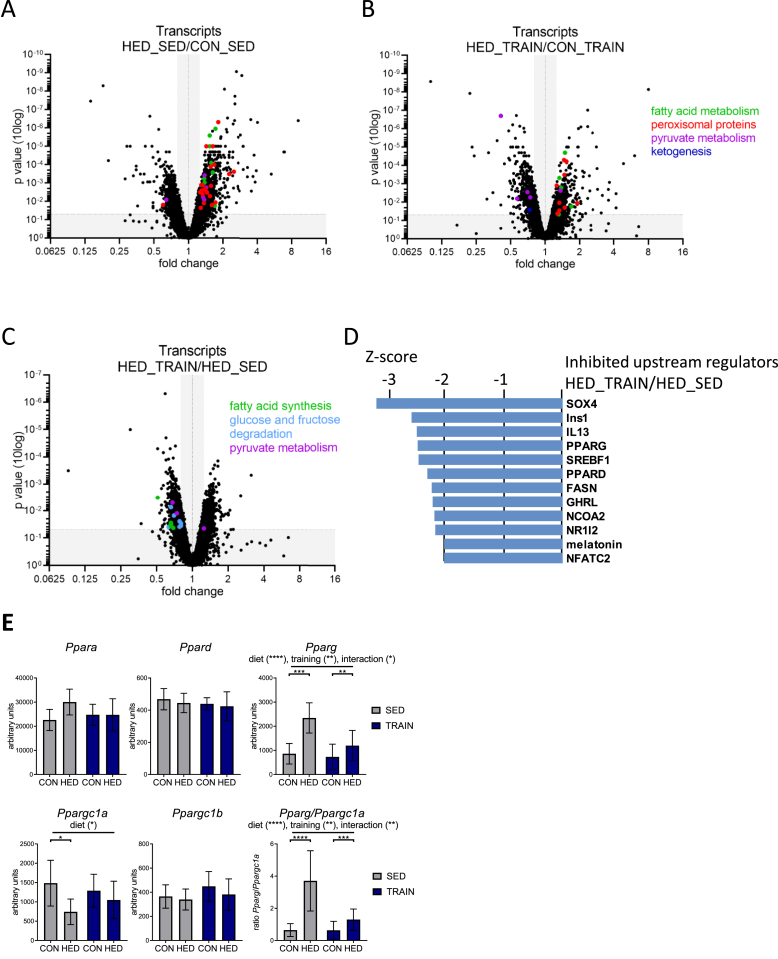
2.4. HED increases hepatic mitochondrial proteins and transcripts related to fatty acid oxidation, ketogenesis, and peroxisomes; training partially mitigates this effect
We applied proteomics to isolated liver mitochondria to investigate alterations in the hepatic mitochondrial proteins that could mediate the partial rescue from metabolic disturbances by training in HED-fed mice. In each of the four groups, proteins with known mitochondrial localization according to Mitocarta version 2.0 [20] accounted for 89% of total protein abundance. Table S2 shows the comparison of the mitochondrial proteomes between all four groups. A STRING pathway analysis revealed a pronounced upregulation of proteins involved in fatty acid oxidation (colored in green), peroxisome function (colored in red), and ketogenesis (colored in blue) when comparing sedentary HED-fed and control-fed mice (Figure 3A and Fig. S2). Proteins of these pathways also increased in the mitochondrial proteome of trained mice receiving the HED, albeit with less conspicuous fold change compared to the trained control fed-group (Figure 3B). A few proteins from these pathways showed significant reduction in the direct comparison of trained and sedentary HED-fed mice (Figure 3C). The heatmaps in Figure 3D,E and Fig. S3A illustrate the differential regulation of the mitochondrial proteins related to mitochondrial fatty acid oxidation, ketogenesis, and peroxisomes in all four groups. Whole genome transcriptome analysis revealed that several of the proteins exhibiting a higher abundance in mitochondria of HED-fed mice were also upregulated on the mRNA level (Figure 3F,G and Supp Fig. S3B). The regulation of several transcripts was modified by training, resulting in a reduced impact of HED feeding. In addition, we observed an enrichment of many proteins involved in peroxisomal fatty acid oxidation and peroxisomal function in isolated mitochondria (Fig. S3A) suggesting a closer direct interaction between mitochondria and peroxisomes, induced by the HED. Proteins located in the endoplasmic reticulum and ribosomes were decreased by HED in both sedentary and trained mice, suggesting reduced contact between mitochondria and endoplasmic reticulum in HED-fed mice (Figure 3A,B). In conclusion, HED increased the amount of enzymes and transport proteins necessary for mitochondrial and peroxisomal fatty acid oxidation as well as for ketogenesis. Training reduced the magnitude of the effect on protein and, in some cases, on transcript levels.
Figure 3.
A high-energy diet increases hepatic mitochondrial proteins and transcripts related to fatty acid oxidation and ketogenesis, and training partially mitigates this effect. A–C: Significantly different proteins in the liver mitochondrial proteome when comparing (A) sedentary HED-fed and sedentary control-fed mice (HED_SED/CON_SED), (B) trained HED-fed and trained control-fed mice (HED_TRAIN/CON_TRAIN), and (C) trained and sedentary HED-fed mice. Data points were colored based on enriched categories of STRING pathways and are plotted by p-values (according to Student's t-test, no correction for multiple testing) versus fold changes between groups. Green = fatty acid oxidation, red = peroxisome, orange = respiratory chain, purple = pyruvate metabolism, light gray = endoplasmic reticulum and ribosomes, and blue = ketogenesis. (A, B: only proteins with fold change <0.85 or >1.15 were colored). See also Table S2. STRING: Search Tool for the Retrieval of Interacting Genes/Proteins. D, E: Significantly different proteins (according to Student's t-test, no correction for multiple testing) related to fatty acid oxidation and ketogenesis in trained or sedentary HED- or control-fed mice. Values were centered to the mean of the respective protein and scaled to unit variance. White color shows values close to the mean and red- and blue-colored values are higher and lower, respectively than the mean. Each column represents one animal; n = 8 per group. ns: not significant. F, G: Transcripts encoding proteins of fatty acid oxidation or ketogenic enzymes increased with HED in mouse liver tissues. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training. n = 6–8, mean ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for diet, training or interaction by 2-way ANOVA as indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for diet by Tukey HSD multiple comparisons test.
In trained HED-fed mice, proteins involved in pyruvate metabolism were reduced compared to both trained control-fed and sedentary HED-fed mice (marked in purple in Figure 3B,C). This is the first indication that pathways related to the mitochondrial conversion of pyruvate to acetyl-CoA were altered by training (see Section 2.6. for further details).
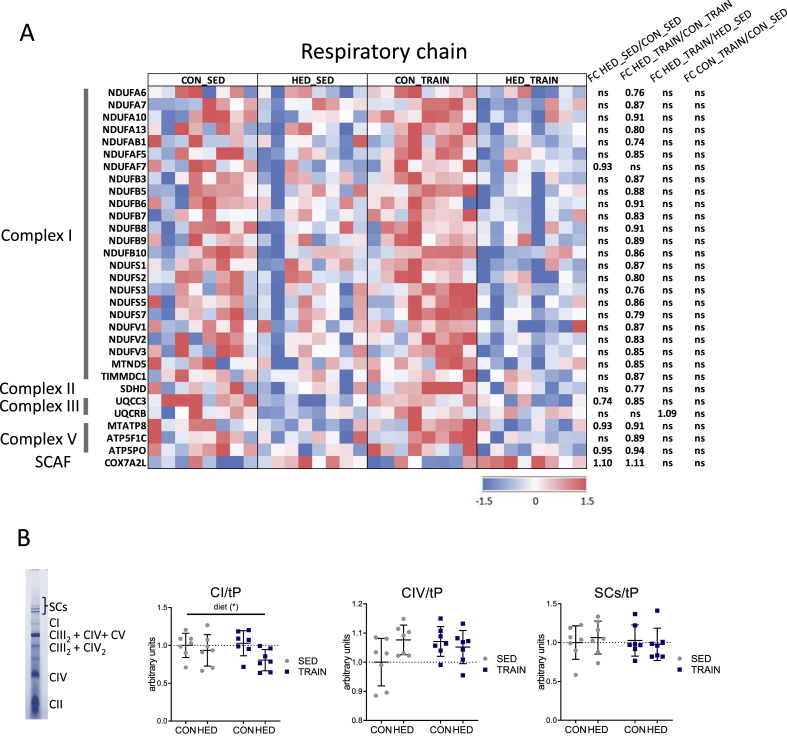
2.5. Trained HED-fed mice have reduced complex I proteins in hepatic mitochondria when compared to trained control-fed mice
Training triggered a significant difference in the mitochondrial content of proteins related to the respiratory chain (Figure 3B, colored in orange). Several of these proteins were reduced in the mitochondrial proteome of trained HED-fed vs. trained control-fed mice. This effect was less pronounced when comparing the two sedentary groups (Figure 4A). No significant effect of training was found in the control-fed mice (Figure 4A). The proteins reduced in mitochondria of trained HED-fed mice relative to trained control-fed mice were mainly components of complex I (Figure 4A). In line with the proteomics data, blue native gel electrophoresis of mitochondrial proteins revealed a lowering of complex I holocomplex by the HED that was most pronounced in the trained HED-fed mice (Figure 4B). No effect was observed on the abundance of the supercomplexes or of complex IV. Complex II was not quantified since no distinct band was evident in the majority of the samples, and complex III was not separated as a single band (Figure 4B). Overall, trained HED-fed mice showed reduced levels of complex I in isolated mitochondria but higher mitochondrial respiration (Figure 1F) than trained control-fed mice.
Figure 4.
Trained high-energy diet-fed mice have reduced complex I proteins in liver mitochondria when compared to trained control-fed mice. A: Significantly different liver mitochondrial proteins (according to Student's t-test, no correction for multiple testing) related to the respiratory chain in trained or sedentary HED- or control-fed mice. Values were centered to the mean of the respective protein and scaled to unit variance. White color shows values close to the mean and red- and blue-colored values are higher and lower, respectively than the mean. Each column represents one animal; n = 8 per group. SCAF, Supercomplex assembly factor. ns: not significant. B: Densitometric quantification of Coomassie-stained native respiratory complexes (exemplarily shown at the left side) in blue native polyacrylamide gel electrophoresis of solubilized liver mitochondria. Signals were normalized to total protein abundance visualized by the Coomassie staining (tP). The mean ± SD values of n = 7 per group are shown. ∗p < 0.05 for diet, training, or interaction by 2-way ANOVA as indicated. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training.
2.6. Training downregulates acetyl-CoA-generating pathways in the liver of HED-fed mice
To elucidate how training alters the connection of mitochondrial metabolism with cytosolic pathways in the livers of trained and sedentary HED-fed mice, we analyzed the whole genome transcriptome followed by a KEGG pathway enrichment analysis on all differentially regulated transcripts (Table S3). In accordance with the proteomics data shown in Figure 3 and Fig. S3, HED-feeding increased transcripts related to fatty acid metabolism and peroxisome, and this effect was more pronounced in sedentary than in trained mice (Figure 5A, B). Trained mice receiving the HED additionally showed a reduction of hepatic transcripts related to pyruvate metabolism compared to trained control-fed mice (Figure 5B). When comparing trained and sedentary HED-fed mice, enriched KEGG pathways were, amongst others, glycolysis/gluconeogenesis (p = 0.002), fructose metabolism (p = 0.006), pentose phosphate pathway (p = 0.029), pyruvate metabolism (p = 0.027), and fatty acid synthesis (p = 0.042) (Table S3). Almost all the corresponding transcripts were reduced in the trained group (Figure 5C). Upstream regulator analysis of all differentially regulated transcripts between the trained and sedentary HED-fed mice suggested reduced activation of the key transcriptional regulators of fatty acid synthesis and oxidation peroxisome proliferator-activated receptor gamma (PPARG), peroxisome proliferator-activated receptor delta (PPARD) and sterol regulatory element binding protein 1 (SREBF1) and of the lipogenic enzyme FASN (Figure 5D). In addition, livers of HED-fed mice showed high levels of the PPARG transcript, which was less induced in trained HED-fed mice (Figure 5E). Ppargc1a transcript levels were reduced by HED only in sedentary mice (Figure 5E). Therefore, training of HED-fed mice normalized the Pparg/Ppargc1a ratio almost to the levels of the control chow-fed mice. Neither diet nor training resulted in any change in the expression of Ppara and Ppard (Figure 5E).
Figure 5.
Training downregulates hepatic transcripts related to glucose oxidation and fatty acid synthesis in high-energy diet-fed mice. A-C: Significantly different transcripts in livers when comparing (A) sedentary HED-fed and sedentary control-fed mice (HED_SED/CON_SED), (B) trained HED-fed and trained control-fed mice (HED_TRAIN/CON_TRAIN), and (C) trained and sedentary HED-fed mice (p < 0.05 according to limma t-test, no correction for multiple testing). Data are plotted by p-values versus fold changes between groups. Transcripts from enriched KEGG categories (Table S3) with fold change <0.8 or >1.3 were colored: Green dots = fatty acid metabolism/fatty acid synthesis, red = peroxisomal proteins, light blue = glycolysis, purple = pyruvate metabolism, dark blue = ketogenesis. A. Two data points with significant increase >16-fold are outside the axis limits (Cyp2b9, Cfd) C. One data point with non-significant fold change is outside the axis limits. D. Upstream regulators inhibited in HED_TRAIN vs. HED_SED based on Ingenuity upstream regulator analysis of differentially regulated transcripts. z-Scores below −2 were considered significant. E. Abundance of peroxisome proliferator-activated receptor (PPAR) transcripts. The mean ± SD values for n = 6–8 are shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for diet, training or interaction by two-way ANOVA as indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for diet by Tukey HSD multiple comparisons test. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training.
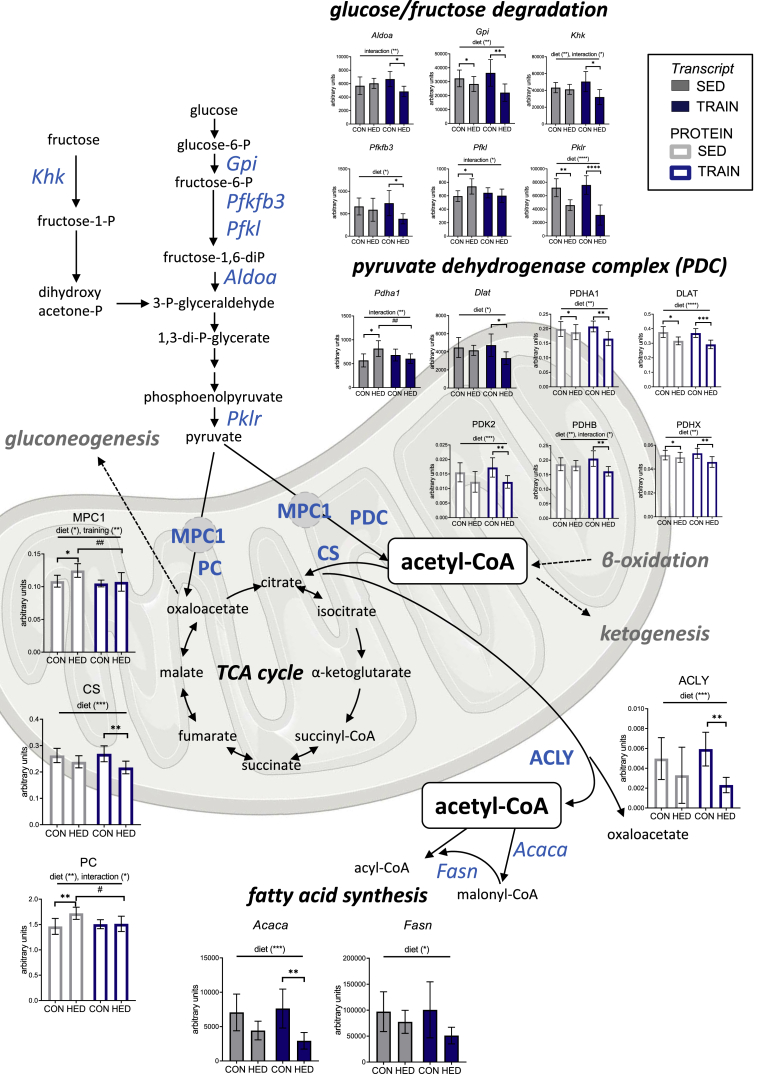
Because several transcripts encoding for enzymes necessary for converting glucose or fructose to pyruvate showed reduced levels by training in HED-fed mice (Figure 5, Figure 6), we closely examined the mitochondrial protein abundance of the enzymes required for pyruvate and acetyl-CoA metabolism. Proteins of the pyruvate dehydrogenase complex, the mitochondrial pyruvate carrier 1 (MPC1), citrate synthase (CS) and pyruvate carboxylase (PC) were all lowered by training, HED-feeding, or the combination of both (Figure 6). Similarly, the amount of ATP citrate lyase (ACLY), the enzyme required to convert citrate to acetyl-CoA to provide acetyl-CoA for fatty acid synthesis in the cytosol was lowered in trained HED-fed mice, together with the transcripts encoding for the cytosolic fatty acid synthesis enzymes Acaca and Fasn (Figure 6). The data are suggestive of a downregulation of the pathways that produce pyruvate and acetyl-CoA, and thus to a potential reduction in the availability of acetyl-CoA for fatty acid and lipid synthesis.
Figure 6.
Training downregulates acetyl-CoA-generatingpathways in the liver of high-energy diet-fed mice. Abundance of transcripts (gray/blue filled bars) and proteins (blank bars) related to glucose/fructose degradation, mitochondrial pyruvate metabolism, acetyl-CoA conversion and fatty acid synthesis. The mean ± SD values for n = 6–8 are shown. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for diet, training or interaction by two-way ANOVA as indicated. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001 for diet, #p < 0.05, ##p < 0.01 for training by Tukey HSD multiple comparisons test. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training.
2.7. A combination of HED and training increases the oxidative capacity of skeletal muscle to a greater extent than training alone
The abovementioned results indicate that in liver mitochondria, training modifies glucose oxidation and acetyl-CoA production, thereby reducing the entry of acetyl-CoA into the TCA cycle and lipogenesis. Training is known to increase the respiratory capacity of the skeletal muscle, which can result in a reduced substrate load for the liver in the HED-fed condition. Thus, we also analyzed respiration and the proteome in mitochondria isolated from skeletal muscles in the four groups. High resolution respirometry revealed a distinct effect of training on pyruvate (POctMp) and succinate-driven (SPOctMp) respiratory capacities in both control- and HED-fed mice (Figure 7A). Similarly, maximal uncoupled phosphorylation (SPOctME) was higher in these mice. Unlike training, HED increased fatty acid oxidation (OctMp) only (Figure 7A). Thus, as opposed to liver mitochondria, training - not diet - had the strongest impact on mitochondrial respiration in skeletal muscle. Trained HED-fed mice also had the highest content of mitochondrial proteins in whole muscle tissue of all groups (Figure 7B). Citrate synthase activity was elevated in sedentary and trained HED-fed mice (Figure 7C). Table S4 shows the comparison of the mitochondrial proteome between all four groups. Proteins with known mitochondrial localization according to the MitoCarta version 2.0 accounted for 96% (CON_TRAIN group 95%) of total protein abundance. Sedentary and trained HED-fed mice demonstrated highly comparable increase in proteins related to fatty acid oxidation, unlike in hepatic mitochondria, where the effect of HED was less in trained than in sedentary mice (Figure 7D and E and Fig. S4A, S4B). Likewise, training did not reduce abundance of proteins of the respiratory chain or citrate synthase in the skeletal muscle mitochondria, neither in control- nor in HED-fed mice (Fig. S4C and D). Pyruvate dehydrogenase kinase 4 (PDK4) was increased to similar levels in both sedentary and trained HED-fed mice (Figure 7D, E, Fig. S4D). PDK4 is the key PDH kinase isoform in skeletal muscle [21]. It phosphorylates PDH leading to its inactivation, thereby reducing glucose oxidation and supporting fatty acid oxidation. Similarly, skeletal muscle mitochondria of sedentary and trained HED-fed mice revealed a comparable increase in the mitochondrial uncoupling protein 3 (UCP3) (Figure 7D and E, Fig. S4D). UCP3 has been shown to enhance fatty acid oxidation and to minimize mitochondrial oxidative damage [23].
Figure 7.
A combination of HED and training increases the oxidative capacity of skeletal muscle to a greater extent than training alone. A: Respiratory analyses of isolated mouse skeletal muscle mitochondria on an Oxygraph-2k. Oxygen fluxes in isolated skeletal muscle mitochondria were normalized for the mitochondrial amount (by protein), dilution throughout the analysis and inter-day variances. Consecutive addition of malate (M), octanoylcarnitine (Oct), ADP, pyruvate (P), and succinate (S), carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) and rotenone (Rot; complex I inhibitor); L = leak respiration (malate and octanoylcarnitine), P = phosphorylating conditions in the presence of ADP, E = uncoupled respiration using the protonophore FCCP. B: Mitochondrial protein yield from skeletal muscle tissue wet weight. C: Citrate synthase activity in skeletal muscle tissue. D–F: Significantly different proteins in the skeletal muscle mitochondrial proteome when comparing (D) sedentary HED-fed and sedentary control-fed mice (HED_SED/CON_SED), (E) trained HED-fed and trained control-fed mice (HED_TRAIN/CON_TRAIN), and (F) trained and sedentary HED-fed mice. Data are plotted by p-value (according to Student's t-test, no correction for multiple testing) versus fold changes between groups. Proteins from the following categories based on enrichment of STRING pathways with fold change <0.85 or >1.15 were colored: Green = fatty acid oxidation, red = peroxisomal proteins, purple = pyruvate metabolism, orange = uncoupling protein, blue = glycolysis. STRING: Search Tool for the Retrieval of Interacting Genes/Proteins. Three data points with a non-significant fold change of HED_TRAIN/CON_TRAIN are outside of the axis limits. See also Fig. S4 and Table S4. CON = control diet, HED = high-energy diet, SED = sedentary, TRAIN = treadmill training. n = 8, (n = 7 for respiratory analyses), mean ± SD. ∗p < 0.05, ∗∗p < 0.01, for diet, training or interaction by two-way ANOVA as indicated. ∗p < 0.05 for diet by Tukey's HSD multiple comparisons test.
We also detected the glycolytic enzymes PKM and ENO3 in the mitochondrial fraction. These enzymes might be associated with the mitochondria, albeit mitochondrial localization has also been reported [22]. In the sedentary HED-fed mice, the mitochondrial association of PKM and ENO3 was clearly diminished compared with that in control diet-fed mice. Only minor changes in the mitochondrial proteome were detected when comparing sedentary and trained HED-fed mice (Figure 7F). Overall, the combination of HED and training increased mitochondrial protein content and mitochondrial substrate oxidation capacity in skeletal muscle, accompanied by an increased abundance of PDK4 and UCP3. This could redirect the substrate overload from liver mitochondria during HED and obesity and help prevent the development of mitochondrial dysfunction, inflammation, and insulin sensitivity in the liver.
3. Discussion
In this study, we used a mouse model of moderate hepatic steatosis to study how regular exercise modulates the early mitochondrial response of the liver to a steatogenic diet, and describe molecular mechanisms that could be relevant for the prevention of NAFLD and comorbidities. Moreover, the results provide a comprehensive resource of exercise-regulated mitochondrial proteins in liver and skeletal muscle of mice fed high-energy diet and control diet.
Expectedly, treadmill training was effective in attenuating weight gain, the increase in fasting insulin and hepatic TG accumulation, and in reversing the glucose intolerance induced by the intake of a sugar- and fat-enriched diet. These metabolic parameters can all be improved by increasing energy consumption. The latter is increased during but also in-between exercise bouts, due to increased skeletal muscle mass and aerobic capacity. However, it has also been suggested that training does more than shift the thermogenic balance of energy intake and consumption [9]. By comprehensively characterizing hepatic transcriptome, mitochondrial proteome, and oxidative capacity, we show that training reduces the mitochondrial acetyl-CoA load caused by overnutrition and decreases the resulting acetyl-CoA flux into lipogenesis, ketogenesis and TCA cycle in the liver. While the oxidative capacity of isolated liver mitochondria was similarly increased when comparing sedentary and trained HED-fed mice, training modified mitochondrial protein abundance and increased mitochondrial mass. Specifically, proteins responsible for the mitochondrial import of pyruvate and the oxidation to acetyl-CoA, and for the entry of pyruvate and acetyl-CoA into the TCA cycle were reduced by the combination of HED-feeding and training. Together with the reduction in transcripts related to glucose/fructose degradation our data strongly suggest that when combined with HED, training decreases hepatic glucose oxidation by concomitantly enhancing fatty acid oxidation. The total fatty acid oxidation capacity in the livers of the trained HED-fed mice was probably higher than that in sedentary HED-fed mice since the former contained more mitochondrial protein. Such mechanisms could promote hepatic fatty acid oxidation without increasing the acetyl-CoA load per mitochondrial unit, while simultaneously decreasing the amount of fatty acids available for re-esterification and TG synthesis. The latter pathway contributes most to hepatic TG accumulation in NAFLD, followed by de-novo lipogenesis from acetyl-CoA [24]. Our data indicate that, in addition to preventing acetyl-CoA accumulation, the addition of training to a HED is effective to reduce the key lipogenic enzymes ACLY, fatty acid synthase (FASN) and acetyl-CoA carboxylase (ACACA). The normalization of the ratio of the lipogenic transcription factor Pparg and the pro-mitochondrial transcriptional co-regulator Ppargc1a by training further points towards a shift in the transcriptome from lipogenesis towards fatty acid oxidation.
Not only can acetyl-CoA be utilized for de-novo lipogenesis, but it can also be utilized in the TCA cycle or for ketogenesis. Notably, the mitochondria of trained mice on HED contained lower amounts of citrate synthase, which could impede the entry of acetyl-CoA into the TCA cycle. In line with this, whole-tissue citrate synthase activity was similar despite a higher content of mitochondria, indicating lower activity per functional mitochondrial unit. Prevention of an increased flux of acetyl-CoA into the TCA cycle may be relevant during the progression of NAFLD, since induction of TCA cycle flux has been linked to increased mitochondrial reactive oxygen species production and gluconeogenesis by virtue of its providing higher amounts of the electron carriers NADH/FADH and oxaloacetate [15,25]. Mitochondria of trained HED-fed mice also contained less pyruvate carboxylase, the enzyme which converts pyruvate to oxaloacetate for TCA cycle anaplerosis or gluconeogenesis. Increased levels of acetyl-CoA can also be disposed of by coupling fatty acid oxidation with ketogenesis. This pathway is physiologically relevant during fasting [26] and is already known to be elevated during the onset of hepatic steatosis in rodents [27]. The higher abundance of transcripts and proteins of key enzymes of ketogenesis in our HED-fed mice indicate that increased ketogenesis can also develop when both sugar and fat are enriched in the diet. The capacity for a ketogenic response to overnutrition seems important, since impaired ketogenesis has been linked to the progression of NAFLD and steatohepatitis [28]. In the current study, this adaptive increase in ketogenic enzymes was weakened by training, probably due to a reduced acetyl-CoA level in the mitochondria.
A recent study also reports how peroxisomal acetyl-CoA production plays a part in the induction of hepatic steatosis by reducing the autophagic degradation of lipid droplets [29]. In this regard, the less pronounced increase in proteins and transcripts related to peroxisomal fatty acid oxidation and markers for hepatic peroxisomal content in the trained HED-fed mice can provide a further mechanistic explanation for the prevention and alleviation of NAFLD by training.
Physical exercise is known to increase mitochondrial substrate oxidation and biogenesis in skeletal muscle, which could have further reduced the energy load in our mice on HED. Treadmill training increased not only the mitochondrial mass in skeletal muscle of HED-fed mice, but also the oxidative capacity per unit of isolated mitochondria. Our data expand the results of a recent study of the skeletal muscle proteome in mice that reported PDK4 and enzymes related to fatty acid oxidation to be induced by both endurance training and a high fat-diet [30]. We show an increase in these enzymes in the skeletal muscle mitochondrial proteome and provide a comprehensive comparison of the regulation of liver and skeletal muscle mitochondrial proteins by training and HED. Another protein highly enriched in skeletal muscle mitochondria of both sedentary and trained HED-fed mice was UCP3. Overexpression of UCP3 in mouse muscle facilitates fatty acid oxidation [31] and mimics the effects of endurance exercise [32]. UCP3 has also been related to the control of reactive oxygen species production and oxidative damage [33,34]. Thus, the increase in UCP3 facilitates higher rates of fatty acid oxidation in skeletal muscle without an increase in oxidative stress, thereby contributing to the protection of liver function by rerouting fatty acid fluxes.
Reversal of the HED-induced lipogenic activity in the liver by training can explain the decreased hepatic content of the DGs containing palmitoyl-chains in the trained HED-fed mice as palmitate is the major product of de-novo lipogenesis. This decrease is highly relevant for the prevention of hepatic insulin resistance. The contribution of different lipid species to disturbances in the hepatic insulin signaling cascade and development of hepatic insulin resistance has been extensively investigated and there is an ongoing debate as to which lipids have the most profound effect and are most relevant for the deleterious effects of chronic overnutrition on insulin action in the liver [35]. DG-mediated down-regulation of the insulin signaling cascade is probably one of the best-studied and understood mechanisms of lipid-induced insulin resistance. DGs are bioactive signaling lipids that are required for the activation of protein kinase C (PKC) isoforms. In particular, activation of the novel PKCε was identified in response to increased DG content in human and rodent liver, a process associated with reduced insulin action [35]. This activation was recently attributed to plasma membrane-bound sn-1, 2-DG species [36]. Only very few studies have investigated the acyl chain composition of the DG species in relation to the development of insulin resistance in NAFLD [37,38]. The results, which are in line with our data, suggest that in particular the fatty acids from de-novo lipogenesis contribute to the increase in DG. The regulation of hepatic DG species containing palmitoyl- and stearoyl-chains in mice that had been subjected to a high fat diet for 8 weeks and to a treadmill training program for the final last 4 weeks was similar to that observed in the current study [39]. However, it remains to be clarified whether distinct DG species are more effective in activating PKCε in the liver than others. Nonetheless, the total hepatic DG content in combination with activation of PKCε, was identified as the best predictor of hepatic insulin resistance in obese patients [40].
The six weeks of HED induced only a very low grade of hepatic inflammation. Nevertheless, trained mice had lower levels of phosphorylated JNK, which is also in line with the aforementioned study in high fat diet-fed mice, where four weeks of exercise lowered the phosphorylation of JNK in the liver [39]. JNK activation, which also inhibits insulin signaling [41], is triggered by saturated fatty acids such as palmitate through Toll-like receptor signaling [42]. Thus, both the attenuated increase in DG species and lower phosphorylation of JNK in the trained mice tally with the training-induced alterations in substrate fluxes and suggest that these training effects can help to preserve hepatic insulin action during high dietary fat and sugar intake.
Notably, training in combination with HED lowered the abundance of complex I proteins and the native complex I holocomplex in our model of fatty liver. Affected proteins were equally related to nicotinamide adenine dinucleotide (NADH) oxidation, ubiquinone reduction, and the proton pumping module, thus not supporting the theory of a subunit-specific effect [43]. Notably, the abundance of supercomplexes remained constant. The fact that supercomplexes detected in liver mitochondria of C57Bl6/6N were all reported to contain complex I [44] pleads in favor of an efficient assembly of complex I to supercomplexes in the trained mice despite a lower content of the single proteins. Supercomplex formation is highly relevant for effective respiration [45]. This may be the reason why the lower abundance of the complex I holocomplex did not weaken the HED-induced increase in oxidative capacity measured in mitochondria isolated from livers of sedentary and trained HED-fed mice. Another explanation is that complex I contributes less than complex II to maximal coupled phosphorylation in isolated liver mitochondria [46]. Supercomplexes are also relevant for low mitochondrial reactive oxygen species production [47]. Lower complex I content and activity have been related to lower mitochondrial hydrogen peroxide release and superoxide production [48,49]. To test the hypothesis that the reduced abundance of complex I in liver mitochondria of trained HED-fed mice can protect them against mitochondrial reactive oxygen species (ROS) production, we studied the respiration-related production of hydrogen peroxide. However, using Amplex red conversion as a measure of hydrogen peroxide production, we obtained high readings in all samples. This was presumably due to the high abundance of carboxylesterases in the liver mitochondria, which promote Amplex red conversion, resulting in high values independent of ROS production in the respiratory chain [50].
Our study focused on mitochondrial adaptations in liver and skeletal muscle to explain the beneficial effect of training on fatty liver and insulin sensitivity. Other mechanisms outside the focus of this study could also have contributed to the prevention of hepatic lipid accumulation. For example, restored peripheral insulin sensitivity in the trained HED-fed mice could have reduced lipolysis in adipose tissue and enhanced glycogen storage in the trained skeletal muscle, two effects lowering the substrate burden on the liver.
A limitation of the study is that we did not directly analyze metabolic fluxes, acetyl-CoA, or hepatic insulin sensitivity. Although we provided functional physiological readouts in combination with proteome, transcriptome and lipidome data, direct proof on a molecular level for the amelioration of oxidative stress or reversal of hepatic insulin action is lacking. Our conclusions are supported by the ipGTT results reflecting whole body glucose tolerance, while twice the number of animals would have been required for an investigation of direct insulin effects on hepatic insulin signaling. We also did not analyze fitness parameters to validate training effects on whole body level. Moreover, treadmill training for five days a week, i.e. more than the three days in our study, has been shown to reduce caloric consumption in mice fed a high-fat diet [51]. This effect could also have contributed to the lower weight and fat storage in our experiment, in addition to an increased energy expenditure caused by treadmill training. Nevertheless, our findings remain relevant as they show differential adaptations in mice receiving a combination of HED and training, particularly on the mitochondrial level, that go beyond a simple normalization towards the sedentary control group. Another open question is the potential long-term effect of training, beyond 6 weeks, on the transition of hepatic steatosis to steatohepatitis and on the development of insulin resistance.
Current approaches for a pharmacological treatment of NAFLD include the development of inhibitors targeting acetyl-CoA carboxylase and fatty acid synthase at the level of fatty acid synthesis, the mitochondrial pyruvate carrier, and ketohexokinase, a key enzyme in the metabolism of fructose, which is a particularly potent driver of de novo lipogenesis [52,53]. By demonstrating that regular exercise can antagonize the upregulation of these factors or reduce their expression in mice fed a steatogenic diet, our results corroborate these intervention strategies. Also, they demonstrate the superiority of physical activity, which targets these and a multitude of additional enzymes and processes simultaneously, for the prevention and treatment of NAFLD. Finally, our results demonstrate that these manifold physical activity-induced molecular alterations can be understood as part of an overarching concept. Collectively, these adaptative processes are crucial for the long-term consequences of high energy intake on liver function.
4. Material and methods
4.1. Animal care, diet and training
The animal experiment was performed in accordance with the Directive 2010/63/EU of the European Union and was approved by the local authorities (Regierungspraesidium Tuebingen). Male C57BL/6N mice were purchased at the age of 9 weeks from Charles River (Sulzfeld, Germany). After two weeks of treadmill acclimatization of all animals, mice were randomized to 4 groups of n = 8 for 6 weeks of dietary intervention and treadmill training: control diet, sedentary (CON_SED); control diet, training (CON_TRAIN); high-energy diet, sedentary (HED_SED); and high-energy diet, training (HED_TRAIN). The high-energy diet (Ssniff, Soest, Germany) E15744-344; 45 kJ% fat, 35 kJ% carbohydrates, 20 kJ% protein; corresponding to Research Diets D12451) contained additional sugar (10 weight% sucrose) and fat (20 weight% lard) and was otherwise similar to the matched control diet (E157453-04/D12450J; 10 kJ% fat, 70 kJ% carbohydrates, 20 kJ% protein). Chow and tap water were provided ad libitum. Treadmill training was conducted for 1 h during the animals’ active phase three times per week and separated by at least one day of rest, as previously described [54]. Treadmill conditions were as follows:
After 5 min warmup, 10 m/min at 12° uphill slope in week 1; 11 m/min, 12° in week 2; 12 m/min, 12° in week 3; 12 m/min, 10° in week 4, 12 m/min, 9° in week 5 and 12 m/min, 8° slope in week 6. After 5 weeks, an i.p. glucose tolerance test (ipGTT) was performed as previously described, after 16h of fasting and with 1.5 g glucose (20% solution; B. Braun, Melsungen, Germany) per kg body weight. After 6 weeks, mice were analgosedated (150 mg ketamine and 10 mg xylazine/kg body weight) and exsanguinated by decapitation before removal of organs, which were immediately processed or flash-frozen in liquid nitrogen. Both glucose tolerance test and organ collection took place 48 h after the preceding training session.
4.2. Isolation of mitochondria
Mitochondria isolation was performed following a previously published protocol [46]. Briefly, 150 mg liver were homogenized in ice-cold STE buffer (250 mM sucrose, 5 mM Tris, 2 mM EGTA, pH 7.4 at 4 °C) + 0.5% BSA and centrifuged at 900 g for 10 min. Pooled muscles from the upper hind limb were used to isolate skeletal muscle mitochondria. Before homogenization and centrifugation as above, 550 mg muscle tissue were digested for 3 min on ice with 33 mg type VIII protease from Bacillus licheniformis (10 U/mg, Sigma-Aldrich) in 5.5 mL STE. The reaction was stopped by adding 10 mL STE + 0.1% BSA, pelletting the muscles by centrifugation and washing three times with STE + 0.1% BSA. The supernatants were centrifuged at 9,000 g for 10 min to yield the crude mitochondrial fraction, which was resuspended in STE +0.1% BSA and centrifuged again at 9,000 g for 10 min. The crude mitochondrial pellet was resuspended in STE. An aliquot was used to determine the protein concentration using Bradford reagent (Carl Roth, Karlsruhe, Germany). 100 μg (per protein) were used for respiration analyses, and additional aliquots stored at −80 °C for blue native PAGE analysis. The remainder of the crude fraction was layered onto 5 ml Percoll (25% in STE) and centrifuged for 20 min at 80,000 g. The lower, mitochondria-rich layer was collected, Percoll was removed by washing, and mitochondria were pelleted and resuspended in PBS. Protein concentration of the purified mitochondrial fraction was determined using a BCA assay (Pierce™ BCA Protein Assay Kit, Pierce Thermo Fisher Scientific, Schwerte, Germany). The aliquots were stored at −80 °C.
4.3. High-resolution respirometry
An Oxygraph-2k (Oroboros Instruments, Innsbruck, Austria) was applied to assess mitochondrial function as previously described [46], using 100 μg (by protein) crude mitochondria. To evaluate electron transport chain capacity and non-mitochondrial oxygen consumption, substrates and inhibitors were added in the following order: Malate (1.28 mM), octanoylcarnitine (0.5 mM), fatty acid oxidation), ADP (2.5 mM, phosphorylating condition), pyruvate (5 mM, complex I respiration), succinate (2.5 mM, complex II respiration), cytochrome c (10 μM, integrity control), FCCP (in 0.5 μM steps to induce the uncoupled state), rotenone (1.25 μM, complex I inhibitor) and antimycin A (5 μM, complex III inhibitor). Mitochondrial membrane integrity was controlled by cytochrome c (20.7 ± 5.8% for liver mitochondria and 3.3 ± 3.1% for muscle mitochondria). Data were corrected for non-mitochondrial background by subtraction of oxygen consumption after antimycin A. Respiratory substrates and inhibitors were purchased from Sigma–Aldrich, ADP was obtained from Calbiochem, Merck (Darmstadt, Germany) and octanoylcarnitine from Tocris Bioscience (Bristol, UK). Respirometry data of the sedentary control diet-fed mice were already published [46].
4.4. Tissue lysates, immunoblots and enzymatic assays
For immunoblots and citrate synthase assays, fresh tissue aliquots were rapidly homogenized at 4 °C in lysis buffer (50 mM HEPES, 150 mM NaCl, 1.5 mM MgCl2 1 mM EGTA, 10% Glycerol, 1% Triton X 100, 100 mM NaF, 10 mM Na4P2O7, 1 mM PMSF, 10 μg/ml Aprotinin, 400 μM Na3VO4) containing protease inhibitors (Complete EDTA-free; Roche, Mannheim, Germany) using a TissueLyser (Qiagen, Hilden, Germany). Immunoblots from 30 μg of protein from tissue lysate were performed as described [55] using IRDye® secondary antibodies (LI-COR Biosciences GmbH, Bad Homburg, Germany). Total protein abundance was visualized by 2, 2, 2-Trichloroethanol (TCE) staining. Antibodies against phospho-JNK (T183/Y185; #9251) and JNK protein (#9252) were from Cell signaling Technology (Danvers, MA, USA). Blue native PAGE analysis of respiratory complexes was performed, as previously described [56] using 100 μg (per protein) mitochondria. The samples were solubilized for 20 min on ice using 800 μg digitonin; centrifuged at 20,000 g, 4 °C for 10 min; 30 μL supernatant were mixed with 200 μg Coomassie G-250, and loaded onto a gradient gel. After electrophoresis with cathode buffers containing high, low, and no Coomassie for 30 min at 150V, 90 min at 250 V and 60 min at 300 V, respectively, the gel was washed in ultrapure water and visualized at 700 nm.
The activity of citrate synthase as quantitative marker for mitochondrial content [57] was recorded for 3 min in a spectrophotometer at 412 nm in a reaction containing 15 μg of protein, 0.25% Triton X-100, 0.31 mM acetyl-CoA, 0.1 mM 5,5′-dithiobis-(2-nitrobenzoic acid) and 0.5 mM oxaloacetate.
Total triacylglycerol content was quantified using an enzymatic assay, as previously described [58], after homogenizing pieces of frozen liver tissue in 0.9% NaCl containing 1% Triton X-100.
4.5. Transcriptome analysis of liver
RNA isolation was performed using the RNeasy kit (Qiagen, Hilden, Germany). The Agilent 2100 Bioanalyzer was applied to assess RNA quality and only high-quality RNA (RIN>7) was used for microarray analysis. Total RNA was amplified with the WT PLUS Reagent Kit (Thermo Fisher Scientific Inc., Waltham, USA) and analyzed on Mouse Clariom S arrays (Thermo Fisher Scientific). Staining and scanning (GeneChip Scanner 3000 7G) were carried out according to manufacturer's instructions. Array data were submitted to the GEO database at NCBI (GSE167046). Expression console (v.1.4.1.46, Affymetrix) was used to obtain annotated normalized SST-RMA gene-level data. Statistical analyses were performed in R (R Development Core Team [59] employing the limma t-test. Two samples were removed from the analysis because of poor-quality data, and one sample was detected as outlier by PCA. To reduce background, gene sets were filtered for fold change >1.3x and for average expression >30 (arbitrary units) in at least one group. Genes were considered to be not expressed if the detection p-value was >0.05 in all samples.
4.6. Proteome analysis of mitochondria
4.6.1. Lysis, carbamidomethylation, on-filter proteolysis and amino acid analysis
Frozen, purified mitochondria were solubilizsed in 50 mM Tris-Cl, 1% SDS, 150 mM NaCl (pH 7.8) containing Complete EDTA-free protease inhibitor cocktail (Roche, Switzerland). After centrifugation at 18,000 g for 30 min, supernatants were transferred to LoBind tubes. Aliquots of 45 μg of protein were reduced (10 mM DTT, 56 °C for 30 min) and alkylated (30 mM iodoacetamide; room temperature for 30 min in the dark). Sample cleanup and on-filter proteolysis were performed using filter-aided sample preparation with minor changes [60,61] commencing with diluting samples in freshly prepared 8 M urea in 100 mM Tris–HCl, pH 8.5 [62], and then using 50 mM NH4HCO3 (pH 7.8) for buffer exchange, followed by incubation with digestion buffer (1:20 (w/w enzyme/substrate) trypsin (sequencing grade modified trypsin, Promega, Madison, USA), 0.2 M guanidinium hydrochloride, 2 mM CaCl2 in 50 mM NH4HCO3 buffer, pH 7.8) at 37 °C for 14 h. Tryptic peptides were recovered by centrifugation followed by flushing first with 50 μL of 50 mM NH4HCO3, then with 50 μL of water, and acidification to pH < 3 with 10% trifluoroacetic acid (TFA). Samples were subsequently desalted by solid phase extraction (C18, 4 mg, Varian, USA) and eluted tryptic peptides dried in a SpeedVac. Peptides were resolubilized in 0.1% TFA and quality was controlled on a monolithic HPLC column, as described previously [63]. Finally, peptides were quantified by amino acid analysis, as previously described [64,65] and stored at −40 °C for subsequent use.
4.6.2. High-pH fractionation and data dependent acquisition (DDA)
A master mix was generated by combining 1.5 μg peptides from eight liver and muscle samples, respectively, dried in a SpeedVac, resolubilized in 10 mM ammonium formate, pH 8.0 and fractionated by reversed-phase chromatography at pH 8.0 on a Biobasic (C18, 0.5 × 150 mm, 5 μm particle size) column using an UltiMate 3000 LC system (both Thermo Scientific, Germany) with buffers A: 10 mM ammonium formate, pH 8.0, and B: 84% ACN in 10 mM ammonium formate, pH 8.0. The flowrate was 12.5 μL/min applying a gradient of 3% B for 10 min, 3–45% B in 40 min, 45–60% B in 5 min, 60–95% B in 5 min, 95% B hold for 5 min, 95%-3% B in 5 min and a final re-equilibration step with 3% B for 20 min. In total 16 fractions were collected at 1 min interval in a concatenation mode, dried in a SpeedVac and stored at −40 °C for later use.
Each fraction was resolubilized in 60 μL of 0.1% TFA. Additionally, 15 μL were spiked with 1.5 μL of diluted iRT standard (Biognosys, Schlieren, Switzerland) and analyzed using an Ultimate 3000 nano RSLC system coupled to a Q Exactive High Field (HF) mass spectrometer (both Thermo Scientific). Peptides were pre-concentrated on a 100 μm × 2 cm C18 trapping column for 5 min using 0.1% TFA with a flow rate of 20 μL/min followed by separation on a 75 μm × 50 cm C18 main column (both Acclaim Pepmap nanoviper, Thermo Scientific) with a 120 min LC gradient ranging from 3 to 35% of B (84% ACN in 0.1% FA) at a flow rate of 250 nL/min. The Q Exactive HF was operated in DDA mode and MS survey scans were acquired from m/z 300 to 1,500 at a resolution of 60,000 using the polysiloxane ion at m/z 371.1012 as a lock mass. The 15 most intense ions were isolated with a 1.2 m/z window and then fragmented by higher energy collisional dissociation with a normalized collision energy of 27%, considering a dynamic exclusion of 20 s. MS/MS spectra were acquired at a resolution of 15,000. Automatic gain control (AGC) target values and maximum fill times were set to 3 × 106 and 120 ms for MS and 5 × 104 and 200 ms for MS/MS.
4.6.3. Data independent acquisition (DIA)
All 64 samples were measured after the DDA runs. Peptides corresponding to 0.5 μg of each sample containing 1.5 μL of iRT standard were analyzed on the same set of LC-MS instruments and chromatographic conditions as mentioned above. The mass spectra on the Q Exactive HF operating in DIA mode were acquired from m/z 300 to 1,201 at a resolution of 60,000. The AGC and fill time values were set to 3 × 106 and 20 ms, respectively. For DIA, the settings were: resolution 30,000; AGC 3 × 106; isolation window 29.1 m/z; DIA segments 32; HCD collision energy: 27% and the maximum fill time was set to auto.
4.6.4. Data analysis for DDA measurements
The DDA MS data acquired from all 16 fractions were processed together with Proteome Discoverer (PD) 1.4 (Thermo Scientific, Germany) and searched against the mouse Uniprot database (downloaded on 22nd of July 2015, containing 16,750 target sequences plus 11 iRT peptide sequences [66]). To maximize the number of peptide spectrum matches, three different search algorithms were included, namely Mascot [67], Sequest [68], and MS Amanda [69] using the same set of parameters, i.e. precursor and fragment ion tolerances of 10 ppm and 0.02 Da for MS and MS/MS, respectively; trypsin as enzyme with a maximum of two missed cleavages; carbamidomethylation of Cys as fixed modification and oxidation of Met as variable modification.
4.6.5. Spectral library generation and DIA based label-free quantitation (LFQ)
The data analysis of all DIA runs was performed with Spectronaut Pulsar software version 11.0.15038.12.33511 (Biognosys). To generate the spectral library, the mouse Uniprot database and the abovementioned PD 1.4 output file were loaded into the Spectronaut software with default settings including 1% false discovery rate (FDR) and search engine rank 1 on the peptide to spectrum match level. A set of 16 DIA raw files belonging to either liver or muscle containing all conditions (CON_SED, HED_SED, CON_TRAIN, HED_TRAIN; n = 4 mice) was then processed independently. For relative LFQ the Spectronaut parameters were: MS1 and MS2 filtering, XIC extraction set to dynamic, calibration and identification set to automatic and dynamic, respectively. Quantification settings were: Major Group Quantity set to Mean peptide quantity; Quantity MS-Level: MS2; Quantity Type set to Area; Min and Max values of Major Group Top N values set to 2 and 4, respectively; Proteotypicity Filter set to Only Proteotypic; Data Filtering set to Qvalue complete (threshold 0.01) and Normalization Strategy set as Global Normalization. Finally, the protein identifications with their respective normalized abundances (PG.Quantity) values were exported from Spectronaut and only those candidates that were commonly quantified across all 16 DIA runs per set were selected for further data analysis. Abundances of individual proteins were normalized to total protein abundance in a given sample. The mass spectrometry proteomics data were deposited in the Proteome Xchange Consortium via the PRIDE partner repository (data set identifier PXD024308) [70].
4.7. Lipid extraction and UHPLC-MS/MS-lipidomics
The lipids were extracted with the combination of MTBE-MeOH-water, as described previously [71] using ∼30 mg of whole liver tissue. Total protein content in the liver extracts was determined using Bradford reagent after solubilizing the pellet retained after MTBE extraction in a buffer containing 5 M urea, 2 mM thiourea, 15 mM DTT and 2% CHAPS. A Waters UHPLC system (Milford, MA, USA) was used for the reversed-phase separation of lipids which was performed on an ACQUITY UHPLC BEH C8 column (100 mm × 2.1 mm × 1.7 μm). The mobile phase consisted of (A) ACN: H2O = 60:40 (v/v) and (B) IPA: ACN = 90:10 (v/v), both containing 10 mM ammonium acetate. Gradient elution was conducted for 20 min at a flow rate of 0.26 ml/min. The gradient elution procedure was as follows: 0–1.5 min, 32% B; 1.5–15.5 min, 32–85% B; 15.5–15.6 min, 85–99% B; 15.6–18 min, 99% B; 18–18.1 min, 99-32% B; 18.1–20 min, 32% B. The column temperature was maintained at 55 °C. The sample tray was maintained at 10 °C throughout the analysis.
A Q Exactive High Field Mass Spectrometer from Thermo Fisher Scientific Inc. (Rockford, IL, USA) was used for lipid detection. Lipids were detected in both positive and negative electrospray ion modes at scan ranges of 400–1300 Da and 200-1 800 Da, respectively. The positive and negative electrospray voltages were 4.0 kV and −3.5 kV, respectively. The ion spray source temperature was 300 °C. The auxiliary gas heater temperature was 350 °C. Sheath gas and auxiliary gas were 45 and 10 arbitrary units, respectively. The S-lens RF level was 50. For full scan, the resolution was 120,000 FWHM, the AGC target was 3 × 106 ion capacity, and maximum IT was 200 ms. For data-dependent MS/MS (dd-MS2), the resolution was 60,000 FWHM, AGC target was 1 × 105, maximum IT was 40 ms, TopN was 15, and isolation window was 1.4 m/z.
Lipidomics data acquisitions were performed using Thermo XCalibur software version 4.2. Lipid identification was carried out on the basis of MS/MS fragmentations, accurate m/z values and relative liquid chromatographic elution time of lipids in combination with the automatic identification output by Thermo Fisher Scientific LipidSearch software (Waltham, USA). The lipidomics data were semi-quantified by normalizing the peak area of detected lipids with those of the appropriate lipid internal standard. The synthetic lipid standards d4-palmitic acid, d4-FFA(16:0), d4-FFA(22:0), CER(d18:1/17:0), d7-DG(15:0/18:1), LPC(15:0), LPC(19:0), PC(15:0)2 PC(19:0)2, PE(17:0)2 SM(d18:1/12:0), TG(15:0)3, CL(14:0)4, were purchased from Avanti Polar Lipids (Alabaster, AL, USA) or Sigma–Aldrich (Munich, Germany). Only data of detected DG and CER species and of the mitochondrial marker lipid tetralinoleoyl cardiolipin were included in this manuscript.
4.8. Statistics
All data in graphs are summarized as mean ± SD. Two-way ANOVA was performed to evaluate the diet, training and interaction effects, followed by Tukey HSD multiple comparison tests. Statistical parameters and significance levels are shown in the figure legends. The statistics for transcriptome data are described in the separate method section. Upstream regulator analyses of differences in gene expression were generated using QIAGEN's Ingenuity Pathway Analysis (IPA®, QIAGEN Redwood City, www.qiagen.com/ingenuity). Enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways associated with differences in gene expression were identified using the InCroMAP platform [72]. Moreover, enrichment analyses of differences in mitochondrial protein abundance were performed with ranked values using STRING 11.0 [73].
Data availability statement
Transcriptome and proteome data are deposited in public databases. Other datasets analyzed during the current study can be obtained from the corresponding author on reasonable request.
Funding
We gratefully acknowledge the financial support from the German Federal Ministry of Education and Research (BMBF) to the German Centre for Diabetes Research (Grant no. 01GI0925), from the Mobility Programme of the Sino-German Center for Research Promotion (M−0257), from the Innovation Program from DICP (DICP I202019), from the National Natural Science Foundation of China (Grant Nos. 21874130 and 22074144), and the support by the “Ministerium für Kultur und Wissenschaft des Landes Nordrhein-Westfalen” and “Der Regierende Bürgermeister von Berlin, Senatskanzlei Wissenschaft und Forschung” (to L. Kollipara and A. Sickmann). This work was partially supported by the Helmholtz Alliance ‘Aging and Metabolic Programming, AMPro’ (to J. Beckers).
Author contributions
M.H., Li.K., R.L., and C.W. designed the study. M.H. carried out the mouse study. Li.K., La.K., C.Hu., M.I., M.H. conducted respirometry, proteomics, transcriptome, and lipidomics experiments and data analysis with timely support from D.B., and C.H. J.B., M.H.A, H.U.H., A.B., A.P., A.S., G.X., R.L., and C.W. contributed to data interpretation and discussion. M.H., Li.K., and C.W. drafted the paper. All authors contributed to the final version of the manuscript.
Acknowledgments
The authors are grateful for the excellent technical support provided by Paul Grubba from the University Hospital Tuebingen, Tuebingen, Germany, and by Mareike Bamberger (Helmholtz Center, Munich).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.molmet.2021.101359.
Conflict of interest
The authors declare no competing interests.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Figure S1-4
Table S1
Table S2
Table S3
Table S4
Multimedia component 6
References
- 1.Paik J.M., Golabi P., Younossi Y., Mishra A., Younossi Z.M. Changes in the global burden of chronic liver diseases from 2012 to 2017: the growing impact of NAFLD. Hepatology. 2020;72(5):1605–1616. doi: 10.1002/hep.31173. [DOI] [PubMed] [Google Scholar]
- 2.Younossi Z.M., Golabi P., de Avila L., Paik J.M., Srishord M., Fukui N., et al. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: a systematic review and meta-analysis. Journal of Hepatology. 2019;71(4):793–801. doi: 10.1016/j.jhep.2019.06.021. [DOI] [PubMed] [Google Scholar]
- 3.Huang D.Q., El-Serag H.B., Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors, and prevention. Nature Reviews Gastroenterology & Hepatology. 2021;18:223–238. doi: 10.1038/s41575-020-00381-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eslam M., Sanyal A.J., George J. MAFLD: a consensus-driven proposed nomenclature for metabolic associated fatty liver disease. Gastroenterology. 2020;158(7):1999–2014. doi: 10.1053/j.gastro.2019.11.312. e1991. [DOI] [PubMed] [Google Scholar]
- 5.EASL-EASD-EASO Clinical Practice Guidelines for the management of the non-alcoholic fatty liver disease. Journal of Hepatology. 2016;64(6):1388–1402. doi: 10.1016/j.jhep.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Younossi Z.M., Corey K.E., Lim J.K. AGA clinical practice update on lifestyle modification using diet and exercise to achieve weight loss in the management of nonalcoholic fatty liver disease: expert review. Gastroenterology. 2021;160(3):912–918. doi: 10.1053/j.gastro.2020.11.051. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi H., Kotani K., Tanaka K., Egucih Y., Anzai K. Therapeutic approaches to nonalcoholic fatty liver disease: exercise intervention and related mechanisms. Frontiers in Endocrinology. 2018;9:588. doi: 10.3389/fendo.2018.00588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hashida R., Kawaguchi T., Bekki M., Omoto M., Matsuse H., Nago T., et al. Aerobic vs. resistance exercise in non-alcoholic fatty liver disease: a systematic review. Journal of Hepatology. 2017;66(1):142–152. doi: 10.1016/j.jhep.2016.08.023. [DOI] [PubMed] [Google Scholar]
- 9.Thyfault J.P., Rector R.S. Exercise combats hepatic steatosis: potential mechanisms and clinical implications. Diabetes. 2020;69(4):517–524. doi: 10.2337/dbi18-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koliaki C., Szendroedi J., Kaul K., Jelenik T., Nowotny P., Jankowiak F., et al. Adaptation of hepatic mitochondrial function in humans with non-alcoholic fatty liver is lost in steatohepatitis. Cell Metabolism. 2015;21(5):739–746. doi: 10.1016/j.cmet.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Fritsch M., Koliaki C., Livingstone R., Phielix E., Bierwagen A., Meisinger M., et al. Time course of postprandial hepatic phosphorus metabolites in lean, obese, and type 2 diabetes patients. American Journal of Clinical Nutrition. 2015;102(5):1051–1058. doi: 10.3945/ajcn.115.107599. [DOI] [PubMed] [Google Scholar]
- 12.Sunny N.E., Parks E.J., Browning J.D., Burgess S.C. Excessive hepatic mitochondrial TCA cycle and gluconeogenesis in humans with nonalcoholic fatty liver disease. Cell Metabolism. 2011;14(6):804–810. doi: 10.1016/j.cmet.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sunny N.E., Bril F., Cusi K. Mitochondrial adaptation in nonalcoholic fatty liver disease: novel mechanisms and treatment strategies. Trends in Endocrinology and Metabolism. 2017;28(4):250–260. doi: 10.1016/j.tem.2016.11.006. [DOI] [PubMed] [Google Scholar]
- 14.Perry R.J., Samuel V.T., Petersen K.F., Shulman G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510(7503):84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Satapati S., Kucejova B., Duarte J.A., Fletcher J.A., Reynolds L., Sunny N.E., et al. Mitochondrial metabolism mediates oxidative stress and inflammation in the fatty liver. Journal of Clinical Investigation. 2015;125(12):4447–4462. doi: 10.1172/JCI82204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cortez-Pinto H., Chatham J., Chacko V.P., Arnold C., Rashid A., Diehl A.M. Alterations in liver ATP homeostasis in human nonalcoholic steatohepatitis: a pilot study. Jama. 1999;282(17):1659–1664. doi: 10.1001/jama.282.17.1659. [DOI] [PubMed] [Google Scholar]
- 17.Perez-Carreras M., Del Hoyo P., Martin M.A., Rubio J.C., Martin A., Castellano G., et al. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology. 2003;38(4):999–1007. doi: 10.1053/jhep.2003.50398. [DOI] [PubMed] [Google Scholar]
- 18.Hu C., Hoene M., Plomgaard P., Hansen J.S., Zhao X., Li J., et al. Muscle-liver substrate fluxes in exercising humans and potential effects on hepatic metabolism. Journal of Clinical Endocrinology & Metabolism. 2020;105(4):1196–1209. doi: 10.1210/clinem/dgz266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Camacho R.C., Donahue E.P., James F.D., Berglund E.D., Wasserman D.H. Energy state of the liver during short-term and exhaustive exercise in C57BL/6J mice. American Journal of Physiology. Endocrinology and Metabolism. 2006;290(3):E405–E408. doi: 10.1152/ajpendo.00385.2005. [DOI] [PubMed] [Google Scholar]
- 20.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Research. 2016;44(D1):D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pilegaard H., Neufer P.D. Transcriptional regulation of pyruvate dehydrogenase kinase 4 in skeletal muscle during and after exercise. Proceedings of the Nutrition Society. 2004;63(2):221–226. doi: 10.1079/pns2004345. [DOI] [PubMed] [Google Scholar]
- 22.Prakasam G., Singh R.K., Iqbal M.A., Saini S.K., Tiku A.B., Bamezai R.N.K. Pyruvate kinase M knockdown-induced signaling via AMP-activated protein kinase promotes mitochondrial biogenesis, autophagy, and cancer cell survival. Journal of Biological Chemistry. 2017;292(37):15561–15576. doi: 10.1074/jbc.M117.791343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson E.J., Yamazaki H., Neufer P.D. Induction of endogenous uncoupling protein 3 suppresses mitochondrial oxidant emission during fatty acid-supported respiration. Journal of Biological Chemistry. 2007;282(43):31257–31266. doi: 10.1074/jbc.M706129200. [DOI] [PubMed] [Google Scholar]
- 24.Donnelly K.L., Smith C.I., Schwarzenberg S.J., Jessurun J., Boldt M.D., Parks E.J. Sources of fatty acids stored in liver and secreted via lipoproteins in patients with nonalcoholic fatty liver disease. Journal of Clinical Investigation. 2005;115(5):1343–1351. doi: 10.1172/JCI23621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fletcher J.A., Deja S., Satapati S., Fu X., Burgess S.C., Browning J.D. Impaired ketogenesis and increased acetyl-CoA oxidation promote hyperglycemia in human fatty liver. JCI Insight. 2019;5(11) doi: 10.1172/jci.insight.127737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balasse E.O. Kinetics of ketone body metabolism in fasting humans. Metabolism. 1979;28(1):41–50. doi: 10.1016/0026-0495(79)90166-5. [DOI] [PubMed] [Google Scholar]
- 27.Satapati S., Sunny N.E., Kucejova B., Fu X., He T.T., Mendez-Lucas A., et al. Elevated TCA cycle function in the pathology of diet-induced hepatic insulin resistance and fatty liver. The Journal of Lipid Research. 2012;53(6):1080–1092. doi: 10.1194/jlr.M023382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cotter D.G., Ercal B., Huang X., Leid J.M., d'Avignon D.A., Graham M.J., et al. Ketogenesis prevents diet-induced fatty liver injury and hyperglycemia. Journal of Clinical Investigation. 2014;124(12):5175–5190. doi: 10.1172/JCI76388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He A., Chen X., Tan M., Chen Y., Lu D., Zhang X., et al. Acetyl-CoA derived from hepatic peroxisomal β-oxidation inhibits autophagy and promotes steatosis via mTORC1 activation. Molecular Cell. 2020;79(1):30–42. doi: 10.1016/j.molcel.2020.05.007. e34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kleinert M., Parker B.L., Jensen T.E., Raun S.H., Pham P., Han X., et al. Quantitative proteomic characterization of cellular pathways associated with altered insulin sensitivity in skeletal muscle following high-fat diet feeding and exercise training. Scientific Reports. 2018;8(1):10723. doi: 10.1038/s41598-018-28540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bezaire V., Spriet L.L., Campbell S., Sabet N., Gerrits M., Bonen A., et al. Constitutive UCP3 overexpression at physiological levels increases mouse skeletal muscle capacity for fatty acid transport and oxidation. The FASEB Journal. 2005;19(8):977–979. doi: 10.1096/fj.04-2765fje. [DOI] [PubMed] [Google Scholar]
- 32.Aguer C., Fiehn O., Seifert E.L., Bézaire V., Meissen J.K., Daniels A., et al. Muscle uncoupling protein 3 overexpression mimics endurance training and reduces circulating biomarkers of incomplete β-oxidation. The FASEB Journal. 2013;27(10):4213–4225. doi: 10.1096/fj.13-234302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goglia F., Skulachev V.P. A function for novel uncoupling proteins: antioxidant defense of mitochondrial matrix by translocating fatty acid peroxides from the inner to the outer membrane leaflet. The FASEB Journal. 2003;17(12):1585–1591. doi: 10.1096/fj.03-0159hyp. [DOI] [PubMed] [Google Scholar]
- 34.Aguer C., Piccolo B.D., Fiehn O., Adams S.H., Harper M.E. A novel amino acid and metabolomics signature in mice overexpressing muscle uncoupling protein 3. The FASEB Journal. 2017;31(2):814–827. doi: 10.1096/fj.201600914R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen M.C., Shulman G.I. Mechanisms of insulin action and insulin resistance. Physiological Reviews. 2018;98(4):2133–2223. doi: 10.1152/physrev.00063.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyu K., Zhang Y., Zhang D., Kahn M., Ter Horst K.W., Rodrigues M.R.S., et al. A membrane-bound diacylglycerol species induces PKC-mediated hepatic insulin resistance. Cell Metabolism. 2020;32(4):654–664. doi: 10.1016/j.cmet.2020.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang M., Hayakawa J., Yang K., Han X. Characterization and quantification of diacylglycerol species in biological extracts after one-step derivatization: a shotgun lipidomics approach. Analytical Chemistry. 2014;86(4):2146–2155. doi: 10.1021/ac403798q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Patterson R.E., Kalavalapalli S., Williams C.M., Nautiyal M., Mathew J.T., Martinez J., et al. Lipotoxicity in steatohepatitis occurs despite an increase in tricarboxylic acid cycle activity. American Journal of Physiology, Endocrinology and Metabolism. 2016;310(7):E484–E494. doi: 10.1152/ajpendo.00492.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jordy A.B., Kraakman M.J., Gardner T., Estevez E., Kammoun H.L., Weir J.M., et al. Analysis of the liver lipidome reveals insights into the protective effect of exercise on high-fat diet-induced hepatosteatosis in mice. American Journal of Physiology, Endocrinology and Metabolism. 2015;308(9):E778–E791. doi: 10.1152/ajpendo.00547.2014. [DOI] [PubMed] [Google Scholar]
- 40.Kumashiro N., Erion D.M., Zhang D., Kahn M., Beddow S.A., Chu X., et al. Cellular mechanism of insulin resistance in nonalcoholic fatty liver disease. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(39):16381–16385. doi: 10.1073/pnas.1113359108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang Q., Vijayakumar A., Kahn B.B. Metabolites as regulators of insulin sensitivity and metabolism. Nature Reviews Molecular Cell Biology. 2018;19(10):654–672. doi: 10.1038/s41580-018-0044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jia L., Vianna C.R., Fukuda M., Berglund E.D., Liu C., Tao C., et al. Hepatocyte Toll-like receptor 4 regulates obesity-induced inflammation and insulin resistance. Nature Communications. 2014;5:3878. doi: 10.1038/ncomms4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Miwa S., Jow H., Baty K., Johnson A., Czapiewski R., Saretzki G., et al. Low abundance of the matrix arm of complex I in mitochondria predicts longevity in mice. Nature Communications. 2014;5:3837. doi: 10.1038/ncomms4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Williams E.G., Wu Y., Jha P., Dubuis S., Blattmann P., Argmann C.A., et al. Systems proteomics of liver mitochondria function. Science. 2016;352(6291):aad0189. doi: 10.1126/science.aad0189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lapuente-Brun E., Moreno-Loshuertos R., Acín-Pérez R., Latorre-Pellicer A., Colás C., Balsa E., et al. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340(6140):1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- 46.Kappler L., Hoene M., Hu C., von Toerne C., Li J., Bleher D., et al. Linking bioenergetic function of mitochondria to tissue-specific molecular fingerprints. American Journal of Physiology, Endocrinology and Metabolism. 2019:E374–E387. doi: 10.1152/ajpendo.00088.2019. [DOI] [PubMed] [Google Scholar]
- 47.Maranzana E., Barbero G., Falasca A.I., Lenaz G., Genova M.L. Mitochondrial respiratory supercomplex association limits the production of reactive oxygen species from complex I. Antioxidants and Redox Signaling. 2013;19(13):1469–1480. doi: 10.1089/ars.2012.4845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verkaart S., Koopman W.J., van Emst-de Vries S.E., Nijtmans L.G., van den Heuvel L.W., Smeitink J.A., et al. Superoxide production is inversely related to complex I activity in inherited complex I deficiency. Biochimica et Biophysica Acta. 2007;1772(3):373–381. doi: 10.1016/j.bbadis.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 49.Lambert A.J., Buckingham J.A., Boysen H.M., Brand M.D. Low complex I content explains the low hydrogen peroxide production rate of heart mitochondria from the long-lived pigeon, Columba livia. Aging Cell. 2010;9(1):78–91. doi: 10.1111/j.1474-9726.2009.00538.x. [DOI] [PubMed] [Google Scholar]
- 50.Miwa S., Treumann A., Bell A., Vistoli G., Nelson G., Hay S., et al. Carboxylesterase converts Amplex red to resorufin: implications for mitochondrial H2O2 release assays. Free Radical Biology and Medicine. 2016;90:173–183. doi: 10.1016/j.freeradbiomed.2015.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Linden M.A., Pincu Y., Martin S.A., Woods J.A., Baynard T. Moderate exercise training provides modest protection against adipose tissue inflammatory gene expression in response to high-fat feeding. Physics Reports. 2014;2(7) doi: 10.14814/phy2.12071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Esler W.P., Bence K.K. Metabolic targets in nonalcoholic fatty liver disease. Cell Mol Gastroenterol Hepatol. 2019;8(2):247–267. doi: 10.1016/j.jcmgh.2019.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gutierrez J.A., Liu W., Perez S., Xing G., Sonnenberg G., Kou K., et al. Pharmacologic inhibition of ketohexokinase prevents fructose-induced metabolic dysfunction. Molecular Metabolism. 2021:101196. doi: 10.1016/j.molmet.2021.101196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoene M., Lehmann R., Hennige A.M., Pohl A.K., Haring H.U., Schleicher E.D., et al. Acute regulation of metabolic genes and insulin receptor substrates in the liver of mice by one single bout of treadmill exercise. Journal of Physiology. 2009;587(Pt 1):241–252. doi: 10.1113/jphysiol.2008.160275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kappler L., Li J., Haring H.U., Weigert C., Lehmann R., Xu G., et al. Purity matters: a workflow for the valid high-resolution lipid profiling of mitochondria from cell culture samples. Scientific Reports. 2016;6:21107. doi: 10.1038/srep21107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wittig I., Braun H.P., Schägger H. Blue native PAGE. Nature Protocols. 2006;1(1):418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
- 57.Larsen S., Nielsen J., Hansen C.N., Nielsen L.B., Wibrand F., Stride N., et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. Journal of Physiology. 2012;590(14):3349–3360. doi: 10.1113/jphysiol.2012.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hoene M., Irmler M., Beckers J., Hrabe de Angelis M., Haring H.U., Weigert C. A vitamin E-enriched antioxidant diet interferes with the acute adaptation of the liver to physical exercise in mice. Nutrients. 2018;10(5):E547. doi: 10.3390/nu10050547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.RCoreTeam . R Foundation for Statistical Computing; Vienna, Austria: 2020. R: a language and environment for statistical computing. [Google Scholar]
- 60.Manza L.L., Stamer S.L., Ham A.-J.L., Codreanu S.G., Liebler D.C. Sample preparation and digestion for proteomic analyses using spin filters. Proteomics. 2005;5(7):1742–1745. doi: 10.1002/pmic.200401063. [DOI] [PubMed] [Google Scholar]
- 61.Wisniewski J.R., Zougman A., Nagaraj N., Mann M. Universal sample preparation method for proteome analysis. Nature Methods. 2009;6(5):359–362. doi: 10.1038/nmeth.1322. [DOI] [PubMed] [Google Scholar]
- 62.Kollipara L., Zahedi R.P. Protein carbamylation: in vivo modification or in vitro artefact? Proteomics. 2013;13(6):941–944. doi: 10.1002/pmic.201200452. [DOI] [PubMed] [Google Scholar]
- 63.Burkhart J.M., Schumbrutzki C., Wortelkamp S., Sickmann A., Zahedi R.P. Systematic and quantitative comparison of digest efficiency and specificity reveals the impact of trypsin quality on MS-based proteomics. Journal of Proteomics. 2012;75(4):1454–1462. doi: 10.1016/j.jprot.2011.11.016. [DOI] [PubMed] [Google Scholar]
- 64.Cohen S.A., Michaud D.P. Synthesis of a fluorescent derivatizing reagent, 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate, and its application for the analysis of hydrolysate amino acids via high-performance liquid chromatography. Analytical Biochemistry. 1993;211(2):279–287. doi: 10.1006/abio.1993.1270. [DOI] [PubMed] [Google Scholar]
- 65.Shindo N., Nojima S., Fujimura T., Taka H., Mineki R., Murayama K. Separation of 18 6-aminoquinolyl-carbamyl-amino acids by ion-pair chromatography. Analytical Biochemistry. 1997;249(1):79–82. doi: 10.1006/abio.1997.2155. [DOI] [PubMed] [Google Scholar]
- 66.Escher C., Reiter L., MacLean B., Ossola R., Herzog F., Chilton J., et al. Using iRT, a normalized retention time for more targeted measurement of peptides. Proteomics. 2012;12(8):1111–1121. doi: 10.1002/pmic.201100463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Perkins D.N., Pappin D.J., Creasy D.M., Cottrell J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis. 1999;20(18):3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 68.Eng J.K., McCormack A.L., Yates J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. Journal of the American Society for Mass Spectrometry. 1994;5(11):976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 69.Dorfer V., Pichler P., Stranzl T., Stadlmann J., Taus T., Winkler S., et al. MS Amanda, a universal identification algorithm optimized for high accuracy tandem mass spectra. Journal of Proteome Research. 2014;13(8):3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Research. 2019;47(D1):D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen S., Hoene M., Li J., Li Y., Zhao X., Haring H.U., et al. Simultaneous extraction of metabolome and lipidome with methyl tert-butyl ether from a single small tissue sample for ultra-high performance liquid chromatography/mass spectrometry. Journal of Chromatography A. 2013;1298:9–16. doi: 10.1016/j.chroma.2013.05.019. [DOI] [PubMed] [Google Scholar]
- 72.Eichner J., Rosenbaum L., Wrzodek C., Haring H.U., Zell A., Lehmann R. Integrated enrichment analysis and pathway-centered visualization of metabolomics, proteomics, transcriptomics, and genomics data by using the InCroMAP software. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2014;966:77–82. doi: 10.1016/j.jchromb.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 73.Szklarczyk D., Gable A.L., Lyon D., Junge A., Wyder S., Huerta-Cepas J., et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Research. 2019;47(D1):D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1-4
Table S1
Table S2
Table S3
Table S4
Multimedia component 6
Data Availability Statement
Transcriptome and proteome data are deposited in public databases. Other datasets analyzed during the current study can be obtained from the corresponding author on reasonable request.