Summary
Background
Vaccines in emergency use are efficacious against COVID-19, yet vaccine-induced prevention against nasal SARS-CoV-2 infection remains suboptimal.
Methods
Since mucosal immunity is critical for nasal prevention, we investigated the efficacy of an intramuscular PD1-based receptor-binding domain (RBD) DNA vaccine (PD1-RBD-DNA) and intranasal live attenuated influenza-based vaccines (LAIV-CA4-RBD and LAIV-HK68-RBD) against SARS-CoV-2.
Findings
Substantially higher systemic and mucosal immune responses, including bronchoalveolar lavage IgA/IgG and lung polyfunctional memory CD8 T cells, were induced by the heterologous PD1-RBD-DNA/LAIV-HK68-RBD as compared with other regimens. When vaccinated animals were challenged at the memory phase, prevention of robust SARS-CoV-2 infection in nasal turbinate was achieved primarily by the heterologous regimen besides consistent protection in lungs. The regimen-induced antibodies cross-neutralized variants of concerns. Furthermore, LAIV-CA4-RBD could boost the BioNTech vaccine for improved mucosal immunity.
Interpretation
Our results demonstrated that intranasal influenza-based boost vaccination induces mucosal and systemic immunity for effective SARS-CoV-2 prevention in both upper and lower respiratory systems.
Funding
This study was supported by the Research Grants Council Collaborative Research Fund, General Research Fund and Health and Medical Research Fund in Hong Kong; Outbreak Response to Novel Coronavirus (COVID-19) by the Coalition for Epidemic Preparedness Innovations; Shenzhen Science and Technology Program and matching fund from Shenzhen Immuno Cure BioTech Limited; the Health@InnoHK, Innovation and Technology Commission of Hong Kong; National Program on Key Research Project of China; donations from the Friends of Hope Education Fund; the Theme-Based Research Scheme.
Keywords: SARS-CoV-2, Receptor binding domain, Mucosal immunity, Nasal prevention, PD1-based DNA vaccine, Live-attenuated influenza-based vaccine
Research in context.
Evidence before this study
Most published vaccines have indicated excellent efficacy in the lungs of vaccinated animals without analysis of SARS-CoV-2-infected cells in the upper respiratory system. Some studies indicated that there were no significant viral load drops in nasal swabs of vaccinated animals after viral challenge, demonstrating that vaccine-induced prevention against nasal SARS-CoV-2 infection remains suboptimal. Considering that vaccine-induced subprotection may underlie asymptomatic transmission, immune-escape viruses and re-infection, an effective vaccine should induce mucosal immunity for nasal prevention of SARS-CoV-2 infection. This is highly relevant to the human situation especially when there are tens of thousands of cases showing vaccine-breakthrough infections.
Added value of this study
In this study, we present the heterologous PD1-RBD-DNA/LAIV-RBD regimen that prevents SARS-CoV-2 infection of upper and lower respiratory systems by the high amount and long-lasting mucosal NAbs and CD8 T cells in two mouse models. Furthermore, intranasal LAIV-RBD boost enhanced humoral and cellular immune responses in mucosal sites when primed with the current approved BioNTech and Sinovac vaccine. These regimens-induced neutralizing antibodies can also cross-neutralize the variants of concern.
Implications of all the available evidence
A booster vaccine that can improve mucosal immunity to prevent asymptomatic transmission and virus spread is urgently needed. LAIV-RBD vaccine has been shown to induce mucosal immunity in pre-clinical studies of SARS-CoV-2 infection. Further studies will be needed to determine whether LAIV-RBD will provide mucosal protection in humans.
Alt-text: Unlabelled box
Introduction
Since the discovery of coronavirus disease 2019 (COVID-19) in human clusters in December 2019,1,2 the causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has led to over 100 million infections globally with nearly 2.1 million deaths after just one year. While the COVID-19 pandemic continues to evolve, hundreds of vaccine candidates have been brought into preclinical and clinical trials at an expedited speed using various platforms of technology.3, 4, 5 Encouragingly, several vaccines have now acquired regulatory approvals for emergency use in various countries, yet few have been evaluated for inducing mucosal protection, especially for preventing robust SARS-CoV-2 infection in nasal turbinate (NT).6,7 NT in the upper respiratory tract (URT) is one of the most important portals of SARS-CoV-2 entry into humans. Ciliated nasal epithelial cells in NT have the highest expression of angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2), supporting rapid and robust SARS-CoV-2 infection.8 Unlike SARS patients who have peak viral load in URT at day 10 after symptom onset,9 COVID-19 patients exhibit the highest viral loads in URT at or soon after the clinical presentation.1,10,11 Multiple SARS-CoV-2 variants can be transmitted simultaneously without a genetic bottleneck.12 Transmitted viruses then quickly impair host innate and adaptive immune responses,12 allowing for robust viral replication and asymptomatic viral spread. These findings indicated that the prevention of SARS-CoV-2 infection in NT is critical for pandemic control.
Current systemic vaccination and passive immunization are less effective or suboptimal for preventing SARS-CoV-2 in NT. Among vaccines authorized for emergency use, the Pfizer-BioNTech BNT162B2 mRNA, the Moderna mRNA-1273, the Oxford-ChAdOx1 and Novavax's NVX-CoV2373 have released phase III results showing the efficacy of 95%, 94.1%, 70.4% and 89.3%, respectively, in preventing COVID-19.13, 14, 15 The efficacy of the Oxford-ChAdOx1 vaccine against asymptomatic SARS-CoV-2 infection was only 3.8% among human vaccinees who received two standard doses.15 For passive immunotherapy, a receptor-binding domain (RBD)-specific human neutralizing antibody (NAb) LY-CoV555 has been evaluated in a phase 2 trial. Of three doses of 700 mg, 2800 mg and 7000 mg tested for monotherapy, only the medium dose of LY-CoV555 appeared to accelerate the natural decline of viral loads in nasopharyngeal swaps by day 11.16 These results indicated that vaccine-induced or passive NAbs are suboptimal to prevent SARS-CoV-2 infection in human NT. Using the animal model, we reported that robust SARS-CoV-2 infection in NT may outcompete passive or vaccine-induced systemic NAbs, revealing a possible mechanism underlying the subprotection against asymptomatic infection.17 Considering that vaccine-induced subprotection may drive immune escape virus variants of concern and allow re-infection,18, 19, 20 we hypothesized that an effective vaccine should induce mucosal immunity for nasal protection to prevent SARS-CoV-2 asymptomatic infection and silent spread. Recently, thousands of vaccine-breakthrough infections in the United States have further indicated the need for priority research on nasal prevention of SARS-CoV-2 infection.21
Methods
Mice
Male and female BALB/c and heterozygous K18-hACE2 mice (imported from The Jackson Laboratory) (aged 6-10 weeks) were obtained from the HKU Laboratory Animal Unit (LAU). The animals were kept in Biosafety Level-2 housing and given access to standard pellet feed and water ad libitum following LAU's standard operational procedures (SOPs). The viral challenge experiments were then conducted in our Biosafety Level-3 animal facility following SOPs strictly.
Cell lines
MDCK (RRID: CVCL_0422), HEK 293T (RRID: CVCL_0063), HEK 293T-hACE2 and Vero E6 cells (RRID: CVCL_0574) (mycoplasma negative) were maintained in Dulbecco's Modified Eagle Medium (DMEM) (Thermo Fisher Scientific) containing 10% fetal bovine serum, 2 mM L-glutamine and 100 U/mL penicillin and were incubated at 37°C in 5% CO2 setting.22 High-level and stable expression of human ACE2 on HEK 293T-hACE2 cell line surface was validated by flow cytometry analysis.
Viruses
Confluent Vero-E6 cells were infected at 0.01 MOI with live SARS-CoV-2 HKU-13 strain (GenBank accession number MT835140). After 3 days incubation, virus supernatant was collected for titration by plaque assay using Vero-E6 cells.
Construction and Generation of PD1-based DNA and live attenuated influenza virus (LAIV)-based vaccine
Codon-optimized SARS-CoV-2 RBD gene was in fusion to a human soluble PD1 domain (PD1-RBD) using the pVAX plasmid as the backbone. To maintain functional domains of the fusion protein, a linker (GGGGS)3 was applied between the PD1 and RBD gene.23 The expression construct contained a human tissue plasminogen activator (tPA) secretory signal sequence to promote antigen secretion. The plasmid DNA transfection into HEK 293T cells was performed using polyethylenimine (PEI), and protein expression was detected by Western blot. The pHW2000-DelNS1-RBD plasmid was constructed by inserting the tPA-linked RBD between the noncoding region (NCR) and autoproteolytic cleavage site (2A) in the pHW2000-DelNS1 plasmid. The V5 tag was added to the C terminal of RBD for better detection of RBD. To rescue the virus, eight pHW2000 plasmids containing the DelNS1-RBD and the other 7 influenza virus genomic segments, together with an NS1 expression plasmid, were transfected into 293T cells using Transit-LT1 (Mirus) according to the manufacturer protocol. After overnight incubation at 33 °C, DNA mix was removed, and MEM supplemented with 1 μg/ml N-tosyl-l-phenylalanine chloromethyl ketone (TPCK)-treated trypsin (Sigma) was added. Virus supernatant was collected 72 hours later and designated passage 0 (P0) virus and was further passaged in chicken embryonated eggs for 48 hours at 33°C. Viruses were aliquoted and titrated by plaque assay using MDCK cells. Two intranasal recombinant LAIV DelNS1-RBD vaccine strains, namely LAIV-CA4-RBD and LAIV-HK68-RBD, were generated using A/CA/04/2009 (H1N1) and A/Hong Kong/1/68 (H3N2) surface proteins (HA and NA) in the A/CA/04/2009 (H1N1) DelNS1 backbone by a reverse genetic procedure.24
Animal immunization and SARS-CoV-2 challenges
All animal experiments were approved by the Committee on the Use of Live Animals in Teaching and Research of the University of Hong Kong (HKU). For vaccine immunization, 6-8 weeks old BALB/c or K18-ACE2 transgenic mice received DNA immunization by intramuscular/electroporation (i.m./EP) with 50 µg PD1-RBD-DNA. The voltage of EP was pre-set 60 V in the TERESA DNA Delivery Device (Shanghai Teresa Healthcare Sci-Tech Co., Ltd). Mice intramuscularly received 1/5 clinical doses of Pfizer/BioNTech or Sinovac vaccine. Mice received LAIV-CA4-RBD or LAIV-HK68-RBD immunization by intranasal inoculation of 106 PFU per mouse at a 3-week interval under anaesthesia by intraperitoneal injection (i.p.) of Ketamine/Xylazine mixture (60 mg/kg and 10 mg/kg, respectively). Blood sera were collected for anti-RBD IgG and neutralization detection. Mice were sacrificed by i.p. infection of Pentobarbitone (150 mg/kg) and cells from lungs and spleen were harvested and subjected to intracellular cytokine staining (ICS) assay. For the SARS-CoV-2 challenge, BALB/c mice were anesthetized and transduced intranasally with 4 × 108 FFU of Ad5-hACE2, kindly provided by Dr. Jincun Zhao,25 in 70 µl DMEM. The transduced BALB/c mice or K18-ACE2 transgenic mice were intranasally infected with live wild type SARS-CoV-2 (HKU clone 13) at a dose of 1 × 104 PFU. Infected animals were sacrificed for endpoint analysis on day 4 post infection (4 dpi). All animal experiments related to SARS-CoV-2 were performed in a biosafety level 3 laboratory in HKU.
Enzyme-linked immunosorbent assay (ELISA)
ELISA was performed to detect SARS-CoV-2 RBD-specific IgG, as previously described.26 In brief, 96-well plates (Costar) were coated with recombinant SARS-CoV-2 RBD antigen (25 ng/well, Sino Biological) at 4°C overnight. After washing with PBST (0.05% Tween-20 in PBS), the plates were blocked with 4% skim milk in PBS for 1 hour at 37°C and incubated with serially diluted patient plasma for 1 hour at 37°C. After washing with PBST, goat anti-mouse IgG (Invitrogen, Cat# 31430, RRID:AB_228307) or anti-mouse IgA (Invitrogen, Cat# 62-6720, RRID: AB_2533951) conjugated with HRP was added, and the whole solution was incubated for 1 hour, followed by washing and the addition of 50 µl HRP chromogenic substrate 3,3’,5,5’-TMB (Sigma, Cat# T4444). Optical density (OD) values were measured at 450 nm using the VARIOSKANTM LUX multimode microplate reader (Thermo Fisher Scientific). The area under the curve (AUC) was measured using GraphPad Prism v8, setting the baseline with the defined endpoint (average of negative control wells + 10 standard deviations) and taking the total peak area as previously described.27
Pseudotyped viral neutralization assay
To determine the neutralizing activity of mouse sera and BAL, specimens were inactivated at 56°C for 30 min before the pseudotyped viral entry assay as previously described.22,28 The results of this assay correlated strongly with that of neutralization assay using replication-competent SARS-CoV or SARS-CoV-2.8,29 The plasmids encoding for D614G, Alpha, Beta and Delta variants were kindly provided by Dr. David D. Ho. In brief, different SARS-CoV-2 pseudotyped viruses were generated through co-transfection of 293T cells with 2 plasmids, pSARS-CoV-2 S and pNL4-3Luc_Env_Vpr, carrying the optimized SARS-CoV-2 S gene and a human immunodeficiency virus type 1 backbone, respectively. At 48 hours post-transfection, viral supernatant was collected and frozen at -80°C. Serially diluted serum samples were incubated with 200 TCID50 of pseudovirus at 37°C for 1 hour. The serum-virus mixtures were then added into pre-seeded HEK 293T-hACE2 cells. After 48 hours, infected cells were lysed, and luciferase activity was measured using Luciferase Assay System kits (Promega) in a Victor3-1420 Multilabel Counter (PerkinElmer). The 50% inhibitory concentrations (IC50) of each specimen were calculated using non-linear regression in GraphPad Prism v8 to reflect anti-SARS-CoV-2 potency.
Surface and intracellular cytokine staining (ICS)
The lung cells of mice were washed one time with staining buffer (PBS contained 2% FBS) followed by staining with anti-mouse antibodies for 30 min at 4 °C, including dead cell dye (Zombie Aqua, Biolegend Cat# 423102), CD19-FITC (Biolegend Cat# 152404, RRID: AB_2629813), CD11b-PerCP/Cy5.5 (Biolegend Cat# 101228, RRID: AB_893232), CD11c-PE-Cy7 (Biolegend Cat# 117318, RRID: AB_493568), Ly6c-APC-Fire750 (Biolegend Cat# 128046, RRID: AB_2616731), F4/80-BV421 (Biolegend Cat# 123137, RRID: AB_2563102), Ly6G-PE (Biolegend Cat# 127608, RRID: AB_1186099), CD103-BV785 (Biolegend Cat# 121439, RRID: AB_2800588) and I-A/I-E-BV605 (Biolegend Cat# 107639, RRID: AB_2565894). To measure antigen-specific T cell response, lymphocytes from mouse lung and spleen were stimulated with 1 µg/mL SARS-CoV-2 RBD peptide pool (15-mer overlapping by 11, spanning the whole RBD sequence at Spike306-543). Cells were incubated at 37°C overnight, and brefeldin A was added at 2 hours post-incubation. PMA/ionomycin stimulation was included as the positive control. After overnight incubation, cells were washed with staining buffer (PBS containing 2% FBS) and surface stained with anti-mouse-CD4-PerCP/Cy5.5 (Biolegend Cat# 116012, RRID: AB_2563023), anti-mouse-CD8-BV785 (Biolegend Cat# 100750, RRID: AB_2562610), anti-mouse CD69-BV711 (Biolegend Cat# 104537, RRID: AB_2566120) and anti-mouse CD103-BV421 (Biolegend Cat# 121422, RRID: AB_2562901). Zombie aqua staining was used to exclude dead cells. For intracellular staining, cells were fixed and permeabilized with BD Cytofix/Cytoperm (BD Biosciences) prior to staining with anti-mouse-IFN-γ-APC (Biolegend Cat# 505810, RRID: AB_315404), anti-mouse-TNF-α-PE (Biolegend Cat# 506306, RRID: AB_315427) and anti-mouse-IL-2-PE-Cy7 (Biolegend Cat# 503832, RRID: AB_2561750). Stained cells were acquired by FACSAriaIII Flow Cytometer (BD Biosciences) and analyzed with FlowJo software (v10.6) (BD Bioscience).
Viral RNA quantification
Half nasal turbinates and lung tissues were homogenized and subjected to viral load determination by quantitative SARS-CoV-2-specific RdRp/Hel reverse-transcription polymerase chain reaction assay.10 Total RNA was extracted using RNAeasy mini kit (Qiagen) and followed by reverse transcription (PrimeScript II 1st Strand cDNA Synthesis Kit). The real-time RT-PCR assay for SARS-CoV-2-RdRp/Hel RNA and nucleocapsid protein (NP) subgenomic RNA detection was performed using One-Step TB Green PrimeScript RT-PCR Kit II according to the manufacture's instruction. Mouse β-actin was used as normalization.
Plaque assay
Infectious virus titration was determined by plaque assay. Confluent Vero-E6 cells in a 12-well plate were incubated with 10-fold serially diluted tissue homogenates for 1 h. The virus supernatant was discarded, and cells were then overlaid with 1% agarose in DMEM and further incubated for 3 days at 37 °C followed by overnight fixation via 4% PFA. Agarose gels were removed, and plaques were visualized by 1 % crystal violet.
Histopathology and Immunofluorescence (IF) Staining
Tissues collected at necropsy were fixed in zinc formalin and then processed into paraffin-embedded tissue blocks. The tissue sections (4 µm) were stained with haematoxylin and eosin (H&E) for light microscopy examination. For identification and localization of SARS-CoV-2 NP in organ tissues, IF staining was performed on deparaffinized and rehydrated tissue sections using rabbit anti-SARS-CoV-2-NP antibody together with AF568-conjugated goat anti-rabbit IgG (Invitrogen, Cat# A-11011, RRID: AB_143157). Briefly, the tissue sections were first treated with antigen unmasking solution (Vector Laboratories) in a pressure cooker. After blocking with 0.1% Sudan black B for 15 min and 1% bovine serum albumin (BSA)/PBS at RT for 30 min, the primary antibody rabbit anti-SARS-CoV-2-N antibody (1:4000 dilution with 1% BSA/PBS) was incubated at 4°C overnight. This step was followed by AF568-conjugated goat anti-rabbit IgG for 30 min and then mounted with 4’,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific, Cat# D1306, RRID:AB_2629482). All tissue sections were examined, the images were captured with a Carl Zeiss LSM780 confocal microscope, and the mean fluorescence intensity (MFI) was further measured by ImageJ v1.53c.
Statistical analysis
Statistical analysis was performed with the GraphPad Prism 8 Software. Data represent mean values or mean values with SEM. Significant differences between the means of multiple groups were tested using a one-way analysis of variance (ANOVA) followed by Tukey's multiple comparisons test. Significant differences between the two groups were performed using the 2-tailed Student's t-test. P < 0.05 was considered statistically significant. Correlation analysis was performed by linear regression using GraphPad Prism 8.0. The animal sample size was determined by the "E" value, where E= Total number of animals-Total number of groups. Based on the ANOVA, the animal size, which lies between 10 and 20, should be considered adequate.30 No randomization, blinding or inclusion/exclusion criteria was performed during the experiment or data analysis.
Animal study approval
All experimental procedures were approved by the Committee on the Use of Live Animals in Teaching and Research (CULATR 5350-20) of the University of Hong Kong.
Role of funding source
The funders of this study had no role in study design, data collection, data analyses, interpretation, or writing of the report.
Results
Construction and characterization of PD1-based DNA and influenza-based vaccines
To address the hypothesis, we sought to study an intramuscular program death 1 (PD1)-based RBD DNA vaccine (PD1-RBD-DNA) and two intranasal live attenuated influenza virus (LAIV)-based vaccines (LAIV-HK68-RBD and LAIV-CA4-RBD). Recently, Y Liu et al. demonstrated that antibodies against the N-terminal domain (NTD) could enhance the binding capacity of the spike protein to ACE2 and infectivity of SARS-CoV-2.31 To focus the immune responses to the most critical region of the Spike protein, we chose RBD as the vaccine immunogen just like similar RBD-based COVID-19 vaccines explored by several groups.32 Since delayed cytotoxic T lymphocyte (CTL) responses are likely associated with COVID-19 severity,12 we constructed the PD1-RBD-DNA vaccine (Figure S1a) that might elicit enhanced antibody and CD8 T cell responses.23 The expression of soluble PD1-RBD protein (∼80 kDa) was readily detected in supernatants of transfected HEK293T cells by Western blot analysis using either anti-SARS-CoV-2 RBD antibody (green lane) or anti-human PD-1 (red lane) antibody (Figure S1b). The released PD1-RBD was able to bind PD-1-ligand 1 (Figure S1c), which might help antigen targeting dendritic cells for cross-presentation.23 Meantime, the NS1-deleted (DelNS1) LAIV was engineered to express the same RBD (Figure S1d), aiming to induce mucosal NAb and T cell immune responses.33 We previously characterized a panel of DelNS1 influenza viruses and evaluated their potential for being used as vaccine vectors.33 Two intranasal recombinant LAIV DelNS1-RBD vaccine strains, namely LAIV-CA4-RBD and LAIV-HK68-RBD, were generated using A/CA/04/2009 (H1N1) and A/Hong Kong/1/68 (H3N2) surface proteins (HA and NA) in the A/CA/04/2009 (H1N1) DelNS1 backbone by a reverse genetic procedure.24 The SARS-CoV-2 RBD and LAIV NP proteins were stably expressed in MDCK cells by Western blot after 5 times of viral passages in MDCK cells (Figure S1e).
Systemic and mucosal antibody responses of vaccine regimens
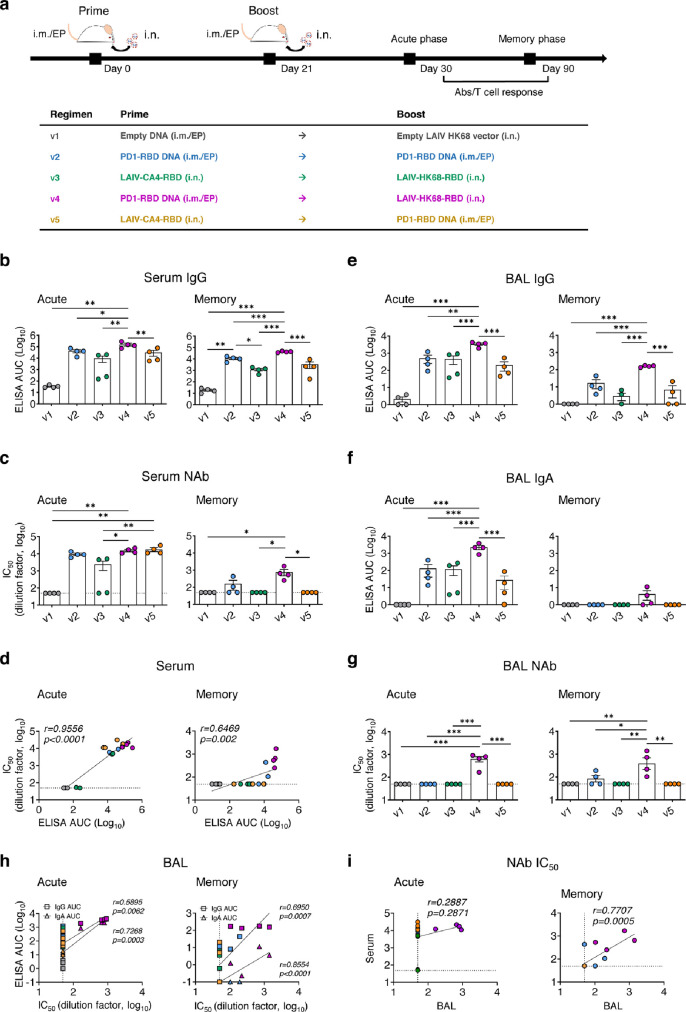
We then chose the immune competent SARS-CoV-2/BALB/c mouse model25 (n=8 per group, 4 mice for acute phase and 4 mice for memory phase), using available antibody reagents to understand the potential correlate of immune protection. Compared with the vector control group (v1) that received 50 µg intramuscular electroporation (i.m./EP) pVAX plasmid prime plus i.n. 106 PFU LAIV-68 vector boost (Figure 1a), we tested groups treated by the homologous 50 µg i.m./EP PD1-RBD-DNA twice (v2), a heterologous i.n. 106 PFU LAIV-CA4-RBD prime plus i.n. 106 PFU LAIV-HK68-RBD boost (v3), a heterologous 50 µg i.m./EP PD1-RBD-DNA prime plus i.n. 106 PFU LAIV-68-RBD boost (v4) and a heterologous i.n. 106 PFU LAIV-CA4-RBD prime plus 50 µg i.m./EP PD1-RBD-DNA boost (v5) at 3-week intervals, consistent with the intervals between dosages of COVID-19 vaccines under emergency use. Vaccine-induced antibody responses were determined on day 9 (acute phase) and day 69 (memory phase) post the second vaccination. Both peripheral blood and bronchoalveolar lavage (BAL) samples were collected from vaccinated mice for antibody detection by ELISA and pseudovirus neutralizing assays. We found that the PD1-RBD-DNA/LAIV-HK68-RBD regimen (v4) elicited and significantly sustained the highest amounts of RBD-specific IgG (acute: mean 5.2, range 4.86-5.45 logs AUC; memory: mean 4.62, range 4.54-4.69 logs AUC) and NAbs (acute: mean 4.19, 4.04-4.34 logs IC50; memory: mean 2.89, 2.4-3.23 logs IC50) in sera during both acute and memory phases as compared with other groups (Figure 1b-c). The RBD-specific IgG titer and NAb IC50 values were positively correlated (Figure 1d), similar to COVID-19 patients’ sera.8 Moreover, v4 animals also developed and sustained significantly higher amounts of RBD-specific mucosal IgG and IgA in BAL during both acute and memory phases as compared with other groups (Figure 1e-f). The amount of BAL NAbs in v4 mice were at mean 2.80 (range 2.22-2.87) and mean 2.59 (range 2.02-3.15) logs IC50 at the acute and memory phases, respectively (Figure 1g). In contrast, v2 and v5 elicited similar amounts of NAb to v4 at the acute phase, yet these responses did not sustain into the memory phase. Moreover, despite heterologous intranasal immunizations twice, the v3 regimen did not induce equally potent and sustained mucosal IgG and IgA as well as NAb responses, compared with the v4 group (Figure 1g). Both BAL IgG and IgA titers correlated positively with the BAL NAb values (Figure 1h) despite the higher amount and better acute/memory of BAL IgG than BAL IgA. Interestingly, serum NAb IC50 values were correlated positively with the BAL NAbs IC50 values at the memory phase but not at the acute phase (Figure 1i). These results indicated that the v4 regimen is likely unique for inducing potent and sustained systemic and mucosal memory IgG/IgA NAb responses.
Figure 1.
Vaccine-induced systemic and mucosal antibody responses. (a) Vaccine immunization schedule and grouping for BALB/c mice (n=8/group). Blood and bronchoalveolar lavage (BAL) were collected and subjected to antibody response analysis on 9 (acute phase, n=4/group) or 69 (memory phase, n=4/group) days post the 2nd immunization, respectively. In sera, (b) RBD-specific IgG titer, (c) NAb IC50 values and (d) positive correlations between RBD-specific IgG titer and NAb IC50 values were analyzed for both acute and memory phases. In BAL, (e) RBD-specific IgG titer, (f) RBD-specific IgA titer, (g) NAb IC50 values and (h) positive correlations between RBD-specific IgG (square) or IgA titers (triangle) and BAL NAb IC50 values were analyzed for both acute and memory phases. (i) Correlate analysis between Peripheral NAb IC50 values and BAL NAb IC50 values at both acute and memory phase. RBD-specific IgG or IgA titers were determined by ELISA at serial dilutions. The area under the curve (AUC) represented the total peak area calculated from ELISA OD values. Neutralization IC50 values were determined against SARS-CoV-2-Spike-pseudovirus infection of 293T-huACE2 cells. Correlation analysis was performed by linear regression using GraphPad Prism 8.0. Each symbol represents an individual mouse with color-coding for corresponding groups. Error bars indicate the standard error of the mean. Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. *p<0.05; **p<0.01.
Acute and memory T cell responses of vaccine regimens
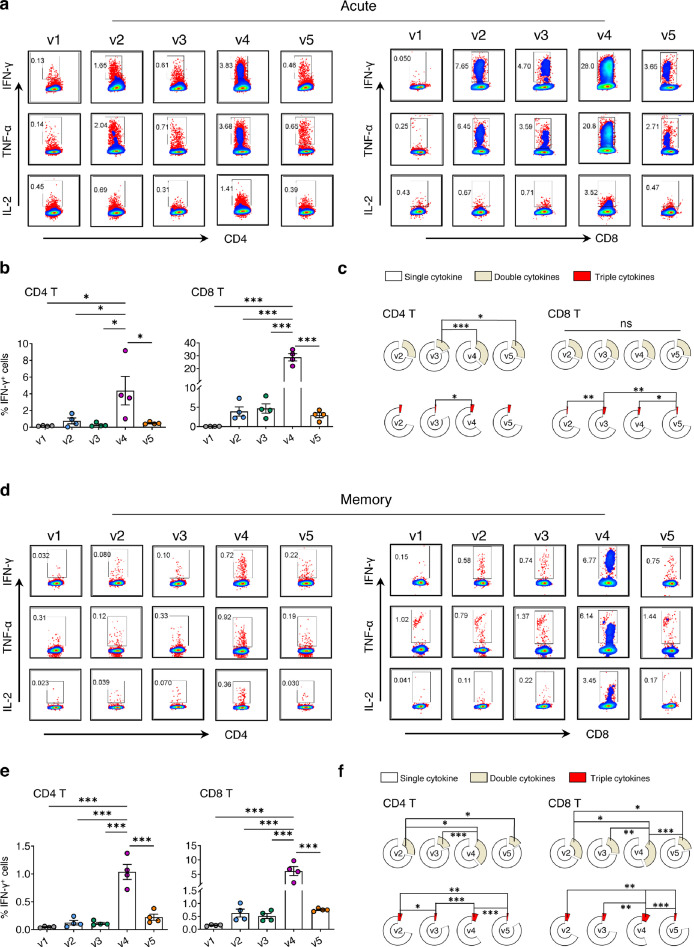
Since SARS-CoV-2-specific T cell responses are essential for control and resolution of viral infection,12,34 we sacrificed five groups of animals to measure vaccine-induced T cell immune responses on day 9 (acute phase) and day 69 (memory phase) post the second vaccination. Lymphocytes were isolated from both lungs (effector site) and spleens (secondary lymph organ) of vaccinated mice for comparison. We found that the v4 regimen elicited and sustained significantly higher frequencies of RBD-specific IFN-γ+ CD8 T cells in lungs and spleens during both acute (Figure 2a-b, Figure S2a-b) and memory (Figure 2d-e, Figure S2d-e) phases as compared with other groups. Similar trends were found with IFN-γ+ CD4 T cells elicited in the v4 regimen but at lower frequencies. At the acute phase, the v4 regimen elicited the highest mean frequency of RBD-specific IFN-γ+ CD8 T cells (mean 28.83%, range 22-34.8%) in lungs (Figure 2b), which was even higher than that in spleens (mean 5.54%, range 3.63-7.06%) (Figure S2b). These cells included the highest frequencies of polyfunctional CTLs with a capacity of releasing two (mean 21.09%, range 13.87-25.31%) or three (mean 2.27%, range 1.02-3%) cytokines (Figure 2c), which was also higher than those in splenic CD8 T cells releasing two (mean 6.68%, range 4.68-9.17%) or three cytokines (mean 0.92%, range 0.7-1.36%) (Figure S2c). At the memory phase, the v4 regimen sustained the highest mean frequency of RBD-specific IFN-γ+ CD8 T cells in lungs (mean 6.11%, range 2.05-9.7%) (Figure 2e) and spleens (mean 2.35%, range 0.7-4.78%) (Figure S2e) as compared with other groups. These cells included the highest frequencies of polyfunctional CTLs with a capacity of releasing two (mean 8.66%, range 2.61-12.41%) or three (mean 2.2, range 0.66-3.09%) cytokines (Figure 2f), which was higher than those in splenic CD8 T cells releasing two (mean 3.59%, range 1.28-6.83%) or three cytokines (mean 0.69%, range 0.31-1.17%) (Figure S2f). These results demonstrated that besides Nabs, the v4 regimen also induced potent and polyfunctional memory CD8 T cell responses, especially in the lungs. Since overall immune responses induced by the heterologous v3 regimen were much weaker than those by the v4 regimen, we also exanimated T cell responses against influenza immunodominant nucleoprotein (NP).35 At acute phase (Figure S3a), the v3 regimen induced the highest frequencies of CD8 T cell response against influenza NP in lungs (mean 19.55%, range 16.2-21.7%) as compared with v1 (mean 8.3%, range 7.77-8.9%) and v5 (mean 3.99%, range 2.55-5.91%). Similar results were observed at the memory phase (Figure S3b). In contrast, the v4 regimen induced significantly lower influenza NP-specific T cell response at both acute (mean 2.26%, range 1.68-2.58%) and memory (mean 0.77%, range 0.49-1.06%) phases. The heterologous prime using PD1-RBD-DNA instead of LAIV-CA4-RBD, therefore, offered an advantage in promoting the RBD immunodominance likely by avoiding anti-vector immune responses.
Figure 2.
Vaccine-induced acute and memory T cell responses in lungs. The vaccine immunization schedule for BALB/c mice was the same as described in Figure 1a. Lung cells were collected and subjected to SARS-CoV-2 RBD-specific T cell responses by ex vivo RBD peptide pool stimulation followed by ICS at both acute (a-c) and memory (d-f) phases. (a, d) Representative dot plots present the gating of IFN-γ+, TNF-α+ or IL-2+ CD4 T (left) and CD8 T (right) against SARS-CoV-2 RBD. (b, e) Quantified results depict the percentage of IFN-γ+ CD4 T (left) and IFN-γ+ CD8 T (right). Each symbol represents an individual mouse. Error bars indicate the standard error of the mean. (c, f) The pie charts indicate the proportion of single or double or triple cytokines produced by CD4 T (left) and CD8 T (right). Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. *p<0.05; **p<0.01; ***p<0.001.
Protective efficacy against intranasal SARS-CoV-2 infection
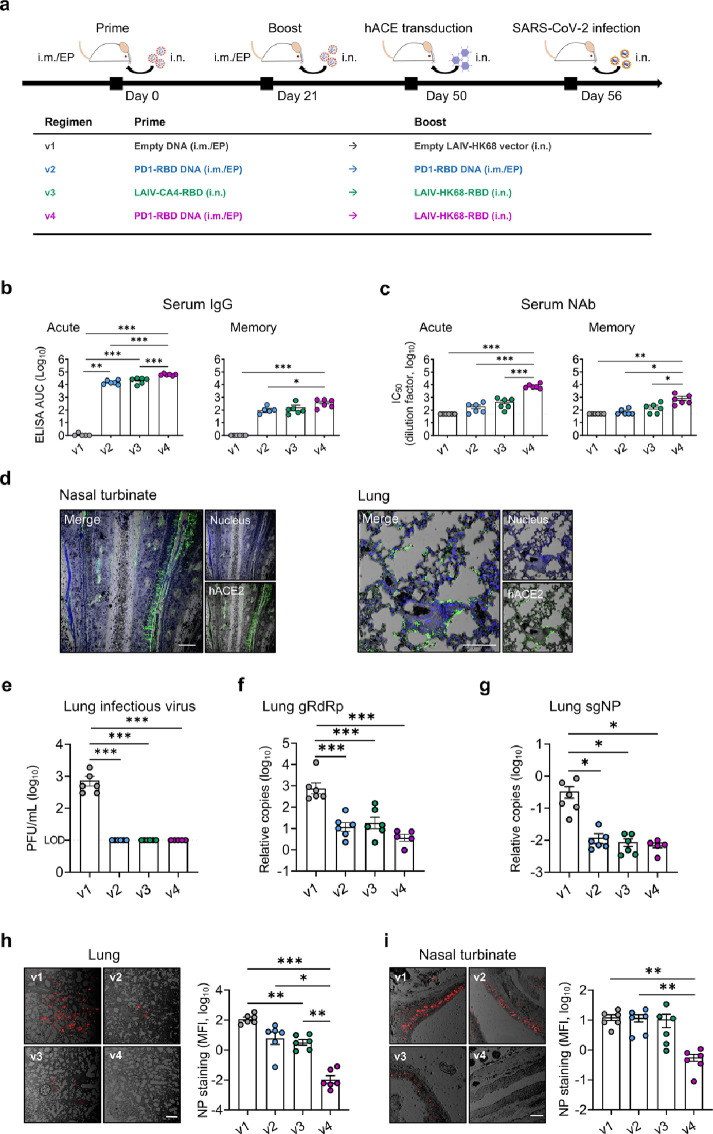
To investigate the efficacy of various vaccine regimens against the live intranasal SARS-CoV-2 challenge, we subsequently immunized additional groups of BALB/c mice (n=6 per group) using the same doses and time interval as described above (Figure 3a). We did not include v5 due to low mucosal immunogenicity and limited space in our animal P3 facility. Sera were collected on day 9 and day 28 after the 2nd immunization to monitor anti-RBD IgG (Figure 3b) and neutralization (Figure 3c). Consistently, the highest IgG (acute: mean 4.79, range 4.7-4.89 logs AUC; memory: mean 2.6, range 2.03-2.81 logs AUC) and neutralizing (acute: mean 3.9, range 3.69-4.14 logs IC50; memory: mean 2.94, range 2.34-3.33 logs IC50) titers were induced in mice by the v4 regimen at both acute and memory phases. Immunized mice were then transduced with Ad5-hACE2 at the memory phase, 29 days post the boost vaccination for expressing human ACE2 in nasal turbinate and lung (Figure 3d), followed by the intranasal SARS-CoV-2 challenge 6 days later as previously described.25 On day 4 after the viral challenge, mice were sacrificed for analysis. Lung specimen was harvested to quantify infectious viruses by plaque assay, viral load by real-time PCR (RT-PCR) and infected cells by immunofluorescence staining (IF). We found that all vaccinated animals had decreased infectious plaque-forming units (PFU) to the limit of detection (10 PFU/mL) in the lungs (Figure 3e). The v4 regimen, however, resulted in the most significant genomic RdRp (gRdRp) drop in the lungs by an average of 2.31 logs compared with 1.81 logs in v2 mice and 1.62 logs in v3 mice (Figure 3f). A similar observation was found in the measurement of nucleocapsid protein (NP) subgenomic RNA (sgNP) (Figure 3g). These findings demonstrated that immune responses induced by v2, v3 and v4 regimens had achieved significant protection in lungs. To determine viral infection in both upper and lower respiratory systems, we further performed immunofluorescence staining of SARS-CoV-2 NP antigen in both lung (Figure 3h) and NT (Figure 3i) tissues. Since murine NT was too small to be sliced for viral load tests, we only used it for the NP staining to maintain the necessary tissue structure. While significantly reduced NP+ cells were observed in the lungs of v2 and v3 mice, infected cells were barely found in the lungs of v4 mice. Furthermore, no significantly reduced NP+ cells were found in NT of v2 and v3 mice as compared with v1 mice, but only a few NP+ cells were seen in v4 mice. Our results demonstrated that while protection was consistently found in lungs of vaccinated animals, significant prevention of robust SARS-CoV-2 infection in NT was only achieved by the v4 regimen.
Figure 3.
Protective efficacy of vaccine regimens in huACE2-transduced mice. (a) Experimental schedule and grouping of BALB/c mice (6 mice/group). On day 29 post the 2nd immunization, mice were transduced to express human ACE2 in vivo by inoculating 4 × 108 Focus-forming units (FFU) Ad5-hACE2 intranasally. Six days later, mice were challenged intranasally with 104 PFU SARS-CoV-2 and sacrificed on day 4 post-infection. (b, c) Serum samples were collected for detection of anti-RBD IgG (b) and neutralizing antibody (c) against pseudovirus, respectively, on day 9 (acute, left) and day 28 (memory, right) post the 2nd immunization. The Area under the curve (AUC) represents the total peak area calculated from ELISA OD values. (d) Human ACE2 expression (green) after the Ad5-hACE2 transduction was evaluated in NT and lung by immunofluorescence (IF) staining using the hACE2-specific antibody on day 6 post-transduction. With nuclei counter staining (blue). Scale bars: 100 µm. (e) A viral plaque assay was used to quantify infectious viruses in lung homogenates. Log10-transformed plaque-forming units (PFU) per mL of tissue extractions were shown for each group. LOD: limit of detection. (f, g) Sensitive RT PCR was used to quantify SARS-CoV-2 RdRp RNA (f) and NP subgenomic RNA (g) copy numbers (normalized by β-actin) in lung homogenates. Confocal images showed SARS-CoV-2 NP positive (red) cells in lungs (h) and NT (i) in the bright field. Scale bars: 200 µm. Mean fluorescent intensities (MFI) of NP+ cells in lung and NT were measured using ImageJ software and plotted with GraphPad prism. Each symbol represents an individual mouse with consistent color-coding. Error bars indicate the standard error of the mean. Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. *p<0.05; **p<0.01; ***p<0.001.
Infection-recalled NAb for correlate of protection
Although safety protocols prevent the removal of tissue specimens for measuring T cell immunity outside the animal P3 laboratory after the SARS-CoV-2 challenge, we determined whether viral infection recalled vaccine-induced NAbs for viral neutralization and clearance.36 By testing RBD-specific IgG and NAb on day 28 (before challenge) and day 39 (also 4 dpi) after the 2nd vaccination, we found that SARS-CoV-2 infection indeed recalled significantly anti-RBD IgG responses in all v2, v3 and v4 animals (Figure S4a). Notably, 62.2-fold and 65.7-fold higher amounts of recalled NAb were found by the v4 regimen (v4, mean 4.64, range 3.22-5.03 logs IC50) than the v2 and v3, respectively (Figure S4b). Furthermore, there were significant negative correlations between viral loads and IgG or NAb IC50 values (Figure S4c-d), as well as between NP+ cells and IgG or NAb IC50 values in lungs (Figure S4e-f) and in NT (Figure S4g-h). Moreover, since the TH1 immune response is likely associated with protective immune responses against COVID-19,37 we also evaluated the ratios of IgG1 and IgG2a in all vaccinated mice. The v4 regimen likely induced a TH1 bias with a higher IgG2a/IgG1 ratio (mean 59.57, range 1.44-279.2) than those induced by v2 (mean 6.72, range 2.63-15.14) and v3 (mean 7.65, range 4.25-12.72) (Figure S4i). Our results demonstrated that the v4 vaccine regimen-induced high amounts of NAbs were correlated with protective efficacy in both NT and lung.
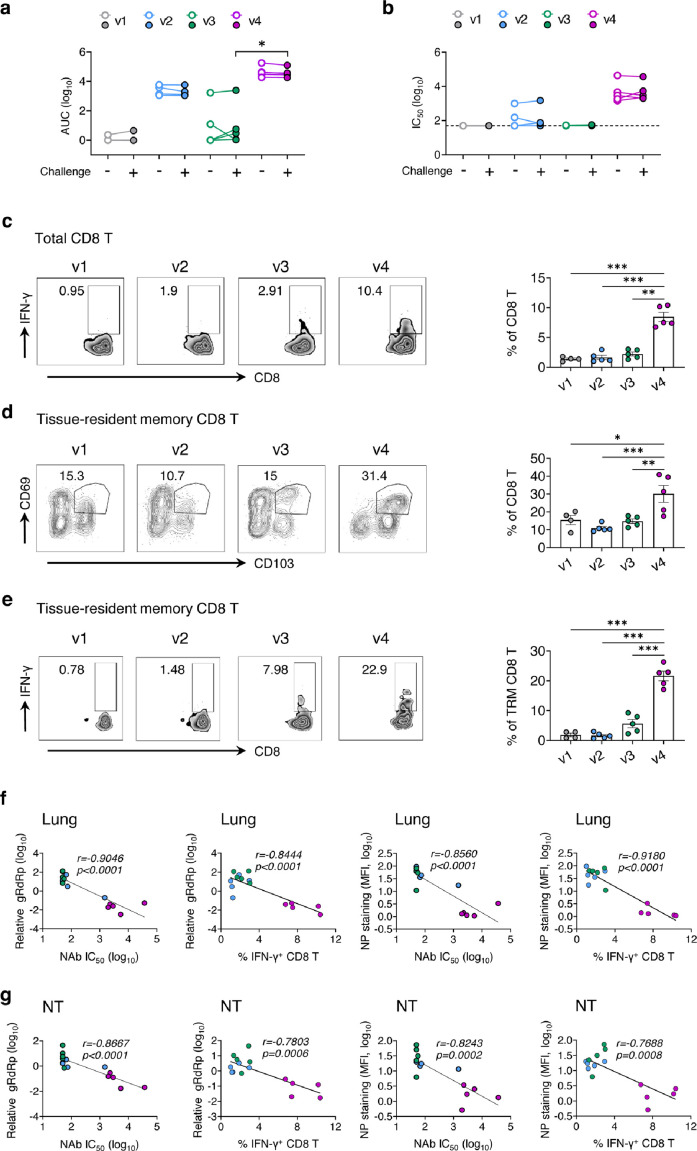
SARS-CoV-2 prevention in both upper and lower respiratory tracts of K18-hACE2 mice
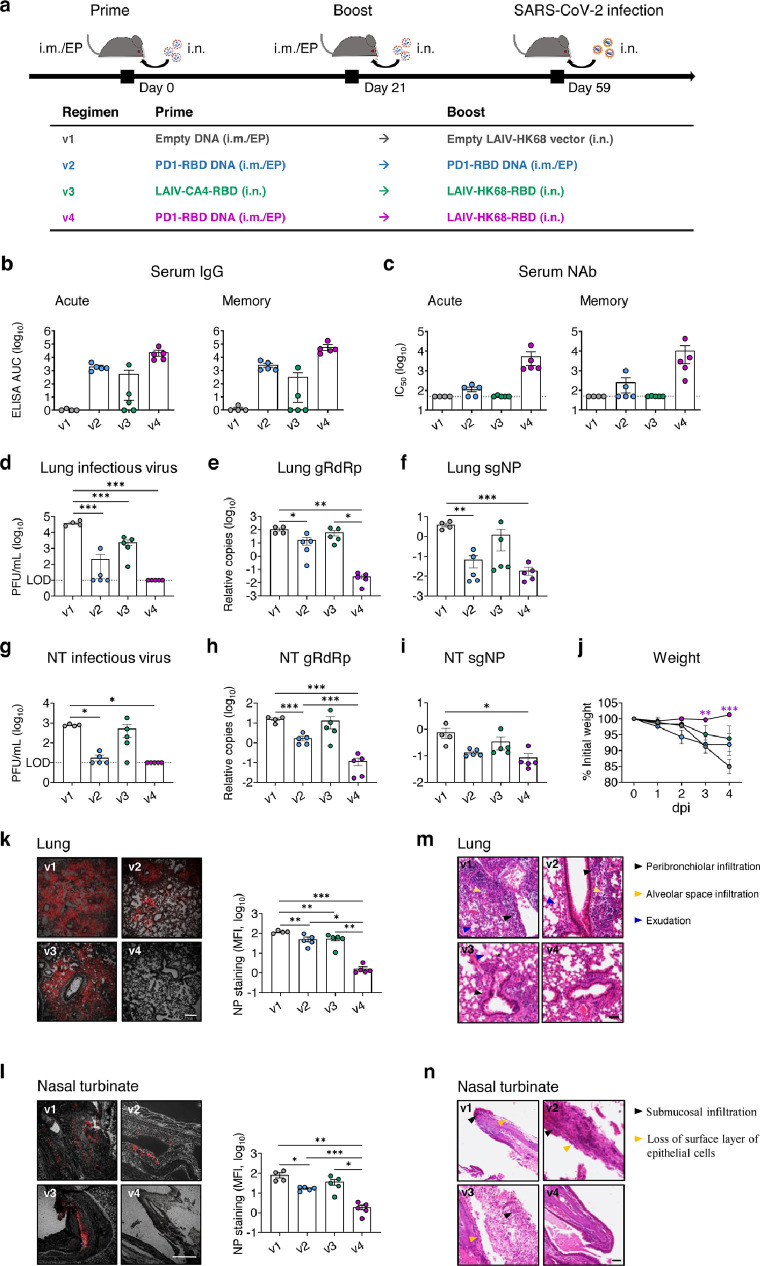
To further determine the vaccine efficacy, we tested the PD1-RBD-DNA/LAIV-HK68-RBD regimen in K18-hACE2 transgenic mice, one of the commonly used animal models for studying COVID-19.38, 39, 40 8-week-old K18-hACE2 mice were vaccinated with various regimens using the doses and time interval described above (4 mice/group for v1 and 5 mice/group for v2-v4) (Figure 4a). The titer of RBD-specific IgG (Figure 4b) and neutralization antibody (Figure 4c) was measured in serum on day 9 (acute phase) and day 33 (memory phase) after the 2nd immunization. Consistently, the PD1-RBD-DNA/LAIV-HK68-RBD regimen (v4) induced and sustained the highest amount of RBD-specific serum IgG (acute: mean 4.38, range 3.81-4.84 logs AUC; memory: mean 4.79, range 4.27-5.25 logs AUC) and Nabs (acute: mean 3.74, range 3.10-4.30 logs IC50; memory: mean 4.02, range 2.48-4.64 logs IC50) during both acute and memory phases as compared with other groups. On day 38 after the 2nd immunization, mice were intranasally challenged with 104 PFU of SARS-CoV-2 (HKU Clone 13). Considering that K18-hACE2 mice can produce anti-SARS-CoV-2 humoral and cellular responses at 6 dpi,39 we sacrificed mice at 4 dpi to measure the viral loads and pathogenesis. In the lungs, no measurable infectious viruses (detection limit: 10 PFU/mL) were found in all v4 mice and 40% v2 mice, whereas infectious viruses were detected in all v1 and v3 mice (Figure 4d). The more sensitive RT-PCR further demonstrated that the v4 regimen resulted in a significant gRdRp drop by an average of 3.57 logs compared with 0.81 logs in v2 mice and 0.25 logs in v3 mice (Figure 4e) at 4 dpi. A similar observation was found with the measurement of NP subgenomic RNA (Figure 4f). Importantly, no measurable infectious viruses were detected in nasal turbinate of v4 mice (Figure 4g), and the v4 regimen significantly suppressed viral genomic/subgenomic RNA (gRdRp and sgNP) (Figure 4h-i). Further IF staining of NP antigen confirmed that significantly reduced NP+ cells were observed in both lungs (Figure 4k) and nasal turbinates (Figure 4l) of v4 mice, compared with other regimens. Daily monitoring of body weight after challenge showed that SARS-CoV-2 infection caused an average of 15%, 8% and 6% weight loss in v1, v2 and v3 mice at 4 dpi, respectively. In contrast, no weight loss was found in v4 mice during the experimental period (Figure 4j). To further characterize the injury of lung and nasal turbinate, we performed pathological analysis on specimens by hematoxylin and eosin (H&E)-staining. Acute lung injury with peribronchiolar infiltration, alveolar space infiltration and exudation were visualized in v1 mice, while v2 and v3 mice showed less alveolar space infiltration. In contrast, only mild peribronchiolar infiltration was observed in v4 mice (Figure 4m). In nasal turbinate, epithelium damage with extensive submucosal immune cell infiltration was observed in v1, v2 and v3 mice, but not in v4 mice (Figure 4n). Consistent results were found in the flow cytometry analysis of immune cell compositions in the lungs. A significant decrease in the frequency of alveolar macrophages as well as reduced CD103+ DCs and CD11b+ DCs (Figure S5a-b) was found in the v4 mice compared with other groups. These results in K18-hACE2 mice, therefore, further demonstrated that the PD1-RBD-DNA/LAIV-HK68-RBD regimen prevents robust SARS-CoV-2 infection not only in the lung but also in NT with minimal infection-associated inflammation and injury.
Figure 4.
PD1-RBD-DNA/LAIV-HK68-RBD regime prevents SARS-CoV-2 infection in the upper and lower respiratory tracts of hACE2 transgenic mice. (a) Experimental schedule and grouping of K18-hACE-2 mice (4 mice in v1 group and 5 mice in other groups). (b, c) Serum samples were collected for detection of anti-RBD IgG (b) and neutralizing antibody (c) against pseudovirus, respectively, on day 9 (acute, left) and day 33 (memory, right) post the 2nd immunization. The area under the curve (AUC) represents the total peak area calculated from ELISA OD values. A viral plaque assay was used to quantify infectious viruses in lung (d) and NT (g) homogenates. Log10-transformed plaque-forming units (PFU) per mL of tissue extractions were shown for each group. LOD: limit of detection. Sensitive RT PCR was used to quantify SARS-CoV-2 RdRp RNA (e, h) and NP subgenomic RNA (f, i) copy numbers (normalized by β-actin) in lung and NT homogenates. (j) Daily body weight was measured after infection. Differences between groups that were given the different vaccine regimes versus PBS were determined using a 2-tailed Student's t-test. Confocal images showed SARS-CoV-2 NP positive (red) cells in lungs (k) and NT (l) in the bright field. Scale bars: 200 µm. Mean fluorescent intensities (MFI) of NP+ cells in lung and NT were measured using ImageJ software and plotted with GraphPad prism. Representative images of animal lung (m) and NT (n) tissues by H&E. Scale bars: 50 µm. Each symbol represents an individual mouse with consistent color-coding. Error bars indicate the standard error of the mean. Statistics were generated using one-way ANOVA followed by Tukey's multiple comparisons test. *p<0.05; **p<0.01; ***p<0.001.
Both NAb and antigen-specific CD8 T cells associated with virus control in K18-hACE2 mice
Compared to the serum collected on day 5 before the challenge, the titer of RBD-specific IgG (Figure 5a) and NAb (Figure 5b) did not increase in sera collected at 4 dpi. We then assessed the T cell responses in lungs by RBD peptide re-stimulation, and a significant increase in IFN-γ producing CD8 T cells (mean 8.45%, range 6.76-10.4%) was observed in the lungs of the mice that received PD1-RBD-DNA prime/LAIV-HK68-RBD boost regimen (Figure 5c). Tissue-resident memory T cells (TRM) is a well-known antiviral T cell type that persists at mucosal sites.41 We found that a higher frequency of TRM cells (CD69+CD103+) (mean 30.1, range 17.6-40.9%) was identified in lung CD8 T cells in v4 mice (Figure 5d). Moreover, a higher frequency of IFN-γ+ TRMs (mean 21.64%, range 17.1-26.3%) was found in v4 mice after RBD peptide re-stimulation (Figure 5e). Correlation analysis showed negative correlations between the NAb titer and viral load (RdRp RNA/NP+ cells) and between the frequency of IFN-γ+ CD8 T cells and viral load in both lungs (Figure 5f) and nasal turbinate (Figure 5g), respectively, indicating their involvements in viral control.
Figure 5.
Robust NAb and antigen-specific T cells were responsible for virus control in K18-hACE2 mice. Blood samples were collected on 33 and 42 days (also 4 dpi) after the 2nd vaccination for analysis. (a, b) The mean ± SEM changes of anti-RBD IgG AUC titer (a) and neutralizing IC50 values (b) were determined by anti-RBD IgG ELISA and pseudovirus assay, respectively. (c) Lung CD8 T cells were assayed for IFN-γ expression by flow cytometry after re-stimulated with the RBD peptide pool. Tissue-resident memory CD8 T cells (TRM) in lungs were phenotyped for expression CD69 and CD103 (d) and the IFN-γ producing TRM were measured by re-stimulated with the RBD peptide pool (e). (f) Viral load (relative gRdRp) in lungs correlated with NAb IC50 titer or percentage of IFN-γ producing CD8 T cells. NP+ cells (MFI) in lungs correlated with NAb IC50 titer or percentage of IFN-γ producing CD8 T cells. (g) Viral load (relative gRdRp) in NT correlated with NAb IC50 titer or percentage of IFN-γ producing CD8 T cells. NP+ cells (MFI) in NT correlated with NAb IC50 titer or percentage of IFN-γ producing CD8 T cells. Correlation analysis was performed by linear regression using GraphPad Prism 8.0. Each color represents a vaccination regimen. Each symbol represents an individual mouse. Statistics were generated using the 2-tailed Student's t test. *p<0.05; **p<0.01; ***p<0.001.
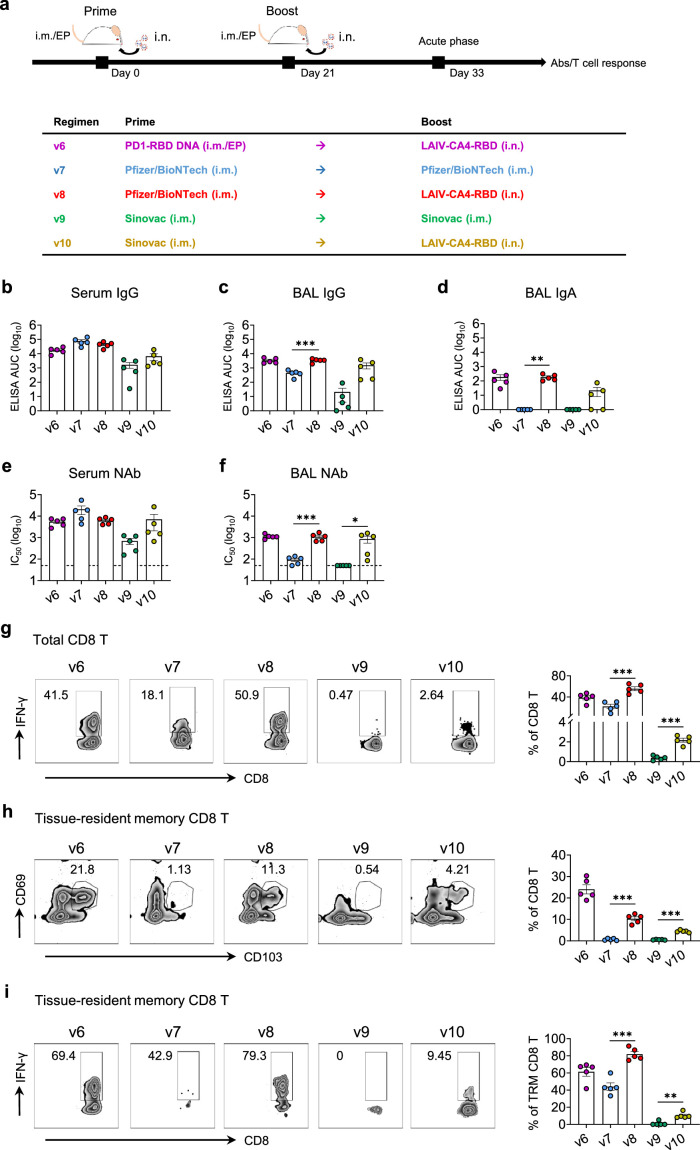
LAIV-CA4-RBD boosted the immunogenicity of vaccines in emergency use
While we were studying the PD1-RBD-DNA/LAIV-HK68-RBD regimen, both the BioNTech mRNA vaccine and the Sinovac inactivated vaccine have been approved for emergency use in Hong Kong. Our LAIV-CA4-RBD vaccine is currently undergoing the phase I clinical trial in Hong Kong (ClinicalTrials.gov Identifier: NCT04809389). Since intramuscular administration of mRNA or inactivated vaccine induces mainly systemic IgG and some T cell immune responses without secretory IgA or tissue-resident memory T cells,42,43 we sought to determine if LAIV-CA4-RBD could boost their mucosal immunogenicity to provide the implication for clinical use. We, therefore, immunized additional groups of BALB/c mice (5 mice per group) as v7, v9, v8, v10 with a homologous i.m. BioNTech (1/5 of clinical dose), a homologous i.m. Sinovac (1/5 of clinical dose), a heterologous i.m. BioNTech plus 106 PFU LAIV-CA4-RBD, a heterologous i.m. Sinovac plus 106 PFU LAIV-CA4-RBD at a 3-week interval, respectively. The selection of the 1/5 of the clinical dose was based on animal studies described previously.44 The control mice in the v6 group were immunized by 50 µg i.m./EP PD1-RBD DNA plus LAIV-CA4-RBD as before (Figure 6a). On day 12 after the 2nd vaccination, serum and BAL were collected and subjected to detection of RBD-specific IgG and IgA as well as neutralization activity. Interestingly, v8 and v10 mice did not show increased titers of IgG and Nab in sera (Figure 6b, e) but had significantly increased titers of IgG (Figure 6c), IgA (Figure 6d) and Nab (Figure 6f) in BAL compared with v7 and v9. We then assessed T cell responses in the lungs by RBD peptide re-stimulation. A significant increase of IFN-γ producing CD8 T cells (mean 55.58%, range 44.9-65.1% in v8 and mean 2.16%, range 1.5-2.64% in v10) was found in mice that received the heterologous boost of LAIV-HK68-RBD than mice received homologous BioNTech or Sinovac (mean 22.14%, range 8.89-30.8% in v7 and mean 0.37, range 0.1-0.66% in v9) (Figure 6g). Furthermore, more total TRMs and RBD-specific TRMs were induced in v8 and v10 compared with v7 and v9 (Figure 6h-i). These results demonstrated that the intranasal LAIV-CA4-RBD boost induced stronger humoral and cellular immune responses in mucosal sites than systemic vaccinations alone.
Figure 6.
Combinations of systemic and mucosal immunization induce robust humoral and cellular immune responses in the effector site. (a) Experimental schedule and grouping of BALB/c mice (5 mice per group). Blood (b, e) and bronchoalveolar lavage (BAL) (c, d, f) were collected and subjected to RBD-specific IgG (b, c) or IgA (d) detection and NAb IC50 activity (e, f). The area under the curve (AUC) represented the total peak area calculated from ELISA OD values. Neutralization IC50 values were determined against SARS-CoV-2-Spike-pseudovirus infection of 293T-huACE2 cells. (f) Lung CD8 T cells were assayed for IFN-γ expression by flow cytometry after re-stimulated with the RBD peptide pool. Tissue-resident memory CD8 T cells (TRM) in lungs were phenotyped for expression CD69 and CD103 (g) and the IFN-γ producing TRM were measured by re-stimulated with the RBD peptide pool (h). Each colour represents a vaccination regimen. Each symbol represents an individual mouse. Statistics were generated using the 2-tailed Student's t test. *p<0.05; **p<0.01; ***p<0.001.
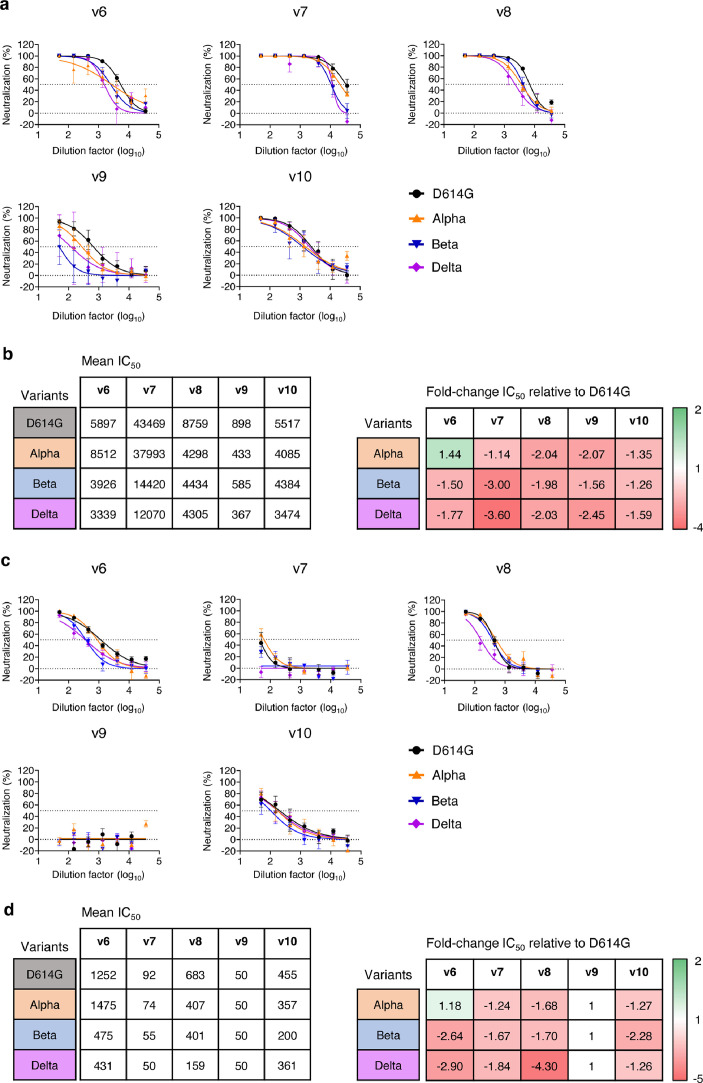
Vaccine-induced NAbs cross-neutralize global SARS-CoV-2 variants of concern
Recently emerged SARS-CoV-2 variants Alpha (B.1.1.7) from the UK, Beta (B.1.351) from South Africa and Delta (B.1.167) from India have become challenges for passive immunotherapy and vaccine-induced protection.19,45 For example, the Novavax vaccine is effective against the wildtype SARS-CoV-2 (95.6%) but provides reduced protection against the variants Alpha (85.6%) and Beta (60%).46 We, therefore, compared the neutralizing activity of immune sera from PD1-RBD-DNA/BioNTech/Sinovac prime and LAIV-CA4-RBD boosted mice (Figure 6) against pseudotyped viruses that contain the D614G, the Alpha, the Beta and the Delta variants (Figure 7a-b) as described previously.19 Compared to the D614G viral strain, v6 sera showed slightly enhanced neutralizing activity against the Alpha variant, while the sera of other groups exhibited reduced neutralization against the Alpha, Beta and Delta variants (Figure 7b). In line with a recent study,19 the Beta and Delta variants were more resistant to neutralization by sera from all vaccine regimens with an average fold reduction of 1.5-1.77 (v6), 3.00-3.60 (v7), 1.98-2.03 (v8), 1.56-2.45 (v9) and 1.26-1.59 (v10) as compared to the D614G strain, respectively (Figure 7b). Although animals in the LAIV-CA4-RBD boost regimens (v6, v8 and v10) showed the mean 1.69-fold reduction against Beta or Delta variants, their mean NAb IC50 titer of 1: 3633 (v6, range 1:1586-1:8250), 1:4370 (v8, range 1:2305-1:9068) and 1:3929 (v10, range 1:189-14393) remained high, respectively, which was superior or comparable to those of homologous-vaccinated mice as well as to the results of clinical vaccines against the wild type virus in murine models.6,47,48 Notably, the BAL from LAIV-CA4-RBD boost groups (v6, v8 and v10) were still able to neutralize the Beta and Delta variants (v6: mean 453, range 117-762; v8: mean 280, range 93-465; v10: mean 281, range 50-760) although they showed the average fold reduction of 2.64-2.90 (v6), 1.7-4.3 (v8) and 1.26-2.28 (v10) as compared to the D614G strain, respectively (Figure 7c-d). These results demonstrated that the systemic prime/LAIV boost regimen-induced high amounts of systemic and mucosal NAbs may confer cross-protection against the variants of concern before the tailor-made vaccines become available.
Figure 7.
Cross-neutralization of vaccine-induced systemic and mucosal NAb against SARS-CoV-2 variants. Neutralization activities of immune sera (a-b) and BAL (c-d) elicited by various vaccine regimens (same as described in Figure 6) were determined against 3 color-coded pseudoviruses in 293T-ACE2 cells. Data showed mean±SEM of 5 mice in each vaccine group. (b and d) Comparison of mean IC50 values against different SARS-CoV-2 variants (left) and fold-change of mean IC50 values in relation to the SARS-CoV-2 D614G strain (right).
Discussion
To control the ongoing COVID-19 pandemic, vaccine-induced protective immune responses should prevent SARS-CoV-2 nasal infection effectively to eliminate viral transmission between humans.17,42 Failure of protection against SARS-CoV-2 in NT may allow asymptomatic viral spread. At the time of writing this manuscript, there have been over 10 thousand cases of vaccine breakthrough infections in the United States.46,49 To date, little is known about the correlation of vaccine-induced mucosal protection for the prevention of intranasal SARS-CoV-2 infection in animal models.15 In this study, we demonstrated that the intranasal LAIV-based vaccination is critical for boosting a systemic PD1-RBD-DNA vaccine prime for effective prevention of SARS-CoV-2 infection in both NT and lungs by inducing high amounts of mucosal IgA/IgG and polyfunctional memory CD8 T cells. Consistent with previous findings that a heterologous prime and boost vaccine regimen may induce stronger immune responses,50,51 our results on effective prevention of intranasal SARS-CoV-2 infection in NT supports further clinical research of PD1-based and LAIV-based COVID-19 vaccines (ClinicalTrials.gov Identifiers: NCT05102643 & NCT04809389). Our findings may have implications for further study of using LAIV-CA4-RBD as a booster for the current vaccines under emergency use, especially the extensively used nucleic acid-based COVID-19 vaccines (e.g., BioNTech mRNA).
The induction of mucosal immune responses is essential for preventing SARS-CoV-2 transmission effectively.40,42 There are, however, some challenges underlying the prevention of SARS-CoV-2 in NT. First, SARS-CoV-2 exhibited a rapid burst of viral replication in NT,1 which has not been previously found with SARS-CoV.9,52 Second, after viral entry, ciliated nasal epithelial cells with the highest expression of ACE2 and TMPRSS2 may facilitate more efficient cell-cell transmission,8 which probably makes it difficult for systemic NAb to block. Third, NAbs by systemic vaccination or passive immunization are less distributed on the surfaces of NT for the prevention of SARS-CoV-2.17 Due to these challenges, it is critical to investigate vaccine-induced mucosal NAbs and CD8 T cells for SARS-CoV-2 prevention in NT. Thus far, most published vaccines have indicated excellent protective efficacy in the lungs of vaccinated animals without detailed analysis in NT.53, 54, 55 Several previous studies had shown no significant viral load drops in nasal swabs or nasal turbinates of vaccinated animals.6,7,44,56,57 One study found that optimal protection was achieved by only the DNA vaccine encoding the full S protein in both the upper and lower respiratory tracts against SARS-CoV-2 in rhesus macaques when vaccinated animals developed pseudovirus neutralizing IC50 titer less than 1:1000.57 Serum NAb titers, as measured by both pseudovirus and live virus neutralization, served as a significant correlate of protection. Similar protection in both the upper and lower respiratory tracts was achieved in another study by the AD26 vaccine encoding the full S protein against SARS-CoV-2 in rhesus macaques in another study.57 Both studies, however, used the macaque model that requires inoculation of 105 TCID50 SARS-CoV-2 into each nare and intratracheal for effective infection, which is not natural and is in great contrast to the robust nature of NT infection in humans. Using a DNA vaccine encoding the full S immunogen, we recently reported that there was significant protection in the lung but not in NT against SARS-CoV-2 in Syrian hamsters even though the vaccinated animals developed pseudovirus neutralizing IC50 titer larger than 1:1000.17 In this study, we consistently found that significant prevention of NT was not achieved by two systemic PD1-RBD-DNA vaccinations in both hACE2-transduced mice and K18-hACE-2 mice.
For direct mucosal vaccination, a single-dose intranasal ChAd vaccine exhibited a significant reduction of viral loads in NT, yet the extent of infected cells in NT was not determined, and its potential for extensive human use remains unclear.40 Here, we showed the highest frequency of RBD-specific tissue-resident memory CD8 T cells in the respiratory system was found by the intranasal boost of LAIV-based vaccine after intramuscularly prime with various systemic vaccines (e.g., PD1-RBD-DNA, Pfizer/BioNTech and Sinovac). Moreover, besides RBD-specific tissue-resident memory CD8 T cells, we also observed the positive correlation between NAb IC50 values and BAL NAb IC50 values, and the negative correlation between NAb IC50 values and NP+ cells, especially in NT. The LAIV-RBD vaccine as a boost vaccination combined with current vaccines in order to achieve mucosal immunity against SARS-CoV-2 warrants further study. Furthermore, the role of the intranasal LAIV-RBD boost route in optimizing the extent and localization of humoral and cellular immune responses in the mucosal site combined with systemic prime immunization is not clear yet and will be determined in the future study. For example, it is unknown whether the intranasal LAIV-RBD boost can potently promote antigen-specific T follicular helper cell responses as well as elicit potent germinal center reaction for neutralizing antibody production in the mucosal site. Secondly, it is crucial to determine how the intranasal LAIV boost develops the antigen-specific tissue resident memory T cells that not only destroy the infected cells but also recruit the innate and adaptive immune cells into the infected tissues via cytokines and chemokines.58 In addition, as K18-hACE2 mice produced anti-SARS-CoV-2 humoral and cellular responses at 6 dpi,39 we sacrificed mice at 4 dpi to measure the viral loads and pathogenesis to avoid the influence of infection-induced immune responses on vaccine-induced immune responses during the challenge experiments. Since high-dose immunization may delay the onset of clinical symptoms, future studies should also evaluate clinical symptoms about 6-8 days after infection.
In summary, we report that the heterologous PD1-RBD-DNA/LAIV-HK68-RBD regimen prevents SARS-CoV-2 nasal infection by inducing sufficient and long-lasting mucosal NAbs and CD8 T cells. Our PD1-RBD-DNA vaccine may prime strong mucosal CD8 T cells responses in addition to inducing potent NAbs. Although intramuscular electroporation delivery has potential limitations for DNA vaccination in large populations, similar approaches have demonstrated safety, tolerability and immunogenicity in clinical trials.59 Future study, however, is needed to develop non-invasive delivery techniques for DNA vaccination in humans. A recent study reported that fully vaccinated individuals with breakthrough infections had similar viral transmission efficiency to unvaccinated cases,60 and thus it is important to further strengthen our study findings on vaccine-mediated prevention of viral transmission in the future.
Declaration of interests
H.C., Z.C. and KY.Y. are co-inventors of PD1-based and LAIV-based COVID-19 vaccine patent. YC. W. and L.L are co-inventors of PD1-based COVID-19 vaccine, P.W. is co-inventors of LAIV-based COVID-19 vaccine. The other authors declare no competing interests. YC. W. is now working at Immuno Cure Holding (HK) Limited.
Acknowledgments
Contributors
Z.C. and H.C. supervised two collaborative teams, conceived and designed the study, and wrote the manuscript. R.Z., P.W., Y.-C.W and H.X. designed the experiments, analysed the data, and prepared the manuscript. R.Z., P.W., Y-.C.W., H.X., S-. Y.L., B. W-.Y.M., K. F. W., H.H., R.C-.Y.T., and S.D. conducted the immunologic and virologic assays. L.L., A.Z. and D.Z. performed the immunofluorescence staining of animal tissue section. A.J.Z. analysed tissue pathology. Q.P., N.L., B.Z., C-.Y.C and D.Y. conducted the ELISA and neutralization assay, Z.D. and K-.K.A. performed the viral RNA measurement. K.-Y.Y. provided critical comments, supports and materials. Z.C. and R.Z. have accessed and verified the underlying data reported in the manuscript. All authors read and approved the final version of the manuscript.
Acknowledgements
This study was supported by the Hong Kong Research Grants Council Collaborative Research Fund (C7156-20G, C1134-20G and C5110-20G), General Research Fund (17107019) and Health and Medical Research Fund (19181052 and 19181012); Outbreak Response to Novel Coronavirus (COVID-19) by the Coalition for Epidemic Preparedness Innovations; Shenzhen Science and Technology Program (JSGG20200225151410198 and JCYJ20210324131610027) and matching fund from the Shenzhen Immuno Cure BioTech Limited; the Hong Kong Health@InnoHK, Innovation and Technology Commission; and the China National Program on Key Research Project (2020YFC0860600, 2020YFA0707500 and 2020YFA0707504); and donations from the Friends of Hope Education Fund. Z.C.’s team was also partly supported by the Hong Kong Theme-Based Research Scheme (T11-706/18-N). The funders of this study had no role in study design, data collection, data analyses, interpretation, or writing of the report.
We sincerely thank Dr. Jincun Zhao for providing the AD5-hACE2 strain, Drs. David D. Ho and Pengfei Wang for the expression plasmids encoding for D614G, Alpha and Beta variants, Dr. Linqi Zhang for the Delta variant plasmid, and Shanghai Teresa Healthcare Sci-Tech Co., Ltd for providing the clinically certified TERESA-EPT-I Drug Delivery Device for DNA vaccination.
Data Sharing Statement
The authors declare that the data supporting the findings of this study are available from the corresponding author upon request.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2021.103762.
Contributor Information
Honglin Chen, Email: hlchen@hku.hk.
Zhiwei Chen, Email: zchenai@hku.hk.
Appendix. Supplementary materials
References
- 1.To K.K., Tsang O.T., Leung W.S., Tam A.R., Wu T.C., Lung D.C., et al. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20(5):565–574. doi: 10.1016/S1473-3099(20)30196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N., Zhang D., Wang W., Li X., Yang B., Song J., et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N Engl J Med. 2020;382(8):727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., et al. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N Engl J Med. 2020;383(25):2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., et al. Immunogenic BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 5.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., et al. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N Engl J Med. 2020;383(25):2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Doremalen N., Lambe T., Spencer A., Belij-Rammerstorfer S., Purushotham J.N., Port J.R., et al. ChAdOx1 nCoV-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586(7830):578–582. doi: 10.1038/s41586-020-2608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu L., To K.K., Chan K.H., Wong Y.C., Zhou R., Kwan K.Y., et al. High neutralizing antibody titer in intensive care unit patients with COVID-19. Emerg Microbes Infect. 2020;9(1):1664–1670. doi: 10.1080/22221751.2020.1791738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peiris J.S., Chu C.M., Cheng V.C., Chan K.S., Hung I.F., Poon L.L., et al. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361(9371):1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan J.F., Yip C.C., To K.K., Tang T.H., Wong S.C., Leung K.H., et al. Improved Molecular Diagnosis of COVID-19 by the Novel, Highly Sensitive and Specific COVID-19-RdRp/Hel Real-Time Reverse Transcription-PCR Assay Validated In Vitro and with Clinical Specimens. J Clin Microbiol. 2020;58(5) doi: 10.1128/JCM.00310-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hung I.F., Cheng V.C., Li X., Tam A.R., Hung D.L., Chiu K.H., et al. SARS-CoV-2 shedding and seroconversion among passengers quarantined after disembarking a cruise ship: a case series. Lancet Infect Dis. 2020;20(9):1051–1060. doi: 10.1016/S1473-3099(20)30364-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wong Y.C., Lau S.Y., Wang To K.K., Mok B.W.Y., Li X., Wang P., et al. Natural transmission of bat-like SARS-CoV-2PRRA variants in COVID-19 patients. Clin Infect Dis. 2020 doi: 10.1093/cid/ciaa953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383(27):2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baden L.R., El Sahly H.M., Essink B., Kotloff K., Frey S., Novak R., et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403–416. doi: 10.1056/NEJMoa2035389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voysey M., Clemens S.A.C., Madhi S.A., Weckx L.Y., Folegatti P.M., Aley P.K., et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397(10269):99–111. doi: 10.1016/S0140-6736(20)32661-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen P., Nirula A., Heller B., Gottlieb R.L., Boscia J., Morris J., et al. SARS-CoV-2 Neutralizing Antibody LY-CoV555 in Outpatients with Covid-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2029849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou D., Chan J.F., Zhou B., Zhou R., Li S., Shan S., et al. Robust SARS-CoV-2 infection in nasal turbinates after treatment with systemic neutralizing antibodies. Cell Host Microbe. 2021 doi: 10.1016/j.chom.2021.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hie B., Zhong E.D., Berger B., Bryson B.. Learning the language of viral evolution and escape. Science. 2021;371(6526):284–288. doi: 10.1126/science.abd7331. [DOI] [PubMed] [Google Scholar]
- 19.Wang P., Nair M.S., Liu L., Iketani S., Luo Y., Guo Y., et al. Antibody Resistance of SARS-CoV-2 Variants B.1.351 and B.1.1.7. Nature. 2021 doi: 10.1038/s41586-021-03398-2. [DOI] [PubMed] [Google Scholar]
- 20.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., et al. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396(10249):467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Birhane M., Bressler S., Chang G., Clark T., Dorough L., Fischer M., et al. COVID-19 Vaccine Breakthrough Infections Reported to CDC — United States, January 1–April 30, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:792–793. doi: 10.15585/mmwr.mm7021e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu L., Wei Q., Lin Q., Fang J., Wang H., Kwok H., et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4(4) doi: 10.1172/jci.insight.123158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou J., Cheung A.K., Tan Z., Wang H., Yu W., Du Y., et al. PD1-based DNA vaccine amplifies HIV-1 GAG-specific CD8+ T cells in mice. J Clin Invest. 2013;123(6):2629–2642. doi: 10.1172/JCI64704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zheng M., Wang P., Song W., Lau S.Y., Liu S., Huang X., et al. An A14U Substitution in the 3′ Noncoding Region of the M Segment of Viral RNA Supports Replication of Influenza Virus with an NS1 Deletion by Modulating Alternative Splicing of M Segment mRNAs. J Virol. 2015;89(20):10273–10285. doi: 10.1128/JVI.00919-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun J., Zhuang Z., Zheng J., Li K., Wong R.L., Liu D., et al. Generation of a Broadly Useful Model for COVID-19 Pathogenesis, Vaccination, and Treatment. Cell. 2020;182(3):734–743. doi: 10.1016/j.cell.2020.06.010. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu X., Guo J., Niu M., An M., Liu L., Wang H., et al. Tandem bispecific neutralizing antibody eliminates HIV-1 infection in humanized mice. J Clin Invest. 2018;128(6):2239–2251. doi: 10.1172/JCI96764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corbett K.S., Flynn B., Foulds K.E., Francica J.R., Boyoglu-Barnum S., Werner A.P., et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in Nonhuman Primates. N Engl J Med. 2020;383(16):1544–1555. doi: 10.1056/NEJMoa2024671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang L., Zhang F., Yu W., He T., Yu J., Yi C.E., et al. Antibody responses against SARS coronavirus are correlated with disease outcome of infected individuals. J Med Virol. 2006;78(1):1–8. doi: 10.1002/jmv.20499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Temperton N.J., Chan P.K., Simmons G., Zambon M.C., Tedder R.S., Takeuchi Y., et al. Longitudinally profiling neutralizing antibody response to SARS coronavirus with pseudotypes. Emerg Infect Dis. 2005;11(3):411–416. doi: 10.3201/eid1103.040906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Charan J., Kantharia N.D.. How to calculate sample size in animal studies? J Pharmacol Pharmaco. 2013;4(4):303–306. doi: 10.4103/0976-500X.119726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Y., Soh W.T., Kishikawa J.I., Hirose M., Nakayama E.E., Li S., et al. An infectivity-enhancing site on the SARS-CoV-2 spike protein targeted by antibodies. Cell. 2021;184(13):3452–3466. doi: 10.1016/j.cell.2021.05.032. e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang S., Li Y., Dai L., Wang J., He P., Li C., et al. Safety and immunogenicity of a recombinant tandem-repeat dimeric RBD-based protein subunit vaccine (ZF2001) against COVID-19 in adults: two randomised, double-blind, placebo-controlled, phase 1 and 2 trials. Lancet Infect Dis. 2021;21(8):1107–1119. doi: 10.1016/S1473-3099(21)00127-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P., Zheng M., Lau S.Y., Chen P., Mok B.W., Liu S., et al. Generation of DelNS1 Influenza Viruses: a Strategy for Optimizing Live Attenuated Influenza Vaccines. mBio. 2019;10(5) doi: 10.1128/mBio.02180-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sette A., Crotty S.. Adaptive immunity to SARS-CoV-2 and COVID-19. Cell. 2021 doi: 10.1016/j.cell.2021.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee L.Y., Ha do L.A., Simmons C., de Jong M.D., Chau N.V., Schumacher R., et al. Memory T cells established by seasonal human influenza A infection cross-react with avian influenza A (H5N1) in healthy individuals. J Clin Invest. 2008;118(10):3478–3490. doi: 10.1172/JCI32460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poland G.A., Ovsyannikova I.G., Kennedy R.B.. SARS-CoV-2 immunity: review and applications to phase 3 vaccine candidates. Lancet. 2020;396(10262):1595–1606. doi: 10.1016/S0140-6736(20)32137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sahin U., Muik A., Derhovanessian E., Vogler I., Kranz L.M., Vormehr M., et al. COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature. 2020;586(7830):594–599. doi: 10.1038/s41586-020-2814-7. [DOI] [PubMed] [Google Scholar]
- 38.Winkler E.S., Bailey A.L., Kafai N.M., Nair S., McCune B.T., Yu J., et al. SARS-CoV-2 infection of human ACE2-transgenic mice causes severe lung inflammation and impaired function. Nat Immunol. 2020;21(11):1327–1335. doi: 10.1038/s41590-020-0778-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng J., Wong L.R., Li K., Verma A.K., Ortiz M.E., Wohlford-Lenane C., et al. COVID-19 treatments and pathogenesis including anosmia in K18-hACE2 mice. Nature. 2021;589(7843):603–607. doi: 10.1038/s41586-020-2943-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hassan A.O., Kafai N.M., Dmitriev I.P., Fox J.M., Smith B.K., Harvey I.B., et al. A Single-Dose Intranasal ChAd Vaccine Protects Upper and Lower Respiratory Tracts against SARS-CoV-2. Cell. 2020;183(1):169–184. doi: 10.1016/j.cell.2020.08.026. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Paik D.H., Farber D.L.. Anti-viral protective capacity of tissue resident memory T cells. Curr Opin Virol. 2021;46:20–26. doi: 10.1016/j.coviro.2020.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krammer F.. SARS-CoV-2 vaccines in development. Nature. 2020;586(7830):516–527. doi: 10.1038/s41586-020-2798-3. [DOI] [PubMed] [Google Scholar]
- 43.Mora J.R., von Andrian U.H.. T-cell homing specificity and plasticity: new concepts and future challenges. Trends Immunol. 2006;27(5):235–243. doi: 10.1016/j.it.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 44.Vogel A.B., Kanevsky I., Che Y., Swanson K.A., Muik A., Vormehr M., et al. BNT162b vaccines protect rhesus macaques from SARS-CoV-2. Nature. 2021 doi: 10.1038/s41586-021-03275-y. [DOI] [PubMed] [Google Scholar]
- 45.Muik A., Wallisch A.K., Sanger B., Swanson K.A., Muhl J., Chen W., et al. Neutralization of SARS-CoV-2 lineage B.1.1.7 pseudovirus by BNT162b2 vaccine-elicited human sera. Science. 2021 doi: 10.1126/science.abg6105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mahase E.. Covid-19: Novavax vaccine efficacy is 86% against UK variant and 60% against South African variant. BMJ. 2021;372:n296. doi: 10.1136/bmj.n296. [DOI] [PubMed] [Google Scholar]
- 47.Tian J.H., Patel N., Haupt R., Zhou H., Weston S., Hammond H., et al. SARS-CoV-2 spike glycoprotein vaccine candidate NVX-CoV2373 immunogenicity in baboons and protection in mice. Nat Commun. 2021;12(1):372. doi: 10.1038/s41467-020-20653-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Graham S.P., McLean R.K., Spencer A.J., Belij-Rammerstorfer S., Wright D., Ulaszewska M., et al. Evaluation of the immunogenicity of prime-boost vaccination with the replication-deficient viral vectored COVID-19 vaccine candidate ChAdOx1 nCoV-19. NPJ Vaccines. 2020;5:69. doi: 10.1038/s41541-020-00221-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gandon S., Mackinnon M.J., Nee S., Read A.F.. Imperfect vaccines and the evolution of pathogen virulence. Nature. 2001;414(6865):751–756. doi: 10.1038/414751a. [DOI] [PubMed] [Google Scholar]
- 50.Liu W., Wong Y.C., Chen S.M.Y., Tang J., Wang H., Cheung A.K.L., et al. DNA prime/MVTT boost regimen with HIV-1 mosaic Gag enhances the potency of antigen-specific immune responses. Vaccine. 2018;36(31):4621–4632. doi: 10.1016/j.vaccine.2018.06.047. [DOI] [PubMed] [Google Scholar]
- 51.Ba L., Yi C.E., Zhang L., Ho D.D., Chen Z.. Heterologous MVA-S prime Ad5-S boost regimen induces high and persistent levels of neutralizing antibody response against SARS coronavirus. Appl Microbiol Biotechnol. 2007;76(5):1131–1136. doi: 10.1007/s00253-007-1073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu L., Wei Q., Nishiura K., Peng J., Wang H., Midkiff C., et al. Spatiotemporal interplay of severe acute respiratory syndrome coronavirus and respiratory mucosal cells drives viral dissemination in rhesus macaques. Mucosal Immunol. 2016;9(4):1089–1101. doi: 10.1038/mi.2015.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang H., Zhang Y., Huang B., Deng W., Quan Y., Wang W., et al. Development of an Inactivated Vaccine Candidate, BBIBP-CorV, with Potent Protection against SARS-CoV-2. Cell. 2020;182(3):713–721. doi: 10.1016/j.cell.2020.06.008. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gao Q., Bao L., Mao H., Wang L., Xu K., Yang M., et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. 2020;369(6499):77–81. doi: 10.1126/science.abc1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang Q., Ji K., Tian S., Wang F., Huang B., Tong Z., et al. A single-dose mRNA vaccine provides a long-term protection for hACE2 transgenic mice from SARS-CoV-2. Nat Commun. 2021;12(1):776. doi: 10.1038/s41467-021-21037-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Routhu N.K., Cheedarla N., Gangadhara S., Bollimpelli V.S., Boddapati A.K., Shiferaw A., et al. A modified vaccinia Ankara vector-based vaccine protects macaques from SARS-CoV-2 infection, immune pathology, and dysfunction in the lungs. Immunity. 2021;54(3):542–556. doi: 10.1016/j.immuni.2021.02.001. e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mercado N.B., Zahn R., Wegmann F., Loos C., Chandrashekar A., Yu J., et al. Single-shot Ad26 vaccine protects against SARS-CoV-2 in rhesus macaques. Nature. 2020;586(7830):583–588. doi: 10.1038/s41586-020-2607-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Morabito K.M., Ruckwardt T., Redwood A., Moin S.M., Price D.A., Graham B.S.. Intranasal administration of RSV antigen-expressing MCMV elicits robust tissue-resident effector and effector memory CD8+T cells in the lung. Mucosal Immunology. 2017;10(2):545–554. doi: 10.1038/mi.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Tebas P., Yang S., Boyer J.D., Reuschel E.L., Patel A., Christensen-Quick A., et al. Safety and immunogenicity of INO-4800 DNA vaccine against SARS-CoV-2: A preliminary report of an open-label, Phase 1 clinical trial. EClinicalMedicine. 2021;31 doi: 10.1016/j.eclinm.2020.100689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Singanayagam A., Hakki S., Dunning J., Madon K.J., Crone M.A., Koycheva A., et al. Community transmission and viral load kinetics of the SARS-CoV-2 delta (B.1.617.2) variant in vaccinated and unvaccinated individuals in the UK: a prospective, longitudinal, cohort study. Lancet Infect Dis. 2021 doi: 10.1016/S1473-3099(21)00648-4. S1473-3099 (21)00648-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.