Abstract
Expression of the bone sialoprotein (BSP) gene, a marker of bone formation, is largely restricted to cells in mineralized tissues. Recent studies have shown that the Cbfa1 (also known as Runx2, AML-3, and PEBP2αA) transcription factor supports commitment and differentiation of progenitor cells to hypertrophic chondrocytes and osteoblasts. This study addresses the functional involvement of Cbfa sites in expression of the Gallus BSP gene. Gel mobility shift analyses with nuclear extracts from ROS 17/2.8 osteoblastic cells revealed that multiple Cbfa consensus sequences are functional Cbfa DNA binding sites. Responsiveness of the 1.2-kb Gallus BSP promoter to Cbfa factors Cbfa1, Cbfa2, and Cbfa3 was assayed in osseous and nonosseous cells. Each of the Cbfa factors mediated repression of the wild-type BSP promoter, in contrast to their well known activation of various hematopoietic and skeletal phenotypic genes. Suppression of BSP by Cbfa factors was not observed in BSP promoters in which Cbfa sites were deleted or mutated. Expression of the endogenous BSP gene in Gallus osteoblasts was similarly downregulated by forced expression of Cbfa factors. Our data indicate that Cbfa repression of the BSP promoter does not involve the transducin-like enhancer (TLE) proteins. Neither coexpression of TLE1 or TLE2 nor the absence of the TLE interaction motif of Cbfa1 (amino acids 501 to 513) influenced repressor activity. However, removal of the C terminus of Cbfa1 (amino acids 362 to 513) relieved suppression of the BSP promoter. Our results, together with the evolutionary conservation of the seven Cbfa sites in the Gallus and human BSP promoters, suggest that suppressor activity by Cbfa is of significant physiologic consequence and may contribute to spatiotemporal expression of BSP during bone development.
Bone sialoprotein (BSP) is a phosphorylated and sulfated 34-kDa protein that represents one of the major noncollagenous, extracellular matrix (ECM) proteins associated with mineralized tissues. In developing rat calvaria, BSP mRNA expression is detected in osteoblasts on day 17 along with type I collagen, osteocalcin (OC), osteonectin, and alkaline phosphatase. BSP expression is also found in hypertrophic chondrocytes (8). BSP, similar to OC, is upregulated during osteoblast differentiation in vitro as the ECM mineralizes. BSP reaches peak levels slightly earlier than OC and is downregulated in mature osteocytes (14, 35, 36, 39, 43). The protein is selectively associated with clusters of needle-like crystals of hydroxyapatite. Various in vitro studies suggest that BSP can either promote mineral nucleation or facilitate mineral growth (22). In other studies the functional role of this protein was assessed by stably transfecting murine osteosarcoma cell lines with avian forms of this gene (15). The results demonstrated that the cell lines expressing the highest levels of BSP displayed the greatest accumulation of calcium in the ECM, consistent with BSP binding to hydroxyapatite (5, 12).
Regulatory elements in the BSP promoter that contribute to its high level of transcription in osteoblasts have not been identified. BSP promoters from several species have been shown to contain a negative vitamin D response element (33), an inverted TATA sequence, a YY1 motif (28), several Sp1 sites, and a homeodomain (engrailed) motif in the proximal promoter (59), but none of these has been demonstrated to mediate tissue-specific expression. The OC promoter, however, has provided examples of several elements that contribute to its osteoblast-specific expression. The OC box I and homeodomain (Msx or Dlx) binding site (20, 21, 56) both restricts and attenuates OC expression in developing osteoblasts. Most significantly, multiple Cbfa (core binding factor α) sites in the various OC promoters support cell type-specific expression, and forced expression of Cbfa factors can confer induction of OC promoter-reporter constructs in both osseous and nonosseous cells (1, 11, 25). Cbfa1 has been shown to be a key regulator of osteogenic differentiation (2, 31, 42) and regulates several osteoblast-related genes.
The Gallus BSP promoter has been sequenced (59) and contains multiple Cbfa consensus motifs. Several important observations prompted examination of the functionality of these putative Cbfa sites. Firstly, the Cbfa1 null mutant revealed loss of BSP expression, along with other osteoblast phenotypic markers (31). Secondly, the similarities between BSP and OC with respect to their high expression levels restricted to mineralized tissues in vivo and in vitro suggest that Cbfa may be a strong enhancer of BSP expression in mature bone cells. However, we report here a series of experimental approaches demonstrating that Cbfa factors mediate repression of the Gallus BSP promoter in rat and Gallus osteoblasts, as well as in nonosseous HeLa cells.
The present studies show that the Gallus BSP promoter contains seven functional Cbfa sites that bind an osteoblast-specific complex comprising Cbfa factors. Cbfa mediates suppression of the BSP promoter by a mechanism that does not involve interaction of the Cbfa VWRPY domain with TLE (the human homologue of Drosophila Groucho), a known suppressor of Cbfa activity. The opposing regulation of the BSP and OC promoters by Cbfa is consistent with distinct hormonal responses and functional properties of the proteins. More importantly, these studies reveal the significance of the promoter context of Cbfa motifs in contributing to transcriptional control.
MATERIALS AND METHODS
Cell cultures.
Osteoblasts were isolated by three sequential trypsin-collagenase treatments of 17-day embryonic chicken (Gallus) calvaria (14). Primary cultures of embryonic chick osteoblasts have been well characterized with respect to their stages of differentiation and reflect bone formation in vivo. Only the cells released during the third digest were used for experiments. Cultures were plated and grown for 2 weeks (until confluence) in minimal essential medium supplemented with 10% fetal bovine serum, with medium changes every 3 days. Upon reaching confluence, the cultures were switched to BGJb medium supplemented with 10% fetal bovine serum. After 2 days this medium was supplemented with 10 mM β-glycerophosphate, and after an additional 2 days the medium was supplemented with 12.5 μg of ascorbic acid per ml. This medium was considered complete medium. All studies were carried out 3 days after switching to complete medium. ROS 17/2.8 cells were maintained in F12 medium supplemented with 5% fetal calf serum (Life Technologies, Inc. [Gibco BRL], Grand Island, N.Y.). HeLa cells were cultured in suspension in Joklik-modified minimal essential medium and plated in Dulbecco modified Eagle medium with 10% fetal calf serum for transfections.
Nuclear extracts.
Nuclear extracts were prepared according to the Dignam method (10). Briefly, 1 × 106 rat osteoblastic cells (ROS 17/2.8) or day 3 Gallus osteoblasts were washed once with 10 ml of ice-cold phosphate-buffered saline (PBS) and collected by centrifuging at 165 × g for 5 min. The cell pellet was resuspended in 400 μl of ice-cold NP-40 lysis buffer (10 mM Tris [pH 7.4], 3 mM MgCl2, 10 mM NaCl, 0.5% NP-40) by gentle pipetting and incubated on ice for 10 min. The nuclei pellet was collected by centrifugation for 30 s and resuspended in 400 μl of cold hypotonic buffer (10 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl). The pellet was again collected by centrifugation at 4,000 × g for 1 min, resuspended in 50 μl of ice-cold extraction buffer (20 mM HEPES [pH 7.9], 1.5 mM MgCl2, 420 mM KCl, 0.2 mM EDTA, 20% glycerol), and vigorously rocked in the cold room for 30 min to 1 h followed by microfuging for 5 min. Aliquots of supernatant containing nuclear proteins were quick-frozen in a dry ice-ethanol bath and stored at −80°C. Protein concentrations were determined using a protein assay reagent (Bradford; Bio-Rad Laboratories, Hercules, Calif.).
Oligonucleotides, probes, and EMSA.
Oligonucleotides representing the wild-type or mutant Cbfa sites in the Gallus BSP promoter are shown in Fig. 1. The upper strands (200 ng) of the oligonucleotides were labeled with 32P for 1 h at 37°C in a 50-μl volume using T4 Polynucleotide Kinase (New England BioLabs, Beverly, Mass.) as indicated by the manufacturer. The reaction was stopped by heat inactivation at 65°C for 1 h. Annealing was performed by addition of a twofold excess amount of bottom strand followed by boiling for 5 min and slow cooling to room temperature. The unincorporated nucleotides were removed using a quick-spin G 25 Sephadex column (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the manufacturer's instructions. Electrophoretic mobility shift assay (EMSA) reaction mixtures were prepared using 50 fmol of probe, 50 mM KCl, 12 mM HEPES, 1 mM EDTA, 1 mM dithiothreitol, 12% glycerol, 2 μg of poly(dI-dC) · poly(dI-dC) and 0 to 10 μg of nuclear protein. After a 20-min incubation at 22°C, unlabeled double-stranded competitor oligonucleotide (12.5-, 25-, 50-, and 100-fold molar excess over probe) was added to the reaction mixtures, and aliquots were loaded onto a 4% nondenaturing polyacrylamide gel. The gels were electrophoresed for 1.5 h at 200 V, dried, and exposed to film for autoradiography.
FIG. 1.
The Cbfa sites are shown in boxed areas; the dotted box indicates a second overlapping Cbfa motif. Mutations of the ACC core are shown in lower case and were selected (ac) to avoid generation of a new DNA recognition motif.
Antibodies.
The affinity-purified polyclonal antibodies (a generous gift from Scott Hiebert [38]), designated anti-Cbfa1, anti-Cbfa2, anti-Cbfa3 and anti-Cbfβ, were raised against purified peptides. Except for anti-Cbfa2 antibody, which recognizes an epitope in the N terminus, all the antibodies recognize epitopes within the C termini. The specificity of each antiserum has previously been demonstrated in detail (38). Anti-Cbfa3 and Cbfa1 are specific for their respective proteins, but anti-Cbfa2 shows a slight cross-reactivity with Cbfa1. For EMSA supershift experiments, after incubation with the DNA probes, 1 μl of undiluted antibody (for each) was added and incubated with either probe alone (as a negative control) or with the reaction mixtures for an additional 20 min at 37°C.
Transient transfection and CAT assay.
Rat osteosarcoma (ROS 17/2.8) or HeLa cells were plated at a density of 0.5 × 106 per ml, 24 h prior to transfection. Cells were transfected at 50% confluency using Superfect transfection reagent (Qiagen Inc., Valencia, Calif.). Briefly, plasmid DNA (2.5 μg of OC or BSP promoter-chloramphenicol acetyltransferase [CAT] constructs, 0.75 μg of either cytomegalovirus vector control or Cbfa and TLE expression plasmid and 0.1 μg of Rous sarcoma virus luciferase construct as an internal control) was added to 100 μl of serum-free medium and mixed with 10 μl of Superfect reagent followed by incubation for 15 min at room temperature. After the addition of 0.4 ml of complete medium the mixture was applied to semiconfluent cultures in 35-mm-diameter culture dishes already washed once with PBS. The cells were incubated with the transfection mixture for 2 h at 37°C, washed with PBS, fed 2 ml of complete medium, and then incubated at 37°C for 30 to 36 h before harvesting.
For reporter assays, cells were first washed twice with ice-cold PBS and lysed with 300 μl of reporter lysis buffer (Promega Corp., Madison, Wis.) for 20 min at room temperature. For CAT assay, cell lysate (50 μl) was incubated with the reaction mixture for 5 h at 37°C. The products were separated by thin-layer chromatography and the amount of incorporated [14C]chloramphenicol was quantified using the Beta-scope (Betagen, Mountain View, Calif.). The data were normalized to luciferase values used as an internal control. For luciferase assay, cell lysate (20 μl) was mixed with luciferase assay buffer (Promega Corp.) and the amount of light units was measured with a Monolith luminometer (Analytical Luminescence Laboratory, San Diego, Calif.). All experiments were repeated four to six times (n = 6 replicates per experiment) using at least three different DNA preparations.
Gallus osteoblasts were transfected with Lipofectamine reagent and 15 μg of plasmid for each assay. Briefly, plasmid DNA was complexed with Lipofectamine reagent in a 1:2 ratio (DNA to lipofectamine) for 30 min at room temperature. Cells cultured for 3 days in complete medium were rinsed once with sterile PBS and overlaid with DNA-lipofectamine complexes in 6 ml of Opti-MEM transfection medium for a total of 5 h. The cells were subsequently rinsed in PBS, fed fresh complete medium, and cultured until harvesting. CAT assays were performed 48 h after transfection on equal aliquots of samples. CAT activity was assayed by liquid scintillation counting (51). The pCAT control vector containing the simian virus 40 promoter and enhancer (Promega Corp.) was used in some experiments to compare the relative activities of the various BSP promoter segments. The final enzyme activities were expressed in counts of [14C]chloramphenicol converted per minute per microgram of protein.
Site-directed mutagenesis and expression constructs.
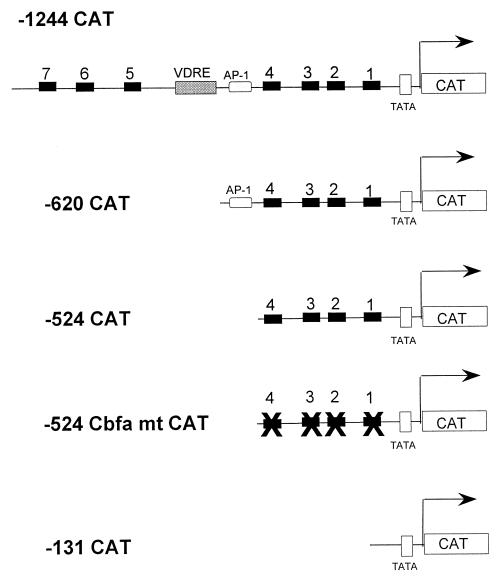
The plasmid construct containing bp −1,239 to +25 of the BSP promoter in the sense direction relative to the CAT coding sequence in the pCAT basic vector (Promega Corp.) was previously described (59). Nested deletions from the 5′ end of the BSP promoter were generated by PCR amplification of selected segments of the promoter from bp −620, −524, or −131 to +25 and have been previously described (58). Site-directed mutagenesis was performed to incorporate 2-nucleotide substitutions into the core binding motif (TGG) of each individual Cbfa site (Fig. 1) in the bp −524 Gallus BSP promoter fragment. Initially the Cbfa site 4 mutant (Fig. 9) was generated by amplifying the bp −524 promoter fragment using the forward primer carrying the mutation (indicated in lower case), 5′TGCCTGCAGCAATTAGAGTacTCTGACCTG3′, and the reverse primer 5′GCTCTAGAGGTGCCACTGGG3′. The PCR product was digested with PstI/XbaI and cloned in similarly digested pCAT vector (Promega Corp.). For sites 3, 2, and 1 the mutant oligonucleotides used as forward and reverse primers were the same as those used for gel shift analyses (Fig. 1). The plasmid bearing mutations in both sites 4 and 3 was generated by using the bp −524 mutant (site 4) promoter as the template. The PCR product was digested with DpnI to destroy the parental DNA, followed by transformation in XL1-Blue competent cells (Stratagene, La Jolla, Calif.). Similar stepwise procedures were followed to generate plasmids carrying mutations in all four Cbfa motifs. Incorporation of the substitution mutations in all constructs was confirmed by sequencing. CMV-driven Cbfa1, Cbfa2, and Cbfa3 expression constructs were described previously (2). The rat OC 5′ promoter fragment (bp −1, 097 to +23) fused to the CAT gene (pOCZCAT) containing three Cbfa-responsive motifs has been reported previously (2, 25). The Cbfa1 MASN isoform expression construct was obtained from James C. Neil (University of Glasgow, Glasgow, United Kingdom). Cbfa1ΔC was generated by digesting the MRIPV isoform with ApaI to remove the part of the cDNA encoding amino acids 362 to 513. Two synthetic oligonucleotides (5′GGGCCCTTGATAACTCGAGGAATTCTTATCAAGGGCCC3′ and 5′GGGCCCTTGATAAGAATTCCTCGAGTTATCAAGGGCCC3′) containing stop codon and unique restriction sites were hybridized to generate a cassette. The cassette was digested with ApaI and ligated into ApaI-digested pCDNA3.1-Cbfa1.
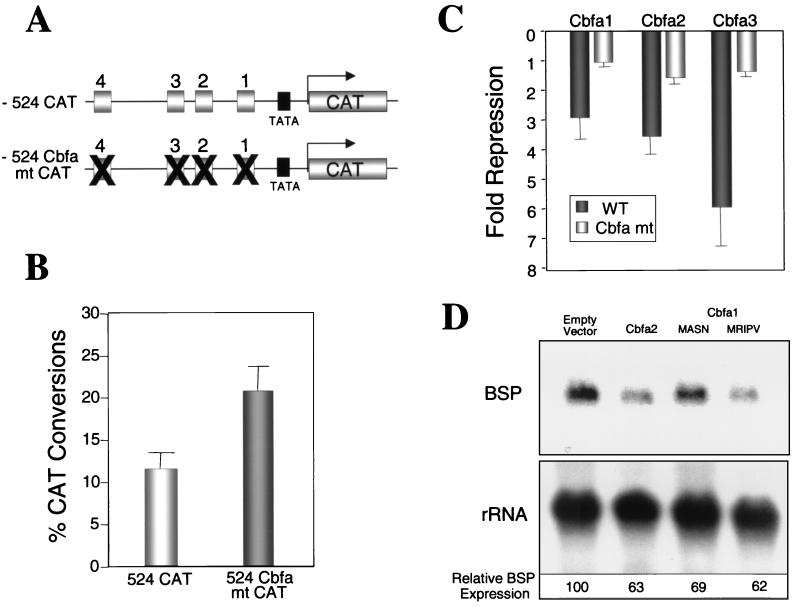
FIG. 9.
Cbfa proteins suppress expression of the BSP gene through interaction with Cbfa motifs. (A) Schematic representation of WT and Cbfa mutant bp −524 promoters. (B) ROS 17/2.8 cells were transfected with 2.5 μg of WT and Cbfa mutant bp −524 promoter to assess basal promoter activity. The cells were assayed for CAT-reporter activity as described in Materials and Methods. n = 6. (C) The WT or mutant promoters were cotransfected with 0.75 μg of empty vector, Cbfa1, Cbfa2, or Cbfa3 expression constructs in HeLa cells. The CAT activity was determined 24 h posttransfection and normalized to RSV luciferase values used as an internal control. The fold suppression data were obtained by comparing Cbfa to empty vector [n = 6]. (D) Primary Gallus osteoblasts were transfected in triplicate with 15 μg of either empty vector, Cbfa2, or two isoforms of Cbfa1. Cells were harvested 24 h later, and total RNA was isolated. A total of 20 μg of RNA was resolved and blots were hybridized with Gallus BSP and rRNA probes. Values for each RNA were determined by radiographic analysis using the STORM Phosphorimager. The normalized values of BSP RNA expression are shown at the bottom.
Cbfa1Δ12 was constructed by digesting the MRIPV isoform with EcoRI-XbaI to remove the part of the cDNA encoding amino acids 501 to 513, followed by blunting and ligation. The sequences of these constructs were confirmed by the dideoxy chain termination method. pBluescript vectors carrying TLE1 or TLE2 cDNAs were obtained from Stefano Stifani, Montreal Neurological Institute, Montreal, Quebec, Canada. TLE1 coding region was amplified by PCR using forward primer 5′GTAATGGATCCAGTATGGGGTTCCCGCAGAGCCGGCACCCG3′ and reverse primer 5′GCCATACTCGAGCTAGTAGATGACTTCATAGACTGTAGC3′. TLE2 was amplified by PCR using forward primer 5′GCAGCGGATCCAGCATGGAATACCCCCAGGGAAGGCAC3′ and reverse primer 5′GCCTGGCTCGAGTCAGTAGACCACCTCATACAC3′. PCR-generated fragments of TLE1 and TLE2 were digested with BamHI-XhoI and cloned into BamHI-XhoI-digested pCDNA3.1 vector (Invitrogen Corporation, San Diego, Calif.). The sequences of these constructs were confirmed by the dideoxy chain termination method.
Isolation of total cellular RNA and Northern blot analysis.
Twenty-four hours after transient transfection, cultures of Gallus osteoblasts were rinsed twice in 1 × PBS and incubated at room temperature for 5 to 10 min in Trizol reagent (Life Technologies, Inc. [Gibco BRL]). (1 ml per 100-mm-diameter plate). Cells were then scraped from the plates into a microfuge tube, and 0.1 ml of 1-bromo, 3-chloropropane (Sigma, St. Louis, Mo.) was added with mixing. After centrifugation at 4°C, the aqueous layer was collected and RNA was precipitated with 0.5 ml of isopropanol. The pellets were washed with 70% ethanol to remove residual salt, briefly air dried, dissolved in diethylpyrocarbonate-treated water, and stored at −70°C.
RNA samples (20 μg) were electrophoresed in 1% formaldehyde-agarose gels and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech, Arlington Heights, Ill.) in 20 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate). Blots were hybridized with randomly primed (Prime-It kit; Stratagene, La Jolla, Calif.), 32P-labeled cDNA probes for Gallus BSP at 42°C overnight. The radiographic signal was quantitated by using a STORM Phosphorimager (Molecular Dynamics, Sunnyvale, Calif.). Blots were then stripped and reprobed for 18S rRNA. The BSP data were normalized to rRNA values for each sample.
Western blot analysis.
Western blots were performed on whole-cell lysates from HeLa and ROS 17/2.8 cells transiently transfected with Cbfa and TLE expression plasmids. For each construct a 100-mm-diameter dish was transfected with 10 μg of either empty vector or expression plasmid in parallel with promoter-reporter transfection studies. After 24 h, cells were washed with 1× PBS and lysed directly with 200 μl of sodium dodecyl sulfate (SDS) lysis buffer (2% SDS, 10 mM dithiothreitol, 10% glycerol, 2 M urea, 1.0 mM phenylmethylsulfonyl fluoride, 10 mM Tris-HCl, 0.002% bromphenol blue, 1× protease inhibitor cocktail). Protein (30 μg/well) was resolved by SDS–10% polyacrylamide gel electrophoresis and transferred to Trans-Blot membrane (Bio-Rad Laboratories). The blots were incubated with 1:5,000 dilution of mouse monoclonal horseradish peroxidase-conjugated anti-Xpress antibody (Invitrogen Corporation) to detect the epitope-tagged Cbfa1 and TLE proteins or with a 1:2,000 dilution of rabbit polyclonal anti-Cbfa2 or -Cbfa3 antibodies. The blot was stripped and reprobed with antibody to lamin B or α-tubulin (Santa Cruz Biotechnology, Inc., Santa Cruz, Calif.) as a control for equal sample loading.
Immunofluorescence analysis.
Primary chick osteoblasts were passed after 6 days in culture onto gelatin-coated glass coverslips (0.5 × 104 cells) and processed for in situ immunofluorescence analysis 36 h after plating. ROS 17/2.8 cells were examined 2 days after plating. Cells were rinsed twice with ice-cold PBS and were fixed for 10 min on ice with 4% formaldehyde. After washing once with PBS, cells were made permeable by incubation for 20 min on ice with 0.1% Triton X-100 in PBS and then incubated at 37°C for 1 h with the appropriate dilution (1:150) of affinity-purified polyclonal primary antibody against Cbfa1. Cells were washed four times with 1 ml of PBSA (0.5% BSA in PBS), followed by incubation with a 1:400 dilution of secondary antibody (fluorescein isothiocyanate goat anti-rabbit) for 1 h at 37°C. Excess secondary antibody was removed by washing the cells four times with PBSA. Cells were stained with DAPI (0.1% 4′, 6-diamidino-2-phenylindole, 0.1% Triton X-100 in PBSA) for 5 min on ice followed by a single rinse with 0.1% Triton X-100 in PBSA. Finally cells were washed twice with ice-cold PBS and were mounted using Vectashield (Vector Laboratories, Inc., Burlingame, Calif.). The coverslips were sealed and stored in the dark at −20°C till microscopy. Cells were viewed in a Zeiss Axioplan2 microscope equipped with epifluorescence filters and a charge-coupled device digital camera. Images were displayed using Metamorph software (Universal Imaging Corporation, West Chester, Pa.) and printed directly from Powerpoint (version 5.1).
RESULTS
Gallus BSP promoter contains functional Cbfa motifs that bind a Cbfa1-containing complex.
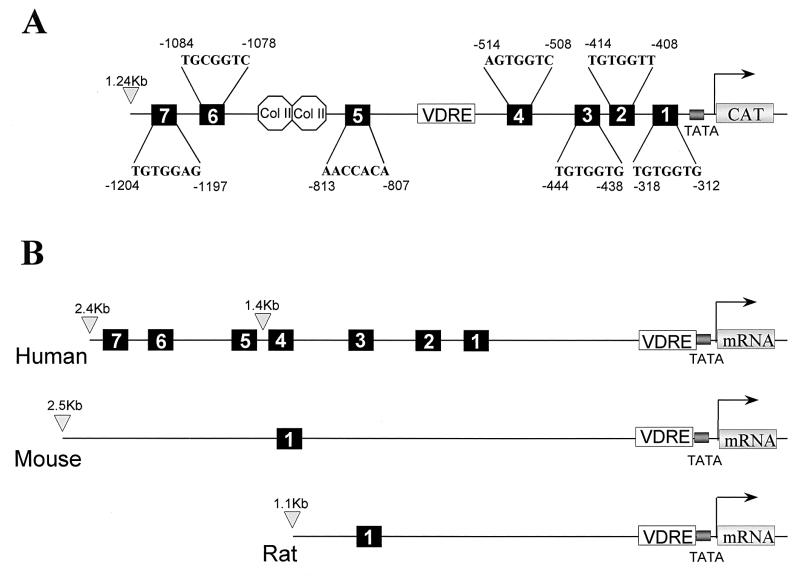
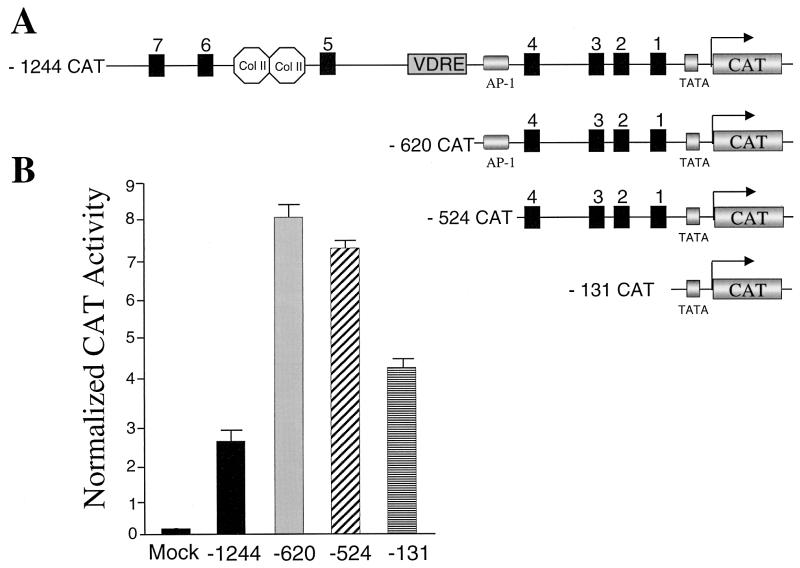
Figure 2 shows the locations of seven consensus Cbfa sites within 1.3 kb of the 5′ flanking sequences of the Gallus BSP gene. The human BSP promoter also contains seven Cbfa recognition motifs, four of which reside within 1.4 kb of 5′ flanking sequences (Fig. 2B). In the Gallus promoter, three Cbfa motifs are distal to the vitamin D response element from kb −1.2 to −0.8, while four sites reside in the proximal promoter from kb −0.5 to −0.3. In contrast, promoters of the rodent species contain either one or two Cbfa consensus sequences (3). In the present studies, we examined the role of the Cbfa elements in transcriptional regulation of the Gallus BSP promoter. We initially assessed which of these Cbfa consensus sites may contribute to BSP transcription by comparing expression of a series of promoter deletion constructs in osteoblasts. We observed both in rat osteoblastic ROS 17/2.8 osteosarcoma cells (Fig. 3) and in Gallus primary osteoblasts (data not shown) that the bp −131 segment lacking Cbfa motifs exhibited a higher level of basal activity than the full-length promoter. This very proximal segment contains several regulatory elements supporting activation (e.g., Sp1 and TATA [29, 59]), and a homeodomain motif shown to mediate enhancer activity of the BSP promoter [4]). Notably, deletion of the upstream promoter segment (from bp −1244 to −620), which includes a silencer motif and three Cbfa sites, resulted in a threefold increase in basal promoter activity. Further deletion of an AP-1 motif (to bp −524) did not significantly change promoter activity. However, removing the four most proximal Cbfa sites decreased activity 44% in ROS 17/2.8 cells and 50% in Gallus osteoblasts. We therefore focused on functional characterization of the Cbfa motifs in the proximal promoter.
FIG. 2.
Gallus and human BSP promoters contain seven Cbfa consensus motifs. (A) Illustration of the positions and sequences of the Cbfa1 sites relative to several conserved regulatory sequences in the Gallus BSP promoter. TATA, TATA box; VDRE, vitamin D response element; Col II-labeled octagons, silencer motifs. (B) Comparative schematic showing the numbers and positions of Cbfa consensus sequences in human, mouse, and rat BSP promoters (not drawn to scale).
FIG. 3.
Analysis of deletion mutants of Gallus BSP promoter in ROS 17/2.8 osteoblastic cells. (A) Schematic illustration of 5′ BSP promoter deletion constructs used in transient transfection assays. Col II-labeled octagons, silencer motifs; VDRE, vitamin D response element. (B) Assay of BSP promoter-CAT reporter constructs in ROS 17/2.8 cells. Cells were transfected with the indicated reporter constructs. Reporter activity was assayed 24 h after transfection and normalized for transfection efficiency (see Materials and Methods). Data are means + the standard errors of the means (SEM), (n = 18 samples); data from three independent experiments were pooled and represent multiple DNA preparations.
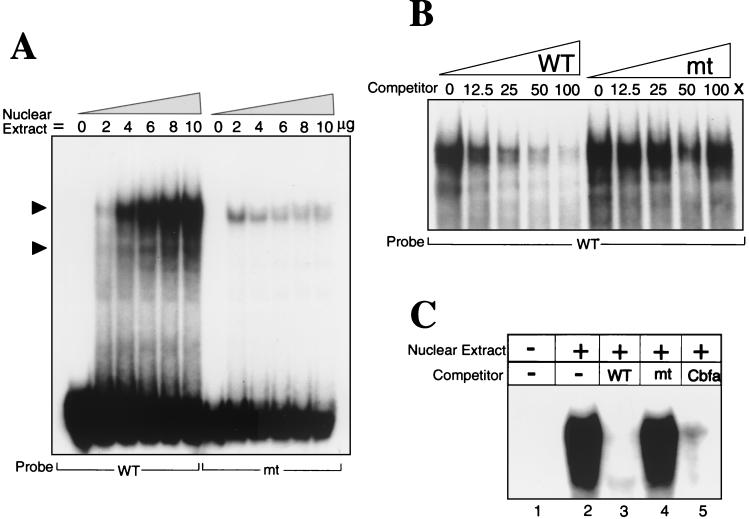
Nuclear extracts from osteoblastic ROS 17/2.8 cells are known to contain a bone-specific DNA binding complex that has been characterized in relation to the Cbfa recognition motifs in the OC promoter (1, 13, 37). Thus, we used these extracts to examine protein-DNA interactions at each of the Cbfa motifs in the BSP proximal promoter. Each of these Cbfa sites (wild type [WT]) forms a major complex that is abolished upon mutation of the core Cbfa motif (Fig. 1 and 4A [representative data shown for site 2]). This complex is not formed at BSP elements using nuclear extracts from HeLa cells (data not shown). Specificity of binding to the putative BSP Cbfa sites was established by competition analyses with oligonucleotides containing wild-type and mutated Cbfa sites (Fig. 4B and data not shown), as well as the Cbfa consensus motif (Fig. 4C). Nuclear extracts from Gallus osteoblasts also formed Cbfa sequence-specific DNA binding complexes at the four Cbfa sites (data not shown). Together, these analyses confirm that each of the Cbfa motifs is a functional Cbfa interaction element.
FIG. 4.
Sequence-specific protein-DNA interactions at the proximal Cbfa site 2 in the Gallus BSP promoter. (A) Oligonucleotides representing wild type (WT) and mutant (mt) sequences for each Cbfa site (Fig. 1) were incubated with increasing concentrations (0 to 10 μg) of nuclear extracts from ROS 17/2.8 cells and examined by gel mobility shift assay Solid arrowheads indicate the formation of different specific complexes. Cbfa-related proteins bind with sequence specificity to all four proximal BSP Cbfa motifs, but representative data are shown only for site 2. The results of competition studies shown in panels B and C demonstrate sequence-specific protein-DNA binding complexes at the BSP Cbfa sites. (B) WT probe was incubated with 6 μg of ROS 17/2.8 nuclear extracts and increasing concentrations (0, 12.5, 25, 50, and 100×) of either WT or mutant (mt) oligonucleotides. (C) Lane 1, WT labeled probe without nuclear extract; lane 2, probe with 6 μg of ROS 17/2.8 nuclear extract; lanes 3, 4, and 5, competition with 100X unlabeled wild-type, mutant, and Cbfa consensus sequence oligonucleotides (Fig. 1), respectively. Only the portion of the gel containing the complexes is shown.
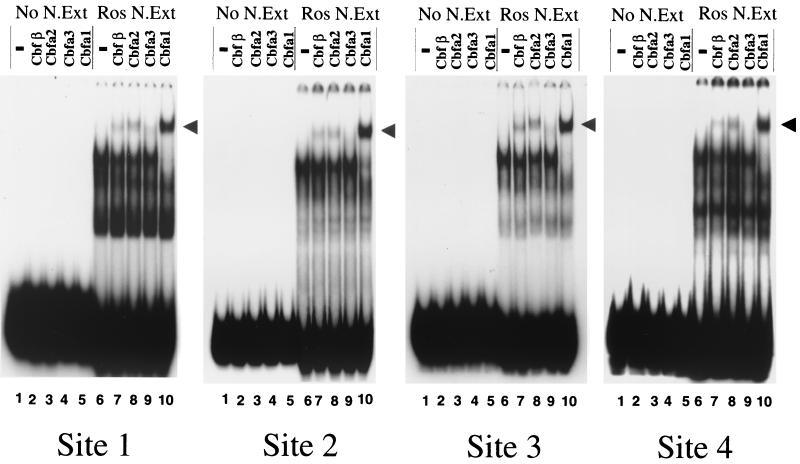
We identified the Cbfa factors involved in complex formation with BSP regulatory elements by antibody supershift analysis using nuclear extracts from ROS 17/2.8 cells. A panel of well characterized, affinity-purified antibodies to the family of Cbfa proteins encoded by distinct genes, Cbfa1, Cbfa2, and Cbfa3 and their non-DNA binding partner, Cbfβ, was used. Figure 5 reveals a complete supershift by the Cbfa1 antibody, as well as a partial supershift with the Cbfa2 antibody which may reflect possible cross-reactivity with Cbfa1 (see Materials and Methods). Cbfβ, a partner protein which enhances Cbfa DNA binding, is also present in the complexes formed at all sites. Additionally, a very weak supershift was detected only on sites 1 and 3 of the BSP promoter with the Cbfa3 antibody. Thus, Cbfa1 is the major component of an osteoblast-derived complex formed on BSP Cbfa motifs, similar to findings for the OC promoter (2, 37).
FIG. 5.
Cbfa1 is the major component of the transcription factor complex binding to all BSP Cbfa sites. Electrophoretic mobility antibody supershift analyses are shown for BSP Cbfa sites 1 to 4. Lanes 1 to 5, antibody controls for each site without nuclear extract, indicating that antisera alone do not react with the probe; lane 6, 6 μg of nuclear extract without antibodies (control); lanes 7 to 10, the supershifts of the Cbfa complex with the indicated antibody. A nearly complete supershift is observed in the presence of antibody specific to Cbfa1 with the complex formed by ROS nuclear extracts at the four proximal sites (1, 2, 3, and 4) in the Gallus BSP promoter. Weak supershifts occurred using antibodies specific to Cbfa2 and Cbfβ, a transactivator partner protein for Cbfa factors.
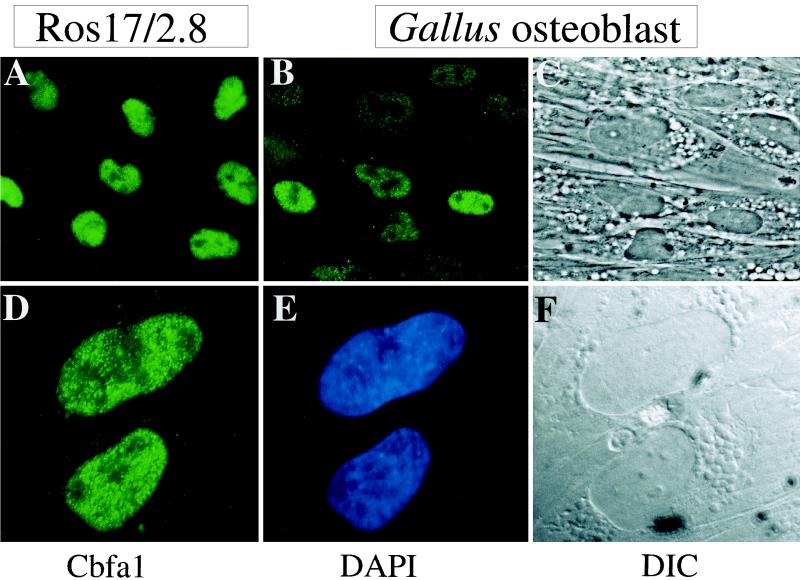
We further confirmed the representation of Cbfa1 in Gallus osteoblasts compared to that in rat osteoblastic ROS 17/2.8 cells by in situ immunofluorescence (Fig. 6). In the ROS 17/2.8 cells, Cbfa1 is predominantly restricted to the nucleus and excluded from nucleoli (Fig. 6A). Cbfa1 is also detected at significant levels in the primary (day 3) Gallus osteoblasts (Fig. 6B and C) by in situ immunofluorescence. The subnuclear distribution of Cbfa1 in Gallus osteoblasts (Fig. 6D to F) is shown at higher magnification. The punctate staining typical of the nuclear matrix association of Cbfa factors is observable (54, 61). Together, these data indicate that Cbfa1 is the major factor in osteoblasts interacting with the multiple Cbfa motifs in the BSP proximal promoter.
FIG. 6.
Immunofluorescence detection of Cbfa1 in osteoblasts. (A) Cellular representation of Cbfa1 in ROS 17/2.8 cells 24 h after plating. (B) Cellular representation of Cbfa1 in day 3 Gallus osteoblasts by immunofluorescence microscopy. Magnification, ×40. Nuclear staining is observed in whole cells incubated with antisera specific to Cbfa1 (as described in Materials and Methods). (C) Corresponding phase-contrast images of the Gallus osteoblast cell layer. (D) Punctate subnuclear distribution of Cbfa1 foci in Gallus osteoblasts. Magnification, ×100. DAPI-stained nuclei. (F) Differential interference contrast (DIC) of the immunopositive cells.
Cbfa factors downregulate BSP promoter activity and expression of the endogenous gene.
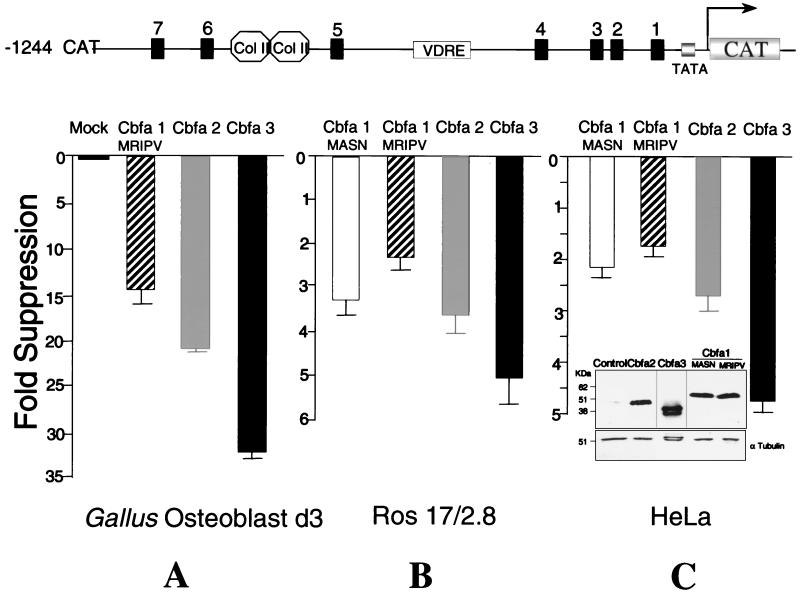
Cbfa1, Cbfa2, and Cbfa3 were previously documented to support enhanced transcriptional activity of the rat OC promoter in both nonosseous cells (1, 11) and the ROS 17/2.8 cell line (2). To assess the consequences of Cbfa protein interactions with Cbfa elements in the Gallus BSP promoter, we transfected the various promoter deletion constructs (from kb −1.2 to −0.1) along with expression plasmids encoding each of the Cbfa proteins into either primary Gallus osteoblasts, ROS 17/2.8 cells, or human HeLa cells, the latter having a zero background of Cbfa factors. As shown in Fig. 7 for the kb −1.2 Gallus BSP promoter, forced expression of the Cbfa factors resulted in suppression of promoter activity in each of the cell types. In the species-homologous Gallus osteoblasts, a highly significant 14.5- to 32-fold suppression was observed with the three Cbfa proteins (Fig. 7A). Notably, in the mammalian cell lines, a greater suppression of the BSP promoter was observed with Cbfa3 than with other family members (Fig. 7B and C). This finding was not the result of significant differences in expression levels, as determined by Western blot analysis of cell layers (Fig. 7C and data not shown). Because of the relatively weaker suppression exhibited by Cbfa1 (MRIPV isoform) on the BSP promoter in HeLa and osteoblast cells, we examined the activity of the bone-related Cbfa1 MASN isoform (52). Only a slightly stronger repression was observed for Cbfa1 (MASN) with all promoter segments in osseous and nonosseous cells (Table 1). Thus, all Cbfa factors and the two Cbfa1 isoforms regulate suppression of BSP promoter activity.
FIG. 7.
Cbfa factors mediate suppression of the Gallus BSP promoter containing seven Cbfa regulatory elements. Various regulatory elements are shown in the diagram at the top of the figure. Gallus (A), ROS 17/2.8 (B), and HeLa (C) cells were transiently transfected with 2.5 μg of −1244 BSP-CAT and 0.75 μg of Cbfa1 (MRIPV), Cbfa1 (MASN), Cbfa2, or Cbfa3 as indicated. CAT activity was quantitated and normalized for transfection efficiency. The data are fold repression values (Cbfa over empty vector). Each bar represents a pool of samples (n = 18) from three independent experiments. Each group showed significant repression (P < 0.001) as tested by analysis of variance. (ANOVA) HeLa cells (100-mm-diameter dishes) were transiently transfected with 10 μg of indicated plasmid. Thirty micrograms of total protein was subjected to SDS–10% PAGE. Blots were incubated with either mouse monoclonal Xpress antibody (to detect tagged Cbfa1 proteins) or rabbit polyclonal anti-Cbfa2 and -Cbfa3 antibodies (see Materials and Methods). Blots were stripped and probed with α-tubulin for equal loading control. The relative mobility of different proteins is indicated. Col II-labeled octagons, silencer motifs; VDRE, vitamin D response element.
TABLE 1.
Summary of repressor effects of Cbfa factors on various segments of the Gallus BSP promoter
| Construct | Mean CAT activity ± SEMa
|
Cell line | |||
|---|---|---|---|---|---|
| Cbfa1 MASN | Cbfa1 MRIPV | Cbfa2 | Cbfa3 | ||
 |
2.1 ± 0.2 | 1.7 ± 0.2 | 2.7 ± 0.3 | 4.7 ± 0.2 | HeLa |
| 3.4 ± 0.4 | 2.4 ± 0.3 | 3.8 ± 0.4 | 5.8 ± 0.6 | ROS 17/2.8 | |
| 6.8 ± 0.8 | 4.0 ± 0.5 | 7.9 ± 1.0 | 11.0 ± 1.5 | HeLa | |
| 7.4 ± 1.2 | 4.9 ± 0.5 | 10.4 ± 1.1 | 14.3 ± 1.3 | ROS 17/2.8 | |
| 6.9 ± 0.8 | 3.9 ± 0.6 | 9.9 ± 1.1 | 13.2 ± 1.7 | HeLa | |
| 8.4 ± 1.3 | 5.5 ± 0.7 | 10.8 ± 1.9 | 16.0 ± 2.4 | ROS 17/2.8 | |
| 1.0 ± 0.1 | 1.6 ± 0.3 | 1.6 ± 0.2 | 1.3 ± 0.2 | HeLa | |
| 1.0 ± 0.1 | 1.3 ± 0.1 | 1.5 ± 0.1 | 1.7 ± 0.3 | ROS 17/2.8 | |
| 1.0 ± 0.1 | 1.2 ± 0.2 | 1.2 ± 0.2 | 1.0 ± 0.2 | HeLa | |
| 1.2 ± 0.1 | 1.1 ± 0.1 | 1.4 ± 0.2 | 1.1 ± 0.1 | ROS 17/2.8 | |
Values shown are normalized CAT activity expressed as fold suppression. n = 18 samples per group for each cell line.
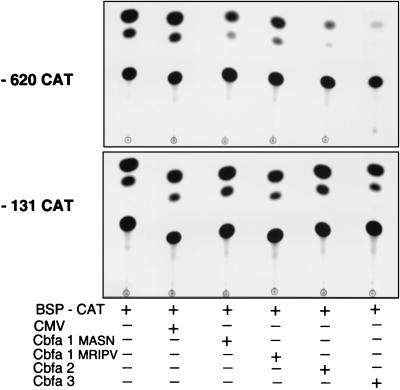
Because the full-length promoter in HeLa and ROS 17/2.8 cells exhibited weaker suppression than in Gallus osteoblasts, we examined Cbfa responsiveness of the bp −620 and −524 promoter deletions which are expressed at higher basal levels and each contain four Cbfa sites (Table 1). The bp −620 promoter construct contains an AP-1 site, which has been deleted from the bp −524 construct. The comparison of these promoters was viewed as necessary since AP-1 proteins can strongly regulate bone-related genes (e.g., those for OC and collagenase 3) and can interact with Cbfa factors (49). The deletion of the AP-1 motif does not influence this suppression. We observe a significant level of repression by Cbfa1 in these shorter segments (from two- to threefold in the kb −1.2 BSP promoter to seven- to eight fold in the bp −620 and −524, promoter fragments). Complete loss of repression was observed with the bp −131 promoter which lacks Cbfa motifs. A representative example of an autoradiogram demonstrating the extent of suppression is shown in Figure 8. In the mammalian cells, we consistently observed a greater fold suppression of the proximal promoter bp −620 and −524 by Cbfa3 and Cbfa2 (from 8- to 16-fold) than by Cbfa1 (four- to eightfold). These differences were observed at several concentrations of expression plasmid (data not shown), are independent of the cell type, and are not a consequence of differences in expression levels, as confirmed by Western blot analysis (Fig. 7C and data not shown). In the same experiment, the kb −1.1 rat OC promoter was similarly examined and the expected upregulation occurred with all three Cbfa expression plasmids (data not shown), ranging from four- to sixfold, as previously reported (2).
FIG. 8.
Representative CAT autoradiogram of Cbfa-mediated suppression of the BSP promoter. ROS 17/2.8 cells were transiently transfected with −620 BSP-CAT containing four Cbfa binding sites (upper panel) and −131 BSP-CAT lacking Cbfa sites (lower panel) and the indicated Cbfa expression constructs. CMV, empty vectors.
In order to establish that suppression of BSP promoter activity is indeed mediated by Cbfa sites, we introduced point mutations (Fig. 1) in each of the four Cbfa sites in the bp −524 promoter (Fig. 9). Interestingly, we observed a twofold increase in basal promoter activity of the mutant promoter (Fig. 9B) and a loss of suppressor activity in response to Cbfa factors (Fig. 9C). These results further confirm the role of Cbfa sites in mediating repressive activity on the BSP promoter. Because suppression of BSP by Cbfa proteins was not anticipated, we examined the effect of Cbfa factors on the expression of the endogenous BSP gene. Gallus primary osteoblasts were transiently transfected with either empty vector or Cbfa expression constructs, and total cellular RNA was isolated 24 h posttransfection. Figure 9D shows a 31 to 39% decrease in BSP mRNA levels in osteoblasts in response to forced expression of Cbfa factors. This decrease in BSP expression, observed in a transient assay, defines the physiological significance of Cbfa suppression of the BSP promoter in osteoblasts.
Cbfa suppression of the BSP promoter is not mediated through Cbfa interactions with the known repressor protein partner TLE.
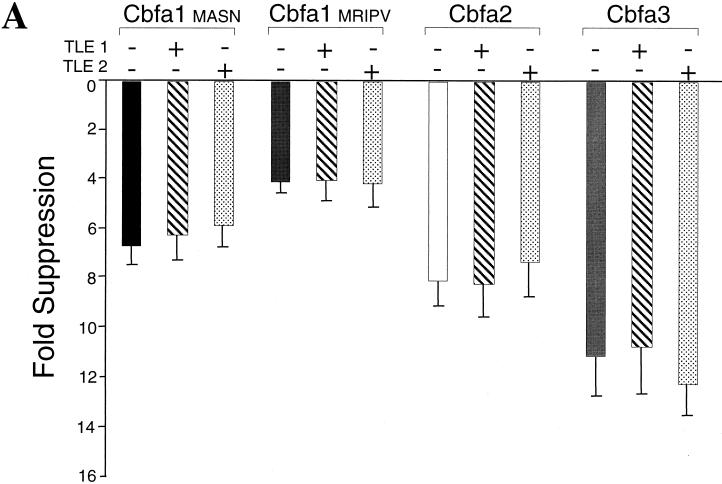
Drosophila Groucho and the related human TLE proteins (TLE1 and TLE2) (53) are well characterized repressors of Cbfa-dependent transcription. The mechanism of this suppression involves a specific protein-protein interaction between TLE proteins and the VWRPY motif in the C terminus of all Cbfa factors. Earlier studies of the OC promoter demonstrated that TLE suppresses Cbfa-mediated transactivation of the OC promoter (18, 55). We anticipated that contransfection of the Cbfa factors with TLE proteins might lead to further suppression or potentially relieve the downregulation of promoter activity by Cbfa. TLE expression plasmids at several concentrations had no effect on activity of the full-length or proximal BSP promoters (data not shown). The effects of forced Cbfa expression in the presence or absence of TLE1 or TLE2 expression on the bp −620 BSP promoter are shown in Figure 10A. The fold suppression mediated by Cbfa factors or Cbfa1 isoforms was not altered by the presence of TLE proteins. Similar results were obtained using the full-length kb −1.2 promoter in osseous and nonosseous cell lines (data not shown). These observations suggested that cellular levels of TLE proteins may be sufficient to provide maximal suppressor activity of Cbfa on the BSP promoter or that suppression is independent of TLE interactions with Cbfa.
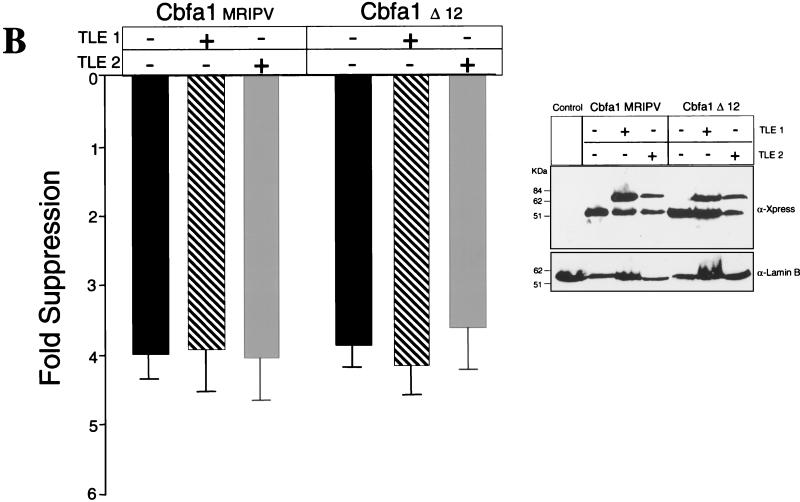
FIG. 10.
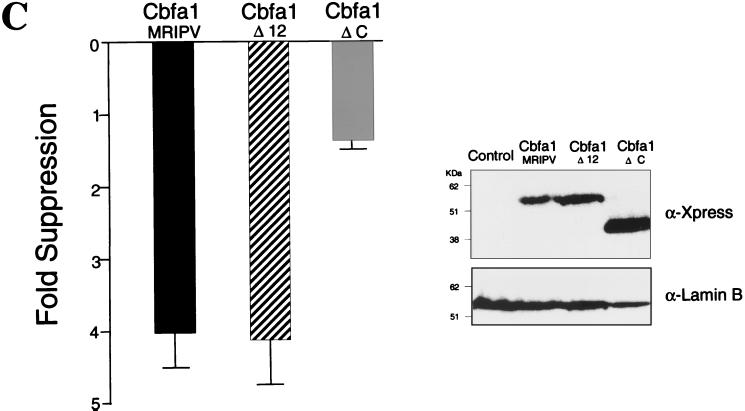
(Continued) TLE repressor proteins do not modulate Cbfa-mediated repression of the Gallus BSP promoter. (A) Coexpression of 0.75 μg of TLE1 or TLE2 with an equal amount of Cbfa expression plasmid with bp −620 Gallus BSP promoter in transiently transfected ROS 17/2.8 cells. The MRIPV and MASN Cbfa1 isoforms are also compared. (B) Cbfa1 Δ12 expression plasmid encodes a mutant which does not include the last 12 amino acids and the conserved VWRPY motif. All groups for panels A and B were assayed in the same experiment, and each bar represents the data for several wells (n = 12). Error bars, SEM. Statistically significant differences were not found by ANOVA. HeLa cells were transiently cotransfected in parallel and assayed for expression levels of these proteins. The blot was incubated with anti-Xpress antibody to detect epitope-tagged TLE and Cbfa proteins. Lamin B was evaluated to assess sample loading. (C) C-terminal domain of Cbfa1 contributes to suppression of the Gallus BSP promoter. The Cbfa1ΔC mutant lacking amino acids 362 to 513 is compared to the WT and to the Cbfa1Δ12 mutant for functional activity on the bp −620 BSP promoter. The inset shows Western blot analysis indicating comparable levels of expression of Cbfa1 proteins.
To address these possibilities, we constructed a series of C-terminal deletion mutations of Cbfa1. The Cbfa1Δ12 expression construct produces a protein that lacks the last 12 amino acids including the conserved VWRPY motif that interacts with TLE proteins. Figure 10B shows that suppressive activity on the BSP promoter by Cbfa1Δ12 is equivalent to that of the wild-type Cbfa1 and that the coexpression of either TLE1 or TLE2 has no effect on this suppression. These results indicate that repression of the BSP promoter by Cbfa1 does not involve TLE interaction. Since the Cbfa1Δ12 exhibited equivalent suppressive activity to (WT)Cbfa1, the repressor activity must reside in another domain of the protein. The C termini of Cbfa factors contain several functional domains, including a 31-amino-acid sequence that is required for trafficking these nuclear factors to subnuclear sites that support transcription (61). Thus, we compared the effect of WT Cbfa1 (from amino acids 1 to 513) and Cbfa1ΔC (from amino acids 1 to 361), lacking the C terminus, on BSP promoter activity. Although DNA binding activity is retained, this mutant protein is not targeted to the nuclear matrix (54). Interestingly, we observed a loss of suppressor function by this mutant protein on BSP promoter activity (Fig. 10C). Taken together, these results indicate that BSP promoter activity and expression (mRNA) in osteoblasts is downregulated by Cbfa1 interaction with Cbfa motifs, but does not involve Cbfa-TLE protein-protein interactions. A different domain in Cbfa1, residing between amino acids 362 and 501, may contribute to Cbfa-mediated suppression of BSP promoter activity.
DISCUSSION
The present studies clearly demonstrate, by several lines of evidence, that the Cbfa family of transcription factors mediate suppression of the Gallus BSP promoter in osseous and nonosseous cell lines. This repression occurred in rodent and human cells (2- to 16-fold) but was most evident (14- to 32-fold) in the species homologous Gallus osteoblasts. Notably, the mouse BSP promoter has been characterized with respect to its responsiveness to Cbfa1 and no consensus Cbfa sequences were defined within the proximal 1.2 kb, nor could the promoter be modulated by forced expression of Cbfa (3). The mouse and rat share similar promoter organizational properties and are in contrast to Gallus and humans, which are more closely related. Thus, downregulation of BSP by Cbfa appears to be more important for regulation in humans and Gallus than for that in rodent species, but the reason why is not clear.
Cbfa factors have been reported to mediate activation of genes related to differentiation of either hematopoietic cells (T- and B-cell differentiation) or osteoblasts, for example, the mouse germ line alpha immunoglobulin promoter (50) and OC promoters (1). Presently, a few gene promoters are known to be downregulated by Cbfa factors (p21 and the multidrug resistance [MDR] genes [34]). However, the suppressive effects on promoter activity were observed only in certain cell types and are either mSin3A- or TLE-dependent (24, 34). In contrast to those studies, we show that suppression of BSP is cell type independent and that both the promoter and endogenous transcripts are downregulated by Cbfa factors. Furthermore, the suppression mechanism does not involve the interaction of TLE proteins. Our findings, which indicate that the seven Cbfa consensus sequences within 1.2 kb of the Gallus BSP promoter mediate a repression of BSP transcription concurrent with the activation of OC in osteoblasts, can be summarized as follows: (i) deletion of 600 bp of distal BSP promoter sequences (from kb −1.2 to −0.6) containing three Cbfa sites raises promoter activity three- to fourfold in osteoblasts and HeLa cells, (ii) forced expression of Cbfa factors in HeLa cells and in rat and Gallus osteoblasts leads to marked downregulation of BSP promoter activity and mRNA levels (31 to 39%) of the BSP gene in osteoblasts, (iii) mutation of Cbfa motifs (bp −524) increases (twofold) basal promoter activity and loss of repression in response to forced expression of Cbfa factors, (iv) competition studies and antibody supershift analyses demonstrate sequence-specific binding of Cbfa factors to the consensus regulatory elements, and, lastly, (v) deletion of the established interaction domain of Cbfa1 with TLE does not affect repressor activity (in contrast, deletion of the entire Cbfa1 C terminus [amino acids 362 to 513] results in loss of suppressor function, suggesting that other regulatory sequences are required for activity). These observations raise several significant questions related to the biology of regulation of this bone-restricted marker of osteoblast differentiation by the Cbfa proteins, as well as the precise molecular mechanism for repressor activity in osteoblasts involving Cbfa1.
The first question raised by our observations is the apparent inconsistency between Cbfa repressor activity on the BSP promoter and the role of Cbfa1 as a critical determinant of osteogenesis and osteoblast differentiation in vivo. Similar to the bone-specific OC gene, BSP mRNA is upregulated in concert with the active deposition of minerals (36, 41, 43, 45, 57). However, BSP is temporally expressed earlier than OC during development of the osteoblast phenotype and is downregulated in mature osteocytes (8, 23, 35, 57, 60). Cbfa1 DNA binding activity is highest in mature osteoblasts (2), when the decline in BSP mRNA is observed (57). This contrasting regulation between the BSP and OC promoters by Cbfa is consistent with other findings. Notably, BSP and OC are quite distinct with respect to their tissue-specific functions and hormonal regulation of expression. For example, vitamin D, dexamethasone, and transforming growth factor β (TGF-β) have opposing effects on transcription of these two genes. BSP is an RGD-containing phosphorylated sialoprotein (5, 17, 40, 60). RGD-containing bone-related proteins (9, 16, 17, 39) have been of considerable interest because they have been shown to interact specifically with the integrin isotypes found on osteoclasts (46, 47). As such, these molecules may be important in the processes of resorption, and, therefore, BSP may require well-controlled transcriptional levels in mature osteoblasts possessing high levels of Cbfa1 in order for bone resorption to be tightly regulated. In this regard, vitamin D, which can promote bone turnover and is a strong enhancer of the OC gene, is a negative regulator of BSP. Thus, OC and BSP genes are similar with respect to bone tissue-specific expression but are distinct with respect to the timing of their expression, their protein properties, their spatial distribution in the bone tissue, and their functions. Thus, it may be necessary to differentially regulate OC and BSP expression through a tissue-restricted transcription factor. Cbfa factors may attenuate BSP levels at certain stages of osteoblast differentiation to accommodate the functional role of BSP in bone.
The question remains as to how BSP is initially activated in osteoblasts, if not by Cbfa1. Studies have shown that Cbfa1 is necessary but not sufficient for osteogenic differentiation (32). For example, both BMP-2 and TGF-β can induce expression of Cbfa1 and homeodomain proteins in the nonosseous myogenic cell line C2C12 (6, 32), but only BMP-2 can support a change to the osteogenic phenotype with concomitant expression of OC and other osteoblast markers (27). Thus Cbfa1 may be inducing or interacting with an unknown factor in response to BMP-2 that functions as a transcriptional activator, either directly as a DNA binding protein or indirectly as a Cbfa partner protein. BMP-2 induces homeodomain proteins (32, 44a) which have been shown to support bone-specific BSP expression and mediate enhancer activity of the BSP promoter (4). Both OC and BSP, as well as collagen type I expressed in bone, have similar homeodomain binding motifs which have been demonstrated to support tissue-restricted expression of OC and collagen in osteoblasts (20, 48, 56).
Several mechanisms may contribute to Cbfa1 suppression of BSP transcription. Our findings indicate that the C terminus of Cbfa1 contains an important regulatory domain specific to repression of the BSP promoter. The mutant CbfaΔC lacking amino acids 362 to 513 (known to interact with a variety of partner proteins) fails to repress the BSP promoter. Thus, a corepressor Cbfa1 interacting partner may participate in transcriptional regulation within the context of BSP Cbfa motifs. Our mutational evidence has proven that Cbfa elements can mediate repression in the BSP promoter, in addition to the activation function that has been shown for other promoters (e.g., OC, TGF-β type 1 receptor, and collagenase 3). Therefore, the context of the Cbfa sequences within a promoter must contribute to the formation of a Cbfa regulatory complex, mediating either repression or activation. While multimers of the Cbfa consensus sequence fused to a reporter result in enhancer activity (13, 55), the multiplicity of Cbfa sites noted within various promoters (rat OC [37] and TGF-β type 1 receptor [26]) may provide one mechanism for context-dependent modulation of activity through structural organization of gene promoters. We have demonstrated for the OC gene that the three Cbfa sites contribute to formation and maintenance of promoter architecture (25). Cbfa factors are known to interact with proteins that influence chromatin architecture (e.g., p300 histone acetyltransferases [30], LEF-ALY [19], and TLE [7, 18, 44]). In the BSP promoter, if Cbfa binding facilitates promoter architecture and chromatin structure, the binding of Cbfa factors may result in secondary interactions allowing accessibility of potential transcription factors that suppress promoter activity.
In conclusion, we have demonstrated promoter context-dependent Cbfa-mediated suppression of the osteoblast-related BSP gene by several lines of evidence. This first demonstration of negative promoter regulation by Cbfa1 for an osteoblast-expressed gene illustrates the necessity for precise control of expression of a gene that is temporally and spatially restricted in bone tissue and during development of the osteoblast phenotype.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (grant no. AR39588, DE12528, AR45689, and HD22400).
REFERENCES
- 1.Banerjee C, Hiebert S W, Stein J L, Lian J B, Stein G S. An AML-1 consensus sequence binds an osteoblast-specific complex and transcriptionally activates the osteocalcin gene. Proc Natl Acad Sci USA. 1996;93:4968–4973. doi: 10.1073/pnas.93.10.4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banerjee C, McCabe L R, Choi J-Y, Hiebert S W, Stein J L, Stein G S, Lian J B. Runt homology domain proteins in osteoblast differentiation: AML-3/CBFA1 is a major component of a bone specific complex. J Cell Biochem. 1997;66:1–8. doi: 10.1002/(sici)1097-4644(19970701)66:1<1::aid-jcb1>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 3.Benson M D, Aubin J E, Xiao G, Thomas P E, Franceschi R T. Cloning of a 2.5 kb murine bone sialoprotein promoter fragment and functional analysis of putative Osf2 binding sites. J Bone Miner Res. 1999;14:396–405. doi: 10.1359/jbmr.1999.14.3.396. [DOI] [PubMed] [Google Scholar]
- 4.Benson M D, Bargeon J L, Xiao G, Thomas P E, Kim A, Cui Y, Franceschi R T. Identification of a homeodomain binding element in the bone sialoprotein gene promoter that is required for its osteoblast-selective expression. J Biol Chem. 2000;275:13907–13917. doi: 10.1074/jbc.275.18.13907. [DOI] [PubMed] [Google Scholar]
- 5.Bianco P, Fisher L W, Young M F, Termine J D, Robey P G. Expression of bone sialoprotein (BSP) in developing human tissues. Calcif Tissue Int. 1991;49:421–426. doi: 10.1007/BF02555854. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Ji X, Harris M A, Feng J Q, Karsenty G, Celeste A J, Rosen V, Mundy G R, Harris S E. Differential roles for bone morphogenetic protein (BMP) receptor type IB and IA in differentiation and specification of mesenchymal precursor cells to osteoblast and adipocyte lineages. J Cell Biol. 1998;142:295–305. doi: 10.1083/jcb.142.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen G, Fernandez J, Mische S, Courey A J. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cowles E A, Derome M E, Pastizzo G, Brailey L L, Gronowicz G A. Mineralization and the expression of matrix proteins during in vivo bone development. Calcif Tissue Int. 1998;62:74–82. doi: 10.1007/s002239900397. [DOI] [PubMed] [Google Scholar]
- 9.Denhardt D T, Guo X. Osteopontin: a protein with diverse functions. FASEB J. 1993;7:1475–1482. [PubMed] [Google Scholar]
- 10.Dignam J D, Lebovitz R M, Roeder R G. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ducy P, Zhang R, Geoffroy V, Ridall A L, Karsenty G. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 1997;89:747–754. doi: 10.1016/s0092-8674(00)80257-3. [DOI] [PubMed] [Google Scholar]
- 12.Fujisawa R, Mizuno M, Nodasaka Y, Kuboki Y. Attachment of osteoblastic cells to hydroxyapatite crystals by a synthetic peptide (Glu7-Pro-Arg-Gly-Asp-Thr) containing two functional sequences of bone sialoprotein. Matrix Biol. 1997;16:21–28. doi: 10.1016/s0945-053x(97)90113-x. [DOI] [PubMed] [Google Scholar]
- 13.Geoffroy V, Ducy P, Karsenty G. A PEBP2α/AML-1-related factor increases osteocalcin promoter activity through its binding to an osteoblast-specific cis-acting element. J Biol Chem. 1995;270:30973–30979. doi: 10.1074/jbc.270.52.30973. [DOI] [PubMed] [Google Scholar]
- 14.Gerstenfeld L C, Chipman S D, Glowacki J, Lian J B. Expression of differentiated function by mineralizing cultures of chicken osteoblasts. Dev Biol. 1987;122:49–60. doi: 10.1016/0012-1606(87)90331-9. [DOI] [PubMed] [Google Scholar]
- 15.Gerstenfeld, L. C., S. S. Shih, C. George, S. Mizunmo, and J. Glowacki. Effect of overexpression of bone sialoprotein or osteocalcin on osteosarcoma tissue growth and mineralization Proc. Am. Acad. Orthoped. Surgeons, in press.
- 16.Gotoh Y, Gerstenfeld L C, Glimcher M J. Identification and characterization of the major chicken bone phosphoprotein. Analysis of its synthesis by cultured embryonic chick osteoblasts. Eur J Biochem. 1990;187:49–58. doi: 10.1111/j.1432-1033.1990.tb15276.x. [DOI] [PubMed] [Google Scholar]
- 17.Gotoh Y, Pierschbacher M D, Grzesiak J J, Gerstenfeld L, Glimcher M J. Comparison of two phosphoproteins in chicken bone and their similarities to the mammalian bone proteins, osteopontin and bone sialoprotein II. Biochem Biophys Res Commun. 1990;173:471–479. doi: 10.1016/s0006-291x(05)81082-4. [DOI] [PubMed] [Google Scholar]
- 18.Guo B, Javed A, Green J, Stein J L, Lian J B, van Wijnen A J, Stein G S. Groucho/TLE proteins associate with the nuclear matrix and repress Cbfa/AML mediated transactivation on osteocalcin promoter, abstr. Bone. 1998;23:S184–S184. [Google Scholar]
- 19.Hernandez-Munain C, Roberts J L, Krangel M S. Cooperation among multiple transcription factors is required for access to minimal T-cell receptor alpha-enhancer chromatin in vivo. Mol Cell Biol. 1998;18:3223–3233. doi: 10.1128/mcb.18.6.3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann H M, Beumer T L, Rahman S, McCabe L R, Banerjee C, Aslam F, Tiro J A, van Wijnen A J, Stein J L, Stein G S, Lian J B. Bone tissue-specific transcription of the osteocalcin gene: role of an activator osteoblast-specific complex and suppressor hox proteins that bind the OC box. J Cell Biochem. 1996;61:310–324. doi: 10.1002/(sici)1097-4644(19960501)61:2<310::aid-jcb14>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 21.Hoffmann H M, Catron K M, van Wijnen A J, McCabe L R, Lian J B, Stein G S, Stein J L. Transcriptional control of the tissue-specific, developmentally regulated osteocalcin gene requires a binding motif for the Msx family of homeodomain proteins. Proc Natl Acad Sci USA. 1994;91:12887–12891. doi: 10.1073/pnas.91.26.12887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hunter G K, Goldberg H A. Nucleation of hydroxyapatite by bone sialoprotein. Proc Natl Acad Sci USA. 1993;90:8562–8565. doi: 10.1073/pnas.90.18.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ibaraki K, Termine J D, Whitson S W, Young M F. Bone matrix mRNA expression in differentiating fetal bovine osteoblasts. J Bone Miner Res. 1992;7:743–754. doi: 10.1002/jbmr.5650070704. [DOI] [PubMed] [Google Scholar]
- 24.Javed A, Guo B, Hiebert S, Choi J-Y, Green J, Zhao S-C, Osborne M A, Stifani S, Stein J L, Lian J B, van Wijnen A J, Stein G S. Groucho/TLE/R-Esp proteins associate with the nuclear matrix and repress RUNX (CBFα/AML/PEBP2α) dependent activation of tissue-specific gene transcription. J Cell Sci. 2000;113:2221–2231. doi: 10.1242/jcs.113.12.2221. [DOI] [PubMed] [Google Scholar]
- 25.Javed A, Gutierrez S, Montecino M, van Wijnen A J, Stein J L, Stein G S, Lian J B. Multiple Cbfa/AML sites in the rat osteocalcin promoter are required for basal and vitamin D responsive transcription and contribute to chromatin organization. Mol Cell Biol. 1999;19:7491–7500. doi: 10.1128/mcb.19.11.7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ji C, Casinghino S, Chang D J, Chen Y, Javed A, Ito Y, Hiebert S W, Lian J B, Stein G S, McCarthy T L, Centrella M. CBFa(AML/PEBP2)-related elements in the TGF-beta type I receptor promoter and expression with osteoblast differentiation. J Cell Biochem. 1998;69:353–363. [PubMed] [Google Scholar]
- 27.Katagiri T, Yamaguchi A, Komaki M, Abe E, Takahashi N, Ikeda T, Rosen V, Wozney J M, Fujisawa-Sehara A, Suda T. Bone morphogenetic protein-2 converts the differentiation pathway of C2C12 myoblasts into the osteoblast lineage. J Cell Biol. 1994;127:1755–1766. doi: 10.1083/jcb.127.6.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kerr J M, Hiscock D R, Grzesik W, Robey P G, Young M F. The human bone sialoprotein gene contains an NF-E1/YY1 cis-acting sequence with putative regulatory activity. Calcif Tissue Int. 1997;60:276–282. doi: 10.1007/s002239900229. [DOI] [PubMed] [Google Scholar]
- 29.Kim R H, Shapiro H S, Li J J, Wrana J L, Sodek J. Characterization of the human bone sialoprotein (BSP) gene and its promoter sequence. Matrix Biol. 1994;14:31–40. doi: 10.1016/0945-053x(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 30.Kitabayashi I, Yokoyama A, Shimizu K, Ohki M. Interaction and functional cooperation of the leukemia-associated factors AML1 and p300 in myeloid cell differentiation. EMBO J. 1998;17:2994–3004. doi: 10.1093/emboj/17.11.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson R T, Gao Y-H, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. doi: 10.1016/s0092-8674(00)80258-5. [DOI] [PubMed] [Google Scholar]
- 32.Lee M H, Javed A, Kim H J, Shin H I, Gutierrez S, Choi J Y, Rosen V, Stein J L, van Wijnen A J, Stein G S, Lian J B, Ryoo H M. Transient upregulation of CBFA1 in response to bone morphogenetic protein-2 and transforming growth factor β1 in C2C12 myogenic cells coincides with suppression of the myogenic phenotype but is not sufficient for osteoblast differentiation. J Cell Biochem. 1999;73:114–125. [PubMed] [Google Scholar]
- 33.Li J J, Kim R H, Zhang Q, Ogata Y, Sodek J. Characteristics of vitamin D3 receptor (VDR) binding to the vitamin D response element (VDRE) in rat bone sialoprotein gene promoter. Eur J Oral Sci. 1998;106:408–417. doi: 10.1111/j.1600-0722.1998.tb02207.x. [DOI] [PubMed] [Google Scholar]
- 34.Lutterbach B, Westendorf J J, Linggi B, Isaac S, Seto E, Hiebert S W. A mechanism of repression by acute myeloid leukemia-1, the target of multiple chromosomal translocations in acute leukemia. J Biol Chem. 2000;275:651–656. doi: 10.1074/jbc.275.1.651. [DOI] [PubMed] [Google Scholar]
- 35.Malaval L, Liu F, Roche P, Aubin J E. Kinetics of osteoprogenitor proliferation and osteoblast differentiation in vitro. J Cell Biochem. 1999;74:616–627. [PubMed] [Google Scholar]
- 36.Manduca P, Palermo C, Caruso C, Brizzolara A, Sanguineti C, Filanti C, Zicca A. Rat tibial osteoblasts III: propagation in vitro is accompanied by enhancement of osteoblast phenotype. Bone. 1997;21:31–39. doi: 10.1016/s8756-3282(97)00037-9. [DOI] [PubMed] [Google Scholar]
- 37.Merriman H L, van Wijnen A J, Hiebert S, Bidwell J P, Fey E, Lian J, Stein J, Stein G S. The tissue-specific nuclear matrix protein, NMP-2, is a member of the AML/CBF/PEBP2/runt domain transcription factor family: interactions with the osteocalcin gene promoter. Biochemistry. 1995;34:13125–13132. doi: 10.1021/bi00040a025. [DOI] [PubMed] [Google Scholar]
- 38.Meyers S, Lenny N, Sun W-H, Hiebert S W. AML-2 is a potential target for transcriptional regulation by the t(8;21) and t(12;21) fusion proteins in acute leukemia. Oncogene. 1996;13:303–312. [PubMed] [Google Scholar]
- 39.Moore M A, Gotoh Y, Rafidi K, Gerstenfeld L C. Characterization of a cDNA for chicken osteopontin: expression during bone development, osteoblast differentiation, and tissue distribution. Biochemistry. 1991;30:2501–2508. doi: 10.1021/bi00223a029. [DOI] [PubMed] [Google Scholar]
- 40.Nagata T, Todescan R, Goldberg H A, Zhang Q, Sodek J. Sulphation of secreted phosphoprotein I (SPPI, osteopontin) is associated with mineralized tissue formation. Biochem Biophys Res Commun. 1989;165:234–240. doi: 10.1016/0006-291x(89)91059-0. [DOI] [PubMed] [Google Scholar]
- 41.Nefussi J R, Brami G, Modrowski D, Oboeuf M, Forest N. Sequential expression of bone matrix proteins during rat calvaria osteoblast differentiation and bone nodule formation in vitro. J Histochem Cytochem. 1997;45:493–503. doi: 10.1177/002215549704500402. [DOI] [PubMed] [Google Scholar]
- 42.Otto F, Thornell A P, Crompton T, Denzel A, Gilmour K C, Rosewell I R, Stamp G W H, Beddington R S P, Mundlos S, Olsen B R, Selby P B, Owen M J. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. doi: 10.1016/s0092-8674(00)80259-7. [DOI] [PubMed] [Google Scholar]
- 43.Owen T A, Soprano D R, Soprano K J. Analysis of the growth factor requirements for stimulation of WI-38 cells after extended periods of density-dependent growth arrest. J Cell Physiol. 1989;139:424–431. doi: 10.1002/jcp.1041390227. [DOI] [PubMed] [Google Scholar]
- 44.Palaparti A, Baratz A, Stifani S. The Groucho/transducin-like enhancer of split transcriptional repressors interact with the genetically defined amino-terminal silencing domain of histone H3. J Biol Chem. 1997;272:26604–26610. doi: 10.1074/jbc.272.42.26604. [DOI] [PubMed] [Google Scholar]
- 44a.Pera E, Stein S, Kessel M. Ectodermal patterning in the avian embryo: epidermis versus neural plate. Development. 1999;126:63–73. doi: 10.1242/dev.126.1.63. [DOI] [PubMed] [Google Scholar]
- 45.Pockwinse S M, Wilming L G, Conlon D M, Stein G S, Lian J B. Expression of cell growth and bone specific genes at single cell resolution during development of bone tissue-like organization in primary osteoblast cultures. J Cell Biochem. 1992;49:310–323. doi: 10.1002/jcb.240490315. [DOI] [PubMed] [Google Scholar]
- 46.Roach H I. Trans-differentiation of hypertrophic chondrocytes into cells capable of producing a mineralized bone matrix. Bone Miner. 1992;19:1–20. doi: 10.1016/0169-6009(92)90840-a. [DOI] [PubMed] [Google Scholar]
- 47.Ross F P, Chappel J, Alvarez J I, Sander D, Butler W T, Farach-Carson M C, Mintz K A, Robey P G, Teitelbaum S L, Cheresh D A. Interactions between the bone matrix proteins osteopontin and bone sialoprotein and the osteoclast integrin alpha v beta 3 potentiate bone resorption. J Biol Chem. 1993;268:9901–9907. [PubMed] [Google Scholar]
- 48.Rossert J A, Chen S S, Eberspaecher H, Smith C N, de Crombrugghe B. Identification of a minimal sequence of the mouse pro-α1(I) collagen promoter that confers high-level osteoblast expression in transgenic mice and that binds a protein selectively present in osteoblasts. Proc Natl Acad Sci USA. 1996;93:1027–1031. doi: 10.1073/pnas.93.3.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Selvamurugan N, Partridge N C. Constitutive expression and regulation of collagenase-3 in human breast cancer cells. Mol Cell Biol Res Commun. 2000;3:218–223. doi: 10.1006/mcbr.2000.0215. [DOI] [PubMed] [Google Scholar]
- 50.Shi M J, Stavnezer J. CBF alpha3 (AML2) is induced by TGF-β1 to bind and activate the mouse germline Ig alpha promoter. J Immunol. 1998;161:6751–6760. [PubMed] [Google Scholar]
- 51.Sleigh M J. A nonchromatographic assay for expression of the chloramphenicol acetyltransferase gene in eucaryotic cells. Anal Biochem. 1986;156:251–256. doi: 10.1016/0003-2697(86)90180-6. [DOI] [PubMed] [Google Scholar]
- 52.Stewart M, Terry A, Hu M, O'Hara M, Blyth K, Baxter E, Cameron E, Onions D E, Neil J C. Proviral insertions induce the expression of bone-specific isoforms of PEBP2αA (CBFA1): evidence for a new myc collaborating oncogene. Proc Natl Acad Sci USA. 1997;94:8646–8651. doi: 10.1073/pnas.94.16.8646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stifani S, Blaumueller C M, Redhead N J, Hill R E, Artavanis-Tsakonas S. Human homologs of a Drosophila enhancer of split gene product define a novel family of nuclear proteins. Nat Genet. 1992;2:119–127. doi: 10.1038/ng1092-119. [DOI] [PubMed] [Google Scholar]
- 54.Tang L, Guo B, Javed A, Choi J-Y, Hiebert S, Lian J B, van Wijnen A J, Stein J L, Stein G S, Zhou G W. Crystal structure of the nuclear matrix targeting signal of the transcription factor AML-1/PEBP2αB/CBFα2. J Biol Chem. 1999;274:33580–33586. doi: 10.1074/jbc.274.47.33580. [DOI] [PubMed] [Google Scholar]
- 55.Thirunavukkarasu K, Mahajan M, McLarren K W, Stifani S, Karsenty G. Two domains unique to osteoblast-specific transcription factor Osf2/Cbfa1 contribute to its transactivation function and its inability to heterodimerize with Cbf β. Mol Cell Biol. 1998;18:4197–4208. doi: 10.1128/mcb.18.7.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Towler D A, Rutledge S J, Rodan G A. Msx-2/Hox 8.1: a transcriptional regulator of the rat osteocalcin promoter. Mol Endocrinol. 1994;8:1484–1493. doi: 10.1210/mend.8.11.7877617. [DOI] [PubMed] [Google Scholar]
- 57.White C, Gardiner E, Eisman J. Tissue specific and vitamin D responsive gene expression in bone. Mol Biol Rep. 1998;25:45–61. doi: 10.1023/a:1006820710966. [DOI] [PubMed] [Google Scholar]
- 58.Yang R, Gerstenfeld L C. Signal transduction pathways mediating parathyroid hormone stimulation of bone sialoprotein gene expression in osteoblasts. J Biol Chem. 1996;271:29839–29846. doi: 10.1074/jbc.271.47.29839. [DOI] [PubMed] [Google Scholar]
- 59.Yang R, Gerstenfeld L C. Structural analysis and characterization of tissue and hormonal responsive expression of the avian bone sialoprotein (BSP) gene. J Cell Biochem. 1997;64:77–93. doi: 10.1002/(sici)1097-4644(199701)64:1<77::aid-jcb11>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 60.Yang R, Gotoh Y, Moore M A, Rafidi K, Gerstenfeld L C. Characterization of an avian bone sialoprotein (BSP) cDNA: comparisons to mammalian BSP and identification of conserved structural domains. J Bone Miner Res. 1995;10:632–640. doi: 10.1002/jbmr.5650100417. [DOI] [PubMed] [Google Scholar]
- 61.Zeng C, van Wijnen A J, Stein J L, Meyers S, Sun W, Shopland L, Lawrence J B, Penman S, Lian J B, Stein G S, Hiebert S W. Identification of a nuclear matrix targeting signal in the leukemia and bone-related AML/CBFα transcription factors. Proc Natl Acad Sci USA. 1997;94:6746–6751. doi: 10.1073/pnas.94.13.6746. [DOI] [PMC free article] [PubMed] [Google Scholar]