Abstract
CIITA is the master regulator of class II major histocompatibility complex gene expression. We present evidence that CIITA can self-associate via two domains: the C terminus (amino acids 700 to 1130) and the GTP-binding domain (amino acids 336 to 702). Heterotypic and homotypic interactions are observed between these two regions. Deletions within the GTP-binding domain that reduce GTP-binding and transactivation function also reduce self-association. In addition, two leucine residues in the C-terminal leucine-rich repeat region are critical for self-association as well as function. This study reveals for the first time a complex pattern of CIITA self-association. These interactions are discussed with regard to the apoptosis signaling proteins, Apaf-1 and Nod1, which share domain arrangements similar to those of CIITA.
Major histocompatibility complex (MHC) class II proteins play a critical role in the initiation of immune responses by presenting peptides from exogenous antigens to T-helper lymphocytes (11, 48). The expression of MHC class II genes is restricted to specific cell types with constitutive expression in B lymphocytes and dendritic cells. In addition, a variety of cell types can be induced to express MHC class II by the proinflammatory cytokine gamma interferon.
The cis-acting elements and the DNA-binding transcription factors that regulate MHC class II expression have been extensively characterized (6, 20). MHC class II gene promoters all contain a well-conserved tripartite arrangement of S, X, and Y sequence elements. The X box can be subdivided into X1 and X2 elements. The class II X1 box binding factor, RFX, has been shown to be defective in a rare immune deficiency, bare lymphocyte syndrome (BLS) (38, 50). Genetic complementation of the defects in these cells resulted in the identification of the components for this DNA-binding transcription factor. RFX is composed of three subunits: RFX5 (56), RFXAP (17), and RFX-B (RFXANK) (43, 46). The X2 element binding protein has recently been identified as CREB (45). While the X box is uniquely present in MHC class II promoters, the Y box is a CCAAT box that is present in a large number of eukaryotic promoters (9, 41). This site is bound by yet another heterotrimeric factor, NF-Y (40, 52).
Although the transcription factors that bind the class II boxes are required for the expression of MHC class II genes, their presence alone is not sufficient for transcriptional activation. A major breakthrough in understanding MHC class II gene expression was provided by complementation studies using the mutant cell line, RJ2.25 (1, 57). The initial isolation of the CIITA cDNA revealed a large protein with an open reading frame encoding 1,130 amino acids (57). CIITA is sufficient for induction of MHC class II gene expression when introduced into cells (10, 14, 58). In certain cases it also has been implicated in the upregulation of MHC class I transcription (21, 42). The N terminus of CIITA contains an acidic domain that can activate transcription when fused to the DNA-binding domain of GAL4 (51, 64). However, CIITA has not been demonstrated to possess any DNA-binding activity. To date, a number of proteins have been described that interact with the amino-terminal sequences of CIITA, including CBP, PCAF, TFIID components, and P-TEFb (18, 19, 31, 36, 39, 54). More recently, it has been shown that CIITA can directly bind to DNA-binding proteins which recognize the MHC class II promoter, including NF-YB, NF-YC, RFX-5, RFX-B (RFXANK), and CREB (16, 22, 65). CIITA interactions are also observed when these factors are organized into an MHC class II promoter-bound enhanceosome complex (44). These interactions support the model of CIITA functioning as a coactivator for MHC class II promoters.
CIITA also has some features unique for a transcriptional activator (24). CIITA was recognized to have sequence similarities to GTP-binding proteins (13), and subsequently CIITA has been demonstrated to be a GTP-binding protein (23). Mutation of the consensus GTP-binding motifs results in loss of function and the accumulation of CIITA in the cytoplasm. A variety of mutations in the C terminus of CIITA have been described in several BLS patient cell lines, underscoring the functional importance of this region (5, 49, 57). In one of these cell lines, defective splicing results in deletion of an exon containing a nuclear import sequence. This results in loss of transactivation and the cytoplasmic localization of CIITA (15).
In this study, we describe the self-association of CIITA. This self-association is complex and involves at least two types of interactions mediated by the peptide sequences within the GTP-binding domain of CIITA and the C terminus (amino acids 700 to 1130), which includes the leucine-rich repeats (LRR). The former is able to interact both with itself and with the LRR contained in the C terminus, while the C terminus also self-associates. Thus, multiple intermolecular, as well as intramolecular, interactions may occur. We discuss the similarities of this interaction with those displayed by Apaf-1 and Nod1, both mediators of apoptosis. CIITA serves a drastically different function as a transcriptional activator and represents the first of its kind to self-associate via a similar mechanism. The significance is discussed in the context of an emerging family of nucleotide-binding domain and LRR domain containing mammalian proteins that display similarity to CIITA.
MATERIALS AND METHODS
Cell lines and culture.
COS-7 and HeLa cells were cultured in Dulbecco modified eagle medium (high glucose) supplemented with 10% fetal bovine serum and 5 mM l-glutamine.
Plasmids.
The FLAG-CIITA expression vector has been described previously (13). The myc-CIITA pcDNA3 expression vector and the C-terminal deletion mutants FLAG-C2 1-335, 1-612, 1-793, and 336-1130 have recently been described (65). The C-terminal deletion mutants FLAG-C2 1-401, 1-650, 1-702, 1-751, and 1-852 were generated using the QuickChange mutagenesis protocol with Pfu Turbo from Stratagene (La Jolla, Calif.). The sense primers used for mutagenesis were as follows: 1-401, 5′-GGCTGAGGTGCTGTaGGCTGCCAAGGAGC-3′; 1-650, 5′-GGCCGTGCAGCCCTCtAgAGCCCCCCCGGGGCC-3′; 1-702, 5′-CCACCGCGGGCCGCAtAGTCCGAGCTGGCC-3′; 1-751, 5′-CCTATGACAACTGGCTGtAGGGCGTGCCACGC-3′; and 1-852, 5′-GGGC AAGGCCTTGtAGGCGGCGGGCCAAGACTTCTCC-3′ (lowercase letters represent mutated bases). The myc-C2 1-852, 1-793, 1-751, and 1-702 expression plasmids were generated by ligating the 1.6-kb SacII/XhoI fragment containing the stop codons into SacII/XhoI-digested myc-CIITA. The myc-C2 1-612 and 1-650 deletion mutants were prepared by using QuickChange mutagenesis to introduce stop codons at positions 613 and 651, respectively.
The expression vector pcDNA3 HisC was obtained from Invitrogen (Carlsbad, Calif.). This expression vector allows for the amino-terminal tagging of CIITA with a hexahistidine tag and an Xpress epitope tag (Xpress-CIITA). Full-length Xpress-CIITA was created by ligating the EcoRI FLAG-CIITA insert into EcoRI-digested pcDNA3 HisC. Xpress-tagged 336-1130 was generated by introduction of a BglII site into CIITA using the QuickChange mutagenesis protocol. The BglII/EcoRI fragment was inserted into BamHI/EcoRI-digested pcDNA3 HisC (Invitrogen). Xpress-CIITA 336-702 was generated after ligating the SacII/XbaI fragment of FLAG-C2 1-702 into SacII/XbaI-digested Xpress-CIITA 336-885.
The internal deletion construct FLAG-C2 1-335:700-1130 was generated by creating an in-frame SacII site at codons 335 and 336 of CIITA using QuickChange mutagenesis. SacII digestion removed the sequences between this designed site and the existing SacII site present at codons 699 and 700 of CIITA. Religation allowed the removal of sequences encoding amino acids 336 to 699. FLAG-C2 700-1130 was created by inserting the SacII/XhoI fragment into a modified pcDNA3 vector that allowed for in-frame fusion of peptide sequence to a FLAG tag using a SacII site. SacII sites were also engineered into CIITA at codons 885 and 886 and codons 938 and 939 using QuickChange mutagenesis to create FLAG-C2 886-1130 and 939-1130 expression plasmids.
Mutations in the LRR domain were generated using the QuickChange mutagenesis protocol. The oligonucleotides used for mutagenesis were as follows: L1007P, 5′-GAGGGTGTCTCGCAGCcCTCAGCCACCTTCCCC-3′; L1035P, 5′-CTGGGTGCCTACAAACcCGCCGAGGCCCTGCCT-3′; L1064P, 5′-GTGGGAGCCGAGAGCccGGCTCGTGTGCTTCCG-3′; L1092P, 5′-GCCGGGGCCCAGCAGCcCGCTGCCAGCCTTCGG-3′; L1007A, 5′-GAGGGTGTCTCGCAGCCCTCAGCCACCTTCCCC-3′; L1035A, 5′-CTGGGTGCCTACAAAgcCGCCGAGGCCCTGCCT-3′; L1064A, 5′-GTGGGAGCCGAGAGCttGGCTCGTGTGCTTCCG-3′; and, L1092A, 5′-GCCGGGGCCCAGCAGgcCGCTGCCAGCCTTCGG-3′. The mutations were transferred into the BamHI/XhoI-digested FLAG-C2 939-1130 expression vector by inserting the BamHI/XhoI insert from the full-length constructs described above. To generate full-length myc-tagged leucine-to-proline mutants of CIITA, SacII/XhoI inserts encoding the site-specific mutations were transferred into SacII/XhoI-digested myc-CIITA expression vector. To generate myc-tagged GTP1Δ, GTP2Δ, and GTP3Δ mutants, NotI inserts encoding the mutation were ligated to NotI-digested and dephosphorylated myc-CIITA. All constructs were verified by sequence analysis. The pcDNA3 expression vector for FLAG-tagged murine p38 was provided by Lee Madrid (laboratory of Albert Baldwin).
Immunoprecipitation and Western blotting.
COS-7 cells were split 2 × 105 cells per well in six-well plates the evening before transfection. Cells were transfected with 500 ng of each expression vector using Fugene 6 (Roche Molecular Biochemicals, Indianapolis, Ind.) according to the standard protocol. At 30 h after transfection, the cells were washed twice with cold phosphate-buffered saline and lysed on the plate with 700 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris, pH 7.4; 200 mM NaCl; 1% Nonidet P-40 [NP-40]; 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 5 mM EDTA, 1 mM dithiothreitol) supplemented with protease inhibitors (Complete EDTA-free; Roche). After centrifugation, 600 μl of the supernatant was transferred to a separate Eppendorf tube and incubated with 1 μg of anti-myc 9E10 or anti-Xpress antibody (Invitrogen) for 1 h at 4°C. Then, 5 μl of prewashed goat anti-mouse M-450 Dynabeads (Dynal, Oslo, Norway) was added, and the complexes were incubated with rotation for 3 h at 4°C. The complexes were washed three times for 10 min each using 500 μl of the lysis buffer. Immunoprecipitated protein was denatured using Laemmli buffer, and the samples were subjected to SDS-polyacrylamide gel electrophoresis. The gels were transferred to nitrocellulose for 1 h at 100 V. For experiments using the FLAG-C2 939-1130 expression vectors polyvinylidene difluoride (Immobilon P; Millipore, Bedford, Mass.) was used, and SDS was omitted from the transfer buffer. The membranes were blocked using 5% milk in Tris-buffered saline–Tween (10 mM Tris, pH 8.0; 150 mM NaCl; 0.05% Tween 20) for 1 h. For detection of FLAG-tagged proteins, anti-FLAG M5 was used (Sigma, St. Louis, Mo.). For detection of myc- and Xpress-tagged proteins in immunoprecipitated complexes, one-quarter of the denatured protein was run on a separate gel and blotted independently of the FLAG-tagged proteins. For detection of myc-tagged proteins, anti-myc 9E10 was used. Secondary detection of the bound primary antibody utilized horseradish peroxidase (HRP)-conjugated goat anti-mouse heavy- and light-chain (κ)-specific antibodies from Southern Biotechnology (Birmingham, Ala.). HRP detection was performed using Supersignal West Pico Chemiluminescent substrate (Pierce, Rockford, Ill.).
In vitro translation.
Plasmids were transcribed and translated in vitro using the TNT T7 Kit (Promega, Madison, Wis.). For each reaction, 1 μg of each indicated plasmid was incubated with 40 μl of the reticulocyte lysate in the presence of 2 μl of [35S]methionine for 90 min at 30°C. Then, 1 μl of the reaction was withheld for detection of the input protein, and the remainder of the reaction was split equally into 500 μl of NP-40 buffer (10 mM Tris, pH 7.4; 140 mM KCl; 20 mM NaCl; 5 mM EDTA; 1% NP-40) and into the same volume of COS-7 cell lysate prepared using the same buffer. Immunoprecipitations were performed using 1.5 μg of the anti-Xpress antibody according to the procedure described above.
Luciferase assays.
HeLa cells were split 2 × 105 cells per well in six-well plates the evening before transfection. A total of 1 μg of each CIITA expression vector was cotransfected with 1 μg of pGL2-DRA 300 (23). Cells were harvested using reporter lysis buffer (Promega) 18 h after transfection. Luciferase activity was normalized to lysate protein content using the Bio-Rad protein assay.
RESULTS
CIITA self-association can be detected in cells but not using in vitro-translated mixtures.
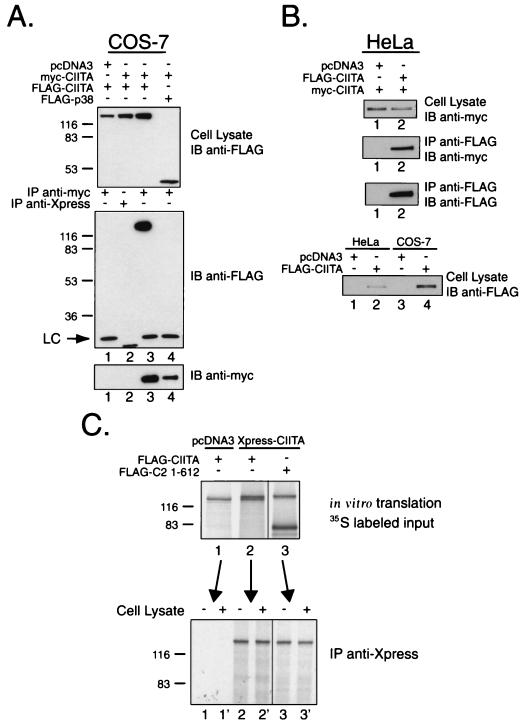
Previous analysis of CIITA indicated that certain mutants had the ability to antagonize wild-type CIITA in a dominant-negative fashion (12, 13). In order to test the hypothesis that this inhibition could be mediated through dimerization of CIITA, COS-7 cells were cotransfected with expression vectors for CIITA tagged with both myc and FLAG epitopes. Cell lysates were prepared using RIPA buffer and immunoprecipitated with anti-myc antibody. Western blots were performed to detect coimmunoprecipitation of the FLAG-tagged CIITA protein. FLAG-CIITA coimmunoprecipitated with myc-CIITA (Fig. 1A, lane 3). Immunoprecipitation with an isotype control, immunoglobulin G1 (IgG1), did not pull down either myc-CIITA or FLAG-CIITA (Fig. 1A, lane 2). As a negative control, an unrelated protein, p38, also failed to coimmunoprecipitate with CIITA (Fig. 1A, lane 4). In addition, we have previously shown that CIITA does not interact with NF-YA (65). Similar results were obtained when using a lysis buffer containing NP-40 as the sole detergent (data not shown). Comparing the signals from total input and immunoprecipitated fractions, it can be estimated that less than 5% of the input FLAG-CIITA coimmunoprecipitated. Since interactions between CIITA molecules with like tags should also occur, less than 15% of the total cellular CIITA would be self-associating in this system.
FIG. 1.
Detection of CIITA self-association using a cellular expression system. (A) FLAG-tagged CIITA coimmunoprecipitates with myc-tagged CIITA in a COS-7 cell expression system. COS-7 cells were cotransfected with pcDNA3 expression vectors encoding the full-length CIITA cDNA (clone 8, amino acids 1 to 1130) with N-terminal FLAG or myc epitope tags. The middle panel shows the results for immunoprecipitation (IP) with anti-myc 9E10 or anti-Xpress as an IgG1 negative control, followed by immunoblotting (IB) with anti-FLAG M5 antibody. The stress-activated protein kinase, p38, served as an irrelevant FLAG-tagged protein control. Expression of FLAG-tagged proteins was verified by performing an anti-FLAG M5 Western blot using 2% of the RIPA lysate (top panel). Expression and immunoprecipitation of the myc-CIITA was confirmed in the bottom panel. Detection of the light chain from the monoclonal antibodies is designated with an “LC.” (B) CIITA self-associates in HeLa cells. Expression and detection of CIITA self-association is performed using the same procedure as in panel A, with immunoprecipitation using anti-FLAG M2-conjugated agarose beads. Expression levels of FLAG-CIITA for the COS-7 and HeLa experiments are shown in the bottom panel. (C) CIITA fails to self-associate when using an in vitro translation system. FLAG-tagged CIITA was cotranslated with an Xpress-tagged CIITA. [35S]methionine-labeled in vitro-translated proteins were incubated with anti-Xpress antibody in the absence or presence of COS-7 cell lysate and immunoprecipitated with goat anti-mouse Dynabeads. No immunoprecipitation was detected for FLAG-CIITA (lanes 2 and 2′) or a C-terminal deletion mutant FLAG-C2 1-612 (lanes 3 and 3′), which was shown to coimmunoprecipitate using the COS-7 expression system (see Fig. 3B). The FLAG-tagged deletion mutant was included because of its obvious size difference from full-length Xpress-CIITA, since full-length FLAG-CIITA and Xpress-CIITA were difficult to distinguish.
To rule out the possibility that the expression of large quantities of CIITA might lead to nonspecific association, we repeated the experiment in HeLa cells. These cells do not harbor the simian virus 40 large-T antigen; thus, they do not amplify the pcDNA3 expression vector. In this experiment, CIITA was immunoprecipitated with anti-FLAG antibody. Myc-tagged CIITA coimmunoprecipitated in HeLa cells (Fig. 1B, lane 2). In three experiments, the level of CIITA expression in HeLa was approximately 1/10 that of COS-7. The transfection efficiency was monitored using green fluorescent protein cotransfection and was equivalent for both cell types (52% for HeLa, 60% for COS-7).
In contrast to the results obtained from the cell expression systems, in vitro-translated Xpress-tagged CIITA failed to coprecipitate FLAG-tagged CIITA. Full-length FLAG-CIITA was cotranslated with Xpress-tagged CIITA in the presence of [35S]methionine. The former has a slightly faster migration pattern than the latter (compare the top bands in Fig. 1C, lanes 1 and 3 which, respectively, represent FLAG-CIITA and Xpress-CIITA). When an anti-Xpress antibody was used for the immunoprecipitation, the slightly smaller FLAG-CIITA failed to coprecipitate. To better distinguish the two differentially tagged molecules, we used a FLAG-CIITA molecule that is significantly smaller and easily distinguishable from the full-length Xpress-CIITA (Fig. 1C, lane 3 [FLAG-C2 1-612]) but readily self-associates in vivo (see Fig. 3B). FLAG-C2 1-612 also did not coprecipitate with Xpress-CIITA. In a previous study, we showed that similarly prepared CIITA coprecipitates NF-YB, NF-YC, and RFX5 (65); thus, in an in vitro translation extract, CIITA, interacts with other proteins but not with itself. The failure to interact could not be rescued by the presence of COS-7 cell lysate (lanes 2′ and 3′), suggesting that the simple presence of a bridging or adapter protein was not the factor resulting in the lack of precipitation. This suggests at least two possibilities: (i) that CIITA undergoes a modification inside cells that promotes self-association and/or (ii) that an adapter molecule, if it exists, has to associate with CIITA in cells. The detection of CIITA self-association exclusively in intact cells prompted us to perform all of the following studies in transfected cells.
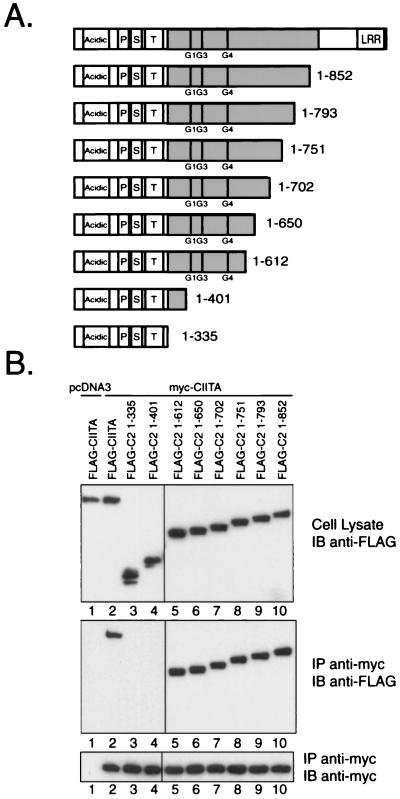
FIG. 3.
N-terminal amino acids 1 to 401 are expendable for interaction with full-length CIITA. (A) Graphical representation of the C-terminal deletion mutants employed for interaction mapping in panel B. The domain representation is the same as that used in Fig. 2A. (B) Full-length myc-tagged CIITA was cotransfected into COS-7 cells with a series of FLAG-tagged C-terminal deletion mutants. A secondary antibody specific for mouse light chain (κ) was used to avoid detection of the heavy chain. Results for the FLAG-C2 1-335, 1-612, and 1-793 deletion mutants have been repeated in excess of 10 times. Similar results are seen when an NP-40 lysis buffer was used. The results for all other deletion mutants have been repeated three times.
Mapping the self-association domains of CIITA.
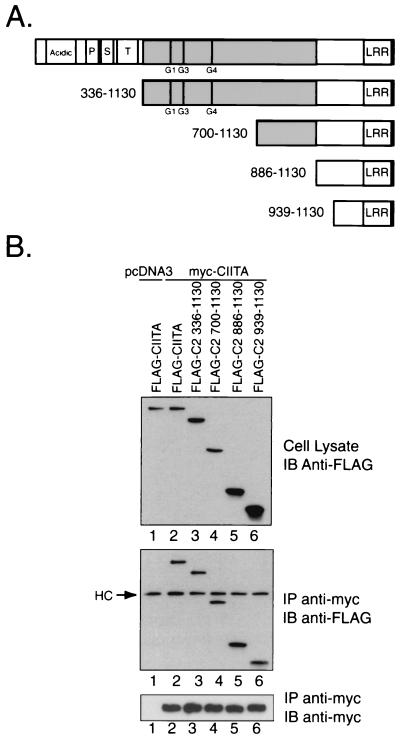
In order to characterize the domains involved in the self-association events of CIITA, we proceeded with the COS-7 cell expression system. For an initial mapping of peptide sequences required for the observed self-association, a series of FLAG-tagged N-terminal deletion mutants were employed. These mutants were coexpressed with myc-tagged full-length CIITA to examine regions required for association. Figure 2A depicts the sequences that are removed in these deletion mutants. Surprisingly, the FLAG-C2 mutants 336-1130, 700-1130, 886-1130, and 939-1130 all interacted with full-length CIITA (Fig. 2B, see lanes 3 through 6). These results provide evidence that the last 192 amino acids of CIITA contain sequences involved in the self-association. This region of CIITA contains six potential LRR, and the data are consistent with the global hypothesis that these repeat structures are frequently involved in mediating protein interactions (34).
FIG. 2.
CIITA amino acids 939 to 1130, which includes the LRR region, interacts with full-length CIITA. (A) Graphical representation of the N-terminal deletion mutants used to map sites required for the CIITA self-association. The N terminus of CIITA contains the acidic domain (30-130), and the proline (P)-, serine (S)-, and threonine (T)-rich regions. Exon 8 encodes the CIITA peptide sequence from amino acids 336 to 886, and this region is shaded in gray. GTP-binding motifs are indicated at their respective positions by G1 (phosphate binding, amino acids 420 to 427), G3 (Mg2+ coordination, amino acids 461 to 464), and G4 (guanine specificity, amino acids 558 to 561). The LRR are located at the extreme C terminus of CIITA (amino acids 957 to 1127). (B) Full-length myc-tagged CIITA was cotransfected into COS-7 with a series of FLAG-tagged N-terminal deletion mutants. An anti-FLAG M5 Western blot on 2% of the RIPA lysate validated expression of the FLAG-tagged deletion constructs. The middle panel shows an anti-FLAG M5 immunoblot after immunoprecipitation (IP) with the anti-myc 9E10 antibody. Heavy chain from the monoclonal anti-myc antibody was detected by the goat anti-mouse secondary antibody and is noted by “HC.” The results were repeated in three experiments using RIPA buffer.
We applied a similar strategy using C-terminal FLAG-tagged deletion mutants to further delineate the self-association sequences. These constructs are shown in Fig. 3A. Deletions of CIITA sequence up to amino acid 612 retained the ability to interact with the full-length myc-tagged CIITA (Fig. 3B). Further deletions beyond the phosphate-binding motif of the GTP-binding domain (G1, amino acids 421 to 428) resulted in two deletion mutants, 1-401 and 1-335, that did not associate with full-length CIITA (Fig. 3B, lanes 3 and 4). An additional deletion mutant, 1-421, was also never observed to bind to full-length CIITA, although the expression level was consistently low and hence the construct was not included in this experiment (data not shown).
The initial interaction mapping with the N- and C-terminal deletions indicates that the amino-terminal sequence of CIITA, which includes the acidic domain, the proline-, serine-, and threonine-rich regions, and the sequences up to the phosphate-binding motif, are not sufficient for CIITA self-association. Additionally, these studies identified two domains involved in self-association: residues 401 to 612, containing the GTP-binding domain, and the C-terminal LRR.
Homotypic self-association between residues 336 and 702 of CIITA.
The data from the deletion mutant analysis suggest that sequences contained in residues 1 to 612 are potentially involved in self-association (Fig. 3), while the N-terminal deletion mutant analysis suggests that the extreme C terminus (residues 939 to 1130) contained sequences that are sufficient for self-association (Fig. 2). Since both of these sets of mutants were tested against full-length CIITA as the associative partner, one explanation is that interactions among multiple domains may occur. To resolve this paradox, the analysis of another nucleotide-binding protein that self-associates provided an interesting model upon which we began to address further mapping of interaction sites. The apoptosis signaling protein, Apaf-1, displays a complex set of interactions that include multimerization of its nucleotide-binding domain (NBD) (55). Similar events are also observed in the C. elegans Apaf-1 homologue, CED-4 (63). Since it was unclear from the prior experiments how the different regions of CIITA were mediating the in vivo interaction, these regions of CIITA were isolated and tested against each other.
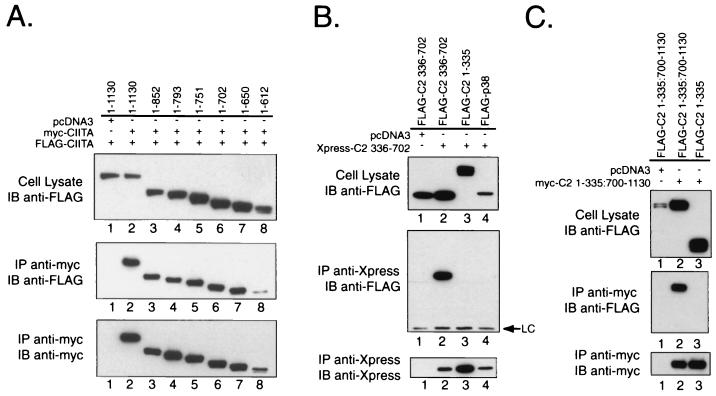
In order to determine whether the GTP-binding domain could interact with itself, two sets of experiments were performed. First, the deletion mutants used in Fig. 3 were tagged with the myc epitope and tested against identical FLAG-tagged deletion mutants. Deletion mutants FLAG-C2 1-852, 1-793, 1-751, 1-702, 1-650, and 1-612 interacted with the corresponding myc-tagged constructs (Fig. 4A), indicating that residues 1 to 612 were sufficient for self-interaction. The expression of the 1-612 construct was lower than the other constructs (Fig. 4A, top and bottom panels). This largely accounts for the lower level of coprecipitated product.
FIG. 4.
Self-association of CIITA is mediated in part by self-interactions between amino acids 336 to 702. (A) The FLAG-tagged C-terminal stop mutants used in Fig. 3 were tested in COS-7 cells for association with a myc-tagged version of the same construct. Self-association of C2 1-612 and 1-793 has been observed using both NP-40 and RIPA buffers. (B) Isolated CIITA sequences from residues 336 to 702 can multimerize. Expression vectors for amino acids 336 to 702 were designed with both FLAG and Xpress N-terminal epitope tags. Coexpression of the differentially tagged constructs from amino acids 336 to 702 in COS-7 cells resulted in coimmunoprecipitation of the FLAG-tagged protein. Coexpression of the FLAG-tagged 1-335 deletion mutant (lane 3) or the irrelevant protein control, FLAG-p38 (lane 4), did not result in coimmunoprecipitation. These results have been verified in four experiments, including one using HeLa cells. (C) CIITA residues encoding amino acids 700 to 1130 can associate with each other. A CIITA construct that deleted the region of residues 336 to 699 was made and designated as 1-335:700-1130. This removes most of the residues from 336 to 702 studied in Fig. 4B but retains the C terminus and the N-terminal region that does not participate in any form of self-association.
To further determine if the GTP-binding domain could self-associate, it was isolated from the N-terminal sequences (residues 1 to 335) and tested against itself in a coimmunoprecipitation assay. Residues 336 to 702 of CIITA were tagged with an Xpress epitope and cotransfected with a FLAG-tagged 336-702 expression vector. This region interacted with itself (Fig. 4B, lane 2) but not with the C-terminal deletion FLAG-C2 1-335 (Fig. 4B, lane 3), which has been shown to lie outside of the self-associative domain (Fig. 2B and 3B). The Xpress-tagged residues 336 to 702 did not interact with a negative control protein, FLAG-tagged p38 (Fig. 4B, lane 4). Cotransfection with the empty pcDNA3 vector also did not result in coprecipitation of the FLAG-C2 336-702 protein, demonstrating the specificity of the anti-Xpress antibody (Fig. 4B, lane 1). Similar findings have been noted in HeLa cells (data not shown).
A second mode of CIITA self-association involves the C-terminal sequences from residues 700 to 1130. An internal deletion within CIITA was constructed deleting residues 336 to 699. This fuses the sequence from residues 700 to 1130 to the N-terminal domain of CIITA. Residues 1 to 335 were included since these allowed better detection of the C-terminal sequences and did not appear to be involved in any self-association. FLAG- and myc-tagged versions of this construct were cotransfected into cells. Figure 4C shows that these two coprecipitated with each other (Fig. 4C, lane 2), whereas there was no interaction with residues 1 to 335 (Fig. 4C, lane 3). This indicates that residues 700 to 1130 may also mediate homotypic self-association.
The LRR sequences in 939-1130 interact with residues 336 to 702.
In addition to homotypic association of the Apaf-1 NBD, heterotypic domain association is observed between the C-terminal WD-40 repeats and the NBD (27). This prompted us to assay for an association between the GTP-binding domain and the LRR region of CIITA. FLAG-C2 939-1130 coprecipitated with the Xpress-tagged 336-702 CIITA sequences, indicating heterotypic association (Fig. 5B, lane 2). Therefore, although CIITA contains LRR as opposed to WD-40 repeats, the complex self-interactions that Apaf-1 displays are mirrored in the interactions detected thus far with CIITA.
FIG. 5.
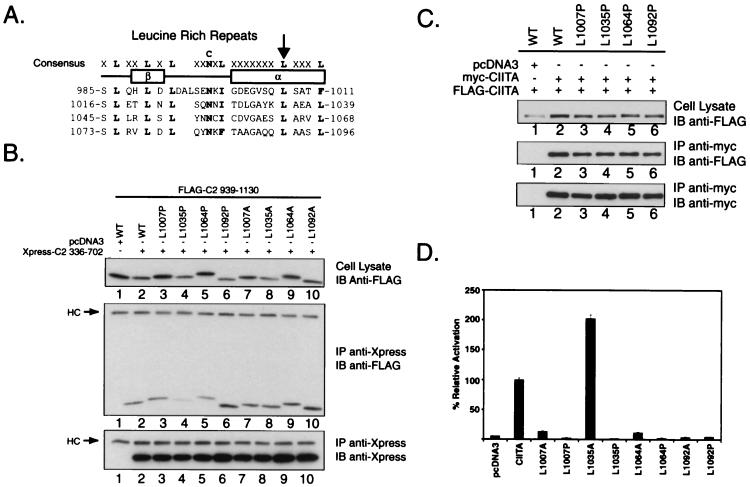
Mutation of leucines in the LRR and effects on self-association and transactivation function. (A) Targeted mutagenesis of the predicted alpha- helix within individual LRR. The consensus sequence for the LRR is shown with alignment of the CIITA motifs shown below. The location of the putative beta-sheet and alpha-helix is indicated under the consensus. The arrowhead indicates the leucine position targeted in the mutagenesis strategy. (B) Mutation of leucine-1035 to proline and, to a lesser extent, leucine-1064 to proline decreases the association of the C-terminal residues from 939 to 1130 with amino acids 336 to 702 of CIITA. FLAG-tagged expression vectors encoding the wild-type amino acid sequence from 939 to 1130 of CIITA or mutant proteins with either leucine-to-proline or leucine-to-alanine substitutions in individual LRR were coexpressed with the Xpress-C2 336-702 expression vector in COS-7 cells. The leucine mutations did not affect the expression of the 939-1130 proteins since expression was detected in each lane (top panel). The proline mutants were tested seven times, and alanine mutants were tested four times. (C) Full-length LRR leucine-to-proline mutations do not interfere with CIITA self-association. LRR mutations were produced in the context of full-length FLAG- and myc-CIITA and assayed for their capacity to self-associate using the conditions described in Fig. 1. (D) Effect of LRR mutations on CIITA transactivation function. Full-length leucine-to-proline and leucine-to-alanine mutants were tested for their ability to activate a DRA promoter luciferase reporter in HeLa cells. Luciferase activity is reported as percent activation relative to wild-type CIITA.
Interactions of mutant LRR with the GTP-binding domain of CIITA.
To explore the functional significance of the LRR in CIITA, a series of mutations were generated using the crystal structure of the LRR-containing protein, Rna1p as a model (25). Leucines within the putative alpha-helical region of the repeat were changed to proline and alanine at residues 1007, 1035, 1064, and 1092 of wild-type CIITA (Fig. 5A). Mutation to proline would be predicted to disrupt the helical component of the repeat, whereas the alanine substitutions should be much less disruptive to the helical structure. When these mutant LRR were tested for their ability to interact with the GTP-binding domain of CIITA, the L1035P mutation consistently showed significantly less interaction compared to the wild type (Fig. 5B, compare lanes 2 and 4). Interaction with the L1064P mutation was also consistently reduced (compare Fig. 5B, top panel, lanes 2 and 5, for the expression level, and the second panel, same lanes for the degree of self-association). Interactions with the two flanking LRR were studied using the L1007P and L1092P mutations; these have little effect on self-association. None of the alanine substitution mutants reduce self-association. These results suggest that the two central LRR are involved in association with the GTP-binding domain. The difference between the proline and alanine substitutions at residues 1035 and 1064 suggests that the helical sequences of these LRR are involved in association.
The effect of the leucine-to-proline mutations on self-association of full-length CIITA was also examined. In contrast to the results seen when we tested the heterotypic interactions between the isolated LRR and GTP-binding domains, none of the leucine-to-proline mutants had a detectable effect on intermolecular self-association when tested within the context of full-length CIITA (Fig. 5C, compare lanes 3 to 6 with lane 2). This result was reproduced in HeLa cells (data not shown). A likely explanation for our inability to detect associative differences between these mutants is that the homotypic interaction occurring between the GTP-binding domains of the two different CIITA molecules can still take place. This homotypic association is likely sufficient to maintain the intermolecular interaction.
To explore the significance of the LRR mutations on the transactivating function of CIITA, these mutants were tested for the capacity to activate an MHC class II promoter. In HeLa cells, all LRR mutants severely abrogated transactivation, with the exception of L1035A (Fig. 5D). Similar results were seen when using the COS-7 and G3A cell lines (J. A. Harton, unpublished results). These results indicate that all the leucine residues are critical for function, yet only two, L1035 and L1064, show a demonstrable effect on the heterotypic association shown in Fig. 5B. The results with L1035 are interesting in that the L1035P mutation affects both association (Fig. 5B) and function (Fig. 5D), whereas the L1035A mutation has no effect in both assays. This correlation of association and function provides support, but not proof, for the contention that association is linked to function. The difference between the proline and alanine mutations of L1035 indicates that conservation of helical structure is important. The role of the other leucines in this heterotypic interaction is unclear at this point. They may have a role in self-association that our approach fails to detect, or they may be critical to other mechanisms not revealed in this study.
Mutations in the GTP-binding domain decrease self-association.
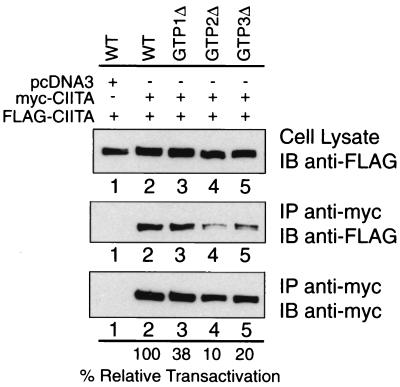
Given the ability of the isolated GTP-binding domain to self-associate (Fig. 4B), we tested whether mutating crucial residues in the GTP-binding domain of full-length CIITA affects intermolecular association. Previously, we have identified sequences corresponding to the G1, G3, and G4 GTP-binding motifs in CIITA (13). CIITA constructs bearing mutations in each of these three motifs are named GTP1Δ, GTP2Δ, and GTP3Δ, respectively (also see the Fig. 6, legend). These mutants display reduced transactivation function and GTP-binding capacity (23). To determine if these mutants display reduced self-association, we initially cotransfected wild-type CIITA together with each of these GTPΔ mutants. Although in selected experiments a small decrease in self-association was observed, these results were not reproducible (data not shown). In contrast, when the myc- and FLAG-tagged CIITA used in the coprecipitation assay both bear mutations in the GTP-binding region, the effect on association was reproducible. GTP1Δ, GTP2Δ, and GTP3Δ mutants were cotransfected in COS-7 cells. Most noteworthy, the GTP2Δ and GTP3Δ mutants displayed reduced self-association (Fig. 6, lanes 4 and 5). The self-association observed between GTP1Δ mutant molecules was similar to that for wild-type CIITA (Fig. 6, lane 3). Interestingly, the reduction observed for self-association parallels the reduction in transactivation potential of these mutants (see the percent relative transactivation values at the bottom of the figure) that we have previously described (13, 23). This strongly suggests that the GTP-binding domain is important in self-association and further shows a correlation between self-association and transactivating function.
FIG. 6.
Mutations within the GTP-binding domain reduce CIITA self-association Expression vectors encoding FLAG- and myc-tagged versions of identical GTP-binding domain mutants were cotransfected into COS-7 cells and tested for association. The consensus GTP-binding motifs are depicted in Fig. 2. GTP1Δ is a three-amino-acid deletion in the G1 box (phosphate binding, Δ419–421). GTP2Δ is a four-amino-acid deletion in the G3 box (Mg2+ coordination, Δ461–464), and GTP3Δ is a four-amino-acid deletion in the G4 box (guanine specificity, Δ558–561). Previously published percent relative transactivation data (13) as measured by a chloramphenicol acetyltransferase assay of each of the mutants are listed below the line at the bottom of the panel.
DISCUSSION
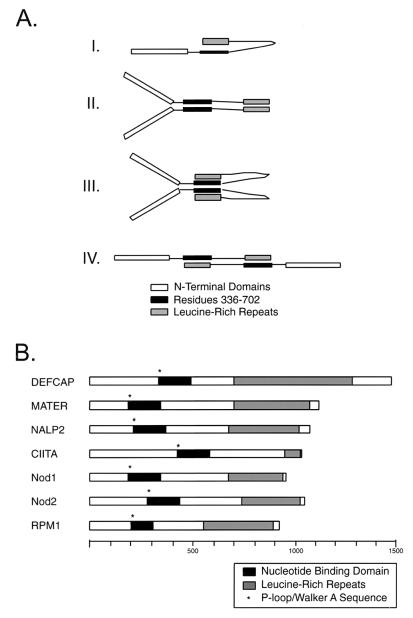
Transcriptional activation involves highly specific interactions among different components of the basal transcription complex, site-specific DNA-binding proteins, coactivators, and chromatin modifiers. Dimerization or multimerization provides an extra layer of regulation and complexity that can allow for multiple signaling outcomes (32). In this study, we provide evidence for a complex set of molecular interactions displayed by the transcriptional coactivator, CIITA. These interactions involve the region comprising residues 336 to 702 and the C terminus of CIITA. The former encompasses the GTP-binding domain and displays the capability for interactions both with itself and with the C-terminal LRR. The data demonstrate a complex pattern of association between distinct domains of CIITA and imply that CIITA occurs as a dimer if not a multimer. Our findings allow for the existence of several binding configurations (Fig. 7A). The simplest of these is a “hairpin” like monomer (Fig. 7A, diagram I), which would result from intramolecular association of the GTP-binding domain with the LRR. However, because our mapping experiments assay only for intermolecular interaction of two differentially tagged CIITA proteins, we can only infer that the intramolecular interaction may take place: The other hypothetical configurations all involve a form of intermolecular interaction (Fig. 7A, diagrams II to IV).
FIG. 7.
Model for self-association and similarity with other mammalian NBD-LRR proteins. (A) Graphical representation of the interactions described in this study. Conformation I would likely alternate with one of the other forms involving homotypic domain interactions (e.g., II or III). Conformation II is a dimeric configuration with homotypic domain interactions. This conformation would also likely alternate with another (e.g., I, III, or IV). Conformation III is a dimeric configuration with homotypic domain interaction (for amino acids 336 to 702) and heterotypic domain interaction (amino acids 336 to 702 with LRR). Multiple copies of this dimeric unit (4-mer, 6-mer, etc.) would allow for all observed interactions to occur simultaneously, obviating the need for additional (or transitional) states. Conformation IV is a dimeric configuration with heterotypic domain interactions. Again, alternating this configuration with other configurations (e.g., II or III) would be likely. (B) Schematic representation of recently described mammalian proteins with similarity to CIITA. Each protein contains a characterized or putative NBD and a series of C-terminal LRR sequences of varying number. NBD-LRR proteins are also common among plant disease resistance genes, as typified by RPM1. The scale shown indicates increments of 100 amino acids.
The initial motivation for these studies was to investigate the trans-dominant-negative function of CIITA mutants (12). One potential mechanism that could explain the results is that dominant-negative mutants could interact directly with wild-type CIITA and block transactivation. Such self-association is shown in this report. In light of the present data, it is possible that the lack of either domain alters the proper balance of self-association to result in the suppression of CIITA function. It is also noteworthy that both the LRR and GTP-binding domains have a role in nuclear translocation (15, 22, 23). It is possible that mutations in these regions cause the protein to assume different configurations, or different associative properties, leading to an effect on import function.
One important observation that arises from these studies is the similarity of these interactions with those that have been described for two other proteins that self-associate, i.e., Apaf-1 and Nod1 (28, 55). Apaf-1 initiates an apoptotic cascade in the presence of cytochrome c and dATP (37). It contains an N-terminal caspase recruitment domain (CARD), an NBD that binds dATP, and C-terminal WD-40 repeats. The NBD of Apaf-1 self-associates (55, 63), and the resultant oligomerization is thought to induce the proximity of procaspase-9 molecules, resulting in cleavage and activation. The C-terminal WD-40 repeats of Apaf-1 interact with the NBD and regulate the activity of Apaf-1 (2, 27, 55). CIITA demonstrates a similar complex pattern of association, although it is unclear what regulates the self-association of CIITA. Based on our data, it is likely that the binding of GTP may represent such a regulatory step. This is supported by the finding that reductions in self-association observed with GTP-binding domain mutants correlates directly with reductions in their transactivation potential. One caveat is that the disruption of the phosphate-binding motif, G1, did not significantly influence the ability of CIITA to self-associate, while this same mutation is known to decrease GTP-binding and transactivation function (23). Thus, we also have to entertain the most conservative interpretation that it is the integrity of the GTP-binding domain that is important for self-association and not the binding of GTP.
The second domain of CIITA that is involved in self-association lies in the C-terminal end, which contains a series of LRR (8). Comparison of the LRR from diverse sources yields the consensus sequence LXXLXLXX(N/C)XL(X)7L(X)3L, although some variation from this consensus can be tolerated as seen in the structure of Rna1p (25). Allowing for such variation, CIITA contains six potential repeats. LRR adopt a horseshoe-like structure composed of alternating beta-sheets and alpha-helices with the beta-sheets lining the interior face and the helices facing outward (33, 34). This motif can be found in a variety of proteins and is thought to be involved in mediating protein-protein interactions (8, 35). This global hypothesis is supported by our results, which show interaction of the LRR with the GTP-binding domain of CIITA.
The results in Fig. 5 indicate that only one of the LRR mutations, L1035P, significantly disrupts binding to the GTP-binding domain, while the L1064P mutation has a lesser effect. The mutation of 1035 is most interesting because, while a proline substitution caused the loss of transactivation function and self-associative properties, an alanine substitution at this site affected neither. This suggests that the helical structure of these repeats, which is most affected by a proline substitution, is actually more significant than an invariant leucine at that site. Aside from these two mutations, mutations of the other LRR motifs produced little effect on self-association, but most caused a dramatic reduction of transactivation activity. The basis for the lack of correlation between self-association and transactivation function may be complicated by other molecular events that require an intact LRR domain. For example, two mutants that self-associate but lack transactivation function, L1007P and L1092P, are exclusively found in the cytoplasm (J. A. Harton et al., unpublished data). Therefore, failure to localize to the nucleus can explain the lack of function observed for these mutants. It is also possible that sequences in addition to the LRR are important for self-association. Along this vein, we have noted that while residues 700 to 1130 exhibit strong homotypic association (Fig. 4C), homotypic associations between residues 939 and 1130 are very weak (data not shown). Finally, we have to consider the possibility that the LRR domain is not only involved in self-association but may also interact with other proteins. Consistent with this hypothesis, the association of CIITA with a 33-kDa protein has been shown to be sensitive to LRR mutagenesis (22).
One crucial question that arises from this study is: why is it necessary for CIITA to self-associate? The answer may lie in recent data which show that CIITA serves as a scaffold for the transcription factors involved in the regulation of MHC class II genes (65). Multimerization of CIITA may enhance the association of these factors in vivo either with each other or with the DRA promoter. This is similar to the induced proximity models discussed for Apaf-1 and more recently for Nod1 and Nod2 (29, 47, 62, 63). In contrast to the apoptosis signaling proteins mentioned above, CIITA does not have a caspase recruitment domain at the N terminus. Instead, the N terminus of CIITA contains a transcriptional activation domain that binds to a number of transcription factors (18, 19, 31, 36, 39). Several factors with enzymatic activity bind in this region, including the acetyltransferase activities of CBP (also called p300) and PCAF (19, 36, 53, 54) and the RNA polymerase II CTD phosphorylation activity of P-TEFb (31). Therefore, it is possible that CIITA could function as a chaperone to induce the proximity of these enzymatic activities at MHC class II promoters.
Although CIITA and Apaf-1 do not display significant primary sequence homology, CIITA has been shown to be similar in sequence to the other apoptotis signaling proteins, Nod1 (also known as CARD4) (4, 28) and Nod2 (47). Sequence similarity with CIITA is strongest in the nucleotide-binding domain and in the C-terminal LRR region. Using a BLAST search of the GenBank database, we have found several other proteins that display similarity to CIITA. These proteins share similarity within their NBDs and are large proteins (typically >1,000 amino acids in length) with LRR at their C termini (Fig. 7B). DEFCAP contains an N-terminal pyrin domain and a C-terminal CARD domain and appears to be yet another NBD-LRR protein capable of signaling apoptosis (26). MATER is an ooplasm-specific maternal effect gene required for embryonic development beyond the two-cell stage (59–61), and its mode of action is ill defined. Finally, some plant disease resistance proteins, such as RPM1, share the NBD-LRR domain arrangement (3, 7, 8). Nod1 and Nod2 have recently been shown to be involved in responses to bacterial products, most notably lipopolysaccharide (30). A thought-provoking possibility in comparing CIITA, Nod1 and -2, and RPM1 is that plants and animals may have adopted a similar molecular strategy to fend off infections.
In summary, this study provides evidence that CIITA self-associates and that this interaction involves the GTP-binding domain and the C terminus, which includes the LRR. The mode of self-association is reminiscent of other NBD-LRR proteins, such as Nod1 and Apaf-1. CIITA is the first example of a transcriptional coactivator that belongs to this group of proteins. It is likely that the mechanisms that mediate self-association among these distinct proteins are similar; however, distinct biological functions are activated depending on the “effector” domain that lies at the N termini. In the case of CIITA, the N terminus contains a classical activation domain, and thus gene transcription is activated. This study has broad implications in the understanding of this emerging family of proteins. Additionally, these findings have specific implications regarding how the biological and molecular function of CIITA is regulated.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (AI29564, AI45580, and AI41751) to J. P.-Y. Ting.
REFERENCES
- 1.Accolla R S, Jotterand-Bellomo M, Scarpellino L, Maffei A, Carra G, Guardiola J. aIr-1, a newly found locus on mouse chromosome 16 encoding a trans-acting activator factor for MHC class II gene expression. J Exp Med. 1986;164:369–374. doi: 10.1084/jem.164.1.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adrain C, Slee E A, Harte M T, Martin S J. Regulation of apoptotic protease activating factor-1 oligomerization and apoptosis by the WD-40 repeat region. J Biol Chem. 1999;274:20855–20860. doi: 10.1074/jbc.274.30.20855. [DOI] [PubMed] [Google Scholar]
- 3.Baker B, Zambryski P, Staskawicz B, Dinesh-Kumar S P. Signaling in plant-microbe interactions. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 4.Bertin J, Nir W J, Fischer C M, Tayber O V, Errada P R, Grant J R, Keilty J J, Gosselin M L, Robison K E, Wong G H, Glucksmann M A, DiStefano P S. Human CARD4 protein is a novel CED-4/Apaf-1 cell death family member that activates NF-kappaB. J Biol Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 5.Bontron S, Steimle V, Ucla C, Eibl M M, Mach B. Two novel mutations in the MHC class II transactivator CIITA in a second patient from MHC class II deficiency complementation group A. Hum Genet. 1997;99:541–546. doi: 10.1007/s004390050403. [DOI] [PubMed] [Google Scholar]
- 6.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 7.Boyes D C, Nam J, Dangl J L. The Arabidopsis thaliana RPM1 disease resistance gene product is a peripheral plasma membrane protein that is degraded coincident with the hypersensitive response. Proc Natl Acad Sci USA. 1998;95:15849–15854. doi: 10.1073/pnas.95.26.15849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchanan S G, Gay N J. Structural and functional diversity in the leucine-rich repeat family of proteins. Prog Biophys Mol Biol. 1996;65:1–44. doi: 10.1016/s0079-6107(96)00003-x. [DOI] [PubMed] [Google Scholar]
- 9.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J Mol Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 10.Chang C H, Fontes J D, Peterlin B M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman H A. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. doi: 10.1016/s0952-7915(98)80038-1. [DOI] [PubMed] [Google Scholar]
- 12.Chin K C, Li G, Ting J P-Y. Activation and transdominant suppression of MHC class II and HLA-DMB promoters by a series of C-terminal class II transactivator deletion mutants. J Immunol. 1997;159:2789–2794. [PubMed] [Google Scholar]
- 13.Chin K C, Li G G, Ting J P-Y. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chin K C, Mao C, Skinner C, Riley J L, Wright K L, Moreno C S, Stark G R, Boss J M, Ting J P-Y. Molecular analysis of G1B and G3A IFN gamma mutants reveals that defects in CIITA or RFX result in defective class II MHC and Ii gene induction. Immunity. 1994;1:687–697. doi: 10.1016/1074-7613(94)90039-6. [DOI] [PubMed] [Google Scholar]
- 15.Cressman D E, Chin K C, Taxman D J, Ting J P-Y. A defect in the nuclear translocation of CIITA causes a form of type II bare lymphocyte syndrome. Immunity. 1999;10:163–171. doi: 10.1016/s1074-7613(00)80017-5. [DOI] [PubMed] [Google Scholar]
- 16.DeSandro A M, Nagarajan U M, Boss J M. Associations and interactions between bare lymphocyte syndrome factors. Mol Cell Biol. 2000;20:6587–6599. doi: 10.1128/mcb.20.17.6587-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durand B, Sperisen P, Emery P, Barras E, Zufferey M, Mach B, Reith W. RFXAP, a novel subunit of the RFX DNA binding complex is mutated in MHC class II deficiency. EMBO J. 1997;16:1045–1055. doi: 10.1093/emboj/16.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fontes J D, Jiang B, Peterlin B M. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Glimcher L H, Kara C J. Sequences and factors: a guide to MHC class-II transcription. Annu Rev Immunol. 1992;10:13–49. doi: 10.1146/annurev.iy.10.040192.000305. [DOI] [PubMed] [Google Scholar]
- 21.Gobin S J, Peijnenburg A, Keijsers V, van den Elsen P J. Site alpha is crucial for two routes of IFN gamma-induced MHC class I transactivation: the ISRE-mediated route and a novel pathway involving CIITA. Immunity. 1997;6:601–611. doi: 10.1016/s1074-7613(00)80348-9. [DOI] [PubMed] [Google Scholar]
- 22.Hake S B, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, In vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol. 2000;20:7716–7725. doi: 10.1128/mcb.20.20.7716-7725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harton J A, Cressman D E, Chin K C, Der C J, Ting J P-Y. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 24.Harton J A, Ting J P. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillig R C, Renault L, Vetter I R, Drell T, Wittinghofer A, Becker J. The crystal structure of rna1p: a new fold for a GTPase-activating protein. Mol Cell. 1999;3:781–791. doi: 10.1016/s1097-2765(01)80010-1. [DOI] [PubMed] [Google Scholar]
- 26.Hlaing, T., R. F. Guo, K. A. Dilley, J. M. Loussia, T. A. Morrish, M. M. Shi, C. Vincenz, and P. A. Ward. Molecular cloning and characterization of DEFCAP-L and -S, two isoforms of a novel member of the mammalian Ced-4 family of apoptosis proteins. J. Biol. Chem., in press. [DOI] [PubMed]
- 27.Hu Y, Ding L, Spencer D M, Nunez G. WD-40 repeat region regulates Apaf-1 self-association and procaspase-9 activation. J Biol Chem. 1998;273:33489–33494. doi: 10.1074/jbc.273.50.33489. [DOI] [PubMed] [Google Scholar]
- 28.Inohara N, Koseki T, del Peso L, Hu Y, Yee C, Chen S, Carrio R, Merino J, Liu D, Ni J, Nunez G. Nod1, an Apaf-1-like activator of caspase-9 and nuclear factor-kappaB. J Biol Chem. 1999;274:14560–14567. doi: 10.1074/jbc.274.21.14560. [DOI] [PubMed] [Google Scholar]
- 29.Inohara N, Koseki T, Lin J, del Peso L, Lucas P C, Chen F F, Ogura Y, Nunez G. An induced proximity model for NF-kappa B activation in the Nod1/RICK and RIP signaling pathways. J Biol Chem. 2000;275:27823–27831. doi: 10.1074/jbc.M003415200. [DOI] [PubMed] [Google Scholar]
- 30.Inohara N, Ogura Y, Chen F F, Muto A, Nunez G. Human Nod1 confers responsiveness to bacterial lipopolysaccharides. J Biol Chem. 2000;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 31.Kanazawa S, Okamoto T, Peterlin B M. Tat competes with CIITA for the binding to P-TEFb and blocks the expression of MHC class II genes in HIV infection. Immunity. 2000;12:61–70. doi: 10.1016/s1074-7613(00)80159-4. [DOI] [PubMed] [Google Scholar]
- 32.Klemm J D, Schreiber S L, Crabtree G R. Dimerization as a regulatory mechanism in signal transduction. Annu Rev Immunol. 1998;16:569–592. doi: 10.1146/annurev.immunol.16.1.569. [DOI] [PubMed] [Google Scholar]
- 33.Kobe B, Deisenhofer J. Crystal structure of porcine ribonuclease inhibitor, a protein with leucine-rich repeats. Nature. 1993;366:751–756. doi: 10.1038/366751a0. [DOI] [PubMed] [Google Scholar]
- 34.Kobe B, Deisenhofer J. The leucine-rich repeat: a versatile binding motif. Trends Biochem Sci. 1994;19:415–421. doi: 10.1016/0968-0004(94)90090-6. [DOI] [PubMed] [Google Scholar]
- 35.Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- 36.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li P, Nijhawan D, Budihardjo I, Srinivasula S M, Ahmad M, Alnemri E S, Wang X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 38.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 39.Mahanta S K, Scholl T, Yang F C, Strominger J L. Transactivation by CIITA, the type II bare lymphocyte syndrome- associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci USA. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999;239:15–27. doi: 10.1016/s0378-1119(99)00368-6. [DOI] [PubMed] [Google Scholar]
- 41.Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin B K, Chin K C, Olsen J C, Skinner C A, Dey A, Ozato K, Ting J P. Induction of MHC class I expression by the MHC class II transactivator CIITA. Immunity. 1997;6:591–600. doi: 10.1016/s1074-7613(00)80347-7. [DOI] [PubMed] [Google Scholar]
- 43.Masternak K, Barras E, Zufferey M, Conrad B, Corthals G, Aebersold R, Sanchez J C, Hochstrasser D F, Mach B, Reith W. A gene encoding a novel RFX-associated transactivator is mutated in the majority of MHC class II deficiency patients. Nat Genet. 1998;20:273–277. doi: 10.1038/3081. [DOI] [PubMed] [Google Scholar]
- 44.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 45.Moreno C S, Beresford G W, Louis-Plence P, Morris A C, Boss J M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 46.Nagarajan U M, Louis-Plence P, DeSandro A, Nilsen R, Bushey A, Boss J M. RFX-B is the gene responsible for the most common cause of the bare lymphocyte syndrome, an MHC class II immunodeficiency. Immunity. 1999;10:153–162. doi: 10.1016/s1074-7613(00)80016-3. [DOI] [PubMed] [Google Scholar]
- 47.Ogura Y, Inohara N, Benito A, Chen F F, Yamaoka S, Nunez G. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NFκB. J Biol Chem. 2000;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 48.Pieters J. MHC class II-restricted antigen presentation. Curr Opin Immunol. 1997;9:89–96. doi: 10.1016/s0952-7915(97)80164-1. [DOI] [PubMed] [Google Scholar]
- 49.Quan V, Towey M, Sacks S, Kelly A P. Absence of MHC class II gene expression in a patient with a single amino acid substitution in the class II transactivator protein CIITA. Immunogenetics. 1999;49:957–963. doi: 10.1007/s002510050579. [DOI] [PubMed] [Google Scholar]
- 50.Reith W, Satola S, Sanchez C H, Amaldi I, Lisowska-Grospierre B, Griscelli C, Hadam M R, Mach B. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 51.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 52.Sinha S, Maity S N, Lu J, de Crombrugghe B. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc Natl Acad Sci USA. 1995;92:1624–1628. doi: 10.1073/pnas.92.5.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sisk T J, Gourley T, Roys S, Chang C H. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J Immunol. 2000;165:2511–2517. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]
- 54.Spilianakis C, Papamatheakis J, Kretsovali A. Acetylation by PCAF enhances CIITA nuclear accumulation and transactivation of major histocompatibility complex class II genes. Mol Cell Biol. 2000;20:8489–8498. doi: 10.1128/mcb.20.22.8489-8498.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Srinivasula S M, Ahmad M, Fernandes-Alnemri T, Alnemri E S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol Cell. 1998;1:949–957. doi: 10.1016/s1097-2765(00)80095-7. [DOI] [PubMed] [Google Scholar]
- 56.Steimle V, Durand B, Barras E, Zufferey M, Hadam M R, Mach B, Reith W. A novel DNA-binding regulatory factor is mutated in primary MHC class II deficiency (bare lymphocyte syndrome) Genes Dev. 1995;9:1021–1032. doi: 10.1101/gad.9.9.1021. [DOI] [PubMed] [Google Scholar]
- 57.Steimle V, Otten L A, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency (or bare lymphocyte syndrome) Cell. 1993;75:135–146. [PubMed] [Google Scholar]
- 58.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 59.Tong Z B, Gold L, Pfeifer K E, Dorward H, Lee E, Bondy C A, Dean J, Nelson L M. Mater, a maternal effect gene required for early embryonic development in mice. Nat Genet. 2000;26:267–268. doi: 10.1038/81547. [DOI] [PubMed] [Google Scholar]
- 60.Tong Z B, Nelson L M. A mouse gene encoding an oocyte antigen associated with autoimmune premature ovarian failure. Endocrinology. 1999;140:3720–3726. doi: 10.1210/endo.140.8.6911. [DOI] [PubMed] [Google Scholar]
- 61.Tong Z B, Nelson L M, Dean J. Mater encodes a maternal protein in mice with a leucine-rich repeat domain homologous to porcine ribonuclease inhibitor. Mamm Genome. 2000;11:281–287. doi: 10.1007/s003350010053. [DOI] [PubMed] [Google Scholar]
- 62.Yang X, Chang H Y, Baltimore D. Autoproteolytic activation of pro-caspases by oligomerization. Mol Cell. 1998;1:319–325. doi: 10.1016/s1097-2765(00)80032-5. [DOI] [PubMed] [Google Scholar]
- 63.Yang X, Chang H Y, Baltimore D. Essential role of CED-4 oligomerization in CED-3 activation and apoptosis. Science. 1998;281:1355–1357. doi: 10.1126/science.281.5381.1355. [DOI] [PubMed] [Google Scholar]
- 64.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 65.Zhu X S, Linhoff M W, Li G, Chin K C, Maity S N, Ting J P. Transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II major histocompatibility complex promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]