Abstract
Cryptococcus neoformans is an opportunistic fungal pathogen that infects the human central nervous system. This pathogen elaborates two specialized virulence factors: the antioxidant melanin and an antiphagocytic immunosuppressive polysaccharide capsule. A signaling cascade controlling mating and virulence was identified. The PKA1 gene encoding the major cyclic AMP (cAMP)-dependent protein kinase catalytic subunit was identified and disrupted. pka1 mutant strains were sterile, failed to produce melanin or capsule, and were avirulent. The PKR1 gene encoding the protein kinase A (PKA) regulatory subunit was also identified and disrupted. pkr1 mutant strains overproduced capsule and were hypervirulent in animal models of cryptococcosis. pkr1 pka1 double mutant strains exhibited phenotypes similar to that of pka1 mutants, providing epistasis evidence that the Pka1 catalytic subunit functions downstream of the Pkr1 regulatory subunit. The PKA pathway was also shown to function downstream of the Gα protein Gpa1 and to regulate cAMP production by feedback inhibition. These findings define a Gα protein-cAMP-PKA signaling pathway regulating differentiation and virulence of a human fungal pathogen.
Cells sense and respond to the environment and communicate with other cells via signal transduction cascades, which allow cells to sense extracellular conditions and appropriately respond. We are interested in the signaling pathways that enable pathogenic fungi to adapt to and survive the dramatically altered environmental conditions they encounter upon infection of the host.
Cryptococcus neoformans is an opportunistic fungal pathogen that causes life-threatening infections of the human central nervous system (CNS) (7, 52). The organism is distributed worldwide, and most individuals have protective immunity and evidence of prior exposure (21, 29). The incidence of cryptococcal meningitis has increased because of immunosuppression as a result of AIDS, cancer chemotherapy, steroid treatment, and organ transplantation. However, even in normal individuals, the infection can be chronic with a state of latency and subsequent reactivation can occur (22, 25). A small percentage of patients with cryptococcal pneumonia, meningitis, or cutaneous infection have no apparent immune system dysfunction, and these patients may be infected with hypervirulent strains (1, 59, 62). In addition, strains of the divergent C. neoformans var. gattii (serotypes B and C) primarily infect hosts with normal immune function (14). Thus, C. neoformans is both an opportunistic and a primary pathogen.
C. neoformans is a basidiomycete with a defined sexual cycle (3, 38, 39, 69). Infection occurs by inhalation of dessicated yeast cells or spores, which then spread hematogenously to the brain (53, 71). Two inducible factors have been linked to virulence: the production of melanin and a polysaccharide capsule (10, 40, 41).
Melanin is not produced under most in vitro culture conditions but can be induced in response to carbon source limitation, which also occurs in the infected host. Melanin is a large polymer that is embedded in the cell wall ensheathing the organism, where it can serve as an antioxidant to protect fungal cells from oxidative and nitrosative challenge by macrophages (31, 77). Melanin synthesis requires the enzyme laccase, which is expressed in vivo and important for full virulence (68). Recent studies reveal that melanin is produced in vivo in infected animals and in the CNS of patients with cryptococcal meningitis (56–58, 66). The substrates for melanin synthesis are diphenolic compounds, such as dopamine and other neurotransmitters. The abundance of these melanin precursors in the CNS may explain the unique tissue tropism of cryptococcal infection.
The polysaccharide capsule is induced in vivo in the infected host by iron limitation and carbon dioxide and surrounds and protects fungal cells from phagocytosis and enhances intracellular survival in macrophages (24, 28, 74). In addition, capsular antigens are shed into the circulation and have potent immunosuppressive activity (18, 19, 32, 75, 76). Mutants lacking either melanin or capsule are avirulent or attenuated in animal models (10, 40, 41, 68), and virtually all clinical isolates produce both melanin and capsule (7).
Recently, the Gα protein Gpa1 was found to regulate melanin and capsule production and virulence of C. neoformans in response to environmental signals (4). gpa1 mutant cells have defects in mating and produce reduced levels of melanin and capsule. As a consequence, gpa1 mutant strains are markedly attenuated for virulence in animal models. Exogenous cyclic AMP (cAMP) restores mating, melanin, and capsule production in gpa1 mutant strains, suggesting that Gpa1 regulates a cAMP signaling pathway (4). We identified here catalytic and regulatory subunits of the cAMP-dependent protein kinase A (PKA) and demonstrate that PKA functions downstream of the Gα protein Gpa1 in a signal transduction cascade that controls mating and virulence of this human pathogen. Related signaling pathways operate to control differentiation in the budding yeast Saccharomyces cerevisiae (37, 46, 61, 65, 67) and in plant fungal pathogens such as Ustilago maydis (23, 26, 27, 35, 64).
MATERIALS AND METHODS
Isolation and disruption of the PKA1 gene.
The PKA1 gene was isolated by degenerate PCR with C. neoformans serotype D cDNA library DNA as template and primers 1883 (5′-ACIYTIGGIACIGGIWSITTYGGIMGIGT), 1886 (5′-TARTCIGGIGTICCRCAIARIGTCCAIGT), and 1887 (5′-TARTCIGGIGTICCRCAIARIGTRTAIGT), where R is A+G, Y is C+T, I is deoxyinosine, W is A+T, M is A+C, and S is G+C. A 410-bp PCR product was cloned, sequenced, and used as a probe to isolate a 12-kb HindIII fragment containing the PKA1 gene from a size-selected genomic library of the serotype A strain H99. The ADE2 gene (2.5-kb BamHI fragment) was inserted at a BglII site in the PKA1 gene and biolistically transformed into the ade2 strain M001 (70). pka1::ADE2 mutants were identified by PCR and confirmed by Southern blot.
Assay of melanin production.
Melanin is produced by the enzyme laccase (CnLac1), which is a phenol oxidase that accepts diphenolic precursors as substrates and oxidizes them, and nonenzymatic polymerization then produces melanin polymers. The activity of CnLac1, the rate-limiting enzyme in melanin production, was assessed by measuring the oxidation of 2,2′-azinobis(3-ethylbenzthiazoline-6-sulfonate) (ABTS) as described elsewhere (6). Wild-type, pka1, gpa1, pka1+PKA1, pkr1, and pkr1 gpa1 strains were incubated at 30°C with shaking for 18 h in minimal asparagine medium with 0.1% glucose. Cells were pelleted, washed twice with water, resuspended in minimal asparagine medium without glucose, and incubated with shaking for 4 h at 30°C. Cells were pelleted and resuspended in 0.1 M sodium acetate buffer (pH 5.0) at 2 × 108 cells/ml. The oxidation of ABTS was assessed spectrophotometrically by measuring the A420 of the supernatant of the cell suspension at 30 min after addition of 0.5 mM ABTS. One unit of enzyme activity was defined as 0.01 absorbance units at 30 min.
Assay of capsule production and measurement of capsule size and volume.
Strains to be assayed for capsule production were grown in low-iron medium (LIM plus EDDHA) for 4 days at 30°C. Cultures were treated with 10% formalin, normalized for cell counts, and added to heparinized Microhematocrit Capillary Tubes (Fisher 22-362-574), and the ends were sealed with clay. Capillary tubes were centrifuged for 10 min in a Microhematocrit Centrifuge model MB (International Equipment Co.). The packed cell volume (PCV) was determined by the following formula: PCV = The length of the packed cells/the length of the total suspension.
The capsule and cell sizes of fungal cells in brain homogenates were determined by photomicroscopy and comparison to images of a 10-μm ruled microscope slide. The cell diameter and capsule thickness were measured for 100 cells each of the pkr1-33 and pkr1-56 strains and for 40 wild-type cells, and the values presented are the mean with the standard deviation. The volume of the capsular shell was calculated by subtracting the volume of the cell (4/3 Πr3) from the volume of the capsule plus the cell.
Animal models of virulence.
The rabbit cryptococcal meningitis model was as described elsewhere (4). In the murine models, BALB/c or A/Jcr mice were infected by tail vein injection or inhalation. Mice were anesthetized by intraperitoneal phenobarbital injections and suspended by the incisors on a silk thread, and 50-μl volumes of the inocula were slowly pipetted into the nares with continued suspension for 10 min. Survival was monitored daily, and moribund animals or those in pain were sacrificed by CO2 inhalation.
Three groups of 10 female A/Jcr mice were infected with a total of 106 yeast cells of serotype A strain H99 (WT) and two pkr1 mutant isolates (pkr1-33 and pkr1-56) via lateral tail vein injection. Three mice from each group were euthanized 3 h, 3 days, or 7 days after infection, exsanguinated by cardiac puncture, and the brain, spleen, and lung tissue were harvested, resuspended in phosphate-buffered saline, and homogenized. Quantitative cultures were performed by plating dilutions of the tissue homogenates on yeast-peptone-dextrose (YPD) medium.
Mouse survival and tissue cryptococcal burden measurements were repeated with BALB/c mice. Three groups of 20 female BALB/c mice were infected with a total of 106 yeast cells of serotype A strain H99 (WT) and two pkr1 mutant isolates (pkr1-33 and pkr1-56) via lateral tail vein injection. The survival of groups of 10 infected mice was determined as described for the A/Jcr mice. The brain cryptococcal burden was also determined as for the A/Jcr mice by sacrificing five mice from each group at 3 or 7 days after infection.
Quantitative mating assay.
Recombinant basidiospore production was measured by mixing serotype A strains (MATα URA5 LYS1) in 100-μl suspensions with the MATa ura5 lys1 serotype D strain JEC53. Then, 5 μl of the cell mix was spotted onto V8 plus uracil-lysine medium and incubated at 25°C. Every 2 days, the mating reaction on one plate was excised, vortexed in 2 ml of water, and plated on 5-fluoroorotic acid medium lacking lysine to detect ura5 LYS1 recombinants. To detect cell fusion, the MATα ura5 serotype A strains were cocultured with the MATa lys1 serotype D strain JEC30 on V8 plus uracil-lysine medium. At 48 and 96 h, cells were removed, resuspended in 2 ml of phosphate-buffered saline, and plated on YNB medium to select heterokaryons and prototrophic recombinants.
Isolation and disruption of the PKR1 gene.
The PKR1 gene was isolated based on a sequence trace from the C. neoformans EST project (University of Oklahoma). Primers 2571 (5′-CGCTCTCGGCCGAAGGACATC) and 2572 (5′-TCAACGATGTAAAAGAAGTCACCG) were used in a PCR reaction with genomic DNA from the serotype A strain H99 as template. The 300-bp PCR product was then used to isolate the PKR1 gene from a size-selected genomic library on a 5-kb HindIII fragment. A 2-kb URA5 gene was inserted into a unique HpaI site in the PKR1 gene, and the serotype A H99 ura5, gpa1 ura5, and pka1 ura5 strains were biolistically transformed. Ura+ transformants were selected on synthetic medium lacking uracil with 1 M sorbitol. pkr1::URA5 mutants were identified by PCR and Southern blot at frequencies of 16.7% in the wild type (10 of 60), 87% in the gpa1 mutant (27 of 31), and 7% in the pka1 mutant (7 of 96).
Two-hybrid assays.
To test if Pka1 and Pkr1 interact in the two hybrid system, a full-length PKA1 cDNA was amplified from a C. neoformans H99 cDNA library with synthetic primers, cleaved with BamHI, and inserted into the two-hybrid plasmid pGBT9 to express a GAL4(BD)-Pka1 fusion protein. The PKR1 cDNA was isolated using primers and the same cDNA template, cleaved with BamHI, and cloned in plasmid pGAD424. Plasmids expressing the GAL4(BD)-Pka1 and GAL4(AD)-Pkr1 fusion proteins supported growth of the two-hybrid strain PJ69-4A on medium lacking adenine or histidine (plus 3-aminotriazole).
cAMP assay.
Cells were precultured overnight at 30°C in YPD medium, inoculated in fresh YPD medium to an optical density at 600 nm (OD600) of 0.05, and then grown under the same conditions for 20 h. Cells were collected by centrifugation and washed twice with water and once with buffer (10 mM morpholineethanesulfonic acid [pH 6.0], 0.1 mM EDTA). Cells were resuspended in buffer and incubated at 30°C with shaking to subject the cells to glucose starvation. After 2 h, glucose was added to a final concentration of 2%. At the time points indicated in Fig. 7, 0.5 ml of the cell suspension was transferred into a tube containing equal volume of ice-cold 10% trichloroacetic acid and 0.3 ml of glass beads and then immediately frozen in liquid nitrogen. Crude cell extracts were prepared by homogenizing with a bead beater at 4°C and were lyophilized. cAMP assays used a cAMP enzyme immunoassay kit (Amersham), as described earlier (47).
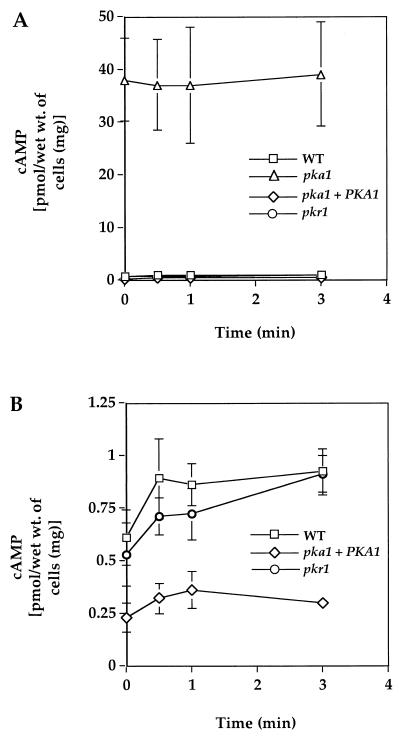
FIG. 7.
PKA negatively regulates cAMP production in C. neoformans. The wild-type, pka1, pka1+PKA1, and pkr1 strains were starved for glucose by growth in YPD medium to stationary phase. Then, 2% glucose was added and, at the indicated times, cells were harvested and crude extracts were prepared. cAMP levels in the extracts were then assessed by enzyme immunoassay. The values shown are the average of triplicate measurements with the standard error of the mean depicted as error bars. The scale of the data shown in panel A has been expanded in panel B to illustrate the levels of cAMP present in the wild type (WT) and in the pkr1 mutant and pka1+PKA1 reconstituted strains.
PKA assay.
Crude cell extracts were prepared from logarithmic-phase (OD600 = 0.6) wild-type, pka1 mutant, and pka1+PKA1 cells as described elsewhere (16). PKA activity was assayed either in the presence or absence of cAMP (5 μM) or with cAMP and the PKA-specific inhibitor (10 μM) using the SignaTECT PKA assay system (Promega).
RESULTS
Identification of the cAMP-dependent protein kinase catalytic subunit Pka1.
To identify the catalytic subunit of PKA, primers were designed to target conserved regions of PKA genes from other fungi. Under reduced stringency conditions, these primers amplified a 410-bp PCR product from C. neoformans pooled cDNA. This PCR product was cloned and sequenced, revealing homology to known PKA catalytic subunits. The PCR product was used as a probe to isolate the full-length PKA1 gene on a 12-kb HindIII fragment from a size-selected genomic library of the serotype A strain H99. Sequence analysis of genomic DNA, 5′ and 3′ RACE (rapid amplification of cDNA ends) products, and cDNA clones was used to define the structure of the PKA1 gene (GenBank accession no. AF288613). The C. neoformans Pka1 protein shares 60% amino acid identity to the Adr1 PKA catalytic subunit from U. maydis, a basidiomycete that infects maize (23, 60). The C. neoformans Pka1 catalytic subunit was more closely related to the S. cerevisiae Tpk2 catalytic subunit that activates filamentous growth than to the Tpk1 and Tpk3 subunits that play a negative regulatory role (61, 65). By Southern blot analysis under conditions of reduced stringency, no genes highly related to PKA1 were detected (data not shown). However, sequence traces to both PKA1 and a gene encoding a second PKA catalytic subunit homolog (PKA2) are present in the Stanford C. neoformans genome sequence database (R. W. Hyman and R. W. Davis, http://www-sequence.stanford.edu/group/C.neoformans/index.html). Our genetic studies suggest that Pka1 plays the predominant role in PKA signaling.
To determine its biological functions, the PKA1 gene was disrupted by transformation and homologous recombination. The ADE2 selectable marker was inserted into a unique BglII site within the PKA1 gene, and the resulting pka1::ADE2 disruption allele was used to replace the wild-type PKA1 gene in the ade2 serotype A strain M001 by biolistic transformation (70). Five pka1 mutant strains were identified from 100 Ade+ transformants. PCR and Southern blot analysis demonstrated that gene replacement without ectopic integration had occurred. The wild-type PKA1 gene linked to the hygromycin B resistance gene was introduced into the pka1 mutant strain by biolistic transformation to produce a pka1+PKA1 reconstituted strain.
To determine the contribution of the PKA1 gene product to PKA activity, cell extracts were prepared from wild-type, pka1 mutant, and pka1+PKA1 reconstituted strains and PKA activity was assayed in vitro. cAMP-dependent protein kinase activity was readily detectable in the wild-type and pka1+PKA1 reconstituted strains, whereas little or no activity was present in extracts from the pka1 mutant strain (Fig. 1A). These findings provide evidence that PKA1 encodes a PKA catalytic subunit that represents the predominant form of the enzyme.
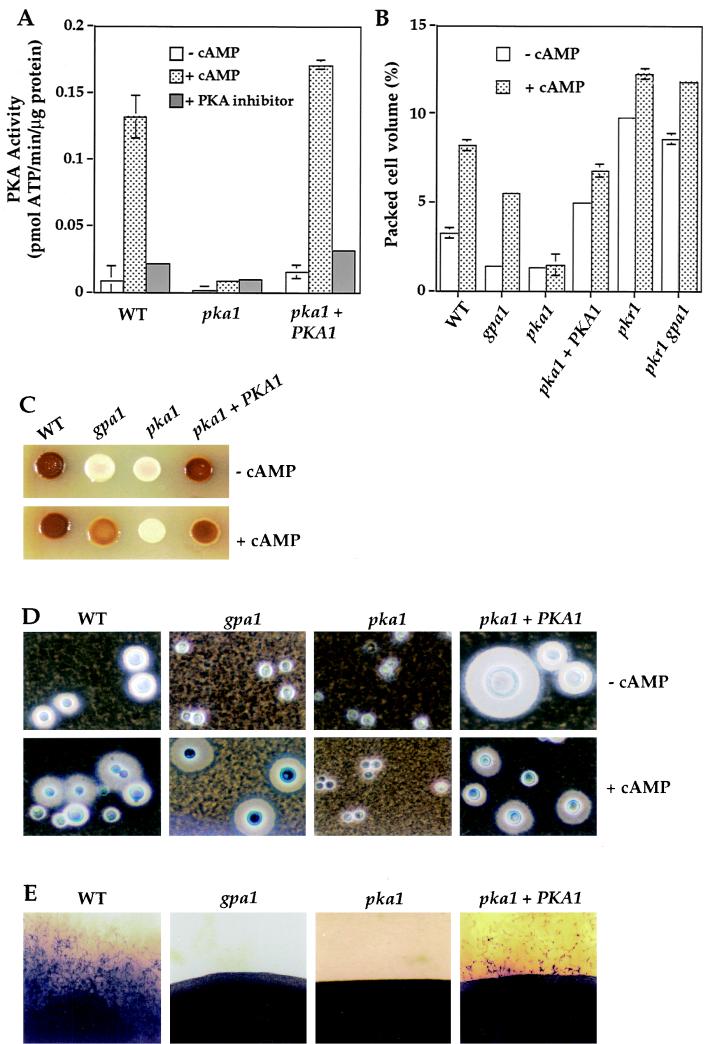
FIG. 1.
PKA regulates melanin and capsule production and mating. (A) Protein extracts were prepared from the wild-type, pka1 mutant, and pka1+PKA1 strains and assayed for PKA activity as described in Materials and Methods. The values shown are the mean of duplicate measurements with the standard error of the mean shown as error bars. In this and other figures, some error bars were too small to be seen. (B) Wild-type, gpa1, pka1, pka1+PKA1, pkr1, and pkr1 gpa1 strains were grown in low-iron medium without or with 10 mM cAMP. Differences in capsule size were quantified by measuring the PCV and are displayed as the mean of triplicate samples with the standard error. (C) Melanin production by the wild-type, gpa1, pka1, and pka1+PKA1 strains was assayed on Niger seed agar without or with 10 mM cAMP. (D) Wild-type, gpa1, pka1, and pka1+PKA1 cells were mixed with India ink, and the capsules, which exclude the ink particles, were photographed. Final magnification, ×360 (E) Mating assays were conducted in which wild-type, gpa1, pka1, and pka1+PKA1 strains were cocultured with serotype D MATa cells on V8 agar for 14 days at 24°C, and the edges of patches of mating cells were photographed (final magnification, ×36). In this assay, a smooth border is produced if no mating occurs, whereas mating results in the production of filaments, basidia, and basidiospores.
pka1 mutant strains exhibit defects in melanin and capsule production.
Similar to gpa1 mutant strains, we found that pka1 mutant strains also exhibited a marked defect in melanization. Melanin production was assayed by culturing on Niger seed (Guizotia abyssinica) medium, on which laccase is induced by limiting glucose and where diphenolic precursors for melanin synthesis are present. The pka1 mutant strain exhibited a severe defect in melanin production, similar to that seen in the gpa1 mutant strain (Fig. 1C). Melanin production was restored in the pka1+PKA1 reconstituted strain. cAMP restored melanin production by the gpa1 mutant strain but not by the pka1 mutant, a result consistent with a model in which Gpa1 regulates cAMP production and PKA is the target of cAMP (Fig. 1C). In a quantitative spectrophotometric assay in which oxidation of diphenolic substrates by cell-associated laccase activity was measured, melanin production was reduced ∼20- to 30-fold by the gpa1 or pka1 mutations compared to wild-type strains (see Materials and Methods and Fig. 4B).
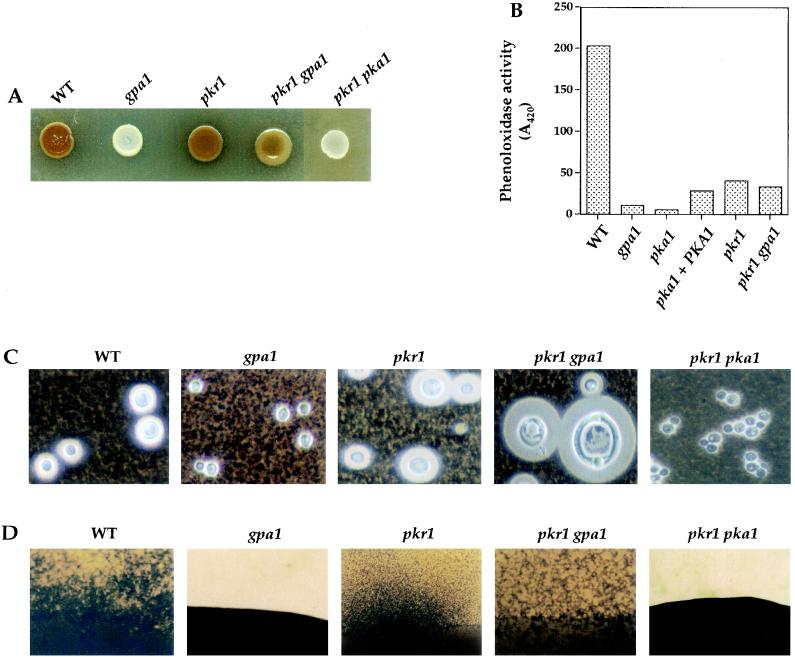
FIG. 4.
PKA regulatory subunit Pkr1 regulates melanin, capsule, and mating and functions downstream of the Gα protein Gpa1. (A) Melanin production in the wild-type, gpa1, pkr1, pkr1 gpa1, and pkr1 pka1 strains was assessed on Niger seed agar following incubation at 37°C for 2 days. (B) Phenoloxidase activity in glucose-starved wild-type, gpa1, pka1, pka1+PKA1, pkr1, and pkr1 gpa1 strains was determined by measuring the oxidation of the diphenolic substrate ABTS (1 U of activity = 0.01 A420 absorbance units in 30 min). The values shown are the average of duplicate samples. (C) The wild-type, gpa1, pkr1, pkr1 gpa1, and pkr1 pka1 strains were grown in low-iron media (LIM plus EDDHA), and the capsule was visualized with India ink and photographed (final magnification, ×324). The packed volumes of these cells are shown in Fig. 1B. (D) Wild-type, gpa1, pkr1, pkr1 gpa1, and pkr1 pka1 strains were cocultured with serotype D MATa cells on V8 agar for 14 days at 24°C and photographed (final magnification, ×32.4).
Pka1 also regulates capsule production. Capsule synthesis was induced by culturing cells in low-iron medium containing an iron chelator (EDDHA). As shown in Fig. 1D, the wild-type and pka1+PKA1 reconstituted strains produced large capsules that excluded India ink particles. In contrast, the pka1 mutant strain exhibited a marked defect in capsule production, a finding similar to that seen with the gpa1 mutant strain. Exogenous cAMP restored capsule production in the gpa1 mutant strain but not in the pka1 mutant strain. cAMP, and also increased Pka1 in the pka1+PKA1 strain, hyperinduced capsule production. These observations were quantified by measuring the packed cell volume of a constant number of fungal cells. The capsule size (and cell volume) of gpa1 and pka1 mutants was reduced ∼3-fold in comparison to wild-type cells (Fig. 1B). The gpa1 and pka1 mutants also produced smaller cells than the wild-type or pka1+PKA1 strains, which may indicate an additional defect in cell size control (Fig. 1D).
Pka1 is required for virulence of C. neoformans.
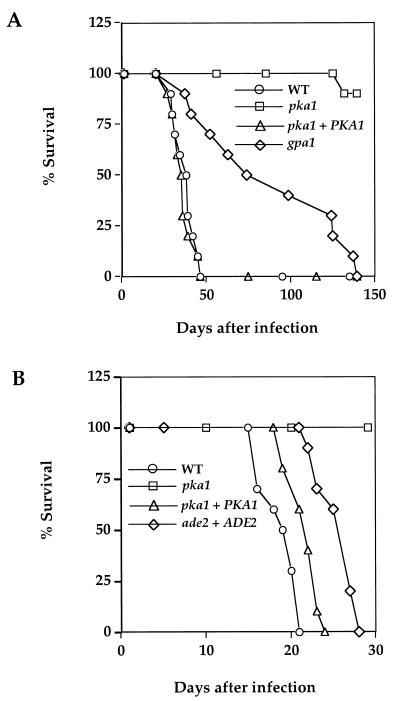
Because pka1 mutant strains have defects in producing melanin and capsule, both established virulence factors, the role of Pka1 in pathogenesis of C. neoformans was assessed in two different animal models. In the first, mice were infected by inhalation with 5 × 104 cells of the wild-type, pka1 mutant, gpa1 mutant, and pka1+PKA1 reconstituted strains. In this model, infection initiates in the lung and spreads hematogenously to other organs. The majority of animals develop severe hydrocephalus and die from cryptococcal meningitis. The average survival of mice infected with the wild-type and reconstituted strains was 36.2 and 34.8 days, respectively (P = 0.57), and both were markedly shorter than the average survival of mice infected with the pka1 (147.5 days, P < 0.0001) or gpa1 mutant strains (86.2 days, P < 0.0001) (Fig. 2A). We note that while 90% of animals infected with the pka1 mutant survived to day 140, 100% of animals infected with the gpa1 mutant succumbed to lethal infection by day 140, although survival was clearly prolonged compared to the wild type (gpa1 versus pka1, P < 0.001). Thus, the phenotype of the pka1 mutant is more severe than that of the gpa1 mutant, a finding consistent with models in which Pka1 functions downstream of Gpa1, and Gpa1 is redundant with other factors regulating adenylyl cyclase (2).
FIG. 2.
PKA regulates virulence of C. neoformans. (A) Groups of 10 female A/Jcr mice were infected with 5 × 104 yeast cells of the wild-type, gpa1, pka1, and pka1+PKA1 strains by nasal inhalation. Survival was monitored over 140 days. (B) Groups of 10 female A/Jcr mice were infected with 5 × 104 yeast cells of the wild-type, pka1, pka1+PKA1, and ade2+ADE2 (M001 ade2 strain restored to prototrophy) strains by nasal inhalation, and survival was monitored for 30 days.
A second virulence experiment in the murine inhalation assay was conducted to compare the pka1 mutant and the pka1+PKA1 reconstituted strain to the ade2 mutant host strain that was used as the transformation recipient to construct these strains (Fig. 2B). In this experiment, the pka1 mutant was again avirulent. Interestingly, the pka1+PKA1 reconstituted strain was modestly increased for virulence compared to the ade2 host strain that was restored to prototrophy by reintroduction of the wild-type ADE2 gene (P = 0.003; Fig. 2B). The ade2 mutant host strain M001 was originally generated by UV irradiation (63, 70) and may differ in virulence potential from the congenic prototrophic parental strain H99. Increased PKA in the pka1+PKA1 reconstituted strain may contribute to enhance virulence beyond the wild-type level.
Virulence of the pka1 mutant was also tested in a rabbit model of cryptococcal meningitis. A total of 108 cells of the wild-type, pka1, and gpa1 mutant strains were injected intrathecally into corticosteroid-immunosuppressed rabbits. The pka1 and gpa1 mutant strains were almost completely cleared from the cerebrospinal fluid (CSF) by day 10 postinfection compared to the isogenic wild-type strain, which persisted at ∼104 cells/ml (data not shown). In summary, the Pka1 catalytic subunit of PKA is required for C. neoformans virulence.
Pka1 regulates mating and filamentation.
To test if Pka1 is required for mating, MATα pka1 mutant cells were cultured with wild-type MATa cells. Like gpa1 mutants, pka1 mutant strains did not produce any mating filaments, basidia, or basidiospores when incubated with MATa cells for 1 to 2 weeks, whereas the wild-type strain produced abundant filaments, basidia, and basidiospores (Fig. 1E). After prolonged incubation (4 weeks), the pka1 mutant did produce some residual filaments. In quantitative assays, cell fusion and recombinant basidiospore production were reduced ∼100-fold with the pka1 mutant compared to the wild type (see Materials and Methods). Mating was restored in the pka1+PKA1 reconstituted strain in both assays (Fig. 1E). Exogenous cAMP restored mating of gpa1 but not pka1 mutants (not shown), indicating Pka1 is required for mating and is the target of cAMP.
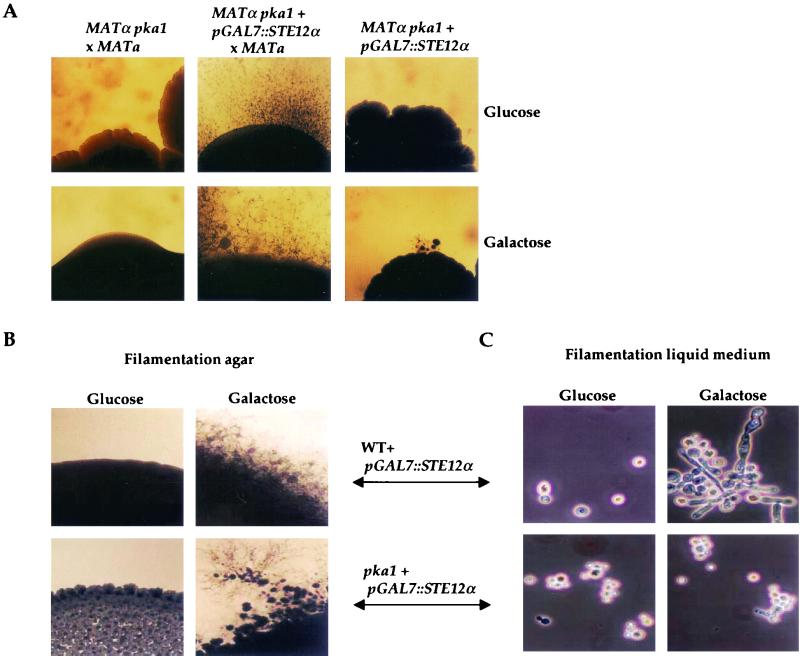
The functions of PKA in mating and filamentous growth were found to be related to the Ste12α transcription factor homolog (13, 79, 82). Overexpression of Ste12α restored mating in pka1 mutant strains, and dikaryotic filaments with fused clamp connections and abundant basidia and basidiospores were produced (Fig. 3A). The pGAL7-regulated STE12α gene restored mating on either galactose (inducing) or glucose (repressing) medium, and expression of the GAL7 gene is known to be less stringently repressed in serotype A strains compared to serotype D strains (17). We also found that overexpression of Ste12α induced haploid filamentous growth not only on nitrogen-limiting solid medium (Fig. 3B), as previously reported (79), but also in nitrogen-limiting liquid medium (Fig. 3C). Pka1 was required for the induction of filamentation by Ste12α in liquid but not solid medium (Fig. 3B and C). The filaments produced in liquid culture in cells overexpressing Ste12α were elongated but failed to produce basidia or basidiospores, indicating that additional environmental signals or factors regulate later steps in filamentous differentiation. The Ste12α proteins from divergent serotype A and D strains of C. neoformans have several conserved PKA consensus sites, and Pka1 was recently found to interact with the Ste12α protein in the two-hybrid system (Y. C. Chang and K. J. Kwon-Chung, personal communication). Ste12α overexpression did not restore melanin or capsule production in pka1 mutant cells (data not shown), suggesting that Ste12α may be one of several downstream targets of PKA.
FIG. 3.
Overexpression of Ste12α restores mating in pka1 mutant cells and induces filamentation in liquid medium in a Pka1-dependent fashion. (A) The MATα pka1 and MATα pka1+pGAL7::STE12α strains were mated with a serotype D MATa PKA1 wild-type strain JEC20 on V8 medium and incubated at 24°C for 14 days. The MATα pka1+pGAL7::STE12α strain was also grown on V8 medium in isolation. The top panels show cultures grown under pGAL7-repressing conditions (glucose), and the bottom panels show cultures grown under pGAL7-inducing conditions (galactose). The edges of the mating patches were photographed (final magnification, ×31.2). (B) The WT+pGAL7::STE12α and pka1+pGAL7::STE12α strains were grown on nitrogen-limiting filamentation agar solid medium under pGAL7-inducing (glucose) or pGAL7-repressing (galactose) conditions. The edges of the colonies were photographed (final magnification, ×31.2). (C) The WT+pGAL7::STE12α and pka1+pGAL7::STE12α strains were grown in nitrogen-limiting filamentation liquid medium with glucose or galactose and photographed (final magnification, ×156).
Identification of the cAMP-dependent protein kinase regulatory subunit Pkr1.
To further analyze PKA function in vivo, the PKR1 gene encoding the PKA regulatory subunit was identified, cloned, and sequenced from the C. neoformans serotype A strain H99 using a probe derived from the Oklahoma EST database (B. A. Roe et al. [http://www.genome.ou.edu/cneo.html]). The 5′ and 3′ RACE products and cDNA clones were isolated and sequenced to establish the structure of the PKR1 gene and to identify the start and stop sites for protein synthesis (GenBank accession no. AF288614). The C. neoformans Pkr1 protein shares sequence identity with many different PKA regulatory subunits, with the highest level of sequence identity to the PKA regulatory subunit from U. maydis (43% identity) (26). The Pkr1 protein was found to interact with the Pka1 catalytic subunit in the yeast two-hybrid assay (see Materials and Methods), further supporting the conclusion that PKR1 encodes the regulatory subunit of cAMP-dependent protein kinase (not shown).
To assess its biological functions, the PKR1 gene was disrupted by inserting the URA5 gene into a unique HpaI site, and the resulting pkr1::URA5 disruption allele was introduced into ura5, ura5 gpa1, and ura5 pka1 C. neoformans strains by biolistic transformation. By Southern blot analysis and PCR with gene-specific primers, the wild-type PKR1 gene was readily replaced with the pkr1::URA5 disruption allele in Ura+ transformants of the wild type (17%), the gpa1 mutant (87%), and the pka1 mutant (7%).
pkr1 mutations increase capsule production and suppress gpa1 mutations.
The effect of the pkr1 mutation on mating and melanin and capsule production was tested (Fig. 4). The pkr1 mutation had no effect on mating in an otherwise wild-type strain (Fig. 4D). The pkr1 mutation suppressed the mating defect conferred by the gpa1 mutation, and mating was restored to a wild-type level in a pkr1 gpa1 double mutant strain (Fig. 4D), providing further evidence that Gpa1 functions upstream in the cAMP-PKA signaling cascade.
Wild-type and pkr1 mutant strains appeared to make similar levels of melanin on Niger seed medium (Fig. 4A). The melanin synthesis defect of the gpa1 mutant was suppressed by the pkr1 mutation, and pkr1 gpa1 double mutants produced increased levels of melanin compared to the gpa1 mutant (Fig. 4A), again indicating that Gpa1 functions upstream of PKA. In a whole-cell assay, laccase activity was found to be decreased ∼5-fold in the pkr1 and pkr1 gpa1 mutant strains compared to the wild type (Fig. 4B). High levels of exogenous cAMP also inhibited melanin production by wild-type strains (not shown). Finally, melanin production was also reduced by PKA overexpression in the pka1+PKA1 reconstituted strain (Fig. 4B). Thus, PKA regulates melanin production, but constitutive or increased PKA activity partially represses melanization.
India ink staining revealed that pkr1 mutant cells produced larger capsules than wild-type cells when they were cultured in low-iron capsule-inducing conditions (Fig. 4C). The capsule production of gpa1 mutant cells was also restored by the pkr1 mutation in the pkr1 gpa1 double mutant strain; in fact, these double mutant cells produced capsules that were significantly larger than wild-type cells and their cell volume was also increased (Fig. 1B and 4C). Increased capsule size in the pkr1 gpa1 mutant cells likely reflects elevated PKA activity, since exogenous cAMP also enhances capsule production (4). We note that the capsules produced by the pkr1 gpa1 mutant cells were larger than those produced by pkr1 single mutant cells, suggesting that Gpa1 may have targets in addition to the PKA pathway and which inhibit capsule production. Previous studies have revealed a similar role for the related Gα protein Gpa2 in S. cerevisiae, which positively regulates cAMP production and pseudohyphal growth and also inhibits the Ime2 kinase to prevent inappropriate entry into meiosis (20, 37, 46). A related Gα protein in Podospora anserina, MOD-D, regulates vegetative growth via a cAMP-dependent pathway and also regulates vegetative incompatibility via a cAMP-independent mechanism (48).
To provide additional evidence that Pka1 is the catalytic subunit and Pkr1 is the regulatory subunit of PKA, pkr1 pka1 double mutant strains were constructed and analyzed. The phenotypes of the pka1 single and the pkr1 pka1 double mutant were indistinguishable (Fig. 4). pkr1 pka1 mutants were sterile and exhibited marked defects in melanin and capsule synthesis. The finding that the pkr1 mutation does not suppress the pka1 mutation is congruent with the observation that cAMP, which inhibits Pkr1 function, also does not suppress the phenotypes of pka1 mutant cells (Fig. 1). This epistasis analysis provides additional evidence that Pka1 functions downstream of Pkr1 and that Pka1 represents the major PKA catalytic subunit.
pkr1 mutations increase virulence and restore virulence of gpa1 mutant strains.
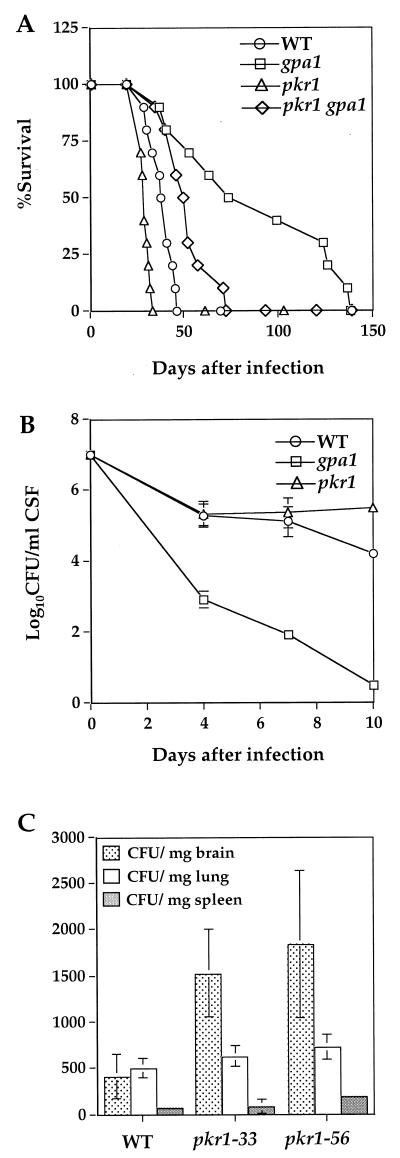
To determine the physiological consequences of constitutive PKA activity and increased capsule production, the virulence of the pkr1 and pkr1 gpa1 mutant strains was assessed in animal models. In the rabbit model of cryptococcal meningitis, the pkr1 mutant was equally or more virulent than the wild type, and viable fungal cells in the CSF of infected animals persisted at >105 CFU/ml for up to 10 days of infection (Fig. 5B).
FIG. 5.
Constitutive activation of PKA enhances virulence. (A) Groups of 10 female A/Jcr mice were infected with 5 × 104 yeast cells of wild-type, and gpa1, pkr1, and pkr1 gpa1 mutant strains by inhalation. The percent survival was plotted for 140 days. (B) Corticosteroid immunosuppressed rabbits were infected with the wild type and gpa1 and pkr1 mutant strains. CSF was withdrawn on days 4, 7, and 10 following infection, and the numbers of viable fungal cells were determined. The values shown are the mean of all cultures for each strain with the standard deviation. (C) Groups of 10 female A/Jcr mice were infected with 106 yeast cells of wild-type or pkr1 mutant strains (pkr1-33 or pkr1-56) by lateral tail vein injections. The brains, lungs, and spleens were harvested on day 7 from three randomly chosen mice from each group. Quantitative cultures were performed by plating the tissue homogenates on YPD medium. The culture data from each sample were averaged and analyzed with a paired Student's t test.
The pkr1 mutant strain was also more virulent than the isogenic wild-type strain in A/Jcr mice (Fig. 5A). A total of 50% of the animals infected by inhalation with the pkr1 mutant strain survived to day 28 postinfection (average survival of 28.3 days), whereas 50% of mice infected with the wild-type strain survived to day 38 postinfection (average survival of 36.2 days; P = 0.043) (Fig. 5A). One-hundred-percent lethality occurred on day 33 with the pkr1 mutant compared to day 46 with the isogenic wild-type strain. In a second independent experiment, the pkr1-33 mutant strain was also found to be more virulent than the H99 ura5 strain used to construct the pkr1-33 mutant and which was restored to prototrophy by transformation with the URA5 gene (P = 0.002; data not shown). In accord with previous studies (4), the virulence of the gpa1 mutant strain was attenuated in mice, and virulence was largely restored in the pkr1 gpa1 double mutant (gpa1 versus pkr1 gpa1; P = 0.035) (Fig. 5A). These findings reveal that the Gpa1-cAMP-PKA signaling cascade plays a central role in regulating virulence of C. neoformans.
A potential mechanism of increased virulence of the pkr1 mutant strain was further assessed in a murine intravenous model of cryptococcosis. In this case, BALB/c mice were infected by tail vein injection with wild-type cells (H99) or two independent pkr1 mutants (pkr1-33 and pkr1-56), and survival was monitored. We found that 50% of the mice infected with the wild-type strain survived to day 15 postinfection (average survival of 15.7 days) (Fig. 6A). In contrast, 50% of the animals infected with the pkr1-33 mutant strain survived to day 9 postinfection (average survival of 10 days; P = 0.007) and 50% infected with the pkr1-56 strain survived to day 11 (average survival of 10.6 days; P = 0.014). One-hundred-percent lethality occurred on day 26 postinfection with the wild type and on days 16 and 18 with the two independent pkr1 mutants (Fig. 6A). These findings support the conclusion that the constitutive activation of PKA results in a hypervirulent phenotype in animal models of cryptococcosis.
FIG. 6.
The virulence of pkr1 mutant strains is increased in BALB/c mice. Groups of 20 female BALB/c mice were infected with 106 yeast cells of wild-type or two independent pkr1 mutants (pkr1-33 and pkr1-56) by lateral tail vein injections. (A) The percent survival of 10 mice from each group was plotted for 26 days. (B) On days 3 and 7 postinfection, five mice from each group were sacrificed, and the brain homogenates were quantitatively cultured on YPD medium. Culture data from each group of five samples were averaged and analyzed with a paired Student's t test. (C) Cells and capsules of the wild-type and two pkr1 mutant strains (pkr1-33 and pkr1-56) grown in capsule-noninducing YPD medium prior to infection (in vitro) and in the brain homogenates of infected animals (in vivo) were stained with India ink and photographed (×900). The scale bar in the middle panels is 10 μm in length and was used to calculate cell and capsule size and volume.
As a second measure of increased virulence, the tissue burden of fungal cells was determined in the brains, lungs, and spleens of A/Jcr mice (Fig. 5C) and in the brains of BALB/c mice after intravenous injection of 106 C. neoformans cells (Fig. 6B). In A/Jcr mice, the CFU of fungal cells per milligram of brain at day 7 after infection were increased fourfold in animals infected with either the pkr1-33 or the pkr1-56 mutant compared to the wild-type strain, whereas little or no difference was apparent in the fungal burden in the lung or spleen (Fig. 5C). Similarly, after 7 days of infection the CFU of fungal cells per milligram of brain were increased five to sixfold in BALB/c mice infected with the pkr1 mutants (P = 0.001384 and 0.000473) compared to the wild-type strain (Fig. 6B). These findings indicate that the fungal burden of cells in the CNS is significantly elevated in animals infected with the pkr1 mutants compared to animals infected with the wild type.
pkr1 mutant strains produce dramatically enlarged capsules in vivo.
The pkr1 mutant strains produced significantly larger capsules in the CNS of infected animals. When cultured in vitro in capsule noninducing YPD medium prior to infection, both the wild type and the pkr1 mutants produced only very limited capsules (Fig. 6C). Notably, following infection, the sizes of capsules of cryptococcal cells in the brain homogenates of animals infected with pkr1 mutants were dramatically increased. The size of the capsule was induced ∼10-fold in wild-type cells and ∼30-fold in the pkr1 mutant cells. Thus, on average the capsules of pkr1 mutant cells were threefold thicker (pkr1-33, 8.1 ± 1.9 μm; pkr1-56, 7.9 ± 2.1 μm; n = 100 cells each) than the capsules surrounding wild-type cells (3.0 ± 0.9 μm; n = 40 cells). This difference is considerably more dramatic when the volume of the capsular sphere is calculated. In this case, the volume of the capsular shell was obtained by subtracting the volume of the cell from the volume of the cell plus the capsule. By this measure, the capsule volume produced by the pkr1 mutants is increased 12- to 13-fold compared to that produced by the wild type (7,910 and 8,030 μm3 for the pkr1-33 and pkr1-56 mutants, respectively, compared to 617 μm3 for the wild type). In addition to the increased capsule volume, the pkr1 mutant cells were larger than the wild-type cells, with a diameter increased by ∼2-fold (pkr1-33, 8.9 ± 2.1 μm; pkr1-56, 9.5 ± 2.1 μm; n = 100 cells each) compared to the wild type (4.9 ± 1.1 μm; n = 40 cells) (Fig. 6C). The dramatic increase in capsule size observed with the pkr1 mutants in the CNS of infected animals provides a plausible molecular mechanism for their increased virulence as measured by survival and the quantitative cell counts and tissue burden in rabbits and mice.
PKA pathway regulates cAMP production.
The PKA signaling pathway was also found to feedback regulate cAMP production. Previous studies in S. cerevisiae have revealed that the PKA signaling pathway participates in a negative feedback loop that represses cAMP production (51, 54). Here we addressed whether PKA regulates cAMP levels via a similar pathway in C. neoformans. When glucose was readded to glucose-starved C. neoformans cells, cAMP production was stimulated similar to previous findings in S. cerevisiae (33, 72, 73) (Fig. 7). Importantly, the basal level of cAMP was dramatically elevated in pka1 mutant cells lacking PKA (Fig. 7A), providing evidence that PKA normally functions to limit the excursions of intracellular cAMP concentrations to a much more modest range. In addition, cAMP levels were found to be modestly reduced in pka1+PKA1 and pkr1 mutant cells with reconstituted or increased PKA activity, respectively (Fig. 7B), providing additional evidence that activation of PKA downregulates cAMP production. These observations indicate that PKA feedback inhibits cAMP production in C. neoformans, a finding analogous to those of previous studies in S. cerevisiae (51, 54).
DISCUSSION
Our studies define the elements of a signal transduction cascade that controls the production of virulence factors and pathogenicity of C. neoformans. The Pka1 catalytic subunit of PKA regulates mating, melanin and capsule production, and virulence. The Pkr1 regulatory subunit of PKA is also a critical component, and mutants lacking Pkr1 overproduced capsule and were hypervirulent by several measures in two different animal models. pkr1 mutant cells also produced dramatically enlarged capsules during infection, and both the larger capsule size and the increased release of immunosuppressive capsular polysaccharides likely contribute to enhanced virulence.
Epistasis analysis further supports the conclusion that the Gα protein Gpa1 is an upstream controlling element for this signaling pathway and that the Ste12α transcription factor may represent one of several downstream targets of PKA that regulate differentiation and virulence. One interesting finding is that mutants with defects in an upstream signaling component (gpa1) were largely restored to virulence by a constitutive downstream mutation (pkr1). These findings indicate that virulence can be uncoupled from the normal regulatory inputs that control the Gpa1-cAMP-PKA signaling pathway in wild-type cells. Similar mutations may be selected against in natural populations in the environment in which upregulated capsule production may have either deleterious effects or divert metabolic potential to unnecessary capsule synthesis. Finally, our findings reveal that the virulence of C. neoformans can be increased by mutations, and this may be occurring in strains that infect individuals with no apparent defects in immune system function.
Our finding that constitutive activation of the PKA signaling pathway results in the production of enlarged cells, increased capsule, and hypervirulence is relevant to previous reports on the association of capsule and cell size with virulence. Two previous reports described the identification of C. neoformans isolates that produced dramatically enlarged cells (40 to 60 μm in diameter) in samples from human lung infection or a brain abscess and also in animal models (15, 49). In one case (15), the isolate formed normal sized yeast cells during in vitro culture, whereas the second isolate formed enlarged yeast cells when cultured at 35°C in brain heart infusion broth (25 μm in diameter) (49). Such clinical isolates could have defects in PKA signaling similar to pkr1 mutants.
Previous studies revealed a correlation between the presence and the relative size of the capsule in the resistance of fungal cells to phagocytosis by macrophages (44, 81). C. neoformans is a facultative intracellular pathogen, and macrophages that have engulfed fungal cells contain multiple intracellular vesicles filled with capsular polysaccharide (24). The capsule likely contributes to virulence by interfering with immune cell function by shed capsular antigen, inhibiting phagocytosis of fungal cells, and enhancing intracellular survival of fungal cells in macrophages. Correspondingly, acapsular mutant strains are avirulent compared to isogenic encapsulated wild-type strains (9–12). Our studies demonstrate that pkr1 mutants with constitutive PKA activity are hypervirulent and produce dramatically enlarged capsules in vivo. Thus, both the presence and the size of the capsule likely contribute to C. neoformans virulence.
Our studies on the hypervirulent pkr1 mutant strain may also be relevant to the relative contributions of melanin and capsule to the virulence of C. neoformans. The pkr1 mutant strains produce enlarged capsules in vitro and in vivo and yet produce ∼20% the level of melanin compared to the wild type. High levels of exogenous cAMP also led to a similar reduction in melanin production. Hence, increased PKA pathway activity hyperinduces capsule production but partially attenuates melanin production. Even so, by several measures the pkr1 mutants are hypervirulent. These findings have several possible implications. First, melanin production may not be limiting during infection, and even a reduced level might be sufficient for virulence. Second, in previous genetic studies, serotype D laccase mutant strains were found to be attenuated for virulence but were still capable of causing lethal infections (68). Thus, melanin production is important but not essential for pathogenesis. The melanin biosynthetic enzyme laccase is expressed in vivo (68), and melanin is produced in vivo in infected animals and in the CNS of patients with cryptococcal meningitis (8, 57, 66). However, fungal cells are not heavily pigmented in vivo (45) and the structure of melanin produced in vivo might differ from that produced in vitro (8). Our findings suggest increased capsule production, even in strains with a reduction in melanin biosynthetic capacity, is the main contributor to hypervirulence in animal models of cryptococcosis.
Previous studies in the yeast S. cerevisiae revealed that the PKA pathway regulates cAMP production via a negative feedback loop (51, 54; reviewed in references 72 and 73). Part of this feedback loop may involve activation of the low-affinity phosphodiesterase Pde1 by PKA-dependent phosphorylation (50). Nonetheless, it has been controversial whether a similar regulatory system operates in other fungi. Our findings provide evidence that a similar regulatory network regulates cAMP production in C. neoformans. Most notably, we found that the basal levels of cAMP were dramatically elevated in strains lacking the PKA catalytic subunit Pka1 (Fig. 7). These findings suggest activation of the PKA pathway normally leads to transient and modest increases in cAMP levels. One reason the cAMP biosynthetic pathway might be regulated in such a fashion is to prevent the inhibition of laccase expression and melanogenesis that occurs in response to hyperactivated PKA signaling. If cAMP levels were not tightly controlled the resulting induction of capsule might also have deleterious consequences. Our findings that the regulation of cAMP production by PKA occurs in a pathogenic basidiomycete that is quite divergent from S. cerevisiae suggests similar regulatory mechanisms likely operate in other fungi.
A related G protein-cAMP-PKA signaling pathway regulates mating, development, and virulence in both model yeasts and plant fungal pathogens (reviewed in references 5, 34, and 43). In budding and fission yeasts, the Gα proteins homologous to C. neoformans Gpa1 (Gpa2 in S. cerevisiae and Gpa2 in S. pombe) are coupled to a nutrient-sensing G-protein-coupled receptor (Gpr1 in S. cerevisiae and Git3 in S. pombe) that regulates cAMP production and PKA activation during mating and filamentation (30, 33, 37, 42, 46, 47, 55, 61, 65, 78, 80).
Mating, filamentous growth, and virulence of the plant fungal pathogen U. maydis are similarly regulated by a related Gα protein (Gpa3), cAMP, and PKA (23, 26, 27, 35, 60, 64). Mutations in the regulatory G protein (gpa3), adenylyl cyclase (uac1), or one of the catalytic subunits of PKA (adr1) abrogate cAMP production or signaling in U. maydis and cause constitutive filamentation (26, 35). Mutations in the gene encoding the PKA regulatory subunit, ubc1, restore normal budding growth in hyperfilamentous gpa3 or uac1 mutants, indicating that Gpa3 regulates the cAMP-PKA signaling pathway similar to the regulation of the Gpa1-cAMP-PKA pathway in C. neoformans (26). PKA hyperactivation also alters virulence of U. maydis (27, 36). Mutant strains expressing a dominant-active Gpa3 mutant allele (Q206L) or lacking the PKA regulatory subunit (Δubc1) colonize maize plants and cause localized symptoms. However, plants infected with Δubc1 mutant dikaryons fail to form tumors (galls) (27), whereas those infected with Gpa3-Q206L mutant strains form smaller tumors (36). Interestingly, U. maydis cells expressing the dominant active Gpa3-Q206L mutant or lacking the PKA regulatory subunit (Δubc1) produce a capsule-like material (36). The nature of this capsule-like material remains to be defined, but these findings are strikingly similar to our finding that hyperactivation of the PKA signaling pathway dramatically enhances capsule production in C. neoformans.
In summary, a conserved Gα protein-cAMP-PKA signal transduction cascade operates during virulence and differentiation of both human and plant fungal pathogens that can be targeted for therapeutic intervention. Hyperactivation of the PKA signaling pathway leads to alterations of virulence in two different fungal pathogens, resulting in defects in gall formation in the plant fungal pathogen U. maydis compared to an enhancement of virulence in the human fungal pathogen C. neoformans. These studies illustrate how a conserved signaling pathway has been coopted to serve related but distinct functions as pathogenic fungi have evolved to adapt to different host environments.
ACKNOWLEDGMENTS
We thank Cristl Arndt and Lora Cavallo for technical assistance, Dana Davis and members of the Heitman lab for discussions, and Yun Chang and June Kwon-Chung for communication of results prior to publication.
This work was supported by NIAID R01 grants AI39115 and AI42159 (to J.H. and J.R.P.), P01 award AI44975 from NIAID to the Duke University Mycology Research Unit, and K08 career development award AI01556 from NIAID (to J.A.A.). Gary Cox is a Burroughs Wellcome New Investigator in Molecular Pathogenic Mycology. Joseph Heitman is a Burroughs Wellcome Scholar in Molecular Pathogenic Mycology and an associate investigator of the Howard Hughes Medical Institute.
REFERENCES
- 1.Aberg J A, Mundy L M, Powderly W G. Pulmonary cryptococcosis in patients without HIV infection. Chest. 1999;115:734–740. doi: 10.1378/chest.115.3.734. [DOI] [PubMed] [Google Scholar]
- 2.Alspaugh J A, Cavallo L M, Perfect J R, Heitman J. RAS1 regulates filamentation, mating and growth at high temperature of Cryptococcus neoformans Mol. Microbiol. 2000;36:352–365. doi: 10.1046/j.1365-2958.2000.01852.x. [DOI] [PubMed] [Google Scholar]
- 3.Alspaugh J A, Davidson R C, Heitman J. Morphogenesis of Cryptococcus neoformans. In: Ernst J F, Schmidt A, editors. Dimorphism in human pathogenic and apathogenic yeasts. 5. Contributions in microbiology. Basel, Switzerland: Karger; 2000. pp. 217–238. [DOI] [PubMed] [Google Scholar]
- 4.Alspaugh J A, Perfect J R, Heitman J. Cryptococcus neoformans mating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Borges-Walmsley M I, Walmsley A R. cAMP signalling in pathogenic fungi: control of dimorphic switching and pathogenicity. Trends Microbiol. 2000;8:133–141. doi: 10.1016/s0966-842x(00)01698-x. [DOI] [PubMed] [Google Scholar]
- 6.Bourbonnais R, Paice M G. Oxidation of non-phenolic substrates. An expanded role for laccase in lignin biodegradation. FEBS Lett. 1990;267:99–102. doi: 10.1016/0014-5793(90)80298-w. [DOI] [PubMed] [Google Scholar]
- 7.Casadevall A, Perfect J R. Cryptococcus neoformans. Washington, D.C.: ASM Press; 1998. [Google Scholar]
- 8.Casadevall A, Rosas A L, Nosanchuk J D. Melanin and virulence in Cryptococcus neoformans Curr. Opin Microbiol. 2000;3:354–358. doi: 10.1016/s1369-5274(00)00103-x. [DOI] [PubMed] [Google Scholar]
- 9.Chang Y C, Cherniak R, Kozel T R, Granger D L, Morris L C, Weinhold L C, Kwon-Chung K J. Structure and biological activities of acapsular Cryptococcus neoformans 602 complemented with the CAP64 gene. Infect Immun. 1997;65:1584–1592. doi: 10.1128/iai.65.5.1584-1592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chang Y C, Kwon-Chung K J. Complementation of a capsule-deficient mutation of Cryptococcus neoformans restores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y C, Kwon-Chung K J. Isolation, characterization, and localization of a capsule-associated gene, CAP10, of Cryptococcus neoformans. J Bacteriol. 1999;181:5636–5643. doi: 10.1128/jb.181.18.5636-5643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang Y C, Penoyer L A, Kwon-Chung K J. The second capsule gene of Cryptococcus neoformans, CAP64, is essential for virulence. Infect Immun. 1996;64:1977–1983. doi: 10.1128/iai.64.6.1977-1983.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chang Y C, Wickes B L, Miller G F, Penoyer L A, Kwon-Chung K J. Cryptococcus neoformans STE12α regulates virulence but is not essential for mating. J Exp Med. 2000;191:871–882. doi: 10.1084/jem.191.5.871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen S, Sorrell T, Nimmo G, Speed B, Currie B, Ellis D, Marriott D, Pfeiffer T, Parr D, Byth K. Epidemiology and host- and variety-dependent characteristics of infection due to Cryptococcus neoformans in Australia and New Zealand. Clin Infect Dis. 2000;31:499–508. doi: 10.1086/313992. [DOI] [PubMed] [Google Scholar]
- 15.Cruickshank J G, Cavill R, Jelbert M. Cryptococcus neoformans of unusual morphology. Appl Microbiol. 1973;25:309–312. doi: 10.1128/am.25.2.309-312.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cruz M C, Poeta M D, Wang P, Wenger R, Zenke G, Quesniaux V F J, Movva N R, Perfect J R, Cardenas M E, Heitman J. Immunosuppressive and nonimmunosuppressive cyclosporin analogs are toxic to the opportunistic fungal pathogen Cryptococcus neoformans via cyclophilin-dependent inhibition of calcineurin. Antimicrob Agents Chemother. 2000;44:143–149. doi: 10.1128/aac.44.1.143-149.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Del Poeta M, Toffaletti D L, Rude T H, Dykstra C C, Heitman J, Perfect J R. Topoisomerase I is essential in Cryptococcus neoformans: role in pathobiology and as an antifungal target. Genetics. 1999;152:167–178. doi: 10.1093/genetics/152.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dong Z M, Murphy J W. Cryptococcal polysaccharides induce L-selectin shedding and tumor necrosis factor receptor loss from the surface of human neutrophils J. Clin Investig. 1996;97:689–698. doi: 10.1172/JCI118466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong Z M, Murphy J W. Effects of the two varieties of Cryptococcus neoformans cells and culture filtrate antigens on neutrophil locomotion. Infect Immun. 1995;63:2632–2644. doi: 10.1128/iai.63.7.2632-2644.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Donzeau M, Bandlow W. The yeast trimeric guanine nucleotide-binding protein α subunit, Gpa2p, controls the meiosis-specific kinase Ime2p activity in response to nutrients. Mol Cell Biol. 1999;19:6110–6119. doi: 10.1128/mcb.19.9.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dromer F, Aucouturier P, Clauvel J P, Saimot G, Yeni P. Cryptococcus neoformans antibody levels in patients with AIDS. Scand J Infect Dis. 1988;20:283–285. doi: 10.3109/00365548809032452. [DOI] [PubMed] [Google Scholar]
- 22.Dromer F, Ronin O, Dupont B. Isolation of Cryptococcus neoformans var. gattii from an Asian patient in France: evidence for dormant infection in healthy subjects. J Med Vet Mycol. 1992;30:395–397. [PubMed] [Google Scholar]
- 23.Dürrenberger F, Wong K, Kronstad J W. Identification of a cAMP-dependent protein kinase catalytic subunit required for virulence and morphogenesis in Ustilago maydis. Proc Natl Acad Sci USA. 1998;95:5684–5689. doi: 10.1073/pnas.95.10.5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformans is a facultative intracellular pathogen in murine pulmonary infection. Infect Immun. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garcia-Hermoso D, Janbon G, Dromer F. Epidemiological evidence for dormant Cryptococcus neoformans infection. J Clin Microbiol. 1999;37:3204–3209. doi: 10.1128/jcm.37.10.3204-3209.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gold S, Duncan G, Barrett K, Kronstad J. cAMP regulates morphogenesis in the fungal pathogen Ustilago maydis. Genes Dev. 1994;8:2805–2816. doi: 10.1101/gad.8.23.2805. [DOI] [PubMed] [Google Scholar]
- 27.Gold S E, Brogdon S M, Mayorga M E, Kronstad J W. The Ustilago maydis regulatory subunit of a cAMP-dependent protein kinase is required for gall formation in maize. Plant Cell. 1997;9:1585–1594. doi: 10.1105/tpc.9.9.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Granger D L, Perfect J R, Durack D T. Virulence of Cryptococcus neoformans: regulation of capsule synthesis by carbon dioxide. J Clin Investig. 1985;76:508–516. doi: 10.1172/JCI112000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henderson D K, Bennett J E, Huber M A. Long-lasting specific immunologic unresponsiveness associated with cryptococcal meningitis. J Clin Investig. 1982;69:1185–1190. doi: 10.1172/JCI110555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isshiki T, Mochizuki N, Maeda T, Yamamoto M. Characterization of a fission yeast gene, gpa2, that encodes a Gα subunit involved in the monitoring of nutrition. Genes Dev. 1992;6:2455–2462. doi: 10.1101/gad.6.12b.2455. [DOI] [PubMed] [Google Scholar]
- 31.Jacobson E S, Tinnell S B. Antioxidant function of fungal melanin. J Bacteriol. 1993;175:7102–7104. doi: 10.1128/jb.175.21.7102-7104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kozel T R, Cazin J J. Nonencapsulated variant of Cryptococcus neoformans. I. Virulence studies and characterization of soluble polysaccharide. Infect Immun. 1971;3:287–294. doi: 10.1128/iai.3.2.287-294.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraakman L, Lemaire K, Ma P, Teunissen A W R H, Donaton M C V, Dijck P V, Winderickx J, De Winde J H, Thevelein J M. A Saccharomyces cerevisiae G-protein coupled receptor, Gpr1, is specifically required for glucose activation of the cAMP pathway during the transition to growth on glucose. Mol Microbiol. 1999;32:1002–1012. doi: 10.1046/j.1365-2958.1999.01413.x. [DOI] [PubMed] [Google Scholar]
- 34.Kronstad J, Maria A D, Funnell D, Laidlaw R D, Lee N, Sá M M d, Ramesh M. Signaling via cAMP in fungi: interconnections with mitogen-activated protein kinase pathways. Arch Microbiol. 1998;170:395–404. doi: 10.1007/s002030050659. [DOI] [PubMed] [Google Scholar]
- 35.Krüger J, Loubradou G, Regenfelder E, Hartmann A, Kahmann R. Crosstalk between cAMP and pheromone signalling pathways in Ustilago maydis. Mol Gen Genet. 1998;260:193–198. doi: 10.1007/s004380050885. [DOI] [PubMed] [Google Scholar]
- 36.Krüger J, Loubradou G, Wanner G, Regenfelder E, Feldbrugge M, Kahmann R. Activation of the cAMP pathway in Ustilago maydis reduces fungal proliferation and teliospore formation in plant tumors. Mol Plant-Microbe Interact. 2000;13:1034–1040. doi: 10.1094/MPMI.2000.13.10.1034. [DOI] [PubMed] [Google Scholar]
- 37.Kübler E, Mösch H U, Rupp S, Lisanti M P. Gpa2p, a G-protein alpha-subunit, regulates growth and pseudohyphal development in Saccharomyces cerevisiae via a cAMP-dependent mechanism. J Biol Chem. 1997;272:20321–20323. doi: 10.1074/jbc.272.33.20321. [DOI] [PubMed] [Google Scholar]
- 38.Kwon-Chung K J. A new genus, Filobasidiella, the perfect state of Cryptococcus neoformans. Mycologia. 1975;67:1197–1200. [PubMed] [Google Scholar]
- 39.Kwon-Chung K J. A new species of Filobasidiella, the sexual state of Cryptococcus neoformans B and C serotypes. Mycologia. 1976;68:943–946. [PubMed] [Google Scholar]
- 40.Kwon-Chung K J, Polacheck I, Popkin T J. Melanin-lacking mutants of Cryptococcus neoformans and their virulence for mice. J Bacteriol. 1982;150:1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kwon-Chung K J, Rhodes J C. Encapsulation and melanin formation as indicators of virulence in Cryptococcus neoformans. Infect Immun. 1986;51:218–223. doi: 10.1128/iai.51.1.218-223.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Landry S, Pettit M T, Apolinario E, Hoffman C S. The fission yeast git5 gene encodes a Gβ subunit required for glucose-triggered adenylate cyclase activation. Genetics. 2000;154:1463–1471. doi: 10.1093/genetics/154.4.1463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lengeler K B, Davidson R C, D'Souza C, Harashima T, Shen W-C, Wang P, Pan X, Waugh M, Heitman J. Signal transduction cascades regulating fungal development and virulence. Microbiol Mol Biol Rev. 2000;64:746–785. doi: 10.1128/mmbr.64.4.746-785.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levitz S M, DiBenedetto D J. Differential stimulation of murine resident peritoneal cells by selectively opsonized encapsulated and acapsular Cryptococcus neoformans. Infect Immun. 1988;56:2544–2551. doi: 10.1128/iai.56.10.2544-2551.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu L, Wakamatsu K, Ito S, Williamson P R. Catecholamine oxidative products, but not melanin, are produced by Cryptococcus neoformans during neuropathogenesis in mice. Infect Immun. 1999;67:108–112. doi: 10.1128/iai.67.1.108-112.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenz M C, Heitman J. Yeast pseudohyphal growth is regulated by GPA2, a G protein α homolog. EMBO J. 1997;16:7008–7018. doi: 10.1093/emboj/16.23.7008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lorenz M C, Pan X, Harashima T, Cardenas M E, Xue Y, Hirsch J P, Heitman J. The G protein-coupled receptor GPR1 is a nutrient sensor that regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Genetics. 2000;154:609–622. doi: 10.1093/genetics/154.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Loubradou G, Begueret J, Turcq B. MOD-D, a Gα subunit of the fungus Podospora anserina, is involved in both regulation of development and vegetative incompatibility. Genetics. 1999;152:519–528. doi: 10.1093/genetics/152.2.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Love G L, Boyd G D, Greer D L. Large Cryptococcus neoformans isolated from brain abscess. J Clin Microbiol. 1985;22:1068–1070. doi: 10.1128/jcm.22.6.1068-1070.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ma P, Wera S, Dijck P V, Thevelein J M. The PDE1-encoded low-affinity phosphodiesterase in the yeast Saccharomyces cerevisiae has a specific function in controlling agonist-induced cAMP signaling. Mol Biol Cell. 1999;10:91–104. doi: 10.1091/mbc.10.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mbonyi K, Aelst L V, Argüelles J C, Jans A W H, Thevelein J M. Glucose-induced hyperaccumulation of cyclic AMP and defective glucose repression in yeast strains with reduced activity of cyclic AMP-dependent protein kinase. Mol Cell Biol. 1990;10:4518–4523. doi: 10.1128/mcb.10.9.4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mitchell T G, Perfect J R. Cryptococcosis in the era of AIDS—100 years after the discovery of Cryptococcus neoformans. Clin Microbiol Rev. 1995;8:515–548. doi: 10.1128/cmr.8.4.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neilson J B, Fromtling R A, Bulmer G S. Cryptococcus neoformans: size range of infectious particles from aerosolized soil. Infect Immun. 1977;17:634–638. doi: 10.1128/iai.17.3.634-638.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nikawa J, Cameron S, Toda T, Ferguson K M, Wigler M. Rigorous feedback control of cAMP levels in Saccharomyces cerevisiae. Genes Dev. 1987;1:931–937. doi: 10.1101/gad.1.9.931. [DOI] [PubMed] [Google Scholar]
- 55.Nocero M, Isshiki T, Yamamoto M, Hoffman C S. Glucose repression of fbp1 transcription in Schizosaccharomyces pombe is partially regulated by adenylate cyclase activation by a G protein α subunit encoded by gpa2 (git8) Genetics. 1994;138:39–45. doi: 10.1093/genetics/138.1.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nosanchuk J D, Rosas A L, Casadevall A. The antibody response to fungal melanin in mice. J Immunol. 1998;160:6026–6031. [PubMed] [Google Scholar]
- 57.Nosanchuk J D, Rosas A L, Lee S C, Casadevall A. Melanisation of Cryptococcus neoformans in human brain tissue. Lancet. 2000;355:2049–2050. doi: 10.1016/S0140-6736(00)02356-4. [DOI] [PubMed] [Google Scholar]
- 58.Nosanchuk J D, Valadon P, Feldmesser M, Casadevall A. Melanization of Cryptococcus neoformans in murine infection. Mol Cell Biol. 1999;19:745–750. doi: 10.1128/mcb.19.1.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Nunez M, Peacock J J E, Chin J R. Pulmonary cryptococcosis in the immunocompetent host. Therapy with oral fluconazole: a report of four cases and a review of the literature. Chest. 2000;118:527–534. doi: 10.1378/chest.118.2.527. [DOI] [PubMed] [Google Scholar]
- 60.Orth A B, Rzhetskaya M, Pell E J, Tien M. A serine (threonine) protein kinase confers fungicide resistance in the phytopathogenic fungus Ustilago maydis. Appl Environ Microbiol. 1995;61:2341–2345. doi: 10.1128/aem.61.6.2341-2345.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pan X, Heitman J. Cyclic AMP-dependent protein kinase regulates pseudohyphal differentiation in Saccharomyces cerevisiae. Mol Cell Biol. 1999;19:4874–4887. doi: 10.1128/mcb.19.7.4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Patel P, Ramanathan J, Kayser M, Baran J J. Primary cutaneous cryptococcosis of the nose in an immunocompetent woman. J Am Acad Dermatol. 2000;43:344–345. doi: 10.1067/mjd.2000.100961. [DOI] [PubMed] [Google Scholar]
- 63.Perfect J R, Toffaletti D L, Rude T H. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformans in cerebrospinal fluid. Infect Immun. 1993;61:4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Regenfelder E, Spellig T, Hartmann A, Lauenstein S, Bölker M, Kahmann R. G proteins in Ustilago maydis: transmission of multiple signals? EMBO J. 1997;16:1934–1942. doi: 10.1093/emboj/16.8.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Robertson L S, Fink G R. The three yeast A kinases have specific signaling functions in pseudohyphal growth. Proc Natl Acad Sci USA. 1998;95:13783–13787. doi: 10.1073/pnas.95.23.13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rosas A L, Nosanchuk J D, Feldmesser M, Cox G M, McDade H C, Casadevall A. Synthesis of polymerized melanin by Cryptococcus neoformans in infected rodents. Infect Immun. 2000;68:2845–2853. doi: 10.1128/iai.68.5.2845-2853.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rupp S, Summers E, Lo H, Madhani H, Fink G. MAP kinase and cAMP filamentation signaling pathways converge on the unusually large promoter of the yeast FLO11 gene. EMBO J. 1999;18:1257–1269. doi: 10.1093/emboj/18.5.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Salas S D, Bennett J E, Kwon-Chung K J, Perfect J R, Williamson P R. Effect of the laccase gene, CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sia R A, Lengeler K B, Heitman J. Diploid strains of the pathogenic basidiomycete Cryptococcus neoformans are thermally dimorphic. Fungal Genet Biol. 2000;29:153–163. doi: 10.1006/fgbi.2000.1192. [DOI] [PubMed] [Google Scholar]
- 70.Sudarshan S, Davidson R C, Heitman J, Alspaugh J A. Molecular analysis of the Cryptococcus neoformans ADE2 gene, a selectable marker for transformation and gene disruption. Fungal Genet Biol. 1999;27:36–48. doi: 10.1006/fgbi.1999.1126. [DOI] [PubMed] [Google Scholar]
- 71.Sukroongreung S, Kitiniyom K, Nilakul C, Tantimavanich S. Pathogenicity of basidiospores of Filobasidiella neoformans var. neoformans. Med Mycol. 1998;36:419–424. [PubMed] [Google Scholar]
- 72.Thevelein J M, Cauwenberg L, Colombo S, De Winde J H, Donaton M, Dumortier F, Kraakman L, Lemaire K, Ma P, Nauwelaers D, Rolland F, Teunissen A, Dijck P V, Versele M, Wera S, Winderickx J. Nutrient-induced signal transduction through the protein kinase A pathway and its role in the control of metabolism, stress resistance, and growth in yeast Enzyme Microb. Technol. 2000;26:819–825. doi: 10.1016/s0141-0229(00)00177-0. [DOI] [PubMed] [Google Scholar]
- 73.Thevelein J M, De Winde J H. Novel sensing mechanisms and targets for the cAMP-protein kinase A pathway in the yeast Saccharomyces cerevisiae. Mol Microbiol. 1999;33:904–18. doi: 10.1046/j.1365-2958.1999.01538.x. [DOI] [PubMed] [Google Scholar]
- 74.Vartivarian S E, Anaissie E J, Cowart R E, Sprigg H A, Tingler M J, Jacobson E S. Regulation of cryptococcal capsular polysaccharide by iron. J Infect Dis. 1993;167:186–190. doi: 10.1093/infdis/167.1.186. [DOI] [PubMed] [Google Scholar]
- 75.Vecchiarelli A, Retini C, Monari C, Tascini C, Bistoni F, Kozel T R. Purified capsular polysaccharide of Cryptococcus neoformans induces interleukin-10 secretion by human monocytes. Infect Immun. 1996;64:2846–2849. doi: 10.1128/iai.64.7.2846-2849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Vecchiarelli A, Retini C, Pietrella D, Monari C, Tascini C, Beccari T, Kozel T R. Downregulation by cryptococcal polysaccharide of tumor necrosis factor alpha and interleukin-1β secretion from human monocytes. Infect Immun. 1995;63:2919–2923. doi: 10.1128/iai.63.8.2919-2923.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang Y, Casadevall A. Susceptibility of melanized and nonmelanized Cryptococcus neoformans to nitrogen- and oxygen-derived oxidants. Infect Immun. 1994;62:3004–3007. doi: 10.1128/iai.62.7.3004-3007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Welton R M, Hoffman C S. Glucose monitoring in fission yeast via the gpa2 Gα, the git5 Gβ and the git3 putative glucose receptor. Genetics. 2000;156:513–521. doi: 10.1093/genetics/156.2.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wickes B L, Edman U, Edman J C. The Cryptococcus neoformans STE12α gene: a putative Saccharomyces cerevisiae STE12 homologue that is mating type specific. Mol Microbiol. 1997;26:951–960. doi: 10.1046/j.1365-2958.1997.6322001.x. [DOI] [PubMed] [Google Scholar]
- 80.Xue Y, Batlle M, Hirsch J P. GPR1 encodes a putative G protein-coupled receptor that associates with the Gpa2p Gα subunit and functions in a Ras-independent pathway. EMBO J. 1998;17:1996–2007. doi: 10.1093/emboj/17.7.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yasuoka A, Kohno S, Yamada H, Kaku M, Koga H. Influence of molecular sizes of Cryptococcus neoformans capsular polysaccharide on phagocytosis. Microbiol Immunol. 1994;38:851–856. doi: 10.1111/j.1348-0421.1994.tb02137.x. [DOI] [PubMed] [Google Scholar]
- 82.Yue C, Cavallo L M, Alspaugh J A, Wang P, Cox G M, Perfect J R, Heitman J. The STE12α homolog is required for haploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]