Abstract
The increasing levels of pesticide resistance in agricultural pests and disease vectors represents a threat to both food security and global health. As insecticide resistance intensity strengthens and spreads, the likelihood of a pest encountering a sub-lethal dose of pesticide dramatically increases. Here, we apply dynamic Bayesian networks to a transcriptome time-course generated using sub-lethal pyrethroid exposure on a highly resistant Anopheles coluzzii population. The model accounts for circadian rhythm and ageing effects allowing high confidence identification of transcription factors with key roles in pesticide response. The associations generated by this model show high concordance with lab-based validation and identifies 44 transcription factors putatively regulating insecticide-responsive transcripts. We identify six key regulators, with each displaying differing enrichment terms, demonstrating the complexity of pesticide response. The considerable overlap of resistance mechanisms in agricultural pests and disease vectors strongly suggests that these findings are relevant in a wide variety of pest species.
Keywords: Insecticide resistance, transcriptional response, transcription factors, Bayesian, time course, Anopheles, pesticide resistance, transcriptomics
Introduction
Insecticides are critical for control of pests in agriculture and disease vectors in public health. The intensive and widespread use of insecticides in each of these settings has led to extensive insecticide resistance (WHO 2020), which poses a threat to both food security and global health. Vector borne diseases account for more than 17% of all infectious diseases annually (WHO 2020), whilst around 35% of crops are lost to pre-harvest pests, underlining the importance of pesticide chemistries in global health and food security (Popp et al., 2013). Malaria control highlights the pivotal role of insecticides in global health with over 80% of the reductions in malaria cases since the turn of the century attributed to their use (Bhatt et al., 2015). Malaria control relies heavily on the distribution and use of insecticide treated bed nets (ITNs), which provide protection to the user and wider community protection through insecticide induced mortality of the adult Anopheles vectors (Hawley et al., 2003, Killeen and Smith, 2007, Killeen et al., 2011). All ITNs currently in use contain the pyrethroid class of insecticide; a fast-acting chemistry that induces immediate knockdown and mortality in susceptible mosquitoes. However, strength of resistance to pyrethroids is now such that populations of Anopheles can survive exposure with minimal effect on their life span (Hughes et al., 2020). Surviving sub-lethal exposures to pesticides is likely to have large and sustained consequences on the biology of the pest species.
Resistance to insecticides both in agricultural pests and disease vectors have been attributed to three characterised mechanisms; changes to the insecticide target site (Weill et al., 2004, Martinez-Torres et al., 1998), thickening of the cuticle to reduce penetrance (Balabanidou et al., 2016) and metabolic clearance through overexpression of detoxification protein families (Müller et al., 2008, Voice et al., 2015, Ingham et al., 2018). Recently, new resistance mechanisms have been reported (Ingham et al., 2018, Ingham et al., 2019) and sub-lethal exposure has been shown to induce large-scale transcriptomic changes, highlighting the complexity of the insects response to insecticides (Ingham et al., 2020).
The demonstration of large-scale changes in transcriptome post-exposure emphasises the importance of transcriptional control in response to insecticide. Despite this, the induction of genes in response to insecticides is poorly studied and the regulatory processes underlying these mechanisms have remained elusive. In most important pests, cis or trans-acting regulatory elements are yet to be identified, and little published research has focused on the role of the non-coding regulatory machinery. Although recent work has identified transcription factors involved in insecticide resistance such as two transcriptional pathways: the Nrf2-cnc pathway in both disease vectors (Ingham et al., 2017, Bottino-Rojas et al., 2018) and agricultural pests (Kalsi and Palli, 2015, Gaddelapati et al., 2018) and the ARNT-AhR in agricultural pests (Peng et al., 2017, Hu et al., 2019), no studies in either setting have examined transcriptional response in a holistic manner. The availability of transcriptomic time-series data from resistant Anopheles coluzzii mosquitoes post-pyrethroid exposure (Ingham et al., 2020) has provided a resource to examine the importance of multiple transcription factors in response to insecticide.
Elucidating complex gene networks from transcriptomic time course data is a fundamental problem in computational systems biology (Delgado and Gómez-Vela, 2019, Thompson et al., 2015, Jackson et al., 2020). Time course data enables measurements of mRNA levels post-perturbation and allows identification of transcripts following similar expression patters over time. Measuring changes in mRNA levels acts as a proxy for protein expression, but regulatory relationships cannot be captured by correlation alone, due to the presence of indirect regulation (gene A regulates gene B which regulates gene C), and post-transcriptional changes. To allow reconstruction of gene regulatory networks, dynamic Bayesian networks have successfully been applied to real-world time course studies (Murphy and Mian, 1999, Dondelinger et al., 2013, Dondelinger and Mukherjee, 2019), allowing identification of key regulatory pathways within a system. These models additionally allow correction for confounding factors. For example, as circadian rhythms can play a significant role in gene expression patterns over short time-scales (Rund et al., 2011), sinusoidal patterns with 24-hour period may be corrected for.
Here, we apply a modified dynamic Bayesian network method to whole-organism microarray data taken at ten time-points post exposure to a pyrethroid insecticide. The method corrects for both circadian patterns and mosquito ageing, which have previously been shown to be important in the insecticide resistance phenotype (Jones et al., 2012, Rund et al., 2016). The Bayesian network approach allows identification of key regulatory factors influencing the expression of transcripts in response to insecticide exposure. Based on validation experiments, we estimate that the inferred network has 70% precision, indicating strong concordance of experimental data to model prediction. The network is made freely available through a ShinyR application, allowing non-bioinformaticians to easily access and visualise the data. Several transcription factors are highlighted as potential key regulators in response to pyrethroid insecticide. This study demonstrates the importance and complexity of transcriptional control of insecticide response, which is likely to have cross species applicability due to relative conservation of transcriptional pathways (Hsia and McGinnis, 2003) and near total overlap of resistance mechanisms.
Results
Identification of transcription factors involved in insecticide resistance
Of the transcripts in the Anopheles microarray, approximately 4% are putative transcription factors, based on FlyMine.org AGAP homologs of Drosophila transcription factors found on FlyTF.org. As exploration of all possible transcription factor/transcript associations was not computationally feasible, the number of transcription factors had to be reduced to <50. Of the 559 total transcription factors, 44 were used in further analyses (Table 1). These transcription factors were selected based on resistance-associated GO term enrichments in transcription factor-transcript clusters (Zhang et al., 2018) found using a previously published library of microarray data comparing resistant and susceptible Anopheles species across Africa (Ingham et al., 2018). A number of these transcription factors have known roles in stress response in Drosophila (Table 1); however, only Maf-S, Met and Dm have previously been linked with insecticide response in mosquitoes (Ingham et al., 2018, Ingham et al., 2017). Of the transcription factors selected for analysis the following have been studied in mosquitoes: p53 has been shown to respond to arboviral infection (Chen et al., 2017); Rbsn-5 has been shown to be involved in egg shell formation (Amenya et al., 2010); l(1)sc is linked with sensory tissue development (Wülbeck and Simpson, 2002); kayak is involved in salivary gland response to arboviral infection through JNK pathway activation (Chowdhury et al., 2020); Hnf4 is linked to ecdysone and Met mediated lipid metabolism (Wang et al., 2017); Cyc controls the circadian ryhthm (Maliti et al., 2016); REL1 and REL2 are involved in immune response (Luna et al., 2006); Kr-h1 is essential for egg development (Fu et al., 2020) and Pan is linked with chromatin changes upon Plasmodium infection (Ruiz et al., 2019).
Table 1.
List of Transcription Factors included in further analysis. 44 Transcription factors included in the analysis with the dynamic Bayesian model, including VectorBase Transcript ID, Drosophila gene name, FBgn identifier, % identity (taken from VectorBase), putative function and network interactor summary KEGG/GO enrichment from this study (See S1 Table 1).
| Transcript ID | Gene Name | Homolog | % Identity | Role in Drosophila | Citation | Network Enrichment |
|---|---|---|---|---|---|---|
| AGAP000057-RA | shaven (sv) | FBgn0005561 | 34.12 | Sensory tissue development | Kavaler et al. 1999 (Kavaler et al., 1999) | None |
| AGAP000066-RA | Sox102F | FBgn0039938 | Neuronal development, behaviour and Wnt signalling | Li et al. 2017 (Li et al., 2017) | mTOR and ECM-receptor interaction | |
| AGAP000141-RA | CG31224 | FBgn0051224 | 17.03 | Unknown | Nuclear-related | |
| AGAP000547-RA | Rbsn-5 | FBgn0261064 | 42.29 | Endosome assembly | Morrison et al 2008 (Morrison et al., 2008) | Polarity, Wingless |
| AGAP000646-RA | Diminuitive (Dm, dMyc) | FBgn0262656 | 13.21 | Glucose and lipid metabolism, development | Parisi et al. 2013 (Parisi et al., 2013) | Sugar Metabolism, Miscellaneous Metabolism |
| AGAP000876-RA | achaete-scute complex (l(1)sc) | FBgn0002561 | 26.42 | Neuronal development, dopaminergic neurons | Stagg et al 2011 (Stagg et al., 2011) | Cuticle-related, Neuroactive ligand-receptor |
| AGAP001093-RA | kayak (kay) | FBgn0001297 | 30.06 | JNK signalling, wound healing, neuronal development | Ramet et al. 2002 (Rämet et al., 2002); Miotto et al. 2006 (Miotto et al., 2006) | RNA/DNA-related Processes |
| AGAP001156-RA | PSEA-binding protein 95kD (Pbp95) | FBgn0037540 | 13.89 | Small nuclear RNA activating complex | Li et al 2004 (Li et al., 2004) | Cytochrome P450s, Signalling Pathways |
| AGAP001388-RA | Doublesex-Mab related 93B (dmrt93B) | FBgn0038851 | 41.61 | Mouth development | Panara et al 2019 (Panara et al., 2019) | Taste/sense-related, Oxidoreductase Activity |
| AGAP001786-RA | Osa | FBgn0261885 | 36.83 | EGFR signalling | Terriente-Feliz and de Celis 2009 (Terriente-Félix and de Celis, 2009) | GSTs |
| AGAP001994-RA | Brahma associated protein 111kD (Bap111) | FBgn0030093 | 40.1 | Chromatin remodelling | Papoulas et al. 2001 (Papoulas et al., 2001) | Miscellaneous Metabolism, Cytochrome P450s, COEs |
| AGAP002082-RA | Squeeze (sqz) | FBgn0010768 | 35.47 | Neuronal development | Terriente-Feliz et al 2007 (Félix et al., 2007) | Ligase Activity |
| AGAP002155-RA | Hepatocyte nuclear factor 4 (Hnf4) | FBgn0004914 | 52.85 | Lipid mobilisation, glucose homeostasis and mitochondrial function | Palanker et al. 2009 (Palanker et al., 2009); Barry and Thummel 2016 (Barry and Thummel, 2016) | Glyoxylate Metabolism, Transcription Coactivator |
| AGAP002352-RB | p53 | FBgn0039044 | 14.2 | Genotoxic stress response | Brodsky et al. 2004 (Brodsky et al., 2004) | Carbon metabolism |
| AGAP002773-RA | Stripe (sr) | FBgn0003499 | Muscle development | Lee et al. 1995 (Lee et al., 1995) | Steroid biosynthesis | |
| AGAP002902-RA | Medea (Med) | FBgn0011655 | 52.42 | Muscle development through BMP and dpp Pathways | Wisotzkey et al. 1998 (Wisotzkey et al., 1998) | Metabolism-related |
| AGAP002920-RA | CG17829 | FBgn0025635 | 17.84 | Unknown | Protein Complex Binding, DNA/RNA processes | |
| AGAP002954-RA | Cell division cycle 5 (Cdc5) | FBgn0035136 | 63.63 | Spliceosome | Herold et al. 2009 (Herold et al., 2009) | Notch Signalling, Apoptosis |
| AGAP003117-RA | Capicua (cic) | FBgn0262582 | 19.37 | EGFR, Torso and TOLL signalling | Astigarraga et al. 2007 (Astigarraga et al., 2007); Papagianni et al.2018 (Papagianni et al., 2018) | Glycan degradation |
| AGAP003449-RA | Rootletin (Root) | FBgn0039152 | 46.08 | Hearing, touch and taste | Chen et al. 2015 (Chen et al., 2015) | Steroid Biosynthesis, Receptor-related activity, Cytochrome P450s |
| AGAP003669-RA | Drop (Dr) | FBgn0000492 | 61.4 | Eye and nerve development | Tearle et al. 1994 (Tearle et al., 1994) | Circadian Pathway |
| AGAP004864-RA | Protein on ecdysone puffs (Pep) | FBgn0004401 | 38.87 | Hsp70 response through hnRNP complex | Hamann et al. 1998 (Hamann and Strätling, 1998) | Response to xenobiotics |
| AGAP004990-RA | Multiprotein bridging factor 1 (mbf1) | FBgn0262732 | 74.15 | Co-activator to induce stress-response genes | Jindra et al. 2004 (Jindra et al., 2004) | Translation-related Processes |
| AGAP005437-RA | Inverted repeat binding protein 18 kDa (Irbp18) | FBgn0036126 | Inhibitor of the conserved stress response protein dATF4/Crc | Blanco et al 2020 (Blanco et al., 2020) | Fatty Acid-related | |
| AGAP005551-RA | Rabaptin-5-associated exchange factor for Rab5 (Rabex-5) | FBgn0262937 | 37.75 | Ras pathway homeostasis | Yan et al. 2010 (Yan et al., 2010) | Apoptosis |
| AGAP005641-RA | CG9705 | FBgn0036661 | 54.78 | Sensory neurons | Iyer et al. 2013 (Iyer et al., 2013) | Protein Sorting, Response to DNA-damage |
| AGAP005655-RA | Cylce (Cyc) | FBgn0023094 | 35.25 | Circadian rhythm | Rutila et al. 1998 (Rutila et al., 1998) | UGTs, Hormone Biosynthesis |
| AGAP006022-RA | Methoprene tolerant (Met) | FBgn0002723 | 21.2 | Juvenile hormone binding | Jindra et al. 2015 (Jindra et al., 2015) | Oxidative Phosphorylation |
| AGAP006061-RA | Ken | FBgn0000286 | 5.92 | JAK/STAT pathway | Arbouzova et al. 2006 (Arbouzova et al., 2006) | GTPase Activity, Vesicle-related, Actin-related |
| AGAP006392-RA | CG4617 | FBgn0029936 | 38.58 | Unknown | Autophagy | |
| AGAP006601-RA | MEP-1 | FBgn0035357 | 31.69 | Chromatin remodelling | Reddy et al. 2010 (Reddy et al., 2010) | Peroxisome, CSPs |
| AGAP006642-RA | Defective proventriculus (dve) | FBgn0020307 | 47.98 | Mitochondrial reactive oxygen species modulator | Baqri et al. 2014 (Baqri et al., 2014) | Behavioural-related, Neuron-related |
| AGAP006736-RA | Sugarbabe (sug) | FBgn0033782 | 28.24 | Regulation of lipid and carbohydrate metabolism | Varghese et al. 2010 (Varghese et al., 2010) | P450, IMD-pathway |
| AGAP006747-RA | Relish (REL2) | FBgn0014018 | 24.12 | Immune response | Dushay et al. 1996 (Dushay et al., 1996) | Transferase, Dendrite-related, CSPs |
| AGAP009444-RA | Suppressor of variegation 205 (Su(var)205) | FBgn0003607 | 23.47 | Hsp70 response through activation of euchromatic genes | Piacentini et al. 2003 (Piacentini et al., 2003) | Ribosome-related, Hippo signalling |
| AGAP009494-RA | Ets at 21C (Ets21C) | FBgn0005660 | 34.25 | Stress inducible transcription factor through JNK | Mundorf et al. 2019 (Mundorf et al., 2019) | Behaviour-related, Neuronal, JAK/STAT |
| AGAP009515-RA | REL1 | FBgn0260632 | 38.96 | Toll pathway | Gross et al. 1999 (Gross et al., 1999) | Vesicle-related Transport, Mitophagy, Toll pathway |
| AGAP009662-RA | Kruppel Homolog 1 (Kr-h1) | FBgn0028420 | 36.47 | 20-hydroxyecdysone linked | Pecasse et al. 2000 (Pecasse et al., 2000) | TCA-cycle |
| AGAP009676-RA | Chameau (chm) | FBgn0028387 | 34.66 | JNK signalling | Miotto et al. 2006 (Miotto et al., 2006) | Transmembrane Signalling, Behavioural-related, Neuronal |
| AGAP009888-RA | CG33695 | FBgn0052831 | 53.3 | Unknown | Hippo Signalling, COEs | |
| AGAP009899-RA | klumpfuss (klu) | FBgn0013469 | 42.86 | Cell death, mitochondrial function, EGFR signalling | Protzer etl al. 2008 (Protzer et al., 2008); Chen et al. 2008 (Chen et al., 2008) | Morphogenesis, Drug Metabolism, UGTs, GSTs |
| AGAP009983-RA | Net | FBgn0002931 | 35.88 | EGFR signalling | Terriente-Feliz and de Celis 2009 (Terriente-Félix and de Celis, 2009) | MAPK/Notch Signalling |
| AGAP010405-RA | Maf-S | FBgn0034534 | 63.7 | Reactive oxygen species stress response | Misra et al. 2011 (Misra et al., 2011) | Respiration-related, Insulin-related |
| AGAP012389-RA | Pangolin (Pan) | FBgn0085432 | 24.47 | Wingless signalling | Brunner et al. 1997 (Brunner et al., 1997) | Wnt-signalling, COEs |
Modelling the insecticide response network
To explore the role of the identified transcription factors in insecticide response, a previously generated time course experiment comparing pyrethroid exposed and unexposed Anopheles coluzzii was used (Ingham et al., 2020). This dataset was then used to model the gene regulatory relationships using a dynamic Bayesian network (DBN) approach (Dondelinger et al., 2013) which infers the regulators of each transcript from the set of selected transcription factors using the time-course of log-fold changes compared to the unexposed baseline measurement, correcting for ageing and circadian rhythms. A Markov chain Monte Carlo (MCMC) algorithm was used to draw samples from the posterior distribution of the network model given the data, and associations were then ranked between target genes and transcription factors using the marginal posterior probability of the corresponding edge (defined as a predicted transcription factor – transcript association) in the network. Since experimental validation of all discovered edges is prohibitively expensive, an important consideration was how many associations needed to be tested in order to establish the validity of the network inference approach. A simulation study was performed under the assumption that the number of genes regulated by each transcription factor follows a Poisson distribution with parameter λ=10. We showed that under some assumptions (see Materials and Methods) testing 4 regulatory relationships for each of 7 transcription factors has a 70% chance of obtaining an estimate of the precision that falls within 10% of the true precision, and a 95% chance of obtaining an estimate that falls within 20% of the true precision. For 5 transcription factors with 4 regulatory relationships, this still gives a 65% chance of an estimate within 10%, and a 90% chance of an estimate within 20% of the true precision.
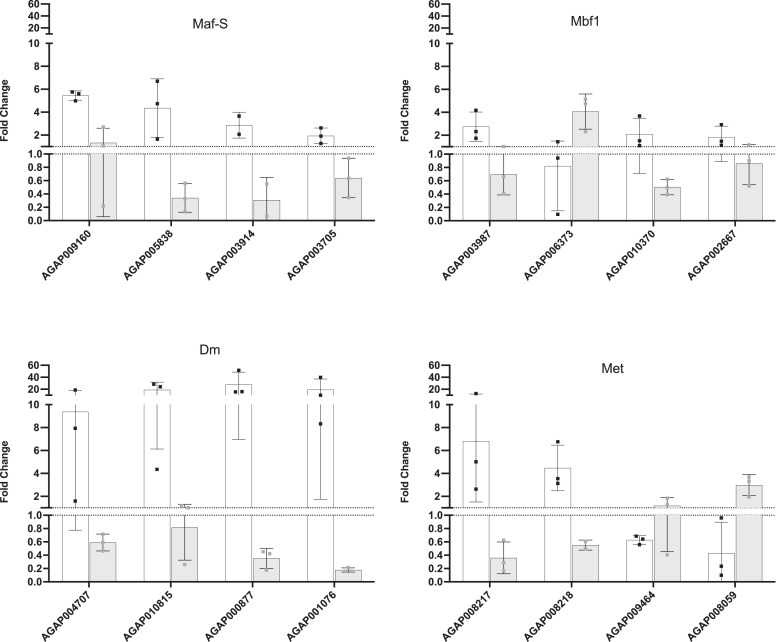
The model was validated using quantitative PCR to confirm the interactions predicted by the model. Successful dsRNA mediated knock down was performed on 5 transcription factors, these showed knock down 48-hours post insecticide exposure (Supplementary Figure 1); the single time point used for model confirmation (Supplementary Figure 1, Supplementary Table 1). Four transcript interactors were chosen randomly for each transcription factor based on a posterior probability of > 0.1. To determine the change in transcript expression post-exposure and to determine whether predicted interactors were influenced by the knock down of the stated transcription factor 2 comparisons were made: (i) GFP-injected exposed vs GFP-injected unexposed and (ii) Exposed transcription factor knockdown compared to exposed GFP-injected for the two comparisons respectively (Supplementary Table 1). Of the 16 interactors (4 transcription factors x 4 interactors), 11 demonstrated concordance with the model, showing a substantial change in expression due to transcription factor knockdown, indicating 69% model precision (Figure 2, Supplementary Table 1).
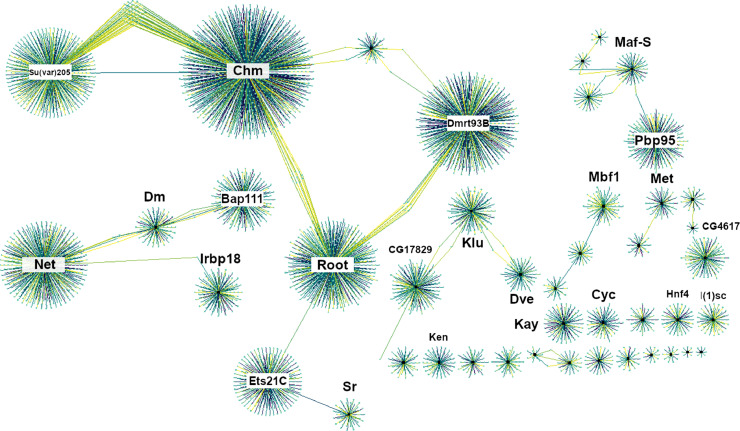
Figure 2.
Network overview. Emboldened black circles represent all 44 transcription factors, with grey nodes representing associated transcripts. Directed edges are coloured on posterior probability gradient from yellow (0.39) through green (0.5) to dark blue (0.97). High posterior probability indicates higher confidence in the interaction. The 23 hub transcription factors, with > 50 associations are labelled.
Network Overview
In order to determine what the optimal cut-off for the marginal posterior probability values should be, a permutation test was performed whereby the observed log-fold values for one of the 44 transcription factors are randomly permuted, so that the original time associations were no longer present (Appendix 1). Any association between this transcription factor and the target gene would then be purely due to chance. This process was then repeated 500 times, inferring the edges for all 44 transcription factors each time. The resulting marginal posterior probability values were then analysed for the randomised transcription factor and showed that a threshold of 0.39 was sufficient to only produce one false positive out of 500 randomizations, or a false positive rate of 0.002 (Appendix 1), which resulted in assignment of 5136 transcripts to the 44 transcription factors.
The complete network using a posterior probability cut-off of 0.39 is displayed in Figure 1. Due to the constraints imposed by this model on number of parent nodes tested, simple network descriptive data was generated only for edges from the selected transcription factors. The average edge count was 118.48±179.62 demonstrating high variance in connectivity as seen in Figure 1 with a range of 8 associations to 951. 23 transcription factors are network hubs, defined as nodes with a high number of associations (>50) (Table 2), including Dm, Met and Maf-S all previously linked with the insecticide resistance phenotype (Ingham et al., 2018, Ingham et al., 2017) and mbf1 a stress response transcription factor (Jindra et al., 2004). To enable the network to be freely accessible an application NetworkVis has been written in ShinyR (Chang et al., 2017) and is available online (https://github.com/VictoriaIngham/NetworkVis_TimeCourse; Supplementary Table 2) with all associated data. Users can manually select a posterior probability cut-off between 0.1-0.8, select and rearrange nodes and edges in the network and identify a priori transcription factors through visual means rather than working with a large text file.
Figure 1.
Model validation. mRNA fold change (y-axis) of each transcript (x-axis) for each transcription factor showing knockdown 48-hours post-deltamethrin exposure. White bars show qPCR results from GFP-injected exposed mosquitoes (48-hours post exposure) compared to GFP-injected unexposed mosquitoes (48-hours post injection) to show induction effect in absence of treatment and grey bars show transcription factor-injected exposed (48 hours post exposure) vs GFP-injected exposed mosquitoes (48-hours) to demonstrate the effect of transcription factor knockdown. Error bars show standard deviation.
Table 2.
Transcription factor hubs. Identifier, gene name and number of associations for 23 transcription factor hubs within the network.
| Transcription Factor | Name | Number of associations |
|---|---|---|
| AGAP009676-RA | Chm | 951 |
| AGAP001388-RA | dmrt93B | 535 |
| AGAP009444-RA | Su(var)205 | 447 |
| AGAP003449-RA | Root | 447 |
| AGAP009983-RA | Net | 399 |
| AGAP009494-RA | Ets21C | 227 |
| AGAP001994-RA | Bap111 | 201 |
| AGAP001156-RA | Pbp95 | 185 |
| AGAP002920-RA | CG17829 | 145 |
| AGAP009899-RA | Klu | 118 |
| AGAP005437-RA | Irbp18 | 113 |
| AGAP006392-RA | CG4617 | 98 |
| AGAP000646-RA | Dm | 91 |
| AGAP001093-RA | Kay | 87 |
| AGAP004990-RA | Mbf1 | 72 |
| AGAP006642-RA | Dve | 69 |
| AGAP005655-RA | Cyc | 66 |
| AGAP010405-RA | Maf-S | 64 |
| AGAP002155-RA | Hnf4 | 60 |
| AGAP006601-RA | Ken | 58 |
| AGAP000876-RA | l(1)sc | 57 |
| AGAP006022-RA | Met | 53 |
| AGAP002773-RA | Sr | 51 |
Enrichment analysis was run for every transcription factor and associated interactors for all GO term categories (Ashburner et al., 2000), KEGG pathways (Kanehisa and Goto, 2000), gene families previously associated with resistance (Balabanidou et al., 2016, Müller et al., 2008, Voice et al., 2015, Ingham et al., 2018, Ingham et al., 2019) and Reactome pathways based on Drosophila homology (Jassal et al., 2020) (Table 1, Supplementary Table 3).
GO enrichments were present for 21/44 of the transcription factors across all ontology categories (Molecular Function, Cellular Component and Biological Process). A large number of GO terms were significant across different transcription factor interactions analysed; however, the terms were largely non-overlapping indicating that the transcription factors are playing differing roles in insecticide response (Supplementary Table 3, Supplementary Figure 2). Seven GO terms (dendrite, dendritic tree, somatodendritic compartment, transmembrane signalling receptor activity, signalling receptor activity, response to drugs) were significant across four transcription factors and relate to terms clearly involved in stress response and associated behavioural changes.
KEGG enrichments were present for 39/44 transcription factors (Supplementary Table 3, Supplementary Figure 3), again there was minimal overlap in the enriched pathways, in agreement with the divergent enriched GO terms. One KEGG pathway was significant for six transcription factor associations (neuroactive ligand-receptor interaction) and two terms were significant for four transcription factor associations (insect hormone biosynthesis, other glycan degradation).
Given our a priori knowledge of insecticide resistance, enrichment analysis was also carried out for detoxification gene families, the cuticular hydrocarbon synthesis pathway and chemosensory proteins; three well described resistance mechanisms (Balabanidou et al., 2016, Müller et al., 2008, Voice et al., 2015, Ingham et al., 2019). Enrichments for these families occurred in 20/44 transcription factors with cytochrome p450s being significantly enriched in eight, GSTs in four, UGTs in three, COEs in eight, chemosensory proteins in two and the cuticular hydrocarbon pathway in three (Supplementary Table 3, Supplementary Figure 4). Reactome enrichment was also carried out, with significance for at least one pathway in 21/44 of the transcription factors (Supplementary Table 3, Supplementary Figure 5).
Taken together, these data indicate that the applied DBN is successfully capturing differing roles of the transcription factors in insecticide exposure response and the enrichment of a large number of a priori detoxification candidates indicates we are successfully capturing transcription factors controlling metabolic response to insecticide exposure.
Key transcriptional regulators of insecticide response
Transcription factors that have previously been implicated in insecticide resistance or stress response and those that have interactors which show a clear functional enrichment from the above analysis are described in greater detail below.
Chameau
Chameau (Chm, AGAP009676-RA) is the transcription factor with the highest number of interactors at 951. Chm interactors are strongly enriched in transmembrane signalling activity (p = 1.32e-8), sensory perception (p = 3.39e-6) and chemosensory behaviour (p = 8.74e-4). 26 other transcription factors interact directly with Chm including Abd-B (AGAP004664-RA) including a known Drosophila interaction and so (AGAP011695-RA), Fer3 (AGAP003756-RA), disco (AGAP01106-RA), C15 (AGAP003674-RA), zfh1 (AGAP000779-RA), hkb (AGAP004517), all known secondary interactors. 14 interactors have posterior probabilities of >0.90, including fringe (AGAP006439-RA) a gene involved in regulating the Notch signalling pathway (Moloney et al., 2000), which is significantly enriched in Chm interactors (p = 0.012) and Roquin (AGAP007114-RA) a protein that translocates to stress granules on chemical induced toxicity (Athanasopoulos et al., 2010, Voßfeldt et al., 2012).
Diminuitive
Diminuitive (Dm, AGAP00646-RA) is a central network hub with 91 interactors and its interactors are enriched in multiple KEGG pathways such as N-glycan biosynthesis, protein processing in the endoplasmic recticulum and starch and sucrose metabolism (Supplementary Table 3) (Martinez-Torres et al., 1998, Ingham et al., 2017, Nagy et al., 2013, Kappes et al., 2011). Previous work has demonstrated that attenuating Dm expression in An. gambiae results in significantly higher mortality post-pyrethroid exposure (Ingham et al., 2018); this role is underlined by significant enrichment of detoxification gene families in this cluster including the COEs (p = 0.031) and ABCs (p = 7.2e-3) (Wilding et al., 2014, Riveron et al., 2014). Interestingly, the ABCs in this network belong to the ABCB family of transporters, which are known as multi-drug transporters and are implicated in insecticide resistance in Drosophila and Anopheles (Gellatly et al., 2015, Pignatelli et al., 2018). Dm also interacts with Bap111, whose network is enriched for fatty acid degradation and cuticular hydrocarbon synthesis and contains the cytochrome p450 CYP4G17, previously linked with cuticular thickening in resistant mosquitoes (Balabanidou et al., 2016). (Balabanidou et al., 2016)
Doublesex-Mab related 93B
Doublesex-Mab related 93B (dmrt93B, AGAP001388-RA) is the second most well-connected node with 535 interactors. Dmrt93B is enriched in multiple GO-terms related to xenobiotic metabolism, including oxidoreductase activity (p = 7.7e-3), heme-binding (p = 2.6e-4) and monooxygenase activity (p = 0.014) as well as being highly enriched in the a priori detoxification gene families; cytochrome p450s (p = 5.53e-6), COEs (p = 0.023) and GSTs (p = 0.029). Taken together, these enrichments indicate that dmrt93B is playing a central role in the response of metabolic transcripts to insecticide exposure. Although not showing enrichment in a related term, 14 cuticular proteins are present in this interactome, one of which CPLCP11 (AGAP009758-RA) has been shown to be up-regulated in resistant mosquitoes compared to susceptible (Balabanidou et al., 2019) and another, CPR133 (AGAP009872-RA), has the highest posterior probability (0.93).
Met and Maf-S
Both Maf-S (AGAP010405-RA) and Met (AGAP006022-RA) have previously been shown to have important roles in insecticide response (Ingham et al., 2018, Ingham et al., 2017). In the absence of insecticide exposure, attenuation of expression of these transcripts demonstrated that both influenced the expression of key pyrethroid metabolisers such as CYP6M2, CYP6Z2, CYP6Z3, CYP6P4, GSTD1 and CYP9K1 (Yunta et al., 2019)(Ingham et al., 2017). Met interacts with CYP6Z2 which is amongst the most strongly induced p450s in the dataset with a marginal posterior probability of 0.88. Interestingly, Maf-S shows enrichment in ABC transporters and terms related to ATP production, indicating Maf-S may play a role in changes in metabolism, which is a striking feature of this dataset. Met shows enrichment in glycolysis, potentially indicated an overlap in the function of these transcription factors, which would be in agreement with the Maf-S knockdown microarray which identified Met as a direct interactor. (Murata et al., 2015, Cornelissen et al., 2018)
Mbf1
Multiprotein bridging factor 1 (mbf1, AGAP004990) has 119 interactors and is enriched for GO terms related to the ribosome (p = 0.026) and RNA binding (p = 0.048) and is highly enriched in the KEGG ribosome (p = 4e-4). The role of mbf1 in Drosophila involved translocation to the nucleus upon cellular stress, where it serves as a co-activator of stress response genes (Jindra et al., 2004); despite this role no enrichment for detoxification transcripts is seen in the predicted mbf1 associations. However, 1 chaperone protein (CCT6) and an oxidative stress sensing protein (AGAP000705-RA) are present in this network. AGAP002667 has the highest posterior probability in the network (0.84) and encodes the homolog of Drosophila Tctp which is necessary for genomic stability under genotoxic stress (Hong and Choi, 2013).
(Taylor-Wells et al., 2015, Zhong and Wu, 2004, Ng and Luo, 2004, Koch et al., 2008, Schaefer et al., 2001, Varghese et al., 2010, Musselman et al., 2018, Bharucha et al., 2008, Beller et al., 2010)
Discussion
In this study, we apply a dynamic Bayesian network approach to whole transcriptome time-course data post-sublethal exposure of An. coluzzii to the pyrethroid insecticide deltamethrin (Ingham et al., 2020). The modified DBN model employed here allows correction for not only circadian rhythms, but also for mosquito ageing, a critical variable in the resistance status (Jones et al., 2012). Interactions predicted by this model were then validated in vivo, demonstrating high model confidence, with 70% precision. The high model precision and the overlapping biological functions with known transcription factors in Drosophila demonstrates the utility of this approach in assigning transcription factor function. Furthermore, this study highlights the potential for use of this methodology across multiple species of interest in which lower resolution time points are more feasible than those seen in model organism studies. Potential applications of this methodology could include exploring transcriptional regulation of pesticide response in other pest species or exposing the same species to additional stressors to distinguish between transcription factors involved in general and insecticide induced stress response.
In this study we highlight 44 transcription factors with putative roles in response to sublethal pesticide exposure, 41 of which have not previously been linked to insecticide resistance. Of the 6585 transcripts differential in the data set used, 5136 transcripts were assigned associations with these 44 transcription factors, using a posterior probability cut-off of >0.39. The assignment of 78% of the overall responsive transcripts is likely due to necessity of reducing the number of transcription factors to less than 50 transcripts and responsive transcripts being regulated by other mechanisms such as non-coding regulatory machinery. The transcription factors selected here for further analysis were identified by applying an SILGGM model (Zhang et al., 2018) to 28 insecticide resistant vs susceptible microarray datasets performed on the Anopheles gambiae species complex collated by Ingham et al. (Ingham et al., 2018) and exploring enrichments of co-correlated transcripts; this represents a confounding aspect of this methodology as these transcripts are constitutively overexpressed and not induced by insecticide exposure due to the nature of the transcriptomic designs.
Of the 44 transcription factors, 3 had previously been linked with insecticide resistance in Anopheles mosquitoes and just 11 had been previously studied in mosquito species in any context (Ingham et al., 2018, Ingham et al., 2017, Ruiz et al., 2019, Chen et al., 2017, Amenya et al., 2010, Wülbeck and Simpson, 2002, Chowdhury et al., 2020, Wang et al., 2017, Maliti et al., 2016, Luna et al., 2006, Fu et al., 2020). All but 4 of these transcription factors have a well-defined role in Drosophila. Using a posterior probability cut off of >0.39, the number of associations showed high levels of variation with an average edge count of 118.48±179.62, potentially demonstrating differential importance in insecticide response, with those transcription factors with a high number of edges or high network connectivity being more important. 23 transcription factors were designated as transcript ‘hubs’ based on high levels of network interconnectivity (>50 edges).
Enrichment analysis was performed for all transcription factors in the network, using GO Terms, KEGG Pathway, Reactome and a priori transcript families with links to resistance. Interestingly, the overlap of enriched terms was low, indicating that each transcription factor may play a differing role in the response to insecticides. 20 transcription factors show enrichments in a priori gene families; this may be an unsurprising feature of this dataset given the obvious change in expression across multiple members of these families within this dataset and their documented importance in insecticide metabolism (Ingham et al., 2020). GO terms enriched across multiple transcription factors include terms expected in an insecticide response, response to drugs, drug metabolism, sensory perception of chemical stimuli and ABC transporters. The former two enrichment terms are in agreement with the well-established dogma that up-regulation of members of the cytochrome p450 class play a direct role in increasing the rate of insecticide metabolism (Ingham et al., 2018, Yunta et al., 2019). (Oliver and Brooke, 2016, Wang et al., 2016). Interestingly, changes to the respiratory pathway through alterations to the oxidative phosphorylation pathway also appears across multiple transcription factors and is a striking feature of this dataset (Ingham et al., 2020).
To cross-validate the function of these transcription factors, their known functions in the model organism Drosophila were explored. Despite the differences in hypotheses explored in this study and the available data in discerning Drosophila pathways, there were clear overlaps in transcription factor roles and associations. For example, Dm is known to play a role in lipid and glucose homeostasis in Drosophila (Parisi et al., 2013) and here, the associations are enriched in the KEGG pathways starch and sucrose metabolism; this is similar to dmrt93B which is involved (Palanker et al., 2009) mouth part development and is enriched in the GO term related to taste receptor activity (Panara et al., 2019). Several further transcription factors show overlap with Drosophila function, including Pep which is involved in stress response through activation of Hsp70 (Hamann and Strätling, 1998, Varghese et al., 2010), dve which is involved in reactive oxygen species modulation (Baqri et al., 2014), Ets21C which is a stress-inducible transcription factor (Mundorf et al., 2019), klumpfuss whose role is related to morphogenesis in the central nervous system (Melcher and Pankratz, 2005), REL1 which is implicated in the TOLL pathway (Gross et al., 1999, Murata et al., 2015) and Chm is a known modulator of the stress responsive JNK pathway with a role in sensory cell fate (Melcher and Pankratz, 2005). (Wang et al., 2017, Ruiz et al., 2019)
This study provides not only previously unreported transcription factors that are involved in the transcriptional response to pesticide exposure but demonstrates the utility of applying a model-based approach to lower-resolution time course data in ascertaining these associations. Here, six transcription factors and their interactomes were delineated as hub transcripts within the network, all of which have either been previously linked to resistance or stress response in Anopheles (Dm, Maf-S and Met) (Ingham et al., 2018, Ingham et al., 2017) or Drosophila (mbf1) (Jindra et al., 2004) or are highly significantly enriched for clear functions (chameau and dmrt93B). These transcription factors are likely to be involved in different facets of insecticide response and represent pathways that should be further explored. The modelling approach taken here, which accounts for both circadian patterns and ageing, two key determinants in pesticide resistance, can be applied widely to other pest or vector species. Using this approach will provide invaluable information on changes to pest biology post-pesticide exposure and will elucidate new pathways to characterise and target to tackle the ongoing threat of pesticide resistance.
Materials and Methods
Microarray Experiments
Microarrays were taken from (Ingham et al., 2020) and consist of deltamethrin exposed mosquitoes compared to unexposed at the following time points post-exposure: 0 minutes, 30 minutes, 1 hour, 2 hours, 4 hours, 8 hours, 12 hours, 24 hours, 48 hours and 72 hours. To account for ageing effects, a twin time course was performed using age matched females that were unexposed to insecticide at the following time points: 8 hours, 12 hours, 24 hours, 48 hours and 72 hours. All mosquitoes within one experimental time course came from the same generation. Experimental data is available on exposure time course (E-MTAB-9422) and ageing time course (E-MTAB-9423). Analysis was performed as previously described.
Transcription factor identification
To identify relevant nodes for the Bayesian analysis, 28 microarray datasets encompassing resistant vs susceptible members of the Anopheles gambiae species complex were used from Ingham et al. 2018 (Ingham et al., 2018). A de-sparsified node-wise scaled lasso (Janková and van de Geer, 2017, Janková and van de Geer, 2018) implemented in the R package SILGGM (Zhang et al., 2018), was used to infer the gene network. This method employs L1-regularisation to preserve sparsity in the estimated network. For the L1-regularisation, the default value of the tuning parameter λ, was used: √log(p)/n, where p is the number of variables and n is the number of samples. The resultant Gaussian graphical model produced a 14079 × 14079 file for every possible interaction in the transcriptome. Each interaction had an associated p-value for precision (Supplementary Table 4). A cut-off value of p ≤ 0.1 was used to filter all interactions to prevent loss of potentially interesting transcription factors due to the differing experimental design of the data set used. Annotated Drosophila transcription factors were downloaded from FlyTF (Adryan and Teichmann, 2006) (http://flytf.gen.cam.ac.uk/) and Anopheles homologs identified using FlyMine (Lyne et al., 2007) (https://www.flymine.org) using the analyse input box, and then selecting An. gambiae homologs, resulting in 559 putative transcription factors; all 559 were then extracted from the inferred network with all associated putative co-correlating transcripts. clusterProfiler (Yu et al., 2012) and AnnotationForge (Carlson and Pagès, 2019) were used to perform GO enrichments using an Anopheles database built from PEST/VectorBase (Giraldo-Calderón et al., 2014) on Biological processes on transcription factors with > 10 interactors. Transcription factors enriched in the following character patterns were extracted: ‘stress’; ‘oxi’; ‘lipid’; ‘behaviour’; ‘response’; ‘fat’; ‘sensory’ and ‘ATP’ leading to 54 transcription factors. The terms were chosen based on previous knowledge of the resistant mechanisms present in An. coluzzii mosquitoes as detailed in the introduction (Balabanidou et al., 2016, Müller et al., 2008, Voice et al., 2015, Ingham et al., 2018, Ingham et al., 2019, Ingham et al., 2020) and ‘ATP’ due to a striking change in metabolism observed in this data set (Ingham et al., 2020). The transcription factors were further filtered on at least 50% of the transcripts in the cluster generated by SILGGM being differentially expressed in at least 1 time point within the time course datasets with an adjusted p value of ≤ 0.05. This procedure resulted in 44 transcription factors being retained. To estimate the impact of transcription factor choice on the network inference, target genes with a marginal posterior probability of >0.75 of having an association with at least one of the transcription factors in the dynamic Bayesian network analysis were selected and the model was re-run using a random selection for 25% of the transcription factors (11/44). The difference in marginal probability of the associations was then analysed. As the majority of differences are <0.2, the 0.39 cut-off used here would still correctly identify the associations with > 0.75 marginal posterior probability in the original analysis (Supplementary Figure 6).
Network reconstruction using Dynamic Bayesian Networks
Dynamic Bayesian networks (DBNs) (Dondelinger et al., 2013) were used to identify directed associations between the transcription factors and putative regulated genes. A dynamic Bayesian network defines a graphical model for the dynamics of time series data, where the gene expression of gene i at time t depends on the gene expression of all transcription factor genes j at time . The relationship can be described by the following auto-regressive linear regression:
where Fistheseto f indices representing the transcription factors. To impose regularisation, we assumed truncated Poisson priors on the number of regression parameters that are non-zero:
where is the maximum number of transcription factors regulating a single gene. We set . Conditional on , the number of non-zero transcription factor-gene associations, the prior on the set of transcription factors for a given gene is simply a uniform distribution.
Inference of the network structure can be done via a Markov Chain Monte Carlo algorithm, with discrete moves allowing for adding and removing edges during the sampling. Convergence was assessed by running each MCMC chain twice from independent starting points and comparing the marginal posterior edge probability estimates. We ran the MCMC algorithm for 500,000 iterations, discarding the first quarter as burn-in, which ensured good convergence across all target genes. For full details on the model and inference procedure, please see Appendix 1. Note that here we employ a simplified version of the model in (Dondelinger et al., 2013) which does not use a changepoint model or information sharing priors.
Prior to applying the network inference model, we pre-processed the log-fold change data by first averaging the values for genes with multiple probes to obtain one measurement per gene. We then employ LOESS estimation (Cleveland et al., 1992), a local regression method which fits low-degree polynomials to subsets of the data, to interpolate the time points at , where we choose hours as the time interval. Interpolation is necessary, as the DBN method requires equal time intervals between each pair of measurements to estimate consistent associations.
We further extend the model to correct for circadian rhythms and ageing effects in the gene expression levels. For the circadian rhythm correction, we assume that all circadian rhythms have a period of 24 hours, and augment the design matrix X = with two additional columns for the sine and cosine functions of a 24-hour periodic signal:
The resulting harmonic regression model with automatically correct for circadian rhythms, including under phase shift, by adding the periodic signal as a parent in the network, while non-periodic genes will remain unconnected to this signal.
Similarly, we add additional columns for the data arising from the ageing controls to correct for the effect of ageing. Note that here we only have data starting from 8 hours, so earlier time points will be uncorrected, and the corresponding values in the design matrix will be set to zero. The final autoregressive model looks as follows:
where is the log-fold change of the ageing controls.
We summarize the results of the DBN analysis using the marginal posterior probability of each transcription factor – target gene association, which can be calculated by obtaining samples from the converged Markov chain and averaging over the presence/absence status of each edge. In order to determine a sensible threshold for the marginal posterior probability that keeps the false discovery rate low, we implement the following permutation test to estimate the posterior probabilities under the null hypothesis of no associations: for each of n=500 iterations, we randomly permute the log-fold changes for one transcription factor. Any associations with the target gene should then be entirely by chance. Taking all n=500 samples of the null distribution obtained in this way, we determine that a threshold of 0.39 was sufficient to only produce one false positive out of 500 randomizations, or a false positive rate of 0.002. Further detail of the model can be found in Appendix 1. The network was displayed using Cytoscape (Shannon et al., 2003).
To estimate how the computational time needed scales with the number of transcription factors, we repeatedly selected 10 target genes and p transcription factors, where p∈(5,100). Network inference using EDISON was then performed on a computational cluster with two Intel Xeon E5-2660 v4s, which have 14 physical cores running @ 2.00GHz each, and 256 GB of RAM, and the resulting computational time is recorded. All MCMC chains for the network inference algorithm are run for 500,000 iterations (Supplementary Figure 7).
NetworkVis App
The NetworkVis app and associated data can be downloaded on Github (https://github.com/VictoriaIngham/NetworkVis_TimeCourse) and installed as described. ShinyR (Chang et al., 2017) was used to create a user interface, both VisNetwork (Almende et al., 2018) and igraph (Csardi and Nepusz, 2006) were used to allow dynamic selection of nodes and edges, and to display the network.
Enrichment analysis
Enrichment analysis was performed using clusterProfiler (Yu et al., 2012) and a custom Anopheles database produced using AnnotationForge (Carlson and Pagès, 2019). GO term and KEGG enrichments were performed using a Benjamini-Hochberg corrected p value cut-off of ≤ 0.05 with transcription factors > 10 interactions. Clusters of each transcription factor were compared using the compareCluster function using default parameters, Benjamini-Hochberg correction and a full background geneset from org.Agambiae.eg.db built from the PEST assembly; these were then displayed using Cytoscape (Shannon et al., 2003). Enrichment analysis on individual gene families were performed using a hypergeometric test with the phyper function in R; significance was considered when p ≤ 0.05. Reactome analysis was also performed using a hypergeometric test with p ≤ 0.05; Drosophila pathway membership was downloaded from Reactome.org (https://reactome.org/) (Jassal et al., 2020) for each pathway of interest, FlyMine (Lyne et al., 2007) was then used to convert these to Anopheles homologs. FlyBase (Consortium, 2003) was used to determine functions of homologs throughout the analysis. We applied the Benjamini-Hochberg correction for multiple testing outputs of the hypergeometric test.
Validation of Network
We first performed a simulation study to determine the number of associations that need to be tested experimentally in order to obtain an accurate estimate of the precision of our network inference method. We made the following assumptions: (i) The mean number of gene regulated by each transcription factor is 10, and the actual number of regulated genes follows a Poisson distribution; (ii) The rate of true positives (correctly predicted associations) of our network is 0.75, and the rate of true negatives (correctly predicted non-associations) is 0.997; this results in a precision of ∼0.56 and a recall of ∼0.72 (Appendix 2); (iii) Transcription factors and regulated genes to test are selected randomly and (iv) The qPCR knockdown test is 100% accurate. The results of the simulation study can be found in Appendix 2. We concluded that testing 4 regulatory relationships for 7 transcription factors has a 70% chance of obtaining an estimate of the precision that falls within 10% of the true precision, and a 95% chance of obtaining an estimate that falls within 20% of the true precision.
In order to choose associations for validation, we then chose interactors by extracting the transcription factor of interest and associated transcripts from the results of the network inference. Transcripts were listed as 1 to n based on posterior probability in descending order. A random number generator was then used to select 4 transcripts for validation from 6 transcription factors chosen based on previous knockdown in the case of Maf-S, Met, Dm or through a random number generator.
Mosquito Rearing
The An. coluzzii VK7 colony reared and profiled at Liverpool School of Tropical Medicine were used for all experiments (Williams et al., 2019). VK7 are a highly pyrethroid resistant population originating from Vallée de Kou, Burkina Faso (Toé et al., 2015). They have been reared at LSTM since 2014 under pyrethroid selection pressure (Williams et al., 2019). All mosquitoes used were reared under standard insectary conditions of 27°C and 70-80% relative humidity under a 12:12 photoperiod and are presumed mated.
dsRNA knockdown
RNAi was performed using 7 transcription factors based on previous publication of knockdown (Maf-s, Met, Dm (Ingham et al., 2018, Ingham et al., 2017)) or through random selection using a random number generator (Med, Pan, l(1)sc, mbf1) (Supplementary Table 5). PCR was performed on 3-day old VK7 unexposed cDNA using Phusion® High-Fidelity DNA Polymerase (Thermo Scientific) following manufacturer's instructions and primer sets with a T7 docking sequence at the 5′ end of both the sense and antisense primers (Supplementary Table 5). Primers were designed as previously described (Ingham et al., 2018). PCR was performed using the following cycles: 98°C for 30s, (98°C 7s, 65°C 10s, 72°C 10s) x35 and 72°C 5 minutes. PCR product was then purified using a Qiagen QIAquick PCR Purification Kit following manufacturers’ instructions. dsRNA was then synthesised using a Megascript® T7 Transcription (Ambion) kit, following manufacturer's instructions (16-hour 37 °C incubation). The dsRNA was cleaned using a MegaClear® Transcription Clear Up (Ambion) kit, with DEPC water, twice heated at 65 °C for 10 min, to elute the sample. The resultant dsRNA product was analysed using a nanodrop spectrometer (Nanodrop Technologies, UK) and subsequently concentrated to 3 μg/μl using a vacuum centrifuge at 35°C. 69nL of dsRNA was subsequently injected into presumed mated, non-blood fed, 3-day old VK7 females immobilised using a CO2 block using a NanoInject II. 50 females were injected with each of the transcription factor dsRNA and 50 with dsGFP as a non-endogenous control.
Insecticide Exposures
25-30 female mosquitoes were exposed to 0.05% deltamethrin impregnated papers for one hour in a standard tube bioassay kit following WHO guidelines. Post-exposure mosquitoes were transferred into holding tubes and maintained on sucrose solution.
RNA extraction and cDNA synthesis
RNA was extracted from 7-10 female mosquitoes in biological triplicate for each experimental group. RNA was extracted from homogenised mosquitoes using a PicoPure RNA isolation kit (Thermo Fisher, UK) following manufacturers’ instructions and treated with DNAase (Qiagen) to remove any DNA contamination. Quality of RNA was checked using a nanodrop spectrophotometer (Nanodrop Technologies UK). 1-4µg of RNA from each experimental set was reversed transcribed using OligoDTT (Invitrogen) and Superscript III (Invitrogen) according to manufacturers’ instructions. The following experimental groups were used: (i) knockdown efficacy for each transcription factor and the GFP control using females 48-hours post RNAi injection and (ii) pathway validation using females 48-hours after they were exposed to 0.05% deltamethrin for 48-hours post-injection for transcription factors and GFP controls.
qPCR validation
Quantitative real-time PCR was performed using SYBR Green Supermix III (Applied Biosystems, UK) using an MX3005 and the associated MxPro software v4.10 (Agilent, UK). Primer Blast (NCBI) was used to design primer pairs. Where possible, primers were designed to span an exon junction (Supplementary Table 5). Each 20µl reaction contained 10µl SYBR Green Supermix, 0.3µM of each primer and 1µl of 4ng/µL cDNA. Standard curves for each primer set were used to calculate efficiency, using five 1:5 dilutions of cDNA to ensure that all primer sets met the MIQE guidelines (90-120% efficiency) (Bustin et al., 2009). qPCR was performed with the following conditions: 3 minutes at 95°C, with 40 cycles of 10 seconds at 95°C and 10 seconds at 60°C. Relative expression was normalised against two housekeeping genes: EF (AGAP005128) and S7 (AGAP010592) and analysed using comparative CT method (Schmittgen and Livak, 2008). qPCR was used to determine the efficacy of transcription factor knockdown by comparing cDNA taken from mosquitoes 48-hours post dsRNA injection for each transcription factor and comparing it to GFP-injected controls all taken from the same mosquito generation. To validate findings in the network, qPCR was performed on dsRNA injected mosquitoes exposed to 0.05% deltamethrin at 48-hours post injection, these mosquitoes were then left for a further 48-hours before harvesting; again, transcription factor injected mosquitoes were compared to the dsGFP injected controls.
Author Contributions
VAI and FD designed and implemented the experiment. SCN performed the SILGGM analysis, FD modified and implemented the dynamic Bayesian network, SE provided rearing, bioassay and molecular biology support, VAI performed the lab-based experiments and analysed all data. VAI and FD drafted the manuscript.
Data Availability
The datasets used in this experiment are available at ArrayExpress under E-MTAB-9422 and E-MTAB-9423. The authors declare that all other data supporting the findings of this study, are available within the article and its Supplementary Information files or are available from the authors upon request.
Code Availability
Code used for analysis in this study is available on GitHub. Network visualisation is available at https://github.com/VictoriaIngham/NetworkVis_TimeCourse, model code is available on the CRAN repository: https://cran.r-project.org/web/packages/EDISON and full analysis is available at https://github.com/FrankD/AnophelesInsecticideExposure.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This work was supported by a Medical Research Council Skills Development Fellowship [MR/R024839/1] to VAI. We thank Hilary Ranson and David Weetman for valuable feedback on the manuscript and Jessica Carson, Marion Morris and Ruth Cowlishaw for insectary support.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.cris.2021.100018.
Contributor Information
Victoria A Ingham, Email: victoria.ingham@med.uni-heidelberg.de.
Sara Elg, Email: sara.elg@lstmed.ac.uk.
Sanjay C Nagi, Email: sanjay.nagi@lstmed.ac.uk.
Frank Dondelinger, Email: fdondelinger.work@gmail.com.
Appendix. Supplementary materials
References
- Adryan B, Teichmann SA. FlyTF: a systematic review of site-specific transcription factors in the fruit fly Drosophila melanogaster. Bioinformatics. 2006;22:1532–1533. doi: 10.1093/bioinformatics/btl143. [DOI] [PubMed] [Google Scholar]
- Almende B V, Thieurmel B, Robert T. visNetwork: Network Visualization using vis. js Library R package version 2.0. 4. 2018.
- Amenya DA, Chou W, Li J, Yan G, Gershon PD, James AA, et al. Proteomics reveals novel components of the Anopheles gambiae eggshell. J Insect Physiol. 2010;56:1414–1419. doi: 10.1016/j.jinsphys.2010.04.013. doi:https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arbouzova NI, Bach EA, Zeidler MP. Ken & Barbie Selectively Regulates the Expression of a Subset of JAK/STAT Pathway Target Genes. Curr Biol. 2006;16:80–88. doi: 10.1016/j.cub.2005.11.033. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25 doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astigarraga S, Grossman R, Díaz-Delfín J, Caelles C, Paroush Z, Jiménez G. A MAPK docking site is critical for downregulation of Capicua by Torso and EGFR RTK signaling. EMBO J. 2007;26:668–677. doi: 10.1038/sj.emboj.7601532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasopoulos V, Barker A, Yu D, Tan AH, Srivastava M, Contreras N, et al. The ROQUIN family of proteins localizes to stress granules via the ROQ domain and binds target mRNAs. FEBS J. 2010;277:2109–2127. doi: 10.1111/j.1742-4658.2010.07628.x. [DOI] [PubMed] [Google Scholar]
- Balabanidou V, Kampouraki A, MacLean M, Blomquist GJ, Tittiger C, Juárez MP, et al. Cytochrome P450 associated with insecticide resistance catalyzes cuticular hydrocarbon production in Anopheles gambiae. Proc Natl Acad Sci. 2016 doi: 10.1073/pnas.1608295113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balabanidou V, Kefi M, Aivaliotis M, Koidou V, Girotti JR, Mijailovsky SJ, et al. Mosquitoes cloak their legs to resist insecticides. Proc R Soc B. 2019;286 doi: 10.1098/rspb.2019.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baqri RM, Pietron A V, Gokhale RH, Turner BA, Kaguni LS, Shingleton AW, et al. Mitochondrial chaperone TRAP1 activates the mitochondrial UPR and extends healthspan in Drosophila. Mech Ageing Dev. 2014 doi: 10.1016/j.mad.2014.09.002. 141–142:35–45doi:https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry WE, Thummel CS. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. Elife. 2016;5:e11183. doi: 10.7554/eLife.11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beller M, Bulankina A V, Hsiao H-H, Urlaub H, Jäckle H, Kühnlein RP. PERILIPIN-Dependent Control of Lipid Droplet Structure and Fat Storage in Drosophila. Cell Metab. 2010;12:521–532. doi: 10.1016/j.cmet.2010.10.001. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Bharucha KN, Tarr P, Zipursky SL. A glucagon-like endocrine pathway in Drosophila modulates both lipid and carbohydrate homeostasis. J Exp Biol. 2008;211 doi: 10.1242/jeb.016451. 3103 LP –3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt S, Weiss DJ, Cameron E, Bisanzio D, Mappin B, Dalrymple U, et al. The effect of malaria control on Plasmodium falciparum in Africa between 2000 and 2015. Nature. 2015;526:207–211. doi: 10.1038/nature15535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco J, Cooper JC, Baker NE. Roles of C/EBP class bZip proteins in the growth and cell competition of Rp (‘Minute’) mutants in Drosophila. Elife. 2020;9:e50535. doi: 10.7554/eLife.50535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottino-Rojas V, Talyuli OAC, Carrara L, Martins AJ, James AA, Oliveira PL, et al. The redox-sensing gene Nrf2 affects intestinal homeostasis, insecticide resistance, and Zika virus susceptibility in the mosquito Aedes aegypti. J Biol Chem. 2018;293:9053–9063. doi: 10.1074/jbc.RA117.001589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky MH, Weinert BT, Tsang G, Rong YS, McGinnis NM, Golic KG, et al. Drosophila melanogaster MNK/Chk2 and p53 Regulate Multiple DNA Repair and Apoptotic Pathways following DNA Damage. Mol Cell Biol. 2004;24 doi: 10.1128/MCB.24.3.1219-1231.2004. 1219 LP –1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Peter O, Schweizer L, Basler K. pangolinencodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature. 1997;385:829–833. doi: 10.1038/385829a0. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Carlson M, Pagès H. AnnotationForge: Tools for building SQLite-Based Annotation Data Packages, 2019. R Packag version. 1.
- Chang W, Cheng J, Allaire J, Xie Y, McPherson J. shiny: Web Application Framework for R. 2017. https://cran.r-project.org/package=shiny.
- Chen J V, Kao L-R, SC Jana, Sivan-Loukianova E, Mendonça S, Cabrera OA, et al. Rootletin organizes the ciliary rootlet to achieve neuron sensory function in Drosophila. J Cell Biol. 2015;211:435–453. doi: 10.1083/jcb.201502032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Shi X, Padmanabhan R, Wang Q, Wu Z, Stevenson SC, et al. Identification of novel modulators of mitochondrial function by a genome-wide RNAi screen in Drosophila melanogaster. Genome Res. 2008;18:123–136. doi: 10.1101/gr.6940108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T-H, Wu Y-J, Hou J-N, Chiu C-H, Chen W-J. The p53 gene with emphasis on its paralogues in mosquitoes. J Microbiol Immunol Infect. 2017;50:747–754. doi: 10.1016/j.jmii.2017.06.006. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Chowdhury A, Modahl CM, Tan ST, Xiang BWW, Missé D, Vial T, et al. JNK pathway restricts DENV, ZIKV and CHIKV infection by activating complement and apoptosis in mosquito salivary glands. bioRxiv. 2020;:2020.03.01.972026. doi:10.1101/2020.03.01.972026. [DOI] [PMC free article] [PubMed]
- Cleveland WS, Grosse E, Shyu WM. Wadsworth Brooks/Cole; Pacific Grove, CA: 1992. Local regression models. Chapter 8 in Statistical models in S; p. 608. JM Chambers and TJ Hastie eds. [Google Scholar]
- Consortium F. The FlyBase database of the Drosophila genome projects and community literature. Nucleic Acids Res. 2003;31:172–175. doi: 10.1093/nar/gkg094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornelissen T, Vilain S, Vints K, Gounko N, Verstreken P, Vandenberghe W. Deficiency of parkin and PINK1 impairs age-dependent mitophagy in Drosophila. Elife. 2018;7:e35878. doi: 10.7554/eLife.35878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csardi G, Nepusz T. The igraph software package for complex network research. InterJournal, complex Syst. 2006;1695:1–9. [Google Scholar]
- Delgado FM, Gómez-Vela F. Computational methods for Gene Regulatory Networks reconstruction and analysis: A review. Artif Intell Med. 2019;95:133–145. doi: 10.1016/j.artmed.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Dondelinger F, Lèbre S, Husmeier D. Non-homogeneous dynamic Bayesian networks with Bayesian regularization for inferring gene regulatory networks with gradually time-varying structure. Mach Learn. 2013;90:191–230. [Google Scholar]
- Dondelinger F, Mukherjee S. Gene regulatory networks. Springer; 2019. Statistical network inference for time-varying molecular data with dynamic bayesian networks; pp. 25–48. [DOI] [PubMed] [Google Scholar]
- Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci. 1996;93 doi: 10.1073/pnas.93.19.10343. 10343 LP –10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix JT, Magariños M, Díaz-Benjumea FJ. Nab controls the activity of the zinc-finger transcription factors Squeeze and Rotund in Drosophila development. Development. 2007;134 doi: 10.1242/dev.003830. 1845 LP –1852. [DOI] [PubMed] [Google Scholar]
- Fu X, Liu P, Dimopoulos G, Zhu J. Dynamic miRNA-mRNA interactions coordinate gene expression in adult Anopheles gambiae. PLOS Genet. 2020;16 doi: 10.1371/journal.pgen.1008765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaddelapati SC, Kalsi M, Roy A, Palli SR. Cap'n’collar C regulates genes responsible for imidacloprid resistance in the Colorado potato beetle, Leptinotarsa decemlineata. Insect Biochem Mol Biol. 2018;99:54–62. doi: 10.1016/j.ibmb.2018.05.006. [DOI] [PubMed] [Google Scholar]
- Gellatly KJ, Yoon KS, Doherty JJ, Sun W, Pittendrigh BR, Clark JM. RNAi validation of resistance genes and their interactions in the highly DDT-resistant 91-R strain of Drosophila melanogaster. Pestic Biochem Physiol. 2015;121:107–115. doi: 10.1016/j.pestbp.2015.01.001. [DOI] [PubMed] [Google Scholar]
- Giraldo-Calderón GI, Emrich SJ, MacCallum RM, Maslen G, Dialynas E, Topalis P, et al. VectorBase: an updated bioinformatics resource for invertebrate vectors and other organisms related with human diseases. Nucleic Acids Res. 2014;43:D707–D713. doi: 10.1093/nar/gku1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross I, Georgel P, Oertel-Buchheit P, Schnarr M, Reichhart J-M. Dorsal-B, a splice variant of the Drosophila factor Dorsal, is a novel Rel/NF-κB transcriptional activator1The sequence described in this paper was deposited in GeneBank under the Accession No. AF053614.1. Gene. 1999;228:233–242. doi: 10.1016/S0378-1119(98)00595-2. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Hamann S, Strätling WH. Specific binding of Drosophila nuclear protein PEP (protein on ecdysone puffs) to hsp70 DNA and RNA. Nucleic Acids Res. 1998;26:4108–4115. doi: 10.1093/nar/26.18.4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawley WA, Phillips-Howard PA, ter Kuile FO, Terlouw DJ, Vulule JM, Ombok M, et al. Community-wide effects of permethrin-treated bed nets on child mortality and malaria morbidity in western Kenya. Am J Trop Med Hyg. 2003;68(4 Suppl):121–127. [PubMed] [Google Scholar]
- Herold N, Will CL, Wolf E, Kastner B, Urlaub H, Lührmann R. Conservation of the Protein Composition and Electron Microscopy Structure of Drosophila melanogaster and Human Spliceosomal Complexes. Mol Cell Biol. 2009;29(281) doi: 10.1128/MCB.01415-08. LP –301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-T, Choi K-W. TCTP directly regulates ATM activity to control genome stability and organ development in Drosophila melanogaster. Nat Commun. 2013;4:1–14. doi: 10.1038/ncomms3986. [DOI] [PubMed] [Google Scholar]
- Hsia CC, McGinnis W. Evolution of transcription factor function. Curr Opin Genet Dev. 2003;13:199–206. doi: 10.1016/S0959-437X(03)00017-0. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Hu B, Huang H, Wei Q, Ren M, Mburu DK, Tian X, et al. Transcription factors CncC/Maf and AhR/ARNT coordinately regulate the expression of multiple GSTs conferring resistance to chlorpyrifos and cypermethrin in Spodoptera exigua. Pest Manag Sci. 2019;75:2009–2019. doi: 10.1002/ps.5316. [DOI] [PubMed] [Google Scholar]
- Hughes A, Lissenden N, Viana M, Toe KH, Ranson H. Anopheles gambiae populations from Burkina Faso show minimal delayed mortality after exposure to insecticide-treated nets. Parasit Vectors. 2020;13:17. doi: 10.1186/s13071-019-3872-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham VA, Brown F, Ranson H. Sub-lethal pyrethroid exposure and ageing lead to pronounced changes in gene expression in insecticide resistance Anopheles coluzziibioRxiv. 2020;:2020.08.14.250852. doi:10.1101/2020.08.14.250852. [DOI] [PMC free article] [PubMed]
- Ingham VA, Pignatelli P, Moore JD, Wagstaff S, Ranson H. The transcription factor Maf-S regulates metabolic resistance to insecticides in the malaria vector Anopheles gambiae. BMC Genomics. 2017;18 doi: 10.1186/s12864-017-4086-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham VA, Wagstaff S, Ranson H. Transcriptomic meta-signatures identified in Anopheles gambiae populations reveal previously undetected insecticide resistance mechanisms. Nat Commun. 2018;9:5282. doi: 10.1038/s41467-018-07615-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham VAVA, Anthousi A, Douris V, Harding NJNJ, Lycett G, Morris M, et al. A sensory appendage protein protects malaria vectors from pyrethroids. Nature. 2019;577 doi: 10.1038/s41586-019-1864-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer EPR, Iyer SC, Sullivan L, Wang D, Meduri R, Graybeal LL, et al. Functional Genomic Analyses of Two Morphologically Distinct Classes of Drosophila Sensory Neurons: Post-Mitotic Roles of Transcription Factors in Dendritic Patterning. PLoS One. 2013;8:e72434. doi: 10.1371/journal.pone.0072434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CA, Castro DM, Saldi G-A, Bonneau R, Gresham D. Gene regulatory network reconstruction using single-cell RNA sequencing of barcoded genotypes in diverse environments. Elife. 2020;9:e51254. doi: 10.7554/eLife.51254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janková J, van de Geer S. Honest confidence regions and optimality in high-dimensional precision matrix estimation. Test. 2017;26:143–162. [Google Scholar]
- Janková J, van de Geer S. Inference in high-dimensional graphical models. arXiv Prepr arXiv180108512. 2018.
- Jassal B, Matthews L, Viteri G, Gong C, Lorente P, Fabregat A, et al. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48:D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindra M, Gaziova I, Uhlirova M, Okabe M, Hiromi Y, Hirose S. Coactivator MBF1 preserves the redox-dependent AP-1 activity during oxidative stress in Drosophila. EMBO J. 2004;23:3538–3547. doi: 10.1038/sj.emboj.7600356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jindra M, Uhlirova M, Charles J-P, Smykal V, Hill RJ. Genetic Evidence for Function of the bHLH-PAS Protein Gce/Met As a Juvenile Hormone Receptor. PLOS Genet. 2015;11 doi: 10.1371/journal.pgen.1005394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones CM, Sanou A, Guelbeogo WM, Sagnon N, Johnson PCD, Ranson H. Aging partially restores the efficacy of malaria vector control in insecticide-resistant populations of Anopheles gambiae s.l. from Burkina Faso. Malar J. 2012;11:24. doi: 10.1186/1475-2875-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi M, Palli SR. Transcription factors, CncC and Maf, regulate expression of CYP6BQ genes responsible for deltamethrin resistance in Tribolium castaneum. Insect Biochem Mol Biol. 2015;65:47–56. doi: 10.1016/j.ibmb.2015.08.002. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kappes G, Deshpande G, Mulvey BB, Horabin JI, Schedl P. The Drosophila Myc gene, diminutive, is a positive regulator of the Sex-lethal establishment promoter, Sxl-Pe. Proc Natl Acad Sci. 2011;108:1543–1548. doi: 10.1073/pnas.1017006108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavaler J, Fu W, Duan H, Noll M, Posakony JW. An essential role for the Drosophila Pax2 homolog in the differentiation of adult sensory organs. Development. 1999;126 doi: 10.1242/dev.126.10.2261. http://dev.biologists.org/content/126/10/2261.abstract 2261 LP –2272. [DOI] [PubMed] [Google Scholar]
- Killeen GF, Okumu FO, N'Guessan R, Coosemans M, Adeogun A, Awolola S, et al. The importance of considering community-level effects when selecting insecticidal malaria vector products. Parasit Vectors. 2011;4:160. doi: 10.1186/1756-3305-4-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Killeen GF, Smith TA. Exploring the contributions of bed nets, cattle, insecticides and excitorepellency to malaria control: a deterministic model of mosquito host-seeking behaviour and mortality. Trans R Soc Trop Med Hyg. 2007;101:867–880. doi: 10.1016/j.trstmh.2007.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch I, Schwarz H, Beuchle D, Goellner B, Langegger M, Aberle H. Drosophila ankyrin 2 is required for synaptic stability. Neuron. 2008;58:210–222. doi: 10.1016/j.neuron.2008.03.019. [DOI] [PubMed] [Google Scholar]
- Lee JC, VijayRaghavan K, Celniker SE, Tanouye MA. Identification of a Drosophila muscle development gene with structural homology to mammalian early growth response transcription factors. Proc Natl Acad Sci. 1995;92 doi: 10.1073/pnas.92.22.10344. 10344 LP –10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A, Hooli B, Mullin K, Tate RE, Bubnys A, Kirchner R, et al. Silencing of the Drosophila ortholog of SOX5 leads to abnormal neuronal development and behavioral impairment. Hum Mol Genet. 2017;26:1472–1482. doi: 10.1093/hmg/ddx051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Harding GA, Parise J, McNamara-Schroeder KJ, Stumph WE. Architectural Arrangement of Cloned Proximal Sequence Element-Binding Protein Subunits on Drosophila U1 and U6 snRNA Gene Promoters. Mol Cell Biol. 2004;24 doi: 10.1128/MCB.24.5.1897-1906.2004. 1897 LP –1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna C, Hoa NT, Lin H, Zhang L, Nguyen HLA, Kanzok SM, et al. Expression of immune responsive genes in cell lines from two different Anopheline species. Insect Mol Biol. 2006;15:721–729. doi: 10.1111/j.1365-2583.2006.00661.x. [DOI] [PubMed] [Google Scholar]
- Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, et al. FlyMine: An Integrated Database for Drosophila and Anopheles Genomics. Genome Biol C7 - R129. 2007;8:1–16. doi: 10.1186/gb-2007-8-7-r129. LA-English. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliti DV, Marsden CD, Main BJ, Govella NJ, Yamasaki Y, Collier TC, et al. Investigating associations between biting time in the malaria vector Anopheles arabiensis Patton and single nucleotide polymorphisms in circadian clock genes: support for sub-structure among An. arabiensis in the Kilombero valley of Tanzania. Parasit Vectors. 2016;9:109. doi: 10.1186/s13071-016-1394-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Torres D, Chandre F, Williamson MS, Darriet F, Berge JB, Devonshire AL, et al. Molecular characterization of pyrethroid knockdown resistance (kdr) in the major malaria vector Anopheles gambiae ss. Insect Mol Biol. 1998;7:179–184. doi: 10.1046/j.1365-2583.1998.72062.x. [DOI] [PubMed] [Google Scholar]
- Melcher C, Pankratz MJ. Candidate gustatory interneurons modulating feeding behavior in the Drosophila brain. PLoS Biol. 2005;3:e305. doi: 10.1371/journal.pbio.0030305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miotto B, Sagnier T, Berenger H, Bohmann D, Pradel J, Graba Y. Chameau HAT and DRpd3 HDAC function as antagonistic cofactors of JNK/AP-1-dependent transcription during Drosophila metamorphosis. Genes Dev. 2006;20:101–112. doi: 10.1101/gad.359506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misra JR, Horner MA, Lam G, Thummel CS. Transcriptional regulation of xenobiotic detoxification in Drosophila. Genes Dev. 2011;25:1796–1806. doi: 10.1101/gad.17280911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moloney DJ, Panin VM, Johnston SH, Chen J, Shao L, Wilson R, et al. Fringe is a glycosyltransferase that modifies Notch. Nature. 2000;406:369–375. doi: 10.1038/35019000. [DOI] [PubMed] [Google Scholar]
- Morrison HA, Dionne H, Rusten TE, Brech A, Fisher WW, Pfeiffer BD, et al. Regulation of early endosomal entry by the Drosophila tumor suppressors Rabenosyn and Vps45. Mol Biol Cell. 2008;19:4167–4176. doi: 10.1091/mbc.e08-07-0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P, Warr E, Stevenson BJ, Pignatelli PM, Morgan JC, Steven A, et al. Field-Caught Permethrin-Resistant Anopheles gambiae Overexpress CYP6P3, a P450 That Metabolises Pyrethroids. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000286. http://dx.doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundorf J, Donohoe CD, McClure CD, Southall TD, Uhlirova M. Ets21c Governs Tissue Renewal, Stress Tolerance, and Aging in the <em>Drosophila</em>Intestine. Cell Rep. 2019;27:3019–3033. doi: 10.1016/j.celrep.2019.05.025. e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata H, Takamatsu H, Liu S, Kataoka K, Huh N, Sakaguchi M. NRF2 Regulates PINK1 Expression under Oxidative Stress Conditions. PLoS One. 2015;10 doi: 10.1371/journal.pone.0142438. https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy K, Mian S. Modelling gene expression data using dynamic Bayesian networks. Citeseer; 1999 [Google Scholar]
- Musselman LP, Fink JL, Maier EJ, Gatto JA, Brent MR, Baranski TJ. Seven-up is a novel regulator of insulin signaling. Genetics. 2018;208:1643–1656. doi: 10.1534/genetics.118.300770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Varga A, Pircs K, Hegedűs K, Juhász G. Myc-driven overgrowth requires unfolded protein response-mediated induction of autophagy and antioxidant responses in Drosophila melanogaster. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng J, Luo L. Rho GTPases regulate axon growth through convergent and divergent signaling pathways. Neuron. 2004;44:779–793. doi: 10.1016/j.neuron.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Oliver S V, Brooke BD. The Role of Oxidative Stress in the Longevity and Insecticide Resistance Phenotype of the Major Malaria Vectors Anopheles arabiensis and Anopheles funestus. PLoS One. 2016;11 doi: 10.1371/journal.pone.0151049. http://dx.doi.org/10.1371%2Fjournal.pone.0151049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanker L, Tennessen JM, Lam G, Thummel CS. <em>Drosophila</em>HNF4 Regulates Lipid Mobilization and β-Oxidation. Cell Metab. 2009;9:228–239. doi: 10.1016/j.cmet.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panara V, Budd GE, Janssen R. Phylogenetic analysis and embryonic expression of panarthropod Dmrt genes. Front Zool. 2019;16:23. doi: 10.1186/s12983-019-0322-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papagianni A, Forés M, Shao W, He S, Koenecke N, Andreu MJ, et al. Capicua controls Toll/IL-1 signaling targets independently of RTK regulation. Proc Natl Acad Sci. 2018;115 doi: 10.1073/pnas.1713930115. 1807 LP –1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoulas O, Daubresse G, Armstrong JA, Jin J, Scott MP, Tamkun JW. The HMG-domain protein BAP111 is important for the function of the BRM chromatin-remodeling complex in vivo. Proc Natl Acad Sci. 2001;98 doi: 10.1073/pnas.091533398. 5728 LP –5733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi F, Riccardo S, Zola S, Lora C, Grifoni D, Brown LM, et al. dMyc expression in the fat body affects DILP2 release and increases the expression of the fat desaturase Desat1 resulting in organismal growth. Dev Biol. 2013;379:64–75. doi: 10.1016/j.ydbio.2013.04.008. doi:https://doi.org/ [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecasse F, Beck Y, Ruiz C, Richards G. Krüppel-homolog, a stage-specific modulator of the prepupal ecdysone response, is essential for Drosophila metamorphosis. Dev Biol. 2000;221:53–67. doi: 10.1006/dbio.2000.9687. [DOI] [PubMed] [Google Scholar]
- Peng T, Chen X, Pan Y, Zheng Z, Wei X, Xi J, et al. Transcription factor aryl hydrocarbon receptor/aryl hydrocarbon receptor nuclear translocator is involved in regulation of the xenobiotic tolerance-related cytochrome P450 CYP6DA2 in Aphis gossypii Glover. Insect Mol Biol. 2017 doi: 10.1111/imb.12311. [DOI] [PubMed] [Google Scholar]
- Piacentini L, Fanti L, Berloco M, Perrini B, Pimpinelli S. Heterochromatin protein 1 (HP1) is associated with induced gene expression in Drosophila euchromatin. J Cell Biol. 2003;161:707–714. doi: 10.1083/jcb.200303012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pignatelli P, Ingham VAA, Balabanidou V, Vontas J, Lycett G, Ranson H. The Anopheles gambiae ATP-binding cassette transporter family: phylogenetic analysis and tissue localization provide clues on function and role in insecticide resistance. Insect Mol Biol. 2018;27:110–122. doi: 10.1111/imb.12351. [DOI] [PubMed] [Google Scholar]
- Popp J, Pető K, Nagy J. Pesticide productivity and food security. A review. Agron Sustain Dev. 2013;33:243–255. doi: 10.1007/s13593-012-0105-x. [DOI] [Google Scholar]
- Protzer CE, Wech I, Nagel AC. Hairless induces cell death by downregulation of EGFR signalling activity. J Cell Sci. 2008;121 doi: 10.1242/jcs.035014. 3167 LP –3176. [DOI] [PubMed] [Google Scholar]
- Rämet M, Lanot R, Zachary D, Manfruelli P. JNK Signaling Pathway Is Required for Efficient Wound Healing in Drosophila. Dev Biol. 2002;241:145–156. doi: 10.1006/dbio.2001.0502. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Reddy BA, Bajpe PK, Bassett A, Moshkin YM, Kozhevnikova E, Bezstarosti K, et al. Drosophila transcription factor Tramtrack69 binds MEP1 to recruit the chromatin remodeler NuRD. Mol Cell Biol. 2010;30:5234–5244. doi: 10.1128/MCB.00266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveron JM, Yunta C, Ibrahim SS, Djouaka R, Irving H, Menze BD, et al. A single mutation in the GSTe2 gene allows tracking of metabolically based insecticide resistance in a major malaria vector. Genome Biol. 2014;15:R27. doi: 10.1186/gb-2014-15-2-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz JL, Yerbanga RS, Lefèvre T, Ouedraogo JB, Corces VG, Gómez-Díaz E. Chromatin changes in Anopheles gambiae induced by Plasmodium falciparum infection. Epigenetics Chromatin. 2019;12:5. doi: 10.1186/s13072-018-0250-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund SSC, Hou TY, Ward SM, Collins FH, Duffield GE. Genome-wide profiling of diel and circadian gene expression in the malaria vector Anopheles gambiae. Proc Natl Acad Sci. 2011;108 doi: 10.1073/pnas.1100584108. E421 LP-E430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rund SSC, O'Donnell AJ, Gentile JE, Reece SE. Daily rhythms in mosquitoes and their consequences for malaria transmission. Insects. 2016;7:14. doi: 10.3390/insects7020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutila JE, Suri V, Le M, So WV, Rosbash M, Hall JC. CYCLE Is a Second bHLH-PAS Clock Protein Essential for Circadian Rhythmicity and Transcription of <em>Drosophila period</em>and <em>timeless</em>. Cell. 1998;93:805–814. doi: 10.1016/S0092-8674(00)81441-5. [DOI] [PubMed] [Google Scholar]
- Schaefer M, Petronczki M, Dorner D, Forte M, Knoblich JA. Heterotrimeric G proteins direct two modes of asymmetric cell division in the Drosophila nervous system. Cell. 2001;107:183–194. doi: 10.1016/s0092-8674(01)00521-9. [DOI] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SB, Guardiola AR, Crews ST. Dual role for Drosophila lethal of scute in CNS midline precursor formation and dopaminergic neuron and motoneuron cell fate. Development. 2011;138 doi: 10.1242/dev.056507. 2171 LP –2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor-Wells J, Brooke BD, Bermudez I, Jones AK. The neonicotinoid imidacloprid, and the pyrethroid deltamethrin, are antagonists of the insect Rdl GABA receptor. J Neurochem. 2015 doi: 10.1111/jnc.13290. n/a-n/a. [DOI] [PubMed] [Google Scholar]
- Tearle R, Tomlinson A, Saint R. The dominant Drop eye mutations of Drosophila melanogaster define two loci implicated in normal eye development. Mol Gen Genet MGG. 1994;244:426–434. doi: 10.1007/BF00286695. [DOI] [PubMed] [Google Scholar]
- Terriente-Félix A, de Celis JF. Osa, a subunit of the BAP chromatin-remodelling complex, participates in the regulation of gene expression in response to EGFR signalling in the Drosophila wing. Dev Biol. 2009;329:350–361. doi: 10.1016/j.ydbio.2009.03.010. [DOI] [PubMed] [Google Scholar]
- Thompson D, Regev A, Roy S. Comparative analysis of gene regulatory networks: from network reconstruction to evolution. Annu Rev Cell Dev Biol. 2015;31:399–428. doi: 10.1146/annurev-cellbio-100913-012908. [DOI] [PubMed] [Google Scholar]
- Toé KH, N'Falé S, Dabiré RK, Ranson H, Jones CM. The recent escalation in strength of pyrethroid resistance in Anopheles coluzzi in West Africa is linked to increased expression of multiple gene families. BMC Genomics. 2015;16:146. doi: 10.1186/s12864-015-1342-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varghese J, Lim SF, Cohen SM. Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev. 2010;24:2748–2753. doi: 10.1101/gad.1995910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voice MW, Kaaz AW, Peet CF, Paine MJ. Recombinant CYP6M2 Inhibition by Insecticides Recommended by WHO for Indoor Residual Spraying Against Malaria Vectors. Drug Metab Rev. 2015;44:56–57. [Google Scholar]
- Voßfeldt H, Butzlaff M, Prüßing K, Chárthaigh R-AN, Karsten P, Lankes A, et al. Large-scale screen for modifiers of ataxin-3-derived polyglutamine-induced toxicity in Drosophila. PLoS One. 2012;7:e47452. doi: 10.1371/journal.pone.0047452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Hou Y, Saha TT, Pei G, Raikhel AS, Zou Z. Hormone and receptor interplay in the regulation of mosquito lipid metabolism. Proc Natl Acad Sci. 2017;114:E2709–E2718. doi: 10.1073/pnas.1619326114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Martínez M-A, Dai M, Chen D, Ares I, Romero A, et al. Permethrin-induced oxidative stress and toxicity and metabolism. A review. Environ Res. 2016;149:86–104. doi: 10.1016/j.envres.2016.05.003. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Weill M, Malcolm C, Chandre F, Mogensen K, Berthomieu A, Marquine M, et al. The unique mutation in ace-1 giving high insecticide resistance is easily detectable in mosquito vectors. Insect Mol Biol. 2004;13:1–7. doi: 10.1111/j.1365-2583.2004.00452.x. [DOI] [PubMed] [Google Scholar]
- WHO . WHO; Geneva: 2020. World Malaria Report. [Google Scholar]
- Wilding CS, Weetman D, Rippon EJ, Steen K, Mawejje HD, Barsukov I, et al. Parallel evolution or purifying selection, not introgression, explains similarity in the pyrethroid detoxification linked GSTE4 of Anopheles gambiae and An. arabiensis. Mol Genet Genomics. 2014:1–15. doi: 10.1007/s00438-014-0910-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams J, Flood L, Praulins G, Ingham VA, Morgan J, Lees RS, et al. Characterisation of Anopheles strains used for laboratory screening of new vector control products. Parasit Vectors. 2019;12:522. doi: 10.1186/s13071-019-3774-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wisotzkey RG, Mehra A, Sutherland DJ, Dobens LL, Liu X, Dohrmann C, et al. Medea is a Drosophila Smad4 homolog that is differentially required to potentiate DPP responses. Development. 1998;125:1433–1445. doi: 10.1242/dev.125.8.1433. [DOI] [PubMed] [Google Scholar]
- Wülbeck C, Simpson P. The expression of pannier and achaete-scute homologues in a mosquito suggests an ancient role of pannier as a selector gene in the regulation of the dorsal body pattern. Development. 2002;129 doi: 10.1242/dev.129.16.3861. http://dev.biologists.org/content/129/16/3861.abstract 3861 LP –3871. [DOI] [PubMed] [Google Scholar]
- Yan H, Jahanshahi M, Horvath EA, Liu H-Y, Pfleger CM. Rabex-5 Ubiquitin Ligase Activity Restricts Ras Signaling to Establish Pathway Homeostasis in <em>Drosophila</em>. Curr Biol. 2010;20:1378–1382. doi: 10.1016/j.cub.2010.06.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Wang L-G, Han Y, He Q-Y. clusterProfiler: an R package for comparing biological themes among gene clusters. Omi a. J Integr Biol. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunta C, Hemmings K, Stevenson B, Koekemoer LL, Matambo T, Pignatelli P, et al. Cross-resistance profiles of malaria mosquito P450s associated with pyrethroid resistance against WHO insecticides. Pestic Biochem Physiol. 2019 doi: 10.1016/j.pestbp.2019.06.007. doi:https://doi.org/ [DOI] [PubMed] [Google Scholar]
- Zhang R, Ren Z, Chen W. SILGGM: An extensive R package for efficient statistical inference in large-scale gene networks. PLoS Comput Biol. 2018;14 doi: 10.1371/journal.pcbi.1006369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong Y, Wu C-F. Neuronal Activity and Adenylyl Cyclase in Environment-Dependent Plasticity of Axonal Outgrowth in Drosophila J Neurosci. 2004 ; 24 :1439 LP – 1445. doi:10.1523/JNEUROSCI.0740-02.2004. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this experiment are available at ArrayExpress under E-MTAB-9422 and E-MTAB-9423. The authors declare that all other data supporting the findings of this study, are available within the article and its Supplementary Information files or are available from the authors upon request.