Abstract
DNA damage checkpoints lead to the inhibition of cell cycle progression following DNA damage. The Saccharomyces cerevisiae Mec1 checkpoint protein, a phosphatidylinositol kinase-related protein, is required for transient cell cycle arrest in response to DNA damage or DNA replication defects. We show that mec1 kinase-deficient (mec1kd) mutants are indistinguishable from mec1Δ cells, indicating that the Mec1 conserved kinase domain is required for all known Mec1 functions, including cell viability and proper DNA damage response. Mec1kd variants maintain the ability to physically interact with both Ddc2 and wild-type Mec1 and cause dominant checkpoint defects when overproduced in MEC1 cells, impairing the ability of cells to slow down S phase entry and progression after DNA damage in G1 or during S phase. Conversely, an excess of Mec1kd in MEC1 cells does not abrogate the G2/M checkpoint, suggesting that Mec1 functions required for response to aberrant DNA structures during specific cell cycle stages can be separable. In agreement with this hypothesis, we describe two new hypomorphic mec1 mutants that are completely defective in the G1/S and intra-S DNA damage checkpoints but properly delay nuclear division after UV irradiation in G2. The finding that these mutants, although indistinguishable from mec1Δ cells with respect to the ability to replicate a damaged DNA template, do not lose viability after UV light and methyl methanesulfonate treatment suggests that checkpoint impairments do not necessarily result in hypersensitivity to DNA-damaging agents.
DNA is prone to alterations, and genomic integrity is ensured by DNA repair systems removing DNA damage and by surveillance mechanisms, known as DNA damage checkpoints, delaying cell cycle progression in response to DNA insults. These mechanisms contribute to the maintenance of genome stability, since they ensure that damaged DNA molecules are neither replicated nor segregated to daughter cells until repaired. Failure to respond properly to DNA damage is a hallmark of cancer cells (reviewed in reference 56).
Cell cycle progression can be transiently arrested by checkpoints at different stages, depending on the cell cycle phase at which DNA alterations occur. In fact, delay of G1/S transition or slowing down of progression through S phase takes place when DNA is damaged in G1 or during DNA synthesis, respectively, thus preventing replication of damaged templates (38, 47). Furthermore, when DNA is damaged in G2 or when DNA replication is incomplete, segregation of damaged or incompletely replicated chromosomes is prevented by delaying nuclear division, thus linking entry into mitosis to proper completion of S phase (58, 59, 60).
Studies of the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae play an important role in the identification of DNA damage checkpoint proteins and in unraveling checkpoint mechanisms. The budding yeast RAD9, RAD24, RAD17, MEC3, and DDC1 genes are necessary for DNA damage checkpoint response and are thought to act early in the DNA damage-induced signaling pathways (reviewed in references 27, 30, and 57). Both Ddc1 and Rad17 are structurally related to the sliding-clamp protein PCNA (proliferating cell nuclear antigen) (50), whose homotrimers form a structure that encircles DNA and tethers DNA polymerase δ to DNA during DNA replication (reviewed in reference 55). This homology and the finding that Ddc1 physically interacts with Rad17 and Mec3 (23, 35) raise the possibility that the Ddc1-Rad17-Mec3 complex may also form clamp-like structures that participate in the recognition and/or processing of damaged DNA.
Central to this signal transduction network is the Mec1 protein, a member of the evolutionarily conserved phosphatidylinositol 3-kinase motif family (6, 19, 21, 62), including S. cerevisiae Tel1 (17, 34), S. pombe Rad3 (4), Drosophila melanogaster Mei-41 (20), and human ATM (45), ATR (4), and DNA-PK (12). MEC1, as well as human ATM and S. pombe Rad3, is required for all known DNA damage checkpoints and for response to incomplete DNA replication. Moreover, the ATM gene is mutated in the familial neural degeneration and cancer-predisposition syndrome ataxia telangiectasia (45). Due to the lack of human ATR mutant cells, the functional role of ATR in the checkpoint pathway is not fully understood. However, overexpression of kinase-defective mutant ATR in wild-type cells abrogates G2/M arrest after exposure to ionizing radiation and increases the sensitivity of cells to ionizing radiation and UV light (9), suggesting some overlapping functions of ATM and ATR.
In addition to its involvement in the checkpoint responses, budding yeast Mec1 is essential for cell viability. However, its essential function, but not its checkpoint functions, can be bypassed by increasing the intracellular concentration of deoxyribonucleotide triphosphates (dNTPs), either by overexpression of RNR genes encoding ribonucleotide reductase (13) or by deletion of the SML1 gene (64), which negatively affects dNTP pools (7).
In S. cerevisiae, several key regulators of the Mec1-dependent signaling pathway, like Rad9, Ddc1, and Ddc2, become phosphorylated in a Mec1-dependent manner in response to DNA damage. The study of the interdependency of these phosphorylation events suggests that Mec1 is implicated in the DNA damage-sensing pathways (14, 35, 48, 49, 53). Moreover, since Mec1 physically interacts with Ddc2 (Lcd1), which is also necessary for all of the DNA integrity checkpoints (36, 42) and undergoes Mec1-dependent DNA damage-induced phosphorylation independently of all of the other known checkpoint proteins, the Mec1-Ddc2 complex might respond to DNA insults independently of the other checkpoint factors (36).
Also, Rad53 and Chk1 undergo Mec1-dependent phosphorylation in response to DNA damage (2, 43, 44) and appear to act downstream of Mec1. Whereas Rad53 is required for all of the DNA integrity checkpoints, Chk1 is specifically required to prevent nuclear division in cdc13 mutants at nonpermissive temperatures, presumably through phosphorylation of the anaphase inhibitor Pds1 (10, 44).
Although phosphorylation of several key regulators in response to DNA damage or a replication block is Mec1 dependent, less is known about the requirement for the Mec1 kinase domain in activation of the DNA damage checkpoints and whether the cell cycle phases at which DNA alterations occur might influence the chance to activate the checkpoint response. To address these points, we generated and characterized two mec1kd alleles specifically altered in the Mec1 conserved kinase domain and searched for new mec1 mutants specifically altered in subsets of DNA damage checkpoint pathways. We show that the Mec1 conserved kinase domain is essential for all of the functions of Mec1. Moreover, overproduction of the Mec1kd mutant forms has a dominant-negative effect specifically on the cell response to DNA damage in G1 or during S phase. We also describe two new hypomorphic mec1 mutants that appear to be completely defective in the G1/S and intra-S checkpoints but proficient in the G2/M checkpoint, suggesting that the Mec1 functions required for response to DNA alterations in the different cell cycle stages are separable.
MATERIALS AND METHODS
Yeast strains and media.
The genotypes of all of the yeast strains used in this study are listed in Table 1. All of the yeast strains were derivatives of W303 (MATa or MATα ade2-1 can1-100 trp1-1 leu2-3,112 his3-11,15 ura3).
TABLE 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| K699 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | 29 |
| K700 | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 | 29 |
| DMP2750.1 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3/ura3 MECI-HA9::LEU2::mec1/MEC1-MYC18::LEU2::mec1 | This study |
| DMP2872/8B | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1-HA2::LEU2::ddc1 mec1kd1 sml1Δ::KanMX4 | This study |
| DMP2872/4A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1-HA2::LEU2::ddc1 sml1Δ::KanMX4 | This study |
| DMP2876/3A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1-HA2::LEU2::ddc1 meclkd2 sml1Δ::KanMX4 | This study |
| DMP2882/2C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1-HA2::LEU2::ddc1 mec1Δ::HIS3 sml1Δ::KanMX4 | This study |
| DMP2885.4 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3/ura3 mec1kd1- HA9::LEU2::mec1/MEC1-MYC18::LEU2::mec1 | This study |
| DMP2893.1 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3/ura3 mec1kd2-HA9::LEU2::mec1/MEC1-MYC18::LEU2::mec1 | This study |
| DMP3048/5B | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1Δ::HIS3 sml1Δ::KanMX4 DDC2-HA3::URA3 | This study |
| DMP3055/8B | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1- MEC1::URA3 DDC2-HA3::URA3 | This study |
| DMP3058/13B | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd1::URA3 DDC2-HA3::URA3 | This study |
| DMP3274/5A | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 chk1Δ::HIS3 | This study |
| DMP3287/2C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-MEC1::URA3 DDC2-HA3::URA3 chk1Δ::HIS3 | This study |
| DMP3288/5A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 chk1Δ::HIS3 | This study |
| DMP3288/8C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd1::URA3 DDC2-HA3::URA3 chk1Δ::HIS3 | This study |
| DMP3295/8B | MATaade2-1 can1-100 his3-11,15 trp1-1 leu2-3,112 ura3 MEC1-HA9::LEU2::mec1 DDC2-MYC18::HIS3 | This study |
| DMP3296/3C | MATaade2-1 can1-100 his3-11,15 trp1-1 leu2-3,112 ura3 mec1kd1-HA9::LEU2::mec1 DDC2-MYC18::HIS3 sml1Δ::KanMX4 | This study |
| DMP3297/6D | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1kd2-HA9::LEU2::mec1 DDC2-MYC18::HIS3 sml1Δ::KanMX4 | This study |
| DMP3343/6C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 mec1-100::LEU2::mec1Δ | This study |
| DMP3344/4A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 mec1-101::LEU2::mec1Δ | This study |
| DMP3412/1A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 CHK1-HA3::URA3 | This study |
| DMP3412/6C | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 CHK1-HA3::URA3 | This study |
| DMP3432/7A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd1::URA3 mec1Δ::HIS3 sml1Δ::KanMX4 DDC2-HA3::URA3 CHK1-HA3::URA3 | This study |
| DMP3455/9A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd1::URA3 DDC2-HA3::URA3 CHK1-HA3::URA3 | This study |
| DMP3459/17C | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-MEC1::URA3 DDC2-HA3::URA3 CHK1-HA3::URA3 | This study |
| YLL334 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC1-HA2::LEU2::ddc1 | 29 |
| YLL447.32 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3/ura3 MEC1/MEC1-MYC18::LEU2::mec1 | This study |
| YLL476.34 | MATa/MATα ade2-1/ade2-1 can1-100/can1-100 his3-11,15/his3-11,15 leu2-3,112/leu2-3,112 trp1-1/trp1-1 ura3/ura3 MEC1/MEC1-HA9::LEU2::mec1 | This study |
| YLL490 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 mec1Δ::HIS3 sml1Δ::KanMX4 | 28 |
| YLL516 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-MEC1::URA3 | This study |
| YLL517 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd1::URA3 | This study |
| YLL518 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd2::URA3 | This study |
| YLL683.8/3D | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 | 36 |
| YLL683.8/4A | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 DDC2-HA3::URA3 | 36 |
| YLL750 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 sml1Δ::KanMX4 mec1-100::LEU2::mec1Δ | This study |
| YLL753 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 sml1Δ::KanMX4 mec1-101::LEU2::mec1Δ | This study |
| YLL769 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3::GAL1-mec1kd1::URA3 pML240 [CEN4 LEU2 GAL1-MEC1] | This study |
| YLL839 | MATaade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3 CHK1-HA3::URA3 | This study |
To obtain strains YLL516, YLL517, and YLL518, carrying, respectively, the GAL1-MEC1, GAL1-mec1kd1, and GAL1-mec1kd2 alleles at the URA3 chromosomal locus, strain K699 was transformed with NcoI-digested plasmids pML236, pML237, and pML238, respectively. Strains DMP3055/8B and DMP3058/13B were derived from crosses of strain DMP683.8/3D with YLL516 and YLL517, respectively. The MEC1 and SML1 deletions (28) and the DDC2-HA3, MEC1-MYC18, and MEC1-HA9 alleles (36) were constructed as previously described. Strains DMP3295/8B, DMP3296/3C, and DMP3297/6D, carrying the DDC2-MYC18 allele at the DDC2 chromosomal locus, and strain YLL839, carrying the CHK1-HA3 allele at the CHK1 chromosomal locus, were generated by the PCR one-step tagging method (22) using, respectively, plasmids 3746 and 3748 (K. Nasmyth, Institute of Molecular Pathology, Vienna, Austria) as templates and oligonucleotides PRP179 (5′-CTT GAG TCA AAA TCA TTC GAT CTA ACC ACA CTA GAG GAG GCC GAT TCA TTA TAT ATC TCA ATG GGA CTG TCC GGT TCT GCT GCT AG-3′) and PRP180 (5′-ATA TAG TTA ATA TTA AGC ATT ACA AGG TTT CTA TAA AGC GTT GAC ATT TTC CCC TTT TGA TTG TTG CCC CTC GAG GCC AGA AGA C-3′) (DDC2-MYC18) or oligonucleotides PRP217 (5′-CTT TAG AAT GGA GAA GAT TGT TCA AGA AAA TTT CAA CTA TCT GTA GGG ATA TTA TCC TAT TCC CAA CTC CGG TTC TGC TGC TAG-3′) and PRP218 (5′-ATA AGT AGA AAG AAT TTT TTT TTT TTT TTG ATC AGT GCA TCT TAA CCC TTC TTT TGT CTC CAT TTT TTC CTC GAG GCC AGA AGA C-3′) (CHK1-HA3) as primers. Strains DMP3412/1A and DMP3412/6C were meiotic segregants from a cross between strains YLL839 and DMP683.8/3D. Strains DMP3455/9A and DMP3459/17C were meiotic segregants from crosses of strains DMP3058/13B and DMP3055/8B with strain DMP3412/6C, respectively. Strain DMP3432/7A was a meiotic segregant from a cross between strains DMP3058/13B and DMP3412/6C, followed by deletion of MEC1 and SML1 as described by Longhese et al. (28). The DDC2-HA3, DDC2-MYC18, MEC1-MYC18, and MEC1-HA9 alleles are fully functional, since strains carrying them at the corresponding chromosomal loci were indistinguishable from the wild type with respect to viability, growth rates at any temperature, and sensitivity to UV radiation, methyl methanesulfonate (MMS), and hydroxyurea (HU). Since CHK1 alterations do not cause obvious phenotypes but do impair Pds1 phosphorylation (44), we verified that DNA damage-induced Pds1 phosphorylation was unaffected in CHK1-HA3 cells.
To generate the CHK1 chromosomal deletion, a chk1Δ::HIS3 cassette was constructed by PCR using the pFA6a-HIS3 plasmid (54) as a template and oligonucleotides PRP190 (5′-TAT CAT AAG TTG CTG TAT ATG GGC AGC ACG TAT TAC TAT GAG TCT CGT ACG CTG CAG GTC GAC-3′) and PRP191 (5′-TGT CTC CAT TTT TTT CAG TTG GGA ATT AGG ATA ATA TCC CTA CAG ATA GTA TCG ATG AAT TCG AGC TCG-3′) as primers, followed by transformation of strain K700 with the PCR product, giving rise to strain DMP3274/5A, where the 1,540 bp of the CHK1 coding region were replaced with the Kluyveromyces lactis HIS3 gene. Strain DMP3287/2C was a meiotic segregant from a cross between strains DMP3274/5A and DMP3055/8B. Strains DMP3288/5A and DMP3288/8C were meiotic segregants from a cross between strains DMP3058/13B and DMP3274/5A. Strain YLL769 was obtained by transforming strain YLL517 with plasmid pML240. Details of strains carrying the mec1kd, mec1-100, and mec1-101 alleles are given in the paragraphs describing the generation of the mutant alleles.
The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR. The standard yeast genetic techniques and media used were described by Rose et al. (41). Cells were grown in YEP medium (1% yeast extract, 2% Bacto Peptone, 50 mg of adenine per liter) supplemented with 2% glucose (YEPD), 2% raffinose (YEP-raf), or 2% raffinose and 1% galactose (YEP-raf-gal). Transformants carrying the KanMX4 cassette were selected on YEPD plates containing G418 (United States Biological) at 400 μg/ml.
Plasmids.
Plasmid pML224, used to generate plasmids pML228.1 and pML229.3 (see next paragraph) and carrying the C-terminal region of MEC1, was originated by inserting into the KpnI-BamHI sites of plasmid YIplac211 (16) the 1,243-bp KpnI-BamHI MEC1 fragment from plasmid pML79 (29). To construct plasmid pML227 (LEU2 CEN4 MEC1), the 8,358-bp XbaI-SpeI fragment containing the whole MEC1 coding region and the 385 bp upstream of the MEC1 ATG codon was cloned into the SpeI site of plasmid YCplac111 (16), followed by excision of the SalI-NarI fragment. To construct plasmid pML236 (YIp5 URA3 GAL1-MEC1), in which a 7,437-bp fragment extending from the MEC1 ATG codon to the SacI site downstream to the MEC1 stop codon is fused to the GAL1 promoter, the SphI-SpeI 8,372-bp fragment from plasmid pML225 (URA3 CEN4 GAL1-MEC1) (36) was cloned into the NheI-SphI sites of plasmid YIp5. Plasmids pML230 and pML231, used to generate plasmids pML237 and pML238, were constructed by cloning, respectively, the 588-bp KpnI-SacII fragment from plasmids pML228.1 and pML229.3 into the KpnI-SacII sites of plasmid pML225. To construct plasmids pML237 (YIp5 URA3 GAL1-mec1kd1) and pML238 (YIp5 URA3 GAL1-mec1kd2), the SphI-SpeI 8,372-bp fragments from plasmids pML230 (URA3 CEN4 GAL1-mec1kd1) and pML231 (URA3 CEN4 GAL1-mec1kd2), respectively, were cloned into the NheI and SphI sites of YIp5. Plasmid pML240 (CEN4 LEU2 GAL1-MEC1) was obtained by cloning a 7,437-bp fragment extending from the MEC1 ATG codon to the SacI site downstream to the MEC1 stop codon into the YCplac111 plasmid. To obtain plasmid pML239, the 8,093-bp SpeI-SpeI fragment containing the whole MEC1 gene from plasmid pML79 was cloned into the XbaI site of plasmid pGEM4.
Generation and transplacement of the mec1kd alleles.
Plasmids pML228.1 and pML229.3, containing, respectively, the base substitutions resulting in the Mec1kd1 D2243E and Mec1kd2 D2224A amino acid changes, were generated by PCR site-directed mutagenesis using plasmid pML224 as a template and oligonucleotides PRP154 (5′-CGG GTA AAG TTC TTC ATG TAG AAT TCG ACT GTT TAT TTG AGA AAG-3′) and PRP155 (5′-CTT TCT CAA ATA AAC AGT CGA ATT CTA CAT GAA GAA CTT TAC CCG-3′) or oligonucleotides PRP152 (5′-GGC CAT ATA TTA GGT CTA GGT GCT AGG CAC TGT GAA AAC ATA TTA-3′) and PRP153 (5′-TAA TAT GTT TTC ACA GTG CCT AGC ACC TAG ACC TAA TAT ATG GCC-3′) as primers to obtain the mec1kd1 or mec1kd2 allele, respectively. The presence of the expected mutations in the above plasmids was assessed by DNA sequencing of the entire PCR fragments.
Transformation of diploid strain W303 with XhoI-digested plasmids pML228.1 and pML229.3 generated MEC1/mec1kd1::URA3 and MEC1/mec1kd2::URA3 heterozygous strains, respectively. Meiotic tetrads from these strains contained only two viable spores carrying the MEC1 allele, while no viable mec1kd1 or mec1kd2 URA3 spores were found. Transformation with the above plasmids of a diploid sml1Δ::KanMX4/SML1 heterozygous strain generated an SML1/ sml1Δ::KanMX4 MEC1/mec1kd::URA3 strain. Several meiotic tetrads from this strain contained more than two viable spores, and viable mec1kd::URA3 sml1Δ::KanMX4 segregants were present with the frequency expected for cosegregation of the two unlinked mec1kd and sml1Δ alleles. Strains DMP2872/8B and DMP2876/3A, in which the MEC1 chromosomal copy was replaced with the mec1kd1 and mec1kd2 alleles, respectively, were obtained by two-step replacement, by transforming a MEC1 DDC1-HA2 sml1Δ strain with XhoI-digested plasmids pML228.1 and pML229.3, followed by excision of the URA3 marker. Similarly, to obtain strains DMP3296/3C and DMP3297/6D, in which the MEC1 chromosomal copy was replaced with the mec1kd1-HA9 and mec1kd2-HA9 alleles, respectively, a MEC1-HA9::LEU2::mec1 DDC2-MYC18::HIS3 sml1Δ strain was transformed with XhoI-digested plasmids pML228.1 and pML229.3, followed by excision of the URA3 marker.
Search for new mec1 mutants.
Mutagenesis of the MEC1 gene was performed by PCR using standard PCR conditions as described by Umezu et al. (52). Primers PRP161 (5′-ATG GAA TCA CAC GTC AAA TAT C-3′) and PRP162 (5′-GAG AAG TGT CTA ATA AAG CAC C-3′) were used to amplify the region between positions +1 and +3307 (gap A), primers PRP163 (5′-CGG AGA AAG CAG ACA GAA AG-3′) and PRP164 (5′-GGG CCA CGT TCA TGT CAA AT-3′) were used to amplify the region between positions +3207 and +6034 (gap B), and primers PRP165 (5′-CAA ACG AGG ATC CAT TAA GGA-3′) and PRP166 (5′-CCA AAA TGG AAG CCA ACC AAT-3′) were used to amplify the region between positions +4747 and +7104 (gap C). PCR mixtures for each set of primers (25 μl) contained 1.25 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.), 10 ng of template DNA (pML239), 1 μM each primer, 200 μM each dNTP, 10 mM Tris-HCl (pH 8.3), 50 mM KCl, and 1.5 mM MgCl2. Twenty independent reaction mixtures were prepared for each set of primers. Strain YLL490 (mec1Δ sml1Δ) was cotransformed with gap A PCR products and the StuI-StuI fragment of pML227 (YCplac111 CEN4 LEU2 MEC1), lacking 2,358 bp between positions +476 and +2834 of the MEC1 coding region, or with gap B or gap C PCR products and the NruI-NotI fragment of pML227, lacking 304 bp between positions +5271 and +5575 of the MEC1 coding region, in order to obtain reconstruction of the whole MEC1 coding region by gap repair. Leu+ transformants were tested for the ability to grow on YEPD plates after UV irradiation (50 J/m2) or in the presence of MMS (0.01%) or HU (100 mM) at 25°C. None of the several mec1 mutants identified were confirmed to be specifically hypersensitive to MMS or UV light, while the mec1-100 and mec1-101 mutants were weakly hypersensitive to HU, but not to MMS and UV light, and were further analyzed. To obtain stable mec1-100 and mec1-101 mutants, the 7,876-bp NcoI-EcoRI fragments from plasmids pML254.1 and pML253.1, containing, respectively, the whole mec1-100 and mec1-101 alleles, were cloned into the SphI-EcoRI sites of the YIplac128 (LEU2) integrative plasmid to generate plasmids pML258.51 and pML266.46, respectively. SpeI digestion was then used to direct the integration of these plasmids into the MEC1 promoter region of mec1Δ sml1Δ strain YLL490, giving rise to strains YLL750 and YLL753, carrying, respectively, the mec1-100 and mec1-101 alleles as the sole complete mec1 alleles at the MEC1 chromosomal locus. SML1 strains DMP3343/6C and DMP3344/4A were meiotic segregants from crosses of strain YLL683.8/3D with strains YLL750 and YLL753, respectively, and their phenotypes were indistinguishable from those of strains YLL750 and YLL753, indicating that the effects of the mec1-100 and mec1-101 alleles are not influenced by SML1.
Other techniques.
Synchronization experiments, immunoprecipitations, Western blot analysis, and kinase assays were performed as previously described (36).
RESULTS
Alteration of the Mec1 conserved kinase domain impairs all Mec1 functions.
In order to investigate whether Mec1 functions as a protein kinase in establishing the DNA integrity checkpoints, we generated the mutations mec1kd1 and mec1kd2, which cause the amino acid changes D2243E and D2224A, respectively, in the Mec1 putative kinase domain (see Materials and Methods). The same amino acid changes in the S. pombe Rad3 lipid kinase domain affected Rad3 function (4). When we analyzed the in vivo consequences of these mutations, we found that both mec1kd alleles resulted in cell lethality that was suppressed by deletion of the SML1 gene (see Materials and Methods), similarly to the MEC1 deletion (64) and to another recently described mec1kd allele (D2224A N2229K) (31). Furthermore, viable mec1kd1 sml1Δ and mec1kd2 sml1Δ strains were as hypersensitive as a mec1Δ sml1Δ strain to UV light, MMS, and HU (Fig. 1A).
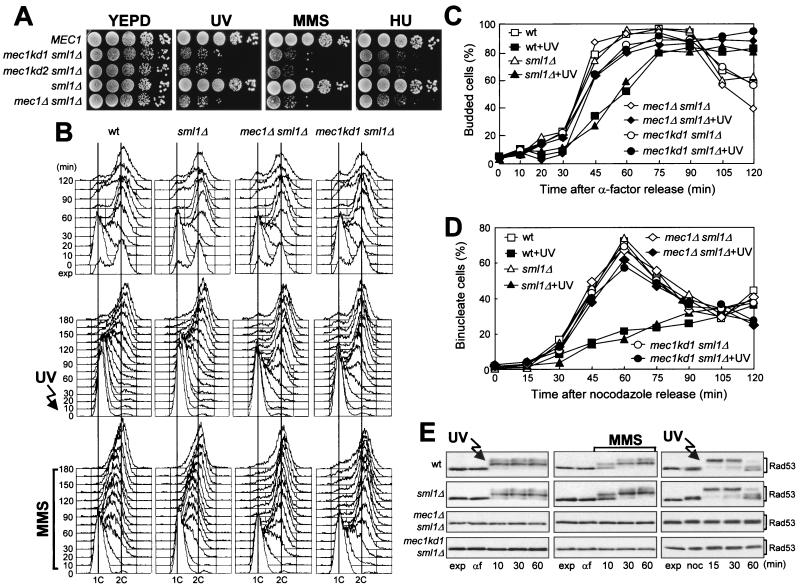
FIG. 1.
mec1 kinase deficiency mutations impair all known DNA damage checkpoints. (A) Serial dilution of cultures of wild-type (wt) YLL334, mec1kd1 sml1Δ DMP2872/8B, mec1kd2 sml1Δ DMP2876/3A, sml1Δ DMP2872/4A, and mec1Δ sml1Δ DMP2882/2C cells growing exponentially in YEPD were spotted on YEPD plates with or without MMS (0.005%) or HU (5 mM). One YEPD plate was UV irradiated (30 J/m2) (UV). (B to E) The strains used were wild-type YLL334, sml1Δ DMP2872/4A, mec1Δ sml1Δ DMP2882/2C, and mec1kd1 sml1Δ DMP2872/8B. (B and C) α-Factor-synchronized cell cultures were UV irradiated (40 J/m2) prior to the release from α-factor in YEPD or were released in YEPD containing 0.02% MMS. (B) Samples of untreated (top), UV-irradiated (middle), or MMS-treated (bottom) cells were taken at the indicated times after release into the cell cycle and analyzed by fluorescence-activated cell sorter. (C) Untreated or UV-irradiated (+UV) cell cultures were scored for the percentage of budded cells at the indicated times. (D) Cell cultures were arrested with nocodazole (noc) and UV irradiated (50 J/m2) prior to the release in YEPD at time zero. Propidium iodide staining was used to directly visualize nuclear division at the indicated times after release from nocodazole in unirradiated and UV-irradiated (+UV) cultures. The survival levels of UV light-treated wild-type, sml1Δ, mec1kd1 sml1Δ, and mec1Δ sml1Δ cells were 78, 90, 8.3, and 7.3%, respectively. (E) Extracts from the above-described G1 UV light-treated (left) or MMS-treated (middle) or G2 UV light-treated (right) cell cultures were analyzed by Western blot assay with anti-Rad53 antibodies. exp, exponentially growing cells.
As shown in Fig. 1, mec1kd1 sml1Δ cells were as defective as mec1Δ sml1Δ cells at all known DNA damage checkpoints. In fact, when mec1kd1 sml1Δ or mec1Δ sml1Δ G1-arrested cells were UV irradiated and then released from the block, both entry into S phase (Fig. 1B, middle) and budding kinetics (Fig. 1C) were much faster and cell survival was much lower (3.4 and 2.5%, respectively) than in wild-type and sml1Δ mutant cell cultures under the same conditions (87 and 89% cell survival, respectively). Furthermore, when α-factor-synchronized mec1kd1 sml1Δ or mec1Δ sml1Δ cells were released from G1 arrest in the presence of MMS, they doubled their DNA content within 30 min and progressively lost viability (both already down to 10% cell survival at 30 min), whereas MMS-treated wild-type and sml1Δ cells progressed through S phase very slowly, completing DNA replication only after 150 min (Fig. 1B, bottom) without losing viability. Finally, mec1kd1 sml1Δ, as well as mec1Δ sml1Δ, cells released from G2 arrest after UV irradiation lost viability and divided nuclei much faster than similarly treated wild-type and sml1Δ cells, which maintained high cell survival and delayed nuclear division compared to unirradiated cells (Fig. 1D).
Since activation of DNA damage checkpoint pathways leads to Mec1-dependent phosphorylation of Rad53 (reviewed in references 27, 30, and 57), we analyzed the Rad53 phosphorylation pattern as a means by which to uncover alterations of Mec1 functions. As shown in Fig. 1E, the inability of mec1kd1 cells to arrest cell cycle progression after DNA damage correlated with impaired Rad53 phosphorylation, since no phosphorylated Rad53 was detectable in mec1kd1 sml1Δ or mec1Δ sml1Δ cells after DNA damage in G1 or G2 or during S phase.
A mec1kd2 sml1Δ strain was also subjected to all of the above-described analyses, and its behavior was always indistinguishable from that of the mec1kd1 sml1Δ strain (data not shown). Thus, the Mec1 kinase domain is required both to sustain cell viability and for proper DNA damage response.
The kinase-deficient Mec1kd1 and Mec1kd2 variants still physically interact with both Ddc2 and wild-type Mec1.
We have previously shown that Mec1 physically interacts with Ddc2 and that its associated kinase activity is capable of phosphorylating Ddc2 in vitro and is impaired by the mec1kd mutations (36). As shown in Fig. 2A, we further confirmed this point, since phosphorylated, Myc-tagged Ddc2 was detected when in vitro kinase assays were performed on immunoprecipitates containing hemagglutinin (HA)-tagged Mec1 but not when the same assays were performed on HA-tagged Mec1kd immunoprecipitates, although similar amounts of Myc-tagged Ddc2 coprecipitated with either the Mec1 or the Mec1kd protein. Thus, both mutations completely abolish the Mec1-associated kinase activity, further strengthening the hypothesis that the kinase is Mec1 itself. Accordingly, Mallory and Petes (31) recently showed that a Mec1 variant with two amino acid substitutions (D2224A and N2229K) in the kinase domain lost the ability to phosphorylate the mammalian protein PHAS-I (phosphorylated heat- and acid-stable protein I) in vitro.
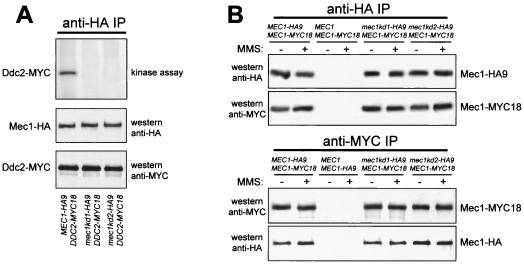
FIG. 2.
Kinase activity and interactions of Mec1kd variants. (A) HA-tagged Mec1 or Mec1kd proteins (Mec1-HA) were immunoprecipitated with anti-HA antibodies (anti-HA IP) from protein extracts prepared from exponentially growing cells concomitantly expressing Mec1-HA9 and Ddc2-MYC18 (DMP3295/8B), Mec1kd1-HA9 and Ddc2-MYC18 (DMP3296/3C), or Mec1kd2-HA9 and Ddc2-MYC18 (DMP3297/6D) from the MEC1 and DDC2 promoters, respectively, as indicated at the bottom. Kinase assays were performed on anti-HA immunoprecipitates, and the results are shown at the top. The same immunoprecipitates were also analyzed by Western blot assay using the antibodies indicated on the right side of the middle and bottom parts of the panel. (B) Immunoprecipitations with anti-HA (anti-HA IP) or anti-MYC (anti-MYC IP) antibodies were performed on extracts from exponentially growing untreated (−) or MMS-treated (+; 0.02% MMS for 1 h) diploid cells with the genotypes indicated in the top part of the panels. Mec1-HA9 and Mec1-MYC18 were then detected by Western blot analysis of the immunoprecipitates by using anti-HA and anti-MYC antibodies. The genotypes of the strains used were MEC1-HA9/MEC1-MYC18 (DMP2750.1), MEC1/MEC1-MYC18 (YLL447.32), MEC1/MEC1-HA9 (YLL476.34), mec1kd1-HA9/MEC1-MYC18 (DMP2885.4), and mec1kd2-HA9/MEC1-MYC18 (DMP2893.1).
Since it was shown that multiple S. pombe Rad3 molecules may be present in complexes (4), we asked whether Mec1 could also form homomeric complexes and whether the Mec1kd variants might still be present in these complexes. To this end, we performed immunoprecipitation assays on protein extracts from untreated and MMS-treated diploid cells carrying fully functional MEC1-HA9 and MEC1-MYC18 alleles at the two MEC1 chromosomal loci. As shown in Fig. 2B, Mec1 molecules can self-associate, since Mec1-MYC18 was specifically recognized by the anti-MYC antibodies in Mec1-HA9 immunoprecipitates, and anti-HA antibodies detected Mec1-HA9 in Mec1-MYC18 immunoprecipitates, independently of DNA damage. Furthermore, when the heterozygous diploid MEC1-MYC18/mec1kd1-HA9 and MEC1-MYC18/mec1kd2-HA9 strains were used in analogous immunoprecipitation assays, Mec1-MYC18 was specifically recognized by the anti-MYC antibodies in both Mec1kd1-HA9 and Mec1kd2-HA9 immunoprecipitates, and anti-HA antibodies detected Mec1kd1-HA9 and Mec1kd2-HA9 in Mec1-MYC18 immunoprecipitates (Fig. 2B), indicating that both Mec1kd inactive forms are still able to interact with wild-type Mec1.
High levels of kinase-deficient Mec1kd1 protein in MEC1 cells cause damage-resistant DNA replication.
Although the mec1kd alleles behave recessively when present in single copy in a mec1kd/MEC1 heterozygous strain (data not shown), the finding that their kinase-deficient gene products are still able to interact in vivo with both wild-type Mec1 and Ddc2 (Fig. 2) led us to ask whether their overexpression might affect the response to DNA damage in the presence of physiological amounts of wild-type Mec1. To address this point, MEC1 strains carrying GAL1-MEC1, GAL1-mec1kd1 and GAL1-mec1kd2 gene fusions at the URA3 locus were first assayed for sensitivity to genotoxic agents under galactose-induced conditions. As shown in Fig. 3A, high levels of inactive Mec1kd proteins in a MEC1 background have a dominant-negative effect, since wild-type cells overproducing Mec1kd1 or Mec1kd2 were more sensitive to HU, MMS, and UV light than otherwise isogenic wild-type or MEC1-overexpressing strains. This hypersensitivity can be suppressed by increasing the level of wild-type Mec1, since MEC1 cells concomitantly expressing the GAL1-mec1kd1 and GAL1-MEC1 fusions were as sensitive as the wild type to HU, MMS, and UV light (Fig. 3A). Therefore, high levels of the kinase-defective variants might determine a dominant defect in the response to DNA damage by competing with wild-type Mec1 molecules.
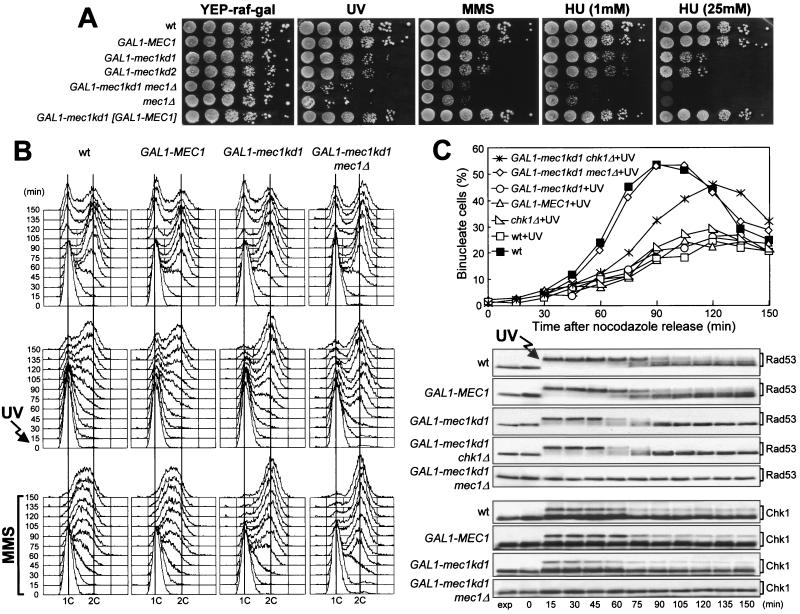
FIG. 3.
Dominant-negative effect of mec1kd1 overexpression. (A) Serial dilutions of exponentially growing (in YEPD) cultures of wild-type (wt) K699, GAL1-MEC1 YLL516, GAL1-mec1kd1 YLL517, GAL1-mec1kd2 YLL518, and GAL1-mec1kd1 [GAL-MEC1] YLL769 cells, all carrying the MEC1 allele at the MEC1 chromosomal locus, and GAL1-mec1kd1 mec1Δ DMP3432/7A and mec1Δ YLL490 cells, both also carrying the sml1Δ allele, were spotted on YEP-raf-gal plates with or without MMS (0.005%) or HU. One YEP-raf-gal plate was UV irradiated (40 J/m2) (UV). (B) Cultures of wild-type DMP3412/1A, GAL1-MEC1 DMP3459/17C, and GAL1-mec1kd1 DMP3455/9A cells, all carrying the MEC1 allele at the MEC1 chromosomal locus, and GAL1-mec1kd1 mec1Δ DMP3432/7A cells, also carrying the sml1Δ allele, logarithmically growing in YEP-raf, were synchronized with α-factor 2.5 h after addition of galactose to 1%. Cell cultures were released from α-factor at time zero into YEP-raf-gal medium with or without 0.02% MMS. One-third of each synchronized culture was UV irradiated (40 J/m2) prior to release. Samples of untreated (top), UV-irradiated (middle), or MMS-treated (bottom) cultures were taken at the indicated times after the release from α-factor and analyzed by fluorescence-activated cell sorter. (C) Cultures of wild-type DMP3412/1A, GAL1-MEC1 DMP3459/17C, GAL1-mec1kd1 DMP3455/9A, GAL1-MEC1 chk1Δ DMP3287/2C, chk1Δ DMP3288/5A, and GAL1-mec1kd1 chk1Δ DMP3288/8C cells, all carrying the MEC1 allele at the MEC1 chromosomal locus, and GAL1-mec1kd1 mec1Δ DMP3432/7A cells, also carrying the sml1Δ allele, logarithmically growing in YEP-raffinose, were synchronized with nocodazole 2 h after addition of 1% galactose and UV irradiated (50 J/m2) prior to release in YEP-raf-gal medium. Nuclear division (top) was directly visualized at the indicated times in untreated and UV light-treated (+UV) cultures by propidium iodide staining. Protein extracts (bottom) from the UV light-treated cell cultures were analyzed by Western blot assay using anti-Rad53 and anti-HA (Chk1) antibodies. exp, exponentially growing cells.
We then analyzed the checkpoint-mediated cell cycle arrest in MEC1 cells overproducing the Mec1kd variants. When G1-arrested, galactose-induced MEC1 GAL1-mec1kd1 cell cultures were UV irradiated prior to release from the block, they not only lost viability (24% survival) but progressed into the cell cycle, reaching a 2C DNA content after 75 min, faster than similarly treated wild-type and MEC1 GAL1-MEC1 cells, which completed DNA replication only after 120 min (Fig. 3B, middle) and maintained high cell survival (79 and 85%, respectively). Furthermore, when G1-arrested MEC1 GAL1-mec1kd1 cells were released from the block in the presence of MMS under galactose-induced conditions, they progressed through S phase much faster than similarly treated wild-type and MEC1 GAL1-MEC1 cells (Fig. 3B, bottom) and progressively lost viability (already down to 30.5% cell survival at 30 min), while the viability of the MMS-treated wild-type and MEC1 GAL1-MEC1 cells was substantially unaffected throughout the experiment. Furthermore, Ddc2 phosphorylation was abolished and Rad53 phosphorylation was severely affected in both of the above-described UV light- and MMS-treated MEC1 GAL1-mec1kd1 cell cultures, compared to similarly treated wild-type and MEC1 GAL1-MEC1 cells (data not shown). Therefore, high levels of the kinase defective Mec1kd1 protein in MEC1 cells have dominant-negative effects on checkpoint response, impairing the ability of cells to regulate DNA replication, as well as to promote Rad53 and Ddc2 phosphorylation when DNA is damaged in G1 or during S phase.
The DNA damage checkpoint defects of MEC1 GAL1-mec1kd1 cells appeared to be less severe than those of mec1Δ cells, suggesting that the presence of physiological amounts of wild-type Mec1 may contribute to partial activation of the DNA damage response in galactose-induced MEC1 GAL1-mec1kd1 cells. Indeed, cells overproducing Mec1kd1 in a mec1Δ sml1Δ background were more sensitive to HU, MMS, and UV light than otherwise isogenic cells overproducing Mec1kd variants in a MEC1 background and were indistinguishable from mec1Δ sml1Δ cells (Fig. 3A). Moreover, when GAL1-mec1kd1 mec1Δ sml1Δ cells were released from G1 arrest after UV irradiation or in the presence of MMS under galactose-induced conditions, they progressed through S phase faster than similarly treated MEC1 GAL1-mec1kd1 cells (Fig. 3B), and DNA damage-induced Rad53 phosphorylation was completely abolished (data not shown). Therefore, the residual activation of the DNA damage response in MEC1 GAL1-mec1kd1 cells was dependent on the presence of wild-type Mec1.
High levels of Mec1kd1 in MEC1 cells do not affect the delay of nuclear division caused by UV irradiation in G2.
The above-described dominant effects of Mec1kd overproduction were limited to the checkpoints controlling S phase entry and progression. In fact, MEC1 GAL1-mec1kd1 galactose-induced cell cultures released from a nocodazole-induced G2 arrest after UV irradiation underwent a delay in nuclear division comparable to that observed in wild-type and MEC1 GAL1-MEC1 cells under the same conditions (Fig. 3C, top), although they showed a premature disappearance of DNA damage-induced Rad53 phosphorylated forms (Fig. 3C, bottom). The activation of the G2/M checkpoint in MEC1 cells overproducing Mec1kd1 was likely due to the presence of wild-type Mec1, since similarly treated GAL1-mec1kd1 mec1Δ sml1Δ cells divided nuclei much faster than did MEC1 GAL1-mec1kd1 cells (Fig. 3C, top) and phosphorylation of Rad53 and Ddc2 was completely abolished (Fig. 3C, bottom), as can be observed in mec1Δ cells under the same conditions (Fig. 1). Therefore, physiological levels of Mec1 in cells overproducing Mec1kd1 might be sufficient to activate Rad53 and/or other proteins specifically required to prevent nuclear division when DNA is damaged in G2. Since Rad53 phosphorylation after UV irradiation in G2 was reduced prematurely in MEC1 cells with high levels of Mec1kd1 (Fig. 3C, bottom), the G2 DNA damage-induced cell cycle arrest of these cells might at least partially depend on proteins acting independently of Rad53. One possible candidate was the Chk1 kinase, which is phosphorylated in a Mec1-dependent manner and is specifically required to prevent anaphase entry in cdc13 mutants at restrictive temperatures, independently of Rad53 (44). Indeed, when galactose-induced cell cultures were released from a nocodazole-induced G2 arrest after UV irradiation, MEC1 GAL1-mec1kd1 chk1Δ cells divided nuclei faster than MEC1 GAL1-mec1kd1 cells, although Rad53 phosphorylation was not further affected (Fig. 3C) and deletion of CHK1 per se was not sufficient to impair either the DNA damage-induced Rad53 phosphorylation or the checkpoint-mediated delay in nuclear division after DNA damage in G2 (Fig. 3C). While the overall amount of Chk1 phosphorylation after UV irradiation was reduced in MEC1 GAL1-mec1kd cells compared to wild-type and GAL1-MEC1 cells during the above-described synchronization experiments (Fig. 3C, bottom), the Chk1 phosphorylated forms persisted until the end of the experiment and were dependent on wild-type Mec1, since they were completely absent in GAL1-mec1kd1 mec1Δ sml1Δ cells under the same conditions (Fig. 3C, bottom).
New mec1 mutants impaired in subsets of DNA integrity checkpoint pathways.
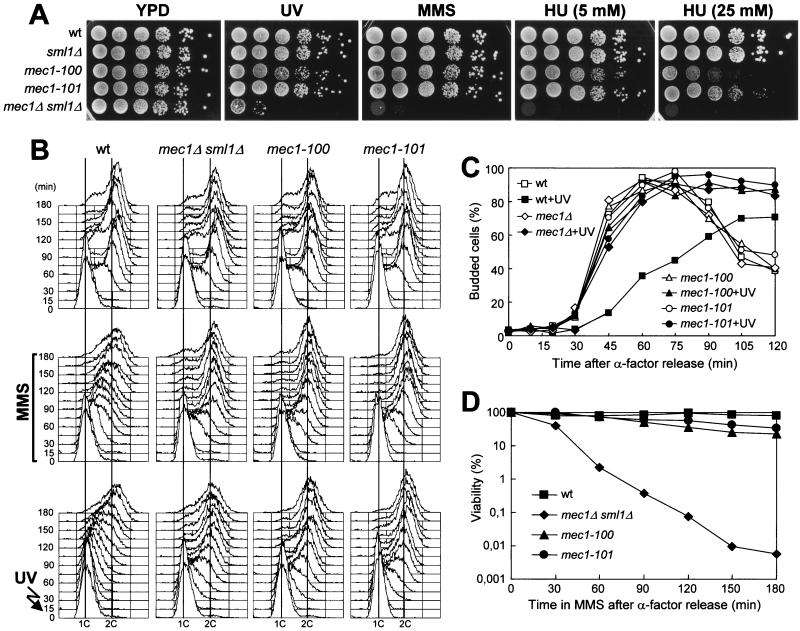
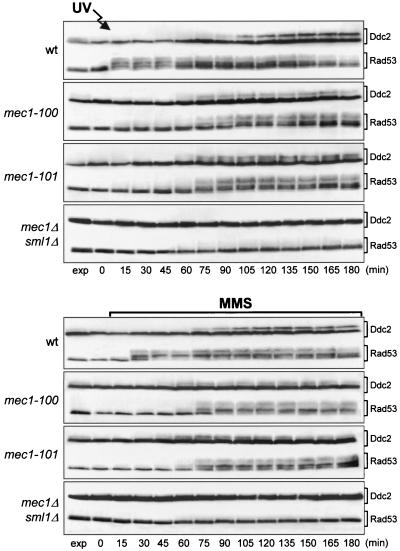
We have shown that an excess of inactive Mec1kd molecules causes dominant DNA damage-resistant DNA replication, but it is not sufficient to abolish the G2/M DNA damage checkpoint, suggesting that specific impairment of Mec1 functions may affect the checkpoint response differently, depending on the cell cycle stages at which DNA alterations occur. If this were the case, it should be possible to isolate mec1 mutants that are defective in slowing down of DNA synthesis but are still able to delay nuclear division in response to DNA damage. Random mutagenesis of the MEC1 gene and screening for mutants that displayed different patterns of sensitivity to genotoxic agents (see Materials and Methods), allowed us to isolate the mec1-100 and mec1-101 mutant alleles. As shown in Fig. 4A, the mec1-100 and mec1-101 mutants did not show hypersensitivity to MMS and UV radiation, while they exhibited a limited sensitivity to HU that was much lower than that of mec1Δ cells. Both mec1-100 and mec1-101 mutants turned out to be completely defective in both the G1/S and intra-S DNA damage checkpoints. In fact, when mec1-100 and mec1-101 G1-arrested cells were UV irradiated and then released into the cell cycle, they entered S phase (Fig. 4B, bottom) and budded (Fig. 4C) much faster than the wild type, similarly to mec1Δ cells, although their cell survival was very similar to that of wild-type cells under the same conditions (75% for mec1-100, 80% for mec1-101, 87% for wild-type, and 3% for mec1Δ sml1Δ cells). Furthermore, when α-factor-synchronized mec1-100 and mec1-101 cells were released from the G1 arrest in the presence of MMS, they doubled their DNA content within 45 min, like mec1Δ cells, whereas MMS-treated wild-type cell cultures progressed through S phase very slowly, completing DNA replication only after 150 min (Fig. 4B, middle). On the contrary, the viability of MMS-treated mec1-100 and mec1-101 mutant cells was much more similar to that of wild-type cells than to that of mec1Δ cells under the same conditions (Fig. 4D). Thus, the new mec1 mutants were completely unable to delay bud emergence and S phase entry and progression when DNA was damaged in G1 or during S phase, although their checkpoint defects did not result in loss of viability. These defective checkpoint responses correlated with defects in the extent and/or timing of Ddc2 and Rad53 phosphorylation. In fact, Rad53 phosphorylation was detectable immediately after UV light and MMS treatment of wild-type cells, while it became detectable in mec1-100 and mec1-101 mutants only at 75 min (Fig. 5), when cells reached late S or G2 phase (Fig. 4B). Furthermore, UV light- and MMS-treated mec1-100 and mec1-101 cells showed reduced amounts of Ddc2 phosphorylated forms that appeared earlier than in wild-type cells (Fig. 5), reflecting the findings that Ddc2 phosphorylation after DNA damage in G1 or during S phase becomes detectable in the wild type only when cells reach the G2 phase (Fig. 4 and 5) (36) and that both mutants reached the G2 phase earlier than the wild type (Fig. 4B).
FIG. 4.
G1/S and intra-S DNA damage checkpoints in mec1-100 and mec1-101 mutants. The strains used were wild-type (wt) YLL683.8/4A, mec1Δ sml1Δ DMP3048/5B, mec1-100 DMP3343/6C, and mec1-101 DMP3344/4A. (A) Serial dilution of exponentially growing (in YEPD) cell cultures were spotted on YEPD plates with or without MMS (0.005%) or HU. One YEPD plate was UV irradiated (40 J/m2) (UV). The data presented in panels B, C, and D all come from the same experiment. (B to D) α-Factor-synchronized cells were released from α-factor at time zero in YEPD (top) or in YEPD containing 0.02% MMS (middle) or were UV irradiated (40 J/m2) prior to the release in YEPD (bottom). (B) Samples of untreated and UV light- and MMS-treated cell cultures were collected at the indicated times after release from α-factor and analyzed by fluorescence-activated cell sorter. (C) Untreated or UV-irradiated (+UV) cell cultures were scored at the indicated times for the percentage of budded cells. (D) Aliquots were removed from the MMS-treated cultures at timed intervals to score for CFU on YEPD plates at 25°C.
FIG. 5.
Rad53 and Ddc2 phosphorylation in mec1-100 and mec1-101 mutants after DNA damage in G1 or during S phase. The strains used were wild-type (wt) YLL683.8/4A, mec1Δ sml1Δ DMP3048/5B, mec1-100 DMP3343/6C, and mec1-101 DMP3344/4A. The data all come from the experiment described in the legend to Fig. 4B, C, and D. Protein extracts from the UV light-treated (top panel) and the MMS-treated (bottom panel) cell cultures were analyzed by Western blot assay using anti-Rad53 and anti-HA (Ddc2) antibodies. exp, exponentially growing cells.
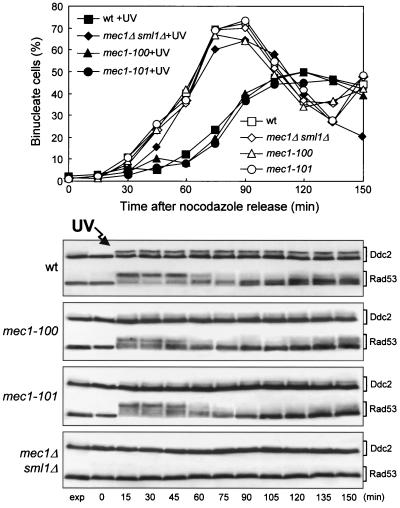
As shown in Fig. 6, the mec1-100 and mec1-101 mutants were not defective in the G2/M DNA damage checkpoint. In fact, when mec1-100 and mec1-101 cell cultures were released from G2 arrest after UV irradiation, they showed a delay in nuclear division comparable to that of wild-type cells under the same conditions, as well as immediate induction of Ddc2 and Rad53 phosphorylation (Fig. 6). Thus, the mec1-100 and mec1-101 mutants are specifically altered only in subsets of the DNA damage checkpoint pathways responding to DNA damage in G1 or during S phase.
FIG. 6.
G2/M DNA damage checkpoint in mec1-100 and mec1-101 mutants. Cultures of wild-type (wt) YLL683.8/4A, mec1Δ sml1Δ DMP3048/5B, mec1-100 DMP3343/6C, and mec1-101 DMP3344/4A cells were arrested with nocodazole and UV irradiated (50 J/m2) prior to release in YEPD. Kinetics of nuclear division were determined as described in the legend to Fig. 1D in untreated and UV light-treated (+UV) cells and are shown at the top. At the bottom is a Western blot analysis of protein extracts from samples of the UV light-treated cell cultures withdrawn at the indicated times. Rad53 and Ddc2 were detected using, respectively, anti-Rad53 and anti-HA (Ddc2) antibodies. exp, exponentially growing cells.
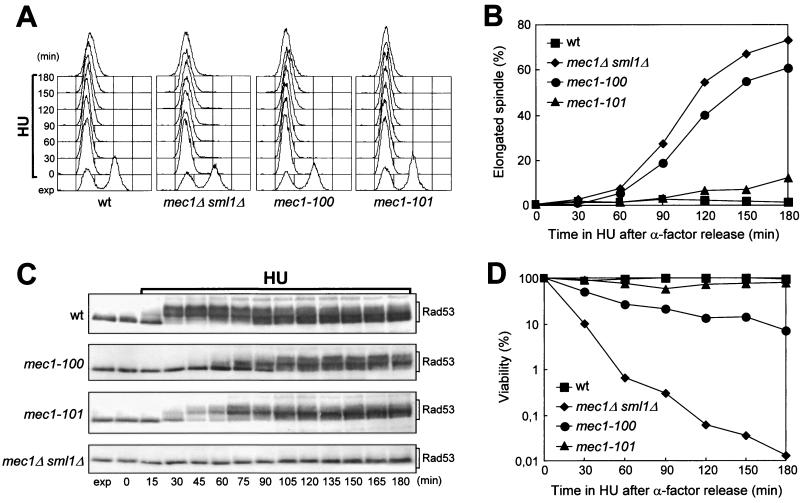
We also asked whether the mec1-100 and mec1-101 mutants were impaired in slowing down of the elongation of mitotic spindles in response to incomplete DNA replication. When cells were released from G1 arrest in the presence of 200 mM HU, all cell cultures arrested DNA synthesis (Fig. 7A) while spindle elongation took place in the mec1-100 mutant, along with aberrant chromosome segregation, with a kinetics only slightly slower than that observed in mec1Δ sml1Δ cells (Fig. 7B). Conversely, the HU-treated mec1-101 cells behaved similarly to HU-treated wild-type cells that, as expected, did not elongate the spindles throughout the experiment (Fig. 7B). Moreover, the viability of wild-type and mec1-101 cells was substantially unaffected by HU, while the mec1-100 mutant lost viability during HU treatment, although to an extent much less than that of mec1Δ sml1Δ cells (Fig. 7D). The differences in the abilities of the two mutants to delay S/M transition in response to incomplete DNA replication correlated with differences in HU-induced Rad53 phosphorylation that was consistently delayed in the mec1-100 mutant compared to wild-type cells, while it was only weakly defective in the mec1-101 mutant (Fig. 7C).
FIG. 7.
Response to HU treatment of mec1-100 and mec1-101 mutants. Cultures of wild-type (wt) YLL683.8/4A, mec1Δ sml1Δ DMP3048/5B, mec1-100 DMP3343/6C, and mec1-101 DMP3344/4A cells were arrested in G1 with α-factor and then released at time zero in YEPD containing 200 mM HU. Cell samples were collected at the indicated times after the release from α-factor. The data presented in panels A to D all come from the same experiment. (A) DNA content was analyzed by fluorescence-activated cell sorter. (B) Cells were stained with antitubulin antibodies to score for the percentage of cells with elongated spindles by indirect immunofluorescence. (C) Protein extracts were analyzed by Western blot assay using anti-Rad53 antibodies. exp, exponentially growing cells. (D) Appropriate dilutions were plated on YEPD at 25°C to score for CFU
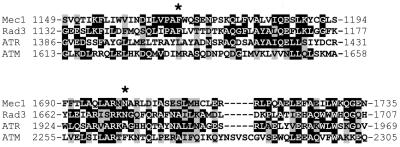
Determination and comparison of the whole wild-type and mutant MEC1 coding sequences revealed that the mec1-100 allele carried two base pair substitutions, resulting in the amino acid changes F1179S and N1700S, while the mec1-101 allele carried three base pair substitutions, leading to the amino acid changes V225G, S552P, and L781S. The contribution of the single amino acid changes to the mutant phenotypes remains to be established. Alignment of the amino acid sequence of Mec1 with those of S. pombe Rad3 and human ATM and ATR indicated that none of the three residues changed by the mec1-101 mutations is conserved among these proteins. On the contrary, both amino acid changes in the mec1-100 gene product involve residues that are identical in Mec1 and Rad3 and belong to regions that also appear to be quite well conserved in human ATM and ATR (Fig. 8).
FIG. 8.
Amino acid residues changed by the mec1-100 mutations. The two Mec1 regions containing the mec1-100-encoded amino acid changes are shown after alignment of the whole Mec1 amino acid sequence with the S. pombe Rad3 and human ATM and ATR amino acid sequences using the ClustalW program. Identical amino acid residues are shaded in black, and similar residues are highlighted in gray. Residues that are changed in the mec1-100 gene product are marked by asterisks.
DISCUSSION
Although Mec1 is necessary to promote all the known phosphorylation events in the DNA damage checkpoint cascade, little is known about the functions and regulation of Mec1 kinase activity in the activation of the DNA damage response in the different cell cycle phases. We previously showed that a kinase activity dependent on an intact Mec1 kinase domain coimmunoprecipitates with Mec1 (36), and recent work by Mallory and Petes (31) further supports this observation. We now demonstrate that the Mec1 conserved kinase domain is essential for all of the functions of Mec1. In fact, two different Mec1kd variants, in which single amino acid residues in the conserved lipid kinase domain are changed to give kinase-deficient proteins, cause the same effects as the lack of Mec1, resulting not only in hypersensitivity to genotoxic agents and SML1-dependent cell lethality but also in a defective DNA damage checkpoint response in all cell cycle phases. Altogether, these data indicate that Mec1 might exert all of its known functions through phosphorylation events. Indeed, we have demonstrated that the kinase activity coprecipitating with Mec1 is able to phosphorylate Ddc2 in vitro (36; this work), thus indicating that Ddc2 may be a target of Mec1 activity in vivo. Both Mec1kd variants completely lose the ability to induce Ddc2 phosphorylation in vitro, although they both still physically interact with Ddc2, indicating that this kinase activity is dependent on the integrity of the Mec1 conserved kinase domain.
Dominant defects caused by mec1kd overexpression.
Similarly to what was observed when kinase-defective Rad3 and ATR mutant proteins were overproduced (4, 9), high levels of Mec1 kinase-deficient variants in wild-type cells cause dominant-negative effects. In fact, MEC1 cells overproducing Mec1kd are hypersensitive to DNA-damaging agents and are defective in the slowing down of S phase entry and progression, as well as in Rad53 and Ddc2 phosphorylation, after DNA damage in G1 or during S phase. Therefore, an excess of Mec1kd proteins in the presence of physiological amounts of wild-type Mec1 may be able to compete for the signals generated by DNA damage, leading to a reduction in the amount of active downstream proteins capable of productively transducing the signal to cell cycle effectors (40). Indeed, we have shown that Mec1 molecules can self-associate and that Mec1-Mec1kd complexes can be formed independently of DNA damage. If Mec1 in vivo functions were dependent on Mec1-Mec1 interaction, Mec1kd overproduction might lead to competition in complex formation, thus reducing the amount of functional Mec1 complexes able to activate downstream effectors like, for example, Rad53. The Mec1kd variants might also titrate Mec1-interacting factors, like Ddc2, into nonfunctional complexes. We found that neither Ddc2 nor Rad53 overproduction can, by itself, suppress the hypersensitivity to DNA-damaging agents or the intra-S checkpoint defect of MEC1 cells overproducing Mec1kd (V. Paciotti et al., unpublished data). Thus, the dominant effect of Mec1kd overproduction likely involves multiple competition events, or if there is a primary target, it does not seem to be either Ddc2 or Rad53. A search for high-copy-number suppressors of the MEC1 GAL1-mec1kd checkpoint defects may help to elucidate this point.
The dominant effects of mec1kd overexpression are limited to the DNA damage checkpoints controlling S phase entry and progression. In fact, MEC1 cells overproducing Mec1kd are still able to activate the G2/M checkpoint, and this depends on the wild-type Mec1 protein, since mec1Δ sml1Δ cells overproducing Mec1kd are completely defective in this response. It is interesting that UV light damage in G2 of MEC1 cells overproducing Mec1kd allows immediate Rad53 phosphorylation, but the Rad53 phosphorylated forms decrease faster in these cells than in wild-type and MEC1 GAL1-MEC1 cells under the same conditions. If this implies that Mec1 activity is continuously needed to both activate and maintain the Rad53-dependent checkpoint response, other factors might be required to maintain the G2 arrest in UV light-treated MEC1 GAL1-mec1kd cells. Our data show that the Chk1 protein kinase is necessary for this checkpoint response. In fact, while inactivation of CHK1 per se is not sufficient to abrogate the UV light-induced G2/M checkpoint in MEC1 cells, it leads to premature nuclear division after UV irradiation of MEC1 cells overexpressing Mec1kd1. Therefore a reduction of Mec1 activity in these cells might uncover the role of Chk1 in this subset of the UV light-induced checkpoint pathways. Conversely, CHK1 deletion is able to promote nuclear division in cdc13 mutants also when Mec1 and Rad53 are fully functional (44), suggesting that the amount and quality of DNA damage might determine the ability of cells to activate the CHK1-dependent checkpoint response.
Mec1 functions required for checkpoint response to DNA damage in different cell cycle phases.
Similar to MEC1 strains overexpressing the mec1kd alleles, both the new mec1-100 and mec1-101 mutants are still able to activate the UV light-induced G2/M checkpoint, while they are completely defective in delaying S phase entry and progression when DNA is damaged in G1 or during S phase. All of the amino acid substitutions in the Mec1-100 and Mec1-101 proteins are located well outside the conserved Mec1 lipid kinase domain. We cannot exclude the possibility that these amino acid changes can directly affect Mec1 kinase activity, and further work is required to address this point. However, we favor the hypothesis that mec1-100 and mec1-101 mutants are defective in interactions with proteins or structures specifically involved in the G1 and intra-S DNA damage responses and that high levels of Mec1kd variants may titrate molecules important for the above responses into nonfunctional complexes. Moreover, the cell cycle phases at which DNA alterations occur and/or are processed might influence the chance to activate the checkpoint pathways. According to this hypothesis, UV irradiation of G2-arrested cells results in immediate Mec1-dependent Ddc2 phosphorylation independently of cell cycle progression, while UV irradiation in G1 is able to induce Ddc2 phosphorylation only when cells are completing S phase or are in G2 (36), although Ddc2 is strictly required to arrest cell cycle progression in response to DNA damage in all of the cell cycle phases (36). It is therefore tempting to speculate that either DNA damage in G2 does not require processing in order to be recognized by Mec1 (36) or specific factors are involved in the processing of DNA damage in G2, allowing easier recognition. If this were the case, it might explain the reduced sensitivity to Mec1 alterations of checkpoint response in G2 compared to G1 or S phase.
It is also worth noting that the mec1-101 mutant that is completely defective in slowing down of S phase progression in response to DNA damage during DNA synthesis is proficient in arresting spindle elongation in the presence of HU, further supporting the hypothesis that the cellular response to DNA replication blocks or to DNA damage during DNA replication involves different Mec1 functions.
DNA damage checkpoint defects and sensitivity to genotoxic agents.
The characterization of the mec1-100 and mec1-101 mutants also provides some new data addressing the important question of whether checkpoint impairment renders cells hypersensitive to genotoxic agents. In fact, while these mutants are indistinguishable from mec1Δ cells with respect to the ability to replicate a damaged DNA template, they do not show hypersensitivity to UV light and MMS, suggesting that a DNA damage checkpoint defect per se does not necessarily cause hypersensitivity to DNA-damaging agents. It is possible that a functional G2 checkpoint can compensate for defects in slowing down of DNA replication in the presence of DNA insults (39). In fact, if a failure to control the replication of damaged template DNA results in genetic instability, activation of the G2/M checkpoint would offer the opportunity to repair strand breaks before sister chromatids are no longer available for repair. If so, the high survival of the mec1-100 and mec1-101 mutants after UV light and MMS treatment may correlate with an increased dependence on DNA damage-induced G2 arrest. However, although MEC1 cells overexpressing mec1kd are still able to activate the G2/M checkpoint, they show hypersensitivity to DNA-damaging agents, indicating that delay of nuclear division is not always sufficient to compensate for the effects of Mec1 impairment on cell survival after DNA damage. Taken together, our results indicate that when DNA replication occurs in the presence of DNA insults, cell lethality of checkpoint mutants might not be purely a cell cycle transition phenomenon, but other processes might be involved. For example, the inability of these mutants to properly carry out chromosomal replication might result in cell lethality, as also suggested by Desany et al. (13). In this view, the high sensitivity to HU treatment of mec1Δ sml1Δ cells might be due to failure of replication structures to recover from the effects of nucleotide depletion, instead of depending on the faster cell cycle progression. In fact, the viability of mec1-100 cells during HU treatment is much higher than that of mec1Δ sml1Δ cells, although the kinetics of spindle elongation in the presence of HU is almost as fast in mec1-100 cells as in mec1Δ sml1Δ cells. Moreover, the sensitivity to genotoxic agents of cells impaired in Mec1 activity may result from the inability to mediate the efficient repair of DNA lesions, leading to a model in which checkpoints are integrated into a larger DNA damage response pathway. Consistent with this hypothesis, recent data have implicated Mec1 in recombination mechanisms. In fact, MEC1 is absolutely required to induce sister chromatid exchange in nucleotide excision repair-deficient cells (H. Neecke et al., unpublished data) and to promote normal meiotic recombination (18, 51). Moreover, phosphorylation of the Rad55, RPA, and Srs2 proteins, all of which are involved in DNA repair and recombination (1, 37), was found to be Mec1 dependent (3, 5, 25). Finally, the implication of the Mec1 human homologue ATM in the control of recombinational repair has been hinted at by various links recently found (8, 11, 46), by the recombinational abnormalities observed in ataxia telangiectasia patients (32), and by the fact that ATM is required for phosphorylation of NBS1 (15, 26, 61, 63) and BRCA1 (11, 24), both of which regulate the repair of double-strand breaks (DSBs) and the proper cellular response to DNA damage. The findings that ATM is required for homologous recombination-mediated repair of DSBs and is a member of the recombinational repair epistasis group (33) clearly indicate that ATM has a role not only in preventing cells from propagating damaged DNA but also in the processing and repair of DSBs. Our data suggest that this is also the case for Mec1, further strengthening the notion of functional conservation between the human and yeast proteins.
ACKNOWLEDGMENTS
We are grateful to Veronica Baldo for computer analysis. We thank J. Diffley and C. Santocanale for the antibody against Rad53, K. Nasmyth for plasmids 3746 and 3748, S. Piatti for helpful suggestions and critical reading of the manuscript, and all of the members of our laboratory for useful discussions and criticisms.
This work was supported by Telethon-Italy (grant E.1247 to M.P.L.), by a grant from the Associazione Italiana Ricerca sul Cancro and Cofinanziamento 1999 MURST-Università di Milano-Bicocca to G.L., and by CNR Target Project on Biotechnology grant CT.97.01180.PF49(F). V.P. was supported by a fellowship from the Fondazione Italiana per la Ricerca sul Cancro.
V.P. and M.C. contributed equally to this work.
REFERENCES
- 1.Aboussekhra A, Chanet R, Zgaga Z, Cassier-Chauvat C, Heude M, Fabre F. RADH, a gene of Saccharomyces cerevisiae encoding a putative DNA helicase involved in DNA repair. Characteristics of radH mutants and sequence of the gene. Nucleic Acids Res. 1989;17:7211–7219. doi: 10.1093/nar/17.18.7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Allen J B, Zhou Z, Siede W, Friedberg E C, Elledge S J. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 1994;8:2416–2428. doi: 10.1101/gad.8.20.2401. [DOI] [PubMed] [Google Scholar]
- 3.Bashkirov V I, King J S, Bashkirova E V, Schmuckli-Maurer J, Heyer W-D. DNA repair protein Rad55 is a terminal substrate of the DNA damage checkpoints. Mol Cell Biol. 2000;20:4393–4404. doi: 10.1128/mcb.20.12.4393-4404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bentley N J, Holtzman D A, Flaggs G, Keegan K S, Demaggio A, Ford J C, Hoekstra M, Carr A M. The Schizosaccharomyces pombe rad3 checkpoint gene. EMBO J. 1996;15:6641–6651. [PMC free article] [PubMed] [Google Scholar]
- 5.Brush G S, Morrow D M, Hieter P, Kelly T J. The ATM homologue MEC1 is required for phosphorylation of replication protein A in yeast. Proc Natl Acad Sci USA. 1996;93:15075–15080. doi: 10.1073/pnas.93.26.15075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carr A M. Control of cell cycle arrest by the Mec1sc/Rad3sp DNA structure checkpoint pathway. Curr Opin Genet Dev. 1997;7:93–98. doi: 10.1016/s0959-437x(97)80115-3. [DOI] [PubMed] [Google Scholar]
- 7.Chabes A, Domkin V, Thelander L. Yeast Sml1, a protein inhibitor of ribonucleotide reductase. J Biol Chem. 1999;274:36679–36683. doi: 10.1074/jbc.274.51.36679. [DOI] [PubMed] [Google Scholar]
- 8.Chen G, et al. Radiation-induced assembly of Rad51 and Rad52 recombination complex requires ATM and c-Abl. J Biol Chem. 1999;274:12748–12752. doi: 10.1074/jbc.274.18.12748. [DOI] [PubMed] [Google Scholar]
- 9.Cliby W A, Roberts C J, Cimprich K A, Stringer C M, Lamb J R, Schreiber S L, Friend S H. Overexpression of a kinase-inactive ATR protein causes sensitivity to DNA-damaging agents and defects in cell cycle checkpoints. EMBO J. 1998;17:159–169. doi: 10.1093/emboj/17.1.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen-Fix O, Koshland D. The anaphase inhibitor of Saccharomyces cerevisiae Pds1p is a target of the DNA damage checkpoint pathway. Proc Natl Acad Sci USA. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cortez D, Wang Y, Quin J, Elledge S J. Requirement of ATM-dependent phosphorylation of Brca1 in the DNA damage response to double-strand breaks. Science. 1999;286:1162–1166. doi: 10.1126/science.286.5442.1162. [DOI] [PubMed] [Google Scholar]
- 12.Critchlow S E, Jackson S P. DNA end-joining: from yeast to man. Trends Biochem Sci. 1998;23:394–398. doi: 10.1016/s0968-0004(98)01284-5. [DOI] [PubMed] [Google Scholar]
- 13.Desany B A, Alcasabas A A, Bachant J B, Elledge S J. Recovery from DNA replicational stress is the essential function of the S-phase checkpoint pathway. Genes Dev. 1998;12:2956–2970. doi: 10.1101/gad.12.18.2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Emili A. MEC1-dependent phosphorylation of Rad9p in response to DNA damage. Mol Cell. 1998;2:183–189. doi: 10.1016/s1097-2765(00)80128-8. [DOI] [PubMed] [Google Scholar]
- 15.Gatei M, Young D, Cerosaletti K M, Desia-Mehta A, Spring K S, Kozlov S, et al. ATM-dependent phosphorylation of nibrin in response to radiation exposure. Nat Genet. 2000;25:115–119. doi: 10.1038/75508. [DOI] [PubMed] [Google Scholar]
- 16.Gietz R D, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six base-pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 17.Greenwell P W, Kronmal S L, Porter S E, Gassenhuber J, Obermaier B, Petes T D. TEL1, a gene involved in controlling telomere length in S. cerevisiae, is homologous to the human ataxia telangiectasia gene. Cell. 1995;82:823–829. doi: 10.1016/0092-8674(95)90479-4. [DOI] [PubMed] [Google Scholar]
- 18.Grushcow J M, Holzen T M, Park K J, Weinert T, Lichten M, Bishop D K. Saccharomyces cerevisiae checkpoint genes MEC1, RAD17 and RAD24 are required for normal meiotic recombination partner choice. Genetics. 1999;153:607–620. doi: 10.1093/genetics/153.2.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Halazonetis T D, Shiloh Y. Many faces of ATM: Eighth International Workshop on Ataxia-Telangiectasia. Biochim Biophys Acta. 1999;1424:R45–R55. doi: 10.1016/s0304-419x(99)00023-2. [DOI] [PubMed] [Google Scholar]
- 20.Hari K L, Santerre A, Sekelsky J J, McKim K S, Boyd J B, Hawley R S. The mei-41 gene of D. melanogaster is a structural and functional homolog of the human ataxia telangiectasia gene. Cell. 1995;82:815–821. doi: 10.1016/0092-8674(95)90478-6. [DOI] [PubMed] [Google Scholar]
- 21.Keith C T, Schreiber S L. PIK-related kinases: DNA repair, recombination, and cell cycle checkpoints. Science. 1995;270:50–51. doi: 10.1126/science.270.5233.50. [DOI] [PubMed] [Google Scholar]
- 22.Knop M, Siegers K, Pereira G, Zachariae W, Winsor B, Nasmyth K, Schiebel E. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast. 1999;15:963–972. doi: 10.1002/(SICI)1097-0061(199907)15:10B<963::AID-YEA399>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 23.Kondo T, Matsumoto K, Sugimoto K. Role of a complex containing Rad17, Mec3, and Ddc1 in the yeast DNA damage checkpoint pathway. Mol Cell Biol. 1999;19:1136–1143. doi: 10.1128/mcb.19.2.1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li S, Ting N S Y, Zheng L, Chen P-L, Ziv Y, Shiloh Y, Lee E Y-H P, Lee W-H. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- 25.Liberi G, Chiolo I, Pellicioli A, Lopes M, Plevani P, Muzi-Falconi M, Foiani M. Srs2 DNA helicase is involved in checkpoint response and its regulation requires a functional Mec1-dependent pathway and Cdk1 activity. EMBO J. 2000;19:5027–5038. doi: 10.1093/emboj/19.18.5027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lim D-S, Kim S-T, Xu B, Maser R S, Lin J, Petrini J H J, Kastan M B. ATM phosphorylates p95/nbs1 in an S-phase checkpoint pathway. Nature. 2000;404:613–617. doi: 10.1038/35007091. [DOI] [PubMed] [Google Scholar]
- 27.Longhese M P, Foiani M, Muzi Falconi M, Lucchini G, Plevani P. DNA damage checkpoint in budding yeast. EMBO J. 1998;17:5525–5528. doi: 10.1093/emboj/17.19.5525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longhese M P, Paciotti V, Neecke H, Lucchini G. Checkpoint proteins influence telomeric silencing and length maintenance in budding yeast. Genetics. 2000;155:1577–1591. doi: 10.1093/genetics/155.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Longhese M P, Paciotti V, Fraschini R, Plevani P, Lucchini G. The novel DNA damage checkpoint protein Ddc1p is phosphorylated periodically during the cell cycle and in response to DNA damage in budding yeast. EMBO J. 1997;16:5216–5226. doi: 10.1093/emboj/16.17.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lowndes N F, Murguia J R. Sensing and responding to DNA damage. Curr Opin Genet Dev. 2000;10:17–25. doi: 10.1016/s0959-437x(99)00050-7. [DOI] [PubMed] [Google Scholar]
- 31.Mallory J C, Petes T D. Protein kinase activity of Tel1p and Mec1p, two Saccharomyces cerevisiae proteins related to the human ATM protein kinase. Proc Natl Acad Sci USA. 2000;97:13749–13754. doi: 10.1073/pnas.250475697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meyn M S. High spontaneous intrachromosomal recombination rates in ataxia-telangiectasia. Science. 1993;260:1327–1330. doi: 10.1126/science.8493577. [DOI] [PubMed] [Google Scholar]
- 33.Morrison C, Sonoda E, Takao N, Shinohara A, Yamamoto K, Takeda S. The controlling role of ATM in homologous recombinational repair of DNA damage. EMBO J. 2000;19:463–471. doi: 10.1093/emboj/19.3.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morrow D M, Tagle D A, Shiloh Y, Collins F S, Hieter P. TEL1, an S. cerevisiae homolog of the human gene mutated in ataxia telangiectasia, is functionally related to the yeast checkpoint gene MEC1. Cell. 1995;82:831–840. doi: 10.1016/0092-8674(95)90480-8. [DOI] [PubMed] [Google Scholar]
- 35.Paciotti V, Lucchini G, Plevani P, Longhese M P. Mec1p is essential for phosphorylation of the yeast DNA damage checkpoint protein Ddc1p, which physically interacts with Mec3p. EMBO J. 1998;17:101–111. doi: 10.1093/emboj/17.14.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paciotti V, Clerici M, Lucchini G, Longhese M P. The checkpoint protein Ddc2, functionally related to S. pombe Rad26, interacts with Mec1 and is regulated by Mec1-dependent phosphorylation in budding yeast. Genes Dev. 2000;14:2046–2059. [PMC free article] [PubMed] [Google Scholar]
- 37.Paques F, Haber J E. Two pathways for removal of nonhomologous DNA ends during double-strand break repair in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paulovich A G, Hartwell L H. A checkpoint regulates the rate of progression through S phase in S. cerevisiae in response to DNA damage. Cell. 1995;82:841–847. doi: 10.1016/0092-8674(95)90481-6. [DOI] [PubMed] [Google Scholar]
- 39.Paulovich A G, Toczyski D P, Hartwell L H. When checkpoints fail. Cell. 1997;88:315–321. doi: 10.1016/s0092-8674(00)81870-x. [DOI] [PubMed] [Google Scholar]
- 40.Perlmutter R M, Alberola I J. The use of dominant-negative mutations to elucidate signal transduction pathways in lymphocytes. Curr Opin Immunol. 1996;8:285–290. doi: 10.1016/s0952-7915(96)80069-0. [DOI] [PubMed] [Google Scholar]
- 41.Rose M D, Winston F, Hieter P. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- 42.Rouse J, Jackson S P. LCD1: an essential gene involved in checkpoint control and regulation of the MEC1 signaling pathway in Saccharomyces cerevisiae. EMBO J. 2000;19:5801–5812. doi: 10.1093/emboj/19.21.5801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sanchez Y, Desany B A, Jones W J, Liu Q, Wang B, Elledge S J. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science. 1996;271:357–360. doi: 10.1126/science.271.5247.357. [DOI] [PubMed] [Google Scholar]
- 44.Sanchez Y, Bachant J, Wang H, Hu F H, Liu D, Tezlaff M, Elledge S J. Control of the DNA damage checkpoint by Chk1 and Rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- 45.Savitsky K, et al. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;286:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- 46.Shafman T, et al. Interaction between ATM protein and c-Abl in response to DNA damage. Nature. 1997;387:520–523. doi: 10.1038/387520a0. [DOI] [PubMed] [Google Scholar]
- 47.Siede W, Friedberg A S, Friedberg E C. RAD9-dependent G1 arrest defines a second checkpoint for damaged DNA in the cell cycle of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1993;90:7985–7989. doi: 10.1073/pnas.90.17.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun Z, Fay D S, Marini F, Foiani M, Stern D F. Spk1/Rad53 is regulated by Mec1-dependent protein phosphorylation in DNA replication and damage checkpoint pathways. Genes Dev. 1996;10:395–406. doi: 10.1101/gad.10.4.395. [DOI] [PubMed] [Google Scholar]
- 49.Sun Z, Hsiao J, Fay D S, Stern D F. Rad53 FHA domain associated with phosphorylated Rad9 in the DNA damage checkpoint. Science. 1998;281:272–274. doi: 10.1126/science.281.5374.272. [DOI] [PubMed] [Google Scholar]
- 50.Thelen M P, Venclovas C, Fidelis K. A sliding clamp model for the Rad1 family of cell cycle checkpoint proteins. Cell. 1999;96:769–770. doi: 10.1016/s0092-8674(00)80587-5. [DOI] [PubMed] [Google Scholar]
- 51.Thompson D A, Stahl F W. Genetic control of recombination partner preference in yeast meiosis: isolation and characterization of mutants elevated for meiotic unequal sister-chromatid recombination. Genetics. 1999;153:621–641. doi: 10.1093/genetics/153.2.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Umezu K, Sugawara N, Chen C, Haber J E, Kolodner R D. Genetic analysis of yeast RPA1 reveals its multiple functions in DNA metabolism. Genetics. 1998;148:989–1005. doi: 10.1093/genetics/148.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vialard J E, Gilbert C S, Green C M, Lowndes N F. The budding yeast Rad9 checkpoint protein is subjected to Mec1/Tel1-dependent hyperphosphorylation and interacts with Rad53 after DNA damage. EMBO J. 1998;17:5679–5688. doi: 10.1093/emboj/17.19.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruption in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- 55.Waga S, Stillman B. The DNA replication fork in eukaryotic cells. Annu Rev Biochem. 1998;67:721–751. doi: 10.1146/annurev.biochem.67.1.721. [DOI] [PubMed] [Google Scholar]
- 56.Weinert T. Yeast checkpoint controls and relevance to cancer. Cancer Surv. 1997;29:109–132. [PubMed] [Google Scholar]
- 57.Weinert T. DNA damage and checkpoint pathways: molecular anatomy and interactions with repair. Cell. 1998;94:555–558. doi: 10.1016/s0092-8674(00)81597-4. [DOI] [PubMed] [Google Scholar]
- 58.Weinert T A, Hartwell L H. The RAD9 gene controls the cell cycle response to DNA damage in Saccharomyces cerevisiae. Science. 1988;241:317–322. doi: 10.1126/science.3291120. [DOI] [PubMed] [Google Scholar]
- 59.Weinert T A, Hartwell L H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics. 1993;134:63–80. doi: 10.1093/genetics/134.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinert T A, Kiser G L, Hartwell L H. Mitotic checkpoint genes in budding yeast and the dependence of mitosis on DNA replication and repair. Genes Dev. 1994;8:652–665. doi: 10.1101/gad.8.6.652. [DOI] [PubMed] [Google Scholar]
- 61.Wu X, Ranganathan V, Weisman D S, Heine W F, Ciccone D N, O'Neill T B, Crick K E, Pierce K A, Lane W S, Rathbun G, Livingston D M, Weaver D T. ATM phosphorylation of Nijmegen breakage syndrome protein is required in a DNA damage response. Nature. 2000;405:477–482. doi: 10.1038/35013089. [DOI] [PubMed] [Google Scholar]
- 62.Zakian V A. ATM-related genes: what do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- 63.Zhao S, Weng Y C, Yuan S-S F, Lin Y-T, Hsu H-C, Lin S-C J, et al. Functional link between ataxia-telangiectasia and Nijmegen breakage syndrome gene products. Nature. 2000;405:473–477. doi: 10.1038/35013083. [DOI] [PubMed] [Google Scholar]
- 64.Zhao X, Muller E G D, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]