Abstract
CDC45 is required for the initiation of DNA replication in Saccharomyces cerevisiae and functions as a DNA polymerase α loading factor in Xenopus, but its role in mammalian DNA replication is unknown. To investigate the genetic and physiological functions of CDC45, we used a gene targeting strategy to generate mice lacking a functional CDC45 gene. Homozygous mutant mice lacking a functional CDC45 gene underwent uterine implantation and induced uterine decidualization but did not develop substantially thereafter. Detailed analysis of CDC45 null embryos cultured in vitro revealed impaired proliferation of the inner cell mass. These findings make CDC45 the only putative replication factor experimentally proven to be essential for mammalian development. The CDC45 gene localizes to human chromosome 22q11.2 in the DiGeorge syndrome critical region (DGCR). Almost 90% of individuals with congenital cardiac and craniofacial defects have a monoallelic deletion in the DGCR that includes CDC45. We report here that heterozygous mutant mice develop into adulthood without any apparent abnormalities, so that it is unlikely that hemizygosity of CDC45 alone is responsible for the cardiac and craniofacial defects in the congenital syndromes.
CDC45 is an essential gene for the initiation of DNA replication in Saccharomyces cerevisiae (7, 17, 30). Yeast CDC45 interacts genetically with the origin recognition complex (ORC) and physically with the minichromosome maintenance (MCM) family members and the yeast replication origins (1, 2, 4, 5, 7, 30, 31). Recent studies indicate that CDC45 functions in late G1 and associates with the prereplicative complex after activation of S-phase-promoting cdk (31). Yeast CDC45 plays an essential role during elongation (24) and is involved in recruiting replication protein A and DNA polymerase α to the DNA (26). It has also been shown that Xenopus CDC45 plays a pivotal role in the loading of DNA polymerase α onto chromatin, a process dependent on S-phase-specific cdk activity (16).
A human homolog of yeast CDC45 has been isolated, and the encoded protein has been shown to associate with human ORC2 protein in cells in culture (22). The human CDC45 protein associates with chromatin in G1 but progressively loses attachment to a nuclear tether as S phase proceeds, such that none of the protein is detected in the nuclear fraction in G2/M. Based on this circumstantial biochemical evidence and the strong conservation of the replication apparatus from yeasts to mammals, the mammalian CDC45 protein is expected to be essential for DNA replication, although this has not been formally proven.
The human CDC45 gene is located in the chromosome region 22q11.2, where interstitial deletions are associated with many developmental disorders, including DiGeorge syndrome (DGS), velocardiofacial syndrome, and conotruncal anomaly face syndrome (3). DGS can be characterized by growth retardation, psychiatric disorders, hypoproliferation of thymus and parathyroid, and outflow tract congenital heart defects, the latter being the most common cause of death (12). A unifying hypothesis is that all these defects might be explained by selective failure of development of the third and fourth branchial arches and pharyngeal pouches that give rise to the conotruncal region of the heart, the thymus, and the parathyroid glands (10, 25). Interestingly, CDC45 is widely expressed in the developing mouse embryo, including the brain, pharyngeal arches, thymus, and kidney, all of which are affected in DGS (23). We know little, however, about the effect of a decrease in CDC45 gene dosage (as seen in hemizygosity) on the proliferation and differentiation of specific lineages, such as neural crest cells.
One patient with DGS has a heterozygous de novo 20-kb microdeletion that removes only exons 1 to 5 of CDC45 and exons 1 to 3 of UFD1 (encoding ubiquitin fusion degradation protein 1) (29). More recently, chromosome-engineering technology was used to model DGS, at least for cardiac anomalies, in mice (13). Hemizygous loss of a 1.2-Mb segment of mouse chromosome 16, syntenic to the human DGS critical region (DGCR), deletes 14 genes, including CDC45 and UFD1, and produces mice with cardiovascular abnormalities of the same type as those seen in DGS. When the hearts of the heterozygous mouse embryos were examined just before birth, a quarter had defects in derivatives of the fourth-branchial-arch artery. Interestingly, mice with a 150- or 550-kb deletion of the proximal region of the DGCR that overlaps with the deleted segment described by Lindsay et al. (13) but does not remove CDC45 show none of the structural abnormalities of DGS (9, 19). Therefore, the genes responsible for the structural defects in DGS map to the distal region of DGCR, where CDC45 maps. Surprisingly, although loss of the entire DGCR produced cardiac anomalies, hemizygosity of just UFD1 did not produce any phenotypic effect (13). Together with the microdeletion reported by Yamagishi et al. (29), where hemizygosity of only UFD1 and CDC45 produced cardiac defects, these results raised the tantalizing possibility that haploinsufficiency of CDC45 alone could contribute to the cardiovascular defects of DGS.
To test whether CDC45 is essential for mammalian DNA replication and to test whether haploinsufficiency of CDC45 alone produces any of the congenital anomalies seen in DGS, we generated mutant mice carrying a disrupted allele by gene targeting in embryonic stem (ES) cells. Analysis of the heterozygous mice shows no anomalies. Further analysis of the progeny from mating of CDC45+/− mice indicates that CDC45 is essential for early embryonic development, consistent with a role of the protein in mammalian DNA replication.
MATERIALS AND METHODS
Construction of the targeting vector.
A mouse bacterial artificial chromosome clone (GenBank accession no. AC005816) derived from mouse strain 129/SvJ and containing the entire mouse CDC45 gene was obtained from Bruce Roe (University of Oklahoma). To delete exons 1 and 2 of CDC45, the 2.4-kb (nucleotides [nt] 30513 to 32970) and 5.3-kb (nt 33431 to 38760) regions were cloned into the targeting vector pKO (Stratagene) with phosphoglycerate kinase I promoter driving the neomycin gene (PGK-neo) for positive selection and polyomavirus enhancer/herpes simplex virus thymidine kinase (MC1) promoter driving the thymidine kinase gene for negative selection (Fig. 1A). Both regions of homology were amplified by PCR, and the identities were confirmed by sequencing.
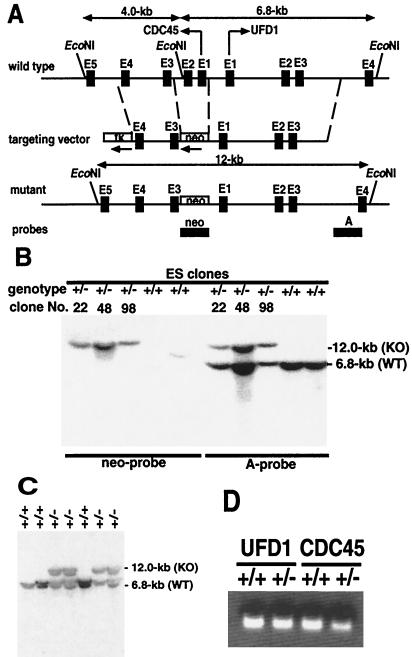
FIG. 1.
Generation of CDC45 mutant mice by gene targeting. (A) The gene structures of the CDC45 and UFD1 loci, the targeting vector, and the targeted allele are shown schematically. Diagnostic restriction fragments indicating the presence of a wild-type (6.8-kb) or targeted (12.0-kb) allele are diagrammed above the wild-type and targeted loci, respectively. The solid boxes and straight lines represent exon and intron sequences, respectively. EcoNI sites are indicated. A 460-bp region of the CDC45 gene, encompassing exons 1 and 2, was replaced with a neo gene. The thymidine kinase (tk) gene was inserted in the 3′ end of the targeting construct. Arrows at 5′ ends or beneath the respective genes indicate the relative orientations of CDC45, UFD1, neo, and tk gene transcripts. (B) Southern blot analysis of the targeted CDC45 allele. Genomic DNA isolated from G418-resistant ES cells, digested with EcoNI, was hybridized with the neo probe (neo gene coding sequence) or probe A (sequence derived from the outside of the targeting vector). The neo probe detects a 12-kb (KO) fragment from a targeted locus. The non-12-kb fragments detected in CDC45+/+ ES cells indicate random integration of the neo gene elsewhere in the genome. Probe A detects a 12-kb (KO) fragment from the targeted CDC45 locus and a 6.8-kb (WT) band from the wild-type CDC45 locus. (C) Genomic DNA isolated from progeny (derived from ES clone 98) of CDC45+/− matings was genotyped with probe A as described for panel B. No homozygous mutant mice were recovered. (D) Levels of CDC45 and UFD1 transcripts were measured by RT-PCR analysis of RNA from 13.5-dpc embryos. The CDC45 transcript level in CDC45+/− mice is roughly half that in CDC45+/+ mice, whereas the UFD1 transcript level is the same in both types of mice.
Gene targeting in ES cells.
ES cells (line J1) were maintained in culture on γ-irradiated primary neo-resistant mouse embryo fibroblast (MEF) feeder cells. The culture medium was supplemented with leukemia inhibitory factor (1,500 U/ml; Gibco-BRL). The NotI-linearized targeting vector (25 μg) was electroporated into 107 ES cells. Targeted clones were selected for 7 to 10 days in the presence of G418 (200 μg/ml) and 1-(2-deoxy-2-fluoro-d-arabinofuranosyl)-5-ioduracil (200 nM) and expanded onto 24-well plates. To screen by Southern blot analysis, candidate clones were grown to confluence on 12-well gelatin-coated plates in the absence of a MEF feeder layer. Individual targeted clones, confirmed by Southern blot analysis, were further expanded for microinjection.
Generation of chimeras.
Chimeric animals were generated by injection of three independent targeted ES cells (clones 22, 48, and 98) into 3.5-day-postcoitus (dpc) C57BL/6 blastocysts by standard procedures (6). After microinjection, the blastocysts were reimplanted into pseudopregnant females. Six- to eight-week-old male progeny with a high percent chimerism (>50%, based on agouti coat color) were bred with C57BL/6 females to produce heterozygous mice capable of transmitting the targeted allele through the germ line. Heterozygous mice were mated with each other to generate homozygous mice.
Genotyping of ES cells, embryos, and animals.
ES cells, embryos, and 2-week-old mice were genotyped by Southern blot or PCR analysis. Genomic DNA was isolated from embryos and tail clipping by digestion overnight at 55°C in lysis buffer (10 mM Tris-HCl [pH 7.5], 100 mM NaCl, 1 mM EDTA, 0.5% sodium dodecyl sulfate [SDS], 100 μg of proteinase K/ml) followed by isopropanol precipitation. Cultured blastocyst outgrowth was frozen in 10 μl of water, heated to 94°C, treated with 1 μl of 2-mg/ml proteinase K for 1 h at 55°C, heated again to 94°C to inactivate the proteinase K, then subjected to amplification using 5 μl of the solubilized cells per reaction. For Southern blot analysis, genomic DNA was digested with EcoNI and resolved on 0.8% agarose gels. The gels were blotted to Nytran membranes (Schleicher & Schuell) by capillary transfer in 10× standard saline citrate (SSC; 1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate). The membranes were prehybridized for 2 h at 42°C in a buffer consisting of 6× SSC, 10× Denhardt's solution (0.02% polyvinylpyrrolidone, 0.02% Ficoll, 0.02% bovine serum albumin), and 50 μg of denatured salmon sperm DNA/ml. Hybridization was overnight at 65°C in a buffer consisting of 6× SSC, 10% dextran sulfate, 1% SDS, and 50 μg of denatured salmon sperm DNA/ml and containing the 32P-labeled DNA probe. The probe, a 1.1-kb DNA fragment (nt 38844 to 39960) that maps upstream of the CDC45 gene (probe A in Fig. 1A), was radiolabeled by random priming using [32P]dCTP. The blots were washed twice for 15 min at 65°C in 3× SSC–0.2% SDS and once for 15 min at 65°C in 0.2× SSC–0.2% SDS before autoradiography.
PCR genotyping of genomic DNA from cultured embryos, yolk sacs, and tail clippings was performed using nested primer sets to detect wild-type and mutant alleles (see Fig. 3A). 5′-ACACTACGTACTTGTCTCACTGTTTGCACT-3′ and 5′-TTGGGTTCCTCACCTCCTGC-3′ were used as forward CDC45 primers (45F1 and 45F2); 5′-TATTGGCTGCTGGCGTGGACCAATCAGAAG-3′ and 5′-TCCGTGTCTCAGCGCCAGTT-3′ were used as reverse CDC45 primers (45R1 and 45R2). 5′-GCCAATATGGGATCGGCCAT-3′ and 5′-GAACAAGATGGATTGCACGC-3′ were forward neo primers (Neo F1 and Neo F2), and 5′-AGCGGCGATACCGTAAAGCA-3′ and 5′-ACAACGTCGAGCACAGCTGC-3′ were reverse neo primers (Neo R1 and Neo R2). The neo primer set (Neo F2 and Neo R2) amplifies a 245-bp fragment from the correctly targeted locus and no fragment from the wild-type CDC45 locus. The wild-type internal primer set (45F2 and 45R2) amplifies a 430-bp DNA fragment from the wild-type CDC45 allele and no product from a Neo-targeted CDC45 allele. PCR products were resolved on 1.0% agarose gels and visualized by UV fluorescence. PCR conditions were 95°C for 3 min, followed by 30 (or 35) cycles of 94°C for 30 s, 58°C for 30 s, and then 72°C for 30 s. After the final cycle, we used a 5-min extension period at 72°C.
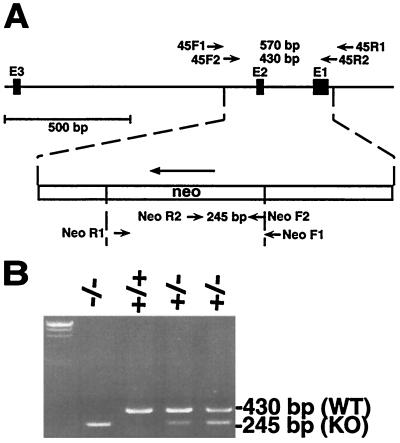
FIG. 3.
PCR analysis of DNAs from embryos resulting from CDC45+/− crosses. (A) PCR strategy for amplification of wild-type and disrupted CDC45 alleles. DNA samples derived from cultured embryos were subjected to PCR amplification using a combination of two sets of nested primers (see Materials and Methods for sequences). Nested PCR of the wild-type CDC45 allele by forward and reverse 45 primers produces a 430-bp DNA fragment, whereas amplification of the disrupted CDC45 allele by forward and reverse Neo primers produces a 245-bp DNA fragment. The arrow indicates the direction of transcription of PGK-neo plus poly(A). (B) Genotype analysis of 5- to 8-day-cultured blastocysts by PCR. Wild-type and targeted CDC45 alleles are amplified as 430-bp (WT) and 245-bp (KO) fragments. λ/HindIII fragments are on the left.
Histological analysis.
For histological examination, samples were fixed in phosphate-buffered saline-buffered 4% formaldehyde, processed, and embedded in paraffin. Sagittal sections (10 μm thick) were cut and stained with hematoxylin and eosin. Embryos (5.5 to 6.5 dpc) obtained from timed matings were dissected from uteri. Serial sections through the whole decidua were mounted onto polyionic slides. Two investigators scored the presence of embryos (or deciduae) in the uterus.
MEF generation and culture.
CDC45+/+ and CDC45+/− embryos (13.5 dpc) were dissected and trypsinized, and MEF were transferred to Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum as described elsewhere (21). γ-Irradiation of the MEF was performed using a 137Cs γ-irradiator (2.12 Gy/s; Gammacell 1000; Atomic Energy of Canada Limited Industrial Products, Mississiagua, Ontario, Canada). Subsequently, 104 cells were plated in triplicate onto six-well plates with 2 ml of medium/well. Viable cells were quantitated by an 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay (Roche) at 5 days after irradiation.
RT-PCR.
Total RNA (10 μg) was prepared from 13.5-dpc embryos and reverse transcribed using oligo(dT) and Superscript II reverse transcriptase (RT; Gibco-BRL). cDNA obtained from 0.5 μg of total RNA was amplified by PCR using the following primer pairs: 5′-CTGAAGCAAGTCAAGCAG-3′ (exon 12) and 5′-CACAAGAGCGTCCAGGAA-3′ (exon 18) as CDC45 primers and 5′-GTGGCGACCTACTCTAAG-3′ (exon 5) and 5′-CCAGGACAAGCTTGATAG-3′ (exon 12) as UFD1 primers. PCR was at 95°C for 3 min, followed by 30 cycles of 94°C for 30 s, 55°C for 30 s, and then 72°C for 30 s. After the final cycle, we used a 5-min extension period at 72°C. The CDC45 and UFD1 primer sets amplify DNA fragments of 723 and 708 bp, respectively.
In vitro culture of preimplantation embryos.
CDC45+/− males and females were intercrossed, and 3.5-dpc embryos were flushed out from the uteri of the plugged females (21). Blastocysts were individually cultured in eight-well chamber slides (Nalge Nunc) in ES medium without leukemia inhibitory factor, in 5% CO2 at 37°C. Photographs of the cultured embryos were taken at 5 or 8 days. After 5 to 8 days in culture, the genotype was determined by nested PCR as described above.
RESULTS
Gene targeting strategy.
Our strategy was to delete exon 1 (including the initiation codon) and exon 2 from the CDC45 gene (Fig. 1A). An internal methionine first occurs in exon 8; thus, at least the first 7 out of 19 exons will be nontranslatable in the mutant. Even if a protein is produced from an internal methionine it will have 199 N-terminal amino acids deleted out of a total of 566 amino acids. These N-terminal 199 residues include acidic patches and nuclear localization signals that are conserved in yeast and Xenopus, so that it is unlikely that such a deletion-containing form of CDC45 would be functional (7, 16). Approximately 170 G418-resistant ES cell clones were screened by Southern blot analysis; six of these were positive for the targeted allele as determined by the presence of a diagnostic 12.0-kb EcoNI fragment (Fig. 1B). Three of the targeted ES cell clones (clones 22, 48, and 98) were microinjected into blastocysts to generate chimeric strains of mice that transmitted the disrupted allele through the germ line. We generated two independent lines from chimeras derived from clones 22 and 98 from a C57BL/6 background. Heterozygous mice generated from each of the independently derived chimeras were identified by Southern blotting and/or PCR analysis of genomic DNA isolated from tail samples of the offspring (Fig. 1C; see also Fig. 3A).
Phenotype of CDC45+/− mice.
No gross anatomical abnormalities have been detected in CDC45 heterozygous mice up to 6 months of age. The mice grow to normal size, are fertile, and do not display any obvious behavioral deficiencies. About 25% of E18.5 embryos hemizygous for a deletion encompassing 22 known genes including CDC45 exhibited various velocardiofacial syndrome- and DGS-associated cardiovascular abnormalities (13). We therefore recovered and examined 18.5-dpc embryos from a cross of CDC45+/− mice. Thirteen CDC45+/− embryos as well as seven wild-type embryos were examined and found to have no abnormal development of the major vessels of the heart (data not shown). RT-PCR analysis of 13.5-dpc embryos revealed that the expression level of CDC45 transcripts in CDC45+/− mice was roughly half that in wild-type mice, whereas levels of UFD1 were the same in CDC45+/− and wild-type mice (Fig. 1D).
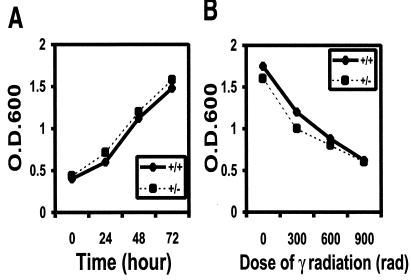
To investigate whether hemizygosity of CDC45 has any phenotypic effect at the cellular level, we prepared MEF from CDC45+/+ and CDC45+/− embryos and measured proliferation rates. Hemizygosity of CDC45 did not have any effect on cell doubling time (Fig. 2A). Since CDC45 is expected to play a role in DNA repair (because of its requirement in the loading of DNA polymerase α), CDC45+/+ and CDC45+/− MEF were γ-irradiated, and the cells surviving radiation was measured. No difference was seen in the radiation sensitivity of CDC45+/+ and CDC45+/− MEF (Fig. 2B).
FIG. 2.
In vitro culture of MEF derived from CDC45+/+ and CDC45+/− embryos. Cell proliferation (A) and the effect of 137Cs γ-irradiation on the number of viable cells (B) were measured by MTT assay, and assays were performed in triplicate. Cell proliferation rates in low serum concentration (0.5%) also showed no difference between CDC45+/+ and CDC45+/− MEF (data not shown). O.D.600, optical density at 600 nm.
Embryonic lethality of the CDC45−/− mutation.
Of 61 live births resulting from crosses between CDC45+/− mice, 39 were heterozygous (CDC45+/−) and 22 were wild-type (CDC45+/+), indicating that disruption of CDC45 results in embryonic lethality (Table 1). The observed ratio of heterozygote to wild-type births is consistent with a predicted ratio of 2:1 and suggests that the CDC45−/− mutation is embryonically lethal.
TABLE 1.
Progeny from CDC45 heterozygotesa
| Age (dpc) | Genotype
|
No. (%) resorbed | Total | ||
|---|---|---|---|---|---|
| +/+ | +/− | −/− | |||
| Neonate | 22 | 39 | 0 | NA | 61 |
| 18.5 | 7 | 13 | 0 | 8 (29) | 28 |
| 13.5 | 6 | 10 | 0 | 9 (36) | 25 |
| 7.5 | 7 | 19 | 0 | 8 (24) | 34 |
| 6.5 | ND | ND | ND | 7 (27) | 26b |
| 5.5 | ND | ND | ND | 6 (25) | 24b |
| 3.5 | 6 | 9 | 3 | NA | 22c |
NA, not available ND, not determined.
All decidua were histologically checked.
Four failed to hatch and produced no PCR products upon genotyping.
To define the stage at which the CDC45−/− animals die, postimplantation embryos (7.5 to 18.5 dpc) from heterozygous matings were surgically explanted from the uterine tissue of pregnant females and genotyped by PCR analysis. At 7.5 dpc, a total of 34 implanted embryos, including 7 resorptions (27%), were found (Table 1). Attempts to genotype the tissue from resorbed embryos were unsuccessful. However, analysis of 62 intact embryos (42 CDC45+/− and 20 CDC45+/+) between 7.5 and 18.5 dpc confirmed the absence of the CDC45−/− genotype (Table 1), suggesting that the CDC45−/− animals die before 7.5 dpc.
To gain more insight into the mutant phenotype, we analyzed fixed paraffin-embedded deciduae from matings of CDC45+/− mice. At 5.5 and 6.5 dpc, we noted two distinct phenotypes. Of a total of 50 deciduae, 74% contained healthy animals (Table 1), characterized by a well-developed embryonic structure (data not shown). However, 24% of the decidua contained no embryos, a number consistent with the possibility that they were derived from the missing CDC45−/− embryos (Table 1). In contrast, out of 19 5.5- and 6.5-dpc deciduas examined from CDC45+/+ × CDC45+/− matings, only 2 were abnormal. These results suggest that CDC45−/− embryos induced the decidual reaction upon implantation but the embryo proper was unable to develop beyond the point of implantation. Even embryos that die or are degraded shortly after attachment stimulate decidualization, a process whereby stromal cells expand, elaborate extracellular matrix, and form tight junctions (28). Since a normal mouse blastocyst attaches to the uterus between 4.5 and 5.0 dpc (20), it is likely that the CDC45−/− mice die between 4.5 and 5.5 dpc.
In vitro growth of blastocysts.
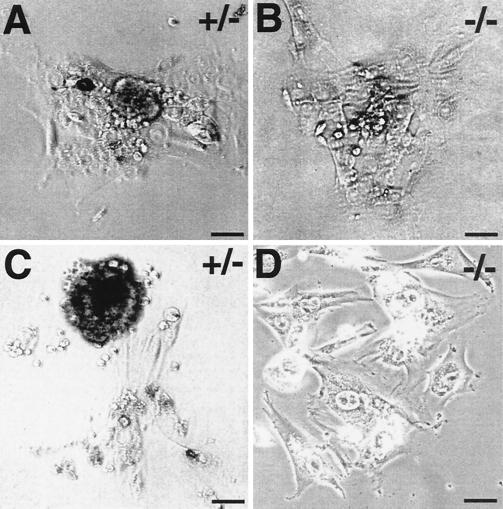
We assessed in vitro growth of blastocysts from heterozygote intercrosses. When 22 blastocysts were individually cultured for 5 to 8 days, 4 did not emerge from the zona pellucida (genotype unknown). Three embryos with trophoblast outgrowths but no proliferation of the inner cell mass (see Fig. 4B and D) were subsequently identified as CDC45−/− (Fig. 3B). The remaining 15 embryos showed trophoblast outgrowths and proliferation of their inner cell mass (Fig. 4A and C); these embryos were found by nested PCR analysis to be either wild type or heterozygous at the CDC45 locus (Fig. 3B). These results strongly suggest that the growth of CDC45−/− embryos is affected at the peri-implantation stage.
FIG. 4.
Morphology of cultured blastocyst outgrowths from crosses of CDC45+/− mice. Cultured heterozygous (A and C) and homozygous (B and D) embryos were photographed using phase microscopy after 5 (A and B) and 8 (C and D) days in culture. The tightly packed inner cell mass is found in heterozygous but not in homozygous embryos. Bars, 100 μm.
DISCUSSION
To the best of our knowledge, this is the first study that genetically demonstrates the requirement of a putative DNA replication factor for normal mammalian development. Because of the difficulty of performing genetic analyses in mammals, most of our knowledge of DNA replication factors in mammals is dependent on biochemical studies in vitro or genetic studies in yeasts and Drosophila. Important roles of CDC45 in the initiation of DNA replication in lower organisms such as yeast (7, 17, 30) as well as Xenopus (16) have been reported. One reasonable explanation for the early lethality of CDC45−/− embryos is that DNA replication initiation is impaired after maternal CDC45 in the embryo is lost. Consistent with this possibility, CDC45 is expressed in multiple tissues from 9.5 to 14.5 days of mouse embryonic development (23). Although evolutionary conservation of core life processes like DNA replication leads us to expect that mammalian homologs of replication factors like CDC45 are essential for viability, this is the first experimental demonstration of this fact. The results reported here eliminate the possibility that mammals have evolved pathways that bypass the requirement of CDC45 in DNA replication.
Maternal stockpiles of replication factors in eggs have been shown to support many rounds of cell division without zygotic transcription. For example, null mutations in ORC2 or ORC3 in Drosophila allow development of the fly to late larval stages, implying that the maternal supply of these proteins can sustain at least 20 cell divisions without any contribution from the embryo (11, 18). Xenopus eggs have a vast excess of ORC proteins, such that more than 99% of the supply has to be immunodepleted before one sees a defect in replication initiation in vitro (27). In light of these observations, it is somewhat surprising that the CDC45−/− mice exhibit such an early embryonic lethality. By 5.5 dpc, there are about 128 to 256 cells in the embryo, so our results suggest that maternal supplies of CDC45 in mammalian eggs can support only about seven to eight cell divisions before lethality. Either the maternal stockpile of CDC45 and other replication factors is low in mouse eggs (relative to that in Xenopus and Drosophila eggs), or CDC45 is unstable in early embryonic cell divisions or the early lethality is an indirect effect of CDC45 deficiency. Since CDC45 is involved during the elongation stage of DNA replication, a decrease in the amount of the protein might lead to defects in replication fork movement well before factors become limiting for replication initiation. Stalled replication forks may lead to DNA damage, so that the early embryonic lethality could be a result of cell cycle arrest due to the activation of cell cycle checkpoints. Examining the phenotypes of CDC45−/− mice that are concurrently defective in p53 and other checkpoint-activating genes will be used to test this hypothesis.
Mice that lack one CDC45 allele develop normally, with no obvious defects in cardiovascular development. We conclude that the loss of one copy of CDC45 alone is not responsible for the congenital defects seen in patients with DGS or in mouse models of the disease. While this paper was under review, three other papers reported that hemizygosity of TBX1, a gene that maps to the distal region of DGCR, was sufficient to produce conotruncal defects in cardiac development in a certain fraction of the mice (8, 14, 15). Homozygous deletion of TBX1 produced more profound defects in cardiac development than seen in the hemizygous mice. Furthermore, TBX1−/− but not TBX1+/− mice had additional features of DGS, such as hypoplasia of the thymus and parathyroid and cleft palate, that could be detected in late-stage embryos (8). The fact that a homozygous deletion of TBX1 is necessary in mice to phenocopy all the defects of DGS (which is seen in humans with hemizygosity of DGCR) might suggest that additional genes in the DGCR contribute to the full spectrum of congenital defects seen in humans.
A hemizygous deletion that removes both TBX1 and CDC45 still does not produce the full spectrum of defects in mice, ruling out CDC45 as a gene that synergizes with TBX1 to produce the congenital defects in mice. We cannot, however, rule out species-specific differences that might allow hemizygosity of the CDC45 and TBX1 to cooperate to produce the full spectrum of defects of DGS in humans but not in mice. It will be necessary to identify DGS patients with mutations only in the TBX1 gene and without any defect in the structure or expression of any other gene in the DGCR (including CDC45) before concluding that TBX1 is solely responsible for all the congenital defects in humans.
The CDC45+/− mice appear to have half the amount of CDC45 mRNA that is found in the CDC45+/+ controls, indicating that both copies of the gene are transcribed in normal development. Although expression of half the normal amount of CDC45 mRNA did not impair cell proliferation or development, this demonstrable decrease in CDC45 transcript levels might leave CDC45+/− cells vulnerable to a similar decrease in levels of another transcript involved in cell proliferation or development. Such a synergistic impairment of cell proliferation in the hemizygous state may be tissue specific if the second gene (like TBX1) has tissue-specific functions. Alternatively, hemizygosity of CDC45 might impair proliferation in specific tissues, as seen with hypomorphic mutations in another gene involved in general cell proliferation, Drosophila ORC3 (18). Future experiments will be directed at generating organisms which are doubly heterozygous for CDC45, a gene involved in cell proliferation, and other genes implicated in normal development or cell proliferation. The results will test whether subtle lesions in different genes, which by themselves have subtle phenotypic effects, interact to produce more profound changes in development or cell proliferation.
ACKNOWLEDGMENTS
We thank the members of our laboratory for helpful discussions, Peter Sicinski for technical advice, and Lina Du for technical assistance.
This work was supported by grant CA60499 from the NIH.
REFERENCES
- 1.Aparicio O M, Weinstein D M, Bell S P. Components and dynamics of DNA replication complexes in S. cerevisiae: redistribution of MCM proteins and Cdc45p during S phase. Cell. 1997;91:59–69. doi: 10.1016/s0092-8674(01)80009-x. [DOI] [PubMed] [Google Scholar]
- 2.Dalton S, Hopwood B. Characterization of Cdc47p-minichromosome maintenance complexes in Saccharomyces cerevisiae: identification of Cdc45p as a subunit. Mol Cell Biol. 1997;17:5867–5875. doi: 10.1128/mcb.17.10.5867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Emanuel B S, Budarf M L, Scambler P J. The genetic basis of conotruncal cardiac defects: the chromosome 22q11.2 deletion. In: Harvey R, Rosenthal N, editors. Heart development. San Diego, Calif: Academic Press; 1999. pp. 463–478. [Google Scholar]
- 4.Hardy C F. Identification of Cdc45p, an essential factor required for DNA replication. Gene. 1997;187:239–246. doi: 10.1016/s0378-1119(96)00761-5. [DOI] [PubMed] [Google Scholar]
- 5.Hennessy K M, Lee A, Chen E, Botstein D. A group of interacting yeast DNA replication genes. Genes Dev. 1991;5:958–969. doi: 10.1101/gad.5.6.958. [DOI] [PubMed] [Google Scholar]
- 6.Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- 7.Hopwood B, Dalton S. Cdc45p assembles into a complex with Cdc46p/Mcm5p, is required for minichromosome maintenance, and is essential for chromosomal DNA replication. Proc Natl Acad Sci USA. 1996;93:12309–12314. doi: 10.1073/pnas.93.22.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jerome L A, Papaioannou V E. DiGeorge syndrome phenotype in mice mutant for the T-box gene, Tbx1. Nat Genet. 2001;27:286–291. doi: 10.1038/85845. [DOI] [PubMed] [Google Scholar]
- 9.Kimber W L, Hsieh P, Hirotsune S, Yuva-Paylor L, Sutherland H F, Chen A, Ruiz-Lozano P, Hoogstraten-Miller S L, Chien K R, Paylor R, Scambler P J, Wynshaw-Boris A. Deletion of 150 kb in the minimal DiGeorge/velocardiofacial syndrome critical region in mouse. Hum Mol Genet. 1999;8:2229–2237. doi: 10.1093/hmg/8.12.2229. [DOI] [PubMed] [Google Scholar]
- 10.Lammer E J, Opitz J M. The DiGeorge anomaly as a developmental field defect. Am J Med Genet. 1986;2(Suppl.):113–127. doi: 10.1002/ajmg.1320250615. [DOI] [PubMed] [Google Scholar]
- 11.Landis G, Kelley R, Spradling A C, Tower J. The k43 gene, required for chorion gene amplification and diploid cell chromosome replication, encodes the Drosophila homolog of yeast origin recognition complex subunit 2. Proc Natl Acad Sci USA. 1997;94:3888–3892. doi: 10.1073/pnas.94.8.3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lindsay E A, Baldini A. Congenital heart defects and 22q11 deletions: which genes count? Mol Med Today. 1998;4:350–357. doi: 10.1016/s1357-4310(98)01302-1. [DOI] [PubMed] [Google Scholar]
- 13.Lindsay E A, Botta A, Jurecic V, Carattini-Rivera S, Cheah Y C, Rosenblatt H M, Bradley A, Baldini A. Congenital heart disease in mice deficient for the DiGeorge syndrome region. Nature. 1999;401:379–383. doi: 10.1038/43900. [DOI] [PubMed] [Google Scholar]
- 14.Lindsay E A, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland H F, Scambler P J, Bradley A, Baldini A. Tbx1 haploinsufficiency in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 15.Merscher S, Funke B, Epstein J A, Heyer J, Puech A, Lu M-M, Xavier R J, Demay M B, Russell R G, Factor S, Tokooya K, St. Jore B, Lopez M, Pandita R K, Lia M, Carrion D, Xu H, Schorle H, Kobler J B, Scambler P, Wynshaw-Boris A, Skoultchi A I, Morrow B E, Kucherlapati R. TBX1 is responsible for cardiovascular defects in velo-cardio-facial/DiGeorge syndrome. Cell. 2001;104:619–629. doi: 10.1016/s0092-8674(01)00247-1. [DOI] [PubMed] [Google Scholar]
- 16.Mimura S, Takisawa H. Xenopus Cdc45-dependent loading of DNA polymerase alpha onto chromatin under the control of S-phase Cdk. EMBO J. 1998;17:5699–5707. doi: 10.1093/emboj/17.19.5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Owens J C, Detweiler C S, Li J J. CDC45 is required in conjunction with CDC7/DBF4 to trigger the initiation of DNA replication. Proc Natl Acad Sci USA. 1997;94:12521–12526. doi: 10.1073/pnas.94.23.12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pinto S, Quintana D G, Smith P, Mihalek R M, Hou Z H, Boynton S, Jones C J, Hendricks M, Velinzon K, Wohlschlegel J A, Austin R J, Lane W S, Tully T, Dutta A. latheo encodes a subunit of the origin recognition complex and disrupts neuronal proliferation and adult olfactory memory when mutant. Neuron. 1999;23:45–54. doi: 10.1016/s0896-6273(00)80752-7. [DOI] [PubMed] [Google Scholar]
- 19.Puech A, Saint-Jore B, Merscher S, Russell R G, Cherif D, Sirotkin H, Xu H, Factor S, Kucherlapati R, Skoultchi A I. Normal cardiovascular development in mice deficient for 16 genes in 550 kb of the velocardiofacial/DiGeorge syndrome region. Proc Natl Acad Sci USA. 2000;97:10090–10095. doi: 10.1073/pnas.97.18.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rinkenberger J L, Cross J C, Werb Z. Molecular genetics of implantation in the mouse. Dev Genet. 1997;21:6–20. doi: 10.1002/(SICI)1520-6408(1997)21:1<6::AID-DVG2>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- 21.Robertson E J. Embryo-derived stem cell lines. In: Robertson E J, editor. Teratocarcinomas and embryonic stem cells. Oxford, United Kingdom: Oxford University Press; 1987. pp. 71–112. [Google Scholar]
- 22.Saha P, Thome K C, Yamaguchi R, Hou Z, Weremowicz S, Dutta A. The human homolog of Saccharomyces cerevisiae CDC45. J Biol Chem. 1998;273:18205–18209. doi: 10.1074/jbc.273.29.18205. [DOI] [PubMed] [Google Scholar]
- 23.Shaikh T H, Gottlieb S, Sellinger B, Chen F, Roe B A, Oakey R J, Emanuel B S, Budarf M L. Characterization of CDC45L: a gene in the 22q11.2 deletion region expressed during murine and human development. Mamm Genome. 1999;10:322–326. doi: 10.1007/s003359900996. [DOI] [PubMed] [Google Scholar]
- 24.Tercero J A, Labib K, Diffley J F. DNA synthesis at individual replication forks requires the essential initiation factor Cdc45p. EMBO J. 2000;19:2082–2093. doi: 10.1093/emboj/19.9.2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Van Mierop L H, Kutsche L M. Cardiovascular anomalies in DiGeorge syndrome and importance of neural crest as a possible pathogenetic factor. Am J Cardiol. 1986;58:133–137. doi: 10.1016/0002-9149(86)90256-0. [DOI] [PubMed] [Google Scholar]
- 26.Walter J, Newport J. Initiation of eukaryotic DNA replication: origin unwinding and sequential chromatin association of Cdc45, RPA, and DNA polymerase alpha. Mol Cell. 2000;5:617–627. doi: 10.1016/s1097-2765(00)80241-5. [DOI] [PubMed] [Google Scholar]
- 27.Walter J, Newport J W. Regulation of replicon size in Xenopus egg extracts. Science. 1997;275:993–995. doi: 10.1126/science.275.5302.993. [DOI] [PubMed] [Google Scholar]
- 28.Welsh A O. Uterine cell death during implantation and early placentation. Microsc Res Tech. 1993;25:223–245. doi: 10.1002/jemt.1070250305. [DOI] [PubMed] [Google Scholar]
- 29.Yamagishi H, Garg V, Matsuoka R, Thomas T, Srivastava D. A molecular pathway revealing a genetic basis for human cardiac and craniofacial defects. Science. 1999;283:1158–1161. doi: 10.1126/science.283.5405.1158. [DOI] [PubMed] [Google Scholar]
- 30.Zou L, Mitchell J, Stillman B. CDC45, a novel yeast gene that functions with the origin recognition complex and Mcm proteins in initiation of DNA replication. Mol Cell Biol. 1997;17:553–563. doi: 10.1128/mcb.17.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zou L, Stillman B. Formation of a preinitiation complex by S-phase cyclin CDK-dependent loading of Cdc45p onto chromatin. Science. 1998;280:593–596. doi: 10.1126/science.280.5363.593. [DOI] [PubMed] [Google Scholar]