Abstract
Variability in disease presentation, progression and treatment response has been a central challenge in medicine. Although variability in host factors and genetics are important, it has become evident that the gut microbiome, with its vast genetic and metabolic diversity, must be considered in moving towards individualized treatment. In this Review, we discuss six broad disease groups: infectious disease, cancer, metabolic disease, cardiovascular disease, autoimmune or inflammatory disease, and allergic and atopic diseases. We highlight current knowledge on the gut microbiome in disease pathogenesis and prognosis, efficacy, and treatment-related adverse events and its promise for stratifying existing treatments and as a source of novel therapies. The Review is not meant to be comprehensive for each disease state but rather highlights the potential implications of the microbiome as a tool to individualize treatment strategies in clinical practice. Although early, the outlook is optimistic but challenges need to be overcome before clinical implementation, including improved understanding of underlying mechanisms, longitudinal studies with multiple data layers reflecting gut microbiome and host response, standardized approaches to testing and reporting, and validation in larger cohorts. Given progress in the microbiome field with concurrent basic and clinical studies, the microbiome will likely become an integral part of clinical care within the next decade.
Inter-individual variability in disease presentation, treatment response and adverse effects of treatment presents a major challenge for the effective management of disease and patient safety. This realization is foundational to precision medicine, which in its simplest form can be described as identifying the right treatment for the right patient without trial and error using an individualized approach. Initial paradigms into the realm of precision medicine have focused on subtyping patients based on host genetics and a range of clinical and laboratory features, for instance, the genetic make-up of tumour tissue1. This process has led to substantial improvements in predicting the efficacy and toxicity of therapies2. However, the host genome represents only part of the genes of the superorganism as the genetic and chemical diversity of the microbial communities in and on our body is greater than that encoded in the human genome3. As individuals harbour a unique and widely variable gut microbiome it is not surprising that it contributes to the heterogeneity in disease phenotype as well as to treatment response4.
An aspect that differentiates the gut microbiome from human genetics is that it represents a dynamic component of our health, continuously interacting with both host and environmental factors through complex networks. Although a potential challenge, the plasticity of the gut microbiome also offers a unique opportunity, making it an appealing target for precision medicine5–8. Several categories and levels of microbiome-based therapeutics currently exist, with variable specificity in targeting the microbiome. Faecal microbiota transplantation (FMT), the least targeted approach, involves the transfer of faeces in its entirety from one individual to another to attempt to repopulate the gut with ‘healthy’ microbiota9. Probiotics (live microorganisms that have presumed health benefits with adequate administration) have potential for targeted therapy, although most current formulations used in clinical practice lack the required specificity10. Prebiotics are compounds that are selectively digested to stimulate the growth or activity of beneficial organisms, thus with much potential for the personalization of therapy11. Synbiotics, which are a mixture of microorganisms and substrates that are selectively utilized by the host to achieve some health benefit, offer even further space for the personalization of microbiome-related therapeutics12. Besides supplementing specific compounds in their purified form, the dietary intake of foods known to be rich in prebiotic compounds is another possible avenue for shifting microbiota populations. The accumulating data on specific microbiota-driven mechanisms underlying disease pathogenesis, prognosis and treatment outcomes has led to the investigation of a multitude of additional approaches, including but not limited to engineering bacteria to perform specific functions, small-molecule drugs and biologic agents that alter microbial function or its interaction with the host, and native or engineered phages to alter microbial composition and function13,14.
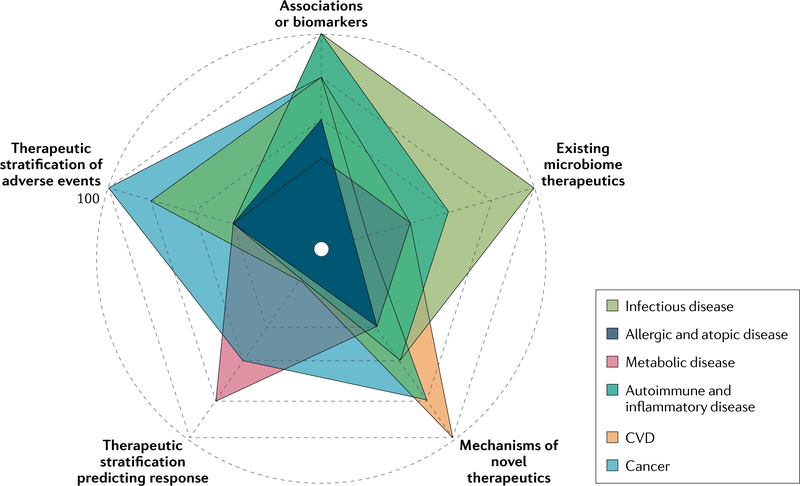
In this Review, we summarize the current evidence in support of using the gut microbiome as a tool for precision medicine and suggest future work needed to incorporate the microbiome as a tool for individualized treatment or interventions. We selected six broad disease groups that have a relatively strong level of evidence for a role for the gut microbiome. Although there are exciting developments in every disease group, the level of promise and maturity when considering clinical impact varies among the different disease groups (FIG. 1; Supplementary Fig. 1). We discuss the disease states from the highest to the lowest maturity of knowledge for clinical practice. Within each section, we aimed to provide information on known taxonomic associations and microbiota mechanisms within the condition, currently used microbiome-directed therapeutics, the role of gut microbiome in predicting treatment response and toxicities, and finishing with an outlook on novel microbiome therapeutics. Given the current differences in evidence between these disease states regarding the gut microbiome, this organization is not entirely uniform across sections. Instead of providing an exhaustive overview of the gut microbiome in each disease state, we aimed to highlight studies that are relevant to the field of precision treatment and synthesize the current status and maturity of this rapidly developing field. The Review provides convincing evidence to support incorporation of the microbiome in clinical practice whilst acknowledging the challenges and outlining a path to get closer to this goal.
Fig. 1 |. Maturity of the gut microbiome as a precision medicine tool.
The advance in microbiome science, while overall remarkable, has been uneven across different disease states. In this figure, we outline the maturity of the microbiome field with regard to advancement to clinical practice in individual disease groups based on a review of available literature in humans. We consider the following parameters within each disease group: presence of robust taxonomic associations or biomarkers; the existence of microbiome therapeutics; knowledge on actionable microbiome mechanisms; use of microbiome in therapeutics stratification; and use of microbiome in prediction of adverse therapeutic effects. A maximal scale of 100 is defined as findings that are currently used in the clinic and the minimal scale of 0 indicates that no progress has been made in this specific aspect. See Supplementary Fig. 1 for each individual disease in a separate plot. CVD, cardiovascular disease.
Infectious disease
The mechanisms involved in antibiotic-induced disruption of the gut microbiome in facilitating opportunistic and nosocomial infections are a topic of intense interest. We use the example of Clostridioides difficile, which is the most common nosocomial diarrhoeal infection15, to highlight microbiome and pathogen-specific features that might explain the inter-individual variability in clinical outcomes. Recurrent C. difficile infection (CDI) has been a central focus of microbiome research from the advent. CDI most commonly follows antibiotic use and, paradoxically, the first-line treatment of CDI also consists of antibiotics16. Although antibiotics are quite effective at the population level, subsets of patients either fail therapy or CDI might recur after successful treatment, which could be related to host features, such as advanced age, or to the use of medications, such as proton pump inhibitors17, combined with features of the specific pathogens in context of the gut microbiome.
In addition to host factors, disruption of gut microbiota has been found to be a key factor underlying CDI. Those with CDI were found to have an elevated relative abundance of Enterococcus, Veillonella, Lactobacillus and Gammaproteobacteria species and lower levels of Bacteroides, Lachnospiraceae and Ruminococcaceae species in comparison to healthy individuals as controls7,18. Moreover, when modelled in germ-free mice, alterations in the gut microbiome driven by a range of host factors increased susceptibility to CDI as a result of elevated amino acid availability, which is a favoured nutrient niche of C. difficile19. Similarly, other studies using mouse models have identified loss of microbiota-derived inhibitory factors as well as an increase in open nutrient niches in CDI, which include decreases in levels of short-chain fatty acids (SCFAs) (specifically valerate) and the secondary bile acid deoxycholic acid and increased levels of the organic acid succinate, sialic acid and amino acids20–23. These microbiome-driven factors that increase susceptibility to CDI vary among individuals and not every individual that develops CDI exhibits all of these abnormalities.
In addition to the gut microbiome, strain to strain variability of C. difficile, such as variation in toxin production, metabolism and biofilm formation capacity, might contribute to different outcomes. A study using whole-genome sequencing of ~400 clinical isolates from patients with CDI found that the majority of disease recurrences are caused by the same strain as the initial infection, which suggests that strain-specific features that enable persistence in the gut might be relevant for recurrent disease24. The organization of C. difficile into multicellular biofilms could allow persistence as biofilms can provide a physical barrier against antibiotics and can interfere with clearance from the gastrointestinal tract25. Hence, diagnostic tests that incorporate whole-genome sequencing along with the metabolic milieu in the gut and the specific microbial taxa will likely provide greater resolution when considering individualized treatment approaches.
The current approach for treating multiple recurrent CDI or treatment failure involves FMT, which has been remarkably effective in treating recurrent CDI, irrespective of the route of administration, the degree of engraftment or specific features of the donor gut microbiome26. The primary feature that predicts FMT failure is continued antibiotic use27. Despite its efficacy, there are concerns about the long-term safety of FMT, which are now being investigated in a long-term registry28. In addition, the question remains as to how to best screen for donor stool. Should this screening be conducted using metagenomics or pathogen whole-genome sequencing to identify the antibiotic resistance profile or is culture or PCR-based screening for pathogens sufficient? The usefulness of a whole-genome sequencing-based approach is illustrated by two clinical trials with cases of bacteraemia with extended-spectrum β-lactamase-producing Escherichia coli linked to donor stool29. This safety concern of FMT is especially relevant for immunocompromised patients.
Beyond FMT, the gut microbiome has also been found to be predictive of treatment outcomes in CDI. At the taxonomic level, the pre-treatment abundance of Ruminococcaceae, Rikenellaceae, Bacteroides and Faecalibacterium has been associated with a positive response to antibiotics in CDI and higher levels of Clostridiaceae, Lachnospiraceae, Blautia, Coprococcus, Streptococcus, Bifidobacterium, Ruminococcus and Actinomyces were associated with non-response in a study including 88 patients with CDI30. Some of these taxa also predicted the risk of recurrent infection30, whereas other taxa have been associated with an overall inability for C. difficile to colonize the gut31 (FIG. 2).
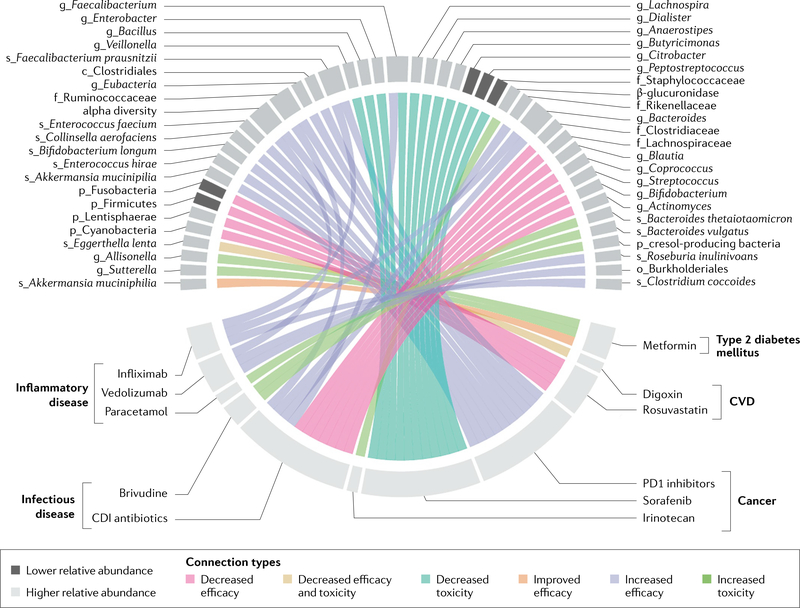
Fig. 2 |. association between the gut microbiome and medication therapy.
Associations between the specific microbial taxa and the efficacy or toxicity of commonly used medications. Only human studies are included and we do not account for differences in the level of confidence in the findings based on association alone or additional biological validation. Prefix indicates phylogenetic level (p_phylum, o_order, f_family, g_genus, s_species). There are a multitude of other medications for which in vitro or animal data suggest associations with outcomes and we expect this field to expand rapidly. CDI, Clostridioides difficile infection; CVD, cardiovascular disease.
The increased understanding of microbiome-driven mechanisms underlying CDI and the prognostic role of the gut microbiome in treatment outcomes, combined with uncertainty regarding long-term risks of FMT, have led to a surge in microbiota-based diagnostics and therapeutics for CDI. The most advanced among these are defined microbial consortia, which showed promise in phase II clinical trials32 and have now been reported to be successful in phase III clinical trials33,34. In addition, there are several approaches utilizing specific bacteria and/or metabolites, prebiotics, and phages that are in early stages of investigation. These narrow range treatment modalities will likely allow for more personalized therapeutics based on host and host microbiome features, potentially further increasing the efficacy and decreasing the risk surrounding treatment of CDI35. This progress provides optimism for the development of future, more precise microbiota-based treatment strategies in CDI (TABLE 1).
Table 1 |.
Current status and outlook for the gut microbiome in disease
| Disease category | Current status | Outlook |
|---|---|---|
| Clostridioides difficile infection | The gut microbiome is an important determinant of susceptibility and response to treatment in C. difficile infection7,18–23,30,31; strain-specific differences in C. difficile might determine the ability to evade antibiotic treatment24,25,264–266 | Although FMT has been effective in treating recurrent C. difficile infection, studies with defined microbial communities have shown promise and could replace FMT32–34 |
| Cancer | Gut microbiota composition can serve as a diagnostics tool to detect early-stage cancer37,40; the gut microbiome can affect cancer development through the production of metabolites and modulation of immune states36,38,41; the gut microbiome can mediate effectiveness and toxicity of chemotherapy and immunotherapy42,45–64 | Novel microbial therapeutics (prebiotics, probiotics, bacterial metabolites) targeting cancer or adverse effects of therapy seem promising37; new tools, such as activity-based protein profiling, can serve as diagnostics for predicting efficacy and adverse events65 |
| Obesity | The gut microbiome is an important determinant of efficacy of dietary, medication and surgical interventions84–93; poor agreement between studies regarding obesity-related microbiome changes has made sub-phenotyping difficult67–72 | Novel microbiome therapeutics show promise in trials82,98–102; clinical studies are needed to validate the stratification of existing therapies based on the microbiome |
| Type 2 diabetes mellitus | Gut microbiota composition can predict glycaemic response to food90,103–109,112; microbial products (SCFAs, BCAAs) play a part in glucose tolerance77,110; efficacy and adverse effects of medications for diabetes are driven in part by the gut microbiome120,121 | Next-generation microbial therapeutics composed of multiple synergistically acting bacteria hold promise for treatment in diabetes122; microbiome-guided dietary and drug therapy can help improve management of diabetes88,89 |
| NAFLD | The gut–liver axis can result in deleterious changes in the liver from microbiota-related mechanisms126; endogenous production of ethanol by bacteria might contribute to steatosis128–130 | Bacteria and bacteriophage-based therapeutics show promise in liver disease and are an area of ongoing investigation133–144 |
| Cardiovascular disease | The association between cardiovascular disease and the microbiome is more related to the functional output of microbiome (metabolites) rather than taxonomic changes148,149,151,152,155–157,159,160; the gut microbiome mediates effectiveness of both dietary and medication-based interventions93,160–163,165–167,169–175 | Novel mechanism-based therapeutics targeting bacterial metabolism are on the horizon152,156 |
| Rheumatoid arthritis | Specific gut and oral bacteria are associated with the development of rheumatoid arthritis179–183; the gut microbiome is an important determinant of efficacy of anti-inflammatory drugs to treat rheumatoid arthritis185,186,188–190,194 | Microbial therapeutics with immune-modulating properties have shown promise in preclinical studies181,199,200 |
| IBD | Feedforward mechanism between the microbiota and host might perpetuate inflammation in IBD184,204–215,217–219; sub-phenotyping based on gut microbiota can enable better stratification to predict disease progression and treatment response220,221 | Microbial therapeutics based on underlying mechanisms and identifying microbiome features that determine success of FMT are active areas of investigation216,223–227 |
| Allergic and atopic diseases | The effect of the gut microbiota early in development is an important risk factor for the development of allergic and atopic diseases, exact mechanisms remain elusive228,233–235,241–243 | Microbial therapy holds promise in allergic conditions by shifting the community to an immuno-tolerant state and degrading allergens237,239,240,250,255,257,258 |
BCAAs, branched-chain amino acids; FMT, faecal microbiota transplantation; IBD, inflammatory bowel disease; NAFLD, non-alcoholic fatty liver disease; SCFAs, short-chain fatty acids.
Cancer
The gut microbiome can inform the field of personalized cancer biology from four different perspectives: inhibiting cancer development, identifying novel treatments, optimization of existing treatments and cancer diagnostics. In regard to cancer development and identifying novel treatments, these issues relate to imperfect functioning of the gut microbiome–immune system axis, which contains both immune inhibitory and immune stimulatory roles for the gut microbiota and the tumour tissue itself36. The optimization of existing treatments relates to reducing adverse effects and predicting response to cancer therapies that range from chemotherapy and radiation therapy to new immunotherapy approaches. Here, the role of the gut microbiome in metabolizing drugs as well as affecting immune cell and cytokine levels can result in changes in therapeutic response and in the development of adverse effects.
Microbiome links to cancer
Mechanistic knowledge of the gut microbiome–immune system axis can be explored both to intercept cancer development and to identify novel treatments. Cancer development and disease progression can be influenced through the oncogenic effects of microorganisms and their products, modulating circulatory metabolite levels that might promote or inhibit tumour growth, and inducing pro-inflammatory and immunosuppressive effects37,38. Localized alterations of microbiota have been identified in association with the development and degree of progression of cancers involving organs that harbour commensal organisms (colorectal, cervical, lung, head and neck)37. The mechanistic connection between the gut microbiome and cancer is most evident in colorectal cancer39. The intricate relationship between microorganisms and their metabolites with the gut mucosa can lead to changes in mucosal permeability increasing localized exposure to a wide array of potential carcinogenic compounds and potentially leading to a chronic inflammatory state39. Another example of a mechanistic link is the presence of E. coli strains that carry the pathogenicity island pks, which encodes genes to synthesize the genotoxic secondary metabolite colibactin and could be used to predict colon cancer risk40. Outside of defined mechanistic features, differences in gut microbial composition and functional attributes have been associated with malignancy in organs with less direct contact37. For example, a study involving 68 patients with pancreatic cancer suggests that the abundance of Pseudoxanthomonas, Saccharopolyspora and Streptomyces in tumour tissue is associated with long-term survival after surgical resection in comparison to those with poorer outcomes41. These protective effects were transferred via faecal transplantation in a mouse model, suggesting FMT as a potential treatment modality that needs further consideration broadly, at least in adjunctive form41.
Microbiome and cancer therapeutics
The response to both chemotherapy and immunotherapy can be influenced by the gut microbiome in regard to both efficacy and toxicity42. This relationship can be due to synergistic effects in antigen presentation, the induction of inflammatory responses and the chemical modification of drugs42. For example, cyclophosphamide, a widely used alkylating agent that induces DNA crosslinks, in part imposes its antitumour effects by modulating the immunological pathways43,44 linked to the generation of specific T helper 17 (TH17) responses after bacterial translocation to secondary lymphoid organs in a mouse model45. In a mouse study on the type 1 immune response-inducing CpG-oligodeoxynucleotides and the DNA crosslinking agent oxaliplatin, therapeutic efficacy was reduced in the presence of antibiotics, which was attributed to reduced tumour necrosis factor (TNF) production by tumour-associated myeloid cells46. This antibiotic-induced reduction of oxaliplatin efficacy is likely multifactorial given that its therapeutic effect is partly driven by the production of reactive oxygen species by the gut microbiota46. Finally, the microbial metabolism of gemcitabine by the action of bacterial cytidine deaminases to generate 2′,2′-difluorodeoxyuridine can reduce its therapeutic effect47.
In addition to chemotherapeutic agents, there is increasing evidence for the importance of the gut microbiota in determining the effectiveness of cancer immunotherapy. In humans, reduced progression-free survival and overall survival has been associated with the use of antibiotics with immunotherapy in multiple cancer types (non-small-cell lung cancer, renal cell cancer and urothelial carcinoma)48,49. This finding has been further investigated using animal models that suggest a role of immune-mediated mechanisms in these negative effects of antibiotics. The efficacy of ipilimumab, a CTLA4 inhibitor, was improved in mice when the Bacteroides species B. fragilis and B. thetaiotaomicron and Burkholderiales were present at increased abundance, which was linked to the upregulation of IL-12-dependent TH1 immune responses50. In a mouse model of colon cancer, efficacy of CTLA4 blockade was enhanced in the presence of Bifidobacterium pseudolongum through immune activation from increased translocation of inosine facilitated by the decreased mucosal barrier function resulting from treatment51. IL-12 has also been found to play a role in CCR9+CXCR3+CD4+ cell recruitment to epithelial tumours in mice, which in turn has been associated with an elevated abundance of Akkermansia muciniphila in the human gut microbiome48. The effect of PD1 and PDL1 checkpoint inhibitors against melanoma can be enhanced in mice with increased abundance of Bifidobacterium species52. Interestingly, Bifidobacterium species might even have anti-melanoma effects in the absence of conventional therapy as its abundance pre-treatment in the same mouse population was associated with tumour growth suppression52. This effect is thought to be related to the upregulation of dendritic cell function, leading to enhanced activity and accumulation of CD8+ T cells in the tumour microenvironment52. The efficacy of PD1 inhibitors in treating melanoma was also enhanced in the presence of a higher abundance of Bifidobacterium longum, Collinsella aerofaciens and Enterococcus faecium in a study of 42 patients with metastatic disease53 whereas, in a separate human cohort of 43 patients with melanoma, increased microbiome diversity was associated with improved cancer survival after PDL1 and PD1 therapy54. Specifically, Ruminococcaceae, Firmicutes, Eubacterium sp., Clostridia, Clostridiales and Faecalibacterium prausnitzii were enriched in patients who were responsive compared with those who were non-responsive (FIG. 2).
The toxicity of chemotherapeutic agents can be a major factor dictating their ability for use. Drug metabolism can be altered by the gut microbiota in several ways, including by competitive inhibition, direct metabolic effects of gut microorganisms and altered host expression of genes involved in metabolic pathways, as seen with the downregulation of xenobiotic detoxifying genes in germ-free mice55,56. Specifically, the toxicity of anticancer agents, such as 5-fluorouracil (5-FU), irinotecan and sorafenib, has been attributed to gut microbial metabolism. The toxicity of the DNA replicator 5-FU when co-administered with the viral DNA polymerase inhibitor sorivudine in a rat model results from reduced metabolism of 5-FU induced by the microbial product of sorivudine transformation, bromovinyl uracil (BVU)42,57,58. Knowledge of the specific biochemical pathways responsible for the formation of BVU could help predict this toxicity to inform alternate treatment options or the development of specific inhibitors of BVU formation. Administration of the topoisomerase inhibitor irinotecan is commonly hindered by the development of severe diarrhoea42, linked to bacterial β-glucuronidase-mediated reactivation of inactive irinotecan metabolites59; antibiotic treatment has been shown to decrease production of the active irinotecan metabolite in vitro60, administration of a probiotic cocktail that lowers β-glucuronidase activity improves diarrhoea in patients with colon cancer and small-molecule inhibitors of glucuronidases have shown promise in preclinical mouse models59,61,62. The tyrosine kinase inhibitor, sorafenib, is another agent whose toxicity might be related to gut microbial activity as both diarrhoea and hand–foot syndrome following sorafenib administration in patients with hepatocellular carcinoma were associated with specific microbial taxa63 (FIG. 2). Specifically, an abundance of Veillonella, Bacillus, Enterobacter, Faecalibacterium, Lachnospira, Dialister and Anaerostipes were protective against hand–foot syndrome, with abundance of Butyricimonas and lower levels of Citrobacter, Peptostreptococcus and Staphylococcaceae associated with less development of diarrhoea. The mechanism underlying this effect might be diminished by enterohepatic recycling of the medication and its metabolites63. A study of patients with metastatic renal cell carcinoma treated with other tyrosine kinase inhibitors (pazopanib and sunitinib) showed improvement in treatment-induced diarrhoea when treated with FMT from healthy donors versus placebo, further implicating these alterations in the microbiome in relation to this adverse effect64. The development of activity-based protein probes to identify the specific microbial pathways responsible for xenobiotic metabolism holds promise as a diagnostic tool and might enable better stratification of treatments65.
Novel therapeutics could use bacterial strains or purified pathogen-associated molecular patterns that function as Toll-like receptor (TLR) agonists to trigger local immune responses in patients with low levels of TLR stimulation37. In addition, levels of faecal and circulatory microbial metabolites that can affect tumour growth (SCFAs, secondary bile acids, vitamins and polyamines) could be used to assess metabolic health status before treatment and, in turn, influence a next generation of biotherapeutics, perhaps in combination with dietary intervention37. A range of immune cell subsets (TH17 cells, T regulatory (Treg) type 1 cells, cytotoxic T lymphocytes, CD4+ cells, CD8+ cells) and cytokine abundances (TNF, IL-12, IL-22 through aryl hydrocarbon receptor (AHR) signalling) are affected by changes in the gut microbiota and might be modulated to influence cancer immunosurveillance. Measurements of such markers can also lead to the assessment of immune health of the patient and provide a target for intervention. These approaches will require detailed personalized multi-omic studies on large cohorts before they can be embraced in the clinic.
The studies thus far provide strong support for a role of the gut microbiota in both the heterogeneity of cancer phenotypes and response to cancer therapy. However, data from human studies are largely associative and still need to be replicated across cohorts. The mechanistic data are obtained primarily from animal studies, raising some concern about translational validity. Despite these concerns, the outlook remains optimistic and integration of the microbiome as a component of treatment strategies in cancer seems inevitable (TABLE 1).
Metabolic disease
Obesity
The number of children and adults with overweight or obesity is growing, with over 40% of adults in the USA now meeting criteria for obesity66. A major challenge in tackling obesity is the complexity of mechanisms resulting from the interplay of genetics, gut microbiome, diet and environment, which result in physiological changes contributing to obesity.
Microbiome links to obesity.
A meta-analysis published in 2016 of curated human microbiome studies found a small but statistically significant association between obesity and lower within-sample alpha diversity metrics of richness and evenness67. Besides alpha diversity, an elevated Firmicutes to Bacteroidetes ratio in obesity has been reported in early microbiota studies both in mouse models and human studies68–72 but was not replicated in this meta-analysis67. Despite the lack of robust compositional markers, a role for gut microbiome in obesity is supported by transplantation experiments showing that germ-free mice colonized by stool from monozygotic twins discordant for obesity exhibit the metabolic phenotype of the donor73. This premise is further strengthened by the observation that weight gain induced by a high-fat diet compared with a low-fat plant polysaccharide diet in humanized mice (germ-free mice colonized with stool from healthy human) can be transmitted to recipient germ-free mice by just transplanting faecal samples from these humanized mice without requiring continued feeding of a high-fat diet74. This finding suggests that the microbiome is important in propagating the obesity phenotype even if it was initiated by other causes. Several mechanisms underlying obesity have been attributed to the gut microbiome such as an increased efficiency of energy extraction from diet, influencing satiety and energy intake, systemic inflammation, and insulin resistance75–78.
The lack of consistent compositional markers in obesity suggests substantial functional redundancy. Indeed, there is less redundancy at the functional level as shifts in several different microbial community configurations can drive changes in production of an array of bioactive factors such as SCFAs, bile acids and lipopolysaccharides (LPS), all of which have been implicated in obesity79. SCFAs play a role in hormonal signalling, such as that of serotonin and peptide YY release, which play a role in satiety, implicating involvement of the gut–brain axis in driving obesity80. Although there have been major advances in the understanding of microbiome-driven mechanisms underlying obesity, we do not yet have sufficient resolution to stratify individuals with obesity based on underlying microbiome-based mechanisms. With accumulating evidence in this area, one can easily envision the microbiome-based stratification of individuals to be part of future personalized strategies for obesity management81. The efficacy of probiotics as a therapeutic regimen for obesity has been suggested in animal models but the results of clinical trials in humans have been mixed and, given the lack of consistency in probiotic formulations used, their role remains unclear at this point82. With regard to the use of prebiotics therapeutically, again there have been encouraging findings in animal models but there is yet to be any clear crossover into humans in clinical trials in terms of durable weight loss83.
Microbiome and obesity therapeutics.
The main therapeutic approaches in the management of obesity are diet, medications (such as glucagon-like peptide 1 (GLP1) agonists, orlistat and phentermine) and bariatric surgery. Diet can affect the gut microbiota both in the short and long term, with alterations occurring in response to brief dietary changes84. More importantly, the therapeutic effect of diet is also dependent on an individual’s microbiome84,85. In simplifying the gut microbiome to 10 microbial strains in germ-free mice it was demonstrated that dietary modifications alter the colonization pattern of the gut and its fermentative capabilities86. One approach to dietary modification in individuals with obesity is increasing the consumption of fruits, vegetables and low-energy density foods whilst concomitantly reducing the intake of foods with high nutrient density; however, the response to such an intervention is quite variable. A pilot study in 26 individuals with overweight or obesity found that a high predicted abundance of glycoside hydrolases in the gut microbiota prior to such a dietary intervention was associated with <5% body weight loss following volumetric dietary intervention, suggesting a potential role for the baseline gut microbiome in predicting outcome87. These findings are in line with inter-individual variability in glycaemic response and lipaemia following meals, which has been attributed to the gut microbiome88–90. Increased calorie intake has been associated with rapid alteration of the gut microbiome in humans within 3 days in a population of 21 healthy individuals, including individuals with normal (between >18.5 and <25) and high (≥30) BMI, showing an increased relative abundance of Firmicutes and depletion of Bacteroidetes91. These changes were associated with increased energy harvest as evidenced by reduced stool caloric content. Hence, reducing caloric intake might exert a beneficial effect by modifying the microbiome in subsets of individuals with obesity76,77,92. The relative abundance of gut bacteria, such as Eubacterium ruminantium and Clostridium felsineum, has also been associated with increased microbiome plasticity in response to several variable dietary interventions in a group of 78 individuals with obesity93. Together, these studies suggest that the concept of some foods being ‘healthy’ for everyone is too simplistic and that dietary choices based on gut microbiome metrics might be beneficial in regard to weight management94.
Few studies have examined the gut microbiome in the context of pharmacological and surgical treatments against obesity. However, the GLP1 agonist liraglutide, which increases insulin release and delays gastric emptying in the setting of elevated blood glucose levels, was found to increase the Firmicutes to Bacteroidetes ratio in rats and might therefore drive weight loss, at least in part, through secondary changes in the gut microbiome95. Having prior Roux-en-Y gastric bypass surgery (a bariatric procedure used to manage patients who fail lifestyle modifications and medications) was associated with a reduced relative abundance of Firmicutes and higher levels of facultative anaerobes, such as Proteobacteria, at 15 months post-surgery96. These changes could play a role in weight loss. An alternative surgery — sleeve gastrectomy — was effective in reducing inflammation and shifted the gut microbiome of 23 patients with pre-surgical obesity closer to that of healthy individuals used as controls, with corresponding recovery of microbially determined plasma glutamate levels as a biomarker of obesity97. Whilst most of the findings need to be validated in larger cohorts and tested in mechanistic models, these studies highlight the utility of microbiome analysis in assessing the efficacy of currently available obesity therapies.
In terms of microbiome-based therapeutics, A. muciniphila is a promising candidate for the treatment of metabolic syndrome and obesity. The consumption of A. muciniphila was protective against weight gain in mice owing to improved intestinal barrier function, diminished endotoxemia and improved glucose tolerance through the activation of TLR signalling as a result of an outer membrane protein98. Pilot data in 32 people suggest the safety and efficacy of A. muciniphila, with modest weight loss and improvement in laboratory markers of obesity over 3 months99. Similarly, Christensenella minuta, noted to be one of the most heritable microbial species and specifically associated with leanness in humans, was also found to be effective in treating obesity in an animal model100 and a randomized controlled trial in humans is planned to start soon101. Although we highlight some ongoing efforts to target the microbiome to decrease intestinally derived inflammatory signals, the alteration of nutrient signalling and modulation of the gut–brain axis can also prove to be effective strategies. These novel approaches of targeting microorganism– host interactions will likely be an important part of preventing and treating obesity102 (TABLE 1).
Insulin resistance or diabetes
Because of the effect on circulatory metabolites and immune status, the gastrointestinal tract and associated gut microbiome can be viewed analogously to an endocrine organ. As such, it is involved in glucose metabolism by affecting insulin signalling.
Microbiome links to altered glycaemic control.
Several studies provide cross-sectional taxonomic changes in the gut microbiome in diabetes with a shift towards a lower abundance of butyrate-producing organisms and overall microbial diversity but findings have not been consistent amongst studies103–105. However, a clear relationship between the gut microbiome and insulin resistance has been elucidated with FMT from donors with metabolic syndrome leading to decreased insulin sensitivity in germ-free mice in comparison to FMT from those with Roux-en-Y gastric bypass106. This was further supported in another human study where insulin sensitivity in individuals with obesity was improved after FMT from lean donors prior to the intervention107. This highlights the potential therapeutic benefit from microbiota-directed therapies in carefully selected patients for whom a shift in the gut microorganisms may lead to more substantial clinical benefit107. A randomized clinical trial in humans showed that increased diversity and abundance of a select group of SCFA-producing strains promoted by dietary fibres led to improvement in haemoglobin A1c levels, which was attributed to increased glucagon-like peptide108. However, other potential SCFA producers were diminished or unchanged, suggesting that not all SCFA producers are created equal and a more targeted restoration of specific microorganisms may be more beneficial. Future studies like this will help identify specific groups of bacteria that are not just capable of a function but in fact work synergistically to restore a key function. Together, these studies support the link between the gut microbiome and diabetes and highlight the promise of using the gut microbiome to optimize therapy (TABLE 1). The mechanisms underlying the role of gut microbiome in insulin resistance or type 2 diabetes mellitus (T2DM) overlap with those identified for obesity, such as low-grade inflammation, changes in gastrointestinal permeability with possible endotoxemia, and decreased SCFA production and absorption, which is consistent with the concept of metabolic syndrome. The changes in SCFAs can in turn affect the production of various metabolic hormones, such as GLP1 and peptide YY, which play a role in insulin secretion77. In addition, elevated branched-chain amino acid (BCAA) levels owing to an altered ratio of microbial BCAA biosynthesis and BCAA degradation have been associated with early insulin resistance in human studies and might be driven by Prevotella copri and Bacteroides vulgatus109. Colonization with P. copri was also associated with insulin resistance in a conventional mouse model109. Another microbial metabolite, imidazole propionate, was found to be elevated in patients with T2DM and can directly impair glucose tolerance and insulin signalling110. These studies highlight how microbiota-derived circulatory metabolites can drive the pathogenesis of diabetes.
Patients with T2DM are primarily managed with diet and medications (such as metformin, sulfonylureas and GLP1 agonists), although they might eventually require insulin replacement therapy and/or surgery. The current approach is to try treatment options sequentially even though there are substantial differences in how individuals respond to each of the treatments and some patients might not respond to diet or medications111. The microbiome offers an important avenue for optimization to determine whether a treatment strategy is better suited to an individual. A seminal study conducted in Israel in 800 non-diabetic individuals outlined the potential for developing personalized dietary recommendations based on an elegant machine learning approach using a combination of measurements, including microbiome and host features as well as blood glucose response to varying diets88,89 (FIG. 1; Supplementary Fig. 1). A subsequent study validated this approach in a US Midwestern population of 327 individuals without diabetes and confirmed that an individual’s microbiome can predict changes in blood glucose in response to different meals112. Interestingly, carbohydrates as a group were still associated with increased glycaemic response, but this approach identifies the major offenders within carbohydrates at an individual level, allowing them to restrict specific carbohydrates rather than a blanket low carbohydrate diet, which often has low adherance94. Another study found that an individual’s microbiome can not only predict changes in blood glucose but also changes in triglycerides in response to different meals90.
Microbiome and diabetes therapeutics.
One of the most commonly used medications for the treatment of diabetes is metformin, which suppresses liver glucose production, increases insulin sensitivity, and enhances muscle and liver glucose uptake113. Metformin efficacy seems to be dependent, at least in part, on the microbiome. Metformin administration in both animal and human studies leads to an increased abundance of A. muciniphila as well as several bacterial species associated with the production of SCFAs (for example, Blautia and Butyricicoccus)114,115 (FIG. 2). A. muciniphila can improve glycaemic control through ileal goblet cell proliferation, a decrease in gastrointestinal permeability with lower endotoxemia and stimulation of TLR signalling as seen in mouse models114,116. The SCFA butyrate is linked to improved energy metabolism in rodents through its beneficial effects on skeletal muscle, brown fat tissue and pancreatic β-cells117. In addition, the SCFA propionate suppresses hepatic gluconeogenesis and reduces appetite and body weight in rodent models118,119. The most common adverse effects of metformin are related to gastrointestinal discomfort such as pain, bloating and nausea. A study including 27 healthy men without diabetes found that abundance of specific genera (Sutterella, Allisonella, Bacteroides and Paraprevotella) in stool prior to initiating metformin was associated with the development of gastrointestinal adverse effects120 (FIG. 2). This finding suggests that, in addition to a role in metformin efficacy, the gut microbiota might also contribute to its gastrointestinal intolerance. Hence, microbiome-based stratification could enable the selection of patients likely to have a favourable response and tolerate therapeutic doses. Data supporting a role for the gut microbiome in other diabetes therapies are sparse but a decrease in the abundance of the phylum Firmicutes upon administration of the GLP1 agonist liraglutide in mice was associated with improvement in glycaemic control121.
Given the data supporting the connection between metformin administration and increased abundance of A. muciniphila and butyrate-producing microorganisms, a multicentre, double blind, randomized placebo-controlled trial studied the administration of these microorganisms in probiotic form in 76 patients with T2DM122. There was a promising trend towards better glycaemic control in those who received a combination therapy synbiotic (A. muciniphila, Clostridium beijerinckii, Clostridium butyricum, Bifidobacterium infantis, Anaerobutyricum hallii and inulin) versus placebo, although the small population and short follow-up (12 weeks) leave it unclear whether this approach might be beneficial in the long-term for T2DM122. Trials to expand on this finding and studies using similar targeted microbiome approaches for diabetes management should help to push treatment of this condition further into precision medicine in the future.
Non-alcoholic fatty liver disease
Non-alcoholic fatty liver disease (NAFLD) is a considerable morbidity associated with metabolic syndrome given the progression to cirrhosis and end-stage liver disease if left unchecked. The gut microbiome is closely linked to the liver through the gut–liver axis123 and detoxification of microbial products is an important function of the liver.
Microbiome links to NAFLD.
Gut microbiota alterations and their consequences observed in metabolic syndrome, such as elevated Firmicutes to Bacteroidetes ratio, increased energy harvest capacity, increased intestinal permeability and low-grade inflammation, have also been reported in NAFLD124–126. The role of the microbiome in NAFLD is supported by the development of steatosis in gnotobiotic mice after the transfer of faeces from human donors with NAFLD127. Other microbiome-mediated mechanisms implicated in NAFLD include microbial bile acid modification and associated effects on liver farnesoid X receptor (FXR) signalling, endotoxemia, and production of uraemic toxins such as methylamines and p-cresyl sulfate126. Although, by definition, NAFLD is not associated with alcohol consumption, the current definition does not account for endogenous production of ethanol. E. coli and other Enterobacteriaceae from the phylum Proteobacteria are able to endogenously produce ethanol; thus, the high metabolic activity of these microorganisms in the gut could conceivably result in elevated levels of ethanol contributing to steatosis in patients considered to have NAFLD128,129. Although extreme cases (referred to as auto-brewery syndrome) are rare, prolonged low levels of ethanol from microbial production might still be a contributing factor130.
Microbiome and NAFLD therapeutics.
Metformin, which is commonly used to treat T2DM98,114,115, is also used in management of NAFLD, with animal studies supporting the efficacy of metformin in this setting131 and human data showing improvement in liver function tests but not in histological response132. As outlined earlier, a successful response to metformin seems to be driven, at least in part, by the gut microbiome. The mechanistic connections between NAFLD and the gut microbiome have led to studies exploring potential microbiota-based therapeutics. FMT has shown promise in animal studies133–135 and preliminary human data also suggest improvement of hepatic steatosis and abnormal intestinal permeability after FMT136,137. Ongoing clinical trials will better clarify the efficacy and safety of these procedures138,139. Probiotic administration has also shown some promise with regard to improving steatosis and markers of liver inflammation. However, the composition of these probiotic formulations have been variable, complicating their clinical recommendation at this point140–143. A study in germ-free mice colonized with faeces from two cytolysin-positive patients with alcoholic hepatitis showed therapeutic benefit with bacteriophages that target cytolytic E. faecalis144 and it remains to be seen whether similar approaches might also be useful in NAFLD. Other novel microbiome therapeutics focused on alleviating sources of liver toxicity from the microbiota seem promising but are in an early stage (TABLE 1).
Cardiovascular disease
Cardiovascular disease (CVD) is the leading cause of death in the USA and continues to rise on a global level, growing by 12.5% between 2005 and 2015 (REF.145). It is estimated that 90% of CVD is preventable through improved lifestyle and diet146. CVD has been repeatedly linked to endotoxemia, increased intestinal permeability and low-grade inflammation, all of which can be driven by the gut microbiome147.
Microbiome links to CVD
An early advance in the field of gut microbiome research was the identification of elevated plasma levels of the metabolite trimethylamine-N-oxide (TMAO) as a risk factor for CVD148–150. TMAO is produced in the liver by action of flavin monooxygenase 3 using the bacterial metabolite trimethylamine (TMA) as a substrate. TMA originates from the bacterial conversion of choline, phosphatidylcholine or l-carnitine148,151. The mechanistic role of TMAO based on animal studies suggests it is likely a major driver of atherosclerotic plaques152 and high plasma l-carnitine levels are associated with decreased event-free survival from cardiovascular events only when there is a concurrent increase in TMAO levels, suggesting that TMAO is the likely driver of cardiovascular risks in humans148,153. The mechanisms by which TMAO contributes to CVD include its effect on foam cells and endothelial cells, vascular inflammation, atherosclerotic lesions, fibrosis, and enhanced platelet aggregation and thrombosis154. Specific gut microorganisms, including Proteus mirabilis, Proteus penneri and Escherichia fergusonii, are known to produce TMA in vitro and in animal models155,156. However, because of considerable inter-strain diversity, the abundance of the genes responsible for TMA production, namely cutC/D or cntA, in the human gut microbiome might have larger predictive promise than levels of specific taxa152,157.
In addition, atherosclerotic CVD in humans was also linked to a reduced functional capacity for microbial fermentation of the gut microbiota as well as to an elevated abundance of the bacterial taxa Enterobacteriaceae and Streptococcus158. Other microbiota-derived metabolites that correlate with lowering of blood pressure are the SCFAs acetate and butyrate159,160. A reduced abundance of SCFA producers, such as Eubacterium rectale, Dorea longicatena, Clostridium clostridioforme and F. prausnitzii, has been associated with the development of heart failure in humans159,160.
Diet is one of the mainstays of prevention and treatment of CVD and, as microbial metabolism of dietary components has a mechanistic role in the pathogenesis of CVD, the gut microbiome might be partly responsible for the effectiveness of dietary interventions. Both a Mediterranean diet and high-fibre diet seem to be protective against CVD and a case–control study in 396 patients with myocardial infarction (MI) and 843 healthy individuals as controls found that absence of P. copri was associated with an 18% lower risk of MI after a Mediterranean diet whereas carriage of P. copri was associated with a non-significant increase in MI following a Mediterranean diet160–162. Adherence to a Mediterranean diet has also been associated with an increased abundance of several gut microorganisms known to metabolize fibre and produce SCFAs such as F. prausnitzii, Eubacterium eligens and Bacteroides cellulosilyticus162. The benefit from increased fibre consumption might be related to increased production of the SCFA acetate by fibre-degrading microorganisms or their interaction partners. Acetate is involved in regulation of the transcription factor Egr1, which in turn regulates cardiac inflammation, fibrosis and hypertrophy in mice163. In addition, elevated abundance of the butyrate producer Clostridium sphenoides prior to various dietary interventions has been associated with a greater decrease in cholesterol levels in individuals with obesity and may also be relevant for CVD93. Contrasting the Mediterranean and high-fibre diets, consumption of a Western diet (high intake of fatty and/or processed meats, saturated fats, salt, sugar and refined grains)164 is associated with an increased risk of CVD, which might be linked to a decreased abundance of gut microorganisms such as Bifidobacterium and Eubacterium spp.163. Interestingly, the TMAO precursors choline, phosphatidylcholine and l-carnitine are prevalent in animal protein, which is a characteristic component of the Western diet. However, the consumption of animal protein such as red meat might only be harmful in a subset of individuals who harbour microorganisms that can generate TMA or other metabolites.
Microbiome and CVD therapeutics
The efficacy and toxicity of several drug treatments directed at CVD is associated with the gut microbiome. A key example of this aspect is the presence of cardiac glycoside reductase genes in Eggerthella lenta that inactivate digoxin, an important drug in the treatment of cardiac arrhythmia that acts through inhibition of the Na+/K+/ATPase at the myocardium165 (FIG. 2). It is likely that this bacterial enzyme activity is due to substrate promiscuity rather than a process evolved in response to environmental exposure of digoxin. This finding provides an example of how the chemical diversity of the gut microbiome can lead to crosstalk with the metabolism of human-designed drugs166. As digoxin has a narrow therapeutic window, determining the presence of this bacterial metabolic pathway before starting treatment could enable more accurate dosing and minimize adverse effects. Interestingly, the genes responsible for digoxin inactivation are repressed in the presence of considerable amounts of the amino acid arginine and a high-protein diet reduced digoxin inactivation in a mouse model165.
Statins, which work through competitive inhibition of HMG-CoA reductase, are the most commonly used medications for CVD-associated hyperlipidaemia, with nearly half the US population aged between 40 and 75 years having an indication for their use167. Interestingly, there is substantial inter-individual variability in response to statins as measured by varying decreases in LDL cholesterol levels168. This variability might originate from the gut microbiome as more robust treatment responses were seen in individuals169,170 and animal models171–174 with higher gut microbial diversity. In addition, having elevated levels of Proteobacteria was associated with decreased efficacy of simvastatin in a study of 100 individuals with total cholesterol levels of 160–400 mg/dl showing variable LDL response170. The efficacy of a different statin, rosuvastatin, was also decreased in the presence of higher levels of Cyanobacteria and Lentisphaerae combined with lower levels of Firmicutes and Fusobacteria in a population of 64 patients with hyperlipidaemia174 (FIG. 2). These studies suggest that statin treatment response can be predicted based on an individual’s gut microbiome. A study published in 2020 found the Bacteroides 2 (Bact2) enterotype, which is associated with obesity, to be less prevalent in patients treated with statins175, which suggests that statins have a microbiome-shaping effect. Whether this finding can be used to predict therapeutic outcomes and direct therapeutic choices in the future remains to be seen.
Given the accumulating evidence in support of the role of gut microbiome in the pathogenesis of CVD, it is not surprising that there are several ongoing clinical trials investigating the role of probiotics in CVD. Two examples include the comparison of the antimicrobial rifaximin and the probiotic Saccharomyces boulardii176 and the effect of Lactobacillus acidophilus on inflammation in patients with heart failure177. Such interventional trials provide a valuable opportunity to study potentially beneficial microbiome rearrangements based on longitudinal information on the gut microbiome collected from patients following treatment. Based on some of these highlighted findings, there seems to be a place for next-generation microbial therapeutics that can drive specific functions such as acetate production or improved barrier function. An alternate approach is the development of small-molecule inhibitors of specific microbial pathways such as the recently described inhibitors of TMA-producing enzymes152,156. These inhibitors might enable more precise therapeutic interventions and can be specifically directed towards patients with high TMAO levels and functional gene levels that indicate a high TMAO production capacity156,178.
In general, the recognition that the gut microbiome plays a role in CVD pathogenesis and treatment is an important advance that opens up novel avenues for disease recognition, stratification and treatment (TABLE 1).
Autoimmune or inflammatory disease
Rheumatoid arthritis
Rheumatoid arthritis (RA) is an autoimmune condition that results in chronic inflammation of the joints. Several studies have described alterations in the gut microbiota in patients with RA and these alterations vary with the stage of disease179.
Microbiome links to RA.
One consistent finding in the literature is that members of the genus Prevotella are associated with progression of disease (P. copri) as well as with disease amelioration (Prevotella histicola) in RA, highlighting that different species and/or strains within the same genus can have divergent effects on host physiology180,181. Hence, it is important to resolve the taxonomic differences at the species or strain level and not to generally label an entire genus as being beneficial or harmful. The potential role of P. copri in the pathogenesis of RA is based on findings in both human and rodent studies. In in vitro studies and in mice, P. copri has been shown to increase TH17 responses, which are in turn associated with increased arthritic bone erosion182. In humans, P. copri 16S rDNA has been found within the synovial fluid of RA-affected joints183. Although P. copri seems to be an important determinant of RA, its levels are reported to be highly variable over time in healthy individuals184; hence, longitudinal studies coupled with assessment of host phenotypes are needed to better understand its role in RA. In addition to the gut microbiome, specific periodontal bacteria and periodontal disease have been associated with an increased risk of RA in humans and mouse models of arthritis. Porphyromonas gingivalis and Aggregatibacter actinomycetemcomitans have both been associated with an increase in autoantibodies against citrullinated peptides and might contribute to the autoimmunity in RA183.
Microbiome and RA therapeutics.
In addition to a role in pathogenesis, the gut microbiota might also play a role in determining response to medications commonly used for RA. These include disease-modifying agents, such as methotrexate and hydroxychloroquine, and anti-inflammatory agents such as sulfasalazine and nonsteroidal anti-inflammatory drugs (NSAIDs). Host factors and genetics have failed to provide a predictive model for response to methotrexate but a higher gut microbial diversity has been associated with methotrexate treatment185–187. A study in 26 drug-naive patients with new-onset RA found distinct microbial taxa and their genes in methotrexate responders and non-responders188. A microbiome-based model developed using machine learning techniques predicted the lack of response to methotrexate in a validation cohort of 21 patients with a high degree of accuracy (AUC 0.84). This finding was attributed to the direct metabolism of methotrexate by gut microbiota given that methotrexate levels following incubation of the drug with distal gut microbiota from patients was predictive of the clinical response188. In another study, the impact of methotrexate treatment on specific microbial taxa and pathways in mouse models led to decreased immune activation and thus decreased disease activity189. These studies suggest that microbial metabolism of methotrexate by gut microbiota may play a role in efficacy of the medication and the effect of methotrexate on the reduction of disease activity is itself driven by modulation of the gut microbiome. Methotrexate, which suppresses immune function through competitive inhibition of dihydrofolate reductase, has been associated with a reduced abundance of Enterobacteriales185 but there remains a lack of clarity as to whether this finding has any effect on medication response185. However, it does further suggest that methotrexate affects the gut microbiome structure and that the microbiome-informed prediction of response to methotrexate could be explored further to guide treatment. The differential gut microbial metabolism of methotrexate itself to inactive or inaccessible forms that remain within bacterial cells is a possible mechanism by which the gut microbiome alters methotrexate efficacy190. Gut microorganisms also play a role in the toxicity of methotrexate; Bacteroides fragilis gavage has been found to be protective against intestinal mucositis, an adverse effect seen in about one-third of patients with methotrexate administration191, following methotrexate treatment in mice192. The efficacy of hydroxychloroquine, which inhibits immune activation via reduced TLR signalling and CD154 expression193, has been associated with gut microbial alpha diversity with a higher pre-treatment diversity favouring greater efficacy, but it is unclear whether it is simply the higher microbial diversity or the increased abundance of specific bacteria that are responsible for this effect186. As with methotrexate, there have been alterations in the gut microbiome associated with the TNF inhibitor etanercept but again without a defined relationship to efficacy in current studies185.
The 5-aminosalicylic acid prodrug sulfasalazine is converted into its active metabolite following acetylation by the enzymatic action of gut microbiota and, as a result, its efficacy is dependent on the gut microbiota194,195. Adverse events related to NSAIDs and paracetamol might be related to the gut microbiota. The activity of bacterial β-glucuronidase can lead to toxicity of NSAIDs and inhibitors of this enzyme decrease NSAID-induced enteropathy in mice196,197. In principle, the measurement of β-glucuronidase activity can help identify individuals who should either avoid NSAIDs or are candidates for co-treatment with specific small-molecule inhibitors of β-glucuronidase. Certain bacteria can produce p-cresol, which competes with paracetamol for enzyme binding in the liver and can lead to production of the hepatotoxic compound NAPQI198 (FIG. 2). Thus, p-cresol levels might be used to guide paracetamol dosing to avoid hepatotoxic adverse effects.
The potential contribution of gut microorganisms to the pathogenesis of RA has led to the exploration of probiotics as a potential therapeutic option. These efforts are primarily focused on modulation of the immune system to counteract changes seen in RA rather than a strategy to potentially replace missing microorganisms or mechanisms. P. histicola decreased the incidence and severity of arthritis in susceptible HLA-DQ8 mice by the suppression of antigen-specific TH17 responses and by stimulating increased transcription of IL-10 (REF.181). Administration of Lactobacillus casei was associated with decreased pro-inflammatory molecules (IL-1β, IL-2, IL-6, IL-12, IL-17, IFNγ, TNF and COX2) by CD4+ T cells in mouse models of collagen-induced arthritis199. In humans, Bacillus coagulans has been studied as a potential adjunctive therapeutic option in 45 adults with RA, with administration leading to improved self-assessment of pain and disability and lower inflammatory markers than placebo200. The differential effect of microbial species on the immune system suggests that immune marker-based subtyping of RA could help select patients most likely to respond to specific microbiota-based therapeutics (TABLE 1).
Inflammatory bowel disease
Inflammatory bowel disease (IBD) is a chronic inflammatory condition that includes Crohn’s disease and ulcerative colitis. The heterogeneous nature of IBD subtypes in terms of disease phenotype, susceptibility, progression and response to treatment has inspired attempts at subtyping them beyond just clinical presentation. The focus has been on host genes and the immune response but the exposome and the microbiome are increasingly being recognized as important determinants of inter-individual variability in IBD.
Microbiome links to IBD.
A central role for gut microbiota in IBD is based on observations such as disease remission following faecal diversion, higher disease burden in areas of the gastrointestinal tract with elevated levels of bacteria, improvement in subsets of patients following antibiotic treatment, and IBD-specific changes in gut microbiota composition and function201–204. Furthermore, enhancement of inflammatory pathways associated with IBD, such as IL-17, was observed after colonization of mice with stool from patients with IBD, highlighting its role in disease184,205.
Both Crohn’s disease and ulcerative colitis are characterized by dramatic shifts in microbial community structure with the most consistent finding being increased relative abundance of the phylum Proteobacteria and, specifically, the family of Enterobacteriaceae. Abundances of specific species have been linked to Crohn’s disease (E. coli, Campylobacter species and Mycobacterium avium)204 and the depletion of specific butyrate-producing bacteria is associated with both Crohn’s disease and ulcerative colitis pathogenesis206,207. Adherent-invasive E. coli has also been identified in the ileal mucosa of patients with Crohn’s disease with an associated increase in TNF secretion but it is unclear whether this bacteria is disease inducing or its presence is a consequence of underlying disease factors208–211. Overall, these community shifts represent an increase in facultative anaerobes, such as Proteobacteria, at the expense of obligate anaerobes211,212. The majority of studies outlined here focus on the luminal microbiome, which, while important in IBD pathogenesis, has lower discriminatory power than the mucosa-associated microbiome in identifying IBD213.
The overall expansion of Enterobacteriaceae in IBD is likely due to changes to the nutritional landscape of the gut such as elevated host production of N-acetyl ethanolamine signalling lipids, which can be exploited by Enterobacteriaceae214–216. In addition, bacterial nitrogen metabolism is linked to Enterobacteriaceae expansion though the production of ureases217 and the availability of nitrate in an inflammatory environment, which can fuel anaerobic respiration of Enterobacteriaceae218. An increased abundance of Proteobacteria might not necessarily be the inciting event in IBD and could result from a combination of host genetic predisposition as well as dietary and environmental exposures. However, Proteobacteria do contain highly immunogenic LPS, which in itself can trigger an inflammatory response. This feedforward mechanism might serve to perpetuate inflammation and allow Proteobacteria to thrive while at the same time excluding bacteria such as F. prausnitzii, which does not fare well in an inflammatory milieu212,219. This hypothesis is supported, in part, by the observation that IBD treatment with TNF inhibitors is associated with restoration of a more diverse gut microbiome in paediatric patients with Crohn’s disease213. Besides LPS, several other bacterial components and metabolites have also been implicated in IBD such as higher levels of polyamines and ATP and lower levels of secondary bile acids and butyrate184.
The microbiome and IBD therapeutics.
The gut microbiome also plays a role in predicting the response to existing IBD treatments. A higher pre-treatment gut microbial alpha diversity is associated with a higher likelihood of remission following treatment with the anti-integrin therapy vedolizumab (α4β7 antagonist), suggesting a potential role for microbial metabolism in determining efficacy220 (FIG. 2). Similarly, specific gut microbiome signatures have been associated with disease recurrence after discontinuation of the anti-TNF treatment infliximab221 (FIG. 2). As the current clinical practice is typically to continue biologic therapies long after achieving remission222, such signatures might allow the selection of patients who could be weaned off therapy. Given the variability in response to biologic medications as well as the cost and morbidity involved, the ability to predict response and persistent remission with microbiome analysis could streamline management of IBD.
These observations have fuelled the development of microbiota-based therapies for treatment of IBD ranging from probiotics and FMT to specific bacterial compounds or metabolites223. F. prausnitzii is one example of a possible probiotic as the reduced abundance of F. prausnitzii in postoperative specimens of patients with Crohn’s disease is associated with increased disease recurrence after resection207. F. prausnitzii has been shown to prevent acute colitis by reducing the secretion of inflammatory cytokines224,225, suggesting it has anti-inflammatory effects, which might be a result of its ability to produce butyrate or through an independent effect on the immune system226. In the future, postoperative profiling of the microbiome could enable the identification of individuals who might benefit from therapeutic strategies such as F. prausnitzii probiotics223.
Similarly, the efficacy of FMT in IBD has been linked to specific features in the donor microbiome. Abundances of the family Lachnospiraceae and the genus Ruminococcus are associated with response, suggesting a role for specific microbial taxa or metabolites in determining response and offering a possible explanation for the variability in response seen in other FMT trials to date227. As we continue to learn more about the role of the gut microbiome in IBD, sub-phenotyping based on the gut microbiome might better stratify patients in terms of predicting progression as well as response to specific treatments and could lead to the development of individualized therapeutic strategies targeting the gut microbiome (TABLE 1).
Allergic and atopic diseases
The critical role of the gut microbiota in immune education makes it an important player in allergic and atopic diseases. The microbiota is most vulnerable during early stages of life and changes during this time can have a long-lasting effect on the immune system. Hence, most microbiome studies in allergic and atopic diseases have focused on early life, with the goal of determining how diet, allergen exposure and microbiome composition of the newborn baby can drive allergic diseases and identify specific targets that can be modulated to prevent these diseases. This potential window of intervention is suggested by the observation that atopy in early life is a risk factor for the development of food allergy and eventually asthma in later life228. This link suggests a shared underlying mechanism classically linked to CD4 TH2 hyperactivation combined with reduced levels of dendritic cell-induced Treg cells, all of which are affected by the microbiome. While the field is still in its infancy, we highlight some areas in which personalized microbiome changes might be relevant.
Food allergies
Food allergies are likely driven by a complex interplay between genetics, diet and the commensal microbiota229,230. In humans, distinct gut microbiota changes have been reported in cohorts with different types of food allergy such as those against eggs, peanuts, soy, wheat and milk231–233. However, the lack of consistency among studies makes it challenging to interpret these changes227,234,235. The role of the gut microbiota is supported by the observations that germ-free mice are sensitive to anaphylactic responses to foods, antibiotics increase allergen sensitization, and transplantation of gut microbiota from healthy infants can protect against food allergies in mice236. In the latter FMT study, the bacterial species Anaerostipes caccae was found to be protective against food allergy response to cow milk. Another FMT study found that bacteriotherapy with the bacterium Subdoligranulum variabile or a consortium of Bacteroides strains was found to be protective to peanut allergy in mice237. Specifically, the Treg cell pathway MyD88–RORγt was found to be important in protecting against food allergies in mice and was identified to be deficient in infants with food allergies237. Treg cell subsets are induced by the microbial metabolite butyrate238 but butyrate was not found to be responsible for the observed effects in this study237.
Besides immune education, the microbiome could be influencing the effective dose of an allergen by either production or degradation. Specific strains of milk-fermenting probiotics, such as Lacticaseibacillus rhamnosus, improved tolerance to milk, supporting a potential role in allergen degradation239,240. Thus, by assessing the capacity of the microbiome to degrade foods, we might be able to predict the spontaneous resolution of an allergy or identify individuals who might benefit from microbiota-based therapies.
Atopy and asthma
Disruption of the gut microbiota in early life, such as that associated with caesarean section delivery and growing up in a ‘clean’ environment with decreased microbial exposure, has been associated with an increased risk of developing atopy and asthma241–243. Although there are no consistent human microbiota signatures associated with atopy and asthma229,244–254, microbial metabolites might be directly involved in disease development. Circulating levels of the SCFA propionate originating from the gut was found to reduce inflammation in the lung in a manner dependent on free fatty acid receptor 3 (FFAR3) and dendritic cell functioning in mice255. Microbial metabolites might be specifically involved in TH2 reprogramming and hyperactivation, even though not all individuals with asthma have elevated TH2 levels256. A microbial metabolite involved in asthma development, the linoleic acid derivative 12,13-diHOME, was identified through a series of metabolomics and microbial genetics studies257. Birth cohorts with an elevated asthma risk were found to have increased faecal levels of 12,13-diHOME, which was linked to reduced anti-inflammatory cytokine and Treg cell levels in the lungs, indicating hampered immune tolerance257. The bacterial epoxide hydroxylase enzyme responsible for the production of 12,13-diHOME may be inhibited as a therapeutic strategy in the subset of patients with elevated enzyme levels.
The supplementation of breast or formula milk with specific strains or communities has been attempted with the aim of reducing the risk of development of atopy or asthma later in life. One such example is the use of L. rhamnosus GG, which led to an increase in microbial species thought to promote immune tolerance in infants at high risk of developing asthma, although the progression to asthma development and severity of disease later in life was not clearly assessed258. In addition, Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possessed strong allergy-protective properties in mice and could be explored in human cohorts250. To better understand the temporal mechanisms underlying the role of gut microbiota in the development of allergic and atopic disease, longitudinal birth cohorts that track clinical outcomes are necessary. Overall, these studies can inform future efforts aimed at personalizing treatment strategies in allergic conditions based on the gut microbiome (TABLE 1).
Future perspectives
There has been much focus on cause-and-effect when considering the microbiome in health and disease, but it is clear that the gut microbiome can contribute to a disease even if it is not the inciting factor. In fact, the gut microbiome is rarely the sole driver of disease and needs to be considered in the context of systems biology involving host genetics, host physiological responses and the environment. We need to understand where the gut microbiome lies in the complex regulatory framework that predisposes an individual to a disease state. Although the gut microbiome has been associated with many diseases, it has been difficult to quantify the relative amount of this contribution compared with other variables such as host (epi)genetics, proteome or transcriptome. A major challenge in delineating the contributions of different host and microbiome factors is due to the difficulty of separating the effect of host and environmental factors on the microbiome from their effect on host biology independent of the microbiome. Vujkovic-Cvijin et al. investigated an array of lifestyle, physiological and dietary factors that might work to confound microbiome-related studies owing to the vast inter-individual variability259. Studies like this one not only help to further the understanding of the microbiome in relation to health and disease but also how to improve the quality of future work to better move towards understanding the power of the microbiome in precision medicine and individualized treatment. Multi-omics approaches in defined model systems, such as gnotobiotic animals, can also help unravel such contributions from different data layers.
Future studies in large cohorts of well-phenotyped patients will need to be multidimensional, incorporating host and microbial multi-omics as well as the exposome to better understand the relative contribution of the gut microbiome and other data levels to a disease state or treatment efficacy. These studies should also encapsulate the temporal and interpersonal variation by using longitudinal sampling. Longitudinal data analysis has been shown to reduce variation as compared with cross-sectional studies260,261 and can be used to make causal links. As an example, the variation that underlies treatment response can be identified through longitudinal monitoring of treatment-naive cohorts to identify relevant microbiome and host factors that could underlie treatment variability. This approach combined with intelligent in vitro and ex vivo approaches can identify novel factors responsible for therapeutic response, adverse effects and drug metabolism5,262. Such studies will also enable us to determine whether the gut microbiome could potentially serve as a readout of other host-associated factors such as diet, genetics, age, and lifestyle and potentially simplify machine learning algorithms for disease classification and treatment stratification (FIG. 3).
Fig. 3 |. the gut microbiome as a readout of host variables in prediction models.
Computational prediction models using artificial intelligence and machine learning hold great promise for the optimization of clinical management. The predictive value of these models depends on the training data, which ideally includes a range of host measurements (left panel). However, determining all these parameters for every patient might be prohibitive in cost and time. As the gut microbiome responds to most of the lifestyle factors it could serve as a valuable abstracted readout that is simple to access and measure. The most relevant microbiome features could be identified in the comprehensive large-scale training cohort (left), allowing the use of the microbiome alone as a predictor of clinical outcomes.
As noted already, describing entire genera as being beneficial or harmful can be misleading and higher resolution techniques should be used when profiling individual microbiomes. A good example of such dichotomy is Prevotella, which has been hailed both as the ‘poison’ as well as the elixir in RA. Microbial functions might be more relevant targets given the generally stronger associations of microbial metabolite levels to disease compared with taxonomic abundances. In addition, functional characteristics of the microbiome are more conserved among individuals. It might therefore be that ‘missing microbial functions’ is a more useful concept for precision treatment than the popularized ‘missing microbes’ concept263. At the same time, it is important to note that more than one microbial function can underlie a disease state, as seen regarding susceptibility to CDI24,25,264–266. Hence, future therapies need to be based on better mechanistic understanding and address a missing function or functions, realizing that there might be different functional abnormalities in different individuals.
As we see continued strong momentum in the field with an increasing number of studies, it can be tempting to run before we can walk. We need to be careful and realize that we are still in the early stages of investigation and there remain major challenges to address before we can translate knowledge of the microbiome into accessible measures that can be used in the clinic to benefit patients. Some of these challenges include a lack of validation of microbiome-based markers across large clinical cohorts, lack of standardization in processing and analysis of microbiome data, and a lack of individualized understanding of microbiome-related mechanisms in different disease states. Despite these shortcomings and challenges, the rapid pace of the field and progress made over the past decade brings optimism that the microbiome will be a part of clinical practice sooner rather than later. We can start to envision how the microbiome can be incorporated in personalized treatment strategies and move beyond the approach of a continuum of treatment strategies for all patients to selecting the best strategy for each patient.
Conclusions
In this Review, we highlight the major advances in our understanding of the role of the gut microbiome in the development, progression and treatment of different disease states. Together, this evidence clearly highlights the gut microbiome’s promise in precision medicine and individualized treatment. While there has been major progress, scientific rigor and the strength of findings supporting a role for the gut microbiome vary between disease states as summarized in this Review (FIG. 1; Supplementary Fig. 1). Although the majority of human studies have focused on compositional analysis to identify microbial biomarkers of response, deeper mechanistic understanding will be required to develop precision therapeutics and stratify treatments. Overall, the field has realized this need and has shifted towards studying the functional aspects of the microbiome. Although a non-targeted ecosystem-based approach shows benefit in CDI, the efficacy of such untargeted measures might not extend to multifactorial chronic diseases in which a multitude of mechanisms are at play.
Supplementary Material
Key poins.
The gut microbiome, with substantially greater genetic diversity than the host, is an important factor in determining the variability in disease development, progression and treatment response.
Tremendous progress has been made in characterizing the microbiome and its influence on biology.
Stratifying to species or strain level is important as microorganisms within the same genus might have a differing effect on the same disease process; the same organism might also have different effects on separate disease processes, making the definition of a universal ‘healthy’ microbiota based on composition alone difficult.
To incorporate the microbiome in the clinic, large patient cohorts with multidimensional and longitudinal analyses are needed to understand the contribution of the microbiome on disease development, progression and treatment in the context of systems biology.
The gut microbiome is an important component in personalized medicine; most of the progress has been in metabolic and cardiovascular disorders as well as in cancer therapies.
The gut microbiome is influenced by numerous factors (including age, diet and host genetics) and hence serves as a readout for those factors, simplifying the input for machine learning-based models in clinical practice being developed to predict disease outcomes or treatment response.
Acknowledgements
The authors thank L. Busby for her secretarial assistance. This work is supported by funding from NIH DK114007, Center for Individualized Medicine and Department of Medicine Mayo Clinic, Rochester, MN, USA.
Footnotes
Competing interests
P.C.K. serves on the advisory board of Novome Biotechnologies and is an ad hoc consultant for Otsuka Pharmaceuticals, Pendulum Therapeutics and IP group. The other authors declare no competing interests.
Peer review information
Nature Reviews Gastroenterology & Hepatology thanks M. Nieuwdorp, O. Pedersen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Supplementary information
The online version contains supplementary material available at https://doi.org/10.1038/s41575-021-00499-1.
References
- 1.Kashyap PC, Chia N, Nelson H, Segal E & Elinav E Microbiome at the frontier of personalized medicine. Mayo Clin. Proc. 92, 1855–1864 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jameson JL & Longo DL Precision medicine–personalized, problematic, and promising. N. Engl. J. Med. 372, 2229–2234 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Gilbert JA et al. Current understanding of the human microbiome. Nat. Med. 24, 392–400 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ejtahed HS, Hasani-Ranjbar S & Larijani B Human microbiome as an approach to personalized medicine. Altern. Ther. Health Med. 23, 8–9 (2017). [PubMed] [Google Scholar]
- 5.Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R & Goodman AL Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature 570, 462–467 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckburg PB et al. Diversity of the human intestinal microbial flora. Science 308, 1635–1638 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schubert AM et al. Microbiome data distinguish patients with Clostridium difficile infection and non-C. difficile-associated diarrhea from healthy controls. mBio 5, e01021–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marchesi JR et al. The gut microbiota and host health: a new clinical frontier. Gut 65, 330–339 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vindigni SM & Surawicz CM Fecal microbiota transplantation. Gastroenterol. Clin. North. Am. 46, 171–185 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Hill C et al. Expert consensus document. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 11, 506–514 (2014). [DOI] [PubMed] [Google Scholar]
- 11.Gibson GR et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 14, 491–502 (2017). [DOI] [PubMed] [Google Scholar]
- 12.Swanson KS et al. The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of synbiotics. Nat. Rev. Gastroenterol. Hepatol. 17, 687–701 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cully M Microbiome therapeutics go small molecule. Nat. Rev. Drug. Discov. 18, 569–572 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Wong AC & Levy M New approaches to microbiome-based therapies. mSystems 4, e00122–19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lessa FC et al. Burden of Clostridium difficile infection in the United States. N. Engl. J. Med. 372, 825–834 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McDonald LC et al. Clinical practice guidelines for clostridium difficile infection in adults and children: 2017 update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA). Clin. Infect. Dis. 66, e1–e48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Song JH & Kim YS Recurrent Clostridium difficile infection: risk factors, treatment, and prevention. Gut Liver 13, 16–24 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antharam V,C. et al. Intestinal dysbiosis and depletion of butyrogenic bacteria in Clostridium difficile infection and nosocomial diarrhea. J. Clin. Microbiol. 51, 2884–2892 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Battaglioli EJ et al. Clostridioides difficile uses amino acids associated with gut microbial dysbiosis in a subset of patients with diarrhea. Sci Transl Med 10, eaam7019 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng KM et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferreyra JA et al. Gut microbiota-produced succinate promotes C. difficile infection after antibiotic treatment or motility disturbance. Cell Host Microbe 16, 770–777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buffie CG et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McDonald JAK et al. Inhibiting growth of Clostridioides difficile by restoring valerate, produced by the intestinal microbiota. Gastroenterology 155, 1495–1507.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho J et al. Clostridioides difficile whole genome sequencing differentiates relapse with the same strain from reinfection with a new strain. Clin. Infect. Dis. 72, 806–813 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Boyd CD & O’Toole GA Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annu. Rev. Cell Dev. Biol. 28, 439–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liubakka A & Vaughn BP Clostridium difficile infection and fecal microbiota transplant. AACN Adv. Crit. Care 27, 324–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tariq R, Saha S, Solanky D, Pardi DS & Khanna S Predictors and management of failed fecal microbiota transplantation for recurrent Clostridioides difficile infection. J. Clin. Gastroenterol. 55, 542–547 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Kelly C, et al R. Fecal microbiota transplant is highly effective in real-world practice: initial results from the FMT National Registry. Gastroenterology 160, 183–192.e3 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeFilipp Z et al. Drug-resistant E. coli bacteremia transmitted by fecal microbiota transplant. N. Engl. J. Med. 381, 2043–2050 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Khanna S. et al. Gut microbiome predictors of treatment response and recurrence in primary Clostridium difficile infection. Aliment. Pharmacol. Ther. 44, 715–727 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seekatz AM & Young VB Clostridium difficile and the microbiota. J. Clin. Invest. 124, 4182–4189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blount KF, Shannon WD, Deych E & Jones C Restoration of bacterial microbiome composition and diversity among treatment responders in a phase 2 trial of RBX2660: an investigational microbiome restoration therapeutic. Open. Forum Infect. Dis. 6, ofz095 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.US National Library of Medicine. ClinicalTrials.gov https://ClinicalTrials.gov/show/NCT03110133. [DOI] [PubMed]
- 34.US National Library of Medicine. ClinicalTrials.gov https://ClinicalTrials.gov/show/NCT03244644. [DOI] [PubMed]
- 35.Gosálbez L The microbiome biotech landscape: an analysis of the pharmaceutical pipeline. Microbiome Times; https://www.microbiometimes.com/the-microbiome-biotech-landscape-an-analysis-of-the-pharmaceutical-pipeline/ (2020). [Google Scholar]
- 36.Zitvogel L, Ayyoub M, Routy B & Kroemer G Microbiome and anticancer immunosurveillance. Cell 165, 276–287 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Zitvogel L, Daillere R, Roberti MP, Routy B & Kroemer G Anticancer effects of the microbiome and its products. Nat. Rev. Microbiol. 15, 465–478 (2017). [DOI] [PubMed] [Google Scholar]
- 38.Kadosh E et al. The gut microbiome switches mutant p53 from tumour-suppressive to oncogenic. Nature 586, 133–138 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Raskov H, Burcharth J & Pommergaard HC Linking gut microbiota to colorectal cancer. J. Cancer 8, 3378–3395 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pleguezuelos-Manzano C et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature 580, 269–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riquelme E et al. Tumor microbiome diversity and composition influence pancreatic cancer outcomes. Cell 178, 795–806.e12 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alexander JL et al. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 14, 356–365 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Schiavoni G et al. Cyclophosphamide synergizes with type I interferons through systemic dendritic cell reactivation and induction of immunogenic tumor apoptosis. Cancer Res. 71, 768–778 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Viaud S et al. Cyclophosphamide induces differentiation of Th17 cells in cancer patients. Cancer Res. 71, 661–665 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Viaud S et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 342, 971–976 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Iida N et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 342, 967–970 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Panebianco C, Andriulli A & Pazienza V Pharmacomicrobiomics: exploiting the drug-microbiota interactions in anticancer therapies. Microbiome 6, 92 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Routy B et al. Gut microbiome influences efficacy of PD-1-based immunotherapy against epithelial tumors. Science 359, 91–97 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Derosa L et al. Negative association of antibiotics on clinical activity of immune checkpoint inhibitors in patients with advanced renal cell and non-small-cell lung cancer. Ann. Oncol. 29, 1437–1444 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vetizou M et al. Anticancer immunotherapy by CTLA-4 blockade relies on the gut microbiota. Science 350, 1079–1084 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mager LF et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 369, 1481–1489 (2020). [DOI] [PubMed] [Google Scholar]
- 52.Sivan A et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 350, 1084–1089 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Matson V et al. The commensal microbiome is associated with anti-PD-1 efficacy in metastatic melanoma patients. Science 359, 104–108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gopalakrishnan V et al. Gut microbiome modulates response to anti-PD-1 immunotherapy in melanoma patients. Science 359, 97–103 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fu ZD, Selwyn FP, Cui JY & Klaassen CD RNA-Seq profiling of intestinal expression of xenobiotic processing genes in germ-free mice. Drug. Metab. Dispos. 45, 1225–1238 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nichols RG, Peters JM & Patterson AD Interplay between the host, the human microbiome, and drug metabolism. Hum. Genomics 13, 27 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Diasio RB Sorivudine and 5-fluorouracil; a clinically significant drug-drug interaction due to inhibition of dihydropyrimidine dehydrogenase. Br. J. Clin. Pharmacol. 46, 1–4 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakayama H et al. Intestinal anaerobic bacteria hydrolyse sorivudine, producing the high blood concentration of 5-(E)-(2-bromovinyl)uracil that increases the level and toxicity of 5-fluorouracil. Pharmacogenetics 7, 35–43 (1997). [DOI] [PubMed] [Google Scholar]
- 59.Wallace BD et al. Alleviating cancer drug toxicity by inhibiting a bacterial enzyme. Science 330, 831–835 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kodawara T et al. The inhibitory effect of ciprofloxacin on the beta-glucuronidase-mediated deconjugation of the irinotecan metabolite SN-38-G. Basic Clin. Pharmacol. Toxicol. 118, 333–337 (2016). [DOI] [PubMed] [Google Scholar]
- 61.Mego M et al. Prevention of irinotecan induced diarrhea by probiotics: a randomized double blind, placebo controlled pilot study. Complement. Ther. Med. 23, 356–362 (2015). [DOI] [PubMed] [Google Scholar]
- 62.Bhatt AP et al. Targeted inhibition of gut bacterial beta-glucuronidase activity enhances anticancer drug efficacy. Proc. Natl Acad. Sci. USA 117, 7374–7381 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yamamoto K et al. Relationship between adverse events and microbiomes in advanced hepatocellular carcinoma patients treated with sorafenib. Anticancer. Res. 40, 665–676 (2020). [DOI] [PubMed] [Google Scholar]
- 64.Ianiro G et al. Faecal microbiota transplantation for the treatment of diarrhoea induced by tyrosine-kinase inhibitors in patients with metastatic renal cell carcinoma. Nat. Commun. 11, 4333 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Whidbey C et al. A probe-enabled approach for the selective isolation and characterization of functionally active subpopulations in the gut microbiome. J. Am. Chem. Soc. 141, 42–47 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hales CM, Carroll MD, Fryar CD & Ogden CL Prevalence of obesity and severe obesity among adults: United States, 2017–2018. NCHS Data Brief 360, 1–8 (2020). [PubMed] [Google Scholar]
- 67.Sze MA & Schloss PD Looking for a signal in the noise: revisiting obesity and the microbiome. mBio 7, e01018–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ley RE et al. Obesity alters gut microbial ecology. Proc. Natl Acad. Sci. USA 102, 11070–11075 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ley RE Obesity and the human microbiome. Curr. Opin. Gastroenterol. 26, 5–11 (2010). [DOI] [PubMed] [Google Scholar]
- 70.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Waldram A et al. Top-down systems biology modeling of host metabotype-microbiome associations in obese rodents. J. Proteome Res. 8, 2361–2375 (2009). [DOI] [PubMed] [Google Scholar]
- 72.Turnbaugh PJ, Backhed F, Fulton L & Gordon JI Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 3, 213–223 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ridaura VK et al. Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341, 1241214 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Turnbaugh PJ et al. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci. Transl Med. 1, 6ra14 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Backhed F et al. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl Acad. Sci. USA 101, 15718–15723 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Turnbaugh PJ et al. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031 (2006). [DOI] [PubMed] [Google Scholar]
- 77.Scheithauer TP, Dallinga-Thie GM, de Vos WM, Nieuwdorp M & van Raalte DH Causality of small and large intestinal microbiota in weight regulation and insulin resistance. Mol. Metab. 5, 759–770 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cani PD et al. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56, 1761–1772 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Belizario JE, Faintuch J & Garay-Malpartida M Gut microbiome dysbiosis and immunometabolism: new frontiers for treatment of metabolic diseases. Mediators Inflamm. 2018, 2037838 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Clarke G et al. Minireview: gut microbiota: the neglected endocrine organ. Mol. Endocrinol. 28, 1221–1238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Davis CD The gut microbiome and its role in obesity. Nutr. Today 51, 167–174 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ejtahed HS, Angoorani P, Soroush AR & Atlasi R Probiotics supplementation for the obesity management; a systematic review of animal studies and clinical trials. Funct. Foods 52, 228–242 (2019). [Google Scholar]
- 83.Cerdo T, Garcia-Santos JA, M, G. B. & Campoy C The role of probiotics and prebiotics in the prevention and treatment of obesity. Nutrients 11, 635 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.David LA et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 505, 559–563 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Smits SA, Marcobal A, Higginbottom S, Sonnenburg JL & Kashyap PC Individualized responses of gut microbiota to dietary intervention modeled in humanized mice. mSystems 1, e00098–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kovatcheva-Datchary P et al. Simplified intestinal microbiota to study microbe-diet-host interactions in a mouse model. Cell Rep. 26, 3772–3783.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Muniz Pedrogo DA et al. Gut microbial carbohydrate metabolism hinders weight loss in overweight adults undergoing lifestyle intervention with a volumetric diet. Mayo Clin. Proc. 93, 1104–1110 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Korem T et al. Bread affects clinical parameters and induces gut microbiome-associated personal glycemic responses. Cell Metab. 25, 1243–1253.e5 (2017). [DOI] [PubMed] [Google Scholar]
- 89.Zeevi D et al. Personalized nutrition by prediction of glycemic responses. Cell 163, 1079–1094 (2015). [DOI] [PubMed] [Google Scholar]
- 90.Berry SE et al. Human postprandial responses to food and potential for precision nutrition. Nat. Med. 26, 964–973 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Jumpertz R et al. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am. J. Clin. Nutr. 94, 58–65 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kolodziejczyk AA, Zheng D & Elinav E Diet-microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 17, 742–753 (2019). [DOI] [PubMed] [Google Scholar]
- 93.Korpela K et al. Gut microbiota signatures predict host and microbiota responses to dietary interventions in obese individuals. PLoS ONE 9, e90702 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zmora N, Suez J & Elinav E You are what you eat: diet, health and the gut microbiota. Nat. Rev. Gastroenterol. Hepatol. 16, 35–56 (2019). [DOI] [PubMed] [Google Scholar]
- 95.Zhao L et al. A glucagon-like peptide-1 receptor agonist lowers weight by modulating the structure of gut microbiota. Front. Endocrinol. 9, 233 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ejtahed HS et al. Adaptation of human gut microbiota to bariatric surgeries in morbidly obese patients: a systematic review. Microb. Pathog. 116, 13–21 (2018). [DOI] [PubMed] [Google Scholar]
- 97.Liu R et al. Gut microbiome and serum metabolome alterations in obesity and after weight-loss intervention. Nat. Med. 23, 859–868 (2017). [DOI] [PubMed] [Google Scholar]
- 98.Plovier H et al. A purified membrane protein from Akkermansia muciniphila or the pasteurized bacterium improves metabolism in obese and diabetic mice. Nat. Med. 23, 107–113 (2017). [DOI] [PubMed] [Google Scholar]
- 99.Depommier C et al. Supplementation with Akkermansia muciniphila in overweight and obese human volunteers: a proof-of-concept exploratory study. Nat. Med. 25, 1096–1103 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Goodrich JK et al. Human genetics shape the gut microbiome. Cell 159, 789–799 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.US National Library of Medicine. ClinicalTrials.gov https://ClinicalTrials.gov/show/NCT04663139. [DOI] [PubMed]
- 102.Choi BS, Daoust L, Pilon G, Marette A & Tremblay A Potential therapeutic applications of the gut microbiome in obesity: from brain function to body detoxification. Int. J. Obes. 44, 1818–1831 (2020). [DOI] [PubMed] [Google Scholar]
- 103.Forslund K et al. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 528, 262–266 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Mandic AD et al. Clostridium ramosum regulates enterochromaffin cell development and serotonin release. Sci. Rep. 9, 1177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Karlsson FH et al. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 498, 99–103 (2013). [DOI] [PubMed] [Google Scholar]
- 106.de Groot P et al. Donor metabolic characteristics drive effects of faecal microbiota transplantation on recipient insulin sensitivity, energy expenditure and intestinal transit time. Gut 69, 502–512 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Vrieze A et al. Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143, 913–916.e7 (2012). [DOI] [PubMed] [Google Scholar]
- 108.Zhao L et al. Gut bacteria selectively promoted by dietary fibers alleviate type 2 diabetes. Science 359, 1151–1156 (2018). [DOI] [PubMed] [Google Scholar]
- 109.Pedersen HK et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 535, 376–381 (2016). [DOI] [PubMed] [Google Scholar]
- 110.Koh A et al. Microbially produced imidazole propionate impairs insulin signaling through mTORC1. Cell 175, 947–961.e17 (2018). [DOI] [PubMed] [Google Scholar]
- 111.Home P., Mant J., Diaz J. & Turner C., Guideline Development Group. Management of type 2 diabetes: summary of updated NICE guidance. BMJ 336, 1306–1308 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Mendes-Soares H et al. Assessment of a personalized approach to predicting postprandial glycemic responses to food among individuals without diabetes. JAMA Netw. Open. 2, e188102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rena G, Hardie DG & Pearson ER The mechanisms of action of metformin. Diabetologia 60, 1577–1585 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Shin NR et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735 (2014). [DOI] [PubMed] [Google Scholar]
- 115.Zhang X et al. Modulation of gut microbiota by berberine and metformin during the treatment of high-fat diet-induced obesity in rats. Sci. Rep. 5, 14405 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee H & Ko G Effect of metformin on metabolic improvement and gut microbiota. Appl. Env. Microbiol. 80, 5935–5943 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Matheus VA, Monteiro L, Oliveira RB, Maschio DA & Collares-Buzato CB Butyrate reduces high-fat diet-induced metabolic alterations, hepatic steatosis and pancreatic beta cell and intestinal barrier dysfunctions in prediabetic mice. Exp. Biol. Med. 242, 1214–1226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Croset M et al. Rat small intestine is an insulin-sensitive gluconeogenic organ. Diabetes 50, 740–746 (2001). [DOI] [PubMed] [Google Scholar]
- 119.De Vadder F et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 156, 84–96 (2014). [DOI] [PubMed] [Google Scholar]
- 120.Bryrup T et al. Metformin-induced changes of the gut microbiota in healthy young men: results of a nonblinded, one-armed intervention study. Diabetologia 62, 1024–1035 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Madsen MSA et al. Metabolic and gut microbiome changes following GLP-1 or dual GLP-1/GLP-2 receptor agonist treatment in diet-induced obese mice. Sci. Rep. 9, 15582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Perraudeau F et al. Improvements to postprandial glucose control in subjects with type 2 diabetes: a multicenter, double blind, randomized placebo-controlled trial of a novel probiotic formulation. BMJ Open Diabetes Res. Care 8, e001319 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Tripathi A et al. The gut-liver axis and the intersection with the microbiome. Nat. Rev. Gastroenterol. Hepatol. 15, 397–411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mouzaki M et al. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 58, 120–127 (2013). [DOI] [PubMed] [Google Scholar]
- 125.Mohammadi Z et al. Fecal microbiota in non-alcoholic fatty liver disease and non-alcoholic steatohepatitis: a systematic review. Arch. Iran. Med. 23, 44–52 (2020). [PubMed] [Google Scholar]
- 126.Leung C, Rivera L, Furness JB & Angus PW The role of the gut microbiota in NAFLD. Nat. Rev. Gastroenterol. Hepatol. 13, 412–425 (2016). [DOI] [PubMed] [Google Scholar]
- 127.Le Roy T et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut 62, 1787–1794 (2013). [DOI] [PubMed] [Google Scholar]
- 128.Zhu L et al. Characterization of gut microbiomes in nonalcoholic steatohepatitis (NASH) patients: a connection between endogenous alcohol and NASH. Hepatology 57, 601–609 (2013). [DOI] [PubMed] [Google Scholar]
- 129.Dawes EA & Foster SM The formation of ethanol in Escherichia coli. Biochim. Biophys. Acta 22, 253–265 (1956). [DOI] [PubMed] [Google Scholar]
- 130.Malik F, Wickremesinghe P & Saverimuttu J Case report and literature review of auto-brewery syndrome: probably an underdiagnosed medical condition. BMJ Open. Gastroenterol. 6, e000325 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Brandt A et al. Metformin attenuates the onset of non-alcoholic fatty liver disease and affects intestinal microbiota and barrier in small intestine. Sci. Rep. 9, 6668 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Y, Liu L, Wang B, Wang J & Chen D Metformin in non-alcoholic fatty liver disease: a systematic review and meta-analysis. Biomed. Rep. 1, 57–64 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma J, Zhou Q & Li H Gut microbiota and nonalcoholic fatty liver disease: insights on mechanisms and therapy. Nutrients 9, 1124 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zhou D et al. Total fecal microbiota transplantation alleviates high-fat diet-induced steatohepatitis in mice via beneficial regulation of gut microbiota. Sci. Rep. 7, 1529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Garcia-Lezana T et al. Restoration of a healthy intestinal microbiota normalizes portal hypertension in a rat model of nonalcoholic steatohepatitis. Hepatology 67, 1485–1498 (2018). [DOI] [PubMed] [Google Scholar]
- 136.Craven L et al. Allogenic fecal microbiota transplantation in patients with nonalcoholic fatty liver disease improves abnormal small intestinal permeability: a randomized control trial. Am. J. Gastroenterol. 115, 1055–1065 (2020). [DOI] [PubMed] [Google Scholar]
- 137.Witjes JJ et al. Donor fecal microbiota transplantation alters gut microbiota and metabolites in obese individuals with steatohepatitis. Hepatol. Commun. 4, 1578–1590 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.US National Library of Medicine. ClinicalTrials.gov https://ClinicalTrials.gov/show/NCT02469272. [DOI] [PubMed]
- 139.US National Library of Medicine. ClinicalTrials.gov https://ClinicalTrials.gov/show/NCT03803540. [DOI] [PubMed]
- 140.Alisi A et al. Randomised clinical trial: The beneficial effects of VSL#3 in obese children with non-alcoholic steatohepatitis. Aliment. Pharmacol. Ther. 39, 1276–1285 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Malaguarnera M et al. Bifidobacterium longum with fructo-oligosaccharides in patients with non alcoholic steatohepatitis. Dig. Dis. Sci. 57, 545–553 (2012). [DOI] [PubMed] [Google Scholar]
- 142.Wong VW et al. Treatment of nonalcoholic steatohepatitis with probiotics. A proof-of-concept study. Ann. Hepatol. 12, 256–262 (2013). [PubMed] [Google Scholar]
- 143.Wong VW et al. Molecular characterization of the fecal microbiota in patients with nonalcoholic steatohepatitis–a longitudinal study. PLoS ONE 8, e62885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Duan Y et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature 575, 505–511 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.GBD 2015 Mortality and Causes of Death Collaborators. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the Global Burden of Disease Study 2015. Lancet 388, 1459–1544 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Capewell S et al. Cardiovascular risk factor trends and potential for reducing coronary heart disease mortality in the United States of America. Bull. World Health Organ. 88, 120–130 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Novakovic M et al. Role of gut microbiota in cardiovascular diseases. World J. Cardiol. 12, 110–122 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Peng J, Xiao X, Hu M & Zhang X Interaction between gut microbiome and cardiovascular disease. Life Sci. 214, 153–157 (2018). [DOI] [PubMed] [Google Scholar]
- 149.Organ CL et al. Choline diet and its gut microbe-derived metabolite, trimethylamine n-oxide, exacerbate pressure overload-induced heart failure. Circ. Heart Fail. 9, e002314 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Tang WH et al. Intestinal microbial metabolism of phosphatidylcholine and cardiovascular risk. N. Engl. J. Med. 368, 1575–1584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Martinez-Del Campo A, Romano KA, Rey FE & Balskus EP The plot thickens: diet microbe interactions may modulate thrombosis risk. Cell Metab. 23, 573–575 (2016). [DOI] [PubMed] [Google Scholar]
- 152.Roberts AB et al. Development of a gut microbe-targeted nonlethal therapeutic to inhibit thrombosis potential. Nat. Med. 24, 1407–1417 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Koeth RA et al. Intestinal microbiota metabolism of L-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 19, 576–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhu Y, Li Q & Jiang H Gut microbiota in atherosclerosis: focus on trimethylamine N-oxide. APMIS 128, 353–366 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Romano KA, Vivas EI, Amador-Noguez D & Rey FE Intestinal microbiota composition modulates choline bioavailability from diet and accumulation of the proatherogenic metabolite trimethylamine-N-oxide. mBio 6, e02481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang Z et al. Non-lethal inhibition of gut microbial trimethylamine production for the treatment of atherosclerosis. Cell 163, 1585–1595 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rath S, Heidrich B, Pieper DH & Vital M Uncovering the trimethylamine-producing bacteria of the human gut microbiota. Microbiome 5, 54 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Jie Z et al. The gut microbiome in atherosclerotic cardiovascular disease. Nat. Commun. 8, 845 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Ahmad AF, Ward NC & Dwivedi G The gut microbiome and heart failure. Curr. Opin. Cardiol. 34, 225–232 (2019). [DOI] [PubMed] [Google Scholar]
- 160.Kamo T et al. Dysbiosis and compositional alterations with aging in the gut microbiota of patients with heart failure. PLoS ONE 12, e0174099 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liyanage T et al. Effects of the Mediterranean diet on cardiovascular outcomes-a systematic review and meta-analysis. PLoS ONE 11, e0159252 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Wang DD et al. The gut microbiome modulates the protective association between a Mediterranean diet and cardiometabolic disease risk. Nat. Med. 27, 333–343 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Marques FZ et al. High-fiber diet and acetate supplementation change the gut microbiota and prevent the development of hypertension and heart failure in hypertensive mice. Circulation 135, 964–977 (2017). [DOI] [PubMed] [Google Scholar]
- 164.Cena H & Calder PC Defining a healthy diet: evidence for the role of contemporary dietary patterns in health and disease. Nutrients 12, 334 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Haiser HJ, Seim KL, Balskus EP & Turnbaugh PJ Mechanistic insight into digoxin inactivation by Eggerthella lenta augments our understanding of its pharmacokinetics. Gut Microbes 5, 233–238 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Spanogiannopoulos P, Bess EN, Carmody RN & Turnbaugh PJ The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat. Rev. Microbiol. 14, 273–287 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Pencina MJ et al. Application of new cholesterol guidelines to a population-based sample. N. Engl. J. Med. 370, 1422–1431 (2014). [DOI] [PubMed] [Google Scholar]
- 168.Iwaki Y, Lee W & Sugiyama Y Comparative and quantitative assessment on statin efficacy and safety: insights into inter-statin and inter-individual variability via dose- and exposure-response relationships. Expert. Opin. Drug Metab. Toxicol. 15, 897–911 (2019). [DOI] [PubMed] [Google Scholar]
- 169.Sun B, Li L & Zhou X Comparative analysis of the gut microbiota in distinct statin response patients in East China. J. Microbiol. 56, 886–892 (2018). [DOI] [PubMed] [Google Scholar]
- 170.Kaddurah-Daouk R et al. Enteric microbiome metabolites correlate with response to simvastatin treatment. PLoS ONE 6, e25482 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.He X et al. Gut microbiota modulation attenuated the hypolipidemic effect of simvastatin in high-fat/cholesterol-diet fed mice. J. Proteome Res. 16, 1900–1910 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Wang L et al. The influence of the intestinal microflora to the efficacy of Rosuvastatin. Lipids Health Dis. 17, 151 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Yoo DH et al. Gut microbiota-mediated drug interactions between lovastatin and antibiotics. Drug. Metab. Dispos. 42, 1508–1513 (2014). [DOI] [PubMed] [Google Scholar]
- 174.Liu Y et al. Gut microbiome associates with lipid-lowering effect of rosuvastatin in vivo. Front. Microbiol. 9, 530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Vieira-Silva S et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature 581, 310–315 (2020). [DOI] [PubMed] [Google Scholar]
- 176.Mayerhofer CCK et al. Design of the GutHeart-targeting gut microbiota to treat heart failure-trial: a phase II, randomized clinical trial. ESC Heart Fail. 5, 977–984 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.US National Library of Medicine. ClinicalTrials.gov https://ClinicalTrials.gov/show/NCT03968549. [DOI] [PubMed]
- 178.Tang WHW, Li DY & Hazen SL Dietary metabolism, the gut microbiome, and heart failure. Nat. Rev. Cardiol. 16, 137–154 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Vaahtovuo J, Munukka E, Korkeamaki M, Luukkainen R & Toivanen P Fecal microbiota in early rheumatoid arthritis. J. Rheumatol. 35, 1500–1505 (2008). [PubMed] [Google Scholar]
- 180.Bodkhe R, Balakrishnan B & Taneja V The role of microbiome in rheumatoid arthritis treatment. Ther Adv Musculoskelet Dis 11, 1759720X19844632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Marietta EV et al. Suppression of inflammatory arthritis by human gut-derived prevotella histicola in humanized mice. Arthritis Rheumatol. 68, 2878–2888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Maeda Y et al. Dysbiosis contributes to arthritis development via activation of autoreactive T cells in the intestine. Arthritis Rheumatol. 68, 2646–2661 (2016). [DOI] [PubMed] [Google Scholar]
- 183.Maeda Y & Takeda K Host-microbiota interactions in rheumatoid arthritis. Exp. Mol. Med. 51, 1–6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Lloyd-Price J et al. Multi-omics of the gut microbial ecosystem in inflammatory bowel diseases. Nature 569, 655–662 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Picchianti-Diamanti A et al. Analysis of gut microbiota in rheumatoid arthritis patients: disease-related dysbiosis and modifications induced by etanercept. Int. J. Mol. Sci. 19, 2938 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Chen J et al. An expansion of rare lineage intestinal microbes characterizes rheumatoid arthritis. Genome Med. 8, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Zhang X et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat. Med. 21, 895–905 (2015). [DOI] [PubMed] [Google Scholar]
- 188.Artach A. et al. The pre-treatment gut microbiome is associated with lack of response to methotrexate in new onset rheumatoid arthritis. Arthritis Rheumatol. 73, 931–942 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nayak RR et al. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host Microbe 29, 362–377.e11 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Sayers E, MacGregor A & Carding SR Drug-microbiota interactions and treatment response: Relevance to rheumatoid arthritis. AIMS Microbiol. 4, 642–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Ince A, Yazici Y, Hamuryudan V & Yazici H The frequency and clinical characteristics of methotrexate (MTX) oral toxicity in rheumatoid arthritis (RA): a masked and controlled study. Clin. Rheumatol. 15, 491–494 (1996). [DOI] [PubMed] [Google Scholar]
- 192.Zhou B et al. Induction and amelioration of methotrexate-induced gastrointestinal toxicity are related to immune response and gut microbiota. EBioMedicine 33, 122–133 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Schrezenmeier E & Dorner T Mechanisms of action of hydroxychloroquine and chloroquine: implications for rheumatology. Nat. Rev. Rheumatol. 16, 155–166 (2020). [DOI] [PubMed] [Google Scholar]
- 194.Scher JU & Abramson SB The microbiome and rheumatoid arthritis. Nat. Rev. Rheumatol. 7, 569–578 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zheng H et al. Modulation of gut microbiome composition and function in experimental colitis treated with sulfasalazine. Front. Microbiol. 8, 1703 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.LoGuidice A, Wallace BD, Bendel L, Redinbo MR & Boelsterli UA Pharmacologic targeting of bacterial beta-glucuronidase alleviates nonsteroidal anti-inflammatory drug-induced enteropathy in mice. J. Pharmacol. Exp. Ther. 341, 447–454 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Saitta KS et al. Bacterial beta-glucuronidase inhibition protects mice against enteropathy induced by indomethacin, ketoprofen or diclofenac: mode of action and pharmacokinetics. Xenobiotica 44, 28–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Clayton TA, Baker D, Lindon JC, Everett JR & Nicholson JK Pharmacometabonomic identification of a significant host-microbiome metabolic interaction affecting human drug metabolism. Proc. Natl Acad. Sci. USA 106, 14728–14733 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.So JS et al. Lactobacillus casei suppresses experimental arthritis by down-regulating T helper 1 effector functions. Mol. Immunol. 45, 2690–2699 (2008). [DOI] [PubMed] [Google Scholar]
- 200.Mandel DR, Eichas K & Holmes J Bacillus coagulans: a viable adjunct therapy for relieving symptoms of rheumatoid arthritis according to a randomized, controlled trial. BMC Complement. Altern. Med. 10, 1 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Lopez J & Grinspan A Fecal microbiota transplantation for inflammatory bowel disease. Gastroenterol. Hepatol. 12, 374–379 (2016). [PMC free article] [PubMed] [Google Scholar]
- 202.Singh S et al. Systematic review with meta-analysis: faecal diversion for management of perianal Crohn’s disease. Aliment. Pharmacol. Ther. 42, 783–792 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Nitzan O, Elias M, Peretz A & Saliba W Role of antibiotics for treatment of inflammatory bowel disease. World J. Gastroenterol. 22, 1078–1087 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Chassaing B & Darfeuille-Michaud A The commensal microbiota and enteropathogens in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140, 1720–1728 (2011). [DOI] [PubMed] [Google Scholar]
- 205.Britton GJ et al. Defined microbiota transplant restores Th17/RORγt+ regulatory T cell balance in mice colonized with inflammatory bowel disease microbiotas. Proc. Natl Acad. Sci. USA 117, 21536–21545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Machiels K et al. A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63, 1275–1283 (2014). [DOI] [PubMed] [Google Scholar]
- 207.Sokol H et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc. Natl Acad. Sci. USA 105, 16731–16736 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kumar M, Garand M & Al Khodor S Integrating omics for a better understanding of inflammatory bowel disease: a step towards personalized medicine. J. Transl Med. 17, 419 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Negroni A et al. Characterization of adherent-invasive Escherichia coli isolated from pediatric patients with inflammatory bowel disease. Inflamm. Bowel Dis. 18, 913–924 (2012). [DOI] [PubMed] [Google Scholar]
- 210.Campos N et al. Macrophages from IBD patients exhibit defective tumour necrosis factor-alpha secretion but otherwise normal or augmented pro-inflammatory responses to infection. Immunobiology 216, 961–970 (2011). [DOI] [PubMed] [Google Scholar]
- 211.Sasaki M et al. Invasive Escherichia coli are a feature of Crohn’s disease. Lab. Invest. 87, 1042–1054 (2007). [DOI] [PubMed] [Google Scholar]
- 212.Zeng MY, Inohara N & Nunez G Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 10, 18–26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Gevers D et al. The treatment-naive microbiome in new-onset Crohn’s disease. Cell Host Microbe 15, 382–392 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Scales BS, Dickson RP & Huffnagle GB A tale of two sites: how inflammation can reshape the microbiomes of the gut and lungs. J. Leukoc. Biol. 100, 943–950 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Garsin DA Ethanolamine utilization in bacterial pathogens: roles and regulation. Nat. Rev. Microbiol. 8, 290–295 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fornelos N et al. Growth effects of N-acylethanolamines on gut bacteria reflect altered bacterial abundances in inflammatory bowel disease. Nat. Microbiol. 5, 486–497 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Ni J et al. A role for bacterial urease in gut dysbiosis and Crohn’s disease. Sci. Transl Med. 9, eaah6888 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Lewis JD et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 22, 247 (2017). [DOI] [PubMed] [Google Scholar]
- 219.Martin R et al. Functional characterization of novel faecalibacterium prausnitzii strains isolated from healthy volunteers: a step forward in the use of F. prausnitzii as a next-generation probiotic. Front. Microbiol. 8, 1226 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Ananthakrishnan AN et al. Gut microbiome function predicts response to anti-integrin biologic therapy in inflammatory bowel diseases. Cell Host Microbe 21, 603–610.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Rajca S et al. Alterations in the intestinal microbiome (dysbiosis) as a predictor of relapse after infliximab withdrawal in Crohn’s disease. Inflamm. Bowel Dis. 20, 978–986 (2014). [DOI] [PubMed] [Google Scholar]
- 222.Jeong DY et al. Induction and maintenance treatment of inflammatory bowel disease: a comprehensive review. Autoimmun. Rev. 18, 439–454 (2019). [DOI] [PubMed] [Google Scholar]
- 223.McIlroy J, Ianiro G, Mukhopadhya I, Hansen R & Hold GL Review article: the gut microbiome in inflammatory bowel disease-avenues for microbial management. Aliment. Pharmacol. Ther. 47, 26–42 (2018). [DOI] [PubMed] [Google Scholar]
- 224.Zhang M et al. Faecalibacterium prausnitzii inhibits interleukin-17 to ameliorate colorectal colitis in rats. PLoS ONE 9, e109146 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Huang XL et al. Faecalibacterium prausnitzii supernatant ameliorates dextran sulfate sodium induced colitis by regulating Th17 cell differentiation. World J. Gastroenterol. 22, 5201–5210 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Zhou L et al. Faecalibacterium prausnitzii produces butyrate to maintain Th17/Treg balance and to ameliorate colorectal colitis by inhibiting histone deacetylase 1. Inflamm. Bowel Dis. 24, 1926–1940 (2018). [DOI] [PubMed] [Google Scholar]
- 227.Moayyedi P et al. Fecal microbiota transplantation induces remission in patients with active ulcerative colitis in a randomized controlled trial. Gastroenterology 149, 102–109.e6 (2015). [DOI] [PubMed] [Google Scholar]
- 228.Hill DA & Spergel JM The atopic march: critical evidence and clinical relevance. Ann. Allergy Asthma Immunol. 120, 131–137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Iweala OI & Nagler CR The microbiome and food allergy. Annu. Rev. Immunol. 37, 377–403 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Huang YJ et al. The microbiome in allergic disease: current understanding and future opportunities-2017 PRACTALL document of the American Academy of Allergy, Asthma & Immunology and the European Academy of Allergy and Clinical Immunology. J. Allergy Clin. Immunol. 139, 1099–1110 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Berin MC & Sampson HA Mucosal immunology of food allergy. Curr. Biol. 23, R389–R400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Zhao W, Ho HE & Bunyavanich S The gut microbiome in food allergy. Ann. Allergy Asthma Immunol. 122, 276–282 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Bunyavanich S et al. Early-life gut microbiome composition and milk allergy resolution. J. Allergy Clin. Immunol. 138, 1122–1130 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Fazlollahi M et al. Early-life gut microbiome and egg allergy. Allergy 73, 1515–1524 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Thompson-Chagoyan OC, Vieites JM, Maldonado J, Edwards C & Gil A Changes in faecal microbiota of infants with cow’s milk protein allergy–a Spanish prospective case-control 6-month follow-up study. Pediatr. Allergy Immunol. 21, e394–e400 (2010). [DOI] [PubMed] [Google Scholar]
- 236.Feehley T et al. Healthy infants harbor intestinal bacteria that protect against food allergy. Nat. Med. 25, 448–453 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Abdel-Gadir A et al. Microbiota therapy acts via a regulatory T cell MyD88/RORγt pathway to suppress food allergy. Nat. Med. 25, 1164–1174 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Furusawa Y et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504, 446–450 (2013). [DOI] [PubMed] [Google Scholar]
- 239.Berni Canani R et al. Specific signatures of the gut microbiota and increased levels of butyrate in children treated with fermented cow’s milk containing heat-killed Lactobacillus paracasei CBA L74. Appl Environ Microbiol 83, e01206–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Berni Canani R et al. Effect of Lactobacillus GG on tolerance acquisition in infants with cow’s milk allergy: a randomized trial. J. Allergy Clin. Immunol. 129, 580–582 (2012). [DOI] [PubMed] [Google Scholar]
- 241.Lynch SV & Boushey HA The microbiome and development of allergic disease. Curr. Opin. Allergy Clin. Immunol. 16, 165–171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Fujimura KE et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat. Med. 22, 1187–1191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Stein MM et al. Innate immunity and asthma risk in Amish and Hutterite farm children. N. Engl. J. Med. 375, 411–421 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Olszak T et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science 336, 489–493 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Arrieta MC et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl Med. 7, 307ra152 (2015). [DOI] [PubMed] [Google Scholar]
- 246.Barcik W et al. Histamine-secreting microbes are increased in the gut of adult asthma patients. J. Allergy Clin. Immunol. 138, 1491–1494.e7 (2016). [DOI] [PubMed] [Google Scholar]
- 247.Ege MJ et al. Exposure to environmental microorganisms and childhood asthma. N. Engl. J. Med. 364, 701–709 (2011). [DOI] [PubMed] [Google Scholar]
- 248.Schuijs MJ et al. Farm dust and endotoxin protect against allergy through A20 induction in lung epithelial cells. Science 349, 1106–1110 (2015). [DOI] [PubMed] [Google Scholar]
- 249.Hammad H et al. House dust mite allergen induces asthma via Toll-like receptor 4 triggering of airway structural cells. Nat. Med. 15, 410–416 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Debarry J et al. Acinetobacter lwoffii and Lactococcus lactis strains isolated from farm cowsheds possess strong allergy-protective properties. J. Allergy Clin. Immunol. 119, 1514–1521 (2007). [DOI] [PubMed] [Google Scholar]
- 251.Frati F et al. The role of the microbiome in asthma: the gut–lung axis. Int. J. Mol. Sci. 20, 123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Penders J et al. Gut microbiota composition and development of atopic manifestations in infancy: the KOALA Birth Cohort Study. Gut 56, 661–667 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.van Nimwegen FA et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J. Allergy Clin. Immunol. 128, 948–955.e1–3 (2011). [DOI] [PubMed] [Google Scholar]
- 254.Bjorksten B, Sepp E, Julge K, Voor T & Mikelsaar M Allergy development and the intestinal microflora during the first year of life. J. Allergy Clin. Immunol. 108, 516–520 (2001). [DOI] [PubMed] [Google Scholar]
- 255.Trompette A et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat. Med. 20, 159–166 (2014). [DOI] [PubMed] [Google Scholar]
- 256.Fujimura KE & Lynch SV Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe 17, 592–602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Levan SR et al. Elevated faecal 12,13-diHOME concentration in neonates at high risk for asthma is produced by gut bacteria and impedes immune tolerance. Nat. Microbiol. 4, 1851–1861 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Durack J et al. Delayed gut microbiota development in high-risk for asthma infants is temporarily modifiable by Lactobacillus supplementation. Nat. Commun. 9, 707 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Vujkovic-Cvijin I et al. Host variables confound gut microbiota studies of human disease. Nature 587, 448–454 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Mars RAT et al. Longitudinal multi-omics reveals subset-specific mechanisms underlying irritable bowel syndrome. Cell 182, 1460–1473.e17 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Poyet M et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat. Med. 25, 1442–1452 (2019). [DOI] [PubMed] [Google Scholar]
- 262.Javdan B et al. Personalized mapping of drug metabolism by the human gut microbiome. Cell 181, 1661–1679.e22 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Blaser MJ Missing microbes: how the overuse of antibiotics is fueling our modern plagues. Emerg. Infect. Dis. 20, 1961 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Vuotto C, Moura I, Barbanti F, Donelli G & Spigaglia P Subinhibitory concentrations of metronidazole increase biofilm formation in Clostridium difficile strains. Pathog. Dis. 74, ftv114 (2016). [DOI] [PubMed] [Google Scholar]
- 265.Maldarelli GA et al. Type IV pili promote early biofilm formation by Clostridium difficile. Pathog. Dis. 74, ftw061 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Ethapa T et al. Multiple factors modulate biofilm formation by the anaerobic pathogen Clostridium difficile. J. Bacteriol. 195, 545–555 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.