Abstract
The coronavirus disease that outbreak in 2019 has caused various health issues. According to the WHO, the first positive case was detected in Bangladesh on 7th March 2020, but while writing this paper in June 2021, the total confirmed, recovered, and death cases were 826922, 766266 and 13118, respectively. Due to the emergence of COVID-19 in Bangladesh, the country is facing a major public health crisis. Unfortunately, the country does not have a comprehensive health policy to address this issue. This makes it hard to predict how the pandemic will affect the population. Machine learning techniques can help us detect the disease's spread. To predict the trend, parameters, risks, and to take preventive measure in Bangladesh; this work utilized the Recurrent Neural Networks based Deep Learning methodologies like LongShort-Term Memory. Here, we aim to predict the epidemic's progression for a period of more than a year under various scenarios in Bangladesh. We extracted the data for daily confirmed, recovered, and death cases from March 2020 to August 2021. The obtained Root Mean Square Error (RMSE) values of confirmed, recovered, and death cases indicates that our result is more accurate than other contemporary techniques. This study indicates that the LSTM model could be used effectively in predicting contagious diseases. The obtained results could help in explaining the seriousness of the situation, also mayhelp the authorities to take precautionary steps to control the situation.
Keywords: COVID-19, LSTM, Recurrent neural networks (RNN), Prediction, RMSE
1. Introduction
In December 2019, several patients in China's Hubei province observed pneumonia that resembled viral pneumonia, and several people quickly increased to fatal sickness and dead results. The International Committee on Taxonomy of Viruses has named it severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). In February 11, 2020, the WHO officially declared the COVID-19 disease a global pandemic. It is the third zoonotic virus of the century to become a global pandemic (Gralinski and Menachery, 2020). Seven types of coronaviruses can contaminate humans only. The first among them was discovered in 1965 (Mcintosh et al., 1970). Symptoms were mild and most commonly was only a common cold. In 2002, people from more than 37 countries were effected by (SARS-CoV). In 2012, the Middle East respiratory syndrome Coronavirus was discovered to have originated from bats (Li et al., 2005). Although it was only confined to the region, it was considered a deadlier variant of the SARS-CoV-2 virus (Sheahan et al., 2020). COVID-19 exhibited the usual symptoms of viral pneumonia, such as fever, difficulty in breathing, and coughing. However, compared to SARS and coronavirus, COVID-19 is a worst contagious disease (Arnold, 2014; de Groot et al., 2013; Guan et al., 2003; Wu et al., 2020; Yangand and Wan, 2020).
The SARS-CoV-2 virus has been spreading globally since it was first spotted in China in 2019. It has been identified as a global threat to the scientific community (Wang et al., 2020). The COVID-19 virus is a positive-sense RNA that can mutate frequently to adapt in different conditions. (Kaneko et al., 2006). It can infect humans for about 6.5 days while patients remain asymptomatic or experience little symptom, hence spreading the virus silently (Lai et al., 2020). In Bangladesh, a relatively high rate of young people were affected, and the rate of recovery for elderly people is lower (Institute of Epidemiology, 2020). According to WHO, the cases are rising daily in Asia, the Americas, and Europe (World Health Organization; Lau et al., 2005), and the preventive measures for COVID-19 are maintaining social distancing, avoiding touching the mouth, nose, face, and washing hands repeatedly.
Bangladesh is one of the densely populated countries in the world. As a result, following the detection of the first cases, it spreads to the whole country within a very short period. Following the confirmation of the first few cases of COVID-19 on March 2020, the government of Bangladesh declared a countrywide lockdown until 30th May 2020, followed by the implementation of zone-based lockdown strategy (Ardabili et al., 2020). At the beginning, only one lab, located in Dhaka city, was engaged in testing of COVID-19 symptoms. However, because of the increasing rate of COVID-19 cases, the number of labs has increased to 37 throughout the country. On the other hand, the country shows a lower recovery rate compared to other countries, which further causes the rise in cumulative way and poses a big challenge to the healthcare facilities in the country. Consequently, the lockdown has not been withdrawn completely but been relaxed or allowed to limit the operation of some sectors including export oriented industries, shopping malls, public transports, etc. In the meantime, the government emphasized on the increase of testing, with the hope that the disease could be mapped more accurately, which may help to take the necessary preventive actions. However, the present testing capacity is not sufficient for a country of 170 million people (Anwar et al., 2020). To overcome this uncertain situation, the authorities need to take decisions that may help to maintain a balance between continuing lockdown and stabilizing the economy. However, to take a viable decision, the necessary information on the transmission characteristics of this virus and the resultant effects on live and livelihood need to be known. Meanwhile, like other countries, many researchers in Bangladesh are working to understand how the virus spreads and find ways to put it under control (Thomas et al., 2020; Yousef et al., 2020).
In this situation, the transmission dynamic of this virus needs to be modeled using publicly available data. Such an effort may help to develop competent public health policy and economic activity guidelines, and planning effective control strategies. Literature shows that machine learning (ML) models not only demonstrates effectiveness in predicting the number of COVID-19 cases (Tuli et al., 2020; Wang et al., 2020) but also the number of deaths caused by COVID-19 (Pathan et al., 2020; Chen & Liu, 2020; Rustam et al., 2020). Some ML based studies have already reported from countries like China (Fanelli and Piazza, 2020; Hu et al., 2020; Li et al., 2020; Liu et al., 2020a; Peng et al., 2020; Roosa et al., 2020), Italy (Grasselli et al., 2020; Wangping et al., 2020; Zhan et al., 2020), France (Fanelli and Piazza, 2020; Massonnaud et al., 2020; Peng et al., 2020), USA (Liu et al., 2020b; Lover and McAndrew, 2020), and South Korea (Suzuki and Suzuki, 2020; Wise et al., 2020). Beside this, the fast growing COVID-19 literature includes different types of modeling tools, such as compartmental deterministic models (Anastassopoulou et al., 2020; Chen et al., 2020; Ivorra et al., 2020; Peirlinck et al., 2020; Yang and Wang, 2020), network models (Peirlinck et al., 2020), stochastic models (Kucharski et al., 2020), and branching process models (Bertozzi et al., 2020; Hellewell et al., 2020).
However, in Bangladesh, the studies are very less till now. No significant research has been found in Bangladesh using the latest data. Observations from the few existing studies in the country are as follows:
-
-
Most studies have explored a limited number of machine learning models, so their forecasting capability is restricted and prediction is less independent.
-
-
Bangladesh needs an adaptable forecasting model for a comprehensive study for pulling things together and planning better for future circumstances.
There is an uncertainty when the outbreak of this pandemic will be ended. At the time of writing this manuscript, in Bangladesh, more than 530,271 were confirmed cases and 7966 death cases. The authority is struggling to provide healthcare facilities to COVID-19 patients. Therefore, some predictive models may predict future cases (Tobías, 2020). The COVID-19 data show nonlinear characteristics, therefore the use of linear methods may not be suitable to predict the COVID-19 dynamics (Chimmula & Zhang, 2020). Similarly, since the data are dynamic in nature, the use of statistical and epidemiological models like Moving Average, Auto-Regressive, Auto-Regressive, Moving Average, etc may provide questionable results (Benvenuto et al., 2020; KrätschmerV, 2006; LiestølK, 2002). To overcome such limitations, this study proposes a model for predicting the outbreak of COVID-19 that fits the real data by using a suitable Artificial Intelligence (AI) based techniques.
Artificial Intelligence and mobile computing evolve as a key player for the success of technology in healthcare systems (PanchTMattie and Celi, 2019). In the world of smart devices, data mining has been done in an unprecedented way than ever before (PanchTMattie and Celi, 2019). Today's world is more connected than ever before w.r.t. the sharing of data between countries in real time. Since COVID-19 is a time series dataset, thus it should be appropriate to use sequential networks to extract the correct patterns (Bandyopadhyay Dutta, 2020, Pathan, Biswas, & Khandaker, 2020; Huang et al., 2020; Pal et al., 2020; TomarA, 2020). In this context, we have adopted data-driven Deep Learning-based LSTM model. The proposed methods approximately fit real data, thus the policymakers can take decision considering the COVID-19 phenomenon in the upcoming days.
In this work, we predicted the COVID-19 outbreak based on the available data for the last 1 year. The coherence of input data was first analyzed to find the key parameters like the number of new cases compared with the previous day reported data. Following the selection of key parameters of the network, several trials were made to predict future infections, recovery rate, and death rate with minimum error. Any biasness of algorithm from the design point of view has been reduced by using LSTM networks. It is expected that with the help of predicted data obtained from real-time forecasting tools, the healthcare providers and frontline staffs in Bangladesh can prepare themselves to handle the COVID-19 crisis. The methodology of this work including the datasets and LSTM models work is presented in Section 2, section 3 comprises the results and discussion, and section 4 addresses the conclusion and future work.
2. Methods and models
2.1. Dataset
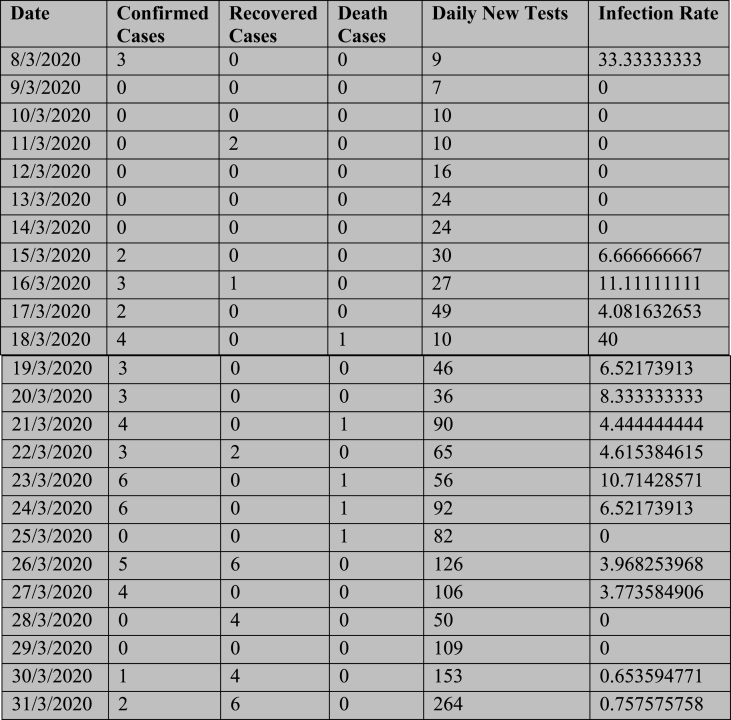
Data related to the SARS – CoV-2 pandemic were released daily by the Health division of the Government of the Republic of Bangladesh. Daily confirmed cases, recovered cases, and death cases of COVID-19 in Bangladesh are collected from the government website. For this study, the data are extracted for the period from March 2020 to August 2021. Dataset is prepared in excel format. This dataset contains 6 columns and 365 rows which is shown in Fig. 1. Starting with the date, the 6 columns include the confirmed cases, recovered cases, death cases, test cases, and infection rate ((test case/confirmed cases)∗ (1/100)). The 365 rows represent 365 days’ data (number of confirmed cases, recovered cases, death cases, test cases, and infection rate).
Fig. 1.
Dataset view.
The dataset is available in the dd-mm-yyyy time series format, and a wavelet transformation (Xu et al., 1994) was applied to mitigate the random noise in the dataset. The conventional function were used to fit the data considering the fundamental point to represent and forecasting the trends. From these data, 70% is used for training the model and 30% is used for testing and validation purposes.
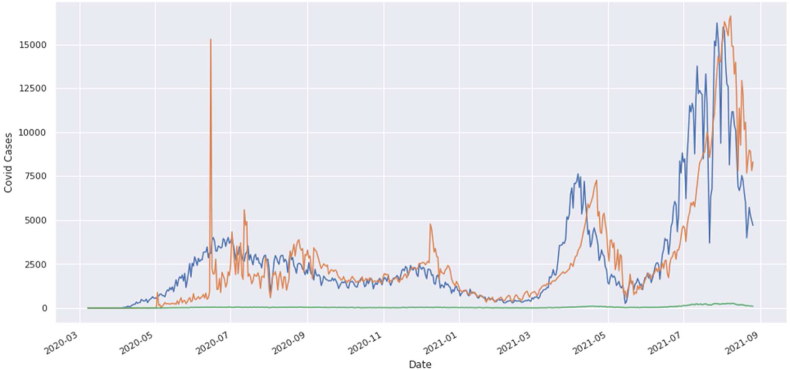
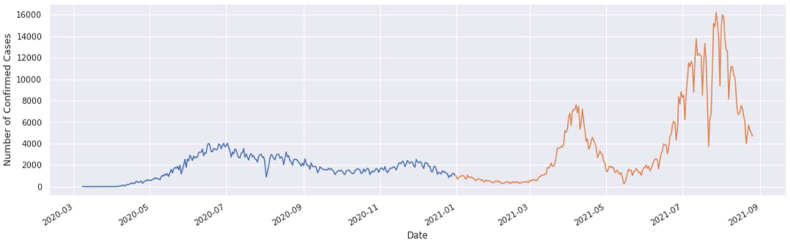
The graphical view of COVID-19 is shown in Fig. 2. In this graph, the green curve indicates the death cases, blue curve indicates the confirmed cases, and the orange curve indicates the recovered cases. This is actually the graphical view of the COVID-19 in Bangladesh until now.
Fig. 2.
Graphical View of COVID-19 data in Bangladesh.
2.2. Methodology
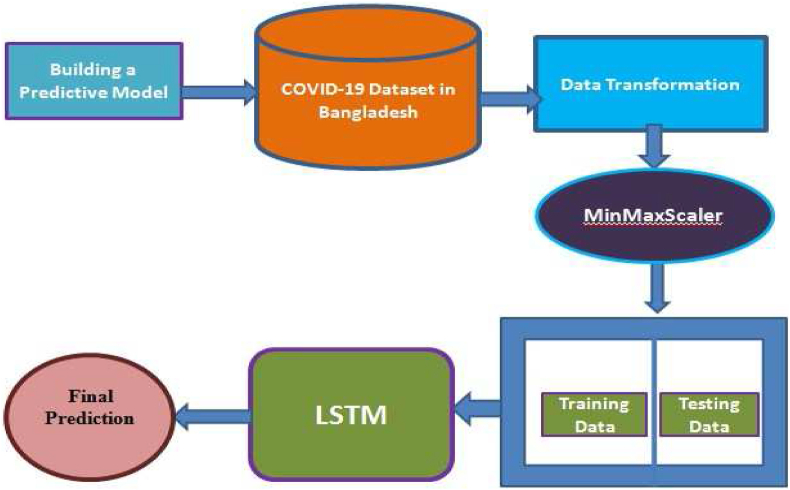
We have used Long Short Term Memory networks for predicting the trend of COVID-19 in Bangladesh. We preprocessed the data using MinMaxScaler. We have added 1 dense layer, 50 hidden layers, and used adam optimizer. For calculating losses, we used Mean_Square_Error. Based on this parameter setting, the LSTM model has been trained using 70% data and the remaining 30% is used for validation. Using the trained model, we have predicted the confirmed cases, recovered cases, and death cases of COVID-19 in Bangladesh. The system architecture is given in Fig. 3.
Fig. 3.
System architecture.
2.3. System implementation
Python programming language has been used to implement the system. Several libraries such as Matlab, Keras, Matplotlib, Numpy are used in this system. Tensorflow was used, because the system is back-end type.
2.4. Time series (TS)
Time Series (TS) is a series of data points over a regular interval of time. It is preferred that the observation is in sequential arrangement in the regression detective modeling approach. Simply, it remains unchanged over time. In TS expressions, the regularity is referred to as the time series being fixed, which represents a constant mean and variance with respect to time. These are significant properties in TS. On the other hand, it can be simply violated by the presence of tendency, seasonality, and inaccuracy. A Time Series is said to have a trend if a certain pattern repeats in regular interval. A nonstationary time series has trend, seasonal effects, therefore, its statistical features may change over time. Due to lockdown, quarantine, social distancing, etc, the TS datasets of COVID-19 may have nonstationary patterns. Therefore, it is essential to know the characteristics of TS before applying for forecasting. This research adopted the Augmented Dickey-Fuller (ADF) test (Cheung & Lai, 1995) to make sure the scenery of TS. ADF is used for a unit root test which helps us to check the impact of trends in a given TS. The outcomes are interpreted by the p-value from the test. The p-value of 1–5% suggests the rejection of the null hypothesis; otherwise, a p-value of above threshold suggests the nonrefusal of the null hypothesis. The p value of >5% indicates a unit root of the input data, hence it is a nonstationary series. Prior to the model architecture, the internal mechanisms of LSTM networks need to be clearly addressed.
2.5. Long Short-Term Memory (LSTM) network
In 1997, the Long Short-Term Memory structural design was first proposed by (Hochreiter and Schmidhuber, 1997a) for recurrent neural networks. LSTM cell was introduced by Hochreiter and Schmidhuber. The LSTM cell has two mechanisms to its condition compared to the RNN cell which are hidden, and internal cell state (Hochreiter and Schmidhuber, 1997b). The hidden state corresponds to the Short-Term Memory (STM) component, and the cell state corresponds to the Long Term Memory (LTM) (Hewamalage et al., 2020; Hochreiter and Schmidhuber, 1997b). LSTM is a unique type of circular neural network construction that can learn the proposed long-term dependence to defeat the problem of slope disappearing (Schuster and Paliwal, 1997). In this context, the LSTM structure replaces the general stress in the circulatory neural network with LSTM cells with little inner memory. These LSTM cells share a common circulatory neural network, serving to memorize multiple levels of error in the cell's internal condition.
LSTM structure facilitates to forget the cell status values and determines how many new input values will be received. The gradient does not finish, and the learning becomes not possible even if this process is recurring. It evaluates the absolute output value throughout the number of hidden sides in a similar way as the standard circulation neural network. In the development of evaluating the number of variables in the hidden layer, gateways are fitted to control the flow of information. As a result, the circulatory neural network using LSTM cells handles even the data in a long sequence of procedures without causing slope loss.
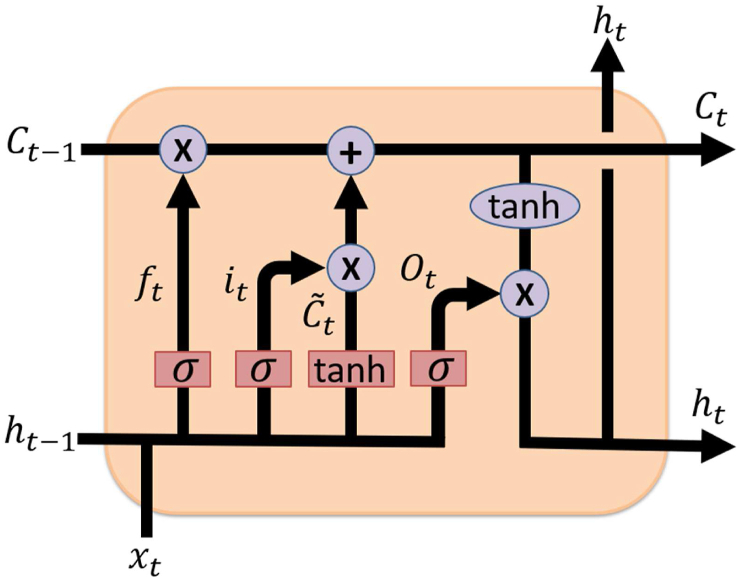
In this study, LSTM is used as a model to forecast the COVID-19 confirmed cases, recovered cases, and death cases. The general structure of LSTM contains of four gates-input gate, forget gate, control gate, and output gate as shown in Fig. 4.
Fig. 4.
Basic LSTM cell structure.
In Fig. 4the, input gate, forget gate, control gate, and output gate are denoted by it, ft, ct, ot, respectively. The details of these four gates are enlightened below:
The input gate is expressed as:
| (1) |
It decides which information can be transferred from the earlier cell to the current cell.
The forget gate is defined by equation (2), and it is used to store the information from the input of previous memory or otherwise.
| (2) |
The control gate controls the update of the cell and it is defined as:
| (3) |
To updates the hidden layer (ht-1) and update the output, finally the output gate is used which is given by the following equations:
| (4) |
In the above equations, xt =input, w = corresponding weight matrix of input, b = corresponding bias of input, Ct-1 = previous block memory, Ct = current block memory, ht-1 = previous block output, ht = current block output. Furthermore, tanh is the hyperbolic tangent function that is used to scale the values in range −1 to 1, and σ is the sigmoid activation function, which gives the output between 0 and 1. The LSTM algorithms were implemented in the following way.
2.6. Performance evaluation criteria: mean square root error
To evaluate the model performance, the Root Mean Square Error (RMSE) is used as a standard statistical metric for getting higher accuracy. It is generally used to evaluate the variation between the predictable value, or the value measured by the model and the value observed in the actual environment. If A is the dataset with the actual value and B is the dataset of predicted values, then the mean square root error can be represented as equation (5) (Benesty et al., 2009; Chai & Draxler, 2014; Gers et al., 1999).
| (5) |
The square root of that value is obtained when the error of the pairs of elements in each set is calculated and then squared to determine the exact difference, averaging the overall error.
3. Results and discussion
In this study, an appropriate model was selected and trained to forecast the three values such as the confirmed case, death case, and recovered case of COVID-19 in Bangladesh. In our review, the LSTM models show a close value to the actual curve by accumulating traffic big data, demonstrating good simulation and forecast effects. Since the lags of unidentified periods could get a position between major events in a time series, thus the LSTM networks appear to be excellent options for classification process and prediction according to time series data. The LSTM program together with Python and Tensorflow are used for the prediction of COVID-19 outbreak. The obtained results are shown in Fig. 5(b), Fig. 5(c), Fig. 6(a), Fig. 6(b), Fig. 6(c), Fig. 7(a), Fig. 7(b), Fig. 7(c), Fig. 8(a), Fig. 8(b), Fig. 8(c). Our study was performed with the most recent data and got more precise predictions compared to the reported results available in the literature.
-
A.
Prediction of Confirmed Cases
Fig. 5(b).
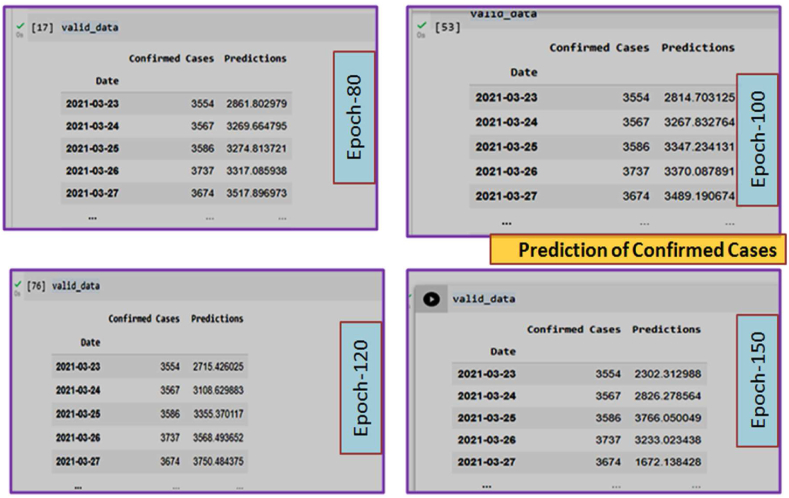
Prediction of Confirmed Cases from March 2020 to August 2021. In this Fig. 5(b), only Epoch-80 has been shown. Other Epoch (Epoch-100, 120, 150) are presented in Supplementary Fig. S1.
Fig. 5(c).
Data validation for confirmed cases.
Fig. 6(a).
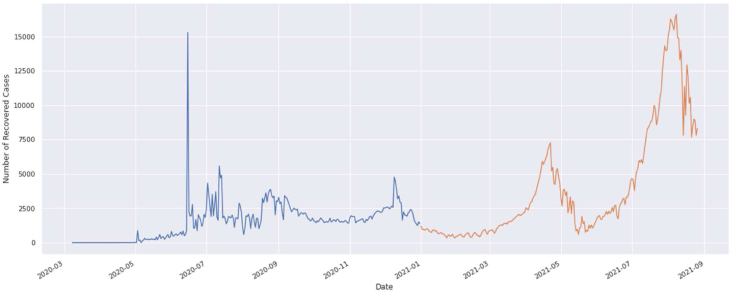
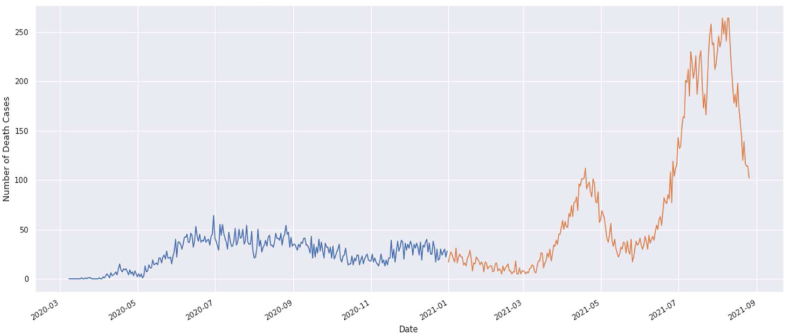
Recovered cases from March 2020 to August 2021.
Fig. 6(b).
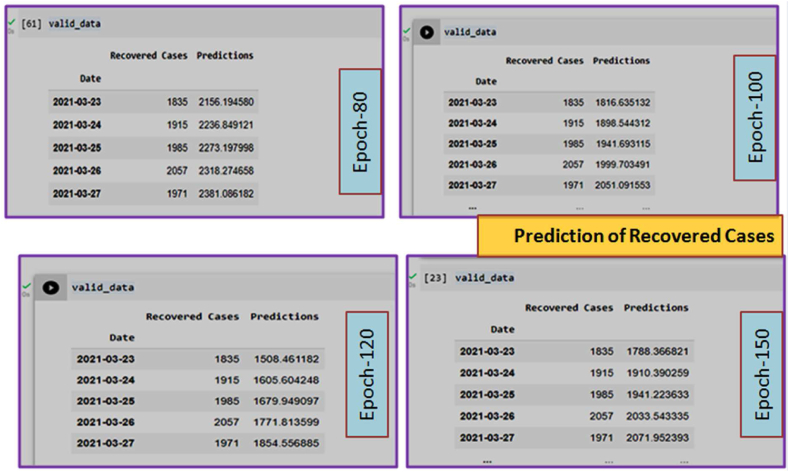
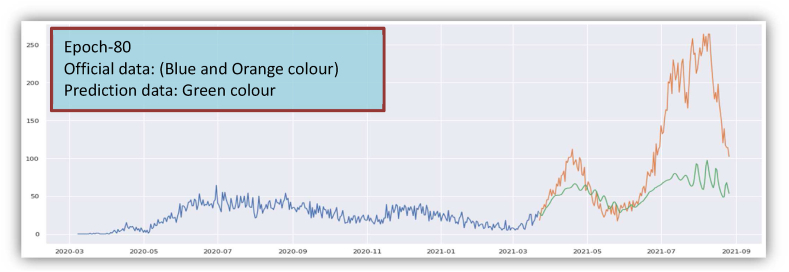
Prediction of Recovered Cases from March 2020 to August 2021. In this Figure, only Epoch-80 has been shown. Other Epoch (Epoch-100, 120, 150) are presented in Supplementary Fig. S2.
Fig. 6(c).
Data validation for recovered cases.
Fig. 7(a).
Prediction of death cases from March 2020 to August 2021.
Fig. 7(b).
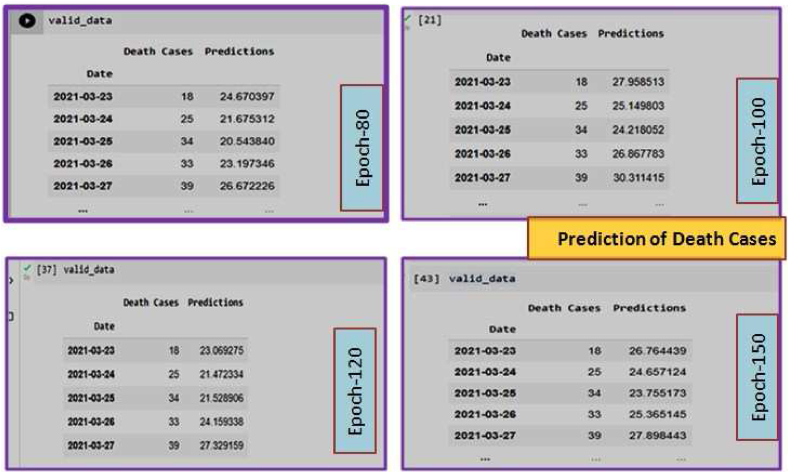
Prediction of Death Cases from March 2020 to August 2021. In this Figure, only Epoch-80 has been shown. Other Epoch (Epoch-100, 120, 150) are presented in Supplementary Fig. S3.
Fig. 7(c).
Data validation for Death Cases.
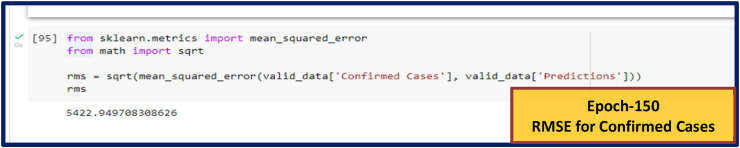
Fig. 8(a).
RMSE for Confirmed Cases. RMSE values for other Epoch (Epoch-80, 100 and 120) are given in Supplementary Fig. S4.
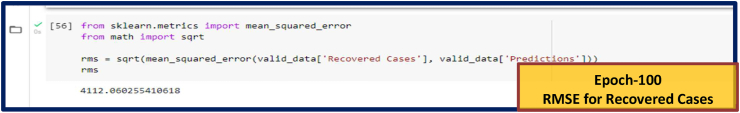
Fig. 8(b).
RMSE for Recovered Cases. RMSE values for other Epoch (Epoch-80, 120 and 150) are given in Supplementary Fig. S5.
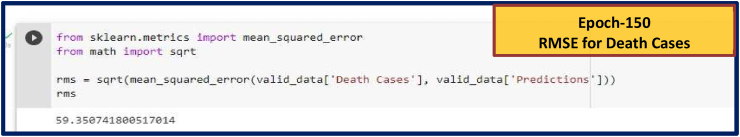
Fig. 8(c).
RMSE for Death Cases. RMSE values for other Epoch (Epoch-80, 100 and 120) are given in Supplementary Fig. S6.
First, we have imported the test set that was used to make the predictions. To predict the confirmed cases, death case, and recovered cases, we were considered a couple of things after loading in the test set:
-
•
Merge the training set and the test set on the 0 axis.
-
•
Set the time step as 60
-
•
Used MinMaxScalar to transform the new dataset
-
•
Reshape the dataset as done previously
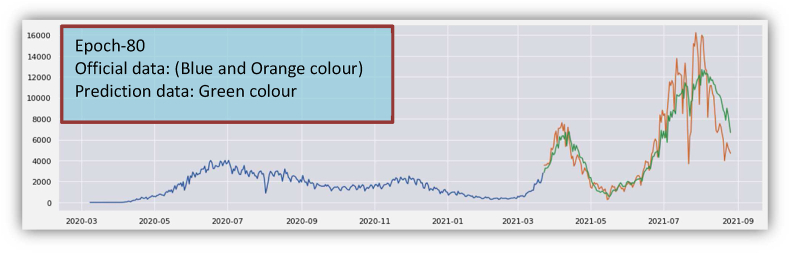
The confirmed cases from March 2020 to August 2021 are shown in Fig. 5(a), where the implementation of prediction of confirmed cases is shown in Fig. 5(b). The blue curve in Fig. 5(b) indicates the training data, the orange curve indicates testing data, and the green curve indicates the predicted data for the confirmed cases.
Fig. 5(a).
Confirmed cases from March 2020 to August 2021.
From Fig. 5(b), it is clear that the official data and the curve of prediction are much closer to each other. Therefore, we conclude that the model has learned the sequential patterns of COVID-19 positive cases successfully. Data validation for confirmed cases is given in Fig. 5(c).
-
B.
Prediction of Recovered Cases
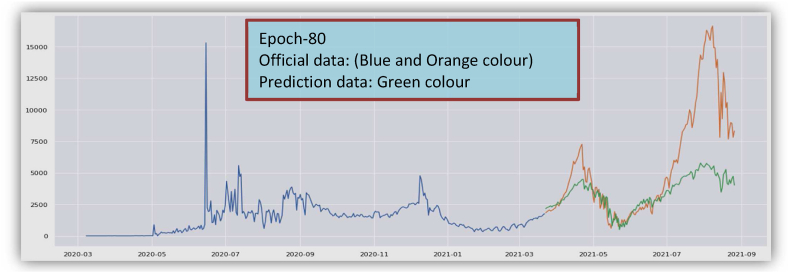
The overall graphical view and prediction results for the recovered cases from March 2020 to August 2021 in Bangladesh are given in Fig. 6(a) and Fig. 6(b). In Fig. 6(b), the blue curve indicates the training data, the orange curve indicates testing data, and the green curve indicates the predicted data for the recovered cases. Data validation for the recovered cases is shown in Fig. 6(c).
-
C.
Prediction of Death Cases
Fig. 7 (a) represents the data from March 2020 to August 2021 for the death cases in our proposed study. Based on these data, we predicted the death case which is shown Fig. 7(b) and the data validation for death cases is given in 7(c).
From the above data validation shown in Fig. 5(c), Fig. 6(c), Fig. 7(c), the difference between the predicted data and the actual data are very low for confirmed cases, recovered cases and death cases respectively. This means that the proposed methods approximately fit real data; thus, the policymakers can use this data to take precautionary steps forCOVID-19 in the upcoming days.
-
D.
Performance Measure
Root Mean Square Error (RMSE) is the standard deviation of the residuals. Residuals are a measure of how far from the regression line data points. In other words, it tells us how concentrated the data is around the line of best fit. RMSE for confirmed cases, recovered cases, and death cases are shown in Fig. 8(a), Fig. 8(b), Fig. 8(c)(a)-8(c).
Epoch is one of the most excellent methods to compare different data for forecasting. We compared the result of our study by a changeable epoches. It represents the highest prediction with the best accuracy at 120 epoches. For measuring the effectiveness of the system, the RMSE has been used. As an example, Hridoy et al. (2020) obtained the RMSE value for Confirmed, Recovered, and Death cases of 593, 1113, and 36, respectively, where our study shows the respective values of 247, 211 and 8 (See in Table 1). Regarding epoch, our approach shows the best result at 120 epochs. If we increase the number of epoches, then the model becomes over fitted and for decreasing the number of epoch, the model predicts a less accurate result.
Table 1.
RMSE comparison with present and previous data for confirmed cases, recovered cases, and death cases.
| Sr. No | No. of Epoch | RMSE |
References | ||
|---|---|---|---|---|---|
| Confirmed Cases | Recovered Cases | Death Cases | |||
| 01 | 80 | 1830.44 | 3872.71 | 77.47 | Present Study (Bangladesh) |
| 100 | 1978.97 | 4112.06 | 91.22 | ||
| 120 | 2801.56 | 3573.72 | 86.18 | ||
| 150 | 5422.95 | 3942.23 | 59.35 | ||
| 02 | – | 593 | 1113 | 36 | Hridoy et al. (2020) |
| 03 | – | 301.99 | 123.8794 | 4.306 | Duttaet al. (2020) |
| 04 | – | 3677.0 | 802.2 | 266.5 | Karim et al. (2019) |
| 05 | – | 2156.2 | 609.7 | 127.2 | |
| 06 | – | 53.38 | 71.2 | 4.97 | Dutta & Bandyopadhyay (2020) |
The forecasted COVID-19 confirmed cases for Bangladesh shows a significant upward trend for some more time in the near future. The actual (blue line) and predicted (green line) data are visualized in Fig. 5(b), Fig. 6(b), Fig. 7(b) having some sudden jump in the forecasted data. Significant downward trend is shown after some time in COVID-19 predicted confirmed cases. The prediction figures show COVID-19 downward trend in three cases, at the end of August 2021. These time-series data are the compiled number of confirmed cases, recovered case, and death cases that are rising high and arriving at a certain coinciding peak curve. LSTM was able to capture the pattern of hypedup growth of the graph with a better correlation value and RMSE.
The combined CNN-LSTM model has a good impact on the confirmation of anticipated findings with respect to actual results, according to the experimental data. Performance measure metrics are used to ensure that all defined cases in relation to COVID-19 are confirmed. The method suggests that those who have been afflicted by the disease may recover, and that a suitable vaccination will be created to minimize the death rate. The proposed technique predicts the illness patterns and assists clinicians in conducting early preventive studies. Clinical doctors have made every effort for success, and it is expected that they will success in the near future. The LSTM has provided more convincing RMSE results. The model is preferable if it has a lower RMSE.
4. Conclusions and future work
A considerable number of studies is in progress in predicting COVID- 19 outbreak since our live and livelihood depends on the outcome of this emergency. We took into consideration of confirmed, recovered, and death cases in Bangladesh to carry out our study ofCOVID-19 prediction. To predict the transmission of epidemic trends, a model based on deep learning approach is applied in this study. LSTM algorithm was used to develop proposed model that shows high performance in time series forecasts, and the results show good performances in the confirmed case, recovered case, and the death cases. The obtained results can be efficiently used to reduce the infectious rate and improve the recovery rate. Since the measure of new cases continue to raise worldwide, our model shows the potential for pandemic curve forecast and hindrance of COVID-19 in new countries. Based on our model simulations, the current COVID-19 pandemic is not expected to be ended in the next few months. Based on the graphical analysis, it can be mentioned that LSTM model shows a great fit for the prediction of COVID-19 because the actual curve and the prediction curve are very close to each other in maximum time. Moreover, the calculated RMSE value indicates the effectiveness of the LSTM model in predicting the COVID-19. It is therefore expected that the LSTM model could be effectively utilized for the prediction of contagious diseases, hence the respective authority may be able to decide what would be the best precautionary steps to prevent further transmissions in the country. In our next work, several other machine learning and deep learning models are planned to be used to predict the trend of COVID-19 and provide the best models in the case of COVID-19 prediction and contemporary issues.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Handling editor: Dr HE DAIHAI HE
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.idm.2021.12.005.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Anastassopoulou C., Russo L., Tsakris A., Siettos C. Data-based analysis, modelling and forecasting of the COVID-19 outbreak. PLoS One. 2020;15:1–21. doi: 10.1371/journal.pone.0230405. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwar S., Nasrullah M., Hosen M.J. COVID-19 and Bangladesh: Challenges and how to address them. Frontiers in Public Health. 2020;8:154. doi: 10.3389/fpubh.2020.00154. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardabili S.F., Mosavi A., Ghamisi P., Ferdinand F., Varkonyi- Koczy A.R., Reuter U., Rabczuk T., Atkinson P.M. 2020. COVID-19 outbreak prediction with machine learning. Available at SSRN 3580188. [Google Scholar]
- Arnold C. Reservoir bats: The invisible enemy. New Scientist. 2014;221(2955):44–47. doi: 10.1016/S0262-4079(14)60285-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S.K., Dutta S. medRxiv; 2020. Machine learning approach for confirmation of COVID-19 cases: Positive, negative, death and release. [Google Scholar]
- Benesty J., Chen J., Huang Y., Cohen I. Noise reduction in speech processing. Springer; Berlin, Heidelberg: 2009. Pearson correlation coefficient; pp. 1–4. [Google Scholar]
- Benvenuto D., Giovanetti M., Vassallo L., Angeletti S., Ciccozzi M. Application of the ARIMA model on the COVID-2019 epidemic dataset. Data in. 2020;brief,:105340. doi: 10.1016/j.dib.2020.105340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertozzi A.L., Franco E., Mohler G., Short M.B., Sledge D. The challenges of modeling and forecasting the spread of COVID-19. Proceedings of the National Academy of Sciences of the United States of America. 2020;117:16732–16738. doi: 10.1073/pnas.2006520117. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai T., Draxler R.R. Root mean square error (RMSE) or mean absolute error (MAE)?–Arguments against avoiding RMSE in the literature. Geoscientific Model Development. 2014;7:1247–1250. [Google Scholar]
- Chen X., Liu Z. medRxiv; 2020. Early prediction of mortality risk among severe COVID-19 patients using machine learning. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T.M., Rui J., Wang Q.P., Zhao Z.Y., Cui J.A., Yin L. A mathematical model for simulating the phase-based transmissibility of a novel coronavirus. Infectious Diseases of Poverty. 2020;9:1–8. doi: 10.1186/s40249-020-00640-3. [CrossRef] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung Y.W., Lai K.S. Lag order and critical values of the augmented dickey—fuller test. Journal of Business & Economic Statistics. 1995;13(3):277–280. [Google Scholar]
- Chimmula V.K.R., Zhang L. Time series forecasting of COVID-19 transmission in Canada using LSTM networks. Chaos, Solitons & Fractals. Jun 2020:135–109864. doi: 10.1016/j.chaos.2020.109864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta, et al. CNN-LSTM model for verifying predictions of COVID-19 cases. AJRCOS. 2020;5(4):25–32. Article no.AJRCOS.57148. [Google Scholar]
- Dutta S., Bandyopadhyay K.S. Machine learning approach for confirmation of COVID-19 cases: Positive, negative, death and release. Iberoamerican Journal of Medicine. 2020;3:172–177. doi: 10.5281/zenodo.3822623. [DOI] [Google Scholar]
- Fanelli D., Piazza F. Analysis and forecast of COVID-19 spreading in China, Italy and France. Chaos, Solitons & Fractals. 2020;134:109761. doi: 10.1016/j.chaos.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gers F.A., Schmidhuber J., Cummins F. Proceedings of the 1999 Ninth International Conference on Artificial neural networks (ICANN) IET; Edinburgh, UK: 1999. Learning to forget: Continual prediction with LSTM; pp. 850–855. [Google Scholar]
- Gralinski L.E., Menachery V.D. Return of the coronavirus: 2019-nCoV. Viruses. 2020;1 12:135. doi: 10.3390/v12020135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grasselli G., Pesenti A., Cecconi M. Critical care utilization for the COVID-19 outbreak in lombardy, Italy: Early experience and forecast during an emergency response. JAMA. 2020;323(16):1545–1546. doi: 10.1001/jama.2020.4031. [DOI] [PubMed] [Google Scholar]
- de Groot R.J., Baker S.C., Baric R.S., Brown C.S., Drosten C., Enjuanes L., Fouchier R.A., Galiano M., Gorbalenya A.E., Memish Z.A., et al. Commentary: Middle east respiratory syndrome coronavirus (merscov): Announcement of the coronavirus study group. Journal of Virology. 2013;87(14):7790–7792. doi: 10.1128/JVI.01244-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Y., Zheng B., He Y., Liu X., Zhuang Z., Cheung C., Luo S., Li P., Zhang L., Guan Y., et al. Isolation and characterization of viruses related to the sars coronavirus from animals in southern China. Science. 2003;302(5643):276–278. doi: 10.1126/science.1087139. [DOI] [PubMed] [Google Scholar]
- Hellewell J., Abbott S., Gimma A., Bosse N.I., Jarvis C.I., Russell T.W., Munday J.D., Kucharski A.J., Edmunds W.J., Sun F., et al. Feasibility of controlling COVID-19 outbreaks by isolation of cases and contacts. The Lancet Infectious Diseases. 2020;8:e488–e496. doi: 10.1016/S2214-109X(20)30074-7. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewamalage H., Bergmeir C., Bandara K. Recurrent neural networks for time series forecasting: Current status and future directions. International Journal of Forecasting. 2020;37:388–427. [Google Scholar]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Computation. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. PMID: 9377276. [DOI] [PubMed] [Google Scholar]
- Hochreiter S., Schmidhuber J. Long short-term memory. Neural Computation. 1997;9:1735–1780. doi: 10.1162/neco.1997.9.8.1735. [DOI] [PubMed] [Google Scholar]
- Huang C.-J., Chen Y.-H., Ma Y., Kuo P.-H. medRxiv; 2020. Multiple-input deep convolutional neural network model for COVID-19 forecasting in China. [Google Scholar]
- Hridoy A.H., Naim M., Tipo I.H. (2020) [DOI]
- Hu Z., Ge Q., Jin L., Xiong M. 2020. Artificial intelligence forecasting of COVID-19 in China. arXiv preprint arXiv:2002.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Institute of Epidemiology DC and R COVID-19. 2020. https://www.iedcr.gov.bd/index.php/component/content/article/73- ncov-2019 Available online at:
- Ivorra B., Ferrández M.R., Vela-Pérez M., Ramos A.M. Mathematical modeling of the spread of the coronavirus disease 2019 (COVID-19) taking into account the undetected infections. The case of China. Communications in Nonlinear Science and Numerical Simulation. 2020;88:105303. doi: 10.1016/j.cnsns.2020.105303. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim F., Majumdar S., Darabi H., Harford S. Multivariate lstm-fcns for time series classification. Neural Networks. 2019;116:237–245. doi: 10.1016/j.neunet.2019.04.014. [DOI] [PubMed] [Google Scholar]
- Krätschmer V. Strong consistency of least-squares estimation in linear regression models with vague concepts. Journal of Multivariate Analysis. 2006;97(3):633–654. [Google Scholar]
- Kucharski A.J., Russell T.W., Diamond C., Liu Y., Edmunds J., Funk S., Eggo R.M., Sun F., Jit M., Munday J.D., et al. Early dynamics of transmission and control of COVID-19 : A mathematical modeling study. The Lancet Infectious Diseases. 2020;20:553–558. doi: 10.1016/S1473-3099(20)30144-4. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): The epidemic and the challenges. International Journal of Antimicrobial Agents. 2020;55(3):105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau S.K., Woo P.C., Li K.S., Huang Y., Tsoi H.W., Wong B.H., Wong S.S., Leung S.Y., Chan K.H., Yuen K.Y. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:14040–14045. doi: 10.1073/pnas.0506735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LiestølK A.P.K. Updating of covariates and choice of time origin in survival analysis: Problems with vaguely defined disease states. Statistics in Medicine. 2002;21(23):3701–3714. doi: 10.1002/sim.1322. [DOI] [PubMed] [Google Scholar]
- Li Q., Feng W., Quan Y.-H. Trend and forecasting of the COVID-19 outbreak in China. Journal of Infection. 2020;80(4):469–496. doi: 10.1016/j.jinf.2020.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Shi Z., Yu M., Ren W., Smith C., Epstein J.H., Wang H., Crameri G., Hu Z., Zhang H., et al. Bats are natural reservoirs of sars-like coronaviruses. Science. 2005;310(5748):676–679. doi: 10.1126/science.1118391. [DOI] [PubMed] [Google Scholar]
- Liu P., Beeler P., Chakrabarty R.K. medRxiv; 2020. COVID-19 progression timeline and effectiveness of response-to-spread interventions across the United States. [Google Scholar]
- Liu Z., Magal P., Seydi O., Webb G. 2020. Predicting the cumulative number of cases for the COVID-19 epidemic in China from early data. arXiv preprint arXiv:2002.12298. [DOI] [PubMed] [Google Scholar]
- Lover A.A., McAndrew T. medRxiv; 2020. Sentinel event surveillance to estimate total sars-cov-2 infections, United States. [Google Scholar]
- Massonnaud C., Roux J., Crépey P. medRxiv; 2020. COVID-19 : Forecasting short term hospital needs in France. [Google Scholar]
- Mcintosh K., Kapikian A.Z., Turner H.C., Hartley J.W., Parrott R.H., Chanock R.M. Seroepidemiologic studies of coronavirus infection in adults and children. American Journal of Epidemiology. 1970;91(6):585–592. doi: 10.1093/oxfordjournals.aje.a121171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaneko Y., Nimmerjahn F., Ravetch J. Anti-inflammatory activity of immunoglobulin G resulting from Fc sialylation. science. Aug 4 2006;313(5787):670–673. doi: 10.1126/science.1129594. [DOI] [PubMed] [Google Scholar]
- Pal R., Sekh A.A., Kar S., Prasad D.K. 2020. Neural network based country wise risk pre- diction of COVID-19. arXiv preprint arXiv:20 040 0959. [Google Scholar]
- PanchT, Mattie H., Celi L.A. The ǣinconvenient truth ǥaboutai in healthcare. Npj Digital Medicine. 2019;2(1):1–3. doi: 10.1038/s41746-019-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathan R.K., Biswas M., Khandaker M.U. Vol. 138. Chaos, Solitons & Fractals; 2020. Time series prediction of COVID-19 by mutation rate analysis using recurrent neural network-based LSTM model. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peirlinck M., Linka K., SahliCostabal F., Kuhl E. Outbreak dynamics of COVID-19 in China and the United states. Biomechanics and Modeling in Mechanobiology. 2020;32342242:1–15. doi: 10.1007/s10237-020-01332-5. [CrossRef] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L., Yang W., Zhang D., Zhuge C., Hong L. 2020. Epidemic analysis of COVID-19 in China by dynamical modeling. arXiv preprint arXiv:2002.06563. [Google Scholar]
- Roosa K., Lee Y., Luo R., Kirpich A., Rothenberg R., Hyman J., Yan P., Chowell G. Real-time forecasts of the COVID-19 epidemic in China from february 5th to february 24th, 2020. Infectious Disease Modelling. 2020;5:256–263. doi: 10.1016/j.idm.2020.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rustam F., Reshi A.A., Mehmood A., Ullah S., On B., Aslam W., Choi G.S. IEEE Access; 2020. Covid 19 future forecasting using supervised machine learning models. [Google Scholar]
- Schuster M., Paliwal K.K. Bidirectional recurrent neural networks. IEEE Transactions on Signal Processing. 1997;45:2673–2681. [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Schäfer A., Won J., Brown A.J., Montgomery S.A., Hogg A., Babusis D., Clarke M.O., et al. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against mers-cov. Nature Communications. 2020;11(1):1–14. doi: 10.1038/s41467-019-13940-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Suzuki A. medRxiv; 2020. Machine learning model estimating number of COVID-19 infection cases over coming 24 days in every province of South Korea (xgboost and multioutputregressor) [Google Scholar]
- Thomas D.M., Sturdivant R., Dhurandhar N.V., Debroy S., Clark N. A primer on COVID-19 mathematical models. Obesity. 2020;28:1375–1377. doi: 10.1002/oby.22881. [CrossRef] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobías A. Evaluation of the lockdowns for the SARS-CoV-2 epidemic in Italy and Spain after one month follow up. Science of the Total Environment. Jul 10 2020:725–138539. doi: 10.1016/j.scitotenv.2020.138539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TomarA G.N. Prediction for the spread of COVID-19 in India and effective- ness of preventive measures. The Science of the Total Environment. 2020:138762. doi: 10.1016/j.scitotenv.2020.138762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuli S., Tuli S., Tuli R., Gill S.S. Internet of Things; 2020. Predicting the growth and trend of COVID-19 pandemic using machine learning and cloud computing; p. 100222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Han D.-w., Li K., Yang X., Yin X.-l., Qiu J., Liang D.-x., Ma Z.-y. 2020. A universal model for prediction of COVID-19 pandemic based on machine learning. [Google Scholar]
- Wang L., Wang Y., Ye D., Liu Q. Review of the 2019 novel coronavirus (SARS-CoV-2) based on current evidence. International journal of antimicrobial agents. 2020;55(6):105948. doi: 10.1016/j.ijantimicag.2020.105948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangping J., Ke H., Yang S., Wenzhe C., Shengshu W., Shanshan Y., Jianwei W., Fuyin K., Penggang T., Jing L., et al. Extended sir prediction of the epidemics trend of COVID-19 in Italy and compared with hunan, China. Frontiers of Medicine. 2020;7:169. doi: 10.3389/fmed.2020.00169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise T., Zbozinek T.D., Michelini G., Hagan C.C., et al. 2020. Changes in risk perception and protective behavior during the first week of the COVID-19 pandemic in the United States. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization www.who.int
- Wu F., Zhao S., Yu B., et al. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., Weaver J.B., Healy D.M., Lu J. Wavelet transform domain filters: A spatially selective noise filtration technique. IEEE Transactions on Image Processing. 1994;3(6):747–758. doi: 10.1109/83.336245. [DOI] [PubMed] [Google Scholar]
- Yangand C., Wan J. A mathematical model for the novel coronavirus epidemic in Wuhan,China. Mathematical Biosciences and Engineering. 2020;17(3):2708–2724. doi: 10.3934/mbe.2020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Wang J. A mathematical model for the novel coronavirus epidemic in Wuhan, China. Mathematical Biosciences and Engineering. 2020;17:2708. doi: 10.3934/mbe.2020148. [CrossRef] [PubMed] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef A., Maryam T., Mojtaba S. The estimate of the basic reproduction number for COVID-19 :A systematic review and meta-analysis. Journal of Preventive Medicine and Public Health. 2020;53:151–157. doi: 10.3961/jpmph.20.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan C., Chi K.T., Lai Z., Hao T., Su J. medRxiv; 2020. Prediction of COVID-19 spreading profiles in South Korea, Italy and Iran by data-driven coding. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.