Abstract
Prostaglandins, pleiotropic immune modulators that induce protein kinase A (PKA), inhibit gamma interferon induction of class II major histocompatibility complex (MHC) genes. We show that phosphorylation of CIITA by PKA accounts for this inhibition. Treatment with prostaglandin E or 8-bromo-cyclic AMP or transfection with PKA inhibits the activity of CIITA in both mouse and human monocytic cell lines. This inhibition is independent of other transcription factors for the class II MHC promoter. These same treatments also greatly reduced the induction of class II MHC mRNA by CIITA. PKA phosphorylation sites were identified using site-directed mutagenesis and phosphoamino acid analysis. Phosphorylation at CIITA serines 834 and 1050 accounts for the inhibitory effects of PKA on CIITA-driven class II MHC transcription. This is the first demonstration that the posttranslational modification of CIITA mediates inhibition of class II MHC transcription.
Class II molecules of the major histocompatibility complex (MHC) play a critical role in T-cell-dependent immunity and the inflammatory response by presenting processed, exogenous antigens to T helper (Th) cells (reviewed in references 13, 42, and 50). These important molecules are commonly used as targets for immune therapies aimed at blocking graft rejection or promoting the recognition of cancer. Although expressed constitutively on “professional” antigen-presenting cells (monocytes/macrophages, B cells, and dendritic cells) class II MHC can be induced in most cell types and tissues in the presence of gamma interferon (IFN-γ) (1, 3, 9–11, 24, 28, 31, 38, 45, 57, 60, 62). Patients lacking class II MHC have reduced numbers of CD4+ Th cells and succumb to repeated bacterial, viral, and protozoal infections, generally dying in early childhood (39). Class II MHC has been implicated as a contributing factor for numerous diseases including diabetes, rheumatoid arthritis, Alzheimer's disease, and multiple sclerosis (7, 39, 49).
The constitutive and inducible expression of nearly all class II MHC and related genes is regulated globally at the level of transcription by the class II transactivator (CIITA) (8, 27, 60). Transient transfection of CIITA into cells which are class II MHC deficient or have low-level expression of class II MHC is sufficient to drive class II MHC transcription and expression (12, 46, 48). In normal tissues, CIITA expression patterns are controlled by as many as four promoters, which allow both constitutive and IFN-γ-inducible expression of CIITA (44, 52). Though not a DNA-binding protein, CIITA is required for the opening and subsequent activation of class II MHC promoters through interactions with the ubiquitously expressed transcription factors RFX and NF-Y, which recognize the X and Y DNA elements of class II and class I MHC promoters (41, 54, 64). CIITA contains an N-terminal acidic domain (residues 1 to 125) which can act as a transcription activation domain when fused to GAL4 DNA-binding sequences (34, 54, 63). A variety of proteins interact with the acidic domain of CIITA, including TFIIB, TAFIIs, CREB-binding protein (CBP), and p300 (22, 23, 34, 40, 58), demonstrating that this domain is coupled to the basal transcription machinery. CIITA also contains a functional GTP-binding domain (26) and a series of C-terminal leucine-rich repeats that are important for nuclear translocation (25). Regulation of CIITA-mediated class II MHC transcription results from specific modulation of the CIITA promoter and/or by modification of the transactivation capacity of CIITA itself. The latter mechanism is illustrated by the ability of human immunodeficiency virus Tat protein to interfere with CIITA-mediated transactivation (61). Little is known regarding posttranslational events that may impact on CIITA.
Prostaglandins are inflammatory mediators produced by the action of cyclo-oxygenase in the arachidonic acid pathway. They contribute to inflammation through vasodilation, potentiating effects on permeability through histamine, bradykinin, and leukoattractants (36). The immunomodulatory effects of prostaglandins on macrophages have been well described. IFN-γ-induced class II MHC expression is inhibited by prostaglandins of the E type (PGE) in macrophages (5, 29, 59, 65). Receptors for PGE activate adenyl cyclase leading to increased intracellular cyclic AMP (cAMP) production and downstream protein kinase A (PKA) activation (21). The effect of PGE on class II MHC expression has been shown to occur at the transcriptional level (2). The mechanism linking PKA to modulation of IFN-γ induction of class II MHC is unknown and is the focus of this study. Since phosphorylation is a frequent form of posttranslational modification in eukaryotic cells and is linked to the control of a multitude of cellular functions, we hypothesized that an effect of PGE, cAMP, and PKA activation might be the direct phosphorylation of CIITA.
In this report, we demonstrate that PGE and cAMP inhibit the function of CIITA. Furthermore, the constitutively active catalytic subunit (α) of PKA (PKAα) can fully reproduce the inhibitory effects of PGE and cAMP on a class II MHC promoter through phosphorylation of CIITA. Through use of the Gal4CIITA fusion and a Gal4-driven reporter, we show that the effects of PGE, cAMP, and PGE on promoter activation by CIITA are independent of specific class II MHC promoter elements. Further, we show that site-specific phosphorylation events are important for PKA-mediated inhibition of CIITA-dependent promoter activation.
MATERIALS AND METHODS
Cell lines and transfection.
Mouse monocyte/macrophage cell lines P388D1 (ATCC TIB 63) and RAW264.7 (ATCC TIB 71) and human monocyte-like cell line U937 (ATCC CRL 1593) were cultured in RPMI 1640 supplemented with 15% fetal calf serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (complete medium). For transfection, cells were harvested in mid-log phase. Transient cotransfection was carried out by electroporation or with the SuperFect (Qiagen) reagent. Briefly, for electroporation, 2 × 106 cells were pelleted by centrifugation, resuspended in 300 μl of complete medium, and transferred into a Gene Pulser cuvette (Bio-Rad Laboratories). The appropriate plasmid DNAs were added in a total volume of 20 μl. For each treatment cells were pulsed at 960 μF and 0.2 kV and transferred into 10 ml of complete medium. Transfections using SuperFect were performed according to the manufacturer's directions. For cells transfected with PKA, 80 μM ZnCl2 was added to ensure PKA kinase activity in vivo. Cells were harvested at 36 h posttransfection. The Gal4DRCAT and Gal4CIITA plasmids were a gift from Jerry Boss.
Analysis of mRNA expression.
P388D1 cells (4 × 106) were transfected by electroporation at 0.24 kV and 960 μF. Total RNA isolation was performed using the Wizard RNA isolation kit (Promega). One microgram of total RNA from each treatment was used for reverse transcription-PCR using class II I-Aβ primers (5′-AAC CAG CCA AGA TCA AAG TGC-3′ and 5′-TGC CGC TCA ACA TCT TGC TCC-3′).
Phosphate labeling in vivo.
For experiments involving [32P]orthophosphate or [33P]orthophosphate labeling of CIITA proteins in vivo, 36 h posttransfection, transfected cells (approximately 2 × 107) were washed once with phosphate-free complete Dulbecco's modified Eagle medium (DMEM) (15% dialyzed fetal calf serum and supplements as for RPMI 1640 above), resuspended in 2.0 ml of phosphate-free complete DMEM, preincubated for 30 min at 37°C in 5% CO2, and incubated for 2 to 4 h with 2 mCi of [32P]orthophosphate or [33P]orthophosphate/ml. At the end of the labeling period, cells were washed once with 10 ml of ice-cold Tris-buffered saline, lysed in 1.0 ml of radioimmunoprecipitation assay (RIPA) buffer (1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 0.01 M sodium phosphate [pH 7.2], 2 mM EDTA, 50 mM NaF, 0.2 mM NaVO4, 0.5 mM phenylmethylsulfonyl fluoride, 1× COMPLETE protease inhibitor cocktail [Roche]), and incubated for 40 min at 4oC. Five microliters of fixed Staphylococcus aureus (Pansorbin; Calbiochem) in RIPA buffer was added if lysates became viscous. The lysates were clarified by centrifugation at 4°C for 10 min in a microcentrifuge at maximum speed (14,000 × g), and the supernatants were used for immunoprecipitation.
Immunoprecipitation and in vitro kinase assay.
For immunoprecipitation of FLAG-tagged CIITA, a 20-μl packed volume of monoclonal anti-Flag M2–agarose affinity gel (A-1205; Sigma, St. Louis, Mo.) was added to cell lysates or the in vitro-translated protein mixture and the resulting mixture was incubated at 4°C for 2 to 4 h. The beads were washed once with RIPA buffer, twice with ice-cold phosphate-buffered saline (PBS), and once with 1× PKA buffer (10 mM Tris [pH 7.0], 5 mM MgCl2). Pellets were incubated with PKAα at a molar ratio of 20:1 in the presence of 10 μCi of [γ-32P]ATP and 100 μM ATP in a total volume of 10 μl of PKA buffer for 10 min at 30°C. Pellets were washed twice with ice-cold PBS containing a mixture of protease and phosphatase inhibitors and boiled for 5 min in 20 μl of 2× SDS sample buffer. Proteins were separated on SDS–8% polyacrylamide gel electrophoresis (PAGE) gels, and phosphorylated bands were visualized by autoradiography.
Enzyme activity assays.
Chloramphenicol acetyltransferase (CAT) and luciferase reporter assays were performed as previously described (15, 51).
Generation of CIITA mutants.
Site-directed mutagenesis was performed using the Transformer (Clontech) site-directed mutagenesis kit according to the manufacturer's instructions. Plasmid p3FGCIITA8 was used as the template in the mutagenesis reaction. All of the primers used in this procedure were 5′ phosphorylated. Selection primer 5′-CCCTGATAAATGCTTCAATGCTAGCGAAAAAGGAAGAGTATGAG-3′ converts an existing SspI site to an NheI site, allowing SspI restriction digest-mediated removal of the parent plasmid. Mutant clones were confirmed by DNA sequence analysis. Site-directed mutation of serine to alanine was achieved with the following mutagenic primers (mutated residues are in boldface): S286A, 5′-GGA GCG AAG GGG CTG GTG GCG CCT GGC CGG TCT GGA GAT GTT GGG-3′; SSS373/374/375AAA, 5′-CCC AGT CCG GGG TGG CCA GTT CCC GCT CCA GGC TCT TGG CGG CGG CCC TCT CCA GCC TGG CCT GCA CCA GAT CCA CCT CC-3′; S674A, 5′-CTG GTC CTC CTG TAG GGT AGC TTG ATG TCT GCG GC-3′; S834A, 5′-GGC GGG TGC CCA GAA AAG CGA GGC GGC CGG GG-3′; SSS942/943/944A, 5′-CCC GAA CAG CAG GGA GCT CCC CAG CTG TGT CTT CCG CGG CAG CTC TCG TCC TCT GAG TCT GCA CAA GCT TTC CCA GG-3′.
Phosphoamino acid analysis and phosphopeptide mapping.
For phosphoamino acid analysis, 32P-labeled CIITA was transferred to a polyvinylidene difluoride membrane (Bio-Rad; 0.2-μm pore size). The transferred product was visualized by autoradiography, excised from the membrane, hydrolyzed in 500 μl of 6 N HCl by heating to 110°C for 60 min, allowed to cool, and microcentrifuged for 2 min at maximum speed. The hydrolysate was dried under vacuum and resuspended in 8 μl of water. Approximately 6,000 cpm of the hydrolysate was spotted on a 20-cm2, 100-μm-thick glass-backed cellulose thin-layer chromatography plate (EM Science). Phosphoamino acid standards (1 μl of a mixture of phosphoserine, phosphothreonine, and phosphotyrosine; 1 mg/ml each) were loaded as described above. The first dimension was resolved by electrophoresis at 1,500 V for 20 min in electrophoresis buffer I (0.58 M formic acid, 1.36 M glacial acetic acid, pH 1.9). Resolution of the second dimension was achieved with electrophoresis at 1,300 V for 16 min in buffer II (0.87 M glacial acetic acid, 0.5% pyridine, 0.5 mM EDTA, pH 3.5). After being dried, plates were sprayed with 0.25% (wt/vol) ninhydrin in acetone and developed at 70°C for 10 min to visualize the phosphoamino acid standards. Autoradiography was performed to visualize labeled CIITA fragments.
For one-dimensional phosphopeptide mapping of CIITA by cyanogen bromide (CNBr), 32P-labeled CIITA was transferred to nitrocellulose (Schleicher & Schuell) and analyzed by autoradiography. A membrane section containing the CIITA band was excised and suspended in degassed 70% (vol/vol) formic acid. After the suspension was flushed with nitrogen, CNBr was added to a concentration of 12 mg/ml and the suspension was incubated under nitrogen at room temperature for 4 h. The sample was diluted 10-fold with water and lyophilized. The sample was dissolved in Laemmli sample buffer, boiled for 5 min, fractionated by SDS–16% PAGE in parallel with prestained molecular weight standards, dried, and visualized by autoradiography.
For two-dimensional phosphopeptide mapping of CIITA using trypsin, polyacrylamide gels (8%) containing CIITA 32P labeled in vitro by PKA were washed three times for 10 min each in 500 ml of sterile deionized water. Following autoradiography, the CIITA band was excised and was placed in a microcentrifuge tube in 200 μl of ice-cold performic acid and incubated for 60 min on ice to achieve oxidation of the 32P-labeled CIITA. The gels were rinsed once with water, and trypsin cleavage was performed as follows. Briefly, 0.1 mg of trypsin (sequencing grade; Boehringer Mannheim)/ml in 1 mM HCl was diluted to 1 to 5 μg of trypsin in 100 μl with digestion buffer (50 mM ammonium bicarbonate, pH 7.8) immediately before use, and sufficient volume to cover each gel slice was used. After 13 h at 37°C, an additional 10-μg aliquot of trypsin was added, and incubation continued for 8 h. Samples were then centrifuged for 30 min, and the supernatants were diluted with 400 μl of water and then lyophilized under vacuum. The tryptic digests were resuspended in 400 μl (pH 1.9) of electrophoresis buffer and lyophilized again. The digests were then dissolved in 5 to 10 μl of electrophoresis buffer, pH 1.9 (15% acetic acid, 5% formic acid). Phosphopeptides were separated in the first dimension by electrophoresis in the same buffer on a thin-layer chromatography cellulose plate (EM Laboratories) at 1,000 V for 65 min using an HTLE 7000 apparatus (C.B.S. Scientific). Plates were then dried and subjected to ascending chromatography in the second dimension for 12 h with 37.5% butanol–25% pyridine–7.5% acetic acid. Radiolabeled phosphopeptides were detected by phosphorimaging.
RESULTS
PGE1, PGE2, 8-bromo-cAMP, and PKA inhibit activation by CIITA.
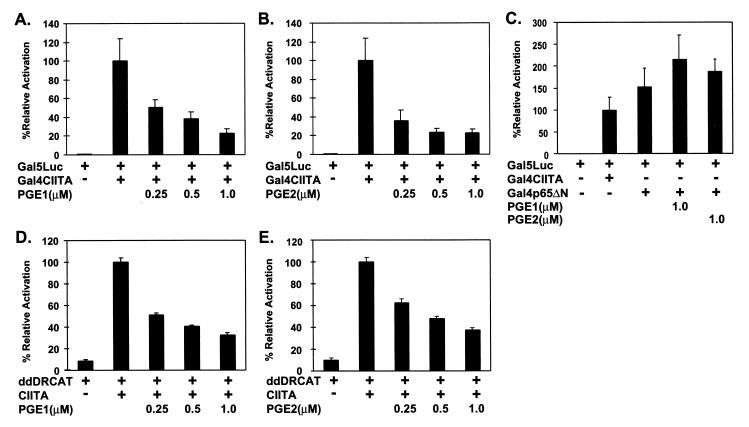
Previous reports have demonstrated the inhibition of constitutive and inducible class II MHC transcription and protein expression by PGE1 and PGE2 (2, 20, 30, 59). As both constitutive and inducible class II MHC transcription is contingent on CIITA, we hypothesized that this inhibition might involve posttranslational modification of CIITA. To investigate the effect of prostaglandin treatment on the ability of CIITA to activate transcription, we coexpressed CIITA fused to the Gal4 DNA-binding domain (Gal4CIITA) and a luciferase reporter containing five upstream Gal4 binding sites (Gal5Luc) in P388D1 cells and treated the cells with 0.25 to 1.0 μM PGE1 or PGE2. The P388D1 cells were chosen because they were used extensively in previous studies analyzing the effects of PGE on class II MHC expression. Use of the Gal5 reporter system is crucial because it allowed us to focus on CIITA-specific effects independent of class II MHC-associated transcription factors. The effect of PGE1 or PGE2 treatment on Gal5Luc reporter activation is shown in Fig. 1. Both PGE1 and PGE2 inhibited the ability of Gal4CIITA to activate transcription in a dose-dependent fashion (Fig. 1A and B). At the highest concentrations of PGE, inhibition is approximately fivefold. The maximal concentrations of PGE had no suppressive effect on activation of the Gal5Luc reporter by an unrelated Gal4–DNA-binding domain fusion containing an N-terminal deletion of the p65 subunit of NF-κB (Gal4p65ΔN) (Fig. 1C). Prostaglandins also inhibit, to a similar extent, class II MHC transcription and promoter function (Fig. 1D and E), consistent with data using the Gal5Luc system. This demonstrates that the ability of CIITA to activate transcription can be suppressed by PGE1- or PGE2-mediated events.
FIG. 1.
PGE1 and PGE2 inhibit CIITA-mediated transactivation. P388D1 cells were transfected with the plasmid DNAs shown and treated with the indicated concentrations of either PGE1 (A, C, and D) or PGE2 (B, C, and E). Luciferase or CAT activity was determined as described in Materials and Methods. Transcriptional activity is expressed as a percentage relative to activity using untreated, wild-type Gal4CIITA (A, B, and C) or CIITA (D and E). Results are presented as the means ± standard deviations of three separate experiments.
The effects of PGE are mediated largely through the activation of adenyl cyclase and subsequent production of second messenger cAMP, which then activates cAMP-dependent PKA (55). Given our observations that PGE1 and PGE2 can suppress transcriptional activation by CIITA, we reasoned that cAMP treatment should have a similar effect. To this end, we analyzed the effect of 8-bromo-cAMP treatment in our system. As anticipated, cAMP treatment of Gal4CIITA-expressing cells also shows suppression of Gal5Luc activation (Fig. 2A). 8-Bromo-cAMP treatment had a similar effect on CIITA-mediated transcription from the DR promoter (Fig. 2B).
FIG. 2.
cAMP inhibits CIITA-mediated transactivation. Transcriptional activation by Gal4CIITA (A) or CIITA (B) was determined using P388D1 cells transfected with the indicated plasmid DNAs and cultured in the absence or presence of increasing concentrations of 8-bromo-cAMP as indicated. Relative activation (as a percentage) is shown as described for Fig. 1. Values are means ± standard deviations from three separate experiments.
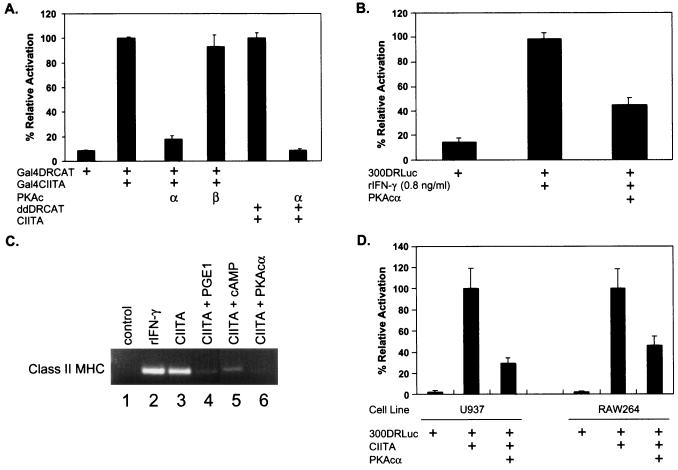
To determine if activation of PKA regulates CIITA activity, we examined the effects of transfected PKA on CIITA activity in vivo using transiently transfected cells. As expected, cells transfected with a Gal4-binding site CAT reporter construct (Gal4DRCAT) (53) alone show only a basal level of CAT acetylation while cotransfection of Gal4DRCAT and Gal4CIITA results in increased reporter activity (Fig. 3A). Significantly, cotransfected constructs encoding the catalytic subunit of PKAα, but not PKAβ, completely suppress DR promoter activity induced by CIITA. This is observed with either Gal4CIITA or the unmodified CIITA construct. To assess if similar treatments might alter class II MHC gene expression in response to IFN-γ, we examined the effect of transfected PKAα on IFN-γ-induced class II MHC promoter activation. As shown in Fig. 3B, transfection of PKAα inhibited promoter activation following treatment with mouse IFN-γ. To further determine if these effects could be extended to the endogenous class II MHC genes, the effects of these treatments on the CIITA-induced expression of class II MHC transcripts were measured. Levels of expression of the induced, endogenous, class II MHC message following either IFN-γ treatment or transfection of CIITA were comparable (Fig. 3C, lanes 1 to 3). Consistent with the reporter assays, PGE1, cAMP, and cotransfected PKAα all had similar effects on expression of class II MHC mRNA in cells transfected with CIITA (Fig. 3C, lanes 4 to 6). PKAα-mediated inhibition of CIITA-induced transcription was also observed in a monocytic cell line of human (U937) and mouse (RAW264) origins (Fig. 3D), indicating that this effect is not specific for the P388D1 cell line. These results demonstrate that PKA can inhibit the ability of CIITA to activate transcription.
FIG. 3.
PKAα inhibits CIITA-mediated transactivation. (A) PKAα inhibits CIITA-mediated promoter activation. Gal4DRCAT and Gal4CIITA (bars 1 to 4) or ddDrCAT and p3FgCIITA (bars 5 and 6) together with plasmids encoding either the PKAα or PKAβ catalytic subunit (c) were cotransfected into P388D1 cells. (B) PKAα represses IFN-γ-induced promoter activity. P388D1 cells were transfected with 300DRLuc with or without cotransfection of PKAα and cultured in the presence or absence of recombinant mouse IFN-γ (rIFN-γ). (C) PKAα inhibits CIITA-mediated transcription of endogenous class II MHC. P388D1 cells were transfected with empty vector or wild-type CIITA and treated as indicated. Cells were treated with IFN-γ (0.8 ng/ml), PGE1 (1.0 μM), or cAMP (1.0 μM) or cotransfected with PKAα as indicated. Expression of mRNA was determined by reverse transcription-PCR (see Materials and Methods). (D) PKAα inhibits CIITA-mediated transactivation in monocytic cell lines. The monocytic cell lines U937 and RAW264 were cotransfected with 300DRLuc and CIITA with or without PKAα using SuperFect (Qiagen). CAT (A) and luciferase (B and D) activities were determined at 36 h posttransfection. The data are the means ± standard deviations of three experiments and are shown normalized to the activity of Gal4CIITA or CIITA on the Gal4DRCAT or 300DRLuc reporter, respectively (100%).
Taken together, the observations that PGE, cAMP, and PKA all inhibit the transactivation function of CIITA suggest that the PGE-, cAMP-, or PKA-mediated inhibition of class II MHC transcription and expression is achieved through modulation of CIITA activity. Furthermore, as the class II MHC transcription factors (i.e., RFX and NF-Y) are not required for activity using the Gal5Luc reporter, this mode of inhibition may be specific for CIITA and independent of class II MHC promoter-specific transcription factors.
Phosphorylation of the CIITA protein by PKA and phosphoamino acid analysis.
As the above experiments implicated the PKA pathway in the modulation of CIITA activity, we examined the phosphorylation state of CIITA in vitro and in vivo (Fig. 4). For in vitro phosphorylation, the Flag-tagged CIITA protein was either expressed in transfected P388D1 cells or in vitro translated. The resulting proteins were immunoprecipitated and incubated with the catalytic subunit of PKAα in the presence of [γ-32P]ATP. CIITA immunoprecipitated from transfected-cell lysate is readily phosphorylated by PKAα (Fig. 4a, lane 1). In vitro-translated CIITA is also subject to phosphorylation by PKAα (lane 2). Phosphorylation was observed in the presence of PKAα (lanes 1 and 2) but not in the presence of a selection of other kinases, including cyclic GMP-dependent protein kinase, protein kinase C, calmodulin-dependent protein kinase II, and growth-associated histone H1 kinase (MPF, cdc2+/CDC28 protein kinase) (lanes 5 to 8). These data demonstrate that CIITA is a substrate for PKA in vitro. Next, we determined whether CIITA could be phosphorylated in vivo. CIITA expressed in transfected P388D1 cells and cultured in the presence of 32Pi reveals a modest level of phosphorylation (Fig. 4, lane 9), which increases markedly when the cells are cotransfected with both CIITA and PKAcα (lane 10). The relatively low level of basal CIITA phosphorylation may indicate the relative inactivity of PKA in P388D1 cells. Alternatively, the observed phosphorylation may also be due to other serine-specific protein kinases (see below). Several lighter bands were also phosphorylated and may represent nonspecific proteins bound by protein A- or G-agarose. However, the observed increase in 32P incorporation in the presence of PKAα demonstrates that CIITA can be phosphorylated in vivo by PKA.
FIG. 4.
CIITA phosphorylation assay in vitro and in vivo with PKA. (a) Lane 1, CIITA immunopurified from transfected-cell lysate; lanes 2 to 4, in vitro-translated and immunopurified CIITA was tested for phosphorylation by PKA with negative controls; lanes 5 through 8, immunopurified CIITA from translation in vitro was assayed for phosphorylation capability with kinases of cGMP-dependent protein kinase, protein kinase C, calmodulin-dependent protein kinase II, and growth-associated histone H1 kinase; lane 9, immunopurified CIITA from transiently transfected cells labeled with [32P]orthophosphate in vivo; lane 10, immunopurified CIITA labeled with [33P]orthophosphate in vivo from P388D1 cells transiently cotransfected with CIITA and PKAα. (b) Two-dimensional phosphoamino acid analysis using [γ-32P]ATP-labeled CIITA in vitro obtained from lane 2. (c) Two-dimensional phosphoamino acid analysis using [33P]orthophosphate-labeled CIITA in vivo obtained from lane 10.
Phosphoamino acid analyses were performed using either in vitro- or in vivo-expressed CIITA phosphorylated by PKA (Fig. 4). In both cases, we observed that phosphorylation occurred exclusively on serine residues. No phosphothreonine or phosphotyrosine was detected in the in vitro sample, even when the thin-layer chromatography plate was exposed to film for up to 2 weeks. This pattern of phosphorylation was identical to that seen with in vivo-expressed CIITA (Fig. 4C). Thus, these data provide strong evidence that any putative PKA phosphorylation sites in CIITA will contain only serine residues as the potential phosphate receptor.
Identification of PKA phosphorylation sites in CIITA.
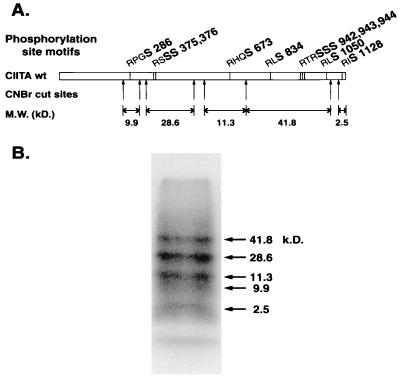
Ten potential PKA phosphorylation site motifs containing serine residues (RXS and RXXS) were identified in CIITA (Ser286; Ser375 and -376; Ser674; Ser834; Ser942, -943, and -944; Ser1050; and Ser1128). To determine which of these residues were likely phosphorylated, the CIITA protein was phosphorylated in vitro with PKA and subjected to CNBr digestion (Fig. 5). Five predicted CNBr fragments encompassed the putative PKA phosphorylation sites. Phosphorylated bands of the predicted molecular weights were observed for each of the five predicted CNBr fragments, suggesting that all 10 putative PKA sites were potentially phosphorylated.
FIG. 5.
Mapping of PKA phosphorylation sites in CIITA. (A) Diagram of CIITA showing potential PKA phosphorylation sites, relative positions of CNBr cleavage sites, and the predicted molecular masses of each cleavage product. Potential PKA phosphorylated serine residues are in boldface. (B) One-dimensional phosphopeptide mapping. Digestion of phosphorylated CIITA was performed with CNBr to generate peptides. Arrows, expected CNBr cleavage peptide fragments; the corresponding predicted molecular masses are shown. Peptides were separated by SDS-PAGE (16%). The observed molecular weights of 32P-labeled fragments are based on migration of molecular weight standards.
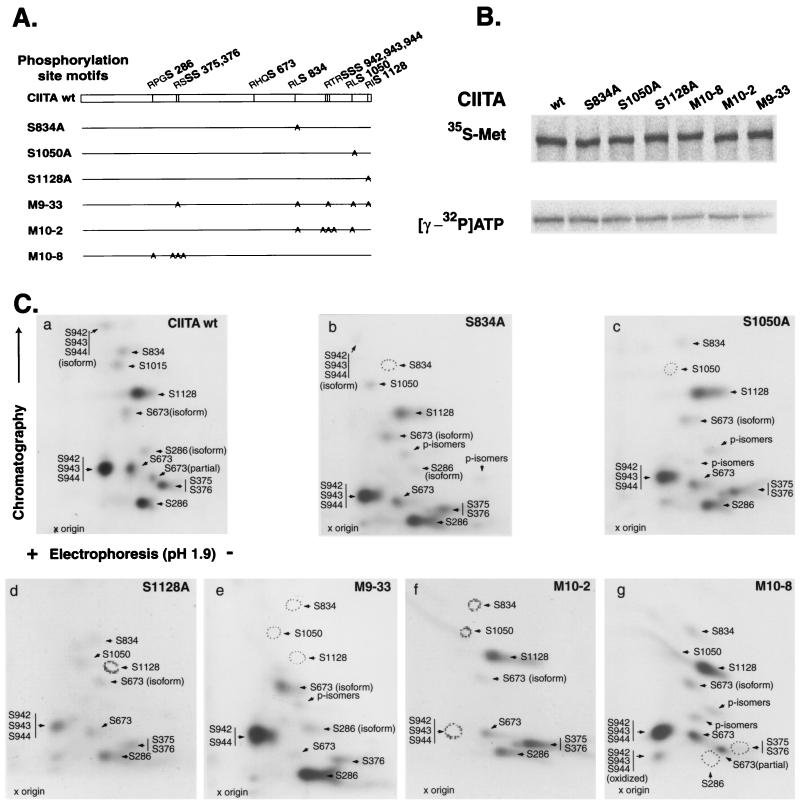
To further analyze CIITA phosphorylation by PKA, we prepared six CIITA mutant constructs in which various groupings of these 10 serine residues were mutated to alanine (Fig. 6A). These included three single mutants, two with five different mutated serine residues, and one with four serine residues mutated. Each of these mutants was translated normally in vitro and in cells, and PKA was able to phosphorylate all of the mutants to various degrees (Fig. 6B); however, mutants with four (M10-8) or five serine mutations (M10-2 and M9-33) showed as much as a 50% decrease in PKA phosphorylation. These observations are consistent with phosphorylation occurring at most if not all the putative PKA sites.
FIG. 6.
CIITA mutants and identification of PKA phosphorylation sites in CIITA. (A) Diagram of CIITA serine mutants. All mutants were sequenced to confirm the absence of deleterious mutations. (B) In vitro translation in the presence of [35S]Met revealed that every mutant yielded a full-length product (top). Bottom, in vitro phosphorylation using [32P]ATP. (C) Two-dimensional phosphopeptide mapping. Wild-type and mutant CIITAs were synthesized in vitro, immunoprecipitated with mouse monoclonal anti-Flag, and phosphorylated with PKA. Labeled proteins were separated by SDS-PAGE and excised for in-gel digestion with trypsin. Phosphopeptides generated by tryptic cleavage were separated by electrophoresis (horizontal dimension; pH 1.9; cathode on right) and thin-layer chromatography (vertical dimension) and detected by autoradiography.
A further delineation of the phosphorylated serine residues was accomplished by two-dimensional tryptic phosphopeptide mapping of wild-type and mutant CIITA proteins translated in vitro (Fig. 6C). A comparison of two-dimensional tryptic phosphopeptide maps from wild-type CIITA and mutant forms of CIITA allowed the assignment of specific serine residues by their absence on autoradiograms. We were able to resolve 10 phosphopeptide spots corresponding to locations that were clearly associated with the PKA consensus phosphorylation site motifs in CIITA. Three of these spots correspond to oxidized phosphopeptides (S286 and S674) or partial digestion products (S674). Two spots correspond to phosphopeptide fragments with multiple putative phosphorylation sites (S942, -943, and -944 and S375 and -376). These findings demonstrate that in vitro CIITA contains phosphorylated serines at the predicted PKA sites. Given the increased spot intensities seen for the two sites with multiple serines, S375 and -376 and S942, -943, and -944, and the decreased phosphorylation of the S375 and -376 spot when S375 is mutated (M9-33), it is probable that S375, S376, and at least two of the three serines from 942 to 944 are phosphorylated.
Mutation of PKA sites in CIITA abolish PKA-mediated inhibition of CIITA.
We have attempted in vivo phosphopeptide analysis of CIITA mutants in the presence of PKA; however these analyses were extremely difficult due to the low level of CIITA in P388D1 cells and the formidable amount of radioisotope required. As an alternative, experiments were performed to test the ability of PKAα to inhibit the transactivation of the HLA-DR promoter by wild-type CIITA or CIITA PKA site mutants in P388D1 cells. Wild-type CIITA or the PKA site mutants were cotransfected together with either a control plasmid or the PKAα plasmid, and activation of the DR promoter-driven CAT reporter was examined (Fig. 7). Without cotransfected PKA, all of the CIITA PKA site mutants retained some ability to activate transcription (approximately 40 to 130% of that of wild-type CIITA). A modest to significant reduction of transactivation in the absence of PKA occurs when PKA sites in the C-terminal portion of CIITA are mutated (S834, S942 to -944, and S1050; mutants S834A, S1050A, M10-2, and M9-33). Mutation of S1128 did not decrease the function of CIITA. Mutation of the more-N-terminal PKA sites S286 and S375 and -376 (M10-8) led to a modest increase in transactivation compared to that for wild-type CIITA (130%). Coexpression of PKAα reduces transcriptional activation by wild-type CIITA to 20% of that seen without PKAα (Fig. 7). Mutants S834A and S1050A are not inhibited by PKAα coexpression at all, indicating that serine residues 834 and 1050 are crucial for PKAα-mediated suppression of CIITA function. In fact, a small but reproducible degree of enhanced transcriptional activation is observed with these two mutants.
FIG. 7.
Suppression of CIITA and phosphorylation mutants by PKAα. (A) P388D1 cells were transiently cotransfected with 5 μg of the indicated plasmids and 2 μg of the DRCAT reporter. CAT activity was assayed and expressed relative to that for wild-type CIITA in the absence of exogenous PKA. The averages of three independent experiments ± standard deviations are shown. (B) Diagram of the observed effect of PGE-signaling events on class II transcription.
In contrast, S1128A and M10-8, mutants that display no decrease in transactivation in the absence of coexpressed PKA, are significantly inhibited in the presence of PKA. The inhibition of these mutants in the presence of PKA is less that of the wild-type CIITA, suggesting that the mutated serines (S286, S374, -375, and -376, and S1128) may play a role in PKA-mediated inhibition of CIITA function but that they are not the key residues involved in this inhibition.
M10-2 and M9-33 display a profile similar to that for S834A and S1050A in that they are no longer susceptible to PKAα-mediated suppression. The latter two constructs contain the S834A and S1050A mutations, consistent with the conclusion that residues 834 and 1050 are crucial. In addition to mutations at S834 and S1050, M10-2 is also mutated at S942 to -944, but the role of these residues is unresolved. Similarly, M9-33 is mutated at S943 and S1128 in addition to S834 and S1050. The characterization of the S1128A mutant described above ruled out a role for this residue in PKA-mediated suppression of CIITA activity. The role of S943 alone is presently unclear. Nonetheless, these data are compatible with the conclusion that the phosphorylation at S834 and S1050 correlates directly with the ability of PKA to inhibit the transactivation of HLA-DR by CIITA. This indicates that the observed action of PKA agonists (PGE, cAMP, forskolin, etc.) on class II MHC genes is likely mediated through phosphorylation of these two residues, although additional serine residues may play a minor role.
DISCUSSION
It has been appreciated for some time that PGE treatment, which increases intracellular cAMP, leads to downregulation of class II MHC surface expression in a transcription-dependent fashion (21, 29, 59). The mechanism by which PGE and cAMP modulate transcription has not been addressed previously. Because the likely targets for such regulation are the transcription factors which regulate class II MHC expression, we examined the class II transactivator (CIITA) as a potential target for PGE- and cAMP-mediated transcriptional repression of class II MHC genes. Here we present compelling evidence that PGE-mediated repression of class II MHC transcription involves the phosphorylation of CIITA by PKA. PGE1, PGE2, 8-bromo-cAMP, and PKA all repress class II MHC transcription and also reduce Gal4CIITA-mediated transcription of a Gal4 binding site containing a promoter lacking W-, X-, and Y-box elements. This suggests that the effects of PGE on class II MHC transcription may be largely independent of the factors that bind these elements. While PKAα is sufficient for these effects, PKAβ is not, indicating the specific activation of PKAα in PGE-mediated modulation of class II MHC expression. In support of these observations PKA can phosphorylate CIITA both in vitro and in vivo, suggesting that direct phosphorylation of CIITA is critical. Four other serine/threonine kinases failed to phosphorylate CIITA in vitro, demonstrating that phosphorylation of CIITA is likely kinase specific. Furthermore, functional characterization and phosphopeptide mapping of PKA site mutations reveal that, despite numerous PKA phosphorylation sites within CIITA, only two individual phosphorylation sites can be unambiguously linked to the repressive effects of PKA. Taken together these results demonstrate a PKA-mediated regulation of CIITA transactivation function and describe a mechanism by which PGEs can modulate transcription of class II MHC genes.
Mechanistic studies prior to this report suggested that elements within the class II MHC promoter were required for the inhibitory effects of cAMP and PKA on class II MHC transcription in B cells (30). S- and X-box sequences appeared to be somewhat important for repression, but the low degree of reporter activity using either mutations in individual promoter elements or deletions made it difficult to interpret the observed loss of repression. With this caveat in mind, mutation of the X2 box, which contains a consensus cAMP response element (CRE), had no effect on PKA-mediated repression but had a substantial effect on constitutive reporter activity. The CRE binding protein (CREB) is activated by PKA and activates many cAMP response promoters through phosphorylation-dependent recruitment of coactivator CBP (16, 32, 35, 47). CREB has been identified as a protein that can bind X2 (43) and interacts with CIITA as part of the class II MHC transcriptional scaffold (64). In murine macrophage cell line P388D1, transcriptional repression of IFN-γ-induced class II MHC by PKA was not affected by mutations in the S, X1, X2, or Y boxes or by complete deletion of S, X1, and X2 (29). However, as in the B-cell study, these mutations did diminish IFN-γ-induced promoter activity from 40 to 70%.
Our identification of CIITA as a target in the PGE/cAMP/PKA pathway is not inconsistent with these previous studies. IFN-γ induces the expression of CIITA, and CIITA is generally required for class II MHC transcription and expression (reviewed in reference 27). Thus CIITA is a critical component of the IFN-γ signaling pathway necessary for class II MHC transcription. At the level of the class II MHC promoter, the W, X, and Y elements are essential for full transcriptional activation. Deletion or mutation of individual sequence elements from class II MHC promoters substantially reduces the induction seen with IFN-γ or CIITA (29, 54, 64). As CIITA-dependent transcription is practically abolished by the introduction of PKA and individual phosphorylation site point mutations in CIITA completely block the effect of PKA, it is highly probable that CIITA is a principal target of the PGE pathway with regard to class II MHC expression in these cells. This hypothesis is supported by our observation that the PGE/cAMP/PKA pathway also inhibits Gal4CIITA activity on a promoter without class II MHC-specific promoter elements. Repression of CIITA activity by PGE is in the range of 2.5- to 5-fold (Fig. 1). In experiments using cAMP, the degree of suppression is similar (Fig. 2). The effects of PGE and cAMP addition may differ, as the effects of added cAMP may be muted due to the cAMP-depleting activity of phosphodiesterases and the lack of a sustained signal (such as PGE) to maintain cAMP generation. PGE, cAMP, and PKA treatment also inhibited transcription of endogenous class II MHC, and the extents of repression by these treatments are comparable. An important caveat in using the Gal4CIITA system is that the contributions of X- and Y-box transcription factors are circumvented by employment of the Gal4 DNA-binding domain such that CIITA is directed to the promoter without RF-X, NF-Y, CREB, etc. However, with this system, modifications that directly impact the ability of CIITA to activate transcription can be observed. Although there is congruence in both our reporter systems (Fig. 1 and 2), we have not ruled out the possibility that disrupted interactions with the essential X- and Y-box binding proteins are another consequence of PKA-mediated CIITA phosphorylation. In addition, PKA phosphorylation of CREB may have other effects on the class II MHC promoter independent of CIITA phosphorylation events. It is also reasonable to consider the possibility that S834 and S1050 of CIITA may be potential targets for other serine kinases with potential roles in regulating class II MHC expression.
By what mechanism does phosphorylation regulate CIITA? This is a key question arising from the results of this study. A priori, the regulatory effects of CIITA phosphorylation could be either positive or negative. Phosphorylation of CIITA could permit binding to the requisite transcription factors, thus positively modulating transactivation. Phosphorylation events leading to protein dimerization followed by productive DNA binding on a promoter are common (e.g., CREB [16] and STATs [14, 17, 18]). This mechanism seems plausible, as the mutation of certain phosphorylation sites in CIITA can downregulate its ability to activate transcription. However, our data indicate that phosphorylation of CIITA by PKA leads to a loss of transactivation ability. This effect is associated with phosphorylation at residues 874 and 1050 of CIITA. Based on our current understanding of CIITA function, there are a number of possibilities that could allow for the observed negative regulation. The simplest is that phosphorylation of S874 and S1050 leads to failed nuclear localization. Nuclear import of yeast transcription factor Pho4 is regulated by a number of serine phosphorylation events (33). Thus far, attempts to visualize CIITA localization by immunofluorescence in the P388D1 cell line have been unsuccessful due to the extremely low levels of CIITA expression (data not shown). In Cos-7 cells, all of the mutants examined in this study had substantial cytoplasmic staining, although nuclear CIITA was detectable in all but M10-8 and M9-33 (data not shown). However, the effects of PKA are apparent even when using a Gal4CIITA fusion that contains Gal4 nuclear localization sequences, strongly suggesting that the effect of PKA is not an alteration of nuclear localization. Another possibility is that phosphorylation of CIITA disrupts a critical protein-protein interaction. Transcription factors RFX5, RFX-ANK, NF-YB, NF-YC, and CREB have been shown to bind CIITA (19, 41, 56, 64), as have several components of the basal transcription machinery, including CBP (23, 34), TFIID (40), and various TATA-binding protein-associated factors (22, 40). Treatment of in vitro-translated CIITA with PKA did not disrupt the binding of CIITA to glutathione S-transferase-CBP fusion proteins in a pull-down assay (data not shown), although this was not surprising as the important serine residues are quite distant from CIITA's N-terminal CBP binding site (23, 34). None of the CIITA-interacting transcription factors mentioned above mapped to a region near amino acid 874 or 1050 of CIITA (64). This suggests that, while disrupting these interactions via phosphorylation is plausible, it would require a mechanism more complex than simple inhibition of a binding site. Our data support this idea to some extent. PKA is able to inhibit transactivation of the Gal5 promoter by Gal4CIITA and transactivation from DR by wild-type CIITA. We conclude that this reflects an independence from the specific promoter elements. The observation also suggests that the effect of phosphorylation is likely to affect the activation domain of CIITA despite its distance from the phosphorylation site. A reasonable hypothesis is that a conformational change alters the accessibility of the activation domain and permits or disallows contact with an important protein. Phosphorylation might affect the folding or conformation of CIITA. Recently, our laboratory has observed that the leucine-rich repeats and sequences in or near the GTP-binding site of CIITA mediate homotypic and heterotypic self-association of CIITA (37). Although the exact role that these associations play is presently unclear, it is conceivable that phosphorylation at S874 and S1050 might influence some presently unknown mechanistic step involving one of more of these self-associations. Another possibility is that phosphorylation targets CIITA for degradation. This has been observed for IkB, the regulator of rel-family transcription factor NF-κB (4), and with c-myb, a transcription factor involved in hematopoiesis (6).
In summary, this work explains the long-observed finding that prostaglandins and cAMP treatment of macrophages cause the inhibition of IFN-γ-induced class II MHC gene expression. This is the first documentation that PGE, cAMP, and PKA can all inhibit CIITA function. This inhibition occurs through the phosphorylation of CIITA, and at least two critical serine residues, S874 and/or S1050, have been identified as the crucial kinase targets. Elucidation of the precise mechanisms involved should prove informative and increase our understanding of CIITA function in macrophage and nonmacrophage cells.
REFERENCES
- 1.Amaldi I, Reith W, Berte C, Mach B. Induction of HLA class II genes by IFN-gamma is transcriptional and requires a trans-acting protein. J Immunol. 1989;142:999–1004. [PubMed] [Google Scholar]
- 2.Askew D, Burger C J, Elgert K D. Tumor-induced modulation of macrophage class II MHC molecule mRNA expression. Mol Immunol. 1993;30:911–920. doi: 10.1016/0161-5890(93)90015-4. [DOI] [PubMed] [Google Scholar]
- 3.Baudeau C, Delarue F, He C J, Nguyen G, Adida C, Peraldi M N, Sraer J D, Rondeau E. Induction of MHC class II molecules HLA-DR, -DP and -DQ and ICAM 1 in human podocytes by gamma-interferon. Exp Nephrol. 1994;2:306–312. [PubMed] [Google Scholar]
- 4.Beg A A, Baldwin A S., Jr The I kappa B proteins: multifunctional regulators of Rel/NF-kappa B transcription factors. Genes Dev. 1993;7:2064–2070. doi: 10.1101/gad.7.11.2064. [DOI] [PubMed] [Google Scholar]
- 5.Berman B, Duncan M R, Smith B, Ziboh V A, Palladino M. Interferon enhancement of HLA-DR antigen expression on epidermal Langerhans cells. J Investig Dermatol. 1985;84:54–58. doi: 10.1111/1523-1747.ep12274691. [DOI] [PubMed] [Google Scholar]
- 6.Bies J, Feikova S, Bottaro D P, Wolff L. Hyperphosphorylation and increased proteolytic breakdown of c-Myb induced by the inhibition of Ser/Thr protein phosphatases. Oncogene. 2000;19:2846–2854. doi: 10.1038/sj.onc.1203613. [DOI] [PubMed] [Google Scholar]
- 7.Bontron S, Ucla C, Mach B, Steimle V. Efficient repression of endogenous major histocompatibility complex class II expression through dominant-negative CIITA mutants isolated by a functional selection strategy. Mol Cell Biol. 1997;17:4249–4258. doi: 10.1128/mcb.17.8.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boss J M. Regulation of transcription of MHC class II genes. Curr Opin Immunol. 1997;9:107–113. doi: 10.1016/s0952-7915(97)80166-5. [DOI] [PubMed] [Google Scholar]
- 9.Brickey W J, Wright K L, Zhu X S, Ting J P. Analysis of the defect in IFN-gamma induction of MHC class II genes in G1B cells: identification of a novel and functionally critical leucine-rich motif (62-LYLYLQL-68) in the regulatory factor X 5 transcription factor. J Immunol. 1999;163:6622–6630. [PubMed] [Google Scholar]
- 10.Brown A M, Wright K L, Ting J P. Human major histocompatibility complex class II-associated invariant chain gene promoter. Functional analysis and in vivo protein/DNA interactions of constitutive and IFN-gamma-induced expression. J Biol Chem. 1993;268:26328–26333. [PubMed] [Google Scholar]
- 11.Campbell I L, Oxbrow L, Koulmanda M, Harrison L. IFN-gamma induces islet cell MHC antigens and enhances autoimmune streptozotocin-induced diabetes in the mouse. J Immunol. 1988;140:1111–1116. [PubMed] [Google Scholar]
- 12.Chang C H, Fontes J D, Peterlin M, Flavell R A. Class II transactivator (CIITA) is sufficient for the inducible expression of major histocompatibility complex class II genes. J Exp Med. 1994;180:1367–1374. doi: 10.1084/jem.180.4.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chapman H A. Endosomal proteolysis and MHC class II function. Curr Opin Immunol. 1998;10:93–102. doi: 10.1016/s0952-7915(98)80038-1. [DOI] [PubMed] [Google Scholar]
- 14.Chatterjee-Kishore M, van den Akker F, Stark G R. Association of STATs with relatives and friends. Trends Cell Biol. 2000;10:106–111. doi: 10.1016/s0962-8924(99)01709-2. [DOI] [PubMed] [Google Scholar]
- 15.Chin K C, Li G G, Ting J P. Importance of acidic, proline/serine/threonine-rich, and GTP-binding regions in the major histocompatibility complex class II transactivator: generation of transdominant-negative mutants. Proc Natl Acad Sci USA. 1997;94:2501–2506. doi: 10.1073/pnas.94.6.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 17.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 18.Decker T, Kovarik P. Transcription factor activity of STAT proteins: structural requirements and regulation by phosphorylation and interacting proteins. Cell Mol Life Sci. 1999;55:1535–1546. doi: 10.1007/s000180050393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSandro A M, Nagarajan U M, Boss J M. Associations and interactions between bare lymphocyte syndrome factors. Mol Cell Biol. 2000;20:6587–6599. doi: 10.1128/mcb.20.17.6587-6599.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fedyk E R, Phipps R P. Prostaglandin E2 receptors of the EP2 and EP4 subtypes regulate activation and differentiation of mouse B lymphocytes to IgE-secreting cells. Proc Natl Acad Sci USA. 1996;93:10978–10983. doi: 10.1073/pnas.93.20.10978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueiredo F, Uhing R J, Okonogi K, Gettys T W, Johnson S P, Adams D O, Prpic V. Activation of the cAMP cascade inhibits an early event involved in murine macrophage Ia expression. J Biol Chem. 1990;265:12317–12323. [PubMed] [Google Scholar]
- 22.Fontes J D, Jiang B, Peterlin B M. The class II trans-activator CIITA interacts with the TBP-associated factor TAFII32. Nucleic Acids Res. 1997;25:2522–2528. doi: 10.1093/nar/25.12.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fontes J D, Kanazawa S, Jean D, Peterlin B M. Interactions between the class II transactivator and CREB binding protein increase transcription of major histocompatibility complex class II genes. Mol Cell Biol. 1999;19:941–947. doi: 10.1128/mcb.19.1.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giacomini P, Fisher P B, Duigou G J, Gambari R, Natali P G. Regulation of class II MHC gene expression by interferons: insights into the mechanism of action of interferon. Anticancer Res. 1988;8:1153–1161. [PubMed] [Google Scholar]
- 25.Hake S B, Masternak K, Kammerbauer C, Janzen C, Reith W, Steimle V. CIITA leucine-rich repeats control nuclear localization, in vivo recruitment to the major histocompatibility complex (MHC) class II enhanceosome, and MHC class II gene transactivation. Mol Cell Biol. 2000;20:7716–7725. doi: 10.1128/mcb.20.20.7716-7725.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harton J A, Cressman D E, Chin K C, Der C J, Ting J P. GTP binding by class II transactivator: role in nuclear import. Science. 1999;285:1402–1405. doi: 10.1126/science.285.5432.1402. [DOI] [PubMed] [Google Scholar]
- 27.Harton J A, Ting J P. Class II transactivator: mastering the art of major histocompatibility complex expression. Mol Cell Biol. 2000;20:6185–6194. doi: 10.1128/mcb.20.17.6185-6194.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hobart M, Ramassar V, Goes N, Urmson J, Halloran P F. IFN regulatory factor-1 plays a central role in the regulation of the expression of class I and II MHC genes in vivo. J Immunol. 1997;158:4260–4269. [PubMed] [Google Scholar]
- 29.Ivashkiv L B, Ayres A, Glimcher L H. Inhibition of IFN-gamma induction of class II MHC genes by cAMP and prostaglandins. Immunopharmacology. 1994;27:67–77. doi: 10.1016/0162-3109(94)90008-6. [DOI] [PubMed] [Google Scholar]
- 30.Ivashkiv L B, Glimcher L H. Repression of class II major histocompatibility complex genes by cyclic AMP is mediated by conserved promoter elements. J Exp Med. 1991;174:1583–1592. doi: 10.1084/jem.174.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kern M J, Stuart P M, Omer K W, Woodward J G. Evidence that IFN-c does not affect MHC class II gene expression at the posttranscriptional level in a mouse macrophage cell line. Immunogenetics. 1989;30:258–265. doi: 10.1007/BF02421329. [DOI] [PubMed] [Google Scholar]
- 32.Kingsley-Kallesen M L, Kelly D, Rizzino A. Transcriptional regulation of the transforming growth factor-beta2 promoter by cAMP-responsive element-binding protein (CREB) and activating transcription factor-1 (ATF-1) is modulated by protein kinases and the coactivators p300 and CREB-binding protein. J Biol Chem. 1999;274:34020–34028. doi: 10.1074/jbc.274.48.34020. [DOI] [PubMed] [Google Scholar]
- 33.Komeili A, O'Shea E K. Roles of phosphorylation sites in regulating activity of the transcription factor Pho4. Science. 1999;284:977–980. doi: 10.1126/science.284.5416.977. [DOI] [PubMed] [Google Scholar]
- 34.Kretsovali A, Agalioti T, Spilianakis C, Tzortzakaki E, Merika M, Papamatheakis J. Involvement of CREB binding protein in expression of major histocompatibility complex class II genes via interaction with the class II transactivator. Mol Cell Biol. 1998;18:6777–6783. doi: 10.1128/mcb.18.11.6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwok R P, Lundblad J R, Chrivia J C, Richards J P, Bachinger H P, Brennan R G, Roberts S G, Green M R, Goodman R H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 36.Larsen G L, Henson P M. Mediators of inflammation. Annu Rev Immunol. 1983;1:335–359. doi: 10.1146/annurev.iy.01.040183.002003. [DOI] [PubMed] [Google Scholar]
- 37.Linhoff M W, Harton J A, Cressman D E, Martin B K, Ting J P-Y. Two distinct domains within CIITA mediate self-association: involvement of the GTP-binding and leucine-rich repeat domains. Mol Cell Biol. 2001;21:3001–3011. doi: 10.1128/MCB.21.9.3001-3011.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y, Ussery G D, Muncaster M M, Gallie B L, Blanck G. Evidence for retinoblastoma protein (RB) dependent and independent IFN-gamma responses: RB coordinately rescues IFN-gamma induction of MHC class II gene transcription in noninducible breast carcinoma cells. Oncogene. 1994;9:1015–1019. [PubMed] [Google Scholar]
- 39.Mach B, Steimle V, Martinez-Soria E, Reith W. Regulation of MHC class II genes: lessons from a disease. Annu Rev Immunol. 1996;14:301–331. doi: 10.1146/annurev.immunol.14.1.301. [DOI] [PubMed] [Google Scholar]
- 40.Mahanta S K, Scholl T, Yang F C, Strominger J L. Transactivation by CIITA, the type II bare lymphocyte syndrome-associated factor, requires participation of multiple regions of the TATA box binding protein. Proc Natl Acad Sci USA. 1997;94:6324–6329. doi: 10.1073/pnas.94.12.6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Masternak K, Muhlethaler-Mottet A, Villard J, Zufferey M, Steimle V, Reith W. CIITA is a transcriptional coactivator that is recruited to MHC class II promoters by multiple synergistic interactions with an enhanceosome complex. Genes Dev. 2000;14:1156–1166. [PMC free article] [PubMed] [Google Scholar]
- 42.McDevitt H O. The role of MHC class II molecules in susceptibility and resistance to autoimmunity. Curr Opin Immunol. 1998;10:677–681. doi: 10.1016/s0952-7915(98)80088-5. [DOI] [PubMed] [Google Scholar]
- 43.Moreno C S, Beresford G W, Louis-Plence P, Morris A C, Boss J M. CREB regulates MHC class II expression in a CIITA-dependent manner. Immunity. 1999;10:143–151. doi: 10.1016/s1074-7613(00)80015-1. [DOI] [PubMed] [Google Scholar]
- 44.Muhlethaler-Mottet A, Otten L A, Steimle V, Mach B. Expression of MHC class II molecules in different cellular and functional compartments is controlled by differential usage of multiple promoters of the transactivator CIITA. EMBO J. 1997;16:2851–2860. doi: 10.1093/emboj/16.10.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohmann H B, Campos M, Lawman M J, Babiuk L A. Induction of MHC class II antigens on bovine cells of nonlymphoid origin by recombinant bovine interferon-gamma and tumor necrosis factor-alpha. J Interferon Res. 1988;8:451–462. doi: 10.1089/jir.1988.8.451. [DOI] [PubMed] [Google Scholar]
- 46.Otten L A, Steimle V, Bontron S, Mach B. Quantitative control of MHC class II expression by the transactivator CIITA. Eur J Immunol. 1998;28:473–478. doi: 10.1002/(SICI)1521-4141(199802)28:02<473::AID-IMMU473>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 47.Parker D, Ferreri K, Nakajima T, LaMorte V J, Evans R, Koerber S C, Hoeger C, Montminy M R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol Cell Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peijnenburg A, Gobin S J, van Eggermond M C, Godthelp B C, van Graafeiland N, van den Elsen P J. Introduction of exogenous class II trans-activator in MHC class II-deficient ABI fibroblasts results in incomplete rescue of MHC class II antigen expression. J Immunol. 1997;159:2720–2727. [PubMed] [Google Scholar]
- 49.Peijnenburg A, van den Berg R, van Eggermond M J, Sanal O, Vossen J M, Lennon A, Alcaide-Loridan C, van den Elsen P J. Defective MHC class II expression in an MHC class II deficiency patient is caused by a novel deletion of a splice donor site in the MHC class II transactivator gene. Immunogenetics. 2000;51:42–49. doi: 10.1007/s002510050007. [DOI] [PubMed] [Google Scholar]
- 50.Pieters J. MHC class II restricted antigen presentation. Curr Opin Immunol. 1997;9:89–96. doi: 10.1016/s0952-7915(97)80164-1. [DOI] [PubMed] [Google Scholar]
- 51.Piskurich J F, Linhoff M W, Wang Y, Ting J P. Two distinct gamma interferon-inducible promoters of the major histocompatibility complex class II transactivator gene are differentially regulated by STAT1, interferon regulatory factor 1, and transforming growth factor beta. Mol Cell Biol. 1999;19:431–440. doi: 10.1128/mcb.19.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Piskurich J F, Wang Y, Linhoff M W, White L C, Ting J P. Identification of distinct regions of 5′ flanking DNA that mediate constitutive, IFN-gamma, STAT1, and TGF-beta-regulated expression of the class II transactivator gene. J Immunol. 1998;160:233–240. [PubMed] [Google Scholar]
- 53.Riley J L, Boss J M. Class II MHC transcriptional mutants are defective in higher order complex formation. J Immunol. 1993;151:6942–6953. [PubMed] [Google Scholar]
- 54.Riley J L, Westerheide S D, Price J A, Brown J A, Boss J M. Activation of class II MHC genes requires both the X box region and the class II transactivator (CIITA) Immunity. 1995;2:533–543. doi: 10.1016/1074-7613(95)90033-0. [DOI] [PubMed] [Google Scholar]
- 55.Samuelsson B, Goldyne M, Granstrom E, Hamberg M, Hammarstrom S, Malmsten C. Prostaglandins and thromboxanes. Annu Rev Biochem. 1978;47:997–1029. doi: 10.1146/annurev.bi.47.070178.005025. [DOI] [PubMed] [Google Scholar]
- 56.Scholl T, Mahanta S K, Strominger J L. Specific complex formation between the type II bare lymphocyte syndrome-associated transactivators CIITA and RFX5. Proc Natl Acad Sci USA. 1997;94:6330–6334. doi: 10.1073/pnas.94.12.6330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siegrist C A, Martinez-Soria E, Kern I, Mach B. A novel antigen-processing-defective phenotype in major histocompatibility complex class II-positive CIITA transfectants is corrected by interferon-gamma. J Exp Med. 1995;182:1793–1799. doi: 10.1084/jem.182.6.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sisk T J, Gourley T, Roys S, Chang C H. MHC class II transactivator inhibits IL-4 gene transcription by competing with NF-AT to bind the coactivator CREB binding protein (CBP)/p300. J Immunol. 2000;165:2511–2517. doi: 10.4049/jimmunol.165.5.2511. [DOI] [PubMed] [Google Scholar]
- 59.Snyder D S, Unanue E R. Corticosteroids inhibit murine macrophage Ia expression and interleukin 1 production. J Immunol. 1982;129:1803. [PubMed] [Google Scholar]
- 60.Steimle V, Siegrist C A, Mottet A, Lisowska-Grospierre B, Mach B. Regulation of MHC class II expression by interferon-gamma mediated by the transactivator gene CIITA. Science. 1994;265:106–109. doi: 10.1126/science.8016643. [DOI] [PubMed] [Google Scholar]
- 61.Tosi G, De Lerma Barbaro A, D'Agostino A, Valle M T, Megiovanni A M, Manca F, Caputo A, Barbanti-Brodano G, Accolla R S. HIV-1 Tat mutants in the cysteine-rich region downregulate HLA class II expression in T lymphocytic and macrophage cell lines. Eur J Immunol. 2000;30:19–28. doi: 10.1002/1521-4141(200001)30:1<19::AID-IMMU19>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 62.Tschickardt M E, Lu Y, Jacim M, Ussery G D, Steimle V, Mach B, Blanck G. RB and a novel E2F-1 binding protein in MHC class II deficient B-cell lines and normal IFN-gamma induction of the class IL transactivator CIITA in class II non-inducible RB-defective tumor lines. Int J Cancer. 1995;62:461–465. doi: 10.1002/ijc.2910620417. [DOI] [PubMed] [Google Scholar]
- 63.Zhou H, Glimcher L H. Human MHC class II gene transcription directed by the carboxyl terminus of CIITA, one of the defective genes in type II MHC combined immune deficiency. Immunity. 1995;2:545–553. doi: 10.1016/1074-7613(95)90034-9. [DOI] [PubMed] [Google Scholar]
- 64.Zhu X S, Linhoff M W, Li G, Chin K C, Maity S N, Ting J P-Y. A transcriptional scaffold: CIITA interacts with NF-Y, RFX, and CREB to cause stereospecific regulation of the class II MHC promoter. Mol Cell Biol. 2000;20:6051–6061. doi: 10.1128/mcb.20.16.6051-6061.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zlotnik A, Shimonkevitz R, Kappler J, Marrack P. Effect of prostaglandin E2 on the gamma-interferon induction of antigen-presenting ability in P388D1 cells and on IL-2 production by T-cell hybridomas. Cell Immunol. 1985;90:154–166. doi: 10.1016/0008-8749(85)90177-7. [DOI] [PubMed] [Google Scholar]