Viruses are diverse and ubiquitous, but many infection models do not recapitulate natural transmission and evolution. Here we bridge natural and experimental systems to study real-time transmission dynamics within and between hosts, identify novel viruses, and infer virus–pathogen relationships.
Abstract
Emerging viruses threaten global health, but few experimental models can characterize the virus and host factors necessary for within- and cross-species transmission. Here, we leverage a model whereby pet store mice or rats—which harbor natural rodent pathogens—are cohoused with laboratory mice. This “dirty” mouse model offers a platform for studying acute transmission of viruses between and within hosts via natural mechanisms. We identified numerous viruses and other microbial species that transmit to cohoused mice, including prospective new members of the Coronaviridae, Astroviridae, Picornaviridae, and Narnaviridae families, and uncovered pathogen interactions that promote or prevent virus transmission. We also evaluated transmission dynamics of murine astroviruses during transmission and spread within a new host. Finally, by cohousing our laboratory mice with the bedding of pet store rats, we identified cross-species transmission of a rat astrovirus. Overall, this model system allows for the analysis of transmission of natural rodent viruses and is a platform to further characterize barriers to zoonosis.
Introduction
Viruses are capable of rapid evolution to both adapt to new hosts and evade host immune responses. Understanding virus population dynamics during intraspecies transmission is particularly important for studying recently emerged viruses. As we have seen with SARS-CoV-2 and influenza viruses, the rise of novel virus variants can have a drastic effect on disease pathogenesis and adaptive immunity (Altman et al., 2018; Hacisuleyman et al., 2021; Wang et al., 2021; Washington et al., 2021). Within a single host, a virus population exists with a wide array of variants. Bottlenecks can have a dramatic impact on virus evolution (McCrone and Lauring, 2018; Zwart and Elena, 2015), but they can be difficult to study in natural settings because it requires access to both reservoir and host. Additionally, meaningful bottleneck analysis is best achieved through natural transmission, but it is difficult to evaluate the role of innate immunity in natural settings. Many laboratory animal models offer an opportunity to study transmission bottlenecks, and transgenic knockout strains are available to characterize the role of specific immune proteins or pathways in controlling bottlenecks. However, few such studies have been completed. Identifying the host factors that constrain transmission bottlenecks is a critical piece in understanding virus evolution.
The global virome is immense, with an estimated 1.6 million viruses capable of infecting mammals and birds (Carroll et al., 2018). Climate change is forcing migration of species into new habitats, resulting in new cross-species interactions, further increasing the odds of pathogens spilling over into new hosts (Olival et al., 2017). This, along with the expanding human population, has led to the ever-increasing threat of a zoonotic event and the devastating consequences of viruses that are able to rapidly adapt and spread in humans. Fortunately, zoonoses are rare. However, this also makes it challenging to study potential spillovers and subsequent transmission chains. Isolating viruses with zoonotic potential for experimental infection in hosts or cells is labor intensive and may not result in infection or pathology. Additionally, growing viruses in vitro and infecting model organisms at a high dose under nonphysiological routes can fail to recapitulate key aspects of disease and virus biology from the native species and often bypasses natural routes of transmission (Gillgrass et al., 2021; Morrison and Diamond, 2017; Muñoz-Fontela et al., 2020). Finally, many cross-species events likely start as dead-end infections, where limited replication and no intra-species transmission occurs until mutational combinations encode for productive infections in the new host (Morens and Fauci, 2020). Dead-end transmission events are invisible to many current techniques. Therefore, new systems to study barriers to viral spillover are of the utmost importance.
Animal models of virus infection and transmission often require genetic modification to express virus receptors and other host-specific infectivity factors. Genomic surveillance of animal populations can identify novel viruses but often lack access to the reservoir species, limiting their utility in assessing transmission (Carroll et al., 2018; Lipkin and Firth, 2013; Williams et al., 2018; Wu et al., 2018; Wu et al., 2012). Models with access to the complete transmission chain are greatly needed to study virus evolution, cross-species transmission, and the role of innate immunity. Rodents are an ideal model to study virus transmission because they live in dense, highly social groups and harbor many pathogens. Rodentia is also diverse, allowing assessment of the impact of evolutionary distance on transmission (Fabre et al., 2012). Furthermore, rodent species are common laboratory animals, and there are readily available tools to study their immune responses.
To evaluate the dynamics of natural virus transmission and evolution within and between species, we housed laboratory mice with pet store mice or the bedding from pet store rats, which harbor myriad natural rodent pathogens. During cohousing, viruses and other microbial species transmit to the laboratory mice via natural routes (Beura et al., 2016). With available knockout mice, we also have the ability to assess the role of antiviral immune pathways in transmission to and dissemination within a host. We identified a wealth of diverse pathogens that transmit between mice, including newly described members of viruses in the Coronaviridae, Astroviridae, Picornaviridae, and Narnaviridae families. Our analyses also uncovered microbes that potentially enhance or inhibit certain virus infections and highlighted the role of IFN in shaping some of these interactions. This model uniquely gives us access to both the reservoir and the new host, allowing us to evaluate genetic alterations in virus populations across the complete transmission chain. Using an amplicon-based approach, we quantified diversity within individual virus populations in both reservoir and host. Finally, we expanded our model to study cross-species transmission using pet store rats. This extension of our cohousing model revealed transmission of a rat astrovirus to laboratory mice in a potential dead-end infection. This model offers a bridge between natural and laboratory settings of virus transmission and evolution studies.
Results
Transmission of microbes from pet store to cohoused laboratory mice reveals role of IFN
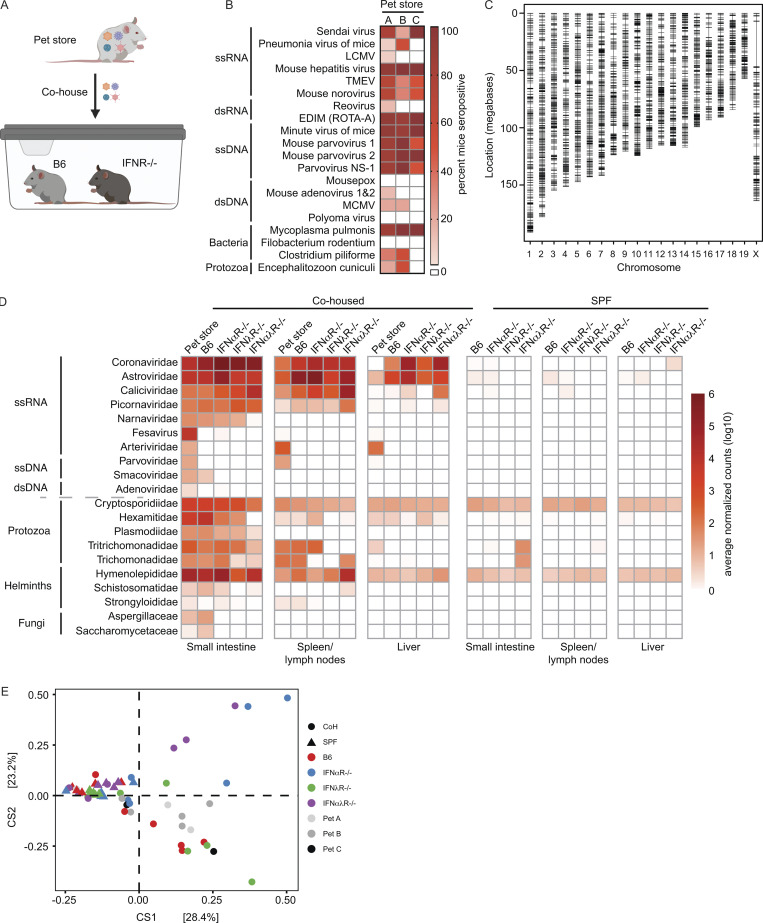
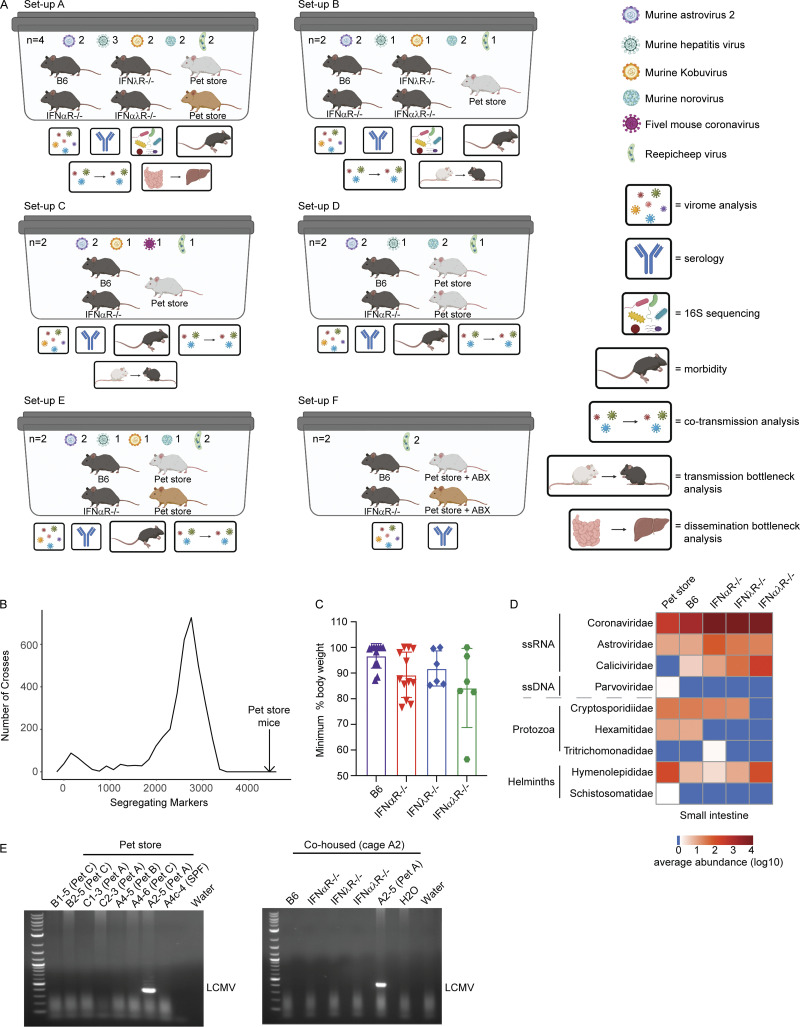
Pet store and specific pathogen–free (SPF) laboratory mice were cohoused for 3–7 d to capture the acute phase of transmission and infection. Housing the animals together in close proximity permits transmission of natural pathogens at physiologic doses and portals of entry from the reservoir animal (pet store) to the new host (cohoused SPF mice). To increase the diversity of pathogens and pathogen combinations, we used several different cohousing models of one to two pet store mice from multiple sources with wild-type C57BL/6 (B6) and laboratory mice deficient in IFN-α receptors (IFNαR−/−), IFN-λ receptors (IFNλR−/−) or IFN-α and -λ receptors (IFNαλR−/−; Figs. 1 A and S1 A and Table S1). Serological testing of the reservoir animals revealed that all pet store mice had been exposed to diverse viral, bacterial, and eukaryotic pathogens (Fig. 1 B). We also detected pet store–specific seropositivity, suggesting that animals from different pet stores have distinct exposure histories (Fig. 1 B). Pet store mice have an unknown genetic ancestry, and therefore we performed genotyping using the mini Mouse Universal Genotyping Array (miniMUGA) assay (Sigmon et al., 2020). All pet store mice used in this study were confirmed to be Mus musculus, and we identified 4,431 genetic markers on the autosomes and X chromosome segregating within our pet store mouse population (Fig. 1 C). Additionally, the number of segregating markers is substantially greater than what can be used to distinguish inbred strains such as C57BL/6 (Fig. S1 B). These data demonstrate a significant degree of diversity within the pool of pet store mice and between pet store mice and inbred mouse strains despite all being M. musculus. Following cohousing, laboratory mice demonstrated clinical signs of disease and morbidity, which were more pronounced in the absence of IFN signaling (Fig. S1 C). To identify viruses, we sequenced polyA-selected RNA from the pet store reservoir and the cohoused SPF hosts. After excluding reads that mapped to the M. musculus genome, we de novo assembled 48,993 transcripts from unmapped reads and assigned prospective taxonomic classifications to 48,735 of these sequences based on nucleotide or amino acid similarity to known sequences. Assessing the small intestine as one portal of entry and liver or spleen as sites of dissemination, we uncovered viruses from ≥10 different families that are not present in un-cohoused SPF mice. We also observed increased transmission and dissemination of viruses in the absence of IFN receptors (Fig. 1 D). While many viruses transferred from pet store mice to cohoused SPF mice, some viruses in the pet store mice failed to transmit. For example, the Arteriviridae member lactate dehydrogenase–elevating virus—which is known to transmit vertically in mice and should therefore not transmit between adults (Zitterkopf et al., 2002)—did not transmit to cohoused mice (Fig. 1 D). In addition to viruses, we detected >10 families of eukaryotic pathogens and uncovered transmission of protozoa, helminths, and fungi (Fig. 1 D). We recovered similar sets of pathogen families when classifying reads with CCMetagen (Marcelino et al., 2020), indicating that our microbial assessments were not strongly biased by using specific sequence databases or metagenomic classification methods (Fig. S1 D).
Figure 1.
Transmission of diverse microbial species to SPF mice during short-term cohousing. (A) Cohousing transmission model. (B) Serology analysis of pet store mouse infection history at the time of sacrifice. (C) Pet store mouse genotyping using the miniMUGA array, identifying a total of 4,431 high-quality markers (black hashes) on the autosomes and X chromosome segregating within the pet store mouse population. (D) Metagenomic classification and quantification of microbes in tissues from pet store, cohoused SPF, and control SPF mice. (E) DPCoA analysis of 16S rRNAseq from the small intestine contents of non-cohoused SPF, cohoused (CoH) SPF, and pet store mice. Data combined from B, n = 14 cages; C, n = 18 pet store mice; D, n = 12 cages; E, n = 6 cages. ds, double-stranded; ss, single-stranded.
Figure S1.
Overview of cohousing setups throughout paper, with expanded pet store genotyping, weight loss, and pathogen discovery data. (A) Models of each cage setup (labeled A–F) used in analyses. Key on the right denotes analyses performed on each cage. See Table S1 for details on which cohousing designs were used for each figure in the manuscript. Cohousing setup F was used only for characterizing pet store mouse serology and assembling the Reepicheep virus genome. (B) Data from miniMUGA genotyping array (Sigmon et al., 2020) that determined the number of segregating markers between 3,653 pairs of inbred strains presented as a frequency distribution (minimum segregating markers = 6, maximum = 3,351, median = 2,654, mean = 2,445). In the 18 pet store mice, the number of segregating markers was 4,431 (black arrow), greatly exceeding these pairwise comparisons of diversity. (C) Maximum weight loss after cohousing. (D) Heatmap of virus reads as quantified using CCMetagen. (E) Specific amplification and identification of arenavirus RNA in pet store (top) and cohoused (bottom) mice. Data combined from D, n = 12 cages. ABX, antibiotics; LCMV, lymphocytic choriomeningitis virus.
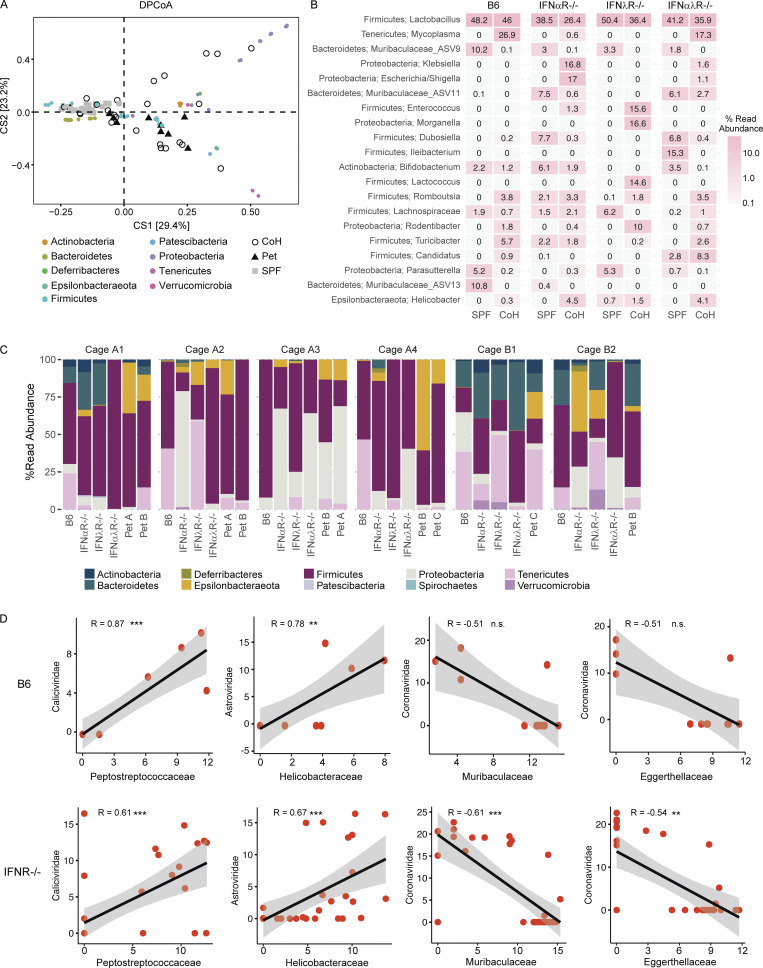
To identify microbial sequences that were not efficiently captured in polyA-selected RNA, we also screened for transmission of bacteria in the small intestine by 16S ribosomal RNA (rRNA) gene sequencing. Double principal coordinate analysis (DPCoA), an ordination approach toward visualizing similarity/dissimilarity between samples, was then used to assess shifts in bacterial community composition after cohousing. While both SPF and pet store mice clustered independently, we observed considerable within-group variation among cohoused mice, some of which was associated with IFN signaling (Fig. 1 E). Variation was also driven by members of Proteobacteria (e.g., Rodentibacter, Klebsiella, Escherichia, and Shigella spp.), Firmicutes (Lactobacillus, Romboutsia, and Turicibacter), and Epsilonbacteraeota (Helicobacter; Fig. S2, A–C), encompassing both natural rodent inhabitants and potential pathogens. To screen for examples of viruses that do not polyadenylate their mRNA in cohoused animals, we prepared cDNA and performed PCR with pan-arenavirus primers to detect lymphocytic choriomeningitis virus (Williams et al., 2018). We identified one positive pet store mouse but did not detect transmission to the cohoused SPF mice (Fig. S1 E). Together, these data demonstrate that this tractable model system can capture the transmission of diverse microorganisms through natural routes and at physiological doses.
Figure S2.
Bacterial abundance and correlations in cohoused mice. (A) DPCoA biplot analysis of small intestine contents from un-cohoused SPF, cohoused SPF, and pet store mice. (B) Mean relative abundance of the 20 most abundant bacterial genera in un-cohoused SPF versus cohoused (CoH) SPF mice. (C) Relative abundance of bacterial phyla in each individual cohoused mouse separated by cage. (D) Correlation of normalized bacterial family read abundance in B6 versus IFNR−/−. Data combined from n = 6 cages (A–C) and n = 12 cages (D). Statistical significance determined by Spearman’s correlation (D). n.s., P > 0.05; **, P < 0.01; *** P < 0.001. Reported P values in D (left to right): 0.00046, 0.0043, 0.11, 0.11 (top) and 0.00018, 2.1 × 10−5, 0.00016, 0.0013 (bottom).
Cotransmission analysis identifies novel virus–microbe interactions
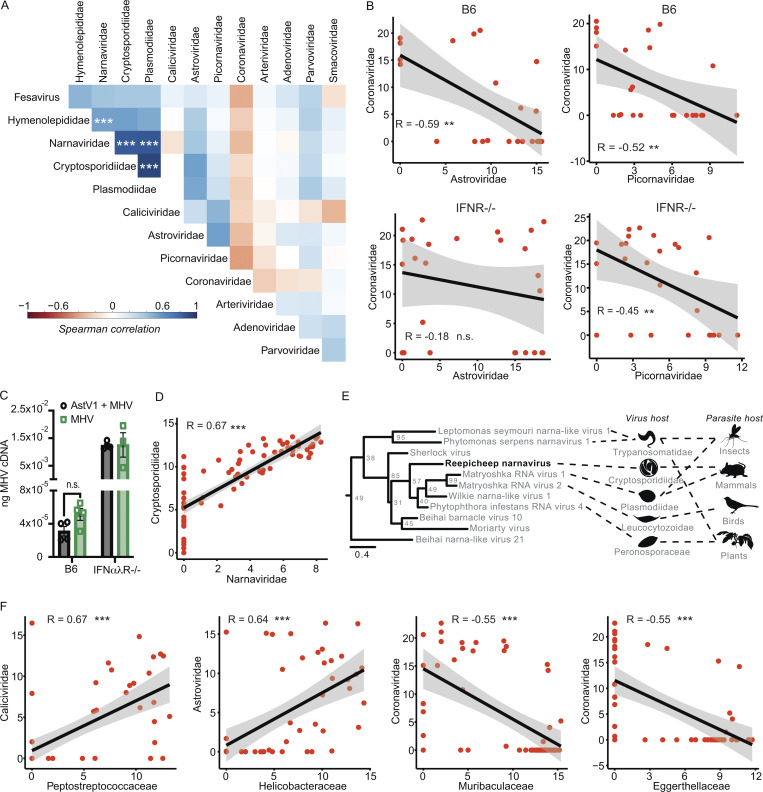
Natural virus transmission occurs in the presence of complex microbial communities that can influence infection and pathology (Desai et al., 2021; Kuss et al., 2011; Nishimoto et al., 2021). For example, the presence of certain bacterial species in mammalian hosts can facilitate enteric virus transmission (Aguilera et al., 2019; Kuss et al., 2011; Robinson et al., 2019). To evaluate potential interactions between the diverse microbes we identified in cohoused mice, we calculated Spearman correlations between microbial family read abundances in the small intestine (Fig. 2 A). Interestingly, we found an anticorrelation between Astroviridae and Coronaviridae reads as well as Coronaviridae and Picornaviridae reads. Evaluating these interactions by host genotype revealed the anticorrelation for Astroviridae and Coronaviridae, but not Coronaviridae and Picornaviridae, was statistically significant in wild-type mice but not IFN-deficient animals. To validate the IFN-dependent anticorrelation of astrovirus and coronavirus, we sequentially infected mice orally with murine astrovirus 1 (AstV1; Cortez et al., 2020) followed by murine hepatitis virus (MHV). We found a reduction in MHV in sequentially infected B6 but not IFNαλR−/− mice (Fig. 2 C). These data demonstrate that the mutual exclusion of astroviruses and coronaviruses in cohoused mice is dependent on IFN signaling. This finding is consistent with a recent report that identified IFN-mediated negative interactions between astroviruses and other enteric viruses in the mouse gut (Ingle et al., 2019).
Figure 2.
Cotransmission analysis identifies potential virus–microbe interactions. (A) Spearman’s correlation of virus and eukaryotic pathogen families. (B) Correlation between Astroviridae versus Coronaviridae reads (left) and Coronaviridae versus Picornaviridae reads (right) in B6 (top) and IFNR−/− (bottom) mice. (C) Sequential infection with murine AstV1 followed by MHV results in reduced MHV loads in B6 but not IFNαλR−/− mice (D) Co-occurrence between Narnaviridae and Cryptosporidiidae. (E) Phylogenetic analysis of Reepicheep narnavirus. (F) Co-occurrences between viral and bacterial pathogens, measured as normalized read counts. In A, B, D, and E, data were combined from n = 12 cages; in C, data from one experiment with n = 3–4 mice per group. Error bars represent SEM. In E, genome assembly combined data from n = 14 cages. Statistical significance determined by Spearman’s correlation (A, B, D, and F) or unpaired t test (C). n.s., P > 0.05; **, P < 0.01; *** P < 0.001. Reported P values (left to right): B, 0.0026 and 0.0097 (top) and 0.38 and 0.022 (bottom); C, 0.0715; D, < 2.2 × 10−16; F, 3.0 × 10−8, 2.1 × 10−7, 1.6 × 10−5, 1.5 × 10−5.
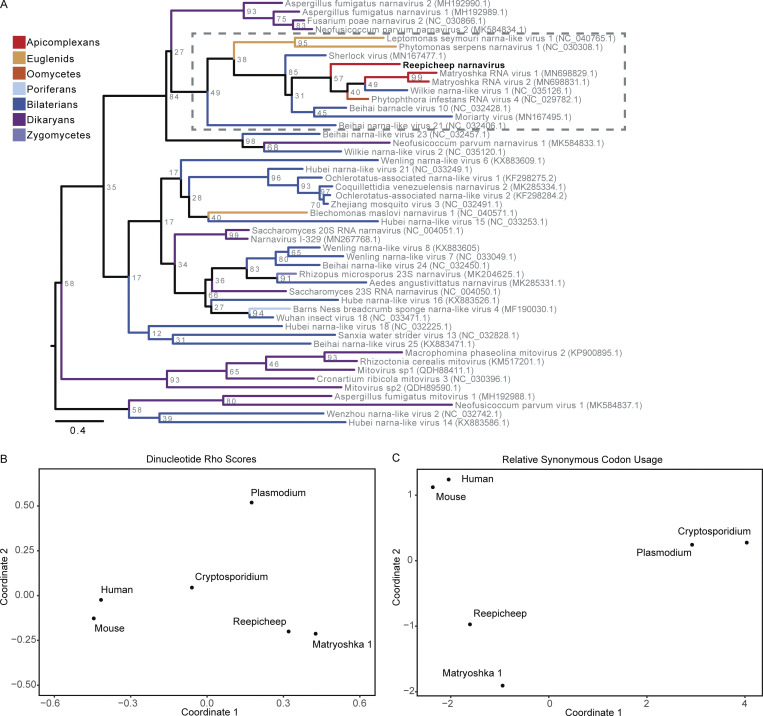
In addition to virus–virus interactions, we observed strong virus–parasite correlations between Narnaviridae and apicomplexans in the Cryptosporidiidae and Plasmodiidae families, but only Cryptosporidiidae abundances tracked strictly with the presence of Narnaviridae (Fig. 2, A and D). This observation may indicate that the narnavirus infects Cryptosporidium, which is an obligate intracellular gastrointestinal parasite that causes gastroenteritis in many vertebrate species including mice (Guérin and Striepen, 2020). Members of Narnaviridae infect diverse nonmetazoan hosts such as fungi, plants, and protists, including apicomplexans (Charon et al., 2019). Our sequence assemblies from cohoused mice include a single new candidate member of Narnaviridae that we named Reepicheep virus (Reepicheep/MN_1/2020, accession no. OK655041), which was most similar to Wilkie narna-like virus 1 (54% amino acid similarity). Phylogenetic analyses with additional potential relatives placed Reepicheep virus within a clade of parasite-infecting narnaviruses (Figs. 2 E and S3 A). While genomic biases in viral genomes such as codon usage have been used as predictors of virus host species (Babayan et al., 2018), this does not appear to be the case for narnaviruses. In analyzing both codon and dinucleotide usage, Reepicheep narnavirus and another closely related narnavirus, Matryoshka RNA virus 1 (MaRNAV-1), cluster with each other but not with their known or putative host, Cryptosporidium and Plasmodium, respectively, suggesting that genomic biases cannot be used to predict narnavirus hosts (Fig. S3, B and C).
Figure S3.
Genetic analysis of Reepicheep narnavirus. (A) Full phylogenetic analysis of Reepicheep narnavirus. (B and C) Multidimensional scaling plots of dinucleotide rho scores (B) and RSCU scores (C) for Reepicheep and MaRNAV-1 narnaviruses against potential hosts. MaRNAV-1, Matryoshka RNA virus 1.
Lastly, we evaluated potential virus–bacteria interactions by merging our RNA and 16S sequencing datasets. We identified patterns of cooccurrence between Caliciviridae and Peptostreptococcaceae as well as Astroviridae and Helicobacteraceae. We also observed negative correlations between Coronaviridae and Eggerthellaceae or Muribaculaceae (Fig. 2 F). However, none of these correlations were impacted by the presence or absence of IFN signaling (Fig. S2 D). Together, these data demonstrate that this model system can identify novel microbial ecology impacting virus transmission.
Discovery of a novel alphacoronavirus
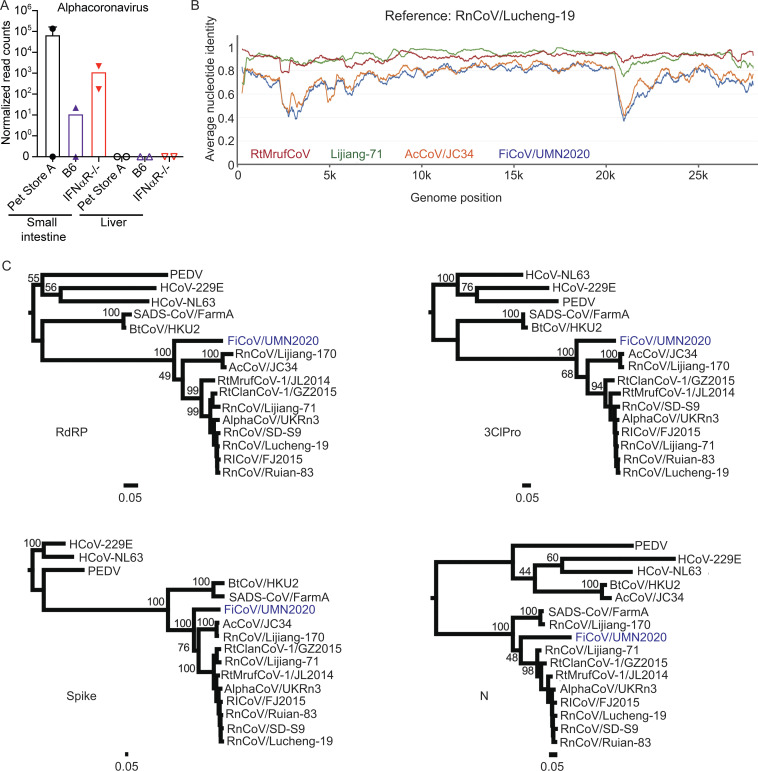
We assembled a 27,663-nt genome of an alphacoronavirus virus with distant similarity to Lucheng Rn rat coronavirus (RnCoV/Lucheng-19; Wang et al., 2015) that we have provisionally named Fievel mouse coronavirus (FiCoV/UMN2020, accession no. OK655840; Fig. 3, A and B). FiCoV was identified in two cages and transmitted from pet store to cohoused IFNαR−/− and wild-type mice (Fig. 3 A). Phylogenetic analysis of different regions of the genome indicate that FiCoV is a new, divergent lineage within a clade of rodent alphacoronaviruses (Fig. 3 C). Furthermore, the closest basic local alignment search tool (BLAST) matches for FiCoV varied across alphacoronaviruses depending on the region of the genome that was queried (Table S2) and exhibited a wide range in percentage identity. In particular, the N, spike, Orf6, and Orf8 genes exhibit significantly lower nucleotide identity to previously described rodent alphacoronaviruses than other parts of the genome, suggesting recombination may have played a role in the evolution of FiCoV. Additionally, FiCoV lacks the betacoronavirus-derived Orf2 gene found in the RnCoV/Lucheng-19 lineage but not the AcCoV/JC34 lineage, indicating the acquisition of this gene was a relatively late event in rodent alphacoronavirus evolution. These data further highlight the utility of evaluating viruses from sources where pathogen burden is not controlled.
Figure 3.
Identification of a novel alphacoronavirus. Contigs assembled using Trinity were entered in NCBI BLASTn to identify closest matches, revealing an alphacoronavirus. (A) Normalized alphacoronavirus reads in tissues of B6 and IFNαR−/− mice. (B) ANI of RtMufCoV, Lijiang-71, AcCoV/JC34, and FiCoV/UMN2020 compared with RnCoV/Lucheng-19 as a reference. (C) Maximum-likelihood phylogenetic trees of RdRP, 3ClPro, Spike, and N genes of FiCoV/UMN2020.
Loose bottlenecks identified during natural transmission of astrovirus
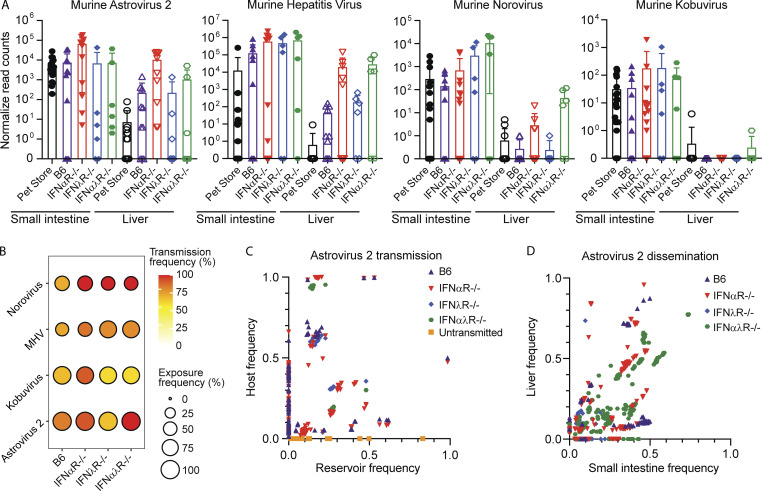
MHV, murine astrovirus 2, norovirus, and Kobuvirus were the most commonly found viruses transmitting from the pet store reservoir to the cohoused SPF hosts. Using mRNA sequencing (mRNAseq) and mixed-effects linear modeling, we tracked levels of replication and dissemination of MHV, astrovirus 2, norovirus, and Kobuvirus in the reservoir and cohoused SPF hosts at the site of transmission (small intestine) and dissemination (liver). We first looked at viral read levels in all cohoused mice, regardless of genotype, and found that only MHV was present at higher levels in the cohoused mice at the acute stage of infection than in the reservoir pet store mouse (Fig. 4 A; t = 3.14, P = 0.002). However, since the reservoir hosts were likely at a different point in the course of acute infection than their cohoused partners, we next compared cohoused mice to each other to determine the effect of specific IFN deficiencies on virus transmission, replication, and dissemination. We did not find any viruses whose transmission was dependent on a lack of IFN signaling, as all four viruses were present in each cohoused genotype to some degree, but we saw differing levels of transmission success when comparing the various IFN deficiencies to B6 mice (Fig. 4 B). For example, transmission frequency of norovirus was 100% in IFN-deficient animals but only 63% in B6 mice, while transmission frequency of Kobuvirus was most successful in IFNαR−/− mice (88% frequency) and equally successful in B6, IFNλR−/−, and IFNαλR−/− mice (58–60% frequency). We found that astrovirus 2, MHV, and norovirus replicated to higher levels in the small intestines of IFN-deficient mice, regardless of genotype, compared with B6 mice (Fig. 4 A; astrovirus 2, t = 2.23, P = 0.03; MHV, t = 2.58, P = 0.01; norovirus, t = 2.29, P = 0.03). There was no difference in replication of Kobuvirus among B6 or IFN-deficient mice. Viral reads at the site of dissemination were higher in IFN-deficient mice only for astrovirus 2 compared with B6 mice (Fig. 4 B; t = 2.44, P = 0.02). In the remaining viruses, replication at the site of dissemination did not differ between B6 and IFN-deficient mice (P > 0.05). Given that these viruses have been coevolving in the same host, they likely all have mechanisms to evade innate immunity. IFN therefore might not be critical for controlling transmission, although it clearly impacts overall viral replication levels and dissemination once established in the new hosts. Our data align with previous reports of IFN-mediated restriction of simian but not murine rotavirus in mice (Lin et al., 2016).
Figure 4.
Evaluating transmission bottlenecks and the impact of IFN responses. (A) Normalized read counts for the indicated viruses in the small intestine and liver of pet store (from pet store A, B, and C pooled from all cages) and cohoused mice. (B) Transmission frequency versus exposure frequency for murine norovirus, MHV, murine Kobuvirus, and murine astrovirus 2 based on the detection of RNAseq reads for the virus of interest. (C and D) Bottleneck analysis of murine astrovirus 2 during transmission and dissemination. Murine astrovirus 2 capsid and RdRP amplicons were generated, and relative variant frequencies were quantified in the reservoir (pet store mice) and host (cohoused mice) to assess transmission bottlenecks (C) and within the small intestine and liver of individual cohoused mice to assess dissemination bottlenecks (D). Data combined from A and B, n = 12 cages; C, n = 3 cages; and D, n = 2 cages.
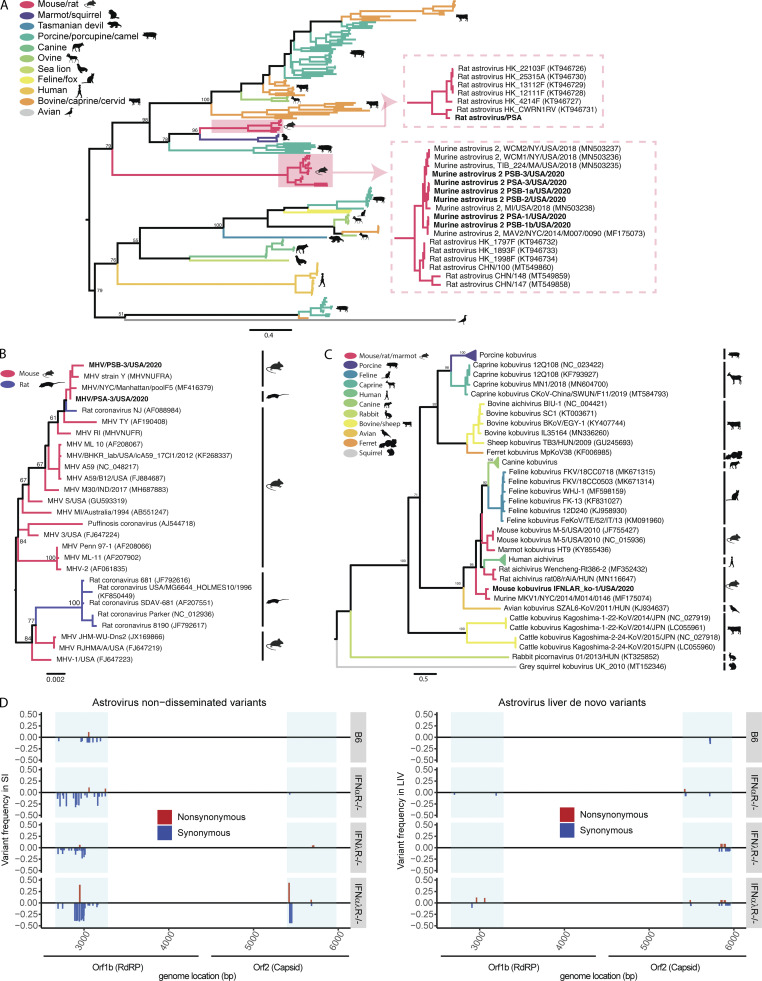
The genetic makeup and mutational capacity of commonly transmitted viruses likely play a strong role in shaping transmission and dissemination. To this end, we examined viral diversity within the model system using phylogenetics. We found that strains of astrovirus 2, Kobuvirus, and MHV were closely related to strains previously identified in murine hosts (Fig. S4, A–C) and captured a range of diversity within the astrovirus 2 sequences found in pet store mice (84–99% shared amino acid identity within the partial ORF2 segments). These phylogenies demonstrated further that our model system gives access to a wide array of virus variation that naturally circulates within reservoir hosts. Our model system further allows access to the complete transmission chain from reservoir to host through natural routes of transmission and at physiological doses. Because immune systems of the host can be manipulated, this model also allows one to study transmission bottlenecks. We chose to interrogate murine astrovirus 2 because of its prevalence in our model system and because astrovirus transmission bottlenecks have never been evaluated. We used amplicon deep sequencing to evaluate variants in the reservoir small intestine, a site of transmission in the new host (small intestine), and a site of dissemination (liver). We evaluated two regions of the viral genome: the RNA-dependent RNA polymerase (RdRP) and the surface capsid (Fig. 4, C and D). Using this approach, we identified variants that did not transmit from the pet store mice to cohoused SPF mice, variants that did transmit with varying efficiencies, and de novo variants that arose in the cohoused SPF mice. Many de novo variants arose in cohoused B6 and IFNαR−/− mice, but few arose in IFNλR−/− or IFNαλR−/− mice. Interestingly, some variants that were at very low frequency (0–0.2) in the pet store mice were present at >0.9 frequency in cohoused B6, IFNαR−/−, and IFNαλR−/− mice, suggesting either a tight bottleneck or strong selection for these variants. No variants present in the IFNλR−/− mice reached this level of saturation. We also observed dissemination of astrovirus 2 to the liver, therefore we can measure bottlenecks from the site of transmission to a distal site. We found many variants present in the small intestine that failed to disseminate. Additionally, variants present in B6 small intestine were less likely to disseminate than variants in the IFN-deficient animals, suggesting that dissemination bottlenecks are predominantly controlled by the IFN response (Fig. 4 C). Interestingly, we found that the variants that failed to disseminate had de novo mutations in the RdRP (Fig. S4 D). These data suggest that the accumulation of potentially deleterious mutations prevented these variants from disseminating. Together, these results demonstrate the utility of this model system to track virus variants from the reservoir to either immunocompetent or -deficient animals and understand how those variants disseminate within the new hosts.
Figure S4.
Astrovirus, MHV, and Kobuvirus phylogeny and variant analyses. (A–C) Phylogenetic relationships of astrovirus ORF2 amino acid sequences (A), MHV nucleocapsid amino acid sequences (B), and Kobuvirus partial whole-genome amino acid sequences (C). Branches are colored by host species; bootstrap percentages (of 500 replicates) are shown at major nodes. (D) Genome position and relative frequency of murine astrovirus 2 SNVs during dissemination. Genome position is shown on the x axis, with blue boxes highlighting areas where amplicons were generated. Relative frequencies are measured as a fraction of the SNV of all nucleotide reads per position. Nonsynonymous SNVs are colored red and shown with a positive frequency, while synonymous SNVs are colored blue and shown with a negative frequency. Data combined from D, n = 2 cages. LIV, liver; SI, small intestine.
Pet store rats reveal cross-species transmission of viruses
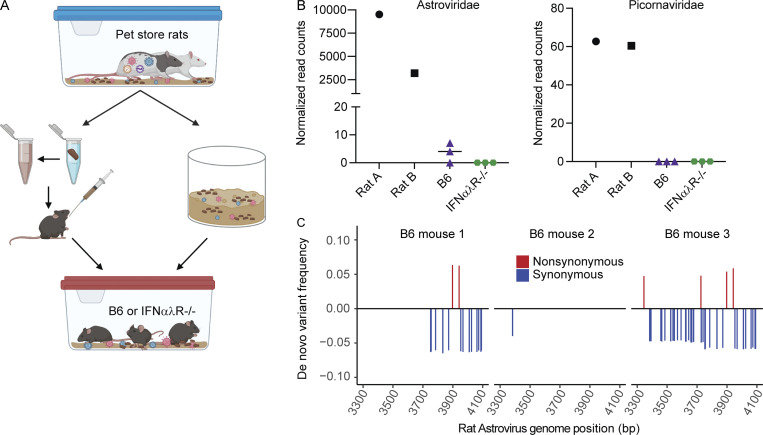
Transmission of viruses between species is the primary catalyst for infectious disease outbreaks, which pose perpetual threats to human health and agriculture (Letko et al., 2020; Marston et al., 2014). Understanding how viruses switch host specificities is critical for preventing or managing new viral outbreaks, but few experimental systems are available for modeling cross-species transmission events. We expanded our rodent cohousing approach to study cross-species transmission of microbes from rats to mice. Rattus and Mus diverged ∼12 million years ago (Kimura et al., 2015; Ramsdell et al., 2008), which likely resulted in significant barriers to the transmission of naturally adapted viruses between these species. To increase the diversity of potential viruses, we obtained two rats from two different pet stores. Rats are natural predators of mice, so animals could not be directly cohoused together. To investigate the potential for cross-species transmission of viruses, we transferred bedding from the rat cage into the mouse cages daily for 7 d. In addition to bedding transfers, we also collected fresh fecal pellets from each of the pet store rats, homogenized the pellets in PBS, and administered the slurry to each mouse via oral gavage daily (Fig. 5 A). We included one cage of B6 mice and one cage of IFNαλR−/− mice. After 7 d, rats and mice were sacrificed, and RNA was extracted and sequenced from the small intestine and liver. Our analysis pipeline identified Astroviridae and Picornaviridae reads in the rat small intestines (Fig. 5 B). Sequence reads assigned to Astroviridae, but not Picornaviridae, were also present in two of the three B6 mouse small intestines, indicating transmission of the rat astrovirus. To assess rat astrovirus replication in the mouse host, we performed amplicon deep sequencing and mutation analysis on the RdRP based on the rat astrovirus contigs assembled from the RNAseq data. We identified variants that were present in the mice but absent from the rats (Fig. 5 C). These variants could have arisen from replicative error or through amplification of otherwise undetectable standing variation in the new host, both of which suggest that the rat astrovirus was able to replicate in the mouse host. Furthermore, these de novo variants are not present in all mice, indicating independent generation of variants in each mouse. These data suggest we are detecting bona fide transmission and replication of a rat astrovirus in mice. The limited replication in mice suggests this was a dead-end transmission, but these astrovirus variants may represent early evolutionary steps toward adaptation in a new host species. Altogether, these data demonstrate the utility of this model for studying rare yet consequential cross-species virus transmission.
Figure 5.
Pet store rats as a platform for cross-species transmission of viruses. (A) Model of rat-to-mouse transmission setup. (B) Normalized RNAseq reads for Astroviridae (left) and Picornaviridae (right) found in rat and mouse small intestine. (C) Relative frequency of rat astrovirus de novo variants present in mice but not rats. Rat astrovirus RdRP amplicons were generated from rat and mouse small intestine to identify SNVs. Data are representative of one experiment. Genome positions are relative to a de novo assembled rat astrovirus contig.
Discussion
Improved virus transmission models are needed to understand factors contributing to within- and between-host virus evolution. Here, we provide a platform to study intra- and interspecies transmission of viruses and other microbial species. We identified transmission of several virus families, including the discovery of two novel viruses. We were also able to evaluate transmission dynamics in the presence and absence of IFN signaling and uncovered new relationships between microbial species during transmission. Finally, we used this model to demonstrate cross-species transmission of a rat astrovirus. This model system complements two other approaches that assess natural mouse pathogens. First, a rewilding approach has been used to expose laboratory mice to natural environments and diverse pathogens (Leung et al., 2018). However, in that system the viral reservoir cannot be identified. Second, a model where laboratory mice are born to wild mice can capture vertical transmission of natural pathogens but generally lacks visibility into acute infections (Rosshart et al., 2019). Our model complements these approaches by allowing access to the viral reservoir and assessment of acute infections.
While the potential coevolution of murine viruses and their hosts limited bottlenecks in our system, we uncovered several interesting trends in the genetic diversity of astroviruses during transmission to B6 and IFN-deficient mice. While understanding the phenotype of virus variants requires isolation or generation of individual variants for direct analysis, our model provides a platform for identifying variants of interest. For example, the majority of de novo variants arose in B6 and IFNαR−/− mice. These variants could contribute to IFN antagonism (B6) or increased replicative fitness in the absence of IFN-I (IFNαR−/−). Additionally, while variants rose to near-saturation in other SPF strains, variants in IFNλR−/− mice never reached >0.7 frequency. The presence of IFN-I may constrain virus replication during early infection, but the absence of IFN-III allows for some genetic diversity. Isolating variants or extending our model to continue the chain of transmission may help resolve the phenotypes associated with specific variants.
This model system exploits animals housed in non-SPF conditions that harbor bacterial, eukaryotic, and viral microorganisms not present in SPF laboratory mice. These animals are outbred, and sourcing from multiple different pet stores led to significant heterogeneity in the viral, eukaryotic, and bacterial carriage. This permitted assessment of transkingdom interactions, which revealed a novel narnavirus putatively replicating in a eukaryotic host. Identifying novel microbial species within complex systems is important for defining how microbial interactions impact transmission and pathogenesis of disease. We additionally uncovered both correlations and anticorrelations between viruses and bacteria, including anticorrelations between viruses driven by the IFN response. Expanding our model to include other immune knockout mice could reveal mechanisms that shape virus–bacterial transmission interactions. Treating mice with antibiotics and recolonizing with specific bacterial species could further define the role of microbial ecology in virus transmission.
Coronaviruses represent a significant zoonotic threat; SARS-CoV-2 is the third emergent coronavirus in less than two decades (Gralinski and Menachery, 2020). Identifying animal sources of coronaviruses and characterizing factors that drive coronavirus evolution are both important steps in preparing for additional emerging viruses. Alphacoronaviruses are known to infect many rodent species, and many viruses can infect a broad range of hosts (Wang et al., 2020). Coinfection of a single host by multiple distinct viruses may potentiate recombination and generation of novel viruses (Su et al., 2016). Models of natural coronavirus transmission, including those where multiple coronaviruses are present in a single host, are important for understanding coronavirus evolution and transmission between species. The novel alphacoronavirus identified here lacks closely related viruses, which makes recombination analysis challenging; however, our data suggest that FiCoV may have a recombinant origin. Continued global identification of rodent coronaviruses will help resolve coronavirus phylogeny and track emergence and evolution of novel species.
While rare, zoonoses are increasing in frequency. There are global efforts to monitor viruses in animal populations from which previous viruses have emerged, including bats and rodents (Carroll et al., 2018; Lipkin and Firth, 2013; Williams et al., 2018; Wu et al., 2018; Wu et al., 2012). These surveillance efforts can help identify viruses capable of crossing species barriers before making the jump to humans. However, this can capture only viruses that are successful, limiting our understanding of factors that drive success or failure in a new host. Our model system overcomes these limitations and recreates important aspects of transmission in natural settings, including diverse microbial ecology and physiological transmission routes, but retains the control of a laboratory setting. Expansion of this model will help uncover additional factors affecting virus transmission within and between host species.
Materials and methods
Cohousing
Wild-type C57BL/6J (B6) and B6(Cg)-Ifnar1tm1.2Ees/J (IFNαR−/−) mice were purchased from The Jackson Laboratory. B6.IL-28RA−/− (IFNλR−/−) and B6.IL- 28RA−/−Ifnar1−/− (IFNαλR−/−; Lin et al., 2016) were generated by Dr. Sergei Kotenko. Pet store mice and rats were purchased from Twin Cities area pet stores. Mice were cohoused in a BSL-3 facility. Age-matched mice housed in SPF facilities served as controls. Male laboratory mice cannot be cohoused with male pet store mice, as this creates significant animal welfare concerns due to fighting, aggression, and social defeat. Therefore, we could ethically only use female mice for cohousing experiments.
Several iterations of cohousing setups were used in these studies, which are illustrated in Fig. S1 with a designated experimental code A–F. Multiple replicates of a cohousing setup were performed, and the associated sequence data (deposited to NCBI under BioProject ID PRJNA775628) are labeled to designate the cage and animal within the cage. For example, A1_1 designates reads from cohousing set-up A, cage 1, animal 1. Full metadata for each animal are available under the BioProject ID PRJNA775628 and Table S4.
Serology
Pet store mice were screened using EZ-spot methods (Charles River Laboratories). Whole blood was collected at the time of sacrifice and submitted as per the Charles River Laboratories guidelines.
Metagenomic classification of RNAseq data from mouse tissues
Indicated tissues were homogenized in a GentleMacs M tube (Miltenyi Biotec) in Buffer RLT Plus (Qiagen) supplemented with 2-mercaptoethanol (10 µl/1 ml) and Reagent DX (0.5% vol/vol; Qiagen). RNA was extracted using the AllPrep DNA/RNA Mini kit (Qiagen). TruSeq mRNA Stranded cDNA libraries were sequenced using NovaSeq (Illumina, 50- or 150-bp PE reads). 12 million to 34 million reads/sample were obtained. Reads were mapped to the M. musculus genome (GRCm38 primary assembly from Ensembl) with STAR v2.7.3a (Dobin et al., 2013). Unmapped reads from all samples were concatenated and assembled de novo using Trinity v2.11.0 (Grabherr et al., 2011). Prospective taxonomic lineages were assigned to each assembled Trinity “gene” by using the best hit per query from BLASTn searches of the nucleotide NCBI database or from BLASTx searches of the nonredundant NCBI database for sequences that did not have significant hits in the nucleotide database. Estimated transcript counts were obtained by mapping reads from each sample to an index of all assembled sequences using Salmon v1.3.0 (Patro et al., 2017) with the validateMappings flag and 20 Gibbs samples. Counts were summed at the taxonomic family level, and normalized counts per family were generated using DESeq2 v.28.1 (Love et al., 2014). Normalized, log10-transformed read levels of astrovirus 2, MHV, norovirus, or Kobuvirus were modeled using mixed-effects linear modeling with the lmerTest package in R. Model 1 looked at replication at the site of transmission (small intestine) in pet store mouse versus cohoused mice. We specified normalized small intestine read counts as an outcome, a two-level fixed effect for mouse condition (pet store versus cohoused SPF), and cage as a random effect. Model 2 looked at replication at the site of transmission in cohoused B6 mice versus IFN-deficient mice, using only data from cohoused mice. Normalized small intestine read counts were modeled as an outcome with a two-level fixed effect for mouse condition (B6 mouse versus IFN-deficient mouse) and cage as a random effect. Model 3 looked at replication at the site of dissemination (liver) in cohoused B6 mice versus IFN-deficient mice, using only data from cohoused mice. Normalized liver read counts were modeled as an outcome with a two-level fixed effect for mouse condition (B6 mouse versus IFN-deficient mouse) and cage as a random effect. t Tests were performed on the fixed-effect estimates using Satterthwaite’s method, and significance was determined at an α value of 0.05.
Phylogenetic analysis
For the narnavirus phylogeny, a single 2,994-bp transcript assembled from the RNAseq data with BLASTx similarity to narnaviruses was translated and used as a query in BLASTp searches to identify all potential relatives. These amino acid sequences were aligned with Clustal Omega (Sievers and Higgins, 2018). Conserved blocks within this alignment were identified with Gblocks v0.91b (Castresana, 2000) using the least stringent settings, which resulted in a multiple sequence alignment of 104 sites. The RtRev+G+I+F model of protein evolution was selected for phylogenetic analyses based on having the highest Akaike information criterion score by Smart Model Selection in PhyML (Lefort et al., 2017). A maximum-likelihood tree with nonparametric bootstrap support was inferred using RAxML-NG, with 1,000 bootstrap replicates (Kozlov et al., 2019).
The metagenomic assemblies described above included partial genome sequences of a coronavirus with distant similarity to alphacoronaviruses. To assemble a full genome sequence, RNA from samples that contained sequences from this virus was used to prepare ribo-depleted libraries for RNAseq. Read processing and de novo assembly were performed as described above, and additional reads were assembled de novo with SPAdes v3.15.0 (Antipov et al., 2020; Goldstein et al., 2021 Preprint) using the coronaSPAdes convenience wrapper (Meleshko et al., 2021). Prospective alphacoronavirus sequences were extracted from Trinity and SPAdes assemblies and manually annotated to construct the full FiCoV genome sequence. The FiCoV genome was aligned to the genome of AcCoV/JC34 using MAFFT and annotated in Geneious. Alignments to RnCoV/Lucheng-19 failed at the Orf1ab/spike boundary due to FiCoV lacking an interceding gene (Lucheng-19 Orf2). Annotated genes, including RdRP and 3ClPro-encoding regions of Orf1ab were subjected to BLASTn and BLASTp analysis to identify closest relatives. The FiCoV genome was then aligned to RnCoV/Lucheng-19, AcCoV/JC34, RtMrufCoV-1/JL2014, and RnCoV/Lijiang-71 with the Orf2 removed where necessary to facilitate the alignment. An average nucleotide identity plot was then generated in IDPlot using this multiple sequence alignment (Antipov et al., 2020).
For FiCoV phylogenetic analysis, a wider diversity of rodent alphacoronaviruses along with divergent alphacoronaviruses were aligned to the indicated Orf1ab regions or 3′ gene. Maximum-likelihood trees for each aligned region with nonparametric bootstrap support were inferred using PhyML with 100 bootstrap replicates (Guindon et al., 2010).
Phylogenies of murine astrovirus 2, MHV, and murine Kobuvirus were built from contigs generated from the RNAseq data. Reads from individual mice were mapped to the mouse genome using STAR. Unmapped reads were de novo assembled with Trinity and BLAST+ was used to query reference sequences for MHV, murine astrovirus 2, and murine Kobuvirus against the contigs. Hits were then mapped to the reference sequences to determine the viral genome position. Contigs were generated from pet store mice for MHV and astrovirus 2 but contigs were only able to be generated from a cohoused IFNαλR−/− mouse for Kobuvirus. Contigs specific to the astrovirus 2 ORF2 capsid gene were used for the astrovirus phylogeny. Astrovirus sequences from GenBank, for which whole-genome sequences were available, were used as reference sequences. The MHV tree was built with nucleocapsid sequences using only MHV reference sequences from GenBank. The Kobuvirus contigs were concatenated and aligned to all whole-genome Kobuvirus sequences available in GenBank. All contigs and reference sequences were translated to their putative amino acid sequence and aligned with MAFFT (Katoh et al., 2002) in Geneious Prime (v2021.1.1, https://www.geneious.com). Maximum-likelihood trees were built using RAxML (Stamatakis, 2014) with the PROTGAMMAAUTO substitution model and 500 bootstrap replicates.
Codon and dinucleotide analyses
Dinucleotide bias was assessed by calculating the rho score for each of the 16 dinucleotides present in Homo sapiens (RefSeq 9606), M. musculus (RefSeq 10090), Plasmodiidae (f.) (RefSeq 5820), Cryptosporidiidae (f.) (RefSeq 5806), Matryoshka RNA Virus 1 (MaRNAV-1; GenBank MN698829.1), and Reepicheep genome sequences. Host genome dinucleotide tables were downloaded from Hive CoCoPUTS (https://hive.biochemistry.gwu.edu/dna.cgi?cmd=tissue_codon_usage&id=586358&mode=cocoputs). The rho function in the seqinr package was used to calculate the rho scores for the narnavirus genomes. For the hosts, the rho score was calculated using the following formula: ρ = fxy/fxfy, where fxy is the frequency of the dinucleotide, and fx and fy are the frequencies of the constituent mononucleotides (Di Giallonardo et al., 2017). Codon usage bias was determined by calculating the relative synonymous codon usage (RSCU) value for each of the 64 codons (Sharp and Li, 1986). For MaRNAV-1 and Reepicheep, the uco function in the seqinr package was used to calculate the RSCU values. Codon usage tables for the hosts were downloaded from HIVE Biochemistry and RSCU values were calculated using the following formulas: RSCU = S × Nc/Na, where S represents the number of synonymous codons encoding the same amino acid, Nc is the frequency of the codon in the genome, and Na is the relative frequency of the codon for that amino acid. For the narnavirus genomes, codon pair score (CPS) was calculated using the CPBias package’s (https://rdrr.io/github/alex-sbu/CPBias/) CPBtable function. For the hosts, codon pair tables were downloaded from HIVE Biochemistry, and CPS scores were calculated using the following formula: CPS = ln{(codon pair[ab] × amino acid[a] × amino acid[b])/(amino acid pair[ab] × codon[a] × codon[b])}.
Amplicon sequencing and variant analysis
Amplicon sequencing was performed based on previously described methods (Grubaugh et al., 2019). Briefly, primers for the murine astrovirus 2 RdRP and capsid genes were designed based on sequences identified in tissue samples from RNAseq of pet store and cohoused mice. Primers for the rat astrovirus RdRP gene were designed based on sequences identified in pet store rat tissue RNAseq samples (see Table S3 for primer information). Primers were designed to amplify 200–600-bp regions of the gene of interest. Virus amplicons were generated using the SuperScript IV One-Step RT-PCR System (Thermo Fisher Scientific). Amplicons were isolated by gel electrophoresis and purified using the NucleoSpin Gel and PCR Clean-up kit (Macherey-Nagel). Paired-end 300-bp reads were generated using the MiSeq v2 platform (Illumina). Each amplicon was independently generated and sequenced in duplicate.
Variant calling and bottleneck analysis
Single nucleotide variants (SNVs) were called in the amplicon sequencing reads based on the iVar pipeline (Grubaugh et al., 2019), which was modified to fit the experimental design. Full details on the pipeline workflow can be found on https://github.com/langloislab/dirty-mouse-virome. Briefly, variants within cohoused SPF mice were called against a consensus sequence generated from the pet store mouse to calculate host frequency. The consensus sequence was generated by mapping reads to a GenBank reference sequence or by de novo assembly where poor reference-based assembly was encountered. To investigate bottlenecks, the pet store mouse reads were also mapped back to the consensus to determine the reservoir variant frequency. Dissemination bottlenecks were determined by generating consensus sequences from small-intestine amplicon reads for each individual mouse. The small-intestine reads and liver reads were separately mapped back to the small-intestine consensus sequence to determine variants present at the sites of both infection and dissemination. In all cases, a cutoff of 3% frequency was used to call variants. Variants present in primer binding regions were discarded, and only variants present in both PCR replicates were included for final analysis.
16S rRNA gene sequencing and analysis
Small-intestine contents were collected and flash frozen at the time of sacrifice. DNA was extracted from frozen content using the AllPrep PowerFecal DNA/RNA Kit (Qiagen) and submitted to the University of Minnesota Genomics Center for 16S rRNA gene library preparation using a two-step PCR protocol (Gohl et al., 2016). The V4 variable region was amplified using V4_515F and V4_806R primers with common adapter sequences as previously described (Gohl et al., 2016). Paired-end 300-bp reads were generated using the MiSeq v2 platform (Illumina). Amplicon sequence analysis was performed in R/4.0.2. Cutadapt/2.1 (Martin, 2011) was used to remove primer and Illumina adapter sequences, with size filtering set to 215 bp (minimum) and 285 bp (maximum). DADA2/1.16 (Callahan et al., 2016) was used to trim sequences and filter sequences. DADA2 corrected sequencing errors by inferring a parametric error model. Reads were dereplicated, paired ends merged, and chimeric reads removed using default options. Genus-level taxonomy was assigned using the RDP Bayesian classifier (Wang et al., 2007) and SILVA-132 taxonomy training set (Quast et al., 2013). Species-level taxonomy was assigned only if an amplicon sequence variant unambiguously matched a sequence in SILVA-132 database. DECIPHER/2.16.1 (Wright, 2016) was used to approximate phylogenetic tree and align sequencing followed by Phangorn/2.5.5 (Schliep, 2011) to construct the phylogenetic tree. Phyla with ambiguous taxonomic assignments, total feature prevalence <10 or mean feature prevalence of 1 were removed, as were amplicon sequence variants with a mean relative abundance <10−4. For downstream ordination, heatmapping, and taxa plots, a rarefied dataset with a depth of 3,000 reads was used.
Pet store mouse genotyping
DNA was extracted from homogenized tissues with the AllPrep DNA/RNA Mini kit (Qiagen). DNA was submitted to TransnetYX for genotyping and MiniMUGA analysis (Sigmon et al., 2020). Data were processed to assign each mouse at each marker a reference (Ref), heterozygous (Het), alternate (Alt), or ambiguous call. We compared these to the genotype calls for all mice from inbred strains and their filial 1s (F1s) presented in Sigmon et al. (2020). We filtered our genotype calls to only those for which genotype calls within an inbred strain were 100% consistent (e.g., all ref or all alt) and F1s had the predicted genotype from the parent inbred strains (e.g., if parent 1 had Ref and parent 2 had Alt, all F1s would be heterozygous). Lastly, we removed single nucleotide polymorphisms for which the ambiguous call rate was >1%. This reduced the marker set to 4,441 markers. We determined the minor allele frequency in our population at each single nucleotide polymorphism by calculating the minimum of #Ref alleles/total # alleles or #Alt alleles/total # alleles. Since all mice were female, for the autosomes and X-chromosome, the denominator is always 36 (18 mice each with 2 chromosomes). See Table S5 for variant calls.
Sequential infection
Mice were infected orally with murine AstV1 as previously described (Cortez et al., 2020). 24 h later, AstV1 infected and uninfected controls were orally inoculated with MHV-Y (Compton et al., 2004). All animals were sacrificed on day 4 after AstV1 infection. Small-intestine content and tissue were processed, and the RNA was extracted as described above. cDNA was generated using SuperScript II Reverse transcription (Thermo Fisher Scientific). Quantitative PCR (qPCR) was performed with iTaq Universal SYBR Green Supermix (Bio-Rad) under the following conditions: 94° for 2 min followed by 40 cycles of 94° for 10 s, 58° for 10 s, and 72° for 30 s on a Bio-Rad CFX96 Real-Time PCR Detection System. Virus was quantified using a standard curve from 10-fold dilutions of a G-block standard (Integrated DNA Technologies). See Table S3 for primer sequences.
Ethics statement
Care and use of the animals were in accordance with the Guide for the Care and Use of Laboratory Animals from the National Research Council and the USDA Animal Care Resource Guide. All experimental protocols involving the use of mice were approved by the Institutional Animal Care and Use Committee at the University of Minnesota.
Online supplementary material
Fig. S1 contains models of each cohousing setup and the analyses performed on each, as well as additional analysis of pet store mouse genotyping and virus read classification using CCMetagen. It also contains the weight loss of all cohoused SPF animal and pet store mouse screening for arenaviruses. Fig. S2 shows additional analysis and results from the 16S rRNAseq of pet store, cohoused SPF, and un-cohoused SPF mice. Fig. S3 contains full phylogenetic analysis of the Reepicheep narnavirus as well as the multidimensional scaling plots of dinucleotide and codon usage for Reepicheep and MaRNAV-1 against their potential hosts. Fig. S4 shows the full phylogenetic relationships for the murine astrovirus, MHV, and murine Kobuvirus in our model based on amino acid identities. It also contains the location and frequency of murine astrovirus 2 de novo variants that did and did not disseminate from the small intestine to the liver. Table S1 provides a detailed list of which Twin Cities pet store the mice originated from for every cage setup as well as the figures and supplemental figures containing data from that setup. Table S2 shows the closest BLAST matches for FiCoV/UMN2020 across the genome. Table S3 lists the primer information and sequences used for amplicon generation and RT-qPCR. Table S4 contains the metadata for the sequencing files associated with each cage setup. Table S5 contains the results from the pet store mouse MiniMUGA genotyping array.
Supplementary Material
describes pet store mice used in all cage setups, and figures containing associated data.
shows the best BLAST matches across the FiCoV/UMN2020 genome.
lists the primers used for lymphocytic choriomeningitis virus screening, amplicon generation, and RT-qPCR.
shows the metadata for sequencing files associated with each cage setup.
shows MiniMUGA mouse genotyping array results for pet store mice.
Acknowledgments
Diagrams created with BioRender.
This work was supported by National Institutes of Health R01 AI132962 and an Academic Investment Research Program grant from the University of Minnesota Medical School to R.A. Langlois. E.J. Fay was supported by National Institutes of Health T32 AI007313. K.M. Balla was supported by National Institutes of Health T32 AI055434-13. S.N. Roach and F.K. Shepherd were supported by National Institutes of Health T32 HL007741. M.T. Ferris was supported by National Institutes of Health U19 AI100625.
Author contributions: E.J. Fay designed, performed, and analyzed experiments. K.M. Balla designed and performed computational analyses. S.N. Roach, F.K. Shepherd, D.S. Putri, A. Tucker, and M. Salnikov performed and analyzed experiments. F.K. Shepherd, M.T. Ferris, S.A. Goldstein, and C.E. Thefaine performed computational and statistical analyses. M. Pierson and D. Masopust maintain, support, and oversee the dirty mouse facility and associated projects. T.D. Wiggen and R.C. Hunter analyzed sequence data. S.V. Kotenko provided IFNR knockout mice. S.R. Compton and V. Cortez provided MHV and murine astrovirus, respectively. R.A. Langlois and N.C. Elde supervised the study. E.J. Fay, K.M. Balla, S.N. Roach, F.K. Shepherd, and R.A. Langlois wrote the manuscript with input from all coauthors.
Data and code availability
Sequence data were deposited and are available as FASTQ files in the NCBI sequence read archive under Bioproject ID PRJNA775628. Fievel virus FiCov/UMN2020 and Reepicheep/MN_1/2020 are deposited under GenBank accession numbers OK655840 and OK655841, respectively.
Previously published software packages and versions used to analyze 16 S rRNA sequence data are cited above. The pipelines used for variant calling and bottleneck analyses are available on https://github.com/langloislab/dirty-mouse-virome.
References
- Aguilera, E.R., Nguyen Y., Sasaki J., and Pfeiffer J.K.. 2019. Bacterial Stabilization of a Panel of Picornaviruses. MSphere. 4:e00183-19. 10.1128/mSphere.00183-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman, M.O., Angeletti D., and Yewdell J.W.. 2018. Antibody Immunodominance: The Key to Understanding Influenza Virus Antigenic Drift. Viral Immunol. 31:142–149. 10.1089/vim.2017.0129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antipov, D., Raiko M., Lapidus A., and Pevzner P.A.. 2020. Metaviral SPAdes: assembly of viruses from metagenomic data. Bioinformatics. 36:4126–4129. 10.1093/bioinformatics/btaa490 [DOI] [PubMed] [Google Scholar]
- Babayan, S.A., Orton R.J., and Streicker D.G.. 2018. Predicting reservoir hosts and arthropod vectors from evolutionary signatures in RNA virus genomes. Science. 362:577–580. 10.1126/science.aap9072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura, L.K., Hamilton S.E., Bi K., Schenkel J.M., Odumade O.A., Casey K.A., Thompson E.A., Fraser K.A., Rosato P.C., Filali-Mouhim A., et al. 2016. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 532:512–516. 10.1038/nature17655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callahan, B.J., McMurdie P.J., Rosen M.J., Han A.W., Johnson A.J., and Holmes S.P.. 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods. 13:581–583. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, D., Daszak P., Wolfe N.D., Gao G.F., Morel C.M., Morzaria S., Pablos-Méndez A., Tomori O., and Mazet J.A.K.. 2018. The Global Virome Project. Science. 359:872–874. 10.1126/science.aap7463 [DOI] [PubMed] [Google Scholar]
- Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540–552. 10.1093/oxfordjournals.molbev.a026334 [DOI] [PubMed] [Google Scholar]
- Charon, J., Grigg M.J., Eden J.S., Piera K.A., Rana H., William T., Rose K., Davenport M.P., Anstey N.M., and Holmes E.C.. 2019. Novel RNA viruses associated with Plasmodium vivax in human malaria and Leucocytozoon parasites in avian disease. PLoS Pathog. 15:e1008216. 10.1371/journal.ppat.1008216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton, S.R., Ball-Goodrich L.J., Johnson L.K., Johnson E.A., Paturzo F.X., and Macy J.D.. 2004. Pathogenesis of enterotropic mouse hepatitis virus in immunocompetent and immunodeficient mice. Comp. Med. 54:681–689. [PubMed] [Google Scholar]
- Cortez, V., Boyd D.F., Crawford J.C., Sharp B., Livingston B., Rowe H.M., Davis A., Alsallaq R., Robinson C.G., Vogel P., et al. 2020. Astrovirus infects actively secreting goblet cells and alters the gut mucus barrier. Nat. Commun. 11:2097. 10.1038/s41467-020-15999-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai, P., Janova H., White J.P., Reynoso G.V., Hickman H.D., Baldridge M.T., Urban J.F. Jr., Stappenbeck T.S., Thackray L.B., and Diamond M.S.. 2021. Enteric helminth coinfection enhances host susceptibility to neurotropic flaviviruses via a tuft cell-IL-4 receptor signaling axis. Cell. 184:1214–1231.e16. 10.1016/j.cell.2021.01.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giallonardo, F., Schlub T.E., Shi M., and Holmes E.C.. 2017. Dinucleotide Composition in Animal RNA Viruses Is Shaped More by Virus Family than by Host Species. J. Virol. 91:e02381-16. 10.1128/JVI.02381-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin, A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., and Gingeras T.R.. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29:15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre, P.H., Hautier L., Dimitrov D., and Douzery E.J.. 2012. A glimpse on the pattern of rodent diversification: a phylogenetic approach. BMC Evol. Biol. 12:88. 10.1186/1471-2148-12-88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillgrass, A., Wessels J.M., Yang J.X., and Kaushic C.. 2021. Advances in Humanized Mouse Models to Improve Understanding of HIV-1 Pathogenesis and Immune Responses. Front. Immunol. 11:617516. 10.3389/fimmu.2020.617516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohl, D.M., Vangay P., Garbe J., MacLean A., Hauge A., Becker A., Gould T.J., Clayton J.B., Johnson T.J., Hunter R., et al. 2016. Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 34:942–949. 10.1038/nbt.3601 [DOI] [PubMed] [Google Scholar]
- Goldstein, S.A., Brown J., Pedersen B.S., Quinlan A.R., and Elde N.C.. 2021. Extensive recombination-driven coronavirus diversification expands the pool of potential pandemic pathogens. bioRxiv (Preprint posted February 4, 2021). 10.1101/2021.02.03.429646 [DOI] [PMC free article] [PubMed]
- Grabherr, M.G., Haas B.J., Yassour M., Levin J.Z., Thompson D.A., Amit I., Adiconis X., Fan L., Raychowdhury R., Zeng Q., et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29:644–652. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralinski, L.E., and Menachery V.D.. 2020. Return of the Coronavirus: 2019-nCoV. Viruses. 12:135. 10.3390/v12020135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubaugh, N.D., Gangavarapu K., Quick J., Matteson N.L., De Jesus J.G., Main B.J., Tan A.L., Paul L.M., Brackney D.E., Grewal S., et al. 2019. An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar. Genome Biol. 20:8. 10.1186/s13059-018-1618-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérin, A., and Striepen B.. 2020. The Biology of the Intestinal Intracellular Parasite Cryptosporidium. Cell Host Microbe. 28:509–515. 10.1016/j.chom.2020.09.007 [DOI] [PubMed] [Google Scholar]
- Guindon, S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., and Gascuel O.. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59:307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Hacisuleyman, E., Hale C., Saito Y., Blachere N.E., Bergh M., Conlon E.G., Schaefer-Babajew D.J., DaSilva J., Muecksch F., Gaebler C., et al. 2021. Vaccine Breakthrough Infections with SARS-CoV-2 Variants. N. Engl. J. Med. 384:2212–2218. 10.1056/NEJMoa2105000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingle, H., Lee S., Ai T., Orvedahl A., Rodgers R., Zhao G., Sullender M., Peterson S.T., Locke M., Liu T.C., et al. 2019. Viral complementation of immunodeficiency confers protection against enteric pathogens via interferon-λ. Nat. Microbiol. 4:1120–1128. 10.1038/s41564-019-0416-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, K., Misawa K., Kuma K., and Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30:3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, Y., Hawkins M.T., McDonough M.M., Jacobs L.L., and Flynn L.J.. 2015. Corrected placement of Mus-Rattus fossil calibration forces precision in the molecular tree of rodents. Sci. Rep. 5:14444. 10.1038/srep14444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov, A.M., Darriba D., Flouri T., Morel B., and Stamatakis A.. 2019. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 35:4453–4455. 10.1093/bioinformatics/btz305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuss, S.K., Best G.T., Etheredge C.A., Pruijssers A.J., Frierson J.M., Hooper L.V., Dermody T.S., and Pfeiffer J.K.. 2011. Intestinal microbiota promote enteric virus replication and systemic pathogenesis. Science. 334:249–252. 10.1126/science.1211057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort, V., Longueville J.E., and Gascuel O.. 2017. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 34:2422–2424. 10.1093/molbev/msx149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letko, M., Seifert S.N., Olival K.J., Plowright R.K., and Munster V.J.. 2020. Bat-borne virus diversity, spillover and emergence. Nat. Rev. Microbiol. 18:461–471. 10.1038/s41579-020-0394-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung, J.M., Budischak S.A., Chung The H., Hansen C., Bowcutt R., Neill R., Shellman M., Loke P., and Graham A.L.. 2018. Rapid environmental effects on gut nematode susceptibility in rewilded mice. PLoS Biol. 16:e2004108. 10.1371/journal.pbio.2004108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J.D., Feng N., Sen A., Balan M., Tseng H.C., McElrath C., Smirnov S.V., Peng J., Yasukawa L.L., Durbin R.K., et al. 2016. Distinct Roles of Type I and Type III Interferons in Intestinal Immunity to Homologous and Heterologous Rotavirus Infections. PLoS Pathog. 12:e1005600. 10.1371/journal.ppat.1005600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipkin, W.I., and Firth C.. 2013. Viral surveillance and discovery. Curr. Opin. Virol. 3:199–204. 10.1016/j.coviro.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcelino, V.R., Clausen P.T.L.C., Buchmann J.P., Wille M., Iredell J.R., Meyer W., Lund O., Sorrell T.C., and Holmes E.C.. 2020. CCMetagen: comprehensive and accurate identification of eukaryotes and prokaryotes in metagenomic data. Genome Biol. 21:103. 10.1186/s13059-020-02014-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston, H.D., Folkers G.K., Morens D.M., and Fauci A.S.. 2014. Emerging viral diseases: confronting threats with new technologies. Sci. Transl. Med. 6:253ps10. 10.1126/scitranslmed.3009872 [DOI] [PubMed] [Google Scholar]
- Martin, M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 17:10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McCrone, J.T., and Lauring A.S.. 2018. Genetic bottlenecks in intraspecies virus transmission. Curr. Opin. Virol. 28:20–25. 10.1016/j.coviro.2017.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meleshko, D., Hajirasouliha I., and Korobeynikov A.. 2021. coronaSPAdes: from biosynthetic gene clusters to RNA viral assemblies. Bioinformatics.:btab597. 10.1093/bioinformatics/btab597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morens, D.M., and Fauci A.S.. 2020. Emerging Pandemic Diseases: How We Got to COVID-19. Cell. 182:1077–1092. 10.1016/j.cell.2020.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison, T.E., and Diamond M.S.. 2017. Animal Models of Zika Virus Infection, Pathogenesis, and Immunity. J. Virol. 91:e00009-17. 10.1128/JVI.00009-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Fontela, C., Dowling W.E., Funnell S.G.P., Gsell P.S., Riveros-Balta A.X., Albrecht R.A., Andersen H., Baric R.S., Carroll M.W., Cavaleri M., et al. 2020. Animal models for COVID-19. Nature. 586:509–515. 10.1038/s41586-020-2787-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimoto, A., Wohlgemuth N., Rosch J., Schultz-Cherry S., Cortez V., and Rowe H.M.. 2021. Transkingdom interactions important for the pathogenesis of human viruses. J. Infect. Dis. 223(12, Suppl 2):S201–S208. 10.1093/infdis/jiaa735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olival, K.J., Hosseini P.R., Zambrana-Torrelio C., Ross N., Bogich T.L., and Daszak P.. 2017. Host and viral traits predict zoonotic spillover from mammals. Nature. 546:646–650. 10.1038/nature22975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro, R., Duggal G., Love M.I., Irizarry R.A., and Kingsford C.. 2017. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods. 14:417–419. 10.1038/nmeth.4197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast, C., Pruesse E., Yilmaz P., Gerken J., Schweer T., Yarza P., Peplies J., and Glöckner F.O.. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41(Database issue, D1):D590–D596. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsdell, C.M., Lewandowski A.A., Glenn J.L., Vrana P.B., O’Neill R.J., and Dewey M.J.. 2008. Comparative genome mapping of the deer mouse (Peromyscus maniculatus) reveals greater similarity to rat (Rattus norvegicus) than to the lab mouse (Mus musculus). BMC Evol. Biol. 8:65. 10.1186/1471-2148-8-65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, C.M., Woods Acevedo M.A., McCune B.T., and Pfeiffer J.K.. 2019. Related Enteric Viruses Have Different Requirements for Host Microbiota in Mice. J. Virol. 93:e01339-19. 10.1128/JVI.01339-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosshart, S.P., Herz J., Vassallo B.G., Hunter A., Wall M.K., Badger J.H., McCulloch J.A., Anastasakis D.G., Sarshad A.A., Leonardi I., et al. 2019. Laboratory mice born to wild mice have natural microbiota and model human immune responses. Science. 365:eaaw4361. 10.1126/science.aaw4361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliep, K.P. 2011. phangorn: phylogenetic analysis in R. Bioinformatics. 27:592–593. 10.1093/bioinformatics/btq706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp, P.M., and Li W.H.. 1986. An evolutionary perspective on synonymous codon usage in unicellular organisms. J. Mol. Evol. 24:28–38. 10.1007/BF02099948 [DOI] [PubMed] [Google Scholar]
- Sievers, F., and Higgins D.G.. 2018. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 27:135–145. 10.1002/pro.3290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigmon, J.S., Blanchard M.W., Baric R.S., Bell T.A., Brennan J., Brockmann G.A., Burks A.W., Calabrese J.M., Caron K.M., Cheney R.E., et al. 2020. Content and Performance of the MiniMUGA Genotyping Array: A New Tool To Improve Rigor and Reproducibility in Mouse Research. Genetics. 216:905–930. 10.1534/genetics.120.303596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis, A. 2014. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 30:1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su, S., Wong G., Shi W., Liu J., Lai A.C.K., Zhou J., Liu W., Bi Y., and Gao G.F.. 2016. Epidemiology, Genetic Recombination, and Pathogenesis of Coronaviruses. Trends Microbiol. 24:490–502. 10.1016/j.tim.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Q., Garrity G.M., Tiedje J.M., and Cole J.R.. 2007. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73:5261–5267. 10.1128/AEM.00062-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Lin X.D., Guo W.P., Zhou R.H., Wang M.R., Wang C.Q., Ge S., Mei S.H., Li M.H., Shi M., et al. 2015. Discovery, diversity and evolution of novel coronaviruses sampled from rodents in China. Virology. 474:19–27. 10.1016/j.virol.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, W., Lin X.D., Zhang H.L., Wang M.R., Guan X.Q., Holmes E.C., and Zhang Y.Z.. 2020. Extensive genetic diversity and host range of rodent-borne coronaviruses. Virus Evol. 6:veaa078. 10.1093/ve/veaa078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, P., Casner R.G., Nair M.S., Wang M., Yu J., Cerutti G., Liu L., Kwong P.D., Huang Y., Shapiro L., and Ho D.D.. 2021. Increased resistance of SARS-CoV-2 variant P.1 to antibody neutralization. Cell Host Microbe. 29:747–751.e4. 10.1016/j.chom.2021.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Washington, N.L., Gangavarapu K., Zeller M., Bolze A., Cirulli E.T., Schiabor Barrett K.M., Larsen B.B., Anderson C., White S., Cassens T., et al. 2021. Emergence and rapid transmission of SARS-CoV-2 B.1.1.7 in the United States. Cell. 184:2587–2594.e7. 10.1016/j.cell.2021.03.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S.H., Che X., Garcia J.A., Klena J.D., Lee B., Muller D., Ulrich W., Corrigan R.M., Nichol S., Jain K., and Lipkin W.I.. 2018. Viral Diversity of House Mice in New York City. MBio. 9:e01354-17. 10.1128/mBio.01354-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright, E. 2016. Using DECIPHER v2. 0 to analyze big biological sequence data in R. R J. 8:352–359. 10.32614/RJ-2016-025 [DOI] [Google Scholar]
- Wu, Z., Ren X., Yang L., Hu Y., Yang J., He G., Zhang J., Dong J., Sun L., Du J., et al. 2012. Virome analysis for identification of novel mammalian viruses in bat species from Chinese provinces. J. Virol. 86:10999–11012. 10.1128/JVI.01394-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Z., Lu L., Du J., Yang L., Ren X., Liu B., Jiang J., Yang J., Dong J., Sun L., et al. 2018. Comparative analysis of rodent and small mammal viromes to better understand the wildlife origin of emerging infectious diseases. Microbiome. 6:178. 10.1186/s40168-018-0554-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zitterkopf, N.L., Haven T.R., Huela M., Bradley D.S., and Cafruny W.A.. 2002. Transplacental lactate dehydrogenase-elevating virus (LDV) transmission: immune inhibition of umbilical cord infection, and correlation of fetal virus susceptibility with development of F4/80 antigen expression. Placenta. 23:438–446. 10.1053/plac.2002.0829 [DOI] [PubMed] [Google Scholar]
- Zwart, M.P., and Elena S.F.. 2015. Matters of Size: Genetic Bottlenecks in Virus Infection and Their Potential Impact on Evolution. Annu. Rev. Virol. 2:161–179. 10.1146/annurev-virology-100114-055135 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
describes pet store mice used in all cage setups, and figures containing associated data.
shows the best BLAST matches across the FiCoV/UMN2020 genome.
lists the primers used for lymphocytic choriomeningitis virus screening, amplicon generation, and RT-qPCR.
shows the metadata for sequencing files associated with each cage setup.
shows MiniMUGA mouse genotyping array results for pet store mice.
Data Availability Statement
Sequence data were deposited and are available as FASTQ files in the NCBI sequence read archive under Bioproject ID PRJNA775628. Fievel virus FiCov/UMN2020 and Reepicheep/MN_1/2020 are deposited under GenBank accession numbers OK655840 and OK655841, respectively.
Previously published software packages and versions used to analyze 16 S rRNA sequence data are cited above. The pipelines used for variant calling and bottleneck analyses are available on https://github.com/langloislab/dirty-mouse-virome.