Summary
Whole-genome sequencing studies applied to large populations or biobanks with extensive phenotyping raise new analytic challenges. The need to consider many variants at a locus or group of genes simultaneously and the potential to study many correlated phenotypes with shared genetic architecture provide opportunities for discovery not addressed by the traditional one variant, one phenotype association study. Here, we introduce a Bayesian model comparison approach called MRP (multiple rare variants and phenotypes) for rare-variant association studies that considers correlation, scale, and direction of genetic effects across a group of genetic variants, phenotypes, and studies, requiring only summary statistic data. We apply our method to exome sequencing data (n = 184,698) across 2,019 traits from the UK Biobank, aggregating signals in genes. MRP demonstrates an ability to recover signals such as associations between PCSK9 and LDL cholesterol levels. We additionally find MRP effective in conducting meta-analyses in exome data. Non-biomarker findings include associations between MC1R and red hair color and skin color, IL17RA and monocyte count, and IQGAP2 and mean platelet volume. Finally, we apply MRP in a multi-phenotype setting; after clustering the 35 biomarker phenotypes based on genetic correlation estimates, we find that joint analysis of these phenotypes results in substantial power gains for gene-trait associations, such as in TNFRSF13B in one of the clusters containing diabetes- and lipid-related traits. Overall, we show that the MRP model comparison approach improves upon useful features from widely used meta-analysis approaches for rare-variant association analyses and prioritizes protective modifiers of disease risk.
Keywords: GWAS, rare variants, aggregation techniques, gene-based analysis
Introduction
Sequencing technologies are quickly transforming human genetic studies of complex traits. It is increasingly possible to obtain whole-genome sequence data on thousands of samples at manageable costs. As a result, the genome-wide study of rare variants (minor allele frequency [MAF] 1%) and their contribution to disease susceptibility and phenotype variation is now feasible.1, 2, 3, 4
In genetic studies of diseases or continuous phenotypes, rare variants are hard to assess individually because of the limited number of observations of each rare variant. Hence, to boost the power to detect a signal, evidence is usually aggregated across variants in blocks. When designing an aggregation method, there are three questions that are usually considered. First, across which biological units should variants be combined (e.g., genes); second, which variants within those units should be included;5 and third, which statistical model should be used?6 Given the widespread observations of shared genetic risk factors across distinct diseases, there is also considerable motivation to use gene discovery approaches that leverage the information from multiple phenotypes jointly. In other words, rather than only aggregating variants that may have effects on a single phenotype, we can also bring together sets of phenotypes for which a single variant or set of variants might have effects.
In this paper, we present a Bayesian multiple rare variants and phenotypes (MRP) model comparison approach for identifying rare-variant associations as an alternative to current, widely used univariate statistical tests. The MRP framework exploits correlation, scale, and/or direction of genetic effects in a broad range of rare-variant association study designs including case-control, multiple diseases and shared controls, a single continuous phenotype, multiple continuous phenotypes, or a mixture of case-control and multiple continuous phenotypes (Figure 1). MRP makes use of Bayesian model comparison whereby we compute a Bayes factor (BF) defined as the ratio of the marginal likelihoods under two models: (1) a null model where all genetic effects are zero and (2) an alternative model where factors such as correlation, scale, and direction of genetic effects are considered. For MRP, the BF represents the statistical evidence for a non-zero effect for a particular group of rare variants on the phenotype(s) of interest and can be used as an alternative to p values from traditional significance testing.
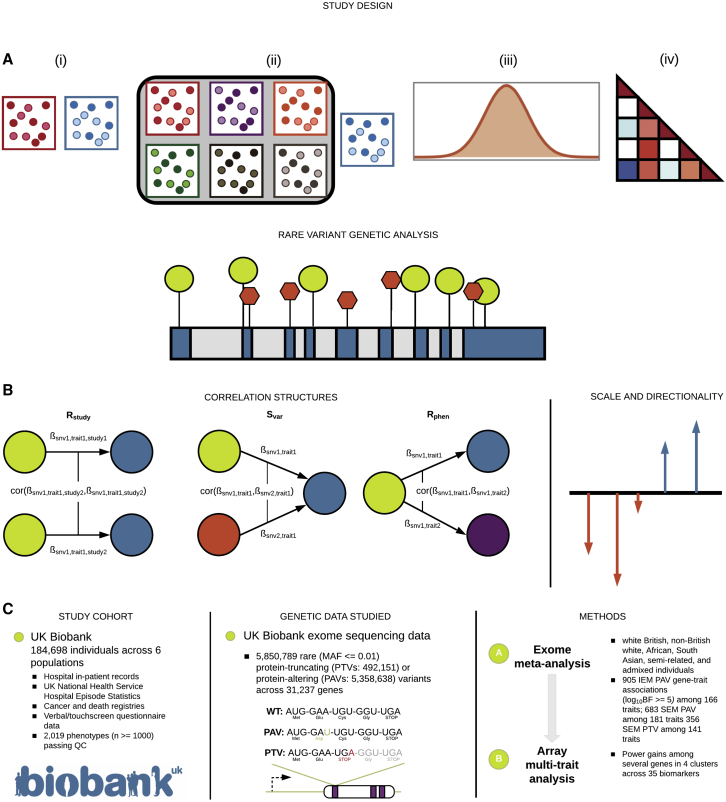
Figure 1.
MRP study overview
(A) MRP is suitable for a broad range of rare variant association study designs, including, from left to right, (i) case-control, (ii) multiple diseases with shared controls, (iii) single quantitative phenotype, and (iv) mixtures of case-control and quantitative phenotypes.
(B) Diagram of factors considered in rare variant association analysis including the correlation matrices: (expected correlation of genetic effects among a group of studies), (expected covariance of genetic effects among a group of variants, potentially accounting for annotation of variants), and (expected correlation of genetic effects among a group of phenotypes). MRP can take into account both scale and direction of effects.
(C) We focused on 184,698 individuals across six ancestry groups in the UK Biobank and analyzed 5,850,789 rare coding variants (492,151 PTVs and 5,358,638 PAVs) in the whole-exome sequencing data via single-trait and multi-trait meta-analyses with a specific focus on 35 biomarker traits.
While many large genetic consortia collect both raw genotype and phenotype data, in practice, sharing of individual genotype and phenotype data across groups is difficult to achieve. To address this, MRP can use summary statistics, such as estimates of effect size and corresponding standard errors from typical single-variant/single-phenotype linear or logistic regressions, as input. Furthermore, we use insights from Liu et al.7 and Cichonska et al.,8 which suggest the use of additional summary statistics such as covariance estimates across variants and studies, respectively, for the lossless ability to detect gene-based association signals with summary statistics alone.
Prior work has explored the use of model comparison and BFs in multi-trait settings. The model comparison in Stephens, 20139 is slightly different in usage. Whereas MRP can be used for meta-analysis and the combining of signal across multiple variants within a block, the method explored in Stephens is used for identifying a distinction between direct and indirect associations. Both have strengths relative to the other.
The multi-trait model comparison approach that is referenced in Pickrell et al., 201610 focuses on the two-phenotype case. In other words, a null (where the SNP is associated to neither trait) is compared to the alternatives of the SNP being associated to one, the other, or both traits. MRP can generalize beyond the two-phenotype case and assumes a more holistic prior across phenotypes by using correlation coefficients.
Aggregation techniques rely on variant annotations to assign variants to groups for analysis. MRP allows for the inclusion of priors on the scale of effect sizes that can be adjusted depending on what type of variants are included in the analysis. For instance, protein-truncating variants (PTVs)11,12 are highly likely to be functional because they often disrupt the normal function of a gene. Additional deleteriousness metrics, such as MPC (a metric that combines subgenic constraints with variant-level data for deleteriousness prediction)13 and pLI (a metric derived from a comparison of the observed number of PTVs in a sample to the number expected in the absence of fitness effects, i.e., under neutrality, given an estimated mutation rate for the gene),14 can further attenuate or accentuate these granular signals. Furthermore, because PTVs typically abolish or severely alter gene function, there is particular interest in identifying protective PTV modifiers of human disease risk that may serve as targets for future therapeutics.15, 16, 17 We therefore demonstrate how the MRP model comparison approach can improve discovery of such protective signals by modeling the direction of genetic effects; this prioritizes variants or genes that are consistent with protecting against disease.
To evaluate the performance of MRP, we use simulations and compare it to other commonly used approaches. Some simple alternatives to MRP include univariate approaches for rare-variant association studies, including the sequence kernel association test (SKAT)18 and the burden test,6 which are special cases of the MRP model comparison when we assign the prior correlation of genetic effects across different variants to be zero or one, respectively.
We apply MRP to summary statistics computed on a tranche of n = 184,698 exomes for thousands of traits in the UK Biobank for which we have exome data for n 1,000 white British individuals, focusing on a meta-analysis context across six UK Biobank subpopulations as defined previously (material and methods).19 We additionally apply multi-phenotype MRP on clusters of biomarker traits within a single-population context (white British individuals). These analyses show that MRP recovers results from single-variant-single-phenotype association analyses while increasing the power to detect new rare-variant associations, including protective modifiers of disease risk.
Material and methods
Description of MRP
In this section, we provide an overview of the MRP model comparison approach. MRP models genome-wide association study (GWAS) summary statistics as being distributed according to one of two models: the null model, where the effect sizes across all studies for a group of variants and a group of phenotypes is zero, and the alternative model, where effect sizes are distributed according to a multivariate normal distribution with a non-zero mean and/or covariance matrix. MRP compares the evidence between the alternative model and the null model with a BF, which is the ratio of the marginal likelihoods under the two models given the observed data.
To define the alternative model, we must specify the prior correlation structure, scale, and direction of the effect sizes. Let N be the number of individuals and K the number of phenotype measurements on each individual. Let M be the number of variants in a testing unit G, where G can be, for example, a gene, a pathway, or a network. Let S be the number of studies from which data is obtained—this data may be in the form of (1) raw genotypes and phenotypes or (2) summary statistics including linkage-disequilibrium (LD) coefficients, effect sizes, and corresponding standard errors. When considering multiple studies (S 1), multiple rare variants (M 1), and multiple phenotypes (K 1), we define the prior correlation structure of the effect sizes as an SMK × SMK matrix, U. In practice, we define U as a Kronecker product of three sub-matrices:
-
•
an S × S matrix containing the correlations of genetic effects among studies that can model the level of heterogeneity in effect sizes between populations;20
-
•
an M × M matrix containing the covariances of genetic effects among genetic variants, which may reflect, e.g., the assumption that all the PTVs in a gene may have the same biological consequence11,12,21 or prior information on scale of the effects obtained through integration of additional functional data;5,22 by assuming zero correlation of genetic effects, MRP becomes a dispersion test similar to C-alpha23,24 and SKAT;18 and
-
•
a K × K matrix containing the correlations of genetic effects among phenotypes, which may be estimated from common variant data.25, 26, 27
The variance-covariance matrix of the effect size estimates may be obtained from readily available summary statistics such as in-study LD matrices, effect size estimates (or log odds ratios), and the standard errors of the effect size estimates.
MRP allows users to specify priors that reflect knowledge of the variants and phenotypes under study. For instance, we can define an independent effects model (IEM) where the effect sizes of different variants are not correlated at all. In this case, is the identity matrix, and MRP behaves similarly to dispersion tests such as C-alpha23,24 and SKAT.18 We can also define a similar effects model (SEM) by setting every value of to 1, where is the correlation matrix corresponding to covariance matrix . This model assumes that all variants under consideration have similar effect sizes (with, possibly, differences in scale, such as in the burden test). Such a model may be appropriate for PTVs, where each variant completely disrupts the function of the gene, leading to a gene knockout. The prior on the scale of effect sizes can be used to denote which variants may have larger effect sizes. For instance, emerging empirical genetic studies have shown that within a gene, PTVs may have stronger effects than missense variants.28 This can be reflected by adjusting the prior variances of effect sizes (σ) for different categories of variants.
Finally, we can utilize a prior on the expected location/direction of effects to specify alternative models where we seek to identify variants with protective effects against disease. By default, we have assumed that the prior mean of genetic effects is zero, which makes it possible to analyze a large number of phenotypes without enumerating the prior mean across all phenotypes. To proactively identify genetic variants that are consistent with a protective profile for a disease, we can include a non-zero vector as a prior mean of genetic effects. For this, we can exploit information from Mendelian randomization studies of common variants, such as recent findings where rare protein-truncating loss-of-function variants in PCSK9 were found to decrease low-density lipoprotein (LDL) and triglyceride levels and decrease coronary artery disease risk,15,29,30 to identify situations where such a prior is warranted.
Applying MRP to variants from a testing unit G yields a BF for that testing unit that describes the evidence that rare variants in that testing unit have a non-zero effect on the traits used in the model. We can turn this evidence into probability via Bayes’ rule. Namely, a multiplication of prior-odds of association by BF transforms the prior-odds to posterior-odds. For example, if our prior probability for one particular gene to be associated with a phenotype is , then an observed BF of means that our posterior probability of association between the gene and the phenotype is over 90%. Although we see advantages in adopting a Bayesian interpretation for MRP, our approach could also be used in a frequentist context by using BF as a test statistic to compute p values.
MRP details
MRP model comparison for association testing
We consider the multivariate linear regression model
where the matrices , , , and describe the phenotype values , copies of minor allele , variant-phenotype effects , and residual errors , for individual i, phenotype k, and variant m.
We assume that each phenotype has been transformed to a standard normal distribution and that the columns of X have been centered, which means that the estimate for the intercept term is 0 and independent of the estimate of B. We use vectorized notation where the rows of B form vector of length .
We define the MRP model comparison as a BF between the alternative model, where at least one variant affects at least one phenotype, and the null model, where all variant-phenotype effects are zero. BF is the ratio of the marginal likelihoods for these two models:
where Data can correspond either to the effect size estimates and the estimated variance-covariance matrix of , , or to the original phenotypes and genotypes, and , and any other covariates that we want to regress out from the phenotypes.
The prior distribution for the null model, , is simply the point mass at .
Likelihood function
A maximum likelihood estimator of B is given by the ordinary least-squares method
which in vectorized form is denoted An estimator of the variance-covariance of is given by
where is the estimated residual variance-covariance matrix of Y given X.
Following Band et al.,20 we approximate the likelihood function of β by a multivariate normal distribution with mean and variance-covariance matrix . Note that by approximating via the trait correlation matrix, this likelihood approximation does not require access to the individual-level data X and Y but only to the summary data of effect sizes , LD-matrix , and a trait correlation estimate.
Prior of β in the alternative model
We construct the prior distribution for the alternative model in three steps, allowing the user to specify correlations between effects of different variants on different traits across different studies.
In a single study, the prior density for β incorporates the expected correlation of genetic effects among a group of variants and among a group of phenotypes . In addition, we incorporate an expected spread of the effect size of each variant by scaling as
where is a diagonal matrix with entries determining the spread of the effect size distribution for each variant . Thus, we can model settings where, e.g., PTVs have larger effect sizes than missense variants . Note that when for all m, then . All in all, our prior density for β under alternative model is
When we have data from multiple studies, we allow for possible differences in genetic effects across ethnicities or populations, extending the approximate BFs of Band et al.20 and the summary statistics approach of RAREMETAL7 from univariate to multivariate phenotypes. Let
where S is the number of studies, M is the number of variants, and K is the number of phenotypes. As with a single study, we incorporate the expected correlation of genetic effects between a pair of variants and a single phenotype by using the matrix , between a variant and a pair of phenotypes by using the matrix , and we introduce the matrix to specify a prior on the similarity in effect sizes across the studies. Thus, the prior is
It is also straightforward to include a non-zero vector μ as a prior mean of genetic effects, in which case the prior is We use this, for example, when screening for protective rare variants that have a pre-specified beneficial profile on a set of risk factors.
The BF is the ratio of the marginal likelihoods between the alternative and the null model. The marginal likelihood for the alternative model is
and the marginal likelihood for the null model is
The BF is given by
When , is an increasing function of the following quadratic form:
Furthermore, this quadratic form is the only part of the that depends on . Thus, by deriving a distribution of under the null model, we can compute a p value (by using the Imhof, Davies, or Farebrother methods) when is used as a test statistic. We include support for computing these p values in the software package for MRP. According to basic properties of quadratic forms of Gaussian variables, , where is an independent sample from a distribution (chi-square with one degree of freedom) and are the eigenvalues of matrix . The distribution function for a mixture of chi-squares can be numerically evaluated by the R package “CompQuadForm,”31 incorporated in the MRP software directly.
To compute the BF
we first consider the term inside the exponential function:
Because and are typically defined through Kronecker products of smaller matrices, their inverses are easier to compute than the inverse of their sum. Hence, we use the Woodbury matrix identity to write
To simplify the determinant calculation, we write
The logarithm of the BF is then
If studies do not share individuals, is a block-diagonal matrix
If studies share individuals, e.g., controls, we can take the approach of Cichonska et al.8 to use summary-level data to estimate the correlation structure of the non-diagonal blocks caused by overlapping individuals.
UK Biobank data
Population definitions
We used a combination of self-reported ancestry (UK Biobank field ID 21000) and principal-component analysis to identify six subpopulations in the study: white British, African, South Asian, non-British white, semi-related, and an admixed population (Table 1). To determine the first four populations, which contain samples not related closer than the third degree, we first used the principal components of the genotyped variants from the UK Biobank, defined thresholds on principal component 1 and principal component 2, and further refined the population definition.19 Semi-related individuals were grouped as individuals whose genetic data (after passing UK Biobank quality control [QC] filters; sufficiently low missingness rates; and genetically inferred sex matching reported sex), via a KING relationship table, were between conditional third and conditional second degrees of relatedness to samples in the first four groups. Admixed individuals were grouped as unrelated individuals who were flagged as “used_in_pca_calculation” by the UK Biobank and were not assigned to any of the other populations.
Table 1.
Number of individuals per population per genotyping platform (exome/array)
| Population | ||
|---|---|---|
| White British | 137,920 | 337,138 |
| Non-British white | 10,432 | 24,905 |
| African | 2,716 | 6,497 |
| South Asian | 3,569 | 7,885 |
| Semi-related | 18,100 | 44,632 |
| Admixed | 11,961 | 28,551 |
| Total | 184,698 | 449,608 |
GWAS summary statistics
We performed genome-wide association analysis on 2,019 UK Biobank traits in the six population subgroups as defined above by using PLINK v2.00a (20 October 2020). We used the “--glm” Firth fallback option in PLINK to apply an additive-effect model across all sites. Quantitative trait values were rank normalized with the “--pheno-quantile-normalize” flag. We used the following covariates in our analysis: age, sex, array type, and the first ten genetic principal components, where array type is a binary variable that represents whether an individual was genotyped with UK Biobank Axiom Array or UK BiLEVE Axiom Array. For variants that were specific to one array and exome data, we did not use array as a covariate.
For the admixed population, we conducted a local ancestry-corrected GWAS. We first assembled a reference panel from 1,380 single-ancestry samples in the 1000 Genomes Project,32 the Human Genome Diversity Project,33 and the Simons Genome Diversity Project,34 choosing appropriate ancestry clusters by running ADMIXTURE35 with the unsupervised setting. Using cross-validation, we identified eight well-supported ancestral population clusters: African, African hunter-gatherer, East Asian, European, Native American, Oceanian, South Asian, and West Asian. We then used RFMix v2.0336 to assign each of the 20,727 windows across the phased genomes to one of these eight ancestry clusters (for all individuals in the UK Biobank). These local ancestry assignments were subsequently used with PLINK2 as local covariates in the GWAS for the admixed individuals for SNPs within those respective windows. PLINK2 allows for the direct input of the RFMix output (the MSP file, which contains the most likely subpopulation assignment per conditional random field [CRF] point) as local covariates with the “--local-cov,” “--local-psam,” and “--local-haps” flags, the “--local-cats0=n” flag (where n is the number of assignments), and the “--local-pos-cols=2,1,2,7” flag (for a typical RFMix MSP output file, see “Association Analysis” page on PLINK website [web resources]).
Variant quality control and metadata generation
For quality control (QC), we ensured that variant-level missingness was less than 10%, that the p value for the Hardy-Weinberg equilibrium test (computed within unrelated individuals of white British ancestry) was greater than , and that the variant was uniquely represented (the “CHROM:POS:REF:ALT” variant string was uniquely identified) in the PLINK dataset file. In total, we removed 195,920 variants that failed to meet all of these criteria, except for 134 variants on the Y chromosome.
For the remainder, we used Variant Effect Predictor (VEP)37 to annotate the most severe consequence, the gene symbol, and HGVSp (the Human Genome Variation Society protein sequence name) of each variant in the UK Biobank exome and array data. We calculated MAFs by using PLINK. MPC13 values (variant-level) and pLI gene memberships14 were annotated from source. To determine LD independence criteria, we used PLINK’s “--indep-pairwise” function with a window size of 1,000 kb, a step size of 1, and an threshold of 0.1 on those variants that pass QC. As our analyses focused on PTVs and protein-altering variants (PAVs), we then performed this same LD independence analysis on only these, overriding assignments in the first analysis if necessary. We provide these essential metadata, which are necessary for MRP, in exome and array tables, available for direct download via the Global Biobank Engine (web resources).38
Prior matrix selection
For exome applications, we chose variants that had MAF 1% and that were LD independent according to the criteria mentioned above. For quantitative traits, we removed variants whose regression effect size had standard error greater than 100, and for binary traits, we removed variants whose regression effect size had standard error greater than 0.2. For array applications, we chose variants with MAF 1% and removed variants whose regression effect size had standard error greater than 0.2. While MRP is capable of handling all variant types (e.g., proximal coding and intronic variants), we included only PAVs and PTVs in both exome and array analyses (exome data feature many more PAVs and thus potential for power gain; Table S1; Figure S1). These sets respectively contain the following consequence annotations:
-
•
PAVs: protein_altering_variant, inframe_deletion, inframe_insertion, splice_region_variant, start_retained_variant, stop_retained_variant, missense_variant
-
•
PTVs: frameshift_variant, splice_acceptor_variant, splice_donor_variant, stop_gained, start_lost, stop_lost
For the exome meta-analysis, we assumed a SEM across studies and an IEM across variants . For example, if we have three studies, e.g., white British, African, and South Asian, assuming a SEM would mean that
The off-diagonals are slightly lower than 1 so as to maintain that the matrix is positive definite and invertible. Likewise, an IEM would imply
The same principle would apply when constructing for a gene with an arbitrary number of variants. We additionally incorporated information about the variant in order to formulate the matrix that is used in the calculation of . For both quantitative and binary traits, PTVs were assigned a σ (standard deviation of prior on effect size) of 0.2, whereas PAVs were assigned a σ value of 0.05. We also incorporated MPC and pLI deleteriousness metrics into our exome analyses. For those PTVs with a pLI of 0.8, we increased σ to 0.5, and for those PAVs with an MPC 1, we set σ = 0.05 × MPC. The matrix was then dotted with a diagonalized version of the σ vector (see MRP details above). These adjustments serve to further granularize and weight MRP results in biologically meaningful ways (Table S2; Figure S2).
We also studied how the application of MRP to multiple phenotypes together would potentially boost power to detect rare-variant associations. We calculated pairwise genetic correlations between 35 biomarker phenotypes19 by using LD score regression27 and then used the “hclust” algorithm in the R stats package39 to generate phenotype clusters. For each of these clusters, using the array data, we performed MRP in the multi-phenotype setting. For this application, we calculated empirically by using correlation coefficients computed with summary statistics from significant (p × ), LD-independent, common (MAF 0.01) variants.
Results
Simulations
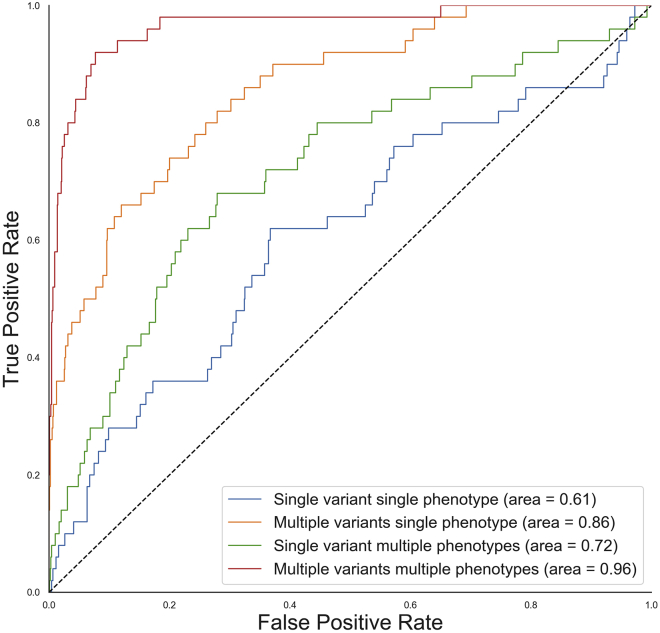
To study the behavior of MRP going from a single phenotype to multiple phenotypes, we conducted a simulation study where we assumed an allelic architecture consistent to that discovered for APOC3 in relation to triglycerides, low-density lipoprotein cholesterol (LDL-C), and high-density lipoprotein cholesterol (HDL-C).30,40,41 We simulated three continuous phenotypes with a total correlation consistent with that observed for triglycerides, LDL-C, and HDL-C. Furthermore, we introduced effects to four variants consistent with the effects observed in four PTVs (approximately 0.35 standard deviations away from the population mean) and to another four variants consistent with the effects observed for missense variants (approximately 0.2 standard deviations away from the population mean), all with MAF of 0.05%. The PTV group of variants had the same effects, whereas out of the missense variants, half had positive and the other half had negative effect sizes. The correlation of effects between the group of phenotypes was set to be directionally consistent with the direction of genetic effects observed for lipid phenotypes and PTVs in APOC3, i.e., proportional effects for triglycerides and LDL-C, and inversely proportional for LDL-C and HDL-C and triglycerides and HDL-C. We simulated 1,000 genes where 50 of the genes contained non-zero effects on the multivariate phenotype. Given we know which of the 1,000 genes contained non-zero effects, we could compute the true positive rates and false positive rates for a given BF threshold. We find MRP with an IEM across variants to be comparable to SKAT and MRP with a SEM across variants to be comparable to the burden test (Figures S3 and S4). We analyze the data as follows: (1) single-variant and single-phenotype, (2) multiple variants and single-phenotype, (3) single-variant and multiple phenotypes, and (4) multiple variants and multiple phenotypes (Figure 2). We find that in some scenarios, analyzing multiple variants and multiple phenotypes jointly improved the ability to detect signals; however, this improvement is only specific to situations in which multiple variants have a true effect and/or the variants have effects on the multiple phenotypes in question (Figures S5 and S6).
Figure 2.
From single-variant and single-phenotype to multiple-variant and multiple-phenotype gene discovery
Receiver operating characteristic (ROC) curves for detecting simulated gene association to any of the phenotypes via single-variant and single-phenotype association (blue) to multiple-variant and multiple-phenotype association (red).
Exome single-phenotype meta-analyses
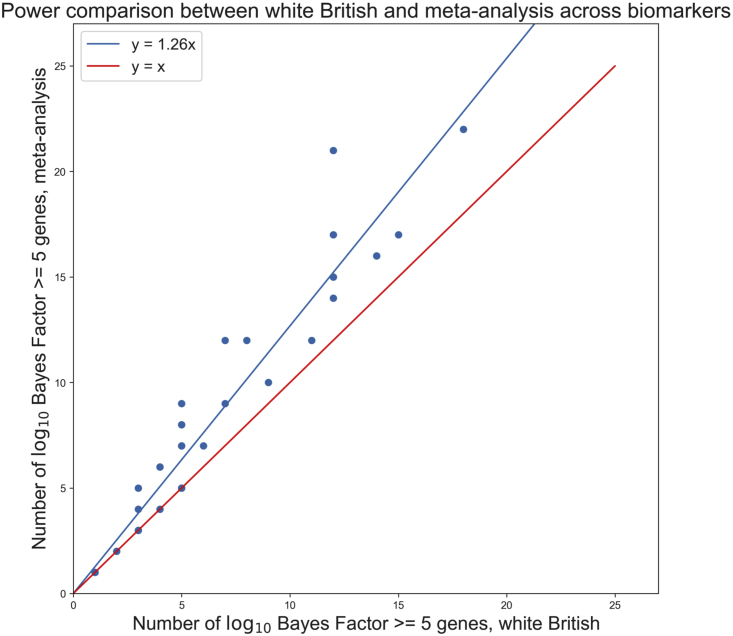
We used MRP to perform exome meta-analysis on 2,019 traits across six UK Biobank populations as described in material and methods. Among the best-powered and -represented traits were a set of 35 biomarkers, the focus of a previous publication.19 We see the number of 5 genes increasing from a single-population to a meta-analysis setting. Because we expect that the meta-analysis over different ancestries cannot be more confounded than the analysis of a single ancestry, we interpret the increase in the number of genes as an increase in the statistical power to detect rare-variant associations (Figure 3).
Figure 3.
From single to multiple populations
Scatterplot showing number of genes with BF 5 for white British population only (x axis) versus meta-analysis (y axis) across 35 biomarkers. Assuming that BFs are correctly calibrated in both analyses and that meta-analysis is not inflated compared to white British-only MRP, this figure suggests a 26% increase in power when incorporating summary statistics across multiple populations.
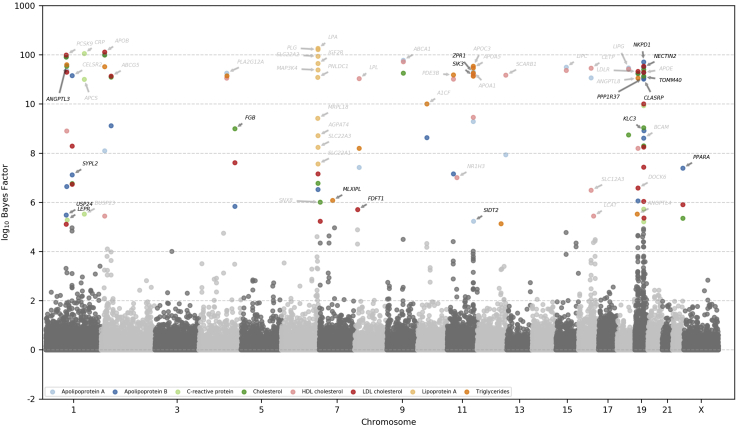
We categorize these biomarkers into six categories as in Sinnott-Armstrong et al.19 (cardiovascular, bone and joint, diabetes, hormone, liver, and renal; Figure 4 and S7–S11), and we recover several known gene-trait associations and discover several others.
Figure 4.
Manhattan plot showing BF under an independent effects variant model among protein-altering variants for cardiovascular phenotypes
Scale is logarithmic after BF 10. Genes found in Sinnott-Armstrong et al.19 are annotated in gray, whereas the other genes are annotated in black.
Among the “bone and joint” biomarkers (alkaline phosphatase, calcium, and vitamin D), we recover associations between CASR and calcium42 and HAL and vitamin D.43 As compared to results from array data as found in Sinnott-Armstrong et al.,19 we also recover exome-specific associations between ALDH5A1 and alkaline phosphatase44 and PDE3B and vitamin D.43
For the “cardiovascular” phenotypes (apolipoprotein A, apolipoprotein B, C-reactive protein, total cholesterol, HDL-C, LDL-C, lipoprotein A, and triglycerides), MRP recovers array associations between the following: PLG, LPA, and lipoprotein A;45 APOC3 and triglycerides;46 ANGPTL3 and triglycerides;47 APOB and apolipoprotein B48 and LDL-C;49 ABCA1 and apolipoprotein A46 and HDL-C;49 PCSK9 and total cholesterol;50 and CRP and C-reactive protein.51 Exome-only signals recover associations such as between ZPR150 and SIK344 and triglycerides.
In the two diabetes-related phenotypes (glucose and HbA1c), we recover associations between G6PC2 and glucose50 as well as PIEZO1 and HbA1c52 and an additional exome association between G6PD and HbA1c.52 Hormonal recoveries include those between SHBG and SHBG and testosterone levels and GH1 and IGF-1 levels.
MRP applied to liver-related phenotypes recover known associations between the following: UGT genes and bilirubins;53 GOT1 and aspartate aminotransferase;54 FCGRT and albumin;44 and GPT and AST-ALT ratio.51 In the exome sequencing, we additionally recover associations between GGT1 and gamma glutamyltransferase,55 TMEM236 and aspartate aminotransferase,44 and SLCO1B3 and bilirubin.56
The renal traits similarly feature a mix of array recoveries and exome discoveries. We recover signal between the following: SLC22A2 and creatinine;57 CST3 and cystatin C;48 COL4A4 and microalbumin;58 TNFRSF13B and non-albumin protein;44 FCGRT and total protein; WDR1, RASGRP2, DRD5, and urate;59,60 and LRP2 and eGFR levels.61 We additionally discover novel gene-trait associations (not found in the NHGRI-EBI catalog or Open Targets Genetics) across these biomarker categories, including the following: GLPD1 and alkaline phosphatase; NKPD1 and apolipoprotein B; RENBP/MAP3K15 and Hba1c; PARPBP and IGF-1; NLGN2 and SHBG; ALB and albumin; ALPL and phosphate; RBM47 and urea; ALDH16A1 and urate; THBD and cystatin C; ITPR3 and phosphate; SLC22A7 and creatinine; and FCGR2B and non-albumin protein.
For the 2,019 traits for which MRP was performed, there were also a considerable number of associations found among non-biomarker traits. We found associations between TUBB1 and platelet distribution width and mean platelet volume;62 IL17RA and monocyte count and percentage;62 OCA2/MC1R and skin color/hair color;63, 64, 65, 66 IQGAP2 and mean platelet volume;62 SLC24A5, HERC2, TCF25, and TYR and skin color;67 SH2B3 and JAK2 and platelet crit68 and count;69 KALRN and mean platelet volume;62 HBB and mean corpuscular volume,70 mean corpuscular hemoglobin,71 and red blood cell count;72 and CXCR2 and neutrophil count.62
The average runtime for these analyses was around 1 day for quantitative traits and 5 h for binary traits with a machine with 16 cores and 200 GB RAM (Table S3).
Array single-population multi-phenotype analyses
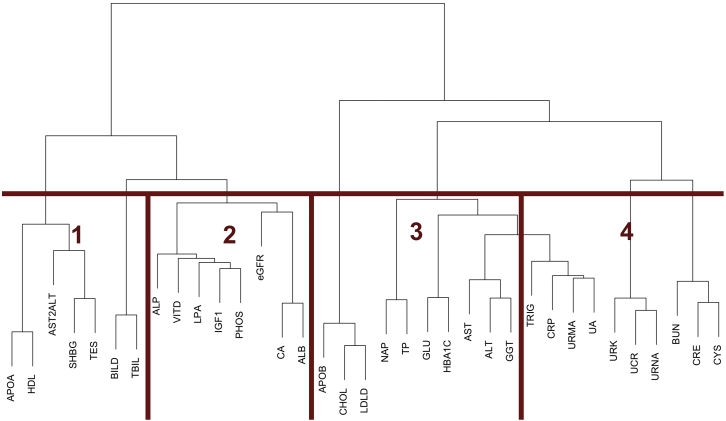
In order to demonstrate the effectiveness of MRP to boost signal in a multi-phenotype context, we used LD-score regression27 to determine genetic correlations between the 35 biomarker traits (Figure S12) that were a focus of a previous paper.19 This correlation matrix was then used for hierarchical clustering followed by dynamic tree cutting, which formed four clusters of between seven and ten traits each (Figure 5). We generated the correlation plots as shown in Figure 6.
Figure 5.
Hierarchical clustering dendrogram and derived clusters from dynamic tree cutting algorithm
Based on genetic correlation derived from an LD-score-regression-based distance matrix between 35 biomarker traits.
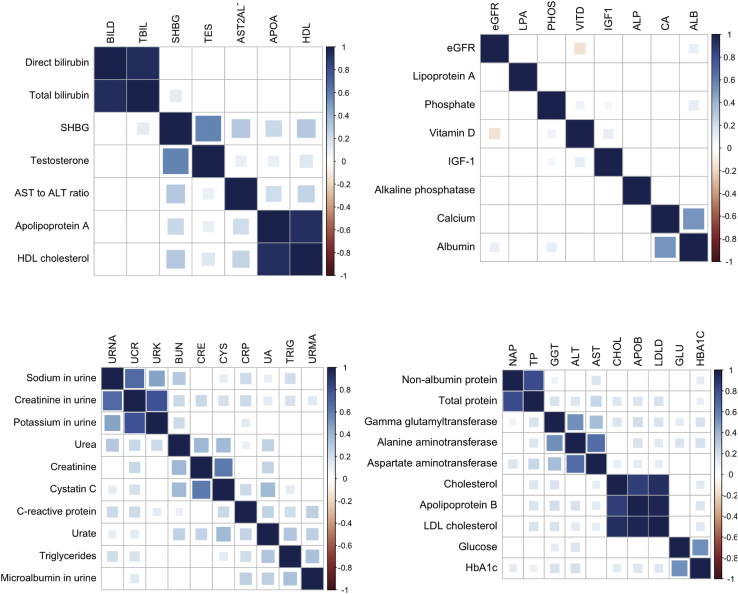
Figure 6.
LD-score-regression-based genetic correlation plots of candidate clusters
Derived from the dendrogram in Figure 5 via a dynamic tree cutting algorithm.
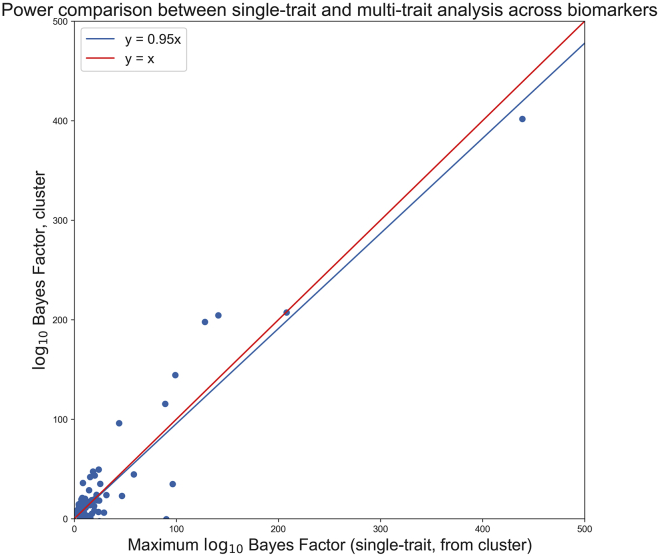
Multi-phenotype MRP results in several substantial power gains throughout the four clusters; one of these clusters is highlighted in Figure 7. As compared to the maximum from the constituent phenotypes, the multi-phenotype analysis generally fares comparably while also highlighting clear targets. We found evidence for association between rare coding variants in several genes and the clusters above; TNFRSF13B ( = 204.5, max = 141.0), APOB ( = 197.9, max = 128.0), and SNX8 ( = 96.0, max = 43.8) receive a boost in of over 50 units for cluster 1 (alanine aminotransferase, aspartate aminotransferase, gamma glutamyltransferase, glucose, HbA1c, total protein, apolipoprotein B, cholesterol, LDL-C, and non-albumin protein). Several other genes that are clearly below 5 (in ) in the single-trait settings become above 5 in the joint setting (e.g., G6PC; = 5.3, max = 1.3). The G6PC gene provides instructions for making the glucose 6-phosphatase enzyme, found on the membrane of the endoplasmic reticulum. The enzyme is expressed in active form in the liver, kidneys, and intestines and is the main regulator of glucose production in the liver; given the traits included in cluster 1, the increase in power may be biologically relevant.73 These results demonstrate that MRP can identify biologically meaningful targets that may be missed by standard GWAS approaches.
Figure 7.
Cluster versus single-trait power analysis
Power comparison of genes with BF 5 in either (i) any of the single-trait analyses of the traits within the cluster or (ii) the multi-trait analysis for a cluster of biomarkers (alanine aminotransferase, aspartate aminotransferase, gamma glutamyltransferase, glucose, HbA1c, total protein, apolipoprotein B, cholesterol, LDL-C, and non-albumin protein). x axis depicts the maximum BF of the gene among any of the constituent single-trait analyses, and y axis depicts the multi-trait result. Multi-trait analyses roughly equal the highest-powered single-trait analyses while also substantially boosting signal in some genes.
The average runtime for these analyses was around 5 h with a machine with 16 cores and 200 GB RAM (Table S3).
Discussion
In this study, we developed MRP, a Bayesian model comparison approach that shares information across variants, phenotypes, and studies to identify rare-variant associations. We used simulations to verify that jointly considering both variants and phenotypes can improve the ability to detect associations. We also applied the MRP model comparison framework in a meta-analysis setting to exome summary statistics across the UK Biobank, identifying strong evidence for the previously described associations between, for example, HAL and vitamin D,43 and discovering several novel associations, such as between GLPD1 and alkaline phosphatase. We made the full results set available on the Global Biobank Engine (web resources).38 We also leveraged MRP to boost signal in a multi-phenotype setting by using the array data (which has many more samples than the exome data), finding genes such as G6PC that do not come up in the single-trait context but show strong evidence in the joint analysis. These results demonstrate the ability of the MRP model comparison approach to leverage information across multiple phenotypes and variants to discover rare-variant associations.
MRP enjoys distinct advantages over prior techniques such as (trans-ancestry) meta-analysis and MultiSKAT in that it allows for both multiple studies as well as multiple phenotypes simultaneously, whereas (trans-ancestry) meta-analysis only allows for multiple studies and not multiple phenotypes and MultiSKAT only allows for multiple phenotypes and not multiple studies. MRP’s closest relative, MetaMultiSKAT,74 allows for these two simultaneously as well. In comparison to MetaMultiSKAT, the novelty in MRP is the flexibility in the models/choices one can make in the priors and the inferential framework. In particular, one can directly compare any two MRP models against each other by comparing their BFs against each other. For example, one can assess the heterogeneity of effects across studies by comparing similar and IEMs for ; if done on a per-variant basis, this can be a useful QC statistic. We can also assess the degree to which variants within a block (genes, annotation categories, etc.) have similar or disparate effects with a proper choice of . Such inferences are not easily available when p values are used as a basis for inference.
Analogous to the work presented in Rivas et al.,11 we see MRP as a natural extension for the analysis of rare variants in gene-based settings. The same advantages we see in using BFs over p values exist here as well. To briefly summarize, studies that follow both discovery and replication phases seek to rank genes for follow-up according to departure from a null. Power in frequentist studies will change with the number of samples and the number of variants included in the analysis, whereas BFs can easily be compared naturally agnostic of the number of variants included as well as the sample size.
We acknowledge some limitations in this study. First, we are pruning variants that are LD dependent. Ancestry misspecification could have a dramatic effect on the computation of LD, especially in the setting of rare variants, whose MAFs could very well be population specific; one could obtain LD estimates that are inaccurate and inapplicable to the study at hand. We propose that if variants in LD are to be used, then LD independence should be computed within the studies that are being meta-analyzed. Many of the rare variants will not be present in external reference panels. Extensions to incorporate in-study LD matrices, as in the work by Feng et al. in RAREMETAL and RAREMETALWORKER,75 are a logical extension to MRP. Second, in the ultra-rare variant setting and/or when case-control numbers are particularly imbalanced, the assumption that effect sizes follow a Gaussian may be invalid.76 Third, removing related individuals as in this study results in a loss of power. Because MRP is agnostic of summary statistic type, though, one can explore models such as SAIGE,76 REGENIE,77 or BOLT-LMM78 that better account for related individuals so as to improve power.
As genetic data linked to high-dimensional phenotype data are increasingly being made available through biobanks, health systems, and research programs, there is a large need for statistical approaches that can leverage information across different genetic variants, phenotypes, and studies to make strong inferences about disease-associated genes. The approach presented here relies only on summary statistics from marginal association analyses, which can be shared with less privacy concerns compared to raw genotype and phenotype data. Combining joint analysis of variants and phenotypes with meta-analysis across studies offers new opportunities to identify gene-disease associations.
Acknowledgments
This research was conducted with the UK Biobank resource under application number 24983, “Generating effective therapeutic hypotheses from genomic and hospital linkage data.” Based on the information provided in protocol 44532, the Stanford IRB has determined that the research does not involve human subjects as defined in 45 CFR 46.102(f) or 21 CFR 50.3(g). All participants in the UK Biobank study provided written informed consent (see “basis of UK Biobank participation” in web resources for more information). Statin adjustment analyses were further conducted via UK Biobank application 7089 with a protocol approved by the Partners HealthCare Institutional Review Board. We thank all the participants in the UK Biobank. We thank members of the Rivas lab for their feedback. M.A.R. is in part supported by the NHGRI of the NIH under award R01HG010140 (M.A.R.) and an NIH Center for Multi- and Trans-ethnic Mapping of Mendelian and Complex Diseases grant (5U01 HG009080). G.R.V. is supported by the National Library of Medicine (NLM) T15 Continuing Education Training Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. Some of the computing for this project was performed on the Sherlock cluster at Stanford University. We would like to thank Stanford University and the Stanford Research Computing Center for providing computational resources and support that contributed to these research results.
Declaration of interests
M.A.R. is on the SAB of 54Gene and Related Sciences; is scientific founder of Broadwing Bio; and has advised BioMarin, Third Rock Ventures, and MazeTx. C.D.B. is the owner and president of C.D.B. Consulting, LTD, and also a director at EdenRoc Sciences, LLC and BigData Bio LLC (LLC) and Etalon DX; founder of Arc Bio LLC (formerly IdentifyGenomics LLC and BigData Bio LLC); and an SAB member of Imprimed, FaunaBio, Columbia Care, and Digitalis Ventures. He is also a venture partner at F-Prime Capital Partners. M.J.D. is a founder of MazeTx.
Published: November 24, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ajhg.2021.11.005.
Contributor Information
Matti Pirinen, Email: matti.pirinen@helsinki.fi.
Manuel A. Rivas, Email: mrivas@stanford.edu.
Web resources
Basis of UK Biobank participation, https://www.ukbiobank.ac.uk/explore-your-participation/basis-of-your-participation
PLINK, https://www.cog-genomics.org/plink/1.9/assoc
Rare variant association analysis of 200k exomes with MRP, https://biobankengine.stanford.edu/RIVAS_HG38/mrpgene/all
Rare variant association analysis of 500k array coding variant data with MRP, https://biobankengine.stanford.edu/RIVAS_HG19/mrpgene/all
UK Biobank Array Metadata for MRP, https://biobankengine.stanford.edu/static/ukb_cal-consequence_wb_maf_gene_ld_indep_mpc_pli.tsv.gz
UK Biobank Exome Metadata for MRP, https://biobankengine.stanford.edu/static/ukb_exm_oqfe-consequence_wb_maf_gene_ld_indep_mpc_pli.tsv.gz
Supplemental information
References
- 1.Abecasis G.R., Altshuler D., Auton A., Brooks L.D., Durbin R.M., Gibbs R.A., Hurles M.E., McVean G.A., 1000 Genomes Project Consortium A map of human genome variation from population scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nejentsev S., Walker N., Riches D., Egholm M., Todd J.A. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rivas M.A., Beaudoin M., Gardet A., Stevens C., Sharma Y., Zhang C.K., Boucher G., Ripke S., Ellinghaus D., Burtt N., et al. Deep resequencing of GWAS loci identifies independent rare variants associated with inflammatory bowel disease. Nat. Genet. 2011;43:1066–1073. doi: 10.1038/ng.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abecasis G.R., Auton A., Brooks L.D., DePristo M.A., Durbin R.M., Handsaker R.E., Kang H.M., Marth G.T., McVean G.A., 1000 Genomes Project Consortium An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majithia A.R., Flannick J., Shahinian P., Guo M., Bray M.-.A., Fontanillas P., Gabriel S.B., Rosen E.D., Altshuler D., NHGRI JHS Rare variants in pparg with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proceedings of the National Academy of Sciences. 2014;111:13127–13132. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee S., Abecasis G.R., Boehnke M., Lin X. Rare-variant association analysis: study designs and statistical tests. Am. J. Hum. Genet. 2014;95:5–23. doi: 10.1016/j.ajhg.2014.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu D.J., Peloso G.M., Zhan X., Holmen O.L., Zawistowski M., Feng S., Nikpay M., Auer P.L., Goel A., Zhang H., et al. Meta-analysis of gene-level tests for rare variant association. Nat. Genet. 2014;46:200–204. doi: 10.1038/ng.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cichonska A., Rousu J., Marttinen P., Kangas A.J., Soininen P., Lehtimäki T., Raitakari O.T., Järvelin M.R., Salomaa V., Ala-Korpela M., et al. metaCCA: summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis. Bioinformatics. 2016;32:1981–1989. doi: 10.1093/bioinformatics/btw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stephens M. A unified framework for association analysis with multiple related phenotypes. PLoS ONE. 2013;8:e65245. doi: 10.1371/journal.pone.0065245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pickrell J.K., Berisa T., Liu J.Z., Ségurel L., Tung J.Y., Hinds D.A. Detection and interpretation of shared genetic influences on 42 human traits. Nat. Genet. 2016;48:709–717. doi: 10.1038/ng.3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rivas M.A., Pirinen M., Neville M.J., Gaulton K.J., Moutsianas L., Lindgren C.M., Karpe F., McCarthy M.I., Donnelly P. Assessing association between protein truncating variants and quantitative traits. Bioinformatics. 2013;29:2419–2426. doi: 10.1093/bioinformatics/btt409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rivas M.A., Pirinen M., Conrad D.F., Lek M., Tsang E.K., Karczewski K.J., Maller J.B., Kukurba K.R., DeLuca D.S., Fromer M., et al. Human genomics. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science. 2015;348:666–669. doi: 10.1126/science.1261877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Samocha K.E., Kosmicki J.A., Karczewski K.J., O’Donnell-Luria A.H., Pierce-Hoffman E., MacArthur D.G., Neale B.M., Daly M.J. Regional missense constraint improves variant deleteriousness prediction. bioRxiv. 2017 doi: 10.1101/148353. [DOI] [Google Scholar]
- 14.Fuller Z.L., Berg J.J., Mostafavi H., Sella G., Przeworski M. Measuring intolerance to mutation in human genetics. Nat. Genet. 2019;51:772–776. doi: 10.1038/s41588-019-0383-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen J., Pertsemlidis A., Kotowski I.K., Graham R., Garcia C.K., Hobbs H.H. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 2005;37:161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 16.Cohen J.C., Boerwinkle E., Mosley T.H., Jr., Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 17.Sullivan D., Olsson A.G., Scott R., Kim J.B., Xue A., Gebski V., Wasserman S.M., Stein E.A. Effect of a monoclonal antibody to PCSK9 on low-density lipoprotein cholesterol levels in statin-intolerant patients: the GAUSS randomized trial. JAMA. 2012;308:2497–2506. doi: 10.1001/jama.2012.25790. [DOI] [PubMed] [Google Scholar]
- 18.Wu M.C., Lee S., Cai T., Li Y., Boehnke M., Lin X. Rare-variant association testing for sequencing data with the sequence kernel association test. Am. J. Hum. Genet. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sinnott-Armstrong N., Tanigawa Y., Amar D., Mars N., Benner C., Aguirre M., Venkataraman G.R., Wainberg M., Ollila H.M., Kiiskinen T., et al. Genetics of 35 blood and urine biomarkers in the UK Biobank. Nat. Genet. 2021;53:185–194. doi: 10.1038/s41588-020-00757-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Band G., Le Q.S., Jostins L., Pirinen M., Kivinen K., Jallow M., Sisay-Joof F., Bojang K., Pinder M., Sirugo G., et al. Imputation-based meta-analysis of severe malaria in three African populations. PLoS Genet. 2013;9:e1003509. doi: 10.1371/journal.pgen.1003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacArthur D.G., Balasubramanian S., Frankish A., Huang N., Morris J., Walter K., Jostins L., Habegger L., Pickrell J.K., Montgomery S.B., et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Findlay G.M., Boyle E.A., Hause R.J., Klein J.C., Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neale B.M., Rivas M.A., Voight B.F., Altshuler D., Devlin B., Orho-Melander M., Kathiresan S., Purcell S.M., Roeder K., Daly M.J. Testing for an unusual distribution of rare variants. PLoS Genet. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Clarke G.M., Rivas M.A., Morris A.P. A flexible approach for the analysis of rare variants allowing for a mixture of effects on binary or quantitative traits. PLoS Genet. 2013;9:e1003694. doi: 10.1371/journal.pgen.1003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cotsapas C., Voight B.F., Rossin E., Lage K., Neale B.M., Wallace C., Abecasis G.R., Barrett J.C., Behrens T., Cho J., et al. Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet. 2011;7:e1002254. doi: 10.1371/journal.pgen.1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Solovieff N., Cotsapas C., Lee P.H., Purcell S.M., Smoller J.W. Pleiotropy in complex traits: challenges and strategies. Nat. Rev. Genet. 2013;14:483–495. doi: 10.1038/nrg3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bulik-Sullivan B., Finucane H.K., Anttila V., Gusev A., Day F.R., Loh P.R., Duncan L., Perry J.R., Patterson N., Robinson E.B., et al. An atlas of genetic correlations across human diseases and traits. Nat. Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Do R., Stitziel N.O., Won H.H., Jørgensen A.B., Duga S., Angelica Merlini P., Kiezun A., Farrall M., Goel A., Zuk O., et al. Exome sequencing identifies rare LDLR and APOA5 alleles conferring risk for myocardial infarction. Nature. 2015;518:102–106. doi: 10.1038/nature13917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Do R., Willer C.J., Schmidt E.M., Sengupta S., Gao C., Peloso G.M., Gustafsson S., Kanoni S., Ganna A., Chen J., et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat. Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Crosby J., Peloso G.M., Auer P.L., Crosslin D.R., Stitziel N.O., Lange L.A., Lu Y., Tang Z.Z., Zhang H., Hindy G., et al. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N. Engl. J. Med. 2014;371:22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duchesne P., Lafaye de Micheaux P. Computing the distribution of quadratic forms: Further comparisons between the liu-tang-zhang approximation and exact methods. Comput. Stat. Data Anal. 2010;54:858–862. [Google Scholar]
- 32.Auton A., Brooks L.D., Durbin R.M., Garrison E.P., Kang H.M., Korbel J.O., Marchini J.L., McCarthy S., McVean G.A., Abecasis G.R., 1000 Genomes Project Consortium A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergström A., McCarthy S.A., Hui R., Almarri M.A., Ayub Q., Danecek P., Chen Y., Felkel S., Hallast P., Kamm J., et al. Insights into human genetic variation and population history from 929 diverse genomes. Science. 2020;367:eaay5012. doi: 10.1126/science.aay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mallick S., Li H., Lipson M., Mathieson I., Gymrek M., Racimo F., Zhao M., Chennagiri N., Nordenfelt S., Tandon A., et al. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature. 2016;538:201–206. doi: 10.1038/nature18964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander D.H., Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics. 2011;12:246. doi: 10.1186/1471-2105-12-246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maples B.K., Gravel S., Kenny E.E., Bustamante C.D. Rfmix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013;93:278–288. doi: 10.1016/j.ajhg.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McLaren W., Gil L., Hunt S.E., Riat H.S., Ritchie G.R.S., Thormann A., Flicek P., Cunningham F. The ensembl variant effect predictor. Genome Biol. 2016;17:1–14. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McInnes G., Tanigawa Y., DeBoever C., Lavertu A., Olivieri J.E., Aguirre M., Rivas M.A. Global Biobank Engine: enabling genotype-phenotype browsing for biobank summary statistics. Bioinformatics. 2019;35:2495–2497. doi: 10.1093/bioinformatics/bty999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murtagh F., Chambers J.M., Gordesch J., Klas A., Lebart L., Sint P.P. Multidimensional clustering algorithms. compstat lect. 4. lect. Comput. Stat. 1985 [Google Scholar]
- 40.Pollin T.I., Damcott C.M., Shen H., Ott S.H., Shelton J., Horenstein R.B., Post W., McLenithan J.C., Bielak L.F., Peyser P.A., et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322:1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jørgensen A.B., Frikke-Schmidt R., Nordestgaard B., Tybjaerg-Hansen A. Loss-of-function mutations in apoc3, remnant cholesterol, ldl cholesterol, and risk of ischemic vascular disease. Atherosclerosis. 2016;252:e251–e252. [Google Scholar]
- 42.Kapur K., Johnson T., Beckmann N.D., Sehmi J., Tanaka T., Kutalik Z., Styrkarsdottir U., Zhang W., Marek D., Gudbjartsson D.F., et al. Genome-wide meta-analysis for serum calcium identifies significantly associated SNPs near the calcium-sensing receptor (CASR) gene. PLoS Genet. 2010;6:e1001035. doi: 10.1371/journal.pgen.1001035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Manousaki D., Mitchell R., Dudding T., Haworth S., Harroud A., Forgetta V., Shah R.L., Luan J., Langenberg C., Timpson N.J., et al. Genome-wide association study for vitamin d levels reveals 69 independent loci. Am. J. Hum. Genet. 2020;106:327–337. doi: 10.1016/j.ajhg.2020.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanai M., Akiyama M., Takahashi A., Matoba N., Momozawa Y., Ikeda M., Iwata N., Ikegawa S., Hirata M., Matsuda K., et al. Genetic analysis of quantitative traits in the Japanese population links cell types to complex human diseases. Nat. Genet. 2018;50:390–400. doi: 10.1038/s41588-018-0047-6. [DOI] [PubMed] [Google Scholar]
- 45.Mack S., Coassin S., Rueedi R., Yousri N.A., Seppälä I., Gieger C., Schönherr S., Forer L., Erhart G., Marques-Vidal P., et al. A genome-wide association meta-analysis on lipoprotein (a) concentrations adjusted for apolipoprotein (a) isoforms. J. Lipid Res. 2017;58:1834–1844. doi: 10.1194/jlr.M076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Richardson T.G., Sanderson E., Palmer T.M., Ala-Korpela M., Ference B.A., Smith G.D., Holmes M.V. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: A multivariable mendelian randomisation analysis. PLoS Med. 2020;17:e1003062. doi: 10.1371/journal.pmed.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kilpeläinen T.O., Bentley A.R., Noordam R., Sung Y.J., Schwander K., Winkler T.W., Jakupović H., Chasman D.I., Manning A., Ntalla I., et al. Multi-ancestry study of blood lipid levels identifies four loci interacting with physical activity. Nat. Commun. 2019;10:376. doi: 10.1038/s41467-018-08008-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Suhre K., Arnold M., Bhagwat A.M., Cotton R.J., Engelke R., Raffler J., Sarwath H., Thareja G., Wahl A., DeLisle R.K., et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8:1–14. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hoffmann T.J., Theusch E., Haldar T., Ranatunga D.K., Jorgenson E., Medina M.W., Kvale M.N., Kwok P.Y., Schaefer C., Krauss R.M., et al. A large electronic-health-record-based genome-wide study of serum lipids. Nat. Genet. 2018;50:401–413. doi: 10.1038/s41588-018-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wojcik G.L., Graff M., Nishimura K.K., Tao R., Haessler J., Gignoux C.R., Highland H.M., Patel Y.M., Sorokin E.P., Avery C.L., et al. Genetic analyses of diverse populations improves discovery for complex traits. Nature. 2019;570:514–518. doi: 10.1038/s41586-019-1310-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nielsen J.B., Rom O., Surakka I., Graham S.E., Zhou W., Roychowdhury T., Fritsche L.G., Gagliano Taliun S.A., Sidore C., Liu Y., et al. Loss-of-function genomic variants highlight potential therapeutic targets for cardiovascular disease. Nat. Commun. 2020;11:6417. doi: 10.1038/s41467-020-20086-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wheeler E., Leong A., Liu C.T., Hivert M.F., Strawbridge R.J., Podmore C., Li M., Yao J., Sim X., Hong J., et al. Impact of common genetic determinants of Hemoglobin A1c on type 2 diabetes risk and diagnosis in ancestrally diverse populations: A transethnic genome-wide meta-analysis. PLoS Med. 2017;14:e1002383. doi: 10.1371/journal.pmed.1002383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gurdasani D., Carstensen T., Fatumo S., Chen G., Franklin C.S., Prado-Martinez J., Bouman H., Abascal F., Haber M., Tachmazidou I., et al. Uganda genome resource enables insights into population history and genomic discovery in africa. Cell. 2019;179:984–1002.e36. doi: 10.1016/j.cell.2019.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moon S., Kim Y.J., Han S., Hwang M.Y., Shin D.M., Park M.Y., Lu Y., Yoon K., Jang H.-M., Kim Y.K., et al. The korea biobank array: design and identification of coding variants associated with blood biochemical traits. Sci. Rep. 2019;9:1382. doi: 10.1038/s41598-018-37832-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seo J.Y., Lee J.-E., Chung G.E., Shin E., Kwak M.-S., Yang J.I., Yim J.Y. A genome-wide association study on liver enzymes in korean population. PLoS ONE. 2020;15:e0229374. doi: 10.1371/journal.pone.0229374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kang T.-W., Kim H.-J., Ju H., Kim J.-H., Jeon Y.-J., Lee H.-C., Kim K.-K., Kim J.-W., Lee S., Kim J.Y., et al. Genome-wide association of serum bilirubin levels in Korean population. Hum. Mol. Genet. 2010;19:3672–3678. doi: 10.1093/hmg/ddq281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Graham S.E., Nielsen J.B., Zawistowski M., Zhou W., Fritsche L.G., Gabrielsen M.E., Skogholt A.H., Surakka I., Hornsby W.E., Fermin D., et al. Sex-specific and pleiotropic effects underlying kidney function identified from GWAS meta-analysis. Nat. Commun. 2019;10:1847. doi: 10.1038/s41467-019-09861-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Casanova F., Tyrrell J., Beaumont R.N., Ji Y., Jones S.E., Hattersley A.T., Weedon M.N., Murray A., Shore A.C., Frayling T.M., et al. A genome-wide association study implicates multiple mechanisms influencing raised urinary albumin-creatinine ratio. Hum. Mol. Genet. 2019;28:4197–4207. doi: 10.1093/hmg/ddz243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gill D., Cameron A.C., Burgess S., Li X., Doherty D.J., Karhunen V., Abdul-Rahim A.H., Taylor-Rowan M., Zuber V., Tsao P.S., et al. Urate, blood pressure, and cardiovascular disease: evidence from mendelian randomization and meta-analysis of clinical trials. Hypertension. 2021;77:383–392. doi: 10.1161/HYPERTENSIONAHA.120.16547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kamatani Y., Matsuda K., Okada Y., Kubo M., Hosono N., Daigo Y., Nakamura Y., Kamatani N. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 61.Gorski M., Van Der Most P.J., Teumer A., Chu A.Y., Li M., Mijatovic V., Nolte I.M., Cocca M., Taliun D., Gomez F., et al. 1000 genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci. Rep. 2017;7:1–11. doi: 10.1038/srep45040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Astle W.J., Elding H., Jiang T., Allen D., Ruklisa D., Mann A.L., Mead D., Bouman H., Riveros-Mckay F., Kostadima M.A., et al. The allelic landscape of human blood cell trait variation and links to common complex disease. Cell. 2016;167:1415–1429.e19. doi: 10.1016/j.cell.2016.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Morgan M.D., Pairo-Castineira E., Rawlik K., Canela-Xandri O., Rees J., Sims D., Tenesa A., Jackson I.J. Genome-wide study of hair colour in uk biobank explains most of the snp heritability. Nat. Commun. 2018;9:1–10. doi: 10.1038/s41467-018-07691-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Adhikari K., Mendoza-Revilla J., Sohail A., Fuentes-Guajardo M., Lampert J., Chacón-Duque J.C., Hurtado M., Villegas V., Granja V., Acuña-Alonzo V., et al. A GWAS in Latin Americans highlights the convergent evolution of lighter skin pigmentation in Eurasia. Nat. Commun. 2019;10:358. doi: 10.1038/s41467-018-08147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu F., Visser M., Duffy D.L., Hysi P.G., Jacobs L.C., Lao O., Zhong K., Walsh S., Chaitanya L., Wollstein A., et al. Genetics of skin color variation in Europeans: genome-wide association studies with functional follow-up. Hum. Genet. 2015;134:823–835. doi: 10.1007/s00439-015-1559-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hysi P.G., Valdes A.M., Liu F., Furlotte N.A., Evans D.M., Bataille V., Visconti A., Hemani G., McMahon G., Ring S.M., et al. Genome-wide association meta-analysis of individuals of European ancestry identifies new loci explaining a substantial fraction of hair color variation and heritability. Nat. Genet. 2018;50:652–656. doi: 10.1038/s41588-018-0100-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lona-Durazo F., Hernandez-Pacheco N., Fan S., Zhang T., Choi J., Kovacs M.A., Loftus S.K., Le P., Edwards M., Fortes-Lima C.A., et al. Meta-analysis of GWA studies provides new insights on the genetic architecture of skin pigmentation in recently admixed populations. BMC Genet. 2019;20:59. doi: 10.1186/s12863-019-0765-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vuckovic D., Bao E.L., Akbari P., Lareau C.A., Mousas A., Jiang T., Chen M.H., Raffield L.M., Tardaguila M., Huffman J.E., et al. The polygenic and monogenic basis of blood traits and diseases. Cell. 2020;182:1214–1231.e11. doi: 10.1016/j.cell.2020.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gieger C., Radhakrishnan A., Cvejic A., Tang W., Porcu E., Pistis G., Serbanovic-Canic J., Elling U., Goodall A.H., Labrune Y., et al. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hodonsky C.J., Jain D., Schick U.M., Morrison J.V., Brown L., McHugh C.P., Schurmann C., Chen D.D., Liu Y.M., Auer P.L., et al. Genome-wide association study of red blood cell traits in Hispanics/Latinos: The Hispanic Community Health Study/Study of Latinos. PLoS Genet. 2017;13:e1006760. doi: 10.1371/journal.pgen.1006760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chen M.H., Raffield L.M., Mousas A., Sakaue S., Huffman J.E., Moscati A., Trivedi B., Jiang T., Akbari P., Vuckovic D., et al. Trans-ethnic and ancestry-specific blood-cell genetics in 746,667 individuals from 5 global populations. Cell. 2020;182:1198–1213.e14. doi: 10.1016/j.cell.2020.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hodonsky C.J., Baldassari A.R., Bien S.A., Raffield L.M., Highland H.M., Sitlani C.M., Wojcik G.L., Tao R., Graff M., Tang W., et al. Ancestry-specific associations identified in genome-wide combined-phenotype study of red blood cell traits emphasize benefits of diversity in genomics. BMC Genomics. 2020;21:228. doi: 10.1186/s12864-020-6626-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hutton J.C., O’Brien R.M. Glucose-6-phosphatase catalytic subunit gene family. J. Biol. Chem. 2009;284:29241–29245. doi: 10.1074/jbc.R109.025544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dutta D., Gagliano Taliun S.A., Weinstock J.S., Zawistowski M., Sidore C., Fritsche L.G., Cucca F., Schlessinger D., Abecasis G.R., Brummett C.M., et al. Meta-MultiSKAT: Multiple phenotype meta-analysis for region-based association test. Genet. Epidemiol. 2019;43:800–814. doi: 10.1002/gepi.22248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Feng S., Liu D., Zhan X., Wing M.K., Abecasis G.R. RAREMETAL: fast and powerful meta-analysis for rare variants. Bioinformatics. 2014;30:2828–2829. doi: 10.1093/bioinformatics/btu367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou W., Nielsen J.B., Fritsche L.G., Dey R., Gabrielsen M.E., Wolford B.N., LeFaive J., VandeHaar P., Gagliano S.A., Gifford A., et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet. 2018;50:1335–1341. doi: 10.1038/s41588-018-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Mbatchou J., Barnard L., Backman J., Marcketta A., Kosmicki J.A., Ziyatdinov A., Benner C., O’Dushlaine C., Barber M., Boutkov B., et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 2021;53:1097–1103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 78.Loh P.R., Tucker G., Bulik-Sullivan B.K., Vilhjálmsson B.J., Finucane H.K., Salem R.M., Chasman D.I., Ridker P.M., Neale B.M., Berger B., et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.