Summary
Obesity is known to affect female reproduction, as evidenced by obese patients suffering from subfertility and abnormal oogenesis. However, the underlying mechanisms by which obesity impairs folliculogenesis are poorly documented. Here, we performed comprehensive single-cell transcriptome analysis in both regular diet (RD) and obese mouse models to systematically uncover how obesity affects ovarian follicle cells and their interactions. We found an increased proportion of Inhbb highly expressed granulosa cells (GCs) among all the GC subpopulations in obese mice. Under obese conditions, excessive androgen secreted from endocrine theca cells (ETCs) may contribute to the imbalanced change of GC subtypes through ETCs-GCs interactions. This is alleviated by enzalutamide, an androgen receptor antagonist. We also identified and confirmed typical GC markers, such as Marcks and Prkar2b, for sensitive evaluation of female fertility in obesity. These data represent a resource for studying transcriptional networks and cell-cell interactions during folliculogenesis under physiological and pathological conditions.
Subject areas: Biological sciences, Endocrinology, Cell biology, Transcriptomics
Graphical abstract
Highlights
-
•
Single-cell transcriptome of folliculogenesis in regular diet-fed and obese mice
-
•
ETCs represent higher activity in androgen synthesis under obese conditions
-
•
Obesity contributes to the disproportion of GC subtypes under excessive androgen
-
•
Enzalutamide alleviates defects in granulosa cells from obese mice
Biological sciences; Endocrinology; Cell biology; Transcriptomics
Introduction
Folliculogenesis is a well-orchestrated process in which the growth of the ovary follicle accompanies and supports oogenetic development. The maturation of the oocyte requires and relies on the synchronous proliferation and differentiation of surrounding somatic cells. In the earliest stage, the primordial follicle comprises a primordial oocyte and a single layer of pregranulosa cells (Coticchio et al., 2015; Li and Albertini, 2013). As the primordial follicle becomes a primary follicle and continues to grow, theca cells are recruited and flattened granulosa cells (GCs) become cube-shaped (Richards et al., 2018). The theca cells are divided into two layers which emerge around the time of birth (Honda et al., 2007; Liu et al., 2015). The outer layer functions in maintaining the structure of the follicle (theca externa), and the endocrine theca cells (ETCs) located at the inner layer express enzymes involved in steroidogenesis, including androgen biosynthesis (Richards et al., 2018; Young and McNeilly, 2010). During folliculogenesis, endocrine theca interna cells first respond to gonadotropin LH surge from pituitary to produce androgen, and Cyp17a1, which is expressed in endocrine theca interna cells, is the rate-limiting enzyme responsible for androgen production. Further, in response to FSH, soluble androgen then diffuses and transfers across the basement transmembrane to GCs where they can be further aromatized to estrogen. Androgen functions in GCs by binding to androgen receptor (Nr3c4), a nuclear receptor belonging to the steroid receptor family (Meinsohn et al., 2019). So, theca cells are the androgen producers while granulosa are the site of aromatase activity; this well-known steroidogenesis pathway in the ovary was coined as the “two-cell, two-gonadotropin theory” (Coticchio et al., 2015; Erickson et al., 1985; Richards et al., 2018). These previous studies showed an intimate interaction between theca cells and GCs which corporately governs folliculogenesis. After the formation of antral follicles, GCs can be further divided into two populations–mural granulosa cells (MGCs) and antral granulosa cells (AGCs), the latter of which will be excreted with the oocyte after follicle rupture. During the whole process, the oocyte produces TGF-β family proteins, such as Gdf9 and Bmp15, to regulate both granulosa cell functions and theca cell development (Li and Albertini, 2013; Monsivais et al., 2017). Therefore, the synchronous development of oocytes and surrounding somatic cells is important for both oogenesis and folliculogenesis.
Obesity is the most common health issue worldwide, and the adverse effects of overweight and obesity on female fertility have attracted broad attention (Hart, 2016; Poston et al., 2016). Previous studies have revealed that obese women, especially polycystic ovary syndrome (PCOS) patients, are at a higher risk of ovulatory dysfunction (Helm et al., 2009; Jain et al., 2007; Santoro et al., 2004), reduced fecundity (Gesink Law et al., 2007; Ramlau-Hansen et al., 2007; van der Steeg et al., 2008; Wise et al., 2010), and poorer outcomes after in vitro fertilization (Marquard et al., 2011; Sermondade et al., 2019; Shah et al., 2011). The impacts of obesity on female reproduction are complicated and multifactorial and include, for example, disrupted meiotic spindle formation, dysfunction of mitochondrial dynamics, cellular impairment caused by excess free fatty acids, altered levels of leptin, and compromised embryo development (Broughton and Moley, 2017). However, despite the serious clinical impacts of obesity, molecular insights into its influences on folliculogenesis await elucidation.
Several classic mouse models have been established for exploring the consequences of obesity in vivo, for instance, the genetically leptin-deficient mice and the leptin-receptor loss-of-function mice (Drel et al., 2006; Kobayashi et al., 2000; Loos and Yeo, 2021). These two strains of mice both manifest increased body weights soon after birth compared with wild-type and regular diet (RD)-fed mice (Latham et al., 2009; Li et al., 2020). In addition, endometrial epithelial cytolipid vacuoles increasingly accumulate in these two mutant mice (Garris, 2004). High-fat diet (HFD)-fed mice are genetically normal but show leptin resistance, and HFD-fed mice also serve as a model of obesity commonly used in research (Chen et al., 2016; Morinaga et al., 2021; Ringel et al., 2020). Occasional studies have tried to reveal the correlation between obesity and impaired function in the ovary by using mouse models (Hohos et al., 2018; Skaznik-Wikiel et al., 2016). However, the molecular alterations in the follicles, which are the basic functional units of the ovary, under obese conditions are still unclear. With the advances in single-cell RNA-seq technology, we can begin to examine the changes in molecular networks and cell-cell interactions within ovarian follicles at single-cell resolution and at the multicellular level in the context of obesity.
In this study, we performed single-cell RNA sequencing of mouse ovarian follicles from RD and HFD mice to uncover obesity-induced effects on murine folliculogenesis. Two genetic obese models, Lep KO and Lepr KO mice, were also included in attempts to identify consistent alterations and mechanisms under obese conditions. We found that obesity would affect genes expression in ETCs, associating with enhanced androgen biosynthesis. Furthermore, hyper androgen contributed to the imbalanced change of GCs subtypes in obese mice. We further identified typical biomarkers and explored the role of androgen receptor antagonist on ETCs-GCs communications in HFD mice. Besides uncovering a regulation of obesity on folliculogenesis, the present investigation could also suggest a promising potential therapeutic approach for alleviating obesity-induced dysfunction in female reproduction.
Results
Single-cell transcriptome profiling of mouse ovarian follicle cells from regular diet and obese mice
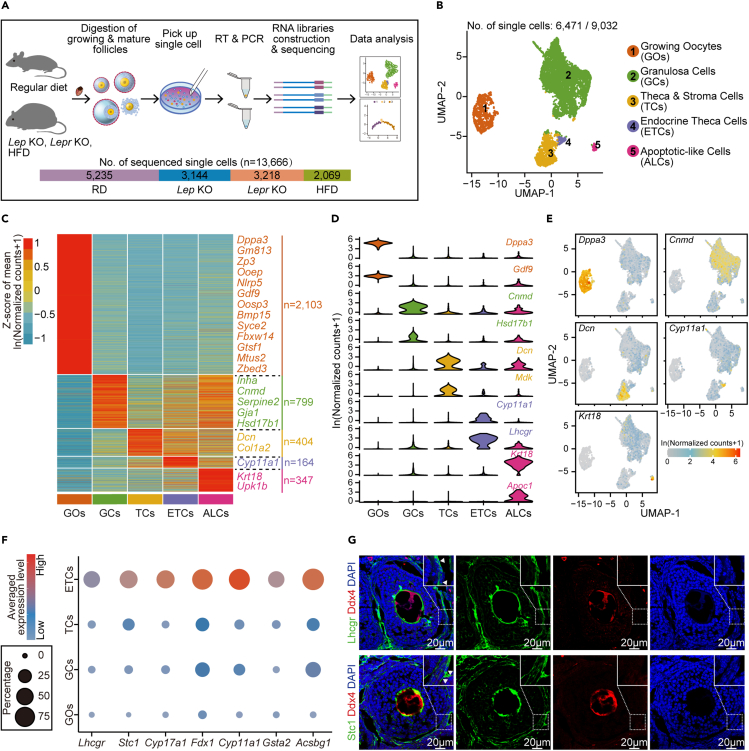
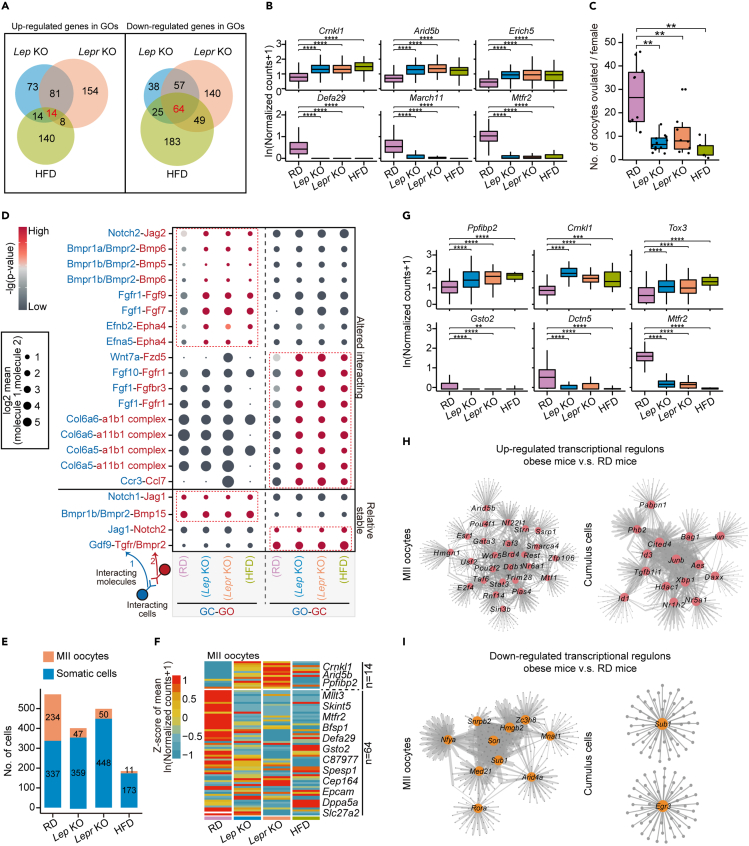
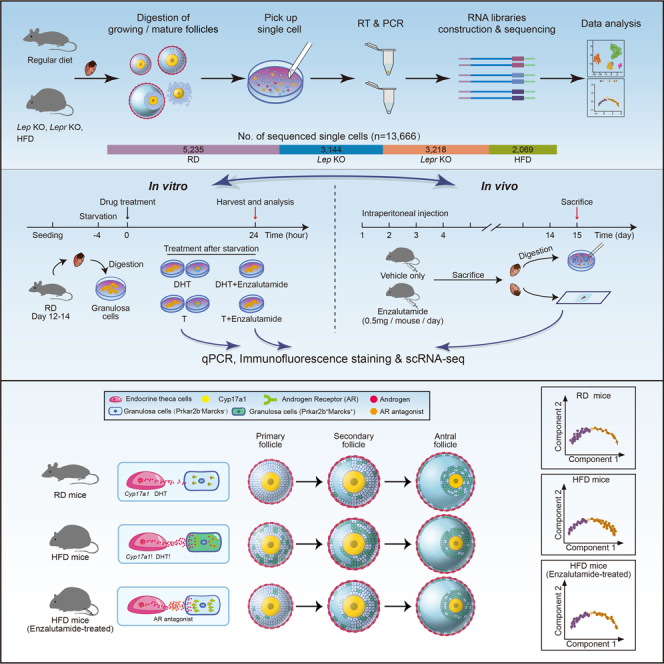
To characterize cell populations and compare the gene expression within ovarian follicles under normal and obesity conditions, we performed single-cell RNA sequencing of mouse follicles from RD and three obese mouse models. We first used two widely corroborated genetic models, leptin knockout (Lep KO) and leptin-receptor knockout (Lepr KO) mice (Figure S1A). Due to leptin deficiency in Lep KO mice or leptin receptor loss-of-function in Lepr KO mice, these two mouse lines developed severe hyperphagia and spontaneous obesity, accompanying the resulting metabolic disorders including insulin-resistance and impaired glucose homeostasis (Chen et al., 2021; Jia et al., 2020; Pan et al., 2021; Tao et al., 2020; Zheng et al., 2021a, 2021b). We also used high-fat diet feeding to obtain HFD mice (Figure S1B). The body weight of each HFD mouse and RD mouse was measured, and the average weight of HFD mice was 1.9-fold higher than that of RD mice (Figure S1C). We collected both growing and mature follicles from RD and obese mice for single-cell RNA sequencing (Figure 1A). The ovaries from 3- to 4-week-old female mice were enriched with secondary follicles, while secondary and antral follicles were enriched in ovaries from 7- to 8-week-old females. Because HFD females were generated by continuous treatment under a high-fat diet for over 16 weeks (Figure S1B), 23- to 24-week-old RD mice were used as controls for HFD female mice. Moreover, we collected MII oocytes and surrounding cumulus cells from both RD and obese mice (Figure 1A) after the injection of pregnant mare serum gonadotropin (PMSG) and human chorionic gonadotropin (hCG). In total, 13,666 single cells, including oocytes and somatic cells, were obtained and analyzed in this study (Figure 1A).
Figure 1.
Cell populations in the ovarian follicles of wild-type and obese mice
(A) Schematic representation of the mouse ovarian follicle single-cell RNA-seq workflow and the downstream analysis (top). The number of sequenced single cells for RD (regular diet), Lep KO, Lepr KO and HFD (high-fat diet) mice are shown as stacked bars (bottom).
(B) UMAP (Uniform Manifold Approximation and Projection) embedding visualization for 6,471 single cells from mouse ovarian follicles showing five major cell populations, including growing oocytes (GOs), granulosa cells (GCs), theca/stroma cells (TCs), endocrine theca cells (ETCs) and apoptotic-like cells (ALCs).
(C) Heatmap depicting DEGs among all identified cell populations derived from natural logarithm-scaled normalized counts based on the single-cell RNA-seq data. Gene expression levels were averaged and scaled.
(D) Violin plots showing expression levels of representative genes that were explicitly expressed in the indicated cell population.
(E) UMAP embedding visualization of cells in (B). Cells are color coded by expression levels of Dppa3, Cnmd, Dcn, Cyp11a1 or Krt18, respectively.
(F) Dot plot depicting the average expression levels of Lhcgr, Stc1, Cyp17a1, Fdx1, Cyp11a1, Gsta2, Acsbg1 in ETCs, TCs, GCs, and GOs, respectively. Averaged expression levels were derived from natural logarithm-scaled normalized counts based on the single-cell RNA-seq data. The percentage of cells expressing each gene is indicated by the size of the dot, and only cells with an expression level of greater than 1 for the indicated gene were counted.
(G) Immunofluorescence staining for Lhcgr or Stc1 in ovarian tissues from RD mice. Tissues were counterstained with Ddx4 and DAPI.
See also Table S1.
We first analyzed 9,032 single cells from growing follicles, including RD and obese mice (Figures 1B and S1D). After stringent quality control (QC) filtering, a total of 6,471 single cells with an average of 118,474 detected unique molecular identifiers (UMIs) and an average of 3,977 detected genes in each cell were retained for the subsequent analysis. By using the Seurat algorithm as previously published (Hafemeister and Satija, 2019; Stuart et al., 2019), these single cells were assigned to four major cell clusters based on the enriched expression of well-known markers (Figure 1B), including growing oocytes (Dpp3a+ and Gdf9+ cluster, GOs), granulosa cells (Cnmd+ and Hsd17b1+ cluster, GCs), theca/stroma cells (Dcn+ and Mdk+ cluster, TCs), and endocrine theca cells (Lhcgr+ and Cyp11a1+ cluster, ETCs) (Fan et al., 2019; Ku et al., 2016; Wagner et al., 2020). Of interest, there was a rare population of apoptotic-like cells (Krt18+ and Apoc1+ cluster, ALCs) (Gaytan et al., 2018; Ku et al., 2016), which were most enriched in Lep KO mice (Figures S1G and S2A). To exclude potential batch effects from sequencing, single cells from different sequencing batches and sample sources were analyzed in each cell cluster, and no obvious sequencing batch effect was observed (Figures S1E and S1F). We next calculated the number of detected genes and UMIs in each cell cluster (Figures S1H and S1I). Because we spiked single-cell lysates with External RNA Controls Consortium (ERCC) mix before reverse transcription (Grun et al., 2014; Risso et al., 2014), the transcript abundance for each cell could also be inferred (Figure S1J). The predicted expression of Lep and Lepr in the RD, Lep KO, Lepr KO and HFD mice was also confirmed at single-cell resolution (Figures S1K and S1L). As expected, there was no Lep detected in follicular cells and deficient Lepr expression in growing oocytes in Lepr KO mice (Figures S1K and S1L). These results together indicated that high-quality single-cell transcriptome data from mouse follicular cells were obtained in this study.
Major cell populations of mouse ovarian follicular cells from RD, Lep KO, Lepr KO and HFD mice
Because genes that are specifically expressed in somatic cells within mouse ovarian follicles have been less studied than those in oocytes, we next compared the differentially expressed genes (DEGs) among each cell cluster (Figures 1C–1E and Table S1). ETCs had much lower cell numbers than GCs and TCs, and little is known about their gene expression profile. Previous studies have identified that the major function of ETCs is the synthesis of androgen, which is then transported into GCs to synthesize estrogen (Richards et al., 2018; Young and McNeilly, 2010). Several marker genes for ETCs have been documented, such as Cyp11a1, Cyp17a1, and Lhcgr (Richards et al., 2018; Young and McNeilly, 2010); these were also confirmed in this study (Figures 1D–1F and Table S1). Furthermore, we uncovered other ETC markers that were not previously validated, such as Fdx1, Stc1, and Acsbg1 (Figure 1F and Table S1). The results of immunofluorescence staining against Lhcgr and Stc1 confirmed our data as well (Figure 1G).
Gene ontology (GO) enrichment analysis was then performed based on the DEGs in each cell cluster (Figure S2B). For the growing oocytes, the DEGs were involved in chromosome segregation and meiotic cell cycle processes, as expected (Figure S2B). GCs and ETCs are known to function in steroid biosynthesis, and their DEGs are associated with the generation of precursor metabolites and energy, hormone metabolic processes, and steroid biosynthetic processes (Figure S2B). For theca/stroma cells, DEGs were related to the positive regulation of cell adhesion and extracellular structure organization (Figure S2B), which reflected the supportive function of those cells.
Comparison of gene expression in granulosa cells between RD and different models of obesity revealed common dysregulated genes in obesity
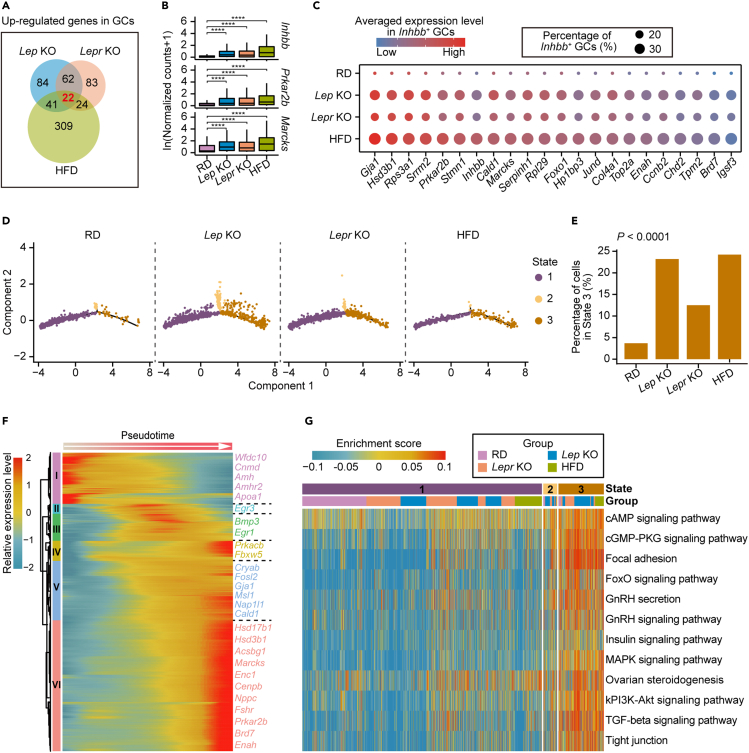
GCs are known to be pivotal in ovarian folliculogenesis, and several studies have reported that obesity may be associated with the alteration of gene expression in GCs (Hamm et al., 2004; Jin et al., 2020; Robker et al., 2011; Xu et al., 2019). However, transcriptome status of GCs from different obese mouse models at single-cell resolution has not yet been studied. We thus analyzed dysregulated genes in GCs between RD and three obese mouse models (Figure S2A). Twenty-two genes were commonly upregulated in Lep KO, Lepr KO, and HFD mice (Figures 2A, 2B, and S2C; Table S2), while 26 genes were commonly downregulated (Figures S3A and S3B; Table S2). Surprisingly, some of these commonly upregulated genes in obese GCs were genes known to be highly expressed in the preovulatory follicle stage, such as Inhbb, Stmn1, and Hsd3b1 (Figures 2B and S2C) (Dou et al., 2016; Elvin et al., 1999; Garcia-Rudaz et al., 2011). Interestingly, we also uncovered genes upregulated in obese GCs, such as Marcks, Prkar2b, and Cald1, that had not previously been documented in GC maturation (Figures 2B and S2C). Because Inhbb, Stmn1, and Hsd3b1 have been reported to be involved in metabolic homeostasis and steroidogenesis in ovarian follicles and are highly expressed in large antral follicles (Dou et al., 2016; Elvin et al., 1999; Garcia-Rudaz et al., 2011), the upregulation of these genes in obese GC population implied a dysregulation in GC differentiation and steroidogenesis.
Figure 2.
Alternations in subgroup composition of ovarian GCs from the obese mice
(A) Venn diagram showing the up-regulated genes in GCs overlapping between Lep KO, Lepr KO, and HFD mice compared with RD mice.
(B) Boxplots showing expression levels of Inhbb, Prkar2b and Marcks in GCs from RD, Lep KO, Lepr KO and HFD mice, respectively. The asterisks above indicate statistical significance. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 (Wilcoxon rank-sum test).
(C) Dot plot depicting the average expression levels of 22 up-regulated genes identified in Figure 2A in Inhbb+ GCs.
(D) Pseudotime trajectory of granulosa cells in Figure 1B. Trajectory was inferred by Monocle 2. Cells are color coded by State inferred in the pseudotime analysis.
(E) Bar plot showing the percentage of GCs in State 3 from RD and the obese mice, respectively. The Chi-squared test was performed.
(F) Heatmap depicting genes with pseudotemporal expression patterns in GCs from RD and the obese mice, which were identified using Monocle2. Gene expression levels were scaled using Monocle2.
(G) Heatmap depicting representative pathways enriched in GCs from different states based on the results of GSVA (gene set variation analysis). Enrichment scores are scaled and color coded. Group and State for GCs are as indicated.
To determine whether the gene expression differences we observed were due to changes of sub-type heterogeneity within the population of GCs, we performed a separate clustering for all the GCs. We classified all GCs (Figure 1B) into four subpopulations, including Ldhb high-expression GCs (Ldhb+ GCs), Htra1+ GCs, Cdkn1b+ GCs and Inhbb+ GCs (Figures S2D–S2G; Table S3). The Ldhb+ GCs highly expressed Ldhb, suggesting they were mainly from preantral follicles as previously reported (Anastacio et al., 2017). Genes involved in steroidogenesis were enriched in both Htra1+ GCs and Inhbb+ GCs, while Inhba, Inhbb, and Fst were with higher expression levels in the Inhbb+ GCs. Consistent with previous reports, Inhbb+ GCs were more mature and were mainly detected in large antral follicles (Wigglesworth et al., 2015), and the Htra1+ GCs could be from small antral follicles (Fenwick et al., 2011). As previously reported, we observed higher expression levels of Gsk3b, Cdkn1b, and Sdc4 in Cdkn1b+ GCs (Figure S2F), inferring that Cdkn1b+ GCs could be from the atretic follicles (Ishiguro et al., 1999; Rajareddy et al., 2007; Wang et al., 2010). The composition of subpopulations we identified in granulosa cells from different mouse models was further examined. The Cdkn1b+ GCs accounted for the lowest proportions among all subpopulations and were most enriched in Lep KO mice (Figure S2E), which was consistent with the previous study (Serke et al., 2012). Interestingly, Inhbb+ GCs highly expressed Marcks, Prkar2b, and Cald1 (Figures S2F and S2G; Table S3) and showed higher proportions in obese mice (Figure S2E), which would contribute to those commonly up-regulated genes expression in all GCs from obese mice. The proportion of Inhbb+ GCs was significantly increased in obese mice; however, representative genes expression levels were similar between RD and obese mice (Figure 2C). These results suggested that elevated proportion of Inhbb+ GCs in obese mice led gene expression changes as represented in Figure 2A.
The Inhbb+GCs exhibited more advanced differentiation status and were enriched in obese mice
To further validate whether those marked genes identified in Inhbb+ GCs were highly expressed in large antral follicles from RD mice, we first analyzed the single-cell transcriptome data of GCs from the preovulatory follicles of RD mice (Figures S3C and S3D). We found that 20 out of 22 (91%) commonly upregulated genes in obese GCs were indeed highly expressed in mature GCs from RD mice (Figure S3D). We next constructed pseudotime trajectories to investigate and compare the differentiation status of GCs in RD and obese mice (Figures 2D, S3E, and S3F). The results of the pseudotime trajectory indicated that cells in State 3 were more advanced in their differentiation status based on the estimated pseudotime (Figures S3E–S3G). Interestingly, the proportion of GCs in State 3 was also significantly increased in obese mice (Figures 2D and 2E). Besides, Inhbb, Marcks, Prkar2b, and Cald1 were highly expressed in GCs from State 3 (Figures 2F and S3H). Interestingly, GCs in State 2 were highly expressed with Cdkn1b and were most enriched in Lep KO mice (Figures 2D and S3H), indicating that GCs from atretic follicles would be hampered in proliferation and differentiation. These results suggested that the Inhbb+ GCs were more advanced in differentiation status among all the GCs.
As controls, GCs from around 10-day-old RD mice whose ovaries were mainly constituted by small secondary follicles and GCs from preovulatory follicles of RD mice were also included (Figures S3G and S3I). GCs from preovulatory follicles of RD mice were mostly assigned to State 3, while most of these GCs from around 10-day-old RD mice were assigned to State 1 (Figures S3G and S3I). In addition, genes with similar pseudotemporal expression patterns were identified (Figures 2F, S3H, and Table S4), such as Inhbb, Prkar2b, and Marcks, which provided potential biomarkers for examining obese-induced alternations in gene expressions in GCs. We further investigated metabolic signaling pathways and processes involved in the differentiation and functions of GCs. Consistently, the activity of these pathways and processes were increasingly enhanced along the pseudotime and were highest in State 3, implying differences in the subtype of GCs among these states (Figure 2G). Besides, we examined correlations in GCs from State 1 and State 3, which showed high correlations among GCs within the same state (Figure S3J). These results indicated that although the proportion of Inhbb+ GCs increased in obese mice, their gene expressions were still similar to those from RD mice.
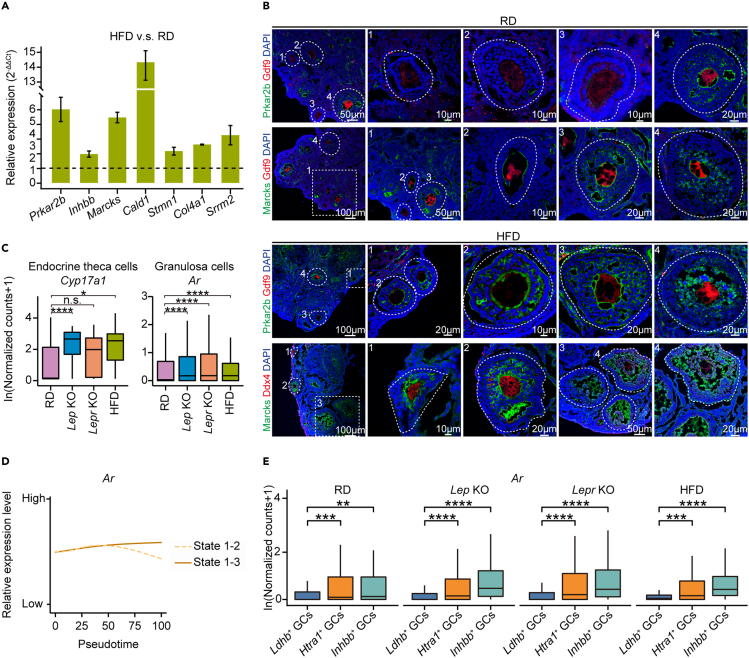
We then validated several genes as biomarkers for obese GCs by using qPCR (Figure 3A), such as Prkar2b, Marcks, and Cald1. Consistent with our single-cell RNA-seq results, these genes were more highly expressed in obese GCs (Figure 3A). Furthermore, we performed immunofluorescence staining for Prkar2b and Marcks to examine their protein signal in situ. Both Prkar2b and Marcks could be readily detected in mature GCs of large antral follicles from RD and obese mice (Figures 3B, S4A, and S4B). These two proteins could only be detected in the GCs of small secondary follicles from obese mice but not RD mice (Figure 3B). Thus, GCs in small growing follicles from obese mice were more advanced in differentiation status, as indicated by the elevated expression levels of Prkar2b, Marcks and Cald1.
Figure 3.
Prkar2b and Marcks were two representative markers for indicating the imbalance of GCs composition in obese mice
(A) qPCR assay of GCs isolated from RD or HFD mouse growing follicles for representative up-regulated genes identified in Figure 2A. Expression levels were normalized to Rps24. Two technical duplicates were performed in individual qPCR experiments, and the representative result of three independent biological replicates is shown. Error bars indicate the mean ± SEM.
(B) Immunofluorescence staining for Prkar2b and Marcks in ovarian tissues from RD and HFD mice. Tissues were counterstained with DAPI and Gdf9 or Ddx4.
(C) Boxplots showing expression levels of Cyp17a1 for ETCs, and Ar for GCs, in RD and the obese mice. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(D) Trend lines showing relative expression levels for Ar in two branches, State 1–2 and State 1–3, identified by using Monocle2.
(E) Boxplots showing expression levels of Ar for Ldhb+ GCs, Htra1+ GCs, and Inhbb+ GCs, in RD and the obese mice. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
Excessive androgen stimulation in endocrine theca cells may contribute to the dysregulation of granulosa cell differentiation in obese mice
GCs and ETCs, which are responsible for ovarian steroidogenesis, play important roles in follicle growth by communicating with each other and coordinating with oocytes (Coticchio et al., 2015). Because the advanced differentiation of GCs in obese mice would give rise to enhanced ovarian steroidogenesis activity (Figure 2G), we next examined expression levels of genes involved in ovarian steroidogenesis in ETCs (Figures S4C and S4D). We found that 17α-hydroxylase-17,20-lyase (Cyp17a1), which converts both pregnenolone and progesterone to androstenedione (Figure S4D), was upregulated in ETCs from obese mice (Figures 3C, S4C, and S4D), implying dysfunction of ovarian steroidogenesis. Interestingly, androgen receptor (Ar), which is located in GCs and delivers androgen to synthesize estrogen (Figure S4D), was also consistently upregulated in GCs from obese mice (Figure 3C). Moreover, we found Ar had a pseudotemporal expression pattern in GCs and was most highly expressed in Inhbb+ GCs (Figures 3D and 3E). These results implied that the enhanced androgen biosynthesis in ETCs may promote the differentiation of GCs in obese mice, which resulted in changes of sub-type heterogeneity within the GC population.
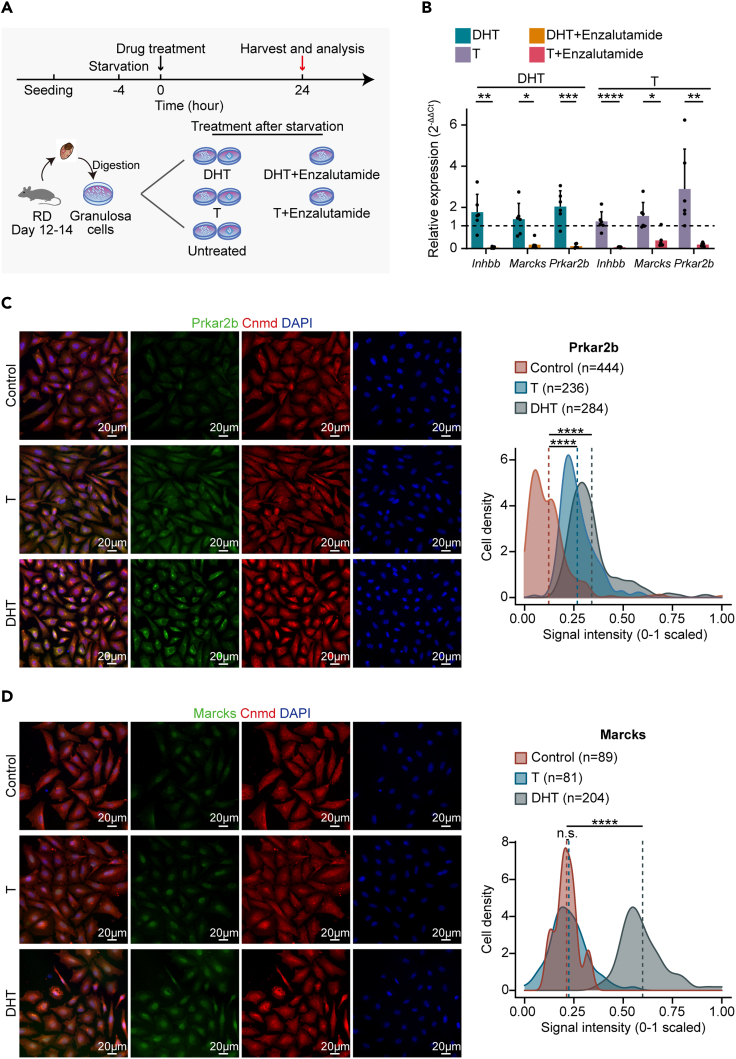
Because testosterone (T, an aromatizable androgen) and dihydrotestosterone (DHT, a nonaromatizable androgen) are more potent than other circulating androgens and DHT has greater affinity than other androgens for the androgen receptor (Burger, 2002; Gao et al., 2005), we isolated immature GCs in small secondary follicles from around 10-day-old RD mice and treated them with T or DHT (Figure 4A). Meanwhile, enzalutamide, an antagonist of the androgen receptor, was used in combination with T or DHT as a control (Figure 4A). The treated and untreated GCs were collected, and expression levels of Inhbb, Prkar2b, and Marcks were quantified by using qPCR (Figure 4B). The expression levels of these three genes were significantly increased in the T- and DHT-treated groups compared with the untreated groups (Figure 4B). More importantly, the activation of these genes by T or DHT was markedly compromised after the addition of enzalutamide (Figure 4B). We further performed immunofluorescence staining against Prkar2b or Marcks for validation at the protein level (Figures 4C and 4D). The signals of both Prkar2b and Marcks were much more readily detectable after the treatment with DHT in GCs (Figures 4C and 4D). These results suggested that excessive androgen production in ETCs may contribute to the dysregulated differentiation of GCs in obese mice.
Figure 4.
Androgens induced expression of Prkar2b and Marcks in vitro in granulosa cells isolated from RD mice
(A) Schematic representation of the androgen stimulation experiments on GCs isolated from 12- to 14-day-old RD mice. DHT, dihydrotestosterone; T, testosterone.
(B) qPCR assay of GCs under different treatments for Inhbb, Marcks, and Prkar2b. Expression levels were normalized to Rps24. Two technical duplicates were performed in individual qPCR experiments. Error bars indicate the mean ± SEM. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(C and D) Immunofluorescence staining for Prkar2b (C) and Marcks (D) in cultured GCs with T or DHT treatment. Tissues were counterstained with DAPI. Scale bars are as indicated. Density plot shows immunofluorescence signal intensity in cells for Prkar2b (C) and Marcks (D) under different treatments. The signal intensity was measured with Zeiss ZEN. The mean signal intensity of specific antibody staining is indicated as a dashed line. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001 and n.s. denotes not significant. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
Altered cell-cell interactions between increased proportion of Inhbb+GCs and growing oocytes in obese mice
Dysfunction in GCs usually affects oogenesis; thus, we investigated the alterations in gene expression in growing oocytes under obesity. Among the calculated DEGs, there were 14 commonly up- and 64 downregulated genes in obese mice compared with RD ones (Figures 5A and 5B and Table S5). Several downregulated genes have been implicated in mitochondrial oxidative metabolism, such as Arid5b and Mtfr2 (Figures 5A and 5B). Although obesity decreased the number of ovulated oocytes, mature MII oocytes could still be obtained (Figure 5C).
Figure 5.
Interactions between oocytes and granulosa cells in RD and obese mice
(A) Venn diagram showing the up-regulated (left) and down-regulated (right) genes in growing oocytes overlapping between Lep KO, Lepr KO, and HFD mice compared with RD mice.
(B) Boxplots showing expression levels of representative up-regulated genes (Crnkl1, Arid5b, Erich5) and down-regulated genes (Defa29, March11, Mtfr2) in growing oocytes in the obese mice compared with RD mice. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(C) Boxplot showing the number of ovulated oocytes in Lep KO (n = 12), Lepr KO (n = 10), and HFD (n = 5) mice is compromised compared with that of RD mice (n = 8). ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(D) Dot plot depicting representative ligand-receptor interactions between GOs and GCs. Interaction strength was measured using log2-scaled means of the average expression level of the ligand in the indicated cell type and receptor in the other cell type. The p values (permutation test) are color coded.
(E) Stacked bar plot showing the number of sequenced single cells in RD, Lep KO, Lepr KO, and HFD mice, grouped by MII oocytes and somatic cells.
(F) Heatmap showing the gene expression patterns in MII oocytes for 78 DEGs identified in Figure 5A.
(G) Boxplots showing expression levels of representative up-regulated genes (Ppfibp2, Crnkl1, Tox3) and down-regulated genes (Gsto2, Dctn5, Mtfr2) in MII oocytes in the obese mice compared with RD mice. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(H and I) Regulatory network of up- (H) and down-regulated (I) regulons in MII oocytes and cumulus cells in obese mice compared with RD mice. Regulons overlapping with DEGs identified in single-cell RNA-seq data (obese mice versus RD mice) are highlighted. The line size indicates the weight of the target regulon.
See also Table S5.
We next explored whether the altered transcriptional network in obese mice was partially contributed by abnormal communication between oocytes and their surrounding GCs. Since we identified three major subpopulations in GCs including the increased Inhbb+ GCs in obese mice (Figure S2E), cell-cell interactions were measured between growing oocytes and Inhbb+ GCs (Figure 5D). Several classic interactions were steady across RD and obese mice, such as Notch1-Jag1, Bmpr1b/Bmpr2-Bmp15, Jag1-Notch2 and Gdf9-Tgfr/Bmpr2 (Figure 5D) (Chang et al., 2016; Juengel and McNatty, 2005; Vanorny and Mayo, 2017). However, several interacting pairs were altered between the GOs and GCs, such as Fgfr1-Fgf9 and Wnt7a-Fzd5 (Figure 5D) (Assou et al., 2006; Boyer et al., 2010), suggesting that the abnormal cell-cell interactions in obese mice may compromise oogenesis and alter gene expression in oocytes.
We next performed single-cell RNA sequencing to examine whether the altered genes expression would persist to the MII oocyte stage (Figures 5E, S5A–S5F, and Table S6). Oocyte-cumulus complexes in RD and obese mice were harvested and sequenced (Figure 5E). The previously identified DEGs between obese and RD mice in growing oocytes were further investigated in MII oocytes, and similar expression patterns were observed indicating that the altered gene expression observed in growing oocytes could persist until the MII oocyte stage (Figures 5F and 5G). To uncover obesity-induced alterations in TF regulons, transcriptional regulatory analysis was performed using SCENIC (Figures S5G and S5H). These results were combined with the single-cell RNA expression data to further construct differential transcriptional regulatory networks for the MII oocytes and cumulus cells (Figures 5H and 5I). For MII oocytes, factors that are known to be involved in the cellular response to stress and DNA repair, such as Hmgb2, were downregulated in obese mice (Figure 5I). However, the specific functions of these downregulated regulons in oocytes remain to be explored.
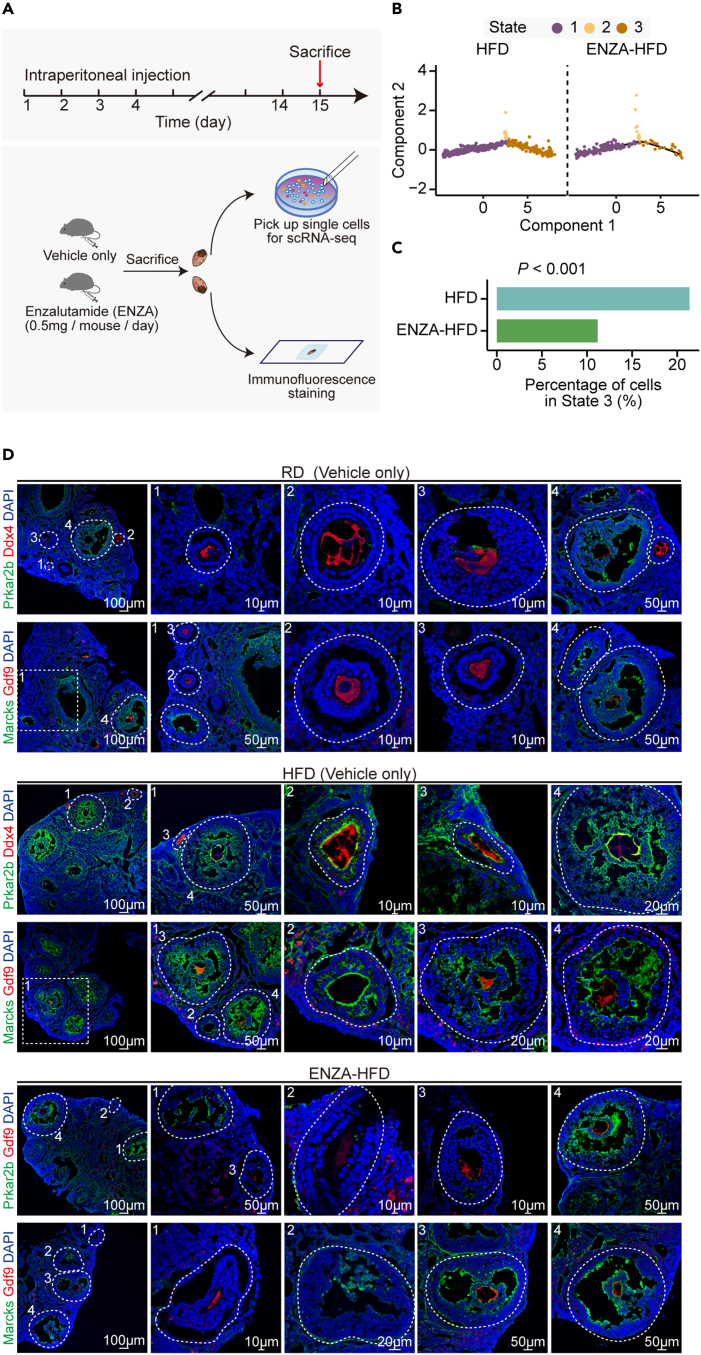
Blocking excessive androgen with an androgen receptor inhibitor alleviated the phenotype of granulosa cells in obese mice
Because enhanced activity in androgen synthesis from obese ETCs may affect the differentiation of the surrounding GCs, as indicated by the in vitro experiment (Figure 4), we asked whether blocking the androgen-estrogen conversion in follicles would delay the marker gene expression in obese GCs. We injected enzalutamide (ENZA) into HFD mice for 14 successive days, followed by single-cell RNA-seq for granulosa cells and immunofluorescence staining of ovarian tissues (Figure 6A). We isolated single cells and randomly picked them for sequencing. After quality control, we combined all granulosa cells and performed pseudotime analysis to compare the differentiation status in GCs from HFD and ENZA-HFD mice (Figures 6B and 6C). Surprisingly, the number of GCs in State 3 was decreased in ENZA-HFD mice (Figures 6B and 6C), suggesting that there were fewer mature GCs after the administration of enzalutamide. We also examined genes with temporal expression patterns along the pseudotime among RD, HFD and ENZA-HFD mice (Figures S6A and S6B) and observed relatively consistent expression patterns in these genes, such as Inhbb, Prkar2b and Marcks.
Figure 6.
Androgen receptor antagonist treatment alleviated the imbalances of the sub-type composition in GCs from obese mice
(A) Schematic representation of the androgen receptor antagonist treatment in vivo experiments on HFD mice.
(B) Pseudotime trajectory of granulosa cells from HFD and ENZA-HFD mice. Trajectory was inferred by Monocle 2. Cells are color coded by State inferred in the pseudotime analysis.
(C) Bar plot showing percentage of cells in State 3 in HFD and ENZA-HFD mice, respectively. The Chi-squared test was performed.
(D) Immunofluorescence staining for Prkar2b and Marcks in ovarian tissues from RD (Vehicle only), HFD (Vehicle only) and ENZA-HFD mice. Tissues were counterstained with DAPI, and Ddx4 or Gdf9.
Moreover, we validated expression levels of Marcks and Prkar2b in ovarian tissues from HFD mice by immunofluorescence staining. As controls, RD mice were also included (Figures 6D and S6B). Importantly, the enzalutamide-treated HFD mice exhibited a similar expression pattern of these genes in their GCs as compared to RD ones (Figures 6D and S6B). Prkar2b and Marcks were detectable in large antral follicles from both RD and HFD mice (Figures 6D and S6B). However, in small secondary follicles that have only two or three layers of GCs around the oocyte, signals for Prkar2b and Marcks could be detected only in the vehicle-treated HFD mice but not in the RD and ENZA-HFD mice (Figures 6D and S6B), suggesting that blocking the androgen-estrogen axis would alleviate the abnormal differentiation of GCs in HFD mice.
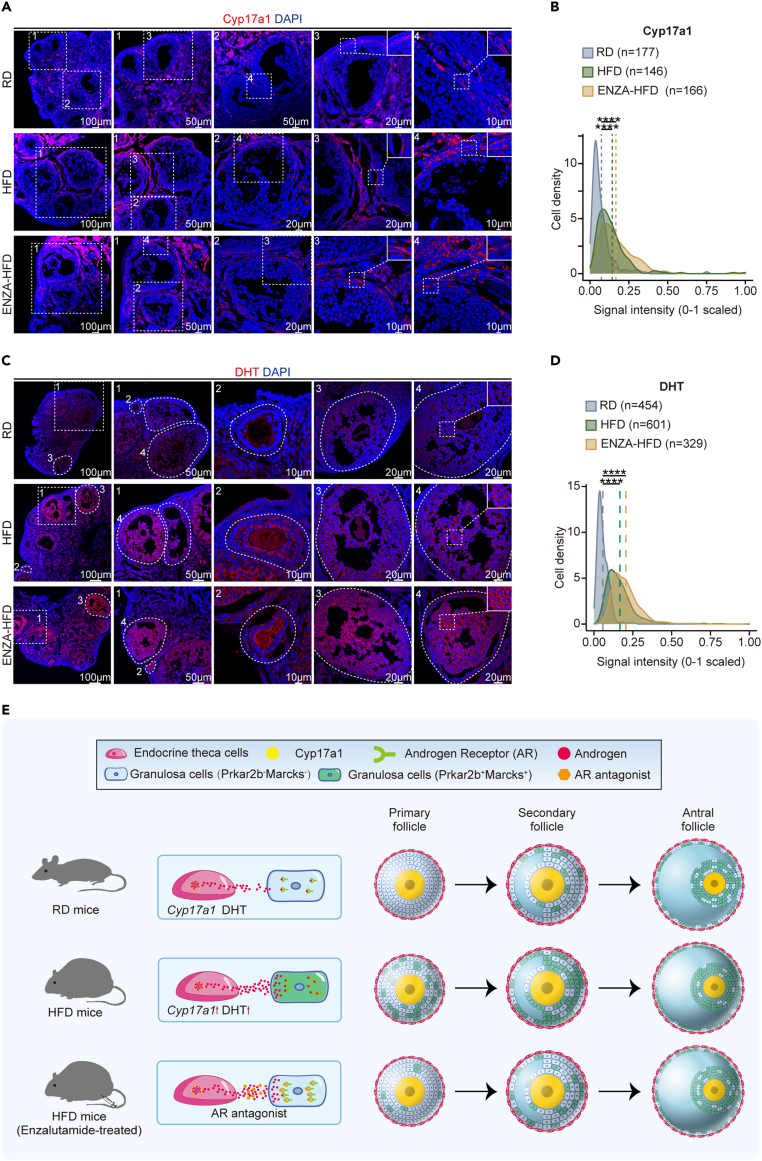
We further explored the effects of enzalutamide's administration in HFD mice. As previously mentioned, Cyp17a1 is a rate-limiting enzyme in androgen biosynthesis. We first examined the expression level of Cyp17a1 in situ in theca cells (Figures 7A, 7B, S7A, and S7B). We observed increased intensity of Cyp17a1 in theca cells from obese mice; however, Cyp17a1 intensity would not decrease in ENZA-HFD mice (Figures 7A, 7B, S7A, and S7B). We further examined distribution of androgen DHT in situ by using immunofluorescence staining (Figures 7C, 7D, S7C, and S7D). Specifically, synthesized in theca interna cells, the locally produced androgen diffuses and transfers across the basement transmembrane to granulosa cells, thus operating in a paracrine manner to orchestrate ovary growth and function. As expected, different from the scene that enzyme Cyp17a1 can be detected specifically within theca interna, DHT hormones are present in several ovarian cell types throughout the follicle (Figure 7C). The signal intensity of DHT was significantly increased in obese mice; however, the intensity of DHT was not decreased after the administration of enzalutamide in HFD mice (Figures 7C, 7D, S7C, and S7D). These results suggested that the enzalutamide functioned in blocking interactions between androgen and its receptor, but not in decreasing androgen levels in ovary microenvironment to alleviate the defects of granulosa cells in HFD mice (Figure 7E).
Figure 7.
Examination of Cyp17a1 and DHT levels in ovarian tissues from RD and HFD mice
(A–D) Immunofluorescence staining for Cyp17a1 (A) and DHT (C) in ovarian tissues from RD, HFD and ENZA-HFD mice. Tissues were counterstained with DAPI. Density plot showing immunofluorescence signal intensity in cells for Cyp17a1 (B) and DHT (D). The signal intensity was measured with Zeiss ZEN. The mean signal intensity of specific antibody staining is indicated as a dashed line. ∗p ≤ 0.05, ∗∗p ≤ 0.01, ∗∗∗p ≤ 0.001, ∗∗∗∗p ≤ 0.0001, n.s. denotes not significant (Wilcoxon rank-sum test).
(E) Model of this study. The increase in Cyp17a1 level may induce excess androgen in growing follicles in HFD mice, resulted in alterations in sub-type composition identified in GCs. However, the defects could be alleviated by using androgen receptor antagonists.
To summarize, these results suggested a feedback response to obesity involving interactions between theca cells and granulosa cells, in which excessive secretion of androgen by theca cells consequently contribute to the defects in differentiation of granulosa cells, represented as the increased subpopulation of Inhbb+ GCs which highly expressed typical markers such as Marcks and Prkar2b (Figure 7E). More importantly, both in vitro and in vivo experiments showed that the androgen receptor antagonist (enzalutamide) could alleviate the phenotype induced by obesity, thus providing a promising therapeutic approach for recovering obesity-induced dysfunction in folliculogenesis (Figure 7E).
Discussion
Here, we have presented the detailed single-cell transcriptome analysis of folliculogenesis in RD and three obese mouse models (Figure 1). We obtained high-quality single-cell transcriptomic data and recovered four major cell types in growing follicles: Growing oocytes (GOs), granulosa cells (GCs), theca/stroma cells (TCs), and endocrine theca cells (ETCs) (Figure 1). Possibly due to the limited cell numbers, little is known about the gene expression network of ETCs in ovarian follicles. In this study, we not only identified additional markers for ETCs but also revealed their potential roles in folliculogenesis under obesity. Furthermore, we systematically analyzed the altered network and regulons in oocytes under obesity and found that the dysregulated gene expression accumulated in GOs would persist to the mature MII stage. Therefore, obesity would impact female fertility in two ways: folliculogenesis, which led to compromised oogenesis in obese mice, and postfertilization development, which requires normal oocyte cytosol (Gosden and Lee, 2010). Notably, the functions of the genes identified as downregulated in oocytes from obese mice require further study.
Although several studies have observed altered gene expression in oocytes from obese mice, the molecular changes in other cell types and their interactions remains to be elucidated (Han et al., 2017, 2018; Hou et al., 2016; Marei et al., 2020). In this study, we uncovered the differences in ovarian follicle cells between RD and obese mouse models. We defined DEGs for each cell cluster among different mouse models of obesity and found that the genes upregulated in GCs common to all three models were affected by androgens produced from ETCs. We also investigated obesity-induced alterations in TF regulons (Figures 5 and S5). Several TFs involved in the cellular response to stress and DNA repair were dysregulated in oocytes from obese mice. In addition, we found that several classic genes involved in cell-cell interactions were consistent across RD and obese mice, such as Bmpr1b/Bmpr2-Bmp15 and Gdf9-Tgfr/Bmpr2 (Chang et al., 2016; Myers et al., 2009; Richani and Gilchrist, 2018; Vanorny and Mayo, 2017). These results suggested that obesity has profound impacts on both somatic cells and oocytes and influences oogenesis through cell–cell interactions in ovarian follicles.
In this study, we examined the potential heterogeneity among GCs based on the subtype clustering or the states inferred by the pseudotime analysis (Figures 2 and S2). Interestingly, the Ldhb+ GCs were in the early differentiation status, while Cdkn1b+ GCs were hampered in differentiation. One important issue is to identify biomarkers for the evaluation of female fertility in obesity. Markers from granulosa cells are most promising because GCs comprise the majority of cells in ovarian follicles and are easier to obtain than oocytes. Nevertheless, possibly due to the heterogeneity in gene expression among GCs, we still lack candidate molecular markers from GCs. We constructed pseudotime trajectories for GCs in RD and obese mice by using Monocle and revealed that the Inhbb+ GCs were enriched in obese mice and exhibited more advanced differentiation status. We identified and confirmed typical markers in GCs for sensitive evaluation of female fertility in obesity, for example, Marcks and Prkar2b. These genes were undetectable in the early-stage follicles from the RD mice but were highly expressed at both the RNA and protein levels in small secondary follicles from the obese mice. More importantly, these genes were sensitive to androgen stimulation and thus could be used as indicators to reflect abnormalities in folliculogenesis at a very early stage.
Previous studies have reported that obese females, especially PCOS patients, show higher androgen levels than healthy ones (Azziz et al., 2016; Escobar-Morreale et al., 2017; Pasquali, 2006). These patients who lose weight could benefit from improved ovulation (Crosignani et al., 2003; Legro et al., 2013; Norman et al., 2007). In addition, similar dysregulation of metabolic phenotype was also observed in a mouse model of PCOS (van Houten et al., 2012). However, the mechanisms and consequences for this high androgen phenotype remain to be explored at the molecular level. Interestingly, we found that Cyp17a1, among all the known androgen synthesis genes, exhibited elevated expression in ETCs from obese mice. Previous studies have confirmed that Cyp17a1 is a key factor in ovarian androgen production and that its overexpression leads to excess androgen synthesis (Richards et al., 2018). We also observed an elevation of signal intensity of DHT in ovarian tissues from obese mice (Figures 7 and S7). Combined with the in vivo and in vitro treatment with exogenous androgen and/or its receptor antagonist, we further revealed that excess androgen could promote the differentiation of GCs. Notably, treatment with an androgen antagonist alleviated the GC phenotype in HFD mice, even though those mice were still fed a high-fat diet. In addition, with single-cell RNA-seq of granulosa cells from HFD and ENZA-HFD mice, we observed that granulosa cells from ENZA-HFD mice were decreased in number of Inhbb+ granulosa cells (Figures 6 and S6). These results could provide a mechanistic insight into the elevated androgen levels observed in obese females and serve as a potential approach for the treatment of obesity-induced fertility problem in females. However, the dosage and interval of androgen antagonist treatment still await optimization. Because Marcks and Prkar2b in GCs could be used as sensitive markers for androgen stimulation, one could use these markers to perform an in-depth evaluation of androgen antagonist treatment in the future.
In summary, we performed single-cell RNA-seq analysis of folliculogenesis in RD and obese mice, providing a regulatory signaling by which obesity impacts murine folliculogenesis. Our high-quality single-cell transcriptome data of ovarian follicle cells and subsequent analysis may be utilized to study folliculogenesis under physiological and pathological conditions in the future.
Limitations of the study
In the present study, detailed single-cell RNA-seq of ovarian follicles in mice suggests an intimate crosstalk between ETCs and GCs, hierarchically mediating impacts of obesity on fertility. Pharmacological blockade of androgen receptor in both in vivo and in vitro settings implies that excessive androgen is one major cause of the obesity-induced defects during folliculogenesis. We have revealed the transcriptional changes in mature oocytes of obese mice, and future work is needed to explore how the disproportioned GCs would contribute to embryonic development via a transcription cascade. Further exploration could be clinically relevant for fertility in women, whether androgen receptor antagonist could be used as an intervention to treat obesity-associated reproductive disorders.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit anti-LECT1 (Cnmd) | Thermo Fisher Scientific | Cat# PA5-76974; RRID: AB_2720701 |
| Rabbit anti-Cyp17a1 | Abcam | Cat# ab134910 |
| Rabbit anti-DHT | Mybiosource | Cat# MBS2042442 |
| Rabbit anti-Ddx4/Mvh | Abcam | Cat# ab13840; RRID: AB_443012 |
| Rabbit anti-Gdf9 | Abcam | Cat# ab254323 |
| Mouse anti-Stc1 | Santa Cruz Biotechnology | Cat# sc-293435 |
| Mouse anti-Lhcgr | Novus | Cat# NBP2-52504; RRID: AB_2783817 |
| Mouse anti-Marcks | Santa Cruz Biotechnology | Cat# sc-100777; RRID: AB_1125958 |
| Mouse anti-Prkar2b | Santa Cruz Biotechnology | Cat# sc-376778 |
| Goat anti-Mouse IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fisher Scientific | Cat# A-11029; RRID: AB_2534088 |
| Goat anti-Rabbit IgG (H+L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 555 | Thermo Fisher Scientific | Cat# A-21429; RRID: AB_2535850 |
| Biological samples | ||
| Mouse ovarian tissue | This study | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| M2 medium | Sigma-Aldrich | Cat# M7167 |
| Penicillin-Streptomycin | GIBCO | Cat# 15140122 |
| Collagenase, Type IV | GIBCO | Cat# 17104019 |
| TrypLE Express Enzyme (1×) | GIBCO | Cat# 12604039 |
| PMSG, Pregnant Mare Serum Gonadotropin | Solarbio | Cat# P9970 |
| hCG, Human Chorionic Gonadotrophin | SANMA | N/A |
| α-MEM | GIBCO | Cat# 12561056 |
| FBS, Fetal Bovine Serum | VisTech | Cat# SE200-ES |
| Insulin-Transferrin-Selenium (ITS -G) (100×) | GIBCO | Cat# 41400045 |
| Nucleosides (100×) | Sigma-Aldrich | Cat# ES-008-D |
| Paraformaldehyde | Sigma-Aldrich | Cat# P6148 |
| Sucrose | Sigma-Aldrich | Cat# V900116 |
| Recombinant RNase Inhibitor | TaKaRa | Cat# 2313B |
| KAPA HiFi HotStart ReadyMix | Roche | Cat# 7958935001 |
| Ampure XP beads | Beckman Coulter | Cat# A63882 |
| dNTP mix | Thermo Fisher Scientific | Cat# R0192 |
| Dynabeads M-280 Streptavidin | Thermo Fisher Scientific | Cat# 11206D |
| Tissue freezing medium (OCT medium) | Leica | Cat# 14020108926 |
| Goat serum | GIBCO | Cat# 16210064 |
| PrimeScript RT Master Mix (Perfect Real Time) | TaKaRa | Cat# RR036A |
| 2× SYBR Green PCR Master Mix | Bimake | Cat# B21202 |
| Dihydrotestosterone, DHT | Solarbio | Cat# ID0310 |
| Enzalutamide (MDV3100) | Selleckchem | Cat# S1250 |
| Testosterone | Solarbio | Cat# IT0110 |
| PEG-400 | KESHI | Cat# 25322-68-22 |
| Tween-80 | Solarbio | Cat# T8360 |
| Triton X-100 | Sigma-Aldrich | Cat# T9284 |
| ProLong Gold Antifade Mountant with DAPI | Thermo Fisher Scientific | Cat# P36935 |
| Critical commercial assays | ||
| Quick-RNA Microprep Kit | Zymo Research | Cat# R1051 |
| DNA Clean & Concentrator-5 (Capped) | Zymo Research | Cat# D4014 |
| NEBNext Ultra II DNA Library Prep Kit for Illumina | New England Biolabs | Cat# E764 |
| Deposited data | ||
| Raw data files and processed data files for ovarian single-cell RNA-seq | This study | GEO: GSE142587 |
| Experimental models: Organisms/strains | ||
| Mouse: C57BL/6J | Vital River | N/A |
| Mouse: BKS-Leprem2Cd479/Gpt | GemPharmatech | Cat# T002407 |
| Mouse: B6/JGpt-Lepem1Cd25/Gpt | GemPharmatech | Cat# T001461 |
| Oligonucleotides | ||
| qPCR primers, see also Table S7 | This paper | N/A |
| Software and algorithms | ||
| ImageJ (version: 1.5.3.c) | NIH | https://imagej.nih.gov/ij/ |
| ZEN | Zeiss | http://www.zeiss.com/microscopy/us/products/microscope-software/zen.html/ |
| Cutadapt (version: 2.3) | Read the Docs | https://cutadapt.readthedocs.io/en/stable/ |
| STAR (version: 2.6.0a) | Github | https://github.com/alexdobin/STAR |
| HTSeq (version: 0.11.2) | Read the Docs | https://htseq.readthedocs.io/en/master/history.html#version-0-11-2 |
| Scater (version: 1.14.6) | Bioconductor | http://bioconductor.org/packages/release/bioc/html/scater.html |
| Seurat (version: 3.2.0) | Satija Lab | https://satijalab.org/seurat/ |
| SCTransform (version: 0.2.1) | Satija Lab | https://satijalab.org/seurat/ |
| clusterProfiler (version: 3.14.3) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html |
| GSVA (version: 1.34.0) | Bioconductor | https://bioconductor.org/packages/release/bioc/html/GSVA.html |
| Monocle 2 (version: 2.14.0) | Bioconductor | http://cole-trapnell-lab.github.io/monocle-release |
| CellPhoneDB (version: 2.14.0) | Github | https://github.com/Teichlab/cellphonedb |
| pySCENIC (version: 0.10.3) | Van de Sande et al. (2020) | https://pyscenic.readthedocs.io/en/latest |
| Cytoscape (version: 3.8.0) | https://cytoscape.org | https://cytoscape.org |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by Lead Contact, Fan Guo (guofan@ioz.ac.cn).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Mice
The animal procedures were performed according to the ethical guidelines of the Laboratory Animal Center of Institute of Zoology, CAS. RD (regular diet), Lep KO, Lepr KO and HFD (high-fat diet) female mice were used in this study. The RD female mice (C57BL/6J) were fed with a regular diet, while the HFD female mice (C57BL/6J) were generated by feeding 6- to 8-week-old mice with high-fat diet (Research Diets, Cat# D12492) and persisted for at least 16 weeks. HFD female mice with weight above 40g were used for the downstream experiments. The Lep KO and Lepr KO female mice were purchased from GemPharmatech (Cat# T002407 and Cat# T001461).
To perform single-cell RNA-seq for ovarian follicles, 3- to 4-week-old and 7- to 8-week-old female were sacrificed to collect secondary follicles and antral follicles in RD, Lep KO, and Lepr KO groups; and because HFD mice used were 23- to 24-week-old, 23- to 24-week-old RD mice were also included as age-matched control to HFD. To perform single-cell RNA-seq for pre-ovulatory granulosa cells and MII oocytes, 8- to 24-week-old adult female were sacrificed in RD, Lep KO, Lepr KO, and HFD groups. To isolate granulosa cells for in vitro culture, 10- to 14-day-old female were sacrificed.
Method details
Isolation of single cells from mouse follicles
Isolation of single cells from mouse follicles was performed as previously described (Gu et al., 2019). Female mice between 10 days and 24 weeks old were sacrificed. To obtain pre-ovulatory follicles, female RD mice were sacrificed 48 hours after PMSG (Pregnant mare serum gonadotropin) injection. Ovaries were dissected and transferred into M2 medium supplemented with 1× penicillin-streptomycin. Then, the ovaries were treated with 1 mg/mL collagenase IV at 37°C for 20 minutes to release follicles. Follicles were manually picked and collected with needles under a stereomicroscope. Then, the follicles were treated with TrypLE Express enzyme at 37°C for 20 minutes. The dissociation solution was neutralized by adding M2 medium. Individual oocytes or somatic cells were picked up under a stereomicroscope.
To obtain ovulatory follicles, female mice were injected with PMSG and hCG (human chorionic gonadotropin). MII oocytes and cumulus cell complexes were collected at 12 hours post hCG injection and treated with hyaluronidase to release single MII oocytes and cumulus cells. The zona pellucida of MII oocytes was removed with Tyrode's solution before collection.
Construction of single-cell RNA-seq library
Single-cell RNA-seq libraries were constructed as previously described (Gu et al., 2019; Li et al., 2017; Yan et al., 2021). Briefly, single cells were manually picked up under a stereomicroscope and transferred into cell lysis buffer. cDNA was synthesized, amplified, and fragmented. RNA-seq libraries were prepared by using the NEBNext Ultra II DNA Library Prep Kit for Illumina as indicated in the instruction manual. The final RNA-seq libraries were pooled together, quality checked and sequenced as 150-bp paired-end reads on an Illumina HiSeq X-Ten instrument (Novogene Co.).
In vitro culture of mouse granulosa cells with or without androgen stimulation
Female C57BL/6J mice were sacrificed between 10 to 14 days of age, and GCs were harvested as described above. After filtration to remove cell debris, cells were centrifuged and resuspended in 1 mL granulosa cell culture medium (GC medium) containing: α-MEM supplemented with 5% FBS, 1× ITS liquid media supplement, 1× nucleosides, 1× penicillin-streptomycin (Gu et al., 2019) for 3 hours and then starved in serum-free medium for 4 hours. To test responses of GCs to androgen stimulation, isolated GCs were treated with 1 μM testosterone or 1 μM dihydrotestosterone for 24 hours before collection. For antagonizing experiments, 100 μM Enzalutamide was introduced to isolated GCs which were treated with either 1 μM testosterone or dihydrotestosterone for 24 hours followed by further analysis.
In vivo treatment of HFD mice with androgen-receptor antagonist
Female HFD mice were randomly picked and divided into two groups, including the control and ENZA-treated group. The ENZA-treated group was injected intraperitoneally in the right lower abdomen area with 0.5 mg of Enzalutamide formulated in 10% DMSO, 30% PEG 400, 5% Tween-80 and 55% physiological saline, while the control group was injected with vehicle only. Mice were monitored and injected daily for 14 days. On the 15th day, mice were weighed and sacrificed, and the ovaries were obtained. One of the ovaries in each mouse was used to obtain GCs as described above for single-cell RNA-seq, and the other one was fixed for immunofluorescence staining.
Immunofluorescence staining of mouse ovarian tissue and somatic cells
Fresh mouse ovarian tissues were fixed in 4% paraformaldehyde at 4°C overnight. Then, the tissues were dehydrated in 20% sucrose solution and were embedded in OCT medium. The embedded tissues were then sectioned into 8-10 μm sections using a cryostat microtome (Leica Co.). Isolated somatic cells from mouse follicles were seeded to wells of a cell culture plate containing coverslips and cultured overnight as described above. Then, the coverslips were rinsed twice with 1× DPBS and fixed in 4% paraformaldehyde for 20 minutes at room temperature.
These sections or coverslips were then rinsed with 1× DPBS and permeabilized in 0.2% Triton X-100 for 20 minutes at room temperature. After rinsing with 1× DPBS, samples were incubated for 30 minutes at room temperature with blocking buffer containing: 1% goat serum, 3% BSA and 0.05% Tween-20 in 1× DPBS, followed by incubating with primary antibodies in blocking buffer at 4°C overnight. After washing with wash buffer (1× DPBS with 0.05% Tween-20), samples were incubated with secondary antibodies in blocking buffer for 1 hour at room temperature. Finally, samples were rinsed with wash buffer, mounted in ProLong Gold Antifade Mountant with DAPI, and then were examined and imaged under a confocal laser scanning microscope (LSM880, Zeiss). For immunofluorescence staining of Cyp17a1 and DHT, the antigen retrieval step was performed before permeabilization. Immunofluorescence signal intensities were measured with either ImageJ software (v1.5.3c) or ZEN software (Zeiss), and at least three independent samples were used for quantification. The signal intensity in each cell was normalized to the intensity of DAPI. The number of measured cells were indicated in corresponding figures.
Total RNA isolation and quantitative real-time PCR
Total RNA was extracted from cultured GCs by using the Quick-RNA Microprep Kit. Reverse transcription of total RNA to cDNA was performed by using PrimeScript RT master mix. The PCR assays were set up in 10 μL volumes consisting of 1 μL of cDNA, 0.2 μL each of forward and reverse 10 μM primers, 5 μL of 2× SYBR Green PCR Master Mix, 0.2 μL of 5× ROX reference dye 2 and 3.4 μL of nuclease-free water. qPCRs were performed in duplicates using the 7500 Real-Time PCR System (Applied Biosystems).
Quantification and statistical analysis
Single-cell RNA-seq data preprocessing
For the processing of raw sequencing reads, the sequencing adapters, template switch oligo (TSO) and poly(A) tail sequences were trimmed using Cutadapt (v2.3) (Martin, 2011). Clean reads were then aligned to the mouse reference genome (Mus musculus, mm9) using STAR (v2.6.0a) (Dobin et al., 2013). Deduplication was further performed on uniquely mapped reads according to UMIs (unique molecular identifiers). Finally, for each barcode observed, gene expression was quantified using HTSeq (v0.11.2) (Anders et al., 2015). To remove low-quality cells, stringent quality control was performed using Scater (v1.14.6) (McCarthy et al., 2017). For oocytes, cells with more than 10,000 detected UMIs and more than 3,000 detected genes were retained for subsequent analysis. For somatic cells, cells were discarded if they did not reach the following criteria: 1) cells with less than 10,000 detected UMIs; 2) cells with abnormally high percentages of mitochondrial gene or ERCC spike-in sequences (value was higher than the median value plus 3 MADs within a specific batch); and 3) cells with abnormally high numbers of detected UMIs or genes (value was higher than the median value plus 3 MADs within a specific batch).
Identifying cell populations in single-cell RNA-seq data
The Seurat R package (v3.2.0) and SCTransform (v0.2.1) R package were used for the downstream analysis of single-cell RNA-seq data (Hafemeister and Satija, 2019; Stuart et al., 2019). Briefly, the expression matrix was processed using SCTransform with default parameters, followed by dimensionality reduction (PCA, principal component analysis) and UMAP embedding using Seurat. Cell clustering was further performed using the FindNeighbors and FindClusters functions in Seurat. Cell populations were defined based on the DEGs identified between clusters. Cells with ambiguous expression patterns were considered contaminants and were removed from this study. For identifying subpopulations of follicle granulosa cells, these genes with high dispersion across all cells in the RD, Lep KO, Lepr KO and HFD mice were used as the features for dimensionality reduction (PCA) and UMAP embedding by following the standard workflow in Seurat.
Identification of differentially expressed genes and Gene Ontology analysis
The DEGs were identified using the function FindAllMarkers or FindMarkers in Seurat based on the natural logarithmic scaled normalized counts (Stuart et al., 2019). Only genes with a natural logarithm-scaled fold change greater than 0.25 and adjusted p-value less than 0.05 were considered as DEGs. The DEGs with adjusted p-value less than 0.001 were used as the input for Gene Ontology analysis using the clusterProfiler R package (v3.14.3) (Yu et al., 2012).
Gene set variation analysis
Gene set variation analysis (GSVA) were performed on the natural logarithm-scaled normalized counts derived from single-cell RNA-seq data according to the recommended procedures. Gene sets for a specific biological process or signaling pathway were retrieved from the KEGG pathway database. GSVA was performed using the GSVA R package (v1.34.0) (Hanzelmann et al., 2013)
Constructing single-cell trajectory and pseudotime analysis
The Monocle2 R package (v2.14.0) was adopted for the single-cell trajectory and pseudotime analyses (Qiu et al., 2017a, 2017b; Trapnell et al., 2014). The genes with high dispersion (dispersion_empirical>= 3 ∗ dispersion_fit) across all cells in the RD, Lep KO, Lepr KO and HFD mice were identified and selected in Monocle2 to construct the trajectory. Cell state identification and pseudotime inference were then performed on the constructed trajectory. Genes with pseudo-temporal expression patterns were identified by using function DifferentialGeneTest in Monocle2. And genes with qval< 0.001 were visualized using Monocle2.
Calculating Pearson correlation for cells in different states
States and pseudotime for cells were identified and inferred in the pseudotime analysis as described above. The log-transformed normalized gene expression levels were used for calculating. For calculating Pearson correlation of cells in different state, cells in the indicated states were ranked by the value of pseudotime. For State 1, the first 50 cells with lower value of pseudotime were selected, while for State 3, the last 50 cells with higher values were selected.
Calculation of ligand-receptor interactions among mouse ovarian follicle cells
The cell-cell communication mediated by ligand-receptor interactions was inferred from single-cell RNA-seq data using CellPhoneDB (v2.14.0) with the default parameters (Efremova et al., 2020). The normalized counts were used as input for CellPhoneDB.
Transcriptional regulatory network analysis
pySCENIC (v0.10.3) was adopted to investigate the alteration in the transcriptional regulatory network for a specific cell type in RD versus the obese mice (Van de Sande et al., 2020). Briefly, the natural logarithm-scaled normalized counts and list of known TFs for mice were used as inputs for network inference (GRNBoost2), followed by candidate regulon generation (cisTarget). After the generation of candidate regulons, the activity score of the regulons in each cell was quantified using AUCell. The activity scores of regulons generated by cisTarget in RD, Lep KO, Lepr KO and HFD mice were averaged, scaled, and visualized via heat map. The network of the regulon and its targets was visualized using Cytoscape (v3.8.0) (Shannon et al., 2003). Only the connections of a specific regulon and its top 50 targets (ranked by interaction weight) were retained.
Statistical analyses
The experimental data were statistically analyzed using unpaired two-tailed t-tests in R to determine whether the means of two groups were equal to each other. The single-cell RNA-seq data were statistically analyzed using the Wilcoxon rank sum test in R. The Chi-squared test was performed in Figures 2E and 6C. Asterisks indicate significance. ∗P <= 0.05, ∗∗P <= 0.01, ∗∗∗P <= 0.001, ∗∗∗∗P <= 0.0001, n.s. denotes for not significant. The error bars in the figures show the mean ± SEM. For box plots in figures, the 25th and 75th percentiles are represented as the lower and upper boundaries of the box, respectively. The median is represented as the line inside the box. The ends of the whiskers represent the sum of the 75th percentiles and 1.5× IQR and the value of the 25th percentiles minus 1.5× IQR.
Acknowledgments
We thank T.X. for mouse care. S.L. and the imaging platform of CAS supported the fluorescence immunoassay. The National Supercomputer in Guangzhou (TianHe-2) supported the bioinformatic analysis of this study. This study was supported by grants from the Ministry of Science and Technology of China (2018YFC1003401, 2018YFA0107701), the National Natural Science Foundation of China (31771590, 31822035), CAS Project for Young Scientists in Basic Research (YSBR-012), and the Strategic Collaborative Research Program of the Ferring Institute of Reproductive Medicine, Ferring Pharmaceuticals and Chinese Academy of Sciences (FIRMC200510).
Author contributions
Fan G. conceived and supervised this study. H.W. and Fei G. helped in revision and discussion. Q.Y., H.Y., J.Q., R.Y., and X.C. performed the experiments. X.L., Q.Z., and C.G. analyzed the bioinformatic data. L.Z. supported this study. Fan G., X.L., and J.Q. wrote the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: January 21, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2021.103627.
Contributor Information
Lin Zhang, Email: zhanglin@scu.edu.cn.
Fan Guo, Email: guofan@ioz.ac.cn.
Supplemental information
Data and code availability
-
•
All raw sequencing data and processed data were deposited in the NCBI GEO database under the accession number GSE142587. Accession numbers are listed in the Key resources table.
-
•
This study did not generate any unique code or algorithm. The algorithm used for the analysis during this study are all publicly available. All code supporting this study is available from the lead contact upon reasonable request.
-
•
Any additional resources are available from the lead contact upon request.
References
- Anastacio A., Rodriguez-Wallberg K.A., Chardonnet S., Pionneau C., Federici C., Almeida Santos T., Poirot C. Protein profile of mouse ovarian follicles grown in vitro. Mol. Hum. Reprod. 2017;23:827–841. doi: 10.1093/molehr/gax056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assou S., Anahory T., Pantesco V., Le Carrour T., Pellestor F., Klein B., Reyftmann L., Dechaud H., De Vos J., Hamamah S. The human cumulus--oocyte complex gene-expression profile. Hum. Reprod. 2006;21:1705–1719. doi: 10.1093/humrep/del065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azziz R., Carmina E., Chen Z., Dunaif A., Laven J.S., Legro R.S., Lizneva D., Natterson-Horowtiz B., Teede H.J., Yildiz B.O. Polycystic ovary syndrome. Nat. Rev. Dis. Prim. 2016;2:16057. doi: 10.1038/nrdp.2016.57. [DOI] [PubMed] [Google Scholar]
- Boyer A., Goff A.K., Boerboom D. WNT signaling in ovarian follicle biology and tumorigenesis. Trends Endocrinol. Metab. 2010;21:25–32. doi: 10.1016/j.tem.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Broughton D.E., Moley K.H. Obesity and female infertility: potential mediators of obesity's impact. Fertil. Steril. 2017;107:840–847. doi: 10.1016/j.fertnstert.2017.01.017. [DOI] [PubMed] [Google Scholar]
- Burger H.G. Androgen production in women. Fertil. Steril. 2002;77(Suppl. 4):S3–S5. doi: 10.1016/s0015-0282(02)02985-0. [DOI] [PubMed] [Google Scholar]
- Chang H.M., Qiao J., Leung P.C. Oocyte-somatic cell interactions in the human ovary-novel role of bone morphogenetic proteins and growth differentiation factors. Hum. Reprod. Update. 2016;23:1–18. doi: 10.1093/humupd/dmw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J., Feng G.H., Peng H., Zhang X., Zhang Y., et al. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- Chen S., Liu X., Peng C., Tan C., Sun H., Liu H., Zhang Y., Wu P., Cui C., Liu C., et al. The phytochemical hyperforin triggers thermogenesis in adipose tissue via a Dlat-AMPK signaling axis to curb obesity. Cell Metab. 2021;33:565–580.e7. doi: 10.1016/j.cmet.2021.02.007. [DOI] [PubMed] [Google Scholar]
- Coticchio G., Dal Canto M., Mignini Renzini M., Guglielmo M.C., Brambillasca F., Turchi D., Novara P.V., Fadini R. Oocyte maturation: gamete-somatic cells interactions, meiotic resumption, cytoskeletal dynamics and cytoplasmic reorganization. Hum. Reprod. Update. 2015;21:427–454. doi: 10.1093/humupd/dmv011. [DOI] [PubMed] [Google Scholar]
- Crosignani P.G., Colombo M., Vegetti W., Somigliana E., Gessati A., Ragni G. Overweight and obese anovulatory patients with polycystic ovaries: parallel improvements in anthropometric indices, ovarian physiology and fertility rate induced by diet. Hum. Reprod. 2003;18:1928–1932. doi: 10.1093/humrep/deg367. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y.D., Zhao H., Huang T., Zhao S.G., Liu X.M., Yu X.C., Ma Z.X., Zhang Y.C., Liu T., Gao X., et al. STMN1 promotes progesterone production via StAR up-regulation in mouse granulosa cells. Sci. Rep. 2016;6:26691. doi: 10.1038/srep26691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drel V.R., Mashtalir N., Ilnytska O., Shin J., Li F., Lyzogubov V.V., Obrosova I.G. The leptin-deficient (ob/ob) mouse: a new animal model of peripheral neuropathy of type 2 diabetes and obesity. Diabetes. 2006;55:3335–3343. doi: 10.2337/db06-0885. [DOI] [PubMed] [Google Scholar]
- Efremova M., Vento-Tormo M., Teichmann S.A., Vento-Tormo R. CellPhoneDB: inferring cell-cell communication from combined expression of multi-subunit ligand-receptor complexes. Nat. Protoc. 2020;15:1484–1506. doi: 10.1038/s41596-020-0292-x. [DOI] [PubMed] [Google Scholar]
- Elvin J.A., Yan C., Wang P., Nishimori K., Matzuk M.M. Molecular characterization of the follicle defects in the growth differentiation factor 9-deficient ovary. Mol. Endocrinol. 1999;13:1018–1034. doi: 10.1210/mend.13.6.0309. [DOI] [PubMed] [Google Scholar]
- Erickson G.F., Magoffin D.A., Dyer C.A., Hofeditz C. The ovarian androgen producing cells: a review of structure/function relationships. Endocr. Rev. 1985;6:371–399. doi: 10.1210/edrv-6-3-371. [DOI] [PubMed] [Google Scholar]
- Escobar-Morreale H.F., Santacruz E., Luque-Ramirez M., Botella Carretero J.I. Prevalence of 'obesity-associated gonadal dysfunction' in severely obese men and women and its resolution after bariatric surgery: a systematic review and meta-analysis. Hum. Reprod. Update. 2017;23:390–408. doi: 10.1093/humupd/dmx012. [DOI] [PubMed] [Google Scholar]
- Fan X., Bialecka M., Moustakas I., Lam E., Torrens-Juaneda V., Borggreven N.V., Trouw L., Louwe L.A., Pilgram G.S.K., Mei H., et al. Single-cell reconstruction of follicular remodeling in the human adult ovary. Nat. Commun. 2019;10:3164. doi: 10.1038/s41467-019-11036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenwick M.A., Mansour Y.T., Franks S., Hardy K. Identification and regulation of bone morphogenetic protein antagonists associated with preantral follicle development in the ovary. Endocrinology. 2011;152:3515–3526. doi: 10.1210/en.2011-0229. [DOI] [PubMed] [Google Scholar]
- Gao W., Bohl C.E., Dalton J.T. Chemistry and structural biology of androgen receptor. Chem. Rev. 2005;105:3352–3370. doi: 10.1021/cr020456u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rudaz C., Dorfman M., Nagalla S., Svechnikov K., Soder O., Ojeda S.R., Dissen G.A. Excessive ovarian production of nerve growth factor elicits granulosa cell apoptosis by setting in motion a tumor necrosis factor alpha/stathmin-mediated death signaling pathway. Reproduction. 2011;142:319–331. doi: 10.1530/REP-11-0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garris D.R. Ultrastructural analysis of progressive endometrial hypercytolipidemia induced by obese (ob/ob) and diabetes (db/db) genotype mutations: structural basis of female reproductive tract involution. Tissue Cell. 2004;36:19–28. doi: 10.1016/j.tice.2003.08.002. [DOI] [PubMed] [Google Scholar]
- Gaytan F., Morales C., Roa J., Tena-Sempere M. Changes in keratin 8/18 expression in human granulosa cell lineage are associated to cell death/survival events: potential implications for the maintenance of the ovarian reserve. Hum. Reprod. 2018;33:680–689. doi: 10.1093/humrep/dey010. [DOI] [PubMed] [Google Scholar]
- Gesink Law D.C., Maclehose R.F., Longnecker M.P. Obesity and time to pregnancy. Hum. Reprod. 2007;22:414–420. doi: 10.1093/humrep/del400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosden R., Lee B. Portrait of an oocyte: our obscure origin. J. Clin.Invest. 2010;120:973–983. doi: 10.1172/JCI41294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grun D., Kester L., van Oudenaarden A. Validation of noise models for single-cell transcriptomics. Nat. Methods. 2014;11:637–640. doi: 10.1038/nmeth.2930. [DOI] [PubMed] [Google Scholar]
- Gu C., Liu S., Wu Q., Zhang L., Guo F. Integrative single-cell analysis of transcriptome, DNA methylome and chromatin accessibility in mouse oocytes. Cell Res. 2019;29:110–123. doi: 10.1038/s41422-018-0125-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafemeister C., Satija R. Normalization and variance stabilization of single-cell RNA-seq data using regularized negative binomial regression. Genome Biol. 2019;20:296. doi: 10.1186/s13059-019-1874-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamm M.L., Bhat G.K., Thompson W.E., Mann D.R. Folliculogenesis is impaired and granulosa cell apoptosis is increased in leptin-deficient mice. Biol. Reprod. 2004;71:66–72. doi: 10.1095/biolreprod.104.027292. [DOI] [PubMed] [Google Scholar]
- Han L., Wang H., Li L., Li X., Ge J., Reiter R.J., Wang Q. Melatonin protects against maternal obesity-associated oxidative stress and meiotic defects in oocytes via the SIRT3-SOD2-dependent pathway. J. Pineal Res. 2017;63:e12431. doi: 10.1111/jpi.12431. [DOI] [PubMed] [Google Scholar]
- Han L., Ren C., Li L., Li X., Ge J., Wang H., Miao Y.L., Guo X., Moley K.H., Shu W., et al. Embryonic defects induced by maternal obesity in mice derive from Stella insufficiency in oocytes. Nat. Genet. 2018;50:432–442. doi: 10.1038/s41588-018-0055-6. [DOI] [PubMed] [Google Scholar]
- Hanzelmann S., Castelo R., Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinf. 2013;14:7. doi: 10.1186/1471-2105-14-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart R.J. Physiological aspects of female fertility: role of the environment, modern lifestyle, and genetics. Physiol. Rev. 2016;96:873–909. doi: 10.1152/physrev.00023.2015. [DOI] [PubMed] [Google Scholar]
- Helm K., Nass R., Evans W., Nass R., Evans W. Yen & Jaffe’s Reproductive Endocrinology; 2009. Physiologic and Pathophysiologic Alterations of the Neuroendocrine Components of the Reproductive axis; pp. 441–488. [Google Scholar]
- Hohos N.M., Cho K.J., Swindle D.C., Skaznik-Wikiel M.E. High-fat diet exposure, regardless of induction of obesity, is associated with altered expression of genes critical to normal ovulatory function. Mol.Cell Endocrinol. 2018;470:199–207. doi: 10.1016/j.mce.2017.10.016. [DOI] [PubMed] [Google Scholar]
- Honda A., Hirose M., Hara K., Matoba S., Inoue K., Miki H., Hiura H., Kanatsu-Shinohara M., Kanai Y., Kono T., et al. Isolation, characterization, and in vitro and in vivo differentiation of putative thecal stem cells. Proc. Natl. Acad. Sci. U S A. 2007;104:12389–12394. doi: 10.1073/pnas.0703787104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou Y.J., Zhu C.C., Duan X., Liu H.L., Wang Q., Sun S.C. Both diet and gene mutation induced obesity affect oocyte quality in mice. Sci. Rep. 2016;6:18858. doi: 10.1038/srep18858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishiguro K., Kojima T., Taguchi O., Saito H., Muramatsu T., Kadomatsu K. Syndecan-4 expression is associated with follicular atresia in mouse ovary. Histochem.Cell Biol. 1999;112:25–33. doi: 10.1007/s004180050388. [DOI] [PubMed] [Google Scholar]
- Jain A., Polotsky A.J., Rochester D., Berga S.L., Loucks T., Zeitlian G., Gibbs K., Polotsky H.N., Feng S., Isaac B., et al. Pulsatile luteinizing hormone amplitude and progesterone metabolite excretion are reduced in obese women. J. Clin. Endocrinol. Metab. 2007;92:2468–2473. doi: 10.1210/jc.2006-2274. [DOI] [PubMed] [Google Scholar]
- Jia L., Jiang Y., Li X., Chen Z. Purβ promotes hepatic glucose production by increasing Adcy6 transcription. Mol. Metab. 2020;31:85–97. doi: 10.1016/j.molmet.2019.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Ma Y., Tong X., Yang W., Dai Y., Pan Y., Ren P., Liu L., Fan H.Y., Zhang Y., et al. Metformin inhibits testosterone-induced endoplasmic reticulum stress in ovarian granulosa cells via inactivation of p38 MAPK. Hum. Reprod. 2020;35:1145–1158. doi: 10.1093/humrep/deaa077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juengel J.L., McNatty K.P. The role of proteins of the transforming growth factor-beta superfamily in the intraovarian regulation of follicular development. Hum. Reprod. Update. 2005;11:143–160. doi: 10.1093/humupd/dmh061. [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Forte T.M., Taniguchi S., Ishida B.Y., Oka K., Chan L. The db/db mouse, a model for diabetic dyslipidemia: molecular characterization and effects of Western diet feeding. Metabolism. 2000;49:22–31. doi: 10.1016/s0026-0495(00)90588-2. [DOI] [PubMed] [Google Scholar]
- Ku N.O., Strnad P., Bantel H., Omary M.B. Keratins: biomarkers and modulators of apoptotic and necrotic cell death in the liver. Hepatology. 2016;64:966–976. doi: 10.1002/hep.28493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham J.R., Pathirathna S., Jagodic M.M., Choe W.J., Levin M.E., Nelson M.T., Lee W.Y., Krishnan K., Covey D.F., Todorovic S.M., et al. Selective T-type calcium channel blockade alleviates hyperalgesia in ob/ob mice. Diabetes. 2009;58:2656–2665. doi: 10.2337/db08-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legro R.S., Arslanian S.A., Ehrmann D.A., Hoeger K.M., Murad M.H., Pasquali R., Welt C.K., Endocrine S. Diagnosis and treatment of polycystic ovary syndrome: an Endocrine Society clinical practice guideline. J. Clin. Endocrinol. Metab. 2013;98:4565–4592. doi: 10.1210/jc.2013-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Albertini D.F. The road to maturation: somatic cell interaction and self-organization of the mammalian oocyte. Nat. Rev. Mol.Cell Biol. 2013;14:141–152. doi: 10.1038/nrm3531. [DOI] [PubMed] [Google Scholar]
- Li L., Dong J., Yan L., Yong J., Liu X., Hu Y., Fan X., Wu X., Guo H., Wang X., et al. Single-cell RNA-seq analysis maps development of human germline cells and gonadal niche interactions. Cell Stem Cell. 2017;20:858–873 e854. doi: 10.1016/j.stem.2017.03.007. [DOI] [PubMed] [Google Scholar]
- Li S., Peng J., Wang H., Zhang W., Brown J.M., Zhou Y., Wu Q. Hepsin enhances liver metabolism and inhibits adipocyte browning in mice. Proc. Natl. Acad. Sci. U S A. 2020;117:12359–12367. doi: 10.1073/pnas.1918445117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Peng J., Matzuk M.M., Yao H.H. Lineage specification of ovarian theca cells requires multicellular interactions via oocyte and granulosa cells. Nat. Commun. 2015;6:6934. doi: 10.1038/ncomms7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loos R.J.F., Yeo G.S.H. The genetics of obesity: from discovery to biology. Nat. Rev. Genet. 2021:1–14. doi: 10.1038/s41576-021-00414-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marei W.F.A., Smits A., Mohey-Elsaeed O., Pintelon I., Ginneberge D., Bols P.E.J., Moerloose K., Leroy J. Differential effects of high fat diet-induced obesity on oocyte mitochondrial functions in inbred and outbred mice. Sci. Rep. 2020;10:9806. doi: 10.1038/s41598-020-66702-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquard K.L., Stephens S.M., Jungheim E.S., Ratts V.S., Odem R.R., Lanzendorf S., Moley K.H. Polycystic ovary syndrome and maternal obesity affect oocyte size in in vitro fertilization/intracytoplasmic sperm injection cycles. Fertil. Steril. 2011;95:2146-9–2149.e1. doi: 10.1016/j.fertnstert.2010.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- McCarthy D.J., Campbell K.R., Lun A.T., Wills Q.F. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinsohn M.C., Smith O.E., Bertolin K., Murphy B.D. The orphan nuclear receptors steroidogenic factor-1 and liver receptor homolog-1: structure, regulation, and essential roles in mammalian reproduction. Physiol. Rev. 2019;99:1249–1279. doi: 10.1152/physrev.00019.2018. [DOI] [PubMed] [Google Scholar]
- Monsivais D., Matzuk M.M., Pangas S.A. The TGF-beta family in the reproductive tract. Cold Spring Harb. Perspect. Biol. 2017;9:a022251. doi: 10.1101/cshperspect.a022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morinaga H., Mohri Y., Grachtchouk M., Asakawa K., Matsumura H., Oshima M., Takayama N., Kato T., Nishimori Y., Sorimachi Y., et al. Obesity accelerates hair thinning by stem cell-centric converging mechanisms. Nature. 2021;595:266–271. doi: 10.1038/s41586-021-03624-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers M., Middlebrook B.S., Matzuk M.M., Pangas S.A. Loss of inhibin alpha uncouples oocyte-granulosa cell dynamics and disrupts postnatal folliculogenesis. Dev. Biol. 2009;334:458–467. doi: 10.1016/j.ydbio.2009.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman R.J., Dewailly D., Legro R.S., Hickey T.E. Polycystic ovary syndrome. Lancet. 2007;370:685–697. doi: 10.1016/S0140-6736(07)61345-2. [DOI] [PubMed] [Google Scholar]
- Pan Q., Lin S., Li Y., Liu L., Li X., Gao X., Yan J., Gu B., Chen X., Li W., et al. A novel GLP-1 and FGF21 dual agonist has therapeutic potential for diabetes and non-alcoholic steatohepatitis. EBioMedicine. 2021;63:103202. doi: 10.1016/j.ebiom.2020.103202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali R. Obesity and androgens: facts and perspectives. Fertil. Steril. 2006;85:1319–1340. doi: 10.1016/j.fertnstert.2005.10.054. [DOI] [PubMed] [Google Scholar]
- Poston L., Caleyachetty R., Cnattingius S., Corvalán C., Uauy R., Herring S., Gillman M.W. Preconceptional and maternal obesity: epidemiology and health consequences. Lancet Diabetes Endocrinol. 2016;4:1025–1036. doi: 10.1016/S2213-8587(16)30217-0. [DOI] [PubMed] [Google Scholar]
- Qiu X., Hill A., Packer J., Lin D., Ma Y.A., Trapnell C. Single-cell mRNA quantification and differential analysis with Census. Nat. Methods. 2017;14:309–315. doi: 10.1038/nmeth.4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X., Mao Q., Tang Y., Wang L., Chawla R., Pliner H.A., Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajareddy S., Reddy P., Du C., Liu L., Jagarlamudi K., Tang W., Shen Y., Berthet C., Peng S.L., Kaldis P., et al. p27kip1 (cyclin-dependent kinase inhibitor 1B) controls ovarian development by suppressing follicle endowment and activation and promoting follicle atresia in mice. Mol. Endocrinol. 2007;21:2189–2202. doi: 10.1210/me.2007-0172. [DOI] [PubMed] [Google Scholar]
- Ramlau-Hansen C.H., Thulstrup A.M., Nohr E.A., Bonde J.P., Sorensen T.I., Olsen J. Subfecundity in overweight and obese couples. Hum. Reprod. 2007;22:1634–1637. doi: 10.1093/humrep/dem035. [DOI] [PubMed] [Google Scholar]
- Richani D., Gilchrist R.B. The epidermal growth factor network: role in oocyte growth, maturation and developmental competence. Hum. Reprod. Update. 2018;24:1–14. doi: 10.1093/humupd/dmx029. [DOI] [PubMed] [Google Scholar]
- Richards J.S., Ren Y.A., Candelaria N., Adams J.E., Rajkovic A. Ovarian follicular theca cell recruitment, differentiation, and impact on fertility: 2017 update. Endocr. Rev. 2018;39:1–20. doi: 10.1210/er.2017-00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringel A.E., Drijvers J.M., Baker G.J., Catozzi A., Garcia-Canaveras J.C., Gassaway B.M., Miller B.C., Juneja V.R., Nguyen T.H., Joshi S., et al. Obesity shapes metabolism in the tumor microenvironment to suppress anti-tumor immunity. Cell. 2020;183:1848–1866.e26. doi: 10.1016/j.cell.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risso D., Ngai J., Speed T.P., Dudoit S. Normalization of RNA-seq data using factor analysis of control genes or samples. Nat. Biotechnol. 2014;32:896–902. doi: 10.1038/nbt.2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robker R.L., Wu L.L., Yang X. Inflammatory pathways linking obesity and ovarian dysfunction. J. Reprod. Immunol. 2011;88:142–148. doi: 10.1016/j.jri.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Santoro N., Lasley B., McConnell D., Allsworth J., Crawford S., Gold E.B., Finkelstein J.S., Greendale G.A., Kelsey J., Korenman S., et al. Body size and ethnicity are associated with menstrual cycle alterations in women in the early menopausal transition: the Study of Women's Health across the Nation (SWAN) Daily Hormone Study. J. Clin. Endocrinol. Metab. 2004;89:2622–2631. doi: 10.1210/jc.2003-031578. [DOI] [PubMed] [Google Scholar]
- Serke H., Nowicki M., Kosacka J., Schroder T., Kloting N., Bluher M., Kallendrusch S., Spanel-Borowski K. Leptin-deficient (ob/ob) mouse ovaries show fatty degeneration, enhanced apoptosis and decreased expression of steroidogenic acute regulatory enzyme. Int. J. Obes. (Lond) 2012;36:1047–1053. doi: 10.1038/ijo.2011.220. [DOI] [PubMed] [Google Scholar]
- Sermondade N., Huberlant S., Bourhis-Lefebvre V., Arbo E., Gallot V., Colombani M., Freour T. Female obesity is negatively associated with live birth rate following IVF: a systematic review and meta-analysis. Hum. Reprod. Update. 2019;25:439–451. doi: 10.1093/humupd/dmz011. [DOI] [PubMed] [Google Scholar]
- Shah D.K., Missmer S.A., Berry K.F., Racowsky C., Ginsburg E.S. Effect of obesity on oocyte and embryo quality in women undergoing in vitro fertilization. Obstet. Gynecol. 2011;118:63–70. doi: 10.1097/AOG.0b013e31821fd360. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaznik-Wikiel M.E., Swindle D.C., Allshouse A.A., Polotsky A.J., McManaman J.L. High-fat diet causes subfertility and compromised ovarian function independent of obesity in mice. Biol. Reprod. 2016;94:108. doi: 10.1095/biolreprod.115.137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao P., Kuang Y., Li Y., Li W., Gao Z., Liu L., Qiang M., Zha Z., Fan K., Ma P., et al. Selection of a full agonist combinatorial antibody that rescues leptin deficiency in vivo. Adv. Sci. (Weinh) 2020;7:2000818. doi: 10.1002/advs.202000818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Cacchiarelli D., Grimsby J., Pokharel P., Li S., Morse M., Lennon N.J., Livak K.J., Mikkelsen T.S., Rinn J.L. The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 2014;32:381–386. doi: 10.1038/nbt.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van de Sande B., Flerin C., Davie K., De Waegeneer M., Hulselmans G., Aibar S., Seurinck R., Saelens W., Cannoodt R., Rouchon Q., et al. A scalable SCENIC workflow for single-cell gene regulatory network analysis. Nat. Protoc. 2020;15:2247–2276. doi: 10.1038/s41596-020-0336-2. [DOI] [PubMed] [Google Scholar]
- van der Steeg J.W., Steures P., Eijkemans M.J., Habbema J.D., Hompes P.G., Burggraaff J.M., Oosterhuis G.J., Bossuyt P.M., van der Veen F., Mol B.W. Obesity affects spontaneous pregnancy chances in subfertile, ovulatory women. Hum. Reprod. 2008;23:324–328. doi: 10.1093/humrep/dem371. [DOI] [PubMed] [Google Scholar]
- van Houten E.L., Kramer P., McLuskey A., Karels B., Themmen A.P., Visser J.A. Reproductive and metabolic phenotype of a mouse model of PCOS. Endocrinology. 2012;153:2861–2869. doi: 10.1210/en.2011-1754. [DOI] [PubMed] [Google Scholar]
- Vanorny D.A., Mayo K.E. The role of Notch signaling in the mammalian ovary. Reproduction. 2017;153:R187–R204. doi: 10.1530/REP-16-0689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner M., Yoshihara M., Douagi I., Damdimopoulos A., Panula S., Petropoulos S., Lu H., Pettersson K., Palm K., Katayama S., et al. Single-cell analysis of human ovarian cortex identifies distinct cell populations but no oogonial stem cells. Nat. Commun. 2020;11:1147. doi: 10.1038/s41467-020-14936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H.X., Li T.Y., Kidder G.M. WNT2 regulates DNA synthesis in mouse granulosa cells through beta-catenin. Biol. Reprod. 2010;82:865–875. doi: 10.1095/biolreprod.109.080903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigglesworth K., Lee K.B., Emori C., Sugiura K., Eppig J.J. Transcriptomic diversification of developing cumulus and mural granulosa cells in mouse ovarian follicles. Biol. Reprod. 2015;92:23. doi: 10.1095/biolreprod.114.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise L.A., Rothman K.J., Mikkelsen E.M., Sorensen H.T., Riis A., Hatch E.E. An internet-based prospective study of body size and time-to-pregnancy. Hum. Reprod. 2010;25:253–264. doi: 10.1093/humrep/dep360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Huang B.Y., Zhan J.H., Liu M.T., Fu Y., Su Y.Q., Sun Q.Y., Wang W.H., Chen D.J., Liu J.Q. Insulin reduces reaction of follicular granulosa cells to FSH stimulation in women with obesity-related infertility during IVF. J. Clin. Endocrinol. Metab. 2019;104:2547–2560. doi: 10.1210/jc.2018-00686. [DOI] [PubMed] [Google Scholar]
- Yan R., Gu C., You D., Huang Z., Qian J., Yang Q., Cheng X., Zhang L., Wang H., Wang P., et al. Decoding dynamic epigenetic landscapes in human oocytes using single-cell multi-omics sequencing. Cell Stem Cell. 2021;28:1641–1656 e1647. doi: 10.1016/j.stem.2021.04.012. [DOI] [PubMed] [Google Scholar]
- Young J.M., McNeilly A.S. Theca: the forgotten cell of the ovarian follicle. Reproduction. 2010;140:489–504. doi: 10.1530/REP-10-0094. [DOI] [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Ding J., Liao M., Qiu Z., Yuan Q., Mai W., Dai Y., Zhang H., Wu H., Wang Y., et al. Immunotherapy against angiotensin II receptor ameliorated insulin resistance in a leptin receptor-dependent manner. FASEB J.: Off. Publ. Fed. Am. Soc. Exp. Biol. 2021;35:e21157. doi: 10.1096/fj.202000300R. [DOI] [PubMed] [Google Scholar]
- Zheng X., Chen T., Jiang R., Zhao A., Wu Q., Kuang J., Sun D., Ren Z., Li M., Zhao M., et al. Hyocholic acid species improve glucose homeostasis through a distinct TGR5 and FXR signaling mechanism. Cell Metab. 2021;33:791–803.e7. doi: 10.1016/j.cmet.2020.11.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
All raw sequencing data and processed data were deposited in the NCBI GEO database under the accession number GSE142587. Accession numbers are listed in the Key resources table.
-
•
This study did not generate any unique code or algorithm. The algorithm used for the analysis during this study are all publicly available. All code supporting this study is available from the lead contact upon reasonable request.
-
•
Any additional resources are available from the lead contact upon request.