Abstract
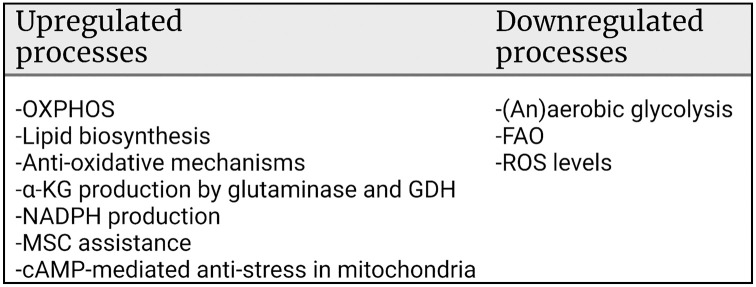
Energy production by means of ATP synthesis in cancer cells has been investigated frequently as a potential therapeutic target in this century. Both (an)aerobic glycolysis and oxidative phosphorylation (OXPHOS) have been studied. Here, we review recent literature on energy production in glioblastoma stem cells (GSCs) and leukemic stem cells (LSCs) versus their normal counterparts, neural stem cells (NSCs) and hematopoietic stem cells (HSCs), respectively. These two cancer stem cell types were compared because their niches in glioblastoma tumors and in bone marrow are similar. In this study, it became apparent that (1) ATP is produced in NSCs and HSCs by anaerobic glycolysis, whereas fatty acid oxidation (FAO) is essential for their stem cell fate and (2) ATP is produced in GSCs and LSCs by OXPHOS despite the hypoxic conditions in their niches with FAO and amino acids providing its substrate. These metabolic processes appeared to be under tight control of cellular regulation mechanisms which are discussed in depth. However, our conclusion is that systemic therapeutic targeting of ATP production via glycolysis or OXPHOS is not an attractive option because of its unwanted side effects in cancer patients.
Keywords: angiogenesis, bone marrow, brain tumors, cancer stem cells, hematopoietic stem cells, leukemia, leukemic stem cells, metabolism, neural stem cells, niches, stem cells, stemness, tumor heterogeneity, tumor immune infiltrate, tumor microenvironment
Introduction
The bulk of malignant cells in cancer patients are rapidly proliferating differentiated cancer cells, and a small fraction of the malignant cells are undifferentiated cancer stem cells (CSCs). CSCs reside in specific microenvironments or niches where they are maintained in a slowly dividing or quiescent state. Quiescence of CSCs protects them from the cytotoxic effects of chemotherapy and radiotherapy, as these therapeutic strategies preferentially target proliferating cells. CSC protection in their niches causes recurrence of cancer demonstrating the importance to develop therapeutic strategies focused on CSCs.1–8
In this review, we discuss targeting of the energy metabolism of CSCs in their niches as therapeutic strategy. We focus on the synthesis of ATP in glioblastoma stem cells (GSCs) in primary brain tumor patients and in leukemic stem cells (LSCs) in the bone marrow of leukemia patients. ATP synthesis in these stem cell types is the focus of our comparison because we have previously shown that GSC niches in glioblastoma tumors and LSC niches in bone marrow are functionally and morphologically similar.9,10 Therefore, energy metabolism may well be regulated in a similar way.
(Cancer) Stem Cell Niches
There has been confusion about types of hematopoietic stem cell (HSC)/LSC niches and GSC niches. In the past, three types of HSC/LSC niches were reported, endosteal, reticular, and perivascular niches, whereas five types of GSC niches, perivascular, extracellular matrix (ECM), periarteriolar, perihypoxic, and peri-immune niches, have been described. We have performed thorough morphological and histological/histochemical analyses of HSC/LSC niches in human bone marrow and GSC niches in human glioblastoma tumors. On the basis of these studies, we came to the conclusion that both in bone marrow and in glioblastoma tumors, only one type of niche exists, the hypoxic periarteriolar HSC/LSC and GSC niche and that the subtypes that have been described in the past are in fact characteristics of that hypoxic periarteriolar niche.11–13 We have analyzed quantitatively in sections of 16 glioblastoma tumors how many arterioles, capillaries, or venules were associated with markers of GSCs and their niches. We found GSC niches around seven arterioles out of 335 investigated or 2% of arterioles were surrounded by a niche, whereas 924 venules and 8085 capillaries were not associated with GSC niche characteristics. 12
In the subventricular zone (SVZ), neural stem cell (NSC) niches are hypoxic but not periarteriolar (see below). Hypoxia induces and maintains the stemness of CSCs and CSCs are well-protected because of their low proliferation rate and their well-developed DNA repair mechanisms and drug efflux pumps. Receptor-chemoattractant interactions are responsible for stem cell maintenance and retention of stem cells in the niches, such as interactions between C-X-C receptor type 4 (CXCR4) and chemokine stromal derived factor-1α (SDF-1α, also named CXCL12), between receptor CD44 and chemokine osteopontin (OPN),9,10,12–15 and between angiopoietin 1 (Ang1) and its tyrosine kinase receptor Tie2.11,16–18 The stemness of CSCs in the present review is considered to be characterized by the expression of stem cell markers.
GSCs are present in GSC niches in glioblastoma tumors and in the SVZ in brains of glioblastoma patients (Fig. 1). 19 The SVZ is a major niche of normal NSCs. The SVZ is morphologically different from GSC and HSC/LSC niches, as the SVZ is not associated with arterioles, but there is also a close association with blood vessels. 18 The SVZ is hypoxic and the CXCR4-SDF1α and CD44-OPN interactions are also associated with homing of GSCs in the SVZ. Thus, GSCs can escape from the effects of cytotoxic therapy in the SVZ as they are maintained in slowly dividing state and are not removed during surgical resection of the tumor. The origin of GSCs is not exactly known. One hypothesis is that GSCs originate from NSCs in the SVZ and have migrated to form a glioblastoma tumor in the brain at distance from the SVZ.20,21 Another hypothesis is that GSCs arise in the glioblastoma tumor, from which they migrate into the SVZ. 22
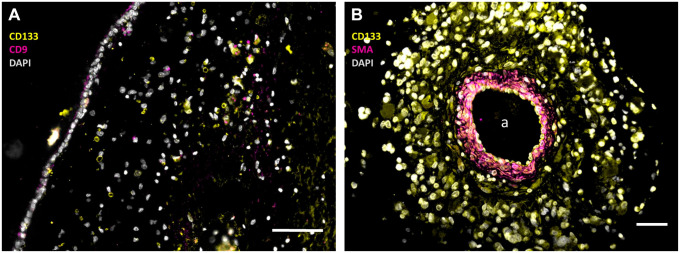
Figure 1.
Immunofluorescence images of paraffin sections of a human SVZ (A) and a human glioblastoma tumor (B) showing NSCs and GSCs in the SVZ (A) and GSCs in a periarteriolar niche in the tumor (B). In the SVZ, NSCs are stained in yellow without pink staining (CD133+and CD9−) and GSCs are stained in yellow and pink (CD133+ and CD9+), whereas cell nuclei are stained in white. In the periarteriolar niche in the glioblastoma tumor, GSCs are stained in yellow with white nuclei. The images were constructed on the basis of monochrome images in green, red, and blue after immunofluorescence staining which are shown in Supplemental Figure 1. The pseudocolor images have been constructed to facilitate clear discrimination of NSCs and GSCs by the color-blind readership according to Jambor et al. 23 a, lumen of arteriole. Bars, 100 µm. Images have been prepared on the basis of images in Hira et al.9,19 Abbreviations: SMA, smooth muscle actin; SVZ, subventricular zone; NSCs, neural stem cells; GSCs, glioblastoma stem cells; DAPI, 4′,6-diamidino-2-phenylindole.
In leukemia patients, LSCs are present in HSC niches (Fig. 2) where they are protected in a similar way as GSCs in their niches in glioblastoma tumors and the SVZ.
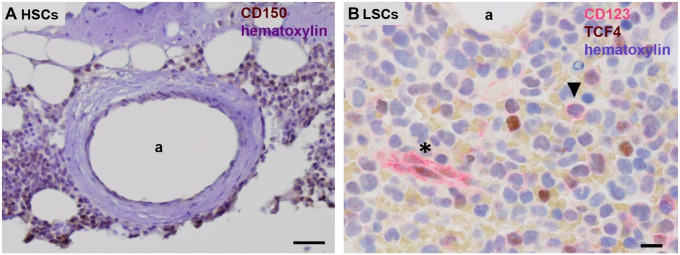
Figure 2.
Chromogenic immunohistochemical images of paraffin sections of human bone marrow. (A) Human CD150-positive HSCs (brown) in their periarteriolar niche in healthy bone marrow adjacent to bone and (B) human CD123-positive LSCs (red, arrow head) in a HSC niche in bone marrow of a leukemia patient. Endothelial cells of capillaries are CD123 positive as well (asterisk). Hematoxylin staining in blue. a, lumen of arteriole. Bars, (A) 100 µm, (B) 25 µm. Tissue sections and staining are prepared according to Hira et al. 9 (A) and El Achi et al. 24 Abbreviations: HSCs, hematopoietic stem cells; LSCs, leukemic stem cells.
Isocitrate Dehydrogenase 1/2 Mutations and ATP Synthesis
Besides the similarity of HSC niches in bone marrow that harbor LSCs in leukemia patients and GSC niches in glioblastoma tumors, there is another remarkable similarity between glioblastoma and leukemia and in particular acute myeloid leukemia (AML). Both types of cancer are induced in a considerable percentage of patients by a distinct mutation in the NADP-dependent isocitrate dehydrogenase (IDH) 1 or 2 genes (10–40% of AML patients and 5% of glioblastoma patients). As IDH1 and IDH2 are both involved in energy metabolism including OXPHOS (for recent reviews, see previous studies25–27), the effects of the IDH1 and IDH2 mutations on ATP production in GSCs and LSCs are discussed as well. The mutation occurs in either the IDH1 or IDH2 gene in similar amounts in AML patients and almost uniquely in IDH1 in glioblastoma, 28 although recently a significant number of IDH2 mutations were found in glioblastoma patients in the Hispanic population in the United States and Mexico. 29
ATP Synthesis Pathways
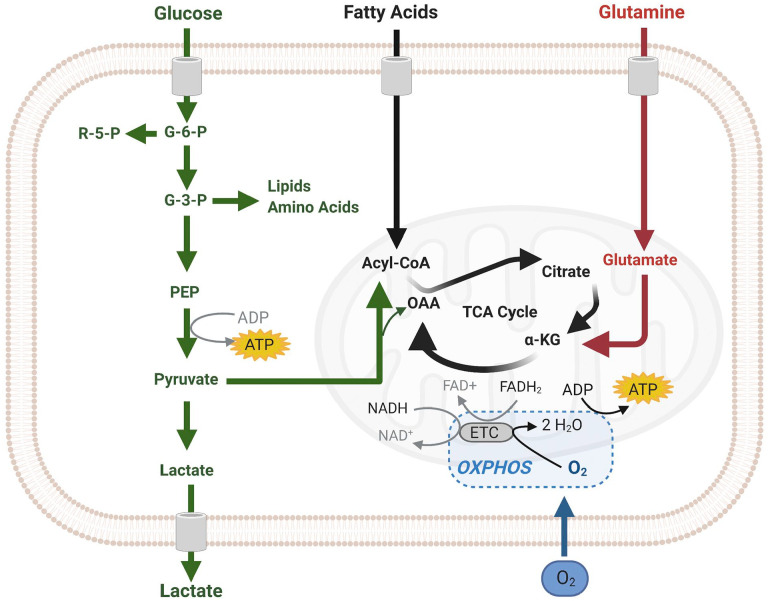
We discuss in the present review ATP synthesis in benign and malignant stem cells in the bone marrow and the brain, HSCs, NSCs, LSCs, and GSCs (Figs. 1 and 2). Two major pathways are available in cells for ATP production (Fig. 3). The most efficient pathway is mitochondrial respiration with the use of oxygen that generates 36-mol ATP per mol glucose, whereas cytoplasmic glycolysis produces 4-mol ATP per mol glucose without the use of oxygen. ATP production by the inefficient glycolytic pathway is a salvage pathway for ATP production in case cells are short of oxygen. In that anaerobic process, lactate is generated from pyruvate and in the early steps of glycolysis, 2-mol ATP per mol glucose are needed to keep glycolysis running, yielding a net 2-mol ATP per mol glucose from glycolysis. When oxygen is available (aerobic conditions), pyruvate is channeled into mitochondria for the more efficient ATP production in the oxidative phosphorylation (OXPHOS) as part of the mitochondrial respiration (Fig. 3).
Figure 3.
Scheme of cellular ATP synthesis via (an)aerobic glycolysis in the cytoplasm (green) and in mitochondria via OXPHOS (black). The image was created using Biorender.com. Abbreviations: OXPHOS, oxidative phosphorylation; TCA, tricarboxylic acid cycle.
It was generally assumed until recently that cancer cells use glycolysis irrespective the presence of oxygen as a consequence of dysfunctional or impaired mitochondria. However, in most tumors, the use of glycolysis for ATP production independently of the presence of oxygen is an essential part of a “selfish” metabolic reprogramming as was explained recently by Vaupel and Multhoff. 30 Therefore, we have investigated at the electron microscopical (EM) level the morphology of mitochondria in differentiated glioblastoma cells and GSCs (Fig. 4). The morphology of mitochondria from the level of individual mitochondrion ultrastructure to the level of the entire mitochondrial network in a cell reflects mitochondrial function. 31 Ultrastructurally, the morphology of cristae can be highly variable in accordance with the metabolic state of mitochondria32,33 and processes that cause their functional decline, such as aging. 34 Generally, more numerous, tightly packed cristae support more efficient OXPHOS because the cristae establish inner mitochondrial compartments and accommodate protein complexes of the electron transport chain and ATP synthase. 35 These protein complexes in turn impact the shape of mitochondrial cristae.36–38 Regarding the mitochondrial network, mitochondrial fusion that produces highly elongated mitochondria supports efficient OXPHOS, whereas mitochondrial fission that results in smaller and rounded mitochondria promotes glycolysis and greater biosynthesis of anabolic precursors.39,40 It appeared that in all glioblastoma cells investigated be it differentiated cells or GSCs, the morphology of mitochondria is deviant from normal cells. Mitochondria in glioblastoma cells are generally small, with very dense matrices and disorganized cristae, which is in line with the assumption that glycolysis is used by cancer cells for ATP production because of defective mitochondria. Similar defective mitochondrial ultrastructure in line with a glycolytic phenotype was reported also by Vallejo et al. 41 for adult and pediatric glioblastoma. Therefore, we are investigating at present the activity of the electron transport chain in mitochondria of glioblastoma cells at the ultrastructural level using enzyme histochemical methods, such as that for demonstrating cytochrome c oxidase activity. 42
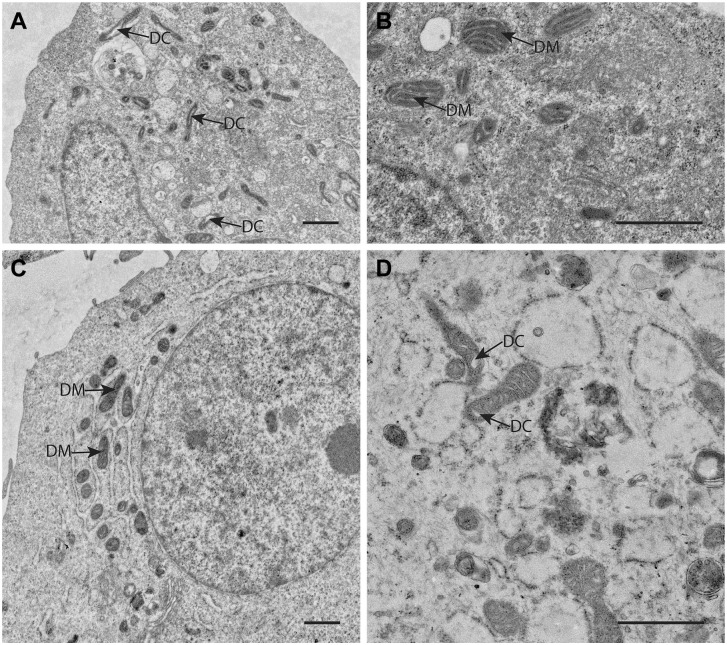
Figure 4.
Electron microscopical (EM) images of mitochondria in human GSCs (A, NCH421 and B, NCH644 primary patient GSCs) and in human differentiated glioblastoma cells (C, U87 cancer cells and D, NIB140 primary patient cancer cells) showing the deviant morphology of mitochondria with relatively few and often dilated cristae (DC) and very electron-dense matrices (DM) in both GSCs and differentiated glioblastoma cells indicating that glycolysis rather than OXPHOS is used by these cells for ATP production. Bars, 1 µm. NCH421 and NCH644 cells are a kind gift of Prof. Dr Christel Herold-Mende, University of Heidelberg, Germany. 43 Cell preparations and EM imaging procedures were performed as described in Bogataj et al. 44 Abbreviations: DC, dilated cristae; GSCs, glioblastoma stem cells; OXPHOS, oxidative phosphorylation.
Nowadays, it is realized that proliferating cells such as differentiated cancer cells predominantly use glucose for ATP production in the cytoplasmic glycolysis instead of OXPHOS independently of the presence of oxygen (the so-called Warburg effect or aerobic glycolysis) because glycolysis enables synthesis of building blocks for macromolecules needed by proliferating cells.30,45,46 Moreover, ample NADPH production is also facilitated in this way for reductive biosynthetic reactions.45,47
Quiescent undifferentiated CSCs have a modest need for building blocks and excessive NADPH production because of their slow proliferation rate and, thus, the question arises what metabolic pathways are used by normal stem cells such as HSCs and NSCs and by CSCs such as LSCs and GSCs for ATP production in their hypoxic niche microenvironment. We discuss the cell biological regulation mechanisms of these metabolic pathways as far as these are presently known. These cell biological regulation mechanisms are explained briefly in texts in Boxes that are separate from the main body of text on ATP production in HSCs, NSCs, LSCs, and GSCs.
Energy Metabolism of HSCs
HSCs are resident in niches in human bone marrow which are hypoxic and periarteriolar and express the chemoattractants SDF1α and OPN and their receptors CXCR4 and CD44, respectively, which are essential for the homing of the slowly dividing or quiescent HSCs.9,10 HSC niches are present in red bone marrow around arterioles adjacent to endosteum and trabecular bone. HSC niches are hypoxic despite the fact that they are uniquely found around arterioles, but arterioles are transport vessels and not exchange vessels like capillaries and thus HSC niches do not receive oxygen from the arteriolar lumen.12,13 HSCs produce low levels of intracellular reactive oxygen species (ROS). 11 HSCs are at the top of the pyramid of blood cells above hematopoietic progenitor cells (HPCs) and differentiated red and white blood cells, including megakaryocytes that fragment into thrombocytes, also named platelets. In healthy humans, one trillion (1012) differentiated red and white blood cells are released into the circulation each day. Under normal conditions, only relatively few HSCs are involved in the formation of new mature red and white blood cells. The vast majority of that trillion red and white blood cells originate from rapidly dividing HPCs which are located in HPC niches in the center of the red bone marrow around the sinusoids and the central vein and are continuous with HSC niches in the periphery of red bone marrow around arterioles. 11 The newly formed differentiating red and white blood cells are released into the sinusoids and leave the bone marrow via the central vein. The HSCs in their hypoxic periarteriolar niches at the periphery of the bone marrow are a life-long reserve to ensure the availability of stem cells during the entire life of individuals and supply HPCs in times of depletion of red and/or white blood cells in cases of blood loss. 48 The HPC niches are also hypoxic but intracellular levels of ROS in HPCs are higher than those in HSCs.11,49–52
Metabolism including ATP synthesis of HSCs, HPCs, and hematopoietic stem and progenitor cells (HSPCs) is probably the best studied metabolic aspect in stem cells because of the therapeutic relevance of metabolism on the ex vivo expansion of phenotypically and functionally defined HSPCs harvested from umbilical cord blood and peripheral blood for transplantation purposes in malignant and non-malignant diseases.48,53–55 The term HSPCs is used for the stem-like cells that are harvested from umbilical cord blood, bone marrow aspirate, and peripheral blood for these purposes. 56 The general consensus is that HSCs are anaerobic glycolytic, whereas during the proliferation phases of HPCs, the energy metabolism is mixed glycolytic and OXPHOS, mainly OXPHOS during the differentiation phase and again glycolytic in the maturation phase.54–58 Anaerobic glycolysis in HSCs is determined by high levels of the transcription factors hypoxia-inducible factor-1 (HIF-1) and HIF-2 (see Box 1).9,48 Moreover, the SDF-1a-CXCR4 axis regulates OXPHOS activity in HSCs as well in a thus far unknown manner. 59
Box 1.
Hypoxia-inducible Factors (HIFs).
| HIF-1, HIF-2, and HIF-3 are heterodimeric transcription factors consisting of an oxygen-labile HIF-1α, HIF-2α, or HIF-3α monomer and a common stable β monomer.60,61 The labile monomers are continuously expressed and in normoxic conditions rapidly oxygen-dependently hydroxylated, ubiquitinated, and proteosomally degraded. They become stabilized in hypoxic conditions.61–64 This complex system enables rapid responses to hypoxic insults without the need for de novo protein synthesis. 61 When HIF-1α, HIF-2α, and HIF-3α become stabilized under hypoxic conditions, they translocate into the cell nucleus and heterodimerize with the stable β monomer (HIF-1β or ARNT) to become HIF-1, HIF-2, and HIF-3, respectively, and then bind to the hypoxia-responsive elements (HREs) of target genes.61–63 HIF-1 and HIF-2 are closely related and both activate HRE-dependent gene transcription. 63 Relatively little is known of the function of HIF-3. It is considered to be a negative regulator of HIF-1 and HIF-2 by competition for HIF-1β60,65 and plays a role in stroke-related diseases. 60 HIF-1 and HIF-2 are closely related but transcription of genes differs and HIF-2 is stable at higher oxygen concentrations than HIF-1. 61 HIF-1 is more acutely responsive to hypoxia, whereas HIF-2 is more chronically responsive.61,66 Moreover, HIF-1 induces preferentially transcription of genes of metabolic (glycolytic) enzymes, whereas HIF-2 induces preferentially angiogenesis-related gene transcription.62,67 Both HIF-1 and HIF-2 promote stemness of cells. 66 Downes et al. 61 analyzed which genes are transcriptionally regulated by HIF-1 and HIF-2 in primary human endothelial cells. HIF-1 induced expression of 700 genes, whereas HIF-2 induced expression of 1450 genes of which 300 genes were transcriptionally activated by both HIF-1 and HIF-2. 61 An alternative explanation for the differences in expression patterns regulated by HIF-1 and HIF-2 is a difference in cell types in which HIF-1 and HIF-2 are active. 63 Indeed, we have observed differences in expression levels of HIF-2α and HIF-1α in GSCs and endothelial cells of arterioles, respectively.9,19 Inhibitors of HIF-1 and HIF-2 have been discussed recently for their potential use to target therapeutically cancer stem cells. 68 |
Abbreviations: ARNT, aryl hydrocarbon receptor nuclear translocator; GSCs, glioblastoma stem cells.
Both HIFs are master switches in cells to adapt to hypoxic conditions 67 and are induced by peroxisome proliferator-activated receptors (PPARs; see Box 2), which are nuclear receptors that act as transcription factors. 69 In fact, intracellular PPAR levels in HSCs are the highest measured in any cell type in the human body. 70 Inhibition of PPARs promotes ex vivo expansion of HSPCs. 54 Moreover, the transcription factor Meis1 (see Box 3), that is expressed in HSCs, transactivates HIF-1α expression by a Meis1-binding motif in the first intron of the HIF-1α gene.55,67 The transcription factor forkhead box O3 (FOXO3; see Box 4) plays a role in many metabolic events in cells, among others protection against ROS, and is a key regulator of HSC quiescence and maintenance of the HSC pool. 48 Finally, autophagy (see Box 5) is also critically important to maintain low mitochondrial metabolic activity and quiescence of HSCs.71,72 Autophagy and stem cell fate are under the control of transcription factor FOXO3 in both normal stem cells and CSCs. 71
Box 2.
Peroxisome Proliferator-activated Receptors (PPARs).
| PPARα, PPARβ/δ, and PPARγ are key members of the nuclear receptor superfamily of transcription factors that sense nutrients and regulate metabolic pathways and in particular FAO.69,73 PPAR β/δ and PPARγ are relevant for HSCs, NSCs, LSCs, and GSCs. 66 The PML-PPARβ/δ-FAO axis regulates the maintenance of HSC stemness via HIF-1 and HIF-2 activity.70,73 PPARγ is expressed in GSCs of the most aggressive subtype of glioblastoma, the mesenchymal subtype 74 and in LSCs. 75 Counterintuitively, activation of PPARγ by agonists is investigated as therapeutic option in GSCs and LSCs because PPARγ upregulation is associated with poor survival of glioblastoma patients. 74 Hua et al. 74 explained this therapeutic effect by PPAR agonists by inhibition of the STAT3 signaling pathway (see Box 8) that prevents a proneural-mesenchymal transition in glioblastoma. Yang et al. 66 explained opposite therapeutic effects of agonists by differential binding to target genes. |
Abbreviations: FAO, fatty acid oxidation; HSCs, hematopoietic stem cells; HIF, hypoxia-inducible factor; STAT3, signal transducer and activator of transcription 3; NSCs, neural stem cells; LSCs, leukemic stem cells; GSCs, glioblastoma stem cells; PML, promyelocytic leukemia protein.
Box 3.
Myeloid Ecotropic Viral Integration Site 1 (Meis1).
| Transcription factor Meis1 was discovered in cancer as a viral integration site and is overexpressed in AML and neuroblastoma76–78 and is associated with therapy resistance. 79 Meis1 is in particular highly expressed in HSCs. 80 Meis1 regulates HSC metabolism and redox state by inducing HIF-1α and HIF-2α expression. 67 Meis1 is a transcription activator-like effector (TALE)-type transcription factor. It cooperates with PBX1 and HOXA9 transcription factors to transactivate target genes such as those of HIF-1α and HIF-2α. 80 Knockdown of Meis1 causes downregulation of HIF-1 and HIF-2 activity with concomitant increased ROS levels and loss of HSC quiescence. 81 Meis1 levels are frequently upregulated in AML.2,55,82 Recently, Turan et al. 80 developed two small molecule Meis1 inhibitors to modulate HSC activity as Meis1 is a key regulator of HSCs. Meis1 inhibition downregulates HIF-1 and HIF-2 activity and reduces HSC quiescence. Therefore, inhibitors of Meis1 may become applicable for transplantation purposes and in anticancer therapy. |
Abbreviations: AML, acute myeloid leukemia; HSCs, hematopoietic stem cells; HIF, hypoxia-inducible factor; ROS, reactive oxygen species.
Box 4.
Forkhead Box O3 (FOXO3).
| Transcription factors FOXO1, FOXO3, FOXO4, and FOXO6 are expressed in mammals in a tissue-specific manner.83–85 FOXO proteins and in particular FOXO3 are longevity genes.
85
FOXO3 is an important transcription factor in stem cells and CSCs in blood and brain and thus for HSCs, NSCs, LSCs, and GSCs,
86
whereas FOXO1 is an important transcription factor in endothelial cells.87,88 The functions of nuclear FOXO1 and FOXO3 are largely overlapping.
89
FOXO3 transcription activity promotes the stemness of stem cells and CSCs and their quiescence,
86
whereas FOXO1 promotes quiescence of mature endothelial cells and is downregulated during angiogenesis.87,88 FOXO1 and FOXO3 promote glycolysis and inhibit OXPHOS and ROS production. As both FOXO1 and FOXO3 promote quiescence of cells, they are considered to be tumor suppressors, but this view may well be too simplistic.85,86,89 FOXOs promote invasion and metastasis of cancer cells by promoting the epithelial–mesenchymal switch and by upregulation of the expression of various extracellular proteases.83,89 Furthermore, FOXO1 and FOXO3 upregulation is associated with poor prognosis of leukemia and glioblastoma patients and therapy resistance.
89
This may well be a consequence of both protection of LSCs and GSCs and promotion of invasion and metastasis. As far as we know, these aspects of transcriptional activity of FOXO1 and especially FOXO3 have not yet been investigated, except for the studies of Firat and Niedermann
84
and Xu et al.
90
The first study demonstrated that FOXO3 is involved in GSC maintenance after irradiation
84
and the second study showed that FOXO3 expression is high in temozolomide-resistant glioblastoma cells.
90
Libby et al. 86 state that it is intriguing that two cancer types (leukemia and glioblastoma) use one and the same protein (FOXO3) to keep the stemness of LSCs and GSCs and the related metabolic shift. Independently, we have concluded that niches of both HSCs and LSCs and of NSCs and GSCs are functionally similar.9,10 |
Abbreviations: OXPHOS, oxidative phosphorylation; ROS, reactive oxygen species; GSCs, glioblastoma stem cells; CSCs, cancer stem cells; HSCs, hematopoietic stem cells; NSCs, neural stem cells; LSCs, leukemic stem cells.
Box 5.
Autophagy.
| In the lysosomes of cells, molecular substrates such as carbohydrates, proteins, lipids, and nucleic acids but also damaged organelles such as mitochondria and peroxisomes are degraded into their building blocks that can be reused in biosynthetic processes and metabolic processes. The acidic environment in lysosomes and the over 60 hydrolases that are active in the lumen of lysosomes are responsible for the degradation. Autophagy, endocytosis, and phagocytosis provide the molecular substrates. 91 Autophagy plays an important role in stem cells because it is a protection mechanism for their quiescence and longevity by preventing cellular stress. 92 However, recently indications have been found that lysosomal regulation of stem cell metabolism is more than just autophagy. Lysosomes seem to play a central role in the regulation of HSC and NSC metabolism and their activation into HPCs and NPCs by coordinating anabolic and catabolic processes by nutrient sensing.91,93–96 Lysosomes are abundant in HSCs and NSCs and scarce in HPCs and NPCs.93,96 Fundamental research and clinical trials are ongoing at the moment to analyze whether inhibition of autophagy has anticancer therapeutic benefits 92 because autophagy in GSCs and LSCs is involved in therapeutic resistance by removal of damaged cellular components. 97 For example, repurposing of nicardipine, a calcium channel antagonist that is FDA approved to treat elevated blood pressure, in combination with temozolomide sensitizes GSCs to temozolomide by inhibiting autophagy. 98 Autophagy protects cells during early steps of carcinogenesis, but once cancer cells are established, autophagy protects cancer cells against therapeutically induced cellular damage.92,97,99 |
Abbreviations: HSCs, hematopoietic stem cells; NSCs, neural stem cells; HPCs, hematopoietic progenitor cells; NPCs, neural progenitor cells; GSCs, glioblastoma stem cells; LSCs, leukemic stem cells; FDA, Food and Drug Administration.
A number of seemingly contradictory aspects is apparent in the energy metabolism of HSCs in their niches. 48 A major issue is that HSCs are anaerobic glycolytic but cannot survive without mitochondria.48,100 This riddle was at least partly solved by demonstrating that PPARs induce both cytoplasmic anaerobic glycolysis and mitochondrial fatty acid oxidation (FAO; Fig. 3).54,70,73 The latter is equally important for maintaining a viable population of HSCs.70,101,102 OXPHOS is inhibited in HSC mitochondria at the level of pyruvate conversion into acetyl-CoA by pyruvate dehydrogenase after its phosphorylation by pyruvate dehydrogenase kinase (PDK).48,70,103 When PDK is inhibited, quiescence of HSCs is lost and levels of ROS are elevated. Thus, pyruvate entry into the mitochondria has a strong impact on HSC fate. 48 This finding shows that the presence of mitochondria does not mean that all their metabolic pathways are active.48,59,69,70,101,102 HSC quiescence can also be interrupted by signaling from differentiated blood cells, for example, by the chemokine (C-C motif) ligand-6 (CCL-6) secreted by eosinophils. 104
It has to be realized that the cellular mechanisms of HSCs as described here are not necessarily relevant for the in vivo situation in human red bone marrow. The studies have been performed either on mouse bone marrow or human ex vivo HSPCs harvested from umbilical cord blood or peripheral blood. For example, low levels of oxygen have been measured in HSC niches in mouse bone marrow. 105 Nevertheless, we think it is safe to conclude that HSCs in their hypoxic periarteriolar niches adjacent to endosteum and trabecular bone are anaerobic glycolytic and only use particular functions of mitochondria, for example FAO, whereas other functions such as OXPHOS are shut down, to meet their metabolic needs as quiescent long-living cells. How HSCs exactly use mitochondria remains to be investigated. 48 HSCs contain relatively many partially inactive mitochondria not only to meet their metabolic needs but also to be able to rapidly switch on mitochondrial activity in case HSCs need to become HPCs. 48
In conclusion, HSCs are dependent on anaerobic glycolysis for their energy supply, whereas FAO is essential for their stem cell fate.
Energy Metabolism of NSCs
Energy metabolism of quiescent NSCs and the drastic alterations in this metabolism associated with the conversion of NSCs into neural progenitor cells (NPCs) are very much the same as those of HSCs and HPCs, as is shown here.
The developing brain is mainly glycolytic and uses FAO, and this energy metabolism shifts toward the use of glucose rather than lipids during adolescence and becomes more and more OXPHOS dependent. 106 In the adult brain, OXPHOS is the major source of ATP in neurons, whereas astrocytes remain glycolytic and produce lactate that is exported into the ECM to be consumed by neurons that take up lactate and convert it to pyruvate to fuel OXPHOS.86,106,107 However, certain areas of the human brain remain glycolytic and contain quiescent NSCs such as the SVZ that lines the lateral ventricles, the subgranular zone (SGZ) between the granule cell layer and the hilus of the dentate gyrus of the hippocampus, and a few areas in the hypothalamus.106,108–111 FOXO3 regulates glycolysis in NSCs and plays a significant role in the maintenance of the stem cell fate of NSCs in combination with the low oxygen levels in SVZ and SGZ in a similar way as in quiescent HSCs.86,108 It also stabilizes HIF-1α and thus regulates the activity of HIF-1 and possibly of HIF-2. 86 Intracellular levels of promyelocytic leukemia protein (PML; see Box 6) that is involved in the formation of nuclear bodies in NSCs as well as in HSCs are also important for their quiescence. Loss of PML induces loss of quiescence and upregulation of proliferation, whereas loss of PML has been found to be associated with decreased levels of PPARγ in HSCs.73,86
Box 6.
Promyelocytic Leukemia Protein (PML).
| PML is a protein that is active in the cell nucleus by the formation of nuclear bodies, the so-called PML-NBs. 112 PML stabilizes the genome and interacts with at least 120 different cellular proteins by physical association with PML-NBs. It is considered to be a tumor suppressor protein because it downregulates cell proliferation, whereas its inactivation and downregulation are associated with cancer. 112 PML induces quiescence of HSCs and NSCs and it stimulates their FAO and PPAR signaling.113,114 For the same reason, it is also important for LSCs and GSCs.86,115 Therefore, PML may be an important player in the protection of LSCs and GSCs in their niches and thus, PML may be a tumor suppressor protein that can also promote cancer as FOXO3 does.114,116 Therapeutic inhibition of PML eradicates LSCs. 116 Furthermore, PML affects the tumor microenvironment as it inhibits angiogenesis via inhibition of mTOR. 117 |
Abbreviations: FAO, fatty acid oxidation; PPAR, peroxisome proliferator-activated receptor; FOXO3, forkhead box O3; HSCs, hematopoietic stem cells; NSCs, neural stem cells; LSCs, leukemic stem cells; GSCs, glioblastoma stem cells; mTOR, mammalian target of rapamycin.
In rodents, NSCs of the SVZ are needed permanently throughout life to differentiate into neurons and astrocytes that migrate toward the olfactory bulb, whereas in humans, this renewal of the neuronal circuits that are responsible for smell hardly occurs or does not occur at all.106,118,119 In humans, it has been observed that NSCs become NPCs that proliferate and differentiate as a consequence of ischemic stroke and that NPCs migrate to the damaged brain tissue for repair.108,120–123
Quiescent NSCs in the SVZ and SGZ are anaerobic glycolytic and need FAO,106,108 exactly like HSCs. The FAO inhibitor etomoxir causes depletion of both quiescent NSCs and HSCs indicating the importance of FAO for their stem cell fate.86,124–126 When NSCs become activated, for example, by an ROS spike and become NPCs, mitochondria switch to OXPHOS127–129 and upregulate the redox state,128,130–132 whereas glycolysis and FAO are downregulated and lipogenesis is elevated.106,108 Upregulation of the redox state involves higher ROS production and more active defense mechanisms against ROS, which is orchestrated by increased expression of the transcription factor erythroid 2-related factor 2 (Nrf2; see Box 7) that senses oxidants and regulates antioxidant defense. 133 Besides the spike of ROS that can activate NSCs into NPCs, branched chain amino acids, the endogenous neuroprotective bile acid tauroursodeoxycholic acid, interferon γ, and signal transducer and activator of transcription 3 (STAT3; see Box 8) have been reported recently to activate human NSCs into NPCs in vitro or in vivo in rodents.134–137 Whether these inducers of switches of NSCs into NPCs are active in vivo in humans is not known yet.
Box 7.
Erythroid 2-related Factor 2 (Nrf2).
| Nrf2 is a transcription factor that is a major regulator of antioxidant defense in cells by checking intracellular levels of ROS and other oxidants. Nrf2 is continuously expressed in cells and at low oxidant levels rapidly ubiquitinated and proteosomally degraded. Nrf2 senses cellular oxidant levels by the oxidation rate of cysteine thiols. When the rate becomes too high, Nrf2 becomes stabilized 133 in a similar way as HIF1α and HIF2α expression is regulated. Nrf2 promotes transcription of genes that are involved in the formation of reduced glutathione, ROS detoxification, and degradation of xenobiotics,138,139 and Nrf2 induces the activity of NADPH-producing dehydrogenases and activates HIFs.140,141 NADPH is a major substrate for cellular detoxifying enzyme systems.27,47 Nrf2 activity is important for HSCs, HPCs, and HSPCs, but it may also be beneficial for cancer cells in relation to therapy resistance. On the other hand, Nrf2 knockout mice are more susceptible to develop cancer. 133 Therefore, it can be concluded that under normal conditions, Nrf2 protects against ROS damage, whereas in CSCs, Nrf2 is an important player in therapy resistance. 139 |
Abbreviations: ROS, reactive oxygen species; HIF, hypoxia-inducible factor; HSCs, hematopoietic stem cells; HPCs, hematopoietic progenitor cells; HSPCs, hematopoietic stem and progenitor cells; CSCs, cancer stem cells.
Box 8.
Signal Transducer and Activator of Transcription 3 (STAT3).
| STAT3 is a transcription factor and transcription activator and its activity is induced by members of the interleukin-6 (IL-6) cytokine family via phosphorylation by Janus family kinases (JAKs) that enables translocation of STAT3 into the cell nucleus.137,142 STAT3 knock out is lethal in embryos, but in adult tissues, conditional knock out results in mild phenotypes and has multiple, sometimes seemingly contradictory, functions. 142 For example, STAT3 has been found to be involved in self-renewal of stem cells and in differentiation of (cancer) stem cells into progenitor cells, or in upregulation of OXPHOS and downregulation of ROS production. STAT3 can also be translocated into mitochondria to promote OXPHOS by binding to complex 1 and it also induces expression of OXPHOS-related genes. 137 Regulatory T cells (Tregs) in GSC niches produce IL-6 and promote GSC stemness via IL-6-STAT3 signaling. 143 Effects of inhibition of STAT3 on stemness of GSCs are presently being investigated.144–147 |
Abbreviations: OXPHOS, oxidative phosphorylation; ROS, reactive oxygen species; GSCs, glioblastoma stem cells.
Finally, the role of glutamine/glutamate in NSCs and NPCs is not exactly known. Recently, a novel methodology to analyze dynamically cell metabolism in time was reported with the example of an extracellular shot of glutamine (15 mM) to both human NSCs and NPCs in vitro. Analysis of metabolites in the cells in time showed that NSCs and NPCs responded in a similar way to the extracellular glutamine shot and it mainly involved levels of amino acids, indicating that the glutamine did not affect energy metabolism very much. 148 It may well be that glutamine/glutamate play their role in tumorigenesis. 149
In conclusion, NSCs are dependent on anaerobic glycolysis for their energy supply, whereas FAO is essential for their stem cell fate. Thus, NSCs and HSCs function in very similar ways for their energy supply.
Energy Metabolism of LSCs
Similarly to their HSC counterparts, LSCs need a hypoxic environment. Therefore, hijacking the hypoxic periarteriolar HSC niches in bone marrow is an attractive option for LSCs.9,10 Metabolic pathways that provide metabolites needed for anabolic cell growth are upregulated in LSCs 150 and mitochondria play an essential role in these pathways in LSCs as has recently been reviewed by Panuzzo et al. 26 Whereas HSCs depend on anaerobic glycolysis for generation of ATP, LSCs produce ATP mainly via OXPHOS.26,151,152 OXPHOS is more efficient as energy producer than glycolysis and sustains LSC energy requirements for survival. 153 Damage by ROS that are generated by OXPHOS is resisted by antioxidative mechanisms. 151 Raffel et al. 152 showed by quantitative proteomics that in comparison with HSCs, LSCs have a profoundly altered mitochondrial metabolism, both at the transcriptional (mRNA) level and at the posttranslational (protein) level. Not only OXPHOS was distinctly different but also FAO, lipid synthesis, and amino acid metabolism.26,152 Glutamine and glutamate are efficiently converted into α-KG via upregulated glutaminase and glutamate dehydrogenase activities.26,154 This pathway produces NADPH which is essential for reduction of glutathione.26,47,155,156 Inhibition of FAO or amino acid uptake reduces OXPHOS activity in LSCs indicating that both pathways supply substrate for OXPHOS.26,154,157 Recently, it was found that the adrenomedullin-calcitonin receptor-like receptor axis 158 and spleen tyrosine kinase 159 both induce OXPHOS activity in LSCs. Inhibition or knock down of both proteins specifically targets LSCs in AML.158,159
Reactive aldehydes that are produced during FAO are effectively detoxified by upregulated fatty aldehyde dehydrogenase, more specifically named aldehyde dehydrogenase 3a2 (ALDH3A2), in LSCs.160,161
There is a strong competition in the niches between HSCs and LSCs. In this competition, LSCs have a greater competitive advantage. 151 This advantage is at least partly caused by assistance of mesenchymal stem cells (MSCs) to LSCs. MSCs are present in niches of HSCs and LSCs (Fig. 5A) and assistance of MSCs enables LSCs to keep their leukemic phenotype and chemotherapy resistance as was demonstrated in an in vivo mouse model of AML. LSCs co-opt energy sources and antioxidant defense mechanisms provided by MSCs in the niches to survive chemotherapy. Assistance of LSCs was proven to be provided at the expense of HSCs. 162 It is not described by Forte et al. 162 how the exchange from MSCs to LSCs takes place but extracellular vesicles (EVs),163,164 tunneling nanotubes (TNTs), and/or tumor microtubes165,166 may well play a role in these exchange processes. Furthermore, extracellular ATP provides resistance of LSCs against chemotherapy by a cAMP-mediated mitochondrial stress response. cAMP is produced by the ectonucleotidase CD39 that converts extracellular ATP into cAMP that is then internalized by LSCs. 167
Figure 5.
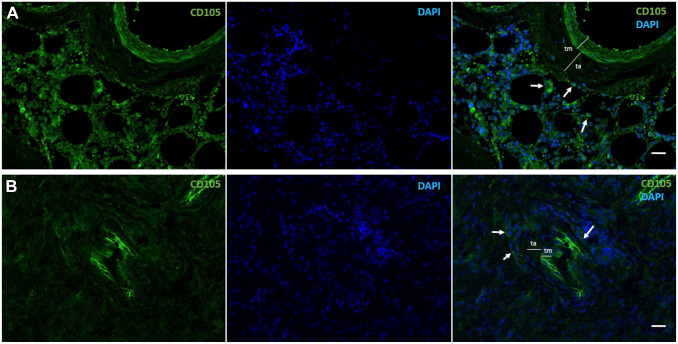
Immunofluorescence images of human CD105-positive MSCs (green, arrows) in a periarteriolar HSC niche in healthy bone marrow adjacent to bone (A) and a periarteriolar GSC niche in a glioblastoma tumor (B). Monochrome images of CD105-positive MSCs in green and DAPI fluorescence (nuclei) in blue and the composition images in green and blue are shown. Smooth muscle cells in the tunica media (tm) of the arterioles are also CD105 positive. ta, Tunica adventitia of the arterioles. Tissue sections and immunofluorescence staining are prepared as described in Hira et al. 9 Bars, 100 µm. Abbreviations: MSCs, mesenchymal stem cells; HSC, hematopoietic stem cell; GSC, glioblastoma stem cell; DAPI, 4′,6-diamidino-2-phenylindole.
All these data have been published recently and indicate that LSCs in hypoxic periarteriolar HSC niches in the bone marrow are different from HSCs with respect to energy metabolism (anaerobic glycolysis versus OXPHOS and altered FAO and amino acid metabolism) and that LSCs in HSC niches are also aided by the microenvironment to survive chemotherapy. This stresses that LSCs have to be removed from HSC niches to optimize the efficiency of anti-LSC therapy, for example, by interfering with their attachment in HSC niches by SDF-1α-CXCR4 with the use of the CXCR4 inhibitor plerixafor.9,10
In a considerable percentage of AML patients (10–20%), cancer cells harbor the IDH1 or IDH2 mutation.25,28,168 The IDH1 and IDH2 mutations occur in similar numbers of AML patients and are mutually exclusive. 28 The effects of IDH1 or IDH2 mutations on the production of ATP [OXPHOS versus (an)aerobic glycolysis] in LSCs do not seem to be profound. The mutations cause metabolic rewiring from aerobic glycolysis to OXPHOS in LSCs and differentiated leukemia cells26,168 independently of their differentiation status. Therefore, we assume that in both IDH1/2-mutated LSCs and differentiated leukemia cells, mitochondrial metabolism including OXPHOS is prevailing. However, as far as we know, this assumption has not been tested experimentally yet.
In conclusion, LSCs depend on OXPHOS for their energy requirements despite the fact that LSCs are hiding in protective hypoxic periarteriolar niches in the bone marrow, whereas FAO and amino acids are needed in the mitochondria to fuel OXPHOS.
Energy Metabolism of GSCs
GSCs play an essential role in glioblastoma tumor development, growth, and aggressiveness. Besides a low proliferation rate that protects GSCs in their hypoxic niches, GSCs possess efficient DNA repair mechanisms and a well-developed drug efflux ABC transporter system.169–171 GSCs express stem cell markers, such as SOX2, NANOG, OLIG2, NESTIN, IDI1, MYC, and MUSHASHI1, and cell surface proteins, such as CD133, CD44, LICAM, and CD15170 and CD9 (Fig. 1) 172 that mediate interactions with their microenvironment of their niches. Furthermore, GSCs interact with differentiated glioblastoma cells and promote malignant progression of glioblastoma. 173 GSCs remain undifferentiated by genetic and epigenetic alterations in signaling pathways such as Notch, bone morphogenetic protein, nuclear factor kappa B, and Wnt.174–177
GSC niches harbor various non-malignant cell types such as immune cells and MSCs (Fig. 5B) that are recruited to the glioblastoma tumor site. The dynamic and complex interactions within niches promote GSC therapeutic resistance and antitumor immune responses. Among the non-cancerous cell types that are present in GSC niches, microglial cells and macrophages prevail. 178 Macrophages are differentiated monocytes from the bone marrow, whereas microglial cells are derived from the yolk sac.179,180 Microglial cells are predominant in newly diagnosed tumors, whereas monocyte-derived macrophages are predominant in recurrent tumors in GSC niches. 181 GSCs recruit macrophages through chemoattractants such as vascular endothelial growth factor, colony stimulating factor 1, SDF-1α, interleukin-6 (IL-6), IL-1b, and the ECM protein periostin, which polarize macrophages to an immune-suppressive and angiogenic phenotype that promotes tumor growth and progression. 182 Not all macrophages are protumoral but for successful application of antitumor macrophages, a better understanding of interactions of GSCs and macrophages is urgently needed. 183
Recently, involvement of cell types and proteins in CSCs and in particular in GSCs has been reviewed, such as a novel cell type, the telocyte, 184 scaffold proteins,185,186 the renin–angiotensin system, 187 and specific proteases and their endogenous inhibitors. 188
MSCs are another cell type that infiltrates glioblastoma tumors and are present in GSC niches, as they are in HSC niches in bone marrow (Fig. 5).9,189 MSCs are important players in glioblastoma because they stimulate proliferation and invasive behavior in differentiated glioblastoma cells and increase the self-renewal capacity of GSCs.9,189–191 MSCs that express CD105 are demonstrated in periarteriolar GSC niches in Fig. 5. Cancer cells, including GSCs, can acquire mitochondria from MSCs in their niches via TNT-mediated intercellular communication. The acquisition of mitochondria increases OXPHOS and ATP production in cancer cells as well as indirectly affects their general metabolism. As a consequence, cancer cells change their invasive and proliferative properties and increase their capacity to develop therapeutic resistance. 192 MSCs also release EVs that contain miRNAs that enhance tumorigenicity of GSCs. 193
Energy metabolism of GSCs is less well studied than that of NSCs, HSCs, and LSCs, but the general consensus is that both IDH1 wild-type and IDH1-mutated GSCs mainly use OXPHOS for the generation of ATP, whereas IDH1wt-differentiated glioblastoma cells use aerobic glycolysis and IDH1-mutated differentiated glioblastoma cells use OXPHOS for ATP production as a consequence of the metabolic rewiring.27,194–197 FAO is essential for GSCs107,198,199 as it is for NSCs, HSCs, and LSCs.
A number of proteins have been described recently that may serve as therapeutic target to reduce OXPHOS activity, especially in GSCs, such as translocator protein (TSPO), insulin-like growth factor 2 mRNA-binding protein 2 (IGF2BP2 or IMP2), oncostatin M, and glycerol-3-phosphate dehydrogenase (GPDI). 27 TSPO is a transmembrane protein in the outer mitochondrial membrane of glial cells 200 and is highly expressed in glioma. 201 Loss of TSPO results in a shift from OXPHOS toward glycolysis. IGF2BP2 is involved in the maintenance of CSCs 202 by delivering electron transport chain subunit-encoding mRNAs to mitochondria and contributing to complex I and IV assembly. 203 Oncostatin M is a cytokine of the IL-6 subfamily and is expressed in brain by various cell types (neurons, astrocytes, and microglia) and is involved in immunosurveillance in the brain. 204 Its receptor is specifically expressed by GSCs in mitochondria and interacts with complex I to promote OXPHOS. Deletion of the oncostatin M receptor reduces OXPHOS activity, but increases ROS levels and sensitizes GSCs to irradiation. 205 GPDI is expressed by GSCs but not by NSCs. 43 GSCs express GPDI in relation with their quiescence and has been proposed as an attractive therapeutic target in glioblastoma.
Promising inhibitors of mitochondrial activity of GSCs have been described as well.107,144,195,206–210 Mudassar et al. 210 proposed to inhibit OXPHOS to increase the low oxygen levels in hypoxic GSC niches to sensitize GSCs to irradiation by repurposing antimalaria drugs such as atovaquone, ivermectin, proguanil, mefloquine, and quinacrine. Lonidamine (LND) is an antiglycolytic drug with limited clinical effects in cancer patients207,211–213 but LND in a mitochondria-targeting form (Mito-LND) appears to be a selective OXPHOS inhibitor with very low toxicity in mice. 213 These characteristics of Mito-LND makes it an attractive candidate to target CSCs in general and GSCs in glioblastoma.
Verteporfin has been Food and Drug Administration (FDA) approved to treat macular degeneration in the eye. It inhibits OXPHOS at complex III and IV and is specifically effective against GSCs and not to differentiated glioblastoma cells or normal cells. 214 Recently, verteporfin was suggested to be a therapeutic agent for epidermal growth factor receptor (EGFR)-amplified and EGFR-mutant glioblastoma on the basis of a study using cultured glioblastoma cells. 215
The FDA-approved antidiabetic drug metformin and the FDA-approved antimalaria drug chloroquine have been tested in a phase IB clinical trial to investigate whether repurposing of these drugs can optimize standard treatment of IDH1-mutated glioblastoma and other IDH1-mutated cancer types, but failed to induce a clinical response.27,197,216 Alternatively, phenformin is a lipophilic analogue of metformin and may reach higher levels in mitochondria of cancer cells and thus may be more effective in the treatment of IDH1-mutated cancer cells. 196 Metformin and phenformin inhibit complex I of OXPHOS, whereas metformin, phenformin, and chloroquine inhibit α-KG production from glutamine and glutamate by glutamate dehydrogenase. 27 Effects of the FAO inhibitor etomoxir were compared with the effects of a ketogenic diet. It appeared that etomoxir prolonged survival of mice, whereas the ketogenic diet did not affect survival or even reduced survival of the mice. It was also found that IDH1 wild-type and IDH1-mutated cells were not differently affected by a ketogenic diet although IDH1-mutated cells use FAO in a different way than IDH1 wild-type cells. 198 In another study of the effects of a ketogenic diet in vitro and in vivo reported modest effects, if any. 217 Therefore, ketogenic diets for glioblastoma patients have to be applied with care to avoid unnecessary negative effects on the quality-of-life of glioblastoma patients because these diets are difficult to maintain.
Repurposing of anti-GSC drugs was reviewed recently. 218
A different approach was proposed by Hira et al.9,10 to sensitize GSCs to radiotherapy and chemotherapy. GSCs are kept in their hypoxic periarteriolar niches in glioblastoma tumors and the SVZ 19 by SDF-1α-CXCR4 interactions in a similar way as HSCs and LSCs are kept in their hypoxic periarteriolar bone marrow niches. Inhibition of CXCR4 by FDA-approved plerixafor is used successfully to remove LSCs out of the bone marrow niches to render them more sensitive to chemotherapy and HSCs in healthy donors to be harvested in the peripheral blood for stem cell transplantation. 10 It is suggested to remove GSCs from their niches in glioblastoma before radiotherapy or chemotherapy.9,10
Concluding Remarks
The present review of the literature shows clearly that quiescent NSCs and HSCs in their niches behave very similar with respect to energy metabolism. The cells are present in a hypoxic environment and depend on anaerobic glycolysis and FAO. When needed, both stem cell types are activated and become progenitor cell types that leave the NSC and HSC niches and switch to OXPHOS and lipid synthesis and downregulate glycolysis and FAO. In both cell types, similar cell biological regulation mechanisms of their stemness and ATP metabolism are involved (Fig. 6). Moreover, mitochondria are present in both quiescent stem cell types but are only partially active (FAO and not OXPHOS) and when needed can switch rapidly to an altered metabolism in proliferating progenitor cells (OXPHOS and lipid biogenesis but not FAO).
Figure 6.
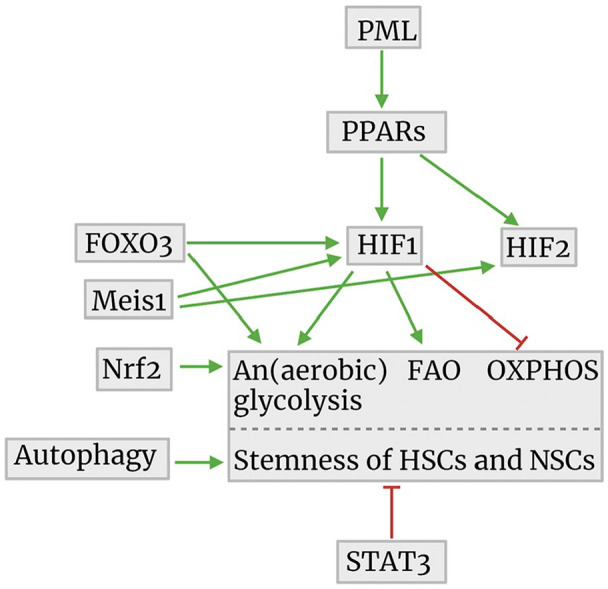
Cell biological regulation mechanisms of ATP synthesis in relationship to the maintenance of stemness of HSCs and NSCs. The image was created using Biorender.com. Abbreviations: PML, promyelocytic leukemia protein; PPARs, peroxisome proliferator-activated receptors; FOXO3, forkhead box O3; FAO, fatty acid oxidation; OXPHOS, oxidative phosphorylation; HSCs, hematopoietic stem cells; STAT3, signal transducer and activator of transcription 3; NSCs, neural stem cells; HIF, hypoxia-inducible factor; Meis1, myeloid ecotropic viral integration site 1; Nrf2, nuclear factor erythroid 2-related factor 2.
GSCs and LSCs appear to be quiescent and keep their stemness in similar ways with FAO and OXPHOS activity for ATP production. So, it seems that inhibition of OXPHOS is a therapeutic option to eradicate GSCs and LSCs to diminish the risk of recurrence of glioblastoma or AML (Fig. 7).
Figure 7.
Metabolic regulation mechanisms in relation to the maintenance of stemness of LSCs and GSCs. The image was created using Biorender.com. Abbreviations: OXPHOS, oxidative phosphorylation; MSC, mesenchymal stem cell; FAO, fatty acid oxidation; ROS, reactive oxygen species; LSCs, leukemic stem cells; GSCs, glioblastoma stem cells; GDH, glutamate dehydrogenase; α-KG, α-ketoglutarate.
However, we argued recently against energy metabolism as target for therapy in cancer. 27 First, inhibition of aerobic glycolysis in differentiated cancer cells has proven in a host of clinical trials to be either ineffective or to cause unwanted side effects.144,211–213 Second, cancer is hypothesized to be a redox disease. 219 Therefore, Watson 220 argued that physical exercise prevents cancer as well as diabetes, dementia, and cardiovascular diseases by producing low levels of ROS that are needed for correctly folding proteins in the endoplasmic reticulum. Generation of low levels of ROS reduces the risk of cancer, 221 prolongs survival of cancer patients and delays recurrence of cancer, 222 and improves quality-of-life of cancer patients including glioblastoma patients.223,224 Because ROS are generated during OXPHOS,27,67 we conclude that systemic treatment of cancer patients with OXPHOS inhibitors is not an effective therapeutic option and leads to adverse effects in patients. Therefore, disruption of the interactions between CSCs and their protective niches seems to be a more promising option as therapeutic strategy because CSCs are then mobilized out of the niches resulting in their differentiation and proliferation and ultimately their sensitization to radiotherapy and chemotherapy.9,10 This approach is currently being investigated by using the FDA-approved drug plerixafor that inhibits CXCR4 to remove LSCs from bone marrow niches before chemotherapeutic treatment in AML and multiple myeloma patients and for harvesting HSCs in patients or healthy donors for HSC transplantation (for a recent review, see Hira et al. 10 ). Clinical trials are ongoing at the moment to investigate whether removal of GSCs from their niches before irradiation or temozolomide chemotherapy of glioblastoma patients is an effective therapeutic option. 10
In conclusion, energy metabolism (OXPHOS) in leukemic and GSCs is not a likely therapeutic avenue because OXPHOS is needed for ROS production in healthy cells and tissues of cancer patients as a first-line anticancer barrier.
Supplemental Material
Supplemental material, sj-pdf-1-jhc-10.1369_00221554211054585 for Cell Biology Meets Cell Metabolism: Energy Production Is Similar in Stem Cells and in Cancer Stem Cells in Brain and Bone Marrow by Cornelis J.F. van Noorden, Barbara Breznik, Metka Novak, Amber J. van Dijck, Saloua Tanan, Miloš Vittori, Urban Bogataj, Noëlle Bakker, Joseph D. Khoury, Remco J. Molenaar and Vashendriya V.V. Hira in Journal of Histochemistry & Cytochemistry
Footnotes
Competing Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Author Contributions: The authors have contributed to this article as follows: conception and design (CJFvN, VVVH), collection and/or assembly of data (NB, MV, UB, JDK), data analysis and interpretation (CJFvN, BB, MN, MV, RJM, VVVH), initial analysis of energy metabolism in glioblastoma stem cells (AJvD) and hematopoietic stem cells (ST), fluorescence image processing (NB), electron microscopical study and imaging (MV, UB), manuscript writing (CJFvN, BB, MN, MV, RJM, VVVH), final approval of manuscript (CJFvN, BB, MN, AJvD, ST, NB, MV, UB, JDK, RJM, VVVH), and supervision of the entire study (CJFvN, BB, MV, VVVH).
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: The Slovenian Research Agency (Project J3-2526; CJFvN, BB, MN, MV, UB) and the Postdoctoral project Z3-1870 (BB), the European Program of Cross-Border Cooperation for Slovenia-Italy Interreg TRANS-GLIOMA (Program 2017; BB, MN), the IVY Interreg Fellowship (VVVH), and the Fondation pour la Recherche Nuovo-Soldati 2019 (RJM).
ORCID iD: Barbara Breznik  https://orcid.org/0000-0003-0247-5811
https://orcid.org/0000-0003-0247-5811
Contributor Information
Cornelis J.F. van Noorden, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia; Department of Medical Biology.
Barbara Breznik, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Metka Novak, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Amber J. van Dijck, Department of Medical Biology
Saloua Tanan, Department of Medical Biology.
Miloš Vittori, Amsterdam UMC Location Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands; Department of Biology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia.
Urban Bogataj, Amsterdam UMC Location Academic Medical Center, University of Amsterdam, Amsterdam, The Netherlands; Department of Biology, Biotechnical Faculty, University of Ljubljana, Ljubljana, Slovenia.
Noëlle Bakker, Department of Medical Biology.
Joseph D. Khoury, Department of Hematopathology, The University of Texas MD Anderson Cancer Center, Houston, Texas
Remco J. Molenaar, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia Department of Medical Oncology.
Vashendriya V.V. Hira, Department of Genetic Toxicology and Cancer Biology, National Institute of Biology, Ljubljana, Slovenia.
Literature Cited
- 1. Gentles AJ, Plevritis SK, Majeti R, Alizadeh AA. Association of a leukemic stem cell gene expression signature with clinical outcomes in acute myeloid leukemia. JAMA. 2010;304(24):2706–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eppert K, Takenaka K, Lechman ER, Waldron L, Nilsson B, van Galen P, Metzeler KH, Poeppl A, Ling V, Beyene J, Canty AJ, Danska JS, Bohlander SK, Buske C, Minden MD, Golub TR, Jurisica I, Ebert BL, Dick JE. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086–93. [DOI] [PubMed] [Google Scholar]
- 3. Ng SW, Mitchell A, Kennedy JA, Chen WC, McLeod J, Ibrahimova N, Arruda A, Popescu A, Gupta V, Schimmer AD, Schuh AC, Yee KW, Bullinger L, Herold T, Görlich D, Büchner T, Hiddemann W, Berdel WE, Wörmann B, Cheok M, Preudhomme C, Dombret H, Metzeler K, Buske C, Löwenberg B, Valk PJ, Zandstra PW, Minden MD, Dick JE, Wang JC. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433–7. [DOI] [PubMed] [Google Scholar]
- 4. Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, McLeod JL, Doedens M, Medeiros JJ, Marke R, Kim HJ, Lee K, McPherson JD, Hudson TJ, Brown AM, Yousif F, Trinh QM, Stein LD, Minden MD, Wang JC, Dick JE. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Auffinger B, Tobias AL, Han Y, Lee G, Guo D, Dey M, Lesniak MS, Ahmed AU. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21(7):1119–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, Parada LF. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Murat A, Migliavacca E, Gorlia T, Lambiv WL, Shay T, Hamou MF, de Tribolet N, Regli L, Wick W, Kouwenhoven MC, Hainfellner JA, Heppner FL, Dietrich PY, Zimmer Y, Cairncross JG, Janzer RC, Domany E, Delorenzi M, Stupp R, Hegi ME. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–24. [DOI] [PubMed] [Google Scholar]
- 8. Pallini R, Ricci-Vitiani L, Banna GL, Signore M, Lombardi D, Todaro M, Stassi G, Martini M, Maira G, Larocca LM, De Maria R. Cancer stem cell analysis and clinical outcome in patients with glioblastoma multiforme. Clin Cancer Res. 2008;14(24):8205–12. [DOI] [PubMed] [Google Scholar]
- 9. Hira VVV, Breznik B, Vittori M, Loncq de Jong A, Mlakar J, Oostra RJ, Khurshed M, Molenaar RJ, Lah T, Van Noorden CJF. Similarities between stem cell niches in glioblastoma and bone marrow: rays of hope for novel treatment strategies. J Histochem Cytochem. 2020;68(1):33–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hira VVV, Van Noorden CJF, Molenaar RJ. CXCR4 antagonists as stem cell mobilizers and therapy sensitizers for acute myeloid leukemia and glioblastoma? Biology. 2020;9(2):31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hira VVV, Van Noorden CJF, Carraway HE, Maciejewski JP, Molenaar RJ. Novel therapeutic strategies to target leukemic cells that hijack compartmentalized continuous hematopoietic stem cell niches. Biochim Biophys Acta Rev Cancer. 2017;1868(1):183–98. [DOI] [PubMed] [Google Scholar]
- 12. Hira VVV, Wormer JR, Kakar H, Breznik B, van der Swaan B, Hulsbos R, Tigchelaar W, Tonar Tonar, Khurshed M, Molenaar RJ, Van Noorden CJF. Periarteriolar glioblastoma stem cell niches express bone marrow hematopoietic stem cell niche proteins. J Histochem Cytochem. 2018;66(3):155–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hira VVV, Aderetti DA, van Noorden CJF. Glioma stem cell niches in human glioblastoma are periarteriolar. J Histochem Cytochem. 2018;66(5):349–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hira VV, Ploegmakers KJ, Grevers F, Verbovšek U, Silvestre-Roig C, Aronica E, Tigchelaar W, Turnšek TL, Molenaar RJ, Van Noorden CJ. CD133+ and nestin+ glioma stem-like cells reside around CD31+ arterioles in niches that express SDF-1α, CXCR4, osteopontin and cathepsin K. J Histochem Cytochem. 2015;63(7):481–93. [DOI] [PubMed] [Google Scholar]
- 15. Goffart N, Lombard A, Lallemand F, Kroonen J, Nassen J, Di Valentin E, Berendsen S, Dedobbeleer M, Willems E, Robe P, Bours V, Martin D, Martinive P, Maquet P, Rogister B. CXCL12 mediates glioblastoma resistance to radiotherapy in the subventricular zone. Neuro Oncol. 2017;19(1):66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Arai F, Hirao A, Ohmura M, Sato H, Matsuoka S, Takubo K, Ito K, Koh GY, Suda T. Tie2/angiopoietin-1 signaling regulates hematopoietic stem cell quiescence in the bone marrow niche. Cell. 2004;118(2):149–61. [DOI] [PubMed] [Google Scholar]
- 17. Saharinen P, Eklund L, Miettinen J, Wirkkala R, Anisimov A, Winderlich M, Nottebaum A, Vestweber D, Deutsch U, Koh GY, Olsen BR, Alitalo K. Angiopoietins assemble distinct Tie2 signalling complexes in endothelial cell-cell and cell-matrix contacts. Nat Cell Biol. 2008;10(5):527–37. [DOI] [PubMed] [Google Scholar]
- 18. Brockman AA, Mobley BC, Ihrie RA. Histological studies of the ventricular-subventricular zone as neural stem cell and glioma stem cell niche. J Histochem Cytochem. Epub 2021. Jul 26. doi: 10.1369/00221554211032003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hira VVV, Molenaar RJ, Breznik B, Lah T, Aronica E, Van Noorden CJF. Immunohistochemical detection of neural stem cells and glioblastoma stem cells in the subventricular zone of glioblastoma patients. J Histochem Cytochem. 2021;69:349–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lee JH, Lee JE, Kahng JY, Kim SH, Park JS, Yoon SJ, Um JY, Kim WK, Lee JK, Park J, Kim EH, Lee JH, Lee JH, Chung WS, Ju YS, Park SH, Chang JH, Kang SG, Lee JH. Human glioblastoma arises from subventricular zone cells with low-level driver mutations. Nature. 2018;560(7717):243–7. [DOI] [PubMed] [Google Scholar]
- 21. Sanai N, Alvarez-Buylla A, Berger MS. Neural stem cells and the origin of gliomas. N Engl J Med. 2005;353(8):811–22. [DOI] [PubMed] [Google Scholar]
- 22. Qin EY, Cooper DD, Abbott KL, Lennon J, Nagaraja S, Mackay A, Jones C, Vogel H, Jackson PK, Monje M. Neural precursor-derived pleiotrophin mediates subventricular zone invasion by glioma. Cell. 2017;170(5):845–59.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jambor H, Antonietti A, Alicea B, Audisio TL, Auer S, Bhardwaj V, Burgess SJ, Ferling I, Gazda MA, Hoeppner LH, Ilangovan V, Lo H, Olson M, Mohamed SY, Sarabipour S, Varma A, Walavalkar K, Wissink EM, Weissgerber TL. Creating clear and informative image-based figures for scientific publications. PLoS Biol. 2021;19(3):e3001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. El Achi H, Dupont E, Paul S, Khoury JD. CD123 as a biomarker in hematolymphoid malignancies: principles of detection and targeted therapies. Cancers. 2020;12(11):3087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Testa U, Castelli G, Pelosi E. Isocitrate dehydrogenase mutations in myelodysplastic syndromes and in acute myeloid leukemias. Cancers. 2020;12(9):2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Panuzzo C, Jovanovski A, Pergolizzi B, Pironi L, Stanga S, Fava C, Cilloni D. Mitochondria: a galaxy in the hematopoietic and leukemic stem cell universe. Int J Mol Sci. 2020;21(11):3928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. van Noorden CJF, Hira VVV, van Dijck AJ, Novak M, Breznik B, Molenaar RJ. Energy metabolism in IDH1 wild-type and IDH1-mutated glioblastoma stem cells: a novel target for therapy? Cells. 2021;10(3):705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Molenaar RJ, Radivoyevitch T, Maciejewski JP, van Noorden CJ, Bleeker FE. The driver and passenger effects of isocitrate dehydrogenase 1 and 2 mutations in oncogenesis and survival prolongation. Biochim Biophys Acta. 2014;1846(2):326–41. [DOI] [PubMed] [Google Scholar]
- 29. McCormack RM, Zhu P, Dono A, Takayasu T, Bhatia A, Blanco AI, Tandon N, Ostrom QT, Gonzales A, Moreno S, Ballester LY, Esquenazi Y. Role of ethnicity and geographic location on glioblastoma IDH1/IDH2 mutations. World Neurosurg. 2021;149:e894–912. [DOI] [PubMed] [Google Scholar]
- 30. Vaupel P, Multhoff G. Revisiting the Warburg effect: historical dogma versus current understanding. J Physiol. 2021;599(6):1745–57. [DOI] [PubMed] [Google Scholar]
- 31. Glancy B, Kim Y, Katti P, Willingham TB. The functional impact of mitochondrial structure across subcellular scales. Front Physiol. 2020;11:541040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hackenbrock CR. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. I. Reversible ultrastructural changes with change in metabolic steady state in isolated liver mitochondria. J Cell Biol. 1966;30(2):269–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hackenbrock CR. Ultrastructural bases for metabolically linked mechanical activity in mitochondria. II. Electron transport-linked ultrastructural transformations in mitochondria. J Cell Biol. 1968;37(2):345–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brandt T, Mourier A, Tain LS, Partridge L, Larsson NG, Kühlbrandt W. Changes of mitochondrial ultrastructure and function during ageing in mice and Drosophila. Elife. 2017;6:e24662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zick M, Rabl R, Reichert AS. Cristae formation-linking ultrastructure and function of mitochondria. Biochim Biophys Acta. 2009;1793(1):5–19. [DOI] [PubMed] [Google Scholar]
- 36. Paumard P, Vaillier J, Coulary B, Schaeffer J, Soubannier V, Mueller DM, Brèthes D, di Rago JP, Velours J. The ATP synthase is involved in generating mitochondrial cristae morphology. Embo J. 2002;21(3):221–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Davies KM, Anselmi C, Wittig I, Faraldo-Gómez JD, Kühlbrandt W. Structure of the yeast F1F0-ATP synthase dimer and its role in shaping the mitochondrial cristae. Proc Natl Acad Sci USA. 2012;109(34):13602–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Davies KM, Strauss M, Daum B, Kief JH, Osiewacz HD, Rycovska A, Zickermann V, Kühlbrandt W. Macromolecular organization of ATP synthase and complex I in whole mitochondria. Proc Natl Acad Sci USA. 2011;108(34):14121–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baum T, Gama V. Dynamic properties of mitochondria during human corticogenesis. Development. 2021;148(4):dev194183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Iwata R, Vanderhaeghen P. Regulatory roles of mitochondria and metabolism in neurogenesis. Curr Opin Neurobiol. 2021;69:231–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Vallejo FA, Shah SS, de Cordoba N, Walters WM, Prince J, Khatib Z, Komotar RJ, Vanni S, Graham RM. The contribution of ketone bodies to glycolytic inhibition for the treatment of adult and pediatric glioblastoma. J Neurooncol. 2020;147(2):317–26. [DOI] [PubMed] [Google Scholar]
- 42. Van Noorden CJ, Frederiks WM. Cerium methods for light and electron microscopical histochemistry. J Microsc. 1993;171(Pt 1):3–16. [DOI] [PubMed] [Google Scholar]
- 43. Rusu P, Shao C, Neuerburg A, Acikgöz AA, Wu Y, Zou P, Phapale P, Shankar TS, Döring K, Dettling S, Körkel-Qu H, Bekki G, Costa B, Guo T, Friesen O, Schlotter M, Heikenwalder M, Tschaharganeh DF, Bukau B, Kramer G, Angel P, Herold-Mende C, Radlwimmer B, Liu HK. GPD1 specifically marks dormant glioma stem cells with a distinct metabolic profile. Cell Stem Cell. 2019;25(2):241–57.e8. [DOI] [PubMed] [Google Scholar]
- 44. Bogataj U, Mrak P, Štrus J, Žnidaršič N. Ultrastructural differentiation of plasma membrane and cell junctions in the hindgut cells is synchronized with key developmental transitions in Porcellio scaber. Arthropod Struct Dev. 2019;50:78–93. [DOI] [PubMed] [Google Scholar]
- 45. Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even Warburg did not anticipate. Cancer Cell. 2012;21(3):297–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Duraj T, García-Romero N, Carrión-Navarro J, Madurga R, Mendivil AO, Prat-Acin R, Garcia-Cañamaque L, Ayuso-Sacido A. Beyond the Warburg effect: oxidative and glycolytic phenotypes coexist within the metabolic heterogeneity of glioblastoma. Cells. 2021;10(2):202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Koehler A, Van Noorden CJ. Reduced nicotinamide adenine dinucleotide phosphate and the higher incidence of pollution-induced liver cancer in female flounder. Environ Toxicol Chem. 2003;22(11):2703–10. [DOI] [PubMed] [Google Scholar]
- 48. Filippi MD, Ghaffari S. Mitochondria in the maintenance of hematopoietic stem cells: new perspectives and opportunities. Blood. 2019;133(18):1943–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Nombela-Arrieta C, Pivarnik G, Winkel B, Canty KJ, Harley B, Mahoney JE, Park SY, Lu J, Protopopov A, Silberstein LE. Quantitative imaging of haematopoietic stem and progenitor cell localization and hypoxic status in the bone marrow microenvironment. Nat Cell Biol. 2013;15(5):533–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ito K, Hirao A, Arai F, Takubo K, Matsuoka S, Miyamoto K, Ohmura M, Naka K, Hosokawa K, Ikeda Y, Suda T. Reactive oxygen species act through p38 MAPK to limit the lifespan of hematopoietic stem cells. Nat Med. 2006;12(4):446–51. [DOI] [PubMed] [Google Scholar]
- 51. Miyamoto K, Araki KY, Naka K, Arai F, Takubo K, Yamazaki S, Matsuoka S, Miyamoto T, Ito K, Ohmura M, Chen C, Hosokawa K, Nakauchi H, Nakayama K, Nakayama KI, Harada M, Motoyama N, Suda T, Hirao A. Foxo3a is essential for maintenance of the hematopoietic stem cell pool. Cell Stem Cell. 2007;1(1):101–12. [DOI] [PubMed] [Google Scholar]
- 52. Tesio M, Golan K, Corso S, Giordano S, Schajnovitz A, Vagima Y, Shivtiel S, Kalinkovich A, Caione L, Gammaitoni L, Laurenti E, Buss EC, Shezen E, Itkin T, Kollet O, Petit I, Trumpp A, Christensen J, Aglietta M, Piacibello W, Lapidot T. Enhanced c-Met activity promotes G-CSF-induced mobilization of hematopoietic progenitor cells via ROS signaling. Blood. 2011;117(2):419–28. [DOI] [PubMed] [Google Scholar]
- 53. Golan K, Vagima Y, Ludin A, Itkin T, Cohen-Gur S, Kalinkovich A, Kollet O, Kim C, Schajnovitz A, Ovadya Y, Lapid K, Shivtiel S, Morris AJ, Ratajczak MZ, Lapidot T. S1P promotes murine progenitor cell egress and mobilization via S1P1-mediated ROS signaling and SDF-1 release. Blood. 2012;119(11):2478–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Guo B, Huang X, Lee MR, Lee SA, Broxmeyer HE. Antagonism of PPAR-γ signaling expands human hematopoietic stem and progenitor cells by enhancing glycolysis. Nat Med. 2018;24(3):360–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kocabas F, Xie L, Xie J, Yu Z, DeBerardinis RJ, Kimura W, Thet S, Elshamy AF, Abouellail H, Muralidhar S, Liu X, Chen C, Sadek HA, Zhang CC, Zheng J. Hypoxic metabolism in human hematopoietic stem cells. Cell Biosci. 2015;5:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Daud H, Browne S, Al-Majmaie R, Murphy W, Al-Rubeai M. Metabolic profiling of hematopoietic stem and progenitor cells during proliferation and differentiation into red blood cells. N Biotechnol. 2016;33(1):179–86. [DOI] [PubMed] [Google Scholar]
- 57. Cabon L, Bertaux A, Brunelle-Navas MN, Nemazanyy I, Scourzic L, Delavallée L, Vela L, Baritaud M, Bouchet S, Lopez C, Quang Van V, Garbin K, Chateau D, Gilard F, Sarfati M, Mercher T, Bernard OA, Susin SA. AIF loss deregulates hematopoiesis and reveals different adaptive metabolic responses in bone marrow cells and thymocytes. Cell Death Differ. 2018;25(5):983–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bertaux A, Cabon L, Brunelle-Navas MN, Bouchet S, Nemazanyy I, Susin SA. Mitochondrial OXPHOS influences immune cell fate: lessons from hematopoietic AIF-deficient and NDUFS4-deficient mouse models. Cell Death Dis. 2018;9(6):581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Messina-Graham S, Broxmeyer H. SDF-1/CXCL12 modulates mitochondrial respiration of immature blood cells in a bi-phasic manner. Blood Cells Mol Dis. 2016;58:13–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gu XX, Tang ZZ, He YL, Zeng ZN, Shi WX, Qiao YC, Wei YS. A functional polymorphism in HIF-3α is related to an increased risk of ischemic stroke. J Mol Neurosci. 2021;71(5):1061–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Downes NL, Laham-Karam N, Kaikkonen MU, Ylä-Herttuala S. Differential but complementary HIF1α and HIF2α transcriptional regulation. Mol Ther. 2018;26(7):1735–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee JW, Ko J, Ju C, Eltzschig HK. Hypoxia signaling in human diseases and therapeutic targets. Exp Mol Med. 2019;51(6):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ratcliffe PJ. HIF-1 and HIF-2: working alone or together in hypoxia? J Clin Invest. 2007;117(4):862–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Zhang Q, Han Z, Zhu Y, Chen J, Li W. Role of hypoxia inducible factor-1 in cancer stem cells (Review). Mol Med Rep. 2021;23(1):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Duan C. Hypoxia-inducible factor 3 biology: complexities and emerging themes. Am J Physiol Cell Physiol. 2016;310(4):C260–9. [DOI] [PubMed] [Google Scholar]
- 66. Yang L, Shi P, Zhao G, Xu J, Peng W, Zhang J, Zhang G, Wang X, Dong Z, Chen F, Cui H. Targeting cancer stem cell pathways for cancer therapy. Signal Transduct Target Ther. 2020;5(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang CC, Sadek HA. Hypoxia and metabolic properties of hematopoietic stem cells. Antioxid Redox Signal. 2014;20(12):1891–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Sun X, Lv X, Yan Y, Zhao Y, Ma R, He M, Wei M. Hypoxia-mediated cancer stem cell resistance and targeted therapy. Biomed Pharmacother. 2020;130:110623. [DOI] [PubMed] [Google Scholar]
- 69. Luchsinger LL, de Almeida MJ, Corrigan DJ, Mumau M, Snoeck HW. Mitofusin 2 maintains haematopoietic stem cells with extensive lymphoid potential. Nature. 2016;529(7587):528–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Ito K, Ito K. Metabolism and the control of cell fate decisions and stem cell renewal. Annu Rev Cell Dev Biol. 2016;32:399–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. El Hout M, Cosialls E, Mehrpour M, Hamaï A. Crosstalk between autophagy and metabolic regulation of cancer stem cells. Mol Cancer. 2020;19(1):27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, Passegué E. Autophagy maintains the metabolism and function of young and old stem cells. Nature. 2017;543(7644):205–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, Pandolfi PP. A PML–PPAR-δ pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hua TNM, Oh J, Kim S, Antonio JM, Vo VTA, Om J, Choi JW, Kim JY, Jung CW, Park MJ, Jeong Y. Peroxisome proliferator-activated receptor gamma as a theragnostic target for mesenchymal-type glioblastoma patients. Exp Mol Med. 2020;52(4):629–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Mrowka P, Glodkowska-Mrowka E. PPARγ agonists in combination cancer therapies. Curr Cancer Drug Targets. 2020;20(3):197–215. [DOI] [PubMed] [Google Scholar]
- 76. Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not PBX1b. Embo J. 1998;17(13):3714–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Spieker N, van Sluis P, Beitsma M, Boon K, van Schaik BD, van Kampen AH, Caron H, Versteeg R. The MEIS1 oncogene is highly expressed in neuroblastoma and amplified in cell line IMR32. Genomics. 2001;71(2):214–21. [DOI] [PubMed] [Google Scholar]
- 78. Jones TA, Flomen RH, Senger G, Nizetić D, Sheer D. The homeobox gene MEIS1 is amplified in IMR-32 and highly expressed in other neuroblastoma cell lines. Eur J Cancer. 2000;36(18):2368–74. [DOI] [PubMed] [Google Scholar]
- 79. Aksoz M, Turan RD, Albayrak E, Kocabas F. Emerging roles of meis1 in cardiac regeneration, stem cells and cancer. Curr Drug Targets. 2018;19(2):181–90. [DOI] [PubMed] [Google Scholar]
- 80. Turan RD, Albayrak E, Uslu M, Siyah P, Alyazici LY, Kalkan BM, Aslan GS, Yucel D, Aksoz M, Tuysuz EC, Meric N, Durdagi S, Gulbas Z, Kocabas F. Development of small molecule MEIS inhibitors that modulate HSC activity. Sci Rep. 2020;10(1):7994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kocabas F, Zheng J, Thet S, Copeland NG, Jenkins NA, DeBerardinis RJ, Zhang C, Sadek HA. Meis1 regulates the metabolic phenotype and oxidant defense of hematopoietic stem cells. Blood. 2012;120(25):4963–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Becker MW, Jordan CT. Leukemia stemness signatures step toward the clinic. Cell Stem Cell. 2011;9(3):185–6. [DOI] [PubMed] [Google Scholar]
- 83. Jiramongkol Y, Lam EW. FOXO transcription factor family in cancer and metastasis. Cancer Metastasis Rev. 2020;39(3):681–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Firat E, Niedermann G. FoxO proteins or loss of functional p53 maintain stemness of glioblastoma stem cells and survival after ionizing radiation plus PI3K/mTOR inhibition. Oncotarget. 2016;7(34):54883–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Calissi G, Lam EW, Link W. Therapeutic strategies targeting FOXO transcription factors. Nat Rev Drug Discov. 2021;20(1):21–38. [DOI] [PubMed] [Google Scholar]
- 86. Libby CJ, McConathy J, Darley-Usmar V, Hjelmeland AB. The role of metabolic plasticity in blood and brain stem cell pathophysiology. Cancer Res. 2020;80(1):5–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wilhelm K, Happel K, Eelen G, Schoors S, Oellerich MF, Lim R, Zimmermann B, Aspalter IM, Franco CA, Boettger T, Braun T, Fruttiger M, Rajewsky K, Keller C, Brüning JC, Gerhardt H, Carmeliet P, Potente M. FOXO1 couples metabolic activity and growth state in the vascular endothelium. Nature. 2016;529(7585):216–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Dumas SJ, García-Caballero M, Carmeliet P. Metabolic signatures of distinct endothelial phenotypes. Trends Endocrinol Metab. 2020;31(8):580–95. [DOI] [PubMed] [Google Scholar]
- 89. Farhan M, Silva M, Xingan X, Huang Y, Zheng W. Role of FOXO transcription factors in cancer metabolism and angiogenesis. Cells. 2020;9(7):1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Xu K, Zhang Z, Pei H, Wang H, Li L, Xia Q. FoxO3a induces temozolomide resistance in glioblastoma cells via the regulation of β-catenin nuclear accumulation. Oncol Rep. 2017;37(4):2391–7. [DOI] [PubMed] [Google Scholar]
- 91. Ghaffari S. Lysosomal regulation of metabolism in quiescent hematopoietic stem cells: more than just autophagy. Cell Stem Cell. 2021;28(3):374–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Zamame Ramirez JA, Romagnoli GG, Kaneno R. Inhibiting autophagy to prevent drug resistance and improve anti-tumor therapy. Life Sci. 2021;265:118745. [DOI] [PubMed] [Google Scholar]
- 93. Liang R, Arif T, Kalmykova S, Kasianov A, Lin M, Menon V, Qiu J, Bernitz JM, Moore K, Lin F, Benson DL, Tzavaras N, Mahajan M, Papatsenko D, Ghaffari S. Restraining lysosomal activity preserves hematopoietic stem cell quiescence and potency. Cell Stem Cell. 2020;26(3):359–76.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Ballabio A, Bonifacino JS. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat Rev Mol Cell Biol. 2020;21(2):101–18. [DOI] [PubMed] [Google Scholar]
- 95. Kobayashi T, Piao W, Takamura T, Kori H, Miyachi H, Kitano S, Iwamoto Y, Yamada M, Imayoshi I, Shioda S, Ballabio A, Kageyama R. Enhanced lysosomal degradation maintains the quiescent state of neural stem cells. Nat Commun. 2019;10(1):5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Leeman DS, Hebestreit K, Ruetz T, Webb AE, McKay A, Pollina EA, Dulken BW, Zhao X, Yeo RW, Ho TT, Mahmoudi S, Devarajan K, Passegué E, Rando TA, Frydman J, Brunet A. Lysosome activation clears aggregates and enhances quiescent neural stem cell activation during aging. Science. 2018;359(6381):1277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Khan I, Baig MH, Mahfooz S, Rahim M, Karacam B, Elbasan EB, Ulasov I, Dong JJ, Hatiboglu MA. Deciphering the role of autophagy in treatment of resistance mechanisms in glioblastoma. Int J Mol Sci. 2021;22(3):1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Shi J, Dong X, Li H, Wang H, Jiang Q, Liu L, Wang L, Dong J. Nicardipine sensitizes temozolomide by inhibiting autophagy and promoting cell apoptosis in glioma stem cells. Aging. 2021;13(5):6820–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Lim SM, Mohamad Hanif EA, Chin SF. Is targeting autophagy mechanism in cancer a good approach? The possible double-edge sword effect. Cell Biosci. 2021;11(1):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Bejarano-García JA, Millán-Uclés Rosado ÁIV, Sánchez-Abarca LI, Caballero-Velázquez T, Durán-Galván MJ, Pérez-Simón JA, Piruat JI. Sensitivity of hematopoietic stem cells to mitochondrial dysfunction by SdhD gene deletion. Cell Death Dis. 2016;7(12):e2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9(4):298–310. [DOI] [PubMed] [Google Scholar]
- 102. Nakamura-Ishizu A, Ito K, Suda T. Hematopoietic stem cell metabolism during development and aging. Dev Cell. 2020;54(2):239–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Takubo K, Nagamatsu G, Kobayashi CI, Nakamura-Ishizu A, Kobayashi H, Ikeda E, Goda N, Rahimi Y, Johnson RS, Soga T, Hirao A, Suematsu M, Suda T. Regulation of glycolysis by PDK functions as a metabolic checkpoint for cell cycle quiescence in hematopoietic stem cells. Cell Stem Cell. 2013;12(1):49–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhang C, Yi W, Li F, Du X, Wang H, Wu P, Peng C, Luo M, Hua W, Wong CC, Lee JJ, Li W, Chen Z, Ying S, Ju Z, Shen H. Eosinophil-derived CCL-6 impairs hematopoietic stem cell homeostasis. Cell Res. 2018;28(3):323–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Spencer JA, Ferraro F, Roussakis E, Klein A, Wu J, Runnels JM, Zaher W, Mortensen LJ, Alt C, Turcotte R, Yusuf R, Côté D, Vinogradov SA, Scadden DT, Lin CP. Direct measurement of local oxygen concentration in the bone marrow of live animals. Nature. 2014;508(7495):269–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Maffezzini C, Calvo-Garrido J, Wredenberg A, Freyer C. Metabolic regulation of neurodifferentiation in the adult brain. Cell Mol Life Sci. 2020;77(13):2483–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Libby CJ, Tran AN, Scott SE, Griguer C, Hjelmeland AB. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochim Biophys Acta Rev Cancer. 2018;1869(2):175–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Navarro Negredo P, Yeo RW, Brunet A. Aging and rejuvenation of neural stem cells and their niches. Cell Stem Cell. 2020;27(2):202–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Bolborea M, Dale N. Hypothalamic tanycytes: potential roles in the control of feeding and energy balance. Trends Neurosci. 2013;36(2):91–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Castinetti F, Davis SW, Brue T, Camper SA. Pituitary stem cell update and potential implications for treating hypopituitarism. Endocr Rev. 2011;32(4):453–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Pellegrino G, Trubert C, Terrien J, Pifferi F, Leroy D, Loyens A, Migaud M, Baroncini M, Maurage CA, Fontaine C, Prévot V, Sharif A. A comparative study of the neural stem cell niche in the adult hypothalamus of human, mouse, rat and gray mouse lemur (Microcebus murinus). J Comp Neurol. 2018;526(9):1419–43. [DOI] [PubMed] [Google Scholar]
- 112. Guan D, Kao HY. The function, regulation and therapeutic implications of the tumor suppressor protein, PML. Cell Biosci. 2015;5:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Cheng X, Guo S, Liu Y, Chu H, Hakimi P, Berger NA, Hanson RW, Kao HY. Ablation of promyelocytic leukemia protein (PML) re-patterns energy balance and protects mice from obesity induced by a Western diet. J Biol Chem. 2013;288(41):29746–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Carracedo A, Weiss D, Leliaert AK, Bhasin M, de Boer VC, Laurent G, Adams AC, Sundvall M, Song SJ, Ito K, Finley LS, Egia A, Libermann T, Gerhart-Hines Z, Puigserver P, Haigis MC, Maratos-Flier E, Richardson AL, Schafer ZT, Pandolfi PP. A metabolic prosurvival role for PML in breast cancer. J Clin Invest. 2012;122(9):3088–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Zhou W, Bao S. PML-mediated signaling and its role in cancer stem cells. Oncogene. 2014;33(12):1475–84. [DOI] [PubMed] [Google Scholar]
- 116. Ito K, Bernardi R, Morotti A, Matsuoka S, Saglio G, Ikeda Y, Rosenblatt J, Avigan DE, Teruya-Feldstein J, Pandolfi PP. PML targeting eradicates quiescent leukaemia-initiating cells. Nature. 2008;453(7198):1072–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Bernardi R, Guernah I, Jin D, Grisendi S, Alimonti A, Teruya-Feldstein J, Cordon-Cardo C, Simon MC, Rafii S, Pandolfi PP. PML inhibits HIF-1alpha translation and neoangiogenesis through repression of mTOR. Nature. 2006;442(7104):779–85. [DOI] [PubMed] [Google Scholar]
- 118. Marshall CA, Suzuki SO, Goldman JE. Gliogenic and neurogenic progenitors of the subventricular zone: who are they, where did they come from, and where are they going? Glia. 2003;43(1):52–61. [DOI] [PubMed] [Google Scholar]
- 119. Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, Song H. In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell. 2011;145(7):1142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Marques BL, Carvalho GA, Freitas EMM, Chiareli RA, Barbosa TG, Di Araújo AGP, Nogueira YL, Ribeiro RI, Parreira RC, Vieira MS, Resende RR, Gomez RS, Oliveira-Lima OC, Pinto MCX. The role of neurogenesis in neurorepair after ischemic stroke. Semin Cell Dev Biol. 2019;95:98–110. [DOI] [PubMed] [Google Scholar]
- 121. Dillen Y, Kemps H, Gervois P, Wolfs E, Bronckaers A. Adult neurogenesis in the subventricular zone and its regulation after ischemic stroke: implications for therapeutic approaches. Transl Stroke Res. 2020;11(1):60–79. [DOI] [PubMed] [Google Scholar]
- 122. Almeida AS, Vieira HLA. Role of cell metabolism and mitochondrial function during adult neurogenesis. Neurochem Res. 2017;42(6):1787–94. [DOI] [PubMed] [Google Scholar]
- 123. Li G, Fang L, Fernández G, Pleasure SJ. The ventral hippocampus is the embryonic origin for adult neural stem cells in the dentate gyrus. Neuron. 2013;78(4):658–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Stoll EA, Makin R, Sweet IR, Trevelyan AJ, Miwa S, Horner PJ, Turnbull DM. Neural stem cells in the adult subventricular zone oxidize fatty acids to produce energy and support neurogenic activity. Stem Cells. 2015;33(7):2306–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Reilly SM, Lee CH. PPAR delta as a therapeutic target in metabolic disease. FEBS Lett. 2008;582(1):26–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Knobloch M, Pilz GA, Ghesquière B, Kovacs WJ, Wegleiter T, Moore DL, Hruzova M, Zamboni N, Carmeliet P, Jessberger S. A fatty acid oxidation-dependent metabolic shift regulates adult neural stem cell activity. Cell Rep. 2017;20(9):2144–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, Ma L, Hamm M, Gage FH, Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128. Khacho M, Slack RS. Mitochondrial activity in the regulation of stem cell self-renewal and differentiation. Curr Opin Cell Biol. 2017;49:1–8. [DOI] [PubMed] [Google Scholar]
- 129. Calvo-Garrido J, Maffezzini C, Schober FA, Clemente P, Uhlin E, Kele M, Stranneheim H, Lesko N, Bruhn H, Svenningsson P, Falk A, Wedell A, Freyer C, Wredenberg A. SQSTM1/p62-directed metabolic reprogramming is essential for normal neurodifferentiation. Stem Cell Reports. 2019;12(4):696–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Mlody B, Lorenz C, Inak G, Prigione A. Energy metabolism in neuronal/glial induction and in iPSC models of brain disorders. Semin Cell Dev Biol. 2016;52:102–9. [DOI] [PubMed] [Google Scholar]
- 132. Yao B, Christian KM, He C, Jin P, Ming GL, Song H. Epigenetic mechanisms in neurogenesis. Nat Rev Neurosci. 2016;17(9):537–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Bifari F, Dolci S, Bottani E, Pino A, Di Chio M, Zorzin S, Ragni M, Zamfir RG, Brunetti D, Bardelli D, Delfino P, Cattaneo MG, Bordo R, Tedesco L, Rossi F, Bossolasco P, Corbo V, Fumagalli G, Nisoli E, Valerio A, Decimo I. Complete neural stem cell (NSC) neuronal differentiation requires a branched chain amino acids-induced persistent metabolic shift towards energy metabolism. Pharmacol Res. 2020;158:104863. [DOI] [PubMed] [Google Scholar]
- 135. Fernandes MB, Costa M, Ribeiro MF, Siquenique S Sá Santos S, Martins J, Coelho AV, Silva MFB, Rodrigues CMP, Solá S. Reprogramming of lipid metabolism as a new driving force behind tauroursodeoxycholic acid-induced neural stem cell proliferation. Front Cell Dev Biol. 2020;8:335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Baumann HJ, Betonio P, Abeywickrama CS, Shriver LP, Leipzig ND. Metabolomic and signaling programs induced by immobilized versus soluble IFN γ in neural stem cells. Bioconjug Chem. 2020;31(9):2125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Su Y, Zhang W, Patro CPK, Zhao J, Mu T, Ma Z, Xu J, Ban K, Yi C, Zhou Y. STAT3 regulates mouse neural progenitor proliferation and differentiation by promoting mitochondrial metabolism. Front Cell Dev Biol. 2020;8:362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lleonart ME, Abad E, Graifer D, Lyakhovich A. Reactive oxygen species-mediated autophagy defines the fate of cancer stem cells. Antioxid Redox Signal. 2018;28(11):1066–79. [DOI] [PubMed] [Google Scholar]
- 139. Choi BH, Kim JM, Kwak MK. The multifaceted role of NRF2 in cancer progression and cancer stem cells maintenance. Arch Pharm Res. 2021;44(3):263–80. [DOI] [PubMed] [Google Scholar]
- 140. Holmström KM, Kostov RV, Dinkova-Kostova AT. The multifaceted role of Nrf2 in mitochondrial function. Curr Opin Toxicol. 2016;1:80–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Hawkins KE, Joy S, Delhove JM, Kotiadis VN, Fernandez E, Fitzpatrick LM, Whiteford JR, King PJ, Bolanos JP, Duchen MR, Waddington SN, McKay TR. NRF2 orchestrates the metabolic shift during induced pluripotent stem cell reprogramming. Cell Rep. 2016;14(8):1883–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Levy DE, Lee CK. What does STAT3 do? J Clin Invest. 2002;109(9):1143–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Liu S, Zhang C, Wang B, Zhang H, Qin G, Li C, Cao L, Gao Q, Ping Y, Zhang K, Lian J, Zhao Q, Wang D, Zhang Z, Zhao X, Yang L, Huang L, Yang B, Zhang Y. Regulatory T cells promote glioma cell stemness through TGF-β-NF-κB-IL6-STAT3 signaling. Cancer Immunol Immunother. 2021;70:2601–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144. Li H, Feng Z, He ML. Lipid metabolism alteration contributes to and maintains the properties of cancer stem cells. Theranostics. 2020;10(16):7053–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145. Mokgautsi N, Wen YT, Lawal B, Khedkar H, Sumitra MR, Wu ATH, Huang HS. An integrated bioinformatics study of a novel niclosamide derivative, NSC765689, a potential GSK3β/β-catenin/STAT3/CD44 suppressor with anti-glioblastoma properties. Int J Mol Sci. 2021;22(5):2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Nie Z, Cai S, Wei Z, Li Y, Bian L, Wang C, Wang X, Wang C. SH3GL3 acts as a novel tumor suppressor in glioblastoma tumorigenesis by inhibiting STAT3 signaling. Biochem Biophys Res Commun. 2021;544:73–80. [DOI] [PubMed] [Google Scholar]
- 147. Liu HW, Su YK, Bamodu OA, Hueng DY, Lee WH, Huang CC, Deng L, Hsiao M, Chien MH, Yeh CT, Lin CM. The disruption of the β-catenin/TCF-1/STAT3 signaling axis by 4-acetylantroquinonol B inhibits the tumorigenesis and cancer stem-cell-like properties of glioblastoma cells, in vitro and in vivo. Cancers. 2018;10(12):491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Vasconcelos ESJ, Simão D, Terrasso AP, Silva MM, Brito C, Isidro IA, Alves PM, Carrondo MJT. Unveiling dynamic metabolic signatures in human induced pluripotent and neural stem cells. PloS Comput Biol. 2020;16(4):e1007780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Bonnay F, Veloso A, Steinmann V, Köcher T, Abdusselamoglu MD, Bajaj S, Rivelles E, Landskron L, Esterbauer H, Zinzen RP, Knoblich JA. Oxidative metabolism drives immortalization of neural stem cells during tumorigenesis. Cell. 2020;182(6):1490–507.e19. [DOI] [PubMed] [Google Scholar]
- 150. Rashkovan M, Ferrando A. Metabolic dependencies and vulnerabilities in leukemia. Genes Dev. 2019;33(21–2):1460–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Chen Y, Liang Y, Luo X, Hu Q. Oxidative resistance of leukemic stem cells and oxidative damage to hematopoietic stem cells under pro-oxidative therapy. Cell Death Dis. 2020;11(4):291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Raffel S, Klimmeck D, Falcone M, Demir A, Pouya A, Zeisberger P, Lutz C, Tinelli M, Bischel O, Bullinger L, Thiede C, Flörcken A, Westermann J, Ehninger G, Ho AD, Müller-Tidow C, Gu Z, Herrmann C, Krijgsveld J, Trumpp A, Hansson J. Quantitative proteomics reveals specific metabolic features of acute myeloid leukemia stem cells. Blood. 2020;136(13):1507–19. [DOI] [PubMed] [Google Scholar]
- 153. Mattes K, Vellenga E, Schepers H. Differential redox-regulation and mitochondrial dynamics in normal and leukemic hematopoietic stem cells: a potential window for leukemia therapy. Crit Rev Oncol Hematol. 2019;144:102814. [DOI] [PubMed] [Google Scholar]
- 154. Jones CL, Stevens BM, D’Alessandro A, Reisz JA, Culp-Hill R, Nemkov T, Pei S, Khan N, Adane B, Ye H, Krug A, Reinhold D, Smith C, DeGregori J, Pollyea DA, Jordan CT. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell. 2018;34(5):724–40.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Jacque N, Ronchetti AM, Larrue C, Meunier G, Birsen R, Willems L, Saland E, Decroocq J, Maciel TT, Lambert M, Poulain L, Hospital MA, Sujobert P, Joseph L, Chapuis N, Lacombe C, Moura IC, Demo S, Sarry JE, Recher C, Mayeux P, Tamburini J, Bouscary D. Targeting glutaminolysis has antileukemic activity in acute myeloid leukemia and synergizes with BCL-2 inhibition. Blood. 2015;126(11):1346–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Gregory MA, Nemkov T, Park HJ, Zaberezhnyy V, Gehrke S, Adane B, Jordan CT, Hansen KC, D’Alessandro A, DeGregori J. Targeting glutamine metabolism and redox state for leukemia therapy. Clin Cancer Res. 2019;25(13):4079–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Pollyea DA, Stevens BM, Jones CL, Winters A, Pei S, Minhajuddin M, D’Alessandro A, Culp-Hill R, Riemondy KA, Gillen AE, Hesselberth JR, Abbott D, Schatz D, Gutman JA, Purev E, Smith C, Jordan CT. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med. 2018;24(12):1859–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Larrue C, Guiraud N, Mouchel PL, Dubois M, Farge T, Gotanègre M, Bosc C, Saland E, Nicolau-Travers ML, Sabatier M, Serhan N, Sahal A, Boet E, Mouche S, Heydt Q, Aroua N, Stuani L, Kaoma T, Angenendt L, Mikesch JH, Schliemann C, Vergez F, Tamburini J, Récher C, Sarry JE. Adrenomedullin-CALCRL axis controls relapse-initiating drug tolerant acute myeloid leukemia cells. Nat Commun. 2021;12(1):422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Polak A, Bialopiotrowicz E, Krzymieniewska B, Wozniak J, Stojak M, Cybulska M, Kaniuga E, Mikula M, Jablonska E, Gorniak P, Noyszewska-Kania M, Szydlowski M, Piechna K, Piwocka K, Bugajski L, Lech-Maranda E, Barankiewicz J, Kolkowska-Lesniak A, Patkowska E, Glodkowska-Mrowka E, Baran N, Juszczynski P. SYK inhibition targets acute myeloid leukemia stem cells by blocking their oxidative metabolism. Cell Death Dis. 2020;11(11):956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Yusuf RZ, Saez B, Sharda A, van Gastel N, Yu VWC, Baryawno N, Scadden EW, Acharya S, Chattophadhyay S, Huang C, Viswanathan V, S’Aulis D, Cobert J, Sykes DB, Keibler MA, Das S, Hutchinson JN, Churchill M, Mukherjee S, Lee D, Mercier F, Doench J, Bullinger L, Logan DJ, Schreiber S, Stephanopoulos G, Rizzo WB, Scadden DT. Aldehyde dehydrogenase 3a2 protects AML cells from oxidative death and the synthetic lethality of ferroptosis inducers. Blood. 2020;136(11):1303–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Will B. No keto for AML stem cells! Blood. 2020;136(11):1219–21. [DOI] [PubMed] [Google Scholar]
- 162. Forte D, García-Fernández M, Sánchez-Aguilera A, Stavropoulou V, Fielding C, Martín-Pérez D, López JA, Costa ASH, Tronci L, Nikitopoulou E, Barber M, Gallipoli P, Marando L, Fernández de, Castillejo CL, Tzankov A, Dietmann S, Cavo M, Catani L, Curti A, Vázquez J, Frezza C, Huntly BJ, Schwaller J, Méndez-Ferrer S. Bone marrow mesenchymal stem cells support acute myeloid leukemia bioenergetics and enhance antioxidant defense and escape from chemotherapy. Cell Metab. 2020;32(5):829–43.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Coumans FAW, Brisson AR, Buzas EI, Dignat-George F, Drees EEE, El-Andaloussi S, Emanueli C, Gasecka A, Hendrix A, Hill AF, Lacroix R, Lee Y, van Leeuwen TG, Mackman N, Mäger I, Nolan JP, van der Pol E, Pegtel DM, Sahoo S, Siljander PRM, Sturk G, de Wever O, Nieuwland R. Methodological guidelines to study extracellular vesicles. Circ Res. 2017;120(10):1632–48. [DOI] [PubMed] [Google Scholar]
- 164. Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, Curry Jr, WT, Carter BS, Krichevsky AM, Breakefield XO. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Resnik N, Erman A, Veranič P, Kreft ME. Triple labelling of actin filaments, intermediate filaments and microtubules for broad application in cell biology: uncovering the cytoskeletal composition in tunneling nanotubes. Histochem Cell Biol. 2019;152(4):311–7. [DOI] [PubMed] [Google Scholar]
- 166. Winkler F, Wick W. Harmful networks in the brain and beyond. Science. 2018;359(6380):1100–1. [DOI] [PubMed] [Google Scholar]
- 167. Aroua N, Boet E, Ghisi M, Nicolau-Travers ML, Saland E, Gwilliam R, de Toni F, Hosseini M, Mouchel PL, Farge T, Bosc C, Stuani L, Sabatier M, Mazed F, Larrue C, Jarrou L, Gandarillas S, Bardotti M, Picard M, Syrykh C, Laurent C, Gotanègre M, Bonnefoy N, Bellvert F, Portais JC, Nicot N, Azuaje F, Kaoma T, Joffre C, Tamburini J, Récher C, Vergez F, Sarry JE. Extracellular ATP and CD39 activate cAMP-mediated mitochondrial stress response to promote cytarabine resistance in acute myeloid leukemia. Cancer Discov. 2020;10(10):1544–65. [DOI] [PubMed] [Google Scholar]
- 168. Stuani L, Sabatier M, Saland E, Cognet G, Poupin N, Bosc C, Castelli FA, Gales L, Turtoi E, Montersino C, Farge T, Boet E, Broin N, Larrue C, Baran N, Cissé MY, Conti M, Loric S, Kaoma T, Hucteau A, Zavoriti A, Sahal A, Mouchel PL, Gotanègre M, Cassan C, Fernando L, Wang F, Hosseini M, Chu-Van E, Le Cam L, Carroll M, Selak MA, Vey N, Castellano R, Fenaille F, Turtoi A, Cazals G, Bories P, Gibon Y, Nicolay B, Ronseaux S, Marszalek JR, Takahashi K, DiNardo CD, Konopleva M, Pancaldi V, Collette Y, Bellvert F, Jourdan F, Linares LK, Récher C, Portais JC, Sarry JE. Mitochondrial metabolism supports resistance to IDH mutant inhibitors in acute myeloid leukemia. J Exp Med. 2021;218(5):e20200924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169. Vitale I, Manic G, De Maria R, Kroemer G, Galluzzi L. DNA damage in stem cells. Mol Cell. 2017;66(3):306–19. [DOI] [PubMed] [Google Scholar]
- 170. Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Codrici E, Enciu AM, Popescu ID, Mihai S, Tanase C. Glioma stem cells and their microenvironments: providers of challenging therapeutic targets. Stem Cells Int. 2016;2016:5728438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Podergajs N, Motaln H, Rajčević U, Verbovšek U, Koršič M, Obad N, Espedal H, Vittori M, Herold-Mende C, Miletic H, Bjerkvig R, Turnšek TL. Transmembrane protein CD9 is glioblastoma biomarker, relevant for maintenance of glioblastoma stem cells. Oncotarget. 2016;7(1):593–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173. Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, Yang K, Morton AR, Zhou W, Zhu Z, Obara EAA, Miller TE, Song A, Lai S, Hubert CG, Jin X, Huang Z, Fang X, Dixit D, Tao W, Zhai K, Chen C, Dong Z, Zhang G, Dombrowski SM, Hamerlik P, Mack SC, Bao S, Rich JN. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell. 2018;22(4):514–28e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Day BW, Stringer BW, Al-Ejeh F, Ting MJ, Wilson J, Ensbey KS, Jamieson PR, Bruce ZC, Lim YC, Offenhäuser C, Charmsaz S, Cooper LT, Ellacott JK, Harding A, Leveque L, Inglis P, Allan S, Walker DG, Lackmann M, Osborne G, Khanna KK, Reynolds BA, Lickliter JD, Boyd AW. EphA3 maintains tumorigenicity and is a therapeutic target in glioblastoma multiforme. Cancer Cell. 2013;23(2):238–48. [DOI] [PubMed] [Google Scholar]
- 175. Rheinbay E, Suvà ML, Gillespie SM, Wakimoto H, Patel AP, Shahid M, Oksuz O, Rabkin SD, Martuza RL, Rivera MN, Louis DN, Kasif S, Chi AS, Bernstein BE. An aberrant transcription factor network essential for Wnt signaling and stem cell maintenance in glioblastoma. Cell Rep. 2013;3(5):1567–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176. Lubanska D, Market-Velker BA, deCarvalho AC, Mikkelsen T, Fidalgo da, Silva E, Porter LA. The cyclin-like protein Spy1 regulates growth and division characteristics of the CD133+ population in human glioma. Cancer Cell. 2014;25(1):64–76. [DOI] [PubMed] [Google Scholar]
- 177. Yan K, Wu Q, Yan DH, Lee CH, Rahim N, Tritschler I, DeVecchio J, Kalady MF, Hjelmeland AB, Rich JN. Glioma cancer stem cells secrete Gremlin1 to promote their maintenance within the tumor hierarchy. Genes Dev. 2014;28(10):1085–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Gutmann DH, Kettenmann H. Microglia/brain macrophages as central drivers of brain tumor pathobiology. Neuron. 2019;104(3):442–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179. Van Hove H, Martens L, Scheyltjens I, De Vlaminck K, Pombo Antunes AR, De Prijck S, Vandamme N, De Schepper S, Van Isterdael G, Scott CL, Aerts J, Berx G, Boeckxstaens GE, Vandenbroucke RE, Vereecke L, Moechars D, Guilliams M, Van Ginderachter JA, Saeys Y, Movahedi K. A single-cell atlas of mouse brain macrophages reveals unique transcriptional identities shaped by ontogeny and tissue environment. Nat Neurosci. 2019;22(6):1021–35. [DOI] [PubMed] [Google Scholar]
- 180. Goldmann T, Wieghofer P, Jordão MJ, Prutek F, Hagemeyer N, Frenzel K, Amann L, Staszewski O, Kierdorf K, Krueger M, Locatelli G, Hochgerner H, Zeiser R, Epelman S, Geissmann F, Priller J, Rossi FM, Bechmann I, Kerschensteiner M, Linnarsson S, Jung S, Prinz M. Origin, fate and dynamics of macrophages at central nervous system interfaces. Nat Immunol. 2016;17(7):797–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Pombo Antunes AR, Scheyltjens I, Lodi F, Messiaen J, Antoranz A, Duerinck J, Kancheva D, Martens L, De Vlaminck K, Van Hove H, Kjølner Hansen SS, Bosisio FM, Van der Borght K, De Vleeschouwer S, Sciot R, Bouwens L, Verfaillie M, Vandamme N, Vandenbroucke RE, De Wever O, Saeys Y, Guilliams M, Gysemans C, Neyns B, De Smet F, Lambrechts D, Van Ginderachter JA, Movahedi K. Single-cell profiling of myeloid cells in glioblastoma across species and disease stage reveals macrophage competition and specialization. Nat Neurosci. 2021;24(4):595–610. [DOI] [PubMed] [Google Scholar]
- 182. Hambardzumyan D, Bergers G. Glioblastoma: defining tumor niches. Trends Cancer. 2015;1(4):252–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183. Squadrito ML, De Palma M. A niche role for periostin and macrophages in glioblastoma. Nat Cell Biol. 2015;17(2):107–9. [DOI] [PubMed] [Google Scholar]
- 184. Rosa I, Marini M, Manetti M. Telocytes: an emerging component of stem cell niche microenvironment. J Histochem Cytochem. Epub 2021. Jun 24. doi: 10.1369/00221554211025489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Rotoli D, Díaz-Flores L, Gutiérrez R, Morales M, Ávila J, Martín-Vasallo P. AmotL2, IQGAP1, and FKBP51 scaffold proteins in glioblastoma stem cell niches. J Histochem Cytochem. Epub 2021. Jun 24. doi: 10.1369/00221554211025480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Rotoli D, Morales M, Maeso MD, Ávila J, Pérez-Rodríguez ND, Mobasheri A, van Noorden CJF, Martín-Vasallo P. IQGAP1, AmotL2, and FKBP51 scaffoldins in the glioblastoma microenvironment. J Histochem Cytochem. 2019;67(7):481–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187. Kilmister EJ, Tan ST. The role of the renin-angiotensin system in the cancer stem cell niche. J Histochem Cytochem. Epub 2021. Jun 24. doi: 10.1369/00221554211026295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Habič A, Novak M, Majc B, Lah Turnšek T, Breznik B. Proteases regulate cancer stem cell properties and remodel their microenvironment. J Histochem Cytochem. Epub 2021. Jul 26. doi: 10.1369/00221554211035192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Shahar T, Rozovski U, Hess KR, Hossain A, Gumin J, Gao F, Fuller GN, Goodman L, Sulman EP, Lang FF. Percentage of mesenchymal stem cells in high-grade glioma tumor samples correlates with patient survival. Neuro Oncol. 2017;19(5):660–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190. Breznik B, Motaln H, Vittori M, Rotter A, Lah Turnšek T. Mesenchymal stem cells differentially affect the invasion of distinct glioblastoma cell lines. Oncotarget. 2017;8(15):25482–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Hossain A, Gumin J, Gao F, Figueroa J, Shinojima N, Takezaki T, Priebe W, Villarreal D, Kang SG, Joyce C, Sulman E, Wang Q, Marini FC, Andreeff M, Colman H, Lang FF. Mesenchymal stem cells isolated from human gliomas increase proliferation and maintain stemness of glioma stem cells through the IL-6/gp130/STAT3 pathway. Stem Cells. 2015;33(8):2400–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Hekmatshoar Y, Nakhle J, Galloni M, Vignais ML. The role of metabolism and tunneling nanotube-mediated intercellular mitochondria exchange in cancer drug resistance. Biochem J. 2018;475(14):2305–28. [DOI] [PubMed] [Google Scholar]
- 193. Figueroa J, Phillips LM, Shahar T, Hossain A, Gumin J, Kim H, Bean AJ, Calin GA, Fueyo J, Walters ET, Kalluri R, Verhaak RG, Lang FF. Exosomes from glioma-associated mesenchymal stem cells increase the tumorigenicity of glioma stem-like cells via transfer of miR-1587. Cancer Res. 2017;77(21):5808–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Vlashi E, Lagadec C, Vergnes L, Matsutani T, Masui K, Poulou M, Popescu R, Della Donna L, Evers P, Dekmezian C, Reue K, Christofk H, Mischel PS, Pajonk F. Metabolic state of glioma stem cells and nontumorigenic cells. Proc Natl Acad Sci USA. 2011;108(38):16062–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Badr CE, Silver DJ, Siebzehnrubl FA, Deleyrolle LP. Metabolic heterogeneity and adaptability in brain tumors. Cell Mol Life Sci. 2020;77(24):5101–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196. Molenaar RJ, Maciejewski JP, Wilmink JW, van Noorden CJF. Wild-type and mutated IDH1/2 enzymes and therapy responses. Oncogene. 2018;37(15):1949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Khurshed M, Molenaar RJ, Lenting K, Leenders WP, van Noorden CJF. In silico gene expression analysis reveals glycolysis and acetate anaplerosis in IDH1 wild-type glioma and lactate and glutamate anaplerosis in IDH1-mutated glioma. Oncotarget. 2017;8(30):49165–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Sperry J, Condro MC, Guo L, Braas D, Vanderveer-Harris N, Kim KKO, Pope WB, Divakaruni AS, Lai A, Christofk H, Castro MG, Lowenstein PR, Le Belle JE, Kornblum HI. Glioblastoma utilizes fatty acids and ketone bodies for growth allowing progression during ketogenic diet therapy. iScience. 2020;23(9):101453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Kant S, Kesarwani P, Prabhu A, Graham SF, Buelow KL, Nakano I, Chinnaiyan P. Enhanced fatty acid oxidation provides glioblastoma cells metabolic plasticity to accommodate to its dynamic nutrient microenvironment. Cell Death Dis. 2020;11(4):253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200. Lee Y, Park Y, Nam H, Lee JW, Yu SW. Translocator protein (TSPO): the new story of the old protein in neuroinflammation. BMB Rep. 2020;53(1):20–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Fu Y, Wang D, Wang H, Cai M, Li C, Zhang X, Chen H, Hu Y, Zhang X, Ying M, He W, Zhang J. TSPO deficiency induces mitochondrial dysfunction, leading to hypoxia, angiogenesis, and a growth-promoting metabolic shift toward glycolysis in glioblastoma. Neuro Oncol. 2020;22(2):240–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Cao J, Mu Q, Huang H. The roles of insulin-like growth factor 2 mRNA-binding protein 2 in cancer and cancer stem cells. Stem Cells Int. 2018;2018:4217259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Janiszewska M, Suvà ML, Riggi N, Houtkooper RH, Auwerx J, Clément-Schatlo V, Radovanovic I, Rheinbay E, Provero P, Stamenkovic I. Imp2 controls oxidative phosphorylation and is crucial for preserving glioblastoma cancer stem cells. Genes Dev. 2012;26(17):1926–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204. Houben E, Hellings N, Broux B, Oncostatin M. An underestimated player in the central nervous system. Front Immunol. 2019;10:1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Sharanek A, Burban A, Laaper M, Heckel E, Joyal JS, Soleimani VD, Jahani-Asl A. OSMR controls glioma stem cell respiration and confers resistance of glioblastoma to ionizing radiation. Nat Commun. 2020;11(1):4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br J Cancer. 2016;114(12):1305–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Sica V, Bravo-San Pedro JM, Stoll G, Kroemer G. Oxidative phosphorylation as a potential therapeutic target for cancer therapy. Int J Cancer. 2020;146(1):10–7. [DOI] [PubMed] [Google Scholar]
- 208. Visweswaran M, Arfuso F, Warrier S, Dharmarajan A. Aberrant lipid metabolism as an emerging therapeutic strategy to target cancer stem cells. Stem Cells. 2020;38(1):6–14. [DOI] [PubMed] [Google Scholar]
- 209. Snyder V, Reed-Newman TC, Arnold L, Thomas SM, Anant S. Cancer stem cell metabolism and potential therapeutic targets. Front Oncol. 2018;8:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. Mudassar F, Shen H, O’Neill G, Hau E. Targeting tumor hypoxia and mitochondrial metabolism with anti-parasitic drugs to improve radiation response in high-grade gliomas. J Exp Clin Cancer Res. 2020;39(1):208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211. Koch K, Hartmann R, Tsiampali J, Uhlmann C, Nickel AC, He X, Kamp MA, Sabel M, Barker RA, Steiger HJ, Hänggi D, Willbold D, Maciaczyk J, Kahlert UD. A comparative pharmaco-metabolomic study of glutaminase inhibitors in glioma stem-like cells confirms biological effectiveness but reveals differences in target-specificity. Cell Death Discov. 2020;6:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Jagust P, de Luxán-Delgado B, Parejo-Alonso B, Sancho P. Metabolism-based therapeutic strategies targeting cancer stem cells. Front Pharmacol. 2019;10:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213. Cheng G, Zhang Q, Pan J, Lee Y, Ouari O, Hardy M, Zielonka M, Myers CR, Zielonka J, Weh K, Chang AC, Chen G, Kresty L, Kalyanaraman B, You M. Targeting lonidamine to mitochondria mitigates lung tumorigenesis and brain metastasis. Nat Commun. 2019;10(1):2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214. Kuramoto K, Yamamoto M, Suzuki S, Sanomachi T, Togashi K, Seino S, Kitanaka C, Okada M. Verteporfin inhibits oxidative phosphorylation and induces cell death specifically in glioma stem cells. FEBS J. 2020;287(10):2023–36. [DOI] [PubMed] [Google Scholar]
- 215. Vigneswaran K, Boyd NH, Oh SY, Lallani S, Boucher A, Neill SG, Olson JJ, Read RD. YAP/TAZ transcriptional coactivators create therapeutic vulnerability to verteporfin in EGFR-mutant glioblastoma. Clin Cancer Res. 2021;27(5):1553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Molenaar RJ, Coelen RJS, Khurshed M, Roos E, Caan MWA, van Linde ME, Kouwenhoven M, Bramer JAM, Bovée J, Mathôt RA, Klümpen HJ, van Laarhoven HWM, van Noorden CJF, Vandertop WP, Gelderblom H, van Gulik TM, Wilmink JW. Study protocol of a phase IB/II clinical trial of metformin and chloroquine in patients with IDH1-mutated or IDH2-mutated solid tumours. BMJ Open. 2017;7(6):e014961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217. Ji CC, Hu YY, Cheng G, Liang L, Gao B, Ren YP, Liu JT, Cao XL, Zheng MH, Li SZ, Wan F, Han H, Fei Z. A ketogenic diet attenuates proliferation and stemness of glioma stem-like cells by altering metabolism resulting in increased ROS production. Int J Oncol. 2020;56(2):606–17. [DOI] [PubMed] [Google Scholar]
- 218. Bahmad HF, Daher D, Aljamal AA, Elajami MK, Oh KS, Alvarez Moreno JC, Delgado R, Suarez R, Zaldivar A, Azimi R, Castellano A, Sackstein R, Poppiti RJ. Repurposing of anticancer stem cell drugs in brain tumors. J Histochem Cytochem. Epub 2021. Jun 24. doi: 10.1369/00221554211025482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219. Molenaar RJ, van Noorden CJ. Type 2 diabetes and cancer as redox diseases? Lancet. 2014;384(9946):853. [DOI] [PubMed] [Google Scholar]
- 220. Watson JD. Type 2 diabetes as a redox disease. Lancet. 2014;383(9919):841–3. [DOI] [PubMed] [Google Scholar]
- 221. Warburton DE, Nicol CW, Bredin SS. Health benefits of physical activity: the evidence. CMAJ. 2006;174(6):801–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222. Irwin ML, Smith AW, McTiernan A, Ballard-Barbash R, Cronin K, Gilliland FD, Baumgartner RN, Baumgartner KB, Bernstein L. Influence of pre- and postdiagnosis physical activity on mortality in breast cancer survivors: the health, eating, activity, and lifestyle study. J Clin Oncol. 2008;26(24):3958–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. Thong MSY, van Noorden CJF, Steindorf K, Arndt V. Cancer-related fatigue: causes and current treatment options. Curr Treat Options Oncol. 2020;21(2):17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Spencer J, Staffileno BA. Exercise intervention: a pilot study to assess the feasibility and impact on cancer-related fatigue and quality of life among patients with high-grade glioma. Clin J Oncol Nurs. 2021;25(2):194–200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-jhc-10.1369_00221554211054585 for Cell Biology Meets Cell Metabolism: Energy Production Is Similar in Stem Cells and in Cancer Stem Cells in Brain and Bone Marrow by Cornelis J.F. van Noorden, Barbara Breznik, Metka Novak, Amber J. van Dijck, Saloua Tanan, Miloš Vittori, Urban Bogataj, Noëlle Bakker, Joseph D. Khoury, Remco J. Molenaar and Vashendriya V.V. Hira in Journal of Histochemistry & Cytochemistry