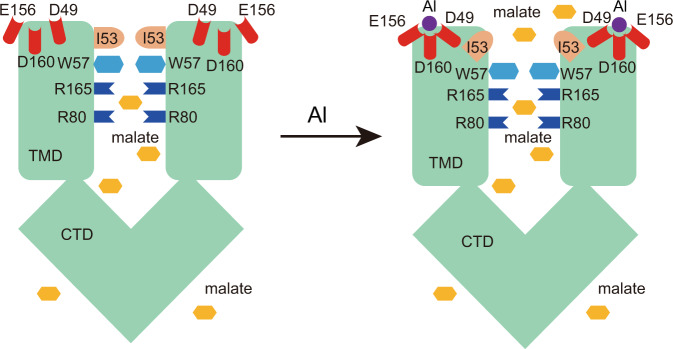
Abstract
The plant aluminum (Al)-activated malate transporter ALMT1 mediates the efflux of malate to chelate the Al in acidic soils and underlies the plant Al resistance. Here we present cryo-electron microscopy (cryo-EM) structures of Arabidopsis thaliana ALMT1 (AtALMT1) in the apo, malate-bound, and Al-bound states at neutral and/or acidic pH at up to 3.0 Å resolution. The AtALMT1 dimer assembles an anion channel and each subunit contains six transmembrane helices (TMs) and six cytosolic α-helices. Two pairs of Arg residues are located in the center of the channel pore and contribute to malate recognition. Al binds at the extracellular side of AtALMT1 and induces conformational changes of the TM1–2 loop and the TM5–6 loop, resulting in the opening of the extracellular gate. These structures, along with electrophysiological measurements, molecular dynamic simulations, and mutagenesis study in Arabidopsis, elucidate the structural basis for Al-activated malate transport by ALMT1.
Subject terms: Cryoelectron microscopy, Plant molecular biology
Introduction
About 40%–50% of the potentially arable lands in the world are covered with acidic soils, where aluminum (Al) toxicity is a major limitation for crop growth.1 Al ions are solubilized at pH below 5 and damage crops by inhibiting root growth and consequently impairing the uptake of water and nutrients.2 To overcome Al toxicity, plants have developed various Al-resistance strategies,3 with the secretion of organic acids from root apices being considered as the predominant means.4,5 The malate, citrate, and oxalate are released into the soil to chelate the Al ions, thus preventing Al uptake into root cells.3,4 The gene Aluminum-activated Malate Transporter (ALMT1), which was first cloned from an Al-resistant wheat (Triticum aestivum) line, encodes an anion channel (TaALMT1) that is responsible for the Al-activated malate extrusion from root tips.6 Soon, ALMT1 from other plant species such as Arabidopsis thaliana (AtALMT1), Brassica napus (BnALMT1), Glycine max (GmALMT1), and Secale cereal (ScALMT1) were also identified as Al-activated malate-permeable channels, suggesting a general Al-resistance mechanism.7–11
The Arabidopsis thaliana ALMT family contains 14 members (Supplementary information, Fig. S1),11 among which only AtALMT1 is involved in Al resistance. Al enhances the malate extrusion by AtALMT1 in two ways. First, it directly interacts with AtALMT1 and induces conformational changes that lead to the increase of transport activity within minutes.7,12 Second, Al up-regulates AtALMT1 expression, via both transcriptional activators (e.g., STOP1) and repressor (e.g., WRKY46).13,14 Because of ALMT1’s essential role in Al resistance, efforts have been made to reveal the molecular basis for the Al activation by electrophysiological, structural, and phylogenetic analyses;5,15,16 however, the Al binding site and its activation mechanism remain largely unknown.
In addition, the ALMT family has been shown to play important roles in the regulation of anion homeostasis,17,18 stomatal aperture,19–23 fruit quality,24 grain filling,25 and γ-aminobutyric acid (GABA) signaling.26,27 For example, in guard cells AtALMT6 and AtALMT9 are localized on the tonoplast and conduct the malate and chloride currents,19,20,22 respectively, whereas AtALMT12 on the plasma membrane mediates the efflux of malate.21,23 The three ALMTs participate in the regulation of stomatal opening and closure by adjusting guard cell volume.11 The ALMT family members share conserved primary sequences (Supplementary information, Fig. S1), implicating similar architectures of anion channels. At present, no structure of ALMT family members has been reported, and our understanding of their overall architecture and structural basis for anion selectivity is limited.
To reveal the structures and Al-activated malate transport mechanisms of ALMT1, we have performed comprehensive structural and functional studies of AtALMT1. We have captured AtALMT1 structures in the apo, malate-bound, and Al-bound states. These studies present structures of a new anion channel, demonstrate the structural basis for malate transport, and decipher the Al activation mechanism.
Results
Functional characterization of AtALMT1 in HEK293 cells
To investigate the electrophysiological properties of AtALMT1, we expressed AtALMT1 in HEK293 cells and measured channel activity by whole-cell patch clamping. In this setting, the pipette solution contains malate at pH 7.4 and the bath solution includes Al ions at pH 4.5. As previously shown, AtALMT1 exhibited basal activity in the absence of Al (Supplementary information, Fig. S2a).7 Extracellular Al increased the inward current density in a dose-dependent manner, with a half-maximal value (EC50) of 98.17 ± 3.53 μM (Supplementary information, Fig. S2a). No similar current was recorded in the cells transfected with the empty vector in the presence of Al (Supplementary information, Fig. S2a). Therefore, the recombinant AtALMT1 expressed in HEK293 cells forms a malate-conducting channel stimulated by extracellular Al.
Structure determination and overall structures of AtALMT1
Under physiological conditions, AtALMT1 is activated by extracellular Al at acidic pH and exports cytosolic malate at neutral pH. So we first determined the apo-state AtALMT1 structures at 3.2 Å resolution at pH 5 (ALMT1apo/pH5) and at 3.4 Å resolution at pH 7.5 (ALMT1apo/pH7.5) (Table 1; Supplementary information, Figs. S3, S4, and Table S1). Both the ALMT1apo/pH5 and ALMT1apo/pH7.5 structures were of high quality with the well-resolved transmembrane domain (TMD) and the cytosolic domain (CTD), allowing us to build de novo models of residues Met1–Glu216 and Asp222–Met405, covering 81% of the full-length protein (Supplementary information, Figs. S3f, S4f). The structures of ALMT1apo/pH5 and ALMT1apo/pH7.5 are essentially the same, with a root mean square deviation (RMSD) value of 0.8 Å over 800 Cα atoms (Supplementary information, Fig. S5a).
Table 1.
Summary of the AtATLM1 structures reported in the study.
| Structure | Ligand | pH | Resolution (Å) | Construct | State |
|---|---|---|---|---|---|
| ALMT1apo/pH5 | apo | 5 | 3.2 | wild type | closed |
| ALMT1apo/pH7.5 | apo | 7.5 | 3.4 | wild type | closed |
| ALMT1malate/pH7.5 | malate | 7.5 | 3.1 | wild type | closed |
| ALMT1Al/pH5 | Al | 5 | 3.6 | wild type | open |
| ALMT1M60A/Al/pH5 | Al | 5 | 3.0 | M60A | open |
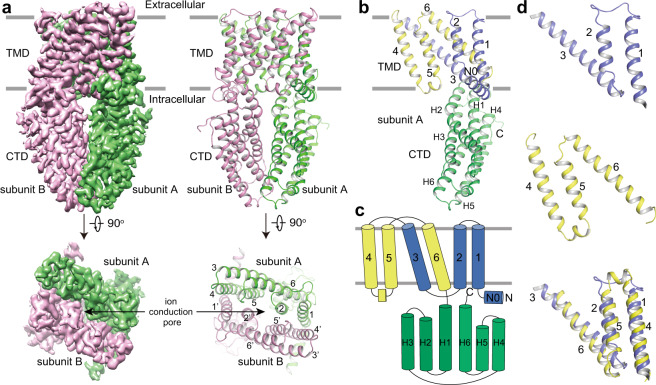
The ALTM1apo/pH5 channel is a homodimer, with dimensions of 75 × 60 × 90 Å (Fig. 1a). Each subunit consists of one N-terminal amphipathic α-helix running at the inner surface of the membrane (N0), six transmembrane helices (TM1–6), and six α-helices (H1–6) in CTD (Fig. 1a–c). Twelve transmembrane helices from two subunits assemble into the ion-conducting pore. TM2 and TM5 form the pore-lining helices at the center while TM1, TM3, TM4, and TM6 are on the periphery (Fig. 1a; Supplementary information, Fig. S5b). In the TMD, an internal structural repeat was identified, relating TM1–3 and TM4–6 by a pseudo-two-fold axis parallel to the membrane (Fig. 1c, d). The Cα atoms of the 80 residues from TM4–6 can be superimposed onto those from TM1–3 by a rotation of about 170°, yielding an RMSD value of 2.9 Å (Fig. 1d), although they only share 10% sequence identity. Following TM6, the H1–6 helices form an α-helix bundle and comprise the CTD (Fig. 1b, c). While most cytosolic helices are straight, H2 bends at Asp241 by about 40°. H2, H3, H6, and the H5–6 loop at the dimer interface contribute to the cytosolic dimerization, which is mediated by extensive hydrophobic interactions, salt bridges, and hydrogen bonds (Supplementary information, Fig. S5c–f).
Fig. 1. The overall structure of ALMT1apo/pH5.
a 3D reconstruction and carton representations of the AtALMT1 anion channel with each subunit individually colored. Numbers indicate transmembrane helices. b Structure of one subunit in the same orientation as the green-colored subunit in a. c Schematic representation of the domain arrangement in one AtALMT1 subunit. Domains are individually colored in b and c. d The internal structural repeat relating TM1–3 and TM4–6 by a pseudo-two-fold axis parallel to the membrane. TM4–6 is aligned to TM1–3 by a rotation of about 170°.
Structural basis for the malate recognition and transport
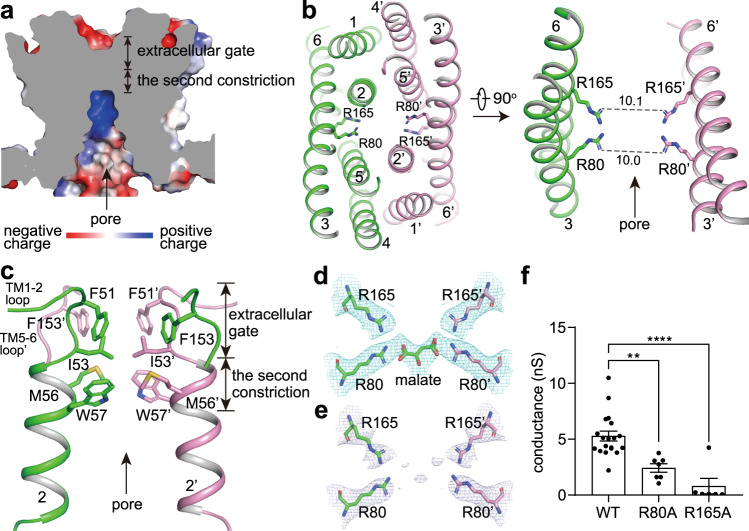
The ion-conducting pore of ALMT1apo/pH5 is funnel-shaped, opening to the cytosolic side with a diagonal atom-to-atom distance of ~14 Å at the inner surface of the membrane (Fig. 2a). Multiple positively charged residues including Lys76 and Arg125 reside on the cytosolic surface of the pore and probably favor the recruitment of malate (Supplementary information, Fig. S5g). Buried deeply in the middle of the membrane, one pair of Arg80 from two TM3 and one pair of Arg165 from two TM6 point to the center of the pore with diagonal atom-to-atom distances of ~10 Å (Fig. 2b). The orientation of the Arg80 side chain is fixed by hydrogen bonds of Arg80–Tyr136 and Arg80–Thr61, whereas the conformation of the Arg165 side chain is maintained by the cation–π interaction between Arg165 and Trp57 (Supplementary information, Fig. S5h). The two pairs of positively charged Arg residues are highly conserved across the ALMT family and they may play an important role in anion binding and conducting (Supplementary information, Fig. S5i). The extracellular gate of ALMT1apo/pH5 is closed mainly by intra- and inter-subunit hydrophobic packing involving residues 51FGI53 in the TM1–2 loop and TM2 N-terminus, and Phe153 in the TM5–6 loop (Fig. 2c). Below the extracellular gate (the first constriction), Met56 and Trp57 in TM2 from the two subunits likely form a second constriction point along the pore axis (Fig. 2c).
Fig. 2. The malate permeation pathway.
a Cross-section of a surface electrostatic potential model of ALMT1apo/pH5 TMD. b Two pairs of Arg residues in the pore center. Numbers on dash lines indicate diagonal atom-to-atom distances (in Å). c The extracellular gate and the second constriction block the extracellular solvent access. d The putative malate binding site in the ALMT1malate/pH7.5 structure. e Densities around the putative malate binding site in the map of ALMT1apo/pH7.5. In d and e, densities are shown at the same contour level of 4.5 σ. f The channel conductance of WT and mutant AtALMT1 measured at –180 mV. Data were analyzed by one-way ANOVA test. **P < 0.01, ****P < 0.0001. Also see Supplementary information, Fig. S2a, c.
To reveal how AtALMT1 recognizes its substrate, we determined the malate-bound AtALMT1 structure at 3.1 Å resolution at pH 7.5 (ALMT1malate/pH7.5) (Table 1; Supplementary information, Fig. S6 and Table S1). While no major conformational differences were observed between ALMT1malate/pH7.5 and ALMT1apo/pH7.5 (Supplementary information, Fig. S6f), strong densities at the contour level of 4 σ were identified between the pair of Arg80 and the pair of Arg165 in the map of ALMT1malate/pH7.5 (Fig. 2d). In comparison, in the map of ALTM1apo/pH7.5, densities at this site were much weaker at the contour level of 4 σ (Fig. 2e). As the shape and size of the densities resemble one malate molecule, we modeled a malate there in one ALMT1malate/pH7.5 channel (Fig. 2d). The putative malate forms interactions with both the pair of Arg80 and the pair of Arg165. Thus, the two pairs of Arg residues stabilize the binding of malate.
The ALMT1malate/pH7.5 map has a C2 symmetry, whereas the malate molecule has no symmetry. During 3D reconstruction, malate molecules in different orientations were averaged, resulting in low-quality densities for malate. To confirm the malate binding site, we performed molecular dynamics simulations in an equilibrium state without an electrochemical gradient, mimicking the structure determination condition (Supplementary information, Fig. S7). Throughout a 200 ns simulation, the deviations of the malate mass center from its original binding site are maintained at ~1.5 Å, suggesting that the malate binds stably between the two pairs of Arg residues (Supplementary information, Fig. S7b).
To verify the function of the two pairs of Arg residues, we recorded the currents of wild-type (WT) and mutant AtALMT1 expressed in HEK293 cells. No detectable current was recorded on the cells expressing the R165A mutant, indicating that the mutant channel lost malate-conducting function (Fig. 2f; Supplementary information, Fig. S2c). While the R80A mutant maintained the Al-sensitivity (Supplementary information, Fig. S2c), its channel conductance was significantly reduced (Fig. 2f; Supplementary information, Fig. S2c). These electrophysiological results support the essential roles of the two Arg residues in transporting the malate.
The Al binding sites
To reveal the Al activation mechanism of ALMT1, we determined the Al-bound AtALMT1 structure at 3.6 Å resolution at pH 5 (ALMT1Al/pH5) (Table 1; Supplementary information, Fig. S8 and Table S1). Strong densities at the contour level of 10 σ were observed between the pair of Met60 in the center of the ion-conducting pore in the map of ALMT1Al/pH5 (Supplementary information, Fig. S9a, b). By contrast, no densities appear at the equivalent site in the map of ALMT1apo/pH5 (Supplementary information, Fig. S9c). Therefore, we modeled one Al ion at this site (site SAl1). The putative Al is coordinated by the sulfhydryl groups of the pair of Met60 (Supplementary information, Fig. S9a, b).
To test whether the Al activates AtALMT1 by binding at the SAl1 site, we recorded whole-cell currents on HEK293 cells expressing AtALMT1 with mutations on the putative Al coordination residue Met60. Both M60A and M60L mutants generated Al-induced currents with concentration-response curves similar to that of WT AtALMT1 (Supplementary information, Fig. S9d, e), indicating that mutations at Met60 do not change the Al activation of the channel. We performed sequence alignment and found that the putative Al coordination residue Met is replaced with a Leu residue in TaALMT1 (Supplementary information, Fig. S9f), which is directly activated by extracellular Al.6 Thus these data suggest that besides SAl1 there is additional Al-binding site(s) in AtALMT1. However, the low quality of densities at the TM1–2 loop and TM5–6 loop in the extracellular region of ALMT1Al/pH5 prevented us from accurately defining the second Al-binding site (Supplementary information, Fig. S10g).
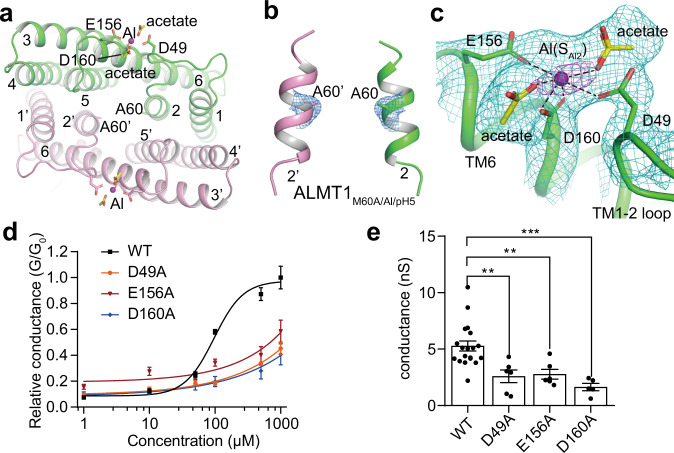
To identify additional Al-binding sites, we further determined the Al-bound structure of the AtALMT1 mutant M60A at 3.0 Å resolution in pH 5 (ALMT1M60A/Al/pH5) (Table 1 and Fig. 3a; Supplementary information, Fig. S10 and Table S1). The structures of ALMT1M60A/Al/pH5 and ALMT1Al/pH5 are essentially the same, except that ALMT1M60A/Al/pH5 loses the SAl1 site due to the substitution of Met60 by an Ala (Fig. 3b), which also validates our assignment of the SAl1 site in WT ALMT1Al/pH5. The ALMT1M60A/Al/pH5 map is of high quality in the extracellular loop regions (Supplementary information, Fig. S10h), allowing us to assign the second Al binding site (SAl2) (Fig. 3c). SAl2 sits between the TM1–2 loop and the N-terminal end of TM6. The Al at SAl2 is surrounded by several negatively charged residues namely Glu156 and Asp160 from TM6, and Asp49 from the TM1–2 loop (Fig. 3c). In addition, the strong nearby densities indicate that the Al is also coordinated by two negatively charged ligands but not Cl- whose densities should be much weaker. We putatively modeled two acetate molecules, the major anion in the protein solution besides Cl− (Fig. 3c).
Fig. 3. The Al binding site SAl2.
a The extracellular view of the SAl2 site in the ALMT1M60A/Al/pH5 structure. b The SAl1 site in the map of ALMT1M60A/Al/pH5. The densities around the SAl1 site and Ala60 are shown at the contour level of 4 σ. No density is observed around the SAl1 site. c Al at the SAl2 site is coordinated by Asp49, Glu156, Asp160, and two putative acetates. The green mesh show densities at the contour level of 5 σ, the magenta mesh shows densities at the contour level of 10 σ. Also see Supplementary information, Fig. S10h. d Concentration-dependent extracellular Al activation of WT and mutant AtALMT1. Data are reported as means ± SEM of at least five independent biological replicates. e The channel conductance of WT and mutant AtALMT1 measured at –180 mV. Data were analyzed by one-way ANOVA test. **P < 0.01, ***P < 0.001. Also see Supplementary information, Fig. S2a, e.
To test whether the SAl2 site is the bona fide Al activation site, we recorded whole-cell currents on HEK293 cells expressing AtALMT1 mutants D49A, E156A, and D160A. Mutations on the three Al coordination residues all significantly reduce the channel’s Al sensitivity (Fig. 3d). In the presence of 1 mM Al at ‒180 mV, the three mutants displayed much lower channel conductance compared with the WT AtALMT1 (Fig. 3e). These data confirm that Al activates AtALMT1 by binding at the SAl2 site. Similarly, in a previous study, the mutation on Glu187 in TaALMT1, which is equivalent to Glu156 in AtALMT1, also reduced the Al sensitivity of TaALMT1.16
The Al activation mechanism
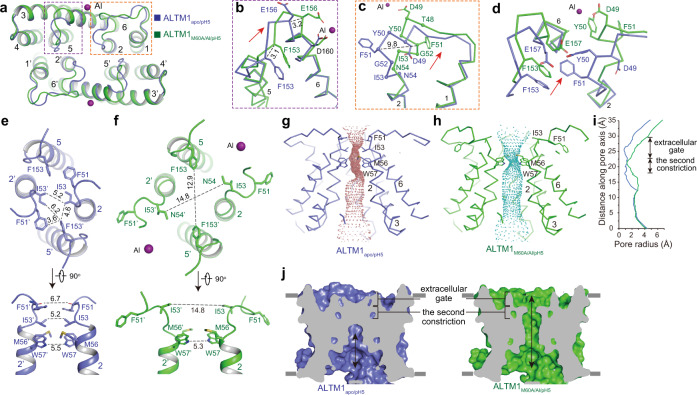
Since the M60A mutant maintains the Al sensitivity (Supplementary information, Fig. S9d, e) and the ALMT1M60A/Al/pH5 structure shares a similar conformation at the extracellular gate with the ALMT1Al/pH5 structure (Supplementary information, Fig. S10g–i), the higher-resolution ALMT1M60A/Al/pH5 structure can be compared with ALMT1apo/pH5 to illustrate the Al activation mechanism. The structural comparison reveals that the Al binding at the SAl2 site induces two major conformational changes involving the TM1–2 loop and TM5–6 loop at the extracellular gate of ALMT1 (Fig. 4a). First, in the ALMT1M60A/Al/pH5 structure, the TM5–6 loop and its connecting TM5 C-terminus and TM6 N-terminus move outwards from the pore central axis by 2–4 Å (Fig. 4b). This movement is induced by the direct chelation of the Al ion at the SAl2 site by Glu156 in TM6. Second, the TM1–2 loop and the TM2 N-terminus undergo considerable structural rearrangements (Fig. 4c). Specifically, the TM2 N-terminus with residues 53IN54 and its preceding β-turn 49DYFG52 lose their secondary structures; meanwhile, another β-turn 48TDYF51 is formed, along with a 5–10 Å outward movement of 49DYFG52 (Fig. 4c). The structural rearrangements of the TM1–2 loop and the TM2 N-terminus are likely caused by the outward movement of the TM5–6 loop, which pushes the β-turn 49DYFG52 outward, away from its original position (Fig. 4d). The Asp49 in the refolded β-turn 48TDYF51 stabilizes the new conformation of the TM1–2 loop by forming direct coordination with the Al ion at the SAl2 site (Fig. 4c).
Fig. 4. The structural comparisons of ALMT1apo/pH5 and ALMT1M60A/Al/pH5.
a The extracellular view of the structural alignment of ALMT1apo/pH5 (blue) and ALMT1M60A/Al/pH5 (green). b Conformational changes at the TM5–6 loop. Numbers on dash lines indicate distances between Cα of two structures (in Å). The red arrow shows the motion direction. c Conformational changes at the TM1–2 loop. Cα atoms of the residues that undergo conformational changes are shown as spheres. d Conformational changes at the interface of TM5–6 loop and TM1–2 loop. e The extracellular and side views of the extracellular gate and the second constriction in the ALMT1apo/pH5 structure. f The extracellular and side views of the extracellular gate and the second constriction in the ALMT1M60A/Al/pH5 structure. g Solvent-accessible pathway along the pore mapped with the HOLE program for ALMT1apo/pH5. Residues located at the extracellular gate and the second constriction are rendered as sticks. h Solvent-accessible pathway along the pore mapped with the HOLE program for ALMT1M60A/Al/pH5. i Comparison of pore radii (calculated with the program HOLE) for ALMT1apo/pH5 (blue) and ALMT1M60A/Al/pH5 (green). j Cross-section of surface-rendered ALMT1apo/pH5 and ALMT1M60A/Al/pH5 at the TMD. ALMT1M60A/Al/pH5 displays an open extracellular gate.
In the ALMT1apo/pH5 structure, the extracellular gate is closed off due to hydrophobic packing by residues 51FGI53 in the TM1–2 loop and TM2 N-terminus, and Phe153 in the TM5–6 loop (Fig. 4e). The shortest diagonal atom-to-atom distance between the pair of Ile53 is ~5 Å (Fig. 4e). In the ALMT1M60A/Al/pH5 structure, upon Al binding at the SAl2 site, the outward movements and structural rearrangements of the TM1–2 loop, TM2 N-terminus, and TM5–6 loop enable the ion-conducting pore to dilate at the extracellular gate (Fig. 4f). The packing interactions between 51FGI53 in the TM1–2 loop and Phe153 in the TM5–6 loop are disrupted, resulting in an enlarged extracellular facing cavity (Fig. 4g–j). The shortest diagonal atom-to-atom distances increase to about 13–15 Å between the pair of Ile53 and the pair of Phe153 (Fig. 4f). Thus, with Al bound at the SAl2 site, the extracellular gate of ALMT1M60A/Al/pH5 opens and is poised to release cytosolic malate (Figs. 4g–j and 5; Supplementary information, Video S1).
Fig. 5. A proposed Al-activated malate transport mechanism of AtALMT1.
Two pairs of Arg residues (R80 and R165) along the ion-conducting pathway are involved in the malate recognition. Under the acidic condition, the Al binds at the extracellular side and is coordinated by three acidic residues (Asp49, Glu156, and Asp160), inducing conformational changes of TM1–2 loop and TM5–6 loop, resulting in the outward movement of Ile53 residues and opening of the extracellular gate. Also see Supplementary information, Video S1.
Upon Al binding at the SAl2 site, the second constriction formed by the pair of Met56 and Trp57 remains almost unchanged. The distance between the amine groups in the Trp57 side chains is about 5–6 Å (Fig. 4e, f). Since the amine group of the Trp57 side chain can be protonated in an acidic solution, it is possible that the malate can pass through this constriction. Alternatively, under physiological conditions, AtALMT1 may need to undergo further conformational changes at the second constriction to conduct the malate.
In vivo validation of key residues in the Al-activated malate transport
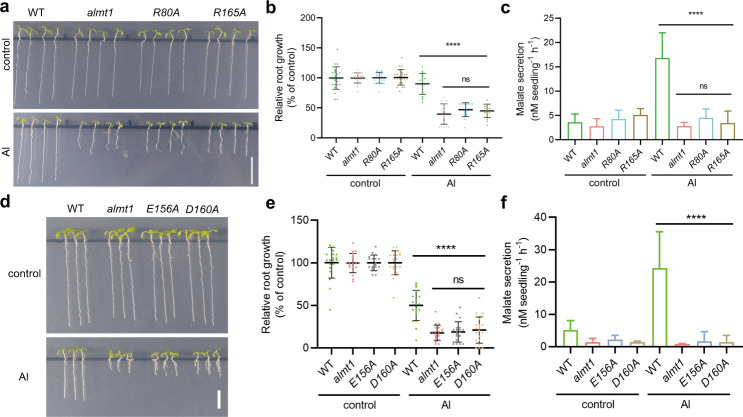
Through structural, computational, and electrophysiological analyses, we reveal the ion-conducting pathway and identify two pairs of Arg residues essential for malate recognition. To confirm the function of two pairs of Arg residues, we generated Arabidopsis thaliana transgenic lines with the AtALMT1 R80A or R165A mutation. The two mutant lines exhibit similar root growth and malate secretion as the WT in the absence of Al (Fig. 6a, b). Under Al treatment, both R80A and R165A lines display significantly reduced root growth, showing a similar phenotype as the almt1 loss-of-function mutant (Fig. 6a, b). Additionally, the Al-induced malate secretion from the R80A or R165A mutant also dramatically decreased (Fig. 6c). These observations thus further validate the essential role of these two Arg residues in the Al-activated malate extrusion in vivo. Similarly, to verify the Al activation site SAl2, we also generated Arabidopsis thaliana transgenic lines with the AtALMT1 mutations on the Al coordination residues Glu156 or Asp160. Both E156A and D160A lines exhibit significantly decreased root growth and malate secretion under Al treatment in comparison with WT Arabidopsis (Fig. 6d–f). Therefore, the in vivo data supports the essential role of the two acidic residues Glu156 and Asp160 in the Al-activated malate extrusion.
Fig. 6. In vivo validation of the Al-activated malate transport by AtALMT1.
a Root growth of WT, almt1, and R80A and R165A mutant lines of Arabidopsis thaliana under control and Al treatment for 10 days. Bar, 1 cm. b Comparison of relative root growth of WT, almt1, and R80A and R165A mutant lines under control and Al treatment. Data were analyzed by one-way ANOVA test. Error bars indicate SD (n = 16–25, ****P < 0.0001, ns, not statistically significant). c Comparison of root malate secretion of WT, almt1, and R80A and R165A mutant lines under control and Al treatment. Data were analyzed by ordinary one-way ANOVA. Error bars indicate SD (n = 4–7, ****P < 0.0001, ns, no significant difference). d Root growth of WT, almt1, and E156A and D160A mutant lines under control and Al treatment for 10 days. Bar, 0.5 cm. e Comparison of relative root growth of WT, almt1, and E156A and D160A mutant lines under control and Al treatment. Data were analyzed by ordinary one-way ANOVA. Error bars indicate SD (n = 20, ****P < 0.0001, ns, no significant difference). f Comparison of root malate secretion of WT, almt1, and E156A and D160A mutant lines under control and Al treatment. Data were analyzed by ordinary one-way ANOVA. Error bars indicate SD (n = 3, ****P < 0.0001).
Discussion
A new anion channel structure
We have elucidated the molecular architecture of AtALMT1, which represents a new anion channel structure. In AtALMT1, one ion-conducting pore is formed by twelve transmembrane helices from two subunits, with an internal structural repeat relating TM1–3 and TM4–6 by a pseudo-two-fold axis parallel to the membrane in each subunit. Along the ion-conducting pathway, two pairs of Arg residues are involved in the malate recognition. These structural features make AtALMT1 different from other anion channels in oligomerization or substrate anion recognition. For example, the dimeric assembly of AtALMT1 differs from the hexameric assembly of volume-regulated anion channel LRRC8A28 and pentameric assembly of type A γ-aminobutyric acid (GABAA) receptor.29 Moreover, in other anion channels such as the CLC chloride channel and Ca2+-activated chloride channel TMEM16A which form homodimers, there is an independent anion conducting pathway in each subunit, instead of one single anion conducting pathway between the two subunits.30–32 In comparison with AtALMT1, the essential role of positively charged pore-lining residues in the anion selectivity is also revealed in LRRC8A,28 but not observed in GABAA receptor,29 CLC,30 or TMEM16A.30–32
Among the five AtALMT1 structures we determined, only the two Al-bound structures, namely ALMT1Al/pH5 and ALMT1M60A/Al/pH5, are in an open state, while the rest three, including ALMT1apo/pH5, ALMT1apo/pH7.5, and ALMT1malate/pH7.5, are in a closed state. AtALMT1 displays basal activity in the absence of extracellular Al (Supplementary information, Fig. S2a).7 It would be interesting to try to capture an open-state or partially open-state structure of AtALMT1 in the absence of extracellular Al at different pH to see whether this constitutively opening mechanism is the same as the Al-activated opening mechanism. These AtALMT1 structures also provide a starting point for a systematic analysis of ion selectivity of the ALMT family. Some ALMT members, such as TaALMT1, ZmALMT2, AtALMT9, and AtALMT6 display selectivity among organic and inorganic anions;17,20,22,33 yet the structural basis for their ion selectivity remain unknown. Whether the conserved two pairs of Arg residues contribute to the anion selectivity of ALMTs awaits further investigation. In the future, high-resolution ALMT structures in complex with different anions, along with electrophysiology and molecular dynamics simulations, will elucidate the structural basis for ion selectivity of ALMTs.
The observation of SAl1 site in vitro
In AtALMT1 structures, we observed two Al binding sites: the SAl1 site sits in the center of the ion-conducting pore and is coordinated by a pair of Met; the SAl2 site is localized at the extracellular side between the TM1–2 loop and the TM6 N-terminus and is surrounded mainly by three acidic residues. Structural and electrophysiological analyses show that SAl2 instead of SAl1 is the Al activation site. Then one may ask whether SAl1 really exists and what its function is. Three lines of evidence support the identification of the SAl1 site. First, the cryo-EM map density at SAl1 is extremely strong, with a decent peak at the contour level above 10 σ, which is unlikely introduced by the C2 symmetry (Supplementary information, Fig. S9b). Second, structural comparison of ALMT1Al/pH5 and ALMT1apo/pH5 shows that no such density exists in the apo-state structure ALMT1apo/pH5 in the absence of Al (Supplementary information, Fig. S9c). Third, in the map of ALMT1M60A/Al/pH5 where the Al coordination residue Met60 at SAl1 is mutated to an Ala, the density corresponding to the Al at SAl1 disappears due to the loss of Al coordination residues (Fig. 3b). Therefore, we verified the observation of the SAl1 site in our ALMT1Al/pH5 structure. But the SAl1 site may only be observed in vitro and Al unlikely binds at this site in vivo. SAl1 site resides in the pore center and theoretically Al binding at SAl1 would inhibit the malate conductance (Supplementary information, Fig. S9a, b). However, the malate conductance of AtALMT1 is not inhibited by the extracellular Al and both mutations of M60A and M60L do not change the Al-activated currents (Supplementary information, Fig. S9d, e), which suggest that Al is unlikely to gain access to the SAl1 site from the extracellular side under our recording condition. In ALMT1Al/pH5 structure, the extracellular gate is closed off by hydrophobic packing and Al can only bind at the SAl1 site from the intracellular side during our sample preparation in vitro. Moreover, the SAl1 site is not conserved for the ALMT family because some Al-sensitive ALMT1 members do not have a Met residue at the equivalent position (Supplementary information, Fig. S9f),6,34 which further tones down the potential function of the SAl1 site.
The CTD of ALMT1
AtALMT1 and TaALMT1 share 42% sequence identity, and both are activated by extracellular Al within minutes.6,7 Previous electrophysiological studies showed that the CTD of TaALMT1 is important for its Al responsiveness in addition to the N-terminal region before the transmembrane domain.15,16 In AtALMT1 structures, the CTD participates in the dimeric assembly of the homodimer channel (Fig. 1a; Supplementary information, Fig. S5c–f), and no Al binding site is observed in the CTD. The CTD may regulate ALMT1 activity by adjusting the dimeric assembly. Mutations in the CTD may disrupt this correct dimeric assembly and further affect the extracellular Al binding site.
Conclusions
In the current study, we have determined the structures of AtALMT1 in the apo closed, malate-bound closed, and Al-bound open states up to 3 Å resolution (Table 1; Supplementary information, Table S1). These structures reveal that the AtALMT1 channel is a homodimer with a molecular mass of ~110 kD. We have identified two pairs of Arg residues in the pore center that are critical for malate recognition and transport. In addition, we have demonstrated that Al binds at the extracellular side between the TM1–2 loop and the TM6 N-terminus, and induces the opening of the extracellular gate (Fig. 5; Supplementary information, Video S1). These mechanistic insights into the molecular basis for the Al-activated malate transport by AtALMT1 can explain the highly efficient malate secretion in Al-resistant plants in response to the Al stress in acidic soil. This study also provides the structural basis for the rational design of the gain-of-function mutants of ALMT1 to enhance the Al-dependent malate secretion and Al-resistance in crops.
Materials and methods
Expression and purification of AtALMT1
The full-length AtALMT1 gene (NCBI: NP_172319.1) that encodes the AtALMT1 transporter was cloned into a pEZT-BM vector with a C-terminal strep-tag.35 AtALMT1 was heterologously expressed in HEK293S GnTI− cells using the BacMam system (Thermo Fisher Scientific). The P3 baculovirus derived from sf9 cells following the standard protocol was used to transfect HEK293S GnTI− cells when the cell density reached 3–3.5 × 106 cells/mL. For 800 mL cultures of HEK293S GnTI− cells, 70 mL P3 virus was added, supplemented with 10 mM sodium butyrate after 8–12 h incubation on an orbital shaker at 37 °C in the presence of 8% CO2.36 Cells were cultured for 48 h before they were harvested by centrifugation at 3000× g for 20 min at 4 °C.
The cells were re-suspended in lysis buffer A (20 mM Tris-HCl, pH 7.5, 150 mM NaCl) supplemented with a protease inhibitor cocktail (2 μg/mL DNase I, 0.5 μg/mL pepstatin, 2 μg/mL leupeptin, 1 μg/mL aprotinin, and 1 mM Phenylmethylsulfonyl fluoride) and homogenized by sonication on ice. The AtALMT1 protein was extracted with 1.5% (w/v) n-Dodecyl-β-D-Maltopyranoside (DDM, Anatrace) by gentle agitation at 100 rpm for 3 h at 4 °C. The supernatant was collected after centrifugation at 46,000× g for 60 min at 4 °C and then incubated with Strep-Tactin Sepharose resin (IBA) at 4 °C for 1.5 h. The collected resin was rinsed with buffer B (buffer A + 0.05% (w/v) DDM) for 10 column volumes. The detergent was then changed to 0.06% (w/v) glyco-diosgenin (GDN) and AtALMT1 protein was eluted with 10 mM desthiobiotin in buffer C (buffer A + 0.06% GDN). The protein sample was further purified by size-exclusion chromatography (SEC) with a Superose 6 10/300 GL column (GE Healthcare) equilibrated with buffer D (50 mM MES-NaOH, pH 5, and 150 mM NaCl) at a flow rate of 0.5 mL/min. The peak fractions were collected and concentrated to 5 mg/mL (ALMTapo/pH5) for single-particle cryo-EM analysis (Supplementary information, Fig. S3a, b).37,38
For the sample of AtALMT1 in pH 7.5, the protein purification followed the same protocol, except that the buffer D in the step of SEC was changed to buffer C. The peak fractions were collected and concentrated to 4.5 mg/mL (ALMTapo/pH7.5) for single-particle cryo-EM analysis (Supplementary information, Fig. S4a, b). For the sample of AtALMT1 bound to malate (ALMT1malate/pH7.5), the peak fractions after SEC in buffer C were mixed with malate to a final concentration of 10 mM. After incubation at 4 °C for 30 min, the mixture was then concentrated to about 4.5 mg/mL for cryo-EM sample preparation (Supplementary information, Fig. S6a). For the sample of AtALMT1 bound to Al3+ (ALMT1Al/pH5), the eluted sample in buffer C was further purified by SEC with buffer E (50 mM NaAc-HAc, pH 5, and 150 mM NaCl), the peak fractions were mixed with AlCl3 to a final concentration of 1 mM. After incubation at 4 °C for 30 min, the mixture sample was concentrated to about 6 mg/mL for cryo-EM sample preparation (Supplementary information, Fig. S8a, b). For the sample of ALMT1M60A/Al/pH5, the protein preparation followed the same protocol of ALMT1Al/pH5 (Supplementary information, Fig. S10a, b).
Cryo-EM data acquisition
The cryo-EM data of AtALMT1 was collected at the Center of Cryo-Electron Microscopy at Zhejiang University. 3 μL aliquots of AtALMT1 were placed on glow-discharged holey carbon grids (Quantifoil Cu R1.2/1.3, 300 mesh), which were blotted for 3 s under 100% humidity at 4 °C before being plunged into liquid ethane using a Mark IV Vitrobot (FEI). The grids were loaded onto a Titan Krios microscope (FEI) operated at 300 kV with a K2 Summit direct electron detector (Gatan). SerialEM software was used for automated data collection following standard procedures. A calibrated magnification of 49,310× was used for imaging, yielding a pixel size of 1.014 Å on images. Data were dose-fractionated to 40 frames with a dose rate of 8 e-/pixel/s and a total exposure time of 8 s, corresponding to a total dose of about 64 e-/Å2.
Image processing
The motion correction was performed using the MotionCorr2 program,39 and the CTF parameters of the micrographs were estimated using the GCTF program.40 All subsequent steps of image processing were carried out with RELION.41
For the sample without any exogenous compound in pH 5, about 1000 particles were manually picked from the micrographs for 2D classification initially. Class averages representing projections of AtALMT1 in different orientations were selected and used as templates for automated particle picking from the full data set of 4026 micrographs. The extracted particles were binned 3 times and subjected to a 2D classification, and a total of 1,692,846 particles were selected for three rounds of 3D classifications using the initial model generated by RELION as the reference. After the third round of 3D classification, 187,470 particles from one 3D class were selected for 3D reconstruction, yielding a 3.2 Å resolution map after 3D refinement with C2 symmetry and particle polishing (ALMT1apo/pH5) (Supplementary information, Fig. S3c). For the samples in pH 7.5 or bound to Al3+ or malate, data processing was performed following similar procedures (Supplementary information, Figs. S4c, S6b, S8c and S10c). The resolutions were estimated by applying a soft mask around the protein density and the gold-standard Fourier shell correlation was set as 0.143 (Supplementary information, Figs. S3e, S4e, S6d, S8e and S10e). RELION was used to calculate the local resolution maps.41
Model building, refinement, and validation
De novo atomic model building was performed in Coot 3.042 based on the 3.2 Å resolution map of the ALMT1apo/pH5. Amino acid residues assignment was achieved based on the clearly defined density for bulky residues (Phe, Trp, Tyr, and Arg). Models were refined against summed maps using phenix.real_space_refine,43 with secondary structure restraints and non-crystallography symmetry applied. The initial EM density map allowed us to construct an AtALMT1 model containing residues Met1–Glu216 and Asp222–Met405, covering 81% of the full-length protein. The ALMT1apo/pH5 model works as a reference for building models with ligands. The statistics for the models’ geometries were generated using MolProbity.44 All the structure figures were prepared in PyMol45 or Chimera.46
Plant materials and growth conditions
Arabidopsis thaliana plants were grown in a controlled environment growth chamber programmed for a 16 h-light/8 h-dark cycle with a daytime temperature of 23 °C and a night temperature of 21 °C. The almt1 knockout mutant (SALK_009629) was kindly provided by Prof. Chao Feng Huang (Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences). ALMT1 mutant lines were generated by transforming the full length of the mutagenized ALMT1 genes under the control of the native promoter (2175 bp) into the almt1 knockout mutant.
For aluminum treatment on root growth, a soaked gel medium was used as described previously with minor modification.47 1/6 Murashige & Skoog (MS) agar medium was first prepared by dissolving MS salts (PhytoTech, M524) in the ddH2O (0.72 g/L) plus 0.5% sucrose and 0.8% agar (Sigma-Aldrich, A7002). The medium was then soaked with a half volume of the same nutrient solution at pH 3.6 for control or with the solution containing 1 mM AlCl3 for Al treatment. After 1 day of soaking, the solution was removed, and seeds were sowed and grown on the agar medium in plates for 7 days. The seedlings were then photographed, and the root lengths were measured. Relative root growth, which was calculated as the percentage of root length with Al treatment divided by that under control treatment, was used to evaluate Al sensitivity.
Determination of malate exudation
For measurement of malate secretion, Arabidopsis seeds were germinated and grown on ½ MS agar medium containing 0.5% sucrose and 0.8% agar. After 7 days’ growth, the seedlings were transferred to a six-well plate (100 seedlings per well) and pretreated with 0.5 mM CaCl2 solution containing 1% sucrose for 2 h at pH 4.5, and then were treated with 4 mL of the same solution with or without 50 µM AlCl3 for 24 h with constant shaking on a rotary shaker (50 rpm) at room temperature in darkness. Malate concentrations in exudation media were determined by a NAD/NADH cycling-coupled enzymatic method as described previously.48
Electrophysiology
The AtALMT1 cDNA was cloned into pEZT vector with a green fluorescent protein (GFP)-strep-tagged at the C-terminus. All single-site mutants were confirmed by DNA sequencing. 0.8 μg of the plasmid was transfected into human embryonic kidney 293 cells containing the SV40 T-antigen (HEK293T) cells that were grown as a mono-layer in 35 mm tissue culture dishes (Corning) (to ~70% confluence) using Lipofectamine 3000 (Thermo Fisher Scientific). 8 h after transfection, cells were dissociated by 0.25% trypsin treatment (Gibco) and kept in a complete serum-containing medium (90% DMEM and 10% FBS) (Sigma) and replated onto microscopy slides (Fisher) and incubated in a cell culture incubator (Thermo Fisher Scientific) with 5% CO2.49–51 Patch-clamp recording was established 24 h after transfection, and a whole-cell configuration was used to measure macroscopic currents of AtALMT1 or its mutants overexpressed in HEK293T cells. The standard bath solution (extracellular side) contained: 150 mM 2-(N-morpholino) ethanesulphonic acid (MES), 1.8 mM CaCl2, 2 mM KCl, 0.1 mM LaCl3, pH 4.5 (adjusted with KOH) and osmolarity at 300 mOsmol (adjusted with 3 M D-sorbitol). To test whether AtALMT1 can be activated by extracellular Al at pH 5, we also measured the currents with bath solution at pH 5 (Supplementary information Fig. S2b). The pipette solution (intracellular side) contained: 105 mM Na2-malate, 10 mM HEPES, pH 7.4 (adjusted with KOH), and osmolarity at 288 mOsmol (adjusted with 3 M D-sorbitol).7
Data were acquired using a HEKA EPC10 amplifier at 10 kHz and filtered offline during data analysis with Igor 5.0 software. Patch pipettes were pulled from borosilicate glass (Sutter) and to a resistance of 3–5 MΩ. After the patch pipette was attached to the cell membrane, a giga seal (> 2 GΩ) was formed by gentle suction. The whole-cell configuration was formed by aspiration. The holding potential was set to 0 mV. The current and voltage relationship (I–V curve) was obtained using voltage pulses ramp from −200 to +80 mV over a 500 ms duration every 5 s. Al3+ was diluted to different concentrations (1, 10, 50, 100, 500, 1000 μM) using standard bath solution from a 50 mM AlCl3 stock solution (pH 4.5). Change of the extracellular solution was performed using an RSC-160 system (Bio-Logic Science Instruments).
All data points shown in Figs. 2f, 3d, e, and Supplementary information, Figs. S2, S9d, e are means ± SEM. of at least five measurements from different patches (n ≥ 5 independent biological replicates).
The current density was calculated by dividing the current amplitude (pA) by the membrane capacitance (pF). To analyze Al3+ activation of wild type and mutant AtALMT1, the currents (I) at −180 mV were used to calculate channel conductance (G) obtained by dividing the current amplitude (pA) by the potential difference (ΔV). The potential difference is referred as the holding potential (Vholding) minus the reversal potential (Vrp).
The true current value is the Al3+-activated currents of various concentrations minus the steady Al3+-free current (the blue curves in Supplementary information, Fig. S2a). The normalized conductance (G/G0) at –180 mV as a function of Al3+ concentration was fitted with the Hill equation:
where G0 is the conductance of wild type AtALMT1 at 1 mM Al, EC50 is the half-maximal effective concentration and n is the Hill coefficient.
Fluorescence imaging
0.8 μg plasmids of wild type and mutant AtALMT1 fused with C-terminal GFP were transfected into HEK293T cells. 24 h after transfection, the bright-field images were directly collected using a halogen lamp and the GFP fluorescence was excited by the mercury lamp light source with a 475/25 nm excitation filter on an inverted fluorescence microscope (Olympus IX83) using a 40× air objective (NA 0.95). The emission was monitored at 510 nm using a CCD camera (HAMAMATSU, C11440). Data were acquired and analyzed using the ImageJ software.
Membrane protein extraction and western blot analysis
25 mL HEK293S GnTI- cells were transiently transfected with 25 μg GFP-strep-tagged WT or mutant AtALMT1 in pEZT-BM vector by 75 ug polyethylenimine when the cell density reached 3.0 × 106 cells/mL. After incubation on an orbital shaker at 37 °C in the presence of 8% CO2 for 4 h, 25 mL complete serum-containing medium (5% FBS) was added, supplemented with 10 mM sodium butyrate to boost protein expression. Cells were cultured for 48 h at 37 °C before they were harvested by centrifugation at 3000× g for 20 min at 4 °C.
Cells were re-suspended in lysis buffer A, and homogenized by sonication on ice. The whole-cell lysate was centrifuged at 8000× g for 10 min at 4 °C and the supernatant was subjected to a second round of centrifugation at 38,000 rpm. (Beckman type 50.2 Ti rotor) for 1 h at 4 °C to pellet the membrane. The membrane fraction was resuspended in buffer A and homogenized with a glass dounce homogenizer. AtALMT1 was extracted with 1% (w/v) DDM (Anatrace) by stirring at 4 °C for 3 h. The supernatant, after extraction, was collected after 40 min centrifugation at 18,000× g at 4 °C. The membrane protein extract was quantified to 10 μg and resolved by SDS-PAGE, transferred to polyvinylidene fluoride membranes, and probed with rabbit anti-strep II antibody (1:2000 dilution; Abcam; ab76949). The secondary antibody used was goat anti-rabbit immunoglobulin G (IgG) (1:5000 dilution; Sangon Biotech; D110058) conjugated to horseradish peroxidase.
Molecular dynamics simulations
The ALMT1malate/pH7.5 structure was subjected to molecular dynamics simulations. The missing residues between E216 and D222 were filled by the protein preparation wizard in Schrodinger (version 2020-1, Schrödinger, Inc.) package. The atoms of protein, lipids, sodium, and TIP3P waters were assigned with CHARMM36m force field.52 CGenFF force field was used for malate molecule, and atom types were determined by both MATCH and paramchem with low charge penalty (< 10).53,54 The whole system was modeled with VMD plugins. The complex structure was solved in a TIP3P water cubic box (> 25 Å buffering for each direction). The coordinates of POPC lipids were assigned according to the detergent density. TIP3P waters in the trans-membrane part, or clashed with lipids and protein were removed. The pore between two subunits of ALMT1 was filled with TIP3P water models. The whole system was neutralized with sodium atoms. For the simulation, a periodic box with dimensions 128 × 128 × 160 Å was generated for all the 207,315 atoms. The lipid tail was firstly relaxed by minimization with 5000 steps, and 0.5 ns molecular dynamics simulation with NVT ensemble, during which other atoms were position restrained with 5 (kcal/mol)/Å2 force constant. Then water, malate, sodium, lipid tail, protein side-chain were relaxed with 1.0 ns simulation with NVT ensemble, while the lipid head, protein backbone were taken as position restraint. Finally, lipid head and protein backbone were released from positions restraint by gradient decreasing the force constant to 0.5 (kcal/mol)/Å2 with a total 7.5 ns simulation with NPT ensemble. The product simulation was run for 200 ns, repeated three times. Structure equilibration and molecular dynamics production were performed with NAMD2 and NAMD3, respectively.55 For each trajectory, 12,500 frames were selected for further analysis by the VMD program. Malate clustering was performed based on the RMSD value (<1.0 Å) of malate heavy atoms by the clustering tool plugin of VMD.
Quantification and statistical analysis
All reported resolutions in cryo-EM studies are based upon the 0.143 Fourier Shell Correlation criterion. Error bars in Figs. 2f, 3d, e, 6b, c, e, f, and Supplementary information, Figs. S2, S9d, e represent the standard error of the mean for at least three independent experiments. Data were analyzed by one-way ANOVA (analysis of variance) test.
Supplementary information
Acknowledgements
Single-particle cryo-EM data were collected at the Center of Cryo-Electron Microscopy at Zhejiang University. We thank Dr. Xing Zhang and Dr. Shenghai Chang for support in facility access and data acquisition. We thank Dr. Chao Feng Huang for kindly providing the almt1 mutant seeds. We thank Dr. Nam Nguyen for his assistance in the paper preparation. This work was supported in part by the Ministry of Science and Technology (2020YFA0908501 and 2018YFA0508100 to J.G., 2016YFA0501100 to X.Z.), the National Natural Science Foundation of China (31870724 to J.G., 82030108, 31872796 and 81571127 to W.Y., 31730006 to S.J.Z., and 31600606 to X.Z.), National Major Special Project on New Drug Innovation of China (2018ZX09711001-004-005 to W.Y.), Zhejiang Provincial Natural Science Foundation (LR19C050002 to J.G., LR16H090001 to W.Y.), and the Fundamental Research Funds for the Central Universities (2021FZZX001-28 to J.G. and S.J.Z.). J.G. is supported by MOE Frontier Science Center for Brain Science & Brain-Machine Integration, Zhejiang University. X.Z. is supported by Guangdong Provincial Key Laboratory of Brain Connectome and Behavior (2017B030301017) and CAS Key Laboratory of Brain Connectome and Manipulation (2019DP173024).
Author contributions
J.G., W.Y., and S.J.Z. conceived and supervised the project. J.W., X.Z., Y.X., and X.L. prepared the samples, performed data acquisition, structure determination, and data analysis. X.Y. and Y.L. performed electrophysiological studies. Z.J.D. and T.Y. performed the in vivo mutagenesis studies. X.X. performed molecular dynamics simulation. All authors participated in the data analysis and paper preparation.
Data availability
The cryo-EM density maps and coordinates of ALMT1apo/pH5, ALMT1apo/pH7.5, ALMT1malate/pH7.5, ALMT1Al/pH5, and ALMT1M60A/Al/pH5 have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-32084, EMD-32085, EMD-32086, EMD-32087, and EMD-32050, and in the RCSB Protein Data Bank (PDB) under accession codes 7VQ3, 7VQ4, 7VQ5, 7VQ7, and 7VOJ.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Jiangqin Wang, Xiafei Yu, Zhong Jie Ding, Xiaokang Zhang.
Contributor Information
Shao Jian Zheng, Email: sjzheng@zju.edu.cn.
Wei Yang, Email: yangwei@zju.edu.cn.
Jiangtao Guo, Email: jiangtaoguo@zju.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41422-021-00587-6.
References
- 1.von Uexküll HR, Mutert E. Global extent, development and economic impact of acid soils. Plant Soil. 1995;171:1–15. [Google Scholar]
- 2.Kochian LV. Cellular mechanisms of aluminum toxicity and resistance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1995;46:237–260. [Google Scholar]
- 3.Ryan PR, Delhaize E, Jones DL. Function and mechanism of organic anion exudation from plant roots. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001;52:527–560. doi: 10.1146/annurev.arplant.52.1.527. [DOI] [PubMed] [Google Scholar]
- 4.Kochian LV, Piñeros MA, Liu J, Magalhaes JV. Plant adaptation to acid soils: the molecular basis for crop aluminum resistance. Annu. Rev. Plant Biol. 2015;66:571–598. doi: 10.1146/annurev-arplant-043014-114822. [DOI] [PubMed] [Google Scholar]
- 5.Ma JF, Chen ZC, Shen RF. Molecular mechanisms of Al tolerance in gramineous plants. Plant Soil. 2014;381:1–12. [Google Scholar]
- 6.Sasaki T, et al. A wheat gene encoding an aluminum-activated malate transporter. Plant J. 2004;37:645–653. doi: 10.1111/j.1365-313x.2003.01991.x. [DOI] [PubMed] [Google Scholar]
- 7.Hoekenga OA, et al. AtALMT1, which encodes a malate transporter, is identified as one of several genes critical for aluminum tolerance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2006;103:9738–9743. doi: 10.1073/pnas.0602868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ligaba A, Katsuhara M, Ryan PR, Shibasaka M, Matsumoto H. The BnALMT1 and BnALMT2 genes from rape encode aluminum-activated malate transporters that enhance the aluminum resistance of plant cells. Plant Physiol. 2006;142:1294–1303. doi: 10.1104/pp.106.085233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang C, et al. Low pH, aluminum, and phosphorus coordinately regulate malate exudation through GmALMT1 to improve soybean adaptation to acid soils. Plant Physiol. 2013;161:1347–1361. doi: 10.1104/pp.112.208934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fontecha G, et al. Candidate gene identification of an aluminum-activated organic acid transporter gene at the Alt4 locus for aluminum tolerance in rye (Secale cereale L.) Theor. Appl. Genet. 2006;114:249–260. doi: 10.1007/s00122-006-0427-7. [DOI] [PubMed] [Google Scholar]
- 11.Sharma T, Dreyer I, Kochian L, Pineros MA. The ALMT family of organic acid transporters in plants and their involvement in detoxification and nutrient security. Front Plant Sci. 2016;7:1488. doi: 10.3389/fpls.2016.01488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang, X., Long, Y., Huang, J. & Xia, J. Molecular mechanisms for coping with Al toxicity in plants. Int. J. Mol. Sci. 20, 1551 (2019). [DOI] [PMC free article] [PubMed]
- 13.Ding ZJ, Yan JY, Xu XY, Li GX, Zheng SJ. WRKY46 functions as a transcriptional repressor of ALMT1, regulating aluminum-induced malate secretion in Arabidopsis. Plant J. 2013;76:825–835. doi: 10.1111/tpj.12337. [DOI] [PubMed] [Google Scholar]
- 14.Iuchi S, et al. Zinc finger protein STOP1 is critical for proton tolerance in Arabidopsis and coregulates a key gene in aluminum tolerance. Proc. Natl. Acad. Sci. USA. 2007;104:9900–9905. doi: 10.1073/pnas.0700117104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Furuichi T, et al. An extracellular hydrophilic carboxy-terminal domain regulates the activity of TaALMT1, the aluminum-activated malate transport protein of wheat. Plant J. 2010;64:47–55. doi: 10.1111/j.1365-313X.2010.04309.x. [DOI] [PubMed] [Google Scholar]
- 16.Ligaba A, et al. Functional, structural and phylogenetic analysis of domains underlying the Al sensitivity of the aluminum-activated malate/anion transporter, TaALMT1. Plant J. 2013;76:766–780. doi: 10.1111/tpj.12332. [DOI] [PubMed] [Google Scholar]
- 17.Ligaba A, Maron L, Shaff JON, Kochian L, PiÑEros M. Maize ZmALMT2 is a root anion transporter that mediates constitutive root malate efflux. Plant Cell Environ. 2012;35:1185–1200. doi: 10.1111/j.1365-3040.2011.02479.x. [DOI] [PubMed] [Google Scholar]
- 18.Piñeros MA, et al. Not all ALMT1-type transporters mediate aluminum-activated organic acid responses: the case of ZmALMT1 - an anion-selective transporter. Plant J. 2007;53:352–367. doi: 10.1111/j.1365-313X.2007.03344.x. [DOI] [PubMed] [Google Scholar]
- 19.De Angeli A, Zhang J, Meyer S, Martinoia E. AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 2013;4:1804. doi: 10.1038/ncomms2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovermann P, et al. The Arabidopsis vacuolar malate channel is a member of the ALMT family. Plant J. 2007;52:1169–1180. doi: 10.1111/j.1365-313X.2007.03367.x. [DOI] [PubMed] [Google Scholar]
- 21.Meyer S, et al. AtALMT12 represents an R-type anion channel required for stomatal movement in Arabidopsis guard cells. Plant J. 2010;63:1054–1062. doi: 10.1111/j.1365-313X.2010.04302.x. [DOI] [PubMed] [Google Scholar]
- 22.Meyer S, et al. Malate transport by the vacuolar AtALMT6 channel in guard cells is subject to multiple regulation. Plant J. 2011;67:247–257. doi: 10.1111/j.1365-313X.2011.04587.x. [DOI] [PubMed] [Google Scholar]
- 23.Sasaki T, et al. Closing plant stomata requires a homolog of an aluminum-activated malate transporter. Plant Cell Physiol. 2010;51:354–365. doi: 10.1093/pcp/pcq016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Angeli A, et al. The vacuolar channel VvALMT9 mediates malate and tartrate accumulation in berries of Vitis vinifera. Planta. 2013;238:283–291. doi: 10.1007/s00425-013-1888-y. [DOI] [PubMed] [Google Scholar]
- 25.Xu M, et al. The barley anion channel, HvALMT1, has multiple roles in guard cell physiology and grain metabolism. Physiol. Plantarum. 2015;153:183–193. doi: 10.1111/ppl.12234. [DOI] [PubMed] [Google Scholar]
- 26.Palmer AJ, Baker A, Muench SP. The varied functions of aluminium-activated malate transporters-much more than aluminium resistance. Biochem. Soc. Trans. 2016;44:856–862. doi: 10.1042/BST20160027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramesh SA, et al. GABA signalling modulates plant growth by directly regulating the activity of plant-specific anion transporters. Nat. Commun. 2015;6:7879. doi: 10.1038/ncomms8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deneka D, Sawicka M, Lam AKM, Paulino C, Dutzler R. Structure of a volume-regulated anion channel of the LRRC8 family. Nature. 2018;558:254–259. doi: 10.1038/s41586-018-0134-y. [DOI] [PubMed] [Google Scholar]
- 29.Laverty D, et al. Cryo-EM structure of the human alpha1beta3gamma2 GABAA receptor in a lipid bilayer. Nature. 2019;565:516–520. doi: 10.1038/s41586-018-0833-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park E, Campbell EB, MacKinnon R. Structure of a CLC chloride ion channel by cryo-electron microscopy. Nature. 2017;541:500–505. doi: 10.1038/nature20812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paulino C, Kalienkova V, Lam AKM, Neldner Y, Dutzler R. Activation mechanism of the calcium-activated chloride channel TMEM16A revealed by cryo-EM. Nature. 2017;552:421–425. doi: 10.1038/nature24652. [DOI] [PubMed] [Google Scholar]
- 32.Dang S, et al. Cryo-EM structures of the TMEM16A calcium-activated chloride channel. Nature. 2017;552:426–429. doi: 10.1038/nature25024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang WH, et al. Characterization of the TaALMT1 protein as an Al3+-activated anion channel in transformed tobacco (Nicotiana tabacum L.) cells. Plant Cell Physiol. 2008;49:1316–1330. doi: 10.1093/pcp/pcn107. [DOI] [PubMed] [Google Scholar]
- 34.Delhaize E, et al. Engineering high-level aluminum tolerance in barley with the ALMT1 gene. Proc. Natl. Acad. Sci. USA. 2004;101:15249–15254. doi: 10.1073/pnas.0406258101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morales-Perez CL, Noviello CM, Hibbs RE. Manipulation of subunit stoichiometry in heteromeric membrane proteins. Structure. 2016;24:797–805. doi: 10.1016/j.str.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goehring A, et al. Screening and large-scale expression of membrane proteins in mammalian cells for structural studies. Nat. Protoc. 2014;9:2574–2585. doi: 10.1038/nprot.2014.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X, et al. Molecular basis for ligand activation of the human KCNQ2 channel. Cell Res. 2020;31:52–61. doi: 10.1038/s41422-020-00410-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu S, Chang S, Han B, Xu L, Guo J. Cryo-EM structures of the human cation-chloride cotransporter KCC1. Science. 2019;366:505–508. doi: 10.1126/science.aay3129. [DOI] [PubMed] [Google Scholar]
- 39.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang K. Gctf: Real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Scheres SHW. RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr. D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schrodinger L. The PyMOL molecular graphics system. Version. 2015;1:0. [Google Scholar]
- 46.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 47.Zhang Y, et al. F-box protein RAE1 regulates the stability of the aluminum-resistance transcription factor STOP1 in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2019;116:319–327. doi: 10.1073/pnas.1814426116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Delhaize E, Ryan PR, Randall PJ. Aluminum tolerance in wheat (Triticum aestivum L.) II. Aluminum-stimulated excretion of malic acid from root apices. Plant Physiol. 1993;103:695–702. doi: 10.1104/pp.103.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luo YH, Yu XF, Ma C, Yang F, Yang W. Effects of calcium-binding sites in the S2-S3 loop on human and Nematostella vectensis TRPM2 channel gating processes. J. Zhejiang Univ. Sci. B. 2019;20:972–982. doi: 10.1631/jzus.B1900477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yu P, et al. Direct Gating of the TRPM2 Channel by cADPR via Specific Interactions with the ADPR Binding Pocket. Cell Rep. 2019;27:3684–3695. doi: 10.1016/j.celrep.2019.05.067. [DOI] [PubMed] [Google Scholar]
- 51.Yuhuan L, Xiafei Y, Cheng M, Jianhong L, Wei Y. Identification of a Novel EF-Loop in the N-terminus of TRPM2 Channel Involved in Calcium Sensitivity. Front. Pharmacol. 2018;9:581. doi: 10.3389/fphar.2018.00581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang J, et al. CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat. Methods. 2016;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vanommeslaeghe K, et al. CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 2009;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yesselman JD, Price DJ, Knight JL, Brooks CL. MATCH: An atom-typing toolset for molecular mechanics force fields. J. Comput. Chem. 2012;33:189–202. doi: 10.1002/jcc.21963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Phillips JC, et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM density maps and coordinates of ALMT1apo/pH5, ALMT1apo/pH7.5, ALMT1malate/pH7.5, ALMT1Al/pH5, and ALMT1M60A/Al/pH5 have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-32084, EMD-32085, EMD-32086, EMD-32087, and EMD-32050, and in the RCSB Protein Data Bank (PDB) under accession codes 7VQ3, 7VQ4, 7VQ5, 7VQ7, and 7VOJ.