Abstract
Lung cancer (LC) is often diagnosed at an advanced stage and conventional treatments for disease management have limitations associated with them. Novel therapeutic targets are thus avidly sought for the effective management of LC. RNA binding proteins (RBPs) have been convincingly established as key players in tumorigenesis, and their dysregulation is linked to multiple cancers, including LC. In this context, we review the role of Human antigen R (HuR), an RBP that is overexpressed in LC, and further associated with various aspects of LC tumor growth and response to therapy. Herein, we describe the role of HuR in LC progression and outline the evidences supporting various pharmacologic and biologic approaches for inhibiting HuR expression and function. These approaches, including use of small molecule inhibitors, siRNAs and shRNAs, have demonstrated favorable results in reducing tumor cell growth, invasion and migration, angiogenesis and metastasis. Hence, HuR has significant potential as a key therapeutic target in LC. Use of siRNA-based approaches, however, have certain limitations that prevent their maximal exploitation as cancer therapies. To address this, in the conclusion of this review, we provide a list of nanomedicine-based HuR targeting approaches currently being employed for siRNA and shRNA delivery, and provide a rationale for the immense potential therapeutic benefits offered by nanocarrier-based HuR targeting and its promise for treating patients with LC.
Keywords: lung cancer, RBP, HuR, siRNA, CRISPR/Cas-9, nanomedicine, drug delivery
Graphical Abstract
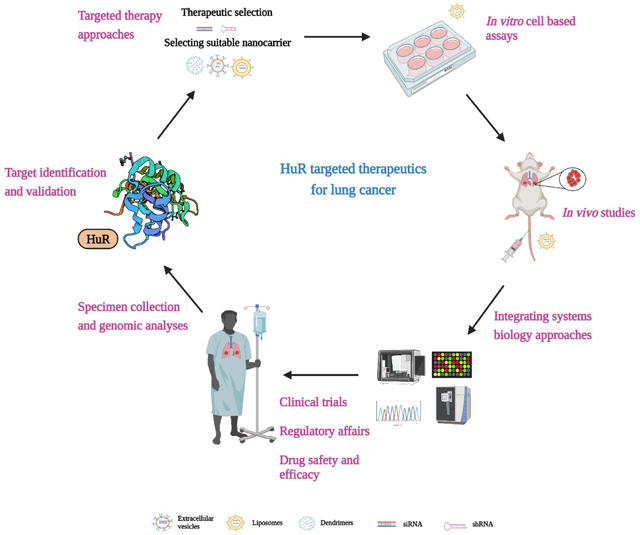
Schematic representation of translating HuR targeted therapeutics from bench to bedside for lung cancer. Image created with BioRender.com
Introduction
Regulation of gene expression occurs at both the transcriptional and post-transcriptional level. In the latter context, RNA Binding Proteins (RBPs), key mediators of RNA metabolism, streamline the flow of genetic material; they are the key players in post-transcriptional regulation of gene expression and as such contribute to growth and development, homeostasis, and effective functioning of the immune system. In addition to directly binding to mRNA, RBPs can bind to non-coding RNAs (ncRNA) and other RBPs, and contribute to regulation of mRNA processing, splicing, polyadenylation, and stability, along with protein translation [1, 2].
The interaction of RBPs with RNA occurs principally via RNA-binding domains (RBD) resulting in the formation of ribonucleotide-protein (RNP) complexes. RBPs can thus be broadly grouped based on their RBDs; over 50 different RBDs are known and include the RNA recognition motif (RRM), which is the most abundant RBD reported in the human genome (e.g. HuR), K homology domain – KH identified from the heterogeneous nuclear riboprotein (hnRNP) (e.g. Nova 2), zinc finger domain (e.g. TTP, MBNL1, ZRANB2), pumilio homology domain (PUF) (e.g. DmPUM, FBF, APUM), double stranded RNA binding domains (dsRBM) (e.g. ADAR2), RGG (Arg-Gly-Gly) box (e.g nucleolin), Piwi/Argonaute/Zwille (PAZ) domain (e.g DICER, Argonaute proteins) and cold shock domains (CSD) (e.g. LIN28A, LIN28B, YBX 1–3) [3–7]. The number of RBD units found in an RBP varies widely, each RBP can either contain multiple repeats of the same RBD unit or can possess multiple different RBDs [8, 9]. RBPs that contain the classic RBDs described above are termed “canonical RBPs”, while those that lack RBDs and rather bind to RNA via intrinsically disordered regions are referred to as “non-canonical” RBPs [10]. Ubiquitously expressed RBPs involved in stabilization and/or destabilization of mRNA are shown in Table 1. Historically, most studies focused on deciphering the role of canonical RBPs in disease processes, however, more recently there is a growing interest in unravelling the mechanistic underpinnings of non-canonical RBPs [11]. In total, 311,571 canonical RBPs and 3,651 non-canonical RBPs have been identified in 162 eukaryotic species in the EuRPDB database [12].
Table 1:
RBP’s in stabilization and destabilization of mRNAs
| S.No. | RBP Name | Stabilization/Destabilization | Target genes | References |
|---|---|---|---|---|
| 1 | hnRNP E1 | Stabilization/Destabilization | eNOS, c-Src, Dab2, ILEI, PRL-3, Inhibin βA | [232–236] |
| 2 | hnRNP E2 | Stabilization/Destabilization | c-Src, Dab2, ILEI, PRL-3, Inhibin βA | [233–236] |
| 3 | HuR | Stabilization/Destabilization | SET, VEGF, COX-2, lncRNA NEAT1, LncRNA-HGBC, p21, cyclin A, cyclin B1, MMP9, GM-CSF, eotaxin, IL-2, c-myc, cyclin E1, LincRNA-p21, lncRNA HOTAIR | [237–245] |
| 4 | LARP1 | Stabilization/Destabilization | Bcl2/BIK | [246] |
| 5 | SRSF1 (SF2/ASF) | Stabilization/Destabilization | CXCL1, PTEN | [247] |
| 6 | CPEB2 | Stabilization | p53 | [248] |
| 7 | eIF4E | Stabilization | PI3-K | [249] |
| 8 | FXR1 | Stabilization | miR17HG, TAL1 | [250] |
| 9 | hnRNP F | Stabilization | Snail1, TCF3, APP | [251–253] |
| 10 | hnRNP H | Stabilization | TCF3 | [252] |
| 11 | hnRNP H1 | Stabilization | APP | [253] |
| 12 | hnRNP I | Stabilization | Insulin | [254] |
| 13 | hnRNP K | Stabilization | c-Src, mPer3 | [233, 255] |
| 14 | hnRNP M | Stabilization | IL-6 | [256] |
| 15 | hnRNP R | Stabilization | CCNB1, CENPF | [257] |
| 16 | hnRNPA2/B1 | Stabilization | BIN1, WWOX, CFLAR, CASP9, CD44, TP53IP2, E-cadherin, Twist, Snail1 | [258] |
| 17 | IGF2BP1 | Stabilization | circXPO1 | [259] |
| 18 | IGF2BP3 (IMP3) | Stabilization | miRlet-7 | [260] |
| 19 | IGFBP2 (IMP2) | Stabilization | miR438–3p, miR151–5p | [261] |
| 20 | Khd1p | Stabilization | MTL1 | [262] |
| 21 | LARP6 | Stabilization | Vimentin | [263] |
| 22 | LARP7 | Stabilization | SIRT1 | [264] |
| 23 | MSI | Stabilization | PTEN, TIMP-3 | [265, 266] |
| 24 | PDCD4 | Stabilization | Bcl-xL | [267] |
| 25 | QKI | Stabilization | RASA1 | [268] |
| 26 | RBM10 | Stabilization | SMN2 | [269] |
| 27 | RBM3 | Stabilization | IGF2, YAP1 | [270] |
| 28 | RBM47 | Stabilization | AXIN1, DKK1 | [271, 272] |
| 29 | SAM68 | Stabilization | lncRNA HOTAIR, miR155HG, | [273] |
| 30 | SMAD | Stabilization | TGF-β1/p53 | [274] |
| 31 | SRSF3 | Stabilization | β-catenin / TCF4 | [275] |
| 32 | TIA-1 | Stabilization | IL-1β | [276] |
| 33 | Wig1 (ZMAT3) | Stabilization | p53 | [277, 278] |
| 34 | CELF1 | Destabilization | BAD, Bax, JunD | [279] |
| 35 | DND1 | Destabilization | CCR4-NOT | [280] |
| 36 | hnRNP D | Destabilization | LDLR | [281] |
| 37 | hnRNPD | Destabilization | p53, mPer3 | [255, 281] |
| 38 | KHSRP | Destabilization | ERRFI1 | [282] |
| 39 | TTP | Destabilization | Nox2 | [283] |
Dysregulation of, and mutations in, RBPs are linked to various disease pathologies, including neurodegenerative diseases [13], cardiovascular diseases [14], and, most importantly for this review, cancer [9]. Manual curation of various high throughput datasets identified about 1,542 RBPs to be expressed in humans samples [15], of which 727 has been implicated in cancers [12]. Significant evidence supports the involvement of RBPs in modulating expression of various oncogenes and tumor suppressor genes. For example, Zhang et al., recently analyzed the expression pattern of RBPs in 16 human cancers and identified aberrant overexpression of 109 RBPs and downregulation of another 41 RBPs; each RBP associated with molecular features and disease prognosis across the various cancer types [16]. Additionally, RBPs are reportedly crucial governors of almost all cancer hallmarks, including cell proliferation (RBM 38, RBM24, IGF2BP1, hnRNP1, HuR), evasion of apoptosis (HuR, eiF4E, TIA-1), angiogenesis (HuR, TIA-1, TTP), epithelial to mesenchymal transition (EMT) and metastasis (HuR, hnRNP, IGF2BP1, KHSRP). All of these RBPsare key regulators of various aspects of tumorigenesis [1, 17], and their expression correlates with disease prognosis and patient survival across multiple cancer types; for example, expression of AUF1 correlates with disease stage and metastasis in both colorectal cancer [18] and esophageal squamous cell carcinoma [19], while elevated expression of Lin 28A and Lin 28B correlates with poor progression free survival and overall survival in multiple malignancies [20]. Many RBPs are also involved in lung cancer (LC) pathogenesis; reported correlations between expression of multiple RBPs and clinico-pathological characteristics of LC tumorigenesis are listed in Table 2.
Table 2:
RBPs and their significance in LC progression
| RBPs | Clinical specimens | Expression level | Assay method | Correlation with histopathological features | References |
|---|---|---|---|---|---|
| HuR | 81 NSCLC tissues and 15 benign lung tissue | Upregulated | IHC | Cytoplasmic HuR correlated with VEGFC-C levels, tumor grade, differentiation status and lymph node metastasis | [59] |
| 236 NSCLC tissues | Upregulated | IHC | Cytoplasmic HuR levels correlated with tumor progression and higher microvessel density, Positive correlation of cytoplasmic and nuclear HuR levels with COX2 | [67] | |
| RBM 5 | 120 tumor and non-tumor NSCLC tissues | Downregulated | RT-PCR and WB | RBM5 protein expression correlated with smoking status, tumor stage and lymph node metastasis of NSCLC | [284] |
| RBM 47 | 175 pairs of NSCLC and adjacent non-cancerous tissues | Upregulated | IHC | RBM47 expression was associated with the pathological type | [285] |
| KHSRP | 75 pairs of cancerous and noncancerous fresh tissues from NSCLC patients | Upregulated | IHC | High expression of KHSRP associated with an advanced TNM stage and metastasis | [286] |
| hnRNP C | 75 pairs of cancerous and noncancerous fresh tissues from NSCLC patients | Upregulated | IHC | No correlation available | [286] |
| IGF2BP1 | Paired samples of NSCLC tissues and adjacent normal tissues | Upregulated | IHC | Upregulation of IGF2BP1 contributed to poorer overall survival in NSCLC | [287] |
| IGF2BP3 | Paired samples of NSCLC tissues and adjacent normal tissues | Upregulated | IHC | Upregulation of IGF2BP3 contributed to poorer overall survival in NSCLC | [287] |
| LARP1 | 84 pairs of primary NSCLC and adjacent non-tumor tissues | Upregulated | qRT-PCR | High LARP1 expression contributes to poorer survival in NSCLC | [288] |
| ESRP1 | 65 cases NSCLC, 20 cases pre-cancerous lesions, 30 benign lung nodules | Upregulated | IHC | Levels of ESRP1 independent prognostic factor for NSCLC | [289] |
| Lin 28 | 69 NSCLC tissue samples | Upregulated | RT-PCR | High levels of Lin-28 correlates with resistance to therapy | [290] |
| SRSF1 | 6 pairs of NSCLC samples and adjacent normal lung tissues | Upregulated | WB | No correlation available | [291] |
| SRSF6 | 5 normal lung and 6 lung tumor tissues | Upregulated | qRT-PCR | No correlation available | [292] |
| hnRNP A1, A2 | 16 tumor tissues from NSCLC patients and normal tissues | Upregulated | IHC, qRT-PCR | High hnRNP A1, A2 levels in combination with low-Tid-1L levels was associated with poor survival of NSCLC patients | [293] |
| PDCD4 | 28 tumor tissues from NSCLC patients and control tissues | Downregulated | qRT-PCR | No correlation available | [294] |
| QKI | 10 adenocarcinoma tissues | Downregulated | WB | No correlation available | [295] |
| MSI-2 | 123 human NSCLC tumor samples and normal lung tissues | Upregulated | TMA | High expression of MSI-2 correlated with extent of disease progression | [296] |
| 40 NSCLC patients and control tissues | Upregulated | IHC | MSI-2 correlated with decreased progression free survival and overall survival | [297] | |
| MSI-1 | 202 TMA specimens | Upregulated | TMA | No correlation available | [298] |
| Kin17 | 97 NSCLC and benign lung lesion tissue specimens | Upregulated | IHC, WB | Kin17 overexpression was associated with tumor grade and lymph node metastasis | [299] |
| eIF4E | 70 cases NSCLC, 19 NSCLC adjacent tissues and 20 normal tissues | Upregulated | TMA, IHC | Higher expression of eIF4E correlates with lymph node metastasis | [300] |
| FXR1 | 292 lung cancer tissues | Upregulated | IHC | Increased expression of FXR1 associated with poor survival of NSCLC | [301] |
| SAM 68 | 208 NSCLC samples | Upregulated | IHC | Increased expression of SAM68 correlates with lymph node metastasis and TNM stage | [302] |
| hnRNP K | 70 lung tissue samples | Upregulated | IHC | No correlation available | [303] |
Since RBPs regulate expression of multiple genes they are attractive targets for cancer management [17]. In a disease like cancer, where multiple genes drive pathogenesis and each of those genes may be regulated via a unique distinct mechanism, definite strategies to mitigate cancer growth are unavailable. A relatively superior approach is potentially offered by modulation of RBP expression, which might in turn lead to global alterations in gene expression of numerous oncogenes and tumor suppressor genes, and thus block cancer progression. Over the last few decades, RBPs have become a prolific field of research and an in-depth knowledge of RBP structure, biology, and function is thus essential to develop them as novel targeted anti-cancer therapies. This review discusses one important and well-studied RBP, Human antigen R (HuR), and describes its profound importance in the context of LC progression, its potential as a therapeutic target for LC, and strategies for targeted delivery of anti-HuR therapies.
HuR
Human Antigen R (HuR), a member of the embryonic lethal abnormal vision like (ELAVL) family of proteins, was first cloned by Ma et al., in 1996 [21]. The ELAVL family of proteins comprises the ubiquitously expressed non-neural protein HuR/HuA (ELAVL1), and the neuronal proteins – HuB (ELAVL2), HuC (ELAVL3), and HuD (ELAVL4). HuR is a 32kDa protein encoded at position 19p13.2 in the human chromosome [22], and contains three characteristic RNA recognition motifs (RRMs i.e. RRM-1, RRM-2 and RRM-3). Early studies, using a poly(A) sepharose beads-based sandwich assay, showed RRM-1 and 2 (the N-terminal domains) bind to AU-rich RNA elements (AREs) in the 3’ untranslated regions (UTR) of mRNAs, while RRM-3 (C-terminal domain) binds the poly(A) tail of mRNAs and 5’U-rich RNA targets [23, 24]. An additional target motif for HuR, specifically a uracil rich RNA motif approximately 17–19 nucleotides in the 3’ UTR, has been reported in various HuR target genes [25]. The RRM3 subunit is implicated in dimerization and RNA binding, and is thought to be pivotal for regulating HuR activity and function; any mutation in RRM3 will result in reduced recognition of target RNAs [24, 26, 27]. There is also a hinge region, the HuR nucleoplasmic shuttling domain (HNS), between the RRM2 and RRM3 subunits which is crucial for the shuttling of HuR between the nucleus and cytoplasm and for its interaction with nuclear proteins [27, 28].
The importance of HuR in biological systems is apparent as early as the embryonic stage. Mouse embryos with genetic ablation of HuR exhibit retarded growth and skeletal deformities and fail to survive beyond mid-gestation [29]. Another study using the Cre-recombination system for HuR deletion showed that HuR is essential for immune and hematopoietic progenitor cell survival [30]. The ubiquitous importance of HuR is underlined by multiple other studies demonstrating its importance in various physiological process including spermatogenesis [31], muscle physiology [32], myogenesis [33], and adipogenesis [34]. Recent studies show that HuR in smooth muscle cells is involved in regulation of blood pressure [35], and HuR knockout (KO) leads to atherosclerosis via increased expression of AMP- α1 and - α2 [36]. Thus, in normal cells HuR is a posttranscriptional regulator modulating expression of genes associated with vital biological processes including homeostasis, organ growth, and development.
Under normal conditions, HuR predominantly localizes to the nucleus; however, when cells are stressed, for example under hypoxia or in presence of proliferative signals, HuR binds to AREs typically located in 3′- UTR of mRNAs (cytokines, growth factors, oncogenes, inflammatory molecules), shuttles to the cytoplasm, and influences the stability and processing of those mRNAs [26]. HuR regulated target mRNAs, including VEGF, COX-2, c-myc, and cyclin B1, among many others, are largely involved in cell growth and proliferation. The known HuR target mRNAs are comprehensively reviewed by Srikantan et al. [37]. Further, the nucleo-cytoplasmic trafficking/shuttling of HuR is largely modulated by the MAPK, AMPK, Chk2, and PKC (α and δ) family of proteins [38]. Both intrinsic (inflammation, oxidative stress, hypoxia) and extrinsic factors (radiation, chemotherapeutic drugs, hormones, viral infections) contribute to the modulation and regulation of HuR function [39–41].
HuR undergoes various post-translational modifications (PTM) such as phosphorylation, methylation, NEDDylation, and ubiquitination; these PTMS largely determine the localization and regulation of HuR function such as binding to mRNA transcripts. For example, the phosphorylation of HuR at S88, S100 (within RRM1), and T118 (within RRM2) by Chk2 and at S318 (within RRM3) by PKC δ influences the ability of HuR to bind target transcripts [27, 42–45]. Another study showed that JAK3-mediated phosphorylation of HuR at Y200 prevents accumulation of HuR in stress granules and lowers its interaction with target mRNAs including siRT1 and VHL [46]. Mutation of HuR at T118 position contributes to reduced interaction between HuR and the target mRNA cPLA2α [47]. Similarly, PTMs and mutations in the HNS region change the nuclear-cytoplasmic localization pattern of HuR. Phosphorylation at S158 and S221 by PKC increases cytoplasmic HuR levels [48], while phosphorylation at S202 by Cdk1 boosts HuR nuclear accumulation [45]. Kim et al., showed that the mutations S242D and S242A led to increased nuclear or cytoplasmic localization of HuR respectively [49]. Furthermore, methylation at R217 correlates with increased cytoplasmic HuR and additionally impacts survival rates of non-small cell lung carcinoma (NSCLC) patients [50]. Other PTMs, such as ubiquitination and NeDDylation at K313 and K326, are prerequisites for HuR RNA binding, protein stabilization, and localization functions [51, 52]. HuR PTMs and their functional effects are comprehensively reviewed by Grammatikakis et al. [53].
HuR plays a central role in the pathogenesis of multiple sclerosis [54], inflammatory diseases [55], diabetes and its associated complications [56], and cancer [39, 40, 57]. Notably, aberrant overexpression of HuR is a characteristic feature of almost all cancer types including LC [59], breast [39], liver [57], and colon [58] (Figure 1A–C). HuR, via its interactions with various cancer-associated mRNAs, mediates the vital processes of cell growth and proliferation, angiogenesis, invasion, and metastasis, and thus contributes to malignancy [39, 40]. The differential expression pattern of HuR between normal and cancerous tissues, along with its involvement in regulation of numerous genes makes HuR an attractive therapeutic target for cancer treatment.
Figure 1.
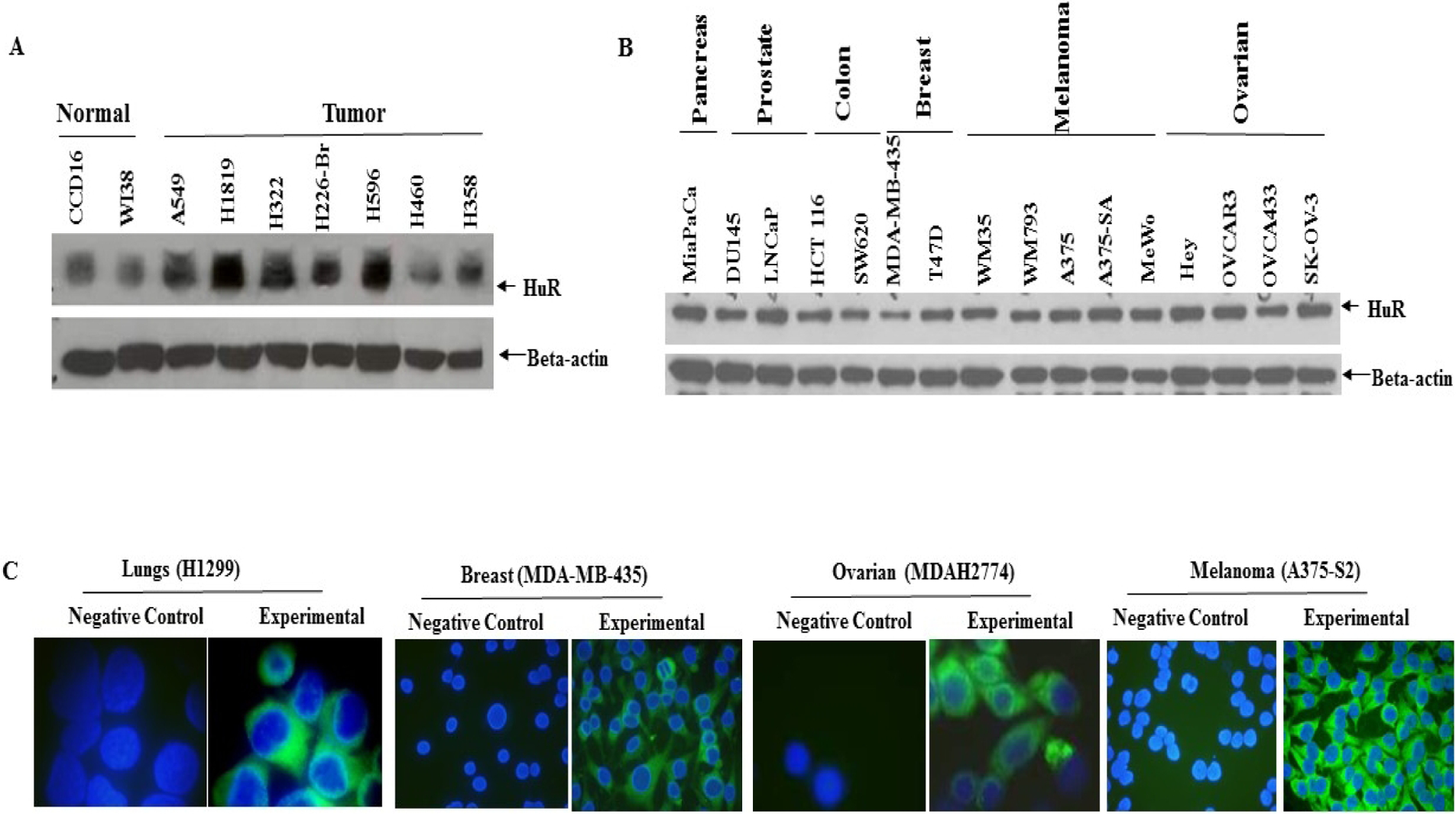
Detection of HuR protein expression by western blotting (A, B) and immunofluorescence (C) in human cancer cell lines. A, HuR expression in human normal lung and tumor cell lines and B, in panel of human cancer cell lines. Beta-actin was used as a loading control C, Immunofluorescence images showing the cytoplasmic localization of HuR in different cancer cell lines. Blue – nuclear DAPI staining; Green – HuR staining.
Lung Cancer (LC)
Due to complex etiology and tumor heterogeneity, delayed diagnosis, and lack of effective treatments, LC remains a leading cause of death in the United States (https://cancerstatisticscenter.cancer.org). The absence of early clinical symptoms also results in diagnosis at late stages (usually III or IV), further limiting treatment options and reducing survival [60]. Additionally, the conventional therapies, namely chemotherapy, radiation, and surgical excision of tumor removal have significant limitations associated with them. Although significant progress, such as molecular targeted therapies [61] and immunotherapy [62], has been made in the management of LC, a clear path to complete tumor remission and cure is still lacking. Thus, there is an urgent need to identify novel mechanisms for drug delivery and disease management in LC.
Clinical significance of HuR expression in LC
As in multiple other cancers types, differential expression of HuR is reported between normal, benign, and tumorigenic lung tissues [63, 64]. Lauriola et al., examined the correlation between HuR levels and age or smoking parameters in stage I and II lung adenocarcinoma (LUAD) patients; and found no correlation between HuR levels and patient dependent factors such as smoking or age [65]. HuR trafficking from nucleus to cytoplasm is further associated with disease pathology (or) oncogenic transformation observed in LC, in contrast to the nuclear HuR expression seen in benign lung tissues, a mixture of both nuclear and cytoplasmic expression is detected in malignant NSCLC tissues [59, 66, 67]. Given this variation in localization patterns, cytoplasmic levels of HuR can be/is used as an indicator of disease prognosis [67]. Furthermore, expression of HuR is correlated with survival rates in patients. The 5-year survival rates of HuR- positive patients was reportedly lower (43%) when compared to HuR-negative patients (85%) [67]. Higher cytoplasmic HuR levels also correlated with risk of death and metastasis in stage I and II LUAD patients [65], and was further associated with clinico-pathological features such as tumor grade (39%, 62% and 68% at stage I, II and IIIA of NSCLC), increased tumor proliferation, micro vessel density, differentiation status, and lymph node metastasis in NSCLC [47, 59, 67, 68]. Additionally, cytoplasmic HuR is associated with higher VEGF-C expression levels [59] and COX-2 levels [67, 68] which in turn could contribute to formation of endothelial cells to support angiogenesis and cancer metastasis in LC. Furthermore, the ratio of nuclear to cytoplasmic HuR is a prognostic factor for evaluating clinical outcomes and metastasis in early stage LUAD patients [65].
Aberrant overexpression of HuR was also seen in the tumorigenic epithelial LC cell lines H1299, A549, HCC827, PC-9 and urethane induced lung tumors in vivo compared to the normal lung cells CCD16, MRC-9 [63, 69, 70] (Figure 1 A). Levels of cytosolic HuR protein were higher in more rapidly growing cell lines derived from papillary tumors (PCC4 and LM2) than in slower growing cell lines derived from solid tumors, supporting modifications in HuR levels concurrent with changes in growth rates/levels (confluence rates) [63].
Based on the clear and pronounced role of HuR in LC, in the current review we aim to summarize the critical role of HuR in LC progression and address the various pharmacological, biological, and genetic approaches for HuR inhibition. Finally, we describe the immense potential of using various novel nanomedicine approaches for targeted therapy of HuR that would benefit multiple cancer types including LC.
Regulation of HuR by non-coding RNAs (ncRNAs)
In the human genome, ncRNAs contribute to regulation of transcriptional and post-transcriptional gene expression and play major roles in biological processes such as splicing, transcription, protein localization and secretion [71]. ncRNAs comprise miRNA, snoRNAs, piRNAs (short ncRNA) and long non-coding RNAs (lncRNA). Multiple studies demonstrate the dysregulated pattern of ncRNAs across various cancer types and discuss avenues for therapeutic targeting of ncRNAs [72, 73]. Various miRNAs have been identified to regulate HuR expression in cancer models; for example, miR-31 was shown to regulate HuR expression in LC cells. Inhibition of miR-31 increased HuR expression which in turn upregulated cyclins and VEGF [74] thereby supporting cell proliferation and angiogenesis. HuR inversely correlates with miR-145 expression in gastric cancer, supporting HuR as a direct target of miR-145 [75]. Similarly, overexpression of miR-22, via the Jun/miR-22/HuR axis, decreased colorectal cancer growth [76].Another miRNA, miR-155 is important for HuR- dependent increase in migration and chemotaxis in colon cancer cells; knockdown of miR-155 decreased cytoplasmic HuR levels and blocked the migratory potential of colon cancer cells [77].
With respect to lncRNAs, the lncRNA MALAT1 binds to HuR, and via the HuR-TIA-1 signaling cascade contributes to the aggressive and metastatic signature of pancreatic cancer cells [78]. Likewise, formation of a ribonucleotide complex encompassing MALAT1 and HuR reportedly regulates CD133 expression and contributes to to EMT in breast cancer [79]. Several ncRNA-based strategies have reached clinical trials (NCT04594720, NCT03830619) and are increasingly recognized for their therapeutic potential. The ncRNAs shown to regulate HuR expression in multiple cancer types are listed in Table 3. Novel strategies employing various ncRNAs that target HuR will thus be an attractive approach in cancer therapeutics.
Table 3:
Regulation of HuR by ncRNAs
| ncRNAs | Mechanism of function | Cancer type | Effect | References |
|---|---|---|---|---|
| circRNAs | ||||
| circAGO2 (hsa_circ_0135889) | activates HuR | Gastric cancer | Progression | [304] |
| circHuR (hsa_circ_0049027) | interacts with CNBP and downregulates HuR | Gastric cancer | Suppression | [305] |
| circDLC1 (hsa_circ_0135718) | Bind to HuR and decreases MMP1 expression | Hepatocellular carcinoma | Suppression | [306] |
| Exosomal circSHKBP1 (hsa_circ_0000936) | Sponged miR-582-3p and increased HuR levels | Gastric cancer | Promotion | [307] |
| circPABPN1 (hsa_circ_0031288) | Binding to HuR lowers translation of PABPN1 | Cervical cancer | Modulator of protein translation | [308] |
| circCCND1 (hsa_circ_0023303) | Physical binding to CCND1 mRNA | Laryngeal squamous cell carcinoma | Promotion | [309] |
| hsa_circ_0074854 | Interacts with HuR and reduces stability | Hepatocellular carcinoma | Suppression | [310] |
| LncRNAs | ||||
| lncRNA NEAT1 | Stabilization by HuR | Ovarian cancer | Progression | [76] |
| lncRNA CASC9 | Binding to HuR | NSCLC | Progression | [64] |
| lncRNA OCC-1 | Destabilizing HuR function via binding | Colorectal cancer | Suppression | [311] |
| lncRNA SNHG-12 | Binds to HuR and modulates YWHAZ/AKT/GS K-3β axis | Gastric cancer | Promotion | [312] |
| lncRNA HGBC | Stabilization by HuR | Gall bladder cancer | Promotion | [237] |
| lncRNA FENDRR | Hinder binding of HuR to MDR-1 | NSCLC | Attenuates stemness | [313] |
| lncRNA HMS | Stabilization by HuR | LUAD | Promotion | [314] |
| lncRNA HOTAIR | Interaction with HuR | Head and neck squamous cell carcinoma | Promotion | [315] |
| lncRNA ASB16-AS1 | Promotes Ubiquitination of HuR | Adrenocortical carcinoma | Promotion | [316] |
| lncRNA LINC00336 | Binding to HuR | LC | Inhibition of ferroptosis | [317] |
| miRNAs | ||||
| miR - 519 | Lowers translation of HuR | Cervical cancer | Suppression | [318] |
| miR- 466i | Interaction with HuR | Th 17 cells | Regulation of GM-CSF expression | [196] |
| mir-22 | Binding to HuR and protein inhibition | Colorectal cancer | Progression | [76] |
| miR-145 | Regulation of HuR expression | Gastric cancer | Suppression | [75] |
| miR-16 | Loss of repression of HuR translation | Breast cancer | Progression | [319] |
| miR-519a | Regulation of HuR expression | Laryngeal squamous cell carcinoma | Suppression | [320] |
Approaches for targeting HuR
As described above, HuR has an established role in tumor progression and drug resistance. Given the ubiquitous expression of HuR in almost all malignancies along with its profound role in post-transcriptional regulation of key genes relevant to tumor progression and survival, HuR is an attractive target for cancer therapy. Thus, it is imperative to identify novel therapeutic strategies to inhibit the biological functions of HuR. Various approaches using inhibitors, siRNAs, and shRNAs, have been proposed and are currently in use to block HuR function; these are discussed in brief in the following sections.
Pharmacologic approaches
a). Inhibitors from natural sources
The most widely used HuR inhibitor, MS-444 (a myosin light chain kinase inhibitor) was identified from microbial culture by Meisner et al., in 1996. MS-444 is reported to interfere with HuR binding, disrupt cytoplasmic trafficking of HuR and its homodimerization, and inhibit interaction of HuR with ARE’s [80]. Colorectal cancer cells treated with MS-444 show decreased growth and increased expression of genes related to apoptosis [81]. The effect of MS-444 was also assayed in glioblastoma cells derived from xenograft models and brain tumor initiating cells. MS-444 induced apoptosis, resulted in cell death, inhibited sphere formation, and the invasive abilities of glioblastoma cells, and decreased the mRNA expression of genes relevant to key cancer hallmarks, namely angiogenesis (VEGF, HIF-1α), evasion of immune response (TGF-β1, TGF-β2, PD-L1, iNOS) and apoptosis (CIAP2, Bcl-2, Bcl-xl, DR5) [82]. Similarly, MS-444 showed promising results in curtailing tumor growth in malignant peripheral nerve sheath tumor (MPNST) models [83]. Programmed death-ligand 1 (PD-L1) is widely overexpressed in tumor cells and via engaging with programmed cell death −1 (PD-1) contributes to immune evasion. Hence, various approaches have been proposed to inhibit the complex PD-1/PD-L1 regulatory pathways [84]. In this regard, few reports support the involvement of HuR in PD-L1 regulation. A recent study showed MS-444 based HuR inhibition upregulated the expression of CMTM6 which consequently led to reduction in the PD-L1 overexpression normally seen under HuR overexpression conditions [85].
Novel inhibitors from natural products also inhibit HuR’s interaction with AU-rich elements. Kaur et al., used a fluorescence polarization assay to identify a compound, azaphilone (AZA-9), from fungal products that blocks HuR–RNA binding residues and inhibits their interaction [86]. However, the efficacy of AZA-9 in blocking LC cell growth and progression requires a thorough assessment. Latrunculin A, a macrolide from sponges, when administered to HepG2 and Huh7 hepatocellular carcinoma cells also reduced cytoplasmic HuR levels, resulting in decreased stability of various HuR-binding mRNAs i.e. COX-2, cyclin A and D1, and subsequently impaired migratory properties and prostaglandin E2 synthesis [87].
Another natural compound, Dihydrotanshione I (DHTS), from Salvia miltiorrhiza (red sage) is widely used in Chinese medicine. DHTS, via stabilizing a closed conformation of HuR [88] reportedly inhibits HuR function and RNA binding activity. Agostino et al., provided supporting evidence that DHTS inhibits breast cancer migratory properties in a HuR-dependent manner [89]. Additionally, DHTS in combination with HuR KO in the HCT 116 colon cancer cell models reduced tumor growth with minimal toxicity to mice [88].
b). Synthetic chemistry-based lead identification:
Fluorescence polarization assays coupled with high throughput screening was employed by Wu et al., to identify small molecule inhibitors that disrupt the interaction of HuR with its target mRNAs. Using this technique, nine compounds from Chemical Methodology and Library Development center (CMLD 1–6) and (NC 1–3) were developed and their ability to disrupt HuR-ARE interactions were tested. Two candidate molecules, CMLD-1 and CMLD-2, showed promising results in inhibiting HuR and its target proteins (Bcl-2 and XIAP) leading to apoptotic cancer cell death [90]. Following this finding, a large number of studies used CMLD-2 to inhibit HuR function in a plethora of cancer cell line models and achieved effective outcomes. CMLD-2, via downregulation of MAD-2, decreased cell viability, increased apoptosis further lowering the migratory and colony forming abilities of thyroid cells [91]. Our own laboratory has assessed the effects of CMLD-2 in NSCLC cells. Consistent with the findings in other cancer types, CMLD-2 treatment resulted in a preferential concentration-dependent cell cytotoxicity selective to tumor cells as characterized by activation of caspases and increased apoptosis. Furthermore, LC cells treated with CMLD-2 exhibited cell cycle arrest at G1 phase consistent with cell death. Therein, we also showed a decrease in the levels of multiple HuR-regulated mRNAs and pro-apoptotic proteins, such as Bcl-2, Cyclin E and Bcl-XL, and increased rates of mitochondrial perturbation in cells treated with CMLD-2 [92].
The newest class of inhibitors potentially inhibiting cytoplasmic dimerization of HuR also showed encouraging results. Filipova et al., combined approaches from medicinal chemistry with high-throughput based firefly-luciferase HuR assays to identify inhibitors (A92, SRI-42127, and SRI-41664) that inhibit cytoplasmic dimerization of HuR. SRI-42127, halted the proliferation and colony forming abilities of patient-derived glioblastoma (PDGx) xenolines. In addition, SRI-42127 reduced the interaction between HuR and Bcl-2 and HuR and Mcl-1 leading to induction of apoptosis and decreased tumor growth. The inhibitor altered various cell signaling pathways and reportedly crossed the blood-brain barrier to decrease glioma growth [93].
Inspired by the functional activity of DHTS against HuR, researchers utilized synthetic chemistry- based approaches to identify various next generation tanshione mimics capable of blocking HuR function. Tanshione mimics 6 and 6a, by keeping HuR in a closed conformation, prevent HuR binding to various target mRNAs [94], leading to reduced tumor cell growth [83]. Additionally, multimerization of HuR (aggregation of 3 domains) is associated with glioma progression, and tanshinone compounds disrupt this HuR multimerization reducing tumor cell proliferation and survival [95]. Similar mechanisms of HuR multimerization in other cancer types warrant further studies, and novel pharmacologic and targeted-therapy based approaches to inhibit HuR multimerization will be beneficial for cancer management.
c). Commercial drugs as HuR inhibitors
Various commercially available drugs are increasingly recognized as HuR inhibitors. For instance, the anthelmintic pyrvinium pamoate reportedly blocks both cytoplasmic accumulation and nucleocytoplasmic translocation of HuR. Furthermore, combinatorial therapy using pyrvinium pamoate and other chemotherapeutic agents resulted in reduced growth of urothelial carcinoma in vitro and in vivo. Pyrvinium pamoate activates the AMPK/importin α1 cascade, thus promoting the nuclear import of HuR, while the nucleocytoplasmic translocation of HuR is blocked by inhibition of the Chk1/Cdk1 pathway [96]. Pyrivinium pamoate also had potent therapeutic effects and led to complete tumor regression in vivo in MPNST models [83]. Similarly, the anti-typanosomal drug suramin, binds to HuR and decreases the expression of cyclins A2 and B1; decreases motility and invasive properties of oral (tongue) carcinoma cells [97]. Eltrombopag, a drug used for treating low platelet count (thrombocytopenia) and anemia, is promising promise as a HuR inhibitor in multiple cancer models. Use of eltrombopag altered HuR levels and decreased mRNA copy numbers of VEGF-A and MMP-9 resulting in reduced microvessel density and impaired angiogenic process [98], providing proof of concept for its anti-tumor and anti-angiogenic effects. Additional data regarding the effect of these drugs on the expression of other HuR regulated mRNAs is necessary to fully realize their potential for cancer therapy.
d). Other inhibitors
Various other small molecule inhibitors have also been tested for their effect on HuR regulation. The microtubule inhibitor MPT0B098, decreases HuR trafficking to the cytoplasm thereby leads to destabilizing HIF-1α mRNA [99]. Additionally, the myosin inhibitor blebbistatin decreases nuclear-cytoplasmic HuR shuttling in hepatocellular carcinoma cells, thereby decreasing destabilized COX-2 and cyclin expression and reducing tumor growth [87]. The anti-proliferative effects of abemaciclib, an established CDK 4/6 inhibitor, were assessed in PDAC cell lines; where induced senescence and cellular apoptosis thus blocking cell cycle progression. The authors showed that abemaciclib resulted in inhibition of HuR and YAP1, which in turn led to downregulation the cyclin D1/CDK4/6 axis thereby leading to the to the aforementioned effects [100].
The information provided in this review on pharmacological approaches of HuR inhibition is not exhaustive, and interested readers are directed to the review by Schultz et al., [101] for more comprehensive information on HuR inhibitors, and their mechanism of action. HuR is a key molecular regulator driving cancer progression in various cancer types, including LC, and the above cited reports provide strong evidence that HuR inhibitors are a promising therapeutic target for disease management. However, more in-depth studies addressing the pharmacokinetics, pharmacodynamics, and regulatory mechanisms of these small molecule inhibitors are essential before they can be tested in the clinical arena as therapeutics for management of cancer.
Biological approaches
a). RNA interference based approach
The process of oncogenesis is tightly controlled by multi-level regulatory networks of genes that dictate cell proliferation and growth. RNA interference (RNAi) is a powerful therapeutic strategy that is being widely used to suppress aberrant overexpression of such disease-causing genes. Gene silencing approaches based on RNAi have an edge over conventional treatment modalities because of their ability to selectively turn off key genes responsible for disease progression. In cancer, RNAi can result in global knockdown of key proteins involved in various cancer-related pathways thus decreasing tumor growth. Furthermore, they are indispensable tools for manipulating “druggable” and, more importantly, “non-druggable” targets that lack any commercially available drugs or small molecule inhibitors. RNAi works via two cellular approaches. In the first, a commercially synthesized small interfering RNA (siRNA) is exogenously added to the biological system; the introduced double-stranded DNA or RNA duplex is cleaved by DICER and converted to shorter siRNA strands, which load into RNA induced silencing complex (RISC). Subsequently the guide strand of the siRNA binds complementarily to the target mRNA of interest followed by mRNA cleavage by Argonaute proteins to result in gene knockdown [102, 103]. siRNAs are delivered into cells via transfection, electroporation, viral vectors and various chemical modifications can also be introduced into the siRNA sequence to improve its stability. The second approach involves use of short hairpin RNA (shRNA). shRNA, widely used for in vivo experimental models, allows target gene knockdown for longer durations. These shRNA systems are also inducible, for example with tetracycline, to allow gene knockdown in a conditional manner. Additionally, the shRNA systems have selectable antibiotic markers which removes untransfected cells from the system [104]. Since their initial development, RNAi based interventions have become increasingly important for management of various diseases including cancer [105]. Currently many RNAi based therapeutics for cancer are being tested in clinical trials (NCT01591356, NCT04995536, NCT02166255). In the following sections, we summarize reports on genetic inhibition of HuR using siRNAs and shRNA, and it effects on various attributes of tumor growth in cancer models, in particular LC.
HuR is extensively studied in the context of cell proliferation. HuR regulates the expression of various pro-survival markers such as COX-2, VEGF, c-Myc, CDK and cyclins, and thus enhances cell growth and survival. High cytoplasmic HuR levels have been correlated with COX2 levels in squamous cell carcinoma [106] and prostate cancer [107], and co-expression of these molecules predicts tumor type and proliferative capacity [68]. Deregulation of the HuR subcellular localization of is further associated with change in the expression of COX-2 [68]. Additionally, a positive correlation between HuR and the DNA replication and proliferation markers MCM-6 and Ki-67, observed in NSCLC, supports the role of HuR in cell proliferation [50]. Higher levels of cytoplasmic HuR were a characteristic feature of rapidly growing cell lines derived from papillary lung tumors (PCC4 and LM2) when compared to slower growing cell lines derived from solid lung tumors. These data support a strong connection between HuR levels and rates of cellular proliferation [63].
Numerous studies show HuR stabilizes the expression of genes (Table 1) responsible for/involved in cancer cell survival, as well as migratory and invasive properties. HuR, via its binding and interaction with CDK3 mRNA promoted apoptosis and decreased proliferation of breast cancer cells [108]. In the same breast cancer cell line models, MCF-7 and MDA-MB-231, HuR knockdown decreased expression of cyclin D1 and MMP-9 leading to stunted growth and impaired invasive abilities [109]. Jimbo et al., attempted a targeted-therapy approach using shRNA and siRNA to knockdown HuR in PDAC cell lines and further establish its role in cell proliferation, invasion, migration and anchorage independent cell growth. The study supported role of HuR knockdown in reducing the malignant phenotype of PDAC cells [110]. Other studies with shRNA- mediated knockdown of HuR in both, in vitro and intracranial in vivo models of glioblastoma also showed decreased tumor growth including anchorage-independent growth and proliferation compared to untreated cells [111].
siRNA and shRNA based-strategies to block HuR in ovarian cancer cells were assessed by Huang et al.,. Transiently and constitutive suppression of HuR in ovarian cancer cells using siRNA and shRNA respectively, led to decreased cell proliferation, colony forming abilities, invasion and migratory potential and suppression of tumor growth in vivo. This was attributed to reduced rates of cellular proliferation rather than induction of tumor cell death. Differential transcriptomic profiles from ovarian cancer A2780 cells with and without HuR knockdown showed that HuR regulated essential processes including cell proliferation, angiogenesis, cell-cycle regulation, apoptosis, and DNA repair [112]. Cancer stemness contributes to self-renewal abilities in cells and tumor progression by giving rise to various lineages of cells. HuR knockdown inhibited expression of Oct4 and Nanog (markers for cancer stemness) in LC cells, suppressing their tumorigenic sphere formation and blocking growth [113]. Since these essential processes are crucial for tumor growth, blocking HuR is beneficial for management of cancer.
Evidence from multiple studies shows that cancer growth to arise from chronic inflammation [114, 115]. HuR, via post transcriptional regulation of inflammation associated cytokines IL-6, IL-8, TNF-α, TGF-β, IFN-γ, and COX-2 is identified to support tumor progression [29, 37, 116–118]. High HuR levels reportedly contribute to an inflammatory microenvironment which in turn fosters fibrosis [119–121], potentially leading to tumorigenesis [122]. Interestingly, HuR expression, function and translocation can be altered by presence of inflammatory factors. For example, one study reported that treatment of A549 LC cells with the pro-inflammatory cytokine IL-1β enhanced the interaction of HuR with cPLA2α mRNA, a molecule implicated in inflammation; a siRNA based HuR knockdown approach in the same cells inhibited IL-1β induced cPLA2α expression [47].
Angiogenesis is a complex process comprising of various steps such as extracellular matrix (ECM) degradation, and migration and proliferation of endothelial cells resulting in the formation of new blood vessels to support tumor growth. Increased angiogenesis is essential for tumorigenesis and metastasis [123]; and HuR is increasingly recognized as a contributor to the process of angiogenesis [124]. Expression of cytoplasmic HuR correlates with angiogenesis and lymphangiogenesis in NSCLC tissues [66]. HuR, via binding to ARE in 3’ UTRs, regulates expression of key molecules in angiogenesis, specifically VEGF, COX-2, MMP-9, HIF-1 and uPA [124–126]. Various studies showed siRNA mediated knockdown of HuR to result in decreased levels of both, VEGF-C [59] and COX-2 [127]. Chang et al., used a selective site specific HuR KO in mouse myeloid cells to establish that expression of HuR in bone marrow macrophages is a prerequisite for stabilizing the expression of genes by binding to 3’UTRs. This study also established the role of miR-200b in regulating the interaction of HuR with VEGF-A, and in doing so modulating angiogenesis [128]. VEGF and COX-2 are crucial mediators of angiogenesis in malignancy, and HuR, by stabilizing VEGF and COX-2 mRNAs, activates the angiogenic phenotype in tumor endothelial cells (TECs) to facilitate angiogenesis. Knockdown of HuR reduces expression of VEGF-A and COX-2 and inhibits motility and tube formation in TECs of both oral carcinoma and melanoma [129]. Additionally, angiotensin has been shown to amplify the cytokine (TNF-α and IL-1β) induced interaction of HuR with COX-2 in glomerular mesangial cells whereas siRNA-based HuR silencing reduced the observed increase in COX-2 and prostaglandin levels seen under stimulated conditions [130].
HuR in combination with another RBP, polypyrimidine tract-binding protein (PTB), binds to HIF-1α mRNA, a master regulator switch for angiogenesis, thus increasing its translation. HIF-1α correlates with VEGF levels [131] as well as with poor survival in LC patients [132]. In breast cancer models, inhibition of HuR expression reduced HIF-1α translation [132] which in turn decreased expression of the transcription factor Hsf1 and reduced angiogenesis [133]. HuR is also implicated in stabilization of the angiogenesis regulator urokinase-type plasminogen activator (uPA) and HuR knockdown using RNAi reduces uPA levels [126]. Collectively, HuR-mediated reduction of HIF-1α or uPA levels would decrease VEGF [126, 131] and constitute a therapeutic approach for targeting angiogenesis.
Cancerous cells, via various discrete mechanisms, have the ability to evade the immune response and thus enhance their survival. HuR is required for survival of hematopoietic stem cells (HSCs) [30], which contributes to generation of various blood cell lineages. Early experiments on mouse spleen cells stimulated with CD3/CD28 showed increased HuR levels [134]. High HuR expression is seen under conditions of B cell activation, further supporting HuR as an elemental factor for B cell growth, differentiation, and antibody production [135]. Additionally, HuR deletion in pro B-cells led to a reduction in the number of immature and mature B cells in the spleen and bone marrow compared to controls; this study also demonstrated that HuR mediates B cell-T cell interactions, further contributing to humoral immunity [136]. Rothamel et al., combined data from multiple high-throughput studies and showed that HuR regulates target mRNAs involved in various pathways of adaptive (Th1 and Th2 cell differentiation) and innate immunity (IL-17 signaling, TNF signaling and TLR signaling) [137]. In terms of tumor progression, glioma models in HuR conditional KO mice showed increased infiltration of CD4+ cells and M1 macrophages contributing to reduced tumor growth and prolonged survival [138]. HuR knockdown also decreased expression of PD-L1, a target for cancer immunotherapy, and a combined approach using KH3 (a small molecule HuR inhibitor) along with an antibody directed at PD-1 (the PD-L1 receptor), decreased tumor growth in breast cancer [139]. Our laboratory has also identified HuR knockdown as contributing to increased PD-L1 expression in LC cells (unpublished data). Additional studies on HuR inhibition and PD-L1 expression are essential to incorporate HuR inhibition to enhance immune cell recognition and benefit cancer immunotherapy. Similarly, another cytokine, granulocyte-macrophage colony stimulating factor (GM-CSF), is gaining prominence in the context of cancer immunotherapy. GM-CSF plays a vital role in immune evasion and tumor progression [140]; GM-CSF upregulates PD-L1 expression and leads to poor prognosis in patients [141]. An association between HuR and GM-CSF has been reported, specifically HuR binds to the 3’UTR of GM-CSF mRNA and regulates its expression; HuR KO in Th17 cells reduces GM-CSF expression [142]. HuR-based regulation of GM-CSF expression, which in turn would modulate PD-L1, would open new avenues for immunotherapy in cancer.
HuR has also been implicated in cancer metastasis. Multiple reports reveal a correlation between HuR levels and its localization pattern with tumor metastasis in both bladder cancer [143] and LC [59, 68]. In addition to playing a role in invasion and migration, HuR, in the presence of increased ATP, increases the stability of MMP-9 mRNA [144] potentially contributing to ECM degradation which is an essential first step in tumor cells intravasating into blood vessels. Furthermore, HuR stabilizes Snail mRNA leading to increased migration [145] and induction of EMT which supports metastasis. HuR-expressing PDAC cells injected into mice preferentially metastasized to the liver and lung compared to HuR-KO mice. The report concluded that HuR KO decreased expression of CTNNB1 and YAP1 mRNAs which led to decreased metastasis [146]. Irigoyen et al., studied the effects of shRNA mediated HuR knockdown on various tumor promoting aspects of MPNSTs; HuR shRNAs decreased cell viability, suppressed MPNST cell growth, decreased clonogenic and anchorage-independent growth, and abrogated tumor formation. The study also examined the influence of HuR depletion in pre-formed tumor models. The authors concluded that HuR depletion altered core biological processes causingdecreased proliferation, increased senescence, G1 phase cell cycle arrest, apoptosis, and necrosis leading to attenuated tumor growth. The HuR depletion also reduced conversion of micro metastasis to macro metastasis, further confirming the role of HuR in cancer metastasis. Pathways downregulated by shHuR included YAP/TAZ (Hippo signaling - responsible for cell survival), Wnt/β-catenin (responsible for cell proliferation, differentiation, and apoptosis), PI3K/AKT/mTOR, and RAS (involved in signal transduction allowing ECM to support tumor growth), alongside altered levels of key transcription factors including MYC, RB, and E2F [83]. Additionally, the HuR inhibitor KH-3 reduced the interaction of HuR and FOXQ1 and inhibited lung metastasis of breast cancer cells [147].
Although various experimental models across multiple cancer types support a role of HuR in cancer metastasis, there is dearth of data revealing pathways mediated by HuR in the context of LC metastasis. Wu et al., identified interactions between CRABP2 and HuR that are mediated by integrin β1/FAK/ERK signaling and lead to metastasis in LC models. They further demonstrated that CRABP2 knockdown with shRNA or siRNA abated the metastatic ability of C10F4 LC cells [148]. Given this preliminary finding, additional studies using preclinical models with LC metastasis are essential to elucidate the regulatory role of HuR in this context.
The goal of cancer therapy is to achieve complete tumor remission and improve over-all patient survival. However, in most cancers, the complex genomic landscape limits the ability to achieve optimal treatment responses and results in therapeutic failure. Drug resistance, largely governed by the tumor microenvironment which contains cancer cells, immune cells and stromal populations, is a multifaceted problem and a principal cause for failure of anti-cancer therapies. The association of HuR with drug resistance has been explored in numerous cancer types, including pancreas, breast, brain, and lung [149], and many studies conclude that HuR post-transcriptionally regulates expression of genes related to multi drug resistance (MDR) further supporting HuR as a therapeutic target. Few studies indeed supported downregulation of HuR as a mechanism contributing to drug resistance in cancer types. One preliminary study reported that HuR knockdown reduced expression of Bim which regulated the resistance to EGFR-TKIs that is common in LC; HuR knockdown reduced apoptosis induced by the EGFR-TKI gefitinib. In contrast, restoring HuR expression levels restored Bim expression and increased sensitivity to gefitinib [70]. EGFR-TKI resistance is widely seen in LC patient and modulation of HuR function has the potential to address this critical issue. HuR, by modulation of Top2A, has also been linked to in vitro resistance of breast cancer cells to doxorubicin [41]. The information provided here regarding the role of HuR in cancer drug resistance is not comprehensive and readers are directed to the review by Zhou et al., for additional information [149].
HuR knockdown has also been reported to sensitize cells to anti-cancer therapies such as chemotherapeutic drugs and ionizing radiation. HuR overexpression, by regulating CDC6, increased proliferation, tumor growth, and resistance to oxiplatin in colorectal cells while HuR knockdown increased sensitivity to oxiplatin [150]. A similar effect of HuR knockdown on the response to epirubicin has been described. Specifically, HuR knockdown increased generation of reactive oxygen species (ROS) and via the ROS-HuR-TGF-β pathway increased sensitivity to epirubicin. Inhibition of galectin-3/β-catenin signaling by siHuR, led to decreased expression of MDR proteins and was identified as a key mechanism for prevention of MDR in colorectal cancer cells [151]. In pancreatic cancer cells, silencing of HuR decreased COX-2 and HO-1 levels thus contributing to increased sensitivity to gemcitabine [152]. In contrast, Constantino et al. concluded that cytoplasmic HuR expression regulates the stabilization of deoxycytidine kinase (dCK) mRNA to serve as a predictor of gemcitabine efficacy [153]. The precise impact of HuR on gemcitabine response is not clearly defined and requires further investigation.
Radiation therapy is a routine treatment modality for cancer management. In a few reports, a link between HuR knockdown and sensitization of cells to ionizing radiation has also been established. HuR, by upregulating caspase-2 and inhibiting thioredoxin reductase, sensitized colorectal [154] and breast cancer cells [155] to radiation therapy. Additionally, studies from our laboratory verified that post-transcriptional regulation of ARID1A by HuR increased resistance to radiation therapy in breast cancer cells [156]. Combining HuR knockdown with radiation therapy could, thus, lead to reduced tumor burden.
In summary, HuR correlates to both therapeutic resistance and sensitivity in cancers. The observed difference in mechanisms reported regarding the role of HuR with respect to resistance and sensitivity may be largely attributable to the complex genomic landscape of different cancers. A more in depth understanding of how HuR regulates treatment response in pre-clinical models and in clinical trials would bridge the missing links and open additional avenues for HuR-based targeted therapy.
b). Genome editing approaches
Mutations confer malignant potential to normal cells and by discrete genetic and epigenetic mechanisms causes tumor cell growth, proliferation, and metastasis. During the carcinogenic process multiple oncogenes are overexpressed and, thus, the goals of cancer therapy are largely to suppress the aberrant expression of oncogenes and in turn profoundly impact therapeutic response and patient survival. Clustered regularly interspaced short palindromic repeats (CRISPR) associated protein (Cas9) is a genome editing tool widely used for genetic manipulation. Using this technology, precise changes, including insertions, deletions and corrections, can be made in a genome in order to study gene function. There is convincing evidence for the potential benefits of CRISPR-Cas9 technology in various disease states, including diabetes [157], obesity [158], and neurodegenerative and eye disorders [159].
Over the last years, CRISPR-Cas9 has gained significant attention in the field of cancer therapy [160]. Some studies have employed CRISPR-Cas9 to better understand HuR regulation and biology in pancreatic and colorectal cancer models [161]. Lal et al., used CRISPR-Cas9 based genome editing to selectively delete the HuR gene in pancreatic (MIAPaCa-2 and Hs766T) and colorectal cancer (HCT116). HuR deletion resulted in a less malignant phenotype, increased apoptosis, and reduced sphere forming abilities in vitro compared to the wild type cells, thus supporting the role of HuR in cell proliferation and growth. Furthermore, MIAPaCa-2, Hs 766T, and HCT 116 mouse models deficient in HuR had reduced tumor volume further substantiating the importance of HuR in regulation of oncogenesis. The authors concluded that deletion of HuR modulated KRAS activity as well as expression of other oncogenic genes to impair tumor growth in mice. In line with this report, Dong et al., and Chand et al., provide evidence supporting CRISPR-Cas9 based HuR deletion to benefit pancreatic cancer therapy. In the first report by Dong et al., HuR KO pancreatic cancer cells exhibited decreased invasion, migration, and spheroid formation compared to controls in vitro, as well as delayed tumor formation in vivo [162]. Chand et al., showed that CRISPR-Cas9-based HuR deletion regulated PARG enhanced sensitivity of PARP inhibitors (olaparib, veliparib and rucaparib), and that HuR silencing combined with PARP inhibitors attenuated tumor growth in PDAC models [163].
While CRISPR-Cas9-based knockdown of HuR has been extensively interrogated in pancreatic cancer models, its effect in other cancers, including lung, remain largely untested. Information on CRISPR-Cas9-based HuR inhibition in LC would advance our understanding of the complex HuR regulatory mechanisms in LC. Given that KRAS mutations are widely reported in LC, it would be worthwhile to study the effect of HuR on KRAS regulation in LC. Likewise, PARP inhibitors are increasingly recognized for management of SCLC [164] and NSCLC (NCT01788332), and integration of HuR silencing with PARP inhibitors may beneficial be for LC management.
In summary, studies from our laboratory and other groups show HuR to be an essential molecular regulator of cancer progression and that HuR inhibition, either transient or stable via use of inhibitors, siRNAs, shRNAs, and CRISPR/Cas9, can strikingly reduce cell viability and tumor growth in vitro and in vivo. Such RNAi, gene silencing, and pharmacologic inhibition based therapies targeting HuR exhibit profound beneficial effects at the preclinical level, however, their successful transition into clinical use to benefit patients is compromised by certain limitations associated with the use of these approaches. Firstly, the efficacy of siRNA delivery largely depends on the route of administration. While few examples of local administration, i.e. intranasal administration of Beclin-1 siRNA for HIV [165] and intravitreal caspase-2 siRNA for improving retinal ganglion cell survival [166] have shown promising results, directed and efficacious delivery to tissues by systemic administration remains a challenge [167]. The transport of siRNAs to a specific target site is primarily limited by their relatively small size and short half-life i.e. faster clearance by the kidneys, the inherent difficulty of penetrating cell membranes due to their negative charge, the unstable nature of siRNAs in the blood stream, and their susceptibility to nuclease degradation [168]. Achieving cell- or tissue-specific delivery of siRNAs is yet another major limiting factor restricting their therapeutic application. There are also significant hurdles to incorporating CRISPR/Cas-9 based genome editing technology into a clinical setting. Constraints pertaining to site specific delivery of CRISPR/Cas-9 systems with minimum off-target effects currently limits its application. Emerging evidence supports incorporating novel nanomedicine-based drug delivery systems to overcome the observed limitations pertaining to degradation, site specific delivery, and off-target effects associated with the use of siRNAs and CRISPR/Cas-9 [169–174]. Developing effective carrier molecules capable of supporting site-specific delivery with minimal cytotoxicity is crucial in order to successfully translate siRNA, shRNA and CRISPR-Cas9 based approaches. These nanomedicine-based approaches are described in the following sections.
Nanomedicine based approaches
The field of nanomedicine, due to its ability to specifically deliver drugs and other biological moieties, such as DNA, RNA, and peptides, to target sites, has gained much attention over the last decade. The term nanomedicine broadly encompasses the use of materials/carriers in the “nano” size range to encapsulate therapeutics and deliver them directly to the diseased site in order to achieve a potent therapeutic effect. Numerous studies report that use of nanomedicine-based approaches offer significant benefits over conventional therapies. Nanomaterials widely used in cancer therapy, especially LC, include polymeric nanoparticles, metallic nanoparticles, dendrimers, liposomes, and solid-lipid nanoparticles [175–177]. The physico-chemical properties such as shape, size, and surface charge largely dictate both, the fate of the nanocarrier within cells and the observed therapeutic effect. Nanoparticles owing to the enhanced permeation and retention (EPR) effect preferentially accumulate in target tissue of interest [178]. Specific features of nanoparticles, i.e. their tunable physico-chemical characteristics, their ease of synthesis and characterization, biocompatibility, and lack of immunogenicity support their application in modulation of cell signaling, drug delivery, imaging, gene therapy and immunotherapy [179]. Their superior and selective tumor-targeting abilities coupled with minimal toxicity to normal healthy cells are major advantages of using nanomedicine-based systems. Nanomedicine-based approaches such as Abraxane, paclitaxel stabilized in a nanoparticle formulation, are approved by the Food and Drug Administration for treating LC [180, 181].
In most cases, nanocarrier-mediated drug delivery depends on the model used for the study i.e. diseased condition and cell or tissue of interest. Every cell type possesses certain specific receptors on their surface, and this characteristic feature can be exploited to target those using nanocarriers. Effective targeting of nanocarrier has been explored in the past by decorating the surface of the nanoparticle with different targeting ligands. Conjugating siRNAs to nanocarriers such as dendrimers and liposomes would prevent degradation of siRNA and help achieve targeted delivery of siRNA and thus effective knockdown of target mRNA [174]. Although, a number of nanomedicine approaches have been reported for targeted therapy in LC [182–184], herein we selectively focus on dendrimers, liposomes, extracellular vesicles (EVs), and anti-sense oligonucleotides (ASOs) utilized for improved delivery of HuR siRNA/shRNA in cancer models. Furthermore, we also discuss a few emerging studies that support incorporation of a similar nanocarrier based approach of HuR knockdown in alleviating pain, diabetes and its associated complications, inflammation, and fibrosis.
Dendrimers
Dendrimers are a category of self-assembled nano-sized polymeric macromolecules [185] with profound importance in cancer drug delivery and therapy. The structure of dendrimers largely consists of a spherical core that is the central element, branched structures or dendrons, and end groups. They are synthesized by both convergent (synthesis begins with dendrons and finally adds them to the central core) and divergent (synthesis starts with the central core and then adds molecules) approaches. The physical, chemical, and biological properties of dendrimers can be modified via functionalization of end groups [186]. These special features such as surface functionalization tailored for specific purposes, controlled monodisperse synthesis of dendrimer particles with modifiable physico-chemical properties, makes them attractive options for use in biomedical applications such as delivery of drugs, gene delivery and as agents for magnetic resonance imaging (MRI) [187]. Chemotherapeutic drugs and other biomolecules and also nanocarriers can be loaded onto end groups attached to the dendrimers and the rate of drug loading and release can be modified by tuning the size of dendrimers.
Dendrimers belonging to the group of non-viral delivery systems have shown immense potential for delivery of nucleic acid therapeutics such as siRNAs and miRNAs [188–190]. The most widely used dendrimers for delivering nucleic acid therapeutics include polyamidoamine (PAMAM) and polypropylene (PPI). Others such as carbosilane dendrimers, triazine dendrimers, polylysine dendrimers, and phosphorous containing dendrimers have also been reported for carrying nucleic acids [188, 191, 192]. Readers are advised to refer to a review by Dzmitruk et al., to gain more information on applications of dendrimers as carrier molecule for siRNA and miRNAs [191].
To enhance the efficiency of drug delivery systems, researchers have also used novel cell-specific receptors (e.g. folate [193], integrins [194], and adhesion molecules [195]) overexpressed in cancerous conditions and designed delivery systems to recognize these cells for targeted delivery of cargoes such as siRNA, drug molecules, and imaging agents. For example, folate receptors are overexpressed in almost all cancer types including ovarian, breast, and lung. As a result, researchers have conjugated ligand (such as folic acid-FA) to a folate receptor in order to deliver siRNAs and anticancer drug to cancer cells using dendrimer- and liposome-based drug delivery systems. This active targeting overcomes a significant limitation of conventional drug delivery, i.e. passive targeting, and delivers the payload selectively to cancer cells which, in turn, offers benefits by increasing efficacy and reducing off target effects. In the context of HuR, a novel Cy3-labeled FA-derivatized DNA dendrimer was utilized by Huang et al., to deliver HuR siRNA to folate receptor overexpressing ovarian cancer cells. To overcome degradation of siRNAs in serum, the authors modified the nucleotide bases to improve stability of siRNA in serum. Intraperitoneal administration of FA-3DNA-siHuR in a mouse model for 4 weeks, reduced tumor growth and minimized chronic inflammation. In addition, survival of FA-3DNA-siHuR mice was prolonged 1.5 fold compared to controls, confirming that nanomedicine-based tumor targeting improves efficacy of targeted siRNA [112].
Dendrimer-based drug delivery can also overcome non-specific toxicities usually seen with systemic administration of chemotherapeutic agents such as cisplatin (cis-dichlorodiammineplatinum; CDDP). Our laboratory used a similar folate receptor-tumor targeting approach to specifically deliver siRNA and drug to LC cells. We developed a chemo-biologic co-delivery system combining CDDP and HuR siRNA with an FA-conjugated-PAMAM dendrimer and polyethyleneimine nanoparticles in order to demonstrate therapeutic efficacy in folate receptor overexpressing LC models [196]. Detailed protocols for synthesis, characterization, surface functionalization as well as the release kinetics of dendrimer-based delivery of CDDP and HuR siRNA to LC cells are described in our publication [197]. Polyethyleneimine was incorporated to increase the efficiency of siRNA complex formation and facilitate endosomal escape of bound siRNA into the cytoplasm. This targeted therapy showed greater therapeutic effect as characterized by increased cytotoxicity to LC cells with minimal cytotoxicity to normal MRC-9 cells [196].
Lipid nanoparticles or liposomes
Liposomes, artificial vesicles created from phospholipids, are a well-established mode of drug delivery [198]. Following the approval of Doxil®, the chemotherapeutic agent doxorubicin hydrochloride encapsulated in liposomes and the first approved liposomal formulation, several other liposomal based formulations (Daunoxome®, Myocet®, Depocyt®) have been approved for human use [199] and even more are undergoing clinical trials (NCT00765973, NCT03088813, NCT04047251). Liposomes offer several advantages including ease of synthesis, improved pharmacokinetics and pharmacodynamics, superior bioavailability, less cytotoxicity, and the ability to load drugs with varying solubility. Hydrophobic drugs are encapsulated in the phospholipid bilayer and hydrophilic drugs are loaded into the aqueous cavity of the liposomes. Furthermore, controlled release of siRNA over an extended time period can be achieved with liposomes [200]. Owing to these features, liposomal-based delivery systems remain the preferred choice for nanomaterial-based therapeutics. Various categories of liposome, such as cationic lipid-based liposomes (DOTAP, DOTMA), neutral lipid-based liposomes (DOPC), solid-lipid nanoparticulate systems, lipoid based formulations, and cholesterol conjugates are currently in use for siRNA delivery and have been extensively reviewed by others [200, 201].
In line with the established aberrant HuR overexpression in LC, our laboratory utilized DOTAP: Cholesterol lipid nanoparticles to selectively downregulate HuR levels and interrogate the effect on modulation of various key oncoproteins. As described above, this system also delivered HuR siRNA selectively to LC cells overexpressing folate receptor. This targeted therapy approach of delivering HuR siRNA via lipid nanoparticles (HuR-FNP) protected the siRNA from degradation and increased cellular uptake. This HuR-FNP formulation in H1299 LC cells induced apoptosis and arrested the cells in G1 phase of cell cycle resulting in reduced cell viability. In addition, HuR knockdown altered expression of cyclin D1, E and Bcl-2, and all major proteins implicated in tumorigenesis [69].
The transferrin receptor (TfR), which binds the iron-carrying protein transferrin for iron import into the cell, is another surface ligand over expressed in NSCLC and expression of which correlates with poor prognosis [202]. We developed a targeted liposomal formulation to deliver HuR siRNA to TfR-overexpressing LC cells. This study using DOTAP: Cholesterol compared the efficacy of targeted (TfR) and non–targeted (i.e. nanoparticles not conjugated to TfR) HuR siRNA delivery (Figure 2). We found greater growth inhibition and HuR knockdown with the TfR-targeted treatment of A549 and HCC827 LC cells, as well as minimal growth inhibition and impact on HuR expression in control MRC-9 cells. In vivo bio-distribution studies further showed preferential accumulation of HuR-TfNP at the tumor site and pronounced reduction in tumor burden [203] (Figure 3).
Figure 2.
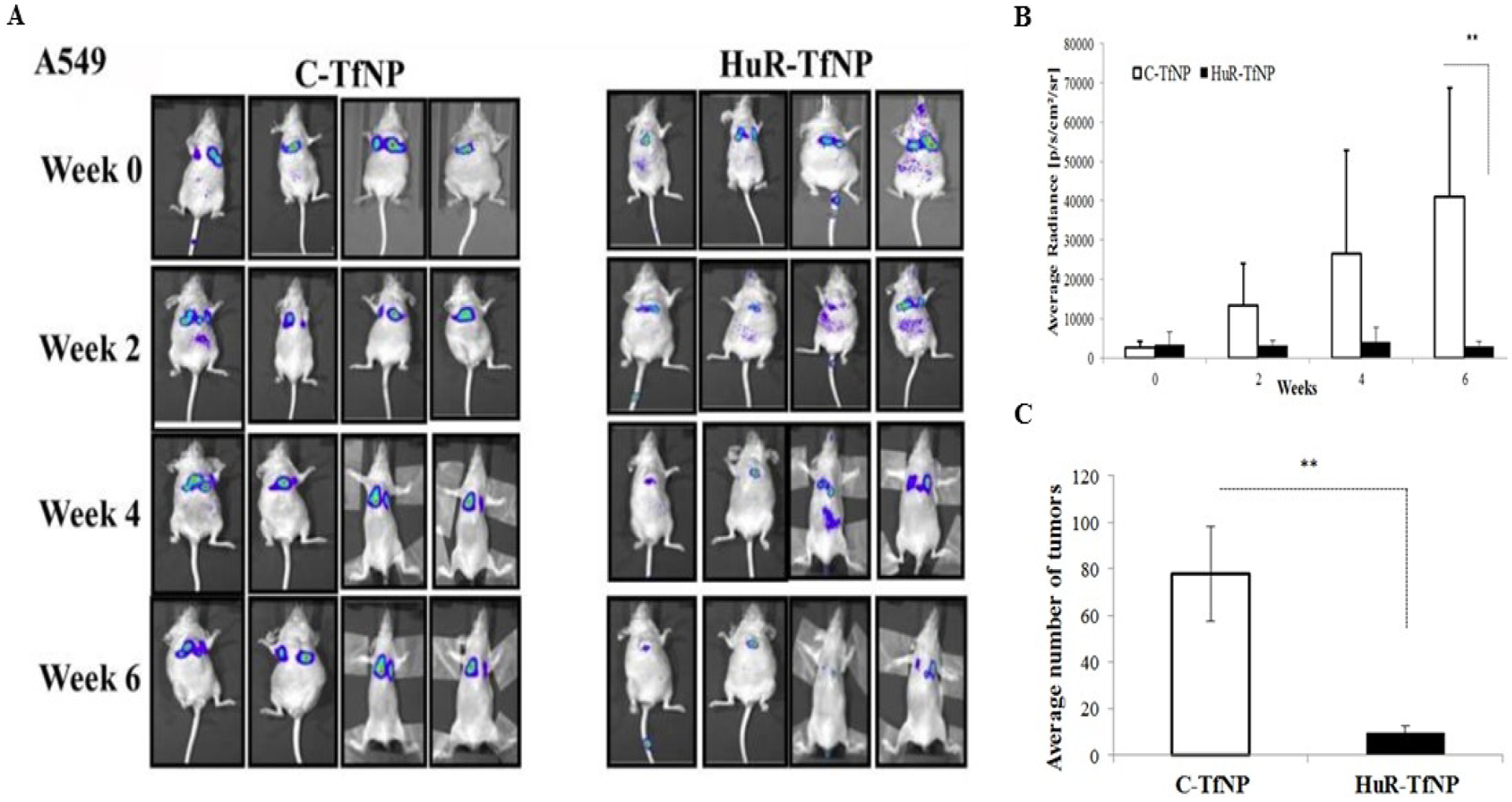
DOTAP: Cholesterol mediated delivery of HuR-TfNP in mice bearing A549-luc experimental lung tumors shows suppression of tumor growth in vivo. A, Representative bioluminescence images showing decreased signal intensity in HuR-TfNP treated mice compared to C-TfNP treatment. B, Average radiance in HuR-TfNP–treated mice compared to C-TfNP (control). C, HuR-TfNP treatment reduced the average number of tumors compared to C-TfNP treatment. Image reproduced from Muralidharan et al., 2017 that is under an open access Creative Commons CC BY 4.0 license.
Figure 3.
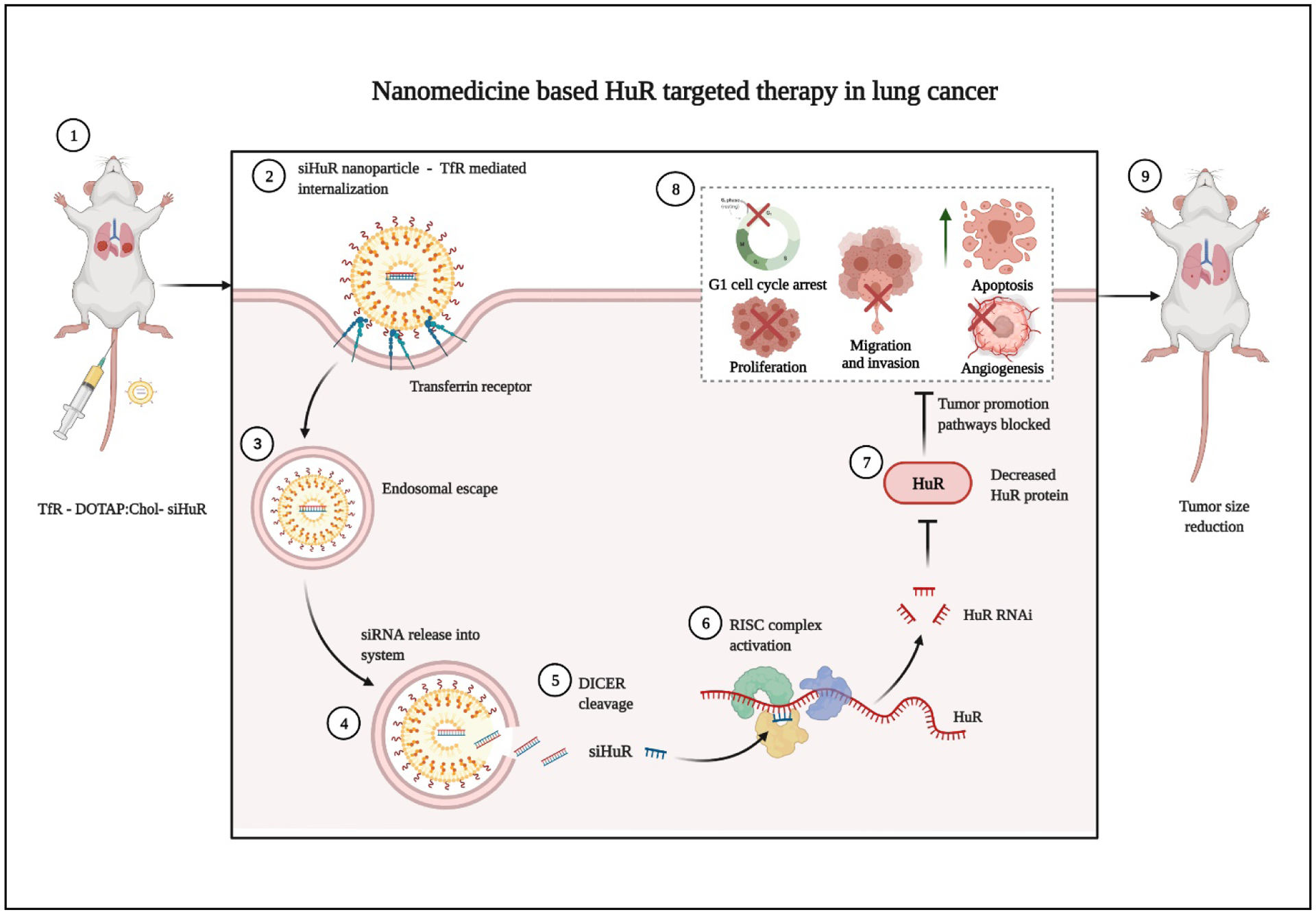
Schematic representation of nanomedicine-based HuR targeted therapy to reduce various aspects of tumor growth in LC. Image created with BioRender.com.
Given the promising results with HuR-FNP-based targeted therapy in LC cells, our laboratory also investigated a combination therapy with HuR siRNA and small-molecule inhibitors in LC cells. Expression of CXCR4, an identified target of HuR [204], correlates with LC invasion and metastasis and predicts poor prognosis in LC. This led us to combine HuR-FNP with AMD3100 (a CXCR4 antagonist) and we foundthe combination more effectively suppressed HuR mRNA expression at 48 h than did HuR-FNP treatment alone. The combination (HuR-FNP +AMD3100) induced cell cycle arrest at G1 phase, decreased cell viability, invasion and migration in H1299 LC cells (Figure 4). Additionally, HuR-FNP+3100 decreased expression of key proteins i.e. HuR, CXCR4, pAKT (Ser473), MMP-2, and MMP-9 implicated in invasion, migration, angiogenesis, and metastasis .hence [205].
Figure 4.
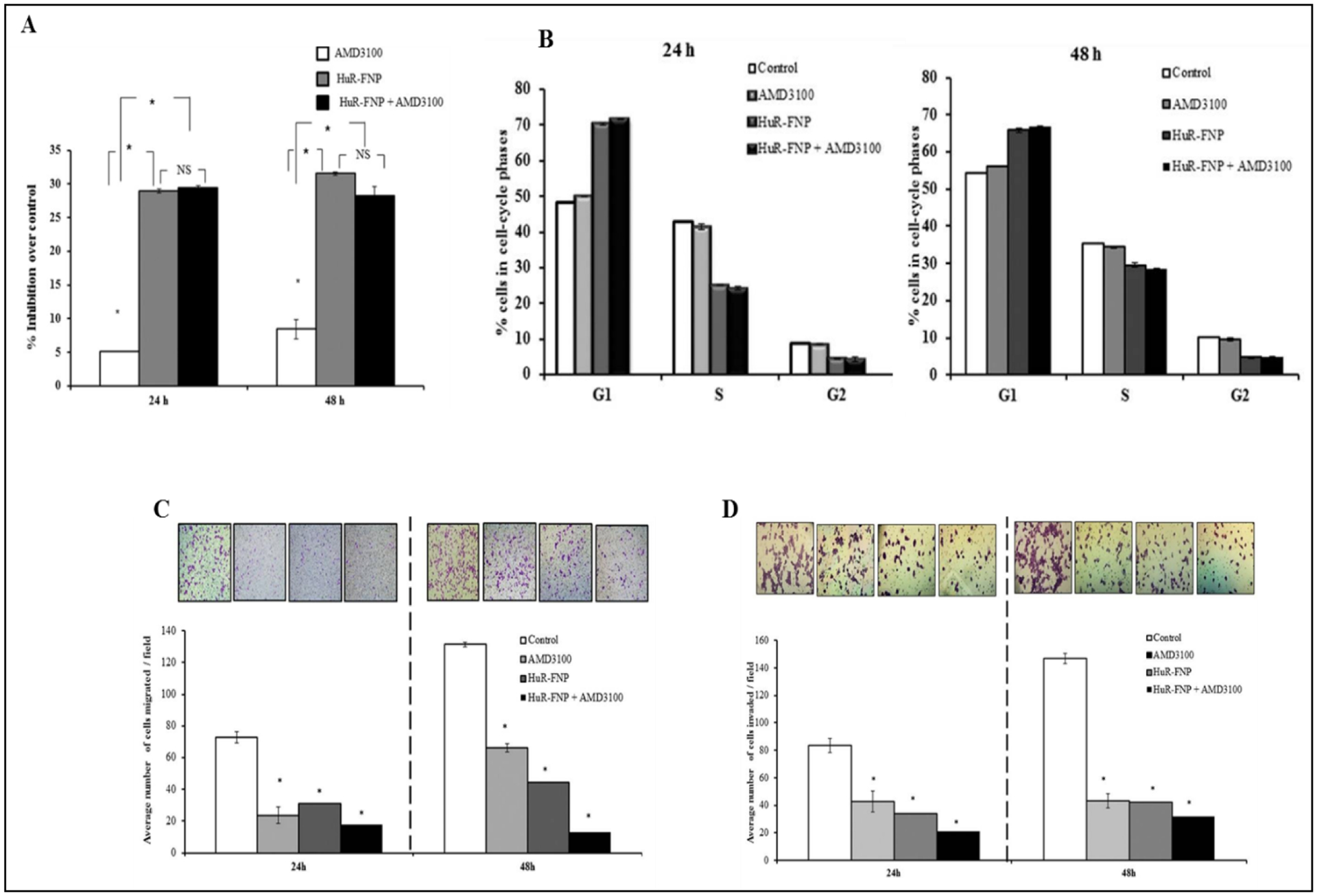
DOTAP:Cholesterol mediated delivery of HuR-FNP + AMD3100 inhibited A, cell proliferation; B, induced cell cycle arrest at G1 phase at 24 h and 48 h after treatment; C, decreased cell migration and D, invasion of H1299 LC cells. * p<0.05. Image reproduced from Muralidharan et al., 2015 under an open access Creative Commons CC BY 4.0 license.
We also extended this combinatorial therapy approach to investigate HuR knockdown in melanoma cells. Inhibition of HuR resulted in reduced cell proliferation and increased cell death characterized by activation of caspase-9 and cleavage of PARP. Furthermore, it led to reduction in expression of various oncoproteins (Cyclin D1, E1, Bcl-2, HIF-1a, VEGF-A, MITF) along with a concordant increase in expression of cell cycle protein p27. Various attributes of tumor growth such as invasion and migration were decreased in HuR siRNA treated melanoma cells compared to control melanocytes. Moreover, a combinatorial therapy of HuR siRNA with a MEK1/2 inhibitor led to greater decrease in MITF levels and p-MEK1/2 Ser 217/221 and increased cell growth inhibition in MITF overexpressing melanoma (MeWo) cell lines [206], establishing the potential for this approach in melanoma management.
While the role of HuR in pathogenesis of cancers is well established, its involvement in other diseases, such as diabetes and its complications (e.g. retinopathy and cardiac diseases), is also a burgeoning research area. For example, the involvement of HuR and activation of the PKCβ/HuR/VEGF pathway in diabetic retinopathy (DR) has been reported. PKCβ functions to activate HuR which in turn binds to VEGF mRNA and increases VEGF protein levels as seen in DR. Furthermore, increased retinal HuR and VEGF levels are seen in experimental STZ-induced diabetes models. To address this, Amadio et al., utilized liposome-siRNA complexes (lipoplexes) to deliver HuR siRNA to rat retinas for management of DR. Liposomal delivery decreased HuR levels, which via post transcriptional regulation of VEGF ameliorated retinal damage induced by STZ treatment [207].
Extracellular Vesicles (EVs)
It is increasingly recognized that EVs, the secreted bodies from cells responsible for cell-cell communication, have great potential for delivery of therapeutics. EVs, a broad term used to describe cell secreted vesicular bodies, largely comprises exosomes, micro vesicles and apoptotic bodies. EVs function to carry cargoes such as DNA, RNA, and other biological moieties and are implicated in various aspects of tumor progression including response to therapy and immune cell regulation [208–210].
Various methods are currently available for synthesis and characterization of EVs. Additionally, the content of EVs largely depends on cell type of origin, and thus EVs can serve as biomarkers or candidates for liquid biopsy of diseased conditions [211–214]. EVs isolated from various cell types, tissues, and bodily fluids (serum/blood, urine, amniotic fluid, saliva and breast milk) have supported their potential for disease diagnosis [214–216]. These natural drug carriers are able to cross biological barriers, including the BBB [217, 218], which conventional drugs cannot, and are key modulators of cell-cell communication. Owing to these attributes EVs constitute an active field of research in cancer diagnosis and therapy.
The advent of nanotechnology has significantly contributed towards exploiting the potential of EVs as drug-delivery vehicles, and this, in turn, has led to an explosion in the number of studies incorporating EVs as carrier molecule for siRNA delivery. Biological moieties when loaded into the lipid bilayer of EVs are protected from nuclease degradation [219], which results in more effective uptake by cells. Furthermore, EVs are an inherent component of cells and as such are neither immunogenic nor cytotoxic. Synthesized EVs can also be functionalized with various targeting ligands towards receptors that are overexpressed in cancer cells for selective delivery of biological molecule.
In this context, the role of HuR in governing levels of exosome secretion in cancer cells has been documented. Overexpression of HuR elevates exosome production four fold in colorectal cancer cells [220]. In contrast, HuR KO reduces exosome secretion in breast cancer cells [139]. EVs are also increasingly recognized as carriers for delivery of RBP therapeutics. Loughlin et al., exploited the therapeutic potential of EVs to deliver lipid (cholesterol)-conjugated HuR siRNA. Various synthesis parameters such as EV: siRNA ratio, incubation time, temperature, and reaction volume were optimized to facilitate efficient uptake of siRNAs. Temperature was a crucial factor dictating the incorporation of siRNA; at 4°C only 12% of cc-siRNA was incorporated into EVs while the efficiency increased to 60% at 22°C, 66% at 37 °C, and 75% at 42°C. The authors reported that 1h incubation at 37°C in 100 μl at a 1:15 ratio of EV: siRNA, resulted in dose-dependent silencing of the target HuR for up to 168 h, further validating the beneficial role of EVs in siRNA delivery strategies [221].
While there only a single study supporting the use of EVs to deliver siRNA to RBPs such as HuR, studies have successfully used EVs to modulate expression of target genes such as VEGF. Yang et al., used exosomes derived from brain endothelial cells to carry fluorescent-labelled VEGF siRNA. This formulation decreased VEGF levels up to 75% in glioblastoma cells (U87-MG). Furthermore, using zebrafish models, the authors showed that the formulation could cross the BBB and successfully deliver the siRNA to brain tumor cells [217]. Our laboratory has used nanosomes (i.e. an exosome/gold nanoparticle-based delivery system) to selectively deliver doxorubicin to LC cells [222]. A similar approach could be used to deliver HuR siRNA to LC cells, by incorporating HuR siRNA into nanosomes. More studies focusing on EVs to inhibit HuR expression along with its target genes are essential to assess if this is a feasible approach for management of LC.
Anti-sense oligonucleotides (ASOs)
ASOs are short stranded synthetic oligonucleotides (18–30 bp in length) which via complementary base pairing bind to mRNA resulting in modification of protein expression [223, 224]. The concept of utilizing antisense oligonucleotides as cancer therapeutics has been described in the context of angiogenesis and drug delivery [225]. Factors such as high specificity and limited toxicity make them attractive options for cancer therapy and a growing number of clinical trials are focused on unravelling the potential of ASOs in targeted cancer therapy (NCT01829113, NCT03101839, NCT00640861). Unmodified ASOs are susceptible to nuclease degradation [226] and thus, various chemical modifications are incorporated to improve stability of ASOs [223]. These include phosphorothioate, phosphorodiamidate, locked nucleic acids, and ribose substitutions at the 2′-O-methyl (2′-OMe) and 2′-O-methoxyethyl (2′-MOE) positions. These modifications greatly improve the stability and target binding features, and result in enhanced uptake of ASOs by various tissues [227], thus improving efficacy [223, 228].
Inflammation is not specific to cancer, but is also associated with various other disorders including spinal cord nerve injury. Peripheral nerve injury contributes to activation of microglia and release of pro-inflammatory factors to result in nerve pain and injury. HuR, via altering expression of key inflammatory molecules such as IL-1β, TNF-α and IL-6, has been implicated in mediating inflammation. As a result, targeted knockdown of HuR using phosphorothioate phosphate- modified ASOs was attempted by Borgonetti et al., to minimize neuropathic pain caused by nerve injury in peripheral mononeuropathy mouse models. The ASOs were incubated with cationic lipid (DOTAP) to enhance cellular uptake and stability, and administered via intranasal and intrathecal routes. Administration of the ASO reduced levels of the pro-inflammatory cytokines IL-1β and iNOS, and increased secretion of IL-10, which converted M1 macrophages to the M2 phenotype and reduced glial cell activation [229].
Studies regarding the role of various ncRNAs in regulation of HuR, and discussion of targeted approaches using ncRNA to modulate HuR, have been described in previous sections of this review. Additionally, the role of lncRNA MALAT1 in the regulation of HuR and its target mRNAs has also been discussed. Use of an ASO-based approach to inhibit lncRNA MALAT1 was described by Amodio et al., [230]; they showed that LNA-gapmeR ASO decreased NRF1 and NRF2 and led to caspase-mediated apoptosis in multiple myeloma cell lines and xenograft models. Although, this report did not provide information on the effect of LNA-gapmeR on HuR levels, we speculate that a HuR-based regulatory mechanism would mediate all of these effects. Follow-up studies on the role of this ASO in HuR function will provide additional essential information. Similar strategies targeting lncRNA MALAT1 and other ncRNAs that regulate HuR are expected to mitigate tumor progression and display therapeutic benefits.
Conclusion
HuR is a pivotal regulator of tumor progression and has profound clinical importance in LC. Increasing evidence, from both experimental and clinical datasets, continues to enhance our knowledge regarding the regulation of key oncogenes and cytokines by HuR. Despite significant advances in understanding the multi-level regulatory networks of HuR in other cancer types, there remains a substantial knowledge gap with respect to LC. The pronounced involvement of HuR in almost every aspect of tumorigenesis, has led researchers to develop strategies to block HuR function. Studies from our laboratory and others have provided direct evidence, using pharmacologic inhibitors, siRNAs, and shRNAs in both in vitro and in vivo models (Figure 5), to support the claim that HuR inhibition is highly beneficial in cancer. However, to date no HuR-based therapy has reached clinical trials. A suitable experimental model using HuR knockout specifically in lung cancer will provide necessary support to progress HuR-based therapies to clinical trials. LC patients have higher incidences of metastasis to bone, liver, and brain, and studying the role of HuR in metastatic LC models could elucidate its specific involvement in metastasis and further enhance development of anti-cancer therapeutics.
Figure 5.
Summary of various pharmacological, biological and nanomedicine-based HuR inhibition strategies available across various cancer models. Image created with BioRender.com
The reports discussed herein convincingly demonstrate the versatility and adaptability of nanotechnology-based approaches (i.e. liposomes, dendrimers, EVs, ASOs) for targeted delivery of cancer therapeutics. However, the advances made in this field remain nascent (Figure 5), and more intense preclinical investigation of these systems is warranted to better utilize and adapt these models for the management of cancer. Given the fact that HuR is required for effective functioning of normal cells and that the global knockdown of HuR in cells is reportedly lethal [30], the use of nanocarriers to selectively suppress HuR in cells and tissues of interest with minimal or no toxicity to normal, healthy cells offers immense potential for clinical application. However, the following points must be carefully considered when seeking to translate these proof-of-concept studies to clinics. First, numerous factors, both intrinsic and extrinsic, have been reported to modulate HuR expression and function, thus a relevant study model to delineate HuR biology is urgently required. Second, efforts to optimize design parameters for nanocarriers must be made in order to deliver equal payloads of siRNA to individual cells. Thirdly, incorporating an exogenous and endogenous stimuli-responsive nanomaterial [231] would provide controlled release of siRNA to the site of interest. Fourth, the route of administration for these biologics must be thoughtfully planned in such a way as to ensure maximum bioavailability and therapeutic effect with minimum cytotoxicity. Finally, the complex intratumoral heterogeneity seen in tumorigenic conditions such as LC cannot be ignored and could potentially result in disparities in HuR levels among different individuals, resulting in failure of therapies. While our advocacy for delivery of siRNA as a standalone therapy may not fulfill all requirements of LC management, combining siRNAs and chemotherapeutic drugs/inhibitors using novel nanomedicine-based drug delivery systems would benefit the field of personalized medicine and provide patient specific siRNA-targeted therapies.
Acknowledgments
This study was supported in part by grants received from the National Cancer Institute (NCI) of the National Institutes of Health (NIH; R01 CA167516, R01 CA233201, R01 CA 254192); from the National Institute of General Medical Sciences (P20 GM103639) of the National Institutes of Health; a Merit Grant (101BX003420A1) from the Department of Veterans Affairs; a Pilot Grant and Seed Grant funded by the National Cancer Institute Cancer Center Support Grant P30CA225520 awarded to the University of Oklahoma Stephenson Cancer Center; from the Department of Defense (DOD) through the Lung Cancer Research Program (LCRP; W81XWH-19-1-0647); a grant (HR18-088) from the Oklahoma Center for Advanced Science and Technology (OCAST); funds received from the Presbyterian Health Foundation Seed Grant, Presbyterian Health Foundation Bridge Grant, Oklahoma Tobacco Settlement Endowment Trust (TSET) awarded to the University of Oklahoma Stephenson Cancer Center, and the Jim and Christy Everest Endowed Chair in Cancer Developmental Therapeutics.
Rajagopal Ramesh is an Oklahoma TSET Research Scholar and holds the Jim and Christy Everest Endowed Chair in Cancer Developmental Therapeutics.
We would like to thank Danny Morton for editorial assistance.
The content is solely the responsibility of the authors. The opinions, interpretations, conclusions and recommendations are those of the authors and not necessarily endorsed by or representative of the official views by the NIH, DOD, Department of Veterans Affairs, or the Oklahoma Tobacco Settlement Endowment Trust.
Abbreviations
- ARE
Adenine rich elements
- ASOs
Anti-sense oligonucleotides
- AZA-9
Azaphilone 9
- BBB
Blood brain barrier
- Cas9
CRISPR- associated protein
- CDDP
Cis-dichlorodiammineplatinum
- circ RNA
Circular RNAs
- CMLD
Chemical Methodology and Library Development center
- CRISPR
Clustered regularly interspaced short palindromic repeats
- DHTS
Dihydrotanshione
- DNA
Deoxyribonucleic acid
- DOTAP
Dioleoyl-3-trimethylammonium propane
- DR
Diabetic retinopathy
- ECM
Extracellular matrix
- EGFR-TKI
Epidermal growth factors receptor - Tyrosine kinase inhibitors
- ELAVL
Embryonic lethal abnormal vision like
- EMT
Epithelial to mesenchymal transition
- EPR
Enhanced permeation and retention
- EVs
Extracellular Vesicles
- FA
Folic acid
- GM-CSF
Granulocyte macrophage colony stimulating factor
- HNS
HuR nucleoplasmic shuttling domain
- HuR
Human Antigen R
- IHC
Immunohistochemistry
- LC
Lung cancer
- lncRNA
Long non-coding RNA
- LUAD
Lung adenocarcinoma
- MDR
Multi drug resistance
- miRNA
MicroRNA
- MPNST
Malignant peripheral nerve sheath tumors
- MRI
Magnetic resonance imaging
- mRNA
Messenger RNA
- ncRNA
Non-coding RNA
- NSCLC
Non-small cell lung carcinoma
- PAMAM
Polyamidoamine
- PDAC
Pancreatic ductal adenocarcinoma
- PDGx
Patient-derived glioblastoma
- PTM
Post-translational modifications
- qRT-PCR
Quantitative reverse transcriptase polymerase chain reaction
- RBD
RNA-binding domain
- RBP
RNA binding proteins
- RISC
RNA induced silencing complex
- RNA
Ribonucleic acid
- RNAi
RNA interference
- ROS
Reactive oxygen species
- RRM
RNA recognition motifs
- RT-PCR
Reverse transcriptase polymerase chain reaction
- SCLC
Small cell lung carcinoma
- shRNA
Short hairpin RNAs
- siRNA
Short interfering RNAs
- STZ
Streptozotocin
- TEC
Tumor endothelial cells
- TfR
Transferrin receptor
- TMA
Tissue microarray
- TNM
Tumor node metastasis
- UTR
Untranslated regions
- WB
Western blotting
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
The authors report no conflicts of interest in this work
References
- [1].Kang D, Lee Y, Lee JS, RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives, Cancers (Basel), 12 (2020) 2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Dreyfuss G, Kim VN, Kataoka N, Messenger-RNA-binding proteins and the messages they carry, Nat Rev Mol Cell Biol, 3 (2002) 195–205. [DOI] [PubMed] [Google Scholar]
- [3].H.-T. AF, Cléry A From structure to function of RNA binding domains, Madame Curie Bioscience Database, Austin (TX), USA, 2000–2013. [Google Scholar]
- [4].Oliveira C, Faoro H, Alves LR, Goldenberg S, RNA-binding proteins and their role in the regulation of gene expression in Trypanosoma cruzi and Saccharomyces cerevisiae, Genet Mol Biol, 40 (2017) 22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, Na H, Irimia M, Matzat LH, Dale RK, Smith SA, Yarosh CA, Kelly SM, Nabet B, Mecenas D, Li W, Laishram RS, Qiao M, Lipshitz HD, Piano F, Corbett AH, Carstens RP, Frey BJ, Anderson RA, Lynch KW, Penalva LO, Lei EP, Fraser AG, Blencowe BJ, Morris QD, Hughes TR, A compendium of RNA-binding motifs for decoding gene regulation, Nature, 499 (2013) 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wang M, Oge L, Perez-Garcia MD, Hamama L, Sakr S, The PUF Protein Family: Overview on PUF RNA Targets, Biological Functions, and Post Transcriptional Regulation, Int J Mol Sci, 19 (2018) 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Heinemann U, Roske Y, Cold-Shock Domains-Abundance, Structure, Properties, and Nucleic-Acid Binding, Cancers (Basel), 13 (2021) 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Lunde BM, Moore C, Varani G, RNA-binding proteins: modular design for efficient function, Nat Rev Mol Cell Biol, 8 (2007) 479–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Qin H, Ni H, Liu Y, Yuan Y, Xi T, Li X, Zheng L, RNA-binding proteins in tumor progression, J Hematol Oncol, 13 (2020) 90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Castello A, Fischer B, Frese CK, Horos R, Alleaume AM, Foehr S, Curk T, Krijgsveld J, Hentze MW, Comprehensive Identification of RNA-Binding Domains in Human Cells, Mol Cell, 63 (2016) 696–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Moore S, Jarvelin AI, Davis I, Bond GL, Castello A, Expanding horizons: new roles for non-canonical RNA-binding proteins in cancer, Curr Opin Genet Dev, 48 (2018) 112–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Liao JY, Yang B, Zhang YC, Wang XJ, Ye Y, Peng JW, Yang ZZ, He JH, Zhang Y, Hu K, Lin DC, Yin D, EuRBPDB: a comprehensive resource for annotation, functional and oncological investigation of eukaryotic RNA binding proteins (RBPs), Nucleic Acids Res, 48 (2020) D307–D313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Cookson MR, RNA-binding proteins implicated in neurodegenerative diseases, Wiley Interdiscip Rev RNA, 8 (2017) 1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Suresh Babu S, Joladarashi D, Jeyabal P, Thandavarayan RA, Krishnamurthy P, RNA-stabilizing proteins as molecular targets in cardiovascular pathologies, Trends Cardiovasc Med, 25 (2015) 676–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Gerstberger S, Hafner M, Tuschl T, A census of human RNA-binding proteins, Nat Rev Genet, 15 (2014) 829–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Zhang B, Babu KR, Lim CY, Kwok ZH, Li J, Zhou S, Yang H, Tay Y, A comprehensive expression landscape of RNA-binding proteins (RBPs) across 16 human cancer types, RNA Biol, 17 (2020) 211–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Mohibi S, Chen X, Zhang J, Cancer the’RBP’eutics-RNA-binding proteins as therapeutic targets for cancer, Pharmacol Ther, 203 (2019) 107390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Tian XY, Li J, Liu TH, Li DN, Wang JJ, Zhang H, Deng ZL, Chen FJ, Cai JP, The overexpression of AUF1 in colorectal cancer predicts a poor prognosis and promotes cancer progression by activating ERK and AKT pathways, Cancer Med, 9 (2020) 8612–8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Gao Y, Wang W, Cao J, Wang F, Geng Y, Cao J, Xu X, Zhou J, Liu P, Zhang S, Upregulation of AUF1 is involved in the proliferation of esophageal squamous cell carcinoma through GCH1, Int J Oncol, 49 (2016) 2001–2010. [DOI] [PubMed] [Google Scholar]
- [20].Zhang J, Xu A, Miao C, Yang J, Gu M, Song N, Prognostic value of Lin28A and Lin28B in various human malignancies: a systematic review and meta-analysis, Cancer Cell Int, 19 (2019) 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ma WJ, Cheng S, Campbell C, Wright A, Furneaux H, Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein, J Biol Chem, 271 (1996) 8144–8151. [DOI] [PubMed] [Google Scholar]
- [22].Ma WJ, Furneaux H, Localization of the human HuR gene to chromosome 19p13.2, Hum Genet, 99 (1997) 32–33. [DOI] [PubMed] [Google Scholar]
- [23].Ma WJ, Chung S, Furneaux H, The Elav-like proteins bind to AU-rich elements and to the poly(A) tail of mRNA, Nucleic Acids Res, 25 (1997) 3564–3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Scheiba RM, de Opakua AI, Diaz-Quintana A, Cruz-Gallardo I, Martinez-Cruz LA, Martinez-Chantar ML, Blanco FJ, Diaz-Moreno I, The C-terminal RNA binding motif of HuR is a multi-functional domain leading to HuR oligomerization and binding to U-rich RNA targets, RNA Biol, 11 (2014) 1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Lopez de Silanes I, Zhan M, Lal A, Yang X, Gorospe M, Identification of a target RNA motif for RNA-binding protein HuR, Proc Natl Acad Sci U S A, 101 (2004) 2987–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Fan XC, Steitz JA, Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs, EMBO J, 17 (1998) 3448–3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Pabis M, Popowicz GM, Stehle R, Fernandez-Ramos D, Asami S, Warner L, Garcia-Maurino SM, Schlundt A, Martinez-Chantar ML, Diaz-Moreno I, Sattler M, HuR biological function involves RRM3-mediated dimerization and RNA binding by all three RRMs, Nucleic Acids Res, 47 (2019) 1011–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fan XC, Steitz JA, HNS, a nuclear-cytoplasmic shuttling sequence in HuR, Proceedings of the National Academy of Sciences, 95 (1998) 15293–15298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Katsanou V, Milatos S, Yiakouvaki A, Sgantzis N, Kotsoni A, Alexiou M, Harokopos V, Aidinis V, Hemberger M, Kontoyiannis DL, The RNA-binding protein Elavl1/HuR is essential for placental branching morphogenesis and embryonic development, Mol Cell Biol, 29 (2009) 2762–2776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ghosh M, Aguila HL, Michaud J, Ai Y, Wu MT, Hemmes A, Ristimaki A, Guo C, Furneaux H, Hla T, Essential role of the RNA-binding protein HuR in progenitor cell survival in mice, J Clin Invest, 119 (2009) 3530–3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chi MN, Auriol J, Jegou B, Kontoyiannis DL, Turner JM, de Rooij DG, Morello D, The RNA-binding protein ELAVL1/HuR is essential for mouse spermatogenesis, acting both at meiotic and postmeiotic stages, Mol Biol Cell, 22 (2011) 2875–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Janice Sanchez B, Tremblay AK, Leduc-Gaudet JP, Hall DT, Kovacs E, Ma JF, Mubaid S, Hallauer PL, Phillips BL, Vest KE, Corbett AH, Kontoyiannis DL, Hussain SNA, Hastings KEM, Di Marco S, Gallouzi IE, Depletion of HuR in murine skeletal muscle enhances exercise endurance and prevents cancer-induced muscle atrophy, Nat Commun, 10 (2019) 4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].von Roretz C, Beauchamp P, Di Marco S, Gallouzi IE, HuR and myogenesis: being in the right place at the right time, Biochim Biophys Acta, 1813 (2011) 1663–1667. [DOI] [PubMed] [Google Scholar]
- [34].Siang DTC, Lim YC, Kyaw AMM, Win KN, Chia SY, Degirmenci U, Hu X, Tan BC, Walet ACE, Sun L, Xu D, The RNA-binding protein HuR is a negative regulator in adipogenesis, Nat Commun, 11 (2020) 213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Liu S, Jiang X, Lu H, Xing M, Qiao Y, Zhang C, Zhang W, HuR (Human Antigen R) Regulates the Contraction of Vascular Smooth Muscle and Maintains Blood Pressure, Arterioscler Thromb Vasc Biol, 40 (2020) 943–957. [DOI] [PubMed] [Google Scholar]
- [36].Liu S, Jiang X, Cui X, Wang J, Liu S, Li H, Yang J, Zhang C, Zhang W, Smooth muscle-specific HuR knockout induces defective autophagy and atherosclerosis, Cell Death Dis, 12 (2021) 385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Srikantan S, Gorospe M, HuR function in disease, Front Biosci (Landmark Ed), 17 (2012) 189–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Doller A, Pfeilschifter J, Eberhardt W, Signalling pathways regulating nucleocytoplasmic shuttling of the mRNA-binding protein HuR, Cell Signal, 20 (2008) 2165–2173. [DOI] [PubMed] [Google Scholar]
- [39].Kotta-Loizou I, Vasilopoulos SN, Coutts RH, Theocharis S, Current Evidence and Future Perspectives on HuR and Breast Cancer Development, Prognosis, and Treatment, Neoplasia, 18 (2016) 674–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang J, Guo Y, Chu H, Guan Y, Bi J, Wang B, Multiple functions of the RNA-binding protein HuR in cancer progression, treatment responses and prognosis, Int J Mol Sci, 14 (2013) 10015–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Latorre E, Tebaldi T, Viero G, Sparta AM, Quattrone A, Provenzani A, Downregulation of HuR as a new mechanism of doxorubicin resistance in breast cancer cells, Mol Cancer, 11 (2012) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Doller A, Schlepckow K, Schwalbe H, Pfeilschifter J, Eberhardt W, Tandem phosphorylation of serines 221 and 318 by protein kinase Cdelta coordinates mRNA binding and nucleocytoplasmic shuttling of HuR, Mol Cell Biol, 30 (2010) 1397–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Schulz S, Doller A, Pendini NR, Wilce JA, Pfeilschifter J, Eberhardt W, Domain-specific phosphomimetic mutation allows dissection of different protein kinase C (PKC) isotype-triggered activities of the RNA binding protein HuR, Cell Signal, 25 (2013) 2485–2495. [DOI] [PubMed] [Google Scholar]
- [44].Abdelmohsen K, Pullmann R Jr., Lal A, Kim HH, Galban S, Yang X, Blethrow JD, Walker M, Shubert J, Gillespie DA, Furneaux H, Gorospe M, Phosphorylation of HuR by Chk2 regulates SIRT1 expression, Mol Cell, 25 (2007) 543–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim HH, Abdelmohsen K, Lal A, Pullmann R Jr., Yang X, Galban S, Srikantan S, Martindale JL, Blethrow J, Shokat KM, Gorospe M, Nuclear HuR accumulation through phosphorylation by Cdk1, Genes Dev, 22 (2008) 1804–1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Yoon JH, Abdelmohsen K, Srikantan S, Guo R, Yang X, Martindale JL, Gorospe M, Tyrosine phosphorylation of HuR by JAK3 triggers dissociation and degradation of HuR target mRNAs, Nucleic Acids Res, 42 (2014) 1196–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Liao WL, Wang WC, Chang WC, Tseng JT, The RNA-binding protein HuR stabilizes cytosolic phospholipase A2alpha mRNA under interleukin-1beta treatment in non-small cell lung cancer A549 Cells, J Biol Chem, 286 (2011) 35499–35508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Doller A, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W, Protein kinase C alpha-dependent phosphorylation of the mRNA-stabilizing factor HuR: implications for posttranscriptional regulation of cyclooxygenase-2, Mol Biol Cell, 18 (2007) 2137–2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Kim HH, Yang X, Kuwano Y, Gorospe M, Modification at HuR(S242) alters HuR localization and proliferative influence, Cell Cycle, 7 (2008) 3371–3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Vigouroux C, Casse JM, Battaglia-Hsu SF, Brochin L, Luc A, Paris C, Lacomme S, Gueant JL, Vignaud JM, Gauchotte G, Methyl(R217)HuR and MCM6 are inversely correlated and are prognostic markers in non small cell lung carcinoma, Lung Cancer, 89 (2015) 189–196. [DOI] [PubMed] [Google Scholar]
- [51].Zhou HL, Geng C, Luo G, Lou H, The p97-UBXD8 complex destabilizes mRNA by promoting release of ubiquitinated HuR from mRNP, Genes Dev, 27 (2013) 1046–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Embade N, Fernandez-Ramos D, Varela-Rey M, Beraza N, Sini M, Gutierrez de Juan V, Woodhoo A, Martinez-Lopez N, Rodriguez-Iruretagoyena B, Bustamante FJ, de la Hoz AB, Carracedo A, Xirodimas DP, Rodriguez MS, Lu SC, Mato JM, Martinez-Chantar ML, Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation, Hepatology, 55 (2012) 1237–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Grammatikakis I, Abdelmohsen K, Gorospe M, Posttranslational control of HuR function, Wiley Interdiscip Rev RNA, 8 (2017) 1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Pistono C, Monti MC, Marchesi N, Boiocchi C, Campagnoli LIM, Morlotti D, Cuccia M, Govoni S, Montomoli C, Mallucci G, Bergamaschi R, Pascale A, Unraveling a new player in multiple sclerosis pathogenesis: The RNA-binding protein HuR, Mult Scler Relat Disord, 41 (2020) 102048. [DOI] [PubMed] [Google Scholar]
- [55].Shang J, Zhao Z, Emerging role of HuR in inflammatory response in kidney diseases, Acta Biochim Biophys Sin (Shanghai), 49 (2017) 753–763. [DOI] [PubMed] [Google Scholar]
- [56].Shang J, Wan Q, Wang X, Duan Y, Wang Z, Wei X, Zhang Y, Wang H, Wang R, Yi F, Identification of NOD2 as a novel target of RNA-binding protein HuR: evidence from NADPH oxidase-mediated HuR signaling in diabetic nephropathy, Free Radic Biol Med, 79 (2015) 217–227. [DOI] [PubMed] [Google Scholar]
- [57].Papatheofani V, Levidou G, Sarantis P, Koustas E, Karamouzis MV, Pergaris A, Kouraklis G, Theocharis S, HuR Protein in Hepatocellular Carcinoma: Implications in Development, Prognosis and Treatment, Biomedicines, 9 (2021) 119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wang J, Zhao W, Guo Y, Zhang B, Xie Q, Xiang D, Gao J, Wang B, Chen Z, The expression of RNA-binding protein HuR in non-small cell lung cancer correlates with vascular endothelial growth factor-C expression and lymph node metastasis, Oncology, 76 (2009) 420–429. [DOI] [PubMed] [Google Scholar]
- [59].Lopez de Silanes I, Fan J, Yang X, Zonderman AB, Potapova O, Pizer ES, Gorospe M, Role of the RNA-binding protein HuR in colon carcinogenesis, Oncogene, 22 (2003) 7146–7154. [DOI] [PubMed] [Google Scholar]
- [60].Gridelli C, Rossi A, Carbone DP, Guarize J, Karachaliou N, Mok T, Petrella F, Spaggiari L, Rosell R, Non-small-cell lung cancer, Nat Rev Dis Primers, 1 (2015) 15009. [DOI] [PubMed] [Google Scholar]
- [61].Hirsch FR, Suda K, Wiens J, Bunn PA Jr., New and emerging targeted treatments in advanced non-small-cell lung cancer, Lancet, 388 (2016) 1012–1024. [DOI] [PubMed] [Google Scholar]
- [62].Corrales L, Scilla K, Caglevic C, Miller K, Oliveira J, Rolfo C, Immunotherapy in Lung Cancer: A New Age in Cancer Treatment, Adv Exp Med Biol, 995 (2018) 65–95. [DOI] [PubMed] [Google Scholar]
- [63].Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD, Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue, Mol Carcinog, 28 (2000) 76–83. [PubMed] [Google Scholar]
- [64].Zhang X, Lian T, Fan W, Zhang G, Chen Z, Gou X, Jha RK, Long-Noncoding RNA CASC9 Promotes Progression of Non-Small Cell Lung Cancer by Promoting the Expression of CDC6 Through Binding to HuR, Cancer Manag Res, 12 (2020) 9033–9043. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [65].Lauriola L, Granone P, Ramella S, Lanza P, Ranelletti FO, Expression of the RNA-binding protein HuR and its clinical significance in human stage I and II lung adenocarcinoma, Histol Histopathol, 27 (2012) 617–626. [DOI] [PubMed] [Google Scholar]
- [66].Wang J, Wang B, Bi J, Zhang C, Cytoplasmic HuR expression correlates with angiogenesis, lymphangiogenesis, and poor outcome in lung cancer, Med Oncol, 28 Suppl 1 (2011) S577–585. [DOI] [PubMed] [Google Scholar]
- [67].Tanaka F, Ogawa E, Wada H, Shanker M, Garcia-Soto A, Branch C, Wistuba I, Roth J, Ramesh R, Clinical significance of HuR expression, an mRNA-binding protein in non-small cell lung cancer (NSCLC), Journal of Clinical Oncology, 24 (2006) 10098–10098. [Google Scholar]
- [68].Giaginis C, Alexandrou P, Tsoukalas N, Sfiniadakis I, Kavantzas N, Agapitos E, Patsouris E, Theocharis S, Hu-antigen receptor (HuR) and cyclooxygenase-2 (COX-2) expression in human non-small-cell lung carcinoma: associations with clinicopathological parameters, tumor proliferative capacity and patients’ survival, Tumour Biol, 36 (2015) 315–327. [DOI] [PubMed] [Google Scholar]
- [69].Muralidharan R, Babu A, Amreddy N, Basalingappa K, Mehta M, Chen A, Zhao YD, Kompella UB, Munshi A, Ramesh R, Folate receptor-targeted nanoparticle delivery of HuR-RNAi suppresses lung cancer cell proliferation and migration, J Nanobiotechnology, 14 (2016) 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Yao Y, Chu H, Wang J, Wang B, Decreased human antigen R expression confers resistance to tyrosine kinase inhibitors in epidermal growth factor receptor-mutant lung cancer by inhibiting Bim expression, Int J Mol Med, 42 (2018) 2930–2942. [DOI] [PubMed] [Google Scholar]
- [71].Anastasiadou E, Jacob LS, Slack FJ, Non-coding RNA networks in cancer, Nat Rev Cancer, 18 (2018) 5–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wang WT, Han C, Sun YM, Chen TQ, Chen YQ, Noncoding RNAs in cancer therapy resistance and targeted drug development, J Hematol Oncol, 12 (2019) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Grillone K, Riillo C, Scionti F, Rocca R, Tradigo G, Guzzi PH, Alcaro S, Di Martino MT, Tagliaferri P, Tassone P, Non-coding RNAs in cancer: platforms and strategies for investigating the genomic “dark matter”, J Exp Clin Cancer Res, 39 (2020) 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Xu H, Ma J, Zheng J, Wu J, Qu C, Sun F, Xu S, MiR-31 Functions as a Tumor Suppressor in Lung Adenocarcinoma Mainly by Targeting HuR, Clin Lab, 62 (2016) 711–718. [DOI] [PubMed] [Google Scholar]
- [75].Li Q, Tong D, Guo C, Wu F, Li F, Wang X, Jiang Q, Wei Y, Liu L, Ni L, Guo B, Huang C, MicroRNA-145 suppresses gastric cancer progression by targeting Hu-antigen R, Am J Physiol Cell Physiol, 318 (2020) C605–C614. [DOI] [PubMed] [Google Scholar]
- [76].Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang C, Cui S, Hong Y, Liang H, Liu M, Zhao C, Ding M, Sun W, Liu Z, Sun F, Zhang C, Zhou Z, Jiang X, Chen X, The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer, Mol Cancer, 17 (2018) 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Al-Haidari A, Algaber A, Madhi R, Syk I, Thorlacius H, MiR-155–5p controls colon cancer cell migration via post-transcriptional regulation of Human Antigen R (HuR), Cancer Lett, 421 (2018) 145–151. [DOI] [PubMed] [Google Scholar]
- [78].Li L, Chen H, Gao Y, Wang YW, Zhang GQ, Pan SH, Ji L, Kong R, Wang G, Jia YH, Bai XW, Sun B, Long Noncoding RNA MALAT1 Promotes Aggressive Pancreatic Cancer Proliferation and Metastasis via the Stimulation of Autophagy, Mol Cancer Ther, 15 (2016) 2232–2243. [DOI] [PubMed] [Google Scholar]
- [79].Latorre E, Carelli S, Raimondi I, D’Agostino V, Castiglioni I, Zucal C, Moro G, Luciani A, Ghilardi G, Monti E, Inga A, Di Giulio AM, Gorio A, Provenzani A, The Ribonucleic Complex HuR-MALAT1 Represses CD133 Expression and Suppresses Epithelial-Mesenchymal Transition in Breast Cancer, Cancer Res, 76 (2016) 2626–2636. [DOI] [PubMed] [Google Scholar]
- [80].Meisner NC, Hintersteiner M, Mueller K, Bauer R, Seifert JM, Naegeli HU, Ottl J, Oberer L, Guenat C, Moss S, Harrer N, Woisetschlaeger M, Buehler C, Uhl V, Auer M, Identification and mechanistic characterization of low-molecular-weight inhibitors for HuR, Nat Chem Biol, 3 (2007) 508–515. [DOI] [PubMed] [Google Scholar]
- [81].Blanco FF, Preet R, Aguado A, Vishwakarma V, Stevens LE, Vyas A, Padhye S, Xu L, Weir SJ, Anant S, Meisner-Kober N, Brody JR, Dixon DA, Impact of HuR inhibition by the small molecule MS-444 on colorectal cancer cell tumorigenesis, Oncotarget, 7 (2016) 74043–74058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Wang J, Hjelmeland AB, Nabors LB, King PH, Anti-cancer effects of the HuR inhibitor, MS-444, in malignant glioma cells, Cancer Biol Ther, 20 (2019) 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Palomo-Irigoyen M, Perez-Andres E, Iruarrizaga-Lejarreta M, Barreira-Manrique A, Tamayo-Caro M, Vila-Vecilla L, Moreno-Cugnon L, Beitia N, Medrano D, Fernandez-Ramos D, Lozano JJ, Okawa S, Lavin JL, Martin-Martin N, Sutherland JD, de Juan VG, Gonzalez-Lopez M, Macias-Camara N, Mosen-Ansorena D, Laraba L, Hanemann CO, Ercolano E, Parkinson DB, Schultz CW, Arauzo-Bravo MJ, Ascension AM, Gerovska D, Iribar H, Izeta A, Pytel P, Krastel P, Provenzani A, Seneci P, Carrasco RD, Del Sol A, Martinez-Chantar ML, Barrio R, Serra E, Lazaro C, Flanagan AM, Gorospe M, Ratner N, Aransay AM, Carracedo A, Varela-Rey M, Woodhoo A, HuR/ELAVL1 drives malignant peripheral nerve sheath tumor growth and metastasis, J Clin Invest, 130 (2020) 3848–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Han Y, Liu D, Li L, PD-1/PD-L1 pathway: current researches in cancer, Am J Cancer Res, 10 (2020) 727–742. [PMC free article] [PubMed] [Google Scholar]
- [85].Liu Y, Li X, Zhang H, Zhang M, Wei Y, HuR up-regulates cell surface PD-L1 via stabilizing CMTM6 transcript in cancer, Oncogene, 40 (2021) 2230–2242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Kaur K, Wu X, Fields JK, Johnson DK, Lan L, Pratt M, Somoza AD, Wang CCC, Karanicolas J, Oakley BR, Xu L, De Guzman RN, The fungal natural product azaphilone-9 binds to HuR and inhibits HuR-RNA interaction in vitro, PLoS One, 12 (2017) e0175471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Doller A, Badawi A, Schmid T, Brauss T, Pleli T, zu Heringdorf DM, Piiper A, Pfeilschifter J, Eberhardt W, The cytoskeletal inhibitors latrunculin A and blebbistatin exert antitumorigenic properties in human hepatocellular carcinoma cells by interfering with intracellular HuR trafficking, Exp Cell Res, 330 (2015) 66–80. [DOI] [PubMed] [Google Scholar]
- [88].Lal P, Cerofolini L, D’Agostino VG, Zucal C, Fuccio C, Bonomo I, Dassi E, Giuntini S, Di Maio D, Vishwakarma V, Preet R, Williams SN, Fairlamb MS, Munk R, Lehrmann E, Abdelmohsen K, Elezgarai SR, Luchinat C, Novellino E, Quattrone A, Biasini E, Manzoni L, Gorospe M, Dixon DA, Seneci P, Marinelli L, Fragai M, Provenzani A, Regulation of HuR structure and function by dihydrotanshinone-I, Nucleic Acids Res, 45 (2017) 9514–9527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].D’Agostino VG, Lal P, Mantelli B, Tiedje C, Zucal C, Thongon N, Gaestel M, Latorre E, Marinelli L, Seneci P, Amadio M, Provenzani A, Dihydrotanshinone-I interferes with the RNA-binding activity of HuR affecting its post-transcriptional function, Sci Rep, 5 (2015) 16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Wu X, Lan L, Wilson DM, Marquez RT, Tsao WC, Gao P, Roy A, Turner BA, McDonald P, Tunge JA, Rogers SA, Dixon DA, Aube J, Xu L, Identification and validation of novel small molecule disruptors of HuR-mRNA interaction, ACS Chem Biol, 10 (2015) 1476–1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Allegri L, Baldan F, Roy S, Aube J, Russo D, Filetti S, Damante G, The HuR CMLD-2 inhibitor exhibits antitumor effects via MAD2 downregulation in thyroid cancer cells, Sci Rep, 9 (2019) 7374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Muralidharan R, Mehta M, Ahmed R, Roy S, Xu L, Aube J, Chen A, Zhao YD, Herman T, Ramesh R, Munshi A, HuR-targeted small molecule inhibitor exhibits cytotoxicity towards human lung cancer cells, Sci Rep, 7 (2017) 9694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Filippova N, Yang X, Ananthan S, Calano J, Pathak V, Bratton L, Vekariya RH, Zhang S, Ofori E, Hayward EN, Namkoong D, Crossman DK, Crowley MR, King PH, Mobley J, Nabors LB, Targeting the HuR Oncogenic Role with a New Class of Cytoplasmic Dimerization Inhibitors, Cancer Res, 81 (2021) 2220–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Manzoni L, Zucal C, Maio DD, D’Agostino VG, Thongon N, Bonomo I, Lal P, Miceli M, Baj V, Brambilla M, Cerofolini L, Elezgarai S, Biasini E, Luchinat C, Novellino E, Fragai M, Marinelli L, Provenzani A, Seneci P, Interfering with HuR-RNA Interaction: Design, Synthesis and Biological Characterization of Tanshinone Mimics as Novel, Effective HuR Inhibitors, J Med Chem, 61 (2018) 1483–1498. [DOI] [PubMed] [Google Scholar]
- [95].Filippova N, Yang X, Ananthan S, Sorochinsky A, Hackney JR, Gentry Z, Bae S, King P, Nabors LB, Hu antigen R (HuR) multimerization contributes to glioma disease progression, J Biol Chem, 292 (2017) 16999–17010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Guo J, Lv J, Chang S, Chen Z, Lu W, Xu C, Liu M, Pang X, Inhibiting cytoplasmic accumulation of HuR synergizes genotoxic agents in urothelial carcinoma of the bladder, Oncotarget, 7 (2016) 45249–45262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kakuguchi W, Nomura T, Kitamura T, Otsuguro S, Matsushita K, Sakaitani M, Maenaka K, Tei K, Suramin, screened from an approved drug library, inhibits HuR functions and attenuates malignant phenotype of oral cancer cells, Cancer Med, 7 (2018) 6269–6280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhu Y, Yang L, Xu J, Yang X, Luan P, Cui Q, Zhang P, Wang F, Li R, Ding X, Jiang L, Lin G, Zhang J, Discovery of the anti-angiogenesis effect of eltrombopag in breast cancer through targeting of HuR protein, Acta Pharm Sin B, 10 (2020) 1414–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Cheng Y-C, Liou J-P, Kuo C-C, Lai W-Y, Shih K-H, Chang C-Y, Pan W-Y, Tseng JT, Chang J-Y, MPT0B098, a Novel Microtubule Inhibitor That Destabilizes the Hypoxia-Inducible Factor-1α mRNA through Decreasing Nuclear–Cytoplasmic Translocation of RNA-Binding Protein HuR, Molecular Cancer Therapeutics, 12 (2013) 1202–1212. [DOI] [PubMed] [Google Scholar]
- [100].Dhir T, Schultz CW, Jain A, Brown SZ, Haber A, Goetz A, Xi C, Su GH, Xu L, Posey J 3rd, Jiang W, Yeo CJ, Golan T, Pishvaian MJ, Brody JR, Abemaciclib Is Effective Against Pancreatic Cancer Cells and Synergizes with HuR and YAP1 Inhibition, Mol Cancer Res, 17 (2019) 2029–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Schultz CW, Preet R, Dhir T, Dixon DA, Brody JR, Understanding and targeting the disease-related RNA binding protein human antigen R (HuR), Wiley Interdiscip Rev RNA, 11 (2020) e1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Meister G, Tuschl T, Mechanisms of gene silencing by double-stranded RNA, Nature, 431 (2004) 343–349. [DOI] [PubMed] [Google Scholar]
- [103].Dana H, Chalbatani GM, Mahmoodzadeh H, Karimloo R, Rezaiean O, Moradzadeh A, Mehmandoost N, Moazzen F, Mazraeh A, Marmari V, Ebrahimi M, Rashno MM, Abadi SJ, Gharagouzlo E, Molecular Mechanisms and Biological Functions of siRNA, Int J Biomed Sci, 13 (2017) 48–57. [PMC free article] [PubMed] [Google Scholar]
- [104].Lambeth LS, Smith CA, Short hairpin RNA-mediated gene silencing, Methods Mol Biol, 942 (2013) 205–232. [DOI] [PubMed] [Google Scholar]
- [105].Mansoori B, Sandoghchian Shotorbani S, Baradaran B, RNA interference and its role in cancer therapy, Adv Pharm Bull, 4 (2014) 313–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Kim GY, Lim SJ, Kim YW, Expression of HuR, COX-2, and survivin in lung cancers; cytoplasmic HuR stabilizes cyclooxygenase-2 in squamous cell carcinomas, Mod Pathol, 24 (2011) 1336–1347. [DOI] [PubMed] [Google Scholar]
- [107].Niesporek S, Kristiansen G, Thoma A, Weichert W, Noske A, Buckendahl AC, Jung K, Stephan C, Dietel M, Denkert C, Expression of the ELAV-like protein HuR in human prostate carcinoma is an indicator of disease relapse and linked to COX-2 expression, Int J Oncol, 32 (2008) 341–347. [PubMed] [Google Scholar]
- [108].Zhang Z, Huang A, Zhang A, Zhou C, HuR promotes breast cancer cell proliferation and survival via binding to CDK3 mRNA, Biomed Pharmacother, 91 (2017) 788–795. [DOI] [PubMed] [Google Scholar]
- [109].Yuan Z, Sanders AJ, Ye L, Wang Y, Jiang WG, Knockdown of human antigen R reduces the growth and invasion of breast cancer cells in vitro and affects expression of cyclin D1 and MMP-9, Oncol Rep, 26 (2011) 237–245. [DOI] [PubMed] [Google Scholar]
- [110].Jimbo M, Blanco FF, Huang YH, Telonis AG, Screnci BA, Cosma GL, Alexeev V, Gonye GE, Yeo CJ, Sawicki JA, Winter JM, Brody JR, Targeting the mRNA-binding protein HuR impairs malignant characteristics of pancreatic ductal adenocarcinoma cells, Oncotarget, 6 (2015) 27312–27331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Filippova N, Yang X, Wang Y, Gillespie GY, Langford C, King PH, Wheeler C, Nabors LB, The RNA-binding protein HuR promotes glioma growth and treatment resistance, Mol Cancer Res, 9 (2011) 648–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Huang YH, Peng W, Furuuchi N, Gerhart J, Rhodes K, Mukherjee N, Jimbo M, Gonye GE, Brody JR, Getts RC, Sawicki JA, Delivery of Therapeutics Targeting the mRNA-Binding Protein HuR Using 3DNA Nanocarriers Suppresses Ovarian Tumor Growth, Cancer Res, 76 (2016) 1549–1559. [DOI] [PubMed] [Google Scholar]
- [113].Zhang Y, Yang L, Ling C, Heng W, HuR facilitates cancer stemness of lung cancer cells via regulating miR-873/CDK3 and miR-125a-3p/CDK3 axis, Biotechnol Lett, 40 (2018) 623–631. [DOI] [PubMed] [Google Scholar]
- [114].Beaugerie L, Svrcek M, Seksik P, Bouvier AM, Simon T, Allez M, Brixi H, Gornet JM, Altwegg R, Beau P, Duclos B, Bourreille A, Faivre J, Peyrin-Biroulet L, Flejou JF, Carrat F, Group CS, Risk of colorectal high-grade dysplasia and cancer in a prospective observational cohort of patients with inflammatory bowel disease, Gastroenterology, 145 (2013) 166–175 e168. [DOI] [PubMed] [Google Scholar]
- [115].Howe LR, Subbaramaiah K, Hudis CA, Dannenberg AJ, Molecular pathways: adipose inflammation as a mediator of obesity-associated cancer, Clin Cancer Res, 19 (2013) 6074–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Gealy C, Denson M, Humphreys C, McSharry B, Wilkinson G, Caswell R, Posttranscriptional suppression of interleukin-6 production by human cytomegalovirus, J Virol, 79 (2005) 472–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhou H, Jarujaron S, Gurley EC, Chen L, Ding H, Studer E, Pandak WM Jr., Hu W, Zou T, Wang JY, Hylemon PB, HIV protease inhibitors increase TNF-alpha and IL-6 expression in macrophages: involvement of the RNA-binding protein HuR, Atherosclerosis, 195 (2007) e134–143. [DOI] [PubMed] [Google Scholar]
- [118].Ge J, Chang N, Zhao Z, Tian L, Duan X, Yang L, Li L, Essential Roles of RNA-binding Protein HuR in Activation of Hepatic Stellate Cells Induced by Transforming Growth Factor-beta1, Sci Rep, 6 (2016) 22141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Green LC, Anthony SR, Slone S, Lanzillotta L, Lorenz J, Tranter M, Pharmacological inhibition of Human Antigen R (HuR) blunts fibroblast activation and cardiac fibrosis, 33 (2019) 817.817–817.817. [Google Scholar]
- [120].Al-Habeeb F, Aloufi N, Traboulsi H, Liu X, Nair P, Haston C, Azuelos I, Huang SK, White ES, Gallouzi IE, Di Marco S, Eidelman DH, Baglole CJ, Human antigen R promotes lung fibroblast differentiation to myofibroblasts and increases extracellular matrix production, J Cell Physiol, 236 (2021) 6836–6851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Woodhoo A, Iruarrizaga-Lejarreta M, Beraza N, Garcia-Rodriguez JL, Embade N, Fernandez-Ramos D, Martinez-Lopez N, Gutierrez-De Juan V, Arteta B, Caballeria J, Lu SC, Mato JM, Varela-Rey M, Martinez-Chantar ML, Human antigen R contributes to hepatic stellate cell activation and liver fibrosis, Hepatology, 56 (2012) 1870–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Peng W, Furuuchi N, Aslanukova L, Huang YH, Brown SZ, Jiang W, Addya S, Vishwakarma V, Peters E, Brody JR, Dixon DA, Sawicki JA, Elevated HuR in Pancreas Promotes a Pancreatitis-Like Inflammatory Microenvironment That Facilitates Tumor Development, Mol Cell Biol, 38 (2018) e00427–00417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Nishida N, Yano H, Nishida T, Kamura T, Kojiro M, Angiogenesis in cancer, Vasc Health Risk Manag, 2 (2006) 213–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Dong R, Yang GD, Luo NA, Qu YQ, HuR: a promising therapeutic target for angiogenesis, Gland Surg, 3 (2014) 203–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Nabors LB, Gillespie GY, Harkins L, King PH, HuR, a RNA stability factor, is expressed in malignant brain tumors and binds to adenine- and uridine-rich elements within the 3’ untranslated regions of cytokine and angiogenic factor mRNAs, Cancer Res, 61 (2001) 2154–2161. [PubMed] [Google Scholar]
- [126].Tran H, Maurer F, Nagamine Y, Stabilization of urokinase and urokinase receptor mRNAs by HuR is linked to its cytoplasmic accumulation induced by activated mitogen-activated protein kinase-activated protein kinase 2, Mol Cell Biol, 23 (2003) 7177–7188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Shen Z, Ye D, Zhang X, Jiang Z, Xiao B, Guo J, Inhibitory effect of HuR gene small interfering RNA segment on laryngeal carcinoma Hep-2 cell growth, J Laryngol Otol, 124 (2010) 1183–1189. [DOI] [PubMed] [Google Scholar]
- [128].Chang SH, Lu YC, Li X, Hsieh WY, Xiong Y, Ghosh M, Evans T, Elemento O, Hla T, Antagonistic function of the RNA-binding protein HuR and miR-200b in post-transcriptional regulation of vascular endothelial growth factor-A expression and angiogenesis, J Biol Chem, 288 (2013) 4908–4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Kurosu T, Ohga N, Hida Y, Maishi N, Akiyama K, Kakuguchi W, Kuroshima T, Kondo M, Akino T, Totsuka Y, Shindoh M, Higashino F, Hida K, HuR keeps an angiogenic switch on by stabilising mRNA of VEGF and COX-2 in tumour endothelium, Br J Cancer, 104 (2011) 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Doller A, Akool el S, Huwiler A, Muller R, Radeke HH, Pfeilschifter J, Eberhardt W, Posttranslational modification of the AU-rich element binding protein HuR by protein kinase Cdelta elicits angiotensin II-induced stabilization and nuclear export of cyclooxygenase 2 mRNA, Mol Cell Biol, 28 (2008) 2608–2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Yohena T, Yoshino I, Takenaka T, Kameyama T, Ohba T, Kuniyoshi Y, Maehara Y, Upregulation of hypoxia-inducible factor-1alpha mRNA and its clinical significance in non-small cell lung cancer, J Thorac Oncol, 4 (2009) 284–290. [DOI] [PubMed] [Google Scholar]
- [132].Lin CS, Liu TC, Lee MT, Yang SF, Tsao TC, Independent Prognostic Value of Hypoxia-inducible Factor 1-alpha Expression in Small Cell Lung Cancer, Int J Med Sci, 14 (2017) 785–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Gabai VL, Meng L, Kim G, Mills TA, Benjamin IJ, Sherman MY, Heat shock transcription factor Hsf1 is involved in tumor progression via regulation of hypoxia-inducible factor 1 and RNA-binding protein HuR, Mol Cell Biol, 32 (2012) 929–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Atasoy U, Watson J, Patel D, Keene JD, ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation, J Cell Sci, 111 (Pt 21) (1998) 3145–3156. [DOI] [PubMed] [Google Scholar]
- [135].Diaz-Munoz MD, Bell SE, Fairfax K, Monzon-Casanova E, Cunningham AF, Gonzalez-Porta M, Andrews SR, Bunik VI, Zarnack K, Curk T, Heggermont WA, Heymans S, Gibson GE, Kontoyiannis DL, Ule J, Turner M, The RNA-binding protein HuR is essential for the B cell antibody response, Nat Immunol, 16 (2015) 415–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].DeMicco A, Naradikian MS, Sindhava VJ, Yoon JH, Gorospe M, Wertheim GB, Cancro MP, Bassing CH, B Cell-Intrinsic Expression of the HuR RNA-Binding Protein Is Required for the T Cell-Dependent Immune Response In Vivo, J Immunol, 195 (2015) 3449–3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Rothamel K, Arcos S, Kim B, Reasoner C, Lisy S, Mukherjee N, Ascano M, ELAVL1 primarily couples mRNA stability with the 3’ UTRs of interferon-stimulated genes, Cell Rep, 35 (2021) 109178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wang J, Leavenworth JW, Hjelmeland AB, Smith R, Patel N, Borg B, Si Y, King PH, Deletion of the RNA regulator HuR in tumor-associated microglia and macrophages stimulates anti-tumor immunity and attenuates glioma growth, Glia, 67 (2019) 2424–2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zhang Q, Wu X, Lan L, Wei L, Xu L, Abstract 1560: Targeting RNA-binding protein HuR to improve anti-PD-1 immunotherapy response in breast cancer, Cancer Research, 81 (2021) 1560–1560. [Google Scholar]
- [140].Yan WL, Shen KY, Tien CY, Chen YA, Liu SJ, Recent progress in GM-CSF-based cancer immunotherapy, Immunotherapy, 9 (2017) 347–360. [DOI] [PubMed] [Google Scholar]
- [141].Rong QX, Wang F, Guo ZX, Hu Y, An SN, Luo M, Zhang H, Wu SC, Huang HQ, Fu LW, GM-CSF mediates immune evasion via upregulation of PD-L1 expression in extranodal natural killer/T cell lymphoma, Mol Cancer, 20 (2021) 80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Chen J, Adamiak W, Huang G, Atasoy U, Rostami A, Yu S, Interaction of RNA-binding protein HuR and miR-466i regulates GM-CSF expression, Sci Rep, 7 (2017) 17233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Miyata Y, Watanabe S, Sagara Y, Mitsunari K, Matsuo T, Ohba K, Sakai H, High expression of HuR in cytoplasm, but not nuclei, is associated with malignant aggressiveness and prognosis in bladder cancer, PLoS One, 8 (2013) e59095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Huwiler A, Akool el S, Aschrafi A, Hamada FM, Pfeilschifter J, Eberhardt W, ATP potentiates interleukin-1 beta-induced MMP-9 expression in mesangial cells via recruitment of the ELAV protein HuR, J Biol Chem, 278 (2003) 51758–51769. [DOI] [PubMed] [Google Scholar]
- [145].Dong R, Lu JG, Wang Q, He XL, Chu YK, Ma QJ, Stabilization of Snail by HuR in the process of hydrogen peroxide induced cell migration, Biochem Biophys Res Commun, 356 (2007) 318–321. [DOI] [PubMed] [Google Scholar]
- [146].McCarthy GA, Jain A, Brown SZ, Yeo CJ, Brody J, Abstract LB-203: The mRNA binding protein Human Antigen R (HuR, ELAVL1) is a novel regulator in pancreatic ductal adenocarcinoma metastatic progression and persistence, Cancer Research, 80 (2020) LB-203–LB-203. [Google Scholar]
- [147].Wu X, Gardashova G, Lan L, Han S, Zhong C, Marquez RT, Wei L, Wood S, Roy S, Gowthaman R, Karanicolas J, Gao FP, Dixon DA, Welch DR, Li L, Ji M, Aube J, Xu L, Targeting the interaction between RNA-binding protein HuR and FOXQ1 suppresses breast cancer invasion and metastasis, Commun Biol, 3 (2020) 193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Wu JI, Lin YP, Tseng CW, Chen HJ, Wang LH, Crabp2 Promotes Metastasis of Lung Cancer Cells via HuR and Integrin beta1/FAK/ERK Signaling, Sci Rep, 9 (2019) 845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [149].Zhou F, Zhang F, Zhou C, Liang M, Cai Z, Lv H, Li W, Wei X, Human antigen R and drug resistance in tumors, Invest New Drugs, 37 (2019) 1107–1116. [DOI] [PubMed] [Google Scholar]
- [150].Cai J, Wang H, Jiao X, Huang R, Qin Q, Zhang J, Chen H, Feng D, Tian X, Wang H, The RNA-Binding Protein HuR Confers Oxaliplatin Resistance of Colorectal Cancer By Upregulating CDC6, Mol Cancer Ther, 18 (2019) 1243–1254. [DOI] [PubMed] [Google Scholar]
- [151].Lin GL, Ting HJ, Tseng TC, Juang V, Lo YL, Modulation of the mRNA-binding protein HuR as a novel reversal mechanism of epirubicin-triggered multidrug resistance in colorectal cancer cells, PLoS One, 12 (2017) e0185625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Jakstaite A, Maziukiene A, Silkuniene G, Kmieliute K, Gulbinas A, Dambrauskas Z, HuR mediated post-transcriptional regulation as a new potential adjuvant therapeutic target in chemotherapy for pancreatic cancer, World J Gastroenterol, 21 (2015) 13004–13019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Costantino CL, Witkiewicz AK, Kuwano Y, Cozzitorto JA, Kennedy EP, Dasgupta A, Keen JC, Yeo CJ, Gorospe M, Brody JR, The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase, Cancer Res, 69 (2009) 4567–4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Badawi A, Hehlgans S, Pfeilschifter J, Rodel F, Eberhardt W, Silencing of the mRNA-binding protein HuR increases the sensitivity of colorectal cancer cells to ionizing radiation through upregulation of caspase-2, Cancer Lett, 393 (2017) 103–112. [DOI] [PubMed] [Google Scholar]
- [155].Mehta M, Basalingappa K, Griffith JN, Andrade D, Babu A, Amreddy N, Muralidharan R, Gorospe M, Herman T, Ding WQ, Ramesh R, Munshi A, HuR silencing elicits oxidative stress and DNA damage and sensitizes human triple-negative breast cancer cells to radiotherapy, Oncotarget, 7 (2016) 64820–64835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Andrade D, Mehta M, Griffith J, Oh S, Corbin J, Babu A, De S, Chen A, Zhao YD, Husain S, Roy S, Xu L, Aube J, Janknecht R, Gorospe M, Herman T, Ramesh R, Munshi A, HuR Reduces Radiation-Induced DNA Damage by Enhancing Expression of ARID1A, Cancers (Basel), 11 (2019) 3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [157].Bevacqua RJ, Dai X, Lam JY, Gu X, Friedlander MSH, Tellez K, Miguel-Escalada I, Bonas-Guarch S, Atla G, Zhao W, Kim SH, Dominguez AA, Qi LS, Ferrer J, MacDonald PE, Kim SK, CRISPR-based genome editing in primary human pancreatic islet cells, Nat Commun, 12 (2021) 2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Crunkhorn S, CRISPR-engineered fat cells prevent obesity, Nat Rev Drug Discov, 19 (2020) 672. [DOI] [PubMed] [Google Scholar]
- [159].Wu W, Tang L, D’Amore PA, Lei H, Application of CRISPR-Cas9 in eye disease, Exp Eye Res, 161 (2017) 116–123. [DOI] [PubMed] [Google Scholar]
- [160].Akram F, Ikram Ul H, Ahmed Z, Khan H, Ali MS, CRISPR-Cas9, A Promising Therapeutic Tool for Cancer Therapy: A Review, Protein Pept Lett, 27 (2020) 931–944. [DOI] [PubMed] [Google Scholar]
- [161].Lal S, Cheung EC, Zarei M, Preet R, Chand SN, Mambelli-Lisboa NC, Romeo C, Stout MC, Londin E, Goetz A, Lowder CY, Nevler A, Yeo CJ, Campbell PM, Winter JM, Dixon DA, Brody JR, CRISPR Knockout of the HuR Gene Causes a Xenograft Lethal Phenotype, Mol Cancer Res, 15 (2017) 696–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [162].Dong R, Chen P, Polireddy K, Wu X, Wang T, Ramesh R, Dixon DA, Xu L, Aube J, Chen Q, An RNA-Binding Protein, Hu-antigen R, in Pancreatic Cancer Epithelial to Mesenchymal Transition, Metastasis, and Cancer Stem Cells, Mol Cancer Ther, 19 (2020) 2267–2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Chand SN, Zarei M, Schiewer MJ, Kamath AR, Romeo C, Lal S, Cozzitorto JA, Nevler A, Scolaro L, Londin E, Jiang W, Meisner-Kober N, Pishvaian MJ, Knudsen KE, Yeo CJ, Pascal JM, Winter JM, Brody JR, Posttranscriptional Regulation of PARG mRNA by HuR Facilitates DNA Repair and Resistance to PARP Inhibitors, Cancer Res, 77 (2017) 5011–5025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Barayan R, Ran X, Lok BH, PARP inhibitors for small cell lung cancer and their potential for integration into current treatment approaches, J Thorac Dis, 12 (2020) 6240–6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [165].Rodriguez M, Lapierre J, Ojha CR, Kaushik A, Batrakova E, Kashanchi F, Dever SM, Nair M, El-Hage N, Intranasal drug delivery of small interfering RNA targeting Beclin1 encapsulated with polyethylenimine (PEI) in mouse brain to achieve HIV attenuation, Sci Rep, 7 (2017) 1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [166].Ahmed Z, Kalinski H, Berry M, Almasieh M, Ashush H, Slager N, Brafman A, Spivak I, Prasad N, Mett I, Shalom E, Alpert E, Di Polo A, Feinstein E, Logan A, Ocular neuroprotection by siRNA targeting caspase-2, Cell Death Dis, 2 (2011) e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].White PJ, Barriers to successful delivery of short interfering RNA after systemic administration, Clin Exp Pharmacol Physiol, 35 (2008) 1371–1376. [DOI] [PubMed] [Google Scholar]
- [168].Babu A, Muralidharan R, Amreddy N, Mehta M, Munshi A, Ramesh R, Nanoparticles for siRNA-Based Gene Silencing in Tumor Therapy, IEEE Trans Nanobioscience, 15 (2016) 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Liu C, Zhang L, Liu H, Cheng K, Delivery strategies of the CRISPR-Cas9 gene-editing system for therapeutic applications, J Control Release, 266 (2017) 17–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].O’Keeffe Ahern J, Lara-Saez I, Zhou D, Murillas R, Bonafont J, Mencia A, Garcia M, Manzanares D, Lynch J, Foley R, Xu Q, Sigen A, Larcher F, Wang W, Non-viral delivery of CRISPR-Cas9 complexes for targeted gene editing via a polymer delivery system, Gene Ther, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Duan L, Ouyang K, Xu X, Xu L, Wen C, Zhou X, Qin Z, Xu Z, Sun W, Liang Y, Nanoparticle Delivery of CRISPR/Cas9 for Genome Editing, Front Genet, 12 (2021) 673286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Cho EY, Ryu JY, Lee HAR, Hong SH, Park HS, Hong KS, Park SG, Kim HP, Yoon TJ, Lecithin nano-liposomal particle as a CRISPR/Cas9 complex delivery system for treating type 2 diabetes, J Nanobiotechnology, 17 (2019) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Shajari N, Mansoori B, Davudian S, Mohammadi A, Baradaran B, Overcoming the Challenges of siRNA Delivery: Nanoparticle Strategies, Curr Drug Deliv, 14 (2017) 36–46. [DOI] [PubMed] [Google Scholar]
- [174].Subhan MA, Torchilin VP, Efficient nanocarriers of siRNA therapeutics for cancer treatment, Transl Res, 214 (2019) 62–91. [DOI] [PubMed] [Google Scholar]
- [175].Caruthers SD, Wickline SA, Lanza GM, Nanotechnological applications in medicine, Curr Opin Biotechnol, 18 (2007) 26–30. [DOI] [PubMed] [Google Scholar]
- [176].Garcia-Fernandez C, Fornaguera C, Borros S, Nanomedicine in Non-Small Cell Lung Cancer: From Conventional Treatments to Immunotherapy, Cancers (Basel), 12 (2020) 1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Sharma P, Mehta M, Dhanjal DS, Kaur S, Gupta G, Singh H, Thangavelu L, Rajeshkumar S, Tambuwala M, Bakshi HA, Chellappan DK, Dua K, Satija S, Emerging trends in the novel drug delivery approaches for the treatment of lung cancer, Chem Biol Interact, 309 (2019) 108720. [DOI] [PubMed] [Google Scholar]
- [178].Greish K, Enhanced permeability and retention (EPR) effect for anticancer nanomedicine drug targeting, Methods Mol Biol, 624 (2010) 25–37. [DOI] [PubMed] [Google Scholar]
- [179].Joo J, Diagnostic and Therapeutic Nanomedicine, Adv Exp Med Biol, 1310 (2021) 401–447. [DOI] [PubMed] [Google Scholar]
- [180].Green MR, Manikhas GM, Orlov S, Afanasyev B, Makhson AM, Bhar P, Hawkins MJ, Abraxane, a novel Cremophor-free, albumin-bound particle form of paclitaxel for the treatment of advanced non-small-cell lung cancer, Ann Oncol, 17 (2006) 1263–1268. [DOI] [PubMed] [Google Scholar]
- [181].Tan H, Hu J, Liu S, Efficacy and safety of nanoparticle albumin-bound paclitaxel in non-small cell lung cancer: a systematic review and meta-analysis, Artif Cells Nanomed Biotechnol, 47 (2019) 268–277. [DOI] [PubMed] [Google Scholar]
- [182].Crintea A, Dutu AG, Samasca G, Florian IA, Lupan I, Craciun AM, The Nanosystems Involved in Treating Lung Cancer, Life (Basel), 11 (2021) 682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Carrasco-Esteban E, Dominguez-Rullan JA, Barrionuevo-Castillo P, Pelari-Mici L, Leaman O, Sastre-Gallego S, Lopez-Campos F, Current role of nanoparticles in the treatment of lung cancer, J Clin Transl Res, 7 (2021) 140–155. [PMC free article] [PubMed] [Google Scholar]
- [184].Mukherjee A, Paul M, Mukherjee S, Recent Progress in the Theranostics Application of Nanomedicine in Lung Cancer, Cancers (Basel), 11 (2019) 597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [185].Tomalia DA, Baker H, Dewald J, Hall M, Kallos G, Martin S, Roeck J, Ryder J, Smith P, A New Class of Polymers: Starburst-Dendritic Macromolecules, Polymer Journal, 17 (1985) 117–132. [Google Scholar]
- [186].Sherje AP, Jadhav M, Dravyakar BR, Kadam D, Dendrimers: A versatile nanocarrier for drug delivery and targeting, Int J Pharm, 548 (2018) 707–720. [DOI] [PubMed] [Google Scholar]
- [187].Abbasi E, Aval SF, Akbarzadeh A, Milani M, Nasrabadi HT, Joo SW, Hanifehpour Y, Nejati-Koshki K, Pashaei-Asl R, Dendrimers: synthesis, applications, and properties, Nanoscale Res Lett, 9 (2014) 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Biswas S, Torchilin VP, Dendrimers for siRNA Delivery, Pharmaceuticals (Basel), 6 (2013) 161–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Zhou K, Nguyen LH, Miller JB, Yan Y, Kos P, Xiong H, Li L, Hao J, Minnig JT, Zhu H, Siegwart DJ, Modular degradable dendrimers enable small RNAs to extend survival in an aggressive liver cancer model, Proc Natl Acad Sci U S A, 113 (2016) 520–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Song C, Xiao Y, Ouyang Z, Shen M, Shi X, Efficient co-delivery of microRNA 21 inhibitor and doxorubicin to cancer cells using core-shell tecto dendrimers formed via supramolecular host-guest assembly, J Mater Chem B, 8 (2020) 2768–2774. [DOI] [PubMed] [Google Scholar]
- [191].Dzmitruk V, Apartsin E, Ihnatsyeu-Kachan A, Abashkin V, Shcharbin D, Bryszewska M, Dendrimers Show Promise for siRNA and microRNA Therapeutics, Pharmaceutics, 10 (2018) 126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Palombarini F, Masciarelli S, Incocciati A, Liccardo F, Di Fabio E, Iazzetti A, Fabrizi G, Fazi F, Macone A, Bonamore A, Boffi A, Self-assembling ferritin-dendrimer nanoparticles for targeted delivery of nucleic acids to myeloid leukemia cells, J Nanobiotechnology, 19 (2021) 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [193].Zwicke GL, Mansoori GA, Jeffery CJ, Utilizing the folate receptor for active targeting of cancer nanotherapeutics, Nano Rev, 3 (2012) 18496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Lasinska I, Mackiewicz J, Integrins as A New Target for Cancer Treatment, Anticancer Agents Med Chem, 19 (2019) 580–586. [DOI] [PubMed] [Google Scholar]
- [195].Simon M, Stefan N, Pluckthun A, Zangemeister-Wittke U, Epithelial cell adhesion molecule-targeted drug delivery for cancer therapy, Expert Opin Drug Deliv, 10 (2013) 451–468. [DOI] [PubMed] [Google Scholar]
- [196].Amreddy N, Babu A, Panneerselvam J, Srivastava A, Muralidharan R, Chen A, Zhao YD, Munshi A, Ramesh R, Chemo-biologic combinatorial drug delivery using folate receptor-targeted dendrimer nanoparticles for lung cancer treatment, Nanomedicine, 14 (2018) 373–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Amreddy N, Ahmed RA, Munshi A, Ramesh R, Tumor-Targeted Dendrimer Nanoparticles for Combinatorial Delivery of siRNA and Chemotherapy for Cancer Treatment, Methods Mol Biol, 2059 (2020) 167–189. [DOI] [PubMed] [Google Scholar]
- [198].Akbarzadeh A, Rezaei-Sadabady R, Davaran S, Joo SW, Zarghami N, Hanifehpour Y, Samiei M, Kouhi M, Nejati-Koshki K, Liposome: classification, preparation, and applications, Nanoscale Res Lett, 8 (2013) 102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Bulbake U, Doppalapudi S, Kommineni N, Khan W, Liposomal Formulations in Clinical Use: An Updated Review, Pharmaceutics, 9 (2017) 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Ozpolat B, Sood AK, Lopez-Berestein G, Liposomal siRNA nanocarriers for cancer therapy, Adv Drug Deliv Rev, 66 (2014) 110–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Xia Y, Tian J, Chen X, Effect of surface properties on liposomal siRNA delivery, Biomaterials, 79 (2016) 56–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [202].Whitney JF, Clark JM, Griffin TW, Gautam S, Leslie KO, Transferrin receptor expression in nonsmall cell lung cancer. Histopathologic and clinical correlates, Cancer, 76 (1995) 20–25. [DOI] [PubMed] [Google Scholar]
- [203].Muralidharan R, Babu A, Amreddy N, Srivastava A, Chen A, Zhao YD, Kompella UB, Munshi A, Ramesh R, Tumor-targeted Nanoparticle Delivery of HuR siRNA Inhibits Lung Tumor Growth In Vitro and In Vivo By Disrupting the Oncogenic Activity of the RNA-binding Protein HuR, Mol Cancer Ther, 16 (2017) 1470–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Al-Souhibani N, Al-Ghamdi M, Al-Ahmadi W, Khabar KS, Posttranscriptional control of the chemokine receptor CXCR4 expression in cancer cells, Carcinogenesis, 35 (2014) 1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Muralidharan R, Panneerselvam J, Chen A, Zhao YD, Munshi A, Ramesh R, HuR-targeted nanotherapy in combination with AMD3100 suppresses CXCR4 expression, cell growth, migration and invasion in lung cancer, Cancer Gene Ther, 22 (2015) 581–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Ahmed R, Muralidharan R, Srivastava A, Johnston SE, Zhao YD, Ekmekcioglu S, Munshi A, Ramesh R, Molecular Targeting of HuR Oncoprotein Suppresses MITF and Induces Apoptosis in Melanoma Cells, Cancers (Basel), 13 (2021) 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [207].Amadio M, Pascale A, Cupri S, Pignatello R, Osera C, V DA, AG DA, Leggio GM, Ruozi B, Govoni S, Drago F, Bucolo C, Nanosystems based on siRNA silencing HuR expression counteract diabetic retinopathy in rat, Pharmacol Res, 111 (2016) 713–720. [DOI] [PubMed] [Google Scholar]
- [208].Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W, Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response, Nature, 560 (2018) 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Srivastava A, Rathore S, Munshi A, Ramesh R, Extracellular Vesicles in Oncology: from Immune Suppression to Immunotherapy, AAPS J, 23 (2021) 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [210].Srivastava A, Amreddy N, Pareek V, Chinnappan M, Ahmed R, Mehta M, Razaq M, Munshi A, Ramesh R, Progress in extracellular vesicle biology and their application in cancer medicine, Wiley Interdiscip Rev Nanomed Nanobiotechnol, 12 (2020) e1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Bebelman MP, Smit MJ, Pegtel DM, Baglio SR, Biogenesis and function of extracellular vesicles in cancer, Pharmacol Ther, 188 (2018) 1–11. [DOI] [PubMed] [Google Scholar]
- [212].O’Brien K, Breyne K, Ughetto S, Laurent LC, Breakefield XO, RNA delivery by extracellular vesicles in mammalian cells and its applications, Nat Rev Mol Cell Biol, 21 (2020) 585–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [213].Wong CH, Chen YC, Clinical significance of exosomes as potential biomarkers in cancer, World J Clin Cases, 7 (2019) 171–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Srivastava A, Moxley K, Ruskin R, Dhanasekaran DN, Zhao YD, Ramesh R, A Noninvasive Liquid Biopsy Screening of Urine-Derived Exosomes for miRNAs as Biomarkers in Endometrial Cancer Patients, AAPS J, 20 (2018) 82. [DOI] [PubMed] [Google Scholar]
- [215].Boukouris S, Mathivanan S, Exosomes in bodily fluids are a highly stable resource of disease biomarkers, Proteomics Clin Appl, 9 (2015) 358–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [216].Kim KU, Kim WH, Jeong CH, Yi DY, Min H, More than Nutrition: Therapeutic Potential of Breast Milk-Derived Exosomes in Cancer, Int J Mol Sci, 21 (2020) 7327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [217].Yang T, Fogarty B, LaForge B, Aziz S, Pham T, Lai L, Bai S, Delivery of Small Interfering RNA to Inhibit Vascular Endothelial Growth Factor in Zebrafish Using Natural Brain Endothelia Cell-Secreted Exosome Nanovesicles for the Treatment of Brain Cancer, AAPS J, 19 (2017) 475–486. [DOI] [PubMed] [Google Scholar]
- [218].Morad G, Carman CV, Hagedorn EJ, Perlin JR, Zon LI, Mustafaoglu N, Park TE, Ingber DE, Daisy CC, Moses MA, Tumor-Derived Extracellular Vesicles Breach the Intact Blood-Brain Barrier via Transcytosis, ACS Nano, 13 (2019) 13853–13865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [219].Akuma P, Okagu OD, Udenigwe CC, Naturally Occurring Exosome Vesicles as Potential Delivery Vehicle for Bioactive Compounds, 3 (2019). [Google Scholar]
- [220].Preet R, Zhuang S, Hung W-T, Christenson LK, Dixon DA, Abstract A41: The RNA binding protein HuR enhances exosome secretion in colorectal cancer, Cancer Research, 76 (2016) A41–A41. [Google Scholar]
- [221].O’Loughlin AJ, Mager I, de Jong OG, Varela MA, Schiffelers RM, El Andaloussi S, Wood MJA, Vader P, Functional Delivery of Lipid-Conjugated siRNA by Extracellular Vesicles, Mol Ther, 25 (2017) 1580–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [222].Srivastava A, Amreddy N, Babu A, Panneerselvam J, Mehta M, Muralidharan R, Chen A, Zhao YD, Razaq M, Riedinger N, Kim H, Liu S, Wu S, Abdel-Mageed AB, Munshi A, Ramesh R, Nanosomes carrying doxorubicin exhibit potent anticancer activity against human lung cancer cells, Sci Rep, 6 (2016) 38541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [223].Bennett CF, Swayze EE, RNA targeting therapeutics: molecular mechanisms of antisense oligonucleotides as a therapeutic platform, Annu Rev Pharmacol Toxicol, 50 (2010) 259–293. [DOI] [PubMed] [Google Scholar]
- [224].Scoles DR, Minikel EV, Pulst SM, Antisense oligonucleotides: A primer, Neurol Genet, 5 (2019) e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [225].Le BT, Raguraman P, Kosbar TR, Fletcher S, Wilton SD, Veedu RN, Antisense Oligonucleotides Targeting Angiogenic Factors as Potential Cancer Therapeutics, Mol Ther Nucleic Acids, 14 (2019) 142–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [226].Eder PS, DeVine RJ, Dagle JM, Walder JA, Substrate specificity and kinetics of degradation of antisense oligonucleotides by a 3’ exonuclease in plasma, Antisense Res Dev, 1 (1991) 141–151. [DOI] [PubMed] [Google Scholar]
- [227].Di Fusco D, Dinallo V, Marafini I, Figliuzzi MM, Romano B, Monteleone G, Antisense Oligonucleotide: Basic Concepts and Therapeutic Application in Inflammatory Bowel Disease, Front Pharmacol, 10 (2019) 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Rinaldi C, Wood MJA, Antisense oligonucleotides: the next frontier for treatment of neurological disorders, Nat Rev Neurol, 14 (2018) 9–21. [DOI] [PubMed] [Google Scholar]
- [229].Borgonetti V, Galeotti N, Intranasal delivery of an antisense oligonucleotide to the RNA-binding protein HuR relieves nerve injury-induced neuropathic pain, Pain, 162 (2021) 1500–1510. [DOI] [PubMed] [Google Scholar]
- [230].Amodio N, Stamato MA, Juli G, Morelli E, Fulciniti M, Manzoni M, Taiana E, Agnelli L, Cantafio MEG, Romeo E, Raimondi L, Caracciolo D, Zuccala V, Rossi M, Neri A, Munshi NC, Tagliaferri P, Tassone P, Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity, Leukemia, 32 (2018) 1948–1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Blum AP, Kammeyer JK, Rush AM, Callmann CE, Hahn ME, Gianneschi NC, Stimuli-responsive nanomaterials for biomedical applications, J Am Chem Soc, 137 (2015) 2140–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Ho JJ, Robb GB, Tai SC, Turgeon PJ, Mawji IA, Man HS, Marsden PA, Active stabilization of human endothelial nitric oxide synthase mRNA by hnRNP E1 protects against antisense RNA and microRNAs, Mol Cell Biol, 33 (2013) 2029–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Messias AC, Harnisch C, Ostareck-Lederer A, Sattler M, Ostareck DH, The DICE-binding activity of KH domain 3 of hnRNP K is affected by c-Src-mediated tyrosine phosphorylation, J Mol Biol, 361 (2006) 470–481. [DOI] [PubMed] [Google Scholar]
- [234].Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH, Identification of an mRNP complex regulating tumorigenesis at the translational elongation step, Mol Cell, 41 (2011) 419–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Wang H, Vardy LA, Tan CP, Loo JM, Guo K, Li J, Lim SG, Zhou J, Chng WJ, Ng SB, Li HX, Zeng Q, PCBP1 suppresses the translation of metastasis-associated PRL-3 phosphatase, Cancer Cell, 18 (2010) 52–62. [DOI] [PubMed] [Google Scholar]
- [236].Howley BV, Hussey GS, Link LA, Howe PH, Translational regulation of inhibin betaA by TGFbeta via the RNA-binding protein hnRNP E1 enhances the invasiveness of epithelial-to-mesenchymal transitioned cells, Oncogene, 35 (2016) 1725–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Hu YP, Jin YP, Wu XS, Yang Y, Li YS, Li HF, Xiang SS, Song XL, Jiang L, Zhang YJ, Huang W, Chen SL, Liu FT, Chen C, Zhu Q, Chen HZ, Shao R, Liu YB, LncRNA-HGBC stabilized by HuR promotes gallbladder cancer progression by regulating miR-502–3p/SET/AKT axis, Mol Cancer, 18 (2019) 167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [238].Lee HH, Son YJ, Lee WH, Park YW, Chae SW, Cho WJ, Kim YM, Choi HJ, Choi DH, Jung SW, Min YJ, Park SE, Lee BJ, Cha HJ, Park JW, Tristetraprolin regulates expression of VEGF and tumorigenesis in human colon cancer, Int J Cancer, 126 (2010) 1817–1827. [DOI] [PubMed] [Google Scholar]
- [239].Young LE, Moore AE, Sokol L, Meisner-Kober N, Dixon DA, The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2, Mol Cancer Res, 10 (2012) 167–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [240].Chai Y, Liu J, Zhang Z, Liu L, HuR-regulated lncRNA NEAT1 stability in tumorigenesis and progression of ovarian cancer, Cancer Med, 5 (2016) 1588–1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [241].Brennan CM, Steitz JA, HuR and mRNA stability, Cell Mol Life Sci, 58 (2001) 266–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [242.Abdelmohsen K Lal A, Kim HH, Gorospe M Posttranscriptional orchestration of an anti-apoptotic program by HuR, Cell Cycle, 6 (2007) 1288–1292. [DOI] [PubMed] [Google Scholar]
- [243].Guo X, Hartley RS, HuR contributes to cyclin E1 deregulation in MCF-7 breast cancer cells, Cancer Res, 66 (2006) 7948–7956. [DOI] [PubMed] [Google Scholar]
- [244].Yoon JH, Abdelmohsen K, Srikantan S, Yang X, Martindale JL, De S, Huarte M, Zhan M, Becker KG, Gorospe M, LincRNA-p21 suppresses target mRNA translation, Mol Cell, 47 (2012) 648–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [245].Yoon JH, Abdelmohsen K, Kim J, Yang X, Martindale JL, Tominaga-Yamanaka K, White EJ, Orjalo AV, Rinn JL, Kreft SG, Wilson GM, Gorospe M, Scaffold function of long non-coding RNA HOTAIR in protein ubiquitination, Nat Commun, 4 (2013) 2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [246].Hopkins TG, Mura M, Al-Ashtal HA, Lahr RM, Abd-Latip N, Sweeney K, Lu H, Weir J, El-Bahrawy M, Steel JH, Ghaem-Maghami S, Aboagye EO, Berman AJ, Blagden SP, The RNA-binding protein LARP1 is a post-transcriptional regulator of survival and tumorigenesis in ovarian cancer, Nucleic Acids Res, 44 (2016) 1227–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [247].Paz S, Ritchie A, Mauer C, Caputi M, The RNA binding protein SRSF1 is a master switch of gene expression and regulation in the immune system, Cytokine Growth Factor Rev, 57 (2021) 19–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [248].Di J, Zhao G, Wang H, Wu Y, Zhao Z, Zhu B, Zhang Y, Zheng J, Liu Y, Hu Y, A p53/CPEB2 negative feedback loop regulates renal cancer cell proliferation and migration, J Genet Genomics, 48 (2021) 606–617. [DOI] [PubMed] [Google Scholar]
- [249].Yu ZJ, Luo HH, Shang ZF, Guan H, Xiao BB, Liu XD, Wang Y, Huang B, Zhou PK, Stabilization of 4E-BP1 by PI3K kinase and its involvement in CHK2 phosphorylation in the cellular response to radiation, Int J Med Sci, 14 (2017) 452–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [250].Cao S, Zheng J, Liu X, Liu Y, Ruan X, Ma J, Liu L, Wang D, Yang C, Cai H, Li Z, Feng Z, Xue Y, FXR1 promotes the malignant biological behavior of glioma cells via stabilizing MIR17HG, J Exp Clin Cancer Res, 38 (2019) 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [251].Li F, Zhao H, Su M, Xie W, Fang Y, Du Y, Yu Z, Hou L, Tan W, HnRNP-F regulates EMT in bladder cancer by mediating the stabilization of Snail1 mRNA by binding to its 3’ UTR, EBioMedicine, 45 (2019) 208–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [252].Yamazaki T, Liu L, Lazarev D, Al-Zain A, Fomin V, Yeung PL, Chambers SM, Lu CW, Studer L, Manley JL, TCF3 alternative splicing controlled by hnRNP H/F regulates E-cadherin expression and hESC pluripotency, Genes Dev, 32 (2018) 1161–1174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [253].Khan MI, Zhang J, Liu Q, HnRNP F and hnRNP H1 regulate mRNA stability of amyloid precursor protein, Neuroreport, 32 (2021) 824–832. [DOI] [PubMed] [Google Scholar]
- [254].Fred RG, Tillmar L, Welsh N, The role of PTB in insulin mRNA stability control, Curr Diabetes Rev, 2 (2006) 363–366. [DOI] [PubMed] [Google Scholar]
- [255].Kim SH, Lee KH, Kim DY, Kwak E, Kim S, Kim KT, Rhythmic control of mRNA stability modulates circadian amplitude of mouse Period3 mRNA, J Neurochem, 132 (2015) 642–656. [DOI] [PubMed] [Google Scholar]
- [256].Masuda K, Ripley B, Nishimura R, Mino T, Takeuchi O, Shioi G, Kiyonari H, Kishimoto T, Arid5a controls IL-6 mRNA stability, which contributes to elevation of IL-6 level in vivo, Proc Natl Acad Sci U S A, 110 (2013) 9409–9414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [257].Chen EB, Qin X, Peng K, Li Q, Tang C, Wei YC, Yu S, Gan L, Liu TS, HnRNPR-CCNB1/CENPF axis contributes to gastric cancer proliferation and metastasis, Aging (Albany NY), 11 (2019) 7473–7491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [258].He Y, Smith R, Nuclear functions of heterogeneous nuclear ribonucleoproteins A/B, Cell Mol Life Sci, 66 (2009) 1239–1256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [259].Huang Q, Guo H, Wang S, Ma Y, Chen H, Li H, Li J, Li X, Yang F, Qiu M, Zhao S, Wang J, A novel circular RNA, circXPO1, promotes lung adenocarcinoma progression by interacting with IGF2BP1, Cell Death Dis, 11 (2020) 1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [260].Jonson L, Christiansen J, Hansen TVO, Vikesa J, Yamamoto Y, Nielsen FC, IMP3 RNP safe houses prevent miRNA-directed HMGA2 mRNA decay in cancer and development, Cell Rep, 7 (2014) 539–551. [DOI] [PubMed] [Google Scholar]
- [261].Dehghani Amirabad A, Ramasamy P, Wierz M, Nordstrom K, Kessler SM, Schulz MH, Simon M, Transgenic expression of the RNA binding protein IMP2 stabilizes miRNA targets in murine microsteatosis, Biochim Biophys Acta Mol Basis Dis, 1864 (2018) 3099–3108. [DOI] [PubMed] [Google Scholar]
- [262].Mauchi N, Ohtake Y, Irie K, Stability control of MTL1 mRNA by the RNA-binding protein Khd1p in yeast, Cell Struct Funct, 35 (2010) 95–105. [DOI] [PubMed] [Google Scholar]
- [263].Challa AA, Stefanovic B, A novel role of vimentin filaments: binding and stabilization of collagen mRNAs, Mol Cell Biol, 31 (2011) 3773–3789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [264].Yu H, Zhang F, Yan P, Zhang S, Lou Y, Geng Z, Li Z, Zhang Y, Xu Y, Lu Y, Chen C, Wang D, Zhu W, Hu X, Wang J, Zhuang T, Zhang Y, Wu G, Liu J, Zeng C, Pu WT, Sun K, Zhang B, LARP7 Protects Against Heart Failure by Enhancing Mitochondrial Biogenesis, Circulation, 143 (2021) 2007–2022. [DOI] [PubMed] [Google Scholar]
- [265].Shin KH, Kim JM, Rho KS, Park KH, Oh JE, Min BM, Inactivation of the PTEN gene by mutation, exonic deletion, and loss of transcript in human oral squamous cell carcinomas, Int J Oncol, 21 (2002) 997–1001. [PubMed] [Google Scholar]
- [266].Brueckl WM, Grombach J, Wein A, Ruckert S, Porzner M, Dietmaier W, Rummele P, Croner RS, Boxberger F, Kirchner T, Hohenberger W, Hahn EG, Jung A, Alterations in the tissue inhibitor of metalloproteinase-3 (TIMP-3) are found frequently in human colorectal tumours displaying either microsatellite stability (MSS) or instability (MSI), Cancer Lett, 223 (2005) 137–142. [DOI] [PubMed] [Google Scholar]
- [267].Pisano A, Ceglia S, Palmieri C, Vecchio E, Fiume G, de Laurentiis A, Mimmi S, Falcone C, Iaccino E, Scialdone A, Pontoriero M, Masci FF, Valea R, Krishnan S, Gaspari M, Cuda G, Scala G, Quinto I, CRL3IBTK Regulates the Tumor Suppressor Pdcd4 through Ubiquitylation Coupled to Proteasomal Degradation, J Biol Chem, 290 (2015) 13958–13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [268].Zhang RL, Yang JP, Peng LX, Zheng LS, Xie P, Wang MY, Cao Y, Zhang ZL, Zhou FJ, Qian CN, Bao YX, RNA-binding protein QKI-5 inhibits the proliferation of clear cell renal cell carcinoma via post-transcriptional stabilization of RASA1 mRNA, Cell Cycle, 15 (2016) 3094–3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [269].Sutherland LC, Thibault P, Durand M, Lapointe E, Knee JM, Beauvais A, Kalatskaya I, Hunt SC, Loiselle JJ, Roy JG, Tessier SJ, Ybazeta G, Stein L, Kothary R, Klinck R, Chabot B, Splicing arrays reveal novel RBM10 targets, including SMN2 pre-mRNA, BMC Mol Biol, 18 (2017) 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [270].Zhu X, Yan J, Bregere C, Zelmer A, Goerne T, Kapfhammer JP, Guzman R, Wellmann S, RBM3 promotes neurogenesis in a niche-dependent manner via IMP2-IGF2 signaling pathway after hypoxic-ischemic brain injury, Nat Commun, 10 (2019) 3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [271].Shen DJ, Jiang YH, Li JQ, Xu LW, Tao KY, The RNA-binding protein RBM47 inhibits non-small cell lung carcinoma metastasis through modulation of AXIN1 mRNA stability and Wnt/beta-catentin signaling, Surg Oncol, 34 (2020) 31–39. [DOI] [PubMed] [Google Scholar]
- [272].Vanharanta S, Marney CB, Shu W, Valiente M, Zou Y, Mele A, Darnell RB, Massague J, Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer, Elife, 3 (2014) e02734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [273].Li N, Hebert S, Song J, Kleinman CL, Richard S, Transcriptome profiling in preadipocytes identifies long noncoding RNAs as Sam68 targets, Oncotarget, 8 (2017) 81994–82005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [274].Samarakoon R, Higgins SP, Higgins CE, Higgins PJ, The TGF-beta1/p53/PAI-1 Signaling Axis in Vascular Senescence: Role of Caveolin-1, Biomolecules, 9 (2019) 341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [275].Goncalves V, Matos P, Jordan P, The beta-catenin/TCF4 pathway modifies alternative splicing through modulation of SRp20 expression, RNA, 14 (2008) 2538–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [276].Ansari MY, Haqqi TM, Interleukin-1beta induced Stress Granules Sequester COX-2 mRNA and Regulates its Stability and Translation in Human OA Chondrocytes, Sci Rep, 6 (2016) 27611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [277].Kim SH, Shahani N, Bae BI, Sbodio JI, Chung Y, Nakaso K, Paul BD, Sawa A, Allele-specific regulation of mutant Huntingtin by Wig1, a downstream target of p53, Hum Mol Genet, 25 (2016) 2514–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [278].Bersani C, Huss M, Giacomello S, Xu LD, Bianchi J, Eriksson S, Jerhammar F, Alexeyenko A, Vilborg A, Lundeberg J, Lui WO, Wiman KG, Genome-wide identification of Wig-1 mRNA targets by RIP-Seq analysis, Oncotarget, 7 (2016) 1895–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [279].Talwar S, Balasubramanian S, Sundaramurthy S, House R, Wilusz CJ, Kuppuswamy D, D’Silva N, Gillespie MB, Hill EG, Palanisamy V, Overexpression of RNA-binding protein CELF1 prevents apoptosis and destabilizes pro-apoptotic mRNAs in oral cancer cells, RNA Biol, 10 (2013) 277–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [280].Yamaji M, Jishage M, Meyer C, Suryawanshi H, Der E, Yamaji M, Garzia A, Morozov P, Manickavel S, McFarland HL, Roeder RG, Hafner M, Tuschl T, DND1 maintains germline stem cells via recruitment of the CCR4-NOT complex to target mRNAs, Nature, 543 (2017) 568–572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [281].Singh AB, Li H, Kan CF, Dong B, Nicolls MR, Liu J, The critical role of mRNA destabilizing protein heterogeneous nuclear ribonucleoprotein d in 3’ untranslated region-mediated decay of low-density lipoprotein receptor mRNA in liver tissue, Arterioscler Thromb Vasc Biol, 34 (2014) 8–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [282].Pan R, Cai W, Sun J, Yu C, Li P, Zheng M, Inhibition of KHSRP sensitizes colorectal cancer to 5-fluoruracil through miR-501–5p-mediated ERRFI1 mRNA degradation, J Cell Physiol, 235 (2020) 1576–1587. [DOI] [PubMed] [Google Scholar]
- [283].Sun X, Zhang H, Xie L, Qian C, Ye Y, Mao H, Wang B, Zhang H, Zhang Y, He X, Zhang S, Tristetraprolin destabilizes NOX2 mRNA and protects dopaminergic neurons from oxidative damage in Parkinson’s disease, FASEB J, 34 (2020) 15047–15061. [DOI] [PubMed] [Google Scholar]
- [284].Liang H, Zhang J, Shao C, Zhao L, Xu W, Sutherland LC, Wang K, Differential expression of RBM5, EGFR and KRAS mRNA and protein in non-small cell lung cancer tissues, J Exp Clin Cancer Res, 31 (2012) 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [285].Li R, Li H, Ge C, Fu Q, Li Z, Jin Y, Tan Q, Zhu Z, Zhang Z, Dong S, Li G, Song X, Increased expression of the RNA-binding motif protein 47 predicts poor prognosis in non-small-cell lung cancer, Oncol Lett, 19 (2020) 3111–3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [286].Yan M, Sun L, Li J, Yu H, Lin H, Yu T, Zhao F, Zhu M, Liu L, Geng Q, Kong H, Pan H, Yao M, RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer, J Exp Clin Cancer Res, 38 (2019) 478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [287].Shi R, Yu X, Wang Y, Sun J, Sun Q, Xia W, Dong G, Wang A, Gao Z, Jiang F, Xu L, Expression profile, clinical significance, and biological function of insulin-like growth factor 2 messenger RNA-binding proteins in non-small cell lung cancer, Tumour Biol, 39 (2017) 1010428317695928. [DOI] [PubMed] [Google Scholar]
- [288].Xu Z, Xu J, Lu H, Lin B, Cai S, Guo J, Zang F, Chen R, LARP1 is regulated by the XIST/miR-374a axis and functions as an oncogene in non-small cell lung carcinoma, Oncol Rep, 38 (2017) 3659–3667. [DOI] [PubMed] [Google Scholar]
- [289].Cui J, Ren P, Li Y, Ma Y, Wang J, Lin C, Jing L, Tong X, Ma S, Chen J, ESRP1 as a prognostic factor of non-small-cell lung cancer is related to the EMT transcription factor of Twist, Thorac Cancer, 12 (2021) 2449–2457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [290].Yin J, Zhao J, Hu W, Yang G, Yu H, Wang R, Wang L, Zhang G, Fu W, Dai L, Li W, Liao B, Zhang S, Disturbance of the let-7/LIN28 double-negative feedback loop is associated with radio- and chemo-resistance in non-small cell lung cancer, PLoS One, 12 (2017) e0172787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [291].Lv Y, Zhang W, Zhao J, Sun B, Qi Y, Ji H, Chen C, Zhang J, Sheng J, Wang T, Dominguez D, Liu H, Liu Q, Meng S, Li X, Wang Y, SRSF1 inhibits autophagy through regulating Bcl-x splicing and interacting with PIK3C3 in lung cancer, Signal Transduct Target Ther, 6 (2021) 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [292].Cohen-Eliav M, Golan-Gerstl R, Siegfried Z, Andersen CL, Thorsen K, Orntoft TF, Mu D, Karni R, The splicing factor SRSF6 is amplified and is an oncoprotein in lung and colon cancers, J Pathol, 229 (2013) 630–639. [DOI] [PubMed] [Google Scholar]
- [293].Chen CY, Jan CI, Pi WC, Wang WL, Yang PC, Wang TH, Karni R, Wang TC, Heterogeneous nuclear ribonucleoproteins A1 and A2 modulate expression of Tid1 isoforms and EGFR signaling in non-small cell lung cancer, Oncotarget, 7 (2016) 16760–16772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [294].Zhen Y, Li D, Li W, Yao W, Wu A, Huang J, Gu H, Huang Y, Wang Y, Wu J, Chen M, Wu D, Lyu Q, Fang W, Wu B, Reduced PDCD4 Expression Promotes Cell Growth Through PI3K/Akt Signaling in Non-Small Cell Lung Cancer, Oncol Res, 23 (2016) 61–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [295].Zong FY, Fu X, Wei WJ, Luo YG, Heiner M, Cao LJ, Fang Z, Fang R, Lu D, Ji H, Hui J, The RNA-binding protein QKI suppresses cancer-associated aberrant splicing, PLoS Genet, 10 (2014) e1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [296].Kudinov AE, Deneka A, Nikonova AS, Beck TN, Ahn YH, Liu X, Martinez CF, Schultz FA, Reynolds S, Yang DH, Cai KQ, Yaghmour KM, Baker KA, Egleston BL, Nicolas E, Chikwem A, Andrianov G, Singh S, Borghaei H, Serebriiskii IG, Gibbons DL, Kurie JM, Golemis EA, Boumber Y, Musashi-2 (MSI2) supports TGF-beta signaling and inhibits claudins to promote non-small cell lung cancer (NSCLC) metastasis, Proc Natl Acad Sci U S A, 113 (2016) 6955–6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [297].Deneka A, Kharin L, Karnaukhov NS, Voloshin M, Ayrapetova TG, Ratner E, Sabirov A, Topchu Y, Mazitova A, Abramova Z, Yugai V, Frantsiyants EM, Kit OI, Boumber Y, Musashi 2 (MSI2) expression as an independent prognostic biomarker in non-small cell lung cancer (NSCLC), Journal of Clinical Oncology, 38 (2020) e21583–e21583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [298].Wang XY, Yu H, Linnoila RI, Li L, Li D, Mo B, Okano H, Penalva LO, Glazer RI, Musashi1 as a potential therapeutic target and diagnostic marker for lung cancer, Oncotarget, 4 (2013) 739–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [299].Zhang Y, Huang S, Gao H, Wu K, Ouyang X, Zhu Z, Yu X, Zeng T, Upregulation of KIN17 is associated with non-small cell lung cancer invasiveness, Oncol Lett, 13 (2017) 2274–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [300].Zhang B, Zhu C, Chen B, Zhang X, Ye M, Lin A, [Expression and its clinical significance of eIF4E in non-small cell lung cancer], Zhongguo Fei Ai Za Zhi, 13 (2010) 1132–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [301].Qian J, Hassanein M, Hoeksema MD, Harris BK, Zou Y, Chen H, Lu P, Eisenberg R, Wang J, Espinosa A, Ji X, Harris FT, Rahman SM, Massion PP, The RNA binding protein FXR1 is a new driver in the 3q26–29 amplicon and predicts poor prognosis in human cancers, Proc Natl Acad Sci U S A, 112 (2015) 3469–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [302].Zhang Z, Xu Y, Sun N, Zhang M, Xie J, Jiang Z, High Sam68 expression predicts poor prognosis in non-small cell lung cancer, Clin Transl Oncol, 16 (2014) 886–891. [DOI] [PubMed] [Google Scholar]
- [303].Tang F, Li W, Chen Y, Wang D, Han J, Liu D, Downregulation of hnRNP K by RNAi inhibits growth of human lung carcinoma cells, Oncol Lett, 7 (2014) 1073–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [304].Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, Li D, Song H, Wang J, Hong M, Wang X, Huang K, Zheng L, Tong Q, Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes, Cell Death Differ, 26 (2019) 1346–1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [305].Yang F, Hu A, Li D, Wang J, Guo Y, Liu Y, Li H, Chen Y, Wang X, Huang K, Zheng L, Tong Q, Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation, Mol Cancer, 18 (2019) 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [306].Liu H, Lan T, Li H, Xu L, Chen X, Liao H, Chen X, Du J, Cai Y, Wang J, Li X, Huang J, Yuan K, Zeng Y, Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR, Theranostics, 11 (2021) 1396–1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [307].Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, Ma P, Jiang H, Wu X, Shu Y, Xu T, Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582–3p/HUR/VEGF axis and suppressing HSP90 degradation, Mol Cancer, 19 (2020) 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [308].Abdelmohsen K, Panda AC, Munk R, Grammatikakis I, Dudekula DB, De S, Kim J, Noh JH, Kim KM, Martindale JL, Gorospe M, Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1, RNA Biol, 14 (2017) 361–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [309].Zang Y, Li J, Wan B, Tai Y, circRNA circ-CCND1 promotes the proliferation of laryngeal squamous cell carcinoma through elevating CCND1 expression via interacting with HuR and miR-646, J Cell Mol Med, 24 (2020) 2423–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [310].Wang Y, Gao R, Li J, Tang S, Li S, Tong Q, Li S, Downregulation of hsa_circ_0074854 Suppresses the Migration and Invasion in Hepatocellular Carcinoma via Interacting with HuR and via Suppressing Exosomes-Mediated Macrophage M2 Polarization, Int J Nanomedicine, 16 (2021) 2803–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [311].Lan Y, Xiao X, He Z, Luo Y, Wu C, Li L, Song X, Long noncoding RNA OCC-1 suppresses cell growth through destabilizing HuR protein in colorectal cancer, Nucleic Acids Res, 46 (2018) 5809–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [312].Zhang T, Beeharry MK, Zheng Y, Wang Z, Li J, Zhu Z, Li C, Long Noncoding RNA SNHG12 Promotes Gastric Cancer Proliferation by Binding to HuR and Stabilizing YWHAZ Expression Through the AKT/GSK-3beta Pathway, Front Oncol, 11 (2021) 645832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [313].Gong F, Dong D, Zhang T, Xu W, Long non-coding RNA FENDRR attenuates the stemness of non-small cell lung cancer cells via decreasing multidrug resistance gene 1 (MDR1) expression through competitively binding with RNA binding protein HuR, Eur J Pharmacol, 853 (2019) 345–352. [DOI] [PubMed] [Google Scholar]
- [314].Priyanka P, Sharma M, Das S, Saxena S, The lncRNA HMS recruits RNA-binding protein HuR to stabilize the 3’-UTR of HOXC10 mRNA, J Biol Chem, 297 (2021) 100997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [315].Xu CZ, Jiang C, Wu Q, Liu L, Yan X, Shi R, A Feed-Forward Regulatory Loop between HuR and the Long Noncoding RNA HOTAIR Promotes Head and Neck Squamous Cell Carcinoma Progression and Metastasis, Cell Physiol Biochem, 40 (2016) 1039–1051. [DOI] [PubMed] [Google Scholar]
- [316].Long B, Yang X, Xu X, Li X, Xu X, Zhang X, Zhang S, Long noncoding RNA ASB16-AS1 inhibits adrenocortical carcinoma cell growth by promoting ubiquitination of RNA-binding protein HuR, Cell Death Dis, 11 (2020) 995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [317].Wang M, Mao C, Ouyang L, Liu Y, Lai W, Liu N, Shi Y, Chen L, Xiao D, Yu F, Wang X, Zhou H, Cao Y, Liu S, Yan Q, Tao Y, Zhang B, Long noncoding RNA LINC00336 inhibits ferroptosis in lung cancer by functioning as a competing endogenous RNA, Cell Death Differ, 26 (2019) 2329–2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [318].Abdelmohsen K, Srikantan S, Kuwano Y, Gorospe M, miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels, Proc Natl Acad Sci U S A, 105 (2008) 20297–20302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [319].Xu F, Zhang X, Lei Y, Liu X, Liu Z, Tong T, Wang W, Loss of repression of HuR translation by miR-16 may be responsible for the elevation of HuR in human breast carcinoma, J Cell Biochem, 111 (2010) 727–734. [DOI] [PubMed] [Google Scholar]
- [320].Shen Z, Zhan G, Deng H, Ren Y, Ye D, Xiao B, Guo J, MicroRNA-519a demonstrates significant tumour suppressive activity in laryngeal squamous cells by targeting anti-carcinoma HuR gene, J Laryngol Otol, 127 (2013) 1194–1202. [DOI] [PubMed] [Google Scholar]


