Abstract
STUDY QUESTION
Are there age-related differences in gene expression during the germinal vesicle (GV) to metaphase II (MII) stage transition in euploid human oocytes?
SUMMARY ANSWER
A decrease in mitochondrial-related transcripts from GV to MII oocytes was observed, with a much greater reduction in MII oocytes with advanced age.
WHAT IS KNOWN ALREADY
Early embryonic development is dependent on maternal transcripts accumulated and stored within the oocyte during oogenesis. Transcriptional activity of the oocyte, which dictates its ultimate developmental potential, may be influenced by age and explain the reduced competence of advanced maternal age (AMA) oocytes compared with the young maternal age (YMA). Gene expression has been studied in human and animal oocytes; however, RNA sequencing could provide further insights into the transcriptome profiling of GV and in vivo matured MII euploid oocytes of YMA and AMA patients.
STUDY DESIGN, SIZE, DURATION
Fifteen women treated for infertility in a single IVF unit agreed to participate in this study. Five GV and 5 MII oocytes from 6, 21–26 years old women (YMA cohort) and 5 GV and 6 MII oocytes from 6, 41–44 years old women (AMA cohort) undergoing IVF treatment were donated. The samples were collected within a time frame of 4 months. RNA was isolated and deep sequenced at the single-cell level. All donors provided either GV or MII oocytes.
PARTICIPANTS/MATERIALS, SETTING, METHODS
Cumulus dissection from donated oocytes was performed 38 h after hCG injection, denuded oocytes were inserted into lysis buffer supplemented with RNase inhibitor. The samples were stored at −80°C until further use. Isolated RNA from GV and MII oocytes underwent library preparation using an oligo deoxy-thymidine (dT) priming approach (SMART-Seq v4 Ultra Low Input RNA assay; Takara Bio, Japan) and Nextera XT DNA library preparation assay (Illumina, USA) followed by deep sequencing. Data processing, quality assessment and bioinformatics analysis were performed using source-software, mainly including FastQC, HISAT2, StringTie and edgeR, along with functional annotation analysis, while scploid R package was employed to determine the ploidy status.
MAIN RESULTS AND THE ROLE OF CHANCE
Following deep sequencing of single GV and MII oocytes in both YMA and AMA cohorts, several hundred transcripts were found to be expressed at significantly different levels. When YMA and AMA MII oocyte transcriptomes were compared, the most significant of these were related to mitochondrial structure and function, including biological processes, mitochondrial respiratory chain complex I assembly and mitochondrial translational termination (false discovery rate (FDR) 6.0E−10 to 1.2E−7). These results indicate a higher energy potential of the YMA MII cohort that is reduced with ageing. Other biological processes that were significantly higher in the YMA MII cohort included transcripts involved in the translation process (FDR 1.9E−2). Lack of these transcripts could lead to inappropriate protein synthesis prior to or upon fertilisation of the AMA MII oocytes.
LARGE SCALE DATA
The RNA sequencing data were deposited in the Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo), under the accession number: GSE164371.
LIMITATIONS, REASONS FOR CAUTION
The relatively small sample size could be a reason for caution. However, the RNA sequencing results showed homogeneous clustering with low intra-group variation and five to six biological replicates derived from at least three different women per group minimised the potential impact of the sample size.
WIDER IMPLICATIONS OF THE FINDINGS
Understanding the effects of ageing on the oocyte transcriptome could highlight the mechanisms involved in GV to MII transition and identify biomarkers that characterise good MII oocyte quality. This knowledge has the potential to guide IVF regimes for AMA patients.
STUDY FUNDING/COMPETING INTEREST(S)
This work was supported by the Medical Research Council (MRC Grant number MR/K020501/1).
Keywords: RNA sequencing, GV oocyte, MII oocyte, ageing, differential gene expression
Introduction
Reproductive ageing is an irreversible biological process that occurs in advance of senescence. Oocyte developmental competence (quality) and fertility rates are reduced upon ageing (Navot et al., 1991). It is widely accepted that increasing chromosome segregation errors lead to significantly higher frequencies of chromosome anomalies, elevated miscarriage rates with advanced age and a significant decrease in pregnancy rate (Duncan et al., 2012; Webster and Schuh, 2017; Gruhn et al., 2019). Other underlying factors that may contribute to reduced oocyte quality with age include the transcriptional activity of the oocyte, which may dictate the developmental potential of the oocyte and the embryo (Song and Wessel, 2005; Stitzel and Seydoux, 2007), along with factors involved in mitochondrial functionality, including mitochondrial gene expression and energy production (Schatten et al., 2014).
Early embryonic cleavage, for example, is governed by maternal RNA stores that accumulate within the oocyte during oogenesis (Cimadomo et al., 2018). Alterations in the mRNA and proteins in the cytoplasm of advanced maternal age (AMA) oocytes and early embryos could therefore partially explain the aetiology of infertility in AMA women (Hamatani et al., 2004). Genes expressed in the oocyte are dramatically degraded upon the transition from the immature germinal vesicles (GVs) to the mature/fertile metaphase II (MII) oocyte in mammals (Su et al., 2007; Reyes et al., 2017). These transcripts are probably important during oogenesis, but may be unnecessary in the final MII oocyte growth phase, while other transcripts are involved in protein synthesis, relying on a combination of the poly(A) tail length and high affinity RNA binding proteins (Kim et al., 2011).
Gene transcripts present in oocytes are mainly involved in meiosis-related processes, as well as energy production and metabolism. Gene expression associated with signalling pathways, appears to be quite stable in the late growth phase of the oocytes, indicating their highly important role in oocyte maturation (Su et al., 2007; Reyes et al., 2017). Oocyte maturation is modulated by a highly regulated programme of gene expression, along with cytoplasmic deadenylation (Reyes et al., 2015). Ageing, however, does not appear to affect GV and MII oocyte gene expression equally, with the latter reporting a much greater number of significantly altered gene transcripts with ageing. Overall, changes in oocyte mitochondrial number and activity combined with epigenetic modifications of the oocyte and altered DNA methylation patterns caused by advancing maternal age, may impact on the health of the offspring (Tarín et al., 2002; Liang et al., 2008, 2011; Khan et al., 2016).
Different studies have investigated the transcriptome derived from mammalian GV and MII oocytes. These studies revealed pathways associated with the ageing process and the transition from GV to MII oocytes, including the study by that used human GV oocytes and in vitro matured (IVM) MII oocytes to investigate the response of young maternal age (YMA; <30 years) and AMA (≥40 years) oocytes to in vitro matured. To the best of our knowledge, our study is the first to investigate the impact of ageing on the transcriptome of individual euploid human oocytes upon the transition of the immature GV to the in vivo matured MII oocyte. The YMA MII oocytes are associated with the greatest potential for successful embryo development. This means that differences identified between the GV and MII oocyte stages and different maternal ages, could improve our understanding of subsequent successful human embryo development and suggest potential molecular biomarkers that are indicative of high oocyte quality.
Materials and methods
Ethical approval
Women presented for infertility treatment at the Reproductive Medicine Unit, Genesis Athens Clinic, Greece, between September and December 2017. All women were eligible to participate in this study. The study was approved by the National Health System A’ Administration of the Health District of Attica, General Children Hospital ‘Aghia Sofia’ (19964/04-09-2014) and the Greek National Authority of Assisted Reproduction. Women participating in the current study provided signed informed consent for oocyte donation. All women were recruited from a single IVF unit.
Sample recruitment and treatment
Fifteen women treated for infertility agreed to participate in this study. Five GV and 5 MII oocytes from 6 women (21–26 years old; YMA cohort) and 5 GV and 6 MII oocytes from 6 women (41–44 years old; AMA cohort) undergoing IVF treatment were donated. The samples were collected within a time frame of 4 months and RNA was isolated and deep sequenced at the single-cell level. Ovarian stimulation was performed according to a short, step down GnRH agonist stimulation protocol with a recombinant FSH (folitropin alpha) (Gonal-F; Merck Serono Europe Ltd., Germany) starting on the second day of the menstrual cycle for final follicular maturation and were followed by oocyte retrieval guided by ultrasound. Doses varied according to women’s’ age and individual infertility profile (range 150–300 IU/day). All women were re-evaluated on Day 5 of stimulation with a transvaginal ultrasound examination and serum oestradiol levels. At this time, adjustments to the dose were made. Triggering was performed with recombinant hCG (Ovitrelle, Merck Serono Europe Ltd., Germany) 36 h prior to transvaginal oocyte collection. Following oocyte collection, cumulus dissection with hyaluronidase (FertiPro, Belgium) from donated oocytes was performed 38 h after hCG administration. Denuded oocytes were assessed for maturation stage and transferred into lysis buffer supplemented with RNase inhibitor.
RNA isolation, library construction and deep sequencing
The SMART-Seq v4 Ultra Low Input RNA method (Takara Bio, Japan) was employed for library construction, according to the manufacturer’s instructions. Following two washing steps in calcium/magnesium free Dulbecco’s Phosphate Buffered Saline (Merck Serono, Germany), the oocytes from all the four cohorts were immediately transferred into the lysis buffer with RNase Inhibitor (Takara Bio, Japan) and frozen at −80°C until required. The first cDNA strand was synthesised using the locked nucleic acid technology and large quantities of full-length cDNA were produced by long-distance PCR. The same number of PCR cycles (16 cycles) were applied to keep constant conditions across the samples. The amplified cDNA was purified by the Agencourt Ampure XP Beads (Beckman Coulter, USA).
Amplified full-length cDNA quantification employed a high sensitivity fluorometric assay (Qubit) (Thermo Fisher Scientific, USA) with 150 pg being processed further using Illumina’s Nextera XT DNA library construction method according to the manufacturer’s instructions (Illumina, USA). Tagging and fragmentation of the full-length cDNA occurred in a single step and a unique combination of i7 and i5 indices per library was used (Illumina, USA). Amplified libraries were purified by Agencourt AMPure XP beads (Beckman Coulter, USA). A high sensitivity fluorometric assay (Qubit) was employed using Qubit 1.0 fluorometer (Thermo Fisher Scientific, USA) together with the 2100 Bioanalyzer system (Agilent Technologies, USA) for evaluating oocyte quantity and quality, respectively. The average length distribution of the fragmented cDNA was calculated using the high sensitivity DNA assay (Agilent, USA), followed by equimolar pooling and deep RNA sequencing (150 base pairs) on a HiSeq 3000 (Illumina, USA).
Bioinformatics analysis
Sequence quality was assessed using FastQC (Andrews, 2010). Trim galore was employed for automated adapter and quality trimming (Krueger, 2015). Reads passing Quality Control (QC) were mapped to the human reference genome (hg38) by the HISAT2 aligner v2.0.4 (Pertea et al., 2016). Samtools v1.3 was used to remove unpaired and unmapped reads (Li et al., 2009) and PCR duplicates were tagged by Picard tools v2.1.1 (Broad Institute (2010). Available online at http://broadinstitute.github.io/picard). StringTie was employed to detect potentially novel transcripts using UCSC hg38 RefSeq annotation of the human transcriptome as a guide (Pertea et al., 2015). To assign and quantify reads mapping to known and predicted transcripts and to cross-check the robustness of differential gene expression (DGE) detection, two approaches were used. The first employed a python script (available online: http://ccb.jhu.edu/software/stringtie/dl/prepDE.py) to extract read counts from the HISAT2/StringTie outputs in a format suitable as input for the Bioconductor/R package edgeR (Robinson and Oshlack, 2010; R Core Team, 2013). The other used the featureCounts function of Rsubread to generate count tables (Liao et al., 2013). Both long coding and long non-coding transcripts were revealed through this approach.
The DaMiRseq R package pipeline was employed to determine molecular biomarkers that could characterise the quality of YMA and AMA oocytes (Chiesa et al., 2018). A correlation cut off of 0.3 was used for normalisation and partial least-square feature selection (FSelect function) to consider gene expression in at least half of the samples of the smaller cohort (i.e. YMA oocytes). We selected the number of surrogate variables that explained at least 90% of the variation. Default values were used for all other functions. Pheatmap and ggplot2 R packages were employed to generate distance matrix and multi-dimensional scaling plots (Kolde and Kolde, 2015; Wickham, 2016).
Statistical analysis
The thresholds for transcript inclusion in the DGE analysis were at least one counts-per-million reads (cpm) present in at least four samples. The calcNormFactors function was used to normalise count data, according to the trimmed mean M value (Robinson and Oshlack, 2010). Gene transcripts represented at significantly different levels among the oocyte cohorts were detected using the generalised linear model (GLM) approach with false discovery rate (FDR) threshold set at 0.05 and fold change greater than 2 (Robinson and Oshlack, 2010).
Functional annotation, aneuploidy analysis and meta-analysis
Ontological analysis of differentially expressed genes was conducted by DAVID (Huang et al., 2009) and goana function in R (Young et al., 2010), including the significantly up-regulated gene transcripts of the YMA and AMA GV and/or MII oocytes. We mainly focused on the biological processes, cellular components and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses. FDR was employed for DGE and ontological analysis. The R package scploid, developed by Griffiths et al. (2017), was employed to determine the ploidy status from YMA and AMA GV and MII oocytes. The RNA sequencing data were deposited in the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo), under the GEO accession number: GSE164371. The current study also considered RNA sequencing data from GV and in vitro matured MII oocytes that were derived from young and advanced age women in the context of meta-analysis (GEO accession: GSE95477).
RNA sequencing validation by qRT-PCR
SMART-Seq v4 ultra-low input RNA method (Takara-Clontech, USA) was employed to validate the RNA-Seq results that derived from single GV and MII oocytes. In brief, reverse transcription was followed by long distance PCR to generate sufficient quantities of cDNA libraries per oocyte. Roche LC480 thermal cycler (Roche, Switzerland) was then used to perform quantitative real-time PCR (qRT-PCR) on three replicates. Annealing temperature was always set at 60°C (Supplementary Table SI). Normalisation was conducted using H3F3B, GAPDH, HNRNPC and ACTB genes as endogenous controls. Gene expression differences between the oocyte groups were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Results
Characterisation of participants
To investigate the molecular transcriptome dynamics involved in maternal ageing, individual GV and in vivo matured MII oocytes were analysed by RNA sequencing from YMA (<30 years old) and AMA (>40 years old) groups. Fifteen women treated for infertility in a single IVF unit agreed to participate in this study by donating oocytes. The mean (SD) ages of the women consisting the YMA GV and MII groups were 22.6 (2.3) years and 21.6 (1.5) years, respectively. The average (SD) age of the women consisting the AMA GV and MII groups were 42.8 (1.1) years and 42 (1.0) years, respectively (Table I). Samples were collected within a 4-month time frame. No morphological differences in the YMA and AMA oocytes were noted. RNA was isolated and deep sequenced.
Table I.
Characteristics of GV and MII oocyte patients.
| GV YMA | MII YMA | GV AMA | MII AMA | |
|---|---|---|---|---|
| Mean age ± SD per oocyte | 22.6 ± 2.3 | 21.6 ± 1.5 | 42.8 ± 1.1 | 42 ± 1.0 |
| Number of women | 4 | 4 | 4 | 3 |
| Number of oocytes | 5 | 5 | 5 | 7 |
| Infertility factor | No (donor) | No (donor) | Unexplained/AMA | Male/AMA |
AMA, advanced maternal age; GV oocyte, germinal vesicle oocyte; MII oocyte, metaphase II oocyte; YMA, young maternal age.
Transcriptome characterisation of individual human oocytes
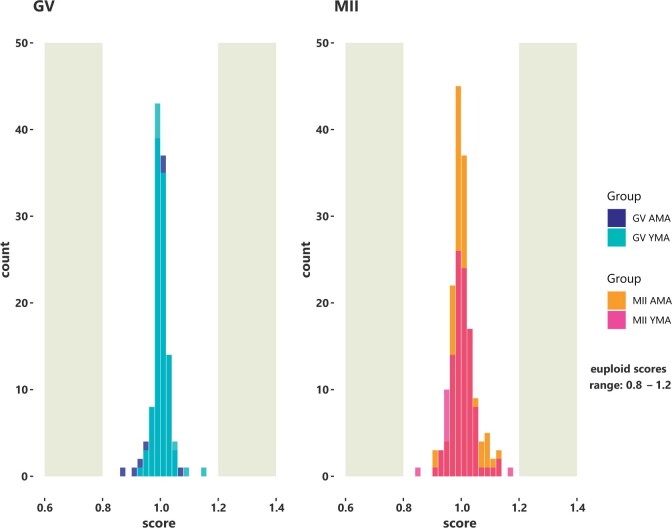
Following library construction and deep sequencing of each oocyte transcriptome, the average aligned reads were 41 (9) and 38 (6) (mean (SD)) millions for the YMA GV and MII oocytes, respectively, and 31 (3) and 30 (5) million reads for the GV and MII oocytes of AMA (Supplementary Table SII). The main genes discussed in this study showed a statistical dispersion of 0.17 and a minimum fold change per comparison of 2.5. Aneuploidy analysis relying on the RNA sequencing data considered only euploid oocytes to participate in the current study. A euploid GV with a normal score range between 0.8 and 1.2 is shown as example (Fig. 1).
Figure 1.
Bell shaped histograms illustrate the score of the average euploid GV and MII oocytes. Normal scores generated by scploid software range between 0.8 and 1.2 (euploidy status). Based on the allele specific gene expression, a score of 1 indicates that no aneuploid chromosomes were present. The results derived from the RNA sequencing data analysis. AMA, advanced maternal age; GV oocyte, germinal vesicle oocyte; MII oocyte, metaphase II oocyte; YMA, young maternal age.
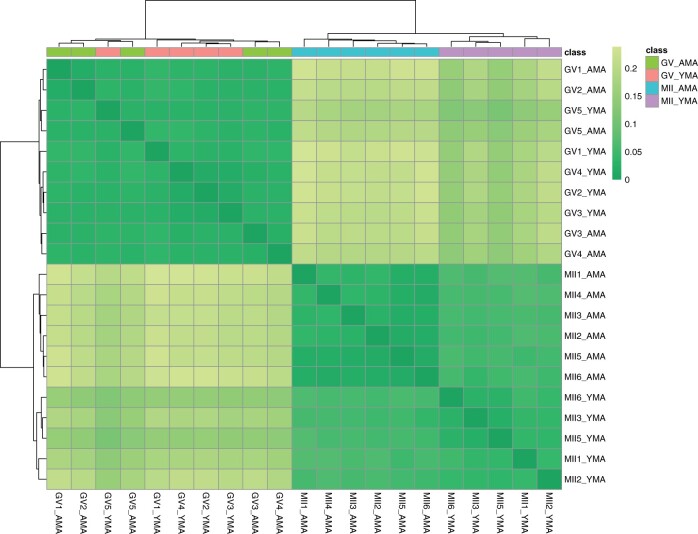
The transcriptome relationships among the four oocyte cohorts were illustrated using a distance matrix, which was generated based on sample-by-sample correlation, highlighting specific clusters related to maternal age (Fig. 2). Unlike the YMA and AMA MII oocytes that resulted in separate clusters following hierarchical clustering, the YMA and AMA, GV oocytes did not show a distinct separation relying on maternal age, which indicates that ageing has a greater effect on the final stage of oocyte differentiation (MII oocytes).
Figure 2.
Heatmap with colours highlighting the distance matrix (Spearman’s correlation). Colour gradient ranges from dark green (minimum distance) to light green. Horizontal bars (on the top of heatmap) represent the maternal age class. Cluster identification is facilitated by the two dendrograms. AMA, advanced maternal age; GV oocyte, germinal vesicle oocyte; MII oocyte, metaphase II oocyte; YMA, young maternal age.
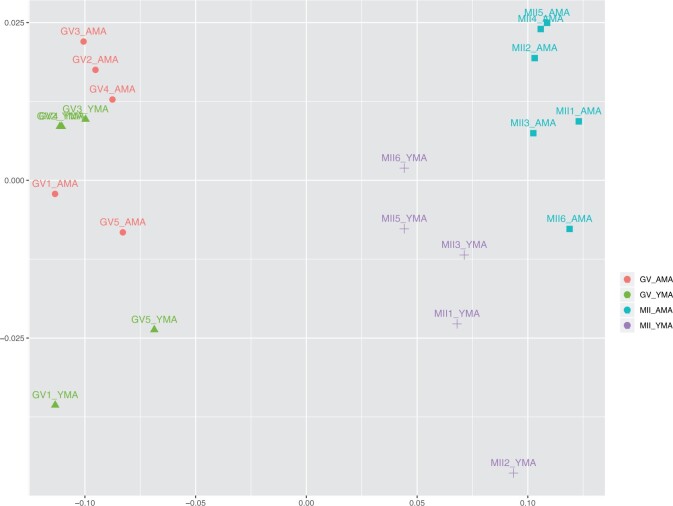
Unsupervised multidimensional scaling plot illustrated oocyte transcriptomes classified based on their developmental stage and maternal age (Fig. 3). GV oocytes are shown on the left side of the diagram with the MII oocytes on the right side. A clear separation between the two developmental stages is illustrated. YMA and AMA GV oocyte transcriptomes lie close with each other sharing an extended overlap. This is also supported by the previous unclear separation of the GV oocytes shown in Fig. 2. A clear transcriptome separation is depicted in the YMA and AMA MII oocytes, indicating a higher number of differentially expressed transcript between the two groups. GV and MII oocytes showed better separation according to the first dimension (X axis), while alterations in both dimensions (dimensions 1 and 2) seem to play a role when ageing is considered (Fig. 3).
Figure 3.
MDS plot illustrating GV and MII oocytes of YMA and AMA categories. The samples are coloured according to the maternal age and the developmental stage of the oocytes. AMA, advanced maternal age; GV oocyte, germinal vesicle oocyte; MDS, multidimensional scaling; MII oocyte, metaphase II oocyte; YMA, young maternal age.
The effect of ageing and developmental stage on the oocyte gene expression
To reveal the genes involved in the ageing process that could play a role in the reduced developmental potential, we investigated the DGE levels of the oocyte cohorts, which were classified based on the maternal age (YMA/AMA) and oocyte developmental stage (GV/MII). The transcriptome relationships among the four oocyte cohorts were assessed using edgeR, along with the ontological enrichment per cohort. No enriched gene ontologies were revealed when YMA and AMA GV oocytes were compared, although individual DGE genes were involved among others in oocyte metabolism (QDPR) and ubiquitin-dependent degradation (FBXO32) (Supplementary Table SIII). These gene expression alterations could illustrate minor differences in the GV oocyte energy production and/or utilisation with ageing.
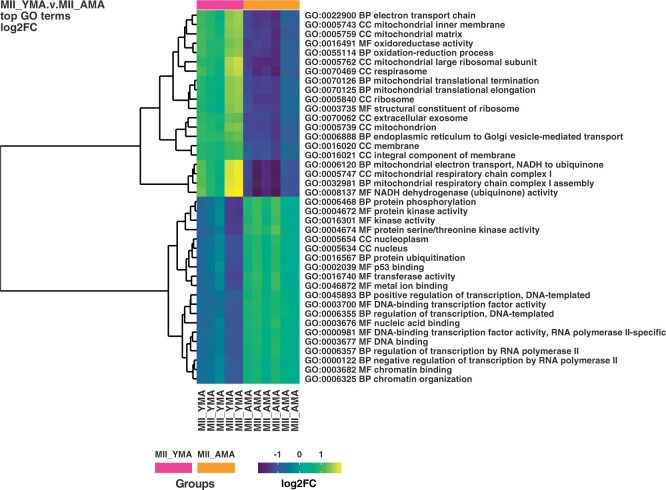
A much greater effect was revealed at the MII oocyte stage, with 1080 genes being significantly higher expressed in the YMA and 111 in the AMA oocytes (Supplementary Table SIV). Although no particular ontologies were significantly enriched in the AMA MII oocytes according to DAVID ontological analysis, the YMA MII oocytes were significantly enriched for mitochondria-related biological processes, including mitochondrial respiratory chain complex I assembly (FDR 4.0E−10) and mitochondrial translational elongation (FDR 3.3E−08). These results were confirmed by the cellular component analysis, with the top gene ontologies including mitochondrial inner membrane (FDR 1.3E−16) and mitochondrion (FDR 6.2E−14). Among other mitochondrial-related ontologies, the extracellular exosome ontology was significantly more highly enriched (FDR 9.6E−07). This ontology consisted of 194 genes or ∼19% of the total number of genes that were significantly more highly expressed in the YMA MII oocytes (Table II). These outcomes are also supported by the ontological analysis performed by goana R package to the YMA and AMA MII oocytes (Fig. 4). Mitochondrial, oxidoreductase activity and translational/post-translational ontological terms were reported, along with the chromatin organisation. The latter could reflect the transition of the nuclear architecture/chromatin structure with ageing, compromising perhaps the developmental potential of the AMA oocytes (Ge et al., 2015; Sun et al., 2018). This process may coincide with the meiotic transcription differences with ageing, due to epigenetic alterations (Navarro-Costa et al., 2016).
Table II.
Illustrates the enriched gene ontology terms of the genes significantly higher expressed in the YMA MII compared with the AMA MII oocytes.
| Gene ontology | Term | Count | % | P-value | FDR |
|---|---|---|---|---|---|
| BP | Mitochondrial respiratory chain complex I assembly | 22 | 2.2 | 2.3E−13 | 4.0E−10 |
| BP | Mitochondrial translational elongation | 23 | 2.3 | 1.9E−11 | 3.3E−8 |
| BP | Mitochondrial electron transport, NADH to ubiquinone | 18 | 1.8 | 2.1E−11 | 3.8E−8 |
| BP | Mitochondrial translational termination | 22 | 2.2 | 1.8E−10 | 3.1E−7 |
| BP | Translation | 28 | 2.8 | 3.6E−5 | 6.4E−2 |
| BP | ER to Golgi vesicle-mediated transport | 20 | 2.0 | 1.2E−4 | 2.2E−1 |
|
| |||||
| Gene ontology | Term | Count | % | P-value | FDR |
| CC | Mitochondrial inner membrane | 70 | 6.9 | 8.7E−20 | 1.3E−16 |
| CC | Mitochondrion | 130 | 12.8 | 4.2E−17 | 6.2E−14 |
| CC | Mitochondrial large ribosomal subunit | 19 | 1.9 | 1.1E−12 | 1.6E−9 |
| CC | Mitochondrial respiratory chain complex I | 18 | 1.8 | 2.1E−11 | 3.0E−8 |
| CC | Extracellular exosome | 194 | 19.1 | 6.6E−10 | 9.6E−7 |
| CC | Mitochondrial matrix | 38 | 3.7 | 3.1E−7 | 4.5E−4 |
| CC | Cytosol | 200 | 19.7 | 9.1E−6 | 1.3E−2 |
| CC | Mitochondrial intermembrane space | 14 | 1.4 | 2.5E−5 | 3.7E−2 |
The number of genes, along with the corresponding P-value and FDR are also reported. The fold-change of the differentially expressed genes has been set at 2.5.
AMA, advanced maternal age; BP, biological process; CC, cellular component; FDR, false discovery rate; MII oocyte; YMA, young maternal age.
Figure 4.
Heatmap illustrating MII YMA and AMA oocyte classification. Colour gradient ranges from dark blue to yellow. Horizontal bars (on the top of heatmap) represent the maternal age class of the oocytes MII YMA (purple) and MII AMA (orange). Ontological cluster identification is facilitated by the dendrogram (left). The ontological term and the type of the ontology terms are illustrated on the right. AMA, advanced maternal age; BP, biological process; CC, cellular component; FC, fold change; GO, gene ontology; MF, molecular function; MII oocyte, metaphase II oocyte; YMA, young maternal age.
When the fold change was set at 2.5, the comparison between different oocyte developmental stages revealed 4681 genes up-regulated in GV YMA and 857 in the MII YMA oocytes. In the AMA oocytes, 6159 genes were up-regulated in the GV and 1368 in the MII cohort. DGE analysis of the GV oocyte transcriptomes with regards to the maternal age, revealed only two up-regulated transcripts, SFXN5 and LINC01755, in the YMA and eight gene transcripts higher in the AMA oocytes, including among others CDA, SLC35A1, NPL, FBXO32 and FGFR2. Unlike the GV oocytes, the MII revealed more differentially expressed genes, with 1080 being up-regulated in the YMA and 111 in the AMA cohort, illustrating a potentially different effect of ageing in the GV to MII oocytes (Table III).
Table III.
Illustrates the differentially expressed genes between GV and MII oocytes upon ageing.
| VS | GV YMA (down) | MII YMA (down) | GV AMA (down) | MII AMA (down) |
|---|---|---|---|---|
| GV YMA (up) | – | 4681 | 2 | – |
| MII YMA (up) | 857 | – | – | 1080 |
| GV AMA (up) | 8 | – | – | 6159 |
| MII AMA (up) | – | 111 | 1358 | – |
The number of up-regulated genes (up) in the different oocyte stages are shown in the first column and the corresponding down-regulated (down) cohorts are shown in the first row (down).
AMA, advanced maternal age; GV, germinal vesicle; MII oocyte; YMA, young maternal age.
Focusing on the developmental stages, a higher total number of genes is expressed in the GV compared with the MII oocytes, irrespectively of the maternal age (Supplementary Table SV). The YMA GV oocytes generally include ontologies enriched for mitochondria and mitochondrial translation (FDR 9.6E−6 to 5.1E−3), proteasome-mediated ubiquitin-dependent degradation (FDR 3.6E−3) and mRNA splicing (FDR 3.7E−3). Among the higher enriched cellular components in the YMA GV oocytes, were mitochondrion and mitochondrial inner membrane ontologies with FDR 2.8E−16 and 5.7E−15, respectively, suggesting that the human GV oocytes express more mitochondrial-related transcripts than the MII oocytes. The YMA MII oocytes showed more highly expressed genes involved in biological processes associated with the preparation of the oocyte for fertilisation and early embryo development. The main biological processes include among others cell and nuclear division (FDR 2.1E−10 to 8.5E−5), cell–cell adhesion (FDR 3.9E−7), RNA metabolism, splicing and post-transcription modifications (FDR 8.4E−9 to 9.3E−3). The majority of the significantly higher expressed MII genes are localised in the nucleoplasm of the MII oocytes with high confidence (FDR 3.1E−56).
When the AMA oocyte gene transcripts are considered, the biological processes enriched for up-regulated AMA GV transcripts include mitochondrial-related ontologies (FDR 8.6E−15 to 1.2E−2), along with alternative splicing (FDR 2.4E−5), proteasome-dependent degradation (FDR 2.1E−4 to 1.5E−2) and vesicle-mediated transport (FDR 3.7E−3 to 3.6E−2). These AMA GV transcripts are mainly localised in mitochondria and the inner mitochondrial membrane with high confidence, reporting FDR of 4.5E−35 and 1.1E−32, respectively (Supplementary Table SVI). The transcripts up-regulated in the AMA MII oocytes were involved in cell division and cell cycle-related ontologies resulting in FDR ranging between 8.6E−20 and 2.1E−2, respectively. Cell–cell adhesion is another significantly enriched ontology in the AMA MII oocytes, with other significant ontologies including DNA repair (FDR 6.9E−12), post-transcription regulation ontologies such as alternative splicing, mRNA export from nucleus, mRNA 3′-end processing (FDR 3.8E−8–1.5E−3), post-translational regulation, including ubiquitin-dependent degradation (FDR 5.1E−8–2.4E−3).
DGE analysis of the RNA-Seq data provided potential molecular biomarkers for subsequent validation by qRT-PCR. RNF123 is an E3 ubiquitin ligase that reported six times higher levels of expression in the MII AMA compared with the MII YMA oocytes (Supplementary Fig. S1). Extracellular exosome-related genes could be good quality markers of MII oocytes with higher likelihood of successful fertilisation. For example, the extracellular-exosome-related gene transcripts CD74 and CD9 were approximately between 4 and 15 times more highly expressed in the MII YMA compared with the MII AMA oocytes (Supplementary Fig. S1), while in some of the MII AMA oocytes the expression levels were extremely low. RAB43 (or RAB11B) belongs to the RAB family and was approximately five times more highly expressed in the MII YMA compared with the MII AMA oocytes. RAB43 is another member of the RAB family that was expressed approximately 4 times higher in the MII YMA compared with the GV YMA oocytes, indicating the importance of this family for the MII YMA group. Finally, the chaperone HSPA6 and the chaperone-like RSAD1, showed a gene expression spectrum from extremely low to almost no expression in the MII YMA oocytes, with significantly higher expression in the GV YMA oocytes. Both the RNA-Seq and qRT-PCR methods showed a good correspondence in the expression levels of the investigated transcripts (Supplementary Fig. S1).
Ageing-related transcriptome alterations during the GV to MII oocyte transition
To investigate the effect of ageing on the GV to MII oocyte transition, we compared the differences of the differentially expressed genes between YMA and AMA cohorts. The majority of these genes (4665) were not affected by ageing (Supplementary Fig. S2). However, a higher number of genes were significantly differentially expressed in the AMA GV/MII transition (2862 genes) compared with the YMA GV/MII transition (873 genes). Considering that AMA MII oocytes correspond to decreased fertilisation rate and lower embryo development success, it is possible that the lower number of YMA transcripts upon the transition from GV to MII oocytes, illustrate a more controlled progression through oogenesis and/or meiotic resumption, unlike the AMA MII oocytes that potentially show a more dysregulated form of expression. Considering that all the GV oocytes showed quite similar gene expression patterns regardless of age, the DGE is more indicative of age effects on the GV/MII transition than on oogenesis per se.
The transcripts that were up-regulated exclusively in the MII oocytes upon the YMA GV/MII transition were not involved in any significant gene ontology, although trends for certain ontologies were noted (Supplementary Table SVII). To better understand the importance of these transcriptome patterns, the genes that were expressed significantly more highly, exclusively in the AMA GV/MII transition, were split based on the oocyte developmental stage. The MII oocyte transcripts with exclusive high gene expression in the AMA GV/MII transition showed a significant enrichment for nucleus and nucleoplasm cellular components. The AMA genes more highly expressed exclusively in GV oocytes, illustrated a high enrichment in several mitochondrial-related biological processes, including mitochondrial electron transport NADH to ubiquinone (FDR 5.1E−5), mitochondrial respiratory chain complex I assembly (FDR 4.4E−4), along with cellular components involved in mitochondria and mitochondrial inner membrane with FDR of 3.9E−9 and 4.4E−9, accordingly (Supplementary Table SVII). As a consequence, these ontologies were significantly down-regulated in the AMA MII oocytes, suggesting that they could play a key role in the decreased oocyte capacity for successful fertilisation and early developmental potential with advancing age.
Molecular biomarkers delineating the effect of ageing on the quality of the MII oocytes
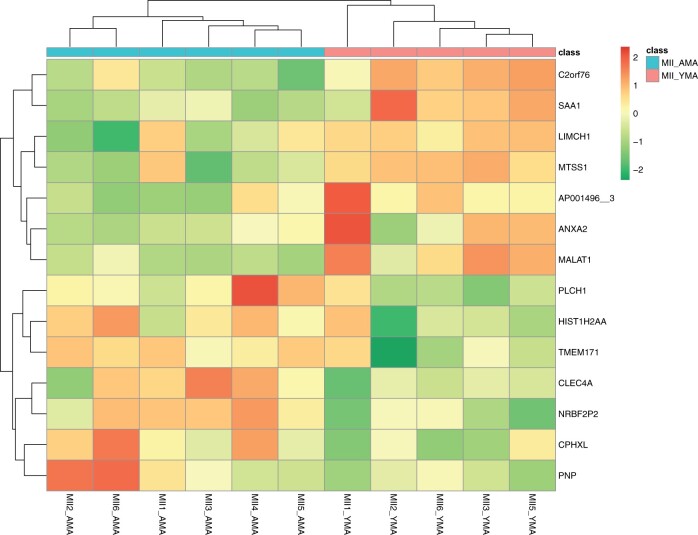
DaMiRseq package in R (Chiesa et al., 2018) was also employed to refine and highlight informative molecular markers that would allow us to increase our confidence in predicting oocyte quality. Read normalisation, filtering and identification of surrogate variables were used to minimise unwanted variation, with a heatmap being generated using Spearman’s correlation, which illustrate the gene expression relationships among cohorts (Fig. 2). This approach used the partial least squares to eliminate class-related features when investigating the transcriptome from the YMA and AMA MII oocytes and resulted in 175 highly important gene features using the FSelect function (Supplementary Table SVIII). To identify a smaller subset of representative gene features that could successfully represent the YMA and AMA MII oocyte groups, DaMirSeq FReduct function was employed. This further reduced the selection of representative biomarkers for each MII oocyte group by excluding the higher correlated genes. Only 14 features were selected using this approach (Fig. 5). These gene features could further support our attempt to classify random oocytes according to their quality, as this was guided by the gene expression patterns of the YMA and AMA oocytes. This methodological approach showed an overlap of 141 (∼81%) gene features with the edgeR DGE analysis, when the FSelect function was employed. The YMA oocytes express genes that could separate them from the AMA oocytes, including the actin-related LIMCH1 and MTSS1 that may be involved in cell migration, along with the TIMP1 metalloprotease that among other functions it is involved in regulating cellular apoptosis, which may contribute to the higher survival rates of YMA oocytes. Histone HIST1H2AA (or H2AC1) could be accumulated in the AMA oocytes, being potentially involved in chromatin organisation (Stefanelli et al., 2018).
Figure 5.
DaMiRseq analysis using the 14 features revealed by the FReduct function. Heatmap with colours highlighting MII YMA and AMA oocytes classification (class). Colour gradient ranges from dark green to red. Horizontal bars (on the top of heatmap) represent the maternal age group. Cluster identification is facilitated by the two dendrograms. AMA, advanced maternal age; MII oocyte, metaphase II oocyte; YMA, young maternal age.
Discussion
The current study employed deep RNA sequencing on YMA and AMA oocytes to reveal the effect of ageing upon the final phase of euploid oocyte maturation in vivo, with a focus on the MII oocytes. This could be invaluable for women undergoing ART, suggesting potential molecular targets that could be used to identify good quality oocytes of greater embryonic development potential. Further to the in vitro matured MII oocytes that participated in the study b, we employed GV and in vivo matured MII oocytes and only analysed oocytes of euploid status, to reflect more accurately the naturally occurring conditions during the late phase of meiotic progression with ageing. The current study reflects results of a study powered at 0.7, given a 2.5-fold change.
While the expression levels of thousands of gene transcripts remained stable upon GV to MII transition, hundreds of transcripts were significantly differentially expressed, highlighting molecular signatures characteristic for each cell type. The MII oocytes are transcriptionally silent and protein synthesis occurs on the existing transcribed genes in the MII oocyte reserve, whereas GV oocytes are transcriptionally active and, if necessary, they could replace important degrading/recycled gene transcripts (Tesařík et al., 1983; Bouniol-Baly et al., 1999). This could partially explain the fact that gene expression patterns between YMA and AMA GV oocytes are quite similar (10 DGE genes), while MII oocytes showed hundreds of differentially expressed genes between YMA and AMA groups (1191 gene transcripts).
The in vivo matured oocytes illustrated lower statistical dispersion compared with the in vitro matured MII oocytes studied by. This may indicate potential differences between the molecular processes involved in in vivo and in vitro oocyte differentiation. For example, the last phase of differentiation may involve a smoother and more targeted homogeneous GV to MII transition when oocyte maturation occurs in the female reproductive system. Taking into account the whole transcriptome, ageing has probably a greater effect in in vivo and in vitro matured MII compared with the GV oocytes, potentially compromising successful MII oocyte and embryo development of AMA women. Finally, inclusion of samples without chromosomal abnormalities, may reduce gene expression variability and increase perhaps the statistical power. Taken together, our findings suggest that the late phase of the GV to MII oocyte transition is likely a sensitive process, highly affected by the maternal age, potentially due to increased cellular stress and accumulated mutations induced as maternal age increases.
During GV to MII oocyte transition, 5538 genes were differentially expressed in the YMA and 7527 in the AMA, respectively, illustrating a 36% increase in the number of differentially expressed genes of the AMA oocytes. Age-induced transcriptome alterations may derive from the dysregulation of gene expression and mRNA instability (Borbolis and Syntichaki, 2015), and could be associated with decreased fertilisation and embryo development rates (Tan et al., 2014; Cimadomo et al., 2018). Furthermore, our study detected ∼8.5% of differentially expressed gene transcripts (2.5-fold change) upon ageing, which is ∼5% in mice (Hamatani et al., 2004). These mouse genes are involved in mitochondrial activity and oxidative stress responses, genome stability, chromatin structure and DNA methylation (Hamatani et al., 2004), with similar molecular processes reported in human (Steuerwald et al., 2007).
The polyadenylated transcripts of this study reported similar abundance in the GV oocytes irrespectively of maternal age. The AMA GV to MII transition of the euploid human oocytes, showed a greater reduction in mitochondrial-related gene transcripts, compared with the YMA oocytes. A similar observation was made in bovine oocytes upon GV to MII transition with ageing, including mitochondrial-related transcripts, involved in energy-related pathways and mitochondrial organisation ontologies (Reyes et al., 2015). This could reflect a lower need for mitochondrial protein synthesis at the MII oocyte stage as the maximum number of mitochondria is reached following the burst of mitochondrial replication associated with oocyte meiotic maturation, with the mitochondrial levels remaining stable during preimplantation embryo development (Cotterill et al., 2013; Tsai and John, 2016). Oxidative phosphorylation and mitochondrial dysfunction were among the processes enriched in the down-regulated transcripts upon GV to in vitro matured MII transition, while in the current study the reduction of mitochondrial-related ontologies was strongly associated with successful GV to MII transition.
The ability of an oocyte to complete meiotic maturation, fertilisation and support early embryo development depends on the mitochondrial number, function and energy production (Reynier et al., 2001; Cummins, 2004; Santos et al., 2006; May-Panloup et al., 2016). According to our study, despite the significant reduction of mitochondrial-related gene expression levels reported in the GV to MII oocyte transition, the YMA MII oocytes maintained significantly higher levels compared with the AMA MII oocytes, potentially affecting mitochondrial activity, through a gradual alteration of the mitochondrial membrane potential associated with ageing (Wilding et al., 2001). The maximum respiratory capacity of the aged oocytes is significantly reduced (Sugimura et al., 2012). Reduced ATP production leads to decreased metabolic activity and could affect fertilisation, embryo development and implantation (Dumollard et al., 2007, 2007; Van Blerkom, 2011; Eichenlaub-Ritter, 2012).
The YMA and AMA GV oocytes reported similar gene expression levels and together with the MII oocytes provided further insights into the oocyte quality. Several gene transcripts were reported in the MII oocyte YMA and AMA DGE analysis, with the majority being significantly up-regulated in the YMA (1080) compared with only 111 genes in the AMA oocytes. Our outcomes indicate dysregulation of gene expression upon ageing, including, for example, peroxiredoxins and cytochrome c oxidases. The Peroxiredoxin-1, PRDX1, has been significantly higher expressed (>2.5 fold-change) in the YMA group. Down-regulation of PRDX1 with ageing, indicates reduced oocyte protection against oxidative stress (Kang et al., 1998). Members of the peroxiredoxin gene family, including PRDX2, PRDX4 and PRDX6, were also higher expressed in the YMA oocytes, indicating AMA oocyte vulnerability to oxidative damage, that could affect the oocyte quality and fertility potential (Mihalas et al., 2017; Sasaki et al., 2019). KEGG pathway analysis revealed 31 genes significantly more highly expressed in the YMA oocytes involved in oxidative phosphorylation (FDR 1.2E−9), including cytochrome c oxidases (COX gene family), such as COX5A, COX7B, COX11, COX17, COX8A, COX8C and COX14, and indicating the importance of this family in energy production and potential effects in MII oocyte physiology and developmental potential (Lord and Aitken, 2013).
Our cellular component analysis revealed 194 genes of the extracellular exosome ontology (FDR 7.0E−8), significantly more highly expressed in the YMA oocytes, including CD44, CD74, CD320 and CD9 that were significantly more highly expressed in the YMA oocytes. CD9 is a tetraspanin protein normally used as exosomal marker and CD9-deficient mouse oocytes do not properly fuse with sperm during fertilisation (Le Naour et al., 2000), suggesting that CD9 gene expression (which was five times lower in the AMA oocytes), could be a critical factor associated with the low fertilisation rate in AMA patients. Similarly, RAB GTPases, including RAB1A, RAB1B, RAB43 (or RAB11B) and RAB11A, act as coordinators of vesicular trafficking (Stenmark, 2009), with RAB11 being one of the few important genes that promote exosome biogenesis and secretion (Blanc and Vidal, 2018). It has been shown that exosomes are transferred from the oocyte prior to sperm-oocyte interaction/fertilisation, potentially inducing the acrosome reaction (Barraud-Lange et al., 2007; Siciliano et al., 2008; Barraud-Lange et al., 2012). Our study suggests that the reduced fertilisation rate observed when conventional insemination is used over ICSI in AMA women (Farhi et al., 2019), could be partially explained by the significantly lower exosome-related transcripts reported in AMA oocytes.
Our pipeline also considered the DaMiRseq methodological approach that could identify molecular targets characterising oocyte quality, potentially in the context of tag-probe labelling for live imaging of the oocyte plasma membrane or exosomal proteins (Yano and Matsuzaki, 2009). For example, the AMA membrane-associated protein ABCA1 that could be involved in ABCA1-mediated cholesterol secretion pathway (Marcil et al., 1999), and/or the YMA membrane-associated KCNB1 and SLC9B2; may indicate easier targets for a diagnostics approach. KCNB1 is a K+ channel that could regulate exocytosis (Albrecht et al., 1993), while SLC9B2 is Na+/H+ antiporter involved in clathrin-mediated endocytosis (Sneitz et al., 2009).
This is the first study that considered the impact of ageing on the transcriptome changes of euploid GV to in vivo matured MII oocytes, revealing various ontologies indicative of the oocyte physiology. Considering that YMA MII oocytes are associated with the highest developmental potential, we focused on the YMA GV to MII oocyte transition, and most importantly the differences between YMA and AMA MII oocytes, revealing factors that could affect fertilisation and early embryo development (Ntostis et al., 2021), supporting also the idea that ICSI is advantageous in AMA women. Using additional methodological approaches, we attempted to confirm our findings and potentially revealed molecular biomarkers that could be used to characterise oocyte quality.
Data availability
RNA sequencing data sets are available at the NCBI Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/) and can be accessed with GSE164371.
Supplementary Material
Acknowledgements
We would like to thank the clinical embryologists, nurses, medical doctors and other staff of Genesis Athens clinic for their support.
Authors’ roles
K.P. and A.P. provided the clinical samples. P.N., G.K., T.V. and K.P. characterised the clinical samples used in the study. G.K. and T.V. conducted the oocyte denuding. P.N. performed the molecular work, optimised and prepared the library construction and RNA sequencing. P.N. and D.I. contributed to the computational analysis of the RNA sequencing data and the interpretation of the results. The manuscript was written collaboratively by P.N., H.P., E.K., G.K., E.K., J.H., K.P., H.M.P. H.M.P. and P.N. directed the data analysis, the conception and design of the study, along with writing and editing of the manuscript. All authors critically reviewed and approved the final version of the manuscript.
Funding
This work was supported by the Medical Research Council (MRC Grant number MR/K020501/1) to H.M.P. and J.H.
Conflict of interest
The authors have no conflict of interest to declare.
References
- Albrecht B, Lorra C, Stocker M, Pongs O.. Cloning and characterization of a human delayed rectifier potassium channel gene. Recept Channels 1993;1:99–110. [PubMed] [Google Scholar]
- Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. http://www.bioinformatics.babraham.ac.uk/projects/fastqc (1 November 2020, date last accessed).
- Barraud-Lange V, Chalas Boissonnas C, Serres C, Auer J, Schmitt A, Lefèvre B, Wolf J-P, Ziyyat A.. Membrane transfer from oocyte to sperm occurs in two CD9-independent ways that do not supply the fertilising ability of Cd9-deleted oocytes. Reproduction 2012;144:53–66. [DOI] [PubMed] [Google Scholar]
- Barraud-Lange V, Naud-Barriant N, Bomsel M, Wolf JP, Ziyyat A.. Transfer of oocyte membrane fragments to fertilizing spermatozoa. FASEB J 2007;21:3446–3449. [DOI] [PubMed] [Google Scholar]
- Blanc L, Vidal M.. New insights into the function of Rab GTPases in the context of exosomal secretion. Small GTPases 2018;9:95–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borbolis F, Syntichaki P.. Cytoplasmic mRNA turnover and ageing. Mech Ageing Dev 2015;152:32–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouniol-Baly C, Hamraoui L, Guibert J, Beaujean N, Szöllösi MS, Debey P.. Differential transcriptional activity associated with chromatin configuration in fully grown mouse germinal vesicle oocytes. Biol Reprod 1999;60:580–587. [DOI] [PubMed] [Google Scholar]
- Chiesa M, Colombo GI, Piacentini L.. DaMiRseq—an R/Bioconductor package for data mining of RNA-Seq data: normalization, feature selection and classification. Bioinformatics 2018;34:1416–1418. [DOI] [PubMed] [Google Scholar]
- Cimadomo D, Fabozzi G, Vaiarelli A, Ubaldi N, Ubaldi FM, Rienzi L.. Impact of maternal age on oocyte and embryo competence. Front Endocrinol (Lausanne) 2018;9:327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotterill M, Harris SE, Collado Fernandez E, Lu J, Huntriss JD, Campbell BK, Picton HM.. The activity and copy number of mitochondrial DNA in ovine oocytes throughout oogenesis in vivo and during oocyte maturation in vitro. Mol Hum Reprod 2013;19:444–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins J. The role of mitochondria in the establishment of oocyte functional competence. Eur J Obstet Gynecol Reprod Biol 2004;115:S23–S29. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J.. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol 2007;77:21–49. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Ward Z, Carroll J, Duchen MR.. Regulation of redox metabolism in the mouse oocyte and embryo. Development 2007;134:455–465. [DOI] [PubMed] [Google Scholar]
- Duncan FE, Hornick JE, Lampson MA, Schultz RM, Shea LD, Woodruff TK.. Chromosome cohesion decreases in human eggs with advanced maternal age. Aging Cell 2012;11:1121–1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U. Oocyte ageing and its cellular basis. Int J Dev Biol 2012;56:841–852. [DOI] [PubMed] [Google Scholar]
- Farhi J, Cohen K, Mizrachi Y, Weissman A, Raziel A, Orvieto R.. Should ICSI be implemented during IVF to all advanced-age patients with non-male factor subfertility? Reprod Biol Endocrinol 2019;17:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Z-J, Schatten H, Zhang C-L, Sun Q-Y.. Oocyte ageing and epigenetics. Reproduction 2015;149:R103–R114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths JA, Scialdone A, Marioni JC.. Mosaic autosomal aneuploidies are detectable from single-cell RNAseq data. BMC Genomics 2017;18:904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruhn JR, Zielinska AP, Shukla V, Blanshard R, Capalbo A, Cimadomo D, Nikiforov D, Chan AC-H, Newnham LJ, Vogel I. et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science 2019;365:1466–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatani T, Falco G, Carter MG, Akutsu H, Stagg CA, Sharov AA, Dudekula DB, VanBuren V, Ko MS.. Age-associated alteration of gene expression patterns in mouse oocytes. Hum Mol Genet 2004;13:2263–2278. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA.. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 2009;37:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SW, Chae HZ, Seo MS, Kim K, Baines IC, Rhee SG.. Mammalian peroxiredoxin isoforms can reduce hydrogen peroxide generated in response to growth factors and tumor necrosis factor-α. J Biol Chem 1998;273:6297–6302. [DOI] [PubMed] [Google Scholar]
- Khan DR, Fournier É, Dufort I, Richard FJ, Singh J, Sirard M-A.. Meta-analysis of gene expression profiles in granulosa cells during folliculogenesis. Reproduction 2016;151:R103–R110. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim J-S, Jeon Y-J, Kim D-W, Yang T-H, Soh Y, Lee HK, Choi N-J, Park S-B, Seo KS. et al. Identification of maturation and protein synthesis related proteins from porcine oocytes during in vitro maturation. Proteome Sci 2011;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolde R. pheatmap: Pretty Heatmaps. R package version 1.0.12. 2019. https://CRAN.R-project.org/package=pheatmap (1 November 2020, date last accessed).
- Krueger F. Trim Galore!: A wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. 2015. http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/.
- Le Naour F, Rubinstein E, Jasmin C, Prenant M, Boucheix C.. Severely reduced female fertility in CD9-deficient mice. Science 2000;287:319–321. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R; 1000 Genome Project Data Processing Subgroup. The sequence alignment/map format and SAMtools. Bioinformatics 2009;25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X-W, Ge Z-J, Wei L, Guo L, Han Z-M, Schatten H, Sun Q-Y.. The effects of postovulatory aging of mouse oocytes on methylation and expression of imprinted genes at mid-term gestation. Mol Hum Reprod 2011;17:562–567. [DOI] [PubMed] [Google Scholar]
- Liang X-W, Zhu J-Q, Miao Y-L, Liu J-H, Wei L, Lu S-S, Hou Y, Schatten H, Lu K-H, Sun Q-Y.. Loss of methylation imprint of Snrpn in postovulatory aging mouse oocyte. Biochem Biophys Res Commun 2008;371:16–21. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W.. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013;41:e108–e108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 2001;25:402–408. [DOI] [PubMed] [Google Scholar]
- Lord T, Aitken RJ.. Oxidative stress and ageing of the post-ovulatory oocyte. Reproduction 2013;146:R217–R227. [DOI] [PubMed] [Google Scholar]
- Marcil M, Brooks-Wilson A, Clee SM, Roomp K, Zhang L-H, Yu L, Collins JA, van Dam M, Molhuizen HO, Loubster O. et al. Mutations in the ABC 1 gene in familial HDL deficiency with defective cholesterol efflux. Lancet 1999;354:1341–1346. [DOI] [PubMed] [Google Scholar]
- May-Panloup P, Boucret L, Chao de la Barca J-M, Desquiret-Dumas V, Ferré-L'Hotellier V, Morinière C, Descamps P, Procaccio V, Reynier P.. Ovarian ageing: the role of mitochondria in oocytes and follicles. Hum Reprod Update 2016;22:725–743. [DOI] [PubMed] [Google Scholar]
- Mihalas BP, Redgrove KA, McLaughlin EA, Nixon B.. Molecular mechanisms responsible for increased vulnerability of the ageing oocyte to oxidative damage. Oxid Med Cell Longev 2017;2017:4015874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro-Costa P, McCarthy A, Prudêncio P, Greer C, Guilgur LG, Becker JD, Secombe J, Rangan P, Martinho RG.. Early programming of the oocyte epigenome temporally controls late prophase I transcription and chromatin remodelling. Nat Commun 2016;7:12331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navot D, Bergh R, Williams MA, Garrisi GJ, Guzman I, Sandler B, Grunfeld L.. Poor oocyte quality rather than implantation failure as a cause of age-related decline in female fertility. Lancet 1991;337:1375–1377. [DOI] [PubMed] [Google Scholar]
- Ntostis P, Swanson G, Kokkali G, Iles D, Huntriss J, Pantou A, Tzetis M, Pantos K, Picton HM, Krawetz SA.. The effects of aging on molecular modulators of human embryo implantation. iScience 2021:102751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL.. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc 2016;11:1650–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL.. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 2015;33:290–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2013. http://www.R-project.org/ (24 December 2013, date last accessed).
- Reyes JM, Chitwood JL, Ross PJ.. RNA‐Seq profiling of single bovine oocyte transcript abundance and its modulation by cytoplasmic polyadenylation. Mol Reprod Dev 2015;82:103–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JM, Silva E, Chitwood JL, Schoolcraft WB, Krisher RL, Ross PJ.. Differing molecular response of young and advanced maternal age human oocytes to IVM. Hum Reprod 2017;32:2199–2208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynier P, May-Panloup P, Chretien M, Morgan C, Jean M, Savagner F, Barriere P, Malthiery Y.. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod 2001;7:425–429. [DOI] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A.. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol 2010;11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos TA, El Shourbagy S, John JCS.. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril 2006;85:584–591. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hamatani T, Kamijo S, Iwai M, Kobanawa M, Ogawa S, Miyado K, Tanaka M.. Impact of oxidative stress on age-associated decline in oocyte developmental competence. Front Endocrinol (Lausanne) 2019;10:811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatten H, Sun Q-Y, Prather R.. The impact of mitochondrial function/dysfunction on IVF and new treatment possibilities for infertility. Reprod Biol Endocrinol 2014;12:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siciliano L, Marcianò V, Carpino A.. Prostasome-like vesicles stimulate acrosome reaction of pig spermatozoa. Reprod Biol Endocrinol 2008;6:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneitz N, Court MH, Zhang X, Laajanen K, Yee KK, Dalton P, Ding X, Finel M.. Human UDP-glucuronosyltransferase UGT2A2: cDNA construction, expression, and functional characterization in comparison with UGT2A1 and UGT2A3. Pharmacogenet Genomics 2009;19:923–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JL, Wessel GM.. How to make an egg: transcriptional regulation in oocytes. Differentiation 2005;73:1–17. [DOI] [PubMed] [Google Scholar]
- Stefanelli G, Azam AB, Walters BJ, Brimble MA, Gettens CP, Bouchard-Cannon P, Cheng H-YM, Davidoff AM, Narkaj K, Day JJ. et al. Learning and age-related changes in genome-wide H2A. Z binding in the mouse hippocampus. Cell Rep 2018;22:1124–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol 2009;10:513–525. [DOI] [PubMed] [Google Scholar]
- Steuerwald NM, Bermúdez MG, Wells D, Munné S, Cohen J.. Maternal age-related differential global expression profiles observed in human oocytes. Reprod Biomed Online 2007;14:700–708. [DOI] [PubMed] [Google Scholar]
- Stitzel ML, Seydoux G.. Regulation of the oocyte-to-zygote transition. Science 2007;316:407–408. [DOI] [PubMed] [Google Scholar]
- Su Y-Q, Sugiura K, Woo Y, Wigglesworth K, Kamdar S, Affourtit J, Eppig JJ.. Selective degradation of transcripts during meiotic maturation of mouse oocytes. Dev Biol 2007;302:104–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura S, Matoba S, Hashiyada Y, Aikawa Y, Ohtake M, Matsuda H, Kobayashi S, Konishi K, Imai K.. Oxidative phosphorylation-linked respiration in individual bovine oocytes. J Reprod Dev 2012;58:636–641. [DOI] [PubMed] [Google Scholar]
- Sun L, Yu R, Dang W.. Chromatin architectural changes during cellular senescence and aging. Genes 2018;9:211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan TY, Lau MSK, Loh SF, Tan HH.. Female ageing and reproductive outcome in assisted reproduction cycles. Singapore Med J 2014;55:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarín JJ, Pérez-Albalá S, Pérez-Hoyos S, Cano A.. Postovulatory aging of oocytes decreases reproductive fitness and longevity of offspring. Biol Reprod 2002;66:495–499. [DOI] [PubMed] [Google Scholar]
- Tesařík J, Trávník P, Kopečný V, Kristek F.. Nucleolar transformations in the human oocyte after completion of growth. Gamete Res 1983;8:267–277. [Google Scholar]
- Tsai T, John JCS.. The role of mitochondrial DNA copy number, variants, and haplotypes in farm animal developmental outcome. Domest Anim Endocrinol 2016;56:S133–S146. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondrial function in the human oocyte and embryo and their role in developmental competence. Mitochondrion 2011;11:797–813. [DOI] [PubMed] [Google Scholar]
- Webster A, Schuh M.. Mechanisms of aneuploidy in human eggs. Trends Cell Biol 2017;27:55–68. [DOI] [PubMed] [Google Scholar]
- Wickham H. ggplot2: elegant Graphics for Data Analysis. New York: Springer, 2016. [Google Scholar]
- Wilding M, Dale B, Marino M, di Matteo L, Alviggi C, Pisaturo ML, Lombardi L, De Placido G.. Mitochondrial aggregation patterns and activity in human oocytes and preimplantation embryos. Hum Reprod 2001;16:909–917. [DOI] [PubMed] [Google Scholar]
- Yano Y, Matsuzaki K.. Tag-probe labeling methods for live-cell imaging of membrane proteins. Biochim Biophys Acta 2009;1788:2124–2131. [DOI] [PubMed] [Google Scholar]
- Young MD, Wakefield MJ, Smyth GK, Oshlack A.. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol 2010;11:R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA sequencing data sets are available at the NCBI Gene Expression Omnibus repository (https://www.ncbi.nlm.nih.gov/geo/) and can be accessed with GSE164371.