Graphical abstract
Abbreviations: NCBI, National Center for Biotechnology Information; OTU, Operational Taxonomic Unit
Keywords: Human gut microbiota, Core microbiota, Enrichment strategies, Metagenomics, Genome databases, Western bias
Abstract
The human gut holds a special place in the study of different microbial environments due to growing evidence that the gut microbiota is related to host health. However, despite extensive research, there is still a lack of knowledge about the core taxa forming the gut microbiota and, moreover, available information is biased towards western microbiomes in both genome databases and most core taxa studies. To tackle these limitations, we tested a database enrichment strategy and analyzed public datasets of whole-genome shotgun data, generated from 545 fecal samples, comprising three gradients of westernization. The NT database was selected as a baseline of biological diversity, subsequently being combined with various studies of interest related to the human microbiota. This enrichment strategy made it possible to improve classification capacity, compared to the original unenriched database, regarding the various lifestyles and populations studied. The effects of incomplete-taxonomy metagenome-assembled genomes on genome database enrichment were also examined, revealing that, while they are helpful, they should be used with caution depending on the taxonomic level of interest. Moreover, in terms of high prevalence, the core analysis revealed a conserved set of bacterial taxa in the healthy human gut microbiota worldwide, despite apparent lifestyle differences. Such taxa show a set of traits, metabolic roles, and ancestral status, making them suitable candidates for a hypothetical phylogenetic core of mutualistic microorganisms co-evolving with the human species.
1. Introduction
The human gut microbiota could be defined as the microbial community that inhabits the human digestive tract. This microbiota represents the overwhelming majority of microorganisms inhabiting the human host, since the bacterial content of the colon surpasses all other body locations by at least two orders of magnitude [1]. Consideration of the microbiota has gone from being virtually ignored by the scientific community a few decades ago to currently monopolizing many scientific studies. This growing interest in microbiota research has a dual motivation. On the one hand, advances in sequencing techniques and bioinformatics have made the field of metagenomics a robust corpus for scientific research [2], [3], [4]. On the other hand, there is growing evidence for the relevance of the microbiota in human health. For instance, a recent study has recollected more than a hundred diseases and disorders associated with changes in gut microbiota composition [5].
One research avenue involves studies attempting to identify a core of taxa shared by most human individuals, focusing on the component of the microbiome found across a considerable proportion of hosts within specific populations and, thus, defining a “Common Core” [6]. This research trend dates back to the Human Microbiome Project (HMP) in 2007 [7]. Since then, many studies have attempted to describe and compare different human-associated taxonomic cores [8], [9], [10], [11], [12]. However, most studies have been conducted with 16S rRNA data gathered using different technologies, and usually at a single taxonomic level, offering less resolution since the 16S rRNA gene is too conserved to obtain species-level identification [13], [14]. An alternative that provides better taxonomic resolution and more accurate abundances is the use of whole-genome shotgun (WGS) data [15], [16]. The few recent examples mentioning a core within the healthy human gut using WGS data [17], [18] do not develop this concept in-depth and present a particular bias towards western populations.
Furthermore, a key factor in any metagenomic study is the use of reference genome/sequence databases. For shotgun metagenomics, the most popular references are NCBI NT and RefSeq databases. Likewise, it is common to use sequences from the GenBank database [4], [19]. However, it is worth pointing out the under-representation of non-western microbiomes for conventional reference databases, which may yield results that cannot be generalized. The microbial diversity of urban western individuals has been the objective of a significant number of studies, and therefore it is better represented in genome databases of known reference organisms [20], [21], [22]. In this respect, applying enrichment strategies using relevant microbiome studies could help to overcome this flaw in conventional databases, approximating read classification levels between different populations.
It is not clear whether a core of microorganisms has evolved with the human species, forming what we might call the phylogenetic component of the human microbiota. The phylogenetic thesis argues that some bacterial taxa have co-evolved with the human species since the beginning [23]. Conversely, the ecological thesis argues that the gut microorganisms of human populations may be highly convergent so that human populations in similar environments would show similar bacterial taxa [24]. Indeed, these two theses are not mutually exclusive but rather complementary. If the phylogenetic thesis were true, we would expect to find highly conserved taxa of an ancestral nature, which would play essential roles in the human microbiota and characteristics that favor the host’s influence over those taxa.
Here, we present a proof-of-concept study focused on addressing the concerns associated with western bias and determining a universal core in the healthy human gut microbiota, based on ubiquitous taxa that are genuinely conserved independently of ecological factors. For this purpose, we used WGS data and an enrichment strategy aiming to account for the under-classification of non-western microbiomes.
2. Materials and methods
2.1. Database enrichment
In brief, for this enrichment strategy, a database widely used in metagenomics was chosen as a baseline of the known biological diversity, and then combined with various studies related to the human microbiota. The NCBI NT database [25] was used as this baseline and then enriched with relevant human microbiome studies, including the HMP [26], FDA-ARGOS [27], BIO-ML [28], CGR [29], HBC [30] and some metagenome-assembled genomes (MAGs) [21], [31] (Supplementary Table 1). All genomes and sequences were downloaded in April 2020.
In the case of studies with MAGs it is not unusual to find genomes with incomplete taxonomies, which may be resolved as species at the assembly level, but are not resolved at the taxonomic level. Therefore, we examined the effects of incomplete-taxonomy MAGs on genome database enrichment. To do so, we filtered these genomes, retaining those that reached a genus level or were Candidatus genomes, thus generating two alternative dataset versions for enrichment (MS and MS-fMAG Sets). In order to minimize redundancies between the NT database and the enrichment datasets we developed a non-redundant filter based on sequence accession. Most of these enrichment genomes belonged to the NCBI, and so a list of their associated sequence identifiers (from RefSeq and GenBank) was generated and used to filter the NT database. Finally, the resulting genomes and sequences were combined, generating two alternative enriched databases. A more detailed description of the entire download and construction process is given in the supplementary materials (Supplementary Document 1; Supplementary Fig. 1).
The improvement of the enriched databases over the original database was assessed in terms of classification capacity (as the percentage of classified sequences) at different taxonomic levels and factors of interest.
2.2. Metagenomic datasets
We collected 545 publicly available metagenomes from the European Nucleotide Archive (ENA), encompassing six different studies [17], [22], [32], [33], [34], [35] (Supplementary Table 2). Technical information was retrieved through the ENA Browser. All available metadata for each sample were retrieved from the ENA API with the mg-toolkit (version 0.6.4; https://pypi.org/project/mg-toolkit/) with further curated inspection of the original publications of each project.
We applied the following criteria for sample selection: (a) human gut samples (stool), (b) healthy individuals or population samples (assuming most individuals will be healthy), (c) only one run accession per sample accession, (d) shotgun metagenomic sequencing and (e) Illumina technology. In this case, different lifestyles were used as a proxy to reflect different levels of westernization: (a) rural populations (reflecting non-westernization), (b) a periurban industrializing shantytown (intermediate level of westernization), and (c) westernized urban populations.
2.3. Quality control
Shotgun reads were quality-trimmed and filtered using the BBDuk tool from BBTools suite (version 38.79) [36]. In the first step, the right end of the reads was quality trimmed to Q15. In a second step, the left ends were quality-trimmed to Q1. Subsequently, we removed the resulting reads containing any ambiguous base (N) or measuring under 60 base pairs (bp) in length after trimming. Quality was checked with FastQC (version 0.11.9) [37] and MultiQC (version 1.8) [38] to study the different datasets and ensure their quality (Supplementary Table 3).
2.4. Metagenomic classification
Kraken 2 software (version 2.0.8-beta) [39] was used to perform classification. The databases were constructed and indexed with default parameters, and classification was performed with a filtering confidence threshold of 0.05 to control the level of false positives.
The Bracken software performed a re-estimation of abundance counts (version 2.5) [40]. Using the bracken-build script, we computed a set of probabilities from the Kraken 2 databases, with a k-mer length of 35 bp (-k 35) and a read length of 60 bp (-l 60) since the minimum read length in our dataset is 60 bp. Subsequently, we used Bracken on the Kraken 2 report results, with a read length of 60 bp (-r 60) and a taxon count threshold of 10 reads (-t 10). Results were obtained at the genus and species levels. The Bracken results were processed with self-made scripts to obtain the corresponding count matrix and taxa table at the genus and species level. Finally, kingdoms Metazoa and Viridiplantae, as well as undefined taxa at the super-kingdom (domain) level were filtered out to perform the subsequent analysis.
2.5. Microbiome analysis
The analyses were carried out mainly with the R programming language (version 4.0.4), using the Microbiome (version 1.12.0) [41], Phyloseq (version 1.34.0) [42], and Vegan (version 2.5–6) [43]] packages.
2.5.1. Defining a universal core
The core microbiome analysis was performed at the genus and species level using the Microbiome package. To avoid the presence of possible spurious low-abundance signals we worked with a 1e − 4 relative abundance threshold (0.01% compositional abundance). When defining cores, the taxa failing to reach this abundance level were considered absent in the sample. Pan-Cores were obtained using a prevalence threshold of 0.9 (presence in at least 90% of samples) for all samples. Additional cores were calculated for each country (El Salvador, Madagascar, Peru, China, Japan, and United States) and lifestyle (rural, periurban shantytown, and urban) using the same parameters. The resulting cores were compared with the UpsetR package (version 1.4.0) [44]. The intersection between the additional cores was used to define universal cores of prevalent taxa. Their prevalence was examined in all samples for different abundance thresholds using the ComplexHeatmap package (version 2.6.2) [45]. Taxonomic relationships among taxa were examined using the Metacoder package (version 0.3.4) [46]. This intersection-based criterion was also used to investigate less prevalent core taxa, using less stringent prevalence thresholds of 0.7 and 0.5, defining medium and soft cores, respectively. Additionally, microbial relative abundances for the most abundant taxa were inspected.
2.5.2. Abundance clustering and pattern analysis
Abundance clusters and differential patterns between groups for these universal core taxa were examined at the genus and species level. These trends were analyzed using relative abundances based on each lifestyle-country combination. Abundance patterns were analyzed employing z-scores. Different groups and taxa were clustered using the k-means clustering algorithm as implemented on the ComplexHeatmap package unless stated otherwise. In these cases, the number of optimal clusters was determined by the Elbow method (Supplementary Figs. 2–3).
2.5.3. Ordination analysis
Inter-sample differences were investigated employing a Principal Component Analysis (PCA) with the Phyloseq package using genus level relative abundances. The scores of the associated taxa were inspected with the Vegan package to investigate the main genera drivers of the differences for the two first components. In addition, Bray-Curtis dissimilarities were also estimated and used with alternative ordination methods, including Principal Coordinates Analysis (PcoA) and Non-Metric Multidimensional Scaling (NMDS).
2.6. Data availability
The accession numbers of the samples and associated metadata used in this work can be consulted in Supplementary Table 4. The scripts used for this work can be found at GitHub (https://github.com/sarpiens/corescripts). The enriched databases will be available on request.
3. Results and discussion
3.1. Considerations on MAG enrichment and comparison of strategies
The Kraken-Bracken tandem was used to verify the improvement provided by the enrichment strategy, which offers good results at the genus and species level, as shown in recent publications [19], [40].
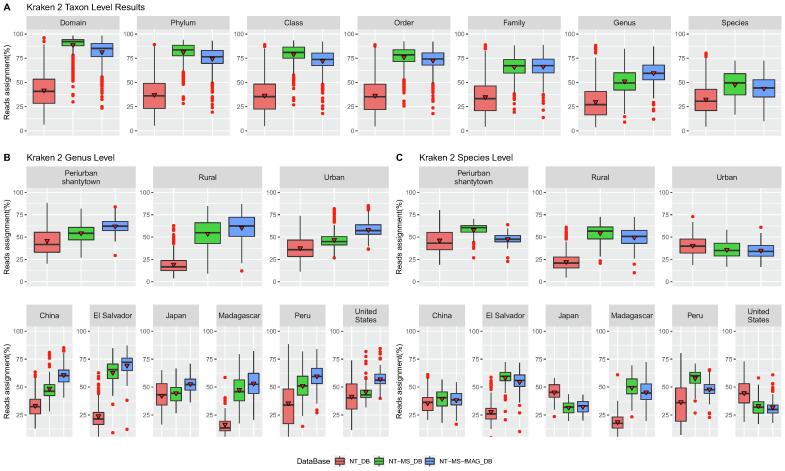
On examining the Kraken 2 results, a general improvement is observed for the enriched databases compared to its original counterpart, which almost doubles its classification capacity from domain to genus levels (Fig. 1A). As we go down the taxonomic tree, we can see how the number of classified reads decreases. The database enriched with the unfiltered-MAG microbiome studies, manages to classify slightly more sequences than its filtered counterpart for taxonomic levels between domain and order. However, this changes for levels between family and genus, where the roles are reversed. The average classification capacity at the genus level is 51.2% for the enriched unfiltered-MAG database and 59.8% for its filtered counterpart. On the contrary, the roles are reversed again at species level, and their values drop to 47.9% and 43.8%, respectively.
Fig. 1.
Kraken 2 results showing a comparison between the enriched and NT original databases. The performance of the databases was examined in terms of classification capacity for all samples as a whole at different taxonomic levels (A) and separating them by lifestyles and countries at genus (B) and species (C) level. In the box plots, the black line within the box marks the median and the red triangle the mean, outliers are presented as red dots. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
In MAG studies, it is not unusual to find genomes with incomplete taxonomies that are resolved as species at the assembly level (usually under the 95% average nucleotide identity criterion), but are not resolved as species at the taxonomic level. In this situation, artefactual species may appear, namely a species-level taxID that presents an incomplete taxonomy and could act as a catch-all taxon comprising assemblies of different species. For example, in the Almeida study, we found 22 assemblies resolved as species with the 95% average nucleotide identity criterion at the assembly level, but they ended up together at the taxonomic level under the taxID “uncultured Bacteroidales bacterium (194843)”. This bacterium has been successfully assigned to the order level to subsequently generate an artefactual species with gaps in its taxonomy. On the one hand, these species would increase the classification capacity at higher taxonomic levels, as far as their taxonomy may allow them. However, this would also imply a decrease at intermediate levels where taxonomic gaps are found. An increase in sequences classified at the species level would also be expected, but to the detriment of identifying more informative species. This would explain the differences observed between the enriched databases.
The emergence of these artefactual species appears to result from a behavioral change in the way MAGs are treated in the prokaryote curation process in the NCBI Taxonomy Database. The practice of assigning specific taxIDs at the species level for MAGs was discontinued in August 2017 because the number of such submissions was expected to rise to a level that would make it impractical to assign individual taxIDs [47]. We believe that this should be taken into account when working with databases that include MAGs, as they could introduce a certain bias and negatively affect the classification capacity depending on the study’s taxonomic level of interest.
Some discrepancies are observed between genus and species level when examining classification capacity based on the variables of interest (lifestyle and location). The general trend in improvement continues for all lifestyles and populations at the genus level, with results that exceed 50% of average classification for the filtered-MAG enriched database. However, the classification capacity decreases for some urban datasets compared to the original database at the species level (Fig. 1B-C). As the number of species in the database increases, so too does the difficulty in estimating species, which is due to large fractions of sequence similarity [40]. This leads to the assignment of many sequences at the genus level or above, but not at the species level since Kraken uses the Last Common Ancestor (LCA) strategy [48] in which lower taxonomic levels will be reached only if the assignment reaches sufficient confidence. This behavior would explain the observed discrepancies, considering the greater representation of urban-western genomes in the databases and the increase in species from the enrichment studies.
Based on these results, we decided to continue using the enriched filtered-MAG database to perform the Bracken re-estimation and all subsequent analyses, as it offers better results at our taxonomic level of interest (genus level). On examining the Bracken results, greater similarities are observed between the genus and species level. The average classification capacity of the NCBI NT database is 45.5% and 82.2% for the enriched filtered-MAG database at the genus level, whereas at the species level, these values are 45.9% and 83.9%, respectively. Classification levels are more similar across lifestyles and locations for the enriched database. In this case, we do not see discrepancies between the results at either the genus or species level. The trend in improvement continues for all lifestyles and populations, with results that exceed 70% average classification (Supplementary Fig. 4). As discussed above, Kraken2 uses the LCA strategy, which implies that in some cases a great number of sequences may be classified at a higher taxonomic level, but not at our level of interest. Bracken applies Bayesian inference to estimate species abundances in a metagenomic sample by probabilistically re-distributing reads on the taxonomic tree. Reads assigned to nodes above the level of interest are distributed down at this level, while reads assigned at lower levels are re-distributed upward to their parent nodes [40]. This would explain the large differences in classification capacity between both tools.
3.2. Defining a core criterion
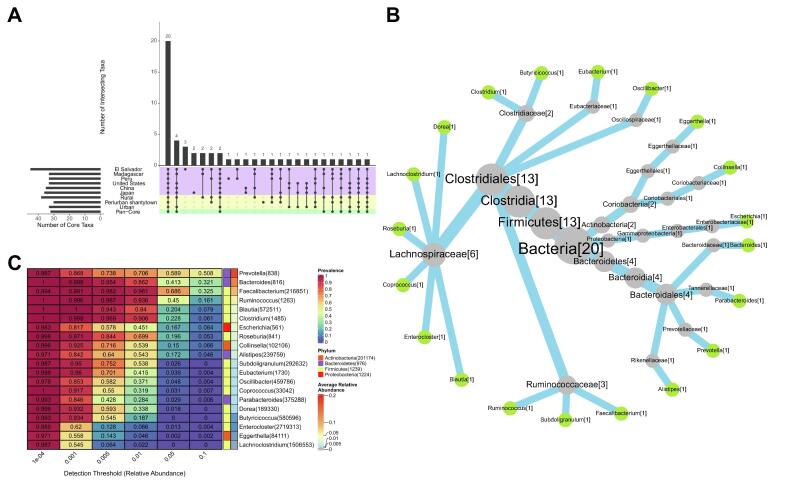
A Pan-Core of 32 genera was defined for all samples using the core analysis approach. Additional cores were generated for each lifestyle and geographic origin (Fig. 2-A; Supplementary Table 5). Among the Pan-Core genera, four members are missing in both the Peru and Periurban Cores (Lachnobacterium, Lachnospira, Solobacterium, and Streptococcus), two in the Peru Core (Anaerobutyricum and Agathobacter), and one more in the Madagascar Core (Bifidobacterium). Other single taxa are missing in multiple cores. It is the case of Anaerostipes (absent in Peru, Madagascar, and Periurban cores), Dialister (absent in Peru, United States, Japan, Periurban and Urban cores), Flavonifractor (absent in Madagascar and Rural cores), Gemmiger (in China, United States, Japan, and Urban cores) and Sutterella (absent in the United States, Japan, and Urban cores). All these missing taxa account for 37.5% of the Pan-Core genera.
Fig. 2.
Universal core genera description. (A) Intersections between cores of interest. (B) Taxonomic relationships between universal core genera. The number of core genera assigned to a particular level is indicated inside the square brackets. (C) Prevalence-Abundance Heatmap. Average relative abundances sort taxa and their NCBI’s taxID is indicated in parentheses.
The differences between the Pan-Core and the rest of the cores of interest reflect that when working with all samples, without accounting for their country of origin or lifestyle, particularities of the different groups are diluted. Consequently, taxa are found that are not truly universal among all the cores of interest. Moreover, sampling differences between groups could explain the presence of some of these taxa, since there may be cases in which a taxon is found in many samples, but not in a condition of interest. For these reasons, rather than defining a Pan-Core, we worked with the intersection of these additional cores in order to better discriminate core taxa that are truly universal, regardless of their country of origin and lifestyle.
3.3. A universal phylogenetic core independent of lifestyle and country of origin
According to this criterion, a total of 20 universal bacterial genera were detected (Fig. 2B). If we study their prevalence, only Bacteroides, Ruminococcus, Blautia, Clostridium, and Coprococcus are prevalent in all samples with a 1e-4 relative abundance threshold, without underrating the rest of the taxa that present values greater than 0.97 prevalence. At higher abundance thresholds, such as 0.01 relative abundance, no taxon was prevalent in all samples, but highly prevalent taxa were still found. Faecalibacterium, Ruminococcus, and Clostridium show the highest values with prevalences above 0.9. Also, Bacteroides and Blautia register high values with prevalences above 0.8, but Coprococcus does not. At even higher abundance thresholds, most taxa show prevalence values below 0.5, except for Prevotella (which is greater at 0.05 and 0.1 relative abundance thresholds) and Faecalibacterium (which is greater at 0.05 relative abundance threshold) (Fig. 2C). These taxa also account for some of the most abundant genera. In terms of average abundance, among these, Prevotella is the most abundant, followed by Bacteroides, Faecalibacterium, Ruminococcus, Blautia, and Clostridium (Supplementary Fig. 4). Thus, these taxa represent some of the most prevalent and abundant genera comprising the human gut microbiota.
On applying this intersection-based core criterion at the species level, a total of 22 universal bacterial species are found (Supplementary Fig. 6A; Supplementary Table 6). Among these, the genus Blautia has the highest number of representatives: uncultured Blautia sp., Blautia obeum, Blautia wexlerae, [Ruminococcus] gnavus and [Ruminococcus] torques. This core comprises uncultured species, but we also find cultured species such as Faecalibacterium prausnitzii, Coprococcus comes, Dorea formicigenerans, Dorea longicatena, Escherichia coli, and [Eubacterium] rectale (Supplementary Fig. 6B). If we study their prevalence at high abundance thresholds, such as 0.01 relative abundance, we find that Faecalibacterium species show the greatest values with prevalences above 0.8 (Supplementary Fig. 6C). These taxa also account for some of the most abundant species (Supplementary Figure 8) and are, thus, the most prevalent and abundant species of the human gut microbiota. The relative abundance of F. prausnitzii in vertebrate animals besides humans [49], [50] would imply it is a functionally important member of the microbiota, with a likely impact on host physiology and health. Indeed, it has been consistently reported as one of the main butyrate producers found in the intestine [51], [52] and butyrate plays a crucial role in gut physiology and host well-being. Furthermore, it is the primary energy source for colonocytes, and has protective properties [53], [54].
Comparing the differences between these two universal cores, we can see the absence of representatives of the core genera Alistipes, Parabacteroides, Eggerthella, Lachnoclostridium, Oscillibacter, and Enterocloster at the species level. In addition, some species are absent at the genus level, such as [Eubacterium] rectale, which has no genus entry in the NCBI Taxonomy database. These differences reinforce the importance of working at various taxonomic levels, as we may encounter species that are less prevalent on their own, but when treated as a whole at higher taxonomic levels, reveal patterns that would otherwise be missed.
All the taxa from these universal cores are bacterial microorganisms, with the majority of them being obligate anaerobes, non-spore-forming, and non-motile [52], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69] (Supplementary Table 7). These characteristics tell us about the environment in which these microorganisms live. Indeed, the intestinal tract is known to constitute an environment largely devoid of oxygen, favoring anaerobic microorganisms [70], [71], [72], whereas other traits such as sporulation and motility, could be related to the process of co-evolution and domestication of the microbiota by its host. Nevertheless, spore-forming and motility characteristics present more variability in members of the Firmicutes phylum. Recent studies show that many members of the gut microbiota can form spores, especially among Firmicutes, which is linked to host-to-host transmission and colonization promotion [30], [73]. Moreover, in the case of the Lachnospiraceae family, only those associated with the human digestive tract possess key sporulation genes [74], [55]. In this respect, we hypothesize that non-motility could be linked to a better spatial control of the host over their microbiota; however, this trait has been related to a greater susceptibility to intestinal expulsion and abundance fluctuations [75].
In accordance with the phylogenetic hypothesis, we could expect the existence of shared taxa with other hominids, if we consider that host phylogenetic relationships have been linked to microbial co-evolution with their primate hosts [23], [76]. Indeed, previous studies have found co-occurrence within the microbiomes of humans and other primates. Moeller et al. [77] studied the gut microbiome at the genus level of gorillas, chimpanzees, bonobos, and different human populations using 16S rRNA amplicon data, defining a “Core Ape Microbiome” that partially overlaps with our core. These genera include Prevotella, Bacteroides, Blautia, Ruminococcus, Clostridium, Roseburia, and Parabacteroides. However, they also found other unclassified taxa that could not be resolved at genus level, such unclassified Enterobacteraceae, unclassified Ruminococcaceae and unclassified Lachnospiraceae, which were better resolved using our core approach. More recent studies, such as Amato et al. [24], have placed special focus on the differences between phylogeny and ecology. They compared the microbiome composition of different human populations, based on 16S rRNA amplicon OTUs (as a proxy of species level), with those of the great apes (which have closer phylogenetic relationships with humans) and distantly related Cercopithecines (a subfamily of Old World monkeys that shares closer ecology traits with humans). Despite the ecological differences between these primate groups, there are some simultaneously shared species which are also present in our core, including Faecalibacterium prausnitzii, Blautia sp., Coprococcus sp., Roseburia sp. and unknown Ruminococcaceae. This highlights the ancestral and phylogenetic importance of these taxa, reinforcing the hypothesis that they have co-evolved with their host since the dawn of the human species, despite ecological differences.
3.4. Different phyla exhibit differential core taxa prevalence
This intersection criterion was also used to further investigate less prevalent core taxa using less stringent prevalence thresholds, defining soft and medium prevalence cores, which were compared with the universal cores, within a prevalence range between 0.5 and 1.
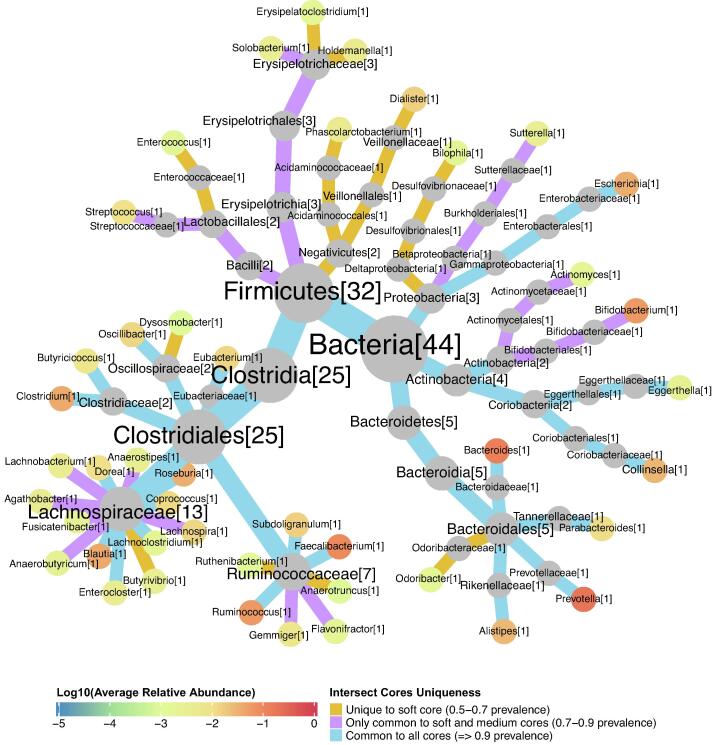
A total of 44 bacterial core genera were detected (Fig. 3; Supplementary Table 5), of which 13 genera are unique to the soft core (prevalence range 0.5–0.7), 11 genera are found only in the soft and medium cores (prevalence range 0.7–0.9) and 20 genera are shared by all three cores (universal core genera, with ≥ 0.9 prevalence). Firmicutes gains most of the new core taxa when relaxing the prevalence threshold (79.17% of the new core taxa), although most of them show lower levels of abundance, and are thus more diverse in terms of prevalence. Within this phylum, the order Clostridiales monopolizes their universal core genera. Proteobacteria gains two more core taxa, also with lower abundances, being equally distributed among the three prevalence ranges studied, with one genus per range. Actinobacteria also gains two new core taxa, being the only phylum to gain a core genus with higher abundance levels, namely the Bifidobacterium genus (which fails to reach the 0.9 threshold in the Madagascar population). This phylum is equally distributed between the intermediate 0.7–0.9 range and the universal core prevalence range (≥0.9 prevalence) with two genera per range. Whereas Bacteroidetes, not only presents members that are generally more prevalent (80% are universal core taxa, with ≥ 0.9 prevalence), but also tend to show higher levels of abundance.
Fig. 3.
Taxonomic relationships between different intersect core genera in a prevalence gradient. Additional intersect cores were computed defining soft and medium prevalence genera cores, which were compared to the corresponding universal genera core, working in a prevalence range between 0.5 and 1.
In applying this criterion at the species level, a total of 78 bacterial and one viral core species are found (Supplementary Fig. 7; Supplementary Table 6), of which 38 species are unique to the soft core, 19 species are found only in the soft and medium cores, while 22 species are shared by all three cores (universal core species). We found that the vast majority of the new core taxa are cultured species, whereas in the universal core, most genera were represented by uncultured ones. Furthermore, the genus Bacteroides is of interest, as we see that although it is one of the most prevalent genera, many of its underlying species are found only in the less prevalent cores. This could indicate that different species of this genus may tend to work together in unison. Indeed, authors have reported the existence of conserved associations between Bacteroides species in healthy human populations, which are designated as a ‘hub’ and ‘bottleneck’ species in association networks [18].
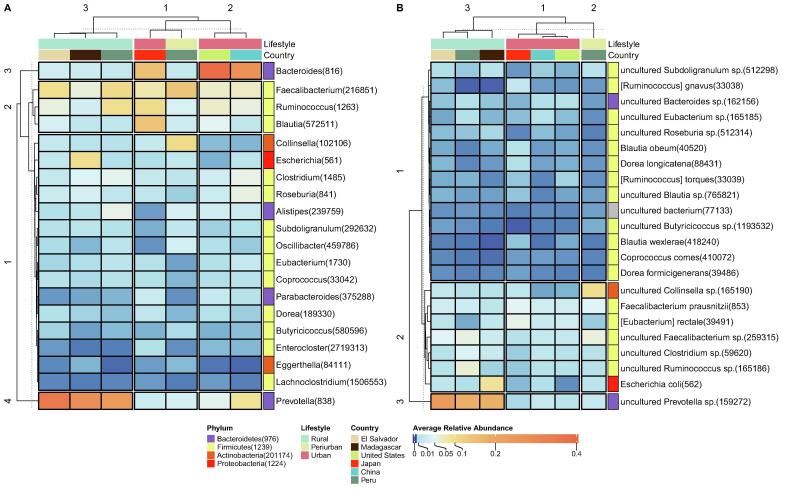
3.5. Abundant universal core taxa are related to crucial roles and ecological adaptation
Four abundance clusters are found when examining average relative abundances at the genus level for the universal core taxa (Fig. 4A). Two abundance clusters, Bacteroides and Prevotella, (clusters 3 and 4), result in dominant bacterial signatures for urban and rural groups, respectively. Bacteroides maintains a complex and generally beneficial relationship with the host when retained in the gut [78]. Carbohydrate fermentation by Bacteroides and other intestinal bacteria produces a pool of volatile fatty acids, which are reabsorbed through the large intestine and utilized by the host as an energy source, providing a significant proportion of the host's daily energy requirement [79]. However, the role of members of the Prevotella genus within the intestinal microbiota and their effects on the host is not fully understood. High prevalence and relative abundance of Prevotella is found in non-westerners who consume a plant-rich diet [80], [81]. In contrast, other studies have associated Prevotella spp. with autoimmune diseases, insulin resistance and diabetes, as well as gut inflammation [82], [83].
Fig. 4.
Abundance clustering analysis for universal core taxa. Average relative abundances for the different group combinations at genus (A) and species (B) level. Groups and taxa were clustered using the k-means algorithm. NCBI’s taxIDs are indicated in parentheses.
We also find another cluster of bacteria that are generally quite abundant in all studied groups (cluster 2), consisting of Faecalibacterium, Ruminococcus, and Blautia. Bacteria of the genus Faecalibacterium, abundant butyric acid-producing bacteria colonizing the human gut, display anti-inflammatory effects and may be used as potential probiotics for treatment of gut inflammation [84], [85]. Ruminococcus members, breakdown cellulose (with the formation of methane) and accumulate a reserve iodophilic polymer of glucose in the cytoplasm [86]. Blautia is widely distributed in mammalian feces and intestines. As a dominant genus in the intestinal microbiota, Blautia, has a significant correlation with host physiological dysfunctions, such as obesity, diabetes, cancer, and various inflammatory and metabolic diseases due to its antibacterial activity against specific microorganisms [87], [88]. Several recent reports have indicated that the composition of and changes in the Blautia population in the intestine are related to factors such as host age, geography, diet, genotype, health, disease state, and other physiological states [89], [90], [91]. Meanwhile, the rest of the universal genera are grouped within the last cluster formed by less abundant taxa (cluster 1). On examining the abundances of individuals for the members of the three main clusters (Supplementary Fig. 9A), we can see that, in general, these taxa maintain high levels of abundance for most individuals, except Prevotella, which is less abundant in urban groups in some cases, which may be related to diet. Clostridium and Roseburia also show stable and relatively abundant levels. Clostridium species ferment a variety of nutrients, like carbohydrate, protein, organic acid and other organics, to produce acetic acid, propionic acid, butyric acid, and some solvents, such as acetone and butanol. In animal and human intestines, Clostridium species mostly utilize indigestible polysaccharide and most of the metabolites they produced afford many benefits to gut health [56]. Roseburia spp. likely play a major role in maintaining gut health and immune defense, such as regulatory T-cell homeostasis, primarily through the production of butyrate. The concomitant decrease in the well-known butyrate-producing bacterial genus Roseburia in many intestinal disorders suggests the potential use of these bacteria as indicators of intestinal health [92].
At the species level, three abundance clusters are found (Fig. 4B). A major abundance cluster (cluster 3), uncultured Prevotella sp., proves to be a dominant bacterial signature of rural groups. Again, we find a cluster formed by bacteria that are generally abundant in all studied groups (cluster 2), with representatives of Faecalibacterium, Clostridium, and Ruminococcus, among others. Finally, we encounter the last cluster containing the remaining less abundant taxa (cluster 1), with representatives of Blautia, Bacteroides, and Roseburia, among others. Examining the abundances across individuals (Supplementary Fig. 9B), Faecalibacterium prausnitzii, uncultured Faecalibacterium sp. and uncultured Clostridium sp. present the most stable and abundant levels. These genera and species are Firmicutes and Bacteroidetes, which are known dominant phyla in the human gut microbiota of healthy individuals [93], [94], [95].
Due to their high abundance, these taxa are of special interest since research suggests that abundant organisms could act as “ecosystem engineers” with the capacity to directly alter their environment [96]. Furthermore, recent findings suggest that abundance is a strong determinant for engaging Horizontal Gene Transfer (HGT) [97], which could provide an ecological adaptation advantage, since transferred gene functions of the microbiome reflect the host’s lifestyle and are driven by niche adaptation [97], [98], [99]. For instance, an interesting case is the transfer of porphyranases and agarases from the marine bacterium Zobellia galactanivorans to the gut bacterium Bacteroides plebeius in the Japanese population, which has been associated with their dietary habits, such as high seaweed consumption [100]. As HGT is a highly frequent process in the human gut, even within single individuals [97], and following the logic that “everything is everywhere, but the environment selects” [99], [101], we hypothesize that certain ancestral organisms which have co-evolved with the human host may present a selective advantage in this ecological adaptation process. This would result from their shared evolutionary history and the associated host’s domestication process.
3.6. Universal core taxa are highly prevalent despite lifestyle and geographic differences
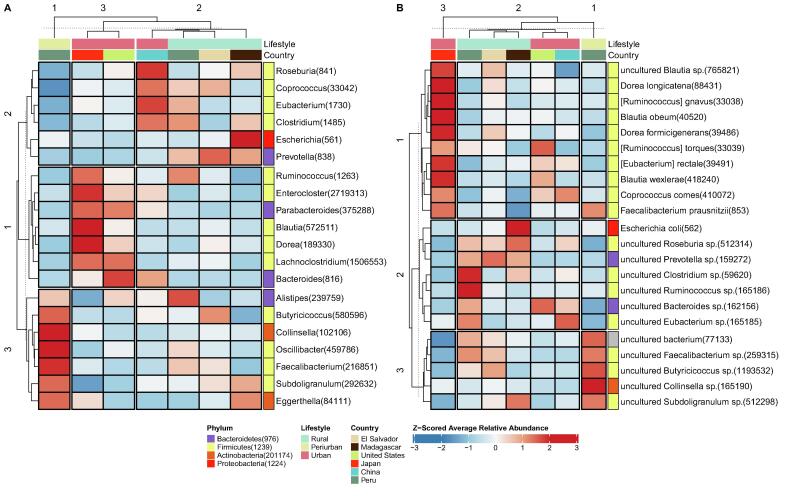
Three pattern clusters are found when examining z-scored average relative abundances at the genus level for the universal core taxa (Fig. 5A). The first cluster presents genera, including Bacteroides, Parabacteroides, and Enterocloster, that are generally more abundant in urban populations. Other country-specific patterns are found, such as Blautia and Dorea in Japan. The second cluster comprises genera that are typically more abundant in China (like Roseburia and Eubacterium) or rural populations (Prevotella is the only one shared by the three rural groups). In addition, the third cluster is formed by genera enriched in the periurban samples, like Faecalibacterium, Oscillibacter, and Collinsella, among others.
Fig. 5.
Patterns analysis for universal core taxa. Z-scored average relative abundances for the different groups at genus (A) and species (B) level. Groups and taxa were clustered using the k-means algorithm. NCBI’s taxIDs are indicated in parentheses.
At the species level, three differential pattern clusters are found (Fig. 5B). In the first cluster, we find more abundant species in urban populations, mainly in Japan and, again, we find species patterns shared by the three urban populations, such as [Eubacterium] rectale and Coprococcus comes. The second cluster is formed by species mainly enriched in rural groups, like uncultured Prevotella sp. and uncultured Roseburia sp. Finally, the third cluster is enriched in periurban samples, with species such as uncultured Collinsella sp. and uncultured Faecalibacterium sp., among others. Interestingly, the urban cluster comprises cultured species, while the rural and periurban clusters are formed by uncultured species, which could be related to the under-representation of non-western microbiomes in conventional reference databases [20], [21], [22]. In general, the aforementioned patterns are maintained across individuals (Supplementary Fig. 10), although they are not completely homogeneous in all cases.
Typically, the differences between urban and rural groups have been explained by their underlying dietary habits associated with these lifestyles. As we have already commented, Prevotella has been associated with rural non-western individuals that present high-fiber and high-carbohydrate diets enriched in plant foods, likely due to their ability to process complex polysaccharides [34], [102]. By contrast, Bacteroides has been related to western urban individuals with high-protein and high-fat diets, which is probably due to their bile-tolerant abilities, which are common in gut environments of consumers of animal-based foods [102], [103]. The same could be said for the related Parabacteroides genus, showing bile-tolerant capabilities [57]. Japan is a special case that shows the highest average lifespan, very low body mass index and a particular microbial uniqueness, with the highest abundance of the genera Blautia [33]. Interestingly, the use of certain traditional Japanese food preparations, acting as prebiotics, has been related to an increase in certain Blautia species [104]. Finally, the lower levels of Prevotella and Bacteroides in the periurban group could be indicative of the transition from a rural area to an industrialized one.
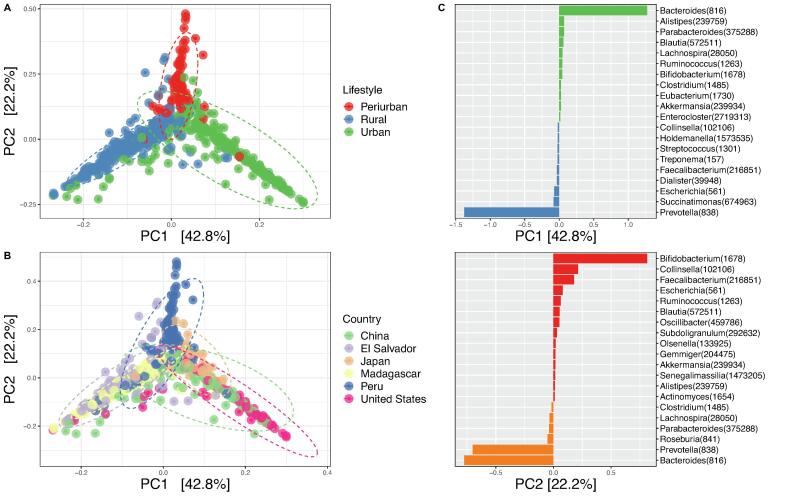
Global differences between samples were examined by means of a PCA at the genus level (Fig. 6). The samples are clearly separated into three groups for the lifestyle variable, each reflecting a different lifestyle. The first component separates the urban and rural samples, whereas the second component separates the periurban samples from the other two groups, with the former being in an intermediate position. These two components manage to explain 65% of the variability between samples. Further analysis using the PcoA and NMDS methods provided similar ordination results (Supplementary Fig. 11). In general, urban populations group together, and rural populations do likewise. We also see that Peru is divided in two, as it has both rural and periurban samples. Altogether, these results reflect differences in community configurations that seem to be associated mainly with lifestyle and the subsequent degree of westernization. Although host factors such as genetics, immune regulation, or age play a role in shaping the gut microbiota [105], [106], [107], environmental factors such as geography, lifestyle, or diet appear to play a dominant role [108], [109], [110]. Moreover, even considering populations that share the same geographical origin, like the Urban Versus Rural Settings in Northern Ecuador of Soto-Girn et al. [111], lifestyle and westernization seem to be the main driver of differences. Furthermore, other findings suggest that the microbial composition of great apes and humans would be more closely related to their host’s lifestyle than to their geography [112].
Fig. 6.
Principal component analysis (PCA). Results are shown by lifestyle (A) and country of origin (B). 95% confidence intervals are represented by ellipses. (C) Top 20 genera scores for the two main components. NCBI’s taxIDs are indicated in parentheses.
The main taxa drivers of the differences of these two first components were investigated (Fig. 6C). In the first component, the main drivers of the differences between urban and rural samples are Bacteroides and Prevotella, respectively. Differences in the human gut composition, especially the Prevotella-Bacteroides exchange between rural and urban populations, have often been explained by their unique dietary traits [20], [108], [109]. Furthermore, these genera have been used to define different community composition types termed enterotypes, which appear to be useful to some degree in attempting to stratify human populations [93], [113]], although with some controversy [114], [115]. On the other hand, for the second component, the main driver towards the periurban samples is Bifidobacterium, and to a lesser degree Collinsella and Faecalibacterium. As stated in the original study from which these samples were taken, diarrheal episodes are frequent in this particular community due to the high prevalence of various infectious agents [32]. The continued presence of pathogenic agents may explain the selection of protective microorganisms. A broad range of beneficial effects on human health has been associated with Bifidobacterium and Faecalibacterium, promoting the use of strains of these genera as probiotics [116], [117], [118], [119]. Probiotics are currently considered to exert anti-diarrheal action through different mechanisms such as homeostasis regulation of the microbiota, immune system activation, manipulation of the intestinal defense barrier, and the production of certain metabolites [120]. For instance, certain Bifidobacterium strains can inhibit growth and adhesion to epithelial cells of some enteropathogens [121]. Likewise, Faecalibacterium is a good example of inhibition by metabolites considering that Faecalibacterium prausnitzii has been considered a major producer of butyrate in the gut, a substance involved in regulating the immune system [119]. By contrast, the presence of Collinsella could have a dietary basis, since it has been associated with low fiber intake [122], [123].
As we can see, even within this core of universal taxa there are differences that are clearly influenced by factors such as geography, lifestyle and their underlying levels of westernization. Their abundances shift according to the different roles they may play depending on these factors, and some of them are even the main drivers of these differences. However, there is no substitution in terms of “replacement”, but rather “displacement”, since they do not disappear completely. Thus, they are highly prevalent in the different studied scenarios, even in spite of apparent differences, which could indicate that these core taxa go beyond ecological factors, playing a vital role in the human gut.
4. Conclusions
Our enrichment strategy improves, and almost double, classification capacity compared to the original NT database. This strategy provides greater similarity of classification levels between the different studied datasets, which compensates for the infra-classification of non-western microbiomes in conventional databases. Furthermore, a MAG filtering strategy was tested and discussed, based on the existence of incomplete-taxonomy MAGs, that could work as catch-all taxa depending on the taxonomic level of interest, illustrating that while these types of genomes are useful, they should be used with caution.
At the same time, we have sought to prove the existence of a phylogenetic core of highly prevalent common taxa in the healthy human gut microbiota worldwide, which we achieved despite clear ecological differences. Giving a leading role to this type of analysis, which is often relegated to a secondary place, and studying it at a greater level of detail thanks to the use of WGS sequence data. Furthermore, the universality of these results was pursued by covering different gradients of westernization, trying to maintain a balanced design between urban and rural datasets, in order to avoid possible bias towards westernized populations. Likewise, the ancestral nature, characteristics, crucial roles and ecological adaptation capacities of these taxa were discussed. The study aims to provide candidates for a hypothetical phylogenetic core of mutualistic microorganisms that has co-evolved with the human species.
Taken together, the results reported here attempt to contribute to the still diffuse knowledge about the use of genome databases in metagenomics and apply it to a problem of great interest, as is the study of the core human gut microbiota.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by grant to AM from the Spanish Ministry of Science and Innovation (PID2019-105969 GB-I00), Generalitat Valenciana (project Prometeo/2018/A/133), Asociación Española contra el Cancer (project AECC 2017-1485) and co-financed by the European Regional Development Fund (ERDF). SP-E was funded by an ACIF fellowship from the Generalitat Valenciana during October 2021 (Reference: ACIF/2021/341), and later on since December 2021 by an FPU fellowship from the Spanish Ministry of Universities (Reference: FPU20/05756).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2021.12.035.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Sender R., Fuchs S., Milo R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016;14(8):e1002533. doi: 10.1371/journal.pbio.1002533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Escobar-Zepeda A., Vera-Ponce de León A., Sanchez-Flores A. The road to metagenomics: from microbiology to DNA sequencing technologies and bioinformatics. Front Genet. 2015;6:348. doi: 10.3389/fgene.2015.00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hiraoka S., Yang C.C., Iwasaki W. Metagenomics and Bioinformatics in Microbial Ecology: Current Status and Beyond. Microbes Environ. 2016;31(3):204–212. doi: 10.1264/jsme2.ME16024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breitwieser F.P., Lu J., Salzberg S.L. A review of methods and databases for metagenomic classification and assembly. Briefings Bioinf. 2019;20(4):1125–1136. doi: 10.1093/bib/bbx120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rojo D., Méndez-García C., Raczkowska B.A. et al. (2017). Exploring the human microbiome from multiple perspectives: factors altering its composition and function. FEMS Microbiology Reviews 41(4):453-478. 10.1093/femsre/fuw046 [DOI] [PMC free article] [PubMed]
- 6.Risely A., Tate A. Applying the core microbiome to understand host–microbe systems. J Anim Ecol. 2020;89(7):1549–1558. doi: 10.1111/1365-2656.13229. [DOI] [PubMed] [Google Scholar]
- 7.Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett C.M., Knight R., Gordon J.I. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E., et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huse S.M., Ye Y., Zhou Y., Fodor A.A., Ahmed N. A core human microbiome as viewed through 16S rRNA sequence clusters. PLoS ONE. 2012;7(6):e34242. doi: 10.1371/journal.pone.0034242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falony G., Joossens M., Vieira-Silva S., Wang J., Darzi Y., Faust K., et al. Population-level analysis of gut microbiome variation. Science. 2016;352(6285):560–564. doi: 10.1126/science.aad3503. [DOI] [PubMed] [Google Scholar]
- 11.Shetty S. A., Hugenholtz F., Lahti L. et al. (2017). Intestinal microbiome landscaping: insight in community assemblage and implications for microbial modulation strategies. FEMS microbiology reviews, 41(2): 182-199. [DOI] [PMC free article] [PubMed]
- 12.Aguirre de Cárcer D. The human gut pan-microbiome presents a compositional core formed by discrete phylogenetic units. Sci Rep. 2018;8(1):1–8. doi: 10.1038/s41598-018-32221-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edgar R.C. Accuracy of taxonomy prediction for 16S rRNA and fungal ITS sequences. PeerJ. 2018;6:e4652. doi: 10.7717/peerj.4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wright E.S., Baum D.A. Exclusivity offers a sound yet practical species criterion for bacteria despite abundant gene flow. BMC Genomics. 2018;19(1):1–12. doi: 10.1186/s12864-018-5099-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ranjan R., Rani A., Metwally A., McGee H.S., Perkins D.L. Analysis of the microbiome: Advantages of whole genome shotgun versus 16S amplicon sequencing. Biochem Biophys Res Commun. 2016;469(4):967–977. doi: 10.1016/j.bbrc.2015.12.083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laudadio I., Fulci V., Palone F., Stronati L., Cucchiara S., Carissimi C. Quantitative assessment of shotgun metagenomics and 16S rDNA amplicon sequencing in the study of human gut microbiome. OMICS. 2018;22(4):248–254. doi: 10.1089/omi.2018.0013. [DOI] [PubMed] [Google Scholar]
- 17.King C.H., Desai H., Sylvetsky A.C., LoTempio J., Ayanyan S., Carrie J., et al. Baseline human gut microbiota profile in healthy people and standard reporting template. PLoS ONE. 2019;14(9):e0206484. doi: 10.1371/journal.pone.0206484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loftus M., Hassouneh S.A.D., Yooseph S. Bacterial associations in the healthy human gut microbiome across populations. Sci Rep. 2021;11(1):1–14. doi: 10.1038/s41598-021-82449-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simon H.Y., Siddle K.J., Park D.J., Sabeti P.C. Benchmarking metagenomics tools for taxonomic classification. Cell. 2019;178(4):779–794. doi: 10.1016/j.cell.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N. Gut microbiome: westernization and the disappearance of intestinal diversity. Curr Biol. 2015;25(14):R611–R613. doi: 10.1016/j.cub.2015.05.040. [DOI] [PubMed] [Google Scholar]
- 21.Nayfach S., Shi Z.J., Seshadri R., Pollard K.S., Kyrpides N.C. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568(7753):505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pasolli E., Asnicar F., Manara S., Zolfo M., Karcher N., Armanini F., et al. Extensive unexplored human microbiome diversity revealed by over 150,000 genomes from metagenomes spanning age, geography, and lifestyle. Cell. 2019;176(3):649–662.e20. doi: 10.1016/j.cell.2019.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ochman H., Worobey M., Kuo C.-H., Ndjango J.-B., Peeters M., Hahn B.H., et al. Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol. 2010;8(11):e1000546. doi: 10.1371/journal.pbio.1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amato K.R., Mallott E.K., McDonald D., Dominy N.J., Goldberg T., Lambert J.E., et al. Convergence of human and Old World monkey gut microbiomes demonstrates the importance of human ecology over phylogeny. Genome Biol. 2019;20(1) doi: 10.1186/s13059-019-1807-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayers E. W., Agarwala R., Bolton E. E. et al. (2019). Database resources of the national center for biotechnology information. Nucleic acids research, 47(Database issue), D23. 10.1093/nar/gky1069 [DOI] [PMC free article] [PubMed]
- 26.Methé B., Nelson K., Pop M., et al. A framework for human microbiome research. Nature. 2012;486(7402):215–221. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sichtig H., Minogue T., Yan Y.i., Stefan C., Hall A., Tallon L., et al. FDA-ARGOS is a database with public quality-controlled reference genomes for diagnostic use and regulatory science. Nat Commun. 2019;10(1) doi: 10.1038/s41467-019-11306-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poyet M., Groussin M., Gibbons S.M., Avila-Pacheco J., Jiang X., Kearney S.M., et al. A library of human gut bacterial isolates paired with longitudinal multiomics data enables mechanistic microbiome research. Nat Med. 2019;25(9):1442–1452. doi: 10.1038/s41591-019-0559-3. [DOI] [PubMed] [Google Scholar]
- 29.Zou Y., Xue W., Luo G., Deng Z., Qin P., Guo R., et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37(2):179–185. doi: 10.1038/s41587-018-0008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forster S.C., Kumar N., Anonye B.O., Almeida A., Viciani E., Stares M.D., et al. A human gut bacterial genome and culture collection for improved metagenomic analyses. Nat Biotechnol. 2019;37(2):186–192. doi: 10.1038/s41587-018-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Almeida A., Mitchell A.L., Boland M., Forster S.C., Gloor G.B., Tarkowska A., et al. A new genomic blueprint of the human gut microbiota. Nature. 2019;568(7753):499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pehrsson E.C., Tsukayama P., Patel S., Mejía-Bautista M., Sosa-Soto G., Navarrete K.M., et al. Interconnected microbiomes and resistomes in low-income human habitats. Nature. 2016;533(7602):212–216. doi: 10.1038/nature17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishijima S., Suda W., Oshima K., Kim S.-W., Hirose Y., Morita H., et al. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016;23(2):125–133. doi: 10.1093/dnares/dsw002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Obregon-Tito A.J., Tito R.Y., Metcalf J., Sankaranarayanan K., Clemente J.C., Ursell L.K., et al. Subsistence strategies in traditional societies distinguish gut microbiomes. Nat Commun. 2015;6(1) doi: 10.1038/ncomms7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X., Zhang D., Jia H., Feng Q., Wang D., Liang D.i., et al. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med. 2015;21(8):895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 36.Bushnell B. (2014). BBTools software package. URL: https://sourceforge.net/projects/bbmap/
- 37.Andrews S. (2010). FastQC: a quality control tool for high throughput sequence data. URL: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
- 38.Ewels P., Magnusson M., Lundin S., Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wood D.E., Lu J., Langmead B. Improved metagenomic analysis with Kraken 2. Genome Biol. 2019;20(1):257. doi: 10.1186/s13059-019-1891-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu J., Breitwieser F.P., Thielen P., et al. Bracken: estimating species abundance in metagenomics data. PeerJ Comput Sci. 2017;3 doi: 10.7717/peerj-cs.104. [DOI] [Google Scholar]
- 41.Lahti L., Shetty S. et al. (2017). Tools for microbiome analysis in R. URL: http://microbiome.github.com/microbiome
- 42.McMurdie P.J., Holmes S., Watson M. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE. 2013;8(4):e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oksanen J., Blanchet F. G., Friendly M. et al. (2019). vegan: Community Ecology Package. R package version 2.5-6. URL: https://CRAN.R-project.org/package=vegan
- 44.Gehlenborg N. UpSetR: A More Scalable Alternative to Venn and Euler Diagrams for Visualizing Intersecting Sets. R package version. 2019;1(4) https://CRAN.R-project.org/package=UpSetR URL. [Google Scholar]
- 45.Gu Z., Eils R., Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32(18):2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 46.Foster Z.S.L., Sharpton T.J., Grünwald N.J., Poisot T. Metacoder: An R package for visualization and manipulation of community taxonomic diversity data. PLoS Comput Biol. 2017;13(2):e1005404. doi: 10.1371/journal.pcbi.1005404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schoch C. L., Ciufo S., Domrachev M. et al. (2020). NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database, 2020. [DOI] [PMC free article] [PubMed]
- 48.Wood D.E., Salzberg S.L. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):1–12. doi: 10.1186/gb-2014-15-3-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castillo M., Skene G., Roca M., Anguita M., Badiola I., Duncan S.H., et al. Application of 16S rRNA gene-targetted fluorescence in situ hybridization and restriction fragment length polymorphism to study porcine microbiota along the gastrointestinal tract in response to different sources of dietary fibre. FEMS Microbiol Ecol. 2007;59(1):138–146. doi: 10.1111/j.1574-6941.2006.00204.x. [DOI] [PubMed] [Google Scholar]
- 50.Nava G.M., Stappenbeck T.S. Diversity of the autochthonous colonic microbiota. Gut Microbes. 2011;2(2):99–104. doi: 10.4161/gmic.2.2.15416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Barcenilla A., Pryde S. E., Martin J. C. et al. (2000). Phylogenetic relationships of butyrate-producing bacteria from the human gut. Applied and environmental microbiology, 66(4): 1654-1661. 10.1128/AEM.66.4.1654-1661.2000 [DOI] [PMC free article] [PubMed]
- 52.Duncan S.H., Hold G.L., Harmsen H.J.M., et al. Growth requirements and fermentation products of Fusobacterium prausnitzii, and a proposal to reclassify it as Faecalibacterium prausnitzii gen. nov., comb. nov. Int J Syst Evol Microbiol. 2002;52(6):2141–2146. doi: 10.1099/00207713-52-6-2141. [DOI] [PubMed] [Google Scholar]
- 53.Christl S. U., Eisner H. D., Dusel G. et al. (1996). Antagonistic effects of sulfide and butyrate on proliferation of colonic mucosa. Digestive diseases and sciences, 41(12): 2477-2481. 10.1007/BF02100146 [DOI] [PubMed]
- 54.Archer S., Meng S., Wu J., Johnson J., Tang R., Hodin R. Butyrate inhibits colon carcinoma cell growth through two distinct pathways. Surgery. 1998;124(2):248–253. doi: 10.1016/S0039-6060(98)70127-8. [DOI] [PubMed] [Google Scholar]
- 55.Egan M., Dempsey E., Ryan C.A., Ross R.P., Stanton C. The Sporobiota of the Human Gut. Gut Microbes. 2021;13(1) doi: 10.1080/19490976.2020.1863134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guo P., Zhang K., Ma X., et al. Clostridium species as probiotics: potentials and challenges. J Anim Sci Biotechnol. 2020;11(1):1–10. doi: 10.1186/s40104-019-0402-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakamoto M., Benno Y. Reclassification of Bacteroides distasonis, Bacteroides goldsteinii and Bacteroides merdae as Parabacteroides distasonis gen. nov., comb. nov., Parabacteroides goldsteinii comb. nov. and Parabacteroides merdae comb. nov. Int J Syst Evol Microbiol. 2006;56(7):1599–1605. doi: 10.1099/ijs.0.64192-0. [DOI] [PubMed] [Google Scholar]
- 58.Trujillo M. E., Dedysh S., DeVos P. et al. (Eds.). (2015). Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley. 10.1002/9781118960608 [DOI]
- 59.Shin Y., Park S.J., Paek J., et al. Bacteroides koreensis sp. nov. and Bacteroides kribbi sp. nov., two new members of the genus Bacteroides. Int J Syst Evol Microbiol. 2017;67(11):4352–4357. doi: 10.1099/ijsem.0.002226. [DOI] [PubMed] [Google Scholar]
- 60.Liu C., Finegold S. M., Song Y. et al. (2008). Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. International Journal of Systematic and Evolutionary Microbiology, 58(8): 1896–1902. 10.1099/ijs.0.65208-0 [DOI] [PubMed]
- 61.Eeckhaut V., Van Immerseel F., Teirlynck E., et al. Butyricicoccus pullicaecorum gen. nov., sp. nov., an anaerobic, butyrate-producing bacterium isolated from the caecal content of a broiler chicken. Int J Syst Evol Microbiol. 2008;58(12):2799–2802. doi: 10.1099/ijs.0.65730-0. [DOI] [PubMed] [Google Scholar]
- 62.Trachsel J., Humphrey S., Allen H.K. Butyricicoccus porcorum sp. nov., a butyrate-producing bacterium from swine intestinal tract. Int J Syst Evol Microbiol. 2018;68(5):1737–1742. doi: 10.1099/ijsem.0.002738. [DOI] [PubMed] [Google Scholar]
- 63.Kageyama A., Benno Y., Nakase T. Phylogenetic evidence for the transfer of Eubacterium lentum to the genus Eggerthella as Eggerthella lenta gen. nov., comb. nov. Int J Syst Evol Microbiol. 1999;49(4):1725–1732. doi: 10.1099/00207713-49-4-1725. [DOI] [PubMed] [Google Scholar]
- 64.Maruo T., Sakamoto M., Ito C., et al. Adlercreutzia equolifaciens gen. nov., sp. nov., an equol-producing bacterium isolated from human faeces, and emended description of the genus Eggerthella. Int J Syst Evol Microbiol. 2008;58(5):1221–1227. doi: 10.1099/ijs.0.65404-0. [DOI] [PubMed] [Google Scholar]
- 65.Kaneuchi C., Watanabe K., Terada A., et al. Taxonomic Study of Bacteroides clostridiiformis subsp. clostridiiformis (Burri and Ankersmit) Holdeman and Moore and of Related Organisms: Proposal of Clostridium clostridiiformis (Burri and Ankersmit) comb. nov. and Clostridium symbiosum (Stevens) comb. nov. Int J Syst Bacteriol. 1976;26(2):195–204. doi: 10.1099/00207713-26-2-195. [DOI] [Google Scholar]
- 66.Haas K.N., Blanchard J.L. Reclassification of the Clostridium clostridioforme and Clostridium sphenoides clades as Enterocloster gen. nov. and Lacrimispora gen. nov., including reclassification of 15 taxa. Int J Syst Evol Microbiol. 2020;70(1):23–34. doi: 10.1099/ijsem.0.003698. [DOI] [PubMed] [Google Scholar]
- 67.Yutin N., Galperin M.Y. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environ Microbiol. 2013;15(10):2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Iino T., Mori K., Tanaka K., et al. Oscillibacter valericigenes gen. nov., sp. nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam. Int J Syst Evol Microbiol. 2007;57(8):1840–1845. doi: 10.1099/ijs.0.64717-0. [DOI] [PubMed] [Google Scholar]
- 69.Le Roy T., Van der Smissen P., Paquot A., et al. Dysosmobacter welbionis gen. nov., sp. nov., isolated from human faeces and emended description of the genus Oscillibacter. Int J Syst Evol Microbiol. 2020;70(9):4851–4858. doi: 10.1099/ijsem.0.003547. [DOI] [PubMed] [Google Scholar]
- 70.Albenberg L., Esipova T.V., Judge C.P., Bittinger K., Chen J., Laughlin A., et al. Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology. 2014;147(5):1055–1063.e8. doi: 10.1053/j.gastro.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zheng L., Kelly C.J., Colgan S.P. Physiologic hypoxia and oxygen homeostasis in the healthy intestine. A review in the theme: cellular responses to hypoxia. American Journal of Physiology-Cell. Physiology. 2015;309(6):C350–C360. doi: 10.1152/ajpcell.00191.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Friedman E.S., Bittinger K., Esipova T.V., et al. Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci. 2018;115(16):4170–4175. doi: 10.1073/pnas.1718635115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Browne H.P., Forster S.C., Anonye B.O., Kumar N., Neville B.A., Stares M.D., et al. Culturing of ‘unculturable’human microbiota reveals novel taxa and extensive sporulation. Nature. 2016;533(7604):543–546. doi: 10.1038/nature17645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Meehan C.J., Beiko R.G. A phylogenomic view of ecological specialization in the Lachnospiraceae, a family of digestive tract-associated bacteria. Genome biology and evolution. 2014;6(3):703–713. doi: 10.1093/gbe/evu050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wiles T.J., Schlomann B.H., Wall E.S., Betancourt R., Parthasarathy R., Guillemin K., et al. Swimming motility of a gut bacterial symbiont promotes resistance to intestinal expulsion and enhances inflammation. PLoS Biol. 2020;18(3):e3000661. doi: 10.1371/journal.pbio.3000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moeller A.H., Caro-Quintero A., Mjungu D., Georgiev A.V., Lonsdorf E.V., Muller M.N., et al. Cospeciation of gut microbiota with hominids. Science. 2016;353(6297):380–382. doi: 10.1126/science.aaf3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moeller A.H., Li Y., Mpoudi Ngole E., Ahuka-Mundeke S., Lonsdorf E.V., Pusey A.E., et al. Rapid changes in the gut microbiome during human evolution. Proc Natl Acad Sci. 2014;111(46):16431–16435. doi: 10.1073/pnas.1419136111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Xu J., Gordon J.I. Honor thy symbionts. Proc Natl Acad Sci. 2003;100(18):10452–10459. doi: 10.1073/pnas.1734063100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hooper L.V., Midtvedt T., Gordon J.I. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev Nutr. 2002;22(1):283–307. doi: 10.1146/annurev.nutr.22.011602.092259. [DOI] [PubMed] [Google Scholar]
- 80.Clemente J.C., Pehrsson E.C., Blaser M.J., Sandhu K., Gao Z., Wang B., et al. The microbiome of uncontacted Amerindians. Sci Adv. 2015;1(3) doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Martínez I., Stegen J., Maldonado-Gómez M., Eren A.M., Siba P., Greenhill A., et al. The gut microbiota of rural papua new guineans: composition, diversity patterns, and ecological processes. Cell reports. 2015;11(4):527–538. doi: 10.1016/j.celrep.2015.03.049. [DOI] [PubMed] [Google Scholar]
- 82.Pedersen H.K., Gudmundsdottir V., Nielsen H.B., Hyotylainen T., Nielsen T., Jensen B.A.H., et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature. 2016;535(7612):376–381. doi: 10.1038/nature18646. [DOI] [PubMed] [Google Scholar]
- 83.Leite A.Z., Rodrigues N.d.C., Gonzaga M.I., Paiolo J.C.C., de Souza C.A., Stefanutto N.A.V., et al. Detection of increased plasma interleukin-6 levels and prevalence of Prevotella copri and Bacteroides vulgatus in the feces of type 2 diabetes patients. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.01107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Quévrain E., Maubert M.A., Michon C., Chain F., Marquant R., Tailhades J., et al. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn’s disease. Gut. 2016;65(3):415–425. doi: 10.1136/gutjnl-2014-307649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sokol H., Pigneur B., Watterlot L., Lakhdari O., Bermudez-Humaran L.G., Gratadoux J.-J., et al. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci. 2008;105(43):16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Falalyeyeva T., Chornenka N., Cherkasova L., Tsyryuk O., Molchek N., Kovalchuk O., et al. Reference Module in Food Science. Elsevier; 2022. Gut Microbiota Interactions With Obesity. [DOI] [Google Scholar]
- 87.Kalyana Chakravarthy S., Jayasudha R., Sai Prashanthi G., Ali M.H., Sharma S., Tyagi M., et al. Dysbiosis in the gut bacterial microbiome of patients with uveitis, an inflammatory disease of the eye. Indian journal of microbiology. 2018;58(4):457–469. doi: 10.1007/s12088-018-0746-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khattab M.S.A., El Tawab A.M.B., Fouad M.T. Isolation and characterization of anaerobic bacteria from frozen rumen liquid and its potential Characterizations. International Journal of Dairy Science. 2017;12(1):47–51. doi: 10.3923/ijds.2017.47.51. [DOI] [Google Scholar]
- 89.Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J.-Z., et al. Age-related changes in gut microbiota composition from newborn to centenarian: a cross-sectional study. BMC Microbiol. 2016;16(1) doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nakayama J., Watanabe K., Jiang J., Matsuda K., Chao S.-H., Haryono P., et al. Diversity in gut bacterial community of school-age children in Asia. Sci Rep. 2015;5(1) doi: 10.1038/srep08397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mao B., Gu J., Li D., Cui S., Zhao J., Zhang H., et al. Effects of different doses of fructooligosaccharides (FOS) on the composition of mice fecal microbiota, especially the bifidobacterium composition. Nutrients. 2018;10(8):1105. doi: 10.3390/nu10081105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tamanai-Shacoori Z., Smida I., Bousarghin L., Loreal O., Meuric V., Fong S.B., et al. Roseburia spp.: a marker of health? Future microbiology. 2017;12(2):157–170. doi: 10.2217/fmb-2016-0130. [DOI] [PubMed] [Google Scholar]
- 93.Arumugam M., Raes J., Pelletier E., Le Paslier D., Yamada T., Mende D.R., et al. Enterotypes of the human gut microbiome. Nature. 2011;473(7346):174–180. doi: 10.1038/nature09944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lozupone C.A., Stombaugh J.I., Gordon J.I., Jansson J.K., Knight R. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489(7415):220–230. doi: 10.1038/nature11550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rinninella E., Raoul P., Cintoni M., Franceschi F., Miggiano G., Gasbarrini A., et al. What is the healthy gut microbiota composition? A changing ecosystem across age, environment, diet, and diseases. Microorganisms. 2019;7(1):14. doi: 10.3390/microorganisms7010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Nemergut D.R., Schmidt S.K., Fukami T., O'Neill S.P., Bilinski T.M., Stanish L.F., et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77(3):342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Groussin M., Poyet M., Sistiaga A., Kearney S.M., Moniz K., Noel M., et al. Elevated rates of horizontal gene transfer in the industrialized human microbiome. Cell. 2021;184(8):2053–2067.e18. doi: 10.1016/j.cell.2021.02.052. [DOI] [PubMed] [Google Scholar]
- 98.Smillie C.S., Smith M.B., Friedman J., Cordero O.X., David L.A., Alm E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature. 2011;480(7376):241–244. doi: 10.1038/nature10571. [DOI] [PubMed] [Google Scholar]
- 99.Brito I.L., Yilmaz S., Huang K., Xu L., Jupiter S.D., Jenkins A.P., et al. Mobile genes in the human microbiome are structured from global to individual scales. Nature. 2016;535(7612):435–439. doi: 10.1038/nature18927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hehemann J.-H., Correc G., Barbeyron T., Helbert W., Czjzek M., Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464(7290):908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- 101.Baas-Becking L. G. M. (1934). Geobiologie; of inleiding tot de milieukunde. WP Van Stockum & Zoon NV.
- 102.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tomova A., Bukovsky I., Rembert E., Yonas W., Alwarith J., Barnard N.D., et al. The effects of vegetarian and vegan diets on gut microbiota. Frontiers in nutrition. 2019;6 doi: 10.3389/fnut.2019.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hamajima H., Matsunaga H., Fujikawa A., et al. Japanese traditional dietary fungus koji Aspergillus oryzae functions as a prebiotic for Blautia coccoides through glycosylceramide: Japanese dietary fungus koji is a new prebiotic. SpringerPlus. 2016;5(1):1–10. doi: 10.1186/s40064-016-2950-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Goodrich J., Waters J., Poole A., Sutter J., Koren O., Blekhman R., et al. Human genetics shape the gut microbiome. Cell. 2014;159(4):789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Salazar N., González S., Nogacka A.M., Rios-Covián D., Arboleya S., Gueimonde M., et al. Microbiota: Current Research and Emerging Trends. Caister Academic Press; 2019. Microbiome: Effects of Ageing and Diet. [DOI] [Google Scholar]
- 107.Woodhams D.C., Bletz M.C., Becker C.G., Bender H.A., Buitrago-Rosas D., Diebboll H., et al. Host-associated microbiomes are predicted by immune system complexity and climate. Genome Biol. 2020;21(1) doi: 10.1186/s13059-019-1908-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Xu Z., Knight R. Dietary effects on human gut microbiome diversity. Br J Nutr. 2015;113(S1):S1–S5. doi: 10.1017/S0007114514004127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Gupta V.K., Paul S., Dutta C. Geography, ethnicity or subsistence-specific variations in human microbiome composition and diversity. Front Microbiol. 2017;8:1162. doi: 10.3389/fmicb.2017.01162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rothschild D., Weissbrod O., Barkan E., Kurilshikov A., Korem T., Zeevi D., et al. Environment dominates over host genetics in shaping human gut microbiota. Nature. 2018;555(7695):210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 111.Soto-Girón M.J., Peña-Gonzalez A., Hatt J.K., Montero L., Páez M., Ortega E., et al. Gut Microbiome Changes with Acute Diarrheal Disease in Urban Versus Rural Settings in Northern Ecuador. The American Journal of Tropical Medicine and Hygiene. 2021;104(6):2275–2285. doi: 10.4269/ajtmh.20-0831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Campbell T.P., Sun X., Patel V.H., Sanz C., Morgan D., Dantas G. The microbiome and resistome of chimpanzees, gorillas, and humans across host lifestyle and geography. The ISME journal. 2020;14(6):1584–1599. doi: 10.1038/s41396-020-0634-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Costea P.I., Hildebrand F., Arumugam M., Bäckhed F., Blaser M.J., Bushman F.D., et al. Enterotypes in the landscape of gut microbial community composition. Nat Microbiol. 2018;3(1):8–16. doi: 10.1038/s41564-017-0072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Knights D., Ward T.L., McKinlay C.E., Miller H., Gonzalez A., McDonald D., et al. Rethinking “enterotypes”. Cell Host Microbe. 2014;16(4):433–437. doi: 10.1016/j.chom.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Gorvitovskaia A., Holmes S.P., Huse S.M. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. 2016;4(1):1–12. doi: 10.1186/s40168-016-0160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Picard C., Fioramonti J., Francois A., et al. bifidobacteria as probiotic agents–physiological effects and clinical benefits. Aliment Pharmacol Ther. 2005;22(6):495–512. doi: 10.1111/j.1365-2036.2005.02615.x. [DOI] [PubMed] [Google Scholar]
- 117.Ku S., Park M.S., Ji G.E., et al. Review on Bifidobacterium bifidum BGN4: functionality and nutraceutical applications as a probiotic microorganism. Int J Mol Sci. 2016;17(9):1544. doi: 10.3390/ijms17091544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Martín R., Bermúdez-Humarán L.G., Langella P. Searching for the bacterial effector: The example of the multi-skilled commensal bacterium Faecalibacterium prausnitzii. Front Microbiol. 2018;9:346. doi: 10.3389/fmicb.2018.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.He X., Zhao S., Li Y. Faecalibacterium prausnitzii: A Next-Generation Probiotic in Gut Disease Improvement. Can J Infect Dis Med Microbiol. 2021;2021:1–10. doi: 10.1155/2021/6666114. [DOI] [Google Scholar]
- 120.Li Y., Xia S., Jiang X., et al. Gut microbiota and diarrhea: an updated review. Front Cell Infect Microbiol. 2021;11 doi: 10.3389/fcimb.2021.625210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Vazquez-Gutierrez P., de Wouters T., Werder J., Chassard C., Lacroix C. High iron-sequestrating bifidobacteria inhibit enteropathogen growth and adhesion to intestinal epithelial cells in vitro. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.01480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Gomez-Arango L.F., Barrett H.L., Wilkinson S.A., Callaway L.K., McIntyre H.D., Morrison M., et al. Low dietary fiber intake increases Collinsella abundance in the gut microbiota of overweight and obese pregnant women. Gut Microbes. 2018;9(3):189–201. doi: 10.1080/19490976.2017.1406584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Sharma A.K., Petrzelkova K., Pafco B., Jost Robinson C.A., Fuh T., Wilson B.A., et al. Traditional human populations and nonhuman primates show parallel gut microbiome adaptations to analogous ecological conditions. Msystems. 2020;5(6) doi: 10.1128/mSystems.00815-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers of the samples and associated metadata used in this work can be consulted in Supplementary Table 4. The scripts used for this work can be found at GitHub (https://github.com/sarpiens/corescripts). The enriched databases will be available on request.