Abstract
Antifungal peptides are effective, biocompatible, and biodegradable, and thus, they are promising to be the next generation of drugs for treating infections caused by fungi. The identification processes of highly active peptides, however, are still time-consuming and labor-intensive. Quantitative structure–activity relationships (QSARs) have dramatically facilitated the discovery of many bioactive drug molecules without a priori knowledge. In this study, we have established an effective QSAR protocol for screening antifungal peptides. The screening protocol integrates an accurate antifungal peptide classification model and four activity prediction models against specified target fungi. A demonstrative application was performed on more than three million candidate peptides, and three outstanding peptides were identified. The whole screening took only a few days, which was much faster than our previous experimental screening works. In conclusion, the protocol is useful and effective for reducing repetitive laboratory efforts in antifungal peptide discovery. The prediction server (antifungal Web server) is freely available at www.chemoinfolab.com/antifungal.
Keywords: Quantitative structure−activity relationship, artificial intelligence, machine learning, large-scale screening, drug discovery, bioactive peptides, antifungal peptides
Identification of bioactive peptides has become more critical than ever on account of the advances in peptide drugs.1 The first peptide therapeutics started in 1922 with the medical use of insulin. At present, there are around 80 peptides on the global markets, more than 150 peptides in clinical development, and 400–600 peptides undergoing preclinical studies.2 Many inherent advantages are associated with bioactive peptides, such as larger surface areas, greater chirality, and more complex spatial structures,2,3 making them sometimes more effective and selective than small molecules.4
Antifungal peptides, as a particular category of bioactive peptides, were regarded as promising therapeutic agents for curing many diseases caused by fungi.5 Antifungal peptides can mimic natural ligands6 and recognize multiple microbial targets to reduce the induced resistance.7 At present, however, isolation, purification, and identification of antifungal peptides are still time-consuming and labor-intensive processes. In general, antifungal peptides can be isolated from plants or animal tissues by using physical and chemical methods and subsequently purified by gel filtration chromatography, ion-exchange chromatography, capillary reverse-phase high-performance liquid chromatography, fast protein liquid chromatography, etc. The above processes may take from several weeks to months. In our previous works,8−10 more than three years were devoted to discover and characterize the antifungal peptide AMP-17. Therefore, development of efficient large-scale screening protocols for antifungal peptide identification is essential.
In silico approaches have greatly facilitated the discovery process of antifungal peptides.11,12 In these methods, quantitative structure–activity relationship (QSAR) provides a new perspective by relating molecular properties to bioactivity.13 Generally, peptides are represented by sequence descriptors that reflect physical or chemical information on molecules,14,15 such as hydrophobicity, bulkiness, charge, and surface energy. Bioactivity data can be retrieved from many specialized databases, such as the DBAAPS,16 APD3,17 DRAMP,18 and CAMP19 databases. Machine learning methods can effectively associate the peptide sequences with their bioactivity values,20−22 and we have proposed many effective algorithms.23−27 However, to the best of our knowledge, few complete works integrate a large-scale screening protocol, successful screening application, chemical synthesis, and bioactivity validation of antifungal peptides.
In this study, an in silico protocol was proposed to select potential antifungal peptides, in which a classification model and four activity prediction models against specified fungi (Candida albicans, Candida krusei, Cryptococcus neoformans, and Candida parapsilosis) were included. Moreover, an antifungal index (AFI) was also proposed to provide a final ranking to these screened peptides. A demonstrative application was conducted on more than three million candidates, and three outstanding sequences were determined. Chemical synthesis and experimental validation were applied, and the results confirmed the prominent antifungal properties of the selected peptides (as in Scheme 1). Antifungal Web server integrating all established models is freely available at www.chemoinfolab.com/antifungal.
Scheme 1. Workflow Scheme of Large-Scale Screening for Antifungal Peptides.
Data set 1 was collected for establishment of the antifungal classification model, in which 5775 antifungal peptides and 5775 negative ones were contained. Figure 1 shows the peptide information in the data set. In Figure 1a, skewed distributions of sequence lengths can be observed, and most are distributed in the range of 11–50 amino acid residues. Figure 1b shows amino acid profiles. Figure 1c depicts the scatter points of charge dense versus isoelectric point (pI), and the points of these two classes of peptides are almost overlapping. In Figure 1d, principal component analysis (PCA) was conducted on the peptide descriptors by resorting to a python package of scikit-learn 0.24.2,28 and the scores of the first two principal components are plotted. The above properties failed to directly separate the antifungal peptides from negatives, suggesting a sophisticated calibration model is needed for an accurate classification.
Figure 1.
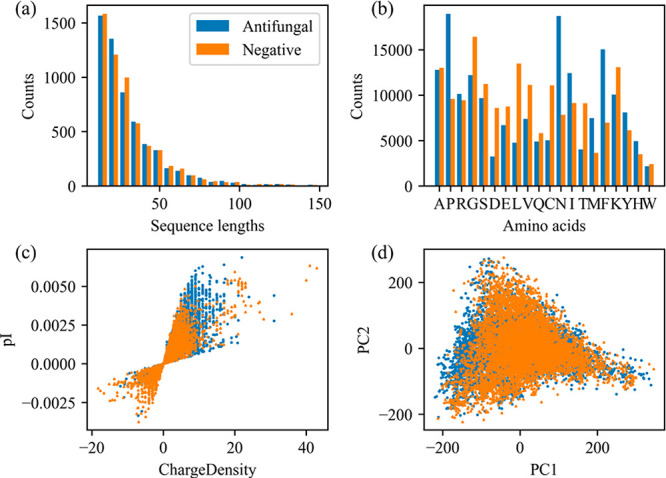
Information of peptides in data set 1 for antifungal peptide classification. Distributions of the sequence lengths (a), amino acid profiles (b), scatter plot of charge density versus isoelectric point (pI) (c), and the PCA scores (d) for peptides in data set 1.
An antifungal peptide classification model was established by utilizing the support vector machine (SVM) method. Hyperparameter C and γ were optimized by 10-fold cross validation in the ranges of 10–1∼105 and 10–8∼102, respectively. Figure 2a presents the mean accuracy of the obtained models with different combinations of C and γ. A large accuracy score represents that an efficient classifier was obtained. When the score reaches the maximum value of 0.91, the optimal C and γ equal 101.08 and 10–4.73, respectively. With the optimized parameters, a classification model was trained. Figure 2b shows the receiver operating characteristic (ROC) curve of the model. The area under the ROC curve (AUC) of the calibration and validation set reaches 0.99 and 0.95, respectively. The results indicate that the obtained model is accurate and robust for antifungal peptide classification. Considering obtained decision values, there are obvious gaps between antifungal peptides and negative ones in the calibration set (Figure 2c) and validation set (Figure 2d), indicating that the trained model is unambiguous for identifying antifungal peptides. The metrics in Table S3 further confirm that the obtained model is effective for identifying antifungal peptides with different lengths.
Figure 2.

Results of antifungal peptide identification. (a) Mean accuracy scores obtained by the models with different combinations of hyperparameters C and γ. (b) ROC curve of the established model for the calibration and validation set. Parts c and d are decision value distributions of peptides in the calibration and validation set, respectively.
Data set 2 was generated for antifungal activity prediction, in which 1583, 95, 275, and 148 activity values (pMIC) were included against C. albicans, C. krusei, C. neoformans, and C. parapsilosis, respectively. Figure 3 displays the information of these peptides. The length distributions of anti-C. albicans, anti-C. krusei, anti-C. neoformans, and anti-C. parapsilosis peptides are displayed in Figure 3a, d, g, and j, respectively. Most of them have sequence lengths smaller than 75 amino acid residues. The distributions of pMIC values against the four target fungi are displayed in Figure 3b, e, h, and k. To indicate the location of the antifungal peptides in principal component space, peptide descriptors in the data set were calculated and projected onto the space spanned in data set 1. The blue dots in Figure 3c, f, i, and l are projections of anti-C. albicans, anti-C. krusei, anti-C. neoformans, and anti-C. parapsilosis peptides, respectively, and the gray points represent the whole antifungal peptide in data set 1. The peptides in this data set account for a very small proportion of data set 1, indicating that only a few antifungal peptides have been measured for a specific antifungal activity.
Figure 3.
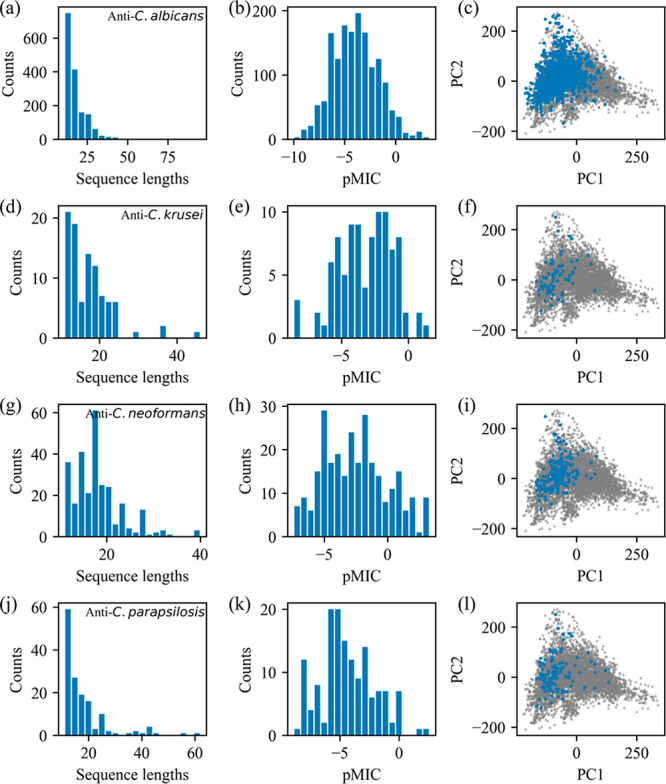
Information of peptides in data set 2 for antifungal activity prediction. Distributions of sequence lengths (a), pMIC values (b), and PCA scores (c) of anti-C. albicans peptides. Subplots d–f, g–i, and j–l are the corresponding distributions of anti-C. krusei, anti-C. neoformans, and anti-C. parapsilosis peptides, respectively. The gray points in parts c, f, i, and l are reference spaces indicating the whole antifungal peptides.
Activity prediction models against C. albicans, C. krusei, C. neoformans, and C. parapsilosis were established by using the support vector regression (SVR) method. Prediction was made on the calibration and validation set, and the results are presented in Figure 4. For C. albicans (Figure 4a), an excellent linear relationship can be observed in terms of the correlation coefficient (R) of 0.93. Figure 4b, c, and d shows the relationships between predicted and experimental pMIC against C. krusei, C. neoformans, and C. parapsilosis, respectively. All models yielded acceptable linearity because all R are larger than 0.90, indicating that accurate models were obtained for predicting antifungal activity.
Figure 4.
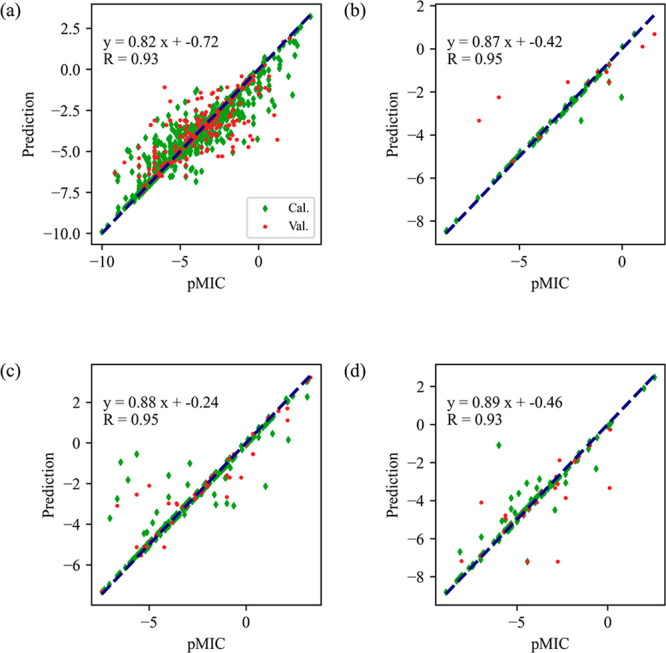
Results of antifungal activity prediction with the targets of C. albicans (a), C. krusei (b), C. neoformans (c), and C. parapsilosis (d).
To further validate the performance of the built models, metrics including RMSE and R2 for the calibration and validation set are listed in Table S4. The RMSEs, as a reflection of the mean prediction error, are close to 1. The results indicate that the prediction error of pMIC is approximately at the experimental level of one broth dilution step (MIC determination method). R2 is another criterion for evaluating the prediction efficiency, and a larger value indicates a more effective model. The R2 values of calibration and validation are in the ranges of 0.90–0.94 and 0.66–0.89, respectively. The results confirm that the built models are efficient for predicting antifungal activity against the four specified fungi.
With the established five accurate models, a multistep screening protocol was proposed to stepwise select potential antifungal peptides without a priori knowledge. Rather than the conventional QSAR method with a single model, the protocol integrates five accurate models. It allows a gradual removal of the most unlikely antifungal peptides in multistep screening, thus giving a relatively high confidence level in the final selected peptide. Moreover, an antifungal index (AFI) was also proposed to provide a final ranking to those selected peptides.
A large-scale screening application was made to evaluate the usefulness of the proposed protocol. A total of 3445312 peptides were collected from the UniProt knowledgebase by restricting sequence lengths in the range of 11–75 amino acid residues. Among these sequences, up to 99.7% of peptides have been only computationally analyzed, and 0.3% have been manually annotated. A demonstrative application was conducted on the unknown peptides, as in Figure 5. After the first step of screening, 491827 antifungal peptides were identified, accounting for just 14% of total sequences. After the second, third, fourth, and fifth steps, only 312327, 306234, 98119, and 61020 peptides with relatively high antifungal activities (MIC < 32 μM) remained, respectively. Finally, three top-ranking antifungal peptides were selected in order of smallest to largest, i.e., KWCFRVCYRGICYRKCR (AFI = 2.11 μM), RRWCFRVCYRGFCYRKCR (AFI = 2.25 μM), and KWCFRVCYRGICYRRCR (AFI = 2.34 μM). For comparison, another 13 reference peptides with moderate antifungal activity (AFI: 4.44–22.71 μM) were also selected by considering both synthesis costs and experiences. Table 1 shows the predictions of the screened 16 peptides. The raw data sets and prediction results are freely available at https://github.com/JinZhangLab/antifungal.
Figure 5.
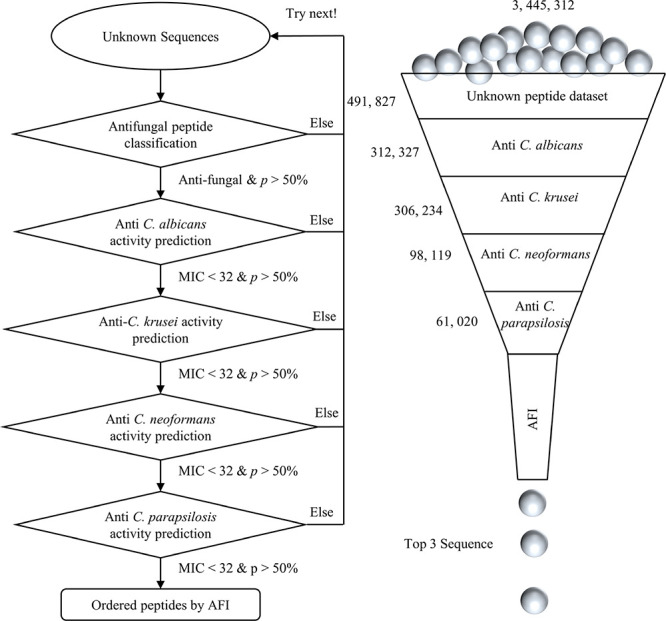
Demonstration of large-scale screening for antifungal peptides.
Table 1. Screened Peptides and the Predicted MIC and AFIa.
| Predicted
MIC/μM (probability/%) |
||||||
|---|---|---|---|---|---|---|
| No. | Sequences | C. albicans | C. krusei | C. neoformans | C. parapsilosis | AFI/μM (probability/%) |
| 1 | KWCFRVCYRGICYRKCR | 1.41(99.1) | 4.25(95.0) | 0.20(63.4) | 16.79(94.4) | 2.11(56.3) |
| 2 | RRWCFRVCYRGFCYRKCR | 1.07(99.2) | 5.22(96.6) | 0.22(72.7) | 20.9(98.7) | 2.25(68.8) |
| 3 | KWCFRVCYRGICYRRCR | 1.87(99.1) | 5.22(95.4) | 0.17(56.0) | 18.25(95.3) | 2.34(50.5) |
| 4 | MRLRKCHKPLTLRLVPWKKQIM | 0.86(99.3) | 9.48(93.1) | 3.70(98.5) | 12.91(96.0) | 4.44(80.5) |
| 5 | MTKILLIVKRLRTVYTKRCLCFRA | 4.15(99.4) | 7.90(78.7) | 1.66(97.7) | 10.87(99.6) | 4.93(74.0) |
| 6 | MYSVFISSIFLFLKIRFKLYPR | 1.18(99.5) | 7.94(91.9) | 5.03(99.3) | 18.98(98.5) | 5.47(81.2) |
| 7 | MRPKWKKKRMRRLKRKRKQRRARYK | 3.59(99.0) | 5.29(70.8) | 4.47(91.7) | 14.5(83.7) | 5.92(49.7) |
| 8 | MNKIFRVIFSKILGRLIVT | 6.94(99.9) | 9.23(97.8) | 1.18(99.1) | 17.65(98.5) | 6.04(88.0) |
| 9 | MTKKQKRKKGIKTKSAGRFGARYGRRIRKAI | 8.71(94.3) | 6.74(67.2) | 4.03(80.0) | 12.52(65.3) | 7.38(29.9) |
| 10 | MKYKKLWALAGIALSCNLLLTA | 1.94(99.5) | 19.59(90.1) | 5.18(99.1) | 16.91(91.8) | 7.60(78.1) |
| 11 | VFQFLGRIIHHVGNFVHGFSHVF | 10.24(99.9) | 8.77(91.1) | 6.63(99.8) | 7.75(98.9) | 8.24(88.9) |
| 12 | MLKKKFGFVFLVCFVIFHSCK | 13.32(99.9) | 8.43(90.8) | 6.74(96.7) | 8.01(98.8) | 8.83(80.1) |
| 13 | YHKIHKVWHIIMKLLAHI | 37.07(99.8) | 9.06(98.3) | 6.51(99.8) | 5.40(96.3) | 10.43(94.3) |
| 14 | YFISCAIHFKILKIRSAAKRREHTKLR | 5.42(97.4) | 10.5(76.9) | 5.33(89.7) | 40.38(63.6) | 10.52(41.4) |
| 15 | MATKKKVVKKAVKKVAKKAPKKAVKKVAKKK | 47.94(92.6) | 8.06(78.3) | 0.84(64.7) | 38.56(68.1) | 10.59(31.9) |
| 16 | PVLKELKSVQKTKSGDTLY | 58.66(99.5) | 29.96(95.0) | 5.79(99.9) | 26.13(97.3) | 22.71(87.2) |
The bold represents the screened top three peptides.
The screened peptides were synthesized and experimentally validated by measuring their antifungal activity. The experimental MIC values against C. albicans SC 5314, C. krusei IFM 56881, C. neoformans H99, and C. parapsilosis ATCC22019 are presented in Table 2. The top three sequences all exhibited excellent antifungal activity. Specifically, the second peptide outperformed most of the reported antifungal peptides in terms of anti-C. neoformans and anti-C. parapsilosis activity. For the remaining 13 screened peptides with moderate activity, nine of them showed moderate antifungal activity.
Table 2. Experimental MIC and AFI of the Screened Peptidesa.
| Experimental
MIC/μM |
||||||
|---|---|---|---|---|---|---|
| No. | Sequences | C. albicans SC 5314 | C. krusei IFM 56881 | C. neoformans H99 | C. parapsilosis ATCC22019 | AFI/μM |
| 1 | KWCFRVCYRGICYRKCR | 14.28 | 28.56 | 1.79 | 7.14 | 8.5 |
| 2 | RRWCFRVCYRGFCYRKCR | 4.07 | 4.07 | 1.02 | 1.02 | 2.0 |
| 3 | KWCFRVCYRGICYRRCR | 14.1 | 28.2 | 3.53 | 7.05 | 10.0 |
| 4 | MRLRKCHKPLTLRLVPWKKQIM | >92.2 | >92.2 | 92.2 | >92.2 | >92.2 |
| 5 | MTKILLIVKRLRTVYTKRCLCFRA | 43.75 | 21.88 | 5.47 | 43.75 | 21.9 |
| 6 | MYSVFISSIFLFLKIRFKLYPR | >92.44 | >92.44 | >92.44 | >92.44 | >92.4 |
| 7 | MRPKWKKKRMRRLKRKRKQRRARYK | >73.77 | >73.77 | 2.31 | 18.44 | >21.9 |
| 8 | MNKIFRVIFSKILGRLIVT | 14.23 | 56.92 | 7.12 | 14.23 | 16.9 |
| 9 | MTKKQKRKKGIKTKSAGRFGARYGRRIRKAI | >70.71 | >70.71 | 8.84 | 4.42 | 21.0 |
| 10 | MKYKKLWALAGIALSCNLLLTA | >105.7 | >105.7 | >105.7 | >105.7 | >105.7 |
| 11 | VFQFLGRIIHHVGNFVHGFSHVF | >94.99 | >94.99 | >94.99 | >94.99 | >95.0 |
| 12 | MLKKKFGFVFLVCFVIFHSCK | >101.5 | >101.5 | >101.5 | >101.5 | >101.5 |
| 13 | YHKIHKVWHIIMKLLAHI | 28.06 | 7.02 | 7.02 | 14.03 | 11.8 |
| 14 | YFISCAIHFKILKIRSAAKRREHTKLR | 77.88 | 19.47 | 38.94 | 77.88 | 46.3 |
| 15 | MATKKKVVKKAVKKVAKKAPKKAVKKVAKKK | >74.87 | >74.87 | 37.44 | >74.87 | >63.0 |
| 16 | PVLKELKSVQKTKSGDTLY | >119.94 | >119.94 | >119.94 | >119.94 | >119.9 |
The bold highlights the MIC values less than 10 μM.
Comparisons were conducted by querying the selected three top-ranking peptides on other online prediction platforms. In Antifp,22 the three top-ranking peptides were predicted to be antifungals with scores of 0.49, 0.56, and 0.52, respectively, but without activity against specific fungi. In DBAASPv3.0,16 the first and second were predicted to be nonantimicrobials, and only the third peptide was an antimicrobial peptide. In CAMPR3,19 the screened peptides were predicted to be antimicrobials with relatively high scores (0.959–1.000) but still without antifungal activity. As mentioned above, approximately 14% of the peptides with appropriate lengths in the Uniprot database are plausible antifungal, but only a few of these peptides have been reported to have specific activity. This implies that the antifungal activity of most peptides may be somewhat less worthy of being reported or studied. The comparison highlights that the activity prediction models are indispensable and the proposed protocol is more comprehensive and valuable for antifungal peptide screening.
Herein, we develop a screening protocol for antifungal peptides, in which an accurate classification model and four activity prediction models against specified fungi were included. Through the protocol, three promising antifungal peptides were screened from three million sequences within a few days in a personal computer. The screened peptides were synthesized and experimentally validated. The results confirmed the outstanding antifungal properties. Compared with the previous experimental identification process, the protocol was fairly efficient for large-scale antifungal peptide screening.
Acknowledgments
This work was supported by National Natural Science Foundation of China (Nos. 22004022, 81760647, and 82060381), Guizhou Science and Technology Department (No. ZK[2021]045), Department of Education of Guizhou Province of China (No. KY[2021]163), Excellent Young Talents Plan of Guizhou Medical University ([2021]104), and Guizhou Medical University (Nos. 19NSP067 and [2019]002).
Glossary
Abbreviations
- MIC
minimum inhibitory concentration
- pMIC
logarithmic transform of MIC
- C. albicans
Candida albicans
- C. krusei
Candida krusei
- C. neoformansm
Cryptococcus neoformans
- C. parapsilosis
Candida parapsilosis
- R
correlation coefficient
- RMSE
root-mean-square error
- R2
coefficient of determination
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsmedchemlett.1c00556.
Text S1–S4: Details about model establishment for antifungal classification, activity prediction, screening protocol, and antifungal peptide screening application. Table S1: Peptide descriptors used in this study. Table S2: Usage of peptide descriptors in antifungal identification. Tables S3 and S4: Results of antifungal peptide classification and activity prediction (PDF)
Mass spectra of all synthesized peptides (ZIP)
Author Contributions
The manuscript was written through contributions of all authors.
The authors declare no competing financial interest.
Supplementary Material
References
- Hamley I. W. Small bioactive peptides for biomaterials design and therapeutics. Chem. Rev. 2017, 117 (24), 14015–14041. 10.1021/acs.chemrev.7b00522. [DOI] [PubMed] [Google Scholar]
- Muttenthaler M.; King G. F.; Adams D. J.; Alewood P. F. Trends in peptide drug discovery. Nat. Rev. Drug Discovery 2021, 20 (4), 309–325. 10.1038/s41573-020-00135-8. [DOI] [PubMed] [Google Scholar]
- Dang T.; Süssmuth R. D. Bioactive peptide natural products as lead structures for medicinal use. Acc. Chem. Res. 2017, 50 (7), 1566–1576. 10.1021/acs.accounts.7b00159. [DOI] [PubMed] [Google Scholar]
- Fosgerau K.; Hoffmann T. Peptide therapeutics: current status and future directions. Drug Discovery Today 2015, 20 (1), 122–128. 10.1016/j.drudis.2014.10.003. [DOI] [PubMed] [Google Scholar]
- Sharma D.; Bisht G. S. Recent Updates on Antifungal Peptides. Mini-Rev. Med. Chem. 2020, 20 (4), 260–268. 10.2174/1389557519666190926112423. [DOI] [PubMed] [Google Scholar]
- Fernández de Ullivarri M.; Arbulu S.; Garcia-Gutierrez E.; Cotter P. D. Antifungal peptides as therapeutic agents. Front. Cell. Infect. Microbiol. 2020, 10, 105. 10.3389/fcimb.2020.00105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautenbach M.; Troskie A. M.; Vosloo J. A. Antifungal peptides: To be or not to be membrane active. Biochimie 2016, 130, 132–145. 10.1016/j.biochi.2016.05.013. [DOI] [PubMed] [Google Scholar]
- Yang L. B.; Guo G.; Zhao X. Y.; Su P. P.; Fu P.; Peng J.; Xiu J. F.; Li B. Y. Antifungal activity and physicochemical properties of a aovel antimicrobial protein AMP-17 from Musca domestica. Pol. J. Microbiol. 2019, 68 (3), 383–390. 10.33073/pjm-2019-041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H. L.; Zhao X. Y.; Yang L. B.; Su P. P.; Fu P.; Peng J.; Yang N.; Guo G. Antimicrobial peptide AMP-17 affects Candida albicans by disrupting its cell wall and cell membrane integrity. Infect. Drug Resist. 2020, 13, 2509–2520. 10.2147/IDR.S250278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G.; Tao R. Y.; Li Y.; Ma H. L.; Xiu J. F.; Fu P.; Wu J. W. Identification and characterization of a novel antimicrobial protein from the housefly Musca domestica. Biochem. Biophys. Res. Commun. 2017, 490 (3), 746–752. 10.1016/j.bbrc.2017.06.112. [DOI] [PubMed] [Google Scholar]
- Das P.; Sercu T.; Wadhawan K.; Padhi I.; Gehrmann S.; Cipcigan F.; Chenthamarakshan V.; Strobelt H.; dos Santos C.; Chen P.-Y.; Yang Y. Y.; Tan J. P. K.; Hedrick J.; Crain J.; Mojsilovic A. Accelerated antimicrobial discovery via deep generative models and molecular dynamics simulations. Nature Biomedical Engineering 2021, 5 (6), 613–623. 10.1038/s41551-021-00689-x. [DOI] [PubMed] [Google Scholar]
- Capecchi A.; Cai X.; Personne H.; Köhler T.; van Delden C.; Reymond J.-L. Machine learning designs non-hemolytic antimicrobial peptides. Chemical Science 2021, 12 (26), 9221–9232. 10.1039/D1SC01713F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bo W. C.; Chen L.; Qin D. Y.; Geng S.; Li J. Q.; Mei H.; Li B.; Liang G. Z. Application of quantitative structure-activity relationship to food-derived peptides: Methods, situations, challenges and prospects. Trends Food Sci. Technol. 2021, 114, 176–188. 10.1016/j.tifs.2021.05.031. [DOI] [Google Scholar]
- Cao D. S.; Xu Q. S.; Liang Y. Z. propy: a tool to generate various modes of Chou’s PseAAC. Bioinformatics 2013, 29 (7), 960–962. 10.1093/bioinformatics/btt072. [DOI] [PubMed] [Google Scholar]
- Müller A. T.; Gabernet G.; Hiss J. A.; Schneider G. modlAMP: Python for antimicrobial peptides. Bioinformatics 2017, 33 (17), 2753–2755. 10.1093/bioinformatics/btx285. [DOI] [PubMed] [Google Scholar]
- Pirtskhalava M.; Amstrong A. A.; Grigolava M.; Chubinidze M.; Alimbarashvili E.; Vishnepolsky B.; Gabrielian A.; Rosenthal A.; Hurt D. E.; Tartakovsky M. DBAASP v3: database of antimicrobial/cytotoxic activity and structure of peptides as a resource for development of new therapeutics. Nucleic Acids Res. 2021, 49 (D1), D288–D297. 10.1093/nar/gkaa991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. s.; Li X.; Wang Z. APD3: the antimicrobial peptide database as a tool for research and education. Nucleic Acids Res. 2016, 44 (D1), D1087–D1093. 10.1093/nar/gkv1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang X. Y.; Dong F. Y.; Shi C.; Liu S. C.; Sun J.; Chen J. X.; Li H. q.; Xu H. M.; Lao X. Z.; Zheng H. DRAMP 2.0, an updated data repository of antimicrobial peptides. Sci. Data 2019, 6 (1), 148. 10.1038/s41597-019-0154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waghu F. H.; Barai R. S.; Gurung P.; Idicula-Thomas S. CAMPR3: a database on sequences, structures and signatures of antimicrobial peptides. Nucleic Acids Res. 2016, 44 (D1), D1094–D1097. 10.1093/nar/gkv1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciociola T.; Magliani W.; De Simone T.; Pertinhez T. A.; Conti S.; Cozza G.; Marin O.; Giovati L. In silico predicted antifungal peptides: In vitro and in vivo anti-Candida activity. J. Fungi 2021, 7 (6), 439. 10.3390/jof7060439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A.; Akbar S.; Khan S.; Hayat M.; Ali F.; Ahmed A.; Tahir M. Deep-AntiFP: Prediction of antifungal peptides using distanct multi-informative features incorporating with deep neural networks. Chemom. Intell. Lab. Syst. 2021, 208, 104214. 10.1016/j.chemolab.2020.104214. [DOI] [Google Scholar]
- Agrawal P.; Bhalla S.; Chaudhary K.; Kumar R.; Sharma M.; Raghava G. P. S. In silico approach for prediction of antifungal peptides. Front Microbiol 2018, 9 (323), 323. 10.3389/fmicb.2018.00323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Li B. Y.; Hu Y.; Zhou L. x.; Wang G. Z.; Guo G.; Zhang Q. H.; Lei S. C.; Zhang A. H. A parameter-free framework for calibration enhancement of near-infrared spectroscopy based on correlation constraint. Anal. Chim. Acta 2021, 1142, 169–178. 10.1016/j.aca.2020.11.006. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Guo C.; Cai W. S.; Shao X. G. Direct non-trilinear decomposition for analyzing high-dimensional data with imperfect trilinearity. Chemom. Intell. Lab. Syst. 2021, 210, 104244. 10.1016/j.chemolab.2021.104244. [DOI] [Google Scholar]
- Zhang J.; Guo C.; Cui X. Y.; Cai W. S.; Shao X. G. A two-level strategy for standardization of near infrared spectra by multi-level simultaneous component analysis. Anal. Chim. Acta 2019, 1050, 25–31. 10.1016/j.aca.2018.11.013. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Cui X. Y.; Cai W. S.; Shao X. G. A variable importance criterion for variable selection in near-infrared spectral analysis. Sci. China: Chem. 2019, 62 (2), 271–279. 10.1007/s11426-018-9368-9. [DOI] [Google Scholar]
- Zhang J.; Cui X. Y.; Cai W. S.; Shao X. G. Combination of heuristic optimal partner bands for variable selection in near-infrared spectral analysis. J. Chemom. 2018, 32 (11), e2971. 10.1002/cem.2971. [DOI] [Google Scholar]
- Pedregosa F.; Varoquaux G.; Gramfort A.; Michel V.; Thirion B.; Grisel O.; Blondel M.; Prettenhofer P.; Weiss R.; Dubourg V. Scikit-learn: Machine learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


