In this study, Pyatnitskaya et al. investigate the mechanism underlying how crossover formation is functionally linked to synaptonemal complex (SC) assembly between homologous chromosomes. They show that Ecm11, a SC central element protein, localizes on both DSB sites and sites that attach chromatin loops to the chromosome axis, which are the starting points of SC formation, in a way that strictly requires the ZMM protein Zip4.
Keywords: aneuploidy, crossing over, homologous recombination, meiosis, chromosome segregation, DSB repair, protein–protein interactions, homologous synapsis
Abstract
Meiotic recombination is triggered by programmed double-strand breaks (DSBs), a subset of these being repaired as crossovers, promoted by eight evolutionarily conserved proteins, named ZMM. Crossover formation is functionally linked to synaptonemal complex (SC) assembly between homologous chromosomes, but the underlying mechanism is unknown. Here we show that Ecm11, a SC central element protein, localizes on both DSB sites and sites that attach chromatin loops to the chromosome axis, which are the starting points of SC formation, in a way that strictly requires the ZMM protein Zip4. Furthermore, Zip4 directly interacts with Ecm11, and point mutants that specifically abolish this interaction lose Ecm11 binding to chromosomes and exhibit defective SC assembly. This can be partially rescued by artificially tethering interaction-defective Ecm11 to Zip4. Mechanistically, this direct connection ensuring SC assembly from CO sites could be a way for the meiotic cell to shut down further DSB formation once enough recombination sites have been selected for crossovers, thereby preventing excess crossovers. Finally, the mammalian ortholog of Zip4, TEX11, also interacts with the SC central element TEX12, suggesting a general mechanism.
Meiosis is a highly conserved process among organisms with sexual development. It produces four haploid gametes from one diploid cell by executing two successive rounds of cell division preceding one round of DNA replication (Hunter 2015). A unique defining feature of meiosis is the pairing/synapsis and homologous recombination between parental chromosomes (homologs). Recombination is initiated by programmed DNA double-strand break (DSB) formation by the topoisomerase-related Spo11 protein together with several meiotic protein partners (Yadav and Claeys Bouuaert 2021). Following DSB formation, the combined action of endo- and exonucleases leads to resection of the DSB 5′ ends, creating 3′ single-strand DNA tails. The strand exchange proteins Rad51 and Dmc1 bind to these tails and form a nucleoprotein filament that invades the homologous chromosome. This results in the formation of a D-loop intermediate that goes through various steps of maturation, leading to two possible outcomes: a crossover (CO) with a physical exchange between chromosomal arms, or a noncrossover (NCO). Meiotic COs can be subdivided into two classes, with class I COs representing ∼85% of total COs formed in budding yeast, mammals, and plants. A characteristic of class I COs is that they are more evenly spaced from each other than would be expected from a random distribution, a phenomenon referred to as “interference” (Berchowitz and Copenhaver 2010). The ZMM group of proteins (Zip1–4, Msh4–5, Mer3, and Spo16) is the major actor promoting class I CO formation (Börner et al. 2004; Pyatnitskaya et al. 2019). Molecularly, these proteins are proposed to act on D-loop recombination intermediates by protecting them against their dismantling by helicases, which would lead to NCO (De Muyt et al. 2012; Zakharyevich et al. 2012). ZMM-protected intermediates are then matured into a particular DNA structure that will be further processed into CO by the endonuclease activity of the MutLγ (Mlh1–Mlh3)–Exo1 complex (Hunter and Kleckner 2001; De Muyt et al. 2012; Zakharyevich et al. 2012). Among the ZMM proteins, the Zip2–Zip4–Spo16 complex plays a predominant role, through its XPF–ERCC1-like module, in specifically binding branched recombination intermediates (De Muyt et al. 2018; Arora and Corbett 2019). In addition, this complex has a scaffolding activity through its Zip4 (originally named Spo22) subunit. Indeed, Zip4 interacts with several other ZMM proteins as well as with Red1, a component of the meiotic chromosome axis (axial element), forming the lateral element of the synaptonemal complex (SC) during homolog synapsis (De Muyt et al. 2018).
The SC appears concomitantly with the maturation of the ZMM-protected recombination intermediates. It is composed of two lateral elements physically maintained together at a precise distance of 100 nm by a central region (Zickler and Kleckner 1999). SC assembly begins with the formation of the axial element along each pair of sister chromatids. Polymerization of axial elements leads to arrays of chromatin loops tethered at their bases to the axial proteins, among which are the meiosis-specific Hop1 and Red1 proteins, and cohesin containing the Rec8 subunit (Smith and Roeder 1997; Klein et al. 1999; Panizza et al. 2011). Homologous chromosomes coalign across their length, and then the central region polymerizes from punctuate sites to progressively connect axial elements of the two homologs until the chromosomes are synapsed along their entire length (Moses 1969; de Boer and Heyting 2006). In budding yeast, the central region is composed of the transverse filament Zip1 and the central element, including Ecm11 and Gmc2, which facilitate Zip1 assembly (Sym et al. 1993; Humphryes et al. 2013; Gao and Colaiácovo 2018).
Whereas in some organisms (C. elegans and D. melanogaster) SC formation is crossover-independent and some (S. pombe and Aspergilus) do meiosis without forming SC, in budding yeast, plants, and mammals, CO formation and SC polymerization are spatially and functionally related (for review, see Pyatnitskaya et al. 2019). Indeed, SC polymerization often initiates from sites called SICs (synapsis initiation complexes), which are enriched in ZMM, and therefore likely represent recombination intermediates, where ZMMs were shown to bind by ChIP-seq approaches (Chua and Roeder 1998; Agarwal and Roeder 2000; Tsubouchi et al. 2006; Shinohara et al. 2008; Serrentino et al. 2013; De Muyt et al. 2018). In Sordaria macrospora, SC also nucleates from recombination nodules, aggregates of active recombination proteins including ZMMs (Dubois et al. 2019). In mammals, whether SC polymerization starts from ZMM-enriched sites is still not fully established. However, a large majority of RNF212, related to the ZMM Zip3 protein, colocalizes with initial stretches of SYCP1, the mouse homolog of Zip1, suggesting that such a mechanism occurs in mammals (Reynolds et al. 2013). Moreover, the absence of ZMM proteins leads to synapsis defects in budding yeast, Sordaria, and mice, suggesting that stabilization of CO precursors is important for correct SC polymerization (for review, see Pyatnitskaya et al. 2019).
On the other hand, the SC is involved in regulating the number and position of crossovers. Whether the SC is involved in mediating crossover interference has been investigated in several model organisms. In budding yeast, this is clearly not the case. A deletion mutant of Zip1, the transverse filament of the SC but also a ZMM protein, is defective in interfering COs. However, in mutants where Zip1 still binds recombination intermediates but does not polymerize, such as the Nter deletion zip1N1 mutant or the central element ecm11Δ and gmc2Δ mutants, COs still interfere, although the strength of interference is slightly but significantly reduced (Voelkel-Meiman et al. 2015, 2016; Lee et al. 2021). These mutants likely preserve the Zip1 “ZMM function” intact, which is independent of its SC assembly function (Börner et al. 2004; Chen et al. 2015; Voelkel-Meiman et al. 2015, 2016). Although SC polymerization is not formally required for the formation of interfering COs, it does seem to play a regulatory role in their distribution. In budding yeast, despite wild-type spore viability, zip1N1, ecm11Δ, and gmc2Δ mutants show increased CO frequency on certain chromosomes, suggesting that the SC could limit ZMM-dependent CO formation (Voelkel-Meiman et al. 2016, 2019; Lee et al. 2021). This may be explained at least in part by recent findings that Ecm11- and Gmc2-dependent SC assembly down-regulates DSB formation by Spo11 (Mu et al. 2020; Lee et al. 2021). Similarly, in plants, mutants of the transverse filament ZEP1 and AtZYP1 in rice and Arabidopsis, respectively, show more COs, indicating that, like in budding yeast, the SC is regulating crossover frequencies. However, contrary to budding yeast, these crossovers lost interference, although they still seem to depend on ZMM (Wang et al. 2010; Capilla-Pérez et al. 2021; France et al. 2021). Similarly, in C. elegans, partial depletion of the synaptonemal complex central region proteins reduces the effective distance over which interference operates, suggesting that synaptonemal complex proteins also limit crossovers in nematodes (Libuda et al. 2013). These apparent differences from fungi deserve further investigation but may stem from the fact that progression through meiosis in plants is not affected by the absence of ZMM proteins.
Despite the temporal and spatial relationships between CO formation and SC assembly, the underlying physical connections between the two processes are elusive. Here, we uncovered a direct interaction between the ZMM protein Zip4 and the central components of the SC Ecm11 and Gmc2 that is essential for the recruitment of the Ecm11 protein to chromosomes and consequently for SC polymerization. We propose a model in which Zip4 brings Ecm11 to recombination sites that are prone to form COs and helps the transverse filament protein Zip1 to nucleate from this location, ensuring a control of recombination starting locally from sites engaged in the crossover repair pathway.
Results
The central element protein Ecm11 interacts with Zip4 and is recruited to DSB and axis attachment sites
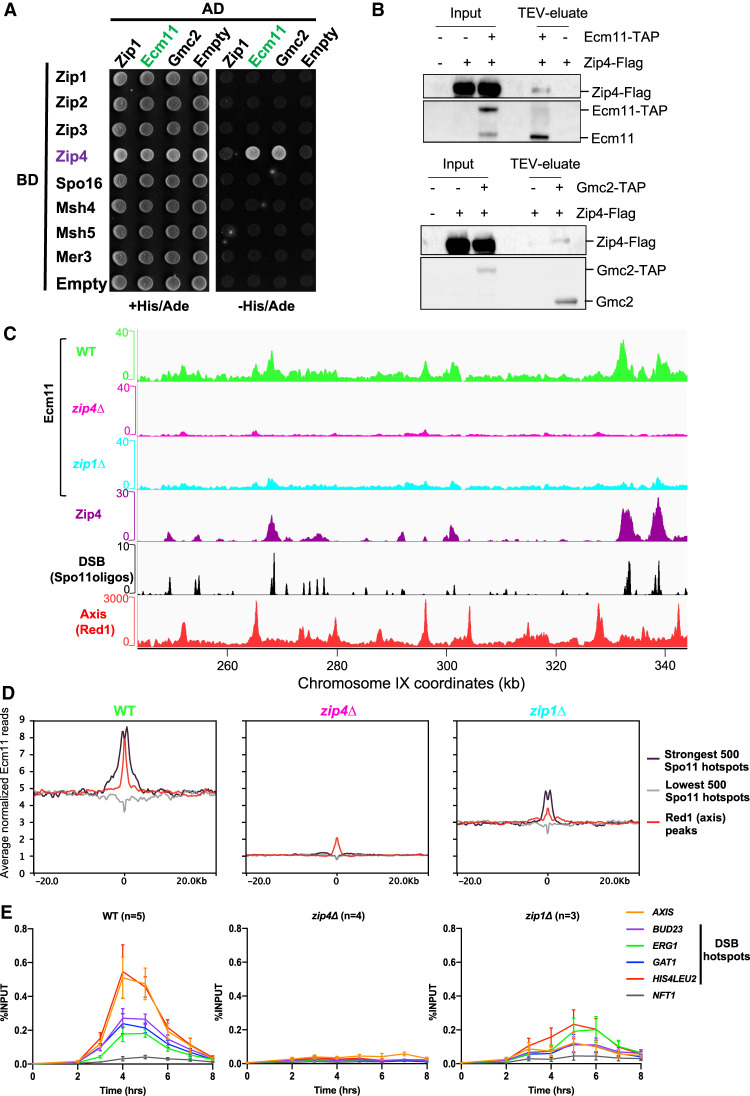
To investigate possible physical connections between crossover formation and synaptonemal complex assembly pathways, we systematically tested by yeast two-hybrid the interactions between ZMM proteins and the known SC components (Fig. 1A). The only interactions were between Zip4 and each of the two known SC central elements: Ecm11 and Gmc2 (Fig. 1A). We confirmed that the endogenous proteins interact in meiotic cells by coimmunoprecipitating Zip4-Flag protein with Ecm11-TAP or Gmc2-TAP (Fig. 1B). Since Zip4 is known to be recruited to recombination sites, we next asked whether Ecm11 shows a similar binding pattern by mapping Ecm11 binding sites at 5 h in meiosis, the expected time of recombination (Hunter and Kleckner 2001), using spike-in calibrated ChIP-seq (Fig. 1C,D; Hu et al. 2015). Strikingly, Ecm11 preferentially localized, like Zip4, at DSB hotspots, indicating that Ecm11 is present at the recombination intermediates. In addition, Ecm11 also preferentially associated to Red1 binding sites, which define the basis of chromatin loops attached to the chromosome axis, where SC polymerizes, consistent with Ecm11 being a component of the SC (Fig. 1C,D; Supplemental Fig. S1). Looking at the kinetics of Ecm11 association with chromatin by ChIP-qPCR revealed that Ecm11 binding to DSB and axis attachment sites was maximum at 4–5 h in meiosis during recombination (Fig. 1E). Then, we sought to find the determinants for Ecm11 association to chromosomes, first by testing whether Zip4 is involved. Indeed, Ecm11 recruitment to chromatin was drastically reduced in a zip4Δ mutant on both DSB and axis sites (Fig. 1C–E; Supplemental Fig. S1). Previous studies have suggested that Zip1 may be important for Ecm11 loading (Voelkel-Meiman et al. 2015, 2016). Interestingly, the recruitment of Ecm11 to DSB hotspots was only partially reduced in the absence of Zip1, while the association with the axis-binding sites was more strongly impaired (Fig. 1C–E; Supplemental Fig. S1). We asked whether the reduced Ecm11 association to chromosomes in zip1Δ may be a consequence of reduced Zip4 binding to chromosomes. Indeed, Zip4 enrichment was strongly reduced in the absence of Zip1, which is likely the reason for reduced Ecm11 binding in zip1Δ (Supplemental Fig. S2). Zip1 therefore seems important for full Ecm11 localization at the SC, likely because Ecm11–Gmc2 copolymerize together with Zip1, but less so for its recruitment to recombination sites.
Figure 1.
Ecm11 localization on DSBs and axis -attachment sites is dependent on Zip4. (A) Yeast two-hybrid interaction analysis between SC components Ecm11 and Gmc2 and the ZMM proteins. Prey and baits are fused with the GAL4 activation domain (GAL4-AD) and with the GAL4 DNA-binding domain (GAL4-BD), respectively. Interaction results in growth on the selective −His/Ade medium. (B) Coimmunoprecipitation between Zip4-Flag and Ecm11-TAP (top panel) or Gmc2-TAP (bottom panel) from meiotic cells at 5 h in meiosis, analyzed by Western blot. The TAP antibody still detects the CBP (calmodulin binding protein) moiety that remains bound to Ecm11 or Gmc2 after TEV cleavage of the TAP tag. (C) ChIP-seq DNA binding of Ecm11-Flag in WT, zip4Δ, and zip1Δ strains. Normalized data were smoothed with a 200-bp window. Zip4 binding profile is also shown (De Muyt et al. 2018). DSB sites are mapped by Spo11 oligos (Zhu and Keeney 2015), and axis attachment sites are mapped by Red1 binding profile (Sun et al. 2015). (D) Average Ecm11 ChIP-seq signal of data shown in A at the indicated features. Alignments were performed on the Spo11 hotspots midpoints from Zhu and Keeney (2015) and Red1 peaks summits from Sun et al. (2015). See also Supplemental Figure S1. (E) ChIP monitoring of Ecm11-Flag association with different chromosomal regions, measured by qPCR using primers that cover the indicated regions. Same strains as in C were used. Values are the mean ± SEM of the indicated number of independent experiments.
Finally, our quantitative Ecm11 ChIP-seq data also revealed relatively uniform Ecm11 binding outside of recombination hotspots and axis sites, which was strongly diminished in the absence of Zip4 (Fig. 1C,D). This was confirmed by qPCR with the enrichment of Ecm11 at the NFT1 site, a locus that shows neither DSB nor detectable axis protein signal (Fig. 1E; Sun et al. 2015; Zhu and Keeney 2015). Such nonspecific binding may reflect, in addition to preferential sites, a mobility of the loop attachment sites to the chromosome axis that may be mediated by constant loop extrusion by cohesin at the basis of these loops, as recently shown in mammalian cells (Fudenberg et al. 2016).
Altogether, we conclude that Ecm11 localizes at recombination sites and along the chromosome axis in a Zip4-dependent manner.
Zip4–Ecm11 interaction is important for normal SC polymerization
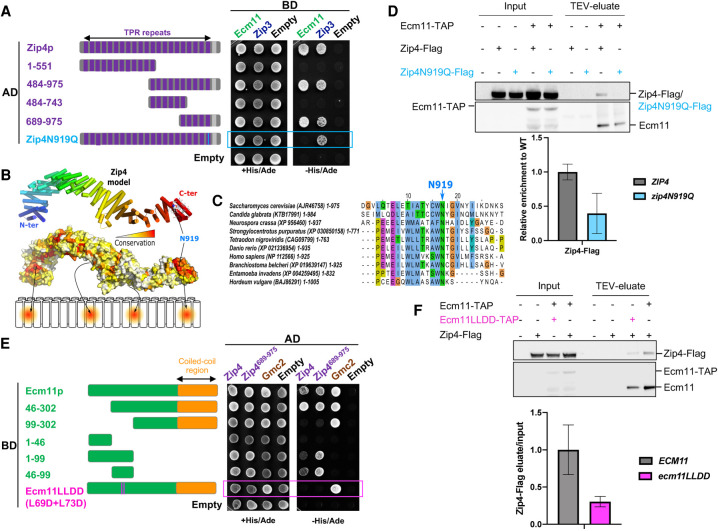
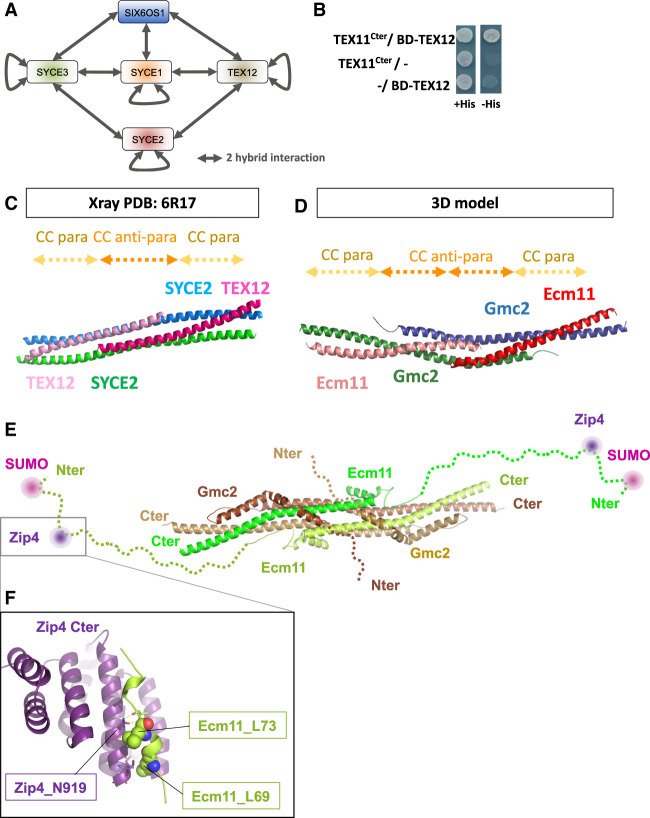
To further investigate the role of the Zip4–Ecm11 interaction in meiosis, we characterized the domains of Zip4 and Ecm11 mediating the interaction. Zip4 encompasses 21 TPR (tetratricopeptide repeat) motifs spanning the whole length of Zip4 and ends with a C-terminal α helix (Fig. 2A; Perry et al. 2005). We generated a 3D model of Zip4, which revealed an extensive surface featuring four distinct conserved patches likely to be involved in protein interactions (Fig. 2B). We used this model to delineate fragments of Zip4 sufficiently long to enable proper folding and maintain interactions without disrupting the conserved patches (Fig. 2A). Yeast two-hybrid experiments showed that Ecm11 interacts with the last C-terminal fragment that contains the most conserved patch of Zip4 (Fig. 2A; Supplemental Fig. S3A). A search for conserved and surface-exposed amino acids potentially involved in protein–protein interactions in this region uncovered a highly conserved aromatic asparagine motif (residues W918–N919 in S. cerevisiae Zip4) (Fig. 2C). This motif is often present in different binding scaffolds, such as in the Armadillo repeats of importin α for interaction with NLS motifs (Fontes et al. 2000). Exposed and conserved asparagine residues in these domains are typically found to mediate specific interactions with the backbone amide groups of the binding partner. Therefore, we mutated this motif by substituting the asparagine 919 with a glutamine (Zip4N919Q), changing only the steric hindrance to have a minimal effect on the rest of the protein. Remarkably, Zip4N919Q completely lost its interaction with Ecm11, as assessed by yeast two-hybrid assay, while keeping its interaction with Zip4's other known partners: Zip3 and Zip2 (Fig. 2A; Supplemental Fig. S3B). Co-IP experiments from meiotic cells also confirmed that the interaction between the Zip4N919Q mutant and Ecm11 was strongly reduced in vivo (Fig. 2D).
Figure 2.
Zip4 specifically interacts with Ecm11. (A) Delineation of the Ecm11-interacting domain in Zip4 by two-hybrid assays. The indicated fragments of Zip4 were fused to GAL4-AD and tested in combination with a GAL4-BD-Ecm11 or GAL4-BD-Zip3 fusion. The blue frame indicates the absence of interaction between Zip4N919Q and Ecm11. (B) 3D model of Zip4 TPR revealing four conserved surface patches. The degree of conservation is shown. (C) Alignment of the Zip4 C-terminal TPR domain. (D) Coimmunoprecipitation between Zip4-Flag, Zip4N919Q-Flag, and Ecm11-TAP from meiotic cells at 5 h in meiosis, analyzed by Western blot. Levels of Zip4-Flag and Zip4N919Q-Flag were quantified relative to the input and normalized by Ecm11-TAP levels. Values are the mean ± SD of two independent experiments. (E) Same assay as in A. Ecm11 domains were fused to GAL4-AD and tested in combination with GAL4-BD-Zip4 or GAL4-BD-Zip4-689–971. The pink frame indicates the loss of interaction between Ecm11LLDD and Zip4. (F) Coimmunoprecipitation of Zip4-Flag with Ecm11-TAP or with Ecm11LLDD-TAP from meiotic cells at 4 h in meiosis, analyzed by Western blot. Levels of Zip4-Flag coimmunoprecipitated with Ecm11-TAP or with Ecm11LLDD-TAP were quantified relative to the input and normalized by Ecm11-TAP or Ecm11LLDD-TAP levels. Values are the mean ± SD of two independent experiments.
To delineate the Ecm11 regions interacting with Zip4, we further analyzed the C-terminal conserved patch of Zip4 in the vicinity of asparagine 919 and identified a set of four exposed apolar residues distributed over the 19th and 20th TPRs (Supplemental Fig. S4A), suggesting that the Ecm11 binding region should contain a significant number of conserved hydrophobic residues to interact with this region. From the multiple sequence alignments of Ecm11 (Supplemental Fig. S4B), sequence analysis predicts the existence of a long disordered N-terminal tail extended by a 70-residue coiled coil in the C terminus. A short stretch spanning residues 68–76 in the disordered tail contains two conserved and hydrophobic positions and a propensity to adopt a helical conformation, making this region a good candidate for interacting with Zip4 in the vicinity of N919. For the interaction between Ecm11 and Gmc2, we exploited a coevolution-based analysis, which suggested that the C-terminal coiled coil of Ecm11 could most likely form antiparallel and parallel coiled coils with Gmc2 (Supplemental Fig. S8). We validated these predictions by Y2H experiments, where the domain 46–99 of Ecm11 was sufficient to interact with Zip4, while the coiled-coil region 212–302 was critical for Gmc2 interaction but not for Zip4 binding (Fig. 2E; Supplemental Fig. S4C). Within the 46–99 region of Ecm11, two well-conserved hydrophobic residues, leucines 69 and 73, are good candidates for Zip4 interaction (Supplemental Fig. S4B). Indeed, their mutation to aspartate (generating the Ecm11L69D-L73D mutant, referred to here as Ecm11LLDD) disrupted the Ecm11–Zip4 interaction, while preserving the Ecm11–Gmc2 interaction in yeast two-hybrid assays (Fig. 2E). This effect was confirmed in vivo where the interaction of Ecm11LLDD with Zip4 was decreased (Fig. 2F). Altogether, these results indicate that Zip4 and Ecm11 interact directly through a region on Ecm11 distinct from the Gmc2-binding region, which establishes a physical connection between CO formation and SC assembly processes.
Disturbing the Zip4–Ecm11 interaction strongly affects Ecm11 recruitment and SC assembly
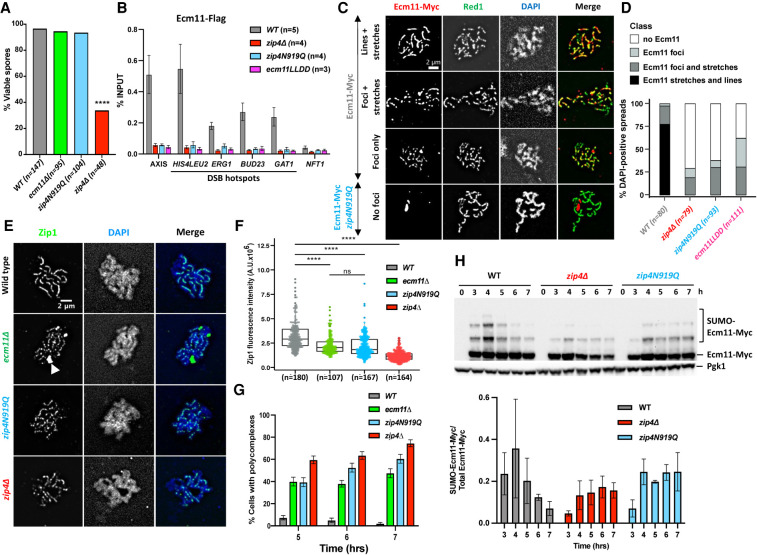
To address the function of the interaction between Zip4 and Ecm11 during meiotic prophase I, we first assessed spore viability and meiotic progression in the interaction mutants. The zip4N919Q and ecm11LLD mutants showed wild-type spore viability, like ecm11Δ but in sharp contrast to zip4Δ (Fig. 3A). In addition, they both showed a shorter delay in meiotic divisions (3 h and 1.5 h, respectively) than zip4Δ (>5 h) (Supplemental Fig. S5A). Together, these data suggest that the Zip4–Ecm11 interaction is not needed for Zip4 ZMM functions in CO formation and that Zip4 fully associates with chromosomes without the need for Ecm11. We noted that the zip4N919Q was slightly more delayed than ecm11Δ (1.5-h delay), which may be related to the lower levels of the Zip4N919Q protein detected during meiosis, affecting the “ZMM” activities of Zip4, whereas Zip4 levels are not affected by ecm11Δ (Supplemental Fig. S5B).
Figure 3.
Synaptonemal complex assembly depends on the interaction of Ecm11 with Zip4. (A) Spore viability assays of strains with the indicated genotype. Numbers of dissected tetrads are indicated. (****) P < 0.0001, Fisher's exact test. (B) Maximum levels of Ecm11-Flag or Ecm11LLDD-Flag in the indicated strains measured by quantitative PCR (qPCR) using primers that cover the indicated regions are shown. Values are the mean ± SEM from at least three independent experiments. The full corresponding time courses are in Figure 1E and Supplemental Figure S5. (C) Ecm11-Myc localization on surface-spread chromosomes in the indicated strains. (Red) Anti-Myc, (green) anti-Red1, (blue) DAPI. Red1-positive spreads were divided into four categories: (1) exhibiting stretches and lines of Ecm11 (synapsis almost complete or complete), (2) exhibiting foci and stretches of Ecm11 (partial synapsis), (3) exhibiting only Ecm11 foci (dotty pattern), and (4) exhibiting no Ecm11. Representative pictures are shown for the indicated strain. The pictures for the other strains are in Supplemental Figure S6. (D) Quantification of the classes shown in C. The number of counted spreads is indicated. (E) Zip1 localization on surface-spread chromosomes in the indicated strains. Only pachytene or pachytene-like stages were considered. (Green) Anti-Zip1, (blue) DAPI (DNA), (white arrow) Zip1 polycomplex. (F) Quantification of total nuclear Zip1 intensity observed in E. Numbers of spreads are indicated for each genotype. (****) P-value < 0.0001, Wilcoxon test. (G) Quantification of DAPI-positive spreads showing a polycomplex. At least 200 spreads were considered for each condition. Values are percentage of cells ± SD of the proportion. (H) Ecm11 SUMOylation in the indicated strains analyzed by Western blot. Quantification is from two independent experiments, with the mean ratio ± SD of SUMOylated versus total Ecm11 protein indicated.
Since the Zip4–Ecm11 interaction itself is not important for Zip4 ZMM function, we next assessed whether it is involved in Ecm11 recruitment to chromatin. Indeed, ChIP-qPCR analyses revealed that Ecm11 was no longer recruited to all tested loci in both zip4N919Q and ecm11LLDD mutants (Fig. 3B; Supplemental Fig. S5C). This loss was further confirmed by Ecm11 and Red1 coimmunostaining of chromosome spreads, where zip4N919Q and ecm11LLDD cells showed no staining or discontinuous Ecm11 pattern, in contrast to wild type, where 75% of meiotic cells showed a continuous Ecm11 pattern (Fig. 3C,D; Supplemental Fig. S6).
We next assessed the consequences of these Ecm11 loading defects on SC assembly by Zip1 immunostaining of meiotic chromosome spreads. In wild-type cells at 5 h (pachytene stage), Zip1 staining was observed along the entire length of the chromosomes (Fig. 3E, top panel). In contrast to wild type, but similar to zip4Δ and ecm11Δ, both zip4N919Q and ecm11LLDD mutants exhibited a discontinuous Zip1 pattern and decrease of the Zip1 fluorescence signal intensity (Fig. 3E,F). In the interaction mutants, Zip1 localization defects were accompanied by the formation of Zip1 aggregates (polycomplexes), like in ecm11Δ (Fig. 3E [arrow], G). Altogether, we showed that the Zip4–Ecm11 interaction is necessary for Ecm11 recruitment to chromosomes and normal SC assembly.
Previous studies have shown that Ecm11 is SUMOylated depending on the Siz1 and Siz2 E3 ligases and that this is required for SC polymerization (Humphryes et al. 2013; Leung et al. 2015). However, we do not know whether SUMOylation is linked to Ecm11 recruitment to chromosomes. In our interaction mutant zip4N919Q, we found that Ecm11 was still SUMOylated (Fig. 3H), clearly indicating that Ecm11 SUMOylation and its association to chromosomes concur independently to allow SC polymerization.
Impaired Zip4–Ecm11 interaction increases homolog nondisjunction
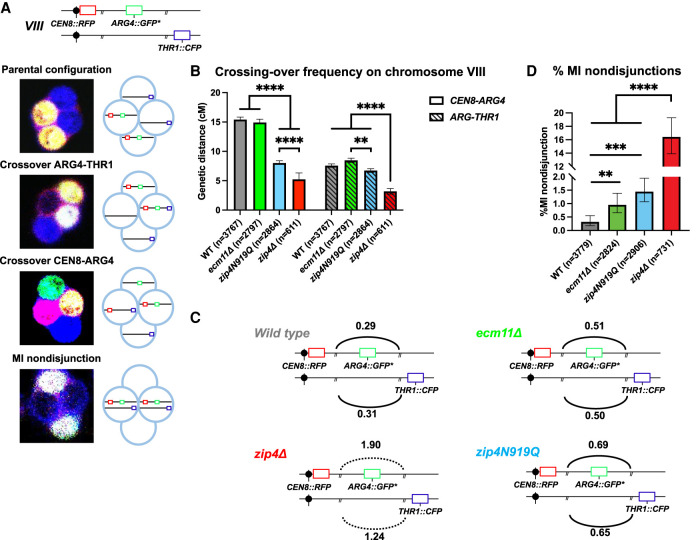
Since Ecm11 and Gmc2 proteins were reported to influence to some extent DSB frequencies and CO distribution (Humphryes et al. 2013; Voelkel-Meiman et al. 2016; Mu et al. 2020; Lee et al. 2021), we investigated the function of the Zip4–Ecm11 interaction in recombination. We first measured CO frequency on two intervals on chromosome VIII (CEN8-ARG4 and ARG4-THR1) by a fluorescent spore-autonomous assay that also allows measuring of homolog missegregation (MI nondisjunction) that can result from recombination defects (Fig. 4A; Thacker et al. 2011). As expected for a zmm mutant known to exhibit reduced CO frequency especially in small chromosomes, CO frequency in zip4Δ was decreased to ∼28%–35% of the wild type in the two intervals (Fig. 4B; Supplemental Table S1). In contrast, ecm11Δ strain showed wild-type CO levels in the ARG4-THR1 interval and a slight but significant CO reduction (95% of wild type) in the CEN8-ARG4 interval, confirming the interval-dependent effect of ecm11Δ. Similarly, the zip4N919Q interaction mutant showed wild-type CO levels in the ARG4-THR1 interval, while it was reduced in the CEN8-ARG4 interval at an intermediate level between wild type and zip4Δ. As mentioned earlier, this slight reduction compared with ecm11Δ may stem from the ZMM function of Zip4N919Q being slightly affected (Supplemental Fig. S5B).
Figure 4.
The effect of the different mutations on meiotic recombination and chromosome segregation. (A) Illustration showing the location of the spore-autonomous reporters on chromosome VIII and the types of tetrads analyzed (Thacker et al. 2011). (B) Crossing-over frequency measured in two genetic intervals—CEN8-ARG4 and ARG4-THR1—on chromosome VIII. Genetic distances are plotted as cM ± SE for the indicated genotypes. (**) P-value < 0.01, (****) P-value < 0.0001, G-test. (C) Interference between the two adjacent CEN8-ARG4 and ARG4-THR1 intervals, calculated based on Malkova et al. (2004), for the indicated genotypes. The solid line indicates that significant interference was observed (P-value < 0.05, G-test). The dotted line indicates the absence of significant interference. (D) MI nondisjunction of chromosome VIII assessed by the spore-autonomous fluorescent reporter assay (see A). The percentage of MI nondisjunction ± 95% CI is plotted. (**) P-value < 0.01, (***) P-value < 0.001, (****) P-value < 0.0001, Fisher's exact test. See also Supplemental Table S1.
We next assessed CO interference between the CEN8-ARG4 and ARG4-THR1 intervals (Fig. 4C). Interference was only slightly diminished in ecm11Δ (0.51 vs. 0.30 in wild type), confirming previous studies (Voelkel-Meiman et al. 2016; Lee et al. 2021). Similarly, interference in the zip4N919Q mutant was slightly reduced (0.67), whereas it was completely abolished in zip4Δ (1.6), as expected for a zmm mutant (Fig. 4C; Supplemental Table S1). Therefore, the Zip4 mutant for interaction with Ecm11 behaves much more like an ecm11Δ mutant than a zip4Δ mutant, confirming the essential role of Ecm11 recruitment by Zip4 for Ecm11's functions in SC assembly and recombination but not for the ZMM functions of Zip4.
Finally, using the spore fluorescent assay, we found that there was a low but significant increase of chromosome MI nondisjunction in both ecm11Δ (0.96%) and zip4N919Q (1.45%) compared with wild type (0.32%), which is much less than that seen in the zip4Δ mutant (16%) (Fig. 4D; Supplemental Table S1). This modest increase in nondisjunction may stem from the altered crossover frequency/distribution in the absence of Ecm11.
Overall, we conclude that impairing the interaction between Zip4 and Ecm11 mimics an ecm11Δ phenotype, confirming that Zip4 is responsible, in addition to its ZMM function, for all of the functions of Ecm11 in SC assembly and recombination control.
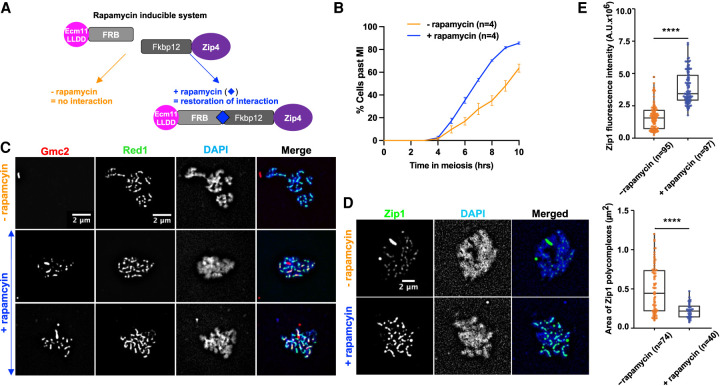
Artificially tethering interaction-deficient Ecm11 to Zip4 reinforces SC polymerization and accelerates meiotic progression
We next tested whether artificially tethering the Ecm11LLDD mutant protein to Zip4 would be sufficient for SC polymerization and meiotic progression. For this, we fused Ecm11LLDD and Zip4 with FRB and FKPB12, respectively, to tether the two proteins upon rapamycin addition at 3.5 h in meiosis, just before the expected time of recombination (Fig. 5A). Interestingly, addition of rapamycin induced a faster meiotic progression, suggesting that facilitating Zip4–Ecm11 interaction may restore SC polymerization, which would result in more rapid shutdown of DSB formation (Fig. 5B; Mu et al. 2020). We thus monitored SC polymerization. First, we checked by surface spreading and staining of meiotic cells that, upon forced interaction between Zip4 and Ecm11, Gmc2 binding was restored and formed lines along chromosomes (Fig. 5C). The recruitment of the Ecm11–Gmc2 was functional, since at all of the time points tested, a strong increase of Zip1 fluorescence signal intensity was observed upon addition of rapamycin compared with the control condition (Fig. 5D,E; Supplemental Fig. S6A,B). In addition, although many cells still contained Zip1 polycomplexes, their size was strongly decreased, consistent with better SC polymerization (Fig. 5D,E; Supplemental Fig. S7C). We conclude that physically tethering Ecm11 to Zip4 is important for the incorporation of Zip1 within the SC and is able to partly compensate for the interaction defects of the Ecm11LLDD mutant. Therefore, our data suggest that rescuing Zip4–Ecm11 association facilitates polymerization of the transverse filament protein Zip1 and accelerates meiotic progression. Unexpectedly, tethering Zip4 to Ecm11 decreased spore viability and genetic distances and increased homolog nondisjunction (Supplemental Fig. S7D–F). However, since meiotic divisions were accelerated upon addition of rapamycin, these phenotypes induced by Zip4–Ecm11 tethering are likely not due to a DSB repair defect but to insufficient DSBs giving rise to crossovers. This may result from reduced DSB frequency, reduced interhomolog–intersister ratio, or reduced crossover/noncrossover ratio. To address this, we monitored DSB formation and found that DSB formation was reduced upon addition of rapamycin (Supplemental Fig. S7G). We therefore propose that the unscheduled, early tethering of Zip4 to Ecm11 triggers premature SC formation, as well as too early inhibition of DSB formation, resulting in too few crossovers.
Figure 5.
Forcing the interaction between Ecm11 and Zip4 is sufficient to restore both Gmc2 recruitment to chromosomes and synaptonemal complex assembly. (A) Strategy to tether Ecm11LLDD fused to the FRB domain to Zip4 fused to the Fkpb12 domain by addition of rapamycin. (B) Meiotic progression as assessed by DAPI staining of nuclei to monitor meiotic divisions, with (+rapamycin) and without (−rapamycin) 1 µM rapamycin added at 3.5 h after meiotic induction. (C) Gmc2 localization on surface-spread chromosomes. Pachytene-stage nuclei are shown. (Red) Gmc2, (green) anti-Red1, (blue) DAPI. (D) Zip1 localization on surface-spread chromosomes. Pachytene-stage nuclei are shown. (Green) Anti-Zip1, (blue) DAPI. (E, top) Quantification of Zip1 intensity observed in D. (Bottom) Quantification of polycomplex areas observed in D. Numbers of spreads are indicated for each condition. (****) P-value < 0.0001, Wilcoxon test.
The mouse Zip4 interacts with TEX12, a component of the SC central element, and Ecm11–Gmc2 show striking structural analogy to TEX12–SYCE2
The whole ZZS complex (TEX11/Zip4–SHOC1/Zip2–SPO16) is present in mammals and is important for CO formation and fertility (Wang et al. 2001; Adelman and Petrini 2008; Yang et al. 2008; Yatsenko et al. 2015; Guiraldelli et al. 2018; Zhang et al. 2018, 2019; Yu et al. 2021). Likewise, the SC overall structure is also conserved between budding yeast and mammals (Zickler and Kleckner 2015). We therefore asked whether the interaction between Zip4 and the SC central element was conserved in mammals by testing the interaction between mouse TEX11 and each of the five known proteins of the mouse SC central element: SYCE1, SYCE2, SYCE3, TEX12, and SIX6OS1 (Fig. 6A; Fraune et al. 2012; Gómez-H et al. 2016). First, we recapitulated all of the previously described interactions among the SC central element proteins by yeast two-hybrid assay, indicating that our constructs are functional for protein–protein interaction (Fig. 6A; Supplemental Table S2). The mouse TEX11 contains an aromatic asparagine motif WN, as the yeast Zip4, in position 857–858 (Fig. 2C). In addition, a recent study in humans showed that the substitution of the Trp by Cys in this WN motif is associated with azoospermia (Sha et al. 2018). We thus generated a truncated TEX11 encompassing the C-terminal part of the protein (residues 637–947), named TEX11Cter, comprising the WN motif. Interestingly, we unveiled an interaction between TEX11Cter and TEX12 (Fig. 6B), reminiscent of the Zip4–Ecm11 interaction in yeast. This suggests that the interaction between the ZMM protein Zip4/TEX11 and the central element of the SC may be conserved, and that TEX12 may be a functional homolog of Ecm11. The three-dimensional structures of human TEX12 and its close interacting partner, SYCE2, have been solved (PDB: 6R17) (Fig. 6C; Davies et al. 2012; Dunce et al. 2021). TEX12 is predicted to be SUMOylated on lysine 8, located at the very N-terminal extremity of the protein (see the Materials and Methods), similar to Ecm11 SUMOylation at lysine 5 (Humphryes et al. 2013). The similarity between TEX12 and Ecm11 is further strengthened by the coevolution patterns that are observed between Gmc2 and Ecm11 on one side and those between SYCE2 and TEX12 on the other side (Supplemental Fig. S8). Strikingly, although no evolutionary relationships could clearly connect the yeast and mammalian systems, their members are both predicted to interact through an antiparallel followed by a parallel coiled coil (Supplemental Fig. S8). This coevolution pattern is fully consistent with the structure of the SYCE2–TEX12 heterotetramer (Fig. 6C; Dunce et al. 2021). Based on this experimental validation that the coevolution patterns for SYCE2–TEX12 are highly meaningful, we used the contacts predicted for the Ecm11–Gmc2 complex to generate a model of how the two proteins could interact with each other, forming a tetrameric bundle likely to further self-assemble through regions flanking the canonical coiled-coil region (Fig. 6D; Supplemental Fig. S8). Interestingly, the C-terminal and N-terminal extremities of TEX12 and SYCE2 appeared to be essential for the complex to make fibers, consistent with a function for SC propagation (Fig. 6E). This prediction is reinforced by the structural model of the Ecm11–Gmc2 tetramer generated with high confidence by the AlphaFold method (Jumper et al. 2021), which predicts the assembly of parallel and antiparallel coiled coils as in the above analysis. Interestingly, the model further predicts that helical motifs upstream of the coiled coils and separated from them by short flexible linkers can wrap on the surface of the coiled-coil bundle proficient for intratetramers but also potentially to mediate intertetramer associations (Fig. 6E).
Figure 6.
Mouse Zip4 (TEX11) interaction with the SC central element and analogies between yeast Ecm11–Gmc2 and mouse SYCE2–TEX12. (A) Illustration showing the SC central element components in mice and the two-hybrid interactions between them (see the text). (B) Yeast two-hybrid interaction analysis between mouse TEX11 and TEX12. (C) Cartoon representation of the crystal structure of the SYCE2–TEX12 coiled coils (PDB: 6R17) (Dunce et al. 2021). SYCE2 is in blue and green, and TEX12 is in dark and light pink. The positions of the antiparallel and parallel coiled-coil stretches are indicated by dashed arrows at the top. (D) 3D model of Ecm11–Gmc2. A model was built using Rosetta to fold the four subunits together under the coevolution constraints. Gmc2 subunits are shown as blue and green cartoons, while Ecm11 is shown as red and salmon cartoons. The locations of the parallel and antiparallel stretches are indicated by dashed arrows at the top (see also Supplemental Fig. S8). (E) Model similar to that in D obtained with AlphaFold integrating the N-terminal regions of Ecm11 and Gmc2, highlighting the SUMOylation (pink circle) and Zip4 (dark-purple circle) interaction sites of Ecm11. (F) 3D model of the interaction between the C terminus of Zip4 (purple cartoon) and the region spanning residues 64–81 of Ecm11 (limon cartoon), predicted using AlphaFold multimer. Ecm11 L69 and L73 residues are indicated as spheres, while N919 is highlighted as sticks.
Discussion
Several studies point to a close relationship between crossover sites and sites of SC nucleation (Pyatnitskaya et al. 2019). However, the connection between these two important processes remained elusive. Here, we describe a direct and functional interaction between the ZMM protein Zip4 and Ecm11, a component of the SC central element, providing a physical link between crossovers and SC polymerization.
Zip4 is an interface protein that integrates signals from both crossover and synapsis promoting factors
Zip4 is a protein with repetitive TPR domains, motifs that are common in scaffold proteins and exhibit a wide range of molecular recognition modes (D'Andrea and Regan 2003; Perez-Riba and Itzhaki 2019). An interesting property of some TPR proteins is their ability to orchestrate different activities by integrating signals from multiple interacting partners. Several pieces of evidence point to such a role for the Zip4 protein. First, on the “ZMM side,” Zip4 interacts directly with its ZMM partners Zip2 and Spo16 to form the ZZS complex. Within this complex, a domain of Zip2 forms with Spo16 an XPF-ERCC1-like module that recognizes DNA joint molecules (De Muyt et al. 2018; Arora and Corbett 2019). The role of Zip4 in this complex is not well understood, but Zip4 is important for Zip2 stability and may act as a chaperone for Zip2 and Spo16, reinforcing their DNA recognition activity (De Muyt et al. 2018). Second, the other ZMM proteins, SUMO/ubiquitin ligase Zip3 and MutSγ, have also been reported to colocalize and interact with Zip4, suggesting that Zip4 integrates multiple ZMM activities to consolidate joint molecule intermediates and promote CO formation (Shinohara et al. 2008; De Muyt et al. 2018).
In addition to ZMMs, Zip4 interacts with components of the SC. In budding yeast, a connection between Zip4 and the synaptonemal complex was first identified via an interaction with Red1, the axial element of the SC (De Muyt et al. 2018). This seems conserved in mammals since the Zip4 ortholog, TEX11, interacts with the SC axial element SYCP2 (Yang et al. 2008). We showed here that Zip4 also binds to the SC central element Ecm11 and Gmc2 proteins, suggesting that Zip4 is tightly connected to SC proteins through multiple interactions. Interestingly, the axial and central elements of the SC are separated from each other by 50 nm, suggesting that Zip4 is present in two different locations within the SC. Based on what is known about the temporal dynamics of recombination intermediates during the successive steps of recombination and the dynamics of ZZS complex association with axis and DSB sites (De Muyt et al. 2018), we envision that Zip4, bound on recombination intermediates through the Zip2–Spo16 module, may first interact with the axial element (via Red1) at an early recombination step, and the ZZS would then be translocated to the future central element location, between the axes, at a later step of recombination to seed SC nucleation via Zip4 interaction with Ecm11. Such dynamics would result in bringing “miniature axes” (or bridges), containing Red1, from parental chromosomes into the interaxis region, as proposed in Sordaria (Dubois et al. 2019).
Several lines of evidence suggest that the SC emerges from ZMM-bound sites. In budding yeast, Sordaria, and mice, SC initiation sites often colocalize with ZMM proteins (Agarwal and Roeder 2000; Tsubouchi et al. 2006; Reynolds et al. 2013; Dubois et al. 2019) and decrease in number in mutants with reduced DSB numbers, while synapsis defects are increased (Tessé et al. 2003, 2017; Henderson and Keeney 2004; Kauppi et al. 2013). This suggests that a minimum number of ZMM/SC nucleation sites is required for full homolog synapsis (Tsubouchi et al. 2006). Since the SC transverse filament protein Zip1 is also a ZMM protein, it was an obvious candidate for the initial recruitment of Ecm11. Moreover, an N-terminal deletion mutant, zip1N1, has a phenotype similar to that of ecm11Δ, and Ecm11–Gmc2 colocalize with Zip1 during synapsis initiation and completion (Tung and Roeder 1998; Humphryes et al. 2013; Voelkel-Meiman et al. 2016). However, we found no evidence of interaction between Zip1 and Ecm11 or Gmc2 in our Y2H experiments. In addition, Ecm11 foci are still visible in a zip1Δ mutant (Humphryes et al. 2013), and Ecm11 still associates with DSB hotspots in our ChIP experiments, implying that Zip1 is not required for the initial SC assembly from the ZMM nucleation sites. Instead, we provide a body of evidence that Zip4, through its direct interaction with Ecm11, plays a pivotal role promoting synapsis at these ZMM binding sites. (1) Ecm11 shows a pattern similar to that of ZMMs, binding both DSB and axis sites. (2) Ecm11 localization at DSB sites strictly requires Zip4 protein, in agreement with the absence of Ecm11 foci in zip4Δ mutant, but not in other tested zmm mutants (Humphryes et al. 2013). (3) Mutations altering the interaction between Zip4 and Ecm11, zip4N919Q and ecm11LLDD, result in defective SC assembly and in polycomplex formation in a manner akin to zip4Δ and ecm11Δ. (4) Tethering Zip4 and a mutated interaction-defective Ecm11 is sufficient to restore SC assembly and faster meiotic progression.
In budding yeast, Ecm11 acts in complex with Gmc2 during SC polymerization (Humphryes et al. 2013). Interestingly, the Ecm11LLDD mutated protein keeps its ability to form a heterodimer with Gmc2. Moreover, Gmc2 also interacts with Zip4 in yeast two-hybrid assays, suggesting that Zip4 may promote SC assembly by depositing a preformed Ecm11–Gmc2 complex. Finally, Zip4 may coordinate signals at the same time through simultaneous interactions between different TPR motifs present throughout its length and its proteins partners (including Zip2, Zip3, Msh5, Red1, Ecm11, and Gmc2). The predicted structural model using AlphaFold trained on a multimer data set is fully consistent with our site-directed mutagenesis experiments (Fig. 6F). An interaction is predicted between the residue N919 side chain and the backbone of Ecm11. Downstream, a short helical stretch in Ecm11 involving residues L69 and L73 is predicted to dock onto the conserved apolar patch exposed at the surface of the Zip4 C terminus. This model provides strong support for the molecular interpretation that we proposed for the designed mutations, and it will be of interest to further identify the role of all of the docking sites connecting Zip4 to its described partners.
Spatio–temporal coupling of crossovers and SC assembly
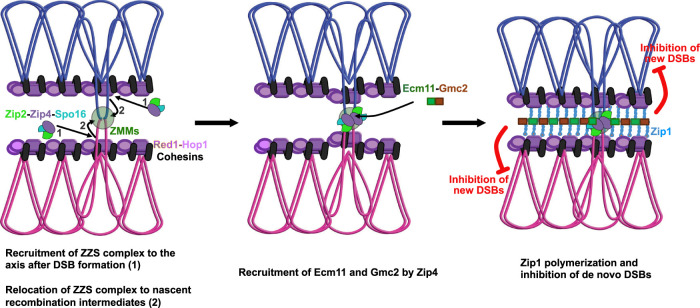
Given the importance of Zip4 in the recognition of DNA joint molecules through the ZZS module and in SC assembly via Ecm11 (and Gmc2) interaction, and to integrate all present and past results, we propose the following model for the Zip4 mechanism of action (Fig. 7): (1) After DSB formation, the ZZS complex associates with the axis component Red1 and with recombination intermediates via the XPF-ERCC1-like DNA recognition module (De Muyt et al. 2018). Other ZMMs, including Zip1, also bind recombination intermediates. (2) Then, still bound on recombination intermediates, the ZZS complex transits from the axis region toward the interaxis region, leading to the formation of chromosomal bridges that progressively align the parental chromosomes (De Muyt et al. 2018; Dubois et al. 2019; Pyatnitskaya et al. 2019). In the meantime, Zip4 helps to bring Ecm11–Gmc2 to these sites by direct protein–protein interaction. The Ecm11–Gmc2 complex then helps initiate the polymerization of the surrounding Zip1. It is at this time that a “synapsis initiation complex” is created, and the SC will start to emanate from this nucleation zone through Zip1 polymerization. Finally, as suggested recently and as we show with our tethering assay, SC polymerization exerts a negative feedback on de novo DSB formation, and therefore locally affects crossover frequencies (Fig. 7; Thacker et al. 2014; Voelkel-Meiman et al. 2016; Mu et al. 2020; Lee et al. 2021). This mechanism of regulation starting from crossover-designated sites would be an elegant way for the cell to fine-tune CO patterning by shutting down DSBs locally through the propagation of the SC along chromosomes.
Figure 7.
Model for the link between crossover sites and SC assembly. The model is based on our study of Zip4–Ecm11 interaction and published studies (see the text). (1) First, the axial element polymerizes, and ZZS is recruited to the axis after DSB formation. (2) Then SICs are formed after the transition of ZMMs (including ZZS) to the interaxis region. the Ecm11–Gmc2 heterodimer is brought to the SIC through its interaction with Zip4, which initiates the polymerization of the TF Zip1. Polymerization of the SC central region inhibits the formation of de novo DSBs, thus avoiding additional break and repair in already synapsed regions.
The relationship between crossovers and SC assembly in other species
Like in budding yeast, in mice and plants, the absence of DSB or efficient interhomolog repair processes leads to synapsis defects, suggesting that synapsis initiation depends on the total number of interhomolog interactions (Mercier et al. 2015; Cahoon and Hawley 2016; Pyatnitskaya et al. 2019). It is currently unknown whether a protein complex similar to the SIC is required for the initiation of SC polymerization at these sites of interhomolog engagement. However, since mouse zmm mutants show synapsis defects, ZMM proteins could participate in the initiation of SC formation, although the different extent of synapsis defects observed among zmm mutants suggests that the absence of some ZMMs might be concealed by a second mechanism based on homology-independent SC extension, known as synapsis adjustment (Zickler and Kleckner 1999). Finally, contrary to budding yeast, whereas ZMM proteins are still detected between homolog axes, the SC central element proteins SYCE1/2/3 and TEX12 are no longer detected on chromosomes in Sycp1−/− (Hamer et al. 2006; Schramm et al. 2011). The central element proteins may have a different mode of recruitment, and/or their abundance is too low to be detected by conventional microscopy, if they form only dots, as in budding yeast.
In plants, SC polymerization seems less dependent on the CO-mediated interhomolog engagement, since zmm mutants do not have apparent synapsis defects (Mercier et al. 2015), but this does not mean that SC polymerization does not initiate from ZMM-bound sites in wild type. In addition, the kinetics of SC assembly and synergistic effects of ZMM mutations have not been thoroughly tested. Indeed, a combination of both zip4 and mer3 mutations leads to severe synapsis defects in rice, suggesting that ZMM proteins might have redundant roles for SC loading in plants (Shen et al. 2012).
Interestingly, the species (like the worm C. elegans or D. melanogaster) that do not rely on recombination for initiating SC polymerization lack many of the ZMM proteins, including Zip4, Zip2, and Spo16, possibly resulting from the absence of selective pressure for CO-designated interhomolog engagement for SC initiation.
Concluding remarks
Recent studies in yeast and plants showed the importance of close homolog juxtaposition by the SC to control recombination frequency and crossover distribution (Mu et al. 2020; Capilla-Pérez et al. 2021; France et al. 2021; Lee et al. 2021). We propose that this control is initiated by the direct interaction between Zip4 and Ecm11. It will be important to understand the interplay between this coupling mechanism and the mechanism of the initial deposition of Zip1, which requires Mek1 phosphorylation, to coordinate SC assembly (Chen et al. 2015). Finally, further investigations of the relationship between the ZMM-dependent CO formation and the SC dynamics in different model organisms will be needed to uncover both their conserved and distinct features and reveal how it could impact human fertility, given the involvement of TEX11 mutations in patients with azoospermia.
Materials and methods
Yeast manipulation
All yeast strains are derivatives of the SK1 background, except those used for two-hybrid experiments and for ChIP-seq spike-in control. Their complete genotypes and their use in different figures are in Supplemental Table S3. All experiments were performed at 30°C. For synchronous meiosis, cells were grown in SPS presporulation medium and transferred to 1% potassium acetate with vigorous shaking at 30°C as described (Murakami et al. 2009). For all strains, spore viability was measured after sporulation on solid sporulation medium for 2 d at 30°C.
Yeast strain construction
Yeast strains were obtained by direct transformation or crossing to obtain the desired genotype. Site-directed mutagenesis and C-terminal deletions were introduced by PCR. All transformants were confirmed using PCR discriminating between correct and incorrect integrations and sequencing for epitope tag insertion or mutagenesis. The functionality of the tagged proteins was measured by spore viability assays. All tagged proteins were functional.
Sequence analyses and modelling of Zip4, Ecm11, and Gmc2 structures
Full-length homologous sequences of Zip4, Ecm11, Gmc2, TEX12, and SYCE2 were retrieved using PSI-BLAST iterations on the nr database, gathering 862, 916, 824, 165, and 184 sequences, respectively. Multiple sequence alignments were generated for these sets of sequences using MAFFT (Katoh and Standley 2013) and represented using Jalview (Waterhouse et al. 2009). Co-MSA for Gmc2–Ecm11 and SYCE2–TEX12 were obtained by selecting a single sequence per species, selecting the hit of lowest e-value, and by concatenating the alignments, resulting in a co-MSA of 451 and 135 sequences, respectively. These alignments were used as input of the RaptorX contact prediction (Wang et al. 2017) to predict the contact maps within and between the pairs of proteins. The structural model of the Gmc2–Ecm11 assembly was generated in a hierarchical manner, assembling first the heterodimeric coiled coil of Ecm11 and Gmc2 using the InterEvDock3 server (Quignot et al. 2021) to take into account the RaptorX coevolution-based contact map in the docking of the two helical stretches of Ecm11 and Gmc2 predicted to form a coiled coil. The tetrameric assembly was subsequently assembled using RosettaRelax (Leman et al. 2020) protocols using standard options and the intermolecular constraints that were not respected in the docking of the Ecm11–Gmc2 heterodimer docking. The 3D model of Zip4 was generated using the latest version of the RoseTTAFold server, combining coevolution and deep learning approaches for the prediction of 3D monomeric structures (Baek et al. 2021). Analyses of the SUMOylation sites were performed using the Jassa server (Beauclair et al. 2015), and those of the coiled coils were performed using PCOILS as implemented in the MPI Bioinformatics Toolkit server (Lupas et al. 1991). As an independent assessment of the predictions that had been generated prior to the AlphaFold publication, the coalignments of Ecm11 and Gmc2 were subjected to the AlphaFold prediction method using the ColabFold interface (Mirdita et al. 2021), allowing for the submission of precomputed alignments and using parameters trained on the monomeric-only structural database (Jumper et al. 2021). For the model of Zip4 and Ecm11, the AlphaFold version trained on a data set of multimers (Evans et al. 2021) was used, since no interaction could be observed with the AlphaFold version trained on monomeric structures.
Yeast two-hybrid analyses
Strains expressing ZMMs are described in De Muyt et al. (2018). ECM11 and GMC2 were PCR-amplified from SK1 genomic DNA. Site-directed mutations were introduced by fusion of PCR products. Full-length mouse Tex11, Tex12, Syce1, Syce2, Syce3, and Six6os1 were PCR-amplified from mouse testis cDNA (a gift from D. Bourc'his). PCR products were cloned in plasmids derived from the two hybrid vectors pGADT7 or pGADCg (GAL4-activating domain) and pGBKT7 or pGBKCg (GAL4-binding domain), creating N-terminal or C-terminal fusions, and transformed in yeast haploid strains Y187 and AH109 (Clontech), respectively. Yeast two-hybrid assays were performed and interactions were scored on selective media exactly as described in Duroc et al. (2017).
Analysis of DSB frequencies
DSBs were analyzed by pulsed-field gel electrophoresis followed by Southern blot with the YHL039W probe and quantified as described previously (Carballo et al. 2013).
Analysis of crossover frequencies
Diploids were sporulated in liquid medium, and recombination between fluorescent markers on chromosome VIII was scored after 24 h of sporulation by microscopy analysis, as described previously (Thacker et al. 2011). Two independent sets of each strain were combined, and at least 730 tetrads were scored for crossovers in two test intervals and for MI nondisjunction events. Genetic distances in the CEN8-ARG4 and ARG4-THR1 intervals were calculated from the distribution of parental ditype (PD), nonparental ditype (NPD), and tetratype (T) tetrads, and genetic distances (cM) were calculated using the Perkins equation cM = [100 (6NPD + T)]/[2(PD + NPD + T)]. SEs of genetic distances were calculated using Stahl laboratory online tools (https://elizabethhousworth.com/StahlLabOnlineTools).
Cytology
For cytology, 1 × 108 cells were harvested at the indicated time point and yeast chromosome spreads were prepared as described in Grubb et al. (2015). Primary antibodies used were mouse monoclonal 9E11 anti-myc antibody (dilution 1:200), rabbit polyclonal anti-Zip1 antibody (dilution 1:100; Santa Cruz Biotechnology sc-33733), rabbit monoclonal anti-Red1 antibody (#16441; dilution 1:200; a gift from N. Hollingsworth), and rabbit anti-Gmc2 antibody (dilution 1:1600; a gift from Amy MacQueen). The secondary antibodies were Alexa488-conjugated goat antirabbit (dilution 1:200; Thermo Fischer Scientific A-11008) and Alexa568-conjugated goat antimouse (dilution 1:200; Thermo Fischer Scientific A-11004). Chromosomal DNA was stained by 4,6-diamidino-2-phenylindole (DAPI). Fluorescence images were visualized and acquired using the Deltavision IX70 system (Applied Precision), 100× objective, and softWoRx imaging software. Images were processed by deconvolution using the constrained iterative deconvolution algorithm within softWoRx. Image analysis and signal quantification were performed using the Fiji software and R scripts. Fluorescence intensity was measured as the average sum of pixel density of Zip1 stretches per nucleus.
TCA extraction and Western blot analysis
Protein extracts were prepared by trichloroacetic acid (TCA) precipitation method. Sprouting cell culture (1.5 mL) was harvested and pellet was immediately frozen in liquid nitrogen. Cells were resuspended in 100 μL of ice-cold NaOH solution (1.85 N NaOH, 7.5% β-mercaptoethanol) and incubated for 10 min on ice. Samples were then mixed with 30 μL of 50% ice-cold TCA and incubated for 10 min on ice. Cell suspension was then harvested at 15000g for 5 min at 4°C and the pellet was resuspended in 100 μL of loading buffer (55 mM Tris at pH 6.8, 6.6 M urea, 4.2% SDS, 0.083 mM EDTA, 0.001% bromophenol blue, 1.5% β-mercaptoethanol). Protein samples were dipped in liquid nitrogen and then incubated for 3 min at 65°C. Samples were centrifuged at 20000g for 5 min and the supernatant was kept at −80°C. Samples were loaded on precast acrylamide gel (4%–12% Bis-Tris gel [Invitrogen]) and transferred to PVDF membrane in MOPS SDS running buffer (Life Technologies). Proteins were detected using mouse monoclonal M2 anti-Flag (dilution 1:1000; Sigma F1804), mouse monoclonal 9E11 anti-myc (dilution 1:500), or rabbit monoclonal anti-TAP antibody (dilution 1:2000; Invitrogen CAB1001). For normalization, mouse monoclonal anti-Pgk1 antibody was used (1:3000;Invitrogen 459250). Image acquisition was performed with Chemidoc system (Bio-Rad). To quantify protein levels, the band intensity in each lane was measured by ImageLab software and divided by the corresponding Pgk1 band intensity in the same lane.
Coimmunoprecipitation
Cells (1.2 × 109) were harvested and 1 mM PMSF was added. Cells were washed once with PBS and lysed in 3 mL of lysis buffer (20 mM HEPES/KOH at pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 10% glycerol, 1 mM MgCl2, 2 mM EDTA, 1 mM PMSF, 1× Complete Mini EDTA-free [Roche], 1× PhosSTOP [Roche]) with 0.5-mm zirconium/silica beads (Biospec Products) three times for 30 sec in a Fastprep instrument (MP Biomedicals). The lysate was incubated for 1 h at 4°C with 125 U/mL benzonase. One-hundred microliters of PanMouse IgG magnetic beads (Thermo Scientific) was washed 1:1 with lysis buffer, preincubated in 100 μg/mL BSA in lysis buffer for 2 h at 4°C, and then washed twice with 1:1 lysis buffer. The lysate was cleared by centrifugation at 13,000g for 5 min and incubated overnight at 4°C with washed PanMouse IgG magnetic beads. The magnetic beads were washed four times with 1 mL of wash buffer (20 mM HEPES/KOH at pH 7.5, 150 mM NaCl, 0.5% Triton X-100, 5% glycerol, 1 mM MgCl2, 2 mM EDTA, 1 mM PMSF, 1× Complete Mini EDTA-free [Roche], 1× Phos-STOP [Roche]). The beads were resuspended in 30 μL of TEV-C buffer (20 mM Tris/HCl at pH 8, 0.5 mM EDTA, 150 mM NaCl, 0.1% NP-40, 5% glycerol, 1 mM MgCl2, 1 mM DTT) with 4 μL of 1 mg/mL TEV protease (which cleaves between the calmodulin binding protein [CBP] and protein A moieties of the TAP tag) and incubated for 2 h at 23°C under agitation to eluate the CBP-tagged protein. The eluate was transferred to a new tube. After washing, beads were resuspended in 25 μL of 2× SDS protein sample buffer. Bead eluate was heated for 3 min at 95°C, loaded onto acrylamide gel (4%–12% Bis-Tris gel [Invitrogen]), and run in MOPS SDS running buffer (Life Technologies). Proteins were then transferred to PVDF membrane using Trans-Blot Turbo transfer system (Bio-Rad) at 2.5 Å constant, up to 25 V for 10 min. Proteins were detected using mouse monoclonal M2 anti-Flag (dilution 1:1000; Sigma F1804) or rabbit monoclonal anti-TAP (dilution 1:2000; Invitrogen CAB1001) antibody. Signal was detected using the SuperSignal West Pico or Femto chemiluminescent substrate (Thermo Fisher). Images were acquired using the Chemidoc system (Bio-Rad). Signal was analyzed with ImageLab software. Results are presented as percentage of input band after subtracting the untagged strain signal and normalizing by TAP-tagged protein level.
Chromatin immunoprecipitation
For each meiotic time point, 2 × 108 cells were processed as described in Duroc et al. (2017), except that before use, magnetic beads were blocked with 5 μg/μL BSA for 4 h at 4°C. Quantitative PCR was performed from the immunoprecipitated DNA or the whole-cell extract using QuantStudio 5 (Applied Biosystems, Thermo Scientific) and analyzed as described (Duroc et al. 2017). Results are expressed as percentage of DNA in the total input present in the immunoprecipitated sample. Primers for GAT1, BUD23, HIS4LEU2, ERG1, AXIS (between RAD18 and SED4 convergent genes), and NFT1 loci have been described (Sanchez et al. 2020).
For ChIP-seq experiments, 1 × 109 cells were processed as described (De Muyt et al. 2018; Sanchez et al. 2020; Sanchez and Borde 2021) except that, for spike-in normalization, 1 × 108 (10%) S. mikatae cells of a single meiotic culture, harvested at 4 h in meiosis and fixed using the same procedure as for S. cerevisiae, were added to each sample before processing.
Illumina sequencing of ChIP DNA and read normalization
Purified DNA was sequenced using an Illumina NovaSeq 6000 instrument following the Illumina TruSeq procedure, generating paired-end 100-bp reads for Ecm11 in wild-type, zip1Δ, zip4Δ, and untagged anti-Flag ChIP. Each experiment was performed in two independent replicates. Reads were aligned to the SaccCer2 S. cerevisiae S288C genome exactly as described (Sanchez et al. 2020), and to the S. mikatae genome assembly (Scannell et al. 2011). Reads that aligned on the S. cerevisiae genome but not on S. mikatae were defined as the experimental reads. For defining the spike-in normalization factor, we then determined the number of reads that did not align on the S.cerevisiae genome but aligned on the S. mikatae genome assembly (Scannell et al. 2011), generating the spike-in reads. The aligned experimental reads from independent replicates were then combined using MergeSamFiles to generate a single Bam file. Next, each Bam file was converted to bigwig format using deepTools bamCoverage, with a bin size of 1, a smoothing window of 200 bp, and a normalization factor 2, obtained as follows: For each sample, the number of experimental reads was first divided by the number of spike-in reads, giving scaling factor 1. Then, the factor 1 of each sample was divided by the mean untagged sample coverage (290), giving scaling factor 2. Finally, for each bigwig file obtained, the scaled untagged sample was subtracted from the scaled tagged sample. These values were used for Figure 1, C and D. Peaks for Red1 ChIP-seq and Spo11 oligonucleotides were from Sun et al. (2015) and Zhu and Keeney (2015), respectively.
Data availability
Sequencing data were deposited at the NCBI Gene Expression Omnibus database with the accession number GSE177033.
Supplementary Material
Acknowledgments
We thank N. Hollingsworth for the anti-Red1 antibody, A. MacQueen for the anti-Gmc2 antibody, A. Hochwagen for the SK1 fpr1 tor1-1 mutant strains, D. Bourc'his for the mouse testis cDNA, and Institut Curie NGS platform, supported by grants from ANR-10-EQPX-03 and ANR10-INBS-09-08 and from the Cancéropôle Ile-de-France. This work was supported by Institut Curie, Centre National de la Recherche Scientifique, Agence Nationale de la Recherche (ANR-15-CE11-0011), Fondation ARC (PJA20171206487), and Ligue Contre le Cancer (5FI13573TAZP). A.P. was a recipient of a predoctoral funding from the Fondation pour la Recherche Médicale.
Author contributions: A.D.M. and V.B. supervised the study. A.P. and A.D.M. performed the experiments. J.A. and R.G. designed all of the protein–protein interaction mutants and performed the structure predictions. A.P., A.D.M., and V.B. wrote the paper, with input from all of the authors.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.348973.121.
Competing interest statement
The authors declare no competing interests.
References
- Adelman CA, Petrini JHJ. 2008. ZIP4H (TEX11) deficiency in the mouse impairs meiotic double strand break repair and the regulation of crossing over. PLoS Genet 4: e1000042. 10.1371/journal.pgen.1000042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal S, Roeder GS. 2000. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 102: 245–255. 10.1016/S0092-8674(00)00029-5 [DOI] [PubMed] [Google Scholar]
- Arora K, Corbett KD. 2019. The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res 47: 2365–2376. 10.1093/nar/gky1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, Wang J, Cong Q, Kinch LN, Schaeffer RD, et al. 2021. Accurate prediction of protein structures and interactions using a three-track neural network. Science 373: 871–876. 10.1126/science.abj8754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beauclair G, Bridier-Nahmias A, Zagury J-F, Saïb A, Zamborlini A. 2015. JASSA: a comprehensive tool for prediction of SUMOylation sites and SIMs. Bioinformatics 31: 3483–3491. 10.1093/bioinformatics/btv403 [DOI] [PubMed] [Google Scholar]
- Berchowitz LE, Copenhaver GP. 2010. Genetic interference: don't stand so close to me. Curr Genomics 11: 91–102. 10.2174/138920210790886835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Börner GV, Kleckner N, Hunter N. 2004. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117: 29–45. 10.1016/S0092-8674(04)00292-2 [DOI] [PubMed] [Google Scholar]
- Cahoon CK, Hawley RS. 2016. Regulating the construction and demolition of the synaptonemal complex. Nat Struct Mol Biol 23: 369–377. 10.1038/nsmb.3208 [DOI] [PubMed] [Google Scholar]
- Capilla-Pérez L, Durand S, Hurel A, Lian Q, Chambon A, Taochy C, Solier V, Grelon M, Mercier R. 2021. The synaptonemal complex imposes crossover interference and heterochiasmy in Arabidopsis. Proc Natl Acad Sci 118: e2023613118. 10.1073/pnas.2023613118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carballo JA, Panizza S, Serrentino ME, Johnson AL, Geymonat M, Borde V, Klein F, Cha RS. 2013. Budding yeast ATM/ATR control meiotic double-strand break (DSB) levels by down-regulating Rec114, an essential component of the DSB-machinery. PLoS Genet 9: e1003545. 10.1371/journal.pgen.1003545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Suhandynata RT, Sandhu R, Rockmill B, Mohibullah N, Niu H, Liang J, Lo H-C, Miller DE, Zhou H, et al. 2015. Phosphorylation of the synaptonemal complex protein Zip1 regulates the crossover/noncrossover decision during yeast meiosis. PLoS Biol 13: e1002329. 10.1371/journal.pbio.1002329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua PR, Roeder GS. 1998. Zip2, a meiosis-specific protein required for the initiation of chromosome synapsis. Cell 93: 349–359. 10.1016/S0092-8674(00)81164-2 [DOI] [PubMed] [Google Scholar]
- D'Andrea LD, Regan L. 2003. TPR proteins: the versatile helix. Trends Biochem Sci 28: 655–662. 10.1016/j.tibs.2003.10.007 [DOI] [PubMed] [Google Scholar]
- Davies OR, Maman JD, Pellegrini L. 2012. Structural analysis of the human SYCE2–TEX12 complex provides molecular insights into synaptonemal complex assembly. Open Biol 2: 120099. 10.1098/rsob.120099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer E, Heyting C. 2006. The diverse roles of transverse filaments of synaptonemal complexes in meiosis. Chromosoma 115: 220–234. 10.1007/s00412-006-0057-5 [DOI] [PubMed] [Google Scholar]
- De Muyt A, Jessop L, Kolar E, Sourirajan A, Chen J, Dayani Y, Lichten M. 2012. BLM helicase ortholog Sgs1 is a central regulator of meiotic recombination intermediate metabolism. Mol Cell 46: 43–53. 10.1016/j.molcel.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Muyt A, Pyatnitskaya A, Andréani J, Ranjha L, Ramus C, Laureau R, Fernandez-Vega A, Holoch D, Girard E, Govin J, et al. 2018. A meiotic XPF–ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev 32: 283–296. 10.1101/gad.308510.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois E, Muyt AD, Soyer JL, Budin K, Legras M, Piolot T, Debuchy R, Kleckner N, Zickler D, Espagne E. 2019. Building bridges to move recombination complexes. Proc Natl Acad Sci 116: 12400–12409. 10.1073/pnas.1901237116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunce JM, Salmon LJ, Davies OR. 2021. Structural basis of meiotic chromosome synaptic elongation through hierarchical fibrous assembly of SYCE2–TEX12. Nat Struct Mol Biol 28: 681–693. 10.1038/s41594-021-00636-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duroc Y, Kumar R, Ranjha L, Adam C, Guérois R, Md Muntaz K, Marsolier-Kergoat M-C, Dingli F, Laureau R, Loew D, et al. 2017. Concerted action of the MutLβ heterodimer and Mer3 helicase regulates the global extent of meiotic gene conversion. Elife 6: e21900. 10.7554/eLife.21900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R, O'Neill M, Pritzel A, Antropova N, Senior A, Green T, Žídek A, Bates R, Blackwell S, Yim J, et al. 2021. Protein complex prediction with AlphaFold-multimer. bioRxiv 10.1101/2021.10.04.463034 [DOI] [Google Scholar]
- Fontes MRM, Teh T, Kobe B. 2000. Structural basis of recognition of monopartite and bipartite nuclear localization sequences by mammalian importin-α. J Mol Biol 297: 1183–1194. 10.1006/jmbi.2000.3642 [DOI] [PubMed] [Google Scholar]
- France MG, Enderle J, Röhrig S, Puchta H, Franklin FCH, Higgins JD. 2021. ZYP1 is required for obligate cross-over formation and cross-over interference in Arabidopsis. Proc Natl Acad Sci 118: e2021671118. 10.1073/pnas.2021671118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraune J, Schramm S, Alsheimer M, Benavente R. 2012. The mammalian synaptonemal complex: protein components, assembly and role in meiotic recombination. Exp Cell Res 318: 1340–1346. 10.1016/j.yexcr.2012.02.018 [DOI] [PubMed] [Google Scholar]
- Fudenberg G, Imakaev M, Lu C, Goloborodko A, Abdennur N, Mirny LA. 2016. Formation of chromosomal domains by loop extrusion. Cell Rep 15: 2038–49. 10.1016/j.celrep.2016.04.085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J, Colaiácovo MP. 2018. Zipping and unzipping: protein modifications regulating synaptonemal complex dynamics. Trends Genet 34: 232–245. 10.1016/j.tig.2017.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez-H L, Felipe-Medina N, Sánchez-Martín M, Davies OR, Ramos I, García-Tuñón I, de Rooij DG, Dereli I, Tóth A, Barbero JL, et al. 2016. C14ORF39/SIX6OS1 is a constituent of the synaptonemal complex and is essential for mouse fertility. Nat Commun 7: 1–16. 10.1038/ncomms13298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb J, Brown MS, Bishop DK. 2015. Surface spreading and immunostaining of yeast chromosomes. J Vis Exp 2015: e53081. 10.3791/53081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guiraldelli MF, Felberg A, Almeida LP, Parikh A, de Castro RO, Pezza RJ. 2018. SHOC1 is a ERCC4-(HhH)2-like protein, integral to the formation of crossover recombination intermediates during mammalian meiosis. PLoS Genet 14: e1007381. 10.1371/journal.pgen.1007381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer G, Gell K, Kouznetsova A, Novak I, Benavente R, Höög C. 2006. Characterization of a novel meiosis-specific protein within the central element of the synaptonemal complex. J Cell Sci 119: 4025–4032. 10.1242/jcs.03182 [DOI] [PubMed] [Google Scholar]
- Henderson KA, Keeney S. 2004. Tying synaptonemal complex initiation to the formation and programmed repair of DNA double-strand breaks. Proc Natl Acad Sci 101: 4519–24. 10.1073/pnas.0400843101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B, Petela N, Kurze A, Chan KL, Chapard C, Nasmyth K. 2015. Biological chromodynamics: a general method for measuring protein occupancy across the genome by calibrating ChIP-seq. Nucleic Acids Res 43: e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphryes N, Leung W-K, Argunhan B, Terentyev Y, Dvorackova M, Tsubouchi H. 2013. The Ecm11-Gmc2 complex promotes synaptonemal complex formation through assembly of transverse filaments in budding yeast. PLoS Genet 9: e1003194. 10.1371/journal.pgen.1003194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. 2015. Meiotic recombination: the essence of heredity. Cold Spring Harb Perspect Biol 7: a016618. 10.1101/cshperspect.a016618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Kleckner N. 2001. The single-end invasion: an asymmetric intermediate at the double-strand break to double-Holliday junction transition of meiotic recombination. Cell 106: 59–70. 10.1016/S0092-8674(01)00430-5 [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Žídek A, Potapenko A, et al. 2021. Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. 10.1038/s41586-021-03819-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauppi L, Barchi M, Lange J, Baudat F, Jasin M, Keeney S. 2013. Numerical constraints and feedback control of double-strand breaks in mouse meiosis. Genes Dev 27: 873–886. 10.1101/gad.213652.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein F, Mahr P, Galova M, Buonomo SBC, Michaelis C, Nairz K, Nasmyth K. 1999. A central role for cohesins in sister chromatid cohesion, formation of axial elements, and recombination during yeast meiosis. Cell 98: 91–103. 10.1016/S0092-8674(00)80609-1 [DOI] [PubMed] [Google Scholar]
- Lee M-S, Higashide MT, Choi H, Li K, Hong S, Lee K, Shinohara A, Shinohara M, Kim KP. 2021. The synaptonemal complex central region modulates crossover pathways and feedback control of meiotic double-strand break formation. Nucleic Acids Res 49: 7537–7553. 10.1093/nar/gkab566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leman JK, Weitzner BD, Lewis SM, Adolf-Bryfogle J, Alam N, Alford RF, Aprahamian M, Baker D, Barlow KA, Barth P, et al. 2020. Macromolecular modeling and design in Rosetta: recent methods and frameworks. Nat Methods 17: 665–680. 10.1038/s41592-020-0848-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung W-K, Humphryes N, Afshar N, Argunhan B, Terentyev Y, Tsubouchi T, Tsubouchi H. 2015. The synaptonemal complex is assembled by a polySUMOylation-driven feedback mechanism in yeast. J Cell Biol 211: 785–793. 10.1083/jcb.201506103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libuda DE, Uzawa S, Meyer BJ, Villeneuve AM. 2013. Meiotic chromosome structures constrain and respond to designation of crossover sites. Nature 502: 703–706. 10.1038/nature12577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. 1991. Predicting coiled coils from protein sequences. Science 252: 1162–1164. 10.1126/science.252.5009.1162 [DOI] [PubMed] [Google Scholar]
- Malkova A, Swanson J, German M, McCusker JH, Housworth EA, Stahl FW, Haber JE. 2004. Gene conversion and crossing over along the 405-kb left arm of Saccharomyces cerevisiae chromosome VII. Genetics 168: 49–63. 10.1534/genetics.104.027961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. 2015. The molecular biology of meiosis in plants. Annu Rev Plant Biol 66: 297–327. 10.1146/annurev-arplant-050213-035923 [DOI] [PubMed] [Google Scholar]
- Mirdita M, Schütze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. 2021. ColabFold—making protein folding accessible to all. bioRxiv 10.1101/2021.08.15.456425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses MJ. 1969. Structure and function of the synaptonemal complex. Genetics 61: 41–51. [PubMed] [Google Scholar]
- Mu X, Murakami H, Mohibullah N, Keeney S. 2020. Chromosome-autonomous feedback down-regulates meiotic DNA break competence upon synaptonemal complex formation. Genes Dev 34: 1605–1618. 10.1101/gad.342873.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami H, Borde V, Nicolas A, Keeney S. 2009. Gel electrophoresis assays for analyzing DNA double-strand breaks in Saccharomyces cerevisiae at various spatial resolutions. Methods Mol Biol 557: 117–142. 10.1007/978-1-59745-527-5_9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panizza S, Mendoza MA, Berlinger M, Huang L, Nicolas A, Shirahige K, Klein F. 2011. Spo11-accessory proteins link double-strand break sites to the chromosome axis in early meiotic recombination. Cell 146: 372–383. 10.1016/j.cell.2011.07.003 [DOI] [PubMed] [Google Scholar]
- Perez-Riba A, Itzhaki LS. 2019. The tetratricopeptide-repeat motif is a versatile platform that enables diverse modes of molecular recognition. Curr Opin Struct Biol 54: 43–49. 10.1016/j.sbi.2018.12.004 [DOI] [PubMed] [Google Scholar]
- Perry J, Kleckner N, Börner GV. 2005. Bioinformatic analyses implicate the collaborating meiotic crossover/chiasma proteins Zip2, Zip3, and Spo22/Zip4 in ubiquitin labeling. Proc Natl Acad Sci 102: 17594–17599. 10.1073/pnas.0508581102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyatnitskaya A, Borde V, De Muyt A. 2019. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma 128: 181–198. 10.1007/s00412-019-00714-8 [DOI] [PubMed] [Google Scholar]
- Quignot C, Postic G, Bret H, Rey J, Granger P, Murail S, Chacón P, Andreani J, Tufféry P, Guerois R. 2021. InterEvDock3: a combined template-based and free docking server with increased performance through explicit modeling of complex homologs and integration of covariation-based contact maps. Nucleic Acids Res 49: W277–W284. 10.1093/nar/gkab358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds A, Qiao H, Yang Y, Chen JK, Jackson N, Biswas K, Holloway JK, Baudat F, de Massy B, Wang J, et al. 2013. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet 45: 269–278. 10.1038/ng.2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A, Borde V. 2021. Methods to map meiotic recombination proteins in Saccharomyces cerevisiae. In Homologous recombination: methods and protocols (ed. Aguilera A, Carreira A), pp. 295–306, Springer US, New York. 10.1007/978-1-0716-0644-5_21 [DOI] [PubMed] [Google Scholar]
- Sanchez A, Adam C, Rauh F, Duroc Y, Ranjha L, Lombard B, Mu X, Wintrebert M, Loew D, Guarné A, et al. 2020. Exo1 recruits Cdc5 polo kinase to MutLγ to ensure efficient meiotic crossover formation. Proc Natl Acad Sci 117: 30577. 10.1073/pnas.2013012117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Zill OA, Rokas A, Payen C, Dunham MJ, Eisen MB, Rine J, Johnston M, Hittinger CT. 2011. The awesome power of yeast evolutionary genetics: new genome sequences and strain resources for the Saccharomyces sensu stricto genus. Adv Genet 1: 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm S, Fraune J, Naumann R, Hernandez-Hernandez A, Höög C, Cooke HJ, Alsheimer M, Benavente R. 2011. A novel mouse synaptonemal complex protein is essential for loading of central element proteins, recombination, and fertility. PLoS Genet 7: e1002088. 10.1371/journal.pgen.1002088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrentino M-E, Chaplais E, Sommermeyer V, Borde V. 2013. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet 9: e1003416. 10.1371/journal.pgen.1003416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha Y, Zheng L, Ji Z, Mei L, Ding L, Lin S, Wang X, Yang X, Li P. 2018. A novel TEX11 mutation induces azoospermia: a case report of infertile brothers and literature review. BMC Med Genet 19: 63. 10.1186/s12881-018-0570-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Tang D, Wang K, Wang M, Huang J, Luo W, Luo Q, Hong L, Li M, Cheng Z. 2012. ZIP4 in homologous chromosome synapsis and crossover formation in rice meiosis. J Cell Sci 125: 2581–2591. [DOI] [PubMed] [Google Scholar]
- Shinohara M, Oh SD, Hunter N, Shinohara A. 2008. Crossover assurance and crossover interference are distinctly regulated by the ZMM proteins during yeast meiosis. Nat Genet 40: 299–309. 10.1038/ng.83 [DOI] [PubMed] [Google Scholar]
- Smith AV, Roeder GS. 1997. The yeast Red1 protein localizes to the cores of meiotic chromosomes. J Cell Biol 136: 957–967. 10.1083/jcb.136.5.957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Huang L, Markowitz TE, Blitzblau HG, Chen D, Klein F, Hochwagen A. 2015. Transcription dynamically patterns the meiotic chromosome-axis interface. Elife 4: e07424. 10.7554/eLife.07424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M, Engebrecht J, Roeder GS. 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. 10.1016/0092-8674(93)90114-6 [DOI] [PubMed] [Google Scholar]
- Tessé S, Storlazzi A, Kleckner N, Gargano S, Zickler D. 2003. Localization and roles of Ski8p protein in Sordaria meiosis and delineation of three mechanistically distinct steps of meiotic homolog juxtaposition. Proc Natl Acad Sci 100: 12865–70. 10.1073/pnas.2034282100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessé S, Bourbon H-M, Debuchy R, Budin K, Dubois E, Liangran Z, Antoine R, Piolot T, Kleckner N, Zickler D, et al. 2017. Asy2/Mer2: an evolutionarily conserved mediator of meiotic recombination, pairing, and global chromosome compaction. Genes Dev 31: 1880–1893. 10.1101/gad.304543.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Lam I, Knop M, Keeney S. 2011. Exploiting spore-autonomous fluorescent protein expression to quantify meiotic chromosome behaviors in Saccharomyces cerevisiae. Genetics 189: 423–439. 10.1534/genetics.111.131326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thacker D, Mohibullah N, Zhu X, Keeney S. 2014. Homologue engagement controls meiotic DNA break number and distribution. Nature 510: 241–246. 10.1038/nature13120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubouchi T, Zhao H, Roeder GS. 2006. The meiosis-specific Zip4 protein regulates crossover distribution by promoting synaptonemal complex formation together with Zip2. Dev Cell 10: 809–819. 10.1016/j.devcel.2006.04.003 [DOI] [PubMed] [Google Scholar]
- Tung K-S, Roeder GS. 1998. Meiotic chromosome morphology and behavior in zip1 mutants of Saccharomyces cerevisiae. Genetics 149: 817–832. 10.1093/genetics/149.2.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Johnston C, Thappeta Y, Subramanian VV, Hochwagen A, MacQueen AJ. 2015. Separable crossover-promoting and crossover-constraining aspects of Zip1 activity during budding yeast meiosis. PLoS Genet 11: e1005335. 10.1371/journal.pgen.1005335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Cheng S-Y, Morehouse SJ, MacQueen AJ. 2016. Synaptonemal complex proteins of budding yeast define reciprocal roles in MutSγ-mediated crossover formation. Genetics 203: 1091–1103. 10.1534/genetics.115.182923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voelkel-Meiman K, Cheng S-Y, Parziale M, Morehouse SJ, Feil A, Davies OR, de Muyt A, Borde V, MacQueen AJ. 2019. Crossover recombination and synapsis are linked by adjacent regions within the N terminus of the Zip1 synaptonemal complex protein. PLoS Genet 15: e1008201. 10.1371/journal.pgen.1008201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang PJ, McCarrey JR, Yang F, Page DC. 2001. An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27: 422–426. 10.1038/86927 [DOI] [PubMed] [Google Scholar]
- Wang M, Wang K, Tang D, Wei C, Li M, Shen Y, Chi Z, Gu M, Cheng Z. 2010. The central element protein ZEP1 of the synaptonemal complex regulates the number of crossovers during meiosis in rice. Plant Cell 22: 417–430. 10.1105/tpc.109.070789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun S, Li Z, Zhang R, Xu J. 2017. Accurate de novo prediction of protein contact map by ultra-deep learning model. PLoS Comput Biol 13: e1005324. 10.1371/journal.pcbi.1005324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics 25: 1189–1191. 10.1093/bioinformatics/btp033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav VK, Claeys Bouuaert C. 2021. Mechanism and control of meiotic DNA double-strand break formation in S. cerevisiae. Front Cell Dev Biol 9: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA, Page DC, Benavente R, Her C, Höög C, McLaughlin KJ, et al. 2008. Meiotic failure in male mice lacking an X-linked factor. Genes Dev 22: 682–691. 10.1101/gad.1613608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko AN, Georgiadis AP, Röpke A, Berman AJ, Jaffe T, Olszewska M, Westernströer B, Sanfilippo J, Kurpisz M, Rajkovic A, et al. 2015. X-Linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N Engl J Med 372: 2097–2107. 10.1056/NEJMoa1406192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X-C, Li M-J, Cai F-F, Yang S-J, Liu H-B, Zhang H-B. 2021. A new TEX11 mutation causes azoospermia and testicular meiotic arrest. Asian J Androl 23: 510–515. 10.4103/aja.aja_8_21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakharyevich K, Tang S, Ma Y, Hunter N. 2012. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell 149: 334–347. 10.1016/j.cell.2012.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Shao J, Fan H-Y, Yu C. 2018. Evolutionarily-conserved MZIP2 is essential for crossover formation in mammalian meiosis. Commun Biol 1: 147. 10.1038/s42003-018-0154-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Ji S-Y, Busayavalasa K, Yu C. 2019. SPO16 binds SHOC1 to promote homologous recombination and crossing-over in meiotic prophase I. Sci Adv 5: eaau9780. 10.1126/sciadv.aau9780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Keeney S. 2015. High-resolution global analysis of the influences of Bas1 and Ino4 transcription factors on meiotic DNA break distributions in Saccharomyces cerevisiae. Genetics 201: 525–542. 10.1534/genetics.115.178293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 1999. Meiotic chromosomes: integrating structure and function. Annu Rev Genet 33: 603–754. 10.1146/annurev.genet.33.1.603 [DOI] [PubMed] [Google Scholar]
- Zickler D, Kleckner N. 2015. Recombination, pairing, and synapsis of homologs during meiosis. Cold Spring Harb Perspect Biol 7: a016626. 10.1101/cshperspect.a016626 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data were deposited at the NCBI Gene Expression Omnibus database with the accession number GSE177033.