Abstract
Abnormal immunological indicators associated with disease severity and mortality in patients with COVID-19 have been reported in several observational studies. However, there are marked heterogeneities in patient characteristics and research methodologies in these studies. We aimed to provide an updated synthesis of the association between immune-related indicators and COVID-19 prognosis. We conducted an electronic search of PubMed, Scopus, Ovid, Willey, Web of Science, Cochrane library, and CNKI for studies reporting immunological and/or immune-related parameters, including hematological, inflammatory, coagulation, and biochemical variables, tested on hospital admission of COVID-19 patients with different severities and outcomes. A total of 145 studies were included in the current meta-analysis, with 26 immunological, 11 hematological, 5 inflammatory, 4 coagulation, and 10 biochemical variables reported. Of them, levels of cytokines, including IL-1β, IL-1Ra, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, TNF-α, IFN-γ, IgA, IgG, and CD4+ T/CD8+ T cell ratio, WBC, neutrophil, platelet, ESR, CRP, ferritin, SAA, D-dimer, FIB, and LDH were significantly increased in severely ill patients or non-survivors. Moreover, non-severely ill patients or survivors presented significantly higher counts of lymphocytes, monocytes, lymphocyte/monocyte ratio, eosinophils, CD3+ T,CD4+T and CD8+T cells, B cells, and NK cells. The currently updated meta-analysis primarily identified a hypercytokinemia profile with the severity and mortality of COVID-19 containing IL-1β, IL-1Ra, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, TNF-α, and IFN-γ. Impaired innate and adaptive immune responses, reflected by decreased eosinophils, lymphocytes, monocytes, B cells, NK cells, T cells, and their subtype CD4+ and CD8+ T cells, and augmented inflammation, coagulation dysfunction, and nonpulmonary organ injury, were marked features of patients with poor prognosis. Therefore, parameters of immune response dysfunction combined with inflammatory, coagulated, or nonpulmonary organ injury indicators may be more sensitive to predict severe patients and those non-survivors.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12016-021-08908-8.
Keywords: COVID-19, Hematological parameters, Immunological indices, Inflammatory responses, Meta-analysis
Introduction
As of 27 September 2021, the outbreak of coronavirus disease 2019 (COVID-19) has affected more than 200 countries, with 231,703,120 confirmed cases and 4,746,620 deaths globally [1]. The disease is caused by infection with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which results in a large number of severe/critical ill patients who require rigorous management in intensive-care units (ICUs) [2–4]. Until now, there has been no consensus on an effective method to eradicate SARS-CoV-2. Prompt recognition and supportive care for potentially severe/critical ill patients are the mainstay treatments to save lives.
Our previous study [5] showed that the counts of lymphocytes, T cell subsets, and eosinophils decreased markedly in severely and fatally ill patients. Non-survivors maintained high levels of, or showed an upward trend in, neutrophil (Neu) counts, interleukin-6 (IL-6), procalcitonin (PCT), serum amyloid A protein (SAA), and C-reactive protein (CRP) levels, while levels of these markers held stable or showed a downward trend in survivors. In addition, studies from other research groups have also investigated the correlation between abnormal immune parameters, including white blood cells (WBC), lymphocytes (Lym), and eosinophil (Eos) counts, infection-related variables, serum inflammatory-cytokine levels, and severity or mortality of the disease [5–7]. Indeed, identifying early and sensitive indicators representative of innate and adaptive immune responses to COVID-19 may help predict the disease progression and potential fatal outcomes.
The evidence of immune abnormalities associated with disease severity and mortality in COVID-19 patients has been widely reported in many published observation clinical studies. However, these studies presented a significant heterogeneity in demographic characteristics, genetic features, and therapeutic approaches before hospital admission. Although previous systematic meta-analyses provided evidence of immune signatures in patients with COVID-19 in the early phase of the disease outbreak [8–11], a number of studies have emerged that offer updated data on the immune abnormality associated with poor clinical outcomes [12–16]. Therefore, we aimed to obtain updated, comprehensive evidence of the immune index alongside hematological, biochemical, inflammatory, and coagulation parameters in either a severity or mortality cohort to present the interplay between impaired immune responses and multi-system abnormality contributing to disease progression.
Materials and Methods
Search Strategy and Selection Criteria
This systematic review was conducted according to the Preferred Reporting in Systematic Reviews and Meta-Analyses (PRISMA) guidelines. We previously registered this meta-analysis in PROSPERO, and the study registration number is CRD42020196272. We searched seven databases, PubMed, Scopus, Ovid, Willey, Web of Science, Cochrane Library and the China National Knowledge Infrastructure (CNKI), using the advanced search mode in the field “Title/Abstract,” the search terms [“COVID-19” OR “SARS-COV-2”] AND [“biomarkers” OR “predictors” OR “parameters”] AND [“severity” OR “mortality”], from January 1, 2020 to August 20, 2021, without any language restrictions. After removing duplicates by Endnote, two reviewers independently assessed the title, abstract, and full text of each article to identify eligibility. Any disagreements were solved by a discussion with a third reviewer to reach a consensus. We included observational studies that consisted of two groups: (a) patients with different severities of COVID-19 and/or (b) patients who died from COVID-19 compared to those who survived. Articles with computable data about immune-related variables, including immunological and/or hematological, coagulation, inflammatory, and biochemical variables, were included in the current meta-analysis. The following results were excluded: reviews and meta-analysis, case reports, editorials, preprints, correspondences and letters, data papers, notes, comments, news, short surveys, erratums and retractions, guidelines, and mathematical models. Moreover, we used the Newcastle–Ottawa Scale (NOS) to evaluate the quality of each included publication.
Data Collection
Based on the classifications of the COVID-19 Diagnosis and Treatment Guideline in China (Interim version 8) [17], the severity of disease was classified as four types: mild, moderate, severe and critical. As the originally reported clinical groups were highly diverse among the included publications, we attempted to combine them into two groups, severe COVID-19 and non-severe COVID-19, for further meta-analysis. The strategy for this combination was as follows: (1) groups consisting of severe or critical cases, cases treated in ICUs, aggravations, refractory disease, and ARDS cases; and (2) groups consisting of non-severe, mild, moderate, common, ordinary, or general cases, cases not treated in ICUs, no aggravations, and cases without ARDS were placed into the non-severe COVID-19 group. Raw published/publicly available data were extracted, verified in duplication, and combined into a single database. In order to present the detailed characteristics of included studies, we extracted basic information of each study, which included the first author, year of publication, country and region, language, original reported groups, combined groups, average age, gender, and sample size of the “case and control groups.” We defined “severe and non-survivors” as “case groups” and “non-severe and survivors” as “control groups.” We also described the collected parameters in each study, including immunological and hematological parameters that are closely associated with immune function, and a few indexes reflecting infection, coagulation, and biochemical status. The final item was the quality score of studies, evaluated by the Newcastle–Ottawa Scale, with a higher score meaning higher quality.
Statistical Analysis
All analyses were performed using R software version 3.6.2 (package: meta/metafor; R Project for Statistical Computing, https://www.r-project.org). We divided studies into two separate cohorts for analysis: a severity cohort and a mortality cohort. For the meta-analysis, we transformed the format of laboratory variables presented as “median [interquartile range (IQR)]” into that of “mean [standard deviation (SD)]” [18, 19]. The value of “mean (SD)” of each included variable in the combined groups was calculated with the raw data from the originally reported groups using the formula proposed by Zhang et al. [20]. Standardized mean differences (SMDs) and 95% confidence intervals (95%CIs) were calculated as the primary metrics for each laboratory variable. Laboratory data was pooled whenever two or more publications reported a given variable. We quantified the variations in observed laboratory variables across studies attributable to heterogeneity using the I2 statistic, a metric ranging from 0% (indicating that all the heterogeneity was spurious) to 100% (indicating that all the heterogeneity was “real” and required further examination or explanation). To probe the sources of heterogeneity, we conducted a meta-regression analysis with three potential factors: the approach of combining disease severity, age, and region. The included variables that presented high heterogeneity (I2 > 50%) and were reported by an adequate number of studies (n ≥ 10) were applied to the analysis. In addition, the robustness of the results was applied by performing leave-one-out sensitivity analysis. The funnel plot method was used to test the publication bias.
Results
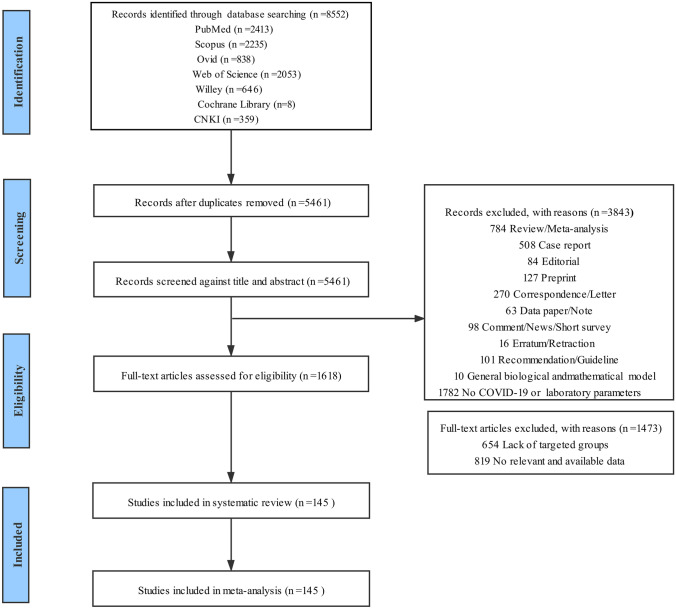
Figure 1 shows the flow diagram of selecting studies according to the PRISMA guidelines. We identified a total of 8552 records by searching seven databases. After removing duplicates, we screened the title and abstract of 5461 articles and excluded ineligible study designs (n = 2061) and unrelated to the topic (n = 1782). Then, we assessed 1618 full-text articles and excluded 1473 publications, mainly owing to no targeted groups (n = 654) and lacking of available and computable laboratory data (n = 819). Ultimately, we included 145 eligible publications in the systematic review and meta-analysis [5, 21–164]. Among the included studies, 91 ones were from China; and 54 studies were from America, Pakistan, Japan, Italy, France, Turkey, Korea, UK, Saudi Arabia, Egypt, India, Serbia, Greece, Libya, Spain, Iran, Mexico, Poland, Germany, and the Netherlands. All studies reported that laboratory variables were measured on admission or early during the hospitalization. There were 137 studies published in English and 8 studies published in Chinese. The characteristics of the included studies are presented in Table 1. Detailed results of the quality assessment of the included studies are presented in Fig. E1.
Fig. 1.
PRISMA flowchart of the study selection process
Table 1.
Characteristics of included studies
| Study | Region | Language | Original reported groups | Combined groups† | Sample size‡ | Case groups | Control groups | Collected parameters | Quality score | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (y) | Male [n (%)] | Age (y) | Male [n (%)] | Immunological | Hematological | Inflammatory | Coagulation | Biochemical | |||||||
| Chen 2020 | Chongqing, China | Chinese | Critical/Severe/Moderate | Severe/ Non severe | 143 (36/107) | 51 | 20 (55.6) | 43 | 52 (48.6) | IL-6 | WBC, Lym, PLT | PCT, CRP | D-dimer | LDH, CK | 8 |
| Li 2020 | Wuhan, China | Chinese | Critical/Severe/Moderate | Severe/Non severe | 62 (44/18) | 55 | 25 (56.8) | 50 | 7 (38.9) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IgA, IgM, IgE, IgG, C3, C4 | WBC, Neu, Lym, PLT | PCT,CRP | APTT, PT | LDH | 8 |
| Ling 2020 | Shanghai, China | Chinese | Severe/ Non severe | Severe/ Non severe | 292 (21/271) | 66 | 19 (90.5) | 49 | 135 (49.8) | CD3+(ab), CD4+(ab), CD8+(ab) | WBC, Neu, Lym | ESR,PCT, CRP | D-dimer, FIB | LDH, CK, cTnI, AST, ALT, CRN | 9 |
| Lu 2020 | Wuhan, China | Chinese | Severe/ Non severe | Severe/Non severe | 101 (34/67) | 61 | 17 (50) | 41 | 17 (25.4) | CD3+(ab), CD4+(ab), CD8+(ab), B(ab), NK(ab), IgA, IgM, IgG, C3, C4 | WBC, Neu, Lym, PLT | CRP | NA | LDH, ALT, AST, CRN, BUN | 7 |
| Xiang 2020 | Jiangxi, China | Chinese | Severe/ Moderate | Severe/Non severe | 49(9/40) | 53 | 8 (88.9) | 41 | 25 (62.5) | CD3+(ab), CD4+(ab), CD8+(ab), B(ab), NK(ab) | WBC, Neu, Lym, Eos, Mono, PLT, HB | PCT, ESR, CRP, SAA | APTT, PT, D-dimer, FIB | LDH, TBIL, ALB, AST, ALT, CRN, CK | 8 |
| Xu 2020 | Hefei,China | Chinese | Severe/ Moderate | Severe/ Non severe | 155 (30/125) | 51 | 20 (66.7) | 40 | 67 (53.6) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, IL-6 | NA | CRP, SAA | APTT | NA | 8 |
| Yang 2020 | Beijing, China | Chinese | Death/ Survival | Non survivors/ Survivors | 94 (13/81) | 77 | 8 (61.5) | 66 | 37 (45.7) | IL-6, IL-8, IL-10, TNF-α | WBC, Neu, Lym | Ferritin | NA | CRN, cTnI | 9 |
| Altschul 2020 | New York, America | English | Died/ Discharged | Non survivors/ Survivors | 2354 (621/ 1733) | 73 | 327 (52.7) | 63 | 771 (44.5) | IL-6 | WBC, Lym, PLT | PCT, CRP, Ferritin | D-dimer | AST, ALT, CRN, BUN, cTnI | 9 |
| Asghar 2021 | Karachi, Pakistan | English | A. ICU/ Ward B. Deceased/Recovered + Home isolation | A. Severe/Non severe B. Non survivors/ Survivors | A. 191 (61/130) B. 191 (44/147) | 53 | NA | NA | NA | NA | LMR, NLR, PLR, HB, PLT | PCT, CRP, Ferritin | NA | LDH | 7 |
| Awano 2020 | Tokyo, Japan | English | Severe/Non severe | Severe/Non severe | 54(21/33) | 66 | 15 (71.4) | 41 | 23 (69.7) | NA | WBC, Lym, Eos | PCT, Ferritin | D-dimer | LDH | 8 |
| Cai 2020 | Shenzhen,China | English | Severe/Non severe | Severe/ Non severe | 298 (58/240) | 61 | 39 (67.2) | 43 | 106 (44.2) | IL-6 | WBC, Neu, Lym, Eos | PCT, ESR, CRP | D-dimer | AST, ALT, CK, BUN, CRN, LDH, TBIL, MYO | 9 |
| Cao 2020 | Wuhan, China | English | Critical/Severe/Moderate | Severe/ Non severe | 244 (153/91) | 64 | 89 (58.2) | 60 | 44 (48.4) | CD4+(ab), CD8+(ab) | WBC, Neu, Lym, HB, PLT | PCT, CRP | NA | cTnI, AST, ALT, CRN, CK, LDH, MYO, TBIL | 8 |
| Chen 2020 | Wuhan, China | English | Severe/ Moderate | Severe/ Non severe | 21 (11/10) | 61 | 10 (90.9) | 50 | 7 (70.0) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), B(ab) | WBC, Neu, Lym, HB, PLT | PCT, CRP, Ferritin | APTT, PT, D-dimer | ALT, AST, BUN, CRN, CK, LDH, TBIL | 8 |
| Chen 2020 | Jiangsu,China | English | Severe or critical/ Mild/ Ordinary | Severe/ Non severe | 598 (31/567) | 61 | 302 (53.3) | 45 | 20 (64.5) | CD3+(ab), CD4+(ab), CD8+(ab), IL-6 | WBC, Neu, Lym, Eos, Mono, PLT, HB | PCT, CRP, ESR | APTT, PT, FIB | AST, ALT, BUN, CRN, LDH, TBIL | 8 |
| Chen 2020 | Wuhan, China | Chinese | Critical/Severe/Moderate | Severe/Non severe | 29 (14/15) | NA | NA | NA | NA | IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α | Lym | CRP | NA | LDH | 7 |
| Chen 2020 | Wuhan, China | English | A. Non survivors/ Survivors B. Critical/Severe/Moderate/Mild | A. Non survivors/ Survivors B. Severe/Non severe | A. 575 (103/445); B. 575 (203/345) | A.67 B.61 | A.69 (67.0)B.131 (64.5) | A.54 B.67 | A.244 (54.8)B.182 (52.8) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, IL-6 | WBC, Neu, Lym, Eos, Mono, Bas, NLR, PLR, PLT, HB | PCT, CRP, Ferritin, SAA | APTT, PT, D-dimer | NA | 8 |
| Chen 2020 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 55 (19/36) | 77 | 16 (84.2) | 72 | 18 (50.0) | IL-6 | WBC, Neu, Lym, PLT | PCT, ESR, CRP | D‐dimer | ALT, AST, ALB, CRN, LDH, CK | 8 |
| Chen 2020 | Wuhan, China | English | Dead/Survive | Non survivors/Survivors | 274(113/161) | 69 | 83 (73.0) | 51 | 171 (62.0) | IL-2R, IL-6, IL-8, IL-10, TNF-α, IgA, IgG, C3, C4 | WBC, Neu, Lym, Mono, HB, PLT | PCT, ESR, CRP,Ferritin | APTT, PT, D-dimer | ALT, AST, ALB, BUN, CRN, CK, LDH, cTnI, TBIL | 9 |
| Chen 2020 | Wuhan, China | English | Critical/Severe/Moderate | Severe/ Non severe | 48 (27/21) | 74 | 24 (88.9) | 53 | 13 (61.9) | IL-6 | WBC, Neu, Lym | PCT | NA | CRN, BUN | 8 |
| Chi 2020 | Nanjing, China | English | Severe/Moderate/Mild | Severe/ Non severe | 66(8/58) | 54 | 5 (63.0) | 42 | 32 (55.2) | IL-1Ra, IL-1β, IL-8, IL-10, IL-18, TNF-α, IFN-r | NA | NA | NA | NA | 8 |
| Ciceri 2020 | Milan, Italy | English | Dead/Discharged | Non survivors/Survivors | 386 (95/291) | 75 | 70 (73.7) | 63 | 207 (71.1) | IL-6 | WBC, Neu, Lym, HB, PLT, NLR | PCT,CRP, Ferritin | D-dimer | LDH, CK, cTnI, AST, CRN, TBIL | 9 |
| Dong 2020 | Wuhan, China | English | Severe/Non severe | Severe/Non severe | 147 (53/94) | 58 | 29 (54.7) | 43 | 34 (36.2) | IL-6, IL-10, TNF-α | WBC, Neu, Lym | CRP, ESR,SAA | D-dimer, FIB | ALT, AST, BUN, CRN | 8 |
| Du 2020 | Wuhan, China | English | Deceased/Survivors | Non survivors/Survivors | 179 (21/158) | 70 | 10 (47.6) | 56 | 87 (55.1) | CD4+(ab), CD8+(ab) | WBC, Neu, Lym | PCT, CRP | APTT, PT, D-dimer | MYO, ALB, TBIL, ALT, AST, CRN, cTnI | 8 |
| Feng 2020 | Wuhan, China | English | Poor outcome/Good outcome | Non survivors/Survivors | 114 (20/94) | 69 | 13 (65.0) | 63 | 58 (61.7) | CD4+(%), CD8+(%), IL-2, IL-4, IL-6, IL-10, IFN-γ | WBC, Neu, Lym, Mono, HB | CRP,Ferritin | APTT, PT, D-dimer, FIB | ALT, AST, BUN, CRN, CK, LDH, cTnI, TBIL | 9 |
| Feng 2020 | China | English | Critical/Severe/Moderate | Severe/Non severe | 476(124/352) | 59 | 81 (65.3) | 50 | 190 (54.0) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), TNF-α, IgA, IgM, IgG | WBC, Neu, Lym, HB, PLT | CRP,PCT, ESR | D-dimer, FIB | TBIL, ALB, MYO, CRN, LDH, CK, BUN | 8 |
| Guo 2020 | Shanghai,China | English | Severe/Non severe | Severe/Non severe | 200 (19/181) 348(68/280) | 57 | 46 (67.6) | 50 | 157 (56.1) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, IL-6, IgA, IgM, IgG | NA | NA | NA | NA | 7 |
| Han 2020 | Anhui, China | English | Severe/Mild | Severe/Non severe | 154 (32/122) | 52 | 23 (71.8) | 40 | 63 (51.6) | CD3+(ab), CD4 + T(ab), CD8 + T(ab), B(ab), NK(ab), IL-6 | WBC, Neu, Lym, PLT, HB | PCT, ESR, CRP, SAA | D-dimer, FIB, PT | ALT, AST, ALB, TBIL, BUN, CRN, CK, LDH | 7 |
| He 2020 | Wuhan, China | English | Severe/Non severe | Severe/Non severe | 204 (69/135) | 62 | 37 (53.6) | 42 | 42 (31.1) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IL-2, IL-4, IL-10, TNF, IFN-γ, IgA, IgM, IgE, IgG, C3, C4 | WBC, Neu, Lym, PLT | PCT, CRP | PT, D-dimer | ALT, AST, CRN, LDH, CK, cTnI | 7 |
| Huang 2020 | Wuhan, China | English | Dead/Alive | Non survivors/Survivors | 50 (10/40) | 38 | 5 (50.0) | 37 | 18 (45.0) | IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α, | Neu, Lym, Eos, Mono | CRP | APTT, PT, D-dimer, FIB | ALT, AST, cTnI, CK, CRN, BUN | 9 |
| Huang 2020 | Wuhan, China | English | Non survivors/ Survivors | Non survivors/Survivors | 151 (15/136) | 76 | 13 (86.7) | 59 | 68 (50.0) | IL-2R, IL-6, IL-8, TNF-α | WBC, Lym | PCT, CRP | NA | AST, ALT, BUN, CRN, MYO | 7 |
| Hue 2020 | Créteil, France | English | Dead/Alive | Non survivors/Survivors | 38 (13/25) | 68 | NA | 57 | NA | IL-6, IL-10 | NA | NA | NA | NA | 9 |
| Jiang 2020 | Wuhan, China | English | Non-survivor/Survivor | Non survivors/Survivors | 215 (72/143) | 70 | 36 (50.0) | 66 | 69 (48.3) | CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, B(ab), NK(ab) | WBC, Neu, Lym, Mono, HB, PLT | PCT, CRP | APTT, PT, D-dimer, FIB | ALT, AST, ALB, TBIL, LDH, cTnI, CRN, BUN | 8 |
| Kazancioglu 2020 | Ankara, Turkey | English | Severe/Non severe | Severe/Non severe | 120 (35/85) | 60 | 20 (57.1) | 44 | 52 (61.2) | IL-6 | WBC, Neu, Lym, Eos, Mono, Bas, HB, PLT | CRP, Ferritin | APTT, PT, D-dimer | ALT, AST, LDH, CK | 7 |
| Lei 2020 | Guangzhou, China | English | Severe & Critical/ Mild & Moderate | Severe/Non severe | 297 (52/245) | 60 | 32 (61.5) | 44 | 111 (45.3) | NA | Lym, Eos | CRP,PCT | NA | TBIL, LDH, ALB | 7 |
| Li 2020 | Shanghai,China | English | Severe/Non severe | Severe/Non severe | 322 (26/296) | 68 | 20 (76.9) | 49 | 147 (49.8) | CD3+(ab), CD4+(ab), CD8+(ab) | WBC, Lym, PLT | PCT, CRP | D-dimer | LDH, AST, ALT, TBIL, BUN, CRN, CK | 7 |
| Li 2020 | Beijing, China | English | Severe/Non severe | Severe/Non severe | 69 (26/43) | 59 | 14 (53.8) | 40 | 26 (60.5) | CD3+(ab), CD4+T(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IL-1β, IL-6, IL-8, TNF-α | WBC, Neu, Lym, Eos, Mono, Bas | PCT, ESR,CRP, Ferritin | FIB, D-dimer | ALB, AST, ALT, LDH | 9 |
| Liao 2020 | Wuhan, China | English | Critical/Severe/Moderate | Severe/Non severe | 380(231/149) | 67 | 137 (59.3) | 55 | 69 (46) | IL-2, IL-4, IL-6, IL-10, TNF-α, IFN-γ | WBC, Neu, Lym, Eos, Mono, Bas, HB, PLT | CRP,Ferritin | APTT, PT, D-dimer, FIB | LDH | 9 |
| Liu 2020 | Wuhan, China | English | Severe/Mild | Severe/Non severe | 140 (33/107) | 77 | 25 (75.8) | 61 | 66 (61.7) | IL-6 | NA | PCT, CRP | NA | NA | 9 |
| Liu 2020 | Wuhan, China | English | Severe/Mild | Severe/Non severe | 40 (13/27) | 60 | 7 (53.8) | 43 | 8 (29.6) | IgA, IgM, IgE, IgG, C3, C4 | WBC, Neu, Lym, Mono, HB, PLT |
CRP,Ferritin, SAA |
APTT, PT, D-Dimer, FIB | TBIL, ALT, AST, LDH, CK, BUN, CRN | 7 |
| Lu 2020 | Shanghai,China | English | Severe and critical/Mild and moderate | Severe/Non severe | 53(9/44) | 68 | 8 (88.9) | 53 | 26 (59.1) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, IgA, IgM, IgG, C3, C4 | WBC, Neu, Lym, Eos, Mono | CRP,ESR | APTT, PT, D-dimer, FIB | ALB, CRN, TBIL, LDH | 8 |
| Luo 2021 | Wuhan, China | English | Non survivors/ Survivors | Non survivors/Survivors | 1018(201/817) | 70 | 133 (66.2) | 56 | 388 (47.5) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, IL-2R, IL-6, IL-8, IL-10, TNF-α | NA | NA | NA | NA | 8 |
| Lv 2020 | Wuhan, China | English | Critical/Severe/Moderate | Severe/Non severe | 354(239/115) | 60 | 117 (49.0) | 54 | 58 (50.4) | IL-2, IL-4, IL-6, IL-10, TNF-α, IFN-γ, IgA, IgM, IgG, C3, C4 | WBC, Neu, Lym | PCT, CRP | D-dimer | BUN, TBIL | 8 |
| Mo 2020 | Wuhan, China | English | Refractory/General | Severe/Non severe | 155(85/70) | 61 | 31 (44.3) | 46 | 55 (64.7) | IL-6 | WBC, Neu, Lym, PLT | PCT, ESR, CRP | D-dimer | ALT, AST, ALB, CRN, CK, LDH | 9 |
| Park 2020 | Daegu, South Korea | English | Fatal cases/Survivor | Non survivors/Survivors | 289 (70/219) | 77 | 42 (60.0) | 70 | 91 (41.6) | NA | WBC, Lym, HB, PLT | CRP,PCT, ESR, Ferritin | PT | CK, AST, ALT, TBIL, BUN, CRN, LDH, ALB | 7 |
| Pei 2020 | Wuhan, China | English | Critical/Severe/Moderate | Severe/Non severe | 333(189/144) | 60 | 115 (60.8) | 51 | 67 (46.5) | IL-2R, IL-6, IL-10, TNF-α | Neu, Lym, Eos, Mono | ESR, CRP | PT, D-dimer | ALT, AST, cTnI,BUN | 9 |
| Qin 2020 | Wuhan, China | English | Severe/Moderate | Severe/Non severe | 452(286/ 166) | 60 | 155 (54.2) | 52 | 80 (48.2) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, B(ab), NK(ab), IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α, IgA, IgM, IgG, C3, C4 | WBC, Neu, Lym, Eos, Mono, Bas | PCT, ESR, CRP, Ferritin | NA | NA | 8 |
| Sinha 2020 | Newport and London, UK | English | Non survivors/Survivors | Non survivors/Survivors | 39 (17/22) | 60 | 14 (82.0) | 52 | 11 (50.0) | IL-6 | WBC, Lym, PLT | CRP,PCT, Ferritin | D-Dimer, FIB | ALB, cTnI, LDH, CRN | 8 |
| Sun 2020 | Jilin, China | English | A. Severe/Non severe B. Died/ Discharged | A. Severe/Non severe B. Non survivors/Survivors | A. 57(45/12) B. 36 (11/25) | 65 | 24 (53.3) | 58 | 5 (41.7) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, B(ab), NK(ab) | WBC, Neu, Mono, Eos, Bas | NA | NA | NA | 8 |
| Sun 2020 | Beijing, China | English | Critical/Severe/Moderate/ Mild | Severe/Non severe | 63 (19/44) | 59 | NA | 42 | NA | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, NK(ab), IL-6 | WBC, Neu, Lym, Eos, Mono, HB, PLT | CRP, ESR, Ferritin | PT, D-dimer, FIB | ALB, TBIL, CRN, ALT, AST, LDH, CK, BUN | 8 |
| Urra 2020 | Spain | English | ICU/Non ICU | Severe/Non severe | 172 (27/145) | 66 | 20 (74.1) | 58 | 84 (57.9) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%) | Neu, Lym, NLR, PLR | CRP, | D-dimer | NA | 8 |
| Wan 2020 | Chong qing, China | English | Severe/Moderate | Severe/Non severe | 123 (21/102) | 61 | NA | 43 | NA | CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IL-4, IL-6, IL-10, TNF-α, IFN-γ | WBC, Neu, Lym | NA | NA | NA | 8 |
| Wang 2020 | Wuhan, China | English | ICU/ Non-ICU | Severe/Non severe | 28(14/14) | 71 | 10 (71.4) | 66 | 11 (78.6) | IL-2R, IL-6, IL-8, IL-10, TNF-α | WBC, Neu, Lym, HB, PLT | PCT, ESR, CRP, Ferritin | PT, APTT, D-dimer | CK, LDH, ALT, AST, ALB, TBIL, cTnI, BUN | 9 |
| Wang 2020 | China | English | Severe/Common | Severe/Non severe | 61(24/37) | 56 | 15 (62.5) | 51 | 16 (43.2) | NA | WBC, Neu, Lym, Mono, PLT, LMR, NLR, PLR | PCT,CRP | PT, D-dimer | AST, LDH, ALB, CRN, CK | 9 |
| Wang 2020 | Wuhan, China | English | Deceased/Alive | Non survivors/Survivors | 119 (16/103) | 72 | 12 (8.2) | 59 | 49 (52.8) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+ | WBC, Neu, Lym, Mono, LMR, NLR | PCT,CRP | PT, APTT, D-dimer, FIB | CK, LDH, ALT, AST, CRN, MYO | 8 |
| Wang 2020 | Beijing, China | English | A. Severe/Moderate B. Non survivors/Survivors | A. Severe/Non severe B. Non survivors/Survivors | A. 199(129/70) B. 199(24/175) | A.65 B.72 | A.70 (54.3)B.16 (66.7) | A.58 B.62 | A.29 (41.4)B.86 (49.1) | IL-1β, IL-2R, IL-8, IL-10, TNF-α | WBC, Neu, Lym, HB, PLT | PCT, CRP, Ferritin | PT, APTT, D-dimer, FIB | AST, ALT, ALB, LDH, TBIL, BUN, CRN | 8 |
| Wang 2020 | Wuhan, China | English | Death/Survival | Nonsurvivors/Survivors | 339 (65/274) | 76 | 39 (60.0) | 69 | 127 (46.4) | CD8+(ab), IL-6 | WBC, Neu, Lym, Mono, HB, PLT | PCT, CRP | PT, APTT, D-dimer | AST, ALT, CRN, CK, cTnI, LDH, BUN | 9 |
| Wang 2020 | Wuhan, China | English | Severe/Non severe | Severe/Non severe | 43(8/35) | 6.81 | 6 (75.0) | 6.93 | 21 (60.0) | CD3+(ab), CD4+(ab), B(ab), NK(ab), IL-2, IL-4, IL-6, IL-10 | WBC, Lym | CRP | D-dimer | LDH, CK, ALT, AST, TBIL | 8 |
| Wang 2020 | Wuhan, China | English | Severe/Mild | Severe/Non severe | 69 (14/55) | 70 | 7 (50.0) | 40 | 25 (45.0) | CD4+(%), CD8+(%), IL-2, IL-4, IL-6, IL-10, TNF-α | WBC, Neu, Lym, Mono, Eos, HB, PLT | PCT, ESR, CRP | NA | AST, ALT, LDH, CRN | 8 |
| Wang 2020 | Wuhan, China | English | Nonsurviving/Surviving | Non survivors/Survivors | 293(116/ 177) | 73 | 65 (56.0) | 50 | 73 (41.2) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, B(ab), NK(ab), NK(%), IgA, IgG, C3, C4 | WBC, Neu, Lym | PCT, CRP | PT, APTT, D-dimer | ALT, AST, ALB, CRN, BUN, CK, LDH, MYO, cTnI, TBIL | 7 |
| Wu 2020 | Wuhan, China | English | A .ARDS/Without ARDS B. Died/Alive | A. Severe/ Non severe B. Non survivors/ Survivors | A. 201 (84/117) B. 84 (44/40) | A.59 B.68 | A.60 (71.4)B.29 (65.9) | A.47 B.49 | A.68 (58.1)B.31 (77.5) | CD3+(ab), CD4+(ab), CD8+(ab), IL-6 | WBC, Neu, Lym, Mono, PLT | ESR, CRP, Ferritin | PT, APTT, D-dimer | TBIL, AST, ALT, ALB, CRN, LDH, BUN | 8 |
| Xie 2020 | Wuhan,China | English | Severe/Non severe | Severe/Non severe | 56 (34/22) | 59 | 6 (27.3) | 53 | 18 (52.9) | CD3+(%), CD4+(%), CD8+(%), CD4+/CD8+ | WBC, Neu, Lym, PLT, HB | PCT, CRP | PT, APTT, D-dimer, FIB | ALT, AST, CRN, cTnI, CK, LDH | 7 |
| Xiong 2020 | Wuhan,China | English | Severe/Non severe | Severe/Non severe | 116 (55/61) | 64 | 38 (69.1) | 52 | 42 (68.9) | CD3+(ab), CD4+(ab), CD8+(ab), IL-6 | WBC, Neu, Lym, Mono, HB, PLT | CRP | PT, APTT, D-dimer | CRN, BUN, AST, ALT, TBIL, LDH, cTnI, MYO | 8 |
| Xu 2020 | Wuhan, China | English | A. Died/Discharged B. Critical/Severe/Mild | A. Non survivors/Survivors B. Severe/Nonsevere | A. 145 (28/117) B. 187 (107/80) | A.73 B.64 | A.17 (60.7)B.73 (68.2) | A.55 B.56 | A.59 (50.4)B.30 (37.5) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IL-1β, IL-6, IL-10, TNF-α | WBC, Neu, Lym, Mono | PCT, CRP, SAA | PT, D-dimer | BUN, CRN, ALT, AST, CK | 8 |
| Yan 2020 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 48(39/9) | 70 | 76 (70.4) | 49 | 38 (44.7) | IL-2R, IL-6, IL-8, TNF-α | WBC, Neu, Lym, HB, PLT | PCT, ESR, CRP, Ferritin | PT, APTT, FIB, D-dimer | ALT, AST, ALB, TBIL, CK, LDH, CRN,cTnI, BUN | 8 |
| Yang 2020 | Wuhan, China | English | Critical/Severe/Mild | Severe/Non severe | 52 (19/33) | NA | NA | NA | NA | CD4+(ab), CD8+(ab), IL-6 | WBC, Neu, Lym | PCT, CRP | D‐dimer | LDH, AST, ALT, CRN, cTnI | 8 |
| Yang 2020 | Shenzhen, China | English | Critical/Severe/Moderate | Severe/Non severe | 50(36/14) | 59 | 22 (61.1) | 50 | 7 (50.0) | CD4+(ab), CD8+(ab) | WBC, Neu, Lym, PLT | PCT, CRP | NA | AST, ALT, CRN, BUN, CK, LDH, TBIL | 8 |
| Yuan 2020 | Shenzhen, China | English | Critical/Severe/Moderate | Severe/Non severe | 214 (92/122) | 58 | 59 (64.1) | 41 | 58 (47.5) | CD4+(ab), IL-6 | WBC, Neu, PLT | CRP | D-Dimer | ALB | 7 |
| Zhang 2020 | China | English | Critical/Severe/Moderate/ Mild | Severe/Non severe | 414 (162/251) | 44 | 34 (57.6) | 42 | 11 (37.9) | NA | WBC, Neu, Lym, Mono, LMR, NLR, PLR | NA | NA | NA | 7 |
| Jun. Zhang 2020 | Wuhan, China | English | Deterioration/Discharge | Non survivors/Survivors | 111 (18/93) | 64 | 14 (77.8) | 38 | 32 (34.4) | IL-2, IL-4, IL-6, IL-10, TNF-α, IFN-γ | WBC, Neu, Lym, Mono, PLT | CRP | NA | CRN, BUN, ALT, AST | 9 |
| Zhang 2020 | Wuhan, China | English | Severe/Non severe | Severe/Non severe | 74 (27/47) | 70 | 18 (66.7) | 61 | 18 (38.3) | CD3+(ab), CD4+(ab), CD8+(ab), B(ab), NK(ab), IL-6, IgM, IgE, IgG | WBC, Neu, Lym, Eos, HB, PLT | PCT, ESR, CRP, SAA | D-dimer | CRN, ALB, AST, ALT, CK, LDH, cTnI | 7 |
| Zhao 2020 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 539 (125/414) | 71 | 71 (56.8) | 52 | 184 (44.4) | CD3+(ab), CD4+(ab), CD8+(ab), B(ab), IL-6, IgA, IgG | WBC, Neu, Lym, HB, PLT | PCT, CRP | NA | NA | 9 |
| Zhao 2020a | Beijing, China | English | Severe/Mild | Severe/Non severe | 71 (18/53) | 64 | 7 (38.9) | 45 | 53 (43.4) | IL-1β, IL-1Ra, IL-2, IL-4, IL-6, IL-10, IL-18, TNF-α | NA | NA | NA | NA | 9 |
| Zheng 2020 | Chengdu,China | English | Critical/Moderate | Severe/Non severe | 99 (32/67) | 64 | NA | 43 | NA | CD4+(ab), CD8+(ab) | WBC, Neu, Lym | CRP | PT, D-dimer | ALT, AST, MYO, cTnI | 7 |
| Zhou 2020 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 191 (54/137) | 69 | 38 (70.0) | 52 | 81 (59.0) | IL-6, | WBC, Lym, HB, PLT | PCT, Ferritin | PT, D-dimer | LDH, ALB, ALT, CK, cTnI | 8 |
| Zhou 2020 | Nanchang, China | English | Aggravation group/Non aggravation group | Severe/Non severe | 17(5/12) | 42 | 0 (0.0) | 42 | 6 (50.0) | CD4+(ab), CD8+(ab) | WBC, Lym | NA | D-dimer | LDH, ALB | 8 |
| Zhu 2020 | Ningbo, China | English | Severe/Non severe | Severe/Non severe | 127 (16/111) | 58 | 9 (56.3) | 50 | 73 (65.8) | IL-2, IL-4, IL-6, IL-10, TNF-α, IFN-γ | WBC, Neu, Lym, NLR, PLR | CRP,ESR | D-dimer, FIB | NA | 8 |
| Abers 2020 | New York, America | English | Critical/Severe/Moderate | Severe/Non severe | 175 (145/30) | NA | NA | NA | NA | IL-1β, IL-1Ra, IL-2, IL-4, IL-8, IL-18, TNF-α | NA | NA | NA | NA | 7 |
| G.Açıksarı 2021 | Turkey | English | Non survivors/Survivors | Non survivors/Survivors | 223 (36/187) | 74 | 15 (12.7) | 57 | 103 (87.3) | NA | WBC, Neu, Lym, Mono, HB, PLT, LMR, NLR, PLR | CRP | NA | NA | 8 |
| Alhumaid 2021 | Alahsa, Saudi Arabia | English | ICU/Non-ICU | Severe/Non severe | 1014(205/809) | 53 | 116 (56.5) | 45 | 466 (57.6) | NA | WBC, Neu, Lym, HB, PLT | CRP,ESR, Ferritin | NA | CK, AST, ALT, LDH, CRN, CK | 8 |
| Aly 2021 | Egypt | English | Critical/Severe/Non severe | Severe/Non severe | 496(311/185) | 57 | 181 (58.2) | 38 | 92 (49.5) | NA | HB, PLT, LMR, NLR, PLR | CRP,Ferritin | D-dimer | NA | 8 |
| Bellan 2021 | Italy | English | Dead/Discharged | Non survivors/Survivors | 664(211/ 453) | 80 | 144 (68.0) | 63 | 260 (57.0) | NA | WBC, Neu, Lym, Eos, NLR, PLT | NA | NA | NA | 7 |
| Bergantini 2021 | Italy | English | Severe/Mild to moderate | Severe/Non severe | 24(10/14) | 65 | 8 (80.0) | 62 | 11 (78.6) | CD4+(%), CD8+(%), IL-6 | WBC, Neu, Lym, Mono, Eos, Bas, PLT | CRP | NA | ALT, AST, LDH | 9 |
| Betti 2021 | Alessandria, Italy | English | Severec & critical/Mild & moderate | Severe/Non severe | 171 (82/89) | 57 | 54 (65.9) | 51 | 50 (56.2) | NA | WBC, Neu, Lym, Eos, HB, PLT | CRP,Ferritin | APTT, PT, D-dimer, FIB | ALT, AST, LDH, TBIL, CRN, cTnI, BUN | 8 |
| Bg 2021 | Davangere, India | English | Non survivors/Survivors | Non survivors/Survivors | 100 (25/75) | 59 | 13 (52.0) | 43 | 44 (58.7) | NA | LMR, NLR, PLR | NA | NA | NA | 8 |
| Cai 2020 | Wuhan, China | English | A. Severe/Non-severe B. Death/Recovery | A. Severe/Non-severe B. Non survivors/Survivors | A. 85(48/37) B. 41(22/19) C. 22(7/15) | A.64 B.67C.70 | A.34 (70.8)B.12 (54.5)C.3 (42.9) | A.55 B.50 C.66 | A.21 (56.8)B.9 (47.4)C.9 (60.0) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, TNF-α, IFN-γ | WBC, Neu, Lym, HB, PLT | CRP | APTT, PT, D-dimer | ALB, BUN, CRN, CK, AST, ALT | 9 |
| Capdevila-Reniu 2021 | Barcelona, Italy | English | Dead/Recovered | Non survivors/Survivors | 159 (53/106) | 86 | 29 (55.0) | 83 | 47 (44.0) | NA | Lym | CRP, Ferritin | D-dimer | LDH | 9 |
| Cekerevac 2021 | Serbia | English | Severe/Moderate/Mild | Severe/Non severe | 127 (70/57) | 61 | 49 (56.3) | 52 | 38 (66.7) | NA | WBC, Lym, HB, PLT | CRP | FIB | LDH, CK | 8 |
| Chen 2020 | Taiwan, China | English | Severe/Non severe | Severe/Non severe | 55 (24/31) | 52.1 | NA | NA | NA | IL-1β, IL-1Ra, IL-6, IL-18, TNF-α, IFN-γ | NA | NA | NA | NA | 7 |
| Conca 2021 | Saudi Arabia | English | Severe/Moderate/Mild | Severe/Non severe | 34 (5/29) | 74 | 5 (100.0) | 44 | 7 (24.1) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab) | WBC | CRP, Ferritin | D-dimer | NA | 7 |
| d’Alessandro 2020 | Siena, Italy | English | Severe/Non severe | Severe/Non severe | 54(14/40) | 65 | 12 (85.7) | 65 | 21 (52.5) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, B(ab), NK(ab) | NA | NA | NA | NA | 8 |
| Deng 2021 | Guangzhou, China | English | Severe & critical/Mild | Severe/Non severe | 166 (17/149) | 59 | 9 (52.9) | 48 | 65 (43.6) | CD3+(%), CD4+(%), CD8+(%), CD4+/CD8+ | HB, PLT | NA | PT, APTT, D-dimer, FIB | LDH, CK, CRN | 8 |
| Eleni 2021 | Greece | English | A. Death/No death B. ICU/No ICU | A. Non survivors/Survivors B. Severe/ Non severe | A. 5 (9/76) B. 85 (17/68) | A. 71 B. 60 | A. 5 (55.6) B. 14 (82.4) | A. 60 B. 60 | A. 44 (57.9) B. 35 (51.5) | NA | Lym, PLT | CRP, Ferritin | D-dimer, FIB | ALT, AST, LDH, cTnI | 8 |
| Elhadi 2021 | Libya | English | Non survivors/Survivors | Non survivors/Survivors | 465(281/184) | 69 | 153 (54.4) | 64 | 87 (47.3) | NA | WBC, Neu, Lym, PLT | CRP,PCT, Ferritin | PT, D-dimer, FIB | cTnI | 8 |
| García de Guadiana-Romualdo 2021 | Spain | English | A. Non survivors/Survivors B. Severe/Non- severe | A. Non survivors/Survivors B. Severe/Non- severe | A. 99 (14/85) B. 99 (25/74) | A. 76 B. 70 | A. 10 (71.4) B. 16 (64.0) | A. 64 B. 65 | A. 51 (60.0) B. 45 (60.8) | IL-6 | WBC, Neu, Lym, NLR, PLT, HB | CRP,PCT, Ferritin | D-dimer | CRN, ALB, ALT, LDH | 9 |
| Huang 2021 | Wuhan, China | English | Critical/Severe/Moderate | Severe/ Non severe | 218 (102/116) | 68 | 57 (49.1) | 56 | 49 (42) | CD3+(ab), CD4+(ab), CD8+(ab), B(ab), NK(ab), IL-6 | Neu, Lym, Mono, Eos, NLR | NA | NA | NA | 8 |
| Karahan 2021 | Turkey | English | A. Severe-Critica/Moderate B. Deceased/Surviving | A. Severe/Non severe B. Non survivors/Survivors | A. 149 (102/47) B. 149 (69/80) | A. 67 B. 68 | A. 58 (56.9) B. 41 (59.4) | A. 56 B. 60 | A. 23 (48.9) B. 40 (50.0) | NA | WBC, Neu, Lym, HB, PLT | CRP | NA | ALB | 8 |
| Karampoor 2021 | Iran | English | ICU/Non ICU | Severe/ Non severe | 120 (63/57) | 61 | 29 (46.0) | 52 | 35 (61.0) | IL-6, IL-10, IL-18 | NA | NA | NA | NA | 8 |
| Li 2021 | Wuhan, China | English | A. Nonsurvivors/Survivors B. ICU/Non ICU | A. Non survivors/Survivors B. Severe/ Non severe | A. 211 (95/116) B. (211/312) | 69 | 56 (58.9) | 56 | 63 (54.3) | IL-6 | WBC, HB | CRP, PCT | NA | BUN, CRN, cTnI, AST, ALT, TBIL, ALB | 8 |
| Li 2021 | Changchun,China | English | Severe or Critical/Nonsevere | Severe/Non severe | 285 (90/164) | 70 | 57 (63.3) | 64 | 73 (44.5) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab), IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α | WBC, Neu, Lym, Mono, PLT, NLR | CRP,PCT, ESR | APTT, PT, D-dimer, FIB | LDH, cTnI, AST, ALT, TBIL, ALB, CRN | 8 |
| Liu 2021 | Wuhan, China | English | Severe Common | Severe/Non severe | 122 (79/43) | 63 | 46 (58.2) | 53 | 26 (60.5) | IL-6 | WBC, Neu, Lym | CRP,PCT, SAA,ESR, Ferritin | APTT, PT, D-dimer, FIB | AST, ALT, CRN, LDH, CK, MYO, cTnI, BUN, TBIL | 8 |
| Liu 2020 | Nanchang,China | English | Severe/ Mild | Severe/ Non severe | 76 (30/46) | NA | NA | NA | NA | CD4+(ab), CD8+(ab), IL-1β, IL-2R, IL-6, IL-8, IL-10 | Lym | NA | APTT, PT, D-dimer, FIB | CK, LDH | 7 |
| López- Escobar 2021 | Madrid, Barcelona and Galicia, Spain | English | Non survivors/Survivors | Non survivors/Survivors | 2088 (321/1767) | 82 | 213(66.4) | 66 | 1032(58.4) | NA | WBC, Neu, Lym, Mono, NLR, PLT | CRP | APTT, PT, D-dimer | AST, ALT, LDH, CRN | 7 |
| Lu 2021 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 77 (40/37) | 60 | 29 (73.0) | 57 | 21 (57.0) | NA | WBC, Neu, Lym | CRP,PCT, ESR, Ferritin | APTT, PT, D-dimer | AST, ALT, CRN, LDH, CK, MYO, cTnI, BUN, TBIL | 9 |
| Marín-Corral 2021 | Spain | English | Critical/Severe/Moderate | Severe/Non severe | 49 (36/13) | 51 | 17 (47.2) | 59 | 7 (53.8) | NA | WBC, Lym | PCT | D-dimer, FIB | LDH | 9 |
| Montrucchio 2021 | Turin, Northern-Italy | English | Non survivors/Survivors | Non survivors/Survivors | 57(31/26) | 66 | 28 (90.3) | 60 | 22 (84.6) | NA | WBC, Lym | CRP,PCT | D-dimer | LDH | 8 |
| Nakamura 2021 | Tokyo, Japan | English | Non survivors/Survivors | Non survivors/Survivors | 32(11/21) | 74 | 10 (91.0) | 67 | 12 (57.0) | NA | WBC, Neu, Lym, HB, PLT | CRP, Ferritin | D-dimer | CRN, ALB, TBIL, LDH | 9 |
| Namendys-Silva 2021 | Mexico | English | Dead/ Alive | Non survivors/Survivors | 164 (85/79) | 57 | 59 (69.4) | 49 | 55 (69.6) | NA | WBC, Neu, Lym, PLT | CRP, Ferritin | D-dimer | CRN, TBIL | 8 |
| Özdemir 2021 | Turkey | English | Deceased/ Surviving | Non survivors/Survivors | 350 (55/295) | 73 | 29 (52.7) | 50 | 165 (55.9) | NA | WBC, Lym, HB | CRP | D-dimer | cTnI, CRN, AST, ALT, ALB | 7 |
| Peiro 2021 | Spain | English | Non survivors/Survivors | Non survivors/Survivors | 196 (37/159) | 76 | 23 (62.2) | 63 | 94 (59.1) | NA | WBC, Lym, HB, PLT | CRP | D-dimer | LDH, cTnI | 8 |
| Provencio 2021 | Spain | English | Died/ Survived | Non survivors/Survivors | 447 (146/301) | 68 | NA | 67 | NA | NA | Neu, Lym, Mono, NLR | CRP | D-dimer | LDH, ALB | 8 |
| Qin 2021 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 262 (23/239) | 69 | 10 (43.5) | 61 | 113 (47.3) | CD3+(ab), CD4+(ab), CD8+(ab), C3, C4 | WBC, Neu, Lym, Mono | CRP | NA | AST, ALT, CRN, LDH, TBIL | 8 |
| Quartuccio 2021 | Udine, Italy | English | Patients with P/F < 300/Patients with P/F ≥ 300 | Severe/ Non severe | 67(22/45) | 58 | 17 (77.3) | 59 | 32 (71.1) | IL-6, IL-18 | WBC, Neu, Lym | CRP | D-dimer | LDH, CK | 9 |
| Sai 2021 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 47 (15/32) | 71 | 7 (46.7) | 70 | 23 (71.9) | IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α | WBC, Neu, Lym, HB, PLT | CRP,PCT | D-dimer | LDH, AST, CRN, cTnI, TBIL | 9 |
| Salto‑Alejandre 2021 | Seville, Spain | English | Severe/ Moderate/ Mild | Severe/ Non severe | 321 (85/236) | 74 | 50 (58.8) | 60 | 119 (50.4) | NA | WBC, Neu, Lym | CRP | D-dimer | CRN, AST, LDH | 8 |
| Scotto 2021 | Italy | English | Unfavourable Outcome/Favourable Outcome | Non survivors/Survivors | 34 (15/19) | NA | NA | NA | NA | IL-6 | WBC, Neu, Lym | CRP | D-dimer | NA | 7 |
| Song 2021 | Hubei, China | English | Critical/Severe/Mild&moderate | Severe/ Non severe | 295 (107/188) | 67 | 67 (62.6) | 51 | 88 (46.8) | IL-6 | WBC, Lym, PLT | NA | D-dimer | ALT, AST, BUN, CRN, TBIL | 7 |
| Sozio 2021 | Italy | English | Death or IOT/Not death and not IOT | Non survivors/Survivors | 111 (28/83) | 64 | 22 (78.6) | 62 | 44 (53.0) | CD4+/CD8+, IL-1β, IL-6, IL-8, TNF-α | WBC, Neu, Lym | CRP,PCT | D-dimer | LDH, CK, CRN | 8 |
| Stachura 2021 | Kraków, Poland | English | Severe/ Non severe | Severe/ Non severe | 100(47/53) | 62.3 | 30 (63.8) | 56.5 | 33 (62.3) | IL-6 | WBC, Neu, Lym | CRP,PCT, Ferritin | APTT, D-dimer | AST, MYO, cTnI, LDH | 9 |
| Tang 2021 | Wuhan, China | English | Critical/Severe/Common | Severe/ Non severe | 100 (44/56) | 49 | 25 (56.8) | 39 | 31 (55.4) | IL-6 | Lym | PCT | NA | NA | 8 |
| Tao 2021 | Wuhan, China | English | Severe/ Non severe | Severe/ Non severe | 222 (20/202) | 68 | 12 (60.0) | 54 | 130 (64.4) | NA | WBC, Neu, Lym, Mono, Eos, Bas, HB, PLT | CRP,PCT, ESR | APTT, PT, D-dimer | MYO, LDH, ALT, AST, TBIL, BUN | 8 |
| Tepasse 2021 | Germany | English | Critical/Severe/Non severe | Severe/ Non severe | 40(31/9) | 59 | 29 (93.5) | 55 | 7 (77.8) | IL-6 | WBC | CRP,PCT, Ferritin | D-dimer | CRN, ALT, ALB | 8 |
| Viana-Llamas 2021 | Spain | English | Deceased /Alive | Non survivors/Survivors | 609 (128/481) | 80 | 85 (66.4) | 66 | 282 (58.6) | NA | WBC, Lym, HB, PLT | CRP, Ferritin | D-dimer, FIB | cTnI, LDH, AST, ALB, CRN | 8 |
| Wang 2021 | Wuhan, China | English | Critical/Severe/Non severe | Severe/ Non severe | A.211 (100/111) B.112(46/66) | 63 | 63 (68.0) | 46 | 38 (34.2) | CD3+(ab), CD3+(%), CD4+(ab), CD4+(%), CD8+(ab), CD8+(%), CD4+/CD8+, B(ab), NK(ab), IL-2, IL-4, IL-6, IL-10, IFN-γ | WBC, Lym | NA | NA | AST, ALT | 8 |
| Wang 2021 | Wuhan, China | English | Non survivors/Survivors | Non survivors/Survivors | 156 (56/100) | 74 | 32 (57.1) | 54 | 44 (44.0) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), IL-6 | WBC, Neu, Lym, HB, PLT | CRP,PCT | D-dimer | CRN, BUN, LDH, AST, ALT, CK, cTnI, TBIL | 8 |
| Waris 2021 | Pakistan | English | Critical/Severe/Moderate/Mild | Severe/ Non severe | 101 (25/76) | 62.1/ 56 | 17 (68.0) | 49.1 /43.24 | 53 (69.7) | NA | WBC, Lym, HB, PLT, LMR, PLR, NLR | NA | NA | NA | 7 |
| Xiong 2021 | China | English | Dead cases/Recovery cases | Non survivors/Survivors | 190 (85/105) | 72 | 53 (62.4) | 59 | 46 (43.8) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+, B(ab), NK(ab) | WBC, Neu, Lym, HB, PLT | CRP,PCT | D-dimer | LDH, CRN, CK, AST, ALT, BUN, TBIL | 7 |
| Xue 2021 | China | English | Severe/ Moderate | Severe/ Non severe | 289 (63/226) | 62 | 31 (49.2) | 54 | 99 (43.8) | NA | WBC, Neu, Lym | CRP | NA | NA | 8 |
| Yang 2021 | Wuhan, China | English | Non survivors/ Survivors | Non survivors/Survivors | 203 (58/145) | 67 | 38 (65.5) | 56 | 77 (53.1) | IL-6 | WBC, Neu, Lym | CRP | APTT, PT, D-dimer | CK, MYO, cTnI, LDH, ALT, AST, TBIL, ALB, CRN, BUN | 8 |
| Zayat 2021 | Heinsberg,German | English | Non survivors/ Survivors | Non survivors/Survivors | 17(8/9) | 57 | 4 (50.0) | 57 | 2 (22.0) | IL-6 | WBC, PLT, HB | CRP,PCT | D-dimer, FIB | LDH, CRN, BUN, CK, ALT | 9 |
| Zhang 2021 | Wuhan, China | English | Died/ Cured | Non survivors/Survivors | 208 (26/182) | 69 | 18 (69.0) | 62 | 111 (61.0) | CD3+(ab), CD3+(%), CD4+(ab), CD8+(ab), B(ab), NK(ab), IL-2R, IL-6, IL-8, IL-10, TNF-α, C3, C4 | WBC, Neu, Lym, Mono, Eos, Bas, HB, PLT | CRP,ESR, Ferritin | PT, D-dimer | AST, ALT, LDH, cTnI, BUN, CRN, TBIL | 9 |
| Zhao 2021b | Wuhan, China | English | Severe/Mild | Severe/ Non severe | 285 (74/211) | 67 | 38 (51.0) | 63 | 96 (45.0) | IL-6 | Neu, Lym, Mono, Eos, NLR, PLR, LMR | CRP | PT, D-dimer | ALT, AST, BUN, CRN | 8 |
| Ahmad 2021 | Northern India | English | Mortality/ Survival | Non survivors/Survivors | 1448(159/1289) | 58 | 128 (12.2) | 47 | 921 (87.8) | NA | HB, WBC, PLT | Ferritin, CRP | D-dimer | ALT, AST, ALB, BUN, CRN, LDH | 7 |
| Akdogan 2021 | Turkey | English | Severe/Non severe | Severe/Non severe | 175(57/118) | 52 | NA | 39 | NA | NA | WBC, Lym | CRP | D-dimer | LDH, AST, ALT, BUN | 7 |
| Berenguer 2020 | Spain | English | Dead/ Alive | Non survivors /Survivors | 4037 (1133/ 2904) | 79 | 1119 (68.5) | 64 | 2868 (58.1) | IL-6 | HB, WBC, Neu, Lym, NLR, PLT | CRP,PCT, Ferritin | D-dimer | ALT, AST, BUN, CRN, ALB, LDH | 7 |
| Albalawi 2021 | Saudi Arabia | English | Non-survivors/Survivors | Non survivors /Survivors | 119 (26/93) | 61 | 14 (53.9) | 53 | 66 (71.0) | NA | WBC, Neu, Lym, HB, PLT | CRP | D-dimer, PT, APTT | AST, ALT, BUN, CRN, LDH, ALB | 7 |
| Arikan 2021 | Turkey | English | Dead/Discharged | Non survivors /Survivor | 225/353 | 71 | 149 (66.2) | 67 | 203 (57.5) | NA | HB, WBC, Neu, Lym, PLT | Ferritin, PCT | FIB, D-dimer | BUN, CRN, AST, ALT, LDH, ALB | 7 |
| Chinnadurai 2020 | Bury, UK | English | Deceased/Alive | Non survivors /Survivors | 215(86/129) | 80 | 51 (59.3) | 68 | 82 (63.5) | NA | HB, Neu, Lym, NLR, PLT | CRP | D-dimer | ALB, ALT | 8 |
| d’Arminio Monforte 2020 | Italy | English | Death/ Survival | Non survivors /Survivors | 541(174/367) | 76 | 117 (67.2) | 61 | 230 (63.0) | NA | HB, WBC, Lym, PLT | CRP,PCT, Ferritin | D-dimer | LDH, CK, ALT, AST, CRN | 8 |
| Gozalbo-Rovira 2020 | Spain | English | ICU/pneumology ward | Severe/ Non severe | 51 (24/27) | 62 | 18 (75.0) | 58 | 14 (52.0) | IL-6 | Lym | CRP, Ferritin | D-dimer | LDH | 8 |
| Gupta 2020 | India | English | ICU/Non ICU | Severe/ Non severe | 200 (32/168) | 51 | 20 (62.4) | 38 | 96 (57.1) | NA | WBC, Lym, HB, PLT | NA | NA | BUN, CRN, AST, ALT | 8 |
| Kaal 2021 | The Netherlands | English | Severe/Non severe | Severe/ Non severe | 142 (41/101) | 69 | 28 (68.3) | 58 | 65 (64.4) | NA | WBC, Lym, PLT | CRP,PCT, Ferritin | NA | CRN, LDH | 8 |
| Duan 2020 | Chongqing, China | English | Severe/Non severe | Severe/ Non severe | 348 (20/328) | 58 | 170 (52.0) | 44 | 14 (70.0) | CD3+(ab), CD4+(ab), CD8+(ab), CD4+/CD8+ | WBC, Lym, NLR, PLT, HB | CRP,PCT | APTT, PT, FIB, D-dimer | ALB, ALT, AST, CRN, BUN, TBIL | 8 |
| Li 2020 | Wuhan, China | English | Non-survivors/Survivor | Non survivors/ Survivors | 102 (15/87) | 68 | 11 (73.0) | 55 | 48 (55.0) | IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α | WBC, Neu, Lym, HB, PLT | CRP,PCT | D-dimer, PT | cTnI, ALB, LDH, ALT, AST, TBIL, CRN, BUN | 8 |
| Li 2021 | Jinan, China | English | Non-survivors/Survivor | Non survivors/ Survivors | 99(9/63) | 72 | 6 (66.7) | 57 | 29 (43.9) | IL-6 | WBC, Lym, PLT | CRP,PCT | D-dimer | BUN, CRN, CK, LDH | 8 |
| Aksel 2021 | Turkey | English | Non-survivors/Survivor | Non survivors/ Survivors | 168 (32/136) | 70 | 17 (53.1) | 62 | 73 (53.7) | NA | WBC, Neu, Lym | CRP | NA | NA | 8 |
Data of age are presened as Mean. NA: not available. Combined groups†: Case group (Non-survivors/Severe)/Control group (Survivors/Non-severe). Sample size‡: Total sample (Case group sample/Control group sample). Quality score*: The Newcastle–Ottawa Scale was used for assessing the quality score of each article, with more stars meaning a higher score
Immunological Results
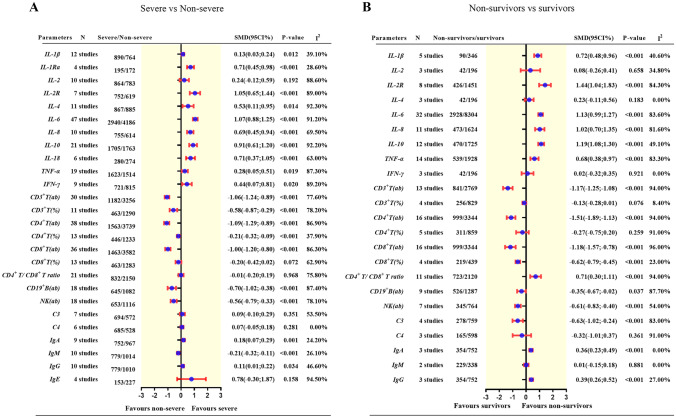
A total of 26 immunological variables were included for comparisons between patients with severe and those with non-severe COVID-19, including IL-1β, IL-1Ra, IL-2, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, tumor necrosis factor-alpha (TNF-α), interferon-γ (IFN-γ), CD3-positive T-lymphocyte absolute count (CD3+ T[ab]), CD3+ T percentage (CD3+T[%]), CD4+T(ab), CD4+T(%), CD8+T(ab), CD8+T(%), CD4+T(ab)/CD8+T(ab) ratio, B-lymphocyte absolute count (B cell[ab]), Natural-killer cell absolute count (NK[ab]), immunoglobulin A (IgA), IgM, IgG, IgE, C3(Complement 3), and C4. Of these, IL-1β, IL-2, IL-2R, IL-4, IL-6, IL-8, IL-10, TNF-α, IFN-γ, CD3+ T(ab), CD3+ T(%), CD4+ T(ab), CD4+T(%), CD8+ T(ab), CD8+T(%), CD4+T(ab)/CD8+T(ab) ratio, B cell(ab), NK cell(ab), IgA, IgM, IgG, C3, and C4 were available for comparisons between non-survivors and survivors infected with SARS-CoV-2. The summarized results are presented in Fig. 2. The detailed forest plots are presented in Fig. E2.
Severe Versus Non-severe COVID-19
Fig. 2.
Summary result of the comparison of immunological parameters between patients with severe COVID-19 and non-severe COVID-19 (A), and between non-survivors and survivors with COVID-19 (B)
IL-1β, IL-1Ra, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, TNF-α, IFN-γ, IgA, and IgG were significantly increased in patients with severe versus those with non-severe COVID-19 (IL-1β = 0.13 [95%CI, 0.03 to 0.24], P = 0.0121, I2 = 39.1%; IL-1Ra = 0.71 [95%CI, 0.45 to 0.98], P < 0.001, I2 = 28.6%; IL-2R = 1.05 [95%CI, 0.65 to 1.44], P < 0.0001, I2 = 89%; IL-4 = 0.53 [95%CI, 0.11 to 0.95], P = 0.014, I2 = 92.3%; IL-6 = 1.07 [95%CI, 0.88 to 1.25], P < 0.001, I2 = 91.2%; IL-8 = 0.69 [95%CI, 0.45 to 0.94], P < 0.0001, I2 = 69.5%; IL-10 = 0.91 [95%CI, 0.61 to 1.20], P < 0.001, I2 = 92.9%; IL-18 = 0.71 [95%CI, 0.37 to 1.05], P < 0.001, I2 = 63%; TNF-α = 0.28 [95%CI, 0.05 to 0.51], P = 0.0186, I2 = 87.3%; IFN-γ = 0.44 [0.07; 0.81], P = 0.0196, I2 = 89.2%; IgA = 0.18 [95%CI, 0.07 to 0.29], P < 0.001, I2 = 24.2%; IgG = 0.11 [95%CI, 0.01 to 0.22], P = 0.0335, I2 = 46.6%); whereas CD3+ T(ab), CD3+ T(%), CD4+ T(ab), CD4+ T(%), CD8+ T(ab), CD8+ T(%), Total B cell(ab), NK cell(ab), and IgM were significantly decreased in patients with severe versus those with non-severe COVID-19 (CD3+ T(ab) = −1.06 [95%CI, −1.24 to −0.89], P < 0.001, I2 = 77.6%; CD3+ T(%) = −0.58[95%CI, −0.87 to −0.29], P < 0.001, I2 = 78.2%; CD4+ T(ab) = −1.09 [95%CI, −1.29 to −0.89], P < 0.001, I2 = 86.9%; CD4+ T(%) = −0.21[95%CI, −0.32 to −0.09], P < 0.001, I2 = 37.9%; CD8+T(ab) = −1.00 [95%CI, −1.20 to −0.80], P < 0.001, I2 = 86.3%; B cell(ab) = −0.70 [95%CI, −1.02 to −0.38], P < 0.001, I2 = 87.4%; NK cell(ab) = −0.56 [95%CI, −0.79 to −0.33], P < 0.001, I2 = 78.1% and IgM = −0.21 [95%CI, −0.32 to −0.11], P < 0.001, I2 = 26.1%). There were no differences in IL-2, CD8+ T(%), CD4+T/CD8 + T ratio, C3, C4, and IgE between the two groups.
-
2.
Non-survivors Versus Survivors of COVID-19
IL-1β, IL-2R, IL-6, IL-8, IL-10, TNF-α and CD4+T/CD8+T ratio, IgA, and IgG were significantly increased in non-survivors versus survivors of COVID-19 (IL-1β = 0.72 [95%CI, 0.48 to 0.96], P < 0.001, I2 = 40.6%; IL-2R = 1.44 [95%CI, 1.04 to 1.83], P < 0.001, I2 = 84.3%; IL-6 = 1.13 [95%CI, 0.99 to 1.27], P < 0.001, I2 = 83.6%; IL-8 = 1.02 [95%CI, 0.70 to 1.35], P < 0.001, I2 = 81.6%; IL-10 = 1.19 [95%CI, 1.08 to 1.30], P < 0.001, I2 = 49.1%; TNF-α = 0.68 [95%CI, 0.38 to 0.97], P < 0.001, I2 = 83.8%; CD4+T/CD8+T ratio = 0.71[95%CI, 0.30 to 1.11], P = 0.0007, I2 = 94%; IgA = 0.36 [95%CI, 0.23 to 0.49], P < 0.001, I2 = 0%; IgG = 0.39[95%CI, 0.26 to 0.52], P < 0.001, I2 = 27%). CD3+ T(ab), CD4+ T(ab), CD8+ T(ab), CD8+ T(%), B cell(ab), NK cell(ab) and C3 were significantly decreased in non-survivors versus survivors (CD3+ T(ab) = −1.51 [95%CI, −1.89 to −1.13], P < 0.001, I2 = 94%; CD4+ T(ab) = −1.12[95%CI, −1.45 to −0.80], P < 0.001, I2 = 94%; CD8+T(ab) = −1.18[95%CI, –1.57 to −0.78], P < 0.001, I2 = 96%; CD8+ T(%) = −0.62 [95%CI, −0.79 to −0.45], P < 0.001, I2 = 23%; B cell(ab) = −0.35 [95%CI, −0.67 to −0.02], P = 0.0367, I2 = 87.7%; NK cell(ab) = −0.61 [95%CI, −0.83 to −0.40], P < 0.001, I2 = 54%; C3 = −0.63[95%CI, -1.02 to −0.24], P = 0.0014, I2 = 83%). There were no differences in IL-2, IL-4, IFN-γ, CD3+ T(%), CD4+ T(%), C4, and IgM between the two groups.
Hematological Results
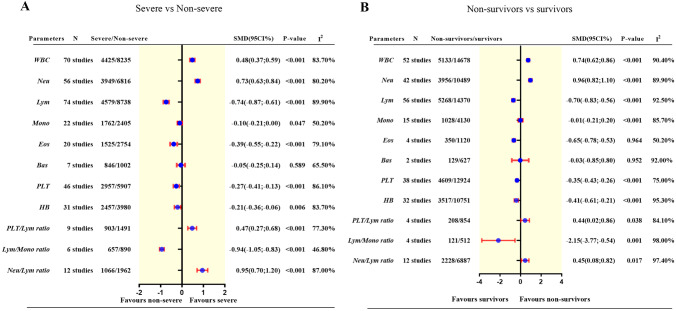
Eleven hematological variables, including WBC, neutrophil (Neu), lymphocyte (Lym), eosinophil (Eos), monocyte (Mono), basophil (Bas) absolute counts and platelet (PLT), hemoglobin (HB), neutrophil/lymphocyte ratio(NLR), lymphocyte/monocyte ratio (LMR), and platelet/lymphocyte ratio (PLR), were included in the meta-analysis for comparisons between patients with severe and non-severe COVID-19. All hematological parameters were available for comparisons between non-survivors and survivors of COVID-19. The summarized results are presented in Fig. 3. The detailed forest plots are presented in Fig. E3.
Severe Versus Non-severe COVID-19
Fig. 3.
Summary result of the comparison of hematological parameters between patients with severe COVID-19 and non-severe COVID-19 (A), and between non-survivors and survivors with COVID-19 (B)
WBC, Neu, NLR, and PLR counts were significantly increased in patients with severe versus those with non-severe COVID-19 (WBC = 0.48 [95%CI, 0.37 to 0.59], P < 0.001, I2 = 83.7%; Neu = 0.73 [95%CI, 0.63 to 0.84], P < 0.001, I2 = 80.2%; NLR = 0.95 [95%CI, 0.70 to 1.20], P < 0.001, I2 = 87%; PLR = 0.47 [95%CI, 0.27 to 0.68], P < 0.001, I2 = 77.3%), whereas Lym, Mono, LMR, Eos, PLT and HB were significantly decreased in patients with severe versus those with non-severe COVID-19 (Lym = −0.74 [95%CI, −0.87 to −0.61], P < 0.001, I2 = 89.9%; Mono = −0.10 [95%CI, −0.21 to −0.00], P = 0.0465, I2 = 50.2%; Eos = −0.39 [95%CI, −0.55 to −0.22], P < 0.001, I2 = 79.1%; LMR = −0.94 [95%CI, −1.05 to −0.83], P < 0.001, I2 = 46.8%; PLT = −0.27 [95%CI, −0.41 to −0.133], P < 0.001, I2 = 86.1%; HB = −0.21 [95%CI, −0.36 to −0.06], P = 0.006, I2 = 83.7%) There was no difference in the Bas count between the two groups.
-
2.
Non-survivors Versus Survivors of COVID-19
Similarly, WBC, Neu, NLR, and PLR were significantly increased in non-survivors versus survivors of COVID-19 (WBC = 0.74 [95%CI, 0.62 to 0.86], P < 0.001, I2 = 90.4%; Neu = 0.96 [95%CI, 0.82 to 1.10], P < 0.001, I2 = 89.9%; NLR = 0.45 [95%CI, 0.08 to 0.82], P = 0.0169, I2 = 97.4%; PLR = 0.44 [95%CI, 0.02 to 0.86], P = 0.038, I2 = 84.1%), whereas Lym, Eos, LMR, PLT and HB were significantly decreased in non-survivors versus survivors (Lym = −0.70 [95%CI, −0.83 to −0.56], P < 0.001, I2 = 92.5%; Eos = −0.65 [95%CI, −0.78 to −0.53], P < 0.001, I2 = 50.2%; LMR = −2.15 [95%CI, −3.77 to −0.54], P = 0.009, I2 = 98%; PLT = −0.35 [95%CI, −0.43 to −0.26], P < 0.001, I2 = 75%; HB = − 0.41 [95%CI, −0.61 to −0.21], P < 0.001, I2 = 95.3%). There were no differences in the Mono and Bas count between the two groups.
Other Abnormal Clinical Parameters Deriving from Immune Dysfunction
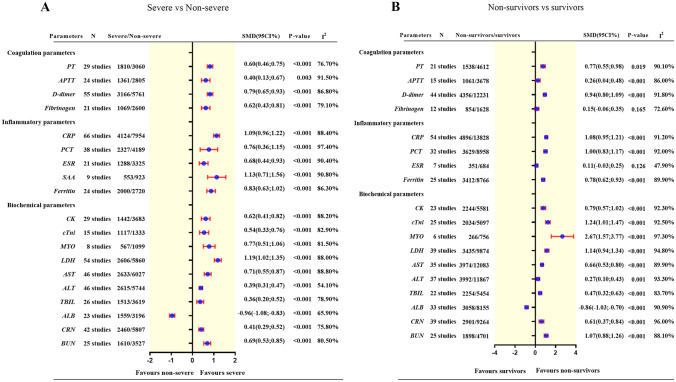
Beyond immunological and hematological cells, cytokines, antibodies and complements, there are some other laboratory parameters that are related to immune dysfunction and reflect the progression of COVID-19 which have been reported in previous studies [165–168]. In the current study, we simultaneously included coagulation parameters (including prothrombin time(PT), activated partial thromboplastin time(APTT), D-dimer and fibrinogen (FIB)), inflammatory parameters (containing C-reactive protein(CRP), procalcitonin(PCT), erythrocyte sedimentation rate(ESR), serum amyloid A(SAA)) and ferritin, biochemical parameters (including cardiac function related ones such as creatine kinase(CK), cardiac troponin I(cTnI), myoglobin (MYO), lactate dehydrogenase (LDH), liver function related ones such as aspartate aminotransferase(AST), alanine aminotransferase (ALT), total bilirubin(TBIL) and kidney function related ones such as creatinine(CRN), albumin(ALB), blood urea nitrogen(BUN). The summarized results are presented in Fig. 4.
Fig. 4.
Summary result of the comparison of coagulation, inflammatory and biochemical parameters between patients with severe COVID-19 and non-severe COVID-19 (A), and between non-survivors and survivors with COVID-19 (B)
Coagulation Results
Four coagulation variables, namely prothrombin time (PT), activated partial thromboplastin time (APTT), D-dimer and fibrinogen (FIB), were included in this study. All coagulation variables were available for comparisons between non-survivors and survivors of COVID-19. The detailed forest plots are presented in Fig. E4.
Severe Versus Non-severe COVID-19
The included four coagulation variables were significantly increased in patients with severe versus those with non-severe COVID-19 (PT = 0.60 [95%CI, 0.46 to 0.75], P < 0.001, I2 = 76.7%; APTT = 0.40 [95%CI, 0.13 to 0.67], P = 0.0033, I2 = 91.5%; D-dimer = 0.79 [95%CI, 0.65 to 0.93], P < 0.001, I2 = 86.8%; FIB = 0.62 [95%CI, 0.43 to 0.81], P < 0.001, I2 = 79.1%).
-
2.
Non-survivors Versus Survivors of COVID-19
Similarly, all the four coagulation variables were significantly increased in non-survivors versus survivors of COVID-19 (PT = 0.77 [95%CI, 0.55 to 0.98], P < 0.001, I2 = 90.1%; APTT = 0.26 [95%CI, 0.04 to 0.48], P = 0.0187, I2 = 86%; D-dimer = 0.94 [95%CI, 0.80; 1.09], P < 0.001, I2 = 91.8%). However, there was no difference in FIB between the two groups.
Inflammatory Results
Five inflammatory variables, C-reactive protein (CRP), procalcitonin (PCT), erythrocyte sedimentation rate (ESR), serum amyloid A (SAA) and ferritin, were included for comparisons between patients with severe and those with non-severe COVID-19, Of these, CRP, PCT, ESR and ferritin were available for comparisons between non-survivors and survivors infected with SARS-CoV-2. The detailed forest plots are presented in Fig. E5.
Severe Versus Non-severe COVID-19
Levels of all five inflammatory variables were significantly increased in patients with severe versus those with non-severe COVID-19 (CRP = 1.09 [95%CI, 0.96 to 1.22], P < 0.001, I2 = 88.4%; PCT = 0.76 [95%CI, 0.36 to 1.15], P < 0.001, I2 = 97.4%; ESR = 0.68 [95%CI, 0.44 to 0.93], P < 0.001, I2 = 90.4%; ferritin = 0.83 [95%CI, 0.63 to 1.02], P < 0.001, I2 = 86.3%; SAA = 1.13 [95%CI, 0.71 to 1.56], P < 0.001, I2 = 90.8%).
-
2.
Non-survivors Versus Survivors of COVID-19
CRP, PCT, and ferritin were significantly increased in non-survivors versus survivors of COVID-19 (CRP = 1.08 [95%CI, 0.95 to 1.21], P < 0.001, I2 = 91.2%; PCT = 1.00 [95%CI, 0.83 to 1.17], P < 0.001, I2 = 92%; ferritin = 0.78 [95%CI, 0.62 to 0.93], P < 0.001, I2 = 89.9%). There was no difference in ESR between the two groups.
Biochemical Results
Ten biochemical variables, namely creatine kinase (CK), cardiac troponin I (cTnI), myoglobin (MYO), lactate dehydrogenase (LDH), aspartate aminotransferase (AST), alanine aminotransferase (ALT), total bilirubin (TBIL), creatinine (CRN), albumin (ALB), and blood urea nitrogen (BUN), were included in this study. All biochemical variables were available for comparisons between non-survivors and survivors of COVID-19. The detailed forest plots are presented in Fig. E6.
Severe Versus Non-severe COVID-19
CK, cTnI, MYO, LDH, AST, ALT, TBIL, CRN, and BUN were significantly increased in patients with severe versus those with non-severe COVID-19 (CK = 0.62 [95%CI, 0.41 to 0.82], P < 0.001, I2 = 88.2%; cTnI = 0.54 [95%CI, 0.33 to 0.76], P < 0.001, I2 = 82.9%; MYO = 0.77 [95%CI, 0.51 to 1.06], P < 0.001, I2 = 81.5%; LDH = 1.19 [95%CI, 1.02 to 1.35], P < 0.001, I2 = 88%; AST = 0.71 [95%CI, 0.55 to 0.87], P < 0.001, I2 = 88.8%; ALT = 0.39 [95%CI, 0.31 to 0.47], P < 0.001, I2 = 54.1%; TBIL = 0.36 [95%CI, 0.20 to 0.52],P < 0.001, I2 = 78.9%; CRN = 0.41 [95%CI, 0.29 to 0.52], P < 0.001, I2 = 75.8%; BUN = 0.69 [95%CI, 0.53 to 0.85], P < 0.001, I2 = 80.5%;), whereas ALB was significantly decreased in patients with severe versus non-severe COVID-19 (ALB = −0.96 [95%CI, −1.08 to −0.83], P < 0.001, I2 = 65.9%).
-
2.
Non-survivors Versus Survivors of COVID-19
Similarly, CK, cTnI, MYO, LDH, AST, ALT, TBIL, CRN, and BUN were significantly increased in non-survivors versus survivors of COVID-19 (CK = 0.79 [95%CI, 0.57 to 1.02], P < 0.001, I2 = 92.3%; cTnI = 1.24 [95%CI, 1.01 to 1.47], P < 0.001, I2 = 92.5%; MYO = 2.67 [95%CI, 1.57 to 3.77], P < 0.001, I2 = 97.3%; LDH = 1.14 [95%CI, 0.94 to 1.34], P < 0.001, I2 = 94.8%; AST = 0.66 [95%CI, 0.53; 0.80], P < 0.001, I2 = 89.9%; ALT = 0.27 [95%CI, 0.10 to 0.43], P = 0.013, I2 = 93.3%; TBIL = 0.47 [95%CI, 0.32 to 0.63], P < 0.001, I2 = 83.7%; CRN = 0.61 [95%CI, 0.37 to 0.84], P < 0.001, I2 = 96%; BUN = 1.07 [95%CI, 0.88 to 1.26], P < 0.001, I2 = 88.1%). In contrast, ALB was significantly decreased in patients with severe versus non-severe COVID-19 (ALB = −0.86 [95%CI, –1.03 to −0.70], P < 0.001, I2 = 90.9%).
Publication Bias
Funnel plots are shown in Figs. E7 and E8. In severe and non-severe patients of COVID-19, the obvious publication bias was presented in B cell (ab), NK cell (ab), IL-1β, IL-4, IL-6, IL-10, TNF-α, NLR, CRP, D-dimer, and cTnI. In contrast, in non-survivors and survivors of COVID-19, obvious publication bias was present in IL-6, IL-8, IL-10, TNF-α, PLT, HB, CRP, Ferritin, ALT, and ALB. Many factors may have led to the publication bias, such as not enough amounts of originally included studies, different characteristics, and the wide ranges of the parameter results.
Sensitivity Analysis
Results of the sensitivity analysis, using the leave-to-out method, showed that most parameters presented good reliability and stability. However, there were also some parameters showed high sensitivity. Detailed results of each parameter are shown in Figs. E9, E10, E11, E12, and E13.
Investigation of Heterogeneity
A majority of included variables in the current review presented significant heterogeneity (I2 > 50%). The heterogeneity might have come from various factors, such as demographic and clinical characteristics of included patients, time of the symptom onset and laboratory parameters measured, and treatment intervention before the admission. Therefore, we conducted a meta-regression analysis with three potential factors, including the approach of combining disease severity, age, and region, to identify the sources of heterogeneity. The included variables presenting high heterogeneity (I2 > 50%) and reported by an adequate number of studies (n ≥ 10) were applied to the analysis. Regarding the approach of combing disease severity, we identified four subgroups in our severe group according to the originally reported disease severity: severe and critical (severe/critical), severe alone, critical alone, and other. The findings showed that the potential heterogeneity of 16 of 39 variables, including CD3+T(%), B cell(ab), NK(ab), IL-4, IL-6, IL-8, Lym, Eos, HB, NLR, CRP, Ferritin, LDH, ALB, CRN, and BUN, were related to the originally reported disease severity. The detailed results are presented in Table E1. Second, based on the available average age of severely ill patients and non-survivors of COVID-19, we classified the studies into six subgroups (average age ≤ 18 years(y), 30 ~ 49y, 50 ~ 59y, 60 ~ 69y, 70 ~ 79y, and ≥ 80y). In severe patients, the findings showed that the potential heterogeneity of 13 of 38 variables, including CD3+T(%), IL-8, PLT, HB, ESR, Ferritin, APTT, PT, FIB, cTnI, ALB, CRN, and BUN, were related to the different ages of the patients in the included studies. The detailed results are presented in Table E2. Similarly, the potential heterogeneity of 14 of 30 variables in non-survivors, including CD3+T(%), CD4+T(ab), CD8+T(ab), Neu, PLT, CRP, PCT, Ferritin, D-dimer, cTnI, AST, ALT, TBIL, and CRN, were related to the ages of the patients in different included studies. The detailed results are presented in Table E3. Moreover, we divided the four subgroups according to the continents (Asia, Europe, North America and Africa). In severe groups, the findings showed that the potential heterogeneity of 28 of 30 variables, including CD3+T(ab), CD3+T(%), CD4+T(ab), CD8+T(ab), B cell(ab), NK(ab), IL-4, IL-6, IL-8, TNF-α, WBC, Neu, Lym, Eos, HB, NLR, PCT, Ferritin, APTT, PT, FIB, CK, cTnI, LDH, AST, ALT, TBIL, and CRN, were related to the region. The detailed results are presented in Table E4. In non-survivors, the findings showed that the potential heterogeneity of 25 of 27 variables, including CD4+T/CD8+T ratio, IL-6, IL-8, TNF-α, WBC, Neu, Lym, PLT, HB, NLR, CRP, PCT, Ferritin, APTT, D-dimer, FIB, CK, cTnI, LDH, AST, ALT, TBIL, ALB, CRN and BUN, were related to the region. The detailed results are presented in Table E5. We considered the major source of heterogeneity as the regional differences among our included studies, while the approach of combining disease severity and the age of patients partially contributed to the marked heterogeneity observed.
Discussion
In the current updated meta-analysis, our synthetic results of 145 included studies identified a hypercytokinemia profile, including IL-1β, IL-1Ra, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, TNF-α, and IFN-γ, which was associated with increased severity and mortality in patients with COVID-19 infection. By contrast, patients with non-severe COVID-19 and survivors exhibited functional innate and adaptive immune responses, presenting by higher levels of eosinophils, lymphocytes, monocytes, B cells, NK cells, T cells and its subset CD4+ T, and CD8+T. Furthermore, in line with an elevated concentration of proinflammatory cytokines, augmented information (indicated by increased WBC, Neu, NLR, PLR, PCT, ESR, CRP, ferritin, or SAA), coagulation dysfunction (indicated by abnormal D-dimer, FIB, APTT and PT) as well as myocardial/liver/renal injury (indicated by elevated CK, cTnI, MYO, LDH, ALT, AST, TBIL, ALB, CRN, and BUN) were the main clinical abnormalities of patients with COVID-19 infection in the severe and fatal cohort.
SARS-CoV-2 infection can initiate a potent immune response, which includes innate immune activation and antiviral immune responses [169, 170]. However, the transition between innate and adaptive immune responses is the core of determining the clinical outcomes and prognosis of COVID-19 infection [171]. Early immune responses against COVID-19 primarily play a protective role in viral clearance, whereas exacerbated and dysregulated immune responses, otherwise known as the “cytokine storm,” can cause tissue damage contributing to poor disease outcomes [172]. An overreactive immune response releases excess pro-inflammatory cytokines and chemokines of which has been well documented [173]. Of these elevated pro-inflammatory cytokines, IL-6 is the most investigated and is a key driver of cytokine dysregulation, which is responsible for the hyper-inflammation in lungs in patients infected with COVID-19 [174]. A recent meta-analysis showed that the anti-IL-6 agent (Tocilizumab) was associated with a lower relative risk of mortality in patients with COVID-19 infection [175]. Other cytokines, such as IL-8 and IL-10, were also proposed to that play a significant role in the inflammatory cascade [176, 177]. We identified an updated abnormal cytokine profile, including IL-1β, IL-1Ra, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, TNF-α, and IFN-γ, relating to severe COVID-19 infection and fatality. It is well known that cytokine storm and the subsequent inflammation cascade relay on a complex cytokine network. Our synthesis results offer updated evidence on revealing the structure of cytokine networks related to the poor clinical outcomes, which helps clarify the underlying complex inflammatory pathways, so we can target new treatment agents.
Current management of COVID-19 is supportive, and respiratory failure from acute respiratory distress syndrome (ARDS) is the leading cause of mortality [178]. Hyperinflammation is the prominent feature of patients with ARDS and those non-survivors. Our previous longitudinal study of 548 revealed that patients who died from COVID-19 infection commonly showed an upward trend for neutrophils, IL-6, and C-reactive protein [5]. Other inflammatory parameters, including WBC, PCT, ESR, and SAA, were also proposed as the predictors of fatality. Our synthesis agrees with the findings of previous studies [179–183]. All patients with COVID-19, regardless of the severity, should be screened for hyperinflammation as precaution for potential ARDS once increases in these indicators are detected. Identification of the early signs of ARDS is critical for early intervention (such as low tidal volumes and prone ventilation) to improve oxygenation and lung compliance. Currently, the rates of bacterial/fungal co-infection reported in patients with COVID-19 appear to be low. Timothy et al. included nine studies and found that only 8% (62/806) of cases of bacterial/fungal co-infection were reported [184]. Nevertheless, our data observed that an increased infectious parameter profile detected on admission was strongly associated with poor clinical outcomes, which suggested that prompt antibiotic therapy should be considered after a comprehensive infectious assessment. Additionally, a combined assessment of using abnormal inflammatory parameters and increased cytokine levels might better identify the subgroup of patients for whom immunosuppression could improve mortality. Beneficial anti-inflammatory effects should be weighed against the potentially detrimental effects of inhibiting anti-viral immunity, thereby delaying virus clearance and perpetuating illness [185].
In addition, we observed substantial decreases in B cells, NK cells, T cells, and its subsets, including CD4+T cells and CD8+ T cells in patients with severe disease, compared to those with non-severe disease. We also found that decreased CD3+T, CD4+T, CD8+ T cells, and higher ratio of CD4+ to CD8+ T cells were associated with a fatal outcome. Our findings were in line with the results from a recent meta-analysis targeting lymphocytes and their subset counts [186] as well as observations from clinical practice. However, the underlying mechanism of observed lymphopenia in severe or fatal COVID-19 patients remains unclear. Based on the current evidence, it is proposed that lymphopenia be relating to the following causes: (1) suppression by cytokine mediation; (2) T cells infected by the virus; (3) T cell exhaustion (4) T cell expansion interfered with by the virus; and (5) organ inflation. Furthermore, our data supported that eosinopenia was associated with both severe disease and a fatal outcome. Our previous study suggested that dynamic changes in blood eosinophil counts might predict COVID-19 progression and recovery [5]. However, the pathophysiology for eosinopenia in COVID-19 remains unclear but is likely multifactorial [187], involving (1) reduced expression of adhesion/chemokine/cytokine, (2) direct eosinophil apoptosis, (3) blockade of eosinophilopoiesis, and (4) inhibition of eosinophil egress from the bone marrow. The finding that eosinophil levels improved in patients before discharge might serve as an indicator of improving clinical status.
The presence of the hypercoagulable state in patients with COVID-19 was another marked clinical feature of patients with increased mortality and a more severe form of the disease. The underlying pathophysiology mechanism was also associated with impaired immune responses [188]. SARS-CoV-2 infects host endothelial cells through ACE2 (an integral membrane protein) [189]. Patients with COVID-19 tend to exhibit greater numbers of ACE2-positive endothelial cells [190]. Therefore, vascular endothelial injury is commonly presented in patients with COVID-19. Vascular endothelial injury caused by COVID-19 infection would lead to the formation of microvascular microthrombi, which would trigger active tissue factor expression on macrophages and endothelial cells [191] Elevated tissue factor levels alongside local hypoxia from COVID-19 induced ARDS create a positive thromboinflammatory feedback loop, also known as a cytokine storm [191] The strong interaction between coagulation cascade activation and the cytokine storm might be responsible for the increased incidence of thrombotic events and aggressive inflammatory reactions. Based on our meta-analysis, increased APTT, PT, D-dimer, and FIB were identified as the indicators of coagulation dysfunction contributing to the unfavorable clinical outcomes. Simultaneously increased coagulation parameters and immune index might imply the interplay between overreactive immune responses and coagulation dysfunction which might serve as a more sensitive predicted index of a poor prognosis of COVID-19 infection. Additionally, we also identified several abnormal biochemical parameters representative of myocardial, liver, or renal injury in the severe and non-survivors cohort, such as CK, cTnI, MYO, LDH, ALT, AST, TBIL, ALB, CRN, and BUN. Although the pathophysiological mechanisms underlying myocardial/liver/renal injury by COVID-19 are not well-known so far, innate dysfunction and adaptive immune systems driving the cytokine storm seem to play a role in non-pulmonary organ damage [192–196], particularly those with comorbidities of cardiovascular, liver, and renal diseases.
The purpose of this meta-analysis is two-fold. First, to provide robust evidence of identifying a series of abnormal immunological indicators early to distinguish patients with poor clinical outcomes and to offer valuable information for exploring the underlying mechanism of COVID-19 progression. Second, to draw a picture of the interaction between immune abnormality and other body system dysfunction, including coagulation, inflammation, and non-pulmonary function. However, our meta-analysis has limitations. In line with the heterogeneity that characterized these observational studies [197, 198], a majority of included variables presented large I2 values, indicating significant variations in terms of outcomes observed. Although we attempted to manage this by performing subgroup analysis and meta-regression by disease severity, the age of included patients, and genetic characteristics, the results could not fully explain the source of heterogeneity. We were confined by the methodologies of the studies included, as well as the heterogeneity in characteristics of included patients, such as comorbidities, the therapeutic approach before hospital admission, and the time of symptom onset, which were not provided in the included studies. However, the observed heterogeneity did not impair our main conclusion that severe COVID-19 and mortality were associated with significant abnormalities in the immunological, hematological, coagulation, inflammatory, and biochemical variables. What the heterogeneity suggests is that these abnormalities might show some variation from one country to another, from one city to another, and from one clinical setting to another.
Conclusions
The currently updated meta-analysis primarily identified a hypercytokinemia profile with the severity and mortality of COVID-19, containing IL-1β, IL-1Ra, IL-2R, IL-4, IL-6, IL-8, IL-10, IL-18, TNF-α, and IFN-γ. Impaired innate and adaptive immune responses, reflected by decreased eosinophils, lymphocytes, monocytes, B cells, NK cells, T cells and their subtype CD4+ and CD8+ T cells, and augmented inflammation, coagulation dysfunction, and nonpulmonary organ injury, were marked features of patients with a poor prognosis. Given the strong interplay between immune response dysfunction, aggressive inflammation, coagulation abnormality, and nonpulmonary organ injury, parameters of immune response dysfunction combined with either inflammatory, coagulated, or nonpulmonary organ injury indicators may be more sensitive to predict outcomes in severe patients versus non-survivors.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file1: Quality assessment of the included studies (Newcastle–Ottawa Scale). +: high quality; –: low quality; ?: unclear quality (PNG 93 KB)
Acknowledgements
We thank LetPub (www.letpub.com) for its linguistic assistance and scientific consultation during the preparation of this manuscript.
Abbreviations
- ALT
Alanine aminotransferase
- APTT
Activated partial thromboplastin time
- AST
Aspartate aminotransferase
- BUN
Blood urea nitrogen
- Bas
Basophil
- COVID-19
Coronavirus disease 2019
- CK
Creatine kinase
- cTnI
Cardiac troponin I
- CNKI
China National Knowledge Infrastructure
- CRP
C-reactive protein
- CRN
Creatinine
- C3
Complement 3
- C4
Complement 4
- CD3+ T(ab)
CD3-positive T-lymphocyte absolute count
- CD3+ T(%)
CD3-positive T-lymphocyte percentage
- CIs
Confidence intervals
- ESR
Erythrocyte sedimentation rate
- Eos
Eosinophil
- FIB
Fibrinogen
- ICUs
Intensive-care units
- IL-6
Interleukin-6
- IFN-γ
Interferon-γ
- IgA
Immunoglobulin A
- IQR
Interquartile range
- 95%CIs
95% Confidence intervals
- LDH
Lactate dehydrogenase
- Lym
Lymphocyte
- LMR
Lymphocyte/monocyte ratio
- Mono
Monocyte
- MYO
Myoglobin
- MERS-CoV
Middle East respiratory syndrome coronavirus
- NLR
Neutrophil/lymphocyte ratio
- NOS
Newcastle–Ottawa Scale
- NK cells
Natural-killer cells
- Neu
Neutrophil
- NETs
Neutrophil extracellular traps
- PCT
Procalcitonin
- PT
Prothrombin time
- PRISMA
Preferred Reporting Items for Systematic Reviews and Meta-Analyses
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- SAA
Serum amyloid A protein
- SMDs
Standardized mean differences
- SD
Standard deviation
- TBIL
Total bilirubin
- TNF-α
Tumor necrosis factor-alpha
- WBC
White blood cell
Author Contribution
All the authors conceived and designed the study project. Li He and Rundong Qin performed the literature search and evaluated the study quality. Li He, Rundong Qin, Xinliu Lin, Renbin Huang, Tian Luo, Yukai Liu, Siyang Yao completed the data extraction. Rundong Qin and Li He performed the statistical analysis. This study was drafted by Rundong Qin, Li He, Zhaowei Yang, Nan Jia, Ruchong Chen, Jiaxing Xie, Wanyi Fu, Hao Chen, Jing Li. It was revised following critical review initially by Jing Li, Rundong Qin, Li He and all the co-authors. All the authors gave final approval of the version to be submitted and agreed to be accountable for the whole paper.
Declarations
Ethics Approval
As the current study did not involve human subjects, the Ethics Committee at the First Affiliated Hospital of Guangzhou Medical University exempted this study from the need for ethical approval.
Study Registration
PROSPERO CRD42020196272.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rundong Qin and Li He contributed equally as co-first authors.
Contributor Information
Mei Jiang, Email: jiangmei927@163.com.
Jing Li, Email: lijing@gird.cn.
References
- 1.World Health Organization (2021) WHO Coronavirus (COVID-19) Dashboard. Available at https://covid19.who.int/
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: A study of a family cluster. Lancet. 2020;395:514–523. doi: 10.1016/s0140-6736(20)30154-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen N, Zhou M, Dong X, Qu J, Gong F, Han Y, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: A descriptive study. Lancet. 2020;395:507–513. doi: 10.1016/S0140-6736(20)30211-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen R, Sang L, Jiang M, Yang Z, Jia N, Fu W, et al. Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J Allergy Clin Immunol. 2020;146:89–100. doi: 10.1016/j.jaci.2020.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu Z, McGoogan JM. Characteristics of and important lessons from the Coronavirus Disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA. 2020;323:1239–1242. doi: 10.1001/jama.2020.2648. [DOI] [PubMed] [Google Scholar]
- 7.Guan WJ, Ni ZY, Hu Y, Liang WH, Ou CQ, He JX, et al. Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu K, Yang T, Peng XF, Lv SM, Ye XL, Zhao TS, et al. A systematic meta-analysis of immune signatures in patients with COVID-19. Rev Med Virol. 2021;31:e2195. doi: 10.1002/rmv.2195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henry BM, de Oliveira MHS, Benoit S, Plebani M, Lippi G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin Chem Lab Med. 2020;58:1021–1028. doi: 10.1515/cclm-2020-0369. [DOI] [PubMed] [Google Scholar]
- 10.Feng X, Li S, Sun Q, Zhu J, Chen B, Xiong M, et al. Immune-inflammatory parameters in COVID-19 cases: A systematic review and meta-analysis. Front Med (Lausanne) 2020;7:301. doi: 10.3389/fmed.2020.00301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jesenak M, Brndiarova M, Urbancikova I, Rennerova Z, Vojtkova J, Bobcakova A, et al. Immune parameters and COVID-19 infection - associations with clinical severity and disease prognosis. Front Cell Infect Microbiol. 2020;10:364. doi: 10.3389/fcimb.2020.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dorgham K, Quentric P, Gökkaya M, Marot S, Parizot C, Sauce D, et al. Distinct cytokine profiles associated with COVID-19 severity and mortality. J Allergy Clin Immunol. 2021;147:2098–2107. doi: 10.1016/j.jaci.2021.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassallo M, Manni S, Pini P, Blanchouin E, Ticchioni M, Seitz-Polski B, et al. Patients with Covid-19 exhibit different immunological profiles according to their clinical presentation. Int J Infect Dis. 2020;101:174–179. doi: 10.1016/j.ijid.2020.09.1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osman M, Faridi RM, Sligl W, Shabani-Rad MT, Dharmani-Khan P, Parker A, et al. Impaired natural killer cell counts and cytolytic activity in patients with severe COVID-19. Blood Adv. 2020;4:5035–5039. doi: 10.1182/bloodadvances.2020002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bolondi G, Russo E, Gamberini E, Circelli A, Meca MCC, Brogi E, et al. Iron metabolism and lymphocyte characterisation during Covid-19 infection in ICU patients: An observational cohort study. World J Emerg Surg. 2020;15:41. doi: 10.1186/s13017-020-00323-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Trigo J, García-Azorín D, Sierra-Mencía Á, Tamayo-Velasco Á, Martínez-Paz P, Tamayo E, et al. Cytokine and interleukin profile in patients with headache and COVID-19: A pilot, CASE-control, study on 104 patients. J Headache Pain. 2021;22:51. doi: 10.1186/s10194-021-01268-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.National Health Commission of the People’s Republic of China (2021) COVID-19 Diagnosis and Treatment Guideline in China (Interim version 8). Available at https://www.gov.cn/zhengce/zhengceku/2020-08/19/5535757/files/da89edf7cc9244fbb34ecf6c61df40bf.pdf
- 18.Luo D, Wan X, Liu J, Tong T. Optimally estimating the sample mean from the sample size, median, mid-range, and/or mid-quartile range. Stat Methods Med Res. 2018;27:1785–1805. doi: 10.1177/0962280216669183. [DOI] [PubMed] [Google Scholar]
- 19.Shi J, Luo D, Weng H, Zeng XT, Lin L, Chu H, et al. Optimally estimating the sample standard deviation from the five-number summary. Res Synth Methods. 2020 doi: 10.1002/jrsm.1429. [DOI] [PubMed] [Google Scholar]
- 20.Zhang BKJ, Chen XM. Methods to combine standard deviations of different subgroups in meta-analysis. Chin J Evid Based Med. 2016;16:851–854. [Google Scholar]
- 21.Altschul DJ, Unda SR, Benton J, de la Garza RR, Cezayirli P, Mehler M, et al. A novel severity score to predict inpatient mortality in COVID-19 patients. Sci Rep. 2020;10:16726. doi: 10.1038/s41598-020-73962-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Asghar MS, Khan NA, Haider Kazmi SJ, Ahmed A, Hassan M, Jawed R, et al. Hematological parameters predicting severity and mortality in COVID-19 patients of Pakistan: A retrospective comparative analysis. J Community Hosp Intern Med Perspect. 2020;10:514–520. doi: 10.1080/20009666.2020.1816276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Awano N, Inomata M, Kuse N, Tone M, Takada K, Muto Y, et al. Serum KL-6 level is a useful biomarker for evaluating the severity of coronavirus disease 2019. Respir Investig. 2020;58:440–447. doi: 10.1016/j.resinv.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Berenguer J, Ryan P, Rodríguez-Baño J, Jarrín I, Carratalà J, Pachón J, et al. Characteristics and predictors of death among 4035 consecutively hospitalized patients with COVID-19 in Spain. Clin Microbiol Infect. 2020;26:1525–1536. doi: 10.1016/j.cmi.2020.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cai Q, Huang D, Ou P, Yu H, Zhu Z, Xia Z, et al. COVID-19 in a designated infectious diseases hospital outside Hubei Province, China. Allergy. 2020;75:1742–1752. doi: 10.1111/all.14309. [DOI] [PubMed] [Google Scholar]
- 26.Cao J, Zheng Y, Luo Z, Mei Z, Yao Y, Liu Z, et al. Myocardial injury and COVID-19: Serum hs-cTnI level in risk stratification and the prediction of 30-day fatality in COVID-19 patients with no prior cardiovascular disease. Theranostics. 2020;10:9663–9673. doi: 10.7150/thno.47980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen G, Wu D, Guo W, Cao Y, Huang D, Wang H, et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J Clin Invest. 2020;130:2620–2629. doi: 10.1172/jci137244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen J, Han T, Huang M, Yang Y, Shang F, Zheng Y, et al. Clinical characteristics of asymptomatic carriers of novel coronavirus disease 2019: A multi-center study in Jiangsu Province. Virulence. 2020;11:1557–1568. doi: 10.1080/21505594.2020.1840122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen L, Liu HG, Liu W, Liu J, Liu K, Shang J, et al. Analysis of clinical features of 29 patients with 2019 novel coronavirus pneumonia. Zhonghua Jie He He Hu Xi Za Zhi. 2020;43:203–208. doi: 10.3760/cma.j.issn.1001-0939.2020.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Chen T, Dai Z, Mo P, Li X, Ma Z, Song S, et al. Clinical characteristics and outcomes of older patients with Coronavirus Disease 2019 (COVID-19) in Wuhan, China: A single-centered, retrospective study. J Gerontol A Biol Sci Med Sci. 2020;75:1788–1795. doi: 10.1093/gerona/glaa089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen T, Wu D, Chen H, Yan W, Yang D, Chen G, et al. Clinical characteristics of 113 deceased patients with coronavirus disease 2019: Retrospective study. BMJ. 2020;368:m1091. doi: 10.1136/bmj.m1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen X, Zhao B, Qu Y, Chen Y, Xiong J, Feng Y, et al. Detectable serum severe acute respiratory syndrome coronavirus 2 viral load (RNAemia) is closely correlated with drastically elevated Interleukin 6 level in critically ill patients with coronavirus disease 2019. Clin Infect Dis. 2020;71:1937–1942. doi: 10.1093/cid/ciaa449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen YJRL, Pang XH, Mou HM, Wang JB, Lang CH, et al. Clinical features of coronavirus disease 2019 in Northeast area of Chongqing: Analysis of 143 cases. J Third Mil Med Univ. 2020;42:549–554. [Google Scholar]
- 34.Chi Y, Ge Y, Wu B, Zhang W, Wu T, Wen T, et al. Serum cytokine and chemokine profile in relation to the severity of coronavirus disease 2019 in China. J Infect Dis. 2020;222:746–754. doi: 10.1093/infdis/jiaa363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chinnadurai R, Ogedengbe O, Agarwal P, Money-Coomes S, Abdurrahman AZ, Mohammed S, et al. Older age and frailty are the chief predictors of mortality in COVID-19 patients admitted to an acute medical unit in a secondary care setting - a cohort study. BMC Geriatr. 2020;20:409. doi: 10.1186/s12877-020-01803-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciceri F, Castagna A, Rovere-Querini P, De Cobelli F, Ruggeri A, Galli L, et al. Early predictors of clinical outcomes of COVID-19 outbreak in Milan. Italy Clin Immunol. 2020;217:108509. doi: 10.1016/j.clim.2020.108509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monforte ADA, Tavelli A, Bai F, Tomasoni D, Falcinella C, Castoldi R, et al. The importance of patients’ case-mix for the correct interpretation of the hospital fatality rate in COVID-19 disease. Int J Infect Dis. 2020;100:67–74. doi: 10.1016/j.ijid.2020.09.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dong Y, Zhou H, Li M, Zhang Z, Guo W, Yu T, et al. A novel simple scoring model for predicting severity of patients with SARS-CoV-2 infection. Transbound Emerg Dis. 2020;67:2823–2829. doi: 10.1111/tbed.13651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Du RH, Liang LR, Yang CQ, Wang W, Cao TZ, Li M et al (2020) Predictors of mortality for patients with COVID-19 pneumonia caused by SARS-CoV-2: A prospective cohort study. Eur Respir J 55. 10.1183/13993003.00524-2020 [DOI] [PMC free article] [PubMed]
- 40.Duan J, Wang X, Chi J, Chen H, Bai L, Hu Q, et al. Correlation between the variables collected at admission and progression to severe cases during hospitalization among patients with COVID-19 in Chongqing. J Med Virol. 2020;92:2616–2622. doi: 10.1002/jmv.26082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng X, Li P, Ma L, Liang H, Lei J, Li W, et al. Clinical characteristics and short-term outcomes of severe patients with COVID-19 in Wuhan, China. Front Med (Lausanne) 2020;7:491. doi: 10.3389/fmed.2020.00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Feng Y, Ling Y, Bai T, Xie Y, Huang J, Li J, et al. COVID-19 with different severities: A multicenter study of clinical features. Am J Respir Crit Care Med. 2020;201:1380–1388. doi: 10.1164/rccm.202002-0445OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gozalbo-Rovira R, Gimenez E, Latorre V, Francés-Gómez C, Albert E, Buesa J, et al. SARS-CoV-2 antibodies, serum inflammatory biomarkers and clinical severity of hospitalized COVID-19 patients. J Clin Virol. 2020;131:104611. doi: 10.1016/j.jcv.2020.104611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guo L, Xiong W, Liu D, Feng Y, Wang P, Dong X, et al. The mNCP-SPI score predicting risk of severe COVID-19 among mild-pneumonia patients on admission. Infect Drug Resist. 2020;13:3593–3600. doi: 10.2147/idr.S263157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gupta N, Ish P, Kumar R, Dev N, Yadav SR, Malhotra N et al (2020) Evaluation of the clinical profile, laboratory parameters and outcome of two hundred COVID-19 patients from a tertiary centre in India. Monaldi Arch Chest Dis 90. 10.4081/monaldi.2020.1507 [DOI] [PubMed]
- 46.Han M, Xu M, Zhang Y, Liu Z, Li S, He T, et al. Assessing SARS-CoV-2 RNA levels and lymphocyte/T cell counts in COVID-19 patients revealed initial immune status as a major determinant of disease severity. Med Microbiol Immunol. 2020;209:657–668. doi: 10.1007/s00430-020-00693-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.He R, Lu Z, Zhang L, Fan T, Xiong R, Shen X, et al. The clinical course and its correlated immune status in COVID-19 pneumonia. J Clin Virol. 2020;127:104361. doi: 10.1016/j.jcv.2020.104361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang H, Zhang M, Chen C, Zhang H, Wei Y, Tian J, et al. Clinical characteristics of COVID-19 in patients with preexisting ILD: A retrospective study in a single center in Wuhan, China. J Med Virol. 2020;92:2742–2750. doi: 10.1002/jmv.26174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang Y, Guo H, Zhou Y, Guo J, Wang T, Zhao X, et al. The associations between fasting plasma glucose levels and mortality of COVID-19 in patients without diabetes. Diabetes Res Clin Pract. 2020;169:108448. doi: 10.1016/j.diabres.2020.108448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hue S, Beldi-Ferchiou A, Bendib I, Surenaud M, Fourati S, Frapard T, et al. Uncontrolled innate and impaired adaptive immune responses in patients with COVID-19 acute respiratory distress syndrome. Am J Respir Crit Care Med. 2020;202:1509–1519. doi: 10.1164/rccm.202005-1885OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang Y, Abudurexiti S, An MM, Cao D, Wei J, Gong P. Risk factors associated with 28-day all-cause mortality in older severe COVID-19 patients in Wuhan, China: A retrospective observational study. Sci Rep. 2020;10:22369. doi: 10.1038/s41598-020-79508-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kazancioglu S, Bastug A, Ozbay BO, Kemirtlek N, Bodur H. The role of haematological parameters in patients with COVID-19 and influenza virus infection. Epidemiol Infect. 2020;148:e272. doi: 10.1017/s095026882000271x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lei C, Lin W, Deng X, Hu F, Chen F, Cai W, et al. Factors associated with clinical outcomes in patients with coronavirus disease 2019 in Guangzhou. China J Clin Virol. 2020;133:104661. doi: 10.1016/j.jcv.2020.104661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li D, Wang M, He B, Xu Y, Zhou XY, Li WJ et al (2020) Laboratory test analysis of sixty-two COVID-19 patients. Med J Wuhan Univ 1–5
- 55.Li K, Chen D, Chen S, Feng Y, Chang C, Wang Z, et al. Predictors of fatality including radiographic findings in adults with COVID-19. Respir Res. 2020;21:146. doi: 10.1186/s12931-020-01411-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li Q, Zhang J, Ling Y, Li W, Zhang X, Lu H, et al. A simple algorithm helps early identification of SARS-CoV-2 infection patients with severe progression tendency. Infection. 2020;48:577–584. doi: 10.1007/s15010-020-01446-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li S, Jiang L, Li X, Lin F, Wang Y, Li B et al (2020) Clinical and pathological investigation of patients with severe COVID-19. JCI Insight 5. 10.1172/jci.insight.138070 [DOI] [PMC free article] [PubMed]
- 58.Liao D, Zhou F, Luo L, Xu M, Wang H, Xia J, et al. Haematological characteristics and risk factors in the classification and prognosis evaluation of COVID-19: A retrospective cohort study. Lancet Haematol. 2020;7:e671–e678. doi: 10.1016/s2352-3026(20)30217-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ling YIY, Qian ZP, Huang D, Zhang DD, Li T et al (2020) Clinical analysis of risk factors for severe patients with novel coronavirus pneumonia. Chin J Infect Dis 193–198. 10.3760/cma.j.cn311365-20200211-00055
- 60.Liu F, Li L, Xu M, Wu J, Luo D, Zhu Y, et al. Prognostic value of interleukin-6, C-reactive protein, and procalcitonin in patients with COVID-19. J Clin Virol. 2020;127:104370. doi: 10.1016/j.jcv.2020.104370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu J, Li S, Liu J, Liang B, Wang X, Wang H, et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. doi: 10.1016/j.ebiom.2020.102763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu Y, Sun K, Guo S, Wang J, Li A, Rong X, et al. Early warning indicators of severe COVID-19: A single-center study of cases from Shanghai. China Front Med (Lausanne) 2020;7:432. doi: 10.3389/fmed.2020.00432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu ZLHR, Jiang WY, Fan T, Geng Q (2020) Clinical characteristics and immune function analysis of COVID-19. Med J Wuhan Univ 41:529–532–546
- 64.Luo M, Liu J, Jiang W, Yue S, Liu H, Wei S (2020) IL-6 and CD8+ T cell counts combined are an early predictor of in-hospital mortality of patients with COVID-19. JCI Insight 5. 10.1172/jci.insight.139024 [DOI] [PMC free article] [PubMed]
- 65.Lv Z, Cheng S, Le J, Huang J, Feng L, Zhang B, et al. Clinical characteristics and co-infections of 354 hospitalized patients with COVID-19 in Wuhan, China: A retrospective cohort study. Microbes Infect. 2020;22:195–199. doi: 10.1016/j.micinf.2020.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mo P, Xing Y, Xiao Y, Deng L, Zhao Q, Wang H et al (2020) Clinical characteristics of refractory COVID-19 pneumonia in Wuhan, China. Clin Infect Dis. 10.1093/cid/ciaa270
- 67.Park JG, Kang MK, Lee YR, Song JE, Kim NY, Kweon YO, et al. Fibrosis-4 index as a predictor for mortality in hospitalised patients with COVID-19: A retrospective multicentre cohort study. BMJ Open. 2020;10:e041989. doi: 10.1136/bmjopen-2020-041989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pei G, Zhang Z, Peng J, Liu L, Zhang C, Yu C, et al. Renal involvement and early prognosis in patients with COVID-19 pneumonia. J Am Soc Nephrol. 2020;31:1157–1165. doi: 10.1681/asn.2020030276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Qin C, Zhou L, Hu Z, Zhang S, Yang S, Tao Y, et al. Dysregulation of immune response in patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin Infect Dis. 2020;71:762–768. doi: 10.1093/cid/ciaa248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinha P, Calfee CS, Cherian S, Brealey D, Cutler S, King C, et al. Prevalence of phenotypes of acute respiratory distress syndrome in critically ill patients with COVID-19: A prospective observational study. Lancet Respir Med. 2020;8:1209–1218. doi: 10.1016/s2213-2600(20)30366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun DW, Zhang D, Tian RH, Li Y, Wang YS, Cao J, et al. The underlying changes and predicting role of peripheral blood inflammatory cells in severe COVID-19 patients: A sentinel? Clin Chim Acta. 2020;508:122–129. doi: 10.1016/j.cca.2020.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sun Y, Dong Y, Wang L, Xie H, Li B, Chang C, et al. Characteristics and prognostic factors of disease severity in patients with COVID-19: The Beijing experience. J Autoimmun. 2020;112:102473. doi: 10.1016/j.jaut.2020.102473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Urra JM, Cabrera CM, Porras L, Ródenas I. Selective CD8 cell reduction by SARS-CoV-2 is associated with a worse prognosis and systemic inflammation in COVID-19 patients. Clin Immunol. 2020;217:108486. doi: 10.1016/j.clim.2020.108486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wan S, Yi Q, Fan S, Lv J, Zhang X, Guo L, et al. Relationships among lymphocyte subsets, cytokines, and the pulmonary inflammation index in coronavirus (COVID-19) infected patients. Br J Haematol. 2020;189:428–437. doi: 10.1111/bjh.16659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang F, Yang Y, Dong K, Yan Y, Zhang S, Ren H, et al. Clinical charactheristics of 28 patients with diabetes and COVID-19 in Wuhan, China. Endocr Pract. 2020;26:668–674. doi: 10.4158/ep-2020-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang H, Xing Y, Yao X, Li Y, Huang J, Tang J et al (2020) Retrospective study of clinical features of COVID-19 in inpatients and their association with disease severity. Med Sci Monit 26:e927674. 10.12659/msm.927674 [DOI] [PMC free article] [PubMed]
- 77.Wang J, Yu H, Hua Q, Jing S, Liu Z, Peng X, et al. A descriptive study of random forest algorithm for predicting COVID-19 patients outcome. PeerJ. 2020;8:e9945. doi: 10.7717/peerj.9945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang J, Zhang H, Qiao R, Ge Q, Zhang S, Zhao Z, et al. Thrombo-inflammatory features predicting mortality in patients with COVID-19: The FAD-85 score. J Int Med Res. 2020;48:300060520955037. doi: 10.1177/0300060520955037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang L, He W, Yu X, Hu D, Bao M, Liu H, et al. Coronavirus disease 2019 in elderly patients: Characteristics and prognostic factors based on 4-week follow-up. J Infect. 2020;80:639–645. doi: 10.1016/j.jinf.2020.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang Y, Zhu F, Wang C, Wu J, Liu J, Chen X, et al. Children hospitalized with severe COVID-19 in Wuhan. Pediatr Infect Dis J. 2020;39:e91–e94. doi: 10.1097/inf.0000000000002739. [DOI] [PubMed] [Google Scholar]
- 81.Wang Z, Yang B, Li Q, Wen L, Zhang R. Clinical features of 69 cases with coronavirus disease 2019 in Wuhan, China. Clin Infect Dis. 2020;71:769–777. doi: 10.1093/cid/ciaa272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wang Z, Ye D, Wang M, Zhao M, Li D, Ye J, et al. Clinical features of COVID-19 patients with different outcomes in Wuhan: A retrospective observational study. Biomed Res Int. 2020;2020:2138387. doi: 10.1155/2020/2138387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wu C, Chen X, Cai Y, Xia J, Zhou X, Xu S, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med. 2020;180:934–943. doi: 10.1001/jamainternmed.2020.0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xiang TXLJ, Xu F, Cheng N, Liu Y, Qian KJ, et al. Analysis of clinical characteristics of 49 patients with coronavirus disease 2019 in Jiangxi. Chin J Resp Crit Care Med. 2020;19:154–160. [Google Scholar]
- 85.Xie J, Ding C, Li J, Wang Y, Guo H, Lu Z, et al. Characteristics of patients with coronavirus disease (COVID-19) confirmed using an IgM-IgG antibody test. J Med Virol. 2020;92:2004–2010. doi: 10.1002/jmv.25930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Xiong S, Liu L, Lin F, Shi J, Han L, Liu H, et al. Clinical characteristics of 116 hospitalized patients with COVID-19 in Wuhan, China: A single-centered, retrospective, observational study. BMC Infect Dis. 2020;20:787. doi: 10.1186/s12879-020-05452-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Xu B, Fan CY, Wang AL, Zou YL, Yu YH, He C, et al. Suppressed T cell-mediated immunity in patients with COVID-19: A clinical retrospective study in Wuhan, China. J Infect. 2020;81:e51–e60. doi: 10.1016/j.jinf.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu J, Han MF, Zhao FD, Zhang T, Ma L (2020) Clinical manifestations and sero-immunological characteristics of 155 patients with COVID-19. Chin J Nosocomiol 2261–2265. 10.11816/cn.ni.2020-200577
- 89.Yan Y, Yang Y, Wang F, Ren H, Zhang S, Shi X et al (2020) Clinical characteristics and outcomes of patients with severe covid-19 with diabetes. BMJ Open Diabetes Res Care 8. 10.1136/bmjdrc-2020-001343 [DOI] [PMC free article] [PubMed]
- 90.Yang F, Shi S, Zhu J, Shi J, Dai K, Chen X. Clinical characteristics and outcomes of cancer patients with COVID-19. J Med Virol. 2020;92:2067–2073. doi: 10.1002/jmv.25972. [DOI] [PubMed] [Google Scholar]
- 91.Yang H, Yang LC, Zhang RT, Ling YP, Ge QG (2020) Risks factors for death among COVID-19 patients combined with hypertension, coronary heart disease or diabetes. J Peking Univ (Health Sci) 52:420–424 [DOI] [PMC free article] [PubMed]
- 92.Yang Y, Shen C, Li J, Yuan J, Wei J, Huang F, et al. Plasma IP-10 and MCP-3 levels are highly associated with disease severity and predict the progression of COVID-19. J Allergy Clin Immunol. 2020;146:119–127.e114. doi: 10.1016/j.jaci.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yuan J, Kou S, Liang Y, Lan J, Li X, Zeng L, et al. Immunological evaluation on potential treatment window for hospitalized COVID-19 patients. J Inflamm Res. 2020;13:985–993. doi: 10.2147/jir.S280331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhang H, Cao X, Kong M, Mao X, Huang L, He P, et al. Clinical and hematological characteristics of 88 patients with COVID-19. Int J Lab Hematol. 2020;42:780–787. doi: 10.1111/ijlh.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang J, Yu M, Tong S, Liu LY, Tang LV. Predictive factors for disease progression in hospitalized patients with coronavirus disease 2019 in Wuhan. China J Clin Virol. 2020;127:104392. doi: 10.1016/j.jcv.2020.104392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang Q, Wei Y, Chen M, Wan Q, Chen X. Clinical analysis of risk factors for severe COVID-19 patients with type 2 diabetes. J Diabetes Complications. 2020;34:107666. doi: 10.1016/j.jdiacomp.2020.107666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zhao Y, Nie HX, Hu K, Wu XJ, Zhang YT, Wang MM, et al. Abnormal immunity of non-survivors with COVID-19: Predictors for mortality. Infect Dis Poverty. 2020;9:108. doi: 10.1186/s40249-020-00723-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhao Y, Qin L, Zhang P, Li K, Liang L, Sun J et al (2020) Longitudinal COVID-19 profiling associates IL-1RA and IL-10 with disease severity and RANTES with mild disease. JCI Insight 5. 10.1172/jci.insight.139834 [DOI] [PMC free article] [PubMed]
- 99.Zheng Y, Xu H, Yang M, Zeng Y, Chen H, Liu R, et al. Epidemiological characteristics and clinical features of 32 critical and 67 noncritical cases of COVID-19 in Chengdu. J Clin Virol. 2020;127:104366. doi: 10.1016/j.jcv.2020.104366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet. 2020;395:1054–1062. doi: 10.1016/s0140-6736(20)30566-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhou Y, Zhang Z, Tian J, Xiong S (2020) Risk factors associated with disease progression in a cohort of patients infected with the 2019 novel coronavirus. Ann Palliat Med 9:428–436. 10.21037/apm.2020.03.26 [DOI] [PubMed]
- 102.Zhu Z, Cai T, Fan L, Lou K, Hua X, Huang Z, et al. Clinical value of immune-inflammatory parameters to assess the severity of coronavirus disease 2019. Int J Infect Dis. 2020;95:332–339. doi: 10.1016/j.ijid.2020.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abers MS, Delmonte OM, Ricotta EE, Fintzi J, Fink DL, de Jesus AAA et al (2021) An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight 6. 10.1172/jci.insight.144455 [DOI] [PMC free article] [PubMed]
- 104.Açıksarı G, Koçak M, Çağ Y, Altunal LN, Atıcı A, Çelik FB, et al. Prognostic value of inflammatory biomarkers in patients with severe COVID-19: A single-center retrospective study. Biomark Insights. 2021;16:11772719211027022. doi: 10.1177/11772719211027022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ahmad S, Kumar P, Shekhar S, Saha R, Ranjan A, Pandey S. Epidemiological, clinical, and laboratory predictors of in-hospital mortality among COVID-19 patients admitted in a tertiary COVID dedicated hospital, Northern India: A retrospective observational study. J Prim Care Community Health. 2021;12:21501327211041486. doi: 10.1177/21501327211041486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Akdogan D, Guzel M, Tosun D, Akpinar O. Diagnostic and early prognostic value of serum CRP and LDH levels in patients with possible COVID-19 at the first admission. J Infect Dev Ctries. 2021;15:766–772. doi: 10.3855/jidc.14072. [DOI] [PubMed] [Google Scholar]
- 107.Aksel G, İslam MM, Algın A, Eroğlu SE, Yaşar GB, Ademoğlu E, et al. Early predictors of mortality for moderate to severely ill patients with Covid-19. Am J Emerg Med. 2021;45:290–296. doi: 10.1016/j.ajem.2020.08.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Albalawi O, Alharbi Y, Bakouri M, Alqahtani A, Alanazi T, Almutairi AZ, et al. Clinical characteristics and predictors of mortality among COVID-19 patients in Saudi Arabia. J Infect Public Health. 2021;14:994–1000. doi: 10.1016/j.jiph.2021.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Alhumaid S, Al Mutair A, Al Alawi Z, Al Salman K, Al Dossary N, Omar A, et al. Clinical features and prognostic factors of intensive and non-intensive 1014 COVID-19 patients: An experience cohort from Alahsa, Saudi Arabia. Eur J Med Res. 2021;26:47. doi: 10.1186/s40001-021-00517-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Aly MM, Meshref TS, Abdelhameid MA, Ahmed SA, Shaltout AS, Abdel-Moniem AE, et al. Can hematological ratios predict outcome of COVID-19 patients? A multicentric study. J Blood Med. 2021;12:505–515. doi: 10.2147/jbm.S316681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Arikan H, Ozturk S, Tokgoz B, Dursun B, Seyahi N, Trabulus S, et al. Characteristics and outcomes of acute kidney injury in hospitalized COVID-19 patients: A multicenter study by the Turkish society of nephrology. PLoS One. 2021;16:e0256023. doi: 10.1371/journal.pone.0256023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bellan M, Azzolina D, Hayden E, Gaidano G, Pirisi M, Acquaviva A, et al. Simple parameters from complete blood count predict in-hospital mortality in COVID-19. Dis Markers. 2021;2021:8863053. doi: 10.1155/2021/8863053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bergantini L, Bargagli E, d'Alessandro M, Refini RM, Cameli P, Galasso L, et al. Prognostic bioindicators in severe COVID-19 patients. Cytokine. 2021;141:155455. doi: 10.1016/j.cyto.2021.155455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Betti M, Bertolotti M, Ferrante D, Roveta A, Pelazza C, Giacchero F, et al. Baseline clinical characteristics and prognostic factors in hospitalized COVID-19 patients aged ≤ 65 years: A retrospective observational study. PLoS One. 2021;16:e0248829. doi: 10.1371/journal.pone.0248829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bg S, Gosavi S, Ananda Rao A, Shastry S, Raj SC, Sharma A, et al. Neutrophil-to-Lymphocyte, Lymphocyte-to-Monocyte, and Platelet-to-Lymphocyte ratios: Prognostic significance in COVID-19. Cureus. 2021;13:e12622. doi: 10.7759/cureus.12622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Cai L, Zhou X, Wang M, Mei H, Ai L, Mu S, et al. Predictive nomogram for severe COVID-19 and identification of mortality-related immune features. J Allergy Clin Immunol Pract. 2021;9:177–184.e173. doi: 10.1016/j.jaip.2020.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Capdevila-Reniu A, Pellice M, Prieto-González S, Ventosa H, Ladino A, Naval J, et al. Clinical characteristics and outcome of patients aged over 80 years with covid-19. Medicine (Baltimore) 2021;100:e24750. doi: 10.1097/md.0000000000024750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cekerevac I, Turnic TN, Draginic N, Andjic M, Zivkovic V, Simovic S, et al. Predicting severity and intrahospital mortality in COVID-19: The place and role of oxidative stress. Oxid Med Cell Longev. 2021;2021:6615787. doi: 10.1155/2021/6615787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen PK, Lan JL, Huang PH, Hsu JL, Chang CK, Tien N, et al. Interleukin-18 is a potential biomarker to discriminate active adult-onset still’s disease from COVID-19. Front Immunol. 2021;12:719544. doi: 10.3389/fimmu.2021.719544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Conca W, Alabdely M, Albaiz F, Foster MW, Alamri M, Alkaff M, et al. Serum β2-microglobulin levels in Coronavirus disease 2019 (Covid-19): Another prognosticator of disease severity? PLoS One. 2021;16:e0247758. doi: 10.1371/journal.pone.0247758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.d’Alessandro M, Bergantini L, Cameli P, Curatola G, Remediani L, Sestini P, et al. Peripheral biomarkers’ panel for severe COVID-19 patients. J Med Virol. 2021;93:1230–1232. doi: 10.1002/jmv.26577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Deng K, Fan Q, Yang Y, Deng X, He R, Tan Y, et al. Prognostic roles of KL-6 in disease severity and lung injury in COVID-19 patients: A longitudinal retrospective analysis. J Med Virol. 2021;93:2505–2512. doi: 10.1002/jmv.26793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Eleni M, Evangelia M, Eleftheria K, Vasilios V, Vana S, Vissaria S, et al. Clinical features and outcomes of hospitalized COVID-19 patients in a low burden region. Pathog Glob Health. 2021;115:243–249. doi: 10.1080/20477724.2021.1893485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Elhadi M, Alsoufi A, Abusalama A, Alkaseek A, Abdeewi S, Yahya M, et al. Epidemiology, outcomes, and utilization of intensive care unit resources for critically ill COVID-19 patients in Libya: A prospective multi-center cohort study. PLoS One. 2021;16:e0251085. doi: 10.1371/journal.pone.0251085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.García de Guadiana-Romualdo L, Calvo Nieves MD, Rodríguez Mulero MD, Calcerrada Alises I, Hernández Olivo M, Trapiello Fernández W, et al. MR-proADM as marker of endotheliitis predicts COVID-19 severity. Eur J Clin Invest. 2021;51:e13511. doi: 10.1111/eci.13511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Huang W, Li M, Luo G, Wu X, Su B, Zhao L, et al. The inflammatory factors associated with disease severity to predict COVID-19 progression. J Immunol. 2021;206:1597–1608. doi: 10.4049/jimmunol.2001327. [DOI] [PubMed] [Google Scholar]
- 127.Kaal A, Snel L, Dane M, van Burgel N, Ottens T, Broekman W, et al. Diagnostic yield of bacteriological tests and predictors of severe outcome in adult patients with COVID-19 presenting to the emergency department. Emerg Med J. 2021;38:685–691. doi: 10.1136/emermed-2020-211027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Karahan S, Katkat F. Impact of serum 25(OH) vitamin D level on mortality in patients with COVID-19 in Turkey. J Nutr Health Aging. 2021;25:189–196. doi: 10.1007/s12603-020-1479-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Karampoor S, Zahednasab H, Farahmand M, Mirzaei R, Zamani F, Tabibzadeh A, et al. A possible pathogenic role of Syndecan-1 in the pathogenesis of coronavirus disease 2019 (COVID-19) Int Immunopharmacol. 2021;97:107684. doi: 10.1016/j.intimp.2021.107684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Li G, Zhou CL, Ba YM, Wang YM, Song B, Cheng XB, et al. Nutritional risk and therapy for severe and critical COVID-19 patients: A multicenter retrospective observational study. Clin Nutr. 2021;40:2154–2161. doi: 10.1016/j.clnu.2020.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Li T, Wang X, Zhuang X, Wang H, Li A, Huang L, et al. Baseline characteristics and changes of biomarkers in disease course predict prognosis of patients with COVID-19. Intern Emerg Med. 2021;16:1165–1172. doi: 10.1007/s11739-020-02560-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Li Z, Jiang N, Li X, Yang B, Jin M, Sun Y et al (2021) Two novel nomograms based on inflammatory cytokines or lymphocyte subsets to differentially diagnose severe or critical and Non-Severe COVID-19. Aging (Albany NY) 13. 10.18632/aging.203307 [DOI] [PMC free article] [PubMed]
- 133.Liu J, Tu C, Zhu M, Wang J, Yang C, Liu W, et al. The clinical course and prognostic factors of severe COVID-19 in Wuhan, China: A retrospective case-control study. Medicine (Baltimore) 2021;100:e23996. doi: 10.1097/md.0000000000023996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Liu Y, Liao W, Wan L, Xiang T, Zhang W. Correlation between relative nasopharyngeal virus RNA load and lymphocyte count disease severity in patients with COVID-19. Viral Immunol. 2021;34:330–335. doi: 10.1089/vim.2020.0062. [DOI] [PubMed] [Google Scholar]
- 135.López-Escobar A, Madurga R, Castellano JM, Ruiz de Aguiar S, Velázquez S, Bucar M, et al. Hemogram as marker of in-hospital mortality in COVID-19. J Investig Med. 2021;69:962–969. doi: 10.1136/jim-2021-001810. [DOI] [PubMed] [Google Scholar]
- 136.Lu Y, Huang Z, Wang M, Tang K, Wang S, Gao P, et al. Clinical characteristics and predictors of mortality in young adults with severe COVID-19: A retrospective observational study. Ann Clin Microbiol Antimicrob. 2021;20:3. doi: 10.1186/s12941-020-00412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Marín-Corral J, Rodríguez-Morató J, Gomez-Gomez A, Pascual-Guardia S, Muñoz-Bermúdez R, Salazar-Degracia A et al (2021) Metabolic signatures associated with severity in hospitalized COVID-19 patients. Int J Mol Sci 22. 10.3390/ijms22094794 [DOI] [PMC free article] [PubMed]
- 138.Montrucchio G, Sales G, Rumbolo F, Palmesino F, Fanelli V, Urbino R, et al. Effectiveness of mid-regional pro-adrenomedullin (MR-proADM) as prognostic marker in COVID-19 critically ill patients: An observational prospective study. PLoS One. 2021;16:e0246771. doi: 10.1371/journal.pone.0246771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nakamura S, Kanemasa Y, Atsuta Y, Fujiwara S, Tanaka M, Fukushima K, et al. Characteristics and outcomes of coronavirus disease 2019 (COVID-19) patients with cancer: A single-center retrospective observational study in Tokyo, Japan. Int J Clin Oncol. 2021;26:485–493. doi: 10.1007/s10147-020-01837-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ñamendys-Silva SA, Alvarado-Ávila PE, Domínguez-Cherit G, Rivero-Sigarroa E, Sánchez-Hurtado LA, Gutiérrez-Villaseñor A, et al. Outcomes of patients with COVID-19 in the intensive care unit in Mexico: A multicenter observational study. Heart Lung. 2021;50:28–32. doi: 10.1016/j.hrtlng.2020.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Özdemir İH, Özlek B, Çetin N. Permanent atrial fibrillation portends poor outcomes in hospitalized patients with COVID-19: A retrospective observational study. J Electrocardiol. 2021;65:113–120. doi: 10.1016/j.jelectrocard.2021.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Peiró ÓM, Carrasquer A, Sánchez-Gimenez R, Lal-Trehan N, Del-Moral-Ronda V, Bonet G, et al. Biomarkers and short-term prognosis in COVID-19. Biomarkers. 2021;26:119–126. doi: 10.1080/1354750x.2021.1874052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Provencio M, Mazarico Gallego JM, Calles A, Antoñanzas M, Pangua C, Mielgo Rubio X, et al. Lung cancer patients with COVID-19 in Spain: GRAVID study. Lung Cancer. 2021;157:109–115. doi: 10.1016/j.lungcan.2021.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Qin W, Bai W, Liu K, Liu Y, Meng X, Zhang K, et al. Clinical course and risk factors of disease deterioration in critically ill patients with COVID-19. Hum Gene Ther. 2021;32:310–315. doi: 10.1089/hum.2020.255. [DOI] [PubMed] [Google Scholar]
- 145.Quartuccio L, Fabris M, Sonaglia A, Peghin M, Domenis R, Cifù A, et al. Interleukin 6, soluble interleukin 2 receptor alpha (CD25), monocyte colony-stimulating factor, and hepatocyte growth factor linked with systemic hyperinflammation, innate immunity hyperactivation, and organ damage in COVID-19 pneumonia. Cytokine. 2021;140:155438. doi: 10.1016/j.cyto.2021.155438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Sai F, Liu X, Li L, Ye Y, Zhu C, Hang Y et al (2021) Clinical characteristics and risk factors for mortality in patients with coronavirus disease 2019 in intensive care unit: A single-center, retrospective, observational study in China. Ann Palliat Med 10:2859–2868. 10.21037/apm-20-1575 [DOI] [PubMed]
- 147.Salto-Alejandre S, Berastegui-Cabrera J, Camacho-Martínez P, Infante-Domínguez C, Carretero-Ledesma M, Crespo-Rivas JC, et al. SARS-CoV-2 viral load in nasopharyngeal swabs is not an independent predictor of unfavorable outcome. Sci Rep. 2021;11:12931. doi: 10.1038/s41598-021-92400-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Scotto R, Pinchera B, Perna F, Atripaldi L, Giaccone A, Sequino D et al (2021) Serum KL-6 could represent a reliable indicator of unfavourable outcome in patients with COVID-19 pneumonia. Int J Environ Res Public Health 18. 10.3390/ijerph18042078 [DOI] [PMC free article] [PubMed]
- 149.Song F, Ma H, Wang S, Qin T, Xu Q, Yuan H, et al. Nutritional screening based on objective indices at admission predicts in-hospital mortality in patients with COVID-19. Nutr J. 2021;20:46. doi: 10.1186/s12937-021-00702-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sozio E, Tascini C, Fabris M, D'Aurizio F, De Carlo C, Graziano E, et al. MR-proADM as prognostic factor of outcome in COVID-19 patients. Sci Rep. 2021;11:5121. doi: 10.1038/s41598-021-84478-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Stachura T, Celejewska-Wójcik N, Polok K, Górka K, Lichołai S, Wójcik K, et al. A clinical profile and factors associated with severity of the disease among Polish patients hospitalized due to COVID-19 - an observational study. Adv Respir Med. 2021;89:124–134. doi: 10.5603/ARM.a2021.0035. [DOI] [PubMed] [Google Scholar]
- 152.Tang J, Lin J, Zhang E, Zhong M, Luo Y, Fu Y, et al. Serum IL-6 and procalcitonin are two promising novel biomarkers for evaluating the severity of COVID-19 patients. Medicine (Baltimore) 2021;100:e26131. doi: 10.1097/md.0000000000026131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Tao Z, Xu J, Chen W, Yang Z, Xu X, Liu L, et al. Anemia is associated with severe illness in COVID-19: A retrospective cohort study. J Med Virol. 2021;93:1478–1488. doi: 10.1002/jmv.26444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tepasse PR, Vollenberg R, Fobker M, Kabar I, Schmidt H, Meier JA et al (2021) Vitamin A plasma levels in COVID-19 patients: A prospective multicenter study and hypothesis. Nutrients 13. 10.3390/nu13072173 [DOI] [PMC free article] [PubMed]
- 155.Viana-Llamas MC, Arroyo-Espliguero R, Silva-Obregón JA, Uribe-Heredia G, Núñez-Gil I, García-Magallón B, et al. Hypoalbuminemia on admission in COVID-19 infection: An early predictor of mortality and adverse events: A retrospective observational study. Med Clin (Barc) 2021;156:428–436. doi: 10.1016/j.medcli.2020.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang M, Fan Y, Chai Y, Cheng W, Wang K, Cao J, et al. Association of clinical and immunological characteristics with disease severity and outcomes in 211 patients with COVID-19 in Wuhan, China. Front Cell Infect Microbiol. 2021;11:667487. doi: 10.3389/fcimb.2021.667487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Wang Z, Wang Z. Identification of risk factors for in-hospital death of COVID - 19 pneumonia – lessions from the early outbreak. BMC Infect Dis. 2021;21:113. doi: 10.1186/s12879-021-05814-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Waris A, Din M, Khalid A, Abbas Lail R, Shaheen A, Khan N, et al. Evaluation of hematological parameters as an indicator of disease severity in Covid-19 patients: Pakistan’s experience. J Clin Lab Anal. 2021;35:e23809. doi: 10.1002/jcla.23809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Xiong L, Zang X, Feng G, Zhao F, Wang S, Zeng W et al (2021) Clinical characteristics and peripheral immunocyte subsets alteration of 85 COVID-19 deaths. Aging (Albany NY) 13:6289–6297. 10.18632/aging.202819 [DOI] [PMC free article] [PubMed]
- 160.Xue M, Zhang T, Chen H, Zeng Y, Lin R, Zhen Y, et al. Krebs Von den Lungen-6 as a predictive indicator for the risk of secondary pulmonary fibrosis and its reversibility in COVID-19 patients. Int J Biol Sci. 2021;17:1565–1573. doi: 10.7150/ijbs.58825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang C, Liu F, Liu W, Cao G, Liu J, Huang S, et al. Myocardial injury and risk factors for mortality in patients with COVID-19 pneumonia. Int J Cardiol. 2021;326:230–236. doi: 10.1016/j.ijcard.2020.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Zayat R, Kalverkamp S, Grottke O, Durak K, Dreher M, Autschbach R, et al. Role of extracorporeal membrane oxygenation in critically Ill COVID-19 patients and predictors of mortality. Artif Organs. 2021;45:E158–e170. doi: 10.1111/aor.13873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zhang J, Wang Z, Wang X, Hu Z, Yang C, Lei P. Risk factors for mortality of COVID-19 patient based on clinical course: A single center retrospective case-control study. Front Immunol. 2021;12:581469. doi: 10.3389/fimmu.2021.581469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhao Y, Yu C, Ni W, Shen H, Qiu M, Zhao Y (2021) Peripheral blood inflammatory markers in predicting prognosis in patients with COVID-19. Some differences with influenza A. J Clin Lab Anal 35:e23657. 10.1002/jcla.23657 [DOI] [PMC free article] [PubMed]
- 165.Elberts SJ, Bateman R, Koutsoubis A, London KS, White JL, Fields JM. The impact of COVID-19 on the sensitivity of D-dimer for pulmonary embolism. Acad Emerg Med. 2021 doi: 10.1111/acem.14348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Liu G, Zhang B, Zhang S, Hu H, Liu T. LDH, CRP and ALB predict nucleic acid turn negative within 14 days in symptomatic patients with COVID-19. Scott Med J. 2021;66:108–114. doi: 10.1177/0036933021994243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Sui J, Noubouossie DF, Gandotra S, Cao L. Elevated plasma fibrinogen is associated with excessive inflammation and disease severity in COVID-19 patients. Front Cell Infect Microbiol. 2021;11:734005. doi: 10.3389/fcimb.2021.734005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Li M, Dong Y, Wang H, Guo W, Zhou H, Zhang Z, et al. Cardiovascular disease potentially contributes to the progression and poor prognosis of COVID-19. Nutr Metab Cardiovasc Dis. 2020;30:1061–1067. doi: 10.1016/j.numecd.2020.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Loske J, Röhmel J, Lukassen S, Stricker S, Magalhães VG, Liebig J, et al. Pre-activated antiviral innate immunity in the upper airways controls early SARS-CoV-2 infection in children. Nat Biotechnol. 2021 doi: 10.1038/s41587-021-01037-9. [DOI] [PubMed] [Google Scholar]
- 170.Choudhary S, Sharma K, Silakari O. The interplay between inflammatory pathways and COVID-19: A critical review on pathogenesis and therapeutic options. Microb Pathog. 2021;150:104673. doi: 10.1016/j.micpath.2020.104673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.García LF. Immune response, inflammation, and the clinical spectrum of COVID-19. Front Immunol. 2020;11:1441. doi: 10.3389/fimmu.2020.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tay MZ, Poh CM, Rénia L, MacAry PA, Ng LFP. The trinity of COVID-19: Immunity, inflammation and intervention. Nat Rev Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ragab D, Salah Eldin H, Taeimah M, Khattab R, Salem R. The COVID-19 cytokine storm; What we know so far. Front Immunol. 2020;11:1446. doi: 10.3389/fimmu.2020.01446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jones SA, Hunter CA. Is IL-6 a key cytokine target for therapy in COVID-19? Nat Rev Immunol. 2021;21:337–339. doi: 10.1038/s41577-021-00553-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Tharmarajah E, Buazon A, Patel V, Hannah JR, Adas M, Allen VB, et al. IL-6 inhibition in the treatment of COVID-19: A meta-analysis and meta-regression. J Infect. 2021;82:178–185. doi: 10.1016/j.jinf.2021.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, et al. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636–1643. doi: 10.1038/s41591-020-1051-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Islam H, Chamberlain TC, Mui AL, Little JP. Elevated Interleukin-10 levels in COVID-19: Potentiation of rro-inflammatory responses or impaired anti-inflammatory action? Front Immunol. 2021;12:677008. doi: 10.3389/fimmu.2021.677008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Acosta MAT, Singer BD (2020) Pathogenesis of COVID-19-induced ARDS: Implications for an ageing population. Eur Respir J 56. 10.1183/13993003.02049-2020 [DOI] [PMC free article] [PubMed]
- 179.Hu R, Han C, Pei S, Yin M, Chen X. Procalcitonin levels in COVID-19 patients. Int J Antimicrob Agents. 2020;56:106051. doi: 10.1016/j.ijantimicag.2020.106051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Chen Z, Xu W, Ma W, Shi X, Li S, Hao M, et al. Clinical laboratory evaluation of COVID-19. Clin Chim Acta. 2021;519:172–182. doi: 10.1016/j.cca.2021.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zhang J, Liu P, Wang M, Wang J, Chen J, Yuan W et al (2020) The clinical data from 19 critically ill patients with coronavirus disease 2019: A single-centered, retrospective, observational study. Z Gesundh Wiss 1–4. 10.1007/s10389-020-01291-2 [DOI] [PMC free article] [PubMed]
- 182.Iwamura APD, Tavares da Silva MR, Hümmelgen AL, Soeiro Pereira PV, Falcai A, Grumach AS, et al. Immunity and inflammatory biomarkers in COVID-19: A systematic review. Rev Med Virol. 2021;31:e2199. doi: 10.1002/rmv.2199. [DOI] [PubMed] [Google Scholar]
- 183.Yang L, Jin J, Luo W, Gan Y, Chen B, Li W. Risk factors for predicting mortality of COVID-19 patients: A systematic review and meta-analysis. PLoS One. 2020;15:e0243124. doi: 10.1371/journal.pone.0243124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Rawson TM, Moore LSP, Zhu N, Ranganathan N, Skolimowska K, Gilchrist M, et al. Bacterial and fungal coinfection in individuals with coronavirus: A rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2020;71:2459–2468. doi: 10.1093/cid/ciaa530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ritchie AI, Singanayagam A. Immunosuppression for hyperinflammation in COVID-19: A double-edged sword? Lancet. 2020;395:1111. doi: 10.1016/s0140-6736(20)30691-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Huang W, Berube J, McNamara M, Saksena S, Hartman M, Arshad T, et al. Lymphocyte subset counts in COVID-19 patients: A meta-analysis. Cytometry A. 2020;97:772–776. doi: 10.1002/cyto.a.24172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Lindsley AW, Schwartz JT, Rothenberg ME. Eosinophil responses during COVID-19 infections and coronavirus vaccination. J Allergy Clin Immunol. 2020;146:1–7. doi: 10.1016/j.jaci.2020.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Kichloo A, Dettloff K, Aljadah M, Albosta M, Jamal S, Singh J, et al. COVID-19 and hypercoagulability: A review. Clin Appl Thromb Hemost. 2020;26:1076029620962853. doi: 10.1177/1076029620962853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Varga Z, Flammer AJ, Steiger P, Haberecker M, Andermatt R, Zinkernagel AS, et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet. 2020;395:1417–1418. doi: 10.1016/s0140-6736(20)30937-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Ackermann M, Verleden SE, Kuehnel M, Haverich A, Welte T, Laenger F, et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl J Med. 2020;383:120–128. doi: 10.1056/NEJMoa2015432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.McGonagle D, O'Donnell JS, Sharif K, Emery P, Bridgewood C. Immune mechanisms of pulmonary intravascular coagulopathy in COVID-19 pneumonia. Lancet Rheumatol. 2020;2:e437–e445. doi: 10.1016/s2665-9913(20)30121-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Oudit GY, Kassiri Z, Jiang C, Liu PP, Poutanen SM, Penninger JM, et al. SARS-coronavirus modulation of myocardial ACE2 expression and inflammation in patients with SARS. Eur J Clin Invest. 2009;39:618–625. doi: 10.1111/j.1365-2362.2009.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Crackower MA, Sarao R, Oudit GY, Yagil C, Kozieradzki I, Scanga SE, et al. Angiotensin-converting enzyme 2 is an essential regulator of heart function. Nature. 2002;417:822–828. doi: 10.1038/nature00786. [DOI] [PubMed] [Google Scholar]
- 194.Oudit GY, Kassiri Z, Patel MP, Chappell M, Butany J, Backx PH, et al. Angiotensin II-mediated oxidative stress and inflammation mediate the age-dependent cardiomyopathy in ACE2 null mice. Cardiovasc Res. 2007;75:29–39. doi: 10.1016/j.cardiores.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 195.Zhao X, Nicholls JM, Chen YG. Severe acute respiratory syndrome-associated coronavirus nucleocapsid protein interacts with Smad3 and modulates transforming growth factor-beta signaling. J Biol Chem. 2008;283:3272–3280. doi: 10.1074/jbc.M708033200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Wong CK, Lam CW, Wu AK, Ip WK, Lee NL, Chan IH, et al. Plasma inflammatory cytokines and chemokines in severe acute respiratory syndrome. Clin Exp Immunol. 2004;136:95–103. doi: 10.1111/j.1365-2249.2004.02415.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Chen DF, Chen Y, Zhong CH, Chen XB, Li SY (2021) Long-term efficacy and safety of the Dumon stent for benign tracheal stenosis: a meta-analysis. J Thorac Dis 13:82–91. 10.21037/jtd-20-2327 [DOI] [PMC free article] [PubMed]
- 198.Alhossan A, Lee CS, MacDonald K, Abraham I. Real-life effectiveness studies of omalizumab in adult patients with severe allergic asthma: Meta-analysis. J Allergy Clin Immunol Pract. 2017;5:1362–1370.e1362. doi: 10.1016/j.jaip.2017.02.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary file1: Quality assessment of the included studies (Newcastle–Ottawa Scale). +: high quality; –: low quality; ?: unclear quality (PNG 93 KB)