Summary
In many bacteria, the stabilities and functions of small regulatory RNAs (sRNAs) that act by base pairing RNA most often are dependent on Hfq or ProQ/FinO-domain proteins, two classes of RNA chaperone proteins. However, while all bacteria appear to have sRNAs, many have neither Hfq nor ProQ/FinO-domain proteins raising the question of whether another factor might act as an sRNA chaperone protein in these organisms. Several recent studies have reported that KH-domain proteins, such as KhpA and KhpB, bind sRNAs. Here we describe what is known about the distribution, structures, RNA binding properties and physiological roles of KhpA and KhpB and discuss evidence for and against these proteins serving as sRNAs chaperones.
Graphical Abstract
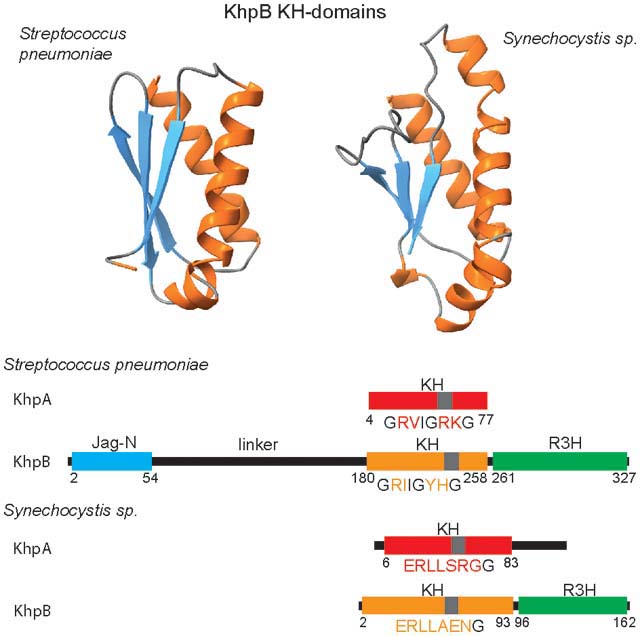
KhpA and KhpB proteins, which are widely distributed together in Gram-positive bacteria, have been found to bind regulatory RNAs, but much remains to be learned about their functions. While KhpA proteins consist of a single KH domain, KhpB proteins are more diverse, consisting of KH and R3H RNA-binding domains, which may or may not be connected to the N-terminal Jag domain by unstructured linkers of various lengths.
Introduction
RNA-binding proteins with KH domains have been found across all kingdoms, with roles in many different processes. The K-homology or KH domain was initially identified in human heterogeneous nuclear ribonucleoprotein K (hnRNP K) and a Xenopus laevis hnRNP K that binds cytidine-rich sequences in pre-mRNAs (Siomi et al., 1993). In KH RNA binding domains, a conserved GXXG amino acid sequence motif located between two α-helices with accompanying β-strands serves to recognize a specific RNA sequence (Nagai, 1996, Nicastro et al., 2015). There are two types of KH domains: type I in eukaryotes where the core KH domain is accompanied by additional α-helix and β-sheet motifs on the C-terminal side, and type II in bacteria where the KH domain is accompanied by additional α-helix and β-sheet motifs on the N-terminal side (Nicastro et al., 2015, Valverde et al., 2008). While eukaryotic proteins often contain multiple KH domains, bacterial proteins typically contain only one or two KH domains (Nicastro et al., 2015).
KH domains have been found in bacterial proteins with a wide range of functions. These include the enzymes polynucleotide phosphorylase (PNPase) (Dendooven et al., 2021, Hardwick et al., 2012) and RNase Y (Shahbabian et al., 2009, Nagata et al., 2008); transcription elongation factor NusA (Gopal et al., 2001, Worbs et al., 2001); GTPases Era, which binds to the 30S ribosomal subunits (Tu et al., 2009, Verstraeten et al., 2011), and Der, which is involved in the assembly of the 50S subunits (Robinson et al., 2002); cold-shock ribosome factor A (RbfA), which assists in the assembly of the 30S subunits (Huang et al., 2003, Verstraeten et al., 2011); and ribosomal protein S3 (Watson et al., 2020, Wimberly et al., 2000).
Two other KH-domain proteins found in multiple bacterial species are KhpA and KhpB (the latter also denoted Jag and EloR). KhpA (KH-domain protein A) initially was identified as a protein involved in cell elongation in Streptococcus pneumoniae (Zheng et al., 2017), while KhpB was first identified as being encoded adjacent to a sporulation gene in Bacillus subtilis, where it was originally named Jag (spoIIIJ-associated gene) (Errington et al., 1992), but later also was shown to be involved in S. pneumoniae cell division (Ulrych et al., 2016, Stamsås et al., 2017, Zheng et al., 2017). Both KhpA and KhpB only have a single KH-domain (Figure 1). The KhpA protein is small and comprised solely of the KH-domain. In contrast, the KhpB protein has a second RNA binding domain, a R3H domain (named for the characteristic spacing of an arginine and a histidine residue) (reviewed in (Grishin, 1998)), on the C-terminus and, in many cases, a Jag-N domain of unknown function on the N-terminus. KhpA proteins have been found to homodimerize as well as form heterodimers with KhpB proteins (Winther et al., 2019).
Fig. 1.
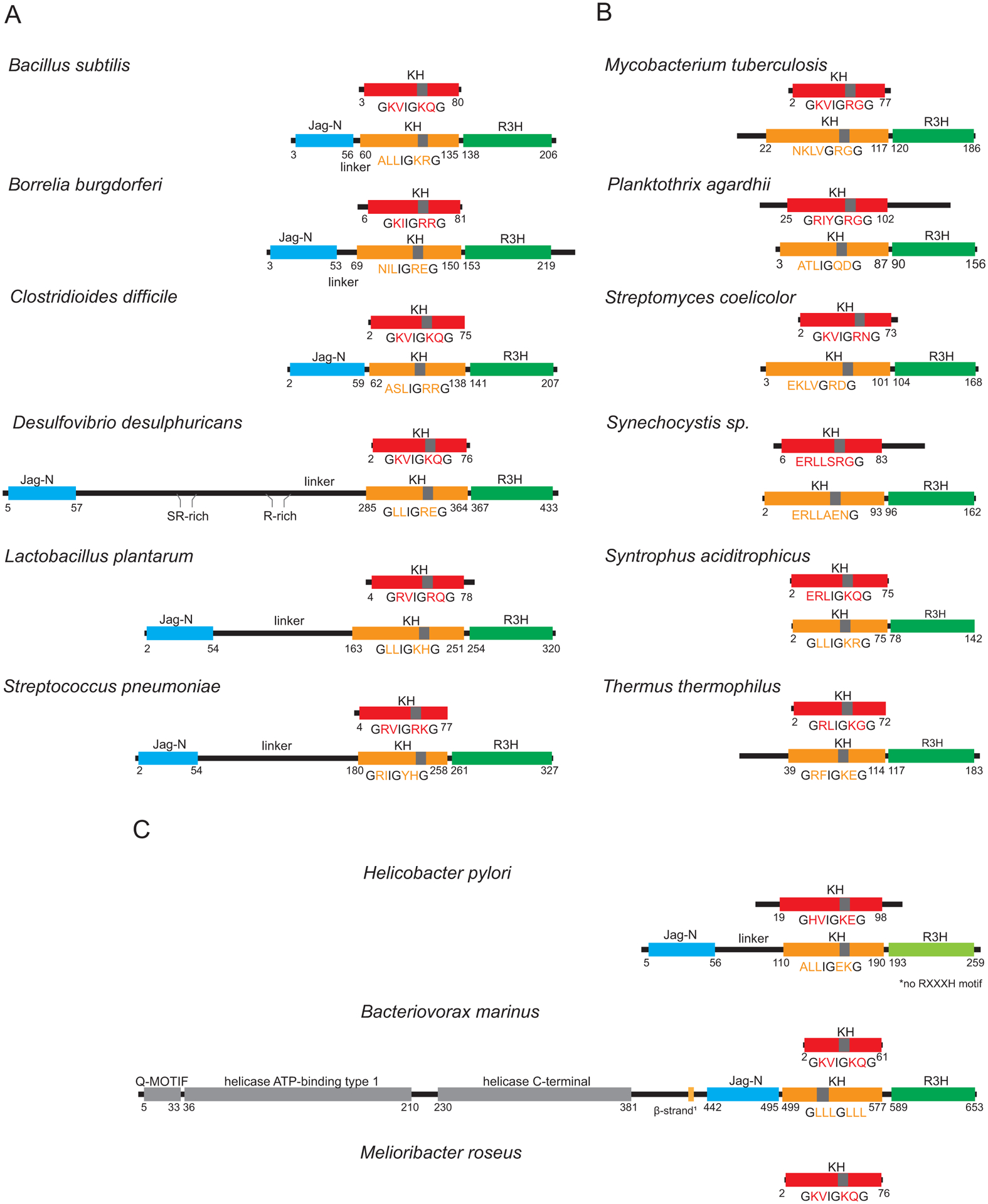
The diversity of structures of KhpA and KhpB proteins for representative organisms. For Figs. 1, S2 and S3, we selected the species that have been the subject of studies on KhpA and KhpB proteins (Errington et al., 1992, Grishin, 1998, Hare et al., 2007, Lamm-Schmidt et al., 2021, Myrbraten et al., 2019, Riediger et al., 2021, Zheng et al., 2017), and additionally included representative species that could illustrate the sequence and structure diversity of KH-domain proteins. Schematic structures of KhpA proteins are presented above the structures of KhpB proteins for each species. The KH domains are colored red in KhpA and orange in KhpB proteins, while the KhpB Jag and R3H domains are colored blue and green, respectively. The atypical R3H domain in H. pylori, which is devoid of an RxxxH sequence, is colored light green, and the additional helicase domains in the N-terminal part of B. marinus KhpB are in grey. The sequence of the GxxIGxxG motif at the junction of the first and second α-helix of the KH domain is provided below each KH domain. The conserved residues in this motif are in black font, while the non-conserved ones are in the color of the KH domain. The position of this sequence in a KH domain is marked with a grey bar. Numbers denote positions of the first and last residue of each domain. To identify domains in protein sequences and assign the domain borders, proteins were folded using ColabFold software (Mirdita et al., 2021) based on AlphaFold 2.0 (Jumper et al., 2021) and MMseqs2 (Steinegger & Soding, 2017). The following bacterial species are represented (the percentages after the species name provide the sequence identity of the KhpA and KhpB proteins, respectively, calculated for homologous regions, relative to the corresponding proteins in S. pneumoniae): (A) KhpA/B sets with KhpB proteins containing the Jag-N domain from Bacillus subtilis (38%, 27%), Borrelia burgdorferi (33%, 19%), Clostridioides difficile (35%, 27%), Desulfovibrio desulphuricans (30%, 18%), Lactobacillus plantarum (46%, 29%), and Streptococcus pneumoniae; (B) KhpA/B sets with KhpB proteins without the Jag-N domain from Mycobacterium tuberculosis (27%, 26%), Planktothrix agardhii (25%, 30%), Streptomyces coelicolor (26%, 25%), Synechocystis sp. PCC 6803 (23%, 20%), Syntrophus aciditrophicus (30%, 24%), and Thermus thermophilus (25%, 22%); (C) unusual KhpA/B sets from Helicobacter pylori (24%, 19%) and Bacteriovorax marinus (35%, 24%), and the KhpA protein from Melioribacter roseus, which does not have a KhpB homolog (30%). The khpA and khpB gene synteny for the above species is shown in Supplemental Figs. S2 and S3, while the phylogeny of KhpA/B proteins is shown in Fig. S1, and the extensive list of KhpA and KhpB homologs is shown in Table S1. For B. marinus β-strand1 denotes a β-strand that interacts with the β-sheet of the KH domain in the ColabFold predicted structure of the KhpB.
Several recent studies have shown that KhpA and KhpB are associated with small regulatory RNAs (sRNAs) (Lamm-Schmidt et al., 2021, Hör et al., 2020, Zheng et al., 2017, Riediger et al., 2021). This is of interest because KhpA and KhpB protein family members are found in several bacteria such as S. pneumoniae, which do not have Hfq and ProQ/FinO-domain RNA chaperone proteins. Hfq and ProQ/FinO-domain proteins stabilize and promote the functions of sRNAs that act by base pairing (reviewed in (Olejniczak & Storz, 2017, Woodson et al., 2018)). These families of proteins are capable of binding both the sRNAs and their base pairing targets consistent with having multiple RNA binding sites and thus help to promote sRNA interactions with target RNAs. For bacteria lacking Hfq and ProQ/FinO-domain proteins altogether or where the deletion of these genes has not resulted in an sRNA phenotype, there has been the unanswered question of whether other proteins facilitate sRNA base pairing.
One source of evidence for KhpA and KhpB binding to sRNAs comes from Grad-seq experiments in which cell extracts are fractionated on gradients and the RNA and protein compositions of the individual gradient fractions are determined by RNA sequencing and mass spectrometry, respectively (Lamm-Schmidt et al., 2021, Riediger et al., 2021, Hör et al., 2020). Co-fractionation of sRNAs and KhpA and KhpB proteins suggest a possible interaction, which in some species has been further verified by tagging the KhpA and KhpB proteins and identifying the RNAs that specifically co-purify with these proteins (Lamm-Schmidt et al., 2021, Zheng et al., 2017).
Here we describe what is known about KhpA and KhpB proteins and discuss their possible roles in facilitating the functions of sRNAs.
Distribution of KhpA and KhpB proteins
We examined the distribution of the KhpA and KhpB proteins based on the presence of the key functional domains of KhpA and KhpB in proteins from the Genome Taxonomy Database (GTDB) predicted by in silico analysis and based on gene synteny (Table S1 and Fig. S1, S2 and S3, 48% of the 45,555 species we analyzed had khpA or khpB or both). These analyses revealed that while khpA and khpB genes are quite prevalent in some phyla such as Actinobacteria and Firmicutes, they are absent in others such as α, β, and γ-Proteobacteria and Bacteroidetes (Prezza et al., 2021). Several other interesting features of these gene families can be noted. First, the genes are only present in single copy in almost all genomes. Second, although the khpA and khpB genes are not physically linked on the chromosome, there is an extremely high co-occurrence of the two genes. More than 80% of all species and more than 90% of the Firmicutes and Actinomycetes species, in which we identified khpA and/or khpB, have both genes (Table S1). There are a limited number of clades where only the khpA gene is present. Third, there are two different categories of KhpB proteins, some of which contain both the Jag-N and R3H domains along with the KH domain (Fig. 1A) and others which only contain KH and R3H domains (Fig. 1B). This observation suggests that the KH and R3H domains form a functional unit that can participate in RNA metabolism, either alone or in connection with the Jag-N domain. Interestingly, very little is known about the function of the Jag-N domain.
Despite similar overall domain composition, KhpA and KhpB from different bacterial species exhibit remarkable sequence diversity. The KH domains of KhpA and KhpB homologs differ in the composition of the variable residues in and around the GxxG motif (Fig. 1). In some species, the conserved G residues even are substituted by other amino acids. Additionally, while the folding of the individual KH, R3H and Jag-N domains of KhpB proteins is similar as judged by ColabFold-based structure predictions, the overall sequence conservation of these domains is low. For example, the sequence identity between the S. pneumoniae KhpB protein and those from B. subtilis or Helicobacter pylori is less than 30% (Fig. 1 legend).
The sequences outside of the conserved domains also vary greatly. Overall, the sequence similarity among KhpA proteins is higher than the similarity among KhpB proteins, but KhpA proteins still vary in the length of adjacent unstructured regions adjacent to the KH domain, resulting in a range in overall size. For instance, Bacteriovorax marinus KhpA is 62 aa and Planktothrix agardhii KhpA is 149 aa. The KhpB proteins that are composed only of the KH and R3H domains are more similar in size ranging from 142 aa for Syntrophus aciditrophicus to 189 aa for T. thermophilus (Fig 1B). In contrast, for KhpB proteins that have Jag-N domains, the length of the linker between the Jag-N and KH domain is remarkably variable resulting in a range of KhpB protein lengths from 208 aa for B. subtilis, and 328 aa for S. pneumoniae to 437 aa for Desulfovibrio desulphuricans (Fig. 1A). It remains to be seen how all the differences affect the functions of KhpA and KhpB in different bacteria.
There also are a few interesting KhpB variants (Fig. 1C). For instance, while the R3H domain of KhpB proteins is defined by an RXXXH motif, this sequence is not present in the corresponding domain of the H. pylori protein. As another example, the KhpB homolog from B. marinus has a helicase domain connected N-terminally of the Jag-N domain. As will be discussed in conjunction with possible KhpA and KhpB functions, there is significant synteny in the genes surrounding khpA and khpB (Fig. S3). The absence of the genes typically found adjacent to khpB suggests that in B. marinus, the khpB gene was fused with another gene at a new genome location.
Structures of KhpA and KhpB proteins
Given that the structures of many KH-domain proteins have been determined, a fair amount is known about the basic structure of the minimal motif comprised of the two α-helices, flanked on either side by one β-strand (Siomi et al., 1993, Valverde et al., 2008). As mentioned above, this minimal KH motif is accompanied by additional α and β structures either on the C-terminus (type I domain architecture) or on the N-terminus (type II domain architecture) (Grishin, 2001). The conserved GxxG sequence motif, located between the two α-helices of the minimal KH motif, together with amino acid residues of the neighboring β-strand, are part of a cleft on the surface of the domain that provides an RNA binding surface (Nicastro et al., 2015, Valverde et al., 2008). While the two middle residues in the GxxG motif are varied, at least one residue is usually positively charged and arginine, lysine or glycine residues are frequent. Double aspartate residues between the glycine residues, on the other hand, have been shown to be detrimental for RNA binding (Hollingworth et al., 2012). In type I and II domains, the additional β-strands are differently engaged with the β-sheet of the minimal KH motif, which also leads to different locations of the variable sequence loop (Valverde et al., 2008).
Previously, the structure of S. pneumoniae KhpA was predicted using i-Tasser (Winther et al., 2019), and the partial structures of Clostridium symbiosum KhpB (pdb: 3GKU) and H. pylori KhpB (pdb: 2PT7) (Hare et al., 2007) were solved using X-ray crystallography. We predicted the structures of KhpA and KhpB proteins from both S. pneumoniae and Clostridioides difficile (Fig. 2) using ColabFold software (Mirdita et al., 2021), which is based on AlphaFold 2.0 (Jumper et al., 2021) and MMseqs2 (Steinegger & Soding, 2017). In the ColabFold predictions the KH domains of KhpA and KhpB can be modeled with high confidence. The comparison of these four predicted KH domain structures shows a fold typical for type II KH proteins with the additional β-strand (β1) on the N-terminus antiparallel to the first β-strand (β2) of the minimal KH domain (Fig. 2). Despite the similarity of the overall fold, the visualization of the electrostatic surface potential shows differences in the locations of charged amino acid side chains, which could affect interactions with RNA molecules (Fig. 2).
Fig. 2.
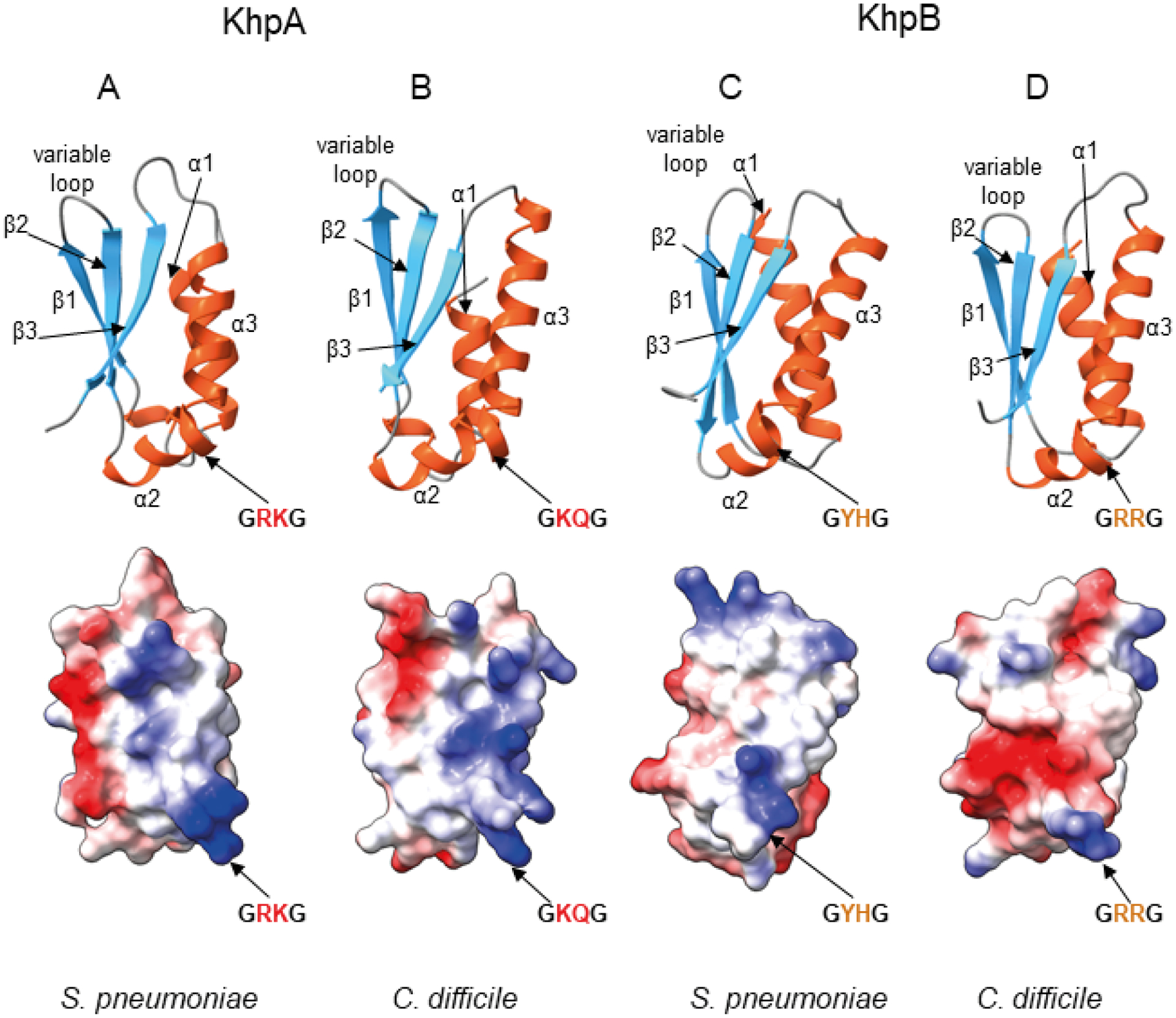
Predicted structures of the KH domains of KhpA and KhpB proteins from S. pneumoniae and C. difficile reveal the same overall fold but different electrostatic surface potential.
A. Predicted structure of KhpA from S. pneumoniae.
B. Predicted structure of KhpA from C. difficile.
C. Predicted structure of KH domain of KhpB protein from S. pneumoniae.
D. Predicted structure of KH domain of KhpB protein from C. difficile. All structures were predicted using ColabFold software (Mirdita et al., 2021) based on AlphaFold 2.0 (Jumper et al., 2021) and MMseqs2 (Steinegger & Soding, 2017), and visualized using ChimeraX (Pettersen et al., 2021). In each pair, a ribbon representation is shown above with α-helices shown in red, and β-strands in blue, and an electrostatic surface potential, calculated using ChimeraX, is shown below. The view of the KH domains is at the face involved in RNA binding.
The structures of some KH-domain proteins other than KhpA and KhpB have been solved in complex with RNA by X-ray crystallography (Fig. 3). In a complex of the Aquifex aeolicus Era GTPase with a 12-nt 3ʹ-terminal fragment of 16S rRNA, the bases and riboses of RNA bind the KH domain using hydrogen bonding along with hydrophobic contacts with the peptide backbone and side chains at and around the GKKG sequence, in the subsequent α-helix, next β-strand, and the variable sequence loop (Tu et al., 2009). In a complex of the Mycobacterium tuberculosis NusA transcription factor, which has two KH domains connected by a six-amino acid linker, with 11-nt RNA fragment of Box C anti-termination sequence, the binding clefts of both KH domains form a continuous binding site for the 11-nt RNA (Beuth et al., 2005). As in other KH domains, each cleft is formed by both α-helices surrounding the GxxG motif (GPMG in the KH1 domain and GKEG in the KH2 domain) and the subsequent β-strand, with additional contacts made with the loops between protein secondary structure motifs. In both the Era and NusA complexes, a stretch of 4–6 nucleotides of RNA fits into the RNA binding cleft of a single KH domain. In the structure of the trimeric Caulobacter crescentus PNPase co-purified with an RNA from E. coli cells, a 12-nt sequence was well resolved in the crystal structure. Here the RNA formed hydrogen bonding contacts with KH domains of each monomer of the trimeric complex via the loops containing GSGG motifs (Hardwick et al., 2012). Hence, while RNA binding occurred at a single KH domain in the Era GTPase, the RNA binding site consisted of more than one KH motif in both NusA and PNPase (Beuth et al., 2005, Hardwick et al., 2012, Tu et al., 2009).
Fig. 3.
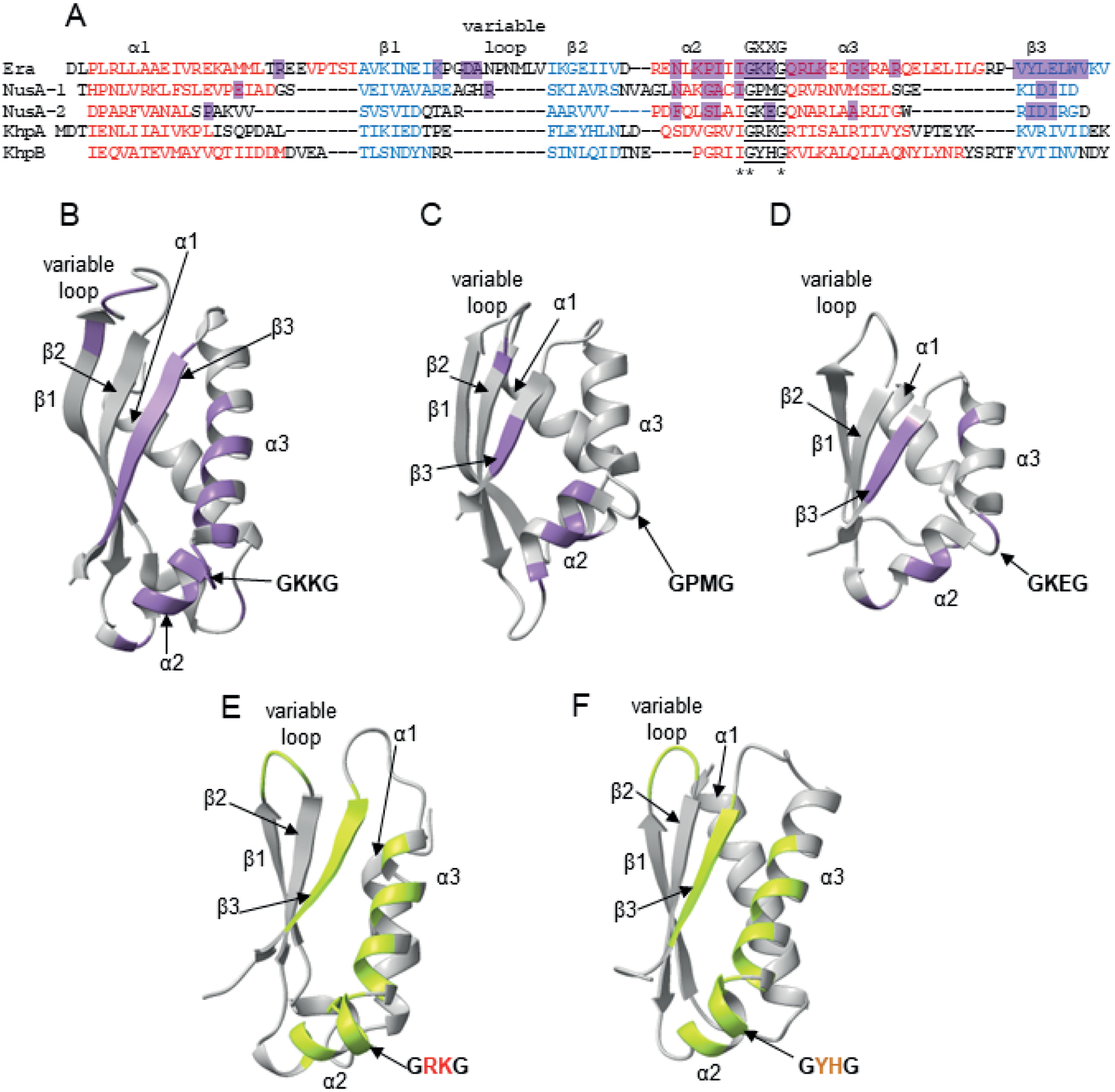
Structurally-determined and predicted RNA binding contacts in KH domains.
A. Alignment of the structurally homologous regions of the KH domain from A. aeolicus Era protein (Tu et al., 2009), the KH domain 1 from M. tuberculosis NusA protein, the KH domain 2 from M. tuberculosis NusA protein (Beuth et al., 2005), and KH domains from Alphafold-predicted S. pneumoniae KhpA and KhpB. For the alignment, homologous sequences were first aligned using Clustal Omega, and then structurally homologous regions were manually aligned based on the Era and NusA structures (Beuth et al., 2005, Tu et al., 2009) and the predicted structures of KhpA and KhpB.
B. The structure of the KH domain of A. aeolicus Era with RNA contacts according to (Tu et al., 2009) marked purple.
C. The structure of the KH 1 domain of M. tuberculosis NusA with RNA contacts according to (Beuth et al., 2005) marked purple.
D. The structure of the KH 2 domain of M. tuberculosis NusA with RNA contacts according to (Beuth et al., 2005) marked purple.
E. The predicted structure of full-length S. pneumoniae KhpA, in which regions that could hypothetically be involved in RNA binding are marked lime-green.
F. The predicted structure of the KH domain of S. pneumoniae KhpB, in which regions that could hypothetically be involved in RNA binding are marked lime-green.
The structures of S. pneumoniae KhpA and KhpB proteins were predicted using ColabFold (Mirdita et al., 2021), and all structures were visualized using Chimera X (Pettersen et al., 2021).
Despite this structural information, the determinants of RNA binding specificity of KH domains are not well understood (Auweter et al., 2006, Corley et al., 2020, Nicastro et al., 2015). It has been proposed that contacts within the RNA binding groove and the shape of the groove determine the RNA binding specificity of individual KH domains (Nicastro et al., 2015). Additionally, the variable sequence loops of KH domains have been proposed to play a role in RNA recognition for KH domains of IMP proteins (Biswas et al., 2019). Furthermore, the glycines in the GXXG motif could be important for RNA binding or just have structural roles as the GXXG loop is located at the bend between two α-helices. Indeed, for Era, one of the glycines contacts RNA, while for NusA, neither of the glycines is directly involved in RNA binding. Nevertheless, while the structures of KhpA and KhpB in complexes with RNA are not yet available, the overlay of amino acid residues contacting RNA in the structures of type II KH domains of M. tuberculosis Era and A. aeolicus NusA onto the homologous sequences of KH domains of KhpA and KhpB suggests regions that could be involved in RNA binding in KhpA and KhpB (Fig. 3).
It should be noted that KhpA and KhpB might bind RNA as homo- or heterodimers, which would result in tandem KH domains (Winther et al., 2019). KhpB also has a second RNA binding domain, R3H (Grishin, 1998, Ciesla et al., 2020). Thus, these proteins might bind RNA regions that are larger than what would fit into a single KH groove. When the binding of isolated KH domains of the FMRP protein to short RNA ligands was measured, the data showed that the RNA binding affinities were very weak (Athar & Joseph, 2020). Hence, it is possible that the tight and specific RNA binding by KH domain-containing proteins requires the cooperation of different KH domains and possibly also other RNA binding domains such as the R3H domain (Dagil et al., 2019, Korn et al., 2021, Schneider et al., 2019).
RNAs bound by KhpA and KhpB proteins
While single-stranded CA-rich sequences and G-rich sequences have been proposed most often as RNA recognition motifs of eukaryotic KH proteins (Nicastro et al., 2015), a U-rich sequence was recently reported as a motif recognized by a KH domain of a DEAD-box helicase (Yadav et al., 2021). Thus, KH domains can bind a wide variety of sequence motifs. It is likely that the amino acid sequence and the exact structure of the recognition motif dictate the binding specificity (Dominguez et al., 2018).
Although there are several datasets for RNAs that co-purify with KhpA and KhpB proteins, no RNA motif that is recognized by either of the two proteins has been reported. RNA immunoprecipitation (RIP) experiments for S. pneumoniae showed that KhpA and KhpB, each tagged with a carboxy terminal 3X FLAG tag, bind the same pool of approximately 170 RNA species, which showed at least a 4-fold enrichment upon immunoprecipitation with either KhpA or KhpB (Zheng et al., 2017). This data set includes mRNAs, two tRNAs and some sRNAs. Another data set corresponds to RNAs that co-purify with KhpB-3xFLAG in C. difficile (Lamm-Schmidt et al., 2021). This study reported enrichment for about 1,400 RNAs. Among these, mRNAs were overrepresented. While there was no enrichment of rRNAs or tRNAs, 12 sRNAs co-purified with KhpB-3xFLAG. It is interesting to note that in C. difficile, which encodes an Hfq protein, some RNAs, including eight of the sRNAs, co-purify with both Hfq and KhpB while other RNAs are only bound by one or the other chaperone suggesting overlapping as well as differing cellular roles. While Hfq predominantly binds to the 5ʹ and 3ʹ ends of mRNAs, the full mRNA and even full operons are enriched for KhpB for C. difficile. Hopefully, further analyses of the S. pneumoniae and C. difficile data sets as well as RNAs that co-purify with KhpA and KhpB from other bacteria will reveal whether the proteins recognize a specific motif(s) or structure(s), that may or may not differ between bacteria, whether these recognition motifs are typically found at a specific location such as the 5ʹ end, middle or 3ʹ end of a transcript, and whether the proteins have binding sites for more than one RNA.
A related question is what occurs to the RNAs upon binding to a KhpA or KhpB monomer or more likely KhpA homodimer or KhpA-KhpB heterodimer. A comparison of the transcriptome of wild-type and a ΔkhpB deletion strain in C. difficile suggests that KhpB might have both positive and negative effects on RNA levels. How this occurs and whether KhpA and KhpB proteins affect the folding of a bound RNA, recruit ribonucleases, or promote base pairing with another RNA remain to be investigated. Spectroscopic studies of NusA binding to a longer 43-nt RNA indicated that this RNA is unfolded upon binding by NusA, suggesting that KH-domain containing proteins could induce changes in RNA structure (Beuth et al., 2005).
Physiological roles of KhpA and KhpB proteins
While the physiological roles of only a limited number of KhpA and KhpB proteins have been examined, a few themes are starting to emerge. The role characterized most extensively is one in cell elongation. KhpA and KhpB may have multiple roles in controlling cell elongation, but in S. pneumoniae, the absence of these RNA-binding proteins clearly leads to increased levels of transcripts in the WalRK regulon, which responds to peptidoglycan stress (Zheng et al., 2017). KhpB similarly has been shown to negatively affect the levels of mRNAs encoding virulence factors in C. difficile (Lamm-Schmidt et al., 2021). The precise mechanisms by which the KH proteins impact the levels of these transcripts are unknown. It is noteworthy that the khpA and khpB genes show conserved synteny with genes encoding proteins involved in RNA processing, protein synthesis, and protein translocation across membranes.
Role in controlling cell elongation
Mutations that inactivate khpA and khpB were identified in two independent screens for suppressors of the growth defect association with the lack of the penicillin binding protein Pbp2b in S. pneumoniae (Stamsås et al., 2017, Tsui et al., 2016, Zheng et al., 2017). Pbp2b is an essential enzyme required for peptidoglycan elongation outward from the midcells of dividing S. pneumoniae cells (reviewed in (Briggs et al., 2021)). The association with elongasome proteins led to the alternate name of EloR (elongasome regulating protein) for KhpB (Stamsås et al., 2017). Consistent with a role in cell elongation, S. pneumoniae strains lacking either khpA or khpB have a reduced length (Zheng et al., 2017, Ulrych et al., 2016, Stamsås et al., 2017) and width (Zheng et al., 2017, Ulrych et al., 2016) compared to wild-type cells and also show slower growth (Zheng et al., 2017, Ulrych et al., 2016, Stamsås et al., 2017). Furthermore, the observation that a ΔkhpA ΔkhpB double mutant has the same phenotype as the single mutants indicates that the two proteins act in the same pathway (Zheng et al., 2017).
Several observations in addition to the co-occurrence of the two genes, indicate that KhpA and KhpB act together (Zheng et al., 2017). The proteins co-localize, diffusing in early divisional cells and enriched at the midcell in dividing cells (Zheng et al., 2017, Winther et al., 2019, Stamsås et al., 2017). Further evidence for an association between the two proteins has come from copurification and bacterial two-hybrid experiments (Winther et al., 2019, Zheng et al., 2017). The studies of different KhpB truncation mutants revealed that KhpA and KhpB heterodimerize via their KH domains and that this interaction is required for the suppression of the Δpbp2b growth defect (Winther et al., 2019). KhpA also can homodimerize, and the α3 helix in the KH domain is required for both homodimerization and heterodimerization with KhpB (Winther et al., 2019).
While the interaction studies revealed that the KH domain is required for oligomerization of KhpA and KhpB, the role of RNA binding in the elongasome is less clear. It is intriguing and consistent with a role in cell division that several mRNAs enriched by co-immunoprecipitation of KhpA or KhpB encode cell division proteins, including the cell division protein FtsA (Zheng et al., 2017). While the relative amount of ftsA-ftsZ mRNA transcript remains nearly the same, the cellular amount of FtsA protein increases in the khp single and double mutant strains. Assays of the effects of different sections of the ftsA gene revealed that the 5ʹ-UTR of ftsAZ mRNA is required for the KhpA and KhpB-dependent down regulation of FtsA levels (Zheng et al., 2017), but it is not known how this post-transcriptional regulation is brought about by the KH-domain proteins. Suggested possibilities include direct down regulation by the KhpA and KhpB proteins or indirect regulation through an sRNA chaperoned by the KH proteins or another protein modulated by KhpA and KhpB.
It is possible that KhpA and KhpB proteins have multiple roles in cell division, only one of which might depend on RNA binding. Several studies showed that S. pneumoniae KhpB is phosphorylated on the linker residue threonine 89 by the StkP kinase that is also part of the elongasome network (Stamsås et al., 2017, Sun et al., 2010, Ulrych et al., 2016). However, phenotypic effects of phosphoablative or phosphomimetic mutations were not observed in all studies, (Zheng et al., 2017, Stamsås et al., 2017), and the threonine 89 residue is not conserved in all KhpB proteins. S. pneumoniae KhpB also has been shown to interact with the peptidoglycan muramidase MpgA (Winther et al., 2021), which was previously called MltG (Tsui et al., 2016, Taguchi et al., 2021). Interestingly, mutations in mpgA, like khpA and khpB mutations, suppress the Δpbp2b phenotype (Tsui et al., 2016). The interaction between KhpB and MpgA involves the Jag-N domain of KhpB and is required for localization of the KhpA-KhpB complex to the midcell (Winther et al., 2021).
While the KhpA and KhpB roles in cell division have been studied most extensively in S. pneumoniae, the phenotypes associated with the lack of these proteins in other bacteria also is consistent with roles in cell division. In Lactobacillus plantarum, a CRISPR knock-down of either khpA or khpB (eloR) results in cell shortening, with the effect being stronger for KhpA (Myrbraten et al., 2019). As the mechanism of cell elongation differs between cocci (peptidoglycan insertion occurs at midcell) and bacilli (peptidoglycan insertion occurs over the full cell length), these observations suggest a very general role for these proteins in cell wall synthesis. However, the KhpA and KhpB may not have a role in cell division in all organisms, given that in C. difficile the size of ΔkhpA cells is like that of wild-type, though ΔkhpB mutants had slightly increased cell length and width (Lamm-Schmidt et al., 2021).
Role in regulating virulence gene expression
In both S. pneumoniae and C. difficile, KhpA and KhpB also bind RNAs that do not encode cell division proteins indicating broader physiological roles of these proteins, including roles in regulating virulence. Among the transcripts bound by KhpB in C. difficile is the tcdA mRNA encoding the clostridial toxin A (Lamm-Schmidt et al., 2021). Consistent with a KhpB role in modulating toxin A production, the levels of both the mRNA and protein increase in a ΔkhpB mutant. However, given only a minimal difference in tdcA mRNA half life after rifampicin treatment when comparing a wild-type and ΔkhpB strain, it is not yet clear how KhpB acts as a negative regulator. The detection of S. pneumoniae khpB in a Tn-seq screen for reduced fitness in a mouse model of pneumonia further supports a KhpB role in regulating virulence gene expression (van Opijnen & Camilli, 2012).
Role in protein synthesis or translocation
Another possible clue to the physiological roles of the KhpA and KhpB proteins comes from the functions of the genes that are syntenic with khpA (Fig. S2) and khpB (Fig. S3). khpA is strongly co-conserved with the rpsP encoding the ribosomal protein S16 as well as rimH encoding a ribosome maturation factor and trmD encoding a tRNA methyltransferase, while khpB appears to be in an operon with yidC (spoIIIJ in B. subtilis) encoding a protein translocase and near rnpA encoding the RNA component of RNase P, among others. All the proteins encoded by syntenic genes directly or indirectly affect translation or protein translocation. Along the same lines, the H. pylori KhpB protein with the noncanonical R3H domain was found to bind to and inhibit the HP0525 inner membrane ATPase which has a role in the transport through type IV secretion systems (Hare et al., 2007). Here the KH and R3H domains contact the ATPase, but again it is not clear if RNA binding is involved.
Outlook
The KhpA and KhpB protein families are clearly broadly distributed in several different phyla, suggesting important roles for these RNA binding proteins. However, even though other KH domains have been studied extensively, relatively little is known about how KhpA and KhpB bind to RNA and the consequences of this binding. Global co-sedimentation approaches and co-purification approaches have revealed binding to sRNAs, but much remains to be learned about the role of this binding. Do KhpA and KhpB promote sRNA pairing with target mRNAs? How is the role in RNA binding connected to the phenotypes related to peptidoglycan synthesis and cell division? Do KhpA and KhpB proteins have different roles in different species? Do the proteins generally act together or also have separate functions? It will be exciting to see what answers future studies will provide to these and other open questions.
Supplementary Material
Acknowledgements
We thank P. Adams, F. Faber, V. Lamm-Schmidt, T. Tsui and M. Winkler for comments. We would also like to thank W. Yan from the Jiang Lab for help with figure generation. Research in the M.O. lab is supported by National Science Centre in Poland [grants No. 2018/31/B/NZ1/02612 and No. 2020/39/O/NZ1/02448]. Research in the X.J. group is supported by the Intramural Research Program of the NIH, National Library of Medicine. Research in the G.S. lab is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development. The authors declare no conflict of interest.
References
- Athar YM, and Joseph S (2020) RNA-binding specificity of the human Fragile X Mental retardation protein. J Mol Biol 432: 3851–3868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auweter SD, Oberstrass FC, and Allain FH (2006) Sequence-specific binding of single-stranded RNA: is there a code for recognition? Nucleic Acids Res 34: 4943–4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuth B, Pennell S, Arnvig KB, Martin SR, and Taylor IA (2005) Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J 24: 3576–3587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas J, Patel VL, Bhaskar V, Chao JA, Singer RH, and Eliscovich C (2019) The structural basis for RNA selectivity by the IMP family of RNA-binding proteins. Nat Commun 10: 4440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs NS, Bruce KE, Naskar S, Winkler ME, and Roper DI (2021) The Pneumococcal divisome: dynamic control of Streptococcus pneumoniae cell division. Front. Microbiol in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesla M, Turowski TW, Nowotny M, Tollervey D, and Boguta M (2020) The expression of Rpb10, a small subunit common to RNA polymerases, is modulated by the R3H domain-containing Rbs1 protein and the Upf1 helicase. Nucleic Acids Res. 48: 12252–12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corley M, Burns MC, and Yeo GW (2020) How RNA-binding proteins interact with RNA: molecules and mechanisms. Mol Cell 78: 9–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagil R, Ball NJ, Ogrodowicz RW, Hobor F, Purkiss AG, Kelly G, Martin SR, Taylor IA, and Ramos A (2019) IMP1 KH1 and KH2 domains create a structural platform with unique RNA recognition and re-modelling properties. Nucleic Acids Res 47: 4334–4348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dendooven T, Sinha D, Roeselova A, Cameron TA, De Lay NR, Luisi BF, and Bandyra KJ (2021) A cooperative PNPase-Hfq-RNA carrier complex facilitates bacterial riboregulation. Mol Cell 81: 2901–2913 e2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez D, Freese P, Alexis MS, Su A, Hochman M, Palden T, Bazile C, Lambert NJ, Van Nostrand EL, Pratt GA, Yeo GW, Graveley BR, and Burge CB (2018) Sequence, structure, and context preferences of human RNA binding proteins. Mol Cell 70: 854–867 e859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Appleby L, Daniel RA, Goodfellow H, Partridge SR, and Yudkin MD (1992) Structure and function of the spoIIIJ gene of Bacillus subtilis: a vegetatively expressed gene that is essential for sG activity at an intermediate stage of sporulation. J. Gen. Microbiol 138: 2609–2618. [DOI] [PubMed] [Google Scholar]
- Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, Colston MJ, and Dodson G (2001) Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 Å resolution. J Mol Biol 314: 1087–1095. [DOI] [PubMed] [Google Scholar]
- Grishin NV (1998) The R3H motif: a domain that binds single-stranded nucleic acids. Trends Biochem. Sci 23: 329–330. [DOI] [PubMed] [Google Scholar]
- Grishin NV (2001) KH domain: one motif, two folds. Nucleic Acids Res 29: 638–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick SW, Gubbey T, Hug I, Jenal U, and Luisi BF (2012) Crystal structure of Caulobacter crescentus polynucleotide phosphorylase reveals a mechanism of RNA substrate channelling and RNA degradosome assembly. Open Biol 2: 120028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare S, Fischer W, Williams R, Terradot L, Bayliss R, Haas R, and Waksman G (2007) Identification, structure and mode of action of a new regulator of the Helicobacter pylori HP0525 ATPase. EMBO J. 26: 4926–4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingworth D, Candel AM, Nicastro G, Martin SR, Briata P, Gherzi R, and Ramos A (2012) KH domains with impaired nucleic acid binding as a tool for functional analysis. Nucleic Acids Res 40: 6873–6886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hör J, Garriss G, Di Giorgio S, Hack LM, Vanselow JT, Förstner KU, Schlosser A, Henriques-Normark B, and Vogel J (2020) Grad-seq in a Gram-positive bacterium reveals exonucleolytic sRNA activation in competence control. EMBO J 39: e103852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJ, Swapna GV, Rajan PK, Ke H, Xia B, Shukla K, Inouye M, and Montelione GT (2003) Solution NMR structure of ribosome-binding factor A (RbfA), a cold-shock adaptation protein from Escherichia coli. J Mol Biol 327: 521–536. [DOI] [PubMed] [Google Scholar]
- Jumper J, Evans R, Pritzel A, Green T, Figurnov M, Ronneberger O, Tunyasuvunakool K, Bates R, Zidek A, Potapenko A, Bridgland A, Meyer C, Kohl SAA, Ballard AJ, Cowie A, Romera-Paredes B, Nikolov S, Jain R, Adler J, Back T, Petersen S, Reiman D, Clancy E, Zielinski M, Steinegger M, Pacholska M, Berghammer T, Bodenstein S, Silver D, Vinyals O, Senior AW, Kavukcuoglu K, Kohli P, and Hassabis D (2021) Highly accurate protein structure prediction with AlphaFold. Nature 596: 583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn SM, Ulshofer CJ, Schneider T, and Schlundt A (2021) Structures and target RNA preferences of the RNA-binding protein family of IGF2BPs: An overview. Structure 29: 787–803. [DOI] [PubMed] [Google Scholar]
- Lamm-Schmidt V, Fuchs M, Sulzer J, Gerovac M, Hör J, Dersch P, Vogel J, and Faber F (2021) Grad-seq identifies KhpB as a global RNA-binding protein in Clostridioides difficile that regulates toxin production. microLife 2: uqab004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirdita M, Ovchinnikov S, and Steinegger M (2021) ColabFold - Making protein folding accessible to all. bioRxiv: doi: 10.1101/2021.1108.1115.456425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myrbraten IS, Wiull K, Salehian Z, Havarstein LS, Straume D, Mathiesen G, and Kjos M (2019) CRISPR interference for rapid knockdown of essential cell cycle genes in Lactobacillus plantarum. mSphere 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai K (1996) RNA-protein complexes. Curr Opin Struct Biol 6: 53–61. [DOI] [PubMed] [Google Scholar]
- Nagata M, Kaito C, and Sekimizu K (2008) Phosphodiesterase activity of CvfA is required for virulence in Staphylococcus aureus. J Biol Chem 283: 2176–2184. [DOI] [PubMed] [Google Scholar]
- Nicastro G, Taylor IA, and Ramos A (2015) KH-RNA interactions: back in the groove. Curr Opin Struct Biol 30: 63–70. [DOI] [PubMed] [Google Scholar]
- Olejniczak M, and Storz G (2017) ProQ/FinO-domain proteins: another ubiquitous family of RNA matchmakers? Mol. Microbiol 104: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Meng EC, Couch GS, Croll TI, Morris JH, and Ferrin TE (2021) UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci 30: 70–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prezza G, Ryan D, Mädler G, Reichardt S, Barquist L, and Westermann AJ (2021) Comparative genomics provides structural and functional insights into Bacteroides RNA biology. Mol. Microbiol In press. [DOI] [PubMed] [Google Scholar]
- Riediger M, Spät P, Bilger R, Voigt K, Maček B, and Hess WR (2021) Analysis of a photosynthetic cyanobacterium rich in internal membrane systems via gradient profiling by sequencing (Grad-seq). Plant Cell 33: 248–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson VL, Hwang J, Fox E, Inouye M, and Stock AM (2002) Domain arrangement of Der, a switch protein containing two GTPase domains. Structure 10: 1649–1658. [DOI] [PubMed] [Google Scholar]
- Schneider T, Hung LH, Aziz M, Wilmen A, Thaum S, Wagner J, Janowski R, Muller S, Schreiner S, Friedhoff P, Huttelmaier S, Niessing D, Sattler M, Schlundt A, and Bindereif A (2019) Combinatorial recognition of clustered RNA elements by the multidomain RNA-binding protein IMP3. Nat Commun 10: 2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbabian K, Jamalli A, Zig L, and Putzer H (2009) RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J 28: 3523–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi H, Matunis MJ, Michael WM, and Dreyfuss G (1993) The pre-mRNA binding K protein contains a novel evolutionarily conserved motif. Nucleic Acids Res 21: 1193–1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamsås GA, Straume D, Ruud Winther A, Kjos M, Frantzen CA, and Havarstein LS (2017) Identification of EloR (Spr1851) as a regulator of cell elongation in Streptococcus pneumoniae. Mol Microbiol 105: 954–967. [DOI] [PubMed] [Google Scholar]
- Steinegger M, and Soding J (2017) MMseqs2 enables sensitive protein sequence searching for the analysis of massive data sets. Nat Biotechnol 35: 1026–1028. [DOI] [PubMed] [Google Scholar]
- Sun X, Ge F, Xiao CL, Yin XF, Ge R, Zhang LH, and He QY (2010) Phosphoproteomic analysis reveals the multiple roles of phosphorylation in pathogenic bacterium Streptococcus pneumoniae. J Proteome Res 9: 275–282. [DOI] [PubMed] [Google Scholar]
- Taguchi A, Page J, Tsui HT, Winkler ME, and Walker S (2021) Biochemical reconstitution defines new functions for membrane-bound glycosidases in assembly of the bacterial cell wall. Proc. Natl. Acad. Sci. USA 118: e2103740118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsui HC, Zheng JJ, Magallon AN, Ryan JD, Yunck R, Rued BE, Bernhardt TG, and Winkler ME (2016) Suppression of a deletion mutation in the gene encoding essential PBP2b reveals a new lytic transglycosylase involved in peripheral peptidoglycan synthesis in Streptococcus pneumoniae D39. Mol Microbiol 100: 1039–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu C, Zhou X, Tropea JE, Austin BP, Waugh DS, Court DL, and Ji X (2009) Structure of ERA in complex with the 3’ end of 16S rRNA: implications for ribosome biogenesis. Proc Natl Acad Sci U S A 106: 14843–14848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulrych A, Holeckova N, Goldova J, Doubravova L, Benada O, Kofronova O, Halada P, and Branny P (2016) Characterization of pneumococcal Ser/Thr protein phosphatase phpP mutant and identification of a novel PhpP substrate, putative RNA binding protein Jag. BMC Microbiol 16: 247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde R, Edwards L, and Regan L (2008) Structure and function of KH domains. FEBS J 275: 2712–2726. [DOI] [PubMed] [Google Scholar]
- van Opijnen T, and Camilli A (2012) A fine scale phenotype-genotype virulence map of a bacterial pathogen. Genome Res. 22: 2541–2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N, Fauvart M, Versees W, and Michiels J (2011) The universally conserved prokaryotic GTPases. Microbiol Mol Biol Rev 75: 507–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson ZL, Ward FR, Meheust R, Ad O, Schepartz A, Banfield JF, and Cate JH (2020) Structure of the bacterial ribosome at 2 Å resolution. Elife 9: e60482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimberly BT, Brodersen DE, Clemons WM Jr., Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, and Ramakrishnan V (2000) Structure of the 30S ribosomal subunit. Nature 407: 327–339. [DOI] [PubMed] [Google Scholar]
- Winther AR, Kjos M, Herigstad ML, Havarstein LS, and Straume D (2021) EloR interacts with the lytic transglycosylase MltG at midcell in Streptococcus pneumoniae R6. J Bacteriol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winther AR, Kjos M, Stamsas GA, Havarstein LS, and Straume D (2019) Prevention of EloR/KhpA heterodimerization by introduction of site-specific amino acid substitutions renders the essential elongasome protein PBP2b redundant in Streptococcus pneumoniae. Sci Rep 9: 3681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodson SA, Panja S, and Santiago-Frangos A (2018) Proteins that chaperone RNA regulation. Microbiol Spectr. 6: RWR-0026–2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worbs M, Bourenkov GP, Bartunik HD, Huber R, and Wahl MC (2001) An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell 7: 1177–1189. [DOI] [PubMed] [Google Scholar]
- Yadav M, Singh RS, Hogan D, Vidhyasagar V, Yang S, Chung IYW, Kusalik A, Dmitriev OY, Cygler M, and Wu Y (2021) The KH domain facilitates the substrate specificity and unwinding processivity of DDX43 helicase. J Biol Chem 296: 100085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng JJ, Perez AJ, Tsui HT, Massidda O, and Winkler ME (2017) Absence of the KhpA and KhpB (JAG/EloR) RNA-binding proteins suppresses the requirement for PBP2b by overproduction of FtsA in Streptococcus pneumoniae D39. Mol Microbiol 106: 793–814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


