Abstract
Chromatin is the epigenomic platform for diverse nuclear processes such as DNA repair, replication, transcription, telomere, and centromere function. In cancer cells, mutations in key processes result in DNA amplification, chromosome translocations, and chromothripsis, severely distorting the natural chromatin state. In normal and diseased states, dozens of chromatin effectors alter the physical integrity and dynamics of chromatin at the level of both single nucleosomes and arrays of nucleosomes folded into 3-dimensional shapes. Integrating these length scales, from the 10 nm sized nucleosome to mitotic chromosomes, whilst jostling within the crowded environment of the cell, cannot yet be achieved by a single technology. In this review, we discuss tools that have proven powerful in the investigation of nucleosome and chromatin fiber dynamics. We also provide a deeper focus into atomic force microscopy (AFM) applications that can bridge diverse length and time scales. Using time course AFM, we observe that chromatin condensation by H1.5 is dynamic, whereas using nano-indentation force spectroscopy we observe that both histone variants and nucleosome binding partners alter material properties of individual nucleosomes. Finally, we demonstrate how high-speed AFM can visualize plasmid DNA dynamics, intermittent nucleosome-nucleosome contacts, and changes in nucleosome phasing along a contiguous chromatin fiber. Altogether, the development of innovative technologies holds the promise of revealing the secret lives of nucleosomes, potentially bridging the gaps in our understanding of how chromatin works within living cells and tissues.
Keywords: epigenetics, nucleosomes, chromatin, chromosome, AFM
Introduction
In eukaryotes, genomic DNA is wrapped around histone cores to form nucleosomes, which in turn form the classic beads-on-a-string structure of chromatin.1–3 Each nucleosome wraps ∼147 bp of DNA and because of histone variants, the composition of nucleosomes is modular.4,5 This modular nucleosomal composition, composed of exchangeable histone dimer units, coupled with dozens of covalent post-translational modifications, makes the chromatin fiber highly plastic and tunable, allowing for tight regulation of access to chromatin factors.6,7 Central questions of outstanding interest include: unveiling the principles by which chromatin domains are organized in the nucleus; dissecting properties they impart to the regulation of the underlying DNA sequence; figuring out whether organization and structure are heritable, dynamic or persistent; and finally, how these phenomena contribute to genome function. In this review, we discuss sophisticated technical advances that have expanded our understanding of the rules of life in the context of the genome.
From Deep Diving into the Genome to Spatial Organization of the Nucleus
The past decade has been dominated by major advances in high throughput sequencing of the genome and super-resolution microscopy of cells. Many studies have focused on sub-nuclear localization of unique histone variants, histone modifications, their protein binding partners and effectors.6,8–15 For example, the Maeshima group used photoactivated localization microscopy (PALM) combined with single-nucleosome tracking to show that nucleosomes form distinct chromatin domains.16,17 High resolution microscopy has also lent critical quantitative insights into the architecture of the human centromere by counting the number of centromeric nucleosomes in vivo.18–20 Genome wide sequencing approaches such as ATAC-seq21 and CUT&Tag22 use the Tn5 transposase to assay which genomic sites are accessible over time; whereas super-resolution microscopy use increasingly sophisticated fluorophores and tracking algorithms to describe the movement of individual molecules in real time.23,24 Thus, such technologies have been informative in figuring out where and how they move in living cells. However, such methods are not yet able to dissect how biophysical properties of epigenetic signatures functionally alter at a given chromatin locus in vivo.
Reconstructing Chromatin Dynamics in Computers and Test Tubes
To complement in vivo studies that capture dynamics (single molecule fluorophore tracking) and sub-nuclear localization (high throughput sequencing, FISH/IF, PALM), powerful in vitro and in silico biophysical tools have been developed to give yield a quantitative understanding of chromatin behavior (Figure 1). Computational modeling uses sophisticated force fields derived from a deep mathematical understanding of the physical relationships between atoms, to create trajectories of macromolecular motion. From these trajectories structural dynamics can be deduced.25–30 In vitro experiments with purified recombinant proteins probe dynamics at high resolution, using elegant tools such as single molecule FRET, magnetic and optical tweezers, DNA nano-curtains, and hydrogen–deuterium exchange mass spectrometry.31–33 Using the optical tweezer technology, invented by Arthur Ashkin,34 the Bustamante group developed a single-molecule transcription assay by clamping one end of a DNA strand with one optical tweezer, and using another optical tweezer to manipulate RNA polymerase 2 (RNAP2).35 With this tour-de-force experiment, it was possible to measure the energy landscape of how RNAP2 transcribes through a nucleosome.35 To understand how chromatin is compacted, the Fierz group utilized single molecule FRET to study how heterochromatin protein HP1α compacts a tetranucleosome array.36 This approached revealed that HP1α transiently stabilized tetranucleosomes in the range of 100 s of milliseconds. Similarly, DNA curtains revealed the dynamics with which condensin I and II compact DNA,37 whereas adding flow to tethered DNA molecules showed that yeast condensin38,39 and human cohesin40,41 forms loops. These latter techniques rely on visualizing DNA with either YOYO-1 or Sytox Orange, which have the capacity to alter DNA topology and dynamics.42–44 Altogether, these various approaches are powerful because they allow for precise measurements of a broad range of chromatin behaviors in vitro, with impressive temporal resolution.
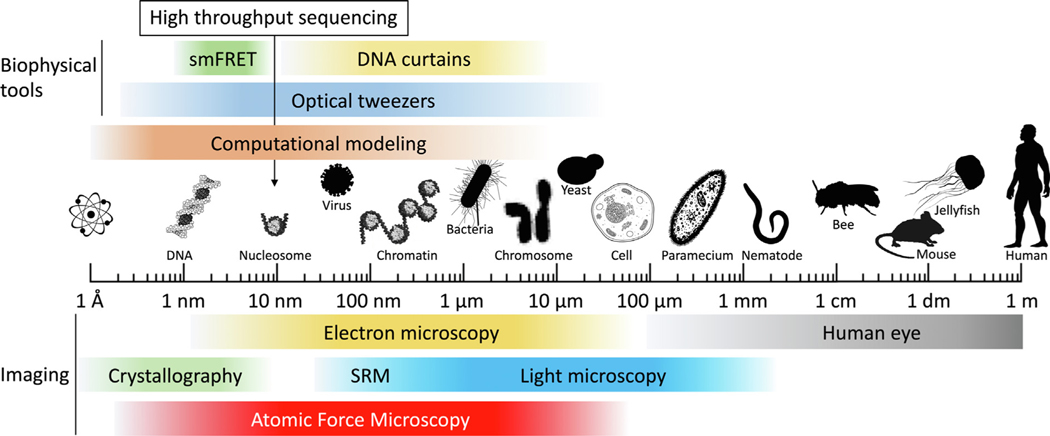
Figure 1.
AFM bridges the gap between crystallography and light microscopy. A logarithmic scale from Ångström to meters highlighting the scale at which various imaging technologies and biophysical tools operate. SRM = super-resolution microscopy. Silhouettes were obtained from PhyloPic (http://phylopic.org/).
Peering into the Structural Heart of Chromatin
Structural studies of chromatin factors rely on imaging techniques like X-ray crystallography, NMR, and cryo-EM. These types of studies have provided a wealth of information linking structure to function.45,46 These studies highlight that despite the static conformation of nucleosomes being remarkably similar irrespective of histone composition, the H2A-H2B acid patch is a preferred binding site for chromatin binding factors,47 whereas internal histone dynamics and DNA site exposure differ quite dramatically. To understand how the histone chaperone FACT is involved in both nucleosome disassembly and reassembly, structural studies provided critical clues. Indeed, FACT bound to a nucleosome forms two types of complexes, which includes interactions with both DNA and all histone variants.48 In addition to these very detailed structural studies, combining super-resolution microscopy with scanning electron microscopy provides intriguing data at the mesoscale of chromatin organization. A recent study that used these approaches showed that chromatin domains act as physically distinct modules that persist after cohesin ablation.49 Combining complementary biophysical methods with structural methods are becoming more common. For instance, to aid the results from the DNA curtain assay, structural understanding was obtained by negative stain electron microscopy.37 Recently, the Narlikar group combined X-ray crystallography with NMR and hydrogen–deuterium exchange mass spectroscopy to show nucleosomes bound by HP1Swi6 altered intra-histone dynamics.50.
The methods described above provide insights in conformational dynamics within the individual nucleosome as well as chromatin structure at large and how these play an essential role in regulating functional aspects of the chromatin fiber. How structural fluctuations facilitate the exposure of DNA sequences and nucleosomal residues,51,52 thereby provide transient access to the nuclear machineries still remain a largely unanswered question. Next, we discuss how atomic force microscopy (AFM), and high-speed AFM (HS-AFM) can help complement high resolution live imaging methodologies to fill critical gaps in our understanding of chromatin spatiotemporal and biophysical dynamics (Figure 1).
AFM as an Imaging and Dissecting Tool
AFM is a topographic imaging technology which allows for the characterization of individual molecules at sub-nanometer resolution, among various other applications such as force spectroscopy and recognition imaging (Figure 1).53–55 One advantage of AFM is that nucleoprotein complexes can be prepared and imaged under gentle conditions, without chemical cross-linking, harsh dehydration, freezing, shadowing or staining.55,56 Furthermore, because every molecule can be directly seen and therefore analyzed, AFM does not require class averaging of thousands of uniformly organized particles for 3D image reconstruction such as in high-resolution techniques like cryo-electron microscopy (cryoEM). As a consequence of class averaging, cryoEM is not bound by the mica surface upon which AFM samples are deposited. It is the presence of this surface that allows samples to be manipulated and assayed in ways that microscopy and cryo-EM cannot do. For instance, determining how elastic nucleosomes are 57,58. These powerful features of AFM permit observation of near-native states of molecules and spontaneous molecular events, such as DNA wrapping and unwrapping of nucleosomes.59–61 AFM can also be used to assess nucleosome distribution62–64 and nucleosome structure.64–66 These studies highlight the variable and dynamic nature of nucleosomes and chromatin. Although DNA sequences can facilitate the positioning of nucleosomes;67 in the nucleus, most nucleosomes are not tightly positioned.23,24,62,63,68,69 AFM has also been combined with a rotaxane molecular reader to assess DNA secondary structures.70 Furthermore, the composition of nucleosomes also impacts their sensitivity to salt concentrations.65 and their linker lengths.64.
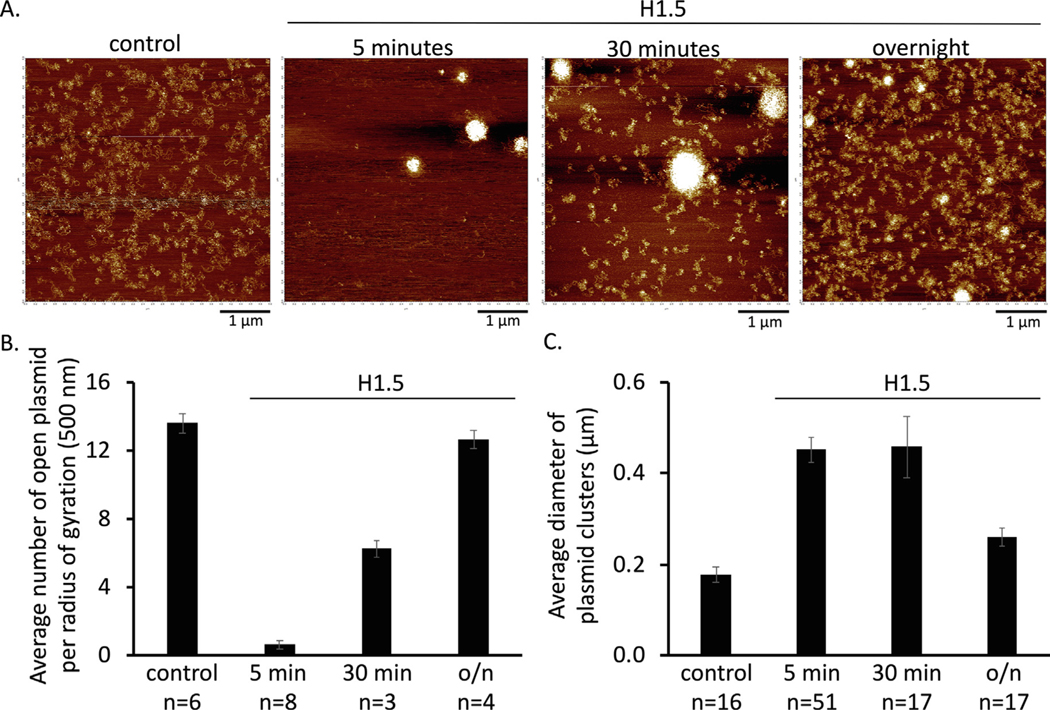
To illustrate how conventional AFM imaging methods can provide temporal information, we performed a time-course of the linker histone H1 compaction of in vitro reconstituted H3 chromatin, as H1 histone proteins are known for compacting chromatin.71–73 We added H1.5 for 5 minutes, 30 minutes, or overnight (Figure 2(a)). From the AFM images, a clear loss of open H3 chromatin is observed within 5 minutes after the addition of H1.5. Interestingly, over the time course, more open H3 chromatin plasmids can be observed (Figure 2(b)), whereas the size of the chromatin clusters decreases over time (Figure 2(c)). These results support the concept that chromatin compaction driven by H1 is dynamic.74,75 As various H1 variants exist76,78 with differing chromatin compaction potential;79 a powerful application of this method is to explore the chromatin compaction dynamics of these H1 variants. These experiments could conceivably be performed also on substrates that contain unique DNA positioning motifs from discrete loci, as well as covalent histone and DNA modifications reported to exist at those loci in vivo. This type of in vitro mimicry of the in vivo epigenetic status of a specific locus may further our mechanistic understanding of the temporal folding and accessibility of chromatin structures.
Figure 2.
Time course of H3 chromatin compaction induced by H1. (a) Representative in air AFM images of reconstituted H3 chromatin without or with H1. (b) Quantification of number of plasmids per radius of gyration as a measure of open chromatin. (c) Quantification of compaction by measuring the diameter of chromatin clusters.
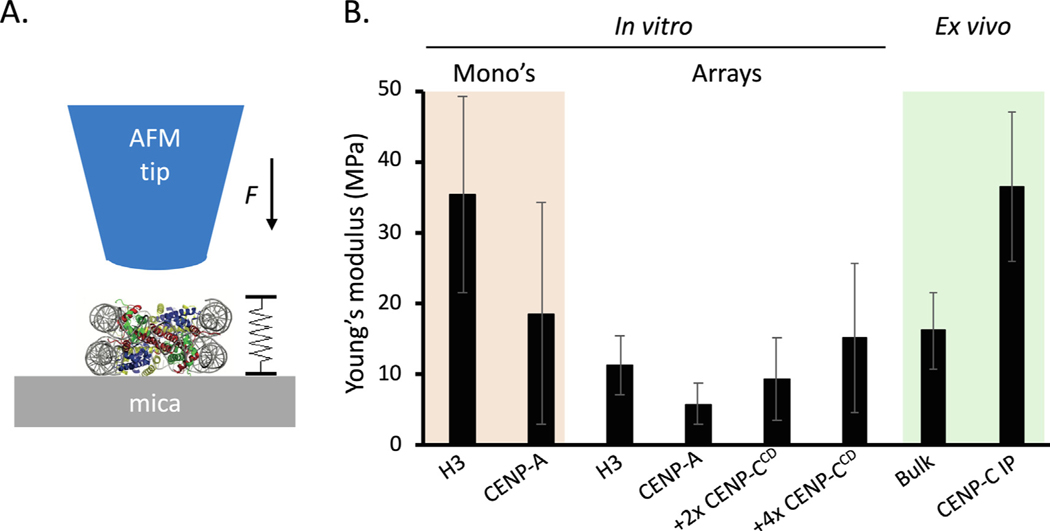
In addition to structural studies, AFM can also be used to determine material properties of molecules and larger structures. Pushing, pulling, sliding and hydrodynamic forces are abound in the crowded internal environment of nuclei, cells and tissues/organs. The response to those forces will rely on inherent elasticity or rigidity of the substrate, which in turn will arise from its constituent components and the environmental context. For example, the nucleus has been subjected to force spectroscopy measurements, wherein nuclei within cells were found to have a higher Young’s modulus (that is, more rigid) than isolated nuclei.80 Mechanical properties of nuclei have also been assayed using micropipettes.81–83 Such a method, while powerful for larger objects, is not well suited for determining material properties of individual macromolecules, such as nucleosomes. In the first measurements of nucleosomes and chromatin using this kind of approach, we used nanoindentation AFM to measure the rigidity (as measured by the Young’s modulus) of reconstituted mononucleosomes and chromatin (Figure 3(a)).58 Interestingly, nucleosomes with the centromere-specific histone H3 variant CENP-A were twice as elastic as canonical H3 nucleosomes (Figure 3 (b)). The primary function of CENP-A nucleosomes is to recruit the kinetochore and the first protein that is recruited is CENP-C.84 Addition of CENP-C resulted in marked rigidification of CENP-A nucleosomes (Figure 3(b)), as well as chromatin clustering.58 We recently applied our nano-indentation AFM protocols to native H3 chromatin and kinetochore complexes purified from human cells.57 Of particular interest from these studies was our observation that H3 chromatin purified from cells was slightly more rigid than in vitro reconstituted H3 chromatin—thus, demonstrating that environmental context does matter (Figure 3(b)).57 These results point to nucleosomal binding proteins (such as H1), or histone modifications, that may alter the rigidity of the chromatin fiber in vivo. Where individual nucleosomes have Young’s moduli in the MPa range, Young’s modulus measurements of whole chromosomes are substantially more elastic (40–400 Pa).85,86 Altogether, an emerging picture is formed that material properties of nucleosomes are inherent but can be modulated. How individual nucleosome dictate the material properties of whole chromosomes will be best studies by focusing on specific loci. Therefore, an outstanding question is how altering material properties of individual nucleosomes could impact chromatin fiber functions at specific loci, such as the speed of DNA repair, accessibility to polymerases during transcription initiation and elongation, replication timing and progression, and mitosis. This becomes particularly pertinent not just in the normal state, where PTMs and histones variants are dynamic, but also in the cancer or aging state, where such proteins are misregulated or even mutated.87–90.
Figure 3.
Young’s modulus varies across different chromatin units (a) Graphical representation how the Young’s modulus is obtained from single molecules such as nucleosomes. (b) Both for mononucleosomes and individual nucleosomes within arrays, CENP-A nucleosomes have a lower Young’s modulus than canonical H3 nucleosomes. Furthermore, CENP-A binding partner CENP-C rigidifies CENP-A nucleosomes. Interestingly, H3 nucleosomes extracted from cells had a larger Young’s modulus than in vitro reconstituted H3 nucleosomes. Data is from Melters et al. and Rakshit et al.58,57
High Speed-AFM to See Chromatin Move and Interact
Structural techniques, such as X-ray crystallography, NMR, and cryo-EM, rely on ensemble averaging. In contrast, AFM creates a topological image of molecules. HS-AFM overcomes the frame rate limitation of conventional AFM, finally breaking through the temporal barrier to visualize the dynamic motions of single molecules in real time. Twelve years ago, the Ando group developed the first generation of custom built HS-AFM.91 In a stunning tour-de-force, they captured the motion of individual myosin V molecules “walking” on an actin filament.92 This landmark paper was quickly followed by the development of commercial HS-AFM systems that can now be purchased for half the cost of an electron microscope, with ambient operation at the benchtop. This is made even more powerful by the fact that even undergraduate and intern researchers can be trained relatively quickly to use these systems, giving them a powerful opportunity to directly peer into the inner workings of macromolecular complexes. With the advent of commercial HS-AFM systems (popular examples are the Cypher VRS from Asylum Research and the JPK NanoRacer by Bruker) and better understanding of tip dynamics,93 researchers have extended the use of HS-AFM for diverse molecular processes. For example, the Nureki group showed how CRISPR-Cas9 cuts a piece of DNA94 and the Dekker group showed that yeast condensin exist in both a closed and open state.95 Expanding the use of HS-AFM to the chromatin field was an inevitable and exciting development. Three pioneering studies, led by the Takeyasu, Dekker, and Lyubchenko groups have imaged nucleosomes with HS-AFM.59,61,96–100 Reconstituted H3 mononucleosomes showed either a one- or two-step spontaneous unfolding event59 as well as sliding.61 In addition, reconstituted CENP-A mononucleosomes also showed extensive loop extrusion and DNA strand transfer.98 A recent study used DNA origami to create a space limited environment for two nucleosomes to interact.101 Approaches as described in this latter study will help in determining how far HS-AFM can be pushed without loss of protein–protein and protein-DNA interactions. Using HS-AFM this study observed that the interaction between two nucleosomes vary considerable over time. Besides the striking visual impact of such movies, especially as a pedagogical tool in the laboratory to novice researchers, these experiments illustrate how rigorously executed HS-AFM experiments can simultaneously capture structural information, while quantifying single molecule dynamics data.
To illustrate applications of HS-AFM for chromatin work in our lab, we first show the dynamic nature of plasmid DNA in fluid (Figure 4 (a), Supplementary Video 1), neatly recapitulating previous work by Lyubchenko and Shlyakhtenko.102 Whereas in air AFM only shows a single conformation per plasmid, HS-AFM shows that the conformation of plasmids changes rather rapidly. We further explore the dynamics of reconstituted H3 nucleosomes on plasmids. Whereas previous studies focused on well positioned mononucleosomes that were seeded at low density or physically confined nucleosomes, we used plasmids containing four human centromeric α-satellite repeats and seeded the reconstituted plasmids to medium density. This permits nucleosomes from one plasmid to interact with a nucleosome of another plasmid. To our surprise, we observed frequent intermittent contact between one nucleosome (Figure 4(b), Supplementary Video 2) and several other nucleosomes. These intermittent contacts are transient, lasting no more than a few seconds. In comparison, when a nucleosome is not in close proximity of another nucleosome it displayed greater mobility. How long inter- or intra-fiber interactions last remains to be determined.103 We therefore speculate that the time scale of intermittent nucleosome-nucleosome contact is within the realm of chromatin inter- or intra-fiber interactions in vivo. Thus HS-AFM should prove powerful in studying how enhancer chromatin loops and interacts with promoters. Next, we illustrate how HS-AFM can shed light on H1-chromatin dynamics in real time. Interestingly, we observed on average three to four nucleosomes would be nicely spaced for several seconds (2.6 ± 2.0 sec; n = 52) (Figure 4 (c), Supplementary Video 3). This observation is in agreement with previously reported nucleosome phasing.104–106 AFM provides multiple sources of data based on how the tip interacts with the surface, which are traditionally reported in distinct channels. For HS-AFM three types of data are recorded: height, amplitude, and phase. The former is commonly reported (Figure 4(a)–(c)), but for HS-AFM, the latter allows for easy distinguishing between DNA and nucleosomes (Figure 4(d), Supplementary Video 4). Altogether, HS-AFM of chromatin has provided tantalizing visual evidence of nucleosome dynamics, both at the mononucleosome and nucleosome array level. Future studies will hopefully provide mechanistic evidence how chromatin machineries, such as RNA polymerase 2 or chromatin remodelers, interact with and modulate nucleosome arrays and how these interactions are altered by nucleosome binding partners.
Figure 4.
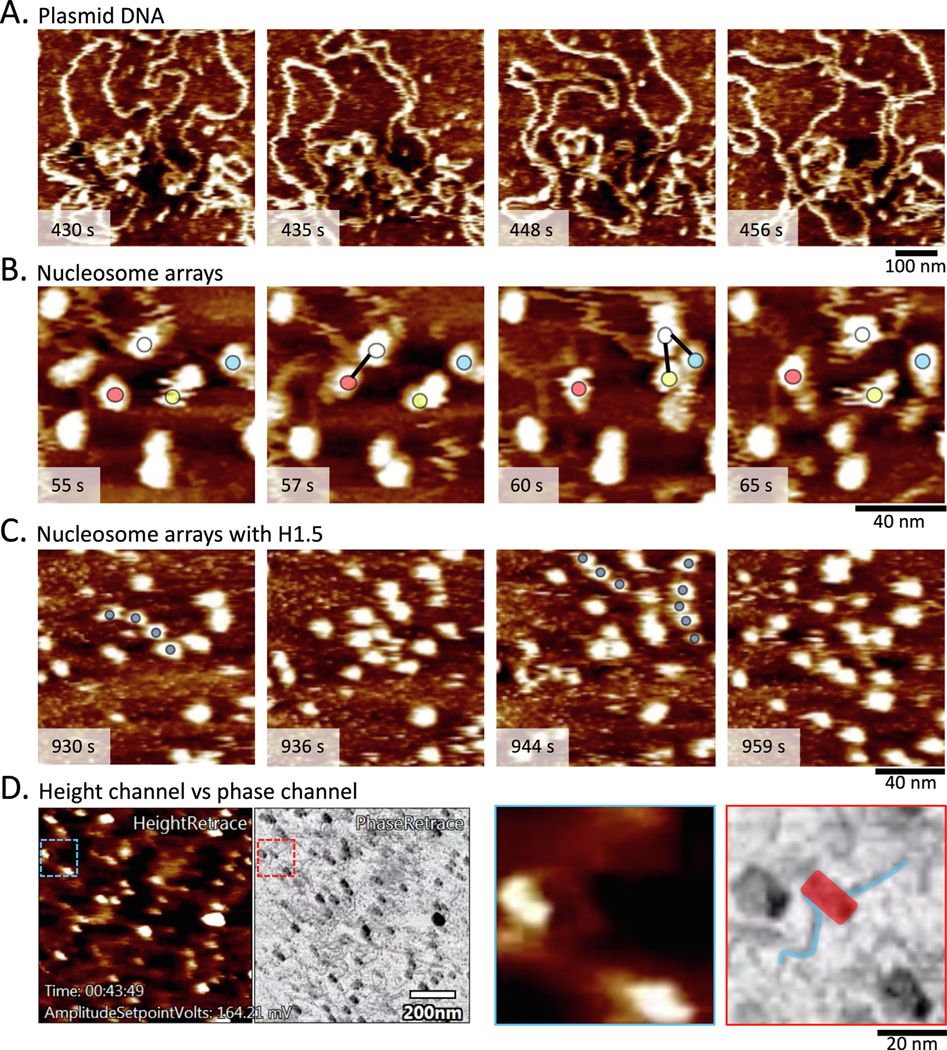
Examples of chromatin sample imaged by HS-AFM. (a) Plasmids in fluid show dynamic behavior (Supplemental Video 1). (b) Tracking an individual nucleosome (white, red, green, and yellow circles), intermittent contact between neighboring nucleosomes were observed (Supplemental Video 2). (c) H3 chromatin compaction induced by H1 shows highly mobile nucleosomes that form temporary nucleosome phasing (dark blue circles; Supplemental Video 3). (d) The height and phase channels provide complementary data. In particular, the phase channel allows for distinguishing lighter DNA from darker nucleosomes (see inset, where DNA (blue) is distinct from nucleosome (red); Supplemental Video 4).
Conclusions and Future Perspective
Recent advances spanning a wide swathe of structural, imaging, and computational methods have opened the door for a more complete understanding of local and global chromatin structure and dynamics. Advances made in cryo-EM has led to the identification of intermediary structures.45,107 Putting these intermediary structures in the correct spatiotemporal order allows for details mechanistic understanding how biomolecules convey their activity. Linking cryo-EM structures with HS-AFM might even provide precise spatiotemporal reconstruction. The development of software, such as DockAFM,108 will facilitate these advances. Another exciting avenue is the potential of HS-AFM in testing predictions made from computational modeling. Indeed, recent biophysical works from our lab, in collaboration with computational modeling from the Papoian lab, of nucleoprotein complexes showcases the interdisciplinary strengths and potential of such partnerships.58,87,109 Another impressive example comes from the Shaban group, which has developed PALM super-resolution microscopy coupled with computational modeling to be able to track chromatin “blobs” and study their dynamics.23 Other sophisticated advances such as Dual-probe AFM allow for the use of two independent AFM probes operating in the same workspace, where one probe can scan and capture data, whilst the other is actively engaged in manipulating the sample,110 in essence, yielding a molecular tweezer and a single molecule imager at the same time. For HS-AFM, development of a fast-wide area scanner would allow for observing morphological dynamics in live cells.111 Coupling HS-AFM to other techniques such as total internal reflection microscopes;112 optical tweezers;113 or immunofluorescence (bioAFM)113 will further expand the AFM tool box to study chromatin dynamics in native samples without need for purification.
In this special issue, experts using a broad range of methods highlighted in our review discuss their latest advances and applications, which provide exciting glimpses into the still somewhat secret life of the nucleus. The biggest challenge remaining, in our view, is a holistic understanding of how a particular epigenetic signature, residing at a specific location in the nucleus, with its inherent set of physical and mechanical properties, and its potential to attract or inhibit unique binding partners, results in a discrete function in vivo. Furthermore, how such functions are turned on or off in a timely manner in response to a changing environment, or at different points in the cell cycle, or over developmental time in different organs, whether such dynamics are conserved or not across different species, remain to be elucidated. Beyond these fundamental basic science questions, analyzing how all these facets of chromatin dynamics are altered in the diseased and aging states is a critical extension. Whether altered chromatin dynamics can be exploited as a therapeutic target is an exciting avenue for biomedical researchers. The next generation of bioengineering and biophysical tools that are currently being developed are precisely those that can bridge these last remaining hurdles. A prescient Dutch proverb (courtesy Cees Dekker of the Kavli Institute for Nanosciences, Delft, the Netherlands) neatly summarizes our views, and resonates with the current challenge facing science and the world: “Wie het kleine niet eert, is het grote niet weerd” (“Who doesn’t appreciate the small things, isn’t worthy of great things”).
Methods
In vitro nucleosome array reconstitution
H3/H4 (cat#16–0008, EpiCypher) and H2A/H2B (cat#15–0311, EpiCypher) were reconstituted on 3 kb plasmid containing four copies of α-satellite sequences as previously described.55,57,66 The linker histone H1.5 was added at a 1–10 molar ratio H1.5 to the reconstituted nucleosome arrays for either 5 minutes, 30 minutes, or overnight.
Conventional atomic force microscopy
Reconstituted chromatin was diluted (67.5 mM NaCl, 2 mM MgCl2) to medium density on APS treated freshly cleared mica.55 Diluted samples were rotated at RT for 30 minutes to gently equilibrate the chromatin in the solution. Next, 10 μL of samples was deposited on mica and incubated for 10 minutes before washing it with 2 × 200 μL ddH2O and subsequently very gently dried with argon gas. The deposited samples were imaged with the Cypher S (Asylum Research) using Olympus cantilevers (OTESPA-R3, Bruker). Subsequent images were analyzed using Gwyddion (http://gwyddion.net/) and R software (https://www.r-project.org/).
High-speed atomic force microscopy
Fresh plasmid DNA or reconstituted chromatin was diluted (67.5 mM NaCl, 2 mM MgCl2) to medium density on 2x APS treated freshly cleaved mica. Mica discs (01900-MB, SPI Supplies) were glued with epoxy on 3 mm sapphire post (cat# 569–029, Asylum Research), which in turn was glued with epoxy on the sapphire grid on the Cypher VRS per manufacturer’s instructions. 10 μL of sample was deposited on top the sapphire post and subsequently allowed to settle on the mica surface for 30 minutes before imaging using the AC10DS probe (Oxford Instruments). Naked plasmids were the same as described above. H3 chromatin was reconstituted as described above. H1.5 was added at 1–10 molar ratio to reconstituted H3 chromatin for 30 minutes before depositing them on the sapphire post. The movies were processed using Asylum’s software.
Supplementary Material
Acknowledgements
We thank Drs. Will Heinz (NCI/NIH AFM core facility), Emilios Dimitriadis (NIBIB/NIH AFM shared facility), and Tatini Rakshit (Bose Institute) for their advice regarding HS-AFM; Drs. Ankita Saha and Craig Mizzen (deceased, University of Illinois, Urbana-Champaign) for the gift of recombinant H1.5 protein; and, CSEM lab members for critical reading of this manuscript. This work was supported by the Intramural Research Program of the Center of Cancer Research of the National Cancer Institute, National Institutes of Health.
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
CRediT authorship contribution statement
Daniël P. Melters: Conceptualization, Methodology, Investigation, Visualization, Writing - original draft, Writing - review & editing. Yamini Dalal: Supervision, Conceptualization, Writing - original draft, Writing - review & editing.
Appendix A. Supplementary material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jmb.2020.11.019.
References
- 1.Kornberg RD, (1974). Chromatin structure: A repeating unit of histones and DNA. Science (80-.), 184, 868–871. 10.1126/science.184.4139.868. [DOI] [PubMed] [Google Scholar]
- 2.Olins AL, Olins DE, (1974). Spheroid chromatin units (m bodies). Science (80-.), 183, 330–332. 10.1126/science.183.4122.330. [DOI] [PubMed] [Google Scholar]
- 3.Woodcock CLF, Safer JP, Stanchfield JE, (1976). Structural repeating units in chromatin: I. Evidence for their general occurrence. Exp. Cell Res,97, 101–110. [DOI] [PubMed] [Google Scholar]
- 4.Melters DP, Nye J, Zhao H, Dalal Y, (2015). Chromatin dynamics in vivo: A game of musical chairs. Genes (Basel), 6, 751–776. 10.3390/genes6031940751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Malik HS, Henikoff S, (2003). Phylogenomics of the nucleosome. Nat. Struct. Biol,10, 882–891. [DOI] [PubMed] [Google Scholar]
- 6.Klemm SL, Shipony Z, Greenleaf WJ, (2019). Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet, 20, 207–220. 10.1038/s41576-018-0089-8. [DOI] [PubMed] [Google Scholar]
- 7.Misteli T, (2020). The self-organizing genome: Principles of genome architecture and function. Cell,. 10.1016/j.cell.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pujals S, Feiner-Gracia N, Delcanale P, Voets I, Albertazzi L, (2019). Super-resolution microscopy as a powerful tool to study complex synthetic materials. Nat. Rev. Chem, 3, 68–84. 10.1038/s41570-018-0070-2. [DOI] [Google Scholar]
- 9.Bonev B, Cavalli G, (2016). Organization and function of the 3D genome. Nat. Rev. Genet, 17, 661–678. 10.1038/nrg.2016.112. [DOI] [PubMed] [Google Scholar]
- 10.Schoenfelder S, Fraser P, (2019). Long-range enhancer–promoter contacts in gene expression control. Nat. Rev. Genet, 20, 437–455. 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 11.Cuvier O, Fierz B, (2017). Dynamic chromatin technologies: from individual molecules to epigenomic regulation in cells. Nat. Rev. Genet, 18, 457–472. 10.1038/nrg.2017.28. [DOI] [PubMed] [Google Scholar]
- 12.Buckwalter JM, Norouzi D, Harutyunyan A, Zhurkin VB, Grigoryev SA, (2017). Regulation of chromatin folding by conformational variations of nucleosome linker DNA. Nucleic Acids Res, 45, 9372–9387. 10.1093/nar/gkx562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Voong LN, Xi L, Wang J-P, Wang X, (2017). Genome-wide mapping of the nucleosome landscape by micrococcal nuclease and chemical mapping. Trends Genet, 33, 495–507. 10.1016/j.tig.2017.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Agbleke AA, Amitai A, Buenrostro JD, Chakrabarti A, Chu L, Hansen AS, Koenig KM, Labade AS, Liu S, Nozaki T, Ovchinnikov S, Seeber A, Shaban HA, Spille J-H, Stephens AD, Su J-H, Wadduwage D, (2020). Advances in chromatin and chromosome research: perspectives from multiple fields. Mol. Cell, 79, 881–901. 10.1016/j.molcel.2020.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shaban HA, Seeber A, (2020). Monitoring the spatiotemporal organization and dynamics of the genome. Nucleic Acids Res, 48, 3423–3434. 10.1093/nar/gkaa135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nozaki T, Imai R, Tanbo M, Nagashima R, Tamura S, Tani T, Joti Y, Tomita M, Hibino K, Kanemaki MT, Wendt KS, Okada Y, Nagai T, Maeshima K, (2017). Dynamic organization of chromatin domains revealed by super-resolution live-cell imaging. Mol. Cell,. 10.1016/j.molcel.2017.06.018. [DOI] [PubMed] [Google Scholar]
- 17.Nagashima R, Hibino K, Ashwin SS, Babokhov M, Fujishiro S, Imai R, Nozaki T, Tamura S, Tani T, Kimura H, Shribak M, Kanemaki MT, Sasai M, Maeshima K, (2019). Single nucleosome imaging reveals loose genome chromatin networks via active RNA polymerase II. J. Cell Biol,. 10.1083/jcb.201811090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bodor DL, Rodríguez MG, Moreno N, Jansen LET, Analysis of protein turnover by quantitative SNAP-based pulse-chase imaging, in: Curr. Protoc. Cell Biol, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2012, p. Unit8.8. 10.1002/0471143030.cb0808s55.. [DOI] [PubMed] [Google Scholar]
- 19.Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE, (2014). The quantitative architecture of centromeric chromatin. Elife,. 10.7554/elife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Milagre I, Pereira C, Oliveira RA, Jansen LET, (2020). Reprogramming of human cells to pluripotency induces CENP-A chromatin depletion. Open Biol, 10, 200227. 10.1098/rsob.200227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Buenrostro JD, Wu B, Chang HY, Greenleaf WJ, (2015). ATAC-seq: A method for assaying chromatin accessibility genome-wide 21.29.1–21.29.9 Curr. Protoc. Mol. Biol, 109 10.1002/0471142727.mb2129s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaya-Okur HS, Wu SJ, Codomo CA, Pledger ES, Bryson TD, Henikoff JG, Ahmad K, Henikoff S, (2019). CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun, 10, 1930. 10.1038/s41467-019-09982-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barth R, Bystricky K, Shaban HA, (2020). Coupling chromatin structure and dynamics by live super-resolution imaging. Sci. Adv, 6, eaaz2196. 10.1126/sciadv.aaz2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu J, Ma H, Jin J, Uttam S, Fu R, Huang Y, Liu Y, (2018). Super-resolution imaging of higher-order chromatin structures at different epigenomic states in single mammalian cells. Cell Rep, 24, 873–882. 10.1016/j.celrep.2018.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bendandi A, Dante S, Zia SR, Diaspro A, Rocchia W, (2020). Chromatin compaction multiscale modeling: a complex synergy between theory, simulation, and experiment. Front. Mol. Biosci, 7, 15. 10.3389/fmolb.2020.00015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pitman M, Melters DP, Dalal Y, (2020). Job opening for nucleosome mechanic: flexibility required. Cells, 9, 580. 10.3390/cells9030580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhuravlev PI, Papoian GA, (2010). Protein functional landscapes, dynamics, allostery: a tortuous path towards a universal theoretical framework. Q. Rev. Biophys, 43, 295–332. 10.1017/S0033583510000119. [DOI] [PubMed] [Google Scholar]
- 28.Moller J, de Pablo JJ, (2020). Bottom-up meets top-down: the crossroads of multiscale chromatin modeling. Biophys. J, 118, 2057–2065. 10.1016/j.bpj.2020.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bascom GD, Myers CG, Schlick T, (2019). Mesoscale modeling reveals formation of an epigenetically driven HOXC gene hub. Proc. Natl. Acad. Sci, 116, 4955–4962. 10.1073/pnas.1816424116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Portillo-Ledesma S, Schlick T, (2020). Bridging chromatin structure and function over a range of experimental spatial and temporal scales by molecular modeling. WIREs Comput. Mol. Sci, 10, e1434. 10.1002/wcms.1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fierz B, Poirier MG, (2019). Biophysics of chromatin dynamics. Annu. Rev. Biophys, 48, 321–345. 10.1146/annurev-biophys-070317-032847. [DOI] [PubMed] [Google Scholar]
- 32.Cherstv VB, Teif Andrey G, Chromatin and epigenetics: current biophysical views, AIMS Biophys. 3 (n.d.) 88–98. 10.3934/biophy.2016.1.88.. [DOI] [Google Scholar]
- 33.Robison AD, Finkelstein IJ, (2014). High-throughput single-molecule studies of protein–DNA interactions. FEBS Lett, 588, 3539–3546. 10.1016/j.febslet.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashkin A, (1997). Optical trapping and manipulation of neutral particles using lasers. Proc. Natl. Acad. Sci, 94, 4853–4860. 10.1073/pnas.94.10.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Z, Gabizon R, Brown AI, Lee A, Song A, Celis CD, Kaplan CD, Koslover EF, Yao T, Bustamante C, (2019). High-resolution and high-accuracy topographic and transcriptional maps of the nucleosome barrier. Elife,. 10.7554/eLife.48281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilic S, Felekyan S, Doroshenko O, Boichenko I, Dimura M, Vardanyan H, Bryan LC, Arya G, Seidel CAM, Fierz B, (2018). Single-molecule FRET reveals multiscale chromatin dynamics modulated by HP1a. Nat. Commun,. 10.1038/s41467-017-02619-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kong M, Cutts EE, Pan D, Beuron F, Kaliyappan T, Xue C, Morris EP, Musacchio A, Vannini A, Greene EC, (2020). Human condensin I and II drive extensive ATP-dependent compaction of nucleosome-bound DNA. Mol. Cell, 79, 99–114.e9. 10.1016/j.molcel.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ganji M, Shaltiel IA, Bisht S, Kim E, Kalichava A, Haering CH, Dekker C, (2018). Real-time imaging of DNA loop extrusion by condensin. Science (80-.), 360, 102–105. 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kim E, Kerssemakers J, Shaltiel IA, Haering CH, Dekker C, (2020). DNA-loop extruding condensin complexes can traverse one another. Nature, 579, 438–442. 10.1038/s41586-020-2067-5. [DOI] [PubMed] [Google Scholar]
- 40.Davidson IF, Bauer B, Goetz D, Tang W, Wutz G, Peters J-M, (2019). DNA loop extrusion by human cohesin. Science (80-.), 366, 1338–1345. 10.1126/science.aaz3418. [DOI] [PubMed] [Google Scholar]
- 41.Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H, (2019). Human cohesin compacts DNA by loop extrusion. Science (80-.), 366, 1345–1349. 10.1126/science.aaz4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Günther K, Mertig M, Seidel R, (2010). Mechanical and structural properties of YOYO-1 complexed DNA. Nucleic Acids Res, 38, 6526–6532. 10.1093/nar/gkq434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biebricher AS, Heller I, Roijmans RFH, Hoekstra TP, Peterman EJG, Wuite GJL, (2015). The impact of DNA intercalators on DNA and DNA-processing enzymes elucidated through force-dependent binding kinetics. Nat. Commun, 6, 7304. 10.1038/ncomms8304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yan X, Habbersett RC, Yoshida TM, Nolan JP, Jett JH, Marrone BL, (2005). Probing the kinetics of SYTOX orange stain binding to double-stranded DNA with implications for DNA analysis. Anal. Chem, 77, 3554–3562. 10.1021/ac050306u. [DOI] [PubMed] [Google Scholar]
- 45.Zhou K, Gaullier G, Luger K, (2019). Nucleosome structure and dynamics are coming of age. Nat. Struct. Mol. Biol,. 10.1038/s41594-018-0166-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou B-R, Bai Y, (2019). Chromatin structures condensed by linker histones. Essays Biochem, 63, 75–87. 10.1042/EBC20180056. [DOI] [PubMed] [Google Scholar]
- 47.Kalashnikova AA, Porter-Goff ME, Muthurajan UM, Luger K, Hansen JC, (2013). The role of the nucleosome acidic patch in modulating higher order chromatin structure. J. R. Soc. Interface,. 10.1098/rsif.2012.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y, Zhou K, Zhang N, Wei H, Tan YZ, Zhang Z, Carragher B, Potter CS, D’Arcy S, Luger K, (2020). FACT caught in the act of manipulating the nucleosome. Nature, 577, 426–431. 10.1038/s41586-019-1820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miron E, Oldenkamp R, Brown JM, Pinto DMS, Xu CS, Faria AR, Shaban HA, Rhodes JDP, Innocent C, de Ornellas S, Hess HF, Buckle V, Schermelleh L, (2020). Chromatin arranges in chains of mesoscale domains with nanoscale functional topography independent of cohesin. Sci. Adv, 6, eaba8811. 10.1126/sciadv.aba8811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ, (2019). HP1 reshapes nucleosome core to promote heterochromatin phase separation. Nature,. 10.1038/s41586-019-1669-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tropberger P, Pott S, Keller C, Kamieniarz-Gdula K, Caron M, Richter F, Li G, Mittler G, Liu ET, Bühler M, Margueron R, Schneider R, (2013). Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell,. 10.1016/j.cell.2013.01.032. [DOI] [PubMed] [Google Scholar]
- 52.Chatterjee N, North JA, Dechassa ML, Manohar M, Prasad R, Luger K, Ottesen JJ, Poirier MG, Bartholomew B, (2015). Histone acetylation near the nucleosome dyad axis enhances nucleosome disassembly by RSC and SWI/SNF. Mol. Cell. Biol,. 10.1128/mcb.00441-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lohr D, Bash R, Wang H, Yodh J, Lindsay S, (2007). Using atomic force microscopy to study chromatin structure and nucleosome remodeling. Methods, 41, 333–341. 10.1016/j.ymeth.2006.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang H, Dalal Y, Henikoff S, Lindsay S, (2008). Single-epitope recognition imaging of native chromatin. Access, 9, 1–9. 10.1186/1756-8935-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Walkiewicz MP, Bui M, Quénet D, Dalal Y, (2014). Tracking histone variant nucleosomes across the human cell cycle using biophysical, biochemical, and cytological analyses. Methods Mol. Biol, 1170, 589–615. 10.1007/978-1-4939-0888-2_34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lyubchenko YL, (2004). DNA structure and dynamics. Cell Biochem. Biophys, 41, 75–98. 10.1385/CBB:41:1:075. [DOI] [PubMed] [Google Scholar]
- 57.Rakshit T, Melters DP, Dimitriadis EK, Dalal Y, (2020). Mechanical properties of nucleoprotein complexes determined by nanoindentation spectroscopy. Nucleus, 11, 264–282. 10.1080/19491034.2020.1816053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Melters DP, Pitman M, Rakshit T, Dimitriadis EK, Bui M, Papoian GA, Dalal Y, (2019). Intrinsic elasticity of nucleosomes is encoded by histone variants and calibrated by their binding partners. Proc. Natl. Acad. Sci. U. S. A, 116, 24066–24074. 10.1073/pnas.1911880116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shlyakhtenko LS, Lushnikov AY, Lyubchenko YL, (2009). Dynamics of nucleosomes revealed by time-lapse atomic force microscopy. Biochemistry, 48, 7842–7848. 10.1021/bi900977t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Filenko NA, Kolar C, West JT, Smith SA, Hassan YI, Borgstahl GEO, Zempleni J, Lyubchenko YL, (2011). The role of histone H4 biotinylation in the structure of nucleosomes. PLoS One, 6, e16299. 10.1371/journal.pone.0016299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miyagi A, Ando T, Lyubchenko YL, (2011). Dynamicsof nucleosomes assessed with time-lapse high-speed atomic force microscopy. Biochemistry, 50, 7901–7908. 10.1021/bi200946z. [DOI] [PubMed] [Google Scholar]
- 62.Yodh JG, Woodbury N, Shlyakhtenko LS, Lyubchenko YL, Lohr D, (2002). Mapping nucleosome locations on the 208–12 by AFM provides clear evidence for cooperativity in array occupation. Biochemistry, 41, 3565–3574. 10.1021/bi011612e. [DOI] [PubMed] [Google Scholar]
- 63.Pisano S, Marchioni E, Galati A, Mechelli R, Savino M, Cacchione S, (2007). Telomeric nucleosomes are intrinsically mobile. J. Mol. Biol, 369, 1153–1162. 10.1016/j.jmb.2007.04.027. [DOI] [PubMed] [Google Scholar]
- 64.Dalal Y, Wang H, Lindsay S, Henikoff S, (2007). Tetrameric structure of centromeric nucleosomes in interphase Drosophila cells. PLoS Biol, 5, e218. 10.1371/journal.pbio.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dimitriadis EK, Weber C, Gill RK, Diekmann S, Dalal Y, (2010). Tetrameric organization of vertebrate centromeric nucleosomes. Proc. Natl. Acad. Sci. U. S. A, 107, 20317–20322. 10.1073/pnas.1009563107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walkiewicz MP, Dimitriadis EK, Dalal Y, (2014). CENP-A octamers do not confer a reduction in nucleosome height by AFM. Nat. Struct. Mol. Biol, 21, 2–3. 10.1038/nsmb.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Widom J, (1998). Structure, dynamics, and function of chromatin in vitro. Annu. Rev. Biophys. Biomol. Struct, 27, 285–327. 10.1146/annurev.biophys.27.1.285. [DOI] [PubMed] [Google Scholar]
- 68.Maehara K, Ohkawa Y, (2016). Exploration of nucleosome positioning patterns in transcription factor function. Sci. Rep, 6, 19620. 10.1038/srep19620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Flores O, Deniz Ö, Soler-López M, Orozco M, (2014). Fuzziness and noise in nucleosomal architecture. Nucleic Acids Res, 42, 4934–4946. 10.1093/nar/gku165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ashcroft BA, Spadola Q, Qamar S, Zhang P, Kada G, Bension R, Lindsay S, (2008). An AFM/rotaxane molecular reading head for sequence-dependent DNA structures. Small, 4, 1468–1475. 10.1002/smll.200800233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Izzo A, Kamieniarz K, Schneider R, The histone H1 family: specific members, specific functions?, Biol. Chem. 389 (n.d.) 333–343. 10.1515/BC.2008.037. [DOI] [PubMed] [Google Scholar]
- 72.Thoma F, Koller T, (1977). Influence of histone H1 onchromatin structure. Cell, 12, 101–107. 10.1016/0092-8674(77)90188-X. [DOI] [PubMed] [Google Scholar]
- 73.Thoma F, Koller T, Klug A, (1979). Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol, 83, 403–427. 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT, (2000). Dynamic binding of histone H1 to chromatin in living cells. Nature, 408, 877–881. 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 75.Bernas T, Brutkowski W, Zarezbski M, Dobrucki J, (2014). Spatial heterogeneity of dynamics of H1 linker histone. Eur. Biophys. J, 43, 287–300. 10.1007/s00249-014-0962-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ponte I, Romero D, Yero D, Suau P, Roque A, (2017). Complex evolutionary history of the mammalian histone H1.1–H1.5 gene family. Mol. Biol. Evol, 34, 545–558. 10.1093/molbev/msw241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kasinsky HE, Lewis JD, Dacks JB, Ausló J, (2001). Origin of H1 linker histones. FASEB J, 15, 34–42. 10.1096/fj.00-0237rev. [DOI] [PubMed] [Google Scholar]
- 78.Hergeth SP, Schneider R, (2015). The H1 linker histones: multifunctional proteins beyond the nucleosomal core particle. EMBO Rep, 16, 1439–1453. 10.15252/embr.201540749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Clausell J, Happel N, Hale TK, Doenecke D, Beato M, (2009). Histone H1 subtypes differentially modulate chromatin condensation without preventing ATP-dependent remodeling by SWI/SNF or NURF. PLoS One,. 10.1371/journal.pone.0007243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu H, Wen J, Xiao Y, Liu J, Hopyan S, Radisic M, Simmons CA, Sun Y, (2014). In situ mechanical characterization of the cell nucleus by atomic force microscopy. ACS Nano, 8, 3821–3828. 10.1021/nn500553z. [DOI] [PubMed] [Google Scholar]
- 81.Maurer M, Lammerding J, (2019). The driving force: nuclear mechanotransduction in cellular function, fate, and disease. Annu. Rev. Biomed. Eng,. 10.1146/annurev-bioeng-060418-052139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rowat AC, Foster LJ, Nielsen MM, Weiss M, Ipsen JH, (2005). Characterization of the elastic properties of the nuclear envelope. J. R. Soc. Interface, 2, 63–69. 10.1098/rsif.2004.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rowat AC, Lammerding J, Ipsen JH, (2006). Mechanical properties of the cell nucleus and the effect of emerin deficiency. Biophys. J, 91, 4649–4664. 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Klare K, Weir JR, Basilico F, Zimniak T, Massimiliano L, Ludwigs N, Herzog F, Musacchio A, (2015). CENP-C is a blueprint for constitutive centromere-associated network assembly within human kinetochores. J. Cell Biol, 210, 11–22. 10.1083/jcb.201412028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nicklas RB, (1983). Measurements of the force produced by the mitotic spindle in anaphase. J. Cell Biol,. 10.1083/jcb.97.2.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Marshall WF, Marko JF, Agard DA, Sedat JW, (2001). Chromosome elasticity and mitotic polar ejection force measured in living Drosophila embryos by four-dimensional microscopy-based motion analysis. Curr. Biol,. 10.1016/S0960-9822(01)00180-4. [DOI] [PubMed] [Google Scholar]
- 87.Bui M, Pitman M, Nuccio A, Roque S, Donlin-Asp PG, Nita-Lazar A, Papoian GA, Dalal Y, (2017). Internal modifications in the CENP-A nucleosome modulate centromeric dynamics. Epigenet. Chromatin, 10, 17. 10.1186/s13072-017-0124-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Athwal RK, Walkiewicz MP, Baek S, Fu S, Bui M, Camps J, Ried T, Sung M-H, Dalal Y, (2015). CENP-A nucleosomes localize to transcription factor hotspots and subtelomeric sites in human cancer cells. Epigenet. Chromatin, 8, 2. 10.1186/1756-8935-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yuen BTK, Knoepfler PS, (2013). Histone H3.3 mutations: A variant path to cancer. Cancer Cell, 24, 567–574. 10.1016/j.ccr.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chen K-Y, Bush K, Klein RH, Cervantes V, Lewis N, Naqvi A, Carcaboso AM, Lechpammer M, Knoepfler PS, (2020). Reciprocal H3.3 gene editing identifies K27M and G34R mechanisms in pediatric glioma including NOTCH signaling. Commun. Biol, 3, 363. 10.1038/s42003-020-1076-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ando T, Uchihashi T, Fukuma T, (2008). High-speed atomic force microscopy for nano-visualization of dynamic biomolecular processes. Prog. Surf. Sci, 83, 337–437. 10.1016/j.progsurf.2008.09.001. [DOI] [Google Scholar]
- 92.Kodera N, Yamamoto D, Ishikawa R, Ando T, (2010). Video imaging of walking myosin V by high-speed atomic force microscopy. Nature, 468, 72–76. 10.1038/nature09450. [DOI] [PubMed] [Google Scholar]
- 93.Strahlendorff T, Dai G, Bergmann D, Tutsch R, (2019). Tip wear and tip breakage in high-speed atomic force microscopes. Ultramicroscopy, 201, 28–37. 10.1016/j.ultramic.2019.03.013. [DOI] [PubMed] [Google Scholar]
- 94.Shibata M, Nishimasu H, Kodera N, Hirano S, Ando T, Uchihashi T, Nureki O, (2017). Real-space and realtime dynamics of CRISPR-Cas9 visualized by high-speed atomic force microscopy. Nat. Commun, 8, 1430. 10.1038/s41467-017-01466-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ryu J-K, Katan AJ, van der Sluis EO, Wisse T, de Groot R, Haering CH, Dekker C, (2020). The condensin holocomplex cycles dynamically between open and collapsed states. Nat. Struct. Mol. Biol,. 10.1038/s41594-020-0508-3. [DOI] [PubMed] [Google Scholar]
- 96.Lyubchenko YL, (2014). Nanoscale nucleosome dynamics assessed with time-lapse AFM. Biophys. Rev, 6, 181–190. 10.1007/s12551-013-0121-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Katan AJ, Vlijm R, Lusser A, Dekker C, (2015). Dynamics of nucleosomal structures measured by high-speed atomic force microscopy. Small, 11, 976–984. 10.1002/smll.201401318. [DOI] [PubMed] [Google Scholar]
- 98.Stumme-Diers MP, Banerjee S, Hashemi M, Sun Z, Lyubchenko YL, (2018). Nanoscale dynamics of centromere nucleosomes and the critical roles of CENP-A. Nucleic Acids Res, 46, 94–103. 10.1093/nar/gkx933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Suzuki Y, Higuchi Y, Hizume K, Yokokawa M, Yoshimura SH, Yoshikawa K, Takeyasu K, (2010). Molecular dynamics of DNA and nucleosomes in solution studied by fast-scanning atomic force microscopy. Ultramicroscopy, 110, 682–688. 10.1016/j.ultramic.2010.02.032. [DOI] [PubMed] [Google Scholar]
- 100.Eeftens JM, Katan AJ, Kschonsak M, Hassler M, de Wilde L, Dief EM, Haering CH, Dekker C, (2016). Condensin Smc2-Smc4 dimers are flexible and dynamic. Cell Rep, 14, 1813–1818. 10.1016/j.celrep.2016.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Endo M, Feng Y, Hashiya F, Hidaka K, Sugiyama H, (2020). Direct observation of dynamic interactions between orientation-controlled nucleosomes in a DNA origami frame. Chem. – Eur. J,. 10.1002/chem.202003071. [DOI] [PubMed] [Google Scholar]
- 102.Lyubchenko YL, Shlyakhtenko LS, (1997). Visualization of supercoiled DNA with atomic force microscopy in situ. Proc. Natl. Acad. Sci, 94, 496–501. 10.1073/pnas.94.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rodriguez J, Larson DR, (2020). Transcription in living cells: molecular mechanisms of bursting. Annu. Rev. Biochem, 89, 189–212. 10.1146/annurevbiochem-011520-105250. [DOI] [PubMed] [Google Scholar]
- 104.Hu J, Gu L, Ye Y, Zheng M, Xu Z, Lin J, Du Y, Tian M, Luo L, Wang B, Zhang X, Weng Z, Jiang C, (2018). Dynamic placement of the linker histone H1 associated with nucleosome arrangement and gene transcription in early Drosophila embryonic development. Cell Death Dis, 9, 765. 10.1038/s41419-018-0819-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fan Y, Nikitina T, Morin-Kensicki EM, Zhao J, Magnuson TR, Woodcock CL, Skoultchi AI, (2003). H1 linker histones are essential for mouse development and affect nucleosome spacing in vivo. Mol. Cell. Biol, 23, 4559–4572. 10.1128/MCB.23.13.4559-4572.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Stein A, Mitchell M, (1988). Generation of different nucleosome spacing periodicities in vitro: Possible origin of cell type specificity. J. Mol. Biol, 203, 1029–1043. 10.1016/0022-2836(88)90127-1. [DOI] [PubMed] [Google Scholar]
- 107.Danev R, Yanagisawa H, Kikkawa M, (2019). Cryo-electron microscopy methodology: current aspects and future directions. Trends Biochem. Sci, 44, 837–848. 10.1016/j.tibs.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 108.Chaves RC, Pellequer J-L, (2013). DockAFM: benchmarking protein structures by docking under AFM topographs. Bioinformatics, 29, 3230–3231. 10.1093/bioinformatics/btt561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zhao H, Winogradoff D, Bui M, Dalal Y, Papoian GA, (2016). Promiscuous histone mis-assembly is actively prevented by chaperones. J. Am. Chem. Soc, 138, 13207–13218. 10.1021/jacs.6b05355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Loganathan M, Al-Ogaidi A, Bristow DA, (2018). Design and control of a dual-probe atomic force microscope. IEEE/ASME Trans. Mechatron, 23, 424–433. 10.1109/TMECH.2017.2779241. [DOI] [Google Scholar]
- 111.Shibata M, Uchihashi T, Ando T, Yasuda R, (2015). Long-tip high-speed atomic force microscopy for nanometer-scale imaging in live cells. Sci. Rep, 5, 8724. 10.1038/srep08724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fukuda S, Uchihashi T, Iino R, Okazaki Y, Yoshida M, Igarashi K, Ando T, (2013). High-speed atomic force microscope combined with single-molecule fluorescence microscope. Rev. Sci. Instrum, 84, 73706. 10.1063/1.4813280. [DOI] [PubMed] [Google Scholar]
- 113.Ando T, (2019). High-speed atomic force microscopy. Curr. Opin. Chem. Biol, 51, 105–112. 10.1016/j.cbpa.2019.05.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.