Abstract
The hidden world of amyloid biology has suddenly snapped into atomic level focus revealing over 80 amyloid protein fibrils, both pathogenic and functional. Unlike globular proteins, amyloid proteins flatten and stack into unbranched fibrils. Stranger still, a single protein sequence can adopt wildly different two-dimensional conformations, yielding distinct fibril polymorphs. Hence, an amyloid protein may define distinct diseases depending on its conformation. At the heart of this conformational variability lies structural frustrations. In functional amyloids, evolution tunes frustration levels to achieve either stability or sensitivity according to the fibril’s biological function, accounting for the vast versatility of the amyloid fibril scaffold.
INTRODUCTION
Amyloid biology is the study of a wide range of pathologies and biological functions performed by proteins in the amyloid fibril state. Amyloid proteins are produced by all three kingdoms of life, and encoded by diverse protein sequences, yet amyloid family members are all unified by a family resemblance arising from a shared amyloid fibril scaffold. In fact, the first observation made 62 years ago(Cohen and Calkins, 1959), revealed the basic features of the amyloid fibril scaffold that define this class of protein assembly to this day. That is, elongated, unbranched morphology, and widths ranging from 50 to 200 nm (Benson et al., 2018),(Sipe and Cohen, 2000).
Members of the amyloid family may be divided into three main branches. Pathological amyloids were the first branch discovered. Its members constitute the pathological hallmarks of amyloid diseases(Chiti and Dobson, 2017) which include the most common degenerative diseases, Alzheimer’s disease (AD), Parkinson’s disease (PD), and type II diabetes. Each disease is associated with fibrils of particular proteins. For example, fibrils of the protein α-synuclein deposit in the brains of PD patients(Baba et al., 1998); fibrils of amyloid β (Aβ) and tau protein deposit in the brains of AD patients(Holtzman et al., 2011)(Jucker and Walker, 2011); and fibrils of the hormone, islet amyloid polypeptide, (IAPP) deposit in pancreatic β-islet cells of type II diabetics (Johnson et al., 1992). The ~35 proteins involved in amyloid diseases share no similarity in sequence or native structure, yet their fibrils share the amyloid family scaffold.
Artificial amyloids were the second branch discovered. Researchers in the 1990s discovered that denaturing conditions can induce familiar globular proteins to form amyloid-like fibrils [Sidebar 1: Amyloid vs. Amyloid-like] (Gustavsson et al., 1991),(Colon and Kelly, 1992),(Guijarro et al., 1998),(Chiti et al., 1999),(Fändrich et al., 2001). We know now that almost any protein can form amyloid fibrils (Goldschmidt et al., 2010) if normally buried amyloid-driving segments become exposed by denaturation(Teng et al., 2012). Indeed, the sequence requirements for amyloid formation were found to be so easily fulfilled that amyloid fibrils could possibly have served as prebiotic enzymes, long before gene-encoded translation(Greenwald et al., 2018),(Rufo et al., 2014).
[Sidebar 1]. Amyloid vs. amyloid-like.
The Nomenclature Committee of the International Society of Amyloidosis recommends that the term “amyloid” (without further explanation) as in “amyloid state” or “functional amyloid” be restricted to pathological deposits. The term “amyloid -like” is recommended for recombinant fibrils of disease-related proteins and of functional “amyloid” proteins(Benson et al., 2018).
Functional amyloids were the latest branch discovered. Nature evolved them to perform a wide range of biological functions in diverse organisms including bacteria, fungi, protozoa, plants, insects, sea slugs, mice and humans(Fowler et al., 2007). For example, they form bacterial biofilms(Chapman et al., 2002), provide scaffolding for melanin synthesis(Fowler et al., 2006), store peptide hormones(Maji et al., 2009), form memories(Li et al., 2016), and facilitate interactions between proteins in subcellular condensates(Kato et al., 2012),(Xiang et al., 2015),(Vogler et al., 2018),(Frey et al., 2006),(Ader et al., 2010).
This diversity of functions seems incompatible with the constraints of a uniform amyloid fibril scaffold. However, amyloid proteins actually vary greatly in conformation. In fact, most proteins are capable of producing several distinct amyloid structures called polymorphs, demonstrating a level of versatility unparalleled in the globular state. Structural variations encode variations in fibril stability. Pathological amyloids are notoriously stable, characterized by resistance to proteolysis, chemical denaturation, and detergent(Balbirnie et al., 2001), whereas functional amyloids range from essentially permanent structures, such as bacterial biofilms to transient barriers such as the pores of nuclear transport receptors(Ader et al., 2010).
Here we review the recent explosion of high-resolution structural information on amyloids to answer three fundamental questions: (1) What features of the amyloid scaffold make it compatible with an enormous number of protein sequences? (2) Why are many pathogenic amyloids subject to polymorphic variations, and how might these variations define distinct diseases? (3) How has evolution tuned functional amyloid fibril structure to disassemble in response to environmental changes? We address these questions in order, after an introduction to amyloid structure.
I. AMYLOID STRUCTURE & ORIGIN OF EXTRAORDINARY STABILITY
Amyloid fibrils assemble on cross-β scaffolds, reinforced by steric zipper motifs
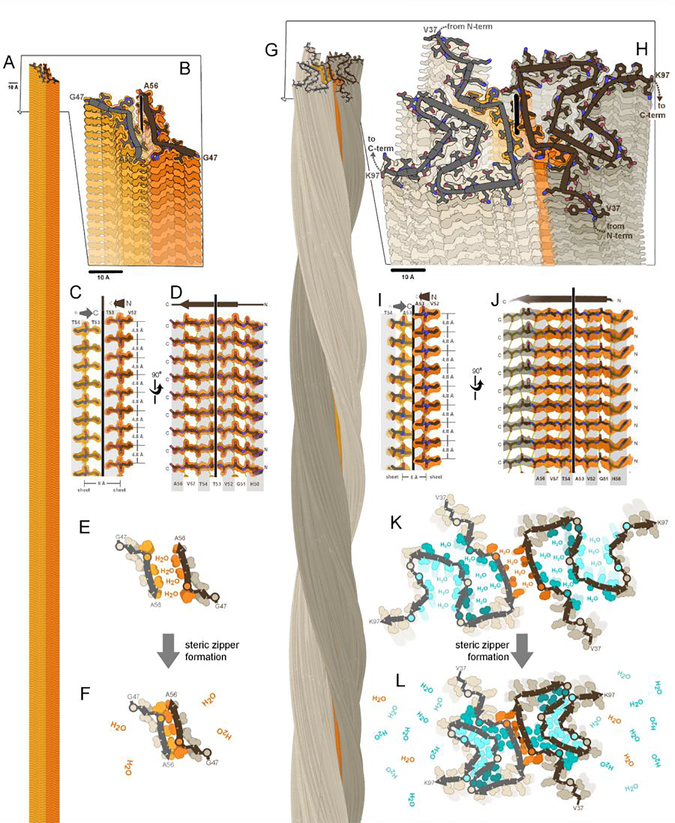
Amyloid fibrils, both functional and pathogenic, display the characteristic “cross-β fiber diffraction pattern”, featuring two orthogonal reflections (Astbury et al., 1935),(Sunde et al., 1997) that signify β-sheets mated together side-by-side, with their β-strands running perpendicular to the fibril axis. The so-called “cross-β motif” was confirmed in atomic resolution detail by crystal structures of fibrils formed by short adhesive segments of amyloid-forming proteins(Nelson et al., 2005; Rodriguez et al., 2015; Sawaya et al., 2007),(Eisenberg and Sawaya, 2017). A prime example is the crystal structure of α-synuclein segment, 47–56 (Figure 1A–F). In the crystal, β-sheets extend microns long by stacking of thousands of β-strands through backbone hydrogen bonding of amide N-H∙∙O groups. Figure 1A captures a short segment of the fibril length. In accord with the cross-β diffraction signature, β-strands are spaced 4.8 Å apart, and the β-sheets are in the range of 8–12 Å apart (Figure 1C–D). Given the variety of protein sequences that form amyloid, it is striking that driven by the power of backbone hydrogen bonding, all sequences conform to this basic cross-β sheet motif, producing the characteristic elongated and unbranched fibril morphology(Cohen and Calkins, 1959),(Sipe and Cohen, 2000).
Figure 1. A fundamental component of amyloid fibrils is the steric zipper motif.
Amyloid fibrils of α-synuclein are associated with Parkinson’s disease and other synucleinopathies. The crystal structure of residues 47–56 offered atomic resolution details of its assembly at 1.4 Å resolution (PDB ID 4ZNN)(Rodriguez et al., 2015). (A) Thousands of copies of this segment stack to form a pair of β-sheets (light and dark orange). (B) A view oblique to the fibril axis reveals the tight fit between side chains of the mated sheets. (C) A view perpendicular to the fibril axis (vertical line) reveals that this zipper-like mating of side chains extends along the entire length of the fibril. (D) The remaining orthogonal view shows how the β-strands stack into β-sheets via main chain hydrogen bonds (dotted lines), parallel and in-register. Amide C=O and N-H groups point up and down, nearly parallel to the fibril axis. (E,F) Steric zipper formation liberates protein-bound water molecules, contributing a hydrophobic effect to fibril stability. (G) As determined by cryoEM at 2.8 resolution (Ni et al., 2019), full-length α-synuclein molecules stack in hydrogen-bonded sheets like the segment, but with a slight twist of each molecule compared to the molecule 4.8 Å below (PDB ID 6osj). Two protofilaments (brown and gray) intertwine to form a fibril. The preNAC region, is colored orange. (H) A view oblique to the fibril axis reveals each molecule is confined to a nearly flat layer. Each chain adopts the same meandering path comprising a series of β-strands and turns which mate together side chains in steric zippers. The steric zipper motif bridging the two protofilaments (orange) is analogous to that of the segment in panel B. (I,J) Views perpendicular to the fibril axis reveal steric zipper interactions and hydrogen bonding patterns analogous to the segment in panels C and D. (K,L) Assembly of 7 steric zippers (orange and cyan shades) liberates water molecules, a process that contributes greatly to amyloid stability.
The cross-β motif is reinforced by interlocking of side chains protruding from the faces of its two mated β-sheets (Figure 1C,F). Formation of this tight, adhesive interface, called a “steric zipper,” liberates water molecules bound to sheet surfaces, thereby producing a favorable gain in entropy that drives amyloid assembly along with the enthalpic contribution from β-sheet hydrogen bonds (Figure1 E–F). The steric zipper motif seen in atomic detail in the α-synuclein segment is a shared feature of the 144 amyloid segment crystals published to date and, as we shall see, lends extraordinary stability to the amyloid fibrils of their full-length parent proteins.
The simplest cross-β scaffold supports a remarkable variety of 2D-protein folds
We now have access to 80 near-atomic-resolution structures of amyloid and amyloid-like fibrils (Figure 2, Suppl. Table 1, and Suppl. Figures 1–9). These structures encompass 20 diverse proteins with core segments ranging from 15 to 139 residues in length. The majority are from the pathogenic branch of the amyloid family and a few are from the functional branch. Most of these fibrils assemble with the simplest type of cross-β scaffold, called parallel, in-register. Here, we survey the range of structural variation among amyloid fibrils and the unique features of this scaffold that enable it to accommodate such variation.
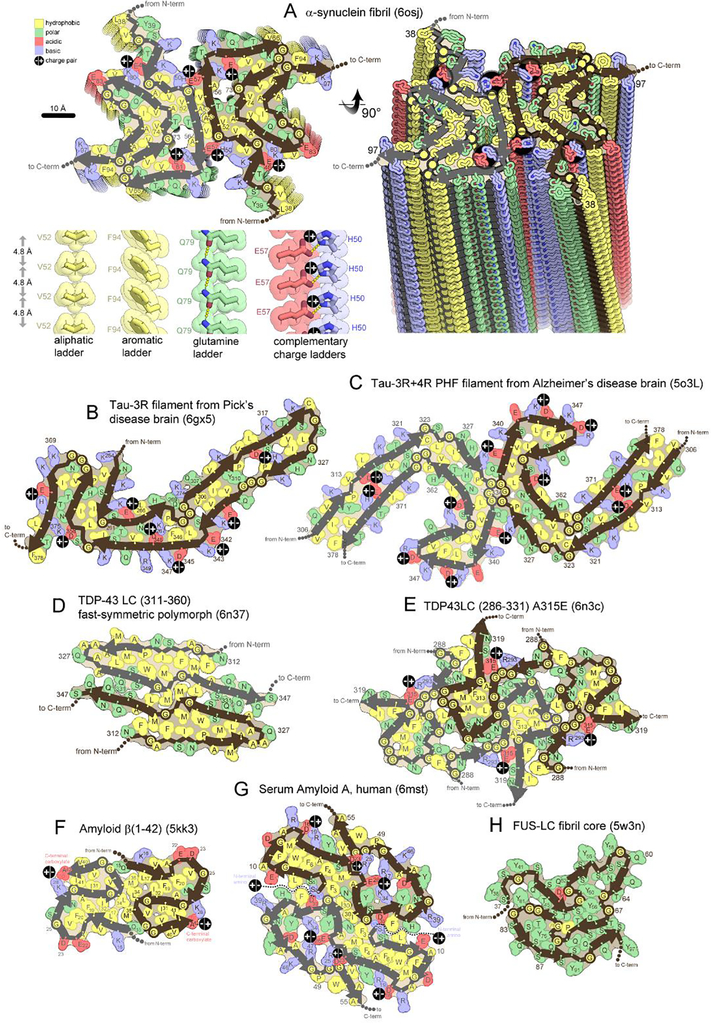
Figure 2. Folding patterns of amyloid proteins in compact, 2D layers.
(A) 2D layers of α-synuclein (upper left) stack into a fibril with a slight twist between layers. Thick lines trace the protein backbone. Brown and gray lines distinguish separate protofilaments. Sidechains are colored according to physical property. A view perpendicular to the fibril axis (right) reveals that identical residues stack in ladders along the fibril axis. Four types of amino acid ladders are depicted in detail (lower left), revealing stabilizing van der Waals contacts and/or hydrogen bonding and/or charge complementation. (B-H) Two dimensional layers of seven diverse amyloid fibrils reveal patterns of side chain association akin to those in globular proteins; hydrophobic residues (yellow) cluster together and tend to be buried. Polar residues also cluster together or reside on the surface. Charged residues associate in complementary pairs (black circle with +/− inscribed) or reside on the surface. β-arches (main chain U-turns) are ubiquitous. Steric zippers (extended β-strands running side-by-side) are evident in all but panel E and H.
Unlike familiar 3-dimensional folds of globular and membrane proteins, amyloid proteins flatten into 2-dimensions (Suppl. Figure 7A) exposing their backbone amides to maximally hydrogen-bond, or “stack” with identical neighboring chains. Thousands of identical, nearly flat protein molecules stack, giving the fibril its length. The structure of an α-synuclein fibril illustrates how a full-length protein molecule “folds” in 2-dimensions (Figure 1H, 2A). Most of the 104-residue-long α-synuclein molecule meanders in a path punctuated by short β-strands and turns. This intricate path is confined to a 4.8 Å thin layer, oriented nearly perpendicular to the fibril axis [Sidebar 2, Warp and Stagger] (Suppl. Figure 10). About 40% of the molecule remains disordered, forming a fuzzy coat on the outside of the fibril.
[Sidebar 2]. Warp and stagger.
The stacked layers of molecules in amyloid fibrils are not entirely flat. Segments of a protein chain rise above or dip below a best-fit plane roughly perpendicular to the fibril axis (Suppl. Figure 10). We describe this atomic deviation from a 2D plane as “warping”. The RMS deviation of a molecule’s α-carbons from a plane (Suppl. Table 1) varies from as little as 0.6 Å for Aβ (1–42) (PDB code 2mxu) to as much as 2.4 Å for Serum Amyloid A (PDB code 6mst). A large degree of warping can give the appearance that one end of the fibril is protruding and the opposite end is receding(Kraus et al., 2021).
Stagger measures the distance between two structural regions along the fibril axis. Stagger is not measured in units of length like angstroms or nanometers. Instead, stagger is measured as the number of molecules (or layers) traversed along the fibril axis to reach from one region of a fibril to another (see Supplement …).
Two-dimensional folds vary markedly among amyloid proteins. For example, molecules of 3R tau are J-shaped in filaments from Pick’s Disease (PiD) patients(Falcon et al., 2018a) (Fig 2B). Molecules of 3R+4R tau are C-shaped in the Paired Helical Filaments (PHFs) from AD patients(Falcon et al., 2018b; Fitzpatrick et al., 2017) (Figure 2C). Molecules of TDP-43 SegA are dagger shaped (Figure 2D, Suppl. Figure 6B–D), and those from TDP-43 SegB are R-shaped(Cao et al., 2019) (Figure 2E). Molecules of Aβ(1–42) are S-shaped(Colvin et al., 2016),(Wälti et al., 2016) (Figure 2F). Molecules of α-synuclein are often amyloid-key-shaped (Figure 2A, Suppl. Figure 4) [Sidebar 3: Hairpins, Arches, and Amyloid Keys]. This alphabet of folds is composed entirely from β-strands and turns. β-strands vary in length, and turns vary in curvature from sharp kinks (Figure 2D) to graceful curves (Figure 2C) in a continuum that defies categorization.
[Sidebar 3]. Hairpins, Arches and Amyloid Keys.
The hairpin-like shapes of side-by-side β-strands found commonly in amyloid fibrils differ from the β-hairpin turns in globular proteins. In β-hairpin turns, the consecutive strands are hydrogen bonded to each other via backbone hydrogen bonds. In the hairpin-like structures of amyloid, consecutive strands interact through side chains rather than backbone hydrogen bonds. To distinguish the bent-hairpin-like structures of amyloid from β-hairpins, the former have been termed “beta arches”(Kajava et al., 2004, 2005).
Greek key patterns prominent in ancient decorative borders are also present in proteins. The Greek key-like arrangements of β-strands found commonly in amyloid fibrils differ from the Greek key-like arrangements in globular proteins. In the Greek keys of globular proteins, the β-strands are hydrogen-bonded through their backbones. However, in the Greek keys of amyloids, the β-strands interact through sidechains, not mainchain hydrogen bonds. To avoid confusion, we refer to the later motif as an amyloid key(Liberta et al., 2019).
Moreover, the cross-β scaffold itself displays variation in propensity to twist and bundle. The stacking of identical molecules along the fibril axis is generally accompanied by a slight twist from the adjacent layer, thereby forming a slowly twisting helix along the fibril length (Figure 1G). Twist angles vary among amyloid fibrils, but typically fall within a few degrees of zero (Suppl. Table 1) [Sidebar 4: Twist]. Hence, the twists observed in full-length proteins are not a great departure from crystalline amyloid segments which lack twists (Figure 1A–B). Many fibrils are composed of a single stack of molecules called a protofilament, exemplified by 3R-tau from PiD (Figure 2B) and FUS (Figure 2H). But, more often, multiple protofilaments bundle together in fibrils. Two protofilaments per fibril is the most common stoichiometry (gray versus brown cartoons in Figure 2A, C–D, F–G), but three (Suppl. Figure 3A–B, 9C), four (Figure 2E) and even 27 protofilaments (Suppl. Figure 6E) have been observed (Suppl. Table 1).
[Sidebar 4]. Twist.
Most fibrils exhibit a slight left-handed twist. Only a few fibrils exhibit a right-handed twist. In all cases, the magnitude of the twist is close to zero. The fibril twist is a manifestation of the twist of its component β-sheets, which run the entire length of the fibril. Thus, the preference for left-handed twist in the β-sheets of amyloid fibrils is consistent with the well demonstrated preference for left-handed twist in β-sheets of globular proteins. The magnitude of the left-handed twist tends be higher in globular folds than in amyloid fibrils. Generally, the longer a β-sheet becomes, the flatter it becomes.
The structural variability described above is remarkable, considering the fibril scaffold is identical in all these examples: parallel, in register. This scaffold aligns protein molecules in parallel (N-terminus aligns with N-terminus, C-terminus aligns with C-terminus), and each residue of a molecule registers (hydrogen-bonds) with the identical residue of adjacent molecules along the fibril axis, 4.8 Å away(Eisenberg and Sawaya, 2017). Few exceptions to parallel, in-register alignment have been observed among amyloid proteins (Suppl. Figure 2J, 9H–I). Just how this one scaffold can be so extraordinarily accommodating of the expansive amyloid family is explained below.
Amino acid ladders zip together in complementary pairs driving folding and stability
Parallel, in-register geometry creates ladders of identical sidechains stacked at 4.8 Å intervals along the fibril length; these ladders can be stabilizing or destabilizing depending on which of the 20 amino acids compose the ladder. Most are stabilizing. For example, when aliphatic or aromatic sidechains stack, they stabilize the fibril through ladders of van der Waals contacts (Val52 in lower left of Figure 2A) or π-π stacking(Hughes et al., 2018; Nelson et al., 2005; Riek and Eisenberg, 2016) (Phe94 or Tyr39 in Figure 2A). Similarly, when asparagine and glutamine sidechains stack, they stabilize the fibril through ladders of sidechain hydrogen bonds termed polar zippers, such as Gln79 in α-synuclein (Figure 2A)(Perutz et al., 1994),(Nelson et al., 2005). However, stacks of identical ionizable residues, such as arginine, lysine, histidine, aspartate or glutamate can be strongly destabilizing due to electrostatic repulsion between like charges spaced only 4.8 Å apart. Parallel, in register geometry has the potential to be the most stable of β-sheet geometries provided that ionizable ladders can be neutralized by hydrogen bonding to water molecules, charged cofactors, or neighboring ladders of opposite charge—conditions which are often observed, as we describe below.
Protein backbones meander in a path that brings together ladders of complementary physical properties. That is, hydrophobic ladders tend to mate with other hydrophobic ladders, generally on the interior of the fibrils, as shown by the streak of hydrophobic residues (yellow-colored residues) crossing the diameter of the Serum Amyloid A fibril (Figure 2G). Polar ladders are often located on the fibril surface. When buried, they tend to mate with other polar ladders, presumably enabling hydrogen bonding networks (cluster of green-colored residues near Gln331 in Figure 2D). Ladders of positive charge tend to mate with ladders of negative charge (juxtaposed blue and pink residues marked by “+/−“). This charge complementation can arise from ladders close in sequence such as Glu342 and Lys343 in 3R-tau from PiD patients (Figure 2B), or distant in sequence like Lys267 and Asp348(Figure 2B), or in separate protofilaments like His50 and Glu57 in α-synuclein (Figure 2A) or Arg293 and Glu315 in TDP-43 SegA (Figure 2E). When an ionizable ladder appears uncomplemented, it is usually located on the surface, where solvent can attenuate electrostatic repulsion. Electrostatic attraction and repulsion between charged ladders may be especially influential in guiding folding patterns since these forces act at a distance(Gaspar et al., 2020). Turns often occur at ladders of glycine or proline, owing to their unique geometric properties.
Mating between complementary ladder types excludes water, thus forming steric zipper motifs like those described above in crystal structures of short amyloid segments (Figure 1E–F). For example, in full-length α-synuclein, aliphatic ladders of Ala53 and Val55 in one protofilament tightly mate with identical ladders in the opposing protofilament (Figure 1K–L). Steric zippers are evident in most amyloid fibril structures and are recognizable wherever sidechains from juxtaposed β-strands contact each other, leaving no intervening solvent accessible gap (Figure 2). Frequently, steric zippers can be spotted within a protofilament, in contiguous strand-turn-strand units called β-arches [Sidebar 4: Hairpins, Arches, and Amyloid Keys]. For example, a β-arch which spans residues 358 to 375 in 3R-tau from PiD patients also hosts a steric zipper composed of its two β-strands (Figure 2B). Even among structures with highly kinked backbones, unpaired segments and cavities are rare (internal white spaces in Figure 2E, 2H). Thus, the formation of tight, dry sheet-sheet interfaces and consequent liberation of surface bound water molecules evidently lend stability to amyloid fibrils.
In summary, amyloid proteins are driven to adopt a structure that can: (1) maximize the number of backbone hydrogen bonds between molecules by adopting favorable β-strand conformation over most residues and stacking in 4.8 Å-thin, nearly flat layers; (2) align aromatic and amide side chains in stabilizing ladders, as occurs when molecules adopt parallel, in-register β-sheet geometry; (3) mate together ladders of complementary amino acid types, as occurs in steric zippers, driving out and liberating solvent molecules; (4) bury hydrophobic sidechains; and (5) expose uncomplemented charges to solvent. Adherence to these principles is evident among pathogenic amyloids (Figure 2, Suppl. Figures 1–8); however, localized violations (frustrations) are common, as we shall discuss. No single fold is capable of satisfying over its entire residue range all these drives towards minimum energy. Instead, as if trying to strike different compromises, most pathogenic amyloid proteins attain multiple alternative amyloid fibril structures called polymorphs.
II. AMYLOID POLYMORPHISM AND ITS ROLE IN DISEASE
Pathogenic proteins each adopt a range of amyloid fibril structures
Polymorphism describes the capacity of a protein sequence to form different structures. A familiar example of a polymorphic protein is hemoglobin. Its uptake and release of oxygen is regulated by distinct structural polymorphs (oxy and deoxy)(Bolton and Perutz, 1970). Similarly, viral capsid proteins take different polymorphic (quasi equivalent) structures to perform distinct structural roles sealing capsid faces and capsid vertices(Caspar and Klug, 1962). A third example is actin fibrils which display at least six distinct polymorphic structures required to perform actin’s diverse biological roles including maintaining cell shape, polarity, and force generation(Galkin et al., 2010). Most disease-related, amyloid-forming protein sequences also perform in a repertoire of polymorphic structures(Fitzpatrick et al., 2017),(Cao et al., 2019),(Li et al., 2018),(Zhang et al., 2019),(Falcon et al., 2019),(Zhang et al., 2020),(Guerrero-Ferreira et al., 2019),(Boyer et al., 2019) [Sidebar 5 What are amyloid protein polymorphs?]. However, rather than having evolved to enhance survival fitness, pathogenic polymorphs are adventitious. They appear more numerous, diverse, and persistent than functional polymorphs. We survey the structural diversity exhibited, and the role of chemical environment in eliciting changes in polymorph populations in vitro and in vivo.
[Sidebar 5]. What are amyloid protein polymorphs?
Amyloid polymorphs, loosely defined, are distinctly different amyloid fibril folds of closely identical amino acid sequences. But more precise definition hinges on the meanings of “distinctly different folds” and “closely identical sequences”. The examples of the protein tau shown in Suppl. Figure 1 reveal the subtleties that must be considered. The C-shaped folds of protein chains with identical sequence of the PHF and SF filaments are essentially identical, but the interfaces of the two protofilaments are distinctly different, justifying their distinction as separate polymorphs (panels A and B). Closer examination shows that there are small rearrangements of sidechain packing in the cores of PHF and SF structures. If these rearrangements are deemed to be significant, then PHF and SF filaments would be distinguished as separate polymorphs based on core-packing differences as well as protofilament interface differences. In contrast PHF filaments and PiD filaments (Panels A and E) with distinctly different folds would not be considered polymorphs because their sequences are not closely identical: the PiD sequence lacks repeat R2 of tau. Deciding on whether a single-residue variant is a polymorphic fold would depend on whether or not the single residue change creates a distinctly different fold. Two amyloid fibrils of different pitch (caused by the twist angle between successive layers) but identical sequence and closely similar chain fold could be classed as polymorphs or not, depending on whether one is concerned with the chain fold or the morphology of the fibril. In short, the term “polymorph” as applied to amyloid fibrils benefits by coupling to further terms that define its geometry.
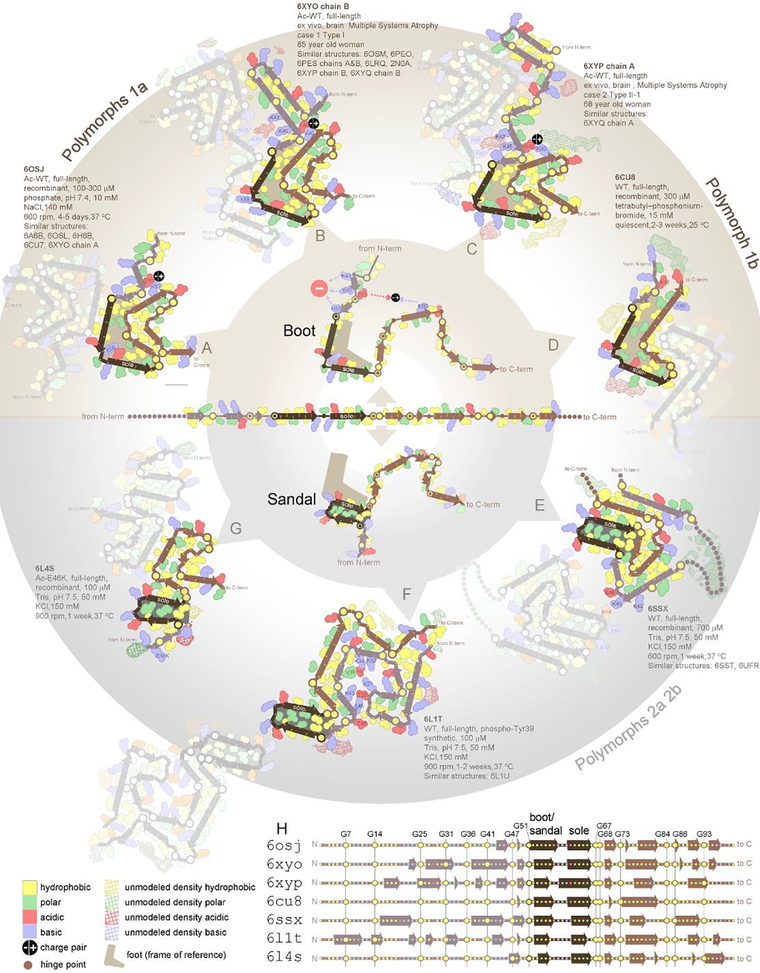
Structural differences between polymorphs vary in magnitude from reorganization of the entire core fold to mere adjustments in side chain conformations. The full gamut is illustrated by 24 distinct α-synuclein structures determined: 18 in vitro and 6 ex vivo (Figure 3). Starting with the most extreme structural differences, not one intramolecular interface is conserved between the folds of polymorph 1a (6osj) (Fig 3A) and polymorph 2a (6ssx) (Fig 3E). Eighteen folds share a small, structurally conserved boot-shaped kernel (residues 50–77)(Boyer et al., 2019) (Figure 3A–D). The remaining six folds share a sandal-shaped core (Figure 3E–G). Some exhibit only local differences in the backbone, such as those that divert the side chain of Lys58 to the interior (6osj) (Figure 3A) or exterior of the kernel (6xyo) (Figure 3B). Lastly, ultrastructural polymorphs maintain identical folds and differ primarily in the arrangement of protofilaments. For example, the H50Q hereditary mutation of α-synuclein displays two polymorphs, narrow (6peo) and wide (6pes), which are composed of single and double protofilaments, respectively (Suppl. Figure 4B–C). Other pathogenic proteins show a similar range of structural diversity, including tau and its isoforms (Suppl. Figure 1), Aβ(1–40) and Aβ(1–42) (Suppl. Fig 2–3), and TDP-43 (Suppl. Fig 6).
Figure 3. A single protein sequence attains multiple amyloid polymorph structures.
At the center, the sequence of α-synuclein is represented in an unfolded state. At the outside of the circle are depicted seven different polymorphs of α-synuclein fibrils (A-G), each obtained from different conditions, ex vivo and in vitro as noted. The seven structures represent distinct groups obtained by structural similarity clustering analysis of 25 structures determined to date. The PDB codes for all members of a group are noted. Each polymorph is composed of a series of β-strands and turns, which are well conserved as evidenced by (H) secondary structure alignment. The turns are most often located near glycine (depicted by prominent yellow spheres centered on backbone), or near clusters of charged residues (pink and blue). Despite the conservation of secondary structure, the diversity of tertiary structure is striking. Some interfaces between strands are conserved between structural groups; however, not one interface is conserved among all seven groups. It appears all polymorphs are built from the same secondary structure building blocks but differ in the angles between them. This point is illustrated by the varied angles observed between strands comprising residues 51–58 and 60–66 (darkest brown color). A 90° angle between these two strands defines the “boot” polymorphs in the upper half of the circle. A 180° angle between these strands defines the “sandal” polymorphs in the lower half of the circle. The alternative paths adopted by the protein chains of distinct polymorphs represent alternative ways to pair hydrophobes together (yellow) and complement opposite charges (pink and blue). Paths are evidently determined by different chemicals in different growth conditions (unmodeled density).
Remarkably, β-strand elements are conserved among polymorphs of α-synuclein (Figure 3H); they are just oriented differently as if the strands were connected by joints (Figure 3 center hub)(Guerrero-Ferreira et al., 2019),(Boyer et al., 2020). Conservation of β-strand elements is also observed among tau polymorphs(Scheres et al., 2020), suggesting these elements are fundamental, modular building blocks of fibril structure.
The full polymorphic potential of a protein is elicited by varying growth conditions. Indeed, the α-synuclein structures illustrated in Figure 3 represent fibrils grown in different salts, different salt concentrations ranging from 5 to 150 mM, and different protein concentrations ranging from 0.3 to 1.0 mM (Table 1 in reference(Guerrero-Ferreira et al., 2019)). Each condition selects a subset of the full polymorphic potential of a protein that is compatible with the particular chemical environment. For example, polyanions select polymorphs 1a and 1b (Figure 3A and D) (Li et al., 2018), whereas a lack of polyanions select polymorphs 2a and 2b (Figure 3E, Suppl. Fig 5E–F) (Guerrero-Ferreira et al., 2019). In other words, which amyloid fibril polymorph emerges depends on environmental conditions, analogous to how the growth of protein crystals is sensitive to crystallization conditions(Jarrett and Lansbury, 1993). The factors influencing fibril polymorphism in vivo are not as well determined due to the complexity of living systems and the years required for amyloid diseases to develop. Interestingly, fibrils extracted from a single patient tend to show less extreme polymorphism than individual in vitro samples cited above. Some evidence suggests that a broad spectrum of polymorphs is present in an individual but only the “fittest” polymorphs (those able to propagate rapidly in that particular environment) are abundant enough to observe by current methods(Oelschlegel and Weissmann, 2013).
Frustrated regions in amyloid fibrils sensitize growth to environmental conditions.
Amyloid fibrils typically exhibit local structural frustrations that, like chinks in armor, impart vulnerability to amyloid fibrils. These frustrations partially offset their extraordinary energetic stability(Bryngelson and Wolynes, 1987), making their growth sensitive to environmental conditions or events that can alleviate these frustrations, such as ligand binding, pH change, mutation, or post translational modification. Understanding frustration and its alleviation lends insights into amyloid pathogenesis and amyloid functionality. Here, we describe three common manifestations of frustration: backbone torsion angle frustrations, charge frustrations, and the formation of cavities or channels (Suppl. Table 2). We illustrate their alleviation and pathogenic consequences.
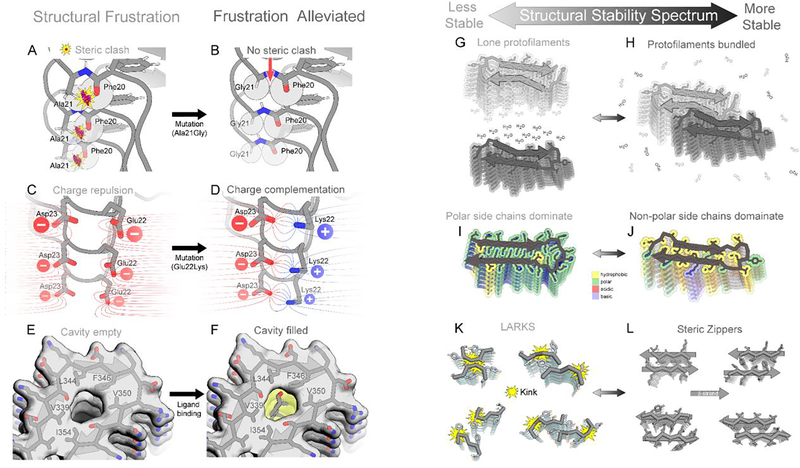
Backbone torsional frustrations occur when the torsion angle between bonded atoms incurs steric clashes (Figure 4A). These clashes weaken structure and delay fibril growth. Such frustration in wild type amyloid fibrils may be alleviated by hereditary mutations with the consequence of facilitating early onset amyloidosis. For example, wild type Aβ(1–42) (PDB ID 2nao(Wälti et al., 2016)) exhibits backbone torsion angle frustrations in the loop region Phe19-Phe20-Ala21-Asp22-Glu23, detected as Ramachandran plot outliers (Suppl. Figure 11A). Hereditary AD mutations such as A21G(Figure 4B), E22G, and E22Δ would eliminate atoms from this crowded loop, potentially alleviating this torsional frustration and accelerating Aβ fibril growth(Wälti et al., 2016). Hence, individuals carrying such frustration-relieving mutations may experience earlier onset of AD compared to individuals carrying wild type Aβ. Similarly, the early-onset hereditary mutation of islet amyloid polypeptide (IAPP), S20G, is proposed to relieve strained backbone torsion angles observed in the wild type structure(Cao et al., 2020).
Figure 4. Structural features that tune amyloid fibril stability.
(A) Torsion angle frustrations manifest as steric clashes. Here, strain in the phi angle of Ala21 produces steric clash between an oxygen atom of Phe20 and hydrogen atom of Ala21 (spheres) in Aβ, (2nao). (B) The Ala21Gly mutation eliminates some steric bulk, so Phe20 atoms do not clash with Gly21 atoms. (C) An example of charge frustration. Close proximity of negatively charged residues Glu22 and Asp23 of Aβ, (2nao) destabilizes the fibril. (D) The Glu22Lys mutation eliminates frustration by introducing a complementary positive charge. (E) A cavity is evident in tau filaments from CTE patients (6nwp) destabilizes the fibril. (F) A complementary ligand fills the cavity and stabilizes the filament. (G, H) A lone filament gains stability when it bundles with other filaments; formation of new interfaces liberates surface bound solvent molecules. (I, J) A fibril gains stability as its polar residues are replaced with hydrophobic residues. (K) Reversible amyloids contain LARKS motifs (Hughes et al., 2018),(Guenther et al., 2018b) which are kinked and exhibit small interfaces, making them less stable than steric zippers (L). Four examples of each are shown here.
Charge frustration weakens molecular contacts in amyloid fibrils by electrostatic repulsion. Charge frustration can be alleviated by a variety of mechanisms, thereby stabilizing fibrils. For example, in wild type fibrils of Aβ(1–42) (PDB ID 2nao(Wälti et al., 2016)), adjacent ladders of negative charge at Glu22 and Asp23 create frustration (Figure 4C)(Suppl. Figure 11A) that may be alleviated by charge-eliminating hereditary mutations E22Q, E22G, E22K (Figure 4D), E22Δ, or D23N(Wälti et al., 2016). The consequence of such frustration-relieving mutations, as noted above, may be early onset of AD. In another example, charge frustration arises from adjacent ladders of positive charge, Lys43 and Lys45, in MSA-derived α-synuclein fibrils(Schweighauser et al., 2020) (PDB ID codes 6xyp, 6xyo), but residual map density suggests it is alleviated by binding a presumably anionic ligand (Figure 3B,C, Suppl. Figure 11B). Growth of this polymorph may be contingent on the presence of such an electrostatically complementary ligand, otherwise charge repulsion would impede fibril growth. A third mechanism for alleviating charge frustration is through charge-altering post-translational modifications such as phosphorylation, acetylation, or ubiquitination. For example, acetylation is proposed to facilitate tau fibrilization in AD and CBD patients by neutralizing some of its abundant lysine residues. The distinct patterns of acetylation observed between AD and CBD patients may account for the distinct structures adopted by tau in the patients of these two diseases(Arakhamia et al., 2020). Similarly, post-translational phosphorylation of α-synuclein residue Tyr39 introduces a −2 charge that alleviates the positive charge frustration incurred by juxtaposition of lysine residues 21, 32, and 34 (Figure 3F)(Zhao et al., 2020).
Cavities incur packing defects in fibrils that may be alleviated by binding ligands, and these ligands may contribute to pathogenesis. For example, a narrow cavity is conspicuous in tau fibrils extracted from CTE patients, bordered by residues Val339, Leu344, Phe346, Val350, and Ile354 (Figure 4E) (Suppl. Figure 1C,D)(PDB ID codes 6nwp, 6nwq)(Falcon et al., 2019). The cryoEM map suggests a ligand is bound there, presumably a slim, non-polar sterol, fatty acid, or short aliphatic chain to complement the shape and hydrophobicity of this cavity (Figure 4F). Fibril formation may be dependent on this ligand considering that water, the alternative void-filling molecule, is incompatible here. In general, the potential of the cell to alleviate any of the types of frustration described above may explain how the cell favors particular polymorphic structures.
The role of polymorphism in defining disease pathogenesis
Distinct fibril polymorphs have been linked to distinct diseases. For example, tau fibrils are deposited in some 25 clinically distinct neurodegenerative and movement disorders called tauopathies. The laboratory of Goedert and Scheres studied the structures of tau fibrils isolated from autopsied brains of multiple patients diagnosed with four diseases: AD (19 patients)(Falcon et al., 2018b), chronic traumatic encephalopathy (CTE) (3 patients)(Falcon et al., 2019), PiD (9 patients)(Falcon et al., 2018c), and CBD (3 patients)(Zhang et al., 2020). Among patients diagnosed with the same disease, no variation in structure was detected using CryoEM and/or immunoreactivity; however, each disease did produce its own distinct structure. Other laboratories observed an analogous pattern among patients of synucleinopathies, whereby structures of brain-derived α-synuclein fibrils were correlated with the particular synucleinopathy diagnosed: Parkinson’s disease (PD) (75 patients)(Shahnawaz et al., 2020), multiple system atrophy (MSA) (5 patients)(Schweighauser et al., 2020), and dementia with Lewy bodies (DLB) (3 patients)(Schweighauser et al., 2020).
Recently two distinct tauopathies have been linked to the same tau polymorph(Shi et al., 2021), demonstrating that each disease may not have its own unique amyloid fibril structure. Nevertheless, each disease appears to be defined by a disease-specific set of polymorphs. Indeed, the structural conformation of a polymorph has been proposed to serve as a basis for classifying amyloid disease. An implication of disease-specific polymorphs is that each disease may require its own diagnostic or therapy to target the deposited fibrils of that particular disease.
Underlying the polymorph-disease relationship is the capability of fibrils to template the growth of new fibrils with structural features faithful to the parent fibril. Indeed, fibril deposits have been observed to spread from cell-to-cell along neural pathways(Goedert, 2015),(Braak and Braak, 1991), and the daughter fibrils propagate true to the original. Fidelity in propagation is evidenced by observation that tau polymorphs are conserved among deposits extracted from seven regions in one AD patient’s brain(Falcon et al., 2018b); similarly, conservation was observed among three regions of one CBD patient’s brain(Zhang et al., 2020). The ability of amyloid fibrils to faithfully self-propagate has long been exploited to produce abundant, homogeneous fibrils in vitro from small amounts of seed material(Petkova et al., 2005),(Saborio et al., 2001). Fibrils grow from their ends(Goldsbury et al., 1999) where molecules are most exposed and accessible to recruit soluble protein to adopt a conformation identical to the seed template. When fibrils break, they produce new seeds(Collins et al., 2004). Cells facilitate fibril spreading by importing and exporting seeds through a variety of mechanisms(Brunello et al., 2020).
The polymorph-disease relationship parallels the more established hypothesis that distinct strains (polymorphs or “conformations”) define distinct prion disease phenotypes(Prusiner, 1998),(Collinge and Clarke, 2007),(Goedert, 2015). Like tauopathies and synucleinopathies, prion diseases are amyloid diseases, but they have the added distinction that fibril seeds are so robust that they spread readily not only between cells of the same individual, but also between individuals. Individuals with prion disease may display different phenotypes. If prion strains are indeed defined by their amyloid fibril conformation, then the various tauopathies may be viewed as strains of tau-associated dementias; similarly, the various synucleinopathies may be viewed as strains of α-synuclein-associated conditions(Peelaerts et al., 2015).
The association of each tauopathy (or synucleinopathy) with its characteristic set of polymorphs raises questions about which comes first, the disease or the polymorph? Does the polymorph emerge first and thereby determine which disease develops? Or do early events in disease progression determine which polymorph emerges? The “polymorph first” hypothesis implies the conditions for fibril nucleation are uniform among individuals, and whichever polymorph stochastically emerges determines the disease. It also implies that distinct polymorphs have the capability to interact differently with cell machinery to bring about different disease phenotypes. Some evidence supports this hypothesis. Distinct α-synuclein polymorphs were found to differentially interact with and inhibit the proteasome, a plausible means of influencing pathology(Suzuki et al., 2020). In contrast, the “disease first” hypothesis suggests biochemical environmental conditions differ among diseases, and that these conditions determine which polymorph emerges. In favor of this hypothesis, evidence of fibril-bound cofactors suggests that cofactor abundance might select the polymorphs particular to AD, CTE, and CBD (previous section). Early events in disease pathogenesis could influence the concentration of cofactors in a given tissue and hence nucleate or select a specific polymorph(Peng et al., 2018), or disease-specific patterns of post-translational modifications like acetylation and ubiquitination could select particular polymorphs of tau(Arakhamia et al., 2020). This hypothesis further holds that structural distinctions between polymorphs are inconsequential to disease phenotype; the mechanism of amyloid pathogenicity is uniform among all polymorphs owing to a shared cross-β scaffold which creates a blunt instrument to damage cells(Engel et al., 2008). A third alternative is a hybrid model in which disease-specific environments select a set of fibril polymorphs which then interacts with cellular components in a disease-specific way to affect phenotype(Prusiner et al., 2015)(Oelschlegel and Weissmann, 2013).
In summary, each protein is prone to adopting a variety of conformations in the amyloid state, known as polymorphs. A particular polymorph is favored by prevailing biochemical conditions that can alleviate its structural frustrations. Specific polymorphs are linked to specific amyloid diseases. Disease-specific conditions might elicit these polymorphs or vice versa.
III. EVOLUTIONARY ADAPTATIONS OF AMYLOID TO PERFORM FUNCTION
Functional and pathogenic amyloids share cross-β scaffolds but differ in activity
Functional amyloids perform biological roles that fall into five broad classes(Perrett et al., 2014),(Otzen and Riek, 2019) (Suppl. Table 3): (i) Structural amyloids maintain shape or support. For example, chaplin fibrils support tubular structures called hyphae in fungi. Curli fibrils support biofilms to anchor and fortify bacterial colonies. PMEL17 fibrils align substrates for polymerization into melanin in humans. (ii) Reservoir amyloids store, protect, and moderate protein activity. For example, β-endorphin fibrilizes as a means to store and protect the hormone from damage. Stress granule proteins such as FUS and hnRNPA2 fibrilize to sequester RNA during times of stress (iii) Information carrier amyloids maintain a particular state over time. For example, Orb2 forms fibrils of sufficient longevity to enable long-term memory in drosophila. HET-s fibrils transmit information to daughter cells regarding compatibility of fusing to other yeast cells (iv) Function-suppressing amyloids reduce the activity of the soluble proteins from which they assemble. For example, pyruvate kinase activity of Cdc19 is reduced when glucose starvation induces Cdc19 fibrilization and (v) Signaling amyloids activate a process. For example, RIPK1/RIPK3 fibrilizes to signal necroptosis. Some amyloids play multiple roles and so belong to multiple classes. Given the importance of their biological roles, nature evolved machinery to regulate assembly of functional amyloid and direct their location in the cell.
To date, seven reportedly functional amyloid fibril structures have been determined: HET-s(Wasmer et al., 2008), Orb2(Hervas et al., 2020), (information carriers), RIPK1-RIPK3(Mompeán et al., 2018) (signaling), FUS(Murray et al., 2017), hnRNPA2(Lu et al., 2020), glucagon(Gelenter et al., 2019), and β-endorphin(Seuring et al., 2020) (reservoirs). These structures show that functional amyloid fibrils assemble on the same cross-β scaffold as their pathogenic cousins.
Despite the structural similarities, functional fibrils show a greater range of stabilities and lifetimes than do pathogenic fibrils, which are highly stable and essentially permanent. Many functional amyloid assemblies evolved to be reversible, their disassembly triggered by a particular cellular stimulus when the fibril is no longer advantageous. For example, β-endorphin fibrils assemble in storage granules at pH 5.5(Maji et al., 2009), but disassemble when free, active hormone is needed, triggered by granule release into the phosphate-free, pH neutral environment of blood(Nespovitaya et al., 2016). PMEL17 fibrils assemble in the melanosome at pH ~4, but exposure to cytosolic conditions (pH ~7) leads to rapid dissolution, which is proposed to protect against cytotoxicity should fibrils escape the melanosome(McGlinchey and Lee, 2017). Other functional fibrils, like yeast prion Sup35 and fungal prion HET-s have greater stability and lifetime, persisting through many generations. Their disassembly requires expenditure of energy; the chaperone Hsp104 hydrolyzes ATP to fuel fibril disassembly(Haslberger et al., 2010). Curli fibrils are even more stable; they enable bacterial colonies to invade hosts, adhere to surfaces, evade immune response, and endure extreme conditions. In contrast to functional amyloids, the majority of pathogenic amyloids occupy only the most stable end of the spectrum. Indeed, stability seems to be a defining feature of pathogenic amyloids(Meersman and Dobson, 2006); if they were unstable, chaperones could clear them and they would not be pathogenic.
Evolution tunes functional amyloid lifespans through structure
Various mechanisms have evolved to impart lability to amyloid structure. Among the seven reportedly functional amyloid fibril structures determined to date, four can be characterized as conditionally labile: FUS(Murray et al., 2017), hnRNPA2(Lu et al., 2020), glucagon(Gelenter et al., 2019), and β-endorphin(Seuring et al., 2020). All four are composed of only a single protofilament, suggesting the absence of protofilament bundling is a feature evolved to facilitate assembly and disassembly (Figure 4G–H). Another mechanism of imparting lability may be evolution of abundant polar residues (Figure 4I–J), as seen in FUS (Figure 1H) and hnRNPA2(Lu et al., 2020) (Suppl. Figure 9E), as well as charged residues as seen in glucagon (Suppl. Figure 9F). The abundance of polar residues helps to shift the equilibrium away from a dehydrated assembly to solvated monomers.
Sensitivity to pH in some amyloids may be facilitated by charge frustration. Fibril structures of β-endorphin, glucagon, and PMEL17 fibrils reveal unpaired charged residues buried in their cores. β-endorphin buries Glu8 (Supp. Fig 9G)(Seuring et al., 2020). Glucagon fibrils bury three aspartic acid residues (Asp9, Aps15, and Asp21) in its core (Suppl. Fig 9F). And structural models of PMEL17 suggest Glu422 is buried in its fibril core; mutagenesis studies confirm Glu422 is critical in controlling the pH sensitivity of PMEL17 fibrils(McGlinchey and Lee, 2017). All three of these fibrils are stable at low pH where the critical buried acid side chains are neutralized, but dissolve upon elevation of pH as the buried acid side chains become charged. Here, nature took advantage of charge frustration to regulate fibril dissolution in response to the presence or absence of protons. In so doing, nature confines these fibrils to their respective acidic granules, but dissolves them upon release into neutral environments where fibrils are no longer needed.
Reversibility in the assembly of subcellular condensates such as stress granules(Molliex et al., 2015),(Yang et al., 2019),(Hennig et al., 2015) may be facilitated additionally by the evolution of smaller and fewer steric zipper motifs and by other structural motifs promoting protein-protein adhesion. These motifs include Low-complexity, Amyloid-like, Reversible, Kinked Segments known as LARKS motifs(Hughes et al., 2018). LARKS were discovered in atomic structures of short segments of low complexity domains (LCDs) [Sidebar 6: IDPs vs. LCDs vs. PrLDs] of proteins known to form reversible condensates, including FUS(Kato et al., 2012), hnRNPA1(Gui et al., 2019), a nuclear porin(Hughes et al., 2018) and TDP-43(Guenther et al., 2018a),(Guenther et al., 2018b). LARKS are similar to steric zippers in that β-sheets mate together through sidechain contacts, but the backbones tend to be kinked so the area buried with the interfaces tends to be smaller (Figure 4K). Computations suggest that the stabilization energy provided by each LARKS is of the order of one half of that of a steric zipper(Hughes et al., 2018)(Figure 4L), consistent with the observation that these fibrils are more readily dissolved than those mated by steric zippers(Guenther et al., 2018b).
[Sidebar 6]. IDPs vs. LCDs vs. PrLDs.
Protein domains that lack well-ordered 3D structures are termed Intrinsically Disordered Proteins (IDPs). Sometimes these are low-complexity domains (LCDs) or Prion-like domains (PrLDs). These terms carry different shades of meaning. Lacking ordered tertiary structures, monomers of Aβ, tau, α-synuclein, and IAPP are unquestionably IDPs. LCDs are characterized by significant bias in amino acid composition, and are usually IDPs. These include, for example, domains from TDP-43, FUS, hnRNPA1 and hnRNPA2, and the yeast prion Sup35. PrLDs in yeast and bacteria are often “low-complexity” domains that typically possess an enrichment in Gln and Asn. These include Sup35 and Rnq1. However, not all PrLDs are LCD or IDPs; mammalian prions are one exception. Numerous proteins containing domains described as IDP, LCD, or PrLDs aggregate into amyloid or amyloid-like fibrils, presumably because they readily expose to nearby identical molecules one or more adhesive, amyloidogenic segments.
Reversible amyloid structures tend to have a lower energetic stability
Energetic evaluation of amyloid structures illuminates why some amyloid assemblies are reversible and others are not. The closer to zero is the free energy of stabilization of a fibril, the more responsive the assembly is to environmental changes. A robust method for estimating the standard free energy of amyloid formation begins with evaluating its solvation energy—the energetic penalty for dissolving a molecule in water(Eisenberg and McLachlan, 1986)(Suppl. Figure 12). By this method, regions of frustration and stability can be identified in energy maps in which each residue of an amyloid structure is colored according to its energetic contribution to fibril stability. Deep red colors indicate the most stabilizing and blue colors the most destabilizing residues (Figure 5). The spatial distribution of deep red residues indicates that the major contribution to stabilization of pathogenic amyloid fibrils arises from the exclusion from solvent of apolar sidechains, as in soluble proteins. These tend to cluster in the core of amyloid folds, in steric zippers (streak of deep red residues crossing the diameter of serum amyloid A Figure 5A). Charged residues may be stabilizing or destabilizing depending on whether the residue is buried or balanced by an adjacent opposite charge. Destabilizing residues include the relatively few charged and polar residues buried away from solvent, such as Glu8 in β-endorphin (6tub) colored blue in Figure 5B, Asp43 in Serum Amyloid A (6mst) (Figure 5A), and Glu66 in transthyretin (6sdz) (Figure 5C). More charge frustration is evident in adjacent negatively charged ladders of Glu22 and Asp23 in Aβ(1–42) (5kk3) (Figure 5D) and adjacent positively charged ladders of Arg17, in glucagon (Figure 5E and Suppl. Figure 9F).
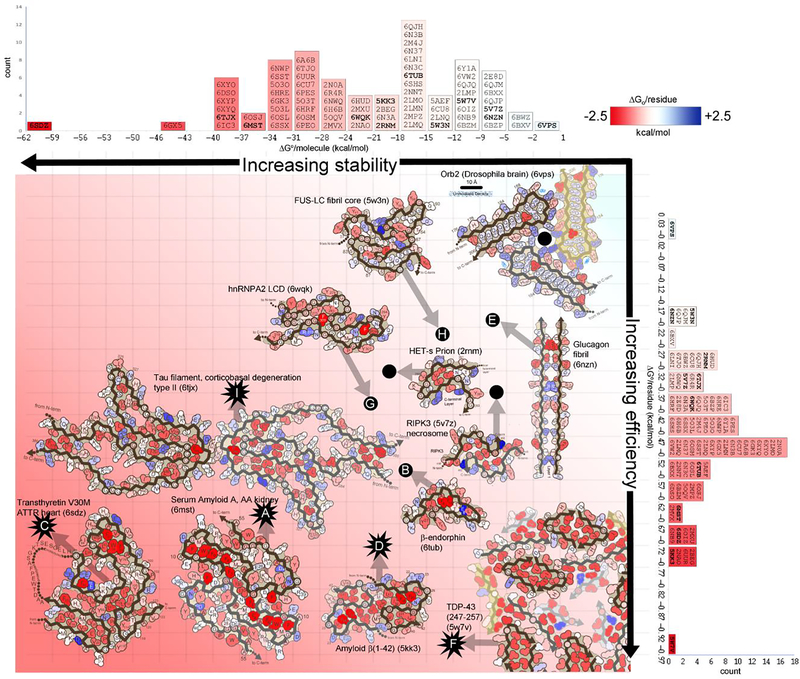
Figure 5. Stabilization energy maps reveal structural features that influence reversibility.
Amyloid fibril structures are colored by energy: strongly stabilizing sidechains are red; destabilizing sidechains are blue. The standard free energies of 75 amyloid structures (indicated by PDB ID codes) are ranked in two dimensions: on a per molecule basis (horizontal histogram) which inform about stability of a molecule, and on a per residue basis (vertical histogram), which inform about energy efficiency (independent of molecule size). Select fibril structures are pictured within the graph. Energy estimates for pathological/irreversible fibrils are indicated with starburst icons. Energy estimates for presumably functional/reversible fibrils are indicated with black dots. Transthyretin V30M (lower left) is evaluated as the most stable structure (62.1 kcal/mol/molecule) and one of the most efficiently stable (−0.68 kcal/mol/residue). It features an abundance of buried hydrophobic residues (deep red). Notably, it is also a pathogenic fibril, extracted from the heart of a patient with hereditary transthyretin amyloidosis. At the opposite extreme, FUS (upper right) is relatively unstable (−12.2 kcal/mol/molecule), and inefficient (−0.20 kcal/mol/residue). It lacks a hydrophobic core. In contrast to transthyretin, FUS aggregation is functional and presumably reversible.
Stabilization energy offers a semi-quantitative means to assess the relative stability of different amyloid fibrils. Some proteins are capable of forming stable amyloid cores with relatively fewer residues. This efficiency in achieving stability with fewer residues is indicated by large negative values of ΔG° per residue (ΔG°resid) (Figure 5, vertical axis) (Suppl. Table 1). Among those most efficient are Aβ polymorphs, exhibiting six of the top ten most negative ΔG°resid values, ranging from −0.76 to −0.67 kcal/mol/residue. The remainder of the top ten are also associated with pathology: TDP-43 segment 247–257 (−0.97 kcal/mol/residue)(Figure 5F), human PrP (−0.74 kcal/mol/residue), transthyretin (−0.68 kcal/mol/residue)(Figure 5C), and serum amyloid A (−0.65 kcal/mol/residue)(Figure 5A). At the bottom end of the spectrum are stress granule-associated fibrils: hnRNPA2(Lu et al., 2020) (−0.38 kcal/mol/residue)(Figure 5G) and FUS(Murray et al., 2017) (−0.20 kcal/mol/residue)(Figure 5H), consistent with the implication that their biological role requires their regulated disassembly. Glucagon fibrils(Gelenter et al., 2019) also may be reversible, and correspondingly exhibit low efficiency (−0.17 kcal/mol/residue)(Figure 5E).
The stability of an amyloid fibril is also dependent on the number of residues in the protein molecule. Small ΔG°resi values are not always correlated with reversibility. Some irreversible, pathogenic amyloid fibrils have small ΔG°resid, but nevertheless are stable. For example, tau CBD type II fibrils occupy the lower end of the spectrum (−0.35 kcal/mol/residue)(Figure 5I). Compensating for this small value, its protofilament core involves 107 residues—over four times more than the most efficient of Aβ fibril cores. Conversely, reversible fibrils of β-endorphin have a relatively high energy efficiency (−0.54 kcal/mol/residue) but only 31 residues contributing to its stability(Figure 5B). Indeed, when we rank amyloid structures by ΔG°/molecule (ΔG°molecule) rather than ΔG°resid, we find the reversible fibrils occupy the lower half of the spectrum, while tau CBD is the eighth most stable fibril by molecule (Figure 5 compare rankings on horizontal and vertical axes). In general, we find that structures of disease-related proteins (Figure 5, starbursts) are more stable (greater negative scores) than structures of functional reversible proteins (Figure 5, filled circles).
Summary
Amyloids have for decades posed biological mysteries only now becoming explained by the recent, technology-driven explosion of amyloid structures. The great number of protein sequences capable of forming amyloid fibrils once seemed inconsistent with the morphological uniformity of the fibrils. Now, we can see that they almost always assemble on the same, uniquely accommodating, parallel, in-register β-sheet scaffold. The extreme polymorphism of amyloid fibrils is enabled by structural frustrations that sensitize fibrils to environmental conditions, such as cofactors or pH. These select which polymorph proliferates, and perhaps define which disease emerges. Pathogenic amyloids are notoriously resistant to degradation; however, some amyloids, like those forming membraneless organelles, must be labile in order to perform their functions. Evolution appears to tune the level of structural frustration, amino acid bias, and presence of LARKS motifs to adjust fibril lifetime. To aid understanding of amyloid biology and development of therapeutics and diagnostics we have constructed a database of amyloid structure and energy (https://people.mbi.ucla.edu/sawaya/amyloidatlas/).
Supplementary Material
Acknowledgements:
We thank our coworkers for discussions and in particular David Boyer for assistance in developing solvation energy maps. We are grateful for support by NIH (AG054022, AG061847, AG070895, AG048120, GM128867), NSF (MCB 1616265), and DOE (DE-FC02-02ER6342).
Footnotes
Declaration of interests
DSE is SAB Chair and an equity holder in ADRx, Inc. JAR is a founder and equity shareholder of Medstruc Inc.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ader C, Frey S, Maas W, Schmidt HB, Görlich D, and Baldus M (2010). Amyloid-like interactions within nucleoporin FG hydrogels. Proc. Natl. Acad. Sci. U.S.A 107, 6281–6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arakhamia T, Lee CE, Carlomagno Y, Duong DM, Kundinger SR, Wang K, Williams D, DeTure M, Dickson DW, Cook CN, et al. (2020). Posttranslational Modifications Mediate the Structural Diversity of Tauopathy Strains. Cell 180, 633–644.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astbury WT, Dickinson S, and Bailey K (1935). The X-ray interpretation of denaturation and the structure of the seed globulins. Biochem. J 29, 2351–2360.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, Trojanowski JQ, and Iwatsubo T (1998). Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson’s disease and dementia with Lewy bodies. Am. J. Pathol 152, 879–884. [PMC free article] [PubMed] [Google Scholar]
- Balbirnie M, Grothe R, and Eisenberg DS (2001). An amyloid-forming peptide from the yeast prion Sup35 reveals a dehydrated beta-sheet structure for amyloid. Proc. Natl. Acad. Sci. U.S.A 98, 2375–2380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson MD, Buxbaum JN, Eisenberg DS, Merlini G, Saraiva MJM, Sekijima Y, Sipe JD, and Westermark P (2018). Amyloid nomenclature 2018: recommendations by the International Society of Amyloidosis (ISA) nomenclature committee. Amyloid 25, 215–219. [DOI] [PubMed] [Google Scholar]
- Berson JF, Theos AC, Harper DC, Tenza D, Raposo G, and Marks MS (2003). Proprotein convertase cleavage liberates a fibrillogenic fragment of a resident glycoprotein to initiate melanosome biogenesis. J Cell Biol 161, 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolton W, and Perutz MF (1970). Three dimensional fourier synthesis of horse deoxyhaemoglobin at 2.8 Angstrom units resolution. Nature 228, 551–552. [DOI] [PubMed] [Google Scholar]
- Boyer DR, Li B, Sun C, Fan W, Sawaya MR, Jiang L, and Eisenberg DS (2019). Structures of fibrils formed by α-synuclein hereditary disease mutant H50Q reveal new polymorphs. Nat. Struct. Mol. Biol 26, 1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer DR, Li B, Sun C, Fan W, Zhou K, Hughes MP, Sawaya MR, Jiang L, and Eisenberg DS (2020). The α-synuclein hereditary mutation E46K unlocks a more stable, pathogenic fibril structure. PNAS 117, 3592–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braak H, and Braak E (1991). Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 82, 239–259. [DOI] [PubMed] [Google Scholar]
- Brunello CA, Merezhko M, Uronen R-L, and Huttunen HJ (2020). Mechanisms of secretion and spreading of pathological tau protein. Cell. Mol. Life Sci 77, 1721–1744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryngelson JD, and Wolynes PG (1987). Spin glasses and the statistical mechanics of protein folding. Proc. Natl. Acad. Sci. U.S.A 84, 7524–7528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Boyer DR, Sawaya MR, Ge P, and Eisenberg DS (2019). Cryo-EM structures of four polymorphic TDP-43 amyloid cores. Nat. Struct. Mol. Biol 26, 619–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Q, Boyer DR, Sawaya MR, Ge P, and Eisenberg DS (2020). Cryo-EM structure and inhibitor design of human IAPP (amylin) fibrils. Nat. Struct. Mol. Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caspar DL, and Klug A (1962). Physical principles in the construction of regular viruses. Cold Spring Harb. Symp. Quant. Biol 27, 1–24. [DOI] [PubMed] [Google Scholar]
- Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, and Hultgren SJ (2002). Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295, 851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, and Dobson CM (2017). Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu. Rev. Biochem 86, 27–68. [DOI] [PubMed] [Google Scholar]
- Chiti F, Webster P, Taddei N, Clark A, Stefani M, Ramponi G, and Dobson CM (1999). Designing conditions for in vitro formation of amyloid protofilaments and fibrils. Proc. Natl. Acad. Sci. U.S.A 96, 3590–3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho Y, Challa S, Moquin D, Genga R, Ray TD, Guildford M, and Chan FK-M (2009). Phosphorylation-Driven Assembly of the RIP1-RIP3 Complex Regulates Programmed Necrosis and Virus-Induced Inflammation. Cell 137, 1112–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Rink R, Jong W. de, Siebring J, Vreugd P. de, Boersma FGH, Dijkhuizen L, and Wösten HAB (2003). A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes Dev. 17, 1714–1726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen AS, and Calkins E (1959). Electron microscopic observations on a fibrous component in amyloid of diverse origins. Nature 183, 1202–1203. [DOI] [PubMed] [Google Scholar]
- Collinge J, and Clarke AR (2007). A general model of prion strains and their pathogenicity. Science 318, 930–936. [DOI] [PubMed] [Google Scholar]
- Collins SR, Douglass A, Vale RD, and Weissman JS (2004). Mechanism of Prion Propagation: Amyloid Growth Occurs by Monomer Addition. PLOS Biology 2, e321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon W, and Kelly JW (1992). Partial denaturation of transthyretin is sufficient for amyloid fibril formation in vitro. Biochemistry 31, 8654–8660. [DOI] [PubMed] [Google Scholar]
- Colvin MT, Silvers R, Ni QZ, Can TV, Sergeyev I, Rosay M, Donovan KJ, Michael B, Wall J, Linse S, et al. (2016). Atomic Resolution Structure of Monomorphic Aβ42 Amyloid Fibrils. J. Am. Chem. Soc 138, 9663–9674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dean DN, and Lee JC (2020). Modulating functional amyloid formation via alternative splicing of the premelanosomal protein PMEL17. J. Biol. Chem 295, 7544–7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D, and McLachlan AD (1986). Solvation energy in protein folding and binding. Nature 319, 199–203. [DOI] [PubMed] [Google Scholar]
- Eisenberg DS, and Sawaya MR (2017). Structural Studies of Amyloid Proteins at the Molecular Level. Annu. Rev. Biochem 86, 69–95. [DOI] [PubMed] [Google Scholar]
- Engel MFM, Khemtémourian L, Kleijer CC, Meeldijk HJD, Jacobs J, Verkleij AJ, Kruijff B. de, Killian JA, and Höppener JWM (2008). Membrane damage by human islet amyloid polypeptide through fibril growth at the membrane. PNAS 105, 6033–6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, and Goedert M (2018a). Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 561, 137–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zhang W, Schweighauser M, Murzin AG, Vidal R, Garringer HJ, Ghetti B, Scheres SHW, and Goedert M (2018b). Tau filaments from multiple cases of sporadic and inherited Alzheimer’s disease adopt a common fold. Acta Neuropathol. 136, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zhang W, Murzin AG, Murshudov G, Garringer HJ, Vidal R, Crowther RA, Ghetti B, Scheres SHW, and Goedert M (2018c). Structures of filaments from Pick’s disease reveal a novel tau protein fold. Nature 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcon B, Zivanov J, Zhang W, Murzin AG, Garringer HJ, Vidal R, Crowther RA, Newell KL, Ghetti B, Goedert M, et al. (2019). Novel tau filament fold in chronic traumatic encephalopathy encloses hydrophobic molecules. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fändrich M, Fletcher MA, and Dobson CM (2001). Amyloid fibrils from muscle myoglobin. Nature 410, 165–166. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick AWP, Falcon B, He S, Murzin AG, Murshudov G, Garringer HJ, Crowther RA, Ghetti B, Goedert M, and Scheres SHW (2017). Cryo-EM structures of tau filaments from Alzheimer’s disease. Nature 547, 185–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Alory-Jost C, Marks MS, Balch WE, and Kelly JW (2006). Functional amyloid formation within mammalian tissue. PLoS Biol 4, e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Koulov AV, Balch WE, and Kelly JW (2007). Functional amyloid--from bacteria to humans. Trends Biochem. Sci 32, 217–224. [DOI] [PubMed] [Google Scholar]
- Frey S, Richter RP, and Görlich D (2006). FG-rich repeats of nuclear pore proteins form a three-dimensional meshwork with hydrogel-like properties. Science 314, 815–817. [DOI] [PubMed] [Google Scholar]
- Galkin VE, Orlova A, Schröder GF, and Egelman EH (2010). Structural polymorphism in F-actin. Nature Structural & Molecular Biology 17, 1318–1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaspar R, Lund M, Sparr E, and Linse S (2020). Anomalous Salt Dependence Reveals an Interplay of Attractive and Repulsive Electrostatic Interactions in α-synuclein Fibril Formation. QRB Discovery 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelenter MD, Smith KJ, Liao S-Y, Mandala VS, Dregni AJ, Lamm MS, Tian Y, Xu W, Pochan DJ, Tucker TJ, et al. (2019). The peptide hormone glucagon forms amyloid fibrils with two coexisting β-strand conformations. Nature Structural & Molecular Biology 26, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goedert M (2015). Alzheimer’s and Parkinson’s diseases: The prion concept in relation to assembled Aβ, tau, and α-synuclein. Science 349. [DOI] [PubMed] [Google Scholar]
- Goldsbury C, Kistler J, Aebi U, Arvinte T, and Cooper GJ (1999). Watching amyloid fibrils grow by time-lapse atomic force microscopy. J. Mol. Biol 285, 33–39. [DOI] [PubMed] [Google Scholar]
- Goldschmidt L, Teng PK, Riek R, and Eisenberg D (2010). Identifying the amylome, proteins capable of forming amyloid-like fibrils. Proc. Natl. Acad. Sci. U.S.A 107, 3487–3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald J, Kwiatkowski W, and Riek R (2018). Peptide Amyloids in the Origin of Life. J. Mol. Biol 430, 3735–3750. [DOI] [PubMed] [Google Scholar]
- Guenther EL, Ge P, Trinh H, Sawaya MR, Cascio D, Boyer DR, Gonen T, Zhou ZH, and Eisenberg DS (2018a). Atomic-level evidence for packing and positional amyloid polymorphism by segment from TDP-43 RRM2. Nat. Struct. Mol. Biol 25, 311–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guenther EL, Cao Q, Trinh H, Lu J, Sawaya MR, Cascio D, Boyer DR, Rodriguez JA, Hughes MP, and Eisenberg DS (2018b). Atomic structures of TDP-43 LCD segments and insights into reversible or pathogenic aggregation. Nat. Struct. Mol. Biol 25, 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero-Ferreira R, Taylor NM, Arteni A-A, Kumari P, Mona D, Ringler P, Britschgi M, Lauer ME, Makky A, Verasdonck J, et al. (2019). Two new polymorphic structures of human full-length alpha-synuclein fibrils solved by cryo-electron microscopy. ELife 8, e48907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gui X, Luo F, Li Y, Zhou H, Qin Z, Liu Z, Gu J, Xie M, Zhao K, Dai B, et al. (2019). Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat Commun 10, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guijarro JI, Sunde M, Jones JA, Campbell ID, and Dobson CM (1998). Amyloid fibril formation by an SH3 domain. Proc. Natl. Acad. Sci. U.S.A 95, 4224–4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O’Donovan K, Fare CM, Diaz Z, Singh N, et al. (2018). Nuclear-Import Receptors Reverse Aberrant Phase Transitions of RNA-Binding Proteins with Prion-like Domains. Cell 173, 677–692.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustavsson A, Engström U, and Westermark P (1991). Normal transthyretin and synthetic transthyretin fragments form amyloid-like fibrils in vitro. Biochem Biophys Res Commun 175, 1159–1164. [DOI] [PubMed] [Google Scholar]
- Haslberger T, Bukau B, and Mogk A (2010). Towards a unifying mechanism for ClpB/Hsp104-mediated protein disaggregation and prion propagation. Biochem. Cell Biol 88, 63–75. [DOI] [PubMed] [Google Scholar]
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al. (2015). Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol 210, 529–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervas R, Rau MJ, Park Y, Zhang W, Murzin AG, Fitzpatrick JAJ, Scheres SHW, and Si K (2020). Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 367, 1230–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman DM, John CM, and Goate A (2011). Alzheimer’s Disease: The Challenge of the Second Century. Sci Transl Med 3, 77sr1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes MP, Sawaya MR, Boyer DR, Goldschmidt L, Rodriguez JA, Cascio D, Chong L, Gonen T, and Eisenberg DS (2018). Atomic structures of low-complexity protein segments reveal kinked β sheets that assemble networks. Science 359, 698–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett JT, and Lansbury PT (1993). Seeding “one-dimensional crystallization” of amyloid: A pathogenic mechanism in Alzheimer’s disease and scrapie? Cell 73, 1055–1058. [DOI] [PubMed] [Google Scholar]
- Johnson KH, O’Brien TD, Betsholtz C, and Westermark P (1992). Islet amyloid polypeptide: mechanisms of amyloidogenesis in the pancreatic islets and potential roles in diabetes mellitus. Lab Invest 66, 522–535. [PubMed] [Google Scholar]
- Jucker M, and Walker LC (2011). Pathogenic Protein Seeding in Alzheimer’s Disease and Other Neurodegenerative Disorders. Ann Neurol 70, 532–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Baxa U, Wickner RB, and Steven AC (2004). A model for Ure2p prion filaments and other amyloids: the parallel superpleated beta-structure. Proc. Natl. Acad. Sci. U.S.A. 101, 7885–7890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kajava AV, Aebi U, and Steven AC (2005). The parallel superpleated beta-structure as a model for amyloid fibrils of human amylin. J. Mol. Biol 348, 247–252. [DOI] [PubMed] [Google Scholar]
- Kato M, Han TW, Xie S, Shi K, Du X, Wu LC, Mirzaei H, Goldsmith EJ, Longgood J, Pei J, et al. (2012). Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell 149, 753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus A, Hoyt F, Schwartz CL, Hansen B, Hughson AG, Artikis E, Race B, and Caughey B (2021). Structure of an infectious mammalian prion. BioRxiv 2021.02.14.431014. [DOI] [PubMed] [Google Scholar]
- Li B, Ge P, Murray KA, Sheth P, Zhang M, Nair G, Sawaya MR, Shin WS, Boyer DR, Ye S, et al. (2018). Cryo-EM of full-length α-synuclein reveals fibril polymorphs with a common structural kernel. Nat Commun 9, 3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao Y-S, Damko E, Moquin D, Walz T, McDermott A, et al. (2012). The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Sanchez CP, Slaughter BD, Zhao Y, Khan MR, Unruh JR, Rubinstein B, and Si K (2016). A Putative Biochemical Engram of Long-Term Memory. Current Biology 26, 3143–3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberta F, Loerch S, Rennegarbe M, Schierhorn A, Westermark P, Westermark GT, Hazenberg BPC, Grigorieff N, Fändrich M, and Schmidt M (2019). Cryo-EM fibril structures from systemic AA amyloidosis reveal the species complementarity of pathological amyloids. Nat Commun 10, 1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Cao Q, Hughes MP, Sawaya MR, Boyer DR, Cascio D, and Eisenberg DS (2020). CryoEM structure of the low-complexity domain of hnRNPA2 and its conversion to pathogenic amyloid. Nature Communications 11, 4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macindoe I, Kwan AH, Ren Q, Morris VK, Yang W, Mackay JP, and Sunde M (2012). Self-assembly of functional, amphipathic amyloid monolayers by the fungal hydrophobin EAS. Proc. Natl. Acad. Sci. U.S.A 109, E804–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maji SK, Perrin MH, Sawaya MR, Jessberger S, Vadodaria K, Rissman RA, Singru PS, Nilsson KPR, Simon R, Schubert D, et al. (2009). Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGlinchey RP, and Lee JC (2017). Reversing the amyloid trend: Mechanism of fibril assembly and dissolution of the repeat domain from a human functional amyloid. Isr. J. Chem 57, 613–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meersman F, and Dobson CM (2006). Probing the pressure–temperature stability of amyloid fibrils provides new insights into their molecular properties. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics 1764, 452–460. [DOI] [PubMed] [Google Scholar]
- Molliex A, Temirov J, Lee J, Coughlin M, Kanagaraj AP, Kim HJ, Mittag T, and Taylor JP (2015). Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell 163, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mompeán M, Li W, Li J, Laage S, Siemer AB, Bozkurt G, Wu H, and McDermott AE (2018). The Structure of the Necrosome RIPK1-RIPK3 Core, a Human Hetero-Amyloid Signaling Complex. Cell 173, 1244–1253.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray DT, Kato M, Lin Y, Thurber KR, Hung I, McKnight SL, and Tycko R (2017). Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 171, 615–627.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson R, Sawaya MR, Balbirnie M, Madsen AØ, Riekel C, Grothe R, and Eisenberg D (2005). Structure of the cross-beta spine of amyloid-like fibrils. Nature 435, 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nespovitaya N, Gath J, Barylyuk K, Seuring C, Meier BH, and Riek R (2016). Dynamic Assembly and Disassembly of Functional β-Endorphin Amyloid Fibrils. J. Am. Chem. Soc 138, 846–856. [DOI] [PubMed] [Google Scholar]
- Oelschlegel AM, and Weissmann C (2013). Acquisition of drug resistance and dependence by prions. PLoS Pathog 9, e1003158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen D, and Riek R (2019). Functional Amyloids. Cold Spring Harb Perspect Biol 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peelaerts W, Bousset L, Van der Perren A, Moskalyuk A, Pulizzi R, Giugliano M, Van den Haute C, Melki R, and Baekelandt V (2015). α-Synuclein strains cause distinct synucleinopathies after local and systemic administration. Nature 522, 340–344. [DOI] [PubMed] [Google Scholar]
- Peng C, Gathagan RJ, Covell DJ, Medellin C, Stieber A, Robinson JL, Zhang B, Pitkin RM, Olufemi MF, Luk KC, et al. (2018). Cellular milieu imparts distinct pathological α-synuclein strains in α-synucleinopathies. Nature 557, 558–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrett S, Pham CLL, Kwan AH, and Sunde M (2014). Functional amyloid: widespread in Nature, diverse in purpose. Essays Biochem 56, 207–219. [DOI] [PubMed] [Google Scholar]
- Perutz MF, Johnson T, Suzuki M, and Finch JT (1994). Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl. Acad. Sci. U.S.A 91, 5355–5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkova AT, Leapman RD, Guo Z, Yau W-M, Mattson MP, and Tycko R (2005). Self-Propagating, Molecular-Level Polymorphism in Alzheimer’s ß-Amyloid Fibrils. Science 307, 262–265. [DOI] [PubMed] [Google Scholar]
- Prusiner SB (1998). Prions. Proc. Natl. Acad. Sci. U.S.A 95, 13363–13383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner SB, Woerman AL, Mordes DA, Watts JC, Rampersaud R, Berry DB, Patel S, Oehler A, Lowe JK, Kravitz SN, et al. (2015). Evidence for α-synuclein prions causing multiple system atrophy in humans with parkinsonism. Proc. Natl. Acad. Sci. U.S.A 112, E5308–5317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riek R, and Eisenberg DS (2016). The activities of amyloids from a structural perspective. Nature 539, 227–235. [DOI] [PubMed] [Google Scholar]
- Rodriguez JA, Ivanova MI, Sawaya MR, Cascio D, Reyes FE, Shi D, Sangwan S, Guenther EL, Johnson LM, Zhang M, et al. (2015). Structure of the toxic core of α-synuclein from invisible crystals. Nature 525, 486–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rufo CM, Moroz YS, Moroz OV, Stöhr J, Smith TA, Hu X, DeGrado WF, and Korendovych IV (2014). Short peptides self-assemble to produce catalytic amyloids. Nat Chem 6, 303–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saad S, Cereghetti G, Feng Y, Picotti P, Peter M, and Dechant R (2017). Reversible protein aggregation is a protective mechanism to ensure cell cycle restart after stress. Nat Cell Biol 19, 1202–1213. [DOI] [PubMed] [Google Scholar]
- Saborio GP, Permanne B, and Soto C (2001). Sensitive detection of pathological prion protein by cyclic amplification of protein misfolding. Nature 411, 810–813. [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJW, McFarlane HT, et al. (2007). Atomic structures of amyloid cross-beta spines reveal varied steric zippers. Nature 447, 453–457. [DOI] [PubMed] [Google Scholar]
- Scheres SH, Zhang W, Falcon B, and Goedert M (2020). Cryo-EM structures of tau filaments. Curr. Opin. Struct. Biol 64, 17–25. [DOI] [PubMed] [Google Scholar]
- Schweighauser M, Shi Y, Tarutani A, Kametani F, Murzin AG, Ghetti B, Matsubara T, Tomita T, Ando T, Hasegawa K, et al. (2020). Structures of α-synuclein filaments from multiple system atrophy. Nature 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seuring C, Verasdonck J, Gath J, Ghosh D, Nespovitaya N, Wälti MA, Maji SK, Cadalbert R, Güntert P, Meier BH, et al. (2020). The three-dimensional structure of human β-endorphin amyloid fibrils. Nat Struct Mol Biol. [DOI] [PubMed] [Google Scholar]
- Shahnawaz M, Mukherjee A, Pritzkow S, Mendez N, Rabadia P, Liu X, Hu B, Schmeichel A, Singer W, Wu G, et al. (2020). Discriminating α-synuclein strains in Parkinson’s disease and multiple system atrophy. Nature 578, 273–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Zhang W, Yang Y, Murzin A, Falcon B, Kotecha A, Beers M. van, Tarutani A, Kametani F, Garringer HJ, et al. (2021). Structure-based Classification of Tauopathies. BioRxiv 2021.05.28.446130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si K, Lindquist S, and Kandel ER (2003). A Neuronal Isoform of the Aplysia CPEB Has Prion-Like Properties. Cell 115, 879–891. [DOI] [PubMed] [Google Scholar]
- Sipe JD, and Cohen AS (2000). Review: History of the Amyloid Fibril. Journal of Structural Biology 130, 88–98. [DOI] [PubMed] [Google Scholar]
- Sunde M, Serpell LC, Bartlam M, Fraser PE, Pepys MB, and Blake CC (1997). Common core structure of amyloid fibrils by synchrotron X-ray diffraction. J. Mol. Biol 273, 729–739. [DOI] [PubMed] [Google Scholar]
- Suzuki G, Imura S, Hosokawa M, Katsumata R, Nonaka T, Hisanaga S-I, Saeki Y, and Hasegawa M (2020). α-synuclein strains that cause distinct pathologies differentially inhibit proteasome. ELife 9, e56825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taneja V, Maddelein M-L, Talarek N, Saupe SJ, and Liebman SW (2007). A Non-Q/N-Rich Prion Domain of a Foreign Prion, [Het-s], Can Propagate as a Prion in Yeast. Molecular Cell 27, 67–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teng PK, Anderson NJ, Goldschmidt L, Sawaya MR, Sambashivan S, and Eisenberg D (2012). Ribonuclease A suggests how proteins self-chaperone against amyloid fiber formation. Protein Sci. 21, 26–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuite MF, and Serio TR (2010). The prion hypothesis: from biological anomaly to basic regulatory mechanism. Nat. Rev. Mol. Cell Biol 11, 823–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villali J, Dark J, Brechtel TM, Pei F, Sindi SS, and Serio TR (2020). Nucleation seed size determines amyloid clearance and establishes a barrier to prion appearance in yeast. Nat. Struct. Mol. Biol 27, 540–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogler TO, Wheeler JR, Nguyen ED, Hughes MP, Britson KA, Lester E, Rao B, Betta ND, Whitney ON, Ewachiw TE, et al. (2018). TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 563, 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälti MA, Ravotti F, Arai H, Glabe CG, Wall JS, Böckmann A, Güntert P, Meier BH, and Riek R (2016). Atomic-resolution structure of a disease-relevant Aβ(1–42) amyloid fibril. Proc. Natl. Acad. Sci. U.S.A 113, E4976–4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wälti MA, Schmidt T, Murray DT, Wang H, Hinshaw JE, and Clore GM (2017). Chaperonin GroEL accelerates protofibril formation and decorates fibrils of the Het-s prion protein. PNAS 114, 9104–9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasmer C, Lange A, Van Melckebeke H, Siemer AB, Riek R, and Meier BH (2008). Amyloid Fibrils of the HET-s(218–289) Prion Form a β Solenoid with a Triangular Hydrophobic Core. Science 319, 1523–1526. [DOI] [PubMed] [Google Scholar]
- Xiang S, Kato M, Wu LC, Lin Y, Ding M, Zhang Y, Yu Y, and McKnight SL (2015). The LC Domain of hnRNPA2 Adopts Similar Conformations in Hydrogel Polymers, Liquid-like Droplets, and Nuclei. Cell 163, 829–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-S, Kato M, Wu X, Litsios A, Sutter BM, Wang Y, Hsu C-H, Wood NE, Lemoff A, Mirzaei H, et al. (2019). Yeast Ataxin-2 Forms an Intracellular Condensate Required for the Inhibition of TORC1 Signaling during Respiratory Growth. Cell 177, 697–710.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Falcon B, Murzin AG, Fan J, Crowther RA, Goedert M, and Scheres SH (2019). Heparin-induced tau filaments are polymorphic and differ from those in Alzheimer’s and Pick’s diseases. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Tarutani A, Newell KL, Murzin AG, Matsubara T, Falcon B, Vidal R, Garringer HJ, Shi Y, Ikeuchi T, et al. (2020). Novel tau filament fold in corticobasal degeneration. Nature 580, 283–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Lim Y-J, Liu Z, Long H, Sun Y, Hu J-J, Zhao C, Tao Y, Zhang X, Li D, et al. (2020). Parkinson’s disease-related phosphorylation at Tyr39 rearranges α-synuclein amyloid fibril structure revealed by cryo-EM. PNAS. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.