Abstract
Persistent inactivity promotes skeletal muscle atrophy, marked by mitochondrial aberrations that affect strength, mobility, and metabolic health leading to the advancement of disease. Mitochondrial quality control (MQC) pathways include biogenesis (synthesis), mitophagy/lysosomal turnover, and the mitochondrial unfolded protein response, which serve to maintain an optimal organelle network. Tumor suppressor p53 has been implicated in regulating muscle mitochondria in response to cellular stress; however, its role in the context of muscle disuse has yet to be explored, and whether p53 is necessary for MQC remains unclear. To address this, we subjected p53 muscle-specific KO (mKO) and WT mice to unilateral denervation. Transcriptomic and pathway analyses revealed dysregulation of pathways pertaining to mitochondrial function, and especially turnover, in mKO muscle following denervation. Protein and mRNA data of the MQC pathways indicated activation of the mitochondrial unfolded protein response and mitophagy–lysosome systems along with reductions in mitochondrial biogenesis and content in WT and mKO tissue following chronic denervation. However, p53 ablation also attenuated the expression of autophagy–mitophagy machinery, reduced autophagic flux, and enhanced lysosomal dysfunction. While similar reductions in mitochondrial biogenesis and content were observed between genotypes, MQC dysregulation exacerbated mitochondrial dysfunction in mKO fibers, evidenced by elevated reactive oxygen species. Moreover, acute experiments indicate that p53 mediates the expression of transcriptional regulators of MQC pathways as early as 1 day following denervation. Together, our data illustrate exacerbated mitochondrial dysregulation with denervation stress in p53 mKO tissue, thus indicating that p53 contributes to organellar maintenance via regulation of MQC pathways during muscle atrophy.
Keywords: skeletal muscle, mitochondria, p53, muscle atrophy, mitochondrial quality control, mitophagy, lysosome, mitochondrial biogenesis, unfolded protein response, transcriptomics
Abbreviations: ATF4, activating transcription factor 4; ATF5, activating transcription factor 5; Atg, autophagy-related protein; cDNA, complementary DNA; CHOP, C/EBP homologous protein; ClpP, caseinolytic mitochondrial matrix peptidase proteolytic subunit; CON, control; COX, cytochrome c oxidase; CtsD, cathepsin D; DEN, denervated; EDL, extensor digitorum longus; GO, Gene Ontology; GSEA, gene set enrichment analysis; HSP, heat shock protein; LC3, microtubule-associated proteins 1a/1b light chain 3; log2FC, log2 fold change; LonP, lon protease homolog, mitochondrial; mKO, muscle-specific KO; MQC, mitochondrial quality control; mtDNA, mitochondrial DNA; p62, sequestosome-1; PGC-1α, peroxisome proliferator–activated receptor gamma coactivator 1 alpha; PINK1, phosphatase and tensin homolog–induced kinase 1; Prkn, Parkin; qPCR, quantitative PCR; REVIGO, reduce and visualize Gene Ontology; ROS, reactive oxygen species; SDH, succinate dehydrogenase; SirT3, sirtuin 3; TA, tibialis anterior; TFE3, transcription factor E3; TFEB, transcription factor EB; UPRmt, mitochondrial unfolded protein response
The influence of skeletal muscle, which accounts for as much as 40% of body mass in lean individuals, has extended implications for mobility, injury, and metabolic diseases, and thus has a major impact on overall quality of life (1, 2, 3, 4). Indeed, a unique and natural feature of aging is the progressive loss of muscle mass, which coincides with diminished aerobic capacity and muscle strength (2, 5, 6). Within muscle, mitochondria are the organelles responsible for the maintenance of metabolic health and contribute to the progression of muscle dysfunction (7). Largely responsible for the provision of ATP through aerobic respiration, mitochondria are also integral to the regulation of oxidative stress and antioxidants, proteostasis, apoptosis, inflammation, and Ca2+ handling (8, 9, 10, 11). Mitochondrial quality control (MQC) involves the coordination of multiple processes that converge on the organelle network to regulate their synthesis (biogenesis), breakdown (mitophagy), and protein handling ability via the mitochondrial unfolded protein response (UPRmt) in order to maintain a healthy mitochondrial pool (7). As such, mitochondrial content and function within muscle is under constant regulation to match the internal metabolic capability with the external demand being placed on the tissue.
Peroxisome proliferator–activated receptor gamma coactivator 1 alpha (PGC-1α) is considered the master regulator of mitochondrial biogenesis, but it has also been implicated in regulating mitochondrial breakdown via the autophagy–lysosome system and through a coordinated regulation with transcription factor EB (TFEB) (12, 13). Thus, this transcriptional regulator plays a dual role in mediating the crosstalk between synthesis and degradation. However, in PGC-1α KO animals subjected to denervation-induced disuse, the coincident induction of autophagy and lysosomal proteins was only partially attenuated (13), suggesting the potential for additional regulators of mitochondrial plasticity following prolonged inactivity. The transcription factor p53 is one such candidate that has been implicated in regulating mitochondria through multiple mechanisms. p53 is a tumor suppressor protein that monitors the cell environment and mounts appropriate action in response to a variety of cellular stressors and is aptly named “the guardian of the genome” (14, 15). Our laboratory has demonstrated that the lack of p53 contributes to diminished exercise performance as well as reduced mitochondrial function, exercise signaling, and mitochondrial DNA (mtDNA) expression in skeletal muscle, implicating its role in moderating mitochondria within skeletal muscle in response to stress (16, 17, 18, 19, 20).
While p53 is identified as a regulator of mitochondrial plasticity with exercise, as well as skeletal muscle atrophy, it has yet been determined whether p53 is required for the regulation of mitochondrial quality, particularly in the context of muscle disuse. In addition, the magnitude of the contribution of p53 to the regulation of mitochondrial content and function in muscle remains up for debate, with others reporting similar mitochondrial profiles in WT muscle, as well as muscle lacking p53 under basal conditions (21). Therefore, our aim was to resolve these issues by performing transcriptomic analysis of over 20,000 genes, along with pathway analysis to determine the transcripts and associated cellular processes that are most affected by the ablation of p53 in muscle, as well as the interaction of p53 ablation with muscle disuse. The transcriptomic data comparing WT and p53 muscle-specific KO (mKO) revealed the differential expression of genes related to mitochondrial remodeling and the discrepant induction of pathways governing MQC. We elaborated on these findings by measuring key markers of mitochondrial biogenesis and content, along with organelle function, and mitochondrial turnover via the mitophagy–lysosome system. We hypothesized that muscle lacking p53 would display further decrements in mitochondrial content and function as a result of impaired induction of MQC pathways. Our data indicate that mitochondrial turnover via the mitophagy–lysosome system is blunted when p53 is absent in denervated (DEN) muscle, and this likely contributes to perturbations in the organelle milieu.
Results
Confirmation of p53 mKO and characterization of the 7-day DEN hindlimb muscle
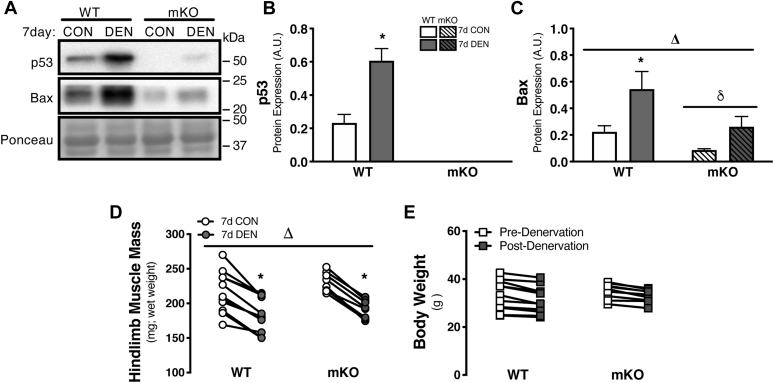
To confirm the effectiveness of our model, we assessed p53 protein expression in WT and mKO animals following 7 days of denervation. It is well documented that p53 is responsive to chronic muscle disuse, which leads to the induction of proapoptotic proteins and thus contributes to muscle atrophy (22, 23). Indeed, we observed a nearly threefold increase in p53 protein expression in WT DEN muscle, whereas p53 protein expression was effectively ablated in the control (CON) and DEN hindlimb of mKO animals (Fig. 1, A and B). Moreover, the proapoptotic protein, and downstream effector of p53, Bax, exhibited a similar threefold induction with denervation in WT tissue, and the overall expression of Bax protein was attenuated in mKO animals, as anticipated (Fig. 1, A and C). We observed no effect of genotype on the collective muscle mass of the hindlimb either prior to the intervention, or brought about by sciatic denervation, as similar 15 to 18% reductions were observed following 7 days of disuse (Fig. 1D). There was also no observable effect on overall body weight in either WT or mKO animals (Fig. 1E).
Figure 1.
Characteristics of p53 mKO with 7 days of hindlimb denervation.A–C, representative whole-muscle Western blots and corresponding graphs for p53 and Bax protein expression following 7 days of denervation. D, hindlimb mass was reduced in both genotypes after chronic muscle disuse. E, body weight was not affected by either genotype or denervation. Bars depict means ± SE; n = 3 to 5 (A–C), n = 8 to 11 (D and E); Δp < 0.05, main effect of denervation; δp < 0.05, main effect of genotype; ∗p < 0.05, DEN versus CON of same genotype by repeated-measures two-way ANOVA. CON, control; DEN, denervated; mKO, muscle-specific KO.
Global transcriptomic signatures of DEN muscle in WT and p53 mKO hindlimbs
To thoroughly characterize the differences in gene expression in the muscle of WT and p53 mKO animals, we performed RNA-Seq analysis to elucidate the divergence in the transcript signatures both basally and following chronic disuse from 7-day DEN tibialis anterior (TA) muscle, along with contralateral CON samples, from both WT and p53 mKO animals. Of the 20,400 transcripts that were measured, 10,416 genes were significantly affected by chronic denervation in WT mice, whereas 10,528 targets were differentially expressed in p53 mKO samples. Moreover, principal component analysis revealed that 95% of the variance was accounted for by the effect of denervation, indicating the pronounced effect that denervation has in altering the expression profile of muscle (Figs. S4 and S5). Surprisingly, only 3% of variance was attributed to the effect of genotype alone. However, the interaction of genotype with denervation revealed discrepancies between WT and muscle lacking p53 in the denervation response at the pathway level (Figs. 2 and 3), which we further explored in the context of mitochondrial regulation and remodeling and describe later.
Figure 2.
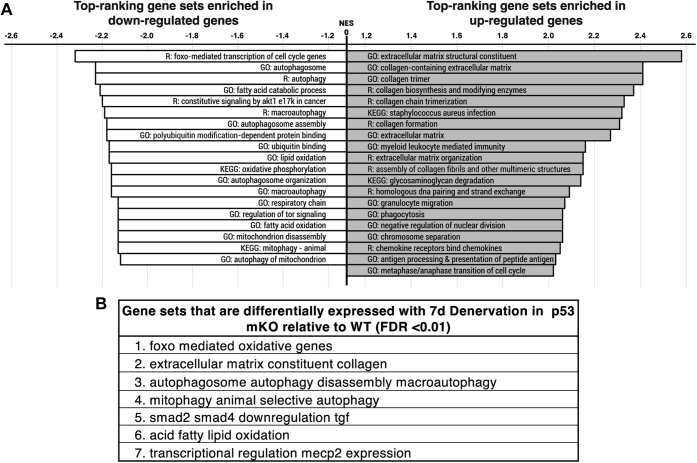
GSEA pathway analysis of 7-day denervated muscle.A, standard GSEA pathway was performed to investigate whether the effects of denervation were modulated by p53 on a more global scale. To address the question of whether the effect of denervation differs across genotypes, an interaction term was included in the statistical model for detection of differentially expressed genes. Normalized enrichment scores (NESs) were determined for the top-ranking gene sets that were enriched in upregulated and downregulated genes based on this interaction term (Table S4). B, gene sets most affected by the interaction of denervation and p53 mKO (FDR <0.01). FDR, false discovery rate; GSEA, gene set enrichment analysis; mKO, muscle-specific KO.
Figure 3.
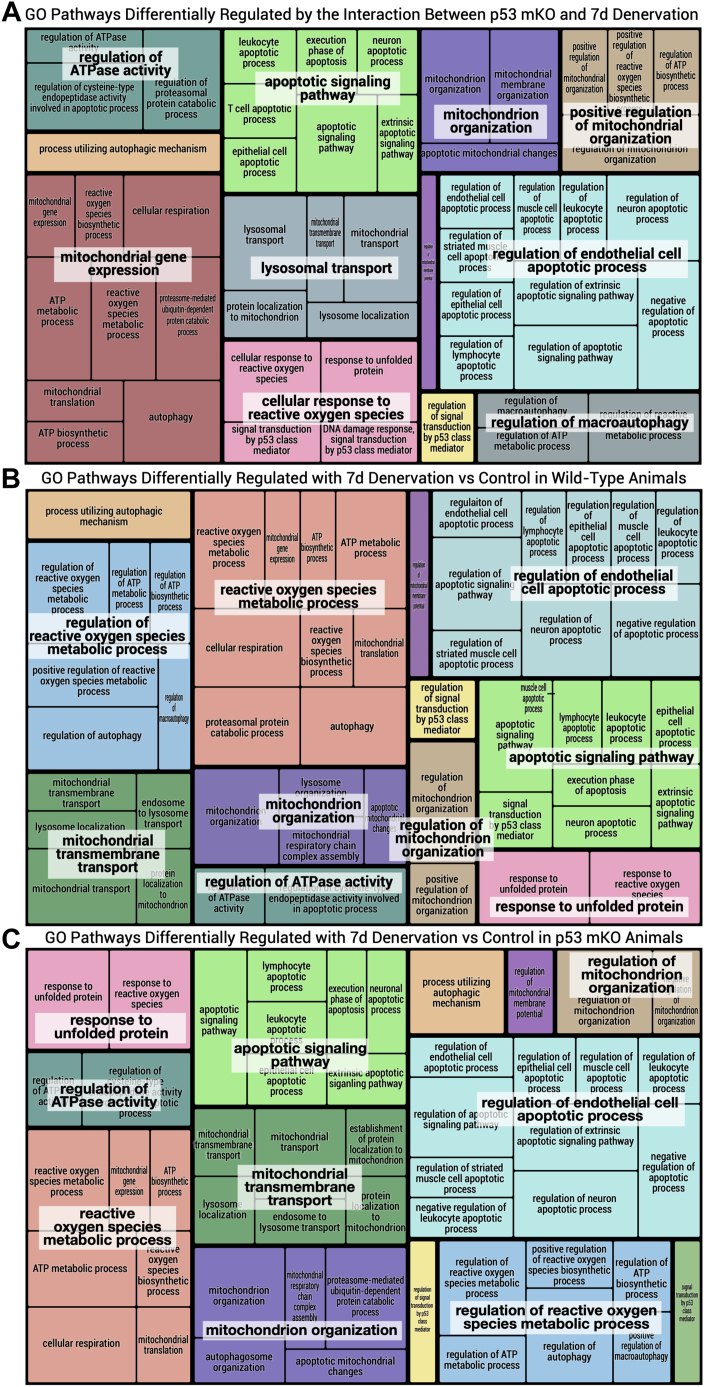
REVIGO (reduce and visualize GO) tree map of GO terms elevated in response to denervation. GO terms pertaining to mitochondrial quality control were selected and used to generate tree maps based on their p value as described previously using the REVIGO web-based application (24). Selection terms for GO pathways included “mitochondria,” “autophagy,” “mitophagy,” “lysosome,” “ROS” or “reactive oxygen species,” “UPR” or “unfolded protein response,” “apoptosis,” “p53,” “protein synthesis,” and “degradation” were used to filter GO terms in order to represent the unique pattern of differentially expressed gene sets in WT versus p53 mKO animals with 7 days of hindlimb denervation. A, tree map representing the GO pathways that are differentially regulated by denervation in p53 mKO as compared with WT animals (based on the analysis of the interaction of genotype and denervation). B and C, individual tree maps of GO pathways that are differentially regulated by chronic disuse in WT animals and p53 mKO animals, respectively. GO, Gene Ontology; mKO, muscle-specific KO.
Chronic denervation–induced differential expression of gene sets in WT and p53 mKO animals
To investigate whether the effects of denervation were modulated by p53 on a more global scale, standard enrichment/pathway analysis was performed. Standard pathway analysis includes GOSeq enrichment (threshold-based method) and gene set enrichment analysis (GSEA) (threshold-free method) methodologies, as each perform independent assessment of the experimental results. However, GOSeq was deemed to not have sufficient power for a meaningful enrichment test, thus only GSEA enrichment analysis was performed. To test whether genotype affected gene expression differentially in the two treatments, an interaction term was included in the statistical model for detection of differentially expressed genes based on the combined effect of genotype with denervation. From this analysis, we compiled a list of the pathways that were most affected by chronic denervation based on genes that were either collectively upregulated or downregulated (Fig. 2A). Most top-ranking gene sets (15 of 19 gene sets) enriched in downregulated genes pertain to regulators of mitochondrial function and turnover, whereas the pathways that were enriched in upregulated genes were not relevant to mitochondrial regulation processes (Fig. 2A). Indeed, when we assessed the most differentially affected pathways without segregating the genes based on whether they were upregulated or downregulated, we determined that four of the seven identified pathways most affected by the absence of p53 with denervation relate to MQC (Fig. 2B). Given the indication that mitochondrial regulatory pathways were particularly susceptible to denervation when p53 is absent from muscle, we then filtered Gene Ontology (GO) terms for specific pathways that govern mitochondria. From the list of MQC-relevant GO terms, tree maps were generated using reduce and visualize Gene Ontology (REVIGO) (24) based on their corresponding p values to visualize the specific pathways that were most different with denervation in WT animals, p53 mKO animals, or as a result of the interaction of mKO with denervation (Fig. 3, A–C). Evidently, WT and p53 mKO muscle display discrepant patterns in their response to 7 days of denervation, when assessing those pathways pertaining to mitochondrial regulation (Fig. 3, B and C). In particular, GO terms related to “mitochondrion gene expression,” “lysosomal transport,” and “regulation of macroautophagy” were uniquely identified in our analysis of pathways affected by the interaction of p53 mKO with denervation (Fig. 3A). Thus, the RNA-Seq data revealed a notable disparity in the MQC response to denervation in the absence of p53.
Effect of p53 ablation on disuse-induced changes in mitochondrial biogenesis, content, and function with chronic disuse
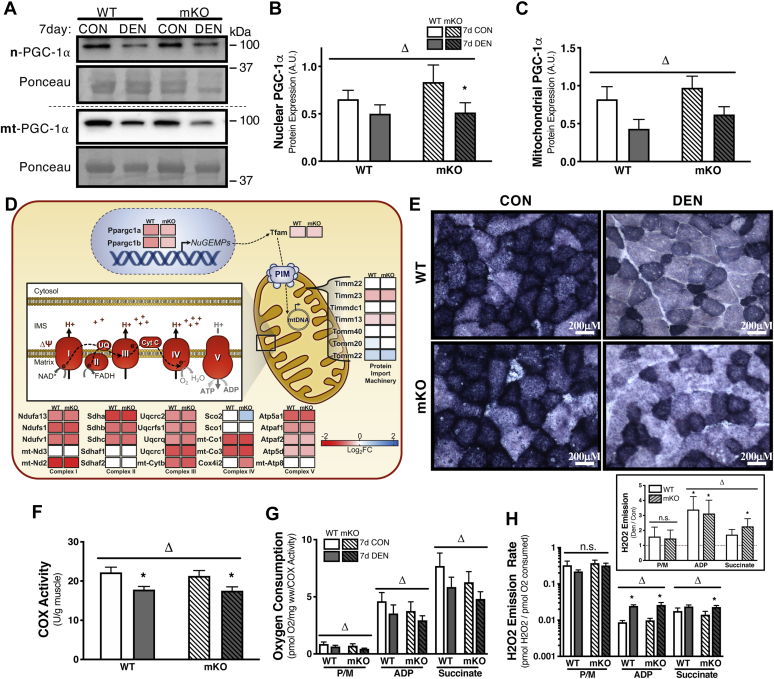
We next wanted to identify whether the altered transcriptome profile within p53 mKO tissue manifests in mitochondrial aberrations. To address this question, our first objective was to compare the mitochondrial phenotype in WT and p53 mKO muscle by measuring markers of mitochondrial biogenesis, content, and function. The transcriptional coactivator, PGC-1α, is widely regarded as the master regulator of mitochondrial biogenesis, which has been localized in both nuclear and mitochondrial compartments, and it has been reported to be regulated by p53. Therefore, we wanted to determine whether the total expression, as well as the subcellular distribution of nuclear and mitochondrial PGC-1α, was affected by the absence of p53. We observed an overall reduction in PGC-1α protein content in both WT and mKO animals following 7 days of disuse (Fig. 4, A–C). However, DEN muscle of mKO animals displayed a more pronounced 33% reduction in nuclear PGC-1α, as compared with the 24% decrease observed in WT DEN tissue (Fig. 4, A and B). Interestingly, this discrepancy in the denervation-induced reduction in PGC-1α content was not observed in the mitochondrial subfraction, as both WT and mKO animals exhibited similar ∼25% decreases in mitochondrial PGC-1α localization following 7 days of disuse (Fig. 4, A and C). To further elucidate the influence of p53 on mitochondrial remodeling with chronic disuse, we assessed the transcript expression profiles of select genes involved in mitochondrial content and biogenesis as determined by RNA-Seq analysis (Fig. 4D and Table S3). Transcriptional regulators of mitochondrial biogenesis, PGC-1α and PGC-1β (Ppargc1a and Ppargc1b), as well as the mtDNA transcriptional regulator, Tfam, were all reduced with 7 days of denervation in both WT and p53 mKO animals; however, PGC-1α and PGC-1β displayed greater reductions in WT relative to p53 mKO tissue (WT, −0.68 log2 fold change [log2FC] versus mKO, −0.44 log2FC; Fig. 4D and Table S3). A subset of genes encoding components of the mitochondrial protein import machinery was also assessed, which revealed similar decreases in the inner membrane import components Timm23 and Timm13 in WT and p53 mKO tissue (Timm23: WT, −0.47 log2FC versus mKO, −0.50 log2FC; Timm13: WT, −0.27 log2FC versus mKO, −0.29 log2FC; Fig. 4D and Table S3). The outer-membrane import receptor, Tomm22, was upregulated in both WT and p53 mKO mice (WT, 0.42 log2FC versus 0.50 log2FC) with chronic denervation, whereas Tomm20 was induced in WT muscle but was not changed in the p53 mKO DEN hindlimb (WT, 0.20 log2FC versus mKO, no change; Fig. 4D and Table S3). In addition, we analyzed a selection of genes encoding subunit proteins of the respiratory chain complexes. Overall, complexes I, II, III, and V displayed a similar pattern of reduction in gene expression with disuse in animals of either genotype (Fig. 4D and Table S3). However, the complex IV subunit Cox4i2 and the assembly factor Sco2 were upregulated and downregulated, respectively, in p53 mKO animals, with no change in their expression observed in WT muscle (Cox4i2: WT, no change versus mKO, −0.90 log2FC; Sco2: WT, no change versus mKO, 0.63 log2FC; Fig. 4D and Table S3).
Figure 4.
Mitochondrial biogenesis, content, and function with chronic disuse.A–C, representative Western blots and corresponding graphs showing nuclear- and mitochondrial-localized PGC-1α subsequent to denervation. D, heat map of DESeq RNA-Seq data showing differential expression of select genes in the context of mitochondrial biogenesis. The data are expressed as log2 fold change following 7 days of denervation as compared with control (reduced gene expression, red; enhanced gene abundance, blue; no change in gene expression, white; FDR < 0.05 and p < 0.05, see Table S3 for expanded data table including both DESeq and EdgeR analysis). E and F, mitochondrial content was measured by cytochrome c oxidase activity and visualized by SDH histochemical stain. G, mitochondrial function was measured in terms of complex I-stimulated (P/M) and complex II-stimulated (succinate) respiration corrected for mitochondrial content (COX activity). H, likewise, ROS production was measured in conjunction with changes in complex I and II respiration. Bars depict means ± SE; n = 5 to 6 (A–C), n = 3 (D), n = 11 (F), n = 8 (G and H); Δp < 0.05, main effect of denervation; ∗p < 0.05, DEN versus CON of same genotype by repeated-measures two-way ANOVA. CON, control; COX, cytochrome c oxidase; DEN, denervated; FDR, false discovery rate; PGC-1α, peroxisome proliferator–activated receptor gamma coactivator 1 alpha; ROS, reactive oxygen species; SDH, succinate dehydrogenase.
Given these observed discrepancies in nuclear genes encoding mitochondrial proteins, we wanted to determine whether there were any observable differences in either mitochondrial content and/or function following denervation in muscle lacking p53 relative to WT tissue. DEN and CON muscle from both WT and p53 mKO animals were cross-sectioned and stained to visualize succinate dehydrogenase (SDH) expression as a representation of mitochondrial content (Fig. 4E). DEN muscle displayed marked reductions in SDH stain intensity in WT and p53 mKO muscle. Furthermore, muscle fiber diameter was reduced in these cross sections, thus underscoring the atrophy that was achieved in these samples with chronic denervation. While histochemistry provided qualitative evidence of diminished mitochondrial content and fiber atrophy with disuse, we corroborated this evidence by measuring cytochrome c oxidase (COX) enzyme activity, which revealed similar ∼20% reductions in mitochondrial content with 7-day denervation in both WT and p53 mKO muscle (Fig. 4F). Given the absence of disparate reductions in mitochondrial content in WT and mKO animals with denervation, we then assessed whether the lack of p53 in muscle would manifest in mitochondrial functional impairments. Deficiencies in organelle function were evident by reductions in respiration rate, measured by oxygen consumption corrected for both tissue weight and mitochondrial content, and the production of reactive oxygen species (ROS) via high-resolution respirometry (Fig. 4, G and H). Similar to our aforementioned findings, denervation induced 22 to 32% reductions in mitochondrial respiration during state 2, as well as state 3 conditions, with no apparent additional effect of p53 ablation. Furthermore, production of ROS was increased during state 3-stimulated respiration following prolonged denervation in both WT and p53 mKO animals. However, following the addition of succinate to stimulate both complex I and II active respiration, a greater 2.3-fold elevation in ROS was observed in the DEN muscle lacking p53, relative to the 1.7-fold induction measured in WT DEN muscle. Thus, the disparate alterations in the gene expression profile between WT and p53 mKO muscle were paralleled by reductions in mitochondrial function despite a similar decrease in mitochondrial content with denervation in both genotypes.
Influence of p53 ablation on the UPRmt in 7-day DEN muscle
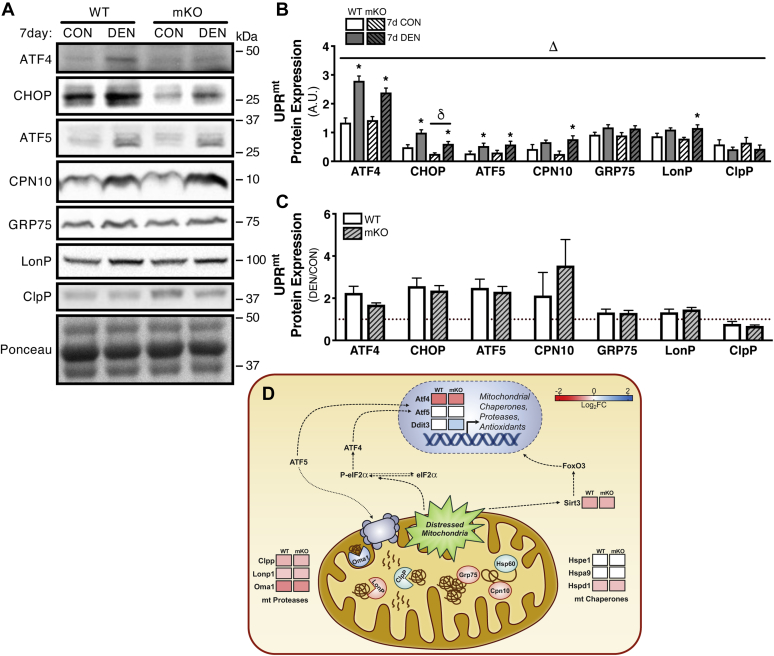
We considered the possibility that an overactivation of the UPRmt may play a role in mediating the mitochondrial remodeling with chronic denervation. The predominant transcriptional regulator of the UPRmt, activating transcription factor 4 (ATF4), along with associated transcription factors, C/EBP homologous protein (CHOP) and activating transcription factor 5 (ATF5), was all significantly upregulated (1.7–2.5-fold) after 7 days of denervation in both WT and p53 mKO animals (Fig. 5, A–C). Protein expression of CHOP was significantly diminished in the absence of p53 in both CON and DEN muscle, whereas genotype had no observable effect on ATF4 or ATF5 (Fig. 5, A–C). Concomitant with the increased expression of these transcriptional regulators, the 10 kDa mitochondrial chaperone displayed a marked 3.5-fold in increase in protein levels in WT muscle, which was further enriched sixfold in p53 mKO samples (Fig. 5, A–C). Another mitochondrial chaperone, glucose-regulated protein 75, was similarly induced 1.3-fold in WT and p53 mKO animals following the chronic disuse stimulus (Fig. 5, A–C). Likewise, mitochondrial proteases (LonP, lon protease homolog, mitochondrial) and caseinolytic mitochondrial matrix peptidase proteolytic subunit (ClpP) were also affected by denervation, with LonP protein levels being enhanced 1.3-fold in WT and 1.45-fold in mKO muscle, whereas ClpP was reduced with denervation by 22% and 32% in WT and mKO muscle, respectively (Fig. 5, A–C).
Figure 5.
Mitochondrial unfolded protein response (UPRmt) subsequent to 7 days of muscle disuse.A–C, representative Western blots and corresponding graphs for ATF4, CHOP, ATF5, CPN10, GRP75, LonP, and ClpP from whole-muscle protein samples from 7-day DEN EDL muscle. D, heat map of DESeq RNA-Seq data showing differential expression of select genes in the context of the UPRmt. The data are expressed as log2 fold change following 7 days of denervation as compared with control (reduced gene expression, red; enhanced gene abundance, blue; no change in gene expression, white; FDR < 0.05 and p < 0.05, see Table S3 for expanded data table including both DESeq and EdgeR analysis). Bars depict means ± SE; n = 4 to 10 (A–C); Δp < 0.05, main effect of denervation; δp < 0.05, main effect of genotype; ∗p < 0.05, DEN versus CON of same genotype by repeated-measures two-way ANOVA. ATF4, activating transcription factor 4; ATF5, activating transcription factor 5; CHOP, C/EBP homologous protein; ClpP, caseinolytic mitochondrial matrix peptidase proteolytic subunit; CON, control; CPN10, 10 kDa mitochondrial chaperone; DEN, denervated; EDL, extensor digitorum longus; FDR, false discovery rate; GRP75, glucose-regulated protein 75; LonP, lon protease homolog, mitochondrial.
Interestingly, RNA-Seq data revealed that, of the genes encoding ATF4, CHOP, and ATF5, only CHOP (Ddit3) was induced with denervation in muscle lacking p53 (0.54 log2FC) with no significant change in the mRNA expression in WT muscle (Fig. 5D and Table S3). ATF4 mRNA was downregulated in both WT and mKO animals following chronic muscle disuse (WT, −0.95 log2FC versus mKO, −0.86 log2FC), whereas ATF5 was not affected in either genotype (Fig. 5D and Table S3). Transcript analysis revealed consistent reductions in the mRNA expression of mitochondrial proteases, Clpp, Lonp1, and Oma1, which was similar across genotypes (Clpp: WT, −0.49 log2FC versus mKO, −0.44 log2FC; Lonp1: WT, −0.34 log2FC versus mKO, −0.39 log2FC; Oma1: WT, −0.82 log2FC versus mKO, −0.75 log2FC; Fig. 5D and Table S3). The expression of genes, heat shock protein family E member 1 (Hspe1) and Hspa9, which encode mitochondrial chaperones 10 kDa mitochondrial chaperone and glucose-regulated protein 75, was not affected by denervation in either WT or mKO samples; however, mRNA expression of another mitochondrial Hspd1 (HSP60) was diminished (WT, −0.40 log2FC versus mKO, −0.41 log2FC) following 7 days of muscle disuse (Fig. 5D and Table S3). The mitochondrial deacetylase, sirtuin 3 (SirT3), has also been implicated in sensing elevations in mitochondrial ROS and appropriately mounting the UPRmt in response to this organelle stress (25). However, we found the mRNA expression of Sirt3 to be downregulated with chronic denervation–induced disuse in animals of both genotypes (WT, −0.55 log2FC versus mKO, −0.51 log2FC; Fig. 5D and Table S3). Collectively, these data suggest that the UPRmt is similarly responsive to chronic denervation in both WT and mKO muscle, which may contribute to either stabilizing the mitochondrial reticulum, or may help promote the recycling of damaged organelles in the presence of disuse-induced stress.
The importance of p53 in the regulation of mitophagy after 7-day disuse
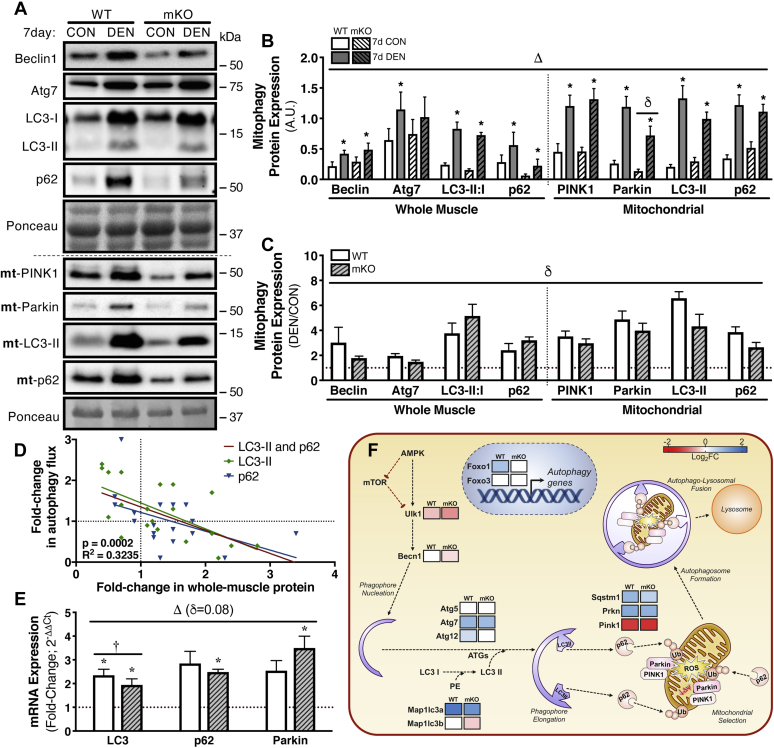
While p53 is classically considered a proapoptotic protein, it is also regarded as a mediator of protein and organelle turnover via the regulation of important components of the autophagy–lysosome system as well as mitophagy (26). Thus, we measured the expression of upstream activators of autophagosomal formation, Beclin1 (enucleation) and Atg7 (autophagy-related protein; phagophore maturation) in WT and mKO animals following the 7-day treatment (Fig. 6, A–C). As anticipated, denervation promoted an increase in the expression of both proteins, regardless of genotype. However, the relative magnitude of induction was blunted in the muscle of p53 mKO animals, as Beclin1 expression increased only 1.8-fold in DEN mKO tissue as compared with a threefold increase in the WT samples. Likewise, Atg7 was upregulated by 1.5-fold in the p53 mKO DEN hindlimb, as opposed to the twofold increase observed in the WT littermates (Fig. 6, A–C). Furthermore, we observed an exacerbated fivefold elevation in the whole-muscle LC3-II:I ratio (microtubule-associated proteins 1a/1b light chain 3) in 7-day DEN p53 mKO tissue relative to the 3.8-fold increase observed in WT samples (Fig. 6, A–C). Likewise, the denervation-induced elevation in whole-muscle sequestosome-1 (p62) expression was also more pronounced in the absence of p53, as mKO animals displayed a 3.2-fold increase in p62 as opposed to the 2.4-fold induction observed in WT tissue (Fig. 6, A–C). These data suggest that the lack of p53 induces a relative impairment in autophagy flux, compared with WT counterparts, as a result of denervation-induced stress. We have validated these commonly used measures of flux by correlating protein levels of p62 and LC3-II to actual changes in flux measured using inhibitors in both cell culture and animal models from several previous studies (Fig. 6D and Table S5).
Figure 6.
Induction of autophagy and mitophagy following 7 days of denervation.A–C, representative Western blots and corresponding graphs showing changes in whole-muscle Beclin1, Atg7, LC3-II:I, and p62, along with mitochondrial-localized PINK1, Parkin, LC3-II, and p62 after 7 days of denervation. D, linear regression analysis of the fold change in whole-muscle LC3-II and p62 protein correlated with the fold change in autophagy flux, derived from previous studies published by our laboratory (Table S5). E, mRNA expression of autophagy transcripts involved in mitochondrial turnover (LC3, p62, and Parkin) measured via real-time quantitative PCR (qPCR). F, heat map of DESeq RNA-Seq data showing differential expression of select genes in the context of autophagy–mitophagy. The data are expressed as log2 fold change following 7 days of denervation as compared with control (reduced gene expression, red; enhanced gene abundance, blue; and no change in gene expression, white; FDR < 0.05 and p < 0.05, see Table S3 for expanded data table including both DESeq and EdgeR analysis). Bars depict means ± SE; n = 5 to 8 (A–C), n = 5 (D and E); Δp < 0.05, main effect of denervation; δp < 0.05, main effect of genotype; †p < 0.05 interaction of genotype and denervation; ∗p < 0.05, DEN versus CON of same genotype by repeated-measures two-way ANOVA. CON, control; DEN, denervated; FDR, false discovery rate; LC3, microtubule-associated proteins 1a/1b light chain 3; p62, sequestosome-1; PINK1, phosphatase and tensin homolog–induced kinase 1.
We next turned our attention to measuring mitophagy-specific protein expression in mitochondrial subcellular fractions. Likewise, we observed an attenuated threefold increase in phosphatase and tensin homolog–induced kinase 1 (PINK1) protein in the DEN muscle from mKO animals, relative to a 3.5-fold increase in WT animals. This corresponded with a similar pattern of Parkin (Prkn) protein expression with denervation, whereby WT muscle exhibited a 4.9-fold induction but increased only fourfold in the absence of p53, and levels were also diminished basally in mKO muscle relative to WT (Fig. 6, A–C). To further assess mitophagy status, we measured LC3-II and p62 protein localized to mitochondria and similarly observed a robust 6.6-fold increase in mitochondrial LC3-II with denervation in WT animals, compared with a blunted 4.3-fold increase in the mKO animals. Likewise, denervation prompted a 3.9-fold increase in p62 within WT muscle, as compared with a 2.6-fold induction in mKO tissue (Fig. 6, A–C). Thus, denervation is a potent inducer of mitophagy proteins, and the absence of p53 consistently leads to a diminished response in the mitophagy machinery at the protein level.
To better ascertain whether the induction in autophagy and mitophagy proteins with denervation and the blunted response observed in p53 mKO muscle were due to enhanced, or impaired protein turnover, we also measured the mRNA expression of LC3, p62, and Prkn (Fig. 6E). Similar to the changes observed at the protein level, transcript expression of all three genes was upregulated by 7-day denervation in both WT and p53 mKO mice, with discrepancies in LC3, p62, and Prkn mRNA levels in p53 mKO animals relative to WT littermates. RNA-Seq data revealed disparate expression of genes involved in the upstream signaling toward autophagy, including Foxo1, which was upregulated (0.68 log2FC) with denervation in WT muscle, but did not change in mKO tissue, as well as Ulk1 and Beclin1, which were both downregulated to a greater extent with disuse in muscle lacking p53 (Ulk1: WT, −0.41 log2FC versus mKO, −0.75 log2FC; Beclin1: WT, no change versus mKO −0.22 log2FC; Fig. 6F and Table S3). Furthermore, RNA-Seq displayed an induction of LC3A (Map1lc3a) that was greater in WT tissue relative to p53 mKO samples (WT, 1.59 log2FC versus mKO, 1.25 log2FC) along with a reduction in LC3B (Map1lc3b; −0.32 log2FC) that was only evident in the DEN muscle lacking p53 (Fig. 6F and Table S3). The lower levels of LC3 subunits revealed by RNA-Seq corroborate our quantitative PCR (qPCR) data in which the interaction of p53 ablation with denervation contributed to a blunted response in LC3 to denervation. In addition to the apparent discrepancies in LC3 gene expression with denervation in the absence of p53, key Atg transcripts responsible for LC3 lipidation and membrane incorporation were also different between genotypes. Atg12 was upregulated in WT muscle (0.38 log2FC) but was not affected in p53 mKO animals, whereas Atg7 was induced similarly regardless of genotype (WT, 0.82 log2FC versus mKO, 0.76 log2FC), and Atg5 was not affected by denervation overall (Fig. 6F and Table S3). PINK1 and Prkn were downregulated and upregulated following the chronic disuse stimulus in both WT and p53 mKO samples, with overall lower levels of these transcripts in the mKO samples (PINK1: WT, −1.48 log2FC versus mKO, −1.59 log2FC; Prkn: WT, 0.97 log2FC versus mKO 0.88 log2FC; Fig. 6F and Table S3). In addition, p62 (Sqstm1) displayed a similar pattern of transcript induction (WT, 0.82 versus mKO 0.51 log2FC; Fig. 6F and Table S3) as observed in the aforementioned qPCR experiments in which mKO muscle had a diminished response to disuse. Thus, the mitophagy machinery is robustly induced in response to chronic denervation as a means to clear damaged organelles; however, the ablation of p53 diminishes this adaptive response in muscle.
p53 ablation on denervation-induced changes in lysosomal content and biogenesis via TFEB/transcription factor E3
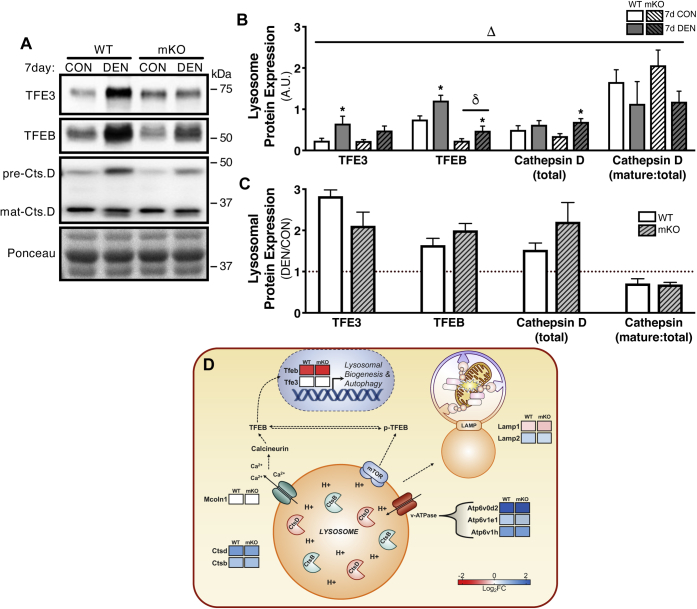
As lysosomes function as the terminal stage of mitochondrial recycling, we naturally wanted to assess lysosomal content and biogenesis following the 7-day denervation stimulus. Our results revealed a similar pattern in the expression of lysosomal markers as observed with autophagy–mitophagy machinery. Whole-muscle transcription factor E3 (TFE3) protein expression increased 2.8-fold in WT muscle, whereas mKO tissue displayed an attenuated 2.1-fold induction (Fig. 7, A–C). Protein expression of TFEB was likewise induced with denervation in both WT and mKO animals by 1.6-2-fold, respectively; however, p53 mKO muscle displayed lower levels in TFEB protein content in both CON and DEN tissue relative to WT littermates (Fig. 7, A–C). We also measured the lysosomal protease, cathepsin D (CtsD), as an indication of lysosomal content, and the ratio of mature:total CtsD as a surrogate measure of lysosomal function. Interestingly, denervation induced a 1.5-fold to 2.2-fold increase in total CtsD of WT and mKO muscle; however, the ratio of mature:total CtsD was similarly diminished ∼30% with chronic denervation in both animal models (Fig. 7, A–C). To corroborate these protein data, we then turned to our RNA-Seq results to determine any changes in transcripts involved in lysosomal biogenesis. Despite the increases in both TFEB and TFE3 protein levels following denervation, mRNA expression of TFEB was markedly reduced in both WT and p53 mKO animals (WT, −1.43 log2FC versus mKO, −1.59 log2FC); however, TFE3 gene expression was not significantly altered (Fig. 7D and Table S3). We also measured transcript levels of key lysosomal constituents. The Mcoln1 gene responsible for encoding the lysosomal cation channel, Mucolipin1, was unaffected by denervation in the presence or the absence of p53 in the muscle (Fig. 8D and Table S3). Conversely, genes encoding various subunits of the lysosomal membrane channel v-ATPase were all induced by denervation (Atp6v0d2: WT, 5.25 log2FC versus mKO, 3.72 log2FC; Atp6v1e1: WT, 0.58 log2FC versus mKO 0.50 log2FC; Atp6v1h: WT, 1.13 log2FC versus mKO, 1.14 log2FC), as were the lysosomal proteases CtsD and CtsB (CtsD: WT, 1.16 log2FC versus mKO, 1.04 log2FC; CtsB: WT, 0.66 log2FC versus mKO, 0.71 log2FC; Fig. 7D and Table S3). Notably, the increased gene expression of the v-ATPase subunit Atp6v0d2 was blunted in p53 mKO muscle as was the induction of CtsD. Paradoxically, the lysosomal membrane–bound Lamp1 and Lamp2, which facilitate lysosomal fusion with the autophagosome, were downregulated and upregulated, respectively, with no discernable difference between animal models (Lamp1: WT, −0.23 log2FC versus mKO, −0.28 log2FC; Lamp2: WT, 0.48 log2FC versus mKO, 0.42 log2FC; Fig. 7D and Table S3). Similar to our observations on the components of mitophagy, lysosomal biogenesis via TFEB–TFE3 signaling appears to be affected by denervation in WT and p53 mKO samples, and the absence of p53 from muscle exacerbates the lysosomal derangements.
Figure 7.
Chronic denervation–induced induction of lysosomal biogenesis and content.A–C, representative Western blots and corresponding graphs from whole-muscle protein extracts showing denervation-induced changes in TFE3, TFEB, total cathepsin along with the ratio of mature:total cathepsin D. D, heat map of DESeq RNA-Seq data showing differential expression of select genes in the context of lysosomal content and biogenesis. The data are expressed as log2 fold change following 7 days of denervation as compared with control (reduced gene expression, red; enhanced gene abundance, blue; and no change in gene expression, white; FDR < 0.05 and p < 0.05, see Table S3 for expanded data table including both DESeq and EdgeR analysis). Bars depict means ± SE; n = 5 to 6 (A–C); Δp < 0.05, main effect of denervation; δp < 0.05, main effect of genotype; ∗p < 0.05, DEN versus CON of same genotype by repeated-measures two-way ANOVA. CON, control; DEN, denervated; FDR, false discovery rate; TFE3, transcription factor E3; TFEB, transcription factor EB.
Figure 8.
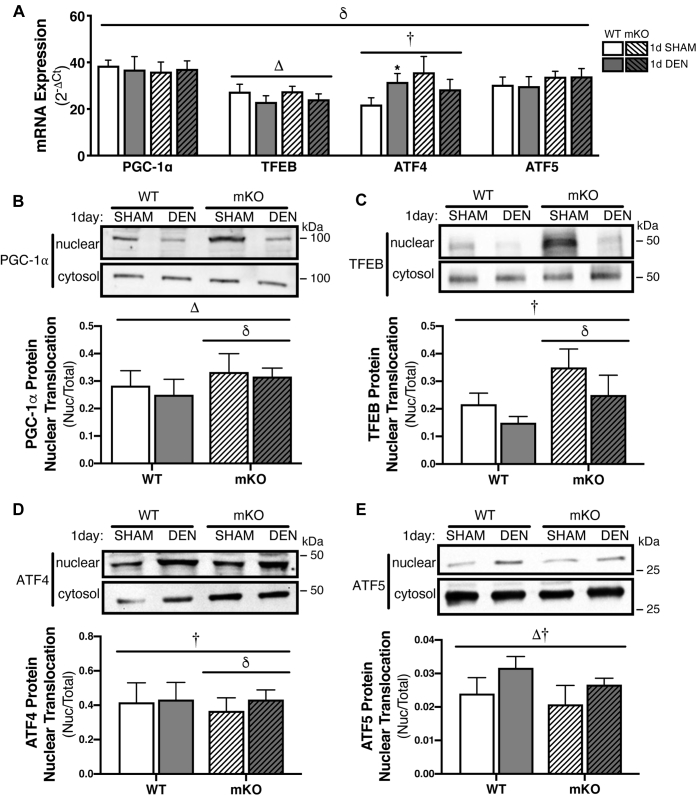
Induction of upstream transcriptional regulators of mitochondrial quality control following 1-day denervation.A, mRNA expression of transcripts involved in mitochondrial quality control pathways measured via real-time quantitative PCR (qPCR) in 1-day DEN and contralateral sham-operated TA muscle. B–E, representative Western blots and corresponding graphs depicting nuclear translocation from the cytosol for transcriptional regulators of mitochondrial biogenesis (PGC-1α), lysosomal biogenesis and autophagy (TFEB), and the UPRmt (ATF4 and ATF5) following 1 day of hindlimb denervation. Subcellular fractions were obtained from gastrocnemius–plantaris muscle. Bars depict means ± SE; n = 6 to 8 (A), n = 5 to 6 (B–E); Δp < 0.05, main effect of denervation; δp < 0.05, main effect of genotype; †p < 0.05 interaction of genotype and denervation; ∗p < 0.05, DEN versus CON of same genotype. mRNA analysis was conducted via repeated-measures two-way ANOVA, whereas nuclear translocation analysis was performed using repeated-measures three-way ANOVA. ATF4, activating transcription factor 4; ATF5, activating transcription factor 5; CON, control; DEN, denervated; PGC-1α, peroxisome proliferator–activated receptor gamma coactivator 1 alpha; TA, tibialis anterior; TFEB, transcription factor EB; UPRmt, mitochondrial unfolded protein response.
Acute effect of p53 ablation on MQC signaling in response to 24 h of denervation-induced disuse
As our data indicate altered MQC responses to 7 days of chronic denervation in WT and mKO tissues, we next wanted to assess whether the effect of p53 ablation had an influence on the acute response to denervation with respect to MQC signaling. Thus, we analyzed the mRNA expression and nuclear translocation of the prominent transcriptional regulators of MQC pathways following 24 h of hindlimb denervation, compared with the contralateral and sham-operated hindlimb muscle. Analysis of the collective mRNA expression for PGC-1α, TFEB, ATF4, and ATF5 suggests a differential expression profile in these regulators of MQC in p53 mKO animals relative to WT littermates; however, only TFEB and ATF4 were affected by acute denervation (Fig. 8A). Specifically, TFEB transcript expression was downregulated 10% in both WT and mKO animals, whereas ATF4 mRNA was upregulated 1.3-fold in the DEN WT hindlimb, while in the absence of p53, ATF4 gene expression was reduced 15% (Fig. 8A). As the proteins encoded by these genes mainly exert their influence on the mitochondrial network as transcriptional regulators via their localization to the nucleus, we also measured their nuclear translocation. Regulators of mitochondrial and lysosomal biogenesis, PGC-1α and TFEB, displayed a similar response to denervation. PGC-1α protein was diminished in the nuclear fraction with denervation in both WT and p53 mKO tissue; however, overall PGC-1α protein content was elevated in the mKO samples (Fig. 8B). Likewise, TFEB nuclear translocation was reduced with denervation regardless of genotype, and p53 mKO tissue displayed higher levels of TFEB protein relative WT (Fig. 8C). Conversely, UPRmt regulators ATF4 and ATF5 were induced in the nuclear fraction following 24 h of hindlimb denervation. Specifically, ATF4 was upregulated in DEN mKO muscle, while no change in protein localization was observed in WT tissue (Fig. 8D), whereas ATF5 nuclear expression was induced with acute denervation in both WT and p53 mKO samples (Fig. 8E). The nuclear expression of all proteins displayed a significantly altered expression pattern in p53 mKO as compared with WT muscle. Evidently, the lack of p53 in muscle affects the acute 1-day response to denervation, which precedes, and likely contributes, to the altered expression of the MQC pathways that we observed following 7 days of hindlimb disuse.
Discussion
The importance of p53 as a regulator of mitochondrial quality and function in muscle has been a focal point of research over several years. We and others have previously described a role for p53 in mediating the mitochondrial adaptive response to acute exercise as well as an endurance training program (17, 18, 19, 20, 27, 28). However, contradictory findings under basal conditions have also been reported (21). Thus, the central focus of this study was to expand previous analyses to determine how p53 regulates the many facets of MQC, given the importance of mitochondrial function in determining skeletal muscle, and indeed whole-body metabolic health. In contrast to our previous work on exercise, in this study, we have used a model of denervation-induced muscle disuse for several reasons. First, this model results in rapid and progressive muscle atrophy and mitochondrial decay, thus providing an accelerated model of the decline in muscle health evident with chronic sedentarism and muscle inactivity, which is pervasive in many developed societies. Understanding the molecular mechanisms of denervation atrophy is also translationally relevant to peripheral neuropathic conditions such as amyotrophic lateral sclerosis and aging itself. Second, denervation induces a robust induction of gene expression signatures relevant to mitochondrial turnover, which should allow for improved analytical power in studying the disuse response in muscle. Third, p53 has been implicated in the maintenance of muscle mass (29, 30), but its role in mediating mitochondrial content and function has not been experimentally evaluated in the context of muscle disuse, as much of the work studying the role of p53 in regulating skeletal muscle mitochondria has focused on basal or exercise-induced differences. Thus, we set out to determine the role of p53 in regulating mitochondrial adaptations following acute (1 day) and chronic (7 days) denervation-induced muscle disuse, and whether these contributed to the impairments observed in the muscle.
Our data reinforce the notion that p53 is important in muscle adaptations following disuse, as we observed an increase in the protein expression of p53 following 7 days of denervation, along with a concomitant increase in the proapoptotic protein Bax, a p53-regulated protein. Our data also reveal that p53 mKO animals experienced similar levels of hindlimb muscle atrophy as WT animals, which differs from a previous report that showed p53 ablation preserved muscle mass following 3 days of hindlimb immobilization (30). However, it is worthwhile to consider the differences in duration and intensity of the disuse stimulus between our experiments and those previously reported. We subjected mice to 7 days of denervation, wherein muscle is no longer able to contract, as compared with the aforementioned study, which utilized hindlimb immobilization for 3 days, a stimulus in which the muscle retains its ability to contract and receive input from the neuromuscular junction. Despite observing no differences in the level of atrophy in WT and mKO tissues in our study, it is important to note that the level of atrophy we observed (∼15–18%) was substantially greater than the levels obtained previously via immobilization (∼4%), which may suggest that the intensity of our disuse intervention exceeds the influence of p53 in attenuating atrophy. Thus, we cannot rule out the possibility that p53 is sufficient to preserve muscle mass during moderate periods of disuse.
The contribution of p53 to the preservation of metabolic function with disuse has not been determined. Therefore, we assessed the transcriptomic alterations associated with both denervation and p53 expression. Principal component analysis revealed the considerable potency of the denervation stimulus in altering the transcript profile of muscle, irrespective of genotype. In contrast, the ablation of p53 alone had considerably less influence in accounting for variability between samples, suggesting that the role of p53 in mediating the regulation of basal mitochondrial function is less than previously thought. However, this does not discount the possibility that the role of p53 may be best understood when it is activated during periods of stress, such as that imposed by denervation, which is a strong stimulus for the induction of mitochondrial turnover. Thus, to better understand how p53 ablation affects skeletal muscle mitochondrial remodeling with denervation, we performed pathway analysis of gene sets most affected by the disuse stimulus (Fig. S6). We evaluated the interaction effect of denervation with genotype, which specifically addressed the question—is the response to denervation different in p53 mKO animals? GSEA of the interaction term revealed that a majority of the gene sets enriched via downregulated genes were specifically related to autophagy–mitophagy machinery, mitochondrial assembly, and oxidation/oxidative phosphorylation, indicating the detrimental effect of p53 ablation on mitochondria during disuse. When considering pathway enrichment based on all differentially expressed genes (upregulated and downregulated combined) based on the interaction of p53 ablation with chronic denervation, four of the seven significantly enriched pathways were likewise related to mitochondrial regulation (Fig. 2B). Moreover, the divergent responses to denervation in mKO and WT muscle is evident in REVIGO-generated tree maps of GO terms, as pathways pertaining to “mitochondrial gene expression,” “lysosomal transport,” and “regulation of macroautophagy” were unique to the interaction term, whereas discrepant patterns and proportions of other MQC-related pathways were evident when comparing the tree maps for WT with mKO animals (Fig. 3C). Thus, since RNA-Seq and enrichment analysis suggested a role for p53 in mediating mitochondrial remodeling following chronic denervation, we next assessed each of the key mitochondrial regulatory pathways individually.
Mitochondrial content is closely related to the expression and activity of PGC-1α, such that with chronic muscle disuse, PGC-1α expression is reduced, contributing to decreases in organelle volume and function (13, 31, 32). In addition to its role as a transcriptional regulator in the nucleus, PGC-1α is also proposed to localize to the mitochondrion to regulate mtDNA transcription. Likewise, p53 also translocates to both the nucleus, where it regulates the expression of PGC-1α, as well as to mitochondria, where it interacts with mitochondrial transcription factor A (Tfam) (16). Indeed, we detected a downregulation of PGC-1α in both compartments following chronic denervation in both WT and mKO animals, indicating reduced drive for mitochondrial biogenesis. Surprisingly, mitochondrial content was not different between genotypes, despite a more significant reduction in nuclear PGC-1α in p53 mKO animals. Since, mitochondrial content does not necessarily equate to function, as dysfunctional organelles may accumulate because of impaired clearance, we measured oxygen consumption rate and ROS production in permeabilized muscle fibers. Our results revealed elevated ROS levels during ADP-stimulated respiration, indicating impaired organelle function with disuse in both WT and mKO animals. Notably, ROS levels were induced to a greater extent in mKO tissue under complex I and complex II-stimulated conditions by the addition of succinate, which corroborates previous reports that showed functional impairments in isolated mitochondria derived from p53 mKO muscle compared with WT tissue (20). In addition, similar levels of respiration in WT and mKO fibers were observed, as well as comparable reductions in respiration with chronic denervation, independent of genotype. Altogether, our respirometry data suggest significant mitochondrial dysfunction resulting from denervation that is magnified in the absence of p53.
The increased levels of mitochondrial ROS observed with muscle disuse can induce the activation of the UPRmt as a means of restoring mitochondrial homeostasis (33, 34). While a link between p53 and the UPRmt requires further investigation (35, 36), we examined the expression of UPRmt proteins with denervation to test the hypothesis that the UPRmt might preserve mitochondria during chronic disuse. We observed an induction of the UPRmt with chronic denervation in both WT and mKO samples, evidenced by increased protein expression of mitochondrial chaperones despite a concomitant decline in mitochondrial content. This was likely driven by the augmented expression of the primary regulators of the UPRmt, the transcription factors ATF4, ATF5, and CHOP, which are otherwise maintained at low levels under basal conditions (37, 38). The fact that ATF4 and CHOP displayed discrepant changes in both their protein and gene expression between WT and mKO samples also implicates p53 as a potential mediator of UPRmt activation. Surprisingly, at the transcript level, several of the genes involved in the UPRmt were downregulated following chronic denervation (Fig. 5D). The increase in protein level in the face of a decrease in their respective transcripts suggests a switch in the CON of UPRmt as denervation time ensues. In this way, the elevations in UPRmt proteins at the 7-day time point reflect an adaptation via the UPRmt pathway at earlier time points to potentially address transient mitochondrial derangements. However, the restorative action of the UPRmt is insufficient to preserve mitochondrial function, despite the elevations in the UPRmt proteins. Our experiments on the early effects of denervation suggest a rapid induction of the UPRmt, evident from increases in ATF4 mRNA and ATF5 translocation to the nucleus in WT muscle. The absence of p53 attenuated this in part, since ATF4 mRNA declined in response to denervation, as did the nuclear localization of ATF5 in p53 mKO tissue. Thus, these data support the possibility that the UPRmt provides a compensatory role in maintaining mitochondrial function following the onset of muscle disuse and indicates that p53 plays a role in regulating this transient stress response; however, following chronic denervation, the UPRmt is insufficient in preserving mitochondrial quality and function and gives way to the mitophagy–lysosome system for the clearance of terminally dysfunctional organelles.
Mitochondrial turnover via the mitophagy–lysosome system is integral for maintaining the quality of the mitochondrial pool. Previous studies have reported that p53 regulates the autophagy–mitophagy and lysosomal machinery and can either promote or suppress the pathway depending on its localization within the cell (39, 40, 41). While denervation exerted a marked increase in the expression of autophagy (Beclin1, Atg7, LC3-II:I, and p62) and mitophagy-specific proteins (mitochondrial-localized PINK1, Prkn, LC3-II, and p62), this induction of mitochondrial turnover was consistently attenuated when p53 was abolished in muscle. In particular, p53 mKO muscle displayed attenuated induction of upstream autophagy regulators, Beclin1 and Atg7 suggesting a diminished initiation of the autophagosome. Likewise, the exacerbated increases in whole-muscle p62 and LC3-II:I ratio provide additional evidence of a reduced autophagosomal turnover in the absence of p53, as the accumulation of these markers indicates reduced autophagy flux. Indeed, mitophagy-specific markers, PINK1 and Prkn, along with mitochondrial-localized LC3-II and p62, corroborate these findings, as their induction in p53 mKO tissue was blunted as compared with WT samples. A similar trend was observed at the mRNA level, assessed using qPCR, thereby substantiating prior reports implicating p53 as a mediator of organelle turnover. In addition, RNA-Seq data further revealed that the induction and maturation of the phagophore membrane was particularly susceptible to the absence of p53. The mRNA expression of the transcriptional regulator Foxo1 (autophagy activation), along with Ulk1 and Beclin1 (phagophore nucleation), was relatively suppressed in the absence of p53 with denervation, thus indicating impaired initiation and activation of the autophagy machinery. Elongation of the phagophore membrane requires the maturation of LC3, which is facilitated by a host of autophagy (Atg) proteins, and RNA-Seq data also revealed that maturation and elongation of the membrane was also disrupted, evidenced by the relatively lower expression of Atg7 and Atg12, as well as LC3 subunits A and B (Map1lc3a/b), in the absence of p53 during denervation.
The availability of functional lysosomes is required for the fusion and digestion of autophagosome-engulfed organelles. Recent evidence suggests a role for p53 in regulating lysosomal biogenesis and autophagy via nuclear translocation of TFEB and TFE3 (42). The influence of TFEB and TFE3 on the transcription of autophagy and lysosomal genes is largely determined by their post-translational modification, which allows their entry into the nucleus. As such, we observed no differences in the mRNA expression of either TFEB or TFE3 with denervation in WT and p53 mKO samples via RNA-Seq; however, p53 mkO muscle displayed a blunted increase in the expression of TFE3 protein with denervation along with overall reduced TFEB protein levels, indicating impaired activation of lysosomal biogenesis. In addition, we measured the ratio of mature CtsD (25 kDa) and its precursor form (37 kDa) as a surrogate measure of lysosomal function, given that CtsD protein maturation requires an acidic lysosomal environment, established by the shuttling of H+ into the lysosomal lumen by the v-ATPase within functional vesicles. Our data suggest a greater increase in total CtsD with denervation in p53 mKO muscle, whereas the mature:total CtsD ratio was reduced, suggesting that lysosomes become dysfunctional with chronic muscle disuse, an effect that was more pronounced in the absence of p53. While RNA-Seq analysis of lysosomal genes displayed similar responses in WT and mKO samples following chronic disuse, the lysosomal membrane protein Lamp1 was reduced to a greater extent in mKO samples than in WT, thus suggesting impairments in the fusion of the lysosome to the autophagosome-engulfed mitochondria and in the formation of the autophagolysosome at the terminal stage of mitophagy. Therefore, we have determined that multiple aspects of mitochondrial turnover, including phagophore formation, mitochondrial selection, as well as lysosomal function and fusion, are impaired in p53 mKO muscle. This attenuated drive for autophagy in p53 mKO muscle may account for the preservation in muscle mass that was previously reported following 3 days of hindlimb immobilization (30). However, during more prolonged stages of muscle disuse, this attenuation may account for the increase in mitochondrial dysfunction observed, resulting for the lack of clearance of poor quality organelles. Thus, while total mitochondrial content may be similar between p53 mKO and WT muscle, the quality of the organelle pool is diminished in the absence of p53.
While our analysis mostly focused on the chronic effect of a 7-day muscle disuse stimulus, we also subjected a subset of WT and p53 mKO animals to 1 day of hindlimb denervation in order to determine whether the early denervation-induced induction of p53 plays a role in initiating the signaling events that dictate the mitochondrial decline observed at later time points of muscle disuse. We noted an effect of genotype on the mRNA expression of PGC-1α, TFEB, ATF4, and ATF5, indicating the influence of p53 on the breadth of the MQC pathways. In addition, we consistently observed an effect of genotype on the nuclear localization of these transcriptional regulators, which reinforces the influence that p53 has on the initiation of mitochondrial regulatory signaling, beginning as an early event following denervation.
It remains to be determined what influence p53 ablation would have at intermediate time points between 1 and 7 days of denervation or in the context of less robust disuse models such as hindlimb suspension or casting. In addition, as p53 has been shown to play a role in exercise-induced mitochondrial biogenesis and remodeling, it would be worthwhile for future studies to examine the influence of p53 ablation on mitochondrial readaptation following muscle reloading. This would not be possible following denervation but would instead require alternative models such as nerve crush, or tetrodotoxin paralysis to remove neural innervation, but allow for its reinstatement following a period of disuse.
In summary, we report that denervation is a potent stimulus for mitochondrial decline in muscle. This is accomplished by promoting decreased mitochondrial biogenesis signaling, along with a substantial induction of stress responses and degradation pathways such as the UPRmt and the mitophagy–lysosome systems. We also highlight a role for p53 in facilitating the acute response to denervation via the regulation of primary MQC transcriptional regulators, as well as mediating mitochondrial remodeling following 7 days of denervation-induced disuse. Mitochondrial turnover is attenuated in muscle lacking p53, which contributes to an accumulation of dysfunctional organelles as indicated by exacerbated ROS. Thus, our data indicate a role for p53 in regulating mitochondrial function, by contributing to the orchestration of the balance between organelle biogenesis and mitophagy, fine-tuned by the UPRmt.
Experimental procedures
Animals
Both WT and muscle-specific p53 KO (mKO) mice were generously provided by Dr Christopher M. Adams (Iowa) and are homozygous for a floxed p53 allele flanked by LoxP restriction sites, whereas mKO mice also express Cre recombinase under control of the muscle creatine kinase promoter (30). Progeny was genotyped postmortem using pectoralis-derived whole-muscle protein extracts separated on an SDS-PAGE gel and immunoblotted for p53 protein using an antibody that was generously provided by Dr Sam Benchimol (York University, Toronto).
Denervation surgery
Male mice aged 5 to 8 months were randomly divided into either 1- or 7-day treatment group. Briefly, mice were anesthetized using isofluorane and had a 2 to 3 mm section of their sciatic nerve surgically dissected from their left hindlimb to induce chronic muscle disuse of the downstream muscles acutely (1 day) or chronically (7 days). Animals in the 1-day group were subjected to sham surgery of the contralateral limb to account for acute effects of surgery itself, whereas the contralateral right hindlimb of 7-day-treated animals served as the intra-animal control. Animals were housed with food and water ad libitum for the duration of the treatment. Following denervation surgery, water supplemented with amoxicillin (0.3 mg/l) was provided, and meloxicam injections (0.05% solution in saline) were administered for 3 days postsurgery for pain management (first dose: 2 μg/g body weight, with each subsequent dose being half of the previous dose). The TA, extensor digitorum longus (EDL), gastrocnemius/plantaris, and soleus tissues from both the CON/sham-operated and DEN leg were collected for analysis. All animal experiments were approved by the York University Animal Care Committee under the auspices of the Canadian Council of Animal Care.
COX activity
COX activity was used as a marker of mitochondrial content. Enzyme extracts were prepared from TA muscle using a Qiagen TissueLyser II and sonicated (3 × 3 s; 30% power). A buffered test solution containing fully reduced horse heart cytochrome c (catalog no.: C-2506; Sigma) was prepared. A multipipette was used to add 240 μl of test solution to 50 μl of whole-muscle homogenate in a 96-well plate. COX enzyme activity was determined spectrophotometrically as the maximal rate of oxidation of fully reduced cytochrome c (catalog no.: C-2506; Sigma), measured by the change in absorbance at 550 nm at 30 °C in a microplate reader (Synergy HT; Bio-Tek Instruments), as previously described (43). For each sample, COX activity was calculated as an average of three trials.
Respiration and ROS production
High-resolution respirometry (Oxygraph-2K; Oroboros Instruments) was used to measure oxygen consumption in permeabilized muscle fibers from the TA muscle (control and DEN) of mice. Briefly, fibers were mechanically separated in ice-cold biopsy preservation solution buffer (2.77 mM CaK2EGTA, 7.23 mM K2EGTA, 7.55 mM Na2ATP, 6.56 mM MgCl2⋅6H2O, 20 mM taurine, 15 mM Na2 phosphocreatine, 20 mM imidazole, 0.5 mM DTT, 50 mM 2-(N-morpholino)ethanesulfonic acid hydrate, and pH 7.1), permeabilized in biopsy preservation solution with 40 μg/μl saponin at 4 °C for 30 min, and washed in buffer Z (105 mM K-2-(N-morpholino)ethanesulfonic acid, 30 mM KCl, 10 mM KH2PO4, 5 mM MgCl2⋅6H2O, 1 mM EGTA, 5 mg/ml bovine serum albumin, and pH 7.4). Fibers were then incubated in the chamber with oxygenated buffer Z supplemented with 10 μM of Amplex-Red to measure ROS production as well as 1 μM blebbistatin (catalog no.: B592500; Toronto Research Chemicals) to prevent tetanus of the muscle and 25 U/ml Cu/Zn SOD1 to convert O2− to H2O2 and 2 mM EGTA. After obtaining background values, substrates were titrated as follows to assess respiration and ROS production: pyruvate–malate (complex I, state 2), ADP (complex I, state 3), and succinate (complex I and II, state 3).
Mitochondrial and nuclear fractionation
Enriched mitochondrial and nuclear cellular subfractions were used to measure nuclear PGC-1α (Fig. 2, A and B) as well as mitochondrial PGC-1α (Fig. 2, A and C), PINK1, Prkn, LC3-II, and p62 (Fig. 3, A–C) from 7-day DEN and CON gastrocnemius muscles, as previously described (44). Briefly, gastrocnemius muscles were minced on ice and homogenized using a Teflon pestle and mortar and suspended in mitochondrial isolation buffer (250 mM sucrose, 20 mM Hepes, 10 mM KCl, 1.5 mM MgCl2, 1 mM EDTA, and 1 mM EGTA) supplemented with protease (Complete, Roche; 1169749801; Roche Diagnostics) and phosphatase inhibitor cocktails (catalog nos.: P5726 and P0044; Cocktail 2 and 3; Sigma). The homogenates were then centrifuged at 900g for 10 min at 4 °C to pellet the nuclei, whereas mitochondrial and cytosolic fractions were contained within the supernate. The supernatant fraction was recentrifuged at 16,000g for 20 min at 4 °C to pellet the mitochondria. The mitochondrial pellet was washed twice and resuspended in a onefold dilution of mitochondrial isolation buffer. Mitochondria were subsequently sonicated 2 × 2 s at 30% power to yield the enriched mitochondrial fraction. Pellets containing nuclei were resuspended in nuclear lysis buffer (1.5 mM MgCl2, 0.2 mM EDTA, 20 mM Hepes, 0.5 M NaCl, 20% glycerol, and 1% Triton X-100), incubated on ice for 30 min, and then sonicated 3 × 10 s followed by a final centrifugation step at 16,000g for 15 min at 4 °C. The supernate was collected to obtain the enriched nuclear fraction. Protein concentrations within the samples were determined using the Bradford method. Fraction purity was confirmed by Western blot analysis (Fig. S1).
Nuclear and cytosolic fractionation
Nuclear and cytosolic fractions were prepared fresh from sham-operated and 1-day DEN gastrocnemius muscles to measure expression and localization of transcription factors PGC-1α, TFEB, ATF4, and ATF5 (Fig. 6, B–E). Fractions were obtained using the NE-PER extraction reagents (catalog no.: 38835; Thermo Fisher Scientific) with minor modifications. Briefly, ∼100 mg of gastrocnemius muscle was minced on ice and homogenized in cytosolic extraction reagent I using a Dounce homogenizer. Homogenates were then vortexed and let to stand on ice for 10 min. Following addition of cytosolic extraction reagent II solution, samples were briefly vortexed and centrifuged at 16,000g for 10 min at 4 °C. The supernatant cytosolic fractions were then collected. The pellets, containing nuclei and cellular debris, were washed in cold 1× PBS and suspended in nuclear extraction buffer. Nuclear fractions were then sonicated three times for ∼3 to 5 s at 30% power and incubated on ice for 40 min. The nuclear fractions were vortexed every 10 min during the incubation and subsequently underwent centrifugation at 16,000g for 10 min at 4 °C. The resulting supernatant nuclear fractions were collected and stored at −80 °C until further analysis. Protein concentrations within the samples were determined using the Bradford method. Fraction purity was confirmed by Western blot analysis (Fig. S2). Paired cytosolic and nuclear samples obtained from the same animals were run and imaged within the same blot for accurate quantification of subcellular translocation.
Whole-muscle protein extraction
Frozen EDL samples (∼10–15 mg) were added to Sakamoto buffer (20 mM Hepes, 2 mM EGTA, 1% Triton X-100, 10% glycerol, 50 mM β-glycerophosphate, 1 mM PMSF, 1 mM DTT, 1 mM sodium orthovanadate, 10 μM leupeptin, 5 μM pepstatin A, and 10 mg/ml aprotinin) and diluted 20-fold. Samples were homogenized using a Qiagen TissueLyser II with steel beads and then centrifuged for 10 min at 12,000g. Supernates were recovered, and protein concentration was determined by Bradford protein assay.
Immunoblotting
Proteins were resolved on 12 to 15% polyacrylamide gels, transferred onto nitrocellulose membranes, and blocked with 5% milk in TBS with Tween-20 for 1 h. Membranes were incubated overnight at 4 °C with 1° antibodies for proteins of interest (Table S1), followed by a 1 h incubation at room temperature with the appropriate 2° antibody coupled to horseradish peroxidase. Membranes were developed with an ECL kit (catalog no.: 170-5061; BIO-RAD) and revealed using enhanced chemiluminescence. Quantification was performed with ImageJ software (the National Institutes of Health).
Histology and cross-sectional area
SDH staining was performed on 10 μm cross sections of EDL muscles as previously described (23). EDL muscles from the CON and DEN hindlimb were excised from the animal, mounted in Cryomatrix (Thermo Fisher Scientific), and frozen in isopentane at the temperature of liquid nitrogen. These muscles were then cryosectioned, transferred to glass slides, and incubated with an SDH-staining solution (0.2 M sodium succinate, 0.2 M phosphate buffer, pH 7.4, and nitro blue tetrazolium) at 37 °C for 20 min. Slides were then rinsed in distilled water, and a microscope cover glass was mounted on slides using DPX mountant for histology (catalog no.: 06522; Sigma). Photos of muscle sections were taken using a Nikon Eclipse 90i camera and QCapture software.
In vitro RNA isolation and reverse transcription
Total RNA was isolated from frozen whole-muscle TA as described previously (45). Briefly, using a Qiagen TissueLyser II, frozen TA muscle samples (∼20–30 mg) were added to TRIzol reagent and mixed with chloroform. Samples were centrifuged at 4 °C at 16,000g for 15 min, and the upper aqueous phase of the sample was transferred to a new tube along with isopropanol and left overnight at −20 °C to precipitate. Samples were once again centrifuged at 4 °C at 16,000g for 10 min. The resultant supernate was discarded, and the pellet was resuspended in 30 μl of molecular-grade sterile water (Wisent Bio Products). The concentration and purity of the RNA were measured using a spectrophotometer (NanoDrop 2000). SuperScript III reverse transcriptase (catalog no.: 18080093; Invitrogen) was used to reverse-transcribe 1.5 μg of total RNA into complementary DNA (cDNA) in a 20 μl reaction.
Real-time qPCR gene expression analysis and qPCR quantification
Sequences from GenBank were used to design primers with Primer 3, version 0.4.0 software (Massachusetts Institute of Technology) for genes of interest (Table S2). Primer specificity was confirmed by OligoAnalyzer 3.1 (Integrated DNA Technologies). mRNA expression was measured with SYBR Green chemistry (PerfeCTa SYBR Green SuperMix, ROX; Quanta BioSciences). Each well contained SYBR Green SuperMix, forward and reverse primers (20 μM), sterile water, and 10 ng of cDNA. All real-time PCR amplifications were detected in a 96-well plate using a StepOnePlus Real-Time PCR System (Applied Biosystems). The final reaction volume of each well was 25 μl. Samples were run in duplicates to ensure accuracy. The PCR program consisted of an initial holding stage (95 °C for 10 min) followed by 40 amplification cycles (60 °C for 1 min and 95 °C for 15 s) and was completed with a final melting stage (95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s). Analysis of melt curves generated by the instrument for SYBR Green analyses was used to control for nonspecific amplification and primer dimers. Negative control wells contained water in place of cDNA. Gene expression was quantitated as follows: first, the average threshold cycle (CT) value of the endogenous reference genes (Gapdh and B2m) was subtracted from the CT value of the target gene: ΔCT = CT(target) − CT(reference avg). Next, the ΔCT value of the control tissue was subtracted from the ΔCT value of the experimental tissue: ΔΔCT = ΔCT(experimental) − ΔCT(control) to be reported as FCs using the ΔΔCT method, calculated as 2−ΔΔCT.
RNA-Seq and pathway analysis
RNA library preparation, sequencing, and pathway analysis were performed by the Toronto Centre for Applied Genomics (The Hospital for Sick Children). NEBNext Ultra II Directional RNA libraries using poly-A selection were prepared. All 12 samples (three WT 7-day DEN with matched controls and three mKO 7-day DEN with matched controls) were multiplexed on one lane of an SP flow cell and sequenced (paired end reads) on the Illumina NovaSeq 6000. Prior to performing RNA-Seq, the resulting DNA-free RNA samples were analyzed for quality on a 2100 Bioanalyzer System (Agilent Technologies). RNA integrity numbers were calculated automatically (Fig. S3). Two-condition differential gene expression analysis was performed with DESeq2 (version 1.26.0s; using R, version 3.6.1) as well as with the edgeR R package (version 3.28.1; using R, version 3.6.1). To investigate whether the effects of denervation were modulated by p53 on a more global scale, standard enrichment/pathway analysis was performed. The standard pathway analysis includes GOSeq enrichment (threshold-based method) and GSEA (threshold-free method) (Table S4). REVIGO was used to summarize and visualize GO terms pertaining to “mitochondria,” “autophagy,” “mitophagy,” “lysosome,” “ROS” or “reactive oxygen species,” “UPR” or “unfolded protein response,” “apoptosis,” “p53,” “protein synthesis,” and “degradation.”
Statistical analysis
Comparisons between DEN and matched control/sham-operated samples from WT and p53 mKO animals were evaluated using two-way ANOVA with repeated measures for all protein, mRNA, respiration, and ROS data. Nuclear translocation data were quantified using three-way ANOVA with repeated measures to compare the effects of genotype, denervation, and fraction localization. Bonferroni post hoc tests were performed when applicable. All values represent the mean ± SE. Data were considered statistically different if p < 0.05.
Data availability
All raw data used to generate the data figures are available upon request from Dr David A. Hood (dhood@yorku.ca).
Supporting information
This article contains supporting information (13, 46, 47, 48, 49, 50).
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Research Council of Canada. We acknowledge the assistance of the Toronto Centre for Applied Genomics for the acquisition of RNA-Seq data and thank Drs Bhooma Thiruv, Roumiana Alexandrova, and Giovanna Pellecchia for their assistance with RNA-Seq and pathway analysis data. We also thank Alisha Rullay for her assistance in preparing experiments and samples.
Author contributions
J. M. M. and D. A. H. conceptualization; J. M. M. and D. A. H. methodology; J. M. M. and D. A. H. validation; J. M. M. and D. A. H. formal analysis; J. M. M. and A. N. O. investigation; J. M. M. and A. N. O. resources; J. M. M. data curation; J. M. M. writing–original draft; J. M. M. and D. A. H. writing–review and editing; J. M. M. visualization; D. A. H. supervision; D. A. H. funding acquisition.
Funding and additional information
D. A. H. is the recipient of a tier I Canada Research Chair in Cell Physiology. J. M. M. is a recipient of the Natural Science and Engineering Research Council Doctoral Canada Graduate Scholarship.
Edited by Qi Qun Tang
Supporting information
References
- 1.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Janssen I., Heymsfield S.B., Ross R. Low relative skeletal muscle mass (sarcopenia) in older persons is associated with functional impairment and physical disability. J. Am. Geriatr. Soc. 2002;50:889–896. doi: 10.1046/j.1532-5415.2002.50216.x. [DOI] [PubMed] [Google Scholar]
- 3.Kelley D.E., Goodpaster B., Wing R.R., Simoneau J.A. Skeletal muscle fatty acid metabolism in association with insulin resistance, obesity, and weight loss. Am. J. Physiol. 1999;277:E1130–E1141. doi: 10.1152/ajpendo.1999.277.6.E1130. [DOI] [PubMed] [Google Scholar]
- 4.Mancini D.M., Walter G., Reichek N., Lenkinski R., McCully K.K., Mullen J.L., Wilson J.R. Contribution of skeletal muscle atrophy to exercise intolerance and altered muscle metabolism in heart failure. Circulation. 1992;85:1364–1373. doi: 10.1161/01.cir.85.4.1364. [DOI] [PubMed] [Google Scholar]
- 5.Bodine S.C. Disuse-induced muscle wasting. Int. J. Biochem. Cell Biol. 2013;45:2200–2208. doi: 10.1016/j.biocel.2013.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Santilli V., Bernetti A., Mangone M., Paoloni M. Clinical definition of sarcopenia. Clin. Cases Miner. Bone Metab. 2014;11:177–180. [PMC free article] [PubMed] [Google Scholar]
- 7.Hood D.A., Memme J.M., Oliveira A.N., Triolo M. Maintenance of skeletal muscle mitochondria in health, exercise, and aging. Annu. Rev. Physiol. 2019;81:19–41. doi: 10.1146/annurev-physiol-020518-114310. [DOI] [PubMed] [Google Scholar]
- 8.Heber D., Ingles S., Ashley J.M., Maxwell M.H., Lyons R.F., Elashoff R.M. Clinical detection of sarcopenic obesity by bioelectrical impedance analysis. Am. J. Clin. Nutr. 1996;64:472S–477S. doi: 10.1093/ajcn/64.3.472S. [DOI] [PubMed] [Google Scholar]
- 9.Kujoth G.C., Hiona A., Pugh T.D., Someya S., Panzer K., Wohlgemuth S.E., Hofer T., Seo A.Y., Sullivan R., Jobling W.A., Morrow J.D., Van Remmen H., Sedivy J.M., Yamasoba T., Tanokura M., et al. Mitochondrial DNA mutations, oxidative stress, and apoptosis in mammalian aging. Science. 2005;309:481–484. doi: 10.1126/science.1112125. [DOI] [PubMed] [Google Scholar]
- 10.Schaap L.A., Pluijm S.M.F., Deeg D.J.H., Visser M. Inflammatory markers and loss of muscle mass (sarcopenia) and strength. Am. J. Med. 2006;119 doi: 10.1016/j.amjmed.2005.10.049. 526.e9-526.e17. [DOI] [PubMed] [Google Scholar]
- 11.Rapizzi E., Pinton P., Szabadkai G., Wieckowski M.R., Vandecasteele G., Baird G., Tuft R.A., Fogarty K.E., Rizzuto R. Recombinant expression of the voltage-dependent anion channel enhances the transfer of Ca2+ microdomains to mitochondria. J. Cell Biol. 2002;159:613–624. doi: 10.1083/jcb.200205091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Erlich A.T., Brownlee D.M., Beyfuss K., Hood D.A. Exercise induces TFEB expression and activity in skeletal muscle in a PGC-1α-dependent manner. Am. J. Physiol. Cell Physiol. 2018;314:C62–C72. doi: 10.1152/ajpcell.00162.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vainshtein A., Desjardins E.M., Armani A., Sandri M., Hood D.A. PGC-1α modulates denervation-induced mitophagy in skeletal muscle. Skelet. Muscle. 2015;5:9. doi: 10.1186/s13395-015-0033-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vogelstein B., Lane D., Levine A.J. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 15.Bartlett J.D., Close G.L., Drust B., Morton J.P. The emerging role of p53 in exercise metabolism. Sports Med. 2014;44:303–309. doi: 10.1007/s40279-013-0127-9. [DOI] [PubMed] [Google Scholar]
- 16.Saleem A., Hood D.A. Acute exercise induces tumour suppressor protein p53 translocation to the mitochondria and promotes a p53-Tfam-mitochondrial DNA complex in skeletal muscle. J. Physiol. 2013;591:3625–3636. doi: 10.1113/jphysiol.2013.252791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saleem A., Carter H., Iqbal S., Hood D.A. Role of p53 within the regulatory network controlling muscle mitochondrial biogenesis. Exerc. Sport Sci. Rev. 2011;39:199–205. doi: 10.1097/JES.0b013e31822d71be. [DOI] [PubMed] [Google Scholar]
- 18.Saleem A., Carter H.N., Hood D.A. P53 is necessary for the adaptive changes in cellular milieu subsequent to an acute bout of endurance exercise. Am. J. Physiol. Cell Physiol. 2014;306:C241–C249. doi: 10.1152/ajpcell.00270.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saleem A., Adhihetty P.J., Hood D.A. Role of p53 in mitochondrial biogenesis and apoptosis in skeletal muscle. Physiol. Genomics. 2009;37:58–66. doi: 10.1152/physiolgenomics.90346.2008. [DOI] [PubMed] [Google Scholar]
- 20.Beyfuss K., Erlich A.T., Triolo M., Hood D.A. The role of p53 in determining mitochondrial adaptations to endurance training in skeletal muscle. Sci. Rep. 2018;8:14710. doi: 10.1038/s41598-018-32887-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stocks B., Dent J.R., Joanisse S., McCurdy C.E., Philp A. Skeletal muscle fibre-specific knockout of p53 does not reduce mitochondrial content or enzyme activity. Front. Physiol. 2017;8:941. doi: 10.3389/fphys.2017.00941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Leary M.F.N., Hood D.A. Effect of prior chronic contractile activity on mitochondrial function and apoptotic protein expression in denervated muscle. J. Appl. Physiol. 2008;105:114–120. doi: 10.1152/japplphysiol.00724.2007. [DOI] [PubMed] [Google Scholar]
- 23.O′Leary M.F.N., Vainshtein A., Carter H.N., Zhang Y., Hood D.A. Denervation-induced mitochondrial dysfunction and autophagy in skeletal muscle of apoptosis-deficient animals. Am. J. Physiol. Cell Physiol. 2012;303:C447–C454. doi: 10.1152/ajpcell.00451.2011. [DOI] [PubMed] [Google Scholar]
- 24.Supek F., Bošnjak M., Škunca N., Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Papa L., Germain D. SirT3 regulates the mitochondrial unfolded protein response. Mol. Cell. Biol. 2014;34:699–710. doi: 10.1128/MCB.01337-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beyfuss K., Hood D.A. A systematic review of p53 regulation of oxidative stress in skeletal muscle. Redox Rep. 2018;23:100–117. doi: 10.1080/13510002.2017.1416773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saleem A., Iqbal S., Zhang Y., Hood D.A. Effect of p53 on mitochondrial morphology, import, and assembly in skeletal muscle. Am. J. Physiol. Cell Physiol. 2015;308:C319–C329. doi: 10.1152/ajpcell.00253.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matoba S., Kang J.-G., Patino W.D., Wragg A., Boehm M., Gavrilova O., Hurley P.J., Bunz F., Hwang P.M. p53 regulates mitochondrial respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 29.Ebert S.M., Dierdorff J.M., Meyerholz D.K., Bullard S.A., Al-Zougbi A., DeLau A.D., Tomcheck K.C., Skopec Z.P., Marcotte G.R., Bodine S.C., Adams C.M. An investigation of p53 in skeletal muscle aging. J. Appl. Physiol. 2019;127:1075–1084. doi: 10.1152/japplphysiol.00363.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fox D.K., Ebert S.M., Bongers K.S., Dyle M.C., Bullard S.A., Dierdorff J.M., Kunkel S.D., Adams C.M. p53 and ATF4 mediate distinct and additive pathways to skeletal muscle atrophy during limb immobilization. Am. J. Physiol. Endocrinol. Metab. 2014;307:E245–E261. doi: 10.1152/ajpendo.00010.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Halling J.F., Pilegaard H. PGC-1α-mediated regulation of mitochondrial function and physiological implications. Appl. Physiol. Nutr. Metab. 2020;45:927–936. doi: 10.1139/apnm-2020-0005. [DOI] [PubMed] [Google Scholar]
- 32.Adhihetty P.J., Uguccioni G., Leick L., Hidalgo J., Pilegaard H., Hood D.A. The role of PGC-1alpha on mitochondrial function and apoptotic susceptibility in muscle. Am. J. Physiol. Cell Physiol. 2009;297:C217–C225. doi: 10.1152/ajpcell.00070.2009. [DOI] [PubMed] [Google Scholar]
- 33.Qureshi M.A., Haynes C.M., Pellegrino M.W. The mitochondrial unfolded protein response: Signaling from the powerhouse. J. Biol. Chem. 2017;292:13500–13506. doi: 10.1074/jbc.R117.791061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He C., Hart P.C., Germain D., Bonini M.G. SOD2 and the mitochondrial UPR: Partners regulating cellular phenotypic transitions. Trends Biochem. Sci. 2016;41:568–577. doi: 10.1016/j.tibs.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourougaa K., Naski N., Boularan C., Mlynarczyk C., Candeias M.M., Marullo S., Fåhraeus R. Endoplasmic reticulum stress induces G2 cell-cycle arrest via mRNA translation of the p53 isoform p53/47. Mol. Cell. 2010;38:78–88. doi: 10.1016/j.molcel.2010.01.041. [DOI] [PubMed] [Google Scholar]
- 36.Bennett C.F., Kaeberlein M. The mitochondrial unfolded protein response and increased longevity: Cause, consequence, or correlation? Exp. Gerontol. 2014;56:142–146. doi: 10.1016/j.exger.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Melber A., Haynes C.M. UPR mt regulation and output: A stress response mediated by mitochondrial-nuclear communication. Cell Res. 2018;28:281–295. doi: 10.1038/cr.2018.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiang D., Cui H., Xie N., Banerjee S., Liu R.-M., Dai H., Thannickal V.J., Liu G. ATF4 mediates mitochondrial unfolded protein response in alveolar epithelial cells. Am. J. Respir. Cell Mol. Biol. 2020;63:478–489. doi: 10.1165/rcmb.2020-0107OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pietrocola F., Izzo V., Niso-Santano M., Vacchelli E., Galluzzi L., Maiuri M.C., Kroemer G. Regulation of autophagy by stress-responsive transcription factors. Semin. Cancer Biol. 2013;23:310–322. doi: 10.1016/j.semcancer.2013.05.008. [DOI] [PubMed] [Google Scholar]
- 40.Goiran T., Duplan E., Rouland L., el Manaa W., Lauritzen I., Dunys J., You H., Checler F., Alves da Costa C. Nuclear p53-mediated repression of autophagy involves PINK1 transcriptional down-regulation. Cell Death Differ. 2018;25:873–884. doi: 10.1038/s41418-017-0016-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang C., Lin M., Wu R., Wang X., Yang B., Levine A.J., Hu W., Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. U. S. A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Z., Wang H., Ding Q., Xing Y., Xu D., Xu Z., Zhou T., Qian B., Ji C., Pan X., Zhong A., Ying Z., Zhou C., Shi M. The tumor suppressor p53 regulates autophagosomal and lysosomal biogenesis in lung cancer cells by targeting transcription factor EB. Biomed. Pharmacother. 2017;89:1055–1060. doi: 10.1016/j.biopha.2017.02.103. [DOI] [PubMed] [Google Scholar]
- 43.Cogswell A.M., Stevens R.J., Hood D.A. Properties of skeletal muscle mitochondria from subsarcolemmal and intermyofibrillar isolated regions. Am. J. Physiol. 1993;264:C383–C389. doi: 10.1152/ajpcell.1993.264.2.C383. [DOI] [PubMed] [Google Scholar]
- 44.Vainshtein A., Kazak L., Hood D.A. Effects of endurance training on apoptotic susceptibility in striated muscle. J. Appl. Physiol. 2011;110:1638–1645. doi: 10.1152/japplphysiol.00020.2011. [DOI] [PubMed] [Google Scholar]
- 45.Ostojic O., O'Leary M.F.N., Singh K., Menzies K.J., Vainshtein A., Hood D.A. The effects of chronic muscle use and disuse on cardiolipin metabolism. J. Appl. Physiol. 2013;114:444–452. doi: 10.1152/japplphysiol.01312.2012. [DOI] [PubMed] [Google Scholar]
- 46.Vainshtein A., Tryon L.D., Pauly M., Hood D.A. Role of PGC-1α during acute exercise-induced autophagy and mitophagy in skeletal muscle. Am. J. Physiol. Cell Physiol. 2015;308:C710–C719. doi: 10.1152/ajpcell.00380.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parousis A., Carter H.N., Tran C., Erlich A.T., Mesbah Moosavi Z.S., Pauly M., Hood D.A. Contractile activity attenuates autophagy suppression and reverses mitochondrial defects in skeletal muscle cells. Autophagy. 2018;14:1886–1897. doi: 10.1080/15548627.2018.1491488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carter H.N., Kim Y., Erlich A.T., Zarrin-khat D., Hood D.A. Autophagy and mitophagy flux in young and aged skeletal muscle following chronic contractile activity. J. Physiol. 2018;596:3567–3584. doi: 10.1113/JP275998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim Y., Triolo M., Erlich A.T., Hood D.A. Regulation of autophagic and mitophagic flux during chronic contractile activity-induced muscle adaptations. Pflügers Arch. 2018;471:431–440. doi: 10.1007/s00424-018-2225-x. [DOI] [PubMed] [Google Scholar]
- 50.Triolo M., Slavin M., Moradi N., Hood D.A. Time-dependent changes in autophagy, mitophagy and lysosomes in skeletal muscle during denervation-induced disuse. J. Physiol. 2022 doi: 10.1113/JP282173. In press. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data used to generate the data figures are available upon request from Dr David A. Hood (dhood@yorku.ca).