Summary
Proteomics analysis of skeletal muscle has recently progressed from whole muscle tissue to single myofibers. Here, we further focus on a specific myofiber domain crucial for force transmission from muscle to tendon, the myotendinous junction (MTJ). To overcome the anatomical constraints preventing the isolation of pure MTJs, we performed in-depth analysis of the MTJ by progressive removal of the muscle component in semitendinosus muscle-tendon samples. Using detergents with increasing stringency, we quantified >3000 proteins across all samples, and identified 112 significantly enriched MTJ proteins, including 24 known MTJ-enriched proteins. Of the 88 novel MTJ markers, immunofluorescence analysis confirmed the presence of tetraspanin-24 (CD151), kindlin-2 (FERMT2), cartilage intermediate layer protein 1 (CILP), and integrin-alpha10 (ITGA10), at the human MTJ. Together, these human data constitute the first detailed MTJ proteomics resource that will contribute to advance understanding of the biology of the MTJ and its failure in pathological conditions.
Subject areas: Biology experimental methods, Molecular physiology, Musculoskeletal anatomy, Proteomics
Graphical abstract
Highlights
-
•
Deep proteome analysis of human skeletal muscle and tendon
-
•
The first proteomic map of human myotendinous junction (MTJ)
-
•
Discovery of 88 novel MTJ-enriched proteins
-
•
Validation of novel MTJ markers by immunofluorescence
Biology experimental methods; Molecular physiology; Musculoskeletal anatomy; Proteomics
Introduction
A major progress in the definition of the protein profile of skeletal muscle, made possible by recent developments in liquid chromatography-mass spectrometry (LCMS)-based approaches, has been the transition from bulk muscle (Deshmukh et al., 2015) to single fiber proteomics (Murgia et al., 2015, 2017, 2021; Deshmukh et al., 2021; Kallabis et al., 2020; Lang et al., 2018). Here, we take a step further in this direction and report the first detailed proteomic analysis of a specific myofiber domain, the myotendinous junction (MTJ). The MTJ is responsible for the transmission of contractile force from muscle to skeleton; thus, knowledge of its molecular composition is essential to understand the process of mechanotransduction at the muscle-tendon interface and its failure under pathological conditions.
The MTJ is highly specialized in both structure and composition to meet the challenges of linking two very different tissues and withstand great mechanical stresses. Structural specialization of the MTJ includes finger-like processes, where collagen fibrils from the tendon interdigitate with extensive folding/invagination in the muscle fiber membrane at the tips of the muscle fibers. These finger-like processes have been estimated to increase the surface contact area between the muscle fibers and tendon by up to 20-fold (Knudsen et al., 2015), thereby reducing the stress, and increasing the strength, at the interface between muscle fibers and tendon (Kojima et al., 2008; Tidball, 1991). Despite the high degree of specialization, the MTJ is the main site of muscle strain injuries. Previous strain injuries, together with aging, are the strongest risk factors for strain injury (Green et al., 2020). Moreover, approximately four out of 5 re-injuries occur at the same site as the original injury (Wangensteen et al., 2016), indicating suboptimal healing of the MTJ after injury.
Despite this important role in injuries, our knowledge of the MTJ composition in general, and the human MTJ in particular, remains limited, mainly because access to human MTJ is restricted to surgically removed tissue. Moreover, the study of the MTJ has been limited to immunohistochemical studies that showed the enrichment of different sarcolemmal proteins at the myofiber tip, which is not surprising given the increased surface area at these sites. The most important finding has been the discovery that collagen XXII is specifically located at the MTJs (Koch et al., 2004). Recently, single-nucleus RNA-seq studies of mouse muscles have identified a specific cluster of MTJ myonuclei with a distinct mRNA profile, including COL22A1 mRNA, supporting the notion of a specialized transcriptional domain at the myofiber tip (Chemello et al., 2020; Dos Santos et al., 2020; Kim et al., 2020; Petrany et al., 2020; Wen et al., 2021). However, an unbiased inventory of the proteins detected at the MTJ based on proteomics analyses is not yet available.
Anatomical constraints do not allow for isolation of pure MTJ from muscle and tendon. Moreover, the proteomic analysis of these specialized tissues is challenging due to their structure and composition (Deshmukh et al., 2015; Sato et al., 2016). Two previous studies have attempted to map MTJ-specific proteins in the rodent muscle. However, the coverage of the proteome was incomplete in one study (Can et al., 2014), in which collagen XXII was not even identified, and the other study was limited to the analysis of extracellular matrix components (Jacobson et al., 2020). Here, we devised an enrichment approach by progressively reducing the muscle component of human semitendinosus muscle/tendon samples and extracting proteins through gradual increase in the stringency of the detergents. Each protein fraction was subsequently analyzed by MS-based proteomics, allowing mapping of the first human MTJ proteome. It is expected that these data will provide a resource and the basis for a better understanding of the intact healthy human MTJ and its remodeling under pathological conditions.
Results
Sequential protein extraction allows for a deep muscle and tendon proteome
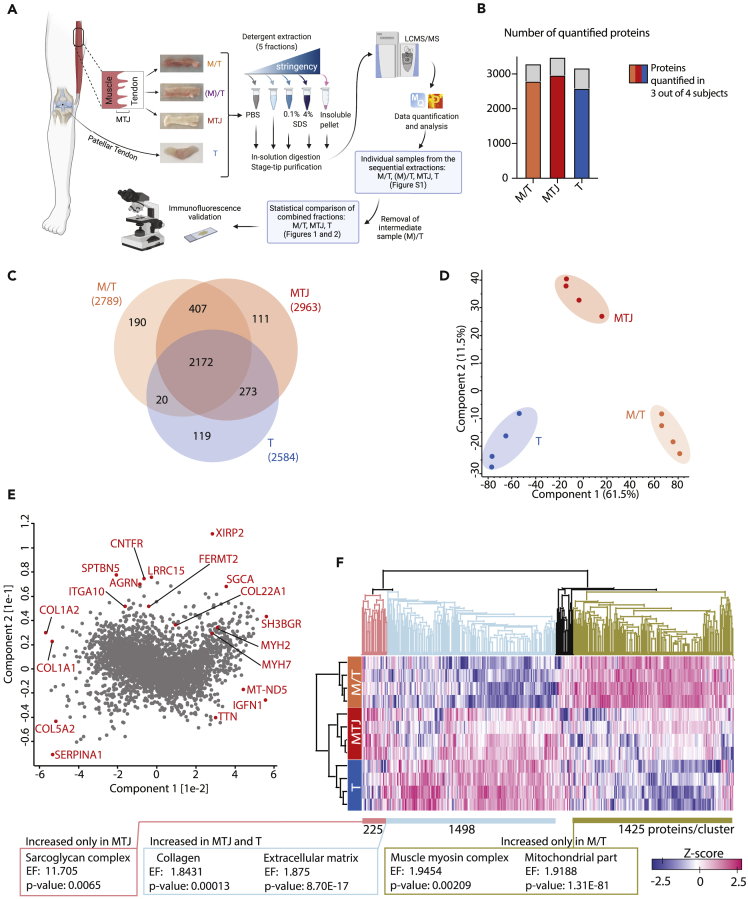
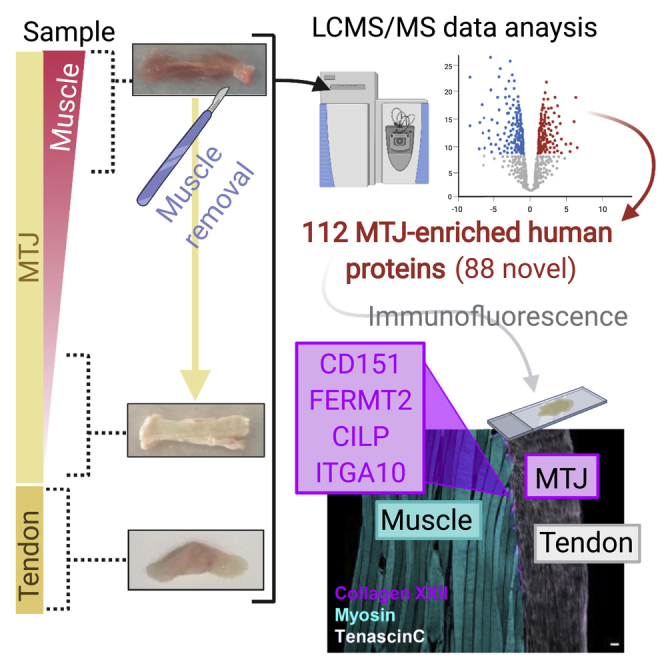
Samples of intact human semitendinosus muscle-tendon complex were collected from four individuals during anterior cruciate ligament (ACL) reconstruction surgery and divided a priori into three sample types by progressively scraping the muscle tissue off the tendon: i) non-scraped samples, consisting of muscle and tendon (muscle/tendon, M/T), ii) samples with the muscle partially removed to obtain less muscle on the tendon (a little muscle with tendon, (M)/T), and iii) samples with all visible muscle scraped away (MTJ) (Figure 1A). In addition, four human patellar tendon biopsies (T), which are not anatomically connected to skeletal muscle tissue, were included as a pure tendon control. The use of buffers with increasing detergent concentration allowed for the sequential extraction of proteins in M/T, (M)/T, MTJ, and T samples, which were then prepared for LCMS-based proteomics (Figure 1A). We first analyzed the individual samples from the sequential protein extractions as independent experiments. This allowed for the study of the accommodation of this method to investigate MTJ proteins (Table S1) as it showed the segregation by extraction buffer (Figure S1A) or sample type (Figure S1B). We observed the highest abundance of the MTJ-marker collagen XXII in the MTJ samples (Table S1). Moreover, Pearson correlation analysis and hierarchical clustering analysis revealed that the (M)/T samples were closely related to M/T and MTJ samples (Figures S1C and S1D). Hence, the intermediate (M)/T sample was removed from the further analysis. Expectedly, removal of the intermediate (M)/T sample resulted in a better segregation between the sample types (Figure S1E). Of note, this segregation was not explained by inter-individual variability among participants (Figure S1F), and the number of proteins detected in each extraction fraction was similar across samples types (Figure S1G).
Figure 1.
Myotendinous junction proteome
(A) Illustration of the workflow and representative images of the four sample types (M/T, (M)/T, MTJ, and T).
(B) Number of quantified proteins in sample types M/T, MTJ, and T after removal of the (M)/T sample, before and after (colored) filtering for proteins quantified in at least three out of four subjects (biological replicates).
(C) Venn diagram showing the overlap between the quantified proteins in each sample type after filtering (as in D).
(D) Principal component analysis (PCA) (fractions/extractions merged per sample type) showing the segregation of the sample types M/T, MTJ, and T, including only proteins quantified in at least three out of four subjects in at least one sample type.
(E) Loading PCA for D, examples of proteins driving the principal components segregation in Figure 1D (plotted furthest from the cluster center) are highlighted (red).
(F) Hierarchical clustering including only proteins quantified in at least three out of four subjects in at least one sample type. Abbreviations: M/T: muscle-tendon sample type; (M)/T: a little muscle with tendon sample type; MTJ: myotendinous junction sample type; T: tendon sample type; PBS: phosphate-buffered saline; NP40: nonidet P-40, non-ionic detergent; SDS: sodium dodecyl sulfate; EF: Enrichment factor.
For the statistical comparison, after removal of the (M)/T sample, proteins quantified in different extraction fractions were combined into their respective sample type. We obtained comprehensive proteome coverage, quantifying 3918 proteins across all samples and on an average > 3000 proteins in each sample type (Table S2). The quantified proteome spanned across six orders of magnitude and included low-abundant proteins such as cytokines (interleukin 33: IL33; C-C-motif chemokine ligand 14 and 18: CCL14 and CCL18), kinases (phosphatidylinositol 4-kinase type 2 alpha: PI4K2A; mechanistic target of rapamycin kinase: MTOR, epidermal growth factor receptor: EGFR), transcription factors (nuclear factor kappa B subunit 1: NFKB1, BCL2-associated transcriptor factor 1: BCLAF1), as well as 121 extracellular matrix (ECM) proteins belonging to the core matrisome (21 collagens, 83 ECM glycoproteins, and 17 proteoglycans) (Naba et al., 2012), which are generally difficult to detect due to their lower solubility (Table S2). In this analysis, we quantified 2789, 2963, and 2584 proteins in M/T, MTJ, and T, respectively (Table S2, Figure 1B). The vast majority of proteins (2172) were quantified in all three sample types in at least three out of four subjects (Figure 1C). A principal component analysis (PCA) showed a clear segregation of each sample type (Figure 1D), and a low and comparable inter-individual variance between subject within each sample type. The segregation of the sample types was driven by different proteins (Figure 1E, in red), such as collagen I and XIRP2 (xin actin-binding repeat-containing protein 2/xinβ), specific proteins for tendon and MTJ, respectively. Additionally, we performed hierarchical clustering of the proteome of the three sample types (Figure 1F), and observed i) the samples separated clearly, with MTJ being closer to tendon (T) than the muscle/tendon (M/T) and ii) three clear expression patterns: a cluster of proteins increased only in MTJ samples (Figure 1F, salmon color), increased in MTJ and T (light blue) and increased only in M/T (golden brown). We then performed the enrichment analysis using GOCC (Gene Ontology Cellular Component) terms within these clusters in order to assess the cellular localization of the majority of proteins (Table S3). Proteins belonging to the sarcoglycan complex were enriched in the MTJ cluster (Figure 1F, salmon); proteins associated with collagen and located in the ECM were over-represented among the proteins increased in the MTJ and T sample types (Figure 1F, light blue), while proteins related to the muscle myosin complex and mitochondria were enriched in the M/T samples (Figure 1F, golden brown). Details regarding the other enriched protein categories for individual clusters can be found in Table S3.
Proteomics analysis revealed known and novel proteins enriched at the MTJ region
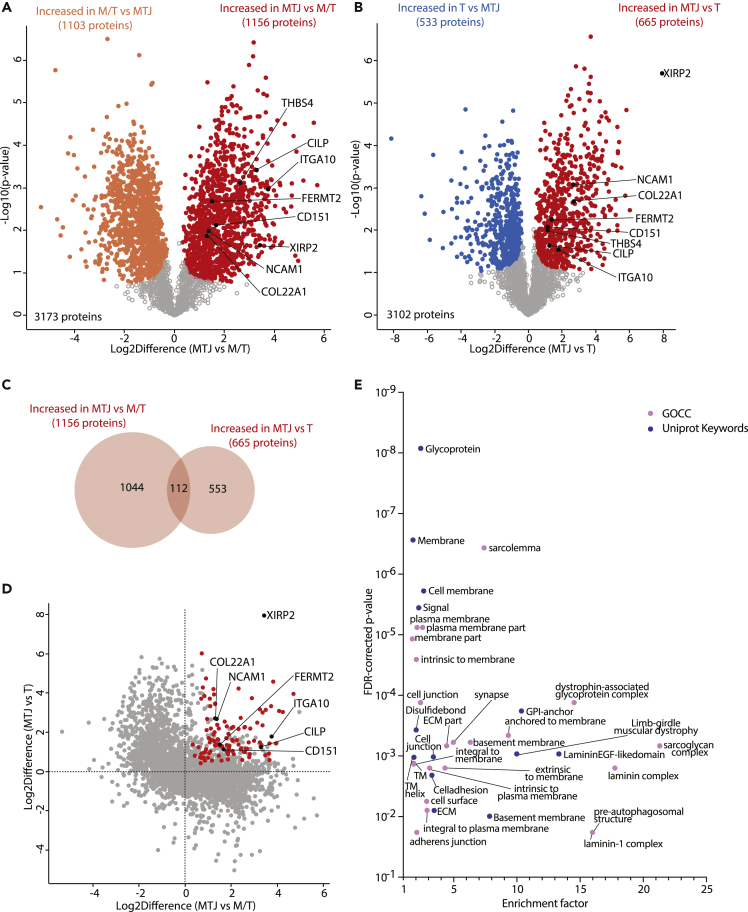
To identify proteins enriched at the MTJ region, we performed pairwise two-samples t test between the proteins in the MTJ vs M/T and MTJ vs T samples (Figures 2A and 2B). In the comparison between MTJ and M/T, 1156 (36.4%) proteins were significantly increased in MTJ (Figure 2A, red, Table S2), whereas in the comparison between MTJ and T, 665 (21.4%) proteins returned which were significantly increased in MTJ (Figure 2B, red, Table S2).
Figure 2.
Comparison of MTJ proteome with muscle and tendon samples
(A) Volcano plot displaying the comparison of M/T vs MTJ proteome. Orange and red indicate differentially down- and upregulated proteins in the MTJ, respectively.
(B) Volcano plot displaying the comparison of T vs MTJ proteome. Blue and red indicate differentially down- and upregulated proteins in the MTJ, respectively.
(C) Venn diagram displaying the overlap between the upregulated proteins in A and B.
(D) Scatter plot showing the fold change of common proteins in both pair-comparisons (A and B). In red, proteins differentially upregulated in both comparisons (also red in A and B).
(E) Fisher‘s exact test enrichment of the 112 upregulated proteins in both comparisons (in red in D) in comparison to all the common proteins (all proteins plotted in D). Student’s T-test (FDR = 0.05, s0 = 0.01) with permutation-based FDR (false discovery rate) correction. Fisher Exact Test cut-off: 0.02. Abbreviations: M/T: muscle-tendon sample type; MTJ: myotendinous junction sample type; T: tendon sample type; GOCC: gene ontology cellular component.
These results were then combined to identify proteins significantly increased in MTJ in both pair comparisons, thus enriched in the MTJ samples, resulting in a final dataset with a total of 112 proteins (Figures 2C and 2D, in red, Table S4). Of these, we can report 88 novel MTJ-enriched proteins, together with 24 proteins known to be enriched at the MTJ in vertebrates (Table S4), including collagen XXII (COL22A1), which is exclusively confined to the MTJ in the skeletal muscle-tendon unit (Jakobsen et al., 2017; Koch et al., 2004), as well as several laminin subunits and 10 proteins belonging to the dystrophin glycoprotein complex (DGC) which is known to be enriched at the MTJ (Charvet et al., 2012; Grady et al., 2003).
To gain further insights into the 112 MTJ-specific proteins, we performed an enrichment analysis within these proteins using GOCC and UniProt Keywords annotations (Figure 2E and Table S5), which revealed an over-representation of cell membrane-related annotations. The two most enriched GOCC terms were the sarcoglycan and the laminin complexes (enrichment factors: 21.3 and 17.8, respectively), and the two most enriched Uniprot Keywords were “lamininEGF-likedomain” (enrichment factor: 13.3) and “GPI-anchor” (enrichment factor: 10.3).
Confirmatory validation of the isolation of the MTJ region with known MTJ-enriched proteins
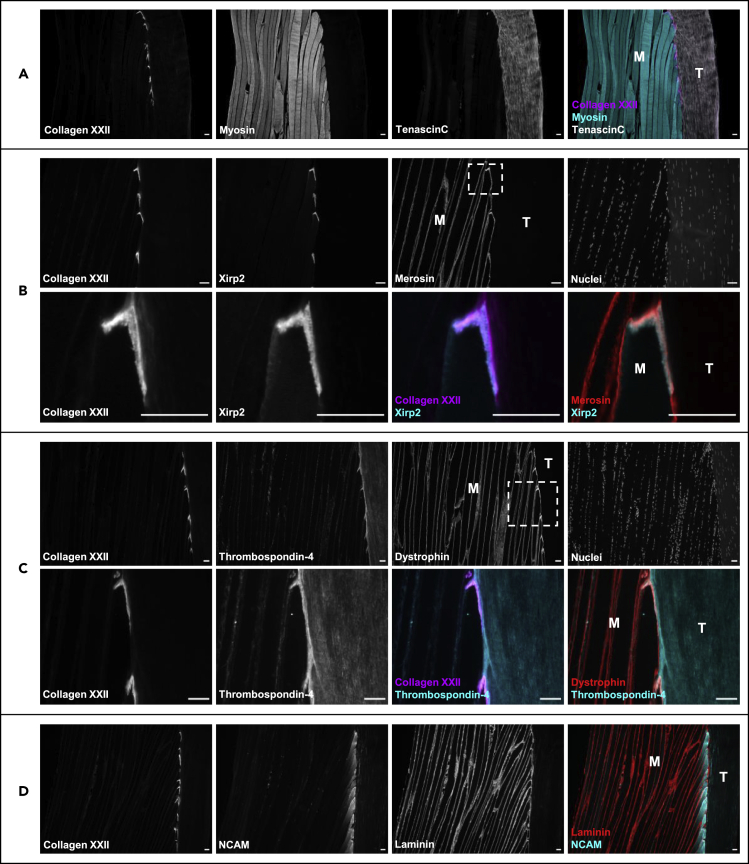
We then performed immunofluorescent staining of selected proteins significantly enriched at the MTJ versus M/T and T, and known to be present in various cellular compartments of the MTJ in vertebrate (XIRP2, thrombospondin-4 (THBS4)) or human (neural cell adhesion molecule 1 (NCAM1)) (Figure 3) to validate the MS-based proteomic data. For illustrative purposes Figure 3A is a representative image of a 10-μm thick MTJ section of human semitendinosus, cut in the longitudinal plane of the muscle fibers, and stained for myosin heavy chain, collagen XXII, and tenascin C, to visualize the muscle fibers, MTJ, and tendon, respectively. A very strong immunofluorescent signal for XIRP2 was evident (Figure 3B), exclusively confined to the MTJ on the muscle fiber side of the collagen XXII staining. In contrast to XIRP2, THBS4 was enriched, but not exclusively confined to the MTJ (Figure 3C), as THBS4 signal was also seen in the tendon together with a vague signal outside the dystrophin-stained sarcolemma along the muscle fibers (Figure 3C). NCAM1 was the only MTJ-enriched protein in the proteome dataset identified from razor peptides only (Table S2), and the immunofluorescent staining showed a concentrated signal in the muscle fiber cytoplasm for a length of 200–300 μm before fiber termination at the MTJ (Figure 3D).
Figure 3.
XIRP2, THBS4, and NCAM at the human MTJ
(A) Illustrative example of a 10-μm thick section of human semitendinosus muscle/tendon tissue from ACL patients. The myotendinous junction is marked with collagen XXII, skeletal muscle fibers “M” are stained for myosin heavy chain, and the tendon “T” is stained with tenascin (C).
(B) Strong XIRP2 (xin actin-binding repeat-containing protein 2) immunofluorescent signal confined to the myotendinous junction (MTJ) on the muscle side of the merosin (LAMA2)-stained basement membrane.
(C) Thrombospondin-4 (THBS4) immunofluorescent signal is enriched at the MTJ, located at the extracellular matrix side of the dystrophin (DMD)-stained sarcolemma.
(D) NCAM1 (neural cell adhesion molecule 1) immunofluorescent signal is concentrated in the muscle fiber cytoplasm at the MTJ. The dotted line boxes indicate areas imaged at higher magnification. Scale bars are 50 μm.
Validation of novel MTJ-enriched proteins
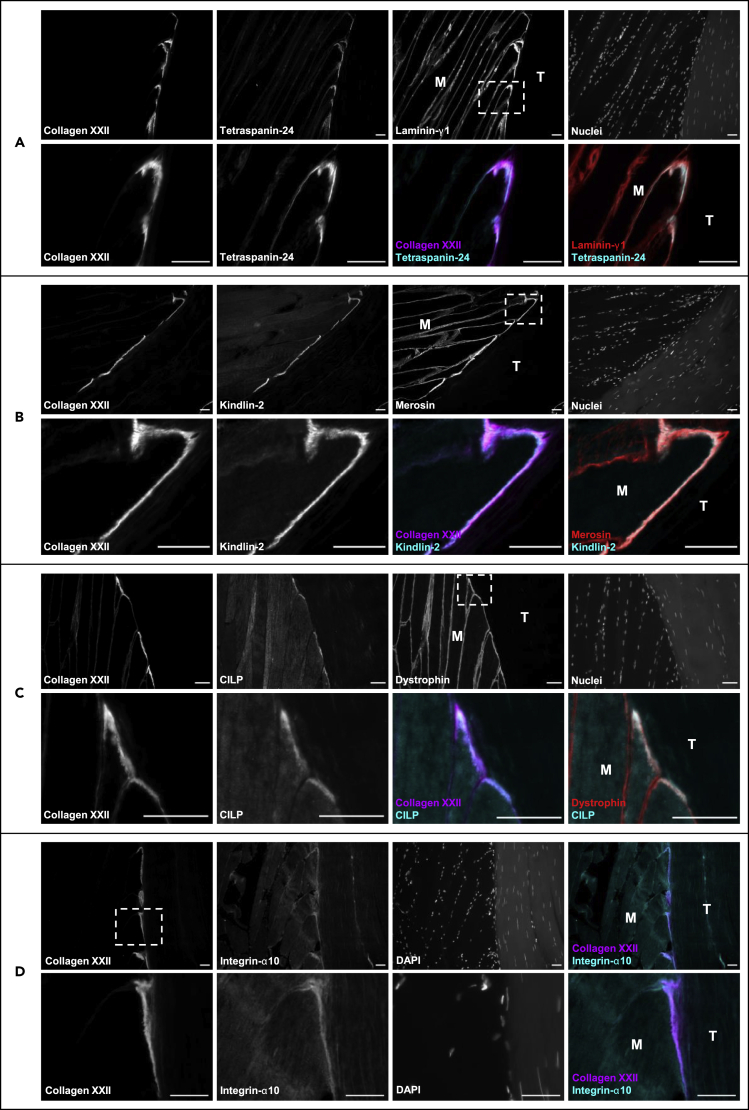
MS-based proteomics and a stringent downstream bioinformatics analysis led to discovery of 88 novel MTJ-enriched proteins, which we aimed to confirm as MTJ-enriched by immunofluorescence (Figure 4). Tetraspanin-24 (CD151/PETA-3, Figure 4A) and kindlin-2 (FERMT2/UNC112B/MIG2, Figure 4B) showed very strong staining exclusively at the MTJ, on the muscle side of collagen XXII, and the laminin-stained basement membrane. A weak tetraspanin-24 signal could also be observed along the muscle fiber membrane in some fibers (Figure 4A). Similarly, cartilage intermediate layer protein 1 (CILP) staining was strong at the muscle fiber tips at the muscle side of collagen XXII and in close proximity to the dystrophin-stained sarcolemma, while a weak CILP reactivity was also seen in the muscle fiber cytoplasm, which appeared stronger in few of the fibers (Figure 4C). In addition, a positive signal was seen for integrin-α10 (ITGA10) at the muscle fiber tips in close proximity to collagen XXII (Figure 4D), although at a lower intensity compared to collagen XXII, XIRP2, tetraspanin-24, kindlin-2, and CILP.
Figure 4.
Tetraspanin-24, kindlin-2, CILP, and integrin-α10 at the human MTJ
All images are from 10-μm thick sections of human semitendinosus muscle/tendon tissue from patients with ACL. Muscle fibers “M” and tendon “T” meet at the myotendinous junction (MTJ), stained with collagen XXII.
(A) Strong tetraspanin-24 (CD151) signal at the MTJ on the muscle side of collagen XXII and the laminin-γ1 (LAMC1)-stained basement membrane.
(B) Strong kindlin-2 (FERMT2) signal at the MTJ on the muscle side of collagen XXII and the merosin (LAMA2) stained basement membrane.
(C) Strong CILP (cartilage intermediate layer protein 1) signal at the MTJ on the muscle side of collagen XXII in close association with the dystrophin (DMD)-stained sarcolemma.
(D) Strong integrin-α10 signal at the MTJ in close proximity with collagen XXII. The dotted line boxes indicate areas imaged at higher magnification. Scale bars are 50 μm.
Discussion
A major recent advance in the proteomic analysis of skeletal muscle has been the development of procedures for single-fiber proteomics. Here, we have zoomed in even further on the MTJ, the specialized domain at the muscle-tendon interface. The MTJ is structurally organized in a complex manner and cannot be anatomically isolated from muscle and tendon, posing a challenge for MS-based proteomics analysis due to presence of high-abundant proteins (Deshmukh et al., 2015; Sato et al., 2016). Moreover, the MTJ consists of proteins belonging to the ECM, which represents a cross-linked and insoluble assembled matrix. It is therefore challenging to extract the proteins from this tissue to study the proteomic composition of the MTJ. To overcome these challenges, we accommodated an experimental approach including scraping the semitendinosus tendon free of muscle tissue before using buffers with increasing stringency to obtain samples enriched in MTJ proteins. The extraction procedure, based on progressively increasing detergent concentration, was essentially similar to that applied by (Schiller et al., 2015) to characterize lung fibrosis. We identified 112 MTJ-enriched proteins of which 24 were known and 88 were novel. Using immunofluorescence, we further confirmed enrichment of the novel MTJ-specific tetraspanin-24, kindlin-2, CILP, and integrin-α10, together representing pathways in force transmission, membrane folding, and extracellular matrix binding. These results represent the first MS-based proteome of the human MTJ and will serve as a valuable resource for developing methods for proteomic studies of specialized domains and the understanding of adaptation and failure of force transmission and regeneration at the MTJ.
The tips of the muscle fibers, where they insert onto the tendon at the MTJ, represent a distinct myocellular domain, characterized by the expression of certain proteins such as Nestin (Vaittinen et al., 1999) and NCAM1 (Daniloff et al., 1989; Jakobsen et al., 2018), not expressed in the rest of the musculotendinous unit. As the muscle fibers (which can be several centimeters long) approach the MTJ, collagen XXII is observed in the muscle fiber basement membrane (Koch et al., 2004). We initially compared semitendinosus muscle-tendon samples with decreasing amounts of muscle by gradually scraping the muscle off the tendon to varying degrees. The greatest enrichment of collagen XXII was detected in the samples that had been most harshly scraped, presumably because the tips of the muscle fibers are deeply embedded in the tendon and the MTJ-enriched proteins are not masked by an excess of muscle proteins. This observation formed the basis of the methodology applied in the present study, where we also included a biopsy of pure patellar tendon (which is not anatomically connected to skeletal muscle and is therefore completely free of muscle- and MTJ-specific proteins) to identify tendon-specific proteins in the MTJ samples. Furthermore, our methodological approach included a sequential protein extraction with buffers of increasing detergent concentration before MS-based proteomics, in combination with a downstream stringent filter that included only those proteins quantified in at least three out of four subjects (biological replicates).
Our methodological approach was validated by an enrichment analysis within the 112 MTJ-enriched proteins that revealed an over-representation of GOCC and UniProt Keywords annotation terms related to “sarcoglycan complex”, “laminin complex”, “cell junction”, “membrane”, “adhesion”, and “extracellular matrix”. These annotations verified that proteins in our list can be located in both the muscle and the tendon side of the MTJ. The high prevalence of proteins belonging to the DGC in the MTJ is not surprising given that DGC proteins are known to be more abundant in the MTJ domain of the myofibers (Charvet et al., 2012; Grady et al., 2003). Moreover, these proteins may have contributed to many of the enriched GOCC terms and UniProt Keywords. Altogether, these enriched annotations reflect that the MTJ is rich in membrane components, in line with the finger-like protrusions that form extensive membrane foldings at the tip of the muscle fibers (Knudsen et al., 2015). The enrichment of annotations such as “cell junction”, “adherens junction”, and “anchored to membrane” further reflect the MTJ-specific requirement of forming a strong link between muscle fibers and tendon.
Several of the 112 MTJ-enriched proteins belonged to the extracellular matrisome (Naba et al., 2012) and have not previously been associated with the MTJ, including but not limited to: Agrin (AGRN), vitrin (VIT), metalloproteinase inhibitor 3 (TIMP3), annexin A3 and A4 (ANXA3, ANXA4), plexin-A1 (PLXNA1), and syndecan-2 (SDC2). Notably, annexin A1 was observed in mouse MTJ with proteomics and immunofluorescence (Can et al., 2014), while our data suggest a role for other members of the annexin family at the human MTJ. Several other novel and potentially interesting MTJ-enriched proteins were observed, such as anthrax toxin receptor 1 (ANTXR1), a transmembrane protein shown to interact with actin and affect cell adhesion; caveolin (CAV2), involved in formation of invaginations to the plasma membrane; the actin binding coactosin-like protein (COTL1); the ephrin type-A receptor 2 (EPHA2), involved in integrin-mediated adhesion and formation of tissue boundaries during embryogenesis; beta-parvin (PARVB), an actin-binding protein with a role in cytoskeleton organization and cell adhesion; and reversion-inducing cysteine-rich protein with Kazal motifs (RECK), a membrane-anchored matrix metalloproteinase-regulator also found in the NMJ (Kawashima et al., 2008). In addition, we also observed enrichment of hemicentin-1 (HMCN1) in MTJ, which is known to link tissues together at the basement membrane and was very recently described at the MTJ (Welcker et al., 2021). The list also contained several proteins located in the Golgi apparatus and/or ER or involved in processes related to endocytosis, autophagy, microtubules, T-cells, enzymatic activity, and the nucleus/transcriptional machinery, together expanding insight into the complex diversity in proteomic composition and cellular processes at the MTJ.
By using immunofluorescence, we confirmed the presence of known components of the MTJ (i.e., XIRP2, THBS4, and NCAM1). XIRP2 and THBS4 have not previously been shown in human MTJ, and the positive immunofluorescent staining of XIRP2, THBS4, and NCAM1 at the MTJ region (Figures 3B–3D) validates our method to detect MTJ-enriched proteins with distinct staining profiles and enrichment scores.
We then performed immunofluorescent stainings for selected targets among the 88 MTJ-enriched proteins in our dataset, which have not been previously reported in the myotendinous location. Kindlin-2 and tetraspanin-24 are both known to associate with the transmembrane integrins and were therefore chosen for further examination by immunofluorescence. Similarly to XIRP2, stainings of tetraspanin-24 (Sterk et al., 2002) and kindlin-2 (Dowling et al., 2008) have been shown at the intercalated disc of the human and murine heart, respectively, as well as at the costameres in cardiac muscle cells and skeletal muscle fibers (Dowling et al., 2008; Sterk et al., 2002). To our knowledge, this is the first report of tetraspanin-24 and kindlin-2 enrichment at the MTJ in skeletal muscle fibers. Although the specific role of these proteins at the MTJ is currently unknown, we find it relevant that tetraspanins are involved in both membrane invagination (Yue et al., 2017) and linking cell membranes via morphogenesis of microextrusions (Huang et al., 2018), whereas kindlin-2 plays a key role in the induction of membrane protrusions and formation of lamellipodia during early isotropic fibroblast cell spreading (Theodosiou et al., 2016). Altogether, this could indicate a role for tetraspanin-24 and kindlin-2 in the extensive folding/invaginations of the tendon into the muscle fiber membrane at the MTJ region.
Some of the novel MTJ-enriched proteins are traditionally associated with cartilage. Given that the MTJ-marker collagen XXII has also been identified in the articular surface of cartilage (Koch et al., 2004), we decided to further examine the location of two cartilage-associated proteins at the MTJ: the integrin-α10 subunit that is expressed together with the β1-subunit in cartilage chondrocytes (Camper et al., 1998) and the cartilage intermediate layer protein 1 (CILP), which is located in the mid-zone of human articular cartilage (Lorenzo et al., 1998). Integrin-α10β1 belongs to a group of collagen-binding integrins (α1β1, α2β1, α10β1, and α11β1), and collagen XXII shows binding affinity for all of them (Zwolanek et al., 2014). At this point, it is not fully clear which muscle fiber membrane proteins collagen XXII binds to at the MTJ, and our data could suggest a role for integrin-α10 as a binding partner for collagen XXII in adult human MTJ.
The localization of CILP near the sarcolemma at the MTJ (Figure 4D) suggests that MTJ-specialized myonuclei may be the source of CILP. In line with this, recent snRNA-seq data in mice showed that CILP gene expression was increased in a sub cluster of MTJ myonuclei (Kim et al., 2020), but also in tenocytes (Petrany et al., 2020) and fibro-adipogenic precursors (Dos Santos et al., 2020). CILP was first discovered in cartilage as an extracellular matrix glycoprotein that increases with aging, progression of osteoarthritis (Lorenzo et al., 1998), and degeneration of intervertebral discs at the lumbar spine (Seki et al., 2005). At the protein level, CILP also shows immunoreactivity in fibrotic regions in human hearts from patients with ischemic cardiomyopathy, but not in healthy controls (Barallobre-Barreiro et al., 2012; Park et al., 2020). As a negative regulator of cardiac fibrosis, it is tempting to speculate that CILP also has an anti-fibrotic function in the MTJ of skeletal muscle, which could be important in a region that is subjected to repeated stress during daily muscle contractions, and also in the accumulation of fibrotic scar tissue after muscle strain injuries (Silder et al., 2008).
In addition to CILP, the pre-mRNAs coding for 19 out of the 112 MTJ-enriched proteins identified in human biopsies were selectively upregulated in MTJ-myonuclei clusters described in four recent snRNA-seq studies of mouse skeletal muscle (Petrany et al., 2020; Dos Santos et al., 2020; Kim et al., 2020; Wen et al., 2021). Only COL22A1, XIRP2, LAMA2, and LAMC1 were found in all four studies; whereas NRAP and APP were found in three studies (Petrany et al., 2020; Dos Santos et al., 2020; Wen et al., 2021); ANXA4, ITGA7, SGCD, and SLC44A2 in two studies (Petrany et al., 2020; Wen et al., 2021); and several transcripts were found in one study: CILP, ITIH5, LAMB1 (Kim et al., 2020); DES, SORBS1 (Dos Santos et al., 2020); FERMT2, LAMB2, NCAM1 (Wen et al., 2021); RRAS2, SGCG (Petrany et al., 2020). Along with CILP, these 19 proteins constitute both known and novel MTJ-enriched proteins. Furthermore, in snRNA-seq data from Ex51 mice (a mouse model of Duchenne’s muscular dystrophy) (Chemello et al., 2020), the gene transcripts for 26 of the 112 MTJ-enriched proteins were differentially regulated (15 upregulated and 11 downregulated). The observation that many of the 112 MTJ-enriched proteins reported here were not found in the snRNA-seq datasets may reflect a species difference (human vs. mouse) and the fact that many proteins at the MTJ do not necessarily originate from MTJ myonuclei, which underlines the specificity and value of measurements at the transcript and protein levels.
To our knowledge, only two studies have pioneered the study of the MTJ proteome in detail (Can et al., 2014; Jacobson et al., 2020). Can et al. used laser capture microdissection of MTJ and pure muscle, but not the tendon, from hindlimb and forelimb muscles in mice. While this approach should give a high enrichment of MTJ-specific proteins, our study demonstrates the limitations of not including a tendon control sample. Another limitation to this time-consuming method was the low total amount of isolated tissue, leading to the need to pool the samples from all biological replicates prior to LCMS analysis and impeding statistical analysis (Can et al., 2014). A different approach was used by Jacobson et al., in which the whole mouse soleus muscle was divided into pure muscle, pure tendon, and a muscle-tendon region that included the MTJ (Jacobson et al., 2020). In that study, the authors used buffers of increasing stringency to extract the insoluble proteins from the ECM and tendon prior to the MS analysis, similarly to the present study. However, our study was adapted to further improve the MTJ proteome coverage by performing an experimental approach where muscle tissue was scraped away to specifically isolate MTJ-enriched region differently than previous studies (Jacobson et al., 2020), therefore avoiding the masking of low-abundant MTJ-specific proteins by high-abundant muscle proteins. Altogether, in addition to the largest MTJ-enriched proteome, we generated the deepest human tendon proteome to date, as well as one of the most comprehensive human muscle proteome (Gonzalez-Freire et al., 2017) which could serve as a valuable resource for the scientific community.
The present study provides the first human MTJ proteome and a deeper understanding of the molecular components of this highly specialized myofiber domain. We combined graded muscle removal to enrich for proteins in the MTJ region, buffers with increasing stringency, and high-resolution MS-based proteomics to deliver the first human MTJ proteome to date. By using this methodology, we obtained a purer MTJ and quantified known MTJ-enriched proteins (i.e. collagen XXII, NCAM), thus demonstrating its fit to also overcome the high dynamic range that could mask low-abundant proteins in this region. The 88 novel MTJ marker proteins identified underline the high degree of specialization of this myofiber domain and, considering their functions in other tissues, suggest an important role in strengthening and repair of the MTJ. Overall, this study provides a valuable proteomics resource with novel insights into the composition of a myofiber domain that is highly specialized to withstand great mechanical forces and, paradoxically, also susceptible to strain injury rupture.
Limitations of the study
One of the limitations of this study is the challenge in obtaining “pure” MTJ tissue, because the MTJ is a combination of muscle fibers and their associated connective tissue on one side, and tendon tissue on the other. While the strong representation of known MTJ proteins in our MTJ fraction validates our approach of progressively removing the muscle tissue, it is likely that the method can be refined further. Remaining work also includes reconciling differences between the MTJ proteins identified in our study and the MTJ genes identified at the transcript level in the mouse.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD151 (Tetraspanin-24), Mouse IgG2b. | Proteintech | Cat# 66567-1-Ig; RRID: AB_2881928 |
| CILP (Cartilage Intermediate Layer Protein 1), Rabbit IgG, Batch/Lot: A20210218198. | MyBioSource | Cat# MBS2006189; RRID: AB_2895588 |
| COL22A1 (Collagen XXII), Guinea pig IgG. | Manuel Koch (Koch et al. 2004) | N/A |
| DMD MANDYS8 (Dystrophin), Mouse IgG2b. | Sigma-Aldrich | Cat# D8168; RRID: AB_259245 |
| FERMT2 (Kindlin-2), Rabbit IgG, Batch/Lot: A106069. | Thermo Fisher Scientific | Cat# PA5-59200; RRID: AB_2643120 |
| ITGA10 (Integrin Subunit Alpha 10), Rabbit IgG. | Abbexa | Cat# abx101874; RRID: AB_2895589 |
| Laminin (all isoforms), Rabbit IgG, Batch/Lot: 20035405. | Dako | Cat# Z0097; RRID: AB_2313665 |
| LAMA2 (Laminin Subunit Alpha 2, Merosin), Mouse IgG1; Batch/Lot: 123108. | Novocastra/Leica Biosystems | Cat# NCL-MEROSIN; RRID: AB_442108 |
| LAMC1 (Laminin Subunit Gamma 1), Mouse IgG2a. | DSHB, deposited by Engvall, E.S. | Cat# 2E8; RRID: AB_528343 |
| Myosin (all isoforms), Mouse IgG2a. | DSHB, deposited by Blau, H.M. | Cat# A4.1025; RRID: AB_528356 |
| CD56 (NCAM1, Neural Cell Adhesion Molecule 1), mouse IgG1. | BD Biosciences | Cat# 347740; RRID: AB_400345 |
| THBS4 (Thrombospondin 4), Rabbit IgG. | Manuel Koch (This Paper) | N/A |
| TNC (Tenascin C), Mouse IgG2b | Novocastra/Leica Biosystems | Cat# NCL-TENAS-C; RRID: AB_564026 |
| XIRP2 (Xin Actin Binding Repeat Containing Protein 2), Rabbit IgG, Batch/Lot: A118323. | Thermo Fischer Scientific | Cat# PA5-57072; RRID: AB_2649681 |
| Goat anti-Guinea Pig IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 647. | Thermo Fischer Scientific | Cat# A-21450; RRID: AB_2735091 |
| Goat anti-Guinea Pig IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568. | Thermo Fischer Scientific | Cat# A-11075; RRID: AB_2534119 |
| Goat anti-Mouse IgG1 Cross-Adsorbed Secondary Antibody, Alexa Fluor 488. | Thermo Fischer Scientific | Cat# A-21121; RRID: AB_2535764 |
| Goat anti-Mouse IgG2a Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fischer Scientific | Cat# A-21131; RRID: AB_2535771 |
| Goat anti-Mouse IgG2a, Cross-Adsorbed Secondary Antibody, Alexa Fluor 680 | Thermo Fischer Scientific | Cat# A31563; RRID: AB_2536177 |
| Goat anti-Mouse IgG2b Cross-Adsorbed Secondary Antibody, Alexa Fluor 488 | Thermo Fischer Scientific | Cat# A-21141; RRID: AB_2535778 |
| Donkey anti-Rabbit IgG (H + L) Highly Cross-Adsorbed Secondary Antibody, Alexa Fluor 568 | Thermo Fischer Scientific | Cat# A10042; RRID: AB_2534017 |
| Alexa Fluor 680 AffiniPure Donkey Anti-Guinea Pig IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 706-625-148; RRID: AB_2340478 |
| Alexa Fluor 594 AffiniPure Donkey Anti-Mouse IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 715-585-151; RRID: AB_2340855 |
| Alexa Fluor 488 AffiniPure Donkey Anti-Rabbit IgG (H + L) | Jackson ImmunoResearch Labs | Cat# 711-545-152; RRID: AB_2313584 |
| Biological samples | ||
| Human Semitendinosus Muscle/Tendon Biopsies | This Paper | N/A |
| Human Patellar Tendon Biopsies | This Paper | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Protease inhibitor cocktail (with EDTA) | Sigma | Cat# S8820 |
| IGEPAL® CA-630 | Sigma | Cat# I8896 |
| MgCl2 | Sigma | Cat# M8266 |
| Benzonase | Merck | Cat# 70746-3 |
| Phosphatase inhibitors | Roche | Cat# 04906837001 |
| Sodium deoxycholate (SDC) | Sigma | Cat# 30970 |
| 10% Sodium dodecyl sulfate (SDS) | Sigma | Cat# 71736 |
| Dithiothreitol (DTT) | Sigma | Cat# D0632 |
| Iodoacetamide (IAA) | Sigma | Cat# I6125 |
| LysC | Wako | Cat# 4548995075888 |
| Trypsin | Promega | Cat# V5111 |
| Trifluoroacetic acid (TFA) | Sigma | Cat# 302031 |
| Acetonitrile (ACN) | Merck | Cat# 100030 |
| Deposited data | ||
| Raw and processed MS data | Proteomics Identifications (PRIDE) | Accession number PXD029307; RRID: SCR_003411 |
| Software and algorithms | ||
| MaxQuant | https://maxquant.org/ | RRID:SCR_014485 |
| Perseus | www.coxdocs.org/doku.php?id=perseus:start | RRID:SCR_015753 |
| Olympus cellSens Software | http://www.olympus-lifescience.com/en/software/cellsens/ | RRID:SCR_014551 |
| ImageJ | https://imagej.net/ | RRID:SCR_003070 |
Resource availability
Lead contact
For further information in relation to resources and reagents in the proteomic analysis, please contact Atul Deshmukh (lead contact) (atul.deshmukh@sund.ku.dk) and for information in relation to the immunofluorescense analysis and antibodies please contact Abigail L. Mackey (abigailmac@sund.ku.dk).
Materials availability
This study did not generate new unique reagents.
Experimental model and subject details
Human subjects and sample collection
All tissue donors provided informed written consent to participate in the study, which was approved by The Research Ethics Committees of the Capital Region of Denmark (ref. H-3-2010–070 and ref. H-2-2010-100), in accordance with the Declaration of Helsinki II.
Human tissue was collected from eight healthy young males (20–44 years old), undergoing either anterior cruciate ligament (ACL) reconstruction surgery (n = 4) or patellar tendon biopsies (n = 4). Uninjured semitendinosus muscle-tendon tissue was collected during ACL surgeries following standard procedures. The surgeon harvested the semitendinosus tendon and secured the tissue needed for the ACL autograft before the excess (most proximal part) of the harvested tissue was placed in PBS in a falcon tube on ice and transferred to the laboratory. The sample consisted of bundles of muscle fibres attached to the semitendinosus tendon and was divided into three parts; M/T, comprised of unaltered muscle-tendon tissue; (M)/T was scraped gently with a scalpel to remove some, but not all, of the muscle from the tendon; MTJ was strongly scraped of all visible muscle tissue to obtain an enrichment of MTJ-proteins together with the tendon. Patellar tendon biopsies (T) were taken with a 14-G needle (Bard Magnum Biopsy Instrument, C.R. Bard, Covington, GA) under local anesthetics (lidocaine, 1%) after sterilization of the skin. All samples for proteomics were placed in cryotubes, frozen in liquid nitrogen and stored at −80°C. In addition, semitendinosus muscle-tendon tissue for histology was carefully aligned and embedded in Tissue-Tek, and frozen in isopentane pre-cooled in liquid nitrogen before storage in cryotubes at −80°C. 10 μm thick sections from human semitendinosus muscle-tendon samples were oriented in the longitudinal plane of the muscle fibers and cut in a cryostat at −20°C. The sections were collected on glass slides and stored at −80°C.
Method details
Proteomics sample preparation
Proteins from the four different types of samples (M/T, (M)/T, MTJ and T) were extracted as described previously with slight modifications (Schiller et al., 2015). Of note, 4% SDS (instead of 1%) was used as the strongest buffer in order to increase the extraction of MTJ-specific proteins. Briefly, ∼40 mg of tissue (wet weight) was homogenized in 10 volumes of PBS (uL), with protease inhibitor cocktail and EDTA using an Ultra-turrax homogenizer. Homogenates were then spun down (20 min, 16000 g, 4°C) and the supernatant with soluble proteins was collected as the PBS fraction. The rest of the proteins were extracted in four steps with 400 uL of buffers with increasing stringency [buffer 1: 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 5% glycerol, 1% IGEPAL® CA-630 (Sigma, #I8896), 1 mM MgCl2, 1× protease inhibitors (+EDTA), 1% benzonase (Merck, #70746-3), 1× phosphatase inhibitors (Roche, #04906837001); buffer 2: 50 mM Tris-HCl (pH 7.5), 5% glycerol, 150 mM NaCl, fresh protease inhibitor tablet (+EDTA), 1.0% IGEPAL® CA-630, 0.5% sodium deoxycholate, 0.1% SDS, 1% benzonase (Merck, #70746-3); and buffer 3:50 mM Tris-HCl (pH 7.5), 5% glycerol, 500 mM NaCl, protease inhibitor tablet (+EDTA), 1.0% IGEPAL® CA-630, 2% sodium deoxycholate, 4% SDS, 1% benzonase (Merck, #70746-3)]. The soluble and insoluble material were separated by centrifugation (20 min, 16000 g) and the soluble material was collected as a new fraction. During each extraction step, insoluble pellets were resuspended in detergent-containing buffers and incubated for 20 min on ice prior centrifugation. Since the buffer 3 contains 4% SDS, the incubation was performed at room temperature. In total, we obtained 4 soluble fractions (PBS, Buffers1-3) and one insoluble pellet. Soluble protein in each fraction was acetone-precipitated: 4 volumes of 100% ice-cold acetone were added to the volume for ∼50 ug of protein and sample were vortexed and stored overnight at −20°C. Samples were centrifuged (10 min, 18000 g, 4°C) and supernatant removed carefully to avoid to dislodge the protein pellet, and then washed twice with 100% acetone and later with 80% acetone. The protein pellet was air-dried after the last wash and resuspended with 40 uL of urea/thiourea (U/T buffer, 6M/2M). The insoluble pellet obtained from the extraction using buffer 3 was resuspended in 300 uL U/T buffer and 200 manual strokes were performed for 2 min continuously with a 2 mL micro-Dounce device. All the samples were then sonicated 5 min (30 s on/off cycles) in a Bioruptor and the insoluble pellet fraction was further tip-sonicated for other 5 min (30 s on, 1 s on, 1 s off, at 50% intensity).
For the LCMSMS analysis, proteins (50 μg protein/sample, or all the sample if less) were digested using in-solution digestion protocol. Samples were reduced with 0.1 M DTT for 30 min, and alkylated with 55 mM of IAA for 20 min in the dark, both steps taking place at room temperature. For the digestion, samples were digested using endoproteinase LysC (1 μg enzyme/50 μg protein) for 1 h at room temperature. The samples were then diluted 1:4 with 25 mM Tris pH 8.5 and further digested with endoproteinase trypsin (1 μg enzyme/50 μg protein) for overnight at 37°C with shaking. For the digestion of insoluble pellet fraction, double amount of enzymes was used and the samples were sonicated (15 min, Bioruptor, 30 s on/off cycles) after the addition of each enzymes. Samples were incubated overnight at 37°C with shaking. Digested proteins were spun down (2000 rpm, 5 min) and the supernatant containing extracted peptides was collected. Samples were acidified by adding 5x volumes of 1% TFA in isopropanol. Peptides were purified using StageTips containing a poly-styrene-divinylbenzene copolymer modified with sulfonic acid groups (SDB-RPS) material (3M, St. Paul, MN, USA). Briefly, peptides were loaded on 3x SDB-RPS discs, washed once with 1% TFA in isopropanol and twice with 0.2%TFA and eluted in a microtiter plate in a single step with 60 uL of 1% ammonia in 80% acetonitrile (ACN). For the washes and elution, the samples were spun down at 700 g for 3-10 min. The elution buffer was evaporated in a concentrator (Eppendorf) and the peptides resuspended in 10 μL of 0.1% TFA in 5% ACN buffer.
LC-MS/MS analysis
Peptides were analyzed on the LCMS instrumentation consisting of an Easy nanoflow UHPLC (Thermo Fisher Scientific) coupled via a nanoelectrospray ion source (Thermo Fisher Scientific) to a Q Exactive mass spectrometer - HFX (Thermo Fisher Scientific). Peptides were separated on 50 cm column with 75 μm inner diameter packed in-house with ReproSil-Pur C18-aq 1.9 μm resin (Dr Maisch). Peptides were loaded in 0.5% formic acid buffer and eluted with a 100 min linear gradient with 80% acetonitrile and 0.5% formic acid (v/v) containing buffer at 350 nL/min. Mass spectra were acquired in a data-dependent manner, with an automatic switch between MS and MS/MS using a top 15 method. MS spectra were acquired in the Orbitrap analyzer with a mass range of 300–1650 m/z and 60 000 resolution at m/z 200. HCD peptide fragments were acquired with normalized collision energy of 27. The maximum ion injection times for the survey scan and the MS/MS scans were 45 ms, and the ion target values were set to 3e6 and 1e4, respectively. Data were acquired using Xcalibur software.
Immunofluorescence experiments
Slides were removed from the freezer and dried at room temperature. Primary antibodies (applied overnight at 4°C) and secondary antibodies (applied the following day for 60 min at room temperature) were diluted in 1% BSA (bovine serum albumin, IgG free) in TBS (tris-buffered saline, tris-base 0.05 mol/L, sodium chloride 0.154 mol/L, pH 7.4–7.6). To optimize staining quality, tests were run for fixation with 4% paraformaldehyde (5 min) or Histofix (12 min) either prior to incubation with primary antibodies (CILP) or after incubation with secondary antibodies. An overview of antibodies is available in the Key Resource Table. Samples were washed for 5 min x3 in TBS between all steps in the protocol. Slides were finally mounted with cover glasses, and DAPI in the mounting medium stained nuclei blue (Molecular Probes ProLong Gold anti-fade reagent, cat. no. P36931). Images were captured with different objectives (10x/0.3NA; 20x/0.5NA; 40x/0.75NA) on an Olympus BX51 microscope with a 0.5x camera (Olympus DP71, Olympus Deutschland GmbH, Hamburg, Germany), controlled by the Olympus cellSens Software (www.olympus-lifescience.com). Images were subsequently viewed and cropped for presentation in ImageJ (version 1.53c; National Institute of Health; USA), and colorblind friendly pseudo colors were applied to composite images.
Quantification and statistical analysis
Computational MSMS data analysis
Mass spectra were analyzed using MaxQuant (version −1.6.01) with the integrated Adromeda search engine (Tyanova et al., 2016a). The initial maximum allowed mass deviation was set to 6 ppm for monoisotopic precursor ions and 20 ppm for MS/MS peaks. Enzyme specificity was set to trypsin, defined as C-terminal to arginine and lysine excluding proline, and a maximum of two missed cleavages were allowed. A minimal peptide length of six amino acids was required. Carbamidomethylcysteine was set as a fixed modification, while N-terminal acetylation and methio-nine oxidation were set as variable modifications. The spectra were searched by the Andromeda search engine against the Human UniProt sequence database combined with 248 common contaminants and concatenated with the reversed versions of all sequences. The false discovery rate (FDR) was set to 1% for peptide and protein identifications. To match the identifications across different LC-MS runs, the “match between runs” option in MaxQuant was enabled with a retention time window of 30 s. In the case of identified peptides that were shared between two or more proteins, these were combined and reported in protein group. Label free protein quantification was performed using built in MaxLFQ algorithm in MaxQuant software. The raw files were processed twice. First, to evaluate the extraction efficiency of different buffers (Table S1, Figures S1A, S1B and S1E)), each raw files were treated as individual experiment. For the statistical comparison (Table S2, Figures 1, 2 ,S1C and S1D), the raw MS files for the proteins extracted with different buffers were treated as individual fractions of respective samples.
Bioinformatics statistical analysis
Global bioinformatics analysis was performed on the label-free quantifications intensities reported by MaxQuant software. The bioinformatics analysis was performed using the Perseus software (version 1.6.5.0) (Tyanova et al., 2016b). Categorical annotations were supplied in the form of Gene Ontology Cellular Component (GOCC) and UniProt Keywords. Contaminants and reverse identification were removed from further data analysis. For the analysis where each fractions were considered as individual experimental sample (Figures S1A, S1B and S1E): proteins quantified in at least 50% of samples were included for further analysis. For the statistical comparison where protein list from the individual fractions was merged into respective sample type (Figures 1, 2, S1C and S1D), proteins quantified in at least 3 out of 4 subjects in at least one of the sample type groups were included in further analysis. The data was imputed to fill missing values by drawing random numbers from Gaussian distribution with 30% of standard deviation in comparison to the standard deviation of the valid values, and one standard deviation down-shifted from the mean to simulate the distribution of low signal values. The number of quantified proteins per group (Figure S1G) are reported without performing imputation.
Principal component analysis and Hierarchical clustering was performed on the imputed data. The paired two sample t-test was performed to compare the MTJ samples with M/T and T samples (FDR = 0.05, s0 = 0.1). Based on the correlation analysis, (M)/T samples were very similar to M/T and MTJ samples (Figure S1C), therefore were excluded from the comparison. Fisher exact tests were performed for the enrichment or depletion of specific protein categories using Benjamini-Hochberg correction with an FDR cutoff of 0.02.
Acknowledgments
The authors would like to thank Rebeca Soria Romero for her assistance with the experimental approach. This work was supported by unconditional donations from the Novo Nordisk Foundation (NNF) to NNF Center for Basic Metabolic Research (NNF18CC0034900) and NNF Center for Protein Research (NNF14CC001); and supported by Nordea Foundation (Center for Healthy Aging Grant), and Novo Nordisk Foundation (NNF0064829) to AM, The Lundbeck Foundation (R303-2018-3427) to AK and by the Deutsche Forschungsgemeinschaft (FOR2722) to Ma.K.
Author contributions
A.S.D. and A.L.M. supervised this work. S.S., Mi.K., A.L.M., and A.S.D. contributed to the first hypothesis generation. All authors contributed to conceptual design. M.R.K., J.R.J., and Ma.K. provided resources. A.K., A.G-F., J.R.J., S.S., A.L.M., and A.S.D. developed the methodology, and A.K. and A.G-F. performed the experiments. A.K. and A.G.F. performed data analysis and visualization. A.K., A.G-F., S.S., A.L.M., and A.S.D. performed analysis and data interpretation. A.K., A.G-F., S.S., A.L.M., and A.S.D. wrote the manuscript. All authors edited and reviewed the manuscript.
Declaration of interest
The authors declare no conflicts of interest.
Published: February 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103836.
Contributor Information
Abigail L. Mackey, Email: abigailmac@sund.ku.dk.
Atul S. Deshmukh, Email: atul.deshmukh@sund.ku.dk.
Supporting citations
The following reference appears in the Supplemental Information: Bao et al., 1993; Feng et al., 2013; Luo et al., 1997; Miner et al., 2006; Subramanian and Schilling, 2014; Subramanian et al., 2007; Tidball, 1992; Tidball et al., 1986; Zhang et al., 2006.
Supplemental information
Filter by proteins with at least 50% of valid values across all fractions and samples
Filter by proteins in at least 3 out of 4 subjects per sample type
Data and code availability
-
•
Data are available via the PRIDE repository (Perez-Riverol et al., 2019) (accession number PXD029307)
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the reported data is available from the lead contact upon request.
References
- Bao Z.Z., Lakonishok M., Kaufman S., Horwitz A.F. Alpha 7 beta 1 integrin is a component of the myotendinous junction on skeletal muscle. J. Cell Sci. 1993;106:579–589. doi: 10.1242/jcs.106.2.579. [DOI] [PubMed] [Google Scholar]
- Barallobre-Barreiro J., Didangelos A., Schoendube F.A., Drozdov I., Yin X., Fernández-Caggiano M., Willeit P., Puntmann V.O., Aldama-López G., Shah A.M., et al. Proteomics analysis of cardiac extracellular matrix remodeling in a porcine model of ischemia/reperfusion injury. Circulation. 2012;125:789–802. doi: 10.1161/CIRCULATIONAHA.111.056952. [DOI] [PubMed] [Google Scholar]
- Camper L., Hellman U., Lundgren-Åkerlund E. Isolation, cloning, and sequence analysis of the integrin subunit α10, a β1-associated collagen binding integrin expressed on chondrocytes. J. Biol. Chem. 1998;273:20383–20389. doi: 10.1074/jbc.273.32.20383. [DOI] [PubMed] [Google Scholar]
- Can T., Faas L., Ashford D.A., Dowle A., Thomas J., O’Toole P., Blanco G. Proteomic analysis of laser capture microscopy purified myotendinous junction regions from muscle sections. Proteome Sci. 2014;12:25. doi: 10.1186/1477-5956-12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charvet B., Ruggiero F., Le Guellec D. The development of the myotendinous junction. A review. Muscles Ligaments Tendons J. 2012;2:53–63. [PMC free article] [PubMed] [Google Scholar]
- Chemello F., Wang Z., Li H., McAnally J.R., Liu N., Bassel-Duby R., Olson E.N. Degenerative and regenerative pathways underlying Duchenne muscular dystrophy revealed by single-nucleus RNA sequencing. Proc. Natl. Acad. Sci. U S A. 2020;117:29691–29701. doi: 10.1073/pnas.2018391117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniloff J.K., Crossin K.L., Pinçon-Raymond M., Murawsky M., Rieger F., Edelman G.M. Expression of cytotactin in the normal and regenerating neuromuscular system. J. Cell Biol. 1989;108:625–635. doi: 10.1083/jcb.108.2.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh A.S., Murgia M., Nagaraj N., Treebak J.T., Cox J., Mann M. Deep proteomics of mouse skeletal muscle enables quantitation of protein isoforms, metabolic pathways, and transcription factors. Mol. Cell Proteomics. 2015;14:841–853. doi: 10.1074/mcp.M114.044222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh A.S., Steenberg D.E., Hostrup M., Birk J.B., Larsen J.K., Santos A., Kjøbsted R., Hingst J.R., Schéele C.C., Murgia M., et al. Deep muscle-proteomic analysis of freeze-dried human muscle biopsies reveals fiber type-specific adaptations to exercise training. Nat. Commun. 2021;12:304. doi: 10.1038/s41467-020-20556-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dos Santos M., Backer S., Saintpierre B., Izac B., Andrieu M., Letourneur F., Relaix F., Sotiropoulos A., Maire P. Single-nucleus RNA-seq and FISH identify coordinated transcriptional activity in mammalian myofibers. Nat. Commun. 2020;11:5102. doi: 10.1038/s41467-020-18789-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling J.J., Gibbs E., Russell M., Goldman D., Minarcik J., Golden J.A., Feldman E.L. Kindlin-2 is an essential component of intercalated discs and is required for vertebrate cardiac structure and function. Circ. Res. 2008;102:423–431. doi: 10.1161/CIRCRESAHA.107.161489. [DOI] [PubMed] [Google Scholar]
- Feng H.-Z., Wang Q., Reiter R.S., Lin J.L.-C., Lin J.J.-C., Jin J.-P. Localization and function of Xinα in mouse skeletal muscle. Am. J. Physiol. Cell Physiol. 2013;304:C1002–C1012. doi: 10.1152/ajpcell.00005.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Freire M., Semba R.D., Ubaida-Mohien C., Fabbri E., Scalzo P., Højlund K., Dufresne C., Lyashkov A., Ferrucci L. The human skeletal muscle proteome project: a reappraisal of the current literature. J. Cachexia Sarcopenia Muscle. 2017;8:5–18. doi: 10.1002/jcsm.12121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grady R.M., Akaaboune M., Cohen A.L., Maimone M.M., Lichtman J.W., Sanes J.R. Tyrosine-phosphorylated and nonphosphorylated isoforms of α-dystrobrevin. J. Cell Biol. 2003;160:741–752. doi: 10.1083/jcb.200209045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green B., Bourne M.N., van Dyk N., Pizzari T. Recalibrating the risk of hamstring strain injury (HSI): a 2020 systematic review and meta-analysis of risk factors for index and recurrent hamstring strain injury in sport. Br. J. Sports Med. 2020;54:1081–1088. doi: 10.1136/bjsports-2019-100983. [DOI] [PubMed] [Google Scholar]
- Huang C., Fu C., Wren J.D., Wang X., Zhang F., Zhang Y.H., Connel S.A., Chen T., Zhang X.A. Tetraspanin-enriched microdomains regulate digitation junctions. Cell Mol. Life Sci. 2018;75:3423–3439. doi: 10.1007/s00018-018-2803-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson K.R., Lipp S., Acuna A., Leng Y., Bu Y., Calve S. Comparative analysis of the extracellular matrix proteome across the myotendinous junction. J. Proteome Res. 2020;19:3955–3967. doi: 10.1021/acs.jproteome.0c00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen J.R., Jakobsen N.R., Mackey A.L., Koch M., Kjaer M., Krogsgaard M.R. Remodeling of muscle fibers approaching the human myotendinous junction. Scand. J. Med. Sci. Sports. 2018;28:1859–1865. doi: 10.1111/sms.13196. [DOI] [PubMed] [Google Scholar]
- Jakobsen J.R., Mackey A.L., Knudsen A.B., Koch M., Kjaer M., Krogsgaard M.R. Composition and adaptation of human myotendinous junction and neighboring muscle fibers to heavy resistance training. Scand. J. Med. Sci. Sports. 2017;27:1547–1559. doi: 10.1111/sms.12794. [DOI] [PubMed] [Google Scholar]
- Kallabis S., Abraham L., Müller S., Dzialas V., Türk C., Wiederstein J.L., Bock T., Nolte H., Nogara L., Blaauw B., et al. High-throughput proteomics fiber typing (ProFiT) for comprehensive characterization of single skeletal muscle fibers. Skelet. Muscle. 2020;10:7. doi: 10.1186/s13395-020-00226-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima S., Imamura Y., Chandana E.P.S., Noda T., Takahashi R., Adachi E., Takahashi C., Noda M. Localization of the membrane-anchored MMP-regulator RECK at the neuromuscular junctions. J. Neurochem. 2008;104:376–385. doi: 10.1111/j.1471-4159.2007.04977.x. [DOI] [PubMed] [Google Scholar]
- Kim M., Franke V., Brandt B., Lowenstein E.D., Schöwel V., Spuler S., Akalin A., Birchmeier C. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat. Commun. 2020;11:6375. doi: 10.1038/s41467-020-20064-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen A.B., Larsen M., Mackey A.L., Hjort M., Hansen K.K., Qvortrup K., Kjaer M., Krogsgaard M.R. The human myotendinous junction: an ultrastructural and 3D analysis study. Scand. J. Med. Sci. Sports. 2015;25 doi: 10.1111/sms.12221. e116–123. [DOI] [PubMed] [Google Scholar]
- Koch M., Schulze J., Hansen U., Ashwodt T., Keene D.R., Brunken W.J., Burgeson R.E., Bruckner P., Bruckner-Tuderman L. A novel marker of tissue junctions, collagen XXII. J. Biol. Chem. 2004;279:22514–22521. doi: 10.1074/jbc.M400536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H., Sakuma E., Mabuchi Y., Mizutani J., Horiuchi O., Wada I., Horiba M., Yamashita Y., Herbert D.C., Soji T., et al. Ultrastructural changes at the myotendinous junction induced by exercise. J. Orthop. Sci. 2008;13:233–239. doi: 10.1007/s00776-008-1211-0. [DOI] [PubMed] [Google Scholar]
- Lang F., Khaghani S., Türk C., Wiederstein J.L., Hölper S., Piller T., Nogara L., Blaauw B., Günther S., Müller S., et al. Single muscle fiber proteomics reveals distinct protein changes in slow and fast fibers during muscle atrophy. J. Proteome Res. 2018;17:3333–3347. doi: 10.1021/acs.jproteome.8b00093. [DOI] [PubMed] [Google Scholar]
- Lorenzo P., Bayliss M.T., Heinegård D. A novel cartilage protein (CILP) present in the mid-zone of human articular cartilage increases with age. J. Biol. Chem. 1998;273:23463–23468. doi: 10.1074/jbc.273.36.23463. [DOI] [PubMed] [Google Scholar]
- Luo G., Zhang J.Q., Nguyen T.P., Herrera A.H., Paterson B., Horowits R. Complete cDNA sequence and tissue localization of N-RAP, a novel nebulin-related protein of striated muscle. Cell Motil Cytoskeleton. 1997;38:75–90. doi: 10.1002/(SICI)1097-0169(1997)38:1<75::AID-CM7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Miner J.H., Go G., Cunningham J., Patton B.L., Jarad G. Transgenic isolation of skeletal muscle and kidney defects in lamininβ 2 mutant mice: implications for Pierson syndrome. Development. 2006;133:967–975. doi: 10.1242/dev.02270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M., Nagaraj N., Deshmukh A.S., Zeiler M., Cancellara P., Moretti I., Reggiani C., Schiaffino S., Mann M. Single muscle fiber proteomics reveals unexpected mitochondrial specialization. EMBO Rep. 2015;16:387–395. doi: 10.15252/embr.201439757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M., Nogara L., Baraldo M., Reggiani C., Mann M., Schiaffino S. Protein profile of fiber types in human skeletal muscle: a single-fiber proteomics study. Skelet Muscle. 2021;11:24. doi: 10.1186/s13395-021-00279-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murgia M., Toniolo L., Nagaraj N., Ciciliot S., Vindigni V., Schiaffino S., Reggiani C., Mann M. Single muscle fiber proteomics reveals fiber-type-specific features of human muscle aging. Cell Rep. 2017;19:2396–2409. doi: 10.1016/j.celrep.2017.05.054. [DOI] [PubMed] [Google Scholar]
- Naba A., Clauser K.R., Hoersch S., Liu H., Carr S.A., Hynes R.O. The matrisome: in silico definition and in vivo characterization by proteomics of normal and tumor extracellular matrices. Mol. Cell Proteomics MCP. 2012;11 doi: 10.1074/mcp.M111.014647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S., Ranjbarvaziri S., Zhao P., Ardehali R. Cardiac fibrosis is associated with decreased circulating levels of full-length CILP in heart failure. JACC Basic Transl Sci. 2020;5:432–443. doi: 10.1016/j.jacbts.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D.J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019;47:D442–D450. doi: 10.1093/nar/gky1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrany M.J., Swoboda C.O., Sun C., Chetal K., Chen X., Weirauch M.T., Salomonis N., Millay D.P. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 2020;11:6374. doi: 10.1038/s41467-020-20063-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N., Taniguchi T., Goda Y., Kosaka H., Higashino K., Sakai T., Katoh S., Yasui N., Sairyo K., Taniguchi H. Proteomic analysis of human tendon and ligament: solubilization and analysis of insoluble extracellular matrix in connective tissues. J. Proteome Res. 2016;15:4709–4721. doi: 10.1021/acs.jproteome.6b00806. [DOI] [PubMed] [Google Scholar]
- Schiller H.B., Fernandez I.E., Burgstaller G., Schaab C., Scheltema R.A., Schwarzmayr T., Strom T.M., Eickelberg O., Mann M. Time- and compartment-resolved proteome profiling of the extracellular niche in lung injury and repair. Mol. Syst. Biol. 2015;11:819. doi: 10.15252/msb.20156123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki S., Kawaguchi Y., Chiba K., Mikami Y., Kizawa H., Oya T., Mio F., Mori M., Miyamoto Y., Masuda I., et al. A functional SNP in CILP, encoding cartilage intermediate layer protein, is associated with susceptibility to lumbar disc disease. Nat. Genet. 2005;37:607–612. doi: 10.1038/ng1557. [DOI] [PubMed] [Google Scholar]
- Silder A., Heiderscheit B.C., Thelen D.G., Enright T., Tuite M.J. MR observations of long-term musculotendon remodeling following a hamstring strain injury. Skeletal Radiol. 2008;37:1101. doi: 10.1007/s00256-008-0546-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterk L.M.T., Geuijen C.A.W., Berg J.G. van den., Claessen N., Weening J.J., Sonnenberg A. Association of the tetraspanin CD151 with the laminin-binding integrinsα 3β1, α6β1, α6β4 and α7β1 in cells in culture and in vivo. J. Cell Sci. 2002;115:1161–1173. doi: 10.1242/jcs.115.6.1161. [DOI] [PubMed] [Google Scholar]
- Subramanian A., Schilling T.F. Thrombospondin-4 controls matrix assembly during development and repair of myotendinous junctions. eLife. 2014;3:e02372. doi: 10.7554/eLife.02372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Wayburn B., Bunch T., Volk T. Thrombospondin-mediated adhesion is essential for the formation of the myotendinous junction in Drosophila. Development. 2007;134:1269–1278. doi: 10.1242/dev.000406. [DOI] [PubMed] [Google Scholar]
- Theodosiou M., Widmaier M., Böttcher R.T., Rognoni E., Veelders M., Bharadwaj M., Lambacher A., Austen K., Müller D.J., Zent R., et al. Kindlin-2 cooperates with talin to activate integrins and induces cell spreading by directly binding paxillin. eLife. 2016;5:e10130. doi: 10.7554/eLife.10130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tidball J.G. Force transmission across muscle cell membranes. J. Biomech. 1991;24:43–52. doi: 10.1016/0021-9290(91)90376-x. [DOI] [PubMed] [Google Scholar]
- Tidball J.G. Desmin at myotendinous junctions. Exp. Cell Res. 1992;199:206–212. doi: 10.1016/0014-4827(92)90425-8. [DOI] [PubMed] [Google Scholar]
- Tidball J.G., O’Halloran T., Burridge K. Talin at myotendinous junctions. J. Cell Biol. 1986;103:1465–1472. doi: 10.1083/jcb.103.4.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016;11:2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Vaittinen S., Lukka R., Sahlgren C., Rantanen J., Hurme T., Lendahl U., Eriksson J.E., Kalimo H. Specific and innervation-regulated expression of the intermediate filament protein Nestin at neuromuscular and myotendinous junctions in skeletal muscle. Am. J. Pathol. 1999;154:591–600. doi: 10.1016/S0002-9440(10)65304-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangensteen A., Tol J.L., Witvrouw E., Van Linschoten R., Almusa E., Hamilton B., Bahr R. Hamstring reinjuries occur at the same location and early after return to sport: a descriptive study of MRI-confirmed reinjuries. Am. J. Sports Med. 2016;44:2112–2121. doi: 10.1177/0363546516646086. [DOI] [PubMed] [Google Scholar]
- Welcker D., Stein C., Feitosa N.M., Armistead J., Zhang J.-L., Lütke S., Kleinridders A., Brüning J.C., Eming S.A., Sengle G., et al. Hemicentin-1 is an essential extracellular matrix component of the dermal-epidermal and myotendinous junctions. Sci. Rep. 2021;11:17926. doi: 10.1038/s41598-021-96824-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y., Englund D.A., Peck B.D., Murach K.A., McCarthy J.J., Peterson C.A. Myonuclear transcriptional dynamics in response to exercise following satellite cell depletion. iScience. 2021;24:102838. doi: 10.1016/j.isci.2021.102838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue S., Zhao K., Erb U., Rana S., Zöller M. Joint features and complementarities of Tspan8 and CD151 revealed in knockdown and knockout models. Biochem. Soc. Trans. 2017;45:437–447. doi: 10.1042/BST20160298. [DOI] [PubMed] [Google Scholar]
- Zhang M., Liu J., Cheng A., Deyoung S.M., Chen X., Dold L.H., Saltiel A.R. CAP interacts with cytoskeletal proteins and regulates adhesion-mediated ERK activation and motility. EMBO J. 2006;25:5284–5293. doi: 10.1038/sj.emboj.7601406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwolanek D., Veit G., Eble J.A., Gullberg D., Ruggiero F., Heino J., Meier M., Stetefeld J., Koch M. Collagen XXII binds to collagen-binding integrins via the novel motifs GLQGER and GFKGER. Biochem. J. 2014;459:217–227. doi: 10.1042/BJ20130642. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Filter by proteins with at least 50% of valid values across all fractions and samples
Filter by proteins in at least 3 out of 4 subjects per sample type
Data Availability Statement
-
•
Data are available via the PRIDE repository (Perez-Riverol et al., 2019) (accession number PXD029307)
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the reported data is available from the lead contact upon request.