Abstract
Transgenic expression of bacterial nitroreductase (NTR) enzymes sensitizes eukaryotic cells to prodrugs such as metronidazole (MTZ), enabling selective cell ablation paradigms that have expanded studies of cell function and regeneration in vertebrates. However, first-generation NTRs required confoundingly toxic prodrug treatments to achieve effective cell ablation, and some cell types have proven resistant. Here, we used rational engineering and cross-species screening to develop a NTR variant, NTR 2.0, which exhibits ~100-fold improvement in MTZ-mediated cell-specific ablation efficacy, eliminating the need for near-toxic prodrug treatment regimens. NTR 2.0 therefore enables sustained cell loss paradigms and ablation of previously resistant cell types. These properties permit enhanced interrogations of cell function, extended challenges to the regenerative capacities of discrete stem cell niches, and novel modeling of chronic degenerative diseases. Accordingly, we have created a series of bipartite transgenic reporter/effector resources to facilitate dissemination of NTR 2.0 to the research community.
Editor’s Summary
An engineered bacterial nitroreductase, NTR 2.0, improves chemically induced cell ablation, facilitating novel sustained ablation paradigms for testing the effects of chronic inflammation on regeneration, and modeling degenerative disease.
INTRODUCTION:
Bacterial nitroreductases (NTRs) are promiscuous enzymes capable of prodrug conversion via reduction of nitro substituents on aromatic rings1–4. This generates genotoxic products that rapidly kill the host cell, a mechanism exploited by anti-cancer and antibiotic prodrugs5. When expressed heterologously, NTRs sensitize vertebrate cells to such prodrugs6. The canonical NTR, Escherichia coli NfsB (NfsB_Ec, “NTR 1.0”), has been widely tested in combination with the anti-cancer prodrug 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB1954) as an enzyme-prodrug therapy for killing tumors1. Transgenic expression of NTR 1.0 in combination with CB1954 was previously advanced as a targeted cell ablation strategy for interrogating cell function in vertebrates7,8. However, CB1954 produces cell-permeable cytotoxins that kill nearby non-targeted cells, i.e., “bystander” cell death9, compromising its use for selective cell ablation. In contrast, the prodrug metronidazole (MTZ) ablates NTR-expressing cells without discernible bystander effects9.
Importantly, fusion proteins between NTR and fluorescent reporters retain MTZ-inducible cell-specific ablation activity10. We therefore adapted the NTR/MTZ ablation system to zebrafish11 to expand studies of cellular regeneration12, reasoning that co-expression of NTR with reporters would enable visualization13,14 and quantification14–17 of MTZ-induced cell loss, and subsequent cell replacement, in vivo. In general, the NTR/MTZ system has proven useful for targeted cell ablation, and has had widespread uptake12. However, the low catalytic efficiency of NTR 1.0 necessitates high concentrations of MTZ (e.g., 10 mM) for effective cell ablation. This precludes sustained ablation paradigms, as MTZ exposures >24 h become increasingly toxic18. In addition, some cell types have proven resistant to NTR/MTZ-mediated ablation (e.g. macrophages and dopaminergic neurons)19,20. To overcome these limitations, we implemented a screening cascade to identify NTR variants exhibiting enhanced MTZ conversion activity.
Using targeted mutagenesis and high-throughput selection, the Searle group previously identified a triple mutant of NTR 1.0 exhibiting a 40- to 80-fold improvement in CB1954 conversion activity (NfsB_Ec T41Q/N71S/F124T, here “NTR 1.1”)21. This variant also enhanced MTZ-induced cell ablation in zebrafish, however, the improvement was only marginal (2- to 3-fold over NTR 1.0)18,22. In contrast, by leveraging a combination of rational engineering and cross-species testing, we identified a rationally engineered NfsB ortholog from the bacterial species Vibrio vulnificus (NfsB_Vv F70A/F108Y, “NTR 2.0”) which improves MTZ-meditated cell ablation efficiency ~100-fold; i.e., robust ablation at 100 μM MTZ versus typical 10 mM treatments. Additional data show that NTR 2.0 will expand the functionality of the NTR/MTZ system by allowing: 1) sustained interrogations of cell function, 2) effective ablation of “resistant” cell types, 3) prolonged cell loss, as novel tests of regenerative capacity, and 4) modeling of degenerative diseases caused by chronic cell loss. Accordingly, we have created a series of bipartite expression vectors and transgenic zebrafish lines co-expressing NTR 2.0 and fluorescent reporters as versatile new toolsets for the research community.
RESULTS
Rational improvement of MTZ-activating NTR variants
We previously compiled an E. coli gene library of 11 nfsB orthologs and used a DNA damage screen in E. coli host cells to monitor activation of SN33623, a PET imaging probe that shares a 5-nitroimidazole core structure with MTZ23. This same library was used here to evaluate ablation efficacy at higher SN33623 doses; relative growth of replicate E. coli cultures was assessed across a dilution series to establish EC50 values. Consistent with the previous DNA damage screen, the six most closely related orthologs of NTR 1.0 (‘NfsB_Ec-like’, >50% amino acid identity with E. coli NfsB) were far less effective at activating SN33623 than the other five enzymes in the panel (Fig. 1a,b). MTZ activation followed the same trend, with one notable exception: the V. vulnificus ortholog (NfsB_Vv), despite being NfsB_Ec-like, was one of the most active MTZ-converting enzymes (Fig. 1c).
Fig. 1: Rationally engineered NfsB-family NTRs for improved activation of MTZ.
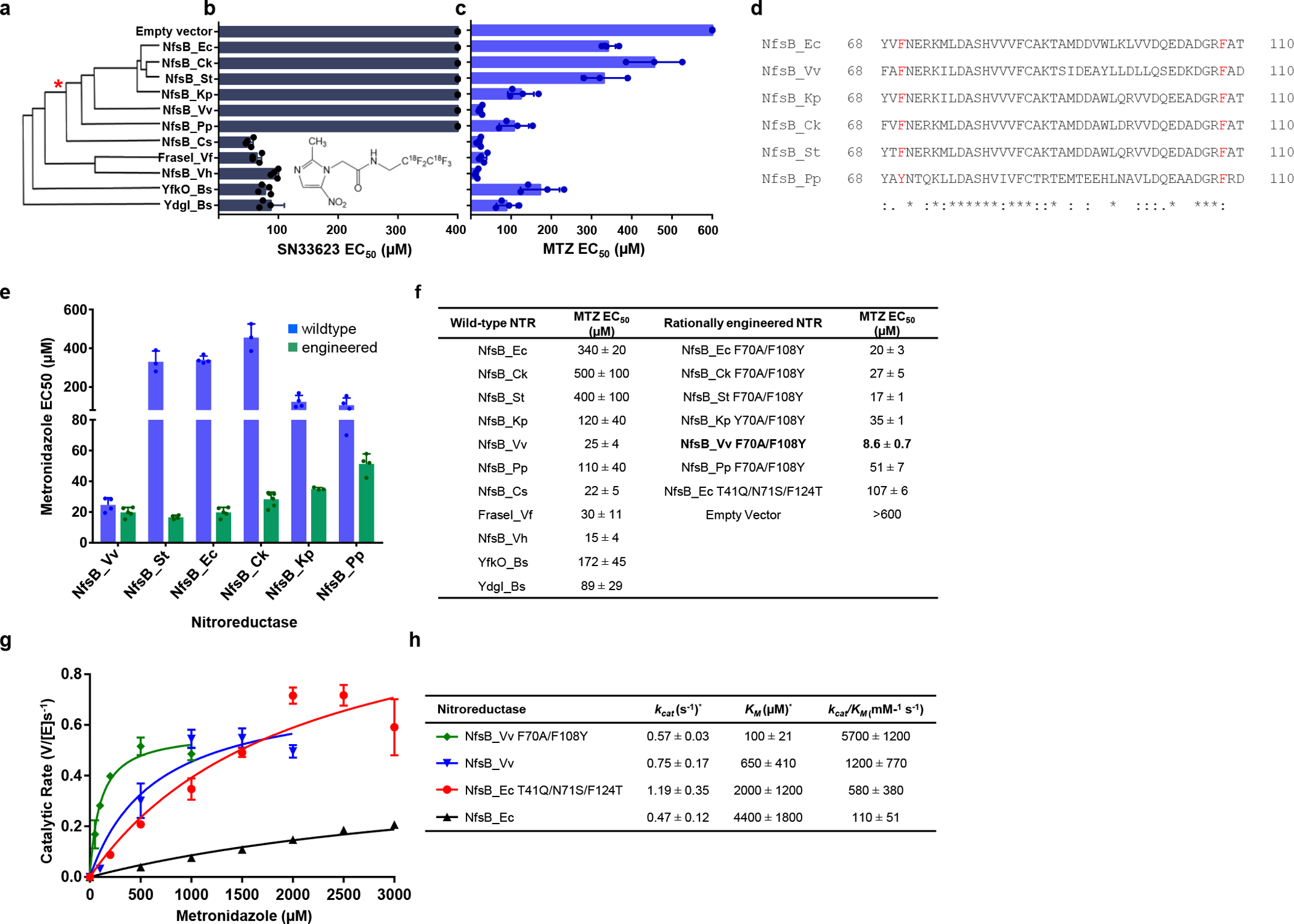
a, Amino acid sequence identity cladogram of eleven NfsB orthologs, grouped according to percent shared amino acid identity with NfsB_Ec. The asterisk (*) marks where other NTR variants diverge from NfsB_Ec-like enzymes. b, E. coli host sensitivity conferred by NfsB variants to the compound SN33623, n=3 biologically independent experiments for all strains except those expressing NfsB_Pp, NfsB_Cs, FraseI_Vf, NfsB_Vh, YfkO_Bs and YdgI_Bs (n=4). c, E. coli host sensitivity conferred by NfsB variants to the compound MTZ, n=4 biologically independent experiments for all strains except those expressing NfsB_Ck and NfsB_St (n=3). b-c, Data are means ± SD, data without error bars indicate host cell sensitivity could not be observed within the tested concentration range. Insets: chemical structures of SN33623 and MTZ. d, Identification of ‘SN33623-blocking’ residues in NfsB_Ec-like NTRs. Partial protein alignment of NfsB_Ec and NfsB_Ec-like enzymes (residues 68 – 110) with ‘SN33623 blocking’ residues highlighted in red. Identical (*), conservative (:), and semi-conservative (.) amino acid differences are indicated. e, E. coli host sensitivity to MTZ conferred by wild-type or rationally engineered NfsB-like enzymes, n=4 biologically independent experiments for all strains except those expressing NfsB_Ck, NfsB_St, and NfsB_Kp Y70A/F108Y (n=3), NfsB_Ec F70A/F108Y (n=5), and NfsB_Ck F70A/F108Y (n=8). Data are means ± SD. f, Summary of MTZ EC50 values for E. coli strains expressing wild-type or rationally engineered NTRs. g, Michaelis-Menten reaction curves of purified NTR variants with MTZ. The indicated NTR enzymes were purified and assayed for MTZ conversion activity across a concentration range spanning from ca. 0.2× to 5× the KM for each variant, n=3 biologically independent experiments. Data presented are means ± SD. h, Michaelis-Menten kinetic parameters for the reduction of MTZ by purified His6-tagged NTRs, monitored at 340 nm, n=3 biologically independent experiments. Data are means ± SD. KM and kcat values were derived using GraphPad Prism 8.0. The asterisk (*) indicates that these are apparent kinetic parameters as measured at 100 μM NADPH.
This was a promising finding, as loss-of-activity and gain-of-activity rational mutagenesis previously demonstrated that the residues F70 and F108 impair 5-nitroimidazole activity in NfsB_Ec23, and these residues are highly conserved in the NfsB_Ec-like enzymes (Fig. 1d). We hypothesized that introducing F70A/F108Y substitutions into NfsB_Vv and the other NfsB_Ec-like enzymes (Y70A/F108Y for NfsB_Pp) would enhance MTZ activity, as was previously found for NfsB_Ec23. This proved true in all cases, with host cells expressing the substituted variants being substantially more sensitive to MTZ. The E. coli strain expressing NfsB_Vv F70A/F108Y was the most sensitive to MTZ, having an EC50 40-fold lower than NfsB_Ec (NTR 1.0) and 12-fold lower than NfsB_Ec T41Q/N71S/F124T (NTR1.1) expressing strains (Fig. 1e,f).To determine whether E. coli EC50 data accurately reflected improved enzymatic reduction of MTZ, the native and F70A/F108Y substituted NfsB_Vv variant were purified as His6-tagged proteins using nickel affinity chromatography, and MTZ conversion activities were compared to the benchmark NTR 1.0 and NTR 1.1 enzymes (Fig. 1g). Relative Michaelis-Menten kinetic parameters were consistent with E. coli sensitivity data; NfsB_Vv F70A/F108Y exhibited the lowest KM and highest catalytic efficiency (kcat/KM), followed by NfsB_Vv, NTR 1.1, and NTR 1.0 respectively (Fig. 1h).
Comparison of NTR variant ablation efficacy: mammalian cells
To assess relative MTZ conversion activities (and expression tolerance) of lead NTR variants in vertebrate cells, NfsB_Vv F70A/F108Y, NfsB_Vv, NTR 1.0 and NTR 1.1 were first stably transfected into human cells (HEK-293). MTZ sensitivity was evaluated using a MTS viability assay24 following a 48 h MTZ dose-response challenge (5 mM to 1 μM, 2-fold dilution series, Fig. 2a). Equivalent MTZ dose-response assays, using the MTT viability assay25, were performed on Chinese hamster ovary (CHO-K1) cell lines expressing either NTR 1.1 or NfsB_Vv F70A/F108Y (Fig. 2b). In HEK-293 cells, NfsB_Vv F70A/F108Y, NfsB_Vv and NTR 1.0 were stably expressed and relative MTZ sensitivities (3, 9 and 1700 μM respectively) were concordant with the E. coli EC50 data (Fig. 2c, compare to Fig. 1f). In contrast, NTR 1.1 expression was lost over time, reflected in the comparatively high MTZ EC50 value (2300 μM). In CHO-K1 cells, EC50 values of 690 μM and 4 μM for NTR 1.1 and NfsB_Vv F70A/F108Y, respectively (Fig. 2c), suggest the improved MTZ activity exhibited by NfsB_Vv F70A/F108Y is retained in heterologous mammalian expression systems.
Fig. 2: Targeted ablation of mammalian cells is enhanced with NsfB_Vv F70A/F108Y.
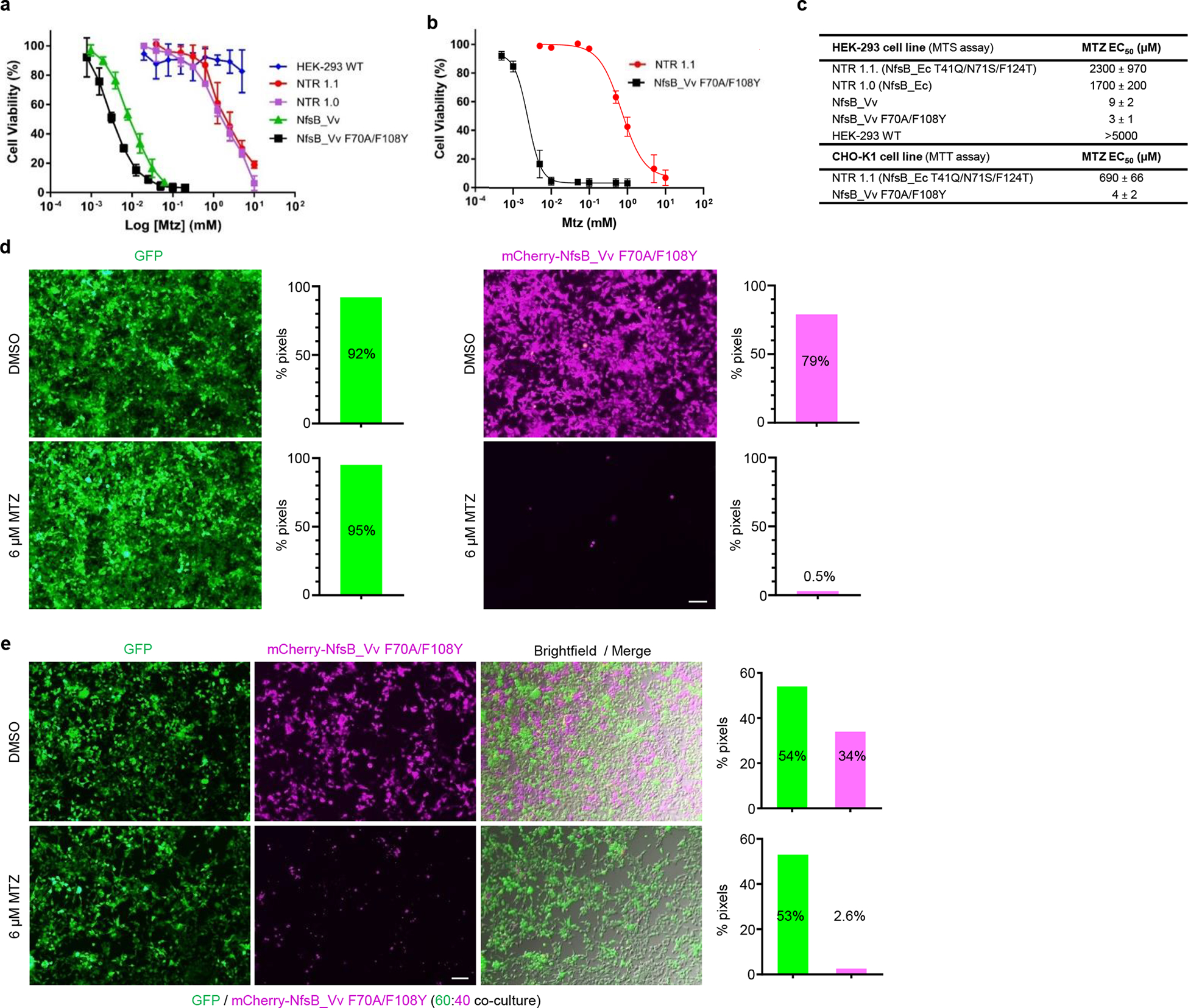
a-b, MTZ dose-response cell viability assays of NTR variants in mammalian cells tested across the indicated MTZ concentrations. a, NTR variants stably expressed in HEK-293, n=5 biologically independent experiments for all cell lines other than wild-type HEK-293 cells (n=4) and those expressing NsfB_Vv F70A/F108Y (n=3) or NfsB_Vv (n=9). b, NTR variants stably expressed in CHO-K1 cells, n=3 biologically independent experiments. Survival rates were measured using MTS (a) or MTT (b) assays and data presented are means ± SD. c, MTZ EC50 values for mammalian HEK-293 and CHO-K1 cell lines stably over-expressing the indicated NTR enzyme variants. Data presented are means ± SD. d-e, Images and quantification of MTZ-induced ablation of transgenic HEK-293 cell lines. Cells expressing GFP or co-expressing mCherry and NfsB_Vv F70A/F108Y were cultured in isolation (d), or as a 60:40 co-culture of both cell lines (e). All cells were treated with 0.01% DMSO or 6 μM MTZ for 48 h and cell viability was assessed qualitatively by fluorescence microscopy and quantitatively by pixel counts (fluorescent pixels/total pixels), n=3 biologically independent experiments per condition. Merged brightfield and fluorescence images (e) confirm loss of NfsB_Vv F70A/F108Y expressing cells as opposed to loss of reporter expression. Scale bars = 100 microns.
Ablation specificity of NfsB_Vv F70A/F108Y: mammalian cells
A key advantage of MTZ is the cell-specific nature of its cytotoxic metabolite, allowing selective ablation of NTR-expressing cells without harming surrounding cells. The targeted nature of NTR/MTZ-induced ablation facilitates precise elimination of discrete cell types, enabling interrogations of cell function during development, regeneration, and other biological processes of interest. To test the specificity of NfsB_Vv F70A/F108Y mediated ablation, stable transgenic HEK-293 cell lines were generated that expressed either green fluorescent protein (GFP) or co-expressed mCherry and NfsB_Vv F70A/F108Y. When single cultures were challenged with 6 μM MTZ or 0.01% (v/v) DMSO (control) for 48 h, cells co-expressing NfsB_Vv F70A/F108Y and mCherry were killed, whereas cells expressing GFP remained healthy (Fig. 2d). Co-cultures of these two cell lines undergoing the same challenge confirmed selective ablation of NfsB_Vv F70A/F108Y-expressing cells (Fig. 2e). The slightly higher survival rate observed in co-cultures appears to be due to dead or dying cells adhering to living cells (Extended Data Fig. 1). Having confirmed enhanced targeted cell ablation in transgenic bacteria and vertebrate cell lines, we dubbed the engineered NfsB_Vv F70A/F108Y variant “NTR 2.0”.
NTR 2.0 cell ablation efficacy in vivo: zebrafish
To determine if NTR 2.0 increases MTZ-induced ablation efficacy in vivo, we generated a UAS reporter/effector transgenic zebrafish line co-expressing yellow fluorescent protein (YFP) and NTR 2.0 in a Gal4 driver-dependent manner, Tg(5xUAS:GAP-tagYFP-P2A-nfsB_Vv F70A/F108Y)jh513 (hereafter, UAS:YFP-NTR2.0). This line was crossed with a neuronally-restricted Gal4 driver line26, Et(2xNRSE-fos:KalTA4)gmc617 (KalTA4 is a zebrafish optimized Gal4/VP16 fusion27), to establish double transgenic larvae co-expressing YFP and NTR 2.0 (hereafter, NRSE:KalTA4;UAS:YFP-NTR2.0) throughout the nervous system (Fig 3a).
Fig. 3: NTR 2.0 enhances cell ablation efficacy in zebrafish.
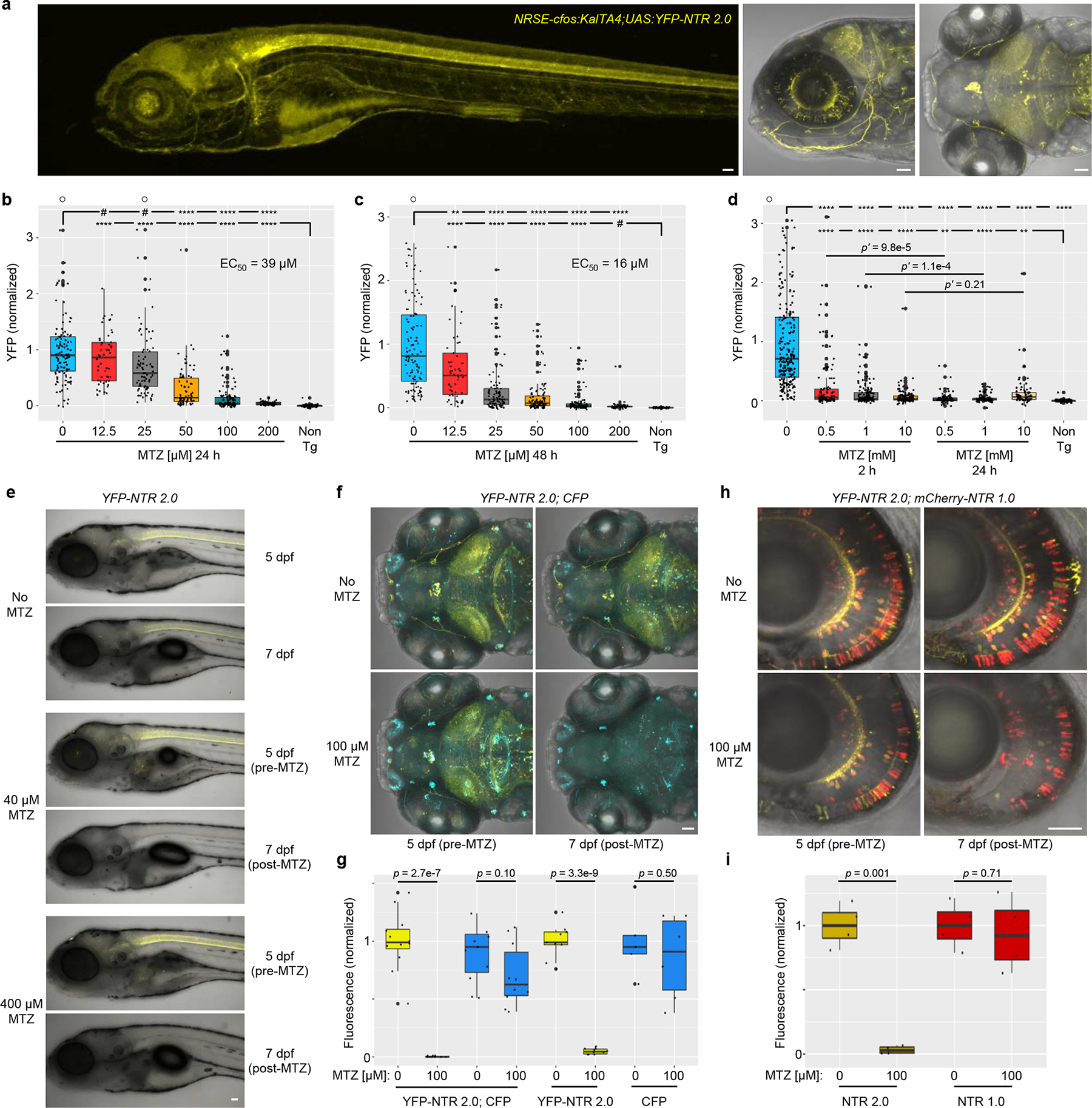
a, Confocal images of 5 dpf NRSE:KalTA4;UAS:YFP-NTR2.0 larvae showing neuronally-restricted YFP expression (co-expressed with NTR 2.0), n=4 biologically independent experiments, 24 larvae examined. b-d, Dose-response tests of MTZ ablation efficacy. 5 dpf NRSE:KalTA4;UAS:YFP-NTR2.0 larvae were exposed to MTZ for 24 h (b), 48 h (c), or either 2 or 24 h (d). Fully detailed statistical comparisons (absolute effect sizes, 95% confidence intervals, Bonferroni-corrected p’-values derived from two-tailed t tests, sample sizes, and the number of biologically independent experiments) between MTZ-treated and control conditions in graphs b-d are provided in Supp. Table 1. e, Representative time series images showing changes in YFP fluorescence in NRSE:KalTA4;UAS:YFP-NTR2.0 larvae treated with 0, 40, or 400 μM MTZ from 5–6 dpf, n=2 biologically independent experiments, 16 larvae imaged per condition. f-g, Test of NTR 2.0 ablation specificity. f, Representative time-series images showing changes in YFP and CFP fluorescence in in NRSE:KalTA4;UAS:YFP-NTR2.0;UAS:CFP larvae treated with 100 μM MTZ from 5–6 dpf, n=4 biologically independent experiments, 24 larvae imaged per condition. g, Imaris-based quantification of changes in YFP and CFP fluorescence in control and MTZ-treated larvae, n=4 biologically independent experiments, dot plots show the number of larvae examined. h-i, Ablation efficacy comparison of NTR 1.0 and NTR 2.0. h, Representative time-series images shows changes in YFP and mCherry fluorescence in nyx:KalTA4; UAS:YFP-NTR2.0;UAS:NTR 1.0-mCherry larvae treated with 100 μM MTZ from 5–6 dpf changes, n=2 biologically independent experiments, 16 larvae imaged per condition. i, Imaris-based quantification of changes in YFP and mCherry fluorescence in control and MTZ-treated larvae, n=2 biologically independent experiments, dot plots show the number of images examined. A two-tailed t test was used to calculate p-values comparing MTZ-treated larvae to corresponding controls per genotype in g and i. All box plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and whiskers = SD, with individual data points (larvae or images) overlaid as a dot plot. Symbols: #p’ > 0.05, *p’ ≤ 0.05, **p’ ≤ 0.01, ***p’ ≤ 0.001, ****p’ ≤0.0001 ○ = outlier data points. Scale bars = 50 microns.
To test NTR 2.0 cell ablation efficacy in vivo, double-transgenic NRSE:KalTA4;UAS:YFP-NTR2.0 larvae were exposed to MTZ across a 2-fold dilution series (from 200 to 12.5 μM) for 24 h (5–6 days post-fertilization, dpf). Using an established plate reader assay15,16, YFP levels were quantified in individual fish at 7 dpf. In the 200 μM MTZ treatment group, YFP levels were reduced 96% relative to non-ablated controls (Fig. 3b; Supp. Table 1). Dose-dependent effects were evident between 25 and 200 μM MTZ with a calculated EC50 of 39 μM. Extending MTZ treatments to 48 h resulted in an estimated EC50 of 16 μM with YFP levels being indistinguishable from non-transgenic controls in the 200 μM MTZ treatment group (Fig. 3c; Supp. Table 1). To determine if NTR 2.0 enabled ablation upon shorter exposures at higher MTZ concentrations, 5 dpf transgenic larvae were incubated with 0.5, 1 or 10 mM MTZ for 2 h or 24 h (control) and YFP levels assessed at 7 dpf. The 2 h incubation with 10 mM MTZ resulted equivalent levels of ablation relative to the 24 h treatment control (Fig. 3d, p’ = 0.21). Visualization by in vivo confocal imaging confirmed effective cell ablation following a one day treatment (5–6 dpf) with either 40 or 400 μM MTZ (Fig. 3e). Together, these data are consistent with NTR 2.0 providing an ~100-fold improvement over NTR 1.011,28 and a ~33-fold improvement over NTR 1.118,22.
Ablation specificity of NTR 2.0 in vivo: zebrafish
As a first test of whether NTR 2.0/MTZ-induced ablation retains the targeted cellular specificity of the NTR 1.0 and NTR 1.1 systems in vivo, we generated triple transgenic fish combining the NRSE:KalTA4;UAS:YFP-NTR2.0 with a second UAS reporter expressing CFP, Tg(5xUAS:GAP-ECFP,he:GAP-ECFP)gmc1913 (i.e., UAS:CFP). Mosaicism of the two UAS reporters led to expression in overlapping and non-overlapping subsets of cells; non-targeted cells expressed CFP only and targeted cells expressed YFP and NTR 2.0 with or without CFP (Fig. 3f). Time-series confocal imaging determined that in the absence of MTZ, mosaic expression of both reporters is maintained from 5 to 7 dpf (No MTZ, Fig. 3f). In contrast, larvae exposed to 100 μM MTZ from 5–6 dpf exhibited loss of cells expressing YFP and NTR 2.0 while cells expressing CFP alone remained (Fig. 3f). Imaris-based quantification of longitudinal changes in fluorescence volume showed significant reductions in YFP relative to pre-treatment levels (99 and 95%, p = 2.7e−7 and p = 3.3e−9, for YFP/CFP and YFP only larvae, respectively). CFP levels also dropped, but only by 30% (YFP/CFP, p = 0.10) and 14% (CFP only, p = 0.50), consistent with some CFP-positive cells co-expressing YFP and NTR 2.0 and a general reduction in CFP detection in older larvae (Fig. 3g). These results suggest that NTR 2.0 maintains the targeted specificity of the original NTR cell ablation systems11. However, mosaic reporter/effector expression somewhat limited our ability to demonstrate this quantitatively. To compare ablation efficacies of NTR 1.0 and NTR 2.0 side-by-side, we generated triple-transgenic fish combining NRSE:KalTA4;UAS:YFP-NTR2.0 with a second UAS reporter expressing a NTR 1.0-mCherry fusion protein, Tg(14xUAS:nfsB_Ec-mCherry)c26429. Mosaicism again led to overlapping and non-overlapping expression of the two UAS transgenes. Time-series imaging showed the expression of both reporters was maintained from 5 to 7 dpf in the absence of MTZ (No MTZ, Fig. 3h). In contrast, cells co-expressing YFP and NTR 2.0 were lost in larvae exposed to 100 μM MTZ from 5–6 dpf, while cells expressing mCherry and NTR 1.0 alone remained (Fig. 3h). Imaris-based quantification (Fig. 3i) showed that YFP dropped by 96% relative to pre-treatment levels (p = 0.001) while mCherry levels dropped only by 7% (p = 0.71), consistent with some mCherry+ cells co-expressing YFP and NTR 2.0. This result suggests NTR 2.0 displays superior ablation efficacy relative to NTR 1.0. However, mosaic expression again complicated the analysis, making it unclear whether NTR 1.0 contributed to any potential enhanced ablation occurring in cells co-expressing NTR 1.0 and NTR 2.0, and disrupting our ability to cleanly demonstrate selective ablation of NTR 2.0-expressing cells.
To assess NTR 2.0/MTZ-induced ablation specificity more directly, selectivity of cell death was compared between neighboring rod (NTR 2.0-expressing) and cone (control) photoreceptor cells. A transgenic line co-expressing YFP and NTR 2.0 in rod cells, Tg(rho:GAP-YFP-2A-nfsB_Vv F70A/F108Y)jh405 (hereafter, rho:YFP-NTR2.0)17, was treated ±MTZ (400 μM) for 24 h (5 to 6 dpf). Retinas were fixed 24 h later (7 dpf), sectioned, and processed for immunohistochemistry with rod (Ab-1-rho, anti-rhodopsin, α-rho, aka 1D1)30 and cone (zpr-1, anti-arrestin 3a, α-arr3a, aka Fret 43)31 labeling antibodies. Confocal images show coincident labeling of YFP-expressing rods (yellow cells) and the rod-specific antibody (α-rho, red cells) in no MTZ controls, and loss of both YFP and α-rho labeling in MTZ-treated retinas (Extended Data Fig. 2a). Manual quantification of both YFP expression and antibody labeling confirmed rod cell loss (Extended Data Fig. 2b). Confocal images of cone antibody labeling showed no appreciable differences between no MTZ controls and MTZ-treated retinas (Extended Data Fig. 2c). Quantification of rods (yellow cells) and cones (red cells) confirmed cone cells were not lost in MTZ-treated retinas, showing instead a slight increase in cone cell number (Extended Data Fig. 2d). It is unclear if this is indicative of ongoing cone cell genesis or cone crowding after rod loss. Nevertheless, these data confirm the absence of a bystander cell death effect when acute MTZ treatments are used to ablate NTR 2.0-expressing cells.
To further quantify the enhanced ablation efficacy of NTR 2.0 in vivo, we used transgenic zebrafish expressing either NTR 1.0 or NTR 2.0 in retinal rod photoreceptor cells, Tg(rho:YFP-nfsB_Ec)gmc500 (hereafter, rho:YFP-NTR1.0) and rho:YFP-NTR2.0, respectively. Expressing each NTR variant in the same cell type controls for differential sensitivity to NTR/MTZ-induced cell death between different cell types. In MTZ dose-response tests (five-fold dilution series, from 5 mM - 0.32 μM, 48 h treatment), treatment of rho:YFP-NTR1.0 fish produced an EC50 of 540 μM MTZ (Fig. 4a), in keeping with prior observations that rod photoreceptors can be ablated with lower MTZ concentrations (2.5 mM)17. Treatment of the rho:YFP-NTR2.0 fish produced an EC50 of 3 μM MTZ, equating to an ~180-fold improvement in ablation efficacy (Fig. 4b; Supp. Table 2). Confocal imaging confirmed these results, showing that a 48 h treatment with 40 μM MTZ (5 to 7 dpf) had no appreciable effect on rho:YFP-NTR1.0 expression (Fig. 4c), but was sufficient for near complete ablation of rho:YFP-NTR2.0-expressing rod cells (Fig. 4d).
Fig. 4: Dose-response test of cell ablation efficacy – NTR 1.0 versus NTR 2.0.
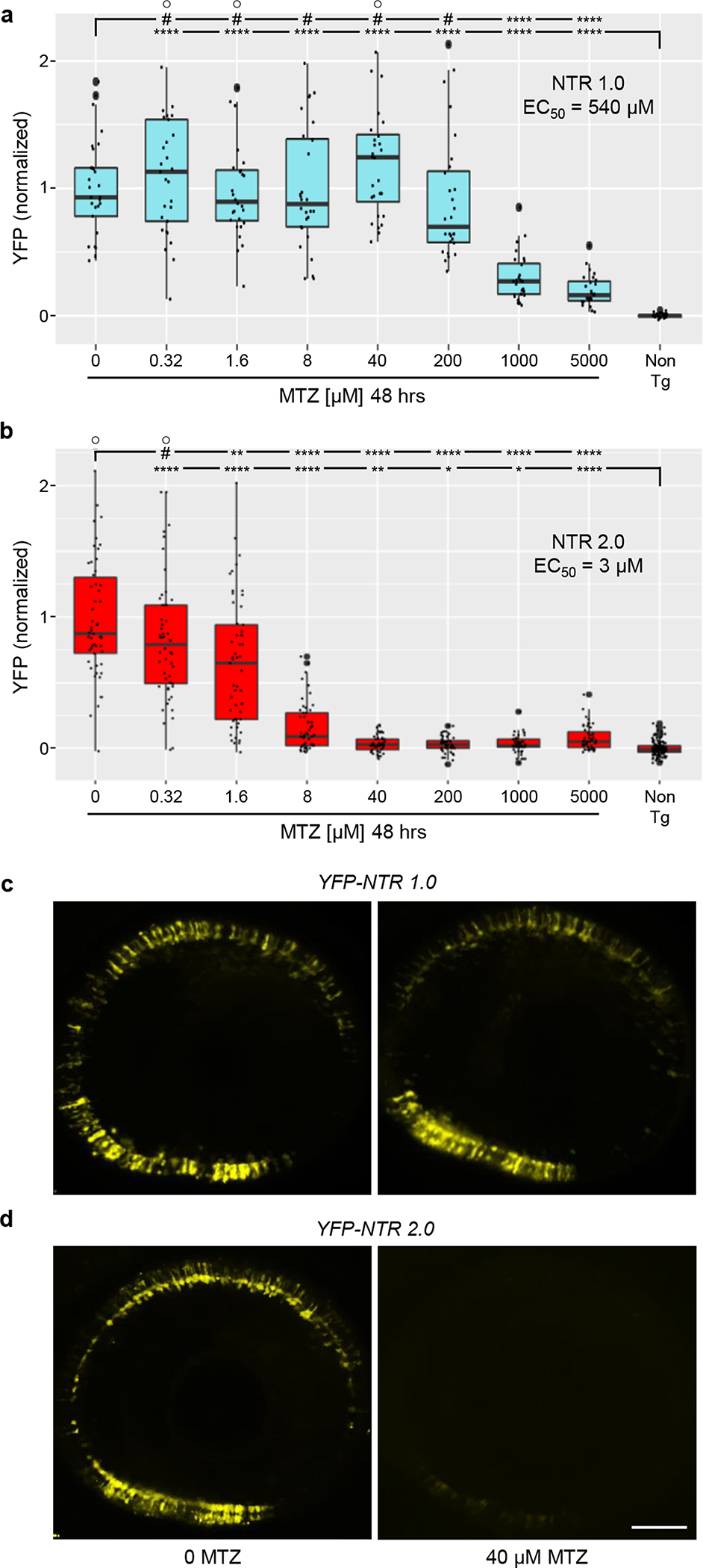
a-b, Transgenic larvae co-expressing either NTR 1.0 and YFP (a,c; rho:YFP-NTR 1.0) or NTR 2.0 and YFP (b,d; rho:YFP NTR 2.0) in rod photoreceptors were treated with MTZ across a 5-fold dilution series (5 mM – 320 nM) for 48 h (5–7 dpf) and YFP levels quantified by plate reader assay (n=4 biologically independent experiments, dot plots show the number of larvae examined). Box plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and whiskers = SD. EC50 values suggest a 180-fold improvement in NTR 2.0-mediated ablation efficacy. Fully detailed statistical comparisons (absolute effect sizes, 95% confidence intervals, Bonferroni-corrected p’-values derived from two-tailed t tests, sample sizes, and the number of biologically independent experiments) between MTZ-treated and control conditions in graphs a-b are provided in Supp. Table 2. Symbols: #p’ > 0.05, *p’ ≤ 0.05, **p’ ≤ 0.01, ***p’ ≤ 0.001, ****p’ ≤ 0.0001; ○ = outlier data points. c-d, Representative confocal images of YFP expression at 7 dpf (post-MTZ) showing differential effects of 40 μM MTZ treatments in NTR 1.0 (c) and NTR 2.0 (d) expressing larvae, n=4 larvae imaged per condition. Scale bar = 50 microns.
Prolonged low-dose MTZ exposures in juvenile zebrafish
A key disadvantage of NTR 1.0 is the near-toxic concentrations of MTZ needed to ablate cells. In addition to potential off-target effects32, near-toxic doses preclude sustained ablation paradigms18. To determine whether zebrafish can tolerate long-term low-dose MTZ exposures, 15 dpf larvae were exposed to 10, 1, 0.1 or 0 mM MTZ for 36 days. As observed previously18, fish maintained in 10 mM MTZ exhibited high rates of lethality with only 6.1% alive after twelve days and none surviving beyond day 34 (Fig. 5a). In contrast, for all other conditions fish survival was equivalent, with ≥92% alive at day 36 (1 mM = 94%, 0.1 mM = 92%, 0 mM = 94%; Fig. 5a).
Fig. 5: Prolonged MTZ treatments are non-toxic and retain targeted ablation specificity in adults.
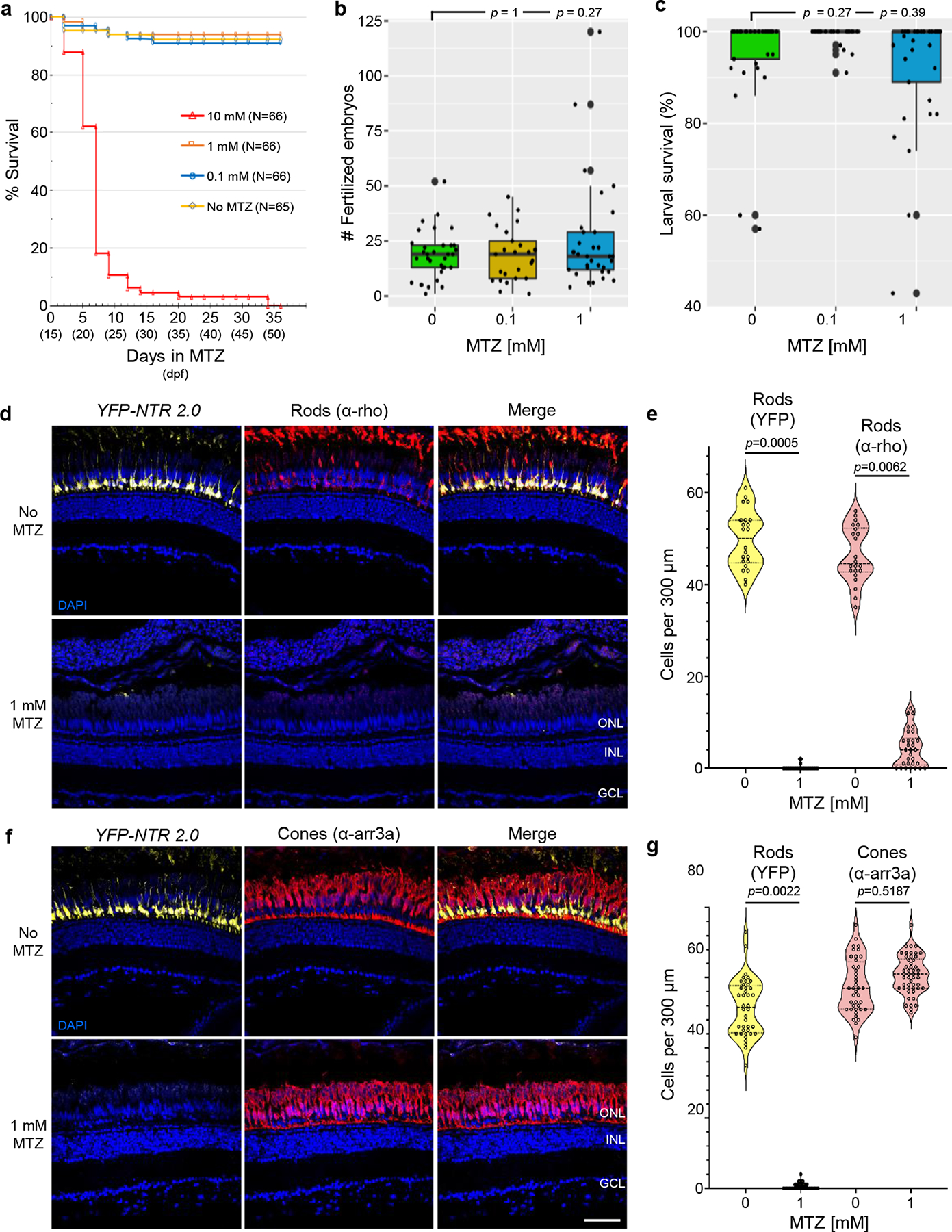
a, Survival of juvenile zebrafish incubated with the indicated concentration of MTZ for 36 days, from 15–51 dpf (n=66 or 65 larvae examined per condition – see inset, n=3 biologically independent experiments). Log-rank (Mantel-Cox) tests and Gehan-Breslow-Wilcoxon tests showed no statistically significant differences between No MTZ controls and 0.1 or 1 mM MTZ conditions. Comparisons between No MTZ controls and the 10 mM condition produced chi-squares of 123 and 97, respectively, and a Bonferroni corrected p’-value of <0.0001 for both tests. b-c, Test of fecundity (b) and offspring survival (c) rates of long-term MTZ exposed fish (n=3 independent mating sessions). Box plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and whiskers = SD, with individual data points (successful matings) overlaid as a dot plot. Fully detailed statistical comparisons (absolute effect sizes, 95% confidence intervals, Bonferroni-corrected p’-values derived from two-tailed t tests, and sample sizes) between MTZ-treated and control conditions in graphs b-c are provided in Supp. Table 3. d-g, Test of NTR 2.0/MTZ-induced ablation efficacy and specificity in adult zebrafish (n=2 zebrafish per condition). Transgenic rho:YFP-NTR2.0 adult zebrafish were treated ±1 mM MTZ for 3 days. Retinas were then fixed, sectioned, and labeled with the nuclear stain DAPI (blue cells) and an antibody specific to rod (α-rho, red cells, d), or cone photoreceptors (α-arr3a, red cells, f). Representative confocal images of YFP-expressing rods (yellow cells, d and f) and antibody labeling show effective ablation of NTR 2.0-expressing rod photoreceptors (d) and maintenance of neighboring cone photoreceptors (f). Manual quantification of YFP-expressing rod cells and either α-rho stained rods (e) or α-arr3a stained cones. Violin plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and the full distribution of the data, with individual data points (number of measurements per condition) overlaid as a dot plot. A two-tailed nested t test (GraphPad, Prism 9) was used to calculate p-values comparing MTZ-treated and control larvae. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bars = 50 microns.
Long-term MTZ exposure has been implicated in rodent male infertility33 and associated (rarely) with neurotoxic effects in patients34. Therefore, we assessed fecundity and offspring survival rates of fish exposed to 1, 0.1, or 0 mM MTZ once they reached sexual maturity. At ~3 months of age, pairs of fish from each MTZ treatment group were naturally mated and the number of egg produced and larval survival rates quantified. No deleterious effects on clutch size or offspring survival rates were observed in MTZ-treated fish compared to untreated controls (Fig. 5b,c; Supp. Table 3; note that roy pigmentation mutants were used throughout this study to facilitate in vivo imaging and quantification of fluorescent reporters, the smaller clutch sizes observed are normal for roy fish at 3 months of age).
Ablation of NTR 2.0-expressing cells in adult zebrafish
To determine if low-dose MTZ treatments were sufficient for ablating NTR 2.0-expressing cells at mature stages, adult rho:YFP-NTR2.0 fish were maintained in 1mM MTZ for 3 days. Retinas were then extracted, fixed, sectioned, and processed for immunohistological labeling. Targeted and bystander cell death was assessed using antibodies specific to either rod (e.g., α-rho)35 or cone (α-arr3a) photoreceptors and confocal imaging (Fig. 5d–g), per corresponding larval-stage assays (see Extended Data Fig. 2). No MTZ controls showed sustained YFP and antibody labeling of NTR 2.0-expressing rod cells, however, MTZ-treated retinas lost both YFP and the rod-specific label (Fig. 5d). Manual quantification showed statistically significant losses of rod cells for both YFP and the rod-specific marker (Fig. 5e, p = 0.0005 and 0.0062, respectively). In contrast, antibody labeling of neighboring cone cells was similar in control and MTZ-treated retinas (Fig. 5f). Manual quantification of YFP labeled rods in the cone stained sections showed a similar MTZ-induced reduction in rod cell numbers (Fig. 5g, YFP, p = 0.0022), but no significant difference in cone cell numbers between control and MTZ-treated larvae (Fig. 5g, α-arr3a, p = 0.5187). Adult-stage rod cell ablation was further validated using 3 and 7 day 1 mM MTZ treatments and both rod-specific antibodies (Ab-4C12 and α-rho, Extended Data Fig. 3). These data confirm that low-dose MTZ treatments are sufficient to ablate NTR 2.0 expressing cells in adult zebrafish. As was seen in larval stage assays, there was no evidence of off-target bystander cell death in neighboring cells. However, as rod cell loss is followed by cone cell loss in retinitis pigmentosa36,37, we anticipate that sustained ablation of rod cells over weeks to months will eventually lead to secondary cone cell loss. Moreover, as secondary or bystander cell death is relevant to other diseases38, development39, and stroke40,41, sustained ablation paradigms may provide novel models to study underlying mechanisms.
NTR 2.0 enables ablation of NTR/MTZ resistant cell types
Another limitation of first-generation NTR resources is that certain cell types have proven difficult to ablate. For example, macrophages co-expressing YFP and NTR 1.1 are resistant to 10 mM (24 h) doses of MTZ14, effective ablation requiring multi-day exposures to 2.5 mM MTZ in adult fish20. To determine if NTR 2.0 enables ablation of “resistant” cell types, a new UAS reporter/effector, Tg(5xUAS:mCherry-2A-NTR 1.1)jh573 (hereafter, UAS:mCherry-NTR 1.1), was established for comparisons to UAS:YFP-NTR 2.0 larvae. Here, a Gal4 driver, Tg(mpeg1.1:Gal4-VP16)gl24 (hereafter, mpeg:Gal4), was used to co-express either mCherry and NTR 1.1 (Fig. 6a,b) or YFP and NTR 2.0 (Fig. 6c,d) specifically in macrophages. To assess relative ablation efficacy, NTR 1.1 and NTR 2.0-expressing larvae were exposed to 0, 0.1, or 10 mM MTZ for 48 h (5– 7 dpf). Confocal time series imaging was used to quantify changes in macrophage numbers per each individual larvae, to control for differences in the number of cells labelled across larvae and between UAS reporter/effectors. MTZ was largely ineffective in ablating cells in UAS:mCherry-NTR 1.1 larvae, with small rounded macrophages surviving in the 10 mM MTZ condition (Fig. 6a,b). In contrast, near complete ablation of NTR 2.0-expressing cells was evident in larvae treated with 100 μM or 10 mM MTZ (Fig. 6c,d). Manual cell counts confirmed differential ablation efficacy of NTR 1.1 and NTR 2.0, with no appreciable change in NTR 1.1-expressing cells (p’ = 1 for both the 100 μM and 10 mM treatments) while NTR 2.0-expressing cells were significantly reduced (p’ = 2.2e−6 and p’ = 2.8e−6 for the 100 μM and 10 mM MTZ treatments, respectively (Fig. 6b,d; Supp. Table 4).
Fig. 6: NTR 2.0 enables ablation of “resistant” cell types.
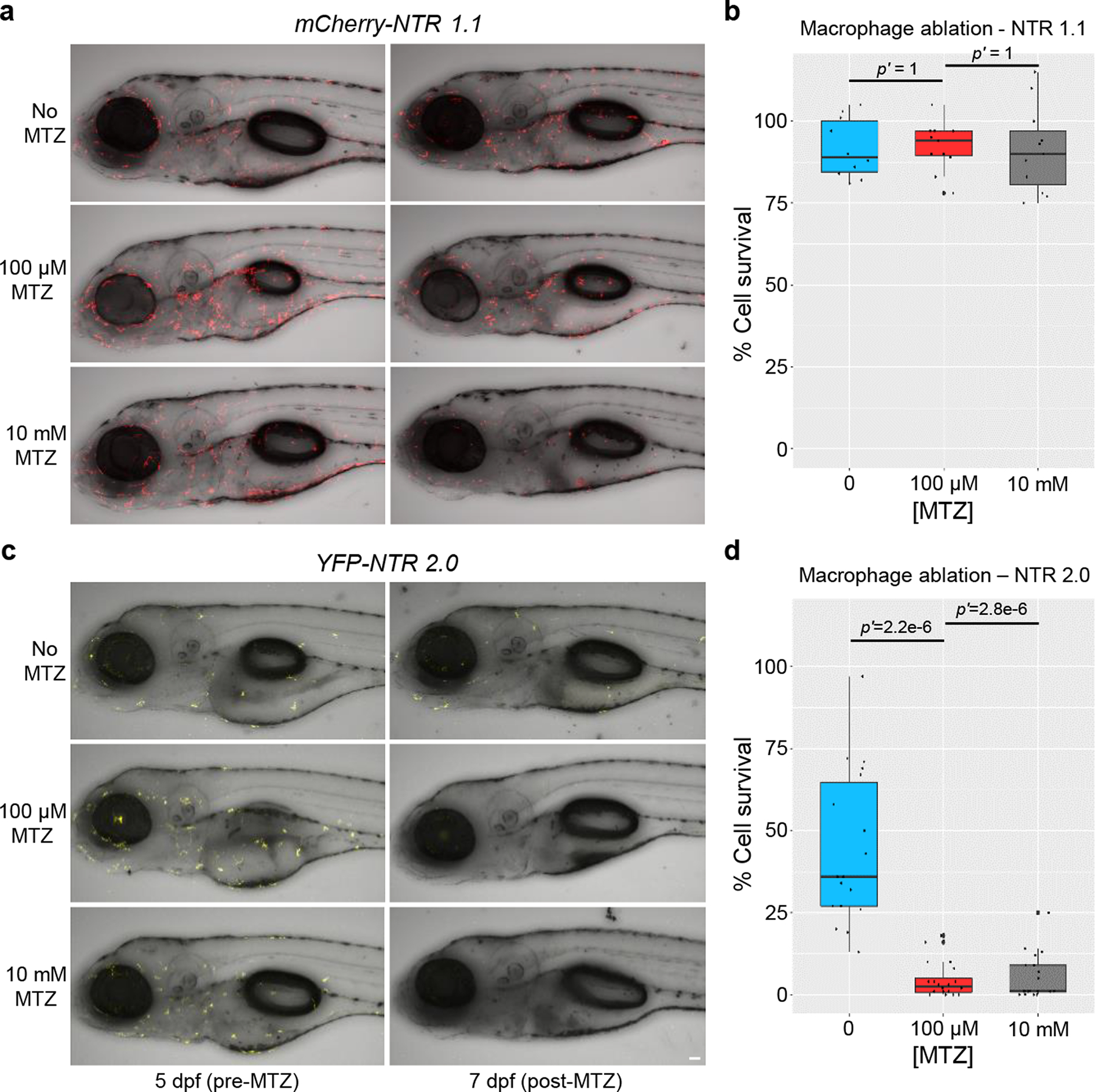
Transgenic larvae co-expressing either mCherry and NTR 1.1 (a,b) or YFP and NTR 2.0 (c,d) in macrophages were treated with 0, 0.1, or 10 mM MTZ from 5–7 dpf, n=3 biologically independent experiments for both assays. a,c, Intravital time series imaging was performed pre-MTZ (5 dpf) and post-MTZ (7 dpf). b,d, Manual counts of macrophage numbers were performed on pre- and post-treatment images. The percent change in cell number was calculated by normalizing day 7 to day 5 image values per each fish. No change in cell number was observed in NTR 1.1 expressing fish (b) due to the persistence of small rounded cells (n=10, 11, and 11 larvae examined, for 0, 0.1, and 10 mM MTZ conditions, respectively). Conversely, both treatment conditions led to near complete ablation of NTR 2.0-expressing macrophages (d; n=18, 20, and 19 larvae examined, for 0, 0.1, and 10 mM MTZ conditions, respectively). Fully detailed statistical comparisons (absolute effect sizes, 95% confidence intervals, Bonferroni-corrected p’-values derived from two-tailed t tests, and sample sizes) between MTZ-treated and control conditions in graphs b and d are provided in Supp. Table 4. Scale bar = 50 microns.
NTR 2.0 does not improve nifurpirinol ablation efficacy
In addition to MTZ, NTR enzymes can convert other prodrug substrates. For instance, a recent study demonstrated robust NTR 1.1-mediated cell ablation following a 24 h exposure to 2.5 μM nifurpirinol (NFP)42. To determine if NTR 2.0 exhibits improved NFP conversion activity, double transgenic larvae co-expressing YFP and NTR 2.0 (NRSE:KalTA4;UAS:YFP-NTR2.0) were exposed to a 1:10 dilution series of nifurpirinol (2.5 μM - 2.5 nM) from 5 to 6 dpf, and YFP quantified at 7 dpf. Contrary to the ~100-fold improvement observed with MTZ, no appreciable enhancement of cell ablation efficacy was observed with nifurpirinol (Extended Data Fig. 4); cell loss being evident only at 2.5 μM NFP, akin to results with NTR 1.142. This suggests that the enhanced prodrug activity of NTR 2.0 is specific to 5-nitroimidazole prodrugs (e.g. MTZ) or not relevant to 5-nitrofuran prodrugs (e.g. NFP). Of note, when higher dose assays were attempted, we observed near complete lethality with 24 h exposure at ≥5 μM NFP. This contrasts with a prior study reporting that lethality initiated at 15 μM NFP for 24 h treatments42.
Bipartite transgenic NTR 2.0 expression resources
To promote dissemination of NTR 2.0 to the research community, we have created UAS and QUAS expression vectors for co-expressing NTR 2.0 with different membrane-tagged fluorescent reporters (e.g., GFP, YFP or mCherry, see list of transgenic lines in Supplementary Information). These plasmids also contain a “tracer” reporter (tagBFP2 for UAS lines, ECFP for QUAS lines) driven by a conserved 5’ element of hatching enzyme (he) homologs to facilitate driver-independent stock maintenance. Existing Gal4 and QF driver lines, respectively, can be combined with these resources to restrict NTR 2.0 and reporter expression to targeted cell types. Plasmids and corresponding transgenic lines will be made available through online depositories, i.e., Addgene (https://www.addgene.org/) and the Zebrafish International Resource Center (ZIRC; https://zebrafish.org/home/guide.php), respectively.
DISCUSSION
NfsB_Ec has received near-exclusive attention as a prodrug-converting NTR for both cancer gene therapy and targeted cell ablation applications1. However, recent evidence suggests this enzyme, dubbed here “NTR 1.0”, is a relatively inefficient NTR compared to orthologous enzymes23,43,44. In particular, we showed that active site residues F70 and F108 impede activity with the 5-nitroimidazole PET probe SN33623 and that rational substitution of these residues yielded marked improvements in 5-nitroimidazole reduction23. Despite also containing F70 and F108 residues, the naturally occurring orthologous enzyme NfsB_Vv was nearly as efficient in activating MTZ as the rationally-engineered NfsB_Ec F70A/F108Y variant. Introducing F70A/F108Y substitutions in NfsB_Vv yielded NTR 2.0, a variant displaying superior MTZ conversion efficiency in all systems tested here: bacteria, rodent cells, human cells, and in zebrafish.
First-generation (NfsB_Ec derived) NTR enzymes have enabled novel cell ablation and regeneration paradigms in the zebrafish liver11, pancreas11,28, kidney45, heart11, brain22, retina46, etc. However, the requirement for near-toxic prodrug doses to achieve effective ablation, and existence of “resistant” cell types, were major drawbacks. NTR 2.0 overcomes these limitations by enhancing MTZ-mediated ablation activity ~100-fold, eliminating the need for toxic MTZ regimens and facilitating effective ablation of resistant cells, e.g., macrophages. By facilitating cell loss at ≤1 mM MTZ, NTR 2.0 also enables prolonged cell ablation. This affords significant advantages to investigations of cell function and will allow regenerative capacities to be challenged with novel paradigms where cell ablation can be maintained for different periods of time and then released. How zebrafish respond to such challenges is unknown since mutant lines exhibiting chronic cell loss have not yet been reversed. However, results from an iterative rapid retinal cell loss paradigm suggest regenerative capacity is maintained, but with decreasing fidelity after successive bouts of cell loss and replacement47. The NTR 2.0/MTZ ablation system, by allowing the extent and duration of cell loss to be precisely controlled, will therefore allow regenerative capacities of discrete stem cell niches to be challenged with conditions that mimic human degenerative diseases; i.e., long-term low rates of cell loss associated with inflammatory signalling cascades.
Although NTR 2.0 exhibited improved MTZ activity across all model systems tested here, some of the other NTR variants evaluated over the course of this study (e.g., the “parental” NfsB_Vv) showed less correlation across species. In addition to the lack of activity presented here for NTR 1.1 in HEK-293 cells, we have previously encountered expression instability issues for the NfsB_Ec F70A/F108Y double mutant and other native or engineered NTR variants in HCT-116 cells23,48–50. Although not reported here, we have also been unable to generate stable transgenic zebrafish lines for several NTR variants that expressed well in both bacteria and HEK-293 cells. This highlights the need to test NTR variants in context when adapting to new models. The reasons for variant/context-specific expression instability of certain NTRs remain unknown, but may stem from NTR substrate promiscuity disrupting host-specific metabolic pathways. Importantly, however, we have not encountered expression stability issues for NTR 2.0 in zebrafish thus far. Nevertheless, these observations suggest heterologous expression of NTR enzymes may need to be attenuated in certain contexts.
Chronic NTR 2.0/MTZ-based cell ablation may also serve as a bona fide means of modeling degenerative diseases at the molecular level. On the face of it, the artificial nature of the system suggests there may be limited relevance to degenerative disease mechanisms. However, our recent data suggests NTR/MTZ cell ablation is mediated by DNA-damage induced cell death pathways broadly implicated in neurodegenerative disease17. Specifically, inhibition of Poly(ADP) ribose polymerase (PARP) protects cells from NTR/MTZ-induced ablation17. PARP1 is a key element in two forms of regulated necroptotic cell death termed parthanatos and cGMP-dependent cell death. The former has been strongly linked to Parkinson’s disease51,52, and the latter in photoreceptor dystrophies53. These data are highly intriguing, emphasizing a need for further investigations of the cell death pathways elicited by the NTR/MTZ system to clarify relevance to degenerative disease mechanisms.
The possibility of applying prolonged low-dose MTZ treatments, enabled by our development of NTR 2.0, necessitates further investigations of the effects of long-term MTZ exposures. Our analysis showed no effects on survival or reproductive health after 36 days of MTZ at 0.1 or 1 mM. However, rare reports of neurotoxic effects in patients34 and infertility post-MTZ in rats33 suggest endogenous prodrug converting activities may be an issue for certain cell types. To date, studies of MTZ effects in zebrafish have been limited. One study showed 10 mM MTZ treatments altered hormone expression in the pituitary gland of larval zebrafish. Importantly, dose-response data from this group suggests treatments of ≤1 mM MTZ would not alter hormone production32. By enabling ablation at micromolar concentrations, NTR 2.0 therefore reduces any potential for unintended effects of MTZ.
Recently nifurpirinol (NFP) has been reported as an alternative prodrug that exhibits less variability in NTR-mediated cell ablation efficacy and is active at micromolar concentrations42. However, toxic effects were observed at 15 μM (24 h) and 10 μM (48 h) treatments42. In our hands, toxic effects arose at 5 μM NFP for 24 h. We do not know the reason for this discrepancy, but differing background strains or NFP source variation are possibilities. Nevertheless, a clear message is that because both MTZ and NFP offer only a small therapeutic index in combination with NfsB_Ec variants―i.e., are toxic at 2 to 3-fold above ablation-promoting doses―potential systemic deleterious effects remain a concern for both prodrugs when paired with first-generation NTR enzymes. While NTR 2.0 showed greatly improved activity with MTZ, it failed to show an increase in activity with NFP. This substrate specificity, however, affords an opportunity to identify NTR variants that exhibit enhanced NFP activity, but are inactive with MTZ. This may allow independent ablation of two cell types, each expressing a different NTR.
Bipartite expression systems such as Gal4/UAS54 and QF/QUAS55, as well as an ever-expanding palette of fluorescent reporters, maximize versatility of zebrafish transgenic resources and accelerate uptake of novel toolsets across the research community. Multi-reporter systems allow the dynamics of cell-cell interactions and signalling pathway activities to be visualized and quantified. Accordingly, we are creating a series of UAS and QUAS reporter/effector plasmids and transgenic lines for co-expressing various fluorescent reporters with NTR 2.0. These resources will facilitate novel NTR 2.0-enabled paradigms for interrogating cell function, exploring regenerative potential, and expanding degenerative disease modeling, thus enhancing the value of NTR targeted cell ablation system for the research community.
METHODS
See the Supplementary Information for methods regarding NTR variant sequences, sequence comparisons, transgenic expression vector cloning, zebrafish husbandry, and zebrafish transgenesis (including a full list of all transgenic resources used or created during these studies).
Bacterial assays - background
Genes encoding NTR candidates were PCR amplified and cloned into the NdeI and SalI sites of two plasmids: pUCX (Addgene #60681), for bacterial overexpression assays, and pET28a(+) (Addgene #69864–3), for His6-tag protein purification. To assess NTR variant properties in the absence of native NTR activity, the E. coli 7NT strain was used, which bears scarless in-frame deletions of seven candidate NTR genes (nfsA, nfsB, azoR, nemA, yieF, ycaK and mdaB) and the efflux pump gene tolC56. For protein purification, the E. coli strain BL21(DE3) was used. All reagents were purchased from Sigma-Aldrich, apart from SN33623, which was custom-synthesized by BOC Sciences.
Bacterial cytotoxicity assays
E. coli 7NT pUCX:nitroreductase cells were used to inoculate wells of a sterile 96 well microplate containing 200 μl LB supplemented with antibiotic and 0.2% (w/v) glucose. Cultures were incubated overnight at 30 °C, 200 rpm. The following morning, cultures were diluted 20-fold in induction medium (LB, antibiotic, 0.2% (w/v) glucose, 50 μM IPTG). Cultures were incubated at 30 °C, 200 rpm for 2.5 h. Culture aliquots (30 μl) were added to sterile 384 well microplates containing 30 μl induction medium ± two-fold the final prodrug concentration. For dose response tests, each culture was exposed, in duplicate, to increasing titrations of prodrug (15 conditions containing 2- or 1.5-fold stepwise increases) and one induction medium-only control. The medium was supplemented with DMSO as appropriate, ensuring that the final DMSO concentration remained <4%. OD600 readings were recorded using an EnSpire™ 2300 Multilabel Reader (PerkinElmer), and initial OD600 values were within the range of 0.12–0.18. Cultures were incubated at 30 °C, 200 rpm for a further 4 h, after which OD600 readings were recorded once more. The increase in OD600 of challenged and unchallenged wells for each strain were compared and used to calculate percent growth at each prodrug concentration.
Mammalian cytotoxicity assays
To generate stably-transfected human embryonic kidney cell lines, plasmid encoding a nitroreductase (F279-V5:ntr, where ntr represents a gene encoding NTR 1.0, NTR 1.1, NfsB_Vv, or NTR 2.0), or a fluorescent protein (pCDNA3-GFP; Addgene plasmid #74165 or mCherry2-N1; Addgene plasmid #54517) was used to transfect HEK-293 cells at 70–90% confluency using Lipofectamine 3000 reagent (Thermo Fisher Scientific) as per the manufacturer’s instructions. After 48 h, cells which had stably integrated plasmid DNA were selected by multiple passage cycles in medium containing escalating concentrations of the selection antibiotic (1–3 μM in the case of puromycin, or 500–900 μg/ml in the case of G418) until cell death was no longer evident. Selection antibiotic concentrations were determined following generation of a dose response curve with wild-type cells.
A standard 48 h MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) cell viability assay was used to evaluate NTR-mediated activation of MTZ and associated cytotoxic activity against HEK-293 cell lines (n ≥ 3 independent experiments with duplicate wells per experiment). Cell lines were treated with MTZ at various concentrations, and a dose−response was generated relative to untreated HEK-293 cells. Cells (100 μL aliquots, 180,000 cells/mL) in RPMI medium/1× glutaMAX/10% fetal calf serum/1% penicillin/streptomycin were seeded into 6.25 mm diameter culture wells and then incubated ~16 h, 37 °C, 5% CO2 before being challenged with 100 μL of RPMI/1× glutaMAX medium ± 2× desired final MTZ concentration. Post-challenge (48 h), 10 μL of CellTitre 96 Aqueous One Solution Cell Proliferation Assay reagent (Promega) was added to all wells and incubated for 60 min at 37 °C, 5% CO2. OD490 was measured with an EnSpire Multilabel plate reader (PerkinElmer), and the absorbance value of the media-only control well subtracted from all other measurements. The baseline-subtracted absorbance of challenged wells was compared to that of unchallenged control wells to determine percent cell viability.
CHO-KI cells were grown in Ham’s F12 medium/10% FBS/penicillin/streptomycin. At 50–60% confluency on 10 cm plates, cells were transfected (Lipofectamine 3000) with either NTR 1.1-mCherry (mCherry-P2A-NfsB_Ec T41Q/N71S/F124T) or NTR 2.0-YFP (tagYFP-P2A- NfsBVv F70A/F108Y). Stable clones were generated by adding G418 (0.5 mg/ml) to the growth medium and selecting resistant mCherry+ or YFP+ colonies. Since not all cells within a colony expressed a given fluorescent reporter, colonies were re-cloned by limiting dilution. Cells were maintained in G418 selection medium until plated for MTZ treatment and MTT (3-[4, 5-dimethylthiazol-2-yl]-2, 5 diphenyl tetrazolium bromide) cell viability assays.
For MTZ dose-response assays, transfected CHO-KI cells were plated into 96-well plates, 10,000 cells/well/100 μl medium, without G418, and allowed to adhere overnight. The following day, the medium was replaced with 100 μl of medium containing varying concentrations of MTZ (≥ 4 wells per MTZ concentration). CHO-KI cells expressing NTR1.1-mCherry were treated with MTZ (0–10 mM) solubilized in water; control wells (0 MTZ) contained 10% water in growth medium. CHO-KI cells expressing NTR 2.0-YFP were treated with MTZ (0–1 mM) solubilized in 0.1% DMSO; control wells (0 MTZ) contained 0.1% DMSO in growth medium. After 24 h of growth in MTZ, an additional 100 μl medium containing the appropriate MTZ concentration was added. Following 48 h total treatment, MTT assays (adapted from Mosmann25) were performed to assess cell viability: 20 μL of 5 mg/ml MTT (Thiazolyl Blue Tetrazolium Bromide, Sigma-Aldrich, M5655) in Dulbecco’s PBS was added to each well. The plates were incubated for 3–4 h, 37 °C, 5% CO2. The supernatant was removed, and 100 μL of DMSO was added. The plates were incubated for 30 min at 37 °C, 5% CO2 and the OD560 measured using a Wallac VICTOR3™ 1420 multilabel plate reader (PerkinElmer). The absorbance values of the DMSO-only controls established the average background absorbance value, which was subtracted from the average absorbance of each MTZ treatment condition. Cell viability (%) was calculated by dividing the average absorbance for each MTZ treatment condition by the absorbance value for cells treated with vehicle alone, multiplied by 100.
Nitroreductase expression and purification
LB/kanamycin medium was inoculated with BL21 (DE3) cells expressing pET28a(+):nitroreductase and grown overnight at 37 °C, 200 rpm. The overnight culture was diluted 100-fold into LB+50 μg/ml kanamycin and incubated at 37 °C, 200 rpm until the OD600 reached 0.6–0.8. The culture was placed on ice for 20 min and IPTG (0.5 mM final concentration) was added to induce nitroreductase expression. The culture was incubated for 24 h, 18 °C and the cells centrifuged (2600 × g, 4 °C, 20 min). The cell pellet was lysed in Bugbuster® (Novagen) and incubated at room temperature for 20 min on an orbital shaker. Following centrifugation (17,000 × g, 4 °C, 15 min), the soluble fractions were decanted and placed on ice. His6-tagged proteins were purified using Ni-NTA His.Bind® Resin (Merck), a peristaltic pump and increasing imidazole buffer washes. Ni-NTA His.Bind® Resin was packed into a Pierce disposable column (Thermo Fisher) and connected to the peristaltic pump. The resin was washed with ddH2O and charged with Charge buffer (50 mM NiSO4). The soluble fraction of the lysed cells was pumped through the column, followed by rinses with Bind buffer (500 mM NaCl, 20 mM Tris-HCl pH 7.9, 5 mM imidazole) and Wash buffer (500 mM NaCl, 20 mM Tris-HCl pH 7.9, 60 mM imidazole). The pump tubing was evacuated and Elution buffer (500 mM NaCl, 20 mM Tris-HCl pH 7.9, 1 M imidazole) run through the column; 500 μl flow through fractions were collected. The three most yellow fractions (indicating highest concentration of FMN-bound nitroreductase) were pooled. Resin beads were washed and stored in Strip buffer (500 mM NaCl, 20 mM Tris-HCl pH 7.9, 100 mM EDTA) at 4 °C. Pooled nickel-purified protein fractions were incubated with excess FMN cofactor (1 mM) for 1 h on ice. Buffer-exchange into 40 mM Tris-Cl pH 7.0 was carried out using HiTrap™ desalting column (GE Healthcare). Only the first 1.5 ml of flow-through was collected to avoid contamination with free FMN.
Steady-state enzyme kinetics
Michaelis-Menten kinetic parameters were derived using NADPH as a cofactor in the presence of an NADPH regenerating system (Bacillus subtilis glucose dehydrogenase + glucose) to maintain constant NADPH levels. Where possible, substrate concentrations were varied from approximately 0.2× KM to 5× KM; however spectroscopic absorption limits prevented concentrations above 3 mM MTZ from being accurately measured. Reactions consisted of 100 μM NADPH, 5 mM glucose, 0.55 μM B. subtilis glucose dehydrogenase, 10 μM NTR, 50 mM sodium phosphate buffer pH 7.0 and varying MTZ concentrations. The OD340 was followed, and mean rates of reduction at each MTZ concentration were obtained from a minimum of triplicate repeats. The extinction coefficient of MTZ reduction at 340 nm was calculated at 1,980 M−1cm−1 by measuring the drop in absorbance of 100 μM MTZ following complete reduction by E. coli NfsA (and subsequent elimination of residual NADPH owing to the intrinsic oxidase activity of NfsA), using a SpectraSuite USB400 UV/Vis spectrophotometer (Ocean Optics).
Fluorescence cultured live cell imaging
For cultured cells, brightfield and fluorescence images were taken on an Olympus IX51 inverted microscope and merged to allow visualization of the ratio of fluorescent to non-fluorescent cells using Olympus’ CellSens software.
Confocal imaging of transgenic zebrafish
All in vivo confocal imaging applied previously detailed methods13,57. Briefly, larval zebrafish were treated with 200 nM PTU at 16 hpf to inhibit pigmentation. Prior to imaging, larvae were anesthetized with tricaine and embedded in 0.8 to 1.5% low-melt agarose. Images were collected on an Olympus FV1000 upright confocal microscope (20× or 60× water immersion objective, NA 0.95 and 1.10, respectively) using Olympus’ FV10-ASW software (v 4.1), except rho:YFP-NTR1.0 and 2.0 fish images which were collected on an Olympus FV3000 Fluoview inverted confocal microscope (30× silicone objective, NA 1.05) using Olympus’ FV31S-SWPAD software.
Automated reporter quantification in vivo (ARQiv) assays
For plate reader-based quantification of YFP-expressing cells in vivo, assays were performed as previously detailed15,16. Briefly, individual zebrafish were aliquoted into wells of 96-well, black, polypropylene, U-bottom plates containing 300 μL of embryo media (+200 nM PTU) and the indicated concentration of MTZ or NFP. On the day of analysis, larvae were anesthetized by addition of 50 μL clove oil (350 ppm) for 15 min. An Infinite M1000 plate reader (Tecan) with iControl software (version 2.0) was used to quantify fluorescence levels in individual fish. Z-dimension focus settings were defined by averaging the maximal z-dimension scan values of five non-ablated controls. Nine regions per well were scanned to account for random orientation of zebrafish using excitation/emission and bandwidth settings of 508 ±5 nm and 528 ±5 nm, respectively, for tagYFP; and 514 ±5 nm and 538 ±10 nm, respectively, for EYFP. The data were processed by first defining a fluorescence “signal” cutoff as the averaged maximal values, plus three standard deviations, of non-transgenic controls. All regional scan values greater than or equal to the signal cutoff were summed to calculate the fluorescence level of each sample. The data were then normalized to a “signal window” bounded by the non-transgenic controls (set at 0%) and 0 MTZ controls (set at 100%) by subtracting the averaged maximal value of the non-transgenic controls from all summed signal values and then dividing the resulting values by the averaged signal from the 0 MTZ controls. Absolute effect sizes for all experimental conditions are expressed as a percentage of the normalized signal window for each assay to facilitate comparisons across biological repeats.
A custom R code package, “ARQiv”16 was used to calculate sample sizes using power analysis. Briefly, to calculate sample sizes, computational iterations randomly sample from large sets of non-ablated (0 mM prodrug) and non-transgenic control data across a range of error rates and effect sizes for both raw and log2-transformed data. Sample sizes for ‘DEFAULT’ effect sizes are calculated at 25, 50, 75, and 100 percent of the positive control. Alternatively, a ‘CUSTOM’ window is available for entering a user-defined effect size. This analysis determined that a sample size of nine larvae was sufficient to detect a 50% loss (EC50) of YFP-expressing cells with the transgenic lines used.
Long-term MTZ exposure
Late larval non-transgenic roy orbison (roya9/a9) fish (15 dpf) were exposed to 0, 0.1, 1 or 10 mM MTZ for 36 days (until 51 dpf). MTZ treatments were initiated at 15 dpf to avoid increases in larval death typical of late larval roy mutant development, i.e., between 10 and 12 dpf. Groups of 30 to 33 larvae were maintained in 1 L of stagnant system water at the indicated concentrations of MTZ, with 750 mL of system water ±MTZ replaced every 2–3 days, and the experiment repeated three times. To assess fecundity post-MTZ treatment, 12–15 pairwise in-crosses of 3 month old fish from each treatment group were setup on three separate occasions. From each successful mating, the number of embryos laid, fertilization rates and offspring survival were recorded until 5 dpf.
Adult zebrafish MTZ exposure
9-month-old transgenic rho:YFP-NTR2.0 zebrafish were maintained for 3 or 7 days with system water ±1 mM MTZ. MTZ was replaced at 48 h for the 3 day treatment and at 48 and 96 h for the 7 day treatment. At 72 or 168 h, eyes were extracted, cryosectioned (10 micron slices), immunostained (detailed below) and imaged using an FV1000 Fluoview confocal microscope (40× oil objective, NA 1.30).
Immunostaining of zebrafish retinal sections
Zebrafish retinas were immunostained as previously described.58 Briefly, 7 dpf larvae or adult zebrafish eyes were extracted, fixed with 4% paraformaldehyde, subjected to five PBS washes, infiltrated with 30% sucrose, embedded in OCT and cryosectioned as dorsa-to-ventral cross sections with a Leica cryostat (CM 3050S) at a thickness of 10 μm. Slides prepared from these sections were washed with PBS and blocked with PBST (PBS/0.5% Triton X-100/ 5% goat serum) for an hour. After PBST removal, sections were incubated in diluted primary antibody overnight. Primary antibody was washed three times with PBST. The secondary antibody was applied for 2 h at room temperature. All slides were washed once with PBST and mounted with VECTASHIELD antifade mounting medium with DAPI (Vector). The primary antibodies and concentrations used in this study were zpr-1 (α-arr3a, 1:200), 1D1 (α-rho, 1:100, aka Ab1-rho), and Ab-4C12 (1:50). The secondary antibody used was a goat anti-mouse Alexa 647 (1:1,000). The zpr-1 monoclonal antibody, which recognizes Arrestin3a, was obtained from the Zebrafish International Resource Center (ZIRC). The 1D1 antibody, which recognizes Rhodopsin, and 4C12 antibody (rod-specific, antigen unknown) was kindly provided by Dr. James M. Fadool. Goat anti-mouse Alexa 647 was obtained from Invitrogen (cat. #C10640, lot #1911332).
Quantification of cells in immunostained zebrafish retinal sections
In 7 dpf larval sections, α-arr3a positive cone cells (zpr-1 antibody) were counted in 100 μm regions of 10 μm thick sections. Because of the unequal distribution of rods in the retina, all YFP-positive or antibody labeled rods present in a single larval section were counted. Per treatment group, 1–4 sections from 10–15 separate retinas were examined. For adult sections, YFP positive, or antibody positive cells were counted in 22–50 300 μm regions of 10 μm retinal sections surrounding the optic nerve from one male and one female zebrafish per condition.
Volumetric Rendering and Quantification with Imaris
Imaris-based volumetric fluorescence quantification of confocal imaging datasets was performed using previously established methods.14 Briefly, confocal z-stacks (Olympus .oib files) of fish expressing YFP, CFP and/or mCherry transgenes were collected using an Olympus FV1000 Fluoview confocal microscope (20× water immersion objective, NA 0.95). Fluorescence signals were calculated for each fluorescence channel across the entire image volume using identical processing parameters (e.g., background subtraction, intensity threshold). Cell surfaces were 3D-rendered and total fluorescence per channel calculated using local background-based volumetric quantification. Statistics for each channel were exported to Microsoft Excel and volume file values summed to calculate channel-specific fluorescence volumes for each fish. To compare pre- and post-treatment images, fluorescence volumes in post-treatment images were normalized to pre-treatment image values for each individual fish.
Microglia/macrophage ablation
Double transgenic mpeg:Gal4;UAS:YFP-NTR2.0 or mpeg:Gal4;UAS:RFP-NTR1.1 larvae were treated with 300 μL of 0, 0.1 or 10 mM MTZ for 48 h from 5–7 dpf in the wells of a 96 well plate. Intravital confocal imaging performed on pre- and post-treatment larvae was acquired with an FV1000 Fluoview confocal microscope (Olympus). Quantification of RFP or YFP positive cell numbers was performed in Imaris (Bitplane, v9.6.1) from 3D rendered z-stacks through the head and trunk. Statistical analyses were performed as described below.
Statistics
For bacterial and mammalian cell culture assays, non-linear regression analysis, Michaelis-Menten curve fitting, and EC50 values (calculated using a dose-response inhibition four–parameter variable slope equation) were generated with GraphPad Prism 8 (GraphPad Software Inc.).
For ARQiv and IMARIS data analysis, pairwise comparisons to either the non-ablated (0 mM prodrug) or non-transgenic controls were performed using Welch’s unequal variances two-tailed t-test (t.test() function). All p-values were adjusted for multiple comparisons using the p.adjust() function, resulting in Bonferroni-corrected p’-values. Standard R code functions were used to calculate absolute effect sizes (mean differences between the treatment group and controls, temp$estimate[1]-temp$estimate[2], where temp is the result of the t.test) and 95% confidence intervals (temp$conf.int, where temp is the result of the t.test). Box plots were created using the function, ggplot()+geom_boxplot()+geom_point().EC50 values were calculated using a dose-response inhibition four-parameter variable slope equation, GraphPad Prism 9.
For zebrafish immunohistology cell count data, control and experimental conditions were compared using a two-tailed nested t test (GraphPad, Prism 9). For long-term MTZ assays, survival data was subjected to Log-rank (Mantel-Cox) tests and Gehan-Breslow-Wilcoxon tests (GraphPad Prism 9) to generate chi-square and p-values. Fecundity and larval survival data was subjected to ANOVA followed by Dunnett’s multiple comparisons test, depending on the outcome of Levene’s test, to generate 95% confidence intervals and p-values. All p-values were subsequently adjusted for multiple comparisons with Bonferroni correction, i.e. p’-values. Absolute effect sizes were calculated as the mean difference between control and experimental conditions. Violin plots were generated with GraphPad, Prism 9.
Code availability
The ARQiv software package is available on the GitHub open-source website (https://github.com/mummlab/ARQiv). All other freely available R code functions used are as listed in the Statistics section.
Extended Data
Extended Data Fig. 1. NfsB_Vv_F70A/F108Y-expressing cells post-MTZ treatment.
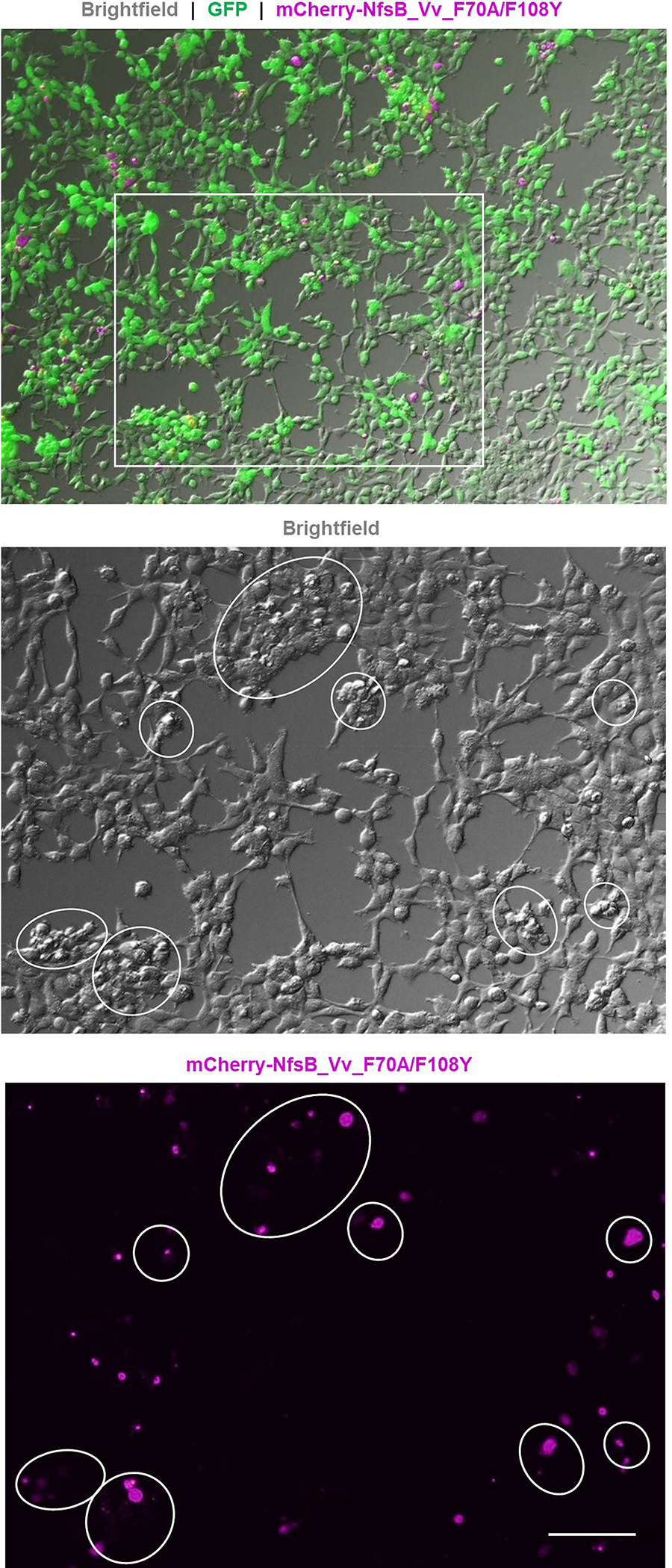
Merged GFP (green), mCherry (magenta), and brightfield image of the MTZ-treated 60:40 co-culture shown in Fig. 2, n=3 biologically independent experiments. Zoomed brightfield and mCherry images of the boxed region show the remaining mCherry fluorescence corresponds to small, round, phase-bright material suggestive of dead or dying cells adhering to healthy GFP-expressing cells. Scale bar = 100 microns.
Extended Data Fig. 2. NTR 2.0/MTZ-induced targeted cell ablation in larval zebrafish.
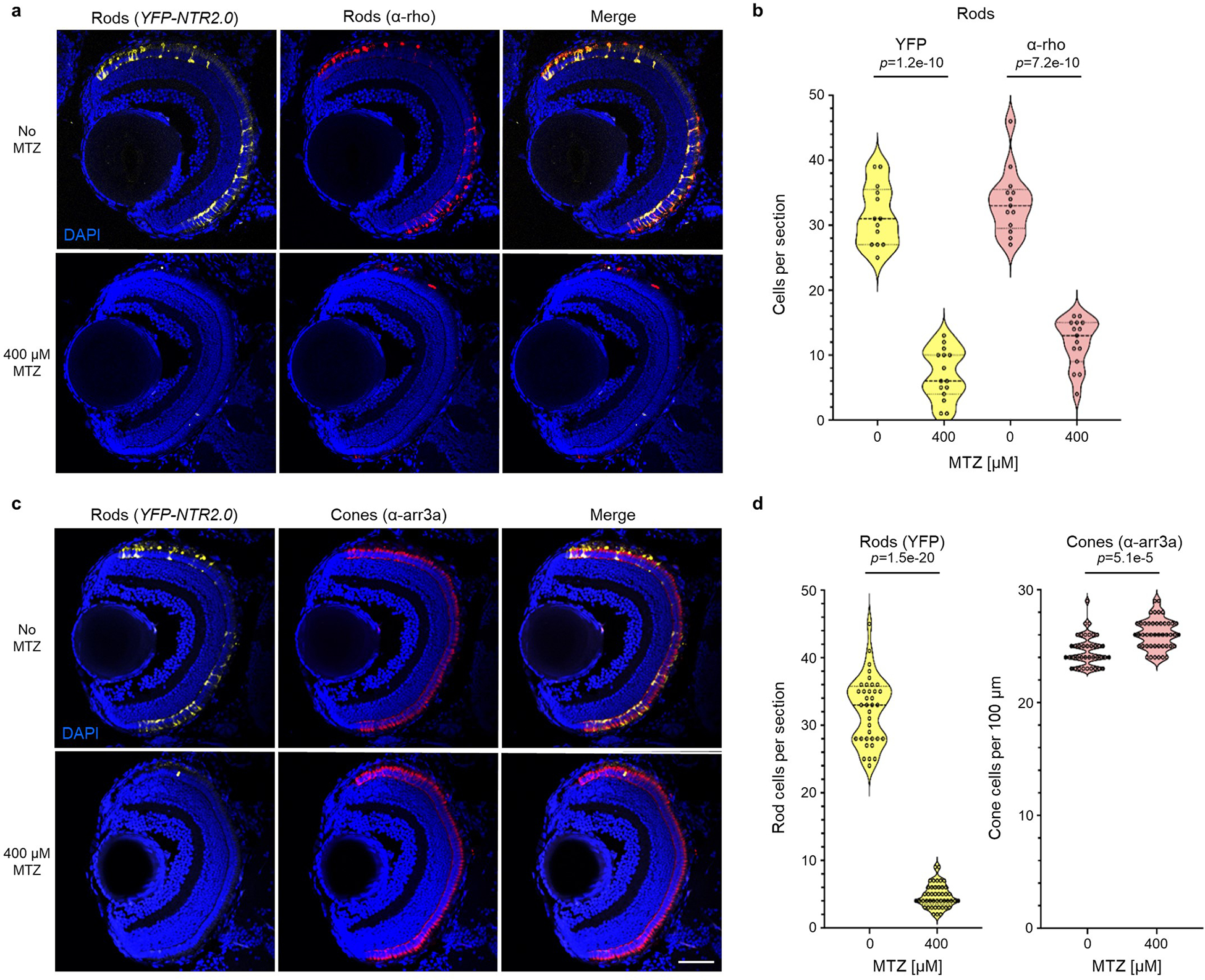
a-d, Transgenic zebrafish larvae co-expressing YFP and NTR 2.0 in rod photoreceptors were treated ±400 μM MTZ for 24 h (5–6 dpf). Retinas were then fixed at 7 dpf, sectioned and labeled with the nuclear stain DAPI (blue cells) and an α-rhodopsin antibody (α-rho, aka 1D1) specific to rods (red cells, a), or an α-arrestin 3a antibody (α-arr3a, aka zpr-1) specific to cones (red cells, c). Representative confocal images of YFP and antibody labeling show effective ablation of NTR 2.0-expressing rod photoreceptors (a, n=10 retinas imaged per condition) and maintenance of neighboring cone photoreceptors (c, n=15 and 14 retinas imaged for the 0 and 400 μM MTZ conditions, respectively). Manual quantification of cell numbers confirmed MTZ-induced loss of rod cells (b) and maintenance of neighboring cones (d). Violin plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and the full distribution of the data, with individual data points (number of measurements per condition) overlaid as a dot plot. A two-tailed nested t test (GraphPad, Prism 9) was used to calculate p-values comparing MTZ-treated and control larvae. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 microns.
Extended Data Fig. 3. NTR 2.0/MTZ-induced rod cell ablation in adult zebrafish.
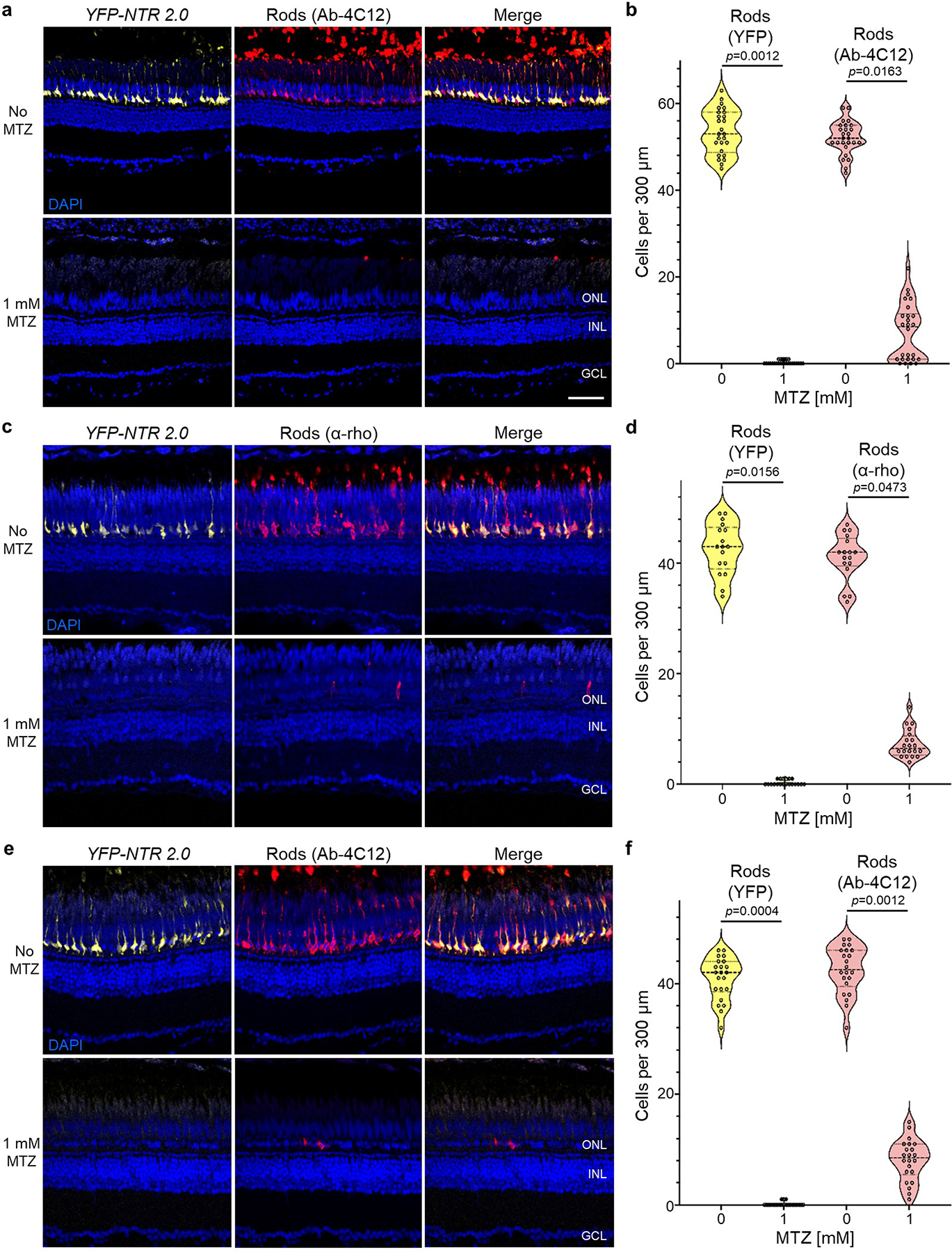
a-f, Transgenic adult zebrafish larvae co-expressing YFP and NTR 2.0 in rod photoreceptors were treated ±1 mM MTZ for 3 (a,b) or 7 days (c-f). Retinas were then fixed, sectioned and labeled with the nuclear stain DAPI (blue cells) and antibodies specific to rods, 4C12 (a and e) or α-rho (c). Representative confocal images of YFP (yellow cells) and antibody labeling (red cells) show effective MTZ-induced ablation of NTR 2.0/YFP-expressing rod photoreceptors and concomitant loss of rod-specific antibody labeling (a, c, and e, n=2 retinas imaged per condition). Manual quantification of cell numbers confirmed MTZ-induced loss of rod cells (b, d, and f. Violin plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and the full distribution of the data, with individual data points (number of measurements per condition) overlaid as a dot plot). A two-tailed nested t test (GraphPad, Prism 9) was used to calculate p-values comparing MTZ-treated and control larvae. Abbreviations: ONL, outer nuclear layer; INL, inner nuclear layer; GCL, ganglion cell layer. Scale bar = 50 microns.
Extended Data Fig. 4. NTR 2.0 does not enhance ablation efficacy with the prodrug nifurpirinol.
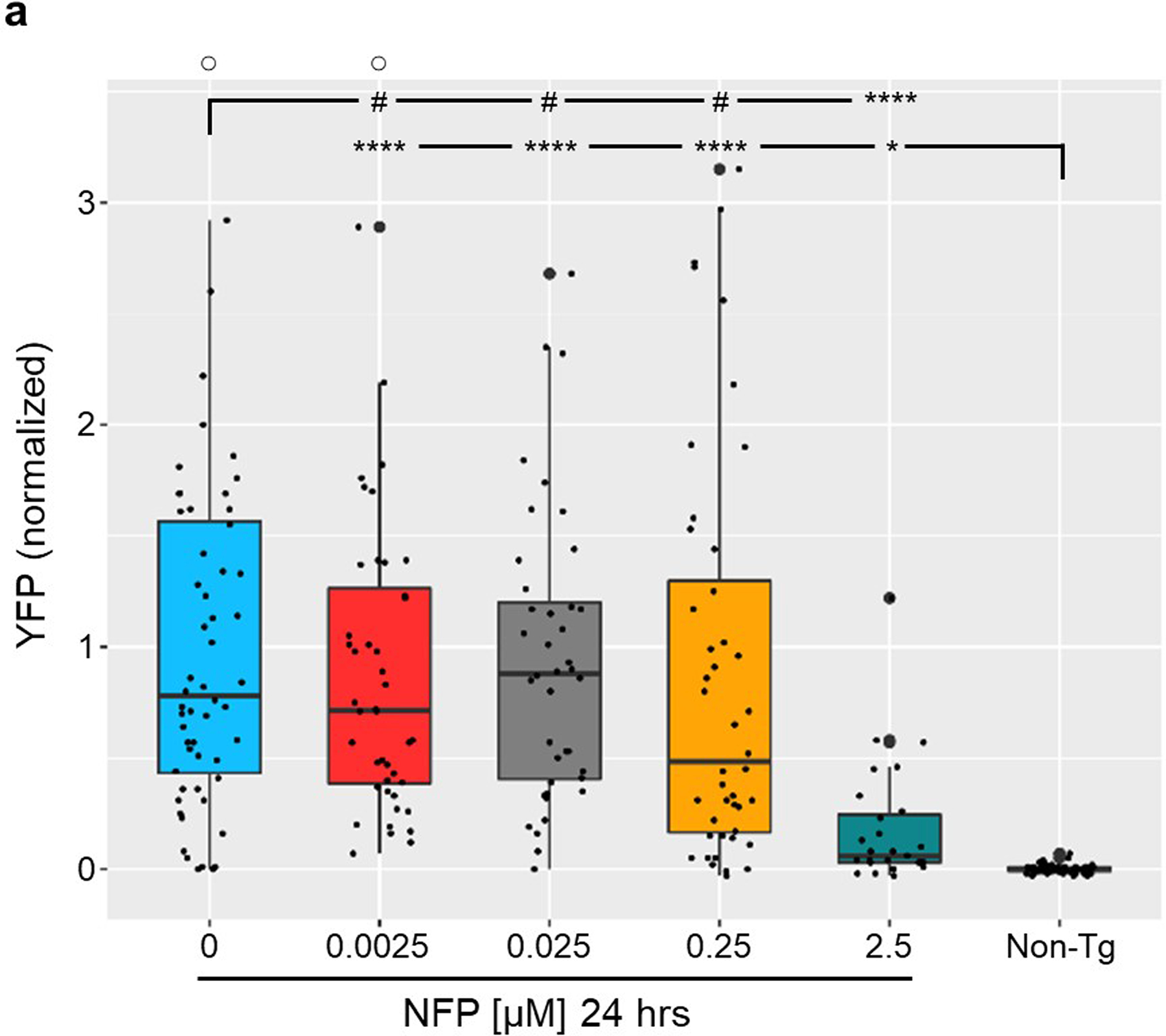
a, Transgenic zebrafish larvae co-expressing YFP and NTR 2.0 in neurons (NRSE:KalTA4;UAS:YFP-NTR2.0) were exposed to the indicated concentrations of NFP for 24 h (5– 6 dpf) and YFP levels were quantified by plate reader at 7 dpf (n=3 biologically independent experiments, dot plots show the number of larvae examined). Box plots show first quartiles (25th percentile), medians, third quartiles (75th percentile), and whiskers = SD, with individual data points (larvae) overlaid as a dot plot. Fully detailed statistical comparisons (absolute effect sizes, 95% confidence intervals, Bonferroni-corrected p’-values derived from two-tailed t tests, and sample sizes) between NFP-treated and control conditions in graphs b and d are provided in Supp. Table 5.Symbols: #p’ > 0.05, *p’ ≤ 0.05, ****p’ ≤ 0.0001; ○ = outlier data points.
Supplementary Material
Acknowledgements
This work was supported by the following grants from the National Institutes of Health, R01OD020376 (JSM and DFA), P30EY001765 (JSM, Animal Module and Imaging Cores), R01NS095355 (ADL and ALC), and R01HG009518 (HJ). Additional consumables support was provided by an HRC Explorer grant (contract 19/750 from the Health Research Council of New Zealand; DFA and JSM). This study was also supported by an unrestricted departmental grant to the Wilmer Eye Institute from Research to Prevent Blindness. The authors wish to thank Makeila Williams, Ben Bich, Noela Lu, and Grace Lee for technical assistance and Drs. Jim Fadool and Marnie Halpern for providing antibodies and a QUAS reporter/effector plasmid, respectively.
Footnotes
Competing Interests Statement
J.S.M. has been awarded patents for the creation (US patent #7,514,595) and use of zebrafish expressing nitroreductase enzymes for gene (US patent #8,071,838) and drug discovery (US patent #8,431,768) applications. M.T.S. is the President and Scientific Director at Luminomics, a biotechnology start-up that offers ARQiv-based screening services. M.T.S. owns stock in Luminomics and J.S.M. serves as a consultant at Luminomics. The remaining authors declare no competing interests.
Data Availability
Source data files used to generate graphs for the following figures are provided in Supplementary information: Figs. 1b, c, e–f, and g–h; 2a–e; 3b–d, g, and i; 4a–b; 5a–c, e, and g; 6b and d; Extended Data Figs. 2b and d. All additional source data files generated during and/or analysed during the current study are available from the corresponding authors upon request.
REFERENCES
- 1.Williams EM et al. Nitroreductase gene-directed enzyme prodrug therapy: insights and advances toward clinical utility. Biochem. J 471, 131–153 (2015). [DOI] [PubMed] [Google Scholar]
- 2.Roldán MD, Pérez-Reinado E, Castillo F & Moreno-Vivián C Reduction of polynitroaromatic compounds: the bacterial nitroreductases. FEMS Microbiol. Rev 32, 474–500 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Venitt S & Crofton-Sleigh C The toxicity and mutagenicity of the anti-tumour drug 5-aziridino-2,4-dinitrobenzamide (CB1954) is greatly reduced in a nitroreductase-deficient strain of E. coli. Mutagenesis 2, 375–81 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Knox RJ et al. The nitroreductase enzyme in Walker cells that activates 5-(aziridin-1-yl)-2,4-dinitrobenzamide (CB 1954) to 5-(aziridin-1-yl)-4-hydroxylamino-2-nitrobenzamide is a form of NAD(P)H dehydrogenase (quinone) (EC 1.6.99.2). Biochem. Pharmacol 37, 4671–7 (1988). [DOI] [PubMed] [Google Scholar]
- 5.Lewis K Platforms for antibiotic discovery. Nat. Rev. Drug Discov 12, 371–387 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Bridgewater JA et al. Expression of the bacterial nitroreductase enzyme in mammalian cells renders them selectively sensitive to killing by the prodrug CB1954. Eur. J. Cancer 31A, 2362–2370 (1995). [DOI] [PubMed] [Google Scholar]
- 7.Drabek D, Guy J, Craig R & Grosveld F The expression of bacterial nitroreductase in transgenic mice results in specific cell killing by the prodrug CB1954. Gene Ther. 4, 93–100 (1997). [DOI] [PubMed] [Google Scholar]
- 8.Clark AJ et al. Selective cell ablation in transgenic mice expression E. coli nitroreductase. Gene Ther. 4, 101–10 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Bridgewater JA, Knox RJ, Pitts JD, Collins MK & Springer CJ The bystander effect of the nitroreductase/CB1954 enzyme/prodrug system is due to a cell-permeable metabolite. Hum. Gene Ther 8, 709–717 (1997). [DOI] [PubMed] [Google Scholar]
- 10.Medico E, Gambarotta G, Gentile A, Comoglio PM & Soriano P A gene trap vector system for identifying transcriptionally responsive genes. Nat. Biotechnol 19, 579–82 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Curado S et al. Conditional targeted cell ablation in zebrafish: a new tool for regeneration studies. Dev. Dyn 236, 1025–35 (2007). [DOI] [PubMed] [Google Scholar]
- 12.White DT & Mumm JS The nitroreductase system of inducible targeted ablation facilitates cell-specific regenerative studies in zebrafish. Methods 62, 232–240 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ariga J, Walker SL & Mumm JS Multicolor time-lapse imaging of transgenic zebrafish: visualizing retinal stem cells activated by targeted neuronal cell ablation. J. Vis. Exp 43, e2093 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White DT et al. Immunomodulation-accelerated neuronal regeneration following selective rod photoreceptor cell ablation in the zebrafish retina. Proc. Natl. Acad. Sci 114, E3719–E3728 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker SL et al. Automated reporter quantification in vivo: High-throughput screening method for reporter-based assays in zebrafish. PLoS One 7, e29916 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White DT et al. ARQiv-HTS, a versatile whole-organism screening platform enabling in vivo drug discovery at high-throughput rates. Nat. Protoc 11, 2432–2453 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L et al. Large-scale phenotypic drug screen identifies neuroprotectants in zebrafish and mouse models of retinitis pigmentosa. Elife 10, e57245 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathias JR, Zhang Z, Saxena MT & Mumm JS Enhanced cell-specific ablation in zebrafish using a triple mutant of Escherichia coli nitroreductase. Zebrafish 11, 85–97 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Godoy R, Noble S, Yoon K, Anisman H & Ekker M Chemogenetic ablation of dopaminergic neurons leads to transient locomotor impairments in zebrafish larvae. J. Neurochem 135, 249–260 (2015). [DOI] [PubMed] [Google Scholar]
- 20.Petrie TA et al. Macrophages modulate adult zebrafish tail fin regeneration. Development 141, 2581–91 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guise CP, Grove JI, Hyde EI & Searle PF Direct positive selection for improved nitroreductase variants using SOS triggering of bacteriophage lambda lytic cycle. Gene Ther 14, 690–8 (2007). [DOI] [PubMed] [Google Scholar]
- 22.Tabor KM et al. Direct activation of the Mauthner cell by electric field pulses drives ultrarapid escape responses. J. Neurophysiol 112, 834–44 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Williams EM et al. Engineering Escherichia coli NfsB To Activate a Hypoxia-Resistant Analogue of the PET Probe EF5 To Enable Non-Invasive Imaging during Enzyme Prodrug Therapy. Biochemistry 58, 3700–3710 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Cory AH, Owen TC, Barltrop JA & Cory JG Use of an Aqueous Soluble Tetrazolium/Formazan Assay for Cell Growth Assays in Culture. Cancer Commun. 3, 207–212 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Mosmann T Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983). [DOI] [PubMed] [Google Scholar]
- 26.Xie X et al. Silencer-delimited transgenesis: NRSE/RE1 sequences promote neural-specific transgene expression in a NRSF/REST-dependent manner. BMC Biol. 10, 93 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Distel M, Wullimann MF & Köster RW Optimized Gal4 genetics for permanent gene expression mapping in zebrafish. Proc. Natl. Acad. Sci. U. S. A 106, 13365–70 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pisharath H, Rhee JM, Swanson M. a, Leach SD & Parsons MJ Targeted ablation of beta cells in the embryonic zebrafish pancreas using E. coli nitroreductase. Mech. Dev 124, 218–29 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davison JM et al. Transactivation from Gal4-VP16 transgenic insertions for tissue-specific cell labeling and ablation in zebrafish. Dev. Biol 304, 811–24 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hyatt GA, Schmitt EA, Fadool JM & Dowling JE Retinoic acid alters photoreceptor development in vivo. Proc Natl Acad Sci U S A 93, 13298–13303 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larison KD & Bremiller R Early onset of phenotype and cell patterning in the embryonic zebrafish retina. Development 109, 567–576 (1990). [DOI] [PubMed] [Google Scholar]
- 32.Cheng X et al. Effects of metronidazole on proopiomelanocortin a gene expression in zebrafish. Gen. Comp. Endocrinol 214, 87–94 (2015). [DOI] [PubMed] [Google Scholar]
- 33.McClain R, Downing JC & Edgcomb JE Effect of metronidazole on fertility and testicular function in male rats. Fundam. Appl. Toxicol 12, 386–396 (1989). [DOI] [PubMed] [Google Scholar]
- 34.Sørensen CG, Karlsson WK, Amin FM & Lindelof M Metronidazole-induced encephalopathy: a systematic review. J. Neurol 267, 1–13 (2020). [DOI] [PubMed] [Google Scholar]
- 35.Morris AC, Schroeter EH, Bilotta J, Wong ROL & Fadool JM Cone survival despite rod degeneration in XOPS-mCFP transgenic zebrafish. Invest. Ophthalmol. Vis. Sci 46, 4762–71 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Narayan DS, Wood JPM, Chidlow G & Casson RJ A review of the mechanisms of cone degeneration in retinitis pigmentosa. Acta Ophthalmologica vol. 94 748–754 (2016). [DOI] [PubMed] [Google Scholar]
- 37.Delyfer M-N et al. Inherited retinal degenerations: therapeutic prospects. Biol. Cell 96, 261–269 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Doughty ML, De Jager PL, Korsmeyer SJ & Heintz N Neurodegeneration in Lurcher mice occurs via multiple cell death pathways. J. Neurosci 20, 3687–3694 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Eroglu M & Derry WB Your neighbours matter-non-autonomous control of apoptosis in development and disease. Cell Death and Differentiation vol. 23 1110–1118 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi DW & Rothman SM The role of glutamate neurotoxicity in hypoxic-ischemic neuronal death. Annual Review of Neuroscience vol. 13 171–182 (1990). [DOI] [PubMed] [Google Scholar]
- 41.Lee JY et al. Central and peripheral secondary cell death processes after transient global ischemia in nonhuman primate cerebellum and heart. in Methods in Molecular Biology vol. 1919 215–225 (Humana Press, New York, NY, 2019). [DOI] [PubMed] [Google Scholar]
- 42.Bergemann D et al. Nifurpirinol: A more potent and reliable substrate compared to metronidazole for nitroreductase-mediated cell ablations. Wound Repair Regen 26, 238–244 (2018). [DOI] [PubMed] [Google Scholar]
- 43.Williams EM et al. A cofactor consumption screen identifies promising NfsB family nitroreductases for dinitrotoluene remediation. Biotechnol. Lett 41, 1155–1162 (2019). [DOI] [PubMed] [Google Scholar]
- 44.Crofts TS et al. Discovery and Characterization of a Nitroreductase Capable of Conferring Bacterial Resistance to Chloramphenicol. Cell Chem. Biol 26, 559–570. e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Huang J et al. A zebrafish model of conditional targeted podocyte ablation and regeneration. Kidney Int. 83, 1193–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Montgomery JE, Parsons MJ & Hyde DR A novel model of retinal ablation demonstrates that the extent of rod cell death regulates the origin of the regenerated zebrafish rod photoreceptors. J. Comp. Neurol 518, 800–814 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ranski AH, Kramer AC, Morgan GW, Perez JL & Thummel R Characterization of retinal regeneration in adult zebrafish following multiple rounds of phototoxic lesion. PeerJ 6, e5646 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Copp JN et al. Engineering a Multifunctional Nitroreductase for Improved Activation of Prodrugs and PET Probes for Cancer Gene Therapy. Cell Chem. Biol. Biol 24, 391–403 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Prosser GA et al. Creation and screening of a multi-family bacterial oxidoreductase library to discover novel nitroreductases that efficiently activate the bioreductive prodrugs CB1954 and PR-104A. Biochem. Pharmacol 85, 1091–103 (2013). [DOI] [PubMed] [Google Scholar]
- 50.Swe PM et al. Targeted mutagenesis of the Vibrio fischeri flavin reductase FRase I to improve activation of the anticancer prodrug CB1954. Biochem. Pharmacol 84, 775–83 (2012). [DOI] [PubMed] [Google Scholar]
- 51.Lee Y et al. Parthanatos mediates AIMP2-activated age-dependent dopaminergic neuronal loss. Nat. Neurosci 16, 1392–1400 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kam T-I et al. Poly(ADP-ribose) drives pathologic α-synuclein neurodegeneration in Parkinson’s disease. Science 362, eaat8407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Power M et al. Cellular mechanisms of hereditary photoreceptor degeneration - Focus on cGMP. Prog. Retin. Eye Res 74, 100772 (2020). [DOI] [PubMed] [Google Scholar]
- 54.Halpern ME et al. Gal4/UAS transgenic tools and their application to zebrafish. Zebrafish 5, 97–110 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Subedi A et al. Adoption of the Q transcriptional regulatory system for zebrafish transgenesis. Methods 66, 433–40 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Copp JN, Hanson-Manful P, Ackerley DF & Patrick WM Error-prone PCR and effective generation of gene variant libraries for directed evolution. Methods Mol. Biol 1179, 3–22 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Mumm JS et al. In vivo imaging reveals dendritic targeting of laminated afferents by zebrafish retinal ganglion cells. Neuron 52, 609–21 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Unal Eroglu A et al. Multiplexed CRISPR/Cas9 Targeting of Genes Implicated in Retinal Regeneration and Degeneration. Front. Cell Dev. Biol 6, 88 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Source data files used to generate graphs for the following figures are provided in Supplementary information: Figs. 1b, c, e–f, and g–h; 2a–e; 3b–d, g, and i; 4a–b; 5a–c, e, and g; 6b and d; Extended Data Figs. 2b and d. All additional source data files generated during and/or analysed during the current study are available from the corresponding authors upon request.


