ABSTRACT
Analysis of the de novo assembled genome of Mammaliicoccus sciuri IMDO-S72 revealed the genetically encoded machinery behind its earlier reported antibacterial phenotype and gave further insight into the repertoire of putative virulence factors of this recently reclassified species. A plasmid-encoded biosynthetic gene cluster was held responsible for the antimicrobial activity of M. sciuri IMDO-S72, comprising genes involved in thiopeptide production. The compound encoded by this gene cluster was structurally identified as micrococcin P1. Further examination of its genome highlighted the ubiquitous presence of innate virulence factors mainly involved in surface colonization. Determinants contributing to aggressive virulence were generally absent, with the exception of a plasmid-associated ica cluster. The native antibiotic resistance genes sal(A) and mecA were detected within the genome, among others, but were not consistently linked with a resistance phenotype. While mobile genetic elements were identified within the genome, such as an untypeable staphylococcal cassette chromosome (SCC) element, they proved to be generally free of virulence- and antibiotic-related genes. These results further suggest a commensal lifestyle of M. sciuri and indicate the association of antibiotic resistance determinants with mobile genetic elements as an important factor in conferring antibiotic resistance, in addition to their unilateral annotation.
IMPORTANCE Mammaliicoccus sciuri has been put forward as an important carrier of virulence and antibiotic resistance genes, which can be transmitted to clinically important staphylococcal species such as Staphylococcus aureus. As a common inhabitant of mammal skin, this species is believed to have a predominant commensal lifestyle, although it has been reported as an opportunistic pathogen in some cases. This study provides an extensive genome-wide description of its putative virulence potential taking into consideration the genomic context in which these genes appear, an aspect that is often overlooked during virulence analysis. Additional genome and biochemical analysis linked M. sciuri with the production of micrococcin P1, gaining further insight into the extent to which these biosynthetic gene clusters are distributed among different related species. The frequent plasmid-associated character hints that these traits can be horizontally transferred and might confer a competitive advantage to its recipient within its ecological niche.
KEYWORDS: Staphylococcus, bacteriocins, genome analysis
INTRODUCTION
With high-throughput sequencing technologies becoming widely used, it became apparent that common techniques, such as phenotypic characterization, ribotyping with EcoRI, internal transcribed spacer (ITS)-PCR fingerprinting, repetitive sequence based PCR (rep-PCR) with (GTG)5 primers, and even phylogenies based on the 16S rRNA gene, often fail to robustly identify species within the Staphylococcaceae (1–4). Recently, genera within the Staphylococcaceae were delineated using core genome phylogenies combined with overall genome-related indices (OGRIs; e.g., average nucleotide identity, average amino acid identity, conserved signature proteins). This triggered a reassignment of the Staphylococcus sciuri species group to a novel genus, i.e., Mammaliicoccus, with Mammaliicoccus sciuri as the type species (5, 6). Also, Mammaliicoccus sciuri itself has been the subject of many taxonomical reclassifications in the past (3, 6). Its current genus name refers to the ecological niche from which these bacteria are typically isolated, being a wide variety of farm and wild mammals, as well as products derived thereof (7–11). Although mostly exhibiting a commensal lifestyle on its animal hosts, M. sciuri has been occasionally identified as an opportunistic pathogen associated with mastitis, dermatitis, and exudative epidermitis (12–15). The species has also been reported as a sporadic colonizer in humans and, to a lesser extent, been isolated from clinical specimens, putting forward questions about its clinical relevance (16–20). It remains unclear, however, if M. sciuri is to be considered a causative agent in infections (9), especially since it is challenging to accurately identify Mammaliicoccus species within polymicrobial infections and distinguish them from closely related staphylococcal species (9).
The sporadic nosocomial appearances have nonetheless triggered interest in Mammaliicoccus species, and M. sciuri in particular, as they could act as a reservoir of resistance and virulence determinants (9, 21). Multiple chromosomally encoded homologues of the methicillin resistance gene mecA have been found but do not seem to confer resistance (22). However, the genetic location of the mecA gene on the chromosomal mobile element staphylococcal cassette chromosome mec (SCCmec) and addition of regulators (mecR1, mecI) turned out to be important in conferring a methicillin-resistant phenotype (23–25). Phenotypical and genome-based in silico analyses have shown that M. sciuri disposes of a substantial repertoire of putative virulence factors (VFs) (9, 21, 26, 27). In general, adhesins and biofilm formation, typically encoded by the ica operon, aiding in the colonization of (a)biotic surfaces are regarded as the most important pathogenic factors for coagulase-negative staphylococci and might be as well for M. sciuri (28–30). Considering that these VFs are needed for both the commensal and pathogenic lifestyles of these bacteria, it remains difficult to define determinants that could differentiate commensal from invasive variants (29, 30).
Being a species isolated from a wide variety of environment-related sources, M. sciuri might also prove an interesting carrier of biosynthetic gene clusters (BGCs) and play a role in the dissemination of the latter between related species. Biological active substances encoded within these gene clusters are mostly found in environmental bacteria, in particular, members of the Streptomyces genus (31). Natural products that originate from a genetic-encoded template and undergo modifications after their ribosomal translation, are referred to as ribosomally synthesized and posttranslationally modified peptides (RiPPs) (32). As a quickly expanding subclass of RiPP antibiotics, thiopeptides have gained much interest for being potent inhibitors of protein synthesis, generally in Gram-positive bacteria (33). They are characterized by a large amount of posttranslational modified heterocycles such as thiazoles, oxazoles, thiazolines, and dehydroamino acids organized in a macrocyclic structure around a central nitrogen-containing six-membered ring (31). Besides Streptomyces, other bacteria have been reported to produce thiopeptides, such as Bacillus spp., Macrococcus caseolyticus, Staphylococcus equorum, and Staphylococcus hominis, all of which share close evolutionary relatedness with Mammaliicoccus spp. (31, 34–37).
The composition of a thiopeptide gene cluster usually includes a gene encoding the structural precursor peptide, which is converted to a biologically active compound by the presence of multiple posttranslational modification enzymes (38). Mechanisms conferring self-immunity to the producing bacterium are typically encoded along the biosynthetic genes necessary for thiopeptide maturation, although this is not always the case (31, 35, 39). The result of thiopeptide activity is the inhibition of protein translation, although the exact mode of action depends on subtle differences in the thiopeptide macrocycle size (33). Thiopeptides containing 26-sized macrocycles, such as micrococcin P1 and thiostrepton, bind at a gap between the ribosomal L11 protein and the 23S rRNA, which are part of the GTPase-associated center (33, 40, 41). Upon binding, these compounds will perturb the association of the elongation factor G (EF-G) with the ribosomal complex, interfering with its GTP hydrolysis, which is necessary for active translation (40). Most resistance mechanisms in native producers involve drug target alterations. Methylation of specific nucleotides in the 23S rRNA or an altered form of the L11 ribosomal protein (or even its complete absence) are common resistance mechanisms reported in thiopeptide-producing microorganisms (31, 35, 42). Nevertheless, for many thiopeptides the adopted resistance mechanisms conferring self-immunity in the native producers are still undetermined and need further scientific attention.
In this study, M. sciuri (strain IMDO-S72) was associated with the production of the thiopeptide micrococcin P1. This strain was originally isolated from fermented meat and attracted interest because of its potent antibacterial activity toward Clostridium botulinum and other foodborne pathogens in previous research (43). The present study aimed at identifying the compound responsible for this antibacterial activity as well as the characterization of the BGC necessary for its production. Additionally, in light of its recent reclassification and putative role as an opportunistic pathogen, a genome-wide virulence analysis was performed to further aid in identifying genetic traits that could discriminate commensal from infectious lifestyles as well as to verify to what extent this species can play a role in the dissemination of antimicrobial resistance genes.
RESULTS AND DISCUSSION
General features of the de novo assembled genome.
The Oxford Nanopore Technologies (ONT) and Illumina read set contained 655,730 reads with an ∼1,600× coverage and 4,048,072 reads with an ∼210× coverage, respectively. The hybrid assembly generated by Unicycler produced a complete assembled genome and did not require further manual intervention. The genome of M. sciuri IMDO-S72 comprises a single circular chromosome of 2,788,517 bp with a G+C content of 32.6% and four plasmid replicons. pIMDO-S72-1 was predicted to be a linear plasmid and was 66,038 bp long with an estimated copy number of 12.2, while pIMDO-S72-2 (21,284 bp), pIMDO-S72-3 (19,094 bp), and pIMDO-S72-4 (3,410 bp) were predicted as circular plasmids with an estimated copy number of 11.0, 9.70, and 104.4, respectively. Based on Prokka, a total of 2,855 coding sequences (CDSs) were predicted, as well as 60 tRNAs and 19 rRNAs. Four isolated CRISPRs, without neighboring cas genes, were detected in the chromosomal DNA, each consisting of two repeats and one spacer region. However, the spacers within these CRISPR arrays were quite conserved among different strains of M. sciuri (results not shown), indicating that these elements might be wrongly identified as CRISPRs (44). A CRISPR-Cas system, similar to a type II-C system, was located on pIMDO-S72-1, containing an array of 4 repeats of 29 bp and 3 spacers with a length of 35 bp, located downstream of a single cas9 gene and 3 putative transactivating CRISPR RNA (tracrRNA)-associated antirepeats (45). Nevertheless, the absence of the universal cas1 and cas2 genes in this system indicates that it concerns a putative nonfunctional remnant of an earlier working CRISPR-Cas system (44, 46).
An intact 51.2-kbp prophage region was present in the chromosome of M. sciuri IMDO-S72 (position 1,060,618 to 1,111,806 bp), with a G+C content of 31.4%. The attachment site attL (5′-TCCCTCACTCTCCG-3′) was located within a tRNASer gene, while attR occurred as a direct repeat at the end of the prophage in an intergenic region. The prophage region contained 77 CDSs, 44 of which encoded hypothetical proteins. The remaining CDSs were phage-associated proteins, among which seven are involved in the tail shaft assembly, and there was one integrase, one terminase, one tail fiber protein, and one baseplate protein. One locus within this region was predicted to be a non-phage-like protein.
The genome also included genetic traits that could reflect the ability of this species to proliferate in meat products. Mammaliicoccus sciuri IMDO-S72 contained a substantial reservoir of genes that could facilitate scavenging of heme iron, as well as coping with the carbohydrate-poor environment by catabolizing peptides/amino acids and purine nucleosides. An iron-sensing determinant (isd) gene cluster was identified in the chromosome of M. sciuri IMDO-S72. IsdC (SSCS72_00397) was predicted as the central conduit of the heme uptake system, containing an NPQTN motif for cell wall anchoring. The isdA gene (SSCS72_00398) encodes a membrane-embedded factor that binds to hemoglobin and transfers its heme group to IsdC. IsdE (SSCS72_00399) is a heme transport lipoprotein that, together with the permease IsdF (SSCS72_00400, SSCS72_02586), most likely enables membrane translocation of the heme into the cytoplasm. The gene locus SSCS72_00401 was not assigned to a specific Isd factor but was predicted to encode an iron complex ATP-binding cassette (ABC) transporter. Sortase B (SSCS72_00402) was encoded within the isd cluster, responsible for covalent anchoring of IsdC to the bacterial cell wall as well as other NPQTN motif-containing surface determinants. The last factor of the Isd system was predicted to encode a heme oxygenase IsdI (SSCS72_00403), catalyzing the oxidative degradation of heme and leading to the release of iron. Extracellular proteolytic enzymes were found as well, such as SspA (SSCS72_00181, SSCS72_01332, SSCS72_02553), ClpX (SSCS72_01289), ClpP (SSCS72_02256, SSCS72_02430), and several putative zinc metalloproteases (SSCS72_00567, SSCS72_01753, SSCS72_02695), which can aid in the degradation of meat tissue with the release of small peptides and/or amino acids. The uptake of such compounds can be facilitated by an oligopeptide permease (opp) operon. The Opp components were ordered within a cluster and consisted of multiple distinct copies of the oligopeptide-binding protein OppA (SSCS72_02071, SSCS72_02072, SSCS72_02073, SSCS72_02074), two membrane-spanning permeases, OppB (SSCS72_02078) and OppC (SSCS72_02077), and two cytoplasmic ATP-binding proteins, OppD (SSCS72_02076) and OppF (SSCS72_02075), that provide energy for the protein-dependent transport system. Two additional opp genes were found to be encoded outside the opp gene cluster, namely, oppC (SSCS72_00103) and oppA (SSCS72_02190). Genes involved in the degradation of purine deoxyribonucleosides (2′-deoxyadenosine and 2′-deoxyguanosine, abundant in meat) were also present in M. sciuri IMDO-S72 (SSCS72_00292, SSCS72_00734, SSCS72_01972) and could improve the survival of this species by catabolizing these compounds for energy production.
While the above-mentioned factors can increase the fitness of this strain within a meat-related environment in view of its potential application as a meat starter culture (43), they can also be regarded as potential VFs within the context of opportunistic pathogen behavior on mammal skin. To widen the view of its ecological functioning, a detailed description of VFs (other than the ones mentioned above) and BGCs involved in bacteriocin production is given below.
Mammaliicoccus sciuri IMDO-S72 contains two bacteriocin biosynthetic gene clusters.
AntiSMASH identified two putative BGCs encoding RiPPs that could exert inhibitory activity toward other (closely related) bacterial species. The first RiPP-like cluster was chromosomally encoded and contained genes that resembled the lactococcin 972 operon (47, 48). Lactococcin 972 was shown to form a homodimer that interferes with cell wall biosynthesis by binding to lipid II (49, 50). The structural gene (SSCS72_02144) encoded a 97-amino acid (aa)-long precursor peptide, containing an N-terminal signal peptide region (aa 1 to 23) which is assumed to be cleaved to yield the mature structural peptide of 74 aa. Next to the structural gene, a bacteriocin-associated integral membrane protein was predicted (SSCS72_02143) with eight membrane-spanning hydrophobic regions. It most likely acts as an immunity protein, although the specific mechanism that confers self-immunity to the producing strain has yet to be elucidated. Sequence alignment analysis using blast revealed that this gene cluster, alongside its upstream and downstream neighboring genes, was rather conserved within different strains of the M. sciuri species. The lactococcin 972 operon has also been identified in different strains of S. equorum (51).
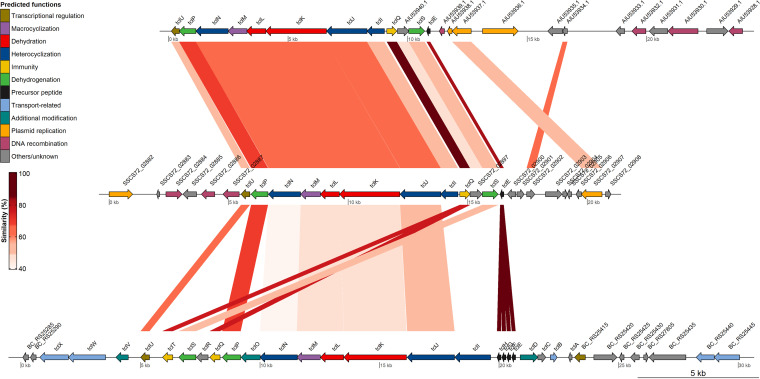
The second BGC was encoded on pIMDO-S72-2 and was associated with thiopeptide production, more specifically, micrococcin P1. A trisubstituted pyridine ring and a 26-membered macrocycle, as predicted features of the core peptide, indicated that it concerned a class d thiopeptide (33). The gene clusters of members of this family that meet the above-mentioned structural features, such as micrococcin P1 and P2 as well as several thiocillins, have been described previously and were used to further annotate the genetic features based on protein homology and to assess to what extent these gene clusters are conserved across species (35, 38, 39, 42). Micrococcin P1 production was also reported in S. equorum WS 2733, but its cognate BGC was never characterized (34). The name allocations of the genes involved in the biosynthetic pathway were done according to the first described thiocillin (tcl) gene cluster (39). Comparison based on protein homology revealed a biosynthetic gene cluster spanning an 11-kb region on pIMDO-S72-2 and containing 12 genes, 11 of which were predicted to have a function in the biosynthetic pathway (Fig. 1). The genetic organization of the micrococcin P1 gene cluster corresponded, in its entirety, with the identified BGC on pBac115 of M. caseolyticus 115 (35). Apart from a homology between a partitioning protein (SSCS72_02907) and a protein with unknown function (SSCS72_02902), the other plasmid regions were generally devoid of any genes showing significant similarity (E value < 0.05). Regardless of being closely related species, this might be an indication that genetic changes in such BGCs are quite restricted by selective constraints on the complex modification enzymes and structural peptide to maintain functionality. Comparison with the chromosomally encoded BGC of Bacillus cereus ATCC 14579, containing 24 genes clustered within a 22-kb region, revealed some more rearrangements. Most genes within the BGC of M. sciuri IMDO-S72 had a homologous counterpart in the gene cluster of B. cereus ATCC 14579, while the latter one contained four copies of the precursor peptide-encoding gene (tclE, tclF, tclG, and tclF) and two copies of the ribosomal protein L11-encoding immunity genes (tclQ and tclT). Interestingly, no homology was found between the tclI genes of M. sciuri IMDO-S72 and B. cereus ATCC 14579, indicating that the translated amino acid sequences differ substantially. Together with putative additional modification enzymes (tclD, tclO, tclV), this might explain why the BGC of B. cereus ATCC 14579 gives rise to eight different thiopeptides instead of one. In general, the precursor peptide (TclE) and ribosomal protein L11 (TclQ and TclT) proved to be the most conserved across the three different BGCs, where TclP showed the highest similarity among the enzymes involved in posttranslational modification. On the other hand, sequence conservation at the nucleotide level between the three BGCs proved to be very low (results not shown). An overview of the complete blastp output can be found in Table S1 in the supplemental material.
FIG 1.
Functional comparison of thiopeptide gene clusters of M. caseolyticus 115 (top), M. sciuri IMDO-S72 (middle), and B. cereus ATCC 14579 (bottom). Homologous regions are indicated with colored connected areas based on protein similarity (i.e., ppos [percentage of positive-scoring matches] values of blastp analysis [see Table S1]). Only blastp hits with an E value of <0.05 were taken into account, except for comparisons with tclE, tclF, tclG, and tclH of B. cereus, which were included for completeness. For M. caseolyticus 115 and M. sciuri IMDO-S72, the complete pBac115 (reverse complement) and pIMDO-S72-2 sequences are depicted; for B. cereus ATCC 14579, a 30-kb region of the chromosome is represented. The figure is drawn to scale.
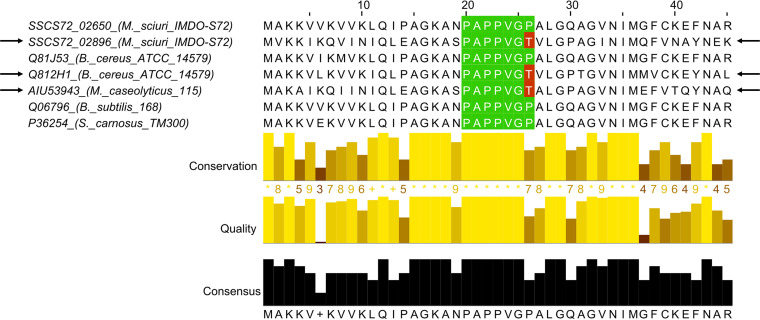
In M. sciuri IMDO-S72, the precursor peptide was encoded by tclE (SSCS72_02899) and contained a 35-aa N-terminal leader peptide together with a conserved 14-aa core peptide that undergoes multiple modifications by the biosynthetic machinery. Genes tclS (SSCS72_02898) and tclP (SSCS72_02889) encode dehydrogenases that are involved in the processing of the C-terminal threonine residue (38, 52). The gene products of tclI, tclJ, and tclN are involved in the heterocyclization of cysteine residues to form thiazoles (SSCS72_02895, SSCS72_02894, and SSCS72_02890, respectively). The TclI protein was predicted to contain a RiPP recognition element (RRE) domain, which typically binds the N-terminal leader region of the precursor peptide, making the core peptide scaffold accessible for posttranslational modification enzymes (53). Cysteine residues are converted to thiazolines by the ATP-dependent action of TclJ, which are further oxidized by the TclN enzyme to thiazole heterocycles. Another characteristic thiopeptide modification is the dehydration of serine and threonine residues to dehydroalanine and dehydrobutyrine, which is most likely performed by the dehydratase-encoding tclK and tclL genes (SSCS72_02892 and SSCS72_02893, respectively). The cyclization reaction, yielding the central pyridine domain and the consequent cleavage of the leader peptide, is facilitated by tclM (SSCS72_02891). Gene tclU encodes a putative transcriptional regulator of the MerR family, containing a helix-turn-helix signature. Multiple sequence alignments of L11 ribosomal proteins (encoded by the rplK gene), encoded within biosynthetic gene clusters and within more distant genomic locations of thiopeptide-producing strains, revealed a single mutation leading to the substitution of a proline residue at position 26 into a threonine within a conserved proline-rich region of the N-terminal domain of L11 (Fig. 2). Probably, self-immunity is achieved by replacement of the native L11 protein within the ribosomal complex by the alternative homologue, thereby significantly decreasing the binding affinity of micrococcin P1 due to conformational changes induced by these specific mutations (40). As micrococcin P1 also interacts with certain residues of the 23S rRNA, specific mutations could also reduce affinity and induce resistance toward the produced thiopeptide (40). Nevertheless, no transitions or transversions were found within the six copies of the 23S rRNA gene of M. sciuri IMDO-S72, supporting the role of tclQ as the sole immunity gene.
FIG 2.
Multiple sequence alignment of the first 45 amino acid residues of the 50S ribosomal protein L11 from M. sciuri IMDO-S72 (SSCS72_02650 and SSCS72_02896), B. cereus ATCC 14579 (GenBank accession numbers Q81J53 and Q812H1), M. caseolyticus 115 (AIU53943), B. subtilis 168 (Q06796) and S. carnosus TM300 (P36254). The latter two have been shown to be susceptible toward micrococcin P1. The conserved proline region in the N-terminal domain of L11 with which micrococcin P1 interacts is indicated in green, while substitutions of Pro-26 by Thr are indicated in red. Ribosomal L11 protein homologs that are encoded within a thiopeptide BGC are indicated with arrows.
Antibacterial activity of Mammaliicoccus sciuri IMDO-S72 originates from a plasmid-encoded thiopeptide gene cluster.
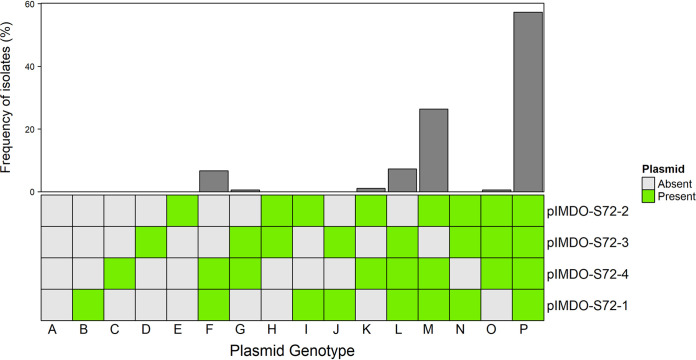
To exclude the possibility of a contributing effect of the chromosomally encoded BGC similar to the lactococcin 972 operon (and other BGCs that were possibly missed during genome mining) toward the antibacterial phenotype of M. sciuri IMDO-S72, plasmid-cured derivatives were obtained by incubating cell cultures at 42°C, arousing stress conditions that mediated plasmid loss. Using multiplex colony PCR, the presence/absence of all four plasmids could be assessed in cells subjected to these conditions. In total, 178 isolates that survived the heat treatment were picked up (representing ∼58% of all surviving isolates) and screened for their plasmid profiles (see Fig. S1). Different plasmid genotypes were found within the 178 isolates, the wild type being the most abundant (plasmid genotype P, n = 102), followed by plasmid genotype M (n = 47) devoid of pIMDO-S72-3, plasmid genotype L (n = 13) where pIMDO-S72-2 was lost, and plasmid genotype F (n = 12) where both pIMDO-S72-2 and pIMDO-S72-3 were absent (Fig. 3). pIMDO-S72-1 was only lost in four isolates, while pIMDO-S72-4 remained present in all the cured derivatives, which can probably be related to its higher estimated copy number (104.4) compared to the other plasmids and, because of its smaller size, a putative lower metabolic burden. The fact that these plasmids were lost under plasmid curing conditions, apart from pIMDO-S72-4, confirmed the prediction of them being separate mobile genetic elements (MGEs) by the final genome assembly generated by Unicycler.
FIG 3.
Frequency of the 16 plasmid genotypes found, based on multiplex colony PCR of 178 screened isolates. The order of plasmid denotation is analogous to the appearance of their amplification products in the gel (see Fig. S1 and S2).
The reassessment of the 178 heat-treated isolates for antibacterial activity revealed a loss of activity that consistently coincided with the absence of the amplified region from pIMDO-S72-2 (894 bp). Indeed, the 26 isolates that lost pIMDO-S72-2 were consistently deficient in the production of an antibacterial compound, providing unequivocal evidence that the thiopeptide BGC encoded on pIMDO-S72-2 is solely responsible for the antibacterial phenotype expressed by the wild-type M. sciuri IMDO-S72. The plasmid profiles of these isolates were reconfirmed with multiplex PCR on their extracted DNA (Fig. S2) and were in full agreement with the PCR profiles generated by multiplex colony PCR.
Purification and structural identification reveal micrococcin P1 as the antibacterial compound.
Initial analysis of fermentation supernatant by liquid chromatography-mass spectrometry (LC-MS) indicated a molecular ion with an m/z ratio of 1,144 (see Fig. S3). To confirm that the observed peak corresponded to micrococcin P1, the extraction procedure was followed by a purification step using semipreparative high-pressure liquid chromatography (HPLC). HPLC analysis and subsequent high-resolution mass spectrometry (HRMS) measurement confirmed that the compound with a retention time (tR) of 3.77 min was found at a [M + H]+ value of 1,144.2173, which is in line with the calculated value of 1,144.2179 for the molecular formula C48H49N13O9S6 (see Fig. S4). Comparison of the obtained 1H NMR spectral data with previously published data (54, 55) confirmed the structure but also revealed the presence of minor impurities (approximately 20%). Assessment of the purified fractions (tR = 3.77) using an agar spot test confirmed antibacterial activity toward the susceptible indicator strain Staphylococcus carnosus IMDO-S5. The nuclear magnetic resonance (NMR) spectral data are represented in Fig. S5 to S9.
Micrococcin P1 follows secondary metabolite production kinetics.
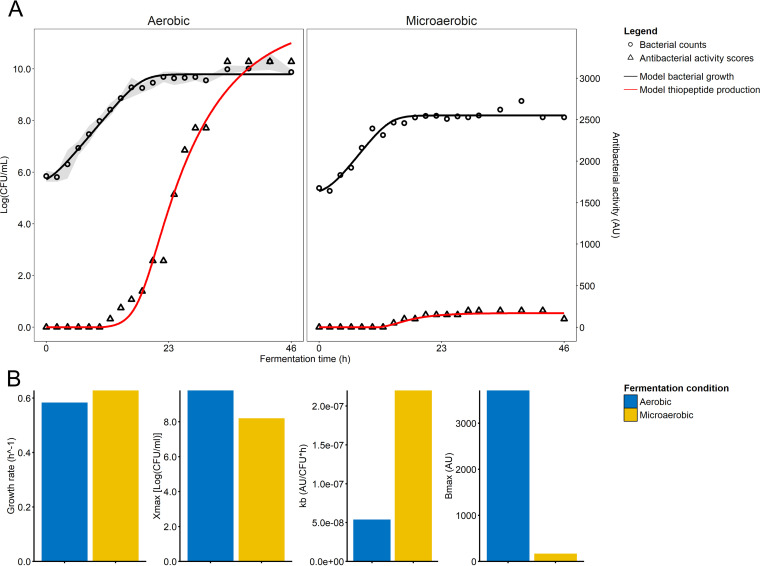
Based on aerobic and microaerobic growth experiments performed with M. sciuri IMDO-S72, micrococcin P1 was marked by a secondary metabolite production kinetics, as an increase was only found in the late-exponential phase (Fig. 4A). In the aerobic experiments, micrococcin P1 production was found after 12 h, reaching a maximum value of 3,200 arbitrary units (AU) in the late stationary phase, where it stayed constant until 46 h. In case of the microaerobic experiment, micrococcin P1 production was found after 14 h, reaching a maximum value of 200 AU in the late stationary phase, which stayed relatively constant until the end of the experiment. Also, when modeling the thiopeptide production, the best fit was obtained using a model without a growth-associated term and with incorporation of a levelling-off toward a certain maximum concentration as follows:
FIG 4.
(A) Growth of M. sciuri IMDO-S72 in MSM under aerobic and microaerobic conditions. Aerobic fermentations were performed in triplicate to confirm biological reproducibility. Average values of the three replicates are shown and were also used for the modeling of the data. Cell counts are represented as log(CFU mL−1), while antibacterial activity is expressed in arbitrary units (AU). Standard deviation of the cell counts is indicated by gray shading. (B) Model parameters obtained from modeling bacterial growth and thiopeptide production, represented by the growth rate (μmax [h−1]), maximum biomass concentration (Xmax [log(CFU mL−1)]), maximum specific production rate (kb [AU (CFU h)−1]), and maximum micrococcin P1 concentration (Bmax [AU]) for aerobic and microaerobic fermentations.
(if B > 0; otherwise dB/dt = 0), with kb being the maximum specific production rate (in AU [CFU h]−1), B the micrococcin P1 concentration (in AU), Bmax the maximum micrococcin P1 concentration (in AU), and X the cell concentration (in CFU ml−1). The maximum specific growth rate of M. sciuri IMDO-S72 (μmax in h−1) was slightly higher under microaerobic conditions, with a value of 0.63 compared to 0.58 h−1 in the case of aerobic conditions (Fig. 4B). Of more importance was the difference in final biomass concentrations that was obtained at the end of the growth experiments, represented by Xmax (in CFU ml−1) (Fig. 4B). The maximum cell population within the aerobic experiments, as simulated by the model, amounted to 9.79 log(CFU mL−1), while under microaerobic conditions 8.20 log(CFU mL−1) was reached. As the thiopeptide production is assumed to be related to the cell concentration based on the equation above, the difference in cell population found between the two conditions could be related to the higher micrococcin P1 production under aerobic growth (i.e., a simulated Bmax of 3,710 AU for aerobic growth compared to a Bmax of 170 AU under microaerobic conditions). This was also confirmed by the maximum specific production rate, kb, which was normalized for cell concentration; the latter was even higher in the microaerobic (2.2 × 10−7 AU [CFU h]−1) than the aerobic experiments (5.4 × 10−8 AU [CFU h]−1). Based on the comparison of these modeling parameters, M. sciuri IMDO-S72 exhibited a generally low specific micrococcin P1 production, requiring high cell densities to reach a substantial increase in concentration. However, because the micrococcin P1-encoding BGC of M. sciuri IMDO-S72 seems to lack any transport-related genes, a characteristic also found inherent in other BGC-producing micrococcin P1, it can be hypothesized that the extracellular release of micrococcin P1 is due to (facilitated) passive diffusion across the cell membrane. Considering its relatively high hydrophobic character, it is not improbable that a part remains within or attached to the producer cells during this process (36). Several studies that focused on micrococcin P1 purification indeed demonstrated that high yields can be obtained from cell pellet extracts (35, 36, 38, 39).
Insights into the virulence potential of Mammaliicoccus sciuri IMDO-S72.
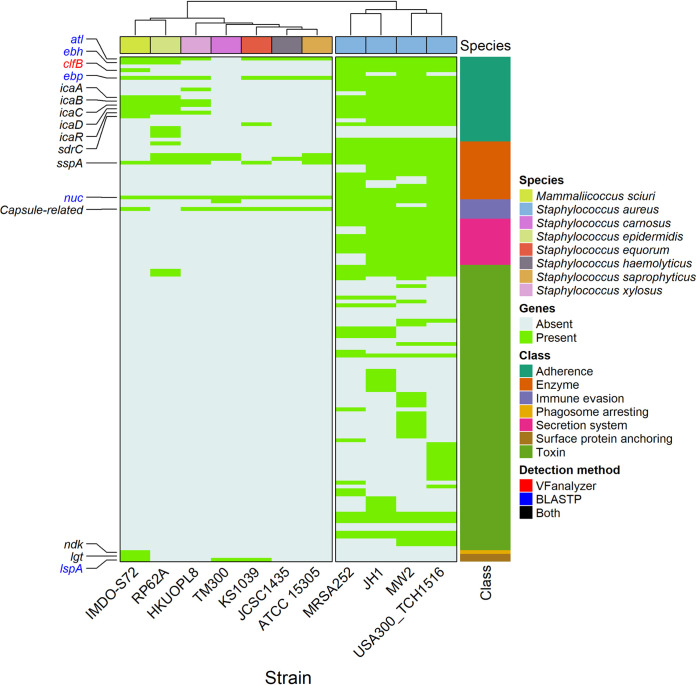
A dedicated genome-wide in silico analysis was performed to chart the virulence potential of M. sciuri IMDO-S72 using different specialized tools and databases in combination with a manual curation. As a first step, VFanalyzer was used to obtain a pathogenomic profile of M. sciuri IMDO-S72 based on preanalyzed reference genomes of well-known staphylococcal (opportunistic) pathogens. Staphylococcus xylosus HKUOPL8, S. carnosus TM300, and S. equorum KS1039, regarded as being less virulent staphylococcal species, were added to extend the comparative analysis. Hierarchical clustering was applied on the output of identified putative VFs, which is represented as a heatmap (Fig. 5).
FIG 5.
Heatmap based on the comparative pathogenomic analysis with VFanalyzer. Genes that had identified protein homologs in M. sciuri IMDO-S72 are indicated on the left-hand side of the figure; their color indicates if they were detected by VFanalyzer, blastp, or both. On the right-hand side, the class of the respective VFs is represented as defined in the VFDB database. Different species included in the comparison are indicated on top of the figure together with their hierarchical clustering, while their corresponding strain names are indicated at the bottom of the figure. The following gene loci of M. sciuri IMDO-S72 were detected as putative VFs: atl (SSCS72_00583, SSCS72_01175, SSCS72_02003, SSCS72_02005, SSCS72_02030), ebh (SSCS72_01534), clfB (SSCS72_00342), ebp (SSCS72_01488), icaA (SSCS72_02865*), icaB (SSCS72_02863*), icaC (SSCS72_00185, SSCS72_01996, SSCS72_02862*), icaD (SSCS72_02864*), icaR (SSCS72_00184, SSCS72_02866*), sdrC (SSCS72_00382), sspA (SSCS72_01332, SSCS72_02553), nuc (SSCS72_02216), capsule-related (SSCS72_00350, SSCS72_00443, SSCS72_00562, SSCS72_00779, SSCS72_01982), ndk (SSCS72_01502), lgt (SSCS72_02264), and lspA (SSCS72_01834). Genes indicated with (*) were located on pIMDO-S72-1.
A distinct separation was obtained between the highly pathogenic S. aureus and other staphylococcal species as well as M. sciuri IMDO-S72. In the latter group, apart from the hlb and hld genes of Staphylococcus epidermidis, no genes were found related to toxin production and secretion systems. The general absence of these genetic traits in M. sciuri IMDO-S72 and the included non-aureus staphylococcal strains hints at a low cytolytic potential compared to S. aureus. In the non-aureus group, two main clusters were found based on their virulence profiles. The first cluster comprised M. sciuri and S. epidermidis, while the second cluster included S. carnosus, S. equorum, Staphylococcus haemolyticus, Staphylococcus saprophyticus, and S. xylosus. Most of the detected VFs of M. sciuri IMDO-S72 and S. epidermidis RP62A fell into the “adherence” class, as defined by the Virulence Factor Database (VFDB), which might indicate a general role of these determinants in the commonly host-related lifestyle of the aforementioned species. These VFs facilitate attachment to abiotic surfaces as well as extracellular matrix proteins and are less likely to be involved in aggressive pathogenesis (29).
In the following paragraphs, an exhaustive description is given of the putative virulon of M. sciuri IMDO-S72 based on the manually curated output of all in silico analyses combined. An overview of all these virulence determinants and their genomic location is given in Fig. 6.
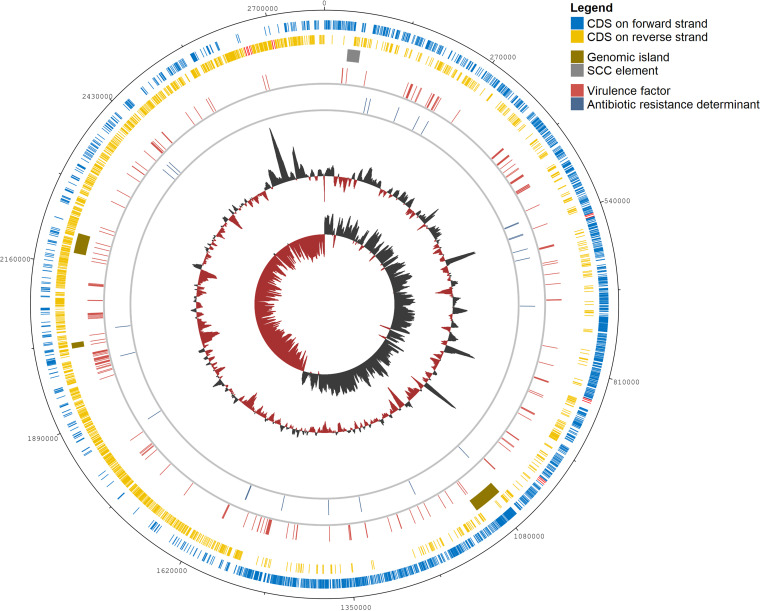
FIG 6.
Genome map of M. sciuri IMDO-S72 (plasmids are not represented). Circle 1 (outer circle), base pair coordinates of the genome; circle 2, coding sequences (CDSs) on the forward strand with rRNA genes indicated in red; circle 3, CDSs on the reverse strand with rRNA genes indicated in red; circle 4, location of putative genomic islands (GIs) and a staphylococcal cassette chromosome (SCC) element; circle 5; genomic location of annotated virulence factors (VFs;165 genes); circle 6, genomic location of annotated antibiotic resistance determinants (25 genes); circle 7, G+C content (black: above average, dark red: below average); circle 8, G+C skew (black: positive values, dark red: negative values).
(i) Adherence. Mammaliicoccus sciuri IMDO-S72 possesses several genes encoding putative bifunctional autolysins similar to atlE (SSCS72_00583, SSCS72_01175, SSCS72_02003, SSCS72_02005, SSCS72_02030, SSCS72_02203), which can facilitate abiotic surface attachment through enhancing hydrophobicity of the cell surface (56). The products of genes SSCS72_00583 and SSCS72_01175 were predicted to be embedded in the membrane. These autolysins have also been shown to interact in a nonspecific manner with matrix proteins such as fibrinogen and fibronectin (56, 57). Additionally, as a typical animal-associated microorganism, M. sciuri IMDO-S72 seems to dispose of a diverse repertoire of so-called microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which enable specific interactions with host-related matrix proteins (29). The following genes with their locus tag were found: SSCS72_00382 is predicted to encode a Ser-Asp-rich fibrinogen-binding protein (Bbp); SSCS72_01488, an elastin-binding protein (EbpS); SSCS72_00151, a serine-rich adhesin for platelets (SraP); SSCS72_01534, a putative fibronectin-binding protein (Ebh); and SSCS72_00342, a protein similar to clumping factor B (ClfB), which is also known to bind fibrinogen. All these MSCRAMMs, except for SSCS72_01488, contain a characteristic LPXTG domain which is cleaved by sortase A (SSCS72_00073 and SSCS72_01792) to covalently anchor these surface proteins to the bacterial cell wall. The product of the gene locus SSCS72_00402 was predicted to encode a sortase B, exerting the same function as SrtA but using a specific NPQTN motif as discussed above.
After primary surface adhesion, the next phase in colonization is the formation of biofilms. These three-dimensional cell clusters embedded in various extracellular biopolymers have a characteristic cell physiology, making them resistant to many host-related defense mechanisms (30). Mammaliicoccus sciuri IMDO-S72 has multiple genetic traits at its disposal that would allow intercellular aggregation, the most important of which is most likely the ica (intercellular adhesion) gene cluster. The complete icaADBC operon was identified on pIMDO-S72-1, which indicates that this trait could be acquired through horizontal gene transfer and is not necessarily a trait specific to the M. sciuri species. This cluster enables the production and secretion of the homopolymer poly-N-acetylglucosamine (PNAG), which is necessary for biofilm formation (29). N-acetylglucosamine transferase IcaA (SSCS72_02865) is responsible for its production, together with the accessory protein IcaD (SSCS72_02864), which is needed for IcaA activity. The IcaB protein (SSCS72_02863) partly deacetylates PNAG, giving it a cationic character, which might further aid in surface adhesion. The membrane-bound IcaC (SSCS72_02862) protein is involved in the transport of PNAG to the extracellular space. The icaR gene (SSCS72_02866) encodes a transcriptional regulator of the biofilm operon. The gene with locus tag SSCS72_02868 located downstream of the ica locus, encoding a putative beta-hexosaminidase, might be involved in the enzymatic degradation of the PNAG exopolymer, leading to biofilm detachment and further dissemination of bacterial cells. Other chromosomally encoded protein homologs were detected for icaR (SSCS72_00184) and icaC (SSCS72_00185, SSCS72_01140, and SSCS72_01996), but as their surrounding genomic region did not contain any other counterparts of the ica operon, it is less likely that these genes contribute to a nonplanktonic mode of growth. Other important structural components of the biofilm matrix include extracellular DNA (eDNA) (30). The competence operon comE, predicted to contain four protein-coding sequences (SSCS72_01365, SSCS72_01366, SSCS72_01367, and SSCS72_01368) might play a role in active secretion of DNA (58).
(ii) Immune evasion. Apart from biofilm formation, containing protective exopolymers such as PNAG shielding bacterial cells from the host immune system, various other mechanisms can contribute to immune evasion. Some of these mechanisms focus on bypassing the antimicrobial action of cationic antimicrobial peptides (CAMPs) that can interact with the negatively charged cell surface via electrostatic attraction (29). One such mechanism is enabled by products of the dltA (SSCS72_02130), dltB (SSCS72_02129), dltC (SSCS72_02128), and dltD (SSCS72_02127) genes from the dlt operon, which are involved in the d-alanylation of (lipo)teichoic acids, increasing their net positive charge. The MprF protein (SSCS72_01620), on the other hand, catalyzes the transfer of a lysyl group to phosphatidylglycerol, producing lysylphosphatidylglycerol (LPG), an important positively charged component of the bacterial cell membrane. In addition to electrostatic repulsion of CAMPs, the ABC transporters VraF (SSCS72_00531) and VraG (SSCS72_00532) are putatively involved in the active export of CAMPs. Upstream of the VraF and VraG transporters, a two-component signal transduction system (TCS) was found to encode a sensor histidine kinase, GraS (SSCS72_00530), and a response regulator, GraR (SSCS72_00529), which is believed to be a CAMP sensory system and regulates the expression of mechanisms that are involved in CAMP resistance (29). Three putative oatA genes were annotated (SSCS72_00235, SSCS72_01983, and SSCS72_01991), encoding an O-acetyltransferase which could facilitate lysozyme resistance via modification of the peptidoglycan backbone (21). Another gene that might be involved in conferring resistance toward host-derived antimicrobials is the speG gene, for which two putative homologs were found (SSCS72_02189, SSCS72_02227). The gene product of the latter concerns a spermidine N-acetyltransferase that could defuse host exogenous polyamines present on the skin and mucous membranes (21, 30).
(iii) Regulation. VFs are conventionally seen as products encoded by accessory genes, i.e., genetic traits that are not essential for in vivo growth but allow microorganisms to circumvent and combat host defense mechanisms (59). The diverse repertoire of these genes is often referred to as the virulon, which is subject to a strict and complex regulatory network that fine-tunes its expression based on different environmental stimuli to meet the biological requirements of the cell. A well-adopted strategy of bacteria is the use of TCSs that down- or upregulate expression of specific genes to adapt to environmental stresses (60). A TCS is typically made up of a membrane-bound sensor histidine kinase, which auto-phosphorylates under the influence of an external stimulus, and in turn, activates a DNA-binding response regulator in the cytoplasm (61). In the genome of M. sciuri IMDO-S72, 15 putative TCSs have been identified (Table 1). Some of them are reported to be involved in the regulation of virulence determinants in other bacterial species, while others are more likely involved in basic cell metabolism.
TABLE 1.
Putative two-component signal transduction systems (TCSs) in the chromosome of Mammaliicoccus sciuri IMDO-S72a
| TCS | Assumed function | Gene(s) | Product | Gene locus |
|---|---|---|---|---|
| AgrA/AgrC | TCS within the QS agr operon (59) | agrA | RR | SSCS72_00884 |
| agrC | HK | SSCS72_00885 | ||
| ArlS/ArlR | Regulation of VFs such as secreted proteins and ica-dependent biofilm formation (59) | arlS | HK | SSCS72_01554 |
| arlR | RR | SSCS72_01553 | ||
| DesK/DesR | Involved in the regulation of membrane fluidity (103) | desK | HK | SSCS72_02396 |
| desR | RR | SSCS72_02395 | ||
| GraS/GraR | Sensing of cAMPs and regulation of a protective response (29) | graS (apsS) | HK | SSCS72_00530 |
| graR (apsR) | RR | SSCS72_00529 | ||
| HssS/HssR | Regulation of heme homeostasis (104) | hssS | HK | SSCS72_01802 |
| hssR | RR | SSCS72_01803 | ||
| LiaS/LiaR | Sensing of cell wall integrity and regulation of cell wall stress responses (105) | liaS (yhcY) | HK | SSCS72_01034 |
| liaR (yhcZ) | RR | SSCS72_01035 | ||
| LytS/LytT | Involved in regulation of cell wall metabolism and biofilm development (106) | lytS | HK | SSCS72_00369 |
| lytT | RR | SSCS72_00370 | ||
| NreB/NreC | Control of nitrate/nitrite reduction in response to oxygen | nreB | HK | SSCS72_00631 |
| nreC | RR | SSCS72_00632 | ||
| PhoR/PhoP | Involved in inorganic phosphate homeostasis (107) | phoR | HK | SSCS72_01273 |
| phoP | RR | SSCS72_01272 | ||
| SaeS/SaeR | Key element in the regulation of VFs such as exoproteins together with the agr QS system (60, 78) | saeS | HK | SSCS72_02479 |
| saeR | RR | SSCS72_02480 | ||
| SrrA/SrrB | Regulation of respiratory metabolism and virulence (108) | srrA (resD) | RR | SSCS72_01482 |
| srrB (resE) | HK | SSCS72_01483 | ||
| WalK/WalR | Regulation of cell wall metabolism (61) | walK (yycG) | HK | SSCS72_00032 |
| walR (yycF) | RR | SSCS72_00033 | ||
| VraS/VraR | Involved in the regulation of cell wall peptidoglycan biosynthesis (78) | vraS | HK | SSCS72_00945 |
| vraR | RR | SSCS72_00946 | ||
| YufL/YufM | Regulation of malate utilization (21) | yufL (dcuS, malK) | HK | SSCS72_00163 |
| yufM (dcuR, malR) | RR | SSCS72_00164 | ||
| NA | Putatively involved in iron homeostasis (21) | NA | HK | SSCS72_00193 |
| NA | RR | SSCS72_00194 |
HK, sensor histidine kinase; RR, response regulator; QS, quorum sensing; cAMP, cationic antimicrobial peptide; NA, not applicable.
(iv) Antibiotic resistance genes. The presence of antibiotic resistance determinants, either acquired or innate, is another important aspect that codefines the clinical importance of a certain species or strain, in conjunction with VFs. Based on CARD and ResFinder, two chromosomally located antibiotic resistance genes were identified, namely, sal(A) (SSCS72_01334) and a mecA gene homologue (SSCS72_00199). The sal(A) gene was shown to be a natural resistance trait within the M. sciuri species, encoding an ATP-binding cassette (ABC) transporter protein conferring moderate resistance toward lincosamides and streptogramin A (62). The penicillin-binding protein (PBP) encoded by the mecA homologue could confer resistance toward methicillin and other β-lactam antibiotics, although it has been shown that this native mecA homologue is not linked with β-lactam resistance unless specific mutations or the presence of an insertion sequence (IS) element in the promoter region allow overexpression of this protein (16, 63, 64). This is in contrast with the mecA gene located on a SCCmec (staphylococcal cassette chromosome mec) element where it is typically flanked by regulatory genes and ISs that control its expression (23, 25, 65). Manual annotation further identified 6 other PBP-like genes (SSCS72_00461, SSCS72_01227, SSCS72_01410, SSCS72_01522, SSCS72_01849, and SSCS72_01990); two of them were also reported as “loose” hits by CARD (SSCS72_01410 and SSCS72_01849), although their involvement in resistance is rather unlikely. The production of a β-lactamase is another mechanism that can facilitate β-lactam resistance by enzymatic inactivation. A putative blaZ gene (SSCS72_00088), encoding a β-lactamase, was identified in M. sciuri IMDO-S72, although it was not located within a typical bla operon, as its neighboring repressor BlaI and signal transducer BlaR seemed to be absent. The relatively low MIC value found for ampicillin indicates that the native mecA gene does indeed not confer β-lactam resistance, and the gene product of blaZ might exert a different function or is not expressed without its cognate regulatory proteins (Table 2). Within the chromosome of M. sciuri IMDO-S72, multiple efflux proteins were identified as well, belonging to the major facilitator superfamily (MFS) and all containing a tetracycline resistance protein signature (SSCS72_00083, SSCS72_00147, SSCS72_00180, SSCS_00488, SSCS72_00490, SSCS72_00524, SSCS72_00543, SSCS72_02051, SSCS72_02427 and SSCS72_02439). One multidrug resistance transporter (SSCS72_00462) was classified under the ABC superfamily and did not contain a tetracycline resistance signature. Apart from SSCS72_00490, SSCS72_00543, SSCS72_02051, and SSCS72_02427, all the other transporter proteins were also reported by CARD as “loose” hits. Regardless of the ubiquitous presence of these multidrug efflux proteins, no resistance phenotype toward tetracycline was found, suggesting the involvement of these proteins in other biological important transport-related functions (Table 2). Although no resistance to kanamycin was found (Table 2), a putative bifunctional AAC/APH enzyme encoded by the aacA-aphD gene (SSCS72_01054) was identified within the chromosome, although it was not associated with any IS or composite transposon. Resistance to aminoglycosides such as kanamycin is indeed typically mediated by Tn4001-like composite transposons which are flanked by ISs (e.g., IS256). These ISs provide strong (hybrid) promoters that modulate the expression of the neighboring aacA-aphD gene captured within the transposable element (66, 67). One antibiotic resistance determinant, mph (SSCS72_02447), was identified that could facilitate resistance toward macrolides such as erythromycin (9). Also here, M. sciuri IMDO-S72 was shown to be susceptible toward this antibiotic. The fact that the presence of intact mph genes does not correspond with phenotypic resistance has been reported before for M. sciuri (68). Both aacA-aphD and mph genes were also detected by CARD as “loose” hits. The absence of antibiotic resistance traits toward chloramphenicol and vancomycin was confirmed by the observed susceptible phenotype during susceptibility testing (Table 2).
TABLE 2.
Overview of in vitro-determined MICs for Mammaliicoccus sciuri IMDO-S72 and their corresponding phenotype based on EUCASTa
| MIC or phenotype | Data for: |
|||||
|---|---|---|---|---|---|---|
| Ampicillin | Chloramphenicol | Erythromycin | Kanamycin | Tetracycline | Vancomycin | |
| MIC (mg/L) | 0.125 | 4 | 0.25 | 0.25 | 0.125 | 1 |
| MIC breakpoint (mg/L)b | NA | R > 8 S ≤ 8 |
R > 2 S ≤ 1 |
R > 8 S ≤ 8 |
R > 2 S ≤ 1 |
R > 4 S ≤ 4 |
| Observed phenotype | NA | S | S | S | S | S |
R, resistant; S, susceptible; NA, not applicable.
MIC breakpoint values based on EUCAST clinical breakpoint tables v. 11.0, valid from 1 January 2021.
The genome of Mammaliicoccus sciuri IMDO-S72 seems to lack virulence- or antibiotic resistance-associated mobile genetic elements.
Next to the mere in silico detection and designation of putative virulence and antibiotic resistance determinants, an assessment of their genomic context can provide more insight into whether these genetic traits are natural or acquired, as well as the probability of their dissemination to other (related) bacterial species. Prediction of MGEs such as plasmids, genomic islands (GIs), transposons, and ISs has become an important aspect of bacterial genome analysis to aid in unravelling the pathogenicity of a certain bacterium (69). After manual refinement of the IslandViewer output, three putative GIs were retained within the chromosome of M. sciuri IMDO-S72 (Fig. 6). The first GI corresponded to the prophage region detected by PHASTER that was described earlier, adding evidence for the horizontal origin of this DNA segment. Phages are known to be an important source of GIs in bacteria, with tRNA genes often acting as a template for their integration (69). The second GI consisted of a 9.8-kbp region comprising 13 genes (position 2,014,458 to 2,024,216 bp), with a G+C content of 30.0%. A 35.1-kbp region with a G+C content of 29.3% defined the third GI (position 2,181,302 to 2,216,433 bp) and contained 49 genes. The delineation of the boundaries of these two last GIs proved more difficult and might need further correction, as their detection most likely solely relied on their sequence composition bias.
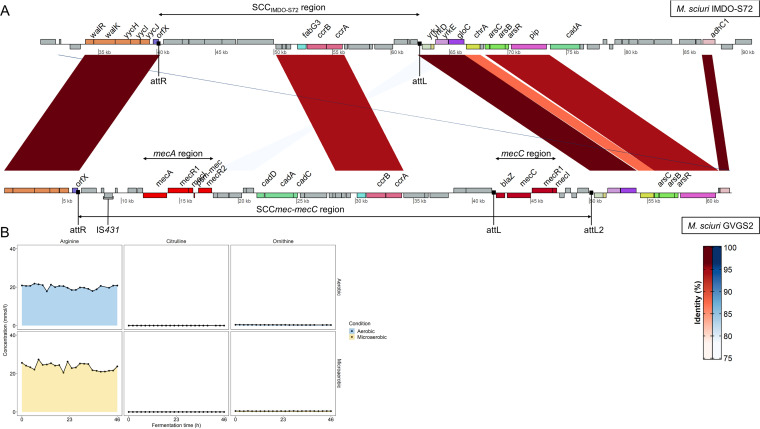
Another important MGE, one that is typically linked with staphylococci and mammaliicocci, is SCCmec (65, 70). It carries the mecA gene (or other mecA homologues, e.g., mecC), which is the core determinant for methicillin resistance, and is acquired by site-specific integration into the 3′ part of the orfX gene (SSCS72_00037) effectuated by cassette chromosome recombinases and encoded by ccr genes (71). Mammaliicoccus sciuri IMDO-S72 contained a nontypeable SCC element of 22.3 kbp, further referred to as SCCIMDO-S72, as it did not contain mecA or any other characteristic genes (Fig. 7A). It was demarcated by two repeat regions, attR (position 40,110 to 40,181 bp) and attL (position 62,292 to 62,362 bp). Sequence alignment analysis using a blast search of the orfX region of M. sciuri IMDO-S72 with the hybrid SCCmec-mecC region of M. sciuri GVGS2 revealed three major homologous regions (23). The first homologous region included the highly conserved walRK operon (walR, walK, walH, walI, and walJ) and the orfX locus (61). SCCIMDO-S72 only showed one homologous region with the hybrid SCCmec-mecC element, comprising four genes encoding hypothetical proteins (SSCS72_00044, SSCS72_00045, SSCS72_00046, SSCS72_00047), a dehydrogenase reductase (SSCS72_00048), and the recombinase genes ccrB (SSCS72_00049) and ccrA (SSCS72_00050), which are an essential part of SCC elements, facilitating their mobilization and integration in the genome. The mecA determinant carried by the SCCmec-mecC element shared 80.48% nucleotide identity with the native mecA gene homologue (SSCS72_00199) located downstream from SCCIMDO-S72, but as discussed above, this natural determinant is generally not linked with β-lactam resistance. Downstream of the attL site of the SCCIMDO-S72 element, another homologous region was found, containing a putative sulfite exporter, YrkJ (SSCS72_00058), a metal-sensitive repressor, YrkD (SSCS72_00059), and a thiosulfate sulfurtransferase, YrkE (SSCS72_00060). Further downstream of these genes, gloC (SSCS72_00061) was found encoding a hydroxyacylglutathione hydrolase, a chromate transporter, chrA (SSCS72_00062), and a putative transcriptional regulator (SSCS72_00063), although the latter gene seemed to be absent in M. sciuri GVGS2. The homologous region was concluded by an arsenic resistance operon, arsCBR (SSCS72_00064, SSCS72_00065, and SSCS72_00066), and a putative membrane-bound phage infection protein (SSCS72_00067). The absence of shared nucleotide identity within the so-called J regions (joining regions) illustrates the structural and genetic diverseness of these MGEs (23, 65, 70). The presence of nontypeable SCC variants has been reported to be common within Mammaliicoccus species (9). For example, the lack of resemblance in nucleotide identity and genomic structure of the orfX region has been shown as well for two different Mammaliicoccus stepanovicii strains (formerly classified as Staphylococcus stepanovicii) (72). In general, the presence of an attachment site (attB) within the conserved orfX locus provides an ideal docking site for SCC elements (typically carrying the second attachment site, attSCC), creating an ideal genomic context for recombination events (73). This facilitates the exchange of genetic information across the boundaries of specific species, in response to variable environmental conditions and the selective pressure created by the intensive use of antibiotics (73). The promiscuous action of Ccr recombinases in recombining attachment sites further adds to the genetic plasticity of SCC elements (74). Different staphylococci and mammaliicocci containing a mec gene complex in the orfX region have been shown to have little homology with known SCCmec types and do not always contain the associated ccr genes (75). The presence of highly similar mec gene complexes within the orfX region devoid of ccr genes, raises the question of whether these complexes can be horizontally transferred independently from SCCmec elements, as they are often associated with transposons (72, 73, 75). Given the high diversity of SCCmec elements, high nucleotide identity between these sequences in different strains or species can therefore be regarded as indicative of horizontal gene transfer (75). In case of M. sciuri, a study showed high conservation of an SCCmec-mecC hybrid element between isolates originating from dairy milk and suggested different independent acquisitions of this element (25). The low host specificity of M. sciuri and its association with SCCmec elements puts this species forward as eminently suited for the dissemination of such elements and the antibiotic resistance genes encoded within.
FIG 7.
(A) Nucleotide sequence alignment comparison between the orfX region of M. sciuri IMDO-S72 containing a putative 22.3-kbp SCC element and the hybrid SCCmec-mecC element of M. sciuri GVGS2 (GenBank accession number HG515014) as reported by Harrison et al. (23). Genomic regions with conserved nucleotide identities are interconnected with red or blue areas. Genes coding for homologous proteins are indicated in the same color; attachment sites are indicated with black squares. Genomic coordinates of att sites for M. sciuri IMDO-S72: attR (40,102 to 40,181 bp); attL (62,292 to 62,362 bp). (B) Metabolite profiles for arginine, citrulline, and ornithine based on UPLC-MS/MS measurements of samples from aerobic and microaerobic fermentations with M. sciuri IMDO-S72.
Next to SCC elements, the arginine catabolic mobile element (ACME) is another GI that is known to use the same attachment site in orfX for its integration (76). This MGE uses the recombination activity of the ccr genes encoded on SCC elements for its mobilization, and therefore they often appear together (77). No ACME element was found in the orfX region of M. sciuri IMDO-S72. Only two native arc genes, arcB (SSCS72_00477) and arcC (SSCS72_00144), were identified within the chromosome, indicating that, in combination with the absence of an ACME element, M. sciuri IMDO-S72 does not possess a functional arginine deiminase pathway, as was confirmed by metabolite measurements (Fig. 7B), which showed no absolute decrease of arginine concentrations.
Joining the identified GIs (including SCCIMDO-S72) together with the genomic location of putative VFs (Fig. 6), it is rather unlikely that these sequences of horizontal origins contribute to a virulent lifestyle for M. sciuri IMDO-S72. While VFs are often disproportionately represented on GIs, in the case of M. sciuri IMDO-S72, on a total of 171 genes that make up the putative virulon, only 8 were located on a GI where they appeared as single isolated genetic traits. The plasmid-associated ica gene cluster described before is therefore the only virulence-related operon that is located on an MGE and might be a genetic determinant of clinical importance to differentiate virulent from commensal strains of this species as has been suggested for S. epidermidis (29). As for the VFs, the same observation was made regarding antibiotic resistance determinants, as none of these genes were associated with the identified GIs, SCCIMDO-S72 element, or plasmids in M. sciuri IMDO-S72. In addition, only a few ISs were identified by ISfinder, three chromosomally encoded ISs belonging to the IS150 subfamily (positions 640,057 to 641,273 bp, 1,663,536 to 1,664,752 bp, and 1,797,195 to 1,798,411 bp) and two ISs from the IS6 family encoded on pIMDO-S72-2 (positions 2319 to 3108 bp and 4759 to 5495 bp). As none of these ISs were in the proximity of antibiotic resistance determinants, it can be assumed that there are no transposon-borne antibiotic resistance genes. These findings can help explain the weak correlation between phenotypic resistance and the presence of antibiotic resistance genes as presented in Table 2, although not all antibiotics were included in this study to cover all reported antibiotic resistance genes, e.g., lincomycin for the sal(A) gene. It is a well-known fact that plasmid- and transposon-associated antibiotic resistance genes confer higher resistance than their chromosomal-encoded homologues that are frequently not linked with their expected resistance due to their lower expression levels or because they encode similar but other functions (9, 70, 78).
Overall, the genome-wide representation of virulence determinants and their lack of association with MGEs indicate that these genes are not acquired but are natural traits in M. sciuri IMDO-S72, performing indigenous functions in the commonly noninfectious lifestyle of this bacterium (9). The benign nature is further underlined by the general absence of highly aggressive toxins that are typically found in pathogenic S. aureus strains (29). A further genome-wide comparison with other M. sciuri strains related to diseases such as bovine mastitis could further contribute to the discovery of genetic traits that can be linked with increased virulence. Apart from the sal(A) gene and mecA gene homologue, there was little evidence for the other annotated antibiotic resistance determinants, indicating that they might encode other functions. As none of these determinants were associated with MGEs, their dissemination via horizontal gene transfer is rather unlikely in the case of M. sciuri IMDO-S72.
Conclusions.
A de novo hybrid assembly of the genome of M. sciuri IMDO-S72 provided insight into the genetic basis behind its antibacterial phenotype. Thiopeptide production was traced back to the presence of a plasmid-encoded biosynthetic gene cluster, the product of which was structurally confirmed as micrococcin P1. The reported presence of such a biosynthetic gene cluster in other related species as well as it being commonly plasmid-based, makes its detection in M. sciuri not unexpected. In M. sciuri IMDO-S72, micrococcin P1 was marked by secondary metabolite production kinetics and a rather low specific production rate, which could hamper future applications of this strain. The usability of genomics was also emphasized in the assessment of the pathogenic potential of this strain. Using a broad approach based on various databases and tools, the virulon of M. sciuri IMDO-S72 was shown to be more related to colonization and survival of this species as a common animal skin-related bacterium, rather than contributing to a pathogenic character. Apart from the plasmid-associated ica cluster, M. sciuri IMDO-S72 was free of virulence- and antibiotic resistance-enriched mobile genetic elements. The discrepancy between the presence of putative antibiotic resistance determinants and the absence of a resistant phenotype underlines the importance of their genomic context, such as association with transposons, plasmids, and SCCmec, which can greatly influence their expression. In addition, unilateral in silico annotations should be interpreted with care, as they remain prone to erroneous functional predictions. The application of whole-genome sequencing could prove very useful in future studies to chart the occurrence of apathogenic strains as well as those carrying aggressive virulence factors and functional antibiotic resistance genes. This would allow further insights into the disease-causing capacity of this species and the genetic factors related to it.
MATERIALS AND METHODS
DNA extraction for Illumina and ONT libraries.
Mammaliicoccus sciuri IMDO-S72 was grown at 30°C for 48 h on mannitol salt phenol-red agar (MSA; Merck, Darmstadt, Germany). A single colony was picked to inoculate brain heart infusion (BHI) broth (Oxoid, Basingstoke, UK) and incubated overnight at 30°C. A bacterial cell pellet was obtained by centrifugation of 15 mL of BHI culture at 16,000 × g for 15 min at 4°C. Genomic DNA was extracted from this pellet using the Genomic-tip G/20 procedure (Qiagen, Düsseldorf, Germany) with some minor modifications. The bacterial pellet was resuspended in 1 mL of buffer B1 (with RNase A [0.2 mg mL−1]) to which 40 μL of both a lysozyme solution (100 mg mL−1; Merck) and a mutanolysin solution was added (12.5 kU mL−1; Sigma), followed by an incubation of 60 min at 37°C. After adding 250 μL of a Qiagen proteinase K stock solution to the bacterial lysate, a second incubation was performed for 2 h at 56°C. Deproteinization was achieved by adding 350 μL of buffer B2 to the lysate with an incubation of 45 min at 50°C. Genomic DNA was further purified by following the remaining steps of the Genomic-tip protocol according to the manufacturer’s instructions. The concentration of the extracted genomic DNA was measured with a Qubit device using the double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit (Thermo Fisher, Waltham, MA, USA), and the identity was confirmed by sequencing of the 16S rRNA and the tuf gene (1).
Whole-genome sequencing and de novo assembly.
Whole-genome sequencing was performed with a combination of long-read and short-read sequencing of the purified genomic DNA. Long reads were generated using the Oxford Nanopore Technologies (ONT) MinION sequencing device; 1 μg of high-molecular-mass DNA was used as input for the ONT library preparation using the ONT ligation sequencing kit (SQK-LSK109) according to the manufacturer’s instructions. To maximize the sequencing read length, the pipette tips used during the library preparation were end-cut to minimize DNA shearing. After loading the final library into an R9.4.1 flow cell, the sequencing run was performed on a MinION MK1b device using MinKNOW software for data acquisition. Base-calling was performed with Guppy v2.3.7 using the configuration file dna_r9.4.1_450bps_flipflop.cfg in high-accuracy GPU-accelerated mode. Short-read sequencing was performed on an Illumina MiniSeq sequencing system (150 bp paired-end) at the KU Leuven Laboratory of Food Microbiology and Leuven Food Science and Nutrition Research Centre (LFoRCe, Leuven, Belgium).
The ONT read set and Illumina read set were combined to generate a hybrid assembly with Unicycler v0.4.7, run in conservative mode (79). The quality of the final assembly was assessed with Bandage v0.8.1 (80).
Genome analysis.
Genome annotation was performed with Prokka v1.12 (81) and was used as the starting point for the subsequent searches of BGCs, VFs, antimicrobial resistance genes, and mobile genetic elements (MGEs). To facilitate the identification of secondary metabolite BGCs, the bacterial version of antiSMASH was used with detection strictness set to “relaxed” (82). Additional blastp analysis (see Table S1) was performed on the antiSMASH output to further assign/confirm predicted functions using already described thiopeptide gene clusters (GenBank accession numbers KM613043.1 and NC_004722.1) (35, 39, 83).
To detect and compare putative VFs, the online platform VFanalyzer of the Virulence Factor Database (VFDB) was used under the default settings (84). At the time of analysis, the genus of the uploaded annotated genome was defined as “Staphylococcus,” as the taxonomic reclassification of Staphylococcus sciuri into M. sciuri was not reported yet. To increase the comparative pathogenomics output of VFanalyzer, a selection of staphylococcal species with an assumed low virulence potential was included in the analysis namely, Staphylococcus carnosus TM300 (GenBank accession number NC_012121.1), Staphylococcus equorum KS1039 (NZ_CP013114.1), and Staphylococcus xylosus HKUOPL8 (NZ_CP007208.1). A manual blastp analysis was performed in parallel, in which all protein sequences of the VFs for the Staphylococcus genus present in the VFDB database were compared with those of M. sciuri IMDO-S72 predicted by Prokka. An overview of this output is given in Table S2. Additionally, the Pathosystems Resource Integration Center database (PATRIC) was used for a proteome comparison of M. sciuri IMDO-S72 against two manually defined feature groups based on representative genomes of the Staphylococcus genus containing VFs of the Victors database and VFDB database, respectively (85).
Identification of putative antimicrobial resistance genes was performed using the Resistance Gene Identifier (RGI) software (v5.1.1) of the Comprehensive Antibiotic Resistance Database (CARD, v3.1.1) as well as ResFinder (v4.1) (86, 87). Screening for putative genomic islands (GIs) was done using IslandViewer 4 (88), PHASTER (89), and SCCmecFinder (v1.2) (90). Detection of insertion sequences (ISs) was facilitated by the use of ISfinder (91). The presence of CRISPR-Cas systems was assessed with CRISPRCasFinder and CRISPRone (44, 92). A manual curation was performed on the output putative VFs and antimicrobial resistance genes by the aforementioned databases/tools using an in-house MySQL database containing different publicly available genome annotation sources as described before in an attempt to correct for possible discrepancies between different annotation sources (93). To visualize the genomic location of VFs, antimicrobial resistance genes, and GIs within the chromosome of M. sciuri IMDO-S72, DNAPlotter was used to create a genome map (94). Graphs depicting the G+C content and G+C skew were plotted using a window size of 10,000 bp and a step size of 200 bp. Visualization of other genome-based comparative analyses was accomplished using the genoPlotR (95) and ComplexHeatmap (96) packages in R (97).
Plasmid curing.
To confirm plasmid-associated thiopeptide production, plasmid-cured derivatives were obtained and screened as follows. Mammaliicoccus sciuri IMDO-S72 was grown on MSA from which a single colony was picked to produce an overnight culture in 10 mL of BHI broth incubated at 30°C. Cells were pelleted (7,500 × g, 15 min at 4°C) and washed with phosphate-buffered saline (PBS; 137 mM NaCl, 10 mM Na2HPO4, 2.7 mM KCl, 2 mM KH2PO4, pH 7) before being diluted into fresh BHI broth at log 4 (CFU mL−1). These cultures were subjected to a heat shock for 24 h at 42°C while being shaken at 40 rpm on a rotator (Stuart, Staffordshire, UK). Afterward, cultures were plated on BHI agar and incubated overnight at 30°C. The surviving isolates were picked and streaked on MSA and stored at −80°C in BHI supplemented with 25% (vol/vol) glycerol. Antimicrobial activity of the heat-treated isolates was assessed by a central-streak assay as described before (43).
Multiplex colony PCR.
To facilitate evaluation of the plasmid profiles from plasmid-cured derivatives, plasmid-specific primers were designed with Primer-BLAST (98) and applied in a multiplex colony PCR (Table 3), taking into account that PCR amplicons needed to differ in size between the different primer sets and avoiding primer dimer formation between the primers. One colony of each isolate obtained after heat treatment was picked up from its MSA plate and dissolved in TE buffer (13 mM Tris base, 1 mM EDTA, pH 8) supplemented with 1% Triton X-100 (Merck). After boiling for 5 min at 100°C, cells were pelleted (13,000 × g for 10 min), and 1 μL of supernatant was used as the template for the PCR. The PCR mixture contained 5 μL of 10× PCR buffer (Roche Diagnostics, Mannheim, Germany), 2.5 μL of bovine serum albumin solution (BSA; Sigma; 0.1 mg mL−1), 0.1 mM deoxynucleotide triphosphate mixture (Sigma), 1.25 U of Taq DNA polymerase (Roche Diagnostics), 0.1 μM each primer (Integrated DNA Technologies, Leuven, Belgium), and 1 μL of template DNA obtained as described above. The multiplex PCR assay comprised an initial denaturation step at 94°C for 2 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 61°C for 1 min, extension at 72°C for 1 min, and a final extension at 72°C for 7 min. The PCR mixtures were subjected to electrophoresis on a 2% agarose gel for 2 h, 30 min at 70V and visualized with ethidium bromide staining to assess successful PCR amplification.
TABLE 3.
Primer sets used in multiplex colony PCR to enable screening for plasmid-cured derivatives (each primer set amplified a specific region of a specific plasmid)
| Target plasmid | Primer | Primer sequence | Position in plasmid (bp) | Amplified fragment (bp) |
|---|---|---|---|---|
| pIMDO-S72-1 | S72_Cas9_F | 5′-ATC AAC CGC TTC AGT GTC CA-3′ | 42194–42213 | 393 |
| S72_Cas9_R | 5′-GTG TTG GAT TTG GTG AGG CG-3′ | 42586–42567 | ||
| pIMDO-S72-2 | S72_ycaO_F | 5′-GGC GCA CTA TTT TCA TCG GA-3′ | 12427–12446 | 894 |
| S72_ycaO_R | 5′-ACC TGG TGT GGG AAG AAC AA-3′ | 13320–13301 | ||
| pIMDO-S72-3 | S72_SAM_F | 5′-GCG AAA TGA GAC CCT GGA TCA-3′ | 13798–13818 | 605 |
| S72_SAM_R | 5′-ATC TTT AGT CCG GGG CAC TG-3′ | 14402–14383 | ||
| pIMDO-S72-4 | S72_PRE_F | 5′-ACC TCT TGA CCT ACA ACC GT-3′ | 2842–2861 | 474 |
| S72_PRE_R | 5′-AGA AAG GGA CGT AGA ACG GG-3′ | 3315–3296 |
Growth experiments and kinetic modeling.
To determine the production kinetics of micrococcin P1, growth experiments were performed with M. sciuri IMDO-S72 in Biostat Cplus bioreactors (Sartorius Stedim Biotech, Göttingen, Germany) under aerobic and microaerobic conditions. To mimic its original isolation source (fermented meat), a meat simulation medium (MSM) was used as described previously with some modifications (99). More information about the MSM composition, inoculum build-up, applied conditions, and semiquantitative determination of antibacterial activity can be found in the supplemental material.
Scoring of antibacterial activity was done in arbitrary units (AU) according to the following formula: AU = 2n × 100, with n being the last dilution still giving rise to a clear inhibition zone (100). Model equations, for both bacterial growth and antibacterial activity, were integrated via the Euler integration method using a stepwise time increment of 0.1 h in Excel. The best model fit was obtained using the Solver function in Excel, minimizing the sum of the squared differences between the model and the experimental data points. For bacterial growth, the model was based on the one proposed by Baranyi and Roberts (101). As such, X being the cell concentration (in CFU mL−1) was modeled as a function of time (in hours). The experimental values used for kinetic modeling are given in Table S3.
Targeted metabolite analysis.
The obtained fermentation supernatants were used to measure extracellular concentrations of arginine, citrulline, and ornithine with ultraperformance liquid chromatography-tandem mass spectrometry (UPLC-MS/MS) using an Acquity UPLC system equipped with an HSS T3 column (column dimensions, 2.1 by 150 mm; particle size, 1.8 μm; pore size, 100 Å) coupled to a triple quadrupole (TQ) tandem mass spectrometer (Waters, Milford, MA, USA). Sample ionization was achieved through positive electrospray ionization. The mobile phase, set at a flow rate of 0.3 mL min−1, consisted of an ultrapure water-acetonitrile mixture (99:1 [vol/vol]) with 0.05% (vol/vol) formic acid and 0.1% (vol/vol) heptafluorobutyric acid (HFBA [Sigma], eluent A) and an ultrapure water-acetonitrile mixture (1:99 [vol/vol]) with 0.05% formic acid and 0.1% HFBA (eluent B). Elution of the compounds was facilitated by applying the following gradient: 0.0 to 1.0 min, isocratic 100% A; 1.0 to 4.0 min, linear from 100% to 50% A and from 0% to 50% B; 4.0 to 5.5 min, isocratic 50% A and 50% B; 5.5 to 5.7 min, linear from 50% A to 100% A and from 50% B to 0% B; 5.7 to 7.0 min, isocratic 100% A. The following mass spectrometric settings were used: capillary voltage, 3.40 kV; source temperature, 150°C; desolvation temperature, 450°C; cone gas flow, 25 L h−1; desolvation gas flow, 800 L h−1. Cone voltage and collision energy were dependent on the detected compound. Detection of the compounds was achieved by scanning for the following selected reaction monitoring (SRM) transitions: arginine, 175.2 > 69.9; citrulline, 176 > 113; and ornithine, 133 > 69.9. Samples were deproteinized by the addition of acetonitrile with 0.15% HFBA in a 1:1 (vol/vol) ratio, briefly vortexed, and centrifuged (13,000 × g, 15 min, 4°C). Before injection into the column (2 μL), deproteinized samples were filtered over H-PTFE filters (Merck). Quantification of the targeted metabolites was achieved through external calibration in triplicate.
Isolation and characterization of micrococcin P1.
Micrococcin P1 was extracted from the fermentation supernatant (10 L) by first reducing the supernatant’s volume to a minimum in vacuo. The residue was partitioned between equal amounts of water and CH2Cl2. The resulting layers were separated, and the water phase was extracted three times with CH2Cl2 (approximately 1 L). The combined organic phases were dried over MgSO4 and filtered, and the solvent was removed in vacuo. The crude mixture was purified using a Gilson semipreparative HPLC system equipped with a Gilson 322 pump, UV detection at 215 nm, and a Grace Vydac 150HC C18 (250 mm by 22 mm, 10 μm) column. The mobile phase used consisted of 0.1% trifluoroacetic acid (TFA; Sigma) in ultrapure water (A) and 0.1% TFA in acetonitrile (B) with a linear gradient from 30% to 100% B over 20 min with a flow rate of 20 mL min−1. Confirmation of the micrococcin P1 structure was initially obtained using an LC-MS setup. Within the LC-MS system, the HPLC unit consisted of a Waters 600 model, combined with a Waters 2487 UV detector at 215 nm, and as stationary phase, an EC 150/2 NUCLEODUR 300-5 C18 ec-column (150 by 2.1 mm, 3 μm, 300 Å) was applied. The solvent system used was 0.1% formic acid in ultrapure water (A) and 0.1% formic acid in acetonitrile (B) with a linear gradient going from 3% to 100% B over 20 min at a flow rate of 0.3 mL min−1. The MS unit, coupled to the HPLC system, was a Waters Micromass Q-Tof micro system. The following mass spectrometric settings were used: capillary voltage, 2.20 kV; source temperature, 90°C; desolvation temperature, 350°C; cone gas flow, 53 L h−1; desolvation gas flow, 450 L h−1. For the high-resolution mass spectrometry (HRMS) analysis, the same MS system was used with reserpine (2 × 10−3 mg mL−1 solution in H2O:CH3CN [1:1]) as the reference. 1H NMR spectra were recorded using a Bruker Avance II 500 spectrometer at 500 MHz and with samples dissolved in dimethyl sulfoxide (DMSO)-d6. The chemical shifts were reported in delta (δ) units in parts per million (ppm) relative to the signal of DMSO-d6 at 2.50 ppm. An overview of the obtained NMR spectra can be found in Fig. S5 to S9.
Antibiotic susceptibility assays.
MICs of ampicillin, chloramphenicol, erythromycin, kanamycin, tetracycline, and vancomycin were determined for M. sciuri IMDO-S72 with 2-fold macrodilutions in Mueller-Hinton broth (Sigma) following the EUCAST recommendations (www.eucast.org) (102). All antibiotics used were obtained from Sigma. MIC determinations were performed in triplicate based on three independent series of experiments, using Staphylococcus aureus NCTC 6571 as the quality control strain. Medium tubes were incubated for 20 h at 37°C. Susceptibility categories (susceptible [S], resistant [R]) were assigned based on the EUCAST breakpoint tables (version 11.0, 2021).
Data availability.
The complete assembled genome of M. sciuri IMDO-S72 has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena/browser/home) and can be retrieved using Project accession number PRJEB46201.
ACKNOWLEDGMENTS
We express our gratitude to Rudy Pelicaen for his help in constructing the MySQL database to improve the manual curation process, as well as to Rafik Benhachemi for providing the Illumina MiniSeq sequence data.
This work was supported by the Research Council of the Vrije Universiteit Brussel (projects SRP7, SRP50, IOF342, and IRP11), the Hercules Foundation (project UABR 09/004), the Research Foundation-Flanders (grant G021518N), and Flanders’ FOOD (Botulinsafe project).
Footnotes
Supplemental material is available online only.
Contributor Information
Frédéric Leroy, Email: frederic.leroy@vub.be.
Danilo Ercolini, University of Naples Federico II.
REFERENCES
- 1.Heikens E, Fleer A, Paauw A, Florijn A, Fluit AC. 2005. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol 43:2286–2290. 10.1128/JCM.43.5.2286-2290.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghebremedhin B, Layer F, Konig W, Konig B. 2008. Genetic classification and distinguishing of staphylococcus species based on different partial gap, 16S rRNA, hsp60, rpoB, sodA, and tuf gene sequences. J Clin Microbiol 46:1019–1025. 10.1128/JCM.02058-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Švec P, Petráš P, Pantůček R, Doškař J, Sedláček I. 2016. High intraspecies heterogeneity within Staphylococcus sciuri and rejection of its classification into S. sciuri subsp. sciuri, S. sciuri subsp. carnaticus and S. sciuri subsp. rodentium. Int J Syst Evol Microbiol 66:5181–5186. 10.1099/ijsem.0.001493. [DOI] [PubMed] [Google Scholar]
- 4.Van Reckem E, De Vuyst L, Leroy F, Weckx S. 2020. Amplicon-based high-throughput sequencing method capable of species-level identification of coagulase-negative staphylococci in diverse communities. Microorganisms 8:897. 10.3390/microorganisms8060897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu X-W, De Meyer S, Trujillo ME. 2018. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466. 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 6.Madhaiyan M, Wirth JS, Saravanan VS. 2020. Phylogenomic analyses of the Staphylococcaceae family suggest the reclassification of five species within the genus Staphylococcus as heterotypic synonyms, the promotion of five subspecies to novel species, the taxonomic reassignment of five Staphylococcus species to Mammaliicoccus gen. nov., and the formal assignment of Nosocomiicoccus to the family Staphylococcaceae. Int J Syst Evol Microbiol 70:5926–5936. 10.1099/ijsem.0.004498. [DOI] [PubMed] [Google Scholar]
- 7.Huber H, Ziegler D, Pflüger V, Vogel G, Zweifel C, Stephan R. 2011. Prevalence and characteristics of methicillin-resistant coagulase-negative staphylococci from livestock, chicken carcasses, bulk tank milk, minced meat, and contact persons. BMC Vet Res 7:6. 10.1186/1746-6148-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhargava K, Zhang Y. 2012. Multidrug-resistant coagulase-negative staphylococci in food animals. J Appl Microbiol 113:1027–1036. 10.1111/j.1365-2672.2012.05410.x. [DOI] [PubMed] [Google Scholar]
- 9.Nemeghaire S, Argudín MA, Feßler AT, Hauschild T, Schwarz S, Butaye P. 2014. The ecological importance of the Staphylococcus sciuri species group as a reservoir for resistance and virulence genes. Vet Microbiol 171:342–356. 10.1016/j.vetmic.2014.02.005. [DOI] [PubMed] [Google Scholar]
- 10.Geniş B, Tuncer Y. 2018. Determination of antibiotic susceptibility and decarboxylase activity of coagulase-negative Staphylococcus and Macrococcus caseolyticus strains isolated from fermented Turkish sausage (sucuk). J Food Process Preserv 42:e13329. 10.1111/jfpp.13329. [DOI] [Google Scholar]
- 11.Ruiz-Ripa L, Gómez P, Alonso CA, Camacho MC, Ramiro Y, de la Puente J, Fernández-Fernández R, Quevedo MÁ, Blanco JM, Báguena G, Zarazaga M, Höfle U, Torres C. 2020. Frequency and characterization of antimicrobial resistance and virulence genes of coagulase-negative staphylococci from wild birds in Spain. Detection of tst-carrying S. sciuri isolates. Microorganisms 8:1317. 10.3390/microorganisms8091317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen S, Wang Y, Chen F, Yang H, Gan M, Zheng SJ. 2007. A highly pathogenic strain of Staphylococcus sciuri caused fatal exudative epidermitis in piglets. PLoS One 2:e147. 10.1371/journal.pone.0000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hauschild T, Wójcik A. 2007. Species distribution and properties of staphylococci from canine dermatitis. Res Vet Sci 82:1–6. 10.1016/j.rvsc.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 14.Frey Y, Rodriguez JP, Thomann A, Schwendener S, Perreten V. 2013. Genetic characterization of antimicrobial resistance in coagulase-negative staphylococci from bovine mastitis milk. J Dairy Sci 96:2247–2257. 10.3168/jds.2012-6091. [DOI] [PubMed] [Google Scholar]
- 15.Lu L, He K, Ni Y, Yu Z, Mao A. 2017. Exudative epidermitis of piglets caused by non-toxigenic Staphylococcus sciuri. Vet Microbiol 199:79–84. 10.1016/j.vetmic.2016.12.016. [DOI] [PubMed] [Google Scholar]
- 16.Couto I, Sanches IS, Sá-Leão R, de Lencastre H. 2000. Molecular characterization of Staphylococcus sciuri strains isolated from humans. J Clin Microbiol 38:1136–1143. 10.1128/JCM.38.3.1136-1143.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stepanović S, Ježek P, Vuković D, Dakić I, Petráš P. 2003. Isolation of members of the Staphylococcus sciuri group from urine and their relationship to urinary tract infections. J Clin Microbiol 41:5262–5264. 10.1128/JCM.41.11.5262-5264.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shittu A, Lin J, Morrison D, Kolawole D. 2004. Isolation and molecular characterization of multiresistant Staphylococcus sciuri and Staphylococcus haemolyticus associated with skin and soft-tissue infections. J Med Microbiol 53:51–55. 10.1099/jmm.0.05294-0. [DOI] [PubMed] [Google Scholar]
- 19.Dakić I, Morrison D, Vuković D, Savić B, Shittu A, Ježek P, Hauschild T, Stepanović S. 2005. Isolation and molecular characterization of Staphylococcus sciuri in the hospital environment. J Clin Microbiol 43:2782–2785. 10.1128/JCM.43.6.2782-2785.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stepanović S, Dakić I, Morrison D, Hauschild T, Ježek P, Petráš P, Martel A, Vuković D, Shittu A, Devriese LA. 2005. Identification and characterization of clinical isolates of members of the Staphylococcus sciuri group. J Clin Microbiol 43:956–958. 10.1128/JCM.43.2.956-958.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Christo-Foroux E, Vallaeys T, Loux V, Dassa E, Deutscher J, Wandersman C, Livernois A, Hot C, Criscuolo A, Dauga C, Clermont D, Chesneau O. 2017. Manual and expert annotation of the nearly complete genome sequence of Staphylococcus sciuri strain ATCC 29059: a reference for the oxidase-positive staphylococci that supports the atypical phenotypic features of the species group. Syst Appl Microbiol 40:401–410. 10.1016/j.syapm.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 22.Monecke S, Müller E, Schwarz S, Hotzel H, Ehricht R. 2012. Rapid microarray-based identification of different mecA alleles in staphylococci. Antimicrob Agents Chemother 56:5547–5554. 10.1128/AAC.00574-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harrison EM, Paterson GK, Holden MTG, Ba X, Rolo J, Morgan FJE, Pichon B, Kearns A, Zadoks RN, Peacock SJ, Parkhill J, Holmes MA. 2014. A novel hybrid SCCmec-mecC region in Staphylococcus sciuri. J Antimicrob Chemother 69:911–918. 10.1093/jac/dkt452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miragaia M. 2018. Factors contributing to the evolution of mecA-mediated β-lactam resistance in staphylococci: update and new insights from whole genome sequencing (WGS). Front Microbiol 9:2723. 10.3389/fmicb.2018.02723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paterson GK. 2020. Genomic epidemiology of methicillin-resistant Staphylococcus sciuri carrying a SCCmec-mecC hybrid element. Infect Genet Evol 79:104148. 10.1016/j.meegid.2019.104148. [DOI] [PubMed] [Google Scholar]
- 26.Stepanović S, Vuković D, Trajković V, Samardžić T, Ćupić M, Švabić-Vlahović M. 2001. Possible virulence factors of Staphylococcus sciuri. FEMS Microbiol Lett 199:47–53. 10.1111/j.1574-6968.2001.tb10649.x. [DOI] [PubMed] [Google Scholar]
- 27.Stepanović S, Dakić I, Opavski N, Ježek P, Ranin L. 2003. Influence of the growth medium composition on biofilm formation by Staphylococcus sciuri. Ann Microbiol 53:63–74. [Google Scholar]
- 28.Zhang Y-Q, Ren S-X, Li H-L, Wang Y-X, Fu G, Yang J, Qin Z-Q, Miao Y-G, Wang W-Y, Chen R-S, Shen Y, Chen Z, Yuan Z-H, Zhao G-P, Qu D, Danchin A, Wen Y-M. 2003. Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol Microbiol 49:1577–1593. 10.1046/j.1365-2958.2003.03671.x. [DOI] [PubMed] [Google Scholar]
- 29.Otto M. 2009. Staphylococcus epidermidis: the “accidental” pathogen. Nat Rev Microbiol 7:555–567. 10.1038/nrmicro2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heilmann C, Ziebuhr W, Becker K. 2019. Are coagulase-negative staphylococci virulent? Clin Microbiol Infect 25:1071–1080. 10.1016/j.cmi.2018.11.012. [DOI] [PubMed] [Google Scholar]
- 31.Bagley MC, Dale JW, Merritt EA, Xiong X. 2005. Thiopeptide antibiotics. Chem Rev 105:685–714. 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 32.Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J, Cotter PD, Craik DJ, Dawson M, Dittmann E, Donadio S, Dorrestein PC, Entian K-D, Fischbach MA, Garavelli JS, Göransson U, Gruber CW, Haft DH, Hemscheidt TK, Hertweck C, Hill C, Horswill AR, Jaspars M, Kelly WL, Klinman JP, Kuipers OP, Link AJ, Liu W, Marahiel MA, Mitchell DA, Moll GN, Moore BS, Müller R, Nair SK, Nes IF, Norris GE, Olivera BM, Onaka H, Patchett ML, Piel J, Reaney MJT, Rebuffat S, Ross RP, Sahl H-G, Schmidt EW, Selsted ME, Severinov K, Shen B, Sivonen K, et al. 2013. Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160. 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Just-Baringo X, Albericio F, Álvarez M. 2014. Thiopeptide antibiotics: retrospective and recent advances. Mar Drugs 12:317–351. 10.3390/md12010317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carnio MC, Höltzel A, Rudolf M, Henle T, Jung G, Scherer S. 2000. The macrocyclic peptide antibiotic micrococcin P1 is secreted by the food-borne bacterium Staphylococcus equorum WS 2733 and inhibits Listeria monocytogenes on soft cheese. Appl Environ Microbiol 66:2378–2384. 10.1128/AEM.66.6.2378-2384.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bennallack PR, Burt SR, Heder MJ, Robison RA, Griffitts JS. 2014. Characterization of a novel plasmid-borne thiopeptide gene cluster in Staphylococcus epidermidis strain 115. J Bacteriol 196:4344–4350. 10.1128/JB.02243-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ovchinnikov KV, Kranjec C, Telke A, Kjos M, Thorstensen T, Scherer S, Carlsen H, Diep DB. 2021. A strong synergy between the thiopeptide bacteriocin micrococcin P1 and rifampicin against MRSA in a murine skin infection model. Front in Immunol 12:2532. 10.3389/fimmu.2021.676534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Liu Y, Du Z, Zhang L, Chen J, Shen Z, Liu Q, Qin J, Lv H, Wang H, He L, Liu J, Huang Q, Sun Y, Otto M, Li M. 2020. Skin microbiota analysis-inspired development of novel anti-infectives. Microbiome 8:85. 10.1186/s40168-020-00866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bennallack PR, Bewley KD, Burlingame MA, Robison RA, Miller SM, Griffitts JS. 2016. Reconstitution and minimization of a micrococcin biosynthetic pathway in Bacillus subtilis. J Bacteriol 198:2431–2438. 10.1128/JB.00396-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brown LCW, Acker MG, Clardy J, Walsh CT, Fischbach MA. 2009. Thirteen posttranslational modifications convert a 14-residue peptide into the antibiotic thiocillin. Proc Natl Acad Sci USA 106:2549–2553. 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CMT, Fucini P. 2008. Translational regulation via L11: molecular switches on the ribosome turned on and off by thiostrepton and micrococcin. Mol Cell 30:26–38. 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 41.Ciufolini MA, Lefranc D. 2010. Micrococcin P1: structure, biology and synthesis. Nat Prod Rep 27:330–342. 10.1039/b919071f. [DOI] [PubMed] [Google Scholar]
- 42.Li C, Kelly WL. 2010. Recent advances in thiopeptide antibiotic biosynthesis. Nat Prod Rep 27:153–164. 10.1039/b922434c. [DOI] [PubMed] [Google Scholar]
- 43.Van der Veken D, Benhachemi R, Charmpi C, Ockerman L, Poortmans M, Van Reckem E, Michiels C, Leroy F. 2020. Exploring the ambiguous status of coagulase-negative staphylococci in the biosafety of fermented meats: the case of antibacterial activity versus biogenic amine formation. Microorganisms 8:167. 10.3390/microorganisms8020167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Q, Ye Y. 2017. Not all predicted CRISPR-Cas systems are equal: isolated cas genes and classes of CRISPR like elements. BMC Bioinform 18:92. 10.1186/s12859-017-1512-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chylinski K, Le Rhun A, Charpentier E. 2013. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol 10:726–737. 10.4161/rna.24321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chylinski K, Makarova KS, Charpentier E, Koonin EV. 2014. Classification and evolution of type II CRISPR-Cas systems. Nucleic Acids Res 42:6091–6105. 10.1093/nar/gku241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martínez B, Suárez JE, Rodríguez A. 1996. Lactococcin 972: a homodimeric lactococcal bacteriocin whose primary target is not the plasma membrane. Microbiology 142:2393–2398. 10.1099/00221287-142-9-2393. [DOI] [PubMed] [Google Scholar]
- 48.Martínez B, Fernández M, Suárez JE, Rodríguez AY. 1999. Synthesis of lactococcin 972, a bacteriocin produced by Lactococcus lactis IPLA 972, depends on the expression of a plasmid-encoded bicistronic operon. Microbiology 145:3155–3161. 10.1099/00221287-145-11-3155. [DOI] [PubMed] [Google Scholar]
- 49.Martínez B, Rodríguez A, Suárez JEY. 2000. Lactococcin 972, a bacteriocin that inhibits septum formation in lactococci. Microbiology 146:949–955. 10.1099/00221287-146-4-949. [DOI] [PubMed] [Google Scholar]
- 50.Martínez B, Böttiger T, Schneider T, Rodríguez A, Sahl H-G, Wiedemann I. 2008. Specific interaction of the unmodified bacteriocin lactococcin 972 with the cell wall precursor lipid II. Appl Environ Microbiol 74:4666–4670. 10.1128/AEM.00092-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee J-H, Heo S, Jeong D-W. 2018. Genomic insights into Staphylococcus equorum KS1039 as a potential starter culture for the fermentation of high-salt foods. BMC Genom 19:136. 10.1186/s12864-018-4532-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bewley KD, Bennallack PR, Burlingame MA, Robison RA, Griffitts JS, Miller SM. 2016. Capture of micrococcin biosynthetic intermediates reveals C-terminal processing as an obligatory step for in vivo maturation. Proc Natl Acad Sci USA 113:12450–12455. 10.1073/pnas.1612161113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kloosterman AM, Shelton KE, van Wezel GP, Medema MH, Mitchell DA. 2020. RRE-Finder: a genome-mining tool for class-Independent RiPP discovery. mSystems 5:e00267-20. 10.1128/mSystems.00267-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Christy MP, Johnson T, McNerlin CD, Woodard J, Nelson AT, Lim B, Hamilton TL, Freiberg KM, Siegel D. 2020. Total synthesis of micrococcin P1 through scalable thiazole forming reactions of cysteine derivatives and nitriles. Org Lett 22:2365–2370. 10.1021/acs.orglett.0c00202. [DOI] [PubMed] [Google Scholar]
- 55.Bycroft BW, Gowland MS. 1978. The structures of the highly modified peptide antibiotics micrococcin P1 and P2. J Chem Soc, Chem Commun (Camb) 1978:256–258. 10.1039/c39780000256. [DOI] [Google Scholar]
- 56.Heilmann C, Hussain M, Peters G, Götz F. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol Microbiol 24:1013–1024. 10.1046/j.1365-2958.1997.4101774.x. [DOI] [PubMed] [Google Scholar]
- 57.Heilmann C, Thumm G, Chhatwal GS, Hartleib J, Uekötter A, Peters G. 2003. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology (Reading) 149:2769–2778. 10.1099/mic.0.26527-0. [DOI] [PubMed] [Google Scholar]
- 58.Rajendran NB, Eikmeier J, Becker K, Hussain M, Peters G, Heilmann C. 2015. Important contribution of the novel locus comEB to extracellular DNA-dependent Staphylococcus lugdunensis biofilm formation. Infect Immun 83:4682–4692. 10.1128/IAI.00775-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novick RP, Geisinger E. 2008. Quorum sensing in staphylococci. Annu Rev Genet 42:541–564. 10.1146/annurev.genet.42.110807.091640. [DOI] [PubMed] [Google Scholar]
- 60.Novick RP, Jiang D. 2003. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology (Reading) 149:2709–2717. 10.1099/mic.0.26575-0. [DOI] [PubMed] [Google Scholar]
- 61.Takada H, Yoshikawa H. 2018. Essentiality and function of WalK/WalR two-component system: the past, present, and future of research. Biosci Biotechnol Biochem 82:741–751. 10.1080/09168451.2018.1444466. [DOI] [PubMed] [Google Scholar]
- 62.Hot C, Berthet N, Chesneau O. 2014. Characterization of sal(A), a novel gene responsible for lincosamide and streptogramin A resistance in Staphylococcus sciuri. Antimicrob Agents Chemother 58:3335–3341. 10.1128/AAC.02797-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Couto I, Wu SW, Tomasz A, de Lencastre H. 2003. Development of methicillin resistance in clinical isolates of Staphylococcus sciuri by transcriptional activation of the mecA homologue native to the species. J Bacteriol 185:645–653. 10.1128/JB.185.2.645-653.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu SW, de Lencastre H, Tomasz A. 2001. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J Bacteriol 183:2417–2424. 10.1128/JB.183.8.2417-2424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Partridge SR, Kwong SM, Firth N, Jensen SO. 2018. Mobile genetic elements associated with antimicrobial resistance. Clin Microbiol Rev 31:e00088-17. 10.1128/CMR.00088-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vandecraen J, Chandler M, Aertsen A, Van Houdt R. 2017. The impact of insertion sequences on bacterial genome plasticity and adaptability. Crit Rev Microbiol 43:709–730. 10.1080/1040841X.2017.1303661. [DOI] [PubMed] [Google Scholar]
- 68.Hauschild T, Schwarz S. 2010. Macrolide resistance in Staphylococcus spp. from free-living small mammals. Vet Microbiol 144:530–531. 10.1016/j.vetmic.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 69.Langille MGI, Hsiao WWL, Brinkman FSL. 2010. Detecting genomic islands using bioinformatics approaches. Nat Rev Microbiol 8:373–382. 10.1038/nrmicro2350. [DOI] [PubMed] [Google Scholar]
- 70.Tsubakishita S, Kuwahara-Arai K, Sasaki T, Hiramatsu K. 2010. Origin and molecular evolution of the determinant of methicillin resistance in staphylococci. Antimicrob Agents Chemother 54:4352–4359. 10.1128/AAC.00356-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang L, Safo M, Archer GL. 2012. Characterization of DNA sequences required for the ccrAB-mediated integration of staphylococcal cassette chromosome mec, a Staphylococcus aureus genomic island. J Bacteriol 194:486–498. 10.1128/JB.05047-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Semmler T, Harrison EM, Lübke-Becker A, Ulrich RG, Wieler LH, Guenther S, Stamm I, Hanssen A-M, Holmes MA, Vincze S, Walther B. 2016. A look into the melting pot: the mecC-harboring region is a recombination hot spot in Staphylococcus stepanovicii. PLoS One 11:e0147150. 10.1371/journal.pone.0147150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lakhundi S, Zhang K. 2018. Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31:e00020-18. 10.1128/CMR.00020-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Misiura A, Pigli YZ, Boyle-Vavra S, Daum RS, Boocock MR, Rice PA. 2013. Roles of two large serine recombinases in mobilizing the methicillin-resistance cassette SCCmec. Mol Microbiol 88:1218–1229. 10.1111/mmi.12253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacFadyen AC, Harrison EM, Ellington MJ, Parkhill J, Holmes MA, Paterson GK. 2018. A highly conserved mecC-encoding SCCmec type XI in a bovine isolate of methicillin-resistant Staphylococcus xylosus. J Antimicrob Chemother 73:3516–3518. 10.1093/jac/dky333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Miragaia M, de Lencastre H, Perdreau-Remington F, Chambers HF, Higashi J, Sullam PM, Lin J, Wong KI, King KA, Otto M, Sensabaugh GF, Diep BA. 2009. Genetic diversity of arginine catabolic mobile element in Staphylococcus epidermidis. PLoS One 4:e7722. 10.1371/journal.pone.0007722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shore AC, Rossney AS, Brennan OM, Kinnevey PM, Humphreys H, Sullivan DJ, Goering RV, Ehricht R, Monecke S, Coleman DC. 2011. Characterization of a novel arginine catabolic mobile element (ACME) and staphylococcal chromosomal cassette mec composite island with significant homology to Staphylococcus epidermidis ACME Type II in methicillin-resistant Staphylococcus aureus genotype ST22-MRSA-IV. Antimicrob Agents Chemother 55:1896–1905. 10.1128/AAC.01756-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jeong D-W, Heo S, Ryu S, Blom J, Lee J-H. 2017. Genomic insights into the virulence and salt tolerance of Staphylococcus equorum. Sci Rep 7:5383. 10.1038/s41598-017-05918-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wick RR, Judd LM, Gorrie CL, Holt KE. 2017. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol 13:e1005595. 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wick RR, Schultz MB, Zobel J, Holt KE. 2015. Bandage: interactive visualization of de novo genome assemblies. Bioinformatics 31:3350–3352. 10.1093/bioinformatics/btv383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 82.Blin K, Shaw S, Steinke K, Villebro R, Ziemert N, Lee SY, Medema MH, Weber T. 2019. antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline. Nucleic Acids Res 47:W81–W87. 10.1093/nar/gkz310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 84.Liu B, Zheng D, Jin Q, Chen L, Yang J. 2019. VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res 47:D687–D692. 10.1093/nar/gky1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Davis JJ, Wattam AR, Aziz RK, Brettin T, Butler R, Butler RM, Chlenski P, Conrad N, Dickerman A, Dietrich EM, Gabbard JL, Gerdes S, Guard A, Kenyon RW, Machi D, Mao C, Murphy-Olson D, Nguyen M, Nordberg EK, Olsen GJ, Olson RD, Overbeek JC, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomas C, VanOeffelen M, Vonstein V, Warren AS, Xia F, Xie D, Yoo H, Stevens R. 2020. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res 48:D606–D612. 10.1093/nar/gkz943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, Edalatmand A, Huynh W, Nguyen A-LV, Cheng AA, Liu S, Min SY, Miroshnichenko A, Tran H-K, Werfalli RE, Nasir JA, Oloni M, Speicher DJ, Florescu A, Singh B, Faltyn M, Hernandez-Koutoucheva A, Sharma AN, Bordeleau E, Pawlowski AC, Zubyk HL, Dooley D, Griffiths E, Maguire F, Winsor GL, Beiko RG, Brinkman FSL, Hsiao WWL, Domselaar GV, McArthur AG. 2020. CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bortolaia V, Kaas RS, Ruppe E, Roberts MC, Schwarz S, Cattoir V, Philippon A, Allesoe RL, Rebelo AR, Florensa AF, Fagelhauer L, Chakraborty T, Neumann B, Werner G, Bender JK, Stingl K, Nguyen M, Coppens J, Xavier BB, Malhotra-Kumar S, Westh H, Pinholt M, Anjum MF, Duggett NA, Kempf I, Nykäsenoja S, Olkkola S, Wieczorek K, Amaro A, Clemente L, Mossong J, Losch S, Ragimbeau C, Lund O, Aarestrup FM. 2020. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother 75:3491–3500. 10.1093/jac/dkaa345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman FS, Simon Fraser University Research Computing Group . 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Arndt D, Grant JR, Marcu A, Sajed T, Pon A, Liang Y, Wishart DS. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res 44:W16–W21. 10.1093/nar/gkw387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kaya H, Hasman H, Larsen J, Stegger M, Johannesen TB, Allesøe RL, Lemvigh CK, Aarestrup FM, Lund O, Larsen AR. 2018. SCCmecFinder, a web-based tool for typing of staphylococcal cassette chromosome mec in Staphylococcus aureus using whole-genome sequence data. mSphere 3:e00612-17. 10.1128/mSphere.00612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res 34:D32–D36. 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Néron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pelicaen R, Gonze D, Teusink B, De Vuyst L, Weckx S. 2019. Genome-scale metabolic reconstruction of Acetobacter pasteurianus 386B, a candidate functional starter culture for cocoa bean fermentation. Front Microbiol 10:2801. 10.3389/fmicb.2019.02801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Carver T, Thomson N, Bleasby A, Berriman M, Parkhill J. 2009. DNAPlotter: circular and linear interactive genome visualization. Bioinformatics 25:119–120. 10.1093/bioinformatics/btn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Guy L, Roat Kultima J, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Gu Z, Eils R, Schlesner M. 2016. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32:2847–2849. 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 97.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation of Statistical Computing, Vienna, Austria. [Google Scholar]
- 98.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinform 13:134. 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ravyts F, Vrancken G, D’Hondt K, Vasilopoulos C, De Vuyst L, Leroy F. 2009. Kinetics of growth and 3-methyl-1-butanol production by meat-borne, coagulase-negative staphylococci in view of sausage fermentation. Int J Food Microbiol 134:89–95. 10.1016/j.ijfoodmicro.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 100.Leroy F, de Vuyst L. 1999. Temperature and pH conditions that prevail during fermentation of sausages are optimal for production of the antilisterial bacteriocin sakacin K. Appl Environ Microbiol 65:974–981. 10.1128/AEM.65.3.974-981.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Baranyi J, Roberts TA. 1994. A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23:277–294. 10.1016/0168-1605(94)90157-0. [DOI] [PubMed] [Google Scholar]
- 102.European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). 2003. Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clin Microbiol Infect 9:1–7. 10.1046/j.1469-0691.2000.00142.x. [DOI] [PubMed] [Google Scholar]
- 103.Albanesi D, Mansilla MC, de Mendoza D. 2004. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol 186:2655–2663. 10.1128/JB.186.9.2655-2663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119. 10.1016/j.chom.2007.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Klinzing DC, Ishmael N, Hotopp JCD, Tettelin H, Shields KR, Madoff LC, Puopolo KM. 2013. The two-component response regulator LiaR regulates cell wall stress responses, pili expression and virulence in group B Streptococcus. Microbiology (Reading) 159:1521–1534. 10.1099/mic.0.064444-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Sharma-Kuinkel BK, Mann EE, Ahn J-S, Kuechenmeister LJ, Dunman PM, Bayles KW. 2009. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol 191:4767–4775. 10.1128/JB.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Santos-Beneit F. 2015. The Pho regulon: a huge regulatory network in bacteria. Front Microbiol 6:402. 10.3389/fmicb.2015.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Yarwood JM, McCormick JK, Schlievert PM. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 183:1113–1123. 10.1128/JB.183.4.1113-1123.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental methods, Tables S1 to S3, and Fig. S1 to S9. Download aem.02088-21-s0001.pdf, PDF file, 3.1 MB (3.1MB, pdf)
Data Availability Statement
The complete assembled genome of M. sciuri IMDO-S72 has been deposited in the European Nucleotide Archive (http://www.ebi.ac.uk/ena/browser/home) and can be retrieved using Project accession number PRJEB46201.